Abstract
Background
Infectious diseases are the leading cause of human morbidity and mortality worldwide. One dramatic issue is the emergence of microbial resistance to antibiotics which is a major public health concern. Surprisingly however, such in vivo adaptive ability has not been reported yet for many intracellular human bacterial pathogens such as Legionella pneumophila.
Methods
We examined 82 unrelated patients with Legionnaire's disease from which 139 respiratory specimens were sampled during hospitalization and antibiotic therapy. We both developed a real time PCR assay and used deep-sequencing approaches to detect antibiotic resistance mutations in L. pneumophila and follow their selection and fate in these samples.
Findings
We identified the in vivo selection of fluoroquinolone resistance mutations in L. pneumophila in two infected patients treated with these antibiotics. By investigating the mutational dynamics in patients, we showed that antibiotic resistance occurred during hospitalization most likely after fluoroquinolone treatment.
Interpretation
In vivo selection of antibiotic resistances in L. pneumophila may be associated with treatment failures and poor prognosis. This hidden resistance must be carefully considered in the therapeutic management of legionellosis patients and in the control of the gradual loss of effectiveness of antibiotics.
Keywords: Legionella pneumophila, Legionellosis, Fluoroquinolone resistance, In vivo selection, Resistance detection, Next generation sequencing
Highlights
-
•
Legionellosis is a pneumonia caused by the inhalation of aerosols containing Legionella, mainly L. pneumophila.
-
•
Its average mortality rate is 10%, despite availability of effective antibiotics such as the macrolides and the fluoroquinolones.
-
•
Using modern molecular approaches, we identified the selection of fluoroquinolone resistance in L. pneumophila in patients under fluoroquinolone therapy.
-
•
This may lead to reduction of treatment efficacy and prognosis worsening.
-
•
Our findings should lead to revised guidelines for therapeutic management and prognosis evaluation of legionellosis.
1. Introduction
Acquired resistance to antibiotics in human bacterial pathogens has been extensively described and is associated with a high public health and economic burden (Martínez, 2008; Skippington and Ragan, 2011; Pendleton et al., 2013). In striking contrast, resistant mutants have been rarely, if ever, detected in humans for many bacterial species with strict or facultative intracellular lifestyles, including Rickettsia spp. (agent of rickettsioses), Coxiella burnetii (Q fever), Ehrlichia and Anaplasma species (ehrlichiosis and anaplasmosis), Chlamydia psittaci (psittacosis), Legionella pneumophila (legionellosis), Francisella tularensis (tularemia), and Brucella melitensis (brucellosis). However, mutants have been easily selected for when these pathogens were grown in vitro (Rolain and Raoult, 2005; Rouli et al., 2012; Maurin et al., 2001; Binet and Maurelli, 2005; Almahmoud et al., 2009; Sutera et al., 2014; Ravanel et al., 2009). These occasional human pathogens thrive in environmental or animal reservoirs, or both, and are not carried by, nor transmitted, among humans.
L. pneumophila is a Gram-negative facultative intracellular bacterium that multiplies in aquatic environments, especially in free-living protozoa and biofilms (Newton et al., 2010; Carratalà and Garcia-Vidal, 2010). Legionellosis is an acute pneumonia caused by inhalation of aerosols containing Legionella sp., with L. pneumophila serogroup (sg) 1 accounting for more than 90% of human infections (Newton et al., 2010; Carratalà and Garcia-Vidal, 2010). The macrolides (especially azithromycin) and the fluoroquinolones are first-line antibiotics for the treatment of legionellosis patients (Chidiac et al., 2012; Jespersen et al., 2010; Yu et al., 2009; Pedro-Botet and Yu, 2009; Burdet et al., 2014). However, treatment failures and relapses may occur despite administration of an appropriate antibiotic therapy (Chidiac et al., 2012; Jespersen et al., 2010; Yu et al., 2009; Pedro-Botet and Yu, 2009; Burdet et al., 2014). The mortality rate of legionellosis is 10% on average, but can be as high as 30% in immunocompromised patients (Chidiac et al., 2012; Jespersen et al., 2010; Yu et al., 2009). Acquired resistance to the macrolides and fluoroquinolones in L. pneumophila has been readily selected for in vitro (Almahmoud et al., 2009; Jonas et al., 2003; Nielsen et al., 2000). In striking contrast, no macrolide resistance (Jespersen et al., 2010; Yu et al., 2009; Pedro-Botet and Yu, 2009; Burdet et al., 2014), and only a single case of fluoroquinolone resistance has been reported in clinical strains (Bruin et al., 2014). In the latter case however, only one patient was studied with only microbiological approaches and, importantly the dynamics of fluoroquinolone-resistance during infection and hospitalization was not investigated. Here, we identified and further characterized, using molecular and genomic approaches, the in vivo selection of fluoroquinolone-resistance mutations in L. pneumophila during a large survey of 82 unrelated legionellosis cases.
2. Methods
2.1. Patients and Clinical Samples
We studied a cohort of 82 unrelated legionellosis patients including 64 males and 18 females (mean age: 55 years, age range: 27–97 years) that were admitted to the Grenoble University Hospital between 2006 and 2011. For diagnostic purposes, 139 lower respiratory tract samples were collected from these patients during hospitalization. After homogenization of the samples (appendix p 3), routine bacteriological tests (bacterial cultures, Legionella detection by culture and PCR) were performed. The homogenized samples were then frozen at − 80 °C. Based on the criteria from the European Centre for Disease Prevention and Control (ECDC, 2012), legionellosis diagnosis was based on the combination of clinical and radiologic findings of pneumonia, and a positive urinary antigen test for L. pneumophila sg1 (Binax Now Legionella, Binax, Portland, ME, USA). In addition, we tried to further confirm diagnosis by isolation of a L. pneumophila strain from respiratory samples and detection of L. pneumophila DNA in the same samples using a previously described quantitative real-time PCR assay (qPCRmip assay) (Maurin et al., 2010).
2.2. Bacterial Strains and Media
We used four reference strains of L. pneumophila sg1 (Paris, Philadelphia, Lens, and Lorraine) (appendix p 5), and 27 strains belonging to 18 Legionella species other than L. pneumophila (appendix p 6), that are all fluoroquinolone-susceptible. We used three fluoroquinolone-resistant mutant strains (LPPI1, LPPI4, and LPPI5) that we previously selected in vitro from the Paris strain (appendix p 5) (Almahmoud et al., 2009). All three have mutations in the gyrA Quinolone Resistance Determinant Region (QRDR). For specificity purposes, we used 33 non-Legionella bacterial strains that were previously isolated from human respiratory samples (appendix p 7). Legionella strains were grown on buffered charcoal yeast extract agar plates (BCYE, Oxoid, Basingstoke, UK) at 37 °C for 48–72 h. The other bacteria were grown on Columbia or chocolate blood agar supplemented with 5% Polyvitex (bioMérieux, Marcy L'Etoile, France).
2.3. Antibiotic Susceptibility Testing
Ciprofloxacin MICs were determined using the E-test method (bioMérieux). A bacterial inoculum of 1 McFarland turbidity standard was prepared in brain–heart infusion (BHI) broth (bioMérieux), and plated on BCYE plates carrying an E-test strip. Plates were incubated for 3–5 days at 35 °C in a 5%-CO2 atmosphere. The MIC is given on the strip at the inhibitory zone edge intersection (Rhomberg and Jones, 1994). We tested the 15 clinical strains of L. pneumophila sg1 sampled from the first respiratory samples of 15 patients (see Results section). The L. pneumophila Paris, Philadelphia, Lens, and Lorraine strains served as ciprofloxacin-susceptible controls, and the LPPI1, LPPI4, and LPPI5 strains as ciprofloxacin-resistant controls.
2.4. Quantitative Real-time PCR Assay Targeting the gyrA QRDR Mutations (qPCRgyrALp)
In gram-negative bacteria, fluoroquinolone resistance is most often related to mutations occurring in the quinolone resistance determining region of gyrA, the gene encoding the sub-unit A of DNA gyrase, especially at the hot-spot codon positions gyrA83 and gyrA87 (Escherichia coli numbering system) (Davies and Davies, 2010; Komp Lindgren et al., 2003; Jacoby, 2005). We previously demonstrated that gyrA83 mutations could occur in vitro in L. pneumophila, and was associated with increased resistance to fluoroquinolones (Almahmoud et al., 2009). Thus, we developed a real-time PCR assay (qPCRgyrALp) targeting mutations in the gyrA QRDR, especially at codon positions gyrA83 and gyrA87.
PCR and real-time PCR experiments were performed with genomic DNA from the relevant strains or from lower respiratory tract samples, after DNA extraction using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations (appendix p 3). We first PCR-amplified and sequenced a 383-bp region of the gyrA QRDR of 31 Legionella strains (appendix pp 3 and 6), as previously described (Almahmoud et al., 2009). We analyzed these sequences to design L. pneumophila-specific primers and probes to elaborate the qPCRgyrALp assay. The sensitivity and specificity of the qPCRgyrALp assay were first checked using a set of 67 strains, including 7 fluoroquinolone-susceptible or -resistant L. pneumophila control strains (appendix p 5), 27 strains belonging to other Legionella species (appendix p 6), and 33 strains belonging to other genera (appendix p 7).
2.5. Next-generation Deep-sequencing of the Legionella gyrA QRDR
We used high-throughput pyrosequencing (appendix p 4, table 1) to confirm and quantify the presence of gyrA QRDR mutated alleles in 11 human respiratory samples, including eight qPCRgyrALp-positive samples from four patients (called patients #1 to #4), and three qPCRgyrALp-negative samples from three patients selected randomly (called control patients).
2.6. Patient and Other Consents
We used respiratory samples and bacterial strains collected in the context of routine care of patients hospitalized at Grenoble University Hospital for severe legionellosis. We have declared this collection of human samples to the French Ministry of Education and Research (DC-2008-677), and we have the authorization to use it for research purposes from our ethics committee (CPP Sud-Est V). In accordance with the French law, patients' information for this type of research is done through a hospital medical booklet and only a non-opposition of the patients is needed.
2.7. Role of the Funding Source
The study was sponsored by the French Centre National de la Recherche Scientifique (CNRS), Université Grenoble Alpes (UGA); and by the Direction de la Recherche Clinique et de l'Innovation (DRCI) Centre Hospitalier Universitaire de Grenoble.
The funding sources had no role in the study design; in data collection, analysis and interpretation; in writing of the manuscript or in the decision to submit it for publication. We did not receive any funds to write this article from a pharmaceutical company or other agencies.
3. Results
A positive urinary antigen test for L. pneumophila sg1 (Binax Now Legionella, Binax, Portland, ME, USA) was found for all 82 investigated pneumonic patients. Legionellosis diagnosis was further confirmed in 15 of the 82 patients by isolation of a L. pneumophila strain from the first collected respiratory sample. These 15 clinical strains of L. pneumophila displayed ciprofloxacin MICs ranging from 0.25 to 0.38 mg/L, whereas MICs were 0.38 mg/L for the susceptible L. pneumophila reference strains (Paris, Philadelphia, Lens, and Lorraine), 1.5 mg/L for the LPPI1 mutant with a single gyrA mutation, and 2 mg/L for the LPPI4 and LPPI5 mutants with two gyrA mutations (appendix p 5). We found no mutation in the gyrA QRDR of the 15 clinical strains. Legionellosis was also confirmed in 61 (74.4%) of the 82 legionellosis patients (including 13 patients with a positive L. pneumophila culture), by direct detection of L. pneumophila DNA in respiratory samples (112 of the 139 collected samples, 80.6%), using the qPCRmip assay (Maurin et al., 2010). The L. pneumophila DNA load in these samples varied from 5.8 × 102 to 6.8 × 107 genome units (GU)/mL of sample. For 19 patients, L. pneumophila culture and PCR assays were negative.
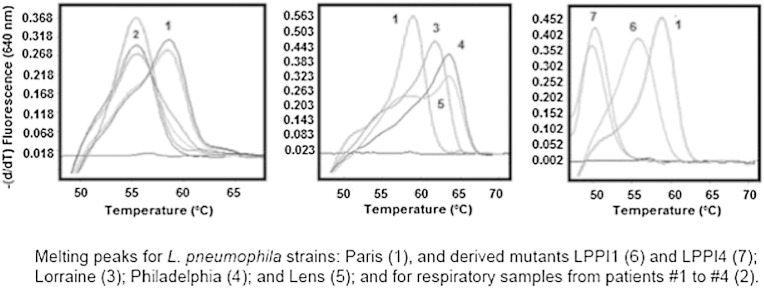
After standardization of the qPCRgyrALp assay, a strong amplification signal of the gyrA QRDR was detected for all tested L. pneumophila strains. Furthermore, this assay could distinguish Legionella versus non-Legionella strains, L. pneumophila versus other Legionella species, fluoroquinolone-susceptible versus resistant mutants of L. pneumophila, and single versus double mutants (Fig. 1; appendix p 3; appendix p 5). When applied to the 139 respiratory samples collected from the 82 legionellosis patients, an amplification signal corresponding to the gyrA QRDR was detected in 114 samples (82%) from 62 patients (75.6%), including 63 and 11 samples collected from patients under and without fluoroquinolone therapy, respectively (Table 1). The qPCRgyrALp assay was positive for 108 (96.4%) of the 112 samples showing a positive qPCRmip assay, but only for 7 of the other 27 qPCRmip-negative samples (25.9%, p < 0.01), suggesting that the DNA load in clinical samples was critical for detection of either the mip or gyrA gene. All qPCRgyrALp-positive respiratory samples (104) collected from 58 patients gave identical results, their melting curve profiles suggesting a reference gyrA QRDR sequence (mean melting peak ± SD, 59.34 ± 0.41 °C) when compared to control strains (Fig. 1, Table 1). For 4 patients however (#1 to #4), we detected melting peaks of 56.35 ± 0.42 °C (three replicates per patient) for 6 of 11 respiratory samples, suggesting the presence of gyrA QRDR mutations.
Fig. 1.
Detection of gyrA QRDR mutations using the qPCRgyrALp assay.
Melting curves and peaks obtained with the qPCRgyrALp assay are shown for the L. pneumophila Paris strain (Martínez, 2008), respiratory samples from patients #1 to #4 (Skippington and Ragan, 2011), the L. pneumophila Lorraine strain (Pendleton et al., 2013), the L. pneumophila Philadelphia strain (Rolain and Raoult, 2005), the L. pneumophila Lens strain (Rouli et al., 2012), and the L. pneumophila Paris mutant strains LPPI1 and LPPI4 harboring one (Maurin et al., 2001), and two (Binet and Maurelli, 2005) gyrA mutations, respectively (appendix p 5). The units of the y axes are expressed as fluorescence levels derived through time. Melting peaks of 59.34 ± 0.41 °C and 56.35 ± 0.42 °C are diagnostic of reference and mutant alleles of the gyrA QRDR, respectively.
Table 1.
Results of the qPCRgyrALp assay with melting temperature (Tm), and presence or absence of the gyrA83/T83I QRDR change determined by PCR amplification and sequencing (PCR-Seq), and Next-Generation Sequencing (NGS, with the percentage of gyrA83 mutant alleles among sequencing reads) for the 139 respiratory samples collected from the 82 patients.
| Patients (n = 82) |
Respiratory samples (n = 139) |
Day of samplinga |
qPCRgyrALp |
T83I GyrA change |
||||
|---|---|---|---|---|---|---|---|---|
| FQ | No FQ | NAb | Total | (Tm) | PCR-Seq | NGS | ||
| n = 20 | 8 | 2 | 10 | 20 | Negative | ND | ||
| n = 58 | 3 | 0 | 1 | 4 | Negative | ND | ||
| 54 | 10c | 40 | 104 | Positive (~ 60 °C) | ND | Yes (< 0.5%) | ||
| #1 | 1 | 1 | D1 | Positive (~ 60 °C) | NPA | No | ||
| 1 | 1 | D3 | Positive (~ 56 °C) | NPA | Yes (< 0.5%) | |||
| 1 | 1 | D7 | Negative | ND | ND | |||
| #2 | 1 | 1 | D0 | Positive (~ 60 °C) | No | Yes (2.9%) | ||
| 1 | 1 | D4 | Positive (~ 56 °C) | Yes | Yes (94%) | |||
| 1 | 1 | D4 | Positive (~ 56 °C) | ND | ND | |||
| #3 | 1 | 1 | D0 | Positive (~ 56 °C) | No | No | ||
| #4 | 1 | 1 | D0 | Positive (~ 60 °C) | No | Yes (1.05%) | ||
| 1 | 1 | D2 | Positive (~ 60 °C) | ND | ND | |||
| 1 | 1 | D3 | Positive (~ 56 °C) | Yes | Yes (75%) | |||
| 1 | 1 | D5 | Positive (~ 56 °C) | Yes | Yes (85%) | |||
| Total | 75 | 13 | 51 | 139 | ||||
Data are given according to the patient's fluoroquinolone (FQ) treatment status at the time of respiratory sample collection. ND: not determined. NPA: no PCR amplification.
Days of sampling are given according to the day of legionellosis diagnosis (D0).
FQ treatment information not available.
Including three D0 samples tested by NGS for the three control patients.
To confirm the presence of gyrA83 or gyrA87 mutations in qPCRgyrALp-positive samples, we tried to PCR-amplify and sequence a 383-bp fragment of the L. pneumophila gyrA QRDR directly from eight respiratory samples of patients #1 to #4 (Table 1). Using next-generation deep sequencing (NGS) technology, we also determined the frequency of these gyrA QRDR mutations by testing the same respiratory samples from patients #1 to #4 and the qPCRgyrALp-negative samples from three control patients (Table 1). We detected no PCR product for samples from patient # 1 collected one and three days (D1 and D3) after the day of legionellosis diagnosis (D0), which may be related to low L. pneumophila DNA load (< 103 GU/mL sample, as determined by the qPCRmip assay). For patient #3, a PCR product was obtained from the D0 sample, in which we identified five mutations none of which affected the gyrA codons 83 or 87. Two silent mutations C219G and C264T (codons 73 and 88, respectively) occurred in the gyrA QRDR but outside the location of the sensor probe of the qPCRgyrALp assay, and thus had no influence on the melting peak. The three others (T278C, A281G, and T285C at codons 93, 94, and 95, respectively) were localized in the region complementary to the anchor probe thereby explaining the observed melting peak decrease. Using the NGS method, we confirmed the lack of gyrA mutation at codons 83 and 87 for sample D0 of patient #3, and the five changes at codons 73 (C219 G), 88 (C264T), 93 (T278C), 94 (A281G), and 95 (T285C), at a frequency of 85.7% of 17,060 reads. More importantly, we detected low levels (0.023% to 0.19% of a total of 3267 to 14,716 reads) of C248T gyrA83 mutations (T83I GyrA change) in samples collected from control legionellosis patients before fluoroquinolone administration, as well as in the D3 sample (0.037% of 5258 reads) from patient #1. These later results suggested that, in some legionellosis patients, the gyrA83 mutants of L. pneumophila were present in the lower respiratory airways before administration of any antibiotic therapy.
For patients #2, a reference gyrA QRDR was detected in the D0 sample, but a C248T mutation resulting in the T83I GyrA change was found in the analyzed D4 sample, suggesting in vivo selection of fluoroquinolone-resistant mutants. Using NGS, we confirmed the presence of the C248T gyrA83 mutation in respiratory samples. It occurred at a frequency of 2.9% of 7205 reads at D0, increasing to 94% of 16,794 reads in the D4 sample after 4 days of treatment with levofloxacin and azithromycin (Table 1). The high proportion of gyrA83 mutants in sample D4 of patients #2 confirmed the melting curve profiles of the qPCRgyrALp assay. Patient #2 recovered from legionellosis under levofloxacin and azithromycin therapy, after 31 days at ICU and a total of 79 days at hospital (Fig. 2).
Fig. 2.
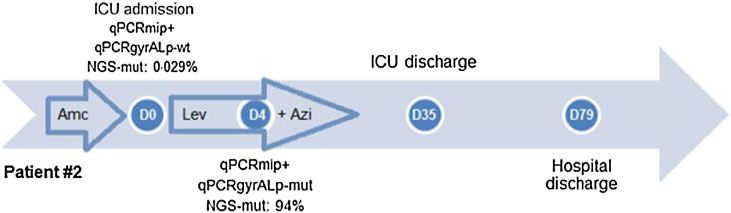
Clinical history and gyrA83 mutational dynamics for patients #2.
The results of qPCRmip, qPCRgyrALp and NGS assays are shown, together with the antibiotic therapy for patients #2 over his hospitalization stay. The number of days for relevant steps during hospitalization is given inside circles according to D0 which corresponds to the day of legionellosis diagnosis as determined by a positive urinary antigen test (UAT +). qPCR-mip + indicates a positive qPCRmip assay in respiratory samples. Results of the qPCRgyrALp assay are indicated as “neg” (no amplification of the gyrA QRDR), “wt” and “mut” (a positive PCR assay with a reference and mutant-type melting peak, respectively). The proportion of the reads corresponding to the T83I gyrA83 mutation, determined by next-generation sequencing, is shown (NGS-mut). Small arrows indicate the periods of antibiotic therapy. Lev: levofloxacin; Cp: ciprofloxacin; Azi: azithromycin; Ery: erythromycin; Amc: amoxicillin-clavulanate; Ca: ceftazidime; Tc: teicoplanin.
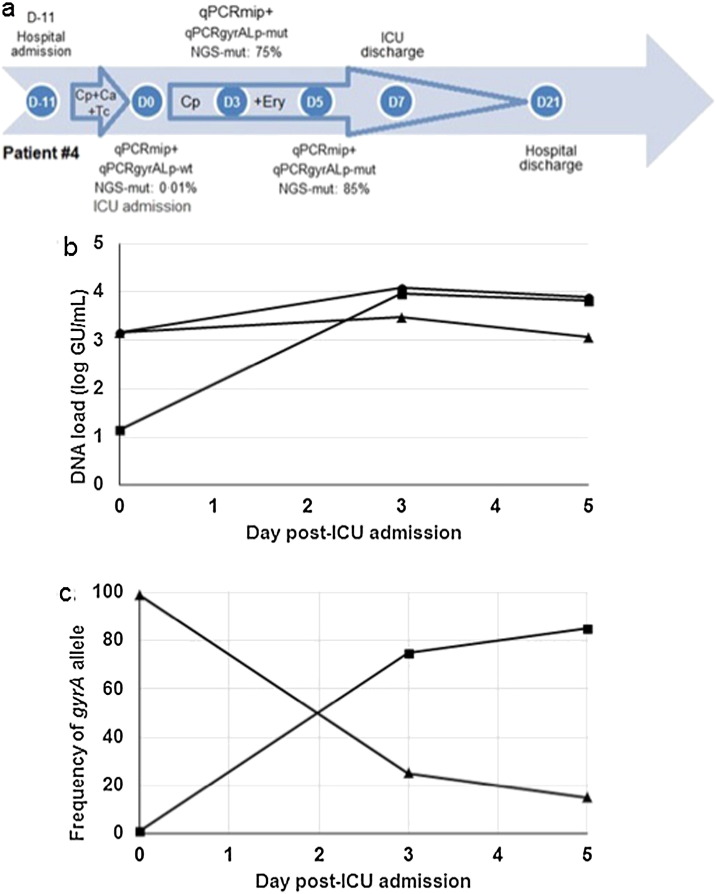
For patients # 4, a reference gyrA QRDR was detected in the D0 sample, but a C248T mutation resulting in the T83I GyrA change was found in both D3 and D5 samples, suggesting in vivo selection of fluoroquinolone-resistant mutants. Using NGS, the C248T gyrA83 mutation was detected in 1.05% of 8063 reads at D0, increasing to 75% of 15,824 reads and 85% of 16,050 reads at D3 and D5, respectively, while the patient was receiving ciprofloxacin and erythromycin in the intensive care unit (ICU) (Table 1). The high proportion of gyrA83 mutants in samples D3 and D5 confirmed the melting curve profiles of the qPCRgyrALp assay. For this patient, we investigated the dynamics of both the DNA load and gyrA83 allele by combining the qPCRmip and NGS approaches. While the global DNA load remained stable in respiratory samples from D0 to D5, the gyrA83 mutated alleles progressively replaced the wild-type alleles, suggesting an increase in the fluoroquinolone-resistant population of L. pneumophila, parallel to the reduction of the susceptible population (Fig. 3). Patient #4 was first admitted to a hematologic ward for severe pneumonia and aplasia, and transferred after 11 days at ICU because of health worsening despite a three-day course of ciprofloxacin, ceftazidime and teicoplanin (Fig. 3). He was cured under ciprofloxacin and erythromycin therapy, with a 32-day hospital stay including 7 days at ICU.
Fig. 3.
Clinical history, and dynamics of L. pneumophila DNA load and gyrA83 mutations in respiratory samples from patient #4.
(a) The results of qPCRmip, qPCRgyrALp and NGS assays are shown, together with the antibiotic therapy for patients #4 over his hospitalization stay (see legend of Fig. 2).
(b) Dynamics of L. pneumophila DNA load, expressed as log GU/mL clinical sample, in respiratory samples of patient # 4 over time after admission in ICU. The total, reference, and mutant gyrA QRDR DNA loads are shown by circles, triangles and squares, respectively. Data were extracted from the qPCRmip assay and NGS experiments.
(c) Relative proportion of the reference (triangles) and mutant (squares) gyrA QRDR allele in respiratory samples of patient # 4 over time after admission in ICU. Data were extracted from the NGS experiments.
We investigated whether a correlation may exist between a positive qPCRgyrALp assay, the presence of gyrA83 mutations in respiratory samples and severe legionellosis evolution (Table 2). A trend was noted, albeit not significant, for more frequent admission to the ICU for patients with a positive qPCRgyrALp assay. Also, patients with a positive qPCRgyrALp assay were more likely to receive a combination of a macrolide and a fluoroquinolone during their hospital stay. No significant difference was observed between the qPCRgyrALp results and the mean global hospital stay or death. A correlation between the presence of gyrA83 mutations in respiratory samples and a worse prognosis was impossible to establish because only 2 patients of 82 displayed such criterion.
Table 2.
Analyses of potential correlations between parameters (qPCRgyrALp assay, qPCRmip assay, antibiotic treatment, duration of stay at hospital and ICU, death) for all 82 patients of the cohort.
| qPCR gyrALp | Patients (n = 82) | qPCRmip |
Legionellosis treatmenta |
Hospital stay (mean days +/− SD) | ICU admission (n) | Death (n) | ||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | FQ only (n) | FQ + MC (n) | |||||
| Positive | 62 | 58 | 4 | 22 | 20 | 18.1 +/− 16.1 | 21 | 5 |
| Negative | 20 | 3 | 17 | 11 | 0 | 14.3 +/− 11.7 | 2 | 0 |
| p < 0.001† | 0.004† | 0.23††† | 0.07†† | 0.32† | ||||
All numbers correspond to patients.
FQ: fluoroquinolone; MC: macrolide.
Fisher exact tests were performed.
Chi2 tests with Yates correction were performed.
t tests were performed.
4. Discussion
It has commonly been assumed that L. pneumophila could not develop fluoroquinolone resistance (Jespersen et al., 2010; Yu et al., 2009; Pedro-Botet and Yu, 2009; Burdet et al., 2014). Very recently however, a fluoroquinolone-resistant strain of L. pneumophila was isolated in a legionellosis patient treated with ciprofloxacin (Bruin et al., 2014). This unique and preliminary observation raises the question of the dynamics and origin of fluoroquinolone resistance in L. pneumophila. It has been speculated that the environmental release of antibiotics may promote the emergence of antibiotic-resistant bacterial lineages (Davies and Davies, 2010), but this has never been demonstrated for Legionella species. Here, using a combination of molecular and genomic approaches, we investigated the infection dynamics of two legionellosis patients in a cohort of 82 during their hospitalization. We demonstrated that infection occurred with a fluoroquinolone-susceptible population of L. pneumophila that evolved in vivo toward a high load of fluoroquinolone-resistant gyrA83 mutants after antibiotic therapy.
We designed a qPCRgyrALp assay that allowed us to detect a signal for the gyrA QRDR in 114 samples from 62 of the 82 patients, including 63 samples collected in patients under fluoroquinolone treatment. We found gyrA83 mutations in two patients (3.1%, patients #2 and #4). We did not seek mutations in either other gyrA regions or other topoisomerase-encoding genes owing to the small amount of biological material, but such mutations may occur in vitro (Almahmoud et al., 2009). Thus, we have likely underestimated the incidence of in vivo selection of fluoroquinolone resistance in L. pneumophila. We investigated the presence of a correlation between a positive qPCRgyrALp assay and higher legionellosis severity in the 82 patients of the cohort. There was a trend for the 62 patients with a positive qPCRgyrALp assay to be more frequently admitted to the ICU, and these patients more frequently received a combination of a macrolide and a fluoroquinolone then those with a negative qPCRgyrALp assay. These findings may reflect higher L. pneumophila bacteria load in their respiratory samples, which we previously correlated with higher severity of the disease using the qPCRmip assay (Maurin et al., 2010). Indeed, there was a significant correlation between results of the qPCRgyrALP and qPCRmip assays.
A gyrA83 mutation leading to the GyrA T83I substitution is likely to be clinically significant for two main reasons: first we previously demonstrated that it produced at least a four-fold increase of ciprofloxacin MIC in L. pneumophila Paris (Almahmoud et al., 2009), and second it is known to be associated with high-level fluoroquinolone resistance in other Gram-negative bacteria (Komp Lindgren et al., 2003; Jacoby, 2005). We detected a rapid increase of gyrA83 mutant alleles under fluoroquinolone treatment to 94% at D4 for patient #2 and 75% and 85% at D3 and D5, respectively, for patient #4. For this latter patient, a fluoroquinolone-susceptible strain of L. pneumophila sg1 was isolated from respiratory samples at D0. During the stay of patient #4 at ICU, we observed a major in vivo shift in the structure of the L. pneumophila population with the replacement of fluoroquinolone-susceptible by resistant cells, confirming that fluoroquinolone therapy selected for in vivo fluoroquinolone resistance during hospitalization. The two patients #2 and #4 suffered from a severe and prolonged course disease, but recovered under combined fluoroquinolone and macrolide treatment. Additional cases will be needed to establish a statistical correlation between the disease severity and the in vivo selection of gyrA QRDR mutant strains of L. pneumophila. We found by NGS a very low percentage (< 0.5%) of mutated alleles in respiratory samples from control patients and in sample D3 from patient #1, and a slightly higher percentage (1.05% and 2.9%, respectively) in sample D0 from patients #2 and #4. These sequences may correspond to false positives introduced during the PCR amplification step of the NGS process itself. Alternatively, they may reflect the spontaneous occurrence of fluoroquinolone-resistance mutations in L. pneumophila, as previously described in other Gram-negative bacteria including E. coli (Komp Lindgren et al., 2003). Therefore, legionellosis patients might have been infected with a complex L. pneumophila population containing low abundance of gyrA83 mutants, with subsequent selection of this population in patients #2 and #4 under fluoroquinolone therapy. An alternative hypothesis could be the in vivo emergence of these mutants. The rapid increase in the proportion of mutated gyrA83 alleles over a few days is more compatible with the former hypothesis. In both cases however, the selective sweeps resulting in the increased abundance of the gyrA83 mutants occurred in vivo during fluoroquinolone treatment. We emphasize that patient #4 already received a three-day course of ciprofloxacin at D0.
We have shown that in vivo selection of gyrA mutations responsible for fluoroquinolone-resistance in L. pneumophila occurs in legionellosis patients during fluoroquinolone treatment. A likely hypothesis is that patients were infected with L. pneumophila populations containing low doses of gyrA83 mutants. If so, it is puzzling that no fluoroquinolone resistance has been so far characterized in environmental strains of L. pneumophila, and only a single fluoroquinolone-resistant strain of this species has been isolated before in a clinical situation (Bruin et al., 2014). It should be emphasized that mutant strains of L. pneumophila have poor ability to propagate outside infected hosts because of the lack of human-to-human transmission and little opportunities to go back to their natural environmental reservoir. However, isolation of L. pneumophila from respiratory samples of legionellosis patients remains challenging, and almost impossible in patients under antibiotic therapy. We recommend the development and use of molecular tools to test the presence of resistance mutations in patients infected by L. pneumophila, especially in the case of poor response to antibiotic treatment. Our strategy should be optimized to allow more specific detection of gyrA83 and gyrA87 mutations, and other previously characterized fluoroquinolone-resistance mutations (Almahmoud et al., 2009; Jonas et al., 2003; Nielsen et al., 2000). This would allow to precisely define the prevalence of fluoroquinolone-resistant mutants in legionellosis patients and their clinical consequences. The molecular tools could also serve to assess the emergence and spread of gyrA83 mutants in environmental L. pneumophila strains. The possibility of in vivo selection for fluoroquinolone-resistance mutations in L. pneumophila fully justifies the current practice of administration of combined antibiotic therapy in severe legionellosis cases (Roig and Rello, 2003). Our study showing in vivo L. pneumophila adaptation to antibiotic pressure paves the way for more intensive investigations of other intracellular bacterial species. This hidden resistance should indeed be intensively explored because it may contribute to the increasing public health threat of the gradual loss of effectiveness of antibiotic treatments.
In conclusion, although fluoroquinolones and macrolides are highly active against Legionella species, death rates of 10–15% are commonly reported in legionellosis patients. No correlation can be found in the literature between treatment failures or relapses and development of antibiotic resistance in Legionella spp. However, fluoroquinolone-resistant strains of L. pneumophila have been selected in vitro, and a fluoroquinolone-resistant isolate was recently obtained from a legionellosis patient. By using state-of-the-art molecular and genomic approaches, we identified, at high frequency, fluoroquinolone resistance mutations in lower respiratory samples from two additional legionellosis patients from a cohort of 82. The detected gyrA83 mutation is associated with a significant increase in fluoroquinolones MICs in L. pneumophila. We demonstrated that selection for fluoroquinolone resistance occurred in vivo during fluoroquinolone therapy. These two patients required ICU admission and prolonged hospitalization, but recovered under combined fluoroquinolone and macrolide treatment. Although the exact clinical impact of our findings could not be statistically evaluated because only two out of 82 legionellosis patients were involved, the possibility of in vivo selection of fluoroquinolone resistance in L. pneumophila should now be considered both for prognostic evaluation and treatment optimization at least in patients with severe legionellosis.
Contributors
SJ, JE, JFT, DS and MM designed the experiments. LS and IA performed the experiments. LS and SL performed the NGS experiments. SJ and JE provided the Legionella strains from species other than L. pneumophila. All authors contributed to literature search. DS and MM wrote the manuscript. LS and MM made the figures. CS and JFT provided all clinical data. All authors had access to data and commented on and approved the final version.
Declaration of Interests
We declare no competing interests.
Acknowledgments
This study was funded by the French Centre National de la Recherche Scientifique, the Université Grenoble Alpes, and the Direction de la Recherche Clinique et de l'Innovation (DRCI) Centre Hospitalier Universitaire de Grenoble. LS thanks to Damascus University (Damascus, Syria) for a Ph.D fellowship. IA thanks Tishreen University (Latakia, Syria) for a Ph.D fellowship. We thank Katia Fusiller and Jeanne-Noëlle Del Bano for technical assistance, and Linda Northrup for English editing. We also thank Roche Diagnostics (Meylan, France) for providing the GS Junior 454 apparatus for next generation deep sequencing experiments.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.07.018.
Appendix A. Supplementary Data
Supplementary material.
References
- Almahmoud I., Kay E., Schneider D., Maurin M. Mutational paths towards increased fluoroquinolone resistance in Legionella pneumophila. J. Antimicrob. Chemother. 2009;64:284–293. doi: 10.1093/jac/dkp173. [DOI] [PubMed] [Google Scholar]
- Binet R., Maurelli A.T. Frequency of spontaneous mutations that confer antibiotic resistance in Chlamydia spp. Antimicrob. Agents Chemother. 2005;49:2865–2873. doi: 10.1128/AAC.49.7.2865-2873.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruin J.P., Koshkolda T., IJzerman E.P.F. Isolation of ciprofloxacin-resistant Legionella pneumophila in a patient with severe pneumonia. J. Antimicrob. Chemother. 2014;69:2869–2871. doi: 10.1093/jac/dku196. [DOI] [PubMed] [Google Scholar]
- Burdet C., Lepeule R., Duval X. Quinolones versus macrolides in the treatment of legionellosis: a systematic review and meta-analysis. J. Antimicrob. Chemother. 2014;69:2354–2360. doi: 10.1093/jac/dku159. [DOI] [PubMed] [Google Scholar]
- Carratalà J., Garcia-Vidal C. An update on Legionella. Curr. Opin. Infect. Dis. 2010;23:152–157. doi: 10.1097/QCO.0b013e328336835b. [DOI] [PubMed] [Google Scholar]
- Chidiac C., Che D., Pires-Cronenberger S. Factors associated with hospital mortality in community-acquired legionellosis in France. Eur. Respir. J. 2012;39:963–970. doi: 10.1183/09031936.00076911. [DOI] [PubMed] [Google Scholar]
- Davies J., Davies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC . 2012. Commission Implementing Decision 2012/506/EU of 8 August 2012, amending Decision 2002/253/EC Laying Down Case Definitions for Reporting Communicable Diseases to the Community Network Under Decision No 2119/98/EC of the European Parliament and of the Council. [Google Scholar]
- Jacoby G.A. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 2005;41(Suppl. 2):120–126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- Jespersen S., Søgaard O.S., Schønheyder H.C., Fine M.J., Ostergaard L. Clinical features and predictors of mortality in admitted patients with community- and hospital-acquired legionellosis: a Danish historical cohort study. BMC Infect. Dis. 2010;10:124. doi: 10.1186/1471-2334-10-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas D., Engels I., Hartung D., Beyersmann J., Frank U., Daschner F.D. Development and mechanism of fluoroquinolone resistance in Legionella pneumophila. J. Antimicrob. Chemother. 2003;51:275–280. doi: 10.1093/jac/dkg054. [DOI] [PubMed] [Google Scholar]
- Komp Lindgren P., Karlsson A., Hughes D. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob. Agents Chemother. 2003;47:3222–3232. doi: 10.1128/AAC.47.10.3222-3232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez J.L. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008;321:365–367. doi: 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- Maurin M., Abergel C., Raoult D. DNA gyrase-mediated natural resistance to fluoroquinolones in Ehrlichia spp. Antimicrob. Agents Chemother. 2001;45:2098–2105. doi: 10.1128/AAC.45.7.2098-2105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurin M., Hammer L., Gestin B. Quantitative real-time PCR tests for diagnostic and prognostic purposes in cases of legionellosis. Clin. Microbiol. Infect. 2010;16:379–384. doi: 10.1111/j.1469-0691.2009.02812.x. [DOI] [PubMed] [Google Scholar]
- Newton H.J., Ang D.K.Y., van Driel I.R., Hartland E.L. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin. Microbiol. Rev. 2010;23:274–298. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K., Bangsborg J.M., Høiby N. Susceptibility of Legionella species to five antibiotics and development of resistance by exposure to erythromycin, ciprofloxacin, and rifampicin. Diagn. Microbiol. Infect. Dis. 2000;36:43–48. doi: 10.1016/s0732-8893(99)00095-4. [DOI] [PubMed] [Google Scholar]
- Pedro-Botet M.L., Yu V.L. Treatment strategies for Legionella infection. Expert. Opin. Pharmacother. 2009;10:1109–1121. doi: 10.1517/14656560902900820. [DOI] [PubMed] [Google Scholar]
- Pendleton J.N., Gorman S.P., Gilmore B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti-Infect. Ther. 2013;11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- Ravanel N., Gestin B., Maurin M. In vitro selection of fluoroquinolone resistance in Brucella melitensis. Int. J. Antimicrob. Agents. 2009;34:76–81. doi: 10.1016/j.ijantimicag.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Rhomberg P.R., Jones R.N. Evaluations of the E-test for antimicrobial susceptibility testing of Legionella pneumophila, including validation of the imipenem and sparfloxacin strips. Diagn. Microbiol. Infect. Dis. 1994;20:159–162. doi: 10.1016/0732-8893(94)90110-4. [DOI] [PubMed] [Google Scholar]
- Roig J., Rello J. Legionnaires' disease: a rational approach to therapy. J. Antimicrob. Chemother. 2003;51:1119–1129. doi: 10.1093/jac/dkg191. [DOI] [PubMed] [Google Scholar]
- Rolain J.M., Raoult D. Genome comparison analysis of molecular mechanisms of resistance to antibiotics in the Rickettsia genus. Ann. N. Y. Acad. Sci. 2005;1063:222–230. doi: 10.1196/annals.1355.035. [DOI] [PubMed] [Google Scholar]
- Rouli L., Rolain J.-M., El Filali A., Robert C., Raoult D. Genome sequence of Coxiella burnetii 109, a doxycycline-resistant clinical isolate. J. Bacteriol. 2012;194:6939. doi: 10.1128/JB.01856-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skippington E., Ragan M.A. Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol. Rev. 2011;35:707–735. doi: 10.1111/j.1574-6976.2010.00261.x. [DOI] [PubMed] [Google Scholar]
- Sutera V., Levert M., Burmeister W.P., Schneider D., Maurin M. Evolution toward high-level fluoroquinolone resistance in Francisella species. J. Antimicrob. Chemother. 2014;69:101–110. doi: 10.1093/jac/dkt321. [DOI] [PubMed] [Google Scholar]
- Yu H., Higa F., Koide M. Lung abscess caused by Legionella species: implication of the immune status of hosts. Int. Med. Tokyo Jpn. 2009;48:1997–2002. doi: 10.2169/internalmedicine.48.2647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.