Abstract
Herein we identify and analyze helical protein interfaces as potential targets for synthetic modulators of protein–protein interactions.
Selective modulation of protein–protein interactions is a grand challenge for chemical biologists and medicinal chemists.1 Protein interfaces are often composed of large shallow surfaces rendering them difficult targets for typical small molecule drugs.2–4 A broad effort to develop new classes of protein–protein interaction inhibitors has focused on the fundamental role played by short folded domains, or protein secondary structures, at protein interfaces.3
α-Helices constitute the largest class of protein secondary structure and mediate many protein interactions.5,6 Helices located within the protein core are vital for the overall stability of protein tertiary structure, whereas exposed α-helices on protein surfaces constitute central bioactive regions for the recognition of numerous proteins, DNAs and RNAs. Fig. 1 displays a selection of complexes in which a helical domain targets a biomolecule. Peptides composed of less than fifteen amino acid residues do not generally form α-helical structures at physiological conditions once excised from the protein environment; much of their ability to specifically bind their intended targets is potentially lost because they adopt an ensemble of conformations rather than the biologically relevant one.7,8 Synthetic strategies that either stabilize short peptides (<15 residues) into α-helical conformations or mimic this domain using non-natural scaffolds are expected to be useful models for the design of bioactive molecules and for studying aspects of protein folding.7–10
Fig. 1.
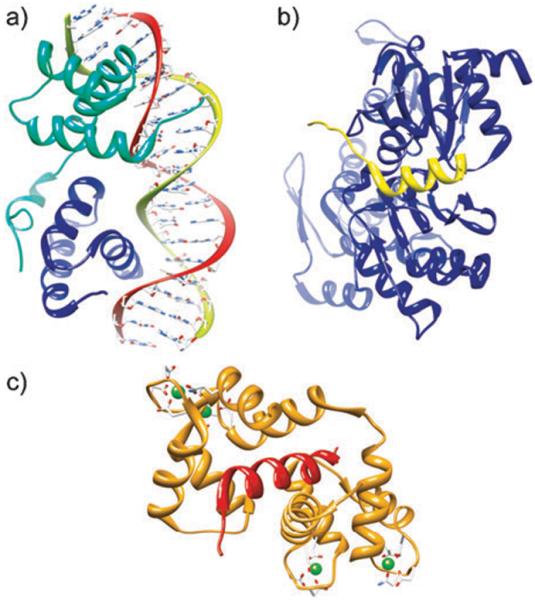
The α-helix is a ubiquitous element in biomolecular recognition. (a) MATa1/MATα2-3A heterodimer bound to DNA (PDB code: 1LE8), (b) complex of the WH2 domain of WAVE with Actin-DNAse I (PDB code: 2A40), (c) endothelial nitric oxide synthase peptide bound to calmodulin (PDB code: 1NIW).
Several classes of helix mimetics have been described by the synthetic organic chemistry community (Fig. 2),7–10 but the use of helix mimetics in biology has been limited to a set of model protein complexes. We attribute the restricted use of these mimetics to the lack of a systematic method for identifying helical protein interfaces that may be targeted by the various classes of stabilized helices and synthetic helix mimetics. In this report, we address this limitation by comprehensively evaluating protein–protein complexes as potential targets for helix mimetics. We expect this study to offer an invaluable starting point for the chemical biology community to develop synthetic inhibitors of protein–protein interactions.
Fig. 2.
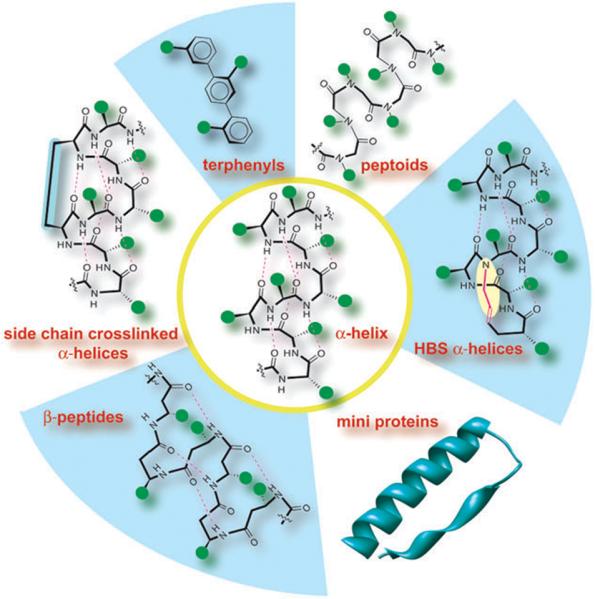
Stabilized helices and non-natural helix mimetics: several strategies that stabilize the R-helical conformation in peptides or mimic this domain with non-natural scaffolds have been described. Efforts include β-peptide helices, terphenyls, miniproteins, peptoids and side-chain and backbone (hydrogen bond surrogate, HBS) crosslinked α-helices.7–10
We were inspired in our undertaking by previous studies to determine the number and class of protein drug targets wherein it was determined that less than 400 druggable domains cover all current drug targets—a number that compares poorly with the projected number of protein families.11 Here we assess the available data on protein–protein complexes with helical interfaces from the Protein Data Bank (PDB, http://www.pdb.org).12 Our endeavor has a dual purpose: to provide a dataset for the chemical biology community representing the variety and number of targets available for helix mimetics, and to examine the nature of helices that appear in interface proteins. Although several examinations of protein–protein interactions have been performed,13 our study is unique in affording a very focused view of interfaces involving a single class of protein secondary structure.
This study identifies a dataset of helical protein–protein complexes (Fig. 3). Atomic coordinates were obtained from the Protein Data Bank for multi-protein complexes. Identification of interface residues and secondary structure was performed using the Rosetta suite of programs.14 Rosetta determines secondary structure by calculating the ϕ and φ angles of the protein backbone. We define a helical segment as one that contains at least four contiguous residues with ϕ and φ angles that are characteristic of an α-helix. An interface residue is defined as (i) a residue that has at least one atom within a 5 Å radius of an atom belonging to a binding partner in the protein complex, or (ii) a residue that becomes significantly buried upon complex formation, as measured by the density of Cβ atoms within a sphere with a radius of 8 Å around the Cβ atom of the residue of interest. We include a detailed explanation of methods used in this study in the ESI.‡ Those structures with helical interfaces in the protein–proteinz (HIPP) interaction were analyzed for helix length and classified according to function.
Fig. 3.
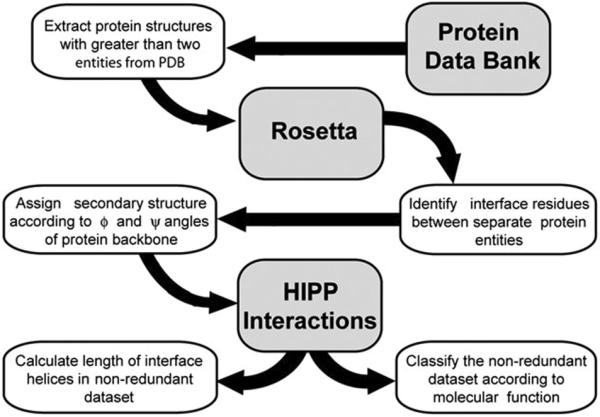
Evaluation of structures from the Protein Data Bank to identify and assess helical interfaces in protein–protein (HIPP) interactions.
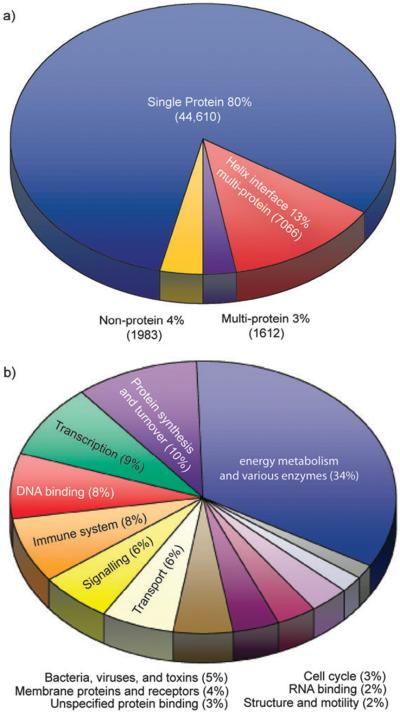
The PDB contains more than 55 000 structures.12 Approximately 80% of these structures contain a single protein entity and 4% contain no protein entities. The remaining 16%, or about 8678 structures, contain more than two separate protein entities and form the dataset for evaluation of HIPP interactions (Fig. 4a). Analysis of this dataset revealed that 13% contained HIPP interactions. These complexes may also contain other secondary motifs, but the current study focuses solely on the helical portions. The 7066 HIPP complexes contain considerable redundancy in sequence and structure owing to the redundancy in the PDB. We removed structures with greater than 95% sequence similarity with the CD-HIT algorithm15 to obtain a better understanding of the types of complexes involved in HIPP interactions. This screen provided a non-redundant dataset of 1658 HIPP interactions for analysis. The PDB codes for interactions from the complete and non-redundant datasets are listed in the ESI.‡
Fig. 4.
(a) Fraction of the current Protein Data Bank involved in helical interfaces. (b) Classification of proteins displaying helical interfaces by function.
We categorized the HIPP interactions in the non-redundant dataset according to function as defined in the PDB (Fig. 4b). Some HIPP interactions could fall into more than one function category, however, we limited each HIPP interaction to one category. Helical interfaces are involved in a wide distribution of functions ranging from enzymatic activity to protein associations. The largest category, energy metabolism and various enzymes, accounts for 34% of HIPP interactions. This category contains many hydrolases, oxidoreductases and transferases, among other enzymes. The protein synthesis and turnover category contains chaperones, proteosomes, ribosomes and other proteins involved in protein synthesis. The transcription category contains proteins that are either involved in transcription regulation, such as activators or repressors, or are part of the transcription machinery, such as those that bind to DNA. The DNA binding category contains proteins that target DNA but are not involved in transcription.
The length of each helix participating at the interface in the complexes in the non-redundant dataset was examined (Fig. 5). Helix length was calculated as the total length of polypeptide chain within the helical segment. Thus, we include the full length of the helix including residues that may not be part of the interface. Our analysis indicates that helices involved in protein interactions range from four residues to 99 residues. The average helix length is fourteen residues with ten residues appearing the most often. The number of helix residues directly engaged in binding has been assessed previously by examining 122 homodimers and 204 protein–protein heterocomplexes.5 This study implicated an average helix length of seven residues in binding.5 Together, these studies emphasize the short length of the helical domain involved in protein interactions.
Fig. 5.
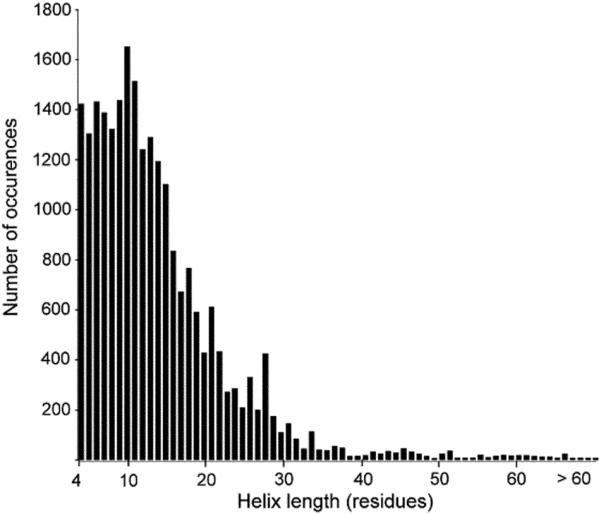
Distribution of helix length at protein interfaces. The present analysis suggests an average length of 14 residues for helices participating in interfaces.
Our study reveals new classes of previously unidentified targets for helix mimetics. Some of these newly identified targets will potentially aid efforts in drug discovery. In particular, it is interesting to note that our study identifies a number of kinases that may be regulated by helix mimetics; a representative list is shown in Table 1. Kinases are an important class of potential drug targets. Typically, kinase inhibitors mimic ATP or substrate conformations. New types of scaffolds that can specifically regulate the function of therapeutically important kinases will fill an important gap in a medicinal chemist's repertoire.16
Table 1.
Representative list of kinases that may be regulated by helix mimetics
| PDB Code | Title |
|---|---|
| 1BLX | P19INK4D/CDK6 complex |
| 1OW6 | Paxillin LD4 motif bound to the focal adhesion targeting domain of the focal adhesion kinase |
| 1WMH | Crystal structure of a PB1 domain complex of protein kinase of CIOTA and PAR6 alpha |
| 1YJ5 | Molecular architecture of mammalian polynucleotide kinase, a DNA repair enzyme |
| 2A19 | PKR kinase domain EIF2 alpha AMP-PNP complex |
| 2CH4 | Complex between bacterial chemotaxis histidine kinase CHEA domains P4 and P5 and adaptor-protein CHEW |
| 2EHB | Structure of the C-terminal domain of the protein kinase ATSOS2 bound to the calcium sensor ATSOS3 |
| 2GIT | Crystal structure of human calmodulin-dependent protein kinase I G |
| 2NPT | Structure of the human mitogen activated protein kinase 5 PHOX domain with protein kinase 2 PHOX domain |
In summary, we have identified and analyzed helical interfaces in protein–protein interactions. We undertook this study to address the significant chasm between the elegant design of helix mimetics and their sporadic use in biology. This study provides an exhaustive list of potential targets for emerging classes of helix mimetics.
Supplementary Material
Acknowledgments
This work was supported by an NIH Grant (GM073943) and an allocation of advanced computing resources supported by the National Science Foundation (CHE090016). The computations were performed in part on the TeraGrid Purdue Steele system.
Footnotes
This article is part of the 2009 Molecular BioSystems `Emerging Investigators' issue: highlighting the work of outstanding young scientists at the chemical- and systems-biology interfaces
Electronic supplementary information (ESI) available: Description of methods, full and non-redundant lists of PDB codes for helical protein interfaces.
References
- 1.Wells JA, McClendon CL. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 2.Argos P. Protein Eng. 1988;2:101–113. doi: 10.1093/protein/2.2.101. [DOI] [PubMed] [Google Scholar]
- 3.Miller S. Protein Eng. 1989;3:77–83. doi: 10.1093/protein/3.2.77. [DOI] [PubMed] [Google Scholar]
- 4.Lo Conte L, Chothia C, Janin J. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 5.Guharoy M, Chakrabarti P. Bioinformatics. 2007;23:1909–1918. doi: 10.1093/bioinformatics/btm274. [DOI] [PubMed] [Google Scholar]
- 6.Jones S, Thornton JM. Prog. Biophys. Mol. Bio. 1995;63:31–65. doi: 10.1016/0079-6107(94)00008-w. [DOI] [PubMed] [Google Scholar]
- 7.Henchey LK, Jochim AL, Arora PS. Curr. Opin. Chem. Biol. 2008;12:692–697. doi: 10.1016/j.cbpa.2008.08.019. and references therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garner J, Harding MM. Org. Biomol. Chem. 2007;5:3577–3585. doi: 10.1039/b710425a. and references therein. [DOI] [PubMed] [Google Scholar]
- 9.Davis JM, Tsou LK, Hamilton AD. Chem. Soc. Rev. 2007;36:326–334. doi: 10.1039/b608043j. [DOI] [PubMed] [Google Scholar]
- 10.Murray JK, Gellman SH. Biopolymers. 2007;88:657–686. doi: 10.1002/bip.20741. [DOI] [PubMed] [Google Scholar]
- 11.Overington JP, Al-Lazikani B, Hopkins AL. Nat. Rev. Drug Discovery. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 12.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keskin Z, Gursoy A, Ma B, Nussinov R. Chem. Rev. 2008;108:1225–1244. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
- 14.Kuhlman B, Baker D. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10383–10388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Jaroszewski L, Godzik A. Bioinformatics. 2001;17:282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 16.Fedorov O, Sundstrom M, Marsden B, Knapp S. Drug Discovery Today. 2007;12:365–372. doi: 10.1016/j.drudis.2007.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.