Abstract
Background and aims
Quercetin is a flavonoid with good antioxidant activity, and exhibits various important pharmacological effects. The aim of the present work was to study the influence of formulation factors on the physicochemical properties of quercetin-loaded polymeric nanoparticles in order to optimize the formulation.
Materials and methods
The nanoparticles were prepared by the nanoprecipitation method. A 3-factor, 3-level Box-Behnken design was employed in this study considering poly(D,L-lactic-co-glycolic) acid (PLGA) concentration, polyvinyl alcohol (PVA) concentration and the stirring speed as independent variables. The responses were particle size, polydispersity index, zeta potential and encapsulation efficiency.
Results
The PLGA concentration seemed to be the most important factor influencing quercetin-nanoparticle characteristics. Increasing PLGA concentration led to an increase in particle size, as well as encapsulation efficiency. On the other hand, it exhibited a negative influence on the polydispersity index and zeta potential. The PVA concentration and the stirring speed had only a slight influence on particle size and polydispersity index. However, PVA concentration had an important negative effect on the encapsulation efficiency. Based on the results obtained, an optimized formulation was prepared, and the experimental values were comparable to the predicted ones.
Conclusions
The overall results indicated that PLGA concentration was the main factor influencing particle size, while entrapment efficiency was predominantly affected by the PVA concentration.
Keywords: nanoparticle, quercetin, experimental design, Box-Behnken, optimization
Introduction
During the past decades, nanoparticles have received considerable attention, due to their potential use as drug delivery systems. Polymeric nanoparticles are colloidal systems ranging in size usually from 10 to 1000 nm. They are formulated from biodegradable polymers in which the active substance can be entrapped, adsorbed or chemically coupled onto the polymer matrix [1].
As opposed to conventional drug delivery systems, nanoparticulate carriers offer several advantages, including high stability, the possibility of incorporating both hydrophilic and hydrophobic substances, protection of the entrapped substance from enzymatic degradation, the ability of delivery by different routes of administration, reduction of frequency of administration, daily doses and side effects [2–4]. Nanoparticles can improve the bioavailability of poorly absorbed drugs, prolong the residence time of these drugs in the body, provide a controlled release, and, last but not least, assure cell targeting [4,5].
The most commonly used polymers for manufacturing polymeric nanoparticles are poly(lactic acid) (PLA), poly(D,L-lactic-co-glycolic) acid (PLGA) and poly(caprolactone) (PCL). These polymeric materials are biocompatible, biodegradable and non-immunogenic [3,6]. Due to its excellent safety profile, PLGA is widely used to obtain nano- and microparticles. PLGA has been approved by the Food and Drug Administration (FDA) and European Medicine Agency (EMA) in several drug delivery systems. Since then, some controlled release PLGA-based products have been licensed for use in humans and have been introduced to the market [6–9]. PLGA consists of two monomers – lactic acid and glycolic acid, formed as a result of the polymer hydrolysis. Both metabolites are endogenous compounds, which are easily metabolized themselves through the body’s Krebs cycle. Consequently, the administration of PLGA nanoparticles is associated with a minimal systemic toxicity [2,9]. PLGA degradation rate depends on its molecular weight and on the molar ratio of the two monomer components. Usually, a higher content of poly glycolic acid leads to faster degradation rates, except for PLGA with a molar ratio of 50:50 poly lactic acid:poly glycolic acid, which exhibits the fastest degradation rate, namely one week [10,11].
Among the materials used to prepare nanoparticles, the stabilizing agents are also included. They act by decreasing the interfacial tension between the hydrophilic and hydrophobic phases, thus by stabilizing the colloidal dispersion. Non-ionic compounds such as polyvinyl alcohol (PVA) or various types of Pluronics are preferred to others, especially for incorporating poorly water soluble substances into nanoparticles [3].
Quercetin (3,3′,4′,5,7-pentahydroxyflavone, QU) is a member of the flavonoid family, commonly found in fruit, vegetables and other plant based food such as apples, onions, berries, red wine, green tea, etc [12]. Several biological properties have been described for QU, among which antioxidant, anti-inflammatory [13], anti-allergic [14], antiviral, anti-proliferative [15], immunomodulatory [16] and anticarcinogenic effects [17]. QU is considered to be one of the best antioxidant flavonoid due to the high number and position of hydroxyl groups, and conjugated π orbitals [18]. However, its clinical use is limited by its low water solubility, high rate of metabolism, short biological half-life and instability in physiological mediums. All these properties result in low bioavailability [15,19,20]. These problems may be overcome by entrapping QU in a nanoscaled delivery system that could improve the drug’s solubility, pharmacokinetic and pharmacological properties [21,22].
The methods frequently used for preparing nanoparticles are nanoprecipitation, emulsification solvent diffusion, solvent evaporation and salting out [4]. Choosing a certain technique depends on the active substance’s solubility [2]. Among these, the nanoprecipitation method is mainly applied for lipophylic compounds that have a limited water solubility, but are easily soluble in organic solvents such as ethanol or acetone [23]. This method involves the addition of an organic solution of the polymer and drug to an aqueous medium, followed by organic solvent evaporation [24]. It is a simple, quick and reproducible method [25], which results in the formation of nanoparticles usually about 200 nm in diameter [26].
The materials used, including the polymer, stabilizing agent and active substance, but also other process parameters, can affect the physicochemical properties of the polymeric nanoparticles such as particle size, polydispersity index (PdI), zeta potential and entrapment efficiency [7]. One goal in the development of drug delivery systems is to incorporate a sufficient amount of drug in order to assure an optimum concentration at the site of action, and thus therapeutic effectiveness. To achieve this, parameters influencing both the nanocarrier and the drug need to be considered during the early stages of development [27].
When developing a complex formulation, traditional experiments have the disadvantage of being time consuming and requiring more effort and materials [28]. Experimental design methodology is a strategy that allows to study different variables simultaneously, the relationship between them and their influence on different experimental responses, by running a small number of experiments [29]. Furthermore, through mathematical models it may determine the optimum level of the variables required for a given response [28]. This technique can be successfully used to optimize nanoparticle preparation conditions [30].
The present study evaluated the influence of three formulation factors on the characteristics of QU-nanoparticles. An experimental design has been used to provide an efficient means to optimize the preparation conditions of the polymeric nanoparticles. This approach involved the analysis of response surfaces in order to establish the relationship between the experimental factors and the output, and also to obtain an appropriate formulation.
Materials and methods
Materials
QU, PVA (average MW 30000–70000) were purchased from Sigma-Aldrich (USA). PLGA (50:50, Resomer RG 502 H, MW 13100) was purchased from Boehringer Ingelheim (Germany). All other chemicals used were of analytical grade and the solvents were of HPLC grade.
Methods
Experimental Design
Prior to elaborating the present experimental design, a prescreening study was preformed (data not presented). Six variables were investigated in the prescreening, namely PLGA, PVA and QU concentration, the volume of organic solvent and aqueous phase and the stirring speed. Based on the results obtained, three formulation factors were selected to further evaluate their influence on the QU-nanoparticles’ properties. In this sense, a three-factor and three-level Box-Behnken Experimental Design was developed using Modde 10 Software (Umetrics, Sweden) [31]. The selected independent variables (formulation factors) were as follows: PLGA concentration (X1), PVA concentration (X2) and stirring speed (X3). Each independent variable in the design was studied at three different levels (−1, 0, 1) (Table I). The dependent variables (responses), were particle size (Y1), PdI (Y2), zeta potential (Y3) and encapsulation efficiency (Y4). The design matrix generated by the software consisted of 15 experiments, of which 3 replicated runs, as shown in Table II. All experiments were carried out in a random order to minimize the effect of unexplainable variability in the observed response [31,32].
Table I.
Independent variables and their levels of variation.
| Independent variable | Symbol | Level of variation | ||
|---|---|---|---|---|
| −1 | 0 | +1 | ||
| PLGA concentration (mg/ml) | X1 | 5 | 12.5 | 20 |
| PVA concentration (%m/V) | X2 | 1 | 2.5 | 4 |
| Stirring speed (rpm) | X3 | 255 | 382.5 | 510 |
Table II.
Design Matrix.
| Experiment Number | Experiment Name | Run Order | X1 | X2 | X3 |
|---|---|---|---|---|---|
| 1 | N1 | 14 | 5 | 1 | 4.5 |
| 2 | N2 | 3 | 20 | 1 | 4.5 |
| 3 | N3 | 7 | 5 | 4 | 4.5 |
| 4 | N4 | 12 | 20 | 4 | 4.5 |
| 5 | N5 | 8 | 5 | 2.5 | 3 |
| 6 | N6 | 4 | 20 | 2.5 | 3 |
| 7 | N7 | 5 | 5 | 2.5 | 6 |
| 8 | N8 | 10 | 20 | 2.5 | 6 |
| 9 | N9 | 1 | 12.5 | 1 | 3 |
| 10 | N10 | 2 | 12.5 | 4 | 3 |
| 11 | N11 | 13 | 12.5 | 1 | 6 |
| 12 | N12 | 6 | 12.5 | 4 | 6 |
| 13 | N13 | 9 | 12.5 | 2.5 | 4.5 |
| 14 | N14 | 15 | 12.5 | 2.5 | 4.5 |
| 15 | N15 | 11 | 12.5 | 2.5 | 4.5 |
X1 – PLGA concentration; X2 – PVA concentration; X3 – stirring speed
Data were fitted by means of partial least squares (PLS) and were analyzed using the statistical module of the Modde 10 Software. In order to establish a statistical correlation between the independent variables and the observed responses and to check the validity of the experimental design, the following statistical parameters were calculated: R2, Q2 and ANOVA. Three dimensional response surface plots that represent the individual and interactive influences of the formulation factors on the responses were generated for a better understanding of these effects.
Preparation of QU-nanoparticles
QU-nanoparticles were prepared by the solvent displacement method, also known as nanoprecipitation method, first described by Fessi et al [24] with slight modifications. Briefly, the corresponding amount of PLGA and 5 mg of QU were dissolved in 7 ml of acetone. The resulting organic solution was added dropwise to an aqueous PVA solution of a certain concentration, which was kept under magnetic stirring at specific rotating speed. The mixture was maintained under continuous magnetic stirring for 4 hours at 40°C in order to completely remove the organic solvent. Subsequently, the QU-nanoparticles were centrifuged at a speed of 25000 rpm for 30 minutes (Sigma, Germany). The supernatant containing the non-entrapped drug was separated from the lower remaining sediment represented by the QU-nanoparticles. The final volume of each dispersion was adjusted to 10 ml by adding double distilled water to the nanoparticles. The dispersion was vortexed for 2 minutes and afterwards sonicated for 10 minutes until homogenization.
Physicochemical characterization of QU-nanoparticles
Particle size and PdI
The particle size and polydispersity of QU-nanoparticles were determined by dynamic light scattering using a Zetasizer Nano-ZS90 (Malvern, UK). 50 μl of nanoparticle suspension were dispersed in double distilled water and then analyzed. Each sample was measured three times. The PdI was calculated based on the distribution of particles.
Zeta potential
The surface charge of the QU-nanoparticles was determined by electrophoretic light scattering with a Zetasizer Nano-ZS90 (Malvern, UK). 300 μl of nanoparticle dispersion were diluted with 50 ml of double distilled water. Measurements were carried out in triplicate.
Encapsulation efficiency
The entrapped QU was assayed by HPLC analysis. 1 ml of QU-nanoparticle dispersion was dissolved in 5 ml of methanol. The obtained solution was further diluted with a solution of acetonitrile:water 75:25 (v/v) in a ratio of 1:1. The samples were then centrifuged at 10000 rpm for 5 minutes. Subsequently, the separated supernatant was introduced in vials for HPLC analysis. The encapsulation efficiency (%EE) was calculated from the ratio of the amount of entrapped QU (Cnp) to that initially added (Ctot), according to the equation: %EE = Cnp/Ctot*100
The quantitative analysis of QU from nanoparticles was performed using an Agilent 1100 series chromatographic system (Agilent Technologies, USA) equipped with a pump, an autosampler and a UV-Vis detector. All measurements were carried out at 25°C using a reverse-phase Gemini C18 column (3 μm). A mixture of acetonitrile and phosphoric acid 0.1% (v/v) 30:70 was used as mobile phase, flowing at a rate of 0.6 ml/min. The injection volume of samples was set at 5 μl and the detection was performed at 370 nm wave length. Data were collected and analyzed using an Agilent ChemStation Software.
Results
Preparation and characterization of QU-nanoparticles
The experimental results concerning particle size, PdI, zeta potential and encapsulation efficiency from all experiments are given in Table III.
Table III.
Results for particle size (Y1), PdI (Y2), zeta potential (Y3) and encapsulation efficiency (Y4).
| Experiment number | Experiment name | Y1 | Y2 | Y3 | Y4 |
|---|---|---|---|---|---|
| 1 | N1 | 135.3±0.472 | 0.100±0.022 | −15.5±0.850 | 26.487 |
| 2 | N2 | 224.8±1.637 | 0.077±0.027 | −27.5±0.721 | 38.577 |
| 3 | N3 | 127.2±0.850 | 0.126±0.007 | −14.6±1.835 | 2.669 |
| 4 | N4 | 257.2±2.662 | 0.086±0.015 | −21.1±0.692 | 13.763 |
| 5 | N5 | 130.9±1.365 | 0.127±0.012 | −14.2±1.069 | 5.100 |
| 6 | N6 | 246.8±2.936 | 0.062±0.021 | −22.0±0.305 | 17.191 |
| 7 | N7 | 122.3±1.400 | 0.115±0.024 | −24.4±4.759 | 5.553 |
| 8 | N8 | 230.1±0.709 | 0.053±0.027 | −26.1±0.900 | 20.854 |
| 9 | N9 | 199.1±0.781 | 0.072±0.016 | −24.0±0.550 | 28.171 |
| 10 | N10 | 228.5±3.426 | 0.074±0.021 | −17.6±0.300 | 8.005 |
| 11 | N11 | 181.1±3.204 | 0.086±0.017 | −23.9±1.514 | 31.210 |
| 12 | N12 | 206.3±1.137 | 0.076±0.023 | −17.8±0.458 | 8.111 |
| 13 | N13 | 196.2±0.321 | 0.072±0.018 | −18.9±0.781 | 13.673 |
| 14 | N14 | 202.7±2.154 | 0.096±0.032 | −19.9±0.346 | 13.203 |
| 15 | N15 | 196.9±0.608 | 0.068±0.009 | −15.8±0.230 | 13.808 |
Data are shown as mean ± standard deviation
Experimental design analysis. Fitting the model
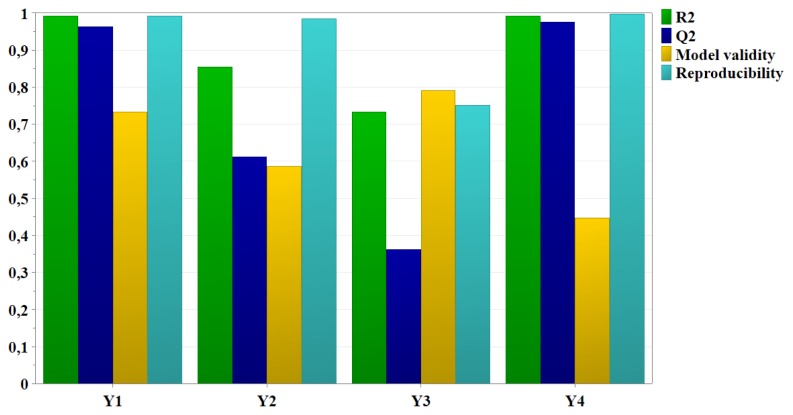
In order to fit the experimental data to the desirable model and to check the validity of the experimental design, R2 and Q2 were calculated (Figure 1) and analysis of variance (ANOVA) was performed. The fitted model is considered adequate if the model is significant (p<0.05) and the lack of fit is not significant (p>0.05).
Figure 1.
Summary of fit for the experimental data.
Experimental design analysis. Regression coefficients analysis
The regression coefficients and their influence on each of the three responses are presented as histograms. A positive value of the regression coefficient indicates a positive effect on the response, while a negative value suggests an inverse relation between the formulation factor and the response [33,34].
To illustrate the influence of the formulation factors on the responses, three-dimensional response surface curves were plotted. The plots were constructed based on the polynomial equations, assessing change in the response surface [34]. These surface plots were used to describe the interaction of two independent variables on the response at one time, while keeping the third variable constant, at its middle level [35]. The regression coefficients and surface plots for each response are shown in Figures 2–5.
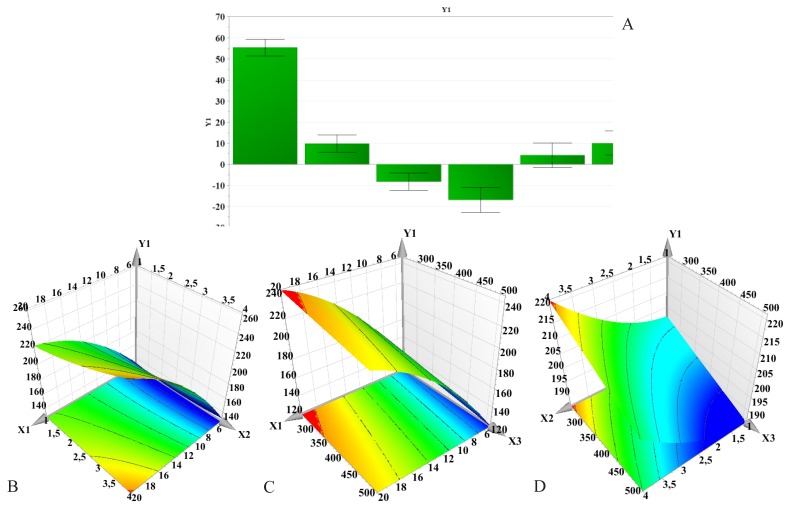
Figure 2.
Regression coefficients (A) and three-dimensional response surface plots showing the effect of formulation factors on particle size (Y1): B – PLGA concentration and PVA concentration effect; C – PLGA concentration and stirring speed effect; D – PVA concentration and stirring speed effect; X1 – PLGA concentration; X2 – PVA concentration; X3 – Stirring speed.
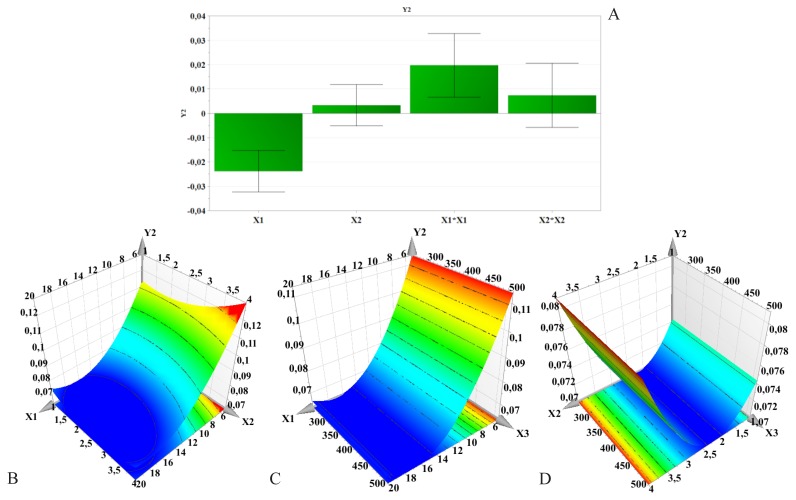
Figure 3.
Regression coefficients (A) and three-dimensional response surface plots showing the effect of formulation factors on PdI (Y2): B – PLGA concentration and PVA concentration effect; C – PLGA concentration and stirring speed effect; D – PVA concentration and stirring speed effect; X1 – PLGA concentration; X2 – PVA concentration; X3 – Stirring speed.
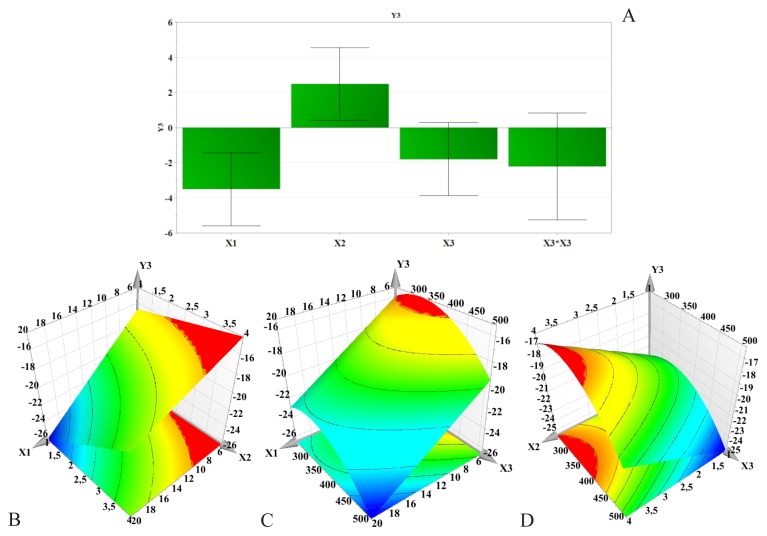
Figure 4.
Regression coefficients (A) and three-dimensional response surface plots showing the effect of formulation factors on zeta potential (Y3): B – PLGA concentration and PVA concentration effect; C – PLGA concentration and stirring speed effect; D – PVA concentration and stirring speed effect; X1 – PLGA concentration; X2 – PVA concentration; X3 – Stirring speed.
Figure 5.
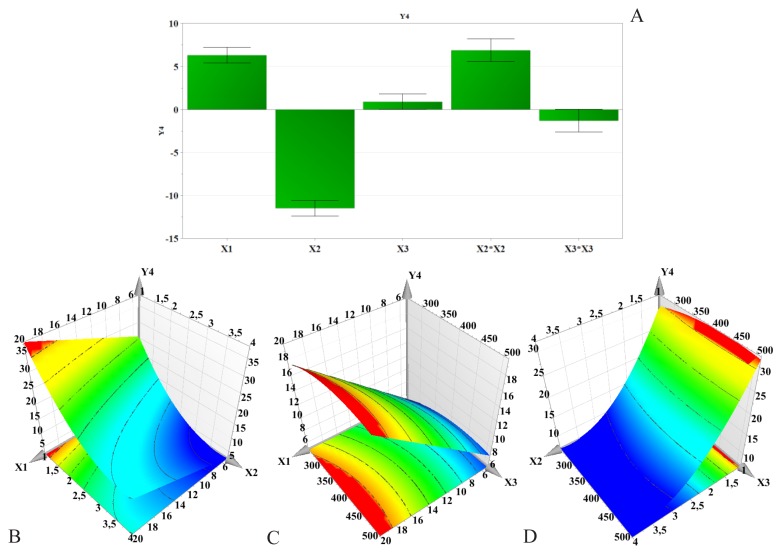
Regression coefficients (A) and three-dimensional response surface plots showing the effect of formulation factors on encapsulation efficiency (Y4): B – PLGA concentration and PVA concentration effect; C – PLGA concentration and stirring speed effect; D – PVA concentration and stirring speed effect; X1 – PLGA concentration; X2 – PVA concentration; X3 – Stirring speed.
Influence of formulation factors on particle size (Y1)
The particle size varied from 122.3±1.400 nm to 257.2±2.662 nm.
According to Figure 2, the PLGA concentration had a significant and positive influence on particle size. Results show that the particle size increased as the amount of polymer increased. The same effect was observed with increasing PVA concentration, but increase in particle size was not as pronounced. In contrast to these findings, the homogenization speed had an opposite effect. Increasing the stirring speed resulted in the formation of smaller particles. As seen in Figures 2, there is an interaction between PLGA concentration (X1) and PVA concentration (X2), which had a somewhat significant effect on particle size. An increase in PLGA and PVA concentrations led to a non-linear increase in particle size when the stirring speed was kept constant.
Influence of formulation factors on PdI (Y2)
Polydispersity indices were low and showed little variability between different samples, ranging from 0.053±0.027 to 0.127±0.012.
The response surfaces for PdI are given in Figure 3. PdI first decreased with increasing PLGA concentration, but higher polymer amounts led to a slight increase of PdI. Also, PVA concentration exhibited a negative influence on PdI.
Influence of formulation factors on zeta potential (Y3)
For all samples, the electric charge was negative, which could be due to the ionized terminal carboxylic groups of PLGA present on the surface of the nanoparticles. The zeta potential ranged between −27.5±0.721 mV and −14.2±1.069 mV, hence, according to literature, all samples are considered to have a poor stability.
Figure 4 reveals the effect of PLGA concentration and PVA concentration on the zeta potential. According to this figure, PLGA concentration seemed to be the main factor influencing the zeta potential. Increase in PLGA concentration led to a decrease of the surface charge. On the contrary, PVA increased zeta potential. The homogenization speed had a rather weak influence on the zeta potential. However, zeta potential absolute values increased with increasing stirring speed.
Influence of formulation factors on encapsulation efficiency (Y4)
Encapsulation efficiency varied on a wide range from a minimum of 2.67% to a maximum of 38.58%.
As illustrated in Figure 5, a positive relationship could be observed between PLGA concentration and the encapsulation efficiency. That is, by increasing PLGA concentration, higher encapsulation efficiency values were obtained. In contrast, the encapsulation efficiency dramatically decreased when PVA concentration increased. The stirring speed had little influence on the encapsulation efficiency, slightly enhancing it.
Optimization
To evaluate the predictive power of the model, QU-nanoparticles were prepared under the optimum conditions suggested by the software. Based on these conditions, the software predicted certain response ranges with target values. The predicted and actual experimental values for the responses are given in Table IV.
Table IV.
Predicted and experimental values of the QU-nanoparticles’ characteristics.
| Response | Target | Predicted value Lower limit | Upper Limit | Experimental value | Bias (%) |
|---|---|---|---|---|---|
| Y1 Particle size (nm) | 214.90 | 207.17 | 222.63 | 222.70 | +3.62 |
| Y2 PdI | 0.067 | 0.054 | 0.081 | 0.065 | −2.98 |
| Y3 Zeta potential (mV) | −24.95 | −28.28 | −21.62 | −25.30 | −1.40 |
| Y3 Encapsulation efficiency (%) | 36.55 | 34.99 | 38.11 | 35.44 | −3.03 |
Discussion
Experimental design analysis. Fitting the model
R2 and Q2 provide the best information on fitting the model. The model validity indicates if the model is appropriate and if the right type of model was chosen from the beginning. Reproducibility reflects a summary of variability [31,36,37]. According to Figure 1, all responses, particle size, PdI, zeta potential and encapsulation efficiency, are well fitted and predicted by the model as R2 has high values. Except for the zeta potential response, Q2 values are above 0.5. Model validity and reproducibility are greater than 0.25 and 0.5, respectively, for each of the four responses. The overall results show that the relationship between the formulation factors and responses was well described by the chosen model, thus indicating a good and valid model with good predictive power.
The analysis of variance (ANOVA) indicates if the variance in the results is due to variations in the formulation factors or is determined by experimental errors [27]. In ANOVA, one of the two F-tests assesses the significance of the regression model, and when p<0.05, the values are considered significant and the test is satisfied. The lack of fit test compares the model error and the replicate error. The lower the model error, then the model shows good fit to the experimental data and has no lack of error. This test is satisfied when p>0.05, therefore the values are considered not significant [36]. p-values for the model were lower than 0.05, while those for lack of fit were greater than 0.05, therefore the model represented the data accurately.
Influence of formulation factors on particle size (Y1)
Particle size is a critical feature for nanoscaled drug delivery systems as it influences the circulating half-life, cellular uptake and biodistribution [38]. As cellular uptake is size dependent, smaller particles could be taken up to a greater extent than bigger particles [39]. The drug’s release kinetics also depends on particle size. Usually, the smaller the particle size, the faster the release rate [32].
The effect of PLGA concentration on particle size can be explained by taking into consideration the viscosity of the organic phase, as well as the number of polymer chains per unit volume of organic solvent [40]. By increasing PLGA concentration, the viscosity of the organic solution increased. A higher viscosity implies a lower net shear stress, therefore leading to the formation of larger size droplets. Furthermore, as a result of increased viscosity, the diffusion of the organic solvent into the aqueous phase is slowed down, causing larger droplets to form, which in turn render larger nanoparticles [5]. On the other hand, higher PLGA concentration favors polymer-polymer interactions, thus more polymer chains remain associated during the solvent’s diffusion into the aqueous medium [40].
PVA can be oriented at the interface between the organic solution and the aqueous medium, thereby reducing the interfacial tension and thus increasing the net shear stress. This in fact would promote the formation of small size particles. Still, by increasing PVA concentration, the viscosity of the aqueous phase increased, hence, as a result of decreased shear stress, the particles’ mean diameter increased [41]. On the other hand, some studies suggest that higher concentrations of PVA promote the coalescence of particles, leading to larger size nanoparticles [4]. The literature reports that a fraction of PVA remains associated with the nanoparticles as it forms an interconnected network with PLGA at the surface [42]. The suggested mechanism involves the interpenetration of PVA and PLGA molecules during nanoparticle formation, particularly during the organic solvent evaporation. Upon entering the organic solution, the hydrophobic segments of PVA remain entrapped in the polymeric matrix [1]. Thus, at higher concentrations, residual PVA could further contribute to increase in particle size. Although these results are contradictory to those reported by most authors [1,39,41], they are in accordance with previous findings published by other groups [4,43].
Results showed that smaller particles were formed when increasing the homogenization speed. This is in agreement with the findings of Kheradmandnia et al and Mehrotra et al [4,44]. As it influences the viscosity of the dispersion, the greater the stirring speed, the lower the generated net shear stress [6]. At the same time, it promotes a rapid diffusion of the organic solvent in the aqueous phase [5].
Influence of formulation factors on PdI (Y2)
PdI is an important property which is used to describe variation of particle size in a population of particles. Most frequently, the size of a population of particles follows a multimodal distribution. When the PdI value is close to 1, the size range is wide. Generally, a value closer to 0 is desired [32].
The PLGA concentration seems to be the most important factor influencing PdI. It could be said that a greater amount of polymer would promote the formation of much more homogenous nanoparticle samples. On the other hand, the homogenization speed had no effect whatsoever on PdI.
Influence of formulation factors on zeta potential (Y3)
The electrostatic potential also referred to as zeta potential is a key feature which offers important information on the stability of colloidal dispersions [45]. It is created by the electric charge present on the nanoparticles’ surface. Nanoparticles having a zeta potential ranging from −10 mV to +10 mV are considered fairly neutral [5]. On the other hand, a zeta potential lower than −30 mV or higher than +30 mV is an indicator of a very stable dispersion [46]. At higher zeta potential values, the repulsive forces between similarly charged particles prevent their aggregation and increase their stability [39].
Different studies suggest that the surface charge of PLGA nanoparticles without any PVA is approximately −45 mV. As mentioned above, this is attributed to the carboxylic end groups of the polymer. PVA is considered a non-ionic stabilizer which forms a protective layer around the nanoparticles, and despite repeated washing it cannot be completely removed from the surface of the particles [3,6]. The less negative zeta potential values seen with increasing PVA concentration are considered to be due to the fact that the PVA coating of the nanoparticles shields the surface charge of PLGA, which is in accordance with the results reported by Sahoo et al [1].
It seems that high stirring speed favors the nanoparticles’ stability. High homogenization speed promotes the formation of small nanoparticles. At the same time, particle size varies inversely with the surface charge.
Influence of formulation factors on encapsulation efficiency (Y4)
A higher entrapment efficiency could be attained by increasing PLGA concentration. This could be due to the fact that higher amounts of polymer led to more viscous organic solutions. One the one hand, as mentioned earlier in this paper, the higher the viscosity, the lower the net shear stress, which would result in larger size particles. Larger nanoparticles provide sufficient surface for QU molecules to be entrapped. On the other hand, an increased viscosity could hinder the drug’s diffusion from the organic phase into the aqueous one, and therefore promote QU’s entrapment [41].
It is suggested that high concentrations of PVA enhance QU’s water solubility [41]. Hence, more drug molecules would pass into the aqueous phase, leaving less QU to be entrapped in the polymeric nanoparticles.
When increasing the homogenization speed, the amount of QU entrapped slightly increased as well. Higher stirring speed causes smaller droplets to form, thus the total surface area of the nanoparticles to increase. This provided additional space for the polymer matrix to accommodate more QU molecules, thereby to improve encapsulation efficiency. These results were similar to those reported by Sapre et al and Narayanan et al [43,47].
Optimization
After establishing the polynomial equations which describe the relationship between the formulation factors and the responses, the optimization process was carried out.
Of the four responses, size and encapsulation efficiency are critical properties of nanoparticles. Theoretically, a minimum particle size and maximum entrapment efficiency are desirable. Therefore, the following criteria were adopted: the particle size (Y1) was minimized, the encapsulation efficiency was maximized (Y4), while PdI (Y2) and zeta potential (Y3) were excluded. The optimum levels of the formulation factors were: a concentration of 18 mg/ml PLGA, a concentration of 1% (%m/v) PVA and a stirring speed of 425 rpm.
The observed response values were comparable to the predicted ones, with low percentage bias (±5%), thus suggesting the optimized formulation is trustworthy and that the model’s prediction ability is quite good.
Conclusions
A nanoprecipitation technique has been successfully employed in this study to obtain QU-loaded polymeric nanoparticles with desirable size and high drug loading. It aimed at evaluating the influence of three formulation factors on the particle size, PdI, zeta potential and encapsulation efficiency of QU-nanoparticles. In this sense, a Box-Behnken experimental design was constructed to study the effects of the variables and to optimize the manufacturing process conditions.
According to the results, the PLGA concentration had a significant effect on all the studied responses, particularly on nanoparticle size. The encapsulation efficiency was mainly influenced by PVA concentration. Based on these findings, an optimized formulation was determined and prepared. Data indicated that a higher amount of PLGA together with a lower PVA concentration and a greater stirring speed were the optimal conditions for the preparation of QU-nanoparticles. The observed response values and the predicted ones were in agreement, therefore confirming the statistical significance of the model and its precision in predicting the optimum preparation conditions for QU-loaded nanoparticles.
In conclusion, a Box-Behnken experimental design was successfully used in order to obtain QU-nanoparticles with optimized characteristics.
References
- 1.Sahoo SK, Panyam J, Prabha S, Labhasetwar V. Residual polyvinyl alcohol associated with poly (D,L-lactide-co-glycolide) nanoparticles affects their physical properties and cellular uptake. J Control Release. 2002;82:105–114. doi: 10.1016/s0168-3659(02)00127-x. [DOI] [PubMed] [Google Scholar]
- 2.Ghasemian E, Vatanara A, Najafabadi AR, Rouini MR, Gilani K, Darabi M. Preparation, characterization and optimization of sildenafil citate loaded PLGA nanoparticles by statistical factorial design. Daru. 2013;21(1):68. doi: 10.1186/2008-2231-21-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sengel Türk CT, Sezgin Bayindir Z, Badilli U. Preparation of polymeric nanoparticles using different stabilizing agents. J Fac Pharm Ankara. 2009;38(4):257–268. [Google Scholar]
- 4.Mehrotra A, Pandit JK. Critical Process Parameters Evaluation of Modified Nanoprecipitation Method on Lomustine Nanoparticles and Cytostatic Activity Study on L132 Human Cancer Cell Line. J Nanomed Nanotechnol. 2012;3:8. [Google Scholar]
- 5.dos Santos KC, da Silva MFGF, Pereira-Filho ER, Fernandes JB, Polikarpov I, Forim MR. Polymeric nanoparticles loaded with the 3,5,3′-triiodothyroacetic acid (Triac), a thyroid hormone: factorial design, characterization, and release kinetics. Nanotechnol Sci Appl. 2012;5:37–48. doi: 10.2147/NSA.S32837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao H, Gagnon J, Häfeli UO. Process and formulation variables in the preparation of injectable and biodegradable magnetic microspheres. Biomagn Res Technol. 2007;5:2. doi: 10.1186/1477-044X-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali H, Kalashnikova I, White MA, Sherman M, Rytting E. Preparation, characterization, and transport of dexamethasone-loaded polymeric nanoparticles across a human placental in vitro model. Int J Pharm. 2013;454(1):149–157. doi: 10.1016/j.ijpharm.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feczkó T, Tóth J, Dósa G, Gyenis J. Optimization of protein encapsulation in PLGA nanoparticles. Chem Eng Prog. 2011;50:757–765. [Google Scholar]
- 9.Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Préat V. PLGA-based nanoparticles: An overview of biomedical applications. J Control Release. 2012;161:505–522. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- 10.Achim M. Micro şi nanoparticule utilizate în terapia la ţintă. Cluj-Napoca: Editura Medicală Universitară „Iuliu Haţieganu”; 2010. [Google Scholar]
- 11.Makadia HK, Siegel SJ. Poly Lactic-co-Glycolic Acid (PLGA) as Biodegradable Controlled Drug Delivery Carrier. Polymers (Basel) 2011;3(3):1377–1397. doi: 10.3390/polym3031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larson AJ, Symons JD, Jalili T. Therapeutic potential of quercetin to decrease blood pressure: review of efficacy and mechanisms. Adv Nutr. 2012;3:39–46. doi: 10.3945/an.111.001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molina MF, Sanchez-Reus I, Iglesias I, Benedi J. Quercetin, a flavonoid antioxidant, prevents and protects against ethanol-induced oxidative stress in mouse liver. Biol Pharm Bull. 2003;26(10):1398–1402. doi: 10.1248/bpb.26.1398. [DOI] [PubMed] [Google Scholar]
- 14.Kumar VD, Verma PRP, Singh SK. Development and evaluation of biodegradable polymeric nanoparticles for the effective delivery of quercetin using a quality by design approach. LWT-Food Sci Technol. 2015;61:330–338. [Google Scholar]
- 15.Shaji J, Iyer S. Novel Double Loaded Quercetin Liposomes: Evidence of Superior Therapeutic Potency Against CCl4 Induced Hepatotoxicity – A Comparative Study. Asian J Pharm Clin Res. 2012;5(2):104–108. [Google Scholar]
- 16.Nday CM, Halevas E, Jackson GE, Salifoglou A. Quercetin encapsulation in modified silica nanoparticles: potential use against Cu(II)-induced oxidative stress in neurodegeneration. J Inorg Biochem. 2015;145:51–64. doi: 10.1016/j.jinorgbio.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Gibellini L, Pinti M, Nasi M, Montagna JP, De Biasi S, Roat E, et al. Quercetin and cancer chemoprevention. Evid Based Complement Alternat Med. 2011;2011:591356. doi: 10.1093/ecam/neq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suntres ZE. Liposomal Antioxidants for Protection against Oxidant-Induced Damage. J Toxicol. 2011;2011:152474. doi: 10.1155/2011/152474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumari A, Yadav SK, Pakade YB, Singh B, Yadav SC. Development of biodegradable nanoparticles for delivery of quercetin. Colloids Surf B Biointerfaces. 2010;80:184–192. doi: 10.1016/j.colsurfb.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 20.Mignet N, Seguin J, Chabot GG. Bioavailability of polyphenol liposomes: a challenge ahead. Pharmaceutics. 2013;5:457–471. doi: 10.3390/pharmaceutics5030457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landi-Librandi AP, Chrysostomo TN, Azzolini AECS, Marzocchi-Machado CM, de Oliveira CA, Lucisano-Valim YM. Study of quercetin-loaded liposomes as potential drug carriers: in vitro evaluation of human complement activation. J Liposome Res. 2012;22(2):89–99. doi: 10.3109/08982104.2011.615321. [DOI] [PubMed] [Google Scholar]
- 22.Morales-Cruz M, Flores-Fernández GM, Morales-Cruz M, Orellano EA, Rodriguez-Martinez JA, Ruiz M, et al. Two-step nanoprecipitation for the production of protein-loaded PLGA nanospheres. Results Pharma Sci. 2012;2:79–85. doi: 10.1016/j.rinphs.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fessi H, Puisieux F, Devissaguet JP, Ammoury N, Benita S. Nanocapsule formation by interfacial polymer deposition following solvent displacement. Int J Pharm. 1989;55(1):R1–R4. [Google Scholar]
- 24.Rao JP, Geckeler KE. Polymer nanoparticles: Preparation techniques and size-control parameters. Prog Polym Sci. 2011;36:887–913. [Google Scholar]
- 25.Pinto Reis CP, Neufeld RJ, Ribeiro AJ, Veiga F. Nanoencapsulation I. Methods for preparation of drug-loaded polymeric nanoparticles. Nanomedicine. 2006;2:8–21. doi: 10.1016/j.nano.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Porfire AS, Tomuţă I, Leucuţa SE, Achim M. Superoxide dismutase loaded liposomes. The influence of formulation factors on enzyme encapsulation and release. Farmacia. 2013;61(5):865–873. [Google Scholar]
- 27.Jain A, Jain SK. Formulation and optimization of temozolomide nanoparticles by 3 factor 2 level factorial design. Biomatter. 2013;3(2):e25102-1–e25102-13. doi: 10.4161/biom.25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez-Rodriguez ML, Barros LB, Palma J, Gonzalez-Rodriguez PL, Rabasco AM. Application of statistical experimental design to study the formulation variables influencing the coating process of lidocaine liposomes. Int J Pharm. 2007;337:336–345. doi: 10.1016/j.ijpharm.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 29.Luo X, Guan R, Chen X, Tao M, Ma J, Zhao J. Optimization on condition of epigallocatechin-3-gallate (EGCG) nanoliposomes by response surface methodology and cellular uptake studies in Caco-2 cells. Nanoscale Res Lett. 2014;9(1):291. doi: 10.1186/1556-276X-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leucuţa SE, Tomuţă I. Planuri experimentale şi optimizarea formulării medicamentelor. Cluj-Napoca: Editura Risoprint; 2011. [Google Scholar]
- 31.Ranjan AP, Mukerjee A, Helson L, Vishwanatha JK. Scale up, optimization and stability analysis of Curcumin C3 complex-loaded nanoparticles for cancer therapy. J Nanobiotechnology. 2012;10:38. doi: 10.1186/1477-3155-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C, Gu C, Peng F, Liu W, Wan J, Xu H, Lam CW, Yang X. Preparation and Optimization of Triptolide-Loaded Solid Lipid Nanoparticles for Oral Delivery with Reduced Gastric Irritation. Molecules. 2013;18:13340–13356. doi: 10.3390/molecules181113340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao J, Fang X, Zhou Y, Wang J, Guo F, Li F, Peng X. Development and optimization of solid lipid nanoparticle formulation for ophthalmic deliver of chloramphenicol using Box-Behnken design. Int J Nanomedicine. 2011;6:683–692. doi: 10.2147/IJN.S17386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varshosaz J, Ghaffari S, Khoshayand MR, Atyabi F, Azarmi S, Kobarfard F. Development and optimization of solid lipid nanoparticles of amikacin by central composite design. J Liposome Res. 2010;20(2):97–104. doi: 10.3109/08982100903103904. [DOI] [PubMed] [Google Scholar]
- 35.Eriksson L, Johansson E, Kettaneh-Wold N, Wikström C, Wold S. Design of Experiments. Principles and Applications. 3rd ed. Umeå: MKS Umetrics AB; 2008. [Google Scholar]
- 36.Muzyka K, Karim K, Guerreiro A, Poma A, Piletsky S. Optimisation of the synthesis of vancomycin-selective molecularly imprinted polymer nanoparticles using automatic photoreactor. Nanoscale Res Lett. 2019;(1):154. doi: 10.1186/1556-276X-9-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie H, Smith JW. Fabrication of PLGA nanoparticles with a fluidic nanoprecipitation system. J Nanobiotechnology. 2010;8:18. doi: 10.1186/1477-3155-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shah U, Joshi G, Sawant K. Improvement in antihypertensive antianginal effects of felodipine by enhanced absorption from PLGA nanoparticles optimized by factorial design. Mater Sci Eng C Mater Biol Appl. 2014;35:153–163. doi: 10.1016/j.msec.2013.10.038. [DOI] [PubMed] [Google Scholar]
- 39.Galindo-Rodriguez S, Allémann E, Fessi H, Doelker E. Physicochemical parameters associated with nanoparticle formation in the salting-out, emulsification-diffusion, and nanoprecipitation methods. Pharmaceut Res. 2004;21(8):1428–1439. doi: 10.1023/b:pham.0000036917.75634.be. [DOI] [PubMed] [Google Scholar]
- 40.Song X, Zhao Y, Hou S, Xu F, Zhao R, He J, Cai Z, Li Y, Chen Q. Dual agents loaded PLGA nanoparticles: Systematic study of particle size and drug entrapment efficiency. Eur J Pharm Biopharm. 2008;69:445–453. doi: 10.1016/j.ejpb.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 41.Kumar MNVR, Bakowsky U, Lehr CM. Preparation and characterization of cationic PLGA nanospheres as DNA carriers. Biomaterials. 2004;25:1771–1777. doi: 10.1016/j.biomaterials.2003.08.069. [DOI] [PubMed] [Google Scholar]
- 42.Narayanan K, Subrahmanyam VM, Rao JV. A Fractional Factorial Design to Study the Effect of Process Variables on the Preparation of Hyaluronidase Loaded PLGA Nanoparticles. Enzyme Res. 2014;2014:162962. doi: 10.1155/2014/162962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kheradmandnia S, Vasheghani-Farahani E, Nosrati M, Atyabi F. The Effect of Process Variables on the Properties of Ketoprofen Loaded Solid Lipid Nanoparticles of Beeswax and Carnauba Wax. Iran J Chem Chem Eng. 2010;29(4):181–187. [Google Scholar]
- 44.Shah R, Eldridge D, Palombo E, Harding I. Optimisation and Stability of Solid Lipid Nanoparticls using Particle Size and Zeta Potential. Journal of Physical Science. 2014;25(1):59–75. [Google Scholar]
- 45.Lasoń E, Sikora E, Ogonowski J. Influence of process parameters on properties of Nanostructured Lipid Carriers (NLC) formulation. Acta Biochim Pol. 2013;60(4):773–777. [PubMed] [Google Scholar]
- 46.Sapre AS, Parikh RK. Design of a buccal mucoadhesive, nanoparticles based delivery system of fluoxetine. JPSBR. 2012;2(3):148–161. [Google Scholar]