Abstract
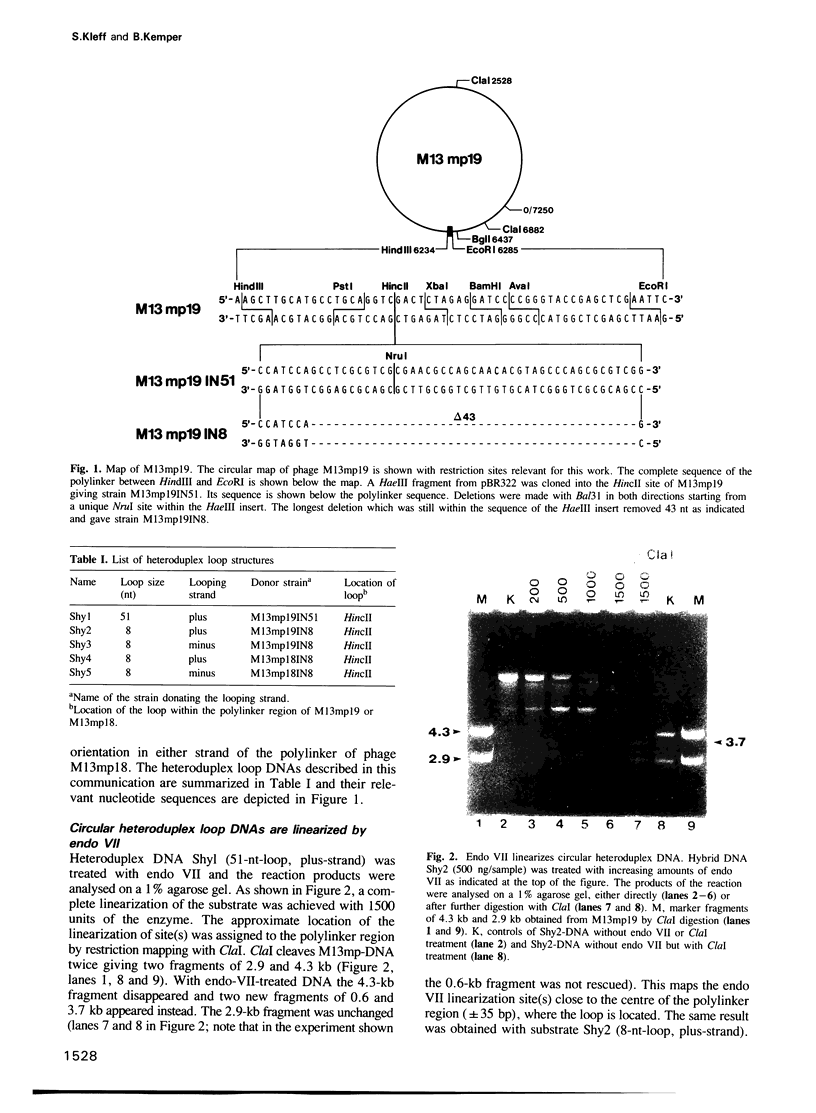
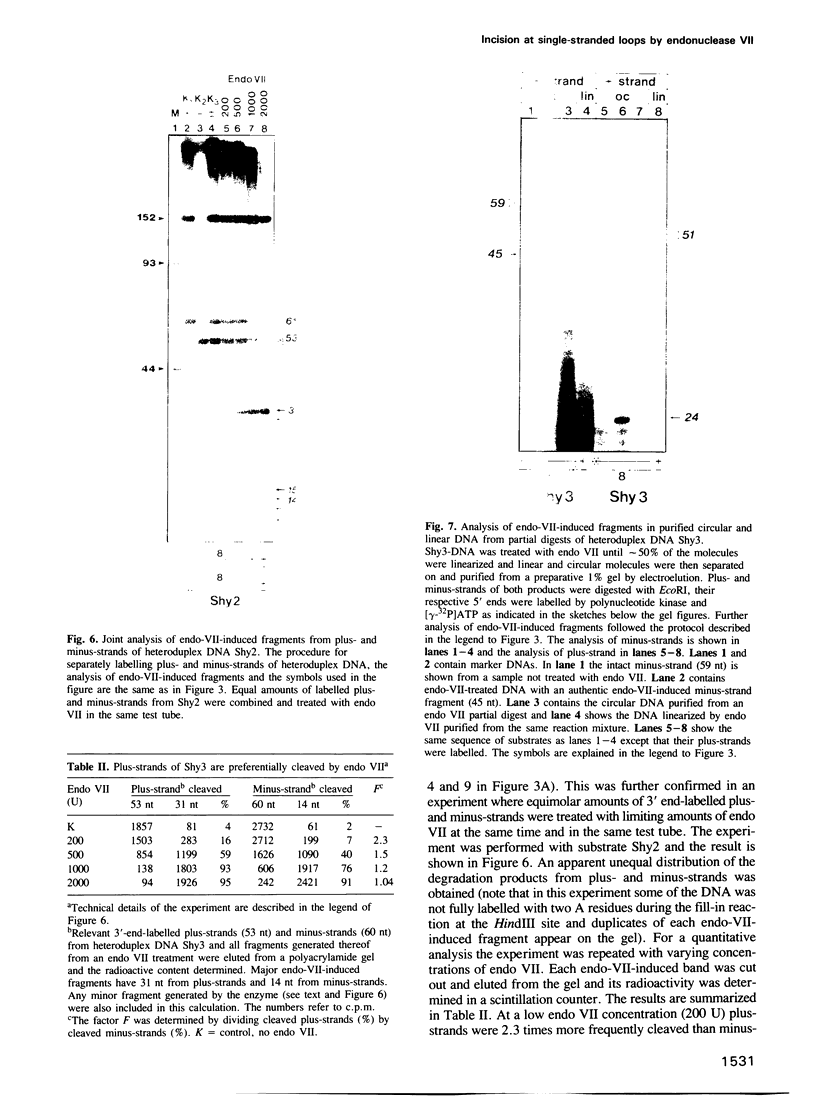
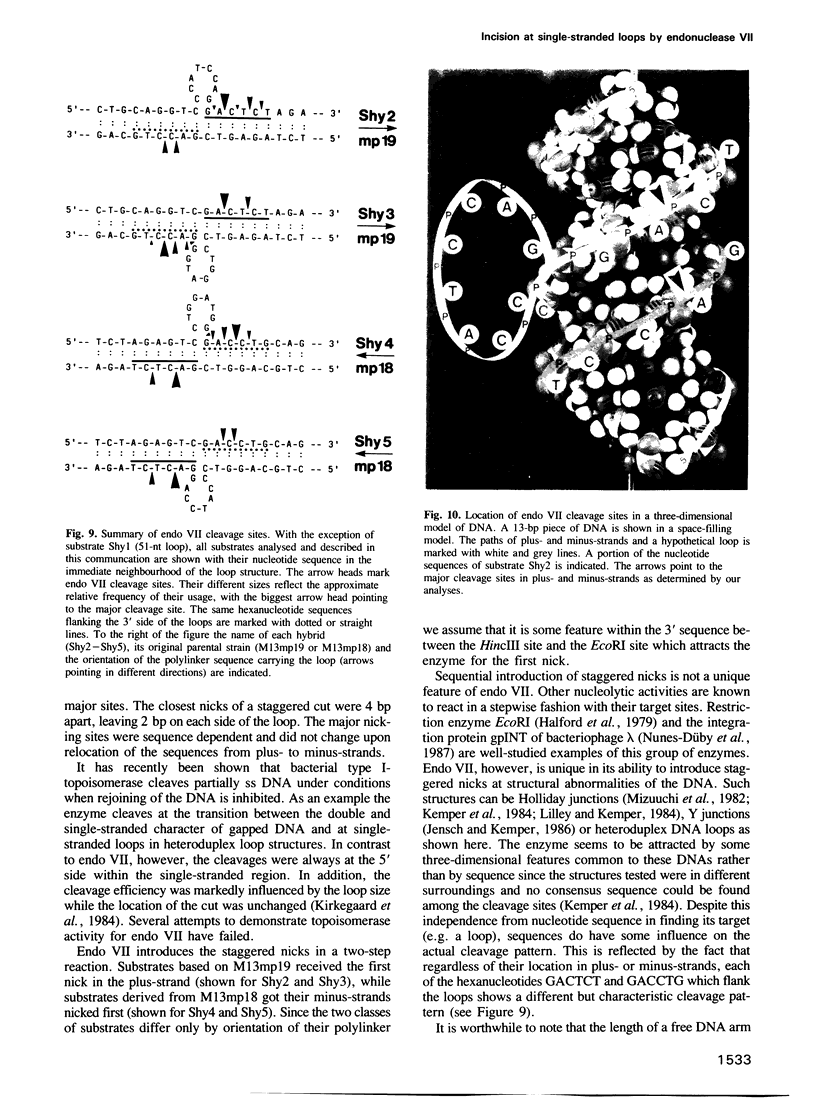
Heteroduplex DNAs with single-stranded loops of 51 nt or 8 nt were constructed in vitro and used in reactions with purified endonuclease VII (endo VII) from phage T4. The enzyme makes double-strand breaks by introducing pairs of staggered nicks flanking the loops. Regardless of loop-size the nicking sites map exclusively at the 3' side of the loop in the looping strand and at the 3' side of the base of the loop in the non-looping strand. The number of potential cleavage sites is small (less than 5) and their distribution depends on DNA sequence. The two closest staggered nicks are 4 bp apart, 2 bp on either side of the loop. Nicking always occurs in the double-stranded part of the molecules; the single-stranded loops are not attacked by endo VII. The nicks are introduced in a stepwise fashion and selection of the strand for the first nick depends on the sequence of 31 base pairs flanking the loops.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ayares D., Ganea D., Chekuri L., Campbell C. R., Kucherlapati R. Repair of single-stranded DNA nicks, gaps, and loops in mammalian cells. Mol Cell Biol. 1987 May;7(5):1656–1662. doi: 10.1128/mcb.7.5.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz W. C., Berger H. Selective allele loss in mixed infections with T4 bacteriophage. Genetics. 1973 Jan;73(1):1–11. doi: 10.1093/genetics/73.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. K., Kolodner R. D. Repair of heteroduplex plasmid DNA after transformation into Saccharomyces cerevisiae. Mol Cell Biol. 1986 Oct;6(10):3401–3409. doi: 10.1128/mcb.6.10.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys J. P., Lacks S. A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986 Jun;50(2):133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohet C., Dzidić S., Wagner R., Radman M. Large non-homology in heteroduplex DNA is processed differently than single base pair mismatches. Mol Gen Genet. 1987 Jan;206(1):181–184. doi: 10.1007/BF00326556. [DOI] [PubMed] [Google Scholar]
- Drake J. W. Heteroduplex heterozygotes in bacteriophage T4 involving mutations of various dimensions. Proc Natl Acad Sci U S A. 1966 Mar;55(3):506–512. doi: 10.1073/pnas.55.3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S. E., Johnson N. P., Grinsted J. The reactions of the EcoRi and other restriction endonucleases. Biochem J. 1979 May 1;179(2):353–365. doi: 10.1042/bj1790353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensch F., Kemper B. Endonuclease VII resolves Y-junctions in branched DNA in vitro. EMBO J. 1986 Jan;5(1):181–189. doi: 10.1002/j.1460-2075.1986.tb04194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper B., Garabett M., Courage U. Studies on T4-head maturation. 2. Substrate specificity of gene-49-controlled endonuclease. Eur J Biochem. 1981 Mar 16;115(1):133–141. [PubMed] [Google Scholar]
- Kemper B., Garabett M. Studies on T4-head maturation. 1. Purification and characterization of gene-49-controlled endonuclease. Eur J Biochem. 1981 Mar 16;115(1):123–131. [PubMed] [Google Scholar]
- Kemper B., Jensch F., von Depka-Prondzynski M., Fritz H. J., Borgmeyer U., Mizuuchi K. Resolution of Holliday structures by endonuclease VII as observed in interactions with cruciform DNA. Cold Spring Harb Symp Quant Biol. 1984;49:815–825. doi: 10.1101/sqb.1984.049.01.092. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Pflugfelder G., Wang J. C. The cleavage of DNA by type-I DNA topoisomerases. Cold Spring Harb Symp Quant Biol. 1984;49:411–419. doi: 10.1101/sqb.1984.049.01.047. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster-Nassal C., Kolodner R. Mismatch correction catalyzed by cell-free extracts of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7618–7622. doi: 10.1073/pnas.83.20.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Matsumoto L., Landy A. Site-specific recombination intermediates trapped with suicide substrates. Cell. 1987 Aug 28;50(5):779–788. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaschewski J., Rüger W. Nucleotide sequence and primary structures of gene products coded for by the T4 genome between map positions 48.266 kb and 39.166 kb. Nucleic Acids Res. 1987 Apr 24;15(8):3632–3633. doi: 10.1093/nar/15.8.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R., Jr, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss U., Wilson J. H. Repair of single-stranded loops in heteroduplex DNA transfected into mammalian cells. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1619–1623. doi: 10.1073/pnas.84.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonesaki T., Minagawa T. T4 phage gene uvsX product catalyzes homologous DNA pairing. EMBO J. 1985 Dec 1;4(12):3321–3327. doi: 10.1002/j.1460-2075.1985.tb04083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]