Abstract
A prolonged QT interval is an important risk factor for ventricular arrhythmias and sudden cardiac death. QT prolongation can be caused by drugs. There are multiple risk factors for drug-induced QT prolongation, including genetic variation. QT prolongation is one of the most common reasons for withdrawal of drugs from the market, despite the fact that these drugs may be beneficial for certain patients and not harmful in every patient. Identifying genetic variants associated with drug-induced QT prolongation might add to tailored pharmacotherapy and prevent beneficial drugs from being withdrawn unnecessarily. In this review, our objective was to provide an overview of the genetic background of drug-induced QT prolongation, distinguishing pharmacokinetic and pharmacodynamic pathways. Pharmacokinetic-mediated genetic susceptibility is mainly characterized by variation in genes encoding drug-metabolizing cytochrome P450 enzymes or drug transporters. For instance, the P-glycoprotein drug transporter plays a role in the pharmacokinetic susceptibility of drug-induced QT prolongation. The pharmacodynamic component of genetic susceptibility is mainly characterized by genes known to be associated with QT interval duration in the general population and genes in which the causal mutations of congenital long QT syndromes are located. Ethnicity influences susceptibility to drug-induced QT interval prolongation, with Caucasians being more sensitive than other ethnicities. Research on the association between pharmacogenetic interactions and clinical endpoints such as sudden cardiac death is still limited. Future studies in this area could enable us to determine the risk of arrhythmias more adequately in clinical practice.
Key Points
| Both pharmacokinetic and pharmacodynamic factors have been identified to influence susceptibility for drug-induced QT interval prolongation, but only few have been consistently associated. |
| Caucasians seem to be more sensitive to drug-induced QT interval prolongation than other ethnicities and therefore ethnicity might be an important factor to take into account when determining risk of ventricular arrhythmias. |
| Future studies on clinical endpoints are needed to find new drug–gene interactions. Identification of more variants influencing susceptibility to drug-induced QT interval prolongation might bring us closer to tailored pharmacotherapy. |
Introduction
The QT interval is measured on the electrocardiogram (ECG) and represents the ventricular depolarization and repolarization. The QT interval is usually corrected for heart rate, often using the formula of Bazett, as heart rate influences the duration of the QT interval. Normally, the average heart rate-corrected QT interval is approximately 410–430 ms for adult men and 420–430 ms for adult women [1]. A prolonged heart rate-corrected QT interval is usually defined as above 450 ms in men, and above 470 ms in women [2]. A prolonged QT interval is a well known risk factor for ventricular arrhythmias and sudden cardiac death [3–6], and can be congenital or acquired [7, 8]. Congenital QT prolongation is caused by mendelian genetic disorders, called the long QT syndromes. The long QT syndromes can be divided into subtypes according to the gene in which the causal mutation is located [9]. Acquired QT prolongation can be caused by cardiac diseases, such as coronary heart disease and heart failure, but it is usually caused by certain drugs [8, 10]. The susceptibility to acquired QT interval prolongation can also be influenced by genetic variation [9]. Two things point to this: first, the heritability of QT interval duration in the general population (excluding congenital long QT syndrome patients) is estimated to be around 35 % [11, 12], and second, first-degree relatives of patients with congenital long QT syndrome have a higher risk of drug-induced QT prolongation than non-related individuals [13]. In genome-wide association studies (GWAS), a great number of genes associated with QT interval duration have been identified [14–16]. The gene with the strongest signal related to QT interval duration is the nitric oxide synthase 1 adaptor protein gene (NOS1AP), located on chromosome 1 [14–20], and is influencing impulse propagation [21]. Other findings from GWAS included polymorphisms within genes known to be mutated in congenital long QT syndromes, genes associated with intracellular calcium handling, as well as genes previously not known to influence cardiac repolarization [9, 15, 16]. Because drug use is the most common cause of acquired QT prolongation [7], QT interval prolongation is an important outcome in drug safety research [5, 6]. In drug safety studies, QT interval prolongation is often combined with torsade de pointes, a rare ventricular arrhythmia associated with QT prolongation. Drugs can lead to a relatively high frequency of torsade de pointes, of up to 5 % [22]. QT interval prolongation is one of the leading causes of drug relabeling or withdrawal of drugs from the market [23, 24]. Drugs that are associated with either QT prolongation or torsade de pointes are mentioned in various lists, such as the University of Arizona list [25, 26]. These lists include antiarrhythmic drugs, such as flecainide and amiodarone, but also non-cardiac drugs [25]. These non-cardiac drugs are a diverse set with various indications and working mechanisms and include antidepressant drugs (e.g., citalopram), antibiotics (e.g., erythromycin and fluoroquinolones), and drugs against nausea (e.g., domperidone) [25]. Regulatory authorities state that these drugs should not be administered to patients already using another drug with QT-prolonging properties or to patients who have an increased risk of QT prolongation [5, 6].
As QT interval duration is influenced by genetic variants, the individual risk of drug-induced QT interval prolongation may also be influenced by genetic factors. Pharmacogenetics describes the role of genetic variation in drug response, which can relate to both pharmacokinetic and pharmacodynamic properties. Pharmacokinetics constitute the effect of the body on the drug, which is usually categorized into effects on absorption, distribution, metabolism, and elimination of the drug. Pharmacodynamics reflect the effect of the drug, in a certain concentration, on the body. Both pharmacokinetics and pharmacodynamics influence efficacy and effectiveness of the drug, and the occurrence of adverse drug reactions. As genotyping has become less expensive over the past years, larger populations can be investigated and more and more variants determining susceptibility to drug-induced QT interval prolongation could possibly be detected. This might bring us closer to the goal of tailored pharmacotherapy [27]. Therefore, our objective was to provide an overview of the pharmacogenetics of drug-induced QT interval prolongation. Hereto, we distinguished pharmacokinetic and pharmacodynamic genetic susceptibility and ethnic differences in QT interval prolongation. We searched PubMed from inception until April 8, 2015 using the search terms (pharmaco)genetic, (pharmaco)genomic, QT/QTc, prolong*, long, drug-induced, drug related, drug, medication, medicine, ethnic, race. We applied a restriction for articles in the English language.
An overview of the pharmacogenetics discussed in this review is shown in Fig. 1 and an overview of the knowledge on ethnic differences in susceptibility to QT prolongation is given in Fig. 2.
Fig. 1.
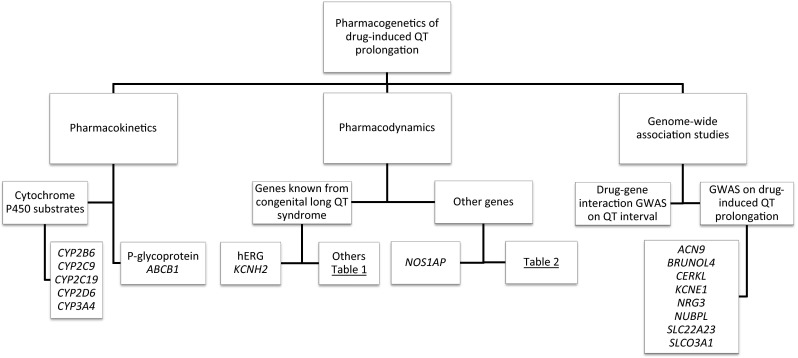
Overview of the genes associated with drug-induced QT interval prolongation
Fig. 2.
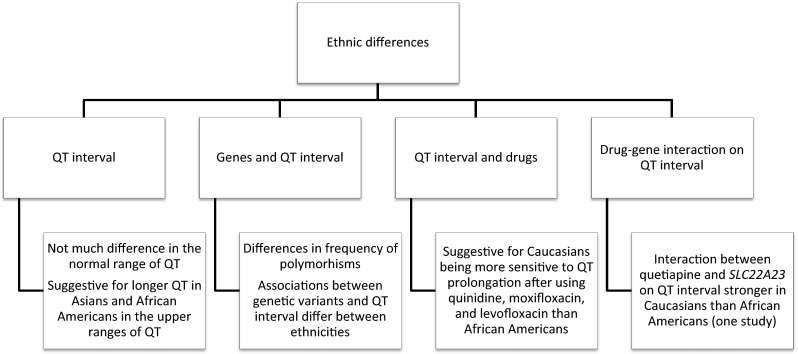
Overview of the available knowledge on ethnic differences in susceptibility to drug-induced QT interval prolongation
Pharmacokinetic Genetic Susceptibility
A description of the pharmacokinetics of all QT-prolonging drugs and interactions with other drugs is beyond the scope of this review. We focused on the influence of genes related to pharmacokinetics on drug-induced QT interval prolongation.
Cytochrome P450 Substrates
Many drugs are metabolized in the liver by cytochrome P450 (CYP), a family of enzymes. These enzymes are encoded by various genes named according to their family (e.g., ‘2’) and subfamily (e.g., ‘C’), based on the amino acid structure. A full code indicates a specific gene, (e.g., CYP2C9). A large proportion of the QT-prolonging drugs are metabolized by CYP2D6, CYP3A4/5, and CYP2C9 [28]. Several cytochrome P450 genes have been investigated in relation to drug-induced QT interval prolongation.
CYP2D6 is the most extensively studied gene of the CYP family in relation to drug-induced QT prolongation. Genetic variation within CYP2D6 was associated with QT prolongation in users of risperidone and iloperidone [29–31]. However, in users of haloperidol and thioridazine, CYP2D6 status was not associated with QT prolongation [32–35]. The proportion of poor metabolizers was not different among users of the antispasmodic terodiline surviving ventricular tachycardia or torsade de pointes than in the total population [36].
Another often studied gene of the CYP family is CYP2C19. CYP2C19 status was a predisposing factor for QT prolongation in antimicrobial (except quinolones) users [37] and a higher proportion of poor metabolizers was found among terodiline users who survived ventricular tachycardia or torsade de pointes compared with the normal population [36]. CYP2C19 status did not influence QT interval in users of nelfinavir or thioridazine [35, 38].
Some other CYP family members were investigated. Methadone users who were CYP2B6 poor metabolizers had an increased QT interval duration and a 4.5-fold increased risk of a prolonged QT interval compared with methadone users who were normal or extensive metabolizers [39]. CYP2C9 was not associated with QT prolongation in risperidone users [29]. CYP3A4 and CYP2C9 variants predisposed to an increased risk of QT prolongation or torsade de pointes induced by antimicrobial agents, with the exception of quinolones [37].
P-Glycoprotein
P-glycoprotein is a transmembrane efflux pump and influences bioavailability of several drugs. P-glycoprotein is encoded by the ATP-binding cassette B1 gene (ABCB1) [40]. The three most frequently investigated polymorphisms within this gene are C1236T, G2677T, and C3435T [41–43]. In a study in 66 schizophrenia patients using risperidone, the QT interval was significantly longer in patients carrying a T allele of the C3435T polymorphism than homozygous C allele carriers [44]. A study on romidepsin, a potentially QT-prolonging anticancer drug, showed that mice lacking P-glycoprotein had higher intracardiac concentrations of the drug. This study also showed that carriers of genetic variants in ABCB1 have less QT interval prolongation following a dose of romidepsin [45]. Thus, P-glycoprotein might also be an effect modifier of other potential QT-prolonging drugs.
Pharmacodynamic Genetic Susceptibility
Genes Known From Congenital Long QT Syndrome
Five to nineteen percent of patients with drug-induced torsade de pointes carry mutations in genes involved in congenital long QT syndrome [46, 47]. These genes are listed in Table 1, which also shows the number of studies that did or did not find an association between these genes and drug-induced QT interval prolongation or torsade de pointes [31, 46–57]. Since the human ether-a-go-go (eag)-related gene (hERG) channel and its associated gene, the potassium channel, voltage gated eag-related subfamily H, member 2 gene (KCNH2), were investigated most frequently and are currently a topic of special interest in drug safety research, it will be discussed separately.
Table 1.
Genes known from congenital long QT syndrome, investigated in drug-induced long QT syndrome or torsade de pointes
| cLQTS type | Gene abbreviation | Gene name [110] | Gene protein product [9] | Association founda | |
|---|---|---|---|---|---|
| Yes | No | ||||
| cLQTS1 | KCNQ1 | Potassium channel, voltage gated KQT-like subfamily Q, member 1 gene | IKs channel alpha-subunit | 3 | 4 |
| cLQTS2 | KCNH2 | Potassium channel, voltage gated eag-related subfamily H, member 2 | IKr channel alpha-subunit | 9 | 4 |
| cLQTS3 | SCN5A | Sodium channel, voltage gated, type V alpha-subunit | INa channel alpha-subunit | 3 | 4 |
| cLQTS4 | ANK2 | Ankyrin 2, neuronal | 1 | 1 | |
| cLQTS5 | KCNE1 | Potassium channel, voltage gated subfamily E regulatory beta-subunit 1 | IKs channel beta-subunit | 4 | 3 |
| cLQTS6 | KCNE2 | Potassium channel, voltage gated subfamily E regulatory beta-subunit 2 | IKr channel beta-subunit | 2 | 6 |
| cLQTS7 | KCNJ2 | Potassium channel, inwardly rectifying subfamily J, member 2 | IKl alpha-subunit | 0 | 3 |
| cLQTS8 | CACNA1C | Calcium channel, voltage-dependent, L type, alpha-1C subunit | ICaL channel alpha-subunit | 2 | 1 |
| cLQTS9 | CAV3 | Caveolin-3 | 0 | 2 | |
| cLQTS10 | SCN4B | Sodium channel, voltage gated, type IV beta-subunit | INa channel beta-subunit 4 | 1 | 1 |
| cLQTS11 | AKAP9 | A kinase anchor protein 9 | 1 | 2 | |
| cLQTS12 | SNTA1 | Syntrophin, acidic 1 | 1 | 1 | |
| cLQTS13 | KCNJ5 | Potassium channel, inwardly rectifying subfamily J, member 5 | IKACh channel alpha-subunit | 0 | 1 |
cLQTS congenital long QT syndrome
aNumber of studies that did or did not find an association with the corresponding gene
hERG Channel Encoded by KCNH2
The hERG channel has received much attention in the past decades. In congenital long QT syndrome type 2, the causal mutation is located in the KCNH2 gene, which encodes this channel [58]. KCNH2 encodes the α subunit of the channel mediating the rapidly activating component of the delayed rectifier K+ (Ikr) current [59]. The hERG channel can be blocked by drugs in two different ways: (1) direct blocking of the channel; (2) indirect blockade by disrupting channel protein trafficking through a reduction of the number of channels in the membrane [58, 60]. The resulting inhibition of the Ikr current in turn leads to an excessive lengthening of the action potential. This can lead to early after depolarizations and subsequently to arrhythmias and sudden cardiac death [61]. Almost all drugs that have been associated with torsade de pointes or QT prolongation block the Ikr current, but not all Ikr-blocking drugs have QT-prolonging properties [62]. In a study of 14 antipsychotics, a significant correlation was reported between the estimated hERG blockade and the increase in QT interval for five Ikr-blocking drugs (haloperidol, olanzapine, risperidone, thioridazine, and ziprasidone) [63]. Several studies identified mutations in the KCNH2 gene in persons with drug-induced prolonged QT interval or torsade de pointes, which were not present in controls [46, 47, 50, 52, 64]. These were KCNH2 mutations not related to congenital long QT syndrome type 2. Several KCNH2 mutations were identified in persons with drug-induced arrhythmias in a database of a spontaneous reporting system for adverse drug reactions [49]. However, in one study, persons with drug-induced torsade de pointes had a similar proportion of hERG mutations as controls [54] and three studies did not find an association between KCNH2 and drug-induced QT prolongation [48, 51, 53]. In a study on dofetilide-induced hERG blockade, it was reported that a blockade of 10 % was associated with a 20-ms [95 % confidence interval (CI) 12–32] increase in QT interval [65]. A study in methadone users showed that persons with a KCNH2 mutation had a 15-ms (95 % CI 5–26) longer QT interval [57].
Other Genes Known from Congenital Long QT Syndrome
Several studies have assessed genes involved in congenital long QT syndrome, to identify variants, other than the causal ones in congenital long QT syndrome, that are associated with drug-induced QT interval prolongation. The design of these studies is very heterogeneous [31, 46–57, 60]. For example, studies included methadone users [57], cases of drug-induced arrhythmias from a database of spontaneous reports of adverse drug reactions [49], and a case report [51]. Results from these studies are ambiguous, as not one gene has consistently been associated with drug-induced QT prolongation. The fact that the study populations were small hampers the possibility to draw firm conclusions.
Other Genes Having Pharmacodynamic Effects
NOS1AP
The NOS1AP gene encodes the nitric oxide synthase 1 activating protein and regulates the enzyme neuronal nitric oxide synthase (nNOS), a regulator of intracellular calcium levels and cardiomyocyte contraction [66, 67]. nNOS inhibits L-type calcium channels and influences cardiac repolarization [68] and impulse propagation [21]. Previous studies have shown that NOS1AP is associated with QT interval duration [14–16, 69], and common variants within this gene have been associated with an increased risk of QT prolongation, induced by amiodarone, sotalol, or diuretics [70]. As the NOS1AP gene is associated with inhibition of L-type calcium channels, which are blocked by certain calcium channel blockers (i.e., diltiazem and verapamil), it can be expected that this gene modifies response to these drugs. Two variants within the NOS1AP gene were associated with a modified effect of the drug on QT interval duration in users of verapamil and, to a lesser degree, diltiazem [71]. NOS1AP has also been associated with antipsychotic-induced QT prolongation [48] and drug-induced torsade de pointes [56]. Heterozygous and homozygous major allele carriers of the rs10494366 variant had an increased risk of a prolonged QT interval following granisetron or dolasetron administration in the perioperative setting, compared with homozygous minor allele carriers [72].
Other Candidate Gene Studies
Several other genes with suspected pharmacodynamic effects were investigated for their association with drug-induced QT prolongation or drug-induced torsade de pointes. Selection of these genes was based on known association with arrhythmias or cardiac diseases or because of a suggestive or significant signal in a GWAS. The results of the candidate gene studies are summarized in Table 2 [48, 52, 53, 56, 73].
Table 2.
Genes investigated in candidate gene studies on drug-induced QT prolongation
| Gene abbreviation | Gene name [110] | Association founda |
|---|---|---|
| AKAP6 | A kinase anchor protein 6 | Yes [52] |
| AKAP7 | A kinase anchor protein 7 | Yes [52] |
| APLP2 | Amyloid beta precursor-like protein 2 | Yes [52] |
| ATP1B1 | ATPase, Na+/K+ transporting, beta 1 polypeptide | No [48] |
| ATP2A2 | ATPase, CA++ transporting, cardiac muscle, slow twitch 2 | Yes [52] |
| – | Beta-adrenergic receptor genes | No [73] |
| BRUNOL4 | Bruno-like 4 | No [48] |
| CACNB2 | Calcium channel, voltage-dependent, beta 2 subunit | Yes [52]/No [53] |
| CALR | Calreticulin | Yes [52] |
| CASQ2 | Calsequestrin 2 | Yes [56] |
| CERKL | Ceramide kinase-like | No [48] |
| CNOT1 | CCR4-NOT transcription complex, subunit 1 | No [48] |
| FKBP1B | FK506 binding protein 1b | No [56] |
| GINS3 | GINS complex subunit 3 | No [48] |
| GPD1L | Glycerol-3-phosphate dehydrogenase 1-like | Yes [52]/No [53, 56] |
| JPH2 | Junctophilin 2 | Yes [52] |
| JPH3 | Junctophilin 3 | Yes [52] |
| KCND3 | Potassium channel, voltage gated Shal related subfamily D, member 3 | Yes [52] |
| KCNN3 | Potassium channel, calcium activated intermediate/small conductance subfamily N alpha, member 3 | Yes [52] |
| LIG3 | Ligase III, DNA, ATP-dependent | No [48] |
| LITAF | Lipopolysaccharide-induced TNF factor | No [48] |
| NDRG4 | NDRG family member 4 | No [48] |
| NRG3 | Neuregulin 3 | No [48] |
| NUBPL | Nucleotide-binding protein-like | Yes [48] |
| PALLD | Palladin, cytoskeletal associated protein | No [48] |
| PLN | Phospholabam | No [48] |
| PPP2R3A | Protein phosphatase 2, regulatory subunit beta, alpha | Yes [52] |
| RNF207 | Ring finger protein 207 | No [48] |
| RYR2 | Ryanodine receptor 2 | Yes [52, 56]/No [53] |
| SCN1B | Sodium channel, voltage gated, type I beta subunit | Yes [56]/No [53] |
| SCN4A | Sodium channel, voltage gated, type IV alpha subunit | Yes [56] |
| SETD6 | SET domain containing 6 | No [48] |
| SLC35F1 | Solute carrier family 35, member F1 | No [48] |
| SLCO3A1 | Solute carrier organic anion transporter family, member 3A1 | No [48] |
| ZFHX3 | Zinc finger homeobox 3 | Yes [52] |
aAssociation, enrichment or predictive value detected for this gene
Genome-Wide Association Studies (GWAS) on Drug-Induced QT Interval Prolongation
In GWAS, associations are studied between a phenotype and an extensive set of polymorphisms throughout the whole genome. Generally, only a subset of all polymorphisms of a person are genotyped, after which the other polymorphisms are imputed with a reference panel. The most frequently used reference panels are HapMap [74, 75] and 1000 Genomes [76–78]. HapMap contains around 2.5 million common polymorphisms; 1000 Genomes contains around 30 million polymorphisms and includes both common and rare variants. The choice of reference panel can influence the number of findings and whether rare variants can be identified. We describe this type of research separately, because both pharmacokinetic and pharmacodynamic polymorphisms can be detected with GWAS. We also describe a whole-exome sequencing study, which is technically not a GWAS, but has a similar aim and methodology.
There are two possible ways to investigate the genetic basis of drug-induced QT prolongation in the framework of GWAS [79]. The first is to investigate the association between the polymorphism and a drug-induced phenotype. The phenotype can be either a dichotomous clinically relevant endpoint (e.g., a heart rate-corrected QT interval above 450 ms in men and above 470 ms in women) [2] or a continuous variable (e.g., the delta QT in ms before and after exposure) [2]. The second is to perform drug–gene interaction GWAS. In these analyses, the interactions between the use of certain drugs and all genetic polymorphisms available in the reference panel are investigated in relation to QT interval as continuous outcome or a prolonged QT interval or torsade de pointes as dichotomous outcome. However, for drug–gene interaction GWAS, very large study populations are required [79]. Moreover, because multiplicative models are tested, biologically relevant interactions, which are usually additive, may be missed.
GWAS on Drug-Induced QT Interval Prolongation
A GWAS in 216 torsade de pointes cases caused by amiodarone, sotalol, and quinidine and 771 controls, using both HapMap and 1000 Genomes imputed data, did not show genome-wide significant associations [80]. In another GWAS in 738 subjects included in a randomized clinical trial using approximately 600,000 polymorphisms, an association was found between the solute carrier family 22 member 23 gene (SLC22A23) and antipsychotic-induced (specifically quetiapine) QT prolongation [48]. SLCO3A1, another member of this family, was associated with QT interval prolongation induced by the antipsychotic iloperidone in 183 subjects included in a randomized trial [81]. This solute carrier family encodes organic ion transporters that shuttle drugs across the cell membrane and are expressed in cardiac tissue [48, 81]. Five other genes were identified in the GWAS on iloperidone-induced QT prolongation using about 300,000 polymorphisms. These included the ceramide kinase-like gene (CERKL), the palladin, cytoskeletal-associated protein gene (PALLD), the bruno-like 4 gene (BRUNOL4), the neuregulin 3 gene (NRG3), and the nucleotide-binding protein-like gene (NUBPL) [81]. CERKL regulates currents of various potassium channels, including the hERG channel. PALLD has been associated with myocardial infarction. BRUNOL4 is expressed in cardiac tissue and has been associated with cardiac structure and function. NRG3 has been found to play a role in cardiac development. The role of NUBPL is unclear [81]. Of these six genes, only the association between NUBPL and drug-induced QT prolongation could be replicated in another study [48]. A whole-exome sequencing study in 65 cases and 148 controls revealed two genes, KCNE1 and ACN9, associated with antiarrhythmic and antipsychotic drug-induced QT interval prolongation [53].
Drug–Gene Interaction GWAS
To our knowledge, there is only one drug–gene interaction GWAS on QT interval duration. This study investigated the interaction between genome-wide genetic variation using HapMap imputed data and four types of exposure (thiazide diuretics, tri/tetracyclic antidepressants, sulfonylurea hypoglycemic agents, and QT-prolonging drugs) in 33,781 participants from ten cohort studies. It did not show a genome-wide significant association with QT interval. There were also no significant interactions with 26 polymorphisms previously reported to be related to QT interval duration [82].
Ethnic Differences in Susceptibility to Drug-Induced QT Interval Prolongation
As ethnic differences ultimately reflect genetic variation, it is useful to study ethnicity with regard to susceptibility to drug-induced QT interval prolongation. However, the role of ethnic differences has not been well established in studies on drug-induced QT interval prolongation. In 20 thorough QT/QTc studies, only 10 % of the total study populations was African American and only 7 % was Asian [83]. We will discuss studies that have investigated the association between ethnicity and QT interval duration, ethnicity and genes associated with QT interval, the influence of ethnicity on associations between drugs and QT interval, and the influence of ethnicity on drug–gene interaction associated with QT interval. This is summarized in Fig. 2.
Ethnic Differences in QT Interval Duration
Data on baseline differences of QT interval between different ethnicities is sparse. The available data indicates that there are no large differences in the normal range [24]. A difference of 6–7 ms was demonstrated for Asian women in the normal range of the QT interval compared with other ethnicities, and this was a difference of 10 ms in the upper range of the QT interval [84]. Among 41 African Americans and 3456 Caucasians with a prolonged QT interval, African Americans had a 29-ms longer heart rate-corrected QT interval [85].
Ethnic Differences in Genetic Variation Associated with QT Interval
The frequency of polymorphisms in genes known from congenital long QT syndrome had varying distributions among ethnic groups [86–89]. A study on SCN5A showed that out of the 39 variants found, 20 were only found in African Americans, seven only in Caucasians, four only in Asians, and three only in Hispanics. Only one polymorphism was found in all four ethnic groups [86, 88]. Of the 49 polymorphisms identified in KCNQ1, KCNH2, KCNE1, and KCNE2, 27 were only found in African Americans, 13 only in Caucasians, two only in Asians, and two only in Hispanics. Only two polymorphisms were identified in all four ethnic groups [87, 89]. A study on KCNH2 in 100 African Americans and 100 Caucasians, showed that the minor alleles of two polymorphisms were more frequent among Caucasians than among African Americans [89]. These studies only described the frequency of variants, but not a potentially causal role of these genes with regard to QT interval prolongation.
Contradictory results were published on the effect of genetic variants on QT between different ethnicities. Among Caucasians, more variants of 28 tagging SNPs of the NOS1AP gene were significantly associated with QT interval than in African Americans, Hispanics, and Chinese [90]. However, other studies reported comparable associations between NOS1AP gene variants and QT interval among Caucasians, Hispanics, and African Americans [91, 92], but also among African Americans of both European and African ancestry [93]. Comparable associations in different ethnic groups were also reported between genetic variants of PLN1, KCNQ1, NDRG4, and ATP1B1 and QT interval [92, 93]. However, one study identified variants within NOS1AP and ATP1B1 among African Americans that had not been reported in Caucasians [92].
Ethnic Differences in Drug Response with Regard to QT Prolongation
African Americans had the highest risk of a prolonged QT interval after acute overdose of QT-prolonging drugs [odds ratio (OR) 2.01; 95% CI 1.03–3.91] while Hispanics had the lowest risk (OR 0.49; 95% CI 0.26–0.92) compared with all other ethnicities [94]. For the other ethnicities, no statistically significant differences were observed [94]. Another study showed that Caucasians using quinidine, a powerful QT-prolonging drug, were more sensitive to QT prolongation than African Americans, even though baseline values of the QT interval were comparable [95, 96]. Moxifloxacin is often used as positive control in thorough QT/QTc studies and is a moderately powerful QT-prolonging drug. Studies showed that there were no ethnic differences in QT prolongation following moxifloxacin and levofloxacin administration [97–100], however the trend suggested that Caucasians may be more sensitive to QT prolongation than Asians [100].
Ethnic Differences in Drug–Gene Interaction on QT Interval
One study compared the interaction between quetiapine use and polymorphisms of the SLC22A23 gene on QT interval and found a stronger effect of this drug–gene interaction in Caucasians than in African Americans [48].
Discussion and Conclusion
Nowadays, QT interval prolongation is one of the most common reasons to withdraw drugs from the market. Many risk factors for QT interval prolongation and associated arrhythmias have been identified, including both non-genetic and genetic risk factors [3, 9, 10]. As QT-prolonging effects might only occur in people with certain genetic variations [24], drugs can be completely withdrawn from the market, while they could be safe and beneficial for certain patients. With more knowledge on the genetic variation associated with drug-induced QT interval prolongation, we might be able to identify which patients would benefit, and which patients would be at risk, and subsequently not have to withdraw drugs with QT-prolonging properties from the market completely [101]. Pharmacovigilance studies focus on drugs that have come onto the market and thus do not have extreme QT-prolonging properties. Therefore, we mostly detect genetic variants with small effects. However, studies into the genetic variation associated with extreme or frequent adverse effects in early-phase studies could tell us more about genetic variants with large effects. Genetic variants associated with large adverse drug effects might be an important aid in risk stratification, give insight into the genes and proteins relevant for drug-induced QT interval prolongation and arrhythmias, or ultimately result in genetic targeted marketing (in accordance with cancer pharmacogenetics) if the effect is fully dependent on a genotype. However, other independent risk factors besides genetic variants play a role in the development of a prolonged QT interval, which are important for risk stratification in individual patients.
GWAS have been instrumental in the discovery of a wide range of associations between genetic variants and many different phenotypes. However, gene–environment interaction GWAS are challenging, because of the large sample size that is required for these kind of studies [79]. Therefore, in the field of pharmacogenetics, candidate gene studies are probably more feasible. These studies are used to elaborate on the possible mechanisms through which drug-induced QT prolongation occurs. They may also shed light on the large interpersonal variability in drug response and therefore add to safer use of many drugs. However, GWAS will remain an important tool for hypothesis generation. Power can be gained by improving the outcome and exposure definition, leading to a reduction in misclassification. Nowadays, QT-prolonging drugs are investigated as one exposure group, often because of the low number of exposed persons; for example, in the drug–gene interaction GWAS by Avery et al. [82]. If the pathways through which these drugs cause QT prolongation comprise the same genetic variants, this approach works. However, if the working mechanisms are heterogeneous, exposure is misclassified and this will reduce power to detect meaningful variants.
Future research could focus on identifying which polymorphisms within a gene have the best predictive value for adverse effects, the so-called fine mapping. With the emergence of whole-genome sequencing and whole-exome sequencing in large cohort studies [102], we will be able to study genotypes in more detail and we will have data available on more rare variants [102]. This might enable us to discover more polymorphisms associated with adverse drug reactions, which might also increase the predictive value of polymorphisms that are important in pharmacogenetic interactions. The increasing availability of expression and methylation data in cohort studies might enable us to determine epigenetic variants influencing the occurrence of disease. Expression and methylation variants might also be associated with the age at which QT prolongation occurs.
Future pharmacogenetic research should also focus on direct clinical endpoints, such as arrhythmias, overall mortality, and sudden cardiac death. This will tell us more about the direct value of pharmacogenetic interactions for daily clinical practice. A research method which is currently emerging is the molecular autopsy [103, 104]. In cases of sudden death, without obvious causes identified with autopsy, the pathologist can only assume that death had an arrhythmogenic origin [103]. In these cases, genotyping family members or the deceased can shed light on the genetic variation which might be the underlying factor that caused the arrhythmia and death [103]. This research method could provide more insight into genetic variations associated with sudden deaths.
Concerns about the value of the QT interval in the prediction of drug-induced arrhythmias and sudden cardiac death have been highlighted in recent years, because some drugs are associated with QT prolongation but not with an increased risk of arrhythmias [105, 106]. In addition to identifying genetic variation contributing to drug-induced QT prolongation, we should also focus on identifying other measurable markers, such as heart-rate variability [107] and QT variability [108], and their genetic determinants of drug-induced arrhythmia in order to improve risk prediction.
Since ethnic differences might be important for individual-specific risk of drug-induced QT prolongation, more research is needed to establish which drugs can be used safely in the different ethnic groups. The studies mentioned thus far suggest that Caucasians are more sensitive to drug-induced QT interval prolongation than other ethnicities. However, studies do not provide consistent results, possibly because other risk factors besides genetic variants play an important role in QT interval prolongation. Shah hypothesizes that given the fact that the differences between ethnicities in these studies are generally small, ethnic differences are probably relatively most important in users of mild QT-prolonging drugs [24]. If a drug has strong QT-prolonging properties in general, it is not likely that the small difference related to ethnicity will make much difference. If a drug has no QT-prolonging properties in general, it is not likely that it will be a cause of strong QT prolongation in one ethnic group. However, if a drug has moderate QT-prolonging properties, the small differences could mean that a drug can be used safely in some ethnic groups, but not in others, because the degree of QT prolongation is either just above or just below the safety cut-off value [24]. The US FDA considers an increase below 5 ms as not clinically relevant [6]. If, for example, a drug prolongs the QT interval with 3 ms in one ethnicity, while it causes an extra 3 ms increase in another ethnicity, the added prolongation is small, but it will be just above the safety cut-off value, meaning that the drug should be contraindicated in one ethnicity, but not in the other. One might question whether such a far-reaching consequence would be justified considering the limited amount of available information. In addition, within each ethnic group, genetic modifiers can be different and pharmacogenetic studies should therefore not be limited to Caucasians only.
As genotyping has become less expensive over the last couple of years, genotyping patients for genetic variants that influence their risk of adverse drug effects has become more feasible. This will allow us to investigate larger populations, enabling us to find more, and also rare, genetic variants. If we know the genetic profile of a patient with regard to adverse drug effects, we might be able to prescribe drugs more safely. This would also allow us to be more accurate in the dosage that a patient needs [101]. However, clinical validity has to be tested for specific drugs, as this has often been done insufficiently to date [109]. Of course, many ethical, legal, and financial problems have to be solved before this can be applied in clinical practice. But as it becomes more and more clear that genetic variation is an important effect modifier when prescribing several drugs, we should continue to identify pharmacogenetic interactions and study their clinical consequences.
In conclusion, we provided an overview of the most important pharmacogenetic background of drug-induced QT interval prolongation, which includes pharmacokinetic and pharmacodynamic variants. GWAS have been proven challenging because of the required large sample size. However, with the availability of genetic data in more and more large study populations, this method remains important for hypothesis generation. Findings from these studies can then be used for candidate gene studies to further elaborate on possible drug–gene interactions. Not much data is available on ethnic differences in susceptibility to drug-induced QT interval prolongation, although Caucasians seem to be more sensitive to QT prolongation than other ethnicities. Future research could focus on fine mapping of genetic regions, epigenetics, clinical endpoints, or use of alternative markers for drug-induced arrhythmias. Knowledge on the genetic variants associated with drug-induced QT interval prolongation is important as this will provide more accurate predictions as to which patients will experience the adverse drug effect and to whom the drugs can be prescribed relatively safely. This may also prevent unnecessary withdrawal of drugs from the market.
Compliance with Ethical Standards
Funding
This work is supported by grants from the Netherlands Organisation for Health Research and Development (ZonMw) (Priority Medicines Elderly 113102005 to ME and PRR; and HTA 80-82500-98-10208 to BHS). The funding sources had no involvement in the collection, analysis, writing, interpretation, or in the decision to submit the paper for publication.
Conflicts of interest
Authors Maartje N. Niemeijer, Marten E. van den Berg, Mark Eijgelsheim, and Bruno H. Stricker have no conflicts of interest that are directly relevant to the content of this study. Author Peter R. Rijnbeek works in a group that has received funding from the European Commission and several pharmaceutical companies. Dr Rijnbeek has no other conflicts of interest that are directly relevant to the content of this study.
References
- 1.Rijnbeek PR, van Herpen G, Bots ML, Man S, Verweij N, Hofman A, et al. Normal values of the electrocardiogram for ages 16–90 years. J Electrocardiol. 2014;47(6):914–21. [DOI] [PubMed]
- 2.Committee for proprietary medicinal products. The assessment of the potential for QT interval prolongation by non-cardiovascular medicinal products. 1997. http://www.fda.gov/ohrms/dockets/ac/03/briefing/pubs/cpmp.pdf. Accessed 13 Nov 2014.
- 3.Zipes DP, Wellens HJ. Sudden cardiac death. Circulation. 1998;98(21):2334–2351. doi: 10.1161/01.CIR.98.21.2334. [DOI] [PubMed] [Google Scholar]
- 4.Roden DM. Drug-induced prolongation of the QT interval. N Engl J Med. 2004;350(10):1013–1022. doi: 10.1056/NEJMra032426. [DOI] [PubMed] [Google Scholar]
- 5.European Medicines Agency. ICH Topic E 14. The clinical evaluation of QT/QTc interval prolongation and proarrhythmic potential for non-arrhythmic drugs. 2005; Available from: http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2009/09/WC500002879.pdf. Cited 2 Mar 2015.
- 6.Food and Drug Administration. International conference on harmonisation; guidance on E14 clinical evaluation of QT/QTc interval prolongation and proarrhythmic potential for non-antiarrhythmic drugs. Fed Regist. 2005;70(202):61134–5. [PubMed]
- 7.Kannankeril PJ, Roden DM. Drug-induced long QT and torsade de pointes: recent advances. Curr Opin Cardiol. 2007;22(1):39–43. doi: 10.1097/HCO.0b013e32801129eb. [DOI] [PubMed] [Google Scholar]
- 8.Kannankeril P, Roden DM, Darbar D. Drug-induced long QT syndrome. Pharmacol Rev. 2010;62(4):760–781. doi: 10.1124/pr.110.003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahida S, Hogarth AJ, Cowan C, Tayebjee MH, Graham LN, Pepper CB. Genetics of congenital and drug-induced long QT syndromes: current evidence and future research perspectives. J Interv Card Electrophysiol. 2013;37(1):9–19. doi: 10.1007/s10840-013-9779-5. [DOI] [PubMed] [Google Scholar]
- 10.Morita H, Wu J, Zipes DP. The QT syndromes: long and short. Lancet. 2008;372(9640):750–763. doi: 10.1016/S0140-6736(08)61307-0. [DOI] [PubMed] [Google Scholar]
- 11.Hong Y, Rautaharju PM, Hopkins PN, Arnett DK, Djousse L, Pankow JS, et al. Familial aggregation of QT-interval variability in a general population: results from the NHLBI Family Heart Study. Clin Genet. 2001;59(3):171–177. doi: 10.1034/j.1399-0004.2001.590305.x. [DOI] [PubMed] [Google Scholar]
- 12.Newton-Cheh C, Larson MG, Corey DC, Benjamin EJ, Herbert AG, Levy D, et al. QT interval is a heritable quantitative trait with evidence of linkage to chromosome 3 in a genome-wide linkage analysis: The Framingham Heart Study. Heart Rhythm. 2005;2(3):277–284. doi: 10.1016/j.hrthm.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Kannankeril PJ, Roden DM, Norris KJ, Whalen SP, George AL, Jr, Murray KT. Genetic susceptibility to acquired long QT syndrome: pharmacologic challenge in first-degree relatives. Heart Rhythm. 2005;2(2):134–140. doi: 10.1016/j.hrthm.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 14.Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38(6):644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 15.Pfeufer A, Sanna S, Arking DE, Muller M, Gateva V, Fuchsberger C, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41(4):407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41(4):399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Post W, Shen H, Damcott C, Arking DE, Kao WH, Sack PA, et al. Associations between genetic variants in the NOS1AP (CAPON) gene and cardiac repolarization in the old order Amish. Hum Hered. 2007;64(4):214–219. doi: 10.1159/000103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aarnoudse AJ, Newton-Cheh C, de Bakker PI, Straus SM, Kors JA, Hofman A, et al. Common NOS1AP variants are associated with a prolonged QTc interval in the Rotterdam Study. Circulation. 2007;116(1):10–16. doi: 10.1161/CIRCULATIONAHA.106.676783. [DOI] [PubMed] [Google Scholar]
- 19.Tobin MD, Kahonen M, Braund P, Nieminen T, Hajat C, Tomaszewski M, et al. Gender and effects of a common genetic variant in the NOS1 regulator NOS1AP on cardiac repolarization in 3761 individuals from two independent populations. Int J Epidemiol. 2008;37(5):1132–1141. doi: 10.1093/ije/dyn091. [DOI] [PubMed] [Google Scholar]
- 20.Lehtinen AB, Newton-Cheh C, Ziegler JT, Langefeld CD, Freedman BI, Daniel KR, et al. Association of NOS1AP genetic variants with QT interval duration in families from the Diabetes Heart Study. Diabetes. 2008;57(4):1108–1114. doi: 10.2337/db07-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapoor A, Sekar RB, Hansen NF, Fox-Talbot K, Morley M, Pihur V, et al. An enhancer polymorphism at the cardiomyocyte intercalated disc protein NOS1AP locus is a major regulator of the QT interval. Am J Hum Genet. 2014;94(6):854–869. doi: 10.1016/j.ajhg.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kannankeril PJ. Understanding drug-induced torsades de pointes: a genetic stance. Expert Opin Drug Saf. 2008;7(3):231–239. doi: 10.1517/14740338.7.3.231. [DOI] [PubMed] [Google Scholar]
- 23.Shah RR. Drug-induced prolongation of the QT interval: why the regulatory concern? Fundam Clin Pharmacol. 2002;16(2):119–124. doi: 10.1046/j.1472-8206.2002.00082.x. [DOI] [PubMed] [Google Scholar]
- 24.Shah RR. Drug-induced QT interval prolongation: does ethnicity of the thorough QT study population matter? Br J Clin Pharmacol. 2013;75(2):347–358. doi: 10.1111/j.1365-2125.2012.04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woosley R. Drugs that prolong the QTc interval and/or induce torsade de pointes; Available from: http://www.crediblemeds.org/everyone/composite-list-all-qtdrugs. Cited 3 Mar 2015.
- 26.De Ponti F, Poluzzi E, Montanaro N. QT-interval prolongation by non-cardiac drugs: lessons to be learned from recent experience. Eur J Clin Pharmacol. 2000;56(1):1–18. doi: 10.1007/s002280050714. [DOI] [PubMed] [Google Scholar]
- 27.Johnson JA, Cavallari LH, Beitelshees AL, Lewis JP, Shuldiner AR, Roden DM. Pharmacogenomics: application to the management of cardiovascular disease. Clin Pharmacol Ther. 2011;90(4):519–531. doi: 10.1038/clpt.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aerssens J, Paulussen AD. Pharmacogenomics and acquired long QT syndrome. Pharmacogenomics. 2005;6(3):259–270. doi: 10.1517/14622416.6.3.259. [DOI] [PubMed] [Google Scholar]
- 29.Llerena A, Berecz R, Dorado P, de la Rubia A. QTc interval, CYP2D6 and CYP2C9 genotypes and risperidone plasma concentrations. J Psychopharmacol. 2004;18(2):189–193. doi: 10.1177/0269881104042618. [DOI] [PubMed] [Google Scholar]
- 30.Cartwright AL, Wilby KJ, Corrigan S, Ensom MH. Pharmacogenetics of risperidone: a systematic review of the clinical effects of CYP2D6 polymorphisms. Ann Pharmacother. 2013;47(3):350–360. doi: 10.1345/aph.1R333. [DOI] [PubMed] [Google Scholar]
- 31.Potkin SG, Preskorn S, Hochfeld M, Meng X. A thorough QTc study of 3 doses of iloperidone including metabolic inhibition via CYP2D6 and/or CYP3A4 and a comparison to quetiapine and ziprasidone. J Clin Psychopharmacol. 2013;33(1):3–10. doi: 10.1097/JCP.0b013e31827c0314. [DOI] [PubMed] [Google Scholar]
- 32.Llerena A, Berecz R, de la Rubia A, Dorado P. QTc interval lengthening is related to CYP2D6 hydroxylation capacity and plasma concentration of thioridazine in patients. J Psychopharmacol. 2002;16(4):361–364. doi: 10.1177/026988110201600411. [DOI] [PubMed] [Google Scholar]
- 33.Desai M, Tanus-Santos JE, Li L, Gorski JC, Arefayene M, Liu Y, et al. Pharmacokinetics and QT interval pharmacodynamics of oral haloperidol in poor and extensive metabolizers of CYP2D6. Pharmacogenomics J. 2003;3(2):105–113. doi: 10.1038/sj.tpj.6500160. [DOI] [PubMed] [Google Scholar]
- 34.Park JY, Shon JH, Kim KA, Jung HJ, Shim JC, Yoon YR, et al. Combined effects of itraconazole and CYP2D6*10 genetic polymorphism on the pharmacokinetics and pharmacodynamics of haloperidol in healthy subjects. J Clin Psychopharmacol. 2006;26(2):135–142. doi: 10.1097/01.jcp.0000203199.88581.c3. [DOI] [PubMed] [Google Scholar]
- 35.Thanacoody RH, Daly AK, Reilly JG, Ferrier IN, Thomas SH. Factors affecting drug concentrations and QT interval during thioridazine therapy. Clin Pharmacol Ther. 2007;82(5):555–565. doi: 10.1038/sj.clpt.6100195. [DOI] [PubMed] [Google Scholar]
- 36.Ford GA, Wood SM, Daly AK. CYP2D6 and CYP2C19 genotypes of patients with terodiline cardiotoxicity identified through the yellow card system. Br J Clin Pharmacol. 2000;50(1):77–80. doi: 10.1046/j.1365-2125.2000.00230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owens RC., Jr QT prolongation with antimicrobial agents: understanding the significance. Drugs. 2004;64(10):1091–1124. doi: 10.2165/00003495-200464100-00005. [DOI] [PubMed] [Google Scholar]
- 38.Damle B, Fosser C, Ito K, Tran A, Clax P, Uderman H, et al. Effects of standard and supratherapeutic doses of nelfinavir on cardiac repolarization: a thorough QT study. J Clin Pharmacol. 2009;49(3):291–300. doi: 10.1177/0091270008329551. [DOI] [PubMed] [Google Scholar]
- 39.Eap CB, Crettol S, Rougier JS, Schlapfer J, Sintra Grilo L, Deglon JJ, et al. Stereoselective block of hERG channel by (S)-methadone and QT interval prolongation in CYP2B6 slow metabolizers. Clin Pharmacol Ther. 2007;81(5):719–728. doi: 10.1038/sj.clpt.6100120. [DOI] [PubMed] [Google Scholar]
- 40.Sakaeda T, Nakamura T, Okumura K. MDR1 genotype-related pharmacokinetics and pharmacodynamics. Biol Pharm Bull. 2002;25(11):1391–1400. doi: 10.1248/bpb.25.1391. [DOI] [PubMed] [Google Scholar]
- 41.Verstuyft C, Schwab M, Schaeffeler E, Kerb R, Brinkmann U, Jaillon P, et al. Digoxin pharmacokinetics and MDR1 genetic polymorphisms. Eur J Clin Pharmacol. 2003;58(12):809–812. doi: 10.1007/s00228-003-0567-5. [DOI] [PubMed] [Google Scholar]
- 42.Kurzawski M, Bartnicka L, Florczak M, Gornik W, Drozdzik M. Impact of ABCB1 (MDR1) gene polymorphism and P-glycoprotein inhibitors on digoxin serum concentration in congestive heart failure patients. Pharmacol Rep. 2007;59(1):107–11. [PubMed]
- 43.Aarnoudse AJ, Dieleman JP, Visser LE, Arp PP, van der Heiden IP, van Schaik RH, et al. Common ATP-binding cassette B1 variants are associated with increased digoxin serum concentration. Pharmacogenet Genomics. 2008;18(4):299–305. doi: 10.1097/FPC.0b013e3282f70458. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki Y, Tsuneyama N, Fukui N, Sugai T, Watanabe J, Ono S, et al. Effect of risperidone metabolism and P-glycoprotein gene polymorphism on QT interval in patients with schizophrenia. Pharmacogenomics J. 2014;14(5):452–456. doi: 10.1038/tpj.2014.6. [DOI] [PubMed] [Google Scholar]
- 45.Sissung TM, Gardner ER, Piekarz RL, Howden R, Chen X, Woo S, et al. Impact of ABCB1 allelic variants on QTc interval prolongation. Clin Cancer Res. 2011;17(4):937–946. doi: 10.1158/1078-0432.CCR-10-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paulussen AD, Gilissen RA, Armstrong M, Doevendans PA, Verhasselt P, Smeets HJ, et al. Genetic variations of KCNQ1, KCNH2, SCN5A, KCNE1, and KCNE2 in drug-induced long QT syndrome patients. J Mol Med (Berl). 2004;82(3):182–188. doi: 10.1007/s00109-003-0522-z. [DOI] [PubMed] [Google Scholar]
- 47.Lehtonen A, Fodstad H, Laitinen-Forsblom P, Toivonen L, Kontula K, Swan H. Further evidence of inherited long QT syndrome gene mutations in antiarrhythmic drug-associated torsades de pointes. Heart Rhythm. 2007;4(5):603–607. doi: 10.1016/j.hrthm.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 48.Aberg K, Adkins DE, Liu Y, McClay JL, Bukszar J, Jia P, et al. Genome-wide association study of antipsychotic-induced QTc interval prolongation. Pharmacogenomics J. 2012;12(2):165–172. doi: 10.1038/tpj.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Bruin ML, van Puijenbroek EP, Bracke M, Hoes AW, Leufkens HG. Pharmacogenetics of drug-induced arrhythmias: a feasibility study using spontaneous adverse drug reactions reporting data. Pharmacoepidemiol Drug Saf. 2006;15(2):99–105. doi: 10.1002/pds.1194. [DOI] [PubMed] [Google Scholar]
- 50.Itoh H, Sakaguchi T, Ding WG, Watanabe E, Watanabe I, Nishio Y, et al. Latent genetic backgrounds and molecular pathogenesis in drug-induced long-QT syndrome. Circ Arrhythm Electrophysiol. 2009;2(5):511–523. doi: 10.1161/CIRCEP.109.862649. [DOI] [PubMed] [Google Scholar]
- 51.Makita N, Horie M, Nakamura T, Ai T, Sasaki K, Yokoi H, et al. Drug-induced long-QT syndrome associated with a subclinical SCN5A mutation. Circulation. 2002;106(10):1269–1274. doi: 10.1161/01.CIR.0000027139.42087.B6. [DOI] [PubMed] [Google Scholar]
- 52.Ramirez AH, Shaffer CM, Delaney JT, Sexton DP, Levy SE, Rieder MJ, et al. Novel rare variants in congenital cardiac arrhythmia genes are frequent in drug-induced torsades de pointes. Pharmacogenomics J. 2013;13(4):325–329. doi: 10.1038/tpj.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weeke P, Mosley JD, Hanna D, Delaney JT, Shaffer C, Wells QS, et al. Exome sequencing implicates an increased burden of rare potassium channel variants in the risk of drug-induced long QT interval syndrome. J Am Coll Cardiol. 2014;63(14):1430–1437. doi: 10.1016/j.jacc.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang P, Kanki H, Drolet B, Yang T, Wei J, Viswanathan PC, et al. Allelic variants in long-QT disease genes in patients with drug-associated torsades de pointes. Circulation. 2002;105(16):1943–1948. doi: 10.1161/01.CIR.0000014448.19052.4C. [DOI] [PubMed] [Google Scholar]
- 55.Sesti F, Abbott GW, Wei J, Murray KT, Saksena S, Schwartz PJ, et al. A common polymorphism associated with antibiotic-induced cardiac arrhythmia. Proc Natl Acad Sci USA. 2000;97(19):10613–10618. doi: 10.1073/pnas.180223197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaab S, Crawford DC, Sinner MF, Behr ER, Kannankeril PJ, Wilde AA, et al. A large candidate gene survey identifies the KCNE1 D85N polymorphism as a possible modulator of drug-induced torsades de pointes. Circ Cardiovasc Genet. 2012;5(1):91–99. doi: 10.1161/CIRCGENETICS.111.960930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hajj A, Ksouda K, Peoc’h K, Curis E, Messali A, Deveaux LL, et al. KCNH2 polymorphism and methadone dosage interact to enhance QT duration. Drug Alcohol Depend. 2014;1(141):34–38. doi: 10.1016/j.drugalcdep.2014.04.027. [DOI] [PubMed] [Google Scholar]
- 58.Vandenberg JI, Perry MD, Perrin MJ, Mann SA, Ke Y, Hill AP. hERG K(+) channels: structure, function, and clinical significance. Physiol Rev. 2012;92(3):1393–1478. doi: 10.1152/physrev.00036.2011. [DOI] [PubMed] [Google Scholar]
- 59.Trudeau MC, Warmke JW, Ganetzky B, Robertson GA. HERG, a human inward rectifier in the voltage-gated potassium channel family. Science. 1995;269(5220):92–95. doi: 10.1126/science.7604285. [DOI] [PubMed] [Google Scholar]
- 60.Rajamani S, Eckhardt LL, Valdivia CR, Klemens CA, Gillman BM, Anderson CL, et al. Drug-induced long QT syndrome: hERG K+ channel block and disruption of protein trafficking by fluoxetine and norfluoxetine. Br J Pharmacol. 2006;149(5):481–489. doi: 10.1038/sj.bjp.0706892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finlayson K, Witchel HJ, McCulloch J, Sharkey J. Acquired QT interval prolongation and HERG: implications for drug discovery and development. Eur J Pharmacol. 2004;500(1–3):129–142. doi: 10.1016/j.ejphar.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 62.Drici MD, Barhanin J. Cardiac K+ channels and drug-acquired long QT syndrome. Therapie. 2000;55(1):185–93. [PubMed]
- 63.Crumb WJ, Jr, Ekins S, Sarazan RD, Wikel JH, Wrighton SA, Carlson C, et al. Effects of antipsychotic drugs on I(to), I (Na), I (sus), I (K1), and hERG: QT prolongation, structure activity relationship, and network analysis. Pharm Res. 2006;23(6):1133–1143. doi: 10.1007/s11095-006-0070-7. [DOI] [PubMed] [Google Scholar]
- 64.Sun Z, Milos PM, Thompson JF, Lloyd DB, Mank-Seymour A, Richmond J, et al. Role of a KCNH2 polymorphism (R1047 L) in dofetilide-induced Torsades de Pointes. J Mol Cell Cardiol. 2004;37(5):1031–1039. doi: 10.1016/j.yjmcc.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 65.Jonker DM, Kenna LA, Leishman D, Wallis R, Milligan PA, Jonsson EN. A pharmacokinetic-pharmacodynamic model for the quantitative prediction of dofetilide clinical QT prolongation from human ether-a-go-go-related gene current inhibition data. Clin Pharmacol Ther. 2005;77(6):572–582. doi: 10.1016/j.clpt.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 66.Barouch LA, Harrison RW, Skaf MW, Rosas GO, Cappola TP, Kobeissi ZA, et al. Nitric oxide regulates the heart by spatial confinement of nitric oxide synthase isoforms. Nature. 2002;416(6878):337–339. doi: 10.1038/416337a. [DOI] [PubMed] [Google Scholar]
- 67.Oceandy D, Cartwright EJ, Emerson M, Prehar S, Baudoin FM, Zi M, et al. Neuronal nitric oxide synthase signaling in the heart is regulated by the sarcolemmal calcium pump 4b. Circulation. 2007;115(4):483–492. doi: 10.1161/CIRCULATIONAHA.106.643791. [DOI] [PubMed] [Google Scholar]
- 68.Splawski I, Timothy KW, Decher N, Kumar P, Sachse FB, Beggs AH, et al. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc Natl Acad Sci USA. 2005;102(23):8089–96 (discussion 6–8). [DOI] [PMC free article] [PubMed]
- 69.Eijgelsheim M, Aarnoudse AL, Rivadeneira F, Kors JA, Witteman JC, Hofman A, et al. Identification of a common variant at the NOS1AP locus strongly associated to QT-interval duration. Hum Mol Genet. 2009;18(2):347–357. doi: 10.1093/hmg/ddn341. [DOI] [PubMed] [Google Scholar]
- 70.Jamshidi Y, Nolte IM, Dalageorgou C, Zheng D, Johnson T, Bastiaenen R, et al. Common variation in the NOS1AP gene is associated with drug-induced QT prolongation and ventricular arrhythmia. J Am Coll Cardiol. 2012;60(9):841–850. doi: 10.1016/j.jacc.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Noord C, Aarnoudse AJ, Eijgelsheim M, Sturkenboom MC, Straus SM, Hofman A, et al. Calcium channel blockers, NOS1AP, and heart-rate-corrected QT prolongation. Pharmacogenet Genomics. 2009;19(4):260–266. doi: 10.1097/FPC.0b013e328324e556. [DOI] [PubMed] [Google Scholar]
- 72.Quraishi SA, Schuler GH, Janicki PK. 5 HT(3)-receptor antagonists and cardiac repolarization time in patients expressing a novel genetic target associated with baseline QTc interval abnormalities. J Clin Anesth. 2011;23(4):297–302. doi: 10.1016/j.jclinane.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 73.Kanki H, Yang P, Xie HG, Kim RB, George AL, Jr, Roden DM. Polymorphisms in beta-adrenergic receptor genes in the acquired long QT syndrome. J Cardiovasc Electrophysiol. 2002;13(3):252–256. doi: 10.1046/j.1540-8167.2002.00252.x. [DOI] [PubMed] [Google Scholar]
- 74.HapMap Project; Available from: http://www.hapmap.org. Cited 4 Mar 2015.
- 75.International HapMap C. The International HapMap Project. Nature. 2003;426(6968):789–96. [DOI] [PubMed]
- 76.1000 Genomes. A deep catalogue of human genetic variation; Available from: http://www.1000genomes.org. Cited 4 Mar 2015.
- 77.Genomes Project Consortium. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Daly AK. Genome-wide association studies in pharmacogenomics. Nat Rev Genet. 2010;11(4):241–246. doi: 10.1038/nrg2751. [DOI] [PubMed] [Google Scholar]
- 80.Behr ER, Ritchie MD, Tanaka T, Kaab S, Crawford DC, Nicoletti P, et al. Genome wide analysis of drug-induced torsades de pointes: lack of common variants with large effect sizes. PLoS One. 2013;8(11):e78511. doi: 10.1371/journal.pone.0078511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Volpi S, Heaton C, Mack K, Hamilton JB, Lannan R, Wolfgang CD, et al. Whole genome association study identifies polymorphisms associated with QT prolongation during iloperidone treatment of schizophrenia. Mol Psychiatry. 2009;14(11):1024–1031. doi: 10.1038/mp.2008.52. [DOI] [PubMed] [Google Scholar]
- 82.Avery CL, Sitlani CM, Arking DE, Arnett DK, Bis JC, Boerwinkle E, et al. Drug-gene interactions and the search for missing heritability: a cross-sectional pharmacogenomics study of the QT interval. Pharmacogenomics J. 2014;14(1):6–13. doi: 10.1038/tpj.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Florian JA, Tornoe CW, Brundage R, Parekh A, Garnett CE. Population pharmacokinetic and concentration—QTc models for moxifloxacin: pooled analysis of 20 thorough QT studies. J Clin Pharmacol. 2011;51(8):1152–1162. doi: 10.1177/0091270010381498. [DOI] [PubMed] [Google Scholar]
- 84.Rautaharju PM, Prineas RJ, Kadish A, Larson JC, Hsia J, Lund B. Normal standards for QT and QT subintervals derived from a large ethnically diverse population of women aged 50 to 79 years [the Women’s Health Initiative (WHI)] Am J Cardiol. 2006;97(5):730–737. doi: 10.1016/j.amjcard.2005.09.108. [DOI] [PubMed] [Google Scholar]
- 85.Fugate T, 2nd, Moss AJ, Jons C, McNitt S, Mullally J, Ouellet G, et al. Long QT syndrome in African–Americans. Ann Noninvasive Electrocardiol. 2010;15(1):73–76. doi: 10.1111/j.1542-474X.2009.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ackerman MJ, Splawski I, Makielski JC, Tester DJ, Will ML, Timothy KW, et al. Spectrum and prevalence of cardiac sodium channel variants among black, white, Asian, and Hispanic individuals: implications for arrhythmogenic susceptibility and Brugada/long QT syndrome genetic testing. Heart Rhythm. 2004;1(5):600–607. doi: 10.1016/j.hrthm.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 87.Ackerman MJ, Tester DJ, Jones GS, Will ML, Burrow CR, Curran ME. Ethnic differences in cardiac potassium channel variants: implications for genetic susceptibility to sudden cardiac death and genetic testing for congenital long QT syndrome. Mayo Clin Proc. 2003;78(12):1479–1487. doi: 10.4065/78.12.1479. [DOI] [PubMed] [Google Scholar]
- 88.Splawski I, Timothy KW, Tateyama M, Clancy CE, Malhotra A, Beggs AH, et al. Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science. 2002;297(5585):1333–1336. doi: 10.1126/science.1073569. [DOI] [PubMed] [Google Scholar]
- 89.Anson BD, Ackerman MJ, Tester DJ, Will ML, Delisle BP, Anderson CL, et al. Molecular and functional characterization of common polymorphisms in HERG (KCNH2) potassium channels. Am J Physiol Heart Circ Physiol. 2004;286(6):H2434–H2441. doi: 10.1152/ajpheart.00891.2003. [DOI] [PubMed] [Google Scholar]
- 90.Shah SA, Herrington DM, Howard TD, Divers J, Arnett DK, Burke GL, et al. Associations between NOS1AP single nucleotide polymorphisms (SNPs) and QT interval duration in four racial/ethnic groups in the Multi-Ethnic Study of Atherosclerosis (MESA) Ann Noninvasive Electrocardiol. 2013;18(1):29–40. doi: 10.1111/anec.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arking DE, Khera A, Xing C, Kao WH, Post W, Boerwinkle E, et al. Multiple independent genetic factors at NOS1AP modulate the QT interval in a multi-ethnic population. PLoS One. 2009;4(1):e4333. doi: 10.1371/journal.pone.0004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Avery CL, Sethupathy P, Buyske S, He Q, Lin DY, Arking DE, et al. Fine-mapping and initial characterization of QT interval loci in African Americans. PLoS Genet. 2012;8(8):e1002870. doi: 10.1371/journal.pgen.1002870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smith JG, Avery CL, Evans DS, Nalls MA, Meng YA, Smith EN, et al. Impact of ancestry and common genetic variants on QT interval in African Americans. Circ Cardiovasc Genet. 2012;5(6):647–655. doi: 10.1161/CIRCGENETICS.112.962787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Manini AF, Stimmel B, Vlahov D. Racial susceptibility for QT prolongation in acute drug overdoses. J Electrocardiol. 2014;47(2):244–50. [DOI] [PMC free article] [PubMed]
- 95.Olatunde A, Price Evans DA. Blood quinidine levels and cardiac effects in white British and Nigerian subjects. Br J Clin Pharmacol. 1982;14(4):513–518. doi: 10.1111/j.1365-2125.1982.tb02022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shin JG, Kang WK, Shon JH, Arefayene M, Yoon YR, Kim KA, et al. Possible interethnic differences in quinidine-induced QT prolongation between healthy Caucasian and Korean subjects. Br J Clin Pharmacol. 2007;63(2):206–215. doi: 10.1111/j.1365-2125.2006.02793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Malik M, Hnatkova K, Schmidt A, Smetana P. Electrocardiographic QTc changes due to moxifloxacin infusion. J Clin Pharmacol. 2009;49(6):674–683. doi: 10.1177/0091270008330984. [DOI] [PubMed] [Google Scholar]
- 98.Kannankeril PJ, Norris KJ, Carter S, Roden DM. Factors affecting the degree of QT prolongation with drug challenge in a large cohort of normal volunteers. Heart Rhythm. 2011;8(10):1530–1534. doi: 10.1016/j.hrthm.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taubel J, Ferber G, Lorch U, Batchvarov V, Savelieva I, Camm AJ. Thorough QT study of the effect of oral moxifloxacin on QTc interval in the fed and fasted state in healthy Japanese and Caucasian subjects. Br J Clin Pharmacol. 2014;77(1):170–179. doi: 10.1111/bcp.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sugiyama A, Nakamura Y, Nishimura S, Adachi-Akahane S, Kumagai Y, Gayed J, et al. Comparison of the effects of levofloxacin on QT/QTc interval assessed in both healthy Japanese and Caucasian subjects. Br J Clin Pharmacol. 2012;73(3):455–459. doi: 10.1111/j.1365-2125.2011.04110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shah RR. Pharmacogenetics in drug regulation: promise, potential and pitfalls. Philos Trans R Soc Lond B Biol Sci. 2005;360(1460):1617–1638. doi: 10.1098/rstb.2005.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kiezun A, Garimella K, Do R, Stitziel NO, Neale BM, McLaren PJ, et al. Exome sequencing and the genetic basis of complex traits. Nat Genet. 2012;44(6):623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tester DJ, Ackerman MJ. The molecular autopsy: should the evaluation continue after the funeral? Pediatr Cardiol. 2012;33(3):461–470. doi: 10.1007/s00246-012-0160-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kamei S, Sato N, Harayama Y, Nunotani M, Takatsu K, Shiozaki T, et al. Molecular analysis of potassium ion channel genes in sudden death cases among patients administered psychotropic drug therapy: are polymorphisms in LQT genes a potential risk factor? J Hum Genet. 2014;59(2):95–99. doi: 10.1038/jhg.2013.125. [DOI] [PubMed] [Google Scholar]
- 105.van Opstal JM, Schoenmakers M, Verduyn SC, de Groot SH, Leunissen JD, van Der Hulst FF, et al. Chronic amiodarone evokes no torsade de pointes arrhythmias despite QT lengthening in an animal model of acquired long-QT syndrome. Circulation. 2001;104(22):2722–2727. doi: 10.1161/hc4701.099579. [DOI] [PubMed] [Google Scholar]
- 106.Thomsen MB, Matz J, Volders PG, Vos MA. Assessing the proarrhythmic potential of drugs: current status of models and surrogate parameters of torsades de pointes arrhythmias. Pharmacol Ther. 2006;112(1):150–170. doi: 10.1016/j.pharmthera.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 107.de Bruyne MC, Kors JA, Hoes AW, Klootwijk P, Dekker JM, Hofman A, et al. Both decreased and increased heart rate variability on the standard 10-second electrocardiogram predict cardiac mortality in the elderly: the Rotterdam Study. Am J Epidemiol. 1999;150(12):1282–1288. doi: 10.1093/oxfordjournals.aje.a009959. [DOI] [PubMed] [Google Scholar]
- 108.Niemeijer MN, van den Berg ME, Eijgelsheim M, van Herpen G, Stricker BH, Kors JA, et al. Short-term QT variability markers for the prediction of ventricular arrhythmias and sudden cardiac death: a systematic review. Heart. 2014;100(23):1831–1836. doi: 10.1136/heartjnl-2014-305671. [DOI] [PubMed] [Google Scholar]
- 109.Wang B, Canestaro WJ, Choudhry NK. Clinical evidence supporting pharmacogenomic biomarker testing provided in US Food and Drug Administration drug labels. JAMA Intern Med. 2014;174(12):1938–1944. doi: 10.1001/jamainternmed.2014.5266. [DOI] [PubMed] [Google Scholar]
- 110.National Center for Biotechnology Information (NCBI) Gene; Available from: http://www.ncbi.nlm.nih.gov/gene/. Cited 10 Mar 2015.


