Significance
Electron transfer is a fundamental process in biology that can be coupled to catalysis within redox enzymes through a careful control of protein conformational movements. Using single-molecule fluorescence resonance energy transfer (FRET) spectroscopy, we find that calmodulin binding to neuronal NO synthase reductase domain (nNOSr) both alters and restricts the distributions of NOSr conformational states and the conformational lifetimes, thereby revealing a physical means by which calmodulin may control electron transfer for catalysis.
Keywords: conformational motion, flavoprotein, domain–domain interaction, FRET, electron transfer
Abstract
Mechanisms that regulate the nitric oxide synthase enzymes (NOS) are of interest in biology and medicine. Although NOS catalysis relies on domain motions, and is activated by calmodulin binding, the relationships are unclear. We used single-molecule fluorescence resonance energy transfer (FRET) spectroscopy to elucidate the conformational states distribution and associated conformational fluctuation dynamics of the two electron transfer domains in a FRET dye-labeled neuronal NOS reductase domain, and to understand how calmodulin affects the dynamics to regulate catalysis. We found that calmodulin alters NOS conformational behaviors in several ways: It changes the distance distribution between the NOS domains, shortens the lifetimes of the individual conformational states, and instills conformational discipline by greatly narrowing the distributions of the conformational states and fluctuation rates. This information was specifically obtainable only by single-molecule spectroscopic measurements, and reveals how calmodulin promotes catalysis by shaping the physical and temporal conformational behaviors of NOS.
Although proteins adopt structures determined by their amino acid sequences, they are not static objects and fluctuate among ensembles of conformations (1). Transitions between these states can occur on a variety of length scales (Å to nm) and time scales (ps to s) and have been linked to functionally relevant phenomena such as allosteric signaling, enzyme catalysis, and protein–protein interactions (2–4). Indeed, protein conformational fluctuations and dynamics, often associated with static and dynamic inhomogeneity, are thought to play a crucial role in biomolecular functions (5–11). It is difficult to characterize such spatially and temporally inhomogeneous dynamics in bulk solution by an ensemble-averaged measurement, especially in proteins that undergo multiple-conformation transformations. In such cases, single-molecule spectroscopy is a powerful approach to analyze protein conformational states and dynamics under physiological conditions, and can provide a molecular-level perspective on how a protein’s structural dynamics link to its functional mechanisms (12–21).
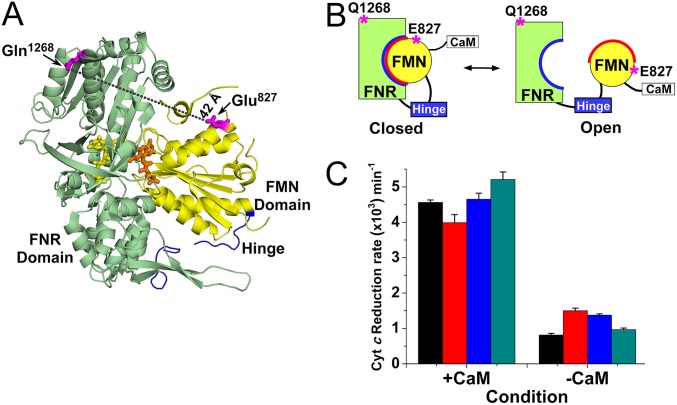
A case in point is the nitric oxide synthase (NOS) enzymes (22–24), whose nitric oxide (NO) biosynthesis involves electron transfer reactions that are associated with relatively large-scale movement (tens of Å) of the enzyme domains (Fig. 1A). During catalysis, NADPH-derived electrons first transfer into an FAD domain and an FMN domain in NOS that together comprise the NOS reductase domain (NOSr), and then transfer from the FMN domain to a heme group that is bound in a separate attached “oxygenase” domain, which then enables NO synthesis to begin (22, 25–27). The electron transfers into and out of the FMN domain are the key steps for catalysis, and they appear to rely on the FMN domain cycling between electron-accepting and electron-donating conformational states (28, 29) (Fig. 1B). In this model, the FMN domain is suggested to be highly dynamic and flexible due to a connecting hinge that allows it to alternate between its electron-accepting (FAD→FMN) or closed conformation and electron-donating (FMN→heme) or open conformation (Fig. 1 A and B) (28, 30–36). In the electron-accepting closed conformation, the FMN domain interacts with the NADPH/FAD domain (FNR domain) to receive electrons, whereas in the electron donating open conformation the FMN domain has moved away to expose the bound FMN cofactor so that it may transfer electrons to a protein acceptor like the NOS oxygenase domain, or to a generic protein acceptor like cytochrome c. In this way, the reductase domain structure cycles between closed and open conformations to deliver electrons, according to a conformational equilibrium that determines the movements and thus the electron flux capacity of the FMN domain (25, 28, 32, 34, 35, 37). A similar conformational switching mechanism is thought to enable electron transfer through the FMN domain in the related flavoproteins NADPH-cytochrome P450 reductase and methionine synthase reductase (38–42).
Fig. 1.
(A) The nNOSr ribbon structure (from PDB: 1TLL) showing bound FAD (yellow) in FNR domain (green), FMN (orange) in FMN domain (yellow), connecting hinge (blue), and the Cy3–Cy5 label positions (pink) and distance (42 Å, dashed line). (B) Cartoon of an equilibrium between the FMN-closed and FMN-open states, with Cy dye label positions indicated. (C) Cytochrome c reductase activity of nNOSr proteins in their CaM-bound and CaM-free states. Color scheme of bar graphs: Black, WT nNOSr unlabeled; Red, Cys-lite (CL) nNOSr unlabeled; Blue, E827C/Q1268C CL nNOSr unlabeled; and Dark cyan, E827C/Q1268C CL nNOSr labeled.
NOS enzymes also contain a calmodulin (CaM) binding domain that is located just before the N terminus of the FMN domain (Fig. 1B), and this provides an important layer of regulation (25, 27). CaM binding to NOS enzymes increases electron transfer from NADPH through the reductase domain and also triggers electron transfer from the FMN domain to the NOS heme as is required for NO synthesis (31, 32). The ability of CaM, or similar signaling proteins, to regulate electron transfer reactions in enzymes is unusual, and the mechanism is a topic of interest and intensive study. It has long been known that CaM binding alters NOSr structure such that, on average, it populates a more open conformation (43, 44). Recent equilibrium studies have detected a buildup of between two to four discreet conformational populations in NOS enzymes and in related flavoproteins, and in some cases, have also estimated the distances between the bound FAD and FMN cofactors in the different species (26, 36, 37, 39, 40), and furthermore, have confirmed that CaM shifts the NOS population distribution toward more open conformations (34, 36, 45). Although valuable, such ensemble-averaged results about conformational states cannot explain how electrons transfer through these enzymes, or how CaM increases the electron flux in NOS, because answering these questions requires a coordinate understanding of the dynamics of the conformational fluctuations. Indeed, computer modeling has indicated that a shift toward more open conformations as is induced by CaM binding to nNOS should, on its own, actually diminish electron flux through nNOS and through certain related flavoproteins (38). Despite its importance, measuring enzyme conformational fluctuation dynamics is highly challenging, and as far as we know, there have been no direct measures on the NOS enzymes or on related flavoproteins, nor studies on how CaM binding might influence the conformational fluctuation dynamics in NOS.
To address this gap, we used single-molecule fluorescence energy resonance transfer (FRET) spectroscopy to characterize individual molecules of nNOSr that had been labeled at two specific positions with Cyanine 3 (Cy3) donor and Cyanine 5 (Cy5) acceptor dye molecules, regarding their conformational states distribution and the associated conformational fluctuation dynamics, which in turn enabled us to determine how CaM binding impacts both parameters. This work provides a unique perspective and a novel study of the NOS enzymes and within the broader flavoprotein family, which includes the mammalian enzymes methionine synthase reductase (MSR) and cytochrome P450 reductase (CPR), and reveals how CaM’s control of the conformational behaviors may regulate the electron transfer reactions of NOS catalysis.
Results
Cys residues in nNOSr that were found to be reactive toward our Cy FRET dyes were changed to Ser using site-directed mutagenesis, to generate a “Cys-lite” version of the enzyme (CLnNOSr) that displayed only residual reactivity toward the FRET dyes (Fig. S1). We then incorporated Cys residues at two surface sites on the enzyme’s FNR and FMN domains (Q1268C and E827C, respectively) (Fig. 1) that are: (i) located outside of the FNR-FMN domain interface, and (ii) expected to fluctuate within a distance range of ∼40–60 Å for the closed and maximally open states, respectively (Fig. 1 A and B), which allows for monitoring of conformational changes by FRET of the Cy3–Cy5 pair, using a corrected Forster radius of Ro, 45 Å that we determined for the dyes when bound to the nNOSr protein. The E827C/Q1268C variant could be labeled with the Cy3 and Cy5 maleimide dyes to near stoichiometry (1.1 and 0.9 mol dye per mol enzyme, respectively) (Fig. S1), and the Cy3–Cy5-labeled enzyme displayed a normal catalytic activity and CaM response compared with native nNOSr enzyme or to the CLnNOSr precursor enzyme (Fig. 1C).
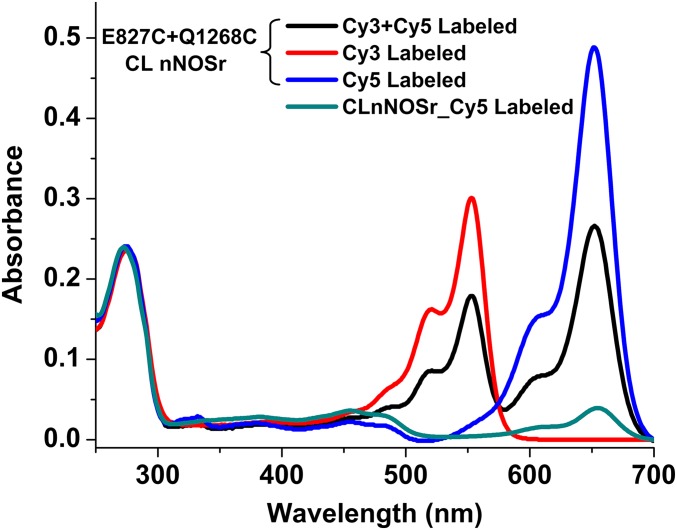
Fig. S1.
UV-vis spectra of Cy3 and Cy5 dye-reacted nNOSr proteins. Two molar equivalents of the donor and acceptor dyes (Cy3 and Cy5, respectively) are bound at ∼1:1 ratio. Extinction coefficients are: nNOSr (457 nm) ε = 22.9 mM−1 cm−1, Cy3 (550 nm) ε = 155 mM−1 cm−1, and Cy 5 (655 nm) ε = 250 mM−1 cm−1. The flavin absorbance is overlapped by Cy3 absorbance. Flavin absorbance is calculated based on the known absorbance ratio 280 nm: 457 nm. Proteins were reacted with the indicated dyes and subject to gel-filtration for further use.
For the FRET spectroscopic experiments, single molecules of dye-labeled nNOSr were entrapped in chambers formed within a buffered 1% agarose gel, then the single-molecule FRET fluctuation time trajectories were recorded under a home-modified single-molecule spectroscopic imaging microscope. A detailed description of the experimental set up and approaches has been published (14, 15).
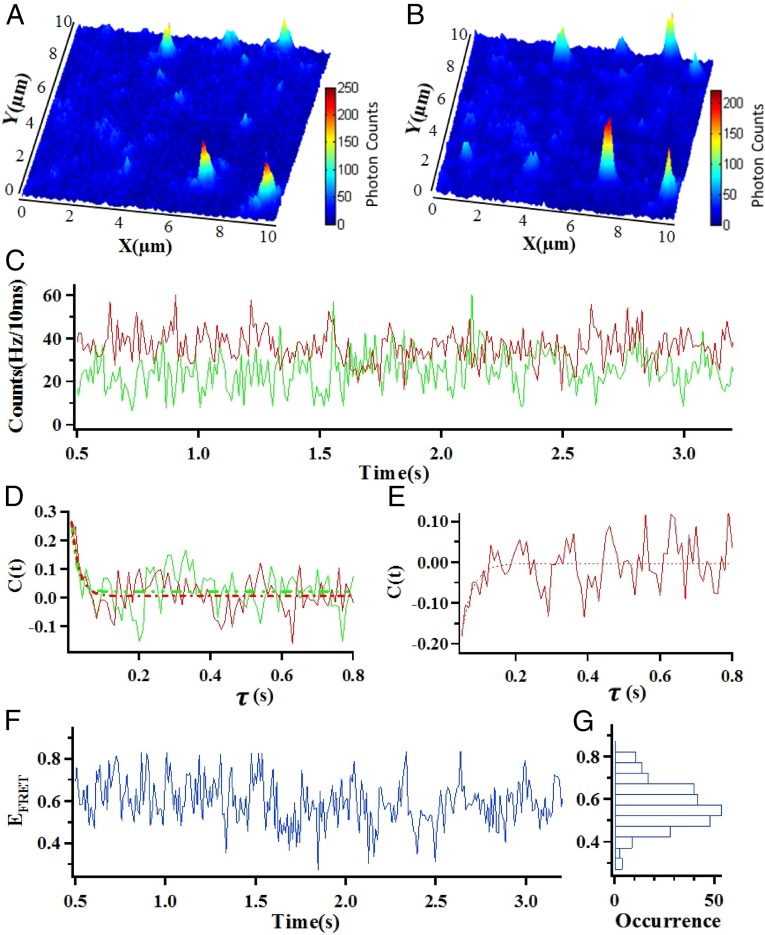
Fig. 2 A and B shows typical donor–acceptor (D–A) two-channel fluorescence images collected for single molecules of Cy3–Cy5 labeled nNOSr within a 10 μm×10 μm laser confocal scanning area in the agarose gel. Each feature in the images is attributed to a single nNOSr molecule; the intensity variation between the molecules is due to FRET and the different longitudinal positions in the light focal depths. Protein conformational changes that cause changes in FRET D–A distance result in coincident and opposite, anticorrelated donor and acceptor fluorescence signal fluctuations (donor decreases, acceptor increases, and vice versa). Fig. 2C shows the intensity vs. time trajectories of the Cy3 donor [Id(t), green]- and Cy5 acceptor [Ia(t), red]-labeled nNOSr under the CaM-free condition. Typically, single-molecule D–A signal fluctuation involves not only FRET-related fluctuation, which shows the anti-correlated relation between the D–A signal fluctuations; but also noncorrelated thermal-related random fluctuation, and measurement noise. To extract the conformational dynamics information from the intensity vs. time trajectories of the Cy3 donor [Id(t), green]- and Cy5 acceptor [Ia(t), red]-labeled nNOSr, we applied autocorrelation and cross-correlation function analysis to characterize the fluctuations due to the FRET energy transfer process. Fig. 2D shows the autocorrelation functions of the donor and acceptor signals, and Fig. 2E shows the cross-correlation function between donor and acceptor signals, all of the correlation functions can be fit with the same decay rate constant within a SD, which confirms that both the donor and acceptor signal fluctuations arise from the same origin, namely the single molecule protein conformational fluctuations.
Fig. 2.
Single-molecule fluorescence images (10 µm × 10 µm) of Cy3/Cy5-labeled E827C/Q1268C nNOSr, the emission is from the FRET dyes. (A) Donor. (B) Acceptor. (C) A portion of a single-molecule intensity vs. time trajectory, green represents donor, and red represents acceptor. (D) Auto-correlation of donor and acceptor from intensity vs. time trajectory. (E) Cross-correlation of donor and acceptor from intensity vs. time trajectory. (F) Single-molecule FRET efficiency vs. time trajectory. (G) The distribution of the FRET efficiencies.
Fig. 2F shows the FRET efficiency vs. time (EFRET∼t) trajectory, calculated as EFRET = Ia/(Id+Ia) from Fig. 2C. The EFRET∼t trajectory reflects the protein conformational motion dynamics, indicating that the Cy3–Cy5 labeled domains of nNOSr fluctuate at certain rate, which provides a statistic result of the protein conformational states. A histogram (Fig. 2G) deduced from the EFRET∼t data of a single nNOSr molecule reflects the distribution of molecular conformations it adopted over the ∼3 s recording period, and shows that they arrange according to a normal distribution, with the most populated conformational states for this molecule corresponding to a EFRET of 0.54.
We then studied how CaM binding impacts the conformational distribution of nNOSr. We analyzed single molecules of CaM-free or CaM-bound nNOSr as described in Fig. 2, determined each molecule’s mean EFRET value (as in Fig. 2 F and G), and then plotted the distributions of these means. The histograms (Fig. 3 A and B) show that the CaM-free and the CaM-bound nNOSr molecules both populate many conformations that have a range of EFRET values. Notably, the CaM-bound molecules populate a tighter distribution of conformational states, which have lower mean FRET efficiency. The mean EFRET value decreased from 0.49 to 0.38 due to CaM binding to nNOSr. Based on the Forster radius of Cy3 and Cy5 and related considerations (supporting information), this corresponds to an increase in the Cy3–Cy5 label mean distance from 4.5 nm to 4.9 nm in the nNOSr, assuming R0 is 4.5 nm. It is important to note that these mean distance values are a statistic result from measures of dynamic structural fluctuations made on single molecules, and the range of structural fluctuation distances that are adopted by any individual molecule is often more extended. The mean values can also fluctuate depending on the dwell times of individual states over the course of the collection time (i.e., shorter-lived conformations may appear more times during the collection period). Nevertheless, our study provides, to our knowledge, the first dynamics-based, single molecule demonstration that under physiologic conditions, CaM binding shifts the conformational distribution of nNOSr toward more open states, consistent with results from ensemble-based studies (26, 27, 34, 37), and we additionally show that CaM narrows the conformational population distribution of nNOSr.
Fig. 3.
(Upper) Histograms of the of FRET efficiency distribution from the EFRET vs. time trajectories (each occurrence is the mean value from histograms as in Fig. 2G) without (A) or with (B) CaM. (Lower) Histograms of the of delay time (τ) distribution from autocorrelation of the intensity vs. time trajectories without (C) or with (D) CaM.
Our single-molecule data can also inform on the conformational fluctuation dynamics of nNOSr and on how CaM binding impacts the dynamics, which are key facets that have not been studied before. Fig. 3 C and D contain histograms of the fluctuation correlation time τ related to the dwell times that we measured for single molecules in any given conformational state, as derived from the autocorrelation functions of the single-molecule FRET intensity ∼ time trajectories. For CaM-free nNOSr, the conformational fluctuation correlation times τ were distributed broadly across 5–300 ms, with an average value of ∼60 ms. These data demonstrate that CaM-free nNOSr molecules adopt a wide range of shorter- and longer-lived conformational lifetimes in solution. Interestingly, the average dwell time for CaM-free nNOSr determined in this single molecule study corresponds fairly well with an ensemble-based estimate of the conformational lifetime for nNOSr open and closed states during catalysis (∼30 ms, based on a two-state conformation model), that was derived indirectly from cytochrome c reductase activity measures and computer modeling (34, 46). Our findings show that CaM-free nNOSr exhibits a wide range of conformational lifetimes, with the more stable conformers (i.e., those with lifetimes exceeding 30 ms) likely helping to limit the steady-state electron flux through the nNOSr during catalysis.
CaM binding to nNOSr caused a dramatic change in the conformational fluctuation correlation times τ (Fig. 3D). CaM shortened the dwell times and also greatly tightened their distribution, such that the enzyme molecules now displayed fluctuation correlation times within the 5–120 ms time window, with most having times less than 50 ms (Fig. 3D). This observation is consistent with CaM binding causing a structural change in nNOSr from a more flexible to a more rigid conformation. More rigid conformational states would have a faster and narrower fluctuation rate distribution than do the CaM-free, more flexible conformational states. An ability of CaM to diminish longer-lived conformational states of nNOSr has not been previously reported, and it likely reveals a mechanism by which CaM binding can increase electron flux through nNOSr during catalysis.
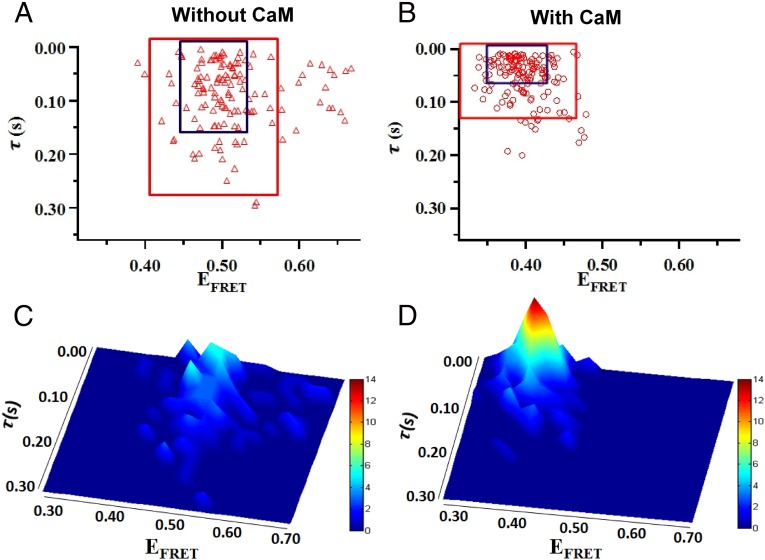
To further illustrate how CaM binding impacts the conformational behaviors of nNOSr, we plotted the pairs of conformational fluctuation correlation times τ and EFRET values that we observed for many individual nNOSr molecules, without and with bound CaM (Fig. 4 A and B). The same data distributions are also plotted as 3D topographical illustrations (Fig. 4 C and D). Several interesting and fundamental concepts emerge from the graphic analysis: First, along with CaM inducing a shift to more open conformations with shorter dwell times, CaM caused a remarkable tightening in the molecular distributions of both parameters. For example, in the absence of CaM (Fig. 4 A and C), the nNOSr molecules distribute broadly in both their conformational space and time dimensions to create a rugged conformational landscape, such that only 83% of the molecules lie within 2 SDs of the mean pair value (the points contained within the red box in Fig. 4A are within 2 SDs). CaM binding caused the nNOSr molecules to distribute much more tightly in both dimensions to create a simpler conformational landscape, such that 91% of the molecules now lie within two SDs of the mean (Fig. 4 B and D). The tighter clustering of paired conformational states and dwell times induced by CaM is such that the rectangles enclosing all CaM-bound nNOSr molecules that lie within 1 or 2 SDs of the mean pair (the blue and red boxes are for 1 and 2 SDs, respectively, Fig. 4B) are each 2.5 times smaller in area than the corresponding boxes enclosing the CaM-free nNOSr molecules that lie within 1 or 2 SDs of the mean pair. This tightening of the distribution is also indicated by the higher central topographic peak in Fig. 4D. Secondly, CaM binding reduced or eliminated the most long-lived and the most closed conformations of nNOSr that are otherwise populated under the CaM-free condition, and whose presence may be expected to retard steady-state electron transfer during catalysis by nNOSr. This effect is manifested by the disappearance of paired points lying beyond τ values of 0.2 and beyond EFRET values of 0.5 in Fig. 4 B and D compared with Fig. 4 A and C. Third, the graphs show that there is no strong correlation apparent between a nNOSr molecule’s conformational state (EFRET) and its conformational dwell time τ in either the CaM-free or CaM-bound condition. This result indicates that longer-lived states of nNOSr do not preferentially populate more closed or more open conformational states, and instead are distributed fairly evenly across the entire range of conformations.
Fig. 4.
Impact of CaM binding on the distribution of single nNOSr molecules regarding their conformational fluctuation correlation times (t) and their associated FRET efficiencies (EFRET). (A and B) Blue and red boxes contain all molecules that lie within 1 and 2 SD from the mean value pair, respectively. (C and D) 3D histograms plotting the molecular pair distributions in A and B. The heights of the peaks and their colors are proportional to the number of molecules, as indicated by the bars.
Discussion
Our single-molecule approach afforded a novel and fundamental study of the conformational dynamics of individual nNOSr molecules, and paired this information with their associated molecular conformational states (EFRET). Because previous ensemble studies could only report on the averaged protein conformational distributions, and could not provide insight into the conformational dynamics (34, 37, 38, 40), our study represents a significant advance and provides an interesting perspective on how CaM may regulate nNOSr electron transfer reactions through its control of protein conformational behavior.
Our data reveal that CaM alters nNOSr conformational behaviors in three ways: (i) It supports faster conformational fluctuations (i.e., shorter dwell times) across the entire conformational landscape. (ii) It tightens the distributions of nNOSr conformational states and dwell times, and diminishes the most closed states and the longest-lived species. (iii) It causes nNOSr to favor more open conformational states. Each of these three CaM effects, on their own, could increase electron flux through a dual-flavin reductase enzyme like nNOSr. However, in the context of nNOSr, the first two CaM effects are likely to be the most important. For example, modeling shows that the surest way to increase electron flux through nNOSr, or through any related dual-flavin reductase enzyme, is to increase the extent of interconversion among the enzyme’s different conformational states (i.e., shortening their dwell times, effect i) (27, 36, 37, 46). CaM’s additional effect of tightening the conformational state and dwell time distributions (effect ii) should also help, particularly because it minimizes populations of closed species which are unable to transfer electrons to an acceptor protein like cytochrome c (or to the nNOS oxygenase domain), and because it minimizes the formation of longer-lived conformational species which may retard the enzyme cycling to support electron flux. These effects are exactly what CaM appears to be doing when it binds to nNOSr. In comparison, CaM’s shifting of the nNOSr conformational equilibrium to favor more open states (effect iii) may not always be useful for improving the electron flux to cytochrome c. This effect is because shifting the conformational equilibrium too far away from unity, or creating extremely open forms of nNOSr (as appears to occur in some cytochrome P450 reductase hinge mutants; refs. 47 and 48) could retard electron flux through the enzyme by making it more difficult for the open nNOSr molecules to close up as is required to continue cycling during catalysis (46). As noted, CaM’s demonstrated ability to increase the interconversion rates among nNOSr conformational species (effect i) could actually make up for, or even overcome, any deleterious shift that CaM may cause in the nNOSr open-closed conformational equilibrium. Thus, our finding that CaM speeds nNOSr conformational dynamics helps to resolve a paradox that kinetic models had uncovered as early as 2009 (27), regarding how CaM’s impact on the open-closed conformational equilibrium could actually increase electron flux through nNOSr.
Our current findings can also inform on how CaM activates heme reduction for NO synthesis in full-length nNOS. Interestingly, CaM’s ability to increase the conformational interconversion rates (effect i) may not be as important in the case of nNOS heme reduction, because the available data and modeling show that the interconversion rates in CaM-free nNOSr are already faster than is the nNOS heme reduction rate, and so should already be sufficient (27, 46). In contrast, CaM’s tightening of the nNOSr conformational distribution (effect ii) may be the more important effect regarding nNOS heme reduction. Consider that cytochrome c is an electron acceptor floating freely in solution, whereas the nNOS oxygenase domain is tethered covalently to nNOSr through the CaM binding site. Thus, restricting the degrees of freedom for FMN domain motion, once it is freed from the FNR domain, could greatly increase the probability of a productive contact between the FMN domain and the nNOS oxygenase domain in unit time, by simply minimizing formation of different open conformational states, most of which are unproductive for nNOS heme reduction but are perfectly fine to achieve cytochrome c reduction. In this context, CaM’s promoting more open conformational states (effect iii) could particularly help to enable nNOS heme reduction, because maintaining a conformational equilibrium setpoint near unity that supports maximal electron flux through nNOSr (as is measured by cytochrome c reduction) is not necessary in the circumstance of NOS heme reduction, nor is it even desired (49).
Although our current study focused on nNOSr, we suspect that similar conformational behavior occurs in other NOS enzymes and in related dual-flavin enzymes like CPR and MSR. In the case of endothelial NOS, previous studies and modeling (34, 38) would predict that it may exhibit significant differences in its conformational states distribution (favoring more closed states relative to nNOSr) and in its conformational dwell times (favoring longer-lived states relative to nNOSr). These differences and related aspects can now be addressed using our single molecule approach.
To conclude, obtaining coordinate information on conformational states and dwell times in a multidomain redox enzyme, at a single molecule level, is a powerful means to define the enzyme’s conformational behavior and to investigate how its conformational behavior relates to catalysis, and in the case of NOS how these facets are regulated by external players such as CaM. Our current results shed new light on how CaM and nNOS function in signal cascades by revealing how their interaction directs the conformational behavior of nNOS, which in turn enables the electron transfer reactions required to support NO synthesis.
Materials and Methods
Expression and Purification of nNOSr Proteins.
nNOSr protein expression was induced at room temperature over 1 or 2 d in E. coli BL21(DE3) that harbored the pACYC-CaM plasmid as described (34). The nNOSr proteins were purified by sequential chromatography on a 2',5′-ADP Sepharose affinity column and CaM-Sepharose resin as reported (34). Purity of each protein was assessed by SDS/PAGE and spectral analysis. For the nNOSr proteins, concentration was determined by using an extinction coefficient of 22,900 M−1 cm−1 at 457 nm for the fully oxidized form (34).
Preparation of the Cy3 + Cy5-Labeled nNOSr.
We used mass spectrometry to identify maleimide dye-reactive Cys residues present in nNOSr, and then used site-directed mutagenesis to convert these to Ser, thus generating a “Cys-lite” version (CLnNOSr). Based on crystal structures, we mutated two cysteine residues at the surface of the FMN and FNR domains (E827C and Q1268C) (28, 47, 50) and labeled them with Cy3 and Cy5 maleimide dyes. Detail has been described in SI Materials and Methods.
Steady-State Cytochrome c Reduction Assay.
The cytochrome c reductase activity was determined at room temperature by monitoring the absorbance increase at 550 nm and using an extinction coefficient ε550 = 21 mM−1 cm−1 as described (34, 38). To overcome a dilution-based dissociation of FMN from the nNOSr, we added 6-OH-FMN, which has much less fluorescence than FMN but still binds within nNOSr and supports its catalysis. 6-OH-FAD was a gift from David Ballou (University of Michigan). 6-OH FMN was generated from 6-OH-FAD using snake venom phosphodiesterase I and then purified as described (51) and the conversion and purity was checked in HPLC. Detail has been described in SI Materials and Methods.
Preparation of Samples for FRET Measurement.
In our experiments, 1 nM nNOSr with oxygen scavenger solution were sandwiched between two clean glass coverslips in 1% agarose gel (in 99% water). In the 1% agarose gel, single nNOSr enzyme molecules can rotate freely (14, 17, 52–54). Detail of sample preparation has been described in SI Materials and Methods.
Single-Molecule Imaging and FRET Measurements.
We used the single-molecule photon stamping approach (16, 17, 52) to record the single-molecule FRET fluctuation time trajectories photon-by-photon for both the donor and acceptor simultaneously. A detailed description of the experimental set up has been published (16, 52). A brief description of measurement has been described in supporting information. Briefly, the fluorescence images and photon-counting trajectories were acquired with an inverted confocal microscope (Axiovert 200, Zeiss). The excitation laser (532 nm CW, Crystalaser) confocal beam was reflected by a dichroic beam splitter (z532rdc, Chroma Technology) and was focused by a high-numerical-aperture objective (1.3 NA, 100×, Zeiss) on the sample at a diffraction limited spot of ∼300 nm diameter. To obtain the fluorescence images and intensity trajectories, the emission signal was split by using a dichroic beam splitter (640dcxr) into two color beams centered at 570 nm and 670 nm representing Cy3 and Cy5 emissions, respectively. The two channels of signal were detected by a pair of Si avalanche photodiode single photon counting modules (SPCM-AQR-16, Perkin-Elmer Optoelectronics). Typical images (10 μm × 10 μm) were acquired by continuously raster-scanning the sample over the laser focus with a scanning speed of 4 ms/pixel, with each image of 100 pixels × 100 pixels. The fluorescence intensity trajectories of the donor (Cy3) and acceptor (Cy5) were recorded by a two-channel Picoharp 300 (PicoQuant) photon-stamping set up. The donor (Cy3) and accepter (Cy5) fluorescence intensity trajectories were typically collected for 50 s.
SI Materials and Methods
Sample Preparation and Characterization
Preparation of the Cy3 + Cy5-Labeled nNOSr.
We used mass spectrometry to identify maleimide dye-reactive Cys residues present in nNOSr, and then used site-directed mutagenesis to convert these to Ser, thus generating a “Cys-lite” version (CLnNOSr). Reacting CLnNOSr with Cy5 maleimide confirmed that only trace reactivity remained (Fig. S1). We then substituted cysteine residues at two locations that lie on the surface of the FMN and FNR domains (E827C and Q1268C) and that are expected to move apart when the protein transitions from the closed conformation to the open conformation, based on available crystal structures (28, 47, 50). The E827C/Q1268C CLnNOSr variant bound Cy3 and Cy5 maleimide dyes equally well, and when the protein was reacted with both dyes approximately equivalent amounts of Cy3 and Cy5 became bound (Fig. S1), indicating it was fit for the single molecule studies. The calibrated Förster distance R0 of the Cy3–Cy5 pair at Cys827 and Cys1268 on the enzyme is ∼45 Å.
Materials and Preparation of the Enzyme Solution.
Calmodulin (CaM) bovine (C4874-10KU, lyophilized powder, ≥98%) and β-Nicotinamide adenine dinucleotide phosphate sodium salt hydrate (NADP, N0505, ≥98% (HPLC) were purchased from Sigma-Aldrich. 6-OH-FAD was a gift from David Ballou (University of Michigan, Ann Arbor). 6-OH FMN was generated from 6-OH-FAD using snake venom phosphodiesterase I and then purified as described (51) and the conversion and purity was checked in HPLC. The buffer solution used for experiments was 40 mM EPPS [4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid] with 10% (vol/vol) glycerol and 150 mM NaCl, pH 7.6.
To overcome a dilution-based dissociation of FMN from the nNOSr, we added 6-OH-FMN, which has much less fluorescence than FMN but still binds within nNOSr and supports its catalysis. The 6-OH-FMN enabled us to avoid a prohibitively high fluorescence background that otherwise became present if FMN was instead added to the diluted enzyme solution. Experimental solutions contained 5 nM dye-labeled nNOSr, 10 µM NADP, and 2 µM 6-OH-FMN in buffer. In some cases, 10 µM CaM and 400 µM CaCl2 were also added.
Preparation of Samples for FRET Measurement.
The samples for single-molecule spectroscopic measurements were prepared by mixing 4 µl enzyme solution described above, with 6 µL of oxygen scavenger solution that contained 0.8% d-glucose, 1 mg/mL glucose oxidase, 0.04 mg/mL catalase, and 1 mM of 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox) and is added to prevent FRET dye molecule photobleaching or blinking due to forming triplet state (53, 54), and with 10 µL of 1% agarose that had been heated just above its gel-transition temperature (26 °C). The mixture was then placed between two glass coverslips to form a sandwich (17, 52). The final concentration of nNOSr is about 1 nM. Once the solution cools it forms a gel containing dispersed single enzyme molecules that are entrapped in chambers large enough to allow the enzyme molecule to freely rotate, and to allow small molecules to diffuse freely. Our previous anisotropy study has demonstrated that confining single enzyme molecules in the agarose gel in this way has no perturbation on the enzyme motions (14).
Single-Molecule Imaging and FRET Measurements
We used the single-molecule photon stamping approach (16, 17, 52) to record the single-molecule FRET fluctuation time trajectories photon-by-photon for both the donor and acceptor simultaneously. A detailed description of the experimental set up has been published elsewhere (16, 52). Briefly, the fluorescence images and photon-counting trajectories were acquired with an inverted confocal microscope (Axiovert 200, Zeiss). The excitation laser (532 nm CW, Crystalaser) confocal beam was reflected by a dichroic beam splitter (z532rdc, Chroma Technology) and was focused by a high-numerical-aperture objective (1.3 NA, 100×, Zeiss) on the sample at a diffraction limited spot of ∼300-nm diameter. To obtain the fluorescence images and intensity trajectories, the emission signal was split by using a dichroic beam splitter (640dcxr) into two color beams centered at 570 nm and 670 nm representing Cy3 and Cy5 emissions, respectively. The two channels of signal were detected by a pair of Si avalanche photodiode single photon counting modules (SPCM-AQR-16, Perkin-Elmer Optoelectronics). Typical images (10 μm × 10 μm) were acquired by continuously raster-scanning the sample over the laser focus with a scanning speed of 4 ms/pixel, with each image of 100 pixels × 100 pixels. The fluorescence intensity trajectories of the donor (Cy3) and acceptor (Cy5) were recorded by a two-channel Picoharp 300 (PicoQuant) photon-stamping set-up. The donor (Cy3) and accepter (Cy5) fluorescence intensity trajectories were typically collected for 50 s.
Acknowledgments
We thank Deb Durrah for technical assistance. The study was supported by National Institutes of Health Grants GM51491 (to D.J.S. and H.P.L.) and HL076491 (to D.J.S.) and support from the Ohio Eminent Scholar Endowment (to H.P.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508829112/-/DCSupplemental.
References
- 1.Fersht A. 1998. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding (W.H. Freeman, New York) 1st Ed.
- 2.Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem Rev. 2006;106(8):3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 3.Boehr DD, McElheny D, Dyson HJ, Wright PE. Millisecond timescale fluctuations in dihydrofolate reductase are exquisitely sensitive to the bound ligands. Proc Natl Acad Sci USA. 2010;107(4):1373–1378. doi: 10.1073/pnas.0914163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zang C, et al. Ultrafast proteinquake dynamics in cytochrome c. J Am Chem Soc. 2009;131(8):2846–2852. doi: 10.1021/ja8057293. [DOI] [PubMed] [Google Scholar]
- 5.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5(11):789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenmesser EZ, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438(7064):117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 7.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450(7172):964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 8.Henzler-Wildman KA, et al. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450(7171):838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 9.Schwans JP, Kraut DA, Herschlag D. Determining the catalytic role of remote substrate binding interactions in ketosteroid isomerase. Proc Natl Acad Sci USA. 2009;106(34):14271–14275. doi: 10.1073/pnas.0901032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watt ED, Shimada H, Kovrigin EL, Loria JP. The mechanism of rate-limiting motions in enzyme function. Proc Natl Acad Sci USA. 2007;104(29):11981–11986. doi: 10.1073/pnas.0702551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyashita O, Onuchic JN, Wolynes PG. Nonlinear elasticity, proteinquakes, and the energy landscapes of functional transitions in proteins. Proc Natl Acad Sci USA. 2003;100(22):12570–12575. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bockenhauer SD, Duncan TM, Moerner WE, Borsch M. 2014. The regulatory switch of F-ATPase studied by single-molecule FRET in the ABEL Trap. Proc. Soc. Photo. Opt. Instrum. Eng 8950:89500H. [DOI] [PMC free article] [PubMed]
- 13.Chung HS, Eaton WA. Single-molecule fluorescence probes dynamics of barrier crossing. Nature. 2013;502(7473):685–688. doi: 10.1038/nature12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y, et al. Probing single-molecule enzyme active-site conformational state intermittent coherence. J Am Chem Soc. 2011;133(36):14389–14395. doi: 10.1021/ja204644y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y, Lu M, Lu HP. Single-molecule photon stamping FRET spectroscopy study of enzymatic conformational dynamics. Phys Chem Chem Phys. 2013;15(3):770–775. doi: 10.1039/c2cp42944f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu R, Hu D, Tan X, Lu HP. Revealing two-state protein-protein interactions of calmodulin by single-molecule spectroscopy. J Am Chem Soc. 2006;128(31):10034–10042. doi: 10.1021/ja057005m. [DOI] [PubMed] [Google Scholar]
- 17.Lu HP, Xun L, Xie XS. Single-molecule enzymatic dynamics. Science. 1998;282(5395):1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 18.Lu HP. Probing single-molecule protein conformational dynamics. Acc Chem Res. 2005;38(7):557–565. doi: 10.1021/ar0401451. [DOI] [PubMed] [Google Scholar]
- 19.Liu S, Bokinsky G, Walter NG, Zhuang X. 2007 Dissecting the multistep reaction pathway of an RNA enzyme by single-molecule kinetic “fingerprinting.” Proc Natl Acad Sci USA 104(31):12634–12639. [Google Scholar]
- 20.Moerner WE. New directions in single-molecule imaging and analysis. Proc Natl Acad Sci USA. 2007;104(31):12596–12602. doi: 10.1073/pnas.0610081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, et al. Protein conformational dynamics probed by single-molecule electron transfer. Science. 2003;302(5643):262–266. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 22.Campbell MG, Smith BC, Potter CS, Carragher B, Marletta MA. Molecular architecture of mammalian nitric oxide synthases. Proc Natl Acad Sci USA. 2014;111(35):E3614–E3623. doi: 10.1073/pnas.1413763111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daff S. NO synthase: Structures and mechanisms. Nitric Oxide. 2010;23(1):1–11. doi: 10.1016/j.niox.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Poulos TL. Heme enzyme structure and function. Chem Rev. 2014;114(7):3919–3962. doi: 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith BC, Underbakke ES, Kulp DW, Schief WR, Marletta MA. Nitric oxide synthase domain interfaces regulate electron transfer and calmodulin activation. Proc Natl Acad Sci USA. 2013;110(38):E3577–E3586. doi: 10.1073/pnas.1313331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobolewska-Stawiarz A, et al. Energy landscapes and catalysis in nitric-oxide synthase. J Biol Chem. 2014;289(17):11725–11738. doi: 10.1074/jbc.M114.548834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuehr DJ, Tejero J, Haque MM. Structural and mechanistic aspects of flavoproteins: electron transfer through the nitric oxide synthase flavoprotein domain. FEBS J. 2009;276(15):3959–3974. doi: 10.1111/j.1742-4658.2009.07120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcin ED, et al. Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J Biol Chem. 2004;279(36):37918–37927. doi: 10.1074/jbc.M406204200. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh DK, Salerno JC. 2003. Nitric oxide synthases: domain structure and alignment in enzyme function and control. Front Biosci 8:d193–d209.
- 30.Daff S. Calmodulin-dependent regulation of mammalian nitric oxide synthase. Biochem Soc Trans. 2003;31(Pt 3):502–505. doi: 10.1042/bst0310502. [DOI] [PubMed] [Google Scholar]
- 31.Dunford AJ, Rigby SEJ, Hay S, Munro AW, Scrutton NS. Conformational and thermodynamic control of electron transfer in neuronal nitric oxide synthase. Biochemistry. 2007;46(17):5018–5029. doi: 10.1021/bi7001339. [DOI] [PubMed] [Google Scholar]
- 32.Feng C, et al. Intraprotein electron transfer in a two-domain construct of neuronal nitric oxide synthase: the output state in nitric oxide formation. Biochemistry. 2006;45(20):6354–6362. doi: 10.1021/bi060223n. [DOI] [PubMed] [Google Scholar]
- 33.Guan ZW, Iyanagi T. Electron transfer is activated by calmodulin in the flavin domain of human neuronal nitric oxide synthase. Arch Biochem Biophys. 2003;412(1):65–76. doi: 10.1016/s0003-9861(03)00009-2. [DOI] [PubMed] [Google Scholar]
- 34.Ilagan RP, et al. Differences in a conformational equilibrium distinguish catalysis by the endothelial and neuronal nitric-oxide synthase flavoproteins. J Biol Chem. 2008;283(28):19603–19615. doi: 10.1074/jbc.M802914200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Persechini A, Tran QK, Black DJ, Gogol EP. Calmodulin-induced structural changes in endothelial nitric oxide synthase. FEBS Lett. 2013;587(3):297–301. doi: 10.1016/j.febslet.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salerno JC, Ray K, Poulos T, Li H, Ghosh DK. Calmodulin activates neuronal nitric oxide synthase by enabling transitions between conformational states. FEBS Lett. 2013;587(1):44–47. doi: 10.1016/j.febslet.2012.10.039. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh DK, Ray K, Rogers AJ, Nahm NJ, Salerno JC. FMN fluorescence in inducible NOS constructs reveals a series of conformational states involved in the reductase catalytic cycle. FEBS J. 2012;279(7):1306–1317. doi: 10.1111/j.1742-4658.2012.08525.x. [DOI] [PubMed] [Google Scholar]
- 38.Haque MM, et al. Distinct conformational behaviors of four mammalian dual-flavin reductases (cytochrome P450 reductase, methionine synthase reductase, neuronal nitric oxide synthase, endothelial nitric oxide synthase) determine their unique catalytic profiles. FEBS J. 2014;281(23):5325–5340. doi: 10.1111/febs.13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang WC, Ellis J, Moody PC, Raven EL, Roberts GC. Redox-linked domain movements in the catalytic cycle of cytochrome p450 reductase. Structure. 2013;21(9):1581–1589. doi: 10.1016/j.str.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pudney CR, Khara B, Johannissen LO, Scrutton NS. Coupled motions direct electrons along human microsomal P450 Chains. PLoS Biol. 2011;9(12):e1001222. doi: 10.1371/journal.pbio.1001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pudney CR, et al. Kinetic and spectroscopic probes of motions and catalysis in the cytochrome P450 reductase family of enzymes. FEBS J. 2012;279(9):1534–1544. doi: 10.1111/j.1742-4658.2011.08442.x. [DOI] [PubMed] [Google Scholar]
- 42.Rigby SE, Lou X, Toogood HS, Wolthers KR, Scrutton NS. ELDOR spectroscopy reveals that energy landscapes in human methionine synthase reductase are extensively remodelled following ligand and partner protein binding. ChemBioChem. 2011;12(6):863–867. doi: 10.1002/cbic.201000630. [DOI] [PubMed] [Google Scholar]
- 43.Craig DH, Chapman SK, Daff S. Calmodulin activates electron transfer through neuronal nitric-oxide synthase reductase domain by releasing an NADPH-dependent conformational lock. J Biol Chem. 2002;277(37):33987–33994. doi: 10.1074/jbc.M203118200. [DOI] [PubMed] [Google Scholar]
- 44.Gachhui R, et al. Characterization of the reductase domain of rat neuronal nitric oxide synthase generated in the methylotrophic yeast Pichia pastoris. Calmodulin response is complete within the reductase domain itself. J Biol Chem. 1996;271(34):20594–20602. doi: 10.1074/jbc.271.34.20594. [DOI] [PubMed] [Google Scholar]
- 45.Arnett DC, Persechini A, Tran QK, Black DJ, Johnson CK. Fluorescence quenching studies of structure and dynamics in calmodulin-eNOS complexes. FEBS Lett. 2015;589(11):1173–1178. doi: 10.1016/j.febslet.2015.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haque MM, Kenney C, Tejero J, Stuehr DJ. A kinetic model linking protein conformational motions, interflavin electron transfer and electron flux through a dual-flavin enzyme-simulating the reductase activity of the endothelial and neuronal nitric oxide synthase flavoprotein domains. FEBS J. 2011;278(21):4055–4069. doi: 10.1111/j.1742-4658.2011.08310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamdane D, et al. Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J Biol Chem. 2009;284(17):11374–11384. doi: 10.1074/jbc.M807868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia C, et al. Conformational changes of NADPH-cytochrome P450 oxidoreductase are essential for catalysis and cofactor binding. J Biol Chem. 2011;286(18):16246–16260. doi: 10.1074/jbc.M111.230532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuehr DJ, Santolini J, Wang ZQ, Wei CC, Adak S. Update on mechanism and catalytic regulation in the NO synthases. J Biol Chem. 2004;279(35):36167–36170. doi: 10.1074/jbc.R400017200. [DOI] [PubMed] [Google Scholar]
- 50.Wang M, et al. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci USA. 1997;94(16):8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sucharitakul J, Chaiyen P, Entsch B, Ballou DP. Kinetic mechanisms of the oxygenase from a two-component enzyme, p-hydroxyphenylacetate 3-hydroxylase from Acinetobacter baumannii. J Biol Chem. 2006;281(25):17044–17053. doi: 10.1074/jbc.M512385200. [DOI] [PubMed] [Google Scholar]
- 52.Chen Y, Hu DH, Vorpagel ER, Lu HP. Probing single-molecule T4 lysozyme conformational dynamics by intramolecular fluorescence energy transfer. J Phys Chem B. 2003;107(31):7947–7956. [Google Scholar]
- 53.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5(6):507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selvin PR, Ha T. Single-Molecule Techniques: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2008. [Google Scholar]