Abstract
Eukaryotic DNA replication terminates when replisomes from adjacent replication origins converge. Termination involves local completion of DNA synthesis, decatenation of daughter molecules, and replisome disassembly. Termination has been difficult to study because termination events are generally asynchronous and sequence non-specific. To overcome these challenges, we paused converging replisomes with a site-specific barrier in Xenopus egg extracts. Upon removal of the barrier, forks underwent synchronous and site-specific termination, allowing mechanistic dissection of this process. We show that DNA synthesis does not slow detectably as forks approach each other and that leading strands pass each other unhindered before undergoing ligation to downstream lagging strands. Dissociation of CMG helicases occurs only after the final ligation step, and is not required for completion of DNA synthesis, strongly suggesting that converging CMGs pass one another and dissociate from double-stranded DNA. This termination mechanism allows rapid completion of DNA synthesis while avoiding premature replisome disassembly
DNA replication occurs in three broad stages: initiation, elongation, and termination. Termination occurs when converging replication forks meet and involves at least four processes, not necessarily in the following order. First, the last stretch of parental DNA between forks is unwound (“dissolution”) and replisomes come into contact; second, any remaining gaps in the daughter strands are filled in and nascent strands are ligated (“ligation”); third, double-stranded DNA intertwinings (i.e. catenanes) are removed (“decatenation”); fourth, the replisome is disassembled. Despite decades of research on termination1, we know little about the order, mechanism, and regulation of the above events, especially during eukaryotic chromosomal replication.
Termination has been most extensively studied in the mammalian DNA tumor virus SV402, where converging replication forks stall during termination1,3,4. Dissolution during SV40 replication requires rotation of the entire fork to produce catenations behind the fork (pre-catenanes)5,6, which are resolved by Topo II6, probably in a manner similar to how Topo IV functions during bacterial termination7,8. The SV40 replicative helicase, large T antigen (T-ag), dissociates from chromatin prior to dissolution, but whether this is required for the completion of replication is unknown9,10. After dissolution, daughter strands retain gaps of ∼60 nucleotides11, which are ultimately filled in by an unknown mechanism in parallel to decatenation12.
Eukaryotic termination has also been investigated. Although convergent forks accumulate at certain replication pause sites in yeast cells lacking 5’-3’ DNA helicases13–15, it is unknown whether forks stall during unperturbed termination. Furthermore, Topo II is not required for dissolution in budding yeast16,17 or during vertebrate termination18,19. Recent work shows that late in S phase, the eukaryotic replicative helicase CMG (CDC45, MCM2–7, GINS)20–23 is removed from chromatin by the ATPase p97 following ubiquitylation of MCM7 (by SCFDia2 in yeast)24,25. While one study implies that DNA replication can go to completion in the absence of CMG unloading24, another reports that tracts of unreplicated DNA remain in the absence of this process25. Given that mis-regulation of bacterial termination can readily trigger re-replication of DNA26,27, a potent driver of genomic instability in mammalian cells28, a better understanding of eukaryotic termination is essential.
Owing to stochastic origin firing29,30 and variable rates of replisome progression31,32, the location and timing of eukaryotic termination is variable30,33, making this process difficult to study. Here, we report that Xenopus egg extracts can be used to induce synchronous and localized termination events. This approach has allowed us to identify and order key events underlying vertebrate termination.
A system to study replication termination
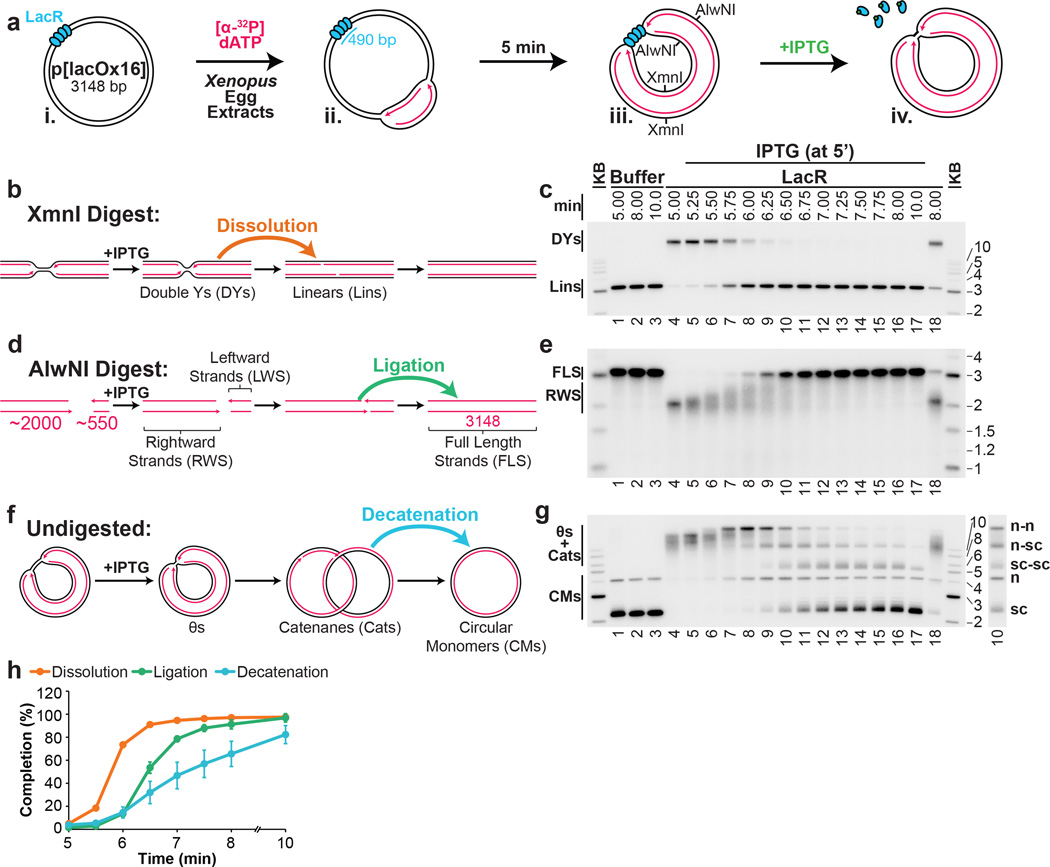
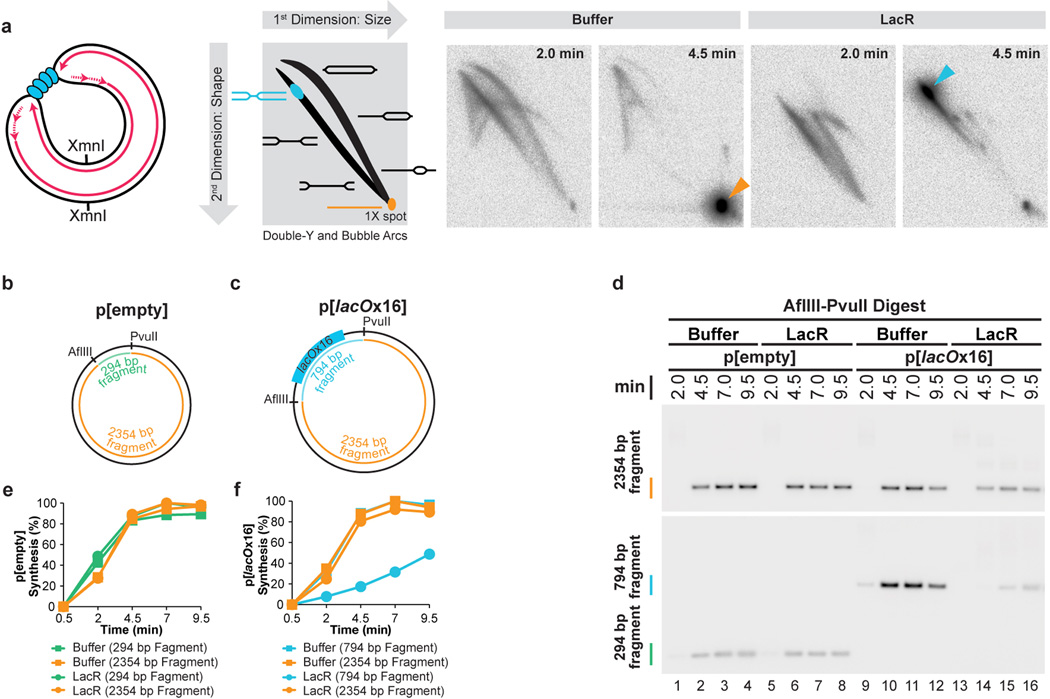
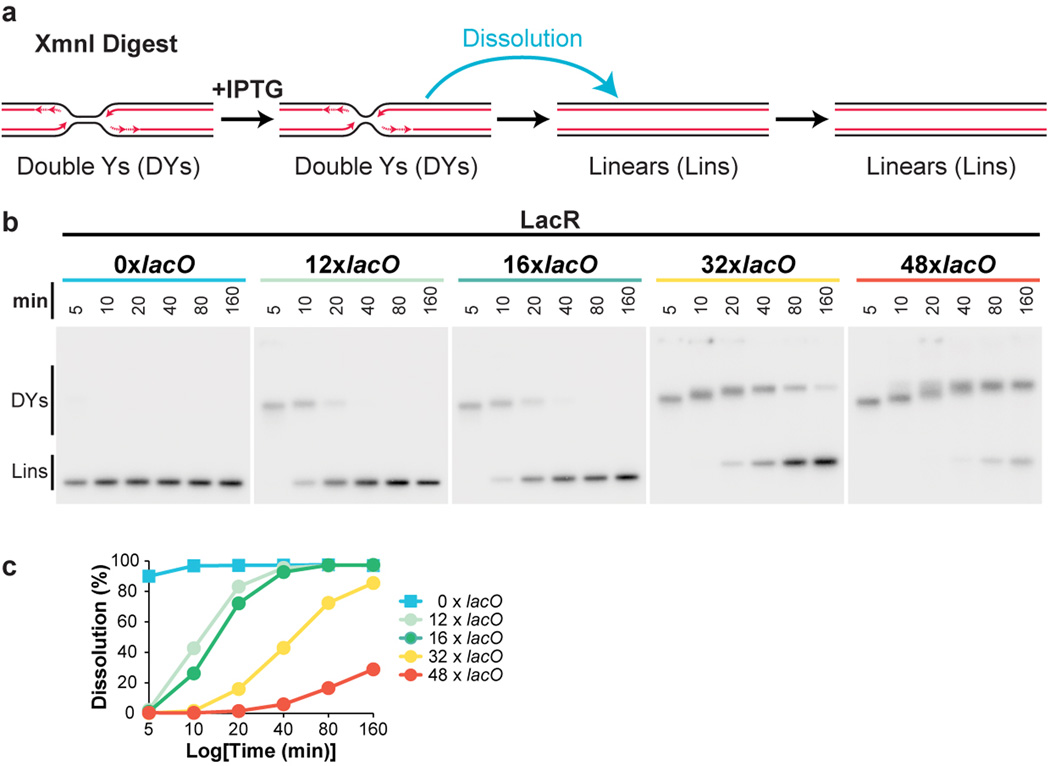
Our strategy was to stall forks on either side of a reversible replication fork barrier (Fig. 1A-i-iii), and subsequently disassemble the barrier to trigger localized and synchronous termination events (Fig. 1A–iv). The barrier we employed consisted of an array of lac repressors (LacRs) bound to lac operators (lacOs)34,35, which can be disrupted by IPTG. We constructed p[lacOx16], which contains 16 tandem copies of lacO (490 basepairs). p[lacOx16] was incubated in nucleus-free Xenopus egg extract, which promotes sequence-non-specific replication initiation on added DNA molecules, followed by a single, complete round of DNA synthesis via a mechanism that appears to reflect events in cells36. To monitor replication, radioactive [α-32P]dATP was included in the reaction. When p[lacOx16] was replicated in the absence of LacR for ∼5 minutes and then cut with XmnI (Fig. 1Aiii), a single linear species representing fully replicated daughter molecules was observed (Fig. 1C, lane 1). In contrast, in the presence of LacR, a slow-mobility product appeared (Fig. 1C, lane 4) that corresponds to a double-Y structure, as shown by 2-D gel electrophoresis (Extended Data Fig. 1A). To confirm that the double-Y resulted from fork stalling at the outer edges of the array, we separately monitored replication in the plasmid backbone and in the lacO array. In the presence of LacR, synthesis of the array was specifically delayed (Extended Data Figure 1F). In contrast, LacR had no effect on replication of a plasmid lacking lacO sites (Extended Data Fig. 1E). These results indicate that replication forks stalled on both sides of the LacR array, consistent with previous findings34,35,37.
Figure 1. A model system to study replication termination.
A. Scheme to induce site-specific termination. Key restriction sites are highlighted.
B. Schematic of the dissolution assay.
C. p[lacOx16] was incubated in buffer or LacR, then replicated in the presence of [α-32P]dATP, before termination was induced by the addition of IPTG. To measure dissolution, radiolabelled termination intermediates were cut with XmnI, separated on a native agarose gel, and analyzed by autoradiography.
D. Schematic of the ligation assay.
E. To measure ligation, replication intermediates were cut with AlwNI and separated on a denaturing agarose gel.
F. Schematic of the decatenation assay
G. To measure decatenation, replication intermediates were separated on a native agarose gel. The additional copy of lane 10 highlights catenated termination intermediates.
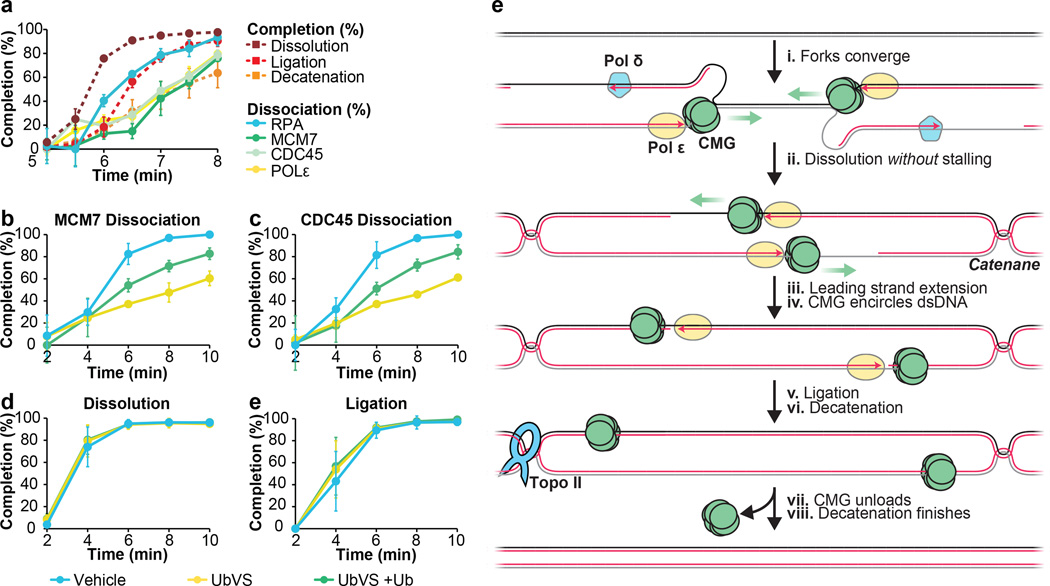
H. Multiple dissolution, ligation, and decatenation assays were quantified. Means±standard deviation (s.d.) are plotted (n=4).
We next addressed whether replication forks stalled by LacR could restart. When IPTG was added to double-Y structures 5 minutes after replication initiation, 90% were converted to unit-sized linear plasmid molecules within a further 1.5 minutes (Fig, 1C, lanes 5–10 and Fig. 1H, yellow circles). In the absence of IPTG, only 21% of double-Y molecules disappeared after 3 minutes (Fig. 1C, lane 18). The conversion of double-Ys to linear species occurs when any remaining parental DNA holding daughter molecules together is unwound (Fig. 1B). This process, which we refer to as “dissolution,” represents a convenient means to measure when converging replisomes meet. Notably, the ATR-Chk1 pathway was not activated above background levels during this procedure (Data not shown).
After dissolution, nascent strands should undergo ligation. To detect the growth and ligation of nascent strands, we digested p[lacOx16] with AlwNI, which cuts the plasmid once, ∼550 nts from the rightward edge of the array and ∼2000 nts from its leftward edge (Fig. 1Aiii and 1D), and we analyzed the products on a denaturing gel. Before IPTG addition, discrete species of ∼2000 nts (Fig. 1E, Lane 4) and ∼550 nts (Extended Data Fig 2A, lane 4) were observed. Upon IPTG addition, both bands grew heterogeneously (Fig. 1E and Extended Data Fig 2A). Since all leading strands were immediately extended upon IPTG addition (Extended Data Fig 2B–C), we infer that the heterogeneity observed resulted because growth of the lagging strand was delayed until ligation of an additional Okazaki fragment. Finally, the nascent strands increased abruptly to the full length of 3100 nts as ligation to downstream lagging strands occurred (Fig. 1E, Lanes 9–13). As expected, dissolution preceded ligation, and there was a ∼45s delay between these two events (Fig. 1H).
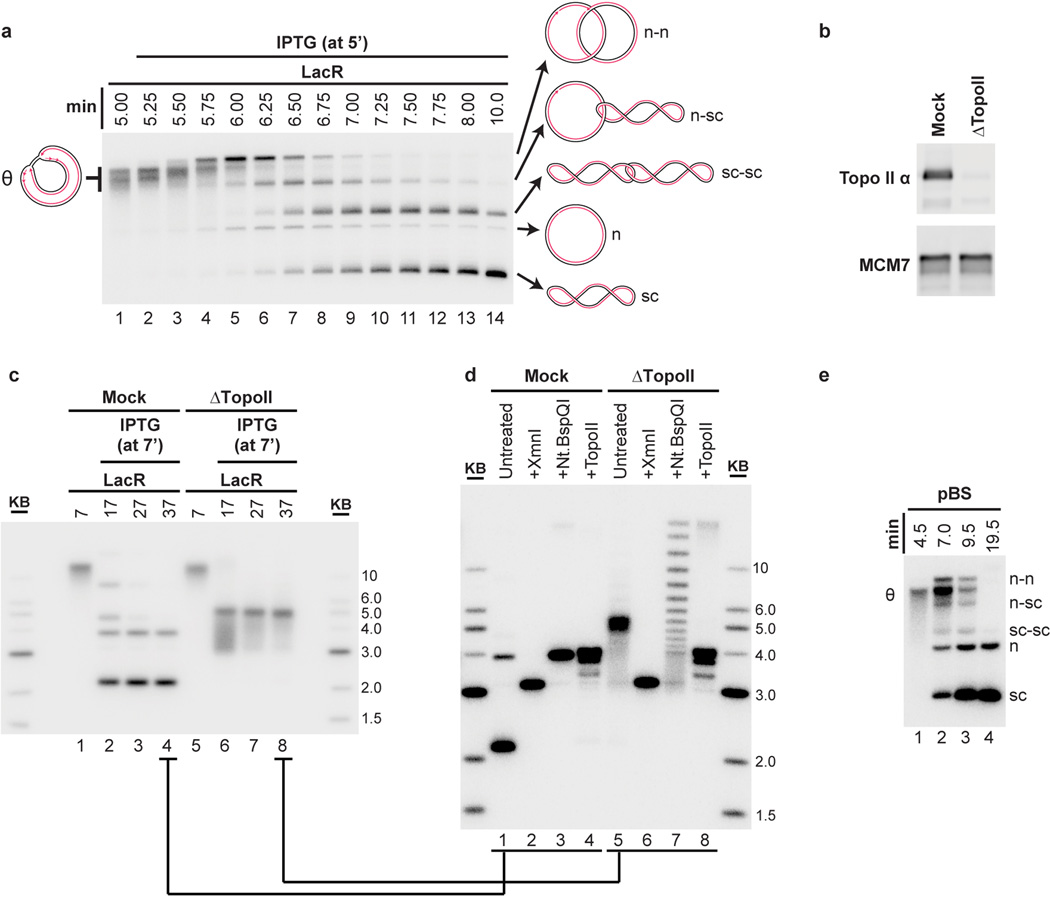
Another important event associated with termination is decatenation of daughter molecules18. To measure this process, we analyzed undigested replication products on native agarose gels (Fig. 1F–G). Before addition of IPTG, when the array had not yet been duplicated, replication products migrated as a compact smear of high molecular weight, θ structures (Fig. 1F and Fig. 1G, lane 4). Upon addition of IPTG, most θs were lost within one minute, and they were successively converted into three types of dimeric catenanes described previously5,18,38: nicked-nicked (n–n), nicked-supercoiled (n-sc), and supercoiled-supercoiled (sc-sc) (Fig. 1G and Extended Data Fig. 3A). n-n catenanes appeared first (Fig. 1G, Lanes 7–8), followed by n-sc (lanes 8–10), and sc-sc (Fig. 1G Lanes 9–12). Supercoiling is the result of nucleosome assembly on closed circular DNA39. Finally, monomeric, supercoiled daughter molecules accumulated (sc, Fig. 1G, Lane 17) dependent on Topoisomerase II (Extended Data Fig 3B–D) as seen in vivo16,17. Topo II was not required for dissolution or ligation (Extended Data Fig 3C–D) suggesting these processes proceed independently of decatenation16,17,19. Like ligation, decatenation began ∼40s after dissolution, but progressed at a slower rate than ligation (Fig. 1H>). The same intermediates were detected in the absence of LacR, but their order of appearance was not well-defined (Extended Data Fig. 3E).
Our results demonstrate that a reversible replication fork barrier allows induction of a synchronous and spatially defined termination event. They also show that soon after forks meet, as measured by dissolution, daughter molecules are quickly ligated and decatenated.
Converging replication forks do not stall
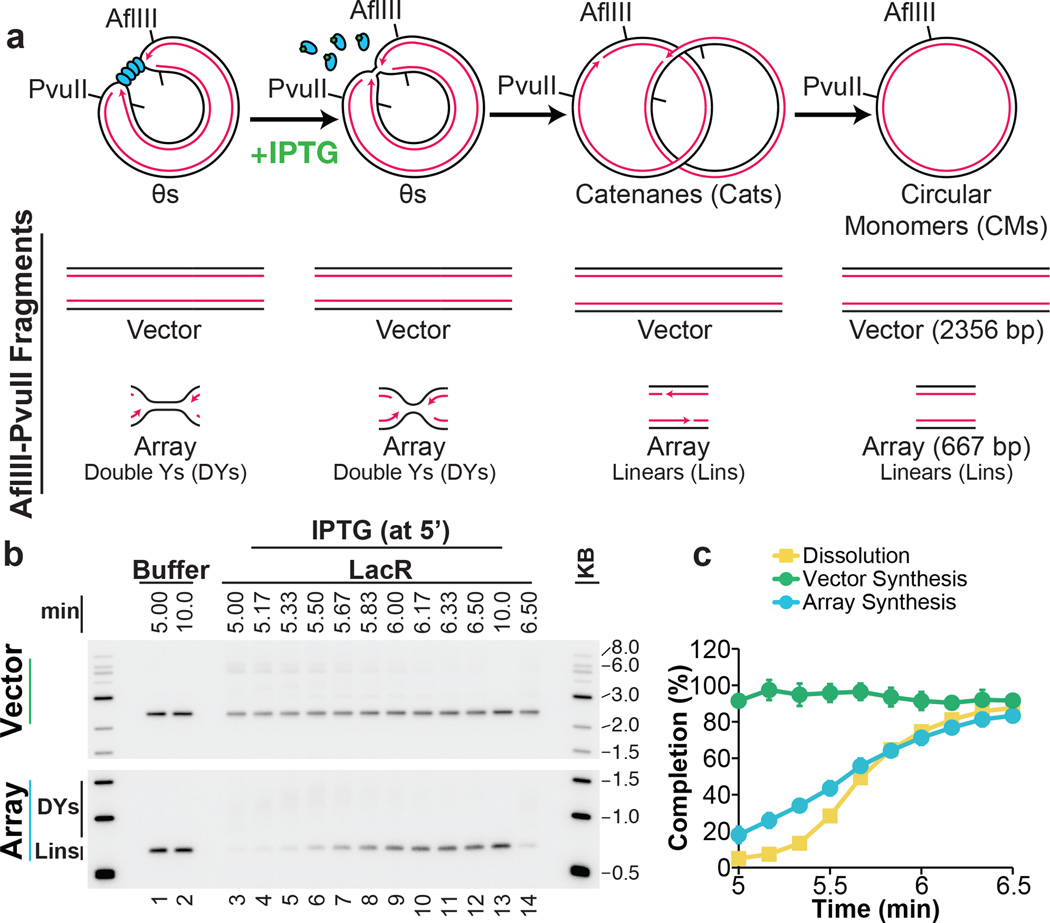
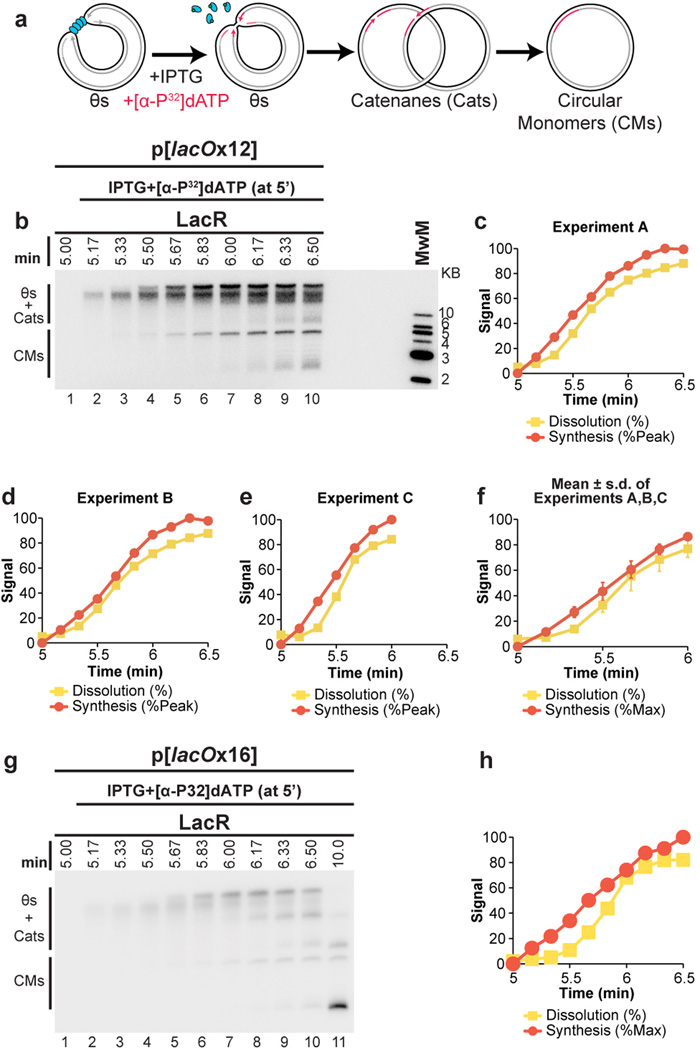
To test the proposal that replication forks slow down or stall during termination1,3,4, we quantified the rate of DNA synthesis as two replisomes converged within the lacO array. To minimize the loss of synchrony among replisomes after IPTG addition, we used a 365 bp array containing only 12 copies of lacO, which was sufficient to prevent dissolution at the 5 minute time point (Extended Data Fig. 4C). We replicated p[lacOx12] in the presence of LacR, added IPTG after 5 minutes, and examined subsequent replication within the array by cutting the plasmid with AflIII and PvuII (Fig. 2A). The rate of DNA synthesis within the array was almost perfectly linear after IPTG addition (Fig. 2B–C) even as dissolution was underway. These data suggest that converging forks do not slow significantly before they meet. A similar conclusion was reached when radiolabelled nucleotides were added at the same time as IPTG and incorporation measured only during the final stage of replication on p[lacOx12] (Extended Data Fig. 5A–F) or p[lacOx16] (Extended Data Fig 5G–H). Moreover, fork rates within the lacO array resembled those previously reported in the same egg extracts (Extended Data Fig. 5F). These results suggest that converging replisomes do not undergo prolonged stalling.
Figure 2. DNA synthesis does not stall during termination.
A. Cartoon depicting the assay for lacO array synthesis.
B. LacR Block-IPTG release was performed on p[lacOx12]. To measure synthesis within the array, termination intermediates were cut with AflIII and PvuII to liberate the array fragment from the vector. Cleaved products were separated by native gel electrophoresis. Different exposures of array and vector fragments are shown (see methods).
C. Array synthesis, vector synthesis, and dissolution were quantified. Means±s.d. are plotted (n=3).
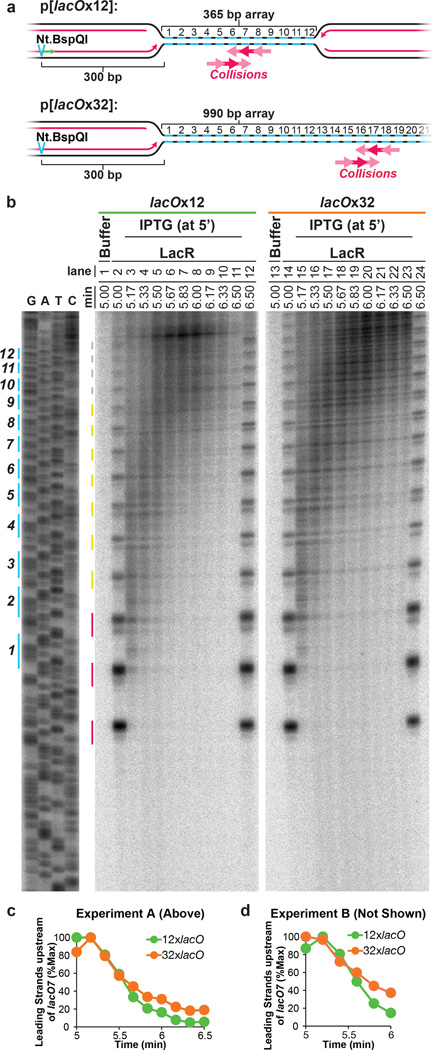
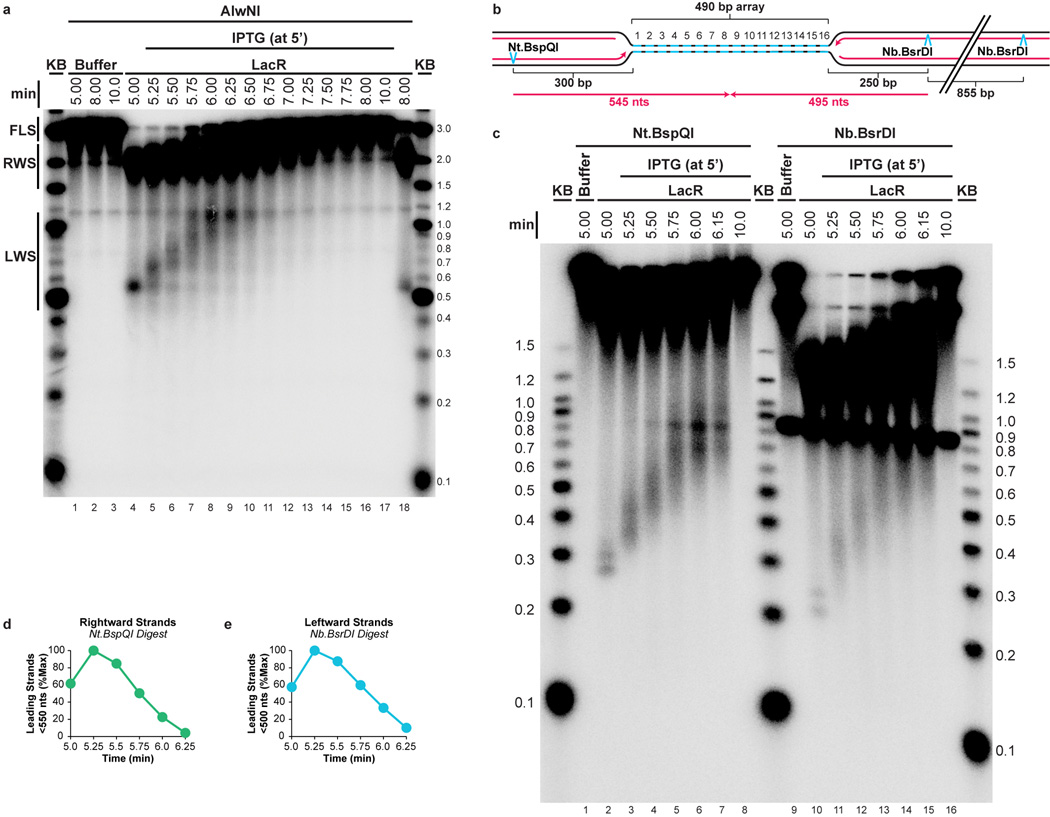
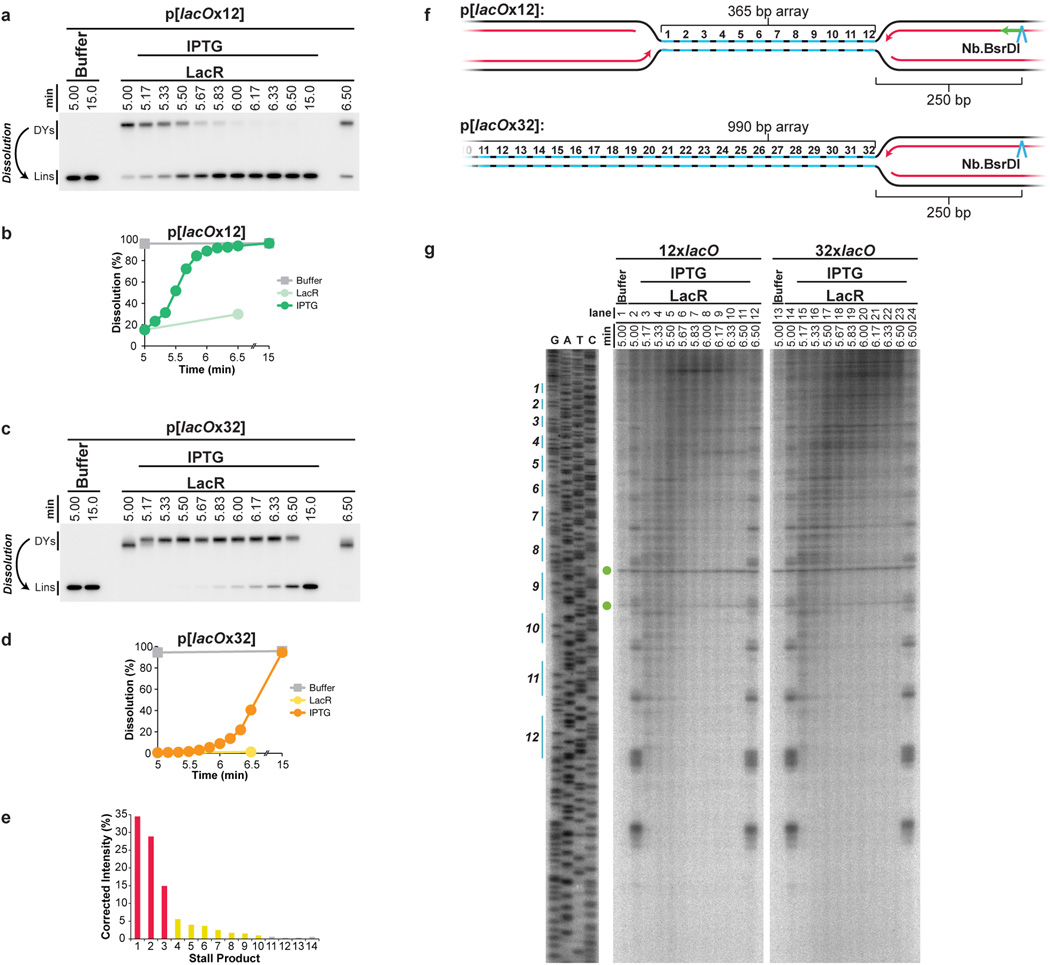
To further evaluate whether forks slow or stall upon encounter with a converging fork, we compared progression of leading strands into arrays containing 12 (12xlacO) or 32 copies (32xlacO) of lacO (Fig. 3A), in which the rightward fork should collide with a converging fork at the 6th or 16th lacO repeats, respectively (Fig. 3A). If converging forks interfere with each other, the rightward leading strand should pause or stall near the 6th repeat in p[lacOx12] but not in p[lacOx32]. As expected, dissolution (Figure 1B) happened much earlier on p[lacOx12] than on p[lacOx32] (Extended Data Fig. 6A–D). To monitor leading strand progression into the array with near-nucleotide resolution, DNA intermediates were purified, digested with the nicking enzyme Nt.BspQI, which released rightward leading strands (Fig 3A), and separated on a denaturing polyacrylamide gel (Fig 3B). Prior to IPTG addition, a discrete ladder of leading strands was seen (Fig 3B, Lanes 2,14), in which the 3’ ends of leading strands stalled ∼29–33 nts from each LacR molecule in the array. This ∼30 nucleotide gap likely corresponds to the footprint of the CMG complex35,40. As shown in Fig. 3B (red lines) and quantified in Extended Data Fig. 6E, 78% of leading strands were stalled at the first 3 lacO sites, indicating that most replisomes were blocked at the outer edges of the array.
Figure 3. Leading strands pass each other unhindered during termination.
A. Schematic of rightward leading strands arrested at 12x and 32xlacO arrays, and the predicted point of fork collision upon IPTG addition.
B. LacR block-IPTG release was performed on p[lacOx12] and p[lacOx32]. Termination intermediates were digested with Nt.BspQI. Nascent strands were separated alongside a sequencing ladder (generated by primer JDO107, green arrow in (A)) on a denaturing polyacrylamide gel and visualized by autoradiography. The lacO sites of p[lacOx12] are indicated in blue. Red, yellow, and grey lines indicate stall products that were quantified (Extended Data Fig. 6E).
C. Leading strands whose 3’ ends were located before lacO7 were quantified (see methods) along with dissolution (Extended Data Fig. 6A–D).
D. Experimental repeat of (C).
Upon addition of IPTG, extension of leading strands resumed immediately (Fig. 3B, lanes 3–11 and 15–23). Importantly, there was no enhanced pausing near the 6th lacO repeat of the 12xlacO array versus the 32xlacO array. By 5.67 minutes, the majority of leading strands had extended beyond the sixth lacO repeat within both arrays (Fig. 3B, lanes 6 and 18 and Extended Fig. 2C). This was also true for the leftward leading strands (Extended Data Fig. 6F–G). Furthermore, leading strands were extended beyond the 6th lacO repeat in the lacOx12 and lacOx32 arrays with similar kinetics (Fig. 3C–D). When leading strands were analyzed on alkaline denaturing gels, we observed that all rightward and leftward leading strands passed the mid-point of the array by 6.25 min (Extended Data Fig 2D–E), indicating that the converging leading strands were readily extended past each other. In summary, we failed to observe detectable slowing or pausing of DNA synthesis during termination, and converging leading strands passed each other unhindered, implying that converging replisomes do not pause or stall significantly.
Lagging strand gaps are rapidly filled in
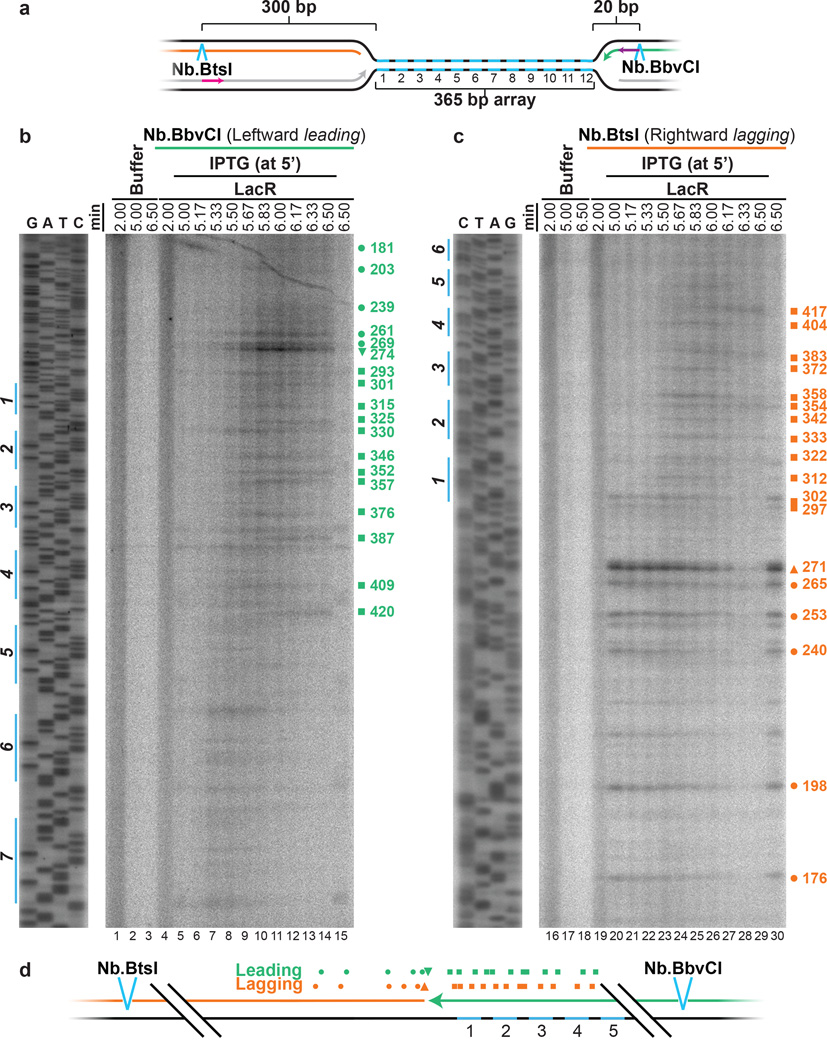
During SV40 replication termination, gaps of ∼60 nucleotides persist following dissolution11. To determine if the appearance of such gaps precedes the ligation step in our system, we mapped the 3’ ends of the leftward leading strands and the 5’ ends of the rightward lagging strands during termination within lacOx12 (Fig. 4A). To this end, we digested DNA intermediates with Nb.BbvCI or Nb.BtsI to release leading or lagging strands, respectively (Fig. 4A>), and separated them on denaturing polyacrylamide gels. Following IPTG addition, we detected a prominent leading strand product beyond the twelfth lacO repeat (species 274 in Fig. 4B; the 3’ and 5’ termini of all leading and lagging strand products, respectively, are mapped relative to the Nb.BtsI site) as seen also in Figure 3B. The 3’ end of this species was located ∼3 nucleotides from the 5’ end of the most abundant lagging strand product of the converging fork (271, Fig. 4C). We observed many other, less prominent leading stra), most of which mapped close to corresponding lagging strand products (176–417, Fig. 4C). The results show that leading strands are generally extended to within ∼3 nucleotides of the lagging strands (Fig. 4D). It is likely that leading strands immediately abut lagging strands and that the ∼3 nucleotide gap reflects imprecise mapping of lagging strands (see methods). In conclusion, we observe no evidence of persistent gaps between leading and lagging strands during replication termination.
Figure 4. Leading strands abut lagging strands of the opposing replisome during termination.
A. Cartoon illustrating the leading and lagging strands released by Nb.BtsI and Nb.BbvCI nicking enzymes. Primers JDO111 (purple arrow) and JDO110 (pink arrow) generated the sequencing ladders in (B) and (C), respectively.
B. LacR block-IPTG release was performed on p[lacOx12]. Termination intermediates were digested with Nb.BbvCI to liberate leftward leading strands, which were separated alongside a sequencing ladder on a denaturing polyacrylamide gel and visualized by autoradiography. Prominent leading strand products are highlighted (green symbols), and their sizes, measured relative to the Nb.BtsI site, are indicated.
C. Same samples as in (B) were digested with Nb.BtsI to liberate rightward lagging strands. The size of prominent lagging strand products (orange symbols), measured relative to the Nb.BtsI site, are indicated.
D. Schematic of the mapped leading (B) and lagging (C) strands.
CMGs dissociate late during termination
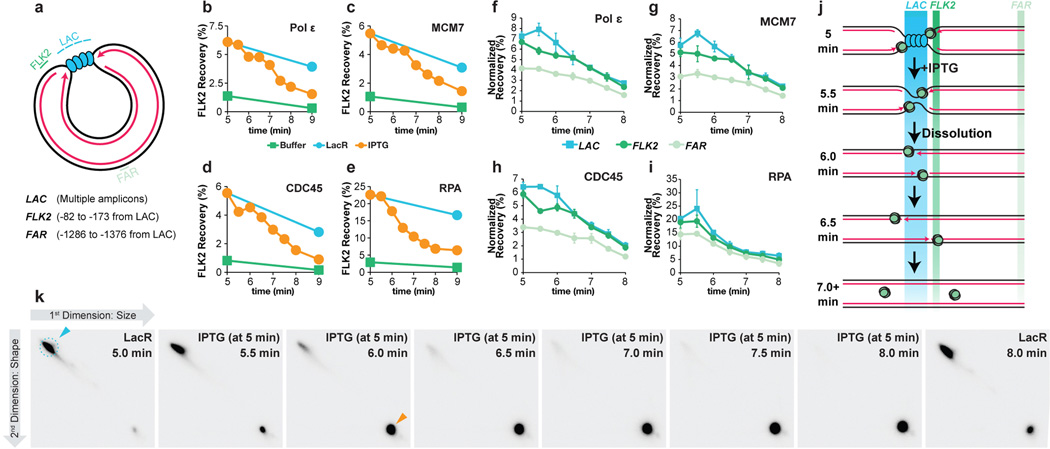
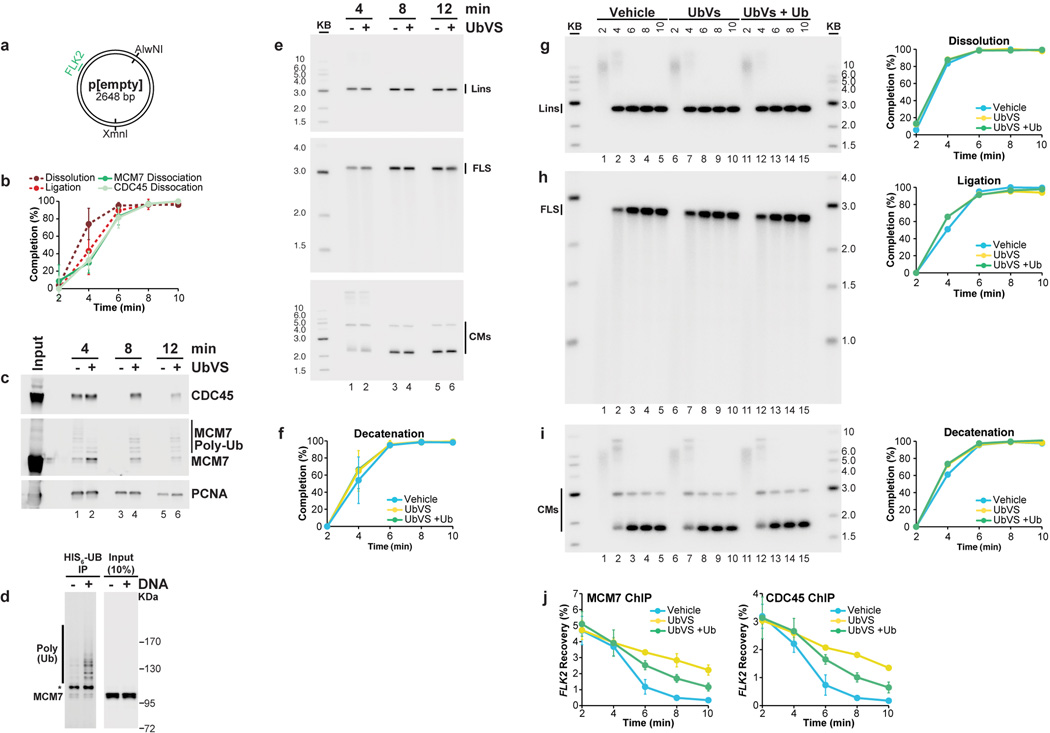
To determine when replisome components dissociate during termination, we monitored MCM7, CDC45, Polε and RPA binding to a site flanking the lacO array using Chromatin ImmunoPrecipitation (ChIP) (FLK2 locus, Extended Data Fig 7A). In parallel, we monitored dissolution, ligation, and decatenation. Prior to IPTG addition, MCM7, CDC45, Polε, and RPA were 4–8-fold enriched at the array in the presence of LacR compared to buffer (Extended Data Fig. 7B–E, Fig. 5 min time point), demonstrating that the ChIP signal reflects replisome stalling at the array. When IPTG was added at 5 minutes, MCM7, CDC45, RPA and Polε largely dissociated by 9 minutes, whereas in the absence of IPTG, they dissociated much more slowly (Extended Data Fig. 7B–E). RPA dissociation correlated well with ligation, as expected, since ligation marks the disappearance of any ssDNA in the termination zone (Figure 5A, compare red squares and blue circles). Strikingly, CDC45, MCM7, and Pol ε dissociated ∼1.5 minutes after dissolution and ∼0.5 minutes after RPA dissociation and ligation (Figure 5A). A time course of ChIP at sequences adjacent to and within the array (Extended Data Fig. 7F–I) was consistent with MCM7, CDC45, and DNA Polε moving into the array and then back out following dissolution (Extended Data Fig. 7J). MCM7 and CDC45 also dissociated after dissolution during replication of plasmid DNA that lacked a lacO array (p[empty], Extended Data Fig. 8A–B). Although the delay between ligation and unloading of MCM7 and CDC45 was not readily detectable on this template (Extended Data Fig. 8B) this was not surprising, given the asynchrony of termination in this setting. Together, the data support a model in which CDC45 and MCM7 dissociate late in termination, long after forks meet (dissolution) and shortly after ligation.
Figure 5. CMGs dissociate after dissolution and ligation.
A. LacR block-IPTG release was followed by MCM7, CDC45, RPA and, Polε ChIP at the indicated times after IPTG addition. Dissolution, ligation, and decatenation were measured in parallel. means±s.d. are plotted (n=3).
B. p[empty] was replicated in extracts treated with Vehicle, Ubiqutin-Vinyl Sulfone (Ub-VS), or Ub-VS and free ubiquitin (UbVS + Ub). Dissociation of MCM7 was measured by ChIP (see methods). mean±s.d. is plotted (n=3).
C. Same as (B), but CDC45 dissociation was measured.
D-E. In parallel to MCM7 and CDC45 dissociation (B, C), dissolution (D) and ligation (E) were measured. mean±s.d. is plotted (n=3). See Extended Data Fig. 8F–I for decatenation measurements and representative gels.
(E) New model of vertebrate replication termination.
If our model is correct, inhibiting CMG unloading should not affect dissolution or ligation. To test this, we inhibited ubiquitin signaling, which is required for chromatin dissociation of CMG24,25,41. p[empty] was replicated in extracts that were incubated with vehicle or the de-ubiquitylating enzyme inhibitor ubiquitin-vinyl-sulfone (Ub-VS), which leads to the depletion of free ubiquitin35,41, and we performed MCM7 and CDC45 ChIP. As shown in Fig. 5B–C, UbVS substantially delayed MCM7 and CDC45 dissociation, and this effect was partially reversed by co-addition of free ubiquitin (Fig 5B–C>). The same inhibitory effect of UbVS on CMG unloading was observed when plasmids were recovered from egg extract and blotted for MCM7 and CDC45 (Extended Data Fig. 8C). This analysis also confirmed prior reports24,25 of MCM7 ubiquitylation during replication. Importantly, dissolution, ligation, and decatenation were not affected by Ub-VS (Fig 5D–E and Extended Data Fig 8F–I). We conclude that defective CMG unloading does not affect dissolution, ligation, or decatenation, strongly supporting our model that CMG unloading is a late event in replication termination.
Discussion
In this study, we present a novel approach to induce synchronous and site-specific replication termination. Using this system, we observe no slowing or pausing of DNA synthesis as forks converge (Fig. 5Ei-ii). Leading strands pass each other unhindered and immediately abut downstream lagging strands before undergoing ligation (Fig. 5E iii-v). CMG remains associated with DNA after dissolution, and it is unloaded only after the leading strand of one fork is ligated to the lagging strand of the opposing fork (Fig. 5E vii). Catenane removal is initiated at the same time as ligation (Fig. 5v–vi). In contrast to models of termination in which replication forks stall1,3,4, our data imply that topological stress between replisomes is handled efficiently and that converging replisomes do not clash or that if they do, any remaining template DNA is immediately reeled into the stalled replisome for duplication (not shown). We previously showed that CMGs encircle the leading strand template at the replication fork42. Therefore, converging CMGs approach each other on opposite strands42,43, which helps explain how they could pass each other. If a fork stalls (e.g. at the ribosomal DNA locus13,14) the same termination mechanism could still operate provided the stalled fork remains stable until a converging fork arrives. We expect this to be the case, given our recent observation that a single fork stalled at an ICL does not collapse or lose its CMG complex34. We speculate that at telomeres the replisome simply runs off the chromosome end. .
Our observations that CMG dissociates after the ligation step (Fig. 5A), and that ligation is not affected when CMG unloading is impaired (Fig 5B–C,E), strongly imply that CMG is unloaded from dsDNA. We propose that when CMG reaches the 5’ end of the opposing fork’s lagging strand, it passes over the ssDNA-dsDNA junction and keeps moving along dsDNA (Fig. 5E), as previously observed for purified MCM2–7 and CMG in vitro (see 23,44 but see also 22). This scenario is appealing, as it would prevent CMG from interfering with ligation of the nascent strands. We propose that CMG ubiquitylation and its removal by p9724,25 is triggered once CMG encircles dsDNA. Such a mechanism would help to avoid inappropriate CMG unloading from active replication forks, where CMG encircles ssDNA. Our results disagree with a recent report, which concluded that inhibition of CMG unloading prevents completion of DNA synthesis25. In contrast, another report that defective CMG unloading does not prevent cell cycle progression24 is consistent with our model. We recently reported that CMG can be unloaded from ssDNA when two replisomes collide with a DNA interstrand cross-link41. However, this process involves a unique, BRCA1-dependent pathway that is not employed during termination41. In conclusion, the termination mechanism described here allows rapid completion of DNA synthesis while minimizing the possibility of premature replisome disassembly.
Methods
Protein Purification
Biotinylated LacR was purified using a protocol adapted from Kenneth Marian’s laboratory (Personal Communication). The LacR open reading frame was fused to a C-terminal AviTag (Avidity, Denver, CO) and expressed from pET11a (pET11a[LacR-Avi]). To biotinylate the AviTag on LacR-Avi, biotin ligase was co-expressed from pBirAcm (Avidity, Denver, CO). To this end, pET11a[LacR-Avi] and pBirAcm were co-transformed into T7 Express cells (New England Biolabs) and grown in the presence of ampicillin (100 µg/ml) and chloramphenicol (17 µg/ml). Expression of LacR-Avi and the biotin ligase was induced by addition of IPTG to a final concentration of 1 mM. Cultures were supplemented with 50 µM biotin (Research Organics, Cleveland, OH) to ensure efficient biotinylation of LacR-Avi.
Cell pellets were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 100 mM NaCl, 1 mM DTT, 10% sucrose (w/v), cOmplete protease inhibitor (Roche, Nutley, NJ). The cells were lysed at room temperature in the presence of 0.2 mg/ml lysozyme and 0.1 % Brij 58. The insoluble, chromatin-containing fraction was isolated by centrifugation at 4°C. Chromatin-bound LacR was then released by sonication (in 50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 1 M NaCl, 1 mM DTT, cOmplete protease inhibitor, 30 mM IPTG). DNA was removed from the soluble fraction by addition of polymin P (final concentration 1%), LacR was precipitated by addition of ammonium sulphate (final concentration 37%). The precipitate was dissolved in wash buffer (50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 2.5 M NaCl, 1 mM DTT, cOmplete protease inhibitor) and then applied to a column of SoftLink avidin resin (Promega, Madison, WI). LacR was eluted (in 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 100 mM NaCl, 1 mM DTT, 5 mM biotin) and dialyzed overnight (against 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1 mM DTT, 38% glycerol (v/v)). Purified LacR was frozen in liquid nitrogen and stored at −80°C. A more detailed purification protocol is available upon request.
Cyclin A was purified as described previously45
Plasmid construction and preparation
pJD82 (Extended Data 9) was created by replacing the SacI-KpnI fragment of pBlueScript II KS- with the sequence:
GAGCTCTCACACCTACAAGGGATGTACATCAATTGTGAGCGGATAACAATTGTTAGGGAGGAATTGTGAGCGGATAACAATTTGGAGTTGATAATTGTGAGCGGATAACAATTGGCTTCAACGTAATTGTGAGCGGATAACAATTTCCGTACGAATGTGCCGAACTTATGGTACC
This contains 4 tandem repeats of the lac operator sequence (AATTGTGAGCGGATAACAATT) interspersed by an average of 10–11 base pairs of random sequence (average 10.33 basepairs). Additional tandem repeats of the BsiWI-BsrGI fragment were then cloned into pJD82, and subsequently derived vectors, to generate arrays of 8, 12, 16, 32 and 48 lacO repeats (Extended Data 9). Recognition sites for nicking enzymes were introduced by QuickChange mutagenesis (Agilent Technologies, Santa Clara, CA) according to the manufacturer’s guidelines.
To propagate lacO plasmid DNA, plasmids were transformed into DH5α cells and grown for a minimal number of passages in the presence of 2 mM IPTG. DNA was prepared using the QIAprep spin kit (Qiagen, Valencia, CA). To eliminate preparations containing genetic rearrangements (typically ∼25%) each preparation was separated by electrophoresis on a 0.8% agarose gel and visualized by ethidium bromide staining. Preparations that were free of rearranged plasmids were then verified by sequencing (Genewiz, Cambridge, MA).
Xenopus egg extracts and DNA replication
Xenopus egg extracts were prepared from Xenopus laevis wild type males and females 2–5 years of age, as approved by the Harvard Medical School Institutional Animal Care and use Committee (IACUC) and as described previously46. For DNA replication, 1 volume of ‘licensing mix’ was prepared by adding plasmid DNA to High Speed Supernatant (HSS) of egg cytoplasm to a final concentration of 7.5–15 ng/µl. Licensing mix was incubated for 30 minutes at room temperature, leading to the formation of pre-replication complexes (pre-RCs). Next, licensing mix was supplemented with 0.1 volumes of Cyclin A to a final volume of 576 nM and incubated a further 10 minutes at room temperature, as previously described45. Cyclin A treatment was performed to achieve highly synchronous DNA replication (Extended Data Fig. 10). Finally, 1.9 volumes of nucleoplasmic extract (NPE) was added to initiate Cdk2-dependent replication at pre-RCs. In all Figures, “0 minutes” represents the time 30 seconds after NPE addition. To radiolabel DNA, NPE was supplemented with [α-32P]dATP. Reactions were stopped with 10 volumes Stop Solution (0.5% SDS, 25 mM EDTA, 50 mM Tris-HCl pH 7.5). DNA in Stop Solution was treated with RNase A (190 ng/µl final concentration) then Proteinase K (909 ng/µl final concentration) before either direct analysis by gel electrophoresis or purification of DNA as described previously40. For UbVS experiments, UbVS (Boston Biochem) was added to final concentration of 20 µM, to HSS 5 minutes prior to addition of plasmid DNA (HSS) and to NPE 5 minutes prior to addition of HSS, with or without 120 µM Ubiquitin (Boston Biochem). Unless otherwise stated in the figure legend, all experiments were performed at least twice and a representative result is shown. Replicate samples were collected from independently assembled replication reactions, and therefore represent biological replicates.
Immunodepletions
To deplete Topo II-α from Xenopus egg extracts one volume of Protein A Sepharose Fast Flow (PAS) (GE Healthcare) was incubated with 4.5 volumes of affinity purified, anti-Topo II-α antibody raised against the C-terminal 20 residues (1 mg/ml). For mock depletion, an equivalent quantity of nonspecific IgGs were used. 5 volumes of pre-cleared HSS or NPE was then mixed with 1 volume of the antibody-bound sepharose and incubated for 45 min at 4°C, and for the NPE this was repeated once. Depleted extracts were collected and used immediately for DNA replication.
Induction of termination
To monitor termination, 0.05 volumes plasmid DNA (150–300 ng/µl) was incubated with 0.1 volumes LacR (54 µM) or dialysis buffer for at least 90 minutes at room temperature to allow formation of LacR arrays on the DNA. Licensing mix was prepared by adding 0.85 volumes of HSS, and DNA was replicated as described above. To induce termination, 0.06 volumes of IPTG was added (to a final concentration of 10 mM) at the time indicated (typically 5 minutes), which triggered dissociation of lacO-bound LacR. To accurately withdraw samples at the times indicated, reactions composed of the same Licensing Mix and NPE were staggered, where necessary.
2D gel electrophoresis
2D gels were performed as described41. Briefly, purified DNA was digested with XmnI (New England BioLabs) and then separated by native-native 2D gel electrophoresis. Samples were separated in the first dimension on a 0.4 % agarose gel at 0.75 V/cm for approximately 40 hours at room temperature. The gel was stained with 0.3 µg/ml ethidium bromide, allowing the 2–8 kilobase pairs size-range to be excised. A second dimension gel containing 1 % agarose and 0.3 µg/ml ethidium bromide was cast over the gel slice from the first dimension. DNA was separated on the second dimension at 4.5 V/cm for 12 hours at 4°C.
Termination assays
To monitor dissolution, 0.25–1.0 ng/µl of purified DNA was incubated in CutSmart Buffer with 0.4 units/µl of XmnI (New England BioLabs) at 37°C for 1 hour. Digested products were separated on a 1.2 % agarose gel at 4 V/cm and detected by autoradiography. Dissolution (%) was calculated as the percentage of total signal in each lane present in the linear products of digestion (Lins, Fig 1C).
To monitor ligation, 0.25–1.0 ng/µl of purified DNA was incubated in CutSmart buffer with 0.2 units/µl of AlwNI (New England BioLabs) at 37°C for 1 hour. Digests were terminated by addition of EDTA to 30 mM, then products were separated on a 1.5 % denaturing alkaline agarose gel at 1.5 V/cm and detected by autoradiography. The percentage of total signal in each lane present in the full-length strands (FLS) was measured (FLS, Fig 1E). During electrophoresis, partial hydrolysis caused signal from the FLS to smear down. To correct for this, a fully ligated plasmid was cleaved and analyzed on the same gel. The percentage of signal in FLS band of the fully ligated plasmid was measured (FLSFL) and used to correct signal in the other lanes to yield an accurate measure of ligation. Ligation (%) was calculated as FLS/FLSFL*100
To monitor decatenation, 0.25–1.0 ng/µl of purified DNA was separated on a 0.8 % agarose gel at 4 V/cm and detected by autoradiography. Decatenation (%) was measured as the percentage of total signal in each lane present in circular monomers (CMs, Fig. 1G).
To monitor DNA synthesis within a lacO array (Figure 2), 0.25–1.0 ng/µl of purified DNA was incubated in Buffer 3.1 with 0.2 units/µl PvuII and 0.2 units/µl AflIII (New England BioLabs) at 37°C for 1 hour. Digested products were separated on a 1.2 % agarose gel at 4 V/cm and detected by autoradiography. To measure array synthesis (SYNARY), the 0.5–1.5 kb region of each lane was quantified (lins and DYs, Fig 2B). To measure vector synthesis (SYNVEC), the 2 to 6 kb region of each lane was quantified, which included the ∼3.0 and ∼6.0 bands that arose when one, or both, lagging strands did not cut, respectively. Total signal in each lane (SYNTOT) was also measured. To correct for differences in efficiency of DNA extraction, total lane signal was also measured in a set of unprocessed samples (SYNUN), which were separated and detected in parallel. Array synthesis (%) was calculated as SYNUN/SYNTOT*SYNARY, vector synthesis was calculated as SYNUN/SYNTOT*SYNVEC and in both cases the 10 minute time point was assigned a value of 100%. The same approach was also used to quantify synthesis of the 294/794 bp fragments (quantified in the same manner as the array) and the 2354 bp fragments (quantified in the same manner as the vector fragments) in Extended Data Figure 1. In Figure 2 and Extended Data Figure 1, a longer exposure of the array fragment is shown because it is less intense than the vector fragment.
To analyze topoisomers (Extended Data Fig 3D) 0.25 ng/µl of radiolabelled DNA was incubated in 1X Buffer A and 1X Buffer B (Topogen) with 0.2 U/µl Human Topo II-α (Topogen) at 37°C for 15 minutes, or in CutSmart Buffer with 0.4 U/µl XmnI or 0.04 U/µl Nt.BspQI (New England Biolabs) for 1 hour.
Nascent strand analysis
To nick rightward leading strands, 1–2 ng/µl of purified DNA was incubated in Buffer 3.1 with 0.4 units/µl Nt.BspQI (New England BioLabs) at 37°C for 1 hour. To nick leftward leading strands, 1–2 ng/µl of purified DNA was incubated in CutSmart buffer with 0.04 units/µl Nb.BsrDI (New England BioLabs) at 65°C for 1 hour. To nick rightward leading strands closer to the lacO array, 1–2 ng/µl of purified DNA was incubated in CutSmart buffer with 0.04 units/µl Nb.BbvCI (New England BioLabs) at 37°C for 1 hour. To nick leftward lagging strands, 1–2 ng/µl of purified DNA was incubated in Buffer 3.1 with 0.04 units/µl Nb.BtsI (New England BioLabs) at 37°C for 1 hour. In all cases, nicking reactions were stopped by the addition of 0.5 volumes of Stop Solution B (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol FF).
Nicked DNA (1.5–2 µl sample) was separated on a 42 cm long, 4 or 5 % polyacrylamide sequencing gel using Model S2 sequencing gel apparatus (Apogee Electrophoresis, Baltimore, MD) according to the manufacturer’s guidelines. To maximize the range of nascent products that could be resolved, gels were cast with a thickness gradient of 0.4 to 1.2 mm, beginning to end, to establish an electrical field gradient during electrophoresis. Sequencing gels were prepared with Rapidgel-XL in 0.8X GTG Buffer (USB Corporation, Cleveland). Sequencing ladders were generated using the Cycle Sequencing Kit (USB Corporation, Cleveland) with primers JDO107, JDO109, JDO110, JDO111 (Extended Data 9) and pJD150 (Extended Data 9) as template DNA.
Mapping and quantification of the Nascent strands in Fig. 3–Fig. 4 was performed as follows. Nascent leading and lagging strands were mapped using the sequencing ladders generated by the primers indicated in Fig. 3A and Fig. 4A (see Extended Data 9 for sequences). Slight discrepancies may exist between mapped and actual lagging strand product sizes (Fig. 4C) since the sequencing ladder (generated by JDO110 Fig. 4A) is complementary to the lagging strands. A fraction of lagging strand products 176–302 were not extended upon IPTG addition, probably because they were reached by the rightward leading strand first. Lagging strand products 312–417 appeared de novo after IPTG addition, and therefore represent growing lagging strands of the leftward fork. To quantify leading strand progression (Fig. 3C–D) leading strands whose 3’ ends were located before lacO7 (in Fig. 3B and data not shown) were quantified, and peak signal was assigned a value of 100 (%Max).
ChIP and quantitative PCR
ChIP and quantitative PCR (qPCR) were performed essentially as described41. Chromatin was withdrawn and crosslinked in the presence of 1% formaldehyde for 10 minutes at room temperature. Crosslinking was then quenched by the addition of 0.1 volumes glycine (1.25 M) for 10 minutes. Samples were then spun through Bio-Spin P-6 Gel (containing Tris Buffer, Bio-Rad) to remove salts and small molecules, before being stored in 10 volumes of sonication buffer (20 mM Tris pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% IGEPAL CA-630 (v/v), 2 mM PMSF, 5 µg/µl aprotinin, 5 µg/µl leupeptin). Samples were then sonicated to shear chromatin into approximately 250 bp fragments.
The antibodies used were described previously21,41. Antibodies were incubated with chromatin overnight at 4°C, then immunoprecipitated by addition of Protein A-Sepharose Fast Flow beads (GE Healthcare) for 2 hours at room temperature. Beads were washed sequentially with sonication buffer, high salt buffer (sonication buffer supplemented with 500 mM NaCl and 100 mM KCl), wash buffer (10 mM Tris pH 7.5, 0.25 M LiCl, 1 mM EDTA, 0.5% NP-40 (v/v), 0.5% SDS (w/v)) and TE (10 mM Tris pH7.5, 1 mM EDTA), before being eluted into elution buffer (50 mM Tris pH7.5, 10 mM EDTA, 1% SDS) at 65°C for 20 minutes. Eluted chromatin, and input samples, were treated with RNase for 30 minutes at 37°C. Finally, proteins were degraded by addition of NaCl (250 mM final) and treatment with Pronase (2 µg/µl final) at 42°C for 6 hours. DNA-peptide crosslinks were reversed by treatment at 70°C for a further 9 hours. DNA was subsequently phenol:chloroform extracted and ethanol precipitated. The absolute amount of DNA recovered from the immunoprecipitated and input samples was measured by quantitative PCR (qPCR) relative to a standard curve. The qPCR primers used are listed in Extended Data 9. Binding was measured as the percentage recovery of immunoprecipitated DNA, relative to the input (EXPREC).
To minimize error in the ChIP process, an internal control was built into all experiments. Xenopus egg extracts were used to separately replicate a different plasmid, pQUANT (see Extended Data 9 for sequences). Mid-way through replication, pQUANT, was crosslinked, quenched, and spun through Bio-Spin P-6 gel (as above) to yield a single pool of heterologous chromatin that was bound by replication proteins. An equal amount of pQUANT chromatin was added to all experimental chromatin samples prior to sonication, and this was carried through the entire ChIP procedure. For each set of immunoprecipitations, the recovery of pQUANT (QNTREC) should be identical between samples. To correct for technical variation in any set of immunopreciptiations, average pQUANT recovery was calculated (QNTAVG) and normalized recovery (%) was calculated as EXPREC*QNTAVG/QNTREC. This ensured that the only sources of technical variation were the crosslinking process and the qPCR. To maximize the reliability of the qPCR, these measurements were performed in quintuplicate and the median value was used. Where three ChIP experiments were combined and plotted as mean±s.d. (Fig 5A–C, Extended Data Figs 7F–I and 9L–M) it was necessary to normalize the data to correct for differences in absolute IP efficiency between experiments. For each protein measured by ChIP, mean recovery across all loci in all samples (meanall) was calculated for each experiment (meanall1, meanall2 and meanall3) and used to generate a correction factor for each experiment (e.g. for experiment 1 the correction factor is [(meanall1+meanall2+meanall3)/3]/meanall1). To measure dissociation (Fig 5A–C), recovery of the FLK2 locus was measured (shown in Extended Data Fig 7F–I and Fig 8L–M) and peak signal was assigned a value of ‘0’, while background signal (measured at 4 or 5 min for Fig 5A, or 10 min for Fig 5B–C) was assigned a value of 100. The experiments shown in Fig 5A and Extended Data Fig. 7F–I were repeated 3 times, once with p[lacOx12] and twice with p[lacOx16].
Plasmid pull downs
Plasmid pull downs were performed essentially as described47, with the following exceptions. Beads were resuspended in buffer supplemented with 4% DMSO and 100 µM NMS-87348 to block further CMG unloading once the samples were withdrawn25. Plasmid-associated proteins from 40–80 ng of plasmid were isolated, and a quarter of the sample was analyzed by Western blotting using previously-described antibodies against CDC45, MCM7, and PCNA41.
HIS6-Ub Immunoprecipitations
Ni-NTA Superflow Resin (Qaigen) was washed three times with Urea Buffer (10mM imidazole, 0.2% NP-40, 8 M urea, 500 mM NaH2PO4, 50 mM Tris HCl, pH 8.0). For each immunoprecipitation, 10 µl of resin was added per tube, and resuspended to 191 µl in Urea buffer. Extracts were supplemented with 100 µM of HIS6-Ubiquitin (Boston Biochem) and replication was carried out as described above. At the indicated time, 9 µl of extract was mixed with the bead mix and samples were incubated for one hour at room temperature, with end-over-end rotation. Resin was washed three times with urea buffer. All residual buffer was removed, and resin was boiled for 5 minutes in 30 μl sample buffer (125 mM Tris-HCl pH 6.8, 20% glycerol, 6.1% SDS, 0.01% bromophenol blue, 10% β-mercaptoethanol). 30 µl of 0.5 M imidazole was added to each sample and HIS6-tagged proteins were eluted off the resin for 60’ at room temperature, with gentle agitation. Resin was spun down at 1000 RCF for 1 minute, and the supernatant was removed. 10 µl of each sample was resolved on an SDS-PAGE gel alongside an input control and analyzed by Western blotting using the previously-described antibody against MCM749. In extended Data Fig. 8D, a longer exposure of the IP lanes is shown, since they are far less intense than the input lanes.
Extended Data
Extended Data Figure 1. Sequence-specific termination can be induced at a LacR array.
(A) To investigate whether a LacR array blocks replication forks, a plasmid containing a tandem array of 16 lac operator (lacO) sequences, p[lacOx16], was incubated with buffer or LacR and then replicated in egg extract containing [α-32P]dATP. Radiolabelled replication intermediates were cleaved with XmnI (far left cartoon) and separated according to size and shape by 2-dimensional gel electrophoresis (see schematic of 2-D gel). As replication neared completion at 4.5 minutes, mainly linear molecules were produced in the presence of buffer (orange arrowhead). In contrast, in the presence of LacR, a discrete spot appeared on the Double Y arc (blue arrowhead), demonstrating that converging replication forks accumulate at a specific locus on p[lacOx16]. These data indicate that 16 copies of LacR block replication forks.
(B–F) To test whether the double-Y structures observed in (A) arose from replication forks stalling at the outer edges of the lacO array, we tested whether LacR specifically inhibited replication of lacO sequences. To this end, p[lacOx16] (C) and the parental plasmid lacking lacO repeats, p[empty] (B), were incubated in the presence of buffer or LacR and replicated using Xenopus egg extracts containing [α-32P]dATP. Radiolabelled replication intermediates were cleaved with AflIII and PvuII to release the 2354 bp plasmid backbone (B and C) and a 294 bp control fragment from p[empty] (B) or a 794 bp lacO fragment from p[lacOx16] (C). The plasmid backbone and the respective inserts were separated on a native gel and detected by autoradiography (D). A longer exposure of the small fragments is shown, since they are less intense than the large fragments. The results in panel (D) were quantified in (E) and (F). Importantly, LacR specifically inhibited replication of the lacO-containing fragment in p[lacOx16] (F, blue circles) but not the control fragment in p[empty] (E, green circles). We conclude that LacR prevents replication of the lacO array and that the double-Y’s in (A) represent forks converged on the outer edges of the array. Importantly, synthesis within the 2354 bp backbone fragment (F, orange circles) of p[lacOx16] was not inhibited in the presence of LacR, indicating that no global structural changes occur that inhibit replication.
Extended Data Figure 2. Supplemental fork progression data.
(A) The gel shown in Figure 1E was overexposed and shown in its entirety so that the smaller leftward strands (LWS, Fig 1D) could be detected. As observed for the rightward strands (RWS, Fig 1E), LWS rapidly increased in size and then disappeared as they were ligated to produce full length strands (FLS, Fig 1E).
(B–E) To determine whether the heterogeneity of LWS (A) and RWS (Fig 1E) was due to delayed extension of lagging strands, or because a significant fraction of leading strands did not restart upon IPTG addition, we specifically monitored leading strand progression upon IPTG addition on p[lacOx16]. To this end, DNA samples were treated with Nt.BspQI or Nb.BsrDI to specifically liberate the rightward or leftward leading strands, respectively (B), and DNAs were separated on a denaturing agarose gel (C). Prior to IPTG addition, discrete leading strand products of the expected size were observed (lanes 2 and 10). The presence of two stall products reflects the fact that at a slow rate, the replisome bypasses LacR (see also Fig 3). Upon IPTG addition, these species rapidly and completely shifted up the gel, indicating that rightward and leftward leading strands restarted efficiently. Therefore, the heterogeneity of the LWS (A) and RWS (Fig. 1E) is probably due to delayed ligation of a new Okazaki fragment to the lagging strands. Quantification of leading strands that had not reached the midpoint of the array (rightward and leftward strands smaller than 550 and 500 nts, respectively, B) revealed that by 6.25 minutes, 90% of rightward and leftward leading strands passed the midpoint of the array (D–E). This demonstrates that leading strands pass each other when forks meet.
Extended Data Figure 3. Topo II-dependent decatenation of p[lacOx16].
(A) The autoradiograph in primary Figure 1G is reproduced with cartoons indicating the structures of the replication and termination intermediates n-n, n-sc, sc-sc, n, and sc (see main text for definitions). The order of appearance of the different catenanes matches previous work5 (n-n, then n-sc, then sc-sc).
(B–D) To determine the role of Topo II during termination within a lacO array, termination was monitored in mock- or Topo II-depleted extracts. To confirm immunodepletion of Topo II, Mock and Topo II-depleted NPE was blotted with MCM7 and Topo II antibodies (B). p[lacOx16] was incubated with LacR, then replicated in either mock- or Topo II-depleted egg extracts in the presence of [α-32P]dATP, and termination was induced with IPTG (at 7’). Untreated DNA intermediates were separated by native gel electrophoresis (C). In the mock-depleted extract, nicked and supercoiled monomers were readily produced (as in (A), albeit with slower kinetics due to nonspecific inhibition of the extracts by the immunodepletion procedure), while in the Topo II-depleted extracts, a discrete species was produced. DNA from the last time point in each reaction (lanes 4 and 8 in (C)) was purified and treated with XmnI, which cuts p[lacOx16] once, or Nt.BspQI, which nicks p[lacOx16] once, or recombinant Topo II, and then separated by native gel electrophoresis (D). Cleavage of the mock- and TopoII-depleted products with XmnI yielded the expected linear 3.15 kb band (lanes 2 and 6), demonstrating that in both extracts, all products were fully dissolved topoisomers of each other. Relaxation of the mock-depleted products by nicking with Nt.BspQI yielded a discrete band corresponding to nicked plasmid (lane 3), while the TopoII-depleted products were converted to a ladder of discrete topoisomers (lane 7), which we infer represent catenated dimers of different linking numbers, since the mobility difference cannot be due to differences in supercoiling. Importantly, the mobility shift following Nt.BspQI treatment (lane 5 vs lane 7) demonstrated that the Topo II-depleted products (lane 5) were covalently closed and thus in the absence of Topo II ligation of the daughter strands still occurred. Treatment of the mock- and Topo II-depleted products with recombinant human Topo II produced the same relaxed monomeric species (lanes 4 and 8), further confirming that the Topo II-depleted products contained catenanes. Collectively, these observations demonstrate that termination within a lacO array in Topo II depleted extracts produces highly catenated supercoiled-supercoiled dimers, as seen in cells lacking Topo II16,17. These data confirm that Topo II is responsible for decatenation and argue that termination within a lacO array reflects physiological termination.
(E) n-n, n-sc, sc-sc, n, and sc products were also detected when plasmid lacking lacO sequences (pBlueScript) was replicated in the absence of LacR without the use of Cyclin A to synchronize replication. Therefore, these intermediates arise in the course of unperturbed DNA replication in Xenopus egg extracts.
Extended Data Figure 4. Inhibition of termination by different size LacR arrays.
(A) Cartoon depicting intermediates detected in the dissolution assay.
(B) To determine the ability of different-sized LacR arrays to inhibit termination, the earliest stage of termination, dissolution (A), was monitored in plasmids containing 0, 12, 16, 32, or 48 lacO repeats. Plasmids were incubated with LacR, and replicated in the presence of [α-32P]dATP. To measure dissolution, radiolabelled termination intermediates were cut with XmnI. Cleaved products were separated on a native agarose gel and detected by autoradiography.
(C) Quantification of dissolution in (B). When 12 or more lacO repeats were present in the array, dissolution was robustly inhibited for at least 5 minutes. Potent inhibition lasted 10 minutes when 32 lacO sequences were present, and 20 minutes in the presence of 48 lacO sequences. In the absence of lacO sequences, dissolution was essentially complete by 5 minutes. Therefore, 12 lacO repeats are sufficient to inhibit termination for 5 minutes.
Extended Data Figure 5. The rate of total DNA synthesis does not slow before dissolution.
(A–C) To further test whether replication stalls or slows prior to dissolution, p[lacOx12] was pre-incubated with LacR and replicated in Xenopus egg extracts. Termination was then induced by addition of IPTG after 5 minutes. Simultaneously, [α-32P]dATP was added to specifically radiolabel DNA synthesized following IPTG addition (A). Radiolabelled DNA was then separated on a native agarose gel and total signal was measured by autoradiography (B). Total signal was quantified, normalized to peak signal, and graphed alongside the rate of dissolution, which was also measured in the same experiment (C). This approach gives a highly sensitive measure of DNA synthesis without manipulation of DNA samples. DNA synthesis should occur primarily within the lacO array (see Extended Data Fig. 1). Upon IPTG addition, there was an approximately linear increase in signal, which plateaued by 5.83 min. Importantly, dissolution was 65% complete by 5.83 min. Therefore, the large majority of dissolution occurs without stalling of DNA synthesis.
(D–E) Experimental repeats of (B–C)
(F) The experiments shown in (C–E) were graphed together with mean±s.d. Synthesis data was normalized so that for each experiment, synthesis at 1 min was assigned a value of 84.4%, since this was the average value from (C–D), where synthesis was allowed to plateau. Given the rate of replication fork progression in these egg extracts (260 bp/minute32) and the size of the array (365 bp), forks should require, on average, 0.7 minutes to converge if no stalling occurs (365 bp ÷ 2 ÷ 260 bp/min = 0.7 min). The time required for dissolution was not appreciably longer than this (dissolution was 50% complete by 0.67 min after IPTG addition, (F)), consistent with a lack of stalling.
(G–H) The experiment shown in (B–C) was repeated using p[lacOx16]. Synthesis was approximately linear until 6.17 minutes, at which point 81% of molecules had dissolved, further demonstrating that the majority of dissolution occurs without stalling of DNA synthesis.
Extended Data Figure 6. Replisome progression through 12x and 32x lacO arrays.
(A–D) To test whether replisomes meet later in a lacOx32 array than a lacOx12 array, we monitored dissolution. LacR Block-IPTG release was performed on p[lacOx12] and p[lacOx32] and radiolabelled termination intermediates were digested with XmnI to monitor the conversion of double-Y molecules to linear molecules (Dissolution). Cleaved molecules were separated on a native agarose gel, detected by autoradiography (A,C), and quantified (B,D). Upon IPTG addition, dissolution was delayed by at least 1 minute within the 32xlacO array compared to the 12xlacO array (B,D). Moreover, by 6 minutes, 92% of forks had undergone dissolution on p[lacOx12] while only 9% had dissolved on p[lacOx32] (B,D).
(E) Stall products within the 12xlacO array (Fig. 3B, Lane 2) were quantified, signal was corrected based on size differences of the products, and the percentage of stall products at each stall point was calculated. 78% of leading strands stalled at the first three arrest points (red columns), 19% stalled at the fourth to tenth arrest points (yellow columns) and the remaining 3% stalled at the tenth to fourteenth arrest points (grey columns). The appearance of fourteen arrest points reproducible and surprising, given that the presence of only twelve lacO sequences was confirmed by sequencing in the very preparation of p[lacOx12] that was used in Fig. 3. The thirteenth and fourteenth arrest points cannot stem from cryptic lacO sites beyond the twelfth lacO site, as this would position the first leftward leading strand stall product ∼90 nucleotides from the lacO array, instead of the observed ∼30 nucleotides (see F-G). At present, we do not understand the origin of these stall products.
(F–G) Progression of leftward leading strands into the array. The same DNA samples used in Fig. 3 were digested with the nicking enzyme Nb.BsrDI, which released leftward leading strands (F), and separated on a denaturing polyacrylamide gel (G). The lacO sites of p[lacOx12] are highlighted in blue on the sequencing ladder (G), which was generated using the primer JDO109 (Green arrow, F). Green circles indicate two non-specific products of digestion. These products arise because nicking enzyme activity varies between experiments, even under the same conditions. There was no significant difference in the pattern of leftward leading strand progression between the 12xlacO and 32xlacO arrays, as seen for the rightward leading strands (Fig. 3B). Specifically, by 5.67 minutes, the majority of leading strands had extended beyond the seventh lacO repeat within lacOx12 (lane 6) and the equivalent region of lacOx32 (lane 18). Therefore, progression of leftward leading strands is unaffected by the presence of an opposing replisome, suggesting that converging replisomes do not stall when they meet.
Extended Data Figure 7. Supplemental ChIP data.
(A) Cartoon depicting the LAC, FLK2 and FAR loci, which were used for ChIP. Their precise locations relative to the leftward edge of the lacO array are indicated. The LAC amplicon is present in four copies distributed across the lacOx16 array and three copies distributed across the lacOx12 array.
(B–E) p[lacOx12] was incubated with buffer or LacR and termination was induced at 5 minutes by IPTG addition. MCM7, RPA, CDC45, and Polε ChIP was performed at different time points after IPTG addition but also in the buffer control and no IPTG control. Recovery of FLK2 was measured as a percentage of input DNA. Upon IPTG addition, ChIP signal declined and by 9 minutes was comparable to the buffer control, demonstrating that unloading of replisomes was induced within 4 minutes of IPTG addition.
(F) To test whether movement of the replisome into and out of the lacO array could be detected upon IPTG addition, termination was monitored within a lacO array, and we performed ChIP of the leading strand polymerase Polε, which was inferred to move into and out of the array based on the behavior of leading strands during termination (Extended Data Fig 2B–E). It was predicted that Polε ChIP at the LAC locus should increase slightly as Polε enters the lacO array and decline again as converging polymerases pass each other, but persist at FLK2 while the polymerases move out of the array. Prior to IPTG addition, Polε was enriched at LAC and FLK2 compared to FAR, consistent with the leading strands being positioned on either side of the lacO array (Extended Data Fig 2C, Fig. 3). Upon IPTG addition, Polε became modestly enriched at LAC compared to FLK2 (5.5 min) but then declined to similar levels at both LAC and FLK2 by 6.5 min. These data are consistent with the leading strand polymerases entering the lacO array and passing each other.
(G–H) To test whether CMG exhibited the same ChIP profile as Polε, MCM7 and CDC45 ChIP was performed using the same samples. Following IPTG addition, MCM7 and CDC45 were enriched at LAC compared to FLK2 (5.5 min), then declined to similar levels at both LAC and FLK2 by 6.5 min, as seen for Polε (F). These data are consistent with a model in which CMGs enter the array and pass each other during termination. A caveat of these experiments is the relatively high recovery of the FAR locus in MCM7, CDC45, and Polε ChIP. Specifically, signal was at most only ∼2-fold enriched at LAC compared to FAR. This was not due to high background binding, because by the end of the experiment (10 minute time point, not shown), we observed a decrease in signal of ∼5–7 fold. Furthermore, we observed ∼5–7 fold enrichment in binding (ChIP) of replisome components to p[lacOx12] that had been incubated in LacR compared to a buffer control (see G-I, below). Instead, the high FAR signal was likely due to poor spatial resolution of the ChIP. Consistent with this, when a plasmid containing a DNA interstrand cross-link (ICL) was replicated, essentially all replisomes converged upon the ICL but the ChIP signal for MCM7 and CDC45 was only ∼3–4 fold enriched at the ICL compared to a control locus41. We speculate that the higher background observed at the control locus in our experiments is due to the decreased distance of the control locus from the experimental locus (1.3 kb for p[lacOx16] and p[lacOx12] vs. 2.4 kb for the ICL plasmid) and possibly due to increased catenation of the parental strands during termination. The high signal at FAR should not complicate interpretation of the MCM7, CDC45 and Polε ChIP (F), as signal at FAR was essentially unaltered between 5 and 6.5 minutes. Further evidence that the high signal seen at the FAR locus emanates from forks stalled near the lacO array is presented in panel (K).
(I) ChIP of RPA was performed on the same chromatin samples used in B-D. As seen for pol ɛ, MCM7, and CDC45, enrichment of RPA at LAC compared to FAR was relatively low, consistent with poor spatial resolution.
(J) Predicted binding of CMGs to the LAC, FLK2 and FAR loci before and after IPTG addition if CMGs converging CMG pass each other.
(K) To determine whether most forks stalled at the array and not elsewhere in the plasmid, we performed a time course in which p[lacOx16] undergoing termination was examined by 2-dimensional gel electrophoresis (2-D gel) at various time points. p[lacOx16] was pre-bound to LacR and replicated in Xenopus egg extract containing [α-32P]dATP. Termination was induced by IPTG addition and samples were withdrawn at different times. Radiolabelled replication intermediates were cleaved with XmnI (as in Extended Data Fig. 1A) and separated according to size and shape on 2-D gels50. A parallel reaction was performed in which samples were analyzed by ChIP, which was one of the repeats analyzed in (B)-(E). In the presence of LacR, a subset of Double Y molecules accumulated (blue arrowhead), demonstrating that 83% of replication intermediates (signal in dashed blue circle) contained two forks converged at a specific locus. Following IPTG addition, linear molecules rapidly accumulated (orange arrow) as dissolution occurred. Importantly, the vast majority of signal was present in the discrete double-Y and linear species (blue and orange arrows), demonstrating that the relatively high ChIP signal observed at FAR in panels F-I was derived from forks present at the lacOx16 array and not elsewhere.
Extended Data Figure 8. Supplemental termination data for p[empty] experiments.
(A) Cartoon depicting the XmnI and AlwNI sites on p[empty], which are used for the dissolution and ligation assays, respectively, and the FLK2 locus, which is used for ChIP.
(B) Plasmid DNA without a lacO array (p[empty]) was replicated and at different times chromatin was subjected to MCM7 and CDC45 ChIP. Percent recovery of FLK2 was quantified and used to measure dissociation of MCM7 and CDC45 (see methods). Dissolution and ligation were also quantified in parallel. mean±s.d. is plotted (n=3). The MCM7 and CDC45 dissociation data is obtained from the vehicle controls in Figure 5B–C, while the dissolution and ligation data are obtained from the vehicle controls in Fig 5D–E.
(C) To seek independent evidence for the conclusions of the ChIP data presented in Figure 5B–C, we used a plasmid pull-down procedure. p[empty] was replicated in egg extracts treated with Vehicle or Ub-VS. At the indicated times, chromatin-associated proteins were captured on LacR-coated beads (which binds DNA independently of lacO sites) and analyzed by Western blotting for CDC45, MCM7, and PCNA. CDC45 and MCM7 dissociated from chromatin by 8’ in the vehicle control, but persisted following UbVS treatment.
(D) To test whether the MCM7 modifications detected in panel (C) represented ubiquitylation, extracts were incubated with HIS6-Ubiquitin in the absence of Cyclin A, and in the absence or presence of plasmid DNA. After 15 minutes, HIS6-tagged proteins were captured by nickel resin pull down and blotted for MCM7. DNA replication greatly increased the levels of ubiquitylated MCM7, with the exception of a single species that was ubiquitylated independently of DNA replication (*). These data show that MCM7 is ubiquitylated during plasmid replication in egg extracts, as observed in yeast and during replication of sperm chromatin following nuclear assembly in egg extracts24,25.
(E) In parallel to the plasmid pull-downs performed in (C), DNA samples were withdrawn for dissolution, ligation, and decatenation assays, none of which were perturbed by UbVS treatment. These data support our conclusion, based on ChIP experiments (Fig. 5), that defective CMG unloading does not affect dissolution, ligation, or decatenation.
(F) Decatenation was measured in the same reactions used to measure Dissolution and Ligation (Fig. 5D–E), mean±s.d. is plotted (n=3).
(G–I) Given the experimental variability at the 4 minute time point in Figures 5D–F, the primary data and quantification for dissolution (G), ligation (H), and decatenation (I) for one of the three experiments summarized in Figure 5D–F is presented. This reveals that Ub-VS does not inhibit dissolution, ligation, or decatenation at the 4 minute time point. The same conclusion applies to two additional repetitions of this experiment (data not shown).
(J) The primary ChIP data used to measure dissociation of MCM7 and CDC45 in Fig 5B–C is shown. Recovery of FLK2 was measured. mean±s.d. is plotted (n=3).
Extended Data Figure 9. Tables of plasmids and oligonucleotides used.
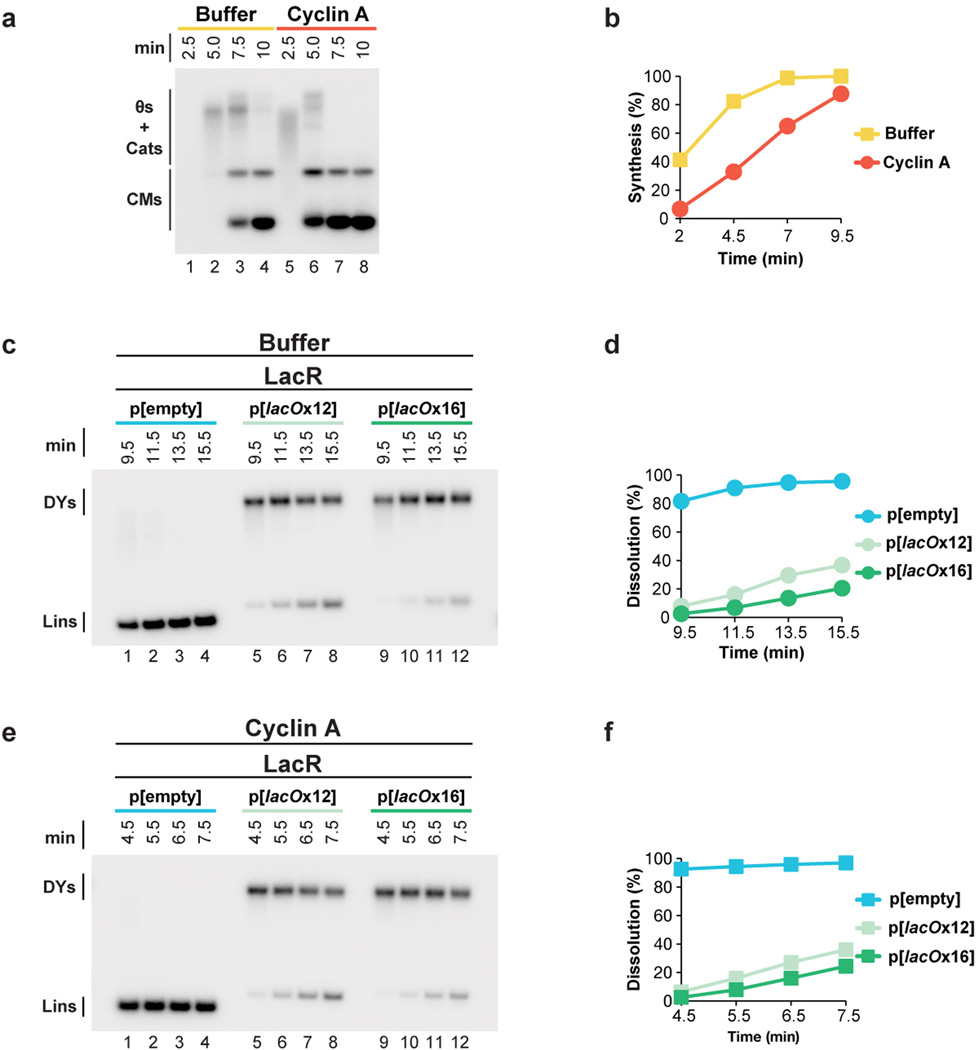
Extended Data Figure 10. Cyclin A treatment synchronizes DNA replication in Xenopus egg extracts.
(A–B) To synchronize DNA replication in Xenopus egg extracts, we treated extracts with Cyclin A, which probably accelerates replication initiation45. Plasmid DNA was incubated in High Speed Supernatant for 20 minutes, then either buffer or Cyclin A was added for a further 20 minutes. NucleoPlasmic extract was added to initiate DNA replication, along with [α-32P]dATP to label replication intermediates. Replication products were separated on a native agarose gel, detected by autoradiography (A), and quantified (B). In the presence of vehicle, replication was not complete by 9.5 minutes, but in the presence of Cyclin A, replication was almost complete by 4.5 minutes (B). Thus, Cyclin A treatment approximately doubles the speed of DNA replication in Xenopus egg extracts.
(C–F) To test whether Cyclin A affects the ability of LacR to inhibit termination, we monitored dissolution of plasmids containing a 12x or 16x LacR array in the presence and absence of Cyclin A. p[lacOx12], p[lacOx16], and the parental control plasmid p[empty] were incubated with LacR, and then treated with buffer or Cyclin A before replication was initiated with NPE in the presence of [α-32P]dATP. Samples were withdrawn when dissolution of p[empty] plateaued (9.5 minutes in the presence of buffer, 4.5 minutes in the presence of Cyclin A). Given that Cyclin A treatment approximately doubles the speed of replication (see B), samples were withdrawn from these reactions twice as frequently as the buffer-treated samples. To measure dissolution, radiolabelled termination intermediates were cut with XmnI to monitor the conversion of double-Y molecules to linear molecules. Cut molecules were separated on a native agarose gel and detected by autoradiography (C,E). By the time the first sample was withdrawn, dissolution of p[empty] was essentially complete, in the absence (9.5 min, D) or presence (4.5 min, F) of Cyclin A. Importantly, dissolution of p[lacOx12] and p[lacOx16] was prevented in the absence (9.5 min, D) or presence (4.5 min, F) of Cyclin A. Moreover, dissolution occurred approximately twice as fast in the presence of Cyclin A (note the similarity between (D) and (F) even though samples are withdrawn twice as frequently in F.) consistent with replication being approximately twice as fast in the presence of Cyclin A. Therefore, Cyclin A does not affect the ability of a LacR array to block replication forks.
Acknowledgements
We thank C. Richardson and members of the Walter laboratory for feedback on the manuscript. We thank K.J Marians and J.T. Yeeles for plasmids and the LacI purification protocol. J.C.W was supported by NIH grant GM62267 and GM80676. J.C.W is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplementary information line:
Supplementary information accompanies this paper on www.nature.com/nature.
Author Contributions
J.M.D and J.C W designed the experiments. J.M.D performed the experiments. M.B. developed methodologies for plasmid pull downs and HIS6-Ub immunoprecipitations. J.M.D and J.C.W interpreted the data and wrote the paper.
The authors declare no competing financial interests.
References
- 1.Levine AJ, Kang HS, Billheimer FE. DNA replication in SV40 infected cells IAnalysis of replicating SV40 DNA. J. Mol. Biol. 1970;50:549–568. doi: 10.1016/0022-2836(70)90211-1. [DOI] [PubMed] [Google Scholar]
- 2.Fanning E, Zhao K. SV40 DNA replication: from the A gene to a nanomachine. Virology. 2009;384:352–359. doi: 10.1016/j.virol.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tapper DP, DePamphilis ML. Discontinuous DNA replication: Accumulation of simian virus 40 DNA at specific stages in its replication. J. Mol. Biol. 1978;120:401–422. doi: 10.1016/0022-2836(78)90427-8. [DOI] [PubMed] [Google Scholar]
- 4.Seidman MM, Salzman NP. Late replicative intermediates are accumulated during simian virus 40 DNA replication in vivo and in vitro. J. Virol. 1979;30:600–609. doi: 10.1128/jvi.30.2.600-609.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sundin O, Varshavsky A. Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell. 1980;21:103–114. doi: 10.1016/0092-8674(80)90118-x. [DOI] [PubMed] [Google Scholar]
- 6.Ishimi Y, Sugasawa K, Hanaoka F, Eki T, Hurwitz J. Topoisomerase II plays an essential role as a swivelase in the late stage of SV40 chromosome replication in vitro. J. Biol. Chem. 1992;267:462–466. [PubMed] [Google Scholar]
- 7.Hiasa H, Marians KJ. Two distinct modes of strand unlinking during theta-type DNA replication. J. Biol. Chem. 1996;271:21529–21535. doi: 10.1074/jbc.271.35.21529. [DOI] [PubMed] [Google Scholar]
- 8.Espeli O, Levine C, Hassing H, Marians KJ. Temporal regulation of topoisomerase IV activity in E. coli. Mol. Cell. 2003;11:189–201. doi: 10.1016/s1097-2765(03)00013-3. [DOI] [PubMed] [Google Scholar]
- 9.Segawa M, Sugano S, Yamaguchi N. Association of simian virus 40 T antigen with replicating nucleoprotein complexes of simian virus 40. J. Virol. 1980;35:320–330. doi: 10.1128/jvi.35.2.320-330.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tack LC, DePamphilis ML. Analysis of simian virus 40 chromosome-T-antigen complexes: T-antigen is preferentially associated with early replicating DNA intermediates. J. Virol. 1983;48:281–295. doi: 10.1128/jvi.48.1.281-295.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen MC, Birkenmeier E, Salzman NP. Simian virus 40 DNA replication: characterization of gaps in the termination region. J. Virol. 1976;17:614–621. doi: 10.1128/jvi.17.2.614-621.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sundin O, Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell. 1981;25:659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- 13.Ivessa AS, Zhou JQ, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 14.Steinacher R, Osman F, Dalgaard JZ, Lorenz A, Whitby MC. The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability. Genes Dev. 2012;26:594–602. doi: 10.1101/gad.184663.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fachinetti D, et al. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol. Cell. 2010;39:595–605. doi: 10.1016/j.molcel.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiNardo S, Voelkel K, Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc. Natl. Acad. Sci. U. S. A. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol. Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 18.Lucas I, Germe T, Chevrier-Miller M, Hyrien O. Topoisomerase II can unlink replicating DNA by precatenane removal. EMBO J. 2001;20:6509–6519. doi: 10.1093/emboj/20.22.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaggioli V, Le Viet B, Germe T, Hyrien O. DNA topoisomerase II controls replication origin cluster licensing and firing time in Xenopus egg extracts. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2–7 helicase by association with Cdc45 and GINS proteins. Mol. Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 22.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang YH, Galal WC, Farina A, Tappin I, Hurwitz J. Properties of the human Cdc45/Mcm2–7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2012 doi: 10.1073/pnas.1203734109. at < http://www.pnas.org/content/early/2012/03/29/1203734109.long> . [DOI] [PMC free article] [PubMed]
- 24.Maric M, Maculins T, De Piccoli G, Labib K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science (80–.) 2014;346:1253596–1253596. doi: 10.1126/science.1253596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Priego Moreno S, Bailey R, Campion N, Herron S, Gambus A. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science (80–.) 2014;346:477–481. doi: 10.1126/science.1253585. [DOI] [PubMed] [Google Scholar]
- 26.Hiasa H, Marians KJ. Tus prevents overreplication of oriC plasmid DNA. J. Biol. Chem. 1994;269:26959–26968. [PubMed] [Google Scholar]
- 27.Rudolph CJ, Upton AL, Stockum A, Nieduszynski Ca, Lloyd RG. Avoiding chromosome pathology when replication forks collide. Nature. 2013;500:608–611. doi: 10.1038/nature12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hook SS, Lin JJ, Dutta A. Mechanisms to control rereplication and implications for cancer. Curr. Opin. Cell Biol. 2007;19:663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Czajkowsky DM, Liu J, Hamlin JL, Shao Z. DNA combing reveals intrinsic temporal disorder in the replication of yeast chromosome VI. J. Mol. Biol. 2008;375:12–19. doi: 10.1016/j.jmb.2007.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGuffee SR, Smith DJ, Whitehouse I. Quantitative, genome-wide analysis of eukaryotic replication initiation and termination. Mol. Cell. 2013;50:123–135. doi: 10.1016/j.molcel.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol. Cell. 2010;40:834–840. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loveland AB, Habuchi S, Walter JC, van Oijen AM. A general approach to break the concentration barrier in single-molecule imaging. Nat. Methods. 2012;9:987–992. doi: 10.1038/nmeth.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santamaría D, et al. Bi-directional replication and random termination. Nucleic Acids Res. 2000;28:2099–2107. doi: 10.1093/nar/28.10.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, et al. DNA interstrand cross-link repair requires replication fork convergence. Nat. Struct. Mol. Biol. In Press doi: 10.1038/nsmb.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duxin JPP, Dewar JMM, Yardimci H, Walter JCC. Replication-coupled repair of a DNA-protein crosslink. Cell. 2014;159:346–357. doi: 10.1016/j.cell.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- 37.Sofueva S, et al. Ultrafine anaphase bridges, broken DNA and illegitimate recombination induced by a replication fork barrier. Nucleic Acids Res. 2011;39:6568–6584. doi: 10.1093/nar/gkr340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charbin A, Bouchoux C, Uhlmann F. Condensin aids sister chromatid decatenation by topoisomerase II. Nucleic Acids Res. 2014;42:340–348. doi: 10.1093/nar/gkt882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laskey RA, Mills AD, Morris NR. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977;10:237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- 40.Räschle M, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long DTT, Joukov V, Budzowska M, Walter JCC. BRCA1 Promotes Unloading of the CMG Helicase from a Stalled DNA Replication Fork. Mol. Cell. 2014;56:174–185. doi: 10.1016/j.molcel.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu YV, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costa A, et al. The structural basis for MCM2–7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaplan DL, Davey MJ, O’Donnell M. Mcm4,6,7 uses a ‘pump in ring’ mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J. Biol. Chem. 2003;278:49171–49182. doi: 10.1074/jbc.M308074200. [DOI] [PubMed] [Google Scholar]
- 45.Prokhorova TA, Mowrer K, Gilbert CH, Walter JC. DNA replication of mitotic chromatin in Xenopus egg extracts. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13241–13246. doi: 10.1073/pnas.2336104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lebofsky R, Takahashi T, Walter JC. DNA replication in nucleus-free Xenopus egg extracts. Methods Mol. Biol. 2009;521:229–252. doi: 10.1007/978-1-60327-815-7_13. [DOI] [PubMed] [Google Scholar]
- 47.Budzowska M, Graham TGW, Sobeck A, Waga S, Walter JC. Regulation of the Rev1-pol ζ complex during bypass of a DNA interstrand cross-link. EMBO J. In Press doi: 10.15252/embj.201490878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magnaghi P, et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat. Chem. Biol. 2013;9:548–556. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- 49.Walter JC, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
Additional Reference
- 50.Long DT, Räschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]