Abstract
Staphylococcus capitis is an opportunistic pathogen of the coagulase negative staphylococci (CoNS). Functional genomic studies of S. capitis have thus far been limited by a lack of available complete genome sequences. Here, we determined the closed S. capitis genome and methylome using Single Molecule Real Time (SMRT) sequencing. The strain, AYP1020, harbors a single circular chromosome of 2.44 Mb encoding 2304 predicted proteins, which is the smallest of all complete staphylococcal genomes sequenced to date. AYP1020 harbors two large mobile genetic elements; a plasmid designated pAYP1020 (59.6 Kb) and a prophage, ΦAYP1020 (48.5 Kb). Methylome analysis identified significant adenine methylation across the genome involving two distinct methylation motifs (1972 putative 6-methyladenine (m6A) residues identified). Putative adenine methyltransferases were also identified. Comparative analysis of AYP1020 and the closely related CoNS, S. epidermidis RP62a, revealed a host of virulence factors that likely contribute to S. capitis pathogenicity, most notably genes important for biofilm formation and a suite of phenol soluble modulins (PSMs); the expression/production of these factors were corroborated by functional assays. The complete S. capitis genome will aid future studies on the evolution and pathogenesis of the coagulase negative staphylococci.
Keywords: coagulase-negative staphylococci, CoNS, SMRT sequencing, genomics, methylation
Introduction
Staphylococcus capitis was first isolated from human skin in 1975 and classified as a species of the coagulase negative staphylococci (CoNS) (Kloos and Schleifer, 1975). S. capitis can be further divided into two subspecies; capitis and ureolyticus based on urease production and maltose fermentation for the latter (Bannerman and Kloos, 1991). Traditionally considered commensals, many CoNS species are now recognized as opportunistic human pathogens. In fact, multiple studies have found CoNS to be the most frequently isolated organisms from bloodstream infections in intensive care units (N.N.I.S, 1999; Wisplinghoff et al., 2004). S. capitis is particularly problematic in neonatal intensive care units causing up to 20% of cases of neonatal sepsis (Van Der Zwet et al., 2002; Rasigade et al., 2012). S. capitis is also occasionally associated with native and prosthetic valve endocarditis, as well as hospital-acquired meningitis (Bandres and Darouiche, 1992). Despite the role of S. capitis in these infections, very little is known about its pathogenicity. In addition, the treatment of infections caused by S. capitis is complicated by the emergence of strains with reduced susceptibility to last line antistaphylococcal agents including vancomycin and linezolid (Cai et al., 2012; Rasigade et al., 2012).
Staphylococcus epidermidis is the most frequently studied CoNS species. S. epidermidis relies heavily on its ability to form robust biofilms for infection and as such, it typically causes infections associated with indwelling medical devices (Von Eiff et al., 2002; Otto, 2009). S. epidermidis produces a range of surface proteins that are important for initial attachment and establishment of biofilm, as well as polysaccharide intercellular adhesin (PIA), which mediates cell to cell adhesion and biofilm accumulation (Mack et al., 1996; Heilmann et al., 1997, 2003). S. epidermidis also produces extracellular peptides termed phenol soluble modulins (PSMs), which are proinflammatory and also contribute to biofilm production (Wang et al., 2011; Peschel and Otto, 2013). Whilst biofilm production also appears to be a virulence determinant for S. capitis (de Silva et al., 2002), the contributing molecular determinants are less well defined for this species.
Whole genome sequencing (WGS) has become a vital tool in understanding the virulence of pathogenic bacteria. WGS has been used to predict the virulence of S. epidermidis RP62a (Gill et al., 2005) and S. epidermidis ATCC 12228 (Zhang et al., 2003) by comparing their genomes to that of the most pathogenic staphylococcal species, S. aureus. These studies gave powerful insights into genes that likely contribute to S. epidermidis virulence, particularly those involved in biofilm formation and immune defense (Zhang et al., 2003; Gill et al., 2005). Subsequently, similar comparative genomics projects have been undertaken for other CoNS species such as S. saprophyticus (Kuroda et al., 2005), S. haemolyticus (Takeuchi et al., 2005), and S. lugdunensis (Heilbronner et al., 2011).
To date, few genome sequences of S. capitis are publically available limiting our capacity to predict genes that may be important for S. capitis virulence. In the present study, we used Single Molecule Real Time (SMRT) sequencing technology to generate the complete S. capitis genome using the clinical bloodstream isolate, AYP1020. SMRT sequencing has emerged as a highly attractive sequencing platform due mainly to its ability to produce long read lengths facilitating efficient and accurate de novo complete genome assembly (Chin et al., 2013; Roberts et al., 2013). SMRT sequencing has the added advantage of allowing for the analysis of DNA methylation (Fang et al., 2012). In prokaryotes, DNA methylation has been shown to be important for critical processes including DNA replication and mismatch repair, as well as controlling gene expression and virulence (Heithoff et al., 1999; Marinus and Casadesus, 2009; Katayama et al., 2010). By comparing the S. capitis genome with that of S. epidermidis RP62a, we have identified factors that likely contribute to S. capitis virulence, including genes important for the establishment and maturation of biofilms and a number of putative PSMs. These genetic attributes were then functionally assessed, which showed that S. capitis AYP1020 produced biofilms on polymeric surfaces and secreted predicted PSMs in culture supernatants.
Methods
Bacterial strains and antibiotic susceptibility testing
S. capitis isolate AYP1020 was originally isolated from blood and confirmed to be S. capitis by Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) and confirmed to be subsp. capitis based on 16S rRNA sequence. After isolation, AYP1020 was cultured on Horse Blood Agar, grown in Heart Infusion Broth (Oxoid) then stored at −80°C in 25% (v/v) Glycerol. S. epidermidis RP62a is a biofilm-producing, methicillin-resistant S. epidermidis (MRSE) strain isolated from a patient with intravascular catheter-associated sepsis (Christensen et al., 1985). It has a fully sequenced genome (accession number NC_002976) (Gill et al., 2005) and was used in this study for comparative genomic analysis. Antibiotic susceptibility testing was performed using broth dilution for penicillin G, gentamicin, methicillin, erythromycin, and streptomycin according to Clinical Laboratory Standards Institute guidelines (C.L.S.I, 2013).
SMRT sequencing, assembly and annotation
S. capitis AYP1020 genomic DNA was prepared using the QIAGEN Blood and Tissue Kit as per manufacturer's instructions following an initial lysis step in TE buffer (pH 8.0) supplemented with 0.4 mg/ml lysostaphin (Sigma-Aldrich). DNA sequencing was performed using the PacBio RS platform (Pacific Biosciences). A 20-kb library was sequenced using P4-C2 chemistry on 2 SMRT cells yielding 565 Mbp from 92,793 reads. The average read length was 6088 bp with a sequencing depth of 164X. The continuous long reads (CLR) were assembled de novo using the PacBio hierarchical genome assembly process (HGAP) and polished using Quiver as described previously (Chin et al., 2013). DNA methylation was determined using the RS_Modification_and_Motif_Analysis protocol within the SMRT Portal v1.3.3. All motifs considered to be modified had a mean modification quality value (QV) > 30 and a mean coverage of >75X. Genome annotation was performed using Prokka (Seemann, 2014). The annotated genome was then manually curated and visualized using Geneious (Biomatters) and Artemis (Rutherford et al., 2000). Prophage were predicted using Phast (Zhou et al., 2011), insertion elements were predicted using ISfinder (Siguier et al., 2006) and putative genomic islands were predicted using Island Viewer (Langille and Brinkman, 2009). The complete genome sequence of S. capitis AYP1020 and its plasmid, pAYP1020 were submitted to GenBank with accession numbers CP007601 and CP007602, respectively.
Comparative genomics
For interspecies comparative analysis, we identified orthologous genes by running a bidirectional BLASTp search between all annotated protein-coding DNA sequences (CDS) for each species. CDS were considered orthologous when amino acid identity was greater than 35 across 75% of the protein, with an e-value limit of 1e-30 (Altschul et al., 1997). To confirm that CDSs predicted to be unique in one strain were not due to differences in annotation or due to the presence of paralogous sequences we ran tBLASTn searches of putative unique gene sequences against the chromosomes. Those genes that hit the chromosome with an e-value less than 1e-30 were considered non-unique. The degree of BLAST identity and the distribution of Clusters of Orthologous Groups (COGs) were visualized as a circular genome representation using CGview (Tatusov et al., 2000; Grant et al., 2012). Phylogenetic analysis was performed using the multilocus sequence typing (MLST) scheme developed for S. epidermidis (Thomas et al., 2007). To compare PSMs, amino acid sequences from S. capitis AYP1020 and S. epidermidis RP62a were clustered using the unweighted pair group method with arithmetic mean and a phylogenetic tree was generated using Geneious (Biomatters). IcaRADBC amino acid sequences were compared using ClustalW (Larkin et al., 2007).
Biofilm analysis
Biofilm formation was assessed using scanning electron microscopy (SEM). AYP1020 was grown overnight in tryptic soya broth (TSB, Oxoid) then diluted 1:100 in TSB with 0.25% glucose in a 24 well microtitre plate containing either a polyurethane or silicon coupon (Biosurface Technologies). Biofilms were allowed to mature for 24 h at 37°C with gentle shaking (75 rpm) then coupons were rinsed three times with phosphate buffered saline (PBS). Samples were fixed with 2.5% glutaraldehyde/1M cacodylate buffer followed by 1% osmium tetroxide, then dehydrated with ethanol and hexamethyldisilazane. Biofilms were coated with gold using a Balzers SCD005 sputter coater then visualized using a Hitachi S570 SEM. Biofilms generated by S. capitis AYP1020 and S. epidermidis RP62a were quantified using 96-well polystyrene microtitre plates (Falcon). Briefly, bacteria were grown as described above, planktonic cells were removed and biofilms were washed three times with PBS. Adherent material was stained using 0.1% crystal violet for 15 min. Crystal violet was emulsified using 96% ethanol and optical densities (570 nm) were determined using an infinite M200 plate reader (Tecan). Biofilms were quantified from four independent experiments and significance was determined by Mann-Whitney U-test with a significance level of P < 0.05.
Detection of PSM peptides using high-performance liquid chromatography (HPLC) and mass spectrometry (MS)
PSM peptides released by S. capitis strain AYP1020 were detected using reverse-phase HPLC and MS as described previously (Gao et al., 2013). Briefly, filtered culture supernatants were analyzed using an Agilent 1200 series HPLC equipped with a reversed-phase analytical column (Agilent Eclipse XDB-C18 4.6 × 150 mm) (Gao et al., 2013). PSM peptides were eluted with a water/acetonitrile (0.1% trifluoroacetic acid) gradient from 0 to 100% acetonitrile over 28 min at 1 mL per min and detected at 214 nm. The identity of PSMs within isolated fractions was determined based on m/z ratios compared to the molecular mass of each predicted PSM using a Waters Micromass ZQ mass spectrometer.
Results and discussion
Genome summary and general features
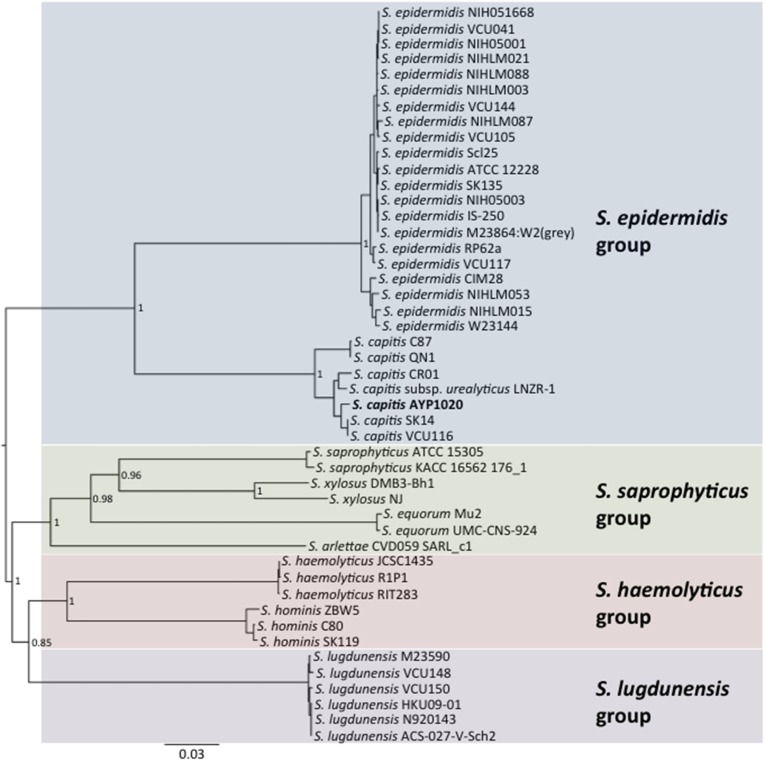
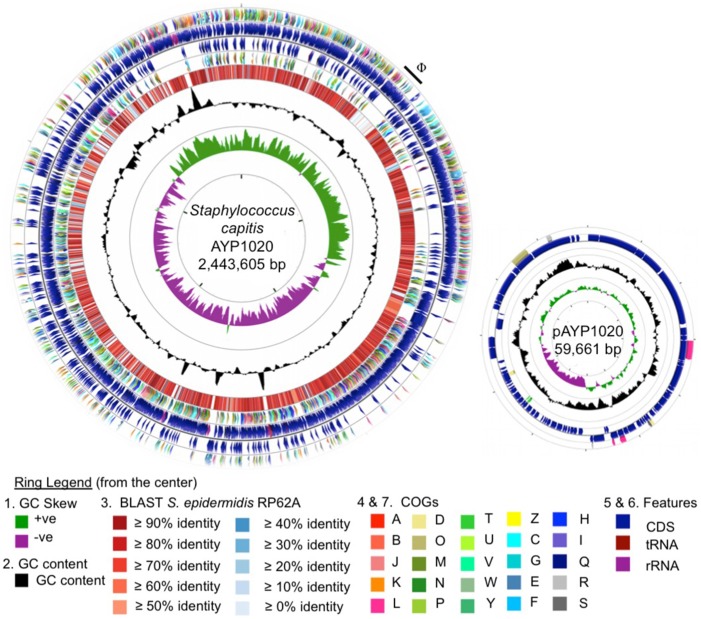
Phylogenetic analysis revealed a close relationship between AYP1020 and other S. capitis strains for which genome sequence information was available, particularly S. capitis SK14 and S. capitis VCU116 (Figure 1). The analysis also shows that S. capitis has closer evolutionary links to S. epidermidis than other clinically relevant coagulase negative staphylococci (Figure 1). S. capitis strain AYP1020 has a 2,443,605 bp chromosome with a GC content of 33.0% (Figure 2). The chromosome is predicted to contain 2304 protein-coding DNA sequences, six rRNA operons, 63 tRNA genes and a single tmRNA (Table 1). S. capitis AYP1020 represents the smallest genome of all closed staphylococcal genomes sequenced to date, with the fewest predicted CDS.
Figure 1.
Neighbor-joining tree showing the relationship between S. capitis AYP1020 and other sequenced CoNS strains. Phylogeny was inferred using the MLST scheme developed for S. epidermidis. S. capitis AYP1020 is represented in bold.
Figure 2.
Circular representation of the S. capitis AYP1020 chromosome and its plasmid, pAYP1020. For the chromosome, the first (innermost) ring indicates the GC skew, followed by the GC content (second ring). The third ring indicates the degree of amino acid identity across all CDS of S. capitis AYP1020 compared to S. epidermidis RP62a, as determined by BLASTp and scaled according to percent identity as indicated in the key (Altschul et al., 1997). Colored arrows on the fourth and seventh rings represent the following COG categories (Tatusov et al., 2000); A, B, J, K, L, information and storage and processing; D, O, M, N, P, T, U, V, W, Y, Z, cellular processes and signaling; C, G, E, F, H, I, Q, metabolism; R and S, poorly characterized, on the reverse and forward strands, respectively. The fifth and sixth rings represent the CDS (blue), tRNA (maroon), and rRNA (purple) on the reverse and forward strands, respectively. For the genome, the scale displayed in the center of the graphic represents 500 kb. For the plasmid, the scale represents 5 kb. The images were generated using CGview (Grant et al., 2012).
Table 1.
General genomic features of S. capitis strain AYP1020 compared with S. epidermidis RP62a.
S. capitis strain AYP1020 has a large plasmid designated pAYP1020. The plasmid is 59.6 Kb in size, has a GC content of 30.3% and is similar to the previously sequenced plasmid, SAP020A (75% coverage, 77% identity) from Staphylococcus sp. CDC3 (GenBank: NC_013373.1). The plasmid contains 71 predicted CDS, the majority of which encode proteins with no assigned function (54 CDS) (Figure 2). The AYP1020 chromosome also has a 48.5 Kb prophage designated ΦAYP1020 (Figure 2), encoding 24 typical phage proteins including integrase, terminase, portal, head, tail, and lysins, attR and attL sites plus an additional 30 hypothetical proteins.
Base modifications
SMRT technology allows for the genome-wide detection of modified nucleotides based on the rate by which DNA polymerase incorporates bases during sequencing (Roberts et al., 2013). Analysis of polymerase kinetic profiles identified 1972 m6A modifications (distribution shown in Figure S1) that were associated with two predicted type I adenine methyltransferase recognition motifs: 5′-CAAN6TGG-3′ and 5′-CTAN7TNYC-3′ (modified bases are underlined and modified bases on the opposite strand are italicized). Adenine modification correlated with the presence of four putative adenine methyltransferases in the AYP1020 genome (AYP1020_0291, AYP1020_0991, AYP1020_1852, and AYP1020_1992). In contrast, no cytosine methylation was detected correlating with the absence of predicted cytosine methyltransferase genes.
The overall distribution of putative methylated sites across the AYP1020 chromosome appeared to be random (Figure S1). However, in line with the predicted involvement of adenine methytransferases, regions of higher GC% contained fewer methylated sites. Aside from regions with higher GC%, methylation did not correlate strongly with predicted regions of horizontally acquired DNA (Figure S1). For example, the ΦAYP1020 prophage was methylated with an apparent equal density to the remainder of the AYP1020 chromosomal sequence. Assuming selective pressure for the accumulation of methylation sites within horizontally acquired DNA over time, this observation may suggest that the rate of sequence amelioration is rapid in AYP1020, or that horizontally acquired DNA was obtained from an organism harboring similar methyltransferases. In contrast to the prophage, the pAYP1020 plasmid sequence contained a low methylation site density (Figure S1), which could indicate that this plasmid is subject to different selective pressures than the host chromosome, or that it was relatively recently acquired within AYP1020.
Comparative genomic analysis of AYP1020 and S. epidermidis RP62a
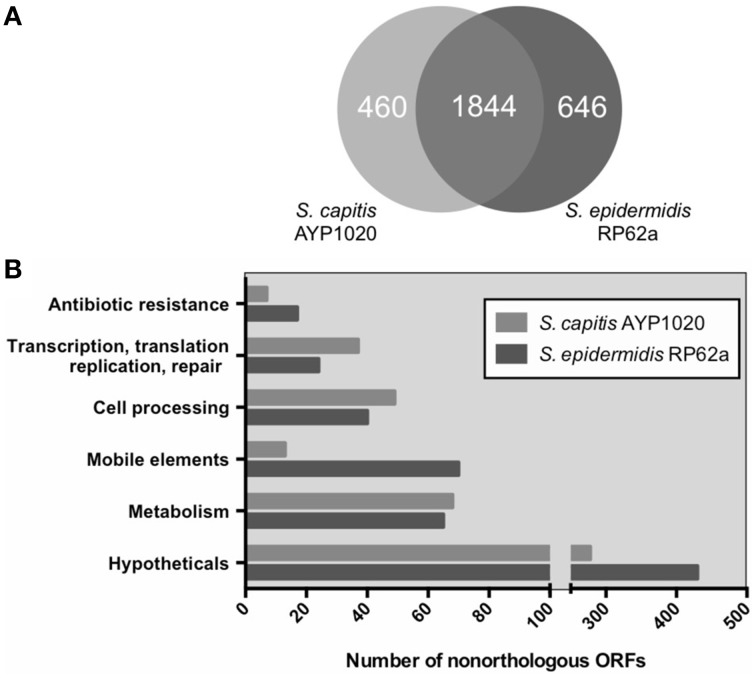
S. epidermidis (RP62a) was chosen for comparative analysis as it is a close relative of S. capitis and it is the most clinically important CoNS (Figure 1) (Poyart et al., 2001; Otto, 2009). Using the Artemis Comparison Tool, we found the degree of synteny between S. capitis AYP1020 and RP62a to be high, with few significant genomic rearrangements detected (Figure S2). We used BLASTp to compare all CDS sequences of AYP1020 with S. epidermidis RP62a and identified a core set of 1844 genes present in both species, which represents 80% of the S. capitis genome (Figure 3A). We identified 480 and 646 non-orthologous genes for S. capitis and S. epidermidis, respectively (Figure 3A). When the non-orthologous genes were categorized based on function, we found that S. epidermidis RP62a had an enhanced repertoire of genes related to mobile genetic elements (Figure 3B), which are largely accounted for by IS elements. The genome of AYP1020 encodes only one predicted IS element compared to the genome of S. epidermidis RP62a that has 64 predicted IS elements (18 intact), as well as five transposons (Gill et al., 2005; Takeuchi et al., 2005). Importantly, key antibiotic resistance determinants are carried on mobile elements in RP62a and these are absent from AYP1020. These include penicillinase encoded by blaZ, as well as SCCmec, which not only harbors determinants important for resistance to methicillin, but also aminoglycosides and macrolides (Table 2) (Gill et al., 2005). The absence of these genes in AYP1020 correlates with the highly antibiotic susceptible phenotype of this S. capitis isolate (Table 2). Multidrug-resistant strains, however, are emerging, with the majority of clinical strains resistant to penicillin and methicillin, and strains with reduced susceptibility to vancomycin also emerging (Ma et al., 2011; Rasigade et al., 2012). Further sequencing projects of multi-drug resistant isolates is required to determine the genetic factors facilitating antibiotic resistance in S. capitis. One such study characterized a novel SCCmec element in pulsotype NRCS-A, which harbored genes not only important for methicillin resistance, but also resistance to cadmium, arsenic, and copper (Martins Simões et al., 2013).
Figure 3.
Orthologous classification of CDS of S. capitis AYP1020 compared to S. epidermidis RP62a. Orthologs were defined by bidirectional BLASTp with an e-value cut off of 10e-30 (A). Non-orthologous CDS were grouped based on COG functional categories (B).
Table 2.
Comparison of antibiotic resistance profiles of S. capitis AYP1020 and S. epidermidis RP62a and their associated mobile elements.
| Antibiotic | Resistance gene (s) | AYP1020 | RP62a | ||||
|---|---|---|---|---|---|---|---|
| Gene | Product | Element | Presence | MIC (μg/mL) | Presence | MIC (μg/mL) | |
| Penicillin | blaZ | β-Lactamase | Plasmid | – | < 0.25 | + | 32 |
| Methicillin | mecA | Penicillin-binding protein 2′ | SCCmec | – | 2 | + | >128 |
| Erythromycin | ermA | rRNA adenine N-6-methyltransferase | Tn554, SCCmec | – | < 0.25 | + | >128 |
| Gentamicin | aacA | 6′-aminoglycoside N-acetyltransferase | Phage | – | < 0.25 | + | 64 |
| Streptomycin | aadE, | Aminoglycoside 6-adenylyltransferase | Plasmid | – | 2 | + | >128 |
| aphA | Aminoglycoside 3′-phosphotransferase | Plasmid | – | + | |||
| spc | Streptomycin 3″-adenylyltransferase | Tn554, SCCmec | – | + | |||
Insights into S. capitis virulence and biofilm production
Importantly, among the CDS orthologs between S. capitis and S. epidermidis, we identified a number of genes encoding for virulence factors that likely contribute to S. capitis pathogenicity (Table 3). CoNS species including S. capitis and S. epidermidis have a lower virulence potential compared to the most virulent staphylococcal species, S. aureus (Massey et al., 2006; Otto, 2009). As such, S. capitis and S. epidermidis do not code for the extensive suite of exotoxins that would be associated with more invasive and severe infections caused by S. aureus. S. capitis does however, encode for a number of factors predicted to be important for biofilm production, persistence and immune evasion.
Table 3.
Orthologous virulence factors of S. capitis strain AYP1020 and S. epidermidis strain RP62a.
| Function | Gene (s) | Product (s) | Locus | |
|---|---|---|---|---|
| S. capitis AYP1020 | S. epidermidis RP62A | |||
| Global regulators | agrADCB | Accessory gene regulator | 1223-6 | 1490-3 |
| sarARVZ | Staphylococcal accessory regulator | 2371 | 0274 | |
| 1469 | 1876 | |||
| 1442 | 1849 | |||
| 1576 | 1979 | |||
| saeRS | SaeRS two-component regulatory system | 0073–4 | 0364–5 | |
| arlRS | ArlRS two-component regulatory system | 0725–6 | 0988–9 | |
| rot | Repressor of toxins | 1043 | 1322 | |
| sigB | RNA polymerase sigma factor B | 1257 | 1677 | |
| Biofilm/PNAG | icaRADBC | Intercellular adhesion proteins | 1906–1910 | 2292–6 |
| PGA | capDACB | Capsule biosynthesis proteins | 1692 | 2103 |
| 1694–6 | 2105–7 | |||
| Exoenyzmes | hlb | Beta Hemolysin | 1985 | 2544 |
| Hemolysin III | 1362 | 1769 | ||
| Hemolysin | – | 2258 | ||
| clpP | Clp protease, proteolytic subunit | 0135 | 0436 | |
| clpBCX | Clp protease, ATP binding subunits | 0244 | 0564 | |
| 2273 | 0165 | |||
| 0957 | 1238 | |||
| sspA | Serine V8 protease | – | 1397 | |
| sspB | Cysteine protease | – | 2390 | |
| sspC | Cysteine protease | – | 2391 | |
| sepA | Metalloprotease elastase | 1844 | 2252 | |
| Zinc metalloprotease | 0571 | 0829 | ||
| Serine protease, putative | 0279 | 0611 | ||
| htrA | HtrA like protease, putative | 1007 | 1292 | |
| splE | Serine protease | 1979 | – | |
| Serine protease | 2166 | 2401 | ||
| lip | Lipase | 1943 | 2336 | |
| geh | Lipase | – | 0018 | |
| geh1 | Lipase | 1911 | 2297 | |
| geh2 | Lipase | 2083 | 2388 | |
| lipA | Lipase/esterase | 0022 | 0309 | |
| Esterase | 1534 | 1941 | ||
| Esterase | 1701 | 2109 | ||
| Pro-inflammatory peptides | psmα | Phenol Soluble modulin alpha | 2169 | 0083 |
| psmδ | Phenol Soluble modulin delta | 2168 | 0082 | |
| psmβ1a | Phenol Soluble modulin beta 1a | 0480 | 0738/9 | |
| psmβ1c | Phenol Soluble modulin beta 1b | 0481 | – | |
| psmβ1d | Phenol Soluble modulin beta 1c | 0482 | – | |
| psmβ2 | Phenol Soluble modulin beta 2 | 0479 | 0737 | |
| psmβ3 | Phenol Soluble modulin beta 3 | – | 0736 | |
| hld | Delta Hemolysin | 1222 | 1489 | |
| Surface proteins/adhesins | fbe | fibronectin binding protein A | 0515 | 0775 |
| atlE | Bifunctional autolysin | 0374 | 0636 | |
| pls | Plasmin-sensitive protein | 2027 | – | |
| aap | Accumulation associated protein | – | 2398 | |
| ebh | Cell wall associated fibronectin binding protein | 0751 | 1011 | |
| sesA | Cell wall surface anchor protein | 1036–7 | 1316 | |
| sesB | Cell wall surface anchor protein | 1776 | 2162 | |
| sesC | Cell wall surface anchor protein | 1864 | 2264 | |
| sesG | Cell wall surface anchor protein | 1214 | 1482 | |
| ebp | Elastin binding protein | – | 1048 | |
| bhp/bap | Cell wall associated biofilm protein | – | 2392 | |
| MSCRAMMs | sdrX | SdrX | 1833 | – |
| sdrZL | SdrZL | 1219 | – | |
| sdrH | SdrH protein | 1487 | ||
| sdrF | SdrF protein | – | 0026 | |
| sdrG | SdrG protein | – | 0207 | |
MSCRAMM, microbial surface component recognizing adhesive molecule; PGA, poly-γ-glutamic acid; PNAG, poly-N-acetlyglucosamine.
aLocus tags AYP1020_
bLocus tages SERP_ (Gill et al., 2005).
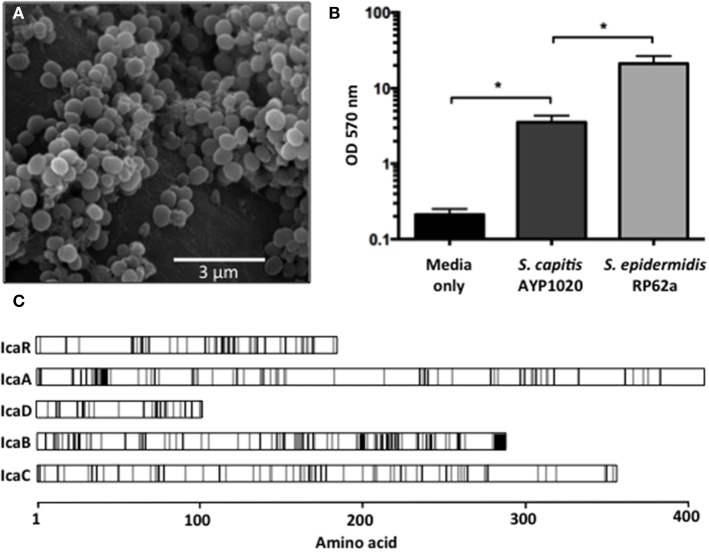
Biofilm formation is highly important for CoNS pathogenicity as they allow bacteria to colonize abiotic surfaces such as indwelling medical devices, which helps to establish infections within the host (Von Eiff et al., 2002). Using SEM, we observed S. capitis AYP1020 biofilms on multiple polymeric surfaces including polyurethane (Figure 4A) and silicone (data not shown). Whilst S. capitis AYP1020 was shown to generate biofilm, the relative production of biofilm was six-fold lower than that of S. epidermidis RP62a (Figure 4B). In S. epidermidis, the bifunctional autolysin, AtlE has been shown to be important for initial biofilm attachment to polymeric surfaces (Heilmann et al., 1997). Further, fibronectin binding proteins Fbe and Ebh are important for binding to host extracellular matrix proteins, which coat medical devices upon implantation (Nilsson et al., 1998; Von Eiff et al., 2002; Williams et al., 2002). The molecular mechanisms contributing to S. capitis surface adhesion are less well defined. A study highlighted the importance of microbial surface component recognizing adhesive molecules (MSCRAMMs) SdrX (Liu et al., 2004) and the “SdrZ like” protein SdrZL, each of which are present in the AYP1020 genome. The homologs of AtlE (70% amino acid identity), Fbe (88% amino acid identity), and Ebh (57% amino acid identity) were also identified. Further studies are required to investigate the role of these proteins in S. capitis adhesion.
Figure 4.
Biofilm formation of S. capitis AYP1020. Scanning electron micrograph of biofilm formed by S. capitis AYP1020 on polyurethane at 10,000X magnification (A). Biofilm was quantified on polystyrene microtitre plates. Data are expressed as mean ± SEM (*P < 0.05) (B). Schematic ClustalW alignment revealed high similarity (71–83% identity) between the IcaRADBC proteins of S. capitis AYP1020 and those of S. epidermidis. White regions represent the same amino acid, gray regions represent similar amino acids and black regions represent non-similar amino acids (C).
Proceeding attachment, biofilm accumulation in staphylococci typically occurs via the production of a poly-N-acetlyglucosamine (PNAG) homopolymer, which facilitates intercellular adhesion (also referred to as polysaccharide intercellular adhesion, PIA) (Mack et al., 1994). PNAG production is dependent on the ica locus, which has been described for S. capitis, and is also present in AYP1020 (Heilmann et al., 1996; de Silva et al., 2002). The genetic arrangement and predicted amino acid sequence for the Ica system was similar between S. capitis AYP1020 and S. epidermidis RP62a (Figure 4C). In addition, a recent study showed that S. capitis biofilms could be disrupted by the addition of proteinase K, suggesting that protein is also important for bacterial accumulation in S. capitis (Greco-Stewart et al., 2013). The plasmin-sensitive protein Pls of AYP1020 is a likely contributor to the proteinaceous nature of S. capitis biofilms as it promotes cell-cell interaction and shares similar domain structure and sequence homology with the accumulation-associated protein (Aap) of S. epidermidis (30% amino acid identity) and SasG of S. aureus (29% amino acid identity), each of which are important for biofilm accumulation (Hussain et al., 1997; Huesca et al., 2002; Geoghegan et al., 2010).
Along with PNAG, the AYP1020 genome also encodes the cap operon, which mediates the production of a second exopolysaccharide, poly-γ-glutamic acid (PGA). In S. epidermidis, PGA has an important role in host immune evasion, more specifically, resistance to host antimicrobial peptides and reduced susceptibility to phagocytosis (Kocianova et al., 2005).
Phenol soluble modulins and exoproteins
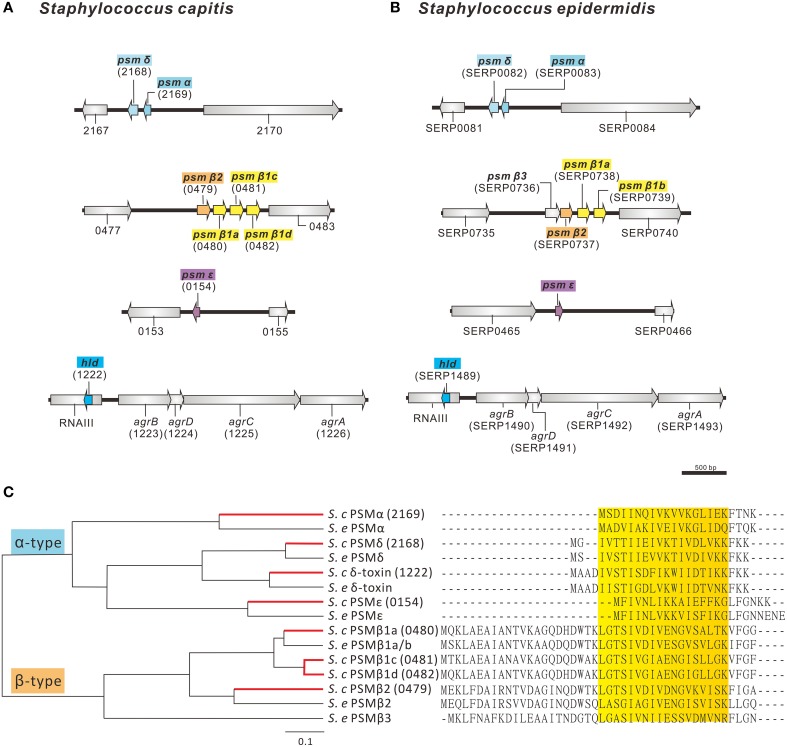
PSMs are secreted amphipathic peptides that appear to have multiple functions in the pathogenicity of staphylococci (Wang et al., 2007b; Peschel and Otto, 2013). PSMs have been shown to be pro-inflammatory and possess cytolytic properties, contribute to biofilm development and have anti-microbial activity, as shown by selective Streptococcus pyogenes killing (Yao et al., 2005; Wang et al., 2007b; Cogen et al., 2010). The AYP1020 genome encodes for a number of peptides with high sequence similarity to the PSMs of S. epidermidis (Figures 5A,B). Like S. epidermidis, S. capitis AYP1020 encodes four α-type PSMs and four β-type PSMs (Figure 5C). The β-type PSMs are present in an operon, whereas the α-type PSMs are found in three distinct genomic locations. The hld gene, encoding the α-type PSM δ-toxin, is present within RNAIII, the key regulatory molecule of the accessory gene regulator system, Agr (Recsei et al., 1986). PSM peptides have an amphipathic α-helical structure, which causes longer retention times during liquid chromatography compared to most other proteins in culture supernatants (Joo and Otto, 2014).
Figure 5.
Predicted phenol-soluble modulin (PSM) genes and amino acid sequences in S. capitis AYP1020. The genetic arrangement of phenol soluble modulin genes in AYP1020 (A) is highly similar when compared to S. epidermidis RP62a (B). In each case, PSMs are found on 4 distinct genetic loci. Predicted PSMs of S. capitis have high amino acid sequence identity when compared to S. epidermidis RP62a (C). AYP1020 has both α- and β-type PSMs, which are classed based on length. Each PSM contains an amphipathic α-helix, which is highlighted in yellow. AYP1020 has three distinct PSM β1 genes (a, c, and d), whereas RP62a has two identical copies of PSM β1 (a and b).
To determine if the predicted PSM peptides were expressed in AYP1020, we analyzed filtered supernatants using reverse phase-HPLC followed by mass spectrometry. The m/z ratios of each peptide peak determined by mass spectrometry were correlated to the predicted mass of each predicted PSM peptide (Table 4). The m/z ratios also indicated that most types of PSMs detected were N-terminal formylated excluding PSMβ2 and PSMε. Given the toxic potential of PSM peptides, future research is required to determine the importance of these peptides to S. capitis physiology and pathogenesis.
Table 4.
Predicted PSMs of S. capitis strain AYP1020 were detected in the culture supernatant using HPLC followed by mass spectrometry.
| PSM | Predicted amino acid sequencea | Molecular Weightb | m/z of peaks (+ H+)c | Retention time (min) |
|---|---|---|---|---|
| α | fMSDIINQIVKVVKGLIEKFTNK | 2547.1 | 1274.2 (+2) | 21.3 |
| 849.8 (+3) | ||||
| 637.4 (+4) | ||||
| β1a | fMQKLAEAIANTVKAGQDHDWTKLGTSIVDIVENGVSALTKVFGG | 4643.3 | 1548.2 (+3) | 20.5 |
| 1161.4 (+4) | ||||
| 929.7 (+5) | ||||
| β1c | fMTKLAEAIANAVKAGQDQDWAKLGTSIVGIAENGISLLGKVFGF | 4563.3 | 1521.9 (+3) | 23.5 |
| 1141.6 (+4) | ||||
| 913.4 (+5) | ||||
| β1d | fMQKLAEAIANTVKAGQDHDWAKLGTSIVGIAENGIGLLGKVFGF | 4599.3 | 1150.3 (+4) | 19.5 |
| 920.3 (+5) | ||||
| β2 | MEKLFDAIRNTVDAGINQDWTKLGTSIVDIVDNGVKVISKFIGA | 4822.5 | 1608.7 (+3) | 25.2 |
| 1206.5 (+4) | ||||
| δ | MGIVTTIIEIVKTIVDLVKKFKK | 2617.3 | 872.5 (+3) | 22.5 |
| 654.6 (+4) | ||||
| fMGIVTTIIEIVKTIVDLVKKFKK | 2646.3 | 1323.7 (+2) | 27 | |
| 882.9 (+3) | ||||
| ε | MFIVNLIKKAIEFFKGLFGNKK | 2586.2 | 862.7 (+3) | 17.5 |
| δ-toxin | MAADIVSTISDFIKWIIDTIKKFKK | 2912.5 | 971.6 (+3) | 19.5 |
| 729.2 (+4) | ||||
| fMAADIVSTISDFIKWIIDTIKKFKK | 2941.5 | 1471.2 (+2) | 25.2 | |
| 981.3 (+3) |
Formylation of the amino-terminus is indicated as f.
The molecular weight is calculated based on predicted amino acid sequence.
The numbers of protons added to the peptides are suggested.
The majority of PSMs have been shown to be regulated by Agr (Wang et al., 2007b). Aside from Agr, AYP1020 encodes a number of systems important for the regulation of virulence in staphylococci including the staphylococcal accessory regulator, Sar, the repressor of toxins, Rot and the two component regulatory systems, SaeRS and ArlRS (Table 3) (Cheung et al., 1992; Giraudo et al., 1994; McNamara et al., 2000; Fournier et al., 2001).
S. capitis AYP1020 also encodes for a suite of exoproteins that likely contribute to infection. These include proteases such as ClpP, which contributes to biofilm formation and SepA, which has been shown to degrade host antimicrobial peptides in CoNS (Wang et al., 2007a; Cheung et al., 2010), as well as hemolysins, lipases, and esterases. These proteins likely facilitate immune evasion, host colonization and persistence, as opposed to acute disease facilitated by typical exoproteins of the more virulent staphylococcal species, S. aureus (Otto, 2009).
In summary, we have generated the closed genome and methylome for S. capitis strain AYP1020 using SMRT sequence technology. Our study focused on genes that are predicted to be important for S. capitis pathogenicity, based on their role in the closely related opportunistic pathogen, S. epidermidis. Having a reliable reference genome invites the sequencing of more diverse S. capitis isolates, which would provide further insights into the evolution of virulence and antibiotic resistance in this emerging pathogen.
Financial support
This work was supported by an Australian Postgraduate Award and a Monash University Postgraduate Publication Award to DC. and an Australian National Health and Medical Research Council (NHMRC) R.D. Wright Career Development Fellowship (APP1047916) and Project Grant (APP1047918) to AP.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Richard J. Maydom for assistance with bioinformatics, Richard Roberts for assistance with REBASE, Paul Stothard for generating images using CGview, Xenia Kostoulias for technical assistance and Joan Clarke from Monash Micro Imaging for scanning electron microscopy.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00980
References
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandres J. C., Darouiche R. O. (1992). Staphylococcus capitis endocarditis: a new cause of an old disease. Clin. Infect. Dis. 14, 366–367. 10.1093/clinids/14.1.366-a [DOI] [PubMed] [Google Scholar]
- Bannerman T. L., Kloos W. E. (1991). Staphylococcus capitis subsp. ureolyticus subsp. nov. from human skin. Int. J. Syst. Bacteriol. 41, 144–147. 10.1099/00207713-41-1-144 [DOI] [PubMed] [Google Scholar]
- Cai J. C., Hu Y. Y., Zhang R., Zhou H. W., Chen G. X. (2012). Linezolid-resistant clinical isolates of meticillin-resistant coagulase-negative staphylococci and Enterococcus faecium from China. J. Med. Microbiol. 61, 1568–1573. 10.1099/jmm.0.043729-0 [DOI] [PubMed] [Google Scholar]
- Cheung A. L., Koomey J. M., Butler C. A., Projan S. J., Fischetti V. A. (1992). Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. U.S.A. 89, 6462–6466. 10.1073/pnas.89.14.6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung G. Y., Rigby K., Wang R., Queck S. Y., Braughton K. R., Whitney A. R., et al. (2010). Staphylococcus epidermidis strategies to avoid killing by human neutrophils. PLoS Pathog. 6:e1001133. 10.1371/journal.ppat.1001133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. S., Alexander D. H., Marks P., Klammer A. A., Drake J., Heiner C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- Christensen G. D., Simpson W. A., Younger J. J., Baddour L. M., Barrett F. F., Melton D. M., et al. (1985). Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C.L.S.I (2013). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. M100-S23. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cogen A. L., Yamasaki K., Sanchez K. M., Dorschner R. A., Lai Y., MacLeod D. T., et al. (2010). Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Invest. Dermatol. 130, 192–200. 10.1038/jid.2009.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva G. D., Kantzanou M., Justice A., Massey R. C., Wilkinson A. R., Day N. P., et al. (2002). The ica operon and biofilm production in coagulase-negative Staphylococci associated with carriage and disease in a neonatal intensive care unit. J. Clin. Microbiol. 40, 382–388. 10.1128/JCM.40.02.382-388.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., Munera D., Friedman D. I., Mandlik A., Chao M. C., Banerjee O., et al. (2012). Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 30, 1232–1239. 10.1038/nbt.2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier B., Klier A., Rapoport G. (2001). The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41, 247–261. 10.1046/j.1365-2958.2001.02515.x [DOI] [PubMed] [Google Scholar]
- Gao W., Cameron D. R., Davies J. K., Kostoulias X., Stepnell J., Tuck K. L., et al. (2013). The RpoB H481Y rifampicin resistance mutation and an active stringent response reduce virulence and increase resistance to innate immune responses in Staphylococcus aureus. J. Infect. Dis. 207, 929–939. 10.1093/infdis/jis772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J. A., Corrigan R. M., Gruszka D. T., Speziale P., O'Gara J. P., Potts J. R., et al. (2010). Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J. Bacteriol. 192, 5663–5673. 10.1128/JB.00628-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. R., Fouts D. E., Archer G. L., Mongodin E. F., Deboy R. T., Ravel J., et al. (2005). Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187, 2426–2438. 10.1128/JB.187.7.2426-2438.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo A. T., Raspanti C. G., Calzolari A., Nagel R. (1994). Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can. J. Microbiol. 40, 677–681. 10.1139/m94-107 [DOI] [PubMed] [Google Scholar]
- Grant J. R., Arantes A. S., Stothard P. (2012). Comparing thousands of circular genomes using the CGView Comparison Tool. BMC Genomics 13:202. 10.1186/1471-2164-13-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco-Stewart V. S., Ali H., Kumaran D., Kalab M., Rood I. G., de Korte D., et al. (2013). Biofilm formation by Staphylococcus capitis strains isolated from contaminated platelet concentrates. J. Med. Microbiol. 62, 1051–1059. 10.1099/jmm.0.050500-0 [DOI] [PubMed] [Google Scholar]
- Heilbronner S., Holden M. T., van Tonder A., Geoghegan J. A., Foster T. J., Parkhill J., et al. (2011). Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors. FEMS Microbiol. Lett. 322, 60–67. 10.1111/j.1574-6968.2011.02339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C., Hussain M., Peters G., Götz F. (1997). Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24, 1013–1024. 10.1046/j.1365-2958.1997.4101774.x [DOI] [PubMed] [Google Scholar]
- Heilmann C., Schweitzer O., Gerke C., Vanittanakom N., Mack D., Götz F. (1996). Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20, 1083–1091. 10.1111/j.1365-2958.1996.tb02548.x [DOI] [PubMed] [Google Scholar]
- Heilmann C., Thumm G., Chhatwal G. S., Hartleib J., Uekötter A., Peters G. (2003). Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology 149, 2769–2778. 10.1099/mic.0.26527-0 [DOI] [PubMed] [Google Scholar]
- Heithoff D. M., Sinsheimer R. L., Low D. A., Mahan M. J. (1999). An essential role for DNA adenine methylation in bacterial virulence. Science 284, 967–970. 10.1126/science.284.5416.967 [DOI] [PubMed] [Google Scholar]
- Huesca M., Peralta R., Sauder D. N., Simor A. E., McGavin M. J. (2002). Adhesion and virulence properties of epidemic Canadian methicillin-resistant Staphylococcus aureus strain 1: identification of novel adhesion functions associated with plasmin-sensitive surface protein. J. Infect. Dis. 185, 1285–1296. 10.1086/340123 [DOI] [PubMed] [Google Scholar]
- Hussain M., Herrmann M., von Eiff C., Perdreau-Remington F., Peters G. (1997). A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 65, 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H. S., Otto M. (2014). The isolation and analysis of phenol-soluble modulins of Staphylococcus epidermidis. Methods Mol. Biol. 1106, 93–100. 10.1007/978-1-62703-736-5_7 [DOI] [PubMed] [Google Scholar]
- Katayama T., Ozaki S., Keyamura K., Fujimitsu K. (2010). Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8, 163–170. 10.1038/nrmicro2314 [DOI] [PubMed] [Google Scholar]
- Kloos W. E., Schleifer K. H. (1975). Isolation and characterization of Staphylococci from human skin II. Descriptions of four new species: Staphylococcus warneri, Staphylococcus capitis, Staphylococcus hominis, and Staphylococcus simulans. Int. J. Syst. Evol. Microbiol. 25, 62–79. 10.1099/00207713-25-1-62 [DOI] [Google Scholar]
- Kocianova S., Vuong C., Yao Y., Voyich J. M., Fischer E. R., Deleo F. R., et al. (2005). Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J. Clin. Invest. 115, 688–694. 10.1172/JCI200523523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Yamashita A., Hirakawa H., Kumano M., Morikawa K., Higashide M., et al. (2005). Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. U.S.A. 102, 13272–13277. 10.1073/pnas.0502950102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M. G., Brinkman F. S. (2009). IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 25, 664–665. 10.1093/bioinformatics/btp030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Liu Y., Ames B., Gorovits E., Prater B. D., Syribeys P., Vernachio J. H., et al. (2004). SdrX, a serine-aspartate repeat protein expressed by Staphylococcus capitis with collagen VI binding activity. Infect. Immun. 72, 6237–6244. 10.1128/IAI.72.11.6237-6244.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. X., Wang E. H., Liu Y., Luo E. J. (2011). Antibiotic susceptibility of coagulase-negative staphylococci (CoNS): emergence of teicoplanin-non-susceptible CoNS strains with inducible resistance to vancomycin. J. Med. Microbiol. 60, 1661–1668. 10.1099/jmm.0.034066-0 [DOI] [PubMed] [Google Scholar]
- Mack D., Fischer W., Krokotsch A., Leopold K., Hartmann R., Egge H., et al. (1996). The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178, 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D., Nedelmann M., Krokotsch A., Schwarzkopf A., Heesemann J., Laufs R. (1994). Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect. Immun. 62, 3244–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Casadesus J. (2009). Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 33, 488–503. 10.1111/j.1574-6976.2008.00159.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins Simões P., Rasigade J. P., Lemriss H., Butin M., Ginevra C., Lemriss S., et al. (2013). Characterization of a novel composite staphylococcal cassette chromosome mec (SCCmec-SCCcad/ars/cop) in the neonatal sepsis-associated Staphylococcus capitis pulsotype NRCS-A. Antimicrob. Agents Chemother. 57, 6354–6357. 10.1128/AAC.01576-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey R. C., Horsburgh M. J., Lina G., Hook M., Recker M. (2006). The evolution and maintenance of virulence in Staphylococcus aureus: a role for host-to-host transmission? Nat. Rev. Microbiol. 4, 953–958. 10.1038/nrmicro1551 [DOI] [PubMed] [Google Scholar]
- McNamara P. J., Milligan-Monroe K. C., Khalili S., Proctor R. A. (2000). Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182, 3197–3203. 10.1128/JB.182.11.3197-3203.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- N.N.I.S . (1999). National Nosocomial Infections Surveillance (NNIS) System Report, Data Summary from January 1990-May 1999, issued June 1999. A report from the NNIS System. Am. J. Infect. Control 27, 520–532. 10.1016/S0196-6553(99)70031-3 [DOI] [PubMed] [Google Scholar]
- Nilsson M., Frykberg L., Flock J. I., Pei L., Lindberg M., Guss B. (1998). A fibrinogen-binding protein of Staphylococcus epidermidis. Infect. Immun. 66, 2666–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2009). Staphylococcus epidermidis–the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7, 555–567. 10.1038/nrmicro2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel A., Otto M. (2013). Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 11, 667–673. 10.1038/nrmicro3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart C., Quesne G., Boumaila C., Trieu-Cuot P. (2001). Rapid and accurate species-level identification of coagulase-negative staphylococci by using the sodA gene as a target. J. Clin. Microbiol. 39, 4296–4301. 10.1128/JCM.39.12.4296-4301.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasigade J. P., Raulin O., Picaud J. C., Tellini C., Bes M., Grando J., et al. (2012). Methicillin-resistant Staphylococcus capitis with reduced vancomycin susceptibility causes late-onset sepsis in intensive care neonates. PLoS ONE 7:e31548. 10.1371/journal.pone.0031548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recsei P., Kreiswirth B., O'Reilly M., Schlievert P., Gruss A., Novick R. P. (1986). Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol. Gen. Genet. 202, 58–61. 10.1007/BF00330517 [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Carneiro M. O., Schatz M. C. (2013). The advantages of SMRT sequencing. Genome Biol. 14:405. 10.1186/gb-2013-14-6-405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein R., Nerz C., Biswas L., Resch A., Raddatz G., Schuster S. C., et al. (2009). Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl. Environ. Microbiol. 75, 811–822. 10.1128/AEM.01982-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M. A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945. 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F., Watanabe S., Baba T., Yuzawa H., Ito T., Morimoto Y., et al. (2005). Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187, 7292–7308. 10.1128/JB.187.21.7292-7308.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. 10.1093/nar/28.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. C., Vargas M. R., Miragaia M., Peacock S. J., Archer G. L., Enright M. C. (2007). Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J. Clin. Microbiol. 45, 616–619. 10.1128/JCM.01934-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Zwet W. C., Debets-Ossenkopp Y. J., Reinders E., Kapi M., Savelkoul P. H., Van Elburg R. M., et al. (2002). Nosocomial spread of a Staphylococcus capitis strain with heteroresistance to vancomycin in a neonatal intensive care unit. J. Clin. Microbiol. 40, 2520–2525. 10.1128/JCM.40.7.2520-2525.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Eiff C., Peters G., Heilmann C. (2002). Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2, 677–685. 10.1016/S1473-3099(02)00438-3 [DOI] [PubMed] [Google Scholar]
- Wang C., Li M., Dong D., Wang J., Ren J., Otto M., et al. (2007a). Role of ClpP in biofilm formation and virulence of Staphylococcus epidermidis. Microbes Infect. 9, 1376–1383. 10.1016/j.micinf.2007.06.012 [DOI] [PubMed] [Google Scholar]
- Wang R., Braughton K. R., Kretschmer D., Bach T. H., Queck S. Y., Li M., et al. (2007b). Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13, 1510–1514. 10.1038/nm1656 [DOI] [PubMed] [Google Scholar]
- Wang R., Khan B. A., Cheung G. Y., Bach T. H., Jameson-Lee M., Kong K. F., et al. (2011). Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J. Clin. Invest. 121, 238–248. 10.1172/JCI42520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. J., Henderson B., Sharp L. J., Nair S. P. (2002). Identification of a fibronectin-binding protein from Staphylococcus epidermidis. Infect. Immun. 70, 6805–6810. 10.1128/IAI.70.12.6805-6810.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisplinghoff H., Bischoff T., Tallent S. M., Seifert H., Wenzel R. P., Edmond M. B. (2004). Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39, 309–317. 10.1086/421946 [DOI] [PubMed] [Google Scholar]
- Yao Y., Sturdevant D. E., Otto M. (2005). Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J. Infect. Dis. 191, 289–298. 10.1086/426945 [DOI] [PubMed] [Google Scholar]
- Zhang Y. Q., Ren S. X., Li H. L., Wang Y. X., Fu G., Yang J., et al. (2003). Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49, 1577–1593. 10.1046/j.1365-2958.2003.03671.x [DOI] [PubMed] [Google Scholar]
- Zhou Y., Liang Y., Lynch K. H., Dennis J. J., Wishart D. S. (2011). PHAST: a fast phage search tool. Nucleic Acids Res. 39, W347–W352. 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.