Abstract
Arsenic is the most prevalent environmental toxic substance and ranks first on the U.S. Environmental Protection Agency’s Superfund List. Arsenic is a carcinogen and a causative agent of numerous human diseases. Paradoxically arsenic is used as a chemotherapeutic agent for treatment of acute promyelocytic leukemia. Inorganic arsenic has two biological important oxidation states: As(V) (arsenate) and As(III) (arsenite). Arsenic uptake is adventitious because the arsenate and arsenite are chemically similar to required nutrients. Arsenate resembles phosphate and is a competitive inhibitor of many phosphate-utilizing enzymes. Arsenate is taken up by phosphate transport systems. In contrast, at physiological pH, the form of arsenite is As(OH)3, which resembles organic molecules such as glycerol. Consequently, arsenite is taken into cells by aquaglyceroporin channels. Arsenic efflux systems are found in nearly every organism and evolved to rid cells of this toxic metalloid. These efflux systems include members of the multidrug resistance protein family and the bacterial exchangers Acr3 and ArsB. ArsB can also be a subunit of the ArsAB As(III)-translocating ATPase, an ATP-driven efflux pump. The ArsD metallochaperone binds cytosolic As(III) and transfers it to the ArsA subunit of the efflux pump. Knowledge of the pathways and transporters for arsenic uptake and efflux is essential for understanding its toxicity and carcinogenicity and for rational design of cancer chemotherapeutic drugs.
1. INTRODUCTION
Environmental arsenic comes primarily from natural geological sources such as volcanoes and hot springs (Inskeep & McDermott, 2005). An arsenic biogeocycle results from biological transformations of arsenic (Bhattacharjee & Rosen, 2007; Rensing & Rosen, 2009; Ye, Rensing, Rosen, & Zhu, in press). The Environmental Protection Agency (EPA) asserts that it pervades our drinking water (Council, 2001) and imperils the safety of our food supply (Stone, 2008). The contamination of groundwater by arsenic in Bangladesh has been called the largest poisoning of a population in history (Smith, Lingas, & Rahman, 2000). As a result of persistent human exposure and health hazard, arsenic ranks first on the Superfund List (http://www.atsdr.cdc.gov/spl/). Arsenic is a carcinogen and a causative agent of cardiovascular and peripheral vascular disease, neurological disorders, diabetes mellitus and various forms of cancer (Abernathy, Thomas, & Calderon, 2003; Tchounwou, Centeno, & Patlolla, 2004; Tseng et al., 2002). Arsenic exposure during pregnancy contributes to low birth weight and fetal loss, as well as delayed infant development (Tofail et al., 2009).
Arsenic is introduced anthropogenically as herbicides and pesticides, wood preservatives, animal feeds and semiconductors. For example, copper–chromium–arsenic (CCA) treatment was used for decades to preserve wood from fungi and insects. Although CCA is no longer used in the United States, playgrounds, household decks, boardwalks, telephone poles and other places where CCA-treated wood was used means that arsenic will contaminate our food and water supplies for the foreseeable future. Inorganic and organic arsenicals have been and are still used for agriculture and animal husbandry. Arsenic acid in the form of herbicides such as Desiccant L-10 by Atochem/Elf Aquitaine was used to defoliate cotton fields so that the next crops could be planted. This arsenic continues to contaminate those former cotton fields in the southern states in the US. Many of those fields are now planted with rice, which is the largest non-seafood source of arsenic in the American diet (Williams et al., 2005; Williams et al., 2007). Arsenic-contaminated brown rice syrup is used in baby food, a serious exposure problem for the most vulnerable (Jackson et al., 2012). Lead arsenate has been one of the most extensively used of arsenical insecticides. It was first used in 1892 against the gypsy moth (Lymantria dispar) (http://soils.tfrec.wsu.edu/leadhistory.htm). According to the EPA, lead arsenate was routinely used as a growth regulator on 17% of the U.S. grapefruit crop in the 1980s. Ten thousand pounds of lead arsenate were also used annually for control of cockroaches, silverfish and crickets (http://pmep.cce.cornell.edu/profiles/insect-mite/fenitrothion-methylpara/lead-arsenate/insect-prof-leadars.html). Even today the arsenic is finding its way into apple juice (http://www.doctoroz.com/videos/arsenic-apple-juice). In addition to inorganic arsenic, organic arsenicals are used in agriculture and animal husbandry. These include salts of methylarsenate [MAs(V)] and dimethylarsenate [DMAs(V)] (for example, one form of Ortho Weed-B-Gone Crabgrass Killer). Although they are no longer being produced, DMAs(V) and MAs(V) are still used as herbicides and fungicides for golf courses in Florida, and runoff from these golf courses introduce these arsenicals into drinking water (http://www.naplesnews.com/news/2006/oct/22/arsenic_levels_get_herbicide_pulled/?local_news). The organic arsenical roxarsone (4-hydroxy-3-nitrophenylarsonic acid) is fed to chickens as feed supplement. In addition to arsenic contamination of the chicken, arsenic-contaminated chicken litter is used as a fertilizer.
Paradoxically, arsenic and the related metalloid antimony have been used for centuries as an herbal remedy for a number of illnesses and more recently as chemotherapeutic agents (Kwong & Todd, 1997). Arsenic- and antimony-containing drugs are currently used for treating acute promyelocytic leukemia (APL) (the arsenical Trisenox) (Liu, Zhou, Chen, & Chen, 2012) and diseases caused by protozoan parasites (the antimonial Pentostam) (Carter & Fairlamb, 1993), and resistance to metalloid drugs is a serious clinical problem.
Inorganic arsenic has two biological important oxidation states: As(V) (arsenate) and As(III) (arsenite). At neutral pH, the arsenate oxyanion resembles phosphate and is a competitive inhibitor of many phosphate-utilizing enzymes. In contrast, As(III) is uncharged As(OH)3 at neutral pH (pKa 9.2). Consequently, As(V) uptake systems are arsenate anion transporters, while the substrate of As(III) transport systems is As(OH)3. This is significant, as discussed below, that most metalloids are taken up by aquaglyceroporin (AQP) channels as neutral species.
Arsenic uptake is by-and-large adventitious—cells have no reason to take up this poisonous metalloid (Bhattacharjee, Rosen, & Mukhopadhyay, 2009). In contrast, arsenic efflux systems are found in nearly every organism and evolved to rid cells of toxic metalloids (Rensing & Rosen, 2009). In bacteria and archaea, the genes for arsenic detoxification are usually encoded by arsenic resistance (ars) operons. Multidrug resistance protein (MRP) orthologues remove arsenite from the cytosol of fungi, plants and animals, including humans, as the reduced glutathione (GSH) conjugate As(GS)3. Knowledge of the pathways and transporters for metalloid uptake and efflux is essential for understanding toxicity and for rational design of metalloid drugs and for treating drug-resistant microorganisms and tumor cells.
2. UPTAKE SYSTEMS FOR ARSENIC
2.1. Arsenate Uptake
The arsenate oxyanion is chemically similar to phosphate, and its toxicity is based primarily on the competitive inhibition of proteins that use phosphate, as do many of the enzymes in intermediary metabolism and oxidative phosphorylation. However, the intracellular concentration of phosphate is usually quite high, so most cells are relatively insensitive to arsenate unless phosphate starved. In most organisms, arsenate is taken up adventitiously by phosphate transporters. For example, in Escherichia coli arsenate is taken up by two phosphate uptake systems (Willsky, Bennett, & Malamy, 1973; Willsky & Malamy, 1980), Pst and Pit. Pst is a high-affinity, low-capacity system induced by phosphate starvation, while Pit is a low-affinity, high-capacity constitutive system that is also the major uptake system for arsenate (Rosenberg, Gerdes, & Chegwidden, 1977).
2.2. Bidirectional Transport of Arsenite by Bacterial AQPs
Trivalent arsenite is considerably more toxic than pentavalent arsenate due to its tendency to react as a soft metal with thiols. Arsenite forms relatively weak bonds with monothiols, and high intracellular concentrations of arsenite can deplete cells of glutathione. It forms strong bonds with dithiols in small molecules such as the lipoic acid cofactor and with vicinal thiols in proteins, leading to inactivation of various enzymes and receptors. Trivalent arsenic is often referred to in the literature as the anion arsenite. But, in contrast to arsenate, which is a negatively charged oxyanion in solution, trivalent inorganic arsenic is, in fact, the neutral undissociated acid As(OH)3 at physiological pH (Ramírez-Solis, Mukopadhyay, Rosen, & Stemmler, 2004). For convenience the terms As(OH)3, As(III) and arsenite will be used interchangeably in this chapter.
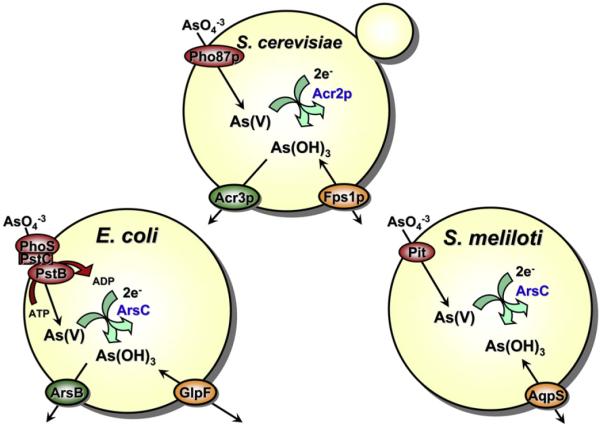
Unlike arsenate, which was long known to be taken up by phosphate transporters, how the more toxic arsenite enters the cell was unknown until the ground-breaking study by Sanders, Rensing, Kuroda, Mitra, and Rosen (1997). Using transposon mutagenesis to identify genes associated with accumulation of metalloids in E. coli, they isolated an Sb(III)-tolerant strain in which the glpF gene was disrupted by insertion of the transposon. Antimony is a related metalloid that shares chemical properties with arsenic. Subsequently, Meng et al. (2004) showed that the glpF mutant also had a significantly reduced rate of arsenic uptake. GlpF is a bacterial glycerol facilitator (Heller, Lin, & Wilson, 1980) that is the first identified member of the AQP or Major intrinsic membrane protein (MIP) superfamily, for which Peter Agre was awarded the Nobel Prize in Chemistry in 2003 (Agre, 2004). There are two main branches in the superfamily: true aquaporins, which are water channels, and AQPs, channels with larger pores that conduct water, glycerol and other small neutral solutes (Agre et al., 2002). These findings suggest that both arsenite and antimonite can be recognized by GlpF as polyol forms related to glycerol. With a pKa of 9.2, arsenite is not an anion but was shown to be uncharged As(OH)3 under physiological conditions (Ramírez-Solis et al., 2004). Although GlpF is responsible for adventitious uptake of As(III) in E. coli, the legume symbiont Sinorhizobium meliloti uses a clever strategy for resistance to arsenate (but not arsenite) using a GlpF orthologue, AqpS, for efflux of As(III) (Fig. 1) (Yang, Cheng, Finan, Rosen, & Bhattacharjee, 2005). When S. meliloti is exposed to environmental arsenate, it is taken up by phosphate transporters and reduced inside the cell to trivalent arsenite by an arsenate reductase, ArsC. The internally generated trivalent arsenite then flows downhill out of the cell through AqpS. Together, AqpS and ArsC constitute a novel arsenate detoxification pathway in S. meliloti. The presence of aqpS and arsC in the meliloti arsenic resistance operon is not an isolated observation, and the combination of AQP and arsenate reductase may confer arsenate tolerance in other bacterial species. The presence of orthologous glycerol facilitators in E. coli and S. meliloti that produce either metalloid sensitivity or resistance demonstrates that bidirectional AQPs can participate in either arsenite uptake or efflux, depending on the direction of the concentration gradient of arsenite.
Figure 1. Pathways of arsenic uptake and efflux.
Arsenate [As(V)] is taken up by phosphate transporters, and As(III) is conducted up by AQPs (GlpF in Escherichia coli, Fps1p in yeast, and AQP7 and AQP9 in mammals). In both E. coli and Saccharomyces cerevisiae, arsenate is reduced to arsenite by arsenate reductases. In E. coli and many other bacteria, arsenite is extruded from the cells by ArsB alone or by the ArsAB ATPase. In many other bacteria, Acr3 replaces ArsB as the plasma membrane arsenite efflux protein. (Some bacteria have both ArsB and Acr3!) Fungi also use Acr3 for arsenite efflux, and yeast Ycf1p, which is a member of the MRP group of the ABC superfamily of drug resistance ATPases, pumps As(GS)3 into the vacuole. The legume symbiotic bacterium Sinorhizobium meliloti is unique in that it uses an AQP for arsenite efflux rather than uptake. In contrast to most bacteria, S. meliloti is very sensitive to arsenite because it has neither ArsB nor Acr3. It is resistant to arsenate, which, as in other organisms, is brought into cells by the phosphate transporters. The first step of detoxification involves reduction of arsenate to arsenite by ArsC. The AqpS channel facilitates downhill transport of internally generated As(III). Thus, this detoxification mechanism functions to confer arsenate but not arsenite resistance in S. meliloti. See the color plate.
2.3. AQPs Are As(III) Transporters in Eukaryotes
Just as prokaryotes use AQPs for As(III) uptake, eukaryotes have AQPs that facilitate the uptake of arsenic and other metalloids (Bhattacharjee, Mukhopadhyay, Thiyagarajan, & Rosen, 2008). For example, the yeast Fps1p is an orthologue of GlpF that is responsible for uptake and efflux of glycerol in Saccharomyces cerevisiae. Deletion of fps1 enhances the tolerance to arsenite and potassium antimonyl tartrate (Liu et al., 2002; Wysocki et al., 2001), suggesting that Fps1p mediates the influx of trivalent metalloid in yeast, so that its absence confers tolerance. Under elevated osmolarity conditions, when the Fps1p channel is closed, wild-type cells show a similar degree of metalloid tolerance as does an fps1 deletion mutant. In contrast, yeast cells expressing a constitutively open form of the Fps1p are extremely sensitive to both arsenite and antimonite. Direct transport assays also indicate that arsenite uptake is mediated by Fps1p. A mitogen-activated protein kinase Hog1p phosphorylates Fps1p on Thr 231 in vivo and regulates influx of arsenic in response to arsenite (Thorsen et al., 2006). The arsenic sensing and signaling events mediated by activation of Hog1p and Fps1p contribute to the tolerance of arsenic acquisition. Wysocki, R., Bobrowicz, P., & Ulaszewski, S. (1997) first showed that Fps1p facilitates efflux of As(III) out of cells and is essential to maintain As(V) resistance (Maciaszczyk-Dziubinska, E., Migocka, M., & Wysocki, R. (2012)). As(V) enters the yeast cell through phosphate transporters (Bun-ya et al., 1996). The arsenate reductase Acr2p converts As(V) to As(III) followed by efflux As(III) out of the cells by both Fps1p and arsenite permease Acr3p (Fig. 1) (Ghosh, Shen, & Rosen, 1999; Liu et al., 2002). This scenario is reminiscent to the As(V) tolerance conferred by the concerted action of AqpS and ArsC in S. meliloti (Yang et al., 2005). It is tempting to speculate that bidirectional As(III) movement by AQPs is involved in both arsenic uptake and efflux in other organisms.
Plants have many AQPs, and a number of these have been shown to facilitate uptake of metalloids, including toxic arsenic and antimony, as well as boron and silicon, which are required in plants, and probably germanium (Bhattacharjee et al., 2008). This has led to the postulate that AQPs may have evolved in the first cells to allow uptake of required metalloids rather than organic polyols, which were not likely to be present in high concentrations in primordial oceans. Vertebrates have fewer; of more than 13 members of AQPs identified in mammals, four (AQP3, AQP7, AQP9 and AQP10) are classical AQPs (Ishibashi et al., 1997, 1998; Ishibashi, Imai, & Sasaki, 2000; Ishibashi, Morinaga, Kuwahara, Sasaki, & Imai, 2002). A yeast Fps1p deletion mutant expressing rat AQP9 was used as a model for studying the function of mammalian AQPs (Liu et al., 2002). The fps1 mutant showed enhanced tolerance to arsenite and antimonite, and had diminished uptake of both metalloids. Complementation of rat AQP9 significantly reversed tolerance and transport of metalloids. Notably, higher rates of metalloid uptake and greater metalloid susceptibility were observed. These reports indicate that AQP9 is a better metalloid channel than Fps1p.
Xenopus laevis oocytes microinjected with either rat AQP7 or AQP9 complementary RNA (cRNA) demonstrated increased uptake of arsenite by AQP7 and a lesser degree by AQP9 (Liu et al., 2002). Likewise, the functions of four known human AQPs hAQP3, hAQP7, fAQP9 and hAQP10 in arsenite transport were determined in X. laevis oocytes (Liu, Carbrey, Agre, & Rosen, 2004). The human AQP9 is a more effective arsenite transporter than that of hAQP7, while little or no uptake was detected in hAQP3 and hAQP10. AQP9 residues Phe64 and Arg219 were predicted to be part of the selectivity filter based on the comparison with the crystal structure of the bacterial homologue GlpF and bovine erythrocyte water channel bAQP1. To investigate the requirement of these residues in the substrate selectivity and conductivity between glycerol and As(OH)3, site-directed mutagenesis was employed. The results from uptake assay showed that a positive-charged lysine residue could substitute for Ala219, which is required at the entry to the AQP9 channel. On the other hand, a hydrophobic phenylalanine residue predicted to position glycerol near the conserved arginine residue is actually dispensable for uptake of substrate (Liu et al., 2004). Taken together, these results suggest that arsenite shares the same translocation pathway with glycerol in AQP9.
The conversion of inorganic arsenic to methylated arsenic is a common metabolic reaction in mammals (Thomas, Waters, & Styblo, 2004). For example, arsenic trioxide is methylated by then enzyme arsenite-S-adenosylmethionine methyltransferase (AS3MT) in mammalian liver. One of the initial products of arsenic methylation is methylarsonous acid, MAs(III), a more toxic and carcinogenic form than inorganic arsenite. Subsequently, these methylated arsenicals are released into circulation and excreted as oxidized species in urine and feces. However, questions on how methylated arsenicals pass through liver cells and translocate to other tissues and the underlying mechanism remain unanswered. Since AQP9 is the major liver isoform, the uptake of methylated arsenicals by GlpF, Fps1p and rat AQP9 was investigated (Liu, Styblo, & Rosen, 2006). Rat AQP9 facilitates uptake of MAs(III) at a higher rate than As(III). In addition, rat AQP9 is a better channel for MAs(III) than that of Fps1p. Mechanistically, the Phe64 is not required, whereas Arg219 is indispensable for MAs(III) translocation. These findings indicate that inorganic and methylated As(III) use the identical translocation pathway in AQP9. Since AQPs have been shown to be bidirectional in bacteria and yeast, As(OH)3 has been proposed to flow down its concentration into hepatocytes, where it is methylated to MAs(OH)2 by AS3MT, and the methylated product then flows out of the cell again by AQP9, where it is excreted in urine either before or after nonenzymatic and spontaneous oxidation to the urinary species MAs(V) (Fig. 2) (Liu et al., 2006).
Figure 2. Uptake and efflux of trivalent arsenic species in liver.
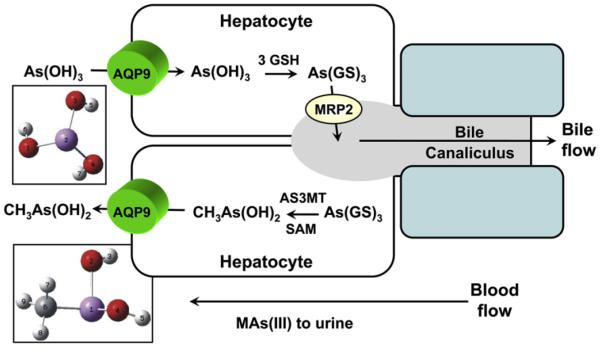
Inorganic As(OH)3 flows down its concentration gradient from blood into hepatocytes through AQP9, the liver AQP isoform. In the cytosol of the hepatocyte, As(III) can be either glutathionylated or methylated to MAs(V), which is reduced to MAs(III). As(GS)3 is pumped into bile by MRP2. Alternatively, As(III) is methylated by the enzyme As(III) SAM methyltransferase (AS3MT) to CH3As(OH)2, which then flows down its concentration gradient via AQP9 into blood, to the kidney and eliminated in the urine. See the color plate.
Since AQP9 is expressed in liver, brain, testes and leukocytes, these tissues may be highly sensitive to arsenic toxicity and carcinogenesis. The susceptibility of AQP9 expressed tissues may be further complicated by a variety of chemical modifications of arsenic, including methylation, oxidation, reduction, and glutathionylation. Carbrey et al. (2003) constructed AQP9-null mice as animal model for the evaluation of arsenic toxicity in vivo. Upon injection of sodium arsenite (NaAsO2), AQP9-null mice display reduced survival. The enhanced accumulation of arsenic is detected in several organs, with the highest in heart and up to 20 times higher than wild-type control. In addition, the AQP9-null mice sustain profound bradycardia within hours after sodium arsenite injection. Despite the arsenic level is detected in multiple organs, the excretion of arsenic in urine and feces is significantly reduced in AQP9-null mice with only one-tenth arsenic level of wild-type mice. The reduced clearance of different arsenicals by AQP9-null mice indicates that AQP9 is involved in the export of multiple arsenic species. Taken together, these results clearly demonstrate that AQP9 facilitates in vivo excretion of arsenicals by liver and suggest a role of AQP9 in partial protection against arsenic toxicity.
The role of AQPs in arsenic transport is an uncharted territory in emerging animal models. Zebra fish (Danio rerio) may develop into a valuable aquatic vertebrate model for investigating the environmental effects of arsenic. Among seven AQPs annotated in the zebra fish genome project, Hamdi et al. (2009) cloned five genes (aqp3, aqp3l, aqp9a, aqp9b and aqp10), which encode homologues of human AQP3, AQP9 and AQP10. Using Reverse transcription polymerase chain reaction (RT-PCR), these AQP genes were detected in multiple tissues and exhibited differential tissue expression. As in humans, liver is the primary organ of arsenic accumulation and detoxification in zebra fish. These zebra fish AQPs show substantial glycerol transport at equivalent rates. In addition, these water channels facilitate the uptake of As(III), MAs(III) and Sb(III). These findings suggest that AQPs are the major pathways for the accumulation of inorganic and methylated arsenicals in zebra fish tissues. However, it is not clear whether AQPs facilitate translocation of pentavalent form of methylated arsenicals such as MAs(V) and DMAs(V), which are the primary forms of organoarsenicals excreted in the urine of mammals. More recently, McDermott, Jiang, Beene, Rosen, and Liu (2010) demonstrated that X. laevis oocytes microinjected with human AQP9 cRNA showed enhanced uptake of both MAs(V) and DMAs(V) at acidic pH (pH 5.5) compared with neutral pH. Hg(II), an aquaporin inhibitor, prevented conduction of arsenicals through AQP9. In contrast, phloretin, an inhibitor of water and glycerol permeation via AQP9, inhibited translocation of pentavalent MAs(V) and DMAs(V) but not trivalent As(III) and MAs(III). These results imply that the translocation pathway for the pentavalent arsenical species is different from that of the trivalent.
2.4. Arsenic and AQPs in Cancer Chemotherapy
In a series of clinical studies evaluating the efficacy of arsenic in treatment of various cancers, intravenous injection of arsenic trioxide (As2O3) was shown to induce a complete remission in patients of newly diagnosed or relapsed APL (Emadi & Gore, 2010). APL is featured with the t(15;17) (q22;q21) chromosomal translocation which fuses the promyelocytic leukemia gene (PML) with the retinoic acid receptor α gene (RAR α) (Au et al., 2011). The PML–RAR fusion gene encodes a chimeric protein that leads to arrest of maturation at the promyelocyte stage of myeloid cell development (Soignet, 2001). Low concentrations of arsenic (0.1–0.5 μM) have been shown to induce differentiation of malignant promyelocyte by inactivation of PML–RAR fusion protein, whereas higher concentration of arsenic (0.5–2.0 μM) causes apoptosis through different signaling pathways (Dilda & Hogg, 2007). These signaling pathways include modulation of redox signaling by arsenic-generated reactive oxygen species (Dai, Weinberg, Waxman, & Jing, 1999) and inactivation of Nuclear factor-kappaB (NF-kB) pathway through binding Inhibitor of nuclear factor-kappaB (IkB) kinase with arsenic (Kapahi et al., 2000). Additionally, arsenic also induces proliferation arrest in several kinds of cancer cells, such as multiple myeloma and myelodysplastic syndromes (Dilda & Hogg, 2007; Emadi & Gore, 2010). To understand the mode of action of the arsenic-based drug, it is essential to delineate the absorption, distribution, metabolism, excretion and toxicity of arsenic drugs in cancer treatment.
Transfection of AQP9 into human leukemia cells renders them hypersensitive to Trisenox (arsenic trioxide) (Bhattacharjee, Carbrey, Rosen, & Mukhopadhyay, 2004). This drug sensitivity is attributed to increased rates of arsenic uptake and proportional to the expression level of AQP9 in different lineages of leukemia cells (Leung, Pang, Yuen, Kwong, & Tse, 2007). HL60 cells treating with vitamin D displayed enhanced sensitivity to arsenic and elevated expression of AQP9. Sensitivity was a consequence of higher rates of arsenic uptake due to elevated levels of AQP9. Similarly, upregulation of AQP9 correlated with increased arsenic uptake in HL60 cells treated with all-trans retinoic acid (ATRA) (Leung et al., 2007). The elevated arsenic uptake led to arsenic-induced cytotoxicity. This may explain the synergism between ATRA and arsenic, where APL patients show improved response upon concomitant treatment with both (Hu et al., 2009). These observations indicate the potential for application of pharmacological agents to enhance AQP9 expression for improving treatment of leukemia.
As chronic exposure of arsenic in drinking water leads to a variety of clinical symptoms, it becomes an emerging epidemic in many regions of the world. Even worse, the clinical symptoms of arsenic poisoning are exacerbated by poor nutritional status. The consequence of arsenic poisoning in populations exposed to high levels of arsenic in the ecosystem is mainly determined by the interaction of multiple factors, including gene expression, environment and nutrition. These factors may dictate the arsenic level in different tissues through modulating the expression level of AQPs. For example, starvation and diabetes mellitus upregulate AQP9 expression up to 20-fold in liver. In addition, AQP9 levels correlate with the nutritional status of the subject and circulating insulin levels (Carbrey et al., 2003). As a result, people with malnutrition are more susceptible to arsenic-induced hepatic toxicity upon drinking arsenic-contaminated water (Agre & Kozono, 2003). Moreover, AQP9 levels are also regulated by steroid hormones (Pastor-Soler et al., 2010) and human chorionic gonadotrophin (Marino, Castro-Parodi, Dietrich, & Damiano, 2010). Further studies on the relationship among AQP expression, arsenic transport and subsequent pharmacological response will elucidate the roles of AQPs in modulating the effects of arsenic in human health and diseases.
3. EFFLUX SYSTEMS FOR ARSENIC
3.1. A Multiplicity of As(III) Efflux Systems
As a consequence of the ubiquity of arsenic, nearly every organism has intrinsic or acquired mechanisms for arsenic detoxification. This includes biotransformations such as oxidation and reduction (Bhattacharjee & Rosen, 2007; Mukhopadhyay & Rosen, 2002), methylation (Ye et al., in press) and demethylation (Yoshinaga, Cai, & Rosen, 2011). But, by far, the most common detoxification mechanism is removal of As(III) from the cytosol, by either efflux out of the cell or sequestration in an organelle (Bhattacharjee & Rosen, 2007). To date, all identified arsenic efflux systems are for trivalent As(III); no As(V) efflux systems have been found. Considering that As(III) is more toxic and carcinogenic than As(V), it is counterintuitive that extrusion systems for As(V) do not exist. We have postulated that one of the earliest challenges of the first cells was to grow in the presence of heavy metals and metalloids that were likely present at high concentrations in primordial oceans (Rosen, 1999). The atmosphere was neutral, and arsenite would have predominated over arsenate, favoring the evolution of transporters that extrude arsenite out of the cell. As the atmosphere became oxidizing, most As(III) would have become oxidized to As(V), so proteins evolved that reduce arsenate to arsenite, the substrate of the existing transport systems.
In many eukaryotes As(IIII) resistance is conferred by members of the MRP group of the Adenosine triphosphate binding cassette (ABC) superfamily of transport ATPases that are physiologically involved in export of GS conjugates such as leukotriene C4 (Haimeur, Conseil, Deeley, & Cole, 2004). MRP1-catalyzed export of glutathione from cells was increased by arsenite, suggesting that MRP1 functions as an ATP-driven As(GS)3 pump (Zaman et al., 1995). In liver, MRP2 extrudes arsenic–glutathione complexes into bile and may be a major route of arsenic detoxification in humans (Fig. 2) (Cui, Kobayashi, Hayakawa, & Hirano, 2004). MRP homologues have been shown to confer arsenic resistance in eukaryotic microbes. Metalloid-containing drugs are still the first-line therapy for trypanosomiasis and leishmaniasis, and clinical resistance is a serious problem in treatment. In arsenite-resistant strains selected in vitro, there is increased expression of pgpA, which encodes an MRP orthologue that pumps arsenite as As(GS)3 (Dey, S., Papadopoulou, B., Haimeur, A., Roy, G., Grondin, K., Dou, D., et al. (1994); Legare et al., 2001). In S. cerevisiae, an MRP orthologue, Ycf1p, confers Cd(II) resistance by pumping Cd(GS)2 into the vacuole (Li et al., 1997). Ycf1p also transports As(GS)3 into the vacuole and confers arsenite resistance in yeast (Ghosh et al., 1999). In addition to Ycf1p, S. cerevisiae has a gene cluster of three ACR genes that also confers arsenic resistance (Bobrowicz, Wysocki, Owsianik, Goffeau, & Ulaszewski, 1997). Acr1p is a transcription factor, and Acr2p is an arsenate reductase (Mukhopadhyay & Rosen, 1998), and Acr3p is an arsenite permease (Ghosh et al., 1999; Wysocki, Bobrowicz, & Ulaszewski, 1997).
3.2. The Acr3 Permease
In bacteria and archaea, arsenic resistance genes are usually organized in ars operons that are controlled by an ArsR As(III)-responsive transcriptional repressor (San Francisco, Hope, Owolabi, Tisa, & Rosen, 1990). Nearly every ars operon has a gene encoding one of two unrelated As(III) efflux proteins, Acr3 or ArsB. The Acr3 permease is a member of the bile/arsenite/riboflavin transporter (BART) superfamily (Mansour, Sawhney, Tamang, Vogl, & Saier, 2007). The Acr3 subfamily includes members found in bacteria, archaea and fungi and is at least as widespread and perhaps more so than members of the ArsB family (Achour, Bauda, & Billard, 2007; Mansour et al., 2007). Unfortunately, the literature is confused by the fact that many members of the Acr3 family are annotated as ArsB even though they exhibit no significant sequence similarity to ArsB. The first identified member of this family is encoded by the ars operon of the skin (sigK intervening) element in the chromosome of Bacillus subtilis (Sato & Kobayashi, 1998).
There are no protein structural data available for Acr3 or related BART proteins, but the transmembrane topology of Acr3 has been explored. The membrane topology of the B. subtilis Acr3 was investigated using translational fusions, but the results could not distinguish between 8 and 10 transmembrane segments (Aaltonen & Silow, 2008). Cysteine scanning mutagenesis and accessibility to thiol-reactive probes were used to determine the transmembrane topology of Acr3-1 from iron-reducing alkaliphile, Alkaliphilus metalliredigens (Fu et al., 2009). The results clearly demonstrate that AmAcr3-1 has 10 transmembrane spanning segments, with the N- and C-termini localized in the cytosol. This method is more likely to yield an accurate topology since it utilizes active membrane proteins. A highly conserved cysteine residue in the P/R-C-T/I-AMV motif of the third transmembrane segment (TM3) of Corynebacterium glutamicum Acr3-1 (CgAcr3-1) and a glutamate in the TM9 may be involved in metalloid translocation. Replacing either Cys129 or Glu305 led to loss of transport activity in CgAcr3-1. The requirement for a thiol suggests that the transport mechanism of Acr3 may be different from that of ArsB, in which no cysteine residues are required for As(III) transport (Chen, Dey, & Rosen, 1996). The role of these residues in mediating transport is still unknown, but it is reasonable to speculate that both residues may serve as selectivity filter for As (III).
The properties of a more distant homologue from Shewanella oneidensis were examined (Xia et al., 2008). The S. oneidensis homologue confers resistance to arsenate but not arsenite. The purified protein binds arsenate, not arsenite, indicating that it is not a true Acr3 orthologue. There is, however, debate over the substrate specificity for Acr3. Fungal members of this family include the ScAcr3p metalloid efflux protein from S. cerevisiae, which was proposed to be selective for As(III) over Sb(III) (Maciaszczyk, Wysocki, Golik, Lazowska, & Ulaszewski, 2004). Recently, ScAcr3p has been suggested to transport Sb(III) in vivo as well (Maciaszczyk-Dziubinska et al., 2012), and yeast plasma membrane vesicles accumulate both As(III) and Sb(III) by exchange with protons (Maciaszczyk-Dziubinska et al., 2012). However, other data indicating that Sb(III) is not transported in yeast plasma membrane vesicles are not in agreement with that conclusion (Ghosh et al., 1999), and bacterial Acr3 orthologues appear to be specific for As(III) and do not transport Sb(III) (Fu et al., 2009; Villadangos et al., 2012).
Cells of E. coli expressing AmAcr3-1 or CgAcr3-1 confer As(III) resistance (Fu et al., 2009). Everted membrane vesicles prepared from C. glutamicum expressing Acr3-1 were shown to transport As(III) but not Sb(III) using Nicotinamide adenine dinucleotide (NADH) as an energy source. However, neither dissipating the positive interior Δψ alone nor the acid interior ΔpH alone had a significant effect on NADH-driven accumulation of As(III) in everted vesicles, indicating that either individual component of the electrochemical proton gradient, Δψ or ΔpH, is capable of energizing As(III) transport via CgAcr3-1. These results demonstrate that Acr3 is an electrophoretic metalloid–proton exchanger with As(OH)3/H+ antiporter activity (Villadangos et al., 2012).
Some plants also have proteins related to Acr3 that confer arsenic tolerance. For example, an Acr3 orthologue was recently identified in the vacuole of the brake fern Pteris vittata and is responsible for arsenic tolerance in the plant (Indriolo, Na, Ellis, Salt, & Banks, 2010). In contrast, rice (Oryza sativa) does not have Acr3, yet arsenic in the rice grain is a serious health hazard (Zhao, McGrath, & Meharg, 2010). To attempt to mitigate this problem, yeast ScAcr3 was expressed in rice, with the result that the arsenic content in the grain was reduced (Duan, Kamiya, Ishikawa, Arao, & Fujiwara, 2012).
3.3. The ArsB Permease and the ArsAB As(III)-Translocating ATPase
Many ars operons have three genes, arsRBC. There are at least three related ArsR transcriptional repressors with As(III)-binding sites in different locations in the protein (Ordóñez et al., 2008; Qin et al., 2007; Shi, Dong, Scott, Ksenzenko, & Rosen, 1996). ArsB, like Acr3, is a As(OH)3/H+ antiporter that extrudes As(III), conferring resistance (Meng et al., 2004). Although it has not been identified in animals, an ArsB homologue exists in plants (Ma et al., 2007). ArsC is an arsenate reductase that converts As(V) to As(III), the substrate of ArsB, extending the range of resistance to include As(V) (Gladysheva, Oden, & Rosen, 1994; Ji & Silver, 1992). Some have two more genes, arsD and arsA, such as the arsRDABC operon in E. coli plasmid R773 (Chen, Misra, Silver, & Rosen, 1986; San Francisco et al., 1990). ArsA and ArsB form the ArsAB complex, a pump utilizing the energy of ATP hydrolysis for As(III) or Sb(III) extrusion (Hsu & Rosen, 1989; Mobley & Rosen, 1982; Rosen, Weigel, Karkaria, & Gangola, 1988). ArsD is a metallochaperone that transfers As(III) or Sb(III) to ArsA (Lin, Walmsley, & Rosen, 2006). Cells expressing the five genes arsRDABC are more resistant to As(V) and As(III) than those expressing only the arsRBC genes (Dey & Rosen, 1995).
3.3.1. The ArsB Permease
Nearly every prokaryote has either an arsB or acr3 gene, which are found in roughly equal frequencies. Often there are multiple copies of either, and sometimes both within a single organism. For example, E. coli has a chromosomal arsB gene as well as plasmids that have additional copy or copies. ArsB from plasmid R773 is a 429-residue membrane protein with 12 transmembrane segments (Tisa & Rosen, 1990; Wu, Tisa, & Rosen, 1992). ArsB is an antiporter that extrudes As(III) or Sb(III) from cells by H+/As(OH)3 exchange coupled to the electrochemical proton gradient (Meng et al., 2004). Many proteins with As(III)-binding sites have groups or pairs of cysteine residues in the binding site. In contrast, ArsB has only a single cysteine residue that could be changed to serine or alanine residues without effect. Thus, unlike most other As(III) detoxification proteins, ArsB does not use soft metal chemistry in As(III) transport.
ArsB is unique in having a dual mode of energy coupling depending on its association with the ArsA ATPase. In vivo studies on the energetics of arsenite extrusion showed that expression of both arsA and arsB genes resulted in ATP-coupled arsenite extrusion independent of the electrochemical proton gradient (Dey & Rosen, 1995). In contrast, in cells expressing only the arsB gene, arsenite extrusion was coupled to electrochemical energy and independent of ATP. Everted membrane vesicles from E. coli cells expressing the ArsA–ArsB complex catalyze ATP-coupled uptake of As(III) (Dey, Dou, Tisa, & Rosen, 1994). On the other hand, everted membrane vesicles prepared from cells expressing only arsB exhibited As(III) uptake coupled to electrochemical energy (Kuroda, Dey, Sanders, & Rosen, 1997). These results suggest that ArsB can be either a secondary antiporter coupled to the proton motive force or an obligatory ATP-coupled pump depending on the subunit composition of the transport complex.
3.3.2. The ArsA As(III)-Activated ATPase
The arsRDABC operon of E. coli plasmid R773 encodes the ArsAB ATPase, a metalloid pump that confers resistance by actively extruding As(III) or Sb(III) from cells (Rosen, 2002). The 583-residue ArsA is a metalloid-activated ATPase that comprises the catalytic subunit of the pump (Hsu & Rosen, 1989). It is normally bound to ArsB (Dey, S., Dou, D., Tisa, L. S., & Rosen, B. P. (1994)), but, in the absence of ArsB, ArsA is found in the cytosol and can be purified as a soluble protein (Hsu & Rosen, 1989). ArsA has two homologous halves, the N-terminal A1 domain (residues 1–288) and the C-terminal A2 domain (residues 314–583), which are connected by a flexible 25-residue linker peptide (residues 289–313). Each half has a consensus nucleotide-binding domain (NBD). At the interface of the two domains is a binding site for As(III) or Sb(III), the metalloid-binding domain (MBD) involved in activation of ATPase activity. Connecting the MBD to the two NBDs are signal transduction domains (STDs) in each half of ArsA. Although the NBDs show considerable sequence similarity, they are not identical. Both are required for resistance, transport and ATPase activity (Karkaria, Chen, & Rosen, 1990; Kaur & Rosen, 1992), and they interact with each other (Li, Liu, & Rosen, 1996; Zhou, Radaev, Rosen, & Gatti, 2000). Mg2+ is required for ArsA ATPase activity (Hsu & Rosen, 1989). Conformational changes at the nucleotide-binding site, as evidenced by an increase in intrinsic tryptophan fluorescence, were observed only on the addition of MgATP, suggesting that Mg2+ binds to ArsA as a complex with ATP (Zhou, Liu, & Rosen, 1995). An aspartic residue in ATP- and GTP-binding proteins serves as Mg2+ ligand. Sequence alignment of ArsA with other ATPase such as nitrogenase iron protein (NifH) (Georgiadis, Komiya, Woo, Kornuc, & Rees, 1992), RecA (Story & Steitz, 1992) and Ras p21 (Pai et al., 1990) suggested that Asp45 might be an Mg2+ ligand in NBD1. Cells expressing Asp45 mutants lost arsenite resistance. Purified D45A and D45N ArsAs were inactive, whereas the D45E enzyme exhibited 5% of wild-type activity, with a five-fold decrease in affinity for Mg2+, demonstrating that Asp45 is part of the Mg2+-binding site.
3.3.2.1. The Nucleotide-Binding Domains
Both NBD1 and NBD2 hydrolyze ATP, with steady state hydrolysis dominated by the activity of NBD1. The overall rate of ATP hydrolysis is slow in the absence of metalloid and is accelerated by metalloid binding. NBD1 and NBD2 appear to play different roles in catalysis. ATP binding and hydrolysis at the individual NBDs were examined by photolabeling with the ATP analogue 8-azido-5′-[α-(32)P]-ATP at 4 °C (Jiang et al., 2005). Metalloid activation correlated with a >10-fold increase in affinity for nucleotide. To investigate the relative contributions of the two NBDs in catalysis, a thrombin site was introduced in the linker between ArsA1 and ArsA2. This allowed discrimination between incorporation of labeled nucleotides into the two halves of ArsA. The results indicate that both NBD1 and NBD2 bind and hydrolyze ATP even in the absence of metalloid. Sb(III) increases the affinity of NBD1 to a greater extent than NBD2. 8-Azido-5′-[γ-(32)P]-ATP was used to measure ATP hydrolysis at 37 °C. Under these catalytic conditions, both nucleotide-binding domains hydrolyze ATP, but hydrolysis in A1 is stimulated to a greater degree by Sb(III) than A2. These results are strong support for the hypothesis that the two homologous halves of the ArsA are functionally nonequivalent.
Other lines of evidence for supporting different roles for the two NBDs came from biochemical studies of the two deviant lysines in the Walker A motifs of NBD1 and NBD2 (Fu, Ajees, Rosen, & Bhattacharjee, 2010). Many ATPases have a Walker A ATP-binding motif (also called the P-loop or phosphate-binding loop), G/AxxxxGKT/S (where x is any amino acid), that makes direct contact with the phosphates of the bound ATP. This family of ATPases has been subdivided into those containing a majority consensus Walker A motif and a subgroup with the “deviant” Walker A motif subgroup, xKGGxxK(T/S) (Koonin, 1993). ArsA is a member of the deviant Walker A motif family, whose members have diverse functions including arsenite transport (ArsA), nitrogen fixation (NifH), DNA segregation (ParA) and spatial regulation of cell division (MinD). An emerging theme for this subgroup is that the role of ATP is to modulate the interaction of the protein with itself and its partner (Lutkenhaus & Sundaramoorthy, 2003; Schindelin, Kisker, Schlessman, Howard, & Rees, 1997). For NifH and ArsA, the dimer is in a more open conformation in the presence of ADP and contains two nucleotide-binding sites at the interface of the two monomers. The binding sites are composed primarily of amino acid residues coming from one of the monomers (Schindelin et al., 1997). Importantly, NifH has also been captured in the transition state, which reveals a more closed conformation, with important new intermonomer contacts. The most important of these is made by the signature “deviant” lysine, which contacts a phosphate of the nucleotide bound to the opposite monomer. To investigate the role of deviant lysines in the Walker A motif of ArsA, Lys16 in NBD1 and Lys335 in NBD2 were mutated for functional analysis (Fu et al., 2010). Phenotypic and biochemical data suggested that Lys335 is necessary for completing NBD1, which is required for ArsA to go through the catalytic cycle by hydrolysis of the ATP in the NBD1. By extrapolation, the role of Lys16 would be to complete NBD2, but the function of NBD2 is not clear.
3.3.2.2. The Metalloid-Binding Domain
In the closed ArsA structure, the two NBDs are located at the interface between A1 and A2, in close proximity to each other (Zhou et al., 2000). Over 20 Å distant from the NBDs is a metalloid-binding domain (MBD), where three Sb(III) or As(III) are bound at the A1–A2 interface. Antimonite or arsenite activates ArsA ATPase activity. In the absence of the metalloids, ArsA has a low level of ATPase activity. Arsenite stimulates ATP hydrolysis by 3- to 5-fold, whereas a 10- to 20-fold stimulation is observed with antimonite as the activator. All other oxyanions tested had no effect on ATPase activity (Hsu & Rosen, 1989). In the crystal structure, one Sb(III) is connected to Cys113 from A1 and Cys422 from the A2 (Site 1), a second to Cys172 from A1 and His-453 from A2 (Site 2), and the third to His-148 from A1 and Ser-420 from A2 (Site 3). Thus, the three metalloid atoms act as molecular glue to bring the A1 and A2 halves of ArsA together, an event that is linked to activation of ATP hydrolysis (Bhattacharjee, Li, Ksenzenko, & Rosen, 1995; Bhattacharjee & Rosen, 1996). ArsA binds a single Sb(III) with high affinity only in the presence of Mg2+-ATPγS. Mutation of the codons for Cys113 and Cys422 eliminated Sb(III) binding to purified ArsA (Ruan, Bhattacharjee, & Rosen, 2006). C113A/C422A ArsA has basal ATPase activity similar to that of the wild type but lacks metalloid-stimulated activity. Cells expressing the arsAC113A/C422AB genes had an intermediate level of metalloid resistance and accumulation between those expressing only arsB alone and those expressing wild-type arsAB genes, indicating that whereas metalloid stimulation of ArsA activity enhances the ability of the pump to reduce the intracellular concentration of metalloid, high-affinity binding of metalloid by ArsA is not obligatory for transport or resistance. However, cells bearing wild-type arsAB replaced cells with mutant arsAC113A/C422AB in mixed populations of cells in a sublethal concentration of arsenite, suggesting that the metalloid-binding site confers an evolutionary advantage. What is the role of the other two metalloids observed in the crystal structure of ArsA, which shows one liganded to Cys172 and His453 and the other liganded to His148 and Ser420? The contribution of those putative metalloid sites was examined (Ruan, Bhattacharjee, & Rosen, 2008). There was little effect of mutagenesis of residues His148 and Ser420 on Sb(III) binding. A C172A ArsA mutant and C172A/H453A double mutant exhibited decreased affinity for Sb(III). Thus, while there appears only a single high-affinity metalloid-binding site in ArsA, the results suggest that Cys172 controls the affinity of this site for metalloid and hence the efficiency of As(III) to activate the ArsAB efflux pump.
3.3.2.3. The Signal Transduction Domain
Connecting the MBD to the two NBDs are the STDs, one in each half of the protein (Zhou & Rosen, 1997). The STDs each have a 12-residue signature sequence D142TAPTGH148TIRLL (STD1) and D447TAPTGH453TLLLL (STD2), which correspond to the Switch II region of other nucleotide-binding proteins and have been proposed to be involved in transmission of the energy of ATP hydrolysis to metalloid transport. ArsA homologues have been found in every sequenced genome of eubacteria, archaea, fungi, plants and animals. This sequence is highly conserved in ArsA homologues from every kingdom, indicating that this common motif has a conserved function. In the structure, the two DTAPTGHT sequences clearly span the spaces between the MgADP-filled NBDs and the MBD (Zhou et al., 2000). During ATP hydrolysis, the carboxy-terminal end of the A1 sequence becomes exposed to a less polar environment, whereas the amino-terminal end becomes exposed to a more hydrophilic environment as the product, ADP, is formed, indicating that this sequence has considerable conformational mobility during the catalytic cycle. From these results the DTAPTGHT sequences have been proposed to be STDs involved in cross talk between the NBDs and the MBDs (Bhattacharjee & Rosen, 2000; Zhou & Rosen, 1997).
3.3.2.4. The Linker Regions
Connecting A1 and A2 is a linker region of 25 residues. In the absence of ATP and metalloid, ArsA is hypersensitive to trypsin (Hsu & Rosen, 1989), with the initial attack within the linker at residue Arg290 (Ramaswamy & Kaur, 1998). Binding of both nucleotide and metalloid provides substantial synergistic protection from protease. The requirement for this linker sequence was examined by the mutagenic insertion of five glycine residues or by the deletion of 5, 10, 15 or 23 residues (Li & Rosen, 2000). Cells expressing arsA with the five-residue insertion had wild-type arsenite resistance. Resistance of cells expressing modified arsA genes with deletions was dependent on the length of the linker but independent of the actual sequence. Cells with 5 or 10 residues deleted exhibited slightly reduced resistance. Further deletions decreased resistance significantly. The purified mutant with the five-residue insertion had the same affinity for ATP and Sb(III) as the wild-type enzyme. Mutants with 10-, 15- or 23-residue deletions exhibited decreased affinity for both Sb(III) and ATP. The enzyme with a 23-residue deletion exhibited only basal ATPase activity and was unable to be activated by Sb(III). These results suggest that the linker brings the two halves of the protein into proper contact with each other, facilitating catalysis.
4. THE ARSD AS(III) METALLOCHAPERONE AND ITS INTERACTION WITH THE ARSA ATPASE
4.1. Properties of Metallochaperones
All cells regulate the intracellular concentration of metals to prevent toxicity. This is necessary because even essential metals become toxic to the cell in excess due to their ability to catalyze cytotoxic reactions. It was estimated that total cytoplasmic concentration of free copper, an essential metal, to be less than 10−18 M, which is several orders of magnitude less than one copper atom per cell (Rae, Schmidt, Pufahl, Culotta, & O’Halloran, 1999). In other words, less than 0.01% of the total cellular copper is ever free in the cytoplasm at any one time. Yet, most copper enzymes bind copper ions with Kd values that are much higher than 10−18 M. For example, the copper- and zinc-dependent enzyme superoxide dismutase (SOD1) binds copper ions with a Kd around 10−15 M in vitro. These considerations suggest that copper enzymes (and perhaps other metalloproteins) rely upon accessory factors for acquiring metals and delivering them to partner proteins without the necessity for free (and toxic) cytosolic metal ions (O’Halloran & Culotta, 2000).
Most identified metallochaperones are for copper, the arsenic chaperone, ArsD, is the first and, to date, only characterized chaperone for the toxic metalloid arsenic (Lin, Walmsley and Rosen, 2006). ArsD sequesters cytosolic As(III) and delivers it to the ArsAB pump for efflux and detoxification. Most ars operons have three genes, arsRBC, but others have two additional genes, arsD and arsA, such as the arsRDABC of plasmid R773. Interestingly, these two genes are nearly always adjacent to each other in the operons and gene clusters, indicating that they were coevolved as a unit. It also suggests that ArsD and ArsA have associated functions in arsenic resistance.
4.2. Properties of the ArsD As(III) Metallochaperone
Cells expressing arsAB are resistant to arsenite even in the absence of an arsD gene (IC50 = 9 mM), and expression of arsD along with arsAB produced only a slight increase in resistance (IC50 = 13 mM) (Lin et al., 2007). So what is the primary physiological role of ArsD? Typical resistance experiments are performed under laboratory conditions, e.g. high concentrations (millimolar) of arsenite for short periods of time (hours). In the environment, bacteria are exposed to low levels (micromolar) of arsenite for long periods of time (days, months, years). In a molecular competition experiment designed to more closely resemble environmental conditions, a mixed culture of cells of E. coli expressing arsDAB and arsAB were grown at 10 μM arsenite, a sub-toxic concentration but one that is found in arsenic-contaminated regions of the world. After a week, the only cells remaining had all three genes, arsDAB. Thus, having an arsD gene gives bacteria a competitive advantage over cells having only arsAB when growing in sub-toxic concentrations of arsenic. Furthermore, cells co-expressing arsD and arsAB genes accumulated less As(III) than those expressing only arsAB genes (Lin et al., 2006).
These results indicate that ArsD acts to increase the efficiency of arsenic extrusion by the ArsAB pump and imply that the two proteins physically interact. Association of ArsD and ArsA was demonstrated by yeast two-hybrid analysis (Lin et al., 2006). Among the four soluble proteins encoded by the R773 arsRDABC operon, ArsD was shown to interact only with ArsA and with itself (because ArsD is a homodimer), but not with the ArsR repressor or the ArsC arsenate [As(V)] reductase. Likewise ArsA only interacts with ArsD. Notably no As(III) or Sb(III) was added to the yeast culture media, indicating that ArsD and ArsA interact to some extent in the absence of metalloid.
4.2.1. Metalloid Binding by ArsD
ArsD is a 120-residue protein that can dimerize, although it is not clear whether the monomer or dimer (or both) interact(s) with ArsA. It has three cysteine pairs, Cys12–Cys13, Cys112–Cys113 and Cys119–120, but only the Cys12–Cys13 pair and a nearby residue, Cys18, are required for ArsD chaperone activity, so most experiments were performed with an ArsD derivate truncated at residue 109 (Lin et al., 2007). The structure of As(III)-binding site was examined by extended X-ray absorption fine structure spectroscopy (EXAFS), which showed that Cys12, Cys13 and Cys18 form three-coordinate binding site in which each cysteine thiolate is 2.24 Å from the centrally bound arsenic atom in what can be predicted to be a pyramidal structure (Yang, Rawat, Stemmler, & Rosen, 2010) (Fig. 3). ArsD has an affinity for Sb(III) of approximately 0.3 μM (Lin et al., 2007). In contrast, its partner, ArsA, has a Kd for Sb(III) approximately 9 μM (Ruan et al., 2006). Thus, ArsA has about 30-fold lower affinity for metalloid than that of ArsD, which raises the question of how metalloid can be transferred quantitatively to ArsA. One possibility is that the process is kinetically, not thermodynamically, driven. The ArsAB ATPase pumps metalloid out of the cells, so, by mass action, metalloid could be pulled from ArsD to the ArsAB pump.
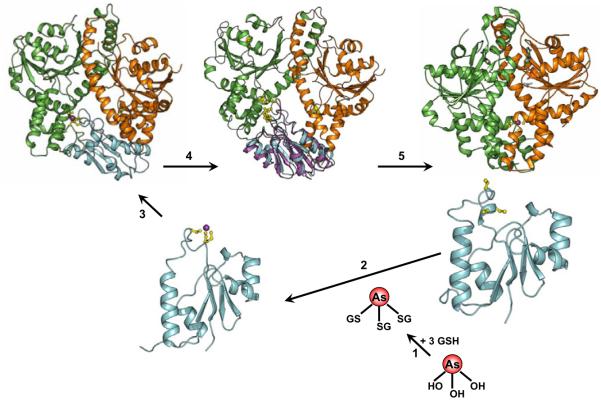
Figure 3. Transfer of As(III) from ArsD to ArsA.
The model shows a hypothetical five-step reaction scheme for transfer of As(III) from ArsD to ArsA for extrusion from the cell. Step 1: Intracellular As(OH)3 complexes with GSH to form As(GS)3. Step 2: ArsD binds As(III) by exchange of the three thiols of As(GS)3 for the three thiols of residues Cys12, Cys13 and Cys18. Step 3: The ArsD–As(III) fits into the cavity of the open form of ArsA, with the three thiols of ArsD juxtaposed to the three thiols of the As(III)-binding site of ArsA. Step 4: In a three-step thiol exchange reaction, As(III) is transferred from ArsD to ArsA. Step 5: As(III) binding to ArsA induces a conformational change that increases the rate of ATP hydrolysis and, consequently, the rate of As(III) extrusion by the ArsAB pump. See the color plate.
Binding of As(III) by ArsD was characterized using tryptophan fluorescence spectroscopy (Yang et al., 2010). ArsD has only two tryptophan residues located distant from the As(III)-binding site. By site-directed mutagenesis, those were changed to tyrosine residues, and single tryptophan derivatives within the As(III)-binding site were constructed by changing either Thr15 or Val17 to tryptophan residues. Either T15W or V17W exhibited quenching of intrinsic protein fluorescence upon binding of As(III) or Sb(III). From the As(III) dependence of quenching, a Kd of approximately 2 μM for arsenite was determined. Mutants lacking any one of the cysteines had 10-fold decreased affinity for As(III). Notably inorganic As(III) quenched fluorescence very slowly. When the glutathione conjugate, As(GS)3, was added, the rate was at least 1000-fold faster. Considering that intracellular GSH concentrations are usually in the millimolar range, nearly all As(III) would be in the form of As(GS1′)3 in vivo, with essentially no free As(III). These results indicate that physiologically glutathione-conjugated arsenic is the species recognized by ArsD (Fig. 3, step 1).
4.2.2. Interactions between ArsD and the ArsA ATPase
A combination of recent biochemical, molecular genetic and structural results have provided insights into the details of the ArsD–ArsA interaction. Interaction of ArsD and ArsA was examined in vitro using purified proteins. This was first demonstrated by reaction with the bifunctional cysteine cross-linking reagent dibromobimane (Lin et al., 2006). Dibromobimane cross-linking was increased by MgATP, suggesting that the two proteins interact during ATP hydrolysis by ArsA. To examine the mechanism of metalloid transfer, an active form of ArsD fused to the maltose-binding protein was constructed. The chimeric protein was loaded with either As(III) or Sb(III), and the metalloid protein bound to amylose column. ArsA was then flowed through the column with a variety of ligands, and, finally, ArsD was eluted with maltose (Lin et al., 2006; Yang et al., 2010). The concentrations of eluted proteins and metalloids were determined, and transfer of metalloid from ArsD to ArsA was compared under catalytic or non-catalytic conditions. The results showed that ATP hydrolysis facilitated metalloid transfer from ArsD to ArsA, suggesting that ArsA is in a transient conformation during the catalytic cycle when transfer takes place.
One important question is whether transfer of As(III) from ArsD to ArsA is mediated by direct interaction of the two proteins that allows for channeling between the thiols of ArsD and ArsA or by a thermodynamic dissociation/reassociation process. The As(III) chelator dimercaptosuccinate was used to differentiate the two mechanisms. The results demonstrated that the As(III) transfer from ArsD to ArsA was insensitive to dimercaptosuccinate, consistent with channeling of metalloid from one to the other, and suggest that ArsD and ArsA form an interface at their metal-binding sites (Yang et al., 2010). At the biochemical level, it appears that transfer of As(III) from ArsD enhances ArsA ATPase activity by increasing its affinity for metalloids (Lin et al., 2006). In the presence of ArsD, the apparent affinity of ArsA for As(III) is increased nearly two orders of magnitude, but the Vmax for ATP hydrolysis remains unchanged. ArsD effectively increases the affinity of ArsA for As(III), making the ArsAB pump more effective at concentrations of As(III) commonly found in the environment.
4.2.3. Structural Basis of ArsD Function
The ArsD structure was recently solved by nuclear magnetic resonance (NMR) and by X-ray crystallization at 1.6 Å (Ye et al., 2010; Ye et al., 2010). The ArsD monomer has a core of four β-strands flanked by four α-helices (Fig. 3). Although the region of ArsD cysteines that form the arsenic-binding site was not visible in that structure, a structure of an oxidized form of an ArsD homologue from Bacteroides vulgatus, in which the cysteines are in disulfide bonds, has been deposited (PDB code: 3KTB; Kim, Y., Tesar, C., Feldmann, B., Joachimiak, A., Midwest Center for Structural Genomics, unpublished data). From superimposition of R773 ArsD and the B. vulgatus orthologue, a model of the metallated form of ArsD109 was constructed (Ye et al., 2010). In the B. vulgatus structure, the Cys12 sulfur is oriented away from the Cys13 and Cys18, which are in a disulfide bond with each other. However, EXAFS data with R773 ArsD indicate that all three sulfur atoms are equidistant to the arsenic atom at 2.24 Å. Consequently, when the unliganded form of ArsD is presented with As(III), it is hypothesized to form an intermediate in which As(III) is weakly bound to the sulfur atoms of Cys13 and Cys18 (Ye et al., 2010). Cys12 could then reorient to become the third ligand, completing the pyramidal high-affinity binding site.
To understand how ArsD functions in solution, an NMR characterization was also performed (Ye et al., 2010). Overall the NMR and crystallographic information were in reasonable agreement. The NMR assignments indicate that the protein is a well-folded homodimer with conserved residues Cys12, Cys13 and Cys18 in an unstructured environment. The unfolded metalloid-binding site is between the structured β-strand 1 and α-helix 1, suggesting that this region of apoArsD is normally flexible and folds around metalloid, allowing the three cysteine residues to form the binding site only when As(III) is present.
ArsA undergoes a number of conformational changes during the catalytic cycle, and, in particular, cycles between an open (unliganded and basal activity) and closed (ligand bound and activated) forms. The crystal structure of a closed form of the R773 ArsA has been solved at 2.1 Å (Zhou et al., 2000). Recently structures of a yeast homologue of ArsA termed Arr4p (Shen, Hsu, Kang, Rosen, & Bhattacharjee, 2003) or Get3 (Auld et al., 2006) that is involved in membrane insertion of tailed-anchored proteins were solved in both closed and open forms (Bozkurt et al., 2009; Mateja et al., 2009). The open form of ArsA was modeled on the open structure of Get3 (Yang, Salam, & Rosen, 2011). The open model of ArsA displays a cavity into which fits ArsD well, and predicts that the ArsD–ArsA interface involves 15 residues from ArsD and 13 from ArsA, with their metalloid-binding sites in close proximity (Yang et al., 2011) (Fig. 3).
A genetic approach was applied to examine the interaction of ArsD and ArsA. In addition to the three conserved N-terminal cysteine residues, 15 ArsD residues were identified as being involved in ArsD function by genetic mapping (Yang et al., 2011). Applying a repressed transactivator yeast two-hybrid selection, nine ArsD mutants, S14R, T20I, D28T, D28V, T31A, Q34R, Q38R and V61A, were identified as increasing interaction with ArsA. Five mutants, V17A, V22A, V27D, Q51H and F55L, were found to decrease ArsA interaction by reverse yeast two-hybrid assays. Two additional mutations in Lys37 and Lys62 were identified by a combination of site-directed mutagenesis and chemical modification. Alanine substitutions in Lys37 and Lys62 were shown to lose interaction with ArsA, and reduce the activity on ArsA ATPase stimulation; however, substitution with arginine at either position was tolerated, suggesting that a positive charge is involved. All 15 residues were mapped on the surface of ArsD, in which four are close to the metal-binding loop and seven are on the outer surface of α-helix 1. Their locations are consistent with the structural model generated by in silico docking. These results strongly point to an interface involves one surface of helix 1 and the metalloid-binding site.
4.3. Do Higher Organisms Have an As(III) Metallochaperone?
To date, the ArsD As(III) metallochaperone has only been identified in prokaryotes. Are there As(III) metallochaperones in eurkaryotes? The ArsD monomer has a thioredoxin fold (Ye et al., 2010). Trx plays an important role in maintaining redox balances in prokaryotic and eukaryotic cells. It includes a group of small proteins of ~12 kDa that have a highly conserved sequence (Cys-Gly-Pro-Cys) that forms a redox-active site of dithiol or disulfide to protect cytosolic proteins from aggregation or inactivation (Arner & Holmgren, 2000). Trx gene expression is induced by As(III) (Hu, Jin, & Snow, 2002). Arsenic–Trx complexes have been detected by liquid chromatography with inductively coupled plasma mass spectrometry (Wang, Zhang, Li, & Le, 2007). Binding stoichiometry for different arsenic species, including inorganic and methylated As(III), were consistent with the available cysteine residues in the protein. It is possible that Trx or another protein with a thioredoxin fold binds and transfers As(III) to other proteins in an analogous fashion as the ArsD As(III) metallochaperone.
ACKNOWLEDGMENTS
The research described in this study was supported by U.S. Public Health Service Grant GM55425 to B.P.R. and Taiwan National Science Council NSC100-2320-B-038-018 to Y.F.L.
REFERENCES
- Aaltonen EK, Silow M. Transmembrane topology of the Acr3 family arsenite transporter from Bacillus subtilis. Biochimica et Biophysica Acta. 2008;1778(4):963–973. doi: 10.1016/j.bbamem.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Abernathy CO, Thomas DJ, Calderon RL. Health effects and risk assessment of arsenic. Journal of Nutrition. 2003;133(5 Suppl. 1):1536S–1538S. doi: 10.1093/jn/133.5.1536S. [DOI] [PubMed] [Google Scholar]
- Achour AR, Bauda P, Billard P. Diversity of arsenite transporter genes from arsenic-resistant soil bacteria. Research in Microbiology. 2007;158(2):128–137. doi: 10.1016/j.resmic.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Agre P. Nobel lecture. Aquaporin water channels. Bioscience Reports. 2004;24(3):127–163. doi: 10.1007/s10540-005-2577-2. [DOI] [PubMed] [Google Scholar]
- Agre P, King LS, Yasui M, Guggino WB, Ottersen OP, Fujiyoshi Y, et al. Aquaporin water channels—from atomic structure to clinical medicine. Journal of Physiology. 2002;542(Pt 1):3–16. doi: 10.1113/jphysiol.2002.020818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agre P, Kozono D. Aquaporin water channels: molecular mechanisms for human diseases. FEBS Letters. 2003;555(1):72–78. doi: 10.1016/s0014-5793(03)01083-4. [DOI] [PubMed] [Google Scholar]
- Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. European Journal of Biochemistry. 2000;267(20):6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- Au WY, Kumana CR, Lee HK, Lin SY, Liu H, Yeung DY, et al. Oral arsenic trioxide-based maintenance regimens for first complete remission of acute promyelocytic leukemia: a 10-year follow-up study. Blood. 2011;118(25):6535–6543. doi: 10.1182/blood-2011-05-354530. [DOI] [PubMed] [Google Scholar]
- Auld KL, Hitchcock AL, Doherty HK, Frietze S, Huang LS, Silver PA. The conserved ATPase Get3/Arr4 modulates the activity of membrane-associated proteins in Saccharomyces cerevisiae. Genetics. 2006;174(1):215–227. doi: 10.1534/genetics.106.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee H, Carbrey J, Rosen BP, Mukhopadhyay R. Drug uptake and pharmacological modulation of drug sensitivity in leukemia by AQP9. Biochemical and Biophysical Research Communications. 2004;322(3):836–841. doi: 10.1016/j.bbrc.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee H, Li J, Ksenzenko MY, Rosen BP. Role of cysteinyl residues in metalloactivation of the oxyanion-translocating ArsA ATPase. The Journal of Biological Chemistry. 1995;270(19):11245–11250. doi: 10.1074/jbc.270.19.11245. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee H, Mukhopadhyay R, Thiyagarajan S, Rosen BP. Aquaglyceroporins: ancient channels for metalloids. Journal of Biology. 2008;7(9):33. doi: 10.1186/jbiol91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee H, Rosen BP. Spatial proximity of Cys113, Cys172, and Cys422 in the metalloactivation domain of the ArsA ATPase. The Journal of Biological Chemistry. 1996;271(40):24465–24470. doi: 10.1074/jbc.271.40.24465. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee H, Rosen BP. Role of conserved histidine residues in metalloactivation of the ArsA ATPase. Biometals. 2000;13(4):281–288. doi: 10.1023/a:1009200215328. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee H, Rosen BP, Simon . Arsenic metabolism in prokaryotic and eukaryotic microbes. In: InNies DHS, editor. Molecular microbiology of heavy metals. Vol. 6. Springer-Verlag; Heidelberg/New York: 2007. 2007. pp. 371–406. [Google Scholar]
- Bhattacharjee H, Rosen BP, Mukhopadhyay R. Aquaglyceroporins and metalloid transport: implications in human diseases. Handbook of Experimental Pharmacology. 2009;190:309–325. doi: 10.1007/978-3-540-79885-9_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowicz P, Wysocki R, Owsianik G, Goffeau A, Ulaszewski S. Isolation of three contiguous genes, ACR1, ACR2 and ACR3, involved in resistance to arsenic compounds in the yeast Saccharomyces cerevisiae. Yeast. 1997;13(9):819–828. doi: 10.1002/(SICI)1097-0061(199707)13:9<819::AID-YEA142>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bozkurt G, Stjepanovic G, Vilardi F, Amlacher S, Wild K, Bange G, et al. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(50):21131–21136. doi: 10.1073/pnas.0910223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bun-ya M, Shikata K, Nakade S, Yompakdee C, Harashima S, Oshima Y. Two new genes, PHO86 and PHO87, involved in inorganic phosphate uptake in Saccharomyces cerevisiae. Current Genetics. 1996;29(4):344–351. [PubMed] [Google Scholar]
- Carbrey JM, Gorelick-Feldman DA, Kozono D, Praetorius J, Nielsen S, Agre P. Aquaglyceroporin AQP9: solute permeation and metabolic control of expression in liver. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(5):2945–2950. doi: 10.1073/pnas.0437994100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter NS, Fairlamb AH. Arsenical-resistant trypanosomes lack an unusual adenosine transporter. Nature. 1993;361(6408):173–176. doi: 10.1038/361173a0. [DOI] [PubMed] [Google Scholar]
- Chen CM, Misra TK, Silver S, Rosen BP. Nucleotide sequence of the structural genes for an anion pump. The plasmid-encoded arsenical resistance operon. The Journal of Biological Chemistry. 1986;261(32):15030–15038. [PubMed] [Google Scholar]
- Chen Y, Dey S, Rosen BP. Soft metal thiol chemistry is not involved in the transport of arsenite by the Ars pump. Journal of Bacteriology. 1996;178(3):911–913. doi: 10.1128/jb.178.3.911-913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Council NR. Arsenic in drinking water: 2001 update. 2001 http://www.nap.edu/books/0309076293/html/ [PubMed]
- Cui X, Kobayashi Y, Hayakawa T, Hirano S. Arsenic speciation in bile and urine following oral and intravenous exposure to inorganic and organic arsenics in rats. Toxicological Sciences. 2004;82(2):478–487. doi: 10.1093/toxsci/kfh265. [DOI] [PubMed] [Google Scholar]
- Dai J, Weinberg RS, Waxman S, Jing Y. Malignant cells can be sensitized to undergo growth inhibition and apoptosis by arsenic trioxide through modulation of the glutathione redox system. Blood. 1999;93(1):268–277. [PubMed] [Google Scholar]
- Dey S, Dou D, Rosen BP. ATP-dependent arsenite transport in everted membrane vesicles of Escherichia coli. The Journal of Biological Chemistry. 1994;269(41):25442–25446. [PubMed] [Google Scholar]
- Dey S, Dou D, Tisa LS, Rosen BP. Interaction of the catalytic and the membrane subunits of an oxyanion-translocating ATPase. Archives of Biochemistry and Biophysics. 1994;311(2):418–424. doi: 10.1006/abbi.1994.1256. [DOI] [PubMed] [Google Scholar]
- Dey S, Papadopoulou B, Haimeur A, Roy G, Grondin K, Dou D, et al. High level arsenite resistance in Leishmania tarentolae is mediated by an active extrusion system. Molecular and Biochemical Parasitology. 1994;67(1):49–57. doi: 10.1016/0166-6851(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Dey S, Rosen BP. Dual mode of energy coupling by the oxyanion-translocating ArsB protein. Journal of Bacteriology. 1995;177(2):385–389. doi: 10.1128/jb.177.2.385-389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilda PJ, Hogg PJ. Arsenical-based cancer drugs. Cancer Treatment Reviews. 2007;33(6):542–564. doi: 10.1016/j.ctrv.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Duan G, Kamiya T, Ishikawa S, Arao T, Fujiwara T. Expressing ScACR3 in rice enhanced arsenite efflux and reduced arsenic accumulation in rice grains. Plant and Cell Physiology. 2012;53(1):154–163. doi: 10.1093/pcp/pcr161. [DOI] [PubMed] [Google Scholar]
- Emadi A, Gore SD. Arsenic trioxide—an old drug rediscovered. Blood Review. 2010;24(4–5):191–199. doi: 10.1016/j.blre.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu HL, Ajees AA, Rosen BP, Bhattacharjee H. Role of signature lysines in the deviant walker a motifs of the ArsA ATPase. Biochemistry. 2010;49(2):356–364. doi: 10.1021/bi901681v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu HL, Meng Y, Ordonez E, Villadangos AF, Bhattacharjee H, Gill JA, et al. Properties of arsenite efflux permeases (Acr3) from Alkaliphilus metalliredigens and Corynebacterium glutamicum. The Journal of Biological Chemistry. 2009;284(30):19887–19895. doi: 10.1074/jbc.M109.011882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis MM, Komiya HPC, Woo D, Kornuc JJ, Rees DC. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science. 1992;257(5077):1653–1659. doi: 10.1126/science.1529353. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Shen J, Rosen BP. Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5001–5006. doi: 10.1073/pnas.96.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladysheva TB, Oden KL, Rosen BP. Properties of the arsenate reductase of plasmid R773. Biochemistry. 1994;33(23):7288–7293. doi: 10.1021/bi00189a033. [DOI] [PubMed] [Google Scholar]
- Haimeur A, Conseil G, Deeley RG, Cole SP. The MRP-related and BCRP/ABCG2 multidrug resistance proteins: biology, substrate specificity and regulation. Current Drug Metabolism. 2004;5(1):21–53. doi: 10.2174/1389200043489199. [DOI] [PubMed] [Google Scholar]
- Hamdi M, Sanchez MA, Beene LC, Liu Q, Landfear SM, Rosen BP, et al. Arsenic transport by zebrafish aquaglyceroporins. BMC Molecular Biology. 2009;10:104. doi: 10.1186/1471-2199-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller KB, Lin EC, Wilson TH. Substrate specificity and transport properties of the glycerol facilitator of Escherichia coli. Journal of Bacteriology. 1980;144(1):274–278. doi: 10.1128/jb.144.1.274-278.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CM, Rosen BP. Characterization of the catalytic subunit of an anion pump. The Journal of Biological Chemistry. 1989;264(29):17349–17354. [PubMed] [Google Scholar]
- Hu J, Liu YF, Wu CF, Xu F, Shen ZX, Zhu YM, et al. Long-term efficacy and safety of all-trans retinoic acid/arsenic trioxide-based therapy in newly diagnosed acute promyelocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(9):3342–3347. doi: 10.1073/pnas.0813280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Jin X, Snow ET. Effect of arsenic on transcription factor AP-1 and NF-kappaB DNA binding activity and related gene expression. Toxicological Letters. 2002;133(1):33–45. doi: 10.1016/s0378-4274(02)00083-8. [DOI] [PubMed] [Google Scholar]
- Indriolo E, Na G, Ellis D, Salt DE, Banks JA. A vacuolar arsenite transporter necessary for arsenic tolerance in the arsenic hyperaccumulating fern Pteris vittata is missing in flowering plants. The Plant Cell. 2010;22(6):2045–2057. doi: 10.1105/tpc.109.069773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inskeep WP, McDermott TR. Geothermal biology and geochemistry in Yellowstone National Park: Proceeding of the Thermal Biology Institute workshop, Yellowstone National Park, WY, October 2003, Bozeman, MT, 2005, 2003. Montana State University Publications; Bozeman, MT: 2003. Thermal biology, I; p. 352. [Google Scholar]
- Ishibashi K, Imai M, Sasaki S. Cellular localization of aquaporin 7 in the rat kidney. Experimental Nephrology. 2000;8(4–5):252–257. doi: 10.1159/000020676. [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Kuwahara M, Gu Y, Kageyama Y, Tohsaka A, Suzuki F, et al. Cloning and functional expression of a new water channel abundantly expressed in the testis permeable to water, glycerol, and urea. The Journal of Biological Chemistry. 1997;272(33):20782–20786. doi: 10.1074/jbc.272.33.20782. [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Kuwahara M, Gu Y, Tanaka Y, Marumo F, Sasaki S. Cloning and functional expression of a new aquaporin (AQP9) abundantly expressed in the peripheral leukocytes permeable to water and urea, but not to glycerol. Biochemical and Biophysical Research Communications. 1998;244(1):268–274. doi: 10.1006/bbrc.1998.8252. [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Morinaga T, Kuwahara M, Sasaki S, Imai M. Cloning and identification of a new member of water channel (AQP10) as an aquaglyceroporin. Biochimica et Biophysica Acta. 2002;1576(3):335–340. doi: 10.1016/s0167-4781(02)00393-7. [DOI] [PubMed] [Google Scholar]
- Jackson BP, Taylor VF, Karagas MR, Punshon T, Cottingham KL. Arsenic, organic foods, and brown rice syrup. Environmental Health Perspectives. 2012 doi: 10.1289/ehp.1104619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji G, Silver S. Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(20):9474–9478. doi: 10.1073/pnas.89.20.9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Bhattacharjee H, Zhou T, Rosen BP, Ambudkar SV, Sauna ZE. Nonequivalence of the nucleotide binding domains of the ArsA ATPase. The Journal of Biological Chemistry. 2005;280(11):9921–9926. doi: 10.1074/jbc.M413391200. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Takahashi T, Natoli G, Adams SR, Chen Y, Tsien RY, et al. Inhibition of NF-kappa B activation by arsenite through reaction with a critical cysteine in the activation loop of Ikappa B kinase. The Journal of Biological Chemistry. 2000;275(46):36062–36066. doi: 10.1074/jbc.M007204200. [DOI] [PubMed] [Google Scholar]
- Karkaria CE, Chen CM, Rosen BP. Mutagenesis of a nucleotide-binding site of an anion-translocating ATPase. The Journal of Biological Chemistry. 1990;265(14):7832–7836. [PubMed] [Google Scholar]
- Kaur P, Rosen BP. Mutagenesis of the C-terminal nucleotide-binding site of an anion-translocating ATPase. The Journal of Biological Chemistry. 1992;267(27):19272–19277. [PubMed] [Google Scholar]
- Koonin EV. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. Journal of Molecular Biology. 1993;229(4):1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Dey S, Sanders OI, Rosen BP. Alternate energy coupling of ArsB, the membrane subunit of the Ars anion-translocating ATPase. The Journal of Biological Chemistry. 1997;272(1):326–331. doi: 10.1074/jbc.272.1.326. [DOI] [PubMed] [Google Scholar]
- Kwong YL, Todd D. Delicious poison: arsenic trioxide for the treatment of leukemia. Blood. 1997;89(9):3487–3488. [PubMed] [Google Scholar]
- Legare D, Richard D, Mukhopadhyay R, Stierhof YD, Rosen BP, Haimeur A, et al. The Leishmania ATP-binding cassette protein PGPA is an intracellular metal-thiol transporter ATPase. The Journal of Biological Chemistry. 2001;276(28):26301–26307. doi: 10.1074/jbc.M102351200. [DOI] [PubMed] [Google Scholar]
- Leung J, Pang A, Yuen WH, Kwong YL, Tse EW. Relationship of expression of aquaglyceroporin 9 with arsenic uptake and sensitivity in leukemia cells. Blood. 2007;109(2):740–746. doi: 10.1182/blood-2006-04-019588. [DOI] [PubMed] [Google Scholar]
- Li J, Liu S, Rosen BP. Interaction of ATP binding sites in the ArsA ATPase, the catalytic subunit of the Ars pump. The Journal of Biological Chemistry. 1996;271(41):25247–25252. doi: 10.1074/jbc.271.41.25247. [DOI] [PubMed] [Google Scholar]
- Li J, Rosen BP. The linker peptide of the ArsA ATPase. Molecular Microbiology. 2000;35(2):361–367. doi: 10.1046/j.1365-2958.2000.01696.x. [DOI] [PubMed] [Google Scholar]
- Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(1):42–47. doi: 10.1073/pnas.94.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YF, Walmsley AR, Rosen BP. An arsenic metallochaperone for an arsenic detoxification pump. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(42):15617–15622. doi: 10.1073/pnas.0603974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YF, Yang J, Rosen BP. ArsD residues Cys12, Cys13, and Cys18 form an As(III)-binding site required for arsenic metallochaperone activity. The Journal of Biological Chemistry. 2007;282(23):16783–16791. doi: 10.1074/jbc.M700886200. [DOI] [PubMed] [Google Scholar]
- Liu JX, Zhou GB, Chen SJ, Chen Z. Arsenic compounds: revived ancient remedies in the fight against human malignancies. Current Opinion in Chemical Biology. 2012 doi: 10.1016/j.cbpa.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Liu Y, Promeneur D, Rojek A, Kumar N, Frokiaer J, Nielsen S, et al. Aquaporin 9 is the major pathway for glycerol uptake by mouse erythrocytes, with implications for malarial virulence. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(30):12560–12564. doi: 10.1073/pnas.0705313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Carbrey JM, Agre P, Rosen BP. Arsenic trioxide uptake by human and rat aquaglyceroporins. Biochemical and Biophysical Research Communications. 2004;316(4):1178–1185. doi: 10.1016/j.bbrc.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Liu Z, Shen J, Carbrey JM, Mukhopadhyay R, Agre P, Rosen BP. Arsenite transport by mammalian aquaglyceroporins AQP7 and AQP9. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(9):6053–6058. doi: 10.1073/pnas.092131899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Styblo M, Rosen BP. Methylarsonous acid transport by aquaglyceroporins. Environmental Health Perspectives. 2006;114(4):527–531. doi: 10.1289/ehp.8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J, Sundaramoorthy M. MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Molecular Microbiology. 2003;48(2):295–303. doi: 10.1046/j.1365-2958.2003.03427.x. [DOI] [PubMed] [Google Scholar]
- Ma JF, Yamaji N, Mitani N, Tamai K, Konishi S, Fujiwara T, et al. An efflux transporter of silicon in rice. Nature. 2007;448(7150):209–212. doi: 10.1038/nature05964. [DOI] [PubMed] [Google Scholar]
- Maciaszczyk E, Wysocki R, Golik P, Lazowska J, Ulaszewski S. Arsenical resistance genes in Saccharomyces douglasii and other yeast species undergo rapid evolution involving genomic rearrangements and duplications. FEMS Yeast Research. 2004;4(8):821–832. doi: 10.1016/j.femsyr.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Maciaszczyk-Dziubinska E, Migocka M, Wysocki R. Acr3p is a plasma membrane antiporter that catalyzes As(III)/H+ and Sb(III)/H+ exchange in Saccharomyces cerevisiae. Biochimica et Biophysica Acta. 2012;1808(7):1855–1859. doi: 10.1016/j.bbamem.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Maciaszczyk-Dziubinska E, Wawrzycka D, Sloma E, Migocka M, Wysocki R. The yeast permease Acr3p is a dual arsenite and antimonite plasma membrane transporter. Biochimica et Biophysica Acta. 2012;1798(11):2170–2175. doi: 10.1016/j.bbamem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Mansour NM, Sawhney M, Tamang DG, Vogl C, Saier MH., Jr. The bile/arsenite/riboflavin transporter (BART) superfamily. FEBS Journal. 2007;274(3):612–629. doi: 10.1111/j.1742-4658.2006.05627.x. [DOI] [PubMed] [Google Scholar]
- Marino GI, Castro-Parodi M, Dietrich V, Damiano AE. High levels of human chorionic gonadotropin (hCG) correlate with increased aquaporin-9 (AQP9) expression in explants from human preeclamptic placenta. Reproductive Science. 2010;17(5):444–453. doi: 10.1177/1933719110361385. [DOI] [PubMed] [Google Scholar]
- Mateja A, Szlachcic A, Downing ME, Dobosz M, Mariappan M, Hegde RS, et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461(7262):361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott JR, Jiang X, Beene LC, Rosen BP, Liu Z. Pentavalent methylated arsenicals are substrates of human AQP9. Biometals. 2010;23(1):119–127. doi: 10.1007/s10534-009-9273-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng YL, Liu Z, Rosen BP. As(III) and Sb(III) uptake by GlpF and efflux by ArsB in Escherichia coli. The Journal of Biological Chemistry. 2004;279:18334–18341. doi: 10.1074/jbc.M400037200. [DOI] [PubMed] [Google Scholar]
- Mobley HL, Rosen BP. Energetics of plasmid-mediated arsenate resistance in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1982;79(20):6119–6122. doi: 10.1073/pnas.79.20.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Rosen BP. Arsenate reductases in prokaryotes and eukaryotes. Environmental Health Perspectives. 2002;110(Suppl. 5):745–748. doi: 10.1289/ehp.02110s5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Rosen BP. The Saccharomyces cerevisiae ACR2 gene encodes an arsenate reductase. FEMS Microbiology Letters. 1998;168:127–136. doi: 10.1111/j.1574-6968.1998.tb13265.x. [DOI] [PubMed] [Google Scholar]
- O’Halloran TV, Culotta VC. Metallochaperones: an intracellular shuttle service for metal ions. The Journal of Biological Chemistry. 2000;275(33):25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- Ordóñez E, Thiyagarajan S, Cook JD, Stemmler TL, Gil JA, Mateos LM, et al. Evolution of metal(loid) binding sites in transcriptional regulators. The Journal of Biological Chemistry. 2008;283(37):25706–25714. doi: 10.1074/jbc.M803209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO Journal. 1990;9(8):2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Soler NM, Fisher JS, Sharpe R, Hill E, Van Hoek A, Brown D, et al. Aquaporin 9 expression in the developing rat epididymis is modulated by steroid hormones. Reproduction. 2010;139(3):613–621. doi: 10.1530/REP-09-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Fu HL, Ye J, Bencze KZ, Stemmler TL, Rawlings DE, et al. Convergent evolution of a new arsenic binding site in the ArsR/SmtB family of metalloregulators. The Journal of Biological Chemistry. 2007;282(47):34346–34355. doi: 10.1074/jbc.M706565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O’Halloran TV. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284(5415):805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Kaur P. Nucleotide binding to the C-terminal nucleotide binding domain of ArsA: studies with an ATP analogue, 5’ -p-fluorosulfonylbenzoyladenosine. The Journal of Biological Chemistry. 1998;273(15):9243–9248. doi: 10.1074/jbc.273.15.9243. [DOI] [PubMed] [Google Scholar]
- Ramírez-Solis A, Mukopadhyay R, Rosen BP, Stemmler TL. Experimental and theoretical characterization of arsenite in water: insights into the coordination environment of As-O. Inorganic Chemistry. 2004;43(9):2954–2959. doi: 10.1021/ic0351592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C, Rosen BP. Heavy metals cycles (arsenic, mercury, selenium, others) In: Schaechter M, editor. Encyclopedia of microbiology. Elsevier; Oxford, U.K.: 2009. pp. 205–219. [Google Scholar]
- Rosen BP. Families of arsenic transporters. Trends in Microbiology. 1999;7:207–212. doi: 10.1016/s0966-842x(99)01494-8. [DOI] [PubMed] [Google Scholar]
- Rosen BP. Biochemistry of arsenic detoxification. FEBS Letters. 2002;529(1):86–92. doi: 10.1016/s0014-5793(02)03186-1. [DOI] [PubMed] [Google Scholar]
- Rosen BP, Weigel U, Karkaria C, Gangola P. Molecular characterization of an anion pump. The arsA gene product is an arsenite(antimonate)-stimulated ATPase. The Journal of Biological Chemistry. 1988;263(7):3067–3070. [PubMed] [Google Scholar]
- Rosenberg H, Gerdes RG, Chegwidden K. Two systems for the uptake of phosphate in Escherichia coli. Journal of Bacteriology. 1977;131(2):505–511. doi: 10.1128/jb.131.2.505-511.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan X, Bhattacharjee H, Rosen BP. Characterization of the metalloactivation domain of an arsenite/antimonite resistance pump. Molecular Microbiology. 2008;67(2):392–402. doi: 10.1111/j.1365-2958.2007.06049.x. [DOI] [PubMed] [Google Scholar]
- Ruan X, Bhattacharjee H, Rosen BP. Cys-113 and Cys-422 form a high affinity metalloid binding site in the ArsA ATPase. The Journal of Biological Chemistry. 2006;281(15):9925–9934. doi: 10.1074/jbc.M600125200. [DOI] [PubMed] [Google Scholar]
- San Francisco MJ, Hope CL, Owolabi JB, Tisa LS, Rosen BP. Identification of the metalloregulatory element of the plasmid-encoded arsenical resistance operon. Nucleic Acids Research. 1990;18(3):619–624. doi: 10.1093/nar/18.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders OI, Rensing C, Kuroda M, Mitra B, Rosen BP. Antimonite is accumulated by the glycerol facilitator GlpF in Escherichia coli. Journal of Bacteriology. 1997;179(10):3365–3367. doi: 10.1128/jb.179.10.3365-3367.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Kobayashi Y. The ars operon in the skin element of Bacillus subtilis confers resistance to arsenate and arsenite. Journal of Bacteriology. 1998;180(7):1655–1661. doi: 10.1128/jb.180.7.1655-1661.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC. Structure of ADP x AIF4(−)-stabilized nitrogenase complex and its implications for signal transduction. Nature. 1997;387(6631):370–376. doi: 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- Shen J, Hsu CM, Kang BK, Rosen BP, Bhattacharjee H. The Saccharomyces cerevisiae Arr4p is involved in metal and heat tolerance. Biometals. 2003;16:369–378. doi: 10.1023/a:1022504311669. [DOI] [PubMed] [Google Scholar]
- Shi W, Dong J, Scott RA, Ksenzenko MY, Rosen BP. The role of arsenic-thiol interactions in metalloregulation of the ars operon. The Journal of Biological Chemistry. 1996;271(16):9291–9297. doi: 10.1074/jbc.271.16.9291. [DOI] [PubMed] [Google Scholar]
- Smith AH, Lingas EO, Rahman M. Contamination of drinking-water by arsenic in Bangladesh: a public health emergency. Bulletin of the World Health Organization. 2000;78(9):1093–1103. [PMC free article] [PubMed] [Google Scholar]
- Soignet SL. Clinical experience of arsenic trioxide in relapsed acute promyelocytic leukemia. Oncologist. 2001;6(Suppl. 2):11–16. doi: 10.1634/theoncologist.6-suppl_2-11. [DOI] [PubMed] [Google Scholar]
- Stone R. Food safety. Arsenic and paddy rice: a neglected cancer risk? Science. 2008;321(5886):184–185. doi: 10.1126/science.321.5886.184. [DOI] [PubMed] [Google Scholar]
- Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355(6358):374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- Tchounwou PB, Centeno JA, Patlolla AK. Arsenic toxicity, mutagenesis, and carcinogenesis—a health risk assessment and management approach. Molecular and Cellular Biochemistry. 2004;255(1–2):47–55. doi: 10.1023/b:mcbi.0000007260.32981.b9. [DOI] [PubMed] [Google Scholar]
- Thomas DJ, Waters SB, Styblo M. Elucidating the pathway for arsenic methylation. Toxicology and Applied Pharmacology. 2004;198(3):319–326. doi: 10.1016/j.taap.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Thorsen M, Di Y, Tangemo C, Morillas M, Ahmadpour D, Van der Does C, et al. The MAPK Hog1p modulates Fps1p-dependent arsenite uptake and tolerance in yeast. Molecular Biology of the Cell. 2006;17(10):4400–4410. doi: 10.1091/mbc.E06-04-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisa LS, Rosen BP. Molecular characterization of an anion pump. The ArsB protein is the membrane anchor for the ArsA protein. The Journal of Biological Chemistry. 1990;265(1):190–194. [PubMed] [Google Scholar]
- Tofail F, Vahter M, Hamadani JD, Nermell B, Huda SN, Yunus M, et al. Effect of arsenic exposure during pregnancy on infant development at 7 months in rural Matlab, Bangladesh. Environmental Health Perspectives. 2009;117(2):288–293. doi: 10.1289/ehp.11670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CH, Tseng CP, Chiou HY, Hsueh YM, Chong CK, Chen CJ. Epidemiologic evidence of diabetogenic effect of arsenic. Toxicological Letters. 2002;133(1):69–76. doi: 10.1016/s0378-4274(02)00085-1. [DOI] [PubMed] [Google Scholar]
- Villadangos AF, Fu HL, Gil JA, Messens J, Rosen BP, Mateos LM. Efflux permease CgAcr3-1 of Corynebacterium glutamicum is an arsenite-specific antiporter. The Journal of Biological Chemistry. 2012;287(1):723–735. doi: 10.1074/jbc.M111.263335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhang H, Li XF, Le XC. Study of interactions between arsenicals and thioredoxins (human and E. coli) using mass spectrometry. Rapid Communications in Mass Spectrometry. 2007;21(22):3658–3666. doi: 10.1002/rcm.3263. [DOI] [PubMed] [Google Scholar]
- Williams PN, Price AH, Raab A, Hossain SA, Feldmann J, Meharg AA. Variation in arsenic speciation and concentration in paddy rice related to dietary exposure. Environmental Science and Technology. 2005;39(15):5531–5540. doi: 10.1021/es0502324. [DOI] [PubMed] [Google Scholar]
- Williams PN, Raab A, Feldmann J, Meharg AA. Market basket survey shows elevated levels of As in South Central U.S. processed rice compared to California: consequences for human dietary exposure. Environmental Science and Technology. 2007;41(7):2178–2183. doi: 10.1021/es061489k. [DOI] [PubMed] [Google Scholar]
- Willsky GR, Bennett RL, Malamy MH. Inorganic phosphate transport in Escherichia coli: involvement of two genes which play a role in alkaline phosphatase regulation. Journal of Bacteriology. 1973;113(2):529–539. doi: 10.1128/jb.113.2.529-539.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsky GR, Malamy MH. Effect of arsenate on inorganic phosphate transport in Escherichia coli. Journal of Bacteriology. 1980;144(1):366–374. doi: 10.1128/jb.144.1.366-374.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Tisa LS, Rosen BP. Membrane topology of the ArsB protein, the membrane subunit of an anion-translocating ATPase. The Journal of Biological Chemistry. 1992;267(18):12570–12576. [PubMed] [Google Scholar]
- Wysocki R, Bobrowicz P, Ulaszewski S. The Saccharomyces cerevisiae ACR3 gene encodes a putative membrane protein involved in arsenite transport. The Journal of Biological Chemistry. 1997;272(48):30061–30066. doi: 10.1074/jbc.272.48.30061. [DOI] [PubMed] [Google Scholar]
- Wysocki R, Chery CC, Wawrzycka D, Van Hulle M, Cornelis R, Thevelein JM, et al. The glycerol channel Fps1p mediates the uptake of arsenite and antimonite in Saccharomyces cerevisiae. Molecular Microbiology. 2001;40(6):1391–1401. doi: 10.1046/j.1365-2958.2001.02485.x. [DOI] [PubMed] [Google Scholar]
- Xia X, Postis VL, Rahman M, Wright GS, Roach PC, Deacon SE, et al. Investigation of the structure and function of a Shewanella oneidensis arsenical-resistance family transporter. Molecular Membrane Biology. 2008;25(8):691–705. doi: 10.1080/09687680802535930. [DOI] [PubMed] [Google Scholar]
- Yang HC, Cheng J, Finan TM, Rosen BP, Bhattacharjee H. Novel pathway for arsenic detoxification in the legume symbiont Sinorhizobium meliloti. Journal of Bacteriology. 2005;187(20):6991–6997. doi: 10.1128/JB.187.20.6991-6997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Rawat S, Stemmler TL, Rosen BP. Arsenic binding and transfer by the ArsD As(III) metallochaperone. Biochemistry. 2010;49(17):3658–3666. doi: 10.1021/bi100026a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Salam AA, Rosen BP. Genetic mapping of the interface between the ArsD metallochaperone and the ArsA ATPase. Molecular Microbiology. 2011;79(4):872–881. doi: 10.1111/j.1365-2958.2010.07494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Ajees AA, Yang J, Rosen BP. The 1.4 Å crystal structure of the ArsD arsenic metallochaperone provides insights into its interaction with the ArsA ATPase. Biochemistry. 2010;49(25):5206–5212. doi: 10.1021/bi100571r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, He Y, Skalicky J, Rosen BP, Stemmler TL. Resonance assignments and secondary structure prediction of the As(III) metallochaperone ArsD in solution. Biomolecular NMR Assignments. 2010;2(3):211–219. doi: 10.1007/s12104-010-9279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rensing C, Rosen BP, Zhu YG. Arsenic biomethylation by photosynthetic organisms. Trends in Plant Science. 2012;17(3):155–162. doi: 10.1016/j.tplants.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga M, Cai Y, Rosen BP. Demethylation of methylarsonic acid by a microbial community. Environmental Microbiology. 2011;13(5):1205–1215. doi: 10.1111/j.1462-2920.2010.02420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman GJ, Lankelma J, van Tellingen O, Beijnen J, Dekker H, Paulusma C, et al. Role of glutathione in the export of compounds from cells by the multidrug-resistance-associated protein. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(17):7690–7694. doi: 10.1073/pnas.92.17.7690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao FJ, McGrath SP, Meharg AA. Arsenic as a food chain contaminant: mechanisms of plant uptake and metabolism and mitigation strategies. Annual Review of Plant Biology. 2010;61:535–559. doi: 10.1146/annurev-arplant-042809-112152. [DOI] [PubMed] [Google Scholar]
- Zhou T, Liu S, Rosen BP. Interaction of substrate and effector binding sites in the ArsA ATPase. Biochemistry. 1995;34(41):13622–13626. doi: 10.1021/bi00041a042. [DOI] [PubMed] [Google Scholar]
- Zhou T, Radaev S, Rosen BP, Gatti DL. Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. EMBO Journal. 2000;19(17):1–8. doi: 10.1093/emboj/19.17.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Rosen BP. Tryptophan fluorescence reports nucleotide-induced conformational changes in a domain of the ArsA ATPase. The Journal of Biological Chemistry. 1997;272(32):19731–19737. doi: 10.1074/jbc.272.32.19731. [DOI] [PubMed] [Google Scholar]