Significance
Sixty billion chickens are produced worldwide each year, and all are at risk from Eimeria, parasites that cause coccidiosis. Control relies widely on chemoprophylaxis, but pressure to reduce drug use in farming urges development of cost-effective vaccines. Antigens such as apical membrane antigen 1 (AMA1) offer promise as anticoccidial vaccine candidates, but experience with related apicomplexans such as Plasmodium, in which pre-existing antigenic diversity and incompatible population structure have undermined vaccine development, tempers confidence. Parasite genotyping identified enormous region-specific variation in haplotype diversity for Eimeria tenella but a contrastingly low level of polymorphism for EtAMA1. Although high levels of polyclonal Eimeria infection and hybridization indicate an ability to disseminate vaccine resistance rapidly, the low level of EtAMA1 diversity promotes vaccine development.
Keywords: Eimeria, coccidiosis, population structure, chickens, food security
Abstract
The phylum Apicomplexa includes serious pathogens of humans and animals. Understanding the distribution and population structure of these protozoan parasites is of fundamental importance to explain disease epidemiology and develop sustainable controls. Predicting the likely efficacy and longevity of subunit vaccines in field populations relies on knowledge of relevant preexisting antigenic diversity, population structure, the likelihood of coinfection by genetically distinct strains, and the efficiency of cross-fertilization. All four of these factors have been investigated for Plasmodium species parasites, revealing both clonal and panmictic population structures with exceptional polymorphism associated with immunoprotective antigens such as apical membrane antigen 1 (AMA1). For the coccidian Toxoplasma gondii only genomic diversity and population structure have been defined in depth so far; for the closely related Eimeria species, all four variables are currently unknown. Using Eimeria tenella, a major cause of the enteric disease coccidiosis, which exerts a profound effect on chicken productivity and welfare, we determined population structure, genotype distribution, and likelihood of cross-fertilization during coinfection and also investigated the extent of naturally occurring antigenic diversity for the E. tenella AMA1 homolog. Using genome-wide Sequenom SNP-based haplotyping, targeted sequencing, and single-cell genotyping, we show that in this coccidian the functionality of EtAMA1 appears to outweigh immune evasion. This result is in direct contrast to the situation in Plasmodium and most likely is underpinned by the biology of the direct and acute coccidian life cycle in the definitive host.
The phylum Apicomplexa contains protozoan parasites of great medical and veterinary importance including Plasmodium, Toxoplasma, and Eimeria species. Cost-effective vaccines and/or drugs are urgently required to control the diseases caused by these pathogens, but their complex life cycles and naturally occurring genetic polymorphism makes the development of such vaccines an extremely difficult task. For Plasmodium species and Toxoplasma gondii, in-depth studies have defined genetic diversity and population structures in many regions and populations, with descriptions ranging from clonality to panmixia (e.g., refs. 1–3). For Eimeria it is widely perceived that species occurrence has been established and that the risks posed are well understood, but population structures within field populations are unclear, and the occurrence of genetic diversity is unexplored (4). Given the recent progress in identifying antigens for inclusion in novel subunit vaccines to control poultry coccidiosis (reviewed in ref. 5), it is now timely to investigate the extent, distribution, and dissemination of genetic diversity in this parasite.
Sustainable food security is a major concern with the global human population set to exceed nine billion by 2050 (6). Poultry are the most efficient source of animal-derived protein (7), and consequentially pathogens that compromise poultry production are reemerging as threats to the global food supply and as factors in human poverty (8, 9). Coccidiosis is a problem in many livestock systems; most notably, in chicken (broiler) production seven different species of Eimeria cause serious intestinal disease that compromises economic productivity and animal welfare, incurring global costs in excess of $3 billion every year (10, 11). Disease control relies on the use of in-feed drugs, but multidrug resistant parasites are ubiquitous, and concerns about residues are widespread (12). Live anticoccidial vaccines have been used in small sectors of the poultry industry for more than 50 y with no evidence of parasite evolution toward resistance/immune escape (11). Examples of naturally occurring genetic diversity have been reliably overcome by the inclusion of more than one “strain” of a single species in some vaccine formulations (11). Each Eimeria species expresses between 6,000 and 9,000 proteins throughout its lifecycle (13), exposing the host to a complex portfolio of antigens. Genetic mapping studies with Eimeria maxima indicated that at least six loci associate absolutely with susceptibility to strong natural immune responses, and additional loci under partial selection also were detectable (14). This breadth of antigenic repertoire is likely to have limited the impact of diversifying selection on the efficacy of live vaccines and explains why resistance has not developed. Unfortunately, these live vaccines must be propagated in chickens, and the low reproductive index of attenuated vaccine parasites makes production costs high and limits capacity (11). More cost-effective subunit vaccines composed of a small number of Eimeria antigens are needed for the mass broiler market; however, these vaccines are likely to induce more focused immune selection, which potentially could drive rapid selection of vaccine-resistant populations. Eimeria tenella is considered one of the most important species because it is widespread and highly pathogenic (13, 15). It also is the best-studied species and therefore was selected as the exemplar in these studies. We used PCR to define the occurrence of E. tenella within fecal samples collected from domestic chickens (Gallus gallus domesticus) across five continents. Positive samples then were genotyped using a custom Sequenom MassARRAY SNP tool, allowing us to define population structure for an Eimeria species for the first time, to our knowledge.
Results
Description of the Farms Sampled and the Occurrence of E. tenella.
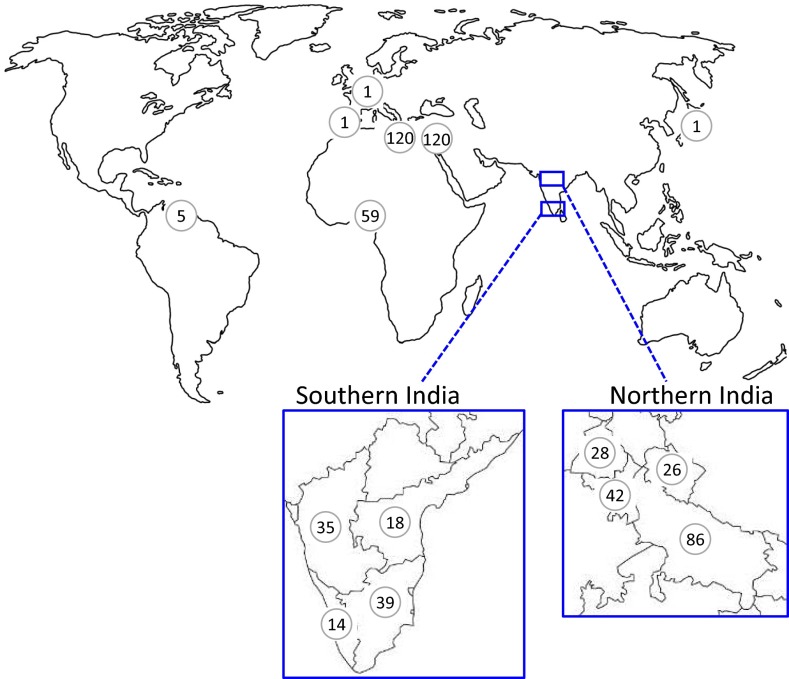
Fecal samples were collected from 595 small- to medium-scale commercial chicken farms (defined here as holding up to 50,000 birds) from eight countries, supplemented by laboratory reference strains and, when combined, representing five continents. Broiler and layer birds were sampled in approximately equal numbers in each region. Each sample comprised several fecal pellets collected from a cage or pen; thus parasites were derived from multiple birds within a single “flock.” PCR screening found E. tenella to be present in all regions that were sampled more than once, most frequently in Nigeria and Southern India (Andhra Pradesh, Karnataka, Kerala, and Tamil Nadu combined), (>80% of farms) and less frequently in Northern India (Haryana, Punjab, Uttarakhand, and Uttar Pradesh combined), Egypt, and Libya (37–66%) (Fig. S1 and Table S1).
Fig. S1.
The geographic origin of chicken fecal samples collected for use during these studies. The number within each node indicates the number of samples collected. The individual states sampled in Northern and Southern India are indicated in the expanded boxes.
Table S1.
The geographic origin of samples and sequences used during these studies
| Continent | Country/region | No. of farms of sampled for species detection by PCR | No. of farms positive for E. tenella (%) | Samples used for Sequenom SNP typing | Clones used for E. tenella AMA-1 cDNA sequencing* |
| Africa | Egypt | 120 | 44 (36.7) | 40 | 2 |
| Libya | 120 | 62 (51.7) | 51 | 5 | |
| Nigeria | 59 | 54 (91.5) | 14 | 31 | |
| Asia | China | 0 | n/a | 0 | 1 (JN032081) |
| Northern India | 182 | 121 (66.5) | 86 | 5 | |
| Southern India | 106 | 85 (80.2) | 53 | ||
| Japan | 1 | 1 (100.0) | 0 | 1 | |
| Europe | Germany | 1 | 1 (100.0) | 0 | 1 |
| Spain | 1 | 0 (0.0) | 0 | 0 | |
| United Kingdom | 0 | N/a | 0 | 3 | |
| North America | United States | 0 | N/a | 0 | 2 |
| South America | Venezuela | 5 | 5 (100.0) | 0 | 5 |
| Total | 595 | 373 (62.6) | 244 | 56 | |
N/a, not applicable.
Accession numbers LN609976-LN610031.
E. tenella Is Genetically Diverse with Evidence of Allopatric Evolution.
Sequence-led studies of genetic diversity within Eimeria species most commonly have focused on the internal transcribed spacer (ITS) sequences 1 and 2 (4). The large number of ITS copies per genome provides a high degree of sensitivity by PCR, although the occurrence of polymorphism between copies, even within single genomes, can result in overestimation of clonal diversity. For more detailed comparisons, we investigated genome-wide genetic diversity among E. tenella samples. First, genetic polymorphism between the Houghton (United Kingdom) reference and a panel of archive E. tenella strains was assessed using amplified fragment-length polymorphism (AFLP) to calculate Jaccard indices of diversity in the absence of more extensive genomic resources (13, 16). Comparison of the Houghton strain with the LCH2 and Weybridge (United Kingdom.), Beltsville and Wisconsin (United States), and Nippon-2 (Nt2; Japan) E. tenella strains revealed Jaccard indices of 0.912, 0.967, 0.956, 0.914, and 0.873, respectively. Using these data, we selected the Wisconsin and Nt2 strains for next-generation sequencing (Illumina), as described previously (13). The sequences are available from the European Nucleotide Archive under the accession number PRJEB4009. Using these resources, we identified SNPs. First, sequences covering a panel of genes with known chromosomal locations were selected to provide an even dispersal of SNPs across the 14 E. tenella chromosomes (Dataset S1) (13, 17). Additional SNPs were selected across chromosomes 1 and 2, including two to four SNPs in each repeat-rich (R) and repeat-poor (P) region (Dataset S1) (13, 18). In total, 55 SNPs were selected, incorporated into two Sequenom MassARRAY multiplexes, and used to genotype 244 field samples found to be PCR positive for E. tenella (Table 1). After SNP genotyping 52 of the 55 SNPs produced informative profiles.
Table 1.
Summary of E. tenella genome-wide genetic data calculated using Sequenom MassARRAY genotyping
| Region | N | SNP haplotypes | Het (±) | IAS | P | |
| Total | Region specific | |||||
| All | 244 | 93 | — | 0.3611 (0.0189) | 0.1238 | <0.0001 |
| Northern India | 86 | 8 | 7 | 0.3331 (0.0254) | 0.1153 | <0.0001 |
| Southern India | 53 | 50 | 49 | 0.3405 (0.0212) | 0.0087 | ns |
| Egypt | 40 | 21 | 13 | 0.1925 (0.0316) | 0.0597 | <0.0001 |
| Libya | 51 | 11 | 3 | 0.1611 (0.0247) | 0.0533 | <0.0001 |
| Egypt + Libya | 91 | 25 | 24 | 0.1789 (0.0275) | 0.0533 | <0.0001 |
| Nigeria | 14 | 14 | 13 | 0.3998 (0.0224) | 0.0012 | ns |
Northern India included samples collected in Haryana, Punjab, Uttarakhand, and Uttar Pradesh. Southern India included samples collected in Andra Pradesh, Karnataka, Kerala, and Tamil Nadu. N, the number of samples tested. The number of SNP haplotypes was calculated using DnaSP v5.10.01. Het, mean genetic diversity; IAS, standardized index of association (calculated using only the first SNP per specific gene-associated sequence or chromosomal repeat type segment to avoid bias as a consequence of close physical linkage), with statistical significance indicated as P, both calculated using LIAN v3.6; ns, not statistically significant.
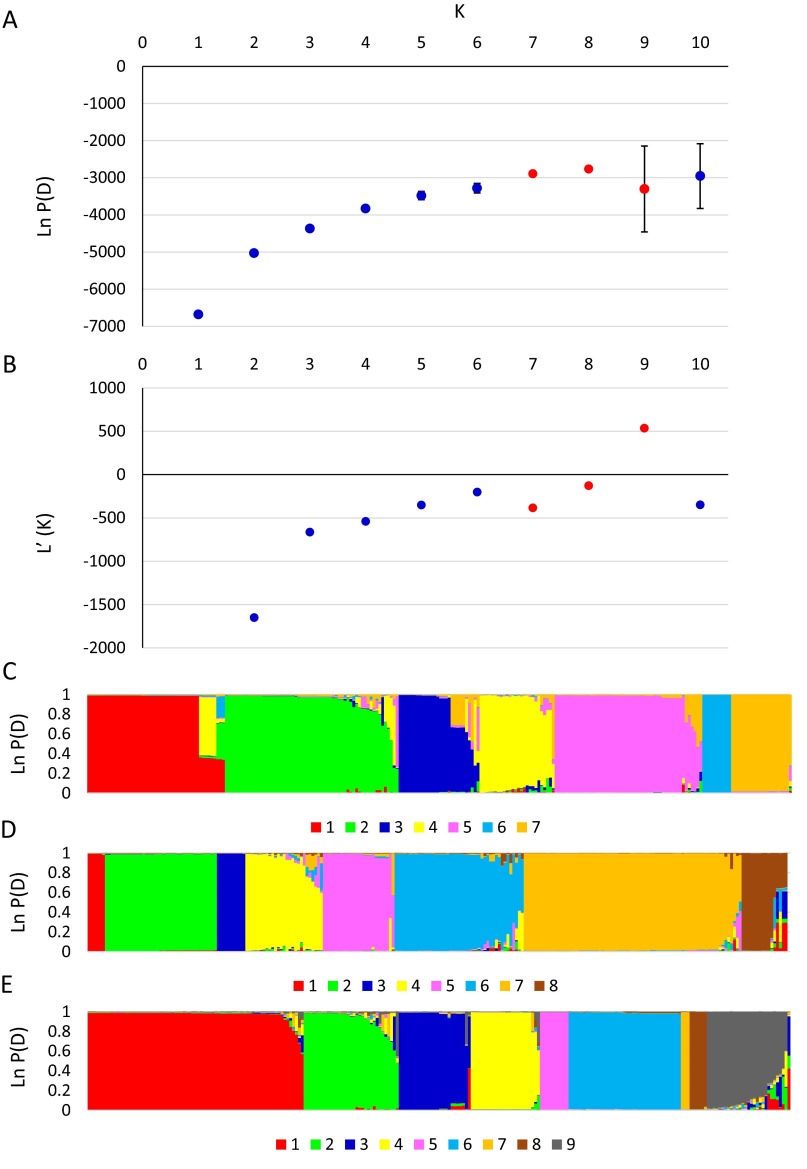
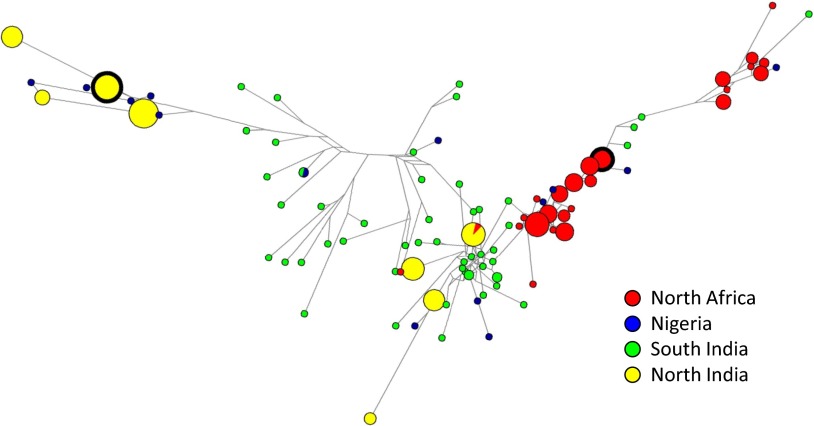
For population genetic analysis, SNP profiles were presented as the major genotype in each sample, determined by the Sequenom SNP frequency of each allelic type in cases of polyclonal populations. Where polyclonal populations were detected the major and minor SNP types were identified, and the major allele was selected for use in haplotype analysis without phasing, because each parasite presented a haploid genome at the time of sampling. The lack of replication per sample and the semiquantitative nature of the assay precluded computational assembly of haplotype complexity within each sample (19). Analysis of all SNP genotypes using DnaSP v5.10.01 (20) identified 93 SNP haplotypes in total, with striking variation in haplotype depth and diversity among regions (Table 1). In Southern India 50 SNP haplotypes were detected from 53 samples, whereas in Northern India only eight were identified from 86 samples (P < 0.005; χ2 test). Comparison between regions revealed that 98% and 87.5% of these haplotypes were unique to their regions, providing evidence of allopatric diversification. In contrast, Egypt and Libya presented an overlapping haplotype profile, so that pooling data for these neighboring countries revealed a similar level of regional specificity. Complementary analysis using STRUCTURE v2.3.4 (21) to assess population structure (K) suggested that eight ancestral genotype clusters best supported the current data, with log likelihood [Ln P(D)] plateauing when K = 7, variation increasing when K = 9, and the mean difference between successive likelihood values of K [L′(K)] peaking at K = 9 (Fig. S2 A and B). STRUCTURE output for K values of 7, 8, and 9 are shown in Fig. S2 C–E, and the input data are provided in Dataset S2. Representation of the data using NETWORK v4.6.1.1 (22) to calculate a median-joining method phylogenetic network illustrated these findings (Fig. 1). A small number of large nodes were calculated for Northern India and North Africa (data from Egypt and Libya combined) distributed across a small number of clusters, indicating multiple occurrences of conserved haplotype profiles. In contrast, Southern India presented a much larger number of smaller nodes in loosely defined clusters, representing the greater haplotype diversity and narrow population structure. The small number of Nigerian samples analyzed revealed the greatest genetic diversity with a broad spread across much of the phylogenetic network (Fig. 1 and Table 1). The distance between clusters was confirmed by calculating the mean genetic diversity (Het) within each region using LIAN v3.6 (23); the regions with more widely distributed clusters presented higher Het (Table 1).
Fig. S2.
Estimate of the ancestral population number (K) for E. tenella produced using STRUCTURE. (A) Plot of the mean log likelihood of the data [Ln P(D)] and associated SDs. Data points colored red were selected for further population structure illustration in C–E. (B) Plot of the mean difference between successive likelihood values of K [L′(K)]. (C–E) Population structure calculated using ancestral population sizes (K) of 7 (C), 8 (D), and 9 (E).
Fig. 1.
Median-joining phylogenetic NETWORK showing the relationships between Sequenom MassARRAY haplotypes for E. tenella samples collected in Northern India (yellow), Southern India (green), Egypt and Libya combined (North Africa, red), and Nigeria (blue). Node size indicates the frequency of haplotype occurrence. Nodes surrounded by a heavy black boundary indicate the haplotypes used in the cross-protection studies.
Linkage disequilibrium (LD) was calculated within the full and regional subpopulations using LIAN v3.6 to test the null hypothesis of linkage equilibrium for all loci. In recognition of the possible bias introduced by SNPs with a close physical linkage, we used the standardized index of association (IAS) using a restricted SNP panel with only the first SNP in each gene-associated sequence or chromosome-specific repeat-type region (the SNPs used are shown in Dataset S1). Statistically significant LD was identified in Northern India and North Africa but not Southern India or Nigeria (Table 1), suggesting that mating and recombination occur freely in the latter but may be restricted in the former populations.
Genomic Diversity Is Associated with Limited Escape from Strain-Specific Immune Killing for E. tenella.
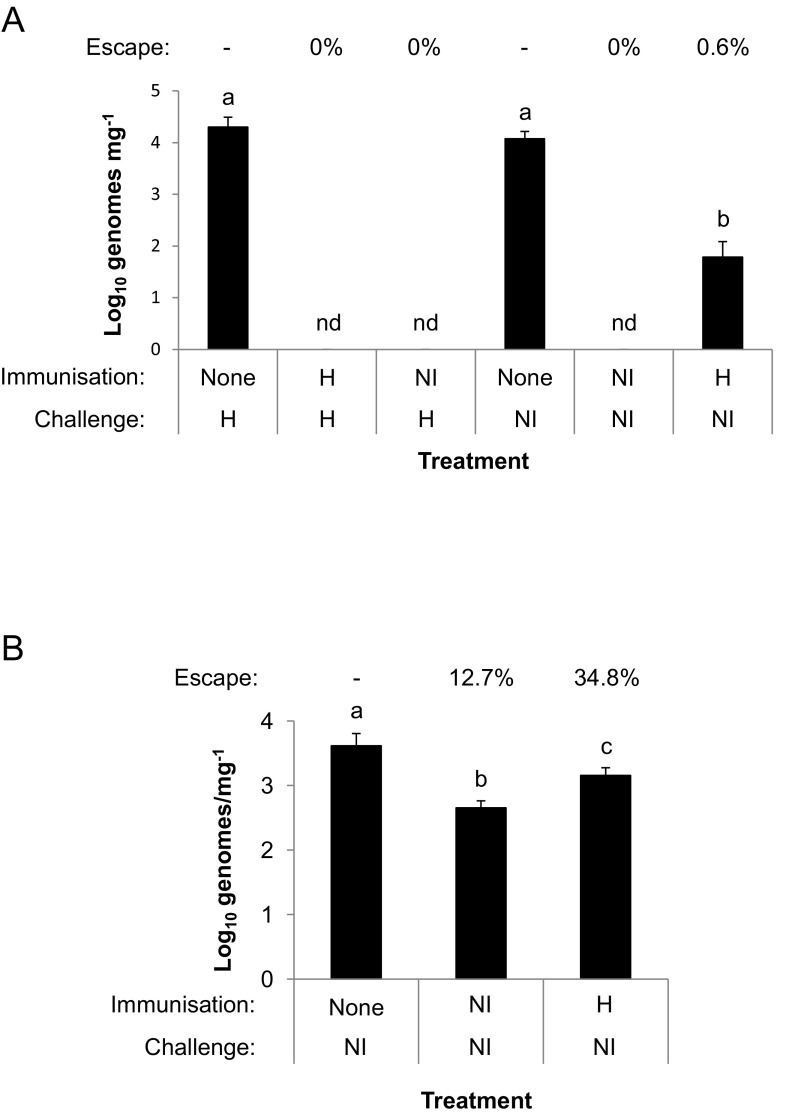
To investigate the likely efficacy of the current generation of anticoccidial vaccines against genetically diverse E. tenella, we tested cross-protective ability between two of the most distantly related strains available. The Houghton reference strain, isolated in the United Kingdom and progenitor for the attenuated E. tenella line included in the Paracox vaccine series, is most closely related to the Libyan LI_01 NETWORK cluster (red node with heavy black boundary in Fig. 1). As a comparison, we chose a clonal line isolated from Northern Indian sample NI_01 (24), which has one of the most distant SNP NETWORK profiles (yellow node with heavy black boundary in Fig. 1). Initially, immunization of outbred Light Sussex chickens at 3 and 4 wk of age used sequential doses of 1,000 and 4,000 sporulated oocysts of either the Houghton or NI strains. Subsequent homologous challenge with 250 sporulated oocysts 2 wk later was blocked, with no detectable oocyst output (Fig. S3A). Heterologous challenge was similarly blocked completely when the Houghton strain was used to challenge birds immunized by the NI strain. However, when the NI strain was used to challenge birds immunized by the Houghton strain, there was a low level of escape, with birds excreting 0.6% of the oocysts produced by unimmunized NI challenge controls. Repeating the trial for only the NI challenge component with a lower-level immunization protocol (sequential doses of 100 and 1,000 sporulated oocysts per bird, administered at 3 and 4 wks of age, respectively) revealed incomplete homologous protection (12.7% oocysts produced compared with the unimmunized control) and a significantly greater escape following heterologous challenge (34.8%; P < 0.05, ANOVA plus Bonferroni post hoc test) (Fig. S3B).
Fig. S3.
Cross-protective ability of European and Asian E. tenella isolates. (A) High-level immunizing dose. Total parasite burden (E. tenella genomes per milligram of cecal tissue, determined using species-specific quantitative PCR normalized against the host genome number 5 d postchallenge) following homologous or heterologous challenge of 6-wk-old birds immunized by infection with 1,000 and then 4,000 sporulated oocysts of the Houghton (H; European) or Northern Indian (NI; Asian) strains at 3 and 4 wk of age, respectively. The challenge dose was 250 sporulated oocysts per bird. Birds given no previous infection were included as a control. nd, no parasite genomes detected (the limit of detection was 17 genomes per gram). Parasite escape was calculated as the average number of parasite genomes detected in the immunized groups as a percentage of the number detected in the challenge strain-matched unimmunized group. (B) Low-level immunizing dose. Total parasite replication (determined as in A) following homologous (NI) or heterologous (H) challenge of 6-wk-old birds immunized by infection with 100 and then 1,000 sporulated oocysts of the NI strain at 3 and 4 wk of age, respectively.
Sequence Diversity Is Limited for the Anticoccidial Vaccine Candidate Apical Membrane Antigen 1.
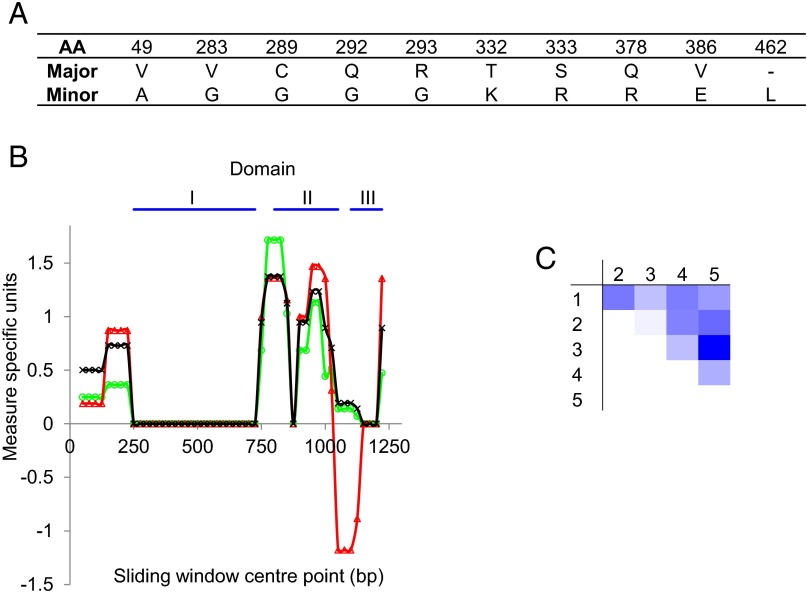
Experimental testing of subunit anticoccidial vaccines containing recombinant antigens such as apical membrane antigen-1 (AMA1) has prompted interest in the extent of naturally occurring genetic diversity in these antigens and has raised the question of whether polymorphism might result in vaccine breakthrough by field parasite populations (14). We sequenced and assembled full-length AMA1-coding sequences for 55 E. tenella samples recovered from four continents (accession nos. LN609976–LN610031) and compared these sequences with the published sequence JN032081 (Table 2 and Table S1). Analysis of the 1,608-bp coding sequence alignment revealed only 13 nucleotide substitutions and one short tandem repeat (STR) variation of the trinucleotide CTG, which is known to be common among Eimeria-coding sequences (13). Ten substitutions were nonsynonymous, resulting in nine amino acid substitutions (two were located within codon 292, glutamine to glycine; Fig. 2A). The STR variation incurred expansion/reduction of a leucine homopolymeric amino acid repeat. The greatest diversity in ama1 was in samples from Asia (China, India, and Japan) and Nigeria as defined by average pairwise difference, nucleotide diversity calculated as π with the Jukes Cantor correction, haplotype number, and haplotype diversity using DnaSP v5.10.01 (Table 2) (20). In contrast to the SNP-based genome-wide polymorphism, only four of the eight E. tenella ama1 haplotypes were region specific.
Table 2.
Summary of AMA1 genetic data
| Region | N | S | dN | dS | k | π Jukes Cantor | H | Hd | H† | Tajima's D | Fu and Li's D* | Fu and Li's F* |
| All | 56 | 13 (13) | 10 | 3 | 4.041 | 0.0032 | 8 | 0.771 | — | 1.259 | 1.513** | 1.688** |
| Asia | 7 | 12 (12) | 9 | 3 | 6 | 0.00475 | 4 | 0.857 | 2 | 1.225 | 1.573‡ | 1.642** |
| Egypt/Libya | 7 | 3 (3) | 0 | 3 | 1.714 | 0.00135 | 2 | 0.571 | 0 | 1.811 | 1.297 | 1.522 |
| Europe/United States | 6 | 4 (4) | 1 | 3 | 2.4 | 0.00189 | 3 | 0.8 | 0 | 1.640 | 1.641 | 1.670 |
| Nigeria | 31 | 11 (3) | 8 | 3 | 4.275 | 0.00338 | 4 | 0.546 | 2 | 1.766 | 1.440** | 1.805** |
| Venezuela | 5 | 2 (2) | 0 | 2 | 1.2 | 0.00095 | 2 | 0.6 | 0 | 1.459 | 1.459 | 1.432 |
All analyses were restricted to the signal peptide cleaved EtAMA1 ectodomain. Tajima’s D and Fu and Li’s D* and F* tests were used to assess of the extent or neutrality of signatures of selection with significance: **P < 0.05; ‡P < 0.02. All measures were calculated using DnaSP v5.10.01. dN, the number of nonsynonymous variant sites; dS, the number of synonymous variant sites; H, the number of sequence haplotypes detected; H†, the number of haplotypes specific to a region; Hd, the haplotype diversity; k, the average number of pairwise differences; N, the number of sequences tested; π, nucleotide diversity, calculated with the Jukes Cantor correction; S, the number of variant sites detected, with the number of parsimony-informative variant sites shown in parentheses.
Fig. 2.
The extent and impact of sequence diversity within the E. tenella AMA11 (EtAMA1) coding sequence. (A) Nonsynonymous polymorphism within the EtAMA1 coding sequence. “Major” denotes the more common sequence type identified; “minor” the less common. (B) Sliding window plot of EtAMA1 coding sequence nucleotide diversity calculated using π with the Jukes Cantor correction (multiplied by 100; green line, open circle markers) and plots of neutrality using Tajima’s D (red, open triangles) and Fu and Li’s F* (black, crosses). The window length was 100 bp with a 25-bp step size. No points were statistically significant. The EtAMA1 domain structure indicated by the blue lines was predicted based on cysteine residue locations. (C) Heat map representing the FST calculated for EtAMA1. Incremental intensity indicates elevated FST. 1, Europe; 2, Nigeria; 3, Asia; 4, the Americas; 5, North Africa.
Signatures of Selection Within E. tenella AMA1.
Antigens under deleterious immune selection commonly exhibit characteristic signatures of balancing selection. We applied Tajima’s D and Fu and Li’s D* and F* tests to the mature EtAMA1 ectodomain coding sequences to assess the occurrence and significance of any departure from neutral evolution. No significant evidence of balancing (or directional) selection was detected using Tajima’s D across the full sequence or using a 100-bp sliding window with a 25-bp step size (Fig. 2B and Table 2). Both Fu and Li’s D* and F* tests showed evidence of a low level of balancing selection acting primarily on domain II, although this finding was statistically significant only when assessed using the full sequence (Fig. 2B and Table 2). Codon-aligned nonsynonymous/synonymous substitutions (dN/dS) analysis of the mature ectodomain reinforced the overall view of low-level selective pressure with a ratio of 0.823, indicating minimal purifying selection. To define global diversity, we calculated Wright’s fixation index (FST) for the full EtAMA1 dataset and each pairwise comparison between sequences pooled as Europe, Nigeria, Asia, the Americas, and North Africa (Fig. 2C). A global figure of 0.083 suggested a high level of parasite interbreeding, although pairwise analysis revealed greater genetic distance between regions, most notably between Asia and North Africa, highlighting the absence of parasite movement and/or strong balancing selection.
Polyclonal Infection Is Common Within E. tenella Field Populations.
The finding of high levels of region-specific genomic diversity in Nigeria and Southern India, but not in Northern India, Egypt and Libya, contrasted sharply with the limited diversity determined within the EtAMA1-coding sequence dataset. Returning to the Sequenom MassARRAY genotyping data, we found 46% of all samples submitted for typing to be polyclonal as determined by a multiple SNP identity at one or more loci. The ratio of polyclonal to monoclonal populations was disproportionately higher in Southern India and Nigeria than in Northern India, Egypt, and Libya (72% and 64% compared with 10%, 12%, and 7%, respectively), likely as a consequence of the higher occurrence and larger number of haplotypes circulating. Nonetheless, opportunities for coinfection, and thus cross-fertilization, were detected in all regions sampled.
Cross-Fertilization Is Common During Polyclonal E. tenella Infection.
Polyclonal infection offers an opportunity for cross-fertilization but does not its confirm occurrence. Historic studies of cross-fertilization within Eimeria species have commonly used lethal drug or immune selection to enrich for hybrid progeny (e.g., refs. 14 and 25), obscuring the efficiency at which cross-fertilization occurs. In response to this knowledge gap, we undertook a series of coinfections between the Houghton and Nt2 E. tenella strains with oocyst ratios of 1:1, 2:1, 5:1, and 10:1 and a total dose of 500 sporulated oocysts per bird (two individual birds per ratio). Single sporocysts isolated from progeny oocysts were SNP-genotyped using Sequenom MassARRAY after FACS and whole-genome DNA amplification. In total, 36 sporocysts were genotyped for each dose ratio (Table 3). The proportion of hybrid sporocysts was highest for the 1:1 dose ratio and decreased as the ratio became more biased toward Houghton; nevertheless, hybrid parasites were detected within every sample. No single hybrid haplotype was detected more than twice, and the proportion of progeny oocysts arising from cross-fertilization always was higher than expected when assuming fertilization to have been a random event (P < 0.05; χ2 test).
Table 3.
The impact of parental ratio during E. tenella coinfection on the likelihood of cross-fertilization (36 sporocysts per ratio)
| Houghton:Nt2 ratio | No. birds | Sporocysts per bird | Self-fertilization | Cross-fertilization | |||
| Expected (Houghton or Nt2) | Observed Houghton | Observed Nt2 | Expected | Observed Houghton or Nt2 | |||
| 1:1 | 2 | 18 | 18 | 8 | 5 | 18 | 23 |
| 2:1 | 2 | 18 | 20 | 11 | 7 | 16 | 18 |
| 5:1 | 2 | 18 | 24 | 20 | 2 | 12 | 14 |
| 10:1 | 2 | 18 | 30 | 24 | 2 | 6 | 10 |
Birds were coinfected with Houghton and Nt2 E. tenella strains. Single-progeny sporocysts were identified as the product of self- or cross-fertilization following FACS purification and whole-genome amplification using Sequenom MassARRAY and comparison with both parental genotypes.
Discussion
Understanding the population structure of Eimeria species parasites is of fundamental importance for explaining the epidemiology of coccidiosis, influencing the development of effective control strategies, and defining the genetic basis of control breakthrough. As recombinant or vectored subunit anticoccidial vaccines become technically more feasible, the regional occurrence of each Eimeria species and the extent of vaccine-targeted antigenic diversity assume greater relevance (5, 14). Under experimental conditions, studies with clonal reference strains of Eimeria have found antigens such as AMA1 to induce good levels of immunoprotection (14, 26). AMA1 also is an effective vaccine candidate in related apicomplexans including Neospora caninum, T. gondii, and several Plasmodium species (27–30), but its value for Plasmodium has been undermined by high levels of naturally occurring antigenic diversity (31, 32). In Eimeria the occurrence and extent of diversity at any antigen-encoding locus is completely unknown. Additionally, the frequency with which cross-fertilization occurs between genetically distinct Eimeria strains under field conditions is unclear, obscuring the relevance of meiotic segregation and recombination to the appearance of hybrid parasite genotypes capable of escaping chemoprophylaxis or vaccination. For novel subunit anticoccidial vaccines to proceed beyond the experimental phase, evidence of likely field efficacy and longevity in the face of immune selection is essential.
Eimeria genomes most commonly have been sampled by sequencing the 18S rDNA and ITS-1 and -2 sequences (e.g., ref. 33). ITS sequences often are used for molecular diagnostics and phylogenetic analyses because of the sensitivity and specificity provided by their high copy number and relatively rapid evolution, although intraclonal polymorphism has proven limiting (33, 34). Here, multiplex SNP genotyping for E. tenella provided far greater genetic power. Comparison of haplotype occurrence and diversity provided strong evidence of the expansion of a limited number of distinct haplotypes with significant LD in Northern India and North Africa, in contrast to Southern India and Nigeria, where no LD was detected. The limited haplotype diversity detected in the Northern regions partially resembles that described for the closely related coccidian T. gondii, for which a small number of dominant clonal genotypes have been described, supplemented by higher levels of genetic diversity in regions such as South America (35). However, comparison between these two genera reveals fundamental biological differences, including an apparently obligate requirement for sexual reproduction and a massive, regularly replaced, immunologically naive pool of potential definitive hosts (chickens) for Eimeria. In Southern India and Nigeria haplotypes were rarely identified more than once, suggesting that mating and recombination occur freely in these populations and that only a fraction of the genetic diversity may have been sampled. Such diversity has been interpreted previously in Trypanosoma cruzi as indicating high levels of parasite transmission, although limited clonal extinction also may be possible (36). Possible explanations for such variation include a higher E. tenella occurrence and elevated levels of poly-haplotype infection in regions found to harbor greater haplotype complexity, indicating more frequent opportunities for hybridization. The lower parasite occurrence in Northern India and North Africa would be expected to reduce opportunities for cross-fertilization, resulting in a more limited expansion of a smaller number of haplotypes with or without true clonal development. The causes of such clear differences in parasite occurrence are not yet apparent, although the greater poultry density in Southern as compared with Northern India does affect the availability of immunologically naive hosts, increasing opportunities for Eimeria replication (37). Larger distances between sampled farms in areas of greater haplotype diversity might have explained the variation detected, indicating greater isolation and opportunity for genetic drift. However, the farm distribution and sampling strategies were the same in Northern and Southern India and are unlikely to explain such distinct population structures. Those regions that were host to higher parasite occurrence and diversity also have been recognized as being more humid [subhumid to humid, compared with semiarid to hyper-arid (38)], potentially supporting higher rates of oocyst survival and sporulation and thus a greater environmental parasite burden.
Comparison of the cross-protective immunizing capacity of live E. tenella parasites from opposing ends of the NETWORK analysis, including the progenitor of the Paracox vaccinal line, revealed significant strain-specific variation and some lack of heterologous immunoprotection, although the breakthrough in parasite replication was at a level unlikely to lead to occurrence of clinical disease (39). Antigenic diversity has been described elsewhere for E. tenella (40), albeit at a much lower level than reported for species such as E. maxima and Eimeria acervulina (14, 41, 42). The relevance of these findings to the other Eimeria species that infect poultry is not currently clear.
Previous genetic-mapping studies have shown AMA1 to be a target of protective anti-Eimeria immunity during secondary infection (14), indicating a likely balancing selective pressure, as has been described for loci encoding many other immunoprotective antigens (e.g., ref. 1). The nature of the immune response stimulated by AMA1 is not currently clear, although T cells and IFN-γ are key components of the anti-E. tenella immune response (43), and anti-recombinant E. tenella AMA1 antibodies have been shown to inhibit the invasion of host cells by sporozoites (26). Analysis of E. tenella AMA1-coding sequences for signatures of selection revealed limited and conflicting evidence. Comparison of dN/dS ratios for the E. tenella AMA1-coding sequences suggested minimal to purifying selection, in agreement with previous cross-Eimeria species Ka/Ks analysis [ratio of the number of nonsynonymous substitutions per nonsynonymous site to the number of synonymous substitutions per synonymous site; gene ETH_00007745, Ka/Ks = 0.12 (13)], implying that conservation of AMA1 protein function may be of greater importance than immune escape. In contrast, application of Fu and Li’s D* and F* tests indicated low levels of balancing selection. Combined, these findings suggest a low level of balancing selection within specific, possibly epitope-coding, regions that is consistent with the incomplete nature of the immune protection attributable to AMA1 revealed when tested as a recombinant protein or DNA vaccine (14, 26). The relatively short and apparently self-limiting in vivo Eimeria lifecycle also is likely to reduce the magnitude of antigen-specific immune selection. Prepatent periods define each Eimeria species with limited variation between strains, even when replicating in immunocompromised hosts such as MHC class I or II−/− knockout mice (11, 44). For E. tenella, microscopy-led studies have consistently identified three rounds of asexual replication before differentiation into gametocytes (45). The ability to select for serotinous lines characterized by extended prepatent periods, additional rounds of schizogony, and enhanced reproductive potential, apparently avoiding significant immune inhibition, indicates a key role for the parasite in the transition from asexual to sexual reproduction and ultimately termination of infection (46, 47). Parasite replication in the presence of minimal or incomplete immunity suggests that the host immune response has a weak role in the evolution of eimerian antigens such as AMA1, whose expression is restricted to the first zoite stage of the lifecycle (45). Alternatively, the relatively recent intensification of poultry production may indicate a similarly recent origin of intensified selection against antigens targeted by the host immune response, limiting any signature of selection. Competing selection incurred by routine chemoprophylaxis may reduce such signatures further. The low level of coding polymorphism detected here for AMA1 across temporally and spatially diverse parasite samples, combined with the cross-protection detected between genetically distinct parasite isolates when tested in outbred chickens, supports the validity of this antigen for use in future vaccine development. Additionally, these data suggest that for Eimeria the dN/dS ratio may not be an effective discriminator for identifying candidate vaccine antigens.
Eimeria are haploid for the majority of their life cycle with the exception of a short diploid phase during sexual reproduction. The appearance of genetic diversity can be accelerated by such a sexual phase, offering an opportunity for cross-fertilization and subsequent independent chromosomal segregation and recombination. The impact of genetic exchange during sexual replication on population structure is largely influenced by the balance between self- and cross-fertilization, which itself will vary by parasite availability as a consequence of survival in the environment and chicken behavior (48). Thus, the frequency at which polyclonal infection occurs is a key determinant. Using a panel of 52 SNPs within an ∼52 Mb genome (13), we found that 46% of all field samples containing E. tenella included at least two distinct genotypes. This finding is almost certainly an underestimate, and the inclusion of more markers or subjecting each sample to next-generation sequencing would likely increase this proportion. Quantitative modeling of SNP proportions at each locus could provide information about the clonal complexity of each sample (19), although this modeling would have required a greater number of replicates. Thus, although it is clear that opportunities for cross-fertilization would have been common for the parasites in our dataset, the proportion of cross-fertilization events per opportunity could not be determined. To shed light on the frequency of cross-fertilization events, we carried out single-sporocyst genotyping on the progeny of a series of experimental biclonal infections and identified multiple hybrid haplotypes from every one of these infections. We used the sporocyst, which contains two identical haploid genomes, as the biological unit for this work to allow FACS isolation of single-clonal units after the completion of meiosis, reducing the risk of allelic drop out (49, 50). Comparison of the proportion of hybrid and parental genotypes revealed higher than expected numbers of hybrid progeny. In the absence of knowledge defining gametocyte ratios during coinfection with the two parasite lines used in the cross, a feature known to vary for the related apicomplexan Plasmodium falciparum (51), it was not possible to assess any bias between cross- and self-fertilization. Nonetheless, it is clear that cross-fertilization was common, highlighting the efficiency with which E. tenella can hybridize and indicating the ease with which vaccine- or drug-resistant alleles may be disseminated and mixed in field parasite populations. Corroboratory data from field parasite populations is scarce, but the large number of singleton haplotypes from Southern India and Nigeria is characteristic of populations with high rates of recombination (52).
Thus, we conclude that Eimeria species that infect chickens are ubiquitous and that at least one species, E. tenella, exhibits variable population structures with a concomitant variation in the probability of genetic exchange. Consequently, when resistance (e.g., to anticoccidial drugs or to possible future single antigen recombinant or subunit vaccines) emerges, it is likely to disseminate rapidly through production systems. However, eimerian antigens such as AMA1 retain promise as anticoccidial vaccine candidates, given the limited genetic and antigenic diversity observed and a conservative signature of selection. The incomplete nature of the protection induced by AMA1 immunization may prove beneficial to vaccine longevity by allowing local field strains to cycle at a level adequate to boost host immune protection and dilute genetic selection for vaccine resistance in a manner comparable to the success of ionophore-based chemoprophylaxis (53). Ultimately, combinations of two or more antigens are likely to be required to control each target Eimeria species.
Materials and Methods
Detailed methods are included in SI Materials and Methods. All primers used were synthesized by Sigma-Genosys and are presented in Table S2.
Table S2.
Details of primers used, their source, annealing temperature, and expected amplicon size
| Target | Name | Sequence (5′–3′) | Reference or source | Annealing | Amplicon size, bp |
| E. tenella | TEN-F | TCGTCTTTGGCTGGCTATTC | Vrba et al., 2010 (63) | 56 °C | 100 |
| gDNA, diagnostic | TEN-R | CAGAGAGTCGCCGTCACAGT | Vrba et al., 2010 (63) | ||
| Eimeria 5S rRNA | 5S_For | TCATCACCCAAAGGGATT | Blake et al., 2006 (64) | 56 °C | ∼110 |
| 5S_Rev | TTCATACTGCGTCTAATGCAC | Blake et al., 2006 (64) | |||
| E. tenella AMA-1 | EtAMA1_fl_F | CACCATGCGGCGGCTTTC | This study | 60 °C | 1,610* |
| cDNA | EtAMA1_fl_R | CCTGGTCCAGCAGCACTTGG | This study |
Amplicon size calculated using cDNA as template.
Ethics Statement.
This study was carried out in strict accordance with the Animals (Scientific Procedures) Act 1986, an Act of Parliament of the United Kingdom. All animal studies and protocols were approved by the Royal Veterinary College Ethical Review Committee and the United Kingdom Government Home Office under the project licenses 30/2545 and 70/7781.
Field Sample Selection.
A panel of 595 fecal samples was collected from eight countries (Table S1). India was most highly represented, with 288 samples collected from the states of Haryana, Punjab, Uttarakhand, and Uttar Pradesh in Northern India and from Andhra Pradesh, Karnataka, Kerala, and Tamil Nadu in Southern India (Fig. S1). Egypt and Libya were equally represented with 120 farms sampled for each, drawn from sites around Alexandria and Cairo and around Benghazi, Zawilah, Misrata, Sirte, and Tripoli, respectively. Every sample was collected from a single pen per farm by walking a predetermined “W” pathway and collecting one fresh dropping every two to five paces (SI Materials and Methods). Thus, each sample represented multiple birds within a single cohort on a single farm. Because of differences in the availability of information, accessibility of farms, and legislation governing the exportation of biological samples, sampling frames using records from veterinary services, poultry suppliers, and farmer organizations were compiled using different approaches in each of the partner countries. Because the aim of the study was to sample for genetic diversity, farms were chosen to maximize parasite diversity, including intensive and extensive broiler and layer systems. Farms less than 100 m away from a previously sampled farm were excluded, as were farms with shared staff or equipment. In countries where 18 or more farms were sampled, those identified were compiled into lists of broiler and layer farms. A panel of farms representing each production type in each country then was randomly selected using Microsoft Excel randomizer (Microsoft Corporation). Samples were collected from one pen per farm where birds older than 3 wk of age were available.
Parasites and Samples.
E. tenella isolates were maintained, purified, and propagated as described previously (11, 54, 55). Genomic DNA was isolated and purified using a QIAamp DNA Stool Mini kit (Qiagen) including an additional physical smashing step (56). Total RNA was prepared from sporulated oocyst field samples using the same approach, followed by purification using a Qiagen RNeasy mini kit as recommended by the manufacturer.
Genetic Crosses: Parasites and Animals, Isolation of Single Sporocysts, and Genomic DNA Preparation.
The E. tenella Houghton and Nt2 strains were used for in vivo coinfection studies (13). Sporulated oocysts recovered from each genetic cross were disrupted mechanically to release sporocysts, which were sorted into 96-well plates using a FACSAria flow cytometer (Becton Dickinson; undertaken at the University of Oxford, Oxford, UK). Single sporocysts were disrupted by five cycles of freeze/thawing alternating between a methanol dry ice bath and the ambient temperature. Disrupted sporocysts were lysed and used as template for whole-genome amplification using a REPLI-g midi kit (Qiagen) as recommended by the manufacturer.
Sequenom SNP Panel Design.
To ensure an even spread of polymorphisms throughout the E. tenella genome, a two-level approach was used in the design of the Sequenom assay SNP panel. The first level of selection focused on gene-specific SNPs. Sequence polymorphisms in chromosomally assigned genes and genes encoding proteins that function at the host/parasite interface were compared for the E. tenella Houghton and Nt2 assemblies (Dataset S1) (13, 17). Gene-specific polymorphisms were selected manually using the local BLAST and sequence comparison tools in CLC Main Workbench (CLC Bio). E. tenella genome and HAPPY (mapping based on the analysis of approximately HAPloid DNA samples using PolYmerase chain reaction) mapping information was used where possible to attribute each gene-specific SNP to a chromosome to give an even dispersal of gene-specific SNPs across the 14 E. tenella chromosomes (13, 17).
The second level of SNP selection was based on intrachromosomal polymorphism in chromosomes 1 and 2. The R and P segmental structure of chromosome 1 and 2 (13, 18) was used as a template for SNP selection by placing a minimum of two SNPs in each R and P region (Dataset S1). R and P regions were designated from published parameters for chromosome 1 (18) and from contigs annotated as R and P regions for chromosome 2 (13). The Genome Analysis Toolkit (GATK, version 2.6-4; Broad Institute) was used to call SNPs for the nucleotides against the Houghton chromosome 1 and 2 assemblies and then to filter for quality (57, 58). SNPs were selected manually by visual comparison in Artemis (version 15; Sanger Institute) for quality across multiple sequence reads and positions within the chromosome to give a uniform distribution within each region (59).
Oligonucleotides for the Sequenom MassARRAY were designed by Sequenom within the left and right 150-bp SNP-flanking regions. Flanking regions for the panel of SNPs were identified manually based on the E. tenella Houghton assembly in CLC Main Workbench and chromosome 1 and 2 using in-house scripts. The MassARRAY genotyping assays were optimized for multiplexing by Sequenom, which separated the panel of SNPs into two separate multiplexes comprising 55 SNPs in total (Dataset S1) (60).
Genetics Analysis.
Software used in these studies to analyze the MassARRAY-generated SNPs included DnaSP v5.10.01 (20), NETWORK v4.6.1.1 (22), STRUCTURE v2.3.4 (21), and LIAN v3.6 (23). Gene-specific population analysis used DnaSP v5.10.01 and SNAP v2.1.1 (61).
SI Materials and Methods
Field Sample Collection.
Fecal samples were collected across each poultry unit following a W-shaped pathway as described previously (62). Briefly, every two to five paces (depending on the size of the unit), one fresh dropping was collected into 50-mL polypropylene conical tubs with screw tops. Samples were fixed in a locally purchased methylated spirit or ethanol solution or were preserved in a 2% (wt/vol) potassium dichromate solution (Sigma Aldrich). Three to five tubes were filled per pen. Each tube then was properly capped, and the contents were mixed thoroughly by vigorous shaking. The samples thus collected were refrigerated at 4 °C before transportation to the United Kingdom under Importation of Animal Pathogens Order (IAPO) permits PATH/71/2010/1 and PATH/71/2011/1–3, 6, 9, 10–13.
Isolation and Identification of Oocysts from Field Samples.
Fecal samples were homogenized by vigorous vortex mixing for ∼1 min. Initially, 1 mL of each fecal sample was added to 9 mL saturated sodium chloride solution (1:10 dilution) for quantification of oocysts per milliliter using a standard McMaster technique (55). Subsequently, additional oocysts were isolated from each sample found to be positive using a modified salt flotation technique. In brief, ∼44 mL saturated salt solution and 1.6 g NaCl were mixed with 6 mL of fecal homogenate. Samples were overlaid with 2 mL of distilled water and were allowed to separate for 10 min before centrifugation (800 × g for 9 min). To purify oocysts, the top layer was transferred using a sterile disposable pastette to microcentrifuge tubes, diluted in distilled water 1:2 (vol/vol), and centrifuged (10,000 × g for 1 min). The supernatant was discarded. Purified oocysts were disrupted in a Mini-BeadBeater-8 (BioSpec) with sterile 0.4- to 0.6-mm glass beads in sterile PBS, and genomic DNA was extracted from the homogenate using a QIAamp DNA Stool Mini kit (Qiagen) (56). Total RNA was prepared from sporulated oocyst field samples using the same physical smashing step followed by purification using a Qiagen RNeasy Mini Kit as recommended by the manufacturer.
Controlled Conditions Experiments: Parasites and Animals.
The E. tenella Houghton and Nt2 strains were used for in vivo coinfection studies (13). Where required, oocysts were propagated in vivo and prepared as described previously (16) using 4-wk-old specific pathogen-free Light Sussex chickens (Pirbright Institute).
PCR.
Standard PCR amplification was completed using Invitrogen Taq DNA polymerase (Life Technologies). Each PCR contained 25 ng template DNA, 20 pmol of relevant forward and reverse primers, 0.5 U Taq DNA polymerase, 10 mM Tris⋅HCl, 1.5 mM MgCl2, 50 mM KCl, and 0.2 mM dNTP. Standard cycle parameters were 1 × (1 min at 95 °C), 30 × (30 s at 94 °C, 1 min at 52–60 °C, and 1 min per kilobase of amplicon at 72 °C), and 1 × (10 min at 72 °C). Where required, cDNA was prepared from total RNA using Invitrogen SuperScript II reverse transcriptase and oligo dT, as described by the manufacturer (Life Technologies). PCR fragments were cloned using pGEM-T Easy (Promega) in XL1-Blue MRF Escherichia coli (Stratagene), miniprepped (Qiagen) and sequenced (GATC Biotech) as described by the respective manufacturers. Sequence assembly, annotation, and interrogation were undertaken using CLC Main Workbench v6.0.2 (CLC Bio) except where otherwise stated.
All primers used were synthesized by Sigma-Genosys and are presented in Table S2. Eimeria occurrence in each field sample was determined initially by PCR targeting the 5S rRNA repeat (6). Samples found to contain Eimeria DNA were screened for species identification using primers specific for E. tenella as described previously (62, 63). E. tenella AMA1-coding sequences were amplified from cDNA prepared from sporulated oocysts using primers targeting the sequences flanking the gene start and stop codons (gene ETH_00007745, located on E. tenella Houghton strain genome contig Eth_scaff19; GenBank accession no. HG674029).
Molecular Quantification of Parasite Replication.
Whole paired cecal tissues collected 5 d after in vivo challenge of chickens by E. tenella were homogenized in the minimum volume of PBS required for sample coverage using a Qiagen TissueRuptor. A 100-µL aliquot was processed for total genomic DNA extraction using a Qiagen DNeasy Blood and Tissue kit as recommended by the manufacturer. The extent of parasite replication was determined using quantitative PCR to calculate the total number of E. tenella genomes per host genome, targeting the E. tenella sequence-characterized amplified regions (SCAR) marker Tn-E03-1161 and the chicken GAPDH coding sequence as described elsewhere (63–65).
In Vivo Experimental Design: Genetic Crosses.
The reference E. tenella Houghton strain was chosen as the first parent for all genetic crosses, with the laboratory strain showing the greatest diversity (calculated using AFLP) selected as the second to maximize polymorphism. Briefly, the LCH2 and Weybridge (Europe), Beltsville and Wisconsin (United States), and Nt2 (Japan) E. tenella laboratory strains were characterized together with the Houghton reference strain using AFLP as described previously with the enzymes EcoRI and MseI and the primer extensions E-C and M-CA or M-CG, respectively (14). Data were used to calculate Jaccard indices of variation compared with the Houghton strain (16). Subsequently, doses of 500 sporulated oocysts (mixtures of the Houghton and Nt2 strains in 1:1, 2:1, 5:1, or 10:1 ratios; two birds per ratio) were administered orally in 0.5 mL distilled water to individually caged 4-wk-old specific pathogen-free Light Sussex chickens (Pirbright Institute). Oocysts were recovered by cecal harvest postmortem and were sporulated and purified following routine protocols (66), keeping samples from each bird separate.
Isolation of Single Sporocysts and Genomic DNA Preparation.
Sporulated oocysts recovered from each genetic cross were disrupted mechanically by vortex mixing in PBS (Sigma) with autoclaved (121 °C for 15 min) glass beads (0.4–0.6 mm, Sigma), checked microscopically under 10×/20× magnification after each 20-s burst. Sporocysts released were purified by running through 0.2- to 0.3-mm glass beads (Sigma) in a 1.0-mL syringe barrel plugged with glass wool (Fisher). Single E. tenella sporocysts were sorted into 96-well plates in 10 µL DNase-free PBS using a FACSAria flow cytometer (Becton Dickinson; undertaken at the University of Oxford, Oxford, UK). Sorted parasites were stored at −80 °C. Single sporocysts were disrupted subsequently by five cycles of freeze/thawing in a methanol dry ice bath and ambient temperature. Disrupted sporocysts were lysed using freshly prepared buffer D2 from a REPLI-g midi kit (Qiagen) incubated on wet ice for 10 min followed by 65 °C for 5 min. Whole-genome amplification then was completed as recommended by the manufacturer. The presence of eimerian genomic DNA was confirmed by PCR amplification of the 5S rRNA gene repeat (64). Whole-genome–amplified DNA then was stored at −80 °C before Sequenom genotyping.
In Vivo Experimental Design: Immunotyping Field Isolates.
The ability of a field isolate to escape immunity induced by previous infection with a European reference strain was assessed in vivo, including six treatment groups of eight specific pathogen-free Light Sussex chickens (Pirbright Institute). All birds were caged in pairs in coccidia-free conditions. Initially, chickens in groups 1 and 2 were mock infected by oral gavage of 1.0 mL sterile PBS at 3 wk of age. In parallel, chickens in groups 3 and 4 were infected by gavage with 1,000 sporulated E. tenella oocysts of the reference Houghton strain (13). Chickens in groups 5 and 6 were infected by equivalent inoculation with the cloned North Indian E. tenella NI strain (24). Infection was repeated using the same groups 1 wk later with a higher dose of 4,000 sporulated oocysts per bird. Six-week-old chickens were challenged with 250 sporulated E. tenella oocysts. Groups 1, 3, and 5 received the E. tenella Houghton strain, and groups 2, 4, and 6 received the E. tenella NI strain. All chickens were killed 5 d post challenge when both caeca were collected from each bird, opened longitudinally, rinsed in sterile PBS and stored in RNAlater (Life Technologies). The NI strain challenge component of the trial was repeated using lower infecting doses (100 and 1,000 oocysts) with group 1 mock infected, group 2 infected with the NI strain, and group 3 infected with the Houghton strain before challenge of all groups by the NI strain. Parasite replication was quantified using quantitative PCR as described above.
Sequenom SNP Panel Design.
To ensure an even spread of polymorphisms throughout the E. tenella genome, a two-level approach was used in the design of the Sequenom assay SNP panel. The first level of selection focused on gene-specific SNPs. Sequence polymorphisms in chromosomally assigned genes and genes encoding proteins that function at the host/parasite interface were compared for the E. tenella Houghton and Nt2 assemblies (Dataset S1) (13, 17). Gene-specific polymorphisms were selected manually using the local BLAST and sequence comparison tools in CLC Main Workbench (CLC Bio). E. tenella genome and HAPPY mapping information was used where possible to attribute each gene-specific SNP to a chromosome to give an even dispersal of gene-specific SNPs across the 14 E. tenella chromosomes (13, 17).
The second level of SNP selection was based on intrachromosomal polymorphism in chromosomes 1 and 2. The R and P segmental structure of chromosome 1 and 2 (13, 18) was used as a template for SNP selection by placing a minimum of two SNPs in each R and P region (Dataset S1). R and P regions were designated from published parameters for chromosome 1 (18) and from contigs annotated as R and P regions for chromosome 2 (13). The Genome Analysis Toolkit (version 2.6-4; Broad Institute) was used to call SNPs for the Nt against the Houghton chromosome 1 and 2 assemblies and then to filter for quality (57, 58). SNPs were selected manually by visual comparison in Artemis (version 15; Sanger Institute) for quality across multiple sequence reads and position within the chromosome to give a uniform distribution within each region (59).
Oligonucleotides for the Sequenom MassARRAY were designed by Sequenom (Sequenom Inc.) within the left and right 150-bp SNP flanking regions. Flanking regions for the panel of SNPs were identified manually based on the E. tenella Houghton assembly in CLC Main Workbench and chromosome 1 and 2 using in-house scripts. The MassARRAY genotyping assays were optimized for multiplexing by Sequenom, which separated the panel of SNPs into two separate multiplexes comprising 55 SNPs in total (Dataset S1).
Sequenom Genotyping.
MassARRAY genotyping was performed by Sequenom (60). Briefly, 5 ng DNA of each sample was air dried for 24 h under ambient conditions and submitted in 384-well plates. MALDI-TOF mass spectrometer-read genotypes were recorded as single allele or major/minor allele SNP profiles.
Genome-Wide Population Genetics Analysis.
In total, 244 samples from Egypt, India, Libya, and Nigeria found to contain E. tenella genomic DNA were submitted to Sequenom for MassARRAY genotyping and used for genome-wide population analysis. Where polyclonal infection was indicated by the presence of multiple alleles, the dominant SNP type was used for genetic analysis. DnaSP v5.10.01 (20) was used to calculate the number of SNP haplotypes (H) in the full dataset and by region, with the number unique to each region confirmed manually. NETWORK v4.6.1.1 was used with the median-joining method to visualize the haplotype network (22). Epsilon was set to zero, and loci were weighted 10:1 in terms of variation. The MP option was included postprocessing to purge superfluous links. Population structure was assessed using STRUCTURE v2.3.4 (21) to assign multilocus genotypes to the optimal number of ancestral clusters. The number of populations (K) was tested from 1 to 10 with 10-fold replication for 100,000 Monte Carlo Markov Chain iterations after a burn-in period of 10,000 (67). The mean log probability of the data [LnP(D)] was plotted, together with the associated SD, to identify the optimal value for K for each population [where LnP(D) plateaued before SD increase]. The mean difference between successive likelihood values of K [L′(K)] was calculated as L(K) − L(K − 1) (35). LD was assessed using LIAN version 3.6 to test the null hypothesis of linkage equilibrium for all loci (23). The Monte Carlo test option was chosen with 100,000 iterations. IAS was calculated as a measure of LD, where 0 = equilibrium, as well as the mean genetic diversity (Het). To minimize possible bias introduced by the Sequenom SNP marker distribution, LD analysis was repeated with a subset of 13 SNP markers including only the first SNP in each gene or those from the ends of the chromosome 1 and 2 sequences (Dataset S1).
Gene-Specific Population Analysis.
Genetic analysis of 56 E. tenella AMA-1 ectodomain sequences included nucleotides 70–1,339 of the coding sequence using DnaSP v5.10.01 (20) after removal of the putative signal and transmembrane sequences (GenBank accession nos. LN609976–LN610031). Analyses were performed on the full sample set and the subsets Asia (representing India, China, and Japan), Egypt plus Libya, Europe/United States, Nigeria, and Venezuela. Analyses included identification of the number of variant sites (S) and numbers of nonsynonymous (dN) or synonymous (dS) substitutions. A description of diversity was provided by calculating average nucleotide differences (k) and nucleotide diversity [π with the Jukes Cantor correction (68)] using (i) the total number of mutations and (ii) a sliding window analysis with a length of 100 bp and a step size of 25 bp. Allelic diversity was defined by calculation of haplotype number (H) and the associated haplotype diversity (Hd) (69). Neutrality was assessed using Tajima’s D (70) and Fu and Li’s D* and F* (71) with the total number of mutations and significance set at P < 0.05. Additionally, Tajima’s D and Fu and Li’s F* were calculated with a sliding window of 100 bp and a step size of 25 bp. Rates of nonsynonymous (dN) and synonymous (dS) substitution and the dN/dS ratio were calculated using SNAP v2.1.1 (61). Population structure as a result of gene flow was assessed using the fixation index (Fst) with pairwise fixation values calculated (72), using 1,000 replicates as a permutation test.
Statistics.
The arithmetic mean and associated SEM for each sample or group were calculated using Excel (Microsoft Excel 2007). Statistical analyses were performed using ANOVA with a Bonferroni post hoc analysis, general linear model regression, or χ2 tests in SPSS Statistics version 20 (IBM). Differences were deemed significant with a P value < 0.05.
Supplementary Material
Acknowledgments
We thank S. Kumar, S. Aarthi, R. Selvabharathi, Dr. Abdul S. Chaudhry, Prof. Olivier Sparagano, Prof. Idris A. Lawal, Prof. J. K. P. Kwaga, and Prof. A. J. Nok for assistance in arranging for the collection of samples used in these studies. The work described here was funded by the UK Department for International Development and the Biotechnology and Biological Sciences Research Council through the Combating Infectious Diseases of Livestock for International Development initiative under Project Reference BB/H009337. This paper has been assigned the reference no. PPB_00975 by the Royal Veterinary College.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. LN609976–LN610031 and LN626984–LN626991).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506468112/-/DCSupplemental.
References
- 1.Amambua-Ngwa A, et al. Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PLoS Genet. 2012;8(11):e1002992. doi: 10.1371/journal.pgen.1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minot S, et al. Admixture and recombination among Toxoplasma gondii lineages explain global genome diversity. Proc Natl Acad Sci USA. 2012;109(33):13458–13463. doi: 10.1073/pnas.1117047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manske M, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487(7407):375–379. doi: 10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck HP, et al. Molecular approaches to diversity of populations of apicomplexan parasites. Int J Parasitol. 2009;39(2):175–189. doi: 10.1016/j.ijpara.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Blake DP, Tomley FM. Securing poultry production from the ever-present Eimeria challenge. Trends Parasitol. 2014;30(1):12–19. doi: 10.1016/j.pt.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 6.O’Neill BC, et al. Global demographic trends and future carbon emissions. Proc Natl Acad Sci USA. 2010;107(41):17521–17526. doi: 10.1073/pnas.1004581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smil V. Worldwide transformation of diets, burdens of meat production and opportunities for novel food proteins. Enzyme Microb Technol. 2002;30(3):305–311. [Google Scholar]
- 8.Godfray HC, et al. Food security: The challenge of feeding 9 billion people. Science. 2010;327(5967):812–818. doi: 10.1126/science.1185383. [DOI] [PubMed] [Google Scholar]
- 9.Perry B, Randolph T, McDermott J, Sones K, Thornton P. 2002. Investing in Animal Health Research to Alleviate Poverty (International Livestock Research Institute), Nairobi, Kenya)
- 10.Dalloul RA, Lillehoj HS. Poultry coccidiosis: Recent advancements in control measures and vaccine development. Expert Rev Vaccines. 2006;5(1):143–163. doi: 10.1586/14760584.5.1.143. [DOI] [PubMed] [Google Scholar]
- 11.Shirley MW, Smith AL, Tomley FM. The biology of avian Eimeria with an emphasis on their control by vaccination. Adv Parasitol. 2005;60:285–330. doi: 10.1016/S0065-308X(05)60005-X. [DOI] [PubMed] [Google Scholar]
- 12.Peek HW, Landman WJ. Coccidiosis in poultry: Anticoccidial products, vaccines and other prevention strategies. Vet Q. 2011;31(3):143–161. doi: 10.1080/01652176.2011.605247. [DOI] [PubMed] [Google Scholar]
- 13.Reid AJ, et al. Genomic analysis of the causative agents of coccidiosis in domestic chickens. Genome Res. 2014;24(10):1676–1685. doi: 10.1101/gr.168955.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blake DP, et al. Genetic mapping identifies novel highly protective antigens for an apicomplexan parasite. PLoS Pathog. 2011;7(2):e1001279. doi: 10.1371/journal.ppat.1001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman HD, Shirley MW. The Houghton strain of Eimeria tenella: A review of the type strain selected for genome sequencing. Avian Pathol. 2003;32(2):115–127. doi: 10.1080/0307945021000071588. [DOI] [PubMed] [Google Scholar]
- 16.Blake DP, et al. Parasite genetics and the immune host: Recombination between antigenic types of Eimeria maxima as an entrée to the identification of protective antigens. Mol Biochem Parasitol. 2004;138(1):143–152. doi: 10.1016/j.molbiopara.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Lim LS, Tay YL, Alias H, Wan KL, Dear PH. Insights into the genome structure and copy-number variation of Eimeria tenella. BMC Genomics. 2012;13(1):389. doi: 10.1186/1471-2164-13-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ling KH, et al. Sequencing and analysis of chromosome 1 of Eimeria tenella reveals a unique segmental organization. Genome Res. 2007;17(3):311–319. doi: 10.1101/gr.5823007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmid-Hempel P, Reber Funk C. The distribution of genotypes of the trypanosome parasite, Crithidia bombi, in populations of its host, Bombus terrestris. Parasitology. 2004;129(Pt 2):147–158. doi: 10.1017/s0031182004005542. [DOI] [PubMed] [Google Scholar]
- 20.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 21.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16(1):37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 23.Haubold B, Hudson RR. LIAN 3.0: Detecting linkage disequilibrium in multilocus data. Linkage Analysis. Bioinformatics. 2000;16(9):847–848. doi: 10.1093/bioinformatics/16.9.847. [DOI] [PubMed] [Google Scholar]
- 24.Kundu K, et al. Cloning and sequencing of beta-tubulin and internal transcribed spacer-2 (ITS-2) of Eimeria tenella isolate from India. J Parasit Dis. 2014 doi: 10.1007/s12639-013-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joyner LP, Norton CC. Transferred drug-resistance in Eimeria maxima. Parasitology. 1975;71(3):385–392. doi: 10.1017/s0031182000047168. [DOI] [PubMed] [Google Scholar]
- 26.Jiang L, et al. Identification and characterization of Eimeria tenella apical membrane antigen-1 (AMA1) PLoS One. 2012;7(7):e41115. doi: 10.1371/journal.pone.0041115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, et al. Neospora caninum: Application of apical membrane antigen 1 encapsulated in the oligomannose-coated liposomes for reduction of offspring mortality from infection in BALB/c mice. Exp Parasitol. 2010;125(2):130–136. doi: 10.1016/j.exppara.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Hodder AN, Crewther PE, Anders RF. Specificity of the protective antibody response to apical membrane antigen 1. Infect Immun. 2001;69(5):3286–3294. doi: 10.1128/IAI.69.5.3286-3294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crewther PE, Matthew ML, Flegg RH, Anders RF. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect Immun. 1996;64(8):3310–3317. doi: 10.1128/iai.64.8.3310-3317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dautu G, et al. Toxoplasma gondii: DNA vaccination with genes encoding antigens MIC2, M2AP, AMA1 and BAG1 and evaluation of their immunogenic potential. Exp Parasitol. 2007;116(3):273–282. doi: 10.1016/j.exppara.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 31.Eisen DP, Saul A, Fryauff DJ, Reeder JC, Coppel RL. Alterations in Plasmodium falciparum genotypes during sequential infections suggest the presence of strain specific immunity. Am J Trop Med Hyg. 2002;67(1):8–16. doi: 10.4269/ajtmh.2002.67.8. [DOI] [PubMed] [Google Scholar]
- 32.Healer J, et al. Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Mol Microbiol. 2004;52(1):159–168. doi: 10.1111/j.1365-2958.2003.03974.x. [DOI] [PubMed] [Google Scholar]
- 33.Schwarz RS, Jenkins MC, Klopp S, Miska KB. Genomic analysis of Eimeria spp. populations in relation to performance levels of broiler chicken farms in Arkansas and North Carolina. J Parasitol. 2009;95(4):871–880. doi: 10.1645/GE-1898.1. [DOI] [PubMed] [Google Scholar]
- 34.Schnitzler BE, Thebo PL, Tomley FM, Uggla A, Shirley MW. PCR identification of chicken Eimeria: A simplified read-out. Avian Pathol. 1999;28(1):89–93. doi: 10.1080/03079459995091. [DOI] [PubMed] [Google Scholar]
- 35.Su C, et al. Globally diverse Toxoplasma gondii isolates comprise six major clades originating from a small number of distinct ancestral lineages. Proc Natl Acad Sci USA. 2012;109(15):5844–5849. doi: 10.1073/pnas.1203190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramírez JD, et al. Contemporary cryptic sexuality in Trypanosoma cruzi. Mol Ecol. 2012;21(17):4216–4226. doi: 10.1111/j.1365-294X.2012.05699.x. [DOI] [PubMed] [Google Scholar]
- 37.USDA Poultry in India. International Egg and Poultry Review. 2013;16(46):1–3. [Google Scholar]
- 38.Deichmann U, Eklundh L. Global Digital Data Sets for Land Degradation Studies: A GIS Approach. United Nations Environment Program/Global Environment Monitoring System and Global and Regional Integrated Data Centers; Nairobi, Kenya: 1991. [Google Scholar]
- 39.McDonald V, Shirley MW, Millard BJ. A comparative study of two lines of Eimeria tenella attenuated either by selection for precocious development in the chicken or by growth in chicken embryos. Avian Pathol. 1986;15(3):323–335. doi: 10.1080/03079458608436296. [DOI] [PubMed] [Google Scholar]
- 40.Awad AM, El-Nahas AF, Abu-Akkada SS. Evaluation of the protective efficacy of the anticoccidial vaccine Coccivac-B in broilers, when challenged with Egyptian field isolates of E. tenella. Parasitol Res. 2013;112(1):113–121. doi: 10.1007/s00436-012-3112-6. [DOI] [PubMed] [Google Scholar]
- 41.Joyner LP. Immunological variation between two strains of Eimeria acervulina. Parasitology. 1969;59(3):725–732. doi: 10.1017/s0031182000031243. [DOI] [PubMed] [Google Scholar]
- 42.Blake DP, et al. EmaxDB: Availability of a first draft genome sequence for the apicomplexan Eimeria maxima. Mol Biochem Parasitol. 2012;184(1):48–51. doi: 10.1016/j.molbiopara.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 43.Hong YH, Lillehoj HS, Lee SH, Dalloul RA, Lillehoj EP. Analysis of chicken cytokine and chemokine gene expression following Eimeria acervulina and Eimeria tenella infections. Vet Immunol Immunopathol. 2006;114(3-4):209–223. doi: 10.1016/j.vetimm.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 44.Smith AL, Hayday AC. Genetic dissection of primary and secondary responses to a widespread natural pathogen of the gut, Eimeria vermiformis. Infect Immun. 2000;68(11):6273–6280. doi: 10.1128/iai.68.11.6273-6280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lal K, et al. Proteomic comparison of four Eimeria tenella life-cycle stages: Unsporulated oocyst, sporulated oocyst, sporozoite and second-generation merozoite. Proteomics. 2009;9(19):4566–4576. doi: 10.1002/pmic.200900305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams RB. “Serotinous”, a descriptor proposed for lines of Eimeria species (Apicomplexa: Eimeriidae) selected for delayed development: An antonym for “precocious”. Vet Parasitol. 2007;145(3-4):388–389. doi: 10.1016/j.vetpar.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Dong H, Suo X, Wang M, Teng K. Characteristics of a line of Eimeria necatrix after 16 successive passages of oocysts collected after peak oocyst production. J Parasitol. 2006;92(6):1229–1234. doi: 10.1645/GE-861R.1. [DOI] [PubMed] [Google Scholar]
- 48.Srinivasa Rao AS, Tomley F, Blake D. Understanding chicken walks on n x n grid: Hamiltonian paths, discrete dynamics, and rectifiable paths. Math Methods Appl Sci. 2014 doi: 10.1002/mma.3301. [DOI] [Google Scholar]
- 49.Shirley MW, Harvey DA. Eimeria tenella: Infection with a single sporocyst gives a clonal population. Parasitology. 1996;112(Pt 6):523–528. doi: 10.1017/s0031182000066099. [DOI] [PubMed] [Google Scholar]
- 50.Nair S, et al. Single-cell genomics for dissection of complex malaria infections. Genome Res. 2014;24(6):1028–1038. doi: 10.1101/gr.168286.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reece SE, Drew DR, Gardner A. Sex ratio adjustment and kin discrimination in malaria parasites. Nature. 2008;453(7195):609–614. doi: 10.1038/nature06954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeo M, et al. Multilocus sequence typing (MLST) for lineage assignment and high resolution diversity studies in Trypanosoma cruzi. PLoS Negl Trop Dis. 2011;5(6):e1049. doi: 10.1371/journal.pntd.0001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chapman HD. Eimeria tenella in chickens: Studies on resistance to the anticoccidial drugs monensin and lasalocid. Vet Parasitol. 1976;2(2):187–196. [Google Scholar]
- 54.Long PL, Millard BJ, Joyner LP, Norton CC. A guide to laboratory techniques used in the study and diagnosis of avian coccidiosis. Folia Vet Lat. 1976;6(3):201–217. [PubMed] [Google Scholar]
- 55. Ministry of Agriculture, Fisheries and Food (1986) Manual of Veterinary Parasitological Laboratory Techniques (Her Majesty's Stationary Office, London)
- 56.Kumar S, et al. An optimised protocol for molecular identification of Eimeria from chickens. Vet Parasitol. 2014;199(1-2):24–31. doi: 10.1016/j.vetpar.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rutherford K, et al. Artemis: Sequence visualization and annotation. Bioinformatics. 2000;16(10):944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 60.Storm N, Darnhofer-Patel B, van den Boom D, Rodi CP. MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol Biol. 2003;212:241–262. doi: 10.1385/1-59259-327-5:241. [DOI] [PubMed] [Google Scholar]
- 61.Korber B. HIV Signature and Sequence Variation Analysis. In: Rodrigo A, Learn G, editors. Computational Analysis of HIV Molecular Sequences. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2000. pp. 55–72. [Google Scholar]
- 62.Fornace KM, et al. Occurrence of Eimeria species parasites on small-scale commercial chicken farms in Africa and indication of economic profitability. PLoS One. 2013;8(12):e84254. doi: 10.1371/journal.pone.0084254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vrba V, Blake DP, Poplstein M. Quantitative real-time PCR assays for detection and quantification of all seven Eimeria species that infect the chicken. Vet Parasitol. 2010;174(3-4):183–190. doi: 10.1016/j.vetpar.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 64.Blake DP, Hesketh P, Archer A, Shirley MW, Smith AL. Eimeria maxima: The influence of host genotype on parasite reproduction as revealed by quantitative real-time PCR. Int J Parasitol. 2006;36(1):97–105. doi: 10.1016/j.ijpara.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 65.Nolan MJ, Tomley FM, Kaiser P, Blake DP. Quantitative real-time PCR (qPCR) for Eimeria tenella replication - Implications for experimental refinement and animal welfare. Parasitol Int. 2015;64(5):464–470. doi: 10.1016/j.parint.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eckert J, Braun R, Shirley M, Coudert P. Guidelines on Techniques In Coccidiosis Research. European Commission; Brussels: 1995. [Google Scholar]
- 67.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol. 2005;14(8):2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 68.Jukes T, Cantor C. Evolution of protein molecules. In: Munro H, editor. Mammalian Protein Metabolism. Academic; New York: 1969. pp. 21–132. [Google Scholar]
- 69.Nei M. Molecular Evolutionary Genetics. Columbia Univ Press; New York: 1987. [Google Scholar]
- 70.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133(3):693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132(2):583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.