Abstract
The gene doublesex, which is placed at the bottom of the sex-determination gene cascade, plays the ultimate discriminatory role for sex determination in insects. In all insects where this gene has been characterized, the dsx premessenger RNA (pre-mRNA) follows a sex-specific splicing pattern, producing male- and female-specific mRNAs encoding the male-DSXM and female-DSXF proteins, which determine male and female development, respectively. This article reports the isolation and characterization of the gene doublesex of dipteran Sciara insects. The Sciara doublesex gene is constitutively transcribed during development and adult life of males and females. Sciara had no sex-specific doublesex mRNAs but the same transcripts, produced by alternative splicing of its primary transcript, were present in both sexes, although their relative abundance is sex specific. However, only the female DSXF protein, but not the male DSXM protein, was produced at similar amounts in both sexes. An analysis of the expression of female and male Sciara DSX proteins in Drosophila showed that these proteins conserved female and male function, respectively, on the control of Drosophila yolk-protein genes. The molecular evolution of gene doublesex of all insects where this gene has been characterized revealed that Sciara doublesex displays a considerable degree of divergence in its molecular organization and its splicing pattern with respect to the rest of dipterans as suggested by its basal position within the doublesex phylogeny. It is suggested that the doublesex gene is involved in Sciara sex determination although it appears not to play the discriminatory role performed in other insects.
Keywords: sex determination, molecular evolution, Sciara, gene doublesex, genetics of sex
MALES and females are different at the morphological, physiological, and behavioral levels. This sexual dimorphism results from the integration of two processes: sex determination and sexual differentiation. Sex determination refers to the developmental program that commits the embryo to either the male or the female pathway. The genes underlying this phenomenon are the sex-determination genes. Sexual differentiation refers to the expression of the sex cytodifferentiation genes (which are controlled by the sex-determination genes), the expression of which gives rise to the formation of the sexually dimorphic structures that characterize the male and female adults.
The animal kingdom possesses a wealth of mechanisms by which sex is decided (Bull 1983; Beukeboom and Perrin 2014). These mechanisms can be classified into three main categories, depending on the origin of the primary sex-determination signal, which can be zygotic, maternal, or environmental. These three primary signals occur and are concatenated in the dipteran Sciara (for details see Sánchez 2010). Briefly, in Sciara, sex determination follows the XX/X0 mechanism: XX zygotes will develop as females and X0 zygotes will develop as males. Yet, the final chromosome constitution of the zygote, which starts as an XXX zygote, depends on a maternal factor. This maternal factor, controlling the number of X chromosomes that are eliminated (either one or two), acts as the primary signal defining the sexual development that the zygote will follow. This implies the existence of a concatenation of two primary signals: the maternal signal preceding and determining the final zygotic signal. This occurs in monogenic species, such as Sciara coprophila (nowadays Bradysia coprophila), where the amount of maternal product deposited in the oocyte is genetically determined and then independent of environmental conditions (Moses and Metz 1928; Metz and Schmuck 1929; Metz 1938). But in other digenic sciarids, such as S. ocellaris, the amount of maternal factor depends on the temperature at which the insects are raised (Metz 1938; Davidheiser 1943; Mori et al. 1979; Nigro et al. 2007). Hence, in these species the environmental signal (temperature) determines the amount of maternal product (maternal signal) placed in the oocytes, which in turn determines the number of X chromosomes to be eliminated in the zygote and then its final chromosome constitution (zygotic signal) that determines its sexual development. Consequently, Sciara is unique in that it offers the possibility of studying the evolutionary relationships between the three main primary signals triggering sexual development (Sánchez 2010). Furthermore, the comparative study between the monogenic and the digenic species will allow study of the cues underlying the reciprocal changes between environmental and genetic systems of determining sex. These are the reasons that prompted us to choose Sciara as an experimental model for exploring the evolution of sex-determination mechanisms.
Drosophila melanogaster has been the paradigm for understanding the genetic and molecular basis underlying sex determination in insects. Briefly, in D. melanogaster, sex determination is under the control of the gene Sex lethal (Sxl) (Cline 1978; reviewed in Penalva and Sánchez 2003). The epistatic relationships between Sxl and the other sex-determination genes transformer (tra), transformer-2 (tra-2), and doublesex (dsx) have revealed that a hierarchical interaction exists between them (Baker and Ridge 1980). Their characterization has shown that sex-specific splicing of their primary transcripts controls their expression during development, the product of one gene controlling the sex-specific splicing of the premessenger RNA (pre-mRNA) of the downstream gene in the cascade (reviewed in Sánchez 2008 and citations therein). Thus, Sxl controls the splicing of the pre-mRNA from the downstream gene tra. This gene is transcribed in both sexes, but its RNA follows sex-specific alternative splicing pathways: in males, a mature mRNA is formed that encodes a truncated, presumably nonfunctional TRA protein, whereas in females, the presence of the SXL protein determines the formation of an mRNA isoform that encodes the functional TRA protein. The TRA product and the product of the constitutively expressed gene tra-2 form a complex, which controls the sex-specific splicing of pre-mRNA from the gene doublesex (dsx). This gene is at the bottom of the sex-determination cascade and is transcribed in both sexes, but gives rise to two different mRNAs, dsxF and dsxM, by sex-specific splicing of its primary transcript. The dsxF and dsxM mRNAs encode, respectively, the female-specific DSXF and the male-specific DSXM proteins. These two DSX proteins are transcription factors that control the activity of the cytodifferentiation genes involved in sexual differentiation. Both proteins share the N-terminal domain, which contains a DNA-binding domain (DM domain). However, they differ in their C-terminal domains, which endow specific functions to these proteins (Burtis and Baker 1989; Hoshijima et al. 1991).
The search for genes orthologous to the sex-determination genes of D. melanogaster has been undertaken in other insects (reviewed in Sánchez 2008; Gempe and Beye 2010; Verhulst et al. 2010). This was a first task of our Sciara project. We have characterized the gene Sxl in the sciarids S. ocellaris (Ruiz et al. 2003), S. coprophila, Rynchosciara americana, and Trichosia pubescens (Serna et al. 2004), as well as the gene tra-2 in S. coprophila and S. ocellaris (Martín et al. 2011). Finally, the isolation and characterization of the gene dsx of dipterans S. coprophila and S. ocellaris are reported.
Outside the drosophilids, the gene dsx has been characterized in the dipterans Megaselia scalaris (Sievert et al. 1997; Kuhn et al. 2000), Musca domestica (Hediger et al. 2004), Lucilia cuprina (Concha et al. 2010), Anopheles gambiae (Scali et al. 2005), and Aedes aegypti (Salvemini et al. 2011); in Bactrocera tryoni (Shearman and Frommer 1998), B. oleae (Lagos et al. 2005), and Ceratitis capitata (Saccone et al. 2008) and in 12 Anastrepha species (Ruiz et al. 2005, 2007); in the lepidopterans Bombyx mori (Ohbayashi et al. 2001; Suzuki et al. 2001), Antheraea assama, and A. mylitta (Shukla and Nagaraju 2010); in the hymenopterans Apis mellifera (Cho et al. 2007) and Nasonia vitripennis (Oliveira et al. 2009); in the coleopteran Tribolium castaneum (Shukla and Palli 2012); and in the crustacean Daphnia magna (Kato et al. 2011) and other cladoceran species (Toyota et al. 2013). The molecular organization of the dsx ORF varies among these organisms, yet in all cases excluding the cladoceran species dsx produces male- and female-specific mRNAs that encode putative male- and female-specific DSX proteins as in Drosophila. In the cladocerans, the regulation of dsx expression does not take place by sex-specific splicing of its pre-mRNA but exhibits sexual differences in the abundance of its transcripts. In Sciara, however, no sex-specific dsx mRNAs but the same transcripts, produced by alternative splicing of its primary transcript, were present in both sexes. Nevertheless, only the female DSXF protein, but not the male DSXM protein, was produced at similar amounts in both sexes. It is suggested that the doublesex gene is involved in Sciara sex determination although it appears not to play the discriminatory role performed in other insects.
Materials and Methods
Fly culture
Drosophila flies were cultured on Equation 4-24 Plain CS Drosophila food from Carolina Biological Supply Company. For the description of the mutant alleles and GAL4 constructs see Lindsley and Zimm (1992) and FlyBase (www.flybase.org). Sciarid flies were raised on “Compost Villacasa” medium for culturing mushrooms. S. coprophila is a monogenic species with two types of females: gynogenic females, which produce only females, and androgenic females, which produce only males; whereas S. ocellaris is a digenic species, where the female has both sex offspring.
Extraction of DNA and RNA
Total genomic DNA was isolated from flies according to Maniatis et al. (1982). Total RNA extracts from frozen specimens were prepared using the Ultraspec-II RNA isolation kit (Biotecx) or Trizol (Invitrogen, Carlsbad, CA). Poly-A+ was isolated using the “mRNA Purification Kit” (GE Healthcare). The GenomeWalker genomic libraries of S. coprophila and S. ocellaris were synthesized using the BD GenomeWalker Universal kit (BD Biosciences). The 3′- and 5′-RACE libraries of S. coprophila and S. ocellaris were synthesized using the “Marathon cDNA Amplification Kit” (Clontech).
PCR and RT-PCR analyses
For long PCR, the “Expand Long Template Enzyme Mix” (Roche) or the “Advantage 2 PCR Enzyme System” (Clontech) was used. Total RNA was reverse transcribed with Superscript II/III (Invitrogen) PCR and RT-PCR products were analyzed by electrophoresis in agarose gels and the amplified fragments subcloned using the TOPO TA-cloning kit (Invitrogen) or the pGEMT-easy vector (Promega, Madison, WI). These subclones were then sequenced using universal forward and reverse primers. In Supporting Information, Table S1 and Figure S1 show the sequences of the primers and their location, respectively. In all cases, PCR on RNA samples was performed to guarantee that they were not contaminated with genomic DNA (negative controls of PCR).
DNA sequencing
Sequencing was performed using an automated 377 DNA sequencer (Applied Biosystems, Foster City, CA) at “Secugen,” Madrid, Spain.
Comparison of DNA and protein sequences
All comparisons were made using Fasta v.3.0t82 (Pearson and Lipman 1988) and ClustalW1.83 software (Thompson et al. 1997).
Production of antibodies against the Sciara DSX proteins
Peptides used to generate the antibodies were synthesized at the Servicio de Proteómica del Centro Nacional de Biotecnología [Consejo Superior de Investigaciones Cientifícas (CSIC)], Madrid, Spain. The sequences of the peptides corresponded to the specific C-terminal domain of ScDSXF protein (YARYNNLNIYDGGELRIDTKRG), to the specific C-terminal domain of ScDSXM protein (QLYHFLNTRVIMNSYANENR), and to part of the common N-terminal region (MVSNDISDWNDIMSDSEVTDT). The peptides were coupled to the KLH carrier protein. Rabbits were immunized by periodic subcutaneous injections of 0.3 mg of purified peptides, following standard procedures (Harlow and Lane 1988). Antibodies were purified using the antigenic peptide coupled to HiTrap NHS-Activated HP columns (GE Healthcare) and ProG 5-µl, 200-µl LTS Tips (Mettler Toledo).
Protein extracts and Western blots
Samples of total proteins from embryos, larvae, and adults of S. coprophila and S. ocellaris and adults of D. melanogaster were prepared by homogenization in STE lysis buffer on ice (10 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA) with protease inhibitor cocktail (Roche). The protein concentration was measured with a Nanodrop 2000 or the Bradford method. Forty micrograms of total protein was separated on SDS–PAGE gels (12%) (Laemmli 1970) and transferred onto nitrocellulose membranes (Towbin et al. 1979). The membranes were blocked for 1 hr at room temperature in PBST (0.05% Tween-20 in PBS) containing 1% BSA and 10% nonfat dried milk and probed with the selected primary antibody [anti-C-terminal DsxM (α-ScDSXM), anti-C-terminal DsxF (α-ScDSXF), and common anti-N-terminal (α-ScDSXC)] overnight at 4°. After washing in PBST, membranes were incubated for 1 hr at room temperature with an anti-rabbit HRP-conjugated secondary antibody (Santa Cruz Biotechnology). Finally, membranes were washed in PBST and developed using the ECL Western blotting detection reagents (GE Healthcare). As a loading control, the membranes were stripped and subsequently reprobed with an anti-α-tubulin monoclonal antibody (T6074, Sigma Aldrich). The above procedure described to prepare the protein extracts was the best one among others that were tested.
Immunoprecipitation
Samples of total proteins from adults of S. coprophila and S. ocellaris were prepared by homogenization in STE lysis buffer on ice (10 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA) with protease inhibitor cocktail (Roche). Immunoprecipitation with the purified α-ScDSXF, α-ScDSXM, and α-ScDSXC antibodies was performed using the “Protein A Sepharose 4 Fast Flow” (GE Heathcare).
Semiquantitative PCR of S. coprophila dsx transcripts
The transcript levels of both female ScdsxF and male ScdsxM mRNA isoforms in Sciara larvae were quantified relative to the level of rpl10 mRNA (encoding the ribosomal protein RPL10) in the two sexes (Ramos-Onsins et al. 1998). The primers used in this analysis were dsxSc1, located in the common exon E1, and either primer dsxMSc (in the male-specific exon E4) to detect the male ScdsxM mRNA or primer dsxFSc (in the female-specific exon E3) to detect the female ScdsxF transcript. RT was carried out on poly-A+ mRNA of male and female larvae, separately, with oligo-dT. The same complementary DNA (cDNA) sample of male and female larvae was used to carry out PCR to detect the male ScdsxM, female ScdsxF, and rpl10 mRNAs. PCR products were loaded in agarose gels and the bands corresponding to the amplicons were measured using the Quantity One program (Bio-Rad, Hercules, CA). A set of PCR was initially performed to decide the number of amplification cycles to prevent saturation effects.
Quantitative analysis of the ScDSXF protein
The amount of ScDSXF protein in male and female larvae was calculated by using tubulin for normalization. The intensity of the bands was measured using the Quantity One program (Bio-Rad), under nonsaturated conditions.
Construction of UAS::ScdsxM-cDNA, UAS::ScdsxF-cDNA, and UAS::DScmg transgenes
For the construction of the UAS::ScdsxM-cDNA (MSc) and UAS::ScdsxF-cDNA (FSc) transgenes, a fragment of 925 and 1118 bp, comprising the whole ORF of ScdsxM and ScdsxF, respectively, was amplified by RT-PCR, using a common primer dsxSccom and a male-specific primer dsxMSc or a female-specific primer dsxFSc at the corresponding 3′-UTR. The amplicon was cloned in the TOPO-TA cloning vector (Invitrogen). The cDNA fragments were then digested with EcoRI and cloned in the pUAST vector. The microinjections for generating the MSc and FSc transgenic D. melanogaster lines were performed by Genetic Services (Sudbury, MA). Standard genetic crosses determined the chromosomal location of the transgenes. To ascertain that each transgenic line was carrying the correct transgene and this was expressed, RT-PCR on total RNA was used to amplify the whole transgene and the amplicons were cloned and sequenced.
For the construction of the UAS::DScmg transgene (Figure S2), four genomic fragments E1I1, I1E2I2, I2E3I3, and I3E4 from the Scdsx gene were generated by PCR on genomic DNA of S. coprophila. The genomic fragment E1I1 (503 bp) that covers the exon E1/intron I1 junction and contains 276 pb from exon E1 plus 227 bp from intron I1 was generated by using the primers e1i1-1 and e1i1-2. The genomic fragment I1E2I2 (770 bp) that is formed by exon E2 (211 bp) plus the adjacent upstream sequences (390 bp) of intron 1 and the adjacent downstream sequences (169 bp) of intron 2 was generated using the primers i1e2i2-1 and i1e2i2-2. The genomic fragment I2E3I3 (1089 bp) that is formed by exon E3 (515 bp) plus the adjacent upstream sequences (451 bp) of intron 2 and the adjacent downstream sequences (123 bp) of intron 3 was generated using the primers i2e3i3-1 and i2e3i3-2. The genomic fragment I3E4 (516 bp) that is formed by the first 144 bp of exon E4 plus the adjacent upstream sequences (372 bp) of intron 3 was generated using the primers i3e4-1 and i3e4-2. Primers were designed to allow the joining of the four fragments in the order E1I1–I1E2I2–I2E3I3–I3E4 and this construct was cloned in the pUAST vector. To do this, the primers contained restriction enzyme sites: e1i1-2 and i1e2i1-1 contained the XbaI sequence, i1e2i2-2 and i2e3i3-1 contained the PstI sequence, i2e3i3-2 and i3e4-1 contained the XhoI sequence, and finally e1i1-1 and i3e4-2 contained, respectively, NotI and KpnI sequences that allowed the cloning of the construct in the pUAST vector in a unique orientation. The amplified fragments were cloned in the pGEM-T easy vector (Promega) and sequenced. Each fragment was then obtained from the vector by digestion with the enzymes whose restriction sites were incorporated in the primers. The pUAST vector was digested with NotI and KpnI. The digestions of the vector and the genomic fragments were resolved on a 0.8% agarose gel and the corresponding bands were cut from the gel and eluted from the agarose, using the “Geneclean Turbo” kit (Qbiogen). Fifty nanograms of each fragment plus 10 ng of the digested pUAST vector were ligated using the “DNA Rapid Ligation” kit (Roche). The presence of the construct in the vector was checked by a colony “cracking” procedure. The cloned constructs were sequenced to ensure the correct orientation.
Semiquantitative PCR of dsx transcripts from the S. coprophila minigene in transgenic Drosophila
The GAL4-UAS system was used to express the DScmg transgene (Brand and Perrimon 1993). The arm-GAL4 driver line, which drives expression ubiquitously, was employed. Reverse transcription of total RNA from male and female Drosophila adults was performed using the primer oligo-dT, while PCR was performed with the primers dsxSc1, located in the common exon E1, and either primer dsxM2Sc (in exon E4) to detect the male ScdsxM mRNA or primer dsxFSc (in exon E3) to detect the female-like ScdsxF transcript. PCR products were loaded in agarose gels and the bands corresponding to the amplicons were measured using the Quantity One program (nonsaturated conditions were used). Normalization was carried out using rp49, encoding the Rp49 ribosomal protein (Ramos-Onsins et al. 1998). The males yw/Y; DScmg/arm-Gal4[w+] and females yw/w; DScmg/arm-Gal4[w+] were the offspring of crosses between females yw; Dscmg and males yw/Y; arm-Gal4[w+]/CyO,Cy.
Comparison of the DSX proteins of Sciara with those of other insects
The putative female ScDSXF and male ScDSXM proteins of S. coprophila (Sciaridae, suborder Nematocera) were compared to the DSXF and DSXM proteins from other dipterans such as A. gambiae and A. aegypti (suborder Nematocera), B. oleae, C. capitata, A. obliqua, D. melanogaster, M. domestica, L. cuprina, and M. scalaris (suborder Brachycera); from lepidopterans B. mori and A. assama; and from the hymenopterans A. mellifera and N. vitripennis and the coleopteran T. castaneum. The proteins were divided into the following regions: N-terminal region, OD1 domain, OD1–OD2 linker region, OD2 domain, and C-terminal region.
Molecular evolutionary analysis
The sequences of male- and female-specific doublesex genes from 21 insect species and 1 outgroup were included in the present work (sequence accession numbers detailed in Table S2). Accession numbers for S. ocellaris DSX proteins are HG934388 (DSXF ) and HG934389 (DSXM ). Sequences corresponding to the gene region common to both sexes were aligned on the basis of the translated amino acid sequences and edited for potential errors, using the BIOEDIT program (Hall 1999). Molecular evolutionary analyses were performed using the computer program MEGA version 6 (Tamura et al. 2011). The phylogeny of doublesex proteins was reconstructed following a maximum-likelihood approach. The models of evolution that best fit the set of sequences analyzed were determined as James-Taylor-Thornton (Jones et al. 1992), considering gamma-distributed variation across sites (G) and invariant sites (I) for the case of proteins, and the Hasegawa-Kishino-Yano model (Hasegawa et al. 1985) with gamma-distributed variation across sites (G) for the case of nucleotide sequences. The reconstructed phylogeny was rooted using the dsx gene from the cladoceran D. magna, diverging from insects ∼369.5 MYA (Hedges and Kumar 2009). The reliability of the resulting topology was assessed through bootstrap analyses (1000 replicates) and further studied using Bayesian analyses implemented in the program BEAST version 1.7 (Drummond et al. 2012). To this end, three independent Markov chain Monte Carlo (MCMC) runs of 10,000,000 generations each were performed, sampling tree topologies every 1000 generations to ensure the independence of successive trees and discarding the first 1000 trees of each run as burn-in.
Results and Discussion
Molecular organization of Sciara doublesex gene
The first step in the isolation of the S. coprophila dsx gene (Scdsx) was to perform RT-PCR on total RNA from female adults. Reverse transcription was performed using the primer oligo-dT, while PCR was performed with two degenerate primers dxsOD3 and dsxOD7 corresponding to the conserved domains OD1 and OD2, respectively, of D. melanogaster, M. scalaris, B. tryoni, B. oleae, M. domestica, and B. mori. An amplicon of 475 bp was amplified, cloned, and sequenced. The conceptual amino acid sequence of this amplicon showed a high degree of similarity with the OD1 (90%) and OD2 (70%) domains of D. melanogaster DSX protein, indicating that a fragment of the putative ScDSX protein was isolated.
Nested PCR was then performed in 3′- and 5′-RACE analyses of males and females, separately. The amplicons were cloned and sequenced. In the 5′-RACE assay, a single fragment of 596 bp was amplified in both sexes. This fragment corresponded to the 5′-UTR region and the N-terminal domain of the putative ScDSX protein. In the 3′-RACE analysis, four amplicons were produced in both sexes. A fragment of 648 bp corresponded to the region orthologous to the OD2 domain plus the C-terminal region of the female DSXF protein of D. melanogaster, as well as the 3′-UTR region. The 739-bp fragment corresponded to the previous fragment containing a 91-bp insertion, which interrupted the sequence encoding the OD2 domain by introducing a translational stop codon that would give rise to a putative truncated version of the ScDSXF protein. The 404- and 572-bp fragments contained the sequence orthologous to the C-terminal region of the male DSXM protein of D. melanogaster, as well as the 3′-UTR region. The dissimilar size of the two fragments was due to the distinct length of their 3′-UTR sequences. None of these two latter fragments contained the sequence corresponding to the OD2 domain common to both DSXF and DSXM proteins of other insects, suggesting that one of the proteins encoded by the Scdsx gene might lack the OD2 domain. To reject the possibility that we were missing downstream male-specific exons, 3′-RACE assays were repeated using a downstream primer corresponding to the 3′ end of the C-terminal domain of the putative ScDSXM protein orthologous to the DSXM protein of D. melanogaster. Two fragments of 284 and 452 bp were amplified that were contained, respectively, within the 404- and 572-bp fragments previously amplified. Thus, no further downstream exons were found.
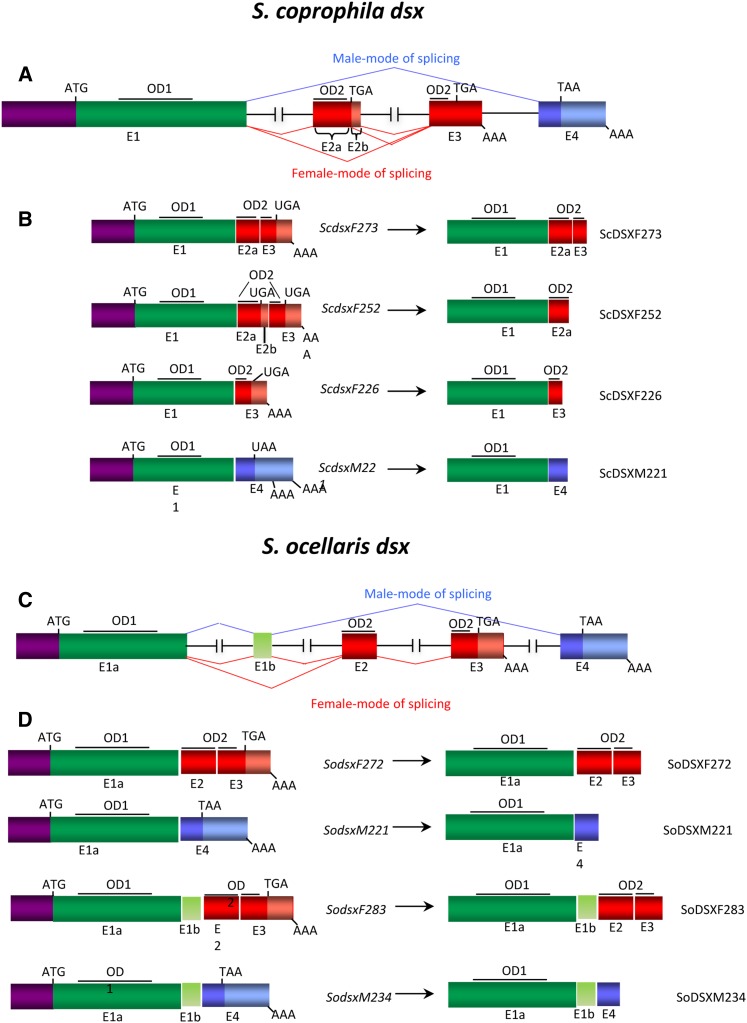
GenomeWalker methodology was next applied to determine the exon–intron junctions on S. coprophila genomic DNA (see Materials and Methods). The sequences of the generated genomic fragments were compared with the S. coprophila cDNA sequences previously found to unambiguously determine the exon–intron junctions. The complete sequence of intron 3 (2106 bp) was confirmed by PCR on genomic DNA with primers flanking the intron. The comparison of the GenomeWalker sequences with those from the 5′-RACE assays indicated that the transcription start site would lie 520 bp upstream of the beginning of the ORF. The molecular organization of Scdsx is shown in Figure 1A.
Figure 1.
(A–D) Molecular organization of the dsx gene of S. coprophila (A) and its transcripts and proteins (B) and dsx of S. ocellaris (C) and its transcripts and proteins (D). Exons (E) (boxes) and introns (lines) are not drawn to scale. The interrupted lines for introns indicate that their lengths remain unknown. The beginning and the end of the ORF are indicated by ATG and TGA (UGA) or TAA (UAA), respectively. AAA stands for poly-A+.
The Scdsx gene is composed of five exons: E1, E2a, E2b, E3, and E4. The primary transcript can follow four modes of splicing (highlighted in red and blue in Figure 1A), producing four types of transcripts (Figure 1B). We followed the nomenclature DSXF and DSXM after comparison of the putative DSX proteins of S. coprophila with those of other insects. Union of exons E1–E2a–E3 produces the ScdsxF273 transcript (hereafter female ScdsxF), which encodes a putative ScDSXF273 protein of 273 amino acids (hereafter female ScDSXF) containing the OD1 and OD2 domains present in the DSXF protein of other insects. Union of exons E1–E2a–E2b–E3 produces the ScdsxF252 transcript, which encodes a putative ScDSXF252 protein of 252 amino acids that is similar to the ScDSXF273 protein except that the 3′ end of OD2 is missing due to the presence of a translational stop codon in exon E2b. Union of exons E1–E3 produces the ScdsxF226 transcript encoding a putative ScDSXF226 protein of 226 amino acids, which shares with the female ScDSXF protein the N-terminal domain carrying the OD1 domain and the 5′ end of the OD2 domain encoded in exon E3. Finally, union of exons E1–E4 produces the ScdsxM221 transcript (hereafter male ScdsxM), which encodes a putative ScDSXM221 protein of 221 amino acids (hereafter male ScDSXM) carrying OD1 but lacking the OD2 domain and having the C-terminal region similar to that in the male DSXM protein of other insects.
To isolate the whole ORF of dsx of S. ocellaris (Sodsx), RT-PCR was performed on total RNA from males and females, individually. GenomeWalking was used to identify the exon–intron junctions. The molecular organization of Sodsx is similar to that of Scdsx, with the exception of a miniexon (exon E1b) between exon 1 and exon 2 (Figure 1C). Exon E1b encodes an extra 11 amino acids in females and 13 amino acids in males. The Sodsx gene produces basically two mRNAs found in both sexes (Figure 1D): SodsxF (composed of exons E1a, E2, and E3) encodes a putative protein of 272 amino acids with a high degree of similarity (95%) to female ScDSXF; and SodsxM (composed of exons E1a, E1b, and E4) encodes a putative protein of 221 amino acids with a high degree of similarity (91%) to male ScDSXM. In both sexes, two further low-abundant dsx mRNAs were found, which encode for putative SoDSXF and SoDSXM proteins containing an additional 11 and 13 amino acids, respectively, corresponding to the incorporation of exon E1b (Figure 1D). In contrast to that observed in S. coprophila, no mRNAs encoding the truncated version of the SoDSXF protein were found; however, a putative splicing site was found downstream of exon 2 that, if used, introduces a stop codon in the mRNA, which would encode a truncated version of SoDSXF protein analogous to S. coprophila.
Expression of dsx in Sciara: the transcripts
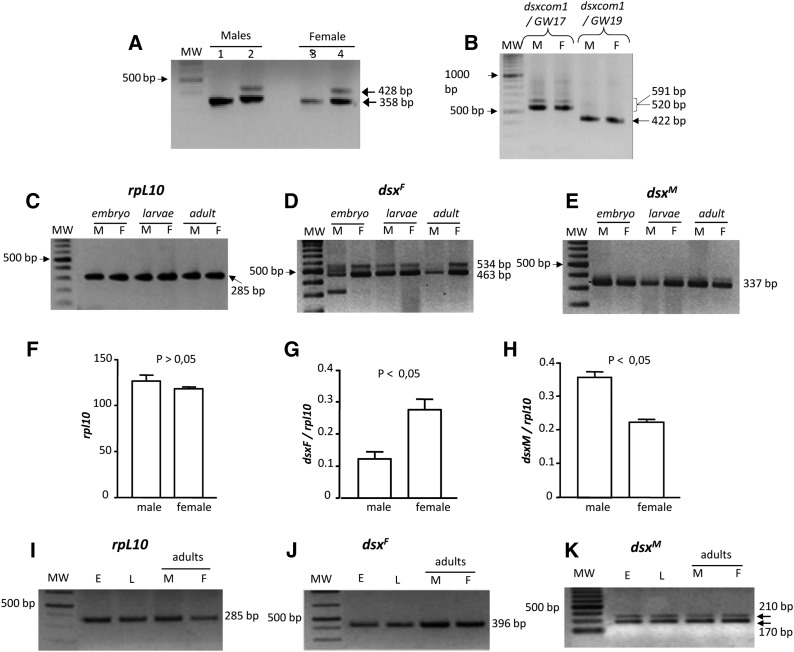
The isolation of the Sciara dsx gene revealed the presence of the same transcripts in both sexes. To confirm this, RT-PCR was performed on poly-A+ mRNA of male and female larvae, separately. As shown in Figure 2A, the female ScdsxF transcript (band of 358 bp in lanes 2 and 4), the female ScdsxF252 transcript (band of 428 bp in lanes 2 and 4), and the male ScdsxM transcript (lanes 1 and 3) were present in both sexes. The ScdsxF226 transcript was visible only after prolonged exposure, indicating that it is present in low amounts (data not shown). The female ScdsxF isoform encoding the full-length putative female ScDSXF protein was more abundant than the female ScdsxF252 isoform encoding the truncated version. Negative controls in all PCR assays produced no amplicons (see Materials and Methods).
Figure 2.
(A–K) Expression analysis of dsx of S. coprophila (A–H) and S. ocellaris (I–K) during development. (A) RT-PCR on poly-A+ of male and female larvae with primers dsxSc1 and either primer dsxM2Sc (lanes 1 and 3) or primer dsxFSc (lanes 2 and 4). (B) RT-PCR on poly-A+ of male and female larvae with primers dsxSc1 and either primer dsxM2Sc (lanes 1 and 3) or primer dsxFSc (lanes 2 and 4). (C) RT-PCR for rpl10 mRNA. (D) RT-PCR for ScdsxF mRNA. (E) RT-PCR for ScdsxM mRNA. (F–H) Semiquantitative PCR for the presence of ScdsxF and ScdsxM mRNAs in male and female larvae, separately. The bars in the histograms represent the 95% confidence intervals corresponding to five replicates. (F) Semiquantitative expression of rpl10 mRNA in male and female larvae. (G) Semiquantitative expression of ScdsxF mRNA in male and female larvae, using rpl10 to normalize. (H) Semiquantitative expression of ScdsxM mRNA in male and female larvae, using rpl10 to normalize. (I) RT-PCR for rpl10 was used as a control. (J) RT-PCR for SodsxF mRNA. (K) RT-PCR for SodsxM mRNA with the two bands corresponding to the absence (band 170 bp) or to the inclusion (band 210 bp) of exon E1b.
The molecular organization of Scdsx, the complete sequencing of intron 3 flanked by exons E3 and E4, and the failure to detect male-specific exons downstream of exon E4 suggested that the four dsx mRNA isoforms were produced by alternative splicing of the same primary transcript. The male ScDSXM protein lacked the OD2 domain, which has been found in all DSXM proteins from dipterans where dsx has been characterized. It could then be argued that a second dipteran-like dsx gene containing the OD2-encoding sequence might exist in Sciara in addition to the described Scdsx transcription unit. If so, an additional amplicon corresponding to the larger size of the male-specific exon in the putative second dipteran-like dsx gene should be expected. This expectation, however, was not observed. RT-PCR was performed on poly-A+ mRNA from male and female larvae separately. Reverse transcription was carried out with a primer anchor-T made by an anchor sequence joined to oligo-dT at its 3′ end. PCR was performed using the anchor primer and a primer dsxcom1 located in the well-conserved N-terminal region in exon E1. Reamplification with primer dsxcom1 and either primer GW17 (located in the female-specific exon E3) or primer GW19 (located in the male-specific exon 4) produced the expected amplicons of sizes 520 and 591 bp in both sexes, corresponding to the female ScdsxF and ScdsxF252 mRNAs, respectively, and the expected amplicon of 422 bp corresponding to the dsxM mRNA (Figure 2B). Cloning and sequencing of the amplicons confirmed their origin. Negative controls for these PCR assays produced no amplicons (see Materials and Methods). Thus, the only amplicons generated were those concurring with the characterized molecular organization of Scdsx shown in Figure 1A.
The Scdsx gene is expressed in both sexes during embryonic and larval development as well as in the adult stage (Figure 2, C–E). The presence of both male- and female-Scdsx mRNA isoforms in the two sexes raised the question about the possible regulation of dsx expression at a quantitative rather than at a qualitative level, that is, whether the two mRNAs would be produced at different amounts in males and females. Thus, a quantitative-PCR assay was attempted to determine the relative abundance of these two transcripts in the two sexes. This investigation failed because it was impossible to find a set of primers that produced neither double amplifications nor dimers. Therefore, a semiquantitative PCR strategy was followed. This consisted of measuring the levels of both female ScdsxF and male ScdsxM mRNA isoforms relative to the level of rpl10 mRNA, which encodes the ribosomal protein RPL10 in the two sexes (Ramos-Onsins et al. 1998). Details of the experimental methodology can be found in Materials and Methods. The results are presented in Figure 2, F–H. The amount of rpl10 mRNA did not significantly vary between males and females (Figure 2F, Student’s t-test, P > 0.05), indicating that the reverse transcriptase reaction was comparable in both sexes so that the amount of rpl10 mRNA was used to normalize the relative abundance of the male ScdsxM and female ScdsxF mRNAs in both sexes. ScdsxF was significantly more abundant than ScdsxM in females (Figure 2G, Student’s t-test, P < 0.05), whereas ScdsxM was significantly more abundant than ScdsxF in males (Figure 2H, Student’s t-test, P < 0.05). Similar results were obtained with other male- and female-specific primers to amplify ScdsxM and ScdsxF, respectively (data not shown). Negative controls for these PCR assays produced no amplicons (see Materials and Methods).
S. ocellaris is a digenic species and each female produces both male and female offspring. Because of this, the individual analysis of Sodsx expression in males and females could be evaluated only at the adult stage. For details see Materials and Methods. Similar to S. coprophila, the dsx gene of S. ocellaris was expressed during development and in adult life. RT-PCR analysis for the control (rpl10) is shown in Figure 2I. The female SodsxF (Figure 2J) and male SodsxM (Figure 2K) mRNA isoforms were detected in embryos and larvae. In adults, both isoforms were expressed in both sexes (Figure 2, J and K), and they were also expressed in ovaries (data not shown). The two bands of 210 and 170 bp observed in the case of SodsxM depended on the inclusion of miniexon E1b. Negative controls for these PCR assays produced no amplicons (see Materials and Methods). To determine the relative abundance of the female SodsxF and male SodsxM mRNAs in the adult males and females, the semiquantitative PCR strategy used for S. coprophila was followed (see Materials and Methods). The SodsxF mRNA was significantly more abundant in female than in male adults, whereas the SodsxM mRNA was more abundant in males than in females, although not significantly (data not shown).
Expression of dsx in Sciara: the proteins
The above results suggested that the regulation of dsx function in Sciara was distinct from that observed in other insects. It is conceivable that the sex-specific function of dsx was implemented by regulating the relative amount of female ScDSXF and male ScDSXM proteins produced in both males and females, as a direct consequence of the different mRNA quantities in the two sexes: a greater amount of ScDSXM than ScDSXF would determine male development, whereas the reverse would cause female development. Alternatively, the sex-specific function of dsx could be implemented by controlling the translation of the male and female mRNAs: in males the mRNA encoding the female ScDSXF protein and in females the mRNA encoding the male ScDSXM protein would not be translated. To explore these two scenarios, an analysis of ScDSXM and ScDSXF protein expression was undertaken. Peptides corresponding to the female-specific and male-specific C-terminal regions of ScDSXF and ScDSXM, respectively, were synthesized and used for immunization of rabbits (see Materials and Methods). To test the specificity of the sera anti-ScDSXF (hereafter α-ScDSXF) and anti-ScDSXM (hereafter α-ScDSXM), an ELISA against the two peptides was performed and results showed that the two sera were specific for the antigens used in immunization (data not shown). Subsequently, Western blotting of purified GST-ScDSXF (full-length ScDSXF fused to GST) and GST-ScDSXM (full-length ScDSXM fused to GST) proteins was carried out and probed with the two sera, demonstrating their specificity; i.e., α-ScDSXF recognizes only ScDSXF (Figure S3A), whereas α-ScDSXM recognizes only ScDSXM (Figure S3B). For construction of these fusion proteins, see legend to Figure S3.
The putative Sciara DSXF female protein is present in both sexes:
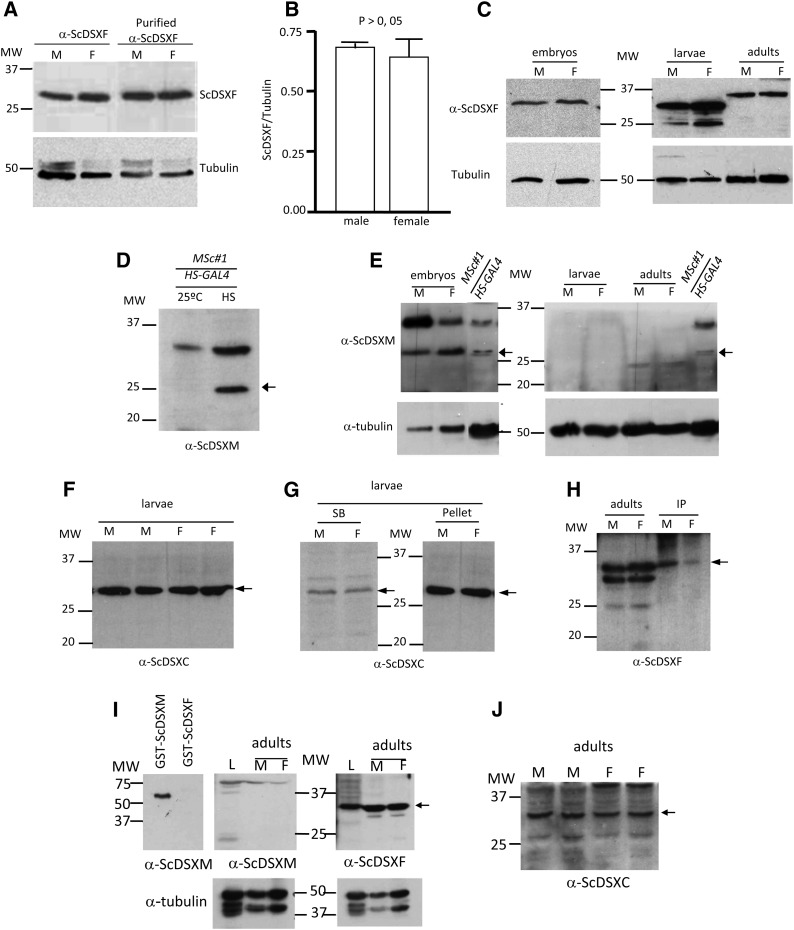
Western blots of total protein extracts from male and female S. coprophila larvae were prepared and incubated with α-ScDSXF serum or with the antigen affinity-purified α-ScDSXF antibody (Figure 3A). A band corresponding to the predicted female ScDSXF protein of ∼30 kDa was observed in both sexes. These results demonstrated that the female ScDSXF protein is present in both males and females.
Figure 3.
Expression analyses of the S. coprophila (A–H) and S. ocellaris (I–J) DSX proteins. (A) Western blot of total protein extracts from male (M) and female (F) larvae probed with the α-ScDSXF serum against the DSXF peptide corresponding to the specific C-terminal domain of the putative ScDSXF protein or with the antigen-affinity-purified α-ScDSXF. (B) Semiquantitative analysis of the amount of ScDSXF protein in male and female larvae, using tubulin for normalization. The bars in the histograms represent the 95% confidence intervals corresponding to five replicates. (C) Expression of the ScDSXF protein during development and in adults of males and females of S. coprophila. (D) Western blot of total protein extracts of transgenic Drosophila flies UAS-ScdsxMSc#1/HS-GAL4 without heat shock (lane 25°) or after heat-shock treatment at 37° (lane HS). The arrow points to the expected band corresponding to the transgenic ScDSXM protein. (E) Western blots of total protein extracts from embryos, larvae, and adults of males (M) and females (F), separately, probed with the purified α-ScDSXM serum. The lane MSc#1/HS-GAL4 carries total protein extracts of Drosophila flies UAS-ScdsxMSc#1/HS-GAL4 after heat-shock treatment at 37° as a positive control for the function of α-ScDSXM (arrow). (F) Western blot of total protein extracts from male (M) larvae and female (F) larvae probed with the purified α-ScDSXC serum. (G) Western blots of soluble (SB) and nonsoluble (Pellet) protein extracts from male (M) and female (F) larvae probed with the purified α-ScDSXC serum. (H) Western blot of total protein extracts from male (M) and female (F) adults (lanes adults) and the corresponding immunoprecipitation protein fraction with the affinity-purified α-ScDSXF serum and probed with this serum (lanes IP). The arrow points to the expected band for the ScDSXF protein. (I) Western blot of total protein extracts from a mixture of male and female larvae (L), and from male and female adults, separately probed with the purified α-ScDSXM or α-ScDSXF. GST-ScDSXM and GST-ScDSXF proteins were used as positive and negative controls, respectively, for the function of α-ScDSXM. (J) Western blot of total protein extracts from male (M) and female (F) adults, separately probed with the affinity-purified α-ScDSXC. The arrow points to the expected band corresponding to the ScDSXF protein.
Next, the amount of female ScDSXF protein was quantified in male and female larvae. Western blots of total protein extracts prepared from male and female larvae, separately, were probed with the affinity-purified α-ScDSXF or a monoclonal antibody against tubulin, which was used as an internal protein-loading control (for experimental details, see Materials and Methods). No differences were found in the ratio of ScDSXF/tubulin in both sexes (Figure 3B), indicating that both males and females have similar amounts of female ScDSXF protein. ScDSXF protein was also observed in embryos and adults of both sexes (Figure 3C), demonstrating that it was present during the development of both males and females. The smaller bands observed in the lanes M and F (Figure 3C) corresponding to larvae might correspond to the predicted truncated ScDSXF221 protein (∼25 kDa) or to the proteolysis of ScDSXF. The different mobility of the ScDSXF band in adults might be due to post-translational modifications in the protein. The alternative possibility that it represented a specific splicing variant in the adults can be rejected since the RT-PCR amplicon corresponding to that putative splicing variant was never found.
The putative Sciara DSXM male protein does not seem to be produced:
The α-ScDSXM serum did not detect the putative Sciara male DSXM protein in Western blots of total protein extracts from male and female S. coprophila larvae (data not shown). This could not be attributed to a lack of activity of the serum since it specifically recognizes the ScDSXM protein in Western blots, using the purified fused GST-ScDSXM protein as antigen (Figure S3B). The possibility that the C-terminal domain (the epitope) of ScDSXM was not accessible to the serum seemed unlikely because of the denaturation conditions used in Western blots. Nevertheless, it was imaginable that the α-ScDSXM serum was working in Western blots of the purified ScDSXM protein but failed to detect this protein when total protein extracts were used. To explore this possibility, we took advantage of Drosophila transgenic lines carrying the ScdsxM-cDNA transgene encoding the male ScDSXM protein. The ScdsxM-cDNA was linked to UAS sequences and the HS-GAL4 driver was used for conditional expression of the transgene (see below). A Western blot of total protein extracts from Drosophila XY transgenic flies is shown in Figure 3D. As expected, the ScDSXM protein was not detected at 25° (lane 25°), whereas the protein was detected after the adult flies were subjected to heat-shock treatments (arrow in lane HS). Thus, the α-ScDSXM serum was also correctly functioning in Western blots of total protein extracts. Then, we performed a Western blot with total protein extracts from S. coprophila male (M) and female (F) embryos, larvae, and adults, separately, and probed this with the antigen-affinity-purified α-ScDSXM serum (Figure 3E). Total protein extract from Drosophila XY adults expressing the transgenic ScDSXM protein was used as a control for α-ScDSXM activity (Figure 3E, MSc#1HS-GAL4 lane). Whereas the ScDSXM protein was detected in the control and in the embryos of both sexes (Figure 3E, arrow), it was not present in the larvae or in the adults of both males and females. It remains to be determined whether the protein detected in embryos is derived from the maternal ScdsxM mRNA (there is expression of Scdsx in ovaries, data not shown) or originates from the expression of the zygotic Scdsx gene. The low-intensity bands below 25 kDa observed in adults were also observed with the secondary antibody alone and thus are probably nonspecific. This failure to detect the ScDSXM protein in larvae and adults cannot be attributed to the method used to prepare the protein extracts since the same method was used with Drosophila adults, as well as with embryos, larvae, and adults of S. coprophila. Since the activity of sera varies from rabbit to rabbit, a new serum was then obtained by immunization of a second rabbit with the same peptide specific for the C-terminal region of the ScDSXM protein. This second serum did not detect the ScDSXM protein in male and female larvae of S. coprophila in spite of the fact that ELISA confirmed its activity (data not shown).
An antibody against a synthetic peptide corresponding to an amino acid sequence from the N-terminal region that is common to both ScDSXF and ScDSXM proteins was prepared. This serum (denoted as α-ScDSXC) was active in ELISA against the common N-terminal peptide but did not recognize the peptides corresponding to the specific C-terminal domain of either ScDSXF or ScDSXM, as expected (data not shown). In Western blots, the α-ScDSXC recognized the purified GST-ScDSXF, GST-ScDSXM, and GST-ScDSXC (the whole N-terminal region common to ScDSXF and ScDSXM fused to GST) (Figure S3C). Western blots of total protein extracts from distinct M and F S. coprophila larvae probed with the antigen-affinity-purified α-ScDSXC serum showed the expected band corresponding to ScDSXF protein (arrow) but not the band (∼25 kDa) corresponding to ScDSXM protein (Figure 3F). Of note, the α-ScDSXC serum was generated against the same epitope present in both ScDSXF and ScDSXM proteins. Hence, the failure to detect the ScDSXM protein cannot be attributed to a lack of activity of the serum.
It could be argued that the male ScDSXM protein was produced but not observed because the epitopes were post-translationally modified, impeding their recognition by α-ScDSXM and α-ScDSXC. If this putative modification was endowing a sex-specific functional role to the ScDSXM protein, this modification should also show sex specificity and consequently ScDSXM would be detected only in one of the sexes. This explanation, however, can be rejected since the protein was observed neither in males nor in females.
Finally, two further approaches were attempted to detect the ScDSXM protein. Total protein extracts from male and female S. coprophila larvae, separately, were fractionated into the soluble and the insoluble portion and analyzed by Western blots probed with the α-ScDSXC serum. Results showed that that ScDSXF but not ScDSXM was present in both fractions of males and females (arrow in Figure 3G). The second approach tested was immunoprecipitation. Thus, total protein extracts from male and female S. coprophila adults, separately, were immunoprecipitated with purified α-ScDSXF; the subsequent Western blot probed with this serum revealed the presence of ScDSXF in males and females (arrows in Figure 3H). The low-intensity bands in the lanes for adults in Figure 3H might correspond to the predicted truncated ScDSXF221 protein (∼25 kDa) or to proteolysis of ScDSXF. Immunoprecipitation with purified α-ScDSXM and subsequent Western blotting with this serum did not indicate the presence of ScDSXM in both sexes (data not shown). Unfortunately, α-ScDSXC did not function in immunoprecipitation
Western blots of total protein extracts from male plus female larvae and from separated male and female adults of S. ocellaris were prepared and probed with α-ScDSXM, α-ScDSXF, and α-ScDSXM. Analogous to S. coprophila, the female SoDSXF protein was detected in both M and F adults (arrow in Figure 3, I and J), whereas the male ScDSXM protein was not detected (Figure 3, I and J). The fusion GST-ScDSXM and GST-ScDSXF proteins were used as positive and negative controls, respectively, for the specific function of α-ScDSXM serum (Figure 3D). Similarly, SoDSXF but not SoDSXM was observed in Western blots of total protein extracts of a mixture of male and female larvae (L in Figure 3I). We repeatedly failed to detect the SoDSXF and SoDSXM proteins in S. ocellaris embryos, for reasons unknown to us. Hence, as found in S. coprophila, the female SoDSXF protein was expressed in both sexes in S. ocellaris, but it appeared that the SoDSXM protein was not produced although its mRNA was expressed.
Collectively, all these results suggested that the Sciara DSXM male protein was not synthesized in Sciara or was synthesized at lower amounts that precluded its detection by Western blotting. Remarkably, this happens in both sexes.
Splicing regulation of the primary transcript of the Sciara dsx gene
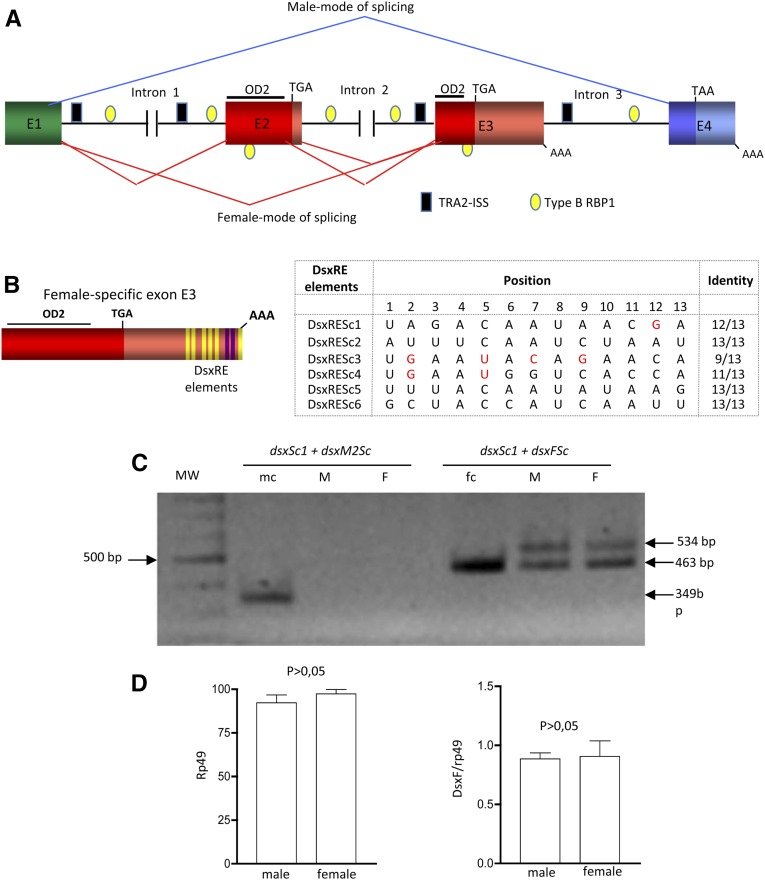
A search for sequences located at the regions involved in the sex-specific splicing regulation of dsx primary transcript in other insects was undertaken in Sciara. Since Sciara is a dipteran, we searched for TRA–TRA2 binding sequences in the Scdsx pre-mRNA that might explain the different amounts of the male- and female-type Scdsx mRNAs in both sexes. Six DsxRE-like sequences were found in the female-specific exon E3 located ∼300 nucleotides upstream of its 3′ end (Figure 4B). In three of the six sequences, there are bases (highlighted in red) in positions never found in the consensus sequence, which is defined in the legend to Figure 4. Two putative PRE elements between the penultimate and final putative TRA–TRA2 binding sequences were also found (marked in purple in Figure 4B). In the dipterans, RBP1, a non-sex-specific SR splicing factor, is also required for female-specific dsx pre-mRNA splicing by binding to target sequences located within the purine-rich polypyrimidine tract of the female-specific exon (Heinrichs and Baker 1995). Two types of RBP1-binding sequences have been identified: DCADCTTTA (type A RBP1) and ATCYNNA (type B RBP1) (Qi et al. 2007). No type A RBP1 sequences were found in Scdsx pre-mRNA and several type B RBP1 sequences were localized in the female-specific exons, one in exon E2 and the other in exon E3, plus numerous copies scattered along all introns (Figure 4A). Salvemini et al. (2011) reported the existence of TRA2-intronic splicing silencer (TRA2-ISS) sequences in Aedes dsx primary transcript in the region involved in its sex-specific splicing regulation. These ISS sequences (CAAGR) are bound by TRA2, causing suppression of splicing (Qi et al. 2007). Several TRA2-ISS sequences were found in Scdsx pre-mRNA: several distributed in intron 1, one in intron 2 and several in intron 3 (Figure 4A). No putative NvdsxRE sites for TRA–TRA2 binding were found. These sequences, which have been found in the hymenopteran Nasonia dsx (Verhulst et al. 2010), have been also observed in the dsx gene of Aedes (Salvemini et al. 2011) and Tribolium (Shukla and Palli 2012). Finally, no BmPSI-exonic splicing suppressor (BmPSI-ESS) sequences were detected in Scdsx pre-mRNA. These ESS sequences are found in dsx from the lepidopterans Bombyx (Suzuki et al. 2001) and Antheraea (Shukla and Nagaraju 2010), which are targets for the sex-specific factor BmPSI that leads to skipping of the female-specific exons 3 and 4 in males (Suzuki et al. 2008).
Figure 4.
Splicing elements of the Scdsx pre-mRNA. (A) Location of the putative TRA–TRA2 binding sequences and TRA2-ISS (black rectangles) and type B RBP1 (yellow ellipses) in Scdsx. (B) Distributions of the DsxREs sequences and the PRE are marked by yellow and purple stripes, respectively, in exon E3. The red letters indicate nucleotides in positions never present in the consensus sequence, which was defined by comparison of putative TRA–TRA2 binding sequences found in dsx of D. melanogaster, M. domestica, B. oleae, B. tryoni, A. obliqua, C. capitata, M. scalaris, A. gambiae, and A. aegypti; in the gene fruitless of D. melanogaster and A. gambiae; and in the gene tra of C. capitata, B. oleae, and A. obliqua. To define the consensus sequence, the nucleotide present in each of the 13 positions independently of its frequency of appearance was considered. (C) Effect of the TRA–TRA2 complex of D. melanogaster on splicing regulation of DScmg pre-mRNA. For details of DScmg construction see Materials and Methods and Figure S1 and its legend; (mc) and (fc) denote the plasmid carrying the ScdsxM or the ScdsxF mRNAs used as positive controls of primer function. (D) Semiquantitative analysis of the amount of female-spliced mRNA isoform from the Dscmg pre-mRNA with respect to rp49 in Drosophila male (M) and female (F) adults. The bars in the histograms represent the 95% confidence intervals corresponding to five replicates.
Since the binding sequences of SR proteins, such as TRA and TRA2, may be quite degenerate (Black 2003), and because the DsxRE sequences of Scdsx showed a high degree of similarity with the consensus TRA–TRA2 binding sequences, a possibility existed that a female-specific TRA–TRA2 complex was involved in the quantitative differential splicing of dsx pre-mRNA in Sciara males and females, with that complex favoring the female against the male type. To test this, a Scdsx minigene (DScmg) was constructed that contained all the complete exons E2, E3, and E4 and 276 bp from the 3′ end of common exon E1, together with their adjacent intron sequences containing the binding sites surrounding the exons described above (see Materials and Methods and Figure S2 and its legend), and DScmg-transgenic Drosophila flies were produced. The idea was to assess the splicing pattern of DScmg pre-mRNA in transgenic Drosophila males (with no TRA–TRA2 complex) and females (with TRA–TRA2 complex). The results are presented in Figure 4C (for details, see Materials and Methods). Only the amplicons corresponding to the female-specific splicing pattern were observed in both males and females. Note the presence of two bands of 463 and 534 bp corresponding to the splicing modes that give rise to female ScdsxF and ScdsxF252 mRNA isoforms, respectively. In the “fc” lane in Figure 4C, there is only one band because it corresponds to the PCR control where the ScdsxF cDNA was used as template. The absence of amplicons generated by the male-splicing pattern in both males and females could not be attributed to a failure of the primers used since these functioned in the PCR control where the ScdsxM cDNA was used as a template (lane “mc” in Figure 4C). Negative controls in these PCR assays produced no amplicons (see Materials and Methods). The amount of rp49 mRNA did not significantly vary between males and females (Student’s t-test, P > 0.05), indicating that the reverse transcription reaction was similarly effective in both sexes (Figure 4D). Thus, the amount of rp49 mRNA was used to normalize the relative abundance of DScmgdsxF mRNA in males and females. Results showed that this is similar in both sexes (Figure 4D, Student’s t-test, P > 0.05). This result cannot be interpreted by considering that the minigene did not carry relevant sequences for splicing regulation as no aberrant splicing amplicons were detected. Furthermore, the binding sequences around the sex-specific exons known to be required for sex-specific splicing regulation of the dipteran dsx primary transcript were present in the DScmg minigene. It is not possible to reject, however, that relevant yet not identified sequences to generate the male isoform were not contained in the minigene construct.
An explanation for the finding that in Drosophila the DScmg pre-mRNA follows the female- but not the male-splicing pattern would be that splicing factors involved in the Sciara dsx pre-mRNA splicing pattern were not present in Drosophila. Whatever the molecular nature of the mechanism underlying dsx pre-mRNA splicing in Sciara and the factors involved, the dipteran sex-specific splicing pattern of dsx pre-mRNA does not apply to Sciara since both dsxF and dsxM transcripts are present in males and in females. Furthermore, the TRA–TRA2 complex appears not to participate in the differential splicing of Sciara dsx pre-mRNA. On the other hand, direct proof of the role of tra-2 in Sciara sex determination is lacking (see Martín et al. 2011).
Effect of the female ScDsxF and male ScDsxM proteins of S. coprophila on the regulation of Drosophila yolk protein genes
The yolk protein (yp) genes of Drosophila are coordinately transcribed in the fat body of adult females under the control of dsx. DSXF and DSXM act as activator and repressor, respectively, by binding to the same regulatory sequences (reviewed in Bownes 1994). These genes are also expressed in the follicle cells of the ovary, yet they are no longer under the control of dsx but instead are regulated by tissue-specific factors present in those cells (Bownes et al. 1990). In loss-of-function dsx mutant XX flies lacking both DSXF and DSXM proteins, basal transcription of the yp genes in the fat body (gonads are not developed) has been reported (Bownes and Nöthiger 1981), although in our dsx1 stock such remnant expression was not observed.
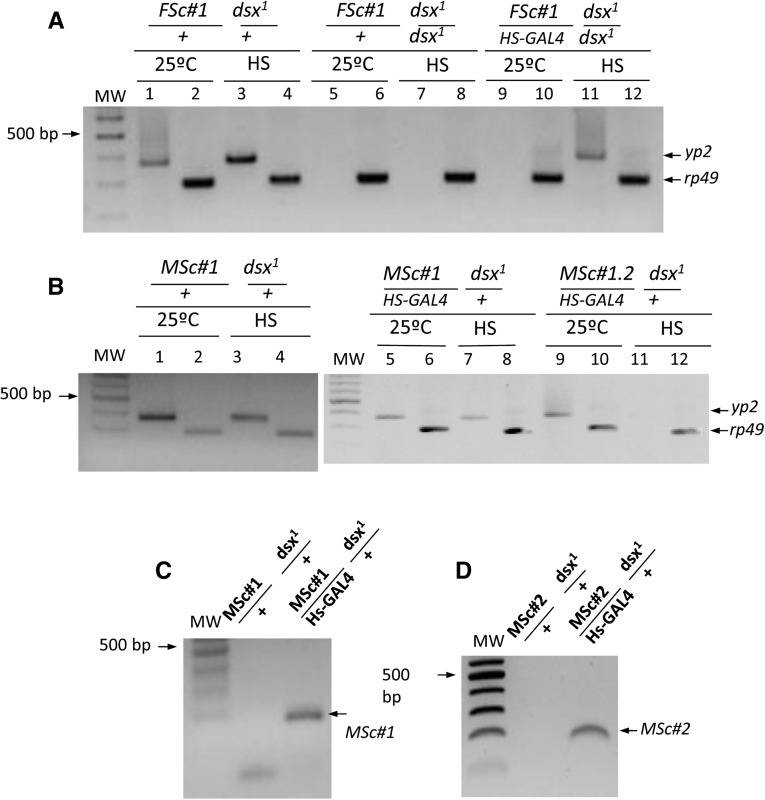
The effect of female ScDSXF protein (FSc transgene) on the regulation of Drosophila yp genes was studied by monitoring the expression of yp2 in transgenic Drosophila XX flies mutant for dsx and either expressing or not the transgenic female ScDSXF protein. As control, the expression of rp49, which codes for the constitutive ribosomal protein 49, was used. For the construction of the FSc and MSc transgenes, see Materials and Methods. The inducible HS-GAL4 driver was used to express the transgenic protein. The results are presented in Figure 5A (for details, see Materials and Methods and legend to Figure 5). As expected, control females (FSc#1/+; dsx1/+) expressed the yp2 gene whereas intersexual flies (FSc#1/+; dsx1/dsx1) did not, whether kept at 25° or subjected to heat shock. The experimental females (FSc#1/HS-GAL4; dsx1/dsx1) did not express the yp2 gene when maintained at 25°, although yp2 was expressed after heat shock (lane 11 in Figure 5A). The three classes of females expressed the control rp49 gene when maintained at 25° and after heat shock. Negative controls in these PCR assays produced no amplicons (see Materials and Methods). These results indicated that the female ScDSXF protein behaved as an activator of the Drosophila yp genes, analogous to DSXF of Drosophila.
Figure 5.
Expression of Drosophila yolk protein 2 gene in Sciara dsx transgenic Drosophila flies. (A) Effect of the ScDsxF protein on the regulation of the Drosophila yp2 gene. (B) Effect of the ScDsxM protein on the regulation of the Drosophila yp2 gene. (C) Heat-shock-dependent expression of MSc#1 transgene. (D) Heat-shock-dependent expression of MSc#2 transgene. yp2 and rp49 stand for the yolk protein 2 and ribosomal protein 49 genes, respectively. The term “25°C” indicates that the flies were maintained at this temperature after eclosion, whereas HS indicates that they were subjected to two 3-hr heat-shock pulses (37°) per day for two consecutive days with recovery at 25° between pulses. PCR amplification of total RNA extracts (without cDNA) yielded no amplification product, indicating that the RNA sample was devoid of DNA. The genotypes in A correspond to the offspring of females yw; FSc#1; dsx1/MKRS,Sb crossed with males w/Y; HS-GAL4[ry+] CyO; dsx1/MKRS,Sb. The genotypes in B correspond to the offspring of females yw; MSc#1; dsx1/MKRS,Sb crossed with males w/Y; HS-GAL4[ry+] CyO; dsx1/MKRS,Sb (B, lanes 1–8) or to the offspring of females yw; MSc#1; Msc#2 crossed with males w/Y; HS-GAL4[ry+] CyO; dsx1/MKRS,Sb (B, lanes 9–12). The genotypes in C correspond to the offspring of females yw; MSc#1 crossed with males w/Y; HS-GAL4[ry+] CyO; dsx1/MKRS,Sb. Finally, the genotypes in D correspond to the offspring of females yw; MSc#2/MKRS,Sb crossed with males w/Y; HS-GAL4[ry+] CyO; dsx1/MKRS,Sb. For the construction of the FSc and MSc transgenes see Materials and Methods.
The same experimental plan described above was followed to determine whether the male ScDSXM protein (MSc transgene) acts as an inhibitor of the yp genes. Both control (MSc#1/+; dsx1/+) and experimental (MSc#1/HS-GAL4; dsx1/+) XX females produced the Drosophila endogenous DSXF protein, while the experimental females additionally produced the transgenic male ScDSXM protein when subjected to heat shock. As expected, the control females expressed the yp2 gene whether maintained at 25° or when subjected to heat shock (Figure 5B). The experimental females also expressed the yp2 gene when maintained at 25° or after being subjected to heat shock (lane 7 in Figure 5B), in spite of the fact that they expressed the transgene MSc#1 (Figure 5C) and the protein was produced as mentioned above (Figure 3D). This absence of effect by the transgenic ScDSXM male protein could be due to its inherent incapacity to successfully compete with the endogenous Drosophila DSXF protein and override its activating effect on expression of yp genes. As described earlier, ScDSXM lacks the OD2 domain found in the male DSXM protein of Drosophila. Alternatively, the lack of effect could be due to an insufficient amount of the transgenic ScDSXM protein to override the endogenous DSXF protein. To address this, the assay was repeated but this time the dsx/+ females carried two copies of the MSc transgene. It was previously verified that the additional copy of the transgene MSc#2 is expressed after the heat-shock treatment (Figure 5D). Therefore, the identical amount of endogenous Drosophila DSXF protein was now confronted with a greater amount of transgenic male ScDSXM protein. In this setting, ScDSXM protein overruled the activating function of endogenous DSXF, causing the repression of the yp genes (lane 11 in Figure 5B). Negative controls in these PCR assays produced no amplicons (see Materials and Methods). These results indicated that the male ScDSXM protein behaved as an inhibitor of the Drosophila yp genes, analogous to DSXM of Drosophila.
Collectively, these results indicated that the Sciara female ScDSXF and male ScDSXM proteins showed conserved female and male sex-determination function, respectively, in Drosophila, at least with respect to the control of yp genes.
Comparison of the DSX proteins of Sciara with those of other insects and dsx phylogeny
The putative female ScDSXF and male ScDSXM proteins of S. coprophila (Sciaridae, suborder Nematocera) were compared to the DSXF and DSXM proteins from dipterans, lepidopterans, hymenopterans, and the coleopterans. For methodological details, see Materials and Methods. The comparisons are shown in Table S3. The higher degree of variation in terms of number of amino acids and similarity was located in the N- and C-terminal regions, as well as in the linker region between the OD1 and OD2 domains.
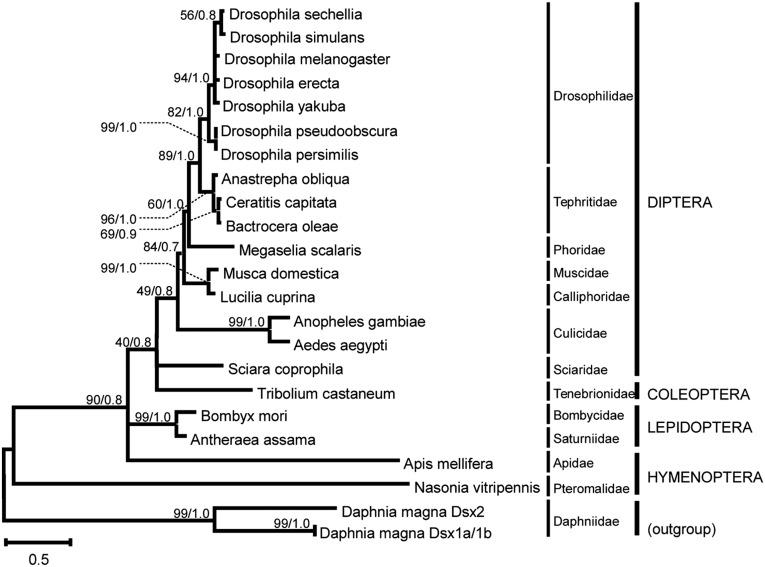
The phylogenetic relationships among DSX proteins were inferred on the basis of the non-sex-specific region of DSX (including the complete oligomerization domain OD1 and part of the OD2 domain), using sequences from 21 insect species and 1 outgroup species (Table S2). For details, see Materials and Methods. The tree was reconstructed by combining maximum-likelihood and Bayesian approaches, producing topologies that were not in conflict (Figure 6). The resulting topology revealed an interesting fact: in spite of the divergent nature of the dsx gene of Sciara in terms of exon composition, splicing regulation, and production of mRNAs with respect to other insects (especially dipterans), the encoded protein is phylogenetically located within the monophyletic group of DSX proteins from dipterans. This result indicates that, in Sciara, the differences observed in gene organization and expression did not result in significant modifications in the DSX protein, probably because of its conserved role as a basal component of the sex-determination cascade (Sánchez 2008). This notion is consistent with the presence of constant evolutionary rates in several sex-determining proteins (including DSX) across insects (Eirín-López and Sánchez 2015).
Figure 6.
Molecular phylogeny of DSX proteins in insect representatives (see Table S2). The tree was reconstructed using maximum-likelihood and Bayesian methods. The numbers for interior branches represent bootstrap probabilities based on 1000 replications followed by the corresponding Bayesian posterior probabilities (shown only when at least one of them is ≥50%). The taxonomic classification of the species used in this analysis is indicated in the right margin.
A possible explanation for the divergence observed in the Sciara dsx lineage may involve a complex pattern of evolution combining purifying selection (Ruiz et al. 2007) with bursts of positive selection as recently described for members of the Anastrepha fraterculus species group (Sobrinho and De Brito 2012). Further analysis will be necessary to decipher the role of positive selection during the evolutionary history of doublesex in insects and its relevance for sex determination.
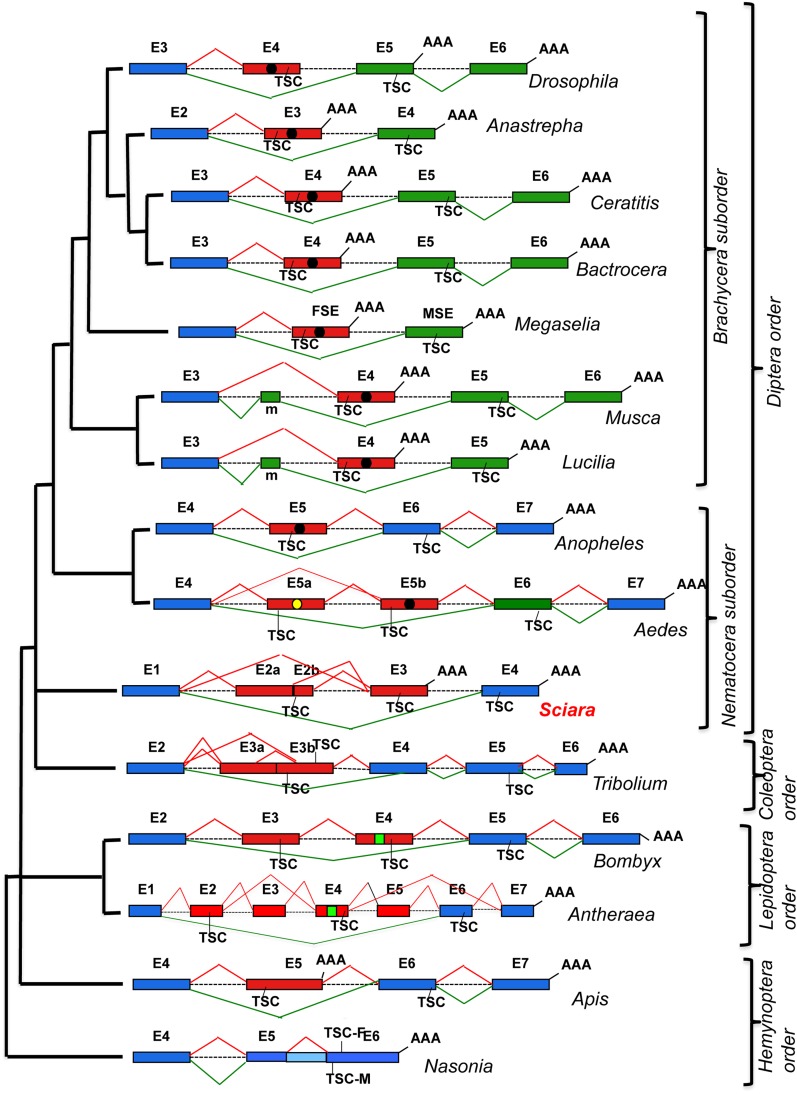
The gene dsx of insects shows a great variability with regard to molecular organization and splicing regulation (Figure 7):
Figure 7.
Molecular evolution of the organization and splicing regulation of gene dsx in insects. It follows the phylogeny shown in Figure 6. Exons (E) (boxes) and introns (lines) are not drawn to scale. The number following E indicates the position of the corresponding exon in the molecular organization of the dsx gene. The dotted lines for introns indicate that their lengths remain unknown. Blue exons are common to both sexes, red exons indicate female-specific exons, and green exons represent male-specific exons. AAA stands for poly-A+. TSC stands for translation stop codon. TSC-F and TSC-M stand for translation stop codon in female and male mRNAs, respectively. Black and yellow circles and green squares within exons represent the binding sites for the corresponding specific splicing factors (see main text).
Exon composition and organization. The dsx gene in all dipterans contains male- and female-specific exons, except in Anopheles (Scali et al. 2005), which has only a female-specific exon and no male-specific exons as in the lepidopterans Bombyx (Ohbayashi et al. 2001; Suzuki et al. 2001) and Antheraea (Shukla and Nagaraju 2010), the hymenopterans Apis (Cho et al. 2007) and Nasonia (Oliveira et al. 2009), and the coleopteran Tribolium (Shukla and Palli 2012). An attribute common to all dsx genes is the location of the female-specific exons upstream of the male-specific exons or to the common exons encoding the C-terminal domain of male and female DSX proteins.
Splicing regulation of dsx pre-mRNA. All dsx genes so far characterized in insects, except Sciara, show sex-specific regulation with the production of mature male- and female-specific mRNAs. This condition corresponds to the existence of sex-specific splicing factors (either activators or inhibitors), whose expression is related to the default splicing regulation of the dsx pre-mRNA, which can be either the male or the female mode of splicing. Strictly speaking, it is not appropriate to talk in terms of default dsx pre-mRNA splicing in Sciara because both male and female dsx mRNAs are simultaneously produced in the two sexes. Sciara could parallel the situation hypothesized in Aedes (Salvemini et al. 2011); that is, putative SR-M and SR-F splicing factors, which would be functional in males and females, respectively, are present in both sexes, but in Sciara these factors would be simultaneously functional in both sexes.
The Dsx proteins. All dsx genes so far characterized in insects, except Sciara, produce male- and female-specific proteins. Sciara dsx seems to produce a single polypeptide, which is reminiscent of the situation encountered in Daphnia (Kato et al. 2011), with the difference being that in the latter the quantity of this protein, which is DSXM, would differ between the two sexes whereas in Sciara, which is DSXF, the quantity would be similar in males and females.
It is proposed that the evolution of dsx would occur by conversion of a single DSX polypeptide-encoding gene into a two DSXF and DSXM polypeptides-encoding gene. The diversity shown by the insect dsx had to be the result of different evolutionary paths undertaken by this gene. These would involve concomitant necessary coevolutionary novelties—sex-specific splicing factors and their target sequences—during insect evolution, allowing the transition toward the production of sex-specific transcripts and their corresponding proteins, thus converting dsx in the ultimately discriminatory gene for sex determination. In this scenario, Sciara might represent a path in dsx evolution characterized by retaining the production of a single protein, a postulated feature of the ancestral gene from which the extant insect dsx genes evolved. The interested reader is addressed to the comprehensive overview of Verhulst and Van De Zande (2015) for a discussion on the evolutionary relationship between gene dsx and its many target genes.
Concluding remarks
The following results indicate that the dsx gene of Sciara was successfully cloned and characterized. First, the putative Sciara proteins showed the highest degree of similarity with DSX proteins from other insects. Second, the putative Sciara female DSXF and male DSXM proteins shared the amino-terminal region and differ in their C-terminal regions, analogous to DSX proteins of other insects. Third, the putative Sciara DSX proteins contained the two domains (OD1 and OD2) required for DNA and protein–protein interactions, respectively, that characterize DSX proteins (An et al. 1996; Cho and Wensink 1997). Moreover, OD1 contained the six amino acids Cys, His, His, Cys, Cys, and Arg that are essential for the binding of DSX proteins to its DNA-target sequences (Erdman and Burtis 1993). Fourth, the putative Sciara DSX proteins were encoded in mRNAs produced by alternative splicing of a single primary transcript. Fifth, the Sciara dsx gene was constitutively transcribed during development and adult life of males and females. Finally, the female DSXF and male DSXM Sciara proteins showed conserved female and male function, respectively, on the control of Drosophila yolk-protein genes. There were, however, three fundamental differences between Sciara and the other insects where dsx has been characterized. First, the putative male DSXM protein contained OD1 but lacked the OD2 domain. Second, Sciara had no bona fide sex-specific dsx mRNAs but the same transcripts were present in both males and females although their relative abundance is sex specific. Third, the female DSXF protein, but not the male DSXM protein, was produced at similar amounts in both sexes.
These findings suggest the existence in Sciara of a control inhibiting the translation of ScdsxM mRNA. Is there any biological reason to justify such translation inhibition? It has been reported that the simultaneous expression of male and female DSX proteins produces sterile intersexual flies (Epper 1981; Nöthiger et al. 1987; Jurnish and Burtis 1993; Alvarez et al. 2009). In this respect, it is noteworthy that the putative DSXM and DSXF proteins of Sciara share the very well-conserved DM/OD1 domain that endows the DSX proteins with the capacity to interact with DNA. In addition, it was shown here that transgenic Sciara DSXM prevented the endogenous Drosophila DSXF protein from activating the yp genes in this species. This result indicates that the Sciara DSXM protein maintains the capacity to interact with DNA such that it can compete with DSXF for the same binding sites. Hence, it is proposed here that a process had to evolve with the simultaneous expression of the female ScdsxF and the male ScdsxM mRNAs in Sciara to prevent the translation of the ScdsxM mRNA; otherwise, the DSXM protein interferes with the function of DSXF and the Sciara zygote would develop as a sterile intersex. In the other insects, such translational inhibition is not required since the sex specificity of the DSX proteins was stipulated by the sex specificity of their mRNAs.
The surprising result of the present study was the finding that the DSXF protein is present at similar amounts in Sciara males and females, whereas the DSXM protein was not detected in the two sexes. This result raises the question of the requirement of dsx in Sciara sex determination, an issue that can be undoubtedly addressed by analyzing the effect of lack of dsx function on Sciara sex determination. It is, however, difficult to provide an answer to this query. First, no dsx mutants are available in Sciara. Second, the application of RNA interference (RNAi) technology to modulate endogenous dsx function is not so straightforward in these insects due to the extreme fragility of their tiny eggs (L. Sánchez and A. L. P. Perondini, unpublished results). Moreover, RNAi would not be appropriate if DSXF, which is expressed throughout the development and in the adult life of Sciara, were continuously required for female development, as in the other insects. Finally, the use of this procedure to analyze the role of DSXF in vitellogenesis (a female-sexual dimorphic feature) is not possible in Sciara because oogenesis occurs only during metamorphosis in this insect (Berry 1941). Hence, direct proof of the role of dsx in Sciara sex determination remains elusive at present. Regardless, it was shown here that the Sciara DSXF and the DSXM proteins exhibited conserved female and male sex-determination function, respectively, in the control of Drosophila yp genes. In addition, the Sciara DSX proteins—mainly DSXF—showed a high degree of conservation with the orthologous proteins of other insects. Because of these observations and due to the fact that the Sciara dsx gene produces only one protein, present at similar amounts in both sexes, it is suggested here that dsx is also involved in Sciara sex determination although it appears not to play the discriminatory role in this process, in contrast to what has been observed in all insects where this gene has been so far characterized.
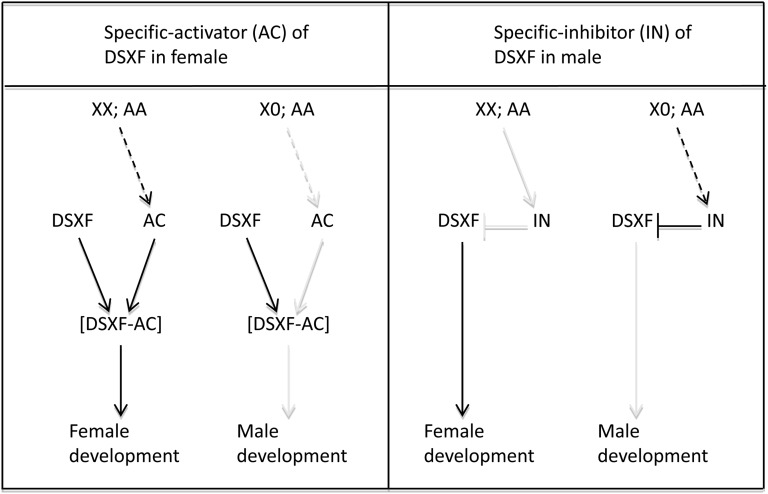
The putative role of dsx in Sciara sex determination is depicted in Figure 8. It is considered that the DSXF protein was needed for female development and was the only one produced by dsx in the two sexes. Two alternative scenarios could be imagined. In the first one, it was assumed that the default state of DSXF was to be inactive, requiring the presence of an activator (AC) that would interact with DSXF, forming the functional DSXF–AC complex that would determine female development. This activator would be produced only in XX;AA individuals so that male development represented the default state. Under this scenario, Sciara DSXF would “behave” as the Intersex protein in Drosophila. This protein is found in both sexes and interacts with DSXF, but not with DSXM, forming a complex to control female development (Chase and Baker 1995; Waterbury et al. 1999; Garrett-Engele et al. 2002). The difference between Drosophila and Sciara would be that DSXF supplies the discriminatory function in the former but not in the latter, where AC, which would be produced only in females, brings the discriminatory function. In the second scenario, the default state of DSXF would be active, requiring the presence of an inhibitor (IN) that would neutralize DSXF and consequently the male development would ensue. This inhibitor would be produced only in X0;AA individuals so that female development represented the default state.
Figure 8.
Scheme showing the putative role of dsx on Sciara sex determination. It was assumed that this gene produces only the female ScDSXF protein at similar amounts in both sexes. Two hypotheses are proposed. The first one assumes that the default state of DSXF protein was to be inactive. In XX;AA specimens, an activator (AC) would be specifically produced (dashed arrow) that forms a complex with DSXF protein, determining the female development. The alternative hypothesis assumes that the default state of DSXF protein was to be active. In X0;AA specimens, an inhibitor (IN) would be specifically synthesized (dashed arrow) that interacts with DSXF, preventing its function, determining male development.
Supplementary Material
Acknowledgments
The authors thank B. Oliver, I. Guerrero, M. Calleja, F. Díaz-Benjumea, E. Sánchez-Herrero, and S. Campuzano for providing Drosophila stocks. L. Sánchez had financial support (BFU2005-03000 and BFU2008-00474) from the Spanish government during the time the group was doing the work presented in this article. However, he had to close his laboratory—with the consequent disappearance of the group—because his grant was not renewed. L. Sánchez thanks very much J. L. Barbero for permitting the authors to finish this work in his laboratory and to be sponsored by his grant. M. Alvarez and F. Sarno were recipients of a predoctoral fellowship from the Spanish Government. J.M.E.-L. was supported by startup funds from the College of Arts and Sciences at Florida International University.
Footnotes
Communicating editor: J. A. Birchler
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.177972/-/DC1.
Literature Cited
- Alvarez M., Ruiz M. F., Sánchez L., 2009. Effect of the gene doublesex of Anastrepha on the somatic sexual development of Drosophila. PLoS ONE 13: e5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An W., Cho S., Ishii H., Wensin P. C., 1996. Sex-specific and non-sex-specific oligomerization domains in both of the Doublesex transcription factors from Drosophila melanogaster. Mol. Cell. Biol. 16: 3106–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B., Ridge K., 1980. Sex and the single cell. I. On the action of the major loci affecting sex determination in Drosophila melanogaster. Genetics 94: 383–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry R. O., 1941. Chromosome behavior in the germ cells and development of the gonads in Sciara ocellaris. J. Morphol. 68: 547–583. [Google Scholar]
- Beukeboom L. W., Perrin N., 2014. The Evolution of Sex Determination. Oxford University Press, Oxford. [Google Scholar]
- Black D. L., 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72: 291–336. [DOI] [PubMed] [Google Scholar]
- Bownes M., 1994. The regulation of the yolk protein genes a family of sex differentiation genes in Drosophila melanogaster. BioEssays 16: 745–752. [DOI] [PubMed] [Google Scholar]
- Bownes M., Nöthiger R., 1981. Sex determining genes and vitellogenin synthesis in Drosophila melanogaster. Mol. Gen. Genet. 182: 222–228. [DOI] [PubMed] [Google Scholar]
- Bownes M., Steinmann-Zwicky M., Nöthiger R., 1990. Differential control of yolk protein gene expression in fat bodies and gonads by the sex-determining gene tra-2 of Drosophila. EMBO J. 9: 3975–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Bull J. B., 1983. Evolution of Sex Determining Mechanisms. Benjamin-Cummings, Menlo Park, CA. [Google Scholar]
- Burtis K. C., Baker B. S., 1989. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56: 997–1010. [DOI] [PubMed] [Google Scholar]
- Chase B. A., Baker B. S., 1995. A genetic analysis of intersex a gene regulating sexual differentiation in Drosophila melanogaster females. Genetics 139: 1649–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S., Wensink P. C., 1997. DNA binding by the male and female doublesex proteins of Drosophila melanogaster. J. Biol. Chem. 272: 3185–3189. [DOI] [PubMed] [Google Scholar]
- Cho S., Huang Z. Y., Zhang J., 2007. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics 177: 1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline T. W., 1978. Two closely-linked mutations in Drosophila melanogaster that are lethal to opposite sexes and interact with daughterless. Genetics 90: 683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha C., Li F., Scott M. J., 2010. Conservation and sex-specific splicing of the doublesex gene in the economically important pest species Lucilia cuprina. J. Genet. 89: 279–285. [DOI] [PubMed] [Google Scholar]
- Davidheiser B., 1943. Inheritance of the X chromosome in exceptional males of Sciara ocellaris (Diptera). Genetics 28: 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Suchard M. A., Xie D., Rambaut A., 2012. Bayesian phylogenetics with BEAUti and the BEAST 17. Mol. Biol. Evol. 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirín-López J. M., Sánchez L., 2015. The comparative study of five sex-determining proteins across insects unveils high rates of evolution at basal components of the sex determination cascade. Dev. Genes Evol. 225: 23–30. [DOI] [PubMed] [Google Scholar]
- Epper F., 1981. Morphological analysis and fate map of the intersexual genital disc of the mutant doublesex-dominant in Drosophila melanogaster. Dev. Biol. 88: 104–114. [DOI] [PubMed] [Google Scholar]
- Erdman E. S., Burtis C. K., 1993. The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 12: 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Engele C. M., Siegal M. L., Manoli D. S., Williams B. C., Li H., et al. , 2002. Intersex a gene required for female sexual development in Drosophila is expressed in both sexes and functions together with doublesex to regulate terminal differentiation. Development 129: 4661–4675. [DOI] [PubMed] [Google Scholar]
- Gempe T., Beye M., 2010. Function and evolution of sex determination mechanisms genes and pathways in insects. BioEssays 33: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98. [Google Scholar]
- Harlow E., Lane D., 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hasegawa M., Kishino K., Yano T., 1985. Dating the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22: 160–174. [DOI] [PubMed] [Google Scholar]
- Hedges S. B., Kumar S., 2009. The TimeTree of Life. Oxford University Press, New York. [Google Scholar]
- Heinrichs V., Baker B. S., 1995. The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J. 14: 3987–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger M., Burghardt G., Siegenthaler C., Buser N., Hilfiker-Kleiner D., et al. , 2004. Sex determination in Drosophila melanogaster and Musca domestica converges at the level of the terminal regulator doublesex. Dev. Genes Evol. 214: 29–42. [DOI] [PubMed] [Google Scholar]
- Hoshijima K., Inoue K., Higuchi I., Sakamoto H., Shimura Y., 1991. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science 252: 833–836. [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M., 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Jurnish V. A., Burtis K., 1993. A positive role in differentiation for the male doublesex protein of Drosophila. Dev. Biol. 155: 235–249. [DOI] [PubMed] [Google Scholar]
- Kato Y., Kobayashi K., Watanabe H., Iguchi T., 2011. Environmental sex determination in the branchiopod crustacean Daphnia magna: deep conservation of a doublesex gene in the sex-determining pathway. PLoS Genet. 7: e1001345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn S., Sievert V., Traut W., 2000. The sex-determining gene doublesex in the fly Megaselia scalaris: conserved structure and sex-specific splicing. Genome 43: 1011–1020. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Lagos D., Ruiz M. F., Sánchez L., Komitopoulou K., 2005. Isolation and characterization of the Bactrocera oleae genes orthologous to the sex determination Sex-lethal and doublesex genes of Drosophila melanogaster. Gene 348: 111–121. [DOI] [PubMed] [Google Scholar]
- Lindsley D. L., Zimm G., 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego. [Google Scholar]
- Maniatis T., Fritsch F., Sambrook J., 1982. Molecular Cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Martín I., M. F. Ruiz, and L. Sánchez, 2011. The gene transformer-2 of Sciara (Diptera Nematocera) and its effect on Drosophila sexual development. BMC Dev. Biol. 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz C. W., 1938. Chromosome behavior, inheritance and sex determination in Sciara. Am. Nat. 72: 485–520. [Google Scholar]
- Metz C. W., Schmuck M. L., 1929. Unisexual progenies and the sex chromosomes mechanism in Sciara. Proc. Natl. Acad. Sci. USA 15: 863–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori L., E. M. Dessen, and A. L. P. Perondini, 1979. A gene that modifies the sex ratio in a bisexual strain of Sciara ocellaris. Heredity 42: 353–357. [DOI] [PubMed] [Google Scholar]
- Moses M. S., Metz C. W., 1928. Evidence that the female is responsible for the sex ratio in Sciara (Diptera). Proc. Natl. Acad. Sci. USA 14: 928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro R. G., C. C. Campos, and A. L. P. Perondini, 2007. Temperature and the progeny sex-ratio in Sciara ocellaris (Diptera, Sciaridae). Genet. Mol. Biol. 30: 152–158. [Google Scholar]
- Nöthiger R., Leuthold M., Andersen N., Gerschwiler P., Gruter A., et al. , 1987. Genetic and developmental analysis of the sex-determining gene doublesex (dsx) of Drosophila melanogaster. Genet. Res. 50: 113–123. [Google Scholar]
- Ohbayashi F., Suzuki M., Mita K., Okano K., Shimada T., 2001. A homologue of the Drosophila doublesex gene is transcribed into sex-specific mRNA isoforms in the silkworm Bombyx mori. Comp. Biochem. Physiol. 128: 145–158. [DOI] [PubMed] [Google Scholar]
- Oliveira C. D., Werren H. J., Verhulst C. E., Giebel D. J., Kamping A., et al. , 2009. Identification and characterization of the doublesex gene of Nasonia. Insect Mol. Biol. 18: 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J., 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85: 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalva L. O. F., Sánchez L., 2003. RNA binding protein sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol. Mol. Biol. Rev. 67: 343–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Su S., Mattox W., 2007. The doublesex splicing enhancer components Tra2 and Rbp1 also repress splicing through an intronic silencer. Mol. Cell. Biol. 27: 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Onsins S., Segarra C., Rozas J., Aguadé M., 1998. Molecular and chromosomal phylogeny in the obscura group of Drosophila inferred from sequences of the rp49 gene region. Mol. Phylogenet. Evol. 9: 33–41. [DOI] [PubMed] [Google Scholar]
- Ruiz M.F., Goday C., González P., Sánchez L., 2003. Molecular analysis and developmental expression of the Sex-lethal gene of Sciara ocellaris (Diptera Order, Nematocera Suborder). Gene Expr. Patt. 3: 341–346. [DOI] [PubMed] [Google Scholar]
- Ruiz M. F., Stefani R. N., Mascarenhas R. O., Perondini A. L. P., Selivon D., et al. , 2005. The gene doublesex of the fruit fly Anastrepha obliqua (Diptera Tephritidae). Genetics 171: 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz M. F., Eirín-López J. M., Stefani R. N., Perondini A. L. P., Selivon D., et al. , 2007. The gene doublesex of Anastrepha fruit flies (Diptera tephritidae) and its evolution in insects. Dev. Genes Evol. 217: 725–731. [DOI] [PubMed] [Google Scholar]
- Saccone G., Salvemini M., Pane A., Polito L. C., 2008. Masculinization of XX Drosophila transgenic flies expressing the Ceratitis capitata Doublesex Misoform. Int. J. Dev. Biol. 52: 1051–1057. [DOI] [PubMed] [Google Scholar]
- Salvemini M., Mauro U., Lombardo F., Milano A., Zazzaro V., et al. , 2011. Genomic organization and splicing evolution of the doublesex gene a Drosophila regulator of sexual differentiation in the dengue and yellow fever mosquito Aedes aegypti. BMC Evol. Biol. 11: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez L., 2008. Sex-determining mechanisms in insects. Int. J. Dev. Biol. 52: 837–856. [DOI] [PubMed] [Google Scholar]
- Sánchez L., 2010. Sciara as an experimental model for studies on the evolutionary relationships between the zygotic, maternal and environmental primary signals for sexual development. J. Genet. 89: 325–331. [DOI] [PubMed] [Google Scholar]
- Scali Ch., Catteruccia F., Li Q., Crisanti A., 2005. Identification of sex-specific transcripts of the Anopheles gambiae doublesex gene. J. Exp. Biol. 208: 3701–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna E., Gorab E., Ruiz M. F., Goday C., Eirín-López J. M., et al. , 2004. The gene Sex-lethal of the Sciaridae family (Order Diptera, Suborder Nematocera) and its phylogeny in dipteran insects. Genetics 168: 907–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman D. C., Frommer A. M., 1998. The Bactrocera tryoni homologue of the Drosophila melanogaster sex-determination gene doublesex. Insect Mol. Biol. 7: 1–12. [DOI] [PubMed] [Google Scholar]
- Shukla J. N., Nagaraju J., 2010. Doublesex: a conserved downstream gene controlled by diverse upstream regulators. J. Genet. 89: 341–356. [DOI] [PubMed] [Google Scholar]
- Shukla J. N., Palli S. R., 2012. Doublesex target genes in the red flour beetle Tribolium castaneum. Sci. Rep. 2: 948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert V., Kuhn S., Traut W., 1997. Expression of the sex determination cascade genes Sex-lethal and doublesex in the phorid fly Megaselia scalaris. Genome 40: 211–214. [DOI] [PubMed] [Google Scholar]
- Sobrinho I. S., Jr, de Brito R. A., 2012. Positive and purifying selection influence the evolution of doublesex in the Anastrepha fraterculus species group. PLoS ONE 7: e33446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. G., Ohbayashi F., Mita K., Shimada T., 2001. The mechanism of sex-specific splicing at the doublesex gene is different between Drosophila melanogaster and Bombyx mori. Insect Biochem. Mol. Biol. 31: 1201–1211. [DOI] [PubMed] [Google Scholar]
- Suzuki M. G., Imanishi S., Dohmae N., Nishimura T., Shimada T., et al. , 2008. Establishment of a novel in vivo sex-specific splicing assay system to identify a trans-acting factor that negatively regulates splicing of Bombyx mori dsx female exons. Mol. Cell. Biol. 28: 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood evolutionary distance and maximum parsimony. Methods Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J., 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76: 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota K., Kato Y., Sato M., Sugiura N., Miyagawa S., et al. , 2013. Molecular cloning of doublesex genes of four cladocera (water flea) species. BMC Genomics 14: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst E. C., van de Zande L., 2015. Double nexus—Doublesex is the connecting element in sex determination. Brief. Funct. Genomics 2015: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst E. C., van de Zande L., Beukeboom L. W., 2010. Insect sex determination: it all evolves around transformer. Curr. Opin. Genet. Dev. 20: 1–8. [DOI] [PubMed] [Google Scholar]
- Waterbury J. A., Jackson L. L., Schedl P., 1999. Analysis of the Doublesex female protein in Drosophila melanogaster: role in sexual differentiation and behavior and dependence on Intersex. Genetics 152: 1653–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.