Two novel flavone synthase enzymes from maize and Arabidopsis integrate flavone metabolism with hormone and biotic stress responses.
Abstract
Flavones are a major group of flavonoids with diverse functions and are extensively distributed in land plants. There are two different classes of FLAVONE SYNTHASE (FNS) enzymes that catalyze the conversion of the flavanones into flavones. The FNSI class comprises soluble Fe2+/2-oxoglutarate-dependent dioxygenases, and FNSII enzymes are oxygen- and NADPH-dependent cytochrome P450 membrane-bound monooxygenases. Here, we describe the identification and characterization of FNSI enzymes from maize (Zea mays) and Arabidopsis (Arabidopsis thaliana). In maize, ZmFNSI-1 is expressed at significantly higher levels in silks and pericarps expressing the 3-deoxy flavonoid R2R3-MYB regulator P1, suggesting that ZmFNSI-1 could be the main enzyme for the synthesis of flavone O-glycosides. We also show here that DOWNY MILDEW RESISTANT6 (AtDMR6), the Arabidopsis homologous enzyme to ZmFNSI-1, has FNSI activity. While dmr6 mutants show loss of susceptibility to Pseudomonas syringae, transgenic dmr6 plants expressing ZmFNSI-1 show similar susceptibility to wild-type plants, demonstrating that ZmFNSI-1 can complement the mutant phenotype. AtDMR6 expression analysis showed a tissue- and developmental stage-dependent pattern, with high expression in cauline and senescing leaves. Finally, we show that Arabidopsis cauline and senescing leaves accumulate apigenin, demonstrating that Arabidopsis plants have an FNSI activity involved in the biosynthesis of flavones. The results presented here also suggest cross talk between the flavone and salicylic acid pathways in Arabidopsis; in this way, pathogens would induce flavones to decrease salicylic acid and, hence, increase susceptibility.
Flavones are a major group of flavonoids that are found extensively in land plants and have diverse physiological functions. These compounds play important physiological roles in UV light protection and in interactions with other organisms (Peters et al., 1986; Mathesius et al., 1998; Schmitz-Hoerner and Weissenböck, 2003; Casati and Walbot, 2005; Kong et al., 2007). In leguminous plants, flavones act as signal molecules for establishing symbiotic relationships with root nodulation bacteria; for example, luteolin induces nod gene expression in Sinorhizobium meliloti, which is essential for root nodulation (Peters et al., 1986). The identification of human cellular targets of apigenin also uncovered roles of this flavone in the regulation of splicing (Arango et al., 2013). Although flavones are generally colorless, they can function as copigments with anthocyanins to alter the color of flowers (Goto and Kondo, 1991; Tanaka et al., 1998; Shiono et al., 2008; Kalisz et al., 2013; Tanaka and Brugliera, 2013) and leaves (Ishikura, 1981; Fossen et al., 2007). On the other hand, flavones as dietary constituents or supplements have beneficial effects, as they have antioxidant properties, they can prevent cancer, they reduce the risks of cardiovascular diseases, they decrease cholesterol levels, and they show antiviral activities (Bontempo et al., 2007; Cai et al., 2007; Liu et al., 2007; Park et al., 2007; Baek et al., 2009; Dharmarajan and Arumugam, 2012; Yarmolinsky et al., 2012; Dajas et al., 2013).
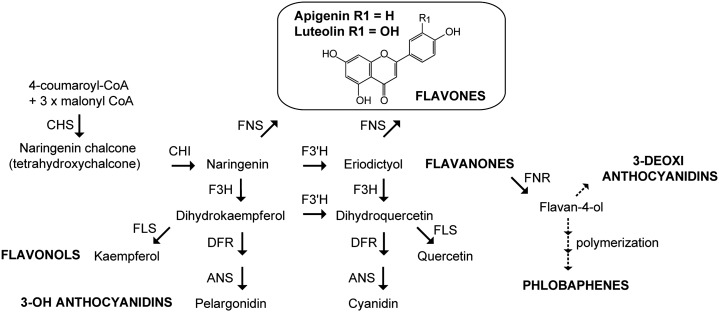
The biosynthesis of flavones begins with flavanones, which are the precursors for all the major flavonoid classes (Fig. 1). There are two different classes of FLAVONE SYNTHASE (FNS) enzymes that catalyze the conversion of the flavanones naringenin and eriodictyol into apigenin and luteolin, respectively. The FNSI class comprises soluble Fe2+ OXOGLUTARATE-DEPENDENT DIOXYGENASEs (2-ODDs), which directly introduce a double bond between C2 and C3 in the flavanone substrates (Martens and Mithöfer, 2005). FNSI shows high sequence identity to FLAVANONE 3-HYDROXYLASE (F3H), another dioxygenase that uses the same flavanones as substrates (Fig. 1; Gebhardt et al., 2007). In contrast, FNSII enzymes are oxygen- and NADPH-dependent cytochrome P450 membrane-bound monooxygenases (Martens and Mithöfer, 2005). All the characterized FNSII enzymes belong to the P450 CYP93 family, and most of these proteins convert flavanones to flavones directly in enzyme assays (Akashi et al., 1999; Martens and Forkmann, 1999; Kitada et al., 2001; Zhang et al., 2007; Fliegmann et al., 2010).
Figure 1.
Simplified scheme of the flavonoid pathway. Representative reactions of each of the major pathway branches are shown. End products are indicated in boldface uppercase letters, and the enzymes catalyzing each step are in uppercase letters. Dotted arrows indicate multiple steps and unidentified enzymes. ANS, Anthocyanidin synthase; CHS, chalcone synthase; CHI, chalcone isomerase; DFR, dihydroflavonol 4-reductase; F3′H, flavonoid-3′-hydroxylase; FLS, flavonol synthase; FNR, flavanone 4-reductase.
In maize (Zea mays), flavonoid biosynthesis is controlled by different classes of regulatory proteins. Anthocyanins are controlled by a MYB domain-containing class (Colored aleurone [C1] or Purple leaf [PL1]; Paz-Ares et al., 1987; Cone et al., 1993) and a basic helix-loop-helix domain-containing class (members of the Red color [R]/Booster [B] gene families; Ludwig et al., 1989). All known anthocyanin genes appear to be coordinately regulated in maize through the concerted action of members of the C1/PL1 and R/B classes of transcription factors (Goff et al., 1992). In addition to 3-hydroxy flavonoids and anthocyanins, maize also accumulates flavones, 3-deoxy flavonoids, and derived pigments, which include the phlobaphenes. The transcription of genes in the biosynthesis of flavones and phlobaphenes is regulated by the Pericarp color1 (P1) regulator, an R2R3-MYB transcription factor similar in the R2R3 MYB domain to C1/PL1 (Grotewold et al., 1991). However, unlike C1/PL1, which requires R/B for function, the P1 regulatory function is independent of R/B (Grotewold and Peterson, 1994).
In general, plants accumulate flavonoids in vacuoles as O-glycoside derivatives, but bryophytes, ferns, gymnosperms, and several angiosperms also produce flavonoid C-glycosides (Harborne, 1993; Rayyan et al., 2005, 2010). Particularly, cereals produce flavonoid C-glycosides such as flavone C-glycosides. In maize, C-glycosyl flavones are involved in the protection against UV-B radiation and in the defense against pathogens (Casati and Walbot, 2005). Maysin, the C-glycosyl flavone predominant in silk tissues of some maize varieties, is a natural insecticide against the corn earworm Helicoverpa zea (McMullen et al., 1998, 2004; Rector et al., 2002). In some maize lines, other flavones, such as the immediate precursor of maysin, rhamnosylisoorientin, can also be present (Gueldner et al., 1989; Snook et al., 1993). The formation of flavone C-glycosides in maize and rice (Oryza sativa) involves the initial hydroxylation of flavanones to the 2-hydroxy derivatives by FLAVANONE 2-HYDROXYLASEs (F2Hs), which are P450 enzymes with very high similarity to FNSII proteins (Brazier-Hicks et al., 2009; Morohashi et al., 2012). Then, the 2-hydroxyflavanones serve as substrates for C-glycosyl transferases to result in flavone 6-C- or 8-C-glucosides (Brazier-Hicks et al., 2009; Falcone Ferreyra et al., 2013). Therefore, enzyme activities that generate 2-hydroxyflavones are necessary for channeling flavanones to flavone C-glycoside formation in cereals. However, flavones accumulate not only as C-glycosides but also as O-linked conjugates in vegetative tissues of grasses, and O-linked modifications are proposed to proceed after the flavone aglycone formation (Brazier-Hicks et al., 2009; Lam et al., 2014). In maize, the presence of flavone O-glycosides, such as apigenin 7-O-glucoside and 6,4-dihydroxy-3-methoxyflavone-7-O-glucoside, has been reported (Ren et al., 2009; Casas et al., 2014; Wen et al., 2014). Thus, maize plants should also have FNS proteins for the biosynthesis of flavone O-glycosides besides the already characterized ZmF2H1 involved in C-glycosyl flavone biosynthesis (Falcone Ferreyra et al., 2013). Therefore, the first aim of this work was to identify and characterize FNS enzymes that could be involved in the synthesis of flavone O-glycosides in maize.
On the other hand, an Arabidopsis (Arabidopsis thaliana) mutant, downy mildew resistant6 (dmr6), was identified in a genetic screen for mutants that showed loss of susceptibility to Hyaloperonospora parasitica (van Damme et al., 2008). dmr6 plants carry a recessive mutation that results in the loss of susceptibility not only to H. parasitica but also to Hyaloperonospora arabidopsidis, Phytophthora capsici, and Pseudomonas syringae, suggesting that AtDMR6 has a role during plant defense (van Damme et al., 2008; Zeilmaker et al., 2015). Interestingly, dmr6 mutants accumulate higher levels of salicylic acid than wild-type plants (Zeilmaker et al., 2015); AtDMR6 expression is also induced by salicylic acid (Arabidopsis eFP Browser; Winter et al., 2007); and because AtDMR6 protein has some amino acid identity with a previously described SALICYLIC ACID 3-HYDROXYLASE (S3H) from Arabidopsis (Zhang et al., 2013), Zeilmaker et al. (2015) suggested that AtDMR6 could be an S3H. Moreover, AtDMR6’s spatial expression was specifically detected at sites that are in direct contact with the pathogen (van Damme et al., 2008). However, two FNSI enzymes described in rice (Kim et al., 2008; Lee et al., 2008) show high amino acid identity with AtDMR6, suggesting that this protein instead has FNSI activity in Arabidopsis plants.
Here, we describe the identification and molecular characterization of FNSI enzymes from maize and Arabidopsis. Transcriptional studies indicate that ZmFNSI-1 could be the main enzyme for the synthesis of flavone O-glycosides in tissues expressing P1 and/or the C1+R and B+PL transcription factors. In addition, Arabidopsis transgenic seedlings expressing ZmFNSI-1 accumulate high levels of apigenin, demonstrating that ZmFNSI-1 is an active enzyme in planta. We show that AtDMR6, the ZmFNSI-1 Arabidopsis homologous enzyme, also has FNS activity in Escherichia coli bioconversion and in vitro activity assays. Correlation analyses between AtDMR6 expression and apigenin accumulation in different Arabidopsis tissues provide additional evidence for the in vivo function of AtDMR6. Furthermore, we here demonstrate that while dmr6 plants show loss of susceptibility to P. syringae, ZmFNSI-1 complements the dmr6 mutant phenotype, restoring the susceptibility of dmr6 plants to P. syringae. Our results also suggest that a cross talk exists between the flavone and salicylic acid pathways; in this way, pathogens would induce flavones to decrease salicylic acid and, hence, increase susceptibility. Together, we provide evidence that AtDMR6 is an active FNS enzyme involved in the synthesis of flavones in specific tissues of Arabidopsis plants.
RESULTS
Flavones and Their Derivatives in Maize Plants
Maysin is the main C-glycosyl flavone predominant in silks of some maize varieties, but O-glycosyl flavones are also present in some maize tissues (Ren et al., 2009; Casas et al., 2014; Wen et al., 2014). To determine the main flavone O-glycosides in maize floral tissues, we analyzed the composition of these compounds in maize floral tissues such as maize silks and 14-d-old pericarps lacking (P1-ww) or accumulating (P1-rr) phlobaphene pigments controlled by the maize P1 gene (Grotewold and Peterson, 1994) by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). This analysis shows that silks accumulate apigenin O-hexosides, while pericarps accumulate both apigenin- and luteolin O-hexosides (Table I; Supplemental Fig. S1). Furthermore, the flavone aglycones apigenin and luteolin, which are the precursors for the synthesis of O-glycosyl flavones (Fig. 1), were also identified in these tissues, showing significantly higher levels in P1-rr than in P1-ww silks and pericarps (Supplemental Fig. S1). Thus, for the synthesis of O-glycosyl flavones, our results indicate that maize plants must have bona fide FNS enzymes.
Table I. Accumulation of flavone aglycones and flavone O-glycosides in maize pericarps (14 DAP) and silks determined by LC-MS/MS.
| Retention Time | Precursor Ion [M+H]+ | Tissue | Compound Assignmenta |
|---|---|---|---|
| min | m/z | ||
| 10.8 | 271 | P1-ww silk, P1-rr silk, P1-ww pericarp, P1-rr pericarp | Apigenin |
| 10 | 287 | P1-rr silk | Luteolin |
| 3.2, 4 | 449 | P1-rr pericarp | Luteolin O-hexoside |
| 3.8 | 433 | P1-rr pericarp, P1-rr silk | Apigenin O-hexoside |
Identification was based on MS/MS fragmentations using standards as references.
Identification of a Putative FNSI Enzyme in Maize
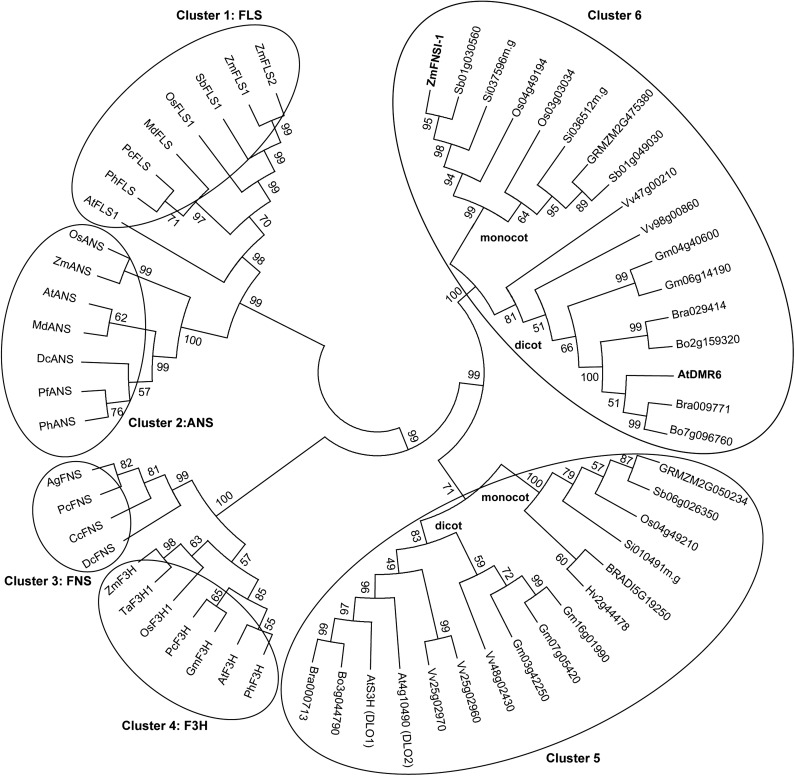
In rice, there are reports of the presence of soluble FNSI enzymes (Kim et al., 2008; Lee et al., 2008). Therefore, since rice is evolutionarily close to maize, we searched for putative maize FNSI enzymes using the two characterized FNSI enzymes from rice in the maize genome sequence using protein sequence homology algorithms. This resulted in the identification of one putative maize FNSI (referred to here as ZmFNSI-1). ZmFNSI-1 sequence alignment with the two rice FNSI proteins showed 83.6% and 78% amino acid identity, suggesting that the maize protein could, in fact, have FNSI activity (Supplemental Fig. S2A). However, comparison of the parsley (Petroselinum crispum) protein and other FNS sequences from Apiaceae species with the maize protein sequence showed that the amino acid identity was only approximately 32% (Supplemental Fig. S2B). Interestingly, the putative maize protein and the two rice FNSI enzymes also show high amino acid identity (59.5% for maize ZmFNSI-1 and 62.2% and 61.9% for the rice proteins) with the DMR6 protein from Arabidopsis (Supplemental Fig. S2A). The sequence of the putative ZmFNSI-1 protein was used in phylogenetic reconstructions with several other plant 2-ODD proteins, primarily involved in phenolic secondary metabolism, including enzymes for flavonoid biosynthesis. The tree shows six well-defined clusters characterized by the enzymatic activity of some of the enzymes included (Fig. 2). Enzymes in cluster 1 are FLS proteins; cluster 2 includes ANS enzymes; clusters 3 and 4 are FNSI and F3H enzymes, respectively. Finally, cluster 5 includes the characterized S3H (DMR6-like oxygenase1 [DLO1]) from Arabidopsis and its paralog DLO2, along with their orthologs in monocot and dicot plants, and cluster 6 groups the two rice FNSI proteins and also their orthologs in monocot and dicot plants. From this analysis, it is clear that ZmFNSI-1 groups with the rice FNSI proteins (Fig. 2) and that this group clusters apart from the FNSI enzymes characterized from the Apiaceae.
Figure 2.
Phylogenetic analysis of 2-oxoglutarate-Fe2+ dioxygenases involved in phenolic secondary metabolism, including enzymes from flavonoid biosynthesis. The phylogenetic tree was constructed with the aligned protein sequences with MEGA (version 5.10; Tamura et al., 2011) using the neighbor-joining method with bootstrap (10,000 replicates). The protein sequences were aligned using ClustalW implemented in MEGA 5.10. Different oxoglutarate-Fe2+ dioxygenases are clustered in circles based on their major demonstrated activities.
ZmFNSI-1 Can Convert Flavanones to Flavones
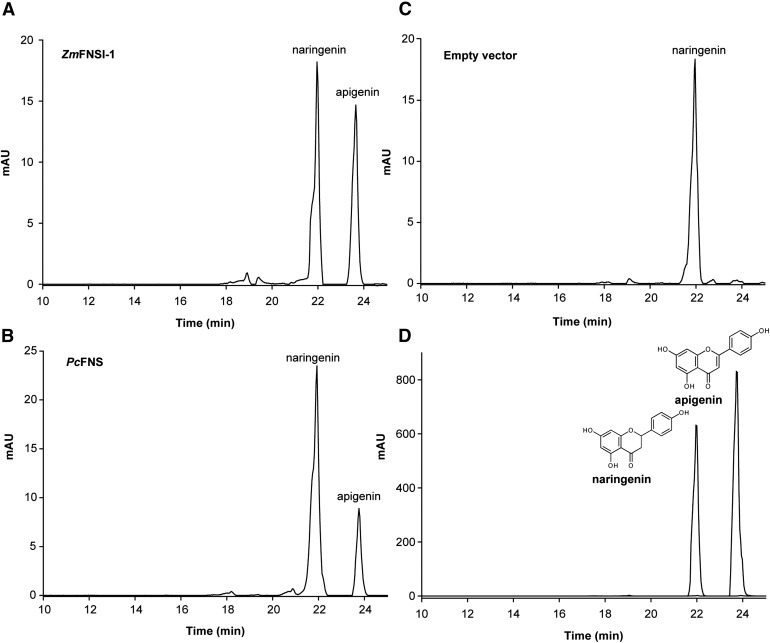
To determine if ZmFNSI-1 encodes a flavone synthase, we investigated its ability to convert flavanones into the respective flavones. For this aim, the full open reading frame was cloned in the pET28a vector, and the protein was expressed in E. coli as an N-terminal fusion protein with a His-6 tag as described in “Materials and Methods.” Activity was assayed by feeding different flavonoids as substrates (as described in Supplemental Table S1) to E. coli cultures expressing ZmFNSI-1. After a 2-d fermentation assay, phenolics were extracted with organic solvent and products were analyzed by liquid chromatography-mass spectrometry (LC-MS). Of all the putative compounds tested as substrates, only naringenin and eriodictyol yielded their corresponding products, apigenin and luteolin (Fig. 3; Supplemental Figs. S3 and S4), which were verified by comparison with commercial standards by LC-MS/MS. The negative control E. coli containing the empty vector did not show the production of any detectable products (Fig. 3; Supplemental Figs. S3 and S4). As a positive control for FNS activity, assays using the recombinant protein PcFNSI were performed (Martens et al., 2001).
Figure 3.
Production of flavones in E. coli expressing ZmFNSI-1. HPLC profiles show FNS activity products in bioconversion assays in E. coli cultures expressing ZmFNSI-1 (A) and PcFNS (B) as a positive control or transformed with the empty vector (C) supplemented with naringenin, showing apigenin production. Naringenin and apigenin standards were used as controls (D). mAU, Milliabsorbance units.
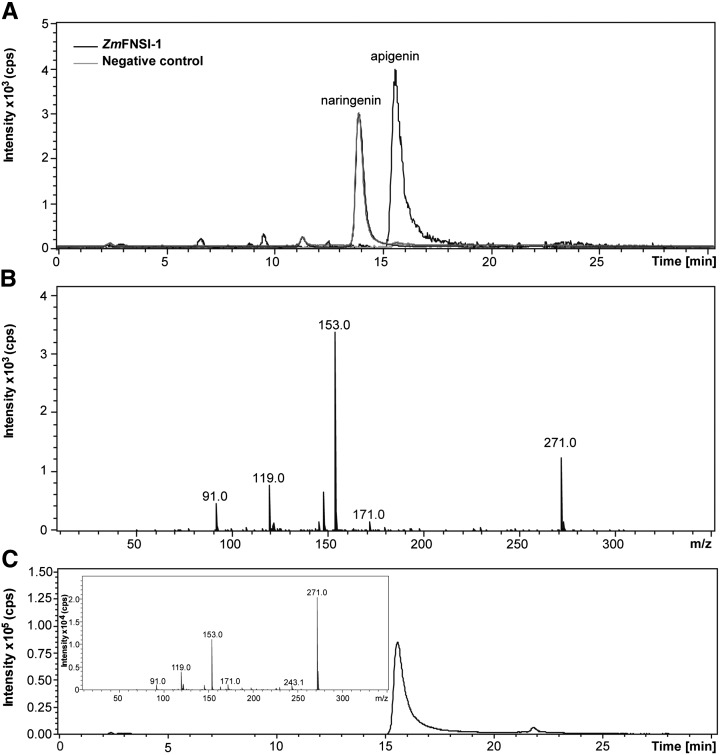
To corroborate the ZmFNSI-1 activity detected in the bioconversion assays in E. coli, the fusion protein was purified using Ni2+-affinity chromatography, and the enzymatic activity of His-6-ZmFNSI-1 was assayed in vitro. The identification of the apigenin product when naringenin was assayed as a substrate by LC-MS analysis using apigenin as a standard confirmed the flavone synthase activity for ZmFNSI-1 (Fig. 4). Moreover, to obtain a comparative estimation of the ZmFNSI-1 activity with the parsley FNS enzyme, we quantified apigenin produced by in vitro enzymatic activity assays with naringenin as a substrate by integration of the peak areas of apigenin in HPLC analysis. Then, the data obtained by integration of the peaks for known amounts of the apigenin standard were compared with the peak areas of the product of enzymatic activities, and these values were used to calculate the specific activity of each enzyme. The PcFNSI enzyme showed a specific activity of 19.45 nmol min−1 mg−1, while the specific activity of ZmFNSI-1 was 15.44 nmol min−1 mg−1 using naringenin as a substrate. On the other hand, when the enzymatic assays were repeated using the different flavonoids described in Supplemental Table S1 as substrates, no product was detected. Together, these results indicate that ZmFNSI-1 is indeed an FNS enzyme with activity comparable to the characterized FNS enzyme from parsley.
Figure 4.
In vitro ZmFNSI-1 activity assayed with naringenin as a substrate. A, LC-MS analysis of purified His-6-ZmFNSI-1 activity. The reaction product generated a molecular ion of mass-to-charge ratio (m/z) = 271 corresponding to apigenin (positive ion chromatogram). A negative control (without protein) did not show the production of any product. B, MS/MS fragmentation profile of the product of the purified His-6-ZmFNSI-1 activity assay. C, Apigenin standard was used as a control. Its MS/MS fragmentation profile corresponds to that of the His-6-ZmFNSI-1 reaction product, which is shown in the inset.
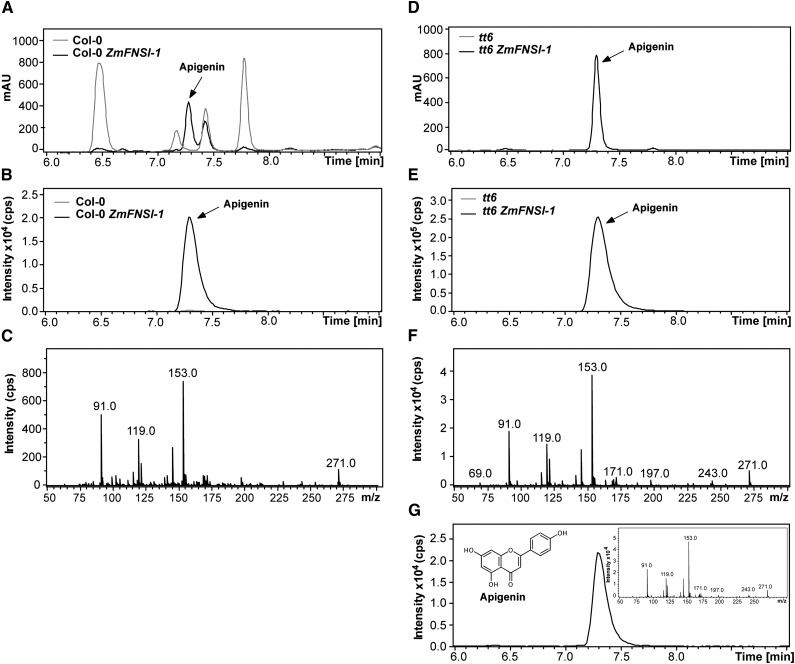
To determine the FNSI activity of ZmFNSI-1 in planta, we transformed Arabidopsis wild-type Columbia-0 (Col-0) and transparent testa6 (tt6) plants, which are mutants in the FLAVANONE 3-HYDROXYLASE gene and accumulate naringenin (Fig. 1), one of the FNS substrates, with ZmFNSI-1 expressed from the constitutive cauliflower mosaic virus 35S promoter (p35S::ZmFNSI-1). Hygromycin-resistant transformed plants were selected, and the presence of the transgene in both types of transgenic plants was examined by PCR analysis of genomic DNA (Supplemental Fig. S5). Accumulation of ZmFNSI-1 mRNA in the transformed seedlings was verified by reverse transcription (RT)-PCR (Supplemental Fig. S5). Then, we investigated flavone accumulation in 15-d-old seedling plants by LC-MS/MS. Apigenin profiles were compared between transgenic plants expressing ZmFNSI-1 (Col-0 ZmFNSI-1 and tt6 ZmFNSI-1), wild type (Col-0), and tt6 mutant plants. While several peaks absorbing in the UV light region were detected in Col-0 plants (Fig. 5A), none of them corresponded to apigenin (Fig. 5B), while tt6 mutants did not show the presence of any detectable UV light-absorbing peaks (Fig. 5D). However, both sets of transgenic plants accumulated apigenin, which was confirmed by comparison of tandem mass spectrometry (MS/MS) fragmentation profiles with the corresponding commercial standard (Fig. 5). It is noteworthy that some peaks (6.5 and 7.8 min; Fig. 5A) present in Col-0 plants with absorbance in the UV light region were not detected in the transgenic plants. Although they were not identified, they likely correspond to phenolic metabolites whose levels are modified by the expression of ZmFNSI-1 in Col-0 plants. Taken together, these results confirm the role of ZmFNSI-1 as a bona fide type I flavone synthase.
Figure 5.
Apigenin determination in Arabidopsis transgenic plants expressing ZmFNSI-1. A, B, D, and E, UV light (A and D) and m/z = 271 ion chromatogram (B and E) profiles of 15-d-old plants: Col-0 (A and B) and tt6 mutants (D and E) and the same plants expressing ZmFNSI-1 (Col-0 ZmFNSI-1 [A and B] and tt6 ZmFNSI-1 [D and E]). C and F, MS/MS fragmentation profiles of the molecular ion m/z = 271 detected in ZmFNSI-1-transformed Arabidopsis plants in Col-0 and tt6 mutant backgrounds. G, Ion chromatogram of the apigenin standard. Its MS/MS fragmentation profile is shown in the inset. mAU, Milliabsorbance units.
Expression Analysis of ZmFNSI-1
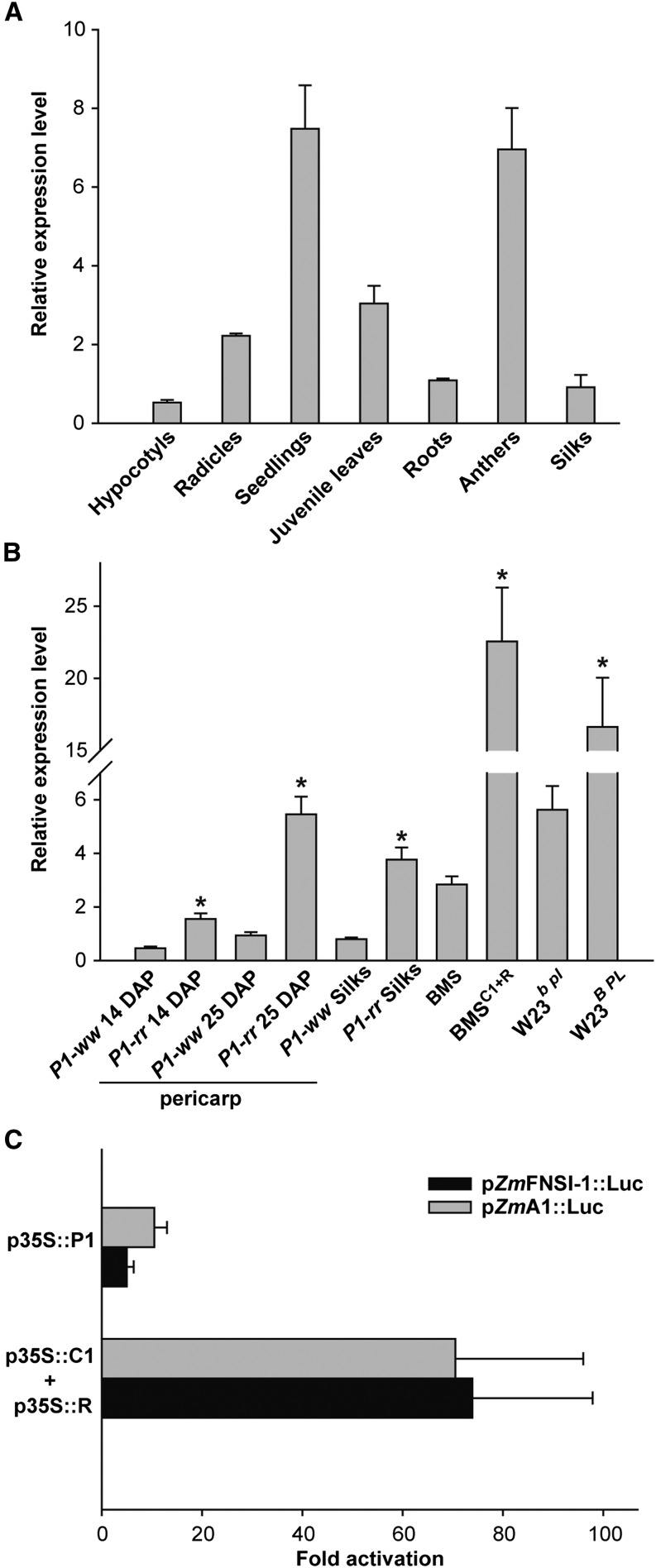
To explore the tissue-specific pattern of expression of ZmFNSI-1, we conducted quantitative reverse transcription (qRT)-PCR on RNA extracted from different maize tissues. Also, the regulation of this transcript by the flavonoid regulators was investigated. For this aim, silks and 14- and 25-d-old pericarps lacking (P1-ww) or accumulating (P1-rr) the phlobaphene pigments controlled by the maize P1 gene (Grotewold and Peterson, 1994), Black Mexican Sweet (BMS) maize cells ectopically expressing the C1+R anthocyanin regulators (BMSC1+R; Grotewold et al., 1998) and untransformed controls (BMS), and leaves from W23 plants expressing or not the B and PL anthocyanin transcription factors in leaves were used.
Transcripts for ZmFNSI-1 were detected in all of the tissues analyzed, with the highest expression found in young seedlings (Fig. 6A). Moreover, ZmFNSI-1 transcripts were present at significantly higher levels in P1-rr compared with P1-ww silks and pericarps, suggesting a regulation of ZmFNS1-1 by P1. More notable was the differential expression between BMS and BMSC1+R and between W23b pl and W23B PL, indicating that ZmFNSI-1 is also under the control of the C1+R and B+PL anthocyanin regulators (Fig. 6B).
Figure 6.
Expression analysis of ZmFNSI-1. A, ZmFNSI-1 relative expression in different maize tissues of the B73 inbred line. B, ZmFNSI-1 relative expression in 15- and 25-d-old pericarps and silks expressing or not the P1 transcription factor (P1-rr and P1-ww), BMS maize cells ectopically expressing the C1+R regulators [BMSC1+R] and untransformed controls (BMS), and juvenile leaves from the W23 line expressing or not the B and PL transcription factors (W23b pl and W23B PL). Asterisks indicate significant (P < 0.05) differences between P1-rr and P1-ww, BMS and BMS C1+R, and W23b pl and W23B PL lines. For all qRT-PCR assays, each reaction was normalized using the cycle threshold values corresponding to the ACTIN1 mRNA (J01238). C, Transient expression following bombardment of BMS cells with p35S::P1 or p35S::C1+p35S::R together with the pZmFNSI-1::Luc reporter construct. A p35S::Renilla construct was included in every bombardment as a normalization control. The pZmA1::Luc construct was used as a positive control. The data were normalized for R. reniformis activity. Each treatment was done in triplicate, and average values of triplicate experiments are shown; the error bars indicate sd.
To better establish the mechanisms by which ZmFNSI-1 expression is modulated, we amplified a 1.5-kb fragment from the translation start codon of ZmFNSI-1 by PCR from B73 genomic DNA (Supplemental Fig. S6) and cloned it upstream of the LUCIFERASE reporter, to generate the pZmFNSI-1::Luc construct. The regulation of pZmFNSI-1::Luc by P1 and C1+R was investigated in BMS cells by bombarding the regulators driven by the p35S promoter (p35S::P1 or p35S::C1+p35S::R) together with the corresponding reporter construct (e.g. pZmFNSI-1::Luc) and the bombardment normalization control, p35S::Renilla. The activation of pZmFNSI-1::Luc was compared with that of pA1::Luc, previously shown to be robustly activated by C1+R and P1 (Grotewold and Peterson, 1994; Sainz et al., 1997; Hernandez et al., 2007; Falcone Ferreyra et al., 2010). As shown in Figure 6C, pZmFNSI-1::Luc is activated by P1 and C1+R at levels similar to pA1::Luc. Consistent with similar results on other flavonoid promoters (Grotewold and Peterson, 1994), R alone did not activate pZmFNSI-1::Luc expression (data not shown). Overall, these results indicate that ZmFNSI-1 is regulated in maize by the flavonoid regulators P1, C1/PL, and R/B.
AtDMR6 Has FNS Activity
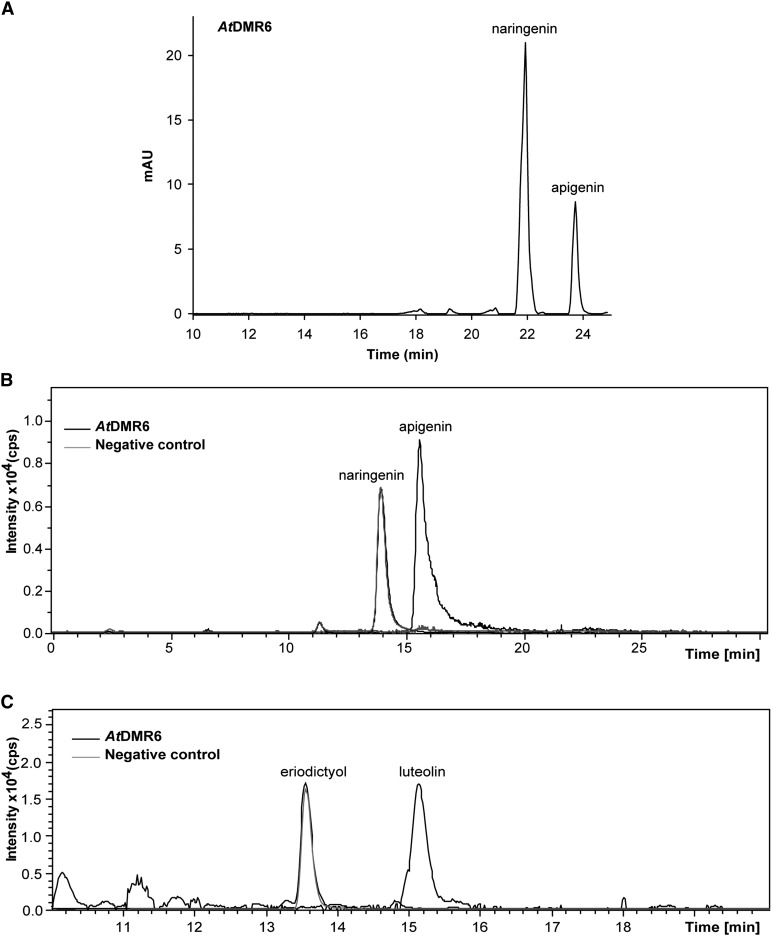
Previously, the Arabidopsis protein AtDMR6 was suggested to have S3H activity, because dmr6 mutants accumulate higher levels of salicylic acid than wild-type plants and because AtDMR6 is 51% identical at its amino acid level with DLO1, a characterized Arabidopsis S3H (van Damme et al., 2008; Zhang et al., 2013; Zeilmaker et al., 2015). Nevertheless, two rice FNSIs show significantly high amino acid identity with AtDMR6 (Kim et al., 2008; Lee et al., 2008). Moreover, ZmFNSI-1 also has high amino acid sequence identity with AtDMR6 (59.5%; Supplemental Fig. S2A). Furthermore, phylogenetic analysis of the proteins involved in phenolic secondary metabolism (Fig. 2) shows that ZmFNSI-1 and AtDMR6 are grouped in the same cluster together with the two rice FNSIs, while AtS3H (DLO1) and DLO2 are clearly grouped in a different cluster, suggesting that AtDMR6 could be an FNSI enzyme. Thus, to investigate the enzymatic properties of AtDMR6, the full open reading frame was cloned in the pET28a vector. The protein was expressed in E. coli as an N-terminal fusion protein with a His-6 tag. Activity was assayed in bioconversion assays by feeding the putative substrates to E. coli cultures expressing AtDMR6 and also in in vitro assays with the purified His-6-AtDMR6 protein using Ni2+-affinity chromatography. Products were analyzed by HPLC (Fig. 7A) and LC-MS/MS (Fig. 7, B and C). As we determined for ZmFNSI-1, from all different compounds tested as putative substrates (Supplemental Table S1), only naringenin and eriodictyol yielded their corresponding products apigenin and luteolin (Fig. 7), while the enzyme failed to metabolize salicylic acid under the conditions tested (Supplemental Fig. S7). The negative E. coli control containing the empty vector did not show the production of any of the compounds (Fig. 7). Furthermore, as described above for ZmFNSI-1, apigenin produced by in vitro enzymatic activity assays with naringenin as substrate was quantified by integration of the peak area of apigenin produced in HPLC analysis, and this value was used to calculate AtDMR6 specific activity. The AtDMR6 enzyme showed a specific activity of 10.86 nmol min−1 mg−1 using naringenin as a substrate, comparable to that of the PcFNSI enzyme (19.45 nmol min−1 mg−1), indicating that AtDMR6 has similar FNS activity to the characterized FNS enzyme from parsley. Hence, by using the recombinant protein in a heterologous system and by in vitro assays, AtDMR6 is able to convert flavanones to flavones.
Figure 7.
AtDMR6 shows FNS activity. A, HPLC profiles of FNS activity in E. coli bioconversion assays. E. coli cultures transformed with the empty vector or expressing AtDMR6 were supplemented with naringenin showing apigenin production. B, In vitro AtDMR6 activity assayed with naringenin as a substrate. LC-MS analysis is shown for purified His-6-AtDMR6 activity; the reaction product generated a molecular ion of m/z = 271 corresponding to apigenin. A negative control (without protein) did not show the production of any product. C, LC-MS analysis of His-6-AtDMR6 activity assayed with eriodictyol as a substrate. The ion chromatogram shows bioconversion in E. coli-expressing AtDMR6 fed with eriodictyol. The reaction product generated a molecular ion of m/z 287 corresponding to luteolin, while E. coli cells transformed with the empty vector did not show the production of the product peak. mAU, Milliabsorbance units.
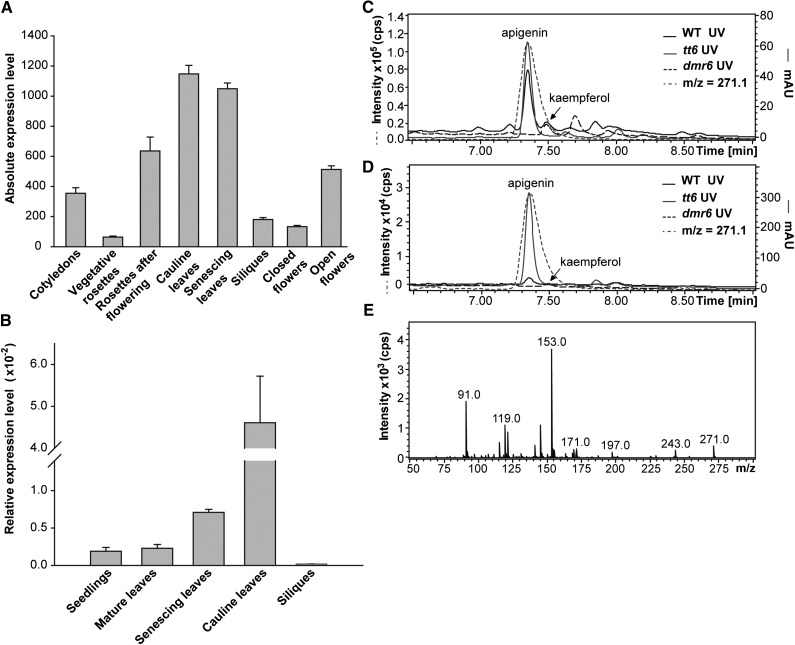
These results led us to investigate whether AtDMR6 could be an active FNS enzyme in certain tissues or under particular developmental stages in Arabidopsis. Therefore, we analyzed the expression pattern of AtDMR6 during different developmental stages using eFP Browser (Fig. 8A; Winter et al., 2007). We established that AtDMR6 has high expression in cauline and senescing leaves. To validate this finding, we further analyzed AtDMR6 expression in different tissues by qRT-PCR. Our results confirmed that AtDMR6 is highly expressed in these leaves (Fig. 8B). Based on these results, we investigated by LC-MS/MS whether Arabidopsis cauline and senescing leaves accumulate apigenin. As presented in Figure 8, C and D, apigenin is in fact accumulated in these organs in both wild-type and tt6 mutant plants, while this compound was not detected in the same organs of the dmr6 mutants. The identity of the apigenin peak was confirmed by comparison of its MS/MS fragmentation profile with the corresponding apigenin commercial standard (Fig. 5G). Furthermore, to gain knowledge about apigenin levels in cauline and senescing leaves from Arabidopsis plants, we compared by LC-MS/MS the amount of this metabolite with that of the flavonol kaempferol, which is usually accumulated in this species (Yonekura-Sakakibara et al., 2008; Stracke et al., 2010). As expected, kaempferol was not detected in tt6 mutants (lacking F3H activity), while this compound was detected in cauline leaves of both the wild type and dmr6 mutants (Fig. 8, C and D; Supplemental Fig. S8). While apigenin was not identified in dmr6 mutants, this flavone concentration in wild-type plants was 5-fold higher than the kaempferol levels. In senescing leaves, we detected kaempferol in dmr6 mutants that do not accumulate apigenin, in contrast to wild-type plants, where apigenin accumulation is notable but kaempferol is almost undetectable. Thus, according to the AtDMR6 expression pattern, FNS activity, and flavone analysis of cauline and senescing leaves, AtDMR6 would be responsible for the synthesis of apigenin in these organs and probably in others where AtDMR6 is expressed, while its low expression in other tissues could explain why apigenin has not been detected previously in Arabidopsis plants.
Figure 8.
AtDMR6 expression analysis and accumulation of apigenin in Arabidopsis. A, AtDMR6 expression levels during plant development. Microarrays data were retrieved from the Arabidopsis eFP Browser (Winter et al., 2007). B, AtDMR6 expression levels in different Arabidopsis tissues determined by RT-quantitative PCR (qPCR). The relative AtDMR6 transcript abundance was analyzed in seedlings (2 weeks old), mature leaves (4 weeks old), senescing leaves (42 d after pollination [DAP]), cauline leaves, and siliques. Each reaction was normalized using the cycle threshold values corresponding to the UBIQUITIN10 (UBQ10) mRNA. The means of the results obtained using three independent RNAs as templates are shown, and the error bars indicate sd. C and D, UV light and ion chromatogram profiles of hydrolyzed flavonoids of cauline (C) and senescing (D) leaves from wild-type (WT), tt6, and dmr6 Arabidopsis plants analyzed by LC-MS/MS showing the m/z = 271 molecular ion in wild-type and tt6 plants that corresponds to apigenin. The peak at 7.5 min that corresponds to kaempferol is also indicated. mAU, Milliabsorbance units. E, MS/MS fragmentation profile of the detected peaks with the molecular ion of m/z = 271.
Overexpression of ZmFNSI-1 Restores the Susceptibility of the dmr6 Mutants to Pathogen Infection
In order to analyze if ZmFNSI-1 can restore the susceptibility of dmr6 mutants to pathogen infection, ZmFNSI-1 was expressed under the control of the 35S promoter and transformed into dmr6-1 mutant plants. To test susceptibility, three independent lines were infected with P. syringae pv tomato DC3000 as described in “Materials and Methods.” While dmr6-1 mutant plants showed resistance to the pathogen, transgenic plants (p35S::ZmFNSI-1) were highly susceptible to the pathogen infection (Zeilmaker et al., 2015; Supplemental Fig. S9), similar to wild-type Landsberg erecta (Ler) plants, indicating that ZmFNSI-1 complements the dmr6 mutant phenotype, restoring susceptibility to P. syringae pv tomato DC3000 infection.
DISCUSSION
Different reports, including this work, have shown that, despite the fact that C-glycosyl flavones are the predominant flavones in maize, this species also accumulates O-glycosyl flavones in different tissues (Ren et al., 2009; Casas et al., 2014; Wen et al., 2014; Table I; Supplemental Fig. S1). We recently demonstrated that C-glycosyl flavone biosynthesis involved 2-hydroxylation of flavanones by ZmF2H1 followed by C-glycosylation by UGT708A6 (Falcone Ferreyra et al., 2013). Moreover, the O-glycosyl flavone biosynthetic pathway was previously demonstrated in rice, showing that the generation of the flavone aglycones occurs first, and then the O-glycosylation takes place (Brazier-Hicks et al., 2009; Lam et al., 2014). Thus, for the synthesis of O-glycosyl flavones, besides the already characterized ZmF2H1, maize plants should also express at least one FNS protein. However, no enzyme of this class was characterized previously in this species. Here, we describe the cloning and molecular characterization of one FNSI enzyme from maize. The only previous reports of such activity in a monocot plant species is in rice, where the presence of two FNSI activities was reported (Kim et al., 2008; Lee et al., 2008); however, in planta, the two putative OsFNSIs failed to produce flavones (Lam et al., 2014). The lack of FNSI activity of the rice proteins in Arabidopsis may be because these two enzymes may fail to interact with other Arabidopsis enzymes involved in flavonoid biosynthesis, being unable to form functional macromolecular complexes required for substrate channeling; alternatively, the affinity for naringenin or its availability could be limiting their in vivo activities. Also, it is possible that low expression levels of the transgenes may result in undetectable levels of apigenin production. Despite this, we here demonstrate that ZmFNSI-1 is capable of converting naringenin and eriodictyol to the corresponding flavones, apigenin and luteolin.
We also here describe that P1-rr pericarps and silks accumulate higher levels of both apigenin and luteolin compared with P1-ww pericarps and silks (Supplemental Fig. S1), suggesting that there must be an FNS activity regulated by P1 in these tissues that could be responsible for the differential accumulation of these flavones. In fact, ZmFNSI-1 transcripts are expressed at significantly higher levels in P1-rr compared with P1-ww silks and pericarps, and transient expression following bombardment of BMS cells with p35S::P1 together with the pZmFNS1-1::Luc reporter construct also showed that ZmFNSI-1 is regulated by P1 (Fig. 6). The results described (Table I; Supplemental Fig. S1) are substantially supported by data recently reported by Casas et al. (2014). Flavone aglycone and flavone O-glycoside contents were quantified both in whole maize kernels expressing or not the P1 transcription factor (P1-rr and P1-ww) as well as in P1-rr kernel mutants in the A1 gene (encoding dihydroflavonol reductase, involved in anthocyanin and flavan 4-ol biosynthesis; P1-rr;a1). Higher levels of flavone and flavone O-glycosides were detected in P1-rr than in P1-ww pericarps, similar to the results presented here (Table I; Supplemental Fig. S1); while P1-rr;a1 kernels showed the highest flavone levels, as this mutant has a higher pool of the intermediate naringenin for flavone formation. These results demonstrated that maize accumulates flavone O-glycosides and the aglycone form, suggesting that there is a bona fide maize flavone synthase that uses flavanones to generate flavones and that have to be expressed in P1-ww but with higher transcript levels in P1-rr. Thus, the expression data of ZmFNSI-1 in pericarps, and its regulation by P1 (Fig. 6) shown in this work, suggest that ZmFNSI-1 is the main active enzyme in maize kernels responsible for flavone O-glycoside formation.
In addition, ZmFNSI-1 is also under the control of the C1+R and B+PL anthocyanin regulators (Fig. 6). It is interesting that ZmFNSI-1 expression is under the control of both P1 and C1+R and B+PL. Flavone biosynthesis in maize has been studied mainly in pericarps and floral tissues like silks, where this pathway is known to be regulated by P1 but not by the anthocyanin regulators (Grotewold et al., 1991, 1998; Morohashi et al., 2012; Casas et al., 2014). However, we previously demonstrated that ZmFLS1 and ZmFLS2, involved in flavonol biosynthesis, are also regulated by both types of transcription factors (Falcone Ferreyra et al., 2012). Complexes between anthocyanins and flavones were found in flowers, leaves, and seeds in different plant species, with flavones acting as copigments (Ishikura, 1981; Goto and Kondo, 1991; Tanaka et al., 1998; Fossen et al., 2007; Shiono et al., 2008; Kalisz et al., 2013; Tanaka and Brugliera, 2013). Consequently, we hypothesize that the regulation of flavone biosynthesis by the same regulators as those for anthocyanin biosynthesis could be necessary to maintain similar levels of both types of flavonoids in particular tissues or conditions, for example, to stabilize anthocyanins, as was suggested in Torenia spp. (Ueyama et al., 2002). Thus, although our results indicate that ZmFNSI-1 could be the main enzyme for the synthesis of O-glycosyl flavones in tissues expressing P1 and/or the C1+R and B+PL transcription factors, we cannot rule out that other enzymes with FNS activity can contribute to the pool of flavones in these and other tissues. Accordingly, we recently found that ZmCYP93G7 (ZmFNSII-1) is also able to convert flavanones into flavones (M.L. Falcone Ferreyra, E.J. Rodriguez, and P. Casati, unpublished data). Nevertheless, the expression of this gene is not P1 regulated (Morohashi et al., 2012) and shows lower transcript levels in pericarps and whole seeds in comparison with ZmFNSI-1 (maize eFP browser; http://bar.utoronto.ca/efp_maize/cgi-bin/efpWeb.cgi; M.L. Falcone Ferreyra and P. Casati, unpublished data), suggesting that ZmCYP93G7 may participate in flavone synthesis in other tissues where P1 is not expressed. Moreover, the ZmFNSI-1 paralogous gene (GRMZM2G475380; Fig. 2), which was not studied in this work, may also contribute to flavone biosynthesis. Finally, it is worth mentioning that, recently, Lan et al. (2015) demonstrated that tricin (a flavone constituted by apigenin with additional modifications such as hydroxylations and methylations) is a structural monomer of lignin polymers in maize. In this way, both ZmFNSI-1 expression and apigenin content in pericarps suggest that ZmFNSI-1 could have a role in lignin biosynthesis in these tissues, providing the structural unit, apigenin.
As described above, there are reports of two different FNSI activities from rice (Kim et al., 2008; Lee et al., 2008). However, in planta, the two putative rice FNSIs failed to produce flavones (Lam et al., 2014). ZmFNSI-1 shows high amino acid similarity to both rice proteins but very low similarity with the characterized PcFNSI from the Apiaceae. However, ZmFNSI-1 has similar enzymatic activity to PcFNSI, determined by bioconversion assays in E. coli, in in vitro assays using the purified enzyme, and in transgenic Arabidopsis plants expressing ZmFNSI-1, suggesting that the evolution of FNSI enzymes in plants may have occurred independently at least twice from a suitable 2-ODD. Interestingly, ZmFNSI-1 also has significant amino acid similarity with AtDMR6 (59.5% identity at the amino acid level). This protein was not characterized previously, and it is required for susceptibility to downy mildew. The Arabidopsis mutant dmr6 carries a recessive mutation that results in the loss of susceptibility to H. parasitica, H. arabidopsidis, P. capsici, and P. syringae, suggesting that AtDMR6 has a role during plant defense (van Damme et al., 2008; Zeilmaker et al., 2015). Moreover, DMR6 spatial expression was specifically detected to the sites that are in direct contact with the pathogen (van Damme et al., 2008). Interestingly, when dmr6-1 mutants from the Ler background were complemented with AtDMR6 from Col-0 plants, this fully restored the susceptibility of Arabidopsis to H. parasitica (van Damme et al., 2008), suggesting that AtDMR6 from different backgrounds of Arabidopsis have the same enzymatic activity. The overexpression of ZmFNSI-1 in drm6-1 mutant plants resulted in reduced resistance to P. syringae; therefore, ZmFNSI-1 is able to restore susceptibility to pathogen infection (Supplemental Fig. S9). In summary, these results demonstrate that ZmFNSI-1 and AtDMR6 have the same activity in planta.
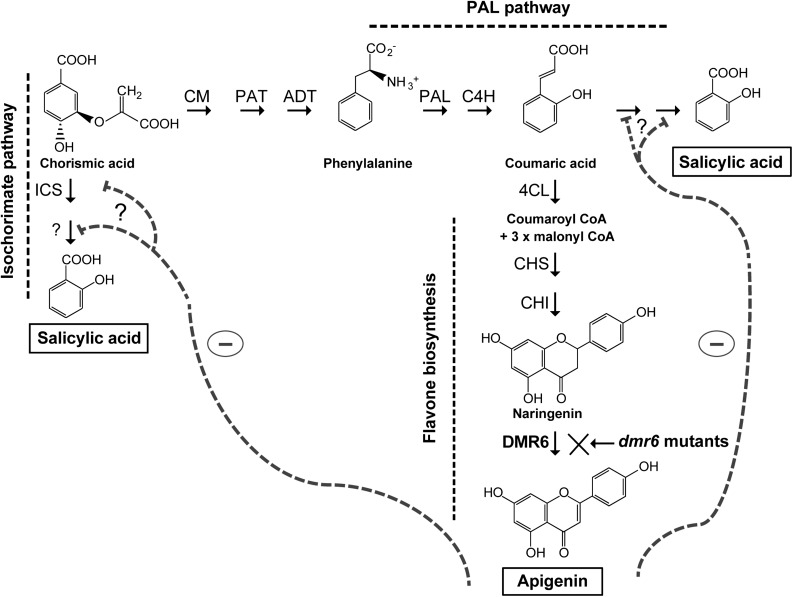
Interestingly, dmr6 mutants accumulate higher levels of salicylic acid than wild-type plants (Zeilmaker et al., 2015), and AtDMR6 expression is also induced by salicylic acid (Arabidopsis eFP Browser; Winter et al., 2007); therefore, Zeilmaker et al. (2015) suggested that AtDMR6 may be an S3H, because it is 51% identical at its amino acid level to a characterized Arabidopsis S3H (Zhang et al., 2013). However, our results failed to detect any such activity in assays using salicylic acid as a substrate (Supplemental Table S1; Supplemental Fig. S7). Moreover, phylogenetic analysis of proteins involved in phenolic specialized metabolism, including enzymes from flavonoid biosynthesis, shows that AtDMR6 groups with ZmFNSI-1 and two rice FNSIs (Fig. 2), and these may represent new types of FNSI enzymes present in both monocot and dicot plants. On the contrary, S3H (DLO1) and DLO2 from Arabidopsis are grouped in a different cluster with other proteins from both monocot and dicot species; this cluster could represent enzymes involved in salicylic acid catabolism. Our hypothesis is that, in the dmr6 mutants, a minor flow through the flavonoid pathway would lead to a higher availability of substrates for salicylic acid biosynthesis (Fig. 9), increasing the levels of salicylic acid and, as a consequence, the loss of susceptibility to pathogens. Interestingly, in dmr6 mutant cauline leaves that do not accumulate apigenin (Fig. 8C), salicylic acid levels are substantially higher than in wild-type Ler cauline leaves that accumulate apigenin (Supplemental Fig. S8D). Moreover, salicylic acid levels in dmr6 cauline leaves are in the same order of magnitude as apigenin levels in wild-type cauline leaves, strengthening the hypothesis that, in the dmr6 mutants, a minor flow through the flavonoid pathway would lead to a higher availability of substrates for salicylic acid biosynthesis (Fig. 9), increasing the levels of salicylic acid and, as a consequence, the loss of susceptibility to pathogens.
Figure 9.
Model for salicylic acid and apigenin accumulation in Arabidopsis plants. Proposed routes for the biosynthesis of salicylic acid via the isochorismatic and Phe ammonium lyase (PAL) pathways, and the flavonoid pathway to apigenin biosynthesis, are shown. In dmr6 mutant plants, a minor flow through the flavonoid pathway would lead to increased availability of substrates for salicylic acid biosynthesis. Apigenin could inhibit the activity of enzymes or the transcription of genes in the salicylic acid biosynthesis pathway (represented by gray dashed lines). Enzymes are abbreviated as follows: arogenate dehydratase (ADT), cinnamate 4-hydroxylase (C4H), 4-coumarate:CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), chorismate mutase (CM), and isochorismate synthase (ICS).
The cross talk between flavonoid and phytohormone pathways has been reported several times. For example, in Arabidopsis, jasmonate biosynthetic genes are highly induced when there is an increased flux through the flavonoid pathway (Pourcel et al., 2013), while mutants in the CHALCONE SYNTHASE gene display an elevated auxin transport in young seedlings, roots, and inflorescences (Buer and Muday, 2004). Thus, it was proposed that flavonoids could function as buffering molecules under biotic and abiotic stress conditions (Pourcel et al., 2013). There are at least two different proposed routes for the biosynthesis of salicylic acid, one via the isochorismatic pathway and the second through the Phe ammonium lyase pathway (Dempsey et al., 2011); the flavonoid pathway for the synthesis of apigenin biosynthesis also derives from Phe through the phenylpropanoid biosynthetic pathway. In this way, in wild-type plants, AtDMR6 probably rewires metabolites into the flavone pathway to lower their availability for salicylic acid in defense responses, making these plants more susceptible to pathogen attack. Moreover, we can hypothesize that apigenin could act as an endogenous modulator of salicylic acid levels; for example, flavones could act as inhibitors of the activity of enzymes in salicylic acid biosynthesis or, alternatively, they could negatively regulate the transcription of genes encoding these enzymes (Fig. 9). Indeed, it was recently reported that the mechanism by which the flavone apigenin exerts its antiinflammatory activity in macrophages is through the down-regulation of microRNA-155 induced by infection (Arango et al., 2015). Interestingly, AtDMR6 is induced by salicylic acid and AtDMR6 is expressed around the infection site during pathogen attack, so a complex feedback inhibition regulation by flavones may exist to balance levels of salicylic acid. Thus, our hypothesis is that a cross talk between the flavone and salicylic acid pathways takes place in Arabidopsis, so pathogens would induce flavones to decrease salicylic acid and, hence, increase susceptibility.
Our data show that AtDMR6 has FNSI activity using both naringenin and eriodictyol as substrates in bioconversion assays in E. coli and in in vitro assays using the purified enzyme, suggesting that this enzyme may also have this role in planta. Our results are controversial, as flavones seemed to be absent in Arabidopsis (Martens and Mithöfer, 2005). However, the presence of two flavone derivatives was recently reported in Col-0 Arabidopsis leaves, apigenin 7-2″,3″-diacetylglucoside and pentamethoxydihydroxy flavone, although they were erroneously grouped as flavonols (Ali and McNear, 2014). In our experiments, we demonstrated that AtDMR6 expression by RT-qPCR is high in cauline and senescing leaves, while its expression is low in seedlings and mature leaves (Fig. 8); in cauline and senescing leaves, this type of flavonoid accumulation has not been analyzed previously. Accordingly, LC-MS/MS analysis of cauline and senescing leaves from wild-type and tt6 mutant plants, which have increased naringenin availability, showed significant apigenin accumulation (Fig. 8). Hence, AtDMR6 may be the enzyme responsible for the synthesis of this compound in some Arabidopsis tissues and under specific conditions, such as after a pathogen attack (Zeilmaker et al., 2015). It is interesting that AtDMR6 expression is induced in senescing leaves where apigenin is accumulated. Anthocyanin biosynthesis also increases during senescence (Hoch et al., 2001; Ougham et al., 2005; Thomas et al., 2009); thus, flavones may stabilize anthocyanins acting as copigments during this process, as found in flowers (Goto and Kondo, 1991; Tanaka et al., 1998; Shiono et al., 2008; Kalisz et al., 2013; Tanaka and Brugliera, 2013). Alternatively, flavones may have a role in chlorophyll degradation, as was reported previously (Yamauchi and Watada, 1994). Future experiments will reveal the role of apigenin in specific tissues as well as under particular situations, for example, after cold, osmotic stress, UV-B radiation, high calcium concentrations, and salicylic acid treatment, all conditions where AtDMR6 gene expression is induced (Arabidopsis eFP Browser; Krinke et al., 2007; Winter et al., 2007; Chan et al., 2008; Sivitz et al., 2008). A close similarity exists between senescence and responses to abiotic and biotic stresses (John et al., 2001; Allu et al., 2014), which is in favor of apigenin playing roles in both processes. Overall, in this work, we were able to identify and characterize an FNSI and detect the product of its activity, apigenin, in Arabidopsis and maize plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Maize (Zea mays) B73 seeds were obtained from the Instituto Nacional de Tecnología Agropecuaria. The two near-isogenic lines that differ in flavonoid phenotype, W23B PL and W23b pl, correspond to those described previously (Casati and Walbot, 2003). The generation and analysis of the BMS cells expressing p35S::C1 and p35S::R were described previously (Grotewold et al., 1998). Pericarps (14 and 25 DAP) from near-isogenic lines containing the P1-rr and P1-ww alleles were described previously (Morohashi et al., 2012). Maize plants were grown in greenhouse conditions with supplemental visible lighting to 1,000 μE m−2 s−1 with 15 h of light and 9 h of dark. Samples were collected from hypocotyls, radicles (3-d-old plants), roots (21-d-old plants), seedlings (7-d-old plants), juvenile leaves (21-d-old plants), and anthers and silks (3 d post emergence).
Arabidopsis (Arabidopsis thaliana) plants from the Col-0 ecotype, the Ler ecotype, dmr6 (dmr6-1; background Ler) and tt6 (tt6-1; background Ler) mutants, and transgenic p35S::ZmFNSI-1 plants were grown in a growth chamber under light (100 μE m−2 s−1) with a 16-h-light/8-h-dark photoperiod after a cold treatment (72 h at 4°C in the dark). Temperature and humidity were maintained at 23°C and 50%, respectively. The tt6 mutant seeds were obtained from the Arabidopsis Biological Resource Center. The dmr6-1 mutant line was described by van Damme et al. (2008). This mutant, generated by ethyl methanesulfonate mutagenesis, has a single point mutation in the second exon (G to A) that causes the change from TGG (Trp codon) to TGA (premature stop codon), so a trunked protein with 141 amino acids is generated, without the essential catalytic domain for its activity. For flavonoid content analysis in transgenic plants, Arabidopsis plants were germinated and grown for 15 d in Murashige and Skoog plant salt mixture and 0.8% (w/v) agar.
Gene Expression Analysis by RT-qPCR
Tissues from three independent biological replicates were frozen in liquid nitrogen and stored at −80°C. Total RNA was extracted following the Trizol protocol (Invitrogen) followed by DNase treatment (Promega). Complementary DNAs (cDNAs) were synthesized from 4 μg of total RNA using SuperScript Reverse Transcription Enzyme II (Invitrogen) with oligo(dT) as a primer. The resulting cDNAs were used as templates for qPCR in the iCycler iQ detection system with the Optical System Software version 3.0a (Bio-Rad), using the intercalation dye SYBR Green I (Invitrogen) as a fluorescent reporter and Platinum Taq Polymerase (Invitrogen). Primers were designed to generate unique 150- to 250-bp fragments using the PRIMER3 software (Rozen and Skaletsky, 2000). Three biological replicates were used for each sample plus a negative control (reaction without reverse transcriptase). To normalize the data, primers for ACTIN1 (J01238) and AtUBQ10 were used for maize and Arabidopsis, respectively (Supplemental Table S2). Amplification conditions were as follows: 2 min of denaturation at 94°C; 40 to 45 cycles at 94°C for 10 s, 57°C for 15 s, and 72°C for 20 s; followed by 5 min at 72°C. Melting curves for each PCR product were determined by measuring the decrease of fluorescence with increasing temperature (from 65°C–95°C). To confirm the sizes of the PCR products and to check that they corresponded to a unique and expected PCR product, the final PCR products were separated on a 2% (w/v) agarose gel, stained with SYBR Green (Invitrogen), and also sequenced. Primers used for ZmFNSI-1 and AtDMR6 are listed in Supplemental Table S2.
Transient Expression Experiments in Maize BMS Cells
The p35S::C1+p35S::R, p35S::P1, p35S::Renilla, p35S::BAR, and pA1::Luc plasmids have all been described previously (Grotewold and Peterson, 1994; Sainz et al., 1997; Hernandez et al., 2004, 2007). To generate the pZmFNSI-1::Luc construct, a 1.5-kb fragment from the translation start codon of ZmFNSI-1 was amplified by PCR from B73 genomic DNA (Supplemental Fig. S6), and it was cloned upstream of the luciferase reporter. Bombardment conditions of maize BMS suspension cells and transient expression assays for luciferase and Renilla reniformis were performed as described previously (Feller et al., 2006; Hernandez et al., 2007). Bombardments were performed in triplicate, and each experiment was repeated at least three times. The assays for firefly (Photinus pyralis) luciferase and R. reniformis luciferase and the normalization of the data were performed as described previously (Hernandez et al., 2007). The fold activation results are expressed as the ratio of arbitrary light units (luciferase) to arbitrary light units (R. reniformis) of the treatment with the transcriptional activator, divided by the ratio of arbitrary light units (luciferase) to arbitrary light units (R. reniformis) of the reporter plasmid in the absence of the regulator.
Cloning of cDNAs and Heterologous Expression
The full-length open reading frame for ZmFNSI-1 (GRMZM2G09967) was amplified by PCR using the primers ZmFNSI-forward and ZmFNSI-reverse1 designed based on the sequence provided by the maize genome sequence (www.maizesequence.org; release 5b.60). The forward primer (ZmFNSI-forward) included the start codon (ZmFNSI-reverse1; for sequences, see Supplemental Table S2). PCR was performed with Platinum Pfx polymerase (Invitrogen) under the following conditions: 1× Pfx buffer, 1× enhancer, 1.5 mm MgSO4, 0.5 mm of each deoxyribonucleotide triphosphate (dNTP), 0.5 mm of each primer, 0.3 units of Platinum Pfx polymerase, cDNA from B73 leaves, and sterile water added to obtain a volume of 20 μL. Cycling conditions were as follows: 30 s of denaturation at 95°C, 30 s of annealing at 65°C, 90 s of amplification at 68°C, with a 1°C decrement of annealing temperature in each cycle until it reached 55°C, followed by 25 cycles of 30 s of denaturation at 95°C, 30 s of annealing at 54°C, and 90 s of amplification at 68°C. PCR product was cloned into pENTR-D-TOPO generating the plasmid pENTR-ZmFNSI-1, sequenced, and recombined into the Gateway site of the pGWB2 binary vector (Karimi et al., 2002), resulting in p35S::ZmFNSI-1.
To express ZmFNSI-1 in Escherichia coli, full-length ZmFNSI cDNA was reamplified by PCR using the pENTR-ZmFNSI vector as a template. The primers ZmFNSI-NdeI-forward and ZmFNSI-BamHI-forward with the NdeI and BamHI restriction sites, respectively, were used for further cloning (Supplemental Table S2). The amplified product was purified, cut with the corresponding NdeI and BamHI restriction enzymes, purified, and cloned into pET28a (Novagen), generating the vector pET28-ZmFNSI.
The full-length open reading frame for AtDMR6 was amplified from cDNA obtained from leaf tissues of wild-type Arabidopsis plants (Col-0). The primers AtDMR6-NdeI-forward and AtDMR6-BamHI-reverse with the NdeI and BamHI restriction sites, respectively, were used for further cloning (Supplemental Table S2). The amplified product was purified, cloned into pGEM-T-Easy vector (Promega), and sequenced. The NdeI-BamHI fragment was further subcloned into pET28a, generating the pET28-AtDMR6 construct. For both amplifications, PCR was performed using GoTaq (Promega) and Pfu polymerases (Invitrogen; 10:1) under the following conditions: 1× buffer, 1.5 mm MgCl2, 0.5 μm of each primer, and 0.5 mm of each dNTP, in 25 μL of final volume.
E. coli BL21-(DE3)-pLys cells were transformed with the construct pET28-ZmFNSI and the empty vector pET28, while recombinant AtDMR6 and PcFNS were expressed in E. coli Rosetta2-(DE3) cells. Cell cultures (Luria-Bertani medium containing 30 mg L−1 kanamycin and 35 mg L−1 chloramphenicol) were grown at 37°C until the optical density at 600 nm reached 0.5 to 0.6, and the recombinant protein expression was achieved by induction with 0.5 mm isopropylthio-β-galactoside for 20 h at 22°C for AtDMR6, whereas ZmFNS1 and PcFNS expression was performed at 30°C for 20 h.
Purification of ZmFNSI-1 and AtDMR6 Proteins
For the purification of His-6-ZmFNSI-1 and His-6-AtDMR6, cells were harvested by centrifugation at 3,000g for 20 min at 4°C. Pellets were resuspended in binding buffer (50 mm sodium phosphate, pH 7.5, 500 mm NaCl, 20 mm imidazole, and 5% [v/v] glycerol) containing 0.1% (v/v) Tween 20, 1 mm phenylmethylsulfonyl fluoride, and complete EDTA-free protease inhibitor cocktail (Thermo). Cells were disrupted by sonication and then centrifuged at 12,000g for 20 min at 4°C to obtain soluble cell extracts. The protein was bound to a nickel-nitrilotriacetic acid agarose resin (Invitrogen) by rocking at 4°C for 1 h, and then the resin was loaded onto a column, washed three times with 15 volumes of binding buffer followed by three washes with 7 volumes of washing buffer (50 mm sodium phosphate, pH 7.5, 500 mm NaCl, 5% [v/v] glycerol, and 40 mm imidazole). Elution was carried out by five sequential additions of 1 mL of elution buffer (50 mm sodium phosphate, pH 7.5, 500 mm NaCl, 5% [v/v] glycerol, and 200 mm imidazole). Finally, the recombinant protein was desalted in desalting buffer (100 mm NaH2PO4, pH 6.8, 10 mm ascorbate, 0.25 mm ferrous sulfate, and 10% [v/v] glycerol) by four cycles of concentration and dilution using Amicon Ultra-15 30K (Millipore) and stored at −80°C. Total protein concentration was determined by the method of Bradford (1976).
Bioconversion Experiments
For in vivo E. coli activity assays, BL21(DE3)-pLys cells harboring pET28-ZmFNSI or empty pET28a plasmids and Rosetta2-(DE3) cells harboring pET28-AtDMR6, pET15-PcFNS, or empty pET28a plasmids were grown at 37°C in Luria-Bertani medium with appropriate antibiotics. Recombinant protein expression was induced by the addition of 0.5 mm isopropylthio-β-galactoside as described above, and cultures were simultaneously supplemented with 80 μg mL−1 of the different flavonoids (Supplemental Table S1). Cultures were grown at 30°C or 22°C for 24 to 48 h and then centrifuged at 15,000g for 5 min. One milliliter of medium aliquots was extracted with ethyl acetate, vacuum dried, and resuspended in methanol for subsequent HPLC and LC-MS analyses. Recombinant PcFNS1 was a gift from Stefan Martens and was used as a positive control in the bioconversion experiments.
In Vitro Activity Assays
The reaction mixture contained 100 mm NaH2PO4, pH 6.8, 10 mm α-ketoglutaric acid (disodium salt), 10 mm ascorbic acid, 0.25 mm ferrous sulfate, 100 μg mL−1 naringenin, and 5 µg of recombinant purified protein in a final volume of 100 µL. The ferrous sulfate solution was prepared in 100 mm sodium acetate, pH 5.5, containing 10 mm ascorbic acid to inhibit the oxidation of Fe2+. Reactions were initiated by the addition of the enzyme and terminated by extraction with ethyl acetate. Activity assays were performed at 30°C for 60 min in open tubes with shaking. S3H activity assays were done as described by Zhang et al. (2013).
HPLC and LC-MS Analyses
HPLC was performed using ÄKTA basic 10/100 equipment (Amersham Bioscience) and a Phenomenex LUNA C18 column (150 × 4.6 mm, 5 μm). Data were collected and analyzed using the UNICORN control system program (version 3.0). Compound separation was by linear gradient elution from 20% (v/v) methanol:80% (v/v) 10 mm ammonium acetate, pH 5.6, to 100% methanol at a flow rate of 0.75 mL min−1. Absorbances were detected at 292 and 340 nm using the UV900 detector (Amersham Bioscience). Retention times of the products analyzed were compared with those of authentic commercial standards (Sigma-Aldrich). The data obtained by integration of the peaks for known amounts of the apigenin standard were compared with the peak areas of the products of each enzymatic activity assay for the quantification of FNS activities.
Reaction products were analyzed by LC-MS using a system consisting of an Agilent 1100 HPLC pump and a Bruker microTOF-Q II mass spectrometer in a positive ion mode-configured LC-MS device with a turbo-ion spray source and collision energy of 25 eV. Samples (10 µL) were chromatographed on a Phenomenex Hypersil GOLD C18 column (3 µm, 2 × 150 mm) at 200 µL min−1 with a linear gradient from 20% (v/v) methanol:80% (v/v) 10 mm ammonium acetate, pH 5.6, to 100% (v/v) methanol over 30 min. Alternatively, to separate substrates from products, a linear gradient from 20% (v/v) acetonitrile to 100% (v/v) in 0.1% (v/v) formic acid over 30 min was used. The eluate was delivered unsplit into the mass spectrometer source. Compounds were identified by comparison of mass spectra with those of authentic commercial standards (Sigma-Aldrich and Indofine). Absorbance units were detected at 295, 330, and 360 nm.
Plant Transformation
The p35S::ZmFNSI-1 construct was transformed into Agrobacterium tumefaciens strain GV3101 by electroporation, and the transformation of Arabidopsis by the resulting bacteria was performed by the floral dip method (Clough and Bent, 1998). Transformed seedlings (T1) were identified by selection on solid Murashige and Skoog plant salt mixture, pH 5.7, and 0.8% (w/v) agar containing hygromycin (30 mg L−1), and the plants were then transferred to soil. The presence of the ZmFNS1 transgene in transformed plants was analyzed by PCR on the genomic DNA from 15-d-old seedlings using the primers ZmFNSI-forward-RT and ZmFNSI-reverse1 (product size of 734 bp). The expression of the ZmFNSI-1 transgene in transformed plants was analyzed in 15-d-old seedlings by RT-PCR using the primers ZmFNSI-forward and ZmFNSI-reverse1 (product size of 1,008 bp; for corresponding primer sequences, see Supplemental Table S2). Primers for CAP-BINDING PROTEIN20 (CBP20) were used as a control. PCR conditions were as follows: 1× buffer GoTaq, 2.5 mm MgCl2, 0.2 mm dNTP, 0.25 μm of each primer, 0.625 units of GoTaq (Promega), and sterile water added to obtain a volume of 25 μL. Cycling conditions were as follows: 2 min of denaturation at 95°C, followed by 35 cycles of 15 s of denaturation at 95°C, 20 s of annealing at 55°C, 1 min of amplification at 72°C, and 7 min of amplification at 72°C. PCR products were separated on a 1% (w/v) agarose gel and stained with SYBR Green (Invitrogen).
Pathogen Infection
Pathogen infection was done as described by Katagiri et al. (2002). Arabidopsis plants used for infection (wild-type Ler, dmr6-1 mutants, and transgenic p35S::ZmFNSI-1 plants in the dmr6-1 background) were grown as described above. Pseudomonas syringae pv tomato DC3000 inoculation was performed on 3-week-old plants (before flowering) by spraying with a bacterial suspension containing 2.5 × 107 colony-forming units mL−1 in water with 0.025% (v/v) Silwet L-77. Samples (four plants per line and three leaves per plant) were collected after inoculation (0 d) and 3 d post inoculation to count colony-forming units. Three independent lines of transgenic plants were analyzed.
Extraction of Total Flavonoid from Maize Silks and Pericarps and Arabidopsis Leaves
Flavonoid extraction from maize tissues was performed as described previously (Casati and Walbot, 2005). Fresh silks and 14- and 25-DAP pericarps were rinsed with water and lyophilized for 1 d. Dry weight was measured and ground to a powder with a mortar and pestle. The powder was extracted for 8 h with 12 volumes of acidic methanol (1% [v/v] HCl in methanol), followed by a second extraction with 12 volumes of chloroform and 6 volumes of distilled water. The extracts were vortexed and centrifuged for 2 min at 3,000g, and organic phases were collected. Flavonoid extracts were analyzed by LC-MS.
Frozen Arabidopsis tissues were homogenized in extraction solvent (50% [v/v] methanol in water) to get a final suspension of 50 mg mL−1 (w/v), and then the samples were sonicated in an ultrasonic water bath at room temperature for 1 h. The resulting extract was centrifuged at 15,000g for 5 min at 4°C, and the supernatant was then hydrolyzed by the addition of an equal volume of 2 n HCl, followed by incubation at 95°C for 1 h. Flavonoid extracts were analyzed by LC-MS as described above.
Cloning of the ZmFNSI-1 Promoter
To amplify the ZmFNSI-1 promoter, primers were designed to amplify a 1.5-kb fragment upstream of the start of the translation codon, as predicted from www.maizesequence.com. Restriction sites, NotI and KpnI, were included in the forward and reverse primers, respectively (ZmFNSI-NotI-prom-F2 and ZmFNSI-Kpn-prom-R2; Supplemental Table S2). Genomic DNA was isolated from leaf tissue using a DNA isolation kit (Qiagen). PCR was performed with PhusionTaq Polymerase (BioLab) in the following conditions: 1× High fidelity or guanine and cytosine-rich buffer, 0.3 mm dimethyl sulfoxide, 1.5 mm MgCl2, 0.5 mm of each primer, 0.5 mm of each dNTP, 100 ng of genomic DNA, and 0.3 units of Phusion Taq polymerase in a volume of 50 μL. Cycling conditions were as follows: 30 s of denaturation at 95°C, 20 s of annealing at 68°C, 90 s of amplification at 72°C, with a 1°C decrement of annealing temperature in each cycle until it reached 58°C, followed by 25 cycles of 30 s of denaturation at 95°C, 20 s of annealing at 58°C, and 90 s of amplification at 72°C. The PCR products were purified from the gels, cut with the corresponding restriction enzymes, and purified. The pA1::Luc construct (pMSZ011; Sainz et al., 1997) was restricted with NotI and KpnI, and the A1 promoter was replaced by the ZmFNSI-1 promoter, resulting in the pZmFNSI::Luc construct.
Phylogenetic Analysis
The tree was constructed using the MEGA 5.1 software with the neighbor-joining method based on ClustalW multiple alignments (Tamura et al., 2011). The following plant 2-ODD sequences were analyzed: PcFLS (parsley [Petroselinum crispum]; AAP57395), PhFLS (Petunia hybrida; CAA80264), MdFLS (Malus domestica; AY965343), AtFLS1 (Arabidopsis; AAB41504), OsFLS1 (rice [Oryza sativa]; BAD17324), SbFLS1 (Sorghum bicolor; EES07584), ZmFLS1 (maize; NP_001140915), PcF3H (AAP57394), PhF3H (CAA43027), AtF3H (TT6; CAD37988), OsF3H1 (NP_001054157), GmF3H (Glycine max; AAU06217), TaF3H1 (Triticum aestivum; ABB20895), ZmF3H (NP_001105695), AgFNS I (Apium graveolens; AXX21537), PcFNS I (AY817680), DcFNS (Daucus carota; AAX21542), CcFNS (Cuminum cyminum; BG78790), OsANS (CAA69252), ZmANS (NP001105074), AtANS (CAD91994), PhANS (P51092), PfANS (Perilla frutescens; BAA20143), and AtDMR6 (NP_197841.1). The search for orthologous genes was performed using the Plaza program (PLAZA 3.0; Proost et al., 2015). Other sequence data for this analysis can be found at http://ensembl.gramene.org/Brassica_oleracea, http://ensembl.gramene.org/Brassica_rapa, http://ensembl.gramene.org/Glycine_max, http://ensembl.gramene.org/Vitis_vinifera, http://ensembl.gramene.org/Arabidopsis_thaliana, http://ensembl.gramene.org/Setaria_italica, http://ensembl.gramene.org/Hordeum_vulgare, http://ensembl.gramene.org/Oryza_sativa, and http://ensembl.gramene.org/Brachypodium_distachyon.
Statistical Analysis
The data presented were analyzed using Student’s t test (P < 0.05) and one-way ANOVA in the Sigma Stat Package. Significant differences are indicated with asterisks or different letters.
Sequence data from this article can be found in the maize genome sequence database (version 3b.60 at maizesequence.org) under accession numbers ZmFNSI-1 (GRMZM2G099467) and ZmACTIN1 (J01238) and in the Arabidopsis Genome Initiative database under accession numbers AtDMR6, At5g24530; CBP20, At5g44200; and AtUBQ10, At4g05320.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. LC-MS/MS analysis of flavone-derived metabolites in maize pericarps (14 DAP) and silks expressing or not the P1 gene (P1-rr and P1-ww).
Supplemental Figure S2. Amino acid sequences alignment of the predicted ZmFNSI-1 with FNS proteins from rice and Arabidopsis plants and Apiaceae.
Supplemental Figure S3. LC-MS analysis of ZmFNSI-1 activity products in E. coli bioconversion assays using naringenin as a substrate.
Supplemental Figure S4. LC-MS analysis of ZmFNSI-1 activity assayed with eriodictyol as a substrate.
Supplemental Figure S5. Presence and expression of the ZmFNSI-1 transgene in transformed Arabidopsis plants.
Supplemental Figure S6. ZmFNSI-1 promoter sequence from the B73 maize line.
Supplemental Figure S7. In vitro AtDMR6 activity assayed with salicylic acid as a substrate.
Supplemental Figure S8. Accumulation of kaempferol, apigenin, and salicylic acid in Arabidopsis.
Supplemental Figure S9. ZmFNSI-1 complements susceptibility of Arabidopsis dmr6-1 mutant plants towards P. syringae pv tomato DC3000 (Pst).
Supplemental Table S1. Substrates tested in bioconversion assays in E. coli expressing ZmFNSI-1 and AtDMR6 and in vitro assays with the recombinant proteins.
Supplemental Table S2. Primers used for cloning, RT-qPCR, and screening.
Supplementary Material
Acknowledgments
We thank Dr. Stefan Martens (Istituto Agrario di San Michele all'Adige, Fondazione Edmund Mach, San Michele all'Adige, Italy) for providing the pET-PcFNS1 construct and for comments on the results; M. Isabel Casas (Molecular Cellular and Developmental Biology, Ohio State University) for providing silks and pericarp maize tissues; Dr. Guido Van den Ackerveken (Department of Biology, Universiteit Utrecht) for providing dmr6-1 mutant seeds; Dr. Adriana Krapp (Instituto de Biología Molecular y Celular de Rosario-Consejo Nacional de Investigaciones Científicas y Técnicas) for providing the P. syringae pv tomato DC3000 strain; and Carla Berosich, a biotechnology student at the Centro de Estudios Fotosintéticos y Bioquímicos, for her contribution to the identification of Arabidopsis transgenic plants and flavonoid extraction from Arabidopsis tissues.
Glossary
- LC-MS/MS
liquid chromatography coupled to tandem mass spectrometry
- LC-MS
liquid chromatography-mass spectrometry
- Col-0
Columbia-0
- RT
reverse transcription
- MS/MS
tandem mass spectrometry
- qRT
quantitative reverse transcription
- Ler
Landsberg erecta
- qPCR
quantitative PCR
- DAP
days after pollination
- cDNA
complementary DNA
- dNTP
deoxyribonucleotide triphosphate
- m/z
mass-to-charge ratio
- BMS
Black Mexican Sweet
Footnotes
This work was supported by Fondo para la Investigación Científica y Tecnológica (grant nos. PICT 2010–00105 and PICT 2013–268 to P.C. and E.G. and grant no. PICT 2013–0082 to M.L.F.F.), by the Agriculture and Food Research Initiative of the U.S. Department of Agriculture National Institute of Food and Agriculture (grant no. 2015–67013–22810), and by the U.S. National Science Foundation (grant no. IOS–1125620 to E.G.).
Articles can be viewed without a subscription.
References
- Akashi T, Fukuchi-Mizutani M, Aoki T, Ueyama Y, Yonekura-Sakakibara K, Tanaka Y, Kusumi T, Ayabe S (1999) Molecular cloning and biochemical characterization of a novel cytochrome P450, flavone synthase II, that catalyzes direct conversion of flavanones to flavones. Plant Cell Physiol 40: 1182–1186 [DOI] [PubMed] [Google Scholar]
- Ali MB, McNear DH Jr (2014) Induced transcriptional profiling of phenylpropanoid pathway genes increased flavonoid and lignin content in Arabidopsis leaves in response to microbial products. BMC Plant Biol 14: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allu AD, Soja AM, Wu A, Szymanski J, Balazadeh S (2014) Salt stress and senescence: identification of cross-talk regulatory components. J Exp Bot 65: 3993–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango D, Diosa-Toro M, Rojas-Hernandez LS, Cooperstone JL, Schwartz SJ, Mo X, Jiang J, Schmittgen TD, Doseff AI (2015) Dietary apigenin reduces LPS-induced expression of miR-155 restoring immune balance during inflammation. Mol Nutr Food Res 59: 763–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango D, Morohashi K, Yilmaz A, Kuramochi K, Parihar A, Brahimaj B, Grotewold E, Doseff AI (2013) Molecular basis for the action of a dietary flavonoid revealed by the comprehensive identification of apigenin human targets. Proc Natl Acad Sci USA 110: E2153–E2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek I, Jeon SB, Song MJ, Yang E, Sohn UD, Kim IK (2009) Flavone attenuates vascular contractions by inhibiting RhoA/Rho kinase pathway. Korean J Physiol Pharmacol 13: 201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontempo P, Mita L, Miceli M, Doto A, Nebbioso A, De Bellis F, Conte M, Minichiello A, Manzo F, Carafa V, et al. (2007) Feijoa sellowiana derived natural flavone exerts anti-cancer action displaying HDAC inhibitory activities. Int J Biochem Cell Biol 39: 1902–1914 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brazier-Hicks M, Evans KM, Gershater MC, Puschmann H, Steel PG, Edwards R (2009) The C-glycosylation of flavonoids in cereals. J Biol Chem 284: 17926–17934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Muday GK (2004) The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16: 1191–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Boocock DJ, Steward WP, Gescher AJ (2007) Tissue distribution in mice and metabolism in murine and human liver of apigenin and tricin, flavones with putative cancer chemopreventive properties. Cancer Chemother Pharmacol 60: 257–266 [DOI] [PubMed] [Google Scholar]
- Casas MI, Duarte S, Doseff AI, Grotewold E (2014) Flavone-rich maize: an opportunity to improve the nutritional value of an important commodity crop. Front Plant Sci 5: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Walbot V (2003) Gene expression profiling in response to ultraviolet radiation in maize genotypes with varying flavonoid content. Plant Physiol 132: 1739–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Walbot V (2005) Differential accumulation of maysin and rhamnosylisoorientin in leaves of high-altitudes landraces of maize after UV-B exposure. Plant Cell Environ 28: 788–799 [Google Scholar]
- Chan CW, Wohlbach DJ, Rodesch MJ, Sussman MR (2008) Transcriptional changes in response to growth of Arabidopsis in high external calcium. FEBS Lett 582: 967–976 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cone KC, Cocciolone SM, Burr FA, Burr B (1993) Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5: 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajas F, Andrés AC, Florencia A, Carolina E, Felicia RM (2013) Neuroprotective actions of flavones and flavonols: mechanisms and relationship to flavonoid structural features. Cent Nerv Syst Agents Med Chem 13: 30–35 [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF (2011) Salicylic acid biosynthesis and metabolism. The Arabidopsis Book 9: e0156, doi/10.1199/tab.0156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmarajan SK, Arumugam KM (2012) Comparative evaluation of flavone from Mucuna pruriens and coumarin from Ionidium suffruticosum for hypolipidemic activity in rats fed with high fat diet. Lipids Health Dis 11: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone Ferreyra ML, Casas MI, Questa JI, Herrera AL, Deblasio S, Wang J, Jackson D, Grotewold E, Casati P (2012) Evolution and expression of tandem duplicated maize flavonol synthase genes. Front Plant Sci 3: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone Ferreyra ML, Rius S, Emiliani J, Pourcel L, Feller A, Morohashi K, Casati P, Grotewold E (2010) Cloning and characterization of a UV-B-inducible maize flavonol synthase. Plant J 62: 77–91 [DOI] [PubMed] [Google Scholar]
- Falcone Ferreyra ML, Rodriguez E, Casas MI, Labadie G, Grotewold E, Casati P (2013) Identification of a bifunctional maize C- and O-glucosyltransferase. J Biol Chem 288: 31678–31688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A, Hernandez JM, Grotewold E (2006) An ACT-like domain participates in the dimerization of several plant basic-helix-loop-helix transcription factors. J Biol Chem 281: 28964–28974 [DOI] [PubMed] [Google Scholar]
- Fliegmann J, Furtwängler K, Malterer G, Cantarello C, Schüler G, Ebel J, Mithöfer A (2010) Flavone synthase II (CYP93B16) from soybean (Glycine max L.). Phytochemistry 71: 508–514 [DOI] [PubMed] [Google Scholar]
- Fossen T, Rayyan S, Holmberg MH, Nimtz M, Andersen ØM (2007) Covalent anthocyanin-flavone dimer from leaves of Oxalis triangularis. Phytochemistry 68: 652–662 [DOI] [PubMed] [Google Scholar]
- Gebhardt YH, Witte S, Steuber H, Matern U, Martens S (2007) Evolution of flavone synthase I from parsley flavanone 3β-hydroxylase by site-directed mutagenesis. Plant Physiol 144: 1442–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Cone KC, Chandler VL (1992) Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev 6: 864–875 [DOI] [PubMed] [Google Scholar]
- Goto T, Kondo T (1991) Structure and molecular stacking of anthocyanins: flower color variation. Angew Chem Int Ed Engl 30: 17–33 [Google Scholar]
- Grotewold E, Athma P, Peterson T (1991) Alternatively spliced products of the maize P gene encode proteins with homology to the DNA-binding domain of myb-like transcription factors. Proc Natl Acad Sci USA 88: 4587–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Chamberlin M, Snook M, Siame B, Butler L, Swenson J, Maddock S, St Clair G, Bowen B (1998) Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10: 721–740 [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Peterson T (1994) Isolation and characterization of a maize gene encoding chalcone flavonone isomerase. Mol Gen Genet 242: 1–8 [DOI] [PubMed] [Google Scholar]
- Gueldner RC, Snook ME, Wiseman BR, Widstrom NW, Himmelsbach DS, Costello CE (1989) Maysin in corn teosinte and centripede grass. In Hedin PA, ed, Naturally Occurring Pest Bioregulators: 197th and 198th National Meetings of the American Chemical Society and the 1989 International Chemical Congress of Pacific Basin Societies. American Chemical Society, Washington, DC, pp 251–263 [Google Scholar]
- Harborne JB. (1993) The Flavonoids: Advances in Research Since 1986. Chapman & Hall, London [Google Scholar]
- Hernandez JM, Feller A, Morohashi K, Frame K, Grotewold E (2007) The basic helix loop helix domain of maize R links transcriptional regulation and histone modifications by recruitment of an EMSY-related factor. Proc Natl Acad Sci USA 104: 17222–17227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JM, Heine GF, Irani NG, Feller A, Kim MG, Matulnik T, Chandler VL, Grotewold E (2004) Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. J Biol Chem 279: 48205–48213 [DOI] [PubMed] [Google Scholar]
- Hoch WA, Zeldin EL, McCown BH (2001) Physiological significance of anthocyanins during autumnal leaf senescence. Tree Physiol 21: 1–8 [DOI] [PubMed] [Google Scholar]
- Ishikura N. (1981) Anthocyanins and flavones in leaves and seeds of Perilla plant. Agric Biol Chem 45: 1855–1860 [Google Scholar]
- John CF, Morris K, Jordan BR, Thomas B, A-H-Mackerness S (2001) Ultraviolet-B exposure leads to up-regulation of senescence-associated genes in Arabidopsis thaliana. J Exp Bot 52: 1367–1373 [PubMed] [Google Scholar]
- Kalisz S, Oszmiański J, Hładyszowski J, Mitek M (2013) Stabilization of anthocyanin and skullcap flavone complexes: investigations with computer simulation and experimental methods. Food Chem 138: 491–500 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He SY (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. The Arabidopsis Book 1: e0039, doi/10.1199/tab.0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Cheon YM, Kim BG, Ahn JH (2008) Analysis of flavonoids and characterization of the OsFNS gene involved in flavone biosynthesis in rice. J Plant Biol 51: 97–101 [Google Scholar]
- Kitada C, Gong Z, Tanaka Y, Yamazaki M, Saito K (2001) Differential expression of two cytochrome P450s involved in the biosynthesis of flavones and anthocyanins in chemo-varietal forms of Perilla frutescens. Plant Cell Physiol 42: 1338–1344 [DOI] [PubMed] [Google Scholar]
- Kong CH, Zhao H, Xu XH, Wang P, Gu Y (2007) Activity and allelopathy of soil of flavone O-glycosides from rice. J Agric Food Chem 55: 6007–6012 [DOI] [PubMed] [Google Scholar]
- Krinke O, Ruelland E, Valentová O, Vergnolle C, Renou JP, Taconnat L, Flemr M, Burketová L, Zachowski A (2007) Phosphatidylinositol 4-kinase activation is an early response to salicylic acid in Arabidopsis suspension cells. Plant Physiol 144: 1347–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PY, Zhu FY, Chan WL, Liu H, Lo C (2014) Cytochrome P450 93G1 is a flavone synthase II that channels flavanones to the biosynthesis of tricin O-linked conjugates in rice. Plant Physiol 165: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Lu F, Regner M, Zhu Y, Rencoret J, Ralph SA, Zakai UI, Morreel K, Boerjan W, Ralph J (2015) Tricin, a flavonoid monomer in monocot lignification. Plant Physiol 167: 1284–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Kim JH, Kim BG, Lim Y, Ahn JH (2008) Characterization of flavone synthase I from rice. BMB Rep 41: 68–71 [DOI] [PubMed] [Google Scholar]
- Liu S, Zhang J, Li D, Liu W, Luo X, Zhang R, Li L, Zhao J (2007) Anticancer activity and quantitative analysis of flavone of Cirsium japonicum DC. Nat Prod Res 21: 915–922 [DOI] [PubMed] [Google Scholar]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR (1989) Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci USA 86: 7092–7096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S, Forkmann G (1999) Cloning and expression of flavone synthase II from Gerbera hybrids. Plant J 20: 611–618 [DOI] [PubMed] [Google Scholar]
- Martens S, Forkmann G, Matern U, Lukacin R (2001) Cloning of parsley flavone synthase I. Phytochemistry 58: 43–46 [DOI] [PubMed] [Google Scholar]
- Martens S, Mithöfer A (2005) Flavones and flavone synthases. Phytochemistry 66: 2399–2407 [DOI] [PubMed] [Google Scholar]
- Mathesius U, Schlaman HRM, Spaink HP, Of Sautter C, Rolfe BG, Djordjevic MA (1998) Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J 14: 23–34 [DOI] [PubMed] [Google Scholar]
- McMullen MD, Byrne PF, Snook ME, Wiseman BR, Lee EA, Widstrom NW, Coe EH (1998) Quantitative trait loci and metabolic pathways. Proc Natl Acad Sci USA 95: 1996–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen MD, Kross H, Snook ME, Cortés-Cruz M, Houchins KE, Musket TA, Coe EH Jr (2004) Salmon silk genes contribute to the elucidation of the flavone pathway in maize (Zea mays L.). J Hered 95: 225–233 [DOI] [PubMed] [Google Scholar]
- Morohashi K, Casas MI, Falcone Ferreyra ML, Mejía-Guerra MK, Pourcel L, Yilmaz A, Feller A, Carvalho B, Emiliani J, Rodriguez E, et al. (2012) A genome-wide regulatory framework identifies maize pericarp color1 controlled genes. Plant Cell 24: 2745–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ougham HJ, Morris P, Thomas H (2005) The colors of autumn leaves as symptoms of cellular recycling and defenses against environmental stresses. Curr Top Dev Biol 66: 135–160 [DOI] [PubMed] [Google Scholar]
- Park HS, Lim JH, Kim HJ, Choi HJ, Lee IS (2007) Antioxidant flavone glycosides from the leaves of Sasa borealis. Arch Pharm Res 30: 161–166 [DOI] [PubMed] [Google Scholar]
- Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H (1987) The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J 6: 3553–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters NK, Frost JW, Long SR (1986) A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233: 977–980 [DOI] [PubMed] [Google Scholar]
- Pourcel L, Irani NG, Koo AJK, Bohorquez-Restrepo A, Howe GA, Grotewold E (2013) A chemical complementation approach reveals genes and interactions of flavonoids with other pathways. Plant J 74: 383–397 [DOI] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inzé D, Mueller-Roeber B, Vandepoele K (2015) PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res 43: D974–D981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayyan S, Fossen T, Andersen ØM (2010) Flavone C-glycosides from seeds of fenugreek, Trigonella foenum-graecum L. J Agric Food Chem 58: 7211–7217 [DOI] [PubMed] [Google Scholar]
- Rayyan S, Fossen T, Andersen ØMJ (2005) Flavone C-glycosides from leaves of Oxalis triangularis. J Agric Food Chem 53: 10057–10060 [DOI] [PubMed] [Google Scholar]
- Rector BG, Snook ME, Widstrom NW (2002) Effect of husk characters on resistance to corn earworm (Lepidoptera: Noctuidae) in high-maysin maize populations. J Econ Entomol 95: 1303–1307 [DOI] [PubMed] [Google Scholar]
- Ren SC, Liu ZL, Ding XL (2009) Isolation and identification of two novel flavone glycosides from corn silk (Stigma maydis). J Med Plant Res 3: 1009–1015 [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Sainz MB, Grotewold E, Chandler VL (1997) Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9: 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Hoerner R, Weissenböck G (2003) Contribution of phenolic compounds to the UV-B screening capacity of developing barley primary leaves in relation to DNA damage and repair under elevated UV-B levels. Phytochemistry 64: 243–255 [DOI] [PubMed] [Google Scholar]
- Shiono M, Matsugaki N, Takeda K (2008) Structure of commelinin, a blue complex pigment from the blue flowers of Commelina communis. Proc Jpn Acad Ser B Phys Biol Sci 84: 452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz AB, Reinders A, Ward JM (2008) Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiol 147: 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snook ME, Gueldner RC, Widstrom NW, Wiseman BR, Himmelsbach DS, Harwood JS, Costello CE (1993) Levels of maysin and maysin analogues in silks of maize germplasm. J Agric Food Chem 41: 1481–1485 [Google Scholar]
- Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B (2010) Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol 188: 985–1000 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Brugliera F (2013) Flower colour and cytochromes P450. Philos Trans R Soc Lond B Biol Sci 368: 20120432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Tsuda S, Kusumi T (1998) Metabolic engineering to modify flower color. Plant Cell Physiol 39: 1119–1126 [Google Scholar]
- Thomas H, Huang L, Young M, Ougham H (2009) Evolution of plant senescence. BMC Evol Biol 9: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueyama Y, Suzuki KI, Fukuchi-Mizutani M, Fukui Y, Miyazaki K, Ohkawa H, Kusumi T, Tanaka Y (2002) Molecular and biochemical characterization of torenia flavonoid 3′-hydroxylase and flavone synthase II and modification of flower color by modulating the expression of these genes. Plant Sci 163: 253–263 [Google Scholar]
- van Damme M, Huibers RP, Elberse J, Van den Ackerveken G (2008) Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J 54: 785–793 [DOI] [PubMed] [Google Scholar]
- Wen W, Li D, Li X, Gao Y, Li W, Li H, Liu J, Liu H, Chen W, Luo J, et al. (2014) Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun 5: 3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi N, Watada AE (1994) Effectiveness of various phenolic compounds in degradation of chlorophyll by in vitro peroxidase-hydrogen peroxide system. J Jpn Soc Hortic Sci 63: 439–444 [Google Scholar]
- Yarmolinsky L, Huleihel M, Zaccai M, Ben-Shabat S (2012) Potent antiviral flavone glycosides from Ficus benjamina leaves. Fitoterapia 83: 362–367 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilmaker T, Ludwig NR, Elberse J, Seidl MF, Berke L, Van Doorn A, Schuurink RC, Snel B, Van den Ackerveken G (2015) DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J 81: 210–222 [DOI] [PubMed] [Google Scholar]
- Zhang J, Subramanian S, Zhang Y, Yu O (2007) Flavone synthases from Medicago truncatula are flavanone-2-hydroxylases and are important for nodulation. Plant Physiol 144: 741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Halitschke R, Yin C, Liu CJ, Gan SS (2013) Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc Natl Acad Sci USA 110: 14807–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.