Abstract
The mammalian MST kinase family, which is related to the Hippo kinase in Drosophila melanogaster, includes five related proteins: MST1 (also called STK4), MST2 (also called STK3), MST3 (also called STK24), MST4, and YSK1 (also called STK25 or SOK1). MST kinases are emerging as key signaling molecules that influence cell proliferation, organ size, cell migration, and cell polarity. Here we review the regulation and function of these kinases in normal physiology and pathologies, including cancer, endothelial malformations, and autoimmune disease.
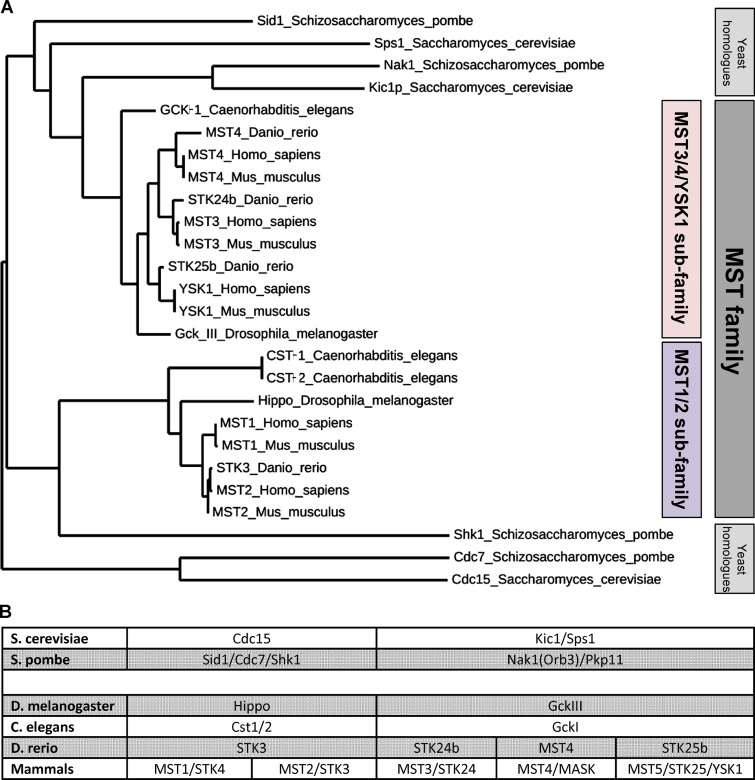
The human kinome features a large branch of the so-called “STE” kinases, named after the yeast Sterile20 kinase. The STE superfamily includes several subfamilies, only one of which is named the “Mammalian Sterile20-like” (MST) family (Creasy and Chernoff, 1995). There are five MST kinases in mammals, and, despite their name, this kinase family is conserved in all metazoans and has homologues in fungi. The five mammalian MST kinases can be broadly divided into two subgroups: MST1 and -2, and MST3/4/YSK1. Representatives of these two subgroups are clearly identifiable in all metazoans, but the homology relationships with yeast kinases are less clear cut. The somewhat confusing nomenclature of these kinases is summarized in Fig. 1. Further confusion can arise from the fact that several other subfamilies of the STE kinases are more closely related to yeast Sterile20 than the MST family itself, for example the PAK family (Fig. S1). In this review, we will use the following nomenclature for the mammalian kinases because it reflects the most common usage: MST1 (STK4), MST2 (STK3), MST3 (STK24), MST4 (STK26), and YSK1 (STK25).
Figure 1.
The MST kinase family. (A) Dendrogram showing the relationship between MST kinases in different model organisms. (B) Table showing the nomenclature of MST kinases in different model organisms. The blank row between the yeast and metazoan genes indicates the imprecise relationship of the kinases across this large evolutionary distance.
Despite millions of years of evolutionary divergence, the functions of MST family kinases are remarkably similar across eukaryotes, with conserved roles in the control of cell polarity and/or the cell division cycle. Another common theme is their regulation by cell architecture and interactions with PP2A complexes, and their regulation of cell and tissue homeostasis. Understanding the function and regulation of these kinases is important given that perturbations in MST kinases are implicated in numerous diseases.
Identification of MST kinases in yeast
Genomic analysis reveals several homologues of metazoan MST kinases in unicellular yeasts. A common theme with these kinases is their role in signal transduction pathways that help control progression of the cell cycle as well as cellular polarity and morphogenesis. Sequence analysis alone does not match the mammalian MST kinases unambiguously to yeast orthologues; however, if sequence and function data are combined then Cdc15 and Sid1 can be considered the kinases most similar to MST1/2, whereas Kic1 and Nak1 are more similar to MST3/4/YSK1 in Saccharomyces cerevisiae and Schizosaccharomyces pombe, respectively. Here we review their discovery, regulation, and function.
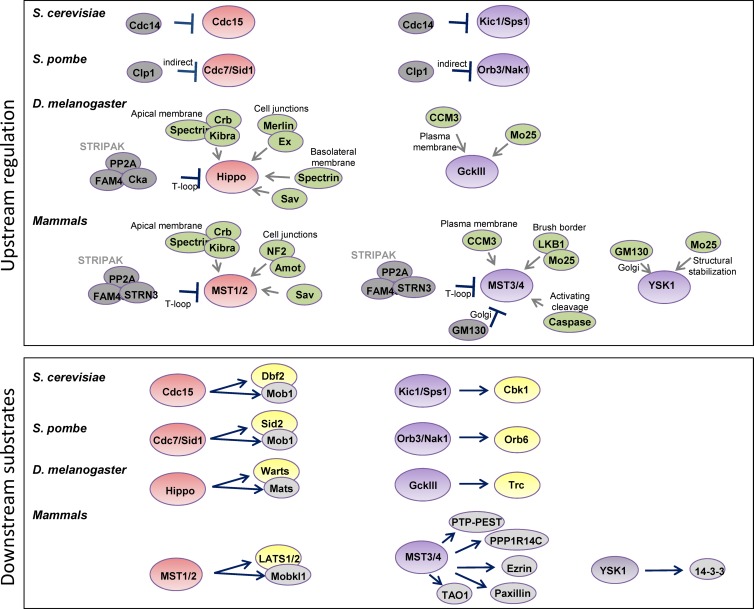
The first MST family kinase to be characterized was Cdc15 in S. cerevisiae (Hartwell et al., 1973; Pringle and Hartwell, 1981; Schweitzer and Philippsen, 1991), which is most similar to MST1/2. Mutants in the Cdc15 gene cause yeast cells to arrest in late mitosis (telophase), unable to complete cytokinesis (Pringle and Hartwell, 1981; Surana et al., 1993; Jaspersen et al., 1998). Cdc15 acts by phosphorylating the NDR/LATS (Nuclear Dbf2-Related/Large Tumour Suppressor)-like kinase Dbf2 (Fig. 2; Xu et al., 2000; Lee et al., 2001b; Mah et al., 2001; Visintin and Amon, 2001; Rock and Amon, 2011). The key downstream effector of Dbf2 is the phosphatase Cdc14, which inactivates the mitotic kinase Cdk1 and allows exit from mitosis and completion of cytokinesis (Surana et al., 1993; Jaspersen et al., 1998). This signaling pathway is named the “MEN,” for “Mitotic Exit Network” (Tóth et al., 2007; for reviews see Bardin and Amon, 2001; Segal, 2011). A similar regulatory network exists in fission yeast: Sid1 and Cdc7, which are similar to the mammalian MST1/2 kinases, are required for the activity of the NDR/LATS family kinase Sid2 to initiate septum formation in cytokinesis (Fig. 2 and Fig. S1 B; Nurse et al., 1976; Sparks et al., 1999). Loss-of-function mutants in the cdc7 or sid2 genes lead to elongated cells with multiple nuclei, due to septation initiation defects (Nurse et al., 1976; Sparks et al., 1999; Hou et al., 2000; Wachowicz et al., 2015). This signaling network was named “SIN,” for “Septation Initiation Network” (for reviews see Bardin and Amon, 2001; Krapp et al., 2004; Krapp and Simanis, 2008).
Figure 2.
Regulation and substrates of MST kinases. (Top) Regulatory inputs into MST kinases in yeast, Drosophila, and mammals. (Bottom) Downstream substrates in yeast, Drosophila, and mammals. MST1/2 kinases are in pink, MST3/4/YSK1 kinases are shown in purple, negative regulators in gray, positive regulators in green, Ndr-related kinase substrates in yellow, and nonkinase substrates in light gray.
Kic1, which is most homologous to the mammalian MST3/4 kinases, phosphorylates the NDR/LATS-like kinase Cbk1 to regulate polarized cell growth and the separation of mother and daughter cells in budding yeast (Fig. 2; Sullivan et al., 1998; Bidlingmaier et al., 2001; Colman-Lerner et al., 2001; Weiss et al., 2002). Loss of either Kic1 or Cbk1 leads to a failure of the F-actin cytoskeleton to polarize, and cells fail to separate, growing as large clusters (Colman-Lerner et al., 2001; Weiss et al., 2002; Nelson et al., 2003). GFP-tagged Kic1 and Cbk1 proteins also localize in a polarized fashion during budding, concentrating at the bud neck during mitosis. In addition to regulating actin polarization, Kic1 and Cbk1 also regulate the Ace2 transcription factor, which controls the daughter cell–specific expression of cell separation genes (Colman-Lerner et al., 2001; Weiss et al., 2002; Nelson et al., 2003; Mazanka et al., 2008).
In fission yeast, Orb3 (also called Nak1, see Fig. 1) is the kinase most similar to mammalian MST3/4. Orb3 is required for polarization of the actin cytoskeleton at the tips of S. pombe cells and for cell separation after cytokinesis (Verde et al., 1995; Leonhard and Nurse, 2005). Orb3 localizes to cell tips, the medial ring, and the spindle pole bodies at various points in the cell cycle (Leonhard and Nurse, 2005). Orb3 appears to act upstream of Orb6, another NDR/LATS-family kinase (Verde et al., 1995; Hou et al., 2003). Loss of function mutations in orb6 causes an orb3-like phenotype, with cells rounding up with unpolarized F-actin and arresting after two to four rounds of cell division. Orb6 also localizes to the growing cell tips and to the middle of dividing cells (Verde et al., 1995; Hou et al., 2003). The Mor2/Cps12 protein, homologous to Drosophila melanogaster Furry, is also involved in this pathway (Hirata et al., 2002). An actin-dependent positive feedback loop has been proposed to localize Orb3 (Leonhard and Nurse, 2005). This signaling network is referred to as the “Morphogenesis” or “MOR” network (Gupta et al., 2013, 2014); however, it might also be useful to think of it as the “Tip Actin Network” (TAN). Thus, the MST acronym could equally stand for MEN-SIN-TAN family kinases, to acknowledge the important contribution of yeast genetics to their discovery and their functional roles.
Metazoan MST kinases
Drosophila has two MST kinases; the MST1/2 homologue is the Hippo (Hpo) kinase, which was discovered in genetic screens for tumor suppressors in the fly eye. Mammals and other tetrapods have two Hpo homologues—MST1 and MST2—that, like Hpo, function to limit cell proliferation. Caenorhabditis elegans has two MST1/2 homologues—Cst-1 and Cst-2—and both Drosophila and C. elegans have one kinase homologous to vertebrate MST3, MST4, and YSK1, confusingly termed GckIII in Drosophila and GCK-I in C. elegans. In mammals, these kinases modulate the several signaling pathways and cellular organization, in particular cell polarity and the actin cytoskeleton. These functions echo the roles of yeast MST homologues in controlling septation and morphogenesis. In the following section we will review the function of these kinases in development and regeneration, and their regulation and effector pathways.
Metazoan MST kinases in development and homeostasis
Regulation of proliferation and tissue size
Loss of Hpo in Drosophila leads to tissue overgrowth in a range of tissues including the eye, wing imaginal disc, gut, and wing. Hippo acts with its cofactor Salvador (Sav) to phosphorylate a key regulatory amino acid in Warts, an NDR/LATS kinase (Kango-Singh et al., 2002; Tapon et al., 2002; Harvey et al., 2003; Pantalacci et al., 2003; Udan et al., 2003; Wu et al., 2003). Warts phosphorylates and inhibits the transcriptional coactivator Yorkie (Yki; homologue of mammalian YAP and TAZ) to repress cell proliferation and promote apoptosis (Huang et al., 2005). Phosphorylation of Yki inhibits its activity by promoting its association with cytoplasmic 14-3-3 proteins (Dong et al., 2007; Oh and Irvine, 2008, 2009). In the nucleus, Yki binds the TEAD-family DNA-binding transcription factor Scalloped (Sd), switching it from a repressor to an activator of transcription (Wu et al., 2008; Zhang et al., 2008; Koontz et al., 2013). Nuclear cofactors of Yki include Mask, Wbp2, Brahma, and possibly Hipk (Chan et al., 2011; Zhang et al., 2011; Chen and Verheyen, 2012; Poon et al., 2012; Jin et al., 2013; Sansores-Garcia et al., 2013; Sidor et al., 2013; Zhu et al., 2015). Important Yki transcriptional target genes include master regulators of proliferation, E2F and myc, and inhibitors of cell death, such as DIAP1. Regulation of these target genes enables the Hippo pathway to regulate cell proliferation and tissue size. Upstream components of the Hippo pathway, such as expanded, are also Yki targets and form part of a negative feedback loop (Hamaratoglu et al., 2006; Nolo et al., 2006; Thompson and Cohen, 2006; Goulev et al., 2008; Neto-Silva et al., 2010; Huang et al., 2014).
Both Hpo and its downstream signaling network are highly conserved in mammals. Both MST1 and -2 can phosphorylate and activate the NDR family kinases LATS1 and LATS2 (Chan et al., 2005; Fig. 2). When active, LATS1 and -2 phosphorylate two transcriptional regulators, YAP and TAZ (homologous to Drosophila Yki). This promotes YAP1 and TAZ interactions with 14-3-3 proteins and the degradation of YAP1 (Zhao et al., 2010); together these mechanisms lead to reduced interaction with TEAD1–4 transcription factors. Similar to Drosophila, some YAP and TAZ target genes are negative regulators of pathway activity (Moroishi et al., 2015). Conditional mouse knockouts for Mst1 and Mst2 revealed a conserved role for these kinases as Hippo pathway components and reinforced the view that YAP and TAZ are the major downstream mediators of MST1/2 function. In the embryo, knocking out both Mst1 and Mst2 caused early lethality. Single knockouts do not yield this phenotype, indicating that the two kinases act redundantly (Oh et al., 2009; Zhou et al., 2009; Song et al., 2010a). In the liver, Mst1/2 double conditional knockouts induced postnatally caused tissue overgrowth and tumor formation (Zhou et al., 2009; Lu et al., 2010; Song et al., 2010a). These liver phenotypes are similar to those of YAP overexpression, Sav knockout, Merlin knockout, or Sav/Merlin double knockout, indicating conservation of the Hippo pathway between Drosophila and mice (Dong et al., 2007; Lee et al., 2010; Lu et al., 2010; Zhang et al., 2010; Yin et al., 2013). In the intestine, Mst1/2 double conditional knockouts cause an expansion of the stem/progenitor cell compartment, again similar to Sav conditional knockouts or overexpression of Yap (Camargo et al., 2007; Cai et al., 2010; Zhou et al., 2011). In the skin, loss of Sav or overexpression of Yap drives proliferation of epithelial stem/progenitor cells (Lee et al., 2008; Schlegelmilch et al., 2011; Zhang et al., 2011). This reflects an emerging body of data showing that YAP and TAZ are key regulators of stem cells. While YAP and TAZ are cytoplasmic in the majority of epithelial cells in the skin or gut, they accumulate in the nucleus in stem cells.
Lats1/2 knockouts might be expected to have similar phenotypes to MST1/2 knockouts. Global mouse knockouts have revealed that Lats1/2 and Yap are involved in cell fate specification in the early mouse embryo. Lats1/2 are required to keep Yap inactive in the future inner cell mass, whereas active YAP helps to specify the trophectoderm (Nishioka et al., 2009; Cockburn et al., 2013; Hirate et al., 2013; Leung and Zernicka-Goetz, 2013). Combined MST1/2 deletion does not yield these phenotypes, suggesting that Lats1/2 may also be regulated by other means. Double conditional knockouts for Lats1/2 have not yet been reported. Interestingly, recent work suggests that the Trc-related kinases Ndr1/2 are essential downstream of Mst1/2 in the mouse intestine (Zhang et al., 2015). This finding is somewhat surprising, as Warts/Lats, rather than Trc, is primarily responsible for regulating Yki in Drosophila. Furthermore, the Lats1 single knockout mice do have tumorigenic phenotypes (St John et al., 1999). Thus, it will be interesting to compare the Lats1/2 and Ndr1/2 double conditional knockouts and the potential for redundancy between these four kinases downstream of MSTs.
Regulation of cell polarity and migration
In addition to regulating Yki, Hippo-Warts signaling can also control polarization of the F-actin cytoskeleton. Both epithelial cells and migrating border cell clusters mutant for hippo or warts up-regulate F-actin at the apical membrane domain (Fernández et al., 2011; Lucas et al., 2013). In border cells, the mislocalized F-actin cytoskeleton in hippo or warts mutants dramatically impairs the collective migration of these clusters (Lucas et al., 2013). This role for Hippo-Warts signaling does not require inhibition of Yki. Instead, the target of Hippo-Warts in polarizing F-actin in Drosophila is the Ena/Capping protein system (Lucas et al., 2013). This dual role of Hippo-Warts in regulating both Yki and F-actin polarization appears to reflect a similar duality in the functions of yeast MST-NDR/LATS kinases in regulating both cell cycle progression and F-actin polarization. Hippo also acts upstream of Warts and other NDR kinases to regulate patterning in the Drosophila nervous system (Zallen et al., 2000; Emoto et al., 2004, 2006). Similar to the border cell migration, this role may also be independent of Yki. MST1 also affects T cell migration and homing to lymph nodes (Katagiri et al., 2009); this may be linked to defects in activation of integrin α4 and LFA-1 (integrin αLβ2; Zhou et al., 2008). MST3 and MST4 regulate actin dynamics in many contexts. In the developing nervous system, MST3 is required for dendritic spine maintenance and limits filopodia formation (Ultanir et al., 2014). MST3 and MST4 also limit actin-dependent protrusions in other cell types. This can result in increased migration on 2D surfaces when they are depleted, but lead to defects in squeezing through gaps in 3D matrices (Lu et al., 2006; Madsen et al., 2015).
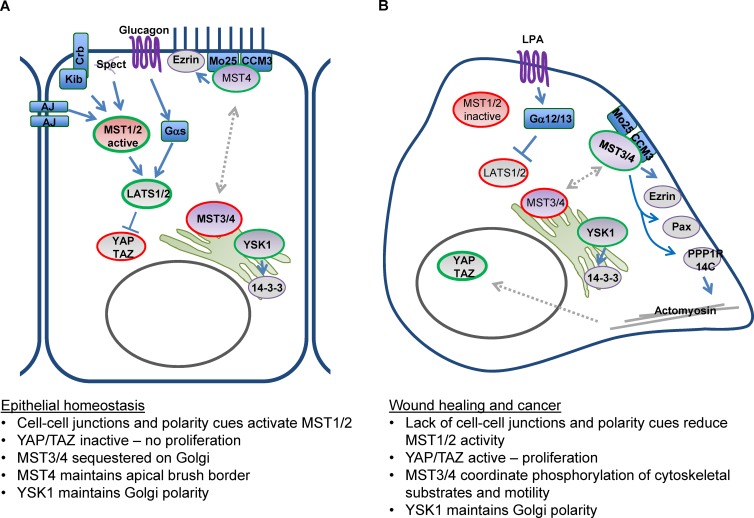
The Drosophila MST3/4 homologue GckIII is required to prevent airway tube dilation along with the CCM3 protein (Song et al., 2013). The related kinase in C. elegans, GCK-1 (Fig. 1), is required to form the excretory canal, possibly by regulating endocytosis and the complex membrane dynamics required for the formation of tubelike structures (Lant et al., 2015). Interestingly, the actin-rich epithelial brush border of intestinal epithelial cells is dependent on MST4 (ten Klooster et al., 2009). This may reflect some conservation of function in regulating the organization of polarized tissues with C. elegans. Mammalian cell culture studies also implicate MST3, MST4, and YSK1 in regulation of cell polarity. MST3, MST4, and YSK1 can all localize to the Golgi apparatus (Preisinger et al., 2004; Lu et al., 2006; ten Klooster et al., 2009). MST4 and YSK1 are recruited to the Golgi apparatus via interaction with GM130. MST3 localization may be through interaction with Striatin proteins. Perturbation of YSK1 function leads to disruption of Golgi organization, and this is believed to trigger a more general loss of cell polarization and migration defects (Preisinger et al., 2004). Interaction with CCM3 or Mo25 can trigger the translocation of MST3 and -4 away from the Golgi apparatus to the plasma membrane (Fig. 2).
Regulation of MST kinases
The regulation of MST kinases is complex and involves the modulation of MST enzymatic activity via T-loop phosphorylation and Mo25 binding, substrate binding, and the regulation of MST localization. In this section we review the major regulatory mechanisms in turn.
Cellular architecture
Recent work has focused on identifying upstream regulator effectors of the MST kinases. Key upstream activators of the Hippo kinase in the fly include the apically localized proteins Crumbs, Expanded, Merlin, Kibra, and apical spectrins, which may constitute a mechanosensory system at the apical domain of epithelial cells (Figs. 2 and 3; Hamaratoglu et al., 2006; Baumgartner et al., 2010; Chen et al., 2010; Genevet et al., 2010; Ling et al., 2010; Yu et al., 2013; Fletcher et al., 2015). There is also likely to be a cell junction–associated complex that activates MST1/2–Angiomotin and Amotl2, which both localize to cell–cell junctions and suppress YAP/TAZ activity (Zhao et al., 2011). In other Drosophila tissues, such as the intestine or ovarian follicle cell epithelium, basolateral spectrins are more important for regulating Hippo signaling, but their mechanism of action remains unknown (Fletcher et al., 2015; Wong et al., 2015). These analyses have led to the idea that MST1/2 activity is regulated by cell and tissue architecture. In homeostatic conditions, MST1/2 are active and therefore cell proliferation is prevented (Fig. 3). It is thought that during tissue growth, MST1/2 activity gradually increases as a result of changes in tissue structure and mechanics; indeed, Rho signaling and actin stress fibers can regulate MST1 and -2 (Densham et al., 2009). The concept of mechanical regulation of MST1/2 is appealing, as physical strain within tissue can increase with size (Mao et al., 2013; Rauskolb et al., 2014). Further, it has been explicitly demonstrated that YAP and TAZ activity are responsive to substrate stiffness, although a role for MST1 and -2 was not found in this study (Dupont et al., 2011). This regulation of YAP and TAZ requires actomyosin function and several regulators of the actin polymerization/depolymerization cycle (Aragona et al., 2013). In response to perturbation of tissue architecture, for example a wound, YAP and TAZ are activated, thereby enabling proliferation of cells to replace the damaged cells in the wound (Lee et al., 2014); this may be linked to reduced MST1/2 activity (Fig. 3). It is likely that this is coordinated with activation of cell migration programs. Intriguingly, mechanical signals may also be integrated with soluble cues at the level downstream of MST1/2. Gα12/13-coupled signals, such as LPA and S1P, can inhibit Lats1 (Yu et al., 2012). In contrast, Gαs-coupled signaling can activate Lats1 (Yu et al., 2012). The latter finding provides a mechanism for hormones that control metabolism, such as glucagon, to be integrated with MST1/2 signals. Metabolic control of MST1/2 signaling may also be achieved by phosphorylation of their adaptor protein Sav by Sik2 (Wehr et al., 2013).
Figure 3.
Role of MST kinases in homeostasis, wound healing, and cancer. (A) Schematic representation of the roles of MST kinases in maintaining epithelial homeostasis. (B) Changes in upstream cues lead to altered regulation of MST kinases and biological consequences. Green outlines indicate functionally active molecules while red outlines indicate inactive molecules. Broken arrows indicate poorly understood regulatory mechanisms.
Several other regulators of the Hippo pathway have been proposed, including the Ds-Ft-Dachs planar polarity system, core apical-basal polarity determinants such as aPKC and Scribble (Skouloudaki et al., 2009; Grzeschik et al., 2010; Verghese et al., 2012), adherens junction components (Bennett and Harvey, 2006; Silva et al., 2006; Willecke et al., 2006), Jnk signaling components (Sun and Irvine, 2013), Src kinases, Echinoid (Yue et al., 2012), and regulators of the F-actin cytoskeleton such as capping proteins, Ajuba, and Zyxin (Fernández et al., 2011; Rauskolb et al., 2011; Sun and Irvine, 2013). However, it remains unclear whether these factors directly influence Hippo-Warts kinase activation or act indirectly through their actions on the cytoskeleton and tissue forces.
Less is known about the regulation of MST3, MST4, and YSK1 by cellular architecture. All three kinases can be localized to the Golgi apparatus through interaction with GM130 (Preisinger et al., 2004; ten Klooster et al., 2009; Fuller et al., 2012). This is believed to keep MST3 and MST4 inactive. In contrast, the adaptor protein CCM3, also called PDCD10, can recruit MST3 to the plasma membrane in both worms and mammals (Figs. 2 and 3). What determines the transition from Golgi to plasma membrane proximal locations remains unclear. In endothelial cells, the interaction with CCM3 may be modulated by HEG1 (Stockton et al., 2010; discussed in more detail later). Unlike MST3 and -4, YSK1 appears to function positively when localized to the Golgi apparatus through its binding to GM130. The localization of active MST3/4 correlates spatially with that of the actomyosin cytoskeleton (Madsen et al., 2015). Further, if the actomyosin function is perturbed, then the localization of MST3 is disrupted, although its biochemical activity is unchanged. This implies that the actin cytoskeleton plays a role in localizing MST3 and has echoes of Orb3 regulation in S. pombe (Leonhard and Nurse, 2005).
Interactions of the SARAH domain
The C-terminal SARAH (Sav, Rassf, Hippo) domain of MST1/2 plays an important role in their regulation. Through its ability to form antiparallel homodimers, the SARAH domain facilitates activating trans-autophosphorylation of the activation loop of MST1 and MST2 (Creasy et al., 1996; Hwang et al., 2007). The SARAH domain also binds to RASSF family proteins and SAV1 (also called WW45 and Salvador in Drosophila). Genetic experiments show that these interactions positively regulate MST1/2 activity (Tapon et al., 2002; Song et al., 2010b), although the mechanistic details are rather unclear (Praskova et al., 2004; Song et al., 2010b; Makbul et al., 2013). Further, the interaction of the MST1 SARAH domain can be modulated by mTORC2-mediated phosphorylation (Sciarretta et al., 2015). It is possible that MST1 or MST2 molecules activated as a result of homodimerization subsequently dissociate and are then targeted to different substrates or subcellular locations by RASSF or SAV1. These latter interactions would help to target MST1/2 to either regulatory complexes or substrates.
PP2A and the STRIPAK complex
A conserved feature of the MST kinases is their association and regulation by PP2A phosphatase. In Drosophila, the PP2A-phosphatase containing the STRIPAK (Striatin Interacting Phosphatase and Kinase) complex can dephosphorylate Hippo (Ribeiro et al., 2010). Interestingly, the Striatin protein that contributes to the name STRIPAK is related to Csc3 in S. pombe, which is part of PP2A phosphatase complex that regulates Sid1 (Singh et al., 2011). In addition, Sid1 is regulated by the phosphatase Clp1 (homologous to S. cerevisiae Cdc14; Trautmann et al., 2001; Wolfe and Gould, 2004; Fig. 2). Proteomic work in mammalian cells has identified MST3, MST4, and YSK1 as components of a large PP2A complex, termed the STRIPAK complex (Glatter et al., 2009; Kean et al., 2011). This complex contains both catalytic and regulatory PP2A components and MST kinases (Filippi et al., 2011). Inhibition of PP2A or depletion of components of the STRIPAK complex increases the phosphorylation of the activation loop of MST3 and MST4 (Madsen et al., 2015). Thus the STRIPAK complex acts as a negative regulator of MST3 and MST4, most likely by directly removing phosphate from the activation loop. However, the regulation of MST3, MST4, and YSK1 by the STRIPAK complex is likely to be more nuanced than a simple negative mechanism. Different splice isoforms of some components have varying abilities to bind to the PP2A catalytic subunits and therefore the ability of the STRIPAK complex to negatively regulate MST kinases may be dependent on its precise molecular makeup (Madsen et al., 2015). It is also unclear where in the cell the complex is located. CCM3 clearly associates with the STRIPAK complex, but smaller “modules” of the STRIPAK complex including CCM3 and the MST kinases may also exist independently of the larger PP2A-containing complex (Goudreault et al., 2009). These may localize differently than the core complex.
Mo25 scaffolds
Another common feature of MST family kinases is their interaction with Mo25 scaffolds. These are armadillo repeat proteins that have an evolutionarily conserved function in binding to members of the larger STE20 family of kinases. The S. cerevisiae homologue of Mo25, Hym1, can interact with Kic1 and regulate the activity of the downstream kinase Cbk1 (Panozzo et al., 2010; Hsu and Weiss, 2013). Similarly, in S. pombe, pMO25 controls the regulation of Nak1-Orb6 (Mendoza et al., 2005; Goshima et al., 2010). Biochemical studies reveal that this interaction can activate kinase activity (Mehellou et al., 2013), although the magnitude of the effect varies greatly (Filippi et al., 2011). It has also been proposed that Mo25 plays a role in subcellular targeting by binding to both MST4 and LKB1. In the absence of M025, LKB1 is not able to regulate MST4 localization and downstream brush border formation. However, the biochemical details of the tertiary complex involving LKB1, Mo25, and MST4 remain to be determined.
Cell stress and death
MST1/2 can be activated by H2O2 redox stress, and in neuronal cells this promotes cell death. MST1, MST2, and MST3 can all be cleaved by caspases during apoptosis (Lee et al., 1998, 2001a). In MST3, the cleavage occurs at amino acid 313 and separates the N-terminal kinase domain from the C-terminal regulatory sequences. This results in nuclear accumulation of the active kinase domain, which can promote apoptosis (Huang et al., 2002; Lee et al., 2004).
MST substrates
We have described many of the biological functions regulated by MST kinases and thereby introduced some of their substrates. Nonetheless it is worth reviewing the direct biochemical substrates on the MST kinases. MST family kinases can autophosphorylate in vitro and this includes phosphorylation of their activation loop (Glantschnig et al., 2002). This autoregulatory event appears to be controlled by the STRIPAK complex that can remove phosphate from the activation loop of MST kinases. The major nonself substrates of MST1 and MST2 are Lats1/2 and Mobkl1a/b (MATS1/2; Fig. 2). MST1/2 phosphorylate the hydrophobic motif of Lats1/2 (S1079 in Lats1) and thereby indirectly promote phosphorylation on their activation loop (S909 in Lats1) and biochemical activity. Phosphorylation of Mob1a/b by MST1/2 promotes their association with Lats kinases and Lats kinase activity. These substrates are sufficient to explain most of the downstream consequences of loss of Hpo in flies and MST1/2 in mammals. Other substrates have been reported under conditions of cell stress, including FOXO transcription factors leading to protective from oxidative stress (Lehtinen et al., 2006), the redox regulator Peroxiredoxin 1 leading to its inhibition (Rawat et al., 2013), and histone H2B in apoptotic cells (Cheung et al., 2003).
In contrast, the situation with MST3, MST4, and YSK1 is more complex. The homologues of MST3/4/YSK1 in S. cerevisiae and S. pombe phosphorylate Ndr family kinases; however, the situation in metazoans is much less certain. In Drosophila it is not clear whether GckIII (the MST3/4/YSK1 homologue) plays a role upstream of the Trc (the Ndr homologue; Fig. 2). The GckIII-related kinase, Misshapen, may be the primary input into Trc in some epithelial tissues (Paricio et al., 1999; Cobreros-Reguera et al., 2010; Horne-Badovinac et al., 2012). There is also evidence for Hippo acting upstream of both Warts and Trc in dendritic tiling and maintenance (Emoto et al., 2004). MST3 can phosphorylate the hydrophobic motif in NDR1 and -2 (Stegert et al., 2005), but evidence that MST4 or YSK1 directly phosphorylate Ndr-related kinases in mammalian systems is currently lacking (Ultanir et al., 2014). MST4 in mammals and GCK-1 in C. elegans have both been reported to modulate the activity of MAPK signaling (Lin et al., 2001; Ma et al., 2007; Schouest et al., 2009); however, the intermediate substrates involved in this regulation are not clear.
Broadly speaking, the kinases within the larger STE20 family are basophilic. Several MST3/4 substrates have been identified that are consistent with this substrate preference. Ezrin can be phosphorylated on its key regulatory site, T567, which enables its binding to both F-actin and the plasma membrane (ten Klooster et al., 2009; Gloerich et al., 2012). This is important for both cell polarity and migration. It is also likely that the Ezrin-related proteins—Moesin and Radixin—are similarly regulated. Other MST3/4 substrates involved in control of cell migration are the adhesion complex molecule paxillin, the tyrosine phosphatase PTP-PEST (Lu et al., 2006), and regulatory subunits of PP1 phosphatase, in particular PPP1R14C (Madsen et al., 2015; Fig. 2). PPP1R14C is phosphorylated by MST3/4 on a key regulatory site T73 that promotes its ability to negatively regulate PP1 complexes. This leads to reduced dephosphorylation of myosin light chain (MLC2/MYL9) and increased actomyosin contractility. Recent work to identify new MST3 substrates has revealed a clear preference for threonine over serine as the phospho-acceptor and a hydrophobic residue immediately C-terminal to this (Ultanir et al., 2014). Interestingly, many cytoskeletal regulators were identified in this study, including the TAO regulators of microtubule stability and the actin regulators EPS8, FMNL2, and Ermin (Ultanir et al., 2014). It will be interesting to determine the role of these substrates downstream of MST3 and -4 in cell migration. The identification of TAO kinases as MST substrates is intriguing, as in Drosophila they can also phosphorylate the activation loop of Hippo (Boggiano et al., 2011), suggesting the possible existence of a regulatory link between MST3 and MST1/2. For regulation of both cell polarity and cell migration, it is likely that locally coordinated phosphorylation of multiple substrates by MST3/4 is required (Madsen et al., 2015). The only well-established YSK-1 substrate is 14-3-3ζ. Phosphorylation of this adaptor protein links YSK1 to Golgi organization and cell polarity (Preisinger et al., 2004).
Metazoan MST kinases in disease
Cancer
Given the potent effects of MST1/2/LATS signaling networks on cell proliferation, these kinases are likely to play important roles in cancer. In particular, a large number of reports have associated up-regulated human YAP and TAZ activity with increased cancer cell proliferation and cancer stem cell function (Lau et al., 2014; Song et al., 2014). Exome sequencing has revealed some mutations and fusions of MST1/2 or LATS1/2 kinases, for example LATS1 fusion in mesothelioma (Miyanaga et al., 2015). However, cancer genome sequencing studies have not revealed high frequencies of mutations, suggesting that other mechanisms must influence either MST signaling or YAP/TAZ activation in cancers. Recent work has indicated that these could include oncogenic Gαq mutations (Feng et al., 2014), or loss of the tumor suppressors NF2 and RASSF family members. Reduced levels of Angiomotin and its close relatives may also contribute increased activity of YAP and TAZ in tumors. YAP also plays a role within the tumor stroma. It is activated in the fibroblastic stroma of breast and squamous cell carcinoma (Calvo et al., 2013). Loss of YAP function in the stroma prevents matrix remodeling and the subsequent invasion of cancer cells. However, the activation of YAP in this context is not associated with reduced MST1/2 activity.
MST3 and MST4 have been implicated in the migration of many cell types. Consistent with this, experimental studies have revealed that they play a positive role in breast cancer metastasis. High levels of MST4 and CCM3 expression are also correlated with more aggressive breast cancer subtypes and worse prognoses. It is likely that CCM3 promotes the activity of MST3 and MST4 at the cell cortex, where they coordinate the phosphorylation of ERM proteins and MLC, enabling cancer cells to squeeze through small gaps (Madsen et al., 2015; Tozluoglu et al., 2015).
Recently, cancer genome sequencing has implicated the STRIPAK complex in cancer. FAM40B is mutated with a high frequency, and the number and type of mutations suggest that it has an oncogenic function (Davoli et al., 2013). Analysis of truncation mutants of FAM40B found in tumors reveals that they are not able to bind to the catalytic subunits of PP2A and may be defective in negatively regulating MST3 and MST4 (Madsen et al., 2015). However, the majority of mutations in FAM40B are point mutations, and these have not yet been analyzed. CCM3 was frequently amplified in many tumor types, especially lung cancer, but the interpretation of these data are complicated because it is very close to the PIK3CA locus, which is a major cancer driver.
Endothelial pathologies
Cerebral cavernous malformation is a common vascular pathology affecting blood vessels in the brain. The malformations are typified by leaky and disordered regions of endothelial cells within the white matter of the brain. Familial forms of the disease are linked to mutations in CCM3 and two other genes, KRIT1/CCM1 and CCM2/OSM. Defective regulation of MST3 and MST4 is implicated in the pathology of endothelial malformations (Stockton et al., 2010; Zheng et al., 2010). Specifically, reduced phosphorylation of ERM proteins leads to increased Rho activity in endothelial cells, and this perturbs endothelial barrier function. Inhibition of the ROCK kinase function downstream of Rho has shown promise in preclinical models of cerebral cavernous malformation. Once again it is interesting to note a role of MST3/4 kinases in the morphogenesis of tubular structures (compare trachea in Drosophila and excretory canal in C. elegans; Song et al., 2013; Lant et al., 2015).
Autoimmunity
During development, MST1/2 act redundantly to restrain the proliferation of epithelial tissues. It has recently been demonstrated that MST1 can play a similar role in T cells. Naive T cells undergo extensive proliferation upon engagement of the T cell receptor (Zhou et al., 2008). This expansion of T cells was partly held in check by MST1 acting in a complex with Nore1B/RAPL. Loss of MST1 leads to hyperproliferation of T cells when stimulated ex vivo (Zhou et al., 2008). The situation in vivo appears more complex; the imbalance in T cell signaling in the absence of MST1 appears to lead to increased apoptosis and fewer T cells (Nehme et al., 2012). This is consistent with reduced T and B cell numbers in humans with an MST1 loss-of-function mutation (Abdollahpour et al., 2012). Further, MST1-defective mice appear less susceptible to experimentally triggered autoimmune encephalitis (Salojin et al., 2014), which may relate to defects in T cell trafficking and extravasation (Katagiri et al., 2009). However, this does not apply to all experimental models. Loss of MST1 renders mice more susceptible to autoimmune disorders including the development of skin lesions around the eye associated with mononuclear cells and splenomegaly. Human studies are also suggestive, but not conclusive. SNPs in MST1 are associated with both Crohn’s disease and colitis (Waterman et al., 2011; Nimmo et al., 2012). Interestingly, MST1 and MST2 do not appear to act redundantly in lymphocytes, possibly reflecting differential expression patterns in immune cells. More work will be required to resolve these issues, including better characterization of the MST1 polymorphisms linked to inflammatory disorders and the targeted loss of MST kinases in leukocyte subsets.
Concluding remarks
MST kinases play a key role in many aspects of biology: MST1/2 couple cellular context within tissues to growth control and can regulate migration, while MST3, MST4, and YSK1 play important roles in highly localized regulation of the cytoskeleton and Golgi apparatus. However, there remains much we do not know. It will be important to learn how MST1/2 integrate the multitude of upstream regulatory mechanisms to achieve such exquisite control over tissue size and structure. Targeted and combined loss-of-function studies of MST3, MST4, and YSK1 will yield insights into the how the localized control of the cytoskeleton influences tissue and organismal level biology in mammals. Finally, the ever increasing analysis of pathological tissue is likely to identify new contexts in which deregulation of these kinases affects human health.
Supplementary Material
Acknowledgements
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Hpo
- Hippo
- Sav
- Salvador
- Yki
- Yorkie
References
- Abdollahpour H., Appaswamy G., Kotlarz D., Diestelhorst J., Beier R., Schäffer A.A., Gertz E.M., Schambach A., Kreipe H.H., Pfeifer D., et al. 2012. The phenotype of human STK4 deficiency. Blood. 119:3450–3457. 10.1182/blood-2011-09-378158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M., Panciera T., Manfrin A., Giulitti S., Michielin F., Elvassore N., Dupont S., and Piccolo S.. 2013. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 154:1047–1059. 10.1016/j.cell.2013.07.042 [DOI] [PubMed] [Google Scholar]
- Bardin A.J., and Amon A.. 2001. Men and sin: what’s the difference? Nat. Rev. Mol. Cell Biol. 2:815–826. 10.1038/35099020 [DOI] [PubMed] [Google Scholar]
- Baumgartner R., Poernbacher I., Buser N., Hafen E., and Stocker H.. 2010. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev. Cell. 18:309–316. 10.1016/j.devcel.2009.12.013 [DOI] [PubMed] [Google Scholar]
- Bennett F.C., and Harvey K.F.. 2006. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr. Biol. 16:2101–2110. 10.1016/j.cub.2006.09.045 [DOI] [PubMed] [Google Scholar]
- Bidlingmaier S., Weiss E.L., Seidel C., Drubin D.G., and Snyder M.. 2001. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:2449–2462. 10.1128/MCB.21.7.2449-2462.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggiano J.C., Vanderzalm P.J., and Fehon R.G.. 2011. Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo-Salvador-Warts tumor suppressor pathway. Dev. Cell. 21:888–895. 10.1016/j.devcel.2011.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Zhang N., Zheng Y., de Wilde R.F., Maitra A., and Pan D.. 2010. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 24:2383–2388. 10.1101/gad.1978810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo F., Ege N., Grande-Garcia A., Hooper S., Jenkins R.P., Chaudhry S.I., Harrington K., Williamson P., Moeendarbary E., Charras G., and Sahai E.. 2013. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 15:637–646. 10.1038/ncb2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo F.D., Gokhale S., Johnnidis J.B., Fu D., Bell G.W., Jaenisch R., and Brummelkamp T.R.. 2007. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17:2054–2060. 10.1016/j.cub.2007.10.039 [DOI] [PubMed] [Google Scholar]
- Chan E.H., Nousiainen M., Chalamalasetty R.B., Schäfer A., Nigg E.A., and Silljé H.H.. 2005. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 24:2076–2086. 10.1038/sj.onc.1208445 [DOI] [PubMed] [Google Scholar]
- Chan S.W., Lim C.J., Huang C., Chong Y.F., Gunaratne H.J., Hogue K.A., Blackstock W.P., Harvey K.F., and Hong W.. 2011. WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ. Oncogene. 30:600–610. 10.1038/onc.2010.438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., and Verheyen E.M.. 2012. Homeodomain-interacting protein kinase regulates Yorkie activity to promote tissue growth. Curr. Biol. 22:1582–1586. 10.1016/j.cub.2012.06.074 [DOI] [PubMed] [Google Scholar]
- Chen C.L., Gajewski K.M., Hamaratoglu F., Bossuyt W., Sansores-Garcia L., Tao C., and Halder G.. 2010. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc. Natl. Acad. Sci. USA. 107:15810–15815. 10.1073/pnas.1004060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W.L., Ajiro K., Samejima K., Kloc M., Cheung P., Mizzen C.A., Beeser A., Etkin L.D., Chernoff J., Earnshaw W.C., and Allis C.D.. 2003. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 113:507–517. 10.1016/S0092-8674(03)00355-6 [DOI] [PubMed] [Google Scholar]
- Cobreros-Reguera L., Fernández-Miñán A., Fernández-Espartero C.H., López-Schier H., González-Reyes A., and Martín-Bermudo M.D.. 2010. The Ste20 kinase misshapen is essential for the invasive behaviour of ovarian epithelial cells in Drosophila. EMBO Rep. 11:943–949. 10.1038/embor.2010.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn K., Biechele S., Garner J., and Rossant J.. 2013. The Hippo pathway member Nf2 is required for inner cell mass specification. Curr. Biol. 23:1195–1201. 10.1016/j.cub.2013.05.044 [DOI] [PubMed] [Google Scholar]
- Colman-Lerner A., Chin T.E., and Brent R.. 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 107:739–750. 10.1016/S0092-8674(01)00596-7 [DOI] [PubMed] [Google Scholar]
- Creasy C.L., and Chernoff J.. 1995. Cloning and characterization of a member of the MST subfamily of Ste20-like kinases. Gene. 167:303–306. 10.1016/0378-1119(95)00653-2 [DOI] [PubMed] [Google Scholar]
- Creasy C.L., Ambrose D.M., and Chernoff J.. 1996. The Ste20-like protein kinase, Mst1, dimerizes and contains an inhibitory domain. J. Biol. Chem. 271:21049–21053. 10.1074/jbc.271.35.21049 [DOI] [PubMed] [Google Scholar]
- Davoli T., Xu A.W., Mengwasser K.E., Sack L.M., Yoon J.C., Park P.J., and Elledge S.J.. 2013. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 155:948–962. 10.1016/j.cell.2013.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Densham R.M., O’Neill E., Munro J., König I., Anderson K., Kolch W., and Olson M.F.. 2009. MST kinases monitor actin cytoskeletal integrity and signal via c-Jun N-terminal kinase stress-activated kinase to regulate p21Waf1/Cip1 stability. Mol. Cell. Biol. 29:6380–6390. 10.1128/MCB.00116-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S.A., Gayyed M.F., Anders R.A., Maitra A., and Pan D.. 2007. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 130:1120–1133. 10.1016/j.cell.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., et al. 2011. Role of YAP/TAZ in mechanotransduction. Nature. 474:179–183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Emoto K., He Y., Ye B., Grueber W.B., Adler P.N., Jan L.Y., and Jan Y.N.. 2004. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 119:245–256. 10.1016/j.cell.2004.09.036 [DOI] [PubMed] [Google Scholar]
- Emoto K., Parrish J.Z., Jan L.Y., and Jan Y.N.. 2006. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 443:210–213. 10.1038/nature05090 [DOI] [PubMed] [Google Scholar]
- Feng X., Degese M.S., Iglesias-Bartolome R., Vaque J.P., Molinolo A.A., Rodrigues M., Zaidi M.R., Ksander B.R., Merlino G., Sodhi A., et al. 2014. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell. 25:831–845. 10.1016/j.ccr.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández B.G., Gaspar P., Brás-Pereira C., Jezowska B., Rebelo S.R., and Janody F.. 2011. Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development. 138:2337–2346. 10.1242/dev.063545 [DOI] [PubMed] [Google Scholar]
- Filippi B.M., de los Heros P., Mehellou Y., Navratilova I., Gourlay R., Deak M., Plater L., Toth R., Zeqiraj E., and Alessi D.R.. 2011. MO25 is a master regulator of SPAK/OSR1 and MST3/MST4/YSK1 protein kinases. EMBO J. 30:1730–1741. 10.1038/emboj.2011.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher G.C., Elbediwy A., Khanal I., Ribeiro P.S., Tapon N., and Thompson B.J.. 2015. The Spectrin cytoskeleton regulates the Hippo signalling pathway. EMBO J. 34:940–954. 10.15252/embj.201489642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller S.J., McGuffin L.J., Marshall A.K., Giraldo A., Pikkarainen S., Clerk A., and Sugden P.H.. 2012. A novel non-canonical mechanism of regulation of MST3 (mammalian Sterile20-related kinase 3). Biochem. J. 442:595–610. 10.1042/BJ20112000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevet A., Wehr M.C., Brain R., Thompson B.J., and Tapon N.. 2010. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell. 18:300–308. 10.1016/j.devcel.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glantschnig H., Rodan G.A., and Reszka A.A.. 2002. Mapping of MST1 kinase sites of phosphorylation. Activation and autophosphorylation. J. Biol. Chem. 277:42987–42996. 10.1074/jbc.M208538200 [DOI] [PubMed] [Google Scholar]
- Glatter T., Wepf A., Aebersold R., and Gstaiger M.. 2009. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol. Syst. Biol. 5:237 10.1038/msb.2008.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloerich M., ten Klooster J.P., Vliem M.J., Koorman T., Zwartkruis F.J., Clevers H., and Bos J.L.. 2012. Rap2A links intestinal cell polarity to brush border formation. Nat. Cell Biol. 14:793–801. 10.1038/ncb2537 [DOI] [PubMed] [Google Scholar]
- Goshima T., Kume K., Koyano T., Ohya Y., Toda T., and Hirata D.. 2010. Fission yeast germinal center (GC) kinase Ppk11 interacts with Pmo25 and plays an auxiliary role in concert with the morphogenesis Orb6 network (MOR) in cell morphogenesis. J. Biol. Chem. 285:35196–35205. 10.1074/jbc.M110.176867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudreault M., D’Ambrosio L.M., Kean M.J., Mullin M.J., Larsen B.G., Sanchez A., Chaudhry S., Chen G.I., Sicheri F., Nesvizhskii A.I., et al. 2009. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell. Proteomics. 8:157–171. 10.1074/mcp.M800266-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulev Y., Fauny J.D., Gonzalez-Marti B., Flagiello D., Silber J., and Zider A.. 2008. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 18:435–441. 10.1016/j.cub.2008.02.034 [DOI] [PubMed] [Google Scholar]
- Grzeschik N.A., Parsons L.M., Allott M.L., Harvey K.F., and Richardson H.E.. 2010. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr. Biol. 20:573–581. 10.1016/j.cub.2010.01.055 [DOI] [PubMed] [Google Scholar]
- Gupta S., Mana-Capelli S., McLean J.R., Chen C.T., Ray S., Gould K.L., and McCollum D.. 2013. Identification of SIN pathway targets reveals mechanisms of crosstalk between NDR kinase pathways. Curr. Biol. 23:333–338. 10.1016/j.cub.2013.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Govindaraghavan M., and McCollum D.. 2014. Cross talk between NDR kinase pathways coordinates cytokinesis with cell separation in Schizosaccharomyces pombe. Eukaryot. Cell. 13:1104–1112. 10.1128/EC.00129-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H., and Halder G.. 2006. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8:27–36. 10.1038/ncb1339 [DOI] [PubMed] [Google Scholar]
- Hartwell L.H., Mortimer R.K., Culotti J., and Culotti M.. 1973. Genetic control of the cell division cycle in yeast: V. Genetic analysis of cdc mutants. Genetics. 74:267–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey K.F., Pfleger C.M., and Hariharan I.K.. 2003. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 114:457–467. 10.1016/S0092-8674(03)00557-9 [DOI] [PubMed] [Google Scholar]
- Hirata D., Kishimoto N., Suda M., Sogabe Y., Nakagawa S., Yoshida Y., Sakai K., Mizunuma M., Miyakawa T., Ishiguro J., and Toda T.. 2002. Fission yeast Mor2/Cps12, a protein similar to Drosophila Furry, is essential for cell morphogenesis and its mutation induces Wee1-dependent G(2) delay. EMBO J. 21:4863–4874. 10.1093/emboj/cdf495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y., Hirahara S., Inoue K., Suzuki A., Alarcon V.B., Akimoto K., Hirai T., Hara T., Adachi M., Chida K., et al. 2013. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr. Biol. 23:1181–1194. 10.1016/j.cub.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne-Badovinac S., Hill J., Gerlach G. II, Menegas W., and Bilder D.. 2012. A screen for round egg mutants in Drosophila identifies tricornered, furry, and misshapen as regulators of egg chamber elongation. G3 (Bethesda). 2:371–378. 10.1534/g3.111.001677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou M.C., Salek J., and McCollum D.. 2000. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr. Biol. 10:619–622. 10.1016/S0960-9822(00)00492-9 [DOI] [PubMed] [Google Scholar]
- Hou M.C., Wiley D.J., Verde F., and McCollum D.. 2003. Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression. J. Cell Sci. 116:125–135. 10.1242/jcs.00206 [DOI] [PubMed] [Google Scholar]
- Hsu J., and Weiss E.L.. 2013. Cell cycle regulated interaction of a yeast Hippo kinase and its activator MO25/Hym1. PLoS ONE. 8:e78334 10.1371/journal.pone.0078334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.Y., Wu Y.M., Hsu C.Y., Lee W.S., Lai M.D., Lu T.J., Huang C.L., Leu T.H., Shih H.M., Fang H.I., et al. 2002. Caspase activation of mammalian sterile 20-like kinase 3 (Mst3). Nuclear translocation and induction of apoptosis. J. Biol. Chem. 277:34367–34374. 10.1074/jbc.M202468200 [DOI] [PubMed] [Google Scholar]
- Huang J., Wu S., Barrera J., Matthews K., and Pan D.. 2005. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 122:421–434. 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Huang H., Li J., Hu L., Ge L., Ji H., Zhao Y., and Zhang L.. 2014. Bantam is essential for Drosophila intestinal stem cell proliferation in response to Hippo signaling. Dev. Biol. 385:211–219. 10.1016/j.ydbio.2013.11.008 [DOI] [PubMed] [Google Scholar]
- Hwang E., Ryu K.S., Pääkkönen K., Güntert P., Cheong H.K., Lim D.S., Lee J.O., Jeon Y.H., and Cheong C.. 2007. Structural insight into dimeric interaction of the SARAH domains from Mst1 and RASSF family proteins in the apoptosis pathway. Proc. Natl. Acad. Sci. USA. 104:9236–9241. 10.1073/pnas.0610716104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Charles J.F., Tinker-Kulberg R.L., and Morgan D.O.. 1998. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell. 9:2803–2817. 10.1091/mbc.9.10.2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Xu J., Yin M.X., Lu Y., Hu L., Li P., Zhang P., Yuan Z., Ho M.S., Ji H., et al. 2013. Brahma is essential for Drosophila intestinal stem cell proliferation and regulated by Hippo signaling. eLife. 2:e00999 10.7554/eLife.00999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kango-Singh M., Nolo R., Tao C., Verstreken P., Hiesinger P.R., Bellen H.J., and Halder G.. 2002. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 129:5719–5730. 10.1242/dev.00168 [DOI] [PubMed] [Google Scholar]
- Katagiri K., Katakai T., Ebisuno Y., Ueda Y., Okada T., and Kinashi T.. 2009. Mst1 controls lymphocyte trafficking and interstitial motility within lymph nodes. EMBO J. 28:1319–1331. 10.1038/emboj.2009.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean M.J., Ceccarelli D.F., Goudreault M., Sanches M., Tate S., Larsen B., Gibson L.C., Derry W.B., Scott I.C., Pelletier L., et al. 2011. Structure-function analysis of core STRIPAK Proteins: a signaling complex implicated in Golgi polarization. J. Biol. Chem. 286:25065–25075. 10.1074/jbc.M110.214486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz L.M., Liu-Chittenden Y., Yin F., Zheng Y., Yu J., Huang B., Chen Q., Wu S., and Pan D.. 2013. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev. Cell. 25:388–401. 10.1016/j.devcel.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A., and Simanis V.. 2008. An overview of the fission yeast septation initiation network (SIN). Biochem. Soc. Trans. 36:411–415. 10.1042/BST0360411 [DOI] [PubMed] [Google Scholar]
- Krapp A., Gulli M.P., and Simanis V.. 2004. SIN and the art of splitting the fission yeast cell. Curr. Biol. 14:R722–R730. 10.1016/j.cub.2004.08.049 [DOI] [PubMed] [Google Scholar]
- Lant B., Yu B., Goudreault M., Holmyard D., Knight J.D., Xu P., Zhao L., Chin K., Wallace E., Zhen M., et al. 2015. CCM-3/STRIPAK promotes seamless tube extension through endocytic recycling. Nat. Commun. 6:6449 10.1038/ncomms7449 [DOI] [PubMed] [Google Scholar]
- Lau A.N., Curtis S.J., Fillmore C.M., Rowbotham S.P., Mohseni M., Wagner D.E., Beede A.M., Montoro D.T., Sinkevicius K.W., Walton Z.E., et al. 2014. Tumor-propagating cells and Yap/Taz activity contribute to lung tumor progression and metastasis. EMBO J. 33:468–481. 10.1002/embj.201386082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.K., Murakawa M., Nishida E., Tsubuki S., Kawashima S., Sakamaki K., and Yonehara S.. 1998. Proteolytic activation of MST/Krs, STE20-related protein kinase, by caspase during apoptosis. Oncogene. 16:3029–3037. 10.1038/sj.onc.1201840 [DOI] [PubMed] [Google Scholar]
- Lee K.K., Ohyama T., Yajima N., Tsubuki S., and Yonehara S.. 2001a MST, a physiological caspase substrate, highly sensitizes apoptosis both upstream and downstream of caspase activation. J. Biol. Chem. 276:19276–19285. 10.1074/jbc.M005109200 [DOI] [PubMed] [Google Scholar]
- Lee S.E., Frenz L.M., Wells N.J., Johnson A.L., and Johnston L.H.. 2001b Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr. Biol. 11:784–788. 10.1016/S0960-9822(01)00228-7 [DOI] [PubMed] [Google Scholar]
- Lee W.S., Hsu C.Y., Wang P.L., Huang C.Y., Chang C.H., and Yuan C.J.. 2004. Identification and characterization of the nuclear import and export signals of the mammalian Ste20-like protein kinase 3. FEBS Lett. 572:41–45. 10.1016/j.febslet.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Lee J.H., Kim T.S., Yang T.H., Koo B.K., Oh S.P., Lee K.P., Oh H.J., Lee S.H., Kong Y.Y., Kim J.M., and Lim D.S.. 2008. A crucial role of WW45 in developing epithelial tissues in the mouse. EMBO J. 27:1231–1242. 10.1038/emboj.2008.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.P., Lee J.H., Kim T.S., Kim T.H., Park H.D., Byun J.S., Kim M.C., Jeong W.I., Calvisi D.F., Kim J.M., and Lim D.S.. 2010. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc. Natl. Acad. Sci. USA. 107:8248–8253. 10.1073/pnas.0912203107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.J., Ran Byun M., Furutani-Seiki M., Hong J.H., and Jung H.S.. 2014. YAP and TAZ regulate skin wound healing. J. Invest. Dermatol. 134:518–525. 10.1038/jid.2013.339 [DOI] [PubMed] [Google Scholar]
- Lehtinen M.K., Yuan Z., Boag P.R., Yang Y., Villén J., Becker E.B., DiBacco S., de la Iglesia N., Gygi S., Blackwell T.K., and Bonni A.. 2006. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 125:987–1001. 10.1016/j.cell.2006.03.046 [DOI] [PubMed] [Google Scholar]
- Leonhard K., and Nurse P.. 2005. Ste20/GCK kinase Nak1/Orb3 polarizes the actin cytoskeleton in fission yeast during the cell cycle. J. Cell Sci. 118:1033–1044. 10.1242/jcs.01690 [DOI] [PubMed] [Google Scholar]
- Leung C.Y., and Zernicka-Goetz M.. 2013. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nat. Commun. 4:2251 10.1038/ncomms3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.L., Chen H.C., Fang H.I., Robinson D., Kung H.J., and Shih H.M.. 2001. MST4, a new Ste20-related kinase that mediates cell growth and transformation via modulating ERK pathway. Oncogene. 20:6559–6569. 10.1038/sj.onc.1204818 [DOI] [PubMed] [Google Scholar]
- Ling C., Zheng Y., Yin F., Yu J., Huang J., Hong Y., Wu S., and Pan D.. 2010. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc. Natl. Acad. Sci. USA. 107:10532–10537. 10.1073/pnas.1004279107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T.J., Lai W.Y., Huang C.Y., Hsieh W.J., Yu J.S., Hsieh Y.J., Chang W.T., Leu T.H., Chang W.C., Chuang W.J., et al. 2006. Inhibition of cell migration by autophosphorylated mammalian sterile 20-like kinase 3 (MST3) involves paxillin and protein-tyrosine phosphatase-PEST. J. Biol. Chem. 281:38405–38417. 10.1074/jbc.M605035200 [DOI] [PubMed] [Google Scholar]
- Lu L., Li Y., Kim S.M., Bossuyt W., Liu P., Qiu Q., Wang Y., Halder G., Finegold M.J., Lee J.S., and Johnson R.L.. 2010. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. USA. 107:1437–1442. 10.1073/pnas.0911427107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas E.P., Khanal I., Gaspar P., Fletcher G.C., Polesello C., Tapon N., and Thompson B.J.. 2013. The Hippo pathway polarizes the actin cytoskeleton during collective migration of Drosophila border cells. J. Cell Biol. 201:875–885. 10.1083/jcb.201210073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zhao H., Shan J., Long F., Chen Y., Chen Y., Zhang Y., Han X., and Ma D.. 2007. PDCD10 interacts with Ste20-related kinase MST4 to promote cell growth and transformation via modulation of the ERK pathway. Mol. Biol. Cell. 18:1965–1978. 10.1091/mbc.E06-07-0608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen C.D., Hooper S., Tozluoglu M., Bruckbauer A., Fletcher G., Erler J.T., Bates P.A., Thompson B., and Sahai E.. 2015. STRIPAK components determine mode of cancer cell migration and metastasis. Nat. Cell Biol. 17:68–80. 10.1038/ncb3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah A.S., Jang J., and Deshaies R.J.. 2001. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA. 98:7325–7330. 10.1073/pnas.141098998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makbul C., Constantinescu Aruxandei D., Hofmann E., Schwarz D., Wolf E., and Herrmann C.. 2013. Structural and thermodynamic characterization of Nore1-SARAH: a small, helical module important in signal transduction networks. Biochemistry. 52:1045–1054. 10.1021/bi3014642 [DOI] [PubMed] [Google Scholar]
- Mao Y., Tournier A.L., Hoppe A., Kester L., Thompson B.J., and Tapon N.. 2013. Differential proliferation rates generate patterns of mechanical tension that orient tissue growth. EMBO J. 32:2790–2803. 10.1038/emboj.2013.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazanka E., Alexander J., Yeh B.J., Charoenpong P., Lowery D.M., Yaffe M., and Weiss E.L.. 2008. The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry. PLoS Biol. 6:e203 10.1371/journal.pbio.0060203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehellou Y., Alessi D.R., Macartney T.J., Szklarz M., Knapp S., and Elkins J.M.. 2013. Structural insights into the activation of MST3 by MO25. Biochem. Biophys. Res. Commun. 431:604–609. 10.1016/j.bbrc.2012.12.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M., Redemann S., and Brunner D.. 2005. The fission yeast MO25 protein functions in polar growth and cell separation. Eur. J. Cell Biol. 84:915–926. 10.1016/j.ejcb.2005.09.013 [DOI] [PubMed] [Google Scholar]
- Miyanaga A., Masuda M., Tsuta K., Kawasaki K., Nakamura Y., Sakuma T., Asamura H., Gemma A., and Yamada T.. 2015. Hippo pathway gene mutations in malignant mesothelioma: revealed by RNA and targeted exon sequencing. J. Thorac. Oncol. 10:844–851. 10.1097/JTO.0000000000000493 [DOI] [PubMed] [Google Scholar]
- Moroishi T., Park H.W., Qin B., Chen Q., Meng Z., Plouffe S.W., Taniguchi K., Yu F.X., Karin M., Pan D., and Guan K.L.. 2015. A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev. 29:1271–1284. 10.1101/gad.262816.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehme N.T., Pachlopnik Schmid J., Debeurme F., André-Schmutz I., Lim A., Nitschke P., Rieux-Laucat F., Lutz P., Picard C., Mahlaoui N., et al. 2012. MST1 mutations in autosomal recessive primary immunodeficiency characterized by defective naive T-cell survival. Blood. 119:3458–3468. 10.1182/blood-2011-09-378364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B., Kurischko C., Horecka J., Mody M., Nair P., Pratt L., Zougman A., McBroom L.D., Hughes T.R., Boone C., and Luca F.C.. 2003. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell. 14:3782–3803. 10.1091/mbc.E03-01-0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto-Silva R.M., de Beco S., and Johnston L.A.. 2010. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev. Cell. 19:507–520. 10.1016/j.devcel.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo E.R., Prendergast J.G., Aldhous M.C., Kennedy N.A., Henderson P., Drummond H.E., Ramsahoye B.H., Wilson D.C., Semple C.A., and Satsangi J.. 2012. Genome-wide methylation profiling in Crohn’s disease identifies altered epigenetic regulation of key host defense mechanisms including the Th17 pathway. Inflamm. Bowel Dis. 18:889–899. 10.1002/ibd.21912 [DOI] [PubMed] [Google Scholar]
- Nishioka N., Inoue K., Adachi K., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R.O., Ogonuki N., et al. 2009. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell. 16:398–410. 10.1016/j.devcel.2009.02.003 [DOI] [PubMed] [Google Scholar]
- Nolo R., Morrison C.M., Tao C., Zhang X., and Halder G.. 2006. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 16:1895–1904. 10.1016/j.cub.2006.08.057 [DOI] [PubMed] [Google Scholar]
- Nurse P., Thuriaux P., and Nasmyth K.. 1976. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146:167–178. 10.1007/BF00268085 [DOI] [PubMed] [Google Scholar]
- Oh H., and Irvine K.D.. 2008. In vivo regulation of Yorkie phosphorylation and localization. Development. 135:1081–1088. 10.1242/dev.015255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., and Irvine K.D.. 2009. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 28:1916–1927. 10.1038/onc.2009.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Reddy B.V., and Irvine K.D.. 2009. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev. Biol. 335:188–197. 10.1016/j.ydbio.2009.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panozzo C., Bourens M., Nowacka A., and Herbert C.J.. 2010. Mutations in the C-terminus of the conserved NDR kinase, Cbk1p of Saccharomyces cerevisiae, make the protein independent of upstream activators. Mol. Genet. Genomics. 283:111–122. 10.1007/s00438-009-0501-3 [DOI] [PubMed] [Google Scholar]
- Pantalacci S., Tapon N., and Léopold P.. 2003. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 5:921–927. 10.1038/ncb1051 [DOI] [PubMed] [Google Scholar]
- Paricio N., Feiguin F., Boutros M., Eaton S., and Mlodzik M.. 1999. The Drosophila STE20-like kinase misshapen is required downstream of the Frizzled receptor in planar polarity signaling. EMBO J. 18:4669–4678. 10.1093/emboj/18.17.4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon C.L., Zhang X., Lin J.I., Manning S.A., and Harvey K.F.. 2012. Homeodomain-interacting protein kinase regulates Hippo pathway-dependent tissue growth. Curr. Biol. 22:1587–1594. 10.1016/j.cub.2012.06.075 [DOI] [PubMed] [Google Scholar]
- Praskova M., Khoklatchev A., Ortiz-Vega S., and Avruch J.. 2004. Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem. J. 381:453–462. 10.1042/BJ20040025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisinger C., Short B., De Corte V., Bruyneel E., Haas A., Kopajtich R., Gettemans J., and Barr F.A.. 2004. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3ζ. J. Cell Biol. 164:1009–1020. 10.1083/jcb.200310061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J.R., and Hartwell L.H.. 1981. The Saccharomyces cerevisiae cell cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Strathern J.N., Jones E.W., and Broach J.R., editors. Cold Spring Harbour Laboratory, Cold Spring Harbour, New York: 97–142. [Google Scholar]
- Rauskolb C., Pan G., Reddy B.V., Oh H., and Irvine K.D.. 2011. Zyxin links fat signaling to the hippo pathway. PLoS Biol. 9:e1000624 10.1371/journal.pbio.1000624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C., Sun S., Sun G., Pan Y., and Irvine K.D.. 2014. Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell. 158:143–156. 10.1016/j.cell.2014.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat S.J., Creasy C.L., Peterson J.R., and Chernoff J.. 2013. The tumor suppressor Mst1 promotes changes in the cellular redox state by phosphorylation and inactivation of peroxiredoxin-1 protein. J. Biol. Chem. 288:8762–8771. 10.1074/jbc.M112.414524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro P.S., Josué F., Wepf A., Wehr M.C., Rinner O., Kelly G., Tapon N., and Gstaiger M.. 2010. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell. 39:521–534. 10.1016/j.molcel.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Rock J.M., and Amon A.. 2011. Cdc15 integrates Tem1 GTPase-mediated spatial signals with Polo kinase-mediated temporal cues to activate mitotic exit. Genes Dev. 25:1943–1954. 10.1101/gad.17257711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salojin K.V., Hamman B.D., Chang W.C., Jhaver K.G., Al-Shami A., Crisostomo J., Wilkins C., Digeorge-Foushee A.M., Allen J., Patel N., et al. 2014. Genetic deletion of Mst1 alters T cell function and protects against autoimmunity. PLoS ONE. 9:e98151 10.1371/journal.pone.0098151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansores-Garcia L., Atkins M., Moya I.M., Shahmoradgoli M., Tao C., Mills G.B., and Halder G.. 2013. Mask is required for the activity of the Hippo pathway effector Yki/YAP. Curr. Biol. 23:229–235. 10.1016/j.cub.2012.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelmilch K., Mohseni M., Kirak O., Pruszak J., Rodriguez J.R., Zhou D., Kreger B.T., Vasioukhin V., Avruch J., Brummelkamp T.R., and Camargo F.D.. 2011. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell. 144:782–795. 10.1016/j.cell.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouest K.R., Kurasawa Y., Furuta T., Hisamoto N., Matsumoto K., and Schumacher J.M.. 2009. The germinal center kinase GCK-1 is a negative regulator of MAP kinase activation and apoptosis in the C. elegans germline. PLoS ONE. 4:e7450 10.1371/journal.pone.0007450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer B., and Philippsen P.. 1991. CDC15, an essential cell cycle gene in Saccharomyces cerevisiae, encodes a protein kinase domain. Yeast. 7:265–273. 10.1002/yea.320070308 [DOI] [PubMed] [Google Scholar]
- Sciarretta S., Zhai P., Maejima Y., Del Re D.P., Nagarajan N., Yee D., Liu T., Magnuson M.A., Volpe M., Frati G., et al. 2015. mTORC2 regulates cardiac response to stress by inhibiting MST1. Cell Reports. 11:125–136. 10.1016/j.celrep.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M. 2011. Mitotic exit control: a space and time odyssey. Curr. Biol. 21:R857–R859. 10.1016/j.cub.2011.09.023 [DOI] [PubMed] [Google Scholar]
- Sidor C.M., Brain R., and Thompson B.J.. 2013. Mask proteins are cofactors of Yorkie/YAP in the Hippo pathway. Curr. Biol. 23:223–228. 10.1016/j.cub.2012.11.061 [DOI] [PubMed] [Google Scholar]
- Silva E., Tsatskis Y., Gardano L., Tapon N., and McNeill H.. 2006. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr. Biol. 16:2081–2089. 10.1016/j.cub.2006.09.004 [DOI] [PubMed] [Google Scholar]
- Singh N.S., Shao N., McLean J.R., Sevugan M., Ren L., Chew T.G., Bimbo A., Sharma R., Tang X., Gould K.L., and Balasubramanian M.K.. 2011. SIN-inhibitory phosphatase complex promotes Cdc11p dephosphorylation and propagates SIN asymmetry in fission yeast. Curr. Biol. 21:1968–1978. 10.1016/j.cub.2011.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skouloudaki K., Puetz M., Simons M., Courbard J.R., Boehlke C., Hartleben B., Engel C., Moeller M.J., Englert C., Bollig F., et al. 2009. Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc. Natl. Acad. Sci. USA. 106:8579–8584. 10.1073/pnas.0811691106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Mak K.K., Topol L., Yun K., Hu J., Garrett L., Chen Y., Park O., Chang J., Simpson R.M., et al. 2010a Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc. Natl. Acad. Sci. USA. 107:1431–1436. 10.1073/pnas.0911409107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Oh S., Oh H.J., and Lim D.S.. 2010b Role of the tumor suppressor RASSF2 in regulation of MST1 kinase activity. Biochem. Biophys. Res. Commun. 391:969–973. 10.1016/j.bbrc.2009.11.175 [DOI] [PubMed] [Google Scholar]
- Song Y., Eng M., and Ghabrial A.S.. 2013. Focal defects in single-celled tubes mutant for Cerebral cavernous malformation 3, GCKIII, or NSF2. Dev. Cell. 25:507–519. 10.1016/j.devcel.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Ajani J.A., Honjo S., Maru D.M., Chen Q., Scott A.W., Heallen T.R., Xiao L., Hofstetter W.L., Weston B., et al. 2014. Hippo coactivator YAP1 upregulates SOX9 and endows esophageal cancer cells with stem-like properties. Cancer Res. 74:4170–4182. 10.1158/0008-5472.CAN-13-3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks C.A., Morphew M., and McCollum D.. 1999. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J. Cell Biol. 146:777–790. 10.1083/jcb.146.4.777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegert M.R., Hergovich A., Tamaskovic R., Bichsel S.J., and Hemmings B.A.. 2005. Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol. Cell. Biol. 25:11019–11029. 10.1128/MCB.25.24.11019-11029.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John M.A., Tao W., Fei X., Fukumoto R., Carcangiu M.L., Brownstein D.G., Parlow A.F., McGrath J., and Xu T.. 1999. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat. Genet. 21:182–186. 10.1038/5965 [DOI] [PubMed] [Google Scholar]
- Stockton R.A., Shenkar R., Awad I.A., and Ginsberg M.H.. 2010. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J. Exp. Med. 207:881–896. 10.1084/jem.20091258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D.S., Biggins S., and Rose M.D.. 1998. The yeast centrin, cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity. J. Cell Biol. 143:751–765. 10.1083/jcb.143.3.751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., and Irvine K.D.. 2013. Ajuba family proteins link JNK to Hippo signaling. Sci. Signal. 6:ra81 10.1126/scisignal.2004324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U., Amon A., Dowzer C., McGrew J., Byers B., and Nasmyth K.. 1993. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 12:1969–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N., Harvey K.F., Bell D.W., Wahrer D.C., Schiripo T.A., Haber D., and Hariharan I.K.. 2002. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 110:467–478. 10.1016/S0092-8674(02)00824-3 [DOI] [PubMed] [Google Scholar]
- ten Klooster J.P., Jansen M., Yuan J., Oorschot V., Begthel H., Di Giacomo V., Colland F., de Koning J., Maurice M.M., Hornbeck P., and Clevers H.. 2009. Mst4 and Ezrin induce brush borders downstream of the Lkb1/Strad/Mo25 polarization complex. Dev. Cell. 16:551–562. 10.1016/j.devcel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Thompson B.J., and Cohen S.M.. 2006. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 126:767–774. 10.1016/j.cell.2006.07.013 [DOI] [PubMed] [Google Scholar]
- Tóth A., Queralt E., Uhlmann F., and Novák B.. 2007. Mitotic exit in two dimensions. J. Theor. Biol. 248:560–573. 10.1016/j.jtbi.2007.06.014 [DOI] [PubMed] [Google Scholar]
- Tozluoglu M., Mao Y., Bates P.A., and Sahai E.. 2015. Cost-benefit analysis of the mechanisms that enable migrating cells to sustain motility upon changes in matrix environments. J. R. Soc. Interface. 12:20141355 10.1098/rsif.2014.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann S., Wolfe B.A., Jorgensen P., Tyers M., Gould K.L., and McCollum D.. 2001. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol. 11:931–940. 10.1016/S0960-9822(01)00268-8 [DOI] [PubMed] [Google Scholar]
- Udan R.S., Kango-Singh M., Nolo R., Tao C., and Halder G.. 2003. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5:914–920. 10.1038/ncb1050 [DOI] [PubMed] [Google Scholar]
- Ultanir S.K., Yadav S., Hertz N.T., Oses-Prieto J.A., Claxton S., Burlingame A.L., Shokat K.M., Jan L.Y., and Jan Y.N.. 2014. MST3 kinase phosphorylates TAO1/2 to enable Myosin Va function in promoting spine synapse development. Neuron. 84:968–982. 10.1016/j.neuron.2014.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F., Mata J., and Nurse P.. 1995. Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J. Cell Biol. 131:1529–1538. 10.1083/jcb.131.6.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese S., Waghmare I., Kwon H., Hanes K., and Kango-Singh M.. 2012. Scribble acts in the Drosophila fat-hippo pathway to regulate warts activity. PLoS ONE. 7:e47173 10.1371/journal.pone.0047173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., and Amon A.. 2001. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol. Biol. Cell. 12:2961–2974. 10.1091/mbc.12.10.2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachowicz P., Chasapi A., Krapp A., Cano Del Rosario E., Schmitter D., Sage D., Unser M., Xenarios I., Rougemont J., and Simanis V.. 2015. Analysis of S. pombe SIN protein association to the SPB reveals two genetically separable states of the SIN. J. Cell Sci. 128:741–754. 10.1242/jcs.160150 [DOI] [PubMed] [Google Scholar]
- Waterman M., Xu W., Stempak J.M., Milgrom R., Bernstein C.N., Griffiths A.M., Greenberg G.R., Steinhart A.H., and Silverberg M.S.. 2011. Distinct and overlapping genetic loci in Crohn’s disease and ulcerative colitis: correlations with pathogenesis. Inflamm. Bowel Dis. 17:1936–1942. 10.1002/ibd.21579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehr M.C., Holder M.V., Gailite I., Saunders R.E., Maile T.M., Ciirdaeva E., Instrell R., Jiang M., Howell M., Rossner M.J., and Tapon N.. 2013. Salt-inducible kinases regulate growth through the Hippo signalling pathway in Drosophila. Nat. Cell Biol. 15:61–71. 10.1038/ncb2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E.L., Kurischko C., Zhang C., Shokat K., Drubin D.G., and Luca F.C.. 2002. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 158:885–900. 10.1083/jcb.200203094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke M., Hamaratoglu F., Kango-Singh M., Udan R., Chen C.L., Tao C., Zhang X., and Halder G.. 2006. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr. Biol. 16:2090–2100. 10.1016/j.cub.2006.09.005 [DOI] [PubMed] [Google Scholar]
- Wolfe B.A., and Gould K.L.. 2004. Fission yeast Clp1p phosphatase affects G2/M transition and mitotic exit through Cdc25p inactivation. EMBO J. 23:919–929. 10.1038/sj.emboj.7600103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K.K., Li W., An Y., Duan Y., Li Z., Kang Y., and Yan Y.. 2015. β-Spectrin regulates the hippo signaling pathway and modulates the basal actin network. J. Biol. Chem. 290:6397–6407. 10.1074/jbc.M114.629493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Huang J., Dong J., and Pan D.. 2003. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 114:445–456. 10.1016/S0092-8674(03)00549-X [DOI] [PubMed] [Google Scholar]
- Wu S., Liu Y., Zheng Y., Dong J., and Pan D.. 2008. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell. 14:388–398. 10.1016/j.devcel.2008.01.007 [DOI] [PubMed] [Google Scholar]
- Xu S., Huang H.K., Kaiser P., Latterich M., and Hunter T.. 2000. Phosphorylation and spindle pole body localization of the Cdc15p mitotic regulatory protein kinase in budding yeast. Curr. Biol. 10:329–332. 10.1016/S0960-9822(00)00382-1 [DOI] [PubMed] [Google Scholar]
- Yin F., Yu J., Zheng Y., Chen Q., Zhang N., and Pan D.. 2013. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 154:1342–1355. 10.1016/j.cell.2013.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.X., Zhao B., Panupinthu N., Jewell J.L., Lian I., Wang L.H., Zhao J., Yuan H., Tumaneng K., Li H., et al. 2012. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 150:780–791. 10.1016/j.cell.2012.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.X., Zhang Y., Park H.W., Jewell J.L., Chen Q., Deng Y., Pan D., Taylor S.S., Lai Z.C., and Guan K.L.. 2013. Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 27:1223–1232. 10.1101/gad.219402.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue T., Tian A., and Jiang J.. 2012. The cell adhesion molecule echinoid functions as a tumor suppressor and upstream regulator of the Hippo signaling pathway. Dev. Cell. 22:255–267. 10.1016/j.devcel.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen J.A., Peckol E.L., Tobin D.M., and Bargmann C.I.. 2000. Neuronal cell shape and neurite initiation are regulated by the Ndr kinase SAX-1, a member of the Orb6/COT-1/warts serine/threonine kinase family. Mol. Biol. Cell. 11:3177–3190. 10.1091/mbc.11.9.3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ren F., Zhang Q., Chen Y., Wang B., and Jiang J.. 2008. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell. 14:377–387. 10.1016/j.devcel.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Bai H., David K.K., Dong J., Zheng Y., Cai J., Giovannini M., Liu P., Anders R.A., and Pan D.. 2010. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell. 19:27–38. 10.1016/j.devcel.2010.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Milton C.C., Poon C.L., Hong W., and Harvey K.F.. 2011. Wbp2 cooperates with Yorkie to drive tissue growth downstream of the Salvador-Warts-Hippo pathway. Cell Death Differ. 18:1346–1355. 10.1038/cdd.2011.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Tang F., Terracciano L., Hynx D., Kohler R., Bichet S., Hess D., Cron P., Hemmings B.A., Hergovich A., and Schmitz-Rohmer D.. 2015. NDR functions as a physiological YAP1 kinase in the intestinal epithelium. Curr. Biol. 25:296–305. 10.1016/j.cub.2014.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Tumaneng K., Wang C.Y., and Guan K.L.. 2010. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCFβ-TRCP. Genes Dev. 24:72–85. 10.1101/gad.1843810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Lu Q., Wang L.H., Liu C.Y., Lei Q., and Guan K.L.. 2011. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25:51–63. 10.1101/gad.2000111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Xu C., Di Lorenzo A., Kleaveland B., Zou Z., Seiler C., Chen M., Cheng L., Xiao J., He J., et al. 2010. CCM3 signaling through sterile 20-like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. J. Clin. Invest. 120:2795–2804. 10.1172/JCI39679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Medoff B.D., Chen L., Li L., Zhang X.F., Praskova M., Liu M., Landry A., Blumberg R.S., Boussiotis V.A., et al. 2008. The Nore1B/Mst1 complex restrains antigen receptor-induced proliferation of naïve T cells. Proc. Natl. Acad. Sci. USA. 105:20321–20326. 10.1073/pnas.0810773105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Conrad C., Xia F., Park J.S., Payer B., Yin Y., Lauwers G.Y., Thasler W., Lee J.T., Avruch J., and Bardeesy N.. 2009. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 16:425–438. 10.1016/j.ccr.2009.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Zhang Y., Wu H., Barry E., Yin Y., Lawrence E., Dawson D., Willis J.E., Markowitz S.D., Camargo F.D., and Avruch J.. 2011. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc. Natl. Acad. Sci. USA. 108:E1312–E1320. 10.1073/pnas.1110428108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Li D., Wang Y., Pei C., Liu S., Zhang L., Yuan Z., and Zhang P.. 2015. Brahma regulates the Hippo pathway activity through forming complex with Yki-Sd and regulating the transcription of Crumbs. Cell. Signal. 27:606–613. 10.1016/j.cellsig.2014.12.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.