Abstract
Although clinical epidemiology lists human enteric viruses to be among the primary causes of acute gastroenteritis in the human population, their circulation in the environment remains poorly investigated. These viruses are excreted by the human population into sewers and may be released into rivers through the effluents of wastewater treatment plants (WWTPs). In order to evaluate the viral diversity and loads in WWTP effluents of the Paris, France, urban area, which includes about 9 million inhabitants (approximately 15% of the French population), the seasonal occurrence of astroviruses and noroviruses in 100 WWTP effluent samples was investigated over 1 year. The coupling of these measurements with a high-throughput sequencing approach allowed the specific estimation of the diversity of human astroviruses (human astrovirus genotype 1 [HAstV-1], HAstV-2, HAstV-5, and HAstV-6), 7 genotypes of noroviruses (NoVs) of genogroup I (NoV GI.1 to NoV GI.6 and NoV GI.8), and 16 genotypes of NoVs of genogroup II (NoV GII.1 to NoV GII.7, NoV GII.9, NoV GII.12 to NoV GII.17, NoV GII.20, and NoV GII.21) in effluent samples. Comparison of the viral diversity in WWTP effluents to the viral diversity found by analysis of clinical data obtained throughout France underlined the consistency between the identified genotypes. However, some genotypes were locally present in effluents and were not found in the analysis of the clinical data. These findings could highlight an underestimation of the diversity of enteric viruses circulating in the human population. Consequently, analysis of WWTP effluents could allow the exploration of viral diversity not only in environmental waters but also in a human population linked to a sewerage network in order to better comprehend viral epidemiology and to forecast seasonal outbreaks.
INTRODUCTION
The epidemiology of viral gastroenteritis is mainly evaluated through the use of clinical data. However, clinical data provide only a partial vision of the gastroenteritis viruses circulating in the human population. Indeed, many viral infections do not require specialized consultations or hospitalizations and so are not directly included in clinical statistics. As a consequence, these data sets offer a limited view of the diversity and occurrence of enteric viruses in the human population.
Human astroviruses (HAstVs) and noroviruses (NoVs) are among the main causes of human acute gastroenteritis worldwide (1, 2). HAstVs and NoVs are nonenveloped viruses and have a positive-sense single-stranded RNA. The genome of HAstV is composed of three open reading frames (ORFs), and ORF2, encoding the capsid protein precursor, allows discrimination of eight genotypes of HAstV, HAstV-1 to HAstV-8 (3). The genome of NoV is also composed of three ORFs, and genotyping based on the genetic similarity of ORF2 and ORF3 allows discrimination of 9 genotypes of NoV genogroup I (NoV GI) and 22 genotypes of NoV genogroup II (NoV GII), which are the most common genogroups associated with human infections.
HAstVs and NoVs are able to infect individuals of all ages and cause a great variety of symptoms, such as vomiting, diarrhea, nausea, abdominal pain, and dehydration (4, 5). A large quantity of viral particles is excreted in the feces of infected people (6, 7), and they are finally circulated through the wastewater network (8, 9). Viruses are generally not eliminated by wastewater treatment plants (WWTPs), and as a consequence, they can be released into rivers at noticeable levels (10–12). Consequently, we assume that the viruses in WWTP effluents could represent the viruses circulating not only in environmental waters but also in the human population.
Recent studies suggested the existence of a relationship between the viruses spread in a sewerage network and the health status of the local population (12). In order to estimate this relationship, seasonal monitoring of the concentration and diversity of HAstV, NoV GI, and NoV GII strains in WWTP effluents from the Paris, France, urban area was performed over 1 year. The viral diversity was assessed by a high-throughput sequencing approach specifically targeting HAstV, NoV GI, and NoV GII. During the same period, the HAstV and NoV genotypes present in fecal clinical samples collected from locations throughout France were determined by the French National Reference Centre for Enteric Viruses. The two data sets were analyzed in order to expand the evaluation of the diversity of the viruses circulating in the sick and healthy virus-infected human population through the evaluation of domestic wastewater. The main goal of this study was to highlight the applicability of monitoring enteric viruses in WWTP effluents by a next-generation sequencing (NGS) strategy in order to appreciate the diversity of viruses circulating in rivers as well as to evaluate it in a consistent human population linked to a sewerage network.
MATERIALS AND METHODS
Sampling campaign.
From May 2013 to May 2014, 4 major WWTPs in the Paris, France, area were sampled in order to determine the concentration and the diversity of HAstV, NoV GI, and NoV II in their effluents. Samples of 10 liters were collected twice a month, and a total of 100 samples were collected: 24 samples in spring, 28 samples in summer, 24 samples in autumn, and 24 samples in winter. Each sample was stored at 4°C for 24 h at the most before concentration of the viral particles. These samples were collected from the WWTPs Seine-Amont (Valenton, France), Seine Centre (Colombes, France), Seine-Aval (Achères, France), and Seine Grésillons (Triel-sur-Seine, France), which treat the wastewater of 263 municipalities with about 9 million inhabitants (approximately 15% of the French population). These WWTPs are designed to eliminate classical pollutants (carbon, nitrogenous compounds, and phosphorus) using primary decantation and biological secondary treatment (details are provided in reference 12). During the same period, 1,395 stool samples were collected from 669 sporadic cases of gastroenteritis and 726 samples were collected from 230 outbreaks of gastroenteritis by the National Reference Centre for Enteric Viruses in Dijon, France, as part of its routine national monitoring.
Concentration of virus from wastewater samples.
Effluent samples were concentrated by three successive filtration/concentration steps as described previously (13). Briefly, 10 liters of sample was filtered using electropositive filters (NanoCeram virus samplers; Argonide, Sanford, FL). The filters were then sonicated at 4°C for 1 h in an elution buffer composed of 1% beef extract (desiccated Bacto beef extract; BD Bioscience, Franklin Lakes, NJ), 0.05 M glycine (Merck, Darmstadt, Germany), 0.1% Tween 80 (Sigma-Aldrich, St. Louis, MO), and 0.1% sodium polyphosphate (Sigma-Aldrich, St. Louis, MO) adjusted to pH 9.5. Then, the viruses were eluted in an inverted flow. The pH was adjusted to 3.5, virus flocculation was allowed to take place with slow magnetic agitation for 1 h, and then the virus suspension was centrifuged at 4,000 × g at 4°C for 2 h. The pellet was resuspended in 1× phosphate-buffered saline (pH 9) and ultracentrifuged on 1 ml of 40% sucrose at 150,000 × g at 4°C for 2 h. Finally, the pellet was resuspended in 1 ml of 40% sucrose. The mean recovery rate by the global detection method in experiments with spiked samples (3 experiments with 10 liters of surface water and 3 experiments with 10 liters of WWTP effluents) ranged from 18 to 42% for an adenovirus type 5 LacZ ΔE1 ΔE3 strain (global control), 31 to 57% for HAstV-1, 40 to 68% for a genotype 4 strain of NoV GI (NoV GI.4), and 42 to 61% for NoV GII.4. Endogenous viral contamination, measured before the samples were spiked, was insignificant compared to the concentration of the spiking solutions and did not affect the estimation of recovery rates.
Extraction of viral nucleic acid from wastewater samples and stool samples.
For the effluent samples, viral nucleic acids from the resuspended pellets were extracted with a MagNA Pure compact extractor and a MagNA Pure compact nucleic acid isolation kit I—large volume (Roche Applied Science, Bâle, Switzerland), which allows the processing of samples up to 1 ml, according to the manufacturer's instructions. The extracted nucleic acids were immediately analyzed, and the remaining material was stored at −80°C.
For the stool samples, viral nucleic acids were extracted from 20% stool suspensions in phosphate-buffered saline using a NucliSENS EasyMAG platform (bioMérieux, Marcy l'Etoile, France), according to the manufacturer's instructions.
Quantification of viral nucleic acid from wastewater samples.
For each effluent sample (n = 100), one-step reverse transcription-quantitative PCR (RT-qPCR) assays for HAstV, NoV GI, and NoV II were performed in a reaction volume of 20 μl containing 5 μl of RNA, primers and probes specific for each virus, and TaqMan fast virus 1-step master mix (Life Technologies, Carlsbad, CA) according to the manufacturer's recommendations. These assays, primers, and probes were described previously (12). Each amplification run included a no-template control and a positive amplification control based on the plasmid used to prepare the standard curves. Results reported for each sample are means for duplicate samples. The raw amplification data were collected with ViiA 7 software (version 1.2.1; Life Technologies, Carlsbad, CA) and then processed with Excel software (Microsoft, Redmond, WA).
Viral concentration controls and analysis validation criteria.
Three controls were included in order to confirm the good recovery rate of the concentration and detection method as described previously (12). A global control consisting of an adenovirus type 5 LacZ ΔE1 ΔE3 strain produced by transfection of the pAD/CMV/V5-GW/LacZ vector (Life Technologies, Carlsbad, CA) in 293A cells was seeded into each water sample at 10,000 genome units before the concentration steps. Bacteriophage MS2, used as an extraction control and as a control for the reverse transcription step, was seeded into each concentrated water sample at 10,000 genome units just before the extraction step. Since the concentration method had a recovery rate below 0.1% for the MS2 phage (data not shown), it was unlikely that the amount of endogenous MS2 phage distorted the assessment of inhibition at the extraction step. A competitive inhibition control for the amplification step, made up of a partial sequence of the human β-actin gene cloned into the pCR2.1-TOPO vector (Life Technologies, Carlsbad, CA) at 1,000 genome units, was added after extraction of total nucleic acids. If the recovery rate of the global control was at least equal to the lower limit determined in the experiments with spiked samples (i.e., 18%) and if no inhibition of the extraction and amplification controls was observed, then the results were validated. If inhibition of the extraction and/or amplification controls (i.e., a shift in the cycle threshold [CT] value of greater than 2 cycles between the water sample and the blank sample) was observed, then a dilution of the extracted nucleic acids was tested to overcome the inhibition. If the recovery rate was not validated and if any inhibition of the two other controls was observed, the sample was excluded from analysis.
Detection of viral nucleic acid from stool samples.
Previously described probes specific for HAstV (14), NoV GI (15), and NoV GII (16) were used to detect the viruses by one-step RT-qPCR with the TaqMan fast virus 1-step master mix (Life Technologies, Carlsbad, CA) according to the manufacturer's recommendations.
Genotyping of viral nucleic acids from wastewater and stool samples.
For wastewater samples, sequencing PCR amplifications were performed independently for HAstV, NoV GI, and NoV GII with each sample that was positive by the RT-qPCR. Then, the PCR products from samples collected in each season were pooled so that each pool consisted of four samples, one from each season. Sequencing was performed with these samples of 4 pooled samples.
A step of reverse transcription at 50°C for 30 min using random primers and SuperScript III reverse transcriptase (Life Technologies, Carlsbad, CA) was performed. Then, the first PCR was performed with an initial denaturation at 94°C for 30 s, followed by 30 cycles of denaturation at 94°C for 30 s, primer annealing at 50°C for 30 s, and an extension reaction at 68°C for 1 min, and then a final extension was performed at 68°C for 5 min. The first PCR was performed using Taq Platinum high-fidelity polymerase (Life Technologies, Carlsbad, CA) in a 50-μl reaction volume with 5 μl of cDNA. The set of primers used for the first PCR for the genotyping of NoV GI was COG1F and G1SKR, and that used for the first PCR for the genotyping of NoV GII was COG2F and G2SKR. A second, seminested PCR was then performed under the same conditions used for the first PCR with the following sets of primers: G1SKF and G1SKR for NoV GI and G2SKF and G2SKR for NoV GII. The primers used for NoV genotyping were previously described (17, 18). Primers MON270 and MON269 were used for the genotyping of HAstV (19), and no seminested PCR was performed. Each sample positive by the RT-qPCR was also positive by the genotyping PCR.
Then, a last PCR step was performed with a forward primer containing the forward primer sequence (MON270, G1SKF, or G2SKF), an adaptor sequence, a bar code sequence, and a 10-bp multiplex identifier (MID). The specific MID was selected in order to discriminate each season. The reverse primer was constructed with the reverse primer sequence (MON269, G1SKR, or G2SKR) and an adaptor sequence. In accordance with the manufacturer's recommendations for the preparation of an amplicon library, amplicons were purified, quantified, and normalized to equimolar concentrations for each sample and then pooled for each season. Emulsion PCR (emPCR) amplification and pyrosequencing were performed with a Lib-L kit and with a GS Junior system, respectively, according to the manufacturer's recommendations (Roche Applied Science, Bâle, Switzerland). Finally, raw sequences and quality files were extracted for data processing.
For stool samples, genotyping of HAstV, NoV GI, and NoV GII from positive samples was performed by sequencing a gene fragment, as described previously (20, 21), on a 3130XL DNA genetic analyzer (Life Technologies, Carlsbad, CA).
Processing of sequencing read data.
All analyses of the sequencing data were performed using QIIME (version 1.9.0) software (22). Only sequences with lengths of between 200 and 1,000 bp were retained, as were sequences with an overall quality score above 25, less than 3 ambiguous bases, and no mismatch with the primer sequences. After the selection of high-quality sequences, sequences were clustered into operational taxonomic units (OTUs) with a 100% similarity threshold. For each virus, reference sequences came from the NCBI database and were aligned using MEGA (version 6) software (23). A minimum of two representative reference sequences for each genotype were used (see Table S1 in the supplemental material). All sequence alignments were performed using the ClustalW algorithm. The aligned sequences were truncated in order to target the same genomic regions from which the sequences obtained by high-throughput sequencing were recovered. A Fasta file containing all OTUs was used to query the file of reference sequences using the BLAST algorithm in order to determine the OTU genotype. OTUs were considered unidentified if the identity was below the threshold of 90% and the E value was >0.0001. A phylogenetic tree was created using the neighbor-joining method with a Kimura two-parameter model in MEGA (version 6) software, and branch support was calculated on the basis of 1,000 bootstrap replicates. The nucleotide sequences of the reference NoV GII.4 variants were collected from the NCBI database.
Statistical analysis.
Statistical analyses were performed with the quantification data only (n = 100), using GraphPad Prism (version 6.01) software (GraphPad, La Jolla, CA). Kruskal-Wallis tests with Dunn's multiple-comparison posttest were performed to highlight the seasonality of the densities of HAstV, NoV GI, and NoV GII in samples of WWTP effluents, and P values of <0.05 were considered significant.
SRA accession number.
All sequences have been deposited in NCBI's Sequence Read Archive (SRA) under accession number SRP061719.
RESULTS
Viral loads and prevalence in WWTP effluents.
For each RT-qPCR assay, all no-template controls were negative and all positive amplification controls were positive (data not shown). For each effluent sample, recovery of the global control allowed us to validate the results (recovery rate, >18%). Moreover, the extraction control and the competitive amplification control did not reveal any significant inhibition, in accordance with the inhibition threshold defined in Materials and Methods. The median viral load and prevalence in all effluent samples (n = 100) were 2.69 × 103 copies per liter and 83%, respectively, for HAstV, 3.97 × 103 copies per liter and 97%, respectively, for NoV GI, and 6.80 × 103 copies per liter and 97%, respectively, for NoV GII (Table 1). The highest values for the viral loads of HAstV, NoV GI, and NoV GII were detected in winter, with 1.39 × 106 copies per liter, 2.04 × 105 copies per liter, and 2.92 × 105 copies per liter, respectively. The viral loads of HAstV and NoV GII were significantly higher in winter. However, only the viral loads of NoV GI measured in winter and spring were significantly higher than those measured in summer and autumn.
TABLE 1.
Seasonal viral loads and prevalence of HAstV, NoV GI, and NoV GII in 100 effluent samples from four WWTPs
| Season | No. of samples analyzed | Viral prevalence (%) |
Median (range) viral load (no. of copies/liter) |
||||
|---|---|---|---|---|---|---|---|
| HAstV | NoV GI | NoV GII | HAstV | NoV GI | NoV GII | ||
| Spring | 24 | 83 | 100 | 100 | 6.50 × 103 (0–4.87 × 105) | 1.65 × 104 (4.04 × 102–1.03 × 105) | 1.11 × 104 (1.69 × 101–1.12 × 105) |
| Summer | 28 | 79 | 93 | 93 | 1.66 × 102 (0–1.04 × 104) | 4.58 × 102 (0–4.88 × 103) | 1.49 × 103 (0–1.13 × 104) |
| Autumn | 24 | 75 | 100 | 96 | 2.03 × 103 (0–2.00 × 105) | 3.82 × 103 (8.79 × 101–3.60 × 104) | 4.40 × 103 (0–1.41 × 105) |
| Winter | 24 | 96 | 96 | 100 | 1.09 × 105 (0–1.39 × 106) | 5.09 × 104 (0–2.04 × 105) | 9.58 × 104 (2.49 × 103–2.92 × 105) |
| Total | 100 | 83 | 97 | 97 | 2.69 × 103 (0–1.39 × 106) | 3.97 × 103 (0–2.04 × 105) | 6.80 × 103 (0–2.92 × 105) |
Viral diversity from WWTP effluents.
A high-throughput sequencing approach was set up in order to determine genotype variations according to season. A total of 5,990, 7,469, and 6,299 sequences were generated from the samples that were positive for HAstV, NoV GI, and NoV GII, respectively. The genotypes identified from WWTP effluent samples are shown in Fig. 1 (left). Four genotypes of HAstV were identified: HAstV-1, HAstV-2, HAstV-5, and HAstV-6. Strains of HAstV-1 represented 76% of the strains identified over the year, and HAstV-1 was clearly the principal genotype recovered regardless of the season (66% of the sequences recovered in spring, 67% recovered in summer, 75% recovered in autumn, and 92% recovered in winter belonged to HAstV-1). HAstV-5 was also identified in every season (3% of the sequences recovered in spring, 25% recovered in summer, 15% recovered in autumn, and 4% recovered in winter belonged to HAstV-5), whereas HAstV-2 was detected only in the autumn (3% of strains) and winter (3% of strains). HAstV-6 was detected only in the winter (<1% of strains).
FIG 1.
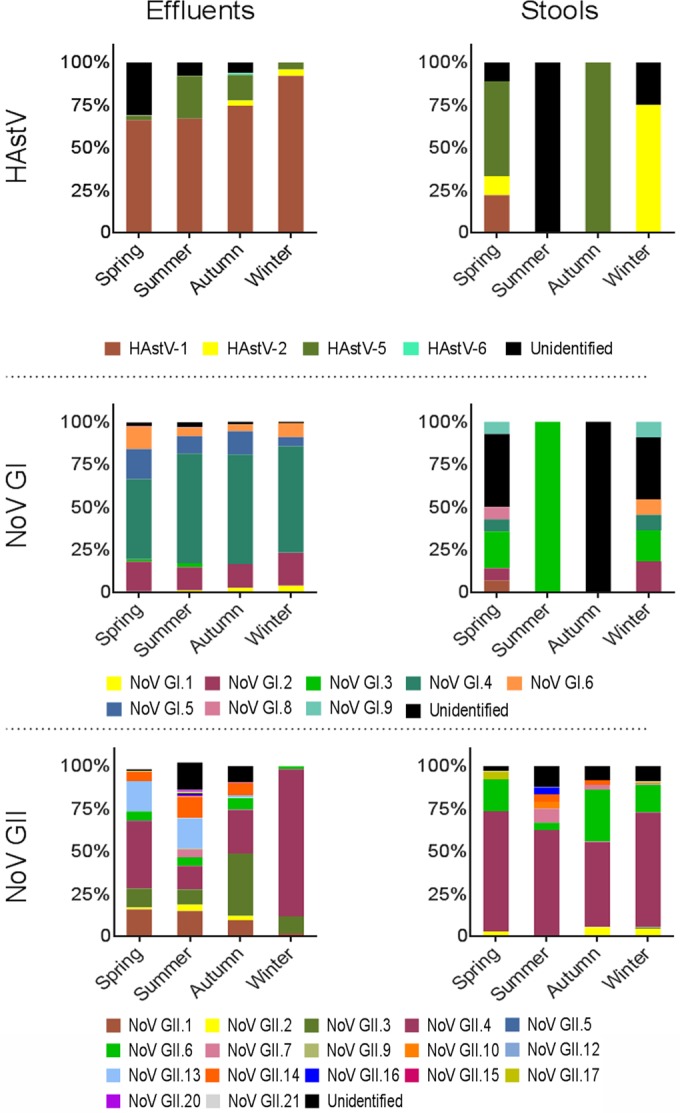
(Left) Genotype diversity of HAstV (n = 5,990), NoV GI (n = 7,469), and NoV GII (n = 6,299) from WWTP effluents according to season between May 2013 and May 2014; (right) genotype diversity of HAstV (n = 15), NoV GI (n = 38), and NoV GII (n = 256) from clinical data collected throughout France according to season between May 2013 and May 2014. Each bar represents the diversity of one type of virus.
Seven human genotypes were present in NoV GI-positive samples, and NoV GI.4 was always the predominant genotype, with detection rates of about 47% in spring and 64% in autumn. The other genotypes were globally detected at rates of 16% for NoV GI.2, 1% for NoV GI.3, 12% for NoV GI.5, and 8% for NoV GI.6. The NoV GI.1 and NoV GI.8 genotypes showed lower rates of occurrence, comprising less than 0.5% of sequences.
For NoV GII, 16 different genotypes (NoV GII.1 to NoV GII.7, NoV GII.9, NoV GII.12 to NoV GII.17, NoV GII.20, and NoV GII.21) of the 19 genotypes known to infect humans were identified. In winter, 7 genotypes could be detected, and among these, the detection rates for NoV GII.1, NoV GII.2, NoV GII.6, NoV GII.13, and NoV GII.14 were below 2% and the detection rate for NoV GII.3 was about 10%. NoV GII.4 strains were present in 86% of samples, and NoV GII.4 was the most prevalent genotype. In autumn, among the 12 identified genotypes, only NoV GII.3 and NoV GII.4 were present at rates above 10%, with their sequences comprising 37% and 26% of the sequences detected, respectively. In spring, the 4 most represented genotypes among the 11 identified genotypes were NoV GII.4 at 40%, NoV GII.13 at 20%, NoV GII.1 at 16%, and NoV GII.3 at 11%. In summer, 14 different genotypes were identified, and the genotype with the highest detection rate was NoV GII.13 (18%).
Viral diversity from stool samples.
During the same period, only 18 strains were detected in the Paris urban area, and these corresponded to 1 HAstV strain and 17 NoV GII strains. In order to have a more relevant vision of the HAstV, NoV GI, and NoV GII strains circulating in the human population and since the population of the study area represented a large part of the French population, the comparison of viral diversity was realized with national data. At the national scale, a total of 309 strains corresponding to 15 HAstV strains, 38 NoV GI strains, and 256 NoV GII strains were identified throughout France (see Table S2 in the supplemental material). The genotypes from each isolated infection and each epidemic event were counted. The highest number of stools positive for HAstV and for NoV GI was observed in spring, with 9 (60%) and 17 (45%), respectively, while 70 (27%) stools were positive for NoV GII. In winter, 4 (27%) HAstV-positive stools, 16 (42%) NoV GI-positive stools, and 119 (46%) NoV GII-positive stools were collected. Both in summer and in autumn, only 1 (about 7%) and 1 (about 7%) stool samples, respectively, were positive for HAstV, 4 (10%) and 1 (about 3%) stool samples, respectively, were positive for NoV GI, and 27 (10%) and 40 (16%) stool samples, respectively, were positive for NoV GII.
The viral diversity found in stool samples is also shown in Fig. 1 (right). In spring, the HAstV-positive samples corresponded to 9 identified gastroenteritis cases related mainly to HAstV-5 (n = 5) and to a lesser extent to HAstV-1 (n = 2) and HAstV-2 (n = 1). In summer and autumn, only one sample in each season was positive for an unidentified genotype and the HAstV-5 genotype, respectively. In winter, three HAstV-5 sequences could be identified among 4 positive cases.
For NoV GI, 45% (17/38) of the reported cases involving 8 different genotypes (NoV GI.1 to NoV GI.4, NoV GI.6, NoV GI.8, NoV GI.9, and NoV GI.f) were identified in spring and 42% (16/38) involving 5 genotypes (NoV GI.2, NoV GI.3, NoV GI.4, NoV GI.6, and NoV GI.9) were identified in winter (see Table S2 in the supplemental material). In opposition, during the summer and autumn periods, this rate was clearly lower, with respective values of 11% (4/38) and <3% (1/38). Among the 4 positive samples obtained in summer, 2 cases of NoV GI.3 and 2 cases of NoV GI.6 could be identified. Moreover, 2 recombinant strains, NoV GI.b/I.6 and NoV GI.b/I.4, were detected in 8 cases and 1 case, respectively (see Table S2 in the supplemental material).
Two hundred fifty-six cases linked to NoV GII were reported between May 2013 and May 2014, and of these, 59% (152/256) were NoV GII.4. The highest rate of detection of epidemic cases, 46% (119/256), was found in winter, and NoV GII.4, which was detected at a rate of 62% (74/119), was the main genotype involved. During this season, 6 genotypes were identified: NoV GII.2 to NoV GII.4, NoV GII.6, NoV GII.7, and NoV GII.17. In the other seasons, NoV GII.4 was still the main genotype identified, and several genotypes were found: 4 (NoV GII.2, NoV GII.4, NoV GII.6, and NoV GII.17) in spring, 6 (NoV GII.4, NoV GII.6, NoV GII.7, NoV GII.10, NoV GII.14, and NoV GII.16) in summer, and 5 (NoV GII.2, NoV GII.4, NoV GII.6, NoV GII.7, and NoV GII.14) in autumn. Additionally, 5 NoV GII recombinants (NoV GII.c/II.1, NoV GII.e/II.2, NoV GII.7/II.6, NoV GII.g/II.12, and NoV GII.21/II.3) were identified (see Table S2 in the supplemental material).
Analysis of NoV GII.4 variants.
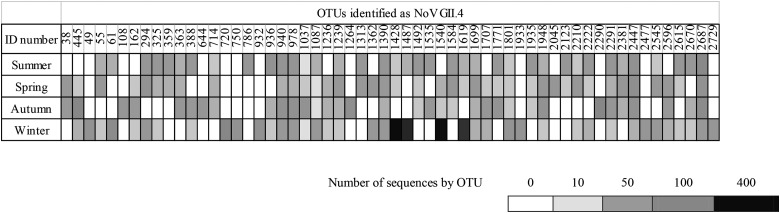
Among the stool samples tested, a total of 152 were positive for NoV GII.4, and these were composed of 4 NoV GII.4 2006.a strains, 3 NoV GII.4 2006b strains, 14 NoV GII.4 2009 strains, and 131 NoV GII.4 2012 strains (see Table S2 in the supplemental material). In the WWTP effluents, the major change in viral diversity was observed in winter, with a significant increase in the number of NoV GII.4 strains, which represented about 86% of the viral diversity for NoV GII, being found (Fig. 1). This change in viral diversity among NoV GII genotypes was primarily linked to an increase in the number of sequences representing 4 specific OTUs which corresponded to GII.4 (Fig. 2). All OTUs identified to be NoV GII.4 were NoV GII.4 2012 variants (data not shown), and consequently, the proportion of sequences representing the 4 specific OTUs greatly increased (see Fig. S1 in the supplemental material).
FIG 2.
Heat map representing the number of OTUs which were identified to be NoV GII.4 in WWTP effluents for each season.
DISCUSSION
Currently, epidemiological studies are essentially based on clinical data, although the majority of infections do not require specialized consultation with health care providers. As a consequence, the prevalence of many circulating enteric viruses is certainly underestimated by these studies. Thus, for the first time, to our knowledge, this study, which used a high-throughput sequencing technology, provides epidemiological data on human viruses detected in the effluents of WWTPs (serving about 9 million inhabitants) over 1 year in order to characterize the seasonal variation in viral loads and the diversity of viruses in WWTP effluents. It also permitted comparison of these environmental data with clinical data obtained by the French National Reference Centre for Enteric Viruses during the same period.
First, the analysis of 100 effluent samples allowed us to highlight the high frequencies of detection and the high loads of HAstV, NoV GI, and NoV GII, which showed an overall consistency with the median viral loads in WWTP effluents observed worldwide, for example, 3.9 × 104 copies per liter for HAstV, 7.1 × 104 copies per liter for NoV GI, and 5.2 × 104 copies per liter for NoV GII in Singapore (9), 2.9 × 103 copies per liter for NoV GI and 2.6 × 103 copies per liter for NoV GII in Japan (8), between 102 and 103 copies per liter for NoV GI and between 102 and 104 copies per liter for NoV GII in New Zealand (24), about 105 copies per liter for NoV GI and NoV GII in Norway (25), about 105 copies per liter for HAstV, NoV GI, and NoV GII in Germany (26), and about 106 copies per liter for NoV GI and NoV GII in France (27). Part of the discrepancy in the detection frequencies could be explained by the differences in the concentration and detection methods and the sample volumes used (10, 11, 28). Our results showed high frequencies of detection and high viral loads in the WWTP effluents, reflecting the high prevalence and high levels of circulation of human HAstV, NoV GI, and NoV GII in the human population. Moreover, viral loads in WWTP effluents greater than 105 copies per liter could be released into the Seine River, which serves as a catchment source for the production of drinking water. When daily water flows (about 1.49 × 106 m3/day in effluents and 3.72 × 107 m3/day in the Seine River) are considered, if all detected genomes were from infectious viruses, it may significantly impair the water resource used by drinking water plants.
Statistical analysis of the viral loads in WWTP effluents showed the significant prevalence of HAstV, NoV GI, and NoV GII in winter and also the significant prevalence of NoV GI in spring. These observations are consistent not only with the data for the clinical samples (where 87% [13/15] of samples were positive for HAstV in winter and spring, 87% [33/38] of samples were positive for NoV GI in winter and spring, and 46% [119/256] of samples were positive for NoV GII in winter) but also with the epidemiological data from different countries with temperate climates (29–33). A seasonal effect in environmental water was also found in Spain (34, 35), China (11), Japan (10), and Ireland (36). Moreover, as in some other studies (12, 24, 37), these results suggest a close link between the health status of the population and the viral loads in WWTP effluents.
A high degree of viral diversity was identified in the WWTP effluent samples, which showed that 4 HAstV genotypes (HAstV-1, HAstV-2, HAstV-5, and HAstV-6), 7 NoV GI genotypes (NoV GI.1 to NoV GI.6 and NoV GI.8), and 16 NoV GII genotypes (NoV GII.1 to NoV GII.7, NoV GII.9, NoV GII.12 to NoV GII.17, NoV GII.20, and NoV GII.21) were indirectly circulating in the resident or working population of the Paris urban area.
The small number of positive stool samples, only 17 for HAstV, NoV GI, and NoV GII in the Paris urban area, highlight the limit of epidemiological data collected by the traditional network. However, a similar global diversity was observed in stools from reported gastroenteritis cases in France, but it is striking to note that the distribution of the genotypes identified was wildly different from that detected in the environmental samples. The predominant HAstV genotype from WWTP effluents was clearly HAstV-1, which was in agreement with the findings of many epidemiological studies (38–40). Previous studies also detected HAstV-2 and/or HAstV-5 in environmental water samples (38, 41); however, to our knowledge, this is the first time that HAstV-6 has been detected in WWTP effluents. In opposition, the predominant genotype from stool samples collected was HAstV-5, following by HAstV-2.
For NoV GI, the most prevalent genotype in WWTP effluents was NoV GI.4, while no NoV GI genotype was clearly prevalent in clinical samples. NoV GI.4 was also reported to be the main genotype of NoV identified in environmental water samples in Spain (34), Japan (10), and South Africa (42). This genotype has often been described to be responsible for foodborne outbreaks (43). Unlike the other two viruses, the predominant NoV GII genotype in WWTP effluents varied according to the season: NoV GII.4 in spring and winter and NoV GII.3 in autumn. Up to 14 different genotypes were found in summer, but no genotype was predominant. In comparison, the majority of stool samples showed NoV GII.4 in all seasons, as in most clinical studies (44, 45). These observations could be explained by the low number of HAstV- or NoV GI-positive stool samples but not NoV GII-positive stool samples collected. They could also reflect, in general, the lower pathogenicity of some circulating strains, which would lead to fewer hospitalizations or medical consultations and to fewer reported cases specifically due to these strains.
Furthermore, all HAstV genotypes from effluent samples except HAstV-6 were also found in the clinical samples; HAstV-6 was rarely found in previous epidemiological studies (46–48). All NoV GI genotypes except NoV GI.5 identified in WWTP effluent samples were also found in clinical samples; NoV GI.5 has frequently been isolated worldwide in both clinical studies (49, 50) and environmental studies (42, 51, 52). The only NoV GII genotype which was found in WWTP effluents and not in stool samples was NoV GII.20, which was also found in previous epidemiological studies (53, 54). The discrepancy between the environmental data and the clinical data is also in agreement with the lack of a systematic analysis of the prevalence of these viruses in feces and the less severe symptoms which are associated with certain genotypes and which thus do not require medical consultation or hospitalization. To our knowledge, no previous study has highlighted the differences in the pathogenicity or virulence of the genotypes of HAstV and NoV. Additional studies will be needed to explain the origin of this discrepancy.
Some genotypes (NoV GI.f and NoV GII.10) were detected only in stool samples. That finding could be explained by the sampling campaign, which was conducted nationwide for stool samples and regionally for WWTP effluents. We were not able to exclude the possibility that some genotypes detected in the environment could present properties conferring higher or lower levels of resistance. This aspect deserves to be studied, because such properties could also confer greater resistance to disinfection treatments.
Despite the weak severity of the infections caused by various classical genotypes, the emergence of new recombinant strains that are more virulent could become a greater public health concern. This high degree of diversity of the viruses circulating in the human population and the potential for coinfection events (due to the ability to infect humans) could promote the emergence of new strains by inter- and intragenotype recombination events and even intergenogroup recombination events (55–58). As such, 2 recombinant strains of Nov GI (NoV GI.b/I.6 and NoV GI.b/I.4) and 5 recombinant strains of NoV GII (NoV GII.c/II.1, NoV GII.e/II.2, NoV GII.7/II.6, NoV GII.g/II.12, and NoV GII.21/II.3) were identified in stool samples. The methodology of this study did not allow evaluation of the rate of occurrence of recombinant strains in WWTP effluents, but it would be interesting to deepen the genotyping analysis, despite the technical limitation of pyrosequencing due to the amplicon length usable by this sequencing technology. However, the methodology allowed the analysis of NoV GII.4 variants in WWTP effluents. All NoV GII.4 strains found were identified to be NoV GII.4 2012 Sydney variants. This observation is consistent with the clinical data, which showed that 86% (131/158) of the NoV GII.4 strains identified in stool samples were NoV GII.4 2012 Sydney variants. It is noteworthy that NoV GII.4 is currently the major genotype contributing to the NoV-related gastroenteritis epidemic. The large number of OTUs in WWTP effluents identified to be NoV GII.4 2012 Sydney variants proved the great variability of the nucleotide sequences coding for the VP1 protein, the major component of the NoV capsid. This variability could contribute to a strategy that allows the virus to escape the adaptive immune response. Thus, identification of the diversity of NoV GII.4 strains in WWTP effluents could be relevant from the perspective of a viral vaccine approach.
Despite the important presence of NoV GII.4, it should be noted that NoV GII.17 was identified in both WWTP effluents and stool samples. In the winter of 2014-2015, one study found that in some Asian countries, NoV GII.17 replaced the previous main NoV GII genotype, NoV GII.4 Sydney 2012 (59). Consequently, it would be interesting to monitor gastroenteritis cases to see if the number of gastroenteritis cases caused by NoV GII.17 strains increases in the years ahead.
Finally, effluent sample analysis could provide deeper information regarding the viral diversity in the population linked to a sewerage network and provide a better comprehension of the viral epidemiology, thereby supplementing the clinical data. Moreover, WWTP effluents are very often released in rivers that can be used for different purposes, such as shellfish farming and catchment to produce drinking water and to irrigate fruits and vegetables (51, 60, 61). This approach would provide a means to identify the circulating viral strains of high or low pathogenicity which could potentially be present in foods.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by Eau de Paris (France), and the Ile-de-France region funded a Ph.D. scholarship associated with this work (DIM R2DS).
We warmly thank the sampling management department of SIAAP (France) for the delivery of all the water samples and the Lariboisiere Hospital (France) for access to its high-throughput sequencing platform.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02076-15.
REFERENCES
- 1.Lopman BA, Hall AJ, Curns AT, Parashar UD. 2011. Increasing rates of gastroenteritis hospital discharges in US adults and the contribution of norovirus, 1996–2007. Clin Infect Dis 52:466–474. doi: 10.1093/cid/ciq163. [DOI] [PubMed] [Google Scholar]
- 2.Jeong AY, Jeong HS, Jo MY, Jung SY, Lee MS, Lee JS, Jee YM, Kim JH, Cheon DS. 2011. Molecular epidemiology and genetic diversity of human astrovirus in South Korea from 2002 to 2007. Clin Microbiol Infect 17:404–408. doi: 10.1111/j.1469-0691.2010.03263.x. [DOI] [PubMed] [Google Scholar]
- 3.Belliot G, Laveran H, Monroe SS. 1997. Detection and genetic differentiation of human astroviruses: phylogenetic grouping varies by coding region. Arch Virol 142:1323–1334. doi: 10.1007/s007050050163. [DOI] [PubMed] [Google Scholar]
- 4.Gallimore CI, Lewis D, Taylor C, Cant A, Gennery A, Gray JJ. 2004. Chronic excretion of a norovirus in a child with cartilage hair hypoplasia (CHH). J Clin Virol 30:196–204. doi: 10.1016/j.jcv.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharya R, Sahoo GC, Nayak MK, Ghosh S, Dutta P, Bhattacharya MK, Mitra U, Gangopadhyay D, Dutta S, Niyogi SK, Saha DR, Naik TN, Bhattacharya SK, Krishnan T. 2006. Molecular epidemiology of human astrovirus infections in Kolkata, India. Infect Genet Evol 6:425–435. doi: 10.1016/j.meegid.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Z, Mitchell DK, Afflerbach C, Jakab F, Walter J, Zhang YJ, Staat MA, Azimi P, Matson DO. 2006. Quantitation of human astrovirus by real-time reverse-transcription-polymerase chain reaction to examine correlation with clinical illness. J Virol Methods 134:190–196. doi: 10.1016/j.jviromet.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, Graham DY. 2008. Norwalk virus shedding after experimental human infection. Emerg Infect Dis 14:1553–1557. doi: 10.3201/eid1410.080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katayama H, Haramoto E, Oguma K, Yamashita H, Tajima A, Nakajima H, Ohgaki S. 2008. One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Res 42:1441–1448. doi: 10.1016/j.watres.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 9.Aw TG, Gin KY. 2010. Environmental surveillance and molecular characterization of human enteric viruses in tropical urban wastewaters. J Appl Microbiol 109:716–730. doi: 10.1111/j.1365-2672.2010.04701.x. [DOI] [PubMed] [Google Scholar]
- 10.Kitajima M, Haramoto E, Phanuwan C, Katayama H, Furumai H. 2012. Molecular detection and genotyping of human noroviruses in influent and effluent water at a wastewater treatment plant in Japan. J Appl Microbiol 112:605–613. doi: 10.1111/j.1365-2672.2012.05231.x. [DOI] [PubMed] [Google Scholar]
- 11.He XQ, Cheng L, Zhang DY, Xie XM, Wang DH, Wang Z. 2011. One-year monthly survey of rotavirus, astrovirus and norovirus in three sewage treatment plants in Beijing, China and associated health risk assessment. Water Sci Technol 63:191–198. doi: 10.2166/wst.2011.032. [DOI] [PubMed] [Google Scholar]
- 12.Prevost B, Lucas FS, Goncalves A, Richard F, Moulin L, Wurtzer S. 2015. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ Int 79:42–50. doi: 10.1016/j.envint.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Wurtzer S, Prevost B, Lucas FS, Moulin L. 2014. Detection of enterovirus in environmental waters: a new optimized method compared to commercial real-time RT-qPCR kits. J Virol Methods 209:47–54. doi: 10.1016/j.jviromet.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Le Cann P, Ranarijaona S, Monpoeho S, Le Guyader F, Ferre V. 2004. Quantification of human astroviruses in sewage using real-time RT-PCR. Res Microbiol 155:11–15. doi: 10.1016/j.resmic.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Lyman WH, Walsh JF, Kotch JB, Weber DJ, Gunn E, Vinjé J. 2009. Prospective study of etiologic agents of acute gastroenteritis outbreaks in child care centers. J Pediatr 154:253–257. doi: 10.1016/j.jpeds.2008.07.057. [DOI] [PubMed] [Google Scholar]
- 16.Loisy F, Atmar RL, Guillon P, Le Cann P, Pommepuy M, Le Guyader FS. 2005. Real-time RT-PCR for norovirus screening in shellfish. J Virol Methods 123:1–7. doi: 10.1016/j.jviromet.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 17.Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, Natori K, Takeda N, Katayama K. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods 100:107–114. doi: 10.1016/S0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- 18.Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Takeda N, Katayama K. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol 41:1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noel JS, Lee TW, Kurtz JB, Glass RI, Monroe SS. 1995. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J Clin Microbiol 33:797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sdiri-Loulizi K, Gharbi-Khelifi H, de Rougemont A, Hassine M, Chouchane S, Sakly N, Pothier P, Guediche MN, Aouni M, Ambert-Balay K. 2009. Molecular epidemiology of human astrovirus and adenovirus serotypes 40/41 strains related to acute diarrhea in Tunisian children. J Med Virol 81:1895–1902. doi: 10.1002/jmv.21586. [DOI] [PubMed] [Google Scholar]
- 21.Sdiri-Loulizi K, Ambert-Balay K, Gharbi-Khelifi H, Sakly N, Hassine M, Chouchane S, Guediche MN, Pothier P, Aouni M. 2009. Molecular epidemiology of norovirus gastroenteritis investigated using samples collected from children in Tunisia during a four-year period: detection of the norovirus variant GGII.4 Hunter as early as January 2003. J Clin Microbiol 47:421–429. doi: 10.1128/JCM.01852-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hewitt J, Leonard M, Greening GE, Lewis GD. 2011. Influence of wastewater treatment process and the population size on human virus profiles in wastewater. Water Res 45:6267–6276. doi: 10.1016/j.watres.2011.09.029. [DOI] [PubMed] [Google Scholar]
- 25.Grøndahl-Rosado R, Yarovitsyna E, Trettenes E, Myrmel M, Robertson L. 2014. A one year study on the concentrations of norovirus and enteric adenoviruses in wastewater and a surface drinking water source in Norway. Food Environ Virol 6:232–245. doi: 10.1007/s12560-014-9161-5. [DOI] [PubMed] [Google Scholar]
- 26.Pusch D, Oh DY, Wolf S, Dumke R, Schröter-Bobsin U, Höhne M, Röske I, Schreier E. 2005. Detection of enteric viruses and bacterial indicators in German environmental waters. Arch Virol 150:929–947. doi: 10.1007/s00705-004-0467-8. [DOI] [PubMed] [Google Scholar]
- 27.da Silva AK, Le Saux J-C, Parnaudeau S, Pommepuy M, Elimelech M, Le Guyader FS. 2007. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl Environ Microbiol 73:7891–7897. doi: 10.1128/AEM.01428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray TY, Mans J, Taylor MB. 2013. Human calicivirus diversity in wastewater in South Africa. J Appl Microbiol 114:1843–1853. doi: 10.1111/jam.12167. [DOI] [PubMed] [Google Scholar]
- 29.Wollants E, De Coster S, Van Ranst M, Maes P. 2015. A decade of norovirus genetic diversity in Belgium. Infect Genet Evol 30:37–44. doi: 10.1016/j.meegid.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Mounts AW, Ando T, Koopmans M, Bresee JS, Noel J, Glass RI. 2000. Cold weather seasonality of gastroenteritis associated with Norwalk-like viruses. J Infect Dis 181(Suppl 2):S284–S287. doi: 10.1086/315586. [DOI] [PubMed] [Google Scholar]
- 31.Liu C, Grillner L, Jonsson K, Linde A, Shen K, Lindell AT, Wirgart BZ, Johansen K. 2006. Identification of viral agents associated with diarrhea in young children during a winter season in Beijing, China. J Clin Virol 35:69–72. doi: 10.1016/j.jcv.2005.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin HC, Kao CL, Chang LY, Hsieh YC, Shao PL, Lee PI, Lu CY, Lee CY, Huang LM. 2008. Astrovirus gastroenteritis in children in Taipei. J Formos Med Assoc 107:295–303. doi: 10.1016/S0929-6646(08)60090-X. [DOI] [PubMed] [Google Scholar]
- 33.Mustafa H, Palombo EA, Bishop RF. 2000. Epidemiology of astrovirus infection in young children hospitalized with acute gastroenteritis in Melbourne, Australia, over a period of four consecutive years, 1995 to 1998. J Clin Microbiol 38:1058–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Sautu U, Sano D, Guix S, Kasimir G, Pinto RM, Bosch A. 2012. Human norovirus occurrence and diversity in the Llobregat River catchment, Spain. Environ Microbiol 14:494–502. doi: 10.1111/j.1462-2920.2011.02642.x. [DOI] [PubMed] [Google Scholar]
- 35.Rusiñol M, Fernandez-Cassi X, Timoneda N, Carratalà A, Abril JF, Silvera C, Figueras MJ, Gelati E, Rodó X, Kay D, Wyn-Jones P, Bofill-Mas S, Girones R. 2015. Evidence of viral dissemination and seasonality in a Mediterranean river catchment: implications for water pollution management. J Environ Manage 159:58–67. doi: 10.1016/j.jenvman.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 36.Flannery J, Keaveney S, Rajko-Nenow P, O'Flaherty V, Doré W. 2012. Concentration of norovirus during wastewater treatment and its impact on oyster contamination. Appl Environ Microbiol 78:3400–3406. doi: 10.1128/AEM.07569-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hellmer M, Paxeus N, Magnius L, Enache L, Arnholm B, Johansson A, Bergstrom T, Norder H. 2014. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl Environ Microbiol 80:6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou N, Lin X, Wang S, Wang H, Li W, Tao Z, Xu A. 2014. Environmental surveillance for human astrovirus in Shandong Province, China in 2013. Sci Rep 4:7539. doi: 10.1038/srep07539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Benedictis P, Schultz-Cherry S, Burnham A, Cattoli G. 2011. Astrovirus infections in humans and animals—molecular biology, genetic diversity, and interspecies transmissions. Infect Genet Evol 11:1529–1544. doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Resque HR, Munford V, Castilho JG, Schmich H, Caruzo TA, Racz ML. 2007. Molecular characterization of astrovirus in stool samples from children in Sao Paulo, Brazil. Mem Inst Oswaldo Cruz 102:969–974. doi: 10.1590/S0074-02762007000800012. [DOI] [PubMed] [Google Scholar]
- 41.Lizasoain A, Tort LFL, García M, Gómez MM, Cristina J, Leite JPG, Miagostovich MP, Victoria M, Colina R. 2015. Environmental assessment of classical human astrovirus in Uruguay. Food Environ Virol 7:142–148. doi: 10.1007/s12560-015-9186-4. [DOI] [PubMed] [Google Scholar]
- 42.Mans J, Netshikweta R, Magwalivha M, Van Zyl WB, Taylor MB. 2013. Diverse norovirus genotypes identified in sewage-polluted river water in South Africa. Epidemiol Infect 141:303–313. doi: 10.1017/S0950268812000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verhoef L, Hewitt J, Barclay L, Ahmed S, Lake R, Hall AJ, Lopman B, Kroneman A, Vennema H. 2015. Norovirus genotype profiles associated with foodborne transmission, 1999-2012. Emerg Infect Dis 21:592. doi: 10.3201/eid2104.141073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan D, Deng L, Wang M, Li X, Ma Y, Liu W. 2015. High prevalence and genetic diversity of noroviruses among children with sporadic acute gastroenteritis in Nanning City, China, 2010-2011. J Med Virol 87:498–503. doi: 10.1002/jmv.24103. [DOI] [PubMed] [Google Scholar]
- 45.Silva PA, Cardoso DD, Schreier E. 2006. Molecular characterization of human astroviruses isolated in Brazil, including the complete sequences of astrovirus genotypes 4 and 5. Arch Virol 151:1405–1417. doi: 10.1007/s00705-005-0704-9. [DOI] [PubMed] [Google Scholar]
- 46.De Grazia S, Platia MA, Rotolo V, Colomba C, Martella V, Giammanco GM. 2011. Surveillance of human astrovirus circulation in Italy 2002-2005: emergence of lineage 2c strains. Clin Microbiol Infect 17:97–101. doi: 10.1111/j.1469-0691.2010.03207.x. [DOI] [PubMed] [Google Scholar]
- 47.Guix S, Caballero S, Villena C, Bartolome R, Latorre C, Rabella N, Simo M, Bosch A, Pinto RM. 2002. Molecular epidemiology of astrovirus infection in Barcelona, Spain. J Clin Microbiol 40:133–139. doi: 10.1128/JCM.40.1.133-139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mendez-Toss M, Griffin DD, Calva J, Contreras JF, Puerto FI, Mota F, Guiscafre H, Cedillo R, Munoz O, Herrera I, Lopez S, Arias CF. 2004. Prevalence and genetic diversity of human astroviruses in Mexican children with symptomatic and asymptomatic infections. J Clin Microbiol 42:151–157. doi: 10.1128/JCM.42.1.151-157.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sang SW, Zhao ZT, Suo JJ, Xing YB, Jia N, Gao Y, Du MM, Xie LJ, Liu BW, Ren SW, Liu YX. 2013. Study on the molecular epidemiological characteristics of norovirus in acute gastroenteritis of Beijing. Zhonghua Liu Xing Bing Xue Za Zhi 34:263–266. (In Chinese.) [PubMed] [Google Scholar]
- 50.Park S, Jung J, Oh S, Jung H, Oh Y, Cho S, Cho S, Cho S, Park H, Jo N, Bae K, Choi S, Kim B, Kim J, Chae Y, Jung H, Cheon D, Kim H. 2012. Characterization of norovirus infections in Seoul, Korea. Microbiol Immunol 56:700–707. doi: 10.1111/j.1348-0421.2012.00494.x. [DOI] [PubMed] [Google Scholar]
- 51.Rajko-Nenow P, Waters A, Keaveney S, Flannery J, Tuite G, Coughlan S, O'Flaherty V, Dore W. 2013. Norovirus genotypes present in oysters and in effluent from a wastewater treatment plant during the seasonal peak of infections in Ireland in 2010. Appl Environ Microbiol 79:2578–2587. doi: 10.1128/AEM.03557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee GC, Kim MJ, Kim JI, Lee CH. 2014. Occurrence and molecular characterization of noroviruses in Korean surface water between 2007 and 2010. J Microbiol Biotechnol 24:556–562. doi: 10.4014/jmb.1311.11089. [DOI] [PubMed] [Google Scholar]
- 53.Yu Y, Yan S, Li B, Pan Y, Wang Y. 2014. Genetic diversity and distribution of human norovirus in China (1999–2011). Biomed Res Int 2014:196169. doi: 10.1155/2014/196169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dove W, Cunliffe NA, Gondwe JS, Broadhead RL, Molyneux ME, Nakagomi O, Hart CA. 2005. Detection and characterization of human caliciviruses in hospitalized children with acute gastroenteritis in Blantyre, Malawi. Biomed Res Int 77:522–527. [DOI] [PubMed] [Google Scholar]
- 55.Nayak MK, Balasubramanian G, Sahoo GC, Bhattacharya R, Vinje J, Kobayashi N, Sarkar MC, Bhattacharya MK, Krishnan T. 2008. Detection of a novel intergenogroup recombinant norovirus from Kolkata, India. Virology 377:117–123. doi: 10.1016/j.virol.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 56.Martella V, Pinto P, Tummolo F, De Grazia S, Giammanco GM, Medici MC, Ganesh B, L'Homme Y, Farkas T, Jakab F, Banyai K. 2014. Analysis of the ORF2 of human astroviruses reveals lineage diversification, recombination and rearrangement and provides the basis for a novel sub-classification system. Arch Virol 159:3185–3196. doi: 10.1007/s00705-014-2153-9. [DOI] [PubMed] [Google Scholar]
- 57.Mans J, Murray TY, Taylor MB. 2014. Novel norovirus recombinants detected in South Africa. Virol J 11:168. doi: 10.1186/1743-422X-11-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verma H, Chitambar SD, Gopalkrishna V. 2010. Astrovirus associated acute gastroenteritis in western India: predominance of dual serotype strains. Infect Genet Evol 10:575–579. doi: 10.1016/j.meegid.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 59.de Graaf M, van Beek J, Vennema H, Podkolzin AT, Hewitt J, Bucardo F, Templeton K, Mans J, Nordgren J, Reuter G. 2015. Emergence of a novel GII.17 norovirus—end of the GII.4 era? Euro Surveill 20(26):pii=21178 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=21178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maunula L, Miettinen IT, von Bonsdorff CH. 2005. Norovirus outbreaks from drinking water. Emerg Infect Dis 11:1716–1721. doi: 10.3201/eid1111.050487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheong S, Lee C, Song SW, Choi WC, Lee CH, Kim SJ. 2009. Enteric viruses in raw vegetables and groundwater used for irrigation in South Korea. Appl Environ Microbiol 75:7745–7751. doi: 10.1128/AEM.01629-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.