Abstract
Core promoter elements play a pivotal role in the transcriptional output, yet they are often detected manually within sequences of interest. Here, we present 2 contributions to the detection and curation of core promoter elements within given sequences. First, the Elements Navigation Tool (ElemeNT) is a user-friendly web-based, interactive tool for prediction and display of putative core promoter elements and their biologically-relevant combinations. Second, the CORE database summarizes ElemeNT-predicted core promoter elements near CAGE and RNA-seq-defined Drosophila melanogaster transcription start sites (TSSs). ElemeNT's predictions are based on biologically-functional core promoter elements, and can be used to infer core promoter compositions. ElemeNT does not assume prior knowledge of the actual TSS position, and can therefore assist in annotation of any given sequence. These resources, freely accessible at http://lifefaculty.biu.ac.il/gershon-tamar/index.php/resources, facilitate the identification of core promoter elements as active contributors to gene expression.
Keywords: BRE, computational tool, core promoter elements/motifs, DPE, initiator, MTE, promoter prediction, RNAP II transcription, TATA box, TCT
Abbreviations
- BRE, TFIIB recognition element
BREd
- BRE downstream of the TATA box
BREu
- BRE upstream of the TATA box
DCE
- downstream core element
DPE
- downstream core promoter element
Inr
- initiator
MTE
- motif 10 element
PWM
- position weight matrix
RNAP II
- RNA Polymerase II
TBP
- TATA box-binding protein
TAFs
- TBP-associated factors
TSS, transcription start site
Introduction
The uniqueness of each cell, as well as the differences between cell types in multicellular organisms, are largely achieved by distinct transcriptional programs. The regulation of transcription initiation is a complex process that is primarily based on the direct interactions between transcription factors and DNA. Transcription initiation occurs at the core promoter region where the RNA Polymerase II (RNAP II) binds, which is often referred to as the ‘gateway to transcription’.1-6 Although it was previously believed that the core promoter is a universal component that works in a similar mechanism for all protein-coding genes, it is nowadays established that core promoters differ in their architecture and function.3,4,7-10 Moreover, distinct core promoter compositions were demonstrated to result in diverse transcriptional outputs.11-15
Transcription initiation is generally thought to occur in either a focused or a dispersed manner with multiple combinations between these modes.4,7 Promoters that exhibit a dispersed initiation pattern typically contain multiple weak transcription start sites (TSSs) within a 50 to 100 bp region and are associated with CpG islands. In vertebrates, dispersed transcription initiation appears to account for the majority of protein-coding genes and is believed to direct the transcription of constitutively-expressed genes. In contrast, focused promoters contain a single predominant TSS or a few TSSs within a narrow region of several nucleotides, and are highly correlated with tightly regulated gene expression.4 The focused core promoter typically spans the region from −40 to +40 relative to the first transcribed nucleotide, which is usually termed “the +1 position.”
The focused core promoter area encompasses distinct DNA sequence motifs, termed core promoter elements or motifs. These elements are recognized by the basal transcription machinery to recruit RNAP II and to form the preinitiation complex.16-18 The TFIID multi-subunit complex is a key basal transcription factor that recognizes the core promoter in the process of transcription initiation.16-19 Distinct TFIID subunits, namely TATA box-binding protein (TBP) and TBP-associated factors (TAFs), recognize specific core promoter sequences.2-4,16,20-23 Table 1 and Figure 1 provide a summary of the characteristics of the known core promoter elements of focused promoters. Remarkably, the MTE, DPE and Bridge elements are exclusively dependent on the presence of a functional initiator with a strict spacing requirement, and are typically enriched in TATA-less promoters.2-4,20,21,23-25
Table 1.
The precisely spaced known core promoter elements within focused promoters
| Name | Position (relative to the TSS) | PWM logo representation | Consensus (in IUPAC characters) | References |
|---|---|---|---|---|
| mammalian Initiator | −2 to +5 |  |
YYANWYY | 70 |
| Drosophila Initiator | −2 to +4 |  |
TCAKTY | |
| TATA box | −30/-31 to -23/-24 |  |
TATAWAAR | 4,71 |
| BREu | Immediately upstream of the TATA box |  |
SSRCGCC | 45 |
| BRE d | Immediately downstream of the TATA box |  |
RTDKKKK | 44 |
| DPE (Inr dependent) | +28 to +33 |  |
DSWYVY (functional range set) | 20,21,24 |
| MTE (Inr dependent) | +18 to +29 |  |
CSARCSSAACGS | 25 |
| Bridge (Inr dependent) | Part I: +18 to +22 Part II: +30 to +33 |  |
Part I: CGANC Part II: WYGT | 23 |
| Drosophila TCT | −2 to +6 |  |
YYCTTTYY | 48 |
| Human TCT | −1 to +6 |  |
YCTYTYY | 48 |
| XCPE1 | −8 to +2 |  |
DSGYGGRASM | 51 |
| XCPE2 | −9 to +2 |  |
VCYCRTTRCMY | 72 |
| DCE | +6 to +11, +16 to +21, +30 to +34 | — | Necessary motifs: CTTC, CTGT, AGC | 73,74 |
The table includes the position (relative to the TSS, +1), motif logo, IUPAC consensus sequence and references for each element.
Figure 1.
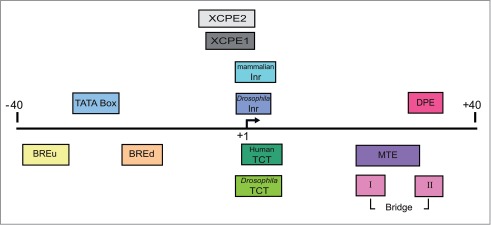
Schematic representation of the major core promoter elements. The region of the core promoter area (−40 to +40 relative to the TSS) is illustrated. The diagram is roughly to scale, and each element is colored according to its color in the output table (see Fig. 2B).
An important aspect of core promoter elements is their synergistic nature. Although the presence of a specific core promoter element is usually sufficient to influence transcription, different combinations of core promoter elements exist, with some shown to act in concert, and, hence, affect the potency of the transcriptional outcome.11,26 It is therefore important to consider all the elements present within the same promoter in order to assess its transcriptional strength.
Prediction of core promoter motifs that affect the transcriptional output, in the absence of experimental validation, is a difficult task. The majority of currently available promoter prediction programs search for over-represented motifs in a given set of promoter sequences (based on annotated TSSs), rather than known core promoter elements.27-29 Most of these programs utilize other features, such as transcription factor binding sites, physical properties of the DNA, DNA accessibility, RNAP II occupancy and various epigenetic markers.29-35 However, even available programs that aim to identify core promoter elements, such as McPromoter36 and Eukaryotic Core Promoter Predictor (YAPP, http://www.bioinformatics.org/yapp/cgi-bin/yapp.cgi), rarely consider the functional constraint of the strict spacing required by the Inr-dependent elements, namely, DPE, MTE, and Bridge.
The selection of promoters that comprise the data set used to predict core promoter elements based on position weight matrices (PWMs) is of pivotal importance, as subtle variations in the sequences may generate completely different PWMs.31 Motif finding algorithms, such as XXmotif, can be used to accurately construct a PWM for over-represented motifs within a given set of sequences.37,38 Unfortunately, even a perfect model that is only based on sequence features, cannot exclusively account for the observed transcriptional activity, as most of the sequence motifs are short and redundant, and can thus be found in many non-transcriptionally active regions of the genome.31 Using experimentally-validated sequences rather than over-represented motifs, can greatly enhance the strength of the prediction program, although it cannot fully guarantee the accuracy of the prediction. Currently, the experimental readout of transcription strength and start sites resulting from mutated promoter sequences is not performed on a high-throughput scale; hence, the currently available experimental results are prone to be biased. Moreover, the known biologically functional sequences may slightly differ from the determined consensus; as a result, a tool for efficient detection of candidate core promoter elements is needed.
Importantly, annotation of individual promoters for the presence of specific core promoter elements can facilitate the discovery of gene groups co-regulated via a common core promoter motif. In a previous study, 205 experimentally-determined Drosophila TSSs were manually annotated for the presence of TATA-box, Initiator and DPE to explore their role and function in gene regulation.24 This annotation facilitated the discovery that the Drosophila Hox gene network is regulated via the DPE.39 A more comprehensive analysis of the whole Drosophila transcriptome revealed that DPE-containing genes are conserved and highly prevalent among the target genes of Dorsal, a key regulator of dorsal-ventral axis formation.12 These examples demonstrate that the comprehensive annotation of core promoter elements in transcripts can greatly advance the understanding of gene expression regulation.
Here we describe 2 contributions in the detection and curation of core promoter elements within sequences of interest, based on experimentally validated sequences. The Elements Navigation Tool (ElemeNT) is a user-friendly web-based, interactive tool for prediction and display of putative core promoter elements and their biologically-relevant combinations in any given sequence, without a need for prior determination of the TSS. The CORE database utilizes the ElemeNT algorithm to annotate putative core promoter elements near CAGE40 and RNA-seq41-defined Drosophila melanogaster TSSs. Together, both the ElemeNT program and the CORE database present new improved tools to assess the presence of core promoter elements within a given DNA sequence.
Methods
Availability
CORE and ElemeNT are freely accessible at http://lifefaculty.biu.ac.il/gershon-tamar/index.php/resources. Each resource is described in a separate page, providing both documentation and resources.
The ElemeNT algorithm
Given a sequence of interest, the algorithm detects in it putative elements whose PWM-similarity to known core promoter elements is above a threshold. For each core promoter element, the user can specify a threshold between 0 and 1 for the presence of the element at a position. Default threshold values were empirically determined for each element, based on known functional sequence elements.
For a PWM matrix P with k columns, the PWM score is calculated for each sub-sequence of length k (k-mer) in the sequence, by multiplying the appropriate values of the PWM for each consecutive position, as follows:
, where is a k-mer starting at position i+1 in sequence S and is the probability for nucleotide x at position j in P, normalized so that for a given j, . The role of this normalization is to guarantee that the final PWM score for every element is between 0 and 1, irrespective of the PWM's parameters. Each sub-sequence with a score exceeding the specified threshold is termed a ‘hit’. The score is calculated for , where n is the length of the input sequence S, and hits are displayed in a list sorted in descending score order for each element. Consensus match scores, which are the number of nucleotide matches of the hit to the motif's consensus (Table 1), are also reported for each hit. The flow diagram of the ElemeNT algorithm is depicted in Figure S1. The PWMs used, as well as their construction processes, are described in File S1.
CORE construction
CORE database construction was based on both CAGE- and RNA-seq-experimentally verified Drosophila TSSs. CAGE-based TSSs were determined based on Hoskins et al.40 For each CAGE peak, the reported probability density functions (PDFs) were used to determine the most probable TSS. If two or more positions at >10 bp distance from each other were assigned with the highest TSS probability, each was considered as a separate TSS. The RNA-seq observed TSSs were reported by Nechaev et al.41 For each determined TSS, the sequence encompassing the TSS ±50 bp was used for downstream analysis by the ElemeNT algorithm, using default score cutoff values.
For each core promoter element, the position relative to the TSS and the corresponding score are reported for all hits within the allowed range (±5 bp relative to the predicted position). All listed positions are with respect to the starting nucleotide of the relevant motif. We list the elements used to construct the CORE, with the relative positions and the cutoff scores provided in parenthesis: BREu (-37, 0.05); TATA box (-30, 0.01); BREd (-24, 0.5); Drosophila Inr (-2, 0.01); Drosophila TCT (-2, 0.1). The Inr-dependent DPE, MTE and Bridge elements were only considered at the precise starting positions Inr+30, Inr+20 and Inr+20, respectively, with cutoff scores of 0.01. A summary of the total numbers of hits of each element within the CAGE and RNA-seq datasets is described in a separate sheet.
GO terms analysis
GO terms enrichment was assessed using the PANTHER classification system42 (http://pantherdb.org/).43 For each examined element (TATA, dInr, DPE, MTE and dTCT), 5 distinct lists were created based on the CORE results- CAGE peaked, CAGE broad, CAGE unclassified, all CAGE tags and RNA-seq. For CAGE data, the classification of promoter types was used as provided with the original data set (see below).40 Each list of genes was analyzed by the PANTHER overrepresentation test (release 20141219) against the Drosophila melanogaster reference list, using GO biological process complete annotation data set (GO ontology database released 2015-04-13). The Bonferroni correction for multiple testing was applied. While enrichment values range between 0.2 and ‘>5', only results with fold enrichment ≥4 are reported.
Results
The elements navigation tool
In order to facilitate the identification of putative core promoter elements and their biologically relevant combinations within a sequence of interest, we developed the Elements Navigation Tool (ElemeNT). ElemeNT is a web-based, interactive tool for rapid and convenient detection of core promoter elements and their combinations within any given sequence. Core promoter elements have been shown to function at a specific distance from the TSS and to affect transcription (e.g., as examined by mutational analysis). ElemeNT scans the input sequences, applying user-specified parameters, for the presence of core promoter elements that are precisely located relative to the TSS (Fig. 1). The elements are represented by PWMs, which were constructed based on validated biologically functional sequences (Table 1, File S1). Notably, for some elements, the PWMs differ from the consensus sequences reported in the literature, reflecting differences in the data sources used to generate these models. The elements that can be searched for are: mammalian initiator, Drosophila initiator, TATA box, MTE, DPE, Bridge, BREu, BREd, human TCT, Drosophila TCT, XCPE1 and XCPE2 (Table 1, Fig. 1). The MTE, DPE and Bridge motifs are only scored at the precise location relative to each detected mammalian/Drosophila initiator, based on the known strict spacing requirement that is crucial for these elements to be functional. The TATA box motif is derived from canonical TATA boxes whose 5’ T is located at -30 or 31 relative to the TSS. Furthermore, the user can search the sequence for any PWM provided by the user. The scores are normalized to a scale of 0 to 1, to allow standardization and comparison between distinct elements. The ElemeNT algorithm is described in the Methods section, and its flow is illustrated in Figure S1.
The output of the program contains the analyzed sequences, a color display of potential combinations of core promoter elements identified, and a table containing the name of each of the detected elements, alongside its position, the sequence, its PWM score and the number of matches with the element's consensus (Fig. 2). Several possible combinations of core promoter elements are displayed, when applicable, in order to indicate potential synergism between elements that may inspire further exploration. Possible combinations considered are one or more of the following: 1) the mammalian/Drosophila initiator and either the MTE, DPE or Bridge motifs; 2) the TATA box and the mammalian/Drosophila initiator; 3) the TATA box and either the BREu or BREd (Fig. 2A).
Figure 2.
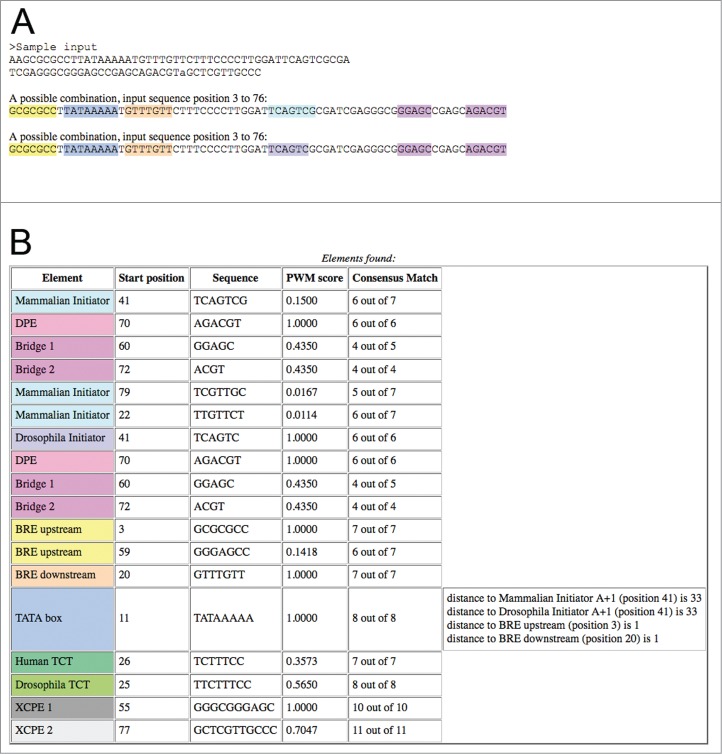
A sample output of the ElemeNT program. (A) The input sequence annotated with the combinations of elements identified in it. ElemeNT detected a TATA box flanked by both a BREu element and a BREd element, Drosophila and mammalian initiator elements and DPE and Bridge elements. The two possible combinations result from a sequence match to both the Drosophila and mammalian initiators, due to the partial sequence redundancy of the 2 elements. (B) A table displaying all the elements identified within the input sequence, their location, PWM and consensus match scores. Note the message displayed for the TATA-box, indicating the presence of mammalian and Drosophila initiators, as well as BREu and BREd, at optimal distances for transcriptional synergy.
In the output table, the elements are ordered by their type and then sorted by PWM scores (Fig. 2B). The MTE, DPE and Bridge motifs, which are strictly dependent on the presence of a functional initiator,2-4,20,21,23,25 are displayed immediately below the corresponding initiator. For TATA box motifs, a message is displayed if the specific TATA box is located 26 to 40 bp upstream of the A+1 of an initiator. In addition, a message is displayed if a BREu or BREd is located in close proximity to the specific TATA box.44-46
To partially assess the performance of the ElemeNT tool, a set of experimentally validated core promoter sequences were analyzed by the tool. The analysis of the Drosophila Inr is presented as an example (Fig. S2). Importantly, ElemeNT detected most of the biologically functional Drosophila initiator motifs among the dataset at cutoff values around 0.01. As expected, lower threshold values used led to detection of a greater number of correct hits, at a cost of a higher false positive rate. False negative hits were scored as well, based on missed motifs. Previously validated sequence variations in core promoter elements resulted in score values of 0.005-0.01, further supporting the defined default cutoffs (data not shown).24
The CORE database
The ElemeNT algorithm was employed to predict core promoter composition of all Drosophila melanogaster transcripts (File S2). TSSs were obtained based on both CAGE40 and RNA-seq41 data-determined Drosophila melanogaster TSSs. The sequence around each TSS was annotated for the presence of core promoter elements near the expected position relative to the TSS. In addition, we summarized the frequencies of the detected elements among the Drosophila transcripts. Importantly, the fraction of promoters containing the distinct elements was similar in the CAGE and RNA-seq data sets. The total analyzed transcripts contained 6-8% TATA box motifs, ∼55% Inr, ∼17% DPE and ∼1% TCT. The CAGE-defined transcripts were previously categorized as peaked, broad and unclassified promoter classes.40 Inr, TATA box and DPE elements were enriched among peaked promoters, as compared to broad and unclassified subsets (Inr- 71% vs. 48% and 54%, TATA box- 14% vs. 6% and 9%, DPE- 32% vs. 11% and 18%, respectively). In contrast, the rare TCT element was slightly more prevalent among the broad promoters class, compared to peaked and unclassified (1.5% vs. 0.5% and 1%, respectively). These results are in the same range as the proportions reported in the original study40 – 70% and 35% Inr, 16% and 4% TATA-box for peaked and broad promoters, respectively.
The distribution of elements found among the allowed positions peaked around the expected relative position (Fig. 3). This peak was observed in both CAGE and RNA-seq data, suggesting that the detected elements are biologically functional. Extending the allowed range around the relative position from ±5 bp to ±10 bp did not reveal additional elements (Fig. S3); hence, a ±5 bp range was used for all downstream analyses. Additionally, the distribution of detected elements among the CAGE defined peaked, broad and unclassified promoters did not differ greatly from the overall distribution (Fig. S4). Reassuringly, the average PWM score also peaked at the biologically relevant positions, although the observed peaks were less profound than the distribution peaks (Fig. 4). Notably, the PWMs were constructed based on completely different data sets obtained by entirely different experimental approaches, as compared to the CAGE and RNA-seq datasets.
Figure 3.
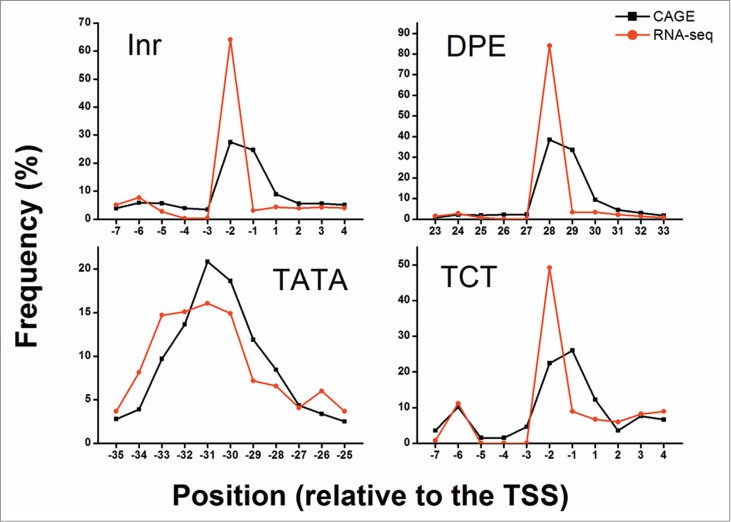
Distribution of core promoter elements’ occurrence at specific positions. The frequency of detected elements (dInr, DPE, TATA, and dTCT) at the allowed positions relative to the determined TSS is presented. The +1 position is the predicted TSS location. Black squares depict the frequency of discovered elements using CAGE whereas red circles depict the frequency of discovered elements using RNA-seq. For both CAGE (black) and RNA-seq (red) data, an enrichment in the frequency of discovered elements is detected at the expected positions (-30 for TATA, -2 for dInr and dTCT and 28 for DPE).
Figure 4.
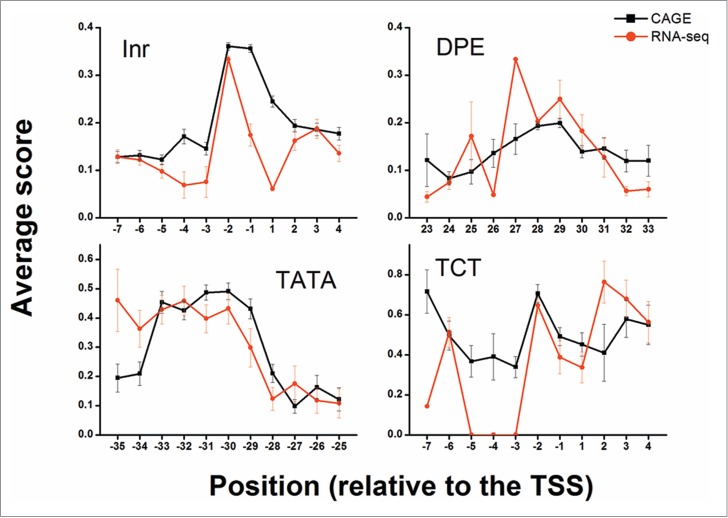
Average PWM score of different core promoter elements at specific positions. The average PWM score of elements (dInr, DPE, TATA and dTCT) at the allowed positions relative to the determined TSS is presented. The +1 position is the predicted TSS location. Black squares depict the average score of discovered elements using CAGE whereas red circles depict the average score of discovered elements using RNA-seq. For both CAGE and RNA-seq data, some enrichment of the mean score is detected at the expected positions (-30 for TATA, -2 for dInr and dTCT and 28 for DPE). Error bars represent the standard errors of the means (SEM).
We also evaluated the CORE database accuracy by GO term analysis of genes sets that were found to contain the same element. We used PANTHER classification system for this aim.42,43 The results (summarized in Table 2, and fully presented in File S3) indicate that distinct GO terms categories are associated with the different core promoter elements. While only a few specific categories were enriched in TATA-containing genes, DPE-containing genes were mostly enriched for development-related gene categories. TCT-containing genes were mostly enriched for translation and ribosomal-related proteins, as well as for structural proteins related to mitosis. Remarkably, the observed enriched categories are in agreement with previous reports, where the DPE was found be associated with developmental genes and TCT with housekeeping and ribosomal genes.4,47-49
Table 2.
Top enriched GO terms categories associated with the analyzed data sets
| TATA | Inr | DPE | TCT | |
|---|---|---|---|---|
| CAGE peak | • chitin-based cuticle development • cuticle development | • branch fusion, open tracheal system • tube fusion • cardiocyte differentiation • ventral cord development • genital disc development | • heart development • circulatory system development • peripheral nervous system development • digestive system development • digestive tract development • reproductive system development • reproductive structure development | • mitotic spindle elongation • centrosome duplication • spindle elongation • centrosome cycle • centrosome organization • microtubule organizing center organization • translation |
| CAGE broad | • chitin-based cuticle development | • NO ENRICHMENT | • negative regulation of molecular function | • translation • cellular macromolecule biosynthetic process • macromolecule biosynthetic process • gene expression • cellular biosynthetic process • organic substance biosynthetic process • biosynthetic process |
| CAGE unclassified | • chitin-based cuticle development | • stem cell fate commitment • regulation of protein localization to nucleus • female meiosis chromosome segregation • regulation of protein import into nucleus | • renal system development • urogenital system development • pigment metabolic process | • Translation • cellular macromolecule biosynthetic process • macromolecule biosynthetic process • gene expression • cellular biosynthetic process • organic substance biosynthetic process • biosynthetic process |
| CAGE all tags | • chitin-based cuticle development • neuropeptide signaling pathway • cuticle development | • NO ENRICHMENT | • cardiocyte differentiation | • translation • cellular macromolecule biosynthetic process • macromolecule biosynthetic process • gene expression • cellular biosynthetic process • organic substance biosynthetic process • biosynthetic process |
| RNA-seq | • cellular modified amino acid metabolic process • glutathione metabolic process • peptide metabolic process • cellular amide metabolic process • sulfur compound metabolic process• cellular amino acid metabolic process • determination of adult lifespan | • NO ENRICHMENT | • heart development • circulatory system development • cardiovascular system development • renal system development • urogenital system development • skeletal muscle organ development • muscle attachment | • translation • mitotic spindle elongation • spindle elongation • cellular macromolecule biosynthetic process • macromolecule biosynthetic process • gene expression |
For each dataset, up to 7 categories that showed significant enrichment (P < 0.05 after Bonferroni corrections) are listed. In case there were more than 7, the top 7 according to the P-value are shown. The different elements are enriched for distinct biological processes categories. The full list of categories along with their P-values is presented in file S3.
Discussion
Core promoter elements, located in the immediate vicinity of the TSSs, have a great effect on the transcriptional output.4,7 The majority of core promoter elements were identified as DNA sequences that are recognized by components of the preinitiation complex.20,44,45,50,51 In addition, overrepresented motifs were discovered in the region around the annotated TSSs.52-54 Some of these motifs affected the transcriptional outcome25 and some were bound by transcription-regulating proteins.55
The uniqueness of the ElemeNT program, as compared to other promoter-prediction software, is its major focus on biologically-functional core promoter elements. This is manifested by 2 major principles adapted in the algorithm. The first is the exclusive use of experimentally validated core promoter motifs, rather than overrepresented motifs, to construct the PWMs used. The second is the obligatory presence of an initiator, and the strict spacing for the downstream promoter elements MTE, DPE, and Bridge, which are crucial for the functionality of the downstream elements. These are overlooked by most of the core promoter elements prediction programs.27,29,32,35,36 Moreover, the identification of combinations of elements, which were experimentally demonstrated to result in synergistic effects,11,25,26 may spark new research directions. In contrast to most of the available promoter prediction programs, the web-based ElemeNT is not designed to produce or analyze a genome-scale data, but is rather intended to narrow down a given region of interest, considering the currently available, experimentally-validated information about core promoter motifs themselves.
The determination of actual TSSs, which influence the motifs discovered in their vicinity, is a critical factor in the prediction of core promoter elements. The TSS of the same gene can vary across the developmental stages, tissues, and time points sampled, which presents a great challenge for integration of the data provided by different studies. To date, a wealth of rapidly evolving high-throughput techniques to identify features and sequences that might affect transcription are available; these include PEAT,56 CAGE,57 FAIRE-seq,58 ChIP-seq,59 and GRO-seq.60 The integrated results will be of utmost importance for re-defining TSSs.
We used the ElemeNT algorithm to annotate Drosophila melanogaster TSSs defined by either CAGE40 or RNA-seq41 for the different core promoter elements. A major contribution of the CORE database for core promoter elements curation is the ability to easily identify all the core promoter elements associated with a specific Drosophila gene, without any previous knowledge.
Generally, CAGE and RNA-seq data showed similar percentages of core promoter elements among the total transcripts. The total frequencies of the TATA box and Inr were in concordance with the numbers reported in the original study.40 However, the original reports on DPE percentages (5% within peaked promoters, 1.5% within broad promoters) are significantly lower than the frequency detected in the CORE database (32% peaked, 11% broad). This discrepancy likely arises from the different approaches taken; while Hoskins et al. searched for a consensus DPE sequence20 within 5 bp of position +26, we have looked for the more biologically relevant functional range set24 located at a precise +28 distance relative to a detected Inr.
Another aspect highlighting the biological relevance of the obtained results is the peak of both the frequency and the average PWM score at the expected positions relative to the TSS (Fig. 3, Fig. 4). The fact that these peaks are clearly evident indicates that both TSS determination and PWM construction have been performed accurately. Further positional constraints apply to the Inr dependent elements—DPE, MTE, and Bridge, as discussed above. Surprisingly, the more strict spacing requirements used in this study yielded a higher proportion of DPE-containing transcripts, thus highlighting the importance of annotation guidelines based on experimentally-validated elements. The TATA box, Inr and DPE elements were enriched among peaked promoters, while the TCT was enriched among the broad promoters class, recapitulating previous observations and highlighting the biological relevance of the obtained results.32,40,49
In addition, GO terms enrichment differed significantly among the gene groups containing distinct core promoter elements (Table 2, File S3), mostly in agreement with the literature.4,7,32,40 The DPE, which was shown to functionally regulate gene expression of developmental gene networks, namely Hox genes39 and mesodermal genes,12 was found to be enriched among circulatory system developmental genes, consistent with the previous findings.13 The Inr element, which is the most abundant motif and is associated with tightly regulated genes, was not found to be enriched for specific gene groups among the total transcripts group. A possible interpretation is that since the Inr is prevalent among most gene groups no enrichment is detected when examining the whole transcriptome. Focused transcription initiation was previously associated with spatiotemporally regulated tissue-specific genes and with canonical core promoter elements that have a positional bias, such as the TATA box, Initiator, MTE and DPE.61,62 However, broad (dispersed) promoters often contain a distinct set of elements with weaker positional biases (as compared to the focused promoters), as Ohler 1, DNA replication element (DRE), Ohler 6, and Ohler 740,62 (a detailed discussion is available in refs4,10). When considering separately the CAGE-defined peaked, broad and unclassified promoter classes, a clear enrichment for developmental processes is evident in the peaked and unclassified subsets. This most probably reflects the DPE-containing Inr fraction, highlighting the major contribution of the DPE motif to transcriptional regulation. The TCT element, which was originally reported to be present among translation and ribosomal-related genes,48 was indeed found to be strongly enriched among these gene groups. In addition, structural processes related to mitosis, such as spindle, microtubule, and centrosome related proteins, were enriched. This highlights the importance of the core promoter elements annotation of individual genes, revealing distinct functions associated with a core promoter element.
The algorithm's performance depends on the accuracy of the constructed models. The redundancy of the core promoter motifs may lead to the identification of sequences that match functionally verified sequences, yet are not functional. Nevertheless, their presence might indicate that the specific genomic locus is transcriptionally active. Based on experience with transcription factors binding motifs,63 sorting out the functionally relevant hits might prove to be a difficult task and will require individual examination. Future improvements of the algorithm will be based on new insights and a better understanding of transcription regulation, obtained by ongoing work of major projects and consortia. These are aimed at dissecting the rules governing transcriptional regulation, and include ENCODE,64 modENCODE,65 and FANTOM5,66 as well as other genome-wide studies.67,68 Importantly, the ElemeNT program can assist in the analysis of sequences from organisms whose TSSs have not yet been comprehensively defined. For example, both the TATA box and the BRE motifs are conserved from Archae to humans69 and many organisms whose transcriptomes have not been annotated, are likely to contain such core promoter elements.
In conclusion, we anticipate that the ElemeNT tool, along with the CORE database, will make the search for specific core promoter elements and their combinations within Drosophila transcripts or any sequence of interest, accessible to scientists and help in elucidating the major role core promoter elements play in gene expression.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Marina Socol, Boris Komraz and Dr. Eli Sloutskin for invaluable assistance in ElemeNT development and web execution. We thank Gal Nuta for assisting with optimization of ElemeNT parameters. We thank Dr. Diana Ideses, Dan Even, Adi Kedmi, Hila Shir-Shapira and Gal Nuta for critical reading of the manuscript.
Funding
This research was supported by grants from the Israel Science Foundation to TJ-G (no. 798/10) and RS (no. 317/13) and the European Union Seventh Framework Programme (Marie Curie International Reintegration Grant) to TJ-G (no. 256491). YO was supported by the Edmond J Safra Center for Bioinformatics at Tel-Aviv University and the Israeli Center for Research Excellence (I-CORE), Gene Regulation in Complex Human Disease, center 41/11.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Heintzman ND, Ren B. The gateway to transcription: identifying, characterizing and understanding promoters in the eukaryotic genome. Cell Mol Life Sci 2007; 64:386-400; PMID:17171231; http://dx.doi.org/10.1007/s00018-006-6295-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Juven-Gershon T, Hsu J-Y, Theisen JWM, Kadonaga JT. The RNA polymerase II core promoter – the gateway to transcription. Curr Opin Cell Biol 2008; 20:253-9; PMID:18436437; http://dx.doi.org/10.1016/j.ceb.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 2010; 339:225-9; PMID:19682982; http://dx.doi.org/10.1016/j.ydbio.2009.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kadonaga JT. Perspectives on the RNA polymerase II core promoter. Wiley Interd Rev Dev Biol 2012; 1:40-51; PMID:23801666; http://dx.doi.org/10.1002/wdev.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smale ST. Core promoters: active contributors to combinatorial gene regulation. Genes Dev 2001; 15:2503-8; PMID:11581155; http://dx.doi.org/10.1101/gad.937701 [DOI] [PubMed] [Google Scholar]
- 6. Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Ann Rev Biochem 2003; 72:449-79; PMID:12651739; http://dx.doi.org/10.1146/annurev.biochem.72.121801.161520 [DOI] [PubMed] [Google Scholar]
- 7. Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat Rev Genet 2012; 13:233-45; PMID:22392219 [DOI] [PubMed] [Google Scholar]
- 8. Muller F, Demeny MA, Tora L. New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J Biol Chem 2007; 282:14685-9; PMID:17395580; http://dx.doi.org/10.1074/jbc.R700012200 [DOI] [PubMed] [Google Scholar]
- 9. Muller F, Tora L. Chromatin and DNA sequences in defining promoters for transcription initiation. Biochim Biophys Acta 2014; 1839:118-28; PMID:24275614; http://dx.doi.org/10.1016/j.bbagrm.2013.11.003 [DOI] [PubMed] [Google Scholar]
- 10. Danino YM, Even D, Ideses D, Juven-Gershon T. The core promoter: at the heart of gene expression. Biochim Biophys Acta 2015; PMID:25934543; http://dx.doi.org/10.1016/j.bbagrm.2015.04.003 [DOI] [PubMed] [Google Scholar]
- 11. Juven-Gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nat Methods 2006; 3:917-22; PMID:17124735; http://dx.doi.org/10.1038/nmeth937 [DOI] [PubMed] [Google Scholar]
- 12. Zehavi Y, Kuznetsov O, Ovadia-Shochat A, Juven-Gershon T. Core promoter functions in the regulation of gene expression of Drosophila dorsal target genes. J Biol Chem 2014; 289:11993-2004; PMID:24634215; http://dx.doi.org/10.1074/jbc.M114.550251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zehavi Y, Sloutskin A, Kuznetsov O, Juven-Gershon T. The core promoter composition establishes a new dimension in developmental gene networks. Nucleus 2014; 5:298-303; PMID:25482118; http://dx.doi.org/10.4161/nucl.29838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Butler JE, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev 2001; 15:2515-9; PMID:11581157; http://dx.doi.org/10.1101/gad.924301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dikstein R. The unexpected traits associated with core promoter elements. Transcription 2011; 2:201-6; PMID:22231114; http://dx.doi.org/10.4161/trns.2.5.17271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 2006; 41:105-78; PMID:16858867; http://dx.doi.org/10.1080/10409230600648736 [DOI] [PubMed] [Google Scholar]
- 17. He Y, Fang J, Taatjes DJ, Nogales E. Structural visualization of key steps in human transcription initiation. Nature 2013; 495:481-6; PMID:23446344; http://dx.doi.org/10.1038/nature11991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grunberg S, Hahn S. Structural insights into transcription initiation by RNA polymerase II. Trends Biochem Sci 2013; 38:603-11; PMID:24120742; http://dx.doi.org/10.1016/j.tibs.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cianfrocco MA, Kassavetis GA, Grob P, Fang J, Juven-Gershon T, Kadonaga JT, Nogales E. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell 2013; 152:120-31; PMID:23332750; http://dx.doi.org/10.1016/j.cell.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burke TW, Kadonaga JT. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev 1996; 10:711-24; PMID:8598298; http://dx.doi.org/10.1101/gad.10.6.711 [DOI] [PubMed] [Google Scholar]
- 21. Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAF(II)60 of Drosophila. Genes Dev 1997; 11:3020-31; PMID:9367984; http://dx.doi.org/10.1101/gad.11.22.3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu CH, Madabusi L, Nishioka H, Emanuel P, Sypes M, Arkhipova I, Gilmour DS. Analysis of core promoter sequences located downstream from the TATA element in the hsp70 promoter from Drosophila melanogaster. Mol Cell Biol 2001; 21:1593-602; PMID:11238896; http://dx.doi.org/10.1128/MCB.21.5.1593-1602.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Theisen JW, Lim CY, Kadonaga JT. Three key subregions contribute to the function of the downstream RNA polymerase II core promoter. Mol Cell Biol 2010; 30:3471-9; PMID:20457814; http://dx.doi.org/10.1128/MCB.00053-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kutach AK, Kadonaga JT. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol Cell Biol 2000; 20:4754-64; PMID:10848601; http://dx.doi.org/10.1128/MCB.20.13.4754-4764.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lim CY, Santoso B, Boulay T, Dong E, Ohler U, Kadonaga JT. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev 2004; 18:1606-17; PMID:15231738; http://dx.doi.org/10.1101/gad.1193404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gershenzon NI, Ioshikhes IP. Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics 2005; 21:1295-300; PMID:15572469; http://dx.doi.org/10.1093/bioinformatics/bti172 [DOI] [PubMed] [Google Scholar]
- 27. Bajic VB, Tan SL, Suzuki Y, Sugano S. Promoter prediction analysis on the whole human genome. Nat Biotechnol 2004; 22:1467-73; PMID:15529174; http://dx.doi.org/10.1038/nbt1032 [DOI] [PubMed] [Google Scholar]
- 28. Frith MC, Valen E, Krogh A, Hayashizaki Y, Carninci P, Sandelin A. A code for transcription initiation in mammalian genomes. Genome Res 2008; 18:1-12; PMID:18032727; http://dx.doi.org/10.1101/gr.6831208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Narlikar L, Ovcharenko I. Identifying regulatory elements in eukaryotic genomes. Brief Funct Genom Proteomics 2009; 8:215-30; PMID:19498043; http://dx.doi.org/10.1093/bfgp/elp014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ohler U, Niemann H. Identification and analysis of eukaryotic promoters: recent computational approaches. Trends Genet 2001; 17:56-60; PMID:11173099; http://dx.doi.org/10.1016/S0168-9525(00)02174-0 [DOI] [PubMed] [Google Scholar]
- 31. Pedersen AG, Baldi P, Chauvin Y, Brunak S. The biology of eukaryotic promoter prediction–a review. Comput Chem 1999; 23:191-207; PMID:10404615; http://dx.doi.org/10.1016/S0097-8485(99)00015-7 [DOI] [PubMed] [Google Scholar]
- 32. Rach EA, Winter DR, Benjamin AM, Corcoran DL, Ni T, Zhu J, Ohler U. Transcription initiation patterns indicate divergent strategies for gene regulation at the chromatin level. PLoS Genet 2011; 7:e1001274; PMID:21249180; http://dx.doi.org/10.1371/journal.pgen.1001274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duran E, Djebali S, Gonzalez S, Flores O, Mercader JM, Guigo R, Torrents D, Soler-Lopez M, Orozco M. Unravelling the hidden DNA structural/physical code provides novel insights on promoter location. Nucleic Acids Res 2013; 41:7220-30; PMID:23761436; http://dx.doi.org/10.1093/nar/gkt511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Abeel T, Saeys Y, Rouze P, Van de Peer Y. ProSOM: core promoter prediction based on unsupervised clustering of DNA physical profiles. Bioinformatics 2008; 24:i24-31; PMID:18586720; http://dx.doi.org/10.1093/bioinformatics/btn172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Datta S, Mukhopadhyay S. A composite method based on formal grammar and DNA structural features in detecting human polymerase II promoter region. PloS One 2013; 8:e54843; PMID:23437045; http://dx.doi.org/10.1371/journal.pone.0054843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ohler U. Identification of core promoter modules in Drosophila and their application in accurate transcription start site prediction. Nucl Acids Res 2006; 34:5943-50; PMID:17068082; http://dx.doi.org/10.1093/nar/gkl608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hartmann H, Guthohrlein EW, Siebert M, Luehr S, Soding J. P-value-based regulatory motif discovery using positional weight matrices. Genome Res 2013; 23:181-94; PMID:22990209; http://dx.doi.org/10.1101/gr.139881.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luehr S, Hartmann H, Soding J. The XXmotif web server for eXhaustive, weight matriX-based motif discovery in nucleotide sequences. Nucleic Acids Res 2012; 40:W104-9; PMID:22693218; http://dx.doi.org/10.1093/nar/gks602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Juven-Gershon T, Hsu J-Y, Kadonaga JT. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev 2008; 22:2823-30; PMID:18923080; http://dx.doi.org/10.1101/gad.1698108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hoskins RA, Landolin JM, Brown JB, Sandler JE, Takahashi H, Lassmann T, Yu C, Booth BW, Zhang D, Wan KH, et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res 2011; 21:182-92; PMID:21177961; http://dx.doi.org/10.1101/gr.112466.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 2010; 327:335-8; PMID:20007866; http://dx.doi.org/10.1126/science.1181421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protocols 2013; 8:1551-66; PMID:23868073; http://dx.doi.org/10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 2013; 41:D377-86; PMID:23193289; http://dx.doi.org/10.1093/nar/gks1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Deng W, Roberts SG. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev 2005; 19:2418-23; PMID:16230532; http://dx.doi.org/10.1101/gad.342405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev 1998; 12:34-44; PMID:9420329; http://dx.doi.org/10.1101/gad.12.1.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deng W, Roberts SG. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma 2007; 116:417-29; PMID:17593382; http://dx.doi.org/10.1007/s00412-007-0113-9 [DOI] [PubMed] [Google Scholar]
- 47. Engstrom PG, Ho Sui SJ, Drivenes O, Becker TS, Lenhard B. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res 2007; 17:1898-908; PMID:17989259; http://dx.doi.org/10.1101/gr.6669607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Parry TJ, Theisen JWM, Hsu J-Y, Wang Y-L, Corcoran DL, Eustice M, Ohler U, Kadonaga JT. The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev 2010; 24:2013-8; PMID:20801935; http://dx.doi.org/10.1101/gad.1951110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 2015; 518:556-9; PMID:25517091; http://dx.doi.org/10.1038/nature13994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chalkley GE, Verrijzer CP. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J 1999; 18:4835-45; PMID:10469661; http://dx.doi.org/10.1093/emboj/18.17.4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tokusumi Y, Ma Y, Song X, Jacobson RH, Takada S. The new core promoter element XCPE1 (X Core Promoter Element 1) directs activator-, mediator-, and TATA-binding protein-dependent but TFIID-independent RNA polymerase II transcription from TATA-less promoters. Mol Cell Biol 2007; 27:1844-58; PMID:17210644; http://dx.doi.org/10.1128/MCB.01363-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. FitzGerald PC, Sturgill D, Shyakhtenko A, Oliver B, Vinson C. Comparative genomics of Drosophila and human core promoters. Genome Biol 2006; 7:R53; PMID:16827941; http://dx.doi.org/10.1186/gb-2006-7-7-r53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ohler U, Liao GC, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome biology 2002; 3:RESEARCH0087; PMID:12537576; http://dx.doi.org/10.1186/gb-2002-3-12-research0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xi H, Yu Y, Fu Y, Foley J, Halees A, Weng Z. Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of YY1. Genome Res 2007; 17:798-806; PMID:17567998; http://dx.doi.org/10.1101/gr.5754707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li J, Gilmour DS. Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor. The EMBO J 2013; 32:1829-41; PMID:23708796; http://dx.doi.org/10.1038/emboj.2013.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ni T, Corcoran DL, Rach EA, Song S, Spana EP, Gao Y, Ohler U, Zhu J. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat Methods 2010; 7:521-7; PMID:20495556; http://dx.doi.org/10.1038/nmeth.1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci USA 2003; 100:15776-81; PMID:14663149; http://dx.doi.org/10.1073/pnas.2136655100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res 2007; 17:877-85; PMID:17179217; http://dx.doi.org/10.1101/gr.5533506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Furey TS. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat Rev Genet 2012; 13:840-52; PMID:23090257; http://dx.doi.org/10.1038/nrg3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 2008; 322:1845-8; PMID:19056941; http://dx.doi.org/10.1126/science.1162228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 2006; 38:626-35; PMID:16645617; http://dx.doi.org/10.1038/ng1789 [DOI] [PubMed] [Google Scholar]
- 62. Rach EA, Yuan HY, Majoros WH, Tomancak P, Ohler U. Motif composition, conservation and condition-specificity of single and alternative transcription start sites in the Drosophila genome. Genome Biol 2009; 10:R73; PMID:19589141; http://dx.doi.org/10.1186/gb-2009-10-7-r73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 2014; 15:272-86; PMID:24614317; http://dx.doi.org/10.1038/nrg3682 [DOI] [PubMed] [Google Scholar]
- 64. ENCODE Project Consortium . The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004; 306:636-40; PMID: 15499007; http://dx.doi.org/10.1126/science.1105136 [DOI] [PubMed] [Google Scholar]
- 65. Washington NL, Stinson EO, Perry MD, Ruzanov P, Contrino S, Smith R, Zha Z, Lyne R, Carr A, Lloyd P, et al. The modENCODE Data Coordination Center: lessons in harvesting comprehensive experimental details. Database (Oxford) 2011; 2011:bar023; PMID:21856757; http://dx.doi.org/10.1093/database/bar023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Lassmann T, Itoh M, Summers KM, Suzuki H, Daub CO, et al. A promoter-level mammalian expression atlas. Nature 2014; 507:462-70; PMID:24670764; http://dx.doi.org/10.1038/nature13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet 2007; 8:424-36; PMID:17486122; http://dx.doi.org/10.1038/nrg2026 [DOI] [PubMed] [Google Scholar]
- 68. Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat Rev Genet 2012; 13:233-45; PMID:22392219 [DOI] [PubMed] [Google Scholar]
- 69. Reeve JN. Archaeal chromatin and transcription. Mol Microbiol 2003; 48:587-98; PMID:12694606; http://dx.doi.org/10.1046/j.1365-2958.2003.03439.x [DOI] [PubMed] [Google Scholar]
- 70. Smale ST, Baltimore D. The "initiator" as a transcription control element. Cell 1989; 57:103-13; PMID:2467742; http://dx.doi.org/10.1016/0092-8674(89)90176-1 [DOI] [PubMed] [Google Scholar]
- 71. Goldberg ML. Ph.D. Thesis. Sequence analysis of Drosophila histone genes. Stanford University 1979. [Google Scholar]
- 72. Anish R, Hossain MB, Jacobson RH, Takada S. Characterization of transcription from TATA-less promoters: identification of a new core promoter element XCPE2 and analysis of factor requirements. PloS One 2009; 4:e5103; PMID:19337366; http://dx.doi.org/10.1371/journal.pone.0005103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lewis BA, Kim TK, Orkin SH. A downstream element in the human beta-globin promoter: evidence of extended sequence-specific transcription factor IID contacts. Proc Natl Acad Sci U S A 2000; 97:7172-7; PMID:10840054; http://dx.doi.org/10.1073/pnas.120181197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee DH, Gershenzon N, Gupta M, Ioshikhes IP, Reinberg D, Lewis BA. Functional characterization of core promoter elements: the downstream core element is recognized by TAF1. Mol Cell Biol 2005; 25:9674-86; PMID:16227614; http://dx.doi.org/10.1128/MCB.25.21.9674-9686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


