Abstract
Individuals within a species vary in their responses to a wide range of stimuli, partly as a result of differences in their genetic makeup. Relatively little is known about the genetic and neuronal mechanisms contributing to diversity of behavior in natural populations. By studying intraspecies variation in innate avoidance behavior to thermal stimuli in the nematode Caenorhabditis elegans, we uncovered genetic principles of how different components of a behavioral response can be altered in nature to generate behavioral diversity. Using a thermal pulse assay, we uncovered heritable variation in responses to a transient temperature increase. Quantitative trait locus mapping revealed that separate components of this response were controlled by distinct genomic loci. The loci we identified contributed to variation in components of thermal pulse avoidance behavior in an additive fashion. Our results show that the escape behavior induced by thermal stimuli is composed of simpler behavioral components that are influenced by at least six distinct genetic loci. The loci that decouple components of the escape behavior reveal a genetic system that allows independent modification of behavioral parameters. Our work sets the foundation for future studies of evolution of innate behaviors at the molecular and neuronal level.
Keywords: quantitative genetics, thermosensation, innate behavior, natural variation
INDIVIDUALS vary widely in their behavioral responses to a given stimulus. Differences in environment, the genetic variation between individuals, or both are thought to contribute to variation in behavior. Although significant progress has been made in understanding the genetic basis of behavior using techniques such as inducing mutations in a single genetic background (Wittenburg and Baumeister 1999; Tracey et al. 2003; Suh et al. 2004; Demir and Dickson 2005; Zhang et al. 2005; Donnelly et al. 2013), much less is known about the genetic basis of behavioral variation in natural populations.
One challenge in understanding the genetic basis of behavior is comprehensively capturing the behavior of interest. Genetic studies of behavior generally focus on a single summary phenotypic metric. However, several distinct motor outputs are integrated over time to give rise to a behavioral response to a given stimulus (Dankert et al. 2009; Brown et al. 2012; Ghosh et al. 2012a; Yemini et al. 2013; Yu et al. 2013). With high-resolution phenotyping, behavioral responses to a particular stimulus can be broken down into simpler components. Diversity in the behavioral responses to a given stimulus in natural populations could result from modifications of the behavioral components through natural selection, as postulated by Darwin ∼150 years ago (Darwin 1909). By quantitatively describing behavioral components and their variation within and between species, Tinbergen and others were able to identify ample heritable variation in behavioral components in multiple species (Tinbergen 1972). These studies laid the framework for understanding the origin and divergence of behaviors within and between species. However, until recently, we did not have the tools with which to ask questions about the genetic basis of behavioral components and the mechanisms through which they are integrated to generate behavioral variation.
The nematode Caenorhabditis elegans is the only organism whose complete neuronal connectivity is known. With well-established genetic and genomic tools and the availability of a large densely genotyped collection of natural isolates (Andersen et al. 2012), C. elegans provides a unique opportunity to study the genetic basis of intraspecies variation in behavior. Tools for dissecting the operation of neuronal circuitry in this organism are also well developed (Leifer et al. 2011; Shipley et al. 2014), thus providing the ideal platform for connecting diversity at the genetic and neuronal circuit level to variation in behavior.
Here, we asked how genetic variation within a population contributes to differences in innate behavioral patterns. Specifically, we focused on the variation in innate escape response of C. elegans to a noxious thermal stimulus, in wild isolates and a recombinant inbred line population. Upon experiencing noxious thermal stimuli, C. elegans animals display a stereotypical behavioral sequence that can be readily broken down into several distinct behavioral components (Wittenburg and Baumeister 1999; Ghosh et al. 2012a). By combining well-developed genetic and genomic resources of C. elegans with a thermal pulse assay (Stephens et al. 2008; Mohammadi et al. 2013), we characterized the genetic basis of natural variation in avoidance responses to thermal stimuli.
We used high-content behavioral phenotyping to capture the avoidance behavior of the global C. elegans population in response to thermal stimuli. Instead of considering a single metric summarizing the avoidance response, we simultaneously quantified multiple aspects of the avoidance response of animals exposed to thermal pulse stimuli. Additionally, we used linkage analysis to identify distinct genetic loci underlying different aspects of the escape behavioral pattern. Our results, consistent with observations in other organisms (Greenwood et al. 2013; Weber et al. 2013), illustrate that genetically separate modules allow for individual variation in behavioral components that together result in behavioral diversity within the C. elegans species. Our results provide novel insights into the genetic mechanisms regulating behavior, the operation of the nervous system, and the evolution of avoidance behaviors.
Materials and Methods
Strains
All strains used in this study were maintained under standard nematode culture conditions. Unless mentioned otherwise, all animals assayed were of the hermaphrodite sex. The strains assayed for the calculation of various thermal response behavioral metrics are listed in Supporting Information, File S5.
Behavioral assays
Unless otherwise mentioned, mid- to late-L4 stage worms were picked on 6-cm standard NGM plates with agar ∼24 hr before the assay. Assay plate preparation: 10-cm plates containing 10 ml of agar medium [17 g of agar (Difco, Detroit, MI), 2.7 g of Bactopeptone (Difco), 0.55 g of Tris base (Sigma, St. Louis, MO), 500 µl of 1 M Tris HCl (Sigma), 2.0 g of NaCl (Fisher Scientific, Pittsburgh, PA), and 1 ml of ethanol containing 5 mg/ml cholesterol (Sigma), per liter H2O]. Plates were stored at 4° and used within 2 weeks.
100µl of Escherichea coli (op50) OD ∼0.6–0.8 was spread evenly on the agar plates at least 16 hr prior to the assay. On the day of the assay, single worms were transferred to the agar plates seeded with bacteria and kept at 20° for at least 10 min. The worms were then subjected to thermal stimulus as described below. We computed a given behavioral metric obtained from 30 (on an average) individual worms per strain. The numbers of worms per strain are depicted in File S5.
Thermal stimulus assay
Worms were imaged using a Leica MZ16APO stereomicroscope and a Basler firewire CMOS camera (A602fm, Basler, Ahrensburg, Germany). A collimated beam with a 1/e diameter of 1.50 mm from a 1440-nm diode laser (FOL1404QQM, Fitel, Peachtree City, GA) was positioned to heat the area covering the worm. The diode laser was driven with a commercial power supply and controller (LDC 210B and TED 200C, Thorlabs, Newton, NJ). A custom program written in LabVIEW (National Instruments, Austin TX) was used to control the firing, power, and duration of the IR laser, while simultaneously recording images of the crawling worm for 15 sec at 14 or 28 Hz. Initially, we had obtained the data at 14 frames per second (fps) (only for the laboratory reference strain). We switched to a higher frame rate (28 fps) for the RIAILs, reference strains, and wild isolates to capture the behavior at higher resolution. However, analysis of the 28 fps data did not reveal any significant behavioral features that could not be extracted when images were obtained at a frame rate of 14 fps. Images were processed offline using custom programs written in LabVIEW and MATLAB (Mathworks, Natick, MA) (Ghosh et al. 2012a). A thermal camera (ICI7320, Infrared Camera Inc., Texas) was used to measure the temperature of the agar when heated by the IR laser. We were able to generate thermal pulses with a temperature change (∆T) ranging from 0.4° to 9.1° with ramp rates ranging from 0.8°/sec to 18°/sec, respectively. These ∆T were measured from the baseline temperature (or room temperature), which was typically ∼20°.
Behavioral quantification
Speed profile of each worm over time was generated by a custom written program in MATLAB and LabVIEW that monitored the position of the center of mass (COM) of the worm image (Ghosh et al. 2012a). From the position of COM in the sequence of captured images, the speed-related traits, namely the speed at every time point, acceleration, and deceleration of the worm, were identified using custom code. All analysis was performed separately on the 14 and 28 fps data. Once processing was complete, the 14 fps data were converted from frames to time (in seconds) and combined with the 28 fps data to produce the final results. A smoothing average of five consecutive time points defined the speeds at 0.18, 0.36, ..., 10 sec and was used in the quantitative trait loci (QTL) analysis. The assay was done for 15 sec, but we used the data for up to 10 sec as in several instances (∼30% for the reference strain, N2) the animal left the field of view after ∼12 sec. Some of the traits, namely the reversal duration, number of body bends during reversals, and probability of responding by reversals and omega turns, were quantified manually from the movies of individual worms. A thermal pulse was applied to a forward-moving worm and any movement opposite to the direction of the movement of the worm after application of the thermal pulse was considered a reversal. The number of body bends and omega turns were assigned following previously published criteria (Gray et al. 2005). We scored 66 behavioral traits. Fifty-six of these were centroid speeds at every 0.18 sec for 10 sec. Additionally, we scored maximum speed after the application of stimulus (MaxS), time to reach minimum speed or pause state after stimulus (TMIN), acceleration (Accn), deceleration (Dec), mean reversal speed (MRS), mean reversal duration (MRD), number of body bends during reversals (W), probability of responding by a reversal (pFR) and omega turn (pRO), and the prestimulus speed (SS) as detailed in File S5 (tab: Phenotype _QTL details).
Genome-wide association studies
Association mapping was carried out with 4690 SNPs across the genome of wild isolate strains of C. elegans using EMMA (Kang et al. 2008). The methodology and the genotyping of these strains are described in more detail elsewhere (Andersen et al. 2012).
QTL mapping
We used 138 advanced intercross recombinant inbred lines (RIAILs) made from N2 and CB4856 strains as described elsewhere (Rockman and Kruglyak 2009) for QTL mapping. The genotype data of these RIAILs are as published in Rockman and Kruglyak (2009). QTL mapping was performed using R/QTL (Broman et al. 2003). Linkage of the escape behavior components (measured by procedures described above) to 1454 markers across the genome was determined under a single-QTL model in an interval-mapping framework. Genome-wide logarithm of odds (LOD) thresholds for mapping a given trait was determined using 1000 random permutations of the data for the RIAILs for the trait in question. Support intervals, 1.5 LOD, were used to determine the confidence intervals for each QTL. To account for large-effect QTL, markers corresponding to these were included as covariates and single-QTL analysis performed again. For multiple QTL mapping of normally distributed traits, we looked for any significant epistatic as well as additive interactions with the markers corresponding to the maximum LOD scores in the chromosomes where the QTL were detected for a given trait in R/QTL. For example, for a trait with two QTL, the model formula was y ∼ Q1 + Q2 + Q1:Q2, where Q1 and Q2 represent the QTL objects corresponding to the markers at the maximum LOD scores of the significant QTL peaks. Q1:Q2 is the interaction term between these two QTL objects. We also performed two-dimensional scans as implemented in R/QTL but failed to detect any epistatic interactions. The fits of these models were used to estimate the contribution of the QTL to the total phenotypic variation in the RIAIL population. We partitioned total phenotypic variation into genotypic variation (VG; i.e., among RIAIL or Wild isolate) and error variation (VE), using a random-effects analysis of variance and the broad-sense heritability was estimated for each trait separately for the RIAIL or the wild isolate populations (Bloom et al. 2013).
NIL construction, sequencing, and genotyping
The sources of the nearly Isogenic Lines (NILs) and the methods use to generate them are outlined in File S5.
NIL sequencing
Genomic DNA was prepared using the DNeasy Blood and Tissue Kit (Qiagen). DNA concentration was determined using the Broad-Range Quant-it dsDNA high-sensitivity DNA quantification kit (Invitrogen). DNA was diluted to 1.7 ng/µl. Libraries were prepared by the Nextera tagmentation procedure using the Nextera HMW buffer according to manufacturer’s (Epicentre) protocol. The transposition reaction was performed for 5 min at 55° and the resulting samples were purified using the MinElute kit (Qiagen). Fifteen microliters of the purified fragmented DNA was PCR amplified and barcoded with custom 5-bp sequences using Ex Taq polymerase (Takara) for 10 PCR cycles. Five µl of each PCR-amplified sample was pooled into one 8- or 16-plex library. The pooled library was loaded on a 2%agarose gel, and the 350- to 550-bp region was excised and gel extracted using QIAquick Gel Extraction Kit (Qiagen). Final libraries were diluted to 3.3 ng/µl and sequenced using the paired-end module on a HiSequation 2000 (Illumina). The strains were sequenced at 12–30× coverage.
NIL genotyping
We used custom R and python code to demultiplex and trim the ends of the barcoded sequencing data. Sequencing reads were assigned to each strain based on the 5-bp barcode at the beginning of each read. The internal 19-bp transposon sequence and 10 bp on the right end of each read were removed. Reads were aligned to the WS210 C. elegans reference genome using the Burrows–Wheeler Aligner using default parameters. Next SAMtools/BCFtools was run to generate pileup and merged vcf files of the NILs for the strains using default parameters. Genotype likelihoods for the N2 and CB4856 alleles for each genotypic variant were extracted from the VCF file as follows.
We ran vcftools to extract variant sites known to be different between N2 and CB4856. We extracted the genotype likelihoods for the homozygous reference and alternate calls. Then we used a hidden Markov model and the Baum–Welch algorithm to calculate the posterior likelihood that a variant is coming from N2 or CB4856. Variants with a higher likelihood of coming from N2 relative to coming from CB4856 were coded as N2 and vice versa. We obtained genotype information of 16853 positions on chromosome X as either belonging to N2 or CB4856 and deduced the breakpoints from the resulting data. We also independently identified breakpoints on chromosome X for the NILs using multiplex shotgun genotyping (MSG) with concordant results. For each strain, two million reads were randomly subsampled to improve the speed of the computation pipeline, which requires very low per-base coverage (Andolfatto et al. 2011). Briefly, reads were mapped to both parental genomes and only ancestry informative markers were used to impute ancestry along the chromosome. Because lines vary in their proportions of each parent on the X chromosome, naïve priors were used (prior parent 1, 0.5; prior parent 2, 0.5). The raw output of the MSG pipeline is posterior probabilities for each genotype (parent1, heterozygous, parent2), and information was obtained at 75430 markers that were used to infer the breakpoints.
Fine mapping using nearly isogenic lines
To phenotype escape behavior, we performed ∼30 replicates per nearly isogenic line to determine the mean speed at 0.18 sec (sp0.18) and the probability of responding by a reversal (pFR). Polymorphism in npr-1, a gene encoding a G-protein-coupled receptor, has been shown to contribute to differences in speed on food between the Bristol and Hawaii strains (de Bono and Bargmann 1998). To determine if background of the NILs or the npr-1 locus has any effect on the speed at 0.18 sec and pFR traits, we performed an analysis of variance using the formula trait ∼ background + npr1 + background * npr1 to identify significant contributors for a given trait. The results of this analysis are shown in Table S2 and Table S3. Next, to narrow the interval encompassing the identified QTL on chromosome X we tested for the association between 15 markers on the X chromosome (spanning intervals ∼4405000–∼6000000 bp) and the traits (sp0.18s and pFR) using a linear model taking into account the effect of the genetic background and the distant QTL identified on X (QTL_Xb). The strength of association (−log 10p) resulting from the fit for each marker is depicted in Figure 5. The significance threshold was determined by Bonferroni correction −log(0.05/15).
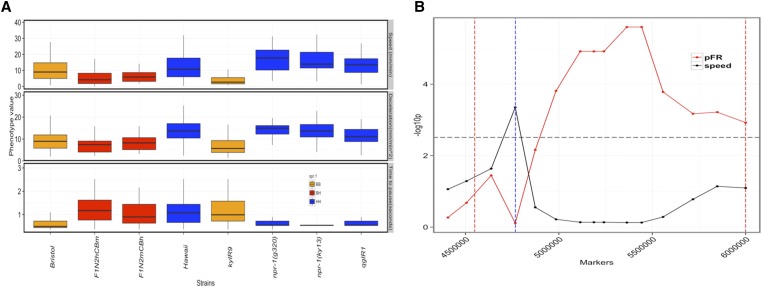
Figure 5.
(A) Distributions of three components of the avoidance behavior for parental, introgression and npr-1 loss-of-function strains. Boxplots of speed at early time points (speed), deceleration (Dec, top), and time-to-pause state (Tmin) for the indicated strains. Bristol and Hawaii are the parental strains. F1N2hCBm: F1 heterozygotes obtained by crossing Bristol hermaphrodites with Hawaiian males. F1N2mCBh: F1 heterozygotes obtained by crossing Bristol males with Hawaiian hermaphrodites. kyIR9 is an introgression line with Bristol like npr-1 in an otherwise Hawaiian background. qgIR1 is an introgression line with Hawaii-like npr-1 in an otherwise Bristol background. The npr-1 status are color coded as follows: orange, Bristol npr-1 allele; blue, Hawaii-like (loss-of-function) npr-1 allele; and BH, F1 heterozygotes from Bristol and Hawaii cross. (B) Speed and probability of responding by reversals are genetically separable. −log10 of P-values (y-axis) resulting from t-test at each marker (x-axis), for the presence of a loci for speed at early time point (speed, black), and probability of responding by reversal (pFR, red) (see Materials and Methods for more details). Black horizontal line: Bonferroni corrected threshold for fourteen markers (black or red dots). Blue vertical dashed line: the position of npr-1. Red vertical dashed lines: the 1.5 LOD drop interval of the proximal QTL on X.
Results
Wild isolates of C. elegans differ in multiple aspects of escape behavior induced by thermal pulse stimulus
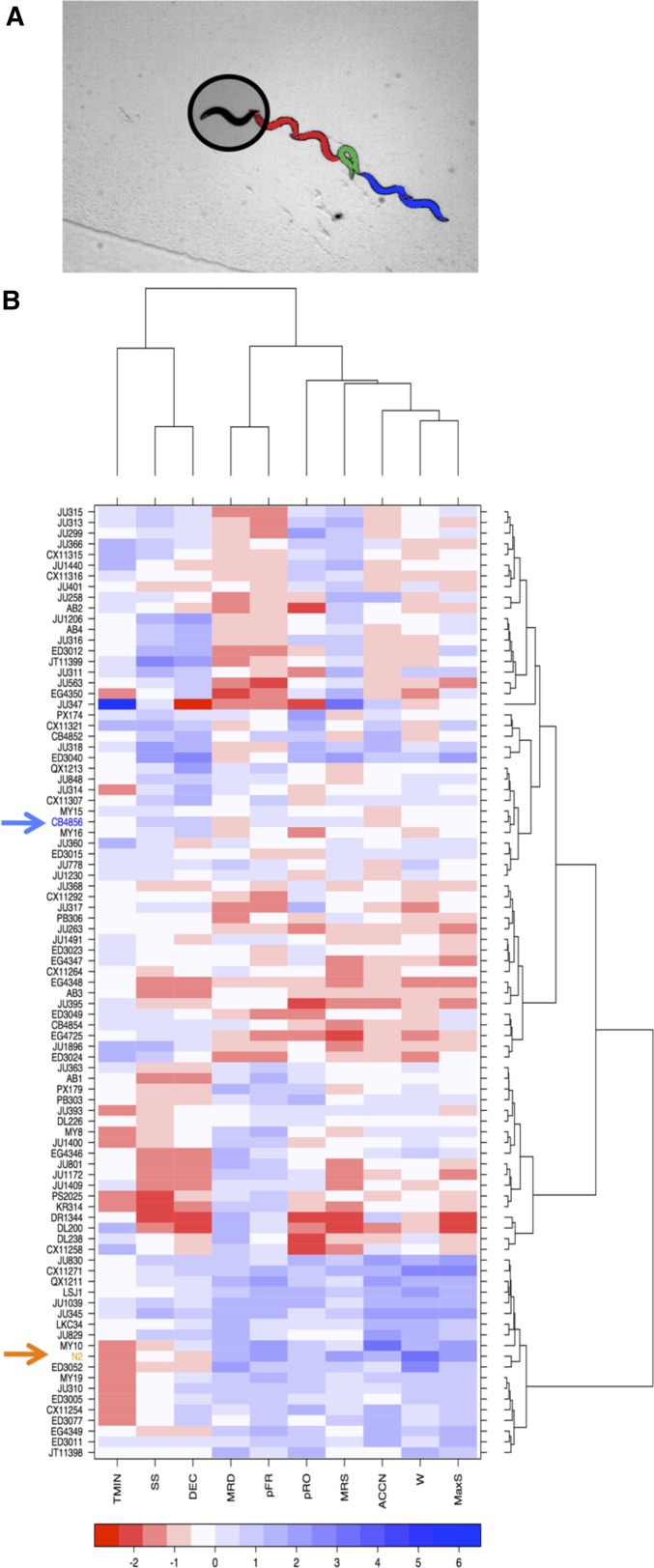
Upon sensation of a noxious thermal stimulus, C. elegans animals typically move backward, turn, and resume forward movement in a new direction (Figure 1A, File S1, File S2, File S3, and File S4). We characterized 66 aspects of the avoidance behavior in response (File S5, Phenotype_QTL details tab) to a thermal pulse that transiently raises the temperature surrounding a forward-moving animal (Ghosh et al. 2012a). Briefly, we recorded the behavior of each animal for 15 sec in response to thermal pulse stimuli of various intensities (Figure 1A) and quantified 66 metrics that characterized the resulting escape behavior (see Materials and Methods) for each thermal pulse stimulus. To investigate the genetic basis of individual variation in response to thermal stimuli, we first surveyed the thermal-pulse-induced behavioral responses of 89 strains of C. elegans isolated from different geographical regions across the globe (Figure 1B) (Andersen et al. 2012). These animals exhibited heritable variation in multiple aspects of their escape behavioral pattern in response to a thermal pulse of 0.4° above the baseline temperature (∆T = 0.4°) (Figure 1B). Depending on the behavioral component analyzed, the broad-sense heritability among the wild-isolate population ranged from 0.04 to 0.21 (Figure S1A). The heritability is in a range consistent with that observed for other behavioral responses to a noxious, stressful stimulus (Mackay et al. 1996; Stirling 2002; Morgan and Mackay 2006). We found no significant correlations among certain escape behavior components in the wild-isolate populations, suggesting a separate genetic basis for these traits (Figure S1B). For example, pRO was not significantly correlated with the speed at an early time point (0.18 sec) in the wild-isolate population. (Figure S1B)
Figure 1.
Variation in components of avoidance behavior among wild isolates of C. elegans in response to thermal pulse. (A) The thermal avoidance assay and response. An infrared laser with a beam width larger than the worm body heats the surrounding area of the animal. The typical responses constitute a reversal (red), an omega turn (green), and forward movement (blue). (B) Profiles of behavioral metrics of 89 wild isolates of C. elegans in response to ∆T ∼ 0.4°. Each behavioral feature is Z-score normalized (See Materials and Methods). TMIN, time to reach pause state; SS, speed at start of the thermal pulse; DEC, deceleration to the pause state; MRD, mean reversal duration; pFR, probability of responding by reversal; pRO, probability of responding by omega; MRS, mean reversal speed; ACCN, acceleration; W, number of body bends during reversals; MaxS, Peak speed after exposure to thermal pulse. In the heat maps, the strains are reordered by the dendrograms resulting from hierarchical clustering. The Bristol strain (N2) and the Hawaiian strain (CB4856) are depicted by arrows and are colored orange and blue, respectively. For simplicity we show only 10 behavioral components. The other 56 behavioral metrics from the centroid speed profiles are not shown.
To determine the genetic architecture underlying diversity in escape behavior in global populations of C. elegans, we conducted genome-wide association mapping of the 66 behavioral metrics that describe the escape response to a thermal stimulus of 0.4° (File S5, tab wild-isolate phenotypes). For the majority of traits, we were unable to detect significant associations. However, loci on chromosomes IV and X contributed to the variation in acceleration, whereas loci on chromosome X contributed to variation in prestimulus speed (Figure S1, C and D), suggesting that these two traits may be genetically separable.
Animals from Hawaii and Bristol differ in their responses to ∆T = 0.4°
Among the strains analyzed for genome-wide association we found that relative to a strain from Hawaii, the laboratory strain (originally from Bristol) differed significantly in multiple aspects of the escape behavioral pattern in response to ∆T = 0.4° (Figure 1B). These two strains exhibited distinct avoidance response behavioral profiles (Figure 1B).
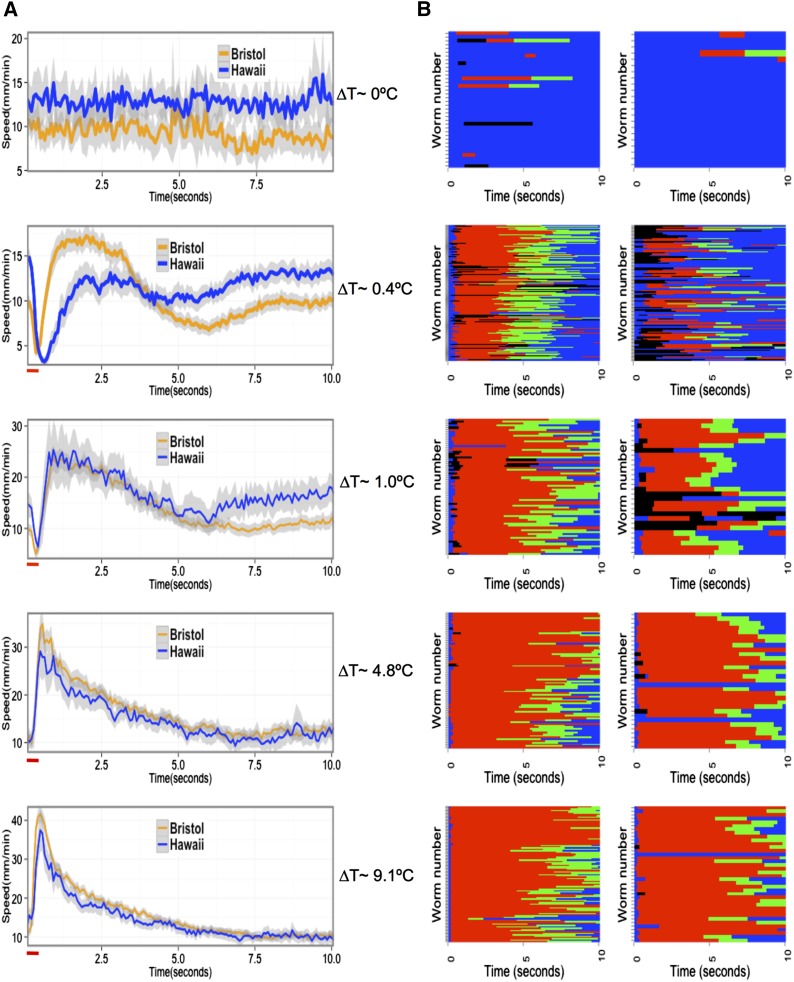
In agreement with previous studies (Stephens et al. 2008), we found that in unstimulated conditions and immediately (<0.2 sec) after exposure to thermal stimuli, Bristol animals move with a significantly lower speed (Welch two sample t-test, P < 0.0001) compared to the Hawaiian animals (Figure 2A, top). Upon experiencing the thermal pulse, both strains decelerate to a pause state and then accelerate away from the stimulus. However, the two strains showed distinct speed profiles during the avoidance response (Figure 2A). Initially (<0.5 sec from application of the thermal pulse), the Hawaiian animals moved forward at a greater speed compared to the Bristol animals, similar to observations under unstimulated conditions (Figure 2A, 2). Subsequently, the Bristol animals moved away from the stimulus significantly faster than the Hawaiian animals. After ∼3 sec, the speed differences between these two strains were similar to the prestimulus state, with the Hawaiian animals once again moving faster (Figure 2A, 2). Bristol and Hawaiian strains also differed in their probability of transition between different behavioral states, as well as in the duration of a given behavioral state (Figure 2B). At stimuli with higher intensities (∆T of 1°, 4.8°, or 9.1°), the differences in avoidance response between these two strains were smaller (Figure 2B). Because the parental strains differed most in their responses elicited by ∆T = 0.4°, we focused our studies of the genetic basis in avoidance behavior at this stimulus intensity.
Figure 2.
Comparison of Hawaii and Bristol strains in responses to thermal stimuli of various intensities. (A) Centroid speed of Bristol (orange) and Hawaii (blue) animals plotted against time for each ∆T. The red horizontal bar indicates duration of the pulse. Temperature increases above the baseline (ΔT) as a result of thermal pulses are indicated. The resulting speed profile corresponds to the indicated ∆T. Shading surrounding the time series represents 95% confidence intervals. (B) Ethogram of different behavioral states of the Bristol (left) and Hawaii (right) animals at indicated ΔT. The behavioral sequence of each animal over the duration of the assay at a given ΔT is shown. Each row represents behavior of a single animal over time. Blue, forward state; red, reversal; black, pause; green, omega turns.
Behavioral differences between Hawaii and Bristol strains are heritable
We characterized 66 components of the escape response (the same as those used for association mapping above) in 138 RIAILs, made from Bristol and Hawaii parent strains (Rockman and Kruglyak 2009). We found substantial phenotypic variation in different behavioral components among the RIAILs. Broad-sense heritability in the RIAIL population ranged from 0.04 to 0.27 for the same set of behavioral metrics measured in the wild isolates. Overall the heritability was higher in the RIAIL population compared to the wild isolates (Figure S1A). To identify loci underlying these traits, we conducted QTL mapping with behavioral metrics extracted from the centroid speed profile (Figure 2A, second from top, see Materials and Methods) and the ethograms (Figure 2B second from top, see Materials and Methods).
Distinct loci contribute to variation in speed profiles
The Bristol and Hawaiian strains differed significantly in their speed profiles in responses to ∆T = 0.4°. We used speed at 0.18-sec increments for 10 sec after the stimulus as a trait in linkage analysis and identified two loci, on chromosomes IV and X, that contributed to variation in the centroid speed profile among the RIAILs (Figure 3A, Figure S2, A–F).
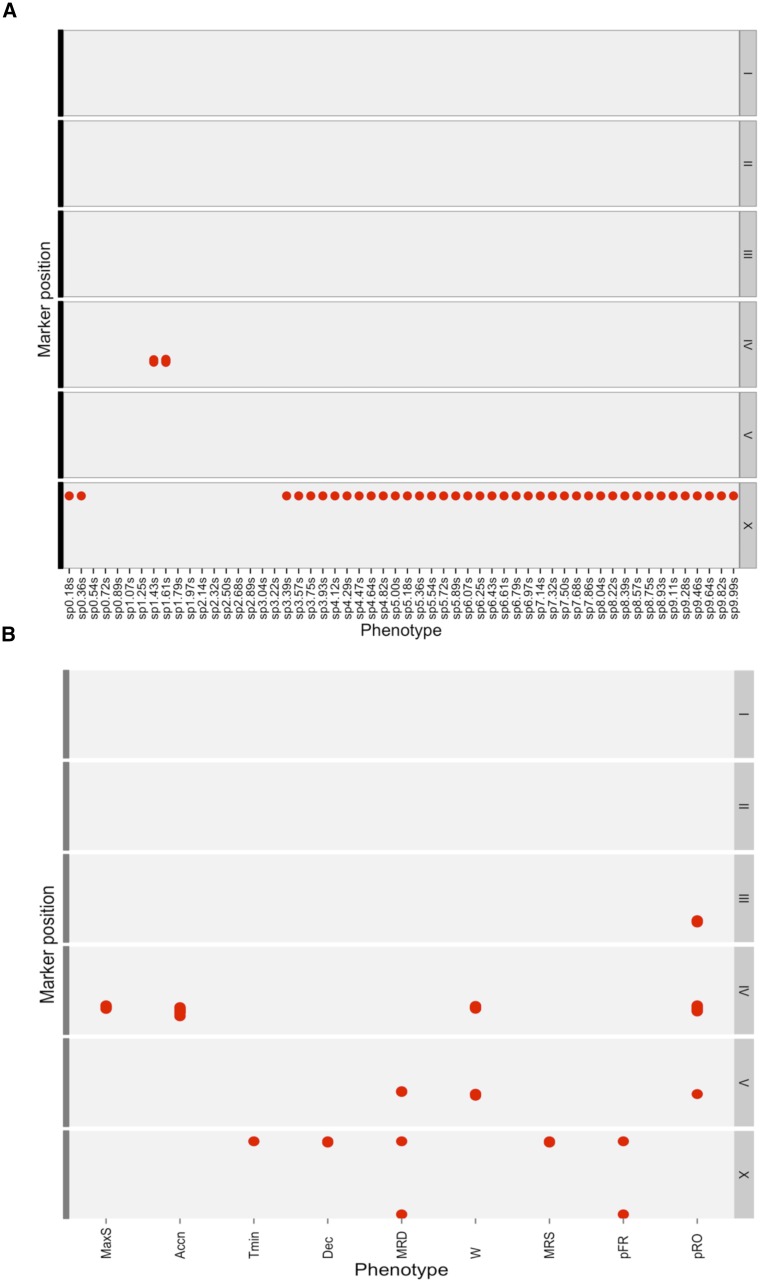
Figure 3.
Genomic intervals contributing to variation in components of escape behavior in 138 RIAILs. (A) The red dot represents position of significant QTL associated with speed at every 0.18 sec (x-axis) at the corresponding marker position plotted on the y-axis. x-axis depicts the traits (the speed profile treated as behavioral metrics) used for linkage mapping. sp*.**s represents speed at *.** sec from start of the assay. y-axis: Marker position for each chromosome. The chromosomes I though X are shown on the left of each facet. (B) The red dots represent position of significant QTL associated with speed at every 0.18 sec (x-axis) at the corresponding marker position plotted on the y-axis (same as A). x-axis depicts the traits used for linkage mapping with 138 RIAILs.
A single significant QTL on chromosome X contributed to variation in speeds at early (<0.5 sec from the thermal pulse) and late (>3.39 sec from the thermal pulse application) time points (Figure 3A, Figure S2, A, C, D, and F). This locus explained ∼47 and 33% of variation in average speeds at 0.18 and 5.0 sec, respectively. The 1.5-LOD support interval of this QTL includes the gene npr-1; polymorphism in this gene has been shown to underlie speed differences between the Bristol and Hawaiian strains. We also identified a QTL on chromosome IV that contributed to variation in speed at 1.43 and 1.61 sec after the thermal pulse (Figure 3A and Figure S2, B and E). This locus explained ∼12% of the variance in speed. We did not detect a significant QTL on chromosome X for speeds at 1.43 and 1.61 sec (Figure 3A, compare Figure S2, E and F), suggesting that the two loci contribute to variation in speed at different phases of the response.
An alternative way to describe the escape behavior is to quantify discrete aspects of the speed profile. Upon exposure to a thermal pulse, forward-moving animals decelerate to a pause state, after which they accelerate away from the stimulus (Figure 2A, 2). Therefore, the centroid speed profile can be divided into an initial deceleration period followed by an acceleration phase. From the speed profiles, we quantified the maximum speed reached after exposure to a thermal pulse, as well as acceleration to maximum speed, defined as the rate of change in the centroid speed from the pause state to the maximum speed. For peak speed and acceleration, we identified a QTL on chromosome IV, at the same location as obtained with speeds at 1.43 and 1.61 sec (Figure 3B, Figure S2, G and H). This locus explained ∼13% of the variation in acceleration and peak speed among the RIAILs. We also quantified the time it takes to reach the pause state as well as deceleration, defined as the rate of change in the centroid speed from the speed at the start of the assay to the pause state. We found that, in contrast to acceleration, overlapping QTL on chromosome X contributed to deceleration and time taken to reach the pause state (Figure 3B, Figure S2, I and J). These loci explained 10.5 and 11.2% of the variation in deceleration and time to minimum speed, respectively, in the RIAIL population. Thus acceleration and deceleration phases during avoidance response behavior are under independent genetic control.
Scanning for additional QTL with markers at the peaks of the two identified QTL used as covariates did not reveal any additional loci. Additionally, a 2D genome-scan failed to find evidence for any significant QTL–QTL interactions. Taken together, these results suggest that polymorphisms at two loci, on chromosomes IV and X, contribute additively to the observed variation in speed during thermal avoidance behavior in the RIAILs.
Distinct loci contribute to variation in different avoidance behavior components
Several aspects of the avoidance behavior could not be fully captured by considering only the centroid of the animal alone. Upon sensation of a thermal stimulus, the posture of an animal changes drastically (Figure 1A) during the reversals and omega turns. We obtained metrics that describe the reversals as well as the transition probabilities among forward, reversal and omega states (see Materials and Methods). We conducted QTL mapping separately for each of these traits and identified unique combinations of five distinct loci contributing to variation in these traits (Figure 3B and Figure S3, A–E).
We detected two distinct loci on chromosome X (QTL_Xa, QTL_Xb) that contributed to variation in the probability of responding by reversal (Figure 3B and Figure S3A) and the average duration of reversals (Figure 3B and Figure S3B). For the average duration of reversals, we detected an additional QTL on chromosome V (QTL_V) (Figure 3B and Figure S3, B) (Figure 3B, Figure S3A, and File S1, tab Phenotype_QTL details).
Two features of the reversal behavior—reversal duration and the number of body bends during reversal—were genetically separable. While the former was significantly influenced by QTL_Xa, QTL_Xb, variation in the latter was attributable to (QTL_IV) (Figure 3B and Figure S3C). QTL_IV contributed to variation in the number of body bends during reversal but not to the duration of reversals, suggesting that the underlying polymorphisms can alter the number of body bends without significantly changing reversal duration. Additionally, we identified QTL_IV, QTL_V, and a QTL on chromosome III as significant contributors to variation in the probability of responding to the thermal stimulus with an omega turn. The QTL on chromosome III was detected only for the probability of responding by omega (Figure 3B, Figure S3D, and File S5, tab Phenotype_QTL details).
We detected QTL at overlapping positions using distinct behavioral measures. For example, QTL_IV, which affected the average number of body bends during reversals and the probability of responding with an omega turn, coincided with the QTL for peak speed and acceleration (Figure 3, A and B). Additionally, we found a significant QTL on chromosome X contributing to mean reversal speed (rate of propagation of body bends during reversal) (Figure 3, A and B, and Figure S3E). This QTL colocalized with the QTL on chromosome X detected for speeds at early and late time points (Figure 3A). Using the peak marker on chromosome X as a covariate for reversal speed, we identified a QTL on chromosome IV at the same position as the QTL for peak speed, acceleration, and speeds at 1.43 and 1.61 sec. This result is not unexpected, as the reversal speed calculated by dividing the number of body bends during reversal by reversal duration includes the acceleration phase of the escape response. We also detected the same QTL_IV for the number of body bends during reversals (Figure 3B). Thus, using distinct behavioral measures, we detected the same loci.
We identified some loci (e.g., QTL_IV) affecting multiple behavioral parameters, whereas others (e.g., QTL_III) affected a single behavioral component of the escape response. Taken together, these data suggest that a modular genetic architecture contributes to diversity in thermal pulse escape response in C. elegans. This suggests that distinct combinations of the detected QTL will result in expected changes in multiple behavioral metrics that together constitute an avoidance behavioral pattern. We next tested this behavioral prediction.
Modular genetic architecture contributes to variation in behavioral responses to a thermal pulse
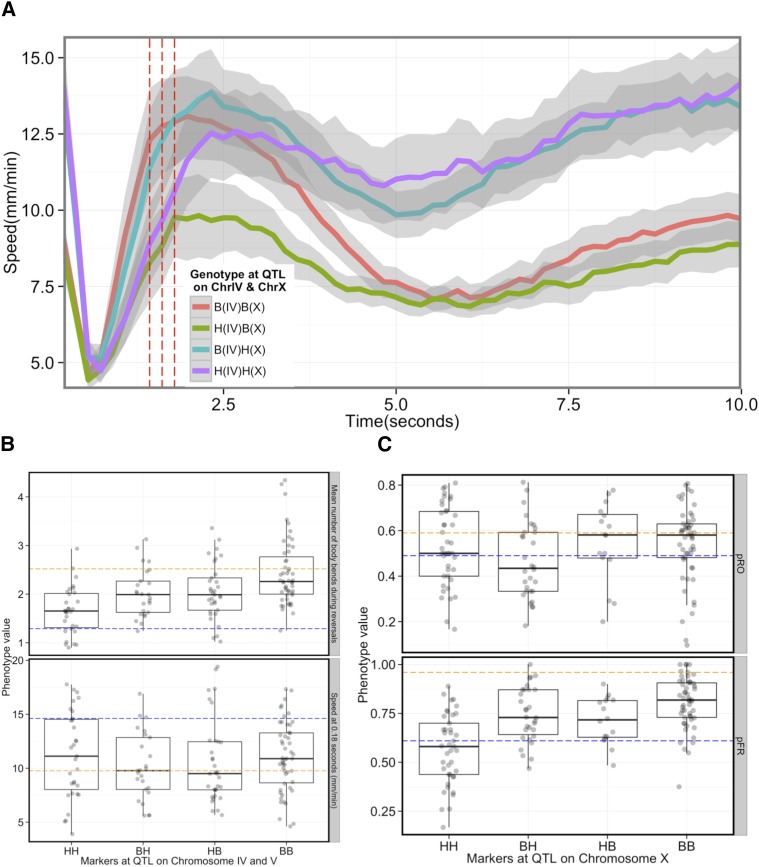
We identified two loci, QTL_IV and QTL_X, contributing to variation in behavioral components quantified from the centroid speed profile (Figure 3A and Figure S3). We next tested whether these two loci were sufficient to generate diversity in speed profiles during avoidance behavior, irrespective of the genetic background. We obtained the average speed profiles of a group of RIAILs that had Bristol alleles at the peak markers at QTL_IV and QTL_X. The speed profile of this group of RIAILs resembled the Bristol parental strain (Figure 4A, red line, compare with Figure 2A, orange line in 2). Similarly, RIAILs with Hawaii alleles at QTL_X and QTL_IV exhibited speed profiles similar to the Hawaii parent (Figure 4A, purple line, compare with blue line in Figure 2A, 2). QTL_IV contributes to variation in speed at intermediate time points, whereas QTL_X contributes at early and late time points. Nonparental combinations of alleles generated centroid profiles expected from a modular genetic architecture for speed. As expected, RIAILs with the Bristol allele of QTL_X and the Hawaii allele of QTL_IV exhibited speed profiles resembling the Bristol parent at early and late time points (Figure 4A, green line) but the Hawaii parent at intermediate time points (Figure 4A, green and purple lines). The reverse was true for RIAILs with the Hawaii allele of QTL_X and the Bristol allele of QTL_IV (Figure 4A, cyan line). Thus the allelic states of QTL_IV and QTL_X define the shape of the centroid speed profile in a predictable manner. These results support the idea that a modular genetic architecture can generate individual variation in behavior, consistent with observations in other organisms (Greenwood et al. 2013; Weber et al. 2013).
Figure 4.
Modular genetic architecture contributes to variation in thermal avoidance behavior. (A) Variation in speed profile is largely explained by two loci. The centroid speed plotted against time (seconds) of four groups of strains in response to ∆T ∼ 0.4°. Strains with Bristol like alleles at the peak marker of the identified QTL on chromosome IV and X are depicted as B (IV) and B (X) whereas strains with Hawaii-like alleles at the same markers are depicted as H(IV) and H(X). The three red dotted lines are the time points, speeds that revealed a QTL on chromosome IV. The shaded area around each line represents the 95% confidence interval. The red horizontal line represents the duration of the laser pulse. (B and C) Additive QTL contributes to variation in escape behavior. (B) Mean number of body bends (top) and speed at 0.18 sec (bottom) in groups of strains with Hawaii (HH) and Bristol (BB) alleles, Hawaii-like allele at peak marker of QTL on chromosome IV and Bristol-like allele at peak marker of QTL on chromosome IV (HB), Bristol-like allele at peak marker of QTL on chromosome IV and Hawaii-like allele at peak marker of QTL on chromosome IV (BH) at peak of detected QTL on chromosomes IV and V for these traits. The blue and orange dotted lines represent the parental Hawaii and Bristol average phenotypes, respectively. (C) Probability of omega turns (top, pRO) and probability of responding by reversals (pFR, bottom) in groups of strains with Hawaii- (HH) and Bristol (BB)-like alleles at peaks of the two QTL detected on chromosome X. BH, Bristol alleles at distal and Hawaii allele at the proximal QTL peak on chromosome X; HB, Hawaii alleles at distal and Bristol allele at the proximal QTL peak on chromosome X. The blue and orange dotted lines represent the parental Hawaii and Bristol average phenotypes, respectively.
QTL contribute additively to variation in escape behavior
We observed no evidence of epistasis for the suite of escape behavior components. The number of body bends during reversals of RIAILs with both Hawaii or both Bristol alleles at QTL_IV and QTL_V resembled that of Hawaii or Bristol parents, respectively (Figure 4B, top). RIAILs with Hawaii allele at QTL_IV and Bristol allele on QTL_V and the alternate allelic combination had intermediate phenotypes. The pattern was absent for speed at 0.18 sec, where we did not detect the QTL on chromosomes IV or V (Figure 4B, bottom).
We detected two additive QTL on chromosome X that contributed to behavioral variation in the probability of responding by reversal, with the proximal QTL (QTL_Xa) explaining ∼18% and the distal QTL (QTL_Xb) explaining ∼7% of the phenotypic variation among the RIAILs. We observed an average probability of reversal of 0.56 (±0.02, N = 41) in RIAILS with Hawaii alleles at these two loci on chromosome X (QTL_Xa and QTL_Xb), whereas the probability of responding by reversals for strains with Bristol alleles at these loci was 0.80 (±0.13, N = 53), similar to the parental values. Strains with Hawaii alleles at QTL_Xa and Bristol alleles at QTL_Xb or the alternate allelic combination responded with an intermediate probability of reversals (Figure 4C, top). The additive pattern was absent for pRO, where we did not detect the QTL, on chromosome X (Figure 4C, top). Additionally, for traits where we detected more than one significant QTL we failed to find any significant effect of interaction through multiple QTL analysis (Table S4, Table S5, Table S6, and Table S7). Taken together, our results suggest that distinct allelic combinations, acting in a largely additive fashion, contribute to variation in escape behavioral metrics.
Fine mapping QTL using nearly isogenic lines
Variation in many avoidance behavior traits was controlled by a locus on chromosome X. This genomic region contributed to variation in speeds at early and late time points, deceleration, time to reach pause state, probability of responding by reversal, and duration of reversals. A single pleiotropic gene or several linked functionally related genes could cause these phenotypic effects. To distinguish between these possibilities, we sought to identify the causative gene(s) at the QTL identified on chromosome X for these traits.
QTL contributing to variation in speed contained npr-1, a gene encoding a G-protein-coupled receptor. A laboratory-derived gain-of-function variant of npr-1 in the Bristol strain background has been previously shown to contribute to variation in several phenotypes (de Bono et al. 2002; Styer et al. 2008; Macosko et al. 2009; McGrath et al. 2009; Reddy et al. 2009; Bendesky et al. 2012; Andersen et al. 2014), including a slower foraging speed relative to the Hawaii strain in the presence of food (de Bono et al. 2002). We found that F1 animals, obtained by reciprocally crossing the Bristol and Hawaii strains (F1N2hCBm and F1N2mCBh in Figure 5A), exhibited speed at the start of the thermal pulse assay resembling Bristol animals, consistent with molecular gain of function in the Bristol strain underlying the observed speed differences (Figure 5A). Consistent with this result and previous reports (de Bono and Bargmann 1998), we found that npr-1 loss of function in an otherwise Bristol background significantly increased the foraging speed of these strains on food [npr-1(g320) and npr-1(ky13) in Figure 5A]. Additionally, nearly isogenic line (NIL) animals with the Bristol npr-1 allele in an otherwise Hawaiian genetic background (kyIR9 in Figure 5A) moved as slowly as the Bristol strain, and NIL animals with Hawaiian npr-1 allele in an otherwise Bristol background moved faster on food, resembling the Hawaiian strain (Figure 5A). Thus npr-1 is likely to be the causative gene underlying the observed speed differences. Using the same set of strains, we also identified npr-1 as the gene contributing to variation in deceleration (Figure 5A), consistent with the strong phenotypic correlation of this trait with speed at early time points (Spearman ρ = 0.9, P < 0.0001).
We detected a significant QTL for mean time to pause state (Tmin) that overlapped with the QTL for speed at early time points and for deceleration. However, unlike with deceleration and speed, we found no evidence for npr-1 contributing to Tmin. Strains with loss-of-function mutations in npr-1 or with the Hawaiian form of npr-1 exhibited Tmin that resembled the Bristol strain, suggesting that this trait is independent of npr-1 (Figure 5A).
Next we focused on fine-mapping QTL that contributed to variation in probability of responding by reversals (pFR). Several lines of evidence suggested that the causative gene(s) underlying pFR is unlikely to be npr-1. Animals with npr-1(lf) mutations or Hawaii-like npr-1 alleles in a Bristol background behaved like the Bristol parent (Table S1). The pFR of a NIL with Hawaii-like npr-1 allele in an otherwise Bristol background was similar to the Bristol parent (Table S1).
To fine map the loci underlying pFR, we generated a panel of 31 X-chromosome introgression lines (NILs) and genotyped them by whole-genome sequencing (Figure S4; see Materials and Methods). We measured pFR and speed at the earliest time point of the assay (speed at 0.18 sec) of these NILs. We examined the effect of npr-1, the genetic background, and QTL_Xb on the variation in speed and pFR in the NIL panel using ANOVA. We found that npr-1 contributed significantly to the variation in speed but not to variation in pFR among these introgression lines (see Materials and Methods, Table S2, and Table S3). Additionally, the genetic background of an introgression line and a distal QTL on chromosome X contributed significantly to variation in pFR but not to speed. Taken together, these data reveal a separate genetic basis for pFR and speed. To further narrow the QTL interval, we next sought to identify markers within the QTL interval that were significant predictors of pFR. We found that markers immediately adjacent to npr-1 were a significant predictor of speed, whereas markers distal to npr-1 significantly contributed to the variation in pFR (Figure 5B). Thus, the gene or genes contributing to variation in pFR are distinct from npr-1 and likely lie within a 750-kb interval distal to npr-1. Taken together, our results suggest that chromosome X harbors distinct QTL in close proximity contributing to variation in different behavioral traits, one of which is the npr-1 gene shaping the variation in foraging speed. Given the significant effects of genetic background and a second distal QTL on chromosome X, further studies with a larger set of introgression lines are needed to localize the causative gene.
Discussion
Most studies investigating the genetic basis of animal behavior have focused on a single genetic background—often the reference laboratory strain. As a result, we have relatively little understanding of the genetic basis of natural variation in behavior. In this study, we sought to identify the genetic mechanisms that generate natural variation in behavioral responses to a thermal stimulus. Our results show that the escape behavior induced by thermal stimuli is composed of simpler behavioral components that are influenced by at least six distinct genetic loci. Our results reveal a genetic system that allows independent modification of behavioral components regulating escape response. Overall, our study reveals the constraints and the flexibility of the genetic mechanisms that generate diversity in C. elegans escape behavior.
We have identified six distinct genetic loci underlying different components of escape behavior and explaining a fraction of the variation that we observe in the RIAIL population (File S5, Table S4, Table S5, Table S6, and Table S7). These genetic loci, acting largely in an additive fashion, contribute to the observed behavioral diversity. For example, we show that it is possible to decouple the genetic mechanisms underlying acceleration and deceleration in response to thermal pulse stimuli. Allelic variation at specific loci can alter some behavioral components but not others, demonstrating that the genetic architecture of the escape response is modular to a large degree. Our findings are similar to those recently published on burrowing patterns in deer mice and schooling behavior in sticklebacks (Greenwood et al. 2013; Weber et al. 2013), providing initial evidence that modular regulation of variation in behavior may be common. These similarities in genetic architecture are striking given the diverse organisms and behaviors studied. Similar genome-wide studies of integration of characters at the genomic level have been conducted for morphological traits (Klingenberg et al. 2004; Miller et al. 2014).
Although we provide strong evidence that distinct genetic loci underlie different components of escape behavior in our RIAIL population, we were less successful in identifying such a set of loci through genome-wide association studies in wild isolates of C. elegans. This could be attributable to a lack of power in the association studies with the relatively small number of available isolates and/or to rare variants contributing to variation in thermal-stimulus-induced escape behavior. Further genetic mapping studies with pairs of wild isolates will elucidate the genetic underpinnings of natural diversity in avoidance behavior and provide novel insights into the genetic basis of a complex behavior.
We further investigated the genetic architecture of escape behavior by focusing on two strains, a wild Hawaii-derived strain, and the commonly used laboratory N2 strain. The RIAIL population we used was constructed from a Hawaiian wild isolate and the laboratory strain N2. It has become increasingly clear that laboratory-derived polymorphisms in a G-protein-coupled receptor gene, npr-1, have large effects on the life history and behavior of N2 (McGrath et al. 2009; Reddy et al. 2009; Bendesky et al. 2011; Andersen et al. 2014). npr-1 has also been implicated in high-temperature (∆T > 9°) avoidance behavior in C. elegans (Glauser et al. 2011). Consistent with previous reports, we observed that npr-1 variation underlies speed differences between the Hawaii and Bristol strains. In addition, npr-1 also contributed to variation in deceleration upon exposure to a thermal pulse stimulus. Studying wild isolates beyond the laboratory strain will provide novel insights masked by the effects of npr-1. This is highlighted by our finding of a locus on chromosome X, distal to the npr-1 locus that contributes to variation in un-stimulated speed in the wild isolates.
Although the QTL associated with time-to-pause state and probability of responding by reversal overlaps with npr-1, our analysis suggests that these are npr-1-independent traits. We found little correlation between the npr-1 genotypes and these traits. Another candidate for decision-making behavior on the X chromosome is the tyramine-gated G-protein-coupled receptor tyra-3. Polymorphisms in tyra-3 have been shown to contribute to variation in decision making between Bristol and Hawaii strains in foraging (Bendesky et al. 2011). However, this gene is not located in the fine-mapped interval and is thus unlikely to regulate escape behavior.
We also detected a QTL on chromosome V contributing to reversal duration and frequency of body bends during reversals. This locus coincides with the location of glc-1, a glutamate-gated chloride channel family gene. Loss-of-function mutations in this gene have been shown to impair reversals and also to cause resistance to the anthelmintic abamectin (Ghosh et al. 2012b). Consistent with this, we found that reversal duration and frequency of body bends during reversals were significantly correlated with abamectin resistance in the RIAILs (R. Ghosh and L. Kruglyak, unpublished results). This implies that glc-1 may integrate reversal duration, number of body bends during reversal state, and probability of omega turns. Different allelic forms of glc-1 may result in differential execution of avoidance behavior in response to thermal stimuli.
Our results from RIAILs provide new insights into the genetic mechanisms of escape behavior in C. elegans. We found that QTL on chromosome V and X contribute to the duration of reversals (‘timer’), whereas QTL on chromosomes IV and V contribute to variation in the number of body bends during reversals (‘counter’). Hence, changing the causative allele(s) on chromosome X or IV can independently modify the timer or the counter during reversals. Changing the causative allele at the QTL on chromosome V will affect both aspects of the reversal and hence may act as a constraint. We also provide evidence that the probability of responding by reversals and the probability of making an omega turn are influenced by unlinked loci, suggesting that there are independent neuronal routes to altering each of these behavioral components. Ultimately, differences in escape behavior at the genetic level will be reflected at the level of neuronal circuitry and/or neuroanatomy. The present study lays the groundwork for future evolutionary, genetic, and neuronal circuitry studies to relate the genetic differences to individual variations in escape behavior.
Supplementary Material
Acknowledgments
The authors are grateful to Frank Albert, Erik Andersen, Alejandro Burga, Meru Sadhu, Sebastian Treusch, Xin Wang and Danny Zeevi for helpful discussions and comments on the manuscript. We also thank Andres Bendesky and Matthew Rockman along with the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources (NCRR), for some nematode strains used in this work. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was funded by a James S. McDonnell Foundation Centennial Fellowship, Howard Hughes Medical Institute, and NIH grant R01-HG004321 (LK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Communicating editor: S. F. Chenoweth
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.178491/-/DC1.
Literature Cited
- Andersen E. C., Gerke J. P., Shapiro J. A., Crissman J. R., Ghosh R., et al. , 2012. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 44: 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E. C., Bloom J. S., Gerke J. P., Kruglyak L., 2014. A variant in the neuropeptide receptor npr-1 is a major determinant of Caenorhabditis elegans growth and physiology. PLoS Genet. 10: e1004156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., Davison D., Erezyilmaz D., Hu T. T., Mast J., et al. , 2011. Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Res. 21: 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendesky A., Tsunozaki M., Rockman M. V., Kruglyak L., Bargmann C. I., 2011. Catecholamine receptor polymorphisms affect decision-making in C. elegans. Nature 472: 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendesky A., Pitts J., Rockman M. V., Chen W. C., Tan M. W., et al. , 2012. Long-range regulatory polymorphisms affecting a GABA receptor constitute a quantitative trait locus (QTL) for social behavior in Caenorhabditis elegans. PLoS Genet. 8: e1003157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. S., Ehrenreich I. M., Loo W. T., Lite T. L., Kruglyak L., 2013. Finding the sources of missing heritability in a yeast cross. Nature 494: 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Brown A. E., Yemini E. I., Grundy L. J., Jucikas T., Schafer W. R., 2012. A dictionary of behavioral motifs reveals clusters of genes affecting Caenorhabditis elegans locomotion. Proc. Natl. Acad. Sci. USA 110: 791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankert H., Wang L., Hoopfer E. D., Anderson D. J., Perona P., 2009. Automated monitoring and analysis of social behavior in Drosophila. Nat. Methods 6: 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin, C. R., 1909 The Origin of Species, Vol. XI, Chap. 8. Collier, New York. [Google Scholar]
- de Bono M., Bargmann C. I., 1998. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94: 679–689. [DOI] [PubMed] [Google Scholar]
- de Bono M., Tobin D. M., Davis M. W., Avery L., Bargmann C. I., 2002. Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature 419: 899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir E., Dickson B. J., 2005. fruitless splicing specifies male courtship behavior in Drosophila. Cell 121: 785–794. [DOI] [PubMed] [Google Scholar]
- Donnelly J. L., Clark C. M., Leifer A. M., Pirri J. K., Haburcak M., et al. , 2013. Monoaminergic orchestration of motor programs in a complex C. elegans behavior. PLoS Biol. 11: e1001529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R., Mohammadi A., Kruglyak L., Ryu W. S., 2012a Multiparameter behavioral profiling reveals distinct thermal response regimes in Caenorhabditis elegans. BMC Biol. 10: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R., Andersen E. C., Shapiro J. A., Gerke J. P., Kruglyak L., 2012b Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans. Science 335: 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauser D. A., Chen W. C., Agin R., Macinnis B. L., Hellman A. B., et al. , 2011. Heat avoidance is regulated by transient receptor potential (TRP) channels and a neuropeptide signaling pathway in Caenorhabditis elegans. Genetics 188: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. M., Hill J. J., Bargmann C. I., 2005. A circuit for navigation in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 102: 3184–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood A. K., Wark A. R., Yoshida K., Peichel C. L., 2013. Genetic and neural modularity underlie the evolution of schooling behavior in threespine sticklebacks. Curr. Biol. 23: 1884–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg C. P., Leamy L. J., Cheverud J. M., 2004. Integration and modularity of quantitative trait locus effects on geometric shape in the mouse mandible. Genetics 166: 1909–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifer A. M., Fang-Yen C., Gershow M., Alkema M. J., Samuel A. D., 2011. Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans. Nat. Methods 8: 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., Hackett J. B., Lyman R. F., Wayne M. L., Anholt R. R., 1996. Quantitative genetic variation of odor-guided behavior in a natural population of Drosophila melanogaster. Genetics 144: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E. Z., Pokala N., Feinberg E. H., Chalasani S. H., Butcher R. A., et al. , 2009. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature 458: 1171–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath P. T., Rockman M. V., Zimmer M., Jang H., Macosko E. Z., et al. , 2009. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron 61: 692–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. T., Glazer A. M., Summers B. R., Blackman B. K., Norman A. R., et al. , 2014. Modular skeletal evolution in sticklebacks is controlled by additive and clustered quantitative trait Loci. Genetics 197: 405–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi A., Byrne Rodgers J., Kotera I., Ryu W. S., 2013. Behavioral response of Caenorhabditis elegans to localized thermal stimuli. BMC Neurosci. 14: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T. J., Mackay T. F., 2006. Quantitative trait loci for thermotolerance phenotypes in Drosophila melanogaster. Heredity 96: 232–242. [DOI] [PubMed] [Google Scholar]
- Reddy K. C., Andersen E. C., Kruglyak L., Kim D. H., 2009. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science 323: 382–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Kruglyak L., 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 5: e1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipley F. B., Clark C. M., Alkema M. J., Leifer A. M., 2014. Simultaneous optogenetic manipulation and calcium imaging in freely moving C. elegans. Front. Neural Circuits 8: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens G. J., Johnson-Kerner B., Bialek W., Ryu W. S., 2008. Dimensionality and dynamics in the behavior of C. elegans. PLOS Comput. Biol. 4: e1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling D. G., Reale D., Roff D. A., 2002. Selection, structure and the heritability of behaviour. J. Evol. Biol. 15: 277–289. [Google Scholar]
- Styer K. L., Singh V., Macosko E., Steele S. E., Bargmann C. I., et al. , 2008. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322: 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh G. S., Wong A. M., Hergarden A. C., Wang J. W., Simon A. F., et al. , 2004. A single population of olfactory sensory neurons mediates an innate avoidance behaviour in Drosophila. Nature 431: 854–859. [DOI] [PubMed] [Google Scholar]
- Tinbergen N., 1972. The Animal in Its World; Explorations of an Ethologist, 1932–1972. Harvard University Press, Cambridge, MA. [Google Scholar]
- Tracey W. D., Jr, Wilson R. I., Laurent G., Benzer S., 2003. painless, a Drosophila gene essential for nociception. Cell 113: 261–273. [DOI] [PubMed] [Google Scholar]
- Weber J. N., Peterson B. K., Hoekstra H. E., 2013. Discrete genetic modules are responsible for complex burrow evolution in Peromyscus mice. Nature 493: 402–405. [DOI] [PubMed] [Google Scholar]
- Wittenburg N., Baumeister R., 1999. Thermal avoidance in Caenorhabditis elegans: an approach to the study of nociception. Proc. Natl. Acad. Sci. USA 96: 10477–10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yemini E., Jucikas T., Grundy L. J., Brown A. E., Schafer W. R., 2013. A database of Caenorhabditis elegans behavioral phenotypes. Nat. Methods 10: 877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Aleman-Meza B., Gharib S., Labocha M. K., Cronin C. J., et al. , 2013. Systematic profiling of Caenorhabditis elegans locomotive behaviors reveals additional components in G-protein Galphaq signaling. Proc. Natl. Acad. Sci. USA 110: 11940–11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lu H., Bargmann C. I., 2005. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438: 179–184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.