Abstract
Despite much attention, history of sheep (Ovis aries) evolution, including its dating, demographic trajectory and geographic spread, remains controversial. To address these questions, we generated 45 complete and 875 partial mitogenomic sequences, and performed a meta-analysis of these and published ovine mitochondrial DNA sequences (n = 3,229) across Eurasia. We inferred that O. orientalis and O. musimon share the most recent female ancestor with O. aries at approximately 0.790 Ma (95% CI: 0.637–0.934 Ma) during the Middle Pleistocene, substantially predating the domestication event (∼8–11 ka). By reconstructing historical variations in effective population size, we found evidence of a rapid population increase approximately 20–60 ka, immediately before the Last Glacial Maximum. Analyses of lineage expansions showed two sheep migratory waves at approximately 4.5–6.8 ka (lineages A and B: ∼6.4–6.8 ka; C: ∼4.5 ka) across eastern Eurasia, which could have been influenced by prehistoric West–East commercial trade and deliberate mating of domestic and wild sheep, respectively. A continent-scale examination of lineage diversity and approximate Bayesian computation analyses indicated that the Mongolian Plateau region was a secondary center of dispersal, acting as a “transportation hub” in eastern Eurasia: Sheep from the Middle Eastern domestication center were inferred to have migrated through the Caucasus and Central Asia, and arrived in North and Southwest China (lineages A, B, and C) and the Indian subcontinent (lineages B and C) through this region. Our results provide new insights into sheep domestication, particularly with respect to origins and migrations to and from eastern Eurasia.
Keywords: wild ancestor, domestication, gene flow, mitogenome, Ovis aries, meta-analysis, colonization simulation
Introduction
As one of the first animals ever domesticated, sheep (Ovis aries) have played an important role in human society and have spread almost globally, following human migrations (Colledge et al. 2005; Chessa et al. 2009). Early evidence implied that modern sheep breeds were first domesticated from Asian mouflon (O. orientalis) in the Fertile Crescent approximately 8–11 thousand years ago (ka) (Ryder 1984). Following domestication, as many as 1,400 sheep breeds have been developed from their wild ancestors after long-term natural and intense artificial selection (Scherf 2000). During this process, human activities have played a significant role in determining the patterns of gene flow among breeds and populations (e.g., Warmuth et al. 2012). Thus, an examination of continent-wide genetic variability among modern native sheep breeds can provide a comprehensive, in-depth understanding of their genetic origins and dispersal, as well as insight into the impact of human activities on sheep throughout history.
In recent decades, remarkable analytical advances in paleontological and molecular genetics have transformed our understanding of the origins and regional expansion of domestic sheep (Poplin 1979; Hiendleder, Mainz, et al. 1998; Pedrosa et al. 2005; Chessa et al. 2009; Meadows et al. 2007, 2011; Kijas et al. 2009, 2012; Demirci et al. 2013). Morphological change and demographic analysis implied that sheep were likely brought under domestication in a region that stretches from northern Zagros to southeastern Anatolia, approximately 10.5–11 ka or perhaps even earlier (Peters et al. 2005). In addition, a recent investigation on endogenous retroviral sequences revealed a remarkable secondary population expansion of improved domestic sheep, most likely out of Southwest Asia (i.e., the Middle East; Chessa et al. 2009). Mitochondrial DNA (mtDNA) sequence analyses have identified a general phenomenon of multiple maternal lineages (i.e., A, B, C, D, and E), some with specific geographic ranges, implying multiple maternal origins and possibly independent domestication events in sheep (Wood and Phua 1996; Hiendleder, Mainz, et al. 1998; Guo et al. 2005; Pedrosa et al. 2005; Tapio et al. 2006; Meadows et al. 2007; Singh et al. 2013).
Estimates from complete and/or partial mtDNA sequences have enabled various divergence time estimates between domestic and wild sheep as well as among the five major maternal lineages of O. aries (e.g., Hiendleder, Mainz, et al. 1998; Pedrosa et al. 2005; Chen et al. 2006; Meadows et al. 2011). In general, the estimated divergence times among the five major lineages have been much earlier than the domestication period inferred from archeological evidence (Bar-Yosef and Meadow 1995; Zeder 2008). For example, the divergence time between the two most common lineages (i.e., A and B) was estimated to be as early as 1.6–1.7 Ma based on cytochrome b (Cyt-b) sequences (Hiendleder, Mainz, et al. 1998). In addition, Pedrosa et al. (2005) and Chen et al. (2006) suggested the divergence time of lineage C from lineages A and B to be approximately 0.42–0.76 Ma and approximately 0.45–0.75 Ma from the analysis of control region and Cyt-b sequences, respectively. However, a more recent study (Meadows et al. 2011) using 12 protein-coding genes from complete mitogenomes implied more recent divergence between the lineages: For example, 0.590 ± 0.17 Ma between A and B and 0.26 ± 0.09 Ma between C and E.
So far, most ovine mtDNA investigations have only focused on one or two segments within Cyt-b gene and the control region (including the hypervariable region; e.g., Pedrosa et al. 2005); nevertheless, high levels of recurrent mutations observed in the short segment within control region in many mammal species may bias dating estimates (e.g., Achilli et al. 2009, 2012; see also the reviews in Torroni et al. 2006; Taberlet et al. 2008). Moreover, previous sheep mtDNA studies have merely included breeds at a regional (e.g., Pedrosa et al. 2005; Chen et al. 2006; Wang et al. 2006; Meadows et al. 2007) or subcontinental scale (e.g., Tapio et al. 2006), whereas maternal lineages of domestic sheep, particularly for breeds in Southwest, Central, East and South Asia, including the Caucasus, Iran, Pakistan, Nepal, Indonesia, Mongolia, China, and India, have been largely excluded from integrated analyses. In addition, the divergence scenarios have not been fully evaluated based on complete mitogenomes either, which could have provided refined phylogenies of maternal lineages and robust estimations of genetic variability and divergence time in domestic animals (see the review in Wang et al. 2014). Therefore, although these early mtDNA studies have provided useful insights into the history of sheep domestication in Eurasia, answers to some basic questions surrounding the domestication process are far from being settled. For example, phylogenetic relationships among wild and domestic sheep (e.g., Hiendleder, Lewalski, et al. 1998; Meadows et al. 2007), divergence times between the major maternal lineages (e.g., Pedrosa et al. 2005; Zeder 2008; Meadows et al. 2011), demographic history and population recolonization (Dobney and Larson 2006; Zeder 2008), and origins of different mtDNA lineages (Tapio et al. 2006; Meadows et al. 2007; Demirci et al. 2013; Singh et al. 2013), as well as the continent-wide patterns of gene flow from the postulated Middle Eastern domestication center to Central, East and South Asia (see, e.g., Tapio et al. 2006, 2010; Cai et al. 2007, 2011) remain provisional or unaddressed.
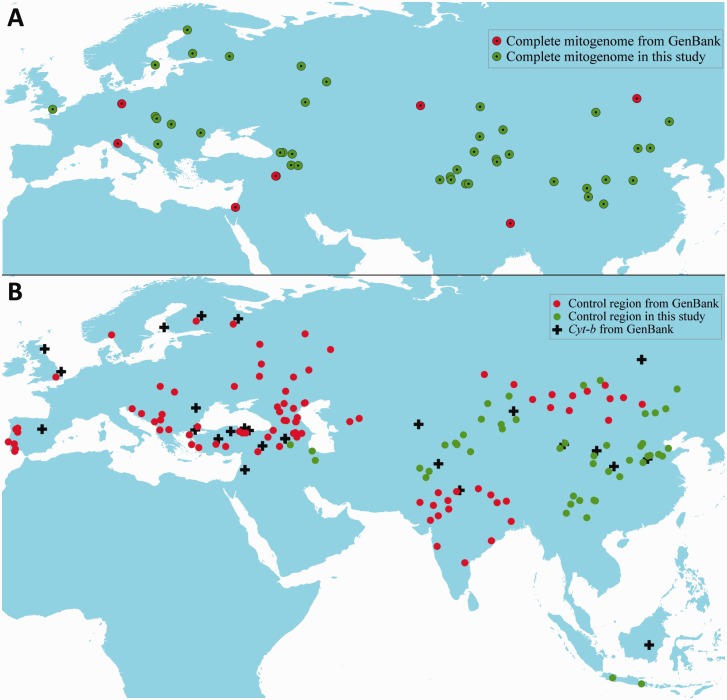
The main objective of our study was to better understand the domestication and expansion of O. aries across Eurasia through a meta-analysis of complete and partial ovine mitogenomic sequences. More specifically, we aimed to refine and challenge existing paradigms on the wild origin, lineage divergence, demographic history and population recolonization of modern sheep, particularly the breeds present in eastern Eurasia. For these purposes, we sequenced the complete mitogenomes of 45 individuals (including O. orientalis, O. vignei, and 42 native breeds of O. aries) and the control region of a total of 875 animals (including 51 native breeds) from eastern Eurasia (fig. 1 and supplementary tables S1 and S2, Supplementary Material online). Together with the sequences retrieved from GenBank, we analyzed 85 complete mitogenomes of domestic sheep including each of the 5 lineages and 10 complete mitogenomes of O. orientalis, O. musimon, O. vignei, O. ammon, and O. canadensis using phylogenetics, molecular-dating, and demographic-reconstruction approaches. Full control region and Cyt-b sequences of seven extant wild sheep species (O. orientalis, O. musimon, O. vignei, O. ammon, O. canadensis, O. dalli, and O. nivicola) were also included in phylogenetic reconstructions. Furthermore, we carried out a meta-analysis and a simulation of colonization (e.g., approximate Bayesian computation, ABC) of mtDNA sequences, including 547 partial Cyt-b and 1,470 partial control region sequences published previously (supplementary tables S2 and S3, Supplementary Material online), from native sheep breeds across Eurasia. We tried to address these questions and test two hypotheses on domestication and migrations of sheep distributed particularly in eastern Eurasia. One is the more recent origin and dispersal of lineage C when compared with those of the two widely distributed lineages A and B (Bruford 2005; Tapio et al. 2006). Another is that the arrival of some Indian sheep from the Middle Eastern domestication center could be through the Mongolian Plateau region, where archeological remains showed an early presence of domestic sheep in ancient history (e.g., Kuo et al. 1999; see also Yang et al. 2015). Our results could help researchers better understand the demographic forces and human practice associated with animal domestication and migration in history (e.g., Hodges 1999; Larson et al. 2007, 2010; Larson and Burger 2013).
Fig. 1.
Geographic distribution of the samples in this and early ovine mtDNA studies.
Results
Geographic Patterns of mtDNA Variation
The 45 complete domestic (GenBank accession numbers KF938317–KF938359) and wild (KF938360–KF938361) sheep mitogenomes (supplementary table S1, Supplementary Material online) sequenced in this study showed considerable sequence variability as well as variation in diversity among different regions (supplementary table S4 and fig. S1, Supplementary Material online). Also, we detected a large number of variable sites in the integrated data of partial Cyt-b and control region (supplementary tables S2 and S3, Supplementary Material online). Full description of the complete mitogenome and partial mtDNA sequence variations is in supplementary information S1, Supplementary Material online.
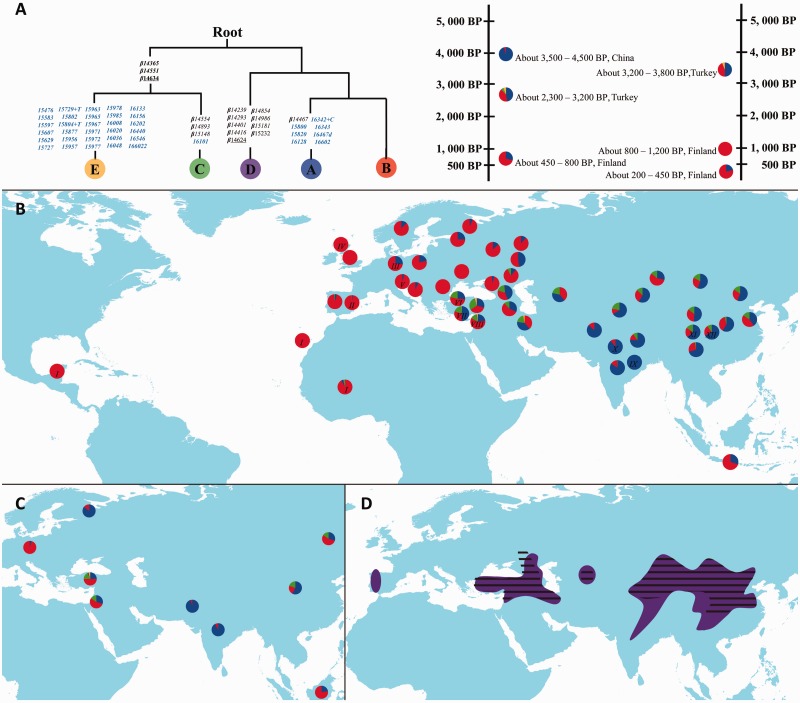
All control region and Cyt-b sequences analyzed in this study can be assigned to the five previously defined lineages (supplementary tables S2 and S3; see also supplementary figs. S2 and S3, Supplementary Material online). The two partial mtDNA fragments displayed similar geographic patterns (fig. 2B and C). For control region sequences, lineages A and B were the most common and most widely distributed, with a mean combined frequency of approximately 89% (fig. 2B and C). Lineage A was extremely frequent (∼77%) in the Indian subcontinent, although its frequency was less than 10% in Europe. In contrast, lineage B was found mostly in Europe, with its highest frequency (>90%) in Southwest Europe (fig. 2B and C). Lineage C occurred mainly in the Middle East, the Caspian Sea region, North China, and the Mongolian Plateau, with a mean frequency of approximately 18% (fig. 2B), whereas a few haplotypes of lineage C were also found in the Iberian Peninsula, India, Nepal, and Southwest China. A majority of the breeds harboring lineage C were fat-tailed (including fat-rump; 73.1%), higher than the proportion of fat-tailed breeds having lineage A (50.8%) or B (44.8%) (supplementary tables S5 and S6, Supplementary Material online). In addition, we found a significantly higher mean frequency of lineage C in fat-tailed breeds than in short-tailed breeds (fat-tailed: fC = 0–0.50, mean fC = 0.19; short-tailed: fC = 0–0.40, mean fC = 0.05; two-sample Kolmogorov–Smirnov test: P < 0.01; supplementary fig. S4, Supplementary Material online). Of the total 149 breeds studied here, 66 are fat-tailed, 78 harbor lineage C, and 57 are fat-tailed sheep carrying lineage C. Compared with the overlap expected by chance, there is a large and significant excess of breeds that are fat-tailed harboring linages C (lineage C: observed n = 57, expected by chance n = 34.65, P < 0.001; supplementary fig. S5; Supplementary Material online). Lineages D and E accounted for approximately 1% of the total samples and were only found in the Middle East (see fig. 2B and C).
Fig. 2.
Geographic distribution of the five major maternal lineages across Eurasia based on sequences obtained in this study and retrieved from GenBank. (A) Phylogenetic tree inferred from partial control region sequences (left) and lineage composition of sheep in different geographic regions at different time points (right) based on ancient specimens (Cai et al. 2007, 2011; Demirci et al. 2013; Niemi et al. 2013); (B) lineage frequency distribution of partial control region sequences; previously reported lineage frequencies in 12 regions (I–XII) are detailed in supplementary table S17, Supplementary Material online; (C) lineage frequency distribution of partial Cyt-b sequences; (D) geographic distribution of fat-tailed native sheep breeds (regions with black lines) and lineage C (region colored in purple). Pie plots show the proportions of the five distinct lineages (A–E) of domestic sheep in the different geographic regions (for the details of the geographic regions, see supplementary tables S5 and S6, Supplementary Material online). In the phylogenetic tree, diagnostic mutations are showed on the branches and are named according to their nucleotide positions relative to the reference sequence AF010406; amino acid replacements are underlined and synonymous replacements are marked in black. Control region mutations (15,437–16,616 bp) are shown in blue. Insertions are indicated by a “+” after the position number and followed by the type of inserted nucleotide(s). Mutations with prefix “β” indicate identical variable sites found in Meadows et al. (2007), which are used to define the five major lineages.
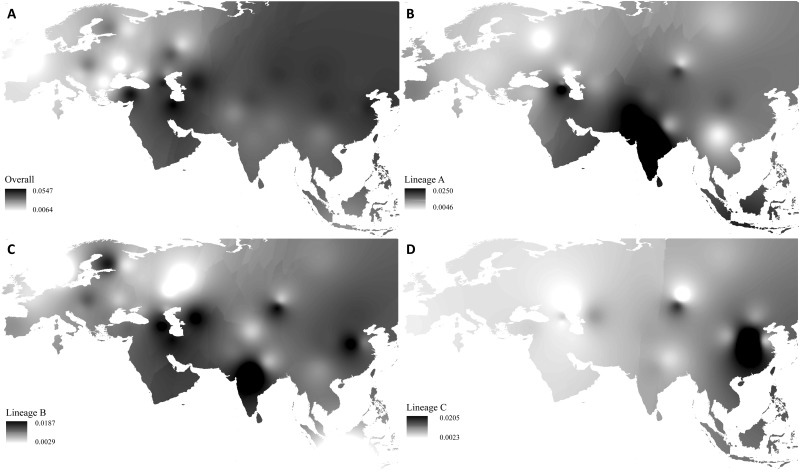
A synthetic map across Eurasia showed that the breeds in the Mongolian Plateau region had the highest genetic variability (π) of control region in Asia (fig. 3A; supplementary table S7, Supplementary Material online). For lineages A and B, a relatively high level of nucleotide diversity was found in the Indian subcontinent (fig. 3B and C). In addition, the synthetic map revealed the highest level of lineage C variability in the breeds of North China, even higher than that of the breeds in the Middle East (fig. 3D), the presumed domestication center of modern sheep (Ryder 1984).
Fig. 3.
Synthetic maps illustrating geographic variation of nucleotide variability for the total lineages and lineages A, B, and C. (A) The total lineages, (B) lineage A, (C) lineage B, and (D) lineage C.
Phylogenetic Relationships
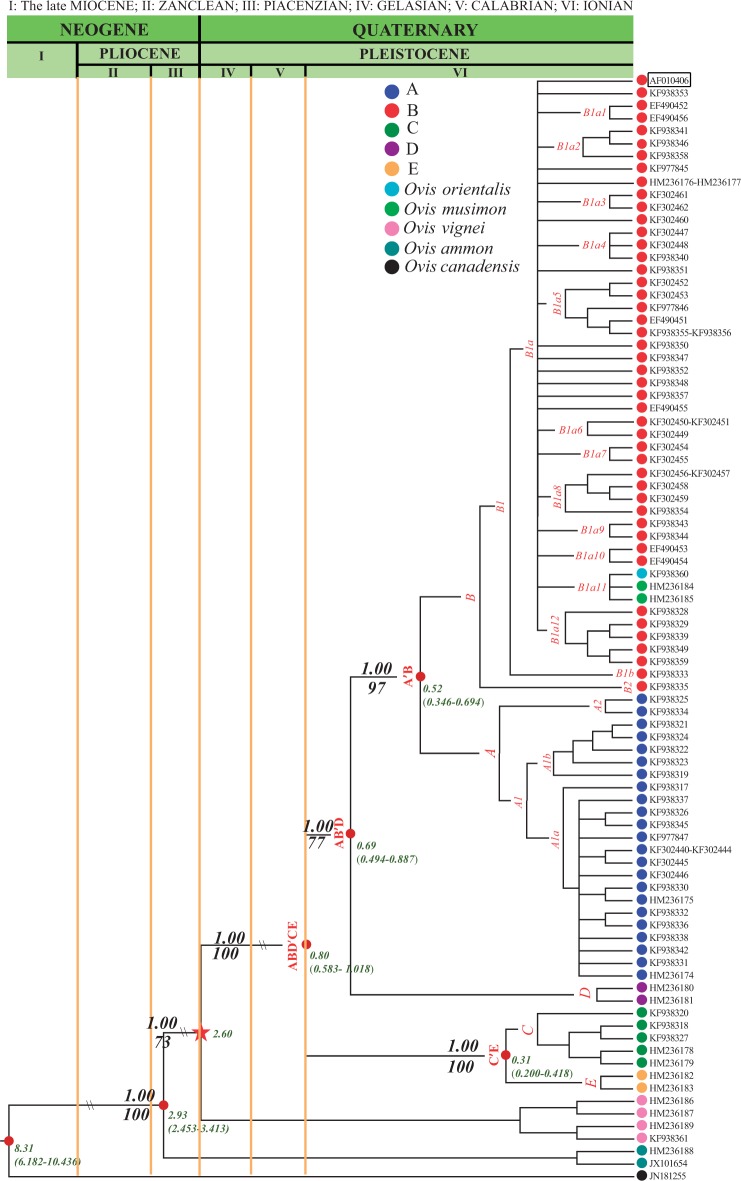
Phylogenetic relationships inferred from all the 95 complete Ovis mitogenomes (supplementary table S1, Supplementary Material online) are shown in figure 4. The 85 complete mitogenomes of O. aries were assigned to five major lineages (fig. 4). Ovis vignei, O. ammon, and O. canadensis clustered into three independent clades separated from O. aries, whereas O. canadensis showed the largest divergence. The clade of O. musimon and O. orientalis was closely related to O. aries. In the phylogenetic trees built from the full control region, Cyt-b, and protein gene sequences of the complete mitogenomes, the four branches of wild sheep agreed with the topology inferred from the complete mitogenomes, but domestic sheep sequences formed an unresolved group rather than the five major lineages (supplementary figs. S6–S9, Supplementary Material online). Additional phylogenetic trees obtained with the full control region and Cyt-b sequences of wild and domestic sheep (supplementary figs. S10 and S11 and tables S8 and S9, Supplementary Material online) showed different topologies from that inferred from the complete mitogenomes (fig. 4 and supplementary fig. S6, Supplementary Material online). Specifically, instead of showing close relationships only to lineage B as inferred from the complete mitogenomes (fig. 4), the haplotypes of O. musimon and O. orientalis clustered with lineages A, B, and C of O. aries control region sequences (supplementary fig. S10, Supplementary Material online), and they even shared some Cyt-b haplotypes of lineages A, B, C, and E (supplementary fig. S11, Supplementary Material online).
Fig. 4.
Phylogeny of domestic and wild sheep inferred from a total of 95 complete mitogenomes (supplementary table S1, Supplementary Material online) using BI and ML methods with posterior probability (the first value) and bootstrap values (the second value) on the nodes, respectively. Divergence times for the lineages (Ma) were estimated only based on the 61 complete mitogenomes of native domestic sheep breeds and wild sheep species (see supplementary table S1, Supplementary Material online).
The reduced median network analysis of partial control region sequences showed several major radiating nodes at a few mutation steps within lineages A and B. Different contributions of breeds to different regions were evident, but none of the major nodes consisted of apparent region-specific haplotypes (supplementary fig. S2, Supplementary Material online). In addition, analysis of molecular variance and pairwise-population FST values indicated genetic differentiation between European and Asian breeds, whereas considerable maternal gene flow was found among the breeds within Asia and Europe, respectively (supplementary figs. S12–S13 and tables S10 and S11, Supplementary Material online).
Selective Pressure on Different Lineages
The log-likelihood values (ln L) under the one-, two-, three- and four-ratio models were ln L = −18,462.81, −18,454.79, −18,419.46 and −18,413.27, respectively (table 1). The ω ratio differed between the branches under the same model and varied for the same branches under different models (table 1). The likelihood ratio tests (LRTs) revealed that the differences between two models for all the pairwise comparisons were significant (P < 0.01) and that the four-ratio model (free-ratio model) best fit the data, which indicated different ω ratios among the lineages. Mean ω values for the lineages were ωA = 0.0457, ωB = 0.0775, ωD = 0.0494, and ωC + E = 0.0496 (supplementary fig. S14, Supplementary Material online); note that these values are all much lower than 1. This observation indicates that the maternal lineages (A, B, D, and C + E) have been under strong but variable intensity of purifying selection: Purifying selection on amino acid changes in lineage B has been slightly weaker than that on the other lineages. Thus, divergence time estimation (see below) based on the protein-coding genes would be biased. Instead, using the synonymous sites might be a better choice for divergence time estimation.
Table 1.
Number of Parameters Fitted, dN/dS Ratios, Log-Likelihood Scores, and Their Differences under Different Models.
| Model | p | ln L | ω | Models Compared | 2Δln L | |
|---|---|---|---|---|---|---|
| A: One ω ratio | ω0 | 102 | −18,462.81 | ω0 = 0.0563 | ||
| B: Two ω ratios | ωB | 103 | −18,454.79 | ωB = 0.0702 | ||
| ω0 | ω0 = 0.0436 | A versus B | 16.04** | |||
| C: Three ω ratios | ωA | 104 | −18,419.46 | ωA = 0.0447 | A versus C | 86.70** |
| ωB | ωB = 0.0744 | A versus D | 99.08** | |||
| ω0 | ω0 = 0.0486 | B versus C | 70.66** | |||
| D: Four ω ratios | ωA | 105 | −18,413.27 | ωA = 0.0457 | B versus D | 83.04** |
| ωB | ωB = 0.0775 | C versus D | 12.38** | |||
| ωD | ωD = 0.0494 | |||||
| ω0 | ω0 = 0.0496 | |||||
Note.—p, number of parameters in the model; ln L, log-likelihood score; ω, the dN/dS ratio for the branches; ωA ωB and ωD are the dN/dS ratios for branches lineages A, B, and D, respectively (see supplementary fig. S14, Supplementary Material online); ω0 is the background dN/dS ratio for the rest branch(es); 2Δln L, twice the log-likelihood difference of the models compared.
**Very significant (P < 0.01).
Divergence Times for the Nodes
The estimated divergence times within the comprehensive evolutionary framework of the Cetartiodactyla are shown in supplementary figure S15, Supplementary Material online. The O. vignei/O. aries split, which is the calibration point applied to estimate the divergence times between extant O. aries lineages, was 2.6 ± 0.9 Ma. That time is far earlier than the most recent common ancestor (TMRCA) of domestic sheep (∼0.79 Ma; 95% CI: 0.64–0.93 Ma; table 2), and even older than the O. ammon/O. aries split (2.13 ± 0.29 Ma) estimated by Meadows et al. (2011). The Capra/Ovis split was estimated to be 14.7 ± 2.1 Ma (supplementary fig. S15, Supplementary Material online), and is much older than the date based on the ungulate fossil record (∼5.00–7.0 Ma; Luikart et al. 2001). Using the calibration point, we obtained a substitution rate of 0.70 × 10−8 substitutions per nucleotide/year for complete mitogenome, 3.12 × 10−8 substitutions per nucleotide/year for control region, and 0.49 × 10−8 per nucleotide/year for Cyt-b without partitions.
Table 2.
Divergence Time Estimated by the Sequences of Complete Mitogenomes and the Protein-Coding Genes (synonymous mutation and the third-codon position) Using ML and BI Methods.
| Method | Data Set | Model | Node | Node 1 (TC/E)Ma | Node 2 (TA/B)Ma | Node 3 (TAB/D)Ma | Node 4 (TABD/CE)Ma | Node 5 (TO.aries/O.vignei)Ma | TO.aries/O.ammonMa | TO.aries/O.canadensisMa |
|---|---|---|---|---|---|---|---|---|---|---|
| ML | Mitogenome | Global | Time | 0.36 | 0.51 | 0.74 | 0.88 | 2.60 | 3.00 | 7.72 |
| 95%(CI) | (0.278–0.439) | (0.402–0.616) | (0.613–0.867) | (0.743–1.013) | — | (2.673–3.323) | (6.567–8.883) | |||
| Local | Time | 0.34 | 0.53 | 0.78 | 0.93 | 2.60 | 3.06 | 8.15 | ||
| 95%(CI) | (0.276–0.472) | (0.397–0.668) | (0.600–0.956) | (0.721–1.131) | — | (2.697–3.419) | (6.635–9.664) | |||
| Synonymous | Global | Time | 0.31 | 0.52 | 0.68 | 0.79 | 2.60 | 2.92 | 8.36 | |
| 95%(CI) | (0.217–0.405) | (0.373–0.661) | (0.536–0.829) | (0.637–0.934) | — | (2.535–3.312) | (6.441–10.286) | |||
| Local | Time | 0.31 | 0.52 | 0.69 | 0.80 | 2.60 | 2.93 | 8.31 | ||
| 95%(CI) | (0.200–0.418) | (0.346–0.694) | (0.494–0.887) | (0.583–1.018) | — | (2.453–3.413) | (6.182–10.436) | |||
| Third codon | Global | Time | 0.29 | 0.50 | 0.64 | 0.73 | 2.60 | 2.81 | 7.47 | |
| 95%(CI) | (0.190–0.390) | (0.361–0.639) | (0.497–0.783) | (0.579–0.881) | — | (2.418–3.202) | (6.157–8.783) | |||
| Local | Time | 0.29 | 0.50 | 0.64 | 0.73 | 2.60 | 2.81 | 7.47 | ||
| 95%(CI) | (0.190–0.390) | (0.359–0.636) | (0.498–0.783) | (0.581–0.882) | — | (2.414–3.200) | (6.158–8.786) | |||
| BI | Mitogenome | Relaxed-molecular clock | Median | 0.35 | 0.55 | 0.85 | 0.92 | 2.60 | 2.68 | 6.13 |
| 95%HPD | (0.130–0.641) | (0.266–0.913) | (0.390–1.413) | (0.464–1.498) | — | (2.462–3.031) | (2.464–11.618) | |||
| Synonymous | Median | 0.41 | 0.61 | 0.96 | 1.06 | 2.60 | 2.62 | 5.89 | ||
| 95%HPD | (0.147–0.772) | (0.291–1.013) | (0.478–1.604) | (0.541–1.716) | — | (2.458–3.338) | (5.456–11.598) | |||
| Third codon | Median | 0.36 | 0.57 | 0.87 | 0.94 | 2.60 | 2.62 | 6.49 | ||
| 95%HPD | (0.142–0.656) | (0.27–0.912) | (0.421–1.428) | (0.472–1.496) | — | (2.461–3.083) | (2.478–12.647) |
Note.—“—,” not available.
The divergence times for each node were mostly concordant under global and local clock models when estimated from the complete mitogenomes, the synonymous mutations or the third-codon positions (table 2). The earliest split was estimated to be approximately 0.73–0.93 Ma for the divergence of C and E from A, B, and D (see the node 4 in table 2), whereas the most recent split was between lineages C and E at approximately 0.29–0.36 Ma (see the node 1 in table 2), greatly predating sheep domestication (∼8–11 ka; Ryder 1984). The time to TMRCA of the two most common lineages (A and B) was estimated to be approximately 0.50–0.53 Ma (see node 2 in table 2). Under the relaxed molecular clock, we also obtained similar estimates of divergence times for the nodes based on different data sets (i.e., complete mitogenomes, synonymous, and third-codon positions; see table 2). However, divergence times for nodes 2, 3, and 4 estimated from synonymous mutations by the Bayesian Inference (BI) approach were significantly (P < 0.05) higher than those by the global and local maximum likelihood (ML) approaches, respectively (supplementary fig. S16, Supplementary Material online).
Prehistoric Population Expansions
Bayesian skyline plot (BSP) reconstructions of historical population expansions using the complete mitogenomes revealed the profile of predomestic change in Ne over large time scales. Based on the estimated TMRCA for the lineages (∼0.79 Ma) from complete mitogenomes, the ovine lineages showed a steep increase in Ne at approximately 20–60 ka (supplementary fig. S17, Supplementary Material online). A prehistoric steep increase in Ne was also identified in the simulations of the partial Cyt-b and control region sequences (supplementary fig. S18, Supplementary Material online). However, population growth was found to have occurred at approximately 50–300 ka, much earlier than the time obtained from simulations of the complete mitogenomes (∼20–60 ka; supplementary fig. S17, Supplementary Material online).
Postdomestic Lineage Expansions
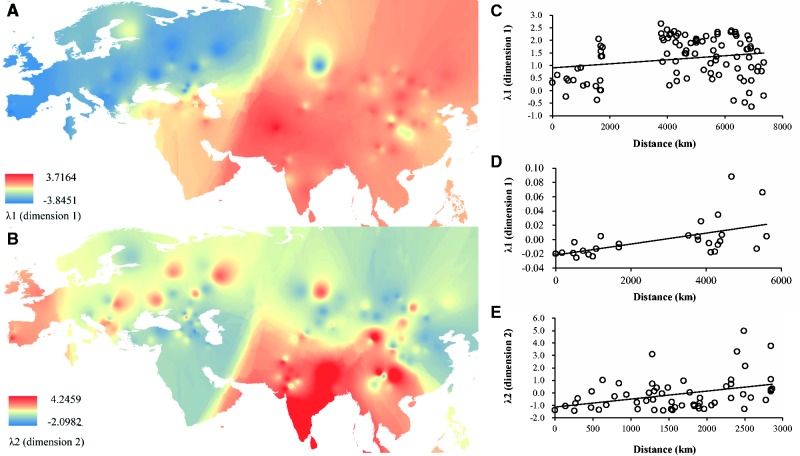
A synthetic map constructed with the use of interpolated λ1 values, the eigenvalues for the first multidimensional scaling (MDS) plot dimension, allows us to examine the gradients of colonization out of the sheep domestication center that peak in the Middle East (fig. 5A; supplementary table S12, Supplementary Material online). λ1 explains 69.3% of the total variation. We observe a significant correlation between the λ1 eigenvalues of Asian populations and their geographic distances from the domestication center (lineages A, B, and C of Central and East Asian populations: r = 0.201; P < 0.05; lineage A of Arabian and Indian populations: r = 0.547; P < 0.01; see fig. 5C and D). This suggests that the major colonization process of the Middle Eastern sheep to eastern Eurasia (including Mongolia, China, and India) was through the Caucasus and Central Asia. The interpolation map of the λ2 eigenvalues suggests that the second MDS dimension could represent genetic influence from the Mongolian Plateau region in China and the Indian subcontinent (fig. 5B). λ2 explains 27.3% of the total variation. Its ranking shows the Mongolian Plateau region at one extreme, whereas the Indian subcontinent at the other extreme. This is supported by a strong and significant correlation observed between geographic distances from the putative region of initial colonization (i.e., the Mongolian Plateau region) and λ2 values across eastern Eurasian populations (r = 0.372; P < 0.01; fig. 5E). Eigenvalues λ1 and λ2 for all the populations are shown in supplementary table S12, Supplementary Material online.
Fig. 5.
Synthetic maps illustrating geographic variation of eigenvalues (λ) for the first two MDS dimensions (λ1 and λ2) and regression of λ versus geographic distance from the putative original site of colonization process. (A) synthetic map for λ1, (B) synthetic map for λ2, (C) regression of λ1 versus geographic distances from the domestication center of sheep (represented by the geographic distance from the Kilis province of Turkey, where ancient domestic sheep are located; Demirci et al. 2013) for Asian populations (r = 0.201; P < 0.05); (D) regression of λ1 (based on lineage A only) versus geographic distances from the domestication center of sheep (represented by the geographic distance from the Kilis province of Turkey, where ancient domestic sheep are located; Demirci et al. 2013) for sheep populations from the Indian subcontinent (r = 0.547; P < 0.01); and (E) regression of λ2 versus geographic distances from a putative “transportation hub” of the Mongolian Plateau region (represented by the geographic distance from the northernmost population [Transbaikal Finewool] sampled) for eastern Eurasian (including China, Mongolia, and India) populations (r = 0.372; P < 0.01).
The star-like median-joining networks (supplementary fig. S2, Supplementary Material online) and mismatch distributions (supplementary fig. S19, Supplementary Material online) revealed genetic signatures of postdomestic demographic population expansions in lineages A, B, and C. The inference was corroborated by Fs (Fu 1997), Tajima’s D (1989), and scaled effective population size statistics (2Nu; N represents the effective population size and u denotes the mutation rate). Both Fu’s Fs and Tajima’s D statistics showed significant (PF < 0.001; PD < 0.001; table 3) departures from neutrality in the three lineages. Additionally, the observed mismatch distributions of the lineages were fitted to the sudden population expansion models with very low values of the sum of squared deviation (SSD ≤ 0.005; table 3) statistic and Harpending’s Raggedness index (Harpending 1994; rH = 0.021–0.031; PR < 0.5; table 3). Furthermore, the estimated pre- and postexpansion scaled effective population sizes (2Nu) indicated an increase in the effective population size for each of the lineages (A: 0.0–77.81; B: 0.04–16.63, and C: 0.00–15.77; table 3). The postdomestic expansion time expressed in twice the number of generations multiplied by the mutation rate (τ = 2 ut) was found to be 6.443 ka (90% CI = 4.279–7.569 ka), 6.811 ka (90% CI: 3.502–9.706 ka), and 4.549 ka (90% CI = 2.402–6.652 ka) for lineages A, B, and C, respectively, when assuming an initial expansion (i.e., lineage A involving European sheep; Tapio et al. 2006) to equal 9 ka (table 3 and fig. 6). Separate analyses for the four major geographic areas (the Middle East, India, East Asia, and Europe) resulted in wider confidence intervals than those for the combined analysis and showed somewhat different estimates of τ (table 3). In particular, the expansion time for lineage C in the Middle East (3.910 ka; 90% CI: 2.818–5.120 ka) was more recent than that in East Asia (4.967 ka; 90% CI: 1.893–7.842 ka), whereas relatively earlier expansions in the Middle East were inferred for lineages A and B (table 3). Additionally, we found much later expansions of lineages A and B in India (lineage A: 4.033 ka: 90% CI: 1.517–23.100 ka; lineage B: 3.393 ka; 90% CI: 0.961–16.311 ka) than those in East Asia (lineage A: 5.877 ka; 90% CI: 5.216–6.681 ka; lineage B: 7.008 ka; 90% CI: 3.030–16.348 ka), respectively.
Table 3.
Sudden Expansion Model Parameters Estimated from the Distribution of Pairwise Differences between Sequences within mtDNA Lineages and Neutrality Tests for Different Lineages.
| Lineages | Areas | τ (90% CI) | θ0 | Θ1 | SSD | PSSD | rH | PR | ka (90% CI) | Tajima’s D |
Fu’s FS |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D | PD | FS | PF | ||||||||||
| A | All | 2.62 (1.740–3.078) | 0.00 | 77.81 | 0.005 | 0.044 | 0.030 | 0.548 | 6.443 (4.279–7.569) | -2.06 | 0.000 | -25.09 | 0.000 |
| India | 1.64 (0.617–9.394) | 4.95 | 99,999.00 | 0.008 | 0.498 | 0.011 | 0.471 | 4.033(1.517–23.100) | -1.08 | 0.103 | -24.68 | 0.000 | |
| East Asiaa | 2.39 (2.121–2.717) | 0.01 | 99,999.00 | 0.000 | 0.437 | 0.039 | 0.246 | 5.877 (5.216–6.681) | -2.33 | 0.000 | -26.50 | 0.000 | |
| Middle East | 2.50 (1.969–3.004) | 0.00 | 99,999.00 | 0.006 | 0.072 | 0.053 | 0.103 | 6.148 (4.842–7.387) | -2.16 | 0.001 | -26.97 | 0.000 | |
| Europe | 3.66 (2.061–5.852) | 0.53 | 41.25 | 0.005 | 0.541 | 0.028 | 0.558 | 9.000 (5.068–14.390) | -1.04 | 0.129 | -4.89 | 0.011 | |
| B | All | 2.77 (1.424–3.947) | 0.04 | 16.63 | 0.001 | 0.783 | 0.021 | 0.759 | 6.811 (3.502–9.706) | -2.24 | 0.000 | -25.48 | 0.000 |
| India | 1.38 (0.391–6.633) | 2.54 | 99,999.00 | 0.006 | 0.474 | 0.020 | 0.594 | 3.393 (0.961–16.311) | -0.83 | 0.190 | -12.88 | 0.001 | |
| East Asiaa | 2.85 (1.232–6.648) | 1.03 | 10.45 | 0.002 | 0.743 | 0.012 | 0.893 | 7.008 (3.030–16.348) | -1.93 | 0.004 | -26.14 | 0.000 | |
| Middle East | 3.00 (1.621–3.949) | 0.00 | 32.66 | 0.001 | 0.673 | 0.023 | 0.685 | 7.377 (3.986–9.711) | -2.28 | 0.000 | -26.66 | 0.000 | |
| Europe | 2.85 (1.232–6.648) | 1.03 | 10.45 | 0.002 | 0.743 | 0.012 | 0.893 | 6.000 (5.383–6.711) | -1.93 | 0.004 | -26.14 | 0.000 | |
| C | All | 1.85 (0.977–2.705) | 0.00 | 15.77 | 0.000 | 0.944 | 0.031 | 0.784 | 4.549 (2.402–6.652) | -2.41 | 0.000 | -27.22 | 0.000 |
| East Asiaa | 2.02 (0.770–3.189) | 0.00 | 11.08 | 0.002 | 0.675 | 0.030 | 0.780 | 4.967 (1.893–7.842) | -2.02 | 0.004 | -24.70 | 0.000 | |
| Middle East | 1.59 (1.146–2.082) | 0.00 | 99,999.00 | 0.000 | 0.829 | 0.052 | 0.406 | 3.910 (2.818–5.120) | -2.24 | 0.001 | -27.33 | 0.000 | |
Note.—τ = 2ut, u is the mutation rate for the haplotypes, t is the time in generations; 90% CI, 90% confidence interval (CI) obtained by the parametric bootstrapping with 1,000 resampling; θ0 = 2uN0, N0 is the initial effective population size; θ1 = 2uN1, N1 is the final effective population size; SSD is the sum of square deviations (SSD) between the observed and the expected mismatch; PSSD, PR, PD, and PF are the corresponding significance values for SSD, rH, Tajima’s D (Tajima 1989), and Fu’s FS (Fu 1997); rH, Harpending’s Raggedness index (Harpending 1994).
aEast Asia includes the regions of Northern China and the Mongolian Plateau (for details, see supplementary table S5, Supplementary Material online).
Fig. 6.
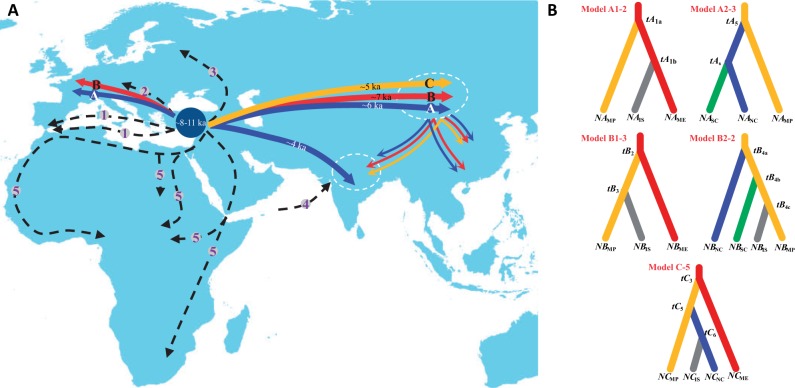
(A) The major migratory events and routes of the major sheep lineages across Eurasia (lineage A, in blue; lineage B, in red; and lineage C, in yellow). Migration routes reported in previous studies have also been included: (1) The Mediterranean routes (Ryder 1984), (2) the Danubian route (Ryder 1984), (3) the northern Europe route (Tapio et al. 2006), (4) the ancient sea trade route to the Indian subcontinent (Singh et al. 2013), and (5) routes of introduction and spread of sheep pastoralism in Africa (Muigai and Hanotte 2013); (B) the optimal population colonization models for A/B/C lineages in DIYABC v.2.0.4 (Cornuet et al. 2014) (supplementary information S5, Supplementary Material online). Colors in the two figures are irrelevant between each other.
ABC analyses based on the control region sequences identified an optimal model for each of the five sets of candidate colonization models (lineage A first-step, lineage A second-step, lineage B first-step, lineage B second-step, and lineage C; supplementary information S4, Supplementary Material online). The optimal models exhibited much higher posterior probability and nonoverlapped 95% CIs as compared with other candidate models (table 4). These optimal models indicated that 1) lineage A first colonized from the Middle East to the Mongolian Plateau region and the Indian subcontinent separately, and later from the Mongolian Plateau region to North China, and then to Southwest China (fig. 6); 2) lineage B first colonized from the Middle East to the Mongolian Plateau region, and then from the Mongolian Plateau region to North and Southwest China and the Indian subcontinent separately (fig. 6); and 3) Lineage C first colonized from the Middle East to the Mongolian Plateau region, and later from the Mongolian Plateau region to North China, and then to the Indian subcontinent (e.g., Nepal) (fig. 6). Taken together, our modeling results provided strong quantitative evidences supporting that the Mongolian Plateau region was acted as a “transportation hub” for sheep migration in Asia.
Table 4.
Results of Model Choice for the Colonization Scenarios of Sheep Lineages A, B, and C Tested in the ABC Analyses.
| Scenarios | Posterior Probability | 95% Confidence Intervals |
|---|---|---|
| The models of lineage A in the first step | ||
| A1‐1 | 0.120 | 0.015 − 0.224 |
| A1‐2 | 0.842 | 0.822 − 0.862 |
| A1‐3 | 0.038 | 0.020 − 0.056 |
| The models of lineage A in the second step | ||
| A2‐1 | 0.215 | 0.168 − 0.263 |
| A2‐2 | 0.060 | 0.016 − 0.105 |
| A2‐3 | 0.725 | 0.707 − 0.742 |
| The models of lineage B in the first step | ||
| B1‐1 | 0.253 | 0.245 − 0.261 |
| B1‐2 | 0.258 | 0.245 − 0.272 |
| B1‐3 | 0.489 | 0.481 − 0.497 |
| The models of lineage B in the second step | ||
| B2‐1 | 0.043 | 0.038 − 0.048 |
| B2‐2 | 0.939 | 0.933 − 0.945 |
| B2‐3 | 0.004 | 0.003 − 0.004 |
| B2‐4 | 0.005 | 0.005 − 0.006 |
| B2‐5 | 0.009 | 0.008 − 0.010 |
| The models of lineage C | ||
| C-1 | 0.158 | 0.149 − 0.167 |
| C-2 | 0.079 | 0.069 − 0.088 |
| C-3 | 0.207 | 0.200 − 0.215 |
| C-4 | 0.138 | 0.128 − 0.147 |
| C-5 | 0.418 | 0.407 − 0.430 |
Note.—The best-supported model with the highest posterior probability is indicated in italics.
Discussion
In this study, we sequenced the largest number of complete ovine mitogenomes and control region sequences in sheep to date. We also performed the first meta-analysis of complete and partial mitogenomic sequences of wild and domestic sheep across Eurasia. Our results clarify the domestication and history of sheep distributed particularly in eastern Eurasia regarding their origins, lineage divergence, demographic history, population recolonization, and evolutionary forces shaping the sheep mitogenomes. Below, we discuss these issues in more details and relate our findings to those that have emerged from early mtDNA studies on the ovine species.
Phylogenies
Phylogenetic analyses of complete mitogenomes showed a high resolution among wild sheep as well as among the major lineages of domestic sheep (fig. 4). The complete mitogenomes of O. orientalis and O. musimon formed a monophyletic group (fig. 4) that was incorporated within lineage B of domestic sheep. However, the analysis of full control region and Cyt-b fragments showed that O. orientalis is also closely related to other lineages (e.g., lineages A, C, and E) of O. aries (supplementary figs. S10 and S11, Supplementary Material online). This difference could be ascribed to the small number (n = 3) of O. musimon and O. orientalis complete mitogenomes available in this study (fig. 4).
Full control region and Cyt-b fragments from the complete mitogenomes produced similar phylogenies with fully resolved phylogenetic relationships of wild sheep, but they failed to define the phylogenetic relationships among the major lineages of domestic sheep (supplementary figs. S7 and S8, Supplementary Material online). Our results suggest that partial fragments of the complete mitogenomes would be problematic when making phylogenetic inferences about domestic sheep. This problem arises due to diagnostic substitutions located elsewhere in the mitogenome (fig. 4; supplementary table S13, Supplementary Material online). Thus, the diagnostic substitutions for species and lineages presented here (fig. 4; supplementary table S13, Supplementary Material online) can serve as an important resource for maternal genetic differentiation between domestic and wild sheep as well as between the lineages within domestic sheep. Also, they might be helpful for addressing certain conflicts described above in future.
Origins and Migrations of Lineage C
Lineage C showed a restricted distribution in semidesert and steppe regions, 30–45°N. Given the limited geographic range, lineage C in domestic sheep may represent a recent genetic introgression from wild ancestors, rather than an independent domestication event. This hypothesis is supported by at least three lines of evidence: 1) lower mean frequency of lineage C (∼10.36%) across the global breeds, and absence or extremely low frequency of lineage C (e.g., due to a recent expansion process; fig. 2B) in native breeds in Europe, the Indian subcontinent (including India and Pakistan), Southwest China, South Asia (including Indonesia), and Africa; 2) the extensive human practice of mating domestic ewes and wild rams documented in Central Asia and North China (Carruthers 1949; Aniwashi et al. 2007); and 3) lineage C shows an even earlier population expansion in East Asia (4.967 ka; 90% CI: 1.893–7.842 ka) than that in the presumed sheep domestication center of the Middle East (3.910 ka; 90% CI: 2.818–5.120 ka). High frequency and genetic variability (h and π) of lineage C were observed in breeds of northern central China (figs. 2B and 4D). These observations suggest that North China could be one of the origin regions for lineage C sheep, where sheep farming has been dated to approximately 5–8 ka at an early Neolithic site (Chen 1990; Cai et al. 2011; Yang et al. 2015). Nevertheless, we note that lineage C was not detected in earlier ancient DNA analyses of archeological sheep remains (∼3.5–4.5 ka; n = 8, Cai et al. 2007; n = 14, Cai et al. 2011; Yang et al. 2015; see also fig. 2A), which could be due to sampling effects.
Interestingly, lineage C co-occurs with indigenous fat-tailed breeds in Eurasia (fig. 2D), and our results suggest that the prevalence of lineage C could be probably linked to geographic range of fat-tailed breeds (see also Bruford 2005; Tapio et al. 2006), as deserving further investigations. Fat-tailed sheep was first recorded on an ancient Uruk II (∼5 ka) and Ur stone vessel and mosaics in Iraq (∼4.4 ka) by archeological evidence (Ryder 1983; see also Muigai and Hanotte 2013). However, we observed very few or not at all lineage C in sheep from Pakistan, India, Southwest China, and Indonesia (fig. 2, supplementary tables S2 and S3, Supplementary Material online). These observations, together with the colonization scenarios of lineage C reconstructed using ABC, suggested that lineage C first colonized from the Middle East to the Mongolian Plateau region through the Caucasus, east of the Caspian Sea and Central Asia, then dispersed to North China and the Qinghai-Tibetan Plateau, and finally arrived in Nepal.
Our results imply that the five maternal lineages evolved at different times before domestication (table 2). Zeder et al. (2006) suggested that independent domestication events in domestic animals might represent the introduction of founders that were subsequently submerged in the recruitment of local wild animals. Larson and Burger (2013) recently proposed that hybridization between local wilds and introduced domestic populations was common in a wide range of plant and animal species. Here, the extremely low occurrence of lineages D and E in domestic sheep and their restricted geographical distributions indicate that these lineages most likely represent additional two introgression events rather than independent domestication events.
Population Expansions and Migrations in Eastern Eurasia
We observed both prehistoric (∼20–60 ka) and postdomestication (∼3.5–9 ka) demographic expansions for the three major lineages (A, B, and C). Thus, the wild ancestors of domestic sheep underwent a major population expansion before the Last Glacial Maximum (∼19.0–26.5 ka). Results of the BSP inferences on demographics should be interpreted with caution due to the confounding effect of population structure (e.g., Grant et al. 2012) and uncertainty in time estimation (e.g., Heller et al. 2013; see also the review by Ho and Shapiro 2011). Nevertheless, the worldwide collection of samples from a large number of populations and the application of multiple approaches in molecular clock calibration should minimize these effects.
We observed high levels of diversity (π) for lineage B in the breeds from Central Asia, with a decreasing gradient to the east (fig. 3C). This geographic pattern could be interpreted as the genetic introgression of Middle Eastern (rather than European) lineage B sheep into East Asian breeds through the Caucasus (Tapio et al. 2006) and Central Asia (fig. 6). This explanation is supported by the similar expansion time of lineage B sheep in the Middle East and East Asia but a later expansion of lineage B in Europe (the Middle East: 7.377 ka, 90% CI: 3.986–9.711 ka; East Asia: 7.008 ka, 90% CI: 3.030–16.348 ka; Europe: 6.000 ka, 90% CI: 5.383–6.711 ka; table 3). We also observed a high level of nucleotide diversity (π) for lineage A in the Caucasus, following a west–east decreasing gradient. In addition, decrease with geographic distance eastwards for the λ1 eigenvalues is significant across East Asian (including China and Mongolia) populations with its peak in the Middle Eastern domestication center (r = 0.201; P < 0.05; fig. 5A and C). Moreover, East Asian breeds showed closer genetic relationships with the breeds in Central Asia, Caucasus, and Turkey than those in Europe, as indicated by the MDS plot and pairwise genetic differentiation (supplementary figs. S12 and S13 and table S12, Supplementary Material online). Thus, the maternal lineages in the Middle Eastern sheep may have been introduced to East Asia through the Caucasus and Central Asia by commercial trade between the West and East (e.g., Cai et al. 2011). Recent archeological studies in North China suggest that the West–East trade contact could be traced back to the prehistoric period through the “Bronze Road” across the vast Eurasian landmass approximately 4.5–5.3 ka (Yi 2004). Also, at this time millet was transported from its domestication site in North China to the Caucasus region approximately 7 ka (Hunt et al. 2008; Jones and Liu 2009). However, regression analysis (fig. 5C) and Bayesian simulations (fig. 6B) of lineage A identified its southeastwards migration from the Middle East to the Indian subcontinent through Arabia (and/or the Arabian Sea; see, e.g., Singh et al. 2013), which could be due to ancient overland (and/or maritime) trade (e.g., the Silk Road) between Arabia and India dated as early as approximately 4.5 ka (e.g., Gauri 2013).
We found high levels of genetic variability in sheep breeds distributed in North China and the Mongolian Plateau (fig. 3A), with a decreasing gradient to the southwest. This finding, together with the extreme ranking of λ2 eigenvalues in these regions (fig. 5B), suggests that the Mongolian Plateau region could be a secondary center of dispersal serving as a “transportation hub” in eastern Eurasia, and sheep have migrated to China (lineages A, B, and C) and the Indian subcontinent (lineages B and C) from the Middle East through this region (fig. 6). This suggestion is corroborated by several pieces of evidence: 1) Bayesian coalescence-based simulations identified two phases of migration in eastern Eurasian sheep: The Middle Eastern sheep first arrived in the Mongolian Plateau region, and then dispersed to other regions including North and Southwest China (lineages A, B, and C; fig. 6) and the Indian subcontinent (lineages B and C; fig. 6); 2) a high and significant correlation was obtained for the southwest dispersal (r = 0.372; P < 0.01; fig. 5E); 3) the expansion of lineage B in the Indian subcontinent was much later than in East Asia (table 4), and lineage C was observed in a few samples from the Indian subcontinent; 4) several animal domestications/early Holocene animal management (e.g., pig, Larson et al. 2010; cattle, Zhang et al. 2013; chicken, Xiang et al. 2014) and crop cultivation events (e.g., millet, Lu et al. 2009; Yang et al. 2012) took place in the middle and lower reaches of the Yellow River in North China; 5) archeological remains showed an early presence of domestic sheep in Inner Mongolia and northern China during the period of Hongshan Culture (∼ 4.2–6.5 ka) (Yang et al. 2015), and archeological sites in Tibet dating to 3.6–5.2 ka are marked by the presence of sheep bones (Chen et al. 2015); and 6) there were several popular ancient trade routes between China and India, by both land (through the Southern Silk Road route and the Tibet–Nepal and Burma–India routes) and sea 3–4 ka (Pe 1959; Shaha 1970). Analyses of genome-wide single nucleotide polymorphisms (SNPs) (∼50 K) in a representative set of 22 native Asian sheep breeds suggest two similar colonization processes of eastern Eurasian sheep domestication as those obtained from mtDNA variability (for the statistical analyses and results of SNPs, see supplementary information S2, table S14, and figs. S20–S23, Supplementary Material online). Also, by using the TreeMix algorithm in the program TreeMix v.1.12 (Pickrell and Pritchard 2012), analyses of the SNP data set reveal strong gene flows between the native breeds of different geographic regions and forward and backward movements as well (supplementary information S3 and fig. S24; Supplementary Material online).
Multiple Forces Acting on the Maternal Genetic Makeup of Eastern Eurasian Sheep
Frequent animal exchange, particularly of breeding ewes, between the breeds in different regions through ancient trade routes (e.g., the Silk Road) could have contributed to the geographic patterns observed in this study (figs. 2 and 3). In addition to the initial population expansion during the Neolithic, sheep maternal lineages could also have spread to East Asia and the Indian subcontinent through the Mongolian invasions. Overall, our findings suggest strong historical human-mediated gene flow between breeds across Eurasia (supplementary figs. S12, S13, and S20–S24, Supplementary Material online; see also, e.g., Kijas et al. 2009, 2012). This hypothesis is also compatible with the high level of genetic admixture revealed by a recent genome-wide analysis of sheep breeds across the world (Kijas et al. 2012).
In addition to the ancient haplotypes (e.g., a1, b1, and c1), we also detected several derived predominant haplotypes (e.g., a2 and b2) composing apparent substructures within the lineages (supplementary fig. S2, Supplementary Material online). During the initial population expansion, a variety of new haplotypes might have appeared in different regions, resulting from local evolutionary dynamics, for example, selective constraints imposed by the regional environments (Zhang et al. 2013). In the Indian subcontinent, we found high levels of nucleotide diversity of lineages A and B (fig. 3B and C). Because the expansion times of lineages A and B in the Middle East greatly predate those in India, this observation implies that some new haplotypes could have arisen under secondary domestication in India (Singh et al. 2013), driven by local population dynamics.
The ω ratios (ω = dN/dS; dN, nonsynonymous substitutions; dS, synonymous substitutions; see Materials and Methods) of lineages A–E ranged from 0.0457 to 0.0775 and were lower than those of other domestic animals such as goat (ω = 0.123–0.387; Nomura et al. 2013), dog (ω = 0.183; Björnerfeldt et al. 2006), and yak (ω = 0.231; Wang et al. 2011). It is reasonable to expect a stronger purifying pressure on sheep than some other domestic animals, as the sheep have often been subject of highly industrialized agricultural practice. Widely variable pastoral environments, such as highland plateau, areas of high precipitations (Joost et al. 2007), and dry desert regions, may have also maintained relatively intense selection pressures on sheep throughout history (e.g., Lv et al. 2014). The low ω values and significant differences between them (table 2) suggest that severe selective pressures have continued to operate on these lineages.
Approaches for Estimating Divergence Time
The use of molecular clock methods to infer date of evolutionary divergence events should be cautioned because a diverse range of available molecular clock methods and models can yield different estimates of ages (e.g., the reviews in Kumar 2005; Ho 2014; Ho and Duchêne 2014). Here, the divergences between domestic lineages dated by BI were much earlier than those by ML (table 2). The difference was consistent with that in an early study on complete mitogenomes from modern horses using external calibration points (Achilli et al. 2012), but contrasted with that of a study on human mtDNA using internal calibration points (Pereira et al. 2010). Achilli et al. (2012) suggested that the BI method behaves better when inferring the ages of nodes for any given lineages using internal calibration points. However, ML should be a more appropriate method by which to estimate the divergence time without a fossil record as internal calibration point. Nomura et al. (2013) argued that the fossil record in general does not always point to the “real” divergence time, but rather that the split of two lineages was older than the age of the first fossil record. In this study, we used a paleontological calibration point (i.e., the divergence time of 2.6 Ma between O. aries and O. vignei) applied to archeological timeframes. Early studies indicated that the paleontological calibration is more reasonable and if this younger fossil age had been used as the calibration point, divergence times would have been grossly underestimated (e.g., Nomura et al 2013; Jiang et al. 2014). Although the approach has been successful in dating interspecies divergence based on mtDNA (e.g., Nomura et al. 2013) and gene sequences (e.g., Jiang et al. 2014), we should be also aware of the uncertainties from such analyses associated with the impact of different timescales (e.g., the reviews in Ho et al. 2011; Ho and Duchêne 2014). The local clock, a more complex model than the global clock model, allows for different molecular clocks for the branches. Because there was genetic differentiation between wild and domestic sheep (p distance = 0.025, data not shown), the evolutionary times estimated under the local clock model with an assumption of different molecular clocks for domestic and wild sheep should be more reasonable (table 2).
Also, several biological and methodological factors can affect time estimation, including natural selection and accurate characterizations of time-dependent molecular evolutionary rate and timescale (e.g., Ho et al. 2011; Ho and Duchêne 2014). We estimated that the oldest split for lineages C and E from A, B, and D in domestic sheep occurred at approximately 0.790 Ma (95% CI: 0.634–0.934 Ma) based on synonymous substitutions, which is more recent than that estimated using a small number of complete mitogenomes in an early study (0.920 Ma, n = 16; Meadows et al. 2011). This may be because molecular clocks at synonymous sites display less time dependence and should be closer to the neutral molecular clock than those at nonsynonymous sites (e.g., the reviews in Kumar 2005; Ho et al. 2011; Ho 2014). Our detection of strong purifying selection on the protein-coding regions of the O. aries lineages (supplementary fig. S14, Supplementary Material online) also suggests that the divergence time estimation based on complete mitogenomes could be biased (see also the reviews in Ho et al. 2011; Ho and Duchêne 2014). Instead, using only synonymous sites might be a better choice for divergence time estimation. Moreover, a variety of molecular evolution rates for different components of mtDNA may also have contributed to the bias in the estimation of divergence time using complete mitogenomes (Torroni et al. 2006; Taberlet et al. 2008; Achilli et al. 2009; see also the reviews in Ho and Shapiro 2011; Ho 2014; Ho and Duchêne 2014). Similar differences were also observed in the mtDNA studies of other domestic animals, such as horse (e.g., Lippold et al. 2011; Achilli et al. 2012) and goat (e.g., Naderi et al. 2008; Nomura et al. 2013).
Conclusions
In conclusion, the first meta-analysis of complete and partial mtDNA sequences of wild and domestic sheep throughout Eurasia reveals that 1) the most common female ancestor of the five O. aries lineages lived at approximately 0.79 Ma (95% CI: 0.637–0.934 Ma) during the Middle Pleistocene, being much earlier than their domestication event (∼8–11 ka); 2) the ancestor of O. aries experienced a rapid increase in Ne immediately before the Last Glacial Maximum (∼19.5–26 ka); 3) two migratory waves (the first wave: lineages A and B; the second wave: lineage C) at approximately 4.5–6.8 ka along different routes created the original maternal genetic makeups of modern sheep across Eurasia, with the influence of prehistoric human activities, and North China could be an origin region for lineage C sheep; 4) the Mongolian Plateau region was a secondary zone/center of sheep dispersal as a “transportation hub” in eastern Eurasia: Sheep from the Middle Eastern domestication center were inferred to have migrated through Caucasus and Central Asia, and arrived in North and Southwest China (lineages A, B, and C) and the Indian subcontinent (lineages B and C) through this region; and 5) lineage A was inferred to have been introduced into the Indian subcontinent from the Middle East through Arabia. The results of this study significantly improve our understanding of sheep domestication across Eurasia, particularly the dispersal to and from eastern Eurasia.
Materials and Methods
Samples, DNA Extraction, mtDNA Sequencing, and Sequence Quality Trimming
Complete mitogenomes of 45 animals representing 42 modern native breeds (O. aries) from Azerbaijan, Moldova, Serbia, Ukraine, Russia, Kazakhstan, Poland, Finland, China, and United Kingdom and two wild species (O. orientalis and O. vignei) from Kazakhstan were sequenced (fig. 1 and supplementary table S1, Supplementary Material online). In addition, the control region of 875 animals representing 51 eastern Eurasian (including 41 Chinese, 3 Iranian, 3 Pakistani, 2 Indonesian, and 2 Siberian [the Republic of Buryatia, Russia]) native breeds was also sequenced (fig. 1 and supplementary tables S2, Supplementary Material online). In all the cases, pedigree information and knowledge of local herdsmen were used to ensure that animals were purebred and unrelated. Summary of sample information, including species/breed names and codes, sites with their geographic coordinates, and sample sizes are detailed in supplementary tables S1 (complete mitogenomes) and S2 (control region), Supplementary Material online. Genomic DNA was extracted from whole blood or marginal ear tissue using a standard phenol–chloroform method (Sambrook and Russell 2001).
Twenty-one primer pairs, including 18 used in Meadows et al. (2011) and three new pairs (supplementary table S15, Supplementary Material online) designed from the O. aries complete mitogenome reference sequence AF010406 (Hiendleder, Mainz, et al. 1998) in this study, were used to amplify the complete mitogenome of domestic and wild sheep. Polymerase chain reaction (PCR) products from the 43 domestic and two wild individuals were produced following Meadows et al. (2007). In addition, a pair of primers, forward primer MSD-F 5′-ACAACACGGACTTCCCACTC-3′ (map positions 15522–15541 bp of AF010406) and reverse primer MSD-R 5′-CCAAGCATCCCCAAAAATTA-3′ (map positions 16299–16318 bp of AF010406), was designed to amplify a 749-bp fragment of the control region. PCR amplification was conducted in 20 µl containing 30 ng of total DNA, 0.4 µM of each primer, and 2 × Es Taq Master Mix (ComWin Biotech, Beijing, China) with an initial denaturation at 95°C for 5 min, followed by 35 cycles of 30 s at 95°C, 30 s at 58°C and 1 min at 72°C, and a final 10-min extension at 72°C. All products were sequenced directly in the forward and reverse directions using the amplification primers on an ABI 3730 capillary sequencer (Applied Biosystems, Life Technologies, NY). Homologous sequences of control region in Indonesian, Iranian, Mongolian, Nepalese, and Pakistani sheep were obtained following Luo et al. (2005).
All the reads were assessed manually and aligned using CLUSTAL_X v.2.1 (Larkin et al. 2007). Complete mitogenomes were assembled and aligned to AF010406 using SeqMan II software in the Lasergene package v.12 (DNASTAR, Inc., Madison), and they were further trimmed to exclude the 75/76 bp tandem repeats (Lancioni et al. 2013) from bp 15650 to 15905. After trimming, the complete mitogenomes were fractionated into four portions, including the concatemers of tRNA, rRNA, 13 protein-coding genes with the ND6 gene readjusted to the same reading direction as the other genes (e.g., Peng and Zhang 2011), and the control region.
mtDNA Data Sets and Sequence Alignment
Fifty complete mitogenomic sequences were retrieved from GenBank, including 42 from O. aries, 2 from O. musimon, 3 from O. vignei, 2 from O. ammon, and 1 from O. canadensis (supplementary table S1, Supplementary Material online). In addition, a total of 2,017 partial O. aries mtDNA sequences (1,470 partial control region sequences from 98 breeds and 547 partial Cyt-b sequences from 50 breeds; supplementary tables S2 and S3, Supplementary Material online) of native Eurasian breeds (covering the Indian subcontinent, the Middle East, the Caucasus, Southwest Asia, Central Asia, Mongolia, and Europe) were extracted from GenBank. We also collected full control region (number of sequences = 346; supplementary table S8, Supplementary Material online) and Cyt-b (number of sequences = 271; supplementary table S9, Supplementary Material online) sequences of domestic and wild sheep (including O. orientalis, O. musimon, O. vignei, O. ammon, O. canadensis, O. dalli, and O. nivicola) from GenBank. All sequences were screened for quality using the approach of Yao et al. (2009) and Shi et al. (2014), and problematic sequences were discarded in downstream analysis.
After quality trimming and filtering, all the complete and partial mitogenomic fragments obtained in this study were compared with the respective sequences collected from GenBank using the nucleotide BLAST (Basic Local Alignment Search Tool) program (Altschul et al. 1997). Common fragments of 506 bp (map positions 14453–14958 of AF010406) for Cyt-b and 292 bp (15541–15654 and 15955–16132 of AF010406) for control region were aligned and edited for analysis using ClustalX v.2.1 (Larkin et al. 2007).
Sequence Variation and Phylogenetic Analyses
Measures of sequence variation, including number and proportion of substitutions (s), transition/transversion ratio, and nucleotide (π) and haplotype diversity (h), were computed using the program DnaSP v.5.1 (Rozas et al. 2003). Estimates of π and h were corrected for sample size using rarefaction. The five major maternal lineages (A–E) of O. aries are defined by specific diagnostic mutations common to the individual lineages (e.g., Meadows et al. 2007, 2011), and the lineage frequencies in each breed/population (or group of breeds/populations) were estimated by counting.
Pairwise-population fixation index (FST) values (Reynolds et al. 1983) and analysis of molecular variation were calculated based on the 2,345 partial control region sequences across Eurasia using Arlequin v.3.5 (Excoffier and Lischer 2010). The obtained genetic distances were used to draw MDS plots in the R package v. 3.1.2 (R Core Team 2014), and eigenvalues (λ) for the first two dimensions (dimensions 1 and 2) were calculated in each breed/population. To visualize the geographic distribution patterns of π and λ, interpolation maps were constructed using the ArcMap program in ArcGIS v.10.0 software. The inverse distance weighted option with a power of 2 was selected for the interpolation of the surface (e.g., Hanotte et al. 2002). Regression of λ1 versus geographic distance from the domestication center of sheep (represented by the geographic distance from the Kilis province of Turkey, where ancient domestic sheep are located; Demirci et al. 2013), and λ2 versus geographic distance from a putative “transportation hub” of the Mongolian Plateau region (represented by the geographic distance from the northernmost population [Transbaikal Finewool] sampled) were conducted separately for populations through various migration routes using SPSS v. 18.0.
Phylogenetic relationships among haplotypes were inferred from the complete mitogenomes using the BI approach within MrBayes v.3.2.2 (Ronquist et al. 2011) and the ML approach within PhyML v.3.0 (Guindon and Gascuel 2003). Hierarchical LRTs were implemented to select a best-fit model of nucleotide substitution in the phylogenetic analysis using the program jModelTest v.2.1.4 (Darriba et al. 2012). Out of 88 candidate models, the best-fit models under the Bayesian information criterion, HKY85 + I + G, HKY + G + I, HKY + G, and TN93 + H + G (for details of the models, see Hasegawa et al. 1985; Tamura and Nei 1993; Posada 2008), were selected for the data sets of protein-coding genes, control region, Cyt-b, and complete sequences of the mitogenomes, respectively. Bootstrap support values for the ML analysis were generated with 1,000 replicates (Felsenstein 1985). The BI analysis was run with four simultaneous Markov chains for 20 million generations, starting from a random tree. The sampling frequency was set to 1/1,000, and the first 20% of trees obtained were discarded as a burn-in. In addition, we reconstructed phylogenetic trees based on the full control region (1,105–1,333 bp) and Cyt-b sequences (1,063–1,143 bp) of wild and domestic sheep (supplementary tables S7 and S8, Supplementary Material online). Phylogenetic trees were inferred using the BI, ML, maximum parsimony (MP), and neighbor-joining methods with the HKY + G model as described in Rezaei et al. (2010). We also constructed median-joining networks from the full control region and Cyt-b sequences of domestic and wild sheep (including the hybrids), and attempted to assign each Cyt-b or control region sequence to the specific lineages defined previously using diagnostic mutations and the near-matching strategy (e.g., Wu et al. 2007; Achilli et al. 2012). Similarly, median-joining networks (Bandelt et al. 1999) were built based on the partial control region or Cyt-b sequences separately.
Detection of Signatures of Selection on Protein-Coding Genes
Signatures of selection on the protein-coding genes of the five lineages were detected using the branch-site models of ω (ω = dN/dS; dN, nonsynonymous substitutions; dS, synonymous substitutions) implemented in the CODEML program of the package PAML v.4.7 (Yang 2007). ω is an indicator of natural selection. Values of ω = 1, ω < 1 and ω > 1 indicate neutral evolution, purifying and diversifying positive selection, respectively (Goldman and Yang 1994; Crandall et al. 1999). Values of ω were estimated for the codons of different branches (lineages A–E) that were assumed to have evolved independently of one another. Four codon-substitution models were used to investigate the signature of selection. The phylogenetic tree for protein-coding genes was constructed using the MP method as implemented in MEGA v.5.2.2 (Tamura et al. 2011).
First, we tested the simplest branch, which assumed the same ω ratio for all the branches in the entire tree. Second, we assumed a two-ratio model that assigned a ω ratio for the tested branch and a background ω ratio for all the other branches. Third, we tested the three-ratio model, which allowed two different ω ratios on the two tested branches and a background ω ratio for the other branches. Finally, we adopted a free-ratio model, which assumed a different ω ratio for each of the four branches in the phylogenetic tree. Log-likelihood scores (ln L) evaluated the quality of the fit of the tested data to the conditions of the model; a higher log-likelihood value (close to zero) was a “better”-fitting model (Yang 1998). LRTs were further employed to compare the fit of the models to the data by comparing twice the log-likelihood difference (2Δln L) to a χ2 distribution with degrees of freedom equal to the difference in the number of parameters between the two models (Yang 1998).
Divergence Time Estimation
Phylogenetic relationships within the genus Ovis (see Results) inferred in previous analyses were used to estimate the divergence times between the major O. aries lineages using PAML v.4.7 (Yang 2007) and BEAST v.1.7.5 (Drummond and Rambaut 2007). Due to the lack of an exact fossil record between Ovis species for the calibration, we used a comprehensive evolutionary framework (see Nomura et al. 2013; Jiang et al. 2014) to estimate the divergence time between O. aries and O. vignei. A phylogenetic tree including 24 species (supplementary table S16, Supplementary Material online) was inferred based on the 13 mtDNA protein-coding genes using the GTR + I + G model in MrBayes v.3.2.2 (Ronquist et al. 2011). The divergence times were estimated based on five fossil calibration points (18.3–28.5 Ma between Bovinae and Caprinae, 52–58 Ma between Cetacea and hippopotamus, >34.1 Ma between baleen and toothed whales, 42.8–63.8 Ma between Caniformia and Feliformia, and 62.3–71.2 Ma between Carnivora and Perissodactyla; see Nomura et al. 2013; Jiang et al. 2014). We applied the obtained O. aries/O. vignei divergence time (2.6 Ma; see Results) and three models to estimate the divergence times between the five O. aries mtDNA lineages. Global and local clock models were implemented using the ML in PAML v.4.7 (Yang 2007) and the uncorrelated relaxed-clock model was implemented using BEAST v.1.7.5 (Drummond and Rambaut 2007).
We only considered the 61 complete mitogenomes of wild sheep species and native breeds of domestic sheep (supplementary table S1; Supplementary Material online) and applied three strategies in the ML analysis: One considered the complete mitogenomes under the TN93 model, the second considered the synonymous mutations under the HKY85 model, and the third considered only the third-codon positions under the HKY85 model. Similarly, Bayesian Markov chain Monte Carlo (MCMC) analysis of molecular sequences was performed by applying the three strategies in the program BEAST v.1.7.5 (Drummond and Rambaut 2007). Parameters of prior distributions, including models of nucleotide substitution and the divergence time between O. aries and O. vignei, were set the same as in the ML analyses described above. Three independent runs were performed with 50 million iterations. Samples were drawn every 5,000 MCMC steps, with the first 25% samples discarded as burn-in. The results of the three independent runs were combined using the LogCombiner program (available at http://beast.bio.ed.ac.uk/LogCombiner, last accessed October 16, 2014) from BEAST v.1.7.5 (Drummond and Rambaut 2007). Convergence was confirmed by effective sampling size (ESS) greater than 200 using the program Tracer v.1.5 (Drummond and Rambaut 2007; available at http://beast.bio.ed.ac.uk/Tracer, last accessed December 26, 2014).
BI of Population Expansions
Based on the divergence time of internal nodes estimated above and the 51 complete mitogenomes of native domestic sheep breeds, we reconstructed the change in Ne of O. aries through time using BSPs (Drummond et al. 2005). The analyses were also performed on the partial Cyt-b and control region sequences. We ran three independent chains in each analysis using BEAST v.1.7.5 (Drummond and Rambaut 2007), with 50 million generations (after discarding the first 10% of sampled generations as burn-in) and samples drawn every 5,000 steps. We applied the HKY85 and TN93 models under relaxed-clock model for complete mitogenomes and partial Cyt-b sequences, respectively. In the analysis of control region sequences, we set similar parameter values of 200 million generations (after discarding the first 10% of sampled generations as burn-in) with samples drawn every 2,000 steps under the HKY85 and relaxed-clock models. The combination of three independent results and checks of convergence were performed following the same procedures as described above.
Signatures of population expansions were examined using Arlequin v.3.5 (Excoffier and Lischer 2010). First, the observed and expected mismatch distributions of pairwise differences between haplotypes were compared using Tajima’s D (Tajima 1989) and Fu’s Fs (Fu 1997) tests of neutrality. Furthermore, we estimated the parameters for the sudden population expansion model (Rogers 1995), and the fit of the data to the sudden population expansion model was tested. Harpending’s raggedness index (rH; Harpending 1994) of the observed mismatch distribution was also calculated. P values of the SSDs test to evaluate the fit and significance for the parameters (rH, D, and Fs) were determined with 1,000 coalescent simulations using Arlequin v.3.5 (Excoffier and Lischer 2010). All the partial control region sequences were included in the calculations.
Further, to corroborate our inference that the Mongolian Plateau region serves as a “transportation hub” in eastern Eurasia (see Results), we distinguished several candidate colonization scenarios for the three main O. aries lineages (A, B, and C) using the ABC (Beaumont et al. 2002) procedure in DIYABC v.2.0.4 (Cornuet et al. 2014). By incorporating all the O. aries control region sequences of a 292-bp-long hypervariable fragment, we tested six, eight, and five colonization models regarding potential migration routes from the Middle Eastern domestication center to different regions in eastern Eurasia (e.g., the Mongolian Plateau region, North China, Southwest China, and the Indian subcontinent) for the lineages A, B, and C, respectively (supplementary fig. S25, Supplementary Material online). Detailed information about the ABC analyses including the candidate colonization models tested was provided in supplementary information S4, Supplementary Material online.
Supplementary Material
Supplementary information S1–S4, figures S1–S25, and tables S1–S19 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank San-Gang He, Ya-Wei Sun, Nurbi Marzanov, Mikhail Ozerov, Maciek Murawski, Tatiana Kiseleva, and the late Mirjana Cinkulov for help in sample collection, Anneli Virta for technical assistance, and Dr Alessandro Achilli (Università di Perugia, Perugia, Italy) for his comments on an earlier version of the manuscript. This work was supported by the 100-talent Program of Chinese Academy of Sciences (CAS), the National High Technology Research and Development Program of China (i.e., 863 Program, grant No. 2013AA102506), the Breakthrough Project of Strategic Priority Program of the Chinese Academy of Sciences (grant No. XDB13000000), the grants from National Natural Science Foundation of China (grants Nos. 31272413 and U1303284), and Academy of Finland (grants Nos. 250633 and 256077) as well as Chinese Government contribution to CAAS-ILRI Joint Laboratory on Livestock and Forage Genetic Resources in Beijing. The paper contributes to the CGIAR Research Program on Livestock and Fish.
References
- Achilli A, Bonfiglio S, Olivieri A, Malusà A, Pala M, Kashani BH, Perego UA, Ajmone-Marsan P, Liotta L, Semino O, et al. 2009. The multifaceted origin of taurine cattle reflected by the mitochondrial genome. PLoS One 4:e5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achilli A, Olivieri A, Soares P, Lancioni H, Kashani BH, Perego UA, Nergadze SG, Carossa V, Santagostino M, Capomaccio S, et al. 2012. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc Natl Acad Sci U S A. 109:2449–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aniwashi J, Jiahan K, Hakaimofu H, Sulaiman Y, Xi S-Y, Du M, Hailati, Tuersenhali, Ayinuer 2007. Study on hybridication of wild argali and Bashibai sheep. Xinjiang Agric Sci. 44:702–705 (in Chinese). [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16:37–48. [DOI] [PubMed] [Google Scholar]
- Bar-Yosef O, Meadow R. 1995. The origins of agriculture in the Near East. In: Price T, Gebauer A-B, editors. Last hunters, first farmers. Santa Fe: School of American Research Press; p. 39–94. [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ. 2002. Approximate Bayesian computation in population genetics. Genetics 162:2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björnerfeldt S, Webster MT, Vilà C. 2006. Relaxation of selective constraint on dog mitochondrial DNA following domestication. Genome Res. 16:990–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruford MW. 2005 Molecular approaches to understanding animal domestication: what have we learned so far? World Poultry Science Association, 4th European Poultry Genetics Symposium. Dubrovnik, Croatia, 6–8 October, 2005.
- Cai DW, Han L, Zhang XL, Zhou H, Zhu H. 2007. DNA analysis of archaeological sheep remains from China. J Archaeol Sci. 34:1347–1355. [Google Scholar]
- Cai DW, Tang ZW, Yu HX, Han L, Ren XY, Zhao XB, Zhu H, Zhou H. 2011. Early history of Chinese domestic sheep indicated by ancient DNA analysis of Bronze Age individuals. J Archaeol Sci. 38:896–902. [Google Scholar]
- Carruthers D. 1949. Beyond the Caspian. A naturalist in Central Asia. Edinburgh: Oliver and Boyd. [Google Scholar]
- Chen FH, Dong GH, Zhang DJ, Liu XY, Jia X, An CB, Ma MM, Xie YW, Barton L, Ren XY, et al. 2015. Agriculture facilitated permanent human occupation of the Tibetan Plateau after 3600 B.P. Science 347:248–250. [DOI] [PubMed] [Google Scholar]
- Chen SY, Duan ZY, Sha T, Xiangyu J, Wu SF, Zhang YP. 2006. Origin, genetic diversity, and population structure of Chinese domestic sheep. Gene 376:216–223. [DOI] [PubMed] [Google Scholar]
- Chen WH. 1990. Index to data of agricultural archaeology-farm-tools. Agric Archaeol. 1:425–427. (in Chinese) [Google Scholar]
- Chessa B, Pereira F, Arnaud F, Amorim A, Goyache F, Mainland I, Kao RR, Pemberton JM, Beraldi D, Stear MJ, et al. 2009. Revealing the history of sheep domestication using retrovirus integrations. Science 324:532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colledge S, Conolly J, Shennan S. 2005. The evolution of Neolithic farming from SW Asian origins to NW European limits. Eur J Archaeol. 8:137–156. [Google Scholar]
- Cornuet J-M, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Marin J-M, Estoup A. 2014. DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30:1187–1189. [DOI] [PubMed] [Google Scholar]
- Crandall KA, Kelsey CR, Imamichi H, Lane HC, Salzman NP. 1999. Parallel evolution of drug resistance in HIV: failure of nonsynonymous/synonymous substitution rate ratio to detect selection. Mol Biol Evol. 16:372–382. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci S, Koban Baştanlar E, Dağtaş ND, Pişkin E, Engin A, Özer F, Yüncü E, Doğan ŞA, Togan İ. 2013. Mitochondrial DNA diversity of modern, ancient and wild sheep (Ovis gmelinii anatolica) from Turkey: new insights on the evolutionary history of sheep. PLoS One 8:e81952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobney K, Larson G. 2006. Genetics and animal domestication: new windows on an elusive process. J Zool. 269:261–271. [Google Scholar]
- Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 22:1185–1192. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 10:564–567. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. [DOI] [PubMed] [Google Scholar]
- Fu Y-X. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauri FN. 2013. Indo-Saudi trade relation. Arabian Journal of Business and Management Review (Nigerian Chapter) 1(2):45–57. [Google Scholar]
- Goldman N, Yang Z. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 11:725–736. [DOI] [PubMed] [Google Scholar]
- Grant WS, Liu M, Gao T, Yanagimoto T. 2012. Limits of Bayesian skyline plot analysis of mtDNA sequences to infer historical demographies in Pacific herring (and other species). Mol Phylogenet Evol. 65:203–212. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704. [DOI] [PubMed] [Google Scholar]
- Guo J, Du LX, Ma YH, Guan WJ, Li HB, Zhao QJ, Li X, Rao SQ. 2005. A novel maternal lineage revealed in sheep (Ovis aries). Anim Genet. 36:331–336. [DOI] [PubMed] [Google Scholar]
- Hanotte O, Bradley DG, Ochieng JW, Verjee Y, Hill EW, Rege JEO. 2002. African pastoralism: genetic imprints of origins and migrations. Science 296:336–339. [DOI] [PubMed] [Google Scholar]
- Harpending H. 1994. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol. 66:591–600. [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T-A. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 22:160–174. [DOI] [PubMed] [Google Scholar]
- Heller R, Chikhi L, Siegismund HR. 2013. The confounding effect of population structure on Bayesian skyline plot inferences of demographic history. PLoS One 8:e62992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiendleder S, Lewalski H, Wassmuth R, Janke A. 1998. The complete mitochondrial DNA sequence of the domestic sheep (Ovis aries) and comparison with the other major ovine haplotype. J Mol Evol. 47:441–448. [DOI] [PubMed] [Google Scholar]
- Hiendleder S, Mainz K, Plante Y, Lewalski H. 1998. Analysis of mitochondrial DNA indicates that domestic sheep are derived from two different ancestral maternal sources: no evidence for contributions from urial and argali sheep. J Hered. 89:113–120. [DOI] [PubMed] [Google Scholar]
- Ho SYW. 2014. The changing face of the molecular evolutionary clock. Trends Ecol Evol. 29:496–503. [DOI] [PubMed] [Google Scholar]
- Ho SYW, Duchêne S. 2014. Molecular-clock methods for estimating evolutionary rates and timescales. Mol Ecol. 23:5947–5965. [DOI] [PubMed] [Google Scholar]
- Ho SYW, Lanfear R, Bromham L, Phillips MJ, Soubrier J, Rodrigo AG, Cooper A. 2011. Time-dependent rates of molecular evolution. Mol Ecol. 20:3087–3101. [DOI] [PubMed] [Google Scholar]
- Ho SYW, Shapiro B. 2011. Skyline-plot methods for estimating demographic history from nucleotide sequences. Mol Ecol Resour. 11:423–434. [DOI] [PubMed] [Google Scholar]
- Hodges J. 1999. Animals and values in society. Livest Res Rural Dev. 11:1–6. [Google Scholar]
- Hunt HV, Vander Linden M, Liu X, Motuzaite-Matuzeviciute G, Colledge S, Jones MK. 2008. Millets across Eurasia: chronology and context of early records of the genera Panicum and Setaria from archaeological sites in the Old World. Veg Hist Archaeobot. 17:5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Xie M, Chen W, Talbot R, Maddox JF, Faraut T, Wu C, Muzny DM, Li Y, Zhang W, et al. 2014. The sheep genome illuminates biology of the rumen and lipid metabolism. Science 344:1168–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MK, Liu X. 2009. Origins of agriculture in East Asia. Science 324:730–731. [DOI] [PubMed] [Google Scholar]
- Joost S, Bonin A, Bruford MW, Despres L, Conord C, Erhardt G, Taberlet P. 2007. A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Mol Ecol. 16:3955–3969. [DOI] [PubMed] [Google Scholar]
- Kijas JW, Lenstra JA, Hayes B, Boitard S, Neto LRP, San Cristobal M, Servin B, McCulloch R, Whan V, Gietzen K, et al. 2012. Genome-wide analysis of the world's sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 10:e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijas JW, Townley D, Dalrymple BP, Heaton MP, Maddox JF, McGrath A, Wilson P, Ingersoll RG, McCulloch R, McWilliam S, et al. 2009. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS One 4:e4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. 2005. Molecular clocks: four decades of evolution. Nat Rev Genet. 6:654–662. [DOI] [PubMed] [Google Scholar]
- Kuo F, Needham J, Chhêng CT. 1999. The history of zoology in China. Beijing (China): Science Press; (in Chinese) [Google Scholar]
- Lancioni H, Di Lorenzo P, Ceccobelli S, Perego UA, Miglio A, Landi V, Antognoni MT, Sarti FM, Lasagna E, Achilli A. 2013. Phylogenetic relationships of three Italian Merino-derived sheep breeds evaluated through a complete mitogenome analysis. PLoS One 8:e73712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- Larson G, Albarella U, Dobney K, Rowley-Conwy P, Schibler J, Tresset A, Vigne J-D, Edwards CJ, Schlumbaum A, Dinu A, et al. 2007. Ancient DNA, pig domestication, and the spread of the Neolithic into Europe. Proc Natl Acad Sci U S A. 104:15276–15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Burger J. 2013. A population genetics view of animal domestication. Trends Genet. 29:197–205. [DOI] [PubMed] [Google Scholar]
- Larson G, Liu R, Zhao X, Yuan J, Fuller D, Barton L, Dobney K, Fan Q, Gu Z, Liu X-H, et al. 2010. Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA. Proc Natl Acad Sci U S A. 107:7686–7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippold S, Matzke NJ, Reissmann M, Hofreiter M. 2011. Whole mitochondrial genome sequencing of domestic horses reveals incorporation of extensive wild horse diversity during domestication. BMC Evol Biol. 11:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zhang J, Liu K-B, Wu N, Li Y, Zhou K, Ye M, Zhang T, Zhang H, Yang X, et al. 2009. Earliest domestication of common millet (Panicum miliaceum) in East Asia extended to 10,000 years ago. Proc Natl Acad Sci U S A. 106:7367–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, Gielly L, Excoffier L, Vigne J-D, Bouvet J, Taberlet P. 2001. Multiple maternal origins and weak phylogeographic structure in domestic goats. Proc Natl Acad Sci U S A. 98:5927–5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y-Z, Cheng S-R, Lkhagva B, Badamdorj D, Hanotte O, Han J-L. 2005. Study on origin and genetic diversity of Mongolian and Chinese sheep. Acta Genet Sin. 32:1256–1265. (in Chinese) [PubMed] [Google Scholar]
- Lv F-H, Agha S, Kantanen J, Colli L, Stucki S, Kijas JW, Joost S, Li M-H, Marsan PA. 2014. Adaptations to climate-mediated selective pressures in sheep. Mol Biol Evol. 31:3324–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JR, Cemal I, Karaca O, Gootwine E, Kijas JW. 2007. Five ovine mitochondrial lineages identified from sheep breeds of the Near East. Genetics 175:1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JRS, Hiendleder S, Kijas JW. 2011. Haplogroup relationships between domestic and wild sheep resolved using a mitogenome panel. Heredity 106:700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muigai AT, Hanotte O. 2013. The origin of African sheep: archaeological and genetic perspectives. Afr Archaeol Rev. 30:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naderi S, Rezaei H-R, Pompanon F, Blum MGB, Negrini R, Naghash H-R, Balkız Ö, Mashkour M, Gaggiotti OE, Ajmone-Marsan P, et al. 2008. The goat domestication process inferred from large-scale mitochondrial DNA analysis of wild and domestic individuals. Proc Natl Acad Sci U S A. 105:17659–17664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi M, Bläuer A, Iso-Touru T, Nyström V, Harjula J, Taavitsainen J-P, Storå J, Lidén K, Kantanen J. 2013. Mitochondrial DNA and Y-chromosomal diversity in ancient populations of domestic sheep (Ovis aries) in Finland: comparison with contemporary sheep breeds. Genet Sel Evol. 45:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K, Yonezawa T, Mano S, Kawakami S, Shedlock AM, Hasegawa M, Amano T. 2013. Domestication process of the goat revealed by an analysis of the nearly complete mitochondrial protein-encoding genes. PLoS One 8:e67775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe H. 1959. G. H. Luce and Pe Maung Tin (ed.): Inscriptions of Burma. Portfolio IV. Down to 702 B.E. (1340 A.D.).—Portfolio V. 703–726 B.E. (1341–1364 A.D.). (University of Rangoon Oriental Studies Publication No. 5, No. 6.) 63 pp., plates 346–462; 38 pp., plates 463–609. Oxford: University Press, 1956. Bull Sch Orient Afr Stud. 22:177. [Google Scholar]
- Pedrosa S, Uzun M, Arranz JJ, Gutierrez-Gil B, San Primitivo F, Bayon Y. 2005. Evidence of three maternal lineages in Near Eastern sheep supporting multiple domestication events. Proc R Soc Lond B Biol Sci. 272:2211–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M-S, Zhang Y-P. 2011. Inferring the population expansions in peopling of Japan. PLoS One 6:e21509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Silva NM, Franco-Duarte R, Fernandes V, Pereira JB, Costa MD, Martins H, Soares P, Behar DM, Richards MB, et al. 2010. Population expansion in the North African Late Pleistocene signalled by mitochondrial DNA haplogroup U6. BMC Evol Biol. 10:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J, von den Driesch A, Helmer D. 2005. The upper Euphrates-Tigris basin: cradle of agro-pastoralism. In: Vigne JD, Peters J, Helmer D, editors. The first steps of animal domestication. Oxford: Oxbow; pp. 96–124. [Google Scholar]
- Pickrell JK, Pritchard JK. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8:e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplin F. 1979. Origine du mouflon de Corse dans une nouvelle perspective paléontologique: par marronnage. Ann Génét Sél Anim. 11:133–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2014. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. 1983. Estimation of the co-ancestry coefficient—basis for a short-term genetic-distance. Genetics 105:767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaei HR, Naderi S, Chintauan-Marquier IC, Taberlet P, Virk AT, Naghash HR, Rioux D, Kaboli M, Pompanon F. 2010. Evolution and taxonomy of the wild species of the genus Ovis (Mammalia, Artiodactyla, Bovidae). Mol Phylogenet Evol. 54:315–326. [DOI] [PubMed] [Google Scholar]
- Rogers AR. 1995. Genetic evidence for a Pleistocene population explosion. Evolution 49:608–615. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck J, Teslenko M. 2011. Draft MrBayes version 3.2 Manual: tutorials and model summaries. Available from: http://mrbayes.sourceforge.net/. [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497. [DOI] [PubMed] [Google Scholar]
- Ryder ML. 1983. Sheep and man. London: Duckworth. [Google Scholar]
- Ryder ML. 1984. Sheep. In: Mason IL, editor. Evolution of domesticated animals. London/New York: Longman; pp. 63–85. [Google Scholar]
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. 3rd ed New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Scherf BD, editor. 2000. World watch list for domestic animal diversity. In: Food and Agriculture Organization of the United Nations. Rome: Food and Agriculture Organization of the United Nations; p. 58. [Google Scholar]
- Shaha R. 1970. Nepal, Tibet and China. J Nepal Council World Aff. 3:13–82. [Google Scholar]
- Shi N-N, Fan L, Yao Y-G, Peng M-S, Zhang Y-P. 2014. Mitochondrial genomes of domestic animals need scrutiny. Mol Ecol. 23:5393–5397. [DOI] [PubMed] [Google Scholar]
- Singh S, Kumar S, Jr, Kolte AP, Kumar S. 2013. Extensive variation and sub-structuring in lineage A mtDNA in Indian sheep: genetic evidence for domestication of sheep in India. PLoS One 8:e77858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet P, Valentini A, Rezaei HR, Naderi S, Pompanon F, Negrini R, Ajmone-Marsan P. 2008. Are cattle, sheep, and goats endangered species? Mol Ecol. 17:275–284. [DOI] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512–526. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapio M, Marzanov N, Ozerov M, Cinkulov M, Gonzarenko G, Kiselyova T, Murawski M, Viinalass H, Kantanen J. 2006. Sheep mitochondrial DNA variation in European, Caucasian, and Central Asian areas. Mol Biol Evol. 23:1776–1783. [DOI] [PubMed] [Google Scholar]
- Tapio M, Ozerov M, Tapio I, Toro MA, Marzanov N, Cinkulov M, Goncharenko G, Kiselyova T, Murawski M, Kantanen J. 2010. Microsatellite-based genetic diversity and population structure of domestic sheep in northern Eurasia. BMC Genet. 11:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Achilli A, Macaulay V, Richards M, Bandelt H-J. 2006. Harvesting the fruit of the human mtDNA tree. Trends Genet. 22:339–345. [DOI] [PubMed] [Google Scholar]
- Wang G-D, Xie H-B, Peng M-S, Irwin D, Zhang Y-P. 2014. Domestication genomics: evidence from animals. Annu Rev Anim Biosci. 2:65–84. [DOI] [PubMed] [Google Scholar]
- Wang X, Ma Y-H, Chen H. 2006. Analysis of the genetic diversity and the phylogenetic evolution of Chinese sheep based on Cyt b gene sequences. Acta Genet Sin. 33:1081–1086. [DOI] [PubMed] [Google Scholar]
- Wang Z, Yonezawa T, Liu B, Ma T, Shen X, Su J, Guo S, Hasegawa M, Liu J. 2011. Domestication relaxed selective constraints on the yak mitochodrial genomes. Mol Biol Evol. 28:1553–1556. [DOI] [PubMed] [Google Scholar]
- Warmuth V, Eriksson A, Bower MA, Barker G, Barrett E, Hanks BK, Li S, Lomitashvili D, Ochir-Goryaeva M, Sizonov GV, et al. 2012. Reconstructing the origin and spread of horse domestication in the Eurasian steppe. Proc Natl Acad Sci U S A. 109:8202–8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood NJ, Phua SH. 1996. Variation in the control region sequence of the sheep mitochondrial genome. Anim Genet. 27:25–33. [DOI] [PubMed] [Google Scholar]
- Wu GS, Yao YG, Qu KX, Ding ZL, Li H, Palanichamy MG, Duan ZY, Li N, Chen YS, Zhang YP. 2007. Population phylogenomic analysis of mitochondrial DNA in wild boars and domestic pigs revealed multiple domestication events in East Asia. Genome Biol. 8:R245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang H, Gao J, Yu B, Zhou H, Cai D, Zhang Y, Chen X, Wang X, Hofreiter M, Zhao X. 2014. Early Holocene chicken domestication in northern China. Proc Natl Acad Sci U S A. 111:17564–17569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Scuderi LA, Wang X, Scuderi LJ, Zhang D, Li H, Forman S, Xu Q, Wang R, Huang W, et al. 2015. Groundwater sapping as the cause of irreversible desertification of Hunshandake Sandy Lands, Inner Mongolia, northern China. Proc Natl Acad Sci U S A. 112:702–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wan Z, Perry L, Lu H, Wang Q, Zhao C, Li J, Xie F, Yu J, Cui T, et al. 2012. Early millet use in northern China. Proc Natl Acad Sci U S A. 109:3726–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 15:568–573. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yao Y-G, Salas A, Logan I, Bandelt H-J. 2009. mtDNA data mining in GenBank needs surveying. Am J Hum Genet. 85:929–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H. 2004. Bronze roads: a introduction to archaic cultural exchange in Eurasia. In: Department of Cultural Heritage and Museum Studies, editor. Antiquities of Eastern Asia. Beijing (China): Cultural Relics Press. (in Chinese) [Google Scholar]
- Zeder MA. 2008. Domestication and early agriculture in the Mediterranean Basin: origins, diffusion, and impact. Proc Natl Acad Sci U S A. 105:11597–11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeder MA, Emshwiller E, Smith BD, Bradley DG. 2006. Documenting domestication: the intersection of genetics and archaeology. Trends Genet. 22:139–155. [DOI] [PubMed] [Google Scholar]
- Zhang H, Paijmans JLA, Chang F, Wu X, Chen G, Lei C, Yang X, Wei Z, Bradley DG, Orlando L, et al. 2013. Morphological and genetic evidence for early Holocene cattle management in northeastern China. Nat Commun. 4:2755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.