ABSTRACT
In hepatitis C virus (HCV)-infected cells, the envelope glycoproteins E1 and E2 assemble as a heterodimer. To investigate potential changes in the oligomerization of virion-associated envelope proteins, we performed SDS-PAGE under reducing conditions but without thermal denaturation. This revealed the presence of SDS-resistant trimers of E1 in the context of cell-cultured HCV (HCVcc) as well as in the context of HCV pseudoparticles (HCVpp). The formation of E1 trimers was found to depend on the coexpression of E2. To further understand the origin of E1 trimer formation, we coexpressed in bacteria the transmembrane (TM) domains of E1 (TME1) and E2 (TME2) fused to reporter proteins and analyzed the fusion proteins by SDS-PAGE and Western blotting. As expected for strongly interacting TM domains, TME1–TME2 heterodimers resistant to SDS were observed. These analyses also revealed homodimers and homotrimers of TME1, indicating that such complexes are stable species. The N-terminal segment of TME1 exhibits a highly conserved GxxxG sequence, a motif that is well documented to be involved in intramembrane protein-protein interactions. Single or double mutations of the glycine residues (Gly354 and Gly358) in this motif markedly decreased or abrogated the formation of TME1 homotrimers in bacteria, as well as homotrimers of E1 in both HCVpp and HCVcc systems. A concomitant loss of infectivity was observed, indicating that the trimeric form of E1 is essential for virus infectivity. Taken together, these results indicate that E1E2 heterodimers form trimers on HCV particles, and they support the hypothesis that E1 could be a fusion protein.
IMPORTANCE HCV glycoproteins E1 and E2 play an essential role in virus entry into liver cells as well as in virion morphogenesis. In infected cells, these two proteins form a complex in which E2 interacts with cellular receptors, whereas the function of E1 remains poorly understood. However, recent structural data suggest that E1 could be the protein responsible for the process of fusion between viral and cellular membranes. Here we investigated the oligomeric state of HCV envelope glycoproteins. We demonstrate that E1 forms functional trimers after virion assembly and that in addition to the requirement for E2, a determinant for this oligomerization is present in a conserved GxxxG motif located within the E1 transmembrane domain. Taken together, these results indicate that a rearrangement of E1E2 heterodimer complexes likely occurs during the assembly of HCV particles to yield a trimeric form of the E1E2 heterodimer. Gaining structural information on this trimer will be helpful for the design of an anti-HCV vaccine.
INTRODUCTION
Hepatitis C virus (HCV) is an enveloped positive-stranded RNA virus that belongs to the genus Hepacivirus in the family Flaviviridae (1). The members of this viral family are classified in three established genera (Flavivirus, Pestivirus, and Hepacivirus) and the new genus Pegivirus (1, 2). The HCV genome encodes a single polyprotein, which is processed by cellular and viral proteases into 10 mature proteins (3). Cleavage of the viral polyprotein by a cellular signal peptidase gives rise to the envelope glycoproteins, E1 and E2, which play a crucial role in HCV entry into host cells (reviewed in reference 4).
The E1 and E2 envelope glycoproteins are two highly glycosylated type I transmembrane (TM) proteins, each with an N-terminal ectodomain of about 160 or 330 amino acids, respectively, and a well-conserved C-terminal TM domain of about 30 amino acids, designated TME1 or TME2, respectively. These hydrophobic domains anchor the envelope proteins to the membrane of the endoplasmic reticulum (ER) and also have a signal peptide-like function (5). Importantly, after signal peptidase cleavage in the ER, there is a dynamic reorientation of the C-terminal segments of these TM domains, leading to a single transmembrane passage topology (6). From a structural point of view, these domains adopt a helical fold with two helical segments connected by a flexible linker (7–9). Moreover, TME1 and TME2 are also involved in E1E2 heterodimerization (7). As a whole, the folding and maturation of individual E1 and E2 glycoproteins, and the formation of E1E2 heterodimers, are slow, interdependent, complex processes that involve the ER chaperone machinery and disulfide bond formation as well as glycosylation (reviewed in references 4 and 10).
Within the E1E2 heterodimer, E2 is currently the better-characterized subunit. Indeed, this glycoprotein is considered the major target of neutralizing antibodies, and it is also the receptor-binding protein, which has been shown to interact with CD81 tetraspanin and scavenger receptor B1 (SRB1), two HCV coreceptors (reviewed in reference 11). Furthermore, the crystal structure of the core of the E2 ectodomain has been determined recently (12, 13). However, contrary to what was suggested previously (14), this protein does not present the expected three-domain organization shared by class II viral fusion proteins but rather shows a globular structure containing many regions with no regular secondary structure (12, 13). These data also indicate that E2 lacks the structural hallmarks of fusion proteins, suggesting that E1 alone or in association with E2 might be responsible for the fusion step (12, 13, 15, 16). However, the structural data concerning the E1 ectodomain are still too limited to support this hypothesis. Indeed, only the crystal structure of the N-terminal region comprising amino acids 1 to 79 has been published recently (17). This partial structure reveals a complex network of covalently linked, intertwined homodimers.
HCV associates with lipoproteins to form lipo-viro-particles (LVP), and this interaction is essential for its infectivity (18–21). In addition to apolipoproteins, LVP also contain viral components, consisting of the RNA genome and core forming the nucleocapsid and the envelope glycoproteins anchored in lipid bilayer patches associated with the nucleocapsid (4, 19). In HCV-infected cells, E1 and E2 interact to form a noncovalent heterodimer, whereas they assemble as large covalent complexes stabilized by disulfide bonds on the viral particle (22). However, the precise organization of functional HCV envelope glycoprotein complexes at the surfaces of LVP remains poorly characterized. Furthermore, during the budding process, the fusion protein is most likely kept in an inactive prefusion state so as to avoid premature fusion with internal membranes during virion release.
Here we investigated the oligomeric state of HCV virion-associated envelope proteins. For this purpose, we analyzed these proteins by SDS-PAGE without thermal denaturation. This revealed the presence of SDS-resistant trimers of E1 on cell culture-derived HCV (HCVcc) as well as on retroviral pseudoparticles harboring HCV envelope glycoproteins (HCVpp), the stability of which depended on a GxxxG motif present in the TM domain of E1. Furthermore, alteration of E1 trimerization by mutation of this motif abrogated HCV infectivity. Our results indicate that a rearrangement of E1E2 complexes occurs during the assembly of HCV particles to yield a trimeric form of the E1E2 heterodimer.
MATERIALS AND METHODS
Cell culture.
293T human embryo kidney cells (HEK293T cells) (ATCC CRL-11268) and Huh-7 human hepatoma cells (23) were grown in Dulbecco's modified essential medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen).
Antibodies.
Anti-HCV monoclonal antibody (MAb) A4 (anti-E1) (24) and MAbs H52 (25) and 3/11 (anti-E2; kindly provided by J. A. McKeating, University of Birmingham, Birmingham, United Kingdom) (25) were produced in vitro by using a MiniPerm apparatus (Heraeus) as recommended by the manufacturer. Human MAb AR3A was kindly provided by M. Law (Scripps Institute, CA). The human anti-E1 monoclonal antibody 1C4 was obtained from Innogenetics, Belgium.
Mutagenesis and production of viruses.
To produce HCVcc, we used the plasmid encoding the JFH1 genome (genotype 2a; GenBank accession number AB237837), kindly provided by T. Wakita (National Institute of Infectious Diseases, Tokyo, Japan) (26) or a modified version. Mutations were introduced in a modified version of the plasmid encoding the full-length JFH1 genome. This virus contains mutations at the C terminus of the core protein leading to amino acid changes F172C and P173S, which have been shown to increase the viral titers (27). Furthermore, the N-terminal E1 sequence encoding residues 196TSSSYMVTNDC has been modified to reconstitute the A4 epitope (SSGLYHVTNDC) as described previously (28). Briefly, HCVcc was produced in Huh-7 cells electroporated with in vitro-transcribed RNA of JFH1. The JFH1-ΔE1E2 plasmid, containing an in-frame deletion in the E1E2 region, and the JFH1-GND replication-defective mutant have been described previously (28, 29). Extracellular and intracellular infectivities were measured in 50% tissue culture infective doses (TCID50) as described previously (30).
HCVpp expressing the green fluorescent protein (GFP) reporter gene were produced in HEK293T cells as described previously (31). Infectivity was determined by flow cytometry analysis of Huh-7 cells 72 h after the addition of HCVpp.
Mutations were constructed by sequential PCR steps as described previously (32), using the Expand High FidelityPLUS PCR system (Roche). The plasmids were verified by sequencing.
HCV core quantification.
The HCV core protein was quantified by a fully automated chemiluminescent microparticle immunoassay according to the manufacturer's instructions (Architect HCVAg test; Abbott, Germany).
Western blotting of HCV envelope glycoproteins.
Viral particles were pelleted by ultracentrifugation and were incubated for 5 min at 37°C or 95°C in Laemmli sample buffer. For the analysis of cell-associated viral envelope glycoproteins, infected cells were lysed in 1× phosphate-buffered saline (PBS) lysis buffer (1% Triton X-100, 20 mM N-ethylmaleimide [NEM], 2 mM EDTA, protease inhibitor cocktail [Roche]). Cell lysates were then precleared by centrifugation at 14,000 × g for 15 min at 4°C. In some experiments, GNA (Galanthus nivalis lectin) pulldown was performed as described previously (22) before Western blotting. Following separation by SDS-PAGE, proteins were transferred to nitrocellulose membranes (Hybond-ECL; GE Healthcare) using a Trans-Blot apparatus (Bio-Rad) and were revealed with specific MAbs. Following incubation with primary antibodies, membranes were incubated with the corresponding peroxidase-conjugated anti-species antibodies. E1 and E2 proteins were revealed by enhanced chemiluminescence (ECL; GE Healthcare) as recommended by the manufacturer.
CD81 pulldown assay.
For the CD81 pulldown assay, glutathione-Sepharose (GS4B) beads were obtained from GE Healthcare Bio-Sciences. Large extracellular loops of recombinant human CD81 in fusion with glutathione S-transferase (GST) were produced as described previously (33). Glutathione-Sepharose beads were first coated with the recombinant CD81 for 2 h at 4°C and washed twice in PBS–1% Triton X-100. They were then further incubated for 2 to 4 h with E1E2-containing lysates and were washed five times in PBS–1% Triton X-100 before the addition of Laemmli loading buffer and Western blot analysis.
Separation of E1E2 complexes on sucrose gradients.
For sucrose gradient analyses, HCV particles were semipurified as described previously (34) and were lysed in TNE (50 mM Tris at pH 7.5, 1 mM EDTA, 100 mM NaCl) containing 1% Triton X-100 and 50 mM dithiothreitol (DTT). Sucrose solutions were made in TNE containing 0.1% Triton X-100 and 5 mM DTT to prepare continuous 5-to-20% sucrose gradients in 13-ml tubes for centrifugation in an SW41 rotor. The virus lysate was laid on top of the gradient. The gradients were spun for 16 h at 210,000 × g in an SW41 rotor (Beckman). Eleven fractions of 1 ml were harvested from the top, and the pellet was resuspended in the gradient buffer. In parallel, a control calibration gradient was run with 250 μg of each of the following proteins: bovine serum albumin (BSA) (66 kDa), β-amylase (200 kDa), and ferritin (440 kDa) (Sigma and GE Healthcare). After similar fractionation, 30 μl of each fraction was analyzed by reducing SDS-PAGE and Coomassie staining to determine the sedimentation pattern of each marker protein.
Immunofluorescence.
Huh-7 cells transfected with HCV RNA were grown on 12-mm glass coverslips. At the indicated time points (Fig. 4 legend), cells were fixed with 3% paraformaldehyde and then permeabilized with 0.1% Triton X-100 in PBS. Both primary- and secondary-antibody incubations were carried out for 30 min at room temperature with PBS containing 10% goat serum. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The coverslips were mounted on slides by using Mowiol 4-88 (Calbiochem)-containing mounting medium. Confocal microscopy was performed with an LSM 780 laser scanning confocal microscope (Zeiss) using a 63× (numerical aperture, 1.4) oil immersion objective. Images were processed using ImageJ and Adobe Photoshop software.
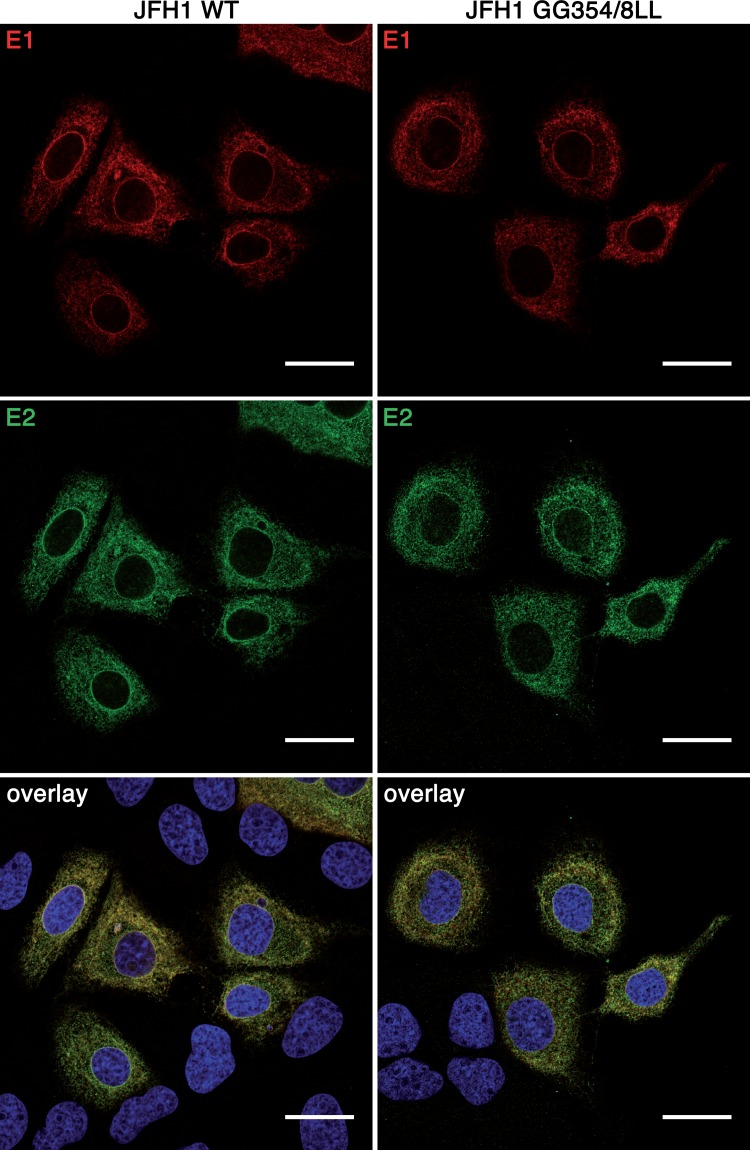
FIG 4.
Mutation of the GxxxG motif in E1 does not affect its subcellular localization. Huh-7 cells were electroporated with HCV RNA transcribed in vitro. At 48 h posttransfection, cells were fixed and were processed for immunofluorescence with MAbs A4 (anti-E1) and AR3A (anti-E2). Bars, 25 μm.
Construction of Trx-TME1 and GST-TME2 fusion protein plasmids.
Genes coding for TME1 and TME2, corresponding, respectively, to amino acids 348 to 383 and 717 to 746 of strain H77 (displayed in Fig. 2), were generated de novo by PCR using optimized codons for expression in Escherichia coli strain Epicurian Coli BL21(DE3)[pLysS] (Stratagene) (35) and were checked by sequencing. Oligonucleotides for PCRs were designed to fuse the TM domains of E1 and E2 in the C-terminal position to the carrier proteins GST and thioredoxin (Trx) as described previously (35) (for details, see below and Table 1). A linker sequence coding for the Asp-Pro dipeptide was inserted between GST or Trx and each TM domain. We have previously reported this insertion to be critical in reducing the toxicity of TME1 and TME2 for their expression in bacteria (35).
FIG 2.
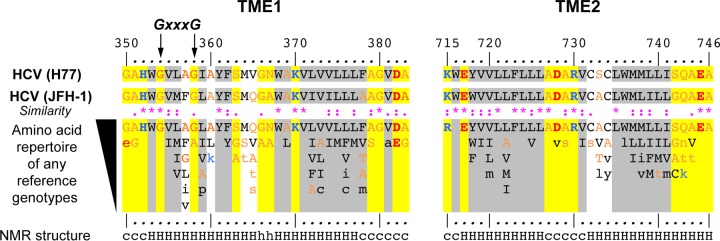
Sequence analyses and NMR structures of the transmembrane domains of HCV E1 and E2. Amino acids of the transmembrane domains of E1 (TME1) (amino acids 350 to 383) and E2 (TME2) (amino acids 715 to 746) are numbered with respect to the HCV polyprotein of the H77 infectious clone (GenBank accession number AF009606) (top). Below the H77 sequence are the sequence of clone JFH-1 (GenBank accession number AB047639) and the amino acid repertoire of the 27 representative TME1 and TME2 sequences from confirmed HCV genotypes and subtypes (listed with accession numbers in Table 1 of reference 74; see the European HCV database for details [https://euhcvdb.ibcp.fr/euHCVdb/] [75]). The degree of amino acid conservation at each position can be inferred from the extent of variability (with the observed amino acids listed in decreasing order of frequency from top to bottom) together with the similarity index according to the CLUSTAL W convention (asterisk, invariant; colon, highly similar; dot, similar) (76). Amino acids observed only once at a given position among the 27 sequences are indicated by lowercase letters. To highlight the variable sequence positions in TME1 and TME2, conserved hydrophilic and hydrophobic positions are highlighted in yellow and gray, respectively. Residues are color-coded according to their hydrophobicity: hydrophobic residues are shown in black (Pro, Cys, Val, Leu, Ile, Met, Phe, Tyr, Trp), polar residues in orange (Gly, Ala, Ser, Thr, Asn, Gln), and positively and negatively charged groups of basic (His, Lys, Arg) and acidic (Glu, Asp) residues in blue and red, respectively. The NMR secondary structure (bottom) shows the conformation of residues determined by nuclear magnetic resonance of the N-terminal part of TME1 in 50% trifluoroethanol (7) (PDB entry 1EMZ) and of recombinant TME1 (9) and TME2 (8) in LPPG [1-palmitoyl-2-hydroxy-sn-glycero-3-phospho-(1′-rac-glycerol)] micelles. Residue conformations are indicated as undetermined (c) or helical (H or h; a lowercase h indicates flexible residues [8]). The glycine residues of the GxxxG motif that were mutated individually (G354L or G358L) or together (G354L G358L) to leucine in the TM domain of E1 are indicated by arrows.
TABLE 1.
Oligonucleotide sequences
| Oligonucleotide | Sequence, 5′ → 3′ |
|---|---|
| baclink | CAGAATTCCTAAGCGTCAACACCAGC |
| EcoTME1 | CAGAATTCCTAAGCGTCAACACCAGC |
| EcoTME2 | CAGAATTCCTAAGCTTCAGCCTGAGAG |
| bacG354L_1 | GTAAGCGATACCAGCCAGAACCAGCCAGTGAGCACCAGCGAT |
| bacG354L_2 | ATCGCTGGTGCTCACTGGCTGGTTCTGGCTGGTATCGCTTAC |
| bacG358L_1 | CAACCATAGAGAAGTAAGCGATCAGAGCCAGAACACCCCAGTG |
| bacG358L_2 | CACTGGGGTGTTCTGGCTCTGATCGCTTACTTCTCTATGGTTG |
| bacG358L+G354L_1 | CAACCATAGAGAAGTAAGCGATCAGAGCCAGAACCAGCCAGTG |
| bacG358L+G354L_2 | CACTGGCTGGTTCTGGCTCTGATCGCTTACTTCTCTATGGTTG |
| bacTrX-TME1_1 | TTCAGTGGCTGTGCATGCAAGGAGATGGCG |
| bacTrX-TME1_2 | TTCAGCCACTGCTAAGCGTCAACACCAGCG |
| eucG354L_1 | TGGTGCTCACTGGCTAGTCCTGGCGGGCATAGCGTATTT |
| eucG354L_2 | CCGCCAGGACTAGCCAGTGAGCACCAGCGATCA |
| eucG358L_1 | GGAGTCCTGGCGCTCATAGCGTATTTCTCCATG |
| eucG358L_2 | AAATACGCTATGAGCGCCAGGACTCCCCAGTG |
| eucG358L+G354L_1 | TGGTGCTCACTGGCTAGTCCTGGCGCTCATAGCGTATTT |
| eucG358L+G354L_2 | GCGCCAGGACTAGCCAGTGAGCACCAGCGATCA |
Thioredoxin-TME1 fusions.
TME1 was fused to the C terminus of Trx encoded by the pET32a(+) plasmid (Novagen). Using PCR, an oligonucleotide hybridizing to the linker sequence (baclink [Table 1]) and an oligonucleotide adding an EcoRI site downstream of the 3′ end of TME1 (EcoTME1 [Table 1]) were used to amplify the linker-TME1 sequence. The resulting PCR fragment was cloned into the MscI and EcoRI restriction sites of pET32a to obtain plasmid Trx-TME1. The G354L and G358L mutations were introduced into Trx-TME1 by site-directed mutagenesis with primer pairs bacG354L_1–bacG354L_2 and bacG358L_1–bacG358L_2, respectively (Table 1). The G358L and G354L double mutation was generated using the pET32Trx-TME1-G354L plasmid as the template and oligonucleotides bacG358L+G354L_1 and bacG358L+G354L_2 (Table 1).
Coexpression of Trx-TME1 and GST-TME2 fusion proteins.
A DNA fragment including the gene coding for Trx-TME1 and its T7 promoter and terminator regions was amplified by PCR using primers bacTrx-TME1_1 and bacTrx-TME1_2 (Table 1). The resulting fragment was first introduced into pCR2.1-Topo (Invitrogen) and then cut out of pCR2.1-Topo by EcoRI and introduced into the unique EcoRI site of pGEXKT-TME2 or pGEXKT-TME2-C731/734A.
Bacterial expression and analysis of fusion proteins.
Trx-TM1 and GST-TM2 chimeras were expressed in BL21-Gold(DE3)[pLysS] (Stratagene) as described in reference 35. Samples were prepared for SDS-PAGE by mixing a 20-μl aliquot of the bacterial cell lysate with an equal volume of lysis buffer (100 mM Tris-Cl [pH 8.0], 1.4 M β-mercaptoethanol, 140 mM SDS, 5 mM EDTA, 8 M urea, and 0.72 mM bromophenol blue). After SDS-PAGE, gels were incubated for 5 min in cold CAPS (N-cyclohexyl-3-aminopropanesulfonic acid) buffer (10 mM CAPS [pH 11.1] [buffered with NaOH], 10% [vol/vol] methanol [MeOH]), and the proteins were blotted onto Immobilon-P membranes with a Bio-Rad Trans-Blot apparatus for 90 min, at 150 V and 250 mA, in cold CAPS buffer. The Immobilon-P membranes were then incubated for 30 min in 20 ml of PBST buffer (90 mM K2HPO4, 10 mM KH2PO4 [pH 7.7], 100 mM NaCl, 0.2% polyoxyethylenesorbitan monolaurate [Tween 20]) containing 5% (wt/vol) dry fatty-acid-free milk. GST was detected by using a horseradish peroxidase-conjugated anti-GST polyclonal antibody (Z-5; catalog no. sc-459; Santa Cruz) and Trx by using a mouse anti-thioredoxin MAb (Invitrogen), each at a 1/2,000 dilution. The blots were then washed three times, for 10 min each time, in 20 ml of PBST buffer and were revealed by chemiluminescence using an ECL kit (GE Healthcare) as recommended by the manufacturer.
Molecular modeling of the TME1 trimer and TME1–TME2 assembly.
The TME1 trimer was built using CNS (Crystallography & NMR System) routines (36). The modeling protocol started with the generation of a random monomer structure with good local geometry, followed by the duplication of the monomeric unit and a rotation of 120° around one of the internal axes to obtain a symmetric trimer. The G354xxxG358 interhelical interactions were encoded by intermolecular restraints between Gly354 residues (G354a-G354b, G354a-G354c, and G354b-G354c) and Gly358 residues (G358a-G358b, G358a-G358c, and G358b-G358c). Intramolecular hydrogen bonds and dihedral angle restraints were extracted from the TME1 monomer coordinates (RCSB Protein Data Bank [PDB] ID code 1EMZ [7]). A pseudoenergy term was used to minimize the root mean square deviation (RMSD) between the different monomeric structures so as to keep the monomers superimposable. For each of the 100 random dimer structures generated, calculations proceeded through three stages: (i) a high-temperature searching phase at 3,000 K (5,000 steps), (ii) an annealing stage from 3,000 to 100 K in temperature steps of 50 K, and (iii) a final gradient minimization consisting of 1,000 cycles of Powell minimization.
A 3-dimensional (3D) model of TME2 was built by setting the ψ/ϕ angle at canonical values for α-helical secondary-structure elements (8) followed by an energy minimization. We then modeled a TME1–TME2 heterodimer assembly using structural restraints based on a Lys370 (TME1)–Asp728 (TME2) salt bridge and an Asn367 (TME1)–Asp728 (TME2) interhelical hydrogen bond as proposed in reference 37. To test whether a trimer of the TME1–TME2 heterodimer [designated (TME1–TME2)3] is structurally and energetically possible, we performed a rigid-body docking starting from three TME1–TME2 heterodimers guided by the GxxxG interhelical restraints, followed by an energy minimization using a noncrystallographic symmetry restraint to reinforce the trimeric symmetry (C3) of the assembly. The resulting models do not show steric clashes and are energetically stable, demonstrating the structural relevance of such a trimer of heterodimers. Due to the lack of experimental restraints between TME1 and TME2 monomers, a conventional high-resolution structure calculation was not conceivable. Therefore, the exact positioning of each TME2 monomer between two TME1 monomers was not determined at the atomic level, although the different TME2 positions relative to the TME1 trimer were all energetically possible.
RESULTS
The E1 glycoprotein is detected as a trimer on HCV particles.
Although the basic unit of HCV envelope proteins has been shown to be an E1E2 heterodimer in infected cells, much larger covalently linked complexes are present at the surface of the viral particle, showing some oligomeric changes during the assembly process (22). To further characterize these envelope proteins in the context of the viral particle, we analyzed E1E2 complexes by sedimentation analysis in sucrose gradients under reducing conditions. Indeed, the presence of intermolecular disulfide bonds leading to the formation of very large complexes, as observed in our previous study (22), does not allow identification of the basic oligomeric state of HCV envelope glycoproteins present on the surface of the virion. We therefore treated HCV envelope glycoproteins with DTT to dissociate intermolecular disulfide bonds in order to identify complexes that are not cross-linked by disulfide bonds. For such analyses, E1 and E2 from semipurified viral preparations were solubilized with Triton X-100. A control gradient with known globular protein standards was spun in parallel to serve as a calibration. Fractions were harvested and were analyzed for the presence of HCV envelope glycoproteins. However, before SDS-PAGE analysis, we first performed a pulldown experiment with Galanthus nivalis lectin (GNA) coupled to agarose, which allows enrichment in E1E2 glycoproteins. As shown in Fig. 1A, HCV envelope glycoproteins showed a rather wide distribution, although a peak was observed at around 200 kDa. These data suggest that the basic E1E2 complex present on the surface of the virion can form complexes larger than heterodimers, for which the expected molecular mass would be about 90 to 100 kDa, including glycans. It should be stressed, however, that this experimental approach cannot yield a reliable estimation of the molecular masses of these complexes because of the presence of numerous glycans (with a higher density than proteins but a larger hydrodynamic size) and the binding of detergent (with a lower density than proteins) to E1 and E2 transmembrane domains.
FIG 1.
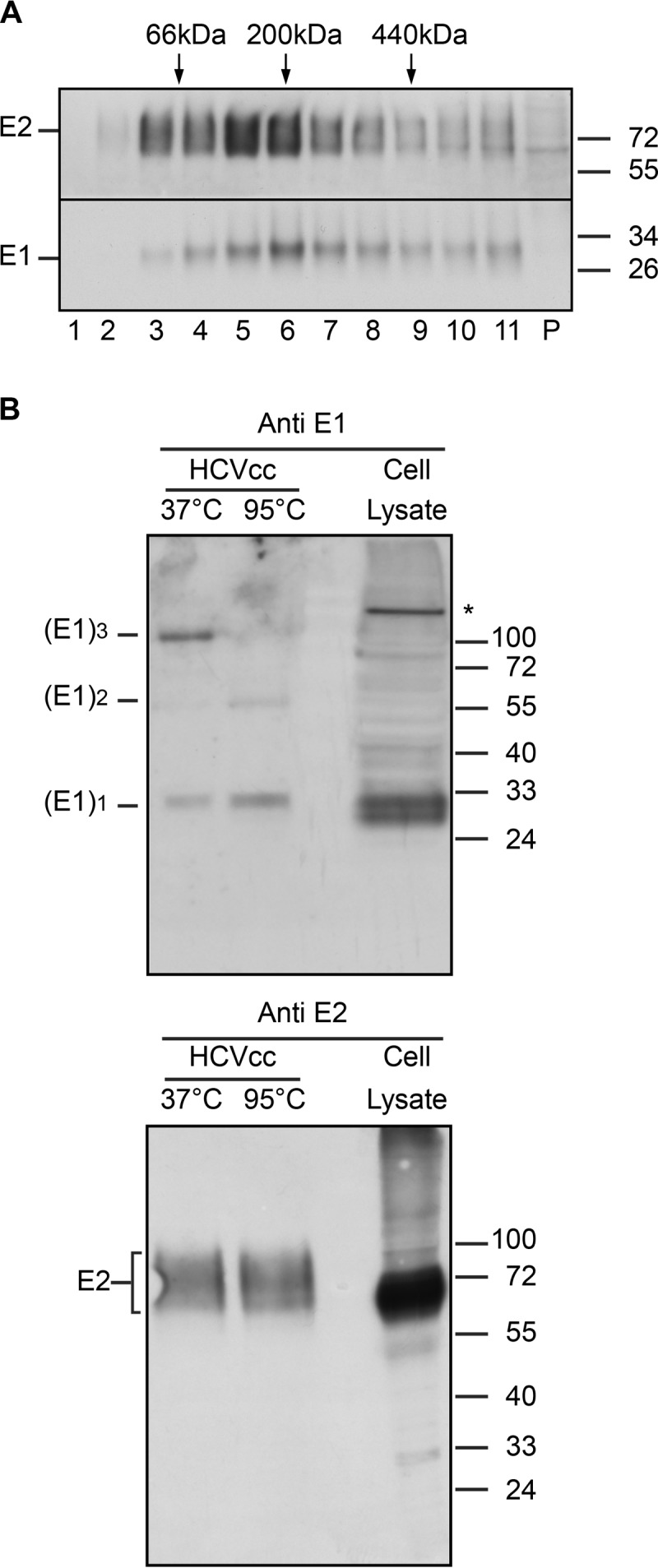
Analysis of HCV envelope glycoproteins associated with viral particles. (A) Separation of HCV envelope glycoproteins in a sucrose density gradient. HCV glycoproteins from a semipurified viral preparation were lysed in 1% Triton X-100 and were separated by sedimentation through a 5-to-20% sucrose gradient in the presence of DTT. Eleven 1-ml fractions and the gradient pellet (P) were harvested. After GNA pulldown, samples were analyzed by reducing SDS-PAGE and Western blotting for the presence of E1 and E2 glycoproteins. The sedimentation profiles of several standard globular proteins in a parallel gradient are indicated above the gel: BSA (66 kDa), β-amylase (200 kDa), and ferritin (440 kDa). (B) Analyses of HCV envelope glycoproteins by SDS-PAGE without heat denaturation. HCVcc particles were lysed in 1% Triton X-100, and HCV envelope glycoproteins were pulled down with a GST-CD81 fusion protein. The proteins were treated with Laemmli sample buffer and were heated for 5 min at 37°C (no thermal denaturation) or 95°C (thermal denaturation) before separation by SDS-PAGE. HCV envelope glycoproteins were revealed by Western blotting with anti-E1 MAb 1C4 (top) and anti-E2 MAb 3/11 (bottom). Lysates of HCV-infected cells treated at 95°C were run in parallel. Molecular mass markers (in kilodaltons) are indicated on the right. The oligomeric forms of E1 are indicated on the left. The asterisk indicates the presence of a nonspecific band.
To further characterize the basic oligomeric state of these envelope proteins in the context of the viral particle, we analyzed them by SDS-PAGE under reducing conditions but without thermal denaturation. A similar treatment has been shown previously to preserve the quaternary structure of the influenza virus spike protein and the trimeric form of the Semliki Forest virus fusion protein (38, 39). To select functional envelope glycoproteins associated with the HCVcc particle, we pulled down E1E2 complexes by the use of a GST-CD81 fusion protein. As shown in Fig. 1B, immunoblotting with anti-E1 and anti-E2 antibodies revealed the presence of SDS-resistant oligomers of E1 when the HCVcc protein samples for SDS-PAGE were pretreated at 37°C instead of 95°C to avoid thermal denaturation. As deduced by comparison with molecular mass markers, the larger fraction of E1 from HCVcc migrated as a trimer. In contrast, protein denaturation by heating the samples at 95°C prior to SDS-PAGE analysis strongly reduced the signal of E1 monomers and some dimers. In contrast to E1, no homo-oligomeric species of E2 (or undetectable levels) were observed. It should be noted that the binding of CD81 to E2 depends on the proper folding of this viral glycoprotein. Furthermore, since the CD81 binding site is located on E2, the pulldown of E1 trimers with CD81 indicates the formation of E1E2 complexes involving E1 trimers. Together, these data indicate that the native form of E1 associated with the viral particle is a stable SDS-PAGE-resistant trimer.
A GxxxG motif is essential for E1 trimer formation.
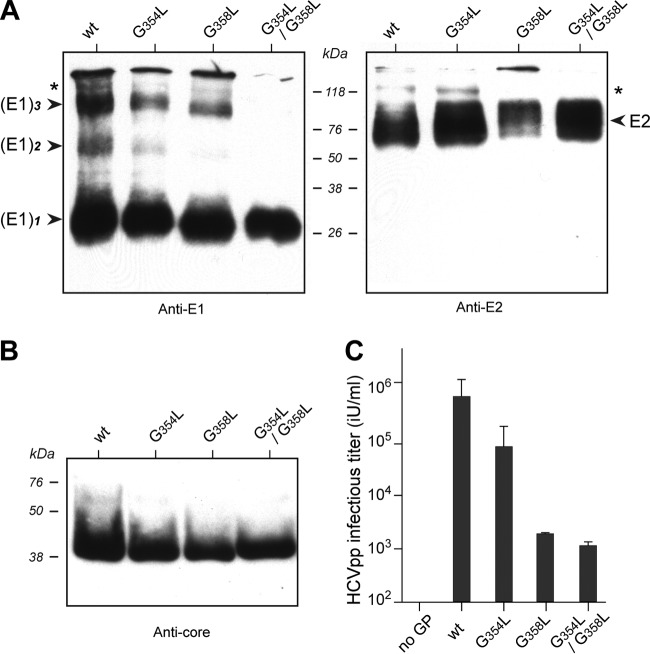
To understand the potential determinants of E1 trimerization, we analyzed the degree of amino acid conservation of the E1 transmembrane sequences of the 27 representative HCV genotypes and subtypes (Fig. 2). The amino acid repertoire revealed that amino acids are strictly conserved in only 24% of the sequence positions (marked with asterisks), underlining the essential role of these residues for the structure and/or function of this domain. However, the apparent variability at most other positions is limited, since the residues observed exhibit similar physicochemical properties, as indicated both by the similarity pattern and by the conservation of the hydropathic character (see the legend to Fig. 2 for details). This indicates that the overall structure of the transmembrane domain of E1 (designated TME1) is conserved among the various HCV genotypes. Of main interest, the GxxxG motif, including residues 354 and 358 (7) belonging to the helical region (7, 9), appears to be almost fully conserved among HCV genotypes. This type of motif is indeed well known to be involved in the homo-oligomerization of TM proteins (reviewed in reference 40). We therefore mutated the GxxxG motif by replacing glycine with leucine, which is a bulky hydrophobic residue able to disrupt potential homo-oligomerization. We then analyzed these mutants under the same SDS-PAGE reducing conditions described above, i.e., samples were treated at 37°C to preserve the trimeric form of the wild-type E1 protein. Due to difficulties in producing large amounts of secreted viral particles in cell culture, we performed our analyses on cell-associated proteins in the context of HCVcc production. We have indeed observed that a fraction of E1 exists as a trimer in HCV-infected cells, together with the E1 dimer (see Fig. 3A). However, to reduce the background and better visualize HCV glycoproteins, we first performed a pulldown experiment with GNA coupled to agarose, which allows for enrichment in E1E2 glycoproteins. The presence of cell-associated E1 dimers and trimers could be due to the presence of cell-associated viral particles. Alternatively, we cannot exclude the possibility that these oligomers are already formed in the prebudding form of HCV envelope glycoproteins. We also observed that E1 trimers present in infected-cell lysates could be pulled down with GST-CD81 (data not shown), confirming that they might represent functional preassembly complexes. As shown in Fig. 3A, replacement of one or both glycine residues within the GxxxG motif abolished the formation and/or stabilization of E1 trimers. In contrast, E1 dimers seemed to be only marginally affected by glycine mutations, indicating that the GxxxG motif is not essential for the formation and/or stabilization of E1 dimers. Since the N-terminal region of E1 is involved in E1E2 heterodimerization (7), we also determined the effect of the glycine mutations on E1E2 interaction by measuring the proportion of E1 protein coprecipitated with E2 in a CD81 pulldown assay. The G354L mutation did not affect E1E2 heterodimerization, and a slight decrease in E1E2 interaction was observed for the G358L mutation (Fig. 3B). However, the double mutant led to a 40% decrease in E1E2 heterodimerization. Importantly, mutated E1 protein still colocalized with E2, as well as with other ER markers, such as calnexin and calreticulin (Fig. 4 and data not shown), indicating that the mutations do not affect the subcellular localization of E1. Together, these data indicate that the GxxxG motif is essential for E1 trimerization and that its mutation had only a limited effect on E1 dimerization and E1E2 heterodimerization.
FIG 3.

Effect of the mutation of the GxxxG motif on E1 trimerization. Glycine residues of the GxxxG motif in the TM domain of E1 were replaced individually (G354L or G358L) or together (G354L/G358L) by leucine in the context of the HCVcc system. Huh-7 cells were electroporated with HCV RNA transcribed in vitro, incubated for 48 h at 37°C, and lysed in 1% Triton X-100. (A) HCV envelope glycoproteins were pulled down with GNA, treated with Laemmli sample buffer, and heated for 5 min at 37°C before separation by SDS-PAGE. HCV envelope glycoproteins were revealed by Western blotting with anti-E1 MAb A4 and anti-E2 MAb 3/11. The oligomeric forms of E1 are indicated on the left and the putative E1E2 heterodimer on the right. The asterisk on the right shows an additional band revealed with the anti-E2 antibody, which likely corresponds to the uncleaved E2p7NS2 precursor. (B) In a parallel experiment, HCV envelope glycoproteins were pulled down with the GST-CD81 fusion protein, treated with Laemmli sample buffer, and heated for 5 min at 70°C before separation by SDS-PAGE. The ratio of coprecipitated E1 glycoprotein to E2 was measured by quantifying the bands by densitometry.
The GxxxG motif is essential for HCV assembly and infectivity.
To further explore the importance of the GxxxG motif in the HCV life cycle, we analyzed the effects of glycine mutations on viral infectivity. As shown in Fig. 5A, single or double mutations abolished the production of infectious viral particles in the supernatants of cells transfected with the corresponding HCV RNA genomes, indicating that the GxxxG motif is essential for the production of infectious viral particles. To determine whether this lack of infectivity is due to a blockade in particle secretion, we also measured intracellular infectivity. However, no infectivity was associated with cells transfected with the mutant RNA molecules (Fig. 5A). This lack of infectivity could potentially be due to the production of defective viral particles or to a defect in virion assembly. To distinguish between these two possibilities, we measured the expression and release of core protein. As a control for assembly defects, we used a mutant in which a large region of the E1E2 sequence was deleted (ΔE1E2). As shown in Fig. 5B, the levels of intracellular core protein in wild-type and mutant viruses were similar. In contrast, the levels of core protein secretion by the mutant viruses were greatly reduced and were similar to that for the mutant with the E1E2 sequence deleted (Fig. 5C). Together, these data indicate that the GxxxG motif is critically involved in particle assembly.
FIG 5.
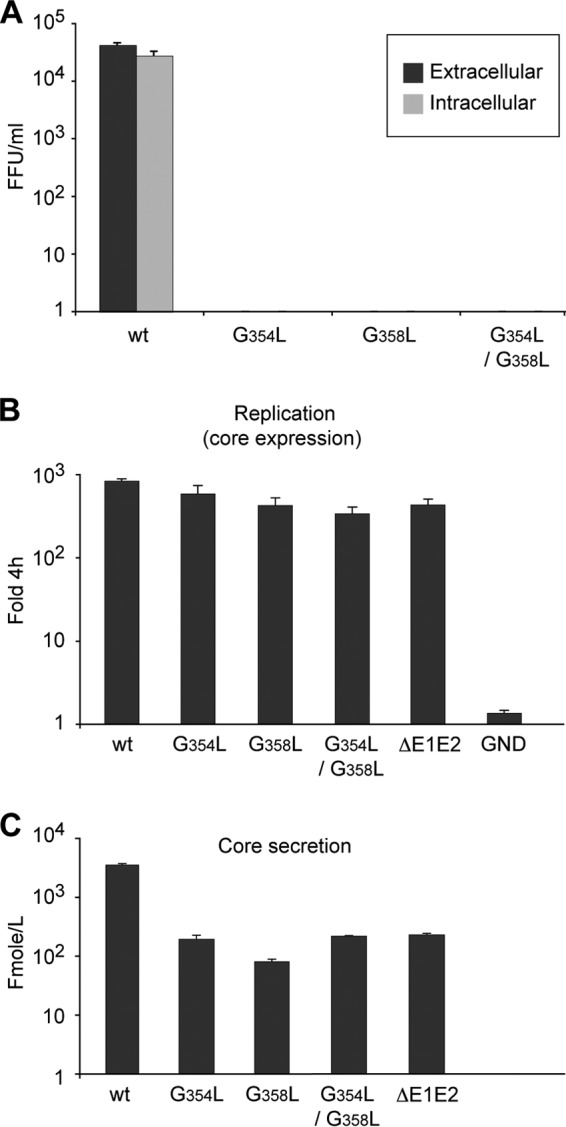
Effects of mutation of the GxxxG motif on HCV infectivity and assembly. In vitro-transcribed RNAs of the GxxxG mutants were electroporated into Huh-7 cells. At 72 h postelectroporation, intracellular and extracellular infectivities (A), expressed in focus-forming units (FFU) per milliliter, were determined by titration, and the amounts of intracellular (B) and extracellular (C) core antigen were measured. Note that the amount of intracellular core protein is expressed as the fold increase in core expression over the level measured at 4 h postelectroporation. The ΔE1E2 mutant, which includes an in-frame deletion in the E1E2 region, and the GND mutant, which includes a mutation in the active site of the viral polymerase, were used as negative controls for virus assembly and replication, respectively.
E2 is essential for E1 trimerization.
An important issue that we were interested in investigating concerns the potential role of E2 in the process of E1 trimer formation. In addition, although we showed that the GxxxG motif is important for viral assembly, we also wanted to check whether this motif could play some role in the entry function of HCV envelope glycoproteins. Since the HCVcc system could not help us to answer these questions, we decided to use the HCVpp system (31). First, we showed that E1 trimers also exist in the context of HCVpp harboring E1-E2 envelope glycoproteins (Fig. 6, left panel, left lane), although some E1 dimer is present and a major fraction of E1 is monomeric. In contrast, no homo-oligomeric form of E2 was observed (Fig. 6, right panel, left lane). We then produced HCVpp containing only E1 in order to determine the potential role of E2 in E1 trimerization. Importantly, no trimer of E1 was observed in the absence of E2 (Fig. 6, left panel, right lane), indicating that E2 plays a major role in E1 trimerization. In contrast, some E1 dimers were observed, indicating that dimer formation is not dependent on the presence of E2. The large amount of E1 monomer observed in the presence of E2 could be attributed to some unassembled E1E2 hetero-oligomers in HCVpp, which are known to be quite heterogeneous (41).
FIG 6.
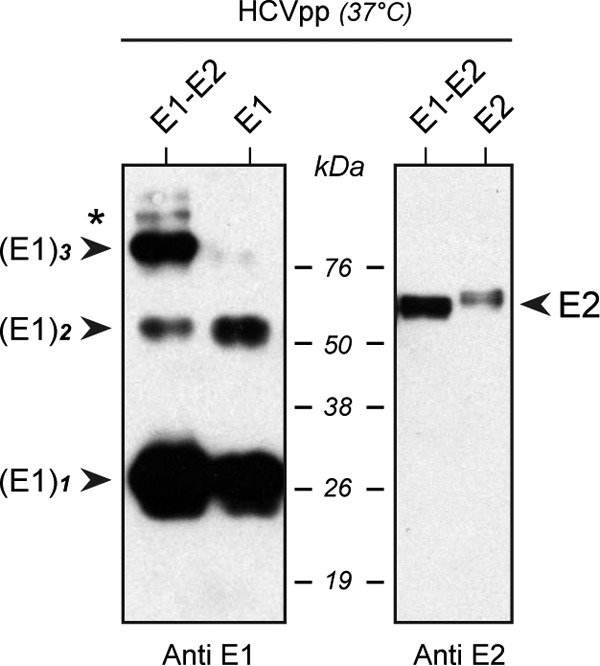
Analysis of HCV envelope glycoproteins associated with HCV pseudoparticles. HCVpp displaying wild-type E1 and E2 complexes, or only E1 or E2, were pelleted through 20% sucrose cushions and were resuspended in PBS. The concentrated viral suspension was treated with Laemmli sample buffer and was heated for 5 min at 37°C prior to separation by SDS-PAGE. HCV envelope glycoproteins were revealed by Western blotting with monoclonal antibodies A4 and H52, directed against E1 and E2, respectively. (Left) The formation of E1 trimers is dependent on the presence of E2 in HCVpp. (Right) The absence of E1 on HCVpp strongly reduces the expression of E2. Homo- and hetero-oligomeric species of E1 and E2 are indicated. The asterisk on the left indicates an additional band that might correspond to the E1E2 heterodimer.
We then tested the effects of glycine mutations in the GxxxG motif in the context of the HCVpp system. As shown in Fig. 7A, individual mutations reduced E1 trimerization, and the double mutant abolished the formation of E1 trimers. Importantly, the mutations had limited effects on the production of proteins and the incorporation of HCV envelope glycoproteins in HCVpp (Fig. 7A and B). We could therefore analyze the entry functions of HCV envelope proteins in the context of these mutations. As shown in Fig. 7C, the glycine mutations had drastic effects on the entry functions of HCV envelope glycoproteins, indicating that the GxxxG motif is essential for these functions.
FIG 7.
The GxxxG motif in TME1 is required for E1 trimerization and HCVpp infectivity. (A) HCVpp displaying wild-type E2 in combination with wild-type E1 (wt) or with E1 displaying the G354L or G358L mutation, or the G354L G358L double mutation, were concentrated on sucrose cushions and resuspended in PBS. The corresponding HCVpp suspensions were treated with Laemmli sample buffer and were heated for 5 min at 37°C prior to analysis by SDS-PAGE and immunoblotting with anti-E1 antibody A4 or anti-E2 antibody H52. The asterisk indicates an additional band that might correspond to the E1E2 heterodimer. (C) The infectivity of HCV pseudoparticles was measured by titrating the unconcentrated viral supernatants onto Huh-7 cells. Virions contained a viral GFP marker gene that leads to the expression of GFP in successfully infected target cells. Titers are expressed as GFP transducing units per milliliter. (B) The amount of viral capsid in each viral supernatant was also quantified by immunoblotting with an anti-capsid antibody.
The TM domain of E1 can form trimers.
To further analyze the role of the TM domain of E1 in the homo-oligomerization of this protein, we constructed a fusion protein containing the TM domain of E1 at the C terminus of the thioredoxin (Trx) reporter protein, and we expressed this chimeric protein (designated Trx-TME1) in E. coli. As shown in Fig. 8, several oligomeric forms of Trx-TME1 resistant to SDS could be observed, including mainly TME1 dimers and trimers. Oligomeric forms of TME1 were found to be associated mainly with E. coli membrane fractions, while inclusion bodies yielded essentially monomeric forms, suggesting that TME1 oligomers are formed exclusively in the membrane context (35). To further confirm the role of the GxxxG motif in TME1 homo-oligomerization, we also mutated glycine residues 354 and 358 in the context of Trx-TME1. Figure 8 (left) shows that single mutation of any glycine in the GxxxG motif strongly reduced the formation and/or stabilization of TME1 trimers, while TME1 dimers appeared to be less sensitive to these mutations. In contrast, the double mutation totally abolished the formation of all TME1 oligomers. Together, these data confirm that the GxxxG motif of the TM domain of E1 plays an essential role in E1 trimerization.
FIG 8.

The TM domain of E1 (TME1) expressed in E. coli forms GxxxG-dependent trimers and TME1–TME2 oligomers. (Left) Bacteria expressing thioredoxin (Trx) fused to wild-type TME1 (Trx-TME1) or to TME1 displaying either the G354L, G358L, or G354L and G358L (G354/358L) mutations were lysed, heated for 5 min at 37°C in Laemmli sample buffer, and subjected to SDS-PAGE, followed by immunoblotting with an anti-Trx antibody. Monomeric, dimeric, and trimeric forms of TME1 are indicated. (Right) Coexpression of Trx-TME1 and GST-TME2 (GST fused to TME2) in E. coli. Bacteria expressing a discistronic construct encoding Trx-TME1 and GST-TME2 were induced and were subjected to SDS-PAGE analysis and immunoblotting with an anti-Trx antibody. Homo- and hetero-oligomeric forms of Trx-TME1 and GST-TME2 are indicated.
Hetero-oligomers are formed when the TM domains of E1 and E2 are coexpressed.
Mutagenesis studies in the context of recombinant HCV envelope glycoproteins have shown that the TM domains of E1 and E2 are involved in E1E2 heterodimerization (5, 7). Therefore, to further understand the complexity of protein-protein interactions at the TM level of HCV envelope glycoproteins, we analyzed whether heterodimerization could also occur in the context of the TM domains devoid of their ectodomains. For this purpose, we coexpressed the Trx-TME1 construct with the GST-TME2 fusion protein, which contains the TM domain of E2 (TME2) at the C terminus of the glutathione S-transferase (GST) protein, in E. coli. Although we observed a majority of homo-oligomeric forms of Trx-TME1 when the Western blot was revealed with an anti-Trx antibody, hetero-oligomeric complexes could also be clearly observed, with molecular masses corresponding to heterodimers or heterotrimers of Trx-TME1 and GST-TME2 (Fig. 8, right). The presence of GST-TME2 in these hetero-oligomeric complexes was confirmed by Western blotting with an anti-GST antibody (data not shown). Together, these data indicate that in addition to Trx-TME1 homo-oligomerization, the TM domains of E1 and E2 can also heterodimerize by themselves.
Molecular modeling of the TME1 trimer and TME1–TME2 assembly.
The obvious implication of the GxxxG motif in the trimerization of E1 enabled us to construct computational molecular models to evaluate whether a trimeric TME1 structure involving a GxxxG motif is possible, taking into account the heterodimerization of TME1–TME2. Figure 9A shows that a TME1 trimer model stabilized by the relative positioning of tightly packed GxxxG motifs can be constructed and is energetically stable, indicating that TME1 can form a chemically plausible three-helix bundle. In this model, glycine residues 354 and 358 point toward each other, ensuring the contact between TME1 subunits. The modeling of TME1–TME2 heterodimer assembly based on a Lys370 (TME1)–Asp728 (TME2) salt bridge and an Asn367 (TME1)–Asp728 (TME2) interhelical hydrogen bond, as proposed by Jusoh, Helms, and coworkers (37), is compatible with this TME1 trimeric assembly. As illustrated in Fig. 9B, several orientations of TME2 relative to TME1 are possible, including the interaction of TME2 either with a single TME1 monomer or with two adjacent TME1 monomers. Although the lack of structural data does not allow one to distinguish between these two possibilities, the TME1 and TME2 interactions reported in this study suggest that the E1 and E2 glycoproteins could form trimers of E1E2 heterodimers at the membrane surfaces of viral particles (Fig. 9C).
FIG 9.
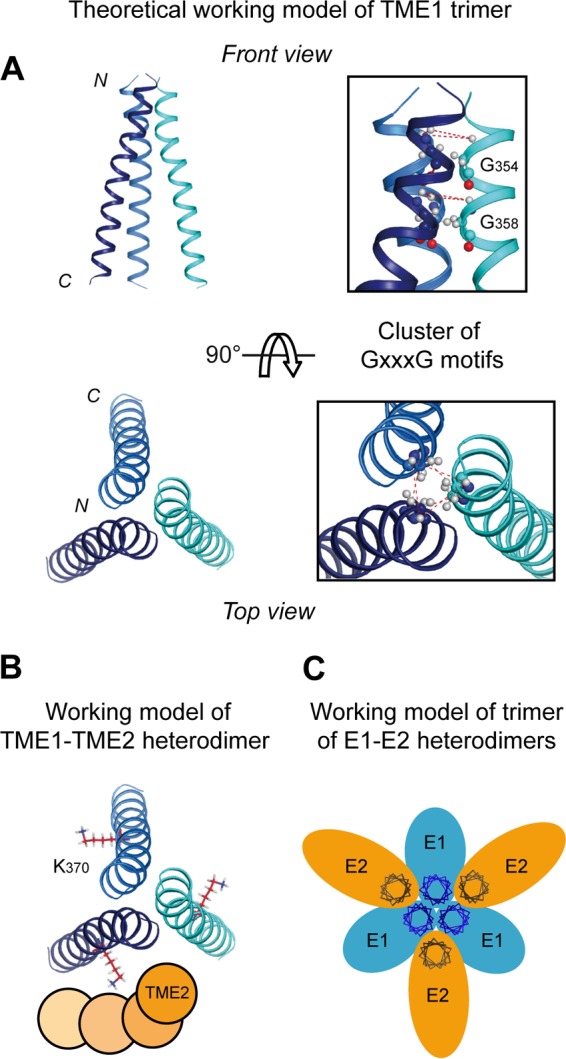
Theoretical models of the TME1 trimer and the trimer of E1E2 heterodimers. (A) Ribbon representation of the trimer model of TME1. Note that, for clarity, TME2 is not represented (see the legend to panel B below). This symmetric model was constructed by assuming the implication of glycine 354 and glycine 358 of each monomer at the trimer interfaces. Dashed red lines indicate interhelical amide proton distances used in the modeling protocol. Glycine residues 354 and 358 forming the GxxxG motifs are shown in ball-and-stick representations. (B) Theoretical model of TME1–TME2 interaction assuming a Lys370 (TME1)–Asp728 (TME2) salt bridge and an Asn367 (TME1)–Asp728 (TME2) interhelical hydrogen bond (as proposed in reference 37). Because numerous possible model solutions were obtained for the relative positioning of TME2 to TME1, TME2 is schematically represented as orange circles at four representative positions for a single TME1–TME2 heterodimer. (C) Schematic representation of a possible organization of the trimer of E1E2 heterodimers at the membrane surface of the viral particle. E1 and E2 ectodomains are represented as blue and yellow ovals, respectively, while TME1 and TME2 are represented as helix projections (i.e., perpendicular to the membrane surface). These models were generated from structure coordinates using VMD (http://www.ks.uiuc.edu/Research/vmd/) and were rendered with POV-Ray (http://www.povray.org/).
DISCUSSION
By being part of the viral particle, HCV envelope glycoproteins play an essential role in virion morphogenesis as well as in HCV entry into liver cells. These two steps necessitate timely and coordinated control of HCV glycoprotein functions, in which protein oligomerization often takes part. Here we investigated the oligomeric state of HCV virion-associated envelope glycoproteins in order to better understand their functions. We demonstrated that E1 forms functional trimers after virion assembly and that E2 is essential for E1 trimer formation, as demonstrated with an HCVpp system, indicating that E2 includes essential structural determinants allowing the formation of trimers of E1E2 heterodimers. Moreover, a main determinant for this trimerization is present in a specific GxxxG motif located within the E1 transmembrane domain. Importantly, alteration of E1 trimerization by mutation of this motif abrogated virion assembly and thus the production of infectious virions. Together, these data indicate that E2-dependent E1 trimerization plays a central role in coordinating the assembly and entry functions of HCV envelope glycoproteins.
The fact that E1 trimers in HCVcc particles have not been detected before the present study is most likely due to the classical 95°C thermal treatment of SDS-PAGE protein samples, which clearly induces the dissociation of E1 trimers into monomers (Fig. 1). As reported previously, for example, for the influenza virus spike protein and the trimeric form of the Semliki Forest virus fusion protein (38, 39), the absence of thermal treatment preserves the quaternary structures of these membrane proteins. Thermal treatment of SDS-PAGE protein samples (which was initially introduced in order to inactivate proteases in complex mixtures of proteins) can disrupt some native oligomeric forms of membrane proteins, while in fact, it is not really necessary to dissociate most protein complexes in the presence of SDS (for discussions of the effects of SDS on protein dissociation and folding, see references 42 and 43).
The results presented in this study establish that TME1 can oligomerize to form trimers thanks to its GxxxG motif, directly mirroring the behavior of full-length E1. The GxxxG motif is a well-known dimerization motif for transmembrane helix-helix association (44–46). The three x amino acids between the glycine residues in a helical structure align the two glycines on the same face of the helix, which creates a flat interaction platform allowing tight interhelical packing. Indeed, the small glycine side chain exposes polar backbone atoms, leading to favorable interactions (47), especially in the hydrophobic membrane environment. In a dimer, the glycine residues of the GxxxG motif face each other, allowing symmetric close packing of the helices. In the present study, mutations of the GxxxG motif had a stronger deleterious effect on the formation and/or stabilization of the trimeric form of E1 than on that of the dimeric form, suggesting that the TME1 GxxxG motif is not a bona fide dimerization motif. Interestingly, it has been reported that the GxxxG motif could be involved in the trimerization of the TM domain of the SARS (severe acute respiratory syndrome) coronavirus spike protein (48). Molecular modeling and molecular dynamics simulations in a hydrated lipid bilayer showed that in this trimeric structure, the glycine residues of the GxxxG motif are facing each other, enhancing helix-helix interactions by allowing for the close positioning of the helices and the stabilization of the trimeric bundle (48). In the present study, a comparable trimeric working model, compatible with TME1–TME2 heterodimerization, could be constructed for TME1 (Fig. 9). As already well documented for the dimerization of TM domains, while the GxxxG motif plays a crucial role in oligomerization, it is often not sufficient by itself to ensure dimerization, and additional neighboring residues are required for the stabilization of the helix bundle (49). This is also likely the case for the trimeric structure of TME1, and further structural studies are required to specify this working model. Interestingly, it has been reported recently that a recombinant ectodomain of HCV E1 protein is oligomeric in solution, with an apparent molecular mass compatible with a trimer when measured by gel filtration (50). This finding not only supports the trimeric assembly of E1 but also indicates that structural determinants for its trimerization are present in its ectodomain, in addition to that identified in its TM domain and in addition to the presence of E2.
Remarkably, the C-terminal TM domains of both the HCV E1 glycoprotein and the SARS coronavirus spike protein play roles in stabilizing the trimeric structures of these proteins, most likely thanks to the direct interactions of their respective GxxxG motifs. One can thus expect that this common structural feature might be shared by other viral envelope proteins containing a GxxxG motif or the so-called “SmallxxxSmall” motifs in their membrane domains (“Small” refers to glycine, alanine, serine, cysteine, or threonine residues). However, because of the difficulties of studying membrane protein domains, only limited structural investigations have been reported to date.
Importantly, the presence of E2 in HCVpp is mandatory for observation of the E1 trimer (Fig. 6), indicating an essential role for E2 in the trimerization of E1. It must be noted that while E1 is observed mainly as a trimer on the HCVcc particle, it is detected mainly as a monomer within infected cells (Fig. 1), most likely associated with E2 as an E1E2 heterodimer, as we reported previously (5, 7). Taken together, these data suggest that the E2-dependent trimerization of E1 occurs by the assembly of preformed E1E2 heterodimers, most likely during the virion assembly step. It is tempting to speculate that the structural transition from E1E2 heterodimers to trimers of heterodimers is a key step in HCV virion envelope assembly, and one can suppose that this process is strictly regulated and probably involves some other viral factors, such as NS2 (51–54).
In this context, one can wonder whether the dimers of E1 observed in cell extracts (Fig. 3) and HCVpp (Fig. 6), but almost absent in HCVcc (Fig. 1B), might be transient molecular species in the process of the formation of trimers of E1E2 heterodimers.
In contrast to what was observed for E1, no homo-oligomeric form of E2 was detected in the present work, and the recombinant ectodomain of E2 has been reported to be monomeric (12, 13). In contrast, the E1E2 heterodimer was repeatedly observed in this study, and in agreement with our previous results using alanine scanning (7), the TME1–TME2 complex observed in bacteria demonstrated that the E1 and E2 transmembrane domains play essential roles in the stabilization of this heterodimer (Fig. 8). Moreover, oligomers including two TME1 molecules and one TME2 molecule were even observed. Interestingly, in this simplified protein coexpression system, TME1 dimers and trimers form nicely even in the absence of TME2, suggesting, again, that some ectodomain determinants might also be important for the regulation of the oligomerization process, which most likely is not triggered only via the TM domains (55). All these features indicate that HCV envelope glycoproteins form a stable trimer of E1E2 heterodimers at the membrane surface of the viral particle, and they have encouraged us to propose a tentative model of E1E2 oligomeric structure based on molecular modeling (Fig. 9). Although this model is compatible with all available structural data, it remains a working model. However, we expect that it will be useful for the design of new structural and functional studies to clarify the structural organization of E1E2 at the virion surface and its role in virus entry.
Increasing evidence favors a major implication of E1 alone or in association with E2 in the fusion process of the HCV virion. Before experimental determination of the structure of E2, similarities in genomic organization between HCV and other members of the family Flaviviridae led to the proposal of E2 as a class II fusion protein (56). However, this hypothesis has been challenged by the publication of a pestivirus E2 glycoprotein structure, which shows no structural homology with class II fusion proteins (57, 58). Furthermore, the recently published structure of the HCV E2 core indicates that this protein clearly does not comply with the class II criteria and also does not resemble its pestivirus homolog (12, 13). These data also indicate that E2 lacks structural hallmarks of fusion proteins. Indeed, the previously proposed fusion peptide (14) is located in secondary-structure elements within the hydrophobic core of the protein and is therefore unlikely to serve as a fusion peptide (12, 13). In addition, this lack of fusion peptide properties has been confirmed recently by a mutagenesis study (59). Finally, structural analyses at low pHs indicate that E2 does not undergo the structural rearrangement needed to play a role in membrane fusion (13). On the other hand, a conserved highly hydrophobic region in E1 (residues 264 to 290) has been proposed previously as a potential HCV fusion loop (60). The importance of this region in viral infectivity has been confirmed by several mutagenesis studies (15, 61, 62). Furthermore, peptides spanning this region have been shown to induce the fusion and disruption of liposomes (63). Finally, very recently, mutations in viruses resistant to an inhibitor of viral fusion have been mapped in the same region (64).
The trimerization of HCV glycoprotein E1 plays a central role in virion assembly and entry. Trimerization is a characteristic feature of viral envelope glycoproteins involved in membrane fusion. The postfusion structures of the class I, II, and III viral envelope glycoproteins reported thus far are indeed trimers (65). Furthermore, prefusion structures of class I and III proteins also form trimers. The case of class II fusion proteins is more complex, since they associate with a companion protein during their biogenesis, and trimers of these heterodimers are formed during virion assembly (reviewed in reference 66). However, after the cleavage of the companion protein, further reorganization can occur for some class II viruses, leading to the formation of homodimers at the surface of the mature virion, as is the case for the flaviviruses. Although they do not belong to class II fusion proteins, HCV envelope glycoproteins also form a heterodimer as observed for the flaviviruses. Due to the close interplay between E1 and E2 during assembly and entry (15, 67, 68), E2 has been proposed to play an accessory role as a molecular scaffold for E1 (16). The functional viral glycoprotein unit involved in viral fusion is therefore likely to be an E1E2 complex, as suggested previously (15). However, as for other envelope glycoproteins, our data indicate that trimerization also plays a central role in the function of HCV envelope glycoproteins. Indeed, the presence of E1 trimers with each E1 subunit linked to an E2 protein strongly suggests that the functional complex is a trimer of E1E2 heterodimers, which is important not only for virion assembly but also for virus entry. A major difference from other viral envelope glycoproteins is the central role played by a TM domain in the homo-oligomerization of the putative fusion protein. A role for the TM domain of E1 in viral fusion had already been suggested previously (69), and here our data suggest that the trimerization of the TM domain could be necessary for E1 to play its role during the fusion process.
As with other enveloped viruses, the HCV fusion process is highly regulated. After endocytosis, the virion needs to be exposed to an acidic compartment in order to initiate membrane fusion (70–72). However, the virus itself is not sensitive to acidic pHs, suggesting that it needs another cue to be able to trigger membrane fusion (72). It has been speculated that the virus needs first to reduce the intermolecular disulfide bonds bridging HCV envelope glycoproteins together on the viral particle (22). E2 interaction with CD81 has also been proposed to prime the virions for the fusion process (73). After priming, as a potential fusion protein, E1 trimers would have to extend, at least transiently, to span the distance between the viral and cellular membranes prior to membrane fusion, which means that the E1 ectodomain would have to adopt an elongated fold as observed for class I, II, and III fusion proteins (16). As with other fusion proteins, the formation of a trimer is likely essential for the destabilization of the host membrane in order to initiate the fusion process. Since E1E2 heterodimers seem to be rather stable, it is likely that E2 plays a role by helping the conformational changes of E1 occurring in the fusion process. However, in the absence of a high-resolution structure of the E1E2 complex, it remains difficult to understand the fusion process induced by this new class of viral fusion protein.
In conclusion, this work demonstrates, for the first time, that the E1 protein is trimeric on HCVcc particles and indicates that the E1 and E2 glycoproteins most likely assemble as stable trimers of E1E2 heterodimers at the surface of the HCV virion. These structural features, together with accumulating evidence, strongly support the notion that E1 plays a major role in the membrane fusion mechanism allowing the delivery of the HCV genome to infected cells. Much structural and functional work remains to be done to further dissect this mechanism. Importantly, these observations are essential for the future design of an efficient vaccine against HCV.
ACKNOWLEDGMENTS
This work was supported by the French National Agency for Research on AIDS and Viral Hepatitis (ANRS) (to J.D., F.P., and F.-L.C.) and a grant from Mapping project ANR-11-BINF-003 (to F.P.). B.A.T. was supported by a Marie Curie Intra European Fellowship within the 7th European Community Framework Programme (PIEF-GA-2009-235147).
We are grateful to Darius Moradpour for scientific advice. We are grateful to Sophana Ung for assistance in the illustrations. We also thank J. McKeating, M. Law, and T. Wakita for providing us with reagents. The fluorescence microscopy data were generated with the help of the Bio-imaging Center Lille Nord-de-France (BICeL).
REFERENCES
- 1.Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. 2011. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol 92:233–246. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams MJ, King AM, Carstens EB. 2013. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2013). Arch Virol 158:2023–2030. doi: 10.1007/s00705-013-1688-5. [DOI] [PubMed] [Google Scholar]
- 3.Moradpour D, Penin F, Rice CM. 2007. Replication of hepatitis C virus. Nat Rev Microbiol 5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 4.Vieyres G, Dubuisson J, Pietschmann T. 2014. Incorporation of hepatitis C virus E1 and E2 glycoproteins: the keystones on a peculiar virion. Viruses 6:1149–1187. doi: 10.3390/v6031149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocquerel L, Wychowski C, Minner F, Penin F, Dubuisson J. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a key role in the processing, subcellular localization, and assembly of these envelope proteins. J Virol 74:3623–3633. doi: 10.1128/JVI.74.8.3623-3633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocquerel L, Op de Beeck A, Lambot M, Roussel J, Delgrange D, Pillez A, Wychowski C, Penin F, Dubuisson J. 2002. Topologic changes in the transmembrane domains of hepatitis C virus envelope glycoproteins. EMBO J 21:2893–2902. doi: 10.1093/emboj/cdf295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Op De Beeck A, Montserret R, Duvet S, Cocquerel L, Cacan R, Barberot B, Le Maire M, Penin F, Dubuisson J. 2000. Role of the transmembrane domains of hepatitis C virus envelope proteins E1 and E2 in the assembly of the noncovalent E1E2 heterodimer. J Biol Chem 275:31428–31437. doi: 10.1074/jbc.M003003200. [DOI] [PubMed] [Google Scholar]
- 8.Shalom-Elazari H, Zazrin-Greenspon H, Shaked H, Chill JH. 2014. Global fold and backbone dynamics of the hepatitis C virus E2 glycoprotein transmembrane domain determined by NMR. Biochim Biophys Acta 1838:2919–2928. doi: 10.1016/j.bbamem.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 9.Zazrin H, Shaked H, Chill JH. 2014. Architecture of the hepatitis C virus E1 glycoprotein transmembrane domain studied by NMR. Biochim Biophys Acta 1838:784–792. doi: 10.1016/j.bbamem.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 10.Lavie M, Goffard A, Dubuisson J. 2007. Assembly of a functional HCV glycoprotein heterodimer. Curr Issues Mol Biol 9:71–86. [PubMed] [Google Scholar]
- 11.Dubuisson J, Helle F, Cocquerel L. 2008. Early steps of the hepatitis C virus life cycle. Cell Microbiol 10:821–827. doi: 10.1111/j.1462-5822.2007.01107.x. [DOI] [PubMed] [Google Scholar]
- 12.Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, Yost SA, Bohannon CD, Jacob J, Grakoui A, Marcotrigiano J. 2014. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB, Wilson IA, Law M. 2013. Hepatitis C virus E2 envelope glycoprotein core structure. Science 342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krey T, d'Alayer J, Kikuti CM, Saulnier A, Damier-Piolle L, Petitpas I, Johansson DX, Tawar RG, Baron B, Robert B, England P, Persson MA, Martin A, Rey FA. 2010. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog 6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavillette D, Pecheur EI, Donot P, Fresquet J, Molle J, Corbau R, Dreux M, Penin F, Cosset FL. 2007. Characterization of fusion determinants points to the involvement of three discrete regions of both E1 and E2 glycoproteins in the membrane fusion process of hepatitis C virus. J Virol 81:8752–8765. doi: 10.1128/JVI.02642-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Modis Y. 2014. A novel membrane fusion protein family in Flaviviridae? Trends Microbiol 22:176–182. doi: 10.1016/j.tim.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Omari K, Iourin O, Kadlec J, Sutton G, Harlos K, Grimes JM, Stuart DI. 2014. Unexpected structure for the N-terminal domain of hepatitis C virus envelope glycoprotein E1. Nat Commun 5:4874. doi: 10.1038/ncomms5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.André P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Brechot C, Paranhos-Baccala G, Lotteau V. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol 76:6919–6928. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartenschlager R, Penin F, Lohmann V, André P. 2011. Assembly of infectious hepatitis C virus particles. Trends Microbiol 19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Catanese MT, Uryu K, Kopp M, Edwards TJ, Andrus L, Rice WJ, Silvestry M, Kuhn RJ, Rice CM. 2013. Ultrastructural analysis of hepatitis C virus particles. Proc Natl Acad Sci U S A 110:9505–9510. doi: 10.1073/pnas.1307527110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merz A, Long G, Hiet MS, Bruegger B, Chlanda P, André P, Wieland F, Krijnse-Locker J, Bartenschlager R. 2011. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem 286:3018–3032. doi: 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vieyres G, Thomas X, Descamps V, Duverlie G, Patel AH, Dubuisson J. 2010. Characterization of the envelope glycoproteins associated with infectious hepatitis C virus. J Virol 84:10159–10168. doi: 10.1128/JVI.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. 1982. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res 42:3858–3863. [PubMed] [Google Scholar]
- 24.Dubuisson J, Hsu HH, Cheung RC, Greenberg HB, Russell DG, Rice CM. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol 68:6147–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flint M, Maidens C, Loomis-Price LD, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating JA. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol 73:6235–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delgrange D, Pillez A, Castelain S, Cocquerel L, Rouillé Y, Dubuisson J, Wakita T, Duverlie G, Wychowski C. 2007. Robust production of infectious viral particles in Huh-7 cells by introducing mutations in hepatitis C virus structural proteins. J Gen Virol 88:2495–2503. doi: 10.1099/vir.0.82872-0. [DOI] [PubMed] [Google Scholar]
- 28.Goueslain L, Alsaleh K, Horellou P, Roingeard P, Descamps V, Duverlie G, Ciczora Y, Wychowski C, Dubuisson J, Rouille Y. 2010. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J Virol 84:773–787. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rocha-Perugini V, Montpellier C, Delgrange D, Wychowski C, Helle F, Pillez A, Drobecq H, Le Naour F, Charrin S, Levy S, Rubinstein E, Dubuisson J, Cocquerel L. 2008. The CD81 partner EWI-2wint inhibits hepatitis C virus entry. PLoS One 3:e1866. doi: 10.1371/journal.pone.0001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alsaleh K, Delavalle PY, Pillez A, Duverlie G, Descamps V, Rouille Y, Dubuisson J, Wychowski C. 2010. Identification of basic amino acids at the N-terminal end of the core protein that are crucial for hepatitis C virus infectivity. J Virol 84:12515–12528. doi: 10.1128/JVI.01393-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartosch B, Dubuisson J, Cosset FL. 2003. Infectious hepatitis C pseudo-particles containing functional E1E2 envelope protein complexes. J Exp Med 197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ausubel FM, Brent R, Kingston RE, Moore AD, Seidman JG, Smith JA, Struhl K. 2000. Current protocols in molecular biology, vol 1 John Wiley and Sons, Inc, New York, NY. [Google Scholar]
- 33.Higginbottom A, Quinn ER, Kuo CC, Flint M, Wilson LH, Bianchi E, Nicosia A, Monk PN, McKeating JA, Levy S. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J Virol 74:3642–3649. doi: 10.1128/JVI.74.8.3642-3649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albecka A, Belouzard S, de Beeck AO, Descamps V, Goueslain L, Bertrand-Michel J, Terce F, Duverlie G, Rouille Y, Dubuisson J. 2012. Role of low-density lipoprotein receptor in the hepatitis C virus life cycle. Hepatology 55:998–1007. doi: 10.1002/hep.25501. [DOI] [PubMed] [Google Scholar]
- 35.Montigny C, Penin F, Lethias C, Falson P. 2004. Overcoming the toxicity of membrane peptide expression in bacteria by upstream insertion of Asp-Pro sequence. Biochim Biophys Acta 1660:53–65. doi: 10.1016/j.bbamem.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 36.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54:905–921. doi: 10.1107/S0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Jusoh SA, Welsch C, Siu SW, Böckmann RA, Helms V. 2010. Contribution of charged and polar residues for the formation of the E1-E2 heterodimer from hepatitis C virus. J Mol Model 16:1625–1637. doi: 10.1007/s00894-010-0672-1. [DOI] [PubMed] [Google Scholar]
- 38.Doms RW, Helenius A. 1986. Quaternary structure of influenza virus hemagglutinin after acid treatment. J Virol 60:833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wahlberg JM, Garoff H. 1992. Membrane fusion process of Semliki Forest virus. I. Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J Cell Biol 116:339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senes A, Engel DE, DeGrado WF. 2004. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol 14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Flint M, Logvinoff C, Rice CM, McKeating JA. 2004. Characterization of infectious retroviral pseudotype particles bearing hepatitis C virus glycoproteins. J Virol 78:6875–6882. doi: 10.1128/JVI.78.13.6875-6882.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong M, Baggetto LG, Falson P, Le Maire M, Penin F. 1997. Complete removal and exchange of sodium dodecyl sulfate bound to soluble and membrane proteins and restoration of their activities, using ceramic hydroxyapatite chromatography. Anal Biochem 247:333–341. doi: 10.1006/abio.1997.2103. [DOI] [PubMed] [Google Scholar]
- 43.Montserret R, McLeish MJ, Bockmann A, Geourjon C, Penin F. 2000. Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding. Biochemistry 39:8362–8373. doi: 10.1021/bi000208x. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Engelman DM, Gerstein M. 2002. Genomic analysis of membrane protein families: abundance and conserved motifs. Genome Biol 3:research0054. doi: 10.1186/gb-2002-3-10-research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russ WP, Engelman DM. 2000. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol 296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 46.Senes A, Gerstein M, Engelman DM. 2000. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol 296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 47.Senes A, Ubarretxena-Belandia I, Engelman DM. 2001. The Cα—H···O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc Natl Acad Sci U S A 98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arbely E, Granot Z, Kass I, Orly J, Arkin IT. 2006. A trimerizing GxxxG motif is uniquely inserted in the severe acute respiratory syndrome (SARS) coronavirus spike protein transmembrane domain. Biochemistry 45:11349–11356. doi: 10.1021/bi060953v. [DOI] [PubMed] [Google Scholar]
- 49.Schneider D, Engelman DM. 2004. Motifs of two small residues can assist but are not sufficient to mediate transmembrane helix interactions. J Mol Biol 343:799–804. doi: 10.1016/j.jmb.2004.08.083. [DOI] [PubMed] [Google Scholar]
- 50.Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, Yost SA, Grakoui A, Marcotrigiano J. 2014. 21st International Symposium on Hepatitis C Virus and Related Viruses, Banff, Canada, abstr O2.03. [Google Scholar]
- 51.Jirasko V, Montserret R, Lee JY, Gouttenoire J, Moradpour D, Penin F, Bartenschlager R. 2010. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Pathog 6:e1001233. doi: 10.1371/journal.ppat.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma Y, Anantpadma M, Timpe JM, Shanmugam S, Singh SM, Lemon SM, Yi M. 2011. Hepatitis C virus NS2 protein serves as a scaffold for virus assembly by interacting with both structural and nonstructural proteins. J Virol 85:86–97. doi: 10.1128/JVI.01070-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Popescu CI, Rouille Y, Dubuisson J. 2011. Hepatitis C virus replication and assembly: a play in one act. Future Virol 6:985–995. doi: 10.2217/fvl.11.69. [DOI] [Google Scholar]
- 54.Stapleford KA, Lindenbach BD. 2011. Hepatitis C virus NS2 coordinates virus particle assembly through physical interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J Virol 85:1706–1717. doi: 10.1128/JVI.02268-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drummer HE, Poumbourios P. 2004. Hepatitis C virus glycoprotein E2 contains a membrane-proximal heptad repeat sequence that is essential for E1E2 glycoprotein heterodimerization and viral entry. J Biol Chem 279:30066–30072. doi: 10.1074/jbc.M405098200. [DOI] [PubMed] [Google Scholar]
- 56.Vaney MC, Rey FA. 2011. Class II enveloped viruses. Cell Microbiol 13:1451–1459. doi: 10.1111/j.1462-5822.2011.01653.x. [DOI] [PubMed] [Google Scholar]
- 57.El Omari K, Iourin O, Harlos K, Grimes JM, Stuart DI. 2013. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep 3:30–35. doi: 10.1016/j.celrep.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Wang J, Kanai R, Modis Y. 2013. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc Natl Acad Sci U S A 110:6805–6810. doi: 10.1073/pnas.1300524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lavie M, Sarrazin S, Montserret R, Descamps V, Baumert TF, Duverlie G, Seron K, Penin F, Dubuisson J. 2014. Identification of conserved residues in hepatitis C virus envelope glycoprotein E2 that modulate virus dependence on CD81 and SRB1 entry factors. J Virol 88:10584–10597. doi: 10.1128/JVI.01402-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Flint M, Thomas JM, Maidens CM, Shotton C, Levy S, Barclay WS, McKeating JA. 1999. Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J Virol 73:6782–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Drummer HE, Boo I, Poumbourios P. 2007. Mutagenesis of a conserved fusion peptide-like motif and membrane-proximal heptad-repeat region of hepatitis C virus glycoprotein E1. J Gen Virol 88:1144–1148. doi: 10.1099/vir.0.82567-0. [DOI] [PubMed] [Google Scholar]
- 62.Russell RS, Kawaguchi K, Meunier JC, Takikawa S, Faulk K, Bukh J, Purcell RH, Emerson SU. 2009. Mutational analysis of the hepatitis C virus E1 glycoprotein in retroviral pseudoparticles and cell-culture-derived H77/JFH1 chimeric infectious virus particles. J Viral Hepat 16:621–632. doi: 10.1111/j.1365-2893.2009.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pérez-Berná AJ, Pabst G, Laggner P, Villalain J. 2009. Biophysical characterization of the fusogenic region of HCV envelope glycoprotein E1. Biochim Biophys Acta 1788:2183–2193. doi: 10.1016/j.bbamem.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 64.Perin PM, Haid S, Brown R, Schulze K, Colpitts CC, Zeilinger C, von Schaewen M, Heller B, Vercauteren K, Luxenburger E, Andonyadis Y, Kirschning A, Schang LM, Muller R, Guzman CA, Randall G, Meuleman P, Ploss A, Pietschmann T. 2014. 21st International Meeting on Hepatitis C Virus and Related Viruses, Banff, Canada, abstr O1.07. [Google Scholar]
- 65.Baquero E, Albertini AA, Vachette P, Lepault J, Bressanelli S, Gaudin Y. 2013. Intermediate conformations during viral fusion glycoprotein structural transition. Curr Opin Virol 3:143–150. doi: 10.1016/j.coviro.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kielian M, Rey FA. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Microbiol 4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Michalak J-P, Wychowski C, Choukhi A, Meunier J-C, Ung S, Rice CM, Dubuisson J. 1997. Characterization of truncated forms of hepatitis C virus glycoproteins. J Gen Virol 78:2299–2306. [DOI] [PubMed] [Google Scholar]
- 68.Patel J, Patel AH, McLauchlan J. 2001. The transmembrane domain of the hepatitis C virus E2 glycoprotein is required for correct folding of the E1 glycoprotein and native complex formation. Virology 279:58–68. doi: 10.1006/viro.2000.0693. [DOI] [PubMed] [Google Scholar]
- 69.Ciczora Y, Callens N, Penin F, Pecheur EI, Dubuisson J. 2007. Transmembrane domains of hepatitis C virus envelope glycoproteins: residues involved in E1E2 heterodimerization and involvement of these domains in virus entry. J Virol 81:2372–2381. doi: 10.1128/JVI.02198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blanchard E, Belouzard S, Goueslain L, Wakita T, Dubuisson J, Wychowski C, Rouille Y. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J Virol 80:6964–6972. doi: 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lavillette D, Bartosch B, Nourrisson D, Verney G, Cosset FL, Penin F, Pecheur EI. 2006. Hepatitis C virus glycoproteins mediate low pH-dependent membrane fusion with liposomes. J Biol Chem 281:3909–3917. doi: 10.1074/jbc.M509747200. [DOI] [PubMed] [Google Scholar]
- 72.Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol 80:1734–1741. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sharma NR, Mateu G, Dreux M, Grakoui A, Cosset FL, Melikyan GB. 2011. Hepatitis C virus is primed by CD81 protein for low pH-dependent fusion. J Biol Chem 286:30361–30376. doi: 10.1074/jbc.M111.263350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, Maertens G, Mizokami M, Murphy D, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin-I T, Stuyver LJ, Thiel H-J, Viazov S, Weiner AJ, Widell A. 2005. Consensus proposals for the unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 75.Combet C, Garnier N, Charavay C, Grando D, Crisan D, Lopez J, Dehne-Garcia A, Geourjon C, Bettler E, Hulo C, Le Mercier P, Bartenschlager R, Diepolder H, Moradpour D, Pawlotsky JM, Rice CM, Trépo C, Penin F, Deléage G. 2007. euHCVdb: the European hepatitis C virus database. Nucleic Acids Res 35:D363–D366. doi: 10.1093/nar/gkl970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]