SUMMARY
The idea that stem cell therapies work only via cell replacement is challenged by the observation of consistent intercellular molecule exchange between the graft and the host. Here we defined a mechanism of cellular signaling by which neural stem/precursor cells (NPCs) communicate with the microenvironment via extracellular vesicles (EVs), and we elucidated its molecular signature and function. We observed cytokine-regulated pathways that sort proteins and mRNAs into EVs. We described induction of interferon gamma (IFN-γ) pathway in NPCs exposed to proinflammatory cytokines that is mirrored in EVs. We showed that IFN-γ bound to EVs through Ifngr1 activates Stat1 in target cells. Finally, we demonstrated that endogenous Stat1 and Ifngr1 in target cells are indispensable to sustain the activation of Stat1 signaling by EV-associated IFN-γ/Ifngr1 complexes. Our study identifies a mechanism of cellular signaling regulated by EV-associated IFN-γ/Ifngr1 complexes, which grafted stem cells may use to communicate with the host immune system.
INTRODUCTION
The systemic injection of neural stem/precursor cells (NPCs) in laboratory animals with immune-mediated experimental CNS demyelination, stroke, or injuries of the spinal cord leads to remarkable neuroprotection and functional recovery (Martino et al., 2011; Uccelli et al., 2011). While a comprehensive understanding of the mechanisms by which stem cell grafts work is still lacking, it is becoming increasingly accepted that they exert some of their therapeutic effects by secreting a complex array of homeostatic molecules (stem cell secretome) with immune regulatory and tissue trophic functions that ultimately reduce tissue damage and/or enhance endogenous repair (Drago et al., 2013).
Partly as drugs and partly as devices, stem cell medicines work like naturally occurring disease-modifying agents that sense signals, migrate to specific areas of the body, make decisions, and execute complex response behaviors—always in the context of specific microenvironments (Fischbach et al., 2013).
Communication between grafted stem cells and the host is delivered via secreted cytokines and/or growth factors or through cellular (Gap) junctional transfer of electrical, metabolic, and immunological information. Furthermore, early work also suggests that extracellular vesicles (EVs) may play a key role when transferred from grafted stem cells to target host neural and nonneural cells (Pluchino and Cossetti, 2013).
EVs are complex membranous structures composed of a lipid bilayer that contain transmembrane proteins and enclose soluble hydrophilic components derived from the cytosol of donor cells. EV is a general term that defines different types of vesicles, including exosomes, microparticles, gesicles (Mangeot et al., 2011), and human endogenous retroviral particles (Balaj et al., 2011). Cells secrete EVs simultaneously, although there are yet no established criteria to distinguish one type of vesicle from another or physical means to separate them once released (Witwer et al., 2013). EVs capture bioactive molecules responsible for direct stimulation (Al-Nedawi et al., 2008) and increased survival of target cells (Frühbeis et al., 2013; Lopez-Verrilli et al., 2013), transmission of infectious agents (Mattei et al., 2009), and horizontal transfer of membrane and/or cargo molecules, which are enriched in specific proteins (Antonyak et al., 2011) and nucleic acids (Mittelbrunn et al., 2011; Valadi et al., 2007).
It is well established that this transfer of information affects the physiology of recipient cells in various ways, from the activation versus suppression of immune responses, to promotion of tissue repair and cancer progression (Breakefield et al., 2011; Théry et al., 2009). Furthermore, experimental therapeutics with either unmodified or functionalized EVs/exosomes collected from mesenchymal stem cells (MSCs) or immune cells are being established as a promising anti-inflammatory (Yu et al., 2013; Zhuang et al., 2011), tissue-protective (Xin et al., 2013), stem cell-free alternative approach for brain repair.
Here, we focused on defining whether the form of communication mediated by EVs exists for NPCs, on elucidating its molecular signature and functional relevance to target cells, and on identifying the key elements responsible for this mechanism of cellular signaling.
We show that NPC EVs primarily consist of exosomes and observe cytokine-regulated pathways that sort proteins and mRNAs into EVs. Moreover, we describe a highly specific induction of the interferon gamma (IFN-γ) pathway in parental NPCs exposed to proinflammatory cytokines that is mirrored in EVs. We determined that activation of Stat1-dependent signaling in target NIH 3T3 cells occurs as a result of the intercellular transfer of IFN-γ bound to interferon gamma receptor 1 (Ifngr1) on the surface of EVs. Finally, we demonstrate that endogenous Stat1 and Ifngr1 in target cells are indispensable to sustain the activation of Stat1 signaling by EV-associated IFN-γ/Ifngr1 complexes.
Our study sheds light on the mechanisms of intercellular information exchange and demonstrates that EV-mediated cytokine signaling is an important mechanism by which NPCs may propagate some of their immune modulatory activities (Pluchino and Cossetti, 2013).
RESULTS
NPCs Secrete EVs
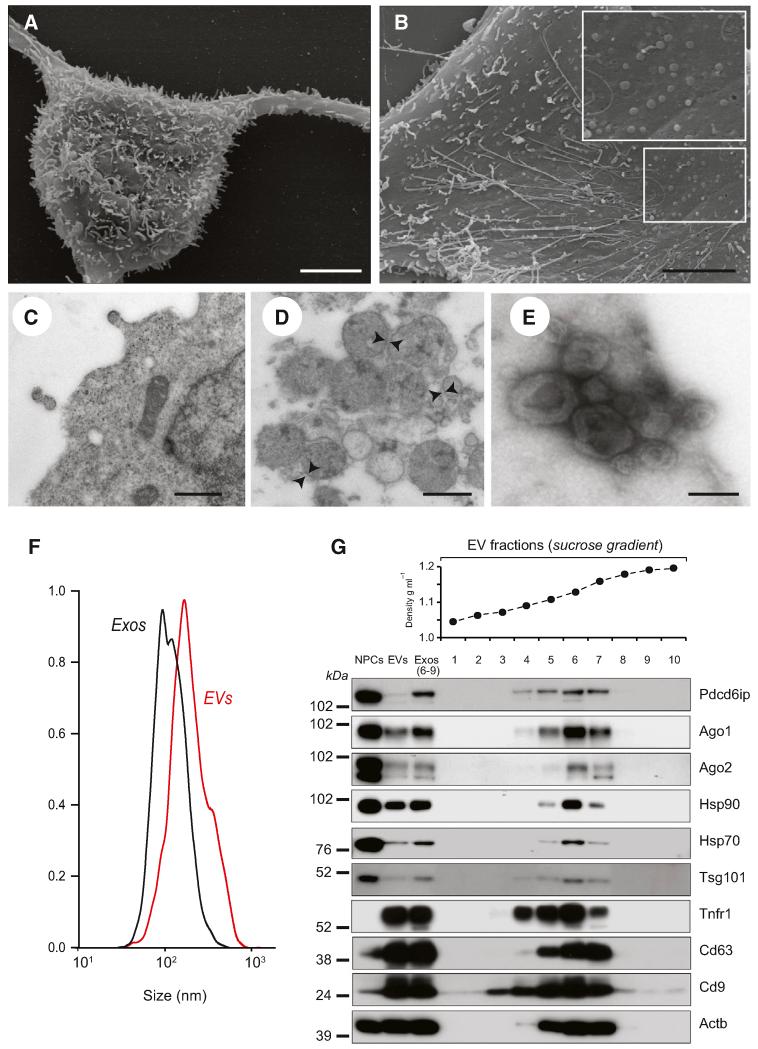
NPCs were established from the subventricular zone (SVZ) of adult mice, as described (Pluchino et al., 2005). Scanning electron microscopy (SEM) of NPC surface revealed polarized membranous structures of small and medium size, which included long nanotubes and round membrane vesicles (Figures 1A and 1B). Transmission electron microscopy (TEM) showed the budding of electron-dense vesicle-like structures directly from the NPC plasma membrane and the presence of cytoplasm electron-transparent vesicles (Figure 1C).
Figure 1. NPCs Secrete EVs.
(A) Scanning electron microscopy (SEM) of a NPC with long adherent expansions, and numerous membrane protrusions on the surface. Scale bars, 5 μm.
(B) SEM of the NPC surface, where membranous nanotubes and circular membrane vesicles are observed (magnified in the inset). Scale bars, 5 μm.
(C) Magnified TEM detail of a NPC secreting two small-sized vesicles. Scale bars, 500 nm.
(D) TEM of NPC EVs. EVs appear as heterogeneous population of differently sized vesicles (range 40–200 nm in diameter) surrounded by a double-layer membrane (arrowheads). Scale bars, 500 nm.
(E) TEM of negative stained EVs, showing cup-shaped vesicles (80–120 nm). Scale bars, 100 nm.
(F) Particle-size distribution of EVs (red line) and Exos (black line) obtained by NTA. Data are represented as mean ± SEM from n = 5 independent replicates and are normalized to 1 for size comparison.
(G) Western blot of exosomal markers in NPCs, EVs, and Exos. Exos correspond to pooled fractions 6–9, having a density between 1.13 and 1.20 g/ml. This panel is representative of n = 3 independent protein preparations showing the same trends.
See also Figure S1.
EVs were isolated from NPC supernatants by differential centrifugation (Théry et al., 2001). We verified the heterogeneity and size of the EVs collected by combination of flow cytometry (FCM) and TEM. FCM analysis showed that the majority of EVs were around 1 μm in size (data not shown), while TEM confirmed the heterogeneity of the EV fraction (Figure 1D). Negative TEM staining indicated the presence of a subfraction of cup-shaped vesicles in the range of 40–120 nm, consistent with previous descriptions of exosomes (Théry et al., 2001) (Figure 1E). Nanoparticle-tracking analysis (NTA) (Dragovic et al., 2011) supported the presence of a multimodal size distribution of EVs, with a major peak corresponding to smaller exosome-like vesicles (diameter 167 ± 2.82 nm) and a smaller peak consisting of larger microparticle-like vesicles (diameter 342.4 ± 36.65 nm) (Figure 1F).
To enrich for exosomes, we applied sucrose gradient density centrifugation and pooled the EV fractions in the range between 1.13 and 1.20 g/ml (Exos), as described previously (Théry et al., 1999). NTA of Exos indicated a strong enrichment in particles with a peak diameter around 100–150 nm, which corresponds to described exosomes (Figure 1F). Biochemical and FCM analyses further confirmed that purified Exos were specifically enriched in several markers associated with exosomes, compared to EVs and NPCs (Théry et al., 2001) (Figure 1G).
Proinflammatory Cytokine Signaling Induces the Export of Specific Protein Cargoes in EVs
To further assess NPC-derived EVs as conveyors of functional (immune) responses (Théry et al., 2009), as well as the potential role of the microenvironment in modulating EV-mediated transfer of information between cells, we examined whether the exposure of NPCs to inflammatory cytokines would affect the protein composition of EVs and Exos.
NPCs were grown in culture media alone (basal) or enriched with cytokine cocktails that mimic a proinflammatory (Th1-like, hereafter referred to as Th1) or anti-inflammatory (Th2-like, hereafter referred to as Th2) microenvironment (Pluchino et al., 2008). Treatment of NPCs with either pro- or anti-inflammatory cytokine cocktails had no effect on expression levels of their cognate receptors, compared to basal (Figure S1, available online). In addition, there was no difference in the EVs/Exos in terms of size, concentration, and total protein content (Figure S2), suggesting that cytokine signaling had minimal impact on the overall protein loading from NPCs into EVs/Exos.
To provide quantitative profiling of the EV proteins, we next performed heavy-light double stable isotope labeling of amino acids in cell culture (SILAC), comparing EVs and Exos collected from NPCs grown in basal, Th1, or Th2 media, as before. Given that we observed a strong (~65%–70%) overlap between the EVs and Exo protein data sets (Table S1), for clarity from here we will only discuss the EV protein content. Data from Exo SILAC are showed in Figure S3.
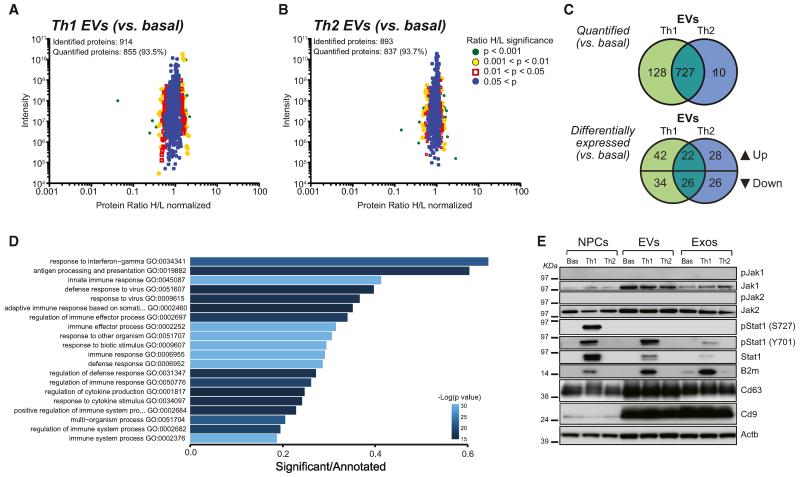
We identified a total of 914 and 893 proteins in Th1 and Th2 EVs, with more than 90% being quantified, and a large (75%) overlap between the two cytokines treatments (727/965), compared to basal (Figures 2A–2C).
Figure 2. Th1 Cytokine Signaling Upregulates the IFN-γ Pathway in NPCs that Is Exported to EVs.
(A–C) SILAC quantification of proteins expressed in Th1 and Th2 EVs. Scatter plots of the identified and quantified proteins in EVs Th1 (A) and Th2 (B) together with a color-coded quantitation significance as provided by MaxQuant software. Protein ratios are plotted against summed peptides intensities. (C) Venn diagrams of the numerical values for the indicated common and unique proteins quantified and differentially expressed in Th1 (green) and Th2 (blue) EVs versus basal EVs. p ≤ 0.05.
(D) Bar chart showing the score of the top 20 most enriched GO terms in Th1 NPCs versus basal from RNA-seq data. Color coding indicates the adjusted p value.
(E) Western blot of the Jak/Stat pathway in NPCs, EVs, and Exos. β-actin (Actb) was used as a loading control. This panel is representative of n = 4 independent protein preparations showing the same trends.
See also Figures S2–S4 and Tables S1 and S2.
We next found that ~12%–15% of proteins were differentially expressed (p ≤ 0.05) in Th1 (124/855) and Th2 (102/837) EVs, with only moderate (27%) overlap between these two differentially expressed data sets (48/178) (Figure 2C). We ultimately identified a set of 76 proteins that were significantly regulated in Th1 EVs only and a smaller set of 54 proteins regulated in Th2 EVs only (Figure 2C). Among the 42 upregulated proteins that were enriched in Th1 EVs, 18 (43%) were categorized as protein metabolism (Table S1).
We then applied Reactome Functional Interaction (FI) Cytoscape plugin to build a pathway-based protein functional interaction subnetwork of the Th1 EV protein metabolism GO terms. From this analysis, we identified that Th1 EVs were enriched for T-complex protein 1 subunit gamma and delta (Cct3 and Cct4) (Figure S3), which have previously been associated with mast cell-derived EVs (Valadi et al., 2007). Reactome pathways enrichment data are available in Table S2.
To examine whether changes in the EV proteome reflect those of the parental NPC protein expression in response to cytokines, we performed SILAC in NPCs as above. We observed a notable difference in NPC protein expression in response to both Th1 and Th2 cytokines compared to basal conditions (Figure S3). However, this did not coincide with the changes previously found in EVs (~10%; 48/482) (Table S1). Interestingly, proteins found in the antigen processing and presentation GO cluster were significantly changed in Th1 NPCs only (p = 0.029), with an upregulation of the two key components of the major histocompatibility complex (MHC): TAP-associated glycoprotein or tapasin (Tapbp) and β2-microglobulin (B2m) (Figure S3).
We concluded that proinflammatory—and to a much lesser extent anti-inflammatory—cytokine signaling pathways regulate the NPC proteome and induce a specific export of protein cargoes in EVs and Exos.
Th1 EVs Are Enriched in mRNAs Coding for the IFN-γ Signaling Pathway
Due to the limitations of SILAC for quantifying low-abundance proteins (Lubec and Afjehi-Sadat, 2007), we next employed RNA sequencing (RNA-seq) on poly(A)-selected RNAs for indepth analysis of the transcriptome of NPCs, EVs, and Exos.
Total RNA levels in NPCs, EVs, and Exos remained constant between basal, Th1, and Th2 conditions (Figure S2); however, we found 686 genes upregulated and 477 genes downregulated (FC > 5) in Th1 NPCs (versus basal; Figure S4). Overall, we found that the transcriptome of both Th1 and Th2 EVs and Exos mirrored that of parental NPCs, with only Th1 EVs and Exos showing remarkable upregulation of mRNAs belonging to inflammatory pathways (Figures 2D and S4).
GO enrichment analysis on the differentially expressed genes revealed that the most altered pathways in Th1 NPCs were response to IFN-γ (p = 3.8 × 10−22) and antigen processing and presentation (p = 2.1 × 10−17; Figure 2D). Th2 NPCs showed differential expression of only a small number of RNAs (Figure S4).
Therefore, proinflammatory cytokine signaling triggered regulation of genes downstream of the IFN-γ pathway in NPCs, which were ultimately exported to EVs and Exos.
We next validated, by western blot analysis, some of the components of the IFN-γ pathway (including Stat1) identified by RNA-seq. The rationale behind this choice was based on three factors: (1) the largely immune-like effects of the proinflammatory cytokine signaling on the NPC, EV, and Exo proteomes (Figure S3 and Table S1); (2) the evidence that EVs traffic proteins and RNAs, often belonging to the very same pathways (Valadi et al., 2007); and (3) the well-known role of the Stat1 pathway in promoting diverse cellular responses to IFN- γ (Ramana et al., 2002).
Th1 NPCs upregulated total and phosphorylated Stat1 (pStat1) at both Y701 and S727 sites, whereas Th1 EVs—and to a much lesser extent Th1 Exos—showed a specific increase in total Stat1 and pStat1 (Y701) only. Th1 NPCs, EVs, and Exos were all enriched with the downstream element of Stat1 pathway B2m (Fellous et al., 1982) (Figure 2E), also confirming a similar enrichment identified by SILAC. Both Jak1 and Jak2 were enriched in EVs, but neither these tyrosine kinase proteins nor their phosphorylated forms underwent upregulation in Th1 samples (Figure 2E), likely reflecting fast activation kinetics of Jaks (Igarashi et al., 1994). We did not observe changes in the expression of either total Stat6 or pStat6 after Th2 treatment (data not shown).
We concluded that proinflammatory cytokine signaling in NPCs activates signal transduction along the IFN-γ/Stat1 pathway and governs the export of specific components of this pathway to EVs and Exos.
Th1 EVs Transfer Specific Signal Transduction along the Stat1 Pathway to Target Cells
To investigate the impact of the transfer of NPC-derived EVs enriched in IFN-γ signaling elements to target cells, we exposed NIH 3T3 cells to increasing quantities of EVs collected from NPCs that expressed either a farnesylated enhanced green fluorescent protein (fEGFP) or the tetraspanin CD63 fused to red fluorescent protein (RFP; Figure 3). We used the NIH 3T3 cell line as a model of target cells, with the key advantages of being robust in terms of reproducibility of outcomes, easy to handle, and very informative for high-throughput approaches investigating the global cellular response to type I and II interferons (Dölken et al., 2008).
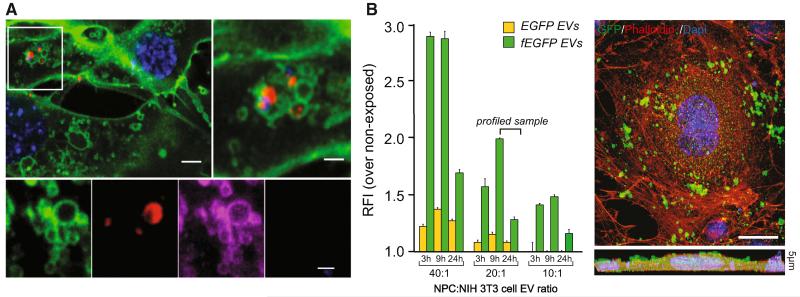
Figure 3. EVs Rapidly Adhere to and Are Incorporated into Target Cells via Plasma Membrane.
(A) CD63-RFP EV uptake in fEGFP target cells (in green) as early as 2 hr after transfer. EVs are in red under confocal and magenta under super-resolution STED microscopy. Scale bars: top left, 5 μm; top right, 2 μm; bottom, 1 μm.
(B) FCM analysis of EGFP internalization by target cells treated with increasing quantities of EVs. Data are represented as mean relative fluorescence intensity ± SEM from n = 3 independent experiments. A representative confocal image of a NIH 3T3 cell (red) exposed to fEGFP EVs (green) is shown. The lower panel is a Z stack of n = 5 optical slices taken at 1 μm intervals. Scale bar, 10 μm.
Nuclei in (A) and (B) were stained with DAPI (blue). See also Movie S1.
We showed, by FCM, confocal, stimulated emission depletion (STED) super-resolution microscopy, and TEM, the rapid adhesion and incorporation of EVs in target cells (peak of fluorescence at 9 hr after exposure; Figure 3 and Movie S1). The lack of association between EVs and the lysosomal-associated membrane protein 1 (LAMP-1) excluded direct or immediate degradation in target cells (data not shown). Whole transcriptome and proteome analyses were carried out on NIH 3T3 cells incubated with EVs (20:1 ratio; ~30 μg of EV proteins/treatment) harvested from NPCs cultured in basal, Th1, or Th2 conditions (Figure 4A).
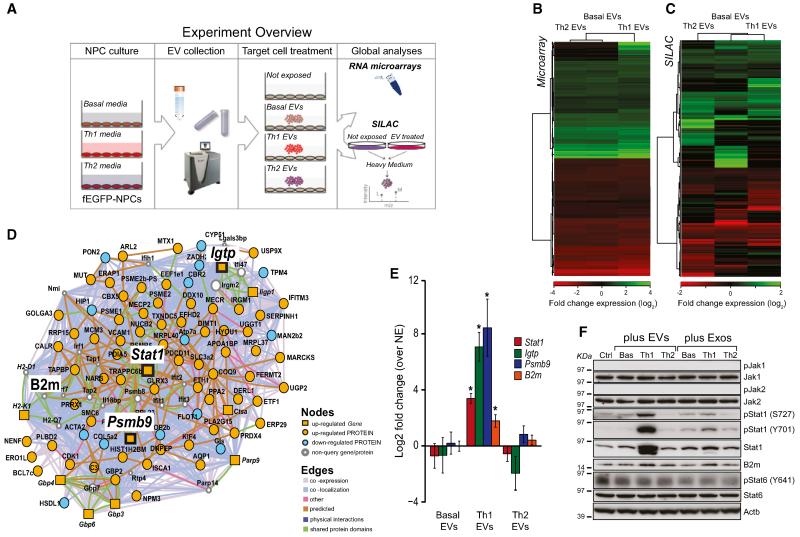
Figure 4. Th1 EVs Activate Signal Transduction along the Stat1 Pathway in Target Cells at Gene and Protein Levels.
(A) Experiment overview of whole transcriptome and proteome analyses in target cells exposed to NPC EVs.
(B) Heatmap of the relative fold changes of genes in target cells exposed to 20:1 NPC:NIH 3T3 ratios of basal, Th1, or Th2 EVs for 24 hr in vitro.
(C) Heatmap of the relative fold changes of proteins in target cells treated as in (B).
(D) Integrated GeneMANIA network of differentially expressed Th1-specific genes and proteins identified using microarray and SILAC, respectively, by comparing target cells exposed to Th1 EVs versus basal EVs.
(E) qPCR analysis of Stat1, Igtp, Psmb9, and B2M expression in target cells treated as in (B). Data are represented as mean log2 fold change ± SEM over target cells not exposed (NE) to EVs from a total of n = 3 independent experiments. *p < 0.05, compared to NE.
(F) Western blot of the Jak/Stat pathway in target cells treated as in (B). This panel is representative of n = 4 independent protein preparations showing the same trends. β-actin (Actb) was used as a loading control.
See also Table S3.
The exposure of target cells to Th1 EVs led to the differential expression of 443 genes (B ≥ 3, compared to NIH 3T3 not exposed [NE]) and 130 proteins (p ≤ 0.01, compared to NIH 3T3 NE). The majority of these genes and proteins were similarly regulated in Th2 and basal EVs with 24 genes and 95 proteins specifically regulated by Th1 EVs only. In contrast, Th2-induced changes were similar to those elicited by basal EVs (Figures 4B and 4C and Table S3).
To understand the functional trends elicited by these EV preparations, we applied GeneMANIA (Mostafavi et al., 2008) and generated an integrated pathway analysis that combined microarray and SILAC data (Table S3).
When generating the GeneMANIA network for the specific response to Th1 EVs (Figure 4D), we found that the most significantly enriched GO term was antigen processing and presentation (p = 3.19 × 10−14). Additionally, we found a smaller, but significant, enrichment of genes/proteins for the GO terms response to interferon beta (p = 2.17 × 10−9), response to interferon gamma (p = 5.26 × 10−7), and response to cytokine stimulus (p = 1.57 × 10−7) (Table S3). Interestingly, these Th1 EV-specific changes broadly overlapped with the changes induced by Th1 cytokines in parental NPCs. On the other hand, we did not find any significant GO enrichment in the Th2-specific GeneMANIA network, which suggested that basal and Th2 EVs elicit broadly similar effects on target cells (Table S3).
Next, we used qPCR to confirm the significant upregulation of the key elements of the Stat1 pathway arising from the Th1-specific GeneMANIA network upon exposure of NIH 3T3 cells to Th1 EVs (Figure 4E). We also found, by western blot analyses, that Th1 EVs specifically induce Stat1, pStat1 (both Y701 and S727), and B2m, but not Stat6 and pStat6 (Y641), in NIH 3T3 cells. As above, neither Jak1 nor Jak2 showed differences in expression, or detectable levels of phosphorylation, in any sample. All changes observed in target cells exposed to EVs were mirrored, although to a lower extent, in cells exposed to Exos (Figure 4F).
Thus, the activation of this Stat1-specific response in NIH 3T3 cells suggests that Th1 EVs/Exos loaded with specific components of the Stat1 pathway may signal either directly via gene (Valadi et al., 2007) and/or protein transfer (Kwon et al., 2014) or indirectly via gene and protein induction (Li et al., 2013).
The EV-Associated IFN- γ/Ifngr1 Complex Activates Signal Transduction along the Stat1 Pathway in Target Cells
To assess the relevance of the direct mRNA and protein transfer via Th1 EVs on target cells, we analyzed the levels of Stat1 and key proteins of the IFN-γ pathway in Stat1−/− target cells treated with wild-type (WT) Th1 EVs. Using a probe specifically designed to detect exogenous Stat1, we observed dose- and time-dependent increase of Stat1 by qPCR. The observed linear correlation between the levels of mRNA and the incubation time (p < 0.001; R2 = 0.7684 and R2 = 0.6061 for 20:1 and 80:1 ratio, respectively) further supported a likely active uptake of EV-derived mRNA by Stat1−/− target cells. However, we failed to detect convincing levels of exogenous Stat1 by western blot, which suggested that the translation of the transferred mRNA and the quantity of exogenous Stat1 from Th1 EVs were both below the detection limits (Figure S5).
These data also suggest a requirement for endogenous Stat1 to sustain the activation of the Stat1 signaling in target cells exposed to Th1 EVs.
Next, to better define the relative contribution of the different components of the Stat1 pathway in the EV-induced activation of signal transduction in target cells, we discriminated between the effects of the Th1 proinflammatory cytokine cocktail and those elicited by IFN-γ alone.
We observed that IFN-γ only induced comparable activation of signal transduction along the Stat1 pathway in NPCs and EVs (Figure S5), as well as in NIH 3T3 cells, when exposed to EVs from NPCs treated with IFN-γ only (Figure 5A).
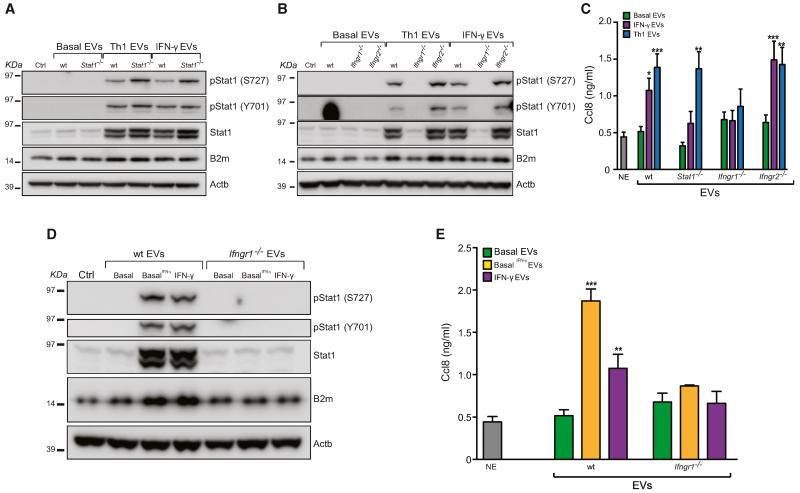
Figure 5. The IFN-γ/Ifngr1 Complex via EVs Activates Signal Transduction along the Stat1 Pathway in Target Cells.
(A and B) Western blot of the Stat1 pathway in target cells treated with WT, Stat1−/− (A) and Ifngr1−/− and Ifngr2−/− (B) EVs as in Figure 4F. These panels are representative of n = 3 independent protein preparations showing the same trends.
(C) Ccl8 release by target cells exposed to EVs as in (A) and (B), as measured by ELISA.
(D) Western blot of the Stat1 pathway in target cells exposed to basal EVs pretreated with 100 ng/ml IFN-γ (basal IFN-γ; same concentration used for NPCs). This panel is representative of n = 3 independent protein preparations showing the same trends.
(E) Ccl8 release by target cells exposed to EVs as in (D), as measured by ELISA.
Data in (C) and (E) are represented as mean ± SEM from a total of n ≤ 3 independent experiments. *p ≤ 0.01, **p ≤ 0.001, ***p ≤ 0.0001, compared to NE target cells not exposed (NE) to EVs. β-actin (Actb) was used as a loading control in (A), (B), and (D). See also Figures S5 and S6.
To clarify which component of the IFN-γ pathway was responsible for the effect of EVs on target cells, we generated NPCs from mice lacking Stat1 (Durbin et al., 1996), Ifngr1 (alpha chain) (Huang et al., 1993), or Ifngr2 (beta chain). The treatment of Stat1−/−, Ifngr1−/−, and Ifngr2−/− NPCs with Th1 cytokines or IFN-γ only failed to activate signal transduction along the Stat1 pathway at both RNA and protein levels (Figure S5). Interestingly, both Stat1−/− and Ifngr2−/− Th1 and IFN-γ EVs were still capable of activating the Stat1 pathway in target cells (Figures 5A and 5B). In contrast, we found that Ifngr1−/− Th1 and IFN-γ EVs failed to elicit the activation of Stat1 in NIH 3T3 (Figure 5B), demonstrating that Ifngr1 is indispensable for the propagation of the IFN-γ-dependent signaling by EVs.
Further, the activation of Stat1 signaling in target cells was also paralleled by secretion of the chemokine monocyte chemoattractant protein 2 (Mcp-2)/C-C motif ligand 8 (Ccl8), as described (Struyf et al., 2009). Again, we did not find any increase in the secretion of Ccl8 by target cells exposed to Ifngr1−/− Th1 or IFN-γ EVs (Figure 5C). These data demonstrated that Ifngr1, but not the enriched mRNAs and proteins in Th1 EVs, is mediating the transfer of proinflammatory signaling to target cells.
To establish whether IFN-γ bound to its cognate receptor on EVs is transferred to target cells, we first measured (by ELISA) the concentration of IFN-γ in the amount of EVs used for the transfer experiments and found no significant differences between WT, Stat1−/−, and Ifngr2−/− Th1 and IFN-γ EVs (range 52.13–210.3 pg/ml, corresponding to ~15 pg of IFN-γ per ~30 μg of EV proteins). Notably, IFN-γ was below the detection limits in both Th1 and IFN-γ Ifngr1−/− EVs (Figure S6). In addition, stimulation of NIH 3T3 cells with ~100-fold more free (soluble) IFN-γ than that estimated by ELISA as being bound to EVs led to lower activation of Stat1-dependent signaling, compared to that induced by IFN-γ bound to EVs via Ifngr1 (Figure S6 and Supplemental Experimental Procedures).
This evidence suggested that IFN-γ bound to its cognate Ifngr1 on EVs has much higher stability compared to the free (soluble) cytokine.
Next, we used EVs treated directly with IFN-γ to confirm that IFN-γ can bind to the Ifngr1 on the surface of EV. We showed that pretreatment of WT, but not Ifngr1−/−, basal EVs with IFN-γ (basal IFN-γ) conferred to basal EVs the very same signaling ability of Th1 or IFN-γ EVs, in terms of both activation of the Stat1 pathway and Ccl8 secretion by target cells (Figures 5D and 5E).
Altogether, these data excluded the possibility that nonspecific IFN-γ binding to EVs plays any role in the observed signaling response of NIH 3T3 and demonstrated that the IFN-γ/Ifngr1 complex on EVs is indispensable to promote intercellular induction of Stat1 signaling.
Ifngr1 on Target Cells Is Required to Sustain the EV-Mediated Activation of the Stat1 Pathway
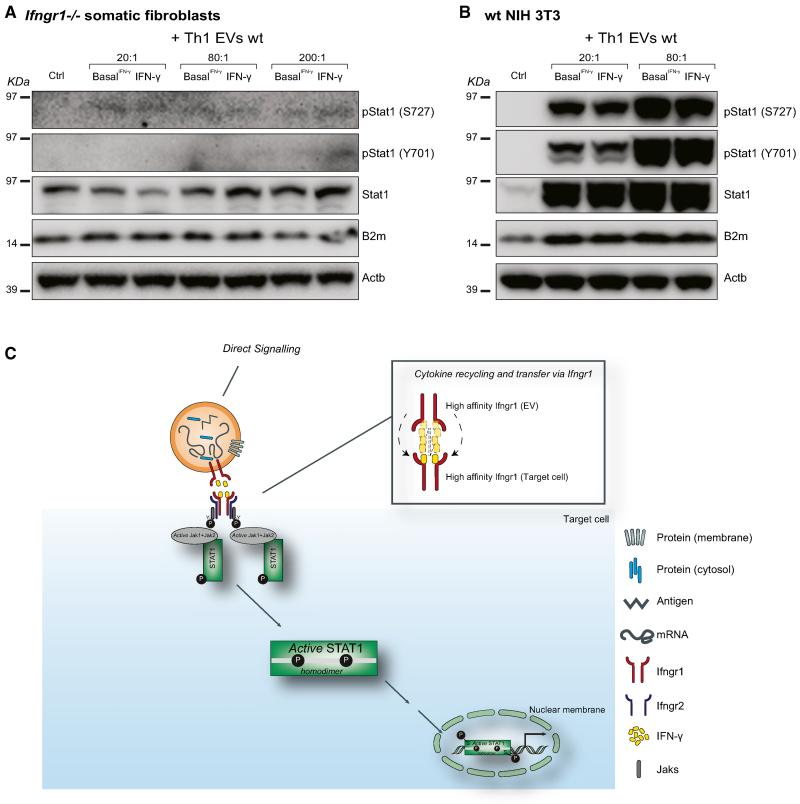
Next, to investigate whether the IFN-γ/Ifngr1 complex on EVs was functionally transferred in target cells, we analyzed the activation of Stat1 signaling in Ifngr1−/− somatic fibroblasts treated with WT IFN-γ and basal IFN-γ EVs (20:1, 80:1, and 200:1 NPC:target cell ratios). We observed a slight and dose-dependent upregulation of total, but not phosphorylated, Stat1 and downstream target B2m by western blot (Figures 6A and 6B). We therefore postulated that Ifngr1 on target cells is required to sustain the activation of the Stat1 pathway.
Figure 6. The EV-Associated IFN-γ/Ifngr1 Complex Requires Ifngr1 on Target Cells to Sustain the Activation of the Stat1 Pathway.
(A and B) Western blot of the Stat1 pathway in Ifngr1−/− (A) or WT (B) target cells treated with different ratios (range 30–300 μg EV protein/treatment) of WT IFN-γ-induced and basal IFN-γ EVs for 24 hr in vitro. Panels are representative of n = 3 independent protein preparations showing the same trends. β-actin (Actb) was used as a loading control.
(C) Proposed model of recycling of soluble (free) IFN-γ by EV-associated Ifngr1 and retargeting to Ifngr1-expressing target cells.
Finally, we wondered how EV-loaded IFN-γ would activate Stat1 signaling in target cells if already bound to a high-affinity receptor. Consulting previously published data on the kinetics of IFN-γ/Ifngr1 complex dissociation, specifically the koff rate, it was possible to estimate that the half-life of the ligand-receptor complex would be 139 s (Sadir et al., 1998). Thus, approximately every 2 min half of the IFN-γ/Ifngr1 complexes dissociates with the now-free IFN-γ allowed to be sequestered by another high-affinity receptor. As IFN-γ-bearing EVs are incubated in the continuous presence of high affinity receptor-expressing target cells for 24 hr, it is plausible to anticipate that within this time frame and experimental set up enough IFN-γ is released from an EV to equilibrate with cell-based IFN-γ receptors (Figure 6C).
All together, our results suggest that the most likely mechanism underlying the EV-mediated activation of the Stat1 pathway in target cells is the direct transfer of IFN-γ from the Ifngr1 associated to EVs to the Ifngr1 exposed on the membrane of target cells.
DISCUSSION
The evidence that secreted EVs provide signals of information that are capable of inducing multiple functional responses in adjacent and distant target cells has only recently emerged (Théry, 2011). EVs are actively secreted by most cell types and have been identified in body fluids working as key players in the regulation of immune responses (Théry et al., 2009).
We show that this form of communication mediated by EVs exists for NPCs, and we elucidate the molecular signature of NPC-derived EVs and their functional relevance on target cells. We also demonstrate that the inflammatory microenvironment, which we mimicked in vitro with two different proinflammatory (Th1-like) and anti-inflammatory (Th2-like) cytokine cocktails, plays a critical role in the regulation of specific RNA and protein sorting toward EVs. We specifically focused on the effect of the cytokine milieu on the function of NPCs as proinflammatory cytokines, such as interferon-gamma (IFN-γ) and tumor necrosis factor alpha (TNF-α), regulate the phenotype, the release of soluble factors, and ultimately the function of stem cells, both in vitro and in vivo (Pluchino and Cossetti, 2013).
Our data show that NPC-derived EVs and Exos are enriched with a composite suite of RNAs and proteins. We observe a significant modification of the EV RNA and protein cohorts under both pro- and anti-inflammatory cytokine treatments. Among the upregulated proteins that were enriched in Th1 EVs only, half were categorized as belonging to the protein metabolism cohort (Valadi et al., 2007). Interestingly, these modifications only minimally overlapped with the changes observed in NPCs in the same conditions. Such a specific enrichment of environment-associated protein cargoes in EVs suggests a link between cytokine receptors, downstream signaling pathways, and a yet to be identified machinery that would sort proteins and eventually regulate the loading of nucleic acids into membrane vesicles, as it has been anticipated in other cellular systems (Villarroya-Beltri et al., 2013). On the other hand, we observed a significant modification of the EV and Exo cohorts in Th1, but not Th2, fractions that mirrored the content of parental NPCs. This category was enriched into genes downstream of the IFN-γ signaling pathway and was paralleled by significant enrichment into proteins of the Stat1 pathway.
While other reports have described that EVs/Exos traffic proteins and RNAs (Balaj et al., 2011; Mittelbrunn et al., 2011; Müller et al., 2011; Valadi et al., 2007), we provide evidence that membrane vesicles broadly sample the parental cellular response to the microenvironment via cytokine receptors and functionally transfer membrane vesicles that include exosomes. Through SILAC and RNA-seq analyses of NPCs, EVs, and exosomes, we have uncovered a comprehensive list of candidate genes and proteins, which may be useful in understanding the mechanisms of EV-mediated cell-to-cell communication via either direct gene (Valadi et al., 2007) and/or protein transfer (Kwon et al., 2014) or indirect gene and protein induction (Li et al., 2013).
We then assessed the ability of NPC-derived EVs to modulate the phenotype and function of target cells. Combining a set of high-sensitivity imaging techniques, we first show the rapid adhesion and incorporation of EVs in target cells.
High-throughput protein and RNA analyses revealed that exposure of NIH 3T3 cells to EVs induced broad changes in both gene and protein expression, comprising significant regulation of the expression of cell-cycle regulators, which was comparable between Basal, Th1, and Th2 EVs. We also observed a Th1 EV-specific response of target cells, broadly overlapping with the changes induced by Th1 mix onto parental NPCs and subsequently exported to EVs and Exos. This Th1 EV-specific modulation of target cell function was also associated with the acquisition of a proinflammatory phenotype of target cells, as suggested by the increased secretion of the chemokine Ccl8 (Struyf et al., 2009).
Specifically, the capacity of Th1 EVs to activate signal transduction in target cells appeared more effective than that of Exos. Whether this implies (1) enrichment of relevant signaling molecules in specific subcellular compartments or (2) a bias in the extraction protocol (e.g., Exos require an extra day to be purified) remains to be fully established.
To clarify the direct contribution of the different key signaling elements of the IFN-γ pathway, we then generated both NPCs and target cells (somatic fibroblasts) from mice lacking Stat1 (Durbin et al., 1996), Ifngr1 (Huang et al., 1993), or Ifngr2.
We demonstrated that the direct transfer of Th1 EV-enriched mRNAs and proteins was not responsible for the activation of Stat1 signaling in target cells, which disagrees with previously published data (Deregibus et al., 2007; Valadi et al., 2007). This may be due to differences in technique sensitivity, absolute versus relative protein abundance, and/or EV uptake by different target cell types. We also cannot exclude that what we show to be just dispensable in somatic fibroblasts, would play a different role when NPC-derived EVs (or EVs from other cell types) are transferred to target cells (or contexts) other than the one studied here.
We show that nonspecific IFN-γ binding to EVs played a negligible role in our system and that EVs use Ifngr1 to recycle free IFN-γ, thus contributing to the perpetuation of the downstream signaling response in target cells. The observation that the direct addition of IFN-γ to target cells at a concentration of ~100-fold more than that bound to EVs led to minimal activation of Stat1-dependent signaling also suggests that IFN-γ bound to its cognate Ifngr1 on EVs has stability much higher than that of the free (soluble) cytokine.
We also establish that both endogenous Stat1 and Ifngr1 in target cells are indispensable to sustain the activation of Stat1 signaling by EV-trafficked IFN-γ/Ifngr1 complexes. The observation that NPCs use EVs to recycle and deliver IFN-γ only to fully equipped target cells highlights the innate target selectivity potential of this mode of intercellular communication. This feature makes EVs capable of delivering functional bioactive ligands in close proximity of cognate receptors on the surface of target cells, thus allowing parental cells to propagate specific and cell-type-restricted signals (Broderick and Hoffman, 2014). These findings may have important implications for the exploitation of either unmodified or functionalized EVs in CNS inflammatory diseases using cell-free technologies (EL Andaloussi et al., 2013).
While our study also demonstrated that Ifngr1 in target cells is indispensable to sustain the responsiveness to EV-bound IFN-γ, more work is required to clarify the relevance of alternative (versus complementary) mechanisms of EV-mediated signaling, as well as its intracellular fate. These include (1) signaling upon membrane-to-membrane interaction or (2) signaling after receptor-mediated endocytosis (Sorkin and von Zastrow, 2009).
Future studies will elucidate whether stem cells possess some intrinsic capacities to engage EV-mediated functional interactions with immune cells—including T lymphocytes, macrophages or microglia, and dendritic cells—and to what extent this type of intercellular communication explains the immune regulatory effects of NPCs when transplanted in laboratory animals affected by experimental CNS inflammatory diseases (Martino et al., 2011; Pluchino and Cossetti, 2013).
In conclusion, our results describe that IFN-γ is exchanged via stem cell-derived EVs and induces specific activation of proinflammatory cytokine signaling in target cells. This work further highlights a surprising role for stem cell-derived EVs in the propagation of cellular signaling, and it represents a significant advance in the understanding the multiple levels of interaction that are established between endogenous versus grafted stem cells and the host immune system (Pluchino and Cossetti, 2013).
EXPERIMENTAL PROCEDURES
NPC Preparations
NPCs were prepared from the subventricular zone (SVZ) of 7- to 12-week-old mice, as described (Pluchino et al., 2008). Conditions for NPC preparation, cell culture, and lentiviral transductions are described in the Supplemental Experimental Procedures.
Collection of EVs and Exos from NPC Media
NPCs were single-cell dissociated and plated overnight at the concentration of 12 × 106 cells/10 ml medium/T75 culture flask. Tissue culture supernatants were collected and centrifuged for 15 min at 1,500 rpm (300 × g) and for 15 min at 2,500 rpm (1,000 × g) to remove cellular debris. EVs were collected by centrifugation at 100,000 × g for 70 min at 4°C using a Beckman Type 70.1 Ti Fixed-Angle Rotor. Pellets were washed with PBS and subjected to an additional centrifugation at 100,000 × g for 1 hr at 4°C using a Beckman Type TLA-55 Fixed-Angle Rotor. For Exo fractionation, pellets of crude EVs were resuspended in 0.5 ml 0.32 M sucrose. The pellet was layered on a 10 ml continuous sucrose density gradient (0.32–2 M sucrose, 5 mM HEPES [pH 7.4]) and centrifuged overnight at 100,000 × g (SW41 rotor; Beckman Coulter). Fractions (1 ml) were collected from the top (low density) to the bottom of the tube (high density) of the sucrose gradient. Fractions ranging from 6 to 9 (density between 1.13 and 1.20 g/ml) from the gradient were harvested, diluted in 0.32 M sucrose, centrifuged at 100,000 × g for 60 min (TLA55 rotor; Beckman Instruments), and then processed for either RNA or protein detection and analyses. Conditions for EV/Exo collection from Th1 versus Th2 cytokine media and transfer experiments are described in the Supplemental Experimental Procedures.
RNA and Protein Analyses
Conditions for RNA and Protein extraction, quantification, and detection from isolated NPCs, EVs, and Exos (Witwer et al., 2013), or from transfer experiments, are described in the Supplemental Experimental Procedures. Gene nomenclature is consistent with that of the Uniprot database (http://www.uniprot.org).
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dolores Di Vizio, Christian Frezza, Tony Green, Claudio Mauro, Jayden A. Smith, Gillian Tannahill, and Aviva Tolkovsky for critically discussing the article and Paula Francis for proof edits. We are grateful to Mario Amendola for his help with lentiviral vector generation; Andrew Malloy for his help with NTA; and Burkhard Becher, Institute of Experimental Immunology, Zürich, Switzerland and Birgit Strobl, University of Veterinary Medicine, Vienna, Austria for providing the Ifngr1−/− and Stat1−/− mice, respectively. We acknowledge the technical assistance of Carla Caddeo, Silvia Coco, Joanna Crawford, Sabine Dietman, Marcel E. Dinger, Giuseppe Filosa, Adaoha Ndibe, Claudio Picchi, Giovanni Pluchino, Gabriella Racchetti, Giuliana Salani, and Umberto Restuccia.
This work was supported by grants from the National Multiple Sclerosis Society (NMSS; RG-4001-A1 to S.P.), the Italian Multiple Sclerosis Foundation (FISM; RG 2010/R/31 to S.P.), the Italian Ministry of Health (GR08/7 to S.P.), the European Research Council (ERC) 2010-StG (RG 260511-SEM_SEM to S.P.), the European Community (EC) 7th Framework Program (FP7/2007-2013; RG 280772-iONE to S.P.), The Evelyn Trust (RG 69865 to S.P.), the Australian Research Council/University of Queensland cosponsored Federation Fellowship (grant FF0561986 to J.S.M.), and the Australian National Health and Medical Research Council Australia Fellowship (grant 631668 to J.S.M.). C.C. received a fellowship (SFRH/BD/15899/2005) from the Fundação para a Ciência e a Tecnologia (FCT). N.I. was supported by a FEBS longterm fellowship, and T.R.M. was supported by the Human Frontiers Science Program. M.P.D. and H.K.S. were supported by a BBSRC grant (BB/J01589X/1) to A.J.E.
Footnotes
ACCESSION NUMBERS
The NCBI Gene Expression Omnibus (GEO) accession number for RNA-seq data is GSE33527. Vesiclepedia accession numbers are 359, 360, 361, 362, 363, and 364.
Supplemental Information includes Supplemental Experimental Procedures, six figures, three tables, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2014.08.020.
REFERENCES
- Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- Antonyak MA, Li B, Boroughs LK, Johnson JL, Druso JE, Bryant KL, Holowka DA, Cerione RA. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc. Natl. Acad. Sci. USA. 2011;108:4852–4857. doi: 10.1073/pnas.1017667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakefield XO, Frederickson RM, Simpson RJ. Gesicles: Microvesicle “cookies” for transient information transfer between cells. Mol. Ther. 2011;19:1574–1576. doi: 10.1038/mt.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick L, Hoffman HM. cASCading specks. Nat. Immunol. 2014;15:698–700. doi: 10.1038/ni.2942. [DOI] [PubMed] [Google Scholar]
- Deregibus MC, Cantaluppi V, Calogero R, Lo Iacono M, Tetta C, Biancone L, Bruno S, Bussolati B, Camussi G. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110:2440–2448. doi: 10.1182/blood-2007-03-078709. [DOI] [PubMed] [Google Scholar]
- Dölken L, Ruzsics Z, Rädle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, Koszinowski UH. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008;14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drago D, Cossetti C, Iraci N, Gaude E, Musco G, Bachi A, Pluchino S. The stem cell secretome and its role in brain repair. Biochimie. 2013;95:2271–2285. doi: 10.1016/j.biochi.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJ, Hole P, Carr B, Redman CW, Harris AL, Dobson PJ, et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine. 2011;7:780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- EL Andaloussi S, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- Fellous M, Nir U, Wallach D, Merlin G, Rubinstein M, Revel M. Interferon-dependent induction of mRNA for the major histocompatibility antigens in human fibroblasts and lymphoblastoid cells. Proc. Natl. Acad. Sci. USA. 1982;79:3082–3086. doi: 10.1073/pnas.79.10.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Bluestone JA, Lim WA. Cell-based therapeutics: the next pillar of medicine. Sci. Transl. Med. 2013;5:ps7. doi: 10.1126/scitranslmed.3005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frühbeis C, Fröhlich D, Kuo WP, Amphornrat J, Thilemann S, Saab AS, Kirchhoff F, Möbius W, Goebbels S, Nave KA, et al. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013;11:e1001604. doi: 10.1371/journal.pbio.1001604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel RM, Aguet M. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Garotta G, Ozmen L, Ziemiecki A, Wilks AF, Harpur AG, Larner AC, Finbloom DS. Interferon-gamma induces tyrosine phosphorylation of interferon-gamma receptor and regulated association of protein tyrosine kinases, Jak1 and Jak2, with its receptor. J. Biol. Chem. 1994;269:14333–14336. [PubMed] [Google Scholar]
- Kwon SH, Liu KD, Mostov KE. Intercellular transfer of GPRC5B via exosomes drives HGF-mediated outward growth. Curr. Biol. 2014;24:199–204. doi: 10.1016/j.cub.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu K, Liu Y, Xu Y, Zhang F, Yang H, Liu J, Pan T, Chen J, Wu M, et al. Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat. Immunol. 2013;14:793–803. doi: 10.1038/ni.2647. [DOI] [PubMed] [Google Scholar]
- Lopez-Verrilli MA, Picou F, Court FA. Schwann cell-derived exosomes enhance axonal regeneration in the peripheral nervous system. Glia. 2013;61:1795–1806. doi: 10.1002/glia.22558. [DOI] [PubMed] [Google Scholar]
- Lubec G, Afjehi-Sadat L. Limitations and pitfalls in protein identification by mass spectrometry. Chem. Rev. 2007;107:3568–3584. doi: 10.1021/cr068213f. [DOI] [PubMed] [Google Scholar]
- Mangeot PE, Dollet S, Girard M, Ciancia C, Joly S, Peschanski M, Lotteau V. Protein transfer into human cells by VSV-G-induced nanovesicles. Mol. Ther. 2011;19:1656–1666. doi: 10.1038/mt.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino G, Pluchino S, Bonfanti L, Schwartz M. Brain regeneration in physiology and pathology: the immune signature driving therapeutic plasticity of neural stem cells. Physiol. Rev. 2011;91:1281–1304. doi: 10.1152/physrev.00032.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei V, Barenco MG, Tasciotti V, Garofalo T, Longo A, Boller K, Löwer J, Misasi R, Montrasio F, Sorice M. Paracrine diffusion of PrP(C) and propagation of prion infectivity by plasma membrane-derived microvesicles. PLoS ONE. 2009;4:e5057. doi: 10.1371/journal.pone.0005057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MA, Bernad A, Sánchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller G, Schneider M, Biemer-Daub G, Wied S. Microvesicles released from rat adipocytes and harboring glycosylphosphatidylinositol-anchored proteins transfer RNA stimulating lipid synthesis. Cell. Signal. 2011;23:1207–1223. doi: 10.1016/j.cellsig.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Pluchino S, Cossetti C. How stem cells speak with host immune cells in inflammatory brain diseases. Glia. 2013;61:1379–1401. doi: 10.1002/glia.22500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluchino S, Zanotti L, Rossi B, Brambilla E, Ottoboni L, Salani G, Martinello M, Cattalini A, Bergami A, Furlan R, et al. Neurosphere-derived multipotent precursors promote neuroprotection by an immunomodulatory mechanism. Nature. 2005;436:266–271. doi: 10.1038/nature03889. [DOI] [PubMed] [Google Scholar]
- Pluchino S, Muzio L, Imitola J, Deleidi M, Alfaro-Cervello C, Salani G, Porcheri C, Brambilla E, Cavasinni F, Bergamaschi A, et al. Persistent inflammation alters the function of the endogenous brain stem cell compartment. Brain. 2008;131:2564–2578. doi: 10.1093/brain/awn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana CV, Gil MP, Schreiber RD, Stark GR. Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol. 2002;23:96–101. doi: 10.1016/s1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- Sadir R, Forest E, Lortat-Jacob H. The heparan sulfate binding sequence of interferon-gamma increased the on rate of the interferon-gamma-interferon-gamma receptor complex formation. J. Biol. Chem. 1998;273:10919–10925. doi: 10.1074/jbc.273.18.10919. [DOI] [PubMed] [Google Scholar]
- Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struyf S, Proost P, Vandercappellen J, Dempe S, Noyens B, Nelissen S, Gouwy M, Locati M, Opdenakker G, Dinsart C, Van Damme J. Synergistic up-regulation of MCP-2/CCL8 activity is counteracted by chemokine cleavage, limiting its inflammatory and anti-tumoral effects. Eur. J. Immunol. 2009;39:843–857. doi: 10.1002/eji.200838660. [DOI] [PubMed] [Google Scholar]
- Théry C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J. Cell Biol. 1999;147:599–610. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- Uccelli A, Laroni A, Freedman MS. Mesenchymal stem cells for the treatment of multiple sclerosis and other neurological diseases. Lancet Neurol. 2011;10:649–656. doi: 10.1016/S1474-4422(11)70121-1. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer KW, Buzás EI, Bemis LT, Bora A, Lässer C, Lötvall J, Nolte-’t Hoen EN, Piper MG, Sivaraman S, Skog J, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Li Y, Cui Y, Yang JJ, Zhang ZG, Chopp M. Systemic administration of exosomes released from mesenchymal stromal cells promote functional recovery and neurovascular plasticity after stroke in rats. J. Cereb. Blood Flow Metab. 2013;33:1711–1715. doi: 10.1038/jcbfm.2013.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Yang F, Jiang L, Chen Y, Wang K, Xu F, Wei Y, Cao X, Wang J, Cai Z. Exosomes with membrane-associated TGF-β1 from gene-modified dendritic cells inhibit murine EAE independently of MHC restriction. Eur. J. Immunol. 2013;43:2461–2472. doi: 10.1002/eji.201243295. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Xiang X, Grizzle W, Sun D, Zhang S, Axtell RC, Ju S, Mu J, Zhang L, Steinman L, et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol. Ther. 2011;19:1769–1779. doi: 10.1038/mt.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.