Abstract
Background:
It is recognised that modulations of the nuclear import of macromolecules have a role in changing cellular phenotypes and carcinogenesis. We and others have noticed that aberrant subcellular localisation of DNA damage response (DDR) proteins in breast cancer (BC) is associated with loss-of-function phenotype. This study aims to investigate the biological and clinical significance of the nucleocytoplasmic transport protein karyopherin α-2 (KPNA2), and its role in controlling DDR proteins subcellular localisation in BC.
Methods:
A large (n=1494) and well-characterised series of early-stage invasive BC with a long-term follow-up was assessed for KPNA2 protein by using immunohistochemistry.
Results:
KPNA2 expression was associated with the subcellular localisation of key DDR proteins that showed cytoplasmic expression including BRCA1, RAD51, SMC6L1, γH2AX, BARD1, UBC9, PIAS1 and CHK1. High level of KPNA2 was associated not only with cytoplasmic localisation of these proteins but also with their low/negative nuclear expression. Positive KPNA2 expression was associated with negative oestrogen receptor and triple-negative phenotype. Survival analysis showed that KPNA2 was associated with poor outcome (P<0.0001), but this effect was not independent of other prognostic variables.
Conclusions:
This study provides further evidence for the complexity of DDR mechanism in BC, and that KNPA2 has a role in the aberrant subcellular localisation of DDR proteins with subsequent impaired function.
Keywords: nucleocytoplasmic transport, breast cancer, tissue microarray, immunohistochemistry, reverse-phase protein array
The mechanisms of nucleocytoplasmic transport have been reported to be associated with several cellular processes, such as gene expression, progression of the cell cycle, apoptosis and signal transduction (Chook and Blobel, 2001). Nuclear transport of macromolecules is also essential for changing cellular phenotypes throughout progression and malignant cell transformation (Poon and Jans, 2005a). Karyopherin α-2 (KPNA2), an adaptor protein, is a member of the karyopherin-α protein family that has a vital role in nucleocytoplasmic transport. Previous studies have demonstrated that karyopherin-α proteins bind to cargo proteins, which contain a classical nuclear localisation signals (NLS), and import macroproteins into the nucleus, hence the name importin (Gorlich et al, 1994). However, the biological function of the different members of karyopherin-α and their role as nuclear transport proteins remain controversial. Some authors have indicated that they mediate the nuclear import of proteins (Moroianu et al, 1995; Zannini et al, 2003; Nishinaka et al, 2004; Huang et al, 2013), others have reported that KPNA2 mediates the export of response molecules to the cytoplasm (Poon and Jans, 2005b). It is also suggested that high nuclear accumulation of KPNA2 leads to cytoplasmic retention of NLS-containing cargo proteins due to defective import: the transporter factor KPNA2 is not recycled back to the cytoplasm to transport the next karyophile into the nucleus leading lack of ‘free' KPNA2 to bind its cargo in the cytoplasm (Gorlich and Mattaj, 1996). Nuclear localisation of KPNA2 in cancer is thought to be due to cellular stress, and that the nuclear retention of KPNA2 in response to cellular stress suppresses the nuclear import (Stochaj et al, 2000a).
Previous studies have demonstrated that nuclear expression of KNPA2 is associated with poor prognosis in patients with oesophageal squamous cell carcinoma (Sakai et al, 2010b), epithelial ovarian carcinomas (Zheng et al, 2010) and melanoma (Winnepenninckx et al, 2006). In breast cancer (BC), expression of KPNA2 is associated with features of aggressive behaviour such as higher tumour grade and positive lymph node (Dankof et al, 2007; Gluz et al, 2008), and poor outcome (Dahl et al, 2006). However, the mechanism of action of KPNA2 and whether its bad prognostic effect in BC is related to its direct function or through modulation of other key driver molecules remain largely unknown. In previous studies, we and others have noted that aberrant subcellular localisation of key proteins including those involved in DNA damage response (DDR) is associated with aggressive behaviour and loss-of-function phenotype (Wilson et al, 1999; Lambie et al, 2003; Rakha et al, 2008; Alshareeda et al, 2012, 2013, 2014). Cytoplasmic location of DDR proteins is also associated with aggressive features in the prostate (Mitra et al, 2009). Subsequently, we hypothesised that an active nucleocytoplasmic transport mechanism contributes to modulation of the subcellular localisation of proteins related to BC development and progression. In this study, KPNA2 protein is assessed in a large series of BC, and its expression is correlated to the subcellular locations of a large panel of relevant proteins and to BC clinicopathological features and outcome.
Materials and Methods
Study cohort
The study cohort was derived from the well-characterised Nottingham Tenovus primary breast carcinoma series. It comprised of 1249 unselected primary operable invasive tumours from female patients presenting between 1989 and 1998. To increase the number of oestrogen receptor (ER)-negative cases, an additional 245 unselected primary operable BC, from patients presenting between 1998 and 2003, were collected. As a control group, a cohort of BC from BRCA1 germline mutation carriers (n=19) was included. Patients' clinicopathological features were obtained including age, menopause status, primary tumour size, tumour type, histological grade, nodal status, lymphovascular invasion and Nottingham Prognostic Index (NPI; Rakha et al, 2006; Alshareeda et al, 2013). Survival data were collected in a prospective way including development of loco-regional and distant recurrences and mortality. BC-specific survival (BCSS) is defined as the interval from the date of primary surgery to the time of death because of BC. Death owing to other causes is considered as a censored event. Distant metastasis (DM) is defined as the interval from the date of primary surgery to the time of DM. Both of these parameters were measured in months.
Tumour characteristics have been considered for patient's managements by selecting NPI and ER status (Rakha et al, 2006). Patients with NPI excellent prognostic group (score ≤3.4) received no adjuvant therapy, but those patients with NPI > 3.4 received Tamoxifen if ER positive (±Zoladex in case the patients were pre-menopausal). On the other hand, classical cyclophosphamide, methotrexate and 5-flurouracil were used if the patients were ER negative and fit to receive chemotherapy. Patients with grade II or III tumours and had node positive were given prophylactic irradiation to the axilla following surgery (Blamey, 2002). Table 1 shows the frequency of the adjuvant treatment received by the patients in the current study.
Table 1. Frequency of treatment received in the study.
| Treatment | N (%) |
|---|---|
|
Chemotherapy | |
| No | 1471 (82.3) |
| Yes | 316 (17.7) |
|
Endocrine treatment | |
| No | 1112 (62.2) |
| Yes | 675 (37.8) |
|
Endocrine and chemotherapy | |
| No | 816 (45.7) |
| Yes | 971 (54.3) |
Abbreviations: N=number of cases; No=did not receive the treatment; Yes=received treatment.
According to the protocol used during the time of this study cohort, patients with oestrogen receptor-positive tumours were not offered systemic therapy, if their Nottingham Prognostic Index score was ≤3.4, and therefore received no adjuvant therapy.
Data on the following biomarkers were available: ER, progesterone receptor (PgR), HER2, DDR proteins (RAD51, PIAS1, BRCA1, BARD1 and CHK1), basal markers cytokeratins (CK5, CK14 and CK17), and the proliferation and cell cycle associated proteins (Ki67 and P53). The immunoreactivity, scoring and categorisation of these markers were defined in this study as previously described (Rakha et al, 2006, 2009; Aleskandarany et al, 2012; Alshareeda et al, 2012, 2013, 2014). In this series, HER2 was assessed using immunohistochemistry (IHC) and dual-colour chromogenic in situ hybridisation as previously published (Aleskandarany et al, 2012). Ki67 labelling index was assessed on whole-tumour tissue sections, and was expressed as the percentage of Ki67-positive cells among a total number of 1000 malignant cells at high power magnification ( × 400) (Aleskandarany et al, 2012). All other markers were assessed using IHC and TMA prepared sections.
This study was approved by Nottingham Research Ethics Committee 2.
KPNA2 IHC
Immunohistochemistry was performed using the Novolink Kit-polymer detection system (Leica, Newcastle, UK). Primary antibody used was KPNA2 (clone Ab84440, Abcam Ltd, Cambridge, UK) with a dilution of 1 : 400 and 60 min incubation. 3-3′Diam-inobenzidine tetrahydrochloride (Novolink DAB substrate buffer plus) was freshly prepared and used as a chromogen. The TMA sections were counterstained with haematoxylin for 6 min (Alshareeda et al, 2012).
Immunohistochemical scoring
Two TMA cores (peripheral or central) were evaluated from each tumour. Only immunostaining of invasive cancer cells within the tissue cores was considered. High-resolution digital images (Nanozoomer; Hamamatsu Photonics, Welwyn Garden City, UK) scanned at × 20 magnification were used to facilitate the manual scoring of the TMA cores via web-based interface (Distiller; Slidepath, Ltd, Dublin, Ireland). Both intensity (scores 0–3 for negative, weak moderate and strong expression, respectively) and percentage of KPNA2 expression (0–100%) were assessed, and H-score was generated (a summation of the percentage of area stained at each intensity level multiplied by the weighted intensity to produce scores from 0 to 300). KPNA2 was categorised based on the frequency histogram distributions. The cutoff point used was chosen based on the median H-score: nuclear KPNA2 (negative/low <35 and positive ⩾35 H-score).
Antibody specificity and reverse-phase protein microarray
To ensure the specificity of the antibody and to confirm the expression of KNPA2 in specific cell lines corresponding to molecular classes of BC, western blotting and reverse-phase protein microarray (RPPA) were performed as previously described (Mannsperger et al, 2010; Aleskandarany et al, 2014; Negm et al, 2014). In this study, two different cell lines were used: luminal phenotype MCF7 cell lines (characterised by positive expression of ER and BRCA1) and MDA-MB-436 (ER- and EGFR+). Cells were grown in RPMI1640 (Sigma Aldrich, Dorset, UK). For western blotting, anti KPNA2 primary antibody was used in a dilution of 1 : 1000 and incubated for 1 h at room temperature. The reaction was developed using enhanced chemiluminescence substrate (GE Healthcare Life Sciences, Buckinghamshire, UK).
For RPPA, in brief; KPNA2 antibody diluted 1 : 500 in diluent with reducing background (DAKO). In addition, β-actin (Sigma Aldrich), diluted 1 : 1000 in the same diluent, was used as a housekeeping protein to control protein loading. Protein signals were determined with background subtraction and normalisation to the internal housekeeping targets using RPPanalyzer, a module within the R statistical language on the CRAN (http://cran.r-project.org/).
Statistical analysis
SPSS 21.0 IBM statistical software (IBM Corporation, New York, NY, USA) was used for different statistical analyses. Categorical variables were analysed using χ2-test. One-way ANOVA was applied to compare the level of the expression among different BC classes (by IHC or cell lines) using post hoc test; Tukey. Associations with outcome were calculated using Kaplan–Meier curves and log-rank test. A two-sided P-value of <0.01 was considered statistically significant.
Results
Expression of KPNA2 in invasive BC
The specificity of KPNA2 primary antibody was validated using western blotting as evident by a single band at the correct protein size (Figure 1). KPNA2 showed nuclear staining, which ranged from negative/weak to strong with no cytoplasmic or membranous staining observed (Figure 2). In sporadic BC, 51% (715 out of 1393) showed nuclear expression compared with 17 out of 19 cases (90%) of the hereditary BRCA1-mutated cases that showed KPNA2 expression (P<0.001).
Figure 1.
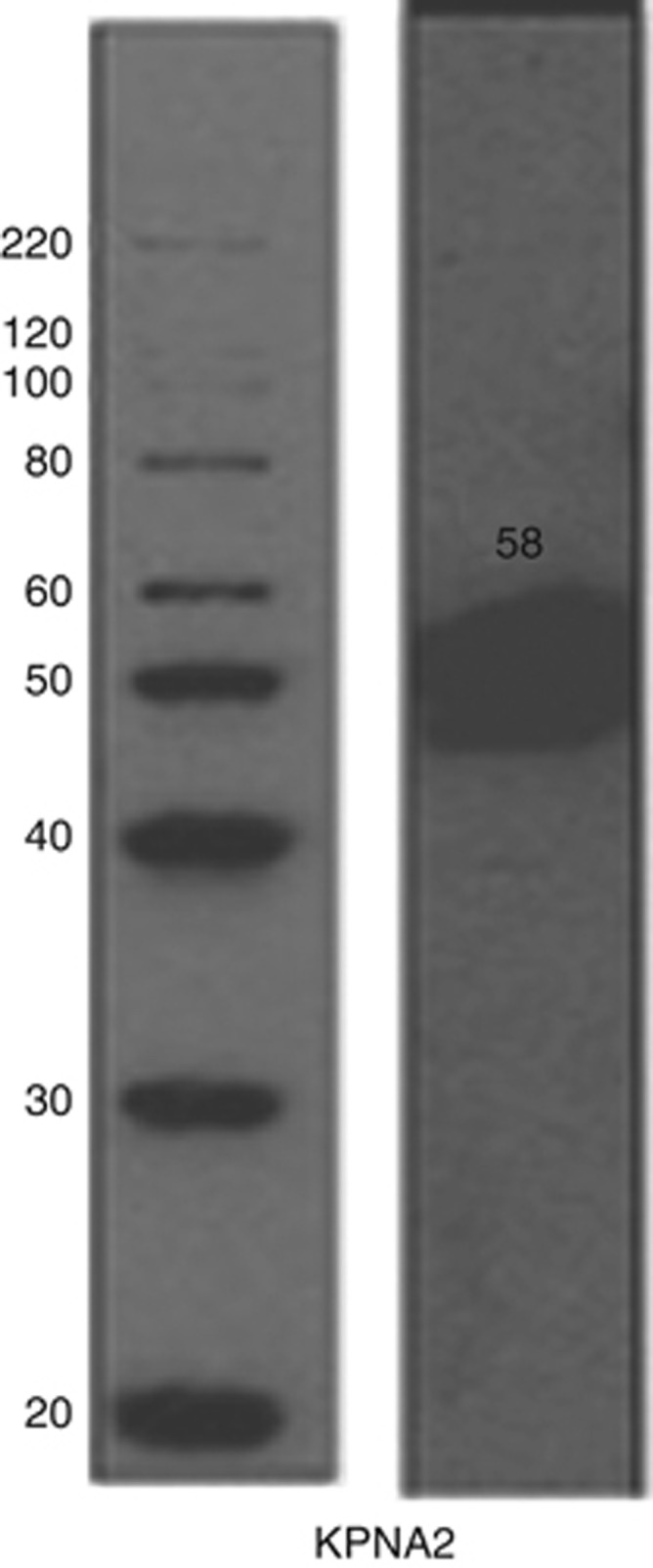
Validation of KPNA2 primary antibody by western blotting. Mixed lysates from MCF7 and MDA-MB-436 cell lines were used.
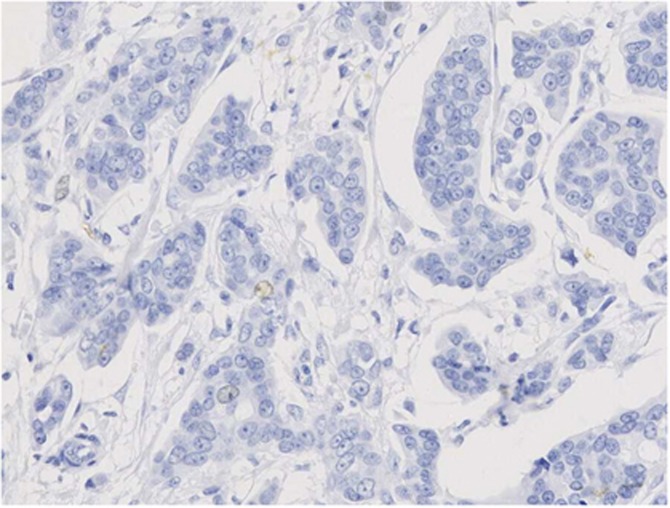
Figure 2.
Nuclear expression of KPNA2 on invasive ductal carcinoma no special type with cytoplasmic staining, which was not considered in the scoring and analysis.
Association between KPNA2 and clinicopathological features
Tables 2 summarises the association between KPNA2 and the various clinicopathological features of BC in the whole series (Table 2A) and in patients who received adjuvant therapy (Table 2B) summarises the association between KPNA2 and the various clinicopathological features of BC. KPNA2 protein expression was associated with features of aggressive behaviour and poor prognosis including younger patient' age, larger tumour size, higher tumour grade (grade III) with marked nuclear pleomorphism, lack of tubular formation and high mitotic counts (P<0.0001).
Table 2A. Relationship between KPNA2 expression and clinicopathological parameters in the whole series.
|
KPNA2 |
||||
|---|---|---|---|---|
| Parameters | Negative, N (%) | Positive, N (%) | χ2 | P-value |
|
Age (years) | ||||
| <50 | 205 (30.5) | 299 (41.9) | 20 | <0.0001 |
| ⩾50 | 468 (69.5) | 414 (58.1) | ||
|
Size (cm) | ||||
| ⩽1.5 | 239 (35.6) | 152 (21.6) | 33 | <0.0001 |
| >1.5 | 432 (64.4) | 551 (78.4) | ||
|
Stage | ||||
| 1 | 434 (64.4) | 404 (56.9) | 10 | 0.007 |
| 2 | 183 (27.2) | 218 (30.7) | ||
| 3 | 57 (8.5) | 88 (12.4) | ||
|
Grade | ||||
| 1 | 145 (21.5) | 18 (2.5) | 326 | <0.0001 |
| 2 | 263 (39) | 84 (11.8) | ||
| 3 | 266 (39.5) | 611 (85.7) | ||
|
Tubules | ||||
| 1 | 40 (6.2) | 7 (1) | 104 | <0.0001 |
| 2 | 251 (38.7) | 133 (18.9) | ||
| 3 | 357 (55.1) | 565 (80.1) | ||
|
Pleomorphism | ||||
| 1 | 15 (2.3) | 3 (0.4) | 220 | <0.0001 |
| 2 | 295 (45.7) | 77 (10.9) | ||
| 3 | 335 (51.9) | 624 (88.6) | ||
|
Mitosis | ||||
| 1 | 287 (44.3) | 57 (8.1) | 316 | <0.0001 |
| 2 | 141 (21.8) | 85 (12.1) | ||
| 3 | 220 (34) | 563 (79.9) | ||
|
NPI | ||||
| Excellent | 88 (13.1) | 10 (1.4) | 214 | <0.0001 |
| Good | 164 (24.5) | 36 (5.1) | ||
| Moderate 1 | 209 (31.2) | 248 (35.4) | ||
| Moderate 2 | 132 (19.7) | 225 (32.1) | ||
| Poor | 61 (9.1) | 136 (19.4) | ||
| Very poor | 16 (2.4) | 46 (6.6) | ||
|
Tumour type | ||||
| Invasive ductal/NST | 363 (55.3) | 586 (82.9) | 169 | <0.0001 |
| Lobular | 47 (7.2) | 11 (1.6) | ||
| Atypical medullary | 7 (1.1) | 31 (4.4) | ||
| aMixed | 206 (31.4) | 68 (9.6) | ||
| bOther | 34 (5.2) | 11 (1.6) | ||
Abbreviations: KPNA2=karyopherin α-2; NPI=Nottingham Prognostic Index; NST=no special type.
Lobular or tubular mixed breast cancers.
Mucinous, alveolar lobular, miscellaneous including metaplastic, adenoid cystic, spindle and tubulolobular.
Table 2B. Relationship between KPNA2 expression and clinicopathological parameters in patients who received adjuvant therapy.
|
KPNA2 |
||||
|---|---|---|---|---|
| Parameters | Negative, N (%) | Positive, N (%) | χ2 | P-value |
|
Age (years) | ||||
| <50 | 83 (36.9) | 142 (63.1) | 7.67 | 0.006 |
| ⩾50 | 186 (48.4) | 198 (51.6) | ||
|
Size (cm) | ||||
| ≤1.5 | 69 (60.5) | 45 (39.5) | 15.03 | <0.001 |
| >1.5 | 201 (40.5) | 295 (59.5) | ||
|
Stage | ||||
| 1 | 97 (37.6) | 161 (62.4) | 11.64 | 0.003 |
| 2 | 142 (51.8) | 132 (48.2) | ||
| 3 | 31 (39.7) | 47 (60.3) | ||
|
Grade | ||||
| 1 | 21 (84.0) | 4 (16.0) | 97.69 | <0.001 |
| 2 | 110 (74.3) | 38 (25.7) | ||
| 3 | 139 (31.8) | 298 (68.2) | ||
|
Tubules | ||||
| 1 | 4 (57.1) | 3 (42.9) | 23.42 | <0.001 |
| 2 | 97 (59.5) | 66 (40.5) | ||
| 3 | 164 (37.7) | 271 (62.3) | ||
|
Pleomorphism | ||||
| 1 | 2 (66.7) | 1 (33.3) | 58.12 | <0.001 |
| 2 | 100 (71.4) | 40 (28.6) | ||
| 3 | 162 (35.1) | 299 (64.9) | ||
|
Mitosis | ||||
| 1 | 88 (81.5) | 20 (18.5) | 117.77 | <0.001 |
| 2 | 62 (65.3) | 33 (34.7) | ||
| 3 | 115 (28.6) | 287 (71.4) | ||
|
NPI | ||||
| Excellent | 2 (100) | 0 | 48.07 | <0.001 |
| Good | 20 (100) | 0 | ||
| Moderate 1 | 105 (51.2) | 100 (48.8) | ||
| Moderate 2 | 92 (42.0) | 127 (58.0) | ||
| Poor | 46 (35.7) | 83 (64.3) | ||
| Very poor | 5 (14.7) | 29 (85.3) | ||
|
Tumour type | ||||
| Invasive ductal/NST | 166 (37.4) | 278 (62.6) | 52.35 | <0.001 |
| Lobular | 22 (78.6) | 6 (21.4) | ||
| Atypical medullary | 2 (12.5) | 14 (87.5) | ||
| aMixed | 75 (65.8) | 39 (34.2) | ||
| bOther | 2 (100) | 0 | ||
Abbreviations: KPNA2=karyopherin α-2; NPI=Nottingham Prognostic Index; NST=no special type.
Lobular or tubular mixed breast cancers.
Mucinous, alveolar lobular, miscellaneous including metaplastic, adenoid cystic, spindle and tubulolobular.
Association between KPNA2 and molecular biomarkers
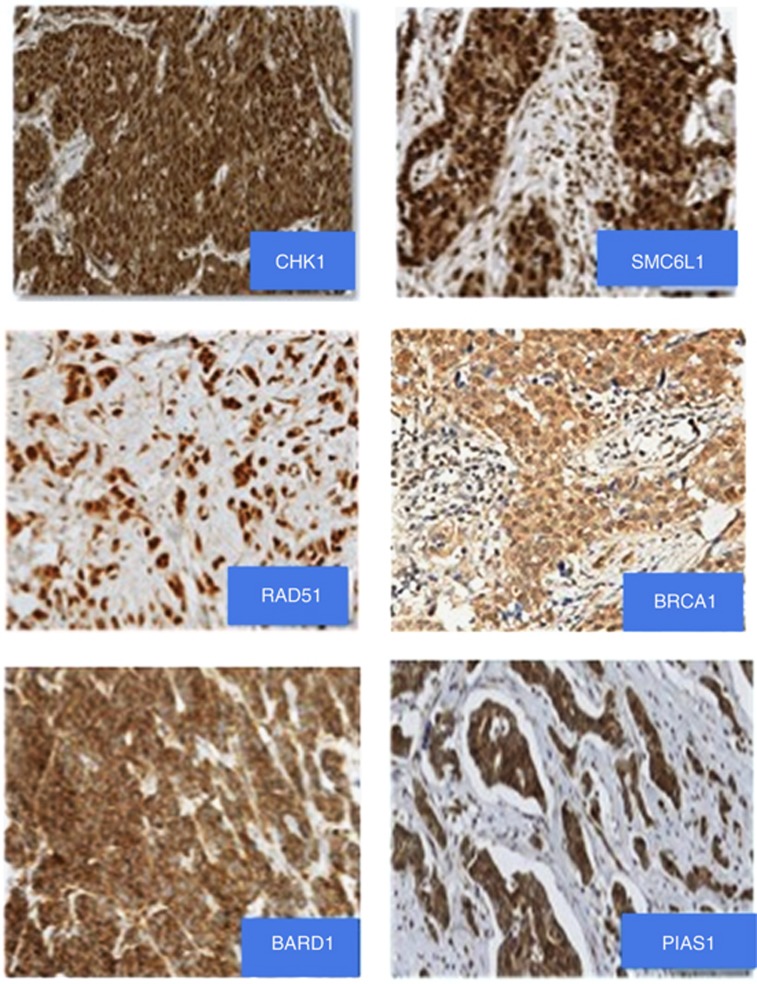
The association between the KPNA2 and other tumour biomarkers are summarised in Table 3. Figure 3 shows some examples of subcellular cytoplasmic and nuclear expression of DDR proteins. There was a significant association between KPNA2 expression and lack of ER and PgR expression, triple-negative phenotype and high expression of the proliferation marker Ki67 (P<0.0001). Regarding DDR proteins, KPNA2 showed an association with expression and subcellular localisation of markers involved in homologous recombination pathway (SMC6L1, BRCA1 and RAD51), DNA signal transducers (CHK1) and SUMOylation (SUMO) markers (PIAS1; P<0.0001). KPNA2 expression was associated with cytoplasmic localisation of these markers with a nuclear-negative/cytoplasmic-positive phenotype (nuclear export function).
Table 3. Relationship between KPNA2 with other tumour biomarkers.
|
KPNA2 |
||||
|---|---|---|---|---|
| Parameters | Negative, N (%) | Positive, N (%) | χ2 | P-value |
|
ER | ||||
| Negative | 153 (23.9) | 447 (63.8) | 215 | <0.0001 |
| Positive | 487 (76.1) | 254 (36.2) | ||
|
PgR | ||||
| Negative | 234 (37.7) | 473 (71.5) | 148 | <0.0001 |
| Positive | 387 (62.3) | 189 (28.5) | ||
|
Triple negative | ||||
| Negative | 533 (85.3) | 373 (55.8) | 134 | <0.0001 |
| Positive | 92 (14.7) | 296 (44.2) | ||
|
HER2 | ||||
| Negative | 585 (90.4) | 521 (76.6) | 46 | <0.0001 |
| Positive | 62 (9.6) | 159 (23.4) | ||
|
Ki67 | ||||
| Negative | 306 (54.2) | 105 (17) | 180 | <0.0001 |
| Positive | 259 (45.8) | 513 (83) | ||
|
CK18 | ||||
| Negative | 44 (8.3) | 109 (24.4) | 47.7 | <0.0001 |
| Positive | 486 (91.7) | 337 (75.6) | ||
|
RAD51 | ||||
| n−c− | 35 (9.6) | 14 (3.2) | 38 | <0.0001 |
| n+c+ | 121 (33.3) | 103 (23.4) | ||
| n−c+ | 187 (51.5) | 313 (71) | ||
| n+c− | 20 (5.5) | 11 (2.5) | ||
|
BRCA1 | ||||
| n−c− | 119 (22.9) | 218 (36.9) | 96 | <0.0001 |
| n+c+ | 105 (20.2) | 64 (10.8) | ||
| n−c+ | 107 (20.6) | 209 (35.4) | ||
| n+c− | 189 (36.3) | 99 (16.8) | ||
|
BARD1 | ||||
| n−c− | 123 (21.1) | 143 (21.5) | 3 | 0.4 |
| n+c+ | 46 (7.9) | 67 (10.1) | ||
| n−c+ | 413 (71) | 455 (68.3) | ||
| n+c− | 0 | 1 (0.2) | ||
|
CHK1 | ||||
| n−c− | 39 (9.2) | 17 (3.7) | 27 | <0.0001 |
| n+c+ | 107 (25.2) | 81 (17.7) | ||
| n−c+ | 259 (60.9) | 348 (76.1) | ||
| n+c− | 20 (4.7) | 11 (2.4) | ||
|
PIAS1 | ||||
| n−c− | 88 (22.4) | 50 (10.9) | 45 | <0.0001 |
| n+c+ | 46 (11.7) | 28 (6.1) | ||
| n−c+ | 239 (60.8) | 373 (81.3) | ||
| n+c− | 20 (5.1) | 8 (1.7) | ||
|
γH2AX | ||||
| n−c− | 9 (1.8) | 11 (1.7) | 47 | <0.0001 |
| n+c+ | 390 (76.5) | 501 (78.2) | ||
| n−c+ | 38 (7.5) | 100 (15.6) | ||
| n+c− | 73 (14.3) | 29 (4.5) | ||
|
SMC6L1 | ||||
| n−c− | 135 (23.3) | 84 (12.8) | 39 | <0.0001 |
| n+c+ | 277 (47.8) | 365 (55.6) | ||
| n−c+ | 78 (13.4) | 138 (21) | ||
| n+c− | 90 (15.5) | 69 (10.5) | ||
|
UBC9 | ||||
| n−c− | 175 (30.1) | 157 (23.2) | 57 | <0.0001 |
| n+c+ | 238 (40.9) | 279 (41.3) | ||
| n−c+ | 104 (17.9) | 217 (32.1) | ||
| n+c− | 65 (11.2) | 23 (3.4) | ||
Abbreviations: c=cytoplasmic; ER=oestrogen receptor; KPNA2=karyopherin α-2; N=number of cases; n=nuclear; PgR=progesterone receptor; ‘−'=negative, ‘+'=positive.
Figure 3.
Immunostaining of key DDR proteins in BC showing subcellular localisation with nuclear and cytoplasmic expressions. Magnification × 20.
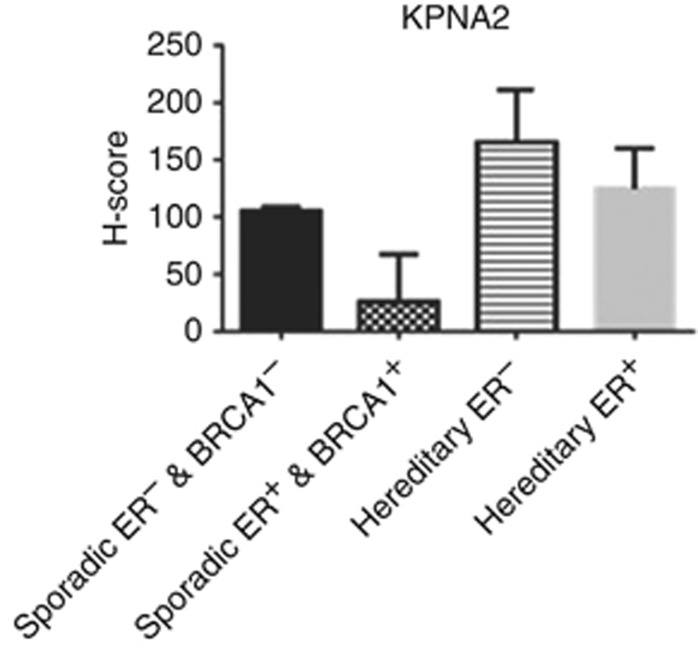
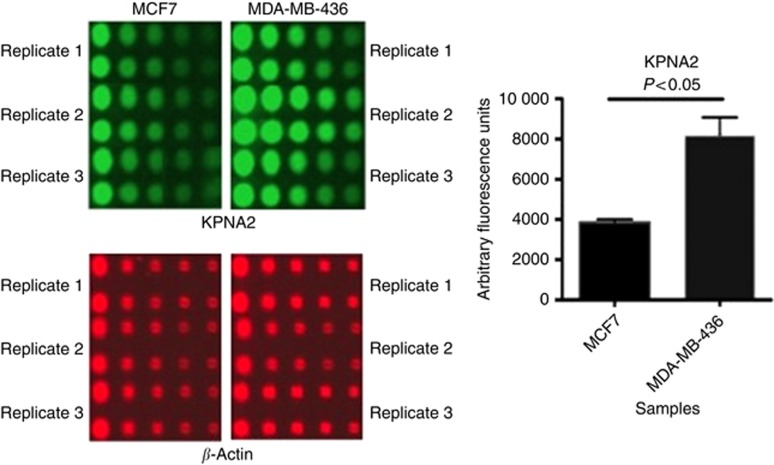
When BC was classified into different molecular classes based on BRCA1 and ER status, high KPNA2 expression was identified in BRCA1-negative/ER-negative phenotype and in BRCA1-mutated compared with BRCA1-positive/ER-positive classes (P<0.0001; Figure 4). RPPA was used to evaluate the expression levels of KPNA2 in the cell lines corresponding to BC molecular classes used in this study. RPPA confirmed the IHC results of KPNA2 and demonstrated lower levels of the expression of KPNA2 in MCF7 cell lines (ER+/BRCA1+) than in MDA-MB-436 (ER−/BRCA1−) cell lines (Figure 5).
Figure 4.
KPNA2 protein level detected by IHC on TMA. Each bar represents different class based on hereditary or sporadic BRCA1 and ER status. Error bars represent mean (s.d.) was created on H-score (range 0–300). A=sporadic cases (ER− and BRCA1−) vs sporadic cases (ER+ and BRCA1+), P<0.0001; B=sporadic cases (ER− and BRCA1−) vs hereditary cases (ER−), P=0.07; C=sporadic cases (ER− and BRCA1−) vs hereditary cases (ER+), P=0.97; D=sporadic cases (ER+ and BRCA1+) vs hereditary cases (ER−), P<0.0001; E=sporadic cases (ER+ and BRCA1+) vs hereditary cases (ER+), P=0.14; and F=hereditary cases (ER−) vs hereditary cases (ER+), P=0.9. ANOVA test was used for each marker within the classes.
Figure 5.
KPNA2 protein levels detected by RPPA in MCF7 and MDA-MB-436 cells. For image of nitrocellulose slide spotted with different cell lysates; the red fluoresence represents detection of β-actin, whereas green fluorsence for KPNA2. Images of scanned nitrocellulose slides printed with extracted protein from cell lines and probed with the antibodies against the target proteins. Five two-fold dilutions of each sample were printed in duplicate. Background was subtracted and the intensity of each spot was normalised to its corresponding β-actin level. Six experimental replicates for each cell linewere used. Error bars represent mean (s.d.). As shown from the graph the expression level of KPNA2 was lower in MCF7 compared with MDA-MB-436 (P<0.05).
Expression of KPNA2 and patient outcome
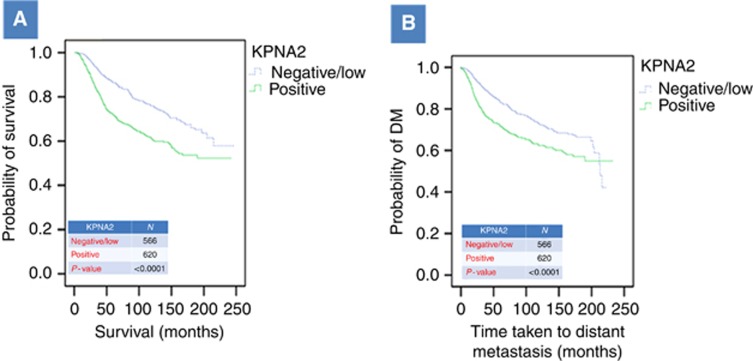
Positive expression of KPNA2 demonstrated an association with poorer outcome in terms of shorter BCSS and shorter DM (P<0.0001; Figure 6A and B respectively). When cases were subclassified according to grade and ER status, KPNA2 maintained its association with outcome in ER-positive group (P<0.001) but not in the ER-negative or in the different tumour grades. Multivariate Cox regression analysis including stage, grade and size indicated that KNPA2 was not independent predictor of outcome (Table 4).
Figure 6.
Association between KPNA2 expression and BCSS (A) and DM (B). N, number of cases. Only patients who died from BC were considered. Cutoff used for KPNA2 was the median of H-score, where negative/low <35 and positive ⩾35.
Table 4. Cox regression analyses for predictors of outcome (BCSS and DM).
|
BCSS |
DM |
|||||||
|---|---|---|---|---|---|---|---|---|
|
95% CI |
95% CI |
|||||||
| Parameters | P-value | HR | Lower | Upper | P-value | HR | Lower | Upper |
| Tumour stage | <0.0001 | 1.92 | 1.7 | 2.2 | <0.0001 | 1.8 | 1.6 | 1.9 |
| Tumour grade | <0.001 | 1.87 | 1.5 | 2.3 | 0.034 | 1.2 | 1.01 | 1.3 |
| Tumour size | 0.001 | 1.07 | 1.03 | 111 | 0.206 | 1.03 | 0.98 | 1.09 |
| KPNA2 | 0.221 | 1.15 | 0.87 | 1.4 | 0.227 | 1.2 | 0.92 | 1.4 |
|
Adjuvant therapy subgroup | ||||||||
| Tumour stage | <0.0001 | 2.05 | 1.7 | 2.5 | <0.0001 | 1.8 | 1.6 | 1.9 |
| Tumour grade | <0.001 | 2.30 | 1.6 | 3.3 | 0.034 | 1.2 | 1.01 | 1.3 |
| Tumour size | 0.087 | 1.18 | 0.98 | 1.3 | 0.206 | 1.03 | 0.98 | 1.09 |
| KPNA2 | 0.300 | 1.13 | 0.87 | 1.6 | 0.227 | 1.2 | 0.92 | 1.4 |
|
No adjuvant therapy subgroup | ||||||||
| Tumour stage | 0.006 | 1.64 | 1.1 | 2.3 | 0.001 | 1.6 | 1.2 | 2.1 |
| Tumour grade | <0.001 | 1.75 | 1.3 | 2.4 | 0.206 | 1.1 | 0.9 | 1.4 |
| Tumour size | <0.001 | 1.58 | 1.2 | 2.0 | 0.004 | 1.3 | 1.1 | 1.6 |
| KPNA2 | 0.826 | 1.05 | 0.7 | 1.7 | 0.168 | 1.3 | 0.9 | 1.8 |
Abbreviations: BCSS=breast cancer-specific survival; CI=confidence interval; DM=distant metastasis; HR=hazard ratio; KPNA2=karyopherin α-2.
Discussion
Studying the localisation of proteins in several subcellular compartments offers valuable insight into understanding state of activation, interaction networks and the biological functions of such proteins (Fu et al, 2010). Understanding the mechanisms underlying aberrant subcellular location of proteins may help in identification of potential targets and drug discovery (Chou and Shen, 2008). The present study investigated the expression of KPNA2 protein as a potential marker involved in the regulation of subcellular location of key proteins in BC. A large panel of biomarkers including DDR proteins was assessed. Although biomarkers involved in DDR pathways are typically located and function in the nucleus, some proteins such as BRCA1, BARD1, PIAS1, RAD51 and CHK1 also show cytoplasmic expression as detected by IHC. In previous studies, we and others noted that cytoplasmic expression of these markers is associated with poor prognostic features, such as higher grade and hormone receptor negativity (Wilson et al, 1999; Lambie et al, 2003; Rakha et al, 2008; Alshareeda et al, 2012, 2013, 2014). The significance and underlying mechanisms of this aberrant subcellular localisation remain to be defined.
It has been reported that overexpression of the nucleocytoplasmic transport marker KPNA2 is associated with progression of BC (Wang et al, 2005; Sotiriou et al, 2006). Gluz et al (2008) have demonstrated that overexpression of nuclear KPNA2 in BCs was significantly associated with aggressive tumour features such as higher grade, negative hormone receptor status, HER2/neu expression, shorter overall survival and DFI, although in this study the effect of KPNA2 on patient's outcome was not an independent prognostic marker. However, the mechanism underlying the association between KPNA2 nuclear expression and aggressive features in BC remains to be determined. To our knowledge, this is the first study to investigate the role of KPNA2 in the subcellular localisation of key proteins related to BC development and progression with particular emphasis on DDR proteins using a large series including different molecular classes.
Our results indicate that KPNA2 nuclear expression is associated with cytoplasmic and not the nuclear expression of the studied DDR proteins. Although there is no direct evidence that links this observation to an export function of KPNA2, it is more likely that high nuclear accumulation of KPNA2 leads to increased amounts of DDR and other cargo proteins in the cytoplasm due to defective import (Gorlich and Mattaj, 1996; Jans et al, 2000). To get across the nuclear membrane and deliver its NLS-containing cargo, KPNA2 may possibly bind importin through its N-terminal IBB domain (Gorlich et al, 1996; Nishinaka et al, 2004). In the nucleus, the components of these complex, including the cargo, end up dissociated, thereby enabling the recycling of KPNA2 returning to the cytoplasm (Moroianu et al, 1996). Lack of ‘free' KPNA2 to bind its cargo in the cytoplasm may be responsible for its cytoplasmic retention. It was also suggested that nuclear retention of KPNA2 in response to cellular stress suppresses nuclear import (Stochaj et al, 2000b). It was also reported that components of the nuclear transport machinery appear to be differentially expressed in malignant cells with alteration of nuclear import/export through different regulatory mechanisms (Poon and Jans, 2005b).
Negative nuclear and positive cytoplasmic expression of different markers such as RAD51, BARD1, BRCA1 CHK1 and PIAS1 was highly associated with KPNA2, and the mechanism inducing cytoplasmic localisation is unclear. Interestingly, in the present study, it is likely that these proteins bind to or interact with another marker, leading to change in their role when expressed in the cytoplasm. Although, ions, small molecules and small proteins (<20 kDa) are able to pass easily by diffusion through NPCs, the macromolecules (>40 kDa) with the appropriate signals are restricted by NPCs (Weis, 2003; Wente and Rout, 2010). However, in the present study, the exclusion of fusion proteins from the nucleus to the cytoplasm is not size dependent, because some markers have >40 kDa such as BRCA1 (∼220 kDa), PIAS1 (71 kDa) and CHK1 (54 kDa), and this can be explained by uncontrol travel of macromolecule from and to the nucleus owing to a defect in NPCs.
In theory, as the nuclear import of the proteins investigated in this study (DDR and SUMO markers) is dependent on interaction with KPNA2, then the expression of KPNA2 is expected to increase and have an effect on their nuclear localisation. Thus, high levels of KPNA2 with other proteins in the cytoplasm may result in their low expression in the nucleus. The expression of these markers (such as RAD51) in the cytoplasm may have a role in the poor prognosis of BC. In this study, there is a strong indication that the poor prognosis of patients is largely associated with the negative nuclear and positive cytoplasmic expression of any marker tested, such as RAD51.
It has been proposed that BRCA1 is one of the proteins whose subcellular distribution is controlled by NLS in direct interaction with KPNA2, and therefore influences DNA repair and cell cycle (Thakur et al, 1997; Narod and Foulkes, 2004). The observation that BRCA1 mutant deficiency of both NLSs can be observed in the nucleus (Wilson et al, 1997; Huber et al, 2001) has resulted in the identification of an alternative process in the importing of BRCA1 (Fabbro et al, 2002). The value of the two alternative pathways continues to be identified. Although, in this study, KPNA2 expression is in association with cytoplasmic localisation of BRCA1 and other key DDR such as RAD51, thereby leading to dysfunction of these nuclear proteins with morphological and immunophenotypical changes, which is similar to that seen in tumours featuring ‘BRCA-ness' phenotype. Although this could suggest that KPNA2 IHC can be used as a surrogate marker for BRCA-ness and subsequently be used as a predictive marker for response to poly-ADP ribose polymerase (PARP) inhibitors, the findings in this study do not provide sufficient evidence to support this suggestion. Previous trials of PARP inhibitors showed significant effect in patients with BRCA1 germline mutation, but no such effect was noted in sporadic tumours even those showing morphological and immunophenotypical similarities to BRCA1-associated tumours (BRCA-ness) (Chen, 2011).
KPNA2 mediates the nucleocytoplasmic transport of some tumour suppressors (Zannini et al, 2003; Nishinaka et al, 2004). KPNA2 also controls both the nuclear localisation of the MRN complexes with the formation of radiation-induced focus (Tseng et al, 2005); therefore, the suppression of KPNA2 prevents the double-stranded breaks (induced by ionising radiation), which will lead to suppression of NSB-mediated DNA repair pathway. Tseng et al (2005) have studied the role of NBS1 NLS in vivo using immunofluorescence, and found that mutation in NBS1 resulted in cytoplasmic redistribution and a decrease of IR-induced nuclear focus formation of NBS1. Therefore, this finding together with the fact that mutation of NLS disrupt the interaction with KPNA2 highlight the value of NBS1 NLS for its binding to KPNA2 and therefore for NBS1 nuclear translocation (Teng et al, 2006).
Poor clinical outcome in BC is linked to overexpression of cell cycle genes due to cell proliferation (Dai et al, 2005). Dai et al (2005) have examined the molecular functions and biological processes of 50 prognostic genes, including KPNA2 and E2F1, and found that the majority of overexpressed genes in tumours with a poor outcome are cell cycle-associated genes (Dai et al, 2005). Interestingly, in the current study, there was a strong correlation between KPNA2 expression and Ki67, and this may highlight the essential role of KPNA2 in the proliferation signalling of BC. Sakai et al (2010a) supported this finding but in oesophageal squamous cell carcinoma. Therefore, the KPNA2 expression could possibly be related to the induction of proliferation and the progression of BC. However, further studies are mandatory to establish this hypothesis. Despite the association between KPNA2 and outcome in univariate analysis, it did not maintain its prognostic value when other well-established prognostic variables were included. This can be explained by the correlation between KPNA2 and other prognostic variables such as histological grade, proliferation, stage and ER status, and that the effect of KPNA2 on patient' outcome is dependent on these variables.
In this study, there was a strong correlation between KPNA2 and patient's age; however, this was mainly a reflection of difference in tumour grade and ER status. When the cohort was stratified based on grade and ER status, the association between KPNA2 and age became insignificant. In gastric adenocarcinoma in which there is no correlation between age and grade or ER status, Li et al (2013) did not find associations between KPNA2 expression and patient' age. In this study, there was no correlation between KPNA2 and BARD1. This can be explained by the predominant cytoplasmic expression of this protein with very small number of cases showing nuclear expression (9%).
Two different methods were used in this study to evaluate the expression level of KPNA2. Interestingly, the RPPA results were in accordance with those results obtained from IHC. The RPPA study confirmed higher expression level in cells deficient in BRCA1 (MDA-MB-436) compared with the BRCA1-proficient cells (MCF7).
In conclusion, KNPA2 nuclear expression is association aggressive features in BC, and this may be related to aberrant cytoplasmic localisation of key nuclear proteins. Further studies of the impact of KPNA2 on the aberrant subcellular localisation of DDR and other key proteins, and identifying alternative mechanism for nuclear import of these proteins in BC, may identify novel therapeutic targets.
References
- Aleskandarany MA, Green AR, Benhasouna AA, Barros FF, Neal K, Reis-Filho JS, Ellis IO, Rakha EA. Prognostic value of proliferation assay in the luminal, HER2-positive, and triple-negative biologic classes of breast cancer. Breast Cancer Res. 2012;14:R3. doi: 10.1186/bcr3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleskandarany MA, Negm OH, Green AR, Ahmed MA, Nolan CC, Tighe PJ, Ellis IO, Rakha EA. Epithelial mesenchymal transition in early invasive breast cancer: an immunohistochemical and reverse phase protein array study. Breast Cancer Res Treat. 2014;145:339–348. doi: 10.1007/s10549-014-2927-5. [DOI] [PubMed] [Google Scholar]
- Alshareeda AT, Negm OH, Albarakati N, Green AR, Nolan C, Sultana R, Madhusudan S, Benhasouna A, Tighe P, Ellis IO, Rakha EA. Clinicopathological significance of KU70/KU80, a key DNA damage repair protein in breast cancer. Breast Cancer Res Treat. 2013;139:301–310. doi: 10.1007/s10549-013-2542-x. [DOI] [PubMed] [Google Scholar]
- Alshareeda AT, Negm OH, Green AR, Nolan C, Tighe P, Albarakati N, Sultana R, Madhusudan S, Ellis IO, Rakha EA. SUMOylation proteins in breast cancer. Breast Cancer Res Treat. 2014;144:519–530. doi: 10.1007/s10549-014-2897-7. [DOI] [PubMed] [Google Scholar]
- Alshareeda AT, Rakha EA, NOLAN CC, Ellis IO, Green AR. Fatty acid binding protein 7 expression and its sub-cellular localization in breast cancer. Breast Cancer Res Treat. 2012;134:519–529. doi: 10.1007/s10549-012-2083-8. [DOI] [PubMed] [Google Scholar]
- Blamey RW. Estimation of prognosis of the individual with primary breast cancer and its applications. Scand J Surg. 2002;91:273–278. doi: 10.1177/145749690209100311. [DOI] [PubMed] [Google Scholar]
- Chen A. PARP inhibitors: its role in treatment of cancer. Chin J Cancer. 2011;30:463–471. doi: 10.5732/cjc.011.10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook Y, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat Protoc. 2008;3:153–162. doi: 10.1038/nprot.2007.494. [DOI] [PubMed] [Google Scholar]
- Dahl E, Kristiansen G, Gottlob K, Klaman I, Ebner E, Hinzmann B, Hermann K, Pilarsky C, Durst M, Klinkhammer-Schalke M, Blaszyk H, Knuechel R, Hartmann A, Rosenthal A, Wild PJ. Molecular profiling of laser-microdissected matched tumor and normal breast tissue identifies karyopherin alpha2 as a potential novel prognostic marker in breast cancer. Clin Cancer Res. 2006;12:3950–3960. doi: 10.1158/1078-0432.CCR-05-2090. [DOI] [PubMed] [Google Scholar]
- Dai H, Van't Veer L, Lamb J, He YD, Mao M, Fine BM, Bernards R, van de Vijver M, Deutsch P, Sachs A, Stoughton R, Friend S. A cell proliferation signature is a marker of extremely poor outcome in a subpopulation of breast cancer patients. Cancer Res. 2005;65:4059–4066. doi: 10.1158/0008-5472.CAN-04-3953. [DOI] [PubMed] [Google Scholar]
- Dankof A, Fritzsche FR, Dahl E, Pahl S, Wild P, Dietel M, Hartmann A, Kristiansen G. KPNA2 protein expression in invasive breast carcinoma and matched peritumoral ductal carcinoma in situ. Virchows Arch. 2007;451:877–881. doi: 10.1007/s00428-007-0513-5. [DOI] [PubMed] [Google Scholar]
- Fabbro M, Rodriguez JA, Baer R, Henderson BR. BARD1 induces BRCA1 intranuclear foci formation by increasing RING-dependent BRCA1 nuclear import and inhibiting BRCA1 nuclear export. J Biol Chem. 2002;277:21315–21324. doi: 10.1074/jbc.M200769200. [DOI] [PubMed] [Google Scholar]
- Fu G, Song XC, Yang X, Peng T, Wang Y, Zhou GW. Protein subcellular localization profiling of breast cancer cells by dissociable antibody microarray staining. Proteomics. 2010;10:1536–1544. doi: 10.1002/pmic.200900585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluz O, Wild P, Meiler R, Diallo-Danebrock R, Ting E, Mohrmann S, Schuett G, Dahl E, Fuchs T, Herr A, Gaumann A, Frick M, Poremba C, Nitz UA, Hartmann A. Nuclear karyopherin alpha2 expression predicts poor survival in patients with advanced breast cancer irrespective of treatment intensity. Int J Cancer. 2008;123:1433–1438. doi: 10.1002/ijc.23628. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Henklein P, Laskey RA, Hartmann E. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Huang L, Wang HY, Li JD, Wang JH, Zhou Y, Luo RZ, Yun JP, Zhang Y, Jia WH, Zheng M. KPNA2 promotes cell proliferation and tumorigenicity in epithelial ovarian carcinoma through upregulation of c-Myc and downregulation of FOXO3a. Cell Death Dis. 2013;4:e745. doi: 10.1038/cddis.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber LJ, Yang TW, Sarkisian CJ, Master SR, Deng CX, Chodosh LA. Impaired DNA damage response in cells expressing an exon 11-deleted murine Brca1 variant that localizes to nuclear foci. Mol Cell Biol. 2001;21:4005–4015. doi: 10.1128/MCB.21.12.4005-4015.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: a key control point in nuclear transport. Bioessays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Lambie H, Miremadi A, Pinder S, Bell J, Wencyk P, Paish E, Macmillan R, Ellis I. Prognostic significance of BRCA1 expression in sporadic breast carcinomas. J Pathol. 2003;200:207–213. doi: 10.1002/path.1348. [DOI] [PubMed] [Google Scholar]
- Li C, Ji L, Ding Z-Y, Zhang Q-D, Huang G-R. Overexpression of KPNA2 correlates with poor prognosis in patients with gastric adenocarcinoma. Tumor Biol. 2013;34:1021–1026. doi: 10.1007/s13277-012-0641-7. [DOI] [PubMed] [Google Scholar]
- Mannsperger HA, Gade S, Henjes F, Beissbarth T, Korf U. RPPanalyzer: analysis of reverse-phase protein array data. Bioinformatics. 2010;26:2202–2203. doi: 10.1093/bioinformatics/btq347. [DOI] [PubMed] [Google Scholar]
- Mitra A, Jameson C, Barbachano Y, Sanchez L, Kote-Jarai Z, Peock S, Sodha N, Bancroft E, Fletcher A, Cooper C. Overexpression of RAD51 occurs in aggressive prostatic cancer. Histopathology. 2009;55:696–704. doi: 10.1111/j.1365-2559.2009.03448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A. The binding site of karyopherin alpha for karyopherin beta overlaps with a nuclear localization sequence. Proc Natl Acad Sci USA. 1996;93:6572–6576. doi: 10.1073/pnas.93.13.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- Negm OH, Mannsperger HA, Mcdermott EM, Drewe E, Powell RJ, Todd I, Fairclough LC, Tighe PJ. A pro-inflammatory signalome is constitutively activated by C33Y mutant TNF receptor 1 in TNF receptor-associated periodic syndrome (TRAPS) Eur J Immunol. 2014;44:2096–2110. doi: 10.1002/eji.201344328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishinaka Y, Masutani H, Oka S, Matsuo Y, Yamaguchi Y, Nishio K, Ishii Y, Yodoi J. Importin alpha1 (Rch1) mediates nuclear translocation of thioredoxin-binding protein-2/vitamin D(3)-up-regulated protein 1. J Biol Chem. 2004;279:37559–37565. doi: 10.1074/jbc.M405473200. [DOI] [PubMed] [Google Scholar]
- Poon IK, Jans DA. Regulation of nuclear transport: central role in development and transformation. Traffic. 2005;6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- Poon IK, Jans DA. Regulation of nuclear transport: central role in development and transformation. Traffic. 2005;6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- Rakha EA, El-Sheikh SE, Kandil MA, El-Sayed ME, Green AR, Ellis IO. Expression of BRCA1 protein in breast cancer and its prognostic significance. Hum Pathol. 2008;39:857–865. doi: 10.1016/j.humpath.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Rakha EA, Elsheikh SE, Aleskandarany MA, Habashi HO, Green AR, Powe DG, El-Sayed ME, Benhasouna A, Brunet JS, Akslen LA, Evans AJ, Blamey R, Reis-Filho JS, Foulkes WD, Ellis IO. Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res. 2009;15:2302–2310. doi: 10.1158/1078-0432.CCR-08-2132. [DOI] [PubMed] [Google Scholar]
- Rakha EA, Putti TC, Abd El-Rehim DM, Paish C, Green AR, Powe DG, Lee AH, Robertson JF, Ellis IO. Morphological and immunophenotypic analysis of breast carcinomas with basal and myoepithelial differentiation. J Pathol. 2006;208:495–506. doi: 10.1002/path.1916. [DOI] [PubMed] [Google Scholar]
- Sakai M, Sohda M, Miyazaki T, Suzuki S, Sano A, Tanaka N, Inose T, Nakajima M, Kato H, Kuwano H. Significance of karyopherin-{alpha} 2 (KPNA2) expression in esophageal squamous cell carcinoma. Anticancer Res. 2010;30:851–856. [PubMed] [Google Scholar]
- Sakai M, Sohda M, Miyazaki T, Suzuki S, Sano A, Tanaka N, Inose T, Nakajima M, Kato H, Kuwano H. Significance of karyopherin-α 2 (KPNA2) expression in esophageal squamous cell carcinoma. Anticancer Res. 2010;30:851–856. [PubMed] [Google Scholar]
- Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, Desmedt C, Larsimont D, Cardoso F, Peterse H, Nuyten D, Buyse M, van de VIjver MJ, Bergh J, Piccart M, Delorenzi M. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- Stochaj U, Rassadi R, Chiu J. Stress-mediated inhibition of the classical nuclear protein import pathway and nuclear accumulation of the small GTPase Gsp1p. FASEB J. 2000;14:2130–2132. doi: 10.1096/fj.99-0751fje. [DOI] [PubMed] [Google Scholar]
- Stochaj U, Rassadi R, Chiu J. Stress-mediated inhibition of the classical nuclear protein import pathway and nuclear accumulation of the small GTPase Gsp1p. FASEB J. 2000;14:2130–2132. doi: 10.1096/fj.99-0751fje. [DOI] [PubMed] [Google Scholar]
- Teng SC, Wu KJ, Tseng SF, Wong CW, Kao L. Importin KPNA2, NBS1, DNA repair and tumorigenesis. J Mol Histol. 2006;37:293–299. doi: 10.1007/s10735-006-9032-y. [DOI] [PubMed] [Google Scholar]
- Thakur S, Zhang HB, Peng Y, Le H, Carroll B, Ward T, Yao J, Farid LM, Couch FJ, Wilson RB, Weber BL. Localization of BRCA1 and a splice variant identifies the nuclear localization signal. Mol Cell Biol. 1997;17:444–452. doi: 10.1128/mcb.17.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SF, Chang CY, Wu KJ, Teng SC. Importin KPNA2 is required for proper nuclear localization and multiple functions of NBS1. J Biol Chem. 2005;280:39594–39600. doi: 10.1074/jbc.M508425200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-Van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CA, Payton MN, Elliott GS, Buaas FW, Cajulis EE, Grosshans D, Ramos L, Reese DM, Slamon DJ, Calzone FJ. Differential subcellular localization, expression and biological toxicity of BRCA1 and the splice variant BRCA1-delta11b. Oncogene. 1997;14:1–16. doi: 10.1038/sj.onc.1200924. [DOI] [PubMed] [Google Scholar]
- Wilson CA, Ramos L, Villasenor MR, Anders KH, Press MF, Clarke K, Karlan B, Chen J-J, Scully R, Livingston D. Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat Genet. 1999;21:236–240. doi: 10.1038/6029. [DOI] [PubMed] [Google Scholar]
- Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, Avril MF, Ortiz Romero PL, Robert T, Balacescu O, Eggermont AM, Lenoir G, Sarasin A, Tursz T, Van Den Oord JJ, Spatz A. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- Zannini L, Lecis D, Lisanti S, Benetti R, Buscemi G, Schneider C, Delia D. Karyopherin-alpha2 protein interacts with Chk2 and contributes to its nuclear import. J Biol Chem. 2003;278:42346–42351. doi: 10.1074/jbc.M303304200. [DOI] [PubMed] [Google Scholar]
- Zheng M, Tang L, Huang L, Ding H, Liao W-T, Zeng M-S, Wang H-Y. Overexpression of karyopherin-2 in epithelial ovarian cancer and correlation with poor prognosis. Obstet Gynecol. 2010;116:884–891. doi: 10.1097/AOG.0b013e3181f104ce. [DOI] [PubMed] [Google Scholar]