Abstract
Restores TBP function 1 (Rtf1) is generally considered to be a subunit of the Paf1 complex (PAF1C), a multifunctional protein complex involved in histone modification and transcriptional or posttranscriptional regulation. Rtf1, however, is not stably associated with the PAF1C in most species except Saccharomyces cerevisiae, and its biochemical functions are not well understood. Here, we show that human Rtf1 is a transcription elongation factor that may function independently of the PAF1C. Rtf1 requires “Rtf1 coactivator” activity, which is most likely unrelated to the PAF1C or DSIF, for transcriptional activation in vitro. A mutational study revealed that the Plus3 domain of human Rtf1 is critical for its coactivator-dependent function. Transcriptome sequencing (RNA-seq) and chromatin immunoprecipitation studies in HeLa cells showed that Rtf1 and the PAF1C play distinct roles in regulating the expression of a subset of genes. Moreover, contrary to the finding in S. cerevisiae, the PAF1C was apparently recruited to the genes examined in an Rtf1-independent manner. The present study establishes a role for human Rtf1 as a transcription elongation factor and highlights the similarities and differences between the S. cerevisiae and human Rtf1 proteins.
INTRODUCTION
Restores TBP (TATA box-binding protein) function 1 (Rtf1) was identified as a suppressor of a TBP mutant in Saccharomyces cerevisiae (1). Subsequent genetic and biochemical studies in yeast have shown that Rtf1 functions as a component of the polymerase-associated factor 1 (Paf1) complex (PAF1C) containing Paf1, Ctr9, Leo1, and Cdc73 (2–5). The PAF1C is a multifunctional protein complex whose primary role is to facilitate histone modifications, such as H2B monoubiquitination at K123 and H3 methylation at K4 and K79 (6–12). The PAF1C also plays important roles in transcription elongation through chromatin, as well as on naked DNA (13–16). Furthermore, the PAF1C is involved in transcription termination and 3′ processing of polyadenylated and nonpolyadenylated Pol II transcripts (17–25).
Paf1, Ctr9, Leo1, Cdc73, and Rtf1 are highly conserved in eukaryotes; however, the subunit compositions of the PAF1C vary among species. Purification of the PAF1C from human HeLa cells yielded a five-subunit complex lacking Rtf1 but containing Ski8 as an additional subunit (22, 26, 27). Similarly, coimmunoprecipitation studies in zebrafish, Drosophila, and Schizosaccharomyces pombe showed that PAF1C homologs in these species lack a stably associated Rtf1 subunit (28–30). At the functional level, however, Rtf1 and other PAF1C subunits have a number of similarities. For example, knockout or knockdown of Rtf1 and other PAF1C subunits results in similar defects in histone modifications in all the species examined (27, 28, 31). At the organismal level, inhibition of Rtf1 and other PAF1C subunits causes similar defects in epidermal morphogenesis in Caenorhabditis elegans and in somitogenesis and cardiomyocyte development in zebrafish (29, 32, 33). Rtf1 and Paf1 colocalize with each other and with RNA polymerase II (RNAPII) in Drosophila polytene chromosomes (28). Thus, in most species except S. cerevisiae, Rtf1 is not stably associated with the PAF1C, although it is functionally similar to the PAF1C in many respects. To indicate the species-specific differences, only Rtf1 from S. cerevisiae is referred to as a PAF1C subunit in this paper.
Little is known about the role of Rtf1 in transcription in higher eukaryotes. In in vitro transcription assays using S. cerevisiae cell extracts and a naked DNA template, Rondon et al. (16) showed that paf1Δ and cdc73Δ cell extracts were defective in elongation, whereas rtf1Δ and leo1Δ cell extracts were not; however, recent papers have shown that S. cerevisiae Rtf1 is critical for the recruitment of the PAF1C (20, 34, 35). The conserved Plus3 domain of Rtf1 interacts with the phosphorylated form of Spt5, the large subunit of DSIF, and thereby recruits the other PAF1C subunits to actively transcribed genes (5, 35–39). In higher eukaryotes, we used a human in vitro transcription system to show that the PAF1C cooperates with DSIF and Tat-SF1 to promote transcription elongation on a naked DNA template (13). The PAF1C lacking Rtf1 was fully active in our transcription assays, and the inclusion of Rtf1 had no discernible effect on the synergistic action of the PAF1C, DSIF, and Tat-SF1 (our unpublished data). In a subsequent study, Kim et al. (14) reconstituted the human PAF1C using recombinant subunits to demonstrate that the PAF1C activates the transcription of a chromatin template in cooperation with TFIIS in vitro. Consistent with our findings, no difference was found between the PAF1C and the PAF1C-Rtf1 complexes in their chromatin transcription assays. Therefore, there is no compelling evidence that metazoan Rtf1 directly controls transcription independently of histone modifications or posttranscriptional regulation.
Our initial goal was to examine the role of human Rtf1 in transcription using in vitro transcription assays. Since efficient transcription elongation was found to be critically dependent on Rtf1 in crude HeLa cell nuclear extracts (NE), we investigated the mechanism of action of Rtf1 and reached an unexpected finding that Rtf1 requires “Rtf1 coactivator” activity, which is most likely unrelated to the PAF1C or DSIF. Mutational studies revealed that the Plus3 domain of human Rtf1 is critical for its coactivator-dependent function. To extend our findings to living cells, gene expression analysis and chromatin immunoprecipitation (ChIP) were performed after knockdown of Rtf1, Paf1, and Ski8. We found that Rtf1 and the PAF1C play distinct roles in the expression of a subset of genes. Moreover, contrary to the findings in S. cerevisiae, the human PAF1C was recruited to the genes examined even when Rtf1 was knocked down. The present study establishes a role for human Rtf1 as a transcription elongation factor that may function independently of the PAF1C. This study also highlights the similarities and differences between the S. cerevisiae and human Rtf1 proteins.
MATERIALS AND METHODS
In vitro transcription assays.
In vitro transcription assays were performed using pSLG402, which contained the adenovirus major late promoter (40), and 2 μl of NE or 6 μl of P1.0 (see below) with or without 0.2 pmol of wild-type or mutant Rtf1. After preincubation of 22-μl reaction mixtures for 40 min, 3 μl of 4 nucleoside triphosphates (NTPs) (30 μM ATP, 300 μM CTP, 300 μM GTP, and 2.5 μM [α-32P]UTP at the respective final concentrations) containing 500 U of RNase T1 was added, and initiation/elongation was allowed to proceed for 20 min unless otherwise stated. The reactions were stopped at the indicated times by the addition of 100 μl of stop buffer (10 mM Tris-HCl, pH 7.5, 0.3 M NaCl, and 5 mM EDTA). The synthesized transcripts were then purified by phenol-chloroform extraction and ethanol precipitation and resolved by 8% urea PAGE. The amounts of radioactivity incorporated were quantified using a Storm 860 phosphorimager (Amersham Pharmacia Biotech).
Primer extension was also performed to analyze transcripts produced from pTF3-6C2AT, the supercoiled plasmid containing the adenovirus E4 promoter, as previously described (41). After the transcription reactions, the purified reaction products were analyzed using one of the following radioactive primers: A, 5′-GAATAATGAGGAAAGGAGAGT-3′; B, 5′-GATGATAGATTTGGGAAATATAA-3′; C, 5′-GGAGAGTAGGGTGGTATAG-3′; or D, 5′-AGGAAACAGCTATGACCATG-3′. The expected lengths of the products were 29 nucleotides (nt), 69 nt, 98 nt, and 432 nt, respectively.
Recombinant proteins.
Human Rtf1 cDNA was prepared by reverse transcription (RT)-PCR from total RNA extracted from HeLa cells and cloned into pFastBac1 with the coding sequence for an N-terminal Flag tag. Coding sequences for full-length Rtf1 and its deletion mutants were subcloned into pET-28c+ for expression of cloned inserts with an N-terminal histidine tag and a C-terminal Flag tag. Recombinant Rtf1 proteins were expressed in Escherichia coli strain BL21 and subjected to tandem-affinity purification using Ni-agarose beads (Qiagen) and M2 Flag-agarose beads (Sigma).
Fractionation of HeLa cell NE.
HeLa cell NE and the phosphocellulose P11 fractions (P.1, P.3, and P1.0) were prepared as previously described (42). P1.0 was dialyzed against HGE (20 mM HEPES, pH 7.9, 20% glycerol, 0.2 mM EDTA, 0.5 mM dithiothreitol) containing 0.1 M KCl (HGE.1; the number following HGE indicates the molar concentration of KCl) and applied to a 50-ml DEAE-Sepharose FF column (Amersham Pharmacia) equilibrated with HGE.1. The column was washed with the same buffer, and proteins were eluted with HGE.225, HGE.3, and HGE1.0. The 0.3 M KCl fraction was dialyzed against HGE.1 before use for in vitro transcription.
Immunological analyses.
Anti-Spt5, anti-Tat-SF1, anti-Leo1, and anti-Cdc73 antibodies were produced in house in rabbits (13). The following commercial antibodies were also used: anti-Rpb1 (Santa Cruz; sc-899), anti-Rpb1 (8WG16), anti-Ctr9 (Bethyl; A301-395A), anti-Rtf1 (c), anti-Paf1 (Bethyl; A300-173A), anti-Paf1 (Abcam; ab20662), anti-Cdc73 (Santa Cruz; sc-33638), anti-Ski8 (Abcam; ab57840), anti-TFIIS (Transduction Laboratories; S84820), anti-H2B (Abcam; ab1790), anti-monoubiquitinated H2B (anti-H2Bub) (Millipore; 05-1312), anti-H3 (Abcam; ab1791), and beta-actin (Abcam; ab6276). Anti-Rpb1 (Santa Cruz; sc-899), anti-Rtf1 (Bethyl; A300-179A), and anti-Paf1 (Abcam; ab20662) antibodies were used for ChIP assays. Immunoprecipitation and immunodepletion were performed as previously described (41, 43).
shRNA-mediated knockdown and RNA analysis.
The following 21-nt sequences were used as short hairpin RNA (shRNA) targets: Rtf1 number 2, 5′-AAGAAUUGAAUCGGGUUCGAU-3′; Ctr9 number 3, 5′-AAGCAGAAGCGGAACAUGAUG-3′; Leo1 number 4, 5′-AAGAGGCAGUGAUAGUGAAGA-3′; Paf1 number 5, 5′-AACCAGUUUGUGGCCUAUUUC-3′; Cdc73 number 1, 5′-AAGUAUAGACAGAAGCGCUCC-3′; and Ski8 number 3, 5′-AGUGGAGCCAUAGAUGGAAUC-3′. Double-stranded oligonucleotides for shRNAs against Rtf1 and Cdc73 were cloned into pBluescript-U6 as previously described (44). After functional validation, cassettes including a mouse U6 promoter were excised and subcloned into pLenti6 (Life Technologies). shRNAs against Ctr9, Leo1, Paf1, and Ski8 were directly cloned into the lentiviral vector pRSI9. Recombinant lentiviruses were produced and concentrated prior to use according to standard procedures. In knockdown experiments, HeLa cells were infected with recombinant lentivirus expressing no shRNA or expressing one of the shRNAs and selected in the presence of 5 μg/ml blasticidin for pLenti6 or 1 μg/ml puromycin for pRSI9. The cells were lysed with high-salt buffer (50 mM Tris-HCl, pH 7.9, 500 mM NaCl, 1% NP-40) for immunoblotting or with Sepasol RNA I Super G (Nacalai Tesque) for RNA analyses.
RNA analyses.
Total RNAs prepared from control HeLa cells and Rtf1, Ski8, and Paf1 knockdown cells were subjected to transcriptome sequencing (RNA-seq) in triplicate. Following the removal of rRNA using the RiboMinus eukaryote kit (Life Technologies), libraries were constructed using the Ion Total RNA-Seq kit and sequenced using the Ion Proton System (Life Technologies). A minimum of 3 million high-quality reads were obtained for each sample. The reads were mapped to the human genome GRch37.p9, and reads per kilobase per million (RPKM) were calculated. The rank product method (RankProd) was used to identify differentially expressed genes using a false-discovery rate (FDR) of 0.05 and a 2.0-fold change as cutoff values. Then, cluster analysis was performed using CLC Genomics Workbench. The average Z scores were calculated for each gene and used to draw a heat map. The heat map (see Fig. 5B) represents the genes differentially expressed in any one of the Rtf1, Ski8, and Paf1 knockdowns. Gene ontology (GO) analysis was performed using DAVID Bioinformatics Resources 6.7 (45).
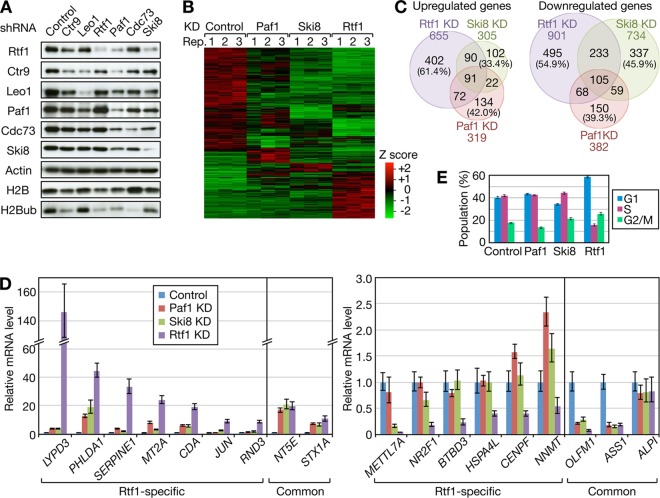
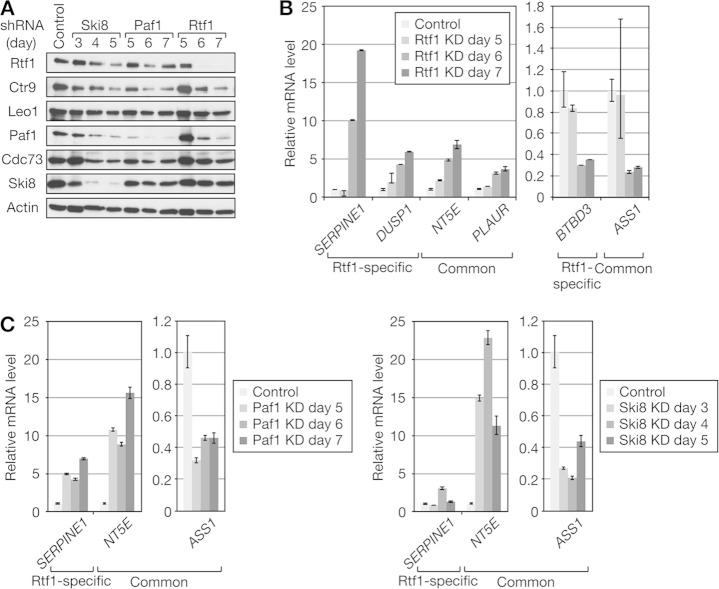
FIG 5.
Comparative analysis of Rtf1- and PAF1C-regulated genes. (A) HeLa cells were infected with lentiviral vectors expressing shRNAs targeting the indicated factors. Four or 7 days postinfection, the cells were harvested for immunoblot analysis. (B) Total RNAs were prepared from Paf1, Ski8, and Rtf1 knockdown (KD) cells and control HeLa cells in triplicate and subjected to RNA-seq. Mean Z scores were calculated for each gene and used to draw the heat map of 2,349 differentially expressed genes. Black indicates the average expression level across 12 samples. Rep, repeat. (C) Venn diagrams comparing the genes affected by Paf1, Ski8, and Rtf1 knockdown. The numbers in parentheses indicate the percentages of genes uniquely regulated by the respective factors. (D) Validation of RNA-seq data. The same set of RNA samples was subjected to qRT-PCR analysis. The averaged expression values for each gene were normalized to the values for GADPH, and the relative expression levels obtained from control knockdown samples were expressed as 1. (E) Knockdown cells were harvested for cell cycle analysis on the same days as RNA-seq analysis. The data are presented as the means ± standard errors of the means (SEM) of the results of 3 independent experiments.
Eighteen genes were selected from the list of differentially expressed genes, and quantitative RT (qRT)-PCR was performed in triplicate using the One Step SYBR PrimeScript Plus RT-PCR kit (TaKaRa). Quantification was performed by the ΔΔCT method using GAPDH (encoding glyceraldehyde-3-phosphate dehydrogenase) as a reference gene.
Cell cycle analysis.
Control and knockdown cells were prepared in triplicate and fixed with 70% cold ethanol in phosphate-buffered saline. After RNase A treatment, the fixed cells were stained with 50 μg/ml propidium iodide. The DNA content measurement and cell cycle analysis were performed using a FACSCalibur flow cytometer and Cell Quest software (Becton Dickinson).
ChIP assays.
ChIP assays were performed as previously described (46). Chromatins were solubilized by micrococcal nuclease digestion, and immunoprecipitation was performed with Dynabeads protein G (Dynal).
RESULTS
Human Rtf1 is a transcription elongation factor.
To characterize human Rtf1, its subcellular localization in HeLa cells was examined by immunostaining. Immunofluorescence signals obtained with anti-Rtf1 antibody perfectly overlapped with those of DAPI (4′,6-diamidino-2-phenylindole) (Fig. 1A), indicating that human Rtf1 is a nuclear protein.
FIG 1.
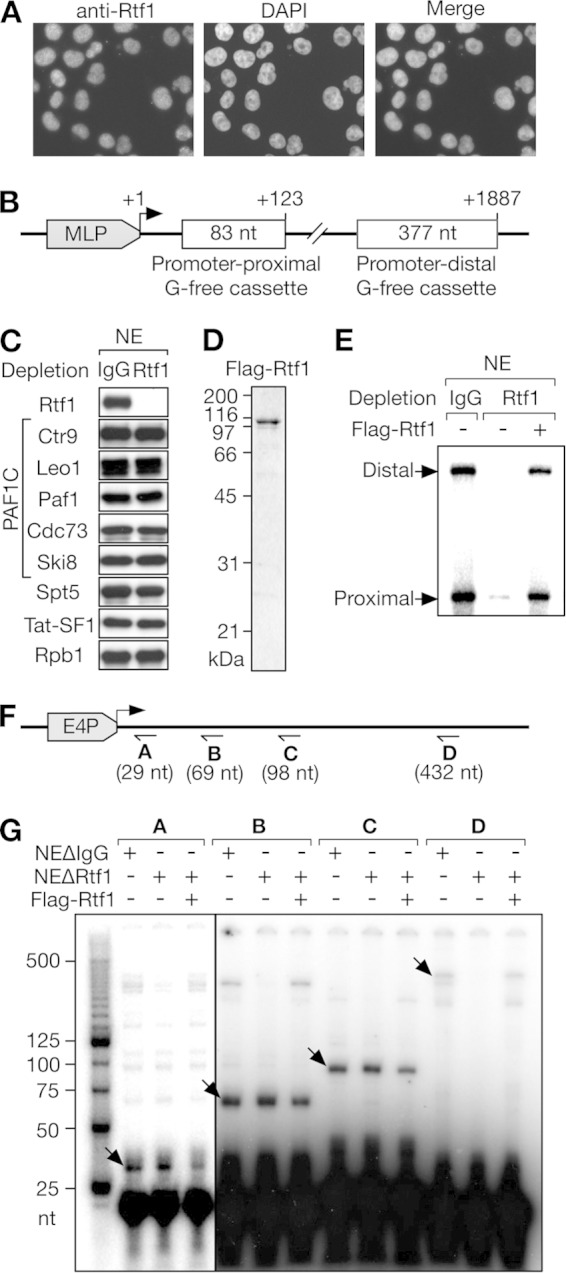
Human Rtf1 is a transcription elongation factor. (A) The subcellular localization of human Rtf1 was examined by immunofluorescence microscopy. HeLa cells were stained with anti-Rtf1 antibody and counterstained with DAPI. (B) Schematic structure of pSLG402, the DNA template containing two G-free cassettes. MLP, adenovirus major late promoter. (C) HeLa cell NE immunodepleted with control IgG or anti-Rtf1 antibody were analyzed by immunoblotting with the indicated antibodies. (D) Coomassie blue-stained Flag-Rtf1 purified from insect cells. (E) In vitro transcription was performed using pSLG402, the NE shown in panel C, and Flag-Rtf1. Promoter-proximal and -distal products are indicated. (F) Schematic structure of the DNA template and the primer-binding sites used in panel G. E4P, adenovirus E4 promoter. (G) Primer extension analysis of the products obtained with the NE immunodepleted with control IgG (NEΔIgG) or anti-Rtf1 (NEΔRtf1). Specific primer extension products are indicated by the arrows.
In vitro transcription assays were performed using crude HeLa cell NE and pSLG402, a naked DNA template containing two G-free cassettes cloned downstream of the adenovirus major late promoter (Fig. 1B). After RNase T1 treatment, 83-nt and 377-nt fragments corresponding to the promoter-proximal and promoter-distal G-free cassettes were generated from the synthesized transcripts. The former represents the efficiency of initiation/early elongation, and the distal-to-proximal ratio represents the efficiency of late elongation. As reported previously by our group (13), HeLa cell NE supported efficient transcription initiation and elongation of pSLG402 (Fig. 1E). To examine the possible role of Rtf1 in transcription, it was depleted from HeLa cell NE using anti-Rtf1 antibody. Rtf1 was efficiently removed from the extract, whereas the levels of all the subunits of the PAF1C, Spt5, Tat-SF1, and Rpb1 were negligibly affected by this procedure (Fig. 1C). Strikingly, Rtf1-depleted NE (NEΔRtf1) exhibited severe transcription defects compared to IgG-depleted control NE (Fig. 1E). Moreover, the addition of Flag-Rtf1 expressed in and purified from insect cells (Fig. 1D) almost fully restored the transcription defects (Fig. 1E). These results demonstrated that human Rtf1 is indispensable for transcription in vitro.
The step of transcription that was affected by Rtf1 depletion was not clear from the results shown in Fig. 1E, because the reduction of promoter-proximal products can be caused by defects in initiation, early elongation, or both. To pinpoint the cause, we analyzed transcripts generated from the adenovirus E4 promoter by primer extension assays as previously described (41). To quantify transcripts of ≥29 nt, ≥69 nt, ≥98 nt, and ≥432 nt, four radiolabeled primers were prepared and hybridized to “cold” runoff products, followed by reverse transcription. As shown in Fig. 1G, the levels of ≥29-nt, ≥69-nt, and ≥98-nt transcripts were negligibly affected by Rtf1 depletion. In contrast, the level of ≥432-nt transcripts was significantly reduced by the immunodepletion and recovered by the addition of Flag-Rtf1 to NEΔRtf1. Since the stimulatory effect of Rtf1 was clearly dependent on transcript length, we concluded that Rtf1 acts only in the elongation phase of transcription in vitro.
Human Rtf1 requires another factor to promote transcription elongation.
To further understand the mechanism of action of Rtf1, we investigated whether Rtf1 requires another factor to promote transcription elongation. To this end, we used a previously described method (13, 41, 42). Briefly, HeLa cell NE was fractionated using a phosphocellulose P11 column into the flowthrough fraction (P0.1), the 0.3 M KCl eluate (P0.3), and the 1.0 M KCl eluate (P1.0) (Fig. 2A). P1.0 contained most of the general transcription factors and Pol II and directed transcription initiation; however, because it lacked several transcription elongation factors, it did not support efficient transcription elongation alone under the conditions used (Fig. 2B and C) (13). Contrary to the observations using NEΔRtf1 (Fig. 1E), the addition of Flag-Rtf1 to P1.0 did not facilitate the efficient synthesis of the promoter-distal region, even at the highest concentration tested (Fig. 2C and 3F), suggesting that another factor is required for Rtf1-mediated transcriptional activation.
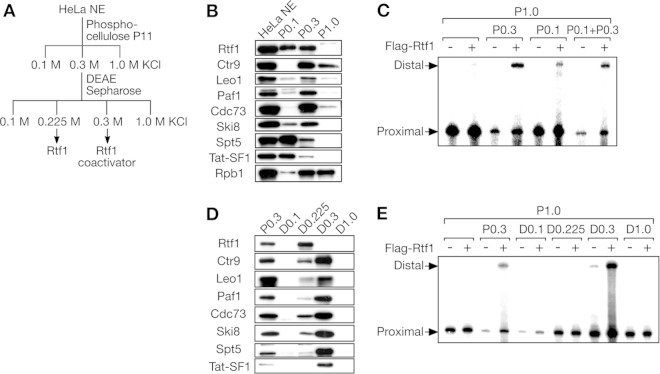
FIG 2.
Human Rtf1 requires another factor to promote transcription elongation. (A) Separation scheme. (B) Immunoblot analysis of HeLa cell NE and phosphocellulose column fractions. (C) In vitro transcription assays of phosphocellulose column fractions. Reaction mixtures containing pSLG402 and the indicated phosphocellulose column fraction(s) were incubated with or without Flag-Rtf1. Promoter-proximal and -distal products are indicated. (D) Immunoblot analysis of DEAE-Sepharose column fractions. P0.3 (input) was analyzed for comparison. (E) In vitro transcription assays of DEAE-Sepharose column fractions. Promoter-proximal and -distal products are indicated.
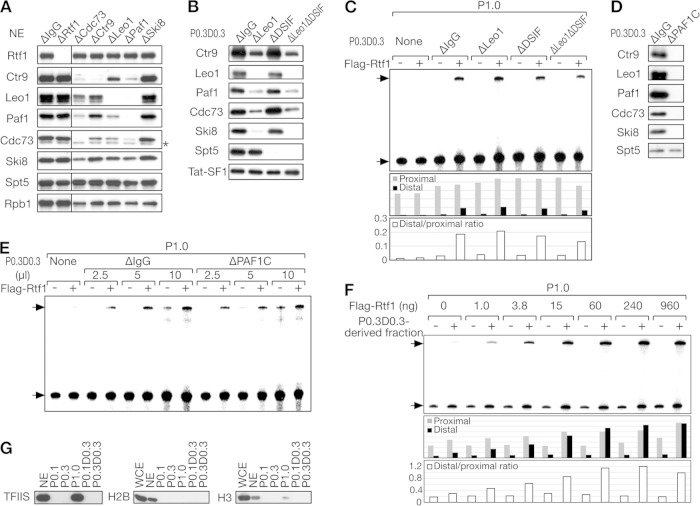
FIG 3.
Human Rtf1 promotes transcription elongation independently of the PAF1C and DSIF. (A) Immunoblot analysis of HeLa cell NE after immunodepletion of one of the indicated factors. Mock-depleted NE (NEΔIgG) were analyzed as a control. The asterisk denotes a nonspecific signal. (B) Immunoblot analysis of the P0.3 D0.3 fraction after immunodepletion of Leo1 and/or DSIF. Mock-depleted P0.3 D0.3 (P0.3 D0.3 ΔIgG) was analyzed as a control. (C) In vitro transcription assays for immunodepleted P0.3 D0.3 fractions. Promoter-proximal and -distal products are indicated by the arrows. The amounts of promoter-proximal and -distal products and their ratios are shown below. (D) Immunoblot analysis of the P0.3 D0.3 fractions immunodepleted with anti-Leo1, anti-Cdc73, and anti-Paf1 antibodies (P0.3 D0.3 ΔPAF1C) or with control IgG. (E) The indicated amounts of P0.3 D0.3 fractions were added with or without Flag-Rtf1 to in vitro transcription assay mixtures. (F) A partially purified Rtf1 coactivator fraction derived from P0.3 D0.3 (P0.3 D0.3 H0.4 S number 5) was added, together with increasing amounts of Flag-Rtf1, to in vitro transcription assay mixtures. The amounts of promoter-proximal and -distal products and their ratios are shown below. (G) Immunoblot analysis of TFIIS and histones H2B and H3.
To search for such an “Rtf1 coactivator,” a limited amount of P0.1 and/or P0.3 was added, together with Flag-Rtf1, to P1.0. As shown in Fig. 2B, Rtf1 was partitioned into P0.1 and P0.3 to similar extents, whereas the PAF1C was largely found in P0.3. In the absence of exogenous Flag-Rtf1, P0.1 and/or P0.3 did not enhance elongation at the concentrations used. On the contrary, P0.3 slightly repressed transcription, possibly because of the enrichment of an inhibitory factor(s) (Fig. 2C). The combination of Flag-Rtf1 and P0.3 significantly increased the promoter-distal products. P0.1 showed a weak coactivation effect with Flag-Rtf1, whereas no further increase was observed in response to the combination of P0.1 and P0.3, suggesting that the same factor was responsible for the effects of P0.1 and P0.3 on Rtf1.
P0.3 was further fractionated on a DEAE-Sepharose column, which yielded the flowthrough fraction (D0.1) and three eluate fractions (D0.225, D.03, and D1.0). At this step, Rtf1 and the PAF1C were largely separated, with Rtf1 exclusively found in D0.225 and the PAF1C mostly detected in D0.3 (Fig. 2D). In regard to other transcription elongation factors, DSIF and Tat-SF1 were largely removed from the active fraction at the phosphocellulose chromatography step (Fig. 2B), whereas a portion of DSIF and Tat-SF1 was cofractionated with the PAF1C and found in D0.3 (Fig. 2D). At the functional level, only D0.3 exhibited strong coactivation with Flag-Rtf1 (Fig. 2E). Incidentally, D0.1 repressed the synthesis of the promoter-proximal region, suggesting that the inhibitory factor(s) found in P0.3 was fractionated into D0.1.
Human Rtf1 promotes transcription elongation independently of the PAF1C and DSIF.
Rtf1 and the Rtf1 coactivator were successfully separated. Based on past studies and the findings shown in Fig. 2, the most likely candidate for the Rtf1 coactivator was the PAF1C. Based on a recent study in S. cerevisiae (35), DSIF might also be a candidate for the Rtf1 coactivator, although DSIF did not comigrate with the coactivator activity at the phosphocellulose step. To explore these possibilities, the PAF1C was immunodepleted from HeLa cell NE using antibodies against the PAF1C. As a result, Cdc73, Ctr9, Leo1, and Paf1, but not Ski8, were efficiently depleted by antibodies against the respective subunits (Fig. 3A). With these antibodies, other subunits of the complex were codepleted to varying degrees, with the Ski8 subunit showing the greatest resistance to codepletion. When anti-Leo1 antibody was used to deplete the PAF1C from the P0.3 D0.3 coactivator fraction, all the PAF1C subunits, including Ski8, were codepleted to a satisfactory level (Fig. 3B). This apparent discrepancy was probably due to the presence of the SKI complex, another Ski8-containing complex. Since the SKI complex was fractionated into P0.1 (data not shown), Ski8 found in P0.3 D0.3 was almost entirely incorporated into the PAF1C and was codepleted with the other PAF1C subunits. With regard to DSIF, it was efficiently and specifically depleted from P0.3 D0.3 using anti-Spt5 antibody (Fig. 3B).
Contrary to our expectations, the coactivator fraction from which Leo1 or DSIF was depleted (P0.3 D0.3 ΔLeo1 or P0.3 D0.3 ΔDSIF) was as efficient as the mock-depleted fraction in stimulating the synthesis of the promoter-distal region (Fig. 3C), suggesting that none of these proteins are responsible for the coactivator activity. To rule out the possibility that the PAF1C and DSIF are functionally redundant and that either can suffice as a coactivator for Rtf1, we codepleted the PAF1C and DSIF from P0.3 D0.3 using a combination of anti-Leo1 and anti-Spt5 antibodies (Fig. 3B). As a result, codepletion of the PAF1C and DSIF did not substantially affect Rtf1-mediated transcriptional activation (Fig. 3C), suggesting that these factors are not responsible for the coactivator activity of P0.3 D0.3.
To eliminate the possibility that residual PAF1C subunits remaining in the depleted fraction supported Rtf1-mediated transcriptional activation, we sought to deplete the PAF1C subunits completely. The combination of anti-Leo1, anti-Cdc73, and anti-Paf1 antibodies resulted in almost complete depletion of the PAF1C from P0.3 D0.3, giving rise to P0.3 D0.3 ΔPAF1C (Fig. 3D). As shown in Fig. 3E, P0.3 D0.3 ΔPAF1C coactivated the synthesis of the promoter-distal region as efficiently as mock-depleted P0.3 D0.3, suggesting that Rtf1 promotes transcription elongation independently of the PAF1C and DSIF in vitro.
To identify the Rtf1 coactivator, P0.3 D0.3 was subjected to heparin-Sepharose chromatography. The resulting active fraction (H0.4) was further resolved on a Mono S column, and the most active fraction (Mono S number 5) was assayed. As shown in Fig. 3F, P0.3 D0.3 H0.4 S number 5 strongly coactivated the synthesis of the promoter-distal region at increasing concentrations of Flag-Rtf1. Moreover, glycerol gradient sedimentation analysis indicated that the apparent molecular mass of the coactivator was 200 kDa (data not shown). Despite various attempts, successful purification and identification of the Rtf1 coactivator have not been achieved.
We performed immunoblot analysis of candidate coactivators. First, Tat-SF1 and TFIIS were investigated because they are known to activate transcription in cooperation with the PAF1C in vitro (13, 14). While a small fraction of Tat-SF1 was found in P0.3 D0.3 (Fig. 2B and D), further purification resulted in separation of the coactivator activity and Tat-SF1 (data not shown). With regard to TFIIS, it was fractionated almost entirely into the P1.0 fraction (Fig. 3G). Since P1.0 was used as a source of the general transcription factors and Pol II in our transcription assays, we cannot exclude the possibility that TFIIS plays a role in Rtf1-dependent activation. It can be said, however, that TFIIS is not responsible for the coactivator activity found in P0.3 D0.3. Considering the tight functional link between Rtf1 and chromatin (9, 14, 34, 38), histones could modulate the transactivation potential of Rtf1 in vitro. Immunoblot analysis showed, however, that P0.3 D0.3 had an undetectable level of histones H2B and H3, and most of the histones were found to be depleted at the phosphocellulose column chromatography step (Fig. 3G). Probably, basic histone molecules were so tightly associated with phosphocellulose that they were barely eluted from the column even at the highest KCl concentration employed. In any case, these results suggest that histones are not involved in the coactivator's function.
Distinct structural requirements for human Rtf1 in H2B monoubiquitination and transcriptional activation in vitro.
Detailed structure-function analysis of S. cerevisiae Rtf1 has shown (i) that its histone modification domain (HMD) is required for H2B monoubiquitination and H3K4 and K79 methylation, (ii) that the Plus3 domain of Rtf1 is required for its interaction with the phosphorylated form of Spt5 and for the recruitment of other PAF1C subunits, and (iii) that the short C-terminal segment of Rtf1 is required for its interaction with the other PAF1C subunits (34). In the present study, we performed similar structure-function analysis of human Rtf1 to understand the structural requirement for its coactivator-dependent function. In addition, we investigated whether the functional domains identified in S. cerevisiae are evolutionarily conserved, because there is at least one major difference between S. cerevisiae and human Rtf1 proteins, i.e., their binding affinities for the PAF1C.
Six deletion mutants of human Rtf1 were constructed for expression in mammalian and bacterial cells (Fig. 4A). HeLa cells were sequentially infected with a lentiviral vector for shRNA targeting Rtf1 and transfected with one of the Rtf1 expression vectors carrying shRNA-resistant mutations. To avoid mislocalization of deletion mutants, the nuclear localization signal from the simian virus 40 (SV40) large T antigen was attached to the N terminus of Rtf1. As expected, knockdown of Rtf1 significantly reduced its expression and the level of H2Bub (Fig. 4B). Overexpression of full-length Rtf1 restored the H2Bub level. Similarly, Rtf1 Δ1, Δ5, and Δ6 restored the H2Bub level, whereas the other Rtf1 mutants did not. Note that the anti-Rtf1 antibody used here recognizes the C terminus of human Rtf1 and hence does not react with the Δ5 and Δ6 mutants. These results are consistent with previous findings in S. cerevisiae (34) and indicate the evolutionarily conserved role of the Rtf1 HMD in histone modifications.
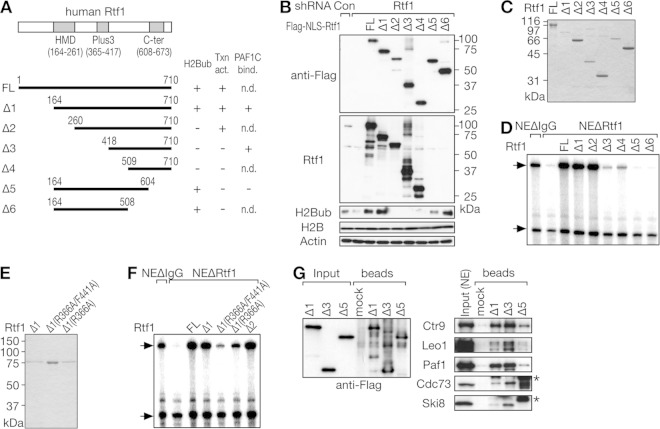
FIG 4.
Distinct structural requirements for human Rtf1 in H2B monoubiquitination and transcriptional activation in vitro. (A) Schematic structures of human Rtf1 deletion mutants. The results shown in panels B to G are summarized on the right. Txn act., transcriptional activation; PAF1C bind., PAF1C binding; n.d., not determined. (B) Knockdown-rescue experiments. HeLa cells were sequentially infected with a lentiviral vector for shRNA targeting Rtf1 and transfected with one of the Flag-NLS-Rtf1 expression vectors carrying shRNA-resistant mutations. Cells were harvested 7 days postinfection (3 days posttransfection) and subjected to immunoblot analysis. (C and E) The purity of full-length (FL) Flag-Rtf1 prepared from insect cells and His-Rtf1-Flag deletion mutants prepared from bacteria was examined by Coomassie blue staining. (D and F) The transactivation potential of Rtf1 mutants was examined by in vitro transcription assays using NEΔRtf1. (G) The indicated Rtf1 mutants were first adsorbed onto anti-Flag-agarose beads and then incubated with HeLa cell NE. Input and bound fractions were analyzed by immunoblotting. The asterisks denote nonspecific signals.
Next, the Rtf1 deletion mutants were expressed in and purified from bacteria by tandem-affinity purification using a histidine tag and a Flag tag (Fig. 4C), and the resulting recombinant proteins were examined by in vitro transcription assays using NEΔRtf1. As shown in Fig. 4D, Rtf1 Δ1 and Δ2 fully restored elongation defects, whereas the other Rtf1 mutants did not, suggesting the possible involvement of the Plus3 domain and the C-terminal segment in transcriptional activation. These findings prompted us to test two previously described Rtf1 point mutants in our assays (35). Single or double mutations of R366A and F441A, which reportedly disrupt the function of the Plus3 domain, were introduced into Rtf1 Δ1 (Fig. 4E). The double point mutation severely impaired the transactivation potential of Rtf1 (Fig. 4F), indicating that the Plus3 domain is critical for the coactivator-dependent function.
To explore the physical interaction between human Rtf1 and the PAF1C, Rtf1 mutants were adsorbed onto anti-Flag-agarose beads and then incubated with HeLa cell NE. Concordant with the results of Kim et al. (14), Rtf1 Δ1 pulled down the PAF1C from HeLa cell NE (Fig. 4G). Δ3, lacking the Plus3 domain, pulled down the PAF1C even more efficiently, but Δ5 did not. Therefore, despite differences in the binding affinities of S. cerevisiae and human Rtf1 proteins for the PAF1C, the same C-terminal segment of Rtf1 is critical for its binding to the PAF1C. Taken together, the results of our mutational study revealed different structural requirements for Rtf1 in H2B monoubiquitination, its coactivator-dependent function in vitro, and the physical interaction with the PAF1C, as summarized in Fig. 4A.
Comparative analysis of Rtf1- and PAF1C-regulated genes.
Based on our findings in vitro, we performed a detailed comparative analysis of Rtf1 and PAF1C knockdown phenotypes. Following lentiviral-vector-mediated expression of shRNAs targeting Rtf1 and individual subunits of the PAF1C in HeLa cells, efficient knockdown of shRNA targets was confirmed by immunoblotting (Fig. 5A). Nontarget PAF1C subunits were codepleted in certain cases; for example, Paf1 knockdown resulted in reduction of all the PAF1C subunits to varying degrees, which was likely due to destabilization of the complex. Moreover, the H2Bub level was reduced in almost all cases.
Paf1 and Ski8 were selected as representative subunits of the PAF1C, and RNA-seq analysis was performed in triplicate to compare the genes affected by Paf1, Ski8, and Rtf1 knockdown (Fig. 5B and C). Since knockdown of any one of Rtf1, Ski8, and Paf1 affected cell growth upon prolonged culture (data not shown), we carefully determined the time course to complete the experiments before the onset of significant growth retardation. For this reason, total RNA was harvested from Ski8 knockdown cells at day 4 and from Rtf1 or Paf1 knockdown cells at day 7. Using an FDR of 0.05 and a 2.0-fold change as cutoff values, 701, 1,039, and 1,556 genes were identified as significantly affected by Paf1, Ski8, and Rtf1 knockdown (data not shown). The number of genes downregulated by the knockdown was slightly larger than the number of upregulated genes (Fig. 5C). In both cases, there was a significant overlap among the genes affected by the knockdown, suggesting functional similarity between Rtf1 and the PAF1C. In addition, many genes were uniquely affected by Rtf1 knockdown (402 upregulated genes and 495 downregulated genes).
To confirm the results of RNA-seq, qRT-PCR analysis was performed (Fig. 5D). For all 18 genes tested, qRT-PCR reproduced the results of RNA-seq. For example, LYPD3 was strongly induced only by Rtf1 knockdown, whereas BTBD3 was selectively repressed by Rtf1 knockdown. In contrast, NT5E and OLFM1 were up- and downregulated, respectively, by Paf1, Ski8, and Rtf1 knockdown to similar extents. These results clearly showed that the expression of certain genes is more sensitive to Rtf1 inhibition than to PAF1C inhibition. The above-mentioned findings also suggest that a significant number of genes are in fact downregulated by Rtf1 and the PAF1C singly or in combination.
To gain insight into the biological significance of the differential gene regulation by Rtf1 and the PAF1C, GO analysis was performed (Table 1). Among the genes commonly affected by Paf1, Ski8, and Rtf1 knockdown, genes involved in nucleosome assembly and cell proliferation were highly enriched. On the other hand, genes involved in the mitotic cell cycle were enriched among the genes uniquely affected by Rtf1 knockdown, suggesting that functionally distinct sets of genes are regulated by Rtf1 alone and by the combination of Rtf1 and the PAF1C.
TABLE 1.
GO analysis of Rtf1-, Paf1-, and Ski8-regulated genes
| GO identification/GO term | Count | FDR |
|---|---|---|
| Genes uniquely affected by Rtf1 knockdown | ||
| GO:0000278/mitotic cell cycle | 42 | 0.0011 |
| GO:0007067/mitosis | 30 | 0.0016 |
| GO:0000280/nuclear division | 30 | 0.0016 |
| GO:0000087/M phase of mitotic cell cycle | 30 | 0.0023 |
| GO:0022403/cell cycle phase | 44 | 0.0034 |
| GO:0048285/organelle fission | 30 | 0.0036 |
| GO:0000279/M phase | 37 | 0.0072 |
| GO:0022402/cell cycle process | 51 | 0.0458 |
| GO:0001836/release of cytochrome c from mitochondria | 8 | 0.0695 |
| GO:0044271/nitrogen compound-biosynthetic process | 34 | 0.0830 |
| Genes affected by Rtf1, Paf1, and Ski8 knockdown in common | ||
| GO:0006334/nucleosome assembly | 21 | 2.14E−08 |
| GO:0031497/chromatin assembly | 21 | 4.28E−08 |
| GO:0065004/protein-DNA complex assembly | 21 | 1.03E−07 |
| GO:0034728/nucleosome organization | 21 | 1.57E−07 |
| GO:0042127/regulation of cell proliferation | 67 | 1.67E−07 |
| GO:0008284/positive regulation of cell proliferation | 45 | 2.06E−07 |
| GO:0006323/DNA packaging | 21 | 1.12E−05 |
| GO:0006333/chromatin assembly or disassembly | 21 | 4.76E−05 |
| GO:0010033/response to organic substance | 56 | 1.90E−04 |
| GO:0009991/response to extracellular stimulus | 25 | 0.0024 |
| GO:0051094/positive regulation of developmental process | 28 | 0.0052 |
| GO:0032101/regulation of response to external stimulus | 20 | 0.0081 |
| GO:0042325/regulation of phosphorylation | 38 | 0.0103 |
| GO:0051174/regulation of phosphorus metabolic process | 39 | 0.0105 |
| GO:0019220/regulation of phosphate metabolic process | 39 | 0.0105 |
| GO:0031667/response to nutrient levels | 22 | 0.0160 |
| GO:0007584/response to nutrient | 18 | 0.0208 |
| GO:0050727/regulation of inflammatory response | 13 | 0.0288 |
| GO:0006979/response to oxidative stress | 19 | 0.0469 |
| GO:0016126/sterol-biosynthetic process | 9 | 0.0483 |
The above-mentioned results prompted us to perform cell cycle analysis of knockdown cells. At the time when RNA-seq analysis was performed, Rtf1 knockdown increased the cell population in G1 phase, whereas Paf1 and Ski8 knockdown had only a weak, if any, effect on the cell cycle (Fig. 5E). Thus, transcriptional changes caused by Rtf1 knockdown were found to be correlated with the phenotypic changes observed.
Time course analysis of knockdown.
shRNA-mediated knockdown of Rtf1, Paf1, and Ski8 all seemed to deplete Rtf1 levels to similar extents (Fig. 5A), making interpretation of the RNA-seq data complicated. In an attempt to differentiate primary and secondary effects of knockdown, we performed time course analysis, based on the assumption that secondary effects should be observed with delayed kinetics. Expression of Rtf1-specific and common genes was negligibly affected on day 5 of Rtf1 knockdown but was activated (PLAUR, DUSP1, SERPINE1, and NT5E) or repressed (BTBD3 and ASS1) on the next day (Fig. 6B). Concordantly, there was no discernible effect on the protein levels of the PAF1C components and Rtf1 on day 5 (Fig. 6A). On the next day, however, the Rtf1 protein almost disappeared, and the levels of the PAF1C components also decreased to various degrees. Expression of common genes was up- or downregulated significantly on day 5 of Paf1 knockdown and on day 3 of Ski8 knockdown (Fig. 6C). On day 5 of Paf1 knockdown, the levels of the PAF1C components decreased to various degrees, with the Paf1 level affected most strongly, whereas there was no apparent reduction of the Rtf1 level (Fig. 6A). On day 3 of Ski8 knockdown, the Ski8 level decreased significantly with only a small effect on the levels of the other PAF1C components, whereas there was no apparent reduction of the Rtf1 level. On the next day, the Ski8 level decreased further, and the levels of the other PAF1C components and Rtf1 also decreased. Collectively, transcriptional defects were observed at the earliest time point when shRNA-mediated knockdown was evident, and the time course of altered gene expression seemed to be correlated well with the time course of knockdown, suggesting that at least several genes studied here are direct targets of Rtf1, Ski8, and Paf1. Moreover, the above-mentioned findings suggest that codepletion of Rtf1 is not responsible, at least in part, for the knockdown phenotype observed following Paf1 or Ski8 knockdown.
FIG 6.
Time course analysis of knockdown. HeLa cells were infected with lentiviral vectors expressing shRNAs targeting Ski8, Paf1, and Rtf1 and harvested for immunoblot analysis (A) and qRT-PCR analysis (B and C) on the indicated days postinfection. qRT-PCR data are presented as the means ± SEM of the results of 3 independent experiments.
Human Rtf1-independent recruitment of the PAF1C.
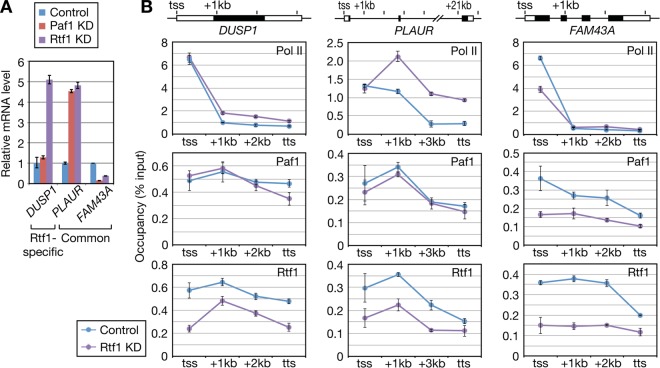
To investigate the mechanisms underlying the differential gene regulation by Rtf1 and the PAF1C, the occupancies of Rtf1 and Paf1 at several gene loci identified in our study were compared. Contrary to our expectation that Rtf1 was more enriched than Paf1 at Rtf1-specific genes, such as LYPD3 and METTL7A, no clear differences in the Rtf1/Paf1 ratio were observed (Fig. 7B and data not shown). Instead, Rtf1 and Paf1 levels were correlated with the level of Pol II over the several genes examined, although Rtf1 and Paf1 were apparently more enriched at the gene body than Pol II. These results suggested that the different outcomes of Rtf1 and PAF1C knockdown arise from the postrecruitment process, i.e., after the factors are recruited to target genes.
FIG 7.
Human Rtf1-independent recruitment of the PAF1C. HeLa cells were infected with lentiviral vectors as for Fig. 5. The infected cells were subjected to qRT-PCR analysis (A) and ChIP (B). ChIP signals are reported as the percentage of input recovered. The data are represented as the means ± SEM of the results of 3 independent experiments. tss, transcription start site.
The functional differences between human Rtf1 and the PAF1C led us to investigate the mechanisms by which these factors find their target genes. In S. cerevisiae, Rtf1 binds to Spt5 and serves as a binding platform for the PAF1C (37–39), and Cdc73 is required for the efficient recruitment of Rtf1 (2, 47). Considering the weak physical interaction between Rtf1 and the PAF1C in metazoans, however, whether such reciprocal recruitment of Rtf1 and the PAF1C is conserved across species is questionable. To address this issue, we selected three genes that gave strong ChIP signals and examined the distributions of Pol II, Paf1, and Rtf1 in response to Rtf1 knockdown (Fig. 7). Compared to the Rtf1 signal, which was significantly reduced, Paf1 occupancy on DUSP1 and PLAUR was negligibly affected by Rtf1 knockdown. FAM43A expression was highly sensitive to Rtf1 knockdown (Fig. 7A). Concordantly, Pol II and Paf1 occupancy on FAM43A was strongly affected by Rtf1 knockdown, whereas the Paf1/Pol II ratio was only modestly affected. These results support the idea that the human PAF1C is recruited to target genes independently of Rtf1. ChIP analysis of Rtf1 after Paf1 knockdown was attempted but was not successful, because Paf1 knockdown resulted in significant growth retardation upon prolonged culture (data not shown).
DISCUSSION
In the present study, we showed for the first time that human Rtf1 directly activates transcription elongation in vitro. Rtf1-mediated transcriptional activation requires coactivator activity, which is most likely unrelated to DSIF or the PAF1C (Fig. 1 to 3). A mutational study showed that the Plus3 domain of human Rtf1, but not its HMD, is required for the coactivator-dependent function (Fig. 4). Consistent with our findings in vitro, we showed that Rtf1 and the PAF1C play distinct roles in the expression of a subset of genes in cultured cells (Fig. 5 and 6). Moreover, the PAF1C was apparently recruited to the genes examined in an Rtf1-independent manner (Fig. 7). Therefore, the present study establishes a role for human Rtf1 as a transcription elongation factor that may function independently of the PAF1C.
PAF1C-dependent and -independent functions of human Rtf1.
Our finding that human Rtf1 can act independently of the PAF1C is consistent with the in vitro and in vivo results of previous studies. Biochemical studies showed that the human PAF1C promotes transcription elongation regardless of the presence of Rtf1 but requires other factors, such as DSIF, Tat-SF1, and TFIIS, for full transcriptional activation in vitro (13, 14). In contrast, the present study showed that human Rtf1 promotes transcription elongation independently of the PAF1C, DSIF, and Tat-SF1. Different requirements for Rtf1- and PAF1C-mediated transcriptional activation indicate that distinct mechanisms underlie these processes. In vivo, Mueller and Jaehning (2) showed that many of the phenotypes associated with paf1Δ or ctr9Δ are not enhanced but, rather, are suppressed by simultaneous deletion of RTF1 in S. cerevisiae. Another recent paper showed that Rtf1 and the PAF1C exert opposing effects on Pol II elongation in S. pombe (30).
Figure 7 suggested that the PAF1C is recruited to some of its target genes independently of Rtf1. This finding is in contrast to the previous finding in S. cerevisiae that Rtf1 serves as a binding platform for the PAF1C (37–39) and raises the question of how the PAF1C is recruited to target genes. As the name suggests, the PAF1C physically interacts with Pol II (3, 4), and a previous study showed that a Paf1-Leo1 subcomplex of the PAF1C is responsible for its association with Pol II (14). Moreover, a few recent papers have revealed that the PAF1C directly interacts with histones (26, 48). In the study by Kim et al. (48), for example, the PAF1C was identified as an isoform-specific interactor of linker histone H1.2. Thus, there seem to be a few independent mechanisms by which the PAF1C is recruited to target genes.
In the present study, we were unable to identify the Rtf1 coactivator. Besides the PAF1C subunits, Rtf1 interacts with several factors, including the transcription elongation factor FACT, the chromodomain-containing protein Chd1, and the 19S proteasome (34, 49–51), and these factors could be considered the prime suspects. Our mutational analysis showed that the HMD of human Rtf1 is dispensable for its coactivator-dependent function in vitro (Fig. 4D). The HMD of S. cerevisiae Rtf1 is necessary and sufficient for Rtf1-mediated histone modification, and the Rtf1 HMDs from various species complement the defects of HMD deletion in S. cerevisiae to varying degrees (9), suggesting its evolutionarily conserved function. Our mutational study of human Rtf1 was consistent with the previous finding in S. cerevisiae (Fig. 4B). Taken together, these results suggest that the coactivator-dependent function of human Rtf1 is independent of Rtf1-mediated histone modification. On the other hand, the Plus3 domain of human Rtf1 was found to be critical for its coactivator-dependent function (Fig. 4D). Originally implicated in single-stranded DNA binding, the Plus3 domain was recently shown to interact with the phosphorylated form of Spt5 (9, 35, 36). The Plus3 domain may serve as a binding platform for not one but multiple factors, including the unidentified Rtf1 coactivator.
PAF1C-controlled, Rtf1-controlled, and H2Bub-controlled genes.
GO analysis showed that genes involved in the mitotic cell cycle were enriched among the genes uniquely affected by Rtf1 knockdown (Table 1). Concordantly, Rtf1 knockdown increased a cell population in G1 phase (Fig. 5E). We were therefore interested in knowing whether transcriptional defects altered the cell cycle or, conversely, whether an altered cell cycle resulted in transcriptional changes. Of the 51 Rtf1-specific genes categorized into GO:0022402 cell cycle process, 44 were downregulated by Rtf1 knockdown while 7 were upregulated by Rtf1 knockdown. A search of Cyclebase 3.0, a cell cycle-dependent gene expression database (52), indicated that 21 of the 44 downregulated genes are highly expressed in S, G2, and/or M phase. These findings are consistent with the idea that the 21 genes, including BUB1, CCNA2, CCNB2, CENPA, and PLK1, were downregulated through an indirect effect of an altered cell cycle. There are, however, many other cell cycle-related genes whose altered expression cannot be explained by an altered cell cycle.
We also compared our RNA-seq data with microarray data of Shema et al. (53), who identified H2Bub-controlled genes in HeLa cells by knocking down RNF20, a component of the E3 ubiquitin ligase complex that mediates H2B monoubiquitination. Meta-analysis of the two data sets revealed that, of 3,469 genes affected by RNF20 knockdown, 538 genes were also affected by the knockdown of Rtf1, Ski8, or Paf1. It is not surprising to us that only a fraction of the genes were identified as coregulated. RNF20 has been reported to target many proteins, such as Syntaxin 1, Ebp1, AP-2alpha, and SREBP1c, for polyubiquitination and degradation (54–57). Hence, it is conceivable that RNF20 knockdown affects the expression of many genes by mechanisms that are independent of H2B monoubiquitination. Similarly, the PAF1C is a multifunctional protein complex that controls transcription in both chromatin-dependent and -independent manners and also affects Pol II termination and 3′ processing (34, 40). Hence, it is plausible that the knockdown of a PAF1C component affects gene expression, in part, in an H2Bub-independent manner.
Functional similarities and differences in Rtf1 proteins among species.
In a recent paper, Wier and colleagues demonstrated that the interaction between the Rtf1 Plus3 domain and phospho-Spt5 is crucial for the recruitment of the PAF1C in S. cerevisiae (35). In S. pombe, however, Ctr9 is recruited independently of Rtf1 (30). Similarly, we found that human Paf1 was recruited to actively transcribed genes, apparently in an Rtf1-independent manner (Fig. 7), suggesting that another mechanism mediates the recruitment of the PAF1C (e.g., via direct interaction between the PAF1C and Pol II). Considering the weak physical interaction between Rtf1 and the PAF1C in many species other than S. cerevisiae, it is reasonable to assume that an additional mechanism could significantly contribute to PAF1C recruitment.
Despite differences in their binding affinities for the PAF1C, the S. cerevisiae and human Rtf1 proteins have similar domain structures. Our mutational analysis showed that the highly conserved HMD of human Rtf1 is critical for H2B monoubiquitination in human cells (Fig. 4B). Moreover, the interaction with the PAF1C subunits was mediated by the Rtf1 C-terminal region of approximately 100 amino acids. These results are consistent with earlier mutational studies in S. cerevisiae (9, 34) and support the fact that the Rtf1 C-terminal region is not well conserved among species; this divergence may underlie the differences in PAF1C-binding affinities among species.
Future perspectives.
Several issues remain to be addressed. The biggest unsolved issue of this work is the identity of the Rtf1 coactivator. Another important question is why some genes are affected only by Rtf1 knockdown despite the recruitment of both Rtf1 and the PAF1C to similar levels. A detailed comparison of the genomic binding sites of human Rtf1 and the PAF1C subunits would be of interest. Identification of the Rtf1 coactivator will be a key step toward the distinction of PAF1C-dependent and -independent mechanisms of action of Rtf1.
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas “Transcription Cycle” from the MEXT, Japan.
We thank Junko Kato for technical assistance.
REFERENCES
- 1.Stolinski LA, Eisenmann DM, Arndt KM. 1997. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol Cell Biol 17:4490–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mueller CL, Jaehning JA. 2002. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol Cell Biol 22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi X, Chang M, Wolf AJ, Chang CH, Frazer-Abel AA, Wade PA, Burton ZF, Jaehning JA. 1997. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol Cell Biol 17:1160–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi X, Finkelstein A, Wolf AJ, Wade PA, Burton ZF, Jaehning JA. 1996. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol Cell Biol 16:669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J 21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11:721–729. doi: 10.1016/S1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 7.Ng HH, Dole S, Struhl K. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem 278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- 8.Ng HH, Robert F, Young RA, Struhl K. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11:709–719. doi: 10.1016/S1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 9.Piro AS, Mayekar MK, Warner MH, Davis CP, Arndt KM. 2012. Small region of Rtf1 protein can substitute for complete Paf1 complex in facilitating global histone H2B ubiquitylation in yeast. Proc Natl Acad Sci U S A 109:10837–10842. doi: 10.1073/pnas.1116994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem 278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 11.Wozniak GG, Strahl BD. 2014. Catalysis-dependent stabilization of Bre1 fine-tunes histone H2B ubiquitylation to regulate gene transcription. Genes Dev 28:1647–1652. doi: 10.1101/gad.243121.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol 25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Yamaguchi Y, Tsugeno Y, Yamamoto J, Yamada T, Nakamura M, Hisatake K, Handa H. 2009. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev 23:2765–2777. doi: 10.1101/gad.1834709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Guermah M, Roeder RG. 2010. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 16.Rondon AG, Gallardo M, García-Rubio M, Aguilera A. 2004. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep 5:47–53. doi: 10.1038/sj.embor.7400045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crisucci EM, Arndt KM. 2011. The roles of the Paf1 complex and associated histone modifications in regulating gene expression. Genet Res Int 2011:707641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaehning JA. 2010. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta 1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan CD, Holland MJ, Winston F. 2005. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J Biol Chem 280:913–922. doi: 10.1074/jbc.M411108200. [DOI] [PubMed] [Google Scholar]
- 20.Nordick K, Hoffman MG, Betz JL, Jaehning JA. 2008. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot Cell 7:1158–1167. doi: 10.1128/EC.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. 2005. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell 20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 22.Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M. 2009. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci U S A 106:755–760. doi: 10.1073/pnas.0812023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheldon KE, Mauger DM, Arndt KM. 2005. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol Cell 20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomson BN, Crisucci EM, Heisler LE, Gebbia M, Nislow C, Arndt KM. 2013. Effects of the Paf1 complex and histone modifications on snoRNA 3′-end formation reveal broad and locus-specific regulation. Mol Cell Biol 33:170–182. doi: 10.1128/MCB.01233-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomson BN, Davis CP, Warner MH, Arndt KM. 2011. Identification of a role for histone H2B ubiquitylation in noncoding RNA 3′-end formation through mutational analysis of Rtf1 in Saccharomyces cerevisiae. Genetics 188:273–289. doi: 10.1534/genetics.111.128645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu X, Qin X, Xu H, Li L, Wang Z, Li F, Xie X, Zhou H, Shen Y, Long J. 2013. Structural insights into Paf1 complex assembly and histone binding. Nucleic Acids Res 41:10619–10629. doi: 10.1093/nar/gkt819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. 2005. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev 19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adelman K, Lis JT. 2006. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol Cell Biol 26:250–260. doi: 10.1128/MCB.26.1.250-260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langenbacher AD, Nguyen CT, Cavanaugh AM, Huang J, Lu F, Chen JN. 2011. The PAF1 complex differentially regulates cardiomyocyte specification. Dev Biol 353:19–28. doi: 10.1016/j.ydbio.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mbogning J, Nagy S, Pagé V, Schwer B, Shuman S, Fisher RP, Tanny JC. 2013. The PAF complex and Prf1/Rtf1 delineate distinct Cdk9-dependent pathways regulating transcription elongation in fission yeast. PLoS Genet 9:e1004029. doi: 10.1371/journal.pgen.1004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenney K, Shilatifard A. 2006. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc Natl Acad Sci U S A 103:11970–11974. doi: 10.1073/pnas.0603620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akanuma T, Koshida S, Kawamura A, Kishimoto Y, Takada S. 2007. Paf1 complex homologues are required for Notch-regulated transcription during somite segmentation. EMBO Rep 8:858–863. doi: 10.1038/sj.embor.7401045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubota Y, Tsuyama K, Takabayashi Y, Haruta N, Maruyama R, Iida N, Sugimoto A. 2014. The PAF1 complex is involved in embryonic epidermal morphogenesis in Caenorhabditis elegans. Dev Biol 391:43–53. doi: 10.1016/j.ydbio.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Warner MH, Roinick KL, Arndt KM. 2007. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol 27:6103–6115. doi: 10.1128/MCB.00772-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wier AD, Mayekar MK, Heroux A, Arndt KM, Vandemark AP. 2013. Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin. Proc Natl Acad Sci U S A 110:17290–17295. doi: 10.1073/pnas.1314754110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Jong RN, Truffault V, Diercks T, Ab E, Daniels MA, Kaptein R, Folkers GE. 2008. Structure and DNA binding of the human Rtf1 Plus3 domain. Structure 16:149–159. doi: 10.1016/j.str.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. 2009. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol 29:4852–4863. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayekar MK, Gardner RG, Arndt KM. 2013. The recruitment of the Saccharomyces cerevisiae Paf1 complex to active genes requires a domain of Rtf1 that directly interacts with the Spt4-Spt5 complex. Mol Cell Biol 33:3259–3273. doi: 10.1128/MCB.00270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou K, Kuo WH, Fillingham J, Greenblatt JF. 2009. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci U S A 106:6956–6961. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JM, Greenleaf AL. 1997. Modulation of RNA Polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J Biol Chem 272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- 41.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41–51. doi: 10.1016/S0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 42.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev 12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. 2007. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell 26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 44.Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. 2006. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell 21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 45.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto J, Hagiwara Y, Chiba K, Isobe T, Narita T, Handa H, Yamaguchi Y. 2014. DSIF and NELF interact with Integrator to specify the correct post-transcriptional fate of snRNA genes. Nat Commun 5:4263. doi: 10.1038/ncomms5263. [DOI] [PubMed] [Google Scholar]
- 47.Amrich CG, Davis CP, Rogal WP, Shirra MK, Heroux A, Gardner RG, Arndt KM, VanDemark AP. 2012. Cdc73 subunit of Paf1 complex contains C-terminal Ras-like domain that promotes association of Paf1 complex with chromatin. J Biol Chem 287:10863–10875. doi: 10.1074/jbc.M111.325647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim K, Lee B, Kim J, Choi J, Kim J-M, Xiong Y, Roeder RG, An W. 2013. Linker histone H1.2 cooperates with Cul4A and PAF1 to drive H4K31 ubiquitylation-mediated transactivation. Cell Rep 5:1690–1703. doi: 10.1016/j.celrep.2013.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krogan NJ, Buratowski S, Greenblatt JF. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan YR, Sun M, Wohlschlegel J, Reed SI. 2013. Cks1 enhances transcription efficiency at the GAL1 locus by linking the Paf1 complex to the 19S proteasome. Eukaryot Cell 12:1192–1201. doi: 10.1128/EC.00151-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J 22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santos A, Wernersson R, Jensen LJ. 2015. Cyclebase 3.0: a multi-organism database on cell-cycle regulation and phenotypes. Nucleic Acids Res 43:D1140–D1144. doi: 10.1093/nar/gku1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, Hublarova P, Moyal L, Gana-Weisz M, Shiloh Y, Yarden Y, Johnsen SA, Vojtesek B, Berger SL, Oren M. 2008. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev 22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chin L-S, Vavalle JP, Li L. 2002. Staring, a novel E3 ubiquitin-protein ligase that targets syntaxin 1 for degradation. J Biol Chem 277:35071–35079. doi: 10.1074/jbc.M203300200. [DOI] [PubMed] [Google Scholar]
- 55.Lee JH, Lee GY, Jang H, Choe SS, Koo S-H, Kim JB. 2014. Ring finger protein20 regulates hepatic lipid metabolism through protein kinase A-dependent sterol regulatory element binding protein1c degradation. Hepatology 60:844–857. doi: 10.1002/hep.27011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Z, Oh S-M, Okada M, Liu X, Cheng D, Peng J, Brat DJ, Sun S-Y, Zhou W, Gu W, Ye K. 2009. Human BRE1 is an E3 ubiquitin ligase for Ebp1 tumor suppressor. Mol Biol Cell 20:757–768. doi: 10.1091/mbc.E08-09-0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ren P, Sheng Z, Wang Y, Yi X, Zhou Q, Zhou J, Xiang S, Hu X, Zhang J. 2014. RNF20 promotes the polyubiquitination and proteasome-dependent degradation of AP-2α protein. Acta Biochim Biophys Sin 46:136–140. doi: 10.1093/abbs/gmt136. [DOI] [PubMed] [Google Scholar]