Abstract
Transcription elongation by RNA polymerase II (RNAP II) involves the coordinated action of numerous regulatory factors. Among these are chromatin-modifying enzymes, which generate a stereotypic and conserved pattern of histone modifications along transcribed genes. This pattern implies a precise coordination between regulators of histone modification and the RNAP II elongation complex. Here I review the pathways and molecular events that regulate co-transcriptional histone modifications. Insight into these events will illuminate the assembly of functional RNAP II elongation complexes and how the chromatin landscape influences their composition and function.
Keywords: histone modifications, transcription elongation, histone H2B ubiquitylation, H3K4 methylation, Cdk9, Cdk7
Introduction
The discovery of the first histone-modifying enzymes solidified the concept of histone posttranslational modifications as instrumental in the activation or repression of transcription.1,2 Subsequent studies unveiled an unanticipated variety of histone modifications. Remarkably, high-resolution mapping of chromatin modifications on a genome-wide scale has revealed that they are generally correlated, either positively or negatively, with gene activity, strongly arguing that transcriptional regulation is the primary determinant of chromatin modification patterns.3
A subset of modifications is primarily associated with the coding regions of transcribed genes, suggesting their involvement in RNAP II elongation. These include histone H3 methylated at lysines 4, 36, and 79 (H3K4me, H3K36me, and H3K79me), and histone H2B monoubiquitylated on lysine 120 (H2Bub1).4 Thus, the known genomic landscape of histone modifications suggests a complex relationship with RNAP II transcription: in some cases, histone modifications are a consequence, rather than a cause, of transcriptional activity.
Such a conceptual shift has broadened the scope of gene regulatory functions thought to be influenced by histone modifications. In particular, co-transcriptional histone modifications have been shown to have roles in directing nucleosome dynamics during transcription, preventing improper transcription initiation within coding regions, regulating RNA processing, and promoting recruitment of DNA repair factors. The ways in which histone modifications influence elongating RNAP II has been the subject of a number of thorough reviews.3-7
The emerging roles of histone modifications during elongation raise the question of how patterns of modifications are generated in concert with the elongation process. Here I review our current knowledge of the molecular links between chromatin-modifying enzymes and the RNAP II elongation complex, highlighting recent advances and outstanding questions.
Landscape of Histone Modifications Within Transcribed Genes
RNAP II elongation is associated with a program of histone modification that is remarkably consistent across genes and species. Signature features of this program are a 5′ peak of trimethyl H3K4 and a 3′ peak of trimethyl H3K36. H2Bub1, H3K79me, and dimethyl forms of both H3K4 and H3K36 are all found broadly distributed across coding regions. In addition, acetylated forms of histones are generally high near gene 5’ ends and low within coding regions.
There is significant regulatory crosstalk between different co-transcriptional histone modifications. For example, formation of H3K79me, and in certain cases H3K4me, requires H2Bub1. The mechanisms underlying these relationships are not fully understood but likely involve direct contact between the ubiquitin and histone methyltransferases, as well as effects on methyltransferase complex recruitment and stability.8 In mammalian cells, these methylation events may also require the monoubiquitylation of H2B at K34, whose roles in elongation have yet to be defined.9
There is also crosstalk between histone methylation (at H3K4 and H3K36) and histone acetylation in gene coding regions, involving methyl-lysine recognition by multiple histone acetyltransferase (HAT) and histone deacetylase (HDAC) complexes. Indeed, H3K4me and H3K36me broadly influence the interactions of regulatory factors with chromatin.10
Points of Contact on the RNAP II Elongation Complex: A Tale of 2 CTDs
Association of regulatory partners with the RNAP II elongation complex generally depends upon Rpb1, the RNAP II large subunit, and Spt5. Both proteins are universally conserved components of the transcriptional machinery. However, eukaryotic variants contain highly unusual C-terminal domains (CTDs) comprising multiple repeats of a short amino acid motif; these CTDs constitute the binding platform on which the RNAP II elongation complex is built.11,12 The canonical Rpb1 CTD motif is Y1S2P3T4S5P6S7 and is nearly invariant across species, although CTDs are typically composed of a combination of canonical and non-canonical repeats. The composition and length of the Spt5 CTD repeat is more variable, both within individual CTD domains and between species. The consensus repeated motif in human Spt5 is GS(R/Q)TP(M/L)Y.13,14 The fission yeast S. pombe Spt5 contains a related but distinct motif of consensus GSKTPAWNS, whereas the consensus repeat in budding yeast Spt5 is highly divergent (SAWGGQ).15-17
The Rpb1 CTD
All of the phosphorylatable residues within the repeated motif are phosphorylated at various stages of the transcription cycle.18 Most phospho-specific interactions with the Rpb1 CTD studied to date, including those related to histone modifications, involve the S2 and S5 positions. Phosphorylation of these sites within heptad repeat motif (S5-P and S2-P) has been studied extensively using phospho-specific antibodies suitable for chromatin immunoprecipitation (ChIP) experiments.18 S5-P and S2-P show highly conserved distribution patterns within gene coding regions: S5-P is preferentially enriched proximal to promoters, whereas S2-P peaks toward gene 3’ ends. Numerous biochemical and structural studies of phospho-CTD interaction domains recognizing S2-P and S5-P have reinforced the notion that S5-P acts early in elongation, whereas S2-P acts later. For example, mRNA capping enzymes, which act on nascent mRNA 5′ ends, are directly engaged and allosterically activated by S5-P. On the other hand, factors involved in mRNA 3′ end cleavage and polyadenylation directly bind to S2-P.12,19
Few direct interactions involving phosphorylation of Y1, T4, or S7 have been characterized, although S7-P plays an important role in snRNA biogenesis through interaction with the Integrator complex.20 The roles of these sites in directing chromatin modifications have not yet been defined.
The Spt5 CTD
Like the Rpb1 CTD, this domain participates in diverse co-transcriptional events, and genetic studies in yeast have revealed significant functional overlap between the Spt5 CTD and that of Rpb1.15,21-23 The Spt5 CTD is typically phosphorylated on threonine of the repeat sequence (the variant Spt5 CTD in budding yeast is phosphorylated on serine).14-17 Interestingly, capping enzyme interactions with the Spt5 CTD are abolished by phosphorylation, whereas binding of the histone modification co-factor Rtf1 is enhanced.13,21,24,25 As such, phosphorylation of the Spt5 CTD is beginning to emerge as an important part of the transition from early to late elongation.
Phosphorylation of the Rpb1 and Spt5 CTDs by Transcription-Associated CDKs
The regulatory pathways coupling histone-modifying enzymes to the RNAP II elongation complex begin with transcription-associated cyclin-dependent kinases (CDKs) that primarily target the Rpb1 and Spt5 CTDs. These include the TFIIH component Cdk7, the positive transcription elongation factor b (P-TEFb) component Cdk9, and Cdk12/13. Current evidence suggests a complex collaboration between these protein kinases in generating the pattern of Rpb1 CTD phosphorylation along genes, as both in vitro and in vivo experiments indicate that they each phosphorylate multiple Rpb1 CTD sites.18 In contrast, the conserved, phosphorylated threonine of the Spt5 CTD is a unique target of Cdk9 in vivo.14,26,27
Cdk7 acts at early stages of transcription elongation. Specific inhibition of Cdk7 in yeast (using “analog-sensitive” alleles that are susceptible to inhibition by bulky ATP analogs) impairs mRNA capping and reduces levels of S5-P near the 5’ ends of genes.28–30 Similar experiments in mammalian cell lines show that Cdk7 activity is necessary for stable association of RNAP II and Spt5 at promoter-proximal pause sites, consistent with a defect in early elongation.31,32
Cdk9 activity is important for formation of stable elongating forms of RNAP II. Cdk9 was originally identified as part of P-TEFb, whose activity relieves promoter-proximal pausing of RNAP II in metazoans.33,34 Pausing is imposed by the unphosphorylated form of Spt5 (as part of the DRB sensitivity inducing factor or DSIF complex) and a negative elongation factor (NELF) complex. Cdk9-dependent phosphorylation of the Rpb1 CTD, the Spt5 CTD, and a subunit of NELF releases RNAP II from the pause.33,34 Phosphorylation of these sites by Cdk9 has positive roles in elongation beyond pause relief per se, since it is also a prominent feature of elongation in yeast where pausing is not generally observed.16,17,26,35
Cdk9 activity has been linked to S2-P on the Rpb1 CTD based on the sensitivity of this modification to DRB and flavopiridol, protein kinase inhibitors with specificity for Cdk9.36,37 However, Cdk9-related kinases Cdk12 and Cdk13 are also sensitive to DRB and flavopiridol and likely contribute to S2-P levels in vivo.38 The orthologs of Cdk12 in Drosophila, in C. elegans, and in yeast (Ctk1 in S. cerevisiae; Lsk1 in S. pombe) are major Rpb1 S2 kinases in these organisms, and S2-P and Ctk1 share important functions in mRNA 3’-end processing.39-43
Co-Transcriptional Histone H2B Ubiquitylation
Given that H2Bub1 helps to orchestrate the co-transcriptional histone modification pattern in transcribed genes, the mechanisms regulating H2Bub1 dynamics during transcription are a key part of the co-transcriptional chromatin modification program (see Fig. 1). H2Bub1 is catalyzed by a complex composed of the E2 ubiquitin conjugating enzyme Rad6 and E3 ubiquitin ligase enzymes related to budding yeast Bre1 (RNF20 and RNF40 in humans).6 The complex also contains a non-catalytic subunit that is not conserved between different eukaryotes but is nonetheless essential for H2Bub1 formation. In the human complex, this subunit, termed WAC, is implicated in interaction of the complex with elongating RNAP II.44 The WAC WW domain interacts with RNAP II in vivo, and depletion of WAC impairs the interaction of RNF20/40 with RNAP II, consistent with a role for WAC in directly coupling RNF20/40 to elongating RNAP II. Purified WAC interacts with the phosphorylated Rpb1 CTD with some preference for the S2-P form.44 An S2A mutant form of RNAP II (in which serine 2 of each Rpb1 CTD repeat is replaced with alanine) is unable to support full levels of H2Bub1 in human cells, consistent with the involvement of S2-P in recruitment of the RNF20/RNF40/WAC complex.45
Figure 1.

Engagement of the phosphorylated Rpb1 and Spt5 CTD domains by H2Bub1 regulators. Dashed arrows indicate unknown interactions between PAF/Rtf1 and the RNF20/40 complex. See text for details.
H2Bub1 requires the activity of P-TEFb/Cdk9, which may directly participate in RNF20/RNF40/WAC recruitment through Rpb1 CTD phosphorylation.45 Cdk9 also promotes H2Bub1 through recruitment of the Polymerase Associated Factor (PAF) complex.46,47 PAF is a necessary cofactor for H2Bub1, and also plays important roles in RNAP II elongation and mRNA 3’ end processing.48 Cdk9 activity drives the recruitment of PAF in part through the phosphorylation of the Spt5 CTD.16,17,24,25,49 The Rtf1 subunit of PAF directly binds to Spt5-P through its conserved Plus 3 domain.13,24,25,49 Structural studies of the Plus 3 domain from human Rtf1 have revealed the molecular basis for this interaction. The Plus 3 domain adopts a fold similar to the Tudor domain, which typically recognizes methyl-arginine and methyl-lysine.13 However, critical aromatic residues required for methyl group recognition are not conserved in Plus 3. A co-crystal structure of the Plus 3-Spt5-P complex (containing a phosphopeptide derived from the CTD of human SPT5) revealed an extended interface containing 2 distinct interaction surfaces bridging the Plus3 domain and Spt5-P. Phosphothreonine recognition involves hydrogen-bonding interactions between conserved arginine side chains on Plus 3 and the phosphate oxygens. The hydrophobic Spt5 CTD residues C-terminal to the phosphosite (a conserved feature of the Spt5 CTD repeat) are accommodated by a hydrophobic surface on the Plus 3 domain. Importantly, mutagenesis studies showed that both parts of the interaction surface are necessary for stable complex formation in vitro.13
Introduction of Plus 3 domain mutations that abrogate the Spt5 interaction into budding yeast results in loss of the entire complex from transcribed genes, confirming the biological importance of this interface.13 However, PAF makes additional contacts with the RNAP II elongation machinery. In fact, both the Ctr9 and Cdc73 subunits of budding yeast PAF have domains that interact with the Rpb1 and Spt5 phospho-CTDs in vitro.49,50 How these domains recognize phospho-CTD is still unclear. Notably, the Cdc73 phospho-CTD interaction domain (PCID) is a highly conserved domain with structural similarity to nucleotide binding domains in Ras family GTPases.51
The existence of multiple mechanisms for PAF complex recruitment to elongating RNAP II may reflect the divergent functions of PAF components in elongation. Many of the biochemical activities of PAF, including stimulation of RNAP II elongation on DNA and chromatin templates, are independent of Rtf1.52–54 In vivo, functional assays demonstrate that Rtf1 is more closely aligned with histone H2B ubiquitylation enzymes than with other PAF components.55,56 Underscoring these differences is the fact that Rtf1 protein isolated from fission yeast or metazoan cells does not co-purify with the other PAF subunits.24,57–59 Thus, the recognition of different phosphosites by Rtf1 or PAF may indicate functionally distinct Cdk9 “pathways” that are differentially utilized during elongation.
The molecular basis for the requirement of PAF/Rtf1 for H2Bub1 remains an open question. A direct interaction between purified human PAF complex and RNF20/40, mediated by the Paf1 subunit, has been reported, but this has not been confirmed in other studies.44,60,61 Human Cdc73 has also been reported to interact with RNF20.62 Remarkably, the N-terminal domain of Rtf1, when overexpressed in yeast, can direct H2Bub1 formation independently of the entire PAF complex.56 The mechanism is not known, but this phenomenon underscores the importance of Rtf1 for the co-transcriptional histone modification program.
Adding to the complexity of Cdk9-dependent mechanisms involved in H2Bub1 formation is the fact that Rad6, the cognate E2 enzyme for H2Bub1, is itself phosphorylated by Cdk9.47,63,64 Rad6 phosphorylation occurs at a conserved serine in proximity to the active site and directly stimulates ubiquitin conjugating activity.47 The relative importance of Cdk9 activity toward Spt5 or Rad6 for H2Bub1 is currently unclear.
Histone Methyltransferases Directly Associate with the Rpb1 CTD
H3K4 methylation by Set1
Set1 enzymes are the catalytic subunits of H3K4 methyltransferase complexes termed Set1C or COMPASS.8 COMPASS-type methyltransferase complexes carry out all H3K4 methylation in yeast. A variety of additional H3K4 methyltransferases have been described in metazoa, including the mixed lineage leukemia (MLL1) protein and its homologs. MLL and COMPASS complexes have a number of shared subunits, suggesting that they have partially overlapping functions.
Consistent with the presence of H3K4me near the 5’ ends of genes, Set1C/COMPASS has been shown to interact specifically with the S5-P form of the Rpb1 CTD (Fig. 2). The S5-P form of Rpb1 is associated with Set1C purified from yeast or mammalian cells.65,66 In vitro studies implicate the Wdr82 subunit of the human Set1C in binding directly to the S5-P CTD.66 This interaction is stimulated by the N-terminal RRM domain of Set1, which is required for Wdr82 interaction with Set1C/COMPASS and, in turn, for association of Set1C with RNAP II in vivo.
Figure 2.
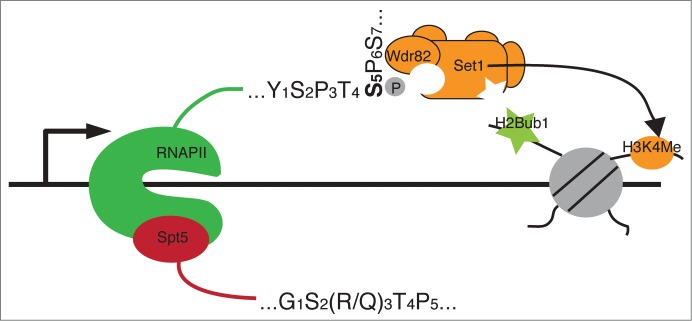
Engagement of the phosphorylated Rpb1 CTD and H2Bub1 by the Set1/COMPASS complex.
The MLL1 complex, which also associates with the S5-P form of RNAP II, does not contain Wdr82 or an RRM domain, suggesting an alternate mode of RNAP II interaction.67 Indeed, PAF1 interacts directly with MLL1, and the PAF complex participates in Hox gene activation by MLL1 fusion proteins.68,69 Direct interaction with PAF is also important for association of COMPASS with the elongation complex, although the nature of this interaction has yet to be defined.65
H3K36 methylation by Set2
H3K36 is methylated within the coding regions of transcribed genes by Set2 family enzymes. In yeast, Set2 is the sole methyltransferase for H3K36 and generates mono-, di-, and trimethylation at this site. This contrasts with the situation in metazoans, in which Set2 homologues (SETD2/HYPB in human) are primarily trimethylases and mono- and di-methyl forms are catalyzed by a variety of other methyltransferases (including nuclear receptor interacting proteins NSD1, 2, and 3).7
Set2 co-purifies with RNAP II and binds directly to the RNAP II CTD through its C-terminal SRI (Set2-Rpb1 interaction) domain70-73 (Fig. 3). The SRI domains of the yeast and human proteins bind weakly to singly phosphorylated S2-P and S5-P CTD repeats, but have a much higher affinity for CTD repeats that are simultaneously phosphorylated on both serines 2 and 5. Longer arrays of doubly phosphorylated repeats result in tighter binding, arguing that Set2 SRI recognizes hyperphosphorylated forms of the Rpb1 CTD.74
Figure 3.
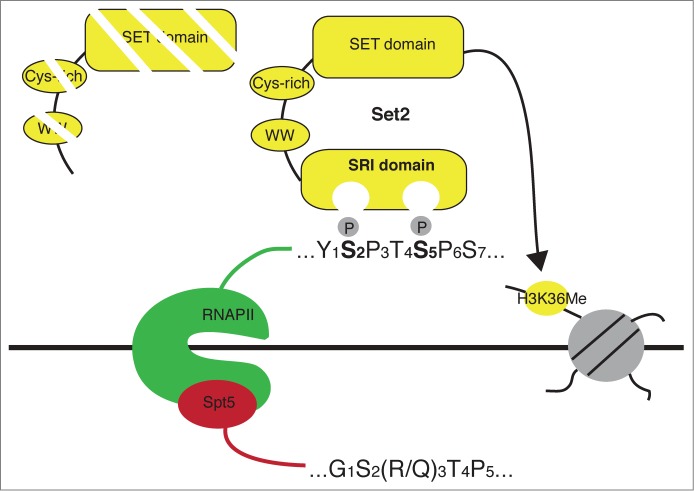
Engagement of the phosphorylated Rpb1 CTD by Set2/HYPB. CTD recognition is mediated by the SRI domain, whereas Set2 protein lacking this domain is targeted for destruction.
Structural studies have shown that the SRI domain, present in Set2 enzymes and in the DNA repair protein RECQ5, is unique among characterized PCID's.74-76 Solution structures of this domain from budding yeast Set2 and the human ortholog HYPB reveal a compact, 3-helix bundle. Phospho-CTD recognition, as inferred from NMR titration and mutagenesis experiments, primarily involves a portion of helix 2 that includes both positively charged and hydrophobic side chains. Although the specific contacts between modified CTD residues and this region are not known, the positive charges are presumed to directly engage the phosphate groups. The hydrophobic residues likely contact tyrosine 1 of the CTD repeat, a property shared by a number of other PCIDs.76
How Set2 is recruited to elongating RNAP II in vivo is still unclear. In budding yeast, deletion of CTK1, encoding the major Rpb1-Ser2 kinase, eliminates H3K36me, consistent with the importance of S2-P for binding of the SRI domain in vitro.70,71 Cdk7 and Cdk9 also promote H3K36me3, suggesting that several kinases may cooperate to generate the dually phosphorylated forms of the Rpb1 CTD recognized by the Set2 SRI domain in vitro.26,31,77,78 However, several lines of evidence point to a more complex regulation of Set2 recruitment. Although deletion of the C-terminal SRI domain in budding yeast Set2 eliminates all forms of H3K36me in vivo, a larger deletion that also removes the adjacent WW and cysteine-rich domains actually leads to a partial restoration of Set2 function.79 This Set2 N-terminal fragment is targeted to gene coding regions and mediates H3K36me2 independently of Ctk1, although H3K36me3 remains absent. This suggests that the SRI domain may also be needed to counteract a negative regulatory role of the WW and cysteine-rich domains. As WW domains typically bind to phosphoserine or phosphothreonine residues,80 negative regulation of Set2 may involve direct recognition of a phosphoprotein. A clue to the nature of this negative regulation comes from recent studies in budding yeast indicating that the WW and/or cysteine-rich domains promote the degradation of Set2 when it is not bound to the phosphorylated Rpb1-CTD.81,82 Regulation of Set2 stability is also proposed to link the PAF complex and the elongation factor Spt6 to H3K36me3, since loss of either of these factors reduces levels of S2-P, Set2, and H3K36me3.70,79,81,83,84 This mechanism represents an attractive strategy for tightly coupling a histone-modifying activity to RNAP II, but how generally it is implemented remains an open question. For example, the mammalian and fission yeast Spt6/Iws1 complexes promote H3K36me3 without affecting Set2 stability, and PAF does not seem to have a major role in formation of H3K36me3 outside of budding yeast.24,57,85,86
Levels of H3K36me in gene coding regions in metazoans are related to mRNA splicing. In mammalian cells, H3K36me is more abundant at intron-containing genes than at intronless genes, suggesting that mRNA splicing is mechanistically linked to H3K36me deposition during transcription.87 Strikingly, small-molecule inhibitors of splicing reduce H3K36me3 levels both globally and at individual genes, and patterns of alternative splicing are correlated with altered patterns of H3K36me3 and HYPB recruitment. Deciphering the molecular basis for this phenomenon is an important goal for future studies.
Targeting Histone H3 Lysine 79 Methylation
Connections between the conserved H3K79 methyltransferase Dot1 and the RNAP II elongation complex remain enigmatic. In metazoans, Dot1 associates with transcription factors AF9, AF10, AF17, and ENL, frequent fusion partners with MLL in mixed-lineage leukemia.88 Interaction of these factors with human DOT1L is of medical interest because it is necessary for the leukemogenic activities of the fusion proteins, and DOT1L is under investigation as a therapeutic target in these cancers.89 Biochemical analyses suggest that DOT1L forms multiple distinct complexes with these factors that contain either AF9 or ENL.90,91 AF9 and ENL can be isolated as components of the Super Elongation Complex (SEC), which also contains P-TEFb and other elongation factors.88 Both proteins contribute to transcription activation by the SEC.91,92 However, interaction of AF9/ENL with the SEC is mutually exclusive with that of DOT1L, and DOT1L has been suggested to negatively regulate SEC activity and elongation.90,91,93 Further study is needed to uncover how DOT1L complexes are recruited to elongating RNAP II and their functions in elongation.
HAT and HDAC Complexes in Transcription Elongation
Histone acetylation (at multiple sites on histones H3 and H4) is strongly associated with transcriptional activity in promoters and gene coding regions and likely facilitates nucleosome remodeling and removal during transcription. Concordantly, HAT complexes promote elongation and localize to coding regions of highly transcribed genes,94-98 but the nature of their interactions with the RNAP II elongation machinery has not been defined.
Although histone acetylation generally correlates with transcription its levels are low in coding regions relative to promoters. This difference likely reflects mechanisms to maintain chromatin structure in the wake of transcribing RNAP II and to prevent transcription of non-coding RNAs from within gene coding regions.4 Multiple HDAC complexes have been shown to act within gene bodies.98-105 The specific contacts between HDACs and the RNAP II elongation complex are mostly unknown. However, a direct interaction has been reported between the Rpd3S HDAC complex and Rpb1 CTD repeat peptides phosphorylated on serines 2 and 5.106 Recognition of these phosphosites fits nicely with the close functional relationship between this complex and Set2, which also binds to multiply phosphorylated CTD repeats. There is also in vivo evidence that proper engagement of Rpd3S with coding regions requires Cdk9-dependent Spt5 phosphorylation, although whether this involves a direct interaction is unknown.107
Feedback Regulation of the RNAP II Elongation Complex by Histone Modifications
A variety of functions has been ascribed to co-transcriptional histone modifications. Interestingly, there is evidence that these functions could involve feedback regulation of the mechanisms that direct their formation. Initial support for this came from studies in budding yeast, which found that loss of function mutations in SET2 and genes encoding components of the Rpd3S HDAC complex bypassed the essential function of Bur1 (the S. cerevisiae ortholog of Cdk9).77,101 Because Bur1 activity is required both for H3K36 methylation by Set2 and for Rpd3S activity, these findings suggested negative feedback regulation of Bur1 by Set2/Rpd3S. Although the molecular basis for these genetic interactions is not yet understood, they suggest that Bur1-dependent H3K36me and Rpd3S action in coding regions serves to counter the positive influence of Bur1 activity on elongation.
Another study in budding yeast demonstrated a negative effect of H2Bub1 on the activity of Ctk1. This was attributed to H2Bub1-mediated inhibition of an interaction between the Ctk1 complex and histone H2A/H2B dimers. Removal of H2Bub1 by the deubiquitylating enzyme Ubp8 was shown to promote Ctk1 activity and S2-P at select genes.108
In S. pombe, an analogous, albeit more complex, interaction was described between Cdk9 and H2Bub1. H2Bub1 and Cdk9-dependent Spt5-P were found to regulate one another through positive feedback. However, genetic interactions actually suggested a negative feedback relationship between H2Bub1 and Cdk9 that presumably involves another Cdk9 target in addition to Spt5.26 Whereas the positive feedback involves Rtf1, the negative feedback relationship involves the PAF complex, suggesting that H2Bub1 participates in multiple aspects of Cdk9 regulation.24 Feedback regulation of Cdk9 by H2Bub1 may be a conserved feature of RNAP II elongation, since a) loss of Rtf1 also bypasses Bur1 mutations in S. cerevisiae,101 and b) positive feedback between Cdk9 and RNF20 has been observed in mammalian cells.109
Perspectives
The basic outline of pathways connecting histone modification to elongating RNAP II has come into view, but a detailed understanding of the underlying interactions is lacking. The histone-modifying enzymes reviewed here are emerging therapeutic targets in human disease, and thus the mechanisms that target them to the RNAP II elongation complex are of key importance.89,110,111 Because of the apparent feedback relationships between the modifying enzymes and the factors that recruit them, further insight into the recruitment mechanisms is also likely to enhance our understanding of the gene regulatory functions of these conserved histone modifications.
The questions to be addressed moving forward reflect the fundamental gaps in our understanding of the RNAP II elongation complex: How do combinations of CTD kinases generate binding sites for histone-modifying enzymes in vivo? What are the relevant phospho-targets and what specific interactions do they engage in? A thorough examination of these issues will involve detailed in vitro and in vivo approaches to define the relevant phosphorylation-dependent interactions and their downstream consequences. There are now tools available in both yeast and metazoan cells for mutational analysis of the chromosomally encoded Rpb1 and Spt5 CTD domains, and for specific inhibition of CTD kinases in vivo.28,39,112-116 These will undoubtedly play invaluable roles in future investigation of these important issues.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
I thank Dr. Terry Hebert, Dr. Robert Fisher, and members of my lab for critical reading of the manuscript. This work was supported by a grant from the Canadian Institutes for Health Research to J.C.T. (MOP-130362). I apologize to colleagues whose work was not cited due to space limitations.
References
- 1. Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 1996; 84:843-51; PMID:8601308; http://dx.doi.org/ 10.1016/S0092-8674(00)81063-6 [DOI] [PubMed] [Google Scholar]
- 2. Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 1996; 272:408-11; PMID:8602529; http://dx.doi.org/ 10.1126/science.272.5260.408 [DOI] [PubMed] [Google Scholar]
- 3. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011; 21:381-95; PMID:21321607; http://dx.doi.org/ 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smolle M, Workman JL. Transcription-associated histone modifications and cryptic transcription. Biochim Biophys Acta 2013; 1829:84-97; PMID:22982198; http://dx.doi.org/ 10.1016/j.bbagrm.2012.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet 2009; 43:559-99; PMID:19886812; http://dx.doi.org/ 10.1146/annurev.genet.032608.103928 [DOI] [PubMed] [Google Scholar]
- 6. Fuchs G, Oren M. Writing and reading H2B monoubiquitylation. Biochim Biophys Acta 2014; 1839:694-701; PMID:24412854; http://dx.doi.org/ 10.1016/j.bbagrm.2014.01.002 [DOI] [PubMed] [Google Scholar]
- 7. Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 2012; 13:115-26; PMID:22266761; http://dx.doi.org/ 10.1038/nrm3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 2012; 81:65-95; PMID:22663077; http://dx.doi.org/ 10.1146/annurev-biochem-051710-134100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu L, Zee BM, Wang Y, Garcia BA, Dou Y. The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in crosstalk with H3 K4 and K79 methylation. Mol Cell 2011; 43:132-44; PMID:21726816; http://dx.doi.org/ 10.1016/j.molcel.2011.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Musselman CA, Kutateladze TG. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res 2011; 39:9061-71; PMID:21813457; http://dx.doi.org/ 10.1093/nar/gkr613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hartzog GA, Fu J. The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim Biophys Acta 2013; 1829:105-115; PMID:22982195; http://dx.doi.org/ 10.1016/j.bbagrm.2012.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 2012; 26:2119-37; PMID:23028141; http://dx.doi.org/ 10.1101/gad.200303.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wier AD, Mayekar MK, Heroux A, Arndt KM, Vandemark AP. Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin. Proc Natl Acad Sci U S A 2013; 110(43):17290-5; PMID:24101474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell 2006; 21:227-37; PMID:16427012; http://dx.doi.org/ 10.1016/j.molcel.2005.11.024 [DOI] [PubMed] [Google Scholar]
- 15. Schneider S, Pei Y, Shuman S, Schwer B. Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD. Mol Cell Biol 2010; 30:2353-64; PMID:20231361; http://dx.doi.org/ 10.1128/MCB.00116-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol 2009; 29:4852-63; PMID:19581288; http://dx.doi.org/ 10.1128/MCB.00609-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou K, Kuo WH, Fillingham J, Greenblatt JF. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci U S A 2009; 106:6956-61; PMID:19365074; http://dx.doi.org/ 10.1073/pnas.0806302106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev 2013; 113:8456-90; PMID:23952966; http://dx.doi.org/ 10.1021/cr400071f [DOI] [PubMed] [Google Scholar]
- 19. Jasnovidova O, Stefl R. The CTD code of RNA polymerase II: a structural view. Wiley Interdiscip Rev RNA 2013; 4:1-16; PMID:23042580; http://dx.doi.org/ 10.1002/wrna.1138 [DOI] [PubMed] [Google Scholar]
- 20. Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 2007; 318:1777-9; PMID:18079403; http://dx.doi.org/ 10.1126/science.1145989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Doamekpor SK, Sanchez AM, Schwer B, Shuman S, Lima CD. How an mRNA capping enzyme reads distinct RNA polymerase II and Spt5 CTD phosphorylation codes. Genes Dev 2014; 28:1323-36; PMID:24939935; http://dx.doi.org/ 10.1101/gad.242768.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev 1999; 13:1774-9; PMID:10421630; http://dx.doi.org/ 10.1101/gad.13.14.1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mayer A, Schreieck A, Lidschreiber M, Leike K, Martin DE, Cramer P. The spt5 C-terminal region recruits yeast 3' RNA cleavage factor I. Mol Cell Biol 2012; 32:1321-31; PMID:22290438; http://dx.doi.org/ 10.1128/MCB.06310-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mbogning J, Nagy S, Page V, Schwer B, Shuman S, Fisher RP, Tanny JC. The PAF complex and Prf1/Rtf1 delineate distinct Cdk9-dependent pathways regulating transcription elongation in fission yeast. PLoS Genet 2013; 9:e1004029; PMID:24385927; http://dx.doi.org/ 10.1371/journal.pgen.1004029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mayekar MK, Gardner RG, Arndt KM. The recruitment of the Saccharomyces cerevisiae Paf1 complex to active genes requires a domain of Rtf1 that directly interacts with the Spt4-Spt5 complex. Mol Cell Biol 2013; 33:3259-73; PMID:23775116; http://dx.doi.org/ 10.1128/MCB.00270-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sanso M, Lee KM, Viladevall L, Jacques PE, Page V, Nagy S, Racine A, St Amour CV, Zhang C, Shokat KM, et al. A positive feedback loop links opposing functions of P-TEFb/Cdk9 and Histone H2B ubiquitylation to regulate transcript elongation in fission yeast. PLoS Genet 2012; 8:e1002822; PMID:22876190; http://dx.doi.org/ 10.1371/journal.pgen.1002822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol 2000; 20:2970-83; PMID:10757782; http://dx.doi.org/ 10.1128/MCB.20.9.2970-2983.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanin EI, Kipp RT, Kung C, Slattery M, Viale A, Hahn S, Shokat KM, Ansari AZ. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc Natl Acad Sci U S A 2007; 104:5812-17; PMID:17392431; http://dx.doi.org/ 10.1073/pnas.0611505104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell 2012; 45:158-70; PMID:22284676; http://dx.doi.org/ 10.1016/j.molcel.2011.11.024 [DOI] [PubMed] [Google Scholar]
- 30. Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, et al. Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol 2010; 17:1154-61; PMID:20802488; http://dx.doi.org/ 10.1038/nsmb.1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol 2009; 29:5455-64; PMID:19667075; http://dx.doi.org/ 10.1128/MCB.00637-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Larochelle S, Amat R, Glover-Cutter K, Sanso M, Zhang C, Allen JJ, Shokat KM, Bentley DL, Fisher RP. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol 2012; 19:1108-15; PMID:23064645; http://dx.doi.org/ 10.1038/nsmb.2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem 2012; 81:119-43; PMID:22404626; http://dx.doi.org/ 10.1146/annurev-biochem-052610-095910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet 2013; 47:483-508; PMID:24050178; http://dx.doi.org/ 10.1146/annurev-genet-110711-155440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Keogh MC, Podolny V, Buratowski S. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol Cell Biol 2003; 23:7005-18; PMID:12972617; http://dx.doi.org/ 10.1128/MCB.23.19.7005-7018.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem 2001; 276:31793-9; PMID:11431468; http://dx.doi.org/ 10.1074/jbc.M102306200 [DOI] [PubMed] [Google Scholar]
- 37. Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell 2004; 13:55-65; PMID:14731394; http://dx.doi.org/ 10.1016/S1097-2765(03)00526-4 [DOI] [PubMed] [Google Scholar]
- 38. Bosken CA, Farnung L, Hintermair C, Merzel Schachter M, Vogel-Bachmayr K, Blazek D, Anand K, Fisher RP, Eick D, Geyer M. The structure and substrate specificity of human Cdk12/Cyclin K. Nat Commun 2014; 5:3505; PMID:24662513; http://dx.doi.org/ 10.1038/ncomms4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Viladevall L, St Amour CV, Rosebrock A, Schneider S, Zhang C, Allen JJ, Shokat KM, Schwer B, Leatherwood JK, Fisher RP. TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol Cell 2009; 33:738-51; PMID:19328067; http://dx.doi.org/ 10.1016/j.molcel.2009.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Karagiannis J, Balasubramanian MK. A cyclin-dependent kinase that promotes cytokinesis through modulating phosphorylation of the carboxy terminal domain of the RNA Pol II Rpb1p sub-unit. PLoS One 2007; 2:e433; PMID:17502918; http://dx.doi.org/ 10.1371/journal.pone.0000433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3' end processing. Mol Cell 2004; 13:67-76; PMID:14731395; http://dx.doi.org/ 10.1016/S1097-2765(03)00492-1 [DOI] [PubMed] [Google Scholar]
- 42. Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 2010; 24:2303-16; PMID:20952539; http://dx.doi.org/ 10.1101/gad.1968210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bowman EA, Bowman CR, Ahn JH, Kelly WG. Phosphorylation of RNA polymerase II is independent of P-TEFb in the C. elegans germline. Development 2013; 140:3703-13; PMID:23903194; http://dx.doi.org/ 10.1242/dev.095778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang F, Yu X. WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol Cell 2011; 41:384-97; PMID:21329877; http://dx.doi.org/ 10.1016/j.molcel.2011.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pirngruber J, Shchebet A, Schreiber L, Shema E, Minsky N, Chapman RD, Eick D, Aylon Y, Oren M, Johnsen SA. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3'-end processing. EMBO Rep 2009; 10:894-900; PMID:19575011; http://dx.doi.org/ 10.1038/embor.2009.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr Biol 2005; 15:1487-93; PMID:16040246; http://dx.doi.org/ 10.1016/j.cub.2005.07.028 [DOI] [PubMed] [Google Scholar]
- 47. Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol Cell 2005; 20:589-99; PMID:16307922; http://dx.doi.org/ 10.1016/j.molcel.2005.09.010 [DOI] [PubMed] [Google Scholar]
- 48. Tomson BN, Arndt KM. The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim Biophys Acta 2013; 1829:116-26; PMID:22982193; http://dx.doi.org/ 10.1016/j.bbagrm.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qiu H, Hu C, Gaur NA, Hinnebusch AG. Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR-independent recruitment of Paf1 complex. Embo J 2012; 31:3494-505; PMID:22796944; http://dx.doi.org/ 10.1038/emboj.2012.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Phatnani HP, Jones JC, Greenleaf AL. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry 2004; 43:15702-19; PMID:15595826; http://dx.doi.org/ 10.1021/bi048364h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Amrich CG, Davis CP, Rogal WP, Shirra MK, Heroux A, Gardner RG, Arndt KM, VanDemark AP. Cdc73 subunit of Paf1 complex contains C-terminal Ras-like domain that promotes association of Paf1 complex with chromatin. J Biol Chem 2012; 287:10863-75; PMID:22318720; http://dx.doi.org/ 10.1074/jbc.M111.325647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 2010; 140:491-503; PMID:20178742; http://dx.doi.org/ 10.1016/j.cell.2009.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rondon AG, Gallardo M, Garcia-Rubio M, Aguilera A. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep 2004; 5:47-53; PMID:14710186; http://dx.doi.org/ 10.1038/sj.embor.7400045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 2006; 125:703-17; PMID:16713563; http://dx.doi.org/ 10.1016/j.cell.2006.04.029 [DOI] [PubMed] [Google Scholar]
- 55. Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell 2011; 42:536-49; PMID:21596317; http://dx.doi.org/ 10.1016/j.molcel.2011.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Piro AS, Mayekar MK, Warner MH, Davis CP, Arndt KM. Small region of Rtf1 protein can substitute for complete Paf1 complex in facilitating global histone H2B ubiquitylation in yeast. Proc Natl Acad Sci U S A 2012; 109:10837-42; PMID:22699496; http://dx.doi.org/ 10.1073/pnas.1116994109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev 2005; 19:1668-73; PMID:16024656; http://dx.doi.org/ 10.1101/gad.1292105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Adelman K, Wei W, Ardehali MB, Werner J, Zhu B, Reinberg D, Lis JT. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol Cell Biol 2006; 26:250-60; PMID:16354696; http://dx.doi.org/ 10.1128/MCB.26.1.250-260.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Langenbacher AD, Nguyen CT, Cavanaugh AM, Huang J, Lu F, Chen JN. The PAF1 complex differentially regulates cardiomyocyte specification. Dev Biol 2011; 353:19-28; PMID:21338598; http://dx.doi.org/ 10.1016/j.ydbio.2011.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 2009; 137:459-71; PMID:19410543; http://dx.doi.org/ 10.1016/j.cell.2009.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell 2005; 20:601-11; PMID:16307923; http://dx.doi.org/ 10.1016/j.molcel.2005.09.025 [DOI] [PubMed] [Google Scholar]
- 62. Hahn MA, Dickson KA, Jackson S, Clarkson A, Gill AJ, Marsh DJ. The tumor suppressor CDC73 interacts with the ring finger proteins RNF20 and RNF40 and is required for the maintenance of histone 2B monoubiquitination. Hum Mol Genet 2012; 21:559-68; PMID:22021426; http://dx.doi.org/ 10.1093/hmg/ddr490 [DOI] [PubMed] [Google Scholar]
- 63. Shchebet A, Karpiuk O, Kremmer E, Eick D, Johnsen SA. Phosphorylation by cyclin-dependent kinase-9 controls ubiquitin-conjugating enzyme-2A function. Cell Cyclel 2012; 11:2122-7; PMID:22592529; http://dx.doi.org/ 10.4161/cc.20548 [DOI] [PubMed] [Google Scholar]
- 64. Sarcevic B, Mawson A, Baker RT, Sutherland RL. Regulation of the ubiquitin-conjugating enzyme hHR6A by CDK-mediated phosphorylation. EMBO J 2002; 21:2009-18; PMID:11953320; http://dx.doi.org/ 10.1093/emboj/21.8.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 2003; 11:721-9; PMID:12667454; http://dx.doi.org/ 10.1016/S1097-2765(03)00091-1 [DOI] [PubMed] [Google Scholar]
- 66. Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol 2008; 28:609-18; PMID:17998332; http://dx.doi.org/ 10.1128/MCB.01356-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci U S A 2005; 102:14765-70; PMID:16199523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, Basrur V, Elenitoba-Johnson KS, Hess JL. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell 2010; 17:609-21; PMID:20541477; http://dx.doi.org/ 10.1016/j.ccr.2010.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Milne TA, Kim J, Wang GG, Stadler SC, Basrur V, Whitcomb SJ, Wang Z, Ruthenburg AJ, Elenitoba-Johnson KS, Roeder RG, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell 2010; 38:853-63; PMID:20541448; http://dx.doi.org/ 10.1016/j.molcel.2010.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 2003; 23:4207-18; PMID:12773564; http://dx.doi.org/ 10.1128/MCB.23.12.4207-4218.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev 2003; 17:654-63; PMID:12629047; http://dx.doi.org/ 10.1101/gad.1055503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol 2005; 25:3305-16; PMID:15798214; http://dx.doi.org/ 10.1128/MCB.25.8.3305-3316.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 2003; 278:8897-903; PMID:12511561; http://dx.doi.org/ 10.1074/jbc.M212134200 [DOI] [PubMed] [Google Scholar]
- 74. Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc Natl Acad Sci U S A 2005; 102:17636-41; PMID:16314571; http://dx.doi.org/ 10.1073/pnas.0506350102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kanagaraj R, Huehn D, MacKellar A, Menigatti M, Zheng L, Urban V, Shevelev I, Greenleaf AL, Janscak P. RECQ5 helicase associates with the C-terminal repeat domain of RNA polymerase II during productive elongation phase of transcription. Nucleic Acids Res 2010; 38:8131-40; PMID:20705653; http://dx.doi.org/ 10.1093/nar/gkq697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vojnic E, Simon B, Strahl BD, Sattler M, Cramer P. Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J Biol Chem 2006; 281:13-5; PMID:16286474; http://dx.doi.org/ 10.1074/jbc.C500423200 [DOI] [PubMed] [Google Scholar]
- 77. Chu Y, Sutton A, Sternglanz R, Prelich G. The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol Cell Biol 2006; 26:3029-38; PMID:16581778; http://dx.doi.org/ 10.1128/MCB.26.8.3029-3038.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Eissenberg JC, Shilatifard A, Dorokhov N, Michener DE. Cdk9 is an essential kinase in Drosophila that is required for heat shock gene expression, histone methylation and elongation factor recruitment. Mol Genet Genomics 2007; 277:101-14; PMID:17001490; http://dx.doi.org/ 10.1007/s00438-006-0164-2 [DOI] [PubMed] [Google Scholar]
- 79. Youdell ML, Kizer KO, Kisseleva-Romanova E, Fuchs SM, Duro E, Strahl BD, Mellor J. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol Cell Biol 2008; 28:4915-26; PMID:18541663; http://dx.doi.org/ 10.1128/MCB.00001-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Salah Z, Alian A, Aqeilan RI. WW domain-containing proteins: retrospectives and the future. Front Biosci (Landmark Ed) 2012; 17:331-48; PMID:22201747; http://dx.doi.org/ 10.2741/3930 [DOI] [PubMed] [Google Scholar]
- 81. Dronamraju R, Strahl BD. A feed forward circuit comprising Spt6, Ctk1 and PAF regulates Pol II CTD phosphorylation and transcription elongation. Nucleic Acids Res 2014; 42:870-81; PMID:24163256; http://dx.doi.org/ 10.1093/nar/gkt1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fuchs SM, Kizer KO, Braberg H, Krogan NJ, Strahl BD. RNA polymerase II carboxyl-terminal domain phosphorylation regulates protein stability of the Set2 methyltransferase and histone H3 di- and trimethylation at lysine 36. J Biol Chem 2012; 287:3249-56; PMID:22157004; http://dx.doi.org/ 10.1074/jbc.M111.273953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell 2004; 14:447-56; PMID:15149594; http://dx.doi.org/ 10.1016/S1097-2765(04)00257-6 [DOI] [PubMed] [Google Scholar]
- 84. Chu Y, Simic R, Warner MH, Arndt KM, Prelich G. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. Embo J 2007; 26:4646-56; PMID:17948059; http://dx.doi.org/ 10.1038/sj.emboj.7601887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev 2008; 22:3422-34; PMID:19141475; http://dx.doi.org/ 10.1101/gad.1720008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. DeGennaro CM, Alver BH, Marguerat S, Stepanova E, Davis CP, Bähler J, Park PJ, Winston F. Spt6 regulates intragenic and antisense transcription, nucleosome positioning, and histone modifications genome-wide in fission yeast. Mol Cell Biol 2013; 33:4779-92; PMID:24100010; http://dx.doi.org/ 10.1128/MCB.01068-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol 2011; 18:977-83; PMID:21792193; http://dx.doi.org/ 10.1038/nsmb.2123 [DOI] [PubMed] [Google Scholar]
- 88. Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev 2011; 25:1345-58; PMID:21724828; http://dx.doi.org/ 10.1101/gad.2057811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 2011; 20:53-65; PMID:21741596; http://dx.doi.org/ 10.1016/j.ccr.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KS, Allis CD, Roeder RG. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc Natl Acad Sci U S A 2011; 108:15751-6; PMID:21896721; http://dx.doi.org/ 10.1073/pnas.1111498108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. He N, Chan CK, Sobhian B, Chou S, Xue Y, Liu M, Alber T, Benkirane M, Zhou Q. Human Polymerase-Associated Factor complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin. Proc Natl Acad Sci U S A 2011; 108:E636-645; PMID:21873227; http://dx.doi.org/ 10.1073/pnas.1107107108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet 2007; 16:92-106; PMID:17135274; http://dx.doi.org/ 10.1093/hmg/ddl444 [DOI] [PubMed] [Google Scholar]
- 93. Cecere G, Hoersch S, Jensen MB, Dixit S, Grishok A. The ZFP-1(AF10)/DOT-1 complex opposes H2B ubiquitination to reduce Pol II transcription. Mol Cell 2013; 50:894-907; PMID:23806335; http://dx.doi.org/ 10.1016/j.molcel.2013.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell 2007; 25:31-42; PMID:17218269; http://dx.doi.org/ 10.1016/j.molcel.2006.11.020 [DOI] [PubMed] [Google Scholar]
- 95. Weake VM, Dyer JO, Seidel C, Box A, Swanson SK, Peak A, Florens L, Washburn MP, Abmayr SM, Workman JL. Post-transcription initiation function of the ubiquitous SAGA complex in tissue-specific gene activation. Genes Dev 2011; 25:1499-509; PMID:21764853; http://dx.doi.org/ 10.1101/gad.2046211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ginsburg DS, Govind CK, Hinnebusch AG. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol Cell Biol 2009; 29:6473-6487; PMID:19822662; http://dx.doi.org/ 10.1128/MCB.01033-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Johnsson A, Durand-Dubief M, Xue-Franzen Y, Ronnerblad M, Ekwall K, Wright A. HAT-HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Rep 2009; 10:1009-14; PMID:19633696; http://dx.doi.org/ 10.1038/embor.2009.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 2009; 138:1019-31; PMID:19698979; http://dx.doi.org/ 10.1016/j.cell.2009.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 2002; 298:1412-4; PMID:12434058; http://dx.doi.org/ 10.1126/science.1077790 [DOI] [PubMed] [Google Scholar]
- 100. Kim T, Xu Z, Clauder-Munster S, Steinmetz LM, Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 2012; 150:1158-69; PMID:22959268; http://dx.doi.org/ 10.1016/j.cell.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 2005; 123:593-605; PMID:16286008; http://dx.doi.org/ 10.1016/j.cell.2005.10.025 [DOI] [PubMed] [Google Scholar]
- 102. Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 2005; 123:581-92; PMID:16286007; http://dx.doi.org/ 10.1016/j.cell.2005.10.023 [DOI] [PubMed] [Google Scholar]
- 103. Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5' transcribed regions. Cell 2009; 137:259-72; PMID:19379692; http://dx.doi.org/ 10.1016/j.cell.2009.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SI. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol 2007; 14:372-80; PMID:17450151; http://dx.doi.org/ 10.1038/nsmb1239 [DOI] [PubMed] [Google Scholar]
- 105. Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 2005; 20:971-8; PMID:16364921; http://dx.doi.org/ 10.1016/j.molcel.2005.11.021 [DOI] [PubMed] [Google Scholar]
- 106. Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell 2010; 39:234-46; PMID:20670892; http://dx.doi.org/ 10.1016/j.molcel.2010.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Drouin S, Laramee L, Jacques PE, Forest A, Bergeron M, Robert F. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet 2010; 6:e1001173; PMID:21060864; http://dx.doi.org/ 10.1371/journal.pgen.1001173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell 2007; 27:275-88; PMID:17643376; http://dx.doi.org/ 10.1016/j.molcel.2007.01.035 [DOI] [PubMed] [Google Scholar]
- 109. Wu L, Li L, Zhou B, Qin Z, Dou Y. H2B ubiquitylation promotes RNA Pol II processivity via PAF1 and pTEFb. Mol Cell 2014; 54:920-31; PMID:24837678; http://dx.doi.org/ 10.1016/j.molcel.2014.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Johnsen SA. The enigmatic role of H2Bub1 in cancer. FEBS Lett 2012; 586:1592-601; PMID:22564770; http://dx.doi.org/ 10.1016/j.febslet.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 111. Kudithipudi S, Jeltsch A. Role of somatic cancer mutations in human protein lysine methyltransferases. Biochim Biophys Acta 2014; 1846:366-79; PMID:25123655 [DOI] [PubMed] [Google Scholar]
- 112. Hsin JP, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3' end processing. Science 2011; 334:683-6; PMID:22053051; http://dx.doi.org/ 10.1126/science.1206034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Hintermair C, Heidemann M, Koch F, Descostes N, Gut M, Gut I, Fenouil R, Ferrier P, Flatley A, Kremmer E, et al. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. Embo J 2012; 31:2784-97; PMID:22549466; http://dx.doi.org/ 10.1038/emboj.2012.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Schwer B, Shuman S. Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell 2011; 43:311-8; PMID:21684186; http://dx.doi.org/ 10.1016/j.molcel.2011.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell 2009; 34:387-93; PMID:19450536; http://dx.doi.org/ 10.1016/j.molcel.2009.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Larochelle S, Batliner J, Gamble MJ, Barboza NM, Kraybill BC, Blethrow JD, Shokat KM, Fisher RP. Dichotomous but stringent substrate selection by the dual-function Cdk7 complex revealed by chemical genetics. Nat Struct Mol Biol 2006; 13:55-62; PMID:16327805; http://dx.doi.org/ 10.1038/nsmb1028 [DOI] [PubMed] [Google Scholar]


