Abstract
Recent advances in experimental and computational methodologies are enabling ultra-high resolution genome-wide profiles of protein-DNA binding events. For example, the ChIP-exo protocol precisely characterizes protein-DNA crosslinking patterns by combining chromatin immunoprecipitation (ChIP) with 5′ → 3′ exonuclease digestion. Similarly, deeply sequenced chromatin accessibility assays (e.g. DNase-seq and ATACseq) enable the detection of protected footprints at protein-DNA binding sites. With these techniques and others, we have the potential to characterize the individual nucleotides that interact with transcription factors, nucleosomes, RNA polymerases, and other regulatory proteins in a particular cellular context. In this review, we explain the experimental assays and computational analysis methods that enable high-resolution profiling of protein-DNA binding events. We discuss the challenges and opportunities associated with such approaches.
Introduction
The central goal of transcriptional regulatory genomics is to understand how regulatory molecules in the nucleus interact with chromatin and each other in order to drive a cell's transcriptional program. Since thousands of distinct proteins, RNAs, and small molecules can be active in the eukaryotic nucleus, it's not surprising that we still understand little about the mechanisms underlying transcriptional regulatory systems. The first step towards generating such understanding is cataloging the activities and genomic binding locations of regulatory actors in transcriptional networks. Characterizing the DNA binding sites of transcription factor (TF) proteins, for example, can provide insight into the genes that they may regulate, or the regulatory proteins with which they may interact. However, we cannot currently predict genomic binding locations from sequence features with any great accuracy, and thus characterizing protein-DNA binding sites remains by necessity experimentally driven.
Over the past fifteen years, assays based on transcriptome profiling, chromatin immunoprecipitation (ChIP), or nuclease digestion (e.g. DNase I or MNase digestion) have enabled genome-wide profiling of genome-associated biochemical processes in a given cell population. The ability of these assays to produce a comprehensive picture of a given biochemical activity has been greatly facilitated by the advent of next generation sequencing technologies. Individual experiments can now tell us the genome-wide distribution of RNA production, chromatin accessibility, DNA methylation, or the localization of various transcription factors, chromatin modifiers, co-activators, RNA polymerases, or histones (and associated post-translational modifications like methylation, acetylation, phosphorylation, ubiquitylation, or citrullination). Sequencing-based assays are even beginning to provide us with insight into the three-dimensional organization of chromatin.
As regulatory genomics assays have proliferated and as access to data has been democratized via databases like GEO and the Short Read Archive (Barrett et al., 2009; Shumway et al., 2010), computational biologists are turning to the challenge of how to integrate disparate data types into cohesive models of regulatory activity. Initial steps in this direction have focused on describing correlative relationships between the genomic distributions of various regulatory processes (Barski et al., 2007; Venters et al., 2011; Dunham et al., 2012; Gerstein et al., 2012), and segmenting the genome into domains that display particular patterns of coordinated activities (Ernst and Kellis, 2010; Hoffman et al., 2012). Such efforts are ultimately motivated by a desire to discover how the various regulatory factors interact with one another, and whether any higher-order patterns of organization can be discerned.
Current models of regulatory organization are hampered by the relatively low spatial resolution of current regulatory genomics assays. Fortunately, recent methodological advances are providing unprecedented high-resolution profiles of protein-DNA binding. New experimental techniques have increased the resolution of particular protein-DNA interaction assays, while improved computational analyses have enabled increased resolution from older assays. In this review, we survey current experimental and computational methods that yield genome-wide protein-DNA occupancy profiles at single base-pair resolution. We also discuss the opportunities and challenges associated with building integrative models of regulatory organization from collections of high-resolution data types.
ChIPing away at the epigenome
Chromatin immunoprecipitation (ChIP) has long been the most popular method for profiling interactions between specific proteins and chromatin (Gilmour and Lis, 1984, 1985; Solomon and Varshavsky, 1985). In ChIP, proteins are covalently crosslinked to DNA in vivo, crosslinked chromatin is lysed and fragmented, and DNA attached to the protein of interest is enriched using an appropriate immobilized antibody. After reversing crosslinks, the resulting DNA can then be identified to assess where the protein is binding on the genome.
ChIP-chip enabled the first genome-wide profiles of ChIP enrichment by hybridizing immunoprecipitated DNA to microarray “chips” composed of DNA probes tiled across the genome (Blat and Kleckner, 1999; Ren et al., 2000; Iyer et al., 2001). The power of ChIP-chip became swiftly apparent, for example by enabling genome-wide occupancy profiles for hundreds of transcription factors in S. cerevisiae (Lieb et al., 2001; Lee et al., 2002; Harbison et al., 2004). However, two aspects of ChIP-chip limit the spatial resolution of profiled protein-DNA binding events. Firstly, the fragmented, immunoprecipitated DNA has a wide range of lengths, typically up to 1 Kbp. A positive hybridization result therefore tells us that a protein-DNA binding event exists in the vicinity of the genomic locus represented by one or more probes, but it does not tell us exactly which nucleotides are bound. Secondly, the number of genomic locations that can be probed using ChIP-chip is inherently limited by microarray design considerations, particularly the number of probes that a given microarray platform can support. While later microarray platforms had sufficient numbers of probes to enable tiling of fungal and smaller invertebrate genomes at 5-40 bp resolution, the application of ChIP-chip to the larger vertebrate genomes had been a compromise of either profiling a small selection of the genome (e.g. tiling of promoter regions or a previously selected set of regions of interest) or using dozens of distinct arrays to profile the entire genome at lower resolution.
The problem of poor genomic coverage was in principle solved by the advent of next-generation sequencing platforms. By directly sequencing the ends of immunoprecipitated fragments, ChIP-seq enables the genome-wide profiling of protein-DNA occupancy in a single experiment (Albert et al., 2007; Barski et al., 2007; Johnson et al., 2007; Mikkelsen et al., 2007). Early ChIP-seq studies demonstrated the assay's ability to profile the distribution of transcription factors, histone modifications, and RNA polymerase across entire genomes (Albert et al., 2007; Barski et al., 2007; Wang et al., 2008; Mikkelsen et al., 2007). The technique was subsequently adopted by the ENCODE and modENCODE projects, resulting in the generation of thousands of ChIP-seq experiments profiling numerous proteins in various human, mouse, worm and fly cell types (Dunham et al., 2012; Gerstein et al., 2010; Roy et al., 2010; Yue et al., 2014). ChIP-seq has thus come to be the dominant assay for characterizing protein-DNA binding on a genomic scale in vivo.
Current sequencing depths and protocol improvements are enabling production-scale processing of ChIP-seq experiments (Blecher-Gonen et al., 2013). However, several technical challenges remain to be optimized in the protocol. For example, ChIP-seq experiments typically contain a large proportion of noise; i.e. sequenced reads that are either produced from non-specific binding events or as the result of imperfect selection during immunoprecipitation. This noise is not evenly distributed over the genome, and may be biased towards accessible regions or highly expressed genes (Teytelman et al., 2013; Park et al., 2013). Similarly, ChIP-seq and other sequencing assays can suffer from GC-content biases (Benjamini and Speed, 2012) and artifactual accumulations of tags over copy-number variants (Rashid et al., 2011). Since accumulations of noise tags may be misinterpreted as true protein-DNA binding events, mitigating the effects of noise would appear to be a critical challenge.
Whilst constituting a vast improvement over ChIP-chip, the spatial resolution of ChIP-seq is still limited. In the standard ChIP-seq protocol, DNA is randomly sheared by sonication to produce DNA fragments in the size range of 200-500 bp. This results in a positional mapping uncertainty of similar magnitude (Figure 1). As a consequence, ChIP-seq data cannot typically resolve individual binding events within binding clusters; for example, if a transcription factor binds to multiple closely spaced motifs, the resulting convolved ChIP-seq signals will often appear as a single ChIP-enriched region. As in ChIP-chip, then, ChIP-seq signal accumulations point to regions that contain protein-DNA binding events, but not the exact locations of the bound nucleotides.
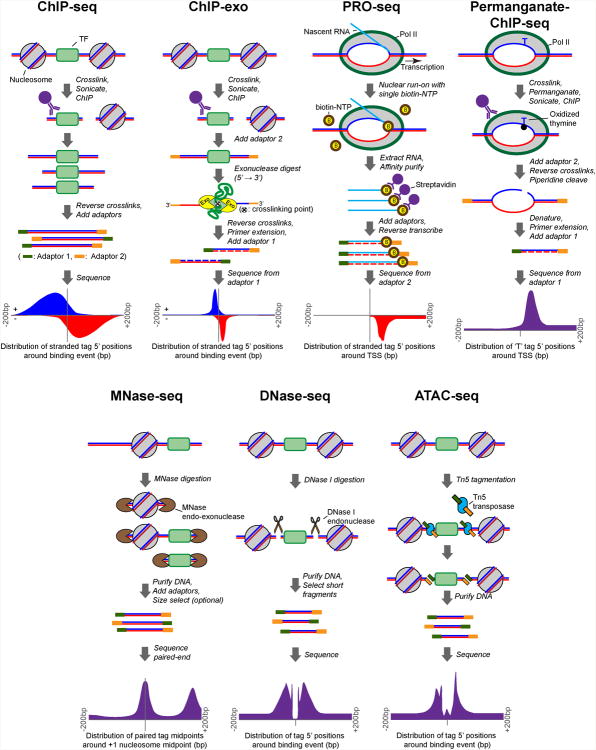
Figure 1.
Outline of seven high-resolution protein-DNA binding assays, summarizing the main protocol steps. Representative tag distribution profiles are taken from the following sources: mouse CTCF ChIP-seq tag 5′ positions (stranded) plotted around midpoints of the CTCF motif (Chen et al., 2008); yeast Reb1 ChIP-exo tag 5′ positions (stranded) plotted around midpoints of the Reb1 motif (Rhee and Pugh, 2011); Drosophila PRO-seq 5′ tag positions (stranded) plotted around annotated gene TSSs (Kwak et al., 2013); Drosophila Pol II permanganate-ChIP-seq 5′ positions of tags that begin with a thymine (unstranded) plotted around annotated gene TSSs (Li et al., 2013); yeast MNase-seq paired-end tag midpoints plotted around +1 nucleosome positions (Whitehouse et al., 2007); human DNase-seq 5′ tag positions (unstranded) plotted around CTCF-occupied motif midpoints (He et al., 2014); human ATAC-seq 5′ tag positions (unstranded) plotted around CTCF-occupied motif midpoints (Buenrostro et al., 2013). A color version of this figure is available online.
The limited resolution in ChIP-seq might be acceptable for some studies. For example, chromatin state analyses seek to characterize the diversity of co-occurring histone modifications in a given cell population (Ernst and Kellis, 2010; Ernst et al., 2011). In these studies, the analysis goal is to capture combinations of epigenomic signatures that occur over the same region (e.g. histone modifications at the same or neighboring nucleosomes), and some analysis approaches actually reduce the effective resolution of the profiled ChIP-seq data by smoothing signals over wider windows (Ernst and Kellis, 2010). Similarly, one may only be interested in listing the promoter and enhancer regions to which a given transcription factor protein binds, or the transcription start sites and gene bodies that contain RNA polymerases, and not the details of which exact nucleotides are directly bound. However, deeper biological insight into the modes and mechanisms of protein-DNA binding and gene regulation are made when we understand the precise interactions between various regulatory proteins and the DNA. The current spatial resolution of ChIP-seq data severely limits this goal.
Improving ChIP-seq's spatial resolution in silico
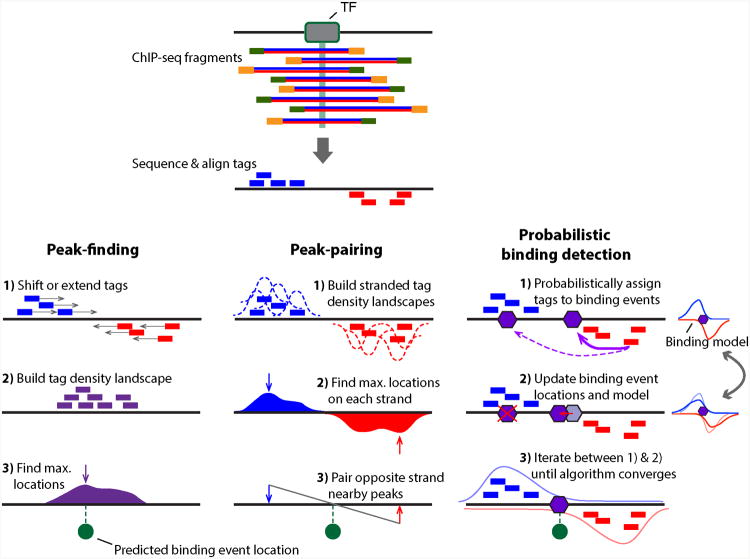
While ChIP-seq-enriched regions are typically several hundred base pairs wide, the underlying protein-DNA binding event locations can be more narrowly determined using computational methods. Intuitively, if fragmentation processes are relatively uniform over the genome, we should expect binding event locations to appear near the center of each ChIP-enriched region. Indeed, the first generation of ChIP-seq “peak-finding” analysis methods used the point of maximum local tag density (i.e. the peak “summit”) within each ChIP-enriched region as the estimated binding event location (Fejes et al., 2008; Zhang et al., 2008; Valouev et al., 2008; Kharchenko et al., 2008) (Figure 2). Other early approaches to defining ChIP-seq binding event locations from tag density information took advantage of the expected bimodal distribution of ChIP-seq tags on opposite strands around binding events (Albert et al., 2007; Kharchenko et al., 2008). In such methods, predicted binding event locations can be defined as the midpoint between paired peak predictions from opposing strands (Albert et al., 2007, 2008) (Figure 2), or relatedly, the position in the centers of ChIP-enriched regions at which the sense and antisense tag densities are most equally weighted (Jothi et al., 2008). More comprehensive discussions of ChIP-seq peak-finding approaches and assessments of their relative performance are available from other sources (Pepke et al., 2009; Park, 2009; Chen et al., 2012; Laajala et al., 2009; Rye et al., 2010).
Figure 2.
Outline of three ChIP-seq binding event detection methods. Peak-finding methods (e.g. MACS (Zhang et al., 2008)) typically either shift the ChIP-seq tag locations in a 3′ direction by half the expected fragment length, or extend the length of the tag in a 3′ direction to be equal to the expected fragment length. Tags from opposite strands are merged to construct an unstranded tag density landscapes, and binding event locations are predicted from the locations with maximum tag coverage within each region that contains a significant enrichment of ChIP-seq tags (i.e. the peak summit). Peak-pairing methods (e.g. GeneTrack (Albert et al., 2008)) build similar tag density landscapes, but retain strandedness information and typically do not shift or extend the tag locations. Peak locations are determined on each strand separately, and nearby peaks in the correct stranded orientation within a given distance are paired together. Binding event locations are predicted from the peak-pair midpoint locations. Probabilistic binding detection methods (e.g. GPS (Guo et al., 2010)) aim to estimate the locations of binding events that could have given rise to the observed ChIP-seq tag locations. These methods begin training with initial guesses of binding event locations and a model of how tags are expected to be distributed around real ChIP-seq binding events. During each training step, every ChIP-seq tag is probabilistically associated with nearby binding events, depending on the distance between the tag and the event location. Given these probabilistic tag assignments, binding event locations are updated to achieve a better fit with their associated tags, and the model of how tags are distributed around binding events is updated to reflect the accumulation of tags around all current binding events. During the training process, binding events with few assigned tags are weeded out of the model, and the process eventually converges to a set of final binding locations. A color version of this figure is available online.
The assumption that tag density information can yield accurate binding positions is justified in highly ChIP-enriched regions (i.e. containing large numbers of ChIP-seq tags) that arise from single protein-DNA binding events. For example, in analyzing the most highly ChIP-enriched regions in mammalian FoxA1 and NRSF transcription factor ChIP-seq datasets, the MACS peak-finder produces a binding event spatial resolution of 10-30 bp (as estimated using the average distance from the predicted binding event location to the nearest cognate motif instance) (Zhang et al., 2008). However, the spatial resolution of standard peak-finders degrades significantly in regions of weaker ChIP-enrichment or in regions containing multiple binding events (where the tag distributions from each binding event overlap and interfere with one another).
Several approaches enable the deconvolution of multiple nearby binding events within ChIP-seq enriched regions, thereby improving the resolution of individual binding event locations (Lun et al., 2009; Guo et al., 2010; Zhang et al., 2011; Wang and Zhang, 2011; Guo et al., 2012; Bardet et al., 2013; Gomes et al., 2014). Such approaches typically aim to estimate a model of binding event locations that would best explain observed ChIP-seq tag positions (Figure 2). For example, the GPS method (developed by one of us – S.M. – and colleagues) conceptually imagines a model containing potential binding events at every nucleotide on the genome (Guo et al., 2010). When GPS analyzes a particular ChIP-seq experiment, the potential binding events in the model compete with each other to take ownership of the observed ChIP-seq tags. Tags are probabilistically assigned to potential binding event locations according to an empirical model of how tags should be bimodally distributed around binding events. The shape of this empirical model is updated throughout the training process, as are the relative strengths of the potential binding events. During numerous iterative cycles, potential binding events that are not supported by sufficient numbers of ChIP-seq tags are weeded out of the model, and GPS analysis results in an accurate set of binding event location predictions. The machine-learning approach behind GPS and similar methods yields significantly higher spatial resolution of individual binding event locations, and can accurately deconvolve neighboring binding events that are at least 100 bp apart in ChIP-seq enriched regions (Guo et al., 2010).
Another advantage of the probabilistic approach to estimating binding event locations is that such methods can incorporate supporting (i.e. prior) information into the search for the precise binding event location. For example, the GEM method (also developed by S.M. & colleagues) incorporates TF binding motif discovery into the discovery of binding event locations, and uses a combination of ChIP-seq tag evidence and motifs to refine predicted binding locations (Guo et al., 2012). In effect, GEM can center transcription factor binding event predictions on appropriate cognate motif instances, depending on which motif instances (if any) are supported by the observed tag evidence. GEM can thus theoretically offer perfect resolution of binding locations for proteins that bind sequence-specifically and in regions that contain recognizable instances of the cognate motif. Of course, it follows that GEM's approach does not improve the resolution of binding events that do not correspond to a recognizable sequence feature, including those bound by histones, chromatin remodelers, transcriptional machinery, and even many sites bound by sequence-specific transcription factors (Afek and Lukatsky, 2012; Afek et al., 2014). However, probabilistic binding event estimation approaches with more limited resolution may still be productively applied to such datasets (e.g. (Zhang et al., 2012)).
ChIP-exo improves on ChIP-seq's spatial resolution
The ChIP-exo protocol (developed by one of us – B.F.P.) aims to experimentally improve upon the resolution of ChIP-seq (Rhee and Pugh, 2011, 2012). ChIP-exo incorporates a step where lambda exonuclease is used to digest immunopurified DNA fragments in a strand-specific 5′ → 3′ direction. The exonuclease either digests background fragments that are not crosslinked to a protein, or digests fragments until blocked by protein-DNA crosslinking (Figure 1). The distribution of the resulting ChIP-exo tag 5′ positions around binding events is much sharper than that of ChIP-seq, thereby leading to more accurate predictions of binding event locations. For example, in analyzing ChIP-exo data for the yeast transcription factor Reb1, the vast majority of predicted binding events were within 5 bp of a cognate motif instance (Rhee and Pugh, 2011).
In one recent demonstration that increased binding resolution can generate increased biological insight, we (B.F.P. & colleagues) used ChIP-exo to characterize the organization of individual histones in the yeast genome (Rhee et al., 2014). The increased resolution of ChIP-exo allowed us to determine that the distribution of histone modifications over the two halves of individual nucleosomes can be asymmetric with respect to the direction of transcription. For example, H3K9ac and H2BK123Ub are preferentially enriched on the promoter-proximal side of the +1 nucleosome, while the histone variant H2A.Z is preferentially enriched on the promoter-distal side. Furthermore, by gaining subnucleosomal resolution on the locations of individual histones, we found evidence for the existence of half nucleosome structures, each containing single copies of the four histones.
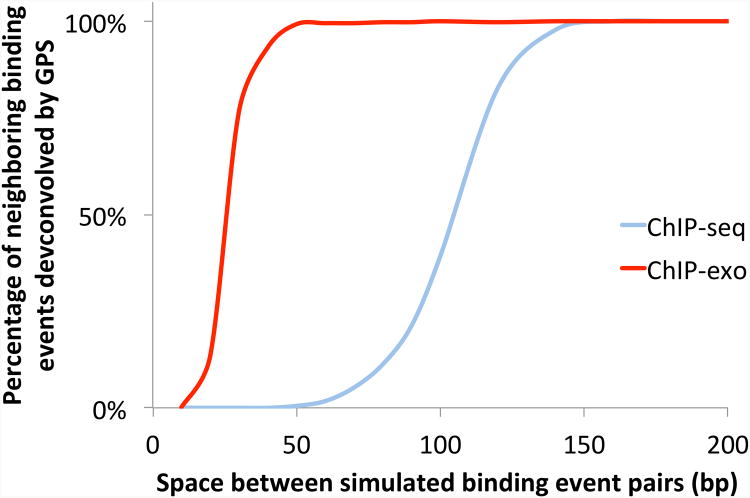
As with ChIP-seq, the resolution of weaker ChIP-exo binding events can be further improved by appropriate computational methods, although relatively few computational analysis methods have yet been designed that are specifically optimized for ChIP-exo analysis (Guo et al., 2012; Bardet et al., 2013; Wang et al., 2014). One immediate benefit of applying binding event deconvolution methods to ChIP-exo data is a greatly increased ability to discriminate between closely spaced binding events (Figure 3), which may be of great relevance for the analysis of densely packed mammalian enhancer elements (Gotea et al., 2010; Hardison and Taylor, 2012).
Figure 3.
GPS can deconvolve more closely spaced binding events in ChIP-exo data compared with ChIP-seq data. GPS was used to detect binding events in synthetic ChIP-seq and ChIP-exo datasets. Synthetic data was generated by randomly simulating tag positions along a genome (90% noise, 10% signal), where signal tags are distributed around predefined binding events of various strengths. Of the simulated binding events, 4,000 were generated as single events (located 50 Kbp away from another event) while 800 were simulated as pairs of events within a certain distance of one another. Simulated signal tags are distributed around binding event locations according to the CTCF ChIP-seq or Reb1 ChIP-exo 5′ tag distributions presented in Figure 1. A color version of this figure is available online.
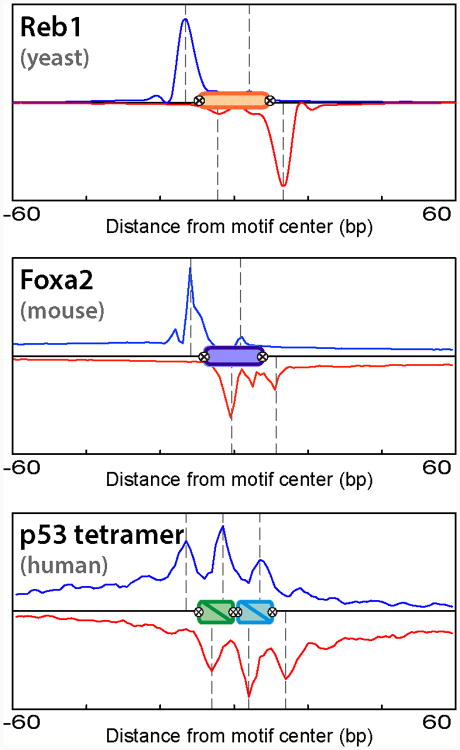
Aside from yielding higher resolution characterization of binding event locations, ChIP-exo may provide types of insight into protein-DNA interactions that are not provided by ChIP-seq. The distribution of ChIP-exo tag 5′ positions around binding events is determined by the protein-DNA crosslinking points that block exonuclease activity, and can therefore be a complex shape that is not necessarily bimodal or symmetric (Figure 4). Careful examination of such distributions (either at individual sites or aggregated across many sites) can provide insight into the organization of protein-DNA complexes. For example, we (B.F.P.) have used analysis of ChIP-exo derived crosslinking patterns to characterize the relative positions of general transcription factors in pre-initiation complexes (Rhee and Pugh, 2012), and the organization of ISW2, SWRC and INO80 chromatin remodeler complexes on nucleosomes (Yen et al., 2012, 2013). Two other recent ChIP-exo studies of glucocorticoid receptor (GR) in human and mouse cells found a distinctive crosslinking pattern at a subset of sites that contain binding motifs for forkhead-family TFs, thus suggesting that GR may bind DNA through FOX proteins at these locations (Starick et al., 2015; Lim et al., 2015).
Figure 4.
ChIP-exo tags have various distributions around TF binding events, depending on the underlying crosslinking pattern. Three examples are shown based on yeast Reb1, mouse FoxA1, and human p53 ChIP-exo datasets. A color version of this figure is available online.
Detailed patterns of crosslinking points within a single binding event can be difficult to interpret in the absence of structural information about the protein. However, when comparison can be made, crosslinking points are often found at the edges of protein-DNA structures and between structures, where there is solvent accessibility (to allow formaldehyde penetration) and transient base pair unstacking (to expose reactive amines). Rarely are crosslinks found inside protein-DNA complexes, except where they induce base unstacking, such as within transcription pre-initation complexes. Crosslinks arising from a single binding event could be spread across a broad region (e.g., 100-200 bp), which may occur if the protein interacts with other neighboring protein-DNA complexes and forms crosslinks with them. This is best exemplified where chromatin remodelers bind to ∼150 bp nucleosomes plus ∼70 bp of adjacent linker/promoter DNA. Such broad regions of crosslinking are difficult to interpret in the context of linear DNA, but take on structural and mechanistic significance when modeled in the context of a 3D structure of a nucleosome.
ChIP-exo has already been used in yeast, mammalian, and bacterial genomes to precisely locate the genome-wide binding locations of various sequence-specific transcription factors (Rhee and Pugh, 2011; Starick et al., 2015; Serandour et al., 2013; Chang et al., 2014; Chen et al., 2014; Wales et al., 2014; Carraro et al., 2014), chromatin remodelers (Yen et al., 2012, 2013), transcriptional machinery components (Rhee and Pugh, 2012), and histones (Rhee et al., 2014). However, some technical limitations have thus far prevented the widespread replacement of ChIP-seq with ChIP-exo. In particular, the additional washes and digestion steps in the ChIP-exo protocol result in less complex DNA libraries compared with ChIP-seq. Consequently, ChIP-exo is currently more prone to redundantly sequencing clonal replicates of DNA molecules than ChIP-seq given the same amount of starting material.
The library complexity issue may be mitigated with further improvements to the ChI-Pexo protocol. For example, the recently described ChIP-nexus protocol ligates both sequencing adaptors onto one end of ChIP fragments in a single step (as opposed to two separate ligation steps in the original ChIP-exo protocol) (He et al., 2015). Exonuclease digestion, DNA self-circularization with circLigase, and restriction enzyme cutting between the two adaptors creates the final library. By removing one of the relatively inefficient ligation steps, ChIP-nexus yields higher complexity libraries with the same resolution as ChIP-exo. One concern is that circLigase activity might have sequence specificity (Kwok et al., 2013), potentially creating bias in the identification of binding locations.
Tracing the footprints of protein-DNA binding interactions
Protection patterns from the actions of non-specific nucleases like micrococcal nuclease (MNase) and deoxyribonuclease I (DNase I) have long been used to characterize protein-DNA binding events with native chromatin (Pirrotta, 1973; Noll, 1974; Galas and Schmitz, 1978). DNase I footprinting, for example, characterizes which nucleotides in a given sequence are protected from DNase I cleavage by a protein-DNA binding event (Galas and Schmitz, 1978). With the advent of high-throughput sequencing technologies, MNase and DNase I have been used to profile genome-wide distributions of nucleosomes and nucleosome-depleted regions, respectively. Now MNase-seq, DNase-seq, and other assays, combined with the high sequencing depths provided by current sequencing platforms, are enabling the detection of subtle protection footprints at individual protein-bound locations across the whole genome. These types of assays differ from ChIP-based assays in that they report on native protein-DNA structures in addition to binding locations, but do not explicitly identify the bound factor. In ChIP assays, ChIP-seq captures locations but not structure. ChIP-exo captures both, to the extent that native crosslinking points reflect native structures.
MNase-ChIP-seq was arguably the first genome-wide high-resolution protein-DNA binding assay, albeit restricted to the characterization of nucleosome positions (Figure 1) (Albert et al., 2007). MNase cleaves unoccupied DNA and so digests linker DNA between proteins (nucleosomes and/or transcription factors). In principle, selection of nucleosome-sized MNase-digested fragments (147 bp), either biochemically or bioinformatically, offers a simple straightforward method of mapping nucleosomes. However, non-nucleosomal complexes may also create MNase-resistant fragments in the same size range, which might erroneously be interpreted as nucleosomes. Histone ChIP, size selection (either biochemically or bioinformatically), along with paired-end sequencing, offers a very precise mapping of stable nucleosome positions (Jiang and Pugh, 2009; Wal and Pugh, 2012). Even higher-resolution characterization of nucleosome locations can be achieved using site-directed hydroxyl radicals to cleave DNA at the center of nucleosomes (Brogaard et al., 2012), although this approach requires the construction of strains that incorporate cysteine into position 47 on histone H4.
In contrast, the DNase-seq assay is typically used to profile regions of accessible chromatin along the genome (Crawford et al., 2006). DNase I preferentially cleaves DNA in nucleosome-depleted regions, resulting in broad accumulations of DNase-seq tags in promoter, enhancer, and insulator regions. A recently proposed assay with a similar effect is ATAC-seq, which profiles transposase-accessible chromatin and which requires much less input material than DNase-seq (Buenrostro et al., 2013). ATAC-seq is based on the action of a Tn5 transposase that simultaneously fragments chromatin and tags ends with sequencing adaptors (also known as “tagmentation” (Adey et al., 2010)). Tn5 preferentially targets accessible chromatin, and thus amplifiable DNA fragments are preferentially located in regulatory regions in a similar distribution to that observed in DNase-seq.
Sufficiently high sequencing depths allow signatures of local enzyme protection (i.e., a lack of mapped tag 5′ ends, surrounded by an enrichment of tags) to be detected over transcription factor binding sites in MNase-seq (Kent et al., 2011; Henikoff et al., 2011), DNase-seq (Hesselberth et al., 2009), and ATAC-seq (Buenrostro et al., 2013) data (Figure 1). When the signals occurring over many binding sites for the same transcription factor are aligned and aggregated, protection patterns often reflect the pattern of protein-DNA contacts. For example, the aggregated DNase I cleavage pattern across hundreds of yeast Reb1 binding sites displays a protection pattern over a contiguous window of approximately 11 bp, centered on the major groove bound by this Myb-domain TF (Hesselberth et al., 2009). While protection footprints are clear in aggregate, they are difficult to detect at individual sites. Therefore, computational methods should be applied to individual footprints to discern binding events from sequence-based DNase-resistance.
Computational analyses of DNase I footprinting are typically reliant on transcription factor binding motifs. In some approaches, footprints are detected as local sites of DNase-seq tag depletion (Hesselberth et al., 2009; Neph et al., 2012; Chen et al., 2010; Boyle et al., 2011). The sequences at predicted footprints are then compared against known TF binding preferences (from databases such as JASPAR (Mathelier et al., 2014), UniPROBE (Hume et al., 2014), or CIS-BP (Weirauch et al., 2014)) in order to predict which proteins might bind at the footprint. An alternative approach first starts with known motifs, and predicts which motif instances on the genome may display the characteristics of a DNase I-protected footprint (Pique-Regi et al., 2011; Sherwood et al., 2014). The reliance of current DNase I footprinting methods on sequence motifs means that analyses are typically focused on sites bound by sequence-specific transcription factors. However, this is far from a narrow application. For example, DNase I footprinting analyses performed by the ENCODE project resulted in 8.4 million predicted footprint sites, many of which were predicted to correspond to known motif instances or instances of hundreds of de novo discovered motifs (Neph et al., 2012).
DNase I footprinting analyses have raised the possibility that nearly all TF binding locations in a given cell type can be characterized in a single experiment. However, recent studies indicate that there may be a substantial false positive rate. One concern centers on the intrinsic sequence biases of DNase I and other nucleases (He et al., 2014; Sung et al., 2014), which may produce false protection footprint signatures over sites with a particular sequence composition. For example, according to one study (He et al., 2014), some of the novel motifs predicted via DNase I footprints by Neph, et al. (Neph et al., 2012) may actually result from such intrinsic DNase I biases. Cleavage biases may be mitigated to some extent by comparative analysis against a control experiment on naked DNA and by using computational methods that account for the sequence bias (He et al., 2014; Yardimci et al., 2014). Differential DNase-seq analysis across conditions may also implicitly control for sequence biases (e.g. (He et al., 2012; Sherwood et al., 2014)). Performing footprinting experiments with multiple distinct nucleases (e.g. benzonase or cyanase) may be an alternative strategy for mitigating biases in the future (He et al., 2014). One further concern with footprinting approaches is that not all TFs may produce detectable footprints. In particular, TFs with short residence times may not produce footprints at bound motifs (Sung et al., 2014).
While highly informative for characterizing a wide swath of regulatory sites in the genome, footprinting approaches cannot provide an unambiguous characterization of the global transcriptional regulatory network. Aside from the methodological caveats noted above, it is difficult to assign a particular TF to individual footprinting sites. Many TFs that share a DNA binding domain structural class will bind to similar motif patterns (Sandelin and Wasserman, 2004; Mahony et al., 2007), and numerous TFs from particular structural classes may be expressed in a given cell type. Therefore, confirming which TF binds to which footprint requires cross-referencing with data from ChIP-based assays.
Characterizing the transcriptional machinery in high-resolution
Characterizing the locations, transcriptional status, and dynamics of RNA polymerase (Pol) II along the genome is critical for developing a complete understanding of gene regulation in a given cell type. ChIP-seq can tell us where Pol II crosslinks to the genome, and antibodies that target specific Pol II carboxy terminal domain (CTD) post-translational modifications can tell us something of Pol II's aggregated transcriptional dynamics over a gene. For example, Pol II CTD serine 5 phosphorylation is correlated with sites of transcriptional initiation, whereas serine 2 phosphorylation is correlated with regions of transcriptional elongation (Palancade and Bensaude, 2003). However, Pol II ChIP-seq cannot definitively tell us if detected sites of Pol II ChIP enrichment represent polymerase that is transcriptionally engaged as opposed to being localized in a non-engaged state, nor can it characterize the direction in which a given transcriptionally-engaged polymerase is traveling. Several assays are enabling the characterization of transcriptionally engaged Pol II and associated transcriptional machinery at high resolution.
One set of approaches for characterizing the locations of transcriptionally engaged Pol II molecules focuses on isolating newly synthesized or nascent RNA transcripts. The 3′ end of a nascent RNA corresponds to the last base added to the transcript, and hence sequencing nascent RNA 3′ ends should provide single base resolution of RNA polymerase locations and the direction in which transcription is proceeding. To isolate nascent RNA, the native elongating transcript sequencing approach (NET-seq (Churchman and Weissman, 2011)) relies on immunoprecipitation of Pol II and sequencing the attached RNA. An alternative approach relies on stable binding of Pol II to the insoluble chromatin, whereby the attached nascent transcript is separated away from the more soluble bulk RNA (Weber et al., 2014). Both approaches have been used to characterize the interplay between nucleosomes and transcriptional pausing, and conclude that the +1 nucleosome is a strong barrier to transcriptional elongation (Churchman and Weissman, 2011; Weber et al., 2014).
NET-seq reveals the locations of chromatin-associated Pol II, but cannot distinguish between arrested and transcriptionally competent polymerases. Precision nuclear run-on sequencing (PRO-seq) enables single base resolution of transcriptionally competent Pol II genome-wide (Kwak et al., 2013) (Figure 1). PRO-seq derives from the global run-on sequencing assay (GRO-seq (Core et al., 2008)). In GRO-seq, newly synthesized transcripts are labeled with bromouridine by incubating cells with bromouridinetriphosphate under conditions that prevent additional Pol II initiation events. PRO-seq achieves single base-pair resolution by incorporating single biotin-labeled nucleoside triphosphates (NTPs), which prevent further elongation of Pol II. After affinity purifying and sequencing the 3′ ends of RNA fragments, tag signals show the positions and traveling directions of transcriptionally competent Pol II. Sequencing from the 5′ ends of 5′-capped RNA fragments enables precise mapping of Pol II initiation locations; relevant protocols include cap analysis gene expression (CAGE (Shiraki et al., 2003)) and variants that derive from GRO-seq (i.e. GRO-cap (Kruesi et al., 2013; Core et al., 2014)) and PRO-seq (i.e. PRO-cap (Kwak et al., 2013)). Kwak, et al. have used PRO-seq to characterize the locations of promoter proximal Pol II pausing, and to demonstrate that Pol II also accumulates at intron-exon junctions and over 3′ polyadenylation sites (Kwak et al., 2013).
As an alternative to profiling Pol II positions via the nascent transcript, the permanganate-ChIP-seq assay (also developed by B.F.P. & colleagues) enables genome-wide detection of transcription bubble locations (Li et al., 2013) (Figure 1). Permanganate oxidizes thymine residues in single stranded DNA (Giardina et al., 1992). In permanganate-ChIP-seq, formaldehyde crosslinked chromatin is treated with permanganate and sheared DNA fragments are immunoprecipitated using a Pol II antibody. By using piperidine to cleave at oxidized thymines and ligating sequencing adaptors to the fragment ends, the 5′ ends of sequencing reads should be located at thymine nucleotides within the transcriptional bubble. Li et al. have used permanganate-ChIP-seq to profile transcription bubbles along the Drosophila genome, determining that thymine reactivity was highly enriched 20-60bp downstream of TSSs at sites of Pol II pausing (Li et al., 2013). The lack of thymine reactivity at the TSS suggested that Pol II rapidly moves into a transcriptionally engaged paused state after PIC assembly.
Gaining a higher-resolution understanding of TF-DNA binding specificity
Current computational approaches cannot accurately predict from sequence features alone which genomic sites will be bound by a particular transcription factor. Standard representations of a given TF's cognate binding motif will typically predict hundreds of thousands of potential high affinity binding sites along a mammalian-scale genome, and yet only a small fraction of these sites appear to be bound under any given cellular context (Wasserman and Sandelin, 2004; Wunderlich and Mirny, 2009). Conversely, for many TFs, a large fraction of ChIP-enriched locations do not appear to contain any match to the TF's cognate motif (Afek and Lukatsky, 2012; Afek et al., 2014; Worsley Hunt and Wasserman, 2014). Part of this lack of predictive power undoubtedly stems from cell-specific interactions with chromatin structure and other regulatory proteins, which restrict some locations from being bound while promoting binding at others (John et al., 2011; Guertin et al., 2012; Mahony et al., 2014). Similarly, not all ChIP-enriched locations represent true direct TF binding sites. However, the models used to represent TF binding preference may also create substantial false positive and false negative binding site predictions. Over the past few years, high-throughput in vitro TF-DNA binding assays have brought our understanding and our representations of TF-DNA binding specificity into sharper focus. While we summarize the main trends here, these themes are more comprehensively surveyed in a recent review by Slattery, et al. (Slattery et al., 2014).
The first high-throughput assay to enable truly comprehensive mapping of a TF's DNA binding preference was the universal protein binding microarray (PBM) (Berger et al., 2006). PBMs rely on microarrays whose probes are cleverly designed to incorporate every possible 8-mer sequence multiple times. By measuring the degree to which a purified TF protein (or just the TF's DNA-binding domain) binds to each probe on the microarray, computational analysis can reconstruct the preference of the TF for every possible 8-mer sequence. Similarly motivated sequencing-based assays that measure a protein's binding preferences in a library of randomized DNA include Bind-n-Seq (Zykovich et al., 2009) and HT-SELEX/SELEX-seq (Zhao et al., 2009; Jolma et al., 2010). Recently, in vitro binding assays have moved to reintroduce the genomic sequence context that a TF reads around its binding sites, either by basing PBM probes on real genomic sequences (the genomic-context PBM (Gordân et al., 2013)) or by immunopurifying and sequencing naked DNA fragments that are bound by the TF (PBseq (Guertin et al., 2012)).
In vitro binding assays have greatly informed our understanding of TF binding specificity. For example, these assays have demonstrated the extent to which highly related TFs from the same structural class can have differing sequence affinities. In vitro binding analysis of numerous homeodomain TFs has shown that while related TFs often share a core high-affinity sequence preference, individual family members can have specific preferences for lower-affinity sites (Berger et al., 2008; Noyes et al., 2008). Related work in the ETS TF family has shown that these minor differences in in vitro binding specificity correlate with binding selectivity in vivo (Wei et al., 2010). In vitro binding assays have also demonstrated that regions flanking the core sequence motif may contain information that strongly contributes to TF binding affinity (Nutiu et al., 2011; Gordân et al., 2013). In particular, the DNA structure (or “shape”) in these flanking regions may be critical for binding recognition for some TFs (Gordân et al., 2013), an observation which has also been made from analysis of solved TF-DNA structures (Rohs et al., 2009).
Given the observed subtleties in TF in vitro binding preferences, it has become clear that our models of TF binding preference are inadequate. The standard representation of TF binding preference is the position weight matrix (PWM), which is a probabilistic representation of the relative occurrence of each nucleotide at each position in an alignment of the TF's observed binding sites (Berg and von Hippel, 1987; Stormo, 2000). PWMs enable an intuitive visualization of the TF's binding preference, and they can be constructed from a small number of observed binding sites. However, it has long been recognized that PWMs are imperfect representations of a TF's DNA binding preferences, particularly since they assume that each DNA position contributes independently to the overall binding energy (Benos et al., 2002). PWMs are often constructed using the most highly occupied sites (as estimated by ChIP enrichment), and thus may not accurately represent low affinity sites that a factor may bind when stabilized through other interactions. Therefore it is not surprising that PWMs do not sufficiently capture a TF's various binding preferences as measured using in vitro binding assays (Weirauch et al., 2013).
Given the shortcomings of PWMs, and given the numbers of training sequences made available by in vitro or ChIP-based in vivo binding assays, several more complex models of TF binding preference have been developed (Weirauch et al., 2013; Zhou and Liu, 2008; Ben-Gal et al., 2005; Sharon et al., 2008; Mathelier and Wasserman, 2013; Hooghe et al., 2012; Agius et al., 2010). These models typically capture higher-order dependencies between positions in the binding sites, and some also make use of additional features like DNA shape (Gordân et al., 2013). The methods used to train and represent these models are varied, and include Bayesian networks (Ben-Gal et al., 2005), Markov networks (Sharon et al., 2008), hidden Markov models (Mathelier and Wasserman, 2013), random forest models (Hooghe et al., 2012), and support vector machines (Gordân et al., 2013; Agius et al., 2010). These models perform better than PWMs in many cases (Weirauch et al., 2013), but can be complex to implement and typically don't lend themselves to intuitive visualization of the features that are important for TF-DNA binding specificity. Therefore, the field as a whole has not yet settled on an alternative to PWMs.
In addition to generating a better understanding of TF binding preferences, in vitro binding assays are complementary to the high-resolution in vivo binding assays discussed in the rest of this review. In particular, genomic context in vitro assays can provide a baseline for where a TF will bind in the absence of any chromatin effects, thus allowing some degree of deconvolution between the sequence and chromatin determinants of binding selectivity (Guertin et al., 2012). Similarly, in vitro binding assays can assess the effects on a TF's binding preference of adding individual co-factors into the environment (Slattery et al., 2011). However, and by definition, only high-resolution in vivo assays can determine where a TF binds given all the regulatory actors in a cellular environment. Therefore, we expect that future studies of TF-DNA selectivity will more closely integrate high-resolution data from both in vivo and in vitro contexts.
Chromatin interactions: adding another dimension to protein-DNA binding
The high-resolution assays discussed thus far provide a one-dimensional view on protein-DNA interactions; i.e. the locations that particular proteins bind along the linear genome. However, the three-dimensional organization of chromatin is also thought to play a key role in transcriptional regulation. For example, enhancers are thought to interact with target promoters that are thousands to millions of base-pairs away in linear sequence space through the creation of loops that bring distal regions into close spatial proximity. Similarly, distal interactions between DNA-bound insulator proteins such as CTCF may lead to the creation of higher-order domains of similarly regulated chromatin. Chromatin interaction assays are providing a new window into the spatial organization of chromatin in the nucleus. These assays do not yet provide high enough resolution to unambiguously characterize the protein-DNA binding locations that mediate chromatin interactions, although protocol advancements may soon enable such insight.
The Hi-C assay combines DNA proximity ligation and paired-end next-generation sequencing to capture chromatin interactions genome-wide (Lieberman-Aiden et al., 2009). The name “Hi-C” reflects that this assay is a genome-wide extension of chromosome conformation capture (3C) assays that used proximity ligation to test chromatin interactions between a small number of loci (Dekker et al., 2002; Dostie et al., 2006). In Hi-C, crosslinked chromatin is cut with restriction enzymes, and the ends of digested DNA fragments are marked with biotin before being ligated to proximal fragments. The assumption underlying this procedure is that pairs of fragments from both sides of a long-range chromatin interaction will be ligated together, as they would be expected to be maintained in proximity to one another in solution via the crosslinked protein-DNA and protein-protein contacts that mediate the chromatin interaction. Due to the low efficiency of ligation, ligated fragments are affinity purified using streptavidin beads before sequencing.
Most Hi-C read pairs represent interactions between DNA fragments that are nearby one another on the linear genome. Sequencing depth therefore limits the degree to which long-distance interactions can be detected with statistical significance. For example, Lieberman-Aiden, et al. limited their analyses to 1 Mbp resolution (i.e. they split the genome into 1 Mbp sized bins, and counted the pairs of Hi-C reads that linked each pair of bins) (Lieberman-Aiden et al., 2009). Even at this very low resolution, the authors were able to demonstrate that the human genome is split into many domains that interact with one another in two distinct compartments (Lieberman-Aiden et al., 2009).
One way to improve the resolution of chromatin interactions is to focus only on interactions that involve a particular protein. The ChIA-PET assay aims to do just this by performing ChIP against a protein of interest before the biotin-labeled ligation step (Fullwood et al., 2009). ChIA-PET can thus assay enhancer-promoter loops if the ChIP step is targeted against Pol II (Li et al., 2012) or an activating transcription factor (Fullwood et al., 2009), or other types of loops mediated by CTCF (Handoko et al., 2011) or cohesin (DeMare et al., 2013). While providing higher resolution than Hi-C for a limited set of chromatin interactions, ChIA-PET does not necessarily provide high resolution on the exact protein-DNA binding events that mediate the interaction. Since chromatin fragments cannot be size-selected after the ChIP step (and before proximity ligation), reads are distributed in 1-2 Kbp windows around binding events.
Other studies have improved the resolution of Hi-C with greater sequencing depths and protocol improvements (Dixon et al., 2012; Jin et al., 2013; Rao et al., 2015; Ma et al., 2015). For example, the reliance on restriction enzymes in the original Hi-C protocol limits the complexity of the ligated products and hence limits the resolution of the method. This issue has recently been addressed by replacing restriction enzymes with DNase I to cut DNA (Ma et al., 2015). Higher resolution Hi-C studies have found a layer of organization at which chromatin is packaged into “topologically associated domains” (TADs) of size ∼1 Mbp (Dixon et al., 2012). A recent higher resolution Hi-C study by Rao, et al. achieved 1 Kbp resolution in one human cell type, and found that chromatin segregated into six subcompartments, each associated with different combinations of histone modifications (Rao et al., 2015). This study also characterized thousands of individual loop interactions directly from Hi-C data. The cost of this increased resolution was in greater sequencing depth requirements; 6.5 billion paired-end reads were required to achieve 1 Kbp resolution on chromatin interactions.
As chromatin interaction assays yield higher resolution characterization of the loops and domains that underlie chromatin organization, attention will inevitably turn towards the protein-DNA binding events that mediate these topological patterns. No existing assay yet enables high-resolution characterization of protein-DNA binding events alongside their associated chromatin interactions. Challenges for the future therefore include asking how best to integrate chromatin interaction assays with the high-resolution (one-dimensional) protein-DNA binding assays described above, or whether computational algorithms can improve the resolution of Hi-C-derived interaction points directly.
High-resolution datasets require retuned analysis methods
Intuitively, as the resolution of protein-DNA binding assays increase, computational identification of binding events from the resulting data should become easier. This is true in many ways; high-resolution assays are typically associated with higher signal-noise ratios, and thus protein-DNA binding events “stick-out” more in such datasets. However, high-resolution assays may also enable detection of subtler protein-DNA binding features, which changes the computational challenges rather than removes them. Furthermore, assumptions useful for analysis of lower resolution assays will not always be appropriate for newer data types, so care should be taken to tune analysis methods to the properties of the particular assay at hand. Here, as an example, we will discuss properties of ChIP-exo that make standard ChIP-seq analyses inappropriate. We expect that similar points also apply to the analysis of other high-resolution datasets.
As we have seen above, ChIP-seq peak-finding often involves detecting some form of tag accumulation midpoint within ChIP-enriched regions. To smooth and amplify local signals, ChIP-seq peak-finding methods and genome browser visualizations typically extend and/or shift mapped ChIP-seq tags, and merge data from positive and negative strands. In higher-resolution ChIP-exo data, only the 5′ position of each mapped tag matters, as this position is typically 6 bp upstream of the crosslinking point that blocked exonuclease digestion. Smoothing or extending ChIP-exo 5′ tag positions in any way obscures the locations of crosslinking points and thus removes some of the resolution of this method. In particular, plotting the entire length of mapped ChIP-exo tags (Serandour et al., 2013) (a common practice in ChIP-seq analysis) lowers the effective resolution of the assay. Merging ChIP-exo data across strands is similarly counter-productive. ChIP-exo binding event analysis has thus far focused on detecting “peak-pairs”; i.e. pairs of sharp tag distributions from alternate strands that are produced at the 5′ borders of crosslinking points (Rhee and Pugh, 2011). Detecting a pair of peak summits at the expected distance and strand orientation adds confidence that a given tag accumulation was generated by a true ChIP crosslinking event, but such analysis is impossible if strand information is thrown away.
A more subtle difference between ChIP-seq and ChIP-exo analyses centers on the interpretation of ChIP-enriched peak summits. In ChIP-seq, the peak summit locations resulting from a given peak-finding analysis method are typically assumed to correspond to predicted binding event locations – i.e. the nucleotides occupied by the assayed protein. In ChIP-exo, the peak-pair midpoints represent crosslinking points, which may or may not involve the actual nucleotides bound in the protein-DNA interaction. Crosslinking points are most likely located near the edge of protein-DNA binding events, where bases and amino acids are more solvent accessible and reactive. However, care should be taken not to conflate ChIP-exo crosslinking points with only the borders of protein-DNA binding events (Wang et al., 2014), as there is not always a 1:1 correspondence between them. For example, the crosslinking position of H3 resides in linker regions between nucleosomes, ∼80 bp from the nucleosome center, where the core of histone H3 actually binds (Rhee et al., 2014). Here, as in most cases, the crosslinking pattern makes clearer sense in the context of a nucleosome structure, where the linker is spatially close to the nucleosome center and the highly regulated amino terminal tail of H3.
A final point that may apply to many high-resolution protein-DNA binding assays concerns the handling of sequencing reads that map to the same genomic coordinate. In a protein-DNA binding assay dataset, there are a number of reasons that we may see multiple identical reads aligned to the same 5′ base position: 1) They may result from expected enrichment during sample purification (i.e. within a ChIP-enriched region); 2) Some DNA fragments may be preferentially amplified by PCR (Aird et al., 2011) or over-enriched by assay-specific biases (Teytelman et al., 2013; Yardimci et al., 2014; Park et al., 2013); 3) Sequencing coverage may exceed the original number of independently obtained DNA fragments (i.e. over-sequencing). Since ChIP-seq reads are spread over a relatively wide window, it is typically assumed that duplicate tags are predominantly caused by artifactual effects (i.e. PCR biases or over-sequencing). Therefore, a common practice in analyses of ChIP-seq and other assays is to ignore all tags that map to a given 5′ position above a certain threshold (Zhang et al., 2008; Jothi et al., 2008; Pepke et al., 2009). As we have seen, the tight distribution of reads around genomic events produced by ChIP-exo, DNase-seq, and other high-resolution assays means that multiple reads should be expected to share the same 5′ position at true biological signals. Therefore, “de-duplicating” tags will throw away the most important signals produced by such assays. Paired-end sequencing can help to clarify which sequencing reads arise from truly identical DNA fragments, although this does not solve the general problem completely. Adding degenerate barcodes to each fragment in the initial library would more clearly resolve the source of duplicate reads, but this approach is not commonly used in practice (He et al., 2015). No computational method yet allows deconvolution of duplicate reads that arise from signals and artifacts, but it may be possible to tackle this issue in the broader context of methods that assign sequenced tags probabilistic weights to correct various biases (Rashid et al., 2011; Yardimci et al., 2014).
Concluding remarks and future directions
As protein-DNA binding assays move towards higher resolutions, it is worth reflecting on the opportunities that such methods will bring. What can we gain from more accurate genomic occupancy profiles of individual proteins in a given cellular population? As discussed earlier, for some applications higher-resolution binding profiles may not yield much further biological insight. For example, if the goal of an analysis is to merely list the genes that a particular protein binds nearby, lower resolution protein-DNA binding assays will suffice. However, high-resolution assays may enable forms of insight that current analyses do not support, and building upon these additional forms of information will be a key future challenge in regulatory genomics.
High-resolution protein-DNA binding assays may enable us to go beyond listing binding locations, and towards characterizing protein-DNA interaction modes and subtypes. In many of the high-resolution protein-DNA binding assays discussed above, the exact distribution of sequenced tags around the binding event is determined by the form of the interaction between the assayed protein and the binding site. For example, the locations of ChIP-exo crosslinking points may vary according to how the protein is interacting with other proteins in its vicinity. Careful analysis of ChIP-exo tag distribution patterns may therefore yield insight into protein-DNA interaction modes as well as binding event locations. However, no current computational analysis methods automatically characterize multiple tag distribution patterns (representing distinct protein-DNA interaction modes) for a single assayed protein using ChIP-exo.
Determining the fine-grained structure of enhancers and other genomic regulatory regions will be another key application enabled by high-resolution assays. Genome segmentation approaches have provided integrative analysis across many regulatory genomic experiments from the same cell type (Ernst and Kellis, 2010; Hoffman et al., 2012, 2013), and in doing so enable an annotation of genomic regions according to shared experimental signatures. However, and as noted earlier, current genome segmentation methods operate at low spatial resolution by design. With higher resolution assays, could genome segmentation methods be adapted to provide more fine-grained annotation of functional region subtypes and the ordering of elements within regions like enhancers? Perhaps, but these state-based methods may not be appropriate for modeling the types of ordering and spacing rules that may potentially underlie enhanceosome organization. Previous approaches to discovering cis-regulatory modules from sequence motif features tried to model ordering and spacing constraints between motif features (e.g. (Zhou and Wong, 2004; Fu et al., 2009)). These methods did not typically uncover syntax rules between the occurrences of TF binding motif instances in enhancers, either because they were overwhelmed with unbound motif instances or because enhancers may not require precise spacing between individual transcription factors that do not directly interact with one another. Could cis-regulatory module discovery techniques be productively applied to high-resolution binding event features instead of sequence features? Alternatively, the most informative models of protein-DNA binding may come from thermodynamically modeling the association between various regulatory actors and chromatin (Wasson and Hartemink, 2009; Zhong et al., 2014), where such models could be parameterized by high-resolution data. Ultimately, determining the best computational approaches to explore and explain high-resolution protein-DNA binding data will inform our understanding of the biophysical processes that guide gene regulation.
Acknowledgments
NIH grant ES013768 to B.F.P.
Footnotes
Declaration of Interests: BFP has a financial interest in Peconic, LLC, which utilizes the ChIP-exo technology implemented in this study and could potentially benefit from the outcomes of this research.
References
- Adey A, Morrison HG, Asan, Xun X, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, Shendure J. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afek A, Lukatsky DB. Nonspecific Protein-DNA Binding Is Widespread in the Yeast Genome. Biophys J. 2012;102:1881–1888. doi: 10.1016/j.bpj.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afek A, Schipper JL, Horton J, Gordân R, Lukatsky DB. Protein-DNA binding in the absence of specific base-pair recognition. Proc Natl Acad Sci U S A. 2014;111:17140–5. doi: 10.1073/pnas.1410569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agius P, Arvey A, Chang W, Noble WS, Leslie C. High resolution models of transcription factor-DNA affinities improve in vitro and in vivo binding predictions. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird D, Ross MG, Chen WS, Danielsson M, Fennell T, Russ C, Jaffe DB, Nusbaum C, Gnirke A. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Albert I, Wachi S, Jiang C, Pugh BF. GeneTrack--a genomic data processing and visualization framework. Bioinformatics. 2008;24:1305–1306. doi: 10.1093/bioinformatics/btn119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardet AF, Steinmann J, Bafna S, Knoblich JA, Zeitlinger J, Stark A. Identification of transcription factor binding sites from ChIP-seq data at high resolution. Bioinformatics. 2013;29:2705–2713. doi: 10.1093/bioinformatics/btt470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Edgar R. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Ben-Gal I, Shani A, Gohr A, Grau J, Arviv S, Shmilovici A, Posch S, Grosse I. Identification of transcription factor binding sites with variable-order Bayesian networks. Bioinformatics. 2005;21:2657–2666. doi: 10.1093/bioinformatics/bti410. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Speed TP. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benos PV, Bulyk ML, Stormo GD. Additivity in protein-DNA interactions: how good an approximation is it? Nucleic Acids Res. 2002;30:4442–4451. doi: 10.1093/nar/gkf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M, Badis G, Gehrke A, Talukder S, Philippakis A, Penacastillo L, Alleyne T, Mnaimneh S, Botvinnik O, Chan E. Variation in Homeodomain DNA Binding Revealed by High-Resolution Analysis of Sequence Preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg OG, von Hippel PH. Selection of DNA binding sites by regulatory proteins.Statistical-mechanical theory and application to operators and promoters. J Mol Biol. 1987;193:723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Blecher-Gonen R, Barnett-Itzhaki Z, Jaitin D, Amann-Zalcenstein D, Lara-Astiaso D, Amit I. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat Protoc. 2013;8:539–554. doi: 10.1038/nprot.2013.023. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Song L, Lee BK, London D, Keefe D, Birney E, Iyer VR, Crawford GE, Furey TS. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogaard K, Xi L, Wang JP, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro N, Matteau D, Luo P, Rodrigue S, Burrus V. The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination. PLoS Genet. 2014;10:e1004714. doi: 10.1371/journal.pgen.1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang GS, Chen XA, Park B, Rhee HS, Li P, Han KH, Mishra T, Chan-Salis KY, Li Y, Hardison RC, Wang Y, Pugh BF. A comprehensive and high-resolution genome-wide response of p53 to stress. Cell Rep. 2014;8:514–527. doi: 10.1016/j.celrep.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, Tjian R, Liu Z. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Hoffman MM, Bilmes JA, Hesselberth JR, Noble WS. A dynamic Bayesian network for identifying protein-binding footprints from single molecule-based sequencing data. Bioinformatics. 2010;26:i334–342. doi: 10.1093/bioinformatics/btq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J. Integration of External Signaling Pathways with the Core Transcriptional Network in Embryonic Stem Cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chen Y, Negre N, Li Q, Mieczkowska JO, Slattery M, Liu T, Zhang Y, Kim TK, He HH, Zieba J, Ruan Y, Bickel PJ, Myers RM, Wold BJ, White KP, Lieb JD, Liu XS. Systematic evaluation of factors influencing ChIP-seq fidelity. Nat Methods. 2012 doi: 10.1038/nmeth.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Martins AL, Danko CG, Waters CT, Siepel A, Lis JT. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, Davis S, Margulies EH, Chen Y, Bernat JA, Ginsburg D, Zhou D, Luo S, Vasicek TJ, Daly MJ, Wolfsberg TG, Collins FS. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- DeMare LE, Leng J, Cotney J, Reilly SK, Yin J, Sarro R, Noonan JP. The genomic landscape of cohesin-associated chromatin interactions. Genome Res. 2013;23:1224–1234. doi: 10.1101/gr.156570.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Dunham I, Ernst J, Furey TS, Gerstein M, Giardine B, Greven M, Hardison RC, Harris RS, Herrero J, Hoffman MM, Iyer S, Kelllis M, Khatun J, Kheradpour P, Kundaje A, Lassmann T, Li Q, Lin X, Marinov GK, Merkel A, Mortazavi A, Parker SCJ, Reddy TE, Rozowsky J, Schlesinger F, Thurman RE, Wang J, Ward LD, Whitfield TW, Wilder SP, Wu W, Xi HS, Yip KY, Zhuang J, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M, Pazin MJ, Lowdon RF, Dillon LAL, Adams LB, Kelly CJ, Zhang J, Wexler JR, Green ED, Good PJ, Feingold EA, Bernstein BE, Birney E, Crawford GE, Dekker J, Elinitski L, Farnham PJ, Gerstein M, Giddings MC, Gingeras TR, Green ED, Guigó R, Hardison RC, Hubbard TJ, Kellis M, Kent WJ, Lieb JD, Margulies EH, Myers RM, Snyder M, Starnatoyannopoulos JA, Tennebaum SA, Weng Z, White KP, Wold B, Khatun J, Yu Y, Wrobel J, Risk BA, Gunawardena HP, Kuiper HC, Maier CW, Xie L, Chen X, Giddings MC, Bernstein BE, Epstein CB, Shoresh N, Ernst J, Kheradpour P, Mikkelsen TS, Gillespie S, Goren A, Ram O, Zhang X, Wang L, Issner R, Coyne MJ, Durham T, Ku M, Truong T, Ward LD, Altshuler RC, Eaton ML, Kellis M, Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Batut P, Bell I, Bell K, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena HP, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Li G, Luo OJ, Park E, Preall JB, Presaud K, Ribeca P, Risk BA, Robyr D, Ruan X, Sammeth M, Sandu KS, Schaeffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Hayashizaki Y, Harrow J, Gerstein M, Hubbard TJ, Reymond A, Antonarakis SE, Hannon GJ, Giddings MC, Ruan Y, Wold B, Carninci P, Guigó R, Gingeras TR, Rosenbloom KR, Sloan CA, Learned K, Malladi VS, Wong MC, Barber GP, Cline MS, Dreszer TR, Heitner SG, Karolchik D, Kent WJ, Kirkup VM, Meyer LR, Long JC, Maddren M, Raney BJ, Furey TS, Song L, Grasfeder LL, Giresi PG, Lee BK, Battenhouse A, Sheffield NC, Simon JM, Showers KA, Safi A, London D, Bhinge AA, Shestak C, Schaner MR, Kim SK, Zhang ZZ, Mieczkowski PA, Mieczkowska JO, Liu Z, McDaniell RM, Ni Y, Rashid NU, Kim MJ, Adar S, Zhang Z, Wang T, Winter D, Keefe D, Birney E, Iyer VR, Lieb JD, Crawford GE, Li G, Sandhu KS, Zheng M, Wang P, Luo OJ, Shahab A, Fullwood MJ, Ruan X, Ruan Y, Myers RM, Pauli F, Williams BA, Gertz J, Marinov GK, Reddy TE, Vielmetter J, Partridge EC, Trout D, Varley KE, Gasper C, Bansal A, Pepke S, Jain P, Amrhein H, Bowling KM, Anaya M, Cross MK, King B, Muratet MA, Antoshechkin I, Newberry KM, McCue K, Nesmith AS, Fisher-Aylor KI, Pusey B, DeSalvo G, Parker SL, Balasubramanian S, Davis NS, Meadows SK, Eggleston T, Gunter C, Newberry JS, Levy SE, Absher DM, Mortazavi A, Wong WH, Wold B, Blow MJ, Visel A, Pennachio LA, Elnitski L, Margulies EH, Parker SCJ, Petrykowska HM, Abyzov A, Aken B, Barrell D, Barson G, Berry A, Bignell A, Boychenko V, Bussotti G, Chrast J, Davidson C, Derrien T, Despacio-Reyes G, Diekhans M, Ezkurdia I, Frankish A, Gilbert J, Gonzalez JM, Griffiths E, Harte R, Hendrix DA, Howald C, Hunt T, Jungreis I, Kay M, Khurana E, Kokocinski F, Leng J, Lin MF, Loveland J, Lu Z, Manthravadi D, Mariotti M, Mudge J, Mukherjee G, Notredame C, Pei B, Rodriguez JM, Saunders G, Sboner A, Searle S, Sisu C, Snow C, Steward C, Tanzer A, Tapanari E, Tress ML, van Baren MJ, Walters N, Washieti S, Wilming L, Zadissa A, Zhengdong Z, Brent M, Haussler D, Kellis M, Valencia A, Gerstein M, Raymond A, Guigó R, Harrow J, Hubbard TJ, Landt SG, Frietze S, Abyzov A, Addleman N, Alexander RP, Auerbach RK, Balasubramanian S, Bettinger K, Bhardwaj N, Boyle AP, Cao AR, Cayting P, Charos A, Cheng Y, Cheng C, Eastman C, Euskirchen G, Fleming JD, Grubert F, Habegger L, Hariharan M, Harmanci A, Iyenger S, Jin VX, Karczewski KJ, Kasowski M, Lacroute P, Lam H, Larnarre-Vincent N, Leng J, Lian J, Lindahl-Allen M, Min R, Miotto B, Monahan H, Moqtaderi Z, Mu XJ, O'Geen H, Ouyang Z, Patacsil D, Pei B, Raha D, Ramirez L, Reed B, Rozowsky J, Sboner A, Shi M, Sisu C, Slifer T, Witt H, Wu L, Xu X, Yan KK, Yang X, Yip KY, Zhang Z, Struhl K, Weissman SM, Gerstein M, Farnham PJ, Snyder M, Tenebaum SA, Penalva LO, Doyle F, Karmakar S, Landt SG, Bhanvadia RR, Choudhury A, Domanus M, Ma L, Moran J, Patacsil D, Slifer T, Victorsen A, Yang X, Snyder M, White KP, Auer T, Centarin L, Eichenlaub M, Gruhl F, Heerman S, Hoeckendorf B, Inoue D, Kellner T, Kirchmaier S, Mueller C, Reinhardt R, Schertel L, Schneider S, Sinn R, Wittbrodt B, Wittbrodt J, Weng Z, Whitfield TW, Wang J, Collins PJ, Aldred SF, Trinklein ND, Partridge EC, Myers RM, Dekker J, Jain G, Lajoie BR, Sanyal A, Balasundaram G, Bates DL, Byron R, Canfield TK, Diegel MJ, Dunn D, Ebersol AK, Ebersol AK, Frum T, Garg K, Gist E, Hansen RS, Boatman L, Haugen E, Humbert R, Jain G, Johnson AK, Johnson EM, Kutyavin TM, Lajoie BR, Lee K, Lotakis D, Maurano MT, Neph SJ, Neri FV, Nguyen ED, Qu H, Reynolds AP, Roach V, Rynes E, Sabo P, Sanchez ME, Sandstrom RS, Sanyal A, Shafer AO, Stergachis AB, Thomas S, Thurman RE, Vernot B, Vierstra J, Vong S, Wang H, Weaver MA, Yan Y, Zhang M, Akey JA, Bender M, Dorschner MO, Groudine M, MacCoss MJ, Navas P, Stamatoyannopoulos G, Kaul R, Dekker J, Stamatoyannopoulos JA, Dunham I, Beal K, Brazma A, Flicek P, Herrero J, Johnson N, Keefe D, Lukk M, Luscombe NM, Sobral D, Vaquerizas JM, Wilder SP, Batzoglou S, Sidow A, Hussami N, Kyriazopoulou-Panagiotopoulou S, Libbrecht MW, Schaub MA, Kundaje A, Hardison RC, Miller W, Giardine B, Harris RS, Wu W, Bickel PJ, Banfai B, Boley NP, Brown JB, Huang H, Li Q, Li JJ, Noble WS, Bilmes JA, Buske OJ, Hoffman MM, Sahu AO, Kharchenko PV, Park PJ, Baker D, Taylor J, Weng Z, Iyer S, Dong X, Greven M, Lin X, Wang J, Xi HS, Zhuang J, Gerstein M, Alexander RP, Balasubramanian S, Cheng C, Harmanci A, Lochovsky L, Min R, Mu XJ, Rozowsky J, Yan KK, Yip KY, Birney E. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes AP, Robertson G, Bilenky M, Varhol R, Bainbridge M, Jones SJM. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EGY, Huang PYH, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KDSA, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RKM, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Ray P, Xing EP. DISCOVER: a feature-based discriminative method for motif search in complex genomes. Bioinformatics. 2009;25:i321–329. doi: 10.1093/bioinformatics/btp230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas DJ, Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, Min R, Alves P, Abyzov A, Addleman N, Bhardwaj N, Boyle AP, Cayting P, Charos A, Chen DZ, Cheng Y, Clarke D, Eastman C, Euskirchen G, Frietze S, Fu Y, Gertz J, Grubert F, Harmanci A, Jain P, Kasowski M, Lacroute P, Leng J, Lian J, Monahan H, O'Geen H, Ouyang Z, Partridge EC, Patacsil D, Pauli F, Raha D, Ramirez L, Reddy TE, Reed B, Shi M, Slifer T, Wang J, Wu L, Yang X, Yip KY, Zilberman-Schapira G, Batzoglou S, Sidow A, Farnham PJ, Myers RM, Weissman SM, Snyder M. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, Alves P, Chateigner A, Perry M, Morris M, Auerbach RK, Feng X, Leng J, Vielle A, Niu W, Rhrissorrakrai K, Agarwal A, Alexander RP, Barber G, Brdlik CM, Brennan J, Brouillet JJ, Carr A, Cheung MS, Clawson H, Contrino S, Dannenberg LO, Dernburg AF, Desai A, Dick L, Dosé AC, Du J, Egelhofer T, Ercan S, Euskirchen G, Ewing B, Feingold EA, Gassmann R, Good PJ, Green P, Gullier F, Gutwein M, Guyer MS, Habegger L, Han T, Henikoff JG, Henz SR, Hinrichs A, Holster H, Hyman T, Iniguez AL, Janette J, Jensen M, Kato M, Kent WJ, Kephart E, Khivansara V, Khurana E, Kim JK, Kolasinska-Zwierz P, Lai EC, Latorre I, Leahey A, Lewis S, Lloyd P, Lochovsky L, Lowdon RF, Lubling Y, Lyne R, MacCoss M, Mackowiak SD, Mangone M, McKay S, Mecenas D, Merrihew G, Miller DM, Muroyama A, Murray JI, Ooi SL, Pham H, Phippen T, Preston EA, Rajewsky N, Rätsch G, Rosenbaum H, Rozowsky J, Rutherford K, Ruzanov P, Sarov M, Sasidharan R, Sboner A, Scheid P, Segal E, Shin H, Shou C, Slack FJ, Slightam C, Smith R, Spencer WC, Stinson EO, Taing S, Takasaki T, Vafeados D, Voronina K, Wang G, Washington NL, Whittle CM, Wu B, Yan KK, Zeller G, Zha Z, Zhong M, Zhou X, modENCODE Consortium. Ahringer J, Strome S, Gunsalus KC, Micklem G, Liu XS, Reinke V, Kim SK, Hillier LW, Henikoff S, Piano F, Snyder M, Stein L, Lieb JD, Waterston RH. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C, Pérez-Riba M, Lis JT. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 1992;6:2190–2200. doi: 10.1101/gad.6.11.2190. [DOI] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT. Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes. Proc Natl Acad Sci U S A. 1984;81:4275–4279. doi: 10.1073/pnas.81.14.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol Cell Biol. 1985;5:2009–2018. doi: 10.1128/mcb.5.8.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes ALC, Abeel T, Peterson M, Azizi E, Lyubetskaya A, Carvalho L, Galagan J. Decoding ChIP-seq with a double-binding signal refines binding peaks to single-nucleotides and predicts cooperative interaction. Genome Res. 2014;24:1686–1697. doi: 10.1101/gr.161711.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordân R, Shen N, Dror I, Zhou T, Horton J, Rohs R, Bulyk ML. Genomic Regions Flanking E-Box Binding Sites Influence DNA Binding Specificity of bHLH Transcription Factors through DNA Shape. Cell Rep. 2013;3:1093–1104. doi: 10.1016/j.celrep.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, Ovcharenko I. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 2010;20:565–77. doi: 10.1101/gr.104471.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin MJ, Martins AL, Siepel A, Lis JT. Accurate prediction of inducible transcription factor binding intensities in vivo. PLoS Genet. 2012;8:e1002610. doi: 10.1371/journal.pgen.1002610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Mahony S, Gifford DK. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput Biol. 2012;8:e1002638. doi: 10.1371/journal.pcbi.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Papachristoudis G, Altshuler RC, Gerber GK, Jaakkola TS, Gifford DK, Mahony S. Discovering homotypic binding events at high spatial resolution. Bioinformatics. 2010;26:3028–3034. doi: 10.1093/bioinformatics/btq590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CWH, Ye C, Ping JLH, Mulawadi F, Wong E, Sheng J, Zhang Y, Poh T, Chan CS, Kunarso G, Shahab A, Bourque G, Cacheux-Rataboul V, Sung WK, Ruan Y, Wei CL. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC, Taylor J. Genomic approaches towards finding cis-regulatory modules in animals. Nat Rev Genet. 2012;13:469–483. doi: 10.1038/nrg3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Chen MW, Jordan VC, Brown M, Liu XS. Differential DNase I Hypersensitivity Reveals Factor-Dependent Chromatin Dynamics. Genome Res. 2012;22:1015–25. doi: 10.1101/gr.133280.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Hu SS, Chen MW, Zang C, Liu Y, Rao PK, Fei T, Xu H, Long H, Liu XS, Brown M. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods. 2014;11:73–78. doi: 10.1038/nmeth.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci. 2011;108:18318–23. doi: 10.1073/pnas.1110731108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Johnston J, Zeitlinger J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat Biotechnol. 2015 doi: 10.1038/nbt.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, Fields S, Stamatoyannopoulos JA. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MM, Buske OJ, Wang J, Weng Z, Bilmes JA, Noble WS. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MM, Ernst J, Wilder SP, Kundaje A, Harris RS, Libbrecht M, Giardine B, Ellenbogen PM, Bilmes JA, Birney E, Hardison RC, Dunham I, Kellis M, Noble WS. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooghe B, Broos S, van Roy F, De Bleser P. A flexible integrative approach based on random forest improves prediction of transcription factor binding sites. Nucleic Acids Res. 2012;40:e106. doi: 10.1093/nar/gks283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume MA, Barrera LA, Gisselbrecht SS, Bulyk ML. UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2014;43:D117–22. doi: 10.1093/nar/gku1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Kivioja T, Toivonen J, Cheng L, Wei G, Enge M, Taipale M, Vaquerizas JM, Yan J, Sillanpää MJ, Bonke M, Palin K, Talukder S, Hughes TR, Luscombe NM, Ukkonen E, Taipale J. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent NA, Adams S, Moorhouse A, Paszkiewicz K. Chromatin particle spectrum analysis: a method for comparative chromatin structure analysis using paired-end mode next-generation DNA sequencing. Nucleic Acids Res. 2011;39:e26. doi: 10.1093/nar/gkq1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. eLife. 2013;2:e00808. doi: 10.7554/eLife.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. Precise Maps of RNA Polymerase Reveal How Promoters Direct Initiation and Pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok CK, Ding Y, Sherlock ME, Assmann SM, Bevilacqua PC. A hybridization-based approach for quantitative and low-bias single-stranded DNA ligation. Anal Biochem. 2013;435:181–186. doi: 10.1016/j.ab.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Laajala T, Raghav S, Tuomela S, Lahesmaa R, Aittokallio T, Elo L. A practical comparison of methods for detecting transcription factor binding sites in ChIP-seq experiments. BMC Genomics. 2009;10:618. doi: 10.1186/1471-2164-10-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong S, Zhang Z, Landt S, Raha D, Euskirchen G, Wei CL, Ge W, Wang H, Davis C, Fisher-Aylor KI, Mortazavi A, Gerstein M, Gingeras T, Wold B, Sun Y, Fullwood MJ, Cheung E, Liu E, Sung WK, Snyder M, Ruan Y. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Rhee HS, Ghosh SKB, Bai L, Pugh BF, Gilmour DS. Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Mol Cell. 2013;50:711–722. doi: 10.1016/j.molcel.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HW, Uhlenhaut NH, Rauch A, Weiner J, Hübner S, Hübner N, Won KJ, Lazar MA, Tuckermann J, Steger DJ. Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Res. 2015 doi: 10.1101/gr.188581.114. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun D, Sherrid A, Weiner B, Sherman D, Galagan J. A blind deconvolution approach to high-resolution mapping of transcription factor binding sites from ChIP-seq data. Genome Biol. 2009;10:R142. doi: 10.1186/gb-2009-10-12-r142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony S, Auron PE, Benos PV. DNA Familial Binding Profiles Made Easy: Comparison of Various Motif Alignment and Clustering Strategies. PLoS Comput Biol. 2007;3:e61. doi: 10.1371/journal.pcbi.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony S, Edwards MD, Mazzoni EO, Sherwood RI, Kakumanu A, Morrison CA, Wichterle H, Gifford DK. An integrated model of multiple-condition ChIP-Seq data reveals predeterminants of Cdx2 binding. PLoS Comput Biol. 2014;10:e1003501. doi: 10.1371/journal.pcbi.1003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Wasserman WW. The Next Generation of Transcription Factor Binding Site Prediction. PLoS Comput Biol. 2013;9:e1003214. doi: 10.1371/journal.pcbi.1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen C, Chou A, Ienasescu H, Lim J, Shyr C, Tan G, Zhou M, Lenhard B, Sandelin A, Wasserman WW. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Ay F, Lee C, Gulsoy G, Deng X, Cook S, Hesson J, Cavanaugh C, Ware CB, Krumm A, Shendure J, Blau CA, Disteche CM, Noble WS, Duan Z. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat Methods. 2015;12:71–78. doi: 10.1038/nmeth.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O/'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, Maurano MT, Humbert R, Rynes E, Wang H, Vong S, Lee K, Bates D, Diegel M, Roach V, Dunn D, Neri J, Schafer A, Hansen RS, Kutyavin T, Giste E, Weaver M, Canfield T, Sabo P, Zhang M, Balasundaram G, Byron R, MacCoss MJ, Akey JM, Bender MA, Groudine M, Kaul R, Stamatoyannopoulos JA. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974;251:249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of Homeodomain Specificities Allows the Family-wide Prediction of Preferred Recognition Sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutiu R, Friedman RC, Luo S, Khrebtukova I, Silva D, Li R, Zhang L, Schroth GP, Burge CB. Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nat Biotechnol. 2011;29:659–664. doi: 10.1038/nbt.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B, Bensaude O. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur J Biochem. 2003;270:3859–3870. doi: 10.1046/j.1432-1033.2003.03794.x. [DOI] [PubMed] [Google Scholar]
- Park D, Lee Y, Bhupindersingh G, Iyer VR. Widespread misinterpretable ChIP-seq bias in yeast. PloS One. 2013;8:e83506. doi: 10.1371/journal.pone.0083506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat Methods. 2009;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Isolation of the operators of phage lambda. Nature New Biol. 1973;244:13–16. doi: 10.1038/newbio244013a0. [DOI] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2015;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid NU, Giresi PG, Ibrahim JG, Sun W, Lieb JD. ZINBA integrates local covariates with DNAseq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 2011;12:R67. doi: 10.1186/gb-2011-12-7-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Bataille AR, Zhang L, Pugh BF. Subnucleosomal Structures and Nucleosome Asymmetry across a Genome. Cell. 2014;159:1377–1388. doi: 10.1016/j.cell.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive Genome-wide Protein-DNA Interactions Detected at Single-Nucleotide Resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, Washietl S, Arshinoff BI, Ay F, Meyer PE, Robine N, Washington NL, Di Stefano L, Berezikov E, Brown CD, Candeias R, Carlson JW, Carr A, Jungreis I, Marbach D, Sealfon R, Tolstorukov MY, Will S, Alekseyenko AA, Artieri C, Booth BW, Brooks AN, Dai Q, Davis CA, Duff MO, Feng X, Gorchakov AA, Gu T, Henikoff JG, Kapranov P, Li R, Macalpine HK, Malone J, Minoda A, Nordman J, Okamura K, Perry M, Powell SK, Riddle NC, Sakai A, Samsonova A, Sandler JE, Schwartz YB, Sher N, Spokony R, Sturgill D, van Baren M, Wan KH, Yang L, Yu C, Feingold E, Good P, Guyer M, Lowdon R, Ahmad K, Andrews J, Berger B, Brenner SE, Brent MR, Cherbas L, Elgin SCR, Gingeras TR, Grossman R, Hoskins RA, Kaufman TC, Kent W, Kuroda MI, Orr-Weaver T, Perrimon N, Pirrotta V, Posakony JW, Ren B, Russell S, Cherbas P, Graveley BR, Lewis S, Micklem G, Oliver B, Park PJ, Celniker SE, Henikoff S, Karpen GH, Lai EC, Macalpine DM, Stein LD, White KP, Kellis M. Identification of Functional Elements and Regulatory Circuits by Drosophila modENCODE. Science. 2010;330:1787–97. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rye MB, Sætrom P, Drabløs F. A manually curated ChIP-seq benchmark demonstrates room for improvement in current peak-finder programs. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Wasserman WW. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J Mol Biol. 2004;338:207–215. doi: 10.1016/j.jmb.2004.02.048. [DOI] [PubMed] [Google Scholar]
- Serandour AA, Brown GD, Cohen JD, Carroll JS. Development of an Illumina-based ChIP-exonuclease method provides insight into FoxA1-DNA binding properties. Genome Biol. 2013;14:R147. doi: 10.1186/gb-2013-14-12-r147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon E, Lubliner S, Segal E. A feature-based approach to modeling protein-DNA interactions. PLoS Comput Biol. 2008;4:e1000154. doi: 10.1371/journal.pcbi.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Hashimoto T, O'Donnell CW, Lewis S, Barkal AA, van Hoff JP, Karun V, Jaakkola T, Gifford DK. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, Fukuda S, Sasaki D, Podhajska A, Harbers M, Kawai J, Carninci P, Hayashizaki Y. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci U S A. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumway M, Cochrane G, Sugawara H. Archiving next generation sequencing data. Nucleic Acids Res. 2010;38:D870–871. doi: 10.1093/nar/gkp1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, Mann RS. Cofactor Binding Evokes Latent Differences in DNA Binding Specificity between Hox Proteins. Cell. 2011;147:1270–1282. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Zhou T, Yang L, Dantas Machado AC, Gordân R, Rohs R. Absence of a simple code: how transcription factors read the genome. Trends Biochem Sci. 2014;39:381–399. doi: 10.1016/j.tibs.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon MJ, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc Natl Acad Sci U S A. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starick SR, Ibn-Salem J, Jurk M, Hernandez C, Love MI, Chung HR, Vingron M, Thomas-Chollier M, Meijsing SH. ChIP-exo signal associated with DNA-binding motifs provide insights into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome Res. 2015 doi: 10.1101/gr.185157.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- Sung MH, Guertin MJ, Baek S, Hager GL. DNase Footprint Signatures Are Dictated by Factor Dynamics and DNA Sequence. Mol Cell. 2014;56:275–85. doi: 10.1016/j.molcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teytelman L, Thurtle DM, Rine J, Oudenaarden Avan. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci. 2013;110:18602–18607. doi: 10.1073/pnas.1316064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, Pugh BF. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wales S, Hashemi S, Blais A, McDermott JC. Global MEF2 target gene analysis in cardiac and skeletal muscle reveals novel regulation of DUSP6 by p38MAPK-MEF2 signaling. Nucleic Acids Res. 2014;42:11349–11362. doi: 10.1093/nar/gku813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wal M, Pugh BF. Genome-wide mapping of nucleosome positions in yeast using high-resolution MNase ChIP-Seq. Methods Enzymol. 2012;513:233–250. doi: 10.1016/B978-0-12-391938-0.00010-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Chen J, Wang C, Uusküla-Reimand L, Chen K, Medina-Rivera A, Young EJ, Zimmermann MT, Yan H, Sun Z, Zhang Y, Wu ST, Huang H, Wilson MD, Kocher JPA, Li W. MACE: model based analysis of ChIP-exo. Nucleic Acids Res. 2014;42:e156. doi: 10.1093/nar/gku846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang X. Pinpointing transcription factor binding sites from ChIP-seq data with SeqSite. BMC Syst Biol. 2011;5(Suppl 2):S3. doi: 10.1186/1752-0509-5-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Wasson T, Hartemink AJ. An ensemble model of competitive multi-factor binding of the genome. Genome Res. 2009;19:2101–2112. doi: 10.1101/gr.093450.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CM, Ramachandran S, Henikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, Yan J, Talukder S, Turunen M, Taipale M, Stunnenberg HG, Ukkonen E, Hughes TR, Bulyk ML, Taipale J. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Cote A, Norel R, Annala M, Zhao Y, Riley TR, Saez-Rodriguez J, Cokelaer T, Vedenko A, Talukder S, Dream5 Consortium. Bussemaker HJ, Morris QD, Bulyk ML, Stolovitzky G, Hughes TR. Evaluation of methods for modeling transcription factor sequence specificity. Nat Biotechnol. 2013;31:126–134. doi: 10.1038/nbt.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, Zheng H, Goity A, van Bakel H, Lozano JC, Galli M, Lewsey MG, Huang E, Mukherjee T, Chen X, Reece-Hoyes JS, Govindarajan S, Shaulsky G, Walhout AJM, Bouget FY, Ratsch G, Larrondo LF, Ecker JR, Hughes TR. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Worsley Hunt R, Wasserman WW. Non-targeted transcription factors motifs are a systemic component of ChIP-seq datasets. Genome Biol. 2014;15:412. doi: 10.1186/s13059-014-0412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich Z, Mirny LA. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet. 2009;25:434–440. doi: 10.1016/j.tig.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardimci GG, Frank CL, Crawford GE, Ohler U. Explicit DNase sequence bias modeling enables high-resolution transcription factor footprint detection. Nucleic Acids Res. 2014;42:11865–11878. doi: 10.1093/nar/gku810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide Nucleosome Specificity and Directionality of Chromatin Remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Pugh BF. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell. 2013;154:1246–1256. doi: 10.1016/j.cell.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, Shen Y, Pervouchine DD, Djebali S, Thurman RE, Kaul R, Rynes E, Kirilusha A, Marinov GK, Williams BA, Trout D, Amrhein H, Fisher-Aylor K, Antoshechkin I, DeSalvo G, See LH, Fastuca M, Drenkow J, Zaleski C, Dobin A, Prieto P, Lagarde J, Bussotti G, Tanzer A, Denas O, Li K, Bender MA, Zhang M, Byron R, Groudine MT, McCleary D, Pham L, Ye Z, Kuan S, Edsall L, Wu YC, Rasmussen MD, Bansal MS, Kellis M, Keller CA, Morrissey CS, Mishra T, Jain D, Dogan N, Harris RS, Cayting P, Kawli T, Boyle AP, Euskirchen G, Kundaje A, Lin S, Lin Y, Jansen C, Malladi VS, Cline MS, Erickson DT, Kirkup VM, Learned K, Sloan CA, Rosenbloom KR, Lacerda de Sousa B, Beal K, Pignatelli M, Flicek P, Lian J, Kahveci T, Lee D, James Kent W, Ramalho Santos M, Herrero J, Notredame C, Johnson A, Vong S, Lee K, Bates D, Neri F, Diegel M, Canfield T, Sabo PJ, Wilken MS, Reh TA, Giste E, Shafer A, Kutyavin T, Haugen E, Dunn D, Reynolds AP, Neph S, Humbert R, Scott Hansen R, De Bruijn M, Selleri L, Rudensky A, Josefowicz S, Samstein R, Eichler EE, Orkin SH, Levasseur D, Papayannopoulou T, Chang KH, Skoultchi A, Gosh S, Disteche C, Treuting P, Wang Y, Weiss MJ, Blobel GA, Cao X, Zhong S, Wang T, Good PJ, Lowdon RF, Adams LB, Zhou XQ, Pazin MJ, Feingold EA, Wold B, Taylor J, Mortazavi A, Weissman SM, Stamatoyannopoulos JA, Snyder MP, Guigo R, Gingeras TR, Gilbert DM, Hardison RC, Beer MA, Ren B The Mouse ENCODE Consortium. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Robertson G, Krzywinski M, Ning K, Droit A, Jones S, Gottardo R. PICS: probabilistic inference for ChIP-seq. Biometrics. 2011;67:151–163. doi: 10.1111/j.1541-0420.2010.01441.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Robertson G, Woo S, Hoffman BG, Gottardo R. Probabilistic inference for nucleosome positioning with MNase-based or sonicated short-read data. PloS One. 2012;7:e32095. doi: 10.1371/journal.pone.0032095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer C, Eeckhoute J, Johnson D, Bernstein B, Nussbaum C, Myers R, Brown M, Li W, Liu XS. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Granas D, Stormo GD. Inferring binding energies from selected binding sites. PLoS Comput Biol. 2009;5:e1000590. doi: 10.1371/journal.pcbi.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Wasson T, Hartemink AJ. Learning protein-DNA interaction landscapes by integrating experimental data through computational models. Bioinformatics. 2014;30:2868–2874. doi: 10.1093/bioinformatics/btu408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Liu JS. Extracting sequence features to predict protein-DNA interactions: a comparative study. Nucleic Acids Res. 2008;36:4137–4148. doi: 10.1093/nar/gkn361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Wong WH. CisModule: de novo discovery of cis-regulatory modules by hierarchical mixture modeling. Proc Natl Acad Sci U S A. 2004;101:12114–12119. doi: 10.1073/pnas.0402858101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zykovich A, Korf I, Segal DJ. Bind-n-Seq: high-throughput analysis of in vitro protein-DNA interactions using massively parallel sequencing. Nucleic Acids Res. 2009;37:e151. doi: 10.1093/nar/gkp802. [DOI] [PMC free article] [PubMed] [Google Scholar]