Abstract
The association between inflammation and endoplasmic reticulum (ER) stress has been observed in many diseases. However, if and how chronic inflammation regulates the unfolded protein response (UPR) and alters ER homeostasis in general, or in the context of chronic disease, remains unknown. Here, we show that, in the setting of obesity, inflammatory input through increased inducible nitric oxide synthase (iNOS) activity causes S-nitrosylation of a key UPR regulator, IRE1α, which leads to a progressive decline in hepatic IRE1α-mediated XBP1 splicing activity in both genetic (ob/ob) and dietary (high-fat diet–induced) models of obesity. Finally, in obese mice with liver-specific IRE1α deficiency, reconstitution of IRE1α expression with a nitrosylation-resistant variant restored IRE1α-mediated XBP1 splicing and improved glucose homeostasis in vivo. Taken together, these data describe a mechanism by which inflammatory pathways compromise UPR function through iNOS-mediated S-nitrosylation of IRE1α, which contributes to defective IRE1α activity, impaired ER function, and prolonged ER stress in obesity.
The endoplasmic reticulum (ER) is the major site for the synthesis and folding of proteins, lipid trafficking, and metabolism, as well as an intracellular store of calcium. The ER is equipped with a robust adaptive response system called the unfolded protein response (UPR), which mitigates stress under many challenging conditions that interfere with folding capacity (1). This adaptive system is mediated by three canonical signaling pathways, initiated by three lumenal sensors—inositol-requiring protein 1 (IRE1α), protein kinase RNA-like ER kinase (PERK), and activating transcription factor 6 (ATF6)—to control a complex network of adaptive responses to restore normal ER function (1, 2). Obesity is associated with ER stress and chronic metabolic inflammation that collectively disrupt systemic glucose homeostasis (2). However, unlike experimental ER stress in which all UPR branches are activated (3), obesity features a disproportionate production of key molecules mediating ER defenses, such as a decline in ATF6 in the presence of sustained PERK activation (4, 5). Here, we investigated the differential regulation of the UPR branches in the liver of obese mice and revealed a mechanism that contributes to the impaired resolution of ER stress and consequent metabolic decline in the setting of obesity.
Obesity results in impaired IRE1α-mediated XBP1 splicing activity
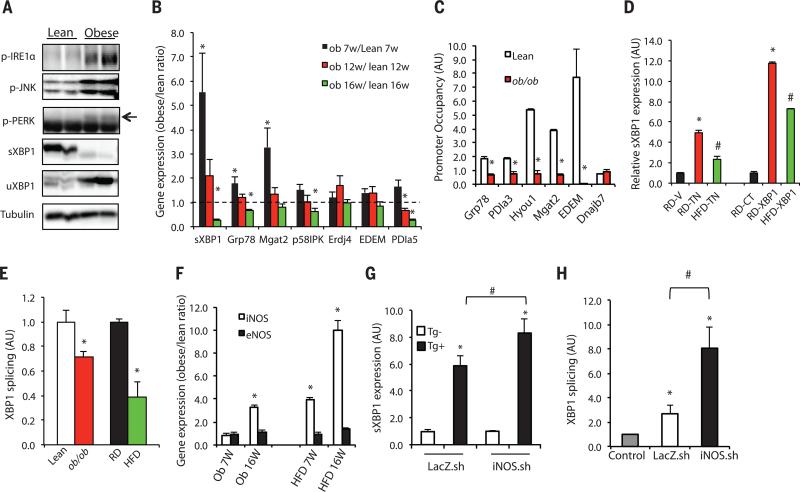
IRE1α plays an important role in maintaining ER homeostasis through initiating unconventional splicing of the mRNA encoding X-box–binding protein 1 (XBP1) to create a translational frame-shift. This produces a potent transcription factor, spliced XBP1 (sXBP1), which regulates expression of genes encoding ER chaperones (6), as well as proteins involved in phospholipid synthesis and de novo lipogenesis (7). As shown in Fig. 1, A and B, and fig. S1A and reported previously in many independent studies (4, 8–10), in the livers of obese mice, we observed sustained activation of the canonical ER stress sensors, indicated by increased PERK and IRE1α phosphorylation, c-Jun N-terminal kinase (JNK) activation, and increased UPR gene expression. However, the level of un-spliced XBP1 protein (uXBP1) was increased in obese liver tissue compared with that of lean controls, in the absence of a corresponding increase in sXBP1, which strongly indicated a defect in the processing of uXBP1 to sXBP1 by IRE1α endoribonuclease activity, despite the robust IRE1α phosphorylation and JNK activation (Fig. 1A). In contrast, in lean mice, most of the uXBP1 was converted into the spliced form. In addition, we detected a marked decline in the expression of hepatic sXBP1, as well as its target ER chaperone genes, at later stages of disease in both genetic (ob/ob) and high-fat diet (HFD)–induced models of obesity (Fig. 1B and fig. S1A).
Fig. 1. Inflammation contributes to defective IRE1α-mediated XBP1 splicing in obese liver.
(A) UPR status was examined in livers of 16-week-old obese (ob/ob) and lean mice by using phosphorylation of IRE1α, PERK, and JNK, as well as expression of sXBP1 and uXBP1 as markers. Data are representative of results from two to three independent cohorts of mice. (B) Expression of hepatic UPR modulators in 7-, 12- and 16-week-old ob/ob mice relative to lean controls (dashed line indicates lean level), n = 4. (C) ChIP assay examining XBP1 target gene occupancy in primary hepatocytes from 16-week-old ob/ob and lean mice. Data were normalized to 2% input, followed by comparison to each immunoglobulin G (IgG) control (n = 4). Data are representative of results from two independent sets of mice. Asterisk (*) indicates statistical significance between lean and obese mice in (B) and (C). Statistical analysis was performed by multiple t tests and significance was determined by the Holm-Šídák method using Prism (B) and Student's t test (C). AU, arbitrary units. (D) sXBP1 was examined in the livers from lean (RD) and obese (HFD-fed) mice injected with vehicle (V), or tunicamycin (TN, 6 hours, 0.5 mg/kg per kg body weight). sXBP1 expression was also examined in the livers from control mice fed RD or HFD transduced with full-length XBP1 (RD-XBP1, HFD-XBP1) or lean controls (RD-XBP1). Asterisk (*) indicates statistical significance between treatments within the control group, and # indicates statistical significance between RD and HFD [one-way analysis of variance (ANOVA) followed by post hoc Tukey's test], n = 6 to 8 mice. (E) In vitro splicing assays measuring the XBP1 splicing efficiency using hepatic IRE1α from mice with dietary (HFD) and genetic (ob/ob) obesity and controls. sXBP1 expression was examined by qRT-PCR. Results are normalized to lean samples. Asterisk (*) indicates statistical significance between lean and obese mice (Student's t test, n = 3). Data are representative of results from two independent sets of mice. (F) iNOS and eNOS mRNAs were examined in livers of 7- and 16-week-old ob/ob or HFD-fed mice and lean controls by qRT-PCR. Asterisk (*) indicates statistical significance between lean and obese mice (Student's t test), n = 4 to 6. (G) sXBP1 expression was examined by qRT-PCR in primary hepatocytes from lean mice transduced with Ad-shiNOS (iNOS.sh) or control virus (LacZ.sh) followed by treatment with thapsigargin (Tg+) for 2 hours, n = 4. (H) In vitro XBP1 splicing assay using IRE1α purified from the livers ob/ob mice after iNOS suppression (normalized to IgG control). Asterisk (*) indicates statistical significance between treatments and controls, and # indicates statistical significance between iNOS.sh group and LacZ.sh group (one-way ANOVA followed by post hoc Tukey's test). All data are shown as means ± SEM. Data are representative of results from two independent sets of mice. *P < 0.05; #P < 0.05.
To determine whether the progressive decline in XBP1 splicing also affected the direct regulation of potential sXBP1 target gene expression, we performed chromatin immunoprecipitation (ChIP) assays in primary hepatocytes from ob/ob mice and matching lean controls. Promoter occupancy of several sXBP1 target genes—including ER chaperones [glucose-regulated protein of 78 kD (Grp78) hypoxia up-regulated 1 (Hyou1 or ORP-150), and protein disulfide isomerase family A member 3 (Pdia3)], as well as genes involved in protein degradation, such as ER degradation-enhancing α-mannosidase (EDEM), and glycosylation, such as mannosyl (α-1,6-)-glycoprotein β-1,2-N-acetylglucosaminyltransferase (Mgat2)—was readily detected in the lean samples but markedly diminished in the ob/ob hepatocytes (Fig. 1C). These results demonstrated that both the expression and activity of sXBP1 are defective in liver cells from obese mice despite phosphorylation and sustained activation of IRE1α.
Next, we examined sXBP1 expression in the livers of HFD-fed mice, as well as lean controls [regular diet (RD)], upon experimentally induced ER stress. As shown in Fig. 1D, injection of the chemical stress inducer tunicamycin acutely induced the production of sXBP1, but this effect was suppressed in the livers of HFD mice. In a second model, HFD or RD mice were transduced with adenovirus-mediated full-length XBP1. As shown in Fig. 1D, in the setting of obesity, the production of sXBP1 was significantly reduced compared with that of lean controls. Next, we asked whether the decrease in sXBP1 expression in obesity was directly related to impaired ribonuclease activity of IRE1α. In an in vitro splicing assay using endogenous IRE1α protein immunopurified from mouse liver, we observed a significant decline in IRE1α-mediated XBP1 processing in samples from obese mice (both ob/ob and HFD) compared with lean controls (Fig. 1E).
Metaflammation is associated with impaired XBP1 splicing
Because IRE1α phosphorylation remained intact in the obese livers but XBP1 splicing activity was markedly diminished, we hypothesized that a phosphorylation-independent, obesity-induced modification of IRE1α might underlie the selective inhibition of its ribonuclease activity. Obesity is characterized by chronic metabolic inflammation, termed metaflammation (11–14), and numerous inflammatory signaling cascades exhibiting aberrant activity in obesity share a common feature: a marked increase in inducible nitric oxide synthase (iNOS) expression (15). Indeed, induction of iNOS and nitric oxide (NO) production is observed in many inflammatory diseases (16, 17). We noted that the decline in the expression of sXBP1 in liver tissue of obese animals coincided with markedly increased iNOS expression in both dietary and genetic obesity models (Fig. 1F), whereas endothelial NOS (eNOS) expression levels were similar between lean and obese tissues, and neuronal NOS mRNA expression was not detectable. To examine whether the nitrosylation-mediated inhibition of the IRE1α ribonuclease activity was a function of iNOS induction as part of metaflammation, we tested the influence of suppression or overexpression of iNOS on XBP1 splicing in primary hepatocytes. Suppression of iNOS expression resulted in enhanced thapsigargin (Tg)–induced XBP1 splicing in primary hepatocytes isolated from lean mice (Fig. 1G). In contrast, reconstitution of iNOS in primary hepatocytes isolated from iNOS-deficient mice resulted in a significant decrease in Tg-induced sXBP1 generation and Grp78 expression (fig. S1, C and D). To examine whether the IRE1α ribonuclease activity was regulated by iNOS induction, we performed in vitro splicing assays in ob/ob liver tissue after in vivo small hairpin RNA (shRNA)–induced suppression of iNOS; expression was reduced more than 75% (fig. S1E). iNOS suppression in vivo led to markedly enhanced IRE1α-mediated XBP1 splicing (Fig. 1H). Consistent with the established role of ER stress in insulin resistance, we observed significantly enhanced hepatic insulin signaling assessed by insulin-induced phosphorylation of insulin receptor and Akt (fig. S1, F and G). There was also a decrease in serum glucose and significantly enhanced systemic glucose tolerance in ob/ob mice after the suppression of hepatic iNOS (fig. S1, H and I). Taken together, these results demonstrate that iNOS is a critical mediator of hepatic IRE1α ribonuclease activity, with consequences for systemic glucose homeostasis.
Nitrosative stress results in IRE1α S-nitrosylation
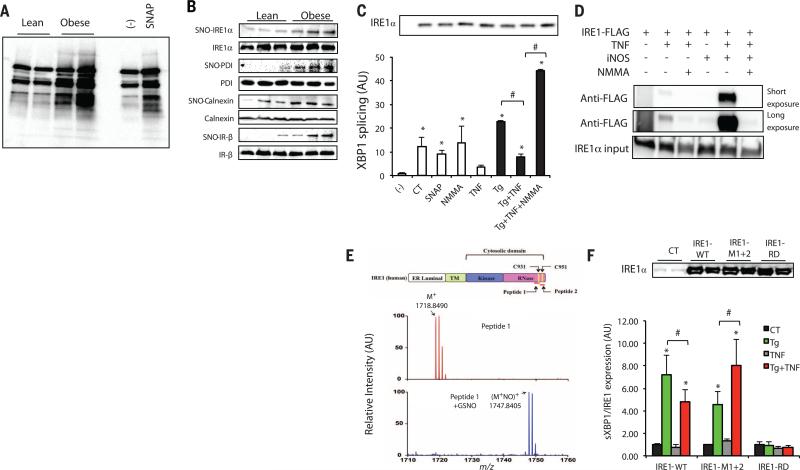
S-Nitrosylation—the covalent attachment of a nitrogen monoxide group to the thiol side chain of cysteine residues—has emerged as a mechanism for dynamic, posttranslational regulation of proteins, including UPR regulators, such as PDI (18). Thus, we next determined whether nitrosylation induced by iNOS-mediated NO production could regulate IRE1α ribonuclease (RNase) activity. For this, we first examined the alterations in general protein S-nitrosylation (SNO) in the livers of lean and obese mice using a biotin switch method (19, 20). There was a significant increase in hepatic protein S-nitrosylation in both dietary and genetic models of obesity (Fig. 2A and fig. S2A). Because protein nitrosylation is highly regulated by glutathione levels (21, 22), we examined the amount of glutathione in its reduced form (GSH) in the livers of ob/ob mice and lean controls and observed a significant decrease in GSH in the livers of ob/ob mice (fig. S2B), consistent with potentially decreased denitrosylation capacity in the obese condition. To determine whether altered nitrosylation status may affect ER function, S-nitrosylated proteins were isolated from the livers of ob/ob mice and lean controls, followed by detection of proteins critical to ER stress and adaptive responses. These experiments demonstrated that several ER chaperones, such as calnexin and protein disulfide isomerase (PDI), exhibited increased nitrosylation (Fig. 2B), as did the insulin receptor (IR), as has been previously reported (23). Of note, we detected significantly enhanced S-nitrosylation of IRE1α in the livers of both ob/ob mice and HFD-induced obese mice compared with lean controls (Fig. 2B and fig. S2C). These data identified IRE1α as a target for S-nitrosylation and raised the possibility that alterations in IRE1α function in the obese liver may result from its modification by this mechanism.
Fig. 2. S-Nitrosylation of IRE1α results in decreased IRE1α RNase activity.
(A) General SNO profile in livers of ob/ob mice (obese) versus lean controls. The S-nitrosylated proteins were purified by a biotin-switch method. SNAP was used as a positive (+) control, and a sample without ascorbic acid treatment served as a negative (−) control. Data are representative of results from four independent cohorts of mice. (B) Specific SNO proteins in the livers of obese (ob/ob) and lean mice. The S-nitrosylated proteins were purified by a biotin-switch method and detected by Western blot analysis. (C) In vitro XBP1 splicing assay in IRE1α−/− MEFs reconstituted with IRE1α. The IRE1α protein levels are shown in the gel (top). NMMA is an iNOS inhibitor. AU, arbitrary units. Asterisk (*) indicates statistical significance compared with control, and # indicates statistically significant differences between treatments (one-way ANOVA followed by post hoc Tukey's test). Data are representative of three individual experiments. (D) IRE1α−/− MEFs reconstituted with IRE1α in the presence or absence of iNOS were treated with 10 ng/ml TNFα overnight. A biotin switch was performed, and samples were blotted with the FLAG-specific antibody. Data are representative of two individual experiments. (E) MS analysis of S-nitrosylated peptide 1, which includes the C931 in the RNase domain of human IRE1α. Shown is unmodified peptide 1, single-charged M+ (m/z 1718.8490), and the peptide after GSNO modification (M+NO)+ (m/z 1747.8405). Nitrosylation of peptide 1 results in a m/z change of +28.9915. (Top) A scheme of IRE1α protein structure is shown. (F) qRT-PCR for sXBP1 in IRE1α-deficient primary hepatocytes reconstituted with WT IRE1α, or nitrosylation-resistant IRE1α (IRE1-M1+2) with mutations in two SNO sites in the RNase domain, or IRE1α with a mutation in its RNase domain (IRE1-RD). The cells were treated with 10 ng/ml TNFα overnight in the presence or absence of Tg (100 nM, 2 hours). Results are presented as sXBP1/IRE1α expression levels in each group normalized to controls. (Top) The IRE1α protein levels are shown. All data are shown as means ± SEM. Asterisk (*) indicates statistical significance compared with control, and # indicates statistically significant differences between treatments (one-way ANOVA followed by post hoc Tukey's test, n = 4). *P < 0.05; #P < 0.05.
To examine the direct effect of NO production on IRE1α-mediated XBP1 splicing, IRE1α−/− mouse embryonic fibroblasts (MEFs) were reconstituted with FLAG-tagged human IRE1α, followed by treatment with tumor necrosis factor-α (TNFα), a key inflammatory mediator in the induction of NO synthesis. TNFα pretreatment significantly dampened Tg-induced production of sXBP1 (fig. S2D). However, the TNFα-mediated suppression of XBP1 splicing was relieved when the experiment was performed in the presence of an iNOS inhibitor, NG-monomethyl-l-arginine (NMMA) (fig. S2D). Reciprocally, NO production by the chemical NO donor S-nitrosoglutathione (GSNO) resulted in decreased ER stress–induced XBP1 splicing (fig. S2E). To confirm that the NO-mediated decrease in XBP1 splicing under ER stress conditions was caused by impaired IRE1α ribonuclease activity, we performed an XBP1 in vitro splicing assay using IRE1α purified from cells treated with Tg and/or TNFα, and in the absence or presence of an iNOS inhibitor. IRE1α from Tg-treated cells mediated robust XBP1 splicing in vitro, whereas this XBP1 splicing activity was significantly inhibited by TNFα treatment and rescued by inhibition of iNOS (Fig. 2C). We further examined the S-nitrosylation state of IRE1α in the presence of NO. First, IRE1α−/− MEFs were reconstituted with FLAG-tagged IRE1α; the cells were then treated with TNFα in the presence or absence of iNOS or iNOS inhibitor, followed by a biotin switch assay. TNFα treatment resulted in S-nitrosylation of IRE1α, which was further enhanced by iNOS overexpression and reduced by iNOS inhibition (Fig. 2D). Furthermore, in vivo, iNOS suppression in liver tissue of ob/ob mice led to decreased S-nitrosylated IRE1α, which was associated with enhanced expression of Grp78 and reduced levels of CHOP expression and IRE1α and JNK phosphorylation, which indicated overall improvement in ER function and reduced ER stress (fig. S2, F and G). These data demonstrated that increased iNOS activity in cells results in IRE1α nitrosylation and inhibition of its ribonuclease activity, which may contribute to the decline in ER function in the presence of an inflammatory signal.
We next sought to identify the relevant S-nitrosylated cysteine residues in human IRE1α, focusing on Cys931 (C931) and Cys951 (C951), the cysteines in the RNase domain. In the presence of a chemical NO donor, GSNO, mass spectrometry (MS) analysis revealed S-nitrosylation of both (C931) and (C951) on human IRE1α peptides (Fig. 2E and fig. S2H). We generated a variant IRE1α with alanine substitutions at these residues to prevent S-nitrosylation (IRE1-M1+2), and introduced this variant into primary hepatocytes isolated from mice with hepatocyte-specific IRE1α deletion (24), along with wild-type (WT) IRE1α and an IRE1α with a mutation in its RNase domain (IRE1-RD). Treatment with Tg induced XBP1 splicing in cells reconstituted with the WT IRE1α, but not in cells expressing IRE1-RD (Fig. 2F). Consistent with our previous findings, in cells expressing the WT IRE1α, TNFα treatment resulted in a significant decrease in Tg-induced XBP1 splicing. However, this inhibitory effect of TNFα on ER stress–induced XBP1 splicing was ameliorated in hepatocytes expressing the IRE1-M1+2 variant (Fig. 2F), which likely reflects an improved adaptive capacity in these cells. We also reproduced these results in IRE1α−/− MEFs reconstituted with WT IRE1α or IRE1α with single substitutions in the same two nitrosylation sites (fig. S2I). Taken together, these experiments in two cellular systems clearly demonstrate that IRE1α S-nitrosylation results in diminished ribonuclease activity and that this inhibition could be prevented by the mutation of target residues in vitro.
S-Nitrosylation impairs IRE1α oligomerization and RNase activity
To gain insights into how S-nitrosylation affects IRE1α RNase activity, we next modeled the relevant residues on the previously published crystal structure of human IRE1α (PDB 3P23) using the “S-nitrosator” Python script (25). As shown in Fig. 3A, C931 and C951 are exposed to a solvent-accessible area of the protein complex, where they are available for S-nitrosylation. Furthermore, because C931 and C951 are equally proximal to the RNA cleavage site and the protein dimerization interface, this modification could potentially prevent binding of another negatively charged molecule (such as XBP1 RNA) or could alter the oligomerization of IRE1α. Either of these outcomes may compromise IRE1α splicing activity (26) and provide a mechanistic explanation for the reduction of XBP1 splicing.
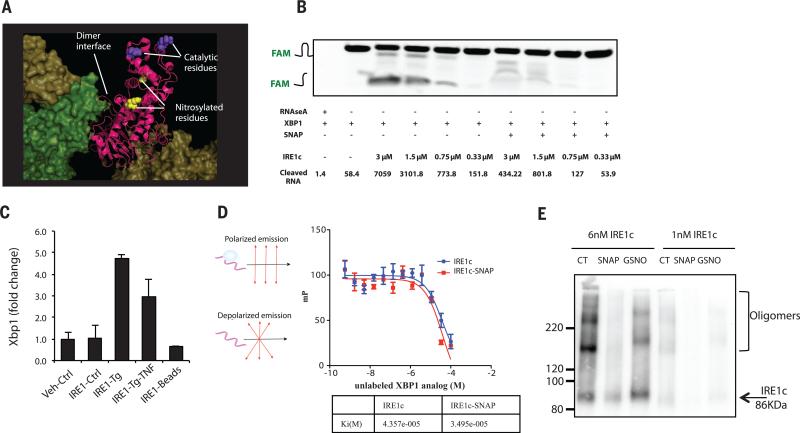
Fig. 3. S-Nitrosylation of IRE1α RNAse domain results in impaired XBP1 processing.
(A) Nitrosylation on C931 and C951 was modeled onto the crystal structure of human IRE1α (PDB 3P23). (B) Urea polyacrylamide gel electrophoresis of sXBP1 substrate (labeled with FAM at the 5′ end) cleaved by different doses of IRE1c in the presence or absence of the chemical NO donor, SNAP (5 mM). RNase A was used as a control. The quantification of cleaved RNA is shown as band intensity (INTmm2). Data are representative of three individual experiments. (C) CLIP assay detecting interaction between IRE1α and unspliced XBP1. IRE1α−/− MEFs were reconstituted with WT IRE1α, followed by treatment with Tg in the absence or presence of TNFα. After UV cross-linking, the IRE1α complexes were immunoprecipitated, and unspliced XBP1 was detected by qRT-PCR. Data are presented as means ± SEM, with n = 6. Data are representative of four individual experiments. (D) Binding of an RNase-resistant analog of XBP1 RNA, HP21 (FAM-dCdCdGdCdAdG) with IRE1c in the absence or presence of NO donor (35) was analyzed by fluorescence polarization assay using a nonfluorescent HP21 as a competitor. The data are presented as units of millipolarization (mP), and the inhibition constant (Ki) is shown (bottom). Data are representative of three individual experiments. (E) IRE1c oligomerization in the absence or presence of NO donors, SNAP (5 mM) or GSNO (0.25 mM). Data are representative of three individual experiments. *P < 0.05; #P < 0.05.
We then performed an in vitro cleavage assay to detect the direct effect of S-nitrosylation on IRE1α-mediated XBP1 processing, using a purified cytosolic portion of the human IRE1α (amino acids 465–977, IRE1c) and a fluorescence resonance energy transfer (FRET)–quenched XBP1 RNA minisubstrate (27). Chemical NO donor treatment impaired IRE1c-mediated XBP1 cleavage in a dose-dependent manner (Fig. 3B and fig. S3, A and B). In addition to the unique, unconventional splicing of XBP1, IRE1α RNase activity is also directed toward other substrates, including a process known as IRE1-dependent decay of mRNA (RIDD) (28). In an in vitro cleavage assay, we found that S-nitrosylation resulted in impaired cleavage of a FRET-RIDD RNA substrate, namely, secreted protein acidic and rich in cysteine (SPARC) (29), which suggests that in vitro, nitrosylation may result in a general impairment in IRE1α RNase enzyme activity (fig. S3C).
We next asked whether S-nitrosylation of IRE1α affected XBP1 recognition using an ultraviolet (UV) cross-linking and immunoprecipitation assay (CLIP) (30). IRE1α−/− MEFs were first reconstituted with WT or S-nitrosylation–resistant IRE1α, followed by treatment with Tg in the absence or presence of TNFα. After UV cross-linking, the IRE1α complexes were immunoprecipitated, followed by RNA isolation, and the unspliced XBP1 abundance was quantified by reverse transcription polymerase chain reaction (qRT-PCR). Tg treatment induced IRE1α interaction with uXBP1, and this was slightly (but not significantly) decreased by TNFα treatment (Fig. 3C and fig. S3D). As a complimentary approach, we used a fluorescence polarization assay (31) to address this same question. In this experimental setting, we assessed the physical interaction between IRE1α and an RNase-resistant RNA analog of HP21 RNA (dCdCdGdCdAdG) (32). NO donor treatment did not alter the C-terminal fragment of IRE1 (IRE1c) binding to HP21 RNA (Fig. 3D). Similarly, nitrosylation did not alter IRE1c recognition of a 2′-deoxy oligonucleotide bearing the RIDD consensus sequence (A.A.A.A.A.U.dG.dC.A.A.A.A.A) (fig. S3E) (33). Taken together, these data did not support the presence of a significant defect in substrate recognition by nitrosylated IRE1α under the experimental conditions tested.
It has been proposed that oligomerization of the ER-lumenal and cytoplasmic domains of IRE1α are important for IRE1α RNase activation (34), the IRE1α RNase output, and consequently the determination of cell fate (35). There is also evidence showing that the IRE1α RNase output can be directed toward XBP1 splicing or RIDD by IRE1α kinase inhibition (36) or alteration of oligomerization (35). As described above, the nitrosylation of cysteine residues could modify oligomerization and could lead to reduced general or substrate-specific IRE1α RNase activity. Indeed, in vitro, NO treatment of IRE1c decreased protein oligomerization (Fig. 3E). In this experiment, we also noted altered IRE1c mobility in the presence of GSNO treatment, which we reason might be caused by additional modification, such as S-glutathionylation, which can also occur after GSNO treatment (37). A similar pattern of reduced higher-molecular-weight oligomers were also observed in obese liver tissue compared with lean controls. Last, it has been suggested that several IRE1α protein partners, such as Bcl2 family proteins and Hsp90 could alter IRE1α-mediated XBP1 splicing (38). However, we did not observe significant changes in these interactions in the presence of NO inducers (fig. S3F). In summary, these data suggest that S-nitrosylation of IRE1α does not impair recognition of substrates or interaction with protein partners, but does alter oligomer formation, ultimately decreasing RNase activity toward XBP1 (fig. S3G).
S-Nitrosylation of IRE1α contributes to the obesity-induced decline in glucose homeostasis
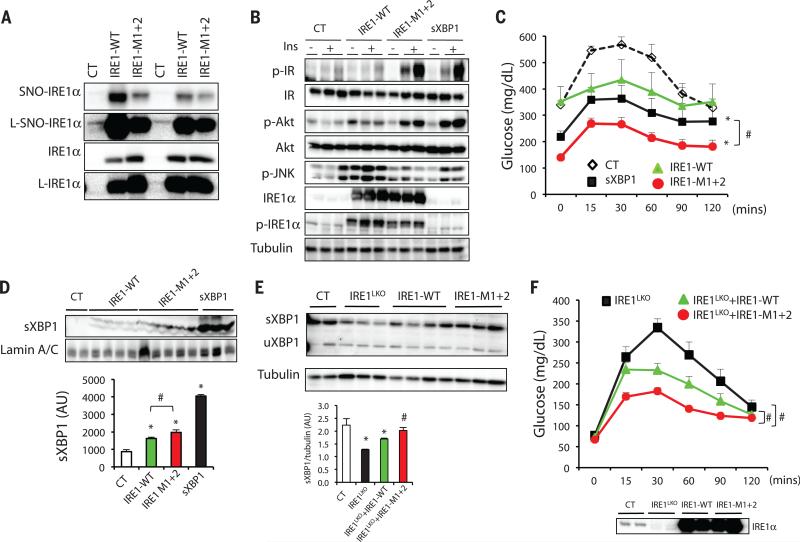
Our experiments demonstrate that inflammation-related nitrosylation impairs IRE1α splicing activity. To more definitively determine whether this mechanism underlies the metabolic decline observed in obesity and whether inhibiting IRE1α nitrosylation could result in metabolic benefit, we exogenously expressed WT IRE1α, as well as the nitrosylation-resistant IRE1-M1+2 variant, in the liver of ob/ob mice. We reasoned that in the obese liver, exogenous WT IRE1α would be subject to nitrosylation and would be compromised in its ability to produce sufficient sXBP1, whereas the IRE1α variant would be resistant to modification by NO and therefore would exhibit positive metabolic activity. We first validated the S-nitrosylation status of the exogenously expressed IRE1α in primary hepatocytes from these mice by performing a biotin switch assay. There was a marked reduction in the S-nitrosylation of IRE1α in hepatocytes expressing IRE1-M1+2 variant compared with those expressing the WT form of IRE1α (Fig. 4A). We found that expression of WT IRE1α did not lead to an improvement in hepatic insulin sensitivity, whereas expression of the S-nitrosylation–resistant IRE1α mutant enhanced hepatic insulin action in ob/ob mice as measured by insulin receptor and Akt phosphorylation (Fig. 4B and fig. S4A). In addition, expression of the nitrosylation-resistant IRE1α mutant resulted in decreased JNK and IRE1α phosphorylation compared with the WT IRE1α, which indicated an improvement in hepatic ER stress in these animals. In glucose tolerance tests, ob/ob mice expressing the S-nitrosylation–resistant form of IRE1α exhibited a marked improvement compared with controls (Fig. 4C). There was also a modest improvement in glucose metabolism in mice expressing the WT IRE1α compared with controls. This effect may reflect residual IRE1α activity resulting from interactions between endogenous and exogenous molecules (34), which could partially overcome the S-nitrosylation–mediated suppression of IRE1α. Although we found a similar increase in hepatic insulin action upon expression of sXBP1 (Fig. 4B), we observed that expression of the nitrosylation-resistant form of IRE1α resulted in a greater improvement in glucose tolerance compared with sXBP1 expression. XBP1 is a dynamically regulated transcription factor involved in many aspects of ER function, and thus, this overexpression system may not recapitulate the physiological regulation and feedback patterns. In addition, expression of nitrosylation-resistant IRE1α may result in changes that are independent of XBP1 splicing. Of note, the level of nuclear sXBP1 was significantly increased in the liver of ob/ob mice overexpressing the mutant IRE1α compared with those expressing the WT IRE1α (Fig. 4D). These experiments clearly demonstrate that expression of a nitrosylation-resistant IRE1α or replenishment with spliced XBP1 can restore insulin action and glucose tolerance in obese animals.
Fig. 4. Effects of IRE1αa S-nitrosylation on insulin action and glucose homeostasis.
(A to D) ob/ob mice were transduced with adenovirus containing sXBP1, WT IRE1α or nitrosylation-resistant IRE1-M1+2. (A) S-Nitrosylation of IRE1α in primary hepatocytes from these mice. The nitrosylation state of IRE1α was examined by biotin switch assay. L, longer exposure time. Data are representative of two individual mouse cohorts. (B) Hepatic insulin action and UPR status. N = 6 to 7, data are representative of two individual cohorts of mice. (C) Glucose tolerance test. All data are presented as means ± SEM, with statistical analysis of the area under the curve (AUC) performed by two-way ANOVA with post hoc Bonferroni test. Asterisk (*) indicates statistical significance compared with control, and # indicates differences between sXBP1 and IRE1-M1+2 expressing mice. Data are representative of two individual cohorts of mice [n = 8 (10-week-old mice)]. (D) sXBP1 expression in the nuclear fraction of liver, with densitometric analysis shown at the bottom of panel. Asterisk (*) indicates statistical significance compared with control, and # indicates statistically significant differences between IRE1-WTand IRE1-M1+2 (one-way ANOVA followed by post hoc Tukey's test). Data are representative of two individual mouse cohorts. (E and F) IRE1α-floxed mice were transduced with Ad-LacZ (Control) or Ad-Cre to delete IRE1α (IRE1LKO), followed by expression of IRE1-WT or the IRE1-M1+2 variant. (E) Hepatic expression of sXBP1 and uXBP1 in the liverof these mice. Quantification of sXBP1/tubulin ratio is shown below. Asterisk (*) indicates statistical significance compared with control, and # indicates statistically significant differences between IRE1-WT and IRE1-M1+2 (one-way ANOVA followed by post hoc Tukey's test). Data are representative of two independent sets of mice. (F) Glucose tolerance test of the mice shown in (E). (Bottom) The IRE1α expression levels are shown [n = 8 (9-week old mice)]. Data are presented as means ± SEM, with statistical analysis of AUC performed by two-way ANOVA with post hoc Bonferroni test. Number sign (#) indicates statistically significant differences comparing IRE1LKO+IRE-M1+2 with IRE1LKO+IRE1-WT, as well as IRE1LKO+IRE1-WT with IRE1LKO. Data are representative of two individual cohorts of mice. *P < 0.05; #P < 0.05.
Finally, considering the possibility that this experimental setting may not rule out the potential influence of endogenous IRE1α expression in obese mice, we tested the function of nitrosylation-resistant IRE1α on XBP1 splicing and glucose metabolism in the absence of endogenous IRE1α in vivo. For this, we first deleted endogenous IRE1α using adenoviral expression of Cre in IRE1α -floxed mice (IRE1LKO), followed by restoration of hepatic IRE1α with either WT IRE1α or the IRE1-M1+2 variant. Whereas overexpression of WT IRE1α resulted in a low level of XBP1 splicing, the expression of sXBP1 and its downstream genes was greatly enhanced in mice expressing the nitrosylation-resistant variant (Fig. 4E and fig. S4B). Notably, in this system, residual endogenous IRE1α expression did contribute to a low level of sXBP1 production in IRE1LKO mice. Unlike the in vitro cleavage assay performed on one synthetic target (fig. S3C), among endogenous RIDD target genes we tested, we did not find significant differences in gene expression when WT IRE1α or the nitrosylation-resistant variant was expressed (fig. S4C), which might be due to the complexity of regulation of these genes in vivo. Deletion of IRE1α in liver resulted in a modest impairment of glucose homeostasis compared with control mice with intact endogenous IRE1α or IRE1α-deficient mice restored with WT IRE1α, as determined by glucose tolerance tests (Fig. 4F). Remarkably, expression of the IRE1-M1+2 variant in liver of IRE1α-deficient mice resulted in the greatest improvement in glucose homeostasis compared with all other groups (Fig. 4F). This improvement was accompanied by decreased JNK phosphorylation and increased expression of chaperones Hsp90 and Grp78 (fig. S4, D and E). Last, we observed an increase in IRE1α oligomerization in the livers from IRE1LKO mice with restoration of the IRE1-M1+2 compared with WT IRE1α (fig. S4F). Although we cannot rule out the possibility of additional effects of the cysteine to alanine mutations in IRE1α, taken together these data demonstrate that prevention of S-nitrosylation of IRE1α at the RNase domain improves hepatic IRE1α-mediated XBP1 splicing and glucose homeostasis.
Discussion
In this study, we have defined a mechanism through which obesity-related chronic inflammation cripples the most conserved branch of the UPR and impairs ER function. Increased hepatic iNOS action results in S-nitrosylation of IRE1α, which in turn results in the inhibition of its ribonuclease, but not kinase, activity and prevents the generation of sufficient levels of sXBP1 while promoting high levels of JNK activity. The data presented here indicate that iNOS-mediated nitrosylation of IRE1α is a critical component contributing to impairment of the UPR and prolonged ER stress in the presence of chronic metabolic and inflammatory stress. Consequently, reversal of this sequence of events, at the level of iNOS, sXBP1 production, or IRE1α nitrosylation, all promote ER homeostasis and result in substantial metabolic benefits. It has been reported that obesity results in decreased sXBP1 nuclear translocation or stability (4, 39, 40); our observations provide a molecular mechanism underlying the defective hepatic XBP1 splicing in obesity, which may be a critical determinant of ER function in metabolic homeostasis.
Previous studies have demonstrated that the N terminus of IRE1α senses stress signals from the ER lumen, which leads to autophosphorylation, which, in turn, activates the C-terminal RNase domain to catalyze site-specific RNA cleavage (1). Furthermore, IRE1α RNase activity can be either up- or down-regulated through selective targeting of its kinase domain by chemical inhibition (1, 41, 42). Ghosh et al. showed that the type II IRE1 kinase inhibitor KIRA6 blocks IRE1α RNAse activity through inhibiting oligomerization, which protects against β cell degeneration and ameliorates hyperglycemia in the Akita mouse model (35). Our findings suggest that S-nitrosylation of IRE1α uncouples kinase and RNase functions of the protein and that inhibition of S-nitrosylation of IRE1α results in enhanced XBP1 splicing, reduced JNK activity, and improved glucose homeostasis in ob/ob mice. Therefore, the regulation of the dual functionalities of IRE1α is complex and multitiered and that the contribution of sXBP1-mediated gene expression, as well as JNK activation, to metabolic homeostasis is fundamentally different between these two mouse models. IRE1α kinase inactivation is predicted to block JNK activation, which may exert a dominant protective effect in the Akita model, which is marked by β cell death (36, 43). On the other hand, preserving the IRE1α-XBP1 axis and, consequently, proper ER homeostasis also leads to suppression of JNK in the liver and recovery of metabolic responses (8, 44).
Finally, our findings indicate that the inflammatory pathways activated in obesity are intimately linked to the UPR and provide strong evidence that inflammatory mechanisms may act upstream of ER dysfunction by compromising the adaptive UPR in conditions of chronic metabolic excess and stress. It will be interesting to examine these interactions in additional tissues that exhibit metaflammation, such as adipocytes in obesity. We suggest that there may be important translational implications of these findings in designing therapeutic interventions against immunometabolic diseases, such as obesity and diabetes, as well as other chronic inflammatory conditions that feature ER dysfunction, to enable the physiological tone of the ER adaptive responses rather than artificially introducing individual components.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Hotamisligil laboratory for their contributions and discussions. We are especially grateful to K. C. Claiborn, A. Tirosh, G. Tuncman, and S. Widenmaier for their critical reading of the manuscript; T. Nakamura (University of Cincinnati) for providing the IRE1-RD construct; and P. Li (NIH) and Y. Lee for scientific discussions and insights. We thank A. Marette (Laval University, Canada) for providing materials for the preliminary experiments and discussions; T. Iwawaki (Gunma University, Japan) for the generous gift of the conditional IRE1α-deletion mouse model; F. Urano (Washington University in St. Louis) for providing the IRE1α−/− MEFs and constructs; R. Prywes (Columbia University) for providing the FLAG-IRE1α construct; and M. Talipov, M. Ivanov, and Q. Timerghazin (Marquette University) for assistance in computational modeling of nitrosylated IRE1α. L.Y. is supported by American Heart Association Postdoctoral Fellowship. This work is supported in part by a grant from NIH to G.S.H. (DK052539).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/349/6247/500/suppl/DC1
Materials and Methods
Figs. S1 to S4
References (45–53)
REFERENCES AND NOTES
- 1.Ron D, Walter P. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 2.Hotamisligil GS. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin JH, et al. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Vera L, Fischer WH, Montminy M. Nature. 2009;460:534–537. doi: 10.1038/nature08111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engin F, et al. Sci. Transl. Med. 2013;5:211ra156. doi: 10.1126/scitranslmed.3006534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidrauski C, Walter P. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 7.Lee AH, Iwakoshi NN, Glimcher LH. Mol. Cell. Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozcan U, et al. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 9.Sreejayan N, Dong F, Kandadi MR, Yang X, Ren J. Obesity (Silver Spring) 2008;16:1331–1337. doi: 10.1038/oby.2008.217. [DOI] [PubMed] [Google Scholar]
- 10.Mao T, et al. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15852–15857. doi: 10.1073/pnas.1107394108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura T, et al. Cell. 2010;140:338–348. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, et al. Cell. 2008;135:61–73. doi: 10.1016/j.cell.2008.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lumeng CN, Saltiel AR. J. Clin. Invest. 2011;121:2111–2117. doi: 10.1172/JCI57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregor MF, Hotamisligil GS. Annu. Rev. Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 15.Kaneki M, Shimizu N, Yamada D, Chang K. Antioxid. Redox Signal. 2007;9:319–329. doi: 10.1089/ars.2006.1464. [DOI] [PubMed] [Google Scholar]
- 16.Noronha BT, Li JM, Wheatcroft SB, Shah AM, Kearney MT. Diabetes. 2005;54:1082–1089. doi: 10.2337/diabetes.54.4.1082. [DOI] [PubMed] [Google Scholar]
- 17.Perreault M, Marette A. Nat. Med. 2001;7:1138–1143. doi: 10.1038/nm1001-1138. [DOI] [PubMed] [Google Scholar]
- 18.Uehara T, et al. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 19.Derakhshan B, Wille PC, Gross SS. Nat. Protoc. 2007;2:1685–1691. doi: 10.1038/nprot.2007.210. [DOI] [PubMed] [Google Scholar]
- 20.Jaffrey SR, Snyder SH. Sci. STKE. 2001;2001:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 21.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 22.Murray CI, Uhrigshardt H, O’Meally RN, Cole RN, Van Eyk JE. Mol. Cell. Proteomics. 2012;11:M111.013441. doi: 10.1074/mcp.M111.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carvalho-Filho MA, et al. Diabetes. 2005;54:959–967. doi: 10.2337/diabetes.54.4.959. [DOI] [PubMed] [Google Scholar]
- 24.Iwawaki T, Akai R, Kohno K. PLOS ONE. 2010;5:e13052. doi: 10.1371/journal.pone.0013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talipov MR, Timerghazin QK. J. Phys. Chem. B. 2013;117:1827–1837. doi: 10.1021/jp310664z. [DOI] [PubMed] [Google Scholar]
- 26.Lee KP, et al. Cell. 2008;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiseman RL, et al. Mol. Cell. 2010;38:291–304. doi: 10.1016/j.molcel.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hollien J, et al. J. Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maurel M, Chevet E, Tavernier J, Gerlo S. Trends Biochem. Sci. 2014;39:245–254. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Tollervey J, Briese M, Turner D, Ule J. Methods. 2009;48:287–293. doi: 10.1016/j.ymeth.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 31.Pagano JM, Clingman CC, Ryder SP. RNA. 2011;17:14–20. doi: 10.1261/rna.2428111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korennykh AV, et al. BMC Biol. 2011;9:47. doi: 10.1186/1741-7007-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han Y, et al. Science. 2014;343:1244–1248. doi: 10.1126/science.1249845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korennykh AV, et al. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh R, et al. Cell. 2014;158:534–548. doi: 10.1016/j.cell.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han D, et al. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martínez-Ruiz A, Lamas S. Cardiovasc. Res. 2007;75:220–228. doi: 10.1016/j.cardiores.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 38.Hetz C. Nat. Rev. Mol. Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 39.Park SW, et al. Nat. Med. 2010;16:429–437. doi: 10.1038/nm.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winnay J, et al. Nat. Med. 2010;16:438–445. doi: 10.1038/nm.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prischi F, Nowak PR, Carrara M, Ali MM. Nat. Commun. 2014;5:3554. doi: 10.1038/ncomms4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, et al. Nat. Chem. Biol. 2012;8:982–989. doi: 10.1038/nchembio.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urano F, et al. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 44.Hirosumi J, et al. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.