Abstract
Metastatic lung cancer is one of the most lethal forms of cancer and molecular pathways driving metastasis are still not clearly elucidated. Metastatic cancer cells undergo an epithelial-mesenchymal transition (EMT) where they lose their epithelial properties and acquire a migratory and invasive phenotype. Here we identify that expression of microRNAs from the miR-200 family and the miR-183~96~182 cluster are significantly co-repressed in non-small cell lung cancer (NSCLC) cell lines and primary tumors from multiple TCGA data sets with high EMT scores. Ectopic expression of the miR-183~96~182 cluster inhibited cancer cell migration and invasion, while its expression was tightly modulated by miR-200. We identified Foxf2 as a common, novel and direct target of both these microRNA families. Foxf2 expression tightly correlates with the transcription factor Zeb1 and is elevated in mesenchymal-like metastatic lung cancer cells. Foxf2 expression induced robust EMT, migration, invasion and metastasis in lung cancer cells, whereas Foxf2 inhibition significantly repressed these phenotypes. We also demonstrated that Foxf2 transcriptionally represses E-Cadherin and miR-200, independent of Zeb1, to form a double negative feedback loop. We therefore identified a novel mechanism whereby the miR-200 family and the miR-183~96~182 cluster inhibit lung cancer invasion and metastasis by targeting Foxf2.
Keywords: miR-200, miR-183-96-182, microRNA, Foxf2, EMT, Metastasis, Zeb1, lung cancer, invasion
Introduction
Lung cancer is the leading cause of cancer related deaths in the world, primarily because of aggressive metastatic progression60. The high mortality for this disease derives partly from limited therapeutic interventions and necessitates an understanding of the molecular pathways driving metastasis. One of the mechanisms contributing to metastasis in epithelial tumors is epithelial-mesenchymal transition (EMT). During this process a subset of cells from the primary tumor acquires invasive and migratory properties with loss of epithelial cell polarity and intercellular adhesion29, 76. A series of transcription factors/repressors regulate EMT by inhibiting epithelial genes like E-Cadherin and miR-200 and inducing mesenchymal markers like vimentin. The Zeb, Snail and Twist families of transcriptional repressors are the most well documented EMT regulators in different cancers69, 71, 72. To elucidate the molecular mechanisms underlying EMT and metastasis our laboratory has developed a syngeneic tumor model using cell lines isolated from a genetically-engineered murine model with mutant KRas and p53 77. Syngeneic tumors in this model vary in their metastatic potential. MicroRNA expression profiling revealed that the microRNA-200 family (miR-200) is repressed in the highly metastatic tumors and exogenous expression of miR-200 in the metastatic cells resulted in their inability to undergo EMT and metastasize 2, 23, 73.
MicroRNAs are 22 nucleotide non-coding RNAs that inhibit target genes by degrading the message or inhibiting translation4, 5, 16, 19, 59. Work from our lab and others has demonstrated that in many tumor types the microRNA-200 family (miR-200) modulates EMT by regulating the Zeb family of transcriptional repressors through a double-negative feedback loop9, 11, 23, 24. Recently the miR-183~96~182 cluster was shown to be repressed by Zeb, in p21 deficient colon cancer cells undergoing EMT and were significantly more invasive and migratory41. miR-183 has been shown to inhibit cancer cell migration and metastasis in breast44 and bone75 cancer cells whereas it induced proliferative and migratory properties in soft tissue sarcomas57 and medulloblastoma68. However a role for the miR-183~96~182 cluster in lung cancer is not defined.
Forkhead-box (Fox) is a super family of transcriptional regulators that have myriad functions from development and proliferation to oncogenesis. Fox proteins have a conserved winged-helix DNA binding domain and less conserved non-Fox domains, which mediate interactions with other transcriptional regulators for specific functions33, 38. The Foxf sub-family consists of two members, of which Foxf1 has been documented to induce EMT and invasion in breast cancers47. Foxf2 is required for gut development and promote extra-cellular matrix production49 but little has been demonstrated on its role in cancer progression or metastasis. Here we provide evidence that the expression of miR-183~96~182 cluster is strongly correlated to miR-200 and EMT in NSCLC and multiple other epithelial tumor types. Both these microRNA families convergently target Foxf2, which can potently regulate EMT, invasion and metastasis in lung cancers by transcriptional repression of E-Cadherin and miR-200.
Results
Expression of miR-183~96~182 cluster is correlated with miR-200 and inhibits cell invasion
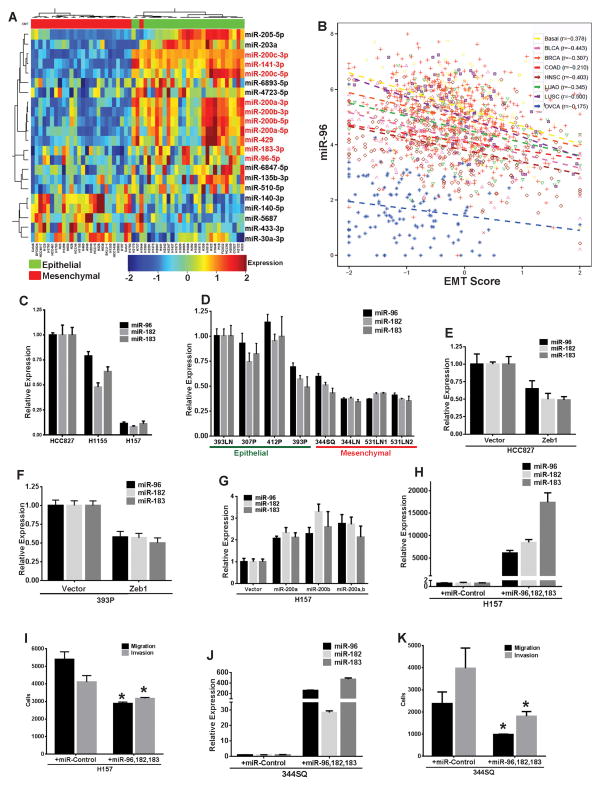
We generated microRNA expression profiles for 55 human NSCLC cell lines and applied a 76 gene EMT signature that we recently described to stratify them into epithelial-like and mesenchymal-like groups based on their gene expression pattern12, 14. In addition to the miR-200 family members, the miR-183~96~182 cluster was significantly repressed in the mesenchymal-like cells lines (Fig. 1A). Like miR-200, the members of the miR-183~96~182 cluster also demonstrated significant negative correlation with the EMT scores12 of samples from multiple TCGA datasets (Fig. 1B and Supplementary. Fig. 1A–B) representing a range of epithelial tumors, thus indicating that both miR-200 and miR-183~96~182 cluster are strongly associated with an epithelial state in different tumor types. Additionally the miR-183~96~182 cluster displayed significant positive correlation with miR-200 for all the tumor datasets (Supplementary Fig. 1C). The expression of the miR-183~96~182 cluster was validated by quantitative RT-PCR in both a panel of human NSCLC cell lines with varying miR-200 levels (Fig. 1C and Supplementary Fig. 1D) and a panel of murine KP lung cancer lines where the miR-183~96~182 cluster expression was significantly lower in the metastatic mesenchymal cell lines (344SQ, 344LN, 531LN1, 531LN2) with low miR-200 expression (as described previously2, 23) than the epithelial non-metastatic cell lines (393LN, 412P, 307P and 393P) with low Zeb1 and high miR-2002 (Fig. 1D). Exogenous Zeb1 expression in epithelial human HCC827 (Supplementary Fig. 1E–H) or murine 393P cell lines resulted in a significant repression of both the miR-200 family2, 23 (Supplementary Fig. 1H) and the miR-183~96~182 cluster (Fig. 1E–F). Conversely, inducible expression of miR-200a and/or -b in mesenchymal human H157 cells14 significantly induced miR-183~96~182 cluster expression (Fig. 1G). To test the functional role of miR-183~96~182 cluster on migration and invasion of NSCLC cells, human or mouse miR-96, 182 and 183 were transfected into mesenchymal human H157 (Fig. 1H) or murine 344SQ cells (Fig. 1J) respectively, which significantly suppressed both migration and invasion (Fig. 1I–K). These results suggest that like miR-200 the expression of the miR-183~96~182 cluster is strongly associated with the epithelial state of cancer cells and directly regulates cancer cell migration and invasion.
Fig. 1. Expression of miR-183~96~182 cluster is correlated with miR-200 and inhibits cell invasion.
(A) Heatmap for microRNA expression profiles of 55 human NSCLC lines, stratified into epithelial and mesenchymal categories based on their EMT score12. (B) Correlation plot for miR-96 expression and EMT scores of human tumors from different TCGA datasets as indicated. (C) Quantitative RT-PCR (qPCR) analysis for relative expression of human miR-183~96~182 cluster in different human NSCLC lines. (D) qPCR analysis for relative expression of mouse miR-183~96~182 cluster in a panel of epithelial-like or mesenchymal-like mouse NSCLC lines. (E) qPCR analysis for relative expression of human miR-183~96~182 cluster in HCC827 cells with stable constitutive expression of either pCDNA vector or Zeb1. (F) qPCR analysis for relative expression of mouse miR-183~96~182 cluster in 393P cells with stable constitutive expression of pCDNA vector or Zeb1. (G) qPCR analysis for relative expression of human miR-183~96~182 cluster in H157 cells with stable inducible expression of either pTRIPZ vector alone, miR-200a, miR-200b or miR-200ab. (H) qPCR analysis for relative expression of human miR-183~96~182 cluster in H157 cells transfected with control miRNA precursor or precursors for human miR-96, miR-182 and miR-183. (I) Trans-well migration or invasion of H157 cells transfected with control miRNA precursor or precursors for human miR-96, miR-182 and miR-183. (J) qPCR analysis for relative expression of mouse miR-183~96~182 cluster in 344SQ cells transfected with control miRNA precursor or precursors for mouse miR-96, miR-182 and miR-183. (K) Trans-well migration or invasion of 344SQ cells transfected with control miRNA precursor or precursors for mouse miR-96, miR-182 and miR-183. (Significance is indicated by * = p≤0.05)
Foxf2 is a novel and direct target of miR-96/182 and miR-200b/c with strong correlation to Zeb1 expression
To investigate the mechanisms by which the mir-200 and the miR-183~96~182 cluster regulate EMT and subsequent cellular migration, invasion and metastasis in NSCLC, we searched for common predicted targets of these microRNA families. First we performed a cross comparison of multiple gene expression datasets from our mouse models of metastasis23. We overlapped 224 genes that were elevated greater than four-fold upon Zeb1 induction in 393P cells, with 210 genes that showed greater than two-fold increase in expression in the metastatic 344SQ cells compared to the non-metastatic 393P cells23 and 143 genes that were repressed to less than 0.5-fold in cells upon exogenous miR-200 expression23 (Fig. 2A). This produced an enriched list of 45 genes that are potential miR-200 targets (Supplementary Table 1). Next we performed an overlap of genes predicted as targets of the miR-200 family (6032 genes) and the miR-183~96~182 cluster (2841 genes), using the microRNA prediction algorithm miRanda (www.microRNA.org) and identified a list of 17 highly conserved common targets (Supplementary Table 2) with a mirSVR7 score less than −6.0 (Fig. 2A). The only 2 genes common in both the overlapping subsets were Zeb1 (ZFHX1A) and Foxf2. We also performed a gene set enrichment analysis on the mRNA profiles from our murine metastatic 344SQ and non-metastatic 393P tumors23 and Foxf2 was again identified in the list of most differentially expressed genes in the metastatic mesenchymal 344SQ tumors (Supplementary Fig. 2A).
Fig. 2. Foxf2 is a novel and direct target of miR-183~96~182 cluster and miR-200 and correlates with Zeb1 expression.
(A) (Left) Heatmaps for gene expression profiles of: mouse 393P cells with stable Zeb1 expression compared to pCDNA vector control cells (genes represented are differentially expressed (p<0.01, fold change>2) with Zeb1), mouse metastatic 344SQ cells compared to non-metastatic 393P cells23, or mouse metastatic 344SQ cells with stable expression of miR-200b~200a~429 cluster compared to control vector cells23. (Right) Venn diagram with the number of genes overlapping for the different datasets as indicated. (B) qPCR analysis for relative expression of mouse Adcy6, Jazf1 or Foxf2 in 344SQ cells transfected with control miRNA precursor or precursors for miR-96, miR-182 and miR-183 (Suppl. Table 7). (C) qPCR analysis for relative expression of human ADCY6, JAZF1 or FOXF2 in H157 cells transfected with control miRNA precursor or precursors for human miR-96, miR-182 and miR-183 (Suppl. Table 7). (D) qPCR analysis for relative expression of mouse Foxf2 in 393 cells with stable expression of miR-200, Zeb1 or miR-200 and Zeb1. (E) qPCR analysis for relative expression of human Foxf2 and Zeb1 in H157 cells with inducible expression of pTRIPZ vector alone, miR-200a, miR-200b or miR-200ab. (F) (Top) Schematic representing reporter constructs of either wild-type mouse Foxf2-3′UTR or mouse Foxf2-3′UTR with a mutated miR-200b/c binding site. (Bottom) Relative luciferase activity from reporter constructs as indicated, with co-expression of either mouse control miRNA precursor or precursors of miR-200a, 200b or 200c in 344SQ cells. (G) (Top) Schematic representing reporter constructs of either wild-type mouse Foxf2-3′UTR or mouse Foxf2-3′UTR with mutation in the miR-96 and miR-182 binding sites. (Bottom) Relative luciferase activity from reporter constructs as indicated, with co-expression of either mouse control miRNA precursor or precursors of miR-96, and miR-182 in 344SQ cells. (H) qPCR analysis for relative expression of mouse Foxf2 and Zeb1 in a panel of epithelial or mesenchymal mouse NSCLC lines. (I) qPCR analysis for relative expression of human Foxf2 and Zeb1 in a panel of epithelial-like or mesenchymal-like human NSCLC lines. (J) Western blot analysis for expression of Zeb1, E-Cadherin, FOXF2 or β-Actin (as a loading control) in a panel of epithelial-like or mesenchymal-like human NSCLC lines. (Significance is indicated by * = p≤0.05)
Foxf2 contains seed sequences for miR-200b/c/429, miR-96, miR-182 and miR-183 in its 3′UTR. Expression of miR-96, miR-182 and miR-183 resulted in significant repression of Foxf2 in both murine (Fig. 2B) and human (Fig. 2C) cells, along with specific down-regulation of ADCY6 and JAZF1, which are two highly predicted targets of the miR-183~96~182 cluster, containing multiple target sequences in their 3′UTR’s6, 7. Stable expression of the miR-200b~200a~429 cluster in 393P murine lung cancer cells significantly inhibited Foxf2 expression, while it was robustly induced when miR-200 was repressed by stable expression of Zeb12 (Fig. 2D). Similarly a significant repression in FOXF2 expression was observed in human H157 cells (Fig. 2E) upon inducible expression of miR-200a and/or -b. In contrast, inhibition of endogenous miR-200a, 200b, 96 and 183 in murine 344SQ cells by anti-miRNA transfection significantly elevated the expression of both Zeb1 and Foxf2 (Supplementary Fig. 2B). To ascertain whether Foxf2 is directly targeted by these microRNAs, we performed luciferase reporter assays with wild-type or miRNA binding site mutant versions of the Foxf2-3′UTR. miR-200b/c significantly repressed luciferase activity of the Foxf2 WT 3′UTR construct, but not the version with a mutated miR-200b/c site (Fig. 2F). Similarly, miR-96 significantly repressed luciferase activity from the WT 3′UTR, which was lost upon mutation of the recognition sites (Fig. 2G). These results indicate that Foxf2 is a novel bonafide target of miR-200b&c and miR-96.
To further test whether Foxf2 expression is triggered by EMT we treated murine or human lung cancer cells with TGFβ and observed a robust induction of Foxf2 (Supplementary Fig. 2C), which was comparable to the increase in Zeb18, 10, 11, 61, 69. We also observed a strong correlation of Foxf2 with Zeb1 expression in our panel of KP mouse (Pearson Correlation: R2 = 0.744, p<0.005) (Fig. 2H) and human (Pearson Correlation: R2 = 0.67, p<0.1) (Fig. 2I–J) lung cancer cell lines.
Foxf2 induces EMT, invasion and metastasis
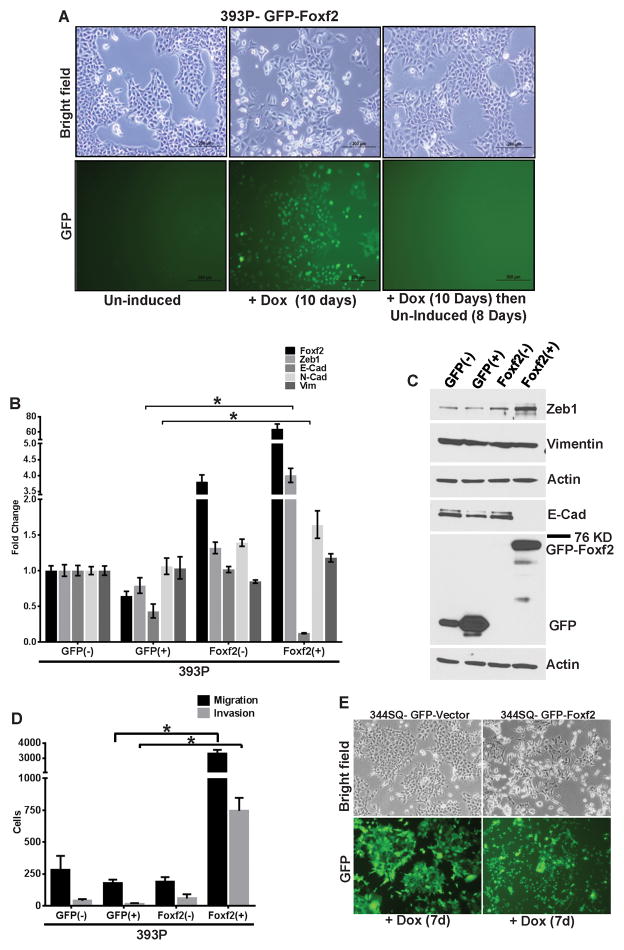
To examine the biological role of Foxf2 in lung cancer, GFP-tagged Foxf2 was exogenously expressed under a doxycycline-inducible promoter in the non-metastatic murine epithelial 393P cells. Foxf2 expression in 393P cells induced a dramatic cellular phenotypic change from a cobble-stone, clustered, epithelial state to a fibroblastic, spindle-shaped, scattered mesenchymal morphology, which reverted upon doxycycline withdrawal (Fig. 3A). Control GFP-expressing cells showed no morphologic change (Supplementary Fig. 3A). This morphologic shift upon 10 days of Foxf2 expression was concordant with a significant induction of Zeb1 and dramatic E-Cadherin repression both at the mRNA (Fig. 3B) and protein (Fig. 3C) levels, but with no significant change in the mesenchymal markers like N-Cadherin or vimentin. 393P-Foxf2(+) cells also exhibited robust increases in cellular migration and invasion in Boyden chambers compared to the control 393P-GFP(+) cells (Fig. 3D and Supplementary Fig. 3B). To determine whether the phenotype produced by Foxf2 is cell line independent, GFP-tagged Foxf2 was inducibly expressed in epithelial murine (531P1) or human (HCC827) lung cancer cells. Foxf2 expression in both 531P1 and HCC827 cells induced an EMT-like morphologic change (data not shown and Supplementary Fig. 3H), with Zeb1 induction and E-Cadherin suppression (Supplementary Fig. 3C–D, 3J). 531P1-Foxf2 cells exhibited pronounced increases in migration and invasion compared to the GFP control cells (Supplementary Fig. 3E–F). The HCC827-FOXF2 cells only demonstrated a modest increase of migration in Boyden chambers (Supplementary Fig. 3I) but when grown in 3D matrigel/collagen I cultures, were more invasive compared to the GFP control cells (Supplementary Fig. 3K). These results demonstrate that Foxf2 induces EMT, cellular migration and invasion in cells that are normally epithelial and non-invasive.
Fig. 3. Foxf2 induces EMT, invasion and metastasis.
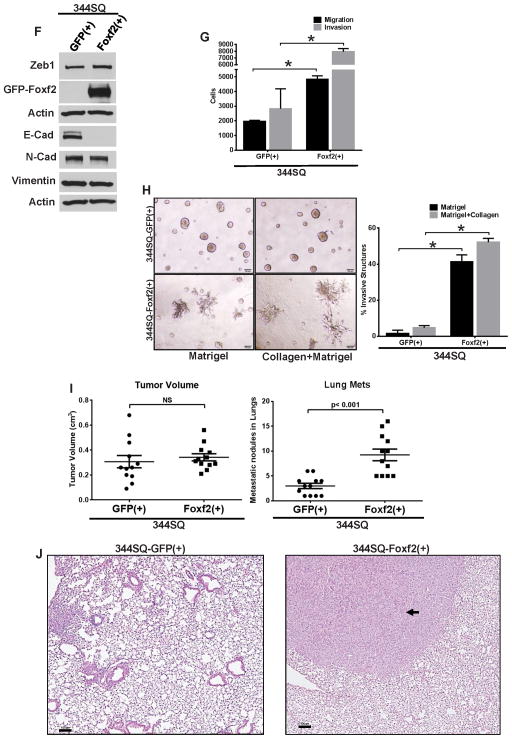
(A) Bright field or GFP fluorescence microscopy images showing morphology of 393P cells, either un-induced (−), induced (+) for GFP-tagged Foxf2 (Foxf2) expression for 10 days or first induced for 10 days and then un-induced for 8 days. (B) qPCR analysis for relative expression of Foxf2 and other EMT markers in 393P cells which are un-induced (−) or induced(+) (10 days) for expression of TRIPZ-GFP control vector (GFP(+)) or GFP-Foxf2 (Foxf2(+)). (C) Western blot analysis for expression of Foxf2 or EMT markers in 393P cells which are un-induced (−) or induced (+) (10 days) for expression of either TRIPZ-GFP control vector (GFP) or GFP-Foxf2 (Foxf2). (D) Trans-well migration or invasion of 393P cells which are un-induced (−) or induced (+) (10 days) for expression of TRIPZ-GFP control vector (GFP) or GFP-Foxf2 (Foxf2). (E) Bright field or GFP fluorescence microscopy images showing morphology of 344SQ cells induced for 7 days for expression of either the TRIPZ-GFP control vector or GFP-Foxf2. (F) Western blot analysis for expression of Foxf2 and other EMT markers in 344SQ cells which are induced for 7 days for expression of either TRIPZ-GFP control vector (GFP(+)) or GFP-Foxf2 (Foxf2(+)) as indicated. (G) Trans-well migration or invasion of 344SQ cells induced for 7 days for expression of TRIPZ-GFP control vector (GFP(+)) or GFP-Foxf2 (Foxf2(+)) as indicated (H) (Left) Bright-field microscopy of 3D spheroids in different matrices as indicated, formed by 344SQ cells induced for 6 days for expression of TRIPZ-GFP control vector (344SQ-GFP(+)) or GFP-Foxf2 (344SQ-Foxf2(+)). (Right) Quantification of invasive structures formed by the respective cell lines in different matrices. (I) (Left) Quantification of tumor volumes of syngeneic mice injected with 344SQ cells induced for expression of TRIPZ-GFP control vector (GFP(+)) or GFP-Foxf2 (Foxf2(+)). (Right) Quantification of metastatic lung surface nodules formed in the mice injected with 344SQ cells induced for expression of either TRIPZ-GFP control vector (GFP(+)) or GFP-Foxf2 (Foxf2(+)). (J) Representative images of H&E sections of lungs from mice injected with 344SQ cells induced for expression of TRIPZ-GFP control vector (344SQ-GFP(+)) or GFP-Foxf2 (344SQ-Foxf2(+)) (Scale bar = 100μM). (Significance is indicated by * = p < 0.05).
To understand the potential role of Foxf2 during invasion and metastasis we wanted to determine whether Foxf2 could modulate these biologic functions in cells that are mesenchymal and have metastatic ability at baseline. For this Foxf2 was inducibly expressed in the murine 344SQ cell line. As in the 393P and 531P1 cells, induction resulted in robust Foxf2 expression, nuclear localization of the protein and an EMT-like change in the cellular morphology when compared to the GFP-expressing cells (Fig. 3E and Supplementary Fig. 4A). Molecularly the 344SQ-Foxf2 cells exhibited a drastic suppression of E-Cadherin, but mesenchymal markers were unchanged (Fig. 3F). This observation was confirmed by IF staining which revealed a complete loss of E-Cadherin from the cell membrane in the Foxf2 expressing cells compared to GFP (control) cells and resulted in re-localization of β-Catenin from the cell surface into the cytoplasm (Supplementary Fig. 4B). The 344SQ-Foxf2 cells also exhibited a significant increase in cellular migration and invasion in Boyden chambers (Fig. 3G and Supplementary Fig. 3G), but no difference in proliferation (Supplementary Fig. 4C). Growth in 3D cultures revealed normal spheroid formation of the control 344SQ-GFP cells and remarkable invasive growth by the 344SQ-Foxf2 cells (Fig. 3H). To determine their in vivo metastatic potencies, 344SQ-Foxf2 or control 344SQ-GFP induced cells were subcutaneously implanted into syngeneic mice. The primary tumor sizes for both the control and the Foxf2 expressing cells were comparable, consistent with no significant difference in cellular proliferation between the tumor types as evident from Ki67 staining (Supplementary Fig. 4D). However, the mice with 344SQ-Foxf2 tumors demonstrated a ~3-fold increase in the number of metastatic lung nodules compared to the control cells (Fig. 3I) within just 4 weeks. This was confirmed by haematoxylin and eosin staining of lung sections from the groups (Fig. 3J). These results establish Foxf2 as a potent suppressor of the epithelial phenotype, which arrests cells in a hyper-invasive state, producing rapid in vivo metastasis.
Foxf2 knockdown suppresses invasion and metastasis
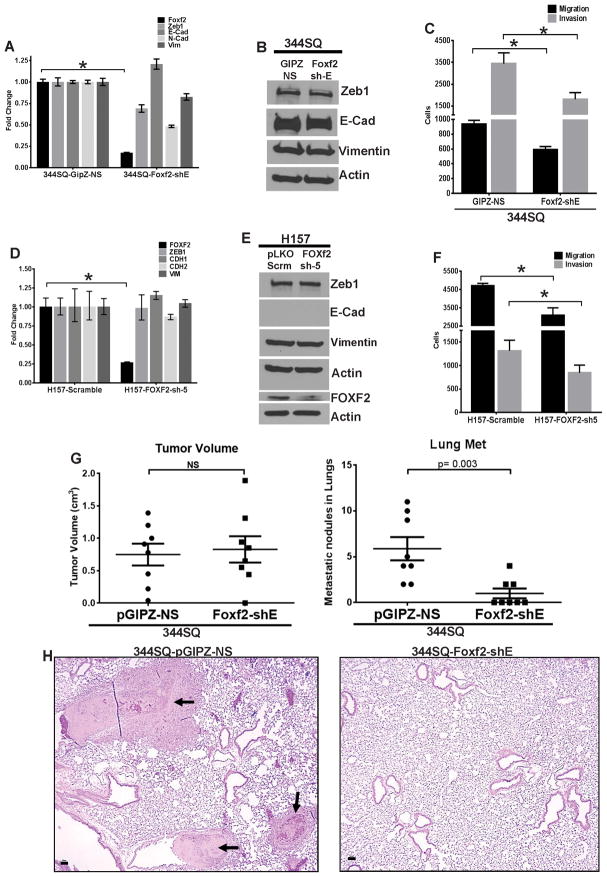
To study the converse effect, we stably knocked down Foxf2 expression in mesenchymal mouse and human cells by shRNA vectors. Foxf2 knockdown in mouse mesenchymal and metastatic 344SQ cells (344SQ-Foxf2-shE) did not result in an apparent change in cell morphology (data not shown), cell proliferation (Supplementary Fig. 4C) or expression of the EMT markers (Fig. 4A–B), but significantly suppressed cellular migration and invasion in Boyden chambers (Fig. 4C and Supplementary Fig. 3L). Similarly in human H157 cells, knockdown of FOXF2 (H157-FOXF2-sh5) did not alter the expression of EMT genes (Fig. 4D–E) but produced significant inhibition of migration and invasion compared to vector controls (Fig. 4F and Supplementary Fig. 3M). To test whether down-regulation of Foxf2 expression could alter the in vivo metastatic potencies, the 344SQ-Foxf2-shE (knockdown) and the 344SQ-pGIPZ-NS (control) cells were injected subcutaneously in syngeneic mice. Both groups formed comparable sized tumors at 8 weeks, with only a slight increase in proliferating cells in the primary tumors formed by the knockdown cells compared to the controls when assayed by Ki-67 staining (Fig. 4G and Supplementary Fig. 4D). In contrast, the Foxf2 knockdown cells exhibited significant repression of lung metastasis (Fig. 4G), which was confirmed by haematoxylin and eosin stained lung sections (Fig. 4H). These results confirm that inhibition of Foxf2 expression could significantly reduce the migratory and invasive capabilities of metastatic cells, abrogating in vivo metastasis. Interestingly by manipulating the levels of Foxf2 in the same (344SQ) cell line we could control the metastatic phenotype of the cells, highlighting the importance of Foxf2 as a metastasis regulator.
Fig. 4. Foxf2 knockdown leads to decreased invasion and metastasis.
(A) qPCR analysis for relative expression of mouse Foxf2 and other EMT markers in 344SQ cells with stable expression of control non-silencing hairpin (344SQ-GipZ-NS) or hairpin targeting Foxf2 (344SQ-Foxf2-shE). (B) Western blot analysis for expression of EMT markers in 344SQ cells with stable expression of either control non silencing hairpin (344SQ-GipZ-NS) or hairpin targeting Foxf2 (344SQ-Foxf2-shE). (C) Trans-well migration or invasion of 344SQ cells with stable expression of either control non silencing hairpin (344SQ-GipZ-NS) or hairpin targeting Foxf2 (344SQ-Foxf2-shE). (D) qPCR analysis for relative expression of human FOXF2 and other EMT markers in H157 cells with stable expression of either control scrambled hairpin (H157-Sramble) or hairpin targeting FOXF2 (H157-FOXF2-sh5). (E) Western blot analysis for human FOXF2 and other EMT markers in H157 cells with stable expression of control scrambled hairpin (H157-Sramble) or hairpin targeting FOXF2 (H157-FOXF2-sh5). (F) Trans-well migration or invasion of H157 cells with stable expression of control scrambled hairpin (H157-Sramble) or hairpin targeting FOXF2 (H157-FOXF2-sh5). (G) (Left) Quantification of tumor volumes or (Right) metastatic lung surface nodules formed in syngeneic mice injected with 344SQ cells with stable expression of control non-silencing hairpin (GipZ-NS) or hairpin targeting Foxf2 (Foxf2-shE). (H) Representative images of H&E sections of lungs from mice in (G) (Scale bar = 50μM). (Significance is indicated by * = p < 0.05).
Foxf2 induces rapid repression of E-cadherin and miR-200 independent of Zeb1
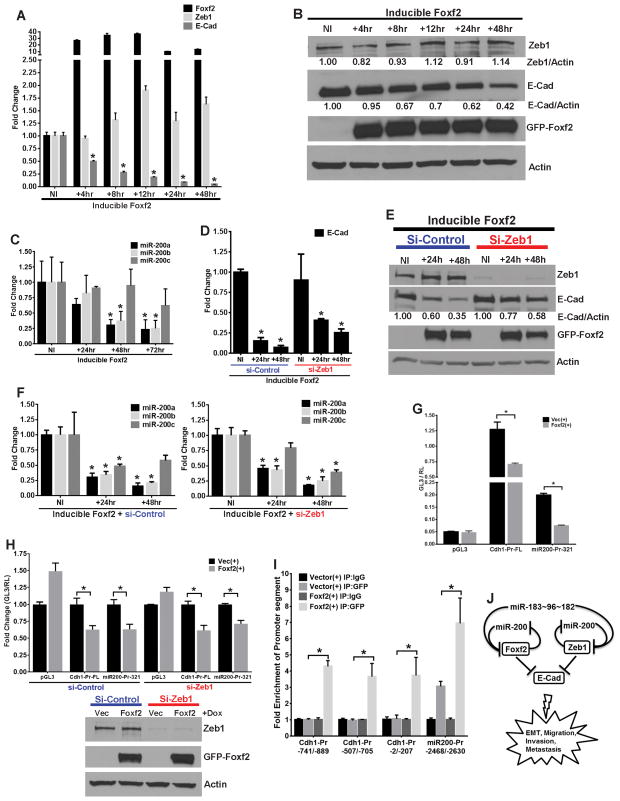
Foxf2 expression induces a strong EMT-like phenotype with increased migration, invasion and metastasis, which is associated with a robust inhibition of E-Cadherin and up-regulation of Zeb1. To understand whether these two changes are a direct and acute consequence of Foxf2 expression, we performed a time course assay to determine the changes in expression of these two markers upon induction of Foxf2 in 393P cells. Upon Foxf2 induction, E-Cadherin was transcriptionally repressed as early as 4 hours (50% at RNA level) and reached its maximum by 48 hours (more than 95% by mRNA and protein level (60%)), whereas Zeb1 protein levels were not considerably elevated (14%) until 48 hours (Fig. 5A–B). Induction of GFP (control vector) did not induce any marker changes (Supplementary Fig. 5A–B). Since the miR-200 family is important regulator of the epithelial phenotype in lung cancer23, we examined whether they are regulated by Foxf2. We observed that mature miR-200a and miR-200b, but not miR-200c, were significantly repressed (70%) within 48 hours of Foxf2 induction (Fig. 5C), whereas no significant changes were observed in the GFP vector cells (Supplementary Fig. 5C). This was confirmed by similar repression of the miR-429 levels (70%) within 72 hours of Foxf2 induction, whereas miR-141 levels were unaltered (Supplementary Fig. 5D). These results suggest that Foxf2 might transcriptionally target only the miR-200b~200a~429 locus and not the miR-141~200c locus.
Fig. 5. Foxf2 induces rapid repression of E-cadherin and miR-200 independent of Zeb1.
(A) qPCR analysis for relative expression and (B) Western blot analysis of mouse Foxf2, Zeb1 and E-Cad in 393P cells induced for Foxf2 expression for the indicated times. Relative band intensity for Zeb1 or E-Cad, normalized to respective actin band intensities are shown for each lane. (C) qPCR analysis for relative expression of mature mouse miR-200a–c in 393P cells induced for Foxf2 expression for the indicated times. (D) qPCR analysis for relative expression and (E) Western blot analysis of mouse E-Cad in 393P cells that were induced for Foxf2 expression for different times as indicated, after transfection of either control siRNA (si-Control) or siRNA targeting Zeb1 (si-Zeb1). Relative band intensity for E-Cad, normalized to respective actin band intensities is shown for each lane. (F) qPCR analysis for relative expression of mature mouse miR-200a-c in 393P cells that were induced for Foxf2 expression for different times as indicated, after transfection of control siRNA (si-Control) (Left) or siRNA targeting Zeb1 (si-Zeb1) (Right). (G) Normalized luciferase activities from reporter constructs of either empty vector (pGL3), full length E-Cad promoter (Cdh1-Pr-FL) or 321 base pair upstream fragment of miR-200b promoter (miR200-Pr-321)9, transfected in 393P cells that were induced for expression of either TRIPZ-GFP control vector (Vec(+)) or GFP-Foxf2 (Foxf2(+)) as indicated. (H) (Top) Relative normalized luciferase activities from reporter constructs; pGL3, Cdh1-Pr-FL and miR200-Pr-3219, transfected in 393P vector control or Foxf2 cells which were pre-transfected with control siRNA (si-Control) or siRNA targeting Zeb1 (si-Zeb1). (Bottom) Western blot analysis demonstrating efficient knockdown of Zeb1 and Foxf2 induction in 393P vector control or Foxf2 cells that were transfected with control siRNA (si-Control) or siRNA targeting Zeb1 (si-Zeb1), which were used in the experiment. (I) Fold-enrichment of promoter segments as indicated, of E-Cadherin and miR-200b~200a~429 promoters after ChIP assays performed in 393P-GFP (Vector) or 393P-GFP-Foxf2 cells, using either an anti-GFP antibody or a mock IgG control antibody. (J) Model representing Foxf2 as a target of miR-200 and miR-183~96~182 cluster, which transcriptionally represses miR-200 and E-Cadherin to induce cancer cell invasion and metastasis.
As the miR-200b~200a~429 cluster and E-cadherin are both known Zeb1 targets 9, 17, 22, we sought to determine whether Foxf2-mediated repression of these two loci is dependent on Zeb1. Control siRNA or siRNA targeting Zeb1 were transfected into control 393P-GFP or 393P-Foxf2 cells and both mRNA and protein were isolated at time points after Foxf2 or GFP induction. Both in the presence and absence of Zeb1, E-Cadherin was significantly repressed upon 48 hours of Foxf2 induction, at the mRNA (>50%) (Fig. 5D) and protein (40%) (Fig. 5E) levels. No change was observed in the 393-GFP vector cells (Supplementary Fig. 5E). To confirm these findings, we generated stable 393P cells with inducible Foxf2 and either scramble (control) hairpin (393P-Foxf2-sh-scramble) or shRNA hairpin targeting Zeb1 (393P-Foxf2-shZeb1), which showed greater than 90% Zeb1 knockdown. Upon Foxf2 induction for 48 hours, E-Cadherin expression was significantly repressed both at RNA (50%) and protein (70%) levels in cells with endogenous Zeb1 expression. Similarly, in cells with stable knockdown of Zeb1, E-Cadherin was significantly repressed both at RNA (50%) and protein (65%) levels after 48 hours of Foxf2 induction (Supplementary Fig. 5G–H). Induction of GFP did not change the levels of E-Cadherin irrespective of Zeb1 status in the control cells (Supplementary Fig. 5F). Similarly, quantitative PCR analysis for mature forms of mir-200a, -b or -c from 393P-Foxf2 (inducible) cells transfected with si-Control or si-Zeb1 revealed that they were significantly repressed upon Foxf2 induction, independent of Zeb1 status (Fig. 5F).
To determine whether E-Cadherin and miR-200 repression are a result of direct inhibition of their promoter activities by Foxf2, we performed luciferase reporter assays using promoter constructs for E-Cadherin and the miR-200b~200a~429 cluster9. We observed a significant repression of activity from both the promoters upon Foxf2 induction (Fig. 5G), which was independent of Zeb1, as demonstrated by use of siRNA targeting Zeb1 versus control siRNA in 393P-GFP (control) or 393P-Foxf2 inducible cells (Fig. 5H). To further assess direct binding of Foxf2 to the endogenous promoters of E-Cadherin and miR-200b~200a~429, we performed ChIP assays with the 393P-GFP (control) or 393P-Foxf2 inducible cells. We observed specific binding and enrichment for Foxf2 at different segments of the E-Cadherin promoter (as indicated) and an unique segment ~2500 bp upstream from the start site of the miR-200b precursor (Fig. 5I). All these enriched segments contain putative Foxf2 binding sites. Taken together these results demonstrate that Foxf2 transcriptionally represses E-Cadherin and miR-200b~200a~429 microRNA cluster, independent of Zeb1, to repress the epithelial phenotype and induce EMT, invasion and metastasis.
Based on our work here and the data in the literature, we propose a model in which the Zeb1-miR-200 axis works in concert with the miR-193~96~182 cluster to convergently target Foxf2 and regulate EMT, invasion and metastasis in lung cancer. In a Zeb1-independent manner, Foxf2 represses E-Cadherin and inhibits miR-200 to form a reinforcing negative feedback loop running in parallel with Zeb1 and miR-200, to regulate invasion and metastasis (Fig. 5J).
Discussion
The molecular underpinnings driving lung cancer metastasis are largely unknown. There is a large body of evidence both in primary human tumors 27, 64, 65 and various mouse models of cancers15, 23, 30, 48, 53, which indicates EMT as a necessary and driving mechanism for metastasis of epithelial tumors. microRNAs are global regulators of gene expression and dysregulation of microRNAs and their signaling networks have been implicated in tumorigenesis and cancer metastasis13, 21, 32, 52. We and others have shown that deregulation of miR-200 by Zeb1 is critical to drive EMT and metastasis11, 23, 24, 37, 51. Metastasis is a complex biological process with compound stages of progression,52, 64 therefore we hypothesized that multiple upstream molecular controllers are deregulated to coordinately control complementary cell functions necessary for initiation and progression of metastasis. We wanted to investigate other microRNAs like miR-200 that act as upstream regulators of EMT and metastasis. By miRNA expression profiling we identified that in addition to miR-200, the miR-183~96~182 cluster shows very strong negative correlation with EMT in a panel of 55 human NSCLC lines and intensive correlative studies in 7 other primary tumor types from TCGA clinical specimens strongly reinforced this observation. This inclusive pan-cancer approach established with high confidence the functional association of the miR-183~96~182 cluster with EMT and metastasis.
MicroRNAs interact with different signaling molecules to produce varied and contrasting outcomes depending on the cell type and the microenvironment1, 18. Similarly, while anti-metastatic function of miR-183~96~182 was observed in certain cancers40, 41, 44, 75, 78, members of this cluster have demonstrated a pro-migratory and invasive role in other tumors25, 26, 31, 39, 58, 63. This suggests that miR-183~96~182 function is regulated contextually in a tissue-dependent manner. To specifically elucidate the molecular targets of miR-200 and miR-183~96~182, we performed a multi-step cross comparison of gene expression profiles of the differently manipulated mouse model cells which overlapped with the list of high confidence common predicted targets of both the miRNA families. This approach gave us the unique advantage to utilize the extensive genomic data from our syngeneic mouse models to discover functionally relevant targets of miR-200 and the miR-183~96~182 cluster with a potential direct role in lung cancer metastasis. Foxf2 is a member of the forkhead box super family of transcriptional regulators which are known to function as oncogenes and tumor suppressors in divergent cancer types26, 33, 36, 38, 67. Other members of the Fox family, namely FOXM1 and FOXC1, have been reported as inducers of EMT, invasion and metastasis56, 70. Foxf1, the other member of the Foxf family has been reported to induce EMT, invasion and metastasis in breast and colorectal cancers43, 47. Foxf2 has been shown to be necessary for gut development and production of the extra cellular matrix46, 66. Two recent reports also demonstrated the association of Foxf2 in a tumor suppressive role in breast36 and prostate26 cancers. Here we established that Foxf2 functions as a robust regulator of EMT, migration, invasion and metastasis in a syngeneic mouse lung cancer model with mutant Kras and p53. It has been reported that constitutive over expression of Foxf2 from a transgenic allele in a mouse model for colorectal carcinogenesis resulted in decreased adenoma formation compared to wild type or Foxf2+/− mice46. This suggests that specific genetic lesions in cancer regulate the activity of Foxf2 as a metastasis driver in a tissue-dependent context.
Multiple groups have convincingly defined the role of miR-200 as a master regulator of EMT and metastasis in different cancer types20, 23, 42, 50, with the double-negative feedback loop between Zeb1/2 and miR-200 as the best documented mechanism of miR-200 regulation11, 23, 24, 37, 51. The importance of this master regulator strongly suggests that there should be multiple layers of control over miR-200 expression. Additional transcription factors and chromatin modifiers have been shown to regulate miR-200 transcription in cancers34, 35, 45, 54, 55. Our data elucidate a novel mechanism where Foxf2 transcriptionally represses E-Cadherin and miR-200, independent of Zeb1 to repress epithelial phenotype. We also identified a novel double-negative feedback loop between Foxf2 and miR-200, thus revealing a parallel axis to the Zeb1-miR-200 loop that controls EMT and metastasis. E-Cadherin and miR-200 loss are nodal events in the onset and progression of EMT and metastasis and independent control in modulating these genes implies a pivotal role for Foxf2 as a parallel regulator of EMT and metastasis. In our miRNA expression profiles on the NSCLC lines we also identified miR-205 and miR-203 which exhibited robust correlation with miR-200 and EMT and further investigation of the functional targets of these miRNAs might reveal new interesting leads in to the regulation of metastasis in lung cancers.
Materials and methods
Plasmids and reagents
Mouse or human Foxf2 cDNA was amplified by RT-PCR as a MfeI/EcoRI – MluI fragment (primers P1-P4, Suppl. Table 4) and cloned in the dox inducible pTRIPZ-GFP vector where the miR-30~RFP cassette was replaced with the GFP cDNA. Mouse Foxf2 shRNA (pGIPZ-Foxf2-shE) construct was purchased from Open-Biosystems/GE-Dharmacon (Cat#RMM4431-200375685/CloneID#V3LMM_518867). Human FOXF2 shRNA (pLKO-FOXF2-sh5) construct was cloned in pLKO vector (Addgene, Cambridge, MA) by ligating annealed oligos (P13, P14, Suppl. Table 4). Reporter construct for Foxf2-3′UTR was generated by RT-PCR amplification of 0.7 KB 3′ UTR region of Foxf2 mRNA, using specific primers (P5, P6, Suppl. Table 4) and cloned in pGL3-Promoter vector (Promega, Madison, WI). miR-200 and miR-96/182 binding site mutants of the Foxf2-3′UTR reporter construct were generated using specific primers (P7-P10, Suppl. Table 4) and Quick change Lightning site directed mutagenesis kit (Agilent, Santa Clara, CA). Reporter construct for E-Cadherin promoter was generated by PCR amplification of 1.6KB promoter region of mouse E-Cadherin using specific primers (P11, P12, Suppl. Table 4) and cloned in pGL3-Basic vector. shRNA construct for Zeb1 (RMM3981-201796052/CloneID#TRCN0000070818) and Scramble control vectors were purchased from Thermo-Scientific. siRNA for Zeb1 (SMARTpool:ON-TARGETplus) was purchased from Dharmacon/GE-Healthcare (Lafayette, CO).
Cell culture and transfections
All cell lines were maintained in RPMI-1640 supplemented with 10% fetal bovine serum. DNA transfections were performed with Lipofectamine-2000 or Lipofectamine-LTX reagents (Life technologies, Grand Island, NY). miRNA precursors and inhibitors (Suppl. Table 7) were transfected at 50nM or 100nM final concentration for 96 hours using Lipofectamin-2000 or Lipofectamine RNAi Max (Invitrogen-Life technologies) and siRNAs were transfected at 25nM final concentration using DharmaFECT-I (Dharmacon/GE-Healthcare) reagents as per manufacturer’s protocols.
Reporter assays
For 3′UTR reporter assays cells were co-transfected with 500 ng of the reporter construct and 50nM miRNA precursors for 24 hours and then assayed for luciferase activity after 24 hours of Doxycycline induction. For promoter-reporter assays cells were first pre-transfected for 72 hours with siRNA followed by transfection for 24 hours with 500ng of the reporter constructs, then assayed for luciferase activity after 24 hours of doxycycline induction. All reporter assays were performed using the Dual-Luciferase reporter assay system (Promega, Madison, WI).
qPCR and Western blot analysis
Total RNA was isolated and RT-PCR was performed using specific primers (Suppl. Table 4) and SYBR® Green PCR Master Mix (Life technologies). SYBR green qPCR analyses were normalized to expression of L32 (60S ribosomal gene). Taq-man assays (Life technologies; Suppl.Table 5) for miRNA qPCR analyses were normalized to miR-16. Western blots were performed with antibodies as listed (Suppl. Table 5).
Analysis of miRNA expression and EMT using TCGA pan-cancer data
Level 3 TCGA pan-cancer data was used including gene expression (RNAseq) and microRNA expression (RNAseq)3, 28. A summary of the samples with matched gene expression and miRNA data was provided in Supplementary Table 6. EMT score was calculated from gene expression data using the EMT signature previously published12. Pearson correlation (r value) was used to quantify the association between miRNA and EMT score.
Analysis of miRNA expression with EMT using human NSCLC cell lines
Affymetrix microRNA 4.0 samples were processed by Affymetrix Expression Console. Same cell line expression levels were averaged for each probe-set. EMT score was calculated from expression data profiled on Illumina WG v3 (GSE32036). There were 55 cell lines with both miRNA expression and EMT score. Two sample t-tests were used to test the difference between Epithelial-like and Mesenchymal-like groups (E is defined as EMT score less than 0 and M otherwise). miRNAs selected at FDR level of 0.05 were used for heatmap.
Analysis of mRNA expression array from Zeb1-transfected cells
Samples were shipped to Asuragen (Austin, TX) for processing and analysis on Affymetrix Mouse Expression Array 430A 2.0 chips as previously described23. Array data have been deposited into the Gene Expression Omnibus (GSE61395).
Migration and Invasion Assays
Trans-well migration (8 μM inserts; BD-Biosciences) and invasion (BD-Biosciences; #354480) assays were performed for 6 hours (human cells) and 16 hours (mouse cells) using standard protocol23. Inserts were stained with crystal violet and migrated or invaded cells were analyzed and counted using Image-J software.
ChIP assays
ChIP was performed as described earlier2, 73 with the modification that cells were fixed first with 5mM DTBP (Pierce-Thermo scientific) and then with 1% formaldehyde. Immuno precipitation was performed using anti-GFP antibody (Abcam ab290) or mock IgG (Santacruz). Promoter segment enrichment was analyzed by qPCR using primers P49-56 (Suppl. Table 4).
3D culture assay
Cells were seeded in a matrix comprising of matrigel (BD-Biosciences; #354230) or a mixture of matrigel and collagen (BD-Biosciences; #354236) and incubated for 6 days with regular replenishment with complete media containing 2% matrigel until formation of spheroids23. Spheroids were imaged and analyzed using an inverted microscope.
In vivo tumor and metastasis experiments
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee at The University of Texas M.D. Anderson Cancer Center. Cells were sub cutaneously injected in the flanks of syngeneic 129/Sv mice of 8–10 weeks age and observed for tumor growth for a period of 4–8 weeks. Upon euthanasia, metastatic nodules on the surface of lung lobes were counted. Lung tissue was fixed in 10% Formalin and then processed for sectioning followed by haematoxylin and eosin staining.
Supplementary Material
(A) Correlation plots for miR-182 and miR-183 expression and EMT scores of human tumors from different TCGA datasets as indicated. (B) Correlation plots for miR-200 expression and EMT scores of human tumors from different TCGA datasets as indicated. (C) Correlation plot for expression of miR-96, miR-182 and miR-183 and miR-200b in human tumors from different TCGA datasets as indicated. (D) qPCR analysis for relative expression of human miR-200b & 200c in panel of human NSCLC lines. (E) Bright field microscopy of HCC827 cells with either pCDNA vector or constitutive expression of Zeb1. (F) Western blot analysis for expression of Zeb1 and other EMT markers in isolated clones of HCC827 constitutively expressing either pCDNA vector of Zeb1. (G) qPCR analysis for relative expression of Zeb1 and other EMT markers in isolated clones of HCC827 constitutively expressing either pCDNA vector of Zeb1. (H) qPCR analysis for relative expression of miR-200a, 200b and 200c in isolated clones of HCC827 constitutively expressing either pCDNA vector of Zeb1.
List of 45 genes that are potential miR-200 targets having a role in the process of EMT and metastasis, generated by overlapping different mouse gene expression datasets as indicated.
List of 17 highly conserved common predicted mouse target genes of mmu-miR-200 and mmu-miR-183~96~182 cluster, using the microRNA prediction algorithms miRanda (www.microRNA.org), with a mirSVR7 score less than -6.0.
List of 48 highly conserved common predicted human target genes of hsa-miR-200 and hsa-miR-183~96~182 cluster, using the microRNA prediction algorithms miRanda (www.microRNA.org), with a mirSVR7 score less than −6.0.
List of primers used for generation of reagents and experiments.
List of miRNA Taq-Man assays and Antibodies.
Summary of samples in TCGA pan-cancer data sets with matched gene expression and miRNA data
(A) Gene set enrichment analysis was performed as described previously62). Gene expression profiles from four syngeneic tumors each, generated from either metastatic 344SQ cells (represented as red squares) or non-metastatic 393P cells (represented as blue squares) were used to perform Gene Set Enrichment Analysis to identify a list of most enriched genes in metastatic cells. The intensity of red or blue colors represent a heat map for magnitude of expression (Red=high ; blue=low). The top genes show Foxf2 as one of the most enriched genes in the metastatic cells compared to the non-metastatic ones. (B) qPCR analysis for relative expression of human Foxf2 and Zeb1 in HCC827 cells that were transfected with either Anti-miRNAs targeting miR-200a, 200b, 96 and 183 or a control Anti-miRNA. (C) (Left) qPCR analysis for relative expression of Zeb1, Foxf2 and other potential miR-200 targets in 344SQ cells treated with TGFβ for 10 days. (Right) qPCR analysis for relative expression of Zeb1 and FOXF2 in human HCC827 cells treated with TGFβ for 10 days.
(A) Bright field or GFP fluorescence microscopy images showing morphology of 393P cells which are either un-induced (−), induced (+) for GFP (control) expression for 10 days or first induced for 10 days and then un-induced for 8 days. (B) Representative images of trans-well chambers for migration and invasion of 393P cells which are un-induced (−) or induced (+) (10 days) for expression of either TRIPZ-GFP control vector (GFP) or GFP-Foxf2 (Foxf2) as indicated. (C) qPCR analysis for relative expression of Foxf2 and other EMT markers in mouse 531P1 cells which are induced(+) (10 days) for expression of either TRIPZ-GFP control vector (GFP(+)) or GFP-Foxf2 (Foxf2(+)). (D) Western Blot analysis for expression of GFP-Foxf2 or EMT markers in mouse 531P1 cells which are induced (+) (10 days) for expression of either TRIPZ-GFP control vector (GFP) or GFP-Foxf2 (Foxf2) as indicated. (E–F) Trans-well migration and invasion of 531P1 cells which are induced (+) (10 days) for expression of either TRIPZ-GFP control vector (GFP), or GFP-Foxf2 (Foxf2), as indicated. (G) Representative images of trans-well chambers for migration and invasion of 344SQ cells which are induced (+) (10 days) for expression of either TRIPZ-GFP control vector (GFP) or GFP-Foxf2 (Foxf2) as indicated. (H) Bright field or GFP fluorescence microscopy images showing morphology of human HCC827 cells which are induced for expression of GFP-vector (control) or GFP-Foxf2 for 10 days. (I) Trans-well migration of human HCC827 cells which are induced (+) (10 days) for expression of either TRIPZ-GFP control vector, or GFP-Foxf2, as indicated. (J) Western Blot analysis for expression of GFP-Foxf2 or EMT markers in human HCC827 cells which are induced (+) (10 days) for expression of either TRIPZ-GFP control vector (GFP) or GFP-Foxf2 (Foxf2) as indicated. (K) Bright-field microscopy and quantitation of 3D spheroids in a matrix of Matrigel:Collagen (1:1), formed by HCC827 cells induced for 10 days for expression of either TRIPZ-GFP control vector (GFP(+)) or GFP-FOX2 (FOXF(+)). (L–M) Representative images of trans-well chambers for migration and invasion of mouse 344SQ or human H157 cells with stable expression of either control non silencing/Scramble hairpins or hairpins targeting mouse or human Foxf2 as indicated.
(A) qPCR analysis for relative expression of mouse Zeb1, Foxf2, E-Cadherin and Vimentin in 344SQ, 344SQ-GFP+ (Vector) and 344SQ-Foxf2+ cells. Gene expressions are normalized to levels in the parental 344SQ cells. (B) Immunofluorescence staining of 344SQ-GFP+ (vector) or 344SQ-Foxf2+ cells was performed for the expression of E-Cadherin or β-Catenin. Secondary antibody used was Alexa-546 labelled (Life technologies). Cells were counter stained with DAPI for nuclear staining. Scale bars are as indicated. Staining was performed as previously described2, 23. (C) Proliferation of indicated cell lines as measured by WST1 reagent (Roche; as per manufacturer’s protocol) when cultured in 2D. (D) (Left) Representative images of IHC stained slides for either Ki67 or no-primary controls for the indicated cells. (Right) Plots representing the quantification of IHC positive cells. 3 independent fields from 2 different tumor sections were acquired and then IHC positive cells were analyzed and counted by image J software. Staining was performed as previously described74. (E) qPCR analysis for relative expression of mouse EMT transcription factors in 393P-Foxf2 cells that were induced for 24 hours for Foxf2 expression. (F) Relative normalized luciferase activity from Zeb1 and Snai1 promoters in 393P-GFP (Vector) or 393P-Foxf2 cells which were induced for GFP or Foxf2 expression respectively. (Asterisks indicate significance p≤0.05)
(A) qPCR analysis for relative expression of mouse Foxf2, Zeb1 and E-Cad in 393P cells that were induced for GFP (vector control) expression for different times as indicated. (B) Western Blot analysis for expression of Zeb1, E-Cad and Foxf2 in 393P cells that were induced for GFP (Vector control) expression for different times as indicated. (C) qPCR analysis for relative expression of mature mouse miR-200a, 200b and 200c in 393P cells that were induced for GFP (vector control) expression for different times as indicated. (D) qPCR analysis for relative expression of mouse miR-141 and miR-429 in 393P-Foxf2 inducible cells that has been induced for different periods of time as indicated. (E) Western Blot analysis for expression of Zeb1, ECad and Foxf2 in 393P cells that were induced for GFP (vector control) expression for different times as indicated, after transfection of either control siRNA (si-Control) or siRNA targeting Zeb1 (si-Zeb1). (F) Western blot analysis for expression of Zeb1, E-Cad and Foxf2 in 393P cells that were induced for GFP (Vector control)for different times, with simultaneous stable expression of scrambled or shRNA hairpin effectively targeting Zeb1. (G) Western blot analysis for expression of Zeb1, E-Cad and Foxf2 in 393P cells that were induced for Foxf2 expression for different times, with simultaneous stable expression of scrambled or shRNA hairpin effectively targeting Zeb1. Relative band intensities for Zeb1 or E-Cad, normalized to respective Actin band intensities are shown for each lanes. (H) qPCR analysis for relative expression of mouse E-Cad in 393P cells that were induced for Foxf2 expression for different times, with simultaneous stable expression of either scrambled or shRNA hairpin effectively targeting Zeb1. (Asterisks indicate significance p≤0.05)
List of miRNA precursors and miRVana miRNA inhibitors.
Acknowledgments
We thank Dr. J. Larner (University of Virginia Health System, VA) for the kind gift of the pTRIPZ-GFP vector. We thank Dr. G. Goodall (University of Adelaide, Australia) for the kind gift of the miR-200 promoter reporter construct9. This work was supported by NCI K08 CA151661 (D.L.G.), an MD Anderson Cancer Center Physician Scientist Award (D.L.G.), Rexanna’s Foundation for Fighting Lung Cancer (D.L.G.), a CDMRP Lung Cancer Research Program award W81XWH-12-10294 (D.L.G. & S.T.K.). D.P. was supported by a CPRIT Graduate Scholar Training Grant (RP140106). L.D., J.W., P.T. are supported by Lung SPORE (P50 CA070907), Cancer Center Support Grant (CCSG CA016672), and Mary K. Chapman Foundation. C.J.C. was supported by CPRIT grant RP120713 and NCI grant CA125123. D.L.G. and L.A.B. are R. Lee Clark Fellows of the University of Texas MD Anderson Cancer Center, supported by the Jeane F. Shelby Scholarship Fund. We would like to thank members of the Gibbons lab for assistance and critical reading of the manuscript.
Footnotes
Conflict of interest.
The authors declare no conflict of interest.
Citations
- 1.Adams BD, Kasinski AL, Slack FJ. Aberrant Regulation and Function of MicroRNAs in Cancer. Current biology : CB. 2014;24:R762–R776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn YH, Gibbons DL, Chakravarti D, Creighton CJ, Rizvi ZH, Adams HP, et al. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. The Journal of clinical investigation. 2012;122:3170–3183. doi: 10.1172/JCI63608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nature communications. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA. org resource: targets and expression. Nucleic acids research. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome biology. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO reports. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer research. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 10.Browne G, Sayan AE, Tulchinsky E. ZEB proteins link cell motility with cell cycle control and cell survival in cancer. Cell cycle. 2010;9:886–891. doi: 10.4161/cc.9.5.10839. [DOI] [PubMed] [Google Scholar]
- 11.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO reports. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nature communications. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derksen PW, Liu X, Saridin F, van der Gulden H, Zevenhoven J, Evers B, et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer cell. 2006;10:437–449. doi: 10.1016/j.ccr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 18.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature reviews Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 19.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nature structural & molecular biology. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 20.Feng X, Wang Z, Fillmore R, Xi Y. MiR-200, a new star miRNA in human cancer. Cancer letters. 2014;344:166–173. doi: 10.1016/j.canlet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annual review of medicine. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 22.Gemmill RM, Roche J, Potiron VA, Nasarre P, Mitas M, Coldren CD, et al. ZEB1-responsive genes in non-small cell lung cancer. Cancer letters. 2011;300:66–78. doi: 10.1016/j.canlet.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes & development. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature cell biology. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 25.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. The Journal of biological chemistry. 2009;284:23204–23216. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirata H, Ueno K, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, et al. MicroRNA-182–5p promotes cell invasion and proliferation by down regulating FOXF2, RECK and MTSS1 genes in human prostate cancer. PloS one. 2013;8:e55502. doi: 10.1371/journal.pone.0055502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirohashi S. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. The American journal of pathology. 1998;153:333–339. doi: 10.1016/S0002-9440(10)65575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Current opinion in cell biology. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, et al. Systemic spread is an early step in breast cancer. Cancer cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Huynh C, Segura MF, Gaziel-Sovran A, Menendez S, Darvishian F, Chiriboga L, et al. Efficient in vivo microRNA targeting of liver metastasis. Oncogene. 2011;30:1481–1488. doi: 10.1038/onc.2010.523. [DOI] [PubMed] [Google Scholar]
- 32.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nature reviews Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh M, Igarashi M, Fukuda H, Nakagama H, Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer letters. 2013;328:198–206. doi: 10.1016/j.canlet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 34.Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. The Journal of experimental medicine. 2011;208:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knouf EC, Garg K, Arroyo JD, Correa Y, Sarkar D, Parkin RK, et al. An integrative genomic approach identifies p73 and p63 as activators of miR-200 microRNA family transcription. Nucleic acids research. 2012;40:499–510. doi: 10.1093/nar/gkr731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong PZ, Yang F, Li L, Li XQ, Feng YM. Decreased FOXF2 mRNA expression indicates early-onset metastasis and poor prognosis for breast cancer patients with histological grade II tumor. PloS one. 2013;8:e61591. doi: 10.1371/journal.pone.0061591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA biology. 2008;5:115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nature reviews Cancer. 2013;13:482–495. doi: 10.1038/nrc3539. [DOI] [PubMed] [Google Scholar]
- 39.Lei R, Tang J, Zhuang X, Deng R, Li G, Yu J, et al. Suppression of MIM by microRNA-182 activates RhoA and promotes breast cancer metastasis. Oncogene. 2014;33:1287–1296. doi: 10.1038/onc.2013.65. [DOI] [PubMed] [Google Scholar]
- 40.Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Targeting of integrin beta1 and kinesin 2alpha by microRNA 183. The Journal of biological chemistry. 2010;285:5461–5471. doi: 10.1074/jbc.M109.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li XL, Hara T, Choi Y, Subramanian M, Francis P, Bilke S, et al. A p21-ZEB1 complex inhibits epithelial-mesenchymal transition through the microRNA 183-96-182 cluster. Molecular and cellular biology. 2014;34:533–550. doi: 10.1128/MCB.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, Wang YK, et al. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Medical oncology. 2012;29:618–626. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 43.Lo PK, Lee JS, Chen H, Reisman D, Berger FG, Sukumar S. Cytoplasmic mislocalization of overexpressed FOXF1 is associated with the malignancy and metastasis of colorectal adenocarcinomas. Experimental and molecular pathology. 2013;94:262–269. doi: 10.1016/j.yexmp.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowery AJ, Miller N, Dwyer RM, Kerin MJ. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC cancer. 2010;10:502. doi: 10.1186/1471-2407-10-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizuguchi Y, Specht S, Lunz JG, 3rd, Isse K, Corbitt N, Takizawa T, et al. Cooperation of p300 and PCAF in the control of microRNA 200c/141 transcription and epithelial characteristics. PloS one. 2012;7:e32449. doi: 10.1371/journal.pone.0032449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nik AM, Reyahi A, Ponten F, Carlsson P. Foxf2 in intestinal fibroblasts reduces numbers of Lgr5(+) stem cells and adenoma formation by inhibiting Wnt signaling. Gastroenterology. 2013;144:1001–1011. doi: 10.1053/j.gastro.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 47.Nilsson J, Helou K, Kovacs A, Bendahl PO, Bjursell G, Ferno M, et al. Nuclear Janus-activated kinase 2/nuclear factor 1-C2 suppresses tumorigenesis and epithelial-to-mesenchymal transition by repressing Forkhead box F1. Cancer research. 2010;70:2020–2029. doi: 10.1158/0008-5472.CAN-09-1677. [DOI] [PubMed] [Google Scholar]
- 48.Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes & development. 1996;10:2462–2477. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 49.Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, et al. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development. 2006;133:833–843. doi: 10.1242/dev.02252. [DOI] [PubMed] [Google Scholar]
- 50.Pacurari M, Addison JB, Bondalapati N, Wan YW, Luo D, Qian Y, et al. The microRNA-200 family targets multiple non-small cell lung cancer prognostic markers in H1299 cells and BEAS-2B cells. International journal of oncology. 2013;43:548–560. doi: 10.3892/ijo.2013.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes & development. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nature cell biology. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 54.Pieraccioli M, Imbastari F, Antonov A, Melino G, Raschella G. Activation of miR200 by c-Myb depends on ZEB1 expression and miR200 promoter methylation. Cell cycle. 2013;12:2309–2320. doi: 10.4161/cc.25405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roy SS, Gonugunta VK, Bandyopadhyay A, Rao MK, Goodall GJ, Sun LZ, et al. Significance of PELP1/HDAC2/miR-200 regulatory network in EMT and metastasis of breast cancer. Oncogene. 2014;33:3707–3716. doi: 10.1038/onc.2013.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer research. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 57.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer research. 2010;70:9570–9580. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 58.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 60.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 61.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer research. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 62.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun Y, Fang R, Li C, Li L, Li F, Ye X, et al. Hsa-mir-182 suppresses lung tumorigenesis through down regulation of RGS17 expression in vitro. Biochemical and biophysical research communications. 2010;396:501–507. doi: 10.1016/j.bbrc.2010.04.127. [DOI] [PubMed] [Google Scholar]
- 64.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes & development. 2013;27:2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vincent-Salomon A, Thiery JP. Host microenvironment in breast cancer development: epithelial-mesenchymal transition in breast cancer development. Breast cancer research : BCR. 2003;5:101–106. doi: 10.1186/bcr578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang T, Tamakoshi T, Uezato T, Shu F, Kanzaki-Kato N, Fu Y, et al. Forkhead transcription factor Foxf2 (LUN)-deficient mice exhibit abnormal development of secondary palate. Developmental biology. 2003;259:83–94. doi: 10.1016/s0012-1606(03)00176-3. [DOI] [PubMed] [Google Scholar]
- 67.Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nature reviews Cancer. 2014;14:233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, et al. Pleiotropic effects of miR-183~96~182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta neuropathologica. 2012;123:539–552. doi: 10.1007/s00401-012-0969-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nature cell biology. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 70.Xia L, Huang W, Tian D, Zhu H, Qi X, Chen Z, et al. Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology. 2013;57:610–624. doi: 10.1002/hep.26029. [DOI] [PubMed] [Google Scholar]
- 71.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 72.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Developmental cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 73.Yang Y, Ahn YH, Gibbons DL, Zang Y, Lin W, Thilaganathan N, et al. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. The Journal of clinical investigation. 2011;121:1373–1385. doi: 10.1172/JCI42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang Y, Ahn YH, Chen Y, Tan X, Guo L, Gibbons DL, et al. ZEB1 sensitizes lung adenocarcinoma to metastasis suppression by PI3K antagonism. The Journal of clinical investigation. 2014;124:2696–2708. doi: 10.1172/JCI72171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao H, Guo M, Zhao G, Ma Q, Ma B, Qiu X, et al. miR-183 inhibits the metastasis of osteosarcoma via downregulation of the expression of Ezrin in F5M2 cells. International journal of molecular medicine. 2012;30:1013–1020. doi: 10.3892/ijmm.2012.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng H, Kang Y. Multilayer control of the EMT master regulators. Oncogene. 2014;33:1755–1763. doi: 10.1038/onc.2013.128. [DOI] [PubMed] [Google Scholar]
- 77.Zheng S, El-Naggar AK, Kim ES, Kurie JM, Lozano G. A genetic mouse model for metastatic lung cancer with gender differences in survival. Oncogene. 2007;26:6896–6904. doi: 10.1038/sj.onc.1210493. [DOI] [PubMed] [Google Scholar]
- 78.Zhu J, Feng Y, Ke Z, Yang Z, Zhou J, Huang X, et al. Down-regulation of miR-183 promotes migration and invasion of osteosarcoma by targeting Ezrin. The American journal of pathology. 2012;180:2440–2451. doi: 10.1016/j.ajpath.2012.02.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Correlation plots for miR-182 and miR-183 expression and EMT scores of human tumors from different TCGA datasets as indicated. (B) Correlation plots for miR-200 expression and EMT scores of human tumors from different TCGA datasets as indicated. (C) Correlation plot for expression of miR-96, miR-182 and miR-183 and miR-200b in human tumors from different TCGA datasets as indicated. (D) qPCR analysis for relative expression of human miR-200b & 200c in panel of human NSCLC lines. (E) Bright field microscopy of HCC827 cells with either pCDNA vector or constitutive expression of Zeb1. (F) Western blot analysis for expression of Zeb1 and other EMT markers in isolated clones of HCC827 constitutively expressing either pCDNA vector of Zeb1. (G) qPCR analysis for relative expression of Zeb1 and other EMT markers in isolated clones of HCC827 constitutively expressing either pCDNA vector of Zeb1. (H) qPCR analysis for relative expression of miR-200a, 200b and 200c in isolated clones of HCC827 constitutively expressing either pCDNA vector of Zeb1.
List of 45 genes that are potential miR-200 targets having a role in the process of EMT and metastasis, generated by overlapping different mouse gene expression datasets as indicated.
List of 17 highly conserved common predicted mouse target genes of mmu-miR-200 and mmu-miR-183~96~182 cluster, using the microRNA prediction algorithms miRanda (www.microRNA.org), with a mirSVR7 score less than -6.0.
List of 48 highly conserved common predicted human target genes of hsa-miR-200 and hsa-miR-183~96~182 cluster, using the microRNA prediction algorithms miRanda (www.microRNA.org), with a mirSVR7 score less than −6.0.
List of primers used for generation of reagents and experiments.
List of miRNA Taq-Man assays and Antibodies.
Summary of samples in TCGA pan-cancer data sets with matched gene expression and miRNA data
(A) Gene set enrichment analysis was performed as described previously62). Gene expression profiles from four syngeneic tumors each, generated from either metastatic 344SQ cells (represented as red squares) or non-metastatic 393P cells (represented as blue squares) were used to perform Gene Set Enrichment Analysis to identify a list of most enriched genes in metastatic cells. The intensity of red or blue colors represent a heat map for magnitude of expression (Red=high ; blue=low). The top genes show Foxf2 as one of the most enriched genes in the metastatic cells compared to the non-metastatic ones. (B) qPCR analysis for relative expression of human Foxf2 and Zeb1 in HCC827 cells that were transfected with either Anti-miRNAs targeting miR-200a, 200b, 96 and 183 or a control Anti-miRNA. (C) (Left) qPCR analysis for relative expression of Zeb1, Foxf2 and other potential miR-200 targets in 344SQ cells treated with TGFβ for 10 days. (Right) qPCR analysis for relative expression of Zeb1 and FOXF2 in human HCC827 cells treated with TGFβ for 10 days.
(A) Bright field or GFP fluorescence microscopy images showing morphology of 393P cells which are either un-induced (−), induced (+) for GFP (control) expression for 10 days or first induced for 10 days and then un-induced for 8 days. (B) Representative images of trans-well chambers for migration and invasion of 393P cells which are un-induced (−) or induced (+) (10 days) for expression of either TRIPZ-GFP control vector (GFP) or GFP-Foxf2 (Foxf2) as indicated. (C) qPCR analysis for relative expression of Foxf2 and other EMT markers in mouse 531P1 cells which are induced(+) (10 days) for expression of either TRIPZ-GFP control vector (GFP(+)) or GFP-Foxf2 (Foxf2(+)). (D) Western Blot analysis for expression of GFP-Foxf2 or EMT markers in mouse 531P1 cells which are induced (+) (10 days) for expression of either TRIPZ-GFP control vector (GFP) or GFP-Foxf2 (Foxf2) as indicated. (E–F) Trans-well migration and invasion of 531P1 cells which are induced (+) (10 days) for expression of either TRIPZ-GFP control vector (GFP), or GFP-Foxf2 (Foxf2), as indicated. (G) Representative images of trans-well chambers for migration and invasion of 344SQ cells which are induced (+) (10 days) for expression of either TRIPZ-GFP control vector (GFP) or GFP-Foxf2 (Foxf2) as indicated. (H) Bright field or GFP fluorescence microscopy images showing morphology of human HCC827 cells which are induced for expression of GFP-vector (control) or GFP-Foxf2 for 10 days. (I) Trans-well migration of human HCC827 cells which are induced (+) (10 days) for expression of either TRIPZ-GFP control vector, or GFP-Foxf2, as indicated. (J) Western Blot analysis for expression of GFP-Foxf2 or EMT markers in human HCC827 cells which are induced (+) (10 days) for expression of either TRIPZ-GFP control vector (GFP) or GFP-Foxf2 (Foxf2) as indicated. (K) Bright-field microscopy and quantitation of 3D spheroids in a matrix of Matrigel:Collagen (1:1), formed by HCC827 cells induced for 10 days for expression of either TRIPZ-GFP control vector (GFP(+)) or GFP-FOX2 (FOXF(+)). (L–M) Representative images of trans-well chambers for migration and invasion of mouse 344SQ or human H157 cells with stable expression of either control non silencing/Scramble hairpins or hairpins targeting mouse or human Foxf2 as indicated.
(A) qPCR analysis for relative expression of mouse Zeb1, Foxf2, E-Cadherin and Vimentin in 344SQ, 344SQ-GFP+ (Vector) and 344SQ-Foxf2+ cells. Gene expressions are normalized to levels in the parental 344SQ cells. (B) Immunofluorescence staining of 344SQ-GFP+ (vector) or 344SQ-Foxf2+ cells was performed for the expression of E-Cadherin or β-Catenin. Secondary antibody used was Alexa-546 labelled (Life technologies). Cells were counter stained with DAPI for nuclear staining. Scale bars are as indicated. Staining was performed as previously described2, 23. (C) Proliferation of indicated cell lines as measured by WST1 reagent (Roche; as per manufacturer’s protocol) when cultured in 2D. (D) (Left) Representative images of IHC stained slides for either Ki67 or no-primary controls for the indicated cells. (Right) Plots representing the quantification of IHC positive cells. 3 independent fields from 2 different tumor sections were acquired and then IHC positive cells were analyzed and counted by image J software. Staining was performed as previously described74. (E) qPCR analysis for relative expression of mouse EMT transcription factors in 393P-Foxf2 cells that were induced for 24 hours for Foxf2 expression. (F) Relative normalized luciferase activity from Zeb1 and Snai1 promoters in 393P-GFP (Vector) or 393P-Foxf2 cells which were induced for GFP or Foxf2 expression respectively. (Asterisks indicate significance p≤0.05)
(A) qPCR analysis for relative expression of mouse Foxf2, Zeb1 and E-Cad in 393P cells that were induced for GFP (vector control) expression for different times as indicated. (B) Western Blot analysis for expression of Zeb1, E-Cad and Foxf2 in 393P cells that were induced for GFP (Vector control) expression for different times as indicated. (C) qPCR analysis for relative expression of mature mouse miR-200a, 200b and 200c in 393P cells that were induced for GFP (vector control) expression for different times as indicated. (D) qPCR analysis for relative expression of mouse miR-141 and miR-429 in 393P-Foxf2 inducible cells that has been induced for different periods of time as indicated. (E) Western Blot analysis for expression of Zeb1, ECad and Foxf2 in 393P cells that were induced for GFP (vector control) expression for different times as indicated, after transfection of either control siRNA (si-Control) or siRNA targeting Zeb1 (si-Zeb1). (F) Western blot analysis for expression of Zeb1, E-Cad and Foxf2 in 393P cells that were induced for GFP (Vector control)for different times, with simultaneous stable expression of scrambled or shRNA hairpin effectively targeting Zeb1. (G) Western blot analysis for expression of Zeb1, E-Cad and Foxf2 in 393P cells that were induced for Foxf2 expression for different times, with simultaneous stable expression of scrambled or shRNA hairpin effectively targeting Zeb1. Relative band intensities for Zeb1 or E-Cad, normalized to respective Actin band intensities are shown for each lanes. (H) qPCR analysis for relative expression of mouse E-Cad in 393P cells that were induced for Foxf2 expression for different times, with simultaneous stable expression of either scrambled or shRNA hairpin effectively targeting Zeb1. (Asterisks indicate significance p≤0.05)
List of miRNA precursors and miRVana miRNA inhibitors.