Summary
Shaping natural killer (NK) cell functions in human immunity and reproduction are diverse killer-cell immunoglobulin-like receptors (KIRs) that recognize polymorphic MHC class I determinants. A survey of placental mammals suggests KIRs serve as variable NK cell receptors only in certain primates and artiodactyls. Divergence of functional and variable KIRs in primates and artiodactyls predates placental reproduction. Among artiodactyls, cattle but not pigs have diverse KIRs. Catarrhine (humans, apes, and Old World monkeys) and platyrrhine (New World monkeys) primates, but not prosimians, have diverse KIRs. Platyrrhine and catarrhine systems of KIR and MHC class I are highly diverged, but within the catarrhines a stepwise co-evolution of MHC class I and KIRs is discerned. In Old World monkeys, diversification focuses on MHC-A and MHC-B and their cognate lineage II KIR. With evolution of C1-bearing MHC-C from MHC-B, as informed by orangutan, the focus changes to MHC-C and its cognate lineage III KIR. Evolution of C2 from C1 and fixation of MHC-C, drove further elaboration of MHC-C-specific KIRs, as exemplified by chimpanzee. In humans, the evolutionary trajectory changes again. Emerging from reorganization of the KIR locus and selective attenuation of KIR avidity for MHC class I are the functionally distinctive KIR A and KIR B haplotypes.
Keywords: NK cells, KIR, MHC class I, co-evolution
Introduction
In comparison to the major histocompatibility complex (MHC) class I genes of other mammalian species, the family of human leukocyte antigen (HLA) class I genes is both simple and tidy. In the human MHC, the HLA complex on chromosome 6, there are only six functional HLA class I genes, HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, and HLA-G, each of which is present on every HLA haplotype. The HLA-A, HLA-B, and HLA-C genes have the unusual characteristic of being highly polymorphic, and this is true for all human populations (1, 2, http://www.ncbi.nlm.nih.gov/projects/gv/mhc/). Antigenic differences between the HLA class I proteins of donors and recipients contribute to the adverse alloreactive immune responses experienced by patients transplanted with allogeneic organs or tissues. In the clinical practice of hematopoietic cell transplantation the severity of alloreactions is reduced by matching the donor and recipient for HLA type. Originally a serological test (3), HLA class I typing is now done by nucleotide sequencing, and in the process has produced a database of around ten thousand different HLA-A, HLA-B, and HLA-C sequences (2). Because HLA class I molecules are critical for the development and functions of both natural killer (NK) cells and killer CD8+ T cells, HLA class I polymorphism is likely to have a modulatory effect on most, if not all, human immune responses.
Unlike almost all other genes, the polymorphic HLA class I genes have numerous alleles, among which there are neither dominant nor wildtype forms. Consequently most individuals in a population are heterozygous for the HLA class I genes and this is true for all human populations. The proteins encoded by the common HLA-A, HLA-B, and HLA-C alleles, called allotypes, differ from each other by multiple amino-acid substitutions. These are concentrated in the antigen recognition site of the α1 and α2 domains. There they modulate the immunological functions of HLA class I molecules by affecting their binding specificity for peptide and lymphocyte receptors. Numerous rare HLA-A, HLA-B, and HLA-C alleles differ from a common allele by single nucleotide substitutions, many of which are silent substitutions or replacement substitutions with little or no functional effect. Contrasting with HLA-A, HLA-B, and HLA-C, the HLA-E, HLA-F, and HLA-G genes are not highly polymorphic and have a pattern of natural variation comparable to that of many other immune-system genes. This typically involves two or three alleles with significant frequency, that exhibit functional differences arising from either a single nucleotide substitution or a single event of insertion, deletion or recombination.
Functions of the conserved HLA class I genes
HLA-E appears to be the ‘oldest’ of the six functional HLA class I genes and is expressed by all cell types that have been examined. The peptide-binding groove of HLA-E has adapted to be specific for peptides cleaved from the leader sequences of other HLA class I polypeptides (4, 5). Binding such a peptide is necessary for HLA-E to leave the endoplasmic reticulum and travel to the cell surface, where it functions as the ligand for the inhibitory CD94:NKG2A and activating CD94:NKG2C receptors of NK (6–8). Consequently, the amount of HLA-E at the surface of a cell reflects the extent of HLA class I synthesis within the cell. By monitoring cell-surface HLA-E, NK cells detect perturbations to HLA class I synthesis caused by infection, malignancy and other forms of cellular stress. The HLA-E, CD94 and NKG2A polypeptides are all conserved (9) and provide an NK-cell response to loss of self HLA class I expression, also called missing-self (10), which is similar in all human individuals (11). CD94:NKG2A, CD94:NKG2C and many other NK cell receptors are also expressed on subpopulations of γ δ T cells and effector/memory αβ T cells (12), but these cells are not a subject for this review.
CD94:NKG2A is an example of a killer cell lectin-like receptor (KLR). The CD94 and NKG2A polypeptides have a similar structure that contains a ligand-binding domain resembling that of a C-type lectin (13). Together the lectin-like domains of CD94 and NKG2A form the site that binds the complex of leader-sequence derived peptide and HLA-E (14). Like other KLRs, CD94:NKG2A binds protein ligands, not carbohydrates as is the case for conventional lectins. The genes for CD94 and NKG2A, as well as those for many other KLR that contribute to innate immunity, are located in a genomic region on human chromosome 12 called the natural killer complex (NKC) (15).
HLA-F is expressed by a range of cell types, including extravillous trophoblast cells, but the purpose of this HLA class I molecule is the least understood (16). Specialized to function in reproduction, HLA-G is expressed only by extravillous trophoblast, the fetal tissue that invades the uterine wall during embryo implantation at the beginning of pregnancy when the placenta is formed (17). This invasion involves a specialized population of maternal NK cells in the uterus (uNK) that interact and cooperate with the extravillous trophoblast cells (18). The binding of trophoblast HLA-E to the CD94:NKG2A expressed by uNK is one molecular interaction that contributes to this cellular co-operation. Another is the binding of trophoblast HLA-G to the inhibitory Leukocyte Immunoglobulin-Like Receptor B1 (LILRB1) expressed by uNK. Although LILRB1 binds to a wide range of HLA class I molecules, its strongest interaction is with HLA-G (19). LILRB1 is one of a family of LILRs, all of which are encoded by genes in a genomic region on human chromosome 19 called the leukocyte receptor complex (LRC) (20). The LRC is densely packed with genes that encode cell surface receptors having ligand-binding sites constructed from immunoglobulin-like domains. Many of these receptors contribute to immune defense or reproduction, and some contribute to both of these vital functions. The NKC and the LRC are thus seen as two separate genomic regions that specialize in leukocyte receptors but make them from totally different protein modules.
Functions of the highly polymorphic HLA class I genes
A major function of the highly polymorphic HLA class I molecules is to bind peptides inside cells and present them on the cell surface to the αβ receptors of killer CD8+ T lymphocytes. Such interactions of HLA class I with αβ T-cell receptors (TCRs) are critical for the development and selection of these cells in the thymus, for establishing tolerance to self MHC class I and for providing effective CD8+ T-cell immunity against unhealthy cells compromised by infection, malignant transformation, or stress.
The high polymorphism of HLA class I confers several benefits to the human species. The individual benefits from having two functionally different allotypic versions of HLA-A, HLA-B, and HLA-C, which can complement each other and serve to increase the breadth of the immune response to any unhealthy cell. Human families benefit, because a rapidly evolving pathogen that adapts to evade the CD8+ T-cell response of one infected family member, cannot similarly evade the response of the other family members it subsequently infects, providing they have different HLA type. High polymorphism also reduces the likelihood that all members of a population or species will succumb during epidemics of acute infectious disease, particularly ones that are new and emerging zoonoses against which the entire population is naive and immunologically inexperienced. HLA-A, HLA-B, and HLA-C polymorphism also prevents the spread of cancer from one person to another, because any transferred tumor cells will trigger and be eliminated by a potent alloreactive CD8+ T-cell response.
Along with their contribution to CD8+ T-cell development and function, some HLA-A and HLA-B allotypes have an analogous involvement in NK cell development, also called NK cell education, and function. Of these, the most important are the HLA-A and HLA-B allotypes that carry the Bw4 epitope, one of the very first antigens described by HLA serologists (3). The Bw4 epitope is carried by ~17% of HLA-A allotypes and ~39% of HLA-B allotypes and is defined by sequence motifs at positions 77-83 of the helix in the α1 domain of the HLA class I molecule (21). The Bw4 epitope is recognized by KIR3DL1 (22), one of the killer cell immunoglobulin-like receptors (KIRs) that are expressed by NK cells and encoded by a multigene family (Fig. 1) adjacent to the LILR genes in the LRC. By educating NK cells to detect perturbations in the expression Bw4-bearing of HLA-A and HLA-B allotypes, the inhibitory KIR3DL1 has an analogous function to CD94:NKG2A. It is also a complementary function, because CD94:NKG2A assesses the overall integrity of HLA-A, HLA-B, and HLA-C expression, whereas KIR3DL1 focuses on particular HLA-A and HLA-B allotypes. In human populations, around 50% of HLA haplotypes encode an HLA-A or an HLA-B allotype that has the Bw4 epitope, and some haplotypes encode both (23). Consequently, ~75% of the human population have the Bw4 epitope, while ~25% lack the Bw4 epitope. Unlike CD94:NKG2A, which is functional in all human individuals, KIR3DL1 cannot function in a minority, but still a large number (~1.8 billion people), of the human population, because in them it has no ligand.
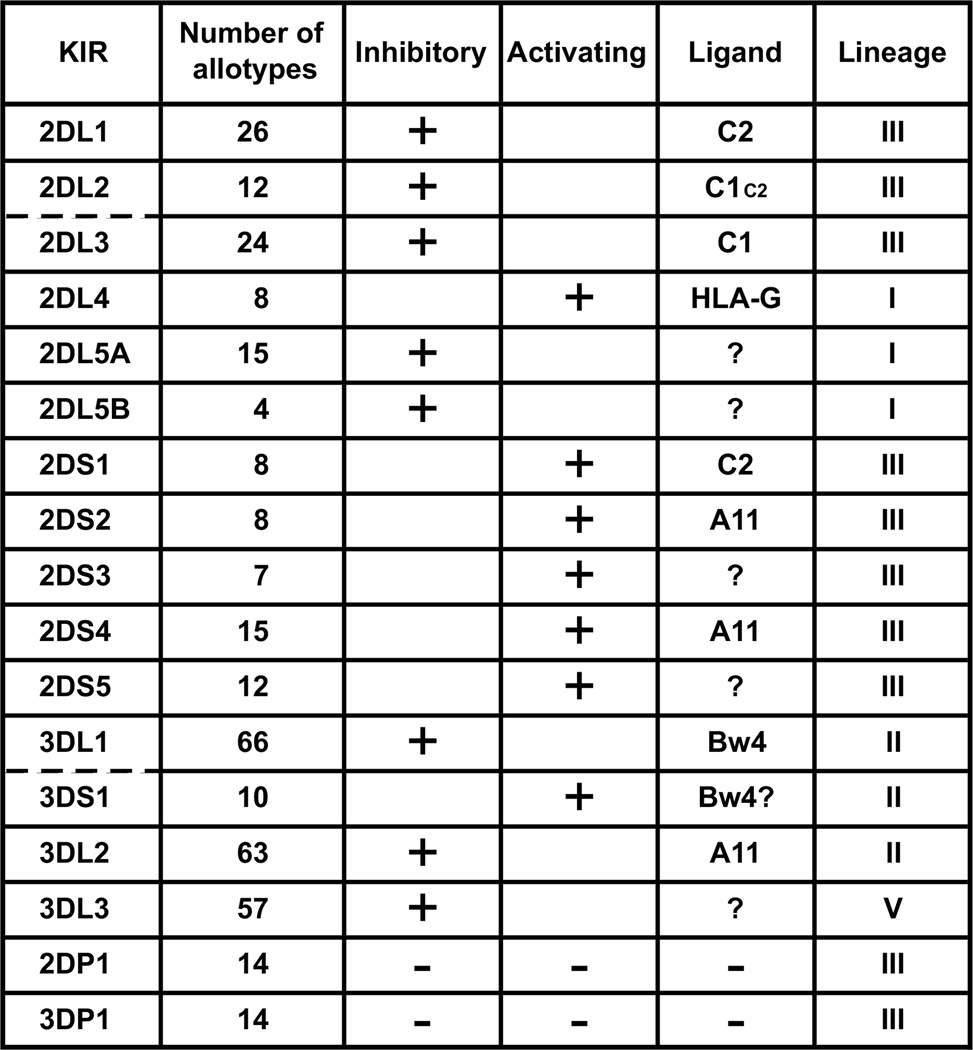
Figure 1. Variability and ligands of human killer cell immunoglobulin-like receptors (KIRs).
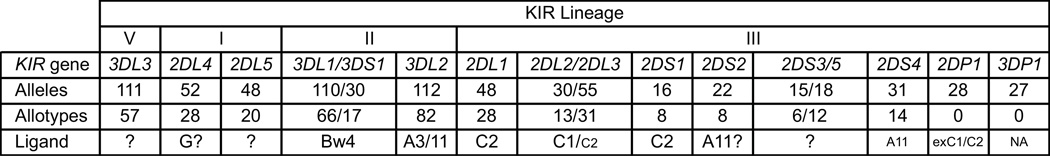
Human KIRs comprise 11 types of expressed KIR and two KIR pseudogenes (2DP1 and 3DP1) that form four phylogenetic lineages: I, II, III, and V. For each KIR, the number of allelic and allotypic variants are given in the rows labeled 'Alleles' and 'Allotypes', respectively. The epitopes of HLA class I molecules that serve as ligands for the various KIR are given in the row labeled 'Ligand'. ? indicates either that there is little or no positive evidence for a ligand (KIR2DL5 and KIR2DS3/5) or that are differing opinions among investigators as to the physiological importance of the ligand (HLA-G/KIR2DL4 and KIR2DS2/A11). C1/C2 denotes a strong reaction with C1 and a weaker cross-reaction with C2. A11 denotes that KIR2DS4 has a weak reaction with A11. exC1/C2 indicates that subsets of allotypes of the 2DP1 pseudogene were likely in the past to have been C1-specific and C2-specific receptors.
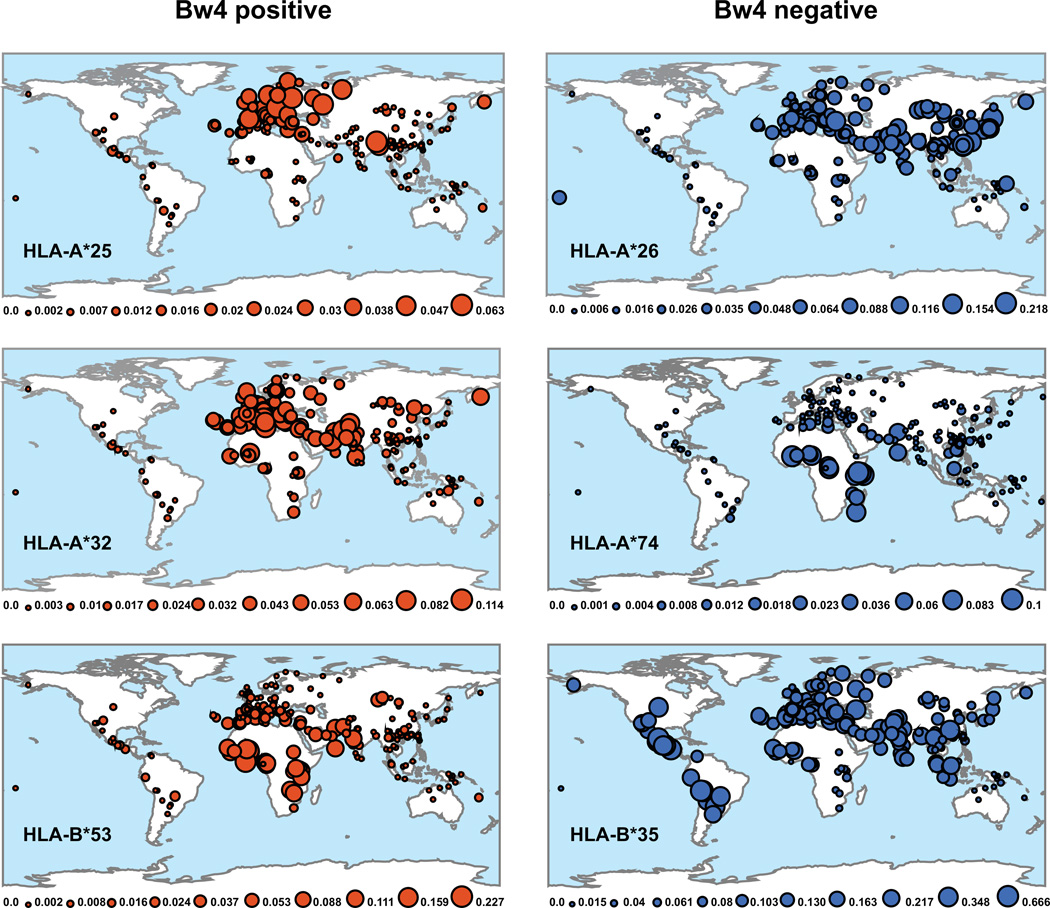
Numerous pairs of HLA-A or HLA-B allotypes differ only at residues 77–83 that determine the presence or absence of the Bw4 epitope (The alternative sequence motifs to Bw4 are called Bw6, another of the first HLA antigens described, but not a ligand for human KIR). The formation of each of these allele pairs required an independent event of recombination that exchanged a Bw6 motif for Bw4 or vice versa and was then followed by natural selection that raised the frequency of the new recombinant allele. HLA-A*25 and its Bw4-bearing partner HLA-A*26 are predominant in Eurasia (Fig. 2, top panels); HLA-A*32 is predominantly in Europe and the Middle East, whereas its Bw4-bearing partner HLA-A*74 is predominantly in Africa (Fig. 2, middle panels). In contrast, HLA-B*53:01, a common allotype of Sub-Saharan African populations, has the Bw4 motif and pairs with HLA-B*35:01, which is widespread and has the Bw6 motif (Fig. 2, bottom panels). HLA-B*53:01 was formed by recombination between HLA-B*35:01 and another HLA-B allele that encodes the Bw4 motif. The likely benefit that caused HLA-B*53:01 to be subject to natural selection is its capacity to confer resistance to childhood cerebral malaria, an endemic disease of tropical Sub-Saharan Africa (24, 25). Although HLA-B*35:01 may also protect against malaria, it is less effective than HLA-B53:01. The protection provided by HLA-B*53:01 could be due to an anti-malarial response of NK cells educated by the Bw4 epitope, or to CD8+ T cells responding to malarial peptides presented by HLA-B*53:01, or to both these arms of the immune response.
Figure 2. Examples of the global distribution of HLA-A and HLA-B allotype pairs differing only in the presence or absence of the Bw4 epitope.
Polymorphisms within residues 77-83 of the α helix of the α1 domain determine whether HLA-A and HLA-B allotypes have or lack the Bw4 epitope. As a consequence of recombination and selection, many pairs of HLA-A and HLA-B allotypes differ only within residues 77-83 and in either having (Bw4 positive) or lacking (Bw4 negative) the Bw4 motif. Three examples are shown here: HLA-A*25/HLA-A*26 (upper panels), HLA-A*32/HLA-A*74 (center panels) and HLA-B*53/HLA-B*35 (lower panels). The maps show the global distribution of each allotype. Each colored circle shows the location of a human population in which the allotypes are present. The size of the circle gives the allele frequency in the population. Given underneath each map is a frequency scale and these differ between the maps. Population frequency data were obtained from http://www.allelefrequencies.net (1)
The HLA-A*03 and HLA-A*11 allotypes carry the A3/11 epitope recognized by a second member of the KIR family, the inhibitory KIR3DL2 receptor. This interaction and its effects on NK cell are not well characterized compared to the interaction of Bw4 with KIR3DL1. Binding of the A3/11 epitope to KIR3DL2 is highly dependent on the peptide bound by HLA-A*03 or HLA-A*11 (26), and this recognition does not appear to educate NK cells in the same way as the Bw4 epitope (27).
Whereas a minority of HLA-A and HLA-B allotypes function as KIR ligands, all HLA-C allotypes have this capacity. Dimorphism at position 80 in the helix of the α1 domain determines if an HLA-C allotype carries the C1 epitope (asparagine 80) or the C2 epitope (lysine 80) (28–30). C1 and C2 are recognized by the inhibitory KIR2DL2/3 and KIR2DL1 receptors, respectively. Both these interactions are effective in educating NK cells to respond to a reduction or loss of HLA-C expression from human cell surfaces. HLA-C is generally less efficient at generating CD8+ T-cell responses than HLA-A and HLA-B (31), but once made they can be effective (32). Correlating with these functional adaptations are distinctive properties of the HLA-C molecule. Adjacent to residues 77-83, the site of the Bw4 motif in HLA-A and -B is, an HLA-C specific ‘KYRV’ motif involving lysine 66, tyrosine 67, arginine 69, and valine 76 (33). The major effect of these residues is to diminish cell-surface expression of HLA-C to one tenth or less that of HLA-A and HLA-B. The KYRV motif reduces the plasticity of the peptide-binding site in HLA-C, making it more selective in binding peptides than HLA-A or HLA-B. This selectivity reduces the efficiency with which HLA-C molecules find a binding peptide in the endoplasmic reticulum and then transit to the cell surface. It also makes them less stable once they are there. Consequently, free HLA-C heavy chains and peptide-free complexes of the heavy chain and β2-microglobulin accumulate inside the cell. Such retention is facilitated by the aspartate 334, serine 336, leucine 337, and isoleucine 338 motif, a dihydrophobic internalization signal that is also specific to HLA-C (34, 35).
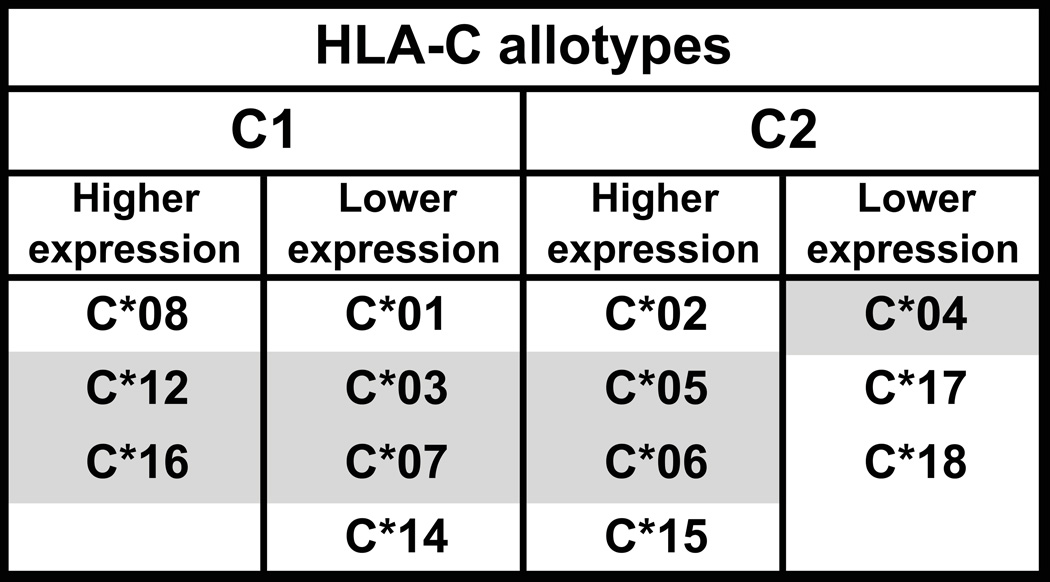
For around one half of the HLA-C alleles, the binding of a microRNA (hsa-miR-148) to the 3′ untranslated region of the HLA-C mRNA prevents translation and further contributes to the low cell-surface expression of HLA-C (36, 37) (Fig. 3). In the other HLA-C alleles, the microRNA binding site is distinguished by the deletion of one nucleotide that abrogates function. Thus the HLA-C allotypes encoded by the latter allele group are expressed at higher cell surface levels than the allotypes encoded by the former allele group (Fig. 3). Although a majority of HLA-B allotypes has asparagine at position 80, they do not have the C1 epitope. Notable exceptions are HLA-B*46 and HLA-B*73, which are recognized by C1-specific KIR (38, 39). These two allotypes are distinguished from all other HLA-B by having valine 76, which is otherwise found only in HLA-C allotypes and in every HLA-C allotype. This shows how this residue of the KYRV motif is critical for making HLA-C a superior ligand for KIR.
Figure 3. HLA-C allotypes differ in their level of expression at cell surfaces.
HLA-C expression level as predicted from polymorphism in the mi-RNA-148 binding site sequence. Allotypes are first grouped according to the presence of the C1 or C2 epitope and then by higher expression, for HLA-C alleles having a nucleotide deletion in the mi-RNA-148 binding site, and lower expression for HLA-C alleles with an intact binding site (37). Expression levels of allotypes in grey-shaded boxes were investigated by Kulkarni et al (36) and conform to these groupings. Analysis by Apps et al (139) gave a hierarchy of cell surface expression that has some differences from the predictions based on the mi-RNA-148 binding site polymorphism. The hierarchy of expression of the C1 containing allotypes was *14>*01>*12>*16>*08>*03>*07 and for the C2 allotypes *18>*06>*15>*04>*02>*05>*17 (139).
Each HLA-A, HLA-B, or HLA-C allotype either carries one of the four epitopes recognized by KIR, A3/11, Bw4, C1, or C2, or has none of these epitopes and does not function as a KIR ligand. No HLA-A, HLA-B, or HLA-C allotype carries more than one of the four epitopes, thus they are mutually exclusive. Crystallographic structures of complexes of various HLA class I and KIR show that the KIR all interact in a similar way with the upper face of the HLA class I molecule and make contact with the two α helices and with the amino acid at position 8 in the bound peptide (28). The footprint of the KIR on the HLA class I molecule overlaps with that of the αβ TCR a further reflection of the many parallel functions of NK cells and T cells.
Human KIR are encoded by a highly variable multigene family
In the human LRC, the human KIR locus comprises a family of 15 KIR genes. Like the HLA class I genes, some of the KIR genes are highly polymorphic and others are conserved or have modest polymorphism (Fig. 4). The KIR locus is flanked on the centromeric side by the LILR gene family and on the telomeric side by the FCAR and NCR1 genes that encode, respectively, the Fc receptor for monomeric IgA and the Natural Cytotoxicity Receptor 1 of NK cells.
Figure 4. Polymorphism and functional variation of human KIR.
Listed for each KIR are the number of allotypes currently defined (http://www.ebi.ac.uk/ipd/kir/) (2), the signaling function of the KIR, its HLA class I ligand, and the phylogenetic lineage to which it belongs. KIR2DL2 and KIR2DL3 are encoded by alleles of the same gene (KIR2DL2/3) but are listed separately because of their functional differences and sequence divergence. Likewise for KIR3DL1 and KIR3DS1, which are both encoded by alleles of the KIR3DL1/S1 gene. ? denotes uncertainty.
Whereas all HLA haplotypes have a fixed set of six functional HLA class I genes, haplotypes of the KIR locus differ in the number of KIR genes they contain (40). This is an important facet of KIR variation, one alternatively called gene-content variation or copy-number variation. In other species, such as macaques (41–44) and seals (45), gene-content variation is a major component of MHC class I gene variation. Conserved features of the KIR locus are the KIR3DL3 and KIR3DL2 genes, which mark the centromeric and telomeric ends of the locus, respectively, and the combination of KIR2DL4 and the KIR3DP1 pseudogene that mark the central part of the locus (46–48). These four genes, collectively called the framework genes, divide the KIR locus into centromeric and telomeric regions, which both vary in the number and nature of the KIR genes they contain. Facilitating this aspect of KIR haplotype variation is the close proximity of adjacent KIR genes and the high sequence similarity of the intervening sequences, as well as the sequence similarity between all the KIR genes. These properties facilitate the process of unequal crossing-over that can produce new recombinant KIR haplotypes with increased or decreased numbers of KIR genes (49). Unequal crossing-over can also produce new recombinant KIR genes, the products of which differ in ligand-binding or signaling functions (23). The inhibitory KIR have long cytoplasmic tails containing immunoreceptor tyrosine-based inhibitory motifs (ITIMs) that engage inhibitory signaling pathways. In contrast, the activating receptors have short cytoplasmic tails that lack ITIMs, but have charged residues in the transmembrane region that engage activating signaling pathways. Further diversification of KIR haplotypes is achieved by the mechanism of equal crossing over. This is facilitated by a distinctive, but highly repetitive sequence in the central part of the KIR locus, which enables centromeric and telomeric gene-content motifs to be recombined in different combinations (48, 50).
The second important aspect of KIR variation is allelic polymorphism (51), which is generally higher for the inhibitory KIR than the activating KIR (Fig. 4). This is exemplified by KIR3DL1/S1 for which there are 66 inhibitory KIR3DL1 allotypes compared to only 17 activating KIR3DS1 allotypes. In part this difference likely stems from the shorter lifetimes, on an evolutionary scale, of the activating receptors (52). Extensive studies of the polymorphism of the inhibitory KIR that recognize the C1, C2, and Bw4 epitopes show that most of the allelic differences have detectable effects on the functions of the cognate KIR2DL2/3, KIR2DL1 and KIR3DL1, respectively. Such effects can alter receptor specificity, avidity, cell-surface expression, or signal transduction (53–59). Many of these functional modulations are caused by substitutions at residues that are not directly involved in contacting MHC class I ligands.
Convergent evolution of variable NK cell receptors for MHC class I in humans and mice
In mice and rats, the functional counterparts of the human KIR are the Ly49 molecules. Before their function as MHC class I receptors was known (60), the gene family encoding Ly49 molecules had been sequenced and shown to specify lectin-like membrane proteins (61, 62). Conversely, the HLA class I specificities of the human NK cell receptors, subsequently named KIR, were described before their primary structures were determined and found to be immunoglobulin-like. Prior to the cloning and sequencing of KIR cDNA (63, 64), the universal expectation was that these variable human NK cell receptors for HLA class I would have similar structure and phylogeny to Ly49. That the human receptors bound MHC class I with immunoglobulin-like domains and had no structural similarity with the Ly49 receptors is an indisputable example of convergent evolution. Under similar selection pressures, but in different circumstance, receptors with comparable function were independently fashioned from two different building blocks. Not only are the binding domains divergent but they interact with completely different sites on MHC class I (28, 65). Although the extracellular parts of Ly49 and KIR are so different, their transmembrane and cytoplasmic tails contain similar functional motifs that enable them to transduce similar signals for inhibiting and activating NK cells. In the human genome the acquisition of a multigene KIR family is accompanied by inactivation of the single human Ly49 gene (66), whereas acquisition of a multigene Ly49 family by the mouse is accompanied with the translocation of the two KIR genes from the LRC to a site on the X chromosome (67).
Distinctive and divergent KIR multigene families in humans and cattle
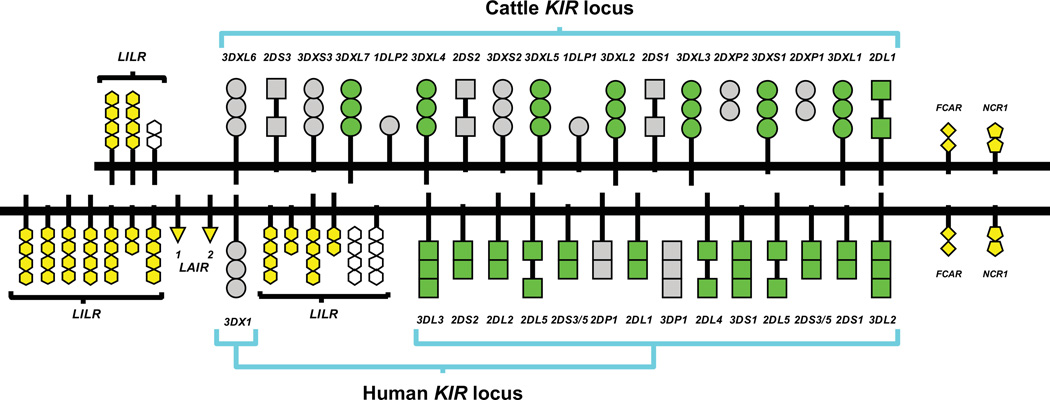
The striking difference between the families of KIR and Ly49 NK cell receptors stimulated study of other mammalian species, to see if they would resemble the mouse model, the human model, or something different again. From examination of species representing five of the 21 orders of placental mammals, functional multigene Ly49 families were found to be present in rodents and in the domestic horse (68, 69) and other equids of the order Perissodactyla (even toed ungulates) (70). The equids also have a single KIR3DL gene that appears to encode a functional inhibitory receptor. Multigene KIR families have been found only in two orders of mammal, the primates and the Artiodactyla (even toed ungulates) (Fig. 5). In primates a functional family of KIR genes has been found in all 23 species of simian primate investigated but not in any prosimian primate. The 23 species of simian primate include 19 that have been reported in the literature (2, 41–44, 71-85) and 4 species for which KIR were identified from analysis of genome assemblies (LAG, unpublished data). Study of artiodactyls has been limited to domestic pig, domestic cattle and aurochs the extinct species from which cattle were originally bred and domesticated (86, 87). The cattle KIR locus consists of 18 genes and pseudogenes (87), whereas the pig KIR locus contains one KIR gene that appears to be the ortholog of the cattle KIR2DL1 gene (86). This is a dramatic difference between these two species of domesticated artiodactyl. In contrast, humans and cattle belong to different mammalian orders but have KIR multigene families of comparable size.
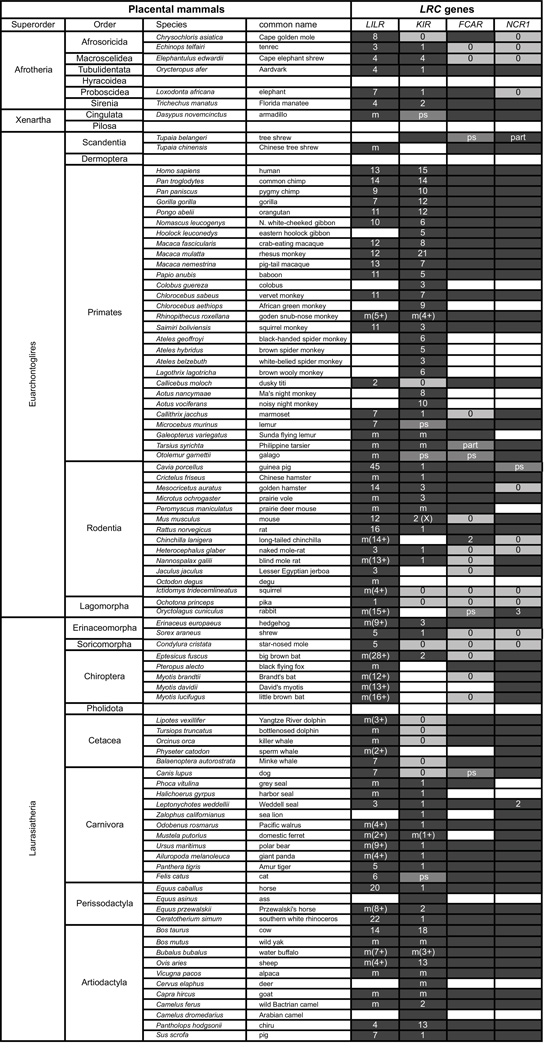
Figure 5. A survey for the presence of KIR and closely-linked LRC genes in species of placental mammal.
For those placental mammals for which genomic sequencing data are available in the Map Viewer at NCBI (http://www.ncbi.nlm.nih.gov/mapview/), we sought evidence for a region syntenic to the human LRC. Each species was analyzed for presence of the LILR, KIR, FCAR, and NCR1 genes. Genes that flank this core LRC region were also assessed, to test initial observations pointing to the absence of LILR, KIR, FCAR, or NCR1. Results were compared with known LRC haplotypes, which have been published and are referenced in the text of this review. The first two columns of the figure give the names for all Superorders and Orders of placental mammals. The third and fourth columns give the species name and common name for those species for which KIR or/and LRC gene content is known. The final four columns indicate presence/absence of the indicated genes and for LILR and KIR the number of genes identified in a species. Dark grey boxes indicate presence of the gene(s). For the medium grey boxes, the label ‘ps’ denotes the presence of a pseudogene and the label ‘part’ denotes that only a partial gene sequence was obtained. Light grey boxes labeled ‘0’ denote absence of the gene. White boxes indicate that insufficient genomic sequence was available to identify the gene. Unless otherwise indicated, single FCAR and NCR1 genes are present. The LILR and KIR genes were assessed in the following ways. For species studied by targeted sequencing, the number of reported KIR genes is listed. For species characterized only by non-targeted whole-genomic sequencing, gene copy number was assessed by counting the number of distinctive LILR or KIR genes identified in the Map Viewer assembly in the region bounded by the flanking markers. Denoted by ‘m’ are incomplete assemblies where multiple genes were identified. Where possible a minimum estimate is given in the brackets and appended with a ’+’.
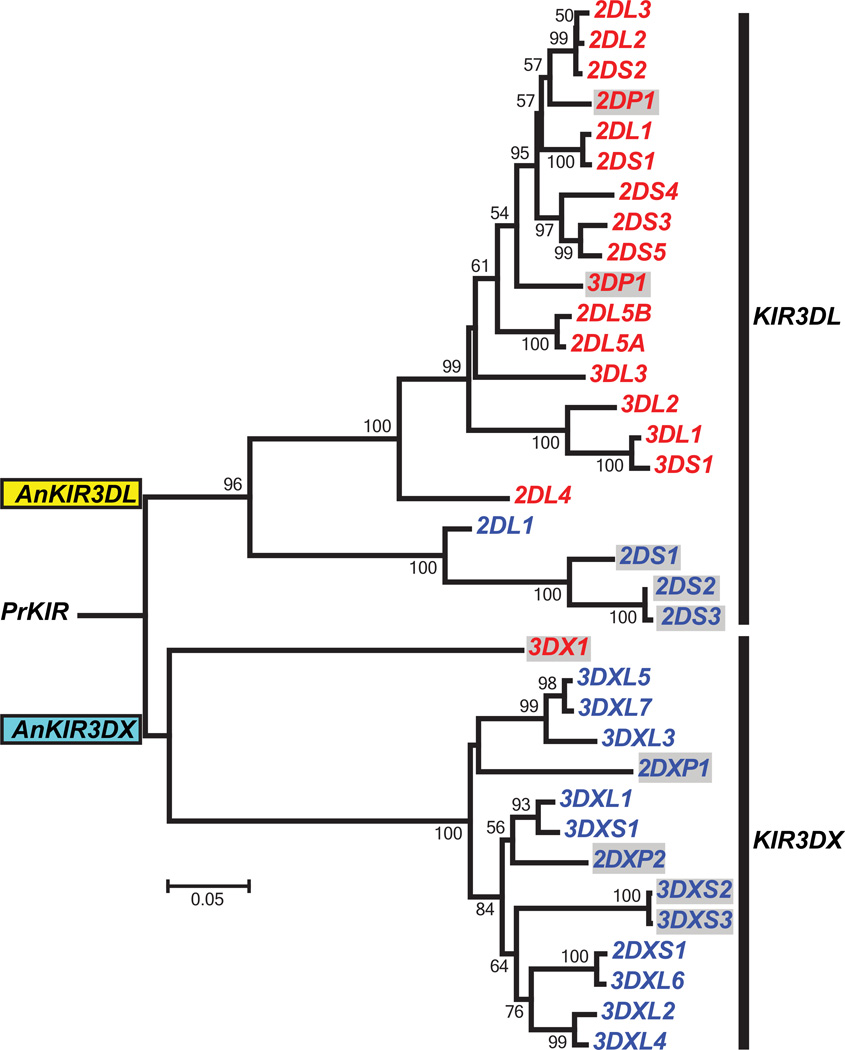
Comparison of the cattle and human KIR loci shows that before the emergence of placental mammals, ~125 million years ago, a primordial KIR gene (PrKIR) duplicated to give two ancestral loci that we shall call AnKIR3DL and AnKIR3DX. In simian primates, the AnKIR3DL gene underwent successive duplications to create a multigene KIR3DL family, whereas the AnKIR3DX gene was not duplicated and became a pseudogene that moved out of the KIR locus and into the neighboring family of LILR genes. Thus simian primates have a family of functional genes derived from AnKIR3DL and one nonfunctional gene derived from AnKIRDLX. By contrast, during the evolution of the cattle KIR gene family, the combination of the AnKIR3DL and AnKIR3DX genes underwent several successive gene duplications to give a mulitigene family that contains an intermingled mixture of KIR3DL and KIR3DX genes (Fig. 6). During or after these duplications there was selective inactivation of all but one of the AnKIR3DL-derived genes. The one functional AnKIR3DL-derived gene in cattle is KIR2DL1, the ortholog of the only gene in the pig KIR locus. Unclear is whether the pig KIR gene is expressed as a functional KIR2DL receptor with D0 and D2 domains (86), or if mutations in what was once an exon encoding the D1 domain cause premature termination and a nonfunctional protein (45). Thus cattle have one functional gene derived from the AnKIR3DL gene and a family of functional genes derived from AnKIR3DX (Fig. 7). Although functions for cattle KIR have yet to be determined, the many functional sequence motifs that they share with human KIR make it likely that cattle KIR function as NK cell receptors for variable determinants of MHC class I molecules. If this assumption is correct, then the primordial KIR is predicted to have been an MHC class I receptor and its AnKIR3DL and AnKIR3DX progeny, MHC class I receptors with different specificities for MHC class I. With each additional duplication of the KIR locus, there would have been potential to increase the repertoire of MHC class I specificities recognized by the KIR. Both pig and cattle have a single functional Ly49 gene (68). The cattle Ly49 gene encodes three divergent allotypes, differing by as many as 16 amino-acid substitutions in the lectin-like domain (88).
Figure 6. Comparison of the genomic organization of the cattle and human KIR loci.
Shown are the KIR genes and flanking LRC genes, comprising LILR and LAIR genes on the telomeric side, FCAR and NCR1 genes on the centromeric side. Each gene is represented by a schematic of the encoded protein in which each symbol represents an Ig-like domain and the type protein in which it is present: circle for KIR3DL, square for KIR3DX, triangle for LAIR, rhombus for FCAR, pentamer for NCR1, and hexagon for LILR. The encoded Ig-like domains of functional genes are colored green for KIR and yellow for other LRC genes. The encoded Ig-like domains of non-functional genes are shaded grey for KIR and white for LILR. This figure is adapted from Fig. 2 in Sanderson et al. (87).
Figure 7. Phylogenetic tree of primate and artiodactyl KIR.
The tree (Neighbor-joining; Tamura-Nei; pairwise deletion; 1000 bootstrap replicates) was constructed from the sequences of exons 1, 2, 3, and 4 from a representative for each of the human (primate) and cattle (artiodactyl) KIR. The tree has two main branches, one comprising the KIR3DL lineage that derives from the AnKIR3DL ancestor and the other comprising the KIR3DX lineage that derives from the AnKIR3DX ancestor. The AnKIR3DL and AnKIR3DX genes arose by duplication of the primordial KIR gene, PrKIR. Human KIR genes have red labels, cattle KIR genes have blue labels. Grey shaded boxes denote non-functional KIR genes. All functional human KIRs are products of KIR3DL lineage genes. Only one functional cattle KIR is the product of a KIR3DL lineage gene, whereas nine are products of KIR3DX lineage genes. Thus human KIR variability is the property of the KIR3DL lineage, whereas cattle KIR variability is the property of the KIR3DX lineage. This figure is adapted from Fig. 4 in Sanderson et al. (87).
The two sequenced cattle KIR haplotypes came from a Holstein-Friesian cow. The two haplotypes have identical KIR gene content and the data from the Aurochs genome project is consistent with the Aurochs KIR haplotype having the same gene-content (87). The comparison of pig and cattle shows that within the same order of placental mammals there can be major differences in the size and functionality of KIR gene families.
Marine carnivores combine conserved KIR and Ly49 with diverse species-specific MHC class I
KIR and Ly49 genes have been examined in four species of marine carnivore: harbor seal, grey seal, Weddell seal and the California sea lion (45). These species all have one KIR gene of AnKIR3DL descent that is flanked by the LILR and FCAR genes. The KIR gene is functional, and encodes an inhibitory, cell-surface expressed KIR3DL in the seals and a KIR2DL with an unexpressed D0 domain in the sea lion. Despite this difference, the gene exhibits 95.8-99.8% sequence similarity even though seals and sea lion are separated by ~33 million years of evolution. Thus in these species, unlike simian primates, the KIR locus has evolved very slowly. Slow evolution also characterizes the single Ly49 gene in these species; it encodes a functional inhibitory receptor and exhibits 94.2-99.9% sequence similarity between species (45).
Contrasting with the KIR and Ly49 genes, there has been extensive evolution of the MHC class I genes in the marine carnivores (89). Of three types of MHC class I gene defined phylogenetically, only one is shared with canines, the terrestrial carnivores to which the seals and sea lions are most closely related. The second type is a pseudogene, and the third type has been elaborated into a diverse and polymorphic multigene family of seal-specific MHC class I genes that have the characteristics of classical MHC class I genes. In the grey seal and the harbor seal, species that separated within the last seven million years, there are 12 and six of these genes, respectively. Some of them are shared by the two species but others are not. Seal MHC class I haplotypes combine gene polymorphism with gene-content diversity.
Contrasting with the marine carnivores are the terrestrial carnivores, as represented by domestic cat and dog. The KIR locus and part of the FCAR gene have been deleted from the dog genome. A critical cysteine residue is missing from the lectin-like domain of protein encoded by the dog Ly49 gene, indicating it is non-functional. The cat has one Ly49 gene that appears functional. The cat’s only KIR gene is flanked by the LILR and FCAR genes, but it cannot function because of a two base pair deletion that causes premature termination in exon 1 (45).
Prosimian and simian primates use different systems of variable MHC class I receptors
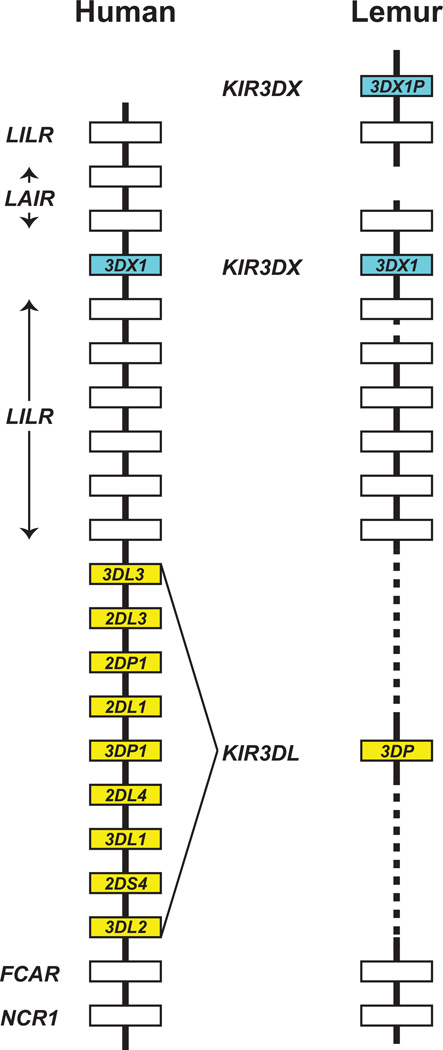
The grey mouse lemur, a prosimian primate, has an LRC with a KIR locus (71). It contains three KIR genes (Fig. 8). Descended from AnKIR3DX is the functional KIR3DX1 gene and the non-functional KIR3DX1P, a pseudogene. Descended from AnKIR3DL is the nonfunctional KIR3DP, also a pseudogene. In short, the lemur has just one functional KIR gene. This situation in a prosimian is unlike that in simian primates, where there is a diversity of functional KIR genes derived from AnKIR3DL. Because the diverse KIR genes of the simian primates are all descended from AnKIR3DL, it is likely and most parsimonious that the common ancestor of all primates had one functional KIR3DX gene and one functional KIR3DL gene, and that the latter was selectively inactivated in the prosimian line (Fig. 8).
Figure 8. Comparison of simian and prosimian KIR loci.
Compared here is the gene content and organization of the human KIR locus (48) and the grey mouse lemur KIR locus (90). Boxes representing genes of the KIR3DL lineage are colored yellow, boxes representing genes of the KIR3DX lineage are colored blue.
In humans, HLA-E and its CD94:NKG2A receptor are highly conserved in structure and function and encoded by single genes. Sequencing CD94 cDNA from six humans and four chimpanzees identified no polymorphism in either species, with the human and chimpanzee sequences differing by only one coding difference and one silent substitution (9). In the grey mouse lemur the condition of CD94 is very different. CD94 is produced from three divergent genes, giving CD94 proteins that differ by 23-24% in amino-acid sequence (71). These CD94 polypeptides randomly associate with five NKG2 polypeptides to give a diversity of 15 different receptors. The diversity is further increased by CD94 gene polymorphism. The qualitative differences between the MHC class I receptors of simians and prosimians is accompanied by equally dramatic differences in their MHC class I molecules, none of which are held in common. The MHC of the grey mouse lemur contains active MHC class II genes, but the four MHC class I genes are inactivated (90, 91). There are six active MHC class I genes, but these were moved out of the MHC to a site on another chromosome. These distinctive properties of the CD94 and MHC class I genes are shared with other prosimian species (71).
As functional studies have yet to be done, it is still in the realm of hypothesis that highly varied CD94:NKG2 receptors are variable NK cell receptors for prosimian MHC class I. But, the circumstantial evidence is impressive. If true then the prosimian and simian groups of primate species are using completely different sets of MHC class I ligands and cognate NK cell receptors.
KIR genes of catarrhine and platyrrhine primates are more divergent than their MHC class I genes
The simian primates divide into two groups: the catarrhines whose nostrils face downward and the platyrrhines whose nostrils face sideways (92). Catarrhine primates comprise the Old World monkeys, lesser apes, greater apes, and humans; platyrrhine primates comprise the New World monkeys. Phylogenetically the catarrhine KIR genes group into four lineages: I, II, III, and V, that are all represented in each species (41–44, 72, 76-78, 81-83, 85, 93). Lineage IV was the name originally given to macaque KIR3D genes, because they formed a distinct phylogenetic group when first compared to human and chimpanzee KIR (83). With the subsequent sequencing of orangutan KIR, and the inclusion of their sequences in phylogenetic analyses, it was realized that the macaque KIR3D belonged to lineage II and so the lineage IV designation was abandoned (47). None of the catarrhine KIR lineages is represented in the platyrrhine primates, who have their own unique lineage VI KIR (73, 75). The sequence and organization of KIR haplotypes from owl monkey and spider monkey, species representing two of the five families of New World monkeys, have been examined (74). The spider monkey haplotype has five functional KIR genes with a pseudogene at the centromeric end of the locus and FCAR flanking the telomeric end. This points to the platyrrhine KIR locus being located in the LRC, at a position syntenic to the catarrhine KIR locus. Distinguishing the spider monkey KIR haplotype from its catarrhine counterparts is an inversion of the KIR genes in the centromeric region of the locus, but not the KIR genes in the telomeric region. This inversion is also a feature of the owl monkey KIR haplotype, which has an additional KIR gene in both the centromeric and telomeric regions. Thus the inversion likely occurred in a common ancestor of the two platyrrhine families ~20 MYA. A novel feature of the owl monkey is a type of KIR having four Ig-like domains. These KIR4DL have two D0 domains. Extensive KIR gene diversity and polymorphism characterize both the owl monkey and spider monkey KIR locus (74, 75).
The common marmoset (Callithrix jacchus: Caja) is the species of New World monkey in which the MHC class I genes have been studied the most (94–96), but unfortunately knowledge of marmoset KIR is currently restricted to the sequence of KIR3DX1 (97), previously named KIR3DL0 (98), and the only KIR found by analysis of the marmoset genome database (Fig. 5). In the marmoset, there is a single MHC-E gene, and it is a strict ortholog of human HLA-E (99). In addition, two large segments of the marmoset MHC class I region have been sequenced. The G/F segment corresponds to the part of the HLA complex containing the HLA-A, HLA-F, and HLA-G genes and has 14 genes related to HLA-G and five related to HLA-F, but none corresponding to HLA-A (94). The B/C segment corresponds to the part of the HLA complex containing the HLA-B and -C genes and contains 9 Caja-B genes, five of which appear functional (95). Of the G/F and B/C MHC class I genes, evidence for transcription has been obtained only for Caja-G genes. These genes exhibit the broad tissue distribution that is characteristic of classical MHC class I genes, not the restricted expression of human HLA-G (96). Consistent with the Caja-G gene family being classical MHC class I genes, individual marmosets express 4-7 different Caja-G variants. The diverse Caja-G genes in the common marmoset appear analogous to the diversity lineage of marine carnivore MHC class I genes (89). Unlike their highly divergent KIR, lineage relationships are clearly evident between the MHC class I genes of catarrhines and platyrrhines. However, as exemplified by MHC-G, the same lineage of MHC class I genes can have distinctive functions in catarrhines and platyrrhines.
Emergence of the Bw4 and Bw6 epitopes drove diversification of lineage II KIR genes
The catarrhine primates all have the same four phylogenetic KIR lineages (I, II, III, and V). In humans, the lineage V gene (KIR3DL3) is at the centromeric end of the locus, a lineage II gene (KIR3DL2) is at the telomeric end and the center of the framework comprises a lineage I gene (KIR2DL4) and a lineage III pseudogene (KIR3DP1). In the variable centromeric and telomeric intervals between the framework genes are variable numbers of lineage I, II, and III genes. This organization is present in the great apes (46, 47, 82, 83, 85), but is less conserved in the lesser apes (84) and Old World monkeys (41–44, 72, 85).
There are two subfamilies of Old World monkeys, the Cercopithecinae and the Colobinae. KIRs have been investigated in four Cercopithecin species [rhesus macaque (41–44, 78), crab-eating macaque (72), pig-tailed macaque (80), and vervet monkey (81)] and two Colobin species [olive baboon and colobus monkey (81)]. As the types of KIR and their variation appear similar, we shall focus on rhesus macaque (Macaca mulatta, the species most studied. At an early stage in this investigation, a rhesus KIR haplotype was sequenced and seen to have only six genes, four framework genes like the human ones, one additional lineage II gene and one additional lineage III gene (85). This initial snapshot of simplicity was not borne out by the subsequent characterization of 22 rhesus KIR genes that are differentially distributed to give KIR haplotypes having from four to fifteen KIR genes (41–44). Some of these haplotypes are lacking one of the framework genes: KIR3DL20, KIR2DL04 and KIR3DL01.
The impressive diversity of rhesus KIR haplotypes is entirely due to the 19 KIR genes of lineage II. By contrast KIR lineages I and V are represented only by one gene each, and KIR lineage III is represented by a pseudogene and KIR1D, neither of which is likely functional. In humans, the lineage II KIR recognize epitopes of HLA-A and HLA-B (KIR3DL1/S1 is specific for the Bw4 epitope of HLA-A and –B and KIR3DL2 is specific for the A3/A11 epitope of HLA-A). Correlating with the expansion of the lineage II KIR genes in Old World monkeys is a corresponding expansion of the MHC-A and MHC-B genes. A sequenced rhesus macaque MHC haplotype has four MHC-A and 19 MHC-B genes (100, 101). Of these many genes only Mamu-A1 is present on every MHC haplotype (102). There is no equivalent to HLA-C in the MHC of Old World monkeys, including rhesus macaque. In addition to haplotypic gene-content diversity the rhesus MHC class I genes exhibit allelic polymorphism and considerable variation in their levels of transcription and cell-surface expression. Thus the number of functional MHC-A and -B class I molecules expressed by each individual may be comparable to the four expressed by most humans (103).
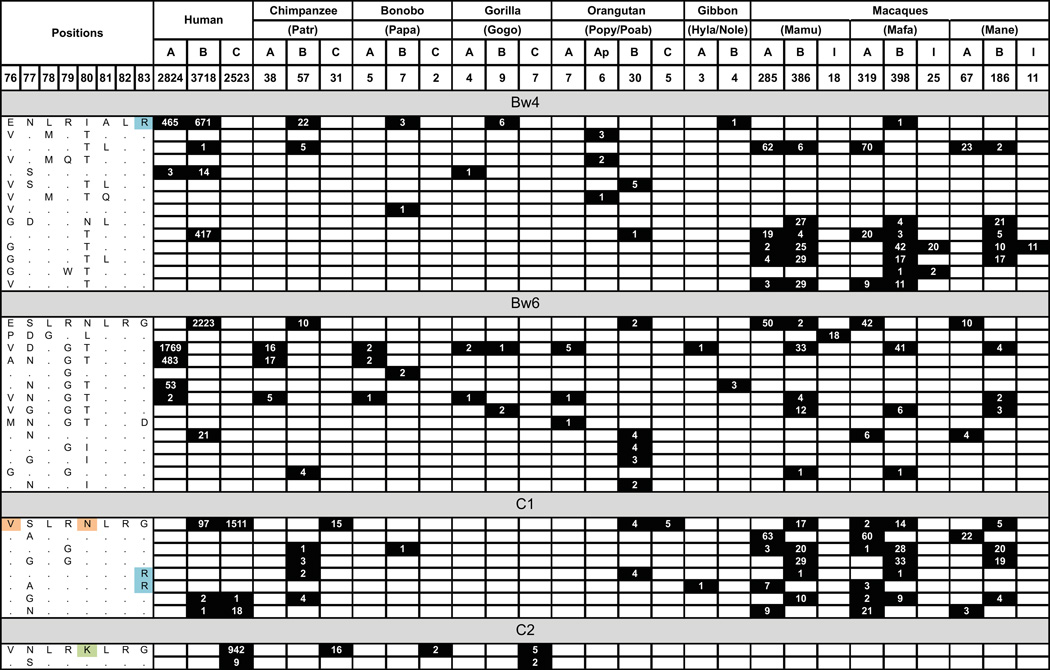
The Bw4 and Bw6 sequence motifs are present in some rhesus MHC-A and MHC-B allotypes (Fig. 9). Only Bw4 is a ligand for human KIR, but both the Bw4 and Bw6 epitopes function as ligands for Old World monkey KIR. Rhesus KIR3DL01 recognizes various Mamu-B allotypes having the Bw4 motif and mutation within the motif abrogates recognition. Other Mamu-B allotypes having the Bw4 motif, however, are not recognized by KIR3DL01 because of substitutions at sites away from the Bw4 motif (104). These characteristics parallel precisely those seen for human KIR3DL1 recognition of Bw4-bearing HLA-B (105). KIR3DL01 is considered the framework gene at the telomeric end of the macaque KIR locus. It is, however, not a component of all KIR haplotypes and is not detectable in ~10% of macaques. Having a complementary specificity to KIR3DL01 is KIR3DLW03, which recognizes Bw4-bearing Mamu-A allotypes. Again the binding is abrogated by mutation that replaces residues of the Bw4 with their counterparts from the Bw6 motif. Rhesus KIR3DLW03 does not recognize Bw4-bearing Mamu-B allotypes nor Mamu-A and -B allotypes lacking a Bw4 motif (106). Thus the Bw4 epitopes on Mamu-A are -B are more diversified and differentiated than those carried by HLA-A and HLA-B.
Figure 9. Distribution of motifs encoding KIR ligands in the sequences of the MHC of catarrhine primate MHC class I molecules.
For each species the number of allotypes analyzed for each MHC class I gene is tabulated at the top of the figure. Ap under orangutan is the orangutan pseudogene orthologous to HLA-A; I under the macaques is a macaque specific MHC-B-related gene. The different sequence motifs at positions 76-83 are given at the left and organized according to whether they have a Bw4, Bw6, C1, or C2 motif. For our purpose, any motif that is not Bw4, C1, or C2 is defined as Bw6. On the right, boxes with black shading denote the genes that encode each sequence motif, and the number of allotypes having the motif are given in white within each black box. Only motifs that were observed in at least 5% of allotypes in at least one species are shown.
Rhesus KIR3DL05 recognizes the common Mamu-A1 allotype, which has the Bw6 motif (107). The strength and specificity of this interaction is modulated by the peptide bound to Mamu-A1 and by the substitutions that distinguish KIR3DL05. This behavior also parallels that of human KIR3DL1 (108). The important difference is that no human KIR recognize HLA class I allotypes with a Bw6 motif, whereas that is not the case for rhesus macaque KIR. It is also the case for a pigtailed macaque KIR that recognizes a broad range of MHC-A and MHC-B allotypes, including ones that have a Bw4 motif, ones that have a Bw6 motif and ones that have neither motif (80).
Testing rhesus KIR3DLW03 against a panel of 95 HLA class I allotypes gave a broad range of reactions with Bw4-bearing HLA-A and HLA-B allotypes, some reactions with Bw6-bearing HLA-B, lesser reactions with HLA-A*11 and weak reactions with other HLA-A (109). Unexpectedly, the strongest reactions were with HLA-C, an MHC class I molecule that is not represented in Old World monkeys. Furthermore, all HLA-C allotypes reacted with the rhesus KIR irrespective of whether they carry the C1 or the C2 epitope. This indicates how the emergence, evolution and adaptation of HLA-C in the hominids involved the selection for changes to the class I molecule that increased its avidity for the existing KIR. In other words the KIR drove the evolution of HLA-C, not vice versa.
At positions 77-83 rhesus Mamu-A and -B molecule have a greater variety of sequence motifs than HLA-A and -B, which includes mixed motifs that combine elements of the Bw4 and Bw6 motifs (Fig. 9). The diversity of the Mamu-A reactive rhesus KIR and the complexity of the binding reactions suggests that a spectrum of KIR ligands are formed by the sequence motifs in residues 77-83 and distinguished by different KIR. Adding to this impression are the properties of the pig-tailed macaque KIR that react with MHC-B having a variety of Bw4, Bw6 and mixed motifs (80). In human this system is replaced by one lineage II KIR, KIR3DL1, and a clear division of the HLA-A and HLA-B allotypes into ones that have a Bw4 motif and are recognized by KIR3DL1, and others that lack a Bw4 motif and are not recognized by KIR3DL1.
The C1 epitope first evolved at an MHC-B gene
Although C1 is usually considered in the context of MHC-C, this epitope is also carried by some MHC-B allotypes (39). The main human C1-bearing HLA-B allotypes are HLA-B*46 and HLA-B*73, both of which have unusual characteristics. HLA-B*46 is the product of a gene conversion between HLA-B and HLA-C that replaced residues 66-76 of the α1 domain helix of an HLA-B*15:01 allotype with the corresponding residues of an HLA-C*01 allotype (38). HLA-B*46 is localized to parts of Southeastern Asia, where it reaches allele frequencies as high as 24% (http://www.allelfrequencies.net). HLA-B*73 differs from other HLA-B allotypes by 54-80 amino-acid substitutions, which are spread throughout the HLA-B sequence of 360 residues, and it thus represents a distinctive second lineage of MHC-B allotypes. HLA-B*73:01 is localized to Western Asia and reaches its highest frequency of 4.9% in Parsis, who now live in Indian and Pakistani cities, but are descended from Zoroastrians who emigrated from Persia (modern day Iran) to the Indian subcontinent some 1100 years ago (110). The extraordinary structure, genetics and genomics of HLA-B*73 all point to this allele having entered the modern human population by introgression from archaic humans (111). In other words, after migrating out of Africa ~45-60 thousand years ago (112) modern humans acquired HLA-B*73 through meeting and mating with the Archaic humans then resident in Western Asia.
Distinguishing HLA-B*46 and HLA-B*73 from other HLA-B allotypes is valine at position 76. Valine 76 in combination with asparagine 80, the most common residue at position 80 of HLA-B, is sufficient to form the C1 epitope recognized by KIR2DL2/3. Residue 80 is within the 77-83 region containing the Bw4/Bw6 motifs and position 76 is adjacent to it. Thus C1 can be seen as a further structural variation on the same theme as Bw4 and Bw6, as is consistent with the mutual exclusivity of the epitopes that function as KIR ligands and the crystallographic studies showing how they interact with KIR (113–116). In addition to HLA-B*46 and HLA-B*73, rare variants of HLA-B*07, HLA-B*08, HLA-B*15, HLA-B*18, HLA-B*35, HLA-B*39, and HLA-B*40 have the valine 76, asparagine 80 motif and are predicted to be ligands for KIR2DL2/3 (46).
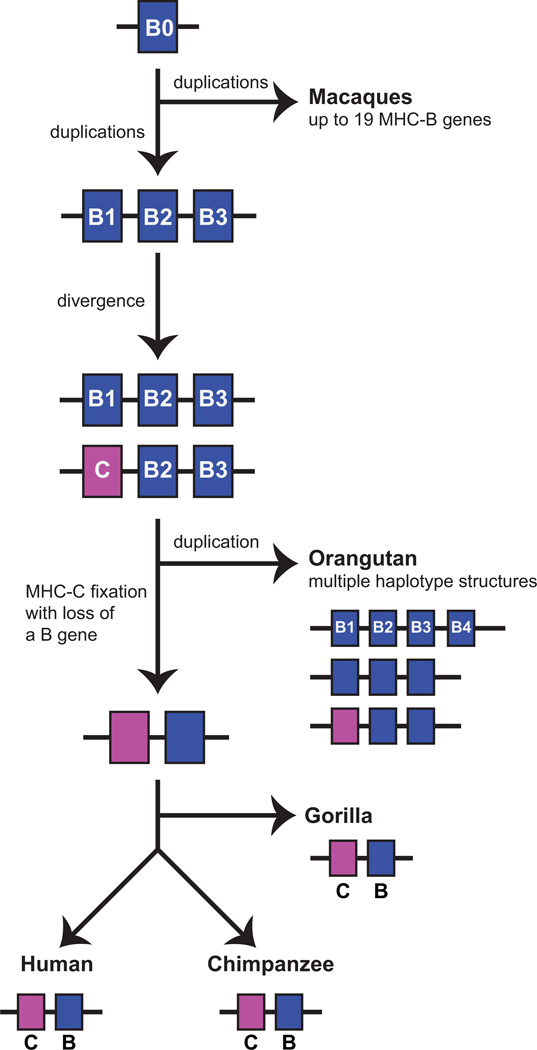
Whereas the human MHC-B allotypes carrying the C1 epitope can be viewed as curiosities, the chimpanzee MHC-B allotypes that have valine 76, asparagine 80 and the C1 epitope are structurally well within the norm and account for around one quarter of the total number of Patr-B allotypes. In orangutan, the species most distant from humans that has MHC-C, a similar fraction of MHC-B allotypes has valine 76 and asparagine 80 and is predicted to carry the C1 epitope. Orangutan MHC haplotypes have 2-4 MHC-B genes and around half of them has an MHC-C gene. Furthermore, the MHC-C gene is syntenic with one of the MHC-B genes on haplotypes lacking MHC-C. These properties are consistent with MHC-C having evolved from an MHC-B gene. That orangutan MHC-C allotypes all carry the C1 epitope, and none have the C2 epitope, argues for MHC-C having evolved from an MHC-B allele that encoded a C1-bearing MHC-B allotype (Fig. 10). Consistent with this hypothesis, phylogenetic analysis, combined with reconstruction of ancestral residues shows that the last common ancestor of MHC-B and MHC-C had valine at position 76 (46). During subsequent evolution, valine 76 was fixed at MHC-C, whereas at MHC-B it has been replaced by glutamate, but to a varying extent in different species. The extent of the replacement has been greatest for human MHC-B.
Figure 10. Proposed model for the evolution of MHC-B and MHC-C.
The primordial MHC-B0 gene was present in the common ancestor of the catarrhine primates. In the macaque lineage of Old World monkeys, MHC-B0 underwent successive duplications to produce at least 19 MHC-B genes. In the hominid lineage, two duplications led to the formation of a smaller number of MHC-B genes: MHC-B1, MHC-B2, and MHC-B3. Under differential selection, an allele of MHC-B1 diverged to become a distinctive MHC-C that was present on some MHC haplotypes but not all. This was the situation in the common ancestor of apes and humans and it is retained in modern orangutans, where MHC haplotypes can have 2-4 MHC-B and 0 or 1 MHC-C genes. In the common ancestor of human and African apes MHC-C was fixed and the number of MHC-B genes was reduced to one, an organization retained in the modern gorilla, chimpanzee, and human species.
The C1 epitope motif, V76 N80, is present among the MHC-A and MHC-B allotypes of Old World monkeys (Fig. 9). So MHC-B molecules with the C1 epitope could have been present in a common catarrhine ancestor where they served as a ligand for lineage II KIR. Under natural selection, and over time, this interaction could have become differentiated from the interactions between other lineage II KIR and the various Bw4 and Bw6 variants. As part of these changes, the gene encoding the C1-bearing MHC-B acquired substitutions that distinguished it from other MHC-B, eventually leading to become what we know now as MHC-C. This evolution of MHC-C from an MHC-B gene is estimated to have occurred ~22.3 million years ago (117, 118), subsequent to the divergence of Old World monkeys and apes.
HLA-C1-mediated expansion of lineage III KIR in hominids
None of the Old World monkey MHC haplotypes has an MHC-C gene. In contrast, all human and chimpanzee MHC haplotypes have MHC-C. This evolution of a fixed MHC-C gene from a single MHC-B progenitor, must have involved numerous intermediate populations of simian primates in which the frequency of MHC haplotypes carrying MHC-C varied from 0-100%. Almost all these intermediate forms died out, leaving the orangutan as the only living species in which MHC-C is present but not fixed (117). During the evolution of the apes from their common ancestor with the Old World monkeys it is likely that the number of class I genes in the MHC was considerably reduced. Complete sequences for orangutan MHC haplotypes have yet to be defined, but the available evidence is consistent with them having a fixed MHC-A gene, a variable number of two to four MHC-B genes and either one or no MHC-C gene. The frequency of orangutan MHC haplotypes containing MHC-C is ~50%, compared to 100% for chimpanzee and human MHC haplotypes. Orangutan MHC-C thus resembles an evolutionary intermediate in the evolution of the MHC-C present in humans and chimpanzees (76).
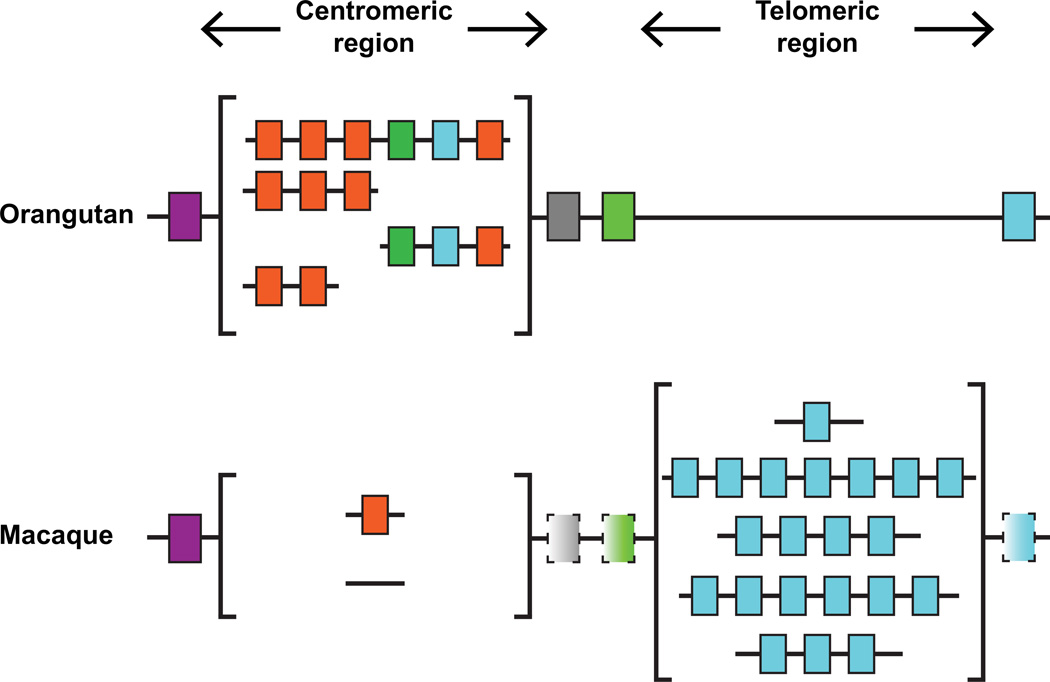
Accompanying the presence of the MHC-C gene in the orangutan MHC are major changes in the KIR locus. The dominance and diversity of lineage II KIR genes that characterizes the Old World monkeys is absent, there being only two lineage II KIR genes in the orangutan KIR locus (Fig. 11). These polymorphic genes encode inhibitory KIR3DL1 and activating KIR3DS1 that are candidate receptors for the Bw4-like and Bw6-like epitopes of orangutan MHC-A and MHC-B. The key feature of the orangutan KIR locus is an increase in the number of lineage III KIR genes to four. These genes encode four types of receptor that recognize MHC-C. Inhibitory KIR2DL10 and activating KIR2DS10, which segregate as alleles, are strong C1-specific receptors that have lysine at position 44 like the human C1-specific receptor KIR2DL2/3 (119). In contrast, inhibitory KIR2DL11 and KIR2DL12 and activating KIR2DS13 have glutamate at position 44, a residue not present in human KIR, and they have a broad C1+C2 specificity for MHC-C that does not discriminate between the C1 and C2 epitopes (119). In the orangutan, which only has C1-bearing MHC-C, these receptors will only engage with the C1 epitope and with an avidity that is less than the C1-specific receptors of other species. Because of their avidity differences the two pairs of orangutan MHC-C reactive KIR receptors are likely to have complementary functions in NK cell education and response. Within each pair of receptors, the activating and inhibitory counterparts have identical avidity associated with complementary signaling functions. Similar to the segregation of KIR2DL2 and KIR2DL3 in humans, orangutan KIR2DS14 behaves as an allele of KIR2DS13. In sequence comparisons, the D1 domain of KIR2DS14 is more similar to the other ape lineage III KIR whereas the other orangutan lineage III KIR have diverged in a species-specific way. Despite the similarity to the other ape KIR and the presence of a lysine at position 44 which should be permissive of binding to MHC, no binding was observed (119). Analysis of sequences provided by Prado-Martinez et al (120) has revealed an additional lineage III gene, 2DS15, which appears to be specific to Bornean orangutans (Pongo pygmaeus). It has lysine at position 44 and based on its similarity with the other orangutan KIR would be predicted to bind to C1 (LAG, unpublished data).
Figure 11. Comparison of the orangutan and macaque KIR loci.
In macaques, which have a diversity of MHC-A and MHC-B genes but no MHC-C gene, there is a diversity of 2-8 lineage II KIR genes (blue colored boxes) that encode receptors recognizing epitopes of MHC-A and MHC-B. In contrast, some macaque KIR haplotypes have only one lineage III KIR gene (orange colored boxes), and others have none. In orangutans, which have several MHC-B genes and an MHC-C gene present on ~50% of haplotypes, the KIR locus is very different from that in macaques. It has only two lineage II KIR genes and multiple lineage III KIR genes that encode MHC-C receptors. The conserved framework of the hominid KIR locus consists of lineage V KIR3DL3 (purple box), a lineage III KIR pseudogene (grey box), lineage I KIR2DL4 (light green box), and a lineage II KIR gene (blue box). These framework genes define centromeric and telomeric regions that are sites for variable KIR gene content. The centromeric lineage V KIR gene is the most conserved feature of macaque KIR haplotypes. The other framework genes, central KIR2DL4 and the pseudogene, and telomeric lineage II KIR3DL, are present only on some macaque KIR haplotypes is indicated by their boxes having a dashed outline and distinctive shading. All macaque KIR haplotypes have a lineage II KIR3DL at the telomeric end and in the majority of haplotypes it is KIR3DL01, as found in the one haplotype sequenced. That ~10% of rhesus macaques lack KIR3DL01 indicates that other KIR3DL genes also occupy this framework position.
The lineage II KIR3D that recognize epitopes of MHC-A and -B have three extracellular Ig-like domains, D0, D1, and D2, that all make contact with the MHC class I molecule and are encoded by separate exons: exons 3, 4, and 5 (116). Lineage III KIR genes have a similar exon-intron organization and could well have evolved from a lineage II KIR gene (76). However, like human lineage III KIR, the orangutan KIR2D receptors for MHC-C have only two extracellular domains and they correspond to the D1 and D2 domains of the lineage II KIR. All lineage III KIR genes encoding KIR2D have an exon 3 encoding a D0 domain but it is not expressed and is therefore called pseudoexon 3 (121). A variety of mechanisms, which differ among the lineage III KIR genes, accounts for the lack of expression of the pseudoexons 3. In the evolution of MHC-C specific lineage III KIR from MHC-A and -B specific lineage II KIR there was clearly some advantage in dispensing with the D0 domain. However, not all lineage III KIR genes have a pseudoexon 3 and in chimpanzee and gorilla there are KIR genes encoding KIR3DL MHC-C receptors that have D0, D1 and D2 domains (82, 83, 93). While the chimpanzee lineage III KIR that encode an expressed D0 domain never had a pseudoexon 3, gorilla KIR3DL7 appears to have had a pseudoexon 3 which subsequently reverted to become an expressed form (83).
As befits its status as an evolutionary intermediate there is flexibility and potential for further evolution of the orangutan KIR as is exemplified by C1-specific KIR2DL10. On testing this orangutan KIR for binding to a panel of almost one hundred human MHC-A, MHC-B, and MHC-C allotypes, its principal specificity was for the C1 epitope, but there were also weak but significant crossreactions with allotypes having the C2, Bw4, Bw6 and HLA-A3/11 epitopes (119). Thus elements of all the KIR specificities of the catarrhine primates are present in this one orangutan KIR. This provides the basis upon which successive natural mutations were able to create KIR specific for each of the epitopes. In the laboratory, extensive mutational analysis indicates that the interaction between KIR and MHC-A, MHC-B, and MHC-C is inherently limited to the known epitopes and specificities.
HLA-C2 mediated expansion of lineage III KIR in hominids
The evolution of the C2 epitope and C2-specific KIR in a common ancestor of humans and the African apes was probably facilitated by the presence of C1+C2 receptors like those still present in the orangutan. Evolving the C2 epitope from C1 would have been a simple matter, requiring only a point substitution in codon 80 that changed the amino acid from asparagine to lysine. That step of creating C2 was absolutely necessary for a system of C2 ligands and C2 receptors to evolve, but it was not sufficient. In addition, the C2 epitope once formed had to be functional and thus subject to a natural selection that would maintain the C2 epitope in the population and increase its frequency there. If the only existing receptors were C1-specific receptors, like orangutan KIR2DL10 and human KIR2DL3, then nascent C2 would not be functional, would not be selected and would not survive. On the other hand, in the presence of existing receptors with C1+C2 specificity and using C1-expressing MHC-B and MHC-C as their ligands, a newly formed C2-bearing MHC-C allotype could from its very inception have been well able to function as a KIR ligand, an educator of NK cells and a regulator of NK cell responses (119).
Once C1+C2 receptors had facilitated the selection and propagation of C2, the stage was set for the emergence of highly specific C2 receptors with methionine 44, like human KIR2DL1. This adaptation of the KIR to the C2 epitope likely involved mutation of lysine to methionine at position 44 in a C1-specific receptor, rather than glutamate to methionine at position 44 in a C1+C2 receptor. The former would have required one nucleotide substitution, whereas the latter would need two. From its inception the C2-specific receptor would encounter C2-bearing MHC-C and be subject to natural selection. The advantage of the C2-specific KIR was such that these receptors complete replaced the C1+C2 receptors with glutamate 44, which are not to be found in humans or African apes (119).
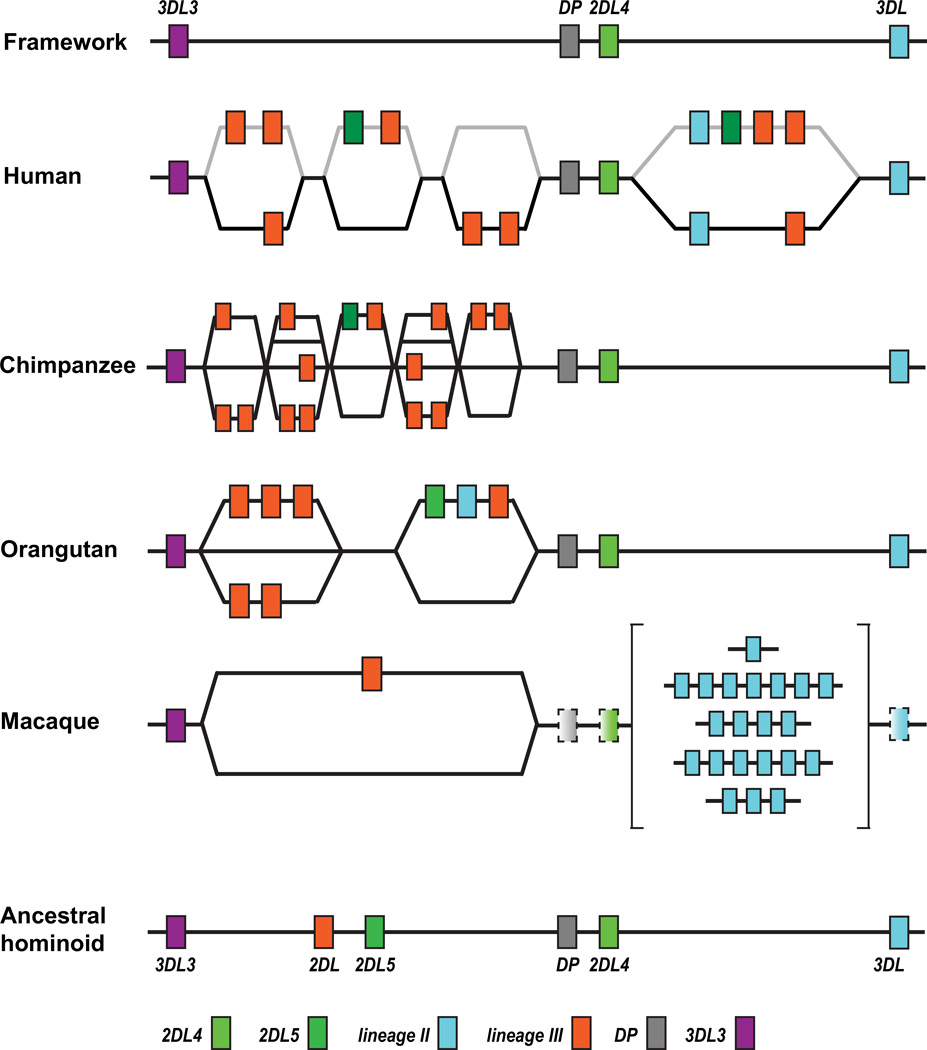
Whereas four lineage III KIR genes are associated with the presence of the C1 epitope in the orangutan, nine chimpanzee lineage III KIR genes are associated with the fixation of MHC-C and the presence of both the C1 and C2 epitopes Fig. 12. Thus the repertoire of MHC-C receptors was doubled, the number of lineage II KIR genes encoding MHC-A and -B receptors was halved from two to one. Eight of the chimpanzee lineage III KIR genes encode high avidity MHC-C receptors, five specific for C2 and three specific for C1 (122). Unlike the orangutan lineage III KIR, four of the eight chimpanzee MHC-C receptors are KIR3DL that have D0, D1 and D2 domains and four are KIR2D that lack the D0 domain, because they are encoded by pseudoexons 3. Removing the D0 domain from the KIR3D had no detectable effect on their function, as tested in vitro (122). The chimpanzee C1- and C2-specific receptors form separate branches in phylogenetic trees, consistent with an initial evolution of the C1 receptors, followed millions of years later by the C2 receptors. Four of the five C2-specific receptors have methionine at position 44 but Pt-KIR2DL9 has glutamate, showing that C2 receptors have evolved form C1 receptors on more than one occasion.
Figure 12. Comparison of the KIR loci in four species of catarrhine primate.
The top line shows the 3DL3 (purple box), DP (grey box), 2DL4 (light green box), and 3DL (blue box) framework genes that are common to human and ape KIR haplotypes. The bottom line gives the predicted structure for the common ancestor of all hominoid (humans and great apes) KIR haplotypes. As well as the framework genes this ancestral haplotype contains lineage I KIR2DL5 (dark green box) and a functional lineage III KIR gene (orange box). For the human, chimpanzee, orangutan and macaque KIR loci, alternative gene-content segments are represented on parallel lines and occur in different combinations to diversify the gene content of KIR haplotypes. For the human KIR haplotypes the boxes for framework genes and genes of the KIR A haplotypes are connected by black lines, whereas the boxes for genes of the KIR B haplotypes are connected by gray lines.
The chimpanzee KIR locus has similar organization to the orangutan KIR locus, in which the telomeric region is conserved and comprises only the framework lineage I (KIR2DL4) and lineage II (KIR3DL1/2) genes (Fig. 12). Pt-KIR3DL1/2, the only chimpanzee lineage II KIR, resembles a common ancestor of KIR3DL1 and KIR3DL2, the two human lineage II KIR. Pt-KIR3DL1/2 recognizes a variety of human and chimpanzee MHC-A and MHC-B allotypes (93), but in a pattern that does not correlate with Bw4 or other known sequence motifs and appears more like that of an Old World monkey KIR than a human KIR. Unequal crossing over within the centromeric region of the chimpanzee KIR locus, the location of the genes for the MHC-C receptors, has produced haplotypes with diverse gene content. The same mechanism also created recombinant receptors in which the same ligand-binding domains are associated with transmembrane and cytoplasmic regions having different signaling capacity (46).
The bonobo (previously called pygmy chimpanzee) is closely related to chimpanzee, having diverged ~2 million years ago (120, 123). Since that time, the system of KIR and MHC class I ligands appears to have diverged significantly. The caveat being that the bonobo has yet to be studied to the depth and resolution that has been achieved for the chimpanzee. The most striking feature is a KIR haplotype that has only the framework genes: lineage V KIR3DL3, lineage I KIR2DL4 and lineage II KIR3DL. In the small population of captive bonobos we studied this haplotype was common (82). Three individuals were homozygotes, including a father of five. These individuals are predicted to lack KIR that recognize MHC-C, but to have an inhibitory KIR3DL receptor for MHC-A and -B. The longest bonobo KIR haplotype contains four additional genes, lineage I KIR2DL5, an additional lineage II KIR3DS, and two lineage III KIR that are orthologous to chimpanzee Pt-KIR3DL4 and Pt-KIR3DL5 and encode inhibitory C2 receptors. There is no candidate for a KIR gene predicted to encode a bonobo C1-specific KIR. Corresponding to the absence of C1-specific KIR, the bonobos studied only had MHC-C allotypes carrying the C2 epitope. Thus our working hypothesis is that the entire C1-mediated arm of NK cell regulation is absent from bonobos and has been replaced by increased use of C2 and MHC-A and -B epitopes that are KIR ligands. Whether this situation is typical of wild bonobo populations is a question needing investigation.
Limited investigation of gorilla KIRs found evidence for one lineage II KIR gene, seven lineage III KIR genes encoding one activating and six inhibitory receptors, KIR2DL4 and KIR2DL5 (83). Similar to the orangutan, six of the seven gorilla lineage III KIR lack the D0 domain, but one does have a D0 domain (83). At position 44, two of the lineage III KIR2D have lysine and two have glutamate, whereas the lineage III KIR3DL has methionine, suggesting a variety of C1 and C2 receptors. It is therefore intriguing that all seven sequenced gorilla MHC-C alleles have the C2 motif. This raises the possibility that gorilla, like bonobo, is reducing its use of the C1 epitope of MHC-C (Fig. 13).
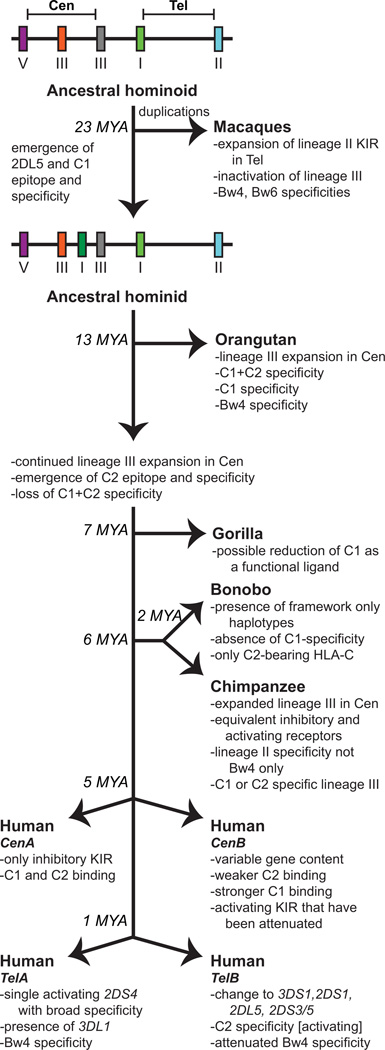
Figure 13. Summary of the co-evolution of MHC class I and KIR in the catarrhine primates.
This schematic diagram shows the distinguishing features of each of the KIR regions in the species shown. Approximate speciation times are indicated at the branch points (MYA, million years ago). Prior to the divergence of Old World monkeys (macaques) and apes, the KIR region minimally contained the framework genes and a lineage III gene as is shown at the top of the figure in the ancestral catarrhine KIR haplotype. Following this speciation event, the macaques expanded the number of lineage II KIR genes through duplication and recombination. In the hominid branch, KIR2DL5 emerged to form the ancestral hominid KIR haplotype. Specific adaptations that occurred subsequently in particular ape species are indicated. For humans, the emergence of distinctive CenA and CenB segments occurred ~5 MYA followed by the formation of distinctive TelA and TelB ~1 MYA. The distinguishing characteristics of these segments are described. Human KIR A haplotypes are defined by a fixed gene content and comprise CenA/TelA. KIR B haplotypes contain at least one of the characteristic B haplotype genes and are much more variable in KIR gene content than the KIR A haplotypes.
Emergence of CenA and CenB KIR haplotypes in hominins
Complete sequences are known for rhesus macaque, orangutan, chimapanzee, and human KIR haplotypes (46–48, 50, 85, 124, 125). A major difference between the species is in the distribution of KIR genes within the centromeric and telomeric regions defined by the framework genes. The expansion of lineage II KIR genes in Old World monkeys occurred in the telomeric region of the KIR locus, whereas the expansion of lineage III KIR genes in apes occurred in the centromeric region. Soon after the divergence of the human and chimpanzee lines, ~7-8 million years ago (126), human ancestors evolved two distinctive gene-content motifs in the centromeric region (50). These were the forbearers of the present day's CenA and CenB motifs (Figure 12). The ancestral motifs from which they derived were probably like the centromeric motifs of chimpanzee KIR haplotypes and contained genes encoding mainly inhibitory receptors for C1 and C2. That remains the characteristic of the CenA motif of the KIR A haplotype, which encodes inhibitory C1 (KIR2DL3) and C2 (KIR2DL1) receptors (50).
The derived motif, which became CenB, encodes KIR2DL1 allotypes that are weaker inhibitory C2 receptors than those encoded by CenA. This attenuation can arise from effects on receptor avidity, signaling capacity, surface expression, and other factors. Most extreme in this regard is the CenB motif that lacks the KIR2DL1 gene. In contrast to KIR2DL1, the KIR2DL2 allotypes encoded by CenB are stronger C1 receptors than the KIR2DL3 allotypes encoded by CenA, and they also exhibit cross-reactivity with C2 (39, 56, 122, 127).
In the centromeric region of the chimpanzee KIR locus, Pt-KIR2DL6 and Pt-KIR3DS6 are a closely linked pair of genes encoding inhibitory and activating receptors with similarly strong avidity for C1 (122). KIR2DL2 and KIR2DS2 are a similar pair of genes in human CenB. Whereas KIR2DL2 binds strongly to C1, KIR2DS2 has no detectable interaction with C1. This failure is due to the unusual substitution of tyrosine for phenylalanine at position 45, which if corrected by in vitro mutagenesis confers strong avidity for C1 on KIR2DS2 (128). This argues for KIR2DS2 having once been a strong C1 receptor but became subject to natural selection that abrogated that function by fixing a variant KIR2DS2 allele encoding tyrosine 45. Although not binding C1, KIR2DS2 is cell-surface expressed (127). The net effect is that CenB encodes a strong inhibitory C1 receptor but no C1-specific activating receptor as a counterbalance. With the combination of strong inhibitory C1 receptor and an attenuated inhibitory C2 receptor, CenB promotes the function of C1 as a KIR ligand and demotes that of C2.
KIR2DS3/5, another activating lineage III KIR encoded by CenB, has threonine at position 44, a substitution that is unique to the human species. Threonine 44 is also encoded by KIR2DP1, a lineage III pseudogene. Like KIR2DS2, KIR2DS3/5 is expressed at the cell surface but does not bind any MHC class I (127). Changing residue 44 of KIR2DL3 from lysine to threonine converts the specificity of the receptor from C1 to C2 (56). When the same change was made to KIR2DL1, it retained C2 specificity but the avidity was reduced by half. This points to KIR2DS3/5 having once encoded an activating C2 receptor, but which became subject to selection against that function, causing the fixation of an allotype that does not bind C2. It appears that a prevailing selection against C2 receptors on CenB has eliminated the C2-binding function of KIR2DS3/5 and made the CenB-encoded KIR2DL1 allotypes weak inhibitory receptors. The final gene that distinguishes CenB from CenA is KIR2DL5, the only non-framework human gene to have orthologs in all the great apes (46, 82, 83, 93). Although KIR2DL5 is surface expressed and delivers inhibitory signals its function remains obscure (129).
Emergence of TelA and TelB KIR haplotypes in hominins
Around one million years ago, the telomeric region of the human KIR locus was reorganized to give two distinctive gene content motifs that became the present day TelA and TelB motifs (50). This involved the importation of four KIR genes that originated in the centromeric region. Of the two human lineage II KIR genes, KIR3DL1/S1 is situated on the telomeric side of KIR2DL4 and KIR3DL2 is placed at the telomeric end of the KIR locus. Situated between KIR3DL1 and KIR3DL2 in TelA is KIR2DS4, which encodes the only activating KIR of the KIR A haplotype (the combination of CenA and TelA) and has an ortholog in the centromeric region of the chimpanzee KIR locus. KIR2DS4 recognizes HLA-A*11, and some HLA-C allotypes in a manner that does not correlate with either the C1 or C2 epitope (130). In the corresponding region of TelB there are three genes that were probably imported en bloc from the centromeric region in an event of unequal cross-over. These three genes comprise KIR2D3/5, KIR2DL5, and KIR2DS1. The latter encodes an activating C2 receptor.
Whereas the chimpanzee genes encoding inhibitory and activating C2 receptors are paired together in the centromeric region, the human counterparts are in different regions and segregate independently of each other in the human population. A major difference between TelA and TelB is in the KIR3DL1/S1 alleles they carry. TelA carries the KIR3DL1 alleles that encode inhibitory Bw4 receptors, whereas TelB carries the homologous KIR3DS1 alleles that encode activating receptors. Like KIR2DS2, the KIR3DS1 proteins have acquired substitutions that reduce Bw4 binding function to where it cannot be detected (131). High polymorphism with functional effect is the property of KIR3DL1. By comparison KIR3DS1 is conserved and KIR3DS1*013 is the most common KIR3DL1/S1 allele (23).
The canonical KIR A haplotype consists of Cen A and Tel A and is a variation on the theme developed in the chimpanzee KIR locus. The canonical KIR B haplotype, consisting of CenB and TelB, is very different from any chimpanzee KIR haplotype. A consequence of the human-specific spreading of KIR variability between the centromeric and telomeric regions is that it enables recombination to generate a spectrum of KIR haplotypes that have CenA combined with Tel B or CenB combined with Tel A. This adds an additional dimension to KIR diversity in humans that is not seen in our closest relatives, the great apes. Although human populations differ in their relative frequencies of CenA, CenB, TelA, and Tel A, all four genomic segments are preserved in all human populations (132). In different ways they must facilitate the survival and propagation of the human species (Fig. 12).
Co-evolution of KIR and HLA class I in modern humans
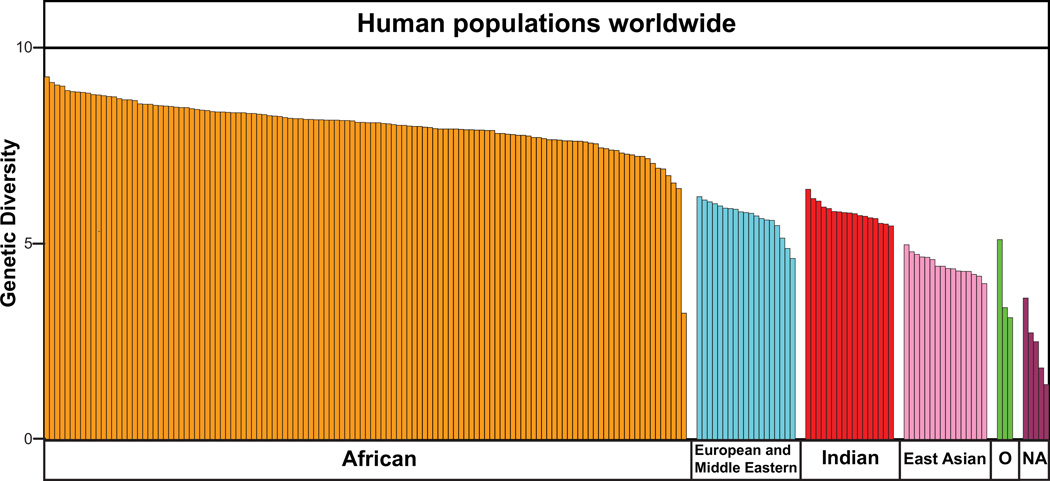
The modern human species originated in East Africa ~200,000 years ago (133) and subsequently extended its range throughout the African continent. Some 75,000 years ago (134), modern humans began to migrate out of Africa and gradually extended the human range to include all the continents, except Antarctica, and many of the offshore and oceanic islands. The migrant populations who left Africa were a small minority of Africans and experienced successive population bottlenecks as they moved further and further away from Africa. Analysis of neutral genetic markers in modern human populations shows that there is an inverse relationship between the overall genetic diversity in a population and the distance travelled from East Africa to reach the place they now inhabit (135, 136) (Fig. 14). Similar correlations were seen when the analysis was done using MHC class I polymorphisms as the genetic markers (135).
Figure 14. The genetic diversity of human populations is inversely correlated with the distance of their place of origin from East Africa.
This diagram shows the genetic diversity assessed from neutral markers of anthropologically well-characterized human populations worldwide. Each bar represents one population and the colors denote their geographical regions of origin. Africans; orange, European/Middle Eastern; blue, Indian; red, East Asian; pink, Oceanian (O); green, and Native American (NA); purple. Adapted with permission from Dr. Sarah Tishkoff from Fig. S2 in Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science. (2009) 324:1035-1044 (136). Reprinted with permission from AAAS.
Illustrating the point is the comparison of MHC class I and KIR diversity in a Sub-Saharan African population, the GaAdangbe of Ghana (25), and a South Amerindian population, the Yucpa of Venezuela (137). The former reside near the site of human origin, while the latter reside towards the end of the longest route of human migration. There is a striking difference in the diversity of HLA class I and KIR haplotypes in these two populations that represent the beginning and the end of the heroic human migrations (Fig. 15). One way such comparisons are informative is in identifying which components of HLA class I and KIR diversity are preserved in populations that have been subject to strong selective pressures and have passed through many population bottlenecks. Because the genes encoding KIR and their MHC class I ligands segregate on different chromosomes, a key issue is the extent to which a population maintains functional ligand-receptor pairs. The Yucpa have maintained the C1 and C2 epitopes of HLA-C as well as Bw4 carried by HLA-A and HLA-B. Absent is the HLA-A3/11 epitope. This evidence from the Yucpa, and other populations, is that C1, C2, and Bw4 epitopes are necessary for long-term human survival, whereas the A3/11 epitope is not.
Figure 15. The HLA and KIR diversity of human populations is inversely correlated with the distance from East Africa of their place of origin.
Compared here is an African population, the Ga-Adangbe from Ghana, and a Native American population, the Yucpa from Venezuela. The upper pie charts show the number and frequencies of allele-level HLA-A,-B,-C haplotypes in the two populations. The lower pie charts show the number and frequencies of allele-level KIR haplotypes in the two populations. The proportions of KIR A and KIR B haplotypes are shown by the curved arrows. Based upon the data of Gendzehhadze et al. (Yucpa) (137) and Norman et al (Ga-Adangbe) (25).
The aspect of the KIR system that is missing from the Yucpa is the centromeric segment containing the KIR2DL5 and KIR2DS3/5 genes. In this population these genes have been completely transferred from the centromeric to the telomeric region of the KIR locus. Thus the centromeric and telomeric regions are more differentiated from each other in the Yucpa than in other populations. This theme of maintaining essentials while limiting redundancy is also evident in the structures of the two dominant and balanced KIR gene-content haplotypes, which have only the framework genes in common. The canonical CenA-TelA haplotype has a frequency of 46%; the CenB-TelB haplotype has a frequency of 48%, and has all the characteristic KIR B genes and lacks KIR2DL1 (137).
An unusual feature of HLA-C in the Yucpa is the 83% frequency of the C1 epitope, one of the highest frequencies observed worldwide. This is mainly due to HLA-C*07:02, a good educator of NK cells (11). As a consequence of their high level of C1, the Yucpa became subject to selection that acted to weaken the KIR2DL3 C1-receptor. In the Yucpa, two new variant alleles were derived from the older KIR2DL3*001 by point mutation. Under selection these two alleles have reached a combined frequency exceeding that of KIR2DL3*001 by four fold. The new variants encode receptors with attenuated function as inhibitory C1 receptors, compared to KIR2DL3*001. Thus the dominating C1 ligand induced a reaction of co-evolution that weakened its cognate inhibitory receptor.
Unlike the Yucpa, the KhoeSan hunter-gather people of Southern Africa have among the highest genetic diversity and a C2 frequency of 64%; again, one of the largest worldwide (138). In this population, natural selection has acted on the KIR2DL1 gene that encodes the inhibitory C2 receptor. Two new KIR2DL1 variant alleles, with reduced function as inhibitory C2 receptors, have risen to higher frequency than the older alleles from which they derived by point mutation (Hilton et al., manuscript submitted). The change in the new KIR2DL1*022 variant was to replace methionine 44 with lysine, which abrogated its function as a C2 receptor but also gave it new function as an inhibitory C1 receptor. Further, the avidity of KIR2DL1*022 for C1 is more than that of any KIR2DL2/3 allotype. The change in the new KIR2DL1*026 variant terminates translation at the end of the transmembrane domain, eliminating the cytoplasmic tail and its ITIMs. This abrogates inhibitory signaling potential and decreases cell surface expression. Thus in the KhoeSan, the high frequency of C2 has induced a reaction that weakened the inhibitory C2 receptors and a strengthened the inhibitory C1 receptors. The Yucpa and the Khoesan are on the extremes of a full spectrum of relative C1 and C2 frequencies (Figure 16). Despite their different demographic histories, the similarity of their response to excess C1 and C2 ligand, suggests that this is a general mechanism of co-evolution that acts to maintain a necessary balance between the C1 and C2 frequencies in human populations.
Figure 16. Frequencies of the C1 and C2 epitopes of HLA-C vary among human populations worldwide.
Frequencies of HLA-C alleles for 153 populations were obtained from http://www.allelefrequencies.net (1). All these populations are anthropologically well defined and are represented by samples of more than 40 individuals. Panels from disease-association studies and transplant cohorts, for which frequency skewing was a possibility, were excluded. Population frequencies were binned in increments of .05 and plotted. The curve approximating the frequency distribution was plotted using Prism 6. Frequencies for C1-bearing HLA-C allotypes are shown on the left; frequencies for C2-bearing HLA-C allotypes are on the right. The upper panels show the frequency distributions for four populations representing Africa (KhoeSan), Europe (Northern Ireland), Asia (Japan), and America (Yucpa) indicated by vertical arrows. The lower panels show the same curves as the upper panels, but they also include the histograms showing the distribution of different population groups. The bars are colored as in Fig. 14: Africans; orange, European/Middle Eastern; blue, Indian; red, East Asian; pink, Oceanian; green, and Native American; purple. Within the global distribution, regional differences can be observed. For example, Africans have higher C2 frequencies than other population groups, whereas East Asians stand out for their higher C1 frequencies.
Acknowledgements
This work was supported by NIH grants AI17892, AI22039, AI24258, and AI31168 to P.P.
Footnotes
The authors declare that they have no conflicts of interest to report
References
- 1.Gonzalez-Galarza FF, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015;43:D784–D788. doi: 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43:D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Rood JJ, Van Leeuwen A. Leukocyte Grouping. A Method and Its Application. J Clin Invest. 1963;42:1382–1390. doi: 10.1172/JCI104822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braud V, Jones EY, McMichael A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur J Immunol. 1997;27:1164–1169. doi: 10.1002/eji.1830270517. [DOI] [PubMed] [Google Scholar]
- 5.O'Callaghan CA, et al. Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA-E. Mol Cell. 1998;1:531–541. doi: 10.1016/s1097-2765(00)80053-2. [DOI] [PubMed] [Google Scholar]
- 6.Brooks AG, et al. Specific recognition of HLA-E, but not classical, HLA class I molecules by soluble CD94/NKG2A and NK cells. J Immunol. 1999;162:305–313. [PubMed] [Google Scholar]
- 7.Lee N, et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci U S A. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vales-Gomez M, Reyburn HT, Erskine RA, Lopez-Botet M, Strominger JL. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J. 1999;18:4250–4260. doi: 10.1093/emboj/18.15.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shum BP, et al. Conservation and variation in human and common chimpanzee CD94 and NKG2 genes. J Immunol. 2002;168:240–252. doi: 10.4049/jimmunol.168.1.240. [DOI] [PubMed] [Google Scholar]
- 10.Ljunggren HG, Karre K. In search of the 'missing self': MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 11.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vivier E, Anfossi N. Inhibitory NK-cell receptors on T cells: witness of the past, actors of the future. Nat Rev Immunol. 2004;4:190–198. doi: 10.1038/nri1306. [DOI] [PubMed] [Google Scholar]
- 13.Weis WI, Taylor ME, Drickamer K. The C-type lectin superfamily in the immune system. Immunol Rev. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Mariuzza RA. Structural basis for recognition of cellular and viral ligands by NK cell receptors. Front Immunol. 2014;5:123. doi: 10.3389/fimmu.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokoyama WM, Seaman WE. The Ly-49 and NKR-P1 gene families encoding lectin-like receptors on natural killer cells: the NK gene complex. Annu Rev Immunol. 1993;11:613–635. doi: 10.1146/annurev.iy.11.040193.003145. [DOI] [PubMed] [Google Scholar]
- 16.Djurisic S, Hviid TV. HLA Class Ib Molecules and Immune Cells in Pregnancy and Preeclampsia. Front Immunol. 2014;5:652. doi: 10.3389/fimmu.2014.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248:220–223. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- 18.Matson BC, Caron KM. Uterine natural killer cells as modulators of the maternal-fetal vasculature. Int J Dev Biol. 2014;58:199–204. doi: 10.1387/ijdb.140032kc. [DOI] [PubMed] [Google Scholar]
- 19.Shiroishi M, et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci U S A. 2003;100:8856–8861. doi: 10.1073/pnas.1431057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torkar M, Norgate Z, Colonna M, Trowsdale J, Wilson MJ. Isotypic variation of novel immunoglobulin-like transcript/killer cell inhibitory receptor loci in the leukocyte receptor complex. Eur J Immunol. 1998;28:3959–3967. doi: 10.1002/(SICI)1521-4141(199812)28:12<3959::AID-IMMU3959>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 21.Wan AM, Ennis P, Parham P, Holmes N. The primary structure of HLA-A32 suggests a region involved in formation of the Bw4/Bw6 epitopes. J Immunol. 1986;137:3671–3674. [PubMed] [Google Scholar]
- 22.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norman PJ, et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 24.Hill AV, et al. Molecular analysis of the association of HLA-B53 and resistance to severe malaria. Nature. 1992;360:434–439. doi: 10.1038/360434a0. [DOI] [PubMed] [Google Scholar]
- 25.Norman PJ, et al. Co-evolution of human leukocyte antigen (HLA) class I ligands with killer-cell immunoglobulin-like receptors (KIR) in a genetically diverse population of sub-Saharan Africans. PLoS Genet. 2013;9:e1003938. doi: 10.1371/journal.pgen.1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansasuta P, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34:1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- 27.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol. 2002;38:1007–1021. doi: 10.1016/s0161-5890(02)00030-5. [DOI] [PubMed] [Google Scholar]
- 29.Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci U S A. 1993;90:12000–12004. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997;158:4026–4028. [PubMed] [Google Scholar]
- 31.Falk CS, Schendel DJ. HLA-C revisited. Ten years of change. Immunol Res. 1997;16:203–214. doi: 10.1007/BF02786363. [DOI] [PubMed] [Google Scholar]
- 32.Honda K, et al. Selection of escape mutant by HLA-C-restricted HIV-1 Pol-specific cytotoxic T lymphocytes carrying strong ability to suppress HIV-1 replication. Eur J Immunol. 2011;41:97–106. doi: 10.1002/eji.201040841. [DOI] [PubMed] [Google Scholar]
- 33.Zemmour J, Parham P. Distinctive polymorphism at the HLA-C locus: implications for the expression of HLA-C. J Exp Med. 1992;176:937–950. doi: 10.1084/jem.176.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sibilio L, et al. A single bottleneck in HLA-C assembly. J Biol Chem. 2008;283:1267–1274. doi: 10.1074/jbc.M708068200. [DOI] [PubMed] [Google Scholar]
- 35.Zipeto D, Beretta A. HLA-C and HIV-1: friends or foes? Retrovirology. 2012;9:39. doi: 10.1186/1742-4690-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulkarni S, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472:495–498. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Huigin C, et al. The molecular origin and consequences of escape from miRNA regulation by HLA-C alleles. Am J Hum Genet. 2011;89:424–431. doi: 10.1016/j.ajhg.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barber LD, et al. The inter-locus recombinant HLA-B*4601 has high selectivity in peptide binding and functions characteristic of HLA-C. J Exp Med. 1996;184:735–740. doi: 10.1084/jem.184.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 40.Uhrberg M, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 41.Blokhuis JH, van der Wiel MK, Doxiadis GG, Bontrop RE. The mosaic of KIR haplotypes in rhesus macaques. Immunogenetics. 2010;62:295–306. doi: 10.1007/s00251-010-0434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blokhuis JH, van der Wiel MK, Doxiadis GG, Bontrop RE. The extreme plasticity of killer cell Ig-like receptor (KIR) haplotypes differentiates rhesus macaques from humans. Eur J Immunol. 2011;41:2719–2728. doi: 10.1002/eji.201141621. [DOI] [PubMed] [Google Scholar]
- 43.Kruse PH, Rosner C, Walter L. Characterization of rhesus macaque KIR genotypes and haplotypes. Immunogenetics. 2010;62:281–293. doi: 10.1007/s00251-010-0433-4. [DOI] [PubMed] [Google Scholar]
- 44.Moreland AJ, et al. Characterization of killer immunoglobulin-like receptor genetics and comprehensive genotyping by pyrosequencing in rhesus macaques. BMC Genomics. 2011;12:295. doi: 10.1186/1471-2164-12-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hammond JA, Guethlein LA, Abi-Rached L, Moesta AK, Parham P. Evolution and survival of marine carnivores did not require a diversity of killer cell Ig-like receptors or Ly49 NK cell receptors. J Immunol. 2009;182:3618–3627. doi: 10.4049/jimmunol.0803026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abi-Rached L, Moesta AK, Rajalingam R, Guethlein LA, Parham P. Human-specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells. PLoS Genet. 2010;6:e1001192. doi: 10.1371/journal.pgen.1001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guethlein LA, Older Aguilar AM, Abi-Rached L, Parham P. Evolution of killer cell Ig-like receptor (KIR) genes: definition of an orangutan KIR haplotype reveals expansion of lineage III KIR associated with the emergence of MHC-C. J Immunol. 2007;179:491–504. doi: 10.4049/jimmunol.179.1.491. [DOI] [PubMed] [Google Scholar]
- 48.Wilson MJ, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting edge: expansion of the KIR locus by unequal crossing over. J Immunol. 2003;171:2192–2195. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- 50.Pyo CW, et al. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS One. 2010;5:e15115. doi: 10.1371/journal.pone.0015115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shilling HG, et al. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168:2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 52.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201:1319–1332. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bari R, et al. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood. 2009;114:5182–5190. doi: 10.1182/blood-2009-07-231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175:5222–5229. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 55.Gardiner CM, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 56.Hilton HG, et al. Mutation at positively selected positions in the binding site for HLA-C shows that KIR2DL1 is a more refined but less adaptable NK cell receptor than KIR2DL3. J Immunol. 2012;189:1418–1430. doi: 10.4049/jimmunol.1100431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. 2003;171:6640–6649. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 58.Steiner NK, Dakshanamurthy S, Nguyen N, Hurley CK. Allelic variation of killer cell immunoglobulin-like receptor 2DS5 impacts glycosylation altering cell surface expression levels. Hum Immunol. 2014;75:124–128. doi: 10.1016/j.humimm.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.VandenBussche CJ, Dakshanamurthy S, Posch PE, Hurley CK. A single polymorphism disrupts the killer Ig-like receptor 2DL2/2DL3 D1 domain. J Immunol. 2006;177:5347–5357. doi: 10.4049/jimmunol.177.8.5347. [DOI] [PubMed] [Google Scholar]
- 60.Karlhofer FM, Ribaudo RK, Yokoyama WM. MHC class I alloantigen specificity of Ly-49+ IL-2-activated natural killer cells. Nature. 1992;358:66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- 61.Yokoyama WM, Jacobs LB, Kanagawa O, Shevach EM, Cohen DI. A murine T lymphocyte antigen belongs to a supergene family of type II integral membrane proteins. J Immunol. 1989;143:1379–1386. [PubMed] [Google Scholar]
- 62.Chan PY, Takei F. Molecular cloning and characterization of a novel murine T cell surface antigen, YE1/48. J Immunol. 1989;142:1727–1736. [PubMed] [Google Scholar]
- 63.Wagtmann N, et al. Molecular clones of the p58 NK cell receptor reveal immunoglobulin-related molecules with diversity in both the extra- and intracellular domains. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 64.Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 65.Deng L, Cho S, Malchiodi EL, Kerzic MC, Dam J, Mariuzza RA. Molecular architecture of the major histocompatibility complex class I-binding site of Ly49 natural killer cell receptors. J Biol Chem. 2008;283:16840–16849. doi: 10.1074/jbc.M801526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Westgaard IH, Berg SF, Orstavik S, Fossum S, Dissen E. Identification of a human member of the Ly-49 multigene family. Eur J Immunol. 1998;28:1839–1846. doi: 10.1002/(SICI)1521-4141(199806)28:06<1839::AID-IMMU1839>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 67.Welch AY, Kasahara M, Spain LM. Identification of the mouse killer immunoglobulin-like receptor-like (Kirl) gene family mapping to chromosome X. Immunogenetics. 2003;54:782–790. doi: 10.1007/s00251-002-0529-6. [DOI] [PubMed] [Google Scholar]
- 68.Gagnier L, Wilhelm BT, Mager DL. Ly49 genes in non-rodent mammals. Immunogenetics. 2003;55:109–115. doi: 10.1007/s00251-003-0558-9. [DOI] [PubMed] [Google Scholar]
- 69.Takahashi T, et al. Natural killer cell receptors in the horse: evidence for the existence of multiple transcribed LY49 genes. Eur J Immunol. 2004;34:773–784. doi: 10.1002/eji.200324695. [DOI] [PubMed] [Google Scholar]
- 70.Futas J, Horin P. Natural killer cell receptor genes in the family Equidae: not only Ly49. PLoS One. 2013;8:e64736. doi: 10.1371/journal.pone.0064736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Averdam A, et al. A novel system of polymorphic and diverse NK cell receptors in primates. PLoS Genet. 2009;5:e1000688. doi: 10.1371/journal.pgen.1000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bimber BN, Moreland AJ, Wiseman RW, Hughes AL, O'Connor DH. Complete characterization of killer Ig-like receptor (KIR) haplotypes in Mauritian cynomolgus macaques: novel insights into nonhuman primate KIR gene content and organization. J Immunol. 2008;181:6301–6308. doi: 10.4049/jimmunol.181.9.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cadavid LF, Lun CM. Lineage-specific diversification of killer cell Ig-like receptors in the owl monkey, a New World primate. Immunogenetics. 2009;61:27–41. doi: 10.1007/s00251-008-0342-y. [DOI] [PubMed] [Google Scholar]
- 74.Cadavid LF, Palacios C, Lugo JS. Bimodal evolution of the killer cell Ig-like receptor (KIR) family in New World primates. Immunogenetics. 2013;65:725–736. doi: 10.1007/s00251-013-0719-4. [DOI] [PubMed] [Google Scholar]
- 75.Garzon-Ospina D, Lopez C, Cadavid LF, Patarroyo ME, Patarroyo MA. Identification and diversity of killer cell Ig-like receptors in Aotus vociferans, a New World monkey. PLoS One. 2013;8:e79731. doi: 10.1371/journal.pone.0079731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guethlein LA, Flodin LR, Adams EJ, Parham P. NK cell receptors of the orangutan (Pongo pygmaeus): a pivotal species for tracking the coevolution of killer cell Ig-like receptors with MHC-C. J Immunol. 2002;169:220–229. doi: 10.4049/jimmunol.169.1.220. [DOI] [PubMed] [Google Scholar]
- 77.Hershberger KL, Kurian J, Korber BT, Letvin NL. Killer cell immunoglobulin-like receptors (KIR) of the African-origin sabaeus monkey: evidence for recombination events in the evolution of KIR. Eur J Immunol. 2005;35:922–935. doi: 10.1002/eji.200425408. [DOI] [PubMed] [Google Scholar]
- 78.Hershberger KL, Shyam R, Miura A, Letvin NL. Diversity of the killer cell Ig-like receptors of rhesus monkeys. J Immunol. 2001;166:4380–4390. doi: 10.4049/jimmunol.166.7.4380. [DOI] [PubMed] [Google Scholar]
- 79.Mager DL, McQueen KL, Wee V, Freeman JD. Evolution of natural killer cell receptors: coexistence of functional Ly49 and KIR genes in baboons. Curr Biol. 2001;11:626–630. doi: 10.1016/s0960-9822(01)00148-8. [DOI] [PubMed] [Google Scholar]
- 80.Maloveste SM, et al. Degenerate recognition of MHC class I molecules with Bw4 and Bw6 motifs by a killer cell Ig-like receptor 3DL expressed by macaque NK cells. J Immunol. 2012;189:4338–4348. doi: 10.4049/jimmunol.1201360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Palacios C, Cuervo LC, Cadavid LF. Evolutionary patterns of killer cell Ig-like receptor genes in Old World monkeys. Gene. 2011;474:39–51. doi: 10.1016/j.gene.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 82.Rajalingam R, Hong M, Adams EJ, Shum BP, Guethlein LA, Parham P. Short KIR haplotypes in pygmy chimpanzee (Bonobo) resemble the conserved framework of diverse human KIR haplotypes. J Exp Med. 2001;193:135–146. doi: 10.1084/jem.193.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rajalingam R, Parham P, Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J Immunol. 2004;172:356–369. doi: 10.4049/jimmunol.172.1.356. [DOI] [PubMed] [Google Scholar]
- 84.Abi-Rached L, et al. A small, variable, and irregular killer cell Ig-like receptor locus accompanies the absence of MHC-C and MHC-G in gibbons. J Immunol. 2010;184:1379–1391. doi: 10.4049/jimmunol.0903016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sambrook JG, et al. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sambrook JG, et al. Identification of a single killer immunoglobulin-like receptor (KIR) gene in the porcine leukocyte receptor complex on chromosome 6q. Immunogenetics. 2006;58:481–486. doi: 10.1007/s00251-006-0110-9. [DOI] [PubMed] [Google Scholar]
- 87.Sanderson ND, et al. Definition of the cattle killer cell Ig-like receptor gene family: comparison with aurochs and human counterparts. J Immunol. 2014;193:6016–6030. doi: 10.4049/jimmunol.1401980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dobromylskyj MJ, Connelley T, Hammond JA, Ellis SA. Cattle Ly49 is polymorphic. Immunogenetics. 2009;61:789–795. doi: 10.1007/s00251-009-0406-7. [DOI] [PubMed] [Google Scholar]
- 89.Hammond JA, Guethlein LA, Norman PJ, Parham P. Natural selection on marine carnivores elaborated a diverse family of classical MHC class I genes exhibiting haplotypic gene content variation and allelic polymorphism. Immunogenetics. 2012;64:915–933. doi: 10.1007/s00251-012-0651-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Averdam A, et al. Sequence analysis of the grey mouse lemur (Microcebus murinus) MHC class II DQ and DR region. Immunogenetics. 2011;63:85–93. doi: 10.1007/s00251-010-0487-3. [DOI] [PubMed] [Google Scholar]
- 91.Flugge P, Zimmermann E, Hughes AL, Gunther E, Walter L. Characterization and phylogenetic relationship of prosimian MHC class I genes. J Mol Evol. 2002;55:768–775. doi: 10.1007/s00239-002-2372-7. [DOI] [PubMed] [Google Scholar]
- 92.Martin RD. Primate Origins and Evolution. Princeton University Press; 1990. [Google Scholar]
- 93.Khakoo SI, et al. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12:687–698. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- 94.Kono A, et al. Genomic sequence analysis of the MHC class I G/F segment in common marmoset (Callithrix jacchus) J Immunol. 2014;192:3239–3246. doi: 10.4049/jimmunol.1302745. [DOI] [PubMed] [Google Scholar]
- 95.Shiina T, et al. Comparative genome analysis of the major histocompatibility complex (MHC) class I B/C segments in primates elucidated by genomic sequencing in common marmoset (Callithrix jacchus) Immunogenetics. 2011;63:485–499. doi: 10.1007/s00251-011-0526-8. [DOI] [PubMed] [Google Scholar]
- 96.van der Wiel MK, Otting N, de Groot NG, Doxiadis GG, Bontrop RE. The repertoire of MHC class I genes in the common marmoset: evidence for functional plasticity. Immunogenetics. 2013;65:841–849. doi: 10.1007/s00251-013-0732-7. [DOI] [PubMed] [Google Scholar]
- 97.Guethlein LA, Abi-Rached L, Hammond JA, Parham P. The expanded cattle KIR genes are orthologous to the conserved single-copy KIR3DX1 gene of primates. Immunogenetics. 2007;59:517–522. doi: 10.1007/s00251-007-0214-x. [DOI] [PubMed] [Google Scholar]
- 98.Sambrook JG, et al. Identification of the ancestral killer immunoglobulin-like receptor gene in primates. BMC Genomics. 2006;7:209. doi: 10.1186/1471-2164-7-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Knapp LA, Cadavid LF, Watkins DI. The MHC-E locus is the most well conserved of all known primate class I histocompatibility genes. J Immunol. 1998;160:189–196. [PubMed] [Google Scholar]
- 100.Shiina T, et al. Rapid evolution of major histocompatibility complex class I genes in primates generates new disease alleles in humans via hitchhiking diversity. Genetics. 2006;173:1555–1570. doi: 10.1534/genetics.106.057034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004;14:1501–1515. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Doxiadis GG, de Groot N, Otting N, Blokhuis JH, Bontrop RE. Genomic plasticity of the MHC class I A region in rhesus macaques: extensive haplotype diversity at the population level as revealed by microsatellites. Immunogenetics. 2011;63:73–83. doi: 10.1007/s00251-010-0486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Doxiadis GG, et al. Haplotype diversity generated by ancient recombination-like events in the MHC of Indian rhesus macaques. Immunogenetics. 2013;65:569–584. doi: 10.1007/s00251-013-0707-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schafer JL, et al. KIR3DL01 recognition of Bw4 ligands in the rhesus macaque: maintenance of Bw4 specificity since the divergence of apes and Old World monkeys. J Immunol. 2014;192:1907–1917. doi: 10.4049/jimmunol.1302883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sanjanwala B, Draghi M, Norman PJ, Guethlein LA, Parham P. Polymorphic sites away from the Bw4 epitope that affect interaction of Bw4+ HLA-B with KIR3DL1. J Immunol. 2008;181:6293–6300. doi: 10.4049/jimmunol.181.9.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rosner C, Kruse PH, Hermes M, Otto N, Walter L. Rhesus macaque inhibitory and activating KIR3D interact with Mamu-A-encoded ligands. J Immunol. 2011;186:2156–2163. doi: 10.4049/jimmunol.1002634. [DOI] [PubMed] [Google Scholar]
- 107.Colantonio AD, et al. KIR polymorphisms modulate peptide-dependent binding to an MHC class I ligand with a Bw6 motif. PLoS Pathog. 2011;7:e1001316. doi: 10.1371/journal.ppat.1001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sharma D, et al. Dimorphic motifs in D0 and D1+D2 domains of killer cell Ig-like receptor 3DL1 combine to form receptors with high, moderate, and no avidity for the complex of a peptide derived from HIV and HLA-A*2402. J Immunol. 2009;183:4569–4582. doi: 10.4049/jimmunol.0901734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Older Aguilar AM, Guethlein LA, Hermes M, Walter L, Parham P. Rhesus macaque KIR bind human MHC class I with broad specificity and recognize HLA-C more effectively than HLA-A and HLA-B. Immunogenetics. 2011;63:577–585. doi: 10.1007/s00251-011-0535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mohyuddin A, Mehdi SQ. HLA analysis of the Parsi (Zoroastrian) population in Pakistan. Tissue Antigens. 2005;66:691–695. doi: 10.1111/j.1399-0039.2005.00507.x. [DOI] [PubMed] [Google Scholar]
- 111.Abi-Rached L, et al. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 2011;334:89–94. doi: 10.1126/science.1209202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Henn BM, Cavalli-Sforza LL, Feldman MW. The great human expansion. Proc Natl Acad Sci U S A. 2012;109:17758–17764. doi: 10.1073/pnas.1212380109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 114.Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat Immunol. 2001;2:452–460. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- 115.Liu J, Xiao Z, Ko HL, Shen M, Ren EC. Activating killer cell immunoglobulin-like receptor 2DS2 binds to HLA-A*11. Proc Natl Acad Sci U S A. 2014;111:2662–2667. doi: 10.1073/pnas.1322052111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vivian JP, et al. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature. 2011;479:401–405. doi: 10.1038/nature10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Adams EJ, Thomson G, Parham P. Evidence for an HLA-C-like locus in the orangutan Pongo pygmaeus. Immunogenetics. 1999;49:865–871. doi: 10.1007/s002510050566. [DOI] [PubMed] [Google Scholar]
- 118.Fukami-Kobayashi K, et al. Genomic evolution of MHC class I region in primates. Proc Natl Acad Sci U S A. 2005;102:9230–9234. doi: 10.1073/pnas.0500770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Older Aguilar AM, Guethlein LA, Adams EJ, Abi-Rached L, Moesta AK, Parham P. Coevolution of killer cell Ig-like receptors with HLA-C to become the major variable regulators of human NK cells. J Immunol. 2010;185:4238–4251. doi: 10.4049/jimmunol.1001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Prado-Martinez J, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vilches C, Pando MJ, Parham P. Genes encoding human killer-cell Ig-like receptors with D1 and D2 extracellular domains all contain untranslated pseudoexons encoding a third Ig-like domain. Immunogenetics. 2000;51:639–646. doi: 10.1007/s002510000184. [DOI] [PubMed] [Google Scholar]
- 122.Moesta AK, Abi-Rached L, Norman PJ, Parham P. Chimpanzees use more varied receptors and ligands than humans for inhibitory killer cell Ig-like receptor recognition of the MHC-C1 and MHC-C2 epitopes. J Immunol. 2009;182:3628–3637. doi: 10.4049/jimmunol.0803401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stone AC, et al. More reliable estimates of divergence times in Pan using complete mtDNA sequences and accounting for population structure. Philos Trans R Soc Lond B Biol Sci. 2010;365:3277–3288. doi: 10.1098/rstb.2010.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hsu KC, Chida S, Geraghty DE, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev. 2002;190:40–52. doi: 10.1034/j.1600-065x.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- 125.Traherne JA, et al. Mechanisms of copy number variation and hybrid gene formation in the KIR immune gene complex. Hum Mol Genet. 2010;19:737–751. doi: 10.1093/hmg/ddp538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Langergraber KE, et al. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc Natl Acad Sci U S A. 2012;109:15716–15721. doi: 10.1073/pnas.1211740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Moesta AK, Graef T, Abi-Rached L, Older Aguilar AM, Guethlein LA, Parham P. Humans differ from other hominids in lacking an activating NK cell receptor that recognizes the C1 epitope of MHC class I. J Immunol. 2010;185:4233–4237. doi: 10.4049/jimmunol.1001951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- 129.Cisneros E, Moraru M, Gomez-Lozano N, Lopez-Botet M, Vilches C. KIR2DL5: An Orphan Inhibitory Receptor Displaying Complex Patterns of Polymorphism and Expression. Front Immunol. 2012;3:289. doi: 10.3389/fimmu.2012.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Graef T, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206:2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gillespie GM, Bashirova A, Dong T, McVicar DW, Rowland-Jones SL, Carrington M. Lack of KIR3DS1 binding to MHC class I Bw4 tetramers in complex with CD8+ T cell epitopes. AIDS Res Hum Retroviruses. 2007;23:451–455. doi: 10.1089/aid.2006.0165. [DOI] [PubMed] [Google Scholar]
- 132.Hollenbach JA, Nocedal I, Ladner MB, Single RM, Trachtenberg EA. Killer cell immunoglobulin-like receptor (KIR) gene content variation in the HGDP-CEPH populations. Immunogenetics. 2012;64:719–737. doi: 10.1007/s00251-012-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Campbell MC, Hirbo JB, Townsend JP, Tishkoff SA. The peopling of the African continent and the diaspora into the new world. Curr Opin Genet Dev. 2014;29:120–132. doi: 10.1016/j.gde.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lippold S, et al. Human paternal and maternal demographic histories: insights from high-resolution Y chromosome and mtDNA sequences. Investig Genet. 2014;5:13. doi: 10.1186/2041-2223-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Prugnolle F, Manica A, Charpentier M, Guegan JF, Guernier V, Balloux F. Pathogen-driven selection and worldwide HLA class I diversity. Curr Biol. 2005;15:1022–1027. doi: 10.1016/j.cub.2005.04.050. [DOI] [PubMed] [Google Scholar]
- 136.Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gendzekhadze K, et al. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci U S A. 2009;106:18692–18697. doi: 10.1073/pnas.0906051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Henn BM, et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci U S A. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Apps R, et al. Influence of HLA-C expression level on HIV control. Science. 2013;340:87–91. doi: 10.1126/science.1232685. [DOI] [PMC free article] [PubMed] [Google Scholar]