Giotopoulos et al. report a novel mouse model that closely mimics the natural progression of human chronic myeloid leukemia to blast crisis, and use this model to identify novel candidate genes and pathways that, in combination with BCR-ABL, drive disease progression.
Abstract
The introduction of highly selective ABL-tyrosine kinase inhibitors (TKIs) has revolutionized therapy for chronic myeloid leukemia (CML). However, TKIs are only efficacious in the chronic phase of the disease and effective therapies for TKI-refractory CML, or after progression to blast crisis (BC), are lacking. Whereas the chronic phase of CML is dependent on BCR-ABL, additional mutations are required for progression to BC. However, the identity of these mutations and the pathways they affect are poorly understood, hampering our ability to identify therapeutic targets and improve outcomes. Here, we describe a novel mouse model that allows identification of mechanisms of BC progression in an unbiased and tractable manner, using transposon-based insertional mutagenesis on the background of chronic phase CML. Our BC model is the first to faithfully recapitulate the phenotype, cellular and molecular biology of human CML progression. We report a heterogeneous and unique pattern of insertions identifying known and novel candidate genes and demonstrate that these pathways drive disease progression and provide potential targets for novel therapeutic strategies. Our model greatly informs the biology of CML progression and provides a potent resource for the development of candidate therapies to improve the dismal outcomes in this highly aggressive disease.
Chronic myeloid leukemia (CML) is a chronic myeloproliferative neoplasm, resulting from a reciprocal translocation between chromosomes 9 and 22, t(9;22)(q34;q11). This lesion was the first recurrent chromosomal abnormality described in cancer (Nowell and Hungerford, 1960; Rowley, 1973) and generates the BCR-ABL oncoprotein, a constitutively activated protein tyrosine kinase (TK; Deininger et al., 2000). Mouse models and human data have demonstrated BCR-ABL expression to be causative in CML (Daley et al., 1990; Heisterkamp et al., 1990; Zhao et al., 2001; Ramaraj et al., 2004; Koschmieder et al., 2005), and this observation has led to the paradigmic development of potent small molecule inhibitors that selectively target ABL enzymatic function and interrupt its oncogenic TK activity. Imatinib mesylate, the prototypic ABL tyrosine kinase inhibitor (TKI), and subsequent second and third generation TKIs, have revolutionized CML treatment (Druker et al., 1996; 2006; Carroll et al., 1997; Heinrich et al., 2000; O’Brien et al., 2003), significantly improving cytogenetic and molecular response rates, keeping the majority of patients in chronic phase, and prolonging overall survival (Druker et al., 2001, 2006; Sawyers et al., 2002; Hughes et al., 2003). However, despite this vast improvement, significant clinical challenges still remain in CML therapy. CML stem cells appear relatively resistant to the effects of TKIs (Copland et al., 2006; Jørgensen et al., 2007; Konig et al., 2008) such that, in the majority of patients, CML is controlled rather than cured. In addition, resistance occurs and this, together with stem cell persistence, facilitates disease transformation. Three distinct phases of the disease have been described. The initial phase, in which ∼85–90% of patients are diagnosed, is the indolent chronic phase (CP), which is readily amenable to treatment. However, without adequate therapy, this almost inevitably progresses to an aggressive acute leukemia of myeloid or lymphoid phenotype (70 and 30%, respectively), termed blast crisis (BC), which may be preceded by an ill-defined intermediate or accelerated phase (AP; during which the levels of myeloblasts in the BM or peripheral blood (PB) are increased but remain <20%). 10–15% of patients present beyond CP and a small percentage of CP cases continue to transform even on TKI therapy. The frequency of transformation is recorded at 3–5% within the first few years of TKI therapy but drops to ∼1% per year thereafter in randomized trials (Druker et al., 2006), although these values have been found to be higher in population-based studies (de Lavallade et al., 2008; Gallipoli et al., 2011). Treatment options for AP and BC are very limited, with response rates to TKIs lower and much less durable. Other options involve highly toxic therapies, such as combination chemotherapy and BM transplantation, and are not available or appropriate for many patients with progression. Therefore, even in the TKI era, the median survival of patients with BC is still dismal at around 6 mo (Hehlmann and Saussele, 2008; Silver et al., 2009), defining it as an unmet clinical need.
Although the chronic phase of CML appears almost entirely dependent on BCR-ABL and CML is regarded as an invaluable model of leukemic evolution, the molecular mechanisms underlying disease progression are still poorly annotated. It is generally accepted that additional mutations cooperate with BCR-ABL during progression to BC (Calabretta and Perrotti, 2004), as is demonstrated by the observation that >75% of BC patients harbor additional cytogenetic abnormalities (Mitelman and Levan, 1978; Radich, 2007). There is also good evidence that the BCR-ABL protein itself contributes to the acquisition of further mutations, through its effects on reactive oxygen species induction, DNA damage, DNA repair, apoptosis, and cellular growth (Perrotti et al., 2010; Nieborowska-Skorska et al., 2012; Bolton-Gillespie et al., 2013), and the levels of BCR-ABL protein can indeed increase in the transition from CP to BC (Gaiger et al., 1995). However, to date, only a small number of mutations in specific pathways have been associated with disease progression in CML. For example, mutations or deletions in TP53, ASXL1, and RUNX1 are commonly described in myeloid BC at frequencies ranging between 3 and 25% (Ahuja et al., 1989; Grossmann et al., 2011), 15 and 20% (Boultwood et al., 2010; Grossmann et al., 2011), and 13 and 33% (Grossmann et al., 2011; Zhao et al., 2012) of cases, respectively. Similarly, mutations or deletions in the CDKN2A/B and IKAROS genes have been reported in up to 50 and 80% of patients in lymphoid BCs, respectively (Sill et al., 1995; Mullighan et al., 2008). Modern sequencing technologies and lowered costs have refined the mutational landscape for many tumors (Pleasance et al., 2010a,b; Curtis et al., 2012; Cancer Genome Atlas Research Network, 2013), but as yet have only been used in a directed fashion in CML (Piccaluga et al., 2009; Boultwood et al., 2010; Grossmann et al., 2011). Therefore, the spectrum of mutations that cooperate with BCR-ABL and the majority of pathways and processes that are corrupted by these mutations during the progression of CML to advanced phases, particularly for myeloid transformation, have yet to be fully described.
Mouse models have greatly informed cancer biology in general and CML in particular. Several models have been previously generated, in which transgenic BCR-ABL expression is driven by several different promoters after either germline or retroviral integration (Hariharan et al., 1989; Castellanos et al., 1997; Honda et al., 1998; Huettner et al., 2000, 2003; Koschmieder et al., 2005). However, many of these models have failed to recapitulate the human disease by either generating predominantly acute lymphoid leukemias that lacked a preceding chronic phase, or a very rapidly fatal myeloproliferative neoplasm (MPN)–like disease not resembling the human counterpart (Daley et al., 1990; Honda et al., 1998; Huettner et al., 2000; Huettner et al., 2003). Models of BC have also been reported, where BCR-ABL expression has been combined with a known second hit, such as p53 or Dok1/Dok2 loss, or NUP98-HOXA9 or Hes1 overexpression (Skorski et al., 1996; Honda et al., 2000; Dash et al., 2002; Yasuda et al., 2004; Neering et al., 2007). Although confirmatory of the cooperation of specific mutations with BCR-ABL, these models have not informed the broader biology of BC due to their directed nature. Previous attempts to model random secondary mutations using retroviral insertional mutagenesis have also proven of limited value, with two reported studies only documenting three common insertions (Notch 1, Zfp423, and BCR-ABL; Mizuno et al., 2008; Miyazaki et al., 2009). Furthermore, the majority of these models have generated lymphoid leukemias, mainly T-ALL, thereby reducing their relevance for human disease.
We therefore set out to generate a novel mouse model of CML progression that would allow us to identify mechanisms of BC progression in an unbiased and tractable manner. Here, we have combined a mouse transposon-based insertional mutagenesis system with a published transgenic mouse model of chronic phase CML (Koschmieder et al., 2005). For the first time, we report a BC model that closely mimics the natural progression of human CML and faithfully recapitulates the cellular and molecular aspects of its biology. We have identified known and novel candidate genes and pathways that, in combination with BCR-ABL, drive disease progression and could act as potential therapeutic targets in BC. Our novel model therefore defines mechanisms of CML progression, identifies therapeutic targets and provides a translational resource to improve clinical outcomes in this aggressive disease.
RESULTS
Generation of a mouse model of CML progression
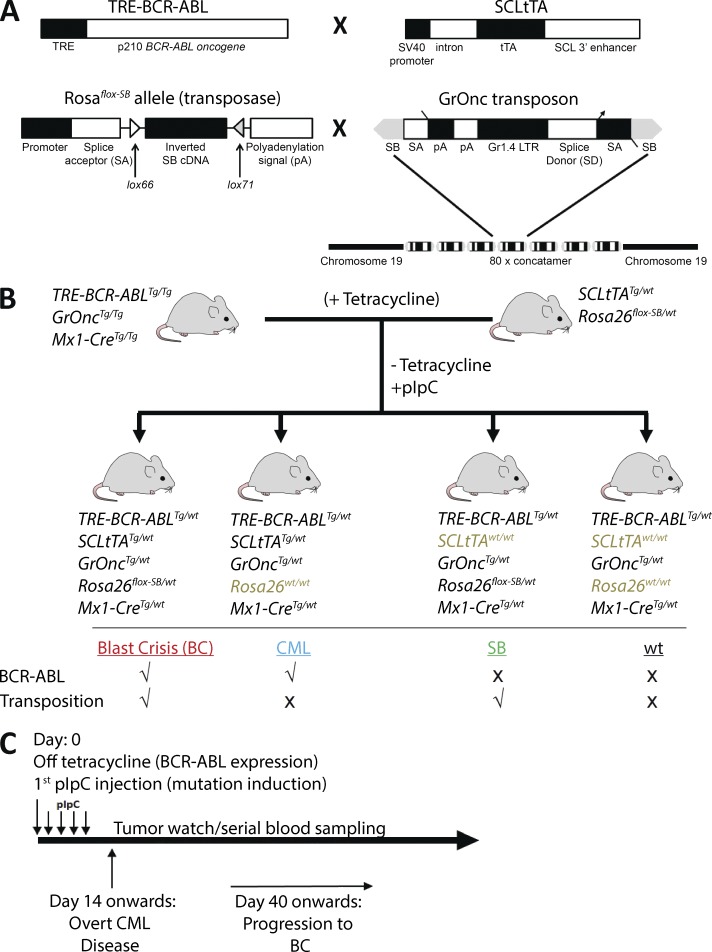
The CML and mutagenesis systems have been previously described (Koschmieder et al., 2005; March et al., 2011; Vassiliou et al., 2011). In brief, BCR-ABL (p210) was expressed in the hematopoietic stem and progenitor cell (HSPC) compartment under the control of the mouse 3′-SCL enhancer (Sánchez et al., 1999) in a tetracycline-dependent manner (Fig. 1 A). Forward mutagenesis was conditionally induced after Mx1-Cre–mediated inversion of the Rosa26flox-SB allele, leading to transposase expression and transposition of GrOnc, a Sleeping Beauty (SB) transposon (Fig. 1 A). To prevent undesired BCR-ABL expression or transposition before the generation of our experimental cohort, SCLtTATg/wt mice were crossbred with Rosa26flox-SB/wt mice to generate SCLtTATg/wt; Rosa26flox-SB/wt mice and TRE-BCR-ABLTg/wt mice were crossbred with Mx1-CreTg/wt; GrOncTg/wt mice to ultimately generate TRE-BCR-ABLTg/Tg; Mx1-CreTg/Tg; GrOncTg/Tg mice (Fig. S1 A). Fig. 1 B illustrates the final cross that generated our four experimental genotypes: TRE-BCR-ABLTg/wt; Mx1-CreTg/wt; GrOncTg/wt; SCLtTATg/wt; Rosa26flox-SB/wt (hereafter BC), which constituted the CML-mutagenesis cohort; TRE-BCR-ABLTg/wt; Mx1-CreTg/wt; GrOncTg/wt; SCLtTATg/wt; Rosa26wt/wt (hereafter CML), which constituted the CML-only cohort; TRE-BCR-ABLTg/wt; Mx1-CreTg/wt; GrOncTg/wt; SCLtTAwt/wt; Rosa26flox-SB/wt (hereafter SB), which constituted the mutagenesis-only cohort; and TRE-BCR-ABLTg/wt; Mx1-CreTg/wt; GrOncTg/wt; SCLtTAwt/wt; Rosa26wt/wt (hereafter wt), which were used as WT controls. The TRE-BCR-ABL and SCLtTA mice were on an FVB/N background, whereas the remaining elements were on a C57BL/6 background (i.e., the experimental cohorts were on a mixed genetic background). To induce expression of BCR-ABL and initiate mutagenesis, respectively, tetracycline was withdrawn from the drinking water and 5–8-wk-old mice were treated with 5 doses of polyinosine-polycyticylic acid (pIpC; Fig. 1 C). Successful expression of BCR-ABL and inversion of the SB transposase and transposon mobilization were confirmed by PCR of PB samples (Fig. 2, B and C; and not depicted).
Figure 1.
Generation of a mouse model of CML progression. (A) Expression of p210 BCR-ABL (TRE-BCR-ABL) is driven by an SCL 3′ enhancer element (SCLtTA transactivator) in a tetracycline-dependent manner (adapted from Koschmieder et al., 2005). Upon pIpC-induced and Mx1-Cre–mediated inversion, the Sleeping Beauty (SB) transposase (Rosa26flox-SB) is expressed and facilitates transposition of the GrOnc transposon. The transposon construct contains a concatamer of 80 U of the Graffi1.4 virus long terminal repeat (Gr1.4 LTR), splice acceptors (SA), bi-directional adenovirus polyadenylation signals (pA), and a splice donor (SD), all flanked by SB inverted repeats (adapted from March et al., 2011; Vassiliou et al., 2011). (B) The parental lines were established as described in Fig. S1. The experimental cohorts were generated by crossing TRE-BCR-ABLTg/Tg, GrOncTg/Tg, Mx1-CreTg/Tg mice with SCL-tTATg/wt, Rosa26flox-SB/wt mice, in the presence of tetracycline. (C) Upon weaning, mice were taken off tetracycline and treated with 5 doses of pIpC, simulating the temporal sequence of human BC transformation where mutagenesis occurs in the presence of BCR-ABL expression, leading to disease progression from a CML to BC phase. A CML phenotype was detected as early as 14 d and BC from as early as 40 d after tetracycline withdrawal.
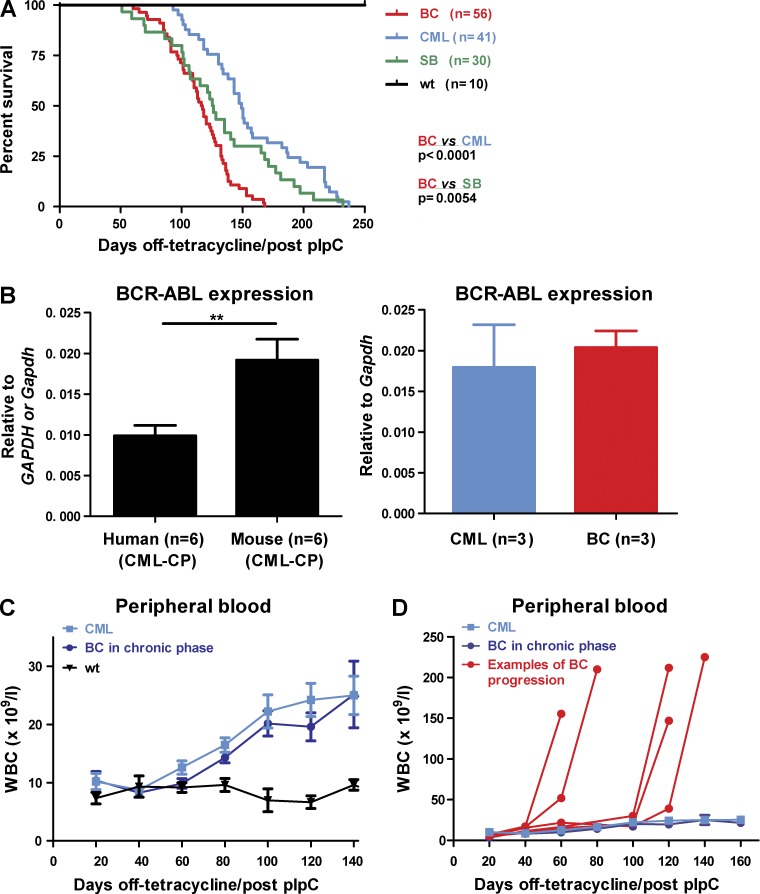
Figure 2.
Transposition leads to decreased survival and CML progression. (A) Kaplan-Meier curves showing survival of BC (n = 56) mice compared with CML (n = 41; P < 0.0001), SB (n = 30; P < 0.0054), and WT (n = 10; P < 0.0001) mice. Log-rank test was used. (B) BCR-ABL expression levels (n = 6 mice) were compared with human CML (n = 6 samples; left). BC (n = 3) and CML (n = 3) mice demonstrated similar levels of BCR-ABL expression throughout the study (right). Student’s t test was used. (C) Disease kinetics between CML and BC mice until the preterminal stages. Genotypic BC (dark blue) and CML mice (light blue) white blood counts (WBC) before transformation, when terminal bleeds for all groups of mice were excluded from the analysis (n = 8–26 mice at each time point). (D) Terminal WBC of representative BC cases (red) in comparison to CML mice (light blue). BC mice (by genotype) that remained in morphological chronic phase (dark blue) before sacrifice are shown for comparison (n = 1–26 mice at each time-point). Data are mean ± SEM. **, P < 0.01.
Transposition in BC mice leads to CML progression and decreased survival
Overall, BC mice demonstrated a decreased survival (median, 116.5 d) compared with both CML (median, 149 d; P < 0.0001) and SB mice (median, 125.5 d; P = 0.0054; Fig. 2 A), a strong indication of a cooperative effect between BCR-ABL and ongoing mutagenesis. The expression levels of BCR-ABL were similar between CML and BC animals and comparable to human CML samples (Fig. 2 B). Upon withdrawal of tetracycline, both CML and BC animals developed a moderate, but persistent (2–3-fold), increase of white blood counts (WBCs), with a marked expansion of the granulocyte compartment, as has been previously reported (Koschmieder et al., 2005). The two groups showed similar early disease kinetics, as assessed by serial WBC, regardless of the length of time from BCR-ABL expression and transposition (Fig. 2 C). However, BC mice demonstrated a dramatic preterminal proliferative change in the kinetics of their disease, with a marked increase in terminal WBC in comparison with CML mice (Fig. 2 D illustrates individual cases and Fig. 3 B averaged terminal WBC). These data demonstrate an explosive alteration in the clinical nature of the leukemia, similar to the usual clinical presentation in patients with AP/BC.
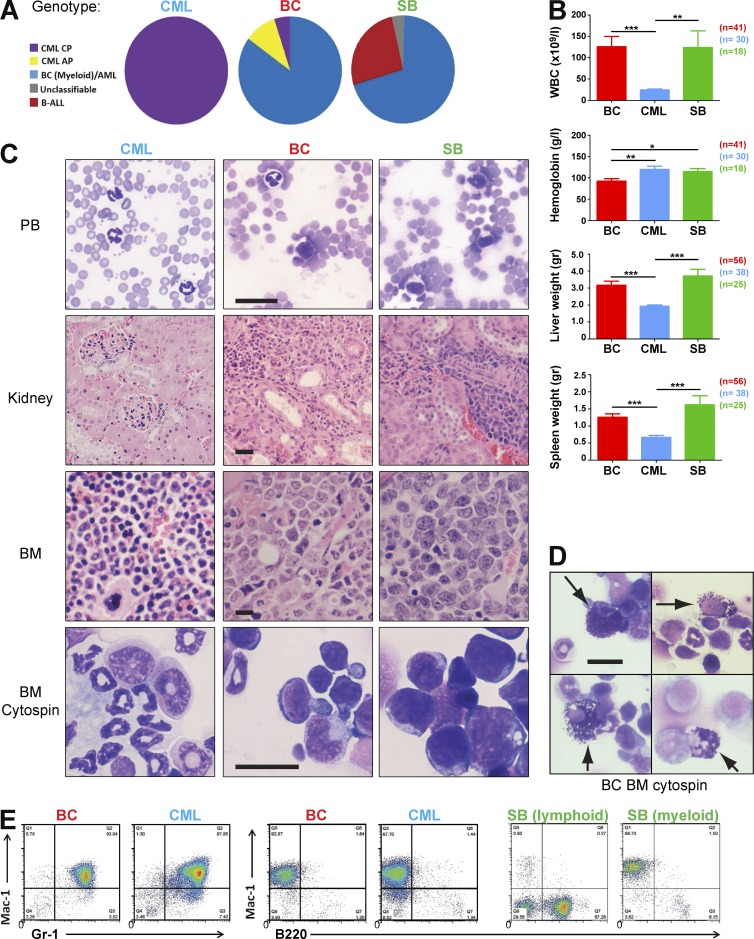
Figure 3.
Transposition leads to myeloid blast crisis. (A) Classification of all leukemias in each experimental cohort by the Bethesda criteria revealed a continuum of myeloid progression in the BC cohort with 85% demonstrating a myeloid transformation. In the SB cohort, 26% of cases were of a lymphoid phenotype. All mice in the CML cohort remained in the chronic phase of the disease. (B) Analysis of WBC, hemoglobin levels, and spleen and liver weights at the terminal endpoints between BC, CML, and SB mice (BC, n = 41; CML, n = 30; and SB, n = 18 for WBC and hemoglobin plots; BC, n = 56; CML, n = 38; and SB, n = 25 for liver and spleen weight plots). Student’s t test was used. (C) Peripheral blood (PB) smears, kidney, and BM histology sections and BM cytospins of representative CML, BC, and SB cases. (D) Cytospin preparations of BC BM from 4 cases showing characteristic abnormal basophils (arrowed), a feature often seen in human BC. (E) Representative flow cytometry profiles of CML, BC, and SB leukemias. Data are representative of at least three independent experiments. Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.0001. Bar, 25 µm.
Transposition generates myeloid BC
In addition to significant differences in disease latency, the experimental cohorts demonstrated marked macroscopic and microscopic differences in leukemia phenotype. All mice underwent macroscopic analysis at necropsy to determine the degree of splenomegaly and liver weight, as well as terminal WBC analysis, cytological analyses of PB smears, and BM cytospins, histology sections, and immunophenotyping by FACS. Individual cases were classified according to the Bethesda criteria for mouse hematological tumors (Kogan et al., 2002; Morse et al., 2002). As expected, all CML animals remained in the chronic phase of the disease. Animals belonging to the SB group developed both lymphoid (26%) and myeloid (70%) acute leukemias. However, in marked contrast to both of these cohorts, an exclusively myeloid and primarily acute leukemia phenotype was seen in the majority of the BC group (85%; Fig. 3 A). A small proportion of BC animals (5%) remained in the chronic phase of the disease and were indistinguishable from CML mice, whereas ∼10% of BC animals developed an AP-like phenotype, distinguishable from CML mice but with <20% blasts. These findings demonstrate a disease continuum in the progression cohort, similar to human CML (Fig. 3 A). Compared with the CML cohort, WBC counts at the terminal endpoint were significantly elevated (Fig. 3 B; P < 0.0001 vs. CML) and hemoglobin levels were decreased in the BC cohort (P < 0.001 vs. CML and P = 0.03 vs. SB). Spleen and liver weight were also significantly increased (P < 0.001 vs. CML) in BC mice. Examination of histological and cytospin preparations of PB, BM, and visceral organs demonstrated increased infiltration of tissues with both granulocytes and immature cells in BC mice, whereas only immature cells were seen in SB mice (Fig. 3 C). Interestingly, disease progression in the BC mice was frequently accompanied by a marked increase in basophils, an extremely rare cell type in mice, but a typical feature of disease progression in the human disease (Fig. 3 D). Assessment by flow cytometry confirmed the absence of lymphoid leukemias in the BC, but not in the SB cohorts (0 vs. 26%; P < 0.0001; Fisher’s exact test, two tailed; Fig. 3 E). These data demonstrate that BCR-ABL synergizes with the GrOnc transposon to promote myeloid but not lymphoid leukemogenesis, recapitulating many clinical and laboratory aspects of human myeloid BC.
Transposition alters the size and function of the HSPC compartment
To characterize alterations in the HSPC compartment after disease progression, we next extended our flow cytometry analyses to this compartment, comparing CML and BC mice. BC mice demonstrated a significant expansion of the lineage-negative (Lin−) BM fraction (Fig. 4 A; P < 0.0001), but no significant differences in total Lin−Sca+c-kit+ (LSK) numbers, long-term and short-term hematopoietic stem cell (HSC), or multipotent progenitor proportions (Fig. 4 A and Fig. S1 B). In contrast, the myeloid progenitor compartment was significantly expanded in the BC mice (Fig. 4 A; P < 0.0024). This expansion of myeloid progenitors, including the GMP compartment (Fig. 4, A and B; P = 0.0116), is in agreement with previous reports in human CML progression, where patients in BC or resistant to Imatinib demonstrated an expansion of the GMP pool (Jamieson et al., 2004). Whereas there was no difference in lymphoid compartment size between BC and CML mice, both demonstrated a marked decrease of lymphoid potential when compared with age-matched WT mice (Fig. 4 A; P < 0.001), with this feature potentially explaining the absence of lymphoid BCs in our model.
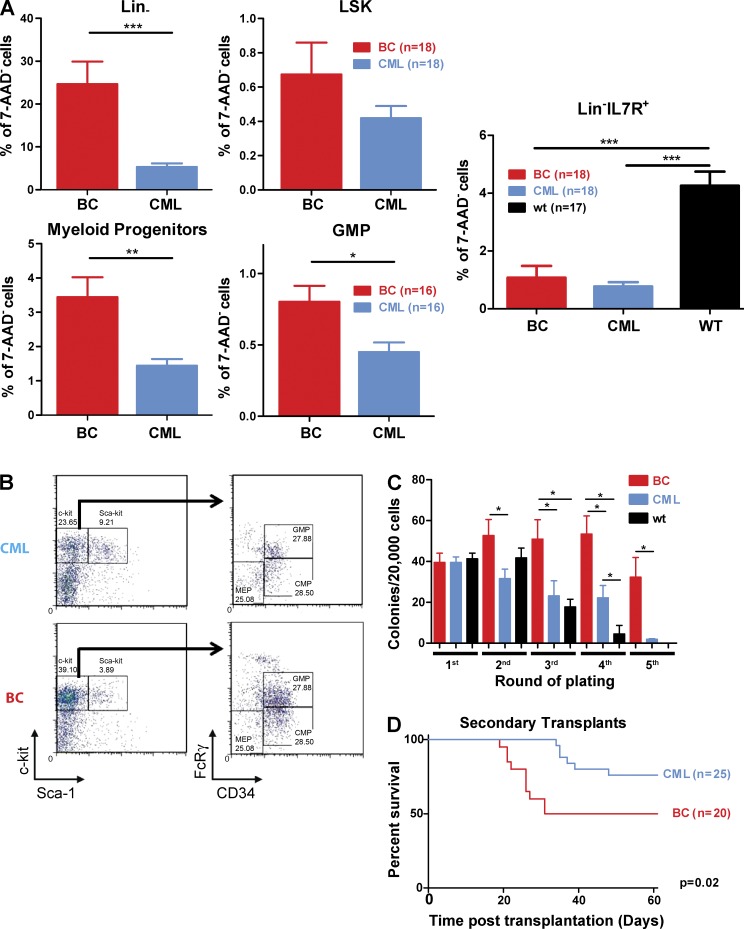
Figure 4.
Progression to blast crisis is accompanied by changes in the frequency and function of hematopoietic stem and progenitor cells. (A) The size/composition of the HSC (LSK) compartment between CML and BC mice (n = 18 for both groups), the myeloid progenitor compartment, including the GMP fraction (n = 16 for both groups) and the Lin−IL7R+ lymphoid progenitor cell (n = 18 for BC and CML and n = 17 for age-matched WT mice) compartments were assessed by flow cytometry. Student’s t test was used. (B) Representative FACS plots of the myeloid progenitor and GMP compartments between BC and SB mice. (C) Mean colony numbers for BC (n = 16), CML (n = 15), and age-matched WT (n = 11) mice in serial replating assays. Student’s t test was used. (D) Kaplan-Meier curves of secondary transplants of BC (n = 20) and CML (n = 25) cases into NOD-SCID recipients (P < 0.05). Log-rank test was used. Data are representative of at least three independent experiments. Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
To assess any alterations in HSPC function upon CML progression, we next performed serial replating assays to assess the self-renewal potential of BC and CML progenitors in vitro. BC cells demonstrated an increased ability to serially replate by comparison with CML and age-matched WT mice (Fig. 4 C). To test their self-renewal in vivo, we performed transplantation experiments using sublethally irradiated NOD-SCID mice as recipients. Both CML and BC cells were able to propagate their respective diseases in vivo, albeit with a compromised efficiency. As shown in Fig. 4 D, a significant increase in self-renewal is demonstrated in BC cases, where the rates of engraftment, disease transfer, and death were twice that of the CML mice. The disease latency was also significantly shorter (median survival 55.5 d for BC and >60 d for CML; P = 0.02). Collectively, these results confirm a dramatic alteration of HSPC regulation and function after transposition and CML progression, with an expansion of the myeloid progenitor compartments and an acquisition of enhanced self-renewal demonstrated both in vitro and in vivo.
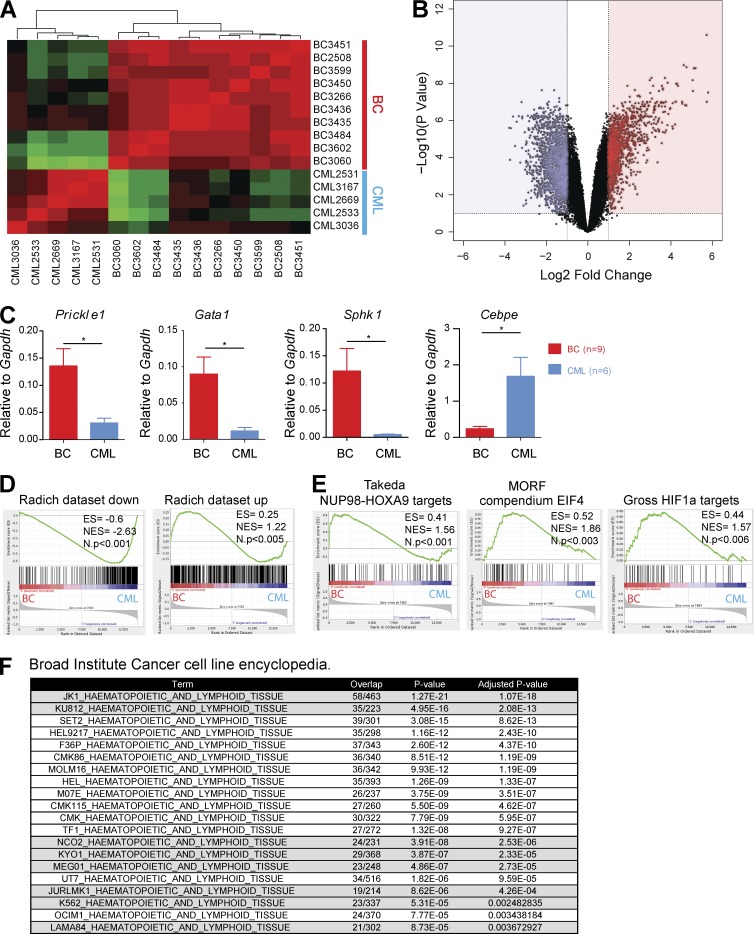
Disease progression induces gene expression changes similar to human BC
To determine molecular drivers of BC, we next examined alterations in gene expression that occurred after CML progression. Microarray analysis revealed distinct differences in gene expression between the CML and BC mice (Fig. 5 A). Hierarchical clustering of gene expression in 5 CML and 10 BC cases demonstrated consistency between gene expression and genotype, with CML and BC mice clustering separately. In total, 2,252 genes were differentially expressed between the two groups (910 up-regulated and 1,342 down-regulated upon disease progression, twofold gene expression difference; Fig. 5 B and Table S1). The expression levels of several genes were validated by qRT-PCR, notably genes that have previously been reported as deregulated in CML progression (Fig. 5 C; Guerrasio et al., 1994; Shimamoto et al., 1995; Radich et al., 2006; Ricci et al., 2009). To test for similarities with human BCs, we then used Gene Set Enrichment Analysis (GSEA; Subramanian et al., 2005) to directly compare our dataset with a widely used human CML-BC dataset (Radich et al., 2006). As illustrated in Fig. 5 D, our expression profiles were significantly enriched in genes reported to be both up- and down-regulated during progression to BC, demonstrating distinct similarities at the gene expression level between our mouse model and the corresponding human disease. Interestingly, additional GSEA analysis, comparing our patterns to MSigDB-curated gene sets, demonstrated other germane gene signatures significantly enriched in the BC cases, including pathways previously reported in human BCs, such as targets of NUP98-HOXA9 (Dash et al., 2002), HIF1a (Ng et al., 2014), Rapamycin, and EIF4E (Lim et al., 2013; Fig. 5 E). Our gene expression profiles were also highly concordant with reported human BC cell line data. In fact, when the list of BC up-regulated genes was compared with up-regulated gene signatures from 947 cell lines in the Cancer Cell Line Encyclopedia (CCLE), using the EnrichR gene set analysis tool (Barretina et al., 2012; Chen et al., 2013a), 8/20 (40%) of the significantly similar human cell lines were generated from patients in BC (P = 1.98 × 10−10; hypergeometric test; Fig. 5 F).
Figure 5.
Progression to BC generates gene expression profiles that are comparable to human blast crisis. (A) Unsupervised hierarchical clustering of gene expression data between CML (n = 5) and BC (n = 10) cases. (B) Volcano plot for BC (n = 10) versus CML (n = 5) samples showing fold change (log2) and p-value significance (−log10) for all genes. (C) Levels of expression for several genes previously identified to be involved in CML progression were validated by qPCR (n = 9 for BC; n = 6 for CML). Data are representative of at least three independent experiments. Student’s t test was used. (D) GSEA plots comparing our gene expression signature to that of a previously published human CML BC dataset. Genes down-regulated during human BC progression (Radich dataset down) and genes up-regulated during human BC progression (Radich dataset up). (E) GSEA plots showing enrichment of previously reported genes/pathways in CML progression including targets of NUP98-HOXA9, HIF1a, and EIF4 in our dataset. (F) The EnrichR tool enabled comparison of our BC up-regulated genes against a gene-set library of genes highly expressed in cancer cell lines from the CCLE database. A significant proportion (40%) of cell lines with a significant overlap of up-regulated genes were CML BC cell lines. Data are mean ± SEM. *, P < 0.05.
Identification of collaborating mutations required for CML progression
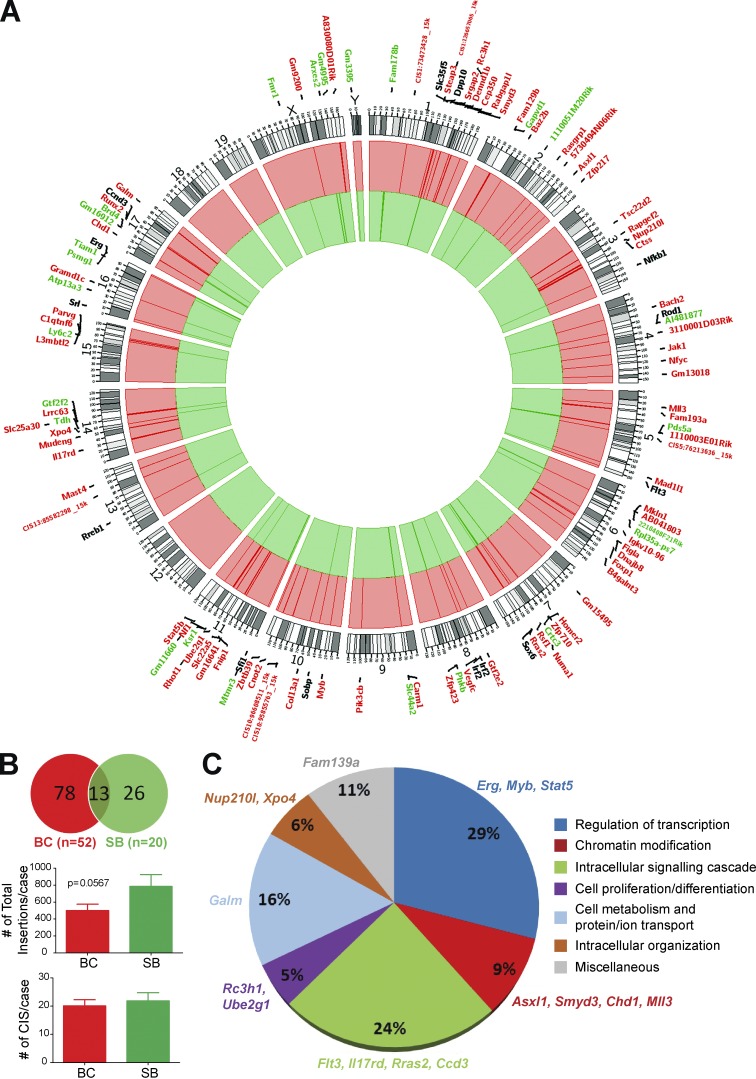
Our model was specifically designed to identify mutations that collaborated with BCR-ABL during CML progression, using ligation-mediated PCR followed by sequencing of transposon insertion sites from individual leukemias (Uren et al., 2009; van der Weyden et al., 2011; Vassiliou et al., 2011). Sequencing generated 534,493 reads, subsequently aligned to the mouse genome with a 44.41% success. These collapsed to 31,739 unique positions in the BC set and 18,301 unique positions in the SB set. 52 BC and 20 SB cases were successfully sequenced and statistically significant common insertion site (CIS) datasets were identified for each cohort. A total of 91 CISs were identified for the BC mice and 39 for the SB, with an average 19.8 CISs/case for the BC mice and 22 CISs/case for the SB mice (Fig. 6, A and B; and Tables S2 and S3). Insertions in chromosome 19 are listed separately to account for local hopping and are not represented in the circos plot, as chromosome 19 harbored the donor transposon concatamer. The number of total insertion events in the two cohorts also differed, with higher insertion rates in the SB (mean = 785.2 insertions/case) compared with the BC mice (mean = 501.1 insertions/case, P = 0.056; Fig. 6 B). These data would be in keeping with the hypothesis that fewer insertional events are required for leukemic transformation in the BC cohort. Of the 91 CISs identified in BC mice, 78 (86%) were specific to this cohort, whereas 26/39 (67%) CISs were unique to the SB cohort (Fig. 6 B), with 13 CISs shared between the two cohorts. Taken as a group and annotated by Gene Ontology (GO) terms, the BC CISs included genes involved in several critical cellular functions, including regulation of transcription, intracellular signaling, chromatin modification, and cellular metabolism (Fig. 6 C). These data demonstrate a high degree of heterogeneity in the pathways that may be affected in BC, very similar to the wide spectrum of dysregulation seen in other acute leukemias.
Figure 6.
Common insertion sites (CISs) in CML BC progression. (A) Circos plot of CISs in the BC (red) and SB (green) cohorts. Genes annotated in black were common to both cohorts. Note: chromosome 19 (the transposon donor) was analyzed separately and is not represented in this plot. (B) Pie chart of the CISs between the BC and SB groups. Bar graphs of the number of CISs per sample and the number of total insertion events between BC mice and SB mice. Student’s t test was used. (C) Functional annotation of the BC CISs according to their gene ontology term. BC (n = 52); SB (n = 20). Data are mean ± SEM. *, P < 0.05; ***, P < 0.0001.
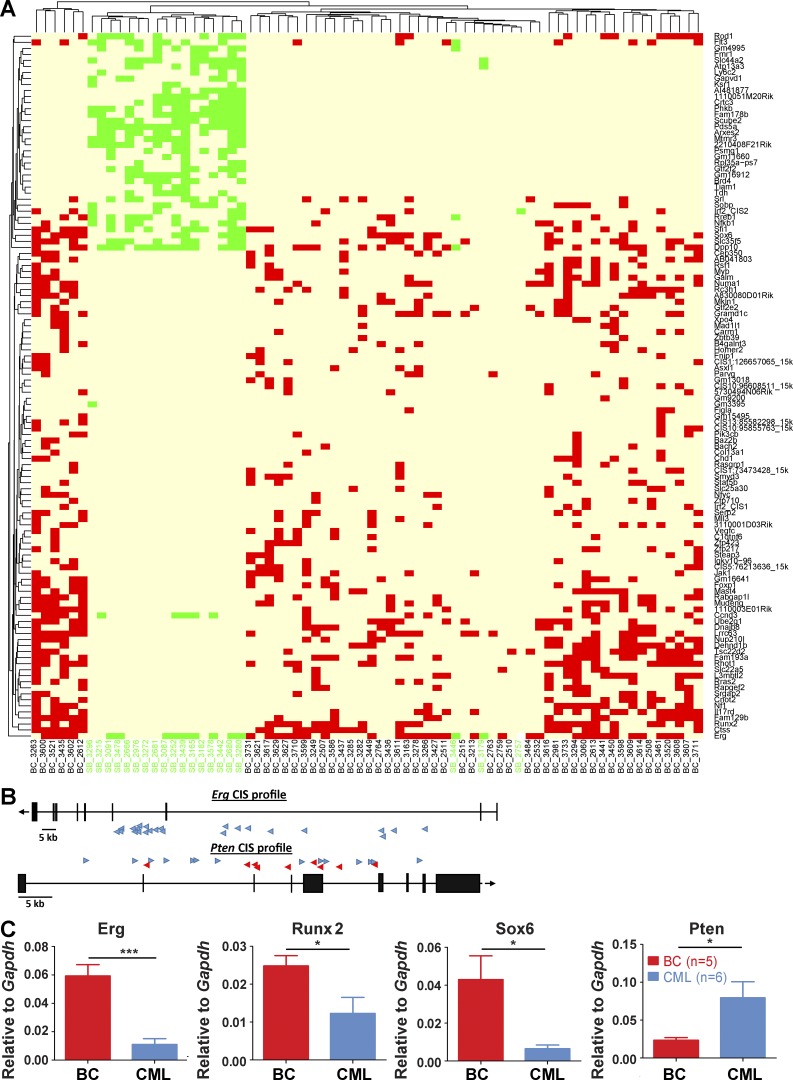
As shown in Fig. 6 A and Table S2, BC CISs include several genes already suggested to be involved in the biology of CML and its progression, including Asxl1, Myb, Stat5b, and Pten (Lidonnici et al., 2008; Boultwood et al., 2010; Peng et al., 2010; Grossmann et al., 2011; Machová Poláková et al., 2011; Makishima et al., 2011; Warsch et al., 2013; Ferri et al., 2014; Gallipoli et al., 2014; Huang et al., 2014). In addition, several novel genes not known to be associated with CML progression, but previously implicated in hematopoiesis or leukemogenesis were also identified, including Jak1, Flt3, Nf1, Erg, and Mll3. At the level of individual cases, BC samples grouped separately from SB samples using unsupervised clustering (Fig. 7 A), with an increased heterogeneity in the identity of their cooperating CISs compared with SB cases. Of interest, the number of cooperating CISs within an individual leukemic sample was also noted to be highly variable for BC cases, ranging from 1–52 insertions (median 18), with SB cases more homogeneous (insertion range 1–29, median 16 insertions; Fig. 7 A).
Figure 7.
Transposition leads to transcriptional deregulation and a distinct mutational signature in the BC cohort. (A) Unsupervised analysis of BC (n = 52) and SB (n = 20) insertion profiles by Euclidean distance, based on insertion number and identity. Each column represents an individual case with BC insertions in red and SB in green. (B) Insertions in the Erg gene were all found in the sense strand (blue arrows), strong indication of activating events. Conversely, insertions on the Pten gene were found in both strands (anti-sense strand in red arrows), suggesting inactivation of this gene. (C) Gene expression levels of selected CIS genes were investigated by qRT-PCR (BC, n = 5; CML, n = 6). Data are representative of at least three independent experiments. Student’s t test was used. Data are mean ± SEM. *, P < 0.05; ***, P < 0.0001.
Mechanisms of action of selected insertion sites and functional validation of cooperativity in CML progression
The GrOnc transposon can activate gene expression through insertion of its strong retroviral enhancer/promoter elements upstream of a gene or can alternatively result in gain or loss of function of genes after intragenic insertion (Ranzani et al., 2013). We documented the insertion location for all CISs and predicted their effects on the function of neighboring genes, as is shown in Fig. 7 (B and C). These included Erg, Runx2, and Sox6, where insertion of the transposon occurred uniquely in the sense strand, either upstream of a predicted transcriptional start site, or in the 5′ portion of the gene, leading to an up-regulation of expression of that gene in BC in comparison to CML (Fig. 7, B and C; and not depicted). Conversely, for other genes, exemplified by the tumor suppressor gene Pten, intragenic insertion of the transposon on either strand led to a down-regulation of gene expression (Fig. 7, B and C).
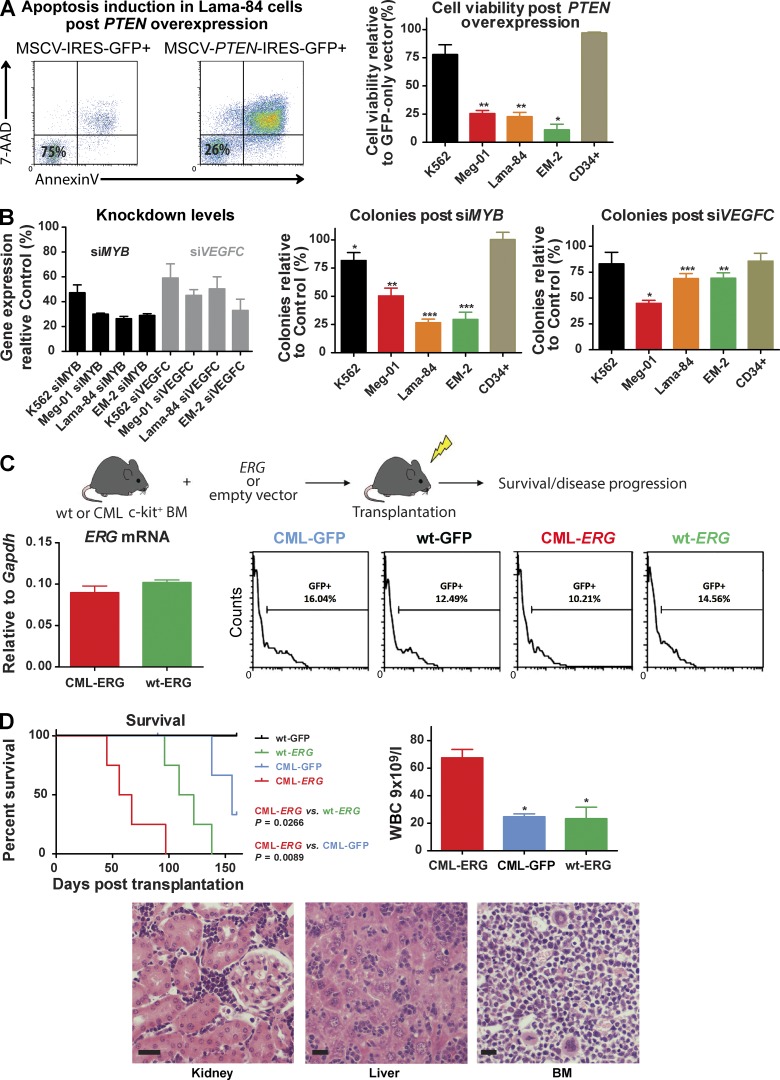
The ability of our model to faithfully recapitulate the cellular and molecular phenotype of BC suggests that our cooperating insertions are relevant for disease progression. To functionally test this hypothesis, we selected 4 candidate genes, Pten, Myb, Vegfc, and Erg for further analysis. Overexpression of PTEN in the human BC cell lines Lama-84, Meg-01, EM-2, and K562 led to an increased apoptotic rate by comparison with overexpression of an empty vector (Fig. 8 A). The rate of apoptosis varied between the cell lines but did not reflect general cellular toxicity, as expression of PTEN in CD34+ progenitor cells demonstrated minimal apoptosis (Fig. 8 A). In addition, modest knockdown of MYB and VEGFC (50–75% of baseline expression), using either pooled siRNA or shRNA, documented a significant decrement in colony formation, as well as increased apoptosis in the same BC cell lines (Fig. 8 B and not depicted). Finally, to provide a functional proof-of-principle for the in vivo cooperation of BCR-ABL with individual CISs, we retrovirally overexpressed ERG, our most commonly occurring CIS, or an empty vector in HSPC from both BCR-ABL–expressing and WT mice, before transplantation into congenic recipients (Fig. 8 C). Although ERG overexpression alone also generated leukemia in recipient mice, as has been previously reported (Goldberg et al., 2013), overexpression of ERG on a CML background resulted in a marked alteration in disease phenotype. Despite very similar transduction efficiencies (Fig. 8 C), CML-ERG mice demonstrated a significantly shortened survival compared with WT-ERG and CML-GFP mice (Fig. 8 D; median survival 61.5 d CML-ERG vs. 115.5 d WT-ERG vs. 156 d CML-GFP; P = 0.0266 CML-ERG vs. wt-ERG; and P = 0.0089 CML-ERG vs. CML-GFP, respectively). The resulting acute leukemias in the CML-ERG cohort phenotypically resembled the original BC cohort, with marked differences in the proportion of primitive cells in comparison to CML-GFP mice and a subtle phenotypic difference, including the WBC, when compared with the WT-ERG cohort (Fig. 8 D). Collectively, these data demonstrate the relevance of our candidate genes to the progression of CML and show that unique individual CISs identified from our cohort may be sufficient, in cooperation with BCR-ABL, to induce myeloid BC.
Figure 8.
Functional validation of CISs candidates. (A) Representative FACS plots and bar charts showing apoptosis induction levels in BC cell lines and normal CD34+ cells, 72 h after retroviral overexpression of PTEN. (B) siRNA-mediated knockdown of MYB or VEGFC effects on colony formation in BC cell lines. (C) Schema of experimental plan. WT or BCR-ABL+ c-kit+ HSPC cells were retrovirally transduced with ERG (or empty vector control) and transplanted into sublethally irradiated congenic recipients. The recipient mice were placed on tumor watch. ERG mRNA levels of CML-ERG and WT-ERG transduced cells (left). Engraftment levels across the four experimental cohorts in the early points after transplantation, as indicated by percentage of GFP+ cells (right). (D) Kaplan-Meier survival curves of CML-ERG (n = 4), CML-GFP (n = 4; P < 0.01) and WT-ERG (n = 4; P < 0.05) mice. Log-rank test was used. Bar charts of terminal white blood counts (Student’s t test was used) and representative photomicrographs of spleen, liver, and BM. Bar, 25 µM. Graphs in A and B represent three independent experiments. Student’s t test was used. Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
Integrated analysis of insertional and transcriptional candidates identify potential therapeutic targets in CML progression
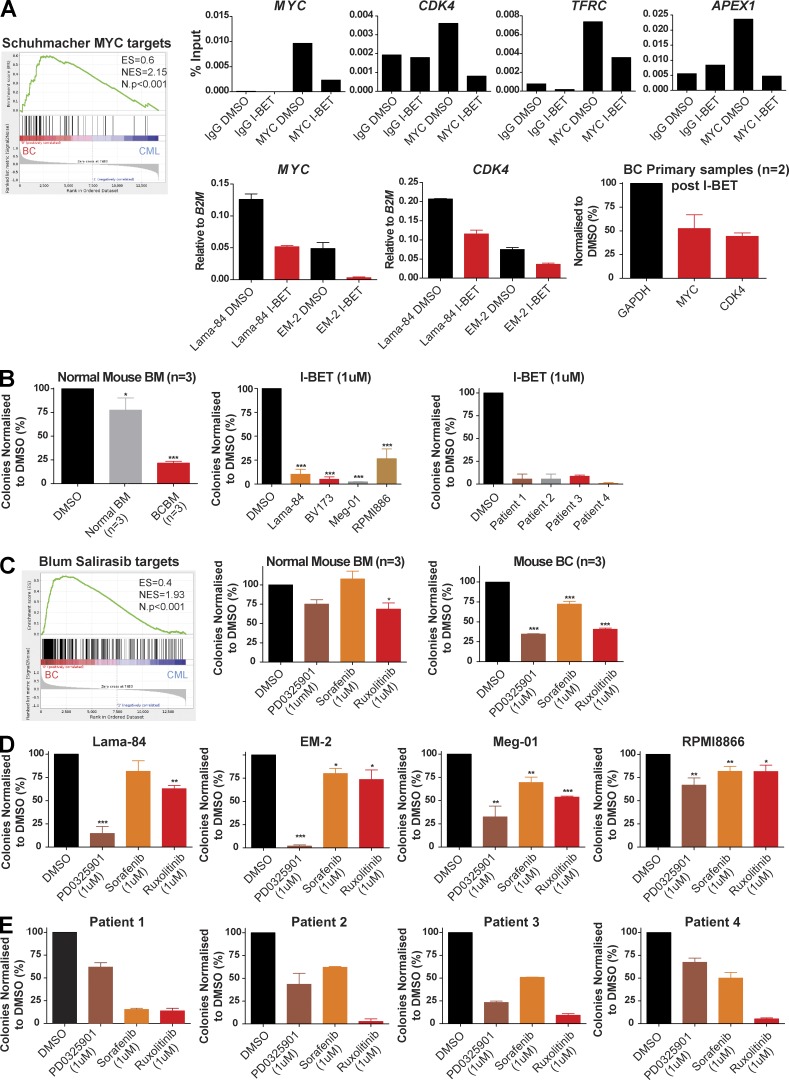
Novel therapies are urgently required for BC and the GSEA and transposon insertional analysis from our model identified several potential therapeutic targets implicated during CML progression. These included (targets of) MYC (Schuhmacher et al., 2001; Fig. 9 A), whose activation was corroborated by the demonstration of increased levels of MYC expression in CML BC samples from both our mouse model and human BC (unpublished data), as well as increased MYC binding at the promoters of known MYC targets (CDK4, TFRC, MYC, and APEX-1; Fig. 9 A). Using an available small molecule inhibitor to the Bromodomain and extra-terminal (BET) proteins, which regulate MYC levels and the MYC transcriptional program (Dawson et al., 2011), and is currently in Phase I trials we were further able to demonstrate that both MYC binding, as well as transcription decreased at these MYC targets in human BC cells upon treatment with I-BET (Fig. 9 A). This correlated with a marked decrement in colony formation for mouse BC cells, BC cell lines (Meg-01, Lama-84, EM-2, and RPMI8866), and samples from BC patients after I-BET treatment (Fig. 9 B and Table S4).
Figure 9.
Identification and in vitro validation of potential therapeutic targets in CML progression. (A) GSEA analysis identified Myc as a potential therapeutic target in BC. Representative ChIP results showing MYC binding at the promoters of known MYC targets (top). Effects of treatment with I-BET (24–48 h) on MYC expression (in Lama-84 and EM-2 cell lines) and mRNA levels of MYC target genes (bottom; in Lama-84, EM-2 cell lines and BC primary samples; n = 2). Data are representative of three independent experiments. (B) Mean colony numbers for control mouse BM (n = 3), BC mice (n = 3), human BC cell lines (n = 4; data represent three independent experiments; Student’s t test) and primary BC samples (n = 4; data represent two independent experiments) in serial replating assays using I-BET. (C) GSEA plot suggesting that the Ras–Raf–Mek–Erk pathway (inhibited by Salirasib) could be a therapeutic target in BC. Mean colony numbers for control mouse BM (n = 3), BC mice (n = 3) and human BC cell lines (n = 4) in serial replating assays using MEK (PD0325901), RAF (Sorafenib) or Jak1/2 (Ruxolitinib) inhibitors. Data represent three independent experiments. Student’s t test was used. (D) Mean colony numbers for four BC cell lines in serial replating assays using MEK (PD0325901), RAF (Sorafenib), or Jak1/2 (Ruxolitinib) inhibitors. Data represent three independent experiments. Student’s t test was used. (E) Mean colony numbers for primary BC patient samples (n = 4) in serial replating assays using MEK (PD0325901), RAF (Sorafenib), or Jak1/2 (Ruxolitinib) inhibitors. Data represent two independent experiments. Data are means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
Insertional and GSEA analysis also identified several other candidates/pathways suggested to be critical mediators of CML progression, including Ras intermediates and the JAK/STAT pathway (Fig. 6 A). We therefore also tested the effects of inhibition of these selected targets on blast-transformed samples, using available small molecule inhibitors. Specifically, Ruxolitinib (targeting the JAK/STAT pathway; Zhou et al., 2014), and PD0325901 and Sorafenib (targeting the MEK and RAF downstream mediators of Ras signaling, respectively; Adnane et al., 2006) were tested against mouse (Fig. 9 C) and human BC (Fig. 9, D and E) samples in similar methylcellulose plating assays. These inhibitors were chosen for their current usage in early phase trials or as established therapies for hematological and other malignancies, to allow rapid repurposing for formal testing in BC. No significant toxicity was demonstrated against normal hematopoietic cells. However, in marked contrast, when the same inhibitors were tested against representative mouse CML BC samples and human CML BC cell lines in similar methylcellulose colony forming assays, all of the inhibitors demonstrated a significant, although somewhat variable, decrement in colony formation, presumably reflecting the molecularly heterogeneous nature of CML BC. Finally, similar sensitivity assays for the inhibitors were performed in available samples from 4 CML BC patients (Fig. 9 E and Table S4). Although the degree of response again varied between patients, a decrease in colony formation was confirmed for all inhibitors. Collectively, these preliminary preclinical data confirm the MYC, JAK/STAT and RAS pathways as potential targets in BC and suggest that other novel insertion sites and expression patterns identified by our model may be important for disease progression and could therefore also be promising targets for treatment.
DISCUSSION
Mouse models have greatly facilitated our understanding of cancer biology, but have been most informative where they faithfully mimic aspects of the human disease. Our model, where both BCR-ABL expression and transposition could be conditionally and independently controlled, allowed us to induce mutagenesis in HSPCs that already express BCR-ABL. This control resulted in a model that recapitulated BC at the clinical and histological level, also modeling alterations in the cellular biology of the HSPC compartment. These alterations included an increase in myeloid progenitor numbers, serial replating in methylcellulose, and an increase in in vivo disease transfer in transplantation experiments for BC mice and mirrored similar studies suggesting an increased self-renewal in human progression (Sirard et al., 1996; Wang et al., 1998; Jamieson et al., 2004). Molecularly, gene expression changes were identified between CML and BC mice that also mirrored alterations seen in patients after progression (Radich et al., 2006). These data demonstrate our model to very closely resemble human myeloid transformation and highlight blocked differentiation, increased self-renewal and proliferation at the cellular and molecular levels as processes required for progression.
Importantly, our BC mice also significantly differed in their disease phenotype from SB mice that also develop acute leukemia but lack BCR-ABL expression. Molecularly, BC and SB mice shared less than a third of their insertion sites. Interestingly, although there was no significant difference in the number of insertions between individual SB and BC leukemias, the insertional spectrum was wider for BC mice with a more heterogeneous pattern of insertions identified (Fig. 7 A). This provides strong functional evidence that BCR-ABL expression provides a selective pressure for specific mutational partners and is more promiscuous in its partner selection. These data also demonstrate that BCR-ABL specifically cooperates with these partners to generate a unique leukemia phenotype. Of note, genes known to be mutated in CML (or previously implicated in CML biology/progression), as well as entirely novel genes, were identified as CISs. Collectively, these CISs formed complementation groups when annotated by GO term. These complementation groups mediated critical cellular functions, including chromatin modification, transcriptional regulation, and intracellular signaling, and the major groupings were highly reminiscent of similar subsets of mutations recently described from a large next generation sequencing (NGS) study in de novo human AML (Cancer Genome Atlas Research Network, 2013).
Several of our identified CISs have previously been implicated in CML biology and progression, particularly those involved in transcriptional regulation of the HSPC compartment. The most commonly identified CIS in our series was located in the Erg locus, with 33/52 (64%) of our BC leukemias demonstrating a 5′ insertion in the forward strand that up-regulated Erg expression (Fig. 7, B and C). ERG has been implicated in normal and leukemia stem cell function (Diffner et al., 2013), is an independent predictor of poor prognosis in normal karypotype AML (Marcucci et al., 2005), amplified in up to 10% of blast-phase transformations of other MPN (Thoennissen et al., 2010), and has been demonstrated to be overexpressed during CML evolution (Kim et al., 2010). Importantly, overexpression of Erg provided functional proof-of-principle that individual CISs identified from the model can cooperate with BCR-ABL to generate an independent BC in retroviral transduction-transplantation experiments (Fig. 8 C and D). The runt-related transcription factor Runx2 was our second most frequent CIS, occurring in half of the BC samples (26/52 cases). Of note, up to one-third of cases of myeloid BC have been reported to carry a mutation in the highly related human paralogue RUNX1, a gene critical for normal HSC activity that is commonly mutated in acute leukemias (Grossmann et al., 2011). Functional redundancy has also previously been demonstrated between Runx1 and Runx2 during mouse skeletal development (Smith et al., 2005), and our data suggest a similar complementation during transformation in our model. Another BC-specific CIS was Asxl1, a gene recurrently mutated or deleted in myeloid malignancies, including in 20% of CML myeloid BC (Boultwood et al., 2010; Grossmann et al., 2011), and an interaction partner of the PRC2 gene repressive complex and negative regulator of HOXA cluster genes (Abdel-Wahab et al., 2012). Similarly, Myb, a critical HSPC transcription factor (Mucenski et al., 1991) that has been described as important for BCR-ABL–mediated transformation, CML leukemia stem cell (LSC) function and a proposed therapeutic target in CP and BC (Ratajczak et al., 1992; Lidonnici et al., 2008), was also a recurrent insertion site in our BC cohort. So was Zfp423, a zinc finger transcription factor previously described as a recurrent insertion site in a retroviral insertional mutagenesis model of BC (Miyazaki et al., 2009).
For many human tumors, our recent understanding of the mechanisms underlying de novo cancer formation and disease progression has been greatly informed by annotation of the mutational landscape in large numbers of patient samples, using NGS technology (Curtis et al., 2012; Cancer Genome Atlas Research Network, 2013). Unfortunately, in CML only a small number of directed (Grossmann et al., 2011) or single case exome NGS studies (Menezes et al., 2013; Huang et al., 2014) have been performed. CISs from our model overlap with these studies and also overlap significantly with mutations described for human BC cell lines in the CCLE, where 13 cell lines with available NGS data exhibited mutations in 14/91 (15%) of our CISs (Table S5). These significant overlaps, along with the strong molecular and cellular similarities to human CML progression, further validate the utility of our model to identify critical regulators of CML progression, and highlight that novel CISs identified in our screen may also be important for CML progression. Further investigation of these novel CISs and comparisons to human disease is warranted.
Integrated analysis of insertion sites and gene expression data from our model identified several specific candidate targets and pathways amenable to therapeutic intervention, particularly those related to chromatin modification and intracellular signaling. Chromatin and epigenetic regulators are recurrently mutated in myeloid malignancies (Shih et al., 2012), and the recent availability of potent, specific small molecule inhibitors of these regulators holds great therapeutic promise (Dawson et al., 2012). GSEA analysis of our BC-associated signature suggested an up-regulation of c-MYC and its targets (Fig. 9 A), which was confirmed by MYC binding and transcriptional activity, demonstrating activation of this pathways during BC progression. This activity was further functionally corroborated, as inhibition of bromodomain and extraterminal (BET) proteins, critical mediators of MYC transcription, with I-BET demonstrated an obvious decrement in MYC binding and transcriptional output, as well as a decrease in clonogenic growth across BC samples from our mouse model, human BC cell lines, and patient samples. In addition, aberrant intracellular signaling is well described as a hallmark of cancer (Hanahan and Weinberg, 2011), and evidence from integration patterns and GSEA also suggested activation of multiple RAS pathway members (Rabgap1l, Rasgrp1, Rapgef2, Rras2, Nf1, Rhot1, and Rreb1; Fig. 9 C) in BC. This prediction proved entirely accurate, with BC samples sensitive to inhibition of downstream MEK (PD0325901) and RAF (Sorafenib) pathways. Another major canonical signaling pathway, the JAK/STAT pathway has recently been implicated in CML LSC maintenance and advocated as a clinical target in CP CML (Chen et al., 2013b; Neviani et al., 2013; Warsch et al., 2013; Gallipoli et al., 2014). Integrations in Jak1 and Stat5b in our model also supported evidence for its up-regulation during CML progression. This association further highlights the important link between mediators of LSC maintenance in CML and pathways up-regulated during progression, with the JAK-STAT pathway, Pten and Myb, all identified from our model, fulfilling this association (Lidonnici et al., 2008; Peng et al., 2010; Gallipoli et al., 2014). The importance of the JAK/STAT pathway to CML progression was demonstrated by the significant growth retardation in BC samples after inhibition with the dual JAK1/JAK2 inhibitor Ruxolitinib. Although the degree of inhibition varied, dependent on the inhibitor and sample tested, all were significant by comparison with control, suggesting the MYC, RAS, and JAK/STAT pathways to be generically activated to a greater or lesser degree in the majority of individual BC cases. Collectively, these data validate the utility of our model to identify targets and pathways critical for progression, to corroborate their role in BC maintenance, and to identify them as potential therapeutic targets. As our choice of inhibitors prioritized agents already in or close to clinical usage, these should be rapidly repurposed to test their efficacy in early phase trials of BC.
In summary, our mouse model carefully recapitulates the process of CML disease progression at the cellular and molecular levels, provides mechanistic detail of pathways and genes involved in this progression, and identifies potential therapeutic targets. For many rare cancers, large human sequencing datasets are difficult or impossible to obtain and indeed, these datasets are not available for BC. Our model is therefore particularly welcome and will form an excellent resource to further our understanding of BC and to identify and model therapeutics for this aggressive disease.
MATERIALS AND METHODS
Mice.
TRE-BCR-ABLTg/wt, SCLtTATg/wt (Koschmieder et al., 2005), and Mx1-CreTg/wt, GrOncTg/wt, and Rosa26flox-SB/wt (March et al., 2011; Vassiliou et al., 2011) were bred as illustrated in Fig. 1 A and Fig. S1 A. To suppress BCR-ABL expression, tetracycline hydrochloride (Sigma-Aldrich) was administered in the drinking water (0.5 g/liter). Mx1-Cre–mediated inversion of the transposase was induced by intraperitoneal injection of 5 doses of pIpC (300 µg/dose; Sigma-Aldrich). Peripheral blood was collected from the saphenous vein into EDTA-coated tubes (Sarstedt). White blood counts (WBCs) were enumerated using a Vet abc counter (Scil Animal Care). All mice were housed in a pathogen-free animal facility and were allowed unrestricted access to food and water. All experiments were conducted in accordance with UK (UK) Home Office regulations, under a UK Home Office project license.
PCR and qPCR.
Total DNA and RNA was extracted using phenol/chloroform (Invitrogen) and TRIzol (Ambion) methods, respectively. cDNA was synthesized using the SuperScript First-Strand RT-PCR kit (Invitrogen). All primers used for mouse genotyping and qPCR can be found in Table S6. Unless otherwise stated, PCR reaction conditions used 35 cycles and a 57°C annealing temperature. qPCR reactions were performed on a Stratagene MX 3000P qPCR system and data were analyzed using MXPro v4.10 software (Stratagene).
Histopathology.
Tissues were fixed in a 10% formalin solution (CellPath Ltd.) and subsequently embedded in paraffin blocks. Bones were decalcified using 0.38 M EDTA (pH7) solution. Tissue sections (4 µm) were stained with Hematoxylin and Eosin (Thermo Fisher Scientific) according to the manufacturer’s protocol. Peripheral blood (PB) and BM cytopsin preparations were fixed in methanol and subsequently stained with Eosin Y and Methylene blue (Rapid Romanowsky Stain; TCS Biosciences Ltd.).
Flow cytometry assays.
BM or spleen single-cell suspensions were prepared as previously described (Chan et al., 2011) and stained with combinations of the following anti–mouse antibodies (BioLegend, unless otherwise stated): CD11b(Mac1) (FITC-conjugated; SouthernBiotech), CD4 (phycoerythrin [PE] conjugated), Ly-6G(Gr-1) (PE-Cy7 conjugated), CD45R(B220) (allophycocyanin [APC]-conjugated; Invitrogen), CD117(c-kit) (PE-Cy7 conjugated), Ly-6A/E(Sca-1) (Pacific Blue [PB] conjugated), CD135(Flt3) (PE conjugated), CD34 (FITC conjugated; BD), CD127(IL-7Ra) (PE conjugated), CD16/32(FcRγ) (PE conjugated), and Mouse Lineage antibody cocktail (APC conjugated; BD). LT-HSC, ST-HSC, MPP, CMP, GMP, and MEP were defined as previously described (Chan et al., 2011). All analyses considered only 7-AAD− (BD) populations. Annexin V (APC conjugated; BD) and 7-AAD were used in cell viability assays according to the manufacturer’s protocol. Flow cytometry was performed on a CyAn ADP FlowCytometer (Dako) or a BD LSRFortessa cell analyzer and all data were analyzed with FlowJo software (Tree Star).
Serial replating assays.
Normal or leukemic mouse BM cells were plated at a concentration of 20,000 cells/plate (in duplicate) using MethoCult GF M3434 (STEMCELL Technologies) methylcellulose medium. Colonies were scored at 7–12 d and equal numbers of cells were replated using the same conditions.
Mouse transplantation experiments.
Unfractionated BM cells (0.5–1 × 106) from primary CML and BC cases were transplanted into sublethally irradiated (2 Gy) NOD-SCID mice, via tail vein injection. Similarly, retrovirally transduced mouse progenitor cells were injected into sublethally irradiated (5 Gy) 8–12-wk-old FVB/N recipients via tail vein injection (106 cells/mouse).
Gene expression profiles and bioinformatic analysis.
Gene expression profiling was performed using a Mouse WG-6 v2.0 expression beadchip kit according to the manufacturer’s protocol (Illumina). Illumina probe sequences were mapped against the mouse reference genome using BLAT. Data analysis was performed using the limma and lumi Bioconductor packages. Batch effects were corrected using COMBAT. Gene set enrichment analysis tools were obtained from The Broad Institute. Default settings and all curated gene sets (c2.All.v4.0.symbols.gmt) were used for the analysis. Gene expression signatures were compared with CCLE (CCLE Broad Institute) human cell lines using the Enrichr tools (Chen et al., 2013a).
Identification of CISs.
Preparation of genomic DNA, splinkerette PCR, and sequencing of barcoded samples have been previously described (Uren et al., 2009; March et al., 2011). In brief, genomic DNA from mouse leukemias underwent restriction enzyme digestion and a splinkerette adaptor was added using ligation-mediated PCR. Leukemia samples were pooled and sequenced on the 454 GS-GLX platform, with each sample identifiable by a unique barcode, allowing deconvolution of the sequencing results for individual cases. Pooled PCR reactions were sequenced on 454-GS-FLX sequencers (Roche). The bioinformatics pipeline used for quality control, sequence annotation, and mapping has been previously described (March et al., 2011). Statistically significant common insertion sites (CISs) were determined using a Gaussian kernel convolution approach (de Ridder et al., 2006; March et al., 2011) and chromosome significance cut-off levels of P < 0.05.
siRNA knockdown assays.
Accell SMARTpool siRNA pools for VEGFC, MYB, as well as a nontargeting pool, were obtained from GE Dharmacon and delivered in human BC cell lines according to the manufacturer’s recommendations. In brief, human BC cell lines were resuspended in Accell Delivery Media at a concentration of 500,000 cells/ml. siRNA pools were resuspended in 1× siRNA buffer and added to the cell suspension at a final concentration of 1 µM. Cells were incubated for 48 h and subsequently plated in methylcellulose. mRNA levels were assessed 72 h after treatment using primer pairs found in Table S6.
Retroviral transduction assays.
TransIT-LT1 transfection reagent (Mirus) was used to transfect the MSCV-ERG3-IRES-GFP vector (Diffner et al., 2013) and the packaging plasmid psiEco into 293T cells according to the manufacturer’s protocol. PTEN (NM_00314) was cloned into an MSCV-IRES-GFP vector and transduced into human cells using TransIT-LT1 and the amphotropic Phoenix 293T-derived cell line. Retroviral supernatants were collected at 48 and 72 h after transfection. BM cells from 8–12-wk-old BCR-ABL–expressing and WT FVB/N mice were selected for cell surface c-kit expression using CD117 MicroBeads (Miltenyi Biotec) according to the manufacturer’s protocol. Cells were spinoculated with retroviruses as previously described (Kvinlaug et al., 2011).
Cell culture and inhibitor assays.
Meg-01, EM-2, Lama-84 and RPMI8866 cells were grown in RPMI-1640 medium supplemented with FBS and penicillin/streptomycin (10–20 and 1% final concentration, respectively; Sigma-Aldrich). Mouse (20,000 cells/plate), human CML BC cell lines (1–5,000 cells/plate), and human CML BC patient primary cells (20–50,000 cells/plate) were plated in duplicate using MethoCult GF M3434, MethoCult H4531, and MethoCult H4435 enriched, respectively, in the presence of a small molecule inhibitor or DMSO. Colonies were scored at 7–12 d.
Chromatin immunoprecipitation (ChIP) and ChIP-PCR assays.
ChIP was performed on human CML BC cells harvested 24–48 h after treatment as previously described (Dawson et al., 2011). In brief, 107 cells were fixed in 1% formaldehyde for 15 min at room temperature. The cross-linking reaction was stopped by addition of glycine (0.125 M final concentration) and additional incubation of 10 min. Cells were washed in PBS and cell pellets were lysed in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.0, and 1 mM sodium orthovanadate and protease inhibitors). Chromatin was sonicated in a Bioruptor (Diagenode) sonicator and precleared for 1 h before immunoprecipitation in equal volumes of protein A and G beads (Dynabeads; Life Technologies). Immunoprecipitation was performed at 4°C overnight in modified RIPA buffer (1% Triton X-100, 0.1% deoxycholate, 0.1% SDS, 90 mM NaCl, 10 mM Tris-HCl pH 8.0, 1 mM sodium orthovanadate, and EDTA-free protease inhibitors) in the presence 2.5 µg of IgG (I5006; Sigma-Aldrich) or c-Myc (sc-764; Santa Cruz Biotechnology) antibodies and A/G beads. DNA was subsequently RNase treated, reverse cross-linked, and purified using QIAquick PCR purification kit (QIAGEN). ChIP-PCR was performed with SYBR Green PCR mastermix using the ABI Prism 7000 system (Applied Biosystems). The primers used can be found in Table S6.
Patient material.
Patient BM or peripheral blood cells (>80% blasts) were obtained after donor/patient consent and under full ethical approval.
Statistical analysis.
Unless otherwise stated, all statistical analyses used Student’s t test on raw data. P-values ≤ 0.05 were considered statistically significant. Survival curves were constructed using the Kaplan-Meier method and statistical significance was determined using log-rank analysis.
Online supplemental material.
Table S1 lists the differentially expressed genes between BC and CML BM samples (adjusted P < 0.05). Table S2 contains the BC CIS list (P < 0.05). Table S3 shows the SB CIS list (P < 0.05). Table S4 contains the CML BC patient details that were used in Fig. 9. Table S5 shows the overlapping mutations between our BC CIS and several human CML BC cell lines (as reported in CCLE). All primer sequences that were used in this study can be found in Table S5. All tables are available as Excel files. Fig. S1 shows the breeding strategy to generate the parental mouse lines and plots of the HSC composition in CML and BC mice as assessed by flow cytometry. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20141661/DC1.
Supplementary Material
Acknowledgments
Work in the Huntly laboratory is funded by CRUK, The European Research Council (ERC), Leukaemia Lymphoma Research, the Kay Kendall Leukaemia Fund, Wellcome Trust, the Medical Research Council (UK), the Leukemia Lymphoma Society America and the Cambridge NIHR Biomedical Research centre. David Adams is funded by Cancer Research UK and Wellcome Trust. Steffen Koschmieder has received funding from Deutsche José Carreras Leukämie-Stiftung (DJCLS; grant 10/23).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AP
- accelerated phase
- BC
- blast crisis
- BET
- bromodomain and extra-terminal (proteins)
- CCLE
- cancer cell line encyclopedia
- CIS
- common insertion site
- CML
- chronic myeloid leukemia
- CP
- chronic phase
- GO
- gene ontology
- GSEA
- gene set enrichment analysis
- HSC
- hematopoietic stem cell
- HSPC
- hematopoietic stem and progenitor cell
- Lin−
- lineage negative
- LSC
- leukemia stem cell
- MPN
- myeloproliferative neoplasm
- NGS
- next generation sequencing
- PB
- peripheral blood
- pIpC
- polyinosine-polycyticylic acid
- SB
- sleeping beauty
- TK
- tyrosine kinase
- TKI
- TK inhibitor
- WBC
- white blood counts
References
- Abdel-Wahab O., Adli M., LaFave L.M., Gao J., Hricik T., Shih A.H., Pandey S., Patel J.P., Chung Y.R., Koche R., et al. 2012. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 22:180–193. 10.1016/j.ccr.2012.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adnane L., Trail P.A., Taylor I., and Wilhelm S.M.. 2006. Sorafenib (BAY 43-9006, Nexavar), a dual-action inhibitor that targets RAF/MEK/ERK pathway in tumor cells and tyrosine kinases VEGFR/PDGFR in tumor vasculature. Methods Enzymol. 407:597–612. 10.1016/S0076-6879(05)07047-3 [DOI] [PubMed] [Google Scholar]
- Ahuja H., Bar-Eli M., Advani S.H., Benchimol S., and Cline M.J.. 1989. Alterations in the p53 gene and the clonal evolution of the blast crisis of chronic myelocytic leukemia. Proc. Natl. Acad. Sci. USA. 86:6783–6787. 10.1073/pnas.86.17.6783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D., et al. 2012. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 483:603–607. 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton-Gillespie E., Schemionek M., Klein H.U., Flis S., Hoser G., Lange T., Nieborowska-Skorska M., Maier J., Kerstiens L., Koptyra M., et al. 2013. Genomic instability may originate from imatinib-refractory chronic myeloid leukemia stem cells. Blood. 121:4175–4183. 10.1182/blood-2012-11-466938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boultwood J., Perry J., Zaman R., Fernandez-Santamaria C., Littlewood T., Kusec R., Pellagatti A., Wang L., Clark R.E., and Wainscoat J.S.. 2010. High-density single nucleotide polymorphism array analysis and ASXL1 gene mutation screening in chronic myeloid leukemia during disease progression. Leukemia. 24:1139–1145. 10.1038/leu.2010.65 [DOI] [PubMed] [Google Scholar]
- Calabretta B., and Perrotti D.. 2004. The biology of CML blast crisis. Blood. 103:4010–4022. 10.1182/blood-2003-12-4111 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368:2059–2074. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll M., Ohno-Jones S., Tamura S., Buchdunger E., Zimmermann J., Lydon N.B., Gilliland D.G., and Druker B.J.. 1997. CGP 57148, a tyrosine kinase inhibitor, inhibits the growth of cells expressing BCR-ABL, TEL-ABL, and TEL-PDGFR fusion proteins. Blood. 90:4947–4952. [PubMed] [Google Scholar]
- Castellanos A., Pintado B., Weruaga E., Arévalo R., López A., Orfao A., and Sánchez-García I.. 1997. A BCR-ABL(p190) fusion gene made by homologous recombination causes B-cell acute lymphoblastic leukemias in chimeric mice with independence of the endogenous bcr product. Blood. 90:2168–2174. [PubMed] [Google Scholar]
- Chan W.I., Hannah R.L., Dawson M.A., Pridans C., Foster D., Joshi A., Göttgens B., Van Deursen J.M., and Huntly B.J.. 2011. The transcriptional coactivator Cbp regulates self-renewal and differentiation in adult hematopoietic stem cells. Mol. Cell. Biol. 31:5046–5060. 10.1128/MCB.05830-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., and Ma’ayan A.. 2013a. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 14:128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Gallipoli P., DeGeer D., Sloma I., Forrest D.L., Chan M., Lai D., Jorgensen H., Ringrose A., Wang H.M., et al. 2013b. Targeting primitive chronic myeloid leukemia cells by effective inhibition of a new AHI-1-BCR-ABL-JAK2 complex. J. Natl. Cancer Inst. 105:405–423. 10.1093/jnci/djt006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copland M., Hamilton A., Elrick L.J., Baird J.W., Allan E.K., Jordanides N., Barow M., Mountford J.C., and Holyoake T.L.. 2006. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood. 107:4532–4539. 10.1182/blood-2005-07-2947 [DOI] [PubMed] [Google Scholar]
- Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y., et al. METABRIC Group. 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 486:346–352. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley G.Q., Van Etten R.A., and Baltimore D.. 1990. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 247:824–830. 10.1126/science.2406902 [DOI] [PubMed] [Google Scholar]
- Dash A.B., Williams I.R., Kutok J.L., Tomasson M.H., Anastasiadou E., Lindahl K., Li S., Van Etten R.A., Borrow J., Housman D., et al. 2002. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc. Natl. Acad. Sci. USA. 99:7622–7627. 10.1073/pnas.102583199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M.A., Prinjha R.K., Dittmann A., Giotopoulos G., Bantscheff M., Chan W.I., Robson S.C., Chung C.W., Hopf C., Savitski M.M., et al. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 478:529–533. 10.1038/nature10509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M.A., Kouzarides T., and Huntly B.J.. 2012. Targeting epigenetic readers in cancer. N. Engl. J. Med. 367:647–657. 10.1056/NEJMra1112635 [DOI] [PubMed] [Google Scholar]
- de Lavallade H., Apperley J.F., Khorashad J.S., Milojkovic D., Reid A.G., Bua M., Szydlo R., Olavarria E., Kaeda J., Goldman J.M., and Marin D.. 2008. Imatinib for newly diagnosed patients with chronic myeloid leukemia: incidence of sustained responses in an intention-to-treat analysis. J. Clin. Oncol. 26:3358–3363. 10.1200/JCO.2007.15.8154 [DOI] [PubMed] [Google Scholar]
- de Ridder J., Uren A., Kool J., Reinders M., and Wessels L.. 2006. Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLOS Comput. Biol. 2:e166 10.1371/journal.pcbi.0020166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger M.W., Goldman J.M., and Melo J.V.. 2000. The molecular biology of chronic myeloid leukemia. Blood. 96:3343–3356. [PubMed] [Google Scholar]
- Diffner E., Beck D., Gudgin E., Thoms J.A., Knezevic K., Pridans C., Foster S., Goode D., Lim W.K., Boelen L., et al. 2013. Activity of a heptad of transcription factors is associated with stem cell programs and clinical outcome in acute myeloid leukemia. Blood. 121:2289–2300. 10.1182/blood-2012-07-446120 [DOI] [PubMed] [Google Scholar]
- Druker B.J., Tamura S., Buchdunger E., Ohno S., Segal G.M., Fanning S., Zimmermann J., and Lydon N.B.. 1996. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat. Med. 2:561–566. 10.1038/nm0596-561 [DOI] [PubMed] [Google Scholar]
- Druker B.J., Sawyers C.L., Kantarjian H., Resta D.J., Reese S.F., Ford J.M., Capdeville R., and Talpaz M.. 2001. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N. Engl. J. Med. 344:1038–1042. 10.1056/NEJM200104053441402 [DOI] [PubMed] [Google Scholar]
- Druker B.J., Guilhot F., O’Brien S.G., Gathmann I., Kantarjian H., Gattermann N., Deininger M.W., Silver R.T., Goldman J.M., Stone R.M., et al. IRIS Investigators. 2006. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 355:2408–2417. 10.1056/NEJMoa062867 [DOI] [PubMed] [Google Scholar]
- Ferri C., Bianchini M., Bengió R., and Larripa I.. 2014. Expression of LYN and PTEN genes in chronic myeloid leukemia and their importance in therapeutic strategy. Blood Cells Mol. Dis. 52:121–125. 10.1016/j.bcmd.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Gaiger A., Henn T., Hörth E., Geissler K., Mitterbauer G., Maier-Dobersberger T., Greinix H., Mannhalter C., Haas O.A., Lechner K., and Lion T.. 1995. Increase of bcr-abl chimeric mRNA expression in tumor cells of patients with chronic myeloid leukemia precedes disease progression. Blood. 86:2371–2378. [PubMed] [Google Scholar]
- Gallipoli P., Shepherd P., Irvine D., Drummond M., and Holyoake T.. 2011. Restricted access to second generation tyrosine kinase inhibitors in the UK could result in suboptimal treatment for almost half of chronic myeloid leukaemia patients: results from a West of Scotland and Lothian population study. Br. J. Haematol. 155:128–130. 10.1111/j.1365-2141.2011.08653.x [DOI] [PubMed] [Google Scholar]
- Gallipoli P., Cook A., Rhodes S., Hopcroft L., Wheadon H., Whetton A.D., Jorgensen H.G., Bhatia R., and Holyoake T.L.. 2014. JAK2/STAT5 inhibition by nilotinib with ruxolitinib contributes to the elimination of chronic phase CML CD34+ cells in vitro and in vivo. Blood. 21:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg L., Tijssen M.R., Birger Y., Hannah R.L., Kinston S.J., Schütte J., Beck D., Knezevic K., Schiby G., Jacob-Hirsch J., et al. 2013. Genome-scale expression and transcription factor binding profiles reveal therapeutic targets in transgenic ERG myeloid leukemia. Blood. 122:2694–2703. 10.1182/blood-2013-01-477133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann V., Kohlmann A., Zenger M., Schindela S., Eder C., Weissmann S., Schnittger S., Kern W., Müller M.C., Hochhaus A., et al. 2011. A deep-sequencing study of chronic myeloid leukemia patients in blast crisis (BC-CML) detects mutations in 76.9% of cases. Leukemia. 25:557–560. 10.1038/leu.2010.298 [DOI] [PubMed] [Google Scholar]
- Guerrasio A., Saglio G., Rosso C., Alfarano A., Camaschella C., Lo Coco F., Biondi A., Rambaldi A., Nicolis S., and Ottolenghi S.. 1994. Expression of GATA-1 mRNA in human myeloid leukemic cells. Leukemia. 8:1034–1038. [PubMed] [Google Scholar]
- Hanahan D., and Weinberg R.A.. 2011. Hallmarks of cancer: the next generation. Cell. 144:646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hariharan I.K., Harris A.W., Crawford M., Abud H., Webb E., Cory S., and Adams J.M.. 1989. A bcr-v-abl oncogene induces lymphomas in transgenic mice. Mol. Cell. Biol. 9:2798–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehlmann R., and Saussele S.. 2008. Treatment of chronic myeloid leukemia in blast crisis. Haematologica. 93:1765–1769. 10.3324/haematol.2008.001214 [DOI] [PubMed] [Google Scholar]
- Heinrich M.C., Griffith D.J., Druker B.J., Wait C.L., Ott K.A., and Zigler A.J.. 2000. Inhibition of c-kit receptor tyrosine kinase activity by STI 571, a selective tyrosine kinase inhibitor. Blood. 96:925–932. [PubMed] [Google Scholar]
- Heisterkamp N., Jenster G., ten Hoeve J., Zovich D., Pattengale P.K., and Groffen J.. 1990. Acute leukaemia in bcr/abl transgenic mice. Nature. 344:251–253. 10.1038/344251a0 [DOI] [PubMed] [Google Scholar]
- Honda H., Oda H., Suzuki T., Takahashi T., Witte O.N., Ozawa K., Ishikawa T., Yazaki Y., and Hirai H.. 1998. Development of acute lymphoblastic leukemia and myeloproliferative disorder in transgenic mice expressing p210bcr/abl: a novel transgenic model for human Ph1-positive leukemias. Blood. 91:2067–2075. [PubMed] [Google Scholar]
- Honda H., Ushijima T., Wakazono K., Oda H., Tanaka Y., Aizawa Si., Ishikawa T., Yazaki Y., and Hirai H.. 2000. Acquired loss of p53 induces blastic transformation in p210(bcr/abl)-expressing hematopoietic cells: a transgenic study for blast crisis of human CML. Blood. 95:1144–1150. [PubMed] [Google Scholar]
- Huang Y., Zheng J., Hu J.D., Wu Y.A., Zheng X.Y., Liu T.B., and Chen F.L.. 2014. Discovery of somatic mutations in the progression of chronic myeloid leukemia by whole-exome sequencing. Genet. Mol. Res. 13:945–953. 10.4238/2014.February.19.5 [DOI] [PubMed] [Google Scholar]
- Huettner C.S., Zhang P., Van Etten R.A., and Tenen D.G.. 2000. Reversibility of acute B-cell leukaemia induced by BCR-ABL1. Nat. Genet. 24:57–60. 10.1038/71691 [DOI] [PubMed] [Google Scholar]
- Huettner C.S., Koschmieder S., Iwasaki H., Iwasaki-Arai J., Radomska H.S., Akashi K., and Tenen D.G.. 2003. Inducible expression of BCR/ABL using human CD34 regulatory elements results in a megakaryocytic myeloproliferative syndrome. Blood. 102:3363–3370. 10.1182/blood-2003-03-0768 [DOI] [PubMed] [Google Scholar]
- Hughes T.P., Kaeda J., Branford S., Rudzki Z., Hochhaus A., Hensley M.L., Gathmann I., Bolton A.E., van Hoomissen I.C., Goldman J.M., and Radich J.P.. International Randomised Study of Interferon versus STI571 (IRIS) Study Group. 2003. Frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukemia. N. Engl. J. Med. 349:1423–1432. 10.1056/NEJMoa030513 [DOI] [PubMed] [Google Scholar]
- Jamieson C.H., Ailles L.E., Dylla S.J., Muijtjens M., Jones C., Zehnder J.L., Gotlib J., Li K., Manz M.G., Keating A., et al. 2004. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 351:657–667. 10.1056/NEJMoa040258 [DOI] [PubMed] [Google Scholar]
- Jørgensen H.G., Allan E.K., Jordanides N.E., Mountford J.C., and Holyoake T.L.. 2007. Nilotinib exerts equipotent antiproliferative effects to imatinib and does not induce apoptosis in CD34+ CML cells. Blood. 109:4016–4019. 10.1182/blood-2006-11-057521 [DOI] [PubMed] [Google Scholar]
- Kim K.I., Park J., Ahn K.S., Won N.H., Kim B.K., Shin W.G., Yoon S.S., and Oh J.M.. 2010. Molecular characterization and prognostic significance of FLT3 in CML progression. Leuk. Res. 34:995–1001. 10.1016/j.leukres.2009.11.008 [DOI] [PubMed] [Google Scholar]
- Kogan S.C., Ward J.M., Anver M.R., Berman J.J., Brayton C., Cardiff R.D., Carter J.S., de Coronado S., Downing J.R., Fredrickson T.N., et al. Hematopathology subcommittee of the Mouse Models of Human Cancers Consortium. 2002. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 100:238–245. 10.1182/blood.V100.1.238 [DOI] [PubMed] [Google Scholar]
- Konig H., Holtz M., Modi H., Manley P., Holyoake T.L., Forman S.J., and Bhatia R.. 2008. Enhanced BCR-ABL kinase inhibition does not result in increased inhibition of downstream signaling pathways or increased growth suppression in CML progenitors. Leukemia. 22:748–755. 10.1038/sj.leu.2405086 [DOI] [PubMed] [Google Scholar]
- Koschmieder S., Göttgens B., Zhang P., Iwasaki-Arai J., Akashi K., Kutok J.L., Dayaram T., Geary K., Green A.R., Tenen D.G., and Huettner C.S.. 2005. Inducible chronic phase of myeloid leukemia with expansion of hematopoietic stem cells in a transgenic model of BCR-ABL leukemogenesis. Blood. 105:324–334. 10.1182/blood-2003-12-4369 [DOI] [PubMed] [Google Scholar]
- Kvinlaug B.T., Chan W.I., Bullinger L., Ramaswami M., Sears C., Foster D., Lazic S.E., Okabe R., Benner A., Lee B.H., et al. 2011. Common and overlapping oncogenic pathways contribute to the evolution of acute myeloid leukemias. Cancer Res. 71:4117–4129. 10.1158/0008-5472.CAN-11-0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidonnici M.R., Corradini F., Waldron T., Bender T.P., and Calabretta B.. 2008. Requirement of c-Myb for p210(BCR/ABL)-dependent transformation of hematopoietic progenitors and leukemogenesis. Blood. 111:4771–4779. 10.1182/blood-2007-08-105072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S., Saw T.Y., Zhang M., Janes M.R., Nacro K., Hill J., Lim A.Q., Chang C.T., Fruman D.A., Rizzieri D.A., et al. 2013. Targeting of the MNK-eIF4E axis in blast crisis chronic myeloid leukemia inhibits leukemia stem cell function. Proc. Natl. Acad. Sci. USA. 110:E2298–E2307. 10.1073/pnas.1301838110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machová Poláková K., Lopotová T., Klamová H., Burda P., Trněný M., Stopka T., and Moravcová J.. 2011. Expression patterns of microRNAs associated with CML phases and their disease related targets. Mol. Cancer. 10:41. 10.1186/1476-4598-10-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima H., Jankowska A.M., McDevitt M.A., O’Keefe C., Dujardin S., Cazzolli H., Przychodzen B., Prince C., Nicoll J., Siddaiah H., et al. 2011. CBL, CBLB, TET2, ASXL1, and IDH1/2 mutations and additional chromosomal aberrations constitute molecular events in chronic myelogenous leukemia. Blood. 117:e198–e206. 10.1182/blood-2010-06-292433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- March H.N., Rust A.G., Wright N.A., ten Hoeve J., de Ridder J., Eldridge M., van der Weyden L., Berns A., Gadiot J., Uren A., et al. 2011. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat. Genet. 43:1202–1209. 10.1038/ng.990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G., Baldus C.D., Ruppert A.S., Radmacher M.D., Mrózek K., Whitman S.P., Kolitz J.E., Edwards C.G., Vardiman J.W., Powell B.L., et al. 2005. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: a Cancer and Leukemia Group B study. J. Clin. Oncol. 23:9234–9242. 10.1200/JCO.2005.03.6137 [DOI] [PubMed] [Google Scholar]
- Menezes J., Salgado R.N., Acquadro F., Gómez-López G., Carralero M.C., Barroso A., Mercadillo F., Espinosa-Hevia L., Talavera-Casañas J.G., Pisano D.G., et al. 2013. ASXL1, TP53 and IKZF3 mutations are present in the chronic phase and blast crisis of chronic myeloid leukemia. Blood Cancer J. 3:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F., and Levan G.. 1978. Clustering of aberrations to specific chromosomes in human neoplasms. III. Incidence and geographic distribution of chromosome aberrations in 856 cases. Hereditas. 89:207–232. [DOI] [PubMed] [Google Scholar]
- Miyazaki K., Yamasaki N., Oda H., Kuwata T., Kanno Y., Miyazaki M., Komeno Y., Kitaura J., Honda Z., Warming S., et al. 2009. Enhanced expression of p210BCR/ABL and aberrant expression of Zfp423/ZNF423 induce blast crisis of chronic myelogenous leukemia. Blood. 113:4702–4710. 10.1182/blood-2007-05-088724 [DOI] [PubMed] [Google Scholar]
- Mizuno T., Yamasaki N., Miyazaki K., Tazaki T., Koller R., Oda H., Honda Z.I., Ochi M., Wolff L., and Honda H.. 2008. Overexpression/enhanced kinase activity of BCR/ABL and altered expression of Notch1 induced acute leukemia in p210BCR/ABL transgenic mice. Oncogene. 27:3465–3474. 10.1038/sj.onc.1211007 [DOI] [PubMed] [Google Scholar]
- Morse H.C. III, Anver M.R., Fredrickson T.N., Haines D.C., Harris A.W., Harris N.L., Jaffe E.S., Kogan S.C., MacLennan I.C., Pattengale P.K., and Ward J.M.. Hematopathology subcommittee of the Mouse Models of Human Cancers Consortium. 2002. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 100:246–258. 10.1182/blood.V100.1.246 [DOI] [PubMed] [Google Scholar]
- Mucenski M.L., McLain K., Kier A.B., Swerdlow S.H., Schreiner C.M., Miller T.A., Pietryga D.W., Scott W.J. Jr., and Potter S.S.. 1991. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 65:677–689. 10.1016/0092-8674(91)90099-K [DOI] [PubMed] [Google Scholar]
- Mullighan C.G., Miller C.B., Radtke I., Phillips L.A., Dalton J., Ma J., White D., Hughes T.P., Le Beau M.M., Pui C.H., et al. 2008. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 453:110–114. 10.1038/nature06866 [DOI] [PubMed] [Google Scholar]
- Neering S.J., Bushnell T., Sozer S., Ashton J., Rossi R.M., Wang P.Y., Bell D.R., Heinrich D., Bottaro A., and Jordan C.T.. 2007. Leukemia stem cells in a genetically defined murine model of blast-crisis CML. Blood. 110:2578–2585. 10.1182/blood-2007-02-073031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neviani P., Harb J.G., Oaks J.J., Santhanam R., Walker C.J., Ellis J.J., Ferenchak G., Dorrance A.M., Paisie C.A., Eiring A.M., et al. 2013. PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. J. Clin. Invest. 123:4144–4157. 10.1172/JCI68951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.P., Manjeri A., Lee K.L., Huang W., Tan S.Y., Chuah C.T., Poellinger L., and Ong S.T.. 2014. Physiologic hypoxia promotes maintenance of CML stem cells despite effective BCR-ABL1 inhibition. Blood. 123:3316–3326. 10.1182/blood-2013-07-511907 [DOI] [PubMed] [Google Scholar]
- Nieborowska-Skorska M., Kopinski P.K., Ray R., Hoser G., Ngaba D., Flis S., Cramer K., Reddy M.M., Koptyra M., Penserga T., et al. 2012. Rac2-MRC-cIII-generated ROS cause genomic instability in chronic myeloid leukemia stem cells and primitive progenitors. Blood. 119:4253–4263. 10.1182/blood-2011-10-385658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell P.C., and Hungerford D.A.. 1960. A minute chromosome in human chronic granulocytic leukemia. Science. 142:1497. [DOI] [PubMed] [Google Scholar]
- O’Brien S.G., Guilhot F., Larson R.A., Gathmann I., Baccarani M., Cervantes F., Cornelissen J.J., Fischer T., Hochhaus A., Hughes T., et al. IRIS Investigators. 2003. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N. Engl. J. Med. 348:994–1004. [DOI] [PubMed] [Google Scholar]
- Peng C., Chen Y., Yang Z., Zhang H., Osterby L., Rosmarin A.G., and Li S.. 2010. PTEN is a tumor suppressor in CML stem cells and BCR-ABL-induced leukemias in mice. Blood. 115:626–635. 10.1182/blood-2009-06-228130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti D., Jamieson C., Goldman J., and Skorski T.. 2010. Chronic myeloid leukemia: mechanisms of blastic transformation. J. Clin. Invest. 120:2254–2264. 10.1172/JCI41246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccaluga P.P., Sabattini E., Bacci F., Agostinelli C., Righi S., Salmi F., Testoni N., Paolini S., Castagnetti F., Martinelli G., et al. 2009. Cytoplasmic mutated nucleophosmin (NPM1) in blast crisis of chronic myeloid leukaemia. Leukemia. 23:1370–1371. 10.1038/leu.2009.95 [DOI] [PubMed] [Google Scholar]
- Pleasance E.D., Cheetham R.K., Stephens P.J., McBride D.J., Humphray S.J., Greenman C.D., Varela I., Lin M.L., Ordóñez G.R., Bignell G.R., et al. 2010a. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 463:191–196. 10.1038/nature08658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance E.D., Stephens P.J., O’Meara S., McBride D.J., Meynert A., Jones D., Lin M.L., Beare D., Lau K.W., Greenman C., et al. 2010b. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 463:184–190. 10.1038/nature08629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radich J.P. 2007. The Biology of CML blast crisis. Hematology (Am Soc Hematol Educ Program). 2007:384–391. 10.1182/asheducation-2007.1.384 [DOI] [PubMed] [Google Scholar]
- Radich J.P., Dai H., Mao M., Oehler V., Schelter J., Druker B., Sawyers C., Shah N., Stock W., Willman C.L., et al. 2006. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc. Natl. Acad. Sci. USA. 103:2794–2799. 10.1073/pnas.0510423103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaraj P., Singh H., Niu N., Chu S., Holtz M., Yee J.K., and Bhatia R.. 2004. Effect of mutational inactivation of tyrosine kinase activity on BCR/ABL-induced abnormalities in cell growth and adhesion in human hematopoietic progenitors. Cancer Res. 64:5322–5331. 10.1158/0008-5472.CAN-03-3656 [DOI] [PubMed] [Google Scholar]
- Ranzani M., Annunziato S., Adams D.J., and Montini E.. 2013. Cancer gene discovery: exploiting insertional mutagenesis. Mol. Cancer Res. 11:1141–1158. 10.1158/1541-7786.MCR-13-0244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak M.Z., Hijiya N., Catani L., DeRiel K., Luger S.M., McGlave P., and Gewirtz A.M.. 1992. Acute- and chronic-phase chronic myelogenous leukemia colony-forming units are highly sensitive to the growth inhibitory effects of c-myb antisense oligodeoxynucleotides. Blood. 79:1956–1961. [PubMed] [Google Scholar]
- Ricci C., Onida F., Servida F., Radaelli F., Saporiti G., Todoerti K., Deliliers G.L., and Ghidoni R.. 2009. In vitro anti-leukaemia activity of sphingosine kinase inhibitor. Br. J. Haematol. 144:350–357. 10.1111/j.1365-2141.2008.07474.x [DOI] [PubMed] [Google Scholar]
- Rowley J.D. 1973. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 243:290–293. 10.1038/243290a0 [DOI] [PubMed] [Google Scholar]
- Sánchez M., Göttgens B., Sinclair A.M., Stanley M., Begley C.G., Hunter S., and Green A.R.. 1999. An SCL 3′ enhancer targets developing endothelium together with embryonic and adult haematopoietic progenitors. Development. 126:3891–3904. [DOI] [PubMed] [Google Scholar]
- Sawyers C.L., Hochhaus A., Feldman E., Goldman J.M., Miller C.B., Ottmann O.G., Schiffer C.A., Talpaz M., Guilhot F., Deininger M.W., et al. 2002. Imatinib induces hematologic and cytogenetic responses in patients with chronic myelogenous leukemia in myeloid blast crisis: results of a phase II study. Blood. 99:3530–3539. 10.1182/blood.V99.10.3530 [DOI] [PubMed] [Google Scholar]
- Schuhmacher M., Kohlhuber F., Hölzel M., Kaiser C., Burtscher H., Jarsch M., Bornkamm G.W., Laux G., Polack A., Weidle U.H., and Eick D.. 2001. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 29:397–406. 10.1093/nar/29.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih A.H., Abdel-Wahab O., Patel J.P., and Levine R.L.. 2012. The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer. 12:599–612. 10.1038/nrc3343 [DOI] [PubMed] [Google Scholar]
- Shimamoto T., Ohyashiki K., Ohyashiki J.H., Kawakubo K., Fujimura T., Iwama H., Nakazawa S., and Toyama K.. 1995. The expression pattern of erythrocyte/megakaryocyte-related transcription factors GATA-1 and the stem cell leukemia gene correlates with hematopoietic differentiation and is associated with outcome of acute myeloid leukemia. Blood. 86:3173–3180. [PubMed] [Google Scholar]
- Sill H., Goldman J.M., and Cross N.C.. 1995. Homozygous deletions of the p16 tumor-suppressor gene are associated with lymphoid transformation of chronic myeloid leukemia. Blood. 85:2013–2016. [PubMed] [Google Scholar]
- Silver R.T., Cortes J., Waltzman R., Mone M., and Kantarjian H.. 2009. Sustained durability of responses and improved progression-free and overall survival with imatinib treatment for accelerated phase and blast crisis chronic myeloid leukemia: long-term follow-up of the STI571 0102 and 0109 trials. Haematologica. 94:743–744. 10.3324/haematol.2009.006999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirard C., Lapidot T., Vormoor J., Cashman J.D., Doedens M., Murdoch B., Jamal N., Messner H., Addey L., Minden M., et al. 1996. Normal and leukemic SCID-repopulating cells (SRC) coexist in the bone marrow and peripheral blood from CML patients in chronic phase, whereas leukemic SRC are detected in blast crisis. Blood. 87:1539–1548. [PubMed] [Google Scholar]
- Skorski T., Nieborowska-Skorska M., Wlodarski P., Perrotti D., Martinez R., Wasik M.A., and Calabretta B.. 1996. Blastic transformation of p53-deficient bone marrow cells by p210bcr/abl tyrosine kinase. Proc. Natl. Acad. Sci. USA. 93:13137–13142. 10.1073/pnas.93.23.13137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N., Dong Y., Lian J.B., Pratap J., Kingsley P.D., van Wijnen A.J., Stein J.L., Schwarz E.M., O’Keefe R.J., Stein G.S., and Drissi M.H.. 2005. Overlapping expression of Runx1(Cbfa2) and Runx2(Cbfa1) transcription factors supports cooperative induction of skeletal development. J. Cell. Physiol. 203:133–143. 10.1002/jcp.20210 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoennissen N.H., Krug U.O., Lee D.H., Kawamata N., Iwanski G.B., Lasho T., Weiss T., Nowak D., Koren-Michowitz M., Kato M., et al. 2010. Prevalence and prognostic impact of allelic imbalances associated with leukemic transformation of Philadelphia chromosome-negative myeloproliferative neoplasms. Blood. 115:2882–2890. 10.1182/blood-2009-07-235119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren A.G., Mikkers H., Kool J., van der Weyden L., Lund A.H., Wilson C.H., Rance R., Jonkers J., van Lohuizen M., Berns A., and Adams D.J.. 2009. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat. Protoc. 4:789–798. 10.1038/nprot.2009.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weyden L., Giotopoulos G., Rust A.G., Matheson L.S., van Delft F.W., Kong J., Corcoran A.E., Greaves M.F., Mullighan C.G., Huntly B.J., and Adams D.J.. 2011. Modeling the evolution of ETV6-RUNX1-induced B-cell precursor acute lymphoblastic leukemia in mice. Blood. 118:1041–1051. 10.1182/blood-2011-02-338848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassiliou G.S., Cooper J.L., Rad R., Li J., Rice S., Uren A., Rad L., Ellis P., Andrews R., Banerjee R., et al. 2011. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat. Genet. 43:470–475. 10.1038/ng.796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.C., Lapidot T., Cashman J.D., Doedens M., Addy L., Sutherland D.R., Nayar R., Laraya P., Minden M., Keating A., et al. 1998. High level engraftment of NOD/SCID mice by primitive normal and leukemic hematopoietic cells from patients with chronic myeloid leukemia in chronic phase. Blood. 91:2406–2414. [PubMed] [Google Scholar]
- Warsch W., Walz C., and Sexl V.. 2013. JAK of all trades: JAK2-STAT5 as novel therapeutic targets in BCR-ABL1+ chronic myeloid leukemia. Blood. 122:2167–2175. 10.1182/blood-2013-02-485573 [DOI] [PubMed] [Google Scholar]
- Yasuda T., Shirakata M., Iwama A., Ishii A., Ebihara Y., Osawa M., Honda K., Shinohara H., Sudo K., Tsuji K., et al. 2004. Role of Dok-1 and Dok-2 in myeloid homeostasis and suppression of leukemia. J. Exp. Med. 200:1681–1687. 10.1084/jem.20041247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R.C., Jiang Y., and Verfaillie C.M.. 2001. A model of human p210(bcr/ABL)-mediated chronic myelogenous leukemia by transduction of primary normal human CD34(+) cells with a BCR/ABL-containing retroviral vector. Blood. 97:2406–2412. 10.1182/blood.V97.8.2406 [DOI] [PubMed] [Google Scholar]
- Zhao L.J., Wang Y.Y., Li G., Ma L.Y., Xiong S.M., Weng X.Q., Zhang W.N., Wu B., Chen Z., and Chen S.J.. 2012. Functional features of RUNX1 mutants in acute transformation of chronic myeloid leukemia and their contribution to inducing murine full-blown leukemia. Blood. 119:2873–2882. 10.1182/blood-2011-08-370981 [DOI] [PubMed] [Google Scholar]
- Zhou T., Georgeon S., Moser R., Moore D.J., Caflisch A., and Hantschel O.. 2014. Specificity and mechanism-of-action of the JAK2 tyrosine kinase inhibitors ruxolitinib and SAR302503 (TG101348). Leukemia. 28:404–407. 10.1038/leu.2013.205 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.