Abstract
Werner syndrome (WS) is a rare autosomal recessive disorder in humans characterized by premature aging and genetic instability. WS is caused by mutations in the WRN gene, which encodes a member of the RecQ family of DNA helicases. Cellular and biochemical studies suggest that WRN plays roles in DNA replication, DNA repair, telomere maintenance, and homologous recombination and that WRN has multiple enzymatic activities including 3′ to 5′ exonuclease, 3′ to 5′ helicase, and ssDNA annealing. The goal of this study was to map and further characterize the ssDNA annealing activity of WRN. Enzymatic studies using truncated forms of WRN identified a C-terminal 79 amino acid region between the RQC and the HRDC domains (aa1072–1150) that is required for ssDNA annealing activity. Deletion of the region reduced or eliminated ssDNA annealing activity of the WRN protein. Furthermore, the activity appears to correlate with DNA binding and oligomerization status of the protein.
Werner syndrome (WS1) is a rare autosomal recessive disorder characterized by early onset aging and genomic instability (1). WS patients carry mutations in the WRN gene, which encodes a member of the RecQ family of DNA helicases. There is growing evidence that WRN plays diverse roles in DNA repair, DNA replication, homologous recombination, and telomere maintenance (2, 3). WRN possesses 3′–5′ helicase, 3′–5′ exonuclease, DNA-dependent ATPase, and ssDNA annealing activities (4, 5). Recent studies also showed that WRN catalyzes DNA strand exchange (6). WRN helicase unwinds a wide variety of DNA substrates in vitro, including Holliday junction and forked DNA duplex structures (7). Additionally, unlike other RecQ helicases, WRN can unwind RNA–DNA heteroduplexes (8).
WRN is the only RecQ helicase with exonuclease activity, which allows it to digest dsDNA with 3′ to 5′ polarity. Preferred WRN DNA substrates include bubble structures, forked DNA duplexes, and Holliday junctions (33). The exonuclease domain is located in the N-terminal region and the helicase domain in the central region of the WRN protein. The C-terminal region includes the RQC (RecQ conserved) and the HRDC (helicase, RNase D Conserved) domains. The RQC domain contains a wing helix (WH) motif, which mediates binding to dsDNA and ssDNA. In addition, WRN contains a nucleolar-targeting sequence (10). In some WS cells, WRN is C-terminally truncated and lacks the nuclear localization signal (NLS). Although previous studies have not mapped any enzymatic function to the WRN C-terminal region, this region mediates both protein–protein interactions and DNA binding. WRN interacts physically and functionally with numerous proteins, including replication protein A (RPA), proliferating cell nuclear antigen (PCNA), Flap endonuclease 1 (Fen 1), DNA polymerase β, and telomere binding protein TRF2 (reviewed in ref 11).
Human cells possess five RecQ helicases: WRN, RecQ1 (RecQL), BLM, RecQ4, and RecQ5 (12). Defects in the activity or expression of BLM and RecQ4 proteins result in Bloom syndrome and Rothmund–Thomson syndrome, respectively. So far, no diseases have been associated with defects in RECQ1 and RECQ5. Helicase activity has been detected biochemically in all human RecQ homologues except RecQ4. All five human RecQ helicases have ssDNA annealing activity (6, 13–17), which in BLM and RecQ5β has been mapped to the poorly conserved C-terminal region. ATP suppresses the annealing activity of RecQ helicases. It has been proposed that ATP binding induces a conformational change in RecQ1, which, in turn, inhibits its ssDNA annealing activity (17).
This study demonstrates that the ssDNA annealing activity of WRN maps to the C-terminal 79 amino acid segment (aa1072–1150) between the RQC and the HRDC domains. Furthermore, the ssDNA annealing activity of WRN correlates with DNA binding and oligomerization status of the protein.
MATERIALS AND METHODS
Cloning and Expression of GST-WRN Fusion Proteins
Recombinant GST-tagged-WRN fragments WRN1–120, WRN239–499, WRN500–946, WRN949–1092, and WRN1072–1432 were constructed as described previously (18, 19). Recombinant 6xHis tagged-WRN fragments, WRN500–1104 (20), WRN1072–1432, and WRN1–368, were cloned and expressed as described previously (18). Fragments of the WRN C-terminal domain were expressed using the Gateway cloning system (Invitrogen). Briefly, the WRN fragments were generated by PCR (20). The PCR product was verified by sequencing and cloned into the pENTR-TEV-D-TOPO vector followed by recombination into bacterial expression plasmid pDEST15. Plasmids were propagated in DH5α during cloning and BL21 cells during expression. Purification was performed as described in ref 21 with some modifications; GST fusion proteins were purified from cell extracts using GST beads and eluted with 10 mM glutathione or 100 units of acTEV protease (Invitrogen) in 100 μL of elution buffer (100 mM Tris at pH 8.0, 120 mM NaCl, 5% glycerol, 0.2% Triton X-100, 300 mM LiSO4, and 5 mM DTT). The cloned human WRN gene was kindly provided by Dr. Junko Oshima (University of Washington Medical School, Seattle, WA).
ssDNA Annealing Activity
ssDNA annealing activity was measured using C80 and G80 oligonucleotides described previously (6), at a concentration of 0.1 nM each, one of which was 5′-32P-end-labeled. Reactions (20 μL) were carried out in 20 mM Tris/HCl at pH 7.5, 2 mM MgCl2, 40 μg/mL BSA, and 1 mM DTT for 15 min at 37 °C. Protein was added in 2-fold serial dilutions at concentrations ranging from 0.5 to 16 nM. Reactions were stopped by the addition of stop buffer (50 mM EDTA, 1% SDS and 50% glycerol) and immediately loaded onto 10% (w/v) native polyacrylamide gel. Gels were run in TBE for 2 h at room temperature at 200 V. Radiolabeled DNA was detected using the Typhoon Imager (GE Healthcare), and percentage of annealed oligonucleotide was quantified using ImageQuant software.
Gel Mobility Shift Assay
Radiolabeled oligonucleotide C80 (1.5 nM) was incubated with WRN in 40 mM TrisHCl at pH 7.5, 20 μg/mL BSA, 8% glycerol, 1 mM EDTA, and 10 mM NaCl for 20 min on ice. The protein–DNA complexes were resolved on a 4% (w/v) native polyacrylamide gel and run in TBE for 2 h at 4 °C at 200V.
Gel Filtration Analysis
A Superdex 200 10/30 GL (GE Healthcare) column was equilibrated in dilution buffer (20 mM Tris/HCl at pH 7.4, 2 mM MgCl2, 100 mM NaCl, and 1 mM DTT) and calibrated with the following protein standards: ferritin, catalase, adolase, albumin, ovalbumin, chymotrypsinogen A, ribonuclease A, and blue dextran 200 (GE healthcare). Samples were diluted to 150 μg/mL in 20 mM Tris/HCl at pH 7.5, 2 mM MgCl2, 100 mM NaCl, and 1 mM DTT and loaded onto the column. Proteins were separated using FPLC-AKTA express (GE healthcare). Column fractions were analyzed by SDS–PAGE followed by silver staining.
RESULTS
Mapping the ssDNA Annealing Activity of WRN
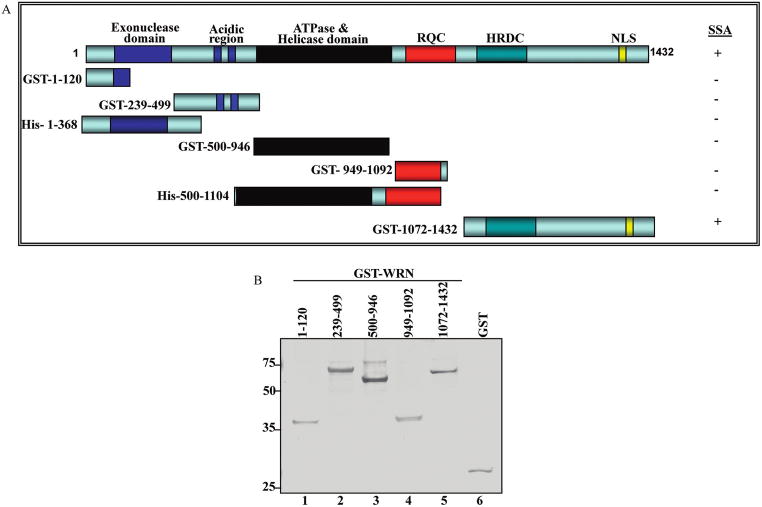
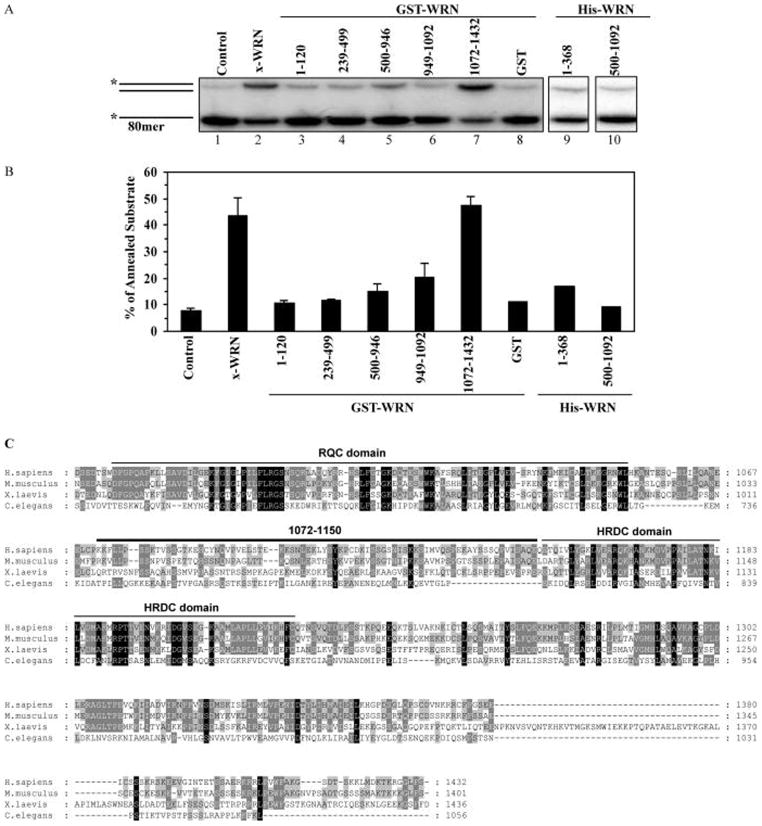
It has been suggested that ssDNA annealing activities map to the C-terminal regions of WRN and BLM proteins and demonstrated that truncation of the C-terminal HRDC domain inhibits the ssDNA annealing activity of BLM (13). Recent studies also showed that WRN and BLM have more efficient ssDNA strand annealing activity than Drosophila melanogaster RecQ5b, most likely because Drosophila’s RecQ5b has a shorter C-terminal region than WRN or BLM (6). This possibility was tested by constructing and characterizing deletion/truncation mutants of WRN tagged with GST or 6X-Histidine (6X-His) (Figure 1A). Selection of WRN fragments was based on proteolytic studies of WRN domain boundaries (19). Initially, five GST- and two 6X-His-tagged WRN fragments were overexpressed, purified, and tested for ssDNA annealing activity (Figures 1 and 2). The purity of the fragments was assessed by silver stain (Figure 1B). The ssDNA annealing activity was significantly above background only for full-length WRN and the GST-WRN fragment containing the C-terminal region, GST-WRN1072–1432. GST was included in the assay as a negative control (Figure 2A, lane 8). This result indicates that the WRN region 1072–1432 (Figure 2A, lanes 2 and 7, and Figure 2B) includes amino acids that are required for ssDNA annealing activity. This region of WRN lies immediately after the RQC domain and includes two conserved motifs: the HRDC domain and an NLS. However, the HRDC domain is not well conserved among RecQ helicases and is present in only two of the five human RECQ helicases, WRN and BLM (reviewed in ref 22). When the WRN amino acid sequence was aligned with RecQ helicases from mouse, Xenopus laevis, and Caenorhabditis elegans using ClustalW (23), weak sequence similarity (17%) was detected in the C-terminal region (Figure 2C). This low level of sequence homology argues against strong evolutionary pressure for functional conservation in the C-terminal region of WRN and other RecQ helicases.
Figure 1.
Recombinant WRN fragments used in this study. (A) Schematic diagram of WRN fragments. Fragments WRN1–368, WRN500–1092, and full length WRN were tagged with 6xHis; the remaining WRN fragments were tagged with GST. (B) GST-WRN fragments (50 ng/lane) were analyzed by SDS–PAGE. Protein bands were detected by silver staining.
Figure 2.
ssDNA annealing activity of WRN fragments. (A) ssDNA annealing assays were carried out as described in Materials and Methods. Full length WRN was used as a positive control, and GST was used as a negative control. (B) ssDNA annealing activity was quantified by PhosphorImager analysis, and values demonstrate the percentage of dsDNA formation. Data values are the mean of at least three independent measurements. (C) Multiple alignment of WRN homologues from Homo sapiens (AAF06121), Mus musculus (AAF64490), Xenopus laevis (O93530), and Caenorhabditis elegans (AAA68410) (C-terminal regions shown). The alignment was computed with clustalW and shaded using BioEdit. Black blocks indicate identity; dark gray, similarities between three sequences; light gray, similarities between two sequences. 1072–1150 region and RQC and HRDC domains are indicated.
Fine-Mapping WRN ssDNA Annealing Activity to a 79-Amino Acid Region between the RQC and HRDC Domains
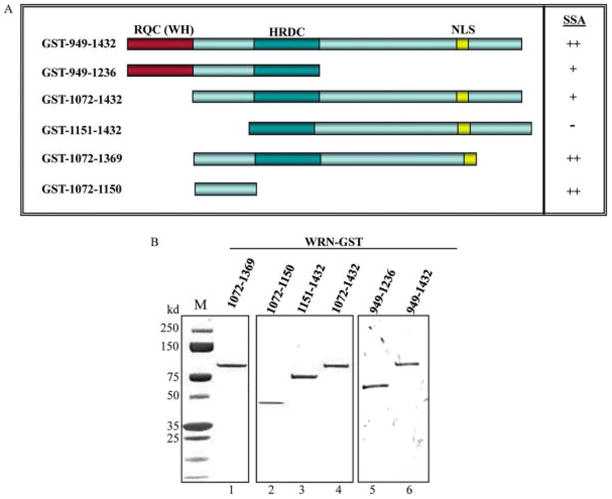
The region required for WRN ssDNA annealing activity was defined more precisely by constructing, overexpressing, and characterizing six additional GST-WRN truncation proteins (Figure 3A). The longest of these variants GST-WRN949–1432 includes the RQC domain, the region between the RQC and HRDC domain, the HRDC domain, and the NLS. Additional WRN variants were truncated C-terminally (aa949–1236), N-terminally (aa1072–1432; aa1151–1432), or both (aa1072–1369; aa1072–1150). The smallest WRN fragment (aa1072–1150) includes only the region between the RQC and HRDC domains. WRN942–1432 and WRN949–1236 fragments include the WH motif from the RQC domain, which has high DNA and protein binding capacities (11). The protein folding patterns of these WRN variants were predicted using the Profile Library Search engine FUGUE (24), and their purity was confirmed using SDS–PAGE followed by staining with SuperBlue reagent (Figure 3B).
Figure 3.
WRN C-terminal fragments. (A) A schematic diagram of WRN C-terminal fragments. All fragments were tagged with GST. The summary of the annealing results from Figure 4 is shown in A (right panel; ++, 50–90% annealing efficiency; +, <50% annealing efficiency; −, no annealing). (B) Purified WRN recombinant fragments were analyzed by SDS–PAGE (1 μg per lane) and visualized by staining with SuperBlue.
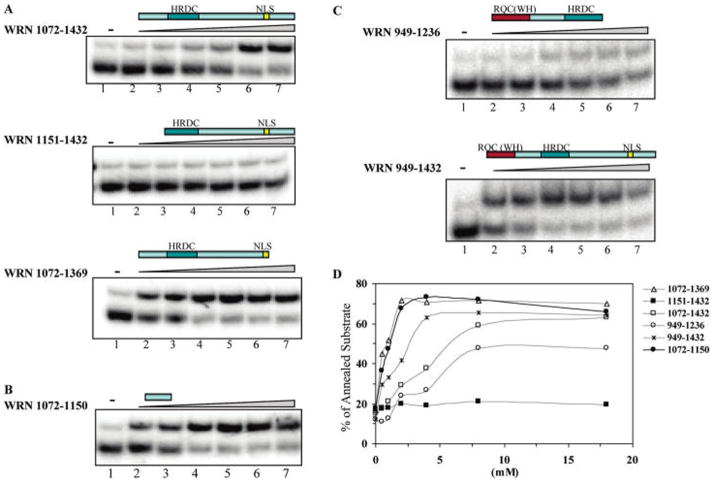
ssDNA annealing assays were carried out with variable amounts of WRN and WRN C-terminal truncations (Figure 4). The results show that WRN1072–1150 is sufficient for ssDNA annealing activity (Figure 4B) and deletion of this region strongly inhibits ssDNA annealing function (Figure 4A, middle panel). WRN1072–1432 (Figure 4A, upper panel) and WRN1072–1369 (Figure 4A, bottom panel) fragments containing that region, but truncated at opposing termini, have annealing activities. The WH motif in the RQC domain together with the intact C-terminal domain had ssDNA annealing activity (Figure 4C, bottom panel), whereas the WRN949–1236 fragment covering the WH motif with the HRDC domain had a slightly inhibitory effect on ssDNA annealing activity (Figure 4C and D). These results suggest that the region required for the ssDNA annealing activity lies between amino acids 1072 and 1150 (Figure 4B and D).
Figure 4.
ssDNA annealing activity of WRN C-terminal fragments. (A, B, and C) ssDNA annealing assays were performed as described in Figure 2, except protein was titrated in 2-fold serial dilutions from 0.5 to 16 nM. (D) ssDNA annealing activity was quantified by PhosphorImager analysis, and the percentage of dsDNA was calculated. Data values are the mean of at least two independent measurements.
Site-directed mutagenesis was used to identify specific amino acids responsible for the ssDNA annealing activity in WRN1072–1150. First, a computer algorithm ConSeq (25) was used to predict putative functionally important amino acids in this region. The amino acids identified were serine 1079, tyrosine 1112, lysine 1113, and lysine 1117. On the basis of the prediction of ConSeq, WRN1072–1432 fragments were generated with mutations of serine 1079 to alanine or leucine, tyrosine 1112 to alanine, lysine 1113 to alanine, and lysine 1117 to alanine. The serine 1079 to leucine variant was constructed because there is a naturally occurring single nucleotide polymorphism (SNP) variant in a human population in sub-Saharan Africa (25). ssDNA annealing assays showed that alanine-substituted and leucine-substituted WRN fragments were fully active (data not shown). Thus, additional studies are needed to identify specific residues or combinations of residues that are required for WRN ssDNA annealing activity.
Amino Acids 1072–1150 Are Required for ssDNA Binding Activity
E. coli and S. cerevisiae RecQ homologues preferentially bind ssDNA via their HRDC domains (26, 27). Previous studies indicate that the WRN C-terminal region binds dsDNA and has lower affinity to ssDNA (19). Here, binding of WRN truncations to an 80-mer ssDNA oligonucleotide (C80) was examined using gel mobility shift assay. The WRN1072–1432 fragment formed a DNA–protein complex with DNA substrate that either did not enter the gel or migrated a small distance into the gel (Figure 5, lanes 5–7). WRN1072–1150 formed smaller amounts of a similar complex (Figure 5, lanes 8–10). In contrast, WRN1151–1432 did not bind the ssDNA substrate (Figure 5, lanes 2–4). These results show that WRN1151–1432 is deficient in both ssDNA binding and ssDNA annealing activity (Figure 4A, middle panel). WRN949–1432 and WRN949–1236 (Figure 5, lanes 12–13 and lanes 14–15, respectively) bind ssDNA with high efficiency forming a protein–DNA complex that has greater electrophoretic mobility than the complexes formed by WRN1072–1432 and WRN1072–1150. This result may indicate that the WH motif of the RQC domain facilitates binding to ssDNA.
Figure 5.
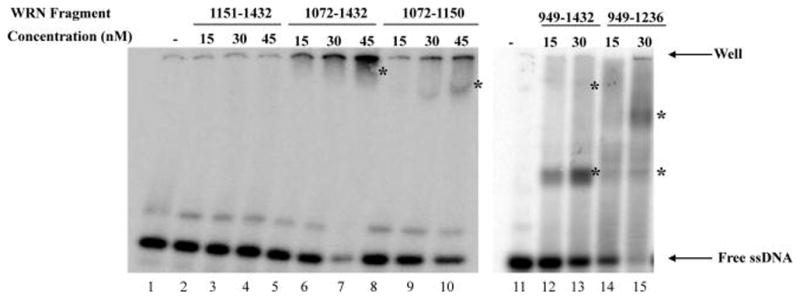
ssDNA binding by gel mobility shift assay. Gel mobility shift assay was performed with 30 fmol radiolabeled C80 oligonucleotide and the indicated amount of WRN protein fragment. Binding assays were analyzed as described in Materials and Methods. Asterisks indicate DNA–protein complexes. The location of the top of the gel (well) is indicated with an arrow.
Oligomerization Status of WRN Fragments
The results of gel mobility shift assays indicate that WRN variants may form protein–DNA complexes of different size/electrophoretic mobility. This could indicate that WRN variants have different aggregation, multimerization, or solubility characteristics. Here, the size and solubility of WRN variants were evaluated using gel filtration chromatography in buffer similar to the buffer for the ssDNA annealing assay. The C-terminal fragment (WRN1072–1432), which has a predicted molecular weight of 66 kDa, eluted in three peaks with molecular weights of 388, 195, and 64 kDa, corresponding to a putative hexamer, trimer, and monomer, respectively. The results also showed that the trimer was the major species formed by the fragment (Figure 6A, upper panel) and that all three peaks contained the WRN1072–1432 polypeptide (Figure 6A, lower panel). WRN1072–1150 had a similar gel filtration profile (data not shown), but WRN1151–1432, which lacks ssDNA annealing activity, eluted primarily as a trimer (Figure 6B). These results and results of the gel mobility shift assays suggest that ssDNA annealing activity may correlate with the oligomerization state of WRN.
Figure 6.
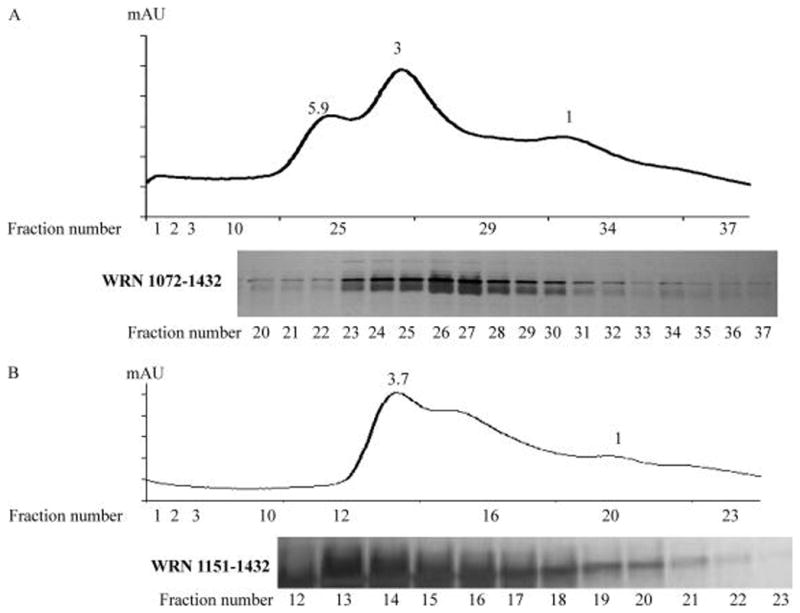
Gel filtration analysis of WRN C-terminal fragments. (A and B) Gel filtration analysis of fragments WRN1072–1432 and WRN1151–1432, respectively. Upper panel is the elution profile from a Superdex 200 10/300 column (GE healthcare). Lower panel is SDS–PAGE analysis of column fractions. Protein bands are visualized by silver staining.
DISCUSSION
Previous studies mapped WRN enzymatic functions and protein–protein interactions to distinct WRN protein domains. In these studies, WRN regions responsible for exonuclease and helicase activities were identified, and the functional properties of the acidic, RQC, and HRDC domains of WRN were characterized. The present study maps the ssDNA annealing activity of WRN to a 79-amino acid region of WRN between residues 1072 and 1150. This region, which lies between the RQC and HRDC domains, is sufficient for ssDNA annealing activity in vitro, and deletion of this region strongly inhibits, but does not completely eliminate, the ssDNA annealing activity of the WRN C-terminal region.
The C-terminal domains of BLM and RecQ5 are also required for ssDNA annealing activities, and the ssDNA annealing activity of BLM was mapped to aa1290–1350 (13). Nevertheless, the C-terminal regions of WRN, BLM, and RecQ5 show little sequence similarity (13, 14). Thus, ssDNA annealing activity does not appear to be an evolutionarily conserved function of RecQ helicases. Although there is weak sequence homology between WRN orthologues in this region, amino acids 1072 to 1150 of WRN are unique, and protein sequences closely related to this region are not found in the protein database. Furthermore, the predicted secondary and tertiary structures of this region lack similarity to any known protein structure.
WRN fragments that retain ssDNA annealing activity form a characteristic protein–DNA complex during gel mobility shift analysis (Figure 5), which correlates with the formation of putative WRN hexamers and trimers during gel filtration (Figure 6). In contrast, WRN1151–1432, which lacks ssDNA annealing activity, exists primarily as a trimer during gel filtration. The oligomerization status of the WRN C-terminal fragment appears to be independent of DNA binding (data not shown), as reported for other proteins with ssDNA annealing activity (28). In addition, previous studies showed that the C-terminal domain of WRN modulates its oligomerization (29) and that the active form of WRN 3′-5′ exonuclease may be a trimer (4). More recently, it was suggested that higher-order protein–DNA complexes of BLM may be active in ssDNA annealing (13). Although BLM also forms a hexamer in solution, the enzymatic properties of the BLM hexamer have not yet been characterized (30).
C-terminal fragments of WRN that include the WH motif of the RQC domain (i.e., WRN949–1432) appear to be more active in ssDNA binding. The WH motif of the RQC domain mediates protein–DNA interaction through the DNA recognition helix of the helix core consisting of three α-helices (20). The recognition helix interacts with the major groove of DNA as in the DNA–protein co-crystal structures of a variety of proteins involved in DNA metabolism (31–33). Therefore, the presence of the WH motif in the WRN RQC domain may offer some explanation as to why the WRN RQC fragments are the strongest DNA binding region followed by the HRDC domain and the exonuclease domain (19). The RQC domain forms a higher oligomer, but the DNA binding does not depend on higher oligomer formation; the ability of higher oligomer formation might correlate with ssDNA activity as suggested for other proteins with ssDNA activity such as Rad52 (28).
RecQ1, BLM, WRN, and RecQ5 catalyze both ssDNA annealing and DNA unwinding in vitro (6, 13, 14, 17). It has been suggested that the balance between RecQ unwinding and annealing functions may be regulated by protein conformation and/or oligomerization (34). In the case of RecQ1, oligomerization is required for ssDNA annealing, and ATP may trigger the switch from ssDNA annealing to DNA unwinding (34). Here, we showed that oligomerization is also required for the ssDNA annealing activity of WRN. The mechanism that regulates WRN ssDNA annealing is not yet known; however, it was previously shown that ATP inhibits ssDNA annealing by WRN, BLM, and RecQ5 (6, 13, 14). It is possible that ATP binding might be one of the factors for the regulation of WRN’s dual function. In addition, WRN oligomerization and its ssDNA annealing activity might be regulated by protein conformation, protein concentration, presence or absence of interacting protein partners, or other as yet unidentified factors.
In future studies, it will be important to determine the in vivo biological significance of the WRN ssDNA annealing activity. This activity may play a role in dsDNA break repair (35), telomere maintenance, resolution of double Holliday junctions, or rescue of stalled or collapsed replication forks, as suggested for BLM (36, 37). Recently, it was shown that yeast and human Dna2 protein, such as WRN and BLM, has ssDNA annealing and DNA strand exchange activities (38) and that it stimulates Fen1 cleavage (39–41). Thus, ssDNA annealing and exchange activities of Dna2, WRN, and BLM may play roles in the processing of Okazaki fragments (38, 42). In support of this hypothesis, we previously reported that the C-terminal domain of WRN (WRN949–1432) interacts with and stimulates Fen1, while WRN ATPase, helicase, and exonuclease activities are not required for the stimulation of Fen1 (21).
In summary, this study clearly demonstrates that the WRN C-terminal domain (aa1072 to 1150) encodes an instrinsic ssDNA annealing activity. This poorly conserved region of WRN may play a role in ssDNA binding and protein oligomerization. Additional studies are needed to understand the in vivo biological role of WRN ssDNA annealing activity.
Acknowledgments
We thank Drs. Sudha Sharma and Mohammed Hedayati for critical reading of the manuscript.
Footnotes
This work was supported by the Intramural Research Program of the NIH, National Institute on Aging.
Abbreviations: WS, Werner syndrome; WRN, Werner protein; RPA, replication protein A; PCNA, proliferating cell nuclear antigen; Fen1, flap endonuclease 1; RQC, RecQ conserved; HRDC, helicase RNaseD conserved; WH, wing helix motif; BLM, Bloom syndrome protein; NLS, nuclear localization signal; GST, glutathione S-transferase.
References
- 1.Salk D, Au K, Hoehn H, Martin GM. Cytogenetics of Werner’s syndrome cultured skin fibroblasts: variegated translocation mosaicism. Cytogenet Cell Genet. 1981;30:92–107. doi: 10.1159/000131596. [DOI] [PubMed] [Google Scholar]
- 2.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 3.Poot M, Hoehn H, Runger TM, Martin GM. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cell lines. Exp Cell Res. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- 4.Huang S, Beresten S, Li B, Oshima J, Ellis NA, Campisi J. Characterization of the human and mouse WRN 3′→5′ exonuclease. Nucleic Acids Res. 2000;28:2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orren DK, Brosh RM, Jr, Nehlin JO, Machwe A, Gray MD, Bohr VA. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machwe A, Xiao L, Groden J, Matson SW, Orren DK. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J Biol Chem. 2005;280:23397–23407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- 7.Brosh RM, Jr, Bohr VA. Roles of the Werner syndrome protein in pathways required for maintenance of genome stability. Exp Gerontol. 2002;37:491–506. doi: 10.1016/s0531-5565(01)00227-3. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki N, Shiratori M, Goto M, Furuichi Y. Werner syndrome helicase contains a 5′→3′ exonuclease activity that digests DNA and RNA strands in DNA/DNA and RNA/DNA duplexes dependent on unwinding. Nucleic Acids Res. 1999;27:2361–2368. doi: 10.1093/nar/27.11.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen JC, Loeb LA. Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res. 2000;28:3260–3268. doi: 10.1093/nar/28.17.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von KC, Harrigan JA, May A, Opresko PL, Dawut L, Cheng WH, Bohr VA. Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol Cell Biol. 2003;23:8601–8613. doi: 10.1128/MCB.23.23.8601-8613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JW, Harrigan J, Opresko PL, Bohr VA. Pathways and functions of the Werner syndrome protein. Mech Ageing Dev. 2005;126:79–86. doi: 10.1016/j.mad.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 13.Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID. The Bloom’s syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33:3932–3941. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machwe A, Lozada EM, Xiao L, Orren DK. Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins. BMC Mol Biol. 2006;7:1. doi: 10.1186/1471-2199-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amsterdam) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM., Jr Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 18.Machwe A, Ganunis R, Bohr VA, Orren DK. Selective blockage of the 3′→5′ exonuclease activity of WRN protein by certain oxidative modifications and bulky lesions in DNA. Nucleic Acids Res. 2000;28:2762–2770. doi: 10.1093/nar/28.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von KC, Thoma NH, Czyzewski BK, Pavletich NP, Bohr VA. Werner syndrome protein contains three structure-specific DNA binding domains. J Biol Chem. 2003;278:52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- 20.Lee JW, Kusumoto R, Doherty KM, Lin GX, Zeng W, Cheng WH, von KC, Brosh RM, Jr, Hu JS, Bohr VA. Modulation of Werner syndrome protein function by a single mutation in the conserved RecQ domain. J Biol Chem. 2005;280:39627–39636. doi: 10.1074/jbc.M506112200. [DOI] [PubMed] [Google Scholar]
- 21.Brosh RM, Jr, von KC, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 25.Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, Ben-Tal N. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–1324. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
- 26.Bernstein DA, Keck JL. Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain. Structure. 2005;13:1173–1182. doi: 10.1016/j.str.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z, Macias MJ, Bottomley MJ, Stier G, Linge JP, Nilges M, Bork P, Sattler M. The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Structure. 1999;7:1557–1566. doi: 10.1016/s0969-2126(00)88346-x. [DOI] [PubMed] [Google Scholar]
- 28.Lloyd JA, Forget AL, Knight KL. Correlation of biochemical properties with the oligomeric state of human rad52 protein. J Biol Chem. 2002;277:46172–46178. doi: 10.1074/jbc.M207262200. [DOI] [PubMed] [Google Scholar]
- 29.Cooper MP, Machwe A, Orren DK, Brosh RM, Ramsden D, Bohr VA. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–912. [PMC free article] [PubMed] [Google Scholar]
- 30.Karow JK, Newman RH, Freemont PS, Hickson ID. Oligomeric ring structure of the Bloom’s syndrome helicase. Curr Biol. 1999;9:597–600. doi: 10.1016/s0960-9822(99)80264-4. [DOI] [PubMed] [Google Scholar]
- 31.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 32.Fogh RH, Ottleben G, Ruterjans H, Schnarr M, Boelens R, Kaptein R. Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy. EMBO J. 1994;13:3936–3944. doi: 10.1002/j.1460-2075.1994.tb06709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gajiwala KS, Chen H, Cornille F, Roques BP, Reith W, Mach B, Burley SK. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000;403:916–921. doi: 10.1038/35002634. [DOI] [PubMed] [Google Scholar]
- 34.Muzzolini L, Beuron F, Patwardhan A, Popuri V, Cui S, Niccolini B, Rappas M, Freemont PS, Vindigni A. Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol. 2007;5:e20. doi: 10.1371/journal.pbio.0050020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan H, McCane J, Toczylowski T, Chen C. Analysis of the Xenopus Werner syndrome protein in DNA double-strand break repair. J Cell Biol. 2005;171:217–227. doi: 10.1083/jcb.200502077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 37.Wu L, Chan KL, Ralf C, Bernstein DA, Garcia PL, Bohr VA, Vindigni A, Janscak P, Keck JL, Hickson ID. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005;24:2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masuda-Sasa T, Polaczek P, Campbell JL. Single strand annealing and ATP-independent strand exchange activities of yeast and human DNA2: possible role in Okazaki fragment maturation. J Biol Chem. 2006;281:38555–38564. doi: 10.1074/jbc.M604925200. [DOI] [PubMed] [Google Scholar]
- 39.Kao HI, Veeraraghavan J, Polaczek P, Campbell JL, Bambara RA. On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J Biol Chem. 2004;279:15014–15024. doi: 10.1074/jbc.M313216200. [DOI] [PubMed] [Google Scholar]
- 40.Sharma S, Sommers JA, Wu L, Bohr VA, Hickson ID, Brosh RM., Jr Stimulation of flap endonuclease-1 by the Bloom’s syndrome protein. J Biol Chem. 2004;279:9847–9856. doi: 10.1074/jbc.M309898200. [DOI] [PubMed] [Google Scholar]
- 41.Sharma S, Otterlei M, Sommers JA, Driscoll HC, Dianov GL, Kao HI, Bambara RA, Brosh RM., Jr WRN helicase and FEN-1 form a complex upon replication arrest and together process branchmigrating DNA structures associated with the replication fork. Mol Biol Cell. 2004;15:734–750. doi: 10.1091/mbc.E03-08-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartos JD, Wang W, Pike JE, Bambara RA. Mechanisms by which Bloom protein can disrupt recombination intermediates of Okazaki fragment maturation. J Biol Chem. 2006;281:32227–32239. doi: 10.1074/jbc.M606310200. [DOI] [PubMed] [Google Scholar]