Abstract
MicroRNAs (miRNAs) are important regulators of neuronal development, network connectivity, and synaptic plasticity. While many neuronal miRNAs were previously shown to modulate neuronal morphogenesis, little is known regarding the regulation of miRNA function. In a large-scale functional screen, we identified two novel regulators of neuronal miRNA function, Nova1 and Ncoa3. Both proteins are expressed in the nucleus and the cytoplasm of developing hippocampal neurons. We found that Nova1 and Ncoa3 stimulate miRNA function by different mechanisms that converge on Argonaute (Ago) proteins, core components of the miRNA-induced silencing complex (miRISC). While Nova1 physically interacts with Ago proteins, Ncoa3 selectively promotes the expression of Ago2 at the transcriptional level. We further show that Ncoa3 regulates dendritic complexity and dendritic spine maturation of hippocampal neurons in a miRNA-dependent fashion. Importantly, both the loss of miRNA activity and increased dendrite complexity upon Ncoa3 knockdown were rescued by Ago2 overexpression. Together, we uncovered two novel factors that control neuronal miRISC function at the level of Ago proteins, with possible implications for the regulation of synapse development and plasticity.
Keywords: Ago2, dendrite, miRNA, Ncoa3, Nova1
See also: F Sananbenesi & A Fischer (September 2015)
Introduction
MicroRNAs (miRNAs) are a large family of small non-coding RNAs that regulate gene expression post-transcriptionally. Over the last 10 years, miRNAs have been implicated in the regulation of virtually every aspect of mammalian nervous system development, including neurogenesis, neuronal differentiation, and plasticity (Fiore et al, 2011; Im & Kenny, 2012). A subset of neuronal miRNAs localizes within the synapto-dendritic compartment, where they constitute a regulatory layer in local mRNA translation and the control of synaptic plasticity (Siegel et al, 2011). Arguably, the best studied synaptic miRNA is miR-134, which (i) is required for activity-dependent dendritogenesis and synaptic scaling by inhibiting Pumilio2 (Fiore et al, 2009, 2014), (ii) mediates spine shrinkage by repressing Lim-domain protein kinase 1 (Schratt et al, 2006), and (iii) impairs synaptic plasticity and memory in the mouse hippocampus in vivo by downregulating CREB (Gao et al, 2010). Other examples of functionally important miRNAs are miR-138, which restricts the growth of dendritic spines by targeting the depalmitoylating enzyme APT1, and the highly conserved let-7, which regulates BDNF-dependent dendritogenesis and axon regeneration (Huang et al, 2012; Zou et al, 2013).
MiRNAs bind the 3′ untranslated region (UTR) of target mRNAs via partial base-pair complementarity, thereby inducing translational repression and/or decay of their targets (Eulalio et al, 2008; Krol et al, 2010). The miRNA-induced silencing complex (miRISC) is a large multi-protein complex that mediates miRNA activity (Tang, 2005). The Argonaute (Ago) family of proteins are the main catalysts of miRNA function as they serve as a platform for miRISC assembly on the target mRNA (Meister, 2013). The mammalian genome comprises four genes encoding for Ago subfamily proteins (Ago1–4), all of which are capable of miRNA-mediated gene silencing. Ago2 is the only member that possesses nuclease (“slicer”) activity (Liu et al, 2004), and therefore is capable of endonucleolytic cleavage of targets in the case of perfect complementarity between the miRNA and the target, which is a rare event in mammals. GW182 (Tnrc6a-c in mammals) proteins represent other core components of miRISC, which help to recruit different effector complexes onto miRNA target mRNAs to induce translational repression and/or mRNA decay (Fabian & Sonenberg, 2012).
miRISC activity is regulated by signal-dependent modification of accessory factors in response to environmental cues. In neurons, activity-dependent degradation of Mov10 was shown to impair miRISC activity, thereby allowing local protein synthesis at synapses to occur (Banerjee et al, 2009). A similar mechanism involving the Mov10 homologue Armitage is further involved in the formation of long-term memories in Drosophila (Ashraf et al, 2006). Finally, dephosphorylation of FMRP in response to elevated activity leads to a release of miRISC from target mRNAs at synapses, thereby allowing synaptic protein synthesis to occur (Muddashetty et al, 2011). In addition, several RNA-binding proteins (RBPs) that do not directly associate with miRISC, but are recruited to the same targeted mRNA, have been previously shown to modulate the effect of miRNAs in non-neuronal cells, including Pumilio (Kedde et al, 2010), HuR (Bhattacharyya et al, 2006), and DND1 (Kedde et al, 2007). However, a comprehensive analysis of miRISC regulation by RBPs in neurons has not been performed.
Here, we interrogated the function of 286 RBPs in neuronal miRISC regulation using siRNA screening in primary mouse cortical neurons. Thereby, we identified Nova1 as a neuron-specific miRISC component and Ncoa3 as a regulator of Ago2 expression. We suggest that neuron-specific modulators of miRISC function such as Nova1 and Ncoa3 could play important roles in activity-dependent gene regulation during synapse development and plasticity.
Results
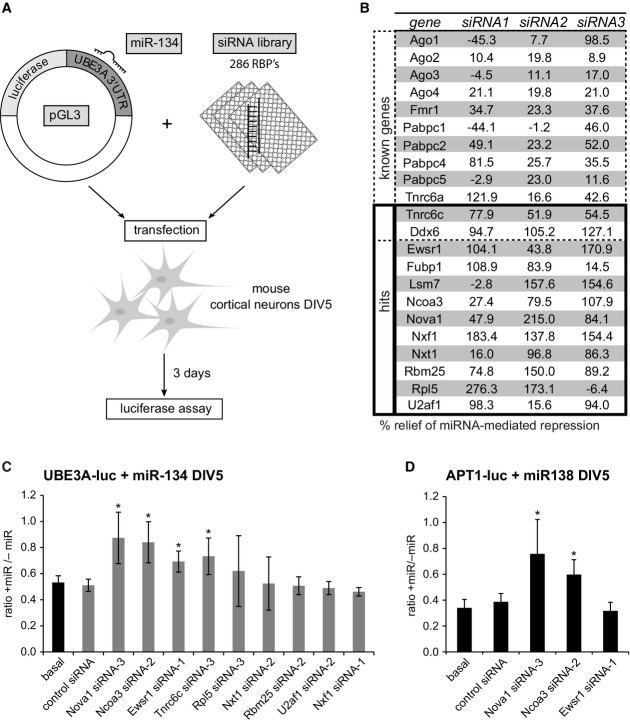
To identify RBPs that modulate the repressive activity of miRNAs in neurons, we combined siRNA-mediated RBP knockdown with a luciferase-based reporter gene readout (Fig1A). Our siRNA library consisted of three different synthetic siRNAs for each of the 286 RBPs which had been shown to be expressed in the postnatal mouse cortex (McKee et al, 2005) (Supplementary Dataset S1). For the screen, each individual siRNA was co-transfected into primary mouse cortical neurons (5 days in vitro DIV) along with a miR-134-responsive luciferase reporter gene (UBE3A-luc) (Valluy et al, 2015) and synthetic miR-134 duplex RNA. miR-134 duplex RNA is directly loaded into miRISC, thereby allowing us to focus on the effects of RBPs downstream of Dicer-dependent pre-miRNA processing. We observed a highly reproducible about 40% repression of UBE3A-luc expression by miR-134 in the absence of additional siRNA. If a given RBP was required for the repressive function of miR-134, the knockdown of the RBP should completely (100%) relieve the miR-134-mediated repression of UBE3A-luc. Because individual siRNAs display different degrees of efficacy, we considered RBPs as positive hits if at least two of three RBP-targeting siRNAs resulted in an at least 50% relief of miR-134-mediated repression averaged over the three independent screen replicates. Results of the analysis using this cutoff are shown in Fig1B (complete study results in Supplementary Dataset S2). Two known miRISC-associated proteins (Tnrc6c, Ddx6) fulfilled our hit selection criteria (Chu & Rana, 2006; Landthaler et al, 2008). In addition, 31 out of 36 siRNAs directed against twelve known regulators of miRNA function (Ago family proteins, Fmr1, Pabpc family members, Tnrc6a) relieved repression of the miR-134 reporter, although the effect was not strong enough to qualify them as hits. This demonstrates that (i) our screen setup allows the identification of miRISC regulatory RBPs; and (ii) our criteria for the selection of hits are very stringent, thereby reducing the number of false positives. In addition to Tnrc6c and Ddx6, ten novel RBPs that had not been previously reported to be involved in miRNA function were identified as hits (Fig1B). These include RBPs implicated in mRNA translation (Rpl5) (Meskauskas & Dinman, 2001), mRNA decay (Lsm7) (Tharun et al, 2000), splicing (Nova1, U2af1, Rbm25) (Jensen et al, 2000; Mollet et al, 2006; Zhou et al, 2008), mRNA nuclear export (Nxf1, Nxt1) (Stutz & Izaurralde, 2003), and transcription (Ewsr1, Fubp1, Ncoa3) (Law et al, 2006; Xu et al, 2009; Zhang & Chen, 2013). Since most of the identified candidates are involved in general RNA metabolism, we considered the possibility that knockdown of these RBPs could affect reporter gene expression independent of miRNAs. Therefore, we conducted a secondary RNAi screen for all the hits where we now also included a condition without miR-134 (“−miR” condition; Fig1C). If the RBP regulates reporter gene expression independent of the transfected miRNA, a similar upregulation of reporter gene expression would be observed in the “+miR” and the “−miR” conditions compared to the respective basal or control siRNA conditions and the ratio “+miR/−miR” would be comparable over all experimental conditions. The validation experiments were performed in rat neurons using siRNAs with conserved target sequences between mouse and rat mRNAs. In addition, an improved reporter gene system (pGL4) was used to further minimize non-specific effects. Using this experimental setup, knockdown of three (Nova1, Ncoa3, Ewsr1) out of the original ten positive hits resulted in a significant increase in the ratio +miR/−miR, suggesting that only these three RBPs specifically affect miR-134-dependent repression of UBE3A-luc. Specific derepression of miRNA target expression by RBP knockdown was further confirmed by our results using a plasmid without miR-134-binding site (Supplementary Fig S1A). The efficacy of the siRNAs targeting Nova1, Ncoa3, and Ewsr1 was validated by Western blot. Each of the three siRNAs led to a specific knockdown of the respective GFP-tagged proteins in HEK293 (Supplementary Fig S1B). Together, we could identify three novel proteins (Nova1, Ncoa3, and Ewsr1) that are specifically required for the repressive function of miR-134. To assess whether these RBPs modulate miRNA activity in a more general fashion, we extended our analysis to another neuronal miRNA, miR-138, and its validated target APT1 (Siegel et al, 2009). Knockdown of Nova1 and Ncoa3 led to a significant derepression of APT1-luc reporter expression (Fig1D) which was comparable to the effects observed with UBE3A-luc. In contrast, the Ewsr1-targeting siRNA had no effect on APT1-luc expression. We conclude that Nova1 and Ncoa3 are required for the repressive function of multiple neuronal miRNAs and therefore decided to focus on these RBPs for further characterization.
Figure 1.
Identification of 12 RNA-binding proteins required for miR-134 repressive function in primary neurons using siRNA-based screening
- Setup of the RNAi screen in mouse primary cortical neurons. Neurons (5 days in vitro (DIV)) were co-transfected with the miR-134-responsive pGL3-UBE3A-3′UTR luciferase reporter (pGL3-UBE3A-luc) together with miR-134 duplex RNA and siRNA (one per condition) directed against 286 RBPs expressed in the mouse cortex (3 siRNAs per RBP). Luciferase activity was determined 3 days after transfection.
- Table of genes which are either known to be involved in the regulation of miRNA activity (“known genes”) or whose knockdown led to an at least 50% relief of miRNA-mediated repression for at least two out of the three siRNAs tested (“hits”). Values are the average of three replicate experiments and represent the percent relief of miRNA-mediated repression for the indicated siRNA compared to a condition without siRNA. Note that two known genes (Tnrc6c, Ddx6) were also present within the 12 hits.
- Luciferase reporter assay in primary rat cortical neurons transfected with pGL4-UBE3A-luc and the indicated siRNAs in the presence or absence of miR-134 duplex RNA. The ratio of normalized luciferase activity from neurons with (“+miR”) to neurons without co-transfected miR-134 (“−miR”) is plotted. Values are the average from three independent experiments ± standard deviation. *P < 0.05 (unpaired t-test compared to control siRNA).
- Luciferase reporter assay as in (C) with pGL4-APT1 3′UTR (APT1-luc) in the presence (“+miR”) or absence (“−miR”) of miR-138. n = 3. *P < 0.05 (unpaired t-test compared to control siRNA).
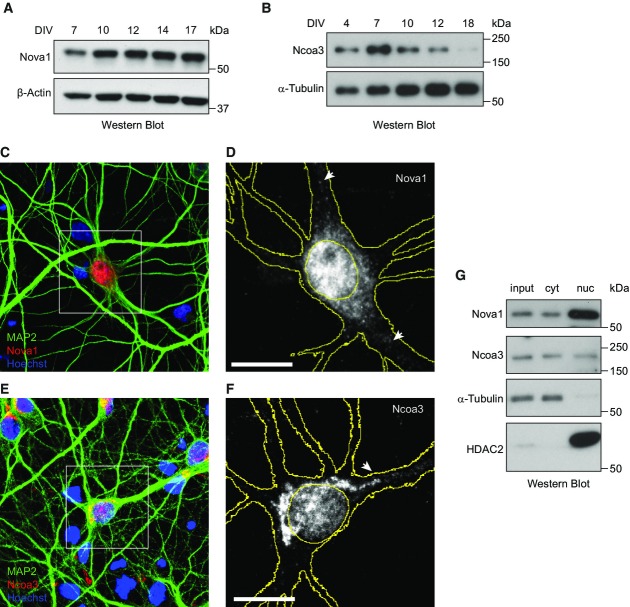
We next studied the expression of Nova1 and Ncoa3 proteins in post-mitotic neurons by Western blot analysis. Nova1 was expressed throughout the in vitro development of primary rat hippocampal (Fig2A) and cortical neurons (Supplementary Fig S2A). The anti-Nova1 antibody was specific, since the Nova1 band was absent when Western blots were performed with total brain lysates from Nova knockout mice (Supplementary Fig S2B). Ncoa3 expression in hippocampal neurons peaked at seven DIV, declined until 14 DIV, and became undetectable at 18 DIV (Fig2B; Supplementary Fig S2C). Using immunocytochemistry (ICC), we observed that both proteins localized to the nuclear and cytoplasmic compartment of hippocampal neurons (14 DIV: Fig2C–F; 7 DIV: Supplementary Fig S2D). The majority of cytoplasmic Nova1 protein was detected as granular structures in the neuronal soma, but some Nova1-positive granules extended into proximal dendrites that could be identified by co-staining for the dendritic marker MAP2 (arrows in Fig2D). Similarly, some of the Ncoa3-positive cytoplasmic granules extended into the proximal part of apical dendrites (arrow in Fig2F), whereas Ncoa3 expression at distal dendrites was generally low. Importantly, the Ncoa3 ICC signal was significantly reduced in cells expressing Ncoa3 shRNA which demonstrates the specificity of the antibody (Supplementary Fig S2E and F). The results obtained by ICC could be confirmed by Western blot analysis of cytoplasmic and nuclear extracts (Fig2G). Taken together, our results demonstrate that Nova1 and Ncoa3 are expressed in both the nuclear and cytosolic compartments of post-mitotic hippocampal neurons at times of synapse development (7–14 DIV).
Figure 2.
Nova1 and Ncoa3 are expressed in developing rat hippocampal neurons in culture
- A, B Western blot analysis of Nova1 (A) and Ncoa3 (B) proteins using whole-cell extracts harvested from FUDR-treated (B) or non-treated (A) rat hippocampal neurons at the indicated days of in vitro (DIV) culture. α-tubulin or β-actin was used as a loading control.
- C–F Immunocytochemistry analysis of Nova1 (C, D) and Ncoa3 (E, F) expression (red) in hippocampal neuron cultures at 14 DIV. MAP2 staining (green) was used to visualize neurons and Hoechst staining (blue) to visualize nuclei. (D, F) Magnification of insert depicted in (C) or (E), respectively. For simplicity, only the Nova1 (D) or Ncoa3 (F) signal is shown in grayscale. Cell and nuclear outlines derived from MAP2 and Hoechst staining, respectively, are shown in yellow. Scale bar = 10 μm.
- G Western blot analysis of Nova1 and Ncoa3 using whole-cell lysates (input), cytoplasmic (cyt) or nuclear (nuc) fractions from FUDR-treated rat hippocampal neurons at seven DIV. α-tubulin was used as a cytoplasmic and HDAC2 as a nuclear marker protein.
Source data are available online for this figure.
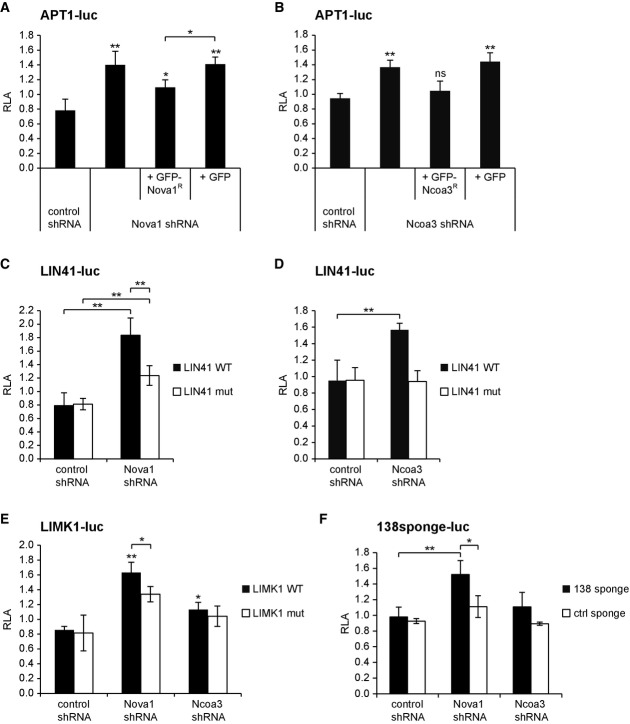
We next investigated whether Nova1 and Ncoa3 are required for the repressive function of endogenously expressed miRNAs in rat hippocampal neurons. To this end, we constructed a series of luciferase reporter plasmids consisting of the 3′UTRs of mRNAs targeted by let-7 (LIN41 3′UTR), miR-138 (APT1 3′UTR), or miR-134 (LIMK1 3′UTR). Importantly, these miRNAs are endogenously expressed in post-mitotic neurons and were previously shown to regulate neuronal morphogenesis via their respective mRNA targets (Schratt et al, 2006; Siegel et al, 2009; Huang et al, 2012). In addition, we cloned luciferase reporters with mutated miRNA-binding sites in order to account for potential miRNA-independent effects. Efficient knockdown of Nova1 and Ncoa3 in rat neurons was achieved by shRNA vectors containing conserved siRNA sequences used in the screen (Supplementary Fig S3A and B). Both Nova1 and Ncoa3 knockdown significantly increased the expression of reporter constructs containing a wild-type APT1 (Fig3A and B), LIN41 (Fig3C and D), and LIMK1 3′UTR (Fig3E). The stimulatory effect of Nova1 and Ncoa3 shRNAs was specific, since reintroduction of shRNA-resistant GFP-fusion proteins at least partially restored basal APT1-luc expression (Fig3A and B). Mutation of the two let-7-binding sites in the context of the LIN41 3′UTR (Fig3C and D) or of the miR-134-binding site in the context of the LIMK1 3′UTR (Fig3E) largely abolished Nova1 and Ncoa3 shRNA-mediated upregulation of the reporters, suggesting that the effects are to a large extent miRNA-dependent. The miR-134 target UBE3A-luc was selectively upregulated by the Nova1, but not by the Ncoa3 shRNA (Supplementary Fig S3C). Similar observations were made for the miR-124 target IQGAP1 (Lim et al, 2005) and another let-7 target, HMGA2 (Nishino et al, 2008) (Supplementary Fig S3D and E). Finally, repression of a reporter construct containing multiple sequential miR-138-binding sites, but lacking a specific 3′UTR context (“miR-138 sponge-luc”), was specifically relieved by Nova1 knockdown (Fig3F). In addition, Nova1 overexpression significantly reduced the expression of APT1 and UBE3A-luc reporters in neurons (Supplementary Fig S3F and G). Collectively, these results suggest that Nova1 is a general stimulator of neuronal miRNA activity, whereas Ncoa3 regulates a specific subset of neuronal miRNA targets depending on the 3′UTR sequence context.
Figure 3.
Nova1 and Ncoa3 are required for the repressive activity of a subset of neuronal miRNAs
- A–F Luciferase reporter gene assays performed in rat cortical neurons (14 DIV) (rat hippocampal neurons in (D)) that had been transfected at 11 or 12 DIV with the indicated 3′UTRs fused to the luc gene of pGL4 together with the given shRNA expression construct. In (A, B), additional shRNA-resistant (R) expression constructs for Nova1 and Ncoa3, respectively, or the empty expression vector (GFP) were co-transfected. Relative luciferase activity (RLA) represents the ratio of firefly reporter activity to Renilla control reporter activity normalized to a basal condition without shRNA expression vector. Values are the mean ± standard deviation from at least three independent experiments. One-way ANOVA: P < 0.0001 (A, C, E), P < 0.005 (B, D, F). Tukey’s HSD (A–D) and unpaired t-test (E, F): *P < 0.05, **P < 0.01.
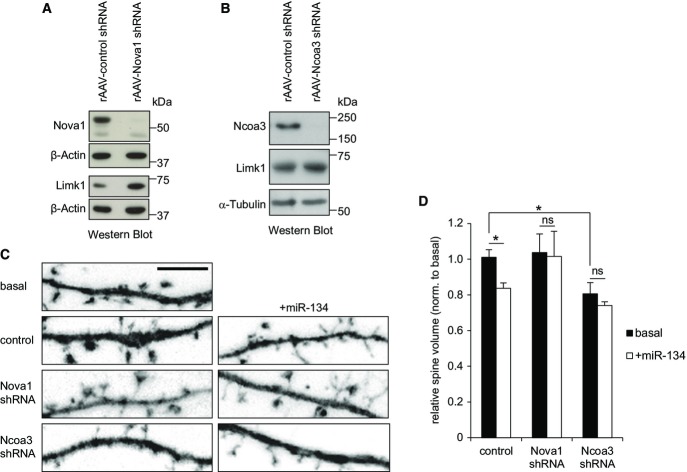
We next examined whether the observed regulation of miRNA reporter genes by Nova1 and Ncoa3 translated into alterations in endogenous miRNA target protein levels in neurons. Therefore, we focused on the regulation of Limk1 by miR-134, which is important during dendritic spine morphogenesis (Schratt et al, 2006). We observed increased levels of Limk1 in hippocampal neurons infected with recombinant adeno-associated virus (rAAV) expressing Nova1 shRNA compared to control shRNA-infected cells (Fig4A; Supplementary Fig S4A and B). Similarly, rAAV-mediated Ncoa3 knockdown was accompanied by significantly elevated Limk1 protein levels (Fig4B; Supplementary Fig S4C–E). Taken together, both Nova1 and Ncoa3 are required for efficient repression of the miR-134 target Limk1, consistent with a positive regulation of miR-134 activity by these RBPs. To directly examine the functional significance of Nova1- and Ncoa3-dependent Limk1 regulation, we investigated whether these proteins are involved in spine shrinkage following miR-134 overexpression (Schratt et al, 2006). As expected, miR-134 overexpression led to a significant reduction in average spine volume in control cells, an effect that was almost completely abolished in the presence of both the Nova1 and Ncoa3 shRNA (Fig4C and D). These results indicate that the positive regulation of miR-134 activity by Nova1 and Ncoa3 is functionally relevant during spine morphogenesis.
Figure 4.
Nova1 and Ncoa3 are required for miR-134-mediated repression of Limk1 and spine morphogenesis
- A, B Western blot analysis of Nova1, Ncoa3, and Limk1 protein in whole-cell extracts from hippocampal neurons (14 DIV) that had been infected with rAAV expressing the indicated shRNAs at four DIV. β-actin or α-tubulin was used as a loading control.
- C Representative dendritic segments with dendritic spines from hippocampal neurons (19 DIV) transfected with the indicated shRNA expression construct (empty plasmid served as a control) in the presence or absence of miR-134 duplex RNA (+miR-134, 20 nM) on 13 DIV. Scale bar = 5 μm.
- D Quantification of the average relative spine volume of hippocampal neurons from (C). Values were normalized to a basal condition without shRNA expression vector and represent averages ± standard deviation from three independent experiments (each including six cells per condition). *P < 0.05 (unpaired t-test).
Source data are available online for this figure.
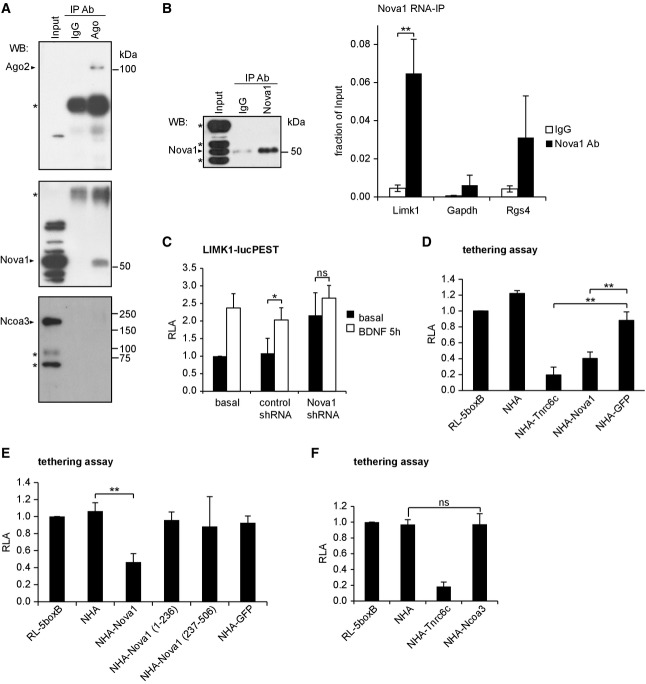
We next investigated the mechanisms underlying the miRNA regulatory function of Nova1 and Ncoa3. Our results from reporter assays suggest that Nova1 regulates the expression of miRNA targets independent of the 3′UTR context (Fig3). This raises the question whether Nova1 is directly associated with the neuronal miRISC. To address this, we studied a potential interaction between Nova1 and Ago using co-immunoprecipitation (co-IP) assays in rat hippocampal lysates. Western blot analysis demonstrated efficient pull-down of Ago proteins using a mouse monoclonal anti-pan-Ago antibody (Fig5A, upper panel). Importantly, we were able to specifically detect Nova1 in pan-Ago IPs, but not in control IgG IPs (Fig5A; Supplementary Fig S5A, middle panels). The interaction of Nova1 with Ago was weak, since only 2% of Nova1 present in the input could be recovered by Ago IP (Supplementary Fig S5B). The amount of co-precipitated Nova1 was unaltered in the presence of RNase (Supplementary Fig S5C and D), suggesting that Nova1 and Ago reside in a protein complex. To further confirm an interaction of Nova1 with endogenous miRNA targets, we performed RNA immunoprecipitation (RIP) experiments (Fig5B). A protocol was chosen that preserves protein–protein interactions, so that indirect Nova1 target mRNA interactions that are mediated by associated RBPs (e.g., Ago) could also be detected. Limk1 mRNA, in contrast to GAPDH mRNA, was significantly enriched in Nova1-RIP samples compared to IgG-RIP controls, suggesting a specific interaction between Nova1 and endogenous Limk1 mRNA (Fig5B). The previously reported Nova1 target Rgs4 served as a positive control (Licatalosi et al, 2008). We have previously shown that the miR-134-dependent repression of Limk1 is reversed by BDNF stimulation (Schratt et al, 2006). Having shown that Nova1 interacts with Ago and Limk1 mRNA, we asked whether Nova1 was involved in BDNF-regulated Limk1 synthesis. We observed that the stimulatory effect of BDNF on a destabilized Limk1-luc reporter (Limk1-lucPEST) was occluded by Nova1 knockdown (Fig5C), consistent with a role of Nova1 in the dynamic regulation of the Limk1-associated miRISC in response to BDNF. To get further mechanistic insight into the gene regulatory function of Nova1 within miRISC, we performed tethering assays (Pillai et al, 2005). Hereby, a protein of interest is artificially recruited to target RNAs by virtue of an N-terminal fusion of the lambda-phage N-peptide that allows binding to stem-loop sequences (boxB) present in the 3′UTR of a reporter RNA. This allowed us to study potential gene regulatory functions of Nova1 on target RNAs independent of the RNA sequence context and the interaction with Ago proteins. When NHA-Nova1 is tethered in this way downstream of a luciferase reporter gene, we observed a significant reduction in luciferase activity compared to a NHA-GFP control construct (Fig5D). The magnitude of repression was comparable to tethering NHA-Tnrc6c, a known miRISC component. Thus, Nova1 is able to repress mRNA translation when recruited to target RNAs, consistent with a function downstream of Ago. The use of deletion constructs further revealed that both the Nova1 N-terminus (containing the NLS, KH1 and KH2 domains) and C-terminus (containing the NES and KH3 domain) are required for the repressive activity of Nova1 (Fig5E). Importantly, all NHA-fusion proteins were expressed to comparable levels and localized in hippocampal neurons as expected (Supplementary Fig S5F and G). Nova1 tethering was further sufficient to reduce reporter gene expression in HEK293 cells (Supplementary Fig S5H). Together, our results suggest that Nova1 is part of neuronal miRISC and possesses the ability to repress mRNA translation independent of its sequence-specific RNA-binding activity.
Figure 5.
Nova1 and Ncoa3 stimulate neuronal miRNA function by different mechanisms
- A Representative co-immunoprecipitation experiment in hippocampal lysates from adult rat using a pan-Ago antibody. Upper panel: anti-Ago2 Western blot demonstrates specific pull-down of Ago2 in pan-Ago IP. Lower panel: anti-Nova1 Western blot demonstrates specific co-precipitation of Nova1 (52 kDa) and Ago proteins. *Non-identified cross-reactive proteins.
- B RNA immunoprecipitation (RIP) in adult rat hippocampus using an anti-Nova1 antibody. Left: specific IP of Nova1 with the applied antibody was confirmed by Western blot analysis. *Non-identified cross-reactive proteins. Right: qPCR for the indicated transcripts is presented as the ratio of mRNA present in Nova1 or control IgG IPs compared to the input material. Data is the average ± standard deviation from three independent experiments. **P < 0.005 (unpaired Student’s t-test).
- C Luciferase assay in cortical neurons (five DIV) transfected with a reporter gene which harbors the Limk1 3′UTR fused to a destabilized firefly ORF (Limk1-lucPEST) and with the indicated shRNAs. Neurons were stimulated with BDNF (100 ng/ml) or mock-treated for 5 h before processing for luminometry. Values were normalized to a co-transfected Renilla reporter and the basal condition without shRNA was set as 1. Data is the average ± standard deviation of four independent experiments. *P < 0.05 (unpaired t-test). ns: not significant.
- D–F Luciferase assay in cortical neurons (five DIV) transfected with RL-5boxB (tethering plasmid) together with plasmids expressing the indicated NHA-fusion proteins. Values were normalized to a co-transfected firefly reporter, and a basal condition without additional NHA protein was set as 1. Data is the average ± standard deviation of four independent experiments. One-way ANOVA: P < 0.0001 (D, F), P < 0.005 (E); Tukey’s HSD test: **P < 0.01; ns: not significant.
Source data are available online for this figure.
In contrast to Nova1, Ncoa3 is involved in the regulation of a specific subset of miRNA target interactions (Fig3). Moreover, Ncoa3 did not interact with Ago based on our co-IP experiments (Fig5A; Supplementary Fig S5A, lower panels), and tethering of Ncoa3 to a target RNA was not sufficient to cause translational repression (Fig5F; Supplementary Fig S5F). Together, these results argue against a direct function of Ncoa3 within neuronal miRISC.
Given that Nova1 and Ncoa3 are required for the function of neuronal miRNAs that play important roles in neuronal morphogenesis (Fig3), we decided to assess their role in dendrite and spine development. Nova1 knockdown in hippocampal neurons (12 DIV) under basal conditions had no significant effect on dendritic complexity or spine morphogenesis (Fig4D; Supplementary Fig S6A and B). In contrast, Ncoa3 knockdown resulted in a highly significant increase in dendritic complexity, which was quantified by Sholl analysis (Fig6A–C). Increased dendritic complexity was rescued by molecular replacement with Ncoa3-GFPR, but not with a control vector (unmodified pEGFP_N1), demonstrating that the phenotype is caused by a specific reduction of Ncoa3 expression (Fig6A–C). Expression and shRNA resistance of Ncoa3R-GFP were confirmed by Western blotting (Supplementary Fig S6C). Therefore, we uncovered a novel function of Ncoa3 in neuronal morphogenesis and decided to investigate the underlying mechanisms in more detail.
Figure 6.
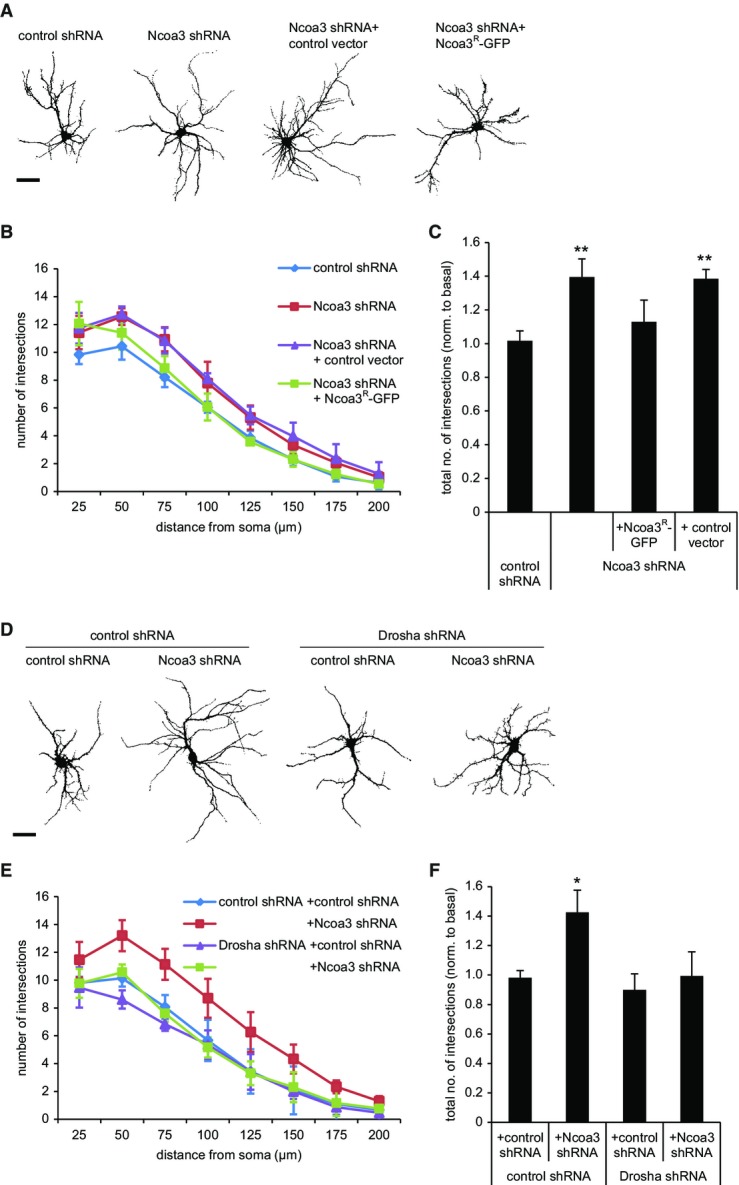
Ncoa3 regulates dendritic complexity in rat hippocampal neurons
- A Representative traces of rat hippocampal pyramidal neurons (12 DIV) that had been transfected with GFP and the indicated shRNA or expression vectors at six DIV. Ncoa3R: Noca3-shRNA resistant. Scale bar = 50 μm.
- B Sholl analysis of hippocampal neurons from (A). The average number of intersections with concentric circles drawn around the cell soma at distances from 25 to 200 μm is shown. Values are the average ± standard deviation from three independent experiments (in each 10–12 cells per condition).
- C Sholl analysis as in (B), except that the total number of intersections was calculated for each condition. Values were normalized to a basal condition without shRNA. Data is the average ± standard deviation of three independent experiments. One-way ANOVA: P = 0.0029, Tukey’s HSD test: **P < 0.01 (to control shRNA condition).
- D Representative traces of rat hippocampal pyramidal neurons (12 DIV) that had been transfected with GFP and the indicated shRNA vectors. Scale bar = 50 μm.
- E, F Results from Sholl analysis performed as in (B, C). Data is the average ± standard deviation of four independent experiments. One-way ANOVA: P = 0.0037; Tukey’s HSD test: *P < 0.05 (to control shRNA condition).
If the observed regulation of miRNA activity by Ncoa3 (Fig3) was functionally important, one would expect that Ncoa3 knockdown should not cause phenotypic alterations in the absence of miRNAs. To test this hypothesis, we combined the Ncoa3 shRNA with knockdown of Drosha, a component of the microprocessor that is crucial for the biogenesis of the vast majority of miRNAs (Gregory et al, 2004). Efficient knockdown of Drosha in neurons was verified by Western blotting (Supplementary Fig S6D) and qPCR (Supplementary Fig S6E). We could also confirm a reduction in the levels of several mature miRNAs upon Drosha knockdown (Supplementary Fig S6F). Using dendritic complexity as readout, we found that Ncoa3 shRNA was no longer able to increase dendritic complexity in the absence of Drosha (Fig6D–F). We therefore conclude that the dendritic phenotype caused by Ncoa3 knockdown is due to aberrant miRNA function in these neurons.
In addition, knockdown of Ncoa3 significantly reduced the average dendritic spine volume (Figs4D and7A and B), which was rescued by co-expression of Ncoa3R-GFP (Fig7A and B). We were interested in whether the morphological phenotypes observed upon Ncoa3 knockdown translated into altered excitatory synapse function. Thus, we measured miniature excitatory postsynaptic currents (mEPSCs) using patch-clamp electrophysiological recordings (Fig7C). Consistent with our previous observation that Ncoa3 knockdown results in reduced spine volume, we detected reduced average mEPSC amplitudes (Fig7D and E), but not frequencies (Fig7F), in Ncoa3 shRNA-transfected neurons compared to control conditions. Therefore, Ncoa3 knockdown affects both morphological and electrophysiological properties of excitatory synapses of hippocampal neurons.
Figure 7.
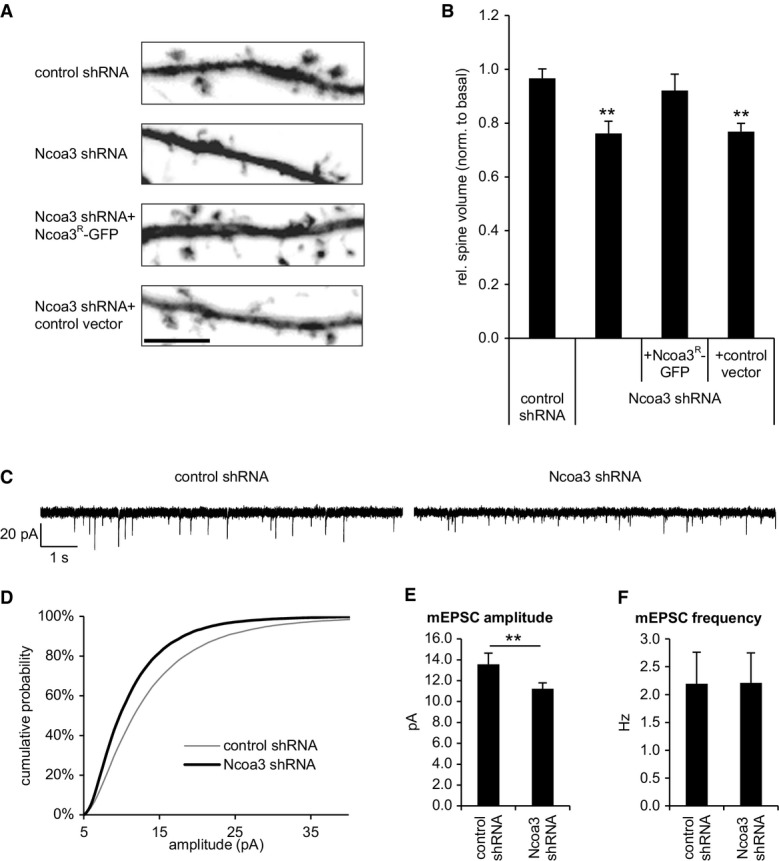
Ncoa3 regulates dendritic spine size and mEPSC amplitudes in rat hippocampal neurons
- A Representative dendritic segments with dendritic spines from hippocampal neurons (19 DIV) transfected with GFP and the indicated shRNA or expression vectors at 13 DIV. Scale bar = 5 μm.
- B Quantification of the average relative spine volume of hippocampal neurons from (A). Values were normalized to a basal condition without shRNA and represent the average ± standard deviation from three independent experiments (in each six cells per condition). One-way ANOVA: P = 0.0009; Tukey’s HSD test: **P < 0.01 (to control shRNA condition).
- C–F Dissociated hippocampal neurons were transfected with indicated shRNA expression vectors on 12–13 DIV, and mEPSCs were measured on 18–21 DIV. (C) Exemplary traces from representative recordings. (D) Cumulative probability plot distribution of mEPSC amplitudes; P < 10−6 (Kolmogorov–Smirnov test). (E) Average mEPSC amplitudes ± standard error, **P < 0.01 (unpaired t-test). (F) Average mEPSC frequency ± standard error. Cells were collected from five independent experiments (control shRNA: n = 18; Ncoa3 shRNA: n = 19; one Ncoa3 shRNA cell was qualified as outlier).
Given the well-documented function of Ncoa3 as transcriptional co-activator, we hypothesized that Ncoa3 promotes the expression of gene(s) that play important roles in miRNA biogenesis and/or activity. To address this hypothesis, we isolated RNA from hippocampal neurons that had been infected with either rAAV expressing Ncoa3 shRNA or a control shRNA (Supplementary Fig S7A). We validated a reduction in Ncoa3 mRNA levels in Ncoa3 shRNA compared to control shRNA-infected neurons (Fig8A). We subsequently subjected isolated RNA to comparative microarray hybridizations using the Affymetrix platform Rat Gene 2.0 ST, which covers a total of 36,685 different transcripts. Hybridization signals from the individual arrays were strongly correlated, demonstrating high reproducibility of the experiments (Supplementary Fig S7B). In total, we identified 100 annotated transcripts that were differentially expressed between Ncoa3 shRNA and control shRNA conditions (cutoff 1.3-fold, P < 0.05, n = 3), among which 68 were downregulated and 32 were upregulated upon Ncoa3 knockdown (Fig8B, complete expression analysis is provided in Supplementary Dataset S3). Intriguingly, among the 15 most downregulated transcripts, we identified the miRISC core component Ago2 and, as expected, Ncoa3 itself (Fig8B, bold). Ncoa3-dependent regulation of Ago2 and three other candidates identified by microarray (Cyb5r3, Map9, and Spast) was independently confirmed by qPCR (Fig8C). In contrast, several other components of miRNA regulatory complexes were not affected by Ncoa3 knockdown (Supplementary Fig S7C). Since downregulation of Ago2 could in principle explain elevated expression of a subset of miRNA targets in Ncoa3 knockdown cells (Fig3; Supplementary Fig S3), we decided to study the regulation of Ago2 in more detail. Using Western blot analysis, we further observed a robust downregulation of Ago2 protein upon infection of neurons with rAAV-expressed Ncoa3 shRNA (Fig8D; Supplementary Fig S7D). To interrogate whether Ncoa3 promotes Ago2 transcription by directly binding to the Ago2 promoter, we performed chromatin immunoprecipitation (ChIP) assays. Ncoa3-ChIP specifically enriched DNA fragments corresponding to the promoter regions of Ago2 and two other Ncoa3-regulated genes (Map9 and Spast) but not Cyb5r3 or the β-globin control (Fig8E; Supplementary Fig S7E and F). The specificity of the Ncoa3-ChIP was further confirmed by the reduced recovery of Ago2, Map9, and Spast promoter fragments from Ncoa3 knockdown neurons. Next, we tested whether Ncoa3 was involved in the regulation of Ago2 transcription using luciferase reporter assays. Therefore, we inserted a fragment of the mouse Ago2 promoter upstream of a minimal promoter driving the luciferase reporter gene. This fragment contains a conserved motif that could serve as a retinoic acid receptor alpha (RXRA)-binding site (JASPAR database; Supplementary Fig S7G). Ncoa3 was required for the induction of this reporter by retinoic acid (RA), suggesting that Ncoa3 functions as a co-activator of RXRA in the context of the Ago2 promoter (Fig8F). Taken together, our experiments establish Ago2 as a direct transcriptional target gene of Ncoa3 in neurons.
Figure 8.
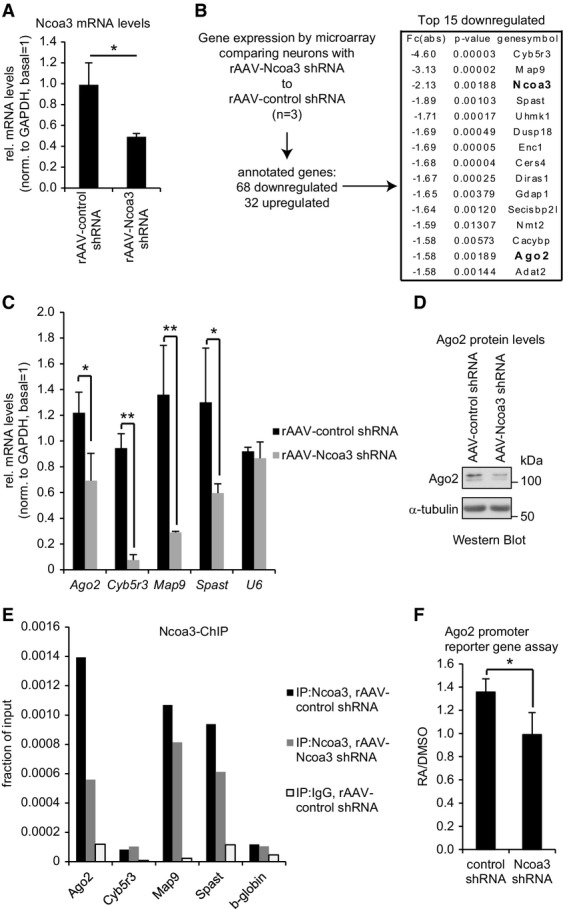
Microarray analysis identifies Ago2 as an Ncoa3-regulated transcript
- qPCR analysis of Ncoa3 mRNA levels in hippocampal neurons (14 DIV) that had been infected with the indicated rAAV-expressed shRNAs at four DIV. The ratio of average Ncoa3 levels compared to the internal GAPDH control is shown after normalization to a non-infected basal condition. Data is the average ± standard deviation of three independent experiments. *P < 0.05 (unpaired t-test).
- Flow chart for the identification of Ncoa3-regulated genes by microarray in hippocampal neurons treated as in (A). Absolute fold changes (Fc(abs)), P-values (t-test), and corresponding gene symbols are given on the right for the top 15 downregulated transcripts. Ncoa3 and Ago2 are highlighted in bold.
- qPCR analysis of indicated Ncoa3-regulated transcripts in hippocampal neurons treated as in (A). The ratio of average candidate mRNA levels compared to the internal GAPDH mRNA control is shown after normalization to a non-infected basal condition. U6 was used as a negative control. Data is the average ± standard deviation of three independent experiments. *P < 0.05, **P < 0.01 (unpaired t-test).
- Western blot analysis of Ago2 protein in whole-cell extracts from hippocampal neurons treated as in (A). α-tubulin was used as a loading control.
- qPCR analysis of indicated Ncoa3-regulated genes from a representative Ncoa3-ChIP experiment performed in either control shRNA (black) or Ncoa3 shRNA (gray) infected neurons. As a specificity control, ChIP with an unrelated IgG was performed in control shRNA-transfected neurons (white). Values are presented as fraction of the respective input DNA used for ChIP.
- Average luciferase activity in retinoic acid (RA)- or mock (DMSO)-treated neurons transfected with an Ago2 promoter reporter gene together with the indicated shRNA constructs. Values are presented as the ratio from RA- to DMSO-treated neurons and represent the average ± standard deviation from three independent experiments. *P < 0.05 (unpaired t-test).
We were then prompted to test whether the functional role of Ncoa3 in miRNA regulation and neuronal morphogenesis is mediated via transcriptional control of Ago2. Therefore, we used a Flag/HA-Ago2 construct which was efficiently expressed in neurons and HEK293 cells as judged by ICC and Western blotting, respectively (Supplementary Fig S8A and B). The quantification of Ago2 levels after Flag/HA-Ago2 overexpression showed that total Ago2 was elevated in control neurons by 1.5-fold and that Ago2 was restored to normal levels in Ncoa3 knockdown neurons (Fig9A; Supplementary Fig S8C). We investigated further the involvement of Ago2 in Ncoa3-mediated miRNA function using the LIN41 3′UTR reporter assay. Importantly, co-expression of Flag/HA-Ago2 in the context of the Ncoa3 shRNA nearly restored reporter expression to basal levels, demonstrating that Ago2 downregulation is required for the loss in let-7 activity in response to Ncoa3 knockdown (Fig9B). Importantly, expression of Flag/HA-Ago2 alone had no repressive activity (Fig9B), suggesting that Ago2 is indeed downstream of Ncoa3 in the regulation of miRNA activity. Finally, we determined the role of Ago2 in Ncoa3-dependent neuronal morphogenesis (Fig9C–E). Neurons expressing Ncoa3 shRNA showed a robust increase in dendritic complexity, which was almost completely rescued upon co-expression of Flag/HA-Ago2, but not the control vector. Similar to the observations from miRNA reporter gene assays, the expression of Flag/HA-Ago2 alone did not significantly affect dendritic complexity. Taken together, our results from miRNA reporter assays and neuromorphological analysis strongly suggest that the transcriptional regulation of Ago2 is responsible for Ncoa3-mediated regulation of miRNA activity and dendritogenesis.
Figure 9.
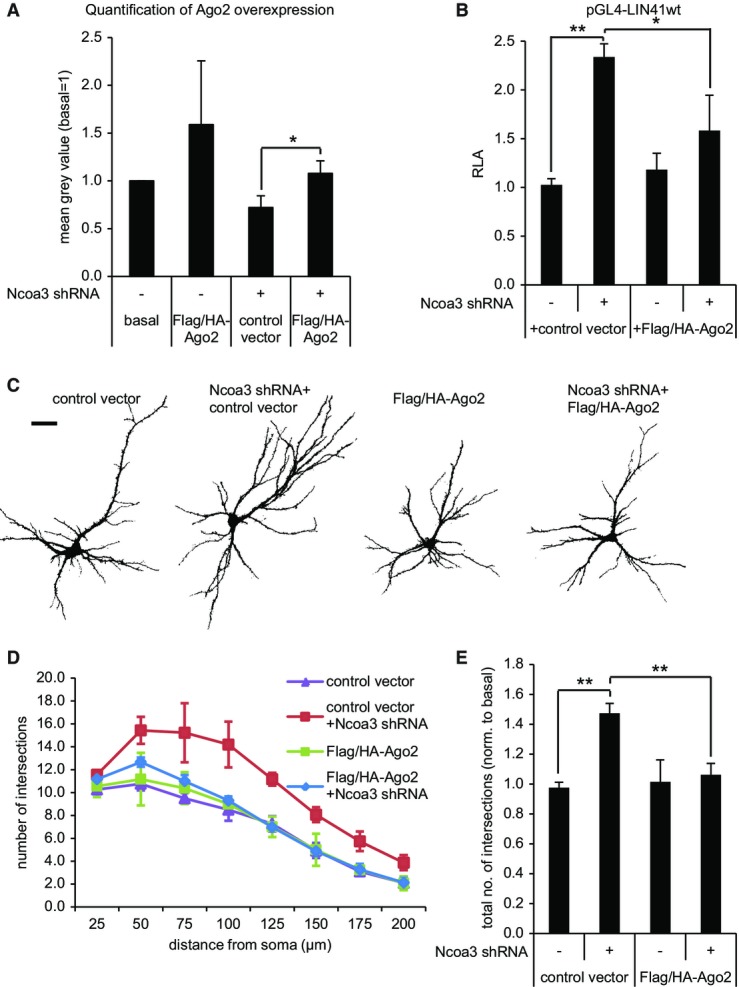
The regulation of Ago2 is required for Ncoa3-dependent miRNA activity and dendritogenesis
- A Quantification of Ago2 overexpression in hippocampal neurons (12 DIV) that have been transfected with GFP, the indicated shRNA expression vectors, and either empty Flag/HA (control vector) or Flag/HA-Ago2 after six DIV by anti-Ago2 immunocytochemistry. Measured was the average signal intensity in the neuronal soma (exemplary images in Supplementary Fig S8C), and values presented are the mean ± standard deviation from three independent experiments (in each 12 cells per condition). *P < 0.05 (pairwise t-test).
- B Reporter gene assay performed in rat hippocampal neurons (14 DIV) that had been transfected with pGL4-LIN41 WT 3′UTR together with the indicated shRNA vectors and either empty Flag/HA (control vector) or Flag/HA-Ago2 three days before. Relative luciferase activity (RLA) represents the ratio of firefly reporter to the internal Renilla control reporter activity normalized to basal condition without additional expression vector. Values are the mean ± standard deviation from three independent experiments. One-way ANOVA: P < 0.0003; Tukey’s HSD test: **P < 0.01.
- C Representative traces of neurons (12 DIV) that have been transfected as described in (A). Scale bar = 50 μm.
- D, E Results from Sholl analysis performed as in Fig6 with neurons treated as in (A). One-way ANOVA: P = 0.0005; Tukey’s HSD test: **P < 0.01.
Discussion
In this study, we present the first large-scale RNAi-based functional screen designed to identify RBPs that modulate miRISC activity in primary neurons. In addition to previously reported miRISC factors (Trnc6c, Ddx6/RCK/p54), we uncovered three novel regulators of miRNA activity: Nova1, Ncoa3, and Ewsr1. The fact that these regulators were missed in previous screens (Chu & Rana, 2006; Eulalio et al, 2007) suggests that neurons employ specific regulatory mechanisms which involve both neuron-specific (such as Nova1) and ubiquitously expressed RBPs (such as Ncoa3 and Ewsr1) to control miRNA activity. In general, our discovery of cell-type specific regulators of miRNA function underscores the need for experiments that address the function and regulation of miRNAs in their natural cellular context.
Nova1 as a novel component of neuronal miRISC
Nova1 was originally identified as an autoantigen in paraneoplastic opsoclonus myoclonus ataxia (POMA), a disorder associated with breast cancer and motor dysfunction (Buckanovich et al, 1993). Later, it was shown that Nova1 is a sequence-specific RNA-binding protein that functions in pre-mRNA splicing (Jensen et al, 2000). Two recent studies indicated that Nova proteins (Nova1 and Nova2) might also have regulatory functions in the cytoplasm. First, CLIP analysis of mouse brain indicated that a fraction of cellular Nova binds to consensus sequence elements within 3′UTR and regulates alternative polyadenylation of target mRNAs (Licatalosi et al, 2008). Second, Nova was shown to co-localize with target RNAs, including GlyRa2 and GIRK2, in dendrites of spinal cord motor neurons (Racca et al, 2010). These reports are consistent with our results from cellular fractionation and immunocytochemistry experiments in hippocampal neurons (Fig2) and with a potential function of Nova1 in the regulation of miRISC activity in the cytoplasm. Surprisingly, although Nova1 associates with the miR-134 target Limk1 mRNA (Fig5B), we found that the modulatory activity toward miRISC does apparently not require sequence-specific interaction of Nova1 with miRNA target RNAs. This conclusion is based on our observations that Nova1 regulates miRNA reporters independent of the 3′UTR context (Fig3) and that artificial tethering of Nova1 to an mRNA is sufficient to induce translational repression (Fig5D). Moreover, Nova1 interacts with Ago in an RNA-independent manner (Supplementary Fig S5C and D), arguing that Nova1 contacts Ago via protein–protein interaction(s), either directly or via additional bridging proteins. Since Nova1 knockdown leads to a residual activation of reporters containing mutated miRNA-binding sites (Fig3), Nova1 might in addition regulate mRNA translation in a miRNA-independent manner. Importantly, Nova1 is also required for miRNA-dependent regulation of neuronal function, in particular miR-134-dependent spine morphogenesis. Since Nova1 deficiency by itself does not cause dendrite or spine abnormalities (Fig4D; Supplementary Fig S6), we speculate that Nova1 function is specifically required for dynamic activity-dependent changes in neuronal morphology, such as those occurring during synaptic plasticity. Our finding that Nova1 is involved in BDNF-dependent Limk1 expression (Fig5C) is consistent with this hypothesis. In this regard, it will be interesting to investigate the potential activity-dependent modifications of the Ago-Nova1 complex.
Ncoa3 as a positive regulator of Ago2 expression
Ncoa3 (SRC-3, AIB-1) is a member of the nuclear receptor co-activator (Ncoa) family of transcriptional co-activators consisting of Ncoa1–3 (also known as SRC1-3) (Leo & Chen, 2000; Dasgupta et al, 2014). In addition to its extensively characterized function in transcription, two reports suggest a post-transcriptional cytosolic function of Ncoa3 in non-neuronal cells (Yu et al, 2007; Long et al, 2010). Using microarray analysis, qPCR, ChIP, and reporter gene assays (Fig8), we could identify the miRISC core factor Ago2 as novel direct transcriptional target gene of Ncoa3. Together with our results from rescue experiments (Fig9), this provides strong evidence that Ncoa3 regulates miRNA function by activating Ago2 transcription in the nucleus. Although Ncoa3 does not interact with Ago (Fig5A) or miRNA target RNAs (data not shown), it is possible that Ncoa3 in addition modulates miRNA function by different mechanisms. Surprisingly, we observed that a relatively subtle reduction of Ago2 levels (30–40%) in Ncoa3 knockdown cells is sufficient to cause profound impairments in miRNA regulation and neuronal morphogenesis, which are rescued by expression of Flag/HA-Ago2. These findings are consistent with previous observations that Ago2 is a rate-limiting factor in RNAi and that titrating endogenous Ago2 levels by excessive supply of small interfering RNAs can affect cellular physiology (Grimm et al, 2006; Borner et al, 2013). In contrast to Nova1, Ncoa3 is only required for the repression of a specific subset of neuronal miRNA targets (Fig3; Supplementary Fig S3). This implies that miRNA targets display a differential susceptibility toward alterations in Ago2 levels. One possible explanation is that some specific miRNAs are preferentially loaded into Ago2. However, there are conflicting data concerning sequence-specific sorting of miRNAs into Ago proteins in mammalian cells (Burroughs et al, 2011; Dueck et al, 2012; Wang et al, 2012). Our observation that mRNAs targeted by the same miRNAs respond differently to Ncoa3 knockdown (e.g., the let-7 targets LIN41 and HMGA2, Fig3D; Supplementary Fig S3D) further argues against this possibility. Alternatively, the miRNA target site context could influence recruitment of specific Ago proteins to mRNAs. In this regard, genome-wide mapping of binding sites for different Ago proteins in conjunction with the interacting miRNAs would be highly informative.
Our results further suggest that the specific pool of miRNA targets affected by the Ncoa3-Ago2 pathway plays an important role in the inhibition of dendrite growth and/or branching. Therefore, interesting candidate Ncoa3-regulated miRNAs might be those with a reported inhibitory role in dendritogenesis, such as miR-137, miR-181a, miR-34a, and miR-375 (Abdelmohsen et al, 2010; Smrt et al, 2010; Agostini et al, 2011; Liu et al, 2013). However, a more systematic analysis of Ncoa3-regulated, Ago2-associated miRNAs and their targets is needed to fully characterize the physiologically important miRNA target interactions that affect neuronal morphogenesis downstream of Ncoa3. The lack of a dendritic phenotype in neurons in which miRNA function is generally perturbed, for example, due to Drosha or Nova1 knockdown, is surprising at first, but could be explained by a perturbed function of both dendrite growth-promoting (e.g., miR-132 (Magill et al, 2010)) and growth-inhibiting (see above) miRNAs. If inhibitory and stimulatory activities balance each other, no net effect on dendritogenesis would be expected upon a global loss of miRNAs.
In summary, we have identified Nova1 and Ncoa3 as novel miRISC regulators that stimulate miRNA activity by different mechanisms converging on Ago proteins. We speculate that signal-dependent regulation of these pathways could be involved in neuronal plasticity and possibly other cellular contexts.
Materials and Methods
Cell culture, transfection, and recombinant AAV production
Dissociated primary hippocampal and cortical neurons from embryonic day (E)18 Sprague–Dawley rats (Charles River Laboratories) were prepared and cultured as described (Schratt et al, 2004). Transfection of neurons was generally performed with Lipofectamine 2000 (Invitrogen) as described (Siegel et al, 2009).
Where indicated, the given amount of plasmid was electroporated to cortical neurons using the NucleofectorTM X-unit (Lonza) as described earlier (Siegel et al, 2009). For the time course experiment of Ncoa3, preparation of cytosolic and nuclear fractions as well as rAAV infections, the culture medium was supplemented with 10 μM of 5-fluorodeoxyuridine (FUDR) to inhibit glial proliferation.
HEK293 cells were cultivated at subconfluent density in MEM containing 10% FCS, 2 mM l-glutamine, and Pen/Strep (all from Gibco). Co-transfection of GFP-fusion constructs and siRNAs was carried out in this medium with Lipofectamine 2000 as described for neurons. For NHA-fusion constructs and rAAV production, HEK293 cells were transfected using the calcium phosphate method as described (Christensen et al, 2010). A titer of 1.8 × 106 IFU/ml was used to infect hippocampal neurons (four DIV).
RNAi screen experiment
Mouse cortical neurons were co-transfected in a 48-well format at five DIV with 62.5 ng of pGL3-UBE3A, 62.5 ng of pGL3-Renilla, 5 pmol of miR-134 duplex RNA, and 7.5 pmol of one synthetic siRNA derived from a custom siRNA library (Ambion) in a total volume of 250 μl using Lipofectamine 2000. Three individual siRNAs for 286 candidate genes were tested. Luciferase assay was performed 3 days after the transfection. Each condition was transfected in duplicates, and the entire screen was repeated three times.
Subcellular fractionation and protein extraction
After one wash with ice-cold PBS, hippocampal cultures were detached in HLB (10 mM KCl, 1 mM EDTA, 1 mM DTT, 10 mM Tris–HCl (pH 7.5), protease inhibitors (Roche)) containing 0.2% NP-40 for 10 min on a shaker at 4°C followed by lysis through 20 strokes with a Teflon potter. After an aliquot was taken as input, nuclei were pelleted at 800 g for 5 min at 4°C and the supernatant was recovered as cytosolic fraction. The nuclei were washed once with HLB containing 0.25 M sucrose. After 10× RIPA (0.5 M Tris pH 8.0, 1.5 M NaCl, 10% Triton X-100, 5% sodiumdeoxycholate, 0.5% SDS, 20 mM EDTA) was diluted to 1× in the input and the nuclear pellet was dissolved in 1× RIPA (containing protease inhibitors), lysis of these two fractions was enforced by sonication. All fractions were subsequently spun down by 16,000 g for 10 min at 4°C, and the respective supernatant was recovered as protein extract. Whole-cell extracts were prepared by lysis in 1× RIPA followed by sonication, centrifugation, and recovery of supernatants as above, or in lysis buffer (150 mM NaCl, 50 mM Tris pH 7,5, 1% Triton) containing protease inhibitors.
Co-immunoprecipitation
Dissected hippocampi from adult brain (E18) were transferred to a 15-ml potter and homogenized on ice by 14 strokes in lysis buffer (20 mM Tris pH 7,5, 150 mM NaCl, 1% NP-40, 2 mM EDTA) containing protease inhibitors (Roche) followed by 10-min centrifugation at 16,000 g at 4°C. For RNase treatment, RNase A (1:20; Ambion) was added to one half of the cell lysate. Lysates were incubated with either mouse anti-pan-Ago (1:200; 2A8, Millipore) or mouse IgG (SCBT) rotating overnight at 4°C. Protein G beads (25 μl per condition; Sigma) were equilibrated in lysis buffer, blocked for 2 h in BSA (1 mg/ml; NEB), washed, resuspended in lysis buffer, and incubated with lysate and antibody for 1 h at 4°C followed by four washes in lysis buffer (last two containing 350 mM NaCl).
RNA immunoprecipitation (RIP)
The hippocampi of an adult female rat were dissected and homogenized in a Dounce potter (20 strokes) in lysis buffer (10 mM Hepes (pH 7.5), 200 mM NaCl, 30 mM EDTA, 0.5% Triton X-100, 0.5 U/μl SUPERase inhibitor (Ambion), complete protease inhibitors (Roche)). Homogenate was then passed five times through a 26-G needle. After centrifugation at 70,000 g (4°C) for 20 min, the supernatant was supplemented with 100 μg/ml yeast tRNA (Sigma-Aldrich) and precleared with Protein G Dynabeads for 30 min at 4°C. After input collection, immunoprecipitation was performed by the addition of 10 μg antibody (rabbit anti-Nova1 (Abcam) or normal rabbit IgG (SCBT)) and 30 μl Protein G Dynabeads for 2 h at 4°C. The beads were washed five times with lysis buffer (0.1 U/ml SUPERase inhibitors and 10 μg/ml yeast tRNA) and once with lysis buffer without additives. Total RNA was extracted using the mirVana Isolation Kit (Ambion) and treated with TURBO DNase (Ambion).
Chromatin immunoprecipitation (ChIP)
Hippocampal neurons were infected with rAAV-expressed Ncoa3 shRNA or rAAV-expressed control shRNA (9 × 105 IFU/well) at four DIV. On 12 DIV, cells were treated with 1% formaldehyde for 10 min followed by the addition of 0.125 M glycine for 5 min. After one wash with cold PBS, cells were lysed (50 mM Hepes–KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.2% Na-deoxycholate, 0.1% SDS, and protease inhibitors (Roche)) by gently shaking at 4°C for 10 min, detached with a cell scraper, and sonicated with a Branson sonifier 250 for 24 cycles with 20 pulses (output 2, 90% duty cycle) and 1-min incubation on ice. After centrifugation at 8,000 g (4°C) for 5 min, the supernatant was precleared for 30 min at 4°C with Protein G Dynabeads (Life Technologies) and used for immunoprecipitation with anti-Ncoa3 (5E11, cell signaling) or normal rabbit IgG (Santa Cruz) antibodies (2 h at 4°C) which were previously coupled to Protein G Dynabeads for 30 min. Beads were washed twice with lysis buffer, four times with high salt buffer (20 mM Tris (pH 8), 1 M NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS), twice with Li wash buffer (10 mM Tris (pH 8), 250 mM LiCl, 1 mM EDTA, 1% NP-40, 1% Na-deoxycholate), and once with TE (20 mM Tris (pH 8), 1 mM EDTA) at 4°C for 5 min. The complexes were eluted in elution buffer (100 mM NaHCO3, 1% SDS) for 15 min at 30°C, and cross-links in the supernatant were reversed by the addition of NaCl (final concentration 370 mM) and incubation at 65°C for 4 h. The input was taken after preclearing, supplemented with proteinase K (10 μg/100 μl lysate from NEB), and incubated at 65°C for 4 h. The DNA was subsequently recovered by phenol/chloroform/isoamyl alcohol extraction (Carl Roth).
Western blot analysis
Western blotting was performed in principle as described (Siegel et al, 2009). Please see Supplementary Materials and Methods for further details.
Immunocytochemistry, microscopy, and image processing
Immunocytochemistry and confocal microscopy were in principle performed as described (Siegel et al, 2009). Please see Supplementary Materials and Methods for further details.
Analysis of neuronal morphology
For analysis of dendritic complexity, random pyramidal neurons (12 DIV) were imaged in epifluorescence mode and analyzed by counting intersections of dendrites with circles of 25 μm incremental radii (range: 25–200 μm) around the soma. The total number of intersections was obtained by summing up intersections between dendrites and circles at each distance. Exemplary traces of analyzed cells are shown. Background signals were removed for clearer presentation. For dendritic spine analysis, confocal images (7 z-stacks at 0.4-μm interval) of pyramidal neurons (19 DIV) were projected to a single-plane image and the individual spine size was quantified by its mean gray value in a 2.2-μm2 circle. More than 100 spines per cell were measured and normalized to the cell’s mean gray values. Analysis was carried out blinded to the experimental conditions.
Luciferase assay
Luciferase assays in primary rat hippocampal or cortical neurons were performed with the Dual-Luciferase Reporter Assay System (Promega). Please see Supplementary Materials and Methods for further details.
DNA constructs and molecular cloning
A detailed description of cloning procedures and primer sequences is provided in the Supplementary Materials and Methods.
RNA extraction and reverse transcription–quantitative PCR (RT–qPCR)
RNA extraction and qPCR were performed according to the manufacturer’s instructions. See Supplementary Materials and Methods for further details.
Microarray
For microarray analysis, total RNA was extracted as described above. RNA quality was confirmed with the Experion System (Bio-Rad). Three independent RNA preparations, each consisting of rAAV-expressed control shRNA and rAAV-expressed Ncoa3 shRNA conditions, were submitted to the GeneCore Facility of EMBL, Heidelberg, Germany, for microarray analysis. Briefly, libraries prepared with the WT Expression Kit (Ambion) were analyzed on the Rat Gene 2.0 ST arrays (Affymetrix). The analysis was performed with Expression Console (Affymetrix) and GeneSpring (Agilent) softwares. In the latter, log2 values below the background control were removed leaving 11,206 transcript IDs for filtering by volcano plot with thresholds of 1.3-fold change and P < 0.05 (moderate t-test). Microarray data are deposited in the GEO database with accession number GSE69131.
Antibodies
A complete list of antibodies used in this study is provided in the Supplementary Materials and Methods.
Statistical analysis
For pairwise comparison, a two-tailed, unpaired Student’s t-test was carried out. For multiple conditions, one-way ANOVA was first applied (http://vassarstats.net/) followed by Tukey’s HSD test for comparing single conditions pairwise.
Acknowledgments
We thank D. Bartel, R. Darnell, W. Filipowicz, G. Meister, R. Pillai, and T. Tuschl for sharing reagents; G. Jarosch, E. Becker, R. Gondrum, U. Beck, and H. Kaiser for outstanding technical support; and A. Antoniou and R. Fiore for critical reading of the manuscript. J. Pistolic assisted with the microarray analysis. B. Honrath, L. Greifenberg, and S. Khudayberdiev generated reagents. This work was funded by grants from the DFG (SFB488, SFB593, SPP1738) and an ERC Starting Grant “Neuromir.”
Author contributions
PHS and JT designed, performed, and analyzed most of the experiments. GaS performed the initial RNAi screen (Fig1) and the Nova1 dendritogenesis assays (Supplementary Fig S5). AAA carried out and analyzed electrophysiological recordings (Fig7). FZ and SS contributed to Supplementary Figs S6 and S8, respectively. GeS supervised the study and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Information
Source Data for Supplementary Figure S1
Source Data for Supplementary Figure S2
Source Data for Supplementary Figure S3
Source Data for Supplementary Figure S4
Source Data for Supplementary Figure S5
Source Data for Supplementary Figure S6
Source Data for Supplementary Figure S7
Source Data for Supplementary Figure S8
Supplementary Dataset S1
Supplementary Dataset S2
Supplementary Dataset S3
Review Process File
Source Data for Figure 2
Source Data for Figure 4
Source Data for Figure 5
References
- Abdelmohsen K, Hutchison ER, Lee EK, Kuwano Y, Kim MM, Masuda K, Srikantan S, Subaran SS, Marasa BS, Mattson MP, Gorospe M. miR-375 inhibits differentiation of neurites by lowering HuD levels. Mol Cell Biol. 2010;30:4197–4210. doi: 10.1128/MCB.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, Forsythe ID, Young KW, Ventura A, Concepcion CP, Han YC, Candi E, Knight RA, Mak TW, Melino G. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci USA. 2011;108:21099–21104. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Borner K, Niopek D, Cotugno G, Kaldenbach M, Pankert T, Willemsen J, Zhang X, Schurmann N, Mockenhaupt S, Serva A, Hiet MS, Wiedtke E, Castoldi M, Starkuviene V, Erfle H, Gilbert DF, Bartenschlager R, Boutros M, Binder M, Streetz K, et al. Robust RNAi enhancement via human Argonaute-2 overexpression from plasmids, viral vectors and cell lines. Nucleic Acids Res. 2013;41:e199. doi: 10.1093/nar/gkt836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Posner JB, Darnell RB. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron. 1993;11:657–672. doi: 10.1016/0896-6273(93)90077-5. [DOI] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Suzuki H, Hayashizaki Y, Daub CO. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8:158–177. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M, Larsen LA, Kauppinen S, Schratt G. Recombinant adeno-associated virus-mediated microRNA delivery into the postnatal mouse brain reveals a role for miR-134 in dendritogenesis in vivo. Front Neural Circuits. 2010;3:16. doi: 10.3389/neuro.04.016.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Lonard DM, O’Malley BW. Nuclear receptor coactivators: master regulators of human health and disease. Annu Rev Med. 2014;65:279–292. doi: 10.1146/annurev-med-051812-145316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueck A, Ziegler C, Eichner A, Berezikov E, Meister G. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 2012;40:9850–9862. doi: 10.1093/nar/gks705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Saba R, Schratt G. MicroRNA function in the nervous system. Prog Mol Biol Transl Sci. 2011;102:47–100. doi: 10.1016/B978-0-12-415795-8.00004-0. [DOI] [PubMed] [Google Scholar]
- Fiore R, Rajman M, Schwale C, Bicker S, Antoniou A, Bruehl C, Draguhn A, Schratt G. MiR-134-dependent regulation of Pumilio-2 is necessary for homeostatic synaptic depression. EMBO J. 2014;33:2231–2246. doi: 10.15252/embj.201487921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Huang YW, Ruiz CR, Eyler EC, Lin K, Meffert MK. Dual regulation of miRNA biogenesis generates target specificity in neurotrophin-induced protein synthesis. Cell. 2012;148:933–946. doi: 10.1016/j.cell.2012.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, Lund AH, Perrakis A, Raz E, Agami R. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law WJ, Cann KL, Hicks GG. TLS, EWS and TAF15: a model for transcriptional integration of gene expression. Brief Funct Genomic Proteomic. 2006;5:8–14. doi: 10.1093/bfgp/ell015. [DOI] [PubMed] [Google Scholar]
- Leo C, Chen JD. The SRC family of nuclear receptor coactivators. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhao Z, Yang F, Gao Y, Song J, Wan Y. microRNA-181a is involved in insulin-like growth factor-1-mediated regulation of the transcription factor CREB1. J Neurochem. 2013;126:771–780. doi: 10.1111/jnc.12370. [DOI] [PubMed] [Google Scholar]
- Long W, Yi P, Amazit L, LaMarca HL, Ashcroft F, Kumar R, Mancini MA, Tsai SY, Tsai MJ, O’Malley BW. SRC-3Delta4 mediates the interaction of EGFR with FAK to promote cell migration. Mol Cell. 2010;37:321–332. doi: 10.1016/j.molcel.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill ST, Cambronne XA, Luikart BW, Lioy DT, Leighton BH, Westbrook GL, Mandel G, Goodman RH. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci USA. 2010;107:20382–20387. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee AE, Minet E, Stern C, Riahi S, Stiles CD, Silver PA. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol. 2005;5:14. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- Meskauskas A, Dinman JD. Ribosomal protein L5 helps anchor peptidyl-tRNA to the P-site in Saccharomyces cerevisiae. RNA. 2001;7:1084–1096. doi: 10.1017/S1355838201001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet I, Barbosa-Morais NL, Andrade J, Carmo-Fonseca M. Diversity of human U2AF splicing factors. FEBS J. 2006;273:4807–4816. doi: 10.1111/j.1742-4658.2006.05502.x. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Racca C, Gardiol A, Eom T, Ule J, Triller A, Darnell RB. The Neuronal Splicing Factor Nova Co-Localizes with Target RNAs in the Dendrite. Front Neural Circuits. 2010;4:5. doi: 10.3389/neuro.04.005.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci. 2004;24:9366–9377. doi: 10.1523/JNEUROSCI.1739-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, Hubel K, Dekker F, Hedberg C, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G, Saba R, Schratt G. microRNAs in neurons: manifold regulatory roles at the synapse. Curr Opin Genet Dev. 2011;21:491–497. doi: 10.1016/j.gde.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, Teng ZQ, Luo Y, Peng J, Bordey A, Jin P, Zhao X. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F, Izaurralde E. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 2003;13:319–327. doi: 10.1016/s0962-8924(03)00106-5. [DOI] [PubMed] [Google Scholar]
- Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Valluy J, Bicker S, Aksoy-Aksel A, Lackinger M, Sumer S, Fiore R, Wust T, Seffer D, Metge F, Dieterich C, Wohr M, Schwarting R, Schratt G. A coding-independent function of an alternative Ube3a transcript during neuronal development. Nat Neurosci. 2015;18:666–673. doi: 10.1038/nn.3996. [DOI] [PubMed] [Google Scholar]
- Wang D, Zhang Z, O’Loughlin E, Lee T, Houel S, O’Carroll D, Tarakhovsky A, Ahn NG, Yi R. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev. 2012;26:693–704. doi: 10.1101/gad.182758.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wu RC, O’Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, York B, Wang S, Feng Q, Xu J, O’Malley BW. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol Cell. 2007;25:765–778. doi: 10.1016/j.molcel.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chen QM. Far upstream element binding protein 1: a commander of transcription, translation and beyond. Oncogene. 2013;32:2907–2916. doi: 10.1038/onc.2012.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Ou AC, Cho A, Benz EJ, Jr, Huang SC. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol Cell Biol. 2008;28:5924–5936. doi: 10.1128/MCB.00560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Chiu H, Zinovyeva A, Ambros V, Chuang CF, Chang C. Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science. 2013;340:372–376. doi: 10.1126/science.1231321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Source Data for Supplementary Figure S1
Source Data for Supplementary Figure S2
Source Data for Supplementary Figure S3
Source Data for Supplementary Figure S4
Source Data for Supplementary Figure S5
Source Data for Supplementary Figure S6
Source Data for Supplementary Figure S7
Source Data for Supplementary Figure S8
Supplementary Dataset S1
Supplementary Dataset S2
Supplementary Dataset S3
Review Process File
Source Data for Figure 2
Source Data for Figure 4
Source Data for Figure 5