Abstract
4,6-α-Glucanotransferase (4,6-α-GTase) enzymes, such as GTFB and GTFW of Lactobacillus reuteri strains, constitute a new reaction specificity in glycoside hydrolase family 70 (GH70) and are novel enzymes that convert starch or starch hydrolysates into isomalto/maltopolysaccharides (IMMPs). These IMMPs still have linear chains with some α1→4 linkages but mostly (relatively long) linear chains with α1→6 linkages and are soluble dietary starch fibers. 4,6-α-GTase enzymes and their products have significant potential for industrial applications. Here we report that an N-terminal truncation (amino acids 1 to 733) strongly enhances the soluble expression level of fully active GTFB-ΔN (approximately 75-fold compared to full-length wild type GTFB) in Escherichia coli. In addition, quantitative assays based on amylose V as the substrate are described; these assays allow accurate determination of both hydrolysis (minor) activity (glucose release, reducing power) and total activity (iodine staining) and calculation of the transferase (major) activity of these 4,6-α-GTase enzymes. The data show that GTFB-ΔN is clearly less hydrolytic than GTFW, which is also supported by nuclear magnetic resonance (NMR) analysis of their final products. From these assays, the biochemical properties of GTFB-ΔN were characterized in detail, including determination of kinetic parameters and acceptor substrate specificity. The GTFB enzyme displayed high conversion yields at relatively high substrate concentrations, a promising feature for industrial application.
INTRODUCTION
Starch is the second-most-abundant carbohydrate on earth and a major dietary carbohydrate for humans; as a storage carbohydrate it is present in seeds, roots, and tubers of plants (1). It consists of α-glucan polymers with α1→4 linkages and a low percentage of α1→6 linkages, in the form of amylose and branched amylopectin (2). Starches are applied in various industrial products such as food, paper, and textiles, often after processing by physical, chemical, or enzymatic treatment (3–6).
Dietary fibers and low-glycemic-index (low-GI) food are considered healthy food contributing to our long-term well-being (7, 8). Of all the nutritional types of starch, slowly digestible starch with low GI has drawn the strongest interest. Annealing/heat-moisture treatment, recrystallization, and enzymatic treatment are recognized approaches to obtain slowly digestible starch (9–11). Slowly digestible starch materials prepared by physical processing suffer losses upon boiling; therefore, structural modifications through enzymatic treatment of starch are more desirable. In the human digestive system, the α1→6 linkages in starch are hydrolyzed at a lower rate than α1→4 linkages (12, 13). Branching enzymes, alone or in combination with β-amylase, are used to increase the percentage of α1→4,6 branches in starches (12–14).
The 4,6-α-glucanotransferase (4,6-α-GTase) enzymes, such as GTFB, GTFW, and GTFML4, of Lactobacillus reuteri strains constitute a subfamily of glycoside hydrolase family 70 (GH70); GH70 mainly consists of glucansucrases (GSes) (http://www.cazy.org). Unlike GSes, 4,6-α-GTases are not sucrose-acting enzymes but rather starch-converting enzymes, capable of converting (1→4)-α-d-gluco-oligosaccharides into dietary fiber isomalto/maltopolysaccharides (IMMPs). It has been proposed that conversion occurs mainly by the stepwise addition of single glucose moieties onto the nonreducing end of an α-glucan, introducing linear chains with α1→6 linkages (15–18). The 4,6-α-GTase enzymes have strong potential for slowly digestible starch or soluble dietary fiber production in the starch industry (19).
The expression yields of all three studied 4,6-α-GTase enzymes in Escherichia coli are rather low, and most of the protein accumulates in inclusion bodies (18, 20); to obtain more active protein, strategies to use denatured refolded GTFB protein, or nonclassical inclusion body preparations, have been tested. The biochemical and catalytic properties of these enzymes, e.g., their hydrolysis and transferase activities, have not been characterized yet because of a lack of suitable quantitative assays. In the present study, the variable N-terminal region of the GTFB enzyme was removed (yielding construct GTFB734–1619), which resulted in increased expression of the soluble and active GTFB-ΔN enzyme in E. coli. In addition, we developed activity assays to characterize the reactions of 4,6-α-GTase enzymes with amylose. These assays were used to define the optimal reaction conditions of GTFB-ΔN and GTFW-ΔN and to quantitatively compare their hydrolysis and transferase activities, as well as to determine key biochemical properties. The developed assays also provide a firm basis for the future characterization of other natural and engineered 4,6-α-GTase enzymes.
MATERIALS AND METHODS
Construction of a truncated GTFB-ΔN mutant.
Based on the alignment of the L. reuteri 121 GTFB with glucansucrase sequences and the crystal structure of L. reuteri 180 GTF180-ΔN glucansucrase (PDB entry 3KLK), the gtfB gene fragment encoding GTFB (UniProt entry Q5SBM0) amino acids 734 to 1619 was amplified by PCR using the High Fidelity PCR enzyme mix (Thermo-Scientific, Landsmeer, The Netherlands) with pET15b-GTFB as the template and the primers CHisFor-dNgtfB (5′-GATGCATCCATGGGCCAGCTCATGAGAAACTTGGTTGCAAAACCTAATA-3′) and CHisRev-dNgtfB (5′-CCTCCTTTCTAGATCTATTAGTGATGGTGATGGTGATGGTTGTTAAAGTTTAATGAAATTGCAGTTGG-3′). A nucleotide sequence encoding a 6×His tag was fused in frame to the 3′ end of the gtfB-ΔN gene using the reverse primer. The resulting PCR product was digested with NcoI and BglII and was ligated into the corresponding site of pET15b. The construct was confirmed by nucleotide sequencing (GATC, Cologne, Germany). Plasmid pET15b-gtfW-ΔN (with the gtfW gene fragment encoding GTFW-ΔN [UniProt entry A5VL73] amino acids 458 to 1363) has been constructed previously (18).
Expression and purification of GTFB, GTFB-ΔN, and 4,6-αGT-W (GTFW-ΔN).
The GTFB protein was produced in E. coli BL21 Star(DE3) carrying the plasmids pRSF-GTFB and pBAD22-GroELS (17). The bacterial inocula were prepared in 0.4-liter Luria-Bertani cultures and grown at 37°C and 220 rpm until the optical density at 600 nm (OD600) had reached 0.4 to 0.5, at which point the inducer 0.4 mM isopropyl-β-d-1-thiogalactopyranoside and l-arabinose (0.02%, wt/vol) were added. The cultures were subsequently incubated at 18°C and 160 rpm for 16 h in an orbital shaker. Cells were harvested by centrifugation (26,000 × g, 40 min) and then washed with 20 ml buffer (20 mM Tris-HCl, pH 8.0, 250 mM NaCl, 1 mM CaCl2). The collected cells were resuspended in 10 ml B-PER protein extraction reagent (Pierce; 100 μg/ml of lysozyme and 5 U/ml of DNase) for 30 min on a rolling device at room temperature, followed by centrifugation (10,000 × g, 30 min). The encoded GTFB protein carrying a C-terminal 6×His tag was purified by His tag affinity chromatography using a 1-ml Hitrap IMAC HP column (GE Healthcare) and anion-exchange chromatography (Resource Q; GE Healthcare). The salt was removed using a 5-ml Hitrap desalting column (GE Healthcare) run with B-PER and 1 mM CaCl2. Purity was checked on 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, and protein concentration was measured with the Bradford reagent (Bio-Rad, Hercules, CA) and bovine serum albumin as the standard.
The L. reuteri DSM 20016 4,6-αGT-W (GTFW-ΔN) and GTFB-ΔN proteins were expressed and purified according to Leemhuis et al. with minor modification (18). E. coli BL21(DE3)/pET15b_gtfW-ΔN/gtfB-ΔN was grown in Luria broth containing 100 mg/liter ampicillin. Protein expression was induced at an OD600 of 0.4 to 0.5 by adding isopropyl-β-d-1-thiogalactopyranoside to 0.1 mM, and cultivation was continued at 18°C and 160 rpm for 16 h. Cells were harvested by centrifugation in Tris-HCl buffer (50 mM, pH 8.0) containing NaCl (250 mM). Cell extracts were made by sonication followed by centrifugation (10,000 × g, 1 h). Purification was performed as described above.
Preparation of amylose V substrate solution.
Amylose V was provided by AVEBE (Veendam, The Netherlands). Note that amylose V is a slightly degraded amylose, as a result of the extraction procedure used to isolate it from regular potato starch. The degree of branching of amylose V was below 0.1%, as reported earlier (21). Amylose V (1%, wt/vol) was prepared as a stock solution. Amylose V (20 mg) was suspended in 1 ml double-distilled H2O (ddH2O) and dissolved by addition of 1 ml 2 M NaOH. The solution was homogenized until clear. Prior to use, the stock solution was neutralized by adding an equal volume of 1 M HCl and the stock was further diluted with reaction buffer.
Enzyme assays.
The optimal pH and temperature of 4,6-α-GTases were determined over the pH range of 3.5 to 7.0 (25 mM sodium acetate [NaAc] buffer [pH 3.5 to 5.5], 25 mM MES [morpholineethanesulfonic acid] buffer [pH 5.5 to 6.5], 25 mM MOPS [morpholinepropanesulfonic acid] buffer [pH 6.5 to 7.0]) at 40°C and temperature range of 30 to 60°C at pH 5.0 with 0.25% (wt/vol) amylose V as the substrate. All other reactions were performed in reaction buffer (25 mM NaAc, pH 5.0, 1 mM CaCl2), with 0.25% (wt/vol) amylose V, 2.5% (wt/vol) maltodextrins (dextrose equivalent = 13 to 17; Sigma-Aldrich, St. Louis, MO), or 10 mM maltoheptaose (Sigma-Aldrich, St. Louis, MO) as the substrate at 40°C. For each reaction, 60 nM enzyme was added. Every 5 min, 100 μl of reaction mixture was taken and the reaction was stopped by addition of 50 μl of 0.4 M NaOH. Afterwards, 50 μl 0.4 M HCl was added to neutralize the reaction mixture. These samples were stored for glucose oxidase/peroxidase (d-glucose assay kit; Megazyme International, Ireland) determination of glucose released, Nelson-Somogyi measurement of reducing power, and (changes in) iodine staining of amylose.
Paselli MD6 maltodextrins (partial hydrolysis product of potato starch with a dextrose equivalent between 5 and 7 and 2.9% α1→6 glycosidic linkages) was provided by AVEBE, and variant concentrations (0.93 to 55.8%, wt/vol) were incubated with GTFB-ΔN (1/1,000 [wt/wt] concentration of substrate) for 72 h at 37°C and pH 5.0.
Amylose-iodine assay.
The iodine staining method was used to quantify (remaining) amylose in the reaction mixture (22). Thus, 0.26 g I2 and 2.6 g KI were dissolved in 10 ml distilled water to prepare the 260-fold-concentrated stock. Prior to using the staining buffer, the stock was diluted 260-fold with distilled water. For each measurement, 150 μl iodine reagent was pipetted into microtiter plate wells, followed by addition of 15 μl enzyme reaction sample solution. The microtiter plate was shaken slowly for 5 min to allow color formation, and the optical density was measured at 660 nm. Amylose V solutions (0 to 0.25%, wt/vol) were used to prepare a calibration curve.
Reducing sugar measurement.
The Nelson-Somogyi assay adapted to microtiter plates was used to measure the reducing power released through hydrolysis activity of the enzymes with amylose V (23). Thus, 40 μl Somogyi copper reagent, consisting of 4 parts KNa tartrate-Na2CO3-Na2SO4-NaHCO3 [1:2:12:1.3] and 1 part CuSO4·5H2O-Na2SO4 (1:9) and 40 μl enzyme reaction sample was mixed and incubated at 90°C for 30 min, after which the mixture was cooled to room temperature. After the microtiter plate was shaken for 1 min to remove CO2, 160 μl arsenomolybdate (25 g ammonium molybdate in 450 ml H2O plus 21 ml 98% H2SO4 plus 3 g Na2HAsO4·7H2O dissolved in 25 ml H2O) was added and mixed by vortexing. Optical densities were read at 500 nm. A calibration curve was made using 0 to 200 μg/ml glucose solutions as the standard.
Glucose measurement.
Glucose was measured using the GOPOD kit, which was adapted for microtiter plates. Five microliters enzyme reaction sample was added to wells containing 195 μl GOPOD reagent, followed by incubation at 40°C for 30 min. Optical densities were read at 510 nm. A calibration curve was made using 0 to 200 μg/ml glucose solutions as the standard.
Definitions of H and total activities, and calculation of T activities.
One unit of hydrolysis activity (H) is defined as the release of 1 mg glucose from amylose V per min in the GOPOD assay mixture. One unit of total activity is defined as the decrease of 1 mg amylose V per min in the assay mixture (as determined by the amylose-iodine staining method).
Amylose V is either converted into free glucose via H or modified by transferase activity (T) introducing linear α1→6-linked glucose chains, resulting in loss of the ability to bind iodine. T was calculated by subtracting H from total activity according to the equation T = total activity − H (all quantities in U/mg). Also, T/H activity ratios can be calculated subsequently.
NMR spectroscopy.
The reaction products were exchanged twice with D2O (99.9 atm% D; Cambridge Isotope Laboratories, Inc.) with intermediate lyophilization and then dissolved in 0.6 ml D2O. Resolution-enhanced 500-MHz one-dimensional (1D) 1H nuclear magnetic resonance (NMR) spectra were recorded with a spectral width of 4,500 Hz in 16k complex data sets and zero filled to 32k in D2O on a Varian Inova spectrometer (NMR Center, University of Groningen) at a probe temperature of 335 K. Suppression of the HOD signal was achieved by applying a WET1D pulse sequence. All spectra were processed using MestReNova 5.3.
RESULTS
Improving expression of soluble GTFB protein.
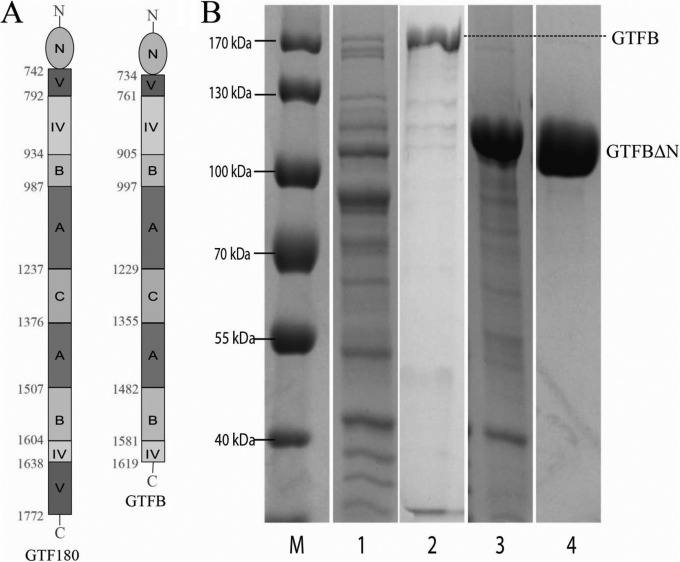
Heterologous expression of L. reuteri 4,6-α-GTase enzymes in E. coli is relatively poor. Optimization of the growth and induction conditions did not improve the soluble expression level of these enzymes, and most of the GTFB and GTFW proteins accumulated in inclusion bodies. Improvement of soluble expression of these enzymes appeared essential in view of their potential industrial application. The 4,6-α-GTase enzymes constitute a subfamily of GH70 showing high similarity with glucansucrases in amino acid sequences. As previously reported, deletion of the N-terminal variable region of GH70 glucansucrases, such as GTFA, did not change the glycosidic linkage types present in the products or the product sizes but significantly improved the expression levels of the enzymes (24–26). Therefore, we applied a similar truncation approach to GTFB. Alignment of the GTFB and glucansucrase protein sequences (27) resulted in identification of amino acids 1 to 733 as the GTFB N-terminal variable region (Fig. 1A). This region contains 5 RDV repeats (sequence R[P/N]DV-X12-SGF-X19–22-R[Y/F]S, where X represents a nonconserved amino acid residue), which were also observed previously in GTF180, GTFA, and GTFML1 (28). Its deletion in GTFB resulted in expression of soluble GTFB-ΔN at a much higher level than the full-length GTFB. Moreover, the purity of GTFB-ΔN protein was improved significantly compared to that of the full-length GTFB enzyme (Fig. 1B). More than 40 mg pure GTFB-ΔN protein was obtained from 1 liter E. coli culture, improving expression 75-fold based on molar calculation. The full-length GTFB and GTFB-ΔN enzymes were compared with regard to their reaction specificities and activities using amylose V and maltoheptaose as the substrates (data not shown). With maltoheptaose (10 mM) as the substrate, the rates of glucose release (GOPOD) and the product profiles of both enzymes were nearly identical. While using amylose V as the substrate, the total and hydrolysis activities measured by the GOPOD and iodine staining assay described below were almost the same. The N-terminally truncated GTFB and the full-length GTFB thus are virtually identical in activity and products.
FIG 1.
(A) Linear schematic representation of the domain organization of full-length GTF180 of L. reuteri 180 and GTFB of L. reuteri 121 (with N-terminal variable region). Structural analysis of the L. reuteri 180 GTF180-ΔN protein has shown that its peptide chain follows a “U” path and that four of its five domains are built up from discontinuous N- and C-terminal parts of the peptide chain. Only the C domain is formed from one continuous stretch of amino acids (27, 40–42). The GTFB protein has a similar organization, except that the C-terminal part of domain V is absent. The amino acid residue numbers indicate the start and end of each domain. (B) SDS-PAGE (8%, Coomassie blue stained) analysis of GTFB and GTFB-ΔN expression. Lane 1, whole-cell extract of E. coli with pET15b-gtfB plasmid; lane 2, purified GTFB (predicted molecular mass, 179 kDa); lane 3, whole-cell extract of E. coli with pET15b-gtfB-ΔN plasmid; lane 4, purified GTFB-ΔN (predicted molecular mass, 99 kDa); lane M, marker proteins.
Products derived from amylose V and maltodextrins by GTFB-ΔN or GTFW-ΔN treatments.
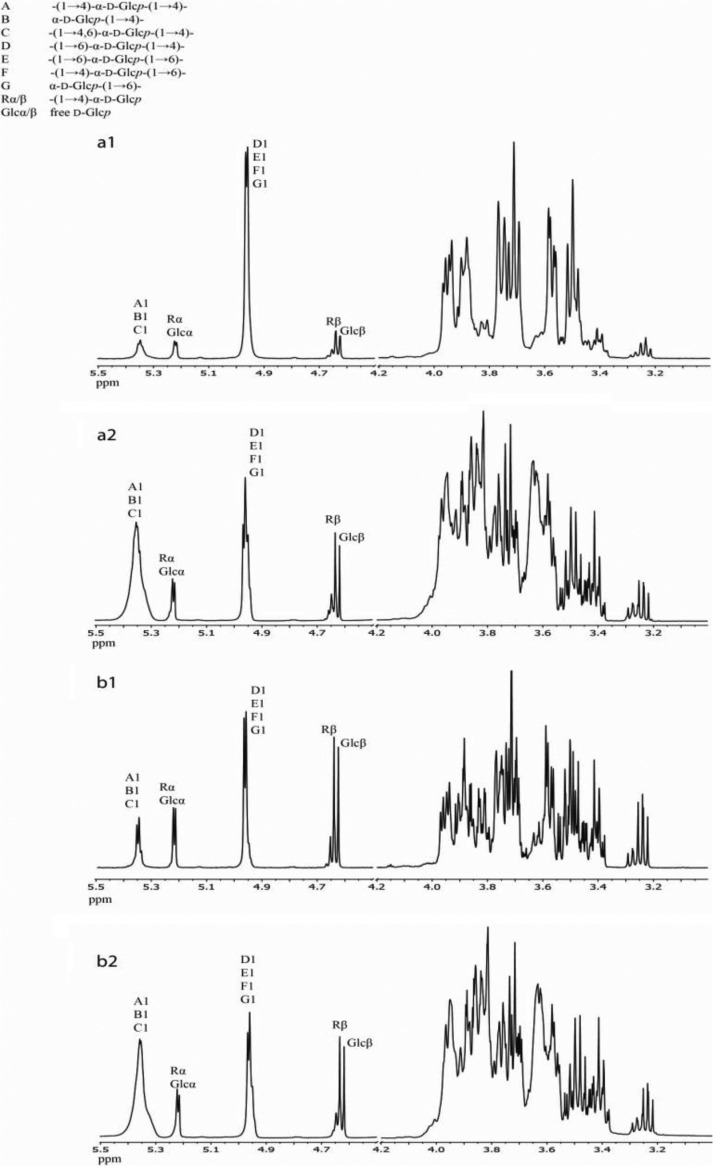
Linear α1→4-linked compounds are preferred substrates for 4,6-α-GTases; therefore, amylose V and short-chain α1→4-linked maltodextrins were compared as donor substrates for the GTFB-ΔN and GTFW-ΔN enzymes. 1H NMR analysis of the products of GTFB-ΔN and GTFW-ΔN incubated with amylose V [(1→4)-α-d-glucan] or maltodextrins [(1→4)-α-d-glucooligosaccharides] for 48 h revealed that both hydrolysis and transferase activities occurred. Besides the presence of α1→4 linkages (H-1, δ ∼5.40), α1→6 linkages (H-1, δ ∼4.97) were newly formed. In the product mixture derived from amylose V, the α1→6/α1→4 linkage ratio increased up to 90:10 (Fig. 2a1) for GTFB-ΔN and 74:26 for GTFW-ΔN (Fig. 2b1) when using identical amounts of purified protein. The spectra also showed the presence of free glucose (Glcα H-1, δ 5.225, Glcβ H-1, δ 4.637), the 4-substituted reducing glucose residues [-(1→4)-α-d-Glcp; Rα H-1, δ 5.225, Rβ H-1, δ 4.652], and a small amount of reducing-end glucose residues which are 6 substituted [-(1→6)-α-d-Glcp; Rα H-1, δ 5.241, Rβ H-1, δ 4.670] (29), suggesting that both GTFB-ΔN and GTFW-ΔN can use glucose as the acceptor substrate, forming maltose, isomaltose, and longer products in the presence of a donor substrate. Comparison of the spectra (Fig. 2a1 and Fig. b1) showed that GTFW-ΔN produced a much higher percentage of free glucose (23%) than GTFB-ΔN (8%) from amylose V. Conceivably, GTFW-ΔN has a higher ratio of hydrolysis to transferase activity than GTFB-ΔN, thereby initially generating more free glucose. In the reactions of both GTFB-ΔN and GTFW-ΔN with maltodextrins that possess branches (Fig. 2a2 and b2), lower conversions (36% for GTFB-ΔN and 32% for GTFW-ΔN) of α1→4 linkages into α1→6 linkages were obtained than in the reactions using amylose V (90% for GTFB-ΔN and 74% for GTFW-ΔN) as the substrate. Long-chain amylose thus appears to be a more efficient substrate for conversion of α1→4 into α1→6 linkages and was therefore used as the standard substrate in subsequent activity assay development and further biochemical analysis.
FIG 2.
One-dimensional 1H NMR spectra (D2O, 335 K) of the products derived from amylose V and maltodextrins via L. reuteri 121 GTFB-ΔN (a1 and a2, respectively) and L. reuteri DSM 20016 GTFW-ΔN (b1 and b2, respectively) treatments.
Measurement of the hydrolysis activity of 4,6-α-GTase enzymes.
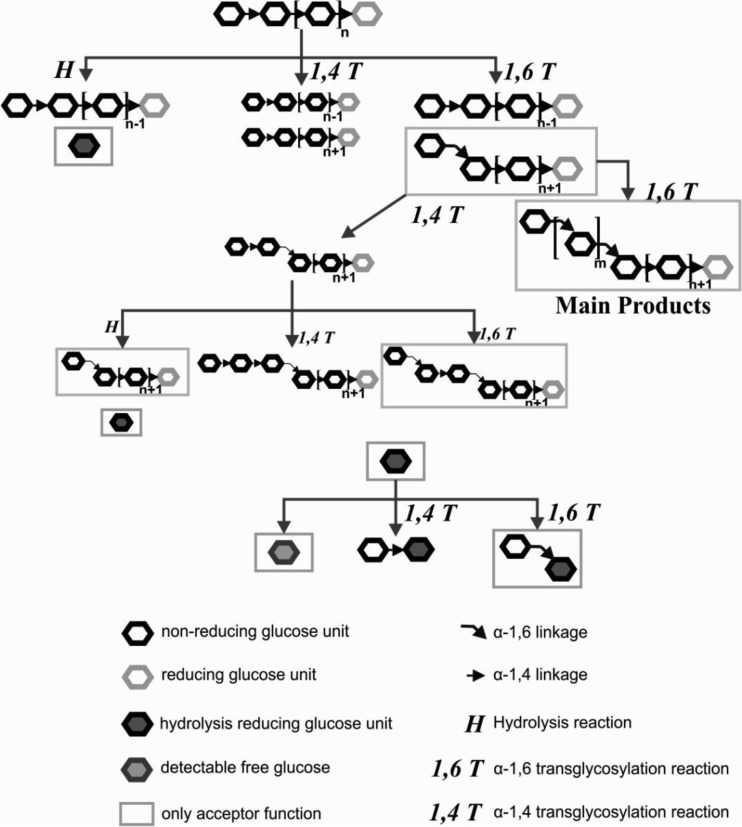
To characterize 4,6-α-GTase enzymes biochemically, assays to determine both their hydrolysis activity (H) and transferase activity (T) are required. All three characterized 4,6-α-GTases synthesize mostly α1→6 linkages and occasionally an α1→4 linkage (Fig. 3) (18). Whereas the 4,6-α-GTase transferase activity is dominant, hydrolysis activity also occurs, resulting in glucose release (Fig. 3). Reducing power and glucose measurements are widely accepted methods to determine H of starch-converting enzymes with either endo- or exohydrolyzing activity, such as α-amylase, α-glucosidase, and cyclodextrin glucanotransferase (30–32). In the case of 4,6-α-GTases, the measurement of free glucose specifically may lead to an underestimation of H. The reducing power assay representing both free glucose and products of the transferase reaction with glucose as the acceptor substrate may give a more precise estimate of H (Fig. 3). Therefore, the Nelson-Somogyi assay measuring reducing power was compared with the GOPOD assay measuring free glucose; the results were the same in the initial stage of the reaction (see Fig. S1 in the supplemental material). Therefore, both assays are equally applicable for measuring the initial rates of hydrolysis activity of 4,6-α-GTase enzymes.
FIG 3.
Schematic diagram of the reactions catalyzed by 4,6-α-GTase as proposed by Leemhuis et al. with minor modifications (18). Reducing ends of sugar molecules are shown in light gray. The molecules with gray frames can be used only as acceptor substrates.
Measurement of the total activity of 4,6-α-GTases.
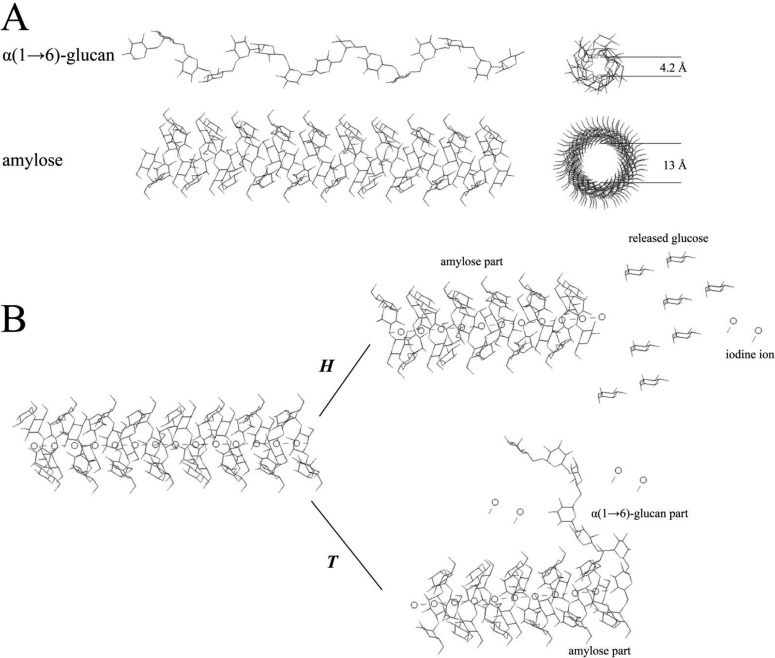
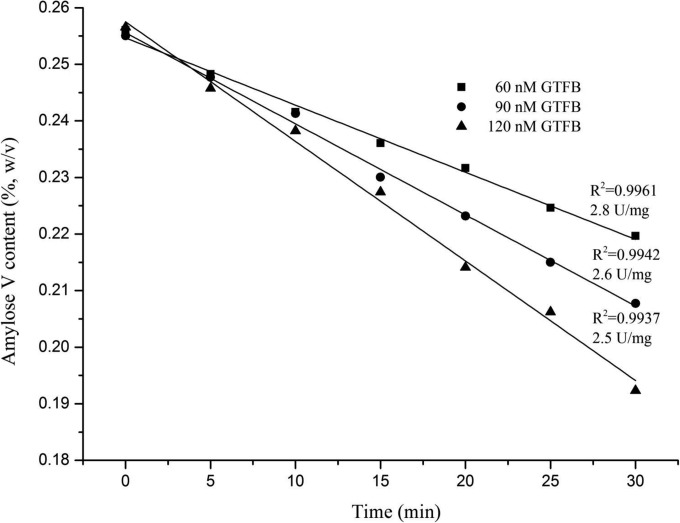
The total activity of 4,6-α-GTases, which includes hydrolysis (H) and transferase (T) activity, was estimated with amylose as the substrate using the iodine staining assay. This assay is based on the fact that iodine stains amylose but not linear (1→6)-α-d-glucan, as can be predicted by the theory of steric hindrance; the corresponding molecular simulations are shown in Fig. 4. The diameter of the iodine atom is 4.2 Å, as measured by Raman spectroscopy (33). The linear (1→6)-α-d-glucan model is based on the results of Genin et al. (34), who showed that five glucose units per rotation in a right-handed helix is the most stable configuration. The inner diameter of such a helix is around 4.1 Å; therefore, the iodine anions cannot be trapped in the cavities of the (1→6)-α-d-glucan due to the steric hindrance. We experimentally verified that dextran does not stain with iodine using 0.5% (wt/vol) dextran T10 (molecular mass: 9 to 11 kDa; Holbaek, Denmark) instead of 0.5% (wt/vol) amylose V in the iodine staining assay as described in Materials and Methods (data not shown). The iodine anions can be entrapped in amylose because of the larger inner diameter (13 Å) and a shorter pitch (8 Å) of each helix (35). H reduces the iodine binding capacity of the amylose substrate since glucose is released gradually from the nonreducing end (Fig. 4) (15). T also impairs the iodine binding capacity of amylose V because it converts the α1→4-linked chains at the nonreducing end into α1→6-linked chains lacking iodine staining capacity. The iodine binding capacity of the amylose V substrate decreased linearly in time during its reaction with GTFB-ΔN (Fig. 5).
FIG 4.
(A) Schematic diagrams of amylose and α(1→6)-glucan; (B) complexes of iodine with amylose and with the amylose-derived products of the 4,6-α-GTase activities, hydrolysis (H) and transglycosylation (T).
FIG 5.
Total activity of the L. reuteri 121 GTFB-ΔN (60, 90, or 120 nM) enzyme with amylose V (0.25%, wt/vol) was followed in time by taking samples and measuring the formation of the iodine-amylose V complex. Resulting total activities (U/mg) are indicated.
Both H and T thus influence the iodine binding capacity of the amylose substrate. Therefore, the iodine assay is applicable for measuring the total activity of 4,6-α-GTases.
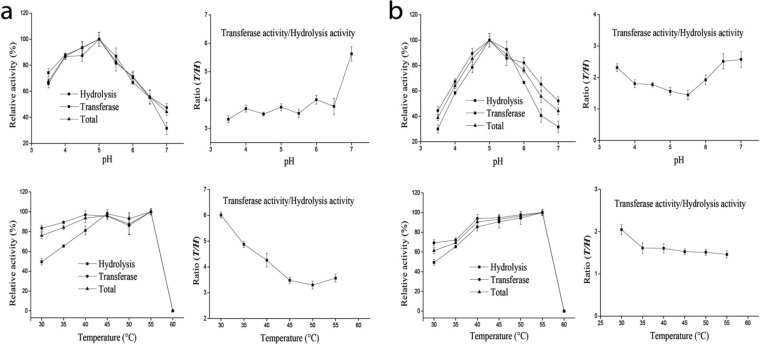
Effects of pH, temperature, and metal ions on activity of GTFB-ΔN and GTFW-ΔN.
The pH optimum for H and T of both GTFB-ΔN and GTFW-ΔN with amylose V was pH 5.0 (Fig. 6). The temperature optimum for H and T of both enzymes at pH 5.0 was 55°C. However, at temperatures of 50 and 55°C, the half-lives of both enzymes are less than 10 min (18, 20). At 40°C, both enzymes were stable for more than 2 h. At lower temperature, the activities of both enzymes remained relatively high (Fig. 6a and b). The transferase activity does not change significantly in the range of 40 to 55°C. Therefore, pH 5.0 and 40°C are the optimal conditions for both enzymes, resulting in T/H ratios of 4.3:1 for GTFB-ΔN and 1.7:1 for GTFW-ΔN.
FIG 6.
The effects of pH and temperature on L. reuteri 121 GTFB-ΔN (a) and L. reuteri DSM 20016 GTFW-ΔN (b) activities, including hydrolysis, transferase, and total activities, using amylose V (0.25%, wt/vol) as the substrate in the presence of 1 mM CaCl2. The experiments were done in triplicate. For pH optimization, values of H, T, and total activities of both GTFB-ΔN (0.6, 2.2, and 2.8 U/mg, respectively) and GTFW-ΔN (1.6, 2.3, and 3.9 U/mg, respectively) determined at pH 5.0 and 40°C were set as 100%. For temperature optimization, values of H, T, and total activities of both GTFB-ΔN (0.6, 2.3, and 2.9 U/mg, respectively) and GTFW-ΔN (1.7, 2.4, and 4.1 U/mg, respectively) determined at pH 5.0 and 55°C were set as 100%.
Aiming to enhance the T/H ratio with the amylose V substrate, most important for industrial application of these enzymes, further variations in incubation temperature and pH were investigated. When the incubation temperature of GTFB-ΔN was decreased from 55 to 30°C, 82% of T and 48% of H remained, resulting in the highest T/H value of 6.0:1 (Fig. 6a). Such higher values of T/H thus result in lower yields of glucose and short-chain oligosaccharides, formed from glucose as the acceptor substrate. Rather, more long-chain polysaccharides (IMMPs), which are considered soluble dietary fiber, are synthesized.
Using the optimal conditions described above (pH 5.0 and 40°C), the H, T, and total activities of both GTFB-ΔN (0.6, 2.2, and 2.8 U/mg, respectively) and GTFW-ΔN (1.6, 2.3, and 3.9 U/mg, respectively) were determined. Compared to GTFB-ΔN, GTFW-ΔN has relatively high H activity, which is in agreement with the NMR results showing more products with reducing ends formed by GTFW-ΔN (Fig. 2).
Both 4,6-α-GTases and glucansucrases (GSes) belong to GH70 and share high protein sequence similarity. GSes are strongly ion dependent and need Ca2+ to stimulate their transferase activity; for example, in the presence of Ca2+ ions, transferase activity of reuteransucrase GTFA of L. reuteri 121 was enhanced 8-fold (24). In the present study, different metal ions had various effects on the hydrolysis and transferase activities of 4,6-α-GTase. As shown in Table 1, Cu2+, Fe2+, and Fe3+ ions strongly inhibited both activities of GTFB-ΔN and GTFW-ΔN. In the presence of Cu2+ ions, no transferase activities were detectable for both enzymes. K+, Na+, and Mg2+ ions had virtually no effect on these activities, especially not on transferase activity, whereas Ca2+ and Mn2+ ions slightly activated both activities of GTFB-ΔN and GTFW-ΔN.
TABLE 1.
Effects of various compounds on GTFB-ΔN and GTFW-ΔN activitya
| Compound (1 mM) | GTFB-ΔN |
GTFW-ΔN |
||
|---|---|---|---|---|
| Hydrolysis activity (%) | Transferase activity (%) | Hydrolysis activity (%) | Transferase activity (%) | |
| None | 100 ± 4 | 405 ± 35 | 100 ± 8 | 180 ± 16 |
| EDTA | 75 ± 5 | 388 ± 51 | 90 ± 7 | 143 ± 14 |
| NaCl | 118 ± 12 | 404 ± 22 | 108 ± 11 | 171 ± 13 |
| CaCl2 | 111 ± 19 | 422 ± 65 | 121 ± 23 | 206 ± 33 |
| MnCl2 | 125 ± 15 | 425 ± 71 | 137 ± 16 | 223 ± 23 |
| MgCl2 | 94 ± 12 | 378 ± 5 | 108 ± 12 | 198 ± 18 |
| KCl | 86 ± 10 | 435 ± 6 | 109 ± 11 | 183 ± 10 |
| CuCl2 | 51 ± 5 | ND | 35 ± 7 | ND |
| FeCl2 | 50 ± 6 | 175 ± 42 | 64 ± 4 | 74 ± 15 |
| FeCl3 | 35 ± 5 | 85 ± 25 | 46 ± 6 | 32 ± 10 |
Incubations were performed at 40°C in a 25 mM NaAc buffer (pH 5.0) with amylose V (0.25%, wt/vol) as the substrate. The hydrolysis activity without additional compound was set at 100% (0.6 U/mg for GTFB-ΔN and 1.3 U/mg for GTFW-ΔN). ND, not detectable. The experiments were done in duplicate. The hydrolysis and transferase activities presented are means ± standard deviations.
Kinetic analysis and acceptor specificity of GTFB-ΔN.
With amylose V as a substrate, GTFB-ΔN displayed Michaelis-Menten type kinetics for hydrolysis, transferase, and total enzyme activities. The kinetic parameters of GTFB-ΔN were calculated from plots of 1/[V] versus 1/[S], where [V] and [S] represent the reaction velocity and concentration of substrate, respectively. The turnover rate (kcat) of the transferase reaction is about 4 times higher than that of the hydrolysis reaction (Table 2). The kcat/Km value for the transferase reaction is 3-fold higher than that for the hydrolysis reaction, indicating that the transferase activity is predominant, resulting mostly in synthesis of modified amylose (IMMP) and in low glucose release.
TABLE 2.
Kinetic parameters of the L. reuteri 121 GTFB-ΔN enzyme with the amylose V substratea
| Activity | Km (g · liter−1) | kcatb (s−1) | kcat/Km (g−1 · s−1 · liter) |
|---|---|---|---|
| Hydrolytic | 0.50 (±0.04) | 9.06 ± 0.18 | 18.5 (±1.8) |
| Transferase | 0.69 (±0.07) | 36.22 ± 1.14 | 53.0 (±7.2) |
| Total | 0.64 (±0.02) | 45.04 ± 1.14 | 70.6 (±4.0) |
A fixed amount of GTFB-ΔN enzyme (60 nM) was used with various concentrations of amylose V (0.025 to 0.4%, wt/vol) at pH 5.0 and 40°C. The values of the kinetic parameters presented are means ± standard deviations.
A molecular mass of 99 kDa (GTFB-ΔN) was used in the calculation of kcat.
To study the acceptor substrate specificity of GTFB-ΔN, the total activities with amylose V in the presence/absence of different acceptor substrates were determined (Table 3). The transglycosylation factor (TF), defined as the ratio of amylose degradation in the presence/absence of small acceptor substrates, was measured. TF reflects the suitability of a certain acceptor substrate for its enzyme (36–38). Addition of these small acceptor substrates increased the reaction rates of GTFB-ΔN with amylose V in all cases. The TFs for α1→4-linked maltose and glucose as acceptor substrates were higher than 1 and similar. The TFs for the α1→6-linked acceptor substrates isomaltose and isomaltotriose clearly were much higher, indicating that isomaltose and isomaltotriose are strong acceptor substrates for the GTFB-ΔN enzyme (Table 3).
TABLE 3.
Amylose V (0.25%, wt/vol) degradation activity (total activity, measured using iodine staining assay) and transglycosylation factors of the L. reuteri 121 GTFB-ΔN enzyme in the presence of different acceptor substratesa
| Acceptor substrate (10 mM) | Amylose degradation (U/mg) | Transglycosylation factorb |
|---|---|---|
| None | 2.8 | 1.0 |
| Glucose | 6.4 | 2.2 |
| Maltose | 7.3 | 2.6 |
| Maltotriose | 7.5 | 2.7 |
| Isomaltose | 13.5 | 4.7 |
| Isomaltotriose | 19.0 | 6.7 |
Experiments were carried out in duplicate, and average data are shown.
The transglycosylation factor is defined as the ratio of amylose degradation rates in the presence/absence of an acceptor substrate.
GTFB is functional at higher substrate concentrations.
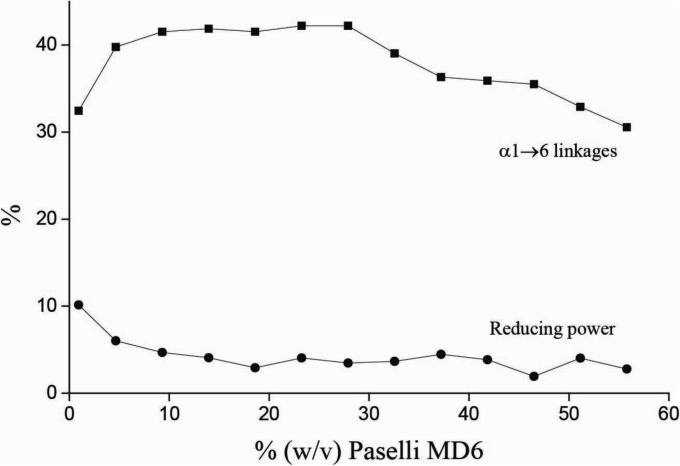
To explore the effects of substrate concentration on the functionality of GTFB, the enzyme was incubated at various Paselli MD6 maltodextrin (partial hydrolysis product of potato starch with a dextrose equivalent between 5 and 7 and 2.9% α1→6 glycosidic linkages) concentrations, keeping the substrate-to-enzyme ratio at 1,000:1 on a mass basis. The results show that GTFB is fully functional at high substrate concentrations. GTFB introduced α1→6 glycosidic linkages at all substrate concentrations, though most efficiently at concentrations between 5 and 30% Paselli MD6, with a maximum of 42.2% α1→6 glycosidic linkages at a 30% concentration (Fig. 7). Moreover, at high substrate concentrations the enzyme has basically no hydrolytic activity, as above 15% (wt/vol) substrate there is no increase in reducing power compared to that for the maltodextrin substrate (3.9% reducing power) used.
FIG 7.
The Paselli MD6 substrate (0.93% to 55.8%, wt/vol) was incubated with Lactobacillus reuteri 121 GTFB-ΔN enzyme (1 to 60 μg/ml). The reducing power of the samples and the percentages of α1→6 glycosidic linkages in the products were determined by 1H NMR. Reactions were performed for 72 h, and reaction mixtures were incubated at 37°C and pH 5.0.
DISCUSSION
The amino acid sequences of 4,6-α-GTases are highly similar to those of glucansucrases, and the 4,6-α-GTases constitute a subfamily of GH70. Most of the glucansucrases characterized from lactobacilli have relatively large N-terminal variable regions, possibly involved in cell wall binding, but a clear functional role has not been established (28). Previous research showed that truncation of the N-terminal variable regions of some glucansucrases had no effect on the size and linkage type distribution of the products formed but resulted in a much higher expression level in E. coli (24–26, 39). The purified truncated GTF180-ΔN and GTFA-ΔN proteins also yielded crystals suitable for X-ray diffraction (40–42). The subsequent elucidation of high-resolution three-dimensional (3D) structures of truncated glucansucrase proteins revealed that their peptide chains follow a “U” path with multiple domains (e.g., glucansucrase GTF180) (40, 42, 43). Multiple-sequence alignment of 4,6-α-GTases with glucansucrases showed that they also have a large N-terminal variable domain (data not shown). In GTFB, this N-terminal domain is formed by residues 1 to 733. Notably, GTFB differs from glucansucrases in lacking the C-terminal polypeptide segment of domain V. When a strategy similar to that used with the glucansucrase was applied, the N-terminal truncation of GTFB (GTFB-ΔN) resulted in a significantly enhanced expression level in E. coli compared to the full-length wild-type GTFB without affecting the activity of the enzyme and products formed.
To biochemically characterize this novel group of 4,6-α-GTase enzymes, several assays have been used, such as high-pH anion-exchange chromatography (HPAEC) and quantitative thin-layer chromatography (TLC) measurement of the saccharides formed from maltoheptaose or the glucose released from maltose as the substrate (16, 18). Both assays have limitations; e.g., they do not allow determination of the specific hydrolysis and transferase activities of these 4,6-α-GTases or a kinetic analysis. Dextran dextrinases (DDases) from Gluconobacter oxydans also catalyze the cleavage of an α1→4-linked glucosyl unit from the nonreducing end of a donor substrate and its subsequent transfer to an acceptor substrate forming an α1→6 linkage (44). The amino acid sequences of these DDase enzymes have not been annotated yet, and further comparison with 4,6-α-GTases is not possible. The dinitrosalicylic acid assay based on changes in reducing power and a viscosity buildup assay based on the changes of rheological properties from maltodextrin to dextran have been used for estimating the activity of DDases (45). However, the poor sensitivity of dinitrosalicylic acid and interference by unreacted maltodextrins and the complex non-Newtonian and time-dependent flow behavior of the maltodextrin-dextran mixture in the reaction mixture severely limit the use of this method. Later, a more reliable assay based on discrete transglycosylation reactions and using maltose as a standard substrate was successfully applied (44). The generated panose was demonstrated to be a valid indicator for transglycosylation activity. A similar assay using p-nitrophenyl-α-d-glucopyranoside (NPG) instead of maltose also provided a reliable estimate of DDase transglycosylation activity. However, these assays are also not applicable for 4,6-α-GTases because of their low or undetectable activity with maltose or NPG. Thus, proper assays for 4,6-α-GTase activities remained to be established.
Starch-converting enzyme activities can be determined with various methods by following amylose degradation/modification (31, 46). Also, 4-α-glucanotransferase enzymes that catalyze both intermolecular (disproportionation) and intramolecular (cyclization) reactions can be studied in this way (36, 47). As shown in our study, amylose also is an efficient substrate for the 4,6-α-GTase-catalyzed transferase and hydrolysis reactions. The data show that the iodine-amylose staining assay is a suitable method to estimate the total activity of 4,6-α-GTases, whereas the GOPOD assay serves to determine hydrolysis (H) activity. The data from both assays can be combined to calculate transferase (T) activity. T/H ratios of 4.3:1 for GTFB-ΔN and 1.7:1 for GTFW-ΔN were obtained under the optimal conditions for both enzymes, indicating that GTFB is less hydrolytic than the GTFW enzyme. Under suboptimal temperature conditions the T/H ratio for GTFB even reached 6.0:1. The data clearly show that GTFB is a proper transglycosylase, with relatively minor hydrolysis activity. T/H ratios have been reported for other transglycosylase enzymes. For glucansucrase GTF180-ΔN from L. reuteri 180 and glucansucrase GTFA-ΔN from L. reuteri 121, these ratios are 2.4:1 and 0.6:1, respectively (41, 48); for cyclodextrin glucanotransferase of Thermoanaerobacterium thermosulfurigenes this ratio is 8.1:1 (49).
Determination of the total activity of GTFB with amylose V in the presence/absence of acceptor substrates clearly showed that isomaltodextrins are preferred over maltodextrins as acceptor substrates. Such information strongly contributes to our understanding of the transglycosylation reaction catalyzed by 4,6-α-GTase enzymes, especially in combination with detailed 3D structural information allowing for identification and characterization of acceptor binding sites. To this end, we are currently aiming to crystallize constructs of GTFB and its homologues (T. Pijning, unpublished data).
To conclude, N-terminal truncation of the L. reuteri GTFB enzyme (GTFB-ΔN) remarkably enhanced the soluble expression level while retaining full activity. Using amylose V as the substrate, the GOPOD assay and the amylose-iodine staining method allowed the characterization of the new 4,6-α-GTase enzymes GTFB and GTFW, including their optimal reaction conditions and hydrolysis and transferase activities. The data show that GTFB is less hydrolytic than GTFW, which is in agreement with the NMR results. Using these assays, various properties of GTFB, the best-characterized 4,6-α-GTase enzyme, were determined, including its kinetic parameters and acceptor substrate specificity. The GTFB enzyme remained active at high substrate concentrations, resulting in high product conversions, a promising feature for industrial applications. Moreover, at high substrate concentrations the hydrolytic activity of GTFB is virtually absent. The developed assays thus provide a firm basis for the future characterization of other natural and engineered 4,6-α-GTase enzymes.
Supplementary Material
ACKNOWLEDGMENTS
The study was financially supported by the Chinese Scholarship Council (to Y.B.), by the University of Groningen, and by the TKI Agri&Food program as coordinated by the Carbohydrate Competence Center (CCC-ABC).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01860-15.
REFERENCES
- 1.Singh N, Singh J, Kaur L, Sodhi NS, Gill BS. 2003. Morphological, thermal and rheological properties of starches from different botanical sources. Food Chem 81:219–231. doi: 10.1016/S0308-8146(02)00416-8. [DOI] [Google Scholar]
- 2.Tester RF, Karkalas J, Qi X. 2004. Starch—composition, fine structure and architecture. J Cereal Sci 39:151–165. doi: 10.1016/j.jcs.2003.12.001. [DOI] [Google Scholar]
- 3.Singh J, Kaur L, McCarthy OJ. 2007. Factors influencing the physico-chemical, morphological, thermal and rheological properties of some chemically modified starches for food applications—a review. Food Hydrocolloid 21:1–22. doi: 10.1016/j.foodhyd.2006.02.006. [DOI] [Google Scholar]
- 4.Guzman-Maldonado H, Paredes-Lopez O, Biliaderis CG. 1995. Amylolytic enzymes and products derived from starch—a review. Crit Rev Food Sci 35:373–403. doi: 10.1080/10408399509527706. [DOI] [PubMed] [Google Scholar]
- 5.van der Maarel MJEC, Leemhuis H. 2013. Starch modification with microbial α-glucanotransferase enzymes. Carbohydr Polym 93:116–121. doi: 10.1016/j.carbpol.2012.01.065. [DOI] [PubMed] [Google Scholar]
- 6.van der Maarel MJEC, van der Veen B, Uitdehaag JCM, Leemhuis H, Dijkhuizen L. 2002. Properties and applications of starch-converting enzymes of the α-amylase family. J Biotechnol 94:137–155. doi: 10.1016/S0168-1656(01)00407-2. [DOI] [PubMed] [Google Scholar]
- 7.Ludwig DDS. 2002. The glycemic index—physiological mechanisms relating to obesity, diabetes, and cardiovascular disease. JAMA 287:2414–2423. doi: 10.1001/jama.287.18.2414. [DOI] [PubMed] [Google Scholar]
- 8.Lattimer JM, Haub MD. 2010. Effects of dietary fiber and its components on metabolic health. Nutrients 2:1266–1289. doi: 10.3390/nu2121266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guraya HS, James C, Champagne ET. 2001. Effect of enzyme concentration and storage temperature on the formation of slowly digestible starch from cooked debranched rice starch. Starke 53:131–139. doi:. [DOI] [Google Scholar]
- 10.Shin SI, Kim HJ, Ha HJ, Lee SH, Moon TW. 2005. Effect of hydrothermal treatment on formation and structural characteristics of slowly digestible non-pasted granular sweet potato starch. Starke 57:421–430. doi: 10.1002/star.200400377. [DOI] [Google Scholar]
- 11.Lee BH, Nichols BL, Hamaker BR. 2013. Enzyme-synthesized highly branched maltodextrins have slow glucogenesis at the mucosal α-glucosidase level and are slowly digestible in vivo. PLoS One 2013:e59745. doi: 10.1371/journal.pone.0059745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kittisuban P, Lee BH, Suphantharika M, Hamaker BR. 2014. Slow glucose release property of enzyme-synthesized highly branched maltodextrins differs among starch sources. Carbohyd Polym 107:182–191. doi: 10.1016/j.carbpol.2014.02.033. [DOI] [PubMed] [Google Scholar]
- 13.Le QT, Lee CK, Kim YW, Lee SJ, Zhang R, Withers SG, Kim YR, Auh JH, Park KH. 2009. Amylolytically-resistant tapioca starch modified by combined treatment of branching enzyme and maltogenic amylase. Carbohyd Polym 75:9–14. doi: 10.1016/j.carbpol.2008.06.001. [DOI] [Google Scholar]
- 14.Lee CK, Le QT, Kim YH, Shim JH, Lee SJ, Park JH, Lee KP, Song SH, Auh JH, Lee SJ, Park KH. 2008. Enzymatic synthesis and properties of highly branched rice starch amylose and amylopectin cluster. J Agric Food Chem 56:126–131. doi: 10.1021/jf072508s. [DOI] [PubMed] [Google Scholar]
- 15.Dobruchowska JM, Gerwig GJ, Kralj S, Grijpstra P, Leemhuis H, Dijkhuizen L, Kamerling JP. 2012. Structural characterization of linear isomalto-/malto-oligomer products synthesized by the novel GTFB 4,6-α-glucanotransferase enzyme from Lactobacillus reuteri 121. Glycobiology 22:517–528. doi: 10.1093/glycob/cwr167. [DOI] [PubMed] [Google Scholar]
- 16.Kralj S, Grijpstra P, van Leeuwen SS, Leemhuis H, Dobruchowska JM, van der Kaaij RM, Malik A, Oetari A, Kamerling JP, Dijkhuizen L. 2011. 4,6-α-Glucanotransferase, a novel enzyme that structurally and functionally provides an evolutionary link between glycoside hydrolase enzyme families 13 and 70. Appl Environ Microbiol 77:8154–8163. doi: 10.1128/AEM.05735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leemhuis H, Dobruchowska JM, Ebbelaar M, Faber F, Buwalda PL, van der Maarel MJ, Kamerling JP, Dijkhuizen L. 2014. Isomalto/malto-polysaccharide, a novel soluble dietary fiber made via enzymatic conversion of starch. J Agric Food Chem 62:12034–12044. doi: 10.1021/jf503970a. [DOI] [PubMed] [Google Scholar]
- 18.Leemhuis H, Dijkman WP, Dobruchowska JM, Pijning T, Grijpstra P, Kralj S, Kamerling JP, Dijkhuizen L. 2013. 4,6-α-Glucanotransferase activity occurs more widespread in Lactobacillus strains and constitutes a separate GH70 subfamily. Appl Microbiol Biotechnol 97:181–193. doi: 10.1007/s00253-012-3943-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dijkhuizen L, van der Maarel MJEC, Kamerling JP, Leemhuis H, Kralj S, Dobruchowska JM. November 2010. Gluco-oligosaccharides comprising (α1→4) and (α1→6) glycosidic bonds, use thereof, and methods for providing them. Patent WO/2010/128859. [Google Scholar]
- 20.Bai Y, van der Kaaij RM, Woortman AJJ, Jin Z, Dijkhuizen L. 2015. Characterization of the 4,6-α-glucanotransferase GTFB enzyme of Lactobacillus reuteri 121 isolated from inclusion bodies. BMC Biotechnol 15:49. doi: 10.1186/s12896-015-0163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palomo M, Pijning T, Booiman T, Dobruchowska JM, van der Vlist J, Kralj S, Planas A, Loos K, Kamerling JP, Dijkstra BW, van der Maarel MJEC, Dijkhuizen L, Leemhuis H. 2011. Thermus thermophilus glycoside hydrolase family 57 branching enzyme crystal structure, mechanism of action, and products formed. J Biol Chem 286:3520–3530. doi: 10.1074/jbc.M110.179515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cochran B, Lunday D, Miskevich F. 2008. Kinetic analysis of amylase using quantitative Benedict's and iodine starch reagents. J Chem Educ 85:401–403. doi: 10.1021/ed085p401. [DOI] [Google Scholar]
- 23.Green F, Clausen CA, Highley TL. 1989. Adaptation of the Nelson-Somogyi reducing-sugar assay to a microassay using microtiter plates. Anal Biochem 182:197–199. doi: 10.1016/0003-2697(89)90578-2. [DOI] [PubMed] [Google Scholar]
- 24.Kralj S, van Geel-Schutten GH, van der Maarel MJEC, Dijkhuizen L. 2004. Biochemical and molecular characterization of Lactobacillus reuteri 121 reuteransucrase. Microbiology 150:2099–2112. doi: 10.1099/mic.0.27105-0. [DOI] [PubMed] [Google Scholar]
- 25.Monchois V, Arguello-Morales M, Russell RRB. 1999. Isolation of an active catalytic core of Streptococcus downei MFe28 GTF-I glucosyltransferase. J Bacteriol 181:2290–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swistowska AM, Wittrock S, Collisi W, Hofer B. 2008. Heterologous hyper-expression of a glucansucrase-type glycosyltransferase gene. Appl Microbiol Biotechnol 79:255–261. doi: 10.1007/s00253-008-1435-0. [DOI] [PubMed] [Google Scholar]
- 27.Leemhuis H, Pijning T, Dobruchowska JM, van Leeuwen SS, Kralj S, Dijkstra BW, Dijkhuizen L. 2013. Glucansucrases: three-dimensional structures, reactions, mechanism, α-glucan analysis and their implications in biotechnology and food applications. J Biotechnol 163:250–272. doi: 10.1016/j.jbiotec.2012.06.037. [DOI] [PubMed] [Google Scholar]
- 28.Kralj S, van Geel-Schutten GH, Dondorff MMG, Kirsanovs S, van der Maarel MJEC, Dijkhuizen L. 2004. Glucan synthesis in the genus Lactobacillus: isolation and characterization of glucansucrase genes, enzymes and glucan products from six different strains. Microbiology 150:3681–3690. doi: 10.1099/mic.0.27321-0. [DOI] [PubMed] [Google Scholar]
- 29.van Leeuwen SS, Leeflang BR, Gerwig GJ, Kamerling JP. 2008. Development of a 1H NMR structural-reporter-group concept for the primary structural characterisation of α-D-glucans. Carbohyd Res 343:1114–1119. doi: 10.1016/j.carres.2008.01.043. [DOI] [PubMed] [Google Scholar]
- 30.Aref HL, Mosbah H, Louati H, Said K, Selmi B. 2011. Partial characterization of a novel amylase activity isolated from Tunisian Ficus carica latex. Pharm Biol 49:1158–1166. doi: 10.3109/13880209.2011.575791. [DOI] [PubMed] [Google Scholar]
- 31.Xiao Z, Storms R, Tsang A. 2006. A quantitative starch-iodine method for measuring α-amylase and glucoamylase activities. Anal Biochem 351:146–148. doi: 10.1016/j.ab.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 32.Kelly RM, Leemhuis H, Dijkhuizen L. 2007. Conversion of a cyclodextrin glucanotransferase into an α-amylase: assessment of directed evolution strategies. Biochemistry 46:11216–11222. doi: 10.1021/bi701160h. [DOI] [PubMed] [Google Scholar]
- 33.Wang DD, Guo WH, Hu JM, Liu F, Chen LS, Du SW, Tang ZK. 2013. Estimating atomic sizes with Raman spectroscopy. Sci Rep 3:1486. doi: 10.1038/srep01486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Genin AL, Banatskaya MI, Nikitin IV, Skokova IF, Vainshtein EF, Kushnerev MY, Kozlov PV. 1983. Criteria of choice of helix type in crystalline dextrane. Polym Sci 25:1717–1722. [Google Scholar]
- 35.Yu XC, Houtman C, Atalla RH. 1996. The complex of amylose and iodine. Carbohyd Res 292:129–141. [Google Scholar]
- 36.Jung JH, Jung TY, Seo DH, Yoon SM, Choi HC, Park BC, Park CS, Woo EJ. 2011. Structural and functional analysis of substrate recognition by the 250s loop in amylomaltase from Thermus brockianus. Proteins 79:633–644. doi: 10.1002/prot.22911. [DOI] [PubMed] [Google Scholar]
- 37.Tang SY, Yang SJ, Cha HJ, Woo EJ, Park C, Park KH. 2006. Contribution of W229 to the transglycosylation activity of 4-α-glucanotransferase from Pyrococcus furiosus. Biochim Biophys Acta 1764:1633–1638. doi: 10.1016/j.bbapap.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 38.Tachibana Y, Takaha T, Fujiwara S, Takagi M, Imanaka T. 2000. Acceptor specificity of 4-α-glucanotransferase from Pyrococcus kodakaraensis KOD1, and synthesis of cycloamylose. J Biosci Bioeng 90:406–409. doi: 10.1016/S1389-1723(01)80009-8. [DOI] [PubMed] [Google Scholar]
- 39.Moulis C, Arcache A, Escalier PC, Rinaudo M, Monsan P, Remaud-Simeon M, Potocki-Veronese G. 2006. High-level production and purification of a fully active recombinant dextransucrase from Leuconostoc mesenteroides NRRL B-512F. FEMS Microbiol Lett 261:203–210. doi: 10.1111/j.1574-6968.2006.00347.x. [DOI] [PubMed] [Google Scholar]
- 40.Pijning T, Vujicic-Zagar A, Kralj S, Dijkhuizen L, Dijkstra BW. 2012. Structure of the α -1,6/α -1,4-specific glucansucrase GTFA from Lactobacillus reuteri 121. Acta Crystallogr F Struct Biol Commun 68:1448–1454. doi: 10.1107/S1744309112044168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pijning T, Vujicic-Zagar A, Kralj S, Eeuwema W, Dijkhuizen L, Dijkstra BW. 2008. Biochemical and crystallographic characterization of a glucansucrase from Lactobacillus reuteri 180. Biocatal Biotransformation 26:12–17. doi: 10.1080/10242420701789163. [DOI] [Google Scholar]
- 42.Vujičić-Žagar A, Pijning T, Kralj S, Lopez CA, Eeuwema W, Dijkhuizen L, Dijkstra BW. 2010. Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes. Proc Natl Acad Sci U S A 107:21406–21411. doi: 10.1073/pnas.1007531107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito K, Ito S, Shimamura T, Weyand S, Kawarasaki Y, Misaka T, Abe K, Kobayashi T, Cameron AD, Iwata S. 2011. Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans. J Mol Biol 408:177–186. doi: 10.1016/j.jmb.2011.02.028. [DOI] [PubMed] [Google Scholar]
- 44.Naessens M, Cerdobbel A, Soetaert W, Vandamme EJ. 2005. Dextran dextrinase and dextran of Gluconobacter oxydans. J Ind Microbiol Biotechnol 32:323–334. doi: 10.1007/s10295-005-0259-5. [DOI] [PubMed] [Google Scholar]
- 45.Naessens M, Vandamme E. 2001. Transglucosylation and hydrolysis activity of Glucanobacter oxydans dextran dextrinase with several donor and acceptor substrates, p 195–203. In Chiellini E, Gil H, Braunegg G, Buchert J, Gatenholm P, van der Zee M (ed), Biorelated polymers: sustainable polymerscience and technology. Kluwer/Plenum, London, United Kingdom. [Google Scholar]
- 46.Cech J, Friedecky B. 1972. Determination of α amylase by a modification of photometric iodine starch method. Cas Lek Cesk 111:309–311. [PubMed] [Google Scholar]
- 47.Terada Y, Fujii K, Takaha T, Okada S. 1999. Thermus aquaticus ATCC 33923 amylomaltase gene cloning and expression and enzyme characterization: production of cycloamylose. Appl Environ Microbiol 65:910–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kralj S, Stripling E, Sanders P, van Geel-Schutten GH, Dijkhuizen L. 2005. Highly hydrolytic reuteransucrase from probiotic Lactobacillus reuteri strain ATCC 55730. Appl Environ Microbiol 71:3942–3950. doi: 10.1128/AEM.71.7.3942-3950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelly RM, Leemhuis H, Rozeboom HJ, van Oosterwijk N, Dijkstra BW, Dijkhuizen L. 2008. Elimination of competing hydrolysis and coupling side reactions of a cyclodextrin glucanotransferase by directed evolution. Biochem J 413:517–525. doi: 10.1042/BJ20080353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.