Abstract
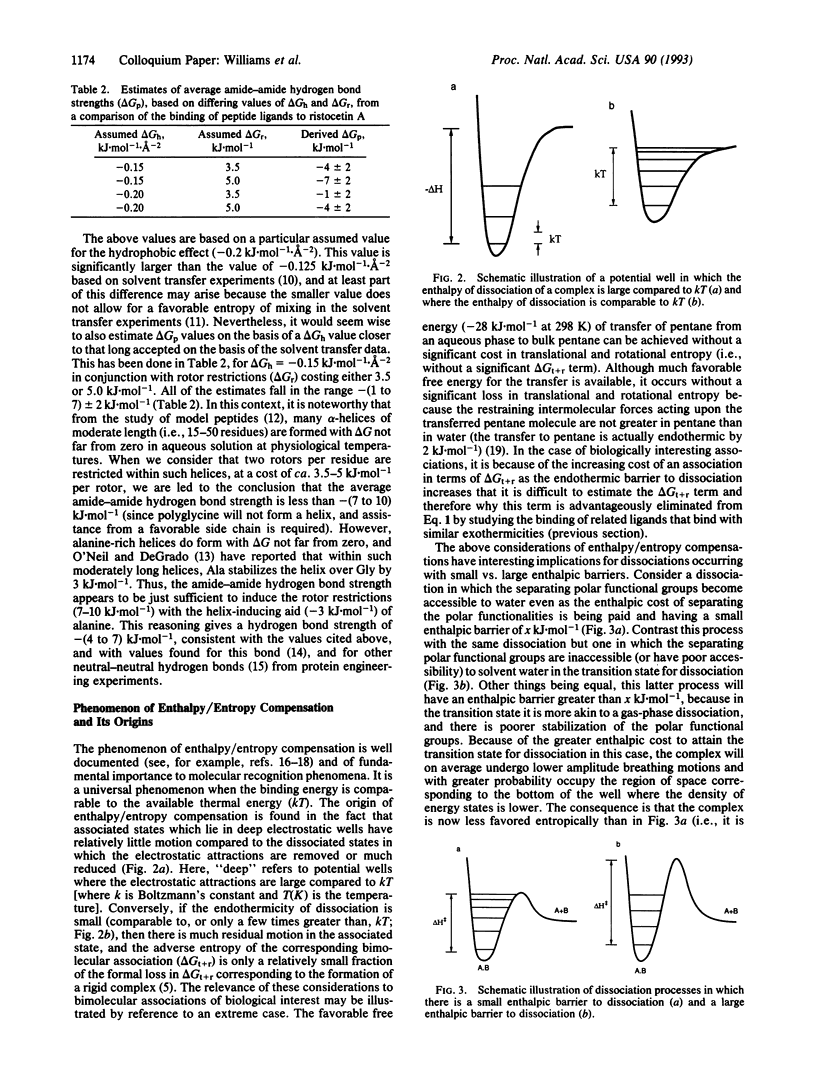
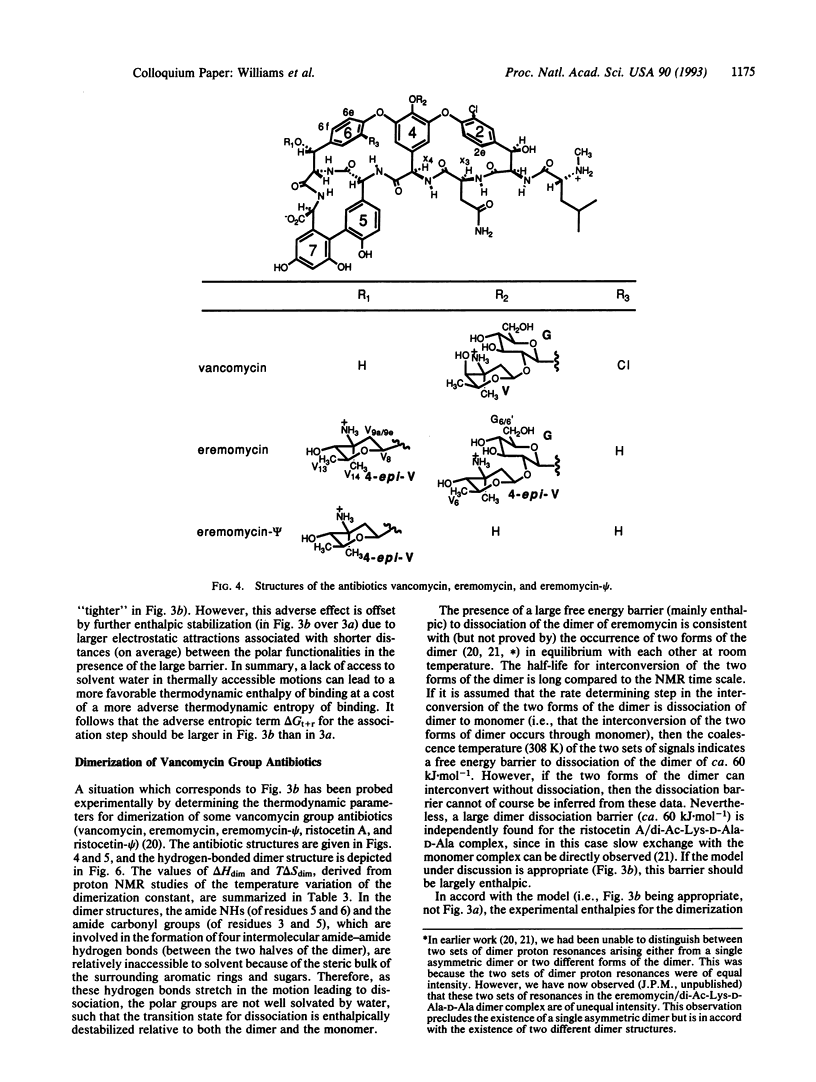
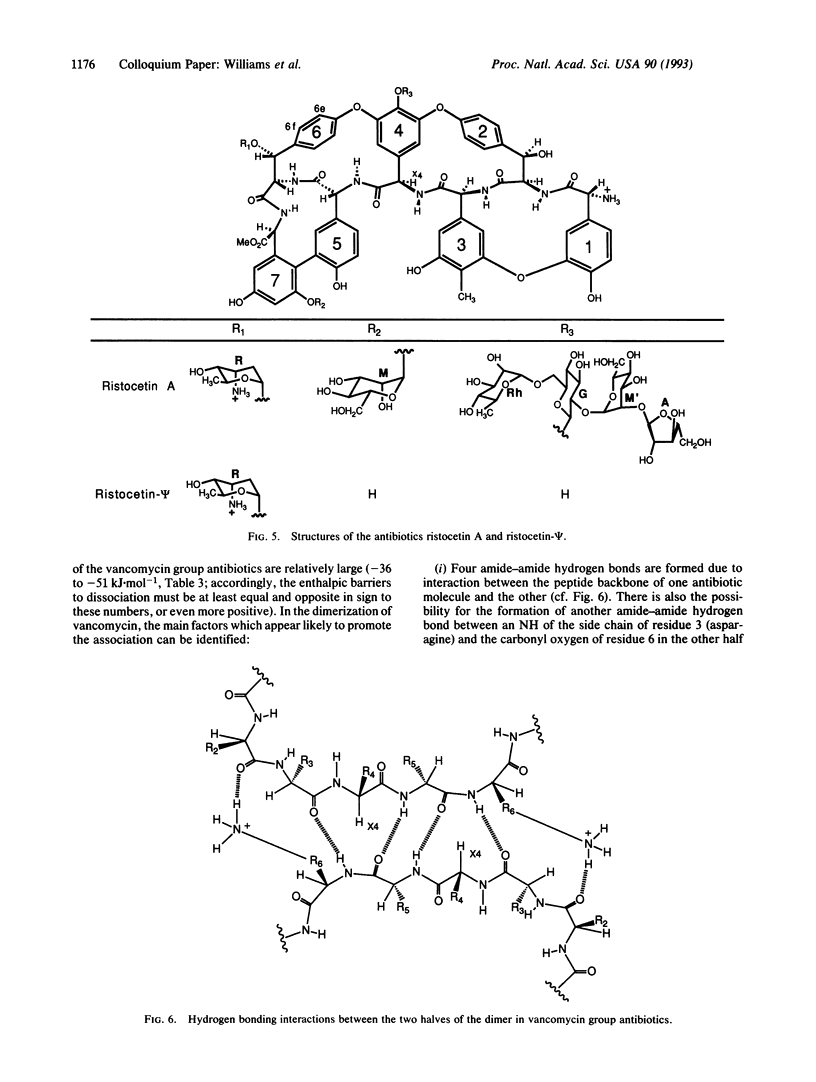
An approach toward the estimation of binding constants for organic molecules in aqueous solution is presented, based upon a partitioning of the free energy of binding. Consideration is given to polar and hydrophobic contributions and to the entropic cost of rotor restrictions and bimolecular associations. Several parameters (derived from an analysis of entropy changes upon the melting of crystals and from the binding of cell wall peptide analogues to the antibiotic ristocetin A) which may be useful guides to a crude understanding of binding phenomena are presented: (i) amide-amide hydrogen bond strengths of -(1 to 7) +/- 2 kJ.mol-1, (ii) a hydrophobic effect of -0.2 +/- 0.05 kJ.mol-1.A-2 of hydrocarbon removed from exposure to water in the binding process, and (iii) free energy costs for rotor restrictions of 3.5-5.0 kJ.mol-1. The validity of the parameters for hydrogen bond strengths is dependent on the validity of the other two parameters. The phenomenon of entropy/enthalpy compensation is considered, with the conclusion that enthalpic barriers to dissociations will result in larger losses in translational and rotational entropy in the association step. The dimerization of some vancomycin group antibiotics is strongly exothermic (-36 to -51 kJ.mol-1) and is promoted by a factor of 50-100 by a disaccharide attached to ring 4 (in vancomycin and eremomycin) and by a factor of ca. 1000 by an amino-sugar attached to the benzylic position of ring 6 in eremomycin. The dimerization process (which, as required for an exothermic association, appears to be costly in entropy) may be relevant to the mode of action of the antibiotics.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin R. L. Temperature dependence of the hydrophobic interaction in protein folding. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8069–8072. doi: 10.1073/pnas.83.21.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C., Janin J. Principles of protein-protein recognition. Nature. 1975 Aug 28;256(5520):705–708. doi: 10.1038/256705a0. [DOI] [PubMed] [Google Scholar]
- Doig A. J., Williams D. H. Is the hydrophobic effect stabilizing or destabilizing in proteins? The contribution of disulphide bonds to protein stability. J Mol Biol. 1991 Jan 20;217(2):389–398. doi: 10.1016/0022-2836(91)90551-g. [DOI] [PubMed] [Google Scholar]
- Good V. M., Gwynn M. N., Knowles D. J. MM 45289, a potent glycopeptide antibiotic which interacts weakly with diacetyl-L-lysyl-D-alanyl-D-alanine. J Antibiot (Tokyo) 1990 May;43(5):550–555. doi: 10.7164/antibiotics.43.550. [DOI] [PubMed] [Google Scholar]
- Jencks W. P. Binding energy, specificity, and enzymic catalysis: the circe effect. Adv Enzymol Relat Areas Mol Biol. 1975;43:219–410. doi: 10.1002/9780470122884.ch4. [DOI] [PubMed] [Google Scholar]
- Jencks W. P. On the attribution and additivity of binding energies. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4046–4050. doi: 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumry R., Rajender S. Enthalpy-entropy compensation phenomena in water solutions of proteins and small molecules: a ubiquitous property of water. Biopolymers. 1970;9(10):1125–1227. doi: 10.1002/bip.1970.360091002. [DOI] [PubMed] [Google Scholar]
- Miklavc A., Kocjan D., Mavri J., Koller J., Hadzi D. On the fundamental difference in the thermodynamics of agonist and antagonist interactions with beta-adrenergic receptors and the mechanism of entropy-driven binding. Biochem Pharmacol. 1990 Aug 15;40(4):663–669. doi: 10.1016/0006-2952(90)90299-z. [DOI] [PubMed] [Google Scholar]
- O'Neil K. T., DeGrado W. F. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science. 1990 Nov 2;250(4981):646–651. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- Page M. I., Jencks W. P. Entropic contributions to rate accelerations in enzymic and intramolecular reactions and the chelate effect. Proc Natl Acad Sci U S A. 1971 Aug;68(8):1678–1683. doi: 10.1073/pnas.68.8.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Tebar A., Vázquez D., Pérez Velázquez J. L., Laynez J., Wadsö I. Thermochemistry of the interaction between peptides and vancomycin or ristocetin. J Antibiot (Tokyo) 1986 Nov;39(11):1578–1583. doi: 10.7164/antibiotics.39.1578. [DOI] [PubMed] [Google Scholar]
- Scholtz J. M., Marqusee S., Baldwin R. L., York E. J., Stewart J. M., Santoro M., Bolen D. W. Calorimetric determination of the enthalpy change for the alpha-helix to coil transition of an alanine peptide in water. Proc Natl Acad Sci U S A. 1991 Apr 1;88(7):2854–2858. doi: 10.1073/pnas.88.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L., Neira J. L., Sancho J., Fersht A. R. Effect of alanine versus glycine in alpha-helices on protein stability. Nature. 1992 Apr 2;356(6368):453–455. doi: 10.1038/356453a0. [DOI] [PubMed] [Google Scholar]
- Sharp K. A., Nicholls A., Fine R. F., Honig B. Reconciling the magnitude of the microscopic and macroscopic hydrophobic effects. Science. 1991 Apr 5;252(5002):106–109. doi: 10.1126/science.2011744. [DOI] [PubMed] [Google Scholar]
- Shirley B. A., Stanssens P., Hahn U., Pace C. N. Contribution of hydrogen bonding to the conformational stability of ribonuclease T1. Biochemistry. 1992 Jan 28;31(3):725–732. doi: 10.1021/bi00118a013. [DOI] [PubMed] [Google Scholar]


