Abstract
In higher eukaryotes the accelerated degradation of mRNAs harboring premature termination codons is controlled by nonsense-mediated mRNA decay (NMD), exon junction complex (EJC), and nuclear cap-binding complex (CBC) factors, but the mechanistic basis for this quality-control system and the specific roles of the individual factors remain unclear. Using Neurospora crassa as a model system, we analyzed the mechanisms by which NMD is induced by spliced 3′-UTR introns or upstream open reading frames and observed that the former requires NMD, EJC, and CBC factors whereas the latter requires only the NMD factors. The transcripts for EJC components eIF4A3 and Y14, and translation termination factor eRF1, contain spliced 3′-UTR introns and each was stabilized in NMD, EJC, and CBC mutants. Reporter mRNAs containing spliced 3′-UTR introns, but not matched intronless controls, were stabilized in these mutants and were enriched in mRNPs immunopurified from wild-type cells with antibody directed against human Y14, demonstrating a direct role for spliced 3′-UTR introns in triggering EJC-mediated NMD. These results demonstrate conclusively that NMD, EJC, and CBC factors have essential roles in controlling mRNA stability and that, based on differential requirements for these factors, there are branched mechanisms for NMD. They demonstrate for the first time autoregulatory control of expression at the level of mRNA stability through the EJC/CBC branch of NMD for EJC core components, eIF4A3 and Y14, and for eRF1, which recognizes termination codons. Finally, these results show that EJC-mediated NMD occurs in fungi and thus is an evolutionarily conserved quality-control mechanism.
Keywords: nonsense-mediated mRNA decay (NMD), RNA stability, exon junction complex (EJC), cap-binding complex (CBC), spliced 3′-UTR intron, post-transcriptional control, ribosome, translation, Neurospora crassa
THE nonsense-mediated mRNA decay (NMD) pathway targets mRNA for degradation through the recognition of translated termination codons determined to be premature (Nicholson et al. 2009; Kervestin and Jacobson 2012; Popp and Maquat 2013). The NMD pathway can degrade mRNAs that contain upstream open reading frames (uORFs) or long 3′-UTRs. The termination-related processes that trigger uORF-mediated or long 3′-UTR-mediated NMD are considered to be similar (Amrani et al. 2004, 2006; Nicholson et al. 2009) in that termination events occurring far from the context created by the mRNA poly(A) tail can result in mRNA degradation (the “faux-UTR” model). In higher eukaryotes, NMD can also act on mRNAs that contain an intron downstream of a termination codon, such as aberrantly spliced transcripts or 3′-UTR intron-containing transcripts (Sauliere et al. 2010; Bicknell et al. 2012). When a spliced intron is positioned at least 50–55 nt downstream of a termination codon, the exon junction complex (EJC) positioned near the exon–exon junction is not displaced by translating ribosomes and its continued association with mRNA targets the mRNA for degradation by NMD (Isken and Maquat 2007). While there are some shared factors, such as the UPF factors, this branch of NMD appears distinct from the faux-UTR branch in that, for NMD to occur, it requires an EJC to remain on the mRNA downstream from the termination event that triggers NMD.
In mammals, the nuclear cap-binding complex (CBC), which is composed of CBP80 and CBP20, is preferentially associated with mRNAs undergoing EJC-targeted NMD (Ishigaki et al. 2001). This has led to a model in which EJC-mediated targeting of mRNAs for NMD occurs during pioneer rounds of translation. During these pioneer rounds, CBC has not yet been replaced by eIF4E, the cytoplasmic cap-binding protein (Maquat et al. 2010). The implication of these studies was that the CBC must also be associated with the mRNA for EJC-mediated NMD to occur. However, more recent data show that EJC-mediated NMD can occur on mRNAs associated with the cytoplasmic cap-binding factor eIF4E (Durand and Lykke-Andersen 2013; Rufener and Muhlemann 2013; Popp and Maquat 2014), and the role of the CBC in NMD, and even whether it is essential for this process (Hosoda et al. 2005; Dzikiewicz-Krawczyk et al. 2008), is not fully understood.
EJC-mediated NMD has been observed in animals and plants, but in the budding yeast Saccharomyces cerevisiae, multiple components of the EJC, and EJC-mediated NMD, are lacking, and in fission yeast Schizosaccharomyces pombe, while EJC components are present, a role for them in determining mRNA stability via NMD-related pathways has not been demonstrated (Wen and Brogna 2010). Thus a comprehensive understanding of the evolutionary significance of EJC and CBC factors in NMD and the means to dissect the functions of the EJC and CBC factors that contribute to NMD in highly tractable model fungal systems remain lacking.
The filamentous fungi generally encode NMD and EJC machinery (Feldbrugge et al. 2008). In Aspergillus nidulans, a role for nmdA, the upf2 homolog, has been identified in the control of mRNA stability (Morozov et al. 2006) and the recognition of premature stop codons through upf1 is associated with additional processes affecting mRNA degradation (Morozov et al. 2012). The genome of the model filamentous fungus Neurospora crassa contains homologs of the NMD components UPF1, UPF2, and UPF3, the core EJC components eIF4A3, Mago, and Y14, and CBC components CBP20 and CBP80. A role for upf1 in N. crassa biology is indicated by identification of alleles causing a circadian rhythm defect (Compton 2003). The molecular basis for this circadian defect—a short period—is not known.
Measurement of mRNA stability is critical for determining whether NMD, EJC, or CBC mutations directly or indirectly affect mRNA levels. Metabolic labeling with 4-thiouracil (4TU) can be used to examine mRNA half-life (Cleary et al. 2005). We developed methods for using metabolic labeling of mRNA with 4TU in pulse–chase analyses to measure mRNA stability in N. crassa. We used this approach to examine the effects of deletions of N. crassa genes specifying NMD, EJC, and CBC factors on the stability of endogenous mRNAs and reporter mRNAs that contain uORFs or 3′-UTR introns. These data show that N. crassa NMD factors can control the stability of mRNAs containing uORFs or spliced 3′-UTR introns and that N. crassa NMD, EJC, and CBC factors can control the stability of mRNAs containing 3′-UTR introns. These data show for the first time that the CBC is essential for NMD of mRNAs induced by spliced introns downstream from termination codons. Importantly, mRNAs demonstrated to be under EJC/CBC control include those encoding the EJC components eIF4A3 and Y14, and the translation termination factor eRF1, indicating that the RNA decay pathways in which these factors are involved have roles in determining cellular levels of these factors through autoregulation of their mRNA levels. These results provide new insight into the mechanistic basis of NMD in eukaryotes and demonstrate for the first time that major branches of NMD are conserved between higher eukaryotes and fungi.
Materials and Methods
Strains
N. crassa strains obtained from the Fungal Genetics Stock Center (FGSC) (McCluskey et al. 2010) included FGSC11230 (ΔNCU04242, Δupf1), FGSC15706 (ΔNCU05267, Δupf2), FGSC11679 (ΔNCU03435, Δupf3), FGSC15492 (ΔNCU03226, Δy14), FGSC13031 (ΔNCU04405, Δmago), FGSC19228 (ΔNCU06678, Δxrn1), FGSC18692 (ΔNCU00210, Δcpb20), FGSC22440 (ΔNCU04187, Δcpb80), FGSC16561 (ΔNCU01446, Δuc-4), and the wild- type (wt) sequenced strain FGSC 2489 (74-OR23-1V A). The deletion mutants were produced by the functional genomics program (Colot et al. 2006).
Homokaryotic isolates with desired genotypes were obtained by selecting appropriate progeny from sexual crosses (Davis and de Serres 1970) or by selecting appropriate culture isolates following purification of asexually produced uninucleate microconidia (Ebbole and Sachs 1990). Purification by microconidiation was performed as described except that microconidia were pelleted in an Eppendorf 5415D centrifuged at 12000 × g for 2 min and, following resuspension and plating, germinated at 30°. Homokaryons were confirmed by PCR analysis of genomic DNA from cultures obtained. A complete list of strains used is given in Supporting Information, Table S5.
The his-3 marker was placed in Δupf1 by crossing RANCR6A (his-3, inl, mat a) (Pratt and Aramayo 2002) with FGSC 11230 (ΔNCU04242, upf1, mat A) and in Δupf2 by crossing FGSC6103 (his-3, mat A) with FGSC15706 (ΔNCU05267, Δupf2, mat a) with the his-3 strain serving as the female parent. To rescue the NMD mutations, plasmids designed for his-3 targeting that contained UPF1, 3XFLAG-tagged UPF1, 3XFLAG-tagged UPF2, or HAT-FLAG-tagged UPF2 were linearized with Spil and used to transform Δupf1, his-3 or Δupf2, his-3 to histidine prototrophy. Transformation of N. crassa by electroporation was accomplished as described previously (Margolin et al. 1997; Wei et al. 2013).
The rescue of Δxrn1, Δy14, Δmago, Δcbp20, and Δcbp80 mutants was accomplished by ectopic integration with the corresponding functional gene using linearized plasmid DNA that also contained the dominant selectable Barr marker. Transformants were selected on the basis of resistance to 250 μg/ml glufosinate (TRC no. G596950) (Pall and Brunelli 1993). Barr homokaryotic strains were obtained by microconidiation and the presence of the rescuing transgene confirmed by Southern or PCR analyses of chromosomal DNA.
Luciferase reporters were placed at specific chromosomal sites for analyses of gene expression. Reporters were integrated at his-3 by transformation of his-3 or his-3 Δupf1 strains. Because y14 (NCU03226, Supercontig 1: 4971616-4972891 +) was closely linked to his-3 (NCU03139, Supercontig 1, 4694797-4697762 +), for analyses of reporter activity in Δy14, a two-step approach was used for placing the reporter at a different chromosomal position in this strain. First, luciferase constructs were placed at csr-1 (NCU00726, Supercontig 1, 7404112-7406331 −) by transformation of the wild-type strain (FGSC 2489) with luc reporters on linearized plasmids designed for targeting to csr-1 (Bardiya and Shiu 2007). Disruption of this locus results in resistance to cyclosporin A. The resulting strains containing luc at csr-1 were then crossed with Δy14 using Δy14 as the male parent; progeny that contained luc and that were either y14+ or Δy14 were used for subsequent analyses. Strains containing luc reporters at csr-1 were also crossed to Δcbp80 to obtain luc reporters in that mutant background; crosses of the resulting strains with Δcbp20 were used to obtain Δcbp20 luc and Δcbp80Δcbp20 luc strains. All progeny were purified by microconidiation and genotypes of strains were confirmed by qPCR and PCR. Confirmation of correct polyadenylation and correct splicing of 3′-UTR introns (where relevant) of endogenous eif4a3, erf1, y14 transcripts and of luc-cox5, luc-eif4a3+I, luc-eif4a3-I reporters were confirmed by 3′-RACE.
Phenotypic analyses
Phenotypic analyses of N. crassa deletion strains and rescued strains were performed as described (Colot et al. 2006) with these modifications. Homokaryotic N. crassa strains were obtained by isolation of microconidia and grown in either 125-ml flasks with 25 ml Vogel’s minimal medium (VM)/1.5% sucrose/2% agar or in 16 × 100-mm glass culture tubes with 3 ml VM/1.5% sucrose/2% agar at 25° with 12:12 hr L:D for 4 days or on 20 ml VM/1.5% sucrose/2% agar or VM/2% yeast extract/1.5% sucrose/2% agar plates at 25° or 37° for 2 days with 12:12 hr L:D. Growth patterns of wt and KO strains were imaged with Canon 50D or 60D digital cameras equipped with Canon EF 50mm f/2.5 macro lenses. Linear growth rates were obtained using race tubes containing 12 ml VM/1.5% sucrose/2% agar at 25° with 12:12 hr L:D for 4 days. Aerial hyphae height was calculated with strains grown with 12:12 hr L:D for 4 days at 25° either with 2 ml VM/1.5% sucrose or VM/2% Yeast Extract/1.5% sucrose. Female sexual development analysis was accomplished by growing strains with 3 ml 1 × SC/1.5% sucrose/2% agar (Westergaard and Mitchell 1947) at 25° with 12:12 hr L:D in 25 ml Falcon tissue culture flasks (Corning Life Sciences DL 353018) with the cap not fully tightened. Scoring for protoperithecia formation and fertilization with FGSC2489 (74A) or FGSC4200 (74a) were accomplished on day 7, perithecia formation was scored on day 14, and ascus formation was scored on day 21. The time-lapse movies were created by incubating plates at room temperature (a detailed protocol is available upon request).
Plasmid construction
Plasmids designed for rescuing N. crassa deletion mutants:
The vectors and oligonucleotides used to create constructs for gene rescue are listed in Table S6. Epitope-tagged genes that could be targeted to the his-3 locus were made using the vectors pCCG::C-3xFLAG (FJ457001) or pCCG::C-HAT::FLAG (FJ457003) (Honda and Selker 2009). Plasmid pZY05 (his-3::upf1P-upf1) has the fragment amplified with oYZ165 and oYZ166 containing 750 bp upstream of upf1 and 3282-bp coding region of upf1 and 526 bp downstream. The NotI/EcoRI-digested upf1 fragment was then inserted to pBM61(Margolin et al. 1997). Plasmid pZY43 (his-3::upf1P-upf1-Gly-3xFLAG) contains the promoter and coding region of upf1 amplified by oYZ165 and oYZ194 inserted into pCCGC–Gly3xFLAG (FJ457001) using NotI and PacI sites. A shuttle plasmid pZY67 based on a modified pSK− vector (containing an additional PacI site between its EcoRI and SmaI sites) contains the promoter of upf2 (970 bp amplified by oZY219 NotI/oYZ220 SpeI) and coding region of upf2 (3422 bp amplified by oYZ221 SpeI/oYZ222 PacI). This plasmid was used to insert a NotI/PacI fragment containing upf2 into pCCGC–Gly3xFLAG (FJ457001) and pCCGC–GlyHATFLAG (FJ457003) to create plasmids pZY68 (his3::upf2P-upf2-Gly3xFLAG) and pYZ70 (his3::upf2P-upf2-GlyHATFLAG), respectively. An MluI/AseI fragment including 972 bp xrn1 promoter, 50-bp 5′-UTR, 4565 bp xrn1 gene, 255-bp 3′-UTR, and 859-bp xrn1downstream region (amplified from N. crassa genomic DNA with oligo-pair oYZ477/oYZ478) was inserted into pBAR (Pall and Brunelli 1993) to create pZY125 (xrn1P-xrn1-[Barr]). An Mlu/NdeI fragment including 450-bp y14 promoter, 174-bp 5′-UTR, 628-bp y14 gene, 474-bp 3′-UTR, and 503 bp y14 downstream (amplified with oligo-pair oYZ435/oYZ436) was inserted into pBAR to create pZY120 (y14P-y14-[Barr]). An Mlu/NdeI fragment including 1304 bp upstream of mago, 59-bp 5′-UTR, 801-bp mago gene, 346-bp 3′-UTR, and 1130 bp downstream of mago (amplified with oligo-pair oYZ437/438) was inserted into pBAR to create pZY121 (magoP-mago-[Barr]). An Mlu/NdeI fragment including 1045 bp upstream of cbp80, 112-bp 5′-UTR, 2766-bp cbp80 gene, 206-bp 3′-UTR, and 1075 bp downstream of cbp80 (amplified with oYZ479/oYZ481) was inserted into pBAR to create pZY126 (cbp80P-cbp80-[Barr]). An Mlu/NdeI fragment including 461 bp upstream of cbp20, 191-bp 5′-UTR, 771-bp cbp20 gene, 481-bp 3′-UTR, and 344 bp downstream of cbp20 (amplified by oYZ501/502) was inserted into pBAR to create pZY128 (cbp20P-cbp20-[Barr]).
Plasmids designed for placing luc reporters in N. crassa:
Vectors and oligonucleotides used to construct luciferase reporters are listed in Table S7. Plasmids pZY78 (his-3::cox-5P\cox-5\luc\) and pZY82 (his-3::cox-5P\cox-5\luc\cox-5) containing codon-optimized firefly luciferase was constructed as described (Wei et al. 2013). pZY92 (his-3::cox-5P\cox-5\luc\eIF4A3+I) and pZY94 (his-3::cox-5P\cox-5\luc\eIF4A3-I) had the same cox-5 promoter as pZY82. The 1486-bp 3′ region of eif4a3 including the intron-containing 3′-UTR was amplified with oYZ330 and oYZ332 from N. crassa genomic DNA template, digested with PacI, and inserted into the pZY78 PacI site. A matched construct containing an intronless 3′-UTR was created by two-step PCR. In the first round, PCR products obtained with (i) oYZ330 and oYZ334 (eIF4A3 intron del Rv), and (ii) oYZ333 (eIF4A3 intron del Fw) and oYZ332 using chromosome DNA as template. In the second-round PCR, the two gel-purified first-round PCR products were used as template and the oYZ330 and oYZ332 primers were used to create the 1264-bp fragment that was digested with PacI to insert into the pZY78 PacI site. pZY84 (his-3::cox-5P\cox-5\luc\eRF1+I) was created by inserting the 1703-bp PacI-digested PCR fragment amplified by oYZ181 and oYZ329 to pZY78. pZY86(his-3::cox-5P\cox-5\luc\eRF1-I) was generated by inserting a PacI–EcoRI fragment of intronless PCR product of oYZ181 and oYZ182 amplified with cDNA as template to pZY84.
For integrating the luc reporters at csr-1, plasmids pZY122 (csr-1::cox-5P\cox-5\luc\cox-5), pZY123 (csr-1::cox-5P\cox-5\luc\eIF4A3+I), and pZY124 (csr-1::cox-5P\cox-5\luc\eIF4A3-I) were created by placing the NotI–ClaI restriction fragments of pYZ82, pZY92, and pZY94, respectively, that contained the reporter genes into the corresponding sites of plasmid pCSR-1 (Bardiya and Shiu 2007).
Culture conditions for metabolic labeling with 4TU
Conidia for labeling experiments were obtained from cultures in 125-ml flasks containing 25 ml VM/2% sucrose/2% agar (Vogel 1956). Cultures were grown at 30° with 12:12 hr L:D for 7 days. Conidia were harvested by suspension in VM/2% sucrose, filtered through cheesecloth, counted with a hemacytometer, and inoculated into 50 ml VM/2% sucrose in a 125-ml flask at a concentration of 107 conidia/ml. Conidia were germinated under constant light at 32° for 6 hr with 150 rpm shaking.
To incorporate 4-thiouracil (4TU), cultures were adjusted to 0.2 mM 4TU (either Sigma 440736-1G or Acros 359930010) from a 1 M stock freshly dissolved in DMSO. After 15 min incubation, uracil (Sigma U0750) was added to a final concentration of 10 mM from a 0.3 M stock prepared in DMSO. Fifteen-milliliter aliquots of each culture were taken at 0, 3, and 10 min and immediately mixed with 7.5 ml VM/2% sucrose/10 mM uracil that had been prefrozen in a 50-ml conical centrifuge tube, and placed on ice. Rapid chilling by mixing with frozen medium stops detectable 4TU incorporation (Figure S3D). Cells were harvested by filtration onto Whatman 541 filter paper, washed with ice-cold sterile water, and cut into ∼0.1-g pieces, and each piece was placed into a 2.0-ml conical screwcap tube (Fisher 02-681-344 tube and VWR 89004-362 cap), quick frozen in liquid nitrogen, and stored at –80° prior to RNA isolation.
RNA isolation
Total RNA from frozen cells was isolated as described (Wei et al. 2013). RNA concentration was determined using a Nanodrop spectrophotometer and quality assessed by denaturing gel electrophoresis in formaldehyde gels and northern analyses (Wei et al. 2013). Poly(A) mRNA was purified using the Poly(A) Purist MAG kit.
3′-RACE
3′-RACE was accomplished as described (Wei et al. 2013) using specific Nest oligos and Nested Universal Primer (NUP) oligo oYZ294 (oligos are listed in Table S8). The second-round PCR products were examined by electrophoresis in 2% TAE agarose gels and the fragments were excised from gels and sequenced.
Southerns and Northerns
Genomic DNA was prepared and Southern analyses were performed as described (Wu et al. 2009). Northerns were performed as described (Sachs and Yanofsky 1991) except that dextran sulfate was omitted from the hybridization buffer. The oligonucleotide sequences and templates used to produce PCR fragments to generate probes are listed in Table S9. Predicted sizes of PCR fragments were computed using SeqBuilder of DNASTAR (v. 11.2.1). 32P-labeled probes were generated from PCR fragments by random priming (Feinberg and Vogelstein 1983) followed by clean-up with Sephadex G-50 Quick spin columns (Roche 11 273 973 001).
Biotinylation and purification of 4TU RNA
Biotinylation of 4TU-labeled total RNA (150-μg input) was performed by modification of the published method (Cleary et al. 2005). RNA (0.2 μg/μl) and biotin–HPDP (Pierce 21314, 0.2 μg/μl) in 10 mM Tris–HCl pH 7.5, 2 mM EDTA were incubated for 1.5 hr at room temperature in foil-wrapped Eppendorf tubes with end-over-end rotation (New Brunswick TC-6 tube rotator, 10 rpm). The reaction mixtures were extracted twice with an equal volume of chloroform at room temperature to remove unreacted biotin–HPDP. The biotinylated RNA was precipitated by adding 1/10 volume 5 M NaCl and an equal volume of isopropanol, followed by freezing at −80° for at least 1 hr and centrifugation at 16100 × g at 4° for 20 min in an Eppendorf 5415R centrifuge. The pellet was washed twice with 80% ethanol and the nucleic acid was resuspended in 100 μl autoclaved and filtered DEPC-treated water.
4TU RNA was purified using Neutr Avidin agarose (Thermo Scientific PI29204). For each 150 μg of biotinylated RNA, Neutr Avidin (150 μl 50% slurry) was equilibrated three times with 250 μl binding buffer (25 mM HEPES–NaOH pH 7.2 and 150 mM NaCl) using a Pierce 89879 spin column and repetitive centrifugations at 500 × g for 1 min at room temperature (Eppendorf 5415R). Then biotinylated RNA (1 μg/μl in binding buffer) was added to the settled resin in the column. After 15 min, the column was centrifuged at 500 × g for 1 min and the eluted material reapplied to the column and incubated another 15 min. The column was centrifuged again, the flow through discarded, and the column containing the biotinylated RNA washed five times with 250 μl Binding Buffer. The 4TU RNA was eluted by two applications of 150 μl 0.1 M DTT in water and centrifugation. All of these steps were at room temperature. Eluted 4TU RNA (300 μl) was precipitated by adding 1/10 volume of 3M sodium acetate and 2.5 volumes of 100% ethanol, freezing at −80° overnight, and pelleting by centrifugation at 4° 16000 × g for 60 min. Pellets were washed twice with 80% ethanol, air dried, and resuspended in 10 μl sterile filtered DEPC treated water. The RNA was quantified using 0.5 μl of material for NanoDrop spectrophotometry. Recovery of 4 TU-RNA was typically 0.5–1% of total RNA input.
ECL detection of biotinylated 4TU-labeled RNA
Biotinylated RNA was either directly dot blotted onto nylon membrane (Bio-Rad Zeta-Probe Blotting membranes no. 162-0159) or was transferred following denaturing agarose-gel electrophoresis. After rinsing the membrane twice for 5 min with PBS, the membrane was incubated with 1000-fold diluted ECL Streptavidin-HRP Conjugate reagent (Amersham RPN 1231V) in PBS buffer for 5 min and washed twice with PBS for 15 min. The excess liquid was removed and the membrane was incubated 2 min with ECL Western blotting detection reagents (Amersham RPN2106) and detected with a UVP Bioimaging system (HAMAMATSU Digital camera C8484-03G).
cDNA synthesis and quantitative PCR from cDNA (RT-qPCR)
For DNase I treatment in a 10-μl reaction volume, 10 μg total RNA or 0.5 μg 4TU RNA in 1× DNase I buffer was incubated at 37° for 30 min with 1 unit DNase I (Ambion AM1907). The reaction was stopped with 1 μl inactivation resin (Ambion) at room temperature for 2 min and the resin-free supernatant obtained by centrifugation according to the manufacturer’s directions. Nucleic acid concentrations were determined by NanoDrop spectrophotometry. DNase I treated total RNA (1 μg) or 4TU RNA (0.2 μg) was used as template to synthesize first-strand cDNA with a combination of oligo(dT)18 and random hexamer primers in a 10-μl final reaction volume as described (Wei et al. 2013). For RT–qPCR, first-strand cDNA (from 8 ng RNA for mRNA measurement and 0.125 ng RNA for 25S rRNA measurement) was used for qPCR as described (Wei et al. 2013). Oligonucleotides used for qPCR are listed in Table S10.
mRNA half-life analysis
Time points were obtained for triplicate cultures. For each individual culture, the 4TU mRNA/25S rRNA ratio at time 0 was set at 1, and the mRNA/25S rRNA ratios at subsequent sampling times normalized to time 0. Half-lives and R2 values were determined by analysis with GraphPad Prism6 software using a nonlinear regression curve-fit for exponential one-phase decay with the plateau value set to 0.
Luciferase assays
Extracts were prepared as described (Luo et al. 1995), quick frozen in aliquots, and stored at −80°. Protein concentration was determined by Bradford assay by mixing 10 μl of serially diluted BSA standards or cell extracts with 150 μl Coomassie Plus Protein Assay Reagent (Thermo Scientific 23236) and absorbance measured with a VICTOR3 V Multilabel Counter (PerkinElmer model 1420) using 96-well plates (Nunclon 167008). Luciferase activity was measured using a VICTOR3 V Multilabel Counter (PerkinElmer model 1420) by mixing 10 μl of samples with 5 μl 5× passive lysis buffer (Promega) and 10 μl firefly luciferase assay reagents (25 mM glyclglycine, 15 mM potassium phosphate solution pH 8.0, 15 mM MgSO4, 4 mM EGTA, 2 mM ATP, 1 mM DTT, 0.1 mM CoA, and 0.075 mM luciferin) (Dyer et al. 2000). Luciferase activity was normalized to protein amount and relative luc mRNA level.
N. crassa cell free translation
N. crassa translation extracts were prepared from wt and used as described (Wu et al. 2007) with N. crassa RNA or synthetic capped and polyadenylated mRNA encoding firefly luciferase (Wei et al. 2012).
Immunopurification of RNA with antibody against human Y14
For each immunopurification, 120 μl (3.6 mg) of a suspended slurry of Dynabeads Protein G (Invitrogen 100.03D) was preequilibrated by washing three times with 200 μl buffer B (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2.5 mM MgC12, 1% NP40) and the beads resuspended with 120 μl of buffer B. N. crassa cell extracts were prepared by adding 200–300 mg of −80° frozen mycelia to 1 g baked (>2 hr at 180°) 0.5-mm zirconia/silica beads (BioSpec 11079105z) and 300 μl of buffer A [10 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.1% Triton-X100, 2.5 mM MgC12, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM vanadyl ribonucleoside complexes (VRC)]; beads and buffer were prechilled on ice in 0.5-ml screwcap tubes (VWR 16466-034). Cells were broken with a Mini-Beadbeater-16 (Biospec) for two cycles of 1 min at full speed with a 1-min rest interval on ice. A hole was poked in the bottom of the tube with a hot needle and the tube placed inside a 2-ml screwcap tube (Fisher 02-681-344) and centrifuged for 1 min at 16,000 × g at 4° and the small tube with the retained beads was discarded. The crude cell extract was then clarified by centrifugation at 4° for 10 min at 16,000 × g. Protein concentrations were measured by Bradford assay and adjusted to 3 mg/ml using buffer A and samples held on ice. Each extract (250 μl) was preincubated with 20 μl preequilibrated Dynabeads at room temperature for 10 min with gentle rotation and the extract without the beads was transferred to a fresh tube containing 100 μl Dynabeads and 5 μl of 0.66 mg/ml anti-Y14, clone 4C4 monoclonal antibody (Millipore 05-1511, Lot no. 05-1511); the Dynabeads/antibody had also been preincubated at room temperature for 10 min with rotation. Extract, beads, and antibody were then incubated for 0.5 hr at 4° with gentle rotation. Beads were collected and washed three times with 200 μl of buffer B per wash; beads were suspended in 0.5 ml RNA extraction buffer. RNA was prepared as described (Wei et al. 2013), dissolved in 10 μl of water, and DNase I treated, and RNA concentrations were determined by NanoDrop spectrophotometry; 150 ng of each RNA was used for cDNA synthesis and the amounts of mRNA relative to input (total RNA extracted from the same cells) were determined by RT–qPCR.
Data analysis
The FunCat database (Ruepp et al. 2004) was used to calculate functional enrichments for the products specified by the 31 identified mRNAs with spliced 3′-UTR introns through queries at http://mips.helmholtz-muenchen.de/funcatDB/ using the N. crassa (TaxId:5141) to p3_p13841_Neu_crass_MIPS containing 10067 annotated genes. Of 31 genes, 28 could be identified in the database; NCU03491 (RNA Splicing factor Pad-1, Broad Funcat as 11.02.03 mRNA synthesis and 11.04.03.01 Splicing), NCU10500 (F-box domain-containing protein), NCU11426 (short chain oxidoreductase) were not identified.
Results
N. crassa NMD mutations increase the levels and stability of mRNAs that are NMD targets
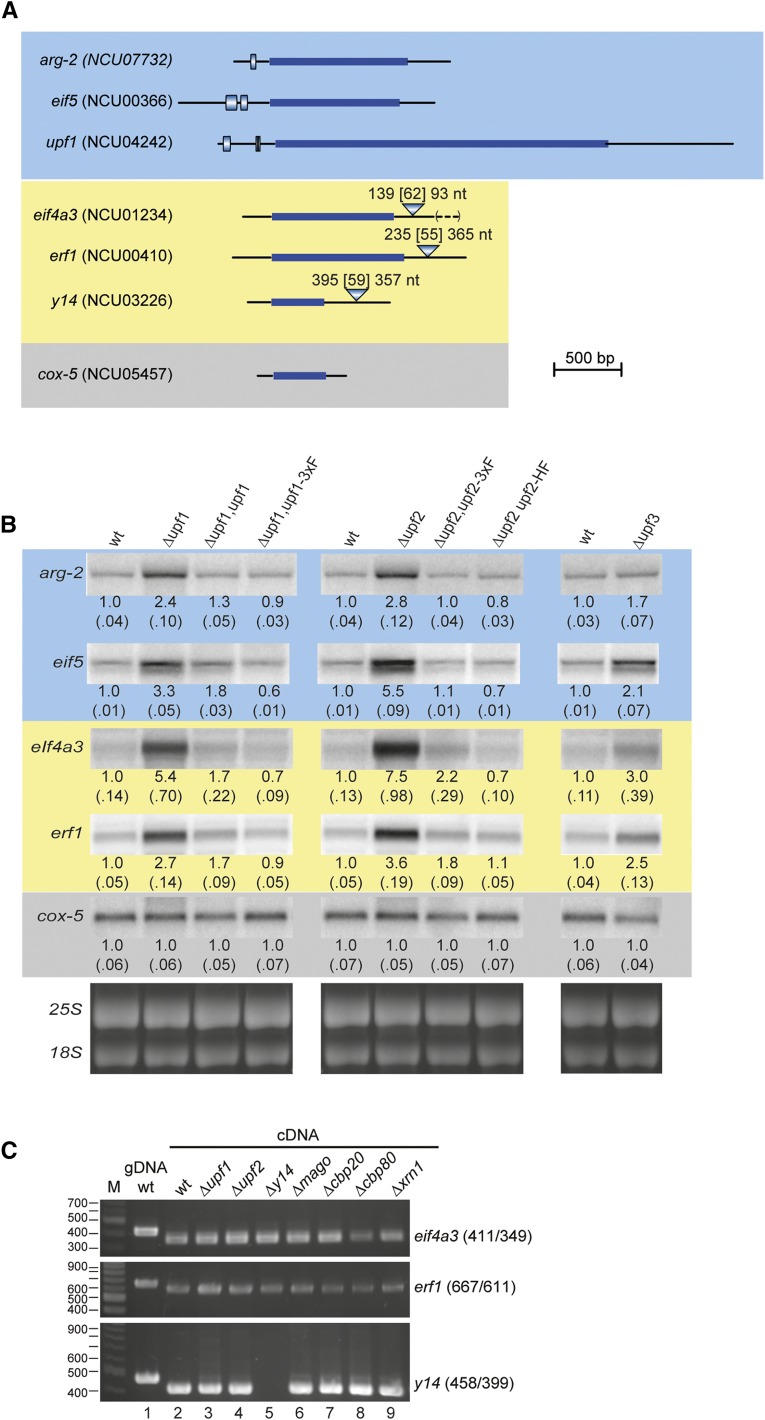
The genes specifying N. crassa UPF1 and UPF2, as well as other factors selected on the basis of their importance for controlling mRNA stability in NMD-related pathways in other organisms, could be identified on the basis of sequence similarities to homologs in other organisms (Table S1 and Figure S1). N. crassa Δupf1 (NCU04242) and Δupf2 (NCU05267) strains had defects in growth and development when cultured in flasks, slants, plates, or race tubes (Figure S2, Table S2, and File S1). The effect of these mutations on mRNA levels for selected genes that had uORFs or spliced 3′-UTR introns (Figure 1A) was examined by Northern analysis (Figure 1B). When compared to the wild-type strain (wt), levels of arg-2, eif5, eif4a3, and erf1 mRNA, but not cox-5 mRNA, increased in Δupf1 and Δupf2. arg-2 contains an evolutionarily conserved uORF specifying the arginine attenuator peptide (AAP) that causes ribosomes to arrest at the uORF stop codon in response to arginine (Wei et al. 2012 and references therein). Ribosome arrest at the homologous S. cerevisiae CPA1 uORF termination codon was demonstrated to trigger NMD (Gaba et al. 2005). N. crassa eif5 contains two uORFs, which by analogy to their functions in the homologous mammalian transcripts are predicted to have roles in eIF5 autoregulation (Loughran et al. 2012). The N. crassa eif4a3, erf1, and y14 transcripts each contain a 3′-UTR intron that was efficiently spliced in wt and all mutants examined here (Figure 1C). Such introns downstream of termination codons could trigger NMD via the EJC if a similar system to that operating in higher eukaryotes were present in N. crassa. The cox-5 transcript lacks features that would be expected to trigger NMD and was used as a negative control. The effects of Δupf1 on growth phenotypes and mRNA levels were corrected by expressing UPF1 (either wild-type UPF1 or UPF1 containing a C-terminal 3XFLAG) from a gene placed at the his-3 locus (Figure 1B, Figure S2, and File S1). Similarly, placing a gene specifying C-terminally tagged UPF2 at his-3 corrected the observed Δupf2 defects (Figure 1B, Figure S2, and File S1). N. crassa Δupf3 (NCU03435) did not show as pronounced effects as Δupf1 or Δupf2 on growth (Table S2) or mRNA levels (Figure 1B). The reason for this is unclear, but the effects of loss of UPF3 activity in mammals on mRNA stability can be different than loss of UPF1 activity (Chan et al. 2007), and this mutant was not examined further.
Figure 1.
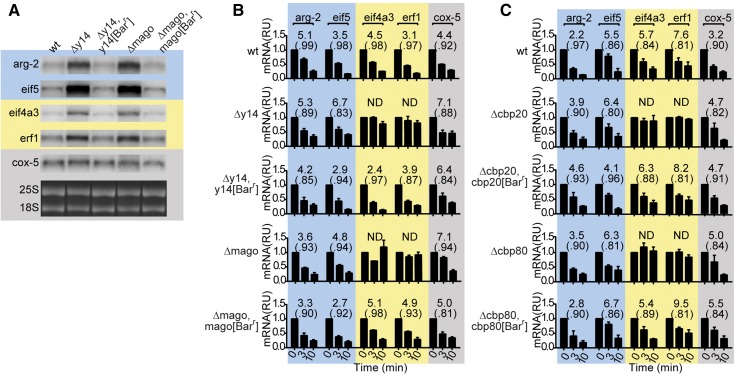
Analyses of uORF-containing and spliced 3′-UTR-containing transcripts. (A) Structures of mRNAs specifying arg-2, eif5, upf1, eif4a3, erf1, y14, and cox-5. The solid bars indicate the genic coding regions; the bars with gradients depict uORFs; the triangles indicate the positions and sizes of 3′-UTR introns (no other intron positions are indicated). The distance between the ORF stop codon and the splice site, the size of the spliced intron, and the distance between the splice site and the major poly(A) site are given [e.g., for eif4a3, 139, (62), 93 nt]. Here and throughout, shading indicates mRNA features: blue, uORFs that trigger NMD; yellow, spliced 3′-UTRs that trigger NMD; gray, no NMD-triggering features. An extended form of the spliced eif4a3 mRNA (Figure S4C) contains an additional 87 nt (indicated by dashed lines between parentheses). (B) Northern analyses of wt, Δupf1, Δupf1 his-3::upf1, Δupf1 his-3::upf1- 3XFLAG, Δupf2, Δupf2 his-3::upf2-3XFLAG, Δupf2, his-3::upf2-HAT-FLAG, and Δupf3. Total RNA (3 μg/lane) was analyzed with radiolabeled probes for arg-2, eif4a3, erf1, eif5, and cox-5. Levels of mRNA were quantified and normalized to the level of cox-5 mRNA. The quantification shown below the bands represents the average and standard deviation from three independent growth experiments. 25S and 18S rRNA bands stained with ethidium bromide from representative gels are shown below the Northerns. The doublet for the intronless eif5 transcript likely represents mRNA isoforms. (C) eif4a3, erf1, and y14 3′-UTR introns are efficiently spliced in all strains examined. M, markers; gDNA, PCR products of genomic DNA template; cDNA, PCR products from cDNA templates. The predicted sizes of PCR products from different templates are indicated. The Δy14 strain lacks y14 mRNA. See also Figure S1 and Figure S2.
To determine whether Δupf1 and Δupf2 directly affected mRNA stability, we first established conditions to metabolically label RNA with 4TU and to perform pulse–chase analyses by pulse-labeling RNA with 4TU and chasing with excess uracil (Figure 2A). 4TU pulse–chase avoids complications from shutting down transcription. 4TU is converted to 4-thiouridine monophosphate by uracil phosphoribosyl transferase [encoded by N. crassa uc-4 (Buxton and Radford 1982)], converted to the triphosphate, and then incorporated into RNA (Cleary et al. 2005). 4TU-RNA can be biotinylated, captured by interaction with avidin, and recovered after reversing linkage to the biotinylation reagent. The capacity to incorporate 4TU into RNA was assessed in wt N. crassa and Δuc-4 (NCU01446). Only wt cells labeled with 4TU gave a strong signal over background (Figure S3, A and B). As expected, new transcription was necessary to obtain 4TU-labeled RNA: preincubation with the general transcription inhibitor thiolutin, which has been shown to decrease transcription in N. crassa (Hoyt et al. 2000; Lee et al. 2009), severely reduced 4TU incorporation, as did chilling cells on ice (Figure S3, C and D). Incorporation of 4TU into RNA rapidly diminished when excess uracil was added to cultures (Figure S3E) and 4TU itself had minimal impact on N. crassa growth under the experimental conditions used (<30 min incubation) or during growth for extended periods (Figure S3F), similar to its minimal effects on growth in S. cerevisiae (Munchel et al. 2011) and S. pombe (Sun et al. 2012). Furthermore, purified 4TU RNA, unlabeled total RNA and unlabeled poly(A) mRNA programmed the synthesis of [35S]Met-labeled polypeptides with similar distributions of masses in N. crassa cell-free translation extracts (Figure S3G). We therefore examined N. crassa mRNA stability by pulse-labeling cells for 15 min with 0.2 mM 4TU added to the growth medium followed by the addition of a 50-fold molar excess of uracil directly to growth medium. This procedure quickly and effectively reduced 4TU-labeling of new RNA for the time-frame of our analyses, streamlined parallel analyses of multiple cultures, and avoided potential complications arising from switching cells from conditioned to fresh medium.
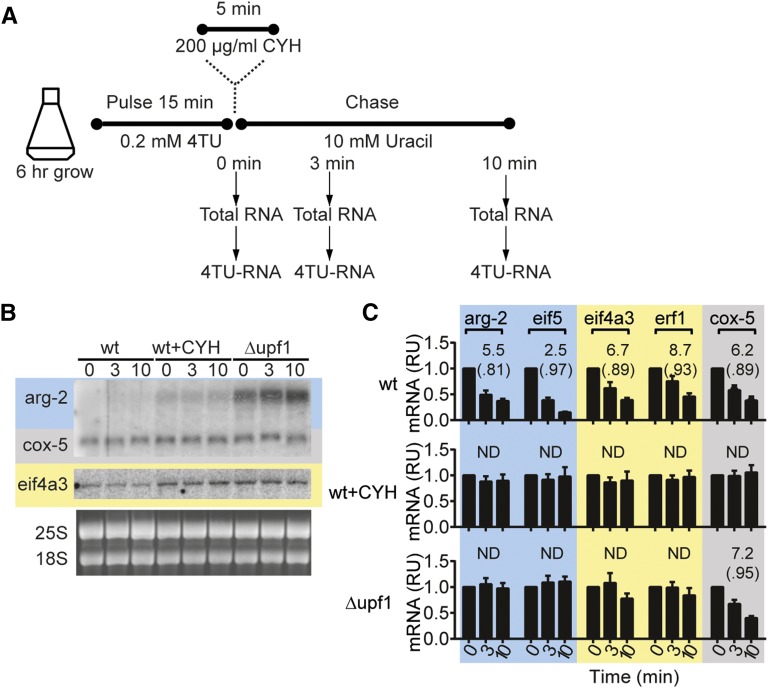
Figure 2.
mRNA levels and mRNA stability are affected by growth in cycloheximide and by Δupf1. (A) Strategy for pulse–chase analyses of mRNA stability. N. crassa conidia were germinated and grown for 6 hr; 0.2 mM 4TU was then added. After 15 min incubation with 4TU, 10 mM uracil was added to the culture, and total RNA and 4TU–RNA was purified from samples taken at 0, 3, and 10 min. In one experiment, cycloheximide (CYH) was added after the 15-min 4TU-pulse; following 5 min of incubation with CYH, the uracil-chase was then performed. (B) Total RNA from wt, Δupf1, and wt cells treated with 200 μg/ml CYH and subjected to 4TU pulse–chase were analyzed by Northern blotting as in Figure 1B; the arg-2 and cox-5 probes were added together. (C) Pulse–chase analyses of 4TU RNA from wt, Δupf1, and CYH-treated wt. 4TU RNA was purified and quantified by RT–qPCR and levels of 4TU mRNAs for arg-2, eif5, eif4a3, erf1, and cox-5 were normalized to levels of 4TU-labeled 25S rRNA in each sample (4TU-labeled 25S rRNA is stable during a 10-min chase) and then normalized to the level at time 0. Differences in expression are shown in relative units (RUs). The results are the average of three independent experiments. The mRNA half-lives (in minutes), calculated by fitting data to first order-decay parameters, are shown, as are R2 values for each calculation (in parentheses). ND, not determined. See also Figure S3.
We examined the effects of Δupf1 and Δupf2 on mRNA levels and stability. Figure 2 compares the level and stability of mRNAs in wt and Δupf1 cells, and wt cells to which cycloheximide (CYH) was added after 4TU labeling but before the uracil chase was started. CYH blocks eukaryotic translation elongation and in previous studies stabilized NMD-targeted mRNAs (Herrick et al. 1990; Carter et al. 1995; Zhang et al. 1997; Saul et al. 2009). In Northern analyses of total RNA in CYH-treated wt cells, levels of arg-2 and eif4a3 increased substantially, and cox-5 increased slightly (Figure 2B). Levels of arg-2 and eif4a3, but not cox-5, increased in Δupf1. Pulse–chase analyses with 4TU showed that arg-2, eif4a3, and cox-5 decayed with first-order kinetics in untreated wt cells (Figure 2C). The stability of arg-2, eif4a3, erf1, eif5, and cox-5 in wt cells increased when cells were treated with CYH (Figure 2C). This indicates that CYH was not affecting NMD substrates specifically, but was acting generally on all mRNAs examined; a mechanism by which it can do so is to inhibit decapping and thus inhibit mRNA degradation (Beelman and Parker 1994). In Δupf1, the stability of arg-2, eif4a3, erf1, and eif5, but not cox-5, increased compared to wt cells as determined by pulse–chase analysis (Figure 2C and Figure 3A). Thus, consistent with studies in other organisms, adding CYH to cultures stabilized mRNA, and the Δupf1 mutation stabilized mRNAs that contained features predicted to make them direct targets of NMD. The effects of Δupf2 on the stability of arg-2, eif4a3, erf1, and eif5 (Figure 3B) were similar to those of Δupf1, indicating that these transcripts are direct targets of NMD. Introduction of functional copies of upf1 and upf2 into the corresponding Δupf1 and Δupf2 strains reduced the stability of arg-2, eif4a3, erf1, and eif5 transcripts to near wild-type levels and largely restored the mutant growth phenotypes, as expected if these phenotypes were direct consequences of the deletion mutations (Figure 3 and Figure S2).
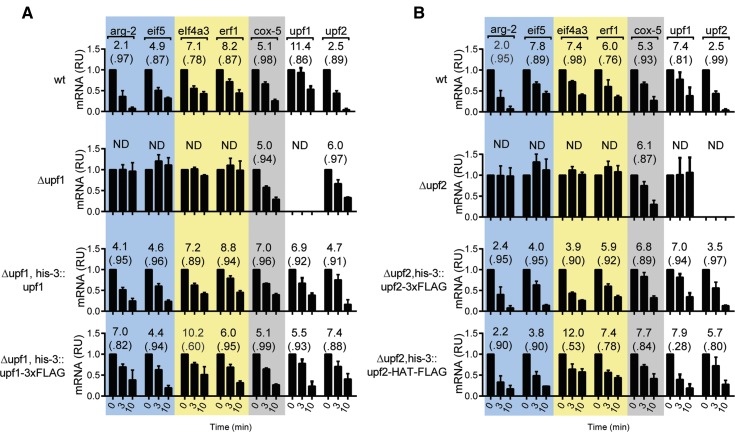
Figure 3.
Effects of Δupf1, Δupf2 mutations and correction of these mutants on the stability of selected mRNAs. 4TU RNA from wt, Δupf1, Δupf1 his-3::upf1, Δupf1 his-3::upf1-3XFLAG strains (A) or Δupf2, Δupf2 his-3::upf2-3XFLAG and Δupf2, his-3::upf2-HAT-FLAG strains (B) were analyzed by pulse–chase for arg-2, cox-5, upf1, upf2, eif4a3, erf1, and eif5 mRNAs using the procedures described in Figure 2. See also Figure S6.
We next examined the consequences of disrupting the predicted N. crassa homolog of XRN1 (NCU06678; see Table S1). XRN1 specifies an evolutionarily conserved 5′–>3′ exonuclease important in general mRNA degradation (Hsu and Stevens 1993; Nagarajan et al. 2013). We examined the Δxrn1 strain to establish whether loss of this 5′–>3′ exonuclease had generalized stabilizing effects on mRNA to provide a necessary context for interpreting subsequent analyses of the functions of NMD, EJC, and CBC components on specific mRNAs. The N. crassa Δxrn1 strain had growth and developmental defects (Figure S2 and Table S2) and stabilized the mRNAs we examined by metabolic labeling with 4TU (Figure 4A). Relative to the wild-type strain, levels of each mRNA were also increased in the mutant (Figure 4B). The growth and mRNA phenotypes were rescued by transforming the Δxrn1 mutant with the wild-type xrn1 gene (Figure S2 and Figure 4). These data indicate that XRN1-dependent mRNA degradation is also important in N. crassa for determining mRNA stability.
Figure 4.
Effects of Δxrn1 and correction of the mutant on the stability of selected mRNAs. (A) 4TU RNA from wt, Δxrn1, and Δxrn1 xrn1[Barr] strains were analyzed by pulse–chase for arg-2, eif5, eif4a3, erf1, and cox-5 mRNAs using the procedures described in Figure 2. (B) Levels of mRNA in total RNA from wt, Δxrn1, and Δxrn1 xrn1[Barr] strains was quantified by RT–qPCR and mRNA levels were normalized to the level of 25S rRNA.
The arg-2 uORF and 3′-UTR introns of eif4a3 and erf1 confer NMD-control to a reporter transcript
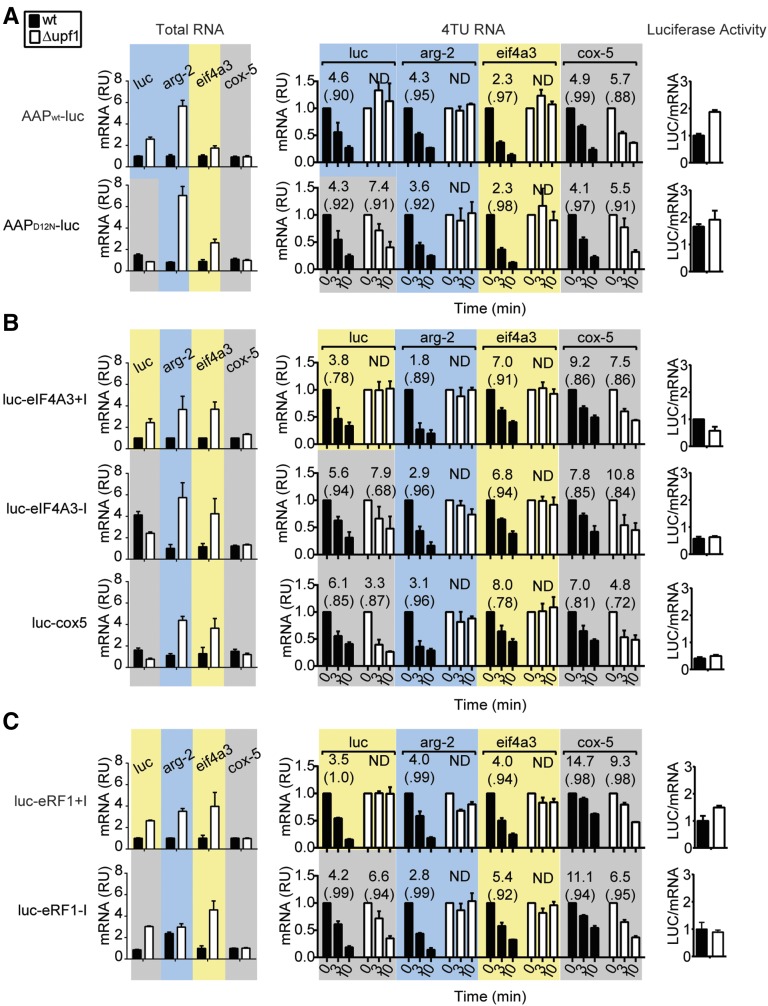
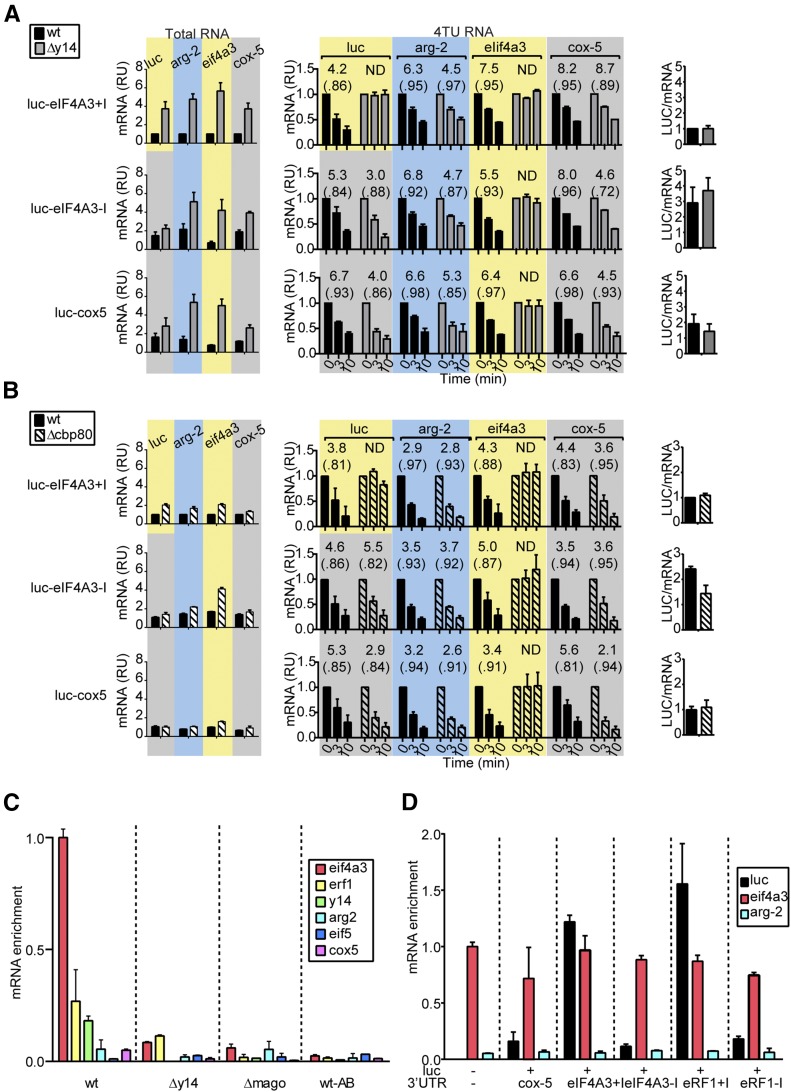
We hypothesized that the arg-2 uORF and the loading of an EJC at the eif4a3 and erf1 3′-UTR introns were responsible for triggering NMD of each respective mRNA. To test this, we introduced these elements into firefly luciferase (luc) reporter constructs. We created wild-type upf1 and Δupf1 strains harboring luc reporters integrated at the his-3 locus (Figure S4A). One matched set of reporters contained either the arg-2 uORF specifying the wild-type AAP (AAPwt–luc) or the mutated D12N AAP (AAPD12N–luc), which eliminates the AAP’s capacity to cause ribosome arrest at the uORF stop codon (Wei et al. 2012 and references therein). Another matched set of reporters produced luc mRNAs with the 3′-UTRs of cox-5, eif4a3, or erf1. Reporters whose 3′-UTRs either contained or lacked the eif4a3 and erf1 3′-UTR introns were constructed (Figure S4A). Analyses of total RNA and 4TU pulse–chase data showed the importance of the arg-2 uORF and the 3′-UTR introns of eif4a3 and erf1 in controlling mRNA stability (Figure 5, A–C). Only the reporter transcripts containing the wt AAP or the spliced eif4a3 or erf1 3′-UTR introns were stabilized in Δupf1 mutant. The 3′-UTRs of reporter mRNAs were mapped by 3′-RACE and showed (i) intron-containing and intron-less reporters were polyadenylated at the sites predicted from the endogenous genes and (ii) the introns were correctly spliced (Figure S4C). The efficiency of translation of each of the mRNAs was compared by measuring LUC enzyme activity in cell extracts and normalizing activities to luc mRNA levels. Neither the 3′-UTR intron of eif4a3 or erf1 substantially affected translation efficiency (Figure 5, B and C).
Figure 5.
Effects of the arg-2 uORF-encoded AAP and the eif4a3 and erf1 3′-UTR introns on the level and stability of luciferase (luc) reporter mRNA in wt and Δupf1 strains. Measurements to analyze the effects on reporter genes containing the arg-2 uORF (A), the eif4a3 3′-UTR intron (B), and the erf1 3′-UTR intron (C) and controls are shown as follows for each: levels of mRNA in total RNA (left), half-lives of mRNA measured by 4TU RNA pulse–chase (middle), and levels of luciferase enzyme activity/mRNA (right). wt (solid bars) or Δupf1 (open bars) strains contained the luc reporters shown in Figure S4A. Measurements of RNA were accomplished as in Figure 4. Luciferase enzyme activity was measured and normalized to luc mRNA levels. See also Figure S4.
Deletions of EJC components increase the levels and stability of mRNAs that contain 3′-UTR introns
Three of the major components of the EJC are eIF4A3, Y14, and Mago (Table S1). The N. crassa Δeif4a3 mutant was not viable: Δy14 grew slowly and had developmental abnormalities; Δmago also displayed growth and development defects but these were not as severe as those of Δy14 (Figure S2, Table S2, and File S1). Overall, mRNA transcript levels in total RNA preparations appeared higher in Δy14 and Δmago than in wt when examined by Northern analysis (Figure 6A) and by RT–qPCR (see Figure 7A). In Δy14 and Δmago, the stability of eif4a3 and erf1 transcripts, which contain 3′-UTR introns, increased, but the stability of arg-2, eif5, and cox-5 transcripts did not (Figure 6B). Thus while increased levels of transcripts in Δy14 and Δmago might reflect altered transcription levels in addition to increased transcript stability, increased RNA stability was observed only for transcripts containing spliced 3′-UTR introns. Reintroduction of functional y14 and mago genes into the respective strains largely restored the observed defects of the mutants (Figure 6, A and B, Figure S2, and File S1).
Figure 6.
Effects of Δy14, Δmago, Δcbp20, and Δcbp80, and correction of these mutants, on the stability of selected mRNAs. (A) Aliquots (3 μg) of total RNA extracted from wt, Δy14, Δy14 y14-Barr, Δmago, and Δmago mago[Barr] were separated on formaldehyde denaturing gels and analyzed as described in Figure 1B with the indicated probes. 4TU RNA from wt, Δy14, Δy14 y14[Barr], Δmago, Δmago mago[Barr] (B) or wt, Δcbp20, Δcbp20 cbp20[Barr], Δcbp80, Δcbp80 cbp80[Barr] (C) was analyzed by pulse–chase for arg-2, eif5, eif4a3, erf1, and cox-5 mRNAs using the procedures described in Figure 2. See also Figure S6.
Figure 7.
Effects of the eif4a3 3′-UTR intron on the level and stability of luc reporter mRNA in wt, Δy14 and Δcpb80 strains. Measurements to analyze the effects on reporter genes containing the eif4a3 3′-UTR intron and controls are shown as follows for each: levels of mRNA in total RNA (left), half-lives of mRNA measured by 4TU RNA pulse–chase (middle), and levels of luciferase enzyme activity/mRNA (right). (A) wt (solid bars) and Δy14 (shaded bars) strains contained the luc reporters shown in Figure S4B. (B) wt (solid bars) and Δcbp80 (hatched bars) strains contained the luc reporters shown in Figure S4B. Measurements were obtained as described in Figure 5. (C) RT–qPCR analysis of eif4a3, erf1, y14, arg-2, eif5, and cox-5 mRNAs purified from wt, Δy14, and Δmago strains by immunopurification of mRNPs with anti-Y14 antibody as described in Materials and Methods. The level of each transcript was normalized to the total amount of purified RNA and then normalized to the level of input transcript. Mock immunopurification from wt without antibody (wt-AB) served as the control. (D) RT–qPCR analyses of luc, eif4a3, and arg-2 mRNAs purified from wt and luc reporter strains by immunopurification of mRNPs with anti-Y14 antibody. mRNA enrichment is shown normalized to the enrichment of eif4a3 in immunopurification from wt cells. See also Figure S5 and Figure S6.
The eIF4A3 3′-UTR intron confers mRNA-stability control through the EJC component Y14
If N. crassa y14 is involved in NMD-mediated control of mRNA stability mediated by an EJC deposited downstream of a termination codon, then the stability of a reporter containing the eif4a3 3′-UTR intron but not a reporter lacking the 3′-UTR intron should be increased in the Δy14 strain. While his-3 is routinely used for site-specific integration of reporters in N. crassa, including the reporters used to study the effects of NMD mutations described above, his-3 and y14 are tightly linked chromosomal loci. We therefore integrated luc reporter constructs at the csr-1 locus, which is not closely linked to y14 (Figure S4B) and crossed these reporter strains to Δy14 to obtain sibling strains containing each reporter and either wild-type y14 or Δy14 alleles to test the impact of Δy14 on the matched reporter constructs.
Δy14 affected the stability of the luc reporter with the intron-containing eif4a3 3′-UTR, but not reporters with the intronless eif4a3 3′-UTR or cox-5 3′-UTR (Figure 7A). In the wild-type strain, the stabilities of all of the reporter mRNAs were similar. Analyses by 3′-RACE showed the reporter transcripts containing the intron were correctly spliced and polyadenylated (Figure S4C). As expected, the stability of the endogenous eif4a3 transcript also was increased in Δy14 but not wt (Figure 7A). Measurements of translational efficiency indicated that the presence of the eif4a3 3′-UTR intron decreased translation efficiency in both the wild-type and Δy14 strains (Figure 7A), arguing against the possibility that increased translation of the spliced mRNA accounts for its instability. These data (Figure 6B and Figure 7A) indicate that Δy14 is specifically affecting the stability of transcripts that contain 3′-UTR introns.
Mutations eliminating nuclear cap-binding complex components CBP20 and CBP80 increase the levels and stability of mRNAs that contain 3′ UTR introns
In higher eukaryotes, CBP20 and CBP80, which form the nuclear CBC, affect the stability of transcripts that contain an intron downstream of the native termination codon (Hwang et al. 2010; Popp and Maquat 2013). Therefore we examined the effects of deleting these factors on the stability of 3′-UTR-intron containing N. crassa transcripts. The Δcbp20 and Δcbp80 strains had mild growth phenotypes (Figure S2, Table S2, and File S1). Analyses of mRNA stability (Figure 6C) showed that deletion of either Δcbp20 or Δcbp80 increased the stability of the 3′-UTR–intron transcripts eif4a3 and erf1 but not arg-2, eifi5, or cox-5. The growth defects and the altered stability of eif4a3 and erf1 transcripts in the mutant strains were corrected when the strains were rescued with wild-type copies of the genes (Figure 6C, Figure S2, and File S1).
We tested whether the effects of these deletions on mRNA stability was due to the presence of a 3′-UTR intron in the affected genes by examining luc reporters with and without the eif4a3 intron (Figure 7B and Figure S5). Only reporters containing the 3′-UTR intron showed increased stability in the Δcbp20, Δcbp80, and the Δcbp20 Δcbp80 double-mutant strains. Measurements of translational efficiency in the Δcbp80 strain (Figure 7B) indicated that the spliced reporter mRNA was translated with less, not more, efficiency than the intronless reporter, arguing against increased translation of the spliced mRNA accounting for its instability.
The y14 mRNA also contains a 3′-UTR intron (Figure 1A). Therefore we examined the half-life of this transcript in Δupf1, Δupf2, Δmago, Δcbp20, and Δcbp80 strains and the corresponding rescued strains (Figure S6, A–C). The effects of these mutations and their corrections on y14 mRNA half-life were similar to the effects observed on eif4a3 and erf1 mRNA half-lives, indicating that the y14 mRNA’s spliced 3′-UTR intron also controls this mRNA’s stability. The levels of y14 mRNA in total RNA in the different strains showed a profile similar to the eif4a3 and erf1mRNAs (Figure S6D). Thus, the y14 mRNA’s spliced 3′-UTR intron appears important for NMD; however, it is also possible that the 752-nt 3′-UTR of the major form of the spliced transcript could itself trigger NMD by virtue of its being a long 3′-UTR.
Immunopurification with anti-Y14 antibody enriches for mRNA containing 3′-UTR introns
The stabilities of N. crassa mRNAs containing spliced 3′-UTR introns appear controlled through the EJC. Thus it is important to obtain direct evidence whether there is binding of the EJC to mRNAs containing spliced 3′-UTR introns. RT–qPCR was used to examine RNAs that were immunopurified from N. crassa extracts using monoclonal antibody directed against human Y14. Enrichment for erf1, eif4a3, and y14 mRNA, relative to arg-2, cox-5, and eif5 mRNA, was seen when wild-type cells were examined (Figure 7C). Such enrichment was not observed in immunopurifications from the Δy14 or Δmago mutants or from mock immunopurifications from the wild-type strain (Figure 7C). Importantly, in wild-type strains containing luc reporters, enrichment was observed for reporters with either eif4a3 or erf1 3′-UTR introns but not from matching reporters lacking 3′-UTR introns (Figure 7D), and enrichment of the endogenous eif4a3 mRNA relative to the arg-2 mRNA is observed in all cases. These data indicate that Y14, which is part of the EJC, is associated with mRNAs containing spliced 3′-UTR introns. Taken together with the RNA stability data, this indicates a direct role for the EJC in governing the stability of mRNAs containing spliced 3′-UTR introns.
Discussion
Here we show that the degradation of mRNA through the NMD pathway in the model filamentous fungus N. crassa has at least two branches, one requiring the NMD factors and the other requiring EJC and CBC factors in addition to NMD factors. We show that this latter pathway controls the stability of spliced 3′-UTR intron containing mRNAs for the EJC components eIF4A3 and Y14, and the nonsense-codon-sensing termination factor eRF1. The absence of evidence for EJC-mediated NMD in fungi has been discussed (Rebbapragada and Lykke-Andersen 2009; Wen and Brogna 2010; Kervestin and Jacobson 2012); this is the first demonstration of EJC-mediated NMD in fungi. Our results thus establish that this quality-control process controlling mRNA stability has evolutionary and functional conservation. While the importance and necessity for CBC factors in EJC-dependent NMD have been debated (Hosoda et al. 2005; Dzikiewicz-Krawczyk et al. 2008; Maquat et al. 2010; Durand and Lykke-Andersen 2013; Rufener and Muhlemann 2013), our data provide the first demonstration in any system that CBC factors are not only important for, but are essential for, the EJC-dependent branch of NMD.
Our results show that N. crassa eIF4A3, eRF1, Y14, and UPF1 are subject to autoregulatory control at the level of mRNA stability through pathways involving NMD, EJC, and CBC factors because mutations that eliminate these factors increase the stability of these transcripts. Specifically, eif4a3 and erf1 mRNA stability increased in Δupf1, Δupf2, Δy14, Δmago, Δcbp20, and Δcbp80 mutants; y14 mRNA stability increased in Δupf1, Δupf2, Δmago, Δcbp20, and Δcbp80 mutants; and upf1 mRNA stability increased in the Δupf2 mutant. The transcripts for two EJC factors (eIF4A3 and Y14) and for termination factor eRF1 each contain a spliced 3′-UTR intron and reporter analyses demonstrated that the spliced eif4a3 and erf1 3′-UTR introns were sufficient to confer control of mRNA stability through NMD-related pathways. Therefore the situation appears analogous to what occurs in higher eukaryotes in which such introns can trigger EJC-mediated NMD. NMD control of N. crassa upf1 mRNA stability might be exerted through recognition of its uORF stop codons as PTCs, as is the case for N. crassa arg-2 (Figure 5A) and S. cerevisiae CPA1 (Gaba et al. 2005), or through its relatively long 3′-UTR (Figure 1A) as is reported for metazoan UPF1 (Huang et al. 2011; Longman et al. 2013), but this remains to be determined.
To our knowledge, this work represents the first demonstration of circuits that enables regulation at the level of mRNA stability for eRF1, eIF4a3, and Y14. Autoregulation of mRNA levels for NMD factors has been reported for metazoans (Huang et al. 2011; Yepiskoposyan et al. 2011; reviewed in Karam et al. 2013; Longman et al. 2013) and autoregulation of NMD and EJC factors has been reported for plants (Nyiko et al. 2013). Measurements of mRNA stability in mammalian cells established that UPF2 and SMG1 (a kinase that phosphorylates UPF1) are stabilized in UPF1-depleted cells and that UPF1 is stabilized in SMG1 depleted cells; furthermore the UPF1 3′-UTR conferred UPF1-dependent mRNA stability to a reporter (Huang et al. 2011). The long 3′-UTRs of mRNAs specifying UPF1, SMG5, and SMG7 (the latter two proteins affect the dephosphorylation of UPF1) reduced the relative stability of an mRNA reporter in mammalian cells (Yepiskoposyan et al. 2011). The transcript levels for Arabidopsis SMG7 and Barentsz/MLN51 (which in higher eukaryotes is another component of the EJC), each contain a spliced 3′-UTR intron (Nyiko et al. 2013). Analyses of reporters containing these spliced 3′-UTR introns showed that their mRNA levels were upregulated in upf1 and upf3 mutants and that the 3′-UTR introns were at least partially responsible for this alteration in transcript levels; in these studies, mRNA stability was not directly assessed (Nyiko et al. 2013).
We confirmed that other filamentous fungi also contained spliced 3′-UTR introns in their eif4a3, y14, and erf1 transcripts, indicating evolutionary conservation of these elements. Specifically, analyses using FungiDB (Stajich et al. 2012) show 3′-UTR introns in erf1 in Fusarium verticillioides (FVEG_08271), A. nidulans (AN8853), and Magnaporthe oryzae (MGG_06266); in eif4a3 in F. oxysporum (FOXG_07544), F. verticillioides (FVEG_04473), M. oryzae (MGG_04885), and y14 in N. discreta (NEUDI_81291), N. tetrasperma (NEUTE1DRAFT_149143), and F. verticillioides (FVEG_03573). The erf1 mRNA of the basidiomycete pathogen Cryptococcus neoformans (CNAG_02948) also has a 3′-UTR intron (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html).
The appearance of spliced 3′-UTR introns in eif4a3, y14, and erf1 transcripts raises the question of their appearance in other transcripts. Gene models from Release 7 of the N. crassa genome indicate that 321 of the 9728 predicted protein coding genes in N. crassa potentially have 3′-UTR introns. For 237, the 3′-UTR-intron was present in all predicted transcript models. We examined these 237 transcripts using our recently published RNA-seq data (Wu et al. 2014). We obtained clear evidence for a spliced 3′-UTR intron in 31 cases (Table S3). FunCat analyses of these 31 genes indicated enrichment in categories related to DNA, RNA, and protein synthesis as well as other several categories (Table S4). Thus the fraction of genes that produce normal transcripts with spliced 3′-UTR introns appears to be quite low in N. crassa, at least in vegetatively growing cultures. In contrast, many N. crassa transcripts contain uORFs; based on EST analyses, 22% of N. crassa mRNAs are estimated to contain uORFs (Galagan et al. 2005).
The N. crassa CBC components CBP20 and CBP80 are crucial for the EJC-mediated NMD processes we observed (Figure 6, Figure 7, and Figure S5). The question of whether mRNA must be associated with the CBC for decay to occur has been debated with respect to mammals (Maquat et al. 2010; Durand and Lykke-Andersen 2013; Rufener and Muhlemann 2013) and while CBC has been considered important for the process, whether it is essential is also unclear. Examination of N. crassa mutants lacking CBP20 and/or CBP80 clearly indicate that the capacity of cells to produce CBC controls the stability of endogenous and reporter transcripts containing a spliced 3′-UTR intron. This directly answers affirmatively the mechanistic question of whether CBC has an essential role for this process. The question of whether in N. crassa the CBC must be associated with the mRNA or can be been exchanged for eIF4E prior to the onset of decay is not yet resolved. Importantly in this regard, N. crassa CBC is not required for controlling the stability of uORF-containing mRNAs such as arg-2 and eif5 whose stability is controlled by NMD factors but not EJC factors. Taken together, these data show that CBC factors are important for producing mRNP that is permissive for EJC-mediated, but not uORF-mediated, NMD. It is increasingly apparent that the nuclear history of a transcript can define its fate in the cytoplasm (Trcek et al. 2011; Haimovich et al. 2013; Zid and O’Shea 2014), and regardless of whether the CBC or eIF4E is associated with the mRNA at the onset of decay, CBC appears essential for EJC-mediated NMD. Thus, the CBC is critical for licensing EJC-mediated NMD.
The levels of RNAs (eif4a3, erf1, and y14) that are stabilized in CBC mutants do not increase as dramatically in the total RNA pools of these mutants as they do in the EJC mutants (Figure 6A, Figure 7, A and B, and Figure S6D). A possible explanation for this is that the CBC mutants leave autoregulatory circuits intact that enable feedback between transcript levels and transcription for eif4a3, erf1, and y14, so that increased stability decreases transcription. With respect to the EJC and NMD mutants, this circuit might be disrupted or there might be additional transcriptional induction independent of an intact feedback circuit. The existence of such feedback circuits that connect RNA stability and transcription has been demonstrated in S. cerevisiae through the functions of XRN1 (Haimovich et al. 2013).
All of the viable deletion mutations affecting NMD, EJC, and CBC components displayed asexual growth phenotypes. Some of these mutants also had defects in their sexual development: the Δy14, Δmago, and Δupf2 strains are female sterile. The formation of protoperithecia, the earliest female-specific developmental structures in N. crassa, is eliminated in Δy14 and Δmago mutants under standard inducing conditions, and Δupf2 reduces protoperithecium formation. These data suggest that Y14, MAGO, and UPF2 have additional roles in the sexual cycle genetically distinguishable from their roles in triggering NMD in asexually growing cells. While the bases for these female-specific phenotypes for this subset of factors involved in NMD remain to be determined, it is interesting in this regard that mago was originally identified in Drosophila melanogaster as a maternal-effect gene affecting oocyte development and subsequent work has established roles for fly Y14 and Mago to correctly position oskar mRNA to enable oocyte development (Boswell et al. 1991; Hachet and Ephrussi 2001, 2004; Mohr et al. 2001; Ghosh et al. 2014). We note here that BLAST comparisons that find close homologs in N. crassa for Y14, Mago, and eIF4A3 do not detect the EJC component Barentsz/MLN51 (Table S1); whether N. crassa contains a functional homolog of Barentsz/MLN51, which has roles in NMD and mRNA positioning (Palacios et al. 2004) remains to be determined.
The N. crassa period-6 (prd-6) gene was identified to harbor a spontaneous mutation that shortens the period of the fungus’s circadian cycle at temperatures above, but not below, 21° (Morgan and Feldman 1997). Unpublished work identified the product of this gene as UPF1 (Compton 2003) and the original prd-6 allele as a frameshift mutation resulting in a premature termination codon in the UPF1 coding region. The basis for the circadian phenotype of this upf1 allele, and whether mutations affecting other components of the NMD pathway similarly affect circadian rhythms in N. crassa, remain to be determined. More generally, the role of NMD in controlling circadian rhythms has not been documented in other organisms, although there is evidence that alternatively spliced transcripts for some Arabidopsis clock genes are degraded by NMD (Kwon et al. 2014) and NMD mutants in this organism show alterations in the way photoperiod-dependent genes are expressed (Shi et al. 2012).
The use of 4TU in N. crassa enabled unprecedented measurements of RNA stability by metabolic labeling. In N. crassa, a good fit to first-order decay kinetics of 4TU-labeled mRNAs is observed immediately upon addition of excess uracil in pulse–chase analyses. This does not appear to be the case when using wild-type strains of S. cerevisiae for similar metabolic labeling analyses. Studies using unlabeled uracil to chase [3H]uracil in S. cerevisiae to measure mRNA half-lives indicate it takes several minutes for a >2000-fold excess of unlabeled uracil to effectively block the incorporation of [3H]uracil into RNA (Crabeel et al. 1990). A study using 4TU to examine mRNA half-life uses a 10-min timepoint following the chase as the initial time point for determining half-life (Munchel et al. 2011) and the measured half-lives of S. cerevisiae mRNAs obtained by this procedure are longer than those determined from other methods. Explanations for this difference between these organisms might be that the addition of excess uracil does not immediately dilute the uracil or 4TU pool in S. cerevisiae because of differences in pyrimidine uptake, compartmentalization, or metabolism between S. cerevisiae and N. crassa.
The conceptual division between NMD-related processes in animals and plants and NMD-related processes in fungi because EJC-linked mRNA instability had not been previously demonstrated in the latter group is bridged by the work described here. Most work on fungal NMD has used S. cerevisiae, whose genome generally lacks introns. For this organism, it is logical to rationalize the absence of EJC-linked NMD because the dearth of introns in its genome could remove selective pressure to retain intron-processing linked quality-control mechanisms. However, the genomes of filamentous fungi are rife with introns and thus this quality-control mechanism would be expected to have important functions in N. crassa and other filamentous fungi. Our data indicate that there is a branch of NMD in N. crassa that corresponds to the faux-UTR branch in that there is no requirement for EJC or CBC factors but there is a requirement for at least UPF1 and UPF2. The stabilities of two uORF-containing mRNAs arg-2 and eif5 appear to be controlled by this branch, and it is likely that other uORF-containing or long 3′-UTR-containing mRNAs will be governed by this pathway.
Our data also demonstrate a second branch of NMD in which an intron downstream of a termination codon elicits NMD and requires EJC and CBC factors as well as at least UPF1 and UPF2. This second branch would have a role in quality control in destroying certain incorrectly or alternatively spliced mRNAs as well as governing levels of mRNAs that normally contain spliced 3′-UTR introns. Determining where there are differences between fungi and higher eukaryotes with respect to specific elements that comprise these mechanisms could provide targets for the development of new therapeutics; for example, the fungal pathogen C. neoformans is avirulent when it lacks XRN1, an exonuclease that has a central role in mRNA degradation (Wollschlaeger et al. 2014). The similarities between fungal systems and higher eukaryotes, and the experimental advantages that fungi offer, will enable experimental approaches for understanding the different branches of NMD and provide new opportunities for insights into fundamental mechanisms governing RNA stability.
Supplementary Material
Acknowledgments
We thank Meray Baştürkmen for initial development of the 4TU protocol, Cheng Wu for assistance with in vitro translation, and Allan Jacobson for helpful discussions. Strains were obtained from the Fungal Genetics Stock Center. This work was supported by National Institutes of Health R01 GM047498 and P01 GM068087. Neither Ying Zhang or Matthew S. Sachs has any financial conflict of interest that might be construed to influence the results or interpretation of this manuscript.
Footnotes
Communicating editor: M. Freitag
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.176743/-/DC1.
Literature Cited
- Amrani N., Ganesan R., Kervestin S., Mangus D. A., Ghosh S., et al. , 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118. [DOI] [PubMed] [Google Scholar]
- Amrani N., Sachs M. S., Jacobson A., 2006. Early nonsense: mRNA decay solves a translational problem. Nat. Rev. Mol. Cell Biol. 7: 415–425. [DOI] [PubMed] [Google Scholar]
- Bardiya N., Shiu P. K., 2007. Cyclosporin A-resistance based gene placement system for Neurospora crassa. Fungal Genet. Biol. 44: 307–314. [DOI] [PubMed] [Google Scholar]
- Beelman C. A., Parker R., 1994. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 269: 9687–9692. [PubMed] [Google Scholar]
- Bicknell A. A., Cenik C., Chua H. N., Roth F. P., Moore M. J., 2012. Introns in UTRs: why we should stop ignoring them. BioEssays 34: 1025–1034. [DOI] [PubMed] [Google Scholar]
- Boswell R. E., Prout M. E., Steichen J. C., 1991. Mutations in a newly identified Drosophila melanogaster gene, mago nashi, disrupt germ cell formation and result in the formation of mirror-image symmetrical double abdomen embryos. Development 113: 373–384. [DOI] [PubMed] [Google Scholar]
- Buxton F. P., Radford A., 1982. Partial characterization of 5-fluoropyrimidine-resistant mutants of Neurospora crassa. Mol. Gen. Genet. 185: 132–135. [Google Scholar]
- Carter M. S., Doskow J., Morris P., Li S., Nhim R. P., et al. , 1995. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 270: 28995–29003. [DOI] [PubMed] [Google Scholar]
- Chan W. K., Huang L., Gudikote J. P., Chang Y. F., Imam J. S., et al. , 2007. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 26: 1820–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary M. D., Meiering C. D., Jan E., Guymon R., Boothroyd J. C., 2005. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat. Biotechnol. 23: 232–237. [DOI] [PubMed] [Google Scholar]
- Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., et al. , 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J., 2003. p. 210 in Advances in Understanding the Molecular Biology of Circadian Rhythms in Neurospora crassa. University of California, Santa Cruz, CA. [Google Scholar]
- Crabeel M., LaValle R., Glansdorff N., 1990. Arginine-specific repression in Saccharomyces cerevisiae: kinetic data on ARG1 and ARG3 mRNA transcription and stability support a transcriptional control mechanism. Mol. Cell. Biol. 10: 1226–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. H., de Serres F. J., 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 27A: 79–143. [Google Scholar]
- Durand S., Lykke-Andersen J., 2013. Nonsense-mediated mRNA decay occurs during eIF4F- dependent translation in human cells. Nat. Struct. Mol. Biol. 20: 702–709. [DOI] [PubMed] [Google Scholar]
- Dyer B. W., Ferrer F. A., Klinedinst D. K., Rodriguez R., 2000. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal. Biochem. 282: 158–161. [DOI] [PubMed] [Google Scholar]
- Dzikiewicz-Krawczyk A., Piontek P., Szweykowska-Kulinska Z., Jarmolowski A., 2008. The nuclear cap-binding protein complex is not essential for nonsense-mediated mRNA decay (NMD) in plants. Acta Biochim. Pol. 55: 825–828. [PubMed] [Google Scholar]
- Ebbole D., Sachs M. S., 1990. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 37: 17–18. [Google Scholar]
- Feinberg A. P., Vogelstein B., 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132: 6–13. [DOI] [PubMed] [Google Scholar]
- Feldbrugge M., Zarnack K., Vollmeister E., Baumann S., Koepke J., et al. , 2008. The posttranscriptional machinery of Ustilago maydis. Fungal Genet. Biol. 45(Suppl. 1): S40–S46. [DOI] [PubMed] [Google Scholar]
- Gaba A., Jacobson A., Sachs M. S., 2005. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell 20: 449–460. [DOI] [PubMed] [Google Scholar]
- Galagan J. E., Calvo S. E., Cuomo C., Ma L. J., Wortman J. R., et al. , 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438: 1105–1115. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Obrdlik A., Marchand V., Ephrussi A., 2014. The EJC binding and dissociating activity of PYM is regulated in Drosophila. PLoS Genet. 10: e1004455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O., Ephrussi A., 2001. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 11: 1666–1674. [DOI] [PubMed] [Google Scholar]
- Hachet O., Ephrussi A., 2004. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 428: 959–963. [DOI] [PubMed] [Google Scholar]
- Haimovich G., Medina D. A., Causse S. Z., Garber M., Millan-Zambrano G., et al. , 2013. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell 153: 1000–1011. [DOI] [PubMed] [Google Scholar]
- Herrick D., Parker R., Jacobson A., 1990. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 2269–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S., Selker E., 2009. Tools for fungal proteomics: multifunctional Neurospora vectors for gene replacement, protein expression and protein purification. Genetics 182:11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda N., Kim Y. K., Lejeune F., Maquat L. E., 2005. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 12: 893–901. [DOI] [PubMed] [Google Scholar]
- Hoyt M. A., Broun M., Davis R. H., 2000. Polyamine regulation of ornithine decarboxylase synthesis in Neurospora crassa. Mol. Cell. Biol. 20: 2760–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. L., Stevens A., 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13: 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Lou C. H., Chan W., Shum E. Y., Shao A., et al. , 2011. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol. Cell 43: 950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J., Sato H., Tang Y., Matsuda D., Maquat L. E., 2010. UPF1 association with the capbinding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol. Cell 39: 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y., Li X., Serin G., Maquat L. E., 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617. [DOI] [PubMed] [Google Scholar]
- Isken O., Maquat L. E., 2007. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 21: 1833–1856. [DOI] [PubMed] [Google Scholar]
- Karam R., Wengrod J., Gardner L. B., Wilkinson M. F., 2013. Regulation of nonsense-mediated mRNA decay: implications for physiology and disease. Biochim. Biophys. Acta 1829: 624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S., Jacobson A., 2012. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 13: 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y. J., Park M. J., Kim S. G., Baldwin I. T., Park C. M., 2014. Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biol. 14: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. C., Chang S. S., Choudhary S., Aalto A. P., Maiti M., et al. , 2009. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459: 274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D., Hug N., Keith M., Anastasaki C., Patton E. E., et al. , 2013. DHX34 and NBAS form part of an autoregulatory NMD circuit that regulates endogenous RNA targets in human cells, zebrafish and Caenorhabditis elegans. Nucleic Acids Res. 41: 8319–8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G., Sachs M. S., Atkins J. F., Ivanov I. P., 2012. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res. 40: 2898–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Freitag M., Sachs M. S., 1995. Translational regulation in response to changes in amino acid availability in Neurospora crassa. Mol. Cell. Biol. 15: 5235–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat L. E., Tarn W. Y., Isken O., 2010. The pioneer round of translation: features and functions. Cell 142: 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin B. S., Freitag M., Selker E. U., 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 44: 34–36. [Google Scholar]
- McCluskey K., Wiest A., Plamann M., 2010. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J. Biosci. 35: 119–126. [DOI] [PubMed] [Google Scholar]
- Mohr S. E., Dillon S. T., Boswell R. E., 2001. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 15: 2886–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan L. W., Feldman J. F., 1997. Isolation and characterization of a temperature-sensitive circadian clock mutant of Neurospora crassa. Genetics 146: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov I. Y., Jones M. G., Gould P. D., Crome V., Wilson J. B., et al. , 2012. mRNA 3′ tagging is induced by nonsense-mediated decay and promotes ribosome dissociation. Mol. Cell. Biol. 32: 2585–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov I. Y., Negrete-Urtasun S., Tilburn J., Jansen C. A., Caddick M. X., et al. , 2006. Nonsense-mediated mRNA decay mutation in Aspergillus nidulans. Eukaryot. Cell 5: 1838–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munchel S. E., Shultzaberger R. K., Takizawa N., Weis K., 2011. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol. Biol. Cell 22: 2787–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan V. K., Jones C. I., Newbury S. F., Green P. J., 2013. XRN 5′→3′ exoribonucleases: structure, mechanisms and functions. Biochim. Biophys. Acta 1829: 590–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P., Yepiskoposyan H., Metze S., Zamudio Orozco R., Kleinschmidt N., et al. , 2009. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell. Mol. Life Sci. 67: 677–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyiko T., Kerenyi F., Szabadkai L., Benkovics A. H., Major P., et al. , 2013. Plant nonsense-mediated mRNA decay is controlled by different autoregulatory circuits and can be induced by an EJC-like complex. Nucleic Acids Res. 41: 6715–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios I. M., Gatfield D., St Johnston D., Izaurralde E., 2004. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427: 753–757. [DOI] [PubMed] [Google Scholar]
- Pall M. L., Brunelli J. P., 1993. A series of six compact fungal transformation vectors containing polylinkers with multiple unique restriction sites. Fungal Genet. Newsl. 40: 58. [Google Scholar]
- Popp M. W., Maquat L. E., 2013. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu. Rev. Genet. 47: 139–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp M. W., Maquat L. E., 2014. The dharma of nonsense-mediated mRNA decay in mammalian cells. Mol. Cells 37: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt R. J., Aramayo R., 2002. Improving the efficiency of gene replacements in Neurospora crassa: a first step towards a large-scale functional genomics project. Fungal Genet. Biol. 37: 56–71. [DOI] [PubMed] [Google Scholar]
- Rebbapragada I., Lykke-Andersen J., 2009. Execution of nonsense-mediated mRNA decay: What defines a substrate? Curr. Opin. Cell Biol. 21: 394–402. [DOI] [PubMed] [Google Scholar]
- Ruepp A., Zollner A., Maier D., Albermann K., Hani J., et al. , 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32: 5539–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufener S. C., Muhlemann O., 2013. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 20: 710–717. [DOI] [PubMed] [Google Scholar]
- Sachs M. S., Yanofsky C., 1991. Developmental expression of genes involved in conidiation and amino acid biosynthesis in Neurospora crassa. Dev. Biol. 148: 117–128. [DOI] [PubMed] [Google Scholar]
- Saul H., Elharrar E., Gaash R., Eliaz D., Valenci M., et al. , 2009. The upstream open reading frame of the Arabidopsis AtMHX gene has a strong impact on transcript accumulation through the nonsense-mediated mRNA decay pathway. Plant J. 60: 1031–1042. [DOI] [PubMed] [Google Scholar]
- Sauliere J., Haque N., Harms S., Barbosa I., Blanchette M., et al. , 2010. The exon junction complex differentially marks spliced junctions. Nat. Struct. Mol. Biol. 17: 1269–1271. [DOI] [PubMed] [Google Scholar]
- Shi C., Baldwin I. T., Wu J., 2012. Arabidopsis plants having defects in nonsense-mediated mRNA decay factors UPF1, UPF2, and UPF3 show photoperiod-dependent phenotypes in development and stress responses. J. Integr. Plant Biol. 54: 99–114. [DOI] [PubMed] [Google Scholar]
- Stajich J. E., Harris T., Brunk B. P., Brestelli J., Fischer S., et al. , 2012. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res. 40: D675–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Schwalb B., Schulz D., Pirkl N., Etzold S., et al. , 2012. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 22: 1350–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T., Larson D. R., Moldon A., Query C. C., Singer R. H., 2011. Single-molecule mRNA decay measurements reveal promoter- regulated mRNA stability in yeast. Cell 147: 1484–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H. J., 1956. A convenient growth medium for Neurospora (Medium N). Microbiol. Genet. Bull. 13: 42–43. [Google Scholar]
- Wei J., Wu C., Sachs M. S., 2012. The arginine attenuator peptide interferes with the ribosome peptidyl transferase center. Mol. Cell. Biol. 32: 2396–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Zhang Y., Ivanov I. P., Sachs M. S., 2013. The stringency of start codon selection in the filamentous fungus Neurospora crassa. J. Biol. Chem. 288: 9549–9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Brogna S., 2010. Splicing-dependent NMD does not require the EJC in Schizosaccharomyces pombe. EMBO J. 29: 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard M., Mitchell H. K., 1947. Neurospora V: a synthetic medium favoring sexual reproduction. Am. J. Bot. 34: 573–577. [Google Scholar]
- Wollschlaeger C., Trevijano-Contador N., Wang X., Legrand M., Zaragoza O., et al. , 2014. Distinct and redundant roles of exonucleases in Cryptococcus neoformans: implications for virulence and mating. Fungal Genet. Biol. 73: 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Amrani N., Jacobson A., Sachs M. S., 2007. The use of fungal in vitro systems for studying translational regulation, pp. 203–225 in Translation Initiation: Extract Systems and Molecular Genetics, edited by Lorsch J. Elsevier, San Diego. [DOI] [PubMed] [Google Scholar]
- Wu C., Kim Y. S., Smith K. M., Li W., Hood H. M., et al. , 2009. Characterization of chromosome ends in the filamentous fungus Neurospora crassa. Genetics 181: 1129–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., F. Yang, K. M. Smith, M. Peterson, R. Dekhang et al., 2014 Genome-wide characterization of light-regulated genes in Neurospora crassa. G3 (Bethesda) 4: 1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yepiskoposyan H., Aeschimann F., Nilsson D., Okoniewski M., Muhlemann O., 2011. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 17: 2108–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Welch E. M., Hogan K., Brown A. H., Peltz S. W., et al. , 1997. Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA 3: 234–244. [PMC free article] [PubMed] [Google Scholar]
- Zid B. M., O’Shea E. K., 2014. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature 514: 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.