Abstract
Even though the Toll-like receptor (TLR) pathway is integral to inflammatory defense mechanisms, its excessive signaling may be devastating. Cells have acquired a cascade of strategies to regulate TLR signaling by targeting protein-protein interactions, or ubiquitin chains, but the details of the inhibition mechanisms are still unclear. Here, we provide the structural basis for the regulation of TLR signaling by constructing architectures of protein-protein interactions. Structural data suggest that 1) Toll/IL-1R (TIR) domain-containing regulators (BCAP, SIGIRR, and ST2) interfere with TIR domain signalosome formation; 2) major deubiquitinases such as A20, CYLD, and DUBA prevent association of TRAF6 and TRAF3 with their partners, in addition to removing K63-linked ubiquitin chains that serve as a docking platform for downstream effectors; 3) alternative downstream pathways of TLRs also restrict signaling by competing to bind common partners through shared binding sites. We also performed in silico mutagenesis analysis to characterize the effects of oncogenic mutations on the negative regulators and to observe the cellular outcome (whether there is/is not inflammation). Missense mutations that fall on interfaces and nonsense/frameshift mutations that result in truncated negative regulators disrupt the interactions with the targets, thereby enabling constitutive activation of the nuclear factor-kappa B, and contributing to chronic inflammation, autoimmune diseases, and oncogenesis.
Introduction
Toll-like receptors (TLRs) play pivotal roles in immune responses against invading pathogens. They signal through major pathways and give rise to inflammation (1,2). Defects in TLR signaling predispose individuals to infections (3). Over-activation can result in chronic inflammatory and autoimmune diseases and contribute to oncogenesis (3–6). To retain a delicate balance between activation and inhibition, and avoid detrimental effects of excessive inflammation, TLR signaling is strictly regulated (7,8). Here, we aim to better understand the mechanisms of negative regulation of the TLR pathway to obtain insights into how the balance is established and the immunological homeostasis is achieved.
TLRs get activated when pathogenic particles, pathogen-associated molecular patterns, as well as endogenous ligands of damaged tissues, damage-associated molecular patterns, bind to their extracellular leucine-rich repeats, resulting in dimerization of their leucine-rich repeats and cytoplasmic Toll/IL-1R (TIR) domains (9). Through their TIR domains, they recruit other TIR domain-containing adaptors, such as Mal, myeloid differentiation primary response protein (MyD88), TRIF-related adaptor molecule (TRAM), and TIR-domain-containing adapter-inducing interferon-β (TRIF) and form TIR domain signalosomes, multimeric protein assemblies. The TIR domain signalosome either comprises TLR/Mal/MyD88 or TLR/TRAM/TRIF (10). Subsequently, MyD88-dependent TIR domain signalosome recruits IRAK4, IRAK2/IRAK1, and TRAF6, which lead to activation of the nuclear factor-kappa B (NF-κB) and mitogen-activated protein kinases (MAPKs), and transcription of proinflammatory cytokines, like interleukin (IL)-1β and tumor necrosis factor (TNF)-α (1). The TRIF-dependent TIR domain signalosome employs TRAF3 to activate interferon-regulatory factors (IRFs) to induce transcription of antiviral interferons (IFNs) and antiinflammatory IL-10 (11,12).
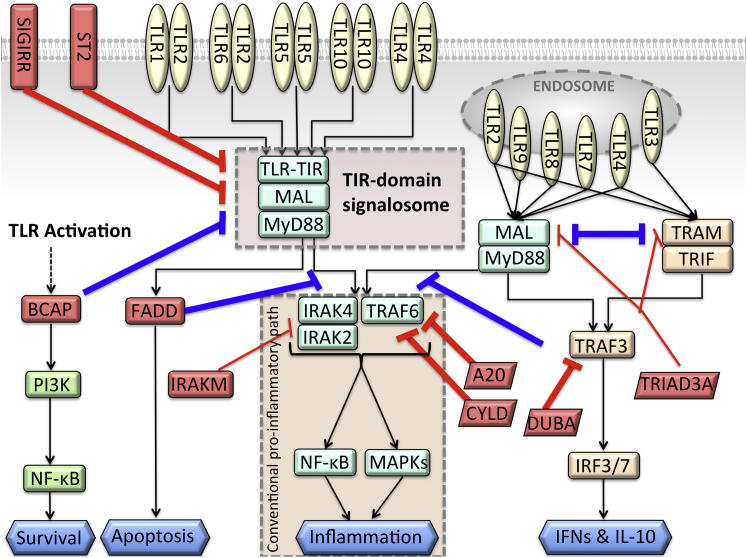
Negative regulation of TLR signaling takes place at multiple steps, ranging from extracellular soluble decoy TLRs, to transmembrane and intracellular inhibitors such as those that contain TIR domain (6,7). At almost each step in the TLR signaling cascade, protein interactions or ubiquitin (Ub) chains are targeted by one or more inhibitors (Fig. 1). The presence of several checkpoints and the redundancy of negative regulators suggest that regulation can be achieved by a cascade of regulators, indicating that a particular inhibitor may be essential but not enough to fine-tune signaling (7), or that different regulators may take action in different tissue types or at different times. Although some inhibitors are constitutively expressed, others are upregulated upon TLR stimulation. TLR-induced apoptosis comes into play when negative regulators fail and the cell becomes overactive (7). Alternative downstream paths of TLR signaling are competitive (10), limiting signaling through the temporally outcompeted pathways. Here, we focus on cytoplasmic and transmembrane regulators to elucidate the mechanisms through which they attenuate TLR signaling and the resulting inflammation.
Figure 1.
Negative regulators take action at almost all steps of TLR signaling. Red-labeled edges show the negative regulators that turn off the TLR signaling. The bolded red edges are the interactions that are modeled here. Blue-labeled edges are negative regulations from the parallel downstream paths to the conventional proinflammatory path. They switch the signaling path, not terminate the signal. To see this figure in color, go online.
Materials and Methods
Building structures of protein-protein interaction complexes
We modeled the architectures of interaction complexes of the negative regulators with the proteins in TLR pathway by exploiting PRISM (protein interactions by structural matching) (13–15). PRISM is a knowledge-based algorithm. It employs prior interface (template) data of known crystal/NMR structures of protein complexes and predicts structures of novel complexes that the target proteins can potentially form. It searches for template interface motifs on the surface of the target proteins, taking into account geometrical complementarities and hotspot conservation. The inputs of PRISM are unbound protein structures and the template interface data set. The outputs are structures of bound protein-protein interaction (PPI) complexes. PRISM structurally aligns the interfaces to query proteins first. Next, there comes a filtering step, where a combination of structural and evolutionary thresholds, i.e., root mean-square deviation and number of matching hotspots, are applied. At the last step, output models are scored and the flexibly refined to relieve possible steric clashes by FiberDock (16). FiberDock assigns a binding energy score by taking van der Waals interactions, partial electrostatics, and hydrogen and disulfide bonds into account (16). An energy score below −10 indicates a favorable interaction (14,17,18). PRISM is much faster than classical docking tools and it achieved almost 100% success in predicting the correct conformation of the complex structure (87 out of 88 cases) (19).
Modeling of proteins with unknown structures
PRISM requires 3D structures of target proteins to model the complex structures of the targets. If the structure of a given target protein is missing from the Protein Data Bank (PDB), we model it by the I-TASSER server (20). TIR domain structures of B-cell adaptor for PI3K (BCAP), single immunoglobulin IL-1R-related molecule (SIGIRR), and ST2 have not been resolved yet. We built a model of BCAP TIR domain (10–144 residues). The template structures used to build the BCAP TIR domain structure are 3h16A (crystal structure of a bacteria TIR domain, PdTIR from Paracoccus denitrificans), 3ub2A (TIR domain of Mal/TIRAP), 4lqcA (the crystal structures of the Brucella protein TcpB and the TLR adaptor protein TIRAP show structural differences in microbial TIR mimicry), and 4c6sA (crystal structure of the TIR domain from the Arabidopsis thaliana disease resistance protein RRS1). Sequence identities, coverage, and normalized Z-scores between the query proteins and the templates are given in Table S1 in the Supporting Material.
The SIGIRR TIR domain structure was also missing in the PDB. We built its model (163–310 residues) and the template used is 1t3gA (TIR domain of human IL-1RAPL).
We also modeled the ST2 TIR domain (375–558 residues) and the templates are 1t3gA, 3j0aA (homology model of human TLR5 fitted into an electron microscopy single-particle reconstruction), and 1fyvA (TIR domain of human TLR1).
Finally, we built a model of the TLR4 TIR domain. The templates are 1fyvA, 3j0aA, and 1fyxA (TLR2 TIR domain).
Mapping clinically observed mutations onto the interfaces of negative regulators and in silico mutagenesis
We obtained clinically observed mutation data from the cBioPortal for Cancer Genomics (The Cancer Genome Atlas, TCGA) (21). The residue numbers in the pdb files are corrected according to the FASTA sequences. We mapped the mutations onto the structures of negative regulators and performed in silico mutagenesis by the MutateAminoAcid function of the Discovery Studio molecular visualizer (Discovery Studio 3.5, Accelrys, San Diego, CA), for the ones that fall onto the interface residues or the first neighbors of interface residues. We identified interface residues and hotspots by the HotRegion database, which uses conservation, accessible surface area and pair potentials (22). We reran PRISM with the mutant protein structures to check whether the mutation abolishes the interaction. If interactions of the mutant structures have a binding energy score higher than that of the wild-type, we then conclude that this mutation is destabilizing. We calculated the number of hydrogen bonds and salt bridges in complexes with both wild-type and mutant structures by using the PDBePISA web server (23).
Results and Discussion
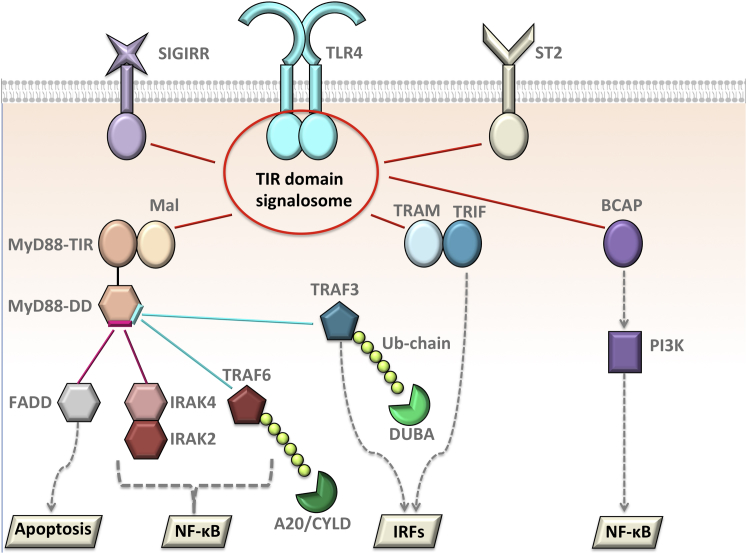
Through cooperative cellular network linkages, multivalent oligomeric proteins display efficient enzymatic reactions and amplify signal (24,25). These large protein assemblies are formed either by direct PPIs or by Ub chains attached to proteins that serve as a dynamic, allosteric scaffold. These scaffolds not only bring proteins together; the shifting landscape that they orchestrate preorganizes the ensembles for subsequent binding and catalytic events (26). The signal that these elicit propagates across the network (27). Cells acquired strategies to block cellular functions such as inflammation by targeting proteins, PPIs, and Ub chains. At almost each step of the TLR signaling cascade, one or more regulators take action to constrain NF-κB activation. In addition, parallel downstream paths also restrict inflammation because they compete with the conventional MyD88-dependent proinflammatory pathway (10) (Fig. 2). Below, we describe the negative regulators focusing on the structural basis of how they forestall signaling.
Figure 2.
The competition in the TLR pathway due to overlapping binding sites. Most of the negative regulators and the proteins that lead to distinct parallel paths of TLRs compete with one another to bind to a common partner. The TIR domain-containing proteins all compete to bind to each other; IRAK4 and FADD compete to bind to overlapping interfaces on DD of MyD88; TRAF6 and TRAF3 have shared interfaces on MyD88-DD. Deubiquitinases, such as A20, CYLD, and DUBA removes Ub chains from TRAF6 and TRAF3. To see this figure in color, go online.
TIR domain-containing negative regulators
Upon stimulation, TLRs dimerize and recruit TIR domain-containing adaptors to form either MyD88- or TRIF-dependent TIR domain signalosomes. In our recent study (10), we have provided structural models for these two signalosomes. We showed that they are competitive and cannot form simultaneously on the same receptor dimer. Apart from Mal, MyD88, TRAM, and TRIF, there are also other TIR domain-containing proteins, which act as negative regulators of TLR signaling. These include BCAP (5), SIGIRR (28), and ST2 (29), which are suggested to interfere with the assembly of TIR domain signalosome. Below, we explain the regulation mechanism of these three TIR domain containing negative regulators structurally and provide models for how they impede TLR signaling.
BCAP
Besides MyD88- and TRIF-dependent downstream paths, the PI3K pathway can also be activated upon TLR stimulation and serve as another parallel pathway of TLRs (5,30). Although the mechanism of TLR-mediated proinflammatory NF-κB and MAPK activation has been investigated extensively, the stimulation mechanism of PI3Kα, a lipid kinase, is still unknown (6). BCAP is a TIR domain-containing adaptor protein that links TLRs to PI3K/Akt pathway (6). BCAP needs to be tyrosine-phosphorylated at its YXXM motif to interact with the SH2 domain of the regulatory subunit (p85) of PI3Kα (3,5). TLR activation increases the phosphorylated BCAP in the membrane, which helps recruit activated PI3K to the membrane where its lipid substrate, phosphoinositide-4,5-bisphosphate, is found (5). If TLR is stimulated and the BCAP level is low, elevated expression of proinflammatory cytokines, such as IL-12 and IL-6, ensues (5–7). Although full-length BCAP (BCAP-L) can bind to both Mal and MyD88 and suppress NF-κB activity, truncated BCAP (BCAP-S) that lacks TIR domain is unable to do so (3,6), suggesting that its TIR domain is critical for TLR regulation. BCAP recruitment and the resulting PI3K activation serve as a checkpoint to limit TLR-induced inflammation (5). BCAP represses MyD88-dependent NF-κB and MAPK activation and there might be two different scenarios for this: in the first, BCAP interferes with the TIR domain signalosome (6,31). In the second, BCAP may allow signalosome formation and function downstream of MyD88 but still interfere with the conventional downstream NF-κB pathway; because Akt is not activated in the absence of MyD88 in macrophages (6) and MyD88 is vital for the recruitment of PI3K p85 regulatory subunit (30).
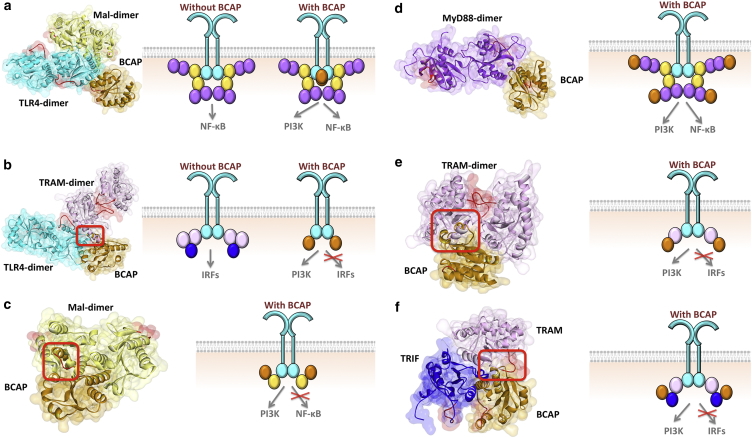
Our results show that the TIR domain of BCAP can form potential complexes with all TIR-domain-containing proteins in TLR signaling, including TLR4, Mal, MyD88, TRAM, and TRIF (Fig. 3). The details of each of the interaction architectures are given in Tables 1 and S2. Some of these interactions interfere with TIR domain signalosome formation, supporting the first scenario. For instance, if BCAP binds to TLR4, it abolishes TLR4-TRAM interaction due to steric clash of BCAP and TRAM, preventing TRIF-dependent signal relay (Fig. 3b). Moreover, if it binds to Mal, due to overlapping binding sites the Mal-BCAP interaction prevents Mal homodimerization, which is vital for MyD88-dependent signalosome formation (32) (Fig. 3c). Similar to MyD88, TRIF can also mediate PI3K activation (33), but the IL-10 level does not change in the absence of BCAP (5). BCAP-TRAM interaction abrogates TRAM homodimerization (Fig. 3e), which facilitates TRIF recruitment by serving as a binding platform (34) and the BCAP-TRIF association disrupts TRAM-TRIF interaction (Fig. 3f).
Figure 3.
BCAP interactions with all TIR domain-containing proteins in TLR signaling. (a) BCAP interaction with TLR4 allows TLR4 dimerization, TLR4-Mal interaction, Mal-dimerization, and the entire MyD88-dependent signalosome formation. (b) BCAP interaction with TLR4 has steric clash with TLR4-TRAM binding therefore these two interactions are mutually exclusive: if BCAP binds to TLR4, TRAM cannot bind to TLR4. (c) BCAP interaction with Mal hinders its dimerization and the assembly of MyD88-dependent signalosome. (d) BCAP-MyD88 interaction enables the signalosome formation. (e) BCAP-TRAM interaction interferes with TRAM-dimerization and prevents TRIF-dependent signalosome assembly. (f) BCAP-TRIF interaction blocks TRIF-TRAM interaction due to overlapping binding sites. Red boxes indicate the location of clash and therefore those complexes are not possible. The red-labeled regions are the BB-loops of the proteins. In all parts, the left-hand side figure shows the structure of PPI complexes obtained by PRISM, and the right-hand side schematic figure shows the TIR domain signalosome and how the negative regulators affect the assembly of signalosome. Circles in the schematics represent the TIR domains: cyan represents TLR, yellow represents Mal, purple represents MyD88, light pink represents TRAM, blue represents TRIF and orange represents BCAP. To see this figure in color, go online.
Table 1.
Details of interactions that are predicted by PRISM
| PPI | Protein1 | Protein2 | Template Interface | Interaction Energy | Reference |
|---|---|---|---|---|---|
| BCAP-TLR4 | model | model | 3dahAB | −46.7 | |
| BCAP-Mal | model | 4lqdA | 3urrAB | −57.21 | (6) |
| BCAP-MyD88 | model | 4eo7A | 3f1rAB | −39.95 | (6,30) |
| BCAP-TRAM | model | 2m1wA | 3bdvAB | −27.05 | |
| BCAP-TRIF | model | 2m1xA | 3dhxAB | −29.97 | (33) |
| SIGIRR-TLR4 | model | model | 3imoBD | −22.7 | (7,38–40) |
| SIGIRR-Mal | model | 4fz5A | 1unhBE | −36.06 | |
| SIGIRR-MyD88 | model | 4domA | 3f13AB | −27.84 | (39,40) |
| SIGIRR-TRAM | model | 2m1wA | 1pzmAB | −11.38 | |
| SIGIRR-TRIF | model | 2m1xA | 1ntcAB | −21.38 | (43,44) |
| ST2L-TLR4 | model | model | 2bq1EF | −16.67 | (40) |
| ST2L-Mal | model | 3ub2A | 3urrAB | −40.66 | (29) |
| ST2L-MyD88 | model | 4eo7A | 2oh1CD | −18.47 | (29,39,40) |
| ST2L-TRAM | model | 2m1wA | 3sf8AB | −43.48 | |
| ST2L-TRIF | model | 2m1xA | 3grzAB | −22.39 | |
| TRAF6-p62 | 2k0bX | 3hctA | 3jygAB | −12.09 | (67) |
| p62-CYLD | 1q02A | 1whmA | 2yvzAB | −20.64 | (61,63,64) |
| TRAF6-CYLD | 1whlA | 3hcuA | 2ekyBD | −21.28 | (62) |
| TRAF6-A20 | 3hcsA | 3zjgB | 1f5qAB | −32.08 | (56) |
| A20 dimer | 2vfjB | 2vfjC | 3dkbCF | −48.57 | (54) |
| TRAF3-DUBA | 3tmp | 1fllA | 2a6aAB | −16.29 | (70) |
In addition to interfering with the formation of TIR-domain signalosome, our results indicate that the second scenario is also plausible. For instance, BCAP-TLR4 enables MyD88-dependent signalosome formation, as it does not abolish TLR4-dimerization or TLR4-Mal interaction (Fig. 3a). Moreover, the interaction of BCAP with MyD88 does not intrude into MyD88-dimerization interface and allow signalosome formation (Fig. 3d). Therefore, if BCAP does not disrupt signalosome formation and functions downstream of MyD88, how does it inhibit NF-κB? Because BCAP is a large protein (811 residues), it was suggested that it may interfere with the formation of the myddosome structure, which is composed of death domains (DDs) of MyD88, IRAK4, and IRAK2/1 (35). Our results indicate that when MyD88 binds to BCAP through its TIR domain, it may not be able to oligomerize with other MyD88 and IRAK molecules through its DD due to steric clash that BCAP may cause. Assembly into large complexes and receptor clustering has been shown to activate signaling cascades even in the absence of essential ligands (36), probably via allosteric activation.
Structural details of the interactions allow us to characterize the mechanisms of oncogenic mutations. We mapped the clinically observed cancer mutations onto interfaces of the PPI architectures that we generated and performed in silico mutagenesis. There are 12 oncogenic mutations on BCAP that fall onto the interface residues (Table 2). Our in silico mutagenesis analysis revealed that seven of these mutations abolish BCAP interactions with either one of five TIR domain-containing proteins (TLR4, Mal, MyD88, TRAM, and TRIF). These mutations are observed in melanoma, breast, lung, head and neck, colorectal and stomach cancers. They do not cluster at a specific location on the BCAP structure; rather they disperse almost over the entire BCAP structure (Fig. S1). A recent study showed that disease-causing mutations often change the number of hydrogen-bonds and salt bridges in comparison to nondisease-causing counterparts (37). In line with this, we found that the E50K mutation on BCAP decreased the number of intermolecular H-bonds by 1 and salt bridges by 2 in the BCAP-TLR4 interaction. Moreover, K139fs frameshift mutation reduced the number of salt bridges from seven to four in the BCAP-TRAM interaction. These decreases in intermolecular bonds may also contribute to loss of the interaction with the mutant BCAP. Additionally, there is also a nonsense mutation (G9∗) on BCAP seen in stomach cancer (Table S3) that causes the loss of BCAP and its interactions. Attenuation of the interactions liberates the conventional TLR downstream pathway that leads to NF-κB and MAPK activation and proinflammatory cytokine production. Without a checkpoint to restrict TLR signaling, constitutive production of inflammatory cytokines leads to chronic inflammation, which promotes tumorigenesis.
Table 2.
Details of clinically observed mutations on negative regulators of TLRs
| Cancer Study | Mutation | Mutation Type | Protein | Interface of PPI | Disrupted PPI | Energy Score | Difference of Energy Scores |
|---|---|---|---|---|---|---|---|
| Melanoma (TCGA) | G48S | missense | BCAP | BCAP-TLR4 | none | −40.33 | 6.37 |
| NCI-60 | E50K | missense | BCAP | BCAP-TLR4 | BCAP-TLR4 | −8.9 | 37.8 |
| Melanoma (Broad) | P82L | missense | BCAP | BCAP-TLR4 | BCAP-TLR4 | −7.7 | 39 |
| Melanoma (Yale) | D113H | missense | BCAP | BCAP-TLR4, BCAP-TRAM | none | −13.0 | 33.7 |
| −16.97 | 10.08 | ||||||
| Breast (TCGA pub) | E21K | missense | BCAP | BCAP-Mal, BCAP-TRIF | BCAP-TRIF | −25.16 | 32.05 |
| −9.26 | 20.71 | ||||||
| Lung squ (TCGA) | L27Q | missense | BCAP | BCAP-Mal, BCAP-TRIF | BCAP-TRIF | −51.4 | 5.81 |
| −1.18 | 28.79 | ||||||
| Uterine (TCGA) | V134M | missense | BCAP | BCAP-Mal | none | −44.24 | 12.97 |
| Head & neck (Broad) | A136D | missense | BCAP | BCAP-Mal | BCAP-Mal | none | none |
| Melanoma (Broad) | L60F | missense | BCAP | BCAP-MyD88 | none | −41.88 | -1.93 |
| Colorectal (Genentech) | C66Y | missense | BCAP | BCAP-MyD88 | BCAP-MyD88 | 12.12 | 52.07 |
| Stomach (TCGA pub) | K139fs | FS del | BCAP | BCAP-TRAM | BCAP-TRAM | −4.87 | 22.18 |
| Head & neck (TCGA) | S39N | missense | BCAP | BCAP-TRIF | none | −11.92 | 18.05 |
| pRCC (TCGA) | L282M | missense | SIGIRR | SIGIRR-Mal, SIGIRR-TRIF | SIGIRR-TRIF | −33.01 | 3.05 |
| −9.84 | 11.54 | ||||||
| Cervical (TCGA) | S297F | missense | SIGIRR | SIGIRR-Mal | none | −22.88 | 11.18 |
| Breast (TCGA) | Y377N | missense | ST2 | ST2-TLR4 | none | −11.99 | 4.68 |
| Melanoma (TCGA) | P384Q | missense | ST2 | ST2-TLR4 | ST2-TLR4 | −5.45 | 11.22 |
| Melanoma (TCGA) | P406L | missense | ST2 | ST2-TLR4 | none | −12.45 | 4.22 |
| Melanoma (Yale) | E410K | missense | ST2 | ST2-TLR4 | none | −14.96 | 1.71 |
| Lung adeno (TCGA pub) | L425Q | missense | ST2 | ST2-TLR4 | ST2-TLR4 | 2.06 | 18.73 |
| Melanoma (TCGA), Melanoma (Yale) | R439Q | missense | ST2 | ST2-TLR4 | none | −19.3 | -2.63 |
| Stomach (TCGA) | E226K | missense | CYLD | CYLD-p62 | none | −20.38 | 0.26 |
| Melanoma (TCGA) | P229S | missense | CYLD | CYLD-p62 | none | −18.34 | 2.3 |
| Head & neck (TCGA) | N300S | missense | CYLD | CYLD-p62 | none | −15.82 | 4.82 |
| Uterine (TCGA) | I303V | missense | CYLD | CYLD-p62 | CYLD-p62 | none | none |
| Lung adeno (TCGA) | A305S | missense | CYLD | CYLD-p62 | none | −16.7 | 3.94 |
| ccRCC (TCGA) | P130T | missense | CYLD | CYLD-TRAF6 | none | −10.35 | 10.93 |
| Uterine (TCGA) | L186R | missense | CYLD | CYLD-TRAF6 | none | −13.92 | 7.36 |
Energy score below −10 indicates favorable interaction. If the difference of energy scores between mutant and wild-type is positive, the mutation is then destabilizing.
SIGIRR
An orphan receptor SIGIRR (also known as TIR8) has a single extracellular immunoglobulin domain, a transmembrane domain, a cytoplasmic TIR domain, and an unusually long tail (95 residues), which is missing in other TIR domain-containing receptors (7,38,39). SIGIRR inhibits NF-κB activation induced by only TIR domain-containing receptors, like TLR and IL-1R1, but not the others such as tumor necrosis factor receptor that do not possess a TIR domain (7). Increased production of inflammatory cytokines and chemokines has been reported in SIGIRR-deficient mice after IL-1 and LPS administration (28). Mice lacking SIGIRR have been shown to be more prone to develop intestinal tumors (39). The mechanism of inhibition of TLRs by SIGIRR remains unclear (40). Although its extracellular immunoglobulin and intracellular TIR domains restrain IL1 signaling by blocking the interaction between IL-1R1 and IL-1RAP, only its TIR domain inhibits TLR signaling (41). In particular, the BB-loop of its TIR domain appears important in association with TLRs, because deletion of the BB-loop abolished the inhibitory role of SIGIRR (39,42). The BB-loop is conserved among TIR domain-containing proteins (34).
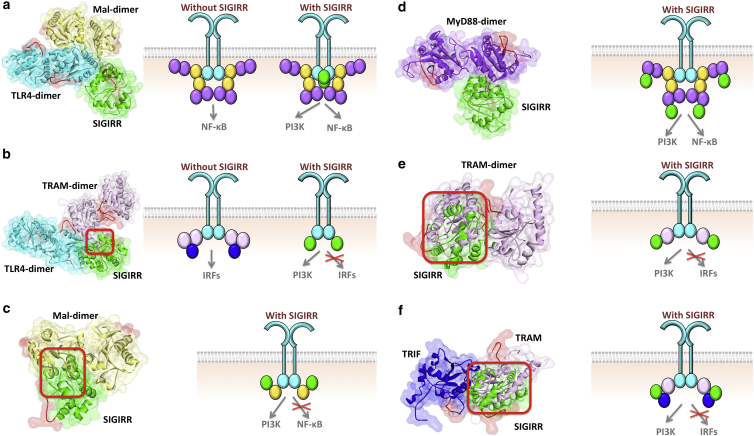
SIGIRR could either interfere with the organization of TIR domain signalosome (39–41) or block the translocation of this complex from the receptor (28). It can form heterodimers with TLR4 (40) and decrease the recruitment of MyD88 and IRAK4 to activated TLR4 (41). We found that SIGIRR can interact with all TIR domain-containing proteins in the TLR pathway (Fig. 4). SIGIRR’s BB-loop is not involved in its interactions with Mal and TRAM, but close to the interface region in the MyD88, TLR4, and TRIF interactions. Similar to BCAP, SIGIRR interactions with Mal, TRAM, and TRIF interfere with Mal-homodimerization (Fig. 4c), TRAM-homodimerization (Fig. 4e), and TRIF-TRAM (Fig. 4f) interactions, respectively; hence, they prevent the signalosome assembly. In addition to the MyD88-dependent pathway, SIGIRR also modulates the TRIF-dependent pathway, because it also inhibits TLR3 signaling (43,44), which signals only through TRIF, but not MyD88. The architectures suggest that the mechanism of this inhibition is by SIGIRR blocking TRAM-homodimerization (Fig. 4e), TLR4-TRAM (Fig. 4b), and TRIF-TRAM (Fig. 4f) interactions.
Figure 4.
SIGIRR interactions with all TIR domain-containing proteins in TLR signaling. (a) SIGIRR interaction with TLR4 allows TLR4 dimerization, TLR4-Mal interaction, Mal-dimerization, and the entire MyD88-dependent signalosome formation. (b) SIGIRR-TLR4 interaction has steric clash with TLR4-TRAM binding therefore these two interactions are mutually exclusive: if SIGIRR binds to TLR4, TRAM cannot bind to TLR4. (c) SIGIRR interaction with Mal hinders its dimerization and the assembly of MyD88-dependent signalosome. (d) SIGIRR-MyD88 interaction enables the signalosome formation. (e) SIGIRR-TRAM interaction interferes with TRAM-dimerization and prevents TRIF-dependent signalosome assembly. (f) SIGIRR-TRIF interaction blocks TRIF-TRAM interaction due to overlapping binding sites. Red boxes indicate the location of clash. The red-labeled regions are the BB-loops of the proteins. In all parts, the left-hand side figure shows the structure of PPI complexes obtained by PRISM, and the right-hand side schematic figure shows the TIR domain signalosome and how the negative regulators affect the assembly of signalosome. Circles in the schematics represent the TIR domains: cyan represents TLR, yellow represents Mal, purple represents MyD88, light pink represents TRAM, blue represents TRIF and green represents SIGIRR. To see this figure in color, go online.
TLR4-SIGIRR (Fig. 4a) and MyD88-SIGIRR (Fig. 4d) architectures are feasible with the MyD88-dependent signalosome formation. Although it has been proposed to hinder MyD88 homodimerization (39,40), which is vital for myddosome formation (45,46), our model suggests that it abolishes neither MyD88-dimerization nor signalosome organization. Therefore, if TLR4 or MyD88 are the proteins that SIGIRR interact with and they do not interfere with the signalosome, how does it hinder the transmission of the signal from TLR4 to NF-κB? The answer could lie in the inhibition of the translocation of this complex from the receptor (28). This strategy is also used by other negative regulators of TLRs, such as IRAK-M, which prevents the dissociation of the myddosome structure from the receptor complex (8,47).
SIGIRR has only two clinically observed oncogenic mutations in the interface (Table 2). Only one of the mutations (L282M) falls on the SIGIRR-Mal and SIGIRR-TRIF interfaces and it abrogated the SIGIRR-TRIF interaction. It also has a nonsense mutation (Q111∗) and a frameshift mutation (P2fs) (Table S3) that produce truncated SIGIRR lacking a TIR domain and abolish its interactions presented here.
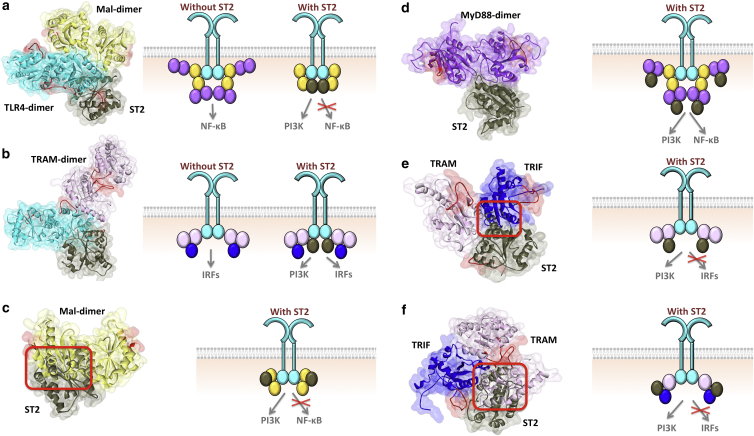
ST2 (ST2L)
ST2 (IL1RL1) is also an orphan receptor with a cytoplasmic TIR domain (7). It inhibits NF-κB activation in response to IL-1R1 and TLR stimulation (29). Overexpression of ST2 prevents activation by TLR4- but not TLR3-induced NF-κB (TLR3 uses only a TRIF-dependent downstream path). In addition, ST2 coimmunoprecipitates with Mal and MyD88, but not with TRIF (29). This suggests that ST2 has inhibitory roles only in the MyD88-dependent pathway of TLRs.
As with other TIR domain-containing regulators, we obtained possible interaction models of ST2 with all TIR domain-containing proteins in TLR signaling (Fig. 5). Although its interactions with TLR4 (Fig. 5a), MyD88 (Fig. 5d), and TRAM (Fig. 5e) allow homodimerization of these proteins, its interactions with Mal and TRIF abolish Mal-homodimerization (Fig. 5c) and TRAM-TRIF interaction (Fig. 5f). Although it may not interact with TRAM or TRIF under physiological conditions (another protein may prevent the association or the proteins may not be coexpressed), PRISM still finds these interactions.
Figure 5.
ST2 interactions with all TIR domain-containing proteins in TLR signaling. (a and b) ST2 interaction with TLR4 allows TLR4 dimerization, Mal-dimerization, and TRAM dimerization and the formation of both MyD88- and TRIF-dependent signalosomes. (c) ST2-Mal interaction hinders its dimerization and the assembly of Myd88-dependent signalosome. (d) ST2-MyD88 interaction enables the signalosome formation. (e) ST2-TRAM interaction interferes with TRAM-TRIF. (f) ST2-TRIF interaction blocks TRIF-TRAM interaction due to overlapping binding sites. Red boxes indicate the location of clash. The red-labeled regions are the BB-loops of the proteins. In all parts, the left-hand side figure shows the structure of PPI complexes obtained by PRISM, and the right-hand side schematic figure shows the TIR domain signalosome and how the negative regulators affect the assembly of signalosome. Circles in the schematics represent the TIR domains: cyan represents TLR, yellow represents Mal, purple represents MyD88, light pink represents TRAM, blue represents TRIF and dark gray represents ST2. To see this figure in color, go online.
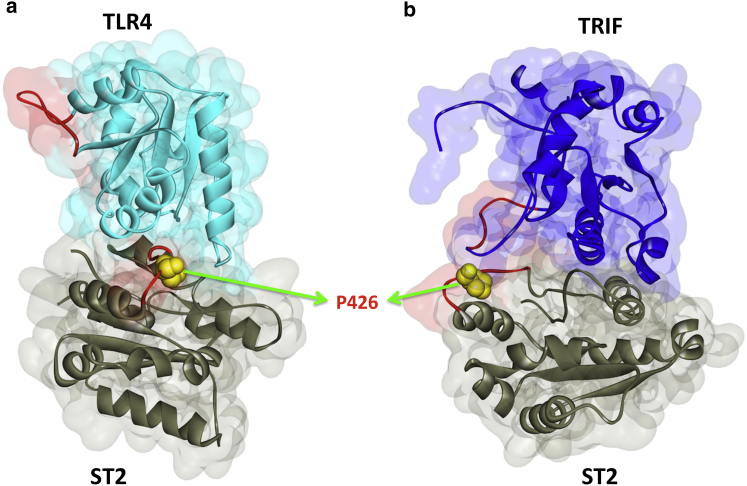
In mice, ST2 loses its inhibitory action in a proline-to-histidine mutation at position 431 of its TIR domain (7,29). This residue corresponds to P426 in the human homolog. P426 is found at the BB-loop of ST2, which is estimated by alignment of the ST2 structure with other TIR domains. This residue is on the TLR4-ST2 and TRIF-ST2 interfaces (Fig. 6) and the P426H mutation abolishes TLR4-ST2 interaction. In addition to this mutation, there are six oncogenic mutations on the interfaces of ST2 (Table 2) and two of them impede the ST2-TLR4 interaction. There are also eight nonsense and frameshift mutations on ST2 (Table S3), leading to loss of the TIR domain and all interactions with the TIR-containing proteins.
Figure 6.
P426 residue of ST2 is at the interface of its interactions with (a) TLR4 and (b) TRIF. P426H mutation abolishes TLR4-ST2 interaction. The red-labeled regions are the BB-loops of the proteins. To see this figure in color, go online.
Ubiquitinases and deubiquitinases as regulators of TLR signaling
Ubiquitination is a reversible posttranslational modification and it is one of the most prevalent in TLR signaling and NF-κB activation (48), like phosphorylation (49,50). Different ubiquitination modes may result in opposing outcomes, like degradation or signaling of TRAF3 (51). Ub chains serve as an anchor, assisting in the assembly of large protein complexes. For instance, the K63-linked Ub chain attached to TRAF6 recruits TAB2/3 and NEMO (IKK-γ); however, these proteins do not associate directly with TRAF6 itself. Because NF-κB activation depends on ubiquitination, evolution developed Ub-dependent regulation to abate inflammation (52). Some of the negative modulators of the TLR pathway are ubiqutinases that add K48-linked Ub chains to essential orchestrators. TRIAD3A catalyzes the K48-linked ubiquitination and leads to proteosomal degradation of Mal and TRIF (48); and Nrdp1 adds K48-Ub to MyD88, inhibits the MyD88-dependent TLR pathway and preferentially enhances TRIF-dependent IFN expression (53). Some others are deubiquitinases (DUBs), such as A20 (also known as TNFAIP3), CYLD (cylindromatosis D), and DUBA (deubiquitinating enzyme A), which negatively modulate TLR signaling by removing the K63-linked Ub chains from proteins, such as TRAF6 and TRAF3 (48). Below we demonstrate that these key DUBs turn off TLR signaling because they prevent the binding of TRAF6 and TRAF3 to their partners in addition to removing their Ub chains.
A20
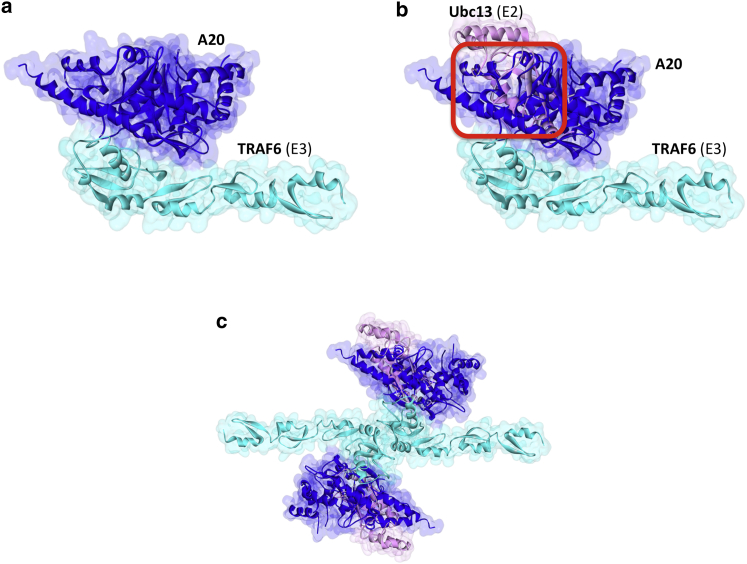
A20 is a pleiotropic Ub editing enzyme: it has an N-terminal ovarian tumor (OTU) domain with DUB activity, and seven C-terminal zinc finger domains (ZF)—the fourth (ZF4) with E3 ligase activity (54,55). Although it is a DUB, it also catalyzes the addition of K48-linked Ub chains (56,57). It antagonizes TLR- and tumor necrosis factor receptor-mediated NF-κB activation (57). TRAF6 is a substrate for A20 in the TLR pathway (48). A20 was also proposed to inhibit the association of TRAF6 (E3) with Ubc13 (E2) and UbcH5c (E2) (56). A20 forms homodimers through the ZF4 domains and its higher-order oligomerization facilitates its deubiquitinase, E3 ligase and Ub-binding functions (54). The A20 homodimer structures in the asymmetric unit of two pdb entries (3dkb_CF and 2vfj_BC) were suggested to be biological because mutation of the interface residues (M15A, R16E, and H351A) suppressed A20-dimerization (54). We obtained similar homodimer with PRISM (Fig. S2 a).
We found interactions between the RING domain of TRAF6 and the OTU domain of A20 (Fig. 7) and this architecture also allows A20 homodimerization (Fig. S2 b). A20-TRAF6 interaction abolishes TRAF6-Ubc13 as proposed earlier (56,57), because they have overlapping binding sites on TRAF6. C103, H256, and T97 residues on A20 were determined to be the catalytic residues (57). The C103A mutation inhibits A20 catalytic function (C103A mutant of A20 was unable to deubiquitinate TRAF6) (57) and TRAF2-Ubc13 interaction (58). However, this residue is not on the interface of TRAF6-A20 that we obtained. This result is expected because C103A mutant of A20 was shown to still bind to TRAF6 (59). The reason for the blockage of TRAF2-Ubc13 interaction may be that the C103 residue is involved in the interface of TRAF2-A20 complex; alternatively, mutation of this residue disrupts the stability of A20 and allosterically modifies its interaction sites. In addition to inhibition of E2-E3 interactions, A20 has also roles in degradation of E2 proteins (e.g., Ubc13) by attaching K48-linked Ub chains to E2, thus affecting E2-dependent E3 function (60). We could not find clinically observed oncogenic mutations on A20 that fall onto A20-TRAF6 interface residues or nonsense/frameshift mutations that lead to truncated A20 without OTU domain the TCGA database.
Figure 7.
A20 interaction with TRAF6. (a) OTU domain of A20 interacts with RING domain of TRAF6. (b) Ubc13 and A20 have almost completely overlapping interfaces on TRAF6 and hence their interactions with TRAF6 are mutually exclusive. Red square indicates the location of clash. TRAF6-Ubc13 interaction is obtained from crystal structure (3hcu_AB). (c) TRAF6-dimer through its RING domain and its interactions with A20 and Ubc13. To see this figure in color, go online.
CYLD
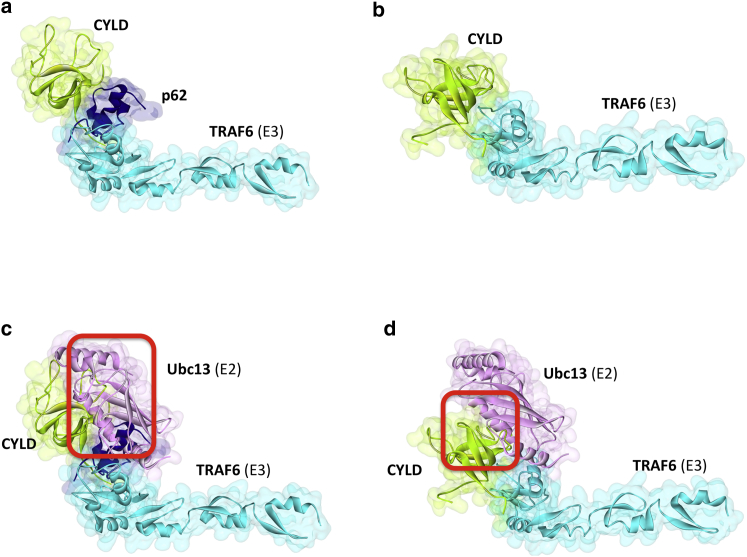
CYLD is a Ub-specific protease with a tumor suppressive role. It removes K63-linked Ub chains from TRAF6 (48) and negatively regulates NF-κB and expression of proinflammatory cytokines (61,62). To date, its NF-κB inhibition mechanism has been unclear. CYLD interacts with TRAF6 (62), but it was also suggested to require some bridging adaptor proteins, like p62 (also known as sequestosome1) to interact with its substrates because its activity is reduced in the absence of p62 (61,63,64).
We found TRAF6 interactions with CYLD either via direct binding or via p62 (Fig. 8, a and b). CYLD has three CAP-Gly (cytoskeleton-associated protein-glycine conserved) domains with similar global folds. The third is suggested to interact with TRAF2 (65,66). As to direct interaction, we found that the first CAP-Gly domain of CYLD binds to the RING domain of TRAF6. However, in the p62 adaptor case, the second CAP-Gly domain of CYLD interacts with p62, which interacts with TRAF6. Residues 225–255 of p62 were already identified as an interface that binds to TRAF6 (67) and this is in agreement with our TRAF6-p62 complex architecture. Both the direct and indirect (through p62) associations sterically hinder the TRAF6-Ubc13 interaction (Fig. 8, c and d). If p62-CYLD or CYLD-itself binds this interface on TRAF6, Ubc13 cannot approach that site and TRAF6 is nonfunctional.
Figure 8.
Architectures of direct and indirect interactions of TRAF6 with CYLD. (a) Indirect association of TRAF6 with CYLD through p62 bridging adaptor. (b) Direct interaction of TRAF6 with CYLD. (c) In the p62 case, both p62 and CYLD has steric clashes with Ubc13. (d) In the direct binding, CYLD has steric clash with Ubc13. Red squares indicate the location of clash. TRAF6-Ubc13 interaction is obtained from crystal structure (3hcu_AB). To see this figure in color, go online.
Why does TLR negative regulation have redundant players with the same function and the same substrate (TRAF6 with K63-Ub chain), such as A20 and CYLD? The timing of their action may address this question: CYLD is constitutively expressed, but A20 is not (61). Alternatively, they may act in distinct cell types, or subcellular locations (68). They may also cooperate to remove the Ub chains from TRAF6.
Seven missense mutations on CYLD correspond to the interface residues, but only one of them causes the loss of its interactions (p62-CYLD) according to our results (Table 2). Additionally, there are also six nonsense or frameshift mutations that lead to truncated protein without CAP-Gly or DUB domains (Table S3). Because these mutations cause loss of the domains that are required for CYLD-TRAF6 or CYLD-p62 interactions, the truncated CYLD can no longer associate with these proteins and deubiquitinate TRAF6. Nonsense and frameshift mutations predominantly constitute the mutations of CYLD in tumors (59,69).
DUBA (OTUD5)
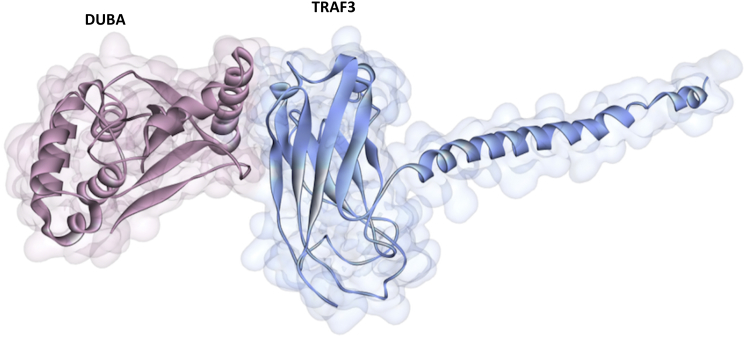
Similar to A20, DUBA (also known as OTUD5) is also an OTU domain family member cysteine protease. It suppresses IFN-I production in response to TLR activation by removing K63-Ub chains from TRAF3 and preventing recruitment of TBK1 and other downstream proteins, which are necessary for NF-κB and IRF activation (48,51,70). Interaction of endogenous TRAF3 and DUBA takes place upon TLR3 stimulation (70). The D221, C224, and H334 are the catalytic residues of DUBA and C224S mutation on DUBA compromised its function (deubiquitinating TRAF3) (70). There is less Ub alteration in TRAF3 lacking a RING domain (70), indicating that the RING domain of TRAF3 is probably the interaction region. However, we could not find interaction between the RING domain of TRAF3 and catalytic OTU domain of DUBA. Instead, we found interaction with concave site on TRAF-C region of TRAF3 (Fig. 9). This concave region is where TRAF3 interacts with CD40 (1fll.pdb) (71), BAFFR (2gkw.pdb) (72), LMP1 (1zms.pdb) (73), Cardif (4ghu.pdb) (74), and also MyD88 (10). If DUBA also binds to TRAF-C region in addition to the RING domain of TRAF3, it attenuates TLR-induced IRF activation not only through its catalytic (DUB) function, but also through interfering with the TRAF3 interactions with other signaling proteins. There are no clinically observed oncogenic mutations on DUBA that fall onto DUBA-TRAF3 interface residues or nonsense/frameshift mutations.
Figure 9.
The architecture of the interaction between the OTU domain of DUBA and TRAF-C domain of TRAF3. To see this figure in color, go online.
Parallel downstream paths of TLRs
The TLR pathway activates different parallel downstream paths. The conventional downstream path of TLRs is the MyD88-dependent path, which involves TRAF6 and IRAKs to activate NF-κB and MAPK and to produce proinflammatory cytokines. There are also TRIF-dependent, TRAF3-dependent, Fas associated death-domain protein (FADD)-dependent, and BCAP-dependent paths, which lead to production of IFNs apoptosis, or activation of the PI3K pathway, respectively. Our recent study (10) suggests that almost all parallel pathways of TLRs compete with each other and thus restrict the activation of one another due to overlapping binding sites on a common binding partner (Fig. 2). These parallel paths only switch the function: while negatively regulating one path, they positively regulate others. The negative regulators covered in this study, except BCAP, cease TLR signaling. Network flow algorithms might be helpful to understand the interplay between parallel paths in TLR pathway (75).
Conclusions
The TLR pathway resembles a double-edged sword: On the one hand, it is vital for triggering proper host immune response against pathogens and its malfunction makes individuals more susceptible to infections. On the other, its overactivation results in excess inflammation, which is detrimental to the host. The cell has evolved several mechanisms to strike a balance between activation and inhibition of TLR signaling at almost each step of the pathway. Although several negative regulators have been identified, their inhibition mechanisms remain elusive. Here, we provide the structural basis for the regulation of TLR signaling. Overall, we observed that negative regulators tend to interfere with the assembly of key signaling complexes, such as TIR-domain signalosome, compete with proteins to bind to their partners, and modify ubiquitination states and target them for proteosomal degradation. Similarly, parallel downstream paths compete with each other and restrain their activation. Several oncogenic mutations on negative modulators tend to disrupt their interactions with the key players in TLR pathway, liberating the conventional downstream path and constitutive activation of NF-κB. Better understanding of negative regulation of TLR signaling holds promise for novel treatment strategies for autoimmune diseases and cancer.
Author Contributions
Conceived and designed research: E.G.M., O.K., A.G., and R.N. Performed research: E.G.M. Analyzed the data: E.G.M. Wrote the article: E.G.M., O.K., A.G., and R.N.
Acknowledgments
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. A.G. and O.K. are members of the Science Academy, Turkey.
Editor: H. Jane Dyson.
Footnotes
Two figures and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00657-8.
Contributor Information
Ozlem Keskin, Email: okeskin@ku.edu.tr.
Ruth Nussinov, Email: nussinor@helix.nih.gov.
Supporting Material
References
- 1.Guven Maiorov E., Keskin O., Nussinov R. The structural network of inflammation and cancer: merits and challenges. Semin. Cancer Biol. 2013;23:243–251. doi: 10.1016/j.semcancer.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Trinchieri G. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu. Rev. Immunol. 2012;30:677–706. doi: 10.1146/annurev-immunol-020711-075008. [DOI] [PubMed] [Google Scholar]
- 3.Matsumura T., Oyama M., Inoue J. Identification of BCAP-(L) as a negative regulator of the TLR signaling-induced production of IL-6 and IL-10 in macrophages by tyrosine phosphoproteomics. Biochem. Biophys. Res. Commun. 2010;400:265–270. doi: 10.1016/j.bbrc.2010.08.055. [DOI] [PubMed] [Google Scholar]
- 4.Chang Z.L. Important aspects of Toll-like receptors, ligands and their signaling pathways. Inflamm. Res. 2010;59:791–808. doi: 10.1007/s00011-010-0208-2. [DOI] [PubMed] [Google Scholar]
- 5.Ni M., MacFarlane A.W., 4th, Hamerman J.A. B-cell adaptor for PI3K (BCAP) negatively regulates Toll-like receptor signaling through activation of PI3K. Proc. Natl. Acad. Sci. USA. 2012;109:267–272. doi: 10.1073/pnas.1111957108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Troutman T.D., Hu W., Pasare C. Role for B-cell adapter for PI3K (BCAP) as a signaling adapter linking Toll-like receptors (TLRs) to serine/threonine kinases PI3K/Akt. Proc. Natl. Acad. Sci. USA. 2012;109:273–278. doi: 10.1073/pnas.1118579109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liew F.Y., Xu D., O’Neill L.A. Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 8.Wang J., Hu Y., Deng W.W., Sun B. Negative regulation of Toll-like receptor signaling pathway. Microbes Infect. 2009;11:321–327. doi: 10.1016/j.micinf.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Pradere J.P., Dapito D.H., Schwabe R.F. The Yin and Yang of Toll-like receptors in cancer. Oncogene. 2014;33:3485–3495. doi: 10.1038/onc.2013.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guven Maiorov, E., O. Keskin, …, R. Nussinov. 2015. The architecture of the TIR domain signalosome in the Toll-like Receptor-4 signaling pathway. Sci. Rep. 10.1038/srep13128. [DOI] [PMC free article] [PubMed]
- 11.Guven-Maiorov E., Acuner-Ozbabacan S.E., Nussinov R. Structural pathways of cytokines may illuminate their roles in regulation of cancer development and immunotherapy. Cancers (Basel) 2014;6:663–683. doi: 10.3390/cancers6020663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acuner-Ozbabacan E.S., Engin B.H., Nussinov R. The structural network of Interleukin-10 and its implications in inflammation and cancer. BMC Genomics. 2014;15(Suppl 4):S2. doi: 10.1186/1471-2164-15-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogmen U., Keskin O., Gursoy A. PRISM: protein interactions by structural matching. Nucleic Acids Res. 2005;33:W331–W336. doi: 10.1093/nar/gki585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuncbag N., Gursoy A., Keskin O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat. Protoc. 2011;6:1341–1354. doi: 10.1038/nprot.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baspinar A., Cukuroglu E., Gursoy A. PRISM: a web server and repository for prediction of protein-protein interactions and modeling their 3D complexes. Nucleic Acids Res. 2014;42:W285–W289. doi: 10.1093/nar/gku397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mashiach E., Nussinov R., Wolfson H.J. FiberDock: flexible induced-fit backbone refinement in molecular docking. Proteins. 2010;78:1503–1519. doi: 10.1002/prot.22668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kar G., Keskin O., Gursoy A. Human proteome-scale structural modeling of E2-E3 interactions exploiting interface motifs. J. Proteome Res. 2012;11:1196–1207. doi: 10.1021/pr2009143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acuner Ozbabacan S.E., Keskin O., Gursoy A. Enriching the human apoptosis pathway by predicting the structures of protein-protein complexes. J. Struct. Biol. 2012;179:338–346. doi: 10.1016/j.jsb.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuncbag N., Keskin O., Gursoy A. Fast and accurate modeling of protein-protein interactions by combining template-interface-based docking with flexible refinement. Proteins. 2012;80:1239–1249. doi: 10.1002/prot.24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E., Gao J., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cukuroglu E., Gursoy A., Keskin O. HotRegion: a database of predicted hot spot clusters. Nucleic Acids Res. 2012;40:D829–D833. doi: 10.1093/nar/gkr929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Ferrao R., Li J., Wu H. Structural insights into the assembly of large oligomeric signalosomes in the Toll-like receptor-interleukin-1 receptor superfamily. Sci. Signal. 2012;5:re3. doi: 10.1126/scisignal.2003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nussinov R., Jang H. Dynamic multiprotein assemblies shape the spatial structure of cell signaling. Prog. Biophys. Mol. Biol. 2014;116:158–164. doi: 10.1016/j.pbiomolbio.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nussinov R., Ma B., Tsai C.J. A broad view of scaffolding suggests that scaffolding proteins can actively control regulation and signaling of multienzyme complexes through allostery. Biochim. Biophys. Acta. 2013;1834:820–829. doi: 10.1016/j.bbapap.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 27.Nussinov R., Tsai C.J., Liu J. Principles of allosteric interactions in cell signaling. J. Am. Chem. Soc. 2014;136:17692–17701. doi: 10.1021/ja510028c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wald D., Qin J., Li X. SIGIRR, a negative regulator of Toll-like receptor-interleukin 1 receptor signaling. Nat. Immunol. 2003;4:920–927. doi: 10.1038/ni968. [DOI] [PubMed] [Google Scholar]
- 29.Brint E.K., Xu D., Liew F.Y. ST2 is an inhibitor of interleukin 1 receptor and Toll-like receptor 4 signaling and maintains endotoxin tolerance. Nat. Immunol. 2004;5:373–379. doi: 10.1038/ni1050. [DOI] [PubMed] [Google Scholar]
- 30.Rhee S.H., Kim H., Pothoulakis C. Role of MyD88 in phosphatidylinositol 3-kinase activation by flagellin/toll-like receptor 5 engagement in colonic epithelial cells. J. Biol. Chem. 2006;281:18560–18568. doi: 10.1074/jbc.M513861200. [DOI] [PubMed] [Google Scholar]
- 31.Troutman T.D., Bazan J.F., Pasare C. Toll-like receptors, signaling adapters and regulation of the pro-inflammatory response by PI3K. Cell Cycle. 2012;11:3559–3567. doi: 10.4161/cc.21572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bovijn C., Desmet A.S., Peelman F. Identification of binding sites for myeloid differentiation primary response gene 88 (MyD88) and Toll-like receptor 4 in MyD88 adapter-like (Mal) J. Biol. Chem. 2013;288:12054–12066. doi: 10.1074/jbc.M112.415810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aksoy E., Vanden Berghe W., Willems F. Inhibition of phosphoinositide 3-kinase enhances TRIF-dependent NF-kappa B activation and IFN-beta synthesis downstream of Toll-like receptor 3 and 4. Eur. J. Immunol. 2005;35:2200–2209. doi: 10.1002/eji.200425801. [DOI] [PubMed] [Google Scholar]
- 34.Enokizono Y., Kumeta H., Inagaki F. Structures and interface mapping of the TIR domain-containing adaptor molecules involved in interferon signaling. Proc. Natl. Acad. Sci. USA. 2013;110:19908–19913. doi: 10.1073/pnas.1222811110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin S.C., Lo Y.C., Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Motshwene P.G., Moncrieffe M.C., Gay N.J. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J. Biol. Chem. 2009;284:25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petukh M., Kucukkal T.G., Alexov E. On human disease-causing amino acid variants: statistical study of sequence and structural patterns. Hum. Mutat. 2015;36:524–534. doi: 10.1002/humu.22770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allavena P., Garlanda C., Mantovani A. Pathways connecting inflammation and cancer. Curr. Opin. Genet. Dev. 2008;18:3–10. doi: 10.1016/j.gde.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Riva F., Bonavita E., Garlanda C. TIR8/SIGIRR is an Interleukin-1 Receptor/Toll Like Receptor Family Member with Regulatory Functions in Inflammation and Immunity. Front. Immunol. 2012;3:322. doi: 10.3389/fimmu.2012.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong J., Wei T., Rössle S.C. Inhibition of Toll-like receptors TLR4 and 7 signaling pathways by SIGIRR: a computational approach. J. Struct. Biol. 2010;169:323–330. doi: 10.1016/j.jsb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 41.Qin J., Qian Y., Li X. SIGIRR inhibits interleukin-1 receptor- and toll-like receptor 4-mediated signaling through different mechanisms. J. Biol. Chem. 2005;280:25233–25241. doi: 10.1074/jbc.M501363200. [DOI] [PubMed] [Google Scholar]
- 42.Lech M., Skuginna V., Anders H.J. Lack of SIGIRR/TIR8 aggravates hydrocarbon oil-induced lupus nephritis. J. Pathol. 2010;220:596–607. doi: 10.1002/path.2678. [DOI] [PubMed] [Google Scholar]
- 43.Garlanda C., Anders H.J., Mantovani A. TIR8/SIGIRR: an IL-1R/TLR family member with regulatory functions in inflammation and T cell polarization. Trends Immunol. 2009;30:439–446. doi: 10.1016/j.it.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Drexler S.K., Kong P., Foxwell B.M. SIGIRR/TIR-8 is an inhibitor of Toll-like receptor signaling in primary human cells and regulates inflammation in models of rheumatoid arthritis. Arthritis Rheum. 2010;62:2249–2261. doi: 10.1002/art.27517. [DOI] [PubMed] [Google Scholar]
- 45.Burns K., Martinon F., Tschopp J. MyD88, an adapter protein involved in interleukin-1 signaling. J. Biol. Chem. 1998;273:12203–12209. doi: 10.1074/jbc.273.20.12203. [DOI] [PubMed] [Google Scholar]
- 46.Fekonja O., Benčina M., Jerala R. Toll/interleukin-1 receptor domain dimers as the platform for activation and enhanced inhibition of Toll-like receptor signaling. J. Biol. Chem. 2012;287:30993–31002. doi: 10.1074/jbc.M112.376186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobayashi K., Hernandez L.D., Flavell R.A. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 48.Bibeau-Poirier A., Servant M.J. Roles of ubiquitination in pattern-recognition receptors and type I interferon receptor signaling. Cytokine. 2008;43:359–367. doi: 10.1016/j.cyto.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 49.Nishi H., Shaytan A., Panchenko A.R. Physicochemical mechanisms of protein regulation by phosphorylation. Front. Genet. 2014;5:270. doi: 10.3389/fgene.2014.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishi H., Demir E., Panchenko A.R. Crosstalk between signaling pathways provided by single and multiple protein phosphorylation sites. J. Mol. Biol. 2015;427:511–520. doi: 10.1016/j.jmb.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Häcker H., Tseng P.H., Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immunol. 2011;11:457–468. doi: 10.1038/nri2998. [DOI] [PubMed] [Google Scholar]
- 52.Wullaert A., Heyninck K., Beyaert R. Ubiquitin: tool and target for intracellular NF-kappaB inhibitors. Trends Immunol. 2006;27:533–540. doi: 10.1016/j.it.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 53.Wang C., Chen T., Cao X. The E3 ubiquitin ligase Nrdp1 ‘preferentially’ promotes TLR-mediated production of type I interferon. Nat. Immunol. 2009;10:744–752. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- 54.Lu T.T., Onizawa M., Ma A. Dimerization and ubiquitin mediated recruitment of A20, a complex deubiquitinating enzyme. Immunity. 2013;38:896–905. doi: 10.1016/j.immuni.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wertz I.E., O’Rourke K.M., Dixit V.M. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 56.Catrysse L., Vereecke L., van Loo G. A20 in inflammation and autoimmunity. Trends Immunol. 2014;35:22–31. doi: 10.1016/j.it.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 57.Lin S.C., Chung J.Y., Wu H. Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20. J. Mol. Biol. 2008;376:526–540. doi: 10.1016/j.jmb.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shembade N., Ma A., Harhaj E.W. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jung S.M., Lee J.H., Park S.H. Smad6 inhibits non-canonical TGF-β1 signalling by recruiting the deubiquitinase A20 to TRAF6. Nat. Commun. 2013;4:2562. doi: 10.1038/ncomms3562. [DOI] [PubMed] [Google Scholar]
- 60.Ma A., Malynn B.A. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat. Rev. Immunol. 2012;12:774–785. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun S.C. CYLD: a tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ. 2010;17:25–34. doi: 10.1038/cdd.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshida H., Jono H., Li J.D. The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling via negative cross-talk with TRAF6 AND TRAF7. J. Biol. Chem. 2005;280:41111–41121. doi: 10.1074/jbc.M509526200. [DOI] [PubMed] [Google Scholar]
- 63.Seibenhener M.L., Babu J.R., Wooten M.W. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell. Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wooten M.W., Geetha T., Moscat J. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J. Biol. Chem. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 65.Saito K., Kigawa T., Yokoyama S. The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKgamma. Structure. 2004;12:1719–1728. doi: 10.1016/j.str.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 66.Zheng C., Yin Q., Wu H. Structural studies of NF-κB signaling. Cell Res. 2011;21:183–195. doi: 10.1038/cr.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wooten M.W., Seibenhener M.L., Moscat J. The atypical protein kinase C-interacting protein p62 is a scaffold for NF-kappaB activation by nerve growth factor. J. Biol. Chem. 2001;276:7709–7712. doi: 10.1074/jbc.C000869200. [DOI] [PubMed] [Google Scholar]
- 68.Veres D.V., Gyurkó D.M., Csermely P. ComPPI: a cellular compartment-specific database for protein-protein interaction network analysis. Nucleic Acids Res. 2015;43:D485–D493. doi: 10.1093/nar/gku1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Komander D., Lord C.J., Barford D. The structure of the CYLD USP domain explains its specificity for Lys-63-linked polyubiquitin and reveals a B box module. Mol. Cell. 2008;29:451–464. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 70.Kayagaki N., Phung Q., Dixit V.M. DUBA: a deubiquitinase that regulates type I interferon production. Science. 2007;318:1628–1632. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- 71.Ni C.Z., Welsh K., Ely K.R. Molecular basis for CD40 signaling mediated by TRAF3. Proc. Natl. Acad. Sci. USA. 2000;97:10395–10399. doi: 10.1073/pnas.97.19.10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ni C.Z., Oganesyan G., Ely K.R. Key molecular contacts promote recognition of the BAFF receptor by TNF receptor-associated factor 3: implications for intracellular signaling regulation. J. Immunol. 2004;173:7394–7400. doi: 10.4049/jimmunol.173.12.7394. [DOI] [PubMed] [Google Scholar]
- 73.Wu S., Xie P., Ely K.R. LMP1 protein from the Epstein-Barr virus is a structural CD40 decoy in B lymphocytes for binding to TRAF3. J. Biol. Chem. 2005;280:33620–33626. doi: 10.1074/jbc.M502511200. [DOI] [PubMed] [Google Scholar]
- 74.Zhang P., Reichardt A., Liu Y. Single amino acid substitutions confer the antiviral activity of the TRAF3 adaptor protein onto TRAF5. Sci. Signal. 2012;5:ra81. doi: 10.1126/scisignal.2003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Isik Z., Ersahin T., Cetin-Atalay R. A signal transduction score flow algorithm for cyclic cellular pathway analysis, which combines transcriptome and ChIP-seq data. Mol. Biosyst. 2012;8:3224–3231. doi: 10.1039/c2mb25215e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.