Abstract
We have previously reported that articular chondrocytes in tissue contain long cytoplasmic arms that physically connect two distant cells. Cell-to-cell communication occurs through connexin channels termed Gap Junction (GJ) channels, which achieve direct cellular communication by allowing the intercellular exchange of ions, small RNAs, nutrients, and second messengers. The Cx43 protein is overexpressed in several human diseases and inflammation processes and in articular cartilage from patients with osteoarthritis (OA). An increase in the level of Cx43 is known to alter gene expression, cell signaling, growth, and cell proliferation. The interaction of proteins with the C-terminal tail of connexin 43 (Cx43) directly modulates GJ-dependent and -independent functions. Here, we describe the isolation of Cx43 complexes using mild extraction conditions and immunoaffinity purification. Cx43 complexes were extracted from human primary articular chondrocytes isolated from healthy donors and patients with OA. The proteomic content of the native complexes was determined using LC-MS/MS, and protein associations with Cx43 were validated using Western blot and immunolocalization experiments. We identified >100 Cx43-associated proteins including previously uncharacterized proteins related to nucleolar functions, RNA transport, and translation. We also identified several proteins involved in human diseases, cartilage structure, and OA as novel functional Cx43 interactors, which emphasized the importance of Cx43 in the normal physiology and structural and functional integrity of chondrocytes and articular cartilage. Gene Ontology (GO) terms of the proteins identified in the OA samples showed an enrichment of Cx43-interactors related to cell adhesion, calmodulin binding, the nucleolus, and the cytoskeleton in OA samples compared with healthy samples. However, the mitochondrial proteins SOD2 and ATP5J2 were identified only in samples from healthy donors. The identification of Cx43 interactors will provide clues to the functions of Cx43 in human cells and its roles in the development of several diseases, including OA.
Direct intercellular communication is accomplished in nearly all tissues and organs by aqueous, low-resistance pores located in the lipid bilayer of the contacted cells. These pores, named gap junctions (GJ)1, are composed of connexins (Cxs) and play critical developmental and functional roles (1, 2). Numerous processes, such as the diffusion of metabolites, nutrients, small RNAs and second messengers, and the rapid transmission of action potentials in heart or neuronal tissue via so-called electrical synapses, are driven by GJ communication (3–7). The junctional channel is composed of two end-to-end hemichannels, each of which is a hexamer of six subunits of Cxs that dock with each other to form a contiguous gap junctional channel. Cxs proteins have a common topology that includes four a-helical transmembrane domains, two extracellular loops, a cytoplasmic loop, and N- and C termini located on the cytoplasmic membrane face. Cx43 (the 43-kDa isoform) is the most widely expressed GJ protein in different cell types (8). Yet, as many as 20 murine and 21 human Cx isoforms have been identified (9).
Cx43 has a relatively large carboxy-terminal domain (CTD) that takes part in multiple proteomic interactions. Increasing evidence indicates that gap-junctional Cx43 is part of a multiprotein complex and that Cx43-interacting proteins are thought to form a dynamic scaffolding protein complex, termed the Nexus, that may function as a platform to localize signaling, structural, and cytoskeletal proteins (10, 11). Indeed many aspects of Cx43 function, for example cellular transport, plaque assembly and stability, and channel conductivity are most likely governed by interactions with regulatory and structural proteins that bind to the cytoplasmic domains of Cx43. These proteins include the tight junction protein zonula occludens-1 (ZO-1) (12–14), 14–3-3 (15), Drebrin (16), β-tubulin (17), c-Src, v-Src (10), and other potential Cx43-interacting proteins that target Cx43 to points of cell–cell contact and regulate gap junctional intercellular communication (GJIC) (10).
Initially, the cellular functions of Cxs were attributed exclusively to direct cell-to-cell diffusion; however, some Cx functions seem to be independent of channel function (18). In fact, several reports have suggested that the Cx43 complex might fulfil functions that are not necessarily related to the control of GJIC. For instance, Cx43 mutants lacking the C-terminal tail that were expressed in transformed cells restored the GJIC but inhibited proliferation (19). Moreover, a Cx43 mutant that does not form GJ has been shown to suppress cell growth (20), and the expression of the C-terminal tail alone was sufficient to reduce proliferation (19, 21). On the other hand, the overexpressed C-terminal tail of Cx43 localized to the nucleus and inhibited cell growth (21, 22). These and other studies raise the possibility that the C-terminal tail of Cx43 can control cell-cycle, gene expression, or different signaling pathways via binding proteins independently of its channel function (18).
The channel-independent effects of Cxs might be explained by the (dys)function of Cx-tail interactions proteins. Our group is interested in investigating the physiopathological mechanism that is associated with osteoarthritis (OA). Recent results suggest a direct role of Cx43 in the development of OA that is not necessary related to its channel function (7, 23, 24). For instance, only a few connexin-interacting proteins have been described to date, and previous studies have used artificial systems, such as yeast two-hybrid screens or Cx43-domain fusion proteins. The aim of this study was to identify Cx43-interacting proteins in primary articular chondrocytes from human adults. In this report, lysates of primary articular chondrocytes were subjected to immunoprecipitation (IP) with an antibody directed against the C-terminal tail of Cx43. Cx43 interactors were identified using liquid chromatography and mass spectrometry (MALDI-TOF/TOF). We have identified >100 Cx43 associated proteins. Among these, we have found out novel functional Cx43 interactors involved in human disease and OA development, which emphasizes the potential importance of Cx43-interactions in normal development and physiology.
EXPERIMENTAL PROCEDURES
Cartilage Collection, Processing, and Primary Culture of Chondrocytes
Human knee and femoral head articular cartilage samples were generously obtained from adult donors undergoing joint surgery. All donors signed an informed consent form, and the institutional Ethics Committee approved the study (Registration Code Galicia CAEIG: 2012/094 - PI13/00591). In situ cartilage samples were frozen immediately in Cryomold® Standard using Tissue-Tek® O.C.T.TM compound and isopentane in liquid nitrogen and stored at −80 °C. Cartilage from healthy persons was obtained in situ from individuals who suffered a knee or hip fracture with no history of joint disease. Healthy samples were assigned on based on their medical record data and histological study. Histological samples (healthy and OA with radiologic diagnosis) were graded using the modified Mankin score (23) (mean age: 71.38 years, range: 63 to 89 years; Mankin score: 0–13 points). The samples (OA patients and age-matched healthy subjects) were divided into four groups: normal/healthy (score = 0–1); early OA (score = 2–3), named grade I; mild OA (score = 4–5), named grade II; moderate OA (score = 6–7), named grade III; and severe OA, named grade IV (score = 8–13). To perform this study, we selected samples from the normal/healthy, early OA and moderate grade III groups. For the isolation and culture of primary chondrocytes, a fresh cartilage was rinsed with saline, and cells were isolated as previously reported (23). The cells (2.5 million) were then seeded into 162-cm2 flasks and incubated at 37 °C in 5% CO2 and air in DMEM supplemented with 100 μg/ml Primocin (In vivo Gen PrimocinTM) and 15% FCS until ∼80–90% confluence was reached. The chondrocytes were used in experiments during the third or fourth weeks of primary culture.
Co-Immunoprecipitation
Co-immunoprecipitation (Co-IP) was performed using primary chondrocytes from human adults as described previously with minor modifications (7). Cells were resuspended in 1000 μl of IP-lysis buffer (50 mm Tris-HCl pH 7.5, 5 mm EDTA pH 8, 0.5% v/v Nonidet P-40, 1.0% v/v Triton-X, 150 mm NaCl) or in IP-lysis buffer containing Sarkosyl and SDS (50 mm Tris-HCl pH 7.5, 5 mm EDTA pH 8, 0.5% v/v Nonidet P-40, 0.5% v/v Sarkosyl, 0.1% SDS v/v, 150 mm NaCl). Both IP buffers contained protease inhibitors (Sigma-Aldrich) and 1 mm PMSF. Lysates were precleared by incubating with 50 μl of Protein G Sepharose (50% slurry). Four micrograms of antiCTD-Cx43 antibody (SC-20, sc-6560 from Santa Cruz Biotechnology, Heidelberg, Germany) followed by Protein G Sepharose beads (50 μl of 50% slurry) were added to the precleared lysates and incubated overnight at 4 °C. A control IP was performed without antibody. The beads were washed four times with the corresponding IP buffer, and the bead pellets were suspended in loading buffer (10% SDS, 0.2 m Tris pH 6.8, 50% glycerol, 0.1% (w/v) bromphenol blue (5x) containing 1.42 m 2-mercaptoethanol) and incubated for 30 min at 37 °C before being boiled for 10 min. Supernatants were stored at −80 °C. Silver stain analysis of immunoprecipitations and Western blotting were used to determine the specificity of the interactions. Silver staining was performed after the electrophoretic separation on polyacrylamide gels. The gels were fixed for 30 min in fixed solution (40% ethanol, 10% acid acetic) and rinsed twice for 10 min in water. After incubating with 0.02% thiosulfate sodium for 1 min, gels were immediately rinsing in water for another min, incubated with silver solution for 30 min (0.2% silver nitrate, 0.075% formalin) and washed for 10–15 s with water. The develop solution contained 3% carbonate potassium, 12.5 mg/ml thiosulfate sodium and 0.025% formalin. The development was stopped by dipping the gel in the stop solution (3% Tris-Buffer, 10% acid acetic) for 15 min. The stained gels were scanned after thoroughly rinsing in water.
For Western blot assays, anti-α-tubulin antibody (T9026) was supplied from Sigma-Aldrich, Schnelldorf, Germany antinucleolin antibody (PAB12541) was from Abnova, Heidelberg, Germany GmbH, the antiranGTP (sc-1156), antiCx43 (sc-6560), antiCalcyclin (sc-271396), and anti-Integrin αV (sc-376156) antibodies were from Santa Cruz Biotechnology, antiAlpha B Crystallin (ab13496) from Abcam. For SDS-PAGE gel electrophoresis, 5 μl of the denatured, immunoprecipitated proteins were loaded and separated on 10% polyacrylamide gels, which were then electro blotted onto polyvinylidene fluoride (PVDF) membrane (Millipore Co., Bedford, MA). The membranes were blocked with 5% milk in PBS and 0.05% Tween-20 (Sigma-Aldrich) and, after probing with antibody, developed using ECL Western blotting Detection reagent from GE Healthcare.
Identification of Proteins Using SDS-PAGE and nanoLC-MALDI-TOF/TOF
Immunoproteins were loaded and separated by SDS-PAGE in 10% acrylamide in-house gels in a Protean® mini-gel system (Bio-Rad, Hercules, CA). Gels were stained and visualized with Coomassie Brilliant blue G250 and the resulting lane was size-fractioned into three sections that were subsequently processed independently. Each section was destained with methanol, diced into small pieces and in-gel digested following a standard procedure (25). Briefly, the samples were desiccated with acetonitrile (26) reduced with 10 mm dithiothreitol (DTT) for 30 min at 56 °C, alkylated with iodoacetamide (50 mm IAA), dehydrated in 100% ACN and trypsin-digested (6.6 ng/μl, Roche Mannheim, Germany) for 16 h at 37 °C. Peptides were then extracted from the gel pieces with 50% ACN/0.1% trifluoroacetic acid (TFA), dried in a speed-vacuum, reconstituted in 0.1% TFA and desalted using homemade stage tips (Empore disk-C18, 47 mm, 3 m O Agilent Technologies, Santa Clara, CA). The peptide fractions were separated using reversed phase chromatography in a Tempo nanoLC system (ABSciex, Madrid, Spain). After precolumn desalting on a trapping column (0.5 × 2 mm, Michrom Bioresources, Auburn, CA) at a flow rate of 15 μl/min for 15 min, tryptic digests were separated on a C18 nanocolumn (Integrafit C18, Proteopep II, 75 μm id, 15 cm, 5 μm, 300 Å; New Objective, Woburn, MA) at a flow rate 350 nl/min, with a 30 min linear gradient of 5 to 50% of buffer B (80% ACN containing 0.1% TFA). The column was washed and regenerated for 5 min using 80% of buffer B. After all nonpolar and nonpeptide materials were removed, the column was re-equilibrated under the initial starting conditions for 15 min. The separated peptides were spotted onto a MALDI plate each 15 s and mixed with α-cyano-4-hydroxycinnamic matrix (3 mg/ml in 0.1% TFA, 70% ACN) at a flow rate of 1.2 μl/min using a Sun Collect MALDI Spotter/Micro-Fraction Collector (SunChrom Wissenschaftliche Geräte Gmbh, Germany). The MS runs for each chromatogram were acquired and analyzed in a MALDI-TOF/TOF instrument (4800 ABSciex, Framingham, MA). MS spectra from m/z 800–4000 were acquired for each fraction and processed with internal calibration. Angiotensin peptide diluted in the matrix (3 fmol/spot) was used for the internal standard calibration at an m/z of 1046.5. After screening of all LC-MALDI sample positions in MS positive reflector mode, the fragmentation of automatically selected precursors was performed at collision energy of 1 kV with CID gas (air). Up to 12 of the most intense ion signals per spot position with signal/noise (S/N) above 80 were selected as precursors for MS/MS acquisition, excluding common trypsin autolysis peaks and matrix ion signals. A second MS/MS was acquired excluding the precursors selected in the previous MS/MS run. Precursors with a S/N above 25–40 were selected to identify proteins that were not identified in the previous run. For protein identification, MS/MS spectra acquired by 4000 Series Explorer software v.3.5.1 were searched with ProteinPilot v.3.0 (ABSciex) against the Uniprot-SwissProt protein database (release version 2012_05; 536,029 entries), using the Paragon Algorithm. The Paragon Algorithm in ProteinPilot software served as the default search program with trypsin as the digestion agent with one missed cleavage allowed, iodoacetamide as a fixed modification of cysteine and biological modifications. Only proteins with a threshold >95% confidence (>1.3 Unused Score) were considered for protein identification. False positive identification rate is around 5%.
Gene Ontology (GO) Analysis
To analyze the MS data and for the representation (protein enrichment) of GO terms, MS data were analyzed using the Protein Information and Knowledge Extractor (PIKE, http://proteo.cnb.csic.es/pike/), which links to the DAVID web system to provide a quality measure of the protein relationships regarding GO (27, 28).
Immunohistochemistry Assays
For immunohistochemistry (IHC), cultured cells on chamber slides were fixed with 4% formaldehyde for 10 min at room temperature as previously described with minor modifications (23). Cells on chamber slides were counterstained with hematoxylin/eosin or counterstained with 4′,6-diamidino-2-phenyindole (DAPI). The antiCx43 antibody (610062) was supplied from BD Transduction Laboratories, antiHSP90β antibody (sc-1057) was from Santa Cruz Biotechnology, and the antivimentin antibody (ab8069) was from Abcam. Negative controls (omitting the primary antibody) were performed to test the specificity of each antibody. For immunofluorescence, cultured cells on chamber slides were fixed with cold methanol for 5 min at −20 °C and permeabilized with 1% Triton-X-100 (v/v) for 5 min at room temperature using the fluorescent-tagged secondary antibodies Polyclonal Rabbit anti-Mouse Immunoglobulins/FITC (F0313, Dako Cytomation), Polyclonal Goat anti-Mouse Immunoglobulins/RPE (R-0480, Dako), and Polyclonal rabbit anti-Goat Immunoglobulins/PE (SC-3755, Santa Cruz Biotechnology). The cells were counterstained with DAPI and mounted in glycergel mounting medium (C0563, DAKO). For IHC, samples were analyzed on an Olympus BX61 microscope using a DP71 digital camera (Olympus, Hamburg, Germany), and calibration and quantification of the images was performed using the AnalySISD 5.0 software (Olympus Biosystems, Hamburg, Germany). Immunofluorescence assays were acquired in a Leica TCS-Sp5 (AOBS) Laser scanning confocal (Leica Microsystems, Wetzlar, Germany) using a 63X HCX PLAPO 1.4 N.A. oil immersion objective and the acquisition software LAS AF 2.6 (Leica Microsystems). Intensity line profile for each channel was performed by using Image J 1.47s (NIH).
RESULTS
Mass Spectrometric Identification of Cx43-Associated Proteins
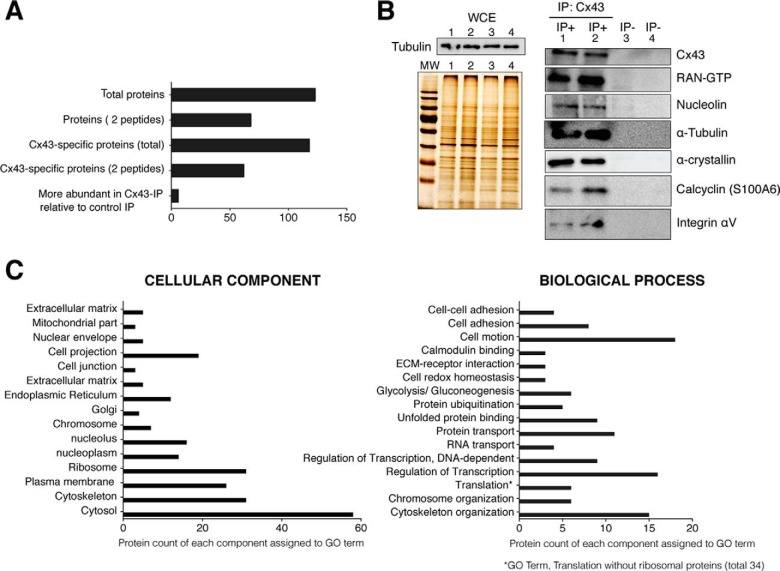
When Cx43-interacting proteins from chondrocytes of healthy donors were analyzed, 123 proteins were identified, of which 68 proteins were represented by at least two unique peptides (Fig. 1A). One hundred and eighteen proteins were specific to the Cx43 immunoprecipitation (IP) and not identified in the control IP, which was performed without antibody (See supplemental Tables S1 and S2). Among these, 62 proteins were represented by at least two unique peptides. We considered a second category called “more abundant proteins” (n = 6) that included proteins such as annexin A2, prelamin-A/C and vimentin. The “more abundant proteins” corresponded to the proteins identified in both, the control IP and Cx43 IP but with more peptides at 95% confidence and with higher coverage and score in the Cx43 IP than in the control IP (supplemental Tables S1 and S2). In all cases, only proteins with >95% confidence and score >1.3 were considered significant.
Fig. 1.
Analysis of Cx43-associated proteins identified using mass-spectrometry. Primary chondrocytes in culture were lysed in the presence of the nonionic surfactants Igepal and Tripton X-100 using a near-physiological-salt concentration of 150 mm NaCl to pull-down the Cx43 (nonjunctional and junctional Cx43). Immunoprecipitated proteins using an antibody that specifically recognized the C-terminal tail of Cx43 were identified using MS. Four different samples of chondrocytes from four healthy donors were analyzed. Overall, proteins were identified with 95% peptide confidence (minimum value 1.3 of unused cutoff) A, Summary of the number of identified proteins. Proteins identified as specific to the Cx43-IP but absent from the control IP (no antibody). The “more abundant proteins” in Cx43-IP relative to control IP correspond to the number of proteins identified as most likely more abundant in the Cx43-IP than in the control-IP (peptide difference ≥2). See supplemental Tables S1 and S2 for a complete list of proteins identified in the Cx43-IP and control IP. B, Whole-cell extracts (WCE) and proteins immunoprecipitated with (IP+) or without (IP−) an antiCTD-Cx43 antibody were analyzed by silver staining and Western blotting. The protein content of the WCE (input, without antibody) (images shown on the left) for each experiment (IP) (images shown on the right) was previously analyzed by silver staining and Western blotting (Tubulin) (image shown on the top-left). Lanes represent 0.5% of the total WCE used for each IP. The numbers (1–4) represent the WCE utilized for the corresponding experiment (IP). For the images shown on the right, lanes represent the 25% of each IP C, Gene Ontology clustering of the protein set. GO analysis (27, 28) for identified proteins reveals a strong enrichment for GO terms associated with cytoskeleton, nuclear localization, translation processes, cell adhesion, and motion and carbohydrate metabolism. See supplemental Tables S1 and S2 for a complete list of the identified proteins.
The Co-IP methodology is the referenced method to investigate interacting proteins or protein complexes. However, this technique has limitations and does not necessary indicate whether interactions are direct. Besides, many protein–protein interactions are temporary and proteins can be found in different cellular compartment. Other limitations include the nonspecific binding to protein A/G and other contaminants. Despite this, the purification strategies used in this report resulted in a low number of proteins identified in the control IP (supplemental Table S2). Nevertheless, a selection of possible Cx43-interacting proteins identified in the MS analysis was further validated by a combination of IP and Western blotting (Fig. 1B). Two IP samples using the Cx43 antibody (IP+) and two IP samples performed without antibody (IP−) confirmed the presence of previously identified proteins in the Cx43 IP. Cx43, nucleolin and the GTP-binding nuclear protein Ran among other interactors were detected in Cx43 IP samples, but not in control IPs performed without antibody. The same number of cells was used for each IP, and the whole-cell extracts that were used for IPs were previously resolved by SDS-PAGE and visualized using silver staining (Fig. 1B).
To explore the functions of the proteins associated with Cx43, we analyzed the GO terms of the specific and more-abundant identified proteins (Fig. 1C). We found significant enrichment for GO processes directly linked with cytoskeleton, ribosome, plasma membrane, nucleus, and cell projection. Cytoskeletal proteins are an important class of contaminant proteins that was identified in a study to bind directly to commonly used immunoprecipitated matrices (30). However, the identified cytoskeletal proteins were enriched in our specific IP, but not in our control IP (supplemental Table S1). Among the 31 identified cytoskeletal-related proteins, 15 were implicated in cytoskeleton organization (Fig. 1C, Table I and supplemental Table S1), and some of the identified cytoskeletal proteins are known to interact with Cx43 (α-/β-tubulin, α-actin, Ezrin, Myosin, Vimentin, or Vinculin).
Table I. Example of Cx43-interacting proteins likely related to Cx trafficking, GJ assembly, GJIC, and turnover (chondrocytes from healthy donors). The cytoskeletal proteins that interact or colocalize with Cxs have a critical functional importance in Cx trafficking and assembly of Cx into GJs. Microfilaments and microtubules are involved in turnover mechanisms that compromise the control of GJIC. The values of peptide probability or score, sequence coverage and number of peptides found are shown in supplementary Fig. S1. Only proteins that show >95% confidence and score >1.3 were considered for this study.
| Protein Group\Name | Gene name |
|---|---|
| α-a and ß-tubulinab | TUBA1B\TUBB\TUBB2A\ TUBB2C |
| α-actina and actin related proteins | ACTB\ACTR3B\ACTN4 |
| Translocon-associated protein subunit delta | SSR4 |
| Myosin motor proteinsa (Myosin ICb) | MYL12B\MYH9\MYO1B |
| Vimentina | VIM |
| Vinculinab | VCL |
| Talin-1b | TLN1 |
| Filamin A, alpha | FLNA |
| Dihydropyrimidinase-related protein 2 | DPYSL2 |
| SEC22 vesicle trafficking protein homolog B | SEC22B |
| Gelsolin | GSN |
| Profilin-1 | PFN1 |
| Erzinab | EZR |
| Radixinb | RDX |
| Moesinb | MSN |
| Annexin A1b\A2b\A5 | ANXA1\ANXA2\ANXA5 |
| Caldesmon | CALD1 |
| Endoplasmin | HSP90B1 |
| Tropomyosin α4 | TPM4 |
| Proteasome subunit beta type-2 | PSMB2 |
| Ubiquitin-40S ribosomal protein S27a | RPS27A |
| Cytoskeleton-associated protein 4 | CKAP4 |
| Thy-1 membrane glycoproteinb | THY1 |
| Protein S100-A6\-A11\-A10 | S100A6\S100A11\S100A10 |
a Previously reported interactions of Cx43.
b Constituent part of the plasma membrane.
The GO term translation (for Biological Process) is caused by the presence of ribosomal proteins and other translation-related proteins in the Cx43 IP (six proteins) compared with the control IP (supplemental Table S1). Note that in this case, the 40S and 60S ribosomal proteins have not been counted in the GO term translation (Fig. 1C). MS results revealed several interactors directly linked to nuclear activity, including several histones and nucleolar-related proteins (Table II and supplemental Table S1). Interestingly, a number of proteins related to metabolic pathways (glycolysis/gluconeogenesis), protein folding and transport, cell adhesion, cell motion, calmodulin binding, and cell-redox homeostasis was also identified to interact with Cx43 (Fig. 1C and Table II).
Table II. Cx43-interacting proteins (represented as gene name) from chondrocytes of healthy donors using the Cx43 IP-based proteomic approach. The values of peptide probability\score, sequence coverage and number of peptides found are shown in supplementary Fig. S1. Only proteins that show >95% confidence and score >1.3 were considered for this study.
| Group\Complex | Gene name |
|---|---|
| Nucleosome. Histones | HIST1H1T, HIST1H2AC, H2AFJ, HIST2H2BF, H3F3A, HIST1H4A |
| Transcription related factors | PTRF, NCL, CAND1, SND1, STAT1, RAN |
| RNA-binding proteins | MVP*, HNRNPA1*, HNRNPA2*, HNRNPA3, HNRNPK, NLC, RAN*, PTRF |
| RNA transport* | |
| RNA splicing | HNRNPA1, HNRNPA2, HNRNPA3, HNRNPK, SNRPD3 |
| Ribosome | RPS16, RPS18, RPS19, RPS24, RPS26, RPS4X, RPS5, RPS7, RPS8, RPS9, RPLP2, RPL11, RPL12, RPL15, RPL17, RPL21, RPL24, RPL26, RPL27A, RPL3, RPL35, RPL35A, RPL36, RPL4, RPL6, RPL7, RPL7A |
| Translation | EEF2, EEF1AL3, EEF1G, EIF5AL1 |
| Endomembrane system (transport within the cell) | KPNB1#, RAN#, LMNA, MVP# |
| Nuclear Pore Complex# | |
| Mitochondrial | SOD2, ATP5J2 |
| Antioxidant activity | PRDX1, SOD2 |
| Generation of precursor metabolites and energy | PKM2, ENO1, GAPDH, LDHA, ALDOA, PYGB, PGK1, SOD2 |
| Protein folding and other-related proteins | CRYAB, HSPA8, HSP90AB1, HSP90B1, PPIB, PPIA, HSPA5, P4HB, PDIA3, HSPB1, SERPINH1 |
Cx43 Nuclear and Perinuclear-Related Interactors
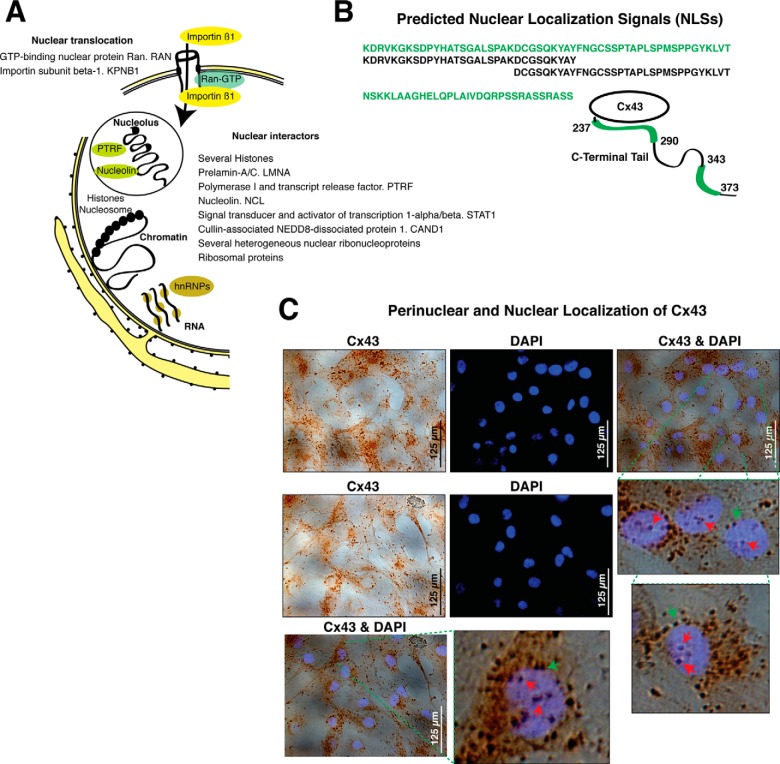
The MS results suggest that Cx43 interacts with two components of the nuclear translocation system: the GTP-binding nuclear protein Ran (RAN) and importin β (KPNB1) (Table II and Fig. 2A). Additionally, using cNLS Mapper to predict nuclear localization signals (NLSs) specific to the importin pathway, we identified two NLSs sites located on the C-Terminal tail of the Cx43 sequence (Fig. 2B). In fact, the images obtained from immunohistochemistry (IHC) experiments using an antibody that specifically recognizes the C-terminal tail of Cx43 and DAPI nuclear staining showed multiple positive spots for Cx43 in the nucleus (Fig. 2C). Cx43 was localized across the nuclear margin and in nuclear foci (one to six per cell).
Fig. 2.
Cx43-nuclear and perinuclear interactors. A, Cx43-associated proteins related to nuclear import and nuclear and perinuclear localization. Schematic view of the nucleus and endoplasmic reticulum; black spots represent ribosomes. The nuclear envelopment is composed of two lipid bilayer membranes, nuclear pore complexes, and the nuclear lamina. The outer nuclear membrane is continuous with the endoplasmic reticulum. Some of the Cx43 nuclear-related interactors specific to Cx43-IP are represented and listed in the schematic representation (unique peptides, 95% confidence). The complete list of identified proteins, number of identified peptides, sequence coverage, and score is found in supplemental Table S1. B, Schematic representation of the C-terminal tail of the Cx43 protein showing the position of the two candidate NLSs that are specific to the importin α/β pathway and identified using cNLS Mapper (93). The amino acid sequence and positions of each NLS are indicated in green. C, DAPI staining and IHC using an antiCx43 antibody on primary human articular chondrocytes in monolayer culture. Merged images and magnifications are shown. Red arrows indicate nuclear staining for Cx43; and green arrows indicate perinuclear Cx43-positive spots.
The identified nuclear-related proteins with a score < 1.3 are listed in supplemental Table S1 and summarized in Fig. 2A and Table II. Among the transcription-related interactors, such as the TBP-interacting protein CAND1, several nucleolus-related proteins were identified. The MS results suggest that Cx43 interacts with the major nucleolar protein nucleolin and with the transcription release factor PTRF, which is required for rRNA transcription and termination.
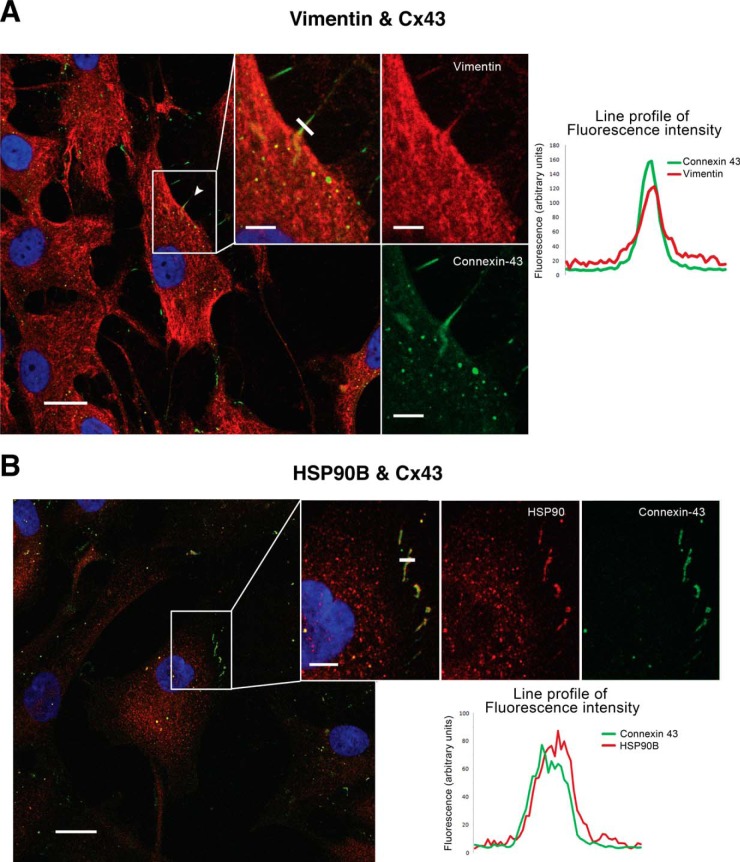
Cx43 Interactors Colocalize with Cx43
To determine whether the Cx43-associated proteins that were identified using MS also associate with Cx43 in situ, we achieved immunofluorescence techniques using antivimentin, antiHSP90B, and monoclonal antiCx43 antibodies to determine if these two candidate proteins colocalize with Cx43 (Fig. 3). Articular chondrocytes in monolayer culture contain high levels of vimentin that is mainly localized in the cytoplasm. Multiple positive spots for Cx43 (green) were found to colocalize with positive staining for vimentin (red) in the cytoplasm, at cell margin (membrane), and in the cellular projections (Fig. 3A). The chaperone HSP90β was also identified by mass spectrometry as a Cx43 interactor, and immunofluorescence assays suggested that this protein colocalized with Cx43 in the cytoplasm and in the plasma membrane (margin of the cells) (Fig. 3B).
Fig. 3.
Colocalization of Cx43 with vimentin and HSP90β in articular chondrocytes. Immunofluorescence confocal microscopy was used to study colocalization of two antigens in one cell. A, Images represent the double staining and DAPI in chondrocytes in monolayer culture that were obtained from healthy human donors. Cx43 is detected in green, and vimentin is in red. B, Colocalization between HSP90β and Cx43 was also studied in human chondrocytes in monolayer culture. Cx43 is shown in green, and HSP90β is in red color. Right panels; colocalization graphs show the fluorescence intensity profile from a line crossing through the cell protrusions, the coincidence of fluorescence intensity peaks, for both red and green, marks the colocalization of both proteins.
Cx43-Associated Proteins Related to OA
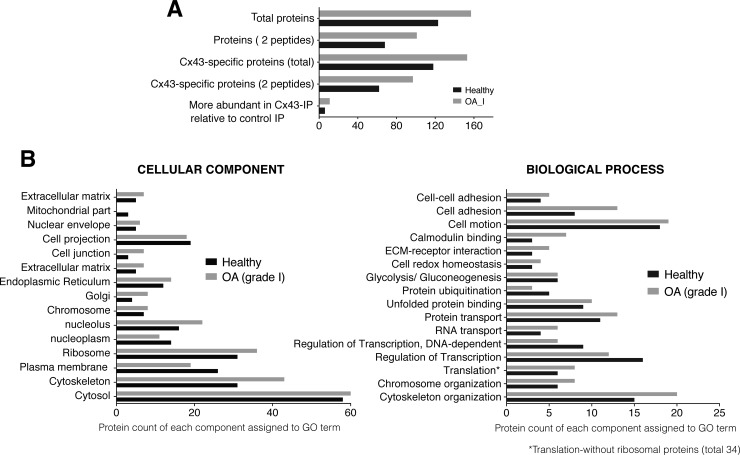
Analysis of the Cx43-associated proteins that were isolated from chondrocyte samples from early phase OA patients (grade 1, following Mankin score, see material and methods) identified 157 proteins, of which 153 were specific to the Cx43 IP and not identified in the control IP performed without antibody (Fig. 4A, supplemental Tables S2 and S3). Eleven proteins, which included annexin A2, glutathione S-transferase and vimentin, were more highly represented in the Cx43 IP than in the control IP (supplemental Table S3); 73 proteins were identified as Cx43-interactors in healthy and OA chondrocyte groups (supplemental Table S3, see peptide difference); and 80 interactors, including the 14-3-3 protein, integrin α-5 and integrin β-1 were exclusively identified in chondrocytes from OA patients (supplemental Table S3, see specific to OA versus healthy).
Fig. 4.
Analysis of Cx43-associated proteins identified in the OA samples. Four different samples of chondrocytes from four OA patients were analyzed. A, A summary of the number of the identified proteins. Gray represents the Cx43 interactors identified in samples from OA grade I. Proteins identified as specific to the Cx43-IP but absent from the control IP (no antibody). The number of proteins identified as most likely more abundant in the Cx43-IP than in the control-IP (peptide difference ≥2). See supplemental Table S3 for a complete list of the proteins identified in the Cx43-IP and control IP. B, GO analysis for the identified proteins. Overall, the proteins were identified with 95% peptide confidence (minimum value 1.3 of unused cutoff). See supplemental Tables S1–S3 for a complete list of the identified proteins. Gray represents the Cx43 interactors that were identified in samples from OA grade I patients.
We next analyzed the GO terms of interactors identified in OA samples and compared them to the results of the proteins identified in healthy samples (Fig. 4B, gray bars). The results showed an enrichment of Cx43 interactors related to cell adhesion, extracellular matrix interaction, calmodulin binding, nucleolus, translation and cytoskeleton-related proteins in OA compared with healthy samples. Critical components for cartilage structure and function, such as collagen XII or collagen VI, which exclusively localize to and delimit the pericellular matrix, were only identified in OA samples (Table III and supplemental Table S3). However, the mitochondrial proteins SOD2 or ATP5J2 were only identified in samples from healthy donors (supplemental Table S1).
Table III. Example of Cx43-interacting proteins identified in this study that were previously reported to be altered in chondrocytes from OA patients (46–53). # Interactors exclusively identified in samples from healthy donors. * Interactors that were only identified in samples from OA patients. The remaining interactors were identified in both samples (healthy and early\moderate OA). Protein Pilot Score, Amino acids sequence coverage for the identified proteins (Cov.) and number of peptides (No. pept.) masses matching\not matching the top hit from MS-Fit PMF. Only proteins that show >95% confidence and score >1.3 were considered for this study.
| Group\Complex | Protein name | Gene name | Healthy OA (g. I\III) | Score | Cov. | No. pept. |
|---|---|---|---|---|---|---|
| Extracellular matrix structural constituent | Collagen alpha-1(I) chain | COL1A1 | Healthy | 6 | 23.9 | 3 |
| OA (g. III) | 8.24 | 50 | 4 | |||
| Collagen alpha-2(I) chain | COL1A2 | Healthy | 4 | 31.3 | 2 | |
| OA (g. III) | 2 | 40.7 | 1 | |||
| Collagen alpha-1(VI) chain* | COL6A1* | OA (g. I\III) | 21.28\10.29 | 57.1\51.3 | 12\6 | |
| Collagen alpha-2(VI) chain* | COL6A2* | OA (g. I\III) | 14.25\6 | 56.1\36.3 | 11\4 | |
| Collagen alpha-3(IV) chain* | COL6A3* | Healthy | 2.84 | 7.4 | 2 | |
| OA (g. I\III) | 95.44\61.62 | 53.9\47.5 | 54\34 | |||
| Collagen alpha-1(XII) chain* | COL12A1* | OA (g. I\III) | 23.48\14.16 | 41.2\32.6 | 11\8 | |
| Carbohydrate metabolic processes | Alpha-enolase | ENO1 | Healthy | 8 | 21 | 4 |
| OA (g. I\III) | 26.89\26.98 | 65\58.1 | 13\14 | |||
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | Healthy | 10.41 | 30.5 | 5 | |
| OA (g. I\III) | 4.04\9.96 | 42.4\31.6 | 2\5 | |||
| Fructose-biphosphate aldolase A | ALDOA | Healthy | 2.01 | 8.8 | 1 | |
| OA (g. I\III) | 2.1\7.56 | 43.7\38.5 | 1\4 | |||
| Cytoskeleton related-proteins | Vimentin | VIM | Healthy | 81.76 | 87.1 | 48 |
| OA (g. I\III) | 124.3\78.96 | 100\94.2 | 158\53 | |||
| Gelsolin | GSN | Healthy | 2 | 6.8 | 1 | |
| OA (g. I\III) | 2\9.81 | 17.4\39.1 | 1\5 | |||
| Vinculin | VCL | Healthy | 5.29 | 20.3 | 3 | |
| OA (g. I\III) | 4.32\13.21 | 40.5\45.3 | 2\7 | |||
| Actin, cytoplasmic 1 | ACTB | Healthy | 25.29 | 53.6 | 16 | |
| OA (g. III) | 29.04 | 62.9 | 21 | |||
| Erzin | EZR1# | Healthy | 2 | 3.9 | 1 | |
| Moesin | MSN | Healthy | 10.09 | 29.5 | 5 | |
| OA (g. I\III) | 10.02\12.35 | 59.1\53 | 6\7 | |||
| Chaperone and Protein folding-related proteins | Alpha-crystallin B chain | CRYAB | Healthy | 4 | 26.3 | 2 |
| OA (g. I\III) | 13.73\4.18 | 65.7\47.4 | 7\2 | |||
| Heat shock cognate 71 KDa protein | HSPA8 | Healthy | 10.72 | 22.5 | 5 | |
| OA (g. I\III) | 3.04\13.06 | 35.8\42.9 | 2\8 | |||
| Heat shock protein HSP 90-beta | HSP90AB1 | Healthy | 7.26 | 17.4 | 4 | |
| OA (g. I\III) | 2.81\16.29 | 28\34.7 | 2\8 | |||
| Endoplasmin | HSP90B1 | Healthy | 5.64 | 5.6 | 3 | |
| OA (g. I\III) | 3.82\22.57 | 31.1\38.5 | 2\11 | |||
| 78 kDa glucose-regulated protein | HSPA5 | Healthy | 22.93 | 30.3 | 11 | |
| OA (g. I\III) | 6.14\32.82 | 53.4\60.6 | 3\17 | |||
| Others | Annexin A1 | ANXA1 | Healthy | 10 | 30.6 | 5 |
| OA (g. I\III) | 10.87\6.15 | 47.4\27.2 | 6\3 | |||
| Annexin A2 | ANXA2 | Healthy | 52.03 | 65.8 | 30 | |
| OA (g. I\III) | 54.59\48.92 | 84.4\86.1 | 40\30 | |||
| Annexin A5 | ANXA5 | Healthy | 2 | 19.7 | 1 | |
| OA (g. I\III) | 2.05\2 | 32.8\20.6 | 1\1 | |||
| Superoxide dismutase [Mn], mitochondrial | SOD2# | Healthy | 2 | 15.3 | 1 | |
| Prelamin-A\C | LMNA | Healthy | 26.38 | 44.7 | 15 | |
| OA (g. I\III) | 39.97\16.48 | 82.7\55.1 | 21\8 | |||
| Eukaryotic translation initation factor 5A-2 | EIF5A2 | OA (g. I) | 2 | 40.5 | 1 | |
| Elongation factor 2 | EEF2 | Healthy | 14.35 | 21.1 | 7 | |
| OA (g. I) | 9.2\19.24 | 35.9\34.4 | 5\9 | |||
| Dihydropyrimidinase-related protein 2 | DPYSL2 | Healthy | 2.37 | 18.9 | 1 | |
| OA (g. III) | 7.31 | 35.1 | 4 | |||
| Major vault protein | MVP | Healthy | 2 | 3.9 | 1 | |
| OA (g. III) | 2.3 | 31 | 1 | |||
| GTP-binding nuclear protein RAN | RAN | Healthy | 4 | 11.6 | 2 | |
| OA (g. I) | 2.01 | 34.7 | 1 | |||
| Integrin alpha-V | ITGAV* | OA (g. I) | 2.03 | 20.9 | 1 | |
| Integrin beta-1 | ITGB1* | OA (g. I\III) | 2\2 | 18.9\8.4 | 1\1 |
The analysis of four different samples from patients with moderate-stage disease (grade 3), revealed a set of 130 Cx43-interactors that were specific to the Cx43 IP (supplemental Table S4). Eighty-eight proteins were identified as Cx43-interactors in both groups in the early and moderate stages of the disease; 64 proteins were identified as interactors in the healthy and moderate stage groups; and 32 interactors, including HMG-1 and Zyxin, were exclusively identified in chondrocytes from patients with moderate-phase disease (supplemental Table 4). The Cx43 interactors that were exclusively found in healthy and OA samples and the common interactors arranged by peptide difference are shown in supplemental Tables S3 and S4 and summarized in Table IV. The proteins were organized into different groups: exclusive to moderate OA, exclusive to early OA, exclusive to healthy, exclusive to OAs (both groups), specific to moderate OA versus early OA and peptide differences between common interactors found in each group.
Table IV. Cx43-interacting proteins exclusively identified in samples from healthy, early OA and moderate OA patients. The table represents some of the Cx43-interacting proteins that were exclusively identified for each group. For the complete list of interactors, see supplementary Table S4. The values of peptide probability\score, sequence coverage and number of peptides found are shown in supplementary figures. Only proteins that show >95% confidence and score >1.3 were considered for this study.
| Protein name | Gene name | Score | Cov. | No. pept. | |
|---|---|---|---|---|---|
| Healthy | ATP synthase subunit f, mitochondrial | ATP5J2 | 2 | 13.8 | 1 |
| Erzin | EZR | 2 | 3.9 | 1 | |
| Protein S100-A10 | S100A10 | 2 | 14.4 | 1 | |
| Protein S100-A11 | S100A11 | 2 | 10.5 | 1 | |
| Protein S100-A6 | S100A6 | 2 | 16.7 | 1 | |
| Ras-related protein Rap-1A | RAP1A | 2 | 7.6 | 1 | |
| Superoxide dismutase [Mn], mitochondrial | SOD2 | 2 | 15.3 | 1 | |
| Thy-1 membrane glycoprotein | THY1 | 4 | 24.2 | 2 | |
| Transforming protein RhoA | RHOA | 2 | 17.6 | 1 | |
| Ubiquitin-40S ribosomal protein S27a | RPS27A | 2.3 | 24.4 | 1 | |
| Vesicle-trafficking protein SEC22b | SEC22B | 2 | 18.1 | 1 | |
| Early OA (grade I) | 14–3-3 protein zeta\delta | YWHAZ | 4 | 28.2 | 2 |
| 1,4-alpha-glucan-branching enzyme | GBE1 | 2 | 13 | 1 | |
| Myosin-10 | MYH10 | 11.36 | 46.5 | 7 | |
| Protein piccolo | PCLO | 1.81 | 19.2 | 1 | |
| Coatomer subunit alpha | COPA | 2 | 28.8 | 1 | |
| Destrin | DSTN | 2.14 | 44.2 | 1 | |
| Integrin alpha-V | ITGAV | 2.03 | 20.9 | 1 | |
| Matrin-3 | MATR3 | 2.05 | 27.4 | 1 | |
| Septin-2 | SEPT2 | 2 | 21.3 | 1 | |
| Transforming growth factor-beta-induced protein ig-h3 | TGFBI | 2.32 | 19.8 | 1 | |
| Vigilin | HDLBP | 3.84 | 34.6 | 2 | |
| Moderate OA (grade III) | 26S proteasome non-ATPase regulatory subunit 2 | PSMD2 | 2 | 7.3 | 1 |
| A-Kinase anchor protein 2 | AKAP2 | 2 | 24.7 | 1 | |
| AP-2 complex subunit alpha-1 | AP2A1 | 2.82 | 16 | 2 | |
| ATP-citrate synthase | ACLY | 3.96 | 18.1 | 2 | |
| Coatomer subunit beta and gamma | COPB1 | 2.03 | 16.8 | 1 | |
| COPG | 6.47 | 24.5 | 4 | ||
| Heterochromatin protein 1-binding protein 3 | HP1BP3 | 2 | 17.9 | 2 | |
| High mobility group protein B1 | HMGB1 | 1.73 | 23.7 | 2 | |
| LIM domain and actin-binding protein 1 | LIMA1 | 3.07 | 28.7 | 2 | |
| LIM domain only protein 7 | LMO7 | 2.53 | 25.9 | 2 | |
| Stress-70 protein, mitochondrial | HSPA9 | 5.26 | 31.1 | 3 | |
| Transgelin | TAGLN | 1.39 | 42.3 | 1 | |
| Zyxin | ZYX | 2.01 | 14.3 | 1 |
The IP and MS methodologies have limitations, and several Cx43-interactors, such as the scaffolding protein zonula occudens-1 (ZO-1) (13, 14) and several phosphatases that are known to interact with Cx43, were not detected in this study. Some of these interactions might depend on the cell cycle stage in which the cells were taken for the IP experiments and the methodology that was used (31, 32). The low expression of tyrosine phosphorylated proteins and other signaling proteins makes these proteins difficult to detect using this methodology. On the other hand, a key factor in maintaining complex formation throughout the steps required for co-IP technique is the lysis (IP) and wash buffers. We used a buffer with low ionic strength and nonionic detergents (Nonidet P-40 and Triton X-100) to maintain the protein–protein interactions during the IP incubations. Using these conditions, we identified more than 100 Cx43-bound proteins (supplemental Tables S2–S4), several of which are a constituent part of the plasma membrane. However, the release of proteins from membranes presents significant challenges. To increase the chance that a particular membrane protein that interacts with Cx43 being detected by MS, we mechanically disrupted the cells using a 25-G needle in the presence of SDS and Sarkosyl. The presence of these ionic detergents favors the solubilization of membrane proteins but affects protein–protein interactions during the IP incubations. Using these conditions, we were only able to identify 32 Cx43-interactors (supplemental Table S5), and 29 were already identified as Cx43 interactors in samples from OA patients. Only Cadherin-23, the four-span transmembrane protein 2 (MS4A7) and Myosin-14 were exclusively identified as Cx43 interactors under these experimental conditions (supplemental Table S5). In addition, the analysis of membrane proteins by MS remains challenging because of the hydrophobicity of transmembrane domains, which are resistant to proteolysis. The resulting hydrophobic peptides are difficult to extract from in-gel digestions and are difficult to solubilize in buffers compatible with MS. These characteristics, together with the poor ionization of hydrophobic peptides, results in the loss of peptides during the process and makes the detection of low-abundance peptides in a complex spectra by MS difficult (33, 34).
DISCUSSION
The mapping of protein interactions with Cx43 have aided in understanding the formation and regulation of gating Cx43-based junctions in molecular detail and contributed to understanding the pathogenesis of diseases such as cardiac disease (35). Biochemical and molecular biology methods, such as GST tag pull-down assays, yeast two-hybrid approaches, in vitro studies, colocalization and co-IP, have been utilized to identify Cx43-interactors (16, 17, 32, 36–45). However, to our knowledge this study is the first, to combine the isolation of human native Cx43 by IP and LC-MS/MS identification. The meta-analysis to determine the degree of overlap between the identified interactors in this study and the identified proteins in previous studies revealed a set of proteins (n = 108 for healthy and n = 129 for chondrocytes from OA patients) that were exclusive to the present study (see Fig. 5) and 15 (for healthy) and 28 (for chondrocytes from OA patients) common interactors. The differences in protein detection between the present and previous studies are most likely because of differences in the cell type, methodology, detection sensitivity, and purification strategies used.
Fig. 5.
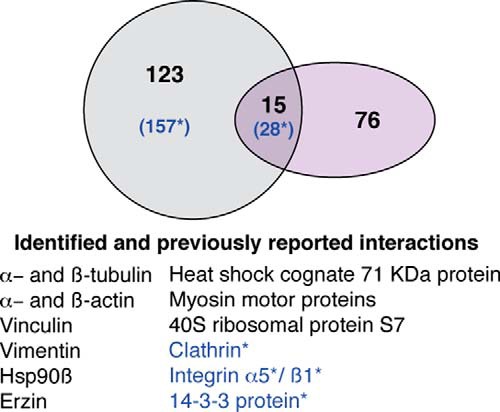
Meta-analysis of Cx43 mass-spectrometric data. Proportional Venn diagram comparing the Cx43-asociated proteins that were identified in the present study (dark gray circle) to those from previous reports (purple circle). Overlap (green) includes proteins present in both groups. Blue letters and asterisk (*) represent Cx43 interactors identified in samples from OA grade I. Some of these previous studies used co-IP, colocalization or affinity techniques and nuclear magnetic resonance using different types of cells and tissues from rat, mouse, human, porcine, goldfish, rabbit, or chick. See the reference list of some of the selected reports and reviews for meta-analysis (16, 17, 32, 36–45). None of the previous studies were performed using cartilage or articular chondrocytes.
Following our methodology, we have identified a large number of known and novel Cx43 interactors, some of which are responsible for the development of human diseases. These include proteins, such as Serpin H1, HnRNP-A2/B1, 78 kDa glucose-regulated protein, collagens, and PPIase B, that are related to connective tissue disorders and others, such as alpha(B)-crystallin, which is responsible for the development of myofibrillar myopathy and cataracts. HspB1 is related to Charcot-Marie-Tooth disease, neuropathy and neurodegeneration; vinculin is related to cardiomyopathy; and Profilin-1 is related to the development of amyotrophic lateral sclerosis and neurodegeneration. Moreover, several interactors that were identified in samples from healthy chondrocytes, such as SOD2, vimentin, vinculin, gelsolin, or alpha-enolase, have been previously reported to be altered in chondrocytes from OA patients (46–53) (Table III) and early to severe stages of the disease contain very high levels of Cx43 (23).
IP experiments were performed in vitro using cultured chondrocytes. Chondrocytes were actively proliferating in an asyncronic culture where cells are in different stages of the cell cycle, whereas they were mainly in a resting stage in normal tissue. This difference, together with physiological conditions and the type of ECM molecules that interact with chondrocytes in tissue, may affect the number of the Cx43-associated proteins identified in this study and in particular identification of Cx43 interactors related with cytoskeletal proteins and proteins involved in cell-matrix adhesion and cell-to-cell communication. Cartilage is very challenging tissue because it is solid and mainly composed of collagen-rich ECM with low cellularity. Extraction of proteins will require the use of chaotropic agents incompatible with protein interactions and IP experiments. These limitations make it impossible to perform IP experiments using tissue as the starting material. Still, the Cx43-associated proteins identified in this and other studies finally depend on the Cx43 amino acid sequence and domains. We have identified a number of multiple Cx43 interacting proteins that might help in understanding the different cellular functions of Cx43. Further studies will be necessary to fully understand the molecular roles of these interactions under physiological conditions and in healthy and disease states.
Especially noteworthy is the identification of several Cx43-interactors related to the generation of precursor metabolites and energy (Table II). Some of the Cx43-interactors identified in this study are enzymes responsible for glycolysis and gluconeogenesis (ALDOA, GAPDH, PGK1, ENO1, and PKM2). Notably, the glycolytic enzymes that were identified as Cx43-binding proteins, except Lactate Dehydrogenase A, which is implicated in the final step of anaerobic glycolysis, are involved in the energy-generation phase of glycolysis, which results in the net synthesis of ATP and NADH molecules. The interactions between Cx43 and these enzymes most likely play an important role in the cellular use of glucose. In fact, the eye lens and articular cartilage of adults are avascular organs, and because no blood supply reaches these tissues, it has been reported that cells in these tissues are maintained by the movement of fluids, ions, nutrients (e.g. glucose), and metabolites through a network of GJs (7, 54, 55). In addition, results from different reports strongly suggest that inhibition of GJIC modifies glucose uptake and that glucose levels modify Cx43 gene expression (56). Reduced levels of Cx43 alters the expression of the glucose transporters GLUT-1 and GLUT-3 and type I and II hexokinases, which are enzymes responsible for the phosphorylation of intracellular glucose to obtain a charged molecule that cannot be transported through the plasma membrane (57, 58). On the other hand, high-glucose conditions have been shown to reduce Cx43 expression and GJIC in normal conditions (59–61). However, increased levels of Cx43 have been detected in pathological conditions, e.g. during wound healing in diabetic rats and humans (62–64). Consequently, cell metabolism and Cx43 appear to be interconnected. It would be interesting to study the role of Cx43 in glucose metabolism in particular chondrocytes because diabetes has been suggested as a possible risk factor for developing OA and OA chondrocytes showed significantly elevated Cx43 protein levels (23).
Recent studies have shown that Cx43 localizes to mitochondria (65–68), forms functional hemichannels, and plays an important role during ischemic preconditioning (65, 66, 68, 69). During prolonged ischemia, ATP levels decrease, leading to lowering of the pH and accumulation of reactive oxygen species (ROS), which ultimately result in cell death. Several studies have shown that mitochondrial Cx43 creates a protective, preconditioning effect during the early stages of cardiac ischemia by most likely favoring ATP and/or glutathione release to generate preconditioning and cell-protective functions. In this report, the MS data revealed that Cx43 might interact with the mitochondrial protein SOD2 and the mitochondrial membrane ATP synthase (ATP5J2). In addition, the Hsp90 chaperone, which has been reported to be involved in the translocation of Cx43 to the mitochondria (67, 70), has been identified in this study and by other authors (67) as a Cx43 binding protein. Importantly, SOD2 and ATP5J2 were only identified as Cx43-interactors in samples from healthy donors, and not in OA chondrocytes (supplemental Table S4). The functional relation between Cx43 and the identified mitochondrial interactors will most likely help in understanding the importance of normal mitochondrial function in degenerative disorders or preconditioning.
Gene array analyses have indicated that Cx43 might represent a central node in the regulation of genome-wide expression (71–73). Full-length Cx43 and its C-terminal tail have been localized to the nucleus, and translocation of the Cx43-CTD into the nucleus modified cell growth, proliferation and gene expression (22). It has been suggested that Cx43 might regulate gene expression through the recruitment of transcription factors to the proximal promoter element termed the connexin response element (CxRE) (74, 75). Yet, how Cx43 can translocate into the nucleus and modify gene expression has not been described. IP and MS analysis has identified two possible Cx43-interactors involved in the RAN-GTP nuclear transport cycle that could be involved in the translocation of Cx43 to the nucleus, the GTP-binding nuclear protein Ran (RAN) and importin β (KPNB1) (Fig. 3A). Additionally, the C-terminal tail of Cx43 contains two potential NLSs that are recognized by the importin α/β complex, which imports proteins to the nucleus through the RAN-GTP cycle (Fig. 3B). However, cNLS Mapper does not predict an importin α-independent NLS. Thus, we cannot rule out that additional sites in the Cx43 sequence might be implicated in the translocation of the protein to the nucleus. The identified Cx43-nuclear interactors include several histones, transcription factors and nucleolar proteins, such as nucleolin or the polymerase I and transcript release factor (PTRF), which suggests that Cx43 or its C-terminal tail might affect chromatin organization and nucleolar activity.
The presence of possible interactors with known functions directly linked to downstream transcription processes, such as RNA splicing, processing, export and translation (Table II), together with the perinuclear localization of Cx43 (Fig. 2C) suggest a potential role of Cx43 in these processes. We have identified several Cx43-interactors involved in post-transcriptional regulation and mRNA processing. Some of these interactors are integrated into the nuclear or endoplasmic reticulum membranes. Together with the identification of several Cx43-binding proteins involved in RNA transport, such as hnRNPs, and translation, which include several ribosomal proteins (Table II), these results suggest a possible role of Cx43 in translation and protein biosynthesis that might be related to RNA species transport across the nuclear and endoplasmic reticulum system. On the other hand, the interaction of Cx43 with the Major Vault Protein (MVP) might be related with the transport of different types of RNA, including mRNA (76, 77), along cytoskeletal elements into dendritic microtubules or the axonal compartment (78–81).
Components of the cell junction and cell projections and proteins related to cell adhesion, motility, and extracellular matrix interactions have been identified using MS as Cx43-associated proteins, and these include several collagens, vinculin, talin, and annexins. The cell adhesion and ECM-receptor interaction proteins collagen VI and XII and integrin alpha-V and beta-I were only detected in samples from OA patients. Moreover, it has been reported that residues 136–158 of the cytoplasmic loop region of Cx43 contains a calmodulin-binding motif (42), and the MS data in this report revealed that calmodulin-interacting proteins, such as caldesmon 1, several types of myosin and a diversity of cytoskeletal proteins, also associate with Cx43 in OA samples. These results suggest that Cx43, particularly in OA chondrocytes, might play a (dys)functional role in cell adhesion and migration. In addition, caldesmon plays important roles in the migration of nonmuscle cells via regulating the actin-myosin system (82), and caldesmon binding to myosin has been reported to result in the enhancement of axon extension (83). Interestingly, we have found that chondrocytes from OA patients contain longer cellular extensions (more than 200 μm in length) when compared with chondrocytes in healthy cartilage (7). Further studies will be necessary to reveal the role of Cx43 in these processes and to discern its function in OA chondrocytes.
Cx43 is a complex protein with multiple functions and cellular localizations. The results reported here suggest that the complex proteomic interactions with Cx43 may play important functions in cell metabolism, gene transcription and translation, cytoskeleton organization, motility, and cell–cell and cell–matrix interactions. It is well documented that Cx43 is up- or down-regulated in a number of diseases such as cancers, inflammation, and disorders such as Alzheimer's disease, Huntington's disease, or OA (18, 23, 84–88). The literature also indicates that mutations in the Cx43 sequence, including single mutations in its C-terminal tail, are responsible for the development of diseases such as oculodentodigital dysplasia (ODDD) (89–92). Many of these observations lack the mechanistic understanding that is required for the development of an effective therapy. The Cx43-interactors identified in this report will serve to gain a better understanding of the cellular functions of Cx43 in health and disease. Although the Cx43 sequence is well conserved from humans to fish, several domains are only highly conserved among primates; therefore, the results presented here can be extended as a guide for further studies of Cx43 interactions in different human cell types.
Supplementary Material
Acknowledgments
We thank Lourdes Sanjurjo and Dolores Salinas Bujan for generously obtaining cartilage samples after surgery; Maria Jose Sanchez Dopico for isolating articular chondrocytes from cartilage for cell culture; Purificación Filgueira-Fernández and Noa Goyanes for collecting and storing cartilage samples, preparing samples and immunohistochemistry and immunofluorescence experiments; members of the Proteomics group (INIBIC) for helpful technical suggestions; Estefania Cives for administrative assistance; thanks to Ada Castro, Purificación Filgueira and Julian Yañez for critical technical input. Marta Varela Eirin for her help in the writing and design of tables. A very special thanks to David Santamaria for his help and critical suggestions.
Footnotes
Author contributions: RG-F designed and performed the experiments and performed data analyses. PC-F performed the experiments. PF-P and JM performed the mass-spec runs and processed the data. DM performed immunofluorescente analysis. BA and EF provided critical input. FJB participated in design of experiments. MDM conceived the study, designed and performed the experiments (IHC analysis), designed and performed the data and bioinformatic analyses (GO, comparisons with other studies) and wrote the paper. All authors discussed the results and commented on the manuscript.
* This work was supported in part through funding from Spanish Society for Rheumatology, SER (FER 2013) (to M.D.M.) and by the National Plan for Scientific Research, Development and Technological Innovation 2013-2016 and funded by the ISCIII-General Subdirection of Assessment and Promotion of the Research - European Regional Development Fund (FEDER) “A way of making Europe”: PI13/00591 to M.D.M. The Fondo Investigación Sanitaria, Madrid, Spain (CIBER-CB06/01/0040; RETIC-RIER-RD12/0009/0018; Proteo-Red/ISCIII) (to F.J.B.); and a Predoctoral fellowship from Xunta de Galicia to Raquel Gago-Fuentes. María D. Mayán was an Isidro Parga Pondal researcher (Xunta de Galicia). The study sponsors had no role in the study design, data collection, data analysis or writing of the manuscript and did not affect the decision to submit the manuscript for publication.
 This article contains supplemental Tables S1 to S5.
This article contains supplemental Tables S1 to S5.
Ethics approval: The study was conducted with the approval of the local ethics committee in Galicia, Spain.
1 The abbreviations used are:
- GJ
- gap junction
- ACN
- acetonitrile
- Co-IP
- co-immunoprecipitation
- CTD
- carboxy-terminal domain
- Cx43
- connexin 43
- DTT
- dithiothreitol
- ECM
- extracellular Matrix
- GJIC
- gap junctional intercellular communication
- GO
- Gene Ontology
- IAA
- iodoacetamide
- IHC
- immunohistochemistry
- IP
- immunoprecipitation
- OA
- osteoarthritis
- TFA
- trifluoroacetic acid.
REFERENCES
- 1. Kar R., Batra N., Riquelme M. A., Jiang J. X. (2012) Biological role of connexin intercellular channels and hemichannels. Arch. Biochem. Biophys. 524, 2–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saez J. C., Berthoud V. M., Branes M. C., Martinez A. D., Beyer E. C. (2003) Plasma membrane channels formed by connexins: their regulation and functions. Physiol. Rev. 83, 1359–1400 [DOI] [PubMed] [Google Scholar]
- 3. Simon A. M., Goodenough D. A., Paul D. L. (1998) Mice lacking connexin40 have cardiac conduction abnormalities characteristic of atrioventricular block and bundle branch block. Curr. Biol. 8, 295–298 [DOI] [PubMed] [Google Scholar]
- 4. Kirchhoff S., Nelles E., Hagendorff A., Kruger O., Traub O., Willecke K. (1998) Reduced cardiac conduction velocity and predisposition to arrhythmias in connexin40-deficient mice. Curr. Biol. 8, 299–302 [DOI] [PubMed] [Google Scholar]
- 5. Alexander D. B., Goldberg G. S. (2003) Transfer of biologically important molecules between cells through gap junction channels. Curr. Med. Chem. 10, 2045–2058 [DOI] [PubMed] [Google Scholar]
- 6. Goldberg G. S., Lampe P. D., Nicholson B. J. (1999) Selective transfer of endogenous metabolites through gap junctions composed of different connexins. Nat. Cell. Biol. 1, 457–459 [DOI] [PubMed] [Google Scholar]
- 7. Mayan M. D., Gago-Fuentes R., Carpintero-Fernandez P., Fernandez-Puente P., Filgueira-Fernandez P., Goyanes N., Valiunas V., Brink P. R., Goldberg G. S., Blanco F. J. (2013) Articular chondrocyte network mediated by gap junctions: role in metabolic cartilage homeostasis. Ann. Rheum. Dis. 74(1), 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Willecke K., Eiberger J., Degen J., Eckardt D., Romualdi A., Guldenagel M., Deutsch U., Sohl G. (2002) Structural and functional diversity of connexin genes in the mouse and human genome. Biol. Chem. 383, 725–737 [DOI] [PubMed] [Google Scholar]
- 9. Sohl G., Willecke K. (2004) Gap junctions and the connexin protein family. Cardiovasc. Res. 62, 228–232 [DOI] [PubMed] [Google Scholar]
- 10. Giepmans B. N. (2004) Gap junctions and connexin-interacting proteins. Cardiovasc. Res. 62, 233–245 [DOI] [PubMed] [Google Scholar]
- 11. Duffy H. S., Fort A. G., Spray D. C. (2006) Cardiac connexins: genes to nexus. Adv. Cardiol. 42, 1–17 [DOI] [PubMed] [Google Scholar]
- 12. Chen J., Pan L., Wei Z., Zhao Y., Zhang M. (2008) Domain-swapped dimerization of ZO-1 PDZ2 generates specific and regulatory connexin43-binding sites. EMBO J. 27, 2113–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Giepmans B. N., Moolenaar W. H. (1998) The gap junction protein connexin43 interacts with the second PDZ domain of the zona occludens-1 protein. Curr. Biol. 8, 931–934 [DOI] [PubMed] [Google Scholar]
- 14. Toyofuku T., Yabuki M., Otsu K., Kuzuya T., Hori M., Tada M. (1998) Direct association of the gap junction protein connexin-43 with ZO-1 in cardiac myocytes. J. Biol. Chem. 273, 12725–12731 [DOI] [PubMed] [Google Scholar]
- 15. Park D. J., Freitas T. A., Wallick C. J., Guyette C. V., Warn-Cramer B. J. (2006) Molecular dynamics and in vitro analysis of Connexin43: A new 14–3-3 mode-1 interacting protein. Protein Sci. 15, 2344–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Butkevich E., Hulsmann S., Wenzel D., Shirao T., Duden R., Majoul I. (2004) Drebrin is a novel connexin-43 binding partner that links gap junctions to the submembrane cytoskeleton. Curr. Biol. 14, 650–658 [DOI] [PubMed] [Google Scholar]
- 17. Giepmans B. N., Verlaan I., Hengeveld T., Janssen H., Calafat J., Falk M. M., Moolenaar W. H. (2001) Gap junction protein connexin-43 interacts directly with microtubules. Curr. Biol. 11, 1364–1368 [DOI] [PubMed] [Google Scholar]
- 18. Zhou J. Z., Jiang J. X. (2014) Gap junction and hemichannel-independent actions of connexins on cell and tissue functions – An update. FEBS Lett. 17; 588(8), 1186–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Omori Y., Yamasaki H. (1999) Gap junction proteins connexin32 and connexin43 partially acquire growth-suppressive function in HeLa cells by deletion of their C-terminal tails. Carcinogenesis 20, 1913–1918 [DOI] [PubMed] [Google Scholar]
- 20. Olbina G., Eckhart W. (2003) Mutations in the second extracellular region of connexin 43 prevent localization to the plasma membrane, but do not affect its ability to suppress cell growth. Mol. Cancer Res. 1, 690–700 [PubMed] [Google Scholar]
- 21. Moorby C., Patel M. (2001) Dual functions for connexins: Cx43 regulates growth independently of gap junction formation. Exp. Cell Res. 271, 238–248 [DOI] [PubMed] [Google Scholar]
- 22. Dang X., Doble B. W., Kardami E. (2003) The carboxy-tail of connexin-43 localizes to the nucleus and inhibits cell growth. Mol. Cell. Biochem. 242, 35–38 [PubMed] [Google Scholar]
- 23. Mayan M. D., Carpintero-Fernandez P., Gago-Fuentes R., Martinez-de-Ilarduya O., Wang H. Z., Valiunas V., Brink P., Blanco F. J. (2013) Human articular chondrocytes express multiple gap junction proteins: differential expression of connexins in normal and osteoarthritic cartilage. Am. J. Pathol. 182, 1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsuchida S., Arai Y., Kishida T., Takahashi K. A., Honjo K., Terauchi R., Inoue H., Oda R., Mazda O., Kubo T. (2013) Silencing the expression of connexin 43 decreases inflammation and joint destruction in experimental arthritis. J. Orthop. Res. 31, 525–530 [DOI] [PubMed] [Google Scholar]
- 25. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 26. Bush P. G., Hall A. C., Macnicol M. F. (2008) New insights into function of the growth plate: clinical observations, chondrocyte enlargement, and a possible role for membrane transporters. J. Bone Joint Surg. Br. 90, 1541–1547 [DOI] [PubMed] [Google Scholar]
- 27. Medina-Aunon J. A., Paradela A., Macht M., Thiele H., Corthals G., Albar J. P. (2010) Protein Information and Knowledge Extractor: discovering biological information from proteomics data. Proteomics 10, 3262–3271 [DOI] [PubMed] [Google Scholar]
- 28. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 29. van der Loos C. M. (2010) Chromogens in multiple immunohistochemical staining used for visual assessment and spectral imaging: the colorful future. J. Histotechnol. 33, 1 [Google Scholar]
- 30. Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., Lamond A. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Solan J. L., Lampe P. D. (2005) Connexin phosphorylation as a regulatory event linked to gap junction channel assembly. Biochim. Biophys. Acta 1711, 154–163 [DOI] [PubMed] [Google Scholar]
- 32. Singh D., Solan J. L., Taffet S. M., Javier R., Lampe P. D. (2005) Connexin 43 interacts with zona occludens-1 and -2 proteins in a cell cycle stage-specific manner. J. Biol. Chem. 280, 30416–30421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu C. C., MacCoss M. J., Howell K. E., Yates J. R., 3rd (2003) A method for the comprehensive proteomic analysis of membrane proteins. Nat. Biotechnol. 21, 532–538 [DOI] [PubMed] [Google Scholar]
- 34. Wu C. C., Yates J. R., 3rd (2003) The application of mass spectrometry to membrane proteomics. Nat. Biotechnol. 21, 262–267 [DOI] [PubMed] [Google Scholar]
- 35. Bruce A. F., Rothery S., Dupont E., Severs N. J. (2008) Gap junction remodelling in human heart failure is associated with increased interaction of connexin43 with ZO-1. Cardiovasc. Res. 77, 757–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Herve J. C., Derangeon M., Sarrouilhe D., Giepmans B. N., Bourmeyster N. (2012) Gap junctional channels are parts of multiprotein complexes. Biochim. Biophys. Acta 1818, 1844–1865 [DOI] [PubMed] [Google Scholar]
- 37. Duffy H. S., Delmar M., Spray D. C. (2002) Formation of the gap junction nexus: binding partners for connexins. J. Physiol. Paris 96, 243–249 [DOI] [PubMed] [Google Scholar]
- 38. Herve J. C., Bourmeyster N., Sarrouilhe D., Duffy H. S. (2007) Gap junctional complexes: from partners to functions. Prog. Biophys. Mol. Biol. 94, 29–65 [DOI] [PubMed] [Google Scholar]
- 39. Olk S., Zoidl G., Dermietzel R. (2009) Connexins, cell motility, and the cytoskeleton. Cell Motil. Cytoskeleton 66, 1000–1016 [DOI] [PubMed] [Google Scholar]
- 40. Giepmans B. N. (2006) Role of connexin43-interacting proteins at gap junctions. Adv. Cardiol. 42, 41–56 [DOI] [PubMed] [Google Scholar]
- 41. Kang E. Y., Ponzio M., Gupta P. P., Liu F., Butensky A., Gutstein D. E. (2009) Identification of binding partners for the cytoplasmic loop of connexin43: a novel interaction with beta-tubulin. Cell Commun. Adhes. 15, 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou Y., Yang W., Lurtz M. M., Ye Y., Huang Y., Lee H. W., Chen Y., Louis C. F., Yang J. J. (2007) Identification of the calmodulin binding domain of connexin 43. J. Biol. Chem. 282, 35005–35017 [DOI] [PubMed] [Google Scholar]
- 43. Singh D., Lampe P. D. (2003) Identification of connexin-43 interacting proteins. Cell Commun Adhes 10, 215–220 [DOI] [PubMed] [Google Scholar]
- 44. Hesketh G. G., Van Eyk J. E., Tomaselli G. F. (2009) Mechanisms of gap junction traffic in health and disease. J. Cardiovasc. Pharmacol. 54, 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hatakeyama T., Dai P., Harada Y., Hino H., Tsukahara F., Maru Y., Otsuji E., Takamatsu T. (2013) Connexin43 functions as a novel interacting partner of heat shock cognate protein 70. Sci. Rep. 3, 2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ruiz-Romero C., Carreira V., Rego I., Remeseiro S., Lopez-Armada M. J., Blanco F. J. (2008) Proteomic analysis of human osteoarthritic chondrocytes reveals protein changes in stress and glycolysis. Proteomics 8, 495–507 [DOI] [PubMed] [Google Scholar]
- 47. Lambrecht S., Verbruggen G., Verdonk P. C., Elewaut D., Deforce D. (2008) Differential proteome analysis of normal and osteoarthritic chondrocytes reveals distortion of vimentin network in osteoarthritis. Osteoarthr. Cartil. 16, 163–173 [DOI] [PubMed] [Google Scholar]
- 48. Ruiz-Romero C., Calamia V., Mateos J., Carreira V., Martinez-Gomariz M., Fernandez M., Blanco F. J. (2009) Mitochondrial dysregulation of osteoarthritic human articular chondrocytes analyzed by proteomics: a decrease in mitochondrial superoxide dismutase points to a redox imbalance. Mol. Cell. Proteomics 8, 172–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ruiz-Romero C., Blanco F. J. (2009) The role of proteomics in osteoarthritis pathogenesis research. Curr. Drug Targets 10, 543–556 [DOI] [PubMed] [Google Scholar]
- 50. Lambrecht S., Dhaenens M., Almqvist F., Verdonk P., Verbruggen G., Deforce D., Elewaut D. (2010) Proteome characterization of human articular chondrocytes leads to novel insights in the function of small heat-shock proteins in chondrocyte homeostasis. Osteoarthr. Cartil. 18, 440–446 [DOI] [PubMed] [Google Scholar]
- 51. Rollin R., Tornero P., Marco F., Camafeita E., Calvo E., López-Durán L., Jover J. A., López J. A., Ramón Lamas J., Fernández-Gutiérrez B. (2008) Differential proteome of articular chondrocytes from patients with osteoarthritis. J. Proteomics Bioinform. 1, 267–280 [Google Scholar]
- 52. Hermansson M., Sawaji Y., Bolton M., Alexander S., Wallace A., Begum S., Wait R., Saklatvala J. (2004) Proteomic analysis of articular cartilage shows increased type II collagen synthesis in osteoarthritis and expression of inhibin betaA (activin A), a regulatory molecule for chondrocytes. J. Biol. Chem. 279, 43514–43521 [DOI] [PubMed] [Google Scholar]
- 53. Wu J., Liu W., Bemis A., Wang E., Qiu Y., Morris E. A., Flannery C. R., Yang Z. (2007) Comparative proteomic characterization of articular cartilage tissue from normal donors and patients with osteoarthritis. Arthritis. Rheum. 56, 3675–3684 [DOI] [PubMed] [Google Scholar]
- 54. Mathias R. T., Rae J. L., Baldo G. J. (1997) Physiological properties of the normal lens. Physiol. Rev. 77, 21–50 [DOI] [PubMed] [Google Scholar]
- 55. Mathias R. T., White T. W., Gong X. (2010) Lens gap junctions in growth, differentiation, and homeostasis. Physiol. Rev. 90, 179–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tabernero A., Medina J. M., Giaume C. (2006) Glucose metabolism and proliferation in glia: role of astrocytic gap junctions. J. Neurochem. 99, 1049–1061 [DOI] [PubMed] [Google Scholar]
- 57. Valle-Casuso J. C., Gonzalez-Sanchez A., Medina J. M., Tabernero A. (2012) HIF-1 and c-Src mediate increased glucose uptake induced by endothelin-1 and connexin43 in astrocytes. PLoS One 7, e32448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Herrero-Gonzalez S., Valle-Casuso J. C., Sanchez-Alvarez R., Giaume C., Medina J. M., Tabernero A. (2009) Connexin43 is involved in the effect of endothelin-1 on astrocyte proliferation and glucose uptake. Glia 57, 222–233 [DOI] [PubMed] [Google Scholar]
- 59. Inoguchi T., Ueda F., Umeda F., Yamashita T., Nawata H. (1995) Inhibition of intercellular communication via gap junction in cultured aortic endothelial cells by elevated glucose and phorbol ester. Biochem. Biophys. Res. Commun. 208, 492–497 [DOI] [PubMed] [Google Scholar]
- 60. Kuroki T., Inoguchi T., Umeda F., Ueda F., Nawata H. (1998) High glucose induces alteration of gap junction permeability and phosphorylation of connexin-43 in cultured aortic smooth muscle cells. Diabetes 47, 931–936 [DOI] [PubMed] [Google Scholar]
- 61. Hills C. E., Bland R., Wheelans D. C., Bennett J., Ronco P. M., Squires P. E. (2006) Glucose-evoked alterations in connexin43-mediated cell-to-cell communication in human collecting duct: a possible role in diabetic nephropathy. Am. J. Physiol. Renal Physiol 291, F1045–F1051 [DOI] [PubMed] [Google Scholar]
- 62. Mendoza-Naranjo A., Cormie P., Serrano A. E., Wang C. M., Thrasivoulou C., Sutcliffe J. E., Gilmartin D. J., Tsui J., Serena T. E., Phillips A. R., Becker D. L. (2012) Overexpression of the gap junction protein Cx43 as found in diabetic foot ulcers can retard fibroblast migration. Cell Biol. Int. 36, 661–667 [DOI] [PubMed] [Google Scholar]
- 63. Becker D. L., Thrasivoulou C., Phillips A. R. (2012) Connexins in wound healing; perspectives in diabetic patients. Biochim. Biophys. Acta 1818, 2068–2075 [DOI] [PubMed] [Google Scholar]
- 64. Wang C. M., Lincoln J., Cook J. E., Becker D. L. (2007) Abnormal connexin expression underlies delayed wound healing in diabetic skin. Diabetes 56, 2809–2817 [DOI] [PubMed] [Google Scholar]
- 65. Li H., Brodsky S., Kumari S., Valiunas V., Brink P., Kaide J., Nasjletti A., Goligorsky M. S. (2002) Paradoxical overexpression and translocation of connexin43 in homocysteine-treated endothelial cells. Am. J. Physiol. Heart Circ. Physiol. 282, H2124–H2133 [DOI] [PubMed] [Google Scholar]
- 66. Boengler K., Dodoni G., Rodriguez-Sinovas A., Cabestrero A., Ruiz-Meana M., Gres P., Konietzka I., Lopez-Iglesias C., Garcia-Dorado D., Di Lisa F., Heusch G., Schulz R. (2005) Connexin 43 in cardiomyocyte mitochondria and its increase by ischemic preconditioning. Cardiovasc. Res. 67, 234–244 [DOI] [PubMed] [Google Scholar]
- 67. Rodriguez-Sinovas A., Boengler K., Cabestrero A., Gres P., Morente M., Ruiz-Meana M., Konietzka I., Miro E., Totzeck A., Heusch G., Schulz R., Garcia-Dorado D. (2006) Translocation of connexin 43 to the inner mitochondrial membrane of cardiomyocytes through the heat shock protein 90-dependent TOM pathway and its importance for cardioprotection. Circ. Res. 99, 93–101 [DOI] [PubMed] [Google Scholar]
- 68. Halestrap A. P. (2006) Mitochondria and preconditioning: a connexin connection? Circ. Res. 99, 10–12 [DOI] [PubMed] [Google Scholar]
- 69. Miro-Casas E., Ruiz-Meana M., Agullo E., Stahlhofen S., Rodriguez-Sinovas A., Cabestrero A., Jorge I., Torre I., Vazquez J., Boengler K., Schulz R., Heusch G., Garcia-Dorado D. (2009) Connexin43 in cardiomyocyte mitochondria contributes to mitochondrial potassium uptake. Cardiovasc. Res. 83, 747–756 [DOI] [PubMed] [Google Scholar]
- 70. Rehling P., Brandner K., Pfanner N. (2004) Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 5, 519–530 [DOI] [PubMed] [Google Scholar]
- 71. Iacobas D. A., Urban-Maldonado M., Iacobas S., Scemes E., Spray D. C. (2003) Array analysis of gene expression in connexin-43 null astrocytes. Physiol. Genomics 15, 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Iacobas D. A., Iacobas S., Urban-Maldonado M., Spray D. C. (2005) Sensitivity of the brain transcriptome to connexin ablation. Biochim. Biophys. Acta 1711, 183–196 [DOI] [PubMed] [Google Scholar]
- 73. Iacobas D. A., Scemes E., Spray D. C. (2004) Gene expression alterations in connexin null mice extend beyond the gap junction. Neurochem. Int. 45, 243–250 [DOI] [PubMed] [Google Scholar]
- 74. Stains J. P., Lecanda F., Screen J., Towler D. A., Civitelli R. (2003) Gap junctional communication modulates gene transcription by altering the recruitment of Sp1 and Sp3 to connexin-response elements in osteoblast promoters. J. Biol. Chem. 278, 24377–24387 [DOI] [PubMed] [Google Scholar]
- 75. Stains J. P., Civitelli R. (2005) Gap junctions regulate extracellular signal-regulated kinase signaling to affect gene transcription. Mol. Biol. Cell 16, 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li J. Y., Volknandt W., Dahlstrom A., Herrmann C., Blasi J., Das B., Zimmermann H. (1999) Axonal transport of ribonucleoprotein particles (vaults). Neuroscience 91, 1055–1065 [DOI] [PubMed] [Google Scholar]
- 77. Herrmann C., Golkaramnay E., Inman E., Rome L., Volknandt W. (1999) Recombinant major vault protein is targeted to neuritic tips of PC12 cells. J. Cell Biol. 144, 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Taneja K. L., Lifshitz L. M., Fay F. S., Singer R. H. (1992) Poly(A) RNA codistribution with microfilaments: evaluation by in situ hybridization and quantitative digital imaging microscopy. J. Cell Biol. 119, 1245–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ferrandon D., Elphick L., Nusslein-Volhard C., St Johnston D. (1994) Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79, 1221–1232 [DOI] [PubMed] [Google Scholar]
- 80. Carson J. H., Worboys K., Ainger K., Barbarese E. (1997) Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil. Cytoskeleton 38, 318–328 [DOI] [PubMed] [Google Scholar]
- 81. Steward O. (1997) mRNA localization in neurons: a multipurpose mechanism? Neuron 18, 9–12 [DOI] [PubMed] [Google Scholar]
- 82. Mayanagi T., Sobue K. (2011) Diversification of caldesmon-linked actin cytoskeleton in cell motility. Cell Adh. Migr. 5, 150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Morita T., Mayanagi T., Sobue K. (2012) Caldesmon regulates axon extension through interaction with myosin II. J. Biol. Chem. 287, 3349–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Loring J. F., Wen X., Lee J. M., Seilhamer J., Somogyi R. (2001) A gene expression profile of Alzheimer's disease. DNA Cell Biol. 20, 683–695 [DOI] [PubMed] [Google Scholar]
- 85. Fushiki S., Perez Velazquez J. L., Zhang L., Bechberger J. F., Carlen P. L., Naus C. C. (2003) Changes in neuronal migration in neocortex of connexin43 null mutant mice. J. Neuropathol. Exp. Neurol. 62, 304–314 [DOI] [PubMed] [Google Scholar]
- 86. Gutmann D. H., Hedrick N. M., Li J., Nagarajan R., Perry A., Watson M. A. (2002) Comparative gene expression profile analysis of neurofibromatosis 1-associated and sporadic pilocytic astrocytomas. Cancer Res. 62, 2085–2091 [PubMed] [Google Scholar]
- 87. Vinken M., Vanhaecke T., Papeleu P., Snykers S., Henkens T., Rogiers V. (2006) Connexins and their channels in cell growth and cell death. Cell Signal. 18, 592–600 [DOI] [PubMed] [Google Scholar]
- 88. Kielian T. (2008) Glial connexins and gap junctions in CNS inflammation and disease. J. Neurochem. 106, 1000–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Paznekas W. A., Boyadjiev S. A., Shapiro R. E., Daniels O., Wollnik B., Keegan C. E., Innis J. W., Dinulos M. B., Christian C., Hannibal M. C., Jabs E. W. (2003) Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am. J. Hum. Genet. 72, 408–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Laird D. W. (2006) Life cycle of connexins in health and disease. Biochem. J. 394, 527–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Laird D. W. (2010) The gap junction proteome and its relationship to disease. Trends Cell Biol. 20, 92–101 [DOI] [PubMed] [Google Scholar]
- 92. Laird D. W. (2014) Syndromic and nonsyndromic disease-linked Cx43 mutations. FEBS Lett. 17; 588(8), 1339–48 [DOI] [PubMed] [Google Scholar]
- 93. Kosugi S., Hasebe M., Tomita M., Yanagawa H. (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. U.S.A. 106, 10171–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.