Abstract
Time or age-dependent accumulation of mitochondrial damage and dysfunction is strongly associated with aging [1]. Thus, a major biomedical goal is to identify and therapeutically manipulate those inherent programs that protect against mitochondrial dysfunction to promote cell survival and organismal health. The mitochondrial unfolded protein response (UPRmt) is such a protective transcriptional response mediated by mitochondrial-to-nuclear signaling that includes mitochondrial proteostasis genes to stabilize mitochondrial function, metabolic adaptations, as well as an innate immunity program. Here, we review the UPRmt and its role during a variety of forms of mitochondrial dysfunction including those caused by mutations in respiratory chain genes as well as upon exposure to pathogens that produce mitochondrial toxins. We also review recent data in support of and against the emerging role of the UPRmt during aging and longevity.
Keywords: Mitochondrial dysfunction, ATFS-1, mitochondrial unfolded protein response, metabolism, innate immunity
1. Introduction
Mitochondria are essential organelles present in nearly all eukaryotic cells best known for their roles in energy metabolism including the tricarboxylic acid cycle (TCA) and oxidative phosphorylation (OxPhos). However, mitochondria also contribute to many other essential cellular processes including nucleotide and amino acid synthesis, iron-sulfur cluster biogenesis, as well as calcium homeostasis and apoptosis. Mitochondria are dynamic double membrane bounded organelles, which divide and fuse throughout their lifetime [2]. Each mitochondrion is comprised of four discrete compartments: the outer membrane, intermembrane space, inner membrane and matrix. The large surface area of the inner membrane allows for cristae formation that accommodates the respiratory chain complexes and ATP synthase promoting the coupling of electron transport and ATP synthesis during OxPhos [3, 4].
Mitochondria are comprised of over 1000 proteins, most of which are encoded by nuclear genes and translated on cytosolic ribosomes prior to import into mitochondria [5]. However, thirteen respiratory chain and ATP synthase components are encoded by the mitochondrial genome (mtDNA) and translated on mitochondrial ribosomes prior to assembly into the OxPhos complexes. The overall architecture of the compartment coupled with the proximity to the reactive oxygen emitting respiratory chain presents considerable challenges to organelle maintenance.
Given the central importance of these organelles in eukaryotic physiology and the challenges in maintaining their optimal function, it is perhaps not surprising that a number of components and signaling pathways have been identified that respond to mitochondrial dysfunction and promote organelle maintenance and recovery, while adapting metabolism to maintain survival. A number of mechanisms are in place to ensure membrane and protein quality including a localized cadre of molecular chaperones, quality control proteases, and anti-oxidant enzymes which promote protein folding and stability while degrading those proteins that fail to fold or assemble [6–8]. In turn, these activities prevent the accumulation of misfolded or potentially toxic damaged proteins maintaining the compartmentalized protein homeostasis essential for mitochondrial function.
A general decline in mitochondrial function has been well documented to occur as organisms age [1]. Those cells that are especially energetic, such as neurons and muscle cells, are most affected [9], but reports of mitochondrial decline have been reported in most cell types. The exact underlying lesion that causes perturbed mitochondrial function are unclear in most cases but mtDNA mutation or deletion accumulation [10], an increase in oxidative damage [11] and/or aggregation of mitochondrial proteins [12], and alterations in mitochondrial morphology have been documented [13]. Presumably, the pathways and machineries in place to protect mitochondrial function are eventually overwhelmed by the prolonged stress leading to mitochondrial dysfunction and the associated pathology including neurodegeneration, muscular and ocular defects as well as metabolic disorders including diabetes [14].
Despite the central importance of mitochondrial function to nearly all cells and the strong link between mitochondrial dysfunction and disease, it has been clearly demonstrated that in a variety of species modest levels of mitochondrial dysfunction leads to increased longevity [15–18]. While the underlying cellular alterations that potentially include metabolic adaptations and mitochondrial maintenance are unclear, increasing evidence points towards a mitochondrial stress response pathway known as the mitochondrial unfolded protein response (UPRmt) having a prominent protective role.
2. Cellular processes affected by UPRmt activation
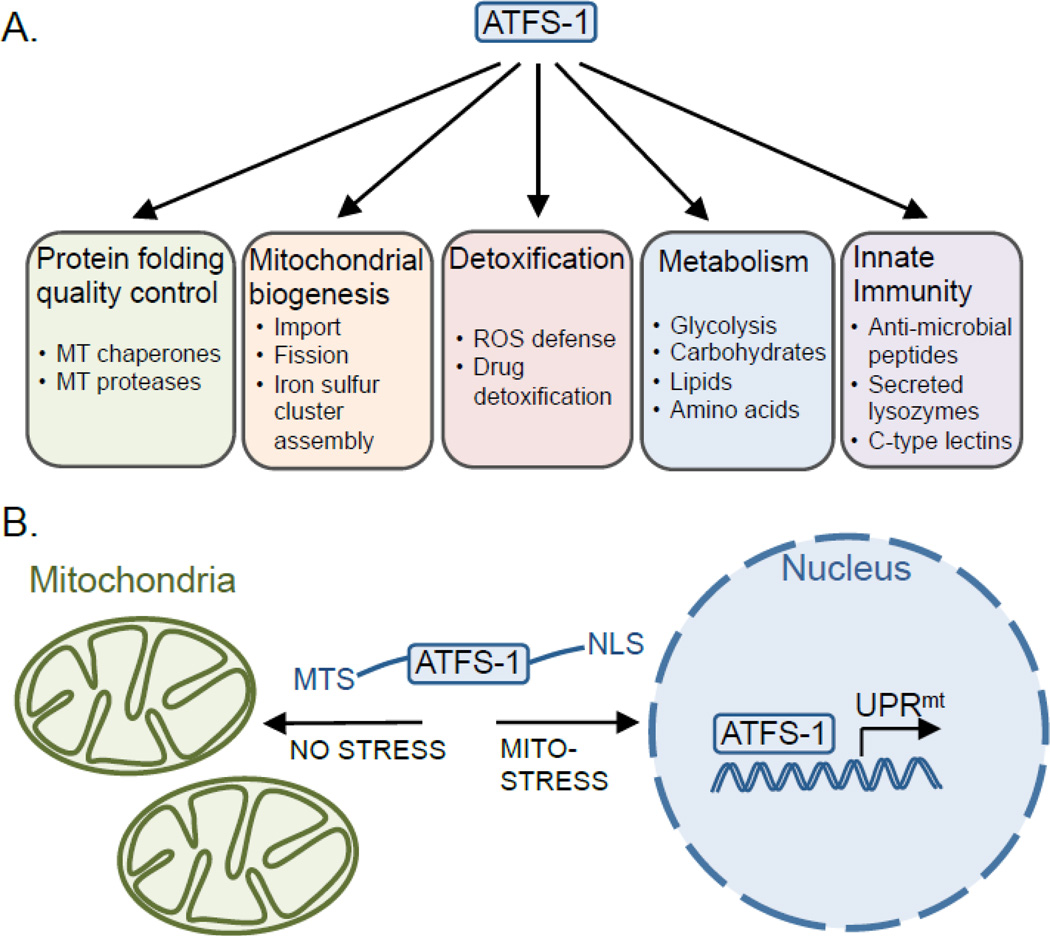
The UPRmt was initially documented in mammalian cell culture as a response to depleted mtDNA or the accumulation of misfolded proteins within the mitochondrial matrix that resulted in increased mitochondrial chaperone and protease transcription to alleviate and promote the recovery from mitochondrial stress [19–21]. More recent work in C. elegans has identified a number of components required for UPRmt activation [22–25], which has suggested a mechanism by which the cell senses mitochondrial dysfunction [26] and for the identification of over 400 genes induced during mitochondrial dysfunction [27, 28], which we have attempted to categorize below (Figure 1A).
Figure 1. UPRmt activation via ATFS-1 regulates a broad transcriptional program.
(A) In response to mitochondrial damage or stress ATFS-1 induces genes involved in mitochondrial repair mechanisms including protein folding and protein quality control, as well as those involved in mitochondrial biogenesis, the detoxification response, metabolism and innate immune gene transcription. (B) The UPRmt is activated during conditions such as mtDNA depletion, respiratory chain dysfunction, increased ROS or increased mitochondrial unfolded proteins, which is regulated by the mitochondrial protein-import efficiency of the transcription factor ATFS-1. In the absence of stress, ATFS-1 localizes to mitochondria via its mitochondria targeting signal (MTS), where it is degraded by the protease Lon. However, during mitochondrial dysfunction or stress, general mitochondrial protein import is attenuated, leading to the accumulation of a portion of ATFS-1 in the cytosol, followed by its translocation to the nucleus via its nuclear localization signal (NLS). In the nucleus, ATFS-1 induces the mitochondrial protective genes described in Figure 1A, which promote survival and recovery from mitochondrial stress.
2.1 Stabilization of mitochondrial function
The increase in the mitochondrial chaperones during stress including the matrix-localized chaperones Hsp60 and mtHsp70 promotes the folding of recoverable proteins, while the increase of proteases including the i-AAA and m-AAA protease removes proteins that fail to fold or assemble. Additionally, the UPRmt includes numerous anti-oxidant genes including a mitochondrial superoxide dismutase and genes involved in glutathione metabolism that limit the protein and membrane perturbations caused by ROS emitted from defective respiratory chains [27]. Increased transcription of protein homeostasis and anti-oxidant genes potentially stabilizes the protein-folding environment to promote organelle function but also prepares for the recovery or regeneration of those salvageable organelles while irreparable organelles are degraded via mitophagy [29–32].
2.2 Metabolic adaptations
Interestingly, the UPRmt also includes multiple glycolysis genes and lactate dehydrogenase suggesting that cells may shift to oxidative glycolysis during respiratory chain and mitochondrial dysfunction [25, 27]. This metabolic adaptation would allow cells to generate ATP from a pathway localized within the cytosol and less likely to be affected by mitochondrial dysfunction. Increased glycolysis allows cells to maintain cellular energy levels to promote normal cellular functions but may also provide the energy required to recover efficient mitochondrial activity. In addition to glycolysis, the UPRmt also includes genes involved in amino acid and additional carbohydrate metabolisms [27] suggesting that considerable and complicated metabolic alterations occur during mitochondrial stress.
2.3 Recovery of mitochondrial function
A separate set of genes induced during mitochondrial stress suggests that UPRmt activation promotes the recovery of mitochondrial function by regenerating and rebuilding the respiratory chain and ATP synthase. Iron-sulfur clusters are essential cofactors synthesized in the mitochondrial matrix [33] required for respiratory chain function but also numerous other intracellular activities such as DNA repair [34]. The UPRmt includes most of the iron-sulfur cluster biogenesis genes suggesting that maintaining iron-sulfur cluster biogenesis is important during mitochondrial stress. In addition to respiratory chain cofactors, the UPRmt also includes multiple respiratory complex assembly factors, which facilitate the assembly of specific respiratory complexes [35, 36] as well as most genes required for ubiquinone biosynthesis, which is involved in electron transport within the respiratory chain [37].
Additionally, the UPRmt includes the mitochondrial RNA polymerase required to transcribe the mtDNA-encoded respiratory chain and ATP synthase genes as well as the core components of the translocase of the inner membrane (TIM), which is required for the import of proteins across the inner mitochondrial membrane [37]. Furthermore, the UPRmt includes most of the mitochondrial fission machinery such as dynamin-related protein (Drp1) but none of the mitochondrial fusion machinery. Mitochondrial fission is required to eliminate severely defective mitochondrial portions [38], but is also required to increase mitochondrial number [2, 39].
Of course, the expression of many additional genes is altered during mitochondrial stress or dysfunction, which are beyond the focus of this review.
2.4 Conditions that activate the UPRmt
A number of mitochondrial perturbations have been found to activate the UPRmt including the above mentioned mtDNA depletion [19, 40] and the accumulation of unfolded proteins within the mitochondrial matrix [21] or intermembrane space [41]. An early RNAi screen demonstrated that inhibition of respiratory chain or ATP synthase components expression was sufficient to trigger the UPRmt [40], which is consistent with activation via mtDNA depletion. More recently, it has been shown that respiratory chain and mitochondrial ribosome gene mutations also activate the UPRmt [6, 27, 42–44] suggesting that an imbalance between mtDNA-encoded and nuclear-encoded OxPhos components is an initiating event in UPRmt activation [44, 45]. Additionally, inhibition of mitochondrial proteases, mitochondrial chaperones, respiratory chain complex assembly factors [46] and mitochondrial tRNA synthetases [47] cause UPRmt activation. Lastly, exposure to a number of respiratory chain inhibitors such as antimycin [48, 49], rotenone and paraquat [27, 40, 49] all activate the UPRmt. The variety of defects within mitochondria that activate the UPRmt suggests that cells likely monitor some aspect of mitochondrial function reliant on multiple mitochondrial activities to initiate the stress response [26, 45, 48].
3. Regulation of the UPRmt
3.1 UPRmt signaling in C. elegans
RNAi screens in C. elegans have identified a number of components required for UPRmt activation [22, 23, 48, 49] allowing the initiation of studies to understand the signaling mechanism by which the status of the mitochondria is transmitted to the nucleus to coordinate adaptive transcription. The only transcription factor identified by screens from three different labs is Activating Transcription Factor associated with Stress-1 or ATFS-1 (originally described as ZC376.7) [24, 48, 49].
Interestingly, ATFS-1 contains a mitochondrial targeting sequence (MTS) in addition to a nuclear localization signal (NLS) within the bZip domain suggesting a unique mechanism of communication between both compartments (Figure 1B). In short, the evaluation of mitochondrial function or dysfunction is based on mitochondrial protein import efficiency; a process that requires mitochondrial chaperones, an efficiently functioning respiratory chain, ATP and intact TIM and TOM (Translocase of the Inner or Outer Membrane) complexes [50]. Like nearly all transcription factors, ATFS-1 has a NLS located near the carboxy-terminus that allows for its nuclear localization, whereas its MTS, located at the amino-terminus, also allows for its import into mitochondria. Normally, the MTS is dominant and ATFS-1 is efficiently imported from the cytosol to the mitochondrial matrix where the MTS is cleaved and it is degraded by the Lon protease. However, if import efficiency is impaired by any of the described mitochondrial defects, a percentage of ATFS-1 accumulates in the cytosol and, because it has a NLS, then traffics to the nucleus to activate the UPRmt [27]. Thus, the cell monitors mitochondrial import efficiency or capacity of the entire cellular pool of organelles. If the capacity is not sufficient to import all ATFS-1, the UPRmt is induced to rectify what is perceived to be deficiency in mitochondrial function [26].
In addition to ATFS-1, a number of additional components have been identified in C. elegans that are required for UPRmt activation including the homeobox transcription factor DVE-1, the ubiquitin-like protein UBL-5, the mitochondrial protease ClpP and the mitochondrial peptide transporter HAF-1, which have been reviewed elsewhere [45, 51, 52]. Understanding how each of these components regulates the UPRmt and how they interact with ATFS-1 is an active area of research. Data from our lab suggests that ClpP and HAF-1 function upstream of ATFS-1 to regulate the UPRmt [24, 27]. HAF-1, which transports peptides from the matrix to the intermembrane space, similarly to the yeast protein Mdl1p [53], functions as a negative regulator of mitochondrial protein import although the mechanism is unknown [26, 27]. Thus, in the absence of HAF-1, ATFS-1 preferentially partitions to mitochondria rather than trafficking to the nucleus during stress. Additional means to adjust mitochondrial import rates including phosphorylation of the protein import machinery [54, 55] as well as the mitochondrial stress-dependent turnover of an essential component of the import channel occur during metabolic stress and may impact UPRmt signaling. It has been suggested that reduced import rates reduce the burden or unfolded protein load on the mitochondrial protein-folding environment in addition to activating the UPRmt [56, 57].
A number of other known factors have also been identified that are required for UPRmt signaling including Tor signaling [6, 23], proteasome and ribosome function [49], as well as mevalonate and ceramide production [48, 58, 59], however the relationship has yet to be resolved.
3.2 UPRmt signaling in mammals
Less is currently known about UPRmt regulation in mammalian cells but the current data suggest that it may be considerably more complicated, potentially receiving inputs from multiple signal transduction pathways. Early studies suggested a role for JNK2 phosphorylation and c-Jun, which binds and activates the promoters of the bZip transcription factor genes CHOP (C/EBP homology protein, also known as GADD153 and DDIT-3) and C/EBPβ to induce mitochondrial chaperone genes during mitochondrial unfolded protein stress [21]. The authors suggest that CHOP-C/EBPβ dimerization is required for the induction of genes containing mitochondrial UPR elements (MUREs) [60]. Interestingly, MUREs exist in the promoters of multiple mitochondrial chaperone and protease genes but the transcription factor that binds the MUREs is currently unclear. Additionally, CHOP is induced by multiple forms of cellular stress [61] and it is therefore currently unclear how CHOP activities integrate with a mitochondrial specific response. Of note, c-Jun was also required for UPRmt activation in flies [17]. Recently, the estrogen receptor and the mitochondrial matrix-localized sirtuin Sirt5 have also been found to play a role in UPRmt regulation in response to unfolded protein accumulation in the intermembrane space [41, 62].
3.3 A role for GCN-2-mediated eIF2α phosphorylation
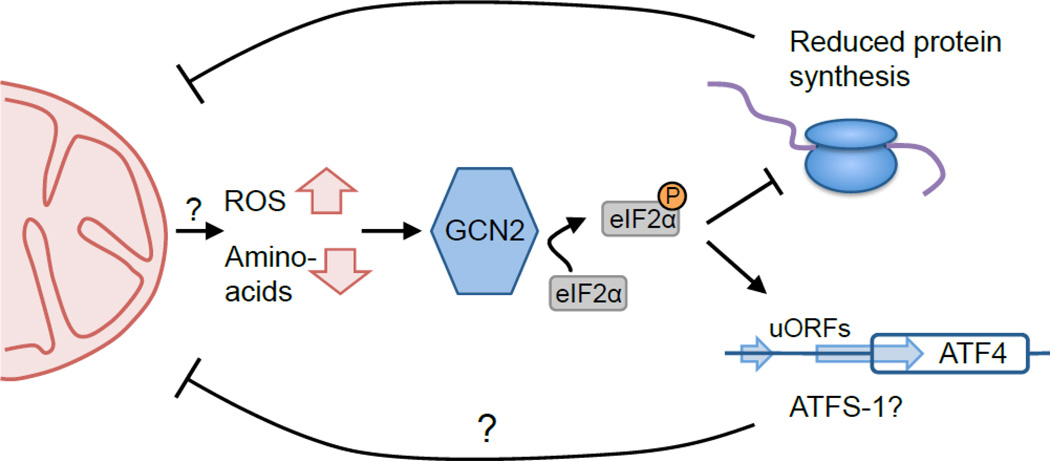
While the transcriptional outputs to mitochondrial stress in worms and mammals are relatively similar, very little mechanistic overlap between mitochondrial-to-nuclear signaling between worms and mammals has been elucidated. In particular, the mammalian ortholog of ATFS-1 is unclear as is the worm ortholog of CHOP; an issue complicated by the high homology amongst the entire bZip family of proteins. Recent work has demonstrated that an increase in eukaryotic translation initiation factor 2 alpha (eIF2α) phosphorylation occurs in response to mitochondrial stress in both worms and mammals [6, 63–65] resulting in the attenuation of global protein synthesis while preferentially translating those mRNAs that contain upstream open reading frames (uORFs) [66]. Four eIF2α kinases exist, but in yeast, C. elegans, and mammals, the kinase general control non-derepressible 2 (GCN2) appears to contribute the most to the induced eIF2α phosphorylation that occurs during mitochondrial stress [6, 63, 64, 67] (Figure 2). However PKR, an eIF2α kinase that responds to double stranded RNA, has also been shown to be required for eIF2α phosphorylation during mitochondrial stress in the mouse intestine suggesting mitochondrial dysfunction activates multiple eIF2α kinases [65].
Figure 2. GCN2 phosphorylates eIF2α during mitochondrial dysfunction.
Dysfunctional mitochondria are major sources of reactive oxygen species (ROS), which accumulate both inside mitochondria and in the cytosol. ROS as well as imbalanced amino acid levels activate the kinase GCN2, which phosphorylates eIF2α to attenuate global mRNA translation. Reduced protein synthesis during mitochondrial stress reduces the burden of unfolded proteins on mitochondria, which facilitates overall recovery. In addition to reducing global protein synthesis, eIF2α phosphorylation selectively increases the translation of mRNAs containing uORFs such as ATF4, which induces transcription of the gene encoding CHOP. Several atfs-1 mRNAs contain uORFs suggesting that it may also be preferentially translated during eIF2α phosphorylation.
GCN2 is activated during amino acid depletion as its tRNA synthetase-like domain binds directly to uncharged tRNAs that accumulate during conditions such as starvation or caloric restriction, leading to kinase activation and eIF2α phosphorylation [66, 68]. GCN2 has also been shown to be activated by ROS and provide resistance to oxidative stress, but the relationship between uncharged tRNA accumulation, ROS and mitochondrial dysfunction is currently unclear [6, 69, 70]. The resultant attenuation of global protein synthesis has been suggested to be protective by reducing the burden on the dysfunctional protein-folding environment in the mitochondria. In addition to a reduction in global protein synthesis, those transcripts with uORFs such as ATF4 are preferentially translated [71] (Figure 2), leading to altered transcriptional outputs. Interestingly, CHOP expression has been shown to increase during mitochondrial dysfunction in an ATF4-dependent manner [64]. However, a protective role for CHOP or ATF4 during mitochondrial dysfunction has yet to be demonstrated. These data generated in both worms and mammals suggest that GCN2-mediated eIF2α phosphorylation may be involved in a UPRmt. And, work from our lab indicates that loss of GCN2 function in C. elegans results in increased ATFS-1 activation consistent with GCN2 and ATFS-1 functioning in separate mitochondrial protective programs [6]. However, it should be noted that multiple atfs-1 transcripts exist, several that contain a single uORF and several that do not. As a single uORF is known to promote translation when eIF2α is phosphorylated [72], it will be interesting to determine if ATFS-1 is preferentially translated in a GCN2-dependent manner during mitochondrial stress.
4. Protective effects mediated by the UPRmt
Transcriptional responses consistent with UPRmt activation have been observed in multiple species, however because the regulatory components have been elucidated in C. elegans, the physiologic roles of the UPRmt are best characterized in this model organism. We primarily focus on the role of ATFS-1, as it is the UPRmt component best understood mechanistically and the transcripts induced during mitochondrial dysfunction requiring ATFS-1 have been characterized (see section 2). Consistent with ATFS-1 being rapidly turned over, atfs-1-deletion affects the expression of very few transcripts in otherwise healthy worms. However, during mitochondrial stress, ATFS-1 is required for the induction of over 400 genes [27]. atfs-1-deletion causes no obvious developmental defects in the soma, but germline defects have been noted [59, 73]. However, the role of ATFS-1 in germline function is unclear.
4.1 Genotoxic respiratory chain defects
C. elegans strains with hypomorphic mutations in respiratory chain and ubiquinone biosynthesis genes including the succinate dehydrogenase component mev-1 (complex II), the cytochrome c reductase component isp-1 (complex III) and the clk-1 gene required for ubiquinone biosynthesis have considerable developmental delays and have been shown to activate the UPRmt [6, 27, 42, 43, 74]. Consistent with a protective role for the UPRmt, the development rate of all three of these mutant worms is further impaired in the absence of ATFS-1 [6, 27, 42]. Similarly, worms lacking GCN2 were also developmentally delayed during mitochondrial dysfunction. And, strains lacking both ATFS-1 and GCN2 are further compromised highlighting the independent effects of both signaling pathways.
4.2 Statin exposure or cholesterol depletion
HMG-CoA reductase is the rate-limiting enzyme in the mevalonate pathway of cholesterol biosynthesis and the target of the cholesterol reducing drugs known as statins. As the mevalonate pathway is also required for the production of coenzyme Q (respiratory chain component, also known as ubiquinone), dolichols (required for protein glycosylation) and isoprenoids (lipid required membrane binding of many small GTPases), statin-mediated inhibition can cause multiple undesirable off-target effects [75]. To understand the compensatory pathways in place to tolerate reduced output of the mevalonate pathway, Rauthan and colleagues performed a mutagenesis screen to isolate C. elegans mutant strains resistant to high levels of statins. From a screen of over 150,000 mutagenized genomes, four resistant strains were isolated. Interestingly, all four mutations were in the atfs-1 gene and caused amino acid substitutions in the MTS [59]. The reduced mitochondrial import efficiency resulted in constitutive activation of the UPRmt, providing protection from statins as well as deletion of the gene encoding HMG-CoA reductase [58]. While these results indicate that transcriptional outputs of the UPRmt are required to tolerate reduced mevalonate pathway output, it is currently not clear which UPRmt-regulated transcripts are required.
4.3 UPRmt-mediated innate immunity
Recent findings suggest a surprising role for the UPRmt during exposure to bacterial pathogens. A relatively benign strain of E. coli is the typical C. elegans food source used experimentally, although its natural habitat contains a large number of species whose metabolic by-products are toxic to mitochondria [76]. These studies evolved from the finding that respiratory toxins produced by bacteria activate the UPRmt including the respiratory chain inhibitors antimycin and cyanide, and the ATP synthase inhibitor oligomycin [48, 49, 73]. And, more recent studies have indicated that pathogen generated siderophores or iron chelators also cause mitochondrial dysfunction and UPRmt activation [73, 77].
Interestingly, in addition to inducing a mitochondrial protective response, ATFS-1 also induces a number of innate immune genes including anti-microbial peptides and secreted lysozymes during mitochondrial dysfunction [27, 73]. Furthermore, ATFS-1 also induced expression of multiple xenobiotic detoxification genes such as the cytochrome P450s [27, 48, 78]. The above findings raised the question as to why an innate immune response would be coupled with a mitochondrial protective response via a single transcription factor activated during mitochondrial stress. Intriguingly, Liu and colleagues examined UPRmt activation when C. elegans were exposed to ~500 natural bacterial isolates. Interestingly, 18% of the strains caused UPRmt activation including species related to human pathogens including Pseusdomonas aeruginosa [48], which is known to produce the respiratory chain inhibitor cyanide [79]. These findings suggested a role for the UPRmt in immunosurveillance; potentially in detecting those bacteria that target mitochondrial function to promote infection (Figure 3).
Figure 3. UPRmt-mediated innate immunity in response to bacterial infection.
In their natural habitat worms are exposed to a variety of bacterial species, many of which cause UPRmt activation [48]. Secreted bacterial toxins such as Pseudomonas aeruginosa-produced cyanide target mitochondria and activate the UPRmt. Cyanide impairs the respiratory chain, thereby perturbing mitochondrial protein import causing ATFS-1 to traffic to the nucleus. In the nucleus, ATFS-1 induces mitochondrial protective genes such as mitochondrial chaperones and proteases, but also innate immune genes such as antimicrobial peptides and secreted lysozymes [73]. Worms with an activated UPRmt have reduced intestinal accumulation of P. aeruginosa and survive longer when exposed to the pathogen [73] indicating that the UPRmt promotes an innate immune response that confers resistance to pathogenic bacteria.
In support of this model, worms lacking ATFS-1 survived for shorter periods of time when raised on P. aeruginosa, demonstrating the importance of the UPRmt when exposed to the pathogen [73]. Interestingly, UPRmt activation caused by P. aeruginosa exposure required the cyanide synthase genes as well the siderophore biosynthesis genes consistent with these compounds perturbing mitochondrial activity of the host. Furthermore, animals with a pre-activated or hyper-activated UPRmt survived longer on P. aeruginosa further demonstrating the protective effects. Most impressively, these worms were able to delay intestinal colonization by P. aeruginosa suggesting the UPRmt mediated a bactericidal activity in addition to a mitochondrial protective response [73]. In sum, these results suggest a role for the UPRmt in immunosurveillance by detecting those pathogens that perturb mitochondrial function and regulating an immune response. Interestingly, mitochondrial stress induces a similar set of innate immune genes in cultured mammalian cells [73], and more recently mitochondrial dysfunction has been shown to induce an anti-viral response in mice, further linking innate immunity and mitochondrial dysfunction [80, 81].
5. Enhanced longevity, mitochondrial stress and the UPRmt
5.1 Mitochondrial stress-associated lifespan extension
Longevity and stress resistance often coincide, as was recently outlined for several well-studied stressors in C. elegans [82]. But, because mitochondrial dysfunction contributes to normal aging as well as multiple devastating diseases [13, 14], the demonstration that moderate mitochondrial perturbation extends lifespan by up to 50% was surprising [83, 84]. Impressively, the lifespan increase associated with mitochondrial dysfunction occurs in yeast [67], worms [83, 84], flies [17] and mice [15, 44]. However, like many treatments or conditions that extend lifespan, it comes at a significant cost to development (as described above), animal size and fecundity [82].
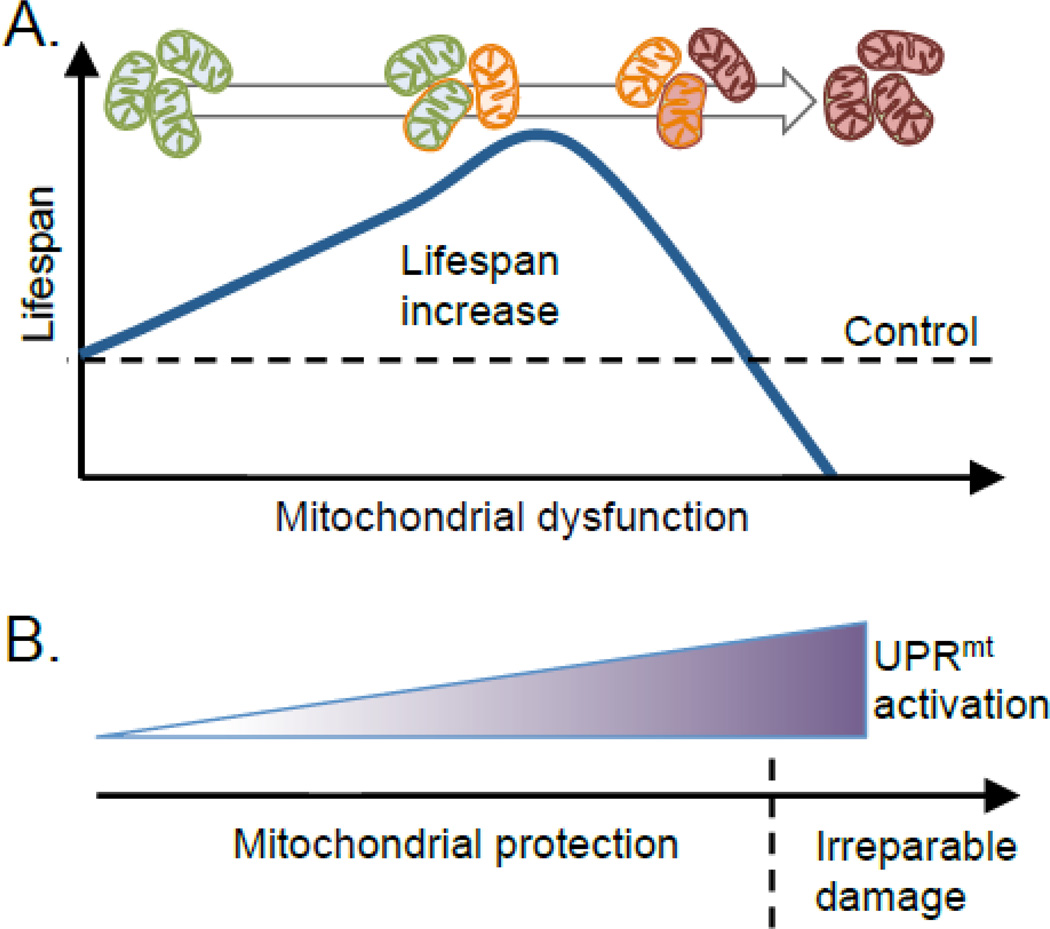
Consistent with both positive and negative effects culminating from dysfunctional mitochondria, Rea and colleagues demonstrated that respiratory chain inhibition has dose-dependent effects on longevity which anti-correlate with developmental rate, fertility and animal size [85] (Figure 4). This “mitochondrial threshold effect” suggests that up to a certain point mitochondria are able to function under compromised conditions and compensate for respiratory chain deficiencies. However, beyond this threshold, compensatory pathways may not be able to offset the severe decline in mitochondrial function, which eventually leads to death [43, 85]. A similar threshold effect was observed when worms were treated with paraquat [86], suggesting that increasing amounts of ROS may cause longevity only up to a certain point, after which the damage becomes detrimental. In sum, these findings suggest that protective effects emanate from defective mitochondria, which may be therapeutically separable from the underlying organelle defects. This section focuses on the role of the UPRmt in the enhanced longevity conferred by modest mitochondrial dysfunction.
Figure 4. Mitochondrial stress and lifespan regulation.
(A) Lifespan increases with moderate levels of mitochondrial dysfunction and is reduced when the damage is too severe. During mild mitochondrial dysfunction (orange), which may affect the entire mitochondrial pool or individuals organelles, the organism is able to promote mitochondrial recovery through physiological alterations that positively affect lifespan. These adaptations include pro-survival metabolic alterations, maintenance of the mitochondrial protein folding environment and resistance to pathogens. (B) The UPRmt is activated in response to mitochondrial dysfunction and promotes mitochondrial repair and metabolic adaptations. At some point, the mitochondrial damage becomes irreparable offsetting the UPRmt-mediated protective effects (dashed line). During severe mitochondrial dysfunction UPRmt activation still occurs, but may not be sufficient to maintain mitochondrial homeostasis and protect survival.
5.2 The UPRmt in longevity
To our knowledge, UPRmt activation occurs in all of the mutant and RNAi-treated worms with increased longevity associated with mitochondrial dysfunction, suggesting the UPRmt promotes longevity [43, 44, 74]. However, accumulating data suggest the relationship between the UPRmt and increased longevity is a complicated one [87].
UPRmt activation in C. elegans occurs only during exposure to mitochondrial stress in development, but not adulthood [40]. Intriguingly, exposure to mitochondrial stress increased lifespan only, if it occurred during development, but not during adulthood [43]. And, ubl-5 was required for the lifespan extension suggesting a requirement for the UPRmt. More recent studies support this findings demonstrating that the level of UPRmt activation correlates with lifespan extension [44], and the requirement for both haf-1 and atfs-1 in the lifespan extension caused by mitochondrial ribosome or respiratory chain perturbation [44, 74]. However, these results are somewhat controversial as a separate study found that atfs-1(RNAi) and a loss-of-function atfs-1-mutation failed to suppress the increased longevity conferred by a complex III defect [42]. It is unclear what accounts for the difference between these studies, but technical differences exist including the use FUDR during the lifespan studies, which limits reproduction by causing germline dysfunction. Regardless, this is clearly an active and rapidly progressing area of research.
Importantly, not all loss-of-function mutations in respiratory chain components promote longevity. For example, defects in complex II shorten lifespan [88], which may be explained by the severity of the defects caused by the mutation or because complex II is the only respiratory complex that also functions in the tricarboxylic acid cycle. Perhaps not surprisingly, the UPRmt is activated in complex II mutants suggesting UPRmt activation is not sufficient to extend lifespan [42, 43]. However, the reduction in lifespan could be reversed by mitochondrial ribosome or complex IV inhibition which both activate the UPRmt to a stronger degree [44]. Another study showed that a gain-of-function mutation in the atfs-1 gene, which causes constitutive UPRmt activation, was not sufficient to induce longevity [59]. Combined, these studies suggest that UPRmt activation is not sufficient to increase longevity but some caution is warranted in addition to the potential toxic side effects associated with complex II defects. For example, the mutation in atfs-1 that causes constitutive UPRmt activation is not well-characterized and may alter other aspects of ATFS-1 activity, as it has been shown that overexpression of a single mitochondrial molecular chaperone is sufficient to confer enhanced longevity [17, 89]. However, these results are also consistent with ATFS-1 and the UPRmt functioning alongside multiple pathways activated during mitochondrial dysfunction (see below). For example, at least three other transcription factors have been shown to be required for the longevity associated with mitochondrial dysfunction including HIF-1 [86], TAF-4 [90], CEH-23 [91] and as well as the kinase GCN2 [6].
5.3 Tissue specificity and extra-cellular communication
Studies in both worms and flies suggest the existence of inter-cellular communication of mitochondrial status or transmission of an extra-cellular UPRmt signal [17, 43, 92]. Interestingly, these studies also suggest a hierarchy for those tissues capable of conferring increased longevity during mitochondrial dysfunction. Inhibition of complex IV specifically in the intestine and neurons, but not muscle cells, was sufficient to confer longevity in C. elegans. Intriguingly, neuronal specific complex IV inhibition resulted in UPRmt activation in the intestine also resulting in increased longevity [43]. An important aspect of these studies is that neuronal-specific impairment of complex IV resulted in less developmental delay, but conferred a similar increase in longevity suggesting the detrimental effects of mitochondrial perturbation can be separated from those that enhance longevity. A similar approach in flies demonstrated that a muscle-specific respiratory chain perturbation in developing animals resulted in UPRmt activation and an increase in longevity. The increase in longevity required an insulin-like growth factor binding protein in addition to UPRmt activation [17, 93].
6. Conclusions, comments and future directions
Considerable progress in understanding which transcripts constitute a UPRmt, and how the stress response is activated and signaled has been made in recent years. The UPRmt is emerging as a regulator of cell survival during a variety of conditions associated with mitochondrial dysfunction including general aging, genotoxic respiratory defects as well as mitochondrial dysfunction associated with pathogen infection. However, many questions remain.
6.1 Identification of ATFS-1 like transcription factors
While it is clear that a UPRmt similar to that which occurs in C. elegans is activated in mammals during mitochondrial stress, it is unclear how it is regulated. It is certainly possible that the mammalian response is regulated in a manner unlike that in C. elegans. However, the present difficulty in identifying the functional ortholog is reminiscent of the difficulties in identifying the functional ortholog of the transcription factor Hac1 that regulates an endoplasmic reticulum stress specific UPR [94]. Hac1 was identified in S. cerevisiae and the functional mammalian ortholog was not identified until six years later in a genetic screen [95] and by induction during ER stress [96] rather than by homology searching. While the homology with Hac1 is quite poor, the mammalian transcription factor known as XBP1 is regulated in nearly the exact same manner and activates transcription of a similar ER-protective program to that identified in yeast, perhaps suggesting a functional mammalian ortholog of ATFS-1 remains to be discovered. Of note, a recent report identified a yeast transcription factor regulated similarly to ATFS-1. Hap1 regulates expression of genes involved in heme biogenesis in response to heme or oxygen levels when Hap1 is in the nucleus. Interestingly, a ribosome profiling experiment showed that the Hap1 mRNA is translated on ribosomes on or near the outer mitochondrial membrane and that the Hap1 protein has a MTS and localizes to mitochondria [97], suggesting it is regulated similarly to ATFS-1. Presumably, a similar approach combined with screening could be used to identify proteins that regulate a UPRmt in mammals.
6.3 Integration of the UPRmt with other mitochondrial protective pathways
Mitochondrial dysfunction is very pleiotropic likely causing dysfunction throughout the cell and activation of multiple protective signaling pathways. A particular intriguing potential interaction is between the UPRmt and the mitochondrial autophagy pathway, which degrades those severely dysfunctional mitochondria that are likely irreparable [98]. Both pathways are regulated by mitochondrial protein import efficiency. Import efficiency of the kinase Pink1 is a major determinant in selecting those severely defective organelles for degradation. Normally, Pink1 is imported into mitochondria, processed and degraded. However, if import is impaired due to the accumulation of misfolded proteins within the mitochondrial matrix or depletion of the inner membrane potential, Pink1 accumulates on the outer membrane where it recruits the ubiquitin ligase Parkin and ultimately the autophagy machinery. Once engulfed by an autophagosome, the defective mitochondrion is delivered to a lysosome where it is ultimately degraded. In addition to import efficiency, a recent screen identified ceramide as an essential regulator of the UPRmt [48]. Intriguingly, ceramide is known to accumulate on damaged mitochondria and is also required for early events in mitophagy [99] suggesting a potential role in coordinating both pathways.
In addition to coordinating with mitochondrial turnover, several interactions with components that regulate translation have been documented. As discussed, the kinase GCN2 is activated concomitantly to the UPRmt and is required for development and lifespan extension during mitochondrial dysfunction [6] but the role reduced translation plays is unclear. Furthermore, the kinase TOR and its upstream regulator Rheb were both found to be required for UPRmt activation [6], but again very little is known regarding the mode of regulation or the physiological significance. Additional interactions likely exist with other metabolic regulators including AMP kinase and the sirtuins [25, 62].
Lastly, as the UPRmt is a transcriptional response that affects over 400 genes controlling many aspects of cell physiology, it will be important to understand which affected activities are the most important in ameliorating the defects associated with mitochondrial dysfunction and potentially the easiest to manipulate. The list of candidates includes but is not limited to mitochondrial repair or regeneration, protein quality control, and metabolic remodeling as suggested by the transcriptional outputs mediated by ATFS-1.
Highlights.
Mitochondrial dysfunction activates a transcriptional response known as the UPRmt
UPRmt activation is regulated by mitochondrial protein import efficiency of ATFS-1
The ATFS-1 -mediated UPRmt includes mitochondrial proteostasis and innate immune genes
The UPRmt also includes a metabolic shift towards glycolysis
Modest mitochondrial dysfunction activates the UPRmt and confers longevity
Acknowledgements
We apologize to those colleagues whose work on mitochondrial stress responses and biology that could not be included due to space limitations. This work was supported by the Deutsche Forschungsgemeinschaft (DFG, SCHU 3023/1-1) to A.M.S. and by the National Institutes of Health (R01AG040061, R01AG047182) to C.M.H.
Abbreviations
- UPRmt
mitochondrial unfolded protein response
- ATFS-1
activating transcription factor associated with stress-1
- mtDNA
mitochondrial DNA
- OxPhos
oxidative phosphorylation
- TCA cycle
tricarboxylic acid cycle
- ATP
adenosine triphosphate
- Hsp
heat shock protein
- TIM
translocase of the inner membrane
- TOM
translocase of the outer membrane
- MTS
mitochondrial targeting sequence
- NLS
nuclear localization sequence
- Tor
target of rapamycin
- uORF
upstream open reading frame
- ROS
reactive oxygen species
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lackner LL. Shaping the dynamic mitochondrial network. BMC biology. 2014;12:35. doi: 10.1186/1741-7007-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheibye-Knudsen M, Fang EF, Croteau DL, Wilson DM, 3rd, Bohr VA. Protecting the mitochondrial powerhouse. Trends in cell biology. 2014 doi: 10.1016/j.tcb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell P. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. 1966. Biochimica et biophysica acta. 2011;1807:1507–1538. doi: 10.1016/j.bbabio.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 5.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker BM, Nargund AM, Sun T, Haynes CM. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS genetics. 2012;8:e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerdes F, Tatsuta T, Langer T. Mitochondrial AAA proteases--towards a molecular understanding of membrane-bound proteolytic machines. Biochimica et biophysica acta. 2012;1823:49–55. doi: 10.1016/j.bbamcr.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Rugarli EI, Langer T. Mitochondrial quality control: a matter of life and death for neurons. The EMBO journal. 2012;31:1336–1349. doi: 10.1038/emboj.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Short KR, Bigelow ML, Kahl J, Singh R, Coenen-Schimke J, Raghavakaimal S, Nair KS. Decline in skeletal muscle mitochondrial function with aging in humans. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5618–5623. doi: 10.1073/pnas.0501559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross JM, Stewart JB, Hagstrom E, Brene S, Mourier A, Coppotelli G, Freyer C, Lagouge M, Hoffer BJ, Olson L, Larsson NG. Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature. 2013;501:412–415. doi: 10.1038/nature12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Souza AD, Parish IA, Krause DS, Kaech SM, Shadel GS. Reducing mitochondrial ROS improves disease-related pathology in a mouse model of ataxia-telangiectasia. Molecular therapy : the journal of the American Society of Gene Therapy. 2013;21:42–48. doi: 10.1038/mt.2012.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS biology. 2010;8:e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes & development. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munkacsy E, Rea SL. The paradox of mitochondrial dysfunction and extended longevity. Experimental gerontology. 2014;56:221–233. doi: 10.1016/j.exger.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yee C, Yang W, Hekimi S. The intrinsic apoptosis pathway mediates the prolongevity response to mitochondrial ROS in C. elegans. Cell. 2014;157:897–909. doi: 10.1016/j.cell.2014.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinus RD, Garth GP, Webster TL, Cartwright P, Naylor DJ, Hoj PB, Hoogenraad NJ. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. European journal of biochemistry / FEBS. 1996;240:98–103. doi: 10.1111/j.1432-1033.1996.0098h.x. [DOI] [PubMed] [Google Scholar]
- 20.Ryan MT, Hoogenraad NJ. Mitochondrial-nuclear communications. Annual review of biochemistry. 2007;76:701–722. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. The EMBO journal. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Developmental cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 24.Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Molecular cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, Guarente L, Auwerx J. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haynes CM, Fiorese CJ, Lin YF. Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends in cell biology. 2013 doi: 10.1016/j.tcb.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Williams EG, Dubuis S, Mottis A, Jovaisaite V, Houten SM, Argmann CA, Faridi P, Wolski W, Kutalik Z, Zamboni N, Auwerx J, Aebersold R. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158:1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashrafi G, Schlehe JS, LaVoie MJ, Schwarz TL. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. The Journal of cell biology. 2014;206:655–670. doi: 10.1083/jcb.201401070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, Kimura M, Komatsu M, Hattori N, Tanaka K. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. The Journal of cell biology. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS biology. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrane J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lill R, Srinivasan V, Muhlenhoff U. The role of mitochondria in cytosolic-nuclear iron-sulfur protein biogenesis and in cellular iron regulation. Current opinion in microbiology. 2014;22C:111–119. doi: 10.1016/j.mib.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 34.Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torraco A, Peralta S, Iommarini L, Diaz F. Mitochondrial Diseases Part I: Mouse models of OXPHOS deficiencies caused by defects on respiratory complex subunits or assembly factors. Mitochondrion. 2015 doi: 10.1016/j.mito.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghezzi D, Zeviani M. Assembly factors of human mitochondrial respiratory chain complexes: physiology and pathophysiology. Advances in experimental medicine and biology. 2012;748:65–106. doi: 10.1007/978-1-4614-3573-0_4. [DOI] [PubMed] [Google Scholar]
- 37.Hirano M, Garone C, Quinzii CM. CoQ(10) deficiencies and MNGIE: two treatable mitochondrial disorders. Biochimica et biophysica acta. 2012;1820:625–631. doi: 10.1016/j.bbagen.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. The EMBO journal. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. Journal of cell science. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 41.Papa L, Germain D. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. Journal of cell science. 2011;124:1396–1402. doi: 10.1242/jcs.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bennett CF, Vander Wende H, Simko M, Klum S, Barfield S, Choi H, Pineda VV, Kaeberlein M. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nature communications. 2014;5:3483. doi: 10.1038/ncomms4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jovaisaite V, Auwerx J. The mitochondrial unfolded protein response-synchronizing genomes. Current opinion in cell biology. 2014;33C:74–81. doi: 10.1016/j.ceb.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pulliam DA, Deepa SS, Liu Y, Hill S, Lin AL, Bhattacharya A, Shi Y, Sloane L, Viscomi C, Zeviani M, Van Remmen H. Complex IV-deficient Surf1(−/−) mice initiate mitochondrial stress responses. The Biochemical journal. 2014;462:359–371. doi: 10.1042/BJ20140291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dogan SA, Pujol C, Maiti P, Kukat A, Wang S, Hermans S, Senft K, Wibom R, Rugarli EI, Trifunovic A. Tissue-specific loss of DARS2 activates stress responses independently of respiratory chain deficiency in the heart. Cell metabolism. 2014;19:458–469. doi: 10.1016/j.cmet.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y, Samuel BS, Breen PC, Ruvkun G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 2014;508:406–410. doi: 10.1038/nature13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Runkel ED, Liu S, Baumeister R, Schulze E. Surveillance-activated defenses block the ROS-induced mitochondrial unfolded protein response. PLoS genetics. 2013;9:e1003346. doi: 10.1371/journal.pgen.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harbauer AB, Zahedi RP, Sickmann A, Pfanner N, Meisinger C. The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell metabolism. 2014;19:357–372. doi: 10.1016/j.cmet.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 51.Jensen MB, Jasper H. Mitochondrial proteostasis in the control of aging and longevity. Cell metabolism. 2014;20:214–225. doi: 10.1016/j.cmet.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pellegrino MW, Nargund AM, Haynes CM. Signaling the mitochondrial unfolded protein response. Biochimica et biophysica acta. 2013;1833:410–416. doi: 10.1016/j.bbamcr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young L, Leonhard K, Tatsuta T, Trowsdale J, Langer T. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science. 2001;291:2135–2138. doi: 10.1126/science.1056957. [DOI] [PubMed] [Google Scholar]
- 54.Harbauer AB, Opalinska M, Gerbeth C, Herman JS, Rao S, Schonfisch B, Guiard B, Schmidt O, Pfanner N, Meisinger C. Mitochondria. Cell cycle-dependent regulation of mitochondrial preprotein translocase. Science. 2014;346:1109–1113. doi: 10.1126/science.1261253. [DOI] [PubMed] [Google Scholar]
- 55.Schmidt O, Harbauer AB, Rao S, Eyrich B, Zahedi RP, Stojanovski D, Schonfisch B, Guiard B, Sickmann A, Pfanner N, Meisinger C. Regulation of mitochondrial protein import by cytosolic kinases. Cell. 2011;144:227–239. doi: 10.1016/j.cell.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 56.Rainbolt TK, Atanassova N, Genereux JC, Wiseman RL. Stress-regulated translational attenuation adapts mitochondrial protein import through Tim17A degradation. Cell metabolism. 2013;18:908–919. doi: 10.1016/j.cmet.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rainbolt TK, Saunders JM, Wiseman RL. YME1L degradation reduces mitochondrial proteolytic capacity during oxidative stress. EMBO reports. 2015;16:97–106. doi: 10.15252/embr.201438976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ranji P, Rauthan M, Pitot C, Pilon M. Loss of HMG-CoA reductase in C. elegans causes defects in protein prenylation and muscle mitochondria. PloS one. 2014;9:e100033. doi: 10.1371/journal.pone.0100033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rauthan M, Ranji P, Aguilera Pradenas N, Pitot C, Pilon M. The mitochondrial unfolded protein response activator ATFS-1 protects cells from inhibition of the mevalonate pathway. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5981–5986. doi: 10.1073/pnas.1218778110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aldridge JE, Horibe T, Hoogenraad NJ. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PloS one. 2007;2:e874. doi: 10.1371/journal.pone.0000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes & development. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Papa L, Germain D. SirT3 regulates the mitochondrial unfolded protein response. Molecular and cellular biology. 2014;34:699–710. doi: 10.1128/MCB.01337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martinez-Reyes I, Sanchez-Arago M, Cuezva JM. AMPK and GCN2-ATF4 signal the repression of mitochondria in colon cancer cells. The Biochemical journal. 2012;444:249–259. doi: 10.1042/BJ20111829. [DOI] [PubMed] [Google Scholar]
- 64.Michel S, Canonne M, Arnould T, Renard P. Inhibition of mitochondrial genome expression triggers the activation of CHOP-10 by a cell signaling dependent on the integrated stress response but not the mitochondrial unfolded protein response. Mitochondrion. 2015 doi: 10.1016/j.mito.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 65.Rath E, Berger E, Messlik A, Nunes T, Liu B, Kim SC, Hoogenraad N, Sans M, Sartor RB, Haller D. Induction of dsRNA-activated protein kinase links mitochondrial unfolded protein response to the pathogenesis of intestinal inflammation. Gut. 2012;61:1269–1278. doi: 10.1136/gutjnl-2011-300767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baird TD, Wek RC. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Advances in nutrition. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delaney JR, Ahmed U, Chou A, Sim S, Carr D, Murakami CJ, Schleit J, Sutphin GL, An EH, Castanza A, Fletcher M, Higgins S, Jelic M, Klum S, Muller B, Peng ZJ, Rai D, Ros V, Singh M, Wende HV, Kennedy BK, Kaeberlein M. Stress profiling of longevity mutants identifies Afg3 as a mitochondrial determinant of cytoplasmic mRNA translation and aging. Aging cell. 2013;12:156–166. doi: 10.1111/acel.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rousakis A, Vlassis A, Vlanti A, Patera S, Thireos G, Syntichaki P. The general control nonderepressible-2 kinase mediates stress response and longevity induced by target of rapamycin inactivation in Caenorhabditis elegans. Aging cell. 2013;12:742–751. doi: 10.1111/acel.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Molecular cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 70.Mascarenhas C, Edwards-Ingram LC, Zeef L, Shenton D, Ashe MP, Grant CM. Gcn4 is required for the response to peroxide stress in the yeast Saccharomyces cerevisiae. Molecular biology of the cell. 2008;19:2995–3007. doi: 10.1091/mbc.E07-11-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. The Journal of cell biology. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Andreev DE, O'Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. Translation of 5' leaders is pervasive in genes resistant to eIF2 repression. eLife. 2015;4 doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pellegrino MW, Nargund AM, Kirienko NV, Gillis R, Fiorese CJ, Haynes CM. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature. 2014;516:414–417. doi: 10.1038/nature13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schieber M, Chandel NS. TOR signaling couples oxygen sensing to lifespan in C. elegans. Cell reports. 2014;9:9–15. doi: 10.1016/j.celrep.2014.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 76.Cohen LB, Troemel ER. Microbial pathogenesis and host defense in the nematode C. elegans. Current opinion in microbiology. 2015;23C:94–101. doi: 10.1016/j.mib.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kirienko NV, Ausubel FM, Ruvkun G. Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:1821–1826. doi: 10.1073/pnas.1424954112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Melo JA, Ruvkun G. Inactivation of conserved C. elegans genes engages pathogen-and xenobiotic-associated defenses. Cell. 2012;149:452–466. doi: 10.1016/j.cell.2012.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gallagher LA, Manoil C. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. Journal of bacteriology. 2001;183:6207–6214. doi: 10.1128/JB.183.21.6207-6214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015 doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rongvaux A, Jackson R, Harman CC, Li T, West AP, de Zoete MR, Wu Y, Yordy B, Lakhani SA, Kuan CY, Taniguchi T, Shadel GS, Chen ZJ, Iwasaki A, Flavell RA. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159:1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shore DE, Ruvkun G. A cytoprotective perspective on longevity regulation. Trends in cell biology. 2013;23:409–420. doi: 10.1016/j.tcb.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 84.Felkai S, Ewbank JJ, Lemieux J, Labbe JC, Brown GG, Hekimi S. CLK-1 controls respiration, behavior and aging in the nematode Caenorhabditis elegans. The EMBO journal. 1999;18:1783–1792. doi: 10.1093/emboj/18.7.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS biology. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Current biology : CB. 2010;20:2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bennett CF, Kaeberlein M. The mitochondrial unfolded protein response and increased longevity: cause, consequence, or correlation? Experimental gerontology. 2014;56:142–146. doi: 10.1016/j.exger.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ishii N, Fujii M, Hartman PS, Tsuda M, Yasuda K, Senoo-Matsuda N, Yanase S, Ayusawa D, Suzuki K. A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature. 1998;394:694–697. doi: 10.1038/29331. [DOI] [PubMed] [Google Scholar]
- 89.Yokoyama K, Fukumoto K, Murakami T, Harada S, Hosono R, Wadhwa R, Mitsui Y, Ohkuma S. Extended longevity of Caenorhabditis elegans by knocking in extra copies of hsp70F, a homolog of mot-2 (mortalin)/mthsp70/Grp75. FEBS letters. 2002;516:53–57. doi: 10.1016/s0014-5793(02)02470-5. [DOI] [PubMed] [Google Scholar]
- 90.Khan MH, Ligon M, Hussey LR, Hufnal B, Farber R, 2nd, Munkacsy E, Rodriguez A, Dillow A, Kahlig E, Rea SL. TAF-4 is required for the life extension of isp-1, clk-1 and tpk-1 Mit mutants. Aging. 2013;5:741–758. doi: 10.18632/aging.100604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Walter L, Baruah A, Chang HW, Pace HM, Lee SS. The homeobox protein CEH-23 mediates prolonged longevity in response to impaired mitochondrial electron transport chain in C. elegans. PLoS biology. 2011;9:e1001084. doi: 10.1371/journal.pbio.1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schinzel R, Dillin A. Endocrine aspects of organelle stress - cell non-autonomous signaling of mitochondria and the ER. Current opinion in cell biology. 2015;33C:102–110. doi: 10.1016/j.ceb.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 93.Droujinine IA, Perrimon N. Defining the interorgan communication network: systemic coordination of organismal cellular processes under homeostasis and localized stress. Frontiers in cellular and infection microbiology. 2013;3:82. doi: 10.3389/fcimb.2013.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 95.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 96.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 97.Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pickrell AM, Youle RJ. The Roles of PINK1, Parkin, and Mitochondrial Fidelity in Parkinson's Disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Selvam SP, Ramshesh VK, Peterson YK, Lemasters JJ, Szulc ZM, Bielawski J, Ogretmen B. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nature chemical biology. 2012;8:831–838. doi: 10.1038/nchembio.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]