Abstract
Yeast cells with DNA damage avoid respiration, presumably because products of oxidative metabolism can be harmful to DNA. We show that DNA damage inhibits the activity of the Snf1 (AMP-activated) protein kinase (AMPK), which activates expression of genes required for respiration. Glucose and DNA damage upregulate SUMOylation of Snf1, catalyzed by the SUMO E3-ligase Mms21, which inhibits SNF1 activity. The DNA damage checkpoint kinases Mec1/ATR and Tel1/ATM, as well as the nutrient sensing protein kinase A (PKA), regulate Mms21 activity towards Snf1. Mec1 and Tel1 are required for two SNF1-regulated processes—glucose sensing and ADH2 gene expression—even without exogenous genotoxic stress. Our results imply that inhibition of Snf1 by SUMOylation is a mechanism by which cells lower their respiration in response to DNA damage. This raises the possibility that activation of DNA damage checkpoint mechanisms could contribute to aerobic fermentation (Warburg effect), a hallmark of cancer cells.
Introduction
Sugars have the potential to cause genotoxic stress for cells because their oxidative metabolism can generate reactive molecules that damage DNA. Indeed, yeast cells with DNA damage seem to avoid respiration: studies of the DNA damage response revealed decreased expression of genes involved in respiration and increased expression of genes involved in fermentation (Caba et al., 2005, Fry et al., 2003, Gasch et al., 2001, Jelinsky and Samson, 1999, Lee et al., 2000, Shalem et al., 2008), and one of the effects of DNA damage in yeast is the reduction of respiration (Kitanovic et al., 2009).
S. cerevisiae has three major glucose sensing pathways: the Gpa1/2-Ras2-PKA pathway that also regulates stress response, the Snf3/Rgt2/Rgt1 (Sensor/Receptor-Repressor, or SRR) pathway that regulates expression of genes encoding hexose transporters (Busti et al., 2010, Johnston and Kim, 2005) required for fermentation (Gamo et al., 1994), and SNF1, a central regulator of carbon metabolism in S. cerevisiae (Ghillebert et al., 2011, Hedbacker et al., 2004) that is the orthologue of the AMP-activated protein kinase (AMPK) of mammalian cells (Hardie et al., 2012, Jiang and Carlson, 1997). SNF1 protein kinase is a heterotrimer composed of the Snf1 catalytic subunit, the Snf4 regulatory subunit, and one of three localizing subunits (Sip1, Sip2 and Gal83). SNF1 stimulates expression of genes involved in respiration, the diauxic shift, ethanol and lactate catabolism, and gluconeogenesis by activating transcriptional activators such as Adr2, the activator of ADH2 (which encodes alcohol dehydrogenase) (Bojunga and Entian, 1999, Hardie et al., 1998, Tachibana et al., 2005, Walther and Schuller, 2001, Young et al., 2003), and by inhibiting the Mig1 repressor of glucose-repressed genes (Vallier and Carlson, 1994). SNF1 also inhibits glucose sensing via the SRR pathway (Gadura et al., 2006, Pasula et al., 2007), which effects degradation of the Mth1 co-repressor of HXT genes encoding glucose transporters. Thus, active SNF1 increases Mth1 levels and thereby reduces HXT expression, and increases Adr1 function, thereby inducing ADH2 expression.
SNF1 is active in glucose-starved yeast cells, in which the Snf1 catalytic subunit is phosphorylated on its activation loop threonine 210 (Elbing et al., 2006, Hedbacker et al., 2004, Hong et al., 2003). Addition of glucose to cells results in a reduction in ADP levels that leads to dephosphorylation of T210 by the Glc7-Reg1 protein phosphatase, thereby inactivating SNF1 (Ludin et al., 1998, Chandrashekarappa et al., 2011, Mayer et al., 2011). The Sit4 and Ptc1 phosphatases also have a role in dephosphorylation of this site (Ruiz et al., 2011, Ruiz et al., 2013). In addition, SNF1 is inhibited by its modification with the Small Ubiquitin-like Modifier protein SUMO, catalyzed by the SUMO E3-ligase Mms21 in response to glucose, which leads to ubiquitinylation and degradation of Snf1 (Simpson-Lavy and Johnston, 2013).
Methyl methanesulfonate (MMS), a DNA-alkylating agent that causes methylation of deoxyguanine and deoxyadenine, activates DNA damage repair pathways and causes cell cycle arrest (Evensen and Seeberg, 1982). The Mec1 protein kinase (orthologue of human ATR), and its homologue Tel1 (orthologue of human ATM), respond to DNA stress by catalyzing phosphorylation of Rad53 (Friedel et al., 2009, Paciotti et al., 2001, Pellicioli and Foiani, 2005) and other proteins involved in the response to genotoxic damage (Morrison et al., 2007, Mallory et al., 2003). Inhibition of SNF1 is involved in the response of cells to MMS, and is required for a concomitant switch in metabolism from respiration to fermentation (Kitanovic et al., 2009), but the basis of that inhibition is unknown. Here, we provide evidence that DNA damage inhibits SNF1 function by inducing its SUMOylation.
Although many targets of SUMO E3-ligases have been identified, the regulation of the ligases themselves remains little understood. To date, the only known phospho-regulation of a SUMO E3-ligase is the regulation of Siz1 stability by the Cdc28 protein kinase via an unidentified mechanism (Takahashi and Kikuchi, 2005). Here we present evidence that suggests that the SUMO (E3) ligase Mms21 is regulated by phosphorylation due to Mec1, Tel1 and PKA activity.
Results
Mms21 is a phosphorylated protein
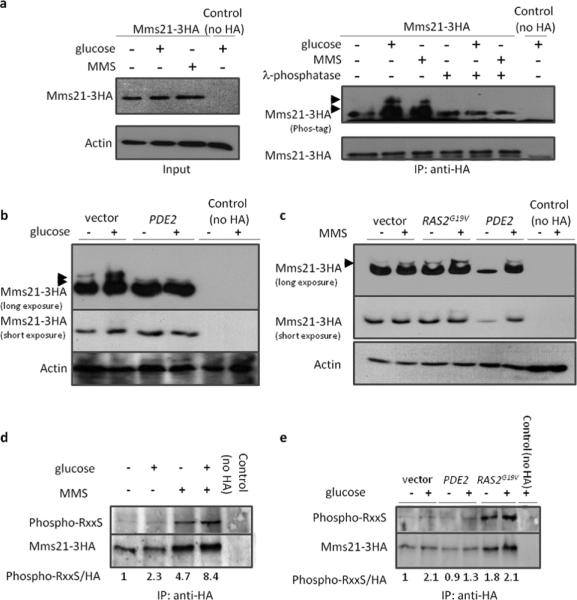
One of the ways glucose inhibits Snf1 activity is by promoting its SUMOylation, catalyzed by the Mms21 SUMO (E3) ligase (Simpson-Lavy and Johnston, 2013). We observed an electrophoretic band-shift of Mms21-3HA that is reversed by λ-phosphatase treatment in cells exposed to either glucose or MMS (Figure 1a). Overexpression of the PDE2 phosphodiesterase (Sass et al., 1986) reduced the amount of this glucose-induced phosphorylation of Mms21 (Figure 1b), suggesting a role for PKA in Mms21 phosphorylation. MMS treatment also results in phosphorylation of Mms21, but this is not inhibited by PDE2 overexpression (Figure 1c). Expression of activated Ras2G19V, which increases PKA activity (Tatchell, 1986) by stimulating adenyl cyclase (Cyr1) (Uno et al., 1985, Uno et al., 1987), increases basal phosphorylation of Mms21, and the level of phosphorylated Mms21 is increased further by MMS treatment (Figure 1c). Thus, both MMS and glucose (via PKA) treatments result in phosphorylation of Mms21.
Figure 1. Mms21 is a phospho-protein.
a. Glucose to 2% (15 minutes) or 0.03% MMS (1 hour) was added to media containing Mms21-3HA cells (or cells lacking the HA tag). Samples were taken for immunoprecipitations using anti-HA. Left: input. Right: Immunopreciptated Mms21-3HA was run on 15% phos-tag and regular gels, and probed with anti-HA. Phosphorylated Mms21 is indicated with arrows. b, c. Glucose to 2% (15 minutes) (b) or 0.03% MMS (1 hour) (c) was added to media containing Mms21-3HA cells (or cells lacking the HA tag) bearing indicated plasmids or empty vector. Cells were processed for immunoblots, and whole extracts loaded onto 15% gels containing phos-tag. A long and a short exposure for anti-HA are shown. After probing for HA, blots were stripped and reprobed for actin (b) or identical amounts of protein extract loaded onto a new gel and the membrane probed for actin (c). Phosphorylated Mms21 is indicated with arrows - only the upper phospho-Mms21 band is regulated by PKA. d, e. Mms21-3HA (or cells lacking the HA tag) were treated as with 4% glucose for 15 minutes and/or 0.3% MMS for 1 hour, and samples taken for immunoprecipitations using anti-HA beads. Immunoprecipated Mms21-3HA was run on 12% gels, and probed with mouse anti-HA and rabbit anti-RxxS* simultaneously using fluorescent secondary antibodies. Ratios of pRxxS/HA were calculated by quantification using ImageJ.
To further characterize this phosphorylation we immunoprecipitated Mms21-3HA from cells treated with glucose or MMS, and probed with an antibody that recognizes phosphorylated serine in the motif RxxS (PKA's consensus site). Phosphorylation of this motif was increased about 2-fold in response to both glucose and MMS (Figure 1d). Expression of Ras2G19V increased basal phosphorylation of this sequence, whereas overexpression of PDE2 attenuated its phosphorylation following addition of glucose (Figure 1e). Mms21 has four RxxS sequences, three in the N-terminal, Smc5-interacting domain, and one in the C-terminal SUMO ligase domain. Examination of the crystal structure of Mms21 bound to Smc5 (Duan et al., 2009) indicates that the former three are either buried or interacting with Smc5, but the site in the SUMO E3 ligase domain is exposed; it is also a more stringent PKA consensus, containing a double basic motif.
Phenotypes of Mms21 phospho-mutants
MMS activates the Mec1/Tel1 protein kinases (Flott et al., 2007, Gasch et al., 2001, Kato and Ogawa, 1994), which phosphorylate their substrates at SQ/TQ sequences (Mallory and Petes, 2000). A cluster of SQ sequence motifs in its C-terminal SUMO E3-ligase domain (residues 227-228, 253-254, 261-262) makes Mms21 a good candidate for a Mec1/Tel1 substrate (Cheung et al., 2012).
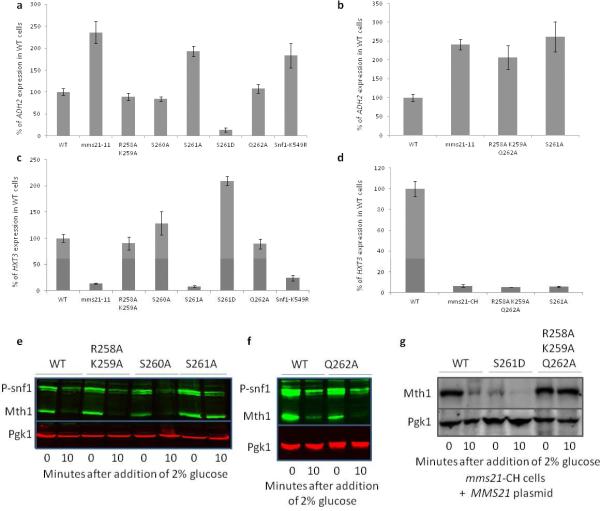
We examined the effect of single potential phospho-site mutations in MMS21. Mutation of a single SQ to AQ does not result in sensitivity to hydroxyurea or MMS (Figure 2a), nor in temperature sensitivity (Figure 2b), which are phenotypes of mms21 mutants (Zhao and Blobel, 2005) (Santa Maria et al., 2007). However, mms21S261A cells grow considerably better than wild-type under strong respiratory conditions (Figure 2c), which suggests this serine may regulate Snf1 mediated processes. Mutation of one of these SQ motifs—S261—but not the other two, to AQ, reduces HXT3 expression (Figure 2d) and increases ADH2 expression (Figure 2e) to the same extent as the mms21-11 mutation; mutation of S261 to the phosphomimicking aspartate has the opposite effect (Figures 2d, e). This is consistent with the idea that S261 mediates regulation of Snf1.
Figure 2. Phenotypes of Mms21 phospho-mutants.
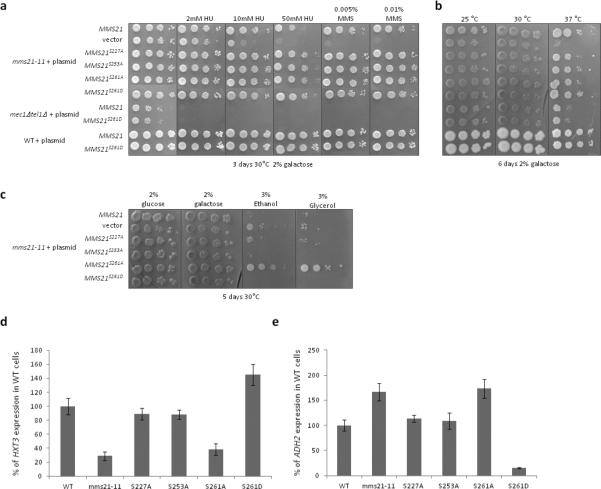
a, b, c. Cells bearing the indicated plasmids were serially diluted tenfold from saturated cultures onto SC-ura plates containing the indicated carbon sources and genotoxins, and grown at the indicated temperatures. Photographs were taken on the indicated days. For the genotoxin and temperature sensitivity assays, similar results were obtained for cells grown on glucose. For the carbon source growth assay, similar results were obtained at 25°C. None of the strains grew on either 3% ethanol or 3% glycerol at 37°C. mec1Δtel1Δ cells are also sml1Δ. d. mms21-11 cells bearing the indicated MMS21 plasmids or empty vector and prHXT3:LacZ were grown overnight in 2% galactose, and glucose added to 2%. HXT3 expression was measured by β-galactosidase activity every half-hour for 90 minutes. The rate of HXT3 expression is normalized to that of wild-type cells. N=3. Error bars are +− 1 standard deviation. e. mms21-11 cells bearing the indicated MMS21 plasmids or empty vector and prADH2:LacZ were grown overnight in 4% glucose. Cells were washed three times with water and resuspended in media containing 3% glycerol. Samples were taken for β-galactosidase assays each hour for four hours. The rate of ADH2 expression is normalized to that of wild-type cells. N=3. Error bars are +− 1 standard deviation.
We changed to alanine residues in the vicinity of Serine 261 to better characterize the sequence required for its phosphorylation, using Mth1 degradation, and HXT3 and ADH2 expression as reporters of SNF1 function. Mutation of serine 261 to alanine results in an increase in ADH2 expression (Figures 3a, b) and a decrease in HXT3 expression (Figures 3c, d) to an extent similar to that caused by the mms21-11 mutation and the non-SUMOylatable Snf1K549R mutation, and impedes Mth1 degradation (Figure 3e). The phosphomimicking S261D mutation has opposite effects, reducing ADH2 expression (Figure 3a) and increasing HXT3 expression (Figure 3c). Mms21S261D cells exhibit a lower level of Mth1 in the absence of glucose, which is further reduced after exposure to glucose (Figure 3g). Mutation of S260A has no effect on ADH2 or HXT3 expression (Figures 3a, c) or on Mth1 degradation (Figure 3e), so the effect of Mms21 phosphorylation on Snf1 functions is specific to S261.
Figure 3. Alanine scan of the C-terminus of Mms21.
a. WT, mms21-11, or cells with the indicated integrated mutations in the MMS21 locus containing a prADH2:LacZ plasmid were grown overnight in 4% glucose. Cells were washed three times with water and resuspended in media containing 3% glycerol. Samples were taken for β-galactosidase assays each hour for four hours. The rate of ADH2 expression is normalized to that of wild-type cells for each experiment. ADH2 expression in snf1Δ cells bearing Snf1K549R is included for comparison. N=3. Error bars are +− 1 standard deviation. b. mms21-11 cells bearing the indicated MMS21 plasmids or empty vector and prADH2:LacZ were grown overnight in 4% glucose. Cells were washed three times with water and resuspended in media containing 3% glycerol. Samples were taken for β-galactosidase assays each hour for four hours. The rate of ADH2 expression is normalized to that of wild-type cells. N=3. Error bars are +− 1 standard deviation. c. WT, mms21-11, or cells with the indicated integrated mutations in the MMS21 locus containing a prHXT3:LacZ plasmid were grown overnight in 2% galactose, and glucose added to 2%. HXT3 expression was measured by β-galactosidase activity each half-hour for 90 minutes. The rate of HXT3 expression is normalized to that of wild-type cells for each experiment. HXT3 expression in snf1Δ cells bearing Snf1K549R is included for comparison. N=3. Error bars are +− 1 standard deviation. d. mms21-CH cells bearing the indicated MMS21 plasmids or empty vector and prHXT3:LacZ were grown overnight in 2% galactose, at 30°C. The temperature was raised to 34°C for one hour, and preheated glucose added to 2%. HXT3 expression was measured by β-galactosidase activity each half-hour for 90 minutes. The rate of HXT3 expression is normalized to that of wild-type cells. N=3. Error bars are +− 1 standard deviation. e, f. Cells bearing the indicated integrated mutations in the MMS21 locus were grown overnight in 2% galactose at 30°C. Glucose was added to 2%, and samples taken for immunoblotting at the indicated times. An extra copy of Snf1 on a CEN plasmid was present which did not affect the experiment. g. mms21-CH cells containing a plasmid with the indicated mutation in MMS21 were grown overnight in 2% galactose at 30°C. The temperature was raised to 34°C for one hour, and preheated glucose added to 2%. Samples were taken for immunoblots at indicated times.
The S261 SQ motif overlaps a potential PKA phosphorylation site (R/K R/K X S*): 258KRSSQ262. Changing to alanine R258 and K259 of the PKA motif, or Q262 of the Mec1/Tel1 motif, does not affect ADH2 (Figure 3a) or HXT3 (Figure 3c) expression or Mth1 degradation (Figure 3e, f). However, cells expressing Mms21R258A, K259A, Q262A show the same increase in ADH2 (Figure 3b) and decrease in HXT3 (Figure 3d) expression as do mms21 E3-ligase domain mutants or non-phosphorylatable Mms21S261A, and are defective for Mth1 degradation in response to glucose (Figure 3g). These results suggest that either PKA or Mec1/Tel1 are capable of phosphorylating S261.
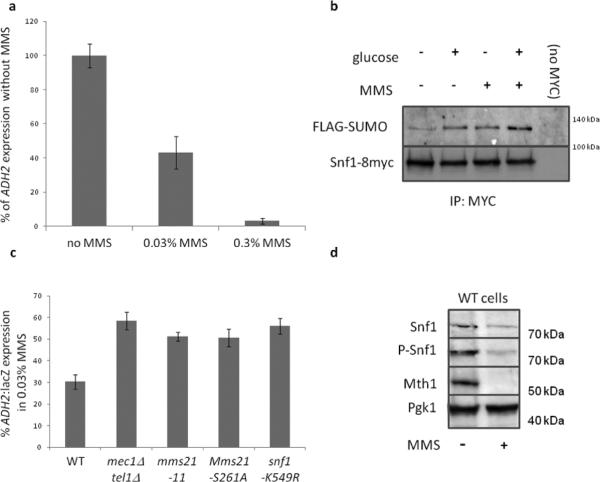
Protein kinases that regulate Mms21
The S261Q262 motif of Mms21 is a possible target of the Mec1 and Tel1 protein kinases. Indeed, deletion of both MEC1 and TEL1 impairs glucose-induced Mth1 degradation (Figure 4a), reduces glucose-induced HXT3 expression (Figure 4b), and enhances ADH2 expression (Figure 4c), all phenotypes expected with loss of Mms21 SUMO E3-ligase activity towards Snf1. Mms21 does not become phosphorylated upon treatment with glucose in mec1Δtel1Δ cells (Figure 4d). MMS21S261D introduced into mec1Δtel1Δ cells restores wild-type Mth1 degradation (Figure 4e) and suppresses the enhanced ADH2 expression of mec1Δtel1Δ cells (Figure 4c). We conclude that promoting phosphorylation of S261 of Mms21 is the sole requirement of Mec1/Tel1 for Mth1 degradation and ADH2 expression. [Note that the effects of Mms21S261D on the mec1Δtel1Δ phenotypes examined are limited to Snf1 regulation: Mms21S261D does not suppress the sensitivity of mec1Δtel1Δ cells to either hydroxyurea, MMS, or elevated temperature (37°C) (Figure 2a)].
Figure 4. Kinases that regulate Mms21 activity towards Snf1 via phosphorylation of S261.
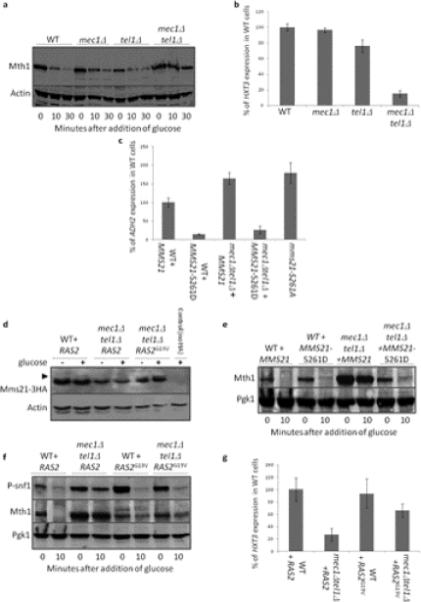
mec1Δ and mec1Δtel1Δ cells are also sml1Δ. a. Cells were grown overnight in 2% galactose at 30°C. Glucose was added to 2% and samples taken for immunoblots at the indicated times. b. Cells were grown overnight in 2% galactose at 30°C. Glucose was added to 2%. HXT3 expression was measured by β-galactosidase activity each half-hour for 90 minutes. The rate of HXT3 expression is normalized to that of wild-type cells. N=3. Error bars are +− 1 standard deviation. c. Indicated cells containing a prADH2:LacZ plasmid and a plasmid with either MMS21WT or MMS21S261D were grown overnight in 4% glucose. Cells were washed three times with water and resuspended in media containing 3% glycerol. Samples were taken for β-galactosidase assays each hour for four hours. The rate of ADH2 expression is normalized to that of wild-type cells. N=3. Error bars are +− 1 standard deviation. d. Indicated cells with an extra copy of plasmid borne Mms21-3HA were grown overnight in 2% galactose at 30°C. Glucose was added to 2% and samples taken after 15 minutes. Cells were processed for immunoblots, and whole extracts loaded onto 15% gels containing phos-tag. After probing for HA, blots were stripped and reprobed for actin. Phosphorylated Mms21 is indicated with arrows. e, f. Indicated cells bearing indicated plasmids were grown overnight in 2% galactose at 30°C. Glucose was added to 2% and samples taken for immunoblots at the indicated times. g. Indicated cells bearing indicated plasmids were grown overnight in 2% galactose at 30°C. Glucose was added to 2%. HXT3 expression was measured by β-galactosidase activity each half-hour for 90 minutes. The rate of HXT3 expression is normalized to that of wild-type cells. N=3. Error bars are +− 1 standard deviation.
Protein kinase A (PKA) affects glucose sensing at multiple levels [inhibition of Snf1 (Barrett et al., 2012), Rgt1 (Kim and Johnston, 2006, Roy et al., 2014) and Mth1 (Ma et al., 2014)], and is activated by Mec1 inhibition of the PKA regulatory subunit Bcy1 (Searle et al., 2011). We observed that phosphorylation of Mms21 in response to glucose (but not MMS) is reduced by overexpression of the PDE2 phosphodiesterase (Figures 1b, c) and that phosphorylation of Mms21 on a PKA consensus motif (RxxS, Figure 1e) is affected by changes in PKA activity. It appears that increased stimulation of the PKA pathway compensates for lack of MEC1 and TEL1 because hyperactive Ras2G19V (McGrath et al., 1984) restores Mms21 phosphorylation (Figure 4d), Mth1 degradation (Figure 4f) and HXT3 expression (Figure 4g) in mec1Δtel1Δ cells.
MMS treatment reduces ADH2 expression via Snf1 SUMOylation
Because MMS treatment of S. cerevisiae cells reduces transcription of genes involved in respiration and increases transcription of genes involved in fermentation (Caba et al., 2005, Fry et al., 2003, Gasch et al., 2001, Jelinsky and Samson, 1999, Lee et al., 2000, Shalem et al., 2008), we monitored the effect of MMS on the expression of ADH2, which is usually induced under respiratory conditions. MMS reduces ADH2 expression (Figure 5a) and increases SUMOylation of Snf1 (Figure 5b). The MMS-induced reduction of ADH2 expression is curtailed to the same extent by deletion of MEC1 and TEL1, or by mutation of mms21, or by preventing Snf1 SUMOylation (Figure 5c). These results suggest that MMS reduces ADH2 expression via Mec1-Tel1 stimulated Mms21-catalyzed SUMOylation of Snf1. We previously showed that glucose-induced SUMOylation of Snf1 leads to its turnover (Simpson-Lavy and Johnston, 2013), and prolonged treatment of cells with a high dose of MMS likewise reduces Snf1 protein levels (Figure 5d).
Figure 5. MMS downregulates ADH2 expression.
a. WT cells were grown overnight in 4% glucose at 30°C. Cells were washed three times with water and resuspended in media containing 3% glycerol and the indicated MMS concentrations. Samples were taken for β-galactosidase assays every hour for four hours. The rate of HXT3 expression is normalized to that of untreated cells. N=3. Error bars are +− 1 standard deviation. mec1Δtel1Δ cells are also sml1Δ. b. ulp1-ts cells (strain 1274 (Wykoff and O'Shea, 2005)) expressing Snf1-8myc (Liu et al., 2011), Snf1K549R-8myc (Simpson-Lavy and Johnston, 2013) or Snf1 (with no tag) (Shirra et al., 2008), together with a plasmid containing GAL:His6-FLAG-SMT3 were grown overnight at 24°C in 2% galactose. The temperature was elevated to 37°C for 4 hours before addition of preheated glucose to 2% for 15 minutes or 0.3% MMS for 1 hour (or both conditions) as indicated. Samples were processed for immunoprecipitations with MYC. c. Cells bearing the indicated mutations were grown overnight in 4% glucose at 30°C. Cells were washed three times with water and resuspended in media containing 3% glycerol with and without 0.03% MMS. Samples were taken for β-galactosidase assays every hour for four hours and the rate of ADH2 expression determined. The rate of ADH2 expression in the presence of 0.03% MMS was normalized against the rate of ADH2 expression for that strain in the absence of MMS (the mutant strains all have dramatically increased ADH2 expression in the absence of MMS (Figures, 2, 3, 4 and (Simpson-Lavy and Johnston, 2013)). Comparing the WT to any of the mutants gave a significant difference at p<0.01. In contrast, the differences between the mutants was not significant (p>0.05). N=3. Error bars are +− 1 standard deviation. d. Wild-type cells were grown overnight in 2% galactose at 30°C. 0.3% MMS was added for two hours. Samples with and without 0.3% MMS treatment for 2 hours were taken for immunoblots.
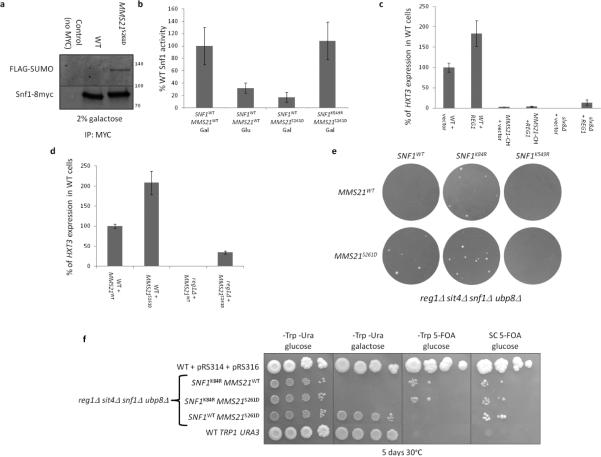
SUMOylation and dephosphorylation regulate Snf1 function
Cells expressing Mms21S261D have an increased amount of SUMOylated Snf1 in galactose-grown cells (Figure 6a). This is consistent with the idea that phosphorylation of S261 of Mms21 activates its SUMO ligase activity. Indeed, the MMS21S261D mutation reduces Snf1 activity in galactose grown cells to levels comparable to those in glucose-grown cells, but not if Snf1 cannot be SUMOylated (Snf1K549R) (Figure 6b).
Figure 6. Snf1 SUMOylation and phosphorylation are independently regulated.
a. ulp1-ts cells (strain 1274 (Wykoff and O'Shea, 2005)) expressing Snf1-8myc (Liu et al., 2011) or Snf1 (with no tag) (Shirra et al., 2008), a plasmid with either MMS21 or MMS21S261D, together with a plasmid containing GAL:His6-FLAG-SMT3 were grown overnight at 24°C in 2% galactose. The temperature was elevated to 37°C for 4 hours before harvesting for anti-myc immunoprecipitations with B480. Blots were probed with anti-myc (for Snf1-8myc) and anti-FLAG (for Smt3). b. Cells containing the indicated Snf1-8myc plasmid and MMS21S261D plasmid where applicable were grown with either 2% galactose or 4% glucose as indicated overnight at 30°C. Cells were harvested and Snf1-8myc immunoprecipated. Cells with untagged Snf1 were used as a control. Protein kinase assays were performed using the ADP-glo kinase assay kit with SAMS peptide as a phosphate acceptor. N=3. c. Indicated cells containing a prHXT3:LacZ plasmid and either a 2μ plasmid overexpressing REG1 or empty vector were grown overnight in 2% galactose at 30°C. The temperature was raised to 34°C for one hour, and preheated glucose added to 2%.HXT3 expression was measured by β-galactosidase activity each half-hour for 90 minutes. The rate of HXT3 expression is normalized to that of wild-type cells. N=3. Error bars are +− 1 standard deviation. d. Indicated cells containing a prHXT3:LacZ plasmid and either a plasmid with MMS21WT or MMS21S261D were grown overnight in 2% galactose. HXT3 expression was measured by β-galactosidase activity each half-hour for 90 minutes. The rate of HXT3 expression is normalized to that of wild-type cells. N=3. Error bars are +− 1 standard deviation. e. reg1Δsit4Δsnf1Δubp8Δ cells were transformed with prs314 (TRP1) bearing either SNF1WT, snf1K84R (kinase dead), or snf1K549R (non-SUMOylatable) and with Ycplac33 (URA3) bearing either MMS21WT or MMS21S261D and plated onto –Trp -Ura 2% glucose plates, and photographed after 5 days incubation at 30°C. f. Wild-type ADE2 cells containing plasmids prs314 (TRP1) and prs316 (URA3) (positive control for growth on 5-FOA containing media), wild-type ADE2 cells with integrated TRP1 and URA3 (negative control for growth on 5-FOA containing media), and the three viable strains from figure 6e were serially tenfold diluted onto the indicated media. Plates were photographed after 5 days incubation at 30°C. The 5-FOA promotes loss of the MMS21S261D plasmid from reg1Δsit4Δubp8Δ SNF1 MMS21S261D cells, resulting in Snf1 toxicity on -Trp 5-FOA media.
What is the relationship of SUMOylation of Snf1 to dephosphorylation of threonine 210? Overexpression of REG1 increases HXT3 expression in wild-type cells [because it reduces SNF1 activity (Tu and Carlson, 1995, Ludin et al., 1998)], but it does not suppress the effects of either an mms21 mutation or a deletion of the SUMO-directed Ubiquitin ligase Slx8 (which targets SUMOylated Snf1 for degradation) (Figure 6c). However, increasing Snf1 SUMOylation via the Mms21S261D mutant increases HXT3 expression, both in the presence and the absence of Reg1 (Figure 6d). This suggests that increasing Snf1 inhibition by SUMOylation can counteract Snf1 activation by T210-phosphorylation, but not vice-versa.
Deletion of both reg1 and sit4 is lethal to S. cerevisiae, even under fermentative conditions. This is due to overactive SNF1, because further deletion of SNF1 restores viability (Ruiz et al., 2011). Introduction of plasmids bearing MMS21S261D and SNF1WT into reg1Δsit4Δsnf1 cells [also carrying ubp8Δ to reduce deubiquitinylation of Snf1 (Wilson et al., 2011)] resulted in viable cells on glucose media (Figure 6e). However, MMS21S261D fails to restore viability when SUMOylation of Snf1 is blocked by the Snf1K549R mutation (Figure 6e). Thus, inhibition of SNF1 by hyperSUMOylation of Snf1 (caused by the MMS21S261D mutation) compensates for lack of Snf1 dephosphorylation. Loss of the MMS21S261D plasmid resulted in their inviability, but viability is restored (to the same extent as in cells expressing kinase-dead Snf1K84R) when selection for the Snf1-bearing plasmid is also relaxed (Figure 6f). These results are consistent with SUMOylation and dephosphorylation of Snf1 occurring independently of each other in response to glucose. However, increasing SUMOylation of Snf1 using Mms21S261D seems to be a more potent inhibitor of Snf1 than REG1 overexpression.
Discussion
Little is known about how the activity of SUMO E3 ligases is regulated. Our results suggest that phosphorylation of serine 261 of Mms21 promoted by the Mec1 and Tel1 protein kinases in response to DNA damage, and also by PKA, induces SUMOylation of Snf1 and thereby inhibits SNF1 function, leading to reduced expression of genes for respiration and to the increased HXT gene expression necessary for fermentation (summarized in Figure 7). Recently, Carlborg et al published an investigation of Mms21 regulation under both ambient conditions and after DNA damage (Carlborg et al., 2015). In accordance with the results presented here, they report that Mms21 is phosphorylated on both S260 and S261. Indeed, we find no basal phosphorylation of Mms21 in mec1Δtel1Δ cells (growing on galactose) (Figure 4d). Their report of increased phosphorylation of S261 following MMS treated is also in agreement with the results of our experiments. Our results, however, do not support a role for S261 phosphorylation in DNA repair, as mms21-11 cells are clearly sensitive to MMS whereas all of the single SQ-AQ mutants exhibit WT growth (Figure 2a). Phosphorylation of Serine261 of Mms21 appears to be specific for SNF1 regulation because changing S261 to A does not affect the sensitivity of cells to the genotoxins hydroxyurea or MMS, or to elevated temperature, nor does Mms21S261D suppress other mec1Δtel1Δ phenotypes. Moreover, growth of cells expressing mms21S261A on ethanol or glycerol is improved compared to wild-type cells; this correlates to increased SNF1 activity in these cells. Since sensitivity to hydroxyurea or MMS or to elevated temperature was not observed for Mms21S261A cells, it is likely that these cells activate other metabolic solutions to genotoxicity, such as the pentose-phosphate pathway, as occurs in humans, where the ATM protein kinase (ortholog of Tel1) activates glucose-6-phosphate dehydrogenase (Zwf1), the rate-limiting enzyme of the pentose phosphate shunt that protects cells from oxidative damage by raising NADPH levels (Cosentino et al., 2011).
Figure 7. Model.
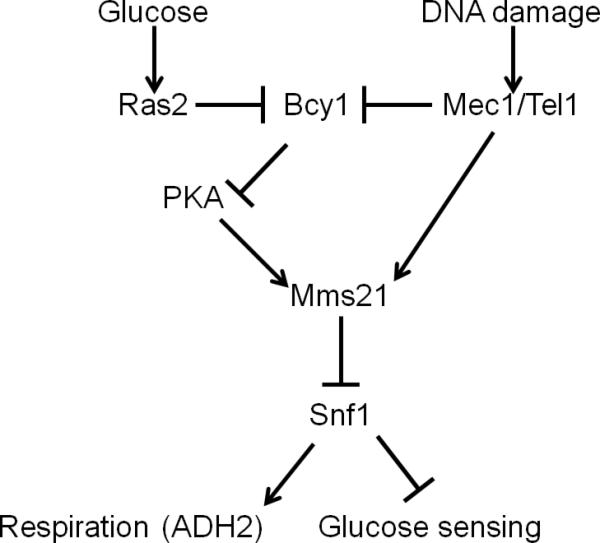
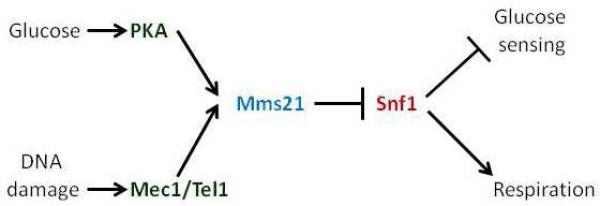
Proposed signaling pathway based on data presented in this article.
SNF1 is involved in regulating many cellular processes, including carbon metabolism, nitrogen availability (Orlova et al., 2006), response to stresses such as high pH (Casamayor et al., 2012), phosphate starvation (Thompson-Jaeger et al., 1991), and sodium ions (Hong and Carlson, 2007). These diverse roles for SNF1 may indicate the existence of different pools of SNF1, which may explain why only a small percentage of Snf1 needs to be SUMOylated to regulate carbon metabolism. Another explanation is that not all of the SNF1 in wild-type cells is active under respiratory conditions, as deletion of multiple phosphatases is lethal (Ruiz et al., 2011, Ruiz et al., 2013), suggesting unutilized Snf1 activity.
We compared the relationship of SUMOylation and dephosphorylation in the regulation of SNF1 activity. Lack of one of these modes of regulation of SNF1 does not prevent the other from occurring ((Simpson-Lavy and Johnston, 2013), Figures 3e, 3f, 6a, 6e, 6f). Hyperinduction of Snf1 SUMOylation with Mms21S261D suppresses the effects of lack of dephosphorylation, as shown by the reduction in SNF1 activity when the Mms21S261D mutant is grown in galactose (Figure 6b), a partial recovery of HXT3 expression in reg1Δ cells (Figure 6d), and viability of reg1Δsit4Δubp8Δ SNF1 Mms21S261D cells (Figures 6e, f).
The consequence of inhibition of SNF1 function by MMS is to throttle respiration, because SNF1 is a central component of the respiration/fermentation switch, inhibiting expression of genes encoding glucose transporters (Gadura et al., 2006, Pasula et al., 2007), which are required for fermentation (Gamo et al., 1994), and activating expression of genes required for respiration (Young et al., 2003). Thus, SUMOylation of Snf1 in response to MMS-induced DNA damage reduces respiration and increases fermentation. We have consistently observed that SUMOylation of Snf1 does not affect its phosphorylation at T210, thus SUMOylation of Snf1 during the DNA damage response downregulates Snf1 irrespective of the available carbon source.
Connections between DNA repair and carbohydrate metabolism are increasingly apparent. MMS treatment of S. cerevisiae decreases transcription of genes involved in respiration and increases transcription of genes involved in fermentation (Caba et al., 2005, Fry et al., 2003, Gasch et al., 2001, Jelinsky and Samson, 1999, Lee et al., 2000, Shalem et al., 2008) (though the effect upon respiration genes in these studies was limited since the experiments were conducted with glucose-grown cells). The requirement of either MEC1 or TEL1 for glucose sensing by the SRR pathway suggests that increased Mec1/Tel1 activity may protect yeast cells from oxidative damage by driving the switch from respiration to fermentation. Thus, Mec1/Tel1 (and possibly ATM/ATR) play important roles in this metabolic switch, in addition to responding the DNA damage.
In mammalian cells p53 promotes respiration in a feed-forward mechanism with AMPK (the ortholog of SNF1), and the absence of p53 in tumor cells is a major contributor to the Warburg effect (the aerobic fermentation exhibited by tumor cells) (Lu et al., 2014, Matoba et al., 2006, Sinthupibulyakit et al., 2010, Wang et al., 2014, Contractor and Harris, 2012, Kim et al., 2013). Our results make us wonder if activation of DNA repair pathways, a hallmark of tumor cells, contributes to the Warburg effect. Our results suggest that the reduction of respiration in response to DNA damage is purposefully controlled, and offer a mechanism by which DNA damage can inhibit respiration by reducing SNF1 activity.
Experimental Procedures
Yeast strains, plasmids, mutagenesis and media
Strains used are listed in Table S1; plasmids used are listed in Table S2. Most experiments were conducted with strains related to W303a (Ma et al., 1987). Snf1 was immunoprecipitated from BY4741/2 (Brachmann et al., 1998) for determining its SUMOylation. Yeasts were transformed with DNA using the frozen lithium acetate method (Knop et al., 1999). To make strains with the mms21-CH mutations (C200A H202A) that abolish its SUMO E3 ligase function, plasmid pMMS21.47 (Santa Maria et al., 2007), which contains the 3’ part of MMS21 with the C200A H202A mutations, was linearized with PvuII to target its integration into the MMS21 locus. Ura+ colonies were isolated and confirmed to carry the mutations by DNA sequencing of a PCR product. Mutations of MMS21 in the vicinity of S261 were made using constructs produced from pYM3 [37] which introduced a 6HA-tag, K.lactis TRP1 marker and mutations in the tail of Mms21 (the primer was codon optimised for codons corresponding to amino-acids 263-267. Primers for tagging MMS21 are listed in Table S3). Mutagenesis was performed using Quickchange (Agilent); all mutations were confirmed by DNA sequencing. To construct the Mms21-3HA plasmid (used in Figure 4d), the 3HA-tag and the G418 resistance marker was cloned out of strain T74-17D (Zhao and Blobel, 2005) together with 244 bases upstream and 213 bases downstream of the STOP codon. This was inserted into plasmid Mms21-46 (Santa Maria et al., 2007) cut with BtrI & ClaI. The resultant plasmid contains Mms21-3HA expressed from its own promoter, and carries selection markers for both URA3 and KAN.
With the exception of snf1Δ cells, which must be grown on 2% glucose, yeasts were grown for HXT3 expression and Mth1 degradation assays in media containing 2% galactose with the appropriate amino acid and nucleotide base supplements. For ADH2 expression assays, cells were grown in 4% glucose, washed 3x with water, and resuspended in media containing 3% glycerol. MMS (Sigma) was added directly to media to the indicated concentration. For serial dilutions, cells were grown overnight in galactose containing media (unless the cells required glucose) to saturation and diluted 10 fold per dilution. The first spotting is a 10-fold dilution from a saturated culture.
β-galactosidase assays
The β-galactosidase assay kit (Pierce, cat. no. 75768) was used in a 96-well plate format. Cell concentration was read at 600nm. Reactions typically proceeded for 5 minutes at room temperature. Additional β-galactosidase assays used a homemade kit, with β-galactosidase activity read at 415nm and cell concentration at 595nm. Rates of gene expression were determined by calculating the increase in LacZ activity per minute (or hour) over the course of the experiment (90 minutes for HXT3 and 4 hours for ADH2 expression) and normalized to that of wild-type for each experiment. All error bars are +− 1 standard deviation.
Immunopreciptations
Cells (40-50ml) were centrifuged for 2 minutes at 4400g in an Eppendorf 5702, resuspended in ice-cold B60 (Hanna et al., 2001), with 480mM KAc and 0.1% NP40 (for Snf1 immunoprecipitation) or 240mM KAc 0.1% NP40 (for Mms21 immunoprecipitation). Lysis buffer also contained 1mM N-ethyl maleimide (NEM), HALT protease and phosphatase inhibitors (Pierce #1861280), and pepstatin A (Sigma #P4265). Cells were vortexed at 4°C for 30 minutes with 1 minute rests every other minute and spun at 13000 rpm for 30 minutes at 4°C. The supernatant was transferred to a new tube and spun for an additional 5 minutes. Protein concentration was determined by BCA (Pierce #23228/1859078). With the exception of Figure 1a, extracts were incubated with Preconjugated EZ-View anti-Myc beads (Sigma #E6654) or anti-HA beads (Sigma #E6779) for 2 hours and washed before eluting by boiling into non-fluorescent sample buffer (Pierce #39001). For Figure 1a, extracts were precleared with a 50/50 mix of protein-A/protein-G beads, and incubated with 2μl anti-HA antibody overnight. A 50/50 mix of protein-A/protein-G beads was added for 3 hours. After the penultimate wash, samples were split and one-half treated with 400 units λ-phosphatase (NEB P0753S) for 40 minutes at 30°C. The beads were washed once more with B240, and proteins extracted by boiling in sample buffer.
Protein kinase assays
Snf1-8myc was immunoprecipitated in B60 with 0.1% Triton X-100 with 1mM NEM, and protease and phosphatase inhibitors as described above. The ADP-Glo protein kinase assay (Promega) kit was used with 1 mM SAMS peptide (Signalchem) as the phosphate acceptor substrate. All error bars are +−1SD.
Immunoblots
To prevent activation of SNF1 by processing of the cells (Wilson et al., 1996), cells were killed before centrifugation by addition of 1ml 100% TCA to 5ml of cells. Cells were vortexed with glass beads, pelleted, and resuspended in non-fluorescent sample buffer (Licor 928-40004). Protein concentration was determined by the Coomassie elution with SDS method (Marbach et al., 2001), except that destaining was done with water. Protein extracts were run on 10% (12% or 15% for Mms21-3HA) TGS gels (Biorad) and transferred to PVDF membranes. Phostag (Waco) was added at 0.15ml/10 mix when required. Membranes were probed with mouse anti-Myc (9E10, Santa Cruz), rabbit anti-S-tag (Abcam), rabbit anti-phospho-RxxS (Cell Signaling), mouse anti-HA (Roche or Santa Cruz), mouse anti-FLAG (M2, Sigma), with all antibodies diluted 1000-fold in blocking buffer (Rockland MB-070). Snf1 phosphorylated on T210 (P-Snf1) was detected using rabbit anti-phospho T172 AMPKα (Cell Signaling). Loading controls were detected with rabbit anti-actin (Epitomics, 500-fold diluted) or mouse anti-Pgk1 (Invitrogen, 10000-fold diluted) antibodies. Secondary antibodies were anti-mouse 680LT and anti-rabbit 800CW (LiCOR) or anti-mouse Dylight 488 and anti-rabbit Dylight 549 (Epitomics). Blots were visualized with a LiCOR odyssey or a Biorad imager. Quantifications were performed using NIH ImageJ.
Supplementary Material
Highlights.
DNA damage and glucose both cause SUMOylation and inactivation of Snf1 by Mms21.
Mec1 (ATR), Tel1 (ATM) and PKA phosphorylate Mms21at Serine 261.
Inactivation of Snf1 by SUMOylation is independent from Snf1 dephosphorylation.
This may contribute to the Warburg effect found in cancer cells
Acknowledgements
This work was supported by National Institutes of Health Grant GM32540 (to M.J.), by the Israel Science Foundation and US-Israel bi-national fund (to M.K.) and by the Israel Cancer Research Fund /MOST Gesher Award (to K.S.-L.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- BARRETT L, ORLOVA M, MAZIARZ M, KUCHIN S. Protein kinase A contributes to the negative control of Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot Cell. 2012;11:119–28. doi: 10.1128/EC.05061-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOJUNGA N, ENTIAN KD. Cat8p, the activator of gluconeogenic genes in Saccharomyces cerevisiae, regulates carbon source-dependent expression of NADP-dependent cytosolic isocitrate dehydrogenase (Idp2p) and lactate permease (Jen1p). Mol Gen Genet. 1999;262:869–75. doi: 10.1007/s004380051152. [DOI] [PubMed] [Google Scholar]
- BRACHMANN CB, DAVIES A, COST GJ, CAPUTO E, LI J, HIETER P, BOEKE JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–32. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- BUSTI S, COCCETTI P, ALBERGHINA L, VANONI M. Glucose signaling-mediated coordination of cell growth and cell cycle in Saccharomyces cerevisiae. Sensors (Basel) 2010;10:6195–240. doi: 10.3390/s100606195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CABA E, DICKINSON DA, WARNES GR, AUBRECHT J. Differentiating mechanisms of toxicity using global gene expression analysis in Saccharomyces cerevisiae. Mutat Res. 2005;575:34–46. doi: 10.1016/j.mrfmmm.2005.02.005. [DOI] [PubMed] [Google Scholar]
- CARLBORG KK, KANNO T, CARTER SD, SJOGREN C. Mec1-dependent phosphorylation of Mms21 modulates its SUMO ligase activity. DNA Repair (Amst) 2015;28:83–92. doi: 10.1016/j.dnarep.2015.01.006. [DOI] [PubMed] [Google Scholar]
- CASAMAYOR A, SERRANO R, PLATARA M, CASADO C, RUIZ A, ARINO J. The role of the Snf1 kinase in the adaptive response of Saccharomyces cerevisiae to alkaline pH stress. Biochem J. 2012;444:39–49. doi: 10.1042/BJ20112099. [DOI] [PubMed] [Google Scholar]
- CHANDRASHEKARAPPA DG, MCCARTNEY RR, SCHMIDT MC. Subunit and domain requirements for adenylate-mediated protection of Snf1 kinase activation loop from dephosphorylation. J Biol Chem. 2011;286:44532–41. doi: 10.1074/jbc.M111.315895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEUNG HC, SAN LUCAS FA, HICKS S, CHANG K, BERTUCH AA, RIBES-ZAMORA A. An S/T-Q cluster domain census unveils new putative targets under Tel1/Mec1 control. BMC Genomics. 2012;13:664. doi: 10.1186/1471-2164-13-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONTRACTOR T, HARRIS CR. p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer Res. 2012;72:560–7. doi: 10.1158/0008-5472.CAN-11-1215. [DOI] [PubMed] [Google Scholar]
- COSENTINO C, GRIECO D, COSTANZO V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J. 2011;30:546–55. doi: 10.1038/emboj.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUAN X, SARANGI P, LIU X, RANGI GK, ZHAO X, YE H. Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol Cell. 2009;35:657–68. doi: 10.1016/j.molcel.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELBING K, MCCARTNEY RR, SCHMIDT MC. Purification and characterization of the three Snf1-activating kinases of Saccharomyces cerevisiae. Biochem J. 2006;393:797–805. doi: 10.1042/BJ20051213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EVENSEN G, SEEBERG E. Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature. 1982;296:773–5. doi: 10.1038/296773a0. [DOI] [PubMed] [Google Scholar]
- FLOTT S, ALABERT C, TOH GW, TOTH R, SUGAWARA N, CAMPBELL DG, HABER JE, PASERO P, ROUSE J. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol Cell Biol. 2007;27:6433–45. doi: 10.1128/MCB.00135-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRIEDEL AM, PIKE BL, GASSER SM. ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol. 2009;21:237–44. doi: 10.1016/j.ceb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- FRY RC, SAMBANDAN TG, RHA C. DNA damage and stress transcripts in Saccharomyces cerevisiae mutant sgs1. Mech Ageing Dev. 2003;124:839–46. doi: 10.1016/s0047-6374(03)00144-1. [DOI] [PubMed] [Google Scholar]
- GADURA N, ROBINSON LC, MICHELS CA. Glc7-Reg1 phosphatase signals to Yck1,2 casein kinase 1 to regulate transport activity and glucose-induced inactivation of Saccharomyces maltose permease. Genetics. 2006;172:1427–39. doi: 10.1534/genetics.105.051698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAMO FJ, LAFUENTE MJ, GANCEDO C. The mutation DGT1-1 decreases glucose transport and alleviates carbon catabolite repression in Saccharomyces cerevisiae. J Bacteriol. 1994;176:7423–9. doi: 10.1128/jb.176.24.7423-7429.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GASCH AP, HUANG M, METZNER S, BOTSTEIN D, ELLEDGE SJ, BROWN PO. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GHILLEBERT R, SWINNEN E, WEN J, VANDESTEENE L, RAMON M, NORGA K, ROLLAND F, WINDERICKX J. The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: structure, function and regulation. FEBS J. 2011;278:3978–90. doi: 10.1111/j.1742-4658.2011.08315.x. [DOI] [PubMed] [Google Scholar]
- HANNA JS, KROLL ES, LUNDBLAD V, SPENCER FA. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol. 2001;21:3144–58. doi: 10.1128/MCB.21.9.3144-3158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARDIE DG, CARLING D, CARLSON M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem. 1998;67:821–55. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- HARDIE DG, ROSS FA, HAWLEY SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–62. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEDBACKER K, HONG SP, CARLSON M. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol Cell Biol. 2004;24:8255–63. doi: 10.1128/MCB.24.18.8255-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HONG SP, CARLSON M. Regulation of snf1 protein kinase in response to environmental stress. J Biol Chem. 2007;282:16838–45. doi: 10.1074/jbc.M700146200. [DOI] [PubMed] [Google Scholar]
- HONG SP, LEIPER FC, WOODS A, CARLING D, CARLSON M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci U S A. 2003;100:8839–43. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JELINSKY SA, SAMSON LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci U S A. 1999;96:1486–91. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG R, CARLSON M. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol Cell Biol. 1997;17:2099–106. doi: 10.1128/mcb.17.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSTON M, KIM JH. Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem Soc Trans. 2005;33:247–52. doi: 10.1042/BST0330247. [DOI] [PubMed] [Google Scholar]
- KATO R, OGAWA H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:3104–12. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM HR, ROE JS, LEE JE, CHO EJ, YOUN HD. p53 regulates glucose metabolism by miR-34a. Biochem Biophys Res Commun. 2013;437:225–31. doi: 10.1016/j.bbrc.2013.06.043. [DOI] [PubMed] [Google Scholar]
- KIM JH, JOHNSTON M. Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J Biol Chem. 2006;281:26144–9. doi: 10.1074/jbc.M603636200. [DOI] [PubMed] [Google Scholar]
- KITANOVIC A, WALTHER T, LORET MO, HOLZWARTH J, KITANOVIC I, BONOWSKI F, VAN BUI N, FRANCOIS JM, WOLFL S. Metabolic response to MMS-mediated DNA damage in Saccharomyces cerevisiae is dependent on the glucose concentration in the medium. FEMS Yeast Res. 2009;9:535–51. doi: 10.1111/j.1567-1364.2009.00505.x. [DOI] [PubMed] [Google Scholar]
- KNOP M, SIEGERS K, PEREIRA G, ZACHARIAE W, WINSOR B, NASMYTH K, SCHIEBEL E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–72. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- LEE SE, PELLICIOLI A, DEMETER J, VAZE MP, GASCH AP, MALKOVA A, BROWN PO, BOTSTEIN D, STEARNS T, FOIANI M, HABER JE. Arrest, adaptation, and recovery following a chromosome double-strand break in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 2000;65:303–14. doi: 10.1101/sqb.2000.65.303. [DOI] [PubMed] [Google Scholar]
- LIU Y, XU X, CARLSON M. Interaction of SNF1 protein kinase with its activating kinase Sak1. Eukaryot Cell. 2011;10:313–9. doi: 10.1128/EC.00291-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU J, TAN M, CAI Q. The Warburg effect in tumor progression: Mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2014 doi: 10.1016/j.canlet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUDIN K, JIANG R, CARLSON M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1998;95:6245–50. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MA H, HAN BK, GUADERRAMA M, ASLANIAN A, YATES JR, 3RD, HUNTER T, WITTENBERG C. Psy2 targets the PP4 family phosphatase Pph3 to dephosphorylate Mth1 and repress glucose transporter gene expression. Mol Cell Biol. 2014;34:452–63. doi: 10.1128/MCB.00279-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MA H, KUNES S, SCHATZ PJ, BOTSTEIN D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–16. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- MALLORY JC, BASHKIROV VI, TRUJILLO KM, SOLINGER JA, DOMINSKA M, SUNG P, HEYER WD, PETES TD. Amino acid changes in Xrs2p, Dun1p, and Rfa2p that remove the preferred targets of the ATM family of protein kinases do not affect DNA repair or telomere length in Saccharomyces cerevisiae. DNA Repair (Amst) 2003;2:1041–64. doi: 10.1016/s1568-7864(03)00115-0. [DOI] [PubMed] [Google Scholar]
- MALLORY JC, PETES TD. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc Natl Acad Sci U S A. 2000;97:13749–54. doi: 10.1073/pnas.250475697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARBACH I, LICHT R, FROHNMEYER H, ENGELBERG D. Gcn2 Mediates Gcn4 Activation in Response to Glucose Stimulation or UV Radiation Not via GCN4 Translation. J Biol Chem. 2001;276:16944–51. doi: 10.1074/jbc.M100383200. [DOI] [PubMed] [Google Scholar]
- MATOBA S, KANG JG, PATINO WD, WRAGG A, BOEHM M, GAVRILOVA O, HURLEY PJ, BUNZ F, HWANG PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–3. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- MAYER FV, HEATH R, UNDERWOOD E, SANDERS MJ, CARMENA D, MCCARTNEY RR, LEIPER FC, XIAO B, JING C, WALKER PA, HAIRE LF, OGRODOWICZ R, MARTIN SR, SCHMIDT MC, GAMBLIN SJ, CARLING D. ADP regulates SNF1, the Saccharomyces cerevisiae homolog of AMP-activated protein kinase. Cell Metab. 2011;14:707–14. doi: 10.1016/j.cmet.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCGRATH JP, CAPON DJ, GOEDDEL DV, LEVINSON AD. Comparative biochemical properties of normal and activated human ras p21 protein. Nature. 1984;310:644–9. doi: 10.1038/310644a0. [DOI] [PubMed] [Google Scholar]
- MORRISON AJ, KIM JA, PERSON MD, HIGHLAND J, XIAO J, WEHR TS, HENSLEY S, BAO Y, SHEN J, COLLINS SR, WEISSMAN JS, DELROW J, KROGAN NJ, HABER JE, SHEN X. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell. 2007;130:499–511. doi: 10.1016/j.cell.2007.06.010. [DOI] [PubMed] [Google Scholar]
- ORLOVA M, KANTER E, KRAKOVICH D, KUCHIN S. Nitrogen availability and TOR regulate the Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:1831–7. doi: 10.1128/EC.00110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PACIOTTI V, CLERICI M, SCOTTI M, LUCCHINI G, LONGHESE MP. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol Cell Biol. 2001;21:3913–25. doi: 10.1128/MCB.21.12.3913-3925.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PASULA S, JOUANDOT D, 2ND, KIM JH. Biochemical evidence for glucose-independent induction of HXT expression in Saccharomyces cerevisiae. FEBS Lett. 2007;581:3230–4. doi: 10.1016/j.febslet.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PELLICIOLI A, FOIANI M. Signal transduction: how rad53 kinase is activated. Curr Biol. 2005;15:R769–71. doi: 10.1016/j.cub.2005.08.057. [DOI] [PubMed] [Google Scholar]
- ROY A, JOUANDOT D, 2ND, CHO KH, KIM JH. Understanding the mechanism of glucose-induced relief of Rgt1-mediated repression in yeast. FEBS Open Bio. 2014;4:105–11. doi: 10.1016/j.fob.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUIZ A, XU X, CARLSON M. Roles of two protein phosphatases, Reg1-Glc7 and Sit4, and glycogen synthesis in regulation of SNF1 protein kinase. Proc Natl Acad Sci U S A. 2011;108:6349–54. doi: 10.1073/pnas.1102758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUIZ A, XU X, CARLSON M. Ptc1 protein phosphatase 2C contributes to glucose regulation of SNF1/AMP-activated protein kinase (AMPK) in Saccharomyces cerevisiae. J Biol Chem. 2013;288:31052–8. doi: 10.1074/jbc.M113.503763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANTA MARIA SR, GANGAVARAPU V, JOHNSON RE, PRAKASH L, PRAKASH S. Requirement of Nse1, a subunit of the Smc5-Smc6 complex, for Rad52-dependent postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:8409–18. doi: 10.1128/MCB.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SASS P, FIELD J, NIKAWA J, TODA T, WIGLER M. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986;83:9303–7. doi: 10.1073/pnas.83.24.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEARLE JS, WOOD MD, KAUR M, TOBIN DV, SANCHEZ Y. Proteins in the nutrient-sensing and DNA damage checkpoint pathways cooperate to restrain mitotic progression following DNA damage. PLoS Genet. 2011;7:e1002176. doi: 10.1371/journal.pgen.1002176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHALEM O, DAHAN O, LEVO M, MARTINEZ MR, FURMAN I, SEGAL E, PILPEL Y. Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol Syst Biol. 2008;4:223. doi: 10.1038/msb.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIRRA MK, MCCARTNEY RR, ZHANG C, SHOKAT KM, SCHMIDT MC, ARNDT KM. A chemical genomics study identifies Snf1 as a repressor of GCN4 translation. J Biol Chem. 2008;283:35889–98. doi: 10.1074/jbc.M805325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMPSON-LAVY KJ, JOHNSTON M. SUMOylation regulates the SNF1 protein kinase. Proc Natl Acad Sci U S A. 2013;110:17432–7. doi: 10.1073/pnas.1304839110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINTHUPIBULYAKIT C, ITTARAT W, ST CLAIR WH, ST CLAIR DK. p53 Protects lung cancer cells against metabolic stress. Int J Oncol. 2010;37:1575–81. doi: 10.3892/ijo_00000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TACHIBANA C, YOO JY, TAGNE JB, KACHEROVSKY N, LEE TI, YOUNG ET. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol Cell Biol. 2005;25:2138–46. doi: 10.1128/MCB.25.6.2138-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKAHASHI Y, KIKUCHI Y. Yeast PIAS-type Ull1/Siz1 is composed of SUMO ligase and regulatory domains. J Biol Chem. 2005;280:35822–8. doi: 10.1074/jbc.M506794200. [DOI] [PubMed] [Google Scholar]
- TATCHELL K. RAS genes and growth control in Saccharomyces cerevisiae. J Bacteriol. 1986;166:364–7. doi: 10.1128/jb.166.2.364-367.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THOMPSON-JAEGER S, FRANCOIS J, GAUGHRAN JP, TATCHELL K. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TU J, CARLSON M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995;14:5939–46. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNO I, MITSUZAWA H, MATSUMOTO K, TANAKA K, OSHIMA T, ISHIKAWA T. Reconstitution of the GTP-dependent adenylate cyclase from products of the yeast CYR1 and RAS2 genes in Escherichia coli. Proc Natl Acad Sci U S A. 1985;82:7855–9. doi: 10.1073/pnas.82.23.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNO I, MITSUZAWA H, TANAKA K, OSHIMA T, ISHIKAWA T. Identification of the domain of Saccharomyces cerevisiae adenylate cyclase associated with the regulatory function of RAS products. Mol Gen Genet. 1987;210:187–94. doi: 10.1007/BF00325683. [DOI] [PubMed] [Google Scholar]
- VALLIER LG, CARLSON M. Synergistic release from glucose repression by mig1 and ssn mutations in Saccharomyces cerevisiae. Genetics. 1994;137:49–54. doi: 10.1093/genetics/137.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WALTHER K, SCHULLER HJ. Adr1 and Cat8 synergistically activate the glucose-regulated alcohol dehydrogenase gene ADH2 of the yeast Saccharomyces cerevisiae. Microbiology. 2001;147:2037–44. doi: 10.1099/00221287-147-8-2037. [DOI] [PubMed] [Google Scholar]
- WANG L, XIONG H, WU F, ZHANG Y, WANG J, ZHAO L, GUO X, CHANG LJ, YOU MJ, KOOCHEKPOUR S, SALEEM M, HUANG H, LU J, DENG Y. Hexokinase 2-mediated Warburg effect is required for PTEN- and p53-deficiency-driven prostate cancer growth. Cell Rep. 2014;8:1461–74. doi: 10.1016/j.celrep.2014.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILSON MA, KOUTELOU E, HIRSCH C, AKDEMIR K, SCHIBLER A, BARTON MC, DENT SY. Ubp8 and SAGA regulate Snf1 AMP kinase activity. Mol Cell Biol. 2011;31:3126–35. doi: 10.1128/MCB.01350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILSON WA, HAWLEY SA, HARDIE DG. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol. 1996;6:1426–34. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- WYKOFF DD, O'SHEA EK. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol Cell Proteomics. 2005;4:73–83. doi: 10.1074/mcp.M400166-MCP200. [DOI] [PubMed] [Google Scholar]
- YOUNG ET, DOMBEK KM, TACHIBANA C, IDEKER T. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J Biol Chem. 2003;278:26146–58. doi: 10.1074/jbc.M301981200. [DOI] [PubMed] [Google Scholar]
- ZHAO X, BLOBEL G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci U S A. 2005;102:4777–82. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.