Abstract
Many cellular functions in eukaryotic pathogens are mediated by the cyclic nucleotide binding (CNB) domain, which senses second messengers such as cyclic AMP and cyclic GMP. Although CNB domain-containing proteins have been identified in many pathogenic organisms, an incomplete understanding of how CNB domains in pathogens differ from other eukaryotic hosts has hindered the development of selective inhibitors for CNB domains associated with infectious diseases. Here, we identify and classify CNB domain-containing proteins in eukaryotic genomes to understand the evolutionary basis for CNB domain functional divergence in pathogens. We identify 359 CNB domain-containing proteins in 31 pathogenic organisms and classify them into distinct subfamilies based on sequence similarity within the CNB domain as well as functional domains associated with the CNB domain. Our study reveals novel subfamilies with pathogen-specific variations in the phosphate-binding cassette. Analyzing these variations in light of existing structural and functional data provides new insights into ligand specificity and promiscuity and clues for drug design. This article is part of a Special Issue entitled: Inhibitors of Protein Kinases.
Keywords: Cyclic nucleotide signaling, Allosteric regulation, Phylogeny, Bioinformatics, Evolution, Pathogenic variation
1. Introduction
Cyclic nucleotide binding (CNB) domains are core components of the cellular machinery that regulate diverse cellular processes in prokaryotes and eukaryotes [1,2] in response to second messenger signals including cyclic AMP (cAMP) and cyclic GMP (cGMP). CNB domains are often linked with a diverse array of functional domains such as ion channels, protein kinases, guanine nucleotide exchange factors and transcription regulators [3,4] and act in conjunction with adenylate/ guanylate cyclases and cyclic nucleotide phosphodiesterases [5] to regulate intracellular cAMP and cGMP levels. An increase in cellular levels of cyclic nucleotides results in activation of cAMP- and cGMP-dependent protein kinases (PKA and PKG), respectively. PKA, often considered the archetype of protein kinases in structural and mechanistic studies, forms a tetrameric complex composed of two individual proteins: the regulatory (PKA-R) and catalytic (PKA-C) subunits. PKA-R contains two CNB domains along with dimerization and localization motifs in the N-terminus (Supplementary Fig. 1). This architecture is conserved across most organisms, with the exception of the docking/ dimerization (D/D) domain, which may be absent in PKA-R in some organisms [6]. PKG, on the other hand, presents a different configuration: the CNB domains are fused to the catalytic kinase domain [7] and this configuration is conserved in some protozoan parasites such as Trypanosoma and Leishmania [8,9].
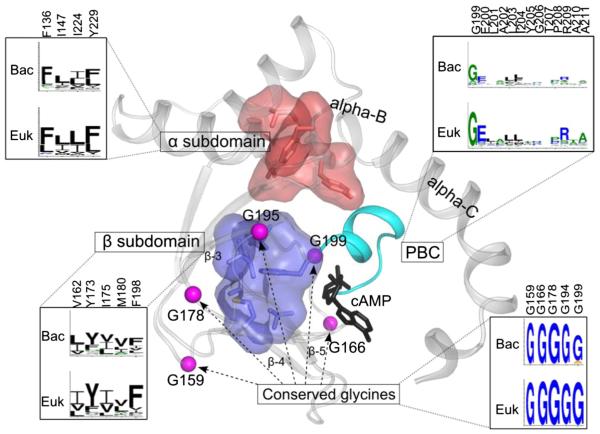
Structurally, CNB domains are composed of a flexible helical subdomain and a stable eight-stranded β sandwich (Fig. 1). Embedded between β-strands 5 and 6 is a small helical motif that is referred to as the phosphate binding cassette (PBC). This signature motif of the CNB domain contains a critical arginine that docks to the phosphate of the cyclic nucleotide. The helical subdomain is made up of two helical motifs in addition to the PBC. At the N-terminus is an N3A motif and following β strand 8 is the B/C helix. In contrast to the beta subdomain, which undergoes minimal conformational change upon cAMP binding, the helical motifs undergo considerable rearrangement [10] upon nucleotide binding and these coordinated conformational changes have been described in detail [3].
Fig. 1.
Key residues and motifs conserved in the CNB domain. The CNB domain of PKA regulatory submit (PDB:1RGS) is used to depict the key residues and motifs. Key regions are labeled and conservation of motifs in eukaryotes (Euk) and bacteria (Bac) is shown as sequence logos (the residue numbers on top are based on pdb id :1RGS). The conserved hydrophobic cores from alpha and beta subunits are shown in surface representation.
Cyclic nucleotide signaling in Plasmodium falciparum-infected red blood cells was shown to be important at different points of parasite intra-erythrocyte development. As the parasite develops into a trophozoite (16–24 h post red blood cell invasion), essential channels in the infected erythrocyte membrane open under the influence of cAMP-dependent protein kinase [11,12]. At the end of the red blood cell cycle (48 h post-invasion), the parasite develops into an invasion-competent form called a merozoite and a major merozoite surface protein (apical membrane antigen 1) becomes phosphorylated at S610 by cAMP-dependent PKA [13]. Even if the role of S610 phosphorylation in invasion has not been clearly demonstrated, PKA-mediated phosphorylation of a wide range of parasite proteins takes place prior to merozoite egress [14]. Merozoite egress and reinvasion of new red blood cells depend on cAMP-dependent PKA [15] and merozoite egress can also be regulated by cGMP-PKG [9,16]. Clearly, both cAMP and cGMP are critical secondary messengers in malaria parasite biology. Therefore, characterizing their corresponding CNB domains present in different parasite proteins is of significant importance for understanding their biological roles and for the possible development of parasite-targeting therapeutics.
CNB domains may serve as attractive drug targets due to their critical regulatory role in essential cellular functions. Since cyclic nucleotide signaling plays a significant role in pathogenic processes, this may serve as a strategy for pathogenic-selective targeting. Cyclic nucleotide analogs were found to have clinical significance for various settings including tumor growth inhibition [17] and at least one compound, 8-Cl-cAMP or tocladesine, has undergone Phase II clinical trials [18]. By defining unique structural features present in pathogenic CNB domains, one can take advantage of divergences between humans and parasites to engineer nucleotide analogs that more closely complement the parasitic CNB domain counterparts. However, an incomplete understanding of how parasitic CNB domains differ from the eukaryotic host has hindered progress in this field.
Previous genome-wide surveys of CNB-domain containing proteins have focused on metagenomes and bacterial genomes [1,3] to understand the evolution of allosteric regulation in the CNB domain. Here, we build upon these studies to identify and classify CNB domain-containing proteins in 31 eukaryotic pathogens (excluding fungi). We identified and classified nearly 13,000 CNB domain-containing sequences into 135 distinct subfamilies, 25 of which are present in pathogen genomes. Through quantitative comparisons of CNB domain sequences and crystal structures, we identify pathogen-specific variations in the phosphate binding cassette and highlight the role of family-specific sequence insertions and deletions in functional adaptation. Implications for inhibitor design are briefiy discussed.
2. Results and discussion
Using profile-based methods, we identified 13,667 CNB domaincontaining proteins in 4272 reference proteomes from Uniprot that span all three kingdoms of life [19] (see Materials and methods). The focus of the present analysis is to understand the functional significance of CNB domains in pathogenic organisms. Towards this end, we focused our analysis on 31 eukaryotic pathogens ranging from unicellular alveolates to multicellular metazoans. In the following sections, we describe the identification and classification of CNB domain-containing proteins in eukaryotic pathogens and discuss key residues and motifs that contribute to the evolutionary divergence of pathogenic CNBs from their eukaryotic homologs.
2.1. Genome-wide survey of CNB-domain containing proteins
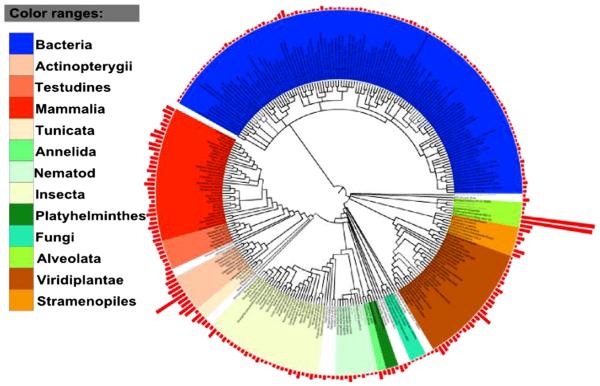
CNB domain-containing proteins are found in organisms spanning major eukaryotic and prokaryotic phyla with significant expansion in chordates (on an average of 50 per organism) (Fig. 2). In addition, freeliving cilliates, Paramecium tetraurelia (427 domains) and Tetrahymena thermophila (417 domains) show expansion of CNB domaincontaining proteins. The observed expansions are functionally significant because ciliary motion in these protozoans is controlled by cAMP and cGMP levels [20]. In Paramecium, for example, swimming speed is controlled by cAMP-mediated regulation of membrane potential [21]. Furthermore, many of the CNB domain-containing proteins such as PKA, PKG and ion channels are involved in sensing and responding to environmental stress in these free living organisms [22].
Fig. 2.
Distribution of CNB domains across diverse phyla. The tree is based on NCBI taxonomy; red bars at each node indicate the number of CNB domains per organism (taxonomy tree generate with itol).
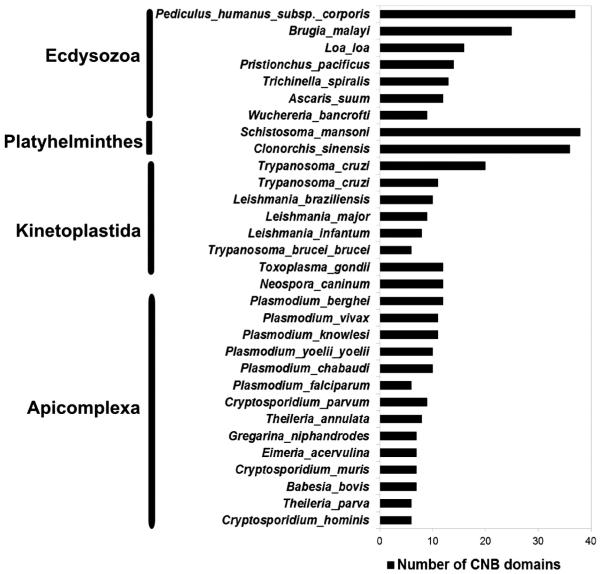
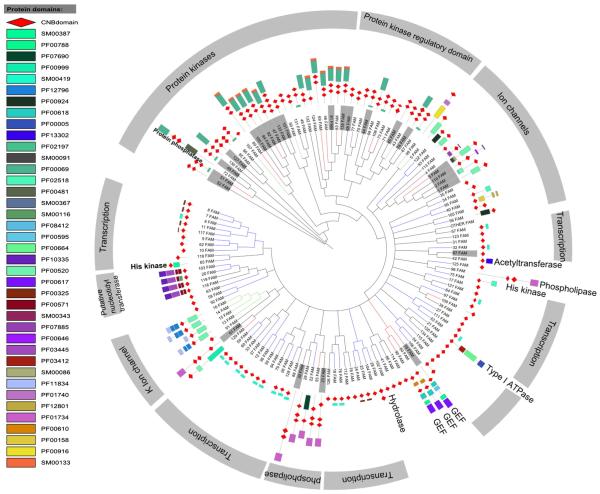
Sequence searches across 31 eukaryotic pathogens revealed 359 CNB domain-containing proteins (Fig. 3), including the regulatory subunit of cAMP-dependent protein kinase A (PKA-R) and cGMP-dependent protein kinase (PKG). In the malaria parasite P. falciparum, cGMP-dependent protein kinase (PfPKG) is first expressed at the ring stage and involved throughout intraerythrocytic development [7,23]. Comparison of PfPKG with its mammalian counterpart revealed the presence of two additional cGMP binding domains, presumably contributing to a unique mode of regulation in P. falciparum [24]. The functional role of PfPKA is also very well established in the malaria parasite [25]. Subtle sequence variations within the CNB domains of the PfPKA regulatory subunit have been noted and such variations offer altered means of regulation via phosphorylation and protein–protein interactions in the parasite [6]. Functions of CNB domain-containing proteins have been noted in other eukaryotic pathogens as well [26–29]. These variations are discussed in later sections in the context of available structural and functional data.
Fig. 3.
Distribution of CNB domains across pathogenic eukaryotes.
2.2. Subfamily organization and distribution of pathogenic organisms
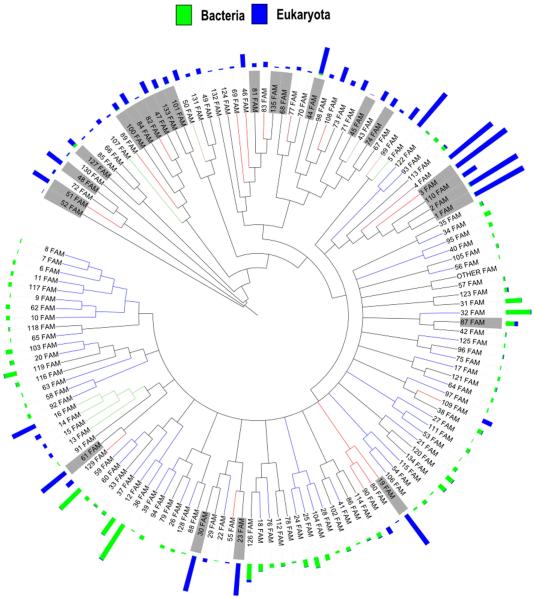
As a starting point for CNB domain functional classification, we clustered 13,667 CNB domain sequences into 135 subfamilies based on similarities and differences in the CNB domain (see Materials and methods for details). The 135 distinct subfamilies, on average, share pairwise sequence identity of 21% (± 3.2) and can be separated based on lineage (eukaryote and prokaryote) (Fig. 4). We also investigated the likely functions associated with each of these subfamilies based on the presence of functional domains associated with the CNB domains (Fig. 5). The eukaryotic subfamilies are predominantly composed of protein kinases, ion channels and guanine nucleotide exchange factors (GEF), while the bacterial subfamilies are predominantly associated with functional domains such as transcription regulators, histidine kinases and hydrolases.
Fig. 4.
Organization of CNB subfamilies. A total of 135 subfamilies identified from 13,667 proteins based on sequence similarity. The blue and green bars indicate the number of eukaryotic and prokaryotic sequences per subfamily, respectively. Nodes highlighted in gray indicate subfamilies which are composed of 10 or more pathogenic sequences.
Fig. 5.
Domain organizations across CNB subfamilies. The most representative domain organization (most frequently occurring domain architecture per subfamily) per subfamily is shown. Nodes are highlighted in gray and indicate subfamilies that are composed of 10 or more pathogenic sequences.
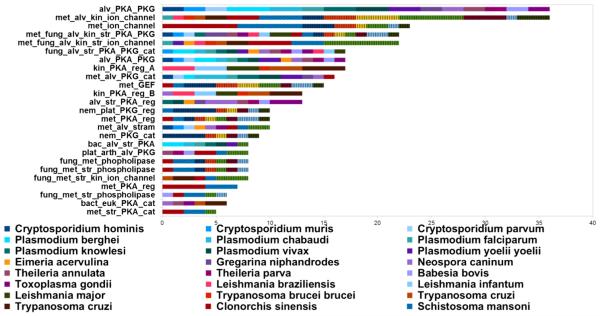
Out of the 135 subfamilies, 25 contain 10 or more sequences from pathogenic organisms (highlighted in gray in Figs. 4 and 5) and the distribution of these subfamilies across 31 eukaryotic pathogens is shown in Fig. 6. Notably, most of the pathogenic sequences cluster with other eukaryotic CNBs. An exception to this was found in two subfamilies of the PKA regulatory subdomain (kin_PKA_reg A & B, Fig. 6), which only consists of pathogenic CNBs from kinetoplastids such as Leishmania. Furthermore, the most represented functional classes among these subfamilies include protein kinases and ion channels (Fig. 6), which are known to play important roles in pathogenesis [6, 30]. P. falciparum, the species responsible for the majority of malariarelated deaths, has three CNB-containing proteins, namely PfPKA-R, PfPKG and Rap guanine nucleotide exchange factor (PfEpac) [15]. P. vivax, P. berghei, P. yoelii, P. knowlesi, P. reichenowi, and P. chabaudi also encode multiple CNB domains in their genomes.
Fig. 6.
Distribution of subfamilies across 31 pathogens. The stacked bar graph shows the number of CNB domains from an organism for each of the subfamilies. The subfamily names are indicated on the y-axis and have been named on the basis of their overall function (based on the domain families associated with CNB domains) and the organism class. Abbreviations for organism classes are as follows: met — metazoan, fung — fungi, bact — bacteria, euk — eukaryotes, str — stramenopiles, alv — alveolates, kin — kinetoplastids, nem — nematode and plat — platyhelminthes.
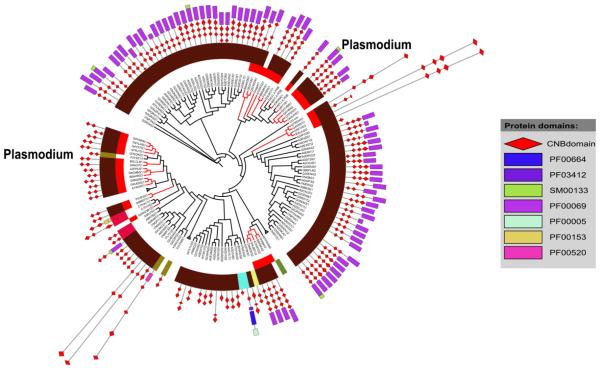
Within individual subfamilies, CNB domains display species-specific variations. This is illustrated for the PKA/PKG subfamily (Fig. 7), where CNB domains from apicomplexans form distinct sub-clusters. The PKA/PKG subfamily predominantly consists of free-living cilliates (80% of the sequences, highlighted in Fig. 7 in maroon) along with apicomplexans (red branches in Fig. 7). The apicomplexan CNBs subcluster into three main regions across the tree, suggesting speciation and functional adaptation. Below, we explore sequence and structural features associated with pathogen-specific evolutionary divergence.
Fig. 7.
Close clustering of pathogenic CNBs within subfamilies. A dendrogram for the cAMP/cGMP dependent protein kinases from alveolates (indicated in maroon) is shown. The subfamily is composed of pathogenic sequences from apicomplexans (red branches). The dendrogram also shows the domain architecture for the individual proteins.
2.3. Structural and sequence variation within pathogenic CNBs from eukaryotes
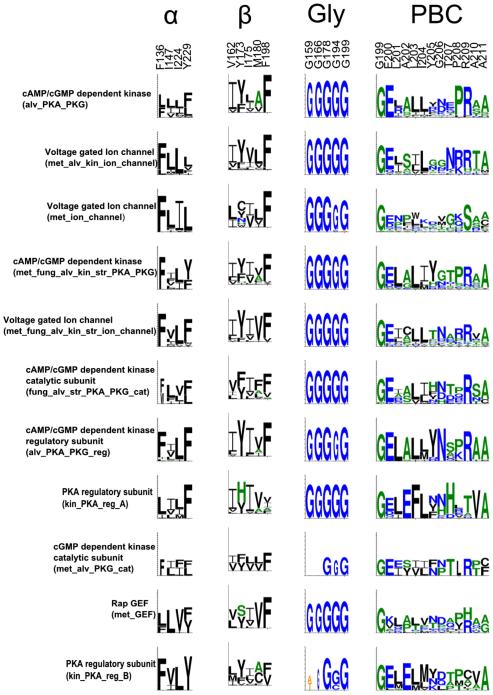
Comparison of pathogenic and non-pathogenic CNB domains indicates key variations in conserved motifs (Fig. 8). Previous evolutionary studies have identified conserved glycine residues in the loops connecting the beta stands in the beta subdomain. In addition, the PBC is characterized by a conserved arginine (R209) and glutamate (E200) that coordinate nucleotide binding [1,3]. These canonical CNB domain residues and motifs are conserved among most pathogenic sequences (Fig. 8). Among the 5 conserved glycine residues, Gly178 in the betasubdomain is the most conserved [1] and this conservation pattern is observed in the CNB of pathogens as well. Likewise, the hydrophobic residues that define the alpha and beta subunit core are also present in pathogen sequences [1,3] (Fig. 8).
Fig. 8.
Conservation of canonical CNB motifs across pathogenic CNBs. The motifs were generated by considering only the pathogenic CNBs from individual subfamilies. The residue numbering on top is based on the first CNB domain in PKA-R (PDB: 1RGS).
Notable variations, however, are observed in the nucleotide binding PBC motif (Fig. 8) in subfamilies such as the CNB-homology domain in voltage-gated potassium channels, which, unlike canonical CNB domains, bind cyclic nucleotides with lower affinity. The low affinity is due, in part, to a unique beta-9 strand in the ligand-binding pocket that occludes binding of the cyclic nucleotide [31–33]. The presence of the beta-9 strand in pathogenic CNB domains suggests variable affinities for cyclic nucleotides. Significant variations are also noted in the kinetoplastid-specific subfamilies of the PKA regulatory subdomain (kin_PKA_reg A & B). Although both families are composed of PKA regulators, they share an overall sequence identify of less than 20% and therefore cluster into two distinct subfamilies. Gly156 and Gly166 are not conserved in the kin_PKA_reg B subfamily. In addition, both subfamilies show variation in the PBC motif. In kin_PKA_reg A, for example, the phosphate-coordinating Arg209 is substituted to a His. Although the functional significance of this variation is not known, it is possible that kin_PKA_reg A has evolved to bind other ligands in the cyclic nucleotide binding pocket as has been demonstrated previously for the heme sensing CNB domains (PDB id: 1FT9) [34].
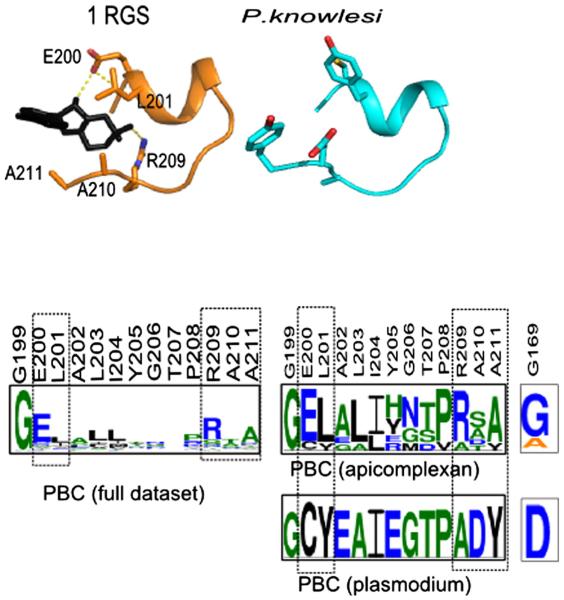
We further investigated pathogen-specific variations within the PBC region by focusing our analysis on the apicomplexan parasites. Fig. 9 shows the conservation of PBC and Gly169 from two subfamilies of PKA/PKG from apicomplexans (met_fung_alv_kin_str_PKA_PKG & alv_str_PKA_reg). Previous sequence analysis revealed strong coconservation of Arg209 with Gly169 [1]. A similar correlation is also observed in apicomplexan CNBs, where lack of conservation of Gly169 is correlated with variations in Arg209 and Glu200. Fig. 9 also shows the PBC region from the modeled structure of a putative cyclic nucleotide-binding domain from P. knowlesi (Uniprot id: B3LAJ2, 4th domain). In the Plasmodium protein, Arg209 and Glu200 are substituted to Ala and Cys, respectively. These variations presumably contribute to Plasmodium-specific functional divergence by facilitating recognition of other small molecule ligands or by altering the affinity for cyclic nucleotides.
Fig. 9.
Variation in the PBC region in Plasmodium. The top panel shows the PBC region from 1RGS and the Plasmodium model structure. The lower panel shows the conservation of the entire PBC region. The residues that are represented as sticks are also highlighted within the motifs. The full dataset of PBCs depicts the canonical PBC motif based on the alignment of all the CNB domains. PBC apicomplexan is based on the alignment of CNBs from apicomplexans only. The P. knowlesi CNB domain sequence is modeled using 1RGS as a template.
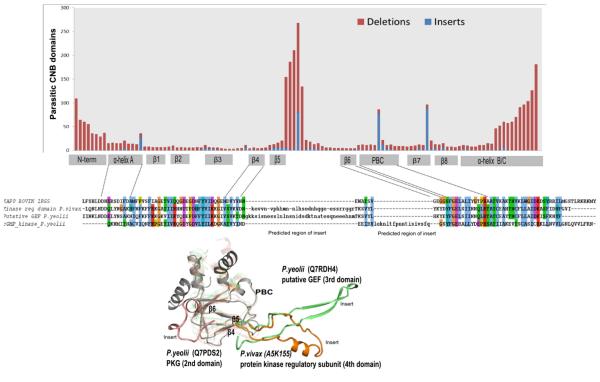
In addition to residue variations in conserved motifs, subfamily-specific insertions and deletions also appear to contribute to CNB domain functional divergence. Fig. 10 shows the distribution of insertions/deletions observed at each position within the CNB domain. A significant number of insertions and deletions occur in the region connecting the beta-4 and beta-6 strands in the beta subdomain. The positional location of the insert is generally conserved in the sequences analyzed, though some exceptions are observed. An example of such variation is illustrated in the PKG CNB domain from P. yeolii (Q7PDS2), where the insert is not conserved in orthologous sequences from closely related apicomplexans (Supplementary Fig. 1a). The variable inserts between the beta-4 and beta-6 strands, however, may be functionally significant because the corresponding regions in some transcription factors [35] are noted to be involved in protein–protein interactions. Moreover, inserts generally provide a framework for evolving new functions from a common scaffold by altering protein regulation and protein–protein interactions [36].
Fig. 10.
Distribution of inserts and deletions within the CNB domain. The stacked bar graph shows the number of proteins with insertions (blue) or deletions (red) at every position of the CNB domain based on an alignment of pathogenic CNB domains with 1RGS as the reference sequence for determining the position of insertions/deletions. The alignment below only represents CNBs from Plasmodium along with the modeled structure highlighting the inserts. All the structures are superimposed on 1RGS (shown in gray, 1st domain).
3. Concluding remarks
Here, we identified and classified CNB domain-containing proteins in eukaryotic pathogens and investigated the sequence and structural basis for pathogen-specific functional adaptation. We identified novel variations in the nucleotide-binding pocket that provide new hypotheses for functional studies. For example, the divergent nature of the PBC motif in Plasmodium can be explored through mutational studies. Like-wise, small molecule screens can potentially identify new high affinity ligands for pathogenic CNBs. In addition to variation in the ligandbinding pocket, our analysis also highlights the presence of pathogen-specific inserts within the CNB domain that can be explored through mutational studies. Several of the identified inserts and deletions are conserved in closely related organisms, suggesting that they play important functional roles. Indeed, insertions/deletions in loop regions have been shown to play important regulatory roles in proteins [36], but to the best of our knowledge the role of these changes in CNB domain functional divergence has not been systematically explored.
Aberrant cyclic nucleotide-mediated signaling is linked to a variety of diseases including cancer, diabetes and cardiovascular diseases [37]. Since CNB domains perform critical roles in cell regulation, numerous cyclic nucleotide analogs were developed and used as probes to uncover CNB-domain functions in signaling pathways. Historically, some of the more commonly used cyclic nucleotide analogs are 8-Br-cAMP [38] and 8-Br-cGMP [30]; and dibutyryl-cAMP [39] and dibutyryl-cGMP [40]. These compounds can bind targets such as PKA and PKG with Ka values in the mid- to low-nM range [41]. However, major limitations of these analogs include susceptibility to hydrolysis by phosphodiesterases (PDEs); lack of target specificity due to the presence of CNB-domains in diverse proteins including HCN, CNG, PKA PKG and EPAC; and limited cell permeability, particularly with the 8-Br-cNMP analogs [42]. Some of these shortcomings have been addressed by synthetic modifications including Rp- and Sp-cAMP analogs, which were found to have increased resistance to PDE hydrolysis [43]. Structure-based design and targeting remains a substantial challenge for CNB domains due to the level of pocket conservation among diverse proteins. Synthetic library screening efforts have afforded the possibility for greatly improved targeting of CNB domains with reduced off-target effects [44–47]. Another successful targeting strategy involves combination treatments of cNMP analogs to synergistically and specifically target particular CNB domains [48,49]. Further bioinformatic and structure-based knowledge of pocket-specific features for the different CNB classes will be crucial for the future development of target-specific inhibitors. In type III PKG, for example, some of the CNB domains fused to a kinase domain are non-functional and cannot bind cGMP. This is deducible from the divergent nature of the PBC (see Table in [8]) and was also recently confirmed experimentally for PfPKG [9]. Thus, the identification of divergent features in the nucleotide-binding pocket as described in our study may aid in the design of pathogen-specific CNB domain inhibitors.
4. Materials and methods
4.1. Dataset and methodology
4.1.1. Identification and alignment of CNB domains
A total of 13,667 CNB domains were identified using a profile based approach [50] from the reference proteomes from Uniprot [19]. The profile consisted of manually curated alignment of the CNB domains from diverse organisms [1] and was used for aligning the 13,667 CNB-domain sequences (Supplementary Table 1 lists all the domains identified in this analysis). The generated alignment was visually inspected for conserved residues and motifs and validated using available crystal structures. Position of large sequence insert segments within the alignment was resolved using crystal structures and predicted secondary structures [51].
4.1.2. Subfamily classification and function annotation
CNB sequences were clustered into subfamilies using a pairwise identity cutoff of 20%. Maximum likelihood tree was constructed using an alignment of consensus sequences obtained from each of the subfamilies with 100 bootstraps using RAXML [52]. The tree generated was used for further clustering of closely related subfamilies. All branches with bootstrap values of 70 or greater were collapsed into a single node; this led to the generation of 135 subfamilies. Subfamilies were named using sequence annotations provided in Uniprot (Supplementary Table 1). Proteins containing CNB domains from pathogenic organisms are listed in Supplementary Table 2 (the list also includes species specific identifiers from EuPathDB). Domain assignments for the sequences were obtained from Interpro [53]. Conserved motifs were generated for the CNB-domain sequence alignment using Weblogo [54]. All the structural models were generated using the SWISS-MODEL [51].
Supplementary Material
Footnotes
This article is part of a Special Issue entitled: Inhibitors of Protein Kinases.
Supplementary data to this article can be found online at http://dx. doi.org/10.1016/j.bbapap.2015.03.012.
Transparency document
The Transparency document associated with this article can be found, in the online version.
References
- [1].Kannan N, Wu J, Anand GS, Yooseph S, Neuwald AF, Venter JC, Taylor SS. Evolution of allostery in the cyclic nucleotide binding module. Genome Biol. 2007;8:R264. doi: 10.1186/gb-2007-8-12-r264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].McCue LA, McDonough KA, Lawrence CE. Functional classification of cNMP-binding proteins and nucleotide cyclases with implications for novel regulatory pathways in Mycobacterium tuberculosis. Genome Res. 2000;10:204–219. doi: 10.1101/gr.10.2.204. [DOI] [PubMed] [Google Scholar]
- [3].Berman HM, Ten Eyck LF, Goodsell DS, Haste NM, Kornev A, Taylor SS. The cAMP binding domain: an ancient signaling module. Proc. Natl. Acad. Sci. U. S. A. 2005;102:45–50. doi: 10.1073/pnas.0408579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shabb JB, Corbin JD. Cyclic nucleotide-binding domains in proteins having diverse functions. J. Biol. Chem. 1992;267:5723–5726. [PubMed] [Google Scholar]
- [5].Lu J, Bao Q, Wu J, Wang H, Li D, Xi Y, Wang S, Yu S, Qu J. CSCDB: the cAMP and cGMP signaling components database. Genomics. 2008;92:60–64. doi: 10.1016/j.ygeno.2008.03.012. [DOI] [PubMed] [Google Scholar]
- [6].Haste NM, Talabani H, Doo A, Merckx A, Langsley G, Taylor SS. Exploring the Plasmodium falciparum cyclic-adenosine monophosphate (cAMP)-dependent protein kinase (PfPKA) as a therapeutic target. Microbes Infect. 2012;14:838–850. doi: 10.1016/j.micinf.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Deng W, Baker DA. A novel cyclic GMP-dependent protein kinase is expressed in the ring stage of the Plasmodium falciparum life cycle. Mol. Microbiol. 2002;44:1141–1151. doi: 10.1046/j.1365-2958.2002.02948.x. [DOI] [PubMed] [Google Scholar]
- [8].Bakkouri ME, Hui R. Diverse cGMP-dependent protein kinases in protozoan parasites and other protist organisms. BBA Proteins and Proteomics. 2015 [Google Scholar]
- [9].Kim JJ, Flueck C, Franz E, Sanabria-Figueroa E, Thompson E, Lorenz R, Bertinetti D, Baker DA, Herberg FW, Kim C. Crystal structures of the carboxyl cGMP binding domain of the Plasmodium falciparum cGMP-dependent protein kinase reveal a novel capping triad crucial for merozoite egress. PLoS Pathog. 2015;11:e1004639. doi: 10.1371/journal.ppat.1004639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kornev AP, Taylor SS, Ten Eyck LF. A generalized allosteric mechanism for cis-regulated cyclic nucleotide binding domains. PLoS Comput. Biol. 2008;4:e1000056. doi: 10.1371/journal.pcbi.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Merckx A, Bouyer G, Thomas SL, Langsley G, Egee S. Anion channels in Plasmodium-falciparum-infected erythrocytes and protein kinase A. Trends Parasitol. 2009;25:139–144. doi: 10.1016/j.pt.2008.12.005. [DOI] [PubMed] [Google Scholar]
- [12].Merckx A, Nivez MP, Bouyer G, Alano P, Langsley G, Deitsch K, Thomas S, Doerig C, Egee S. Plasmodium falciparum regulatory subunit of cAMP-dependent PKA and anion channel conductance. PLoS Pathog. 2008;4:e19. doi: 10.1371/journal.ppat.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Leykauf K, Treeck M, Gilson PR, Nebl T, Braulke T, Cowman AF, Gilberger TW, Crabb BS. Protein kinase a dependent phosphorylation of apical membrane antigen 1 plays an important role in erythrocyte invasion by the malaria parasite. PLoS Pathog. 2010;6:e1000941. doi: 10.1371/journal.ppat.1000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lasonder E, Green JL, Camarda G, Talabani H, Holder AA, Langsley G, Alano P. The Plasmodium falciparum schizont phosphoproteome reveals extensive phosphatidylinositol and cAMP-protein kinase A signaling. J. Proteome Res. 2012;11:5323–5337. doi: 10.1021/pr300557m. [DOI] [PubMed] [Google Scholar]
- [15].Dawn A, Singh S, More KR, Siddiqui FA, Pachikara N, Ramdani G, Langsley G, Chitnis CE. The central role of cAMP in regulating Plasmodium falciparum merozoite invasion of human erythrocytes. PLoS Pathog. 2014;10:e1004520. doi: 10.1371/journal.ppat.1004520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Collins CR, Hackett F, Strath M, Penzo M, Withers-Martinez C, Baker DA, Blackman MJ. Malaria parasite cGMP-dependent protein kinase regulates blood stage merozoite secretory organelle discharge and egress. PLoS Pathog. 2013;9:e1003344. doi: 10.1371/journal.ppat.1003344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cho-Chung YS, Clair T, Tagliaferri P, Ally S, Katsaros D, Tortora G, Neckers L, Avery TL, Crabtree GW, Robins RK. Site-selective cyclic AMP analogs as new biological tools in growth control, differentiation, and proto-oncogene regulation. Cancer Invest. 1989;7:161–177. doi: 10.3109/07357908909038282. [DOI] [PubMed] [Google Scholar]
- [18].Tortora G, Ciardiello F, Pepe S, Tagliaferri P, Ruggiero A, Bianco C, Guarrasi R, Miki K, Bianco AR. Phase I clinical study with 8-chloro-cAMP and evaluation of immunological effects in cancer patients. Clin. Cancer Res. 1995;1:377–384. [PubMed] [Google Scholar]
- [19].Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bonini NM, Nelson DL. Differential regulation of Paramecium ciliary motility by cAMP and cGMP. J. Cell Biol. 1988;106:1615–1623. doi: 10.1083/jcb.106.5.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bonini NM, Gustin MC, Nelson DL. Regulation of ciliary motility by membrane potential in Paramecium: a role for cyclic AMP. Cell Motil. Cytoskeleton. 1986;6:256–272. doi: 10.1002/cm.970060303. [DOI] [PubMed] [Google Scholar]
- [22].Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Jr., Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA, Krieger CJ, del Toro C, Ryder HF, Williamson SC, Barbeau RA, Hamilton EP, Orias E. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Taylor HM, McRobert L, Grainger M, Sicard A, Dluzewski AR, Hopp CS, Holder AA, Baker DA. The malaria parasite cyclic GMP-dependent protein kinase plays a central role in blood-stage schizogony. Eukaryot. Cell. 2010;9:37–45. doi: 10.1128/EC.00186-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Deng W, Parbhu-Patel A, Meyer DJ, Baker DA. The role of two novel regulatory sites in the activation of the cGMP-dependent protein kinase from Plasmodium falciparum. Biochem. J. 2003;374:559–565. doi: 10.1042/BJ20030474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wurtz N, Chapus C, Desplans J, Parzy D. cAMP-dependent protein kinase from Plasmodium falciparum: an update. Parasitology. 2011;138:1–25. doi: 10.1017/S003118201000096X. [DOI] [PubMed] [Google Scholar]
- [26].Gurnett AM, Liberator PA, Dulski PM, Salowe SP, Donald RG, Anderson JW, Wiltsie J, Diaz CA, Harris G, Chang B, Darkin-Rattray SJ, Nare B, Crumley T, Blum PS, Misura AS, Tamas T, Sardana MK, Yuan J, Biftu T, Schmatz DM. Purification and molecular characterization of cGMP-dependent protein kinase from apicomplexan parasites. A novel chemotherapeutic target. J. Biol. Chem. 2002;277:15913–15922. doi: 10.1074/jbc.M108393200. [DOI] [PubMed] [Google Scholar]
- [27].Laxman S, Beavo JA. Cyclic nucleotide signaling mechanisms in trypanosomes: possible targets for therapeutic agents. Mol. Interv. 2007;7:203–215. doi: 10.1124/mi.7.4.7. [DOI] [PubMed] [Google Scholar]
- [28].Treptau T, Piram P, Cook PF, Rodriguez PH, Hoffmann R, Jung S, Thalhofer HP, Harris BG, Hofer HW. Comparison of the substrate specificities of cAMP-dependent protein kinase from bovine heart and Ascaris suum muscle. Biol. Chem. Hoppe Seyler. 1996;377:203–209. doi: 10.1515/bchm3.1996.377.3.203. [DOI] [PubMed] [Google Scholar]
- [29].Saito-Ito A, He S, Kimura M, Matsumura T, Tanabe K. Cloning and structural analysis of the gene for cAMP-dependent protein kinase catalytic subunit from Plasmodium yoelii. Biochim. Biophys. Acta. 1995;1269:1–5. doi: 10.1016/0167-4889(95)00119-d. [DOI] [PubMed] [Google Scholar]
- [30].Nelböck M, Michal G, Weimann G. Analogues of cyclic AMP and their physiological response. Pure Appl. Chem. 1973;35:411–437. [Google Scholar]
- [31].Brelidze TI, Carlson AE, Sankaran B, Zagotta WN. Structure of the carboxyterminal region of a KCNH channel. Nature. 2012;481:530–533. doi: 10.1038/nature10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Brelidze TI, Gianulis EC, DiMaio F, Trudeau MC, Zagotta WN. Structure of the C-terminal region of an ERG channel and functional implications. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11648–11653. doi: 10.1073/pnas.1306887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Marques-Carvalho MJ, Sahoo N, Muskett FW, Vieira-Pires RS, Gabant G, Cadene M, Schonherr R, Morais-Cabral JH. Structural, biochemical, and functional characterization of the cyclic nucleotide binding homology domain from the mouse EAG1 potassium channel. J. Mol. Biol. 2012;423:34–46. doi: 10.1016/j.jmb.2012.06.025. [DOI] [PubMed] [Google Scholar]
- [34].Lanzilotta WN, Schuller DJ, Thorsteinsson MV, Kerby RL, Roberts GP, Poulos TL. Structure of the CO sensing transcription activator CooA. Nat. Struct. Biol. 2000;7:876–880. doi: 10.1038/82820. [DOI] [PubMed] [Google Scholar]
- [35].Joyce MG, Levy C, Gabor K, Pop SM, Biehl BD, Doukov TI, Ryter JM, Mazon H, Smidt H, van den Heuvel RH, Ragsdale SW, van der Oost J, Leys D. CprK crystal structures reveal mechanism for transcriptional control of halorespiration. J. Biol. Chem. 2006;281:28318–28325. doi: 10.1074/jbc.M602654200. [DOI] [PubMed] [Google Scholar]
- [36].Birkholtz LM, Wrenger C, Joubert F, Wells GA, Walter RD, Louw AI. Parasite-specific inserts in the bifunctional S-adenosylmethionine decarboxylase/ornithine decarboxylase of Plasmodium falciparum modulate catalytic activities and domain interactions. Biochem. J. 2004;377:439–448. doi: 10.1042/BJ20030614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gold MG, Gonen T, Scott JD. Local cAMP signaling in disease at a glance. J. Cell Sci. 2013;126:4537–4543. doi: 10.1242/jcs.133751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Meyer RB, Jr., Miller JP. Analogs of cyclic AMP and cyclic GMP: general methods of synthesis and the relationship of structure to enzymic activity. Life Sci. 1974;14:1019–1040. doi: 10.1016/0024-3205(74)90228-8. [DOI] [PubMed] [Google Scholar]
- [39].Steinberg RA, Agard DA. Turnover of regulatory subunit of cyclic AMP-dependent protein kinase in S49 mouse lymphoma cells. Regulation by catalytic subunit and analogs of cyclic AMP. J. Biol. Chem. 1981;256:10731–10734. [PubMed] [Google Scholar]
- [40].Tuganowski W, Kopec P, Gorczyca J, Tarnowski W, Grudzien E. The influence of dibutyryl cGMP on the rabbit auricle. Pol. J. Pharmacol. Pharm. 1978;30:73–75. [PubMed] [Google Scholar]
- [41].Schwede F, Maronde E, Genieser H, Jastorff B. Cyclic nucleotide analogs as bio-chemical tools and prospective drugs. Pharmacol. Ther. 2000;87:199–226. doi: 10.1016/s0163-7258(00)00051-6. [DOI] [PubMed] [Google Scholar]
- [42].Poppe H, Rybalkin SD, Rehmann H, Hinds TR, Tang XB, Christensen AE, Schwede F, Genieser HG, Bos JL, Doskeland SO, Beavo JA, Butt E. Cyclic nucleotide analogs as probes of signaling pathways. Nat. Methods. 2008;5:277–278. doi: 10.1038/nmeth0408-277. [DOI] [PubMed] [Google Scholar]
- [43].Eckstein F, Simonson LP, Bar HP. Adenosine 3′,5′-cyclic phosphorothioate: synthesis and biological properties. Biochemistry. 1974;13:3806–3810. doi: 10.1021/bi00715a029. [DOI] [PubMed] [Google Scholar]
- [44].Enserink JM, Christensen AE, de Rooij J, van Triest M, Schwede F, Genieser HG, Doskeland SO, Blank JL, Bos JL. A novel Epac-specific cAMP analogue demonstrates independent regulation of Rap1 and ERK. Nat. Cell Biol. 2002;4:901–906. doi: 10.1038/ncb874. [DOI] [PubMed] [Google Scholar]
- [45].Kang G, Joseph JW, Chepurny OG, Monaco M, Wheeler MB, Bos JL, Schwede F, Genieser HG, Holz GG. Epac-selective cAMP analog 8-pCPT-2′-O-Me-cAMP as a stimulus for Ca2+-induced Ca2+ release and exocytosis in pancreatic beta-cells. J. Biol. Chem. 2003;278:8279–8285. doi: 10.1074/jbc.M211682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bertinetti D, Schweinsberg S, Hanke SE, Schwede F, Bertinetti O, Drewianka S, Genieser HG, Herberg FW. Chemical tools selectively target components of the PKA system. BMC Chem. Biol. 2009;9:3. doi: 10.1186/1472-6769-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Almahariq M, Tsalkova T, Mei FC, Chen H, Zhou J, Sastry SK, Schwede F, Cheng X. A novel EPAC-specific inhibitor suppresses pancreatic cancer cell migration and invasion. Mol. Pharmacol. 2013;83:122–128. doi: 10.1124/mol.112.080689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Beebe SJ, Blackmore PF, Chrisman TD, Corbin JD. Use of synergistic pairs of site-selective cAMP analogs in intact cells. Methods Enzymol. 1988;159:118–139. doi: 10.1016/0076-6879(88)59013-4. [DOI] [PubMed] [Google Scholar]
- [49].Van Sande J, Lefort A, Beebe S, Roger P, Perret J, Corbin J, Dumont JE. Pairs of cyclic AMP analogs, that are specifically synergistic for type I and type II cAMP-dependent protein kinases, mimic thyrotropin effects on the function, differentiation expression and mitogenesis of dog thyroid cells. Eur. J. Biochem. 1989;183:699–708. doi: 10.1111/j.1432-1033.1989.tb21101.x. [DOI] [PubMed] [Google Scholar]
- [50].Neuwald AF. Rapid detection, classification and accurate alignment of up to a million or more related protein sequences. Bioinformatics. 2009;25:1869–1875. doi: 10.1093/bioinformatics/btp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- [52].Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, de Castro E, Coggill P, Corbett M, Das U, Daugherty L, Duquenne L, Finn RD, Fraser M, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, McMenamin C, Mi H, Mutowo-Muellenet P, Mulder N, Natale D, Orengo C, Pesseat S, Punta M, Quinn AF, Rivoire C, Sangrador-Vegas A, Selengut JD, Sigrist CJ, Scheremetjew M, Tate J, Thimmajanarthanan M, Thomas PD, Wu CH, Yeats C, Yong SY. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.