Abstract
Eisosomes are among the few remaining eukaryotic cellular differentations that lack a defined function(s). These trough-shaped invaginations of the plasma membrane have largely been studied in Saccharomyces cerevisiae, in which their associated proteins, including two BAR domain proteins, have been identified, and homologues have been found throughout the fungal radiation. Using quick-freeze deep-etch electron microscopy to generate high-resolution replicas of membrane fracture faces without the use of chemical fixation, we report that eisosomes are also present in a subset of red and green microalgae as well as in the cysts of the ciliate Euplotes. Eisosome assembly is closely correlated with both the presence and the nature of cell walls. Microalgal eisosomes vary extensively in topology and internal organization. Unlike fungi, their convex fracture faces can carry lineage-specific arrays of intramembranous particles, and their concave fracture faces usually display fine striations, also seen in fungi, that are pitched at lineage-specific angles and, in some cases, adopt a broad-banded patterning. The conserved genes that encode fungal eisosome-associated proteins are not found in sequenced algal genomes, but we identified genes encoding two algal lineage-specific families of predicted BAR domain proteins, called Green-BAR and Red-BAR, that are candidate eisosome organizers. We propose a model for eisosome formation wherein (i) positively charged recognition patches first establish contact with target membrane regions and (ii) a (partial) unwinding of the coiled-coil conformation of the BAR domains then allows interactions between the hydrophobic faces of their amphipathic helices and the lipid phase of the inner membrane leaflet, generating the striated patterns.
INTRODUCTION
In a pioneering 1963 paper employing their newly developed freeze fracture electron microscopy (EM) technique, Moor and Mühlethaler (1) documented “rod-like invaginations,” averaging 300 nm long, 20 to 30 nm wide, and 50 nm deep, in the Saccharomyces cerevisiae plasma membrane. Such invaginations, also called furrows or troughs, have since been identified in additional freeze fracture EM studies of S. cerevisiae (2–6) and other fungi (7–13).
In 2002, Young et al. (14) reported that green fluorescent protein (GFP)-tagged Sur7p, a tetraspanning plasma membrane protein of S. cerevisiae, localized to stable punctate domains of the yeast membrane; in 2004, a tagged proton-arginine transporter, Can1p, was found to colocalize with Sur7p (15); and in 2006, two tagged cytoplasmic proteins, Pil1p and Lsp1p, were localized to these domains (16). These observations were brought together by Strádalová et al. (5), who showed that the membrane compartment occupied by Can1p (MCC) localized to the invaginations seen with freeze fracture EM, and by Karotki et al. (6), who showed that Pil1p localizes to the cytoplasmic surfaces of these invaginations. Additional proteins also associate with these punctate domains, in some cases in a transient fashion (17), and the MCC component is reportedly enriched in ergosterol (18) and influenced by phosphoinositide (6, 19, 20) and sphingolipid (14, 17, 21–24) levels. Hence, the current yeast model (25, 26) proposes that Pil1p and Lsp1p, which contain membrane curvature-inducing BAR domains (6, 27, 28), together with Seg1p (29, 30), form a submembrane complex (6) reportedly influenced by Pil1p phosphorylation (25); this complex then either creates or associates with the MCC domains, presumably inducing an inward curvature. The MCC protein Nce102p has also been implicated in generating curvature (5, 31).
Walther et al. (16) proposed the name eisosome (eis, Gr., into or portal) for the Pil1p/Lsp1p heteromer because their experiments indicated that it was associated with endocytosis, an interpretation that has since been challenged (29, 32–35; but see reference 36). Formally, the full membrane/submembrane complex of yeast proteins should be denoted “MCC/eisosomes,” and the invaginations should be denoted “furrows” (25). However, the term “eisosome” has entered the published literature and is used in this report to designate this class of membrane differentiation.
Several published studies of unicellular green algae have included freeze fracture images of rod-like plasma membrane furrows that are morphologically homologous to fungal eisosomes (3, 13, 37–39). During the course of a recent quick-freeze deep-etch electron microscopic (QFDEEM) (40) survey of several dozen unicellular algae deemed to be candidate producers of triacylglycerols for biodiesel, we obtained high-resolution images of eisosomes from members of several red and green microalgal radiations, including lichen photobionts. Here we document lineage-specific features of their freeze fracture ultrastructure and large-scale topology. No homologues of identified fungal eisosome-specific proteins were detected in microalgae with sequenced genomes. However, BLAST searches identified two additional families of BAR proteins—designated Green-BAR and Red-BAR—whose presence largely correlates with an eisosomal endowment. It is possible that these proteins serve Pil1p/Lsp1p-related functions in the green and red algal lineages. We present a model of eisosome formation that integrates some of our findings with those from previous publications.
MATERIALS AND METHODS
When not otherwise specified, organisms were grown in their source labs and transported or overnight shipped to Washington University for immediate freezing. Organisms were obtained from the following sources: Penicillium sp. isolate K17, collected from desert rock varnish in New Mexico by Diana Northup, University of New Mexico; Saccharomyces cerevisiae, David Drubin, University of California, Berkeley; Candida albicans, Peter Lipke, Brooklyn College; Cryptococcus neoformans, Tamara L. Doering, Washington University School of Medicine; Schizosaccharomyces pombe, David Kovar, University of Chicago; Candelaria concolor, collected from a tree branch in Missouri by U. Goodenough; Cladonia grayi, Daniele Armaleo, Duke University (solo cultures were grown in the Goodenough lab); Cyanidioschizon YNP 1A and Galdieria sulfuraria CCMEE 5587.1, Peter Lammers, currently at Arizona State University, obtained from R. W. Castenholz, University of Oregon, Culture Collection of Microorganisms from Extreme Environments (CCMEE), now at the Pacific Northwest National Laboratory under the direction of S. L. Cady; Auxenochlorella protothecoides UTEX 25, Shayani Pieris, Missouri Baptist University, and Richard Sayre, currently at Los Alamos National Laboratory; Chlamydomonas monoica, Karen VanWinkle-Swift, Northern Arizona University (cultures were grown in the Goodenough lab); Chlamydomonas reinhardtii, J.-H. Lee, currently at the University of British Columbia (cultures were grown in the Goodenough lab); Polytomella parva strain SAG 63-3, Robert W. Lee, Dalhousie University (cultures were grown in the Goodenough lab with the permission of SAG); Haematococcus sp. strain Haema001, collected from a bird bath in California by Jeurgen Polle, Brooklyn College (cultures were grown in the Goodenough lab); Borodinellopsis texensis, collected from a dry roadside ditch in New Mexico by Jeurgen Polle (cultures were grown in the Goodenough lab); and Euplotes sp., collected from pond water in Kyoto, Japan, by J. E. Heuser.
Using previously described procedures (40), living organisms were gently pelleted, snap-frozen without fixation at liquid-helium temperatures, and then subjected to fracture, deep etching, and replication at the Deep Etch EM Facility (http://www.heuserlab.wustl.edu/services/index.shtml) of the Department of Cell Biology and Physiology, Washington University School of Medicine. The replicas were examined with a model JEM 1400 JEOL electron microscope equipped with an AMTV601 digital camera.
The following programs and servers were used for gene and protein analysis: for BAR domain (PF03144) analysis, HMMER (v3.0) (http://hmmer.janelia.org); for α-helical predictions, Heliquest (http://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParamsV2.py); for three-dimensional (3D) template searches and modeling, SWISS-MODEL (http://swissmodel.expasy.org); and for 3D structure visualization, Swiss Pdb Viewer (v4.1) (http://spdbv.vital-it.ch). All the BAR candidates listed in Table S2 in the supplemental material were confirmed to have structural similarity to verified 3D models of BAR domain proteins posted on the Protein Data Bank (PDB) site.
RESULTS
Freeze fracture deep-etch characteristics of eisosomes.
When a frozen plasma membrane is fractured, the result is either a cross-fracture, equivalent to a cross section, or the fracture plane travels within the membrane lipid bilayer, generating two faces, called E (ectoplasmic; the leaflet adjacent to the cell exterior) and P (protoplasmic; the leaflet adjacent to the cell interior). In the QFDEEM procedure employed in this study (40), the exposed membrane faces are then etched under a vacuum (surface water is allowed to sublime) and rotary shadowed (platinum is deposited uniformly), during which the more protuberant entities are coated with more platinum and hence appear whiter than the less protuberant entities in the negative images shown. The platinum replica is then cleaned of cellular material and examined by transmission EM.
Eisosomes, which invaginate into the cytoplasm, display two curved fracture faces: the cytoplasmic leaflet, carried by the P face, appears concave, and its reciprocal, carried by the E face, appears convex. Several figures (e.g., see Fig. S1 in the supplemental material) show 3D anaglyphs that display these curvatures unambiguously; in the cropped images used for the text figures, they may be more difficult to discern. The eisosomal concave face is either amorphous or variously striated, whereas in algae, the convex face often carries globular entities resembling intramembrane particles (IMPs), which are traditionally interpreted as representing the transmembrane domains of membrane-spanning proteins (41).
Eisosomes of free-living fungi.
To our knowledge, no wild-type fungus has been reported to lack eisosomes. In current studies of free-living (in contrast to lichen-forming) fungi, eisosomes are usually visualized by fluorescence microscopy of tagged resident proteins. Figure 1 and Fig. S1 in the supplemental material show representative QFDEEM images.
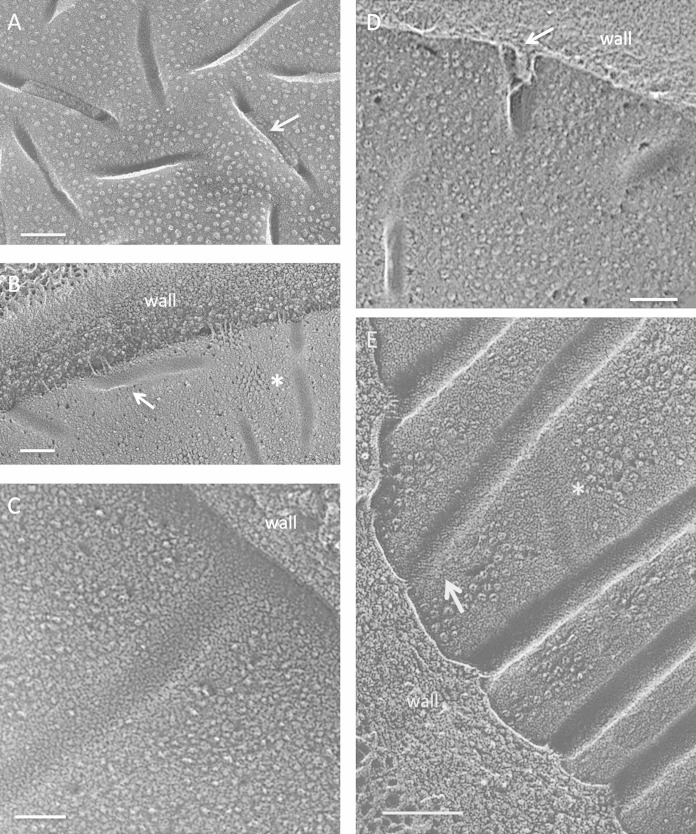
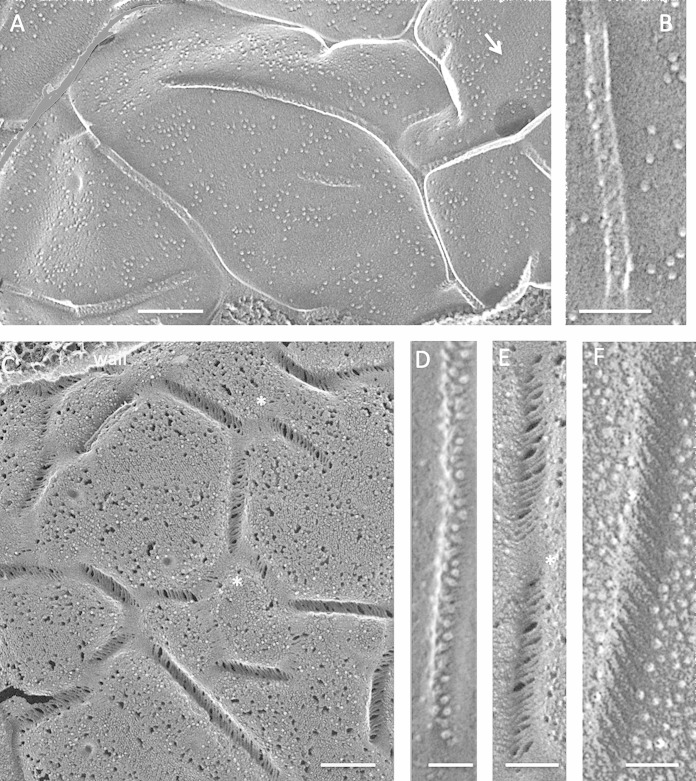
FIG 1.
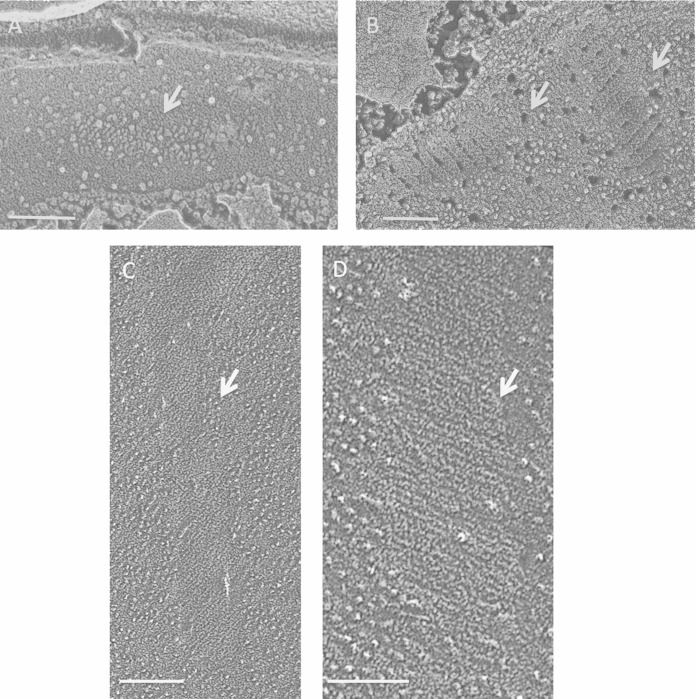
Fungi. (A) Punctate concave eisosomal faces, some containing wall remnants (arrow), from an unidentified ascomycete bread mold. Bar, 100 nm. (B) Saccharomyces cerevisiae punctate concave eisosomal faces. Arrow, diagonal striations; asterisk, cluster of Pma1p proton pumps in the membrane compartment occupied by Pma1p (MCP). Bar, 100 nm. (C) Cryptococcus neoformans elongated concave eisosomal face with a uniform granular texture. Bar, 50 nm. (D) Candida albicans punctate convex eisosomal faces, with the wall entering at the arrow. Bar, 100 nm. (E) Schizosaccharomyces pombe elongated concave eisosomal faces. Arrow, diagonal striations; asterisk, eisosome with reduced depth and at right angles to others. Bar, 100 nm.
Two large-scale eisosomal topologies are encountered in the fungi studied to date. The most common, which we designate “punctate,” entails short individual eisosomes that are sparsely or densely arrayed in what appear to be random orientations, as seen in an unidentified bread mold (Fig. 1A), S. cerevisiae (Fig. 1B) (1–3), Cryptococcus neoformans (Fig. 1C) (8), and Candida albicans (Fig. 1D) (10). The alternative, which we designate “elongated,” entails long eisosomes that are oriented either randomly, as in Penicillium sp. isolate K17 (42) (see Fig. S1 in the supplemental material) and Melampsore lini (7), or in parallel or anastomosing rows, as in S. pombe (Fig. 1E) (9, 12, 43) and Malassezia (12). Interestingly, the punctate eisosomes of S. cerevisiae and C. albicans adopt an elongated anastomosing configuration when the cells are converted to protoplasts (6, 44, 45), and C. albicans protoplasts restore the punctate configuration when allowed to regrow new walls (45). Additional examples of such topological variability were encountered in the present study (see below).
The concave eisosomal fracture faces of fungi have a fine-grained and uniform texture (Fig. 1C; see Fig. S1B in the supplemental material) which, in some instances, is organized into thin striations pitched at ∼30° with respect to the long axis of the furrow (Fig. 1B and E, arrows), mirroring the pitch displayed by in vitro polymers of Pil1p/Lsp1p (6). The convex fracture faces of fungal eisosomes are either smooth (Fig. 1D) or roughened (see Fig. S1A) and are IMP-free.
Eisosomes of lichen-forming fungi of the lichens Candelaria concolor and Cladonia grayi.
Figure 2 shows eisosomes of the fungal (mycobiont) partner in the foliose lichen Candelaria concolor. In some cases, the hyphae display sparse, punctate, and straight eisosomes (Fig. 2A; see Fig. S2A in the supplemental material), but more commonly, they are sparse, punctate, and curved (Fig. 2B; see Fig. S2B and C). Curved eisosomes have also been reported for the mycobiont of Myelochroa leucotyliza (13), whereas the mycobionts of several Lichenaceae species display only straight punctate eisosomes (46). Particularly dramatic is a single example wherein the eisosomes are highly dense, elongated, and sinuous (Fig. 2C).
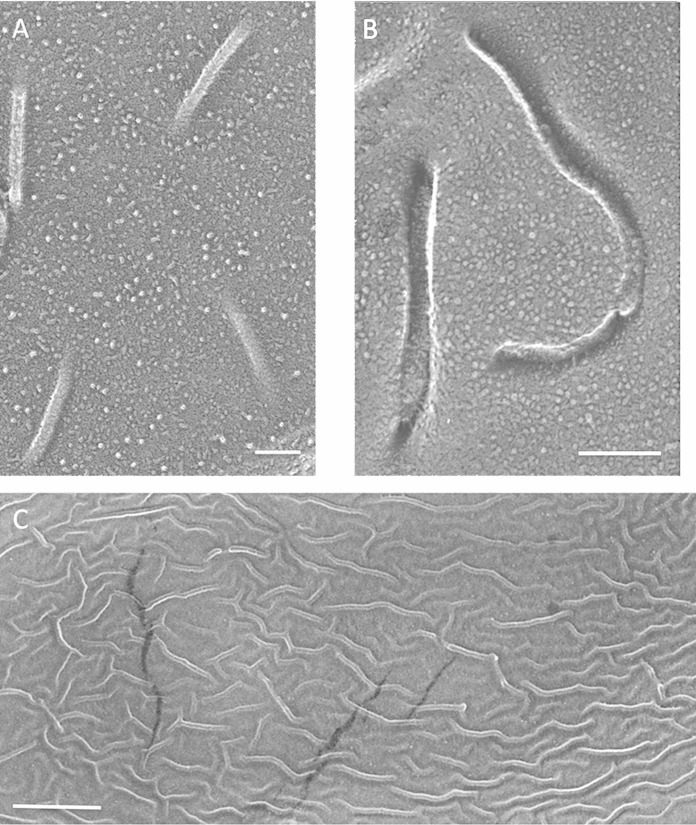
FIG 2.
Lichenized Candelaria concolor mycobiont. (A) Sparse, straight, punctate convex faces. Bar, 100 nm. (B) Sparse, curved, punctate convex faces. Bar, 100 nm. (C) High-density elongated sinuous convex faces. Bar, 500 nm.
The fungus of the fruticose lichen Cladonia grayi has been visualized both in the lichenized state and in solo culture on agar plates (47). In the lichen, its hyphae display only sparse punctate eisosomes (see Fig. S3 in the supplemental material), and some hyphae continue this sparse pattern on plates (see Fig. S4A and B). The majority, however, display a high-density, elongated, and sinuous population (Fig. 3) similar to the one example of this topology that we encountered in lichenized C. concolor (Fig. 2C). The concave faces of these sinuous eisosomes are also distinctive, with irregular alternating strands and gaps (Fig. 3C) at right angles to the long axis of the furrow.
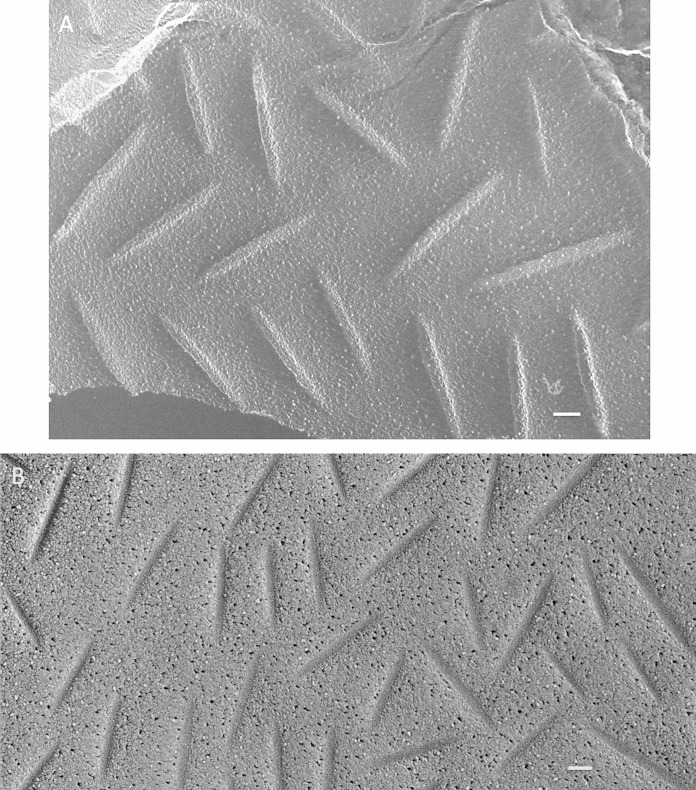
FIG 3.
Nonlichenized Cladonia grayi (solo). (A) Hyphal eisosomes in both cross-fracture and planar fracture convex views, showing a concentration of eisosomes in the region of wall curvature (arrows) and their absence in the relatively flat domain. Bar, 200 nm. (B) High-density elongated sinuous convex faces. Bar, 100 nm. (C) High-density elongated sinuous concave faces carrying struts alternating with gaps. Arrow, wall entering eisosome. Bar, 100 nm.
Microalgal overview.
When frozen microalgae are fractured, numerous and extensive views of plasma membrane interiors are generated; hence, it is possible to ascertain the presence versus absence of eisosomes in a given cell type with considerable confidence. Table S1 in the supplemental material lists the numerous microalgae examined that do not display eisosomes during vegetative growth. Considered below are the 10 microalgae in our survey that produce eisosomes either as vegetative cells or as cysts.
Eisosomes of the red alga Cyanidioschizon YNP 1A.
The extensively studied extremophilic red alga Cyanidioschizon merolae (48–50), isolated from hot springs near Naples, Italy, lacks a cell wall and carries no eisosomes in its plasma membrane. A recently isolated red alga, Cyanidioschizon YNP 1A, obtained from hot springs in Yellowstone National Park, WY, has 18S rRNA and RubisCO large subunit gene sequences nearly identical to those of C. merolae (51), and both species have very similar cellular morphologies (see Fig. S5 in the supplemental material). However, YNP 1A possesses a cell wall (51) and also displays abundant eisosomes.
The elongated eisosomes of YNP 1A can be regularly spaced (Fig. 4A to C) and can span the entire circumference of some cells (see Fig. S6B and S7B in the supplemental material). In other cases, notably in daughter cells, the spacing can be more irregular (see Fig. S6A and S7A). Elements connecting parallel eisosomes are occasionally present (Fig. 4C, asterisk). Cell wall material fills the invaginations (Fig. 4A and B). The convex fracture face has a higher density of IMPs than the surrounding membrane (see Fig. S8A), while the concave face either is amorphous or displays fine striations pitched ∼30° with respect to the long axis (Fig. 4D; see Fig. S8B).
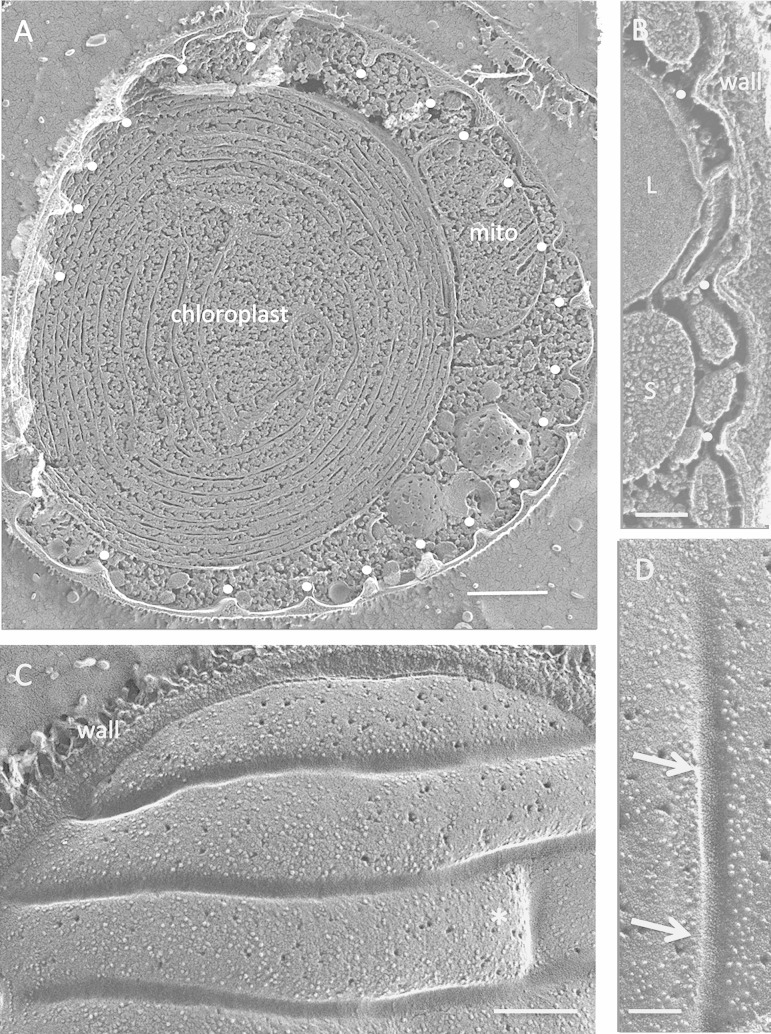
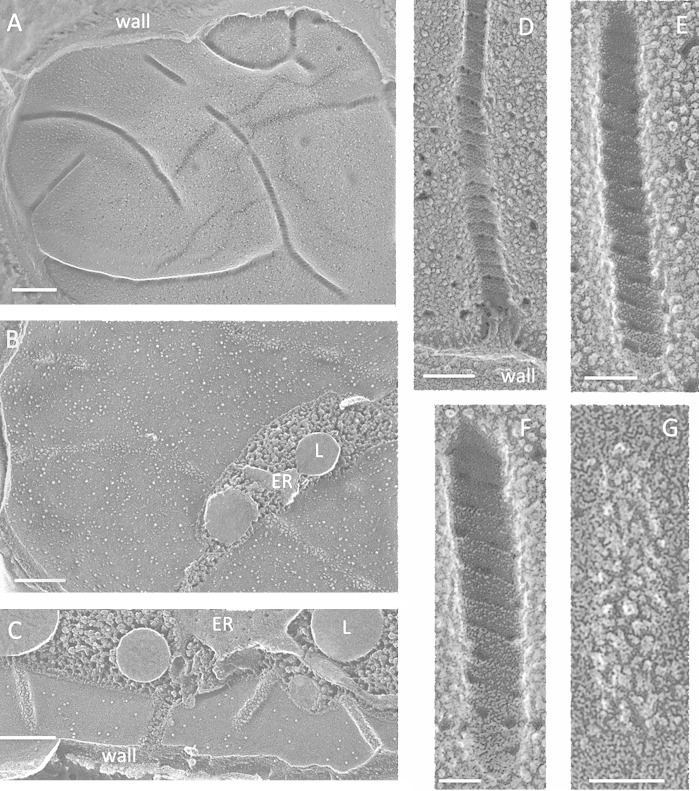
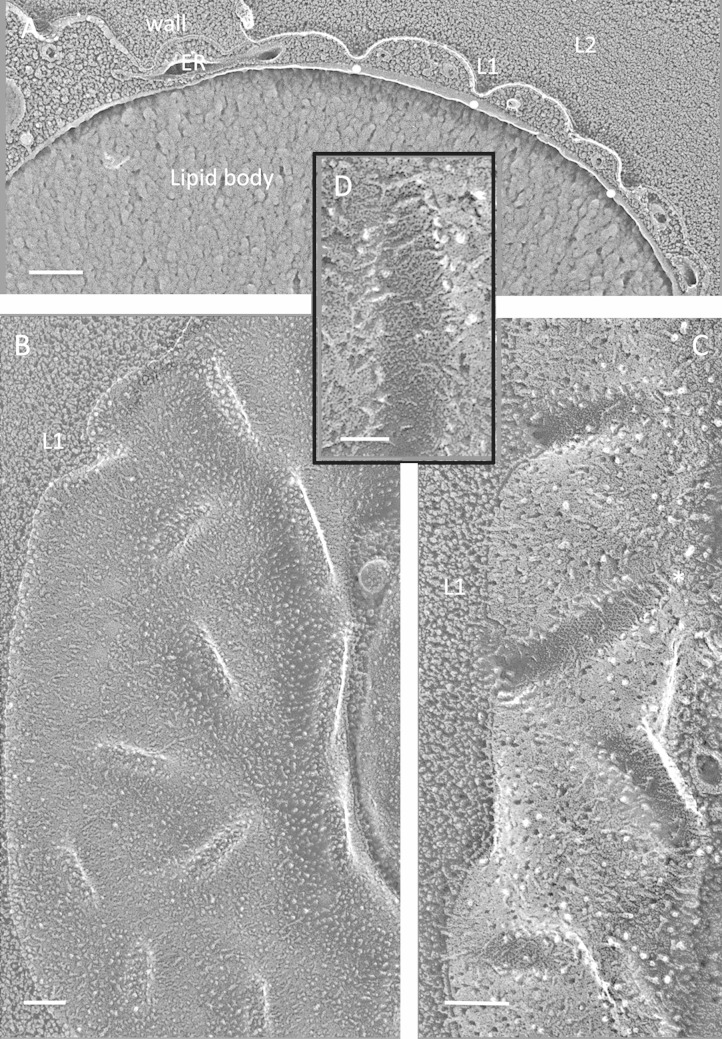
FIG 4.
Cyanidioschizon YNP 1A. (A) Cell survey with indented eisosome cross-fractures marked with white dots. Bar, 500 nm. (B) Eisosome indentations (white dots). L, lipid body; S, starch. Bar, 100 nm. (C) Elongated parallel eisosomes with a cross-segment at the asterisk (field enlarged from Fig. S6B in the supplemental material). Bar, 200 nm. (D) Concave face with regions of angled striations (arrows). Bar, 100 nm.
Eisosomes of the red alga Galdieria sulfuraria.
Galdieria sulfuraria is estimated to have diverged from C. merolae ∼1 billion years ago (BYA) and shares only 42% of its genes (52), yet both are adapted to similar habitats (low pH and high sulfur and temperature). The G. sulfuraria strain examined here was isolated from hot springs in Yellowstone Park (51) but has properties that are very different from those of YNP 1A: it is larger, has a different intracellular organization (compare Fig. S9 and S5B in the supplemental material), and can grow on a wide range of carbon sources (51–53), whereas YNP 1A and its sister C. merolae are obligate phototrophs.
The plasma membrane of G. sulfuraria is unique among the algae examined in this study in carrying flat rectangular domains (Fig. 5A, arrow; see Fig. S10 in the supplemental material) and circular “islands” of IMPs (see Fig. S10A and S11A) that are reminiscent of the Pma1p proton pump clusters in S. cerevisiae (Fig. 1B). Its elongated eisosomes, apparently independent of these features, form anastomosing networks (Fig. 5A and C) rather than the circumferential bands seen in YNP 1A. Their convex fracture faces can display an angled striated patterning, pitched at ∼40° (Fig. 5B), that is usually obscured by an abundant population of IMPs (Fig. 5D). Their concave faces display alternating bands of material, reminiscent of the solo C. grayi concave face (Fig. 3C), that are also pitched at ∼40° but are more regularly spaced (Fig. 5C and E). Short segments of the concave faces, often at intersecting nodes, lack the bands (Fig. 5C and E, asterisks).
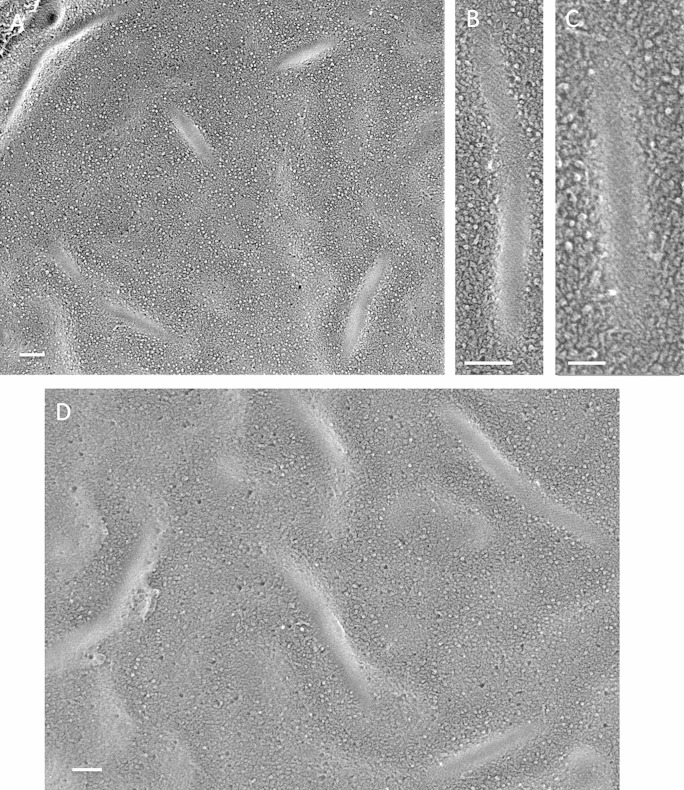
FIG 5.
Galdieria sulfuraria. (A) Elongated anastomosing eisosomes, convex face. Arrow, rectangular membrane feature (see Fig. S10 in the supplemental material). Bar, 250 nm. (B) Convex face with few IMPs, displaying a diagonal pitch. Bar, 100 nm. (C) Long anastomosing concave eisosomes. Asterisks, domains lacking bands. Bar, 200 nm. (D) Convex face festooned with IMPs. Bar, 50 nm. (E) Concave face, banded. Bar, 100 nm. (F) Concave face, nonetched, banded. Bar, 50 nm.
The irregular gaps between the bands in Fig. 5C and E proved to be generated during the deep-etching process. As shown in Fig. 5F and in Fig. S11 in the supplemental material, when G. sulfuraria eisosomes are fractured but not etched, the concave face is regularly corrugated. Hence, the native structure presumably alternates between relatively etchable (watery or fragile) and nonetchable domains; this may also be the case for solo C. grayi eisosomes (Fig. 3C). Figures S12 and S13 in the supplemental material show 3D anaglyphs of the two nonetched faces of G. sulfuraria eisosomes.
Eisosomes of the trebouxiophyte green alga Auxenochlorella protothecoides.
Auxenochlorella protothecoides, which is in the Chlorella lineage clade (54) and has a published genome (55), displays eisosomes during both its growth phase and its thicker-walled stationary phase. The cell wall has a granular inner layer that penetrates the furrow and a denser outer layer that does not (see Fig. S14 in the supplemental material). The eisosomes can adopt an elongated topology with occasional anastomoses, as in S. pombe and the red alga YNP 1A (Fig. 6A and B; see Fig. S15), but can also be organized as oblong punctate units, as in most fungi (Fig. 6C; see Fig. S16). The convex face is densely covered with IMPs (Fig. 6B, C, and G), and the concave face is highly distinctive, with broad bands pitched at a shallow, ∼20° angle (see Fig. S17), regularly alternating with narrow gaps (Fig. 6D to F; see Fig. S15C and D), gaps that may have been created during etching as documented earlier for G. sulfuraria (Fig. 5F; see Fig. S11).
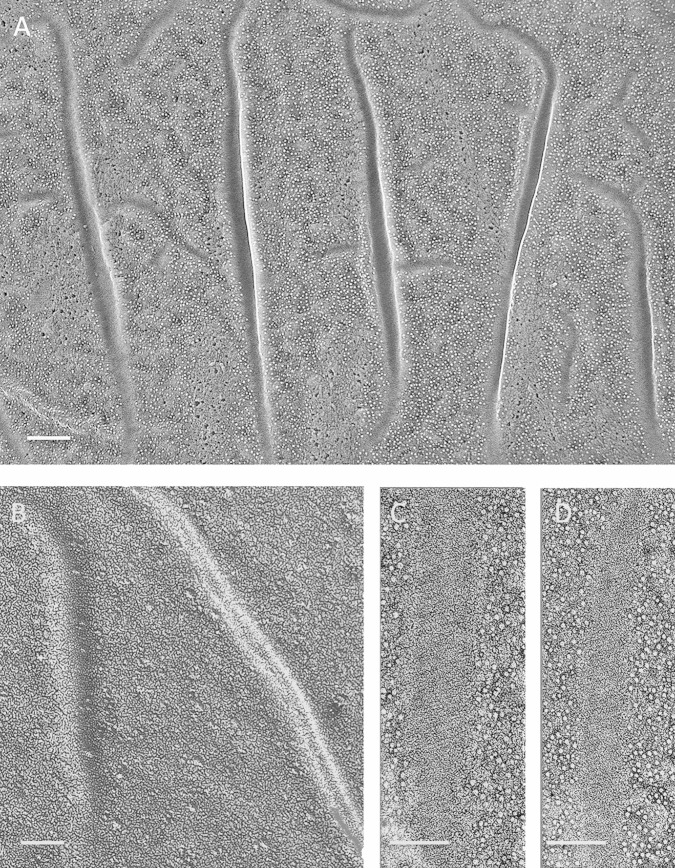
FIG 6.
Auxenochlorella protothecoides. (A) Elongated eisosomes that occasionally intersect, concave face. Bar, 250 nm. (B) Elongated parallel eisosomes, convex face. L, lipid body in cytoplasm. Bar, 200 nm. (C) Convex face. Bar, 200 nm. (D) Concave face. Bar, 100 nm. (E) Concave face. Bar, 50 nm. (F) Concave face. Bar, 50 nm. (G) Convex face. Bar, 50 nm.
Eisosomes of trebouxiophyte green algal photobionts of Candelaria concolor and Cladonia grayi lichens.
Lichens, which are named for their ∼20,000 different fungal (mycobiont) species, harbor trebouxiophyte microalgae (photobionts) in just two genera: Trebouxia (56) and Asterochloris (57). Our survey includes Trebouxia jamesii from the Candelaria concolor lichen (58) and Asterochloris sp. from the Cladonia grayi lichen (47).
When lichenized, T. jamesii has abundant eisosomes (Fig. 7A and B), as also reported for Trebouxia sp. in the lichen Myelochroa leucotyliza (13), that are uniformly organized as punctate oblong units rather than elongated systems. The convex faces carry various populations of IMPs. The concave face is finely granular, with no evidence of striations.
FIG 7.
Trebouxia jamesii. (A) High-density punctate eisosomes, convex face. Bar, 100 nm. (B) Concave face. Bar, 100 nm.
No eisosomes were encountered in numerous images of lichenized Asterochloris sp.; in contrast, they were often encountered when the alga was grown alone on agar medium. They are less dense and more sinuous than the eisosomes of T. jamesii (Fig. 8A and D), their convex faces are smooth, and their concave faces carry prominent diagonal striations pitched at ∼30° (Fig. 8B and C).
FIG 8.
Asterochloris sp. (A) Punctate concave eisosomal faces. Bar, 100 nm. (B) Concave face with diagonal striations. Bar, 100 nm. (C) Concave face with diagonal striations. Bar, 50 nm. (D) Sinuous concave faces. Bar, 100 nm.
Eisosomes of the chlorophyte green alga Chlamydomonas monoica.
The chlorophyte Chlamydomonas monoica (59), one of an interfertile group that includes Chlamydomonas noctigama and Chlamydomonas geitleri, is a member of the Moewusii clade that also includes Chlamydomonas eugametos; this lineage is only distantly related to the Reinhardtinia clade that includes the well-studied species Chlamydomonas reinhardtii (60–62). The eisosomes of C. eugametos have been imaged previously (37, 38).
The eisosomes of both C. eugametos and C. monoica are punctate and abundant (Fig. 9A). Strikingly, in both species, there has been a reversal of the orientation of the two faces: whereas in all other organisms examined the concave fracture face (cytoplasmic leaflet; P face) is amorphous, finely striated, or banded, in C. monoica and C. eugametos it is densely populated with IMPs (Fig. 9C and D); conversely, the convex fracture face (external leaflet; E face) is finely striated (Fig. 9E and F; see Fig. S18 in the supplemental material), with an ∼30° pitch. Figures S19 to S21 in the supplemental material show stereo views of this reversed orientation (compare with Fig. S1, S12, and S13). The convex face also carries a variable number of irregularly shaped and distributed particles (Fig. 9E and F, arrows; see Fig. S18 and S20) that are absent from the amorphous/striated faces of other organisms. These look more like postfracture condensations than like IMPs (see Fig. S20); if this is the case, then these are artifacts that for some reason are restricted to these domains in this alga.
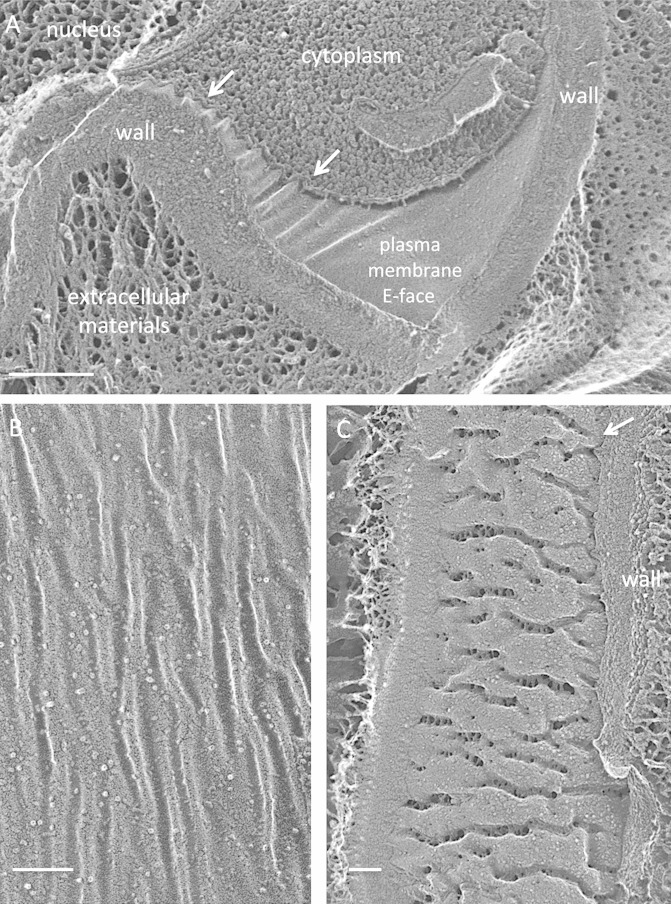
FIG 9.
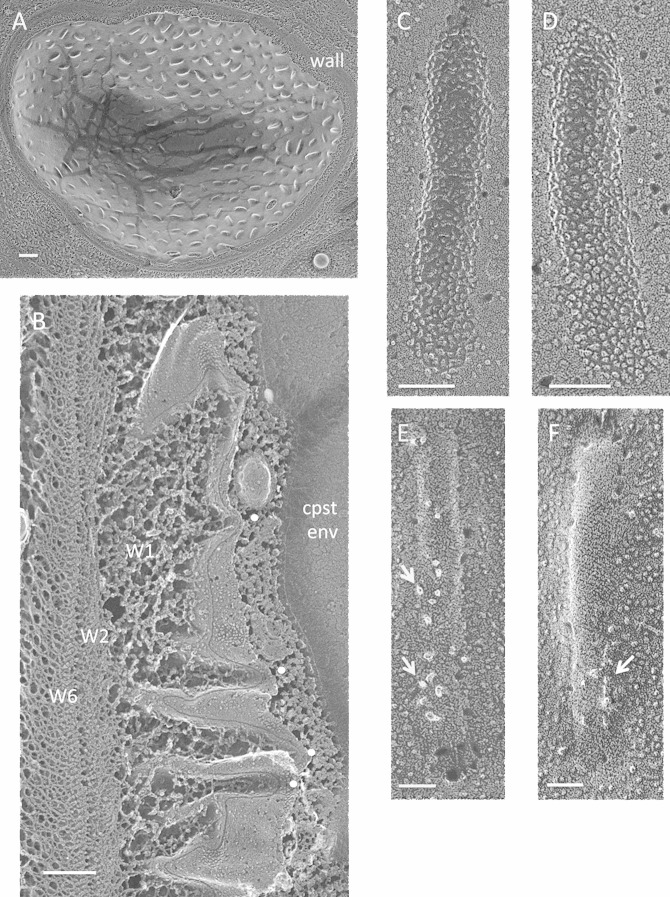
Chlamydomonas monoica. (A) Cell survey showing high-density punctate eisosomal topology. Bar, 500 nm. (B) The trabecular W1 layer of the wall associates with eisosomal invaginations (white dots); the dense W2 to W6 layers do not. Cpst env, chloroplast envelope. Bar, 250 nm. (C and D) Concave faces. Bars, 100 nm. (E and F) Convex faces. Arrows, irregularly shaped and distributed particles. Bars, 50 nm.
A second C. monoica/C. eugametos distinction lies in the eisosomal relationship to the cell wall. In all other organisms examined, the wall layer that enters and occupies the eisosomal furrow is dense and directly associated with the rest of the compact cell wall (Fig. 1D, 3A and C, 4B, and 6C; see Fig. S9 in the supplemental material, and see additional examples in cysts [described below]). The eisosomes of C. monoica/C. eugametos, in contrast, are not in direct contact with the dense W2 to W6 layers of the wall (38) but rather with an open trabecular layer designated W1 (38) (Fig. 9B; see Fig. S21 and S22).
Eisosomes of chlorophyte green algal species in zygotes and cysts.
The four remaining chlorophyte species examined in this study lack eisosomes as vegetative cells but produce them either as diploid sexual zygotes or as asexual cysts.
Chlamydomonas reinhardtii.
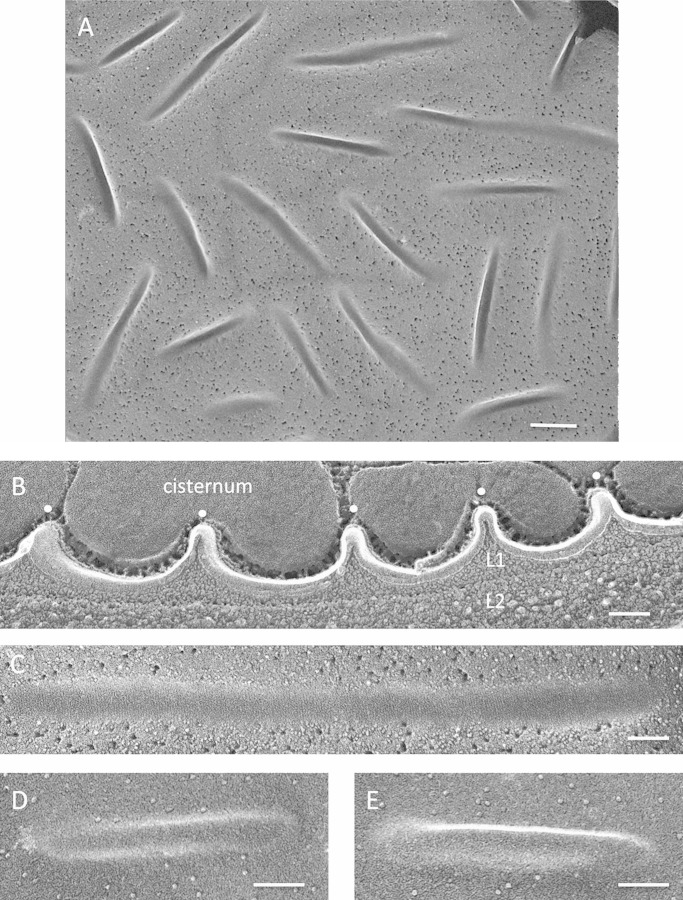
The mature sexual zygotes of C. reinhardtii (62, 63) display abundant punctate eisosomes, with an apparently random orientation (Fig. 10A), that extend into the cytoplasm between a system of cisternae (Fig. 10B; see Fig. S23 in the supplemental material) that we posit to contain the orange pigment produced by these cells. Reminiscent of the case in A. protothecoides (see Fig. S14), the finely granular innermost wall layer (L1) (Fig. 10B) directly enters the invaginations, whereas the denser layer beneath it (L2) does not. Fine striations, at right angles to the eisosome axis, mark some but not all of the concave faces (Fig. 10C; see Fig. S24A). The smooth convex face carries no IMPs or other structural features (Fig. 10D and E; see Fig. S24B).
FIG 10.
Chlamydomonas reinhardtii zygotes. (A) High-density punctate eisosomes, concave faces. Bar, 250 nm. (B) Eisosome cross-fractures (white dots). L1, penetrant granular wall layer; L2, denser wall layer. Bar, 100 nm. (C) Concave face. Bar, 100 nm. (D and E) Convex faces. Bars, 100 nm.
Polytomella parva.
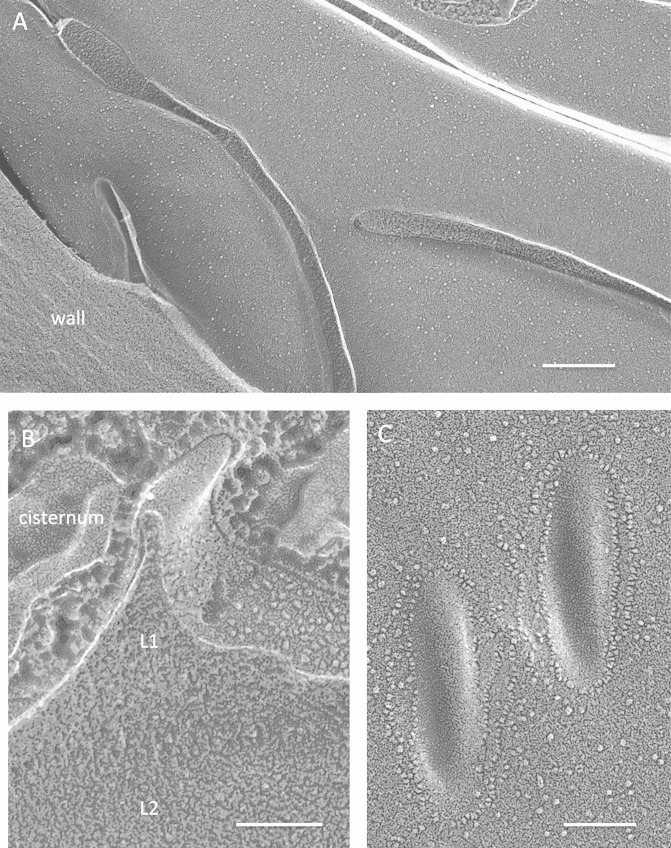
P. parva, a close relative of C. reinhardtii (64, 65), is a colorless quadriflagellate that is cell wall-less during vegetative growth but forms an asexual cyst in the stationary phase (66). The P. parva cyst eisosomes can be either elongated (Fig. 11A) or punctate (Fig. 11C). The smooth convex face is unique to our samples in being surrounded by linear chains of IMPs (Fig. 11C). As in the C. reinhardtii zygote, a granular wall layer (L1) enters the eisosome, while a denser sublayer (L2) does not (Fig. 11B; see Fig. S25 in the supplemental material). The cysts also display subplasmalemmal cisternae (Fig. 11B; see Fig. S25).
FIG 11.
Polytomella parva cysts. (A) Elongated parallel eisosomes. Bar, 200 nm. (B) Eisosome cross-fracture (white dot). L1, granular wall layer; L2, denser wall layer. Bar, 100 nm. (C) Convex faces of punctate eisosomes encircled with chains of IMPs. Bar, 100 nm.
Haematococcus sp.
The Haematococcus sp. strain examined has been assigned morphologically to the Chlorogonium clade (Haematococcus pluvialis/Haematococcus lacustris) (67; J. Polle, personal communication). The walled green vegetative cells lack eisosomes, whereas the red asexual cysts have a punctate eisosome distribution (Fig. 12A) very similar to that in the C. reinhardtii zygote (Fig. 10A), where, again, a granular wall layer (L1) enters the eisosome and a denser layer (L2) does not (Fig. 12B and C). However, no striations are evident on the concave face (Fig. 12D), and no subplasmalemmal cisternae are present. Hagen et al. (39) provided an image of the smooth convex face of eisosomes in H. pluvialis cysts.
FIG 12.
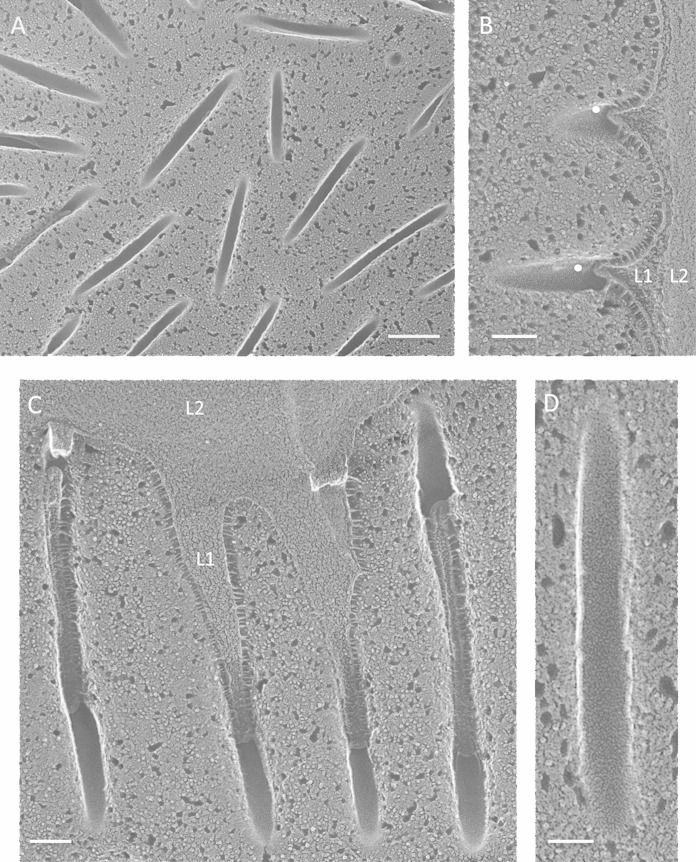
Haematococcus sp. cysts. (A) High-density punctate eisosomes, concave faces. Bar, 200 nm. (B) Cross-fractured eisosomes (white dots). L1, penetrant granular wall layer; L2, denser wall layer. Bar, 100 nm. (C) Parallel invaginations filled with L1 wall material, concave faces. Bar, 100 nm. (D) Concave face. Bar, 50 nm.
Borodinellopsis texensis.
B. texensis is a member of the Sphaeropleales clade of the chlorophytes. Green B. texensis vegetative cells, although walled, display no eisosomes, whereas eisosomes are numerous in the orange asexual cysts (Fig. 13). Their convex faces are IMP-rich, in contrast to the smooth convex faces in the five other cysts examined, and their concave faces are unique in carrying a system of rivulets that can adopt a honeycomb configuration (Fig. 13C and D). Again, a granular inner wall layer (L1) enters the eisosome (Fig. 13A; see Fig. S26 in the supplemental material).
FIG 13.
Borodinellopsis texensis cysts. (A) Cross-fractured eisosomes (white dots) containing the L1 but not the L2 wall layer. Bar, 200 nm. (B) Punctate convex faces. Bar, 100 nm. (C) Concave faces. Bar, 100 nm. (D) Enlargement of eisosome denoted with asterisk in panel C. Bar, 50 nm.
Eisosomes of the ciliate Euplotes sp. cysts.
Given the presence of eisosomes in the four algal zygotes/cysts examined, we searched for eisosomes in the spores of Dictyostelium discoideum (Amoebozoa) and the cysts of Naegleria gruberi (Discoba), and none were encountered. In contrast, abundant eisosomes are produced by the cysts of the ciliate Euplotes sp. (Alveolata) (68) (Fig. 14; see Fig. S27 in the supplemental material). These are more elongated (Fig. 14A; see Fig. S27B) than those encountered in algal zygotes/cysts, with the exception of a subset in P. parva (Fig. 11A), and they display a smooth convex face (Fig. 14A and B). The concave face carries diagonal striations pitched at ∼20° (Fig. 14C and D).
FIG 14.
Euplotes cysts. (A) Elongated parallel eisosomes, convex faces. Bar, 100 nm. (B) Smooth convex faces. Bar, 50 nm. (C and D) Concave faces with diagonal striations. Bar, 50 nm.
Flat eisosomes.
Infrequently, differentiated domains of microalgal plasma membranes are encountered that have the dimensions and ultrastructure of eisosomes but are coplanar with the membrane, showing no inward curvature. Figure 15 shows examples from A. protothecoides (Fig. 15A and B) and C. monoica (Fig. 15C and D), in which their IMP and striated features are indistinguishable from their furrowed counterparts (Fig. 6D to F and Fig. 9F). “Flat” eisosomes have also been described for the mycobiont of the lichen Myelochroa leucotyliza (13).
FIG 15.
Flat eisosomes (arrows). (A) Flat eisosome comparable to the convex face in Auxenochlorella protothecoides (c.f. Fig. 6C and G). Bar, 100 nm. (B) Flat eisosome comparable to the concave face in A. protothecoides (c.f. Fig. 6E and F). Bar, 100 nm. (C) Flat eisosome comparable to the convex face in Chlamydomonas monoica (c.f. Fig. 9E and F). Bar, 100 nm. (D) Flat eisosome comparable to the convex face in C. monoica (c.f. Fig. 9E and F). Bar, 100 nm.
Images at the resolution of Fig. 15D permitted the striation periodicity to be calculated as ∼10 nm, equivalent to the 10-nm period of the 30° repeats in the 3D maps of in vitro-polymerized Lsp1 (6) and the perpendicular repeats of the in vitro membrane-associated animal BAR domain proteins amphiphysin (69) and formin-binding protein 17 (70).
Search for algal genes homologous to genes encoding fungal eisosomal BAR domain proteins.
Homologues of genes encoding Pil1p and Lsp1p, the cytoplasmic proteins that dominate S. cerevisiae eisosomes (16), are restricted to the ascomycetes and basidiomycetes (28, 33; our BLAST results), a result that has generated the speculation that eisosomes are confined to the fungal kingdom (27).
Since Pil1p and Lsp1p contain N-terminal BAR domains—three coiled-coil α-helices that dimerize and participate in generating membrane curvature (6, 27, 28)—we went on to search for additional BAR domain-containing protein genes in sequenced microalgal genomes. The results are shown in Table S2 and Fig. S28 to S30 and S33 in the supplemental material.
As is the case in animals and fungi, most algal BAR domain protein genes encode proteins homologous to proteins involved with cytoplasmic trafficking (e.g., carrying ArfGAP [AGD] or sorting nexin [SNX] domains) (see Table S2 in the supplemental material).
Of interest in the green lineage is a family of genes, homologous to the Cre16.g653450 gene in C. reinhardtii, that encode ∼400-amino-acid proteins that contain N-terminal BAR domains but lack identified trafficking motifs (see Table S2 in the supplemental material). With one exception, representatives of this family are found in all sequenced green algae, including the cyst-forming multicellular charophyte Klebsormidium flaccidum (see Table S2 and Fig. S28); the family is therefore designated Green-BAR. The one exception is the eisosome-free Mamiellales group of prasinophytes, which instead carries a distinctive gene family (Prasino-BAR) encoding proteins with a BAR domain located at the C terminus (see Table S2 and Fig. S29).
Green-BAR homologues are absent from red algal genomes, but the red alga G. sulfuraria carries eight genes that encode a fourth distinctive family of BAR domain proteins lacking trafficking motifs. Homologues of these genes are absent from fungi and green algae but are found, with one exception, in all sequenced red algal genomes, including the cyst-forming multicellular organism Chondrus crispus (see Table S2 and Fig. S30 in the supplemental material); the family is therefore designated Red-BAR. The one exception is the eisosome-free red alga C. merolae, whose two short BAR domain sequences show no homology to any other proteins.
Red-BAR homologues are also present in the genomes of four algae that have not yet been examined by QFDEEM: (i) the walled glaucophyte Cyanophora paradoxa, a close relative of the red algae; (ii) the phaeophyte Ectocarpus siliculosus, a multicellular secondary endosymbiont with a red alga-derived plastid and several encysted stages in its life cycle; (iii) the nucleomorph-containing cryptophyte Guillardia theta, which harbors a red alga-derived plastid; and (iv) the nucleomorph-containing chlorarachniophyte Bigelowiella natans, a wall-less genus that forms cysts and harbors a green alga-derived plastid (see Table S2 in the supplemental material). Red-BAR genes are unexpectedly found in two species of the eustigmatophyte Nannochloropsis (see Table S2) that do not form eisosomes as vegetative cells by QFDEEM and have not been reported to form cysts. Red-BAR genes are absent from all available ciliate genomes (see Table S3), although Euplotes, which does not have a published genome, produces morphological eisosomes (Fig. 14); it is possible that the ciliates harbor a distinctive BAR domain family, but we have not pursued this question. Table S3 in the supplemental material lists other queried genomes in which Red-BAR and Green-BAR homologues are not found.
Figures S28 and S30 in the supplemental material show alignments of the Green-BAR and Red-BAR families. The proteins are most closely related to the N-BAR family typified by the amphiphysins (71). In animals, amphiphysins bind to clathrin proteins at the plasma membrane (71, 72) and in the trans-Golgi network (73) via LLDLDFDP and PWDLW motifs, but no such sequences, nor any D or W clusters, are found in any of the Green-BAR or Red-BAR proteins. The Green-BAR proteins resemble amphiphysin in having a short amphipathic α-helix N-terminal to the BAR domain, and the Red-BAR proteins in secondary endosymbionts resemble amphiphysin in having C-terminal SH3 domains (see Fig. S28 and S30). The three α-helices of the amphiphysin and the Lsp1/Pil1 BAR domains are predicted to be amphipathic (see Fig. S31 and S32), as expected for domains that adopt a coiled-coil tertiary structure (74); this is also the case for Red-BAR proteins (see Fig. S30) and for two of the three α-helices of Green-BAR proteins (see Fig. S28). No homology is evident in the unstructured C-terminal domains of either algal protein family.
Figure S33 in the supplemental material compares our constructed space-filling model (Gbar) of the BAR domain from a Chlamydomonas Green-BAR monomer (Cre16.g653450) with PDB files of crystal structures for the BAR domains of eisosomal Lsp1p from yeast (PDB entry 3plt), amphiphysin from Drosophila (PDB entry 1uru), and a Red-BAR protein from G. sulfuraria (PDB entry 3caz). The three α-helices of the Lsp1 BAR monomer are coplanar, generating a relatively wide structure, whereas in amphiphysin one helix is located behind the other two, generating a narrower structure. Like amphiphysin, the Green-Bar and Red-Bar monomers are narrow. In all four monomers, the amphipathic α-helical configurations generate patches or strips of hydrophobic residues (white regions in Fig. S33 in the supplemental material; also see Fig. 2A of reference 75). Arrows in Fig. S28 and S32 indicate the N-terminal amphipathic α-helices found in amphiphysin and in the Green-BAR proteins.
Taken together, our genome searches indicate that the Green-BAR and Red-BAR proteins are possible candidates as eisosome components in their respective lineages. With that said, there is no evidence for a “universal” set of eisosome-associated BAR proteins in eukaryotes.
Search for algal genes homologous to genes encoding fungal eisosomal non-BAR-domain proteins.
The cytoplasmic protein Seg1p, reported to control the length of eisosomes in S. cerevisiae (30), is found only within the budding yeasts (Saccharomycetales); its functional homologue, the “Seg1-like” Sle1p protein in S. pombe (30), shares sequence homology only in the polybasic C-terminal region. Not surprisingly, no algal Seg1 homologues were found.
The transmembrane proteins Can1p, Sur7p, and Nce102p colocalize with S. cerevisiae eisosomes and are considered members of the MCC domain (25, 26). Can1p is an arginine-proton cotransporter (76). Of the two CAN1 homologues in C. reinhardtii, AOC5 is strongly upregulated in eisosome-free vegetative cells with nitrogen starvation (77), as are other arginine transporters, whereas AOC6 is expressed at constant low levels (∼10 reads per kilobase per million mapped reads); expression levels in eisosome-forming C. reinhardtii zygotes have not been evaluated. Sur7p fails to colocalize with eisosomes in S. pombe, whereas Nce102p/Fhn1 is important for their integrity (43), as is also the case in S. cerevisiae (31). Sequences homologous to Sur7p and to the conserved Marvel domain of Nce102p were found in fungi and metazoa but not in any algae. Thus, there is no evidence as yet for a “universal” set of transmembrane eisosome-associated proteins in eukaryotes.
DISCUSSION
Eisosome distribution in eukaryotic microalgae.
The first conclusion of this report is that whereas the ubiquity of eisosomes throughout the fungal kingdom presumably indicates that eisosomes perform one or more selectable functions during fungal life cycles, they are evidently optional for microalgae writ large: as listed in Table S1 in the supplemental material, most of the microalgae investigated do not form eisosomes during vegetative growth. Importantly, while we can state with confidence that the microalgae listed in Table S1 are devoid of eisosomes under the growth conditions employed, it is possible that their assembly is induced under other conditions and/or during nonsampled stages of their life cycles.
While Table S1 in the supplemental material documents that the presence of cell walls per se does not dictate the formation of algal eisosomes, eisosomes prove to be absent from all wall-less microalgae examined to date. This correlation is particularly striking in the case of the wall-less species Cyanidioschizon merolae and the walled species Cyanidioschizon YNP 1A. Both species are nearly identical in their probed gene sequences, ultrastructure, and habitat, with the one known exception being that YNP 1A has walls and eisosomes, whereas C. merolae has neither. The genome sequence of YNP 1A, which is unavailable as yet, may document eisosomal gene candidates that are absent from the sequenced C. merolae genome.
In fungi, wall-less protoplasts of S. cerevisiae (11, 15) and C. albicans (45) retain their eisosomes, indicating that whereas walls may be involved in fungal eisosome assembly, they are not necessary for eisosome stability. In both S. cerevisiae and S. pombe, eisosomes are actively excluded from cellular domains where new cell wall is being laid down, such as small buds, growing tips, and mitotic cleavage planes (9, 14, 43, 78, 79), suggesting that they may actually interfere with early fungal wall formation but participate in wall maturation.
Another comparison of interest is between the vegetative cells of Chlamydomonas reinhardtii (Reinhardtinia clade) (60) and Chlamydomonas monoica (Moewusii clade) (61), since both construct similar vegetative cell walls by using a related group of hydroxyproline-rich glycoproteins (38, 80, 81). A proposed function of eisosomes is to anchor the wall to the cell membrane (25); this concept is framed in the context of the dense cell walls of fungi. In the case of C. monoica, however, the W1 wall layer with which the eisosomes are in contact is loosely fibrillar (Fig. 9C; see Fig. S21 and S22 in the supplemental material), with the more dense layers located at the periphery. Moreover, the C. reinhardtii plasma membrane associates with a similarly organized W1 wall layer yet forms no eisosomes (3, 81). Other members of the Reinhardtinia clade—Chlamydomonas komma/C. debaryana and Volvox carteri-–also lack eisosomes as vegetative cells (3; U. Goodenough and J. E. Heuser, unpublished data).
Eisosome organization: fracture face ultrastructure.
Prior to this report, knowledge of eisosome ultrastructure has largely been restricted to fungi, all of which display similar membrane interiors: a smooth or slightly roughened convex fracture face and an amorphous or finely striated concave fracture face. Our QFDEEM images of free-living fungi largely confirm these findings (Fig. 1; see Fig. S1 in the supplemental material), but our images of lichen-forming fungi and of microalgae reveal that eisosomes can adopt a far wider range of ultrastructural features.
The convex eisosomal face is smooth in most fungi and most cysts, where “smooth” represents a differentiated state compared with the surrounding IMP-rich plasma membrane. In contrast, in microalgal vegetative cells and in Borodinellopsis texensis cysts, the convex face carries a population of discrete IMPs ranging in density from moderate to abundant. This description also applies to C. monoica, except that its IMP-rich fracture face is concave rather than convex (Fig. 9). Ordinarily, the two fracture faces of cell membranes are expected to be complementary to one another, with IMPs on one face and corresponding pits—where the particles have pulled out—on the opposite face (82). However, the particle-free faces of microalgal eisosomes lack complementary pits. Such an outcome could arise if the IMPs were restricted to one leaflet rather than being transmembrane or if the lipids in the corresponding half-membrane were capable of rapidly sealing any pits after fracture and prior to etching. Notably, several proteins with transmembrane domains have been localized to yeast eisosomes, yet neither IMPs nor pits are observed on either fracture face. Clearly, the eisosome interior has unique properties compared to other biological membranes.
The concave faces of microalgal and lichen-forming fungal eisosomes display considerable diversity in their ultrastructure. They are usually finely granular, with the granules often aligned in fine striations at right angles (Fig. 10C) or at various angles of tilt with respect to the long axis of the furrows (Fig. 4D, 8B and C, and 9F). Three concave faces are particularly distinctive. (i) In the green alga Borodinellopsis texensis cyst, the striations form a honeycomb pattern (Fig. 13C and D). (ii) In the red alga G. sulfuraria (Fig. 5) and the green alga A. protothecoides (Fig. 6), the concave face regularly alternates between broad and narrow bands, with one set of bands being more sensitive to etching than the other. (iii) In the lichen-forming fungus C. grayi, concave faces display an irregularly banded organization (Fig. 3C).
It is our working assumption that the striated and banded patterns are the consequence of an “embossing” process wherein cytosol-localized proteins that associate with eisosomes are able to order the inner leaflet lipids into regular configurations. Support for this assumption comes (i) from the concordance between the ∼30° pitch of the S. cerevisiae striations and the ∼30° pitch adopted by in vitro polymers of Pil1p/Lsp1p (6) and (ii) from the 10-nm period displayed both by the striations (Fig. 15D) and by membrane-associated animal BAR domains in vitro (69, 70). To our knowledge, such an ordered patterning of membrane lipids has been described only for eisosomes. Eisosomal striations in all the fungi examined are pitched at an ∼30° angle, and all fungi queried carry Pil1p/Lsp1p homologues, whereas the striations or bands in algae and the ciliate Euplotes, which lack Pil1p/Lsp1p homologues, can be at right angles to the long axis of the furrow (C. reinhardtii) or pitched at ∼20° (Auxenochlorella and Euplotes), ∼30° (YNP 1A, Asterochloris sp., and C. monoica), or ∼40° (Galdieria). Hence, the embossment hypothesis posits that the systems organizing the eisosomal lipids in various lineages are both similar in their general features and diverse in their fine-grained features, where the elements that organize the broad-banded examples are expected to be particularly complex. Any model of eisosome architecture will need to include an explanation of the striking topological reversal of membrane organization in C. monoica and C. eugametos, in which the embossed leaflet faces the exterior.
An interesting feature of the finely striated embossments in both fungi and microalgae is that they are not uniformly present. This is illustrated in Fig. 4D, in which two striated regions are denoted by arrows but no striations are evident elsewhere. In some cases, a membrane may carry a mix of striated and nonstriated eisosomes; in other cases, one cell will be uniformly amorphous, while an adjacent cell is uniformly striated. We have ruled out the possibility that this phenomenon is an artifact of platinum replication. One possibility is that striation variability may reflect a switch in “levels of engagement” between the eisosomal lipids and their underlying organizing systems. If this is the case, the variability could be a morphological reflection of (some of) the signal transduction properties attributed to eisosomes (19, 20, 22).
Walther and colleagues (30, 83) have suggested analogies between eisosomes and caveolae. Caveolae, found in certain animal cell types, start out as flat, cholesterol-rich lipid rafts that are induced to form spherical invaginations via the association of accessory cholesterol-binding proteins, called caveolins, which hairpin into but do not span the plasma membrane, and the cavins, peripheral membrane proteins thought to control caveolin oligomerization. However, no “embossed” ultrastructural patterning is observed in either of the membrane fracture faces of caveolae (84; and J. E. Heuser, unpublished data).
Flat eisosomes have occasionally been encountered in fungi (13) and algae (Fig. 15); since confocal images of fungal eisosome/MCC domains are unable to distinguish whether tagged domains are flat or furrowed, it is possible that certain mutant strains will be found to be capable of the former configuration but not the latter. Flat eisosomes carry the same “embossed” patterns as their curved counterparts, indicating that the establishment of the membrane domains and the posited interactions that emboss the lipids can be independent of the generation of curvature. Flat eisosomes may also be remnants of furrows that have lost their curvature, and indeed, there might be furrow-flat-furrow cycles that would be expected to modulate cell surface area and/or wall interactions.
Eisosome organization: large-scale topology.
In many non-lichen-forming fungi, the eisosomes have a uniform short size, are independent, and are relatively equidistant (Fig. 1A to D), and some algae adopt an analogous punctate topology (Fig. 7, 8A to C, 9A, 10A, 12A, and 13B). Other fungi, such as S. pombe and Penicillium sp., form elongated long meshworks that may anastomose (Fig. 1E; see Fig. S1 in the supplemental material), a pattern also seen in several algae (Fig. 4C, 5A and B, 6A and B, and 11A) and in Euplotes cysts (Fig. 14). These topological alternatives are evidently not conferred by the BAR domain proteins per se: when the S. pombe PIL1 gene is introduced into pil1 mutants of S. cerevisiae, punctate eisosomes are formed; reciprocally, S. pombe mutants rescued with S. cerevisiae PIL1 form elongated eisosomes (43, 85). Protoplast fungi have been observed to switch from punctate to elongated and back again (6, 44, 45), the eisosomes of S. cerevisiae are punctate in growing cells but elongated in ascospores (86), and deletion of the ypr050c gene in S. cerevisiae leads to elongated eisosomes in growing cells (5). Lichenizing fungi and algae also display several eisosomal topologies (see below). In several microalgae (e.g., A. protothecoides [Fig. 6]), cells with either punctate or elongated topologies are encountered, possibly because they are in different states of differentiation. Most of the organisms analyzed in this report were observed only under a limited set of growth conditions/states of differentiation, so it is possible that such switches are even more common than suggested by our sampling. Presumably these large-scale topological features have functional correlates.
Our observations specifically encourage research into the role that eisosomal topology might play in the biology of lichens. In Cladonia, eisosomes are sparse in both the mycobiont and the photobiont (Asterochloris) when the organisms are lichenized, and hence interacting and growth restricted, whereas they are abundant when the organisms are cultured separately on agar and engaging in rapid growth (Fig. 3C and D and 8). Arakawa-Kobayashi and Kanaseki (13) also noted differences in the eisosomes of the Myelochroa leucotyliza mycobiont when it was lichenized versus growing alone. Within the Candelaria lichen, no variability is evident in the photobiont Trebouxia (Fig. 7), but several configurations are encountered in the mycobiont. In both Cladonia and Candelaria, the eisosomes are often curved (Fig. 2B and 3A), a configuration not seen in nonlichenizing fungi but prominent as well in Myelochroa leucotyliza (13). These variations may be elicited by local environmental parameters (e.g., levels of intralichen humidity or proximity to symbionts or bacteria) and/or reflect alternative states of differentiation.
Particularly dramatic is a system of densely packed sinuous eisosomes encountered once in a Candelaria lichen (Fig. 2C) and often in Cladonia grown on agar (Fig. 3B and C) but in no other fungi. The eisosomes of free-living Asterochloris, the photobiont in Cladonia, are also sinuous (Fig. 8), although less densely packed and with fine striations (Fig. 8B and C) rather than bands (Fig. 3C) on their concave faces. This sinuous class of eisosome conceivably plays a role in the mysterious process wherein specific solo fungi and microalgae recognize one another and initiate lichen formation.
Conserved versus nonconserved eisosomal parameters.
Our studies point to the following six eisosomal features that are encountered across several phyla: (i) a curved, elongated membrane indentation of uniform width (∼30 to 50 nm) and depth (∼50 nm) that fails to pinch off into vesicles; (ii) the presence of cell wall material in the furrow; (iii) a large-scale topology that can be either punctate or elongated, with the capacity to convert between the two configurations; (iv) “embossed” lipid configurations and IMP endowments that differ from the surrounding plasma membrane; (v) fracture faces that are noncomplementary; and (vi) “flat” versions of the intramembranous domains. In addition, fungal eisosomes tagged with GFP-labeled components are observed to be immobile (16, 21, 29, 35, 43), albeit they exhibit subunit turnover (87); the uniform distribution of punctate eisosomes in algae (e.g., see Fig. 7, 9, 10, and 12) suggests that this immobility feature is also conserved.
Given these conserved features, we anticipated that we would find algal homologues of the set of proteins identified as eisosome-localized proteins in S. cerevisiae and other fungi. However, no homologues were found in the available sequenced genomes of eisosome-forming microalgae. Thus, the assembly of apparently homologous eisosomal furrows can be achieved using a variety of protein components, including complements of algal proteins that have yet to be identified. With that said, the common ancestor to the fungal, algal, and ciliate radiations is presumed to be close to the last eukaryotic common ancestor (LECA), indicating that the capacity to form eisosomal membrane furrows represents an ancient adaptation.
Although no fungal eisosomal protein homologues were identified, genes encoding two distinct families of BAR domain proteins were found in all known eisosome-forming green and red or red-derived algae examined and were absent from all known eisosome-free algae except Nannochloropsis (see Table S2 and Fig. S28 to S30 and S33 in the supplemental material). These families, which we call Green-BAR and Red-BAR, represent candidate Pil1p/Lsp1p alternatives, and the obvious future experiment will be to test their localization using fluorophore-tagged proteins. If eisosome associations are documented, then at least three BAR domain families—Pil1p/Lsp1p, Green-BAR, and Red-BAR—will have been shown to participate in eisosome construction, with the sequence differences within the Green- and Red-BAR families (see Fig. S28 and S30) perhaps contributing to the ultrastructural variability documented in this report.
Eisosomes also have within-group taxonomic potential, as illustrated by the following two examples. (i) Among the chlorophytes, C. monoica and C. eugametos are closely related members of the Moewusii clade and share a “reversed” eisosomal organization: the IMP-dense leaflet abuts the cytoplasm, while the striated leaflet faces the exterior (Fig. 9) (37, 38). In contrast, the eisosomes of two members of the branching Chloromonas clade (61, 88), Chlamydomonas nivalis/augusta and Chlamydomonas yellowstoniensis, display a standard, nonreversed orientation (3). (ii) Among the trebouxiophytes, Auxenochlorella protothecoides, in the Chlorella lineage clade (54), has a broad-banded eisosomal concave face (Fig. 6), whereas Oocystis apiculata, in the branching Oocystis lineage clade (54), displays narrow striations (89). Hence, eisosomal ultrastructure could serve as a diagnostic marker for related radiations.
Functional speculations.
A recent review of fungal eisosomes (90) concludes: “Thus, we have a complex and strongly conserved cellular apparatus, which has even been called an organelle, without an obvious physiological function.” Knockouts of eisosome-associated proteins in fungi generate a range of mutant phenotypes, from undetectable (33, 35, 90) to mild (14, 16) to substantial (25, 29, 91, 92). Douglas and Konopka (25) reviewed in detail the numerous functions that have been proposed for fungal eisosomes. Our observations on algal eisosomes have led us to the following additional perspectives.
First, as noted above, it seems clear that a strong relationship exists between eisosomes and cell walls and that this relationship can be independent of a membrane-associated actin cytoskeleton, since microalgae lack such a cytoskeleton. This conclusion carries the important caveat that although no wall-less algae have been found to carry eisosomes, there are numerous algae that have walls but no eisosomes (see Table S1 in the supplemental material). Therefore, the relationship holds for only a subset of cases. Wall “rigidity” does not appear to be a common denominator since, for example, Nannochloropsis and Botryococcus have robust cell walls (93, 94) but no eisosomes. A chemical analysis of the walls of eisosome-forming organisms to determine whether they share any common constituents would be valuable. Wang et al. (92) propose, for example, that eisosomes are needed for proper synthesis of the β-1,3-glucan component of the Candida cell wall.
For organisms that engage in an eisosome-wall relationship, outcomes of this relationship could include the generation of wall curvature, as suggested by Fig. 3A, or the regulation of stresses such as osmotic imbalance, freezing, and drying. The presence of eisosomes in all examined algal cysts and in Euplotes cysts is consonant with a role in such activities, albeit our observation that eisosomes are absent from Dictyostelium spores and Naegleria cysts indicates that the mediation of such stresses can be organized in other ways.
Finally, studies of fungi document that eisosomes accumulate a subset of transmembrane proteins (e.g., Can1p, Sur7p, and Nce102p) in some but not all species. Extending this finding, we suggest that once such a differentiated domain of the plasma membrane is established, perhaps initially to serve a role(s) in membrane-wall relationships, it may accumulate additional, lineage-specific roles in docking and segregating particular proteins and lipids for specific functions. If this is the case, then it follows that eisosomes may represent platforms or scaffolds for the acquisition of diverse differentiated traits during the radiation of a subset of eukaryotic microbes.
Model of BAR domain-membrane interactions and eisosome formation.
The three α-helices of all BAR domain proteins are amphipathic, generating nonpolar patches or strips that mediate the adoption of the coiled-coil configuration (74, 75) (see Fig. S28 to S33 in the supplemental material). Yu and Schulten (95) performed molecular dynamic simulations of the interactions between animal F-BAR proteins and membrane liposomes made of 33% phosphatidylserine (as 1,2-dioleoyl-sn-glycero-3-phosphoserine [DOPS]) and 67% phosphatidylcholine (as 1,2-dioleoyl-sn-glycero-3-phosphocholine [DOPC]). They reported that following the electrostatic interactions that establish the initial protein-membrane binding, there occurs a partial unwinding of the domain's 3-helix bundle structure. We suggest that this uncoiling might be driven by interactions between the nonpolar residues within the coiled coil and the hydrophobic interior of the membrane. At such a close range, such interactions would be expected to be energetically favorable, particularly given that the electrostatic potential would presumably have been neutralized during the initial binding.
Ergosterol is an abundant component of yeast plasma membranes (∼50% of total lipid [96]), and yeast eisosomes have been shown to be dramatically enriched in ergosterol compared with the rest of the membrane (18). Since embossed patterns have not been observed in (ergo)sterol-poor membranes (e.g., Golgi vesicles) that also associate with BAR proteins, the embossed patterns of yeast eisosomes suggest that ergosterol may play a role. High sterol concentrations increase the viscosity of membrane interiors (97, 98). We therefore suggest that once yeast eisosomal BAR proteins are tethered to plasma membranes by their electrostatic targeting signals (6, 28), the uncoiling and the posited association of their nonpolar domains with the membrane interior can generate regular striations in the ergosterol-rich phase of the membrane, an ordering that may not readily be imposed on a more fluid fatty acid-rich phase.
Since ergosterol-rich domains are absent in Pil1p deletion mutants (18), we further suggest that Pil1p/Lsp1p may recruit ergosterol to the eisosomal membrane, perhaps preferentially to its inner leaflet. A membrane with an ergosterol-rich inner leaflet may be prone to bending (99) but incapable of fusing, explaining why the furrows do not pinch off as vesicles or tubules in vivo. This proposal might also explain why ergosterol-free liposomes incubated with Pil1p/Lsp1p in vitro form closed tubules rather than displaying ridge-like deformations (6). The regulation of Pil1p/Lsp1p abundance and/or membrane affinity would be expected, in this scenario, to influence plasma membrane fluidity by sequestering or releasing ergosterol moieties.
Red, green, and red-derived algae also produce ergosterol or ergosterol-related sterols (100, 101). Hence, these speculations may also apply to other eisosome-constructing organisms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carrie Goodson for cell culture and Yu Hang and Klaus Schulten (NIH Center for Macromolecular Modeling & Informatics, UI Urbana) for valuable discussions.
Support for this work was provided as follows. The Goodenough lab was supported by contract DE-EE0003046 via the National Alliance for Advanced Biofuels and Bioproducts (NAABB) from the U.S. Department of Energy (DOE) and by grant DE-SC0006873 from the DOE Office of Science (BER). The J.-H. Lee lab was supported by grant 2013M1A8A1056300 from the Korea CCS R&D Center (KCRC), Korean Ministry of Science, and by Discovery Grant 418471-12 from the Natural Sciences and Engineering Research Council (NSERC) of Canada. The Northrup lab was supported by NSF grants EAR031193 and EAR0311930D and Kenneth Ingham Consulting, with technical support from the University of New Mexico’s Molecular Biology Facility supported by NIH grant P20RR18754. The Doering lab was supported by National Institutes of Health (NIH) grants R01 AI071007 and AI07879. The Kovar lab was supported by an NIH postdoctoral fellowship in Tom Pollard's lab at Yale University. The Lammers lab was supported by DOE contract DE-EE0006316 for the Realization of Algae Potential and by National Science Foundation award IIA-1301346 Energize New Mexico (EPSCoR). The R. W. Lee lab was supported by discovery grant 9599-12 from the Natural Sciences and Engineering Research Council (NSERC) of Canada. The Lipke lab was supported by NIH grant R01 GM 098616. The Polle lab was supported by the NAABB. The Sayre lab was supported by the NAABB. The VanWinkle-Swift lab was supported by NIH grant 2R15GM071374.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00106-15.
REFERENCES
- 1.Moor H, Mühlethaler K. 1963. Fine structure in frozen-etched yeast cells. J Cell Biol 17:609–628. doi: 10.1083/jcb.17.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gross H, Kuebler O, Bas E, Moor H. 1978. Decoration of specific sites on freeze-fractured membranes. J Cell Biol 79:646–656. doi: 10.1083/jcb.79.3.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke KJ, Leeson EA. 1985. Plasmalemma structure in freezing tolerant unicellular algae. Protoplasma 129:120–126. doi: 10.1007/BF01279909. [DOI] [Google Scholar]
- 4.Den Hertog AL, van Marle J, van Veen HA, Van't Hof W, Bolscher JGM, Veerman ECI, Niew Amerongen AV. 2005. Candidacidal effects of two antimicrobial peptides: histatin 5 causes small membrane defects, but LL-37 causes massive disruption of the cell membrane. Biochem J 388:689–695. doi: 10.1042/BJ20042099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strádalová V, Stahlschmidt W, Grossman G, Blažĺková M, Rachel R, Tanner W, Malinsky J. 2009. Furrow-like invaginations of the yeast plasma membrane correspond to membrane compartment of Can1. J Cell Sci 122:2887–2894. doi: 10.1242/jcs.051227. [DOI] [PubMed] [Google Scholar]
- 6.Karotki L, Huiskonen JT, Stefan CJ, Ziółkowska NE, Roth R, Surma MA, Krogan NJ, Emr SD, Heuser J, Grünewald K, Walther TC. 2011. Eisosome proteins assemble into a membrane scaffold. J Cell Biol 195:889–902. doi: 10.1083/jcb.201104040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Littlefield LJ, Bracker CE. 1972. Ultrastructural specialization at the host-pathogen interface in rust-infected flax. Protoplasma 74:271–305. doi: 10.1007/BF01282533. [DOI] [Google Scholar]
- 8.Takeo K, Uesaka I, Uehira K, Nishiura M. 1973. Fine structure of Cryptococcus neoformans grown in vitro as observed by freeze-etching. J Bacteriol 113:1442–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeo K. 1984. Lack of invaginations of the plasma membrane during budding and cell division of Saccharomyces cerevisiae and Schizosaccharomyces pombe. FEMS Microbiol Lett 22:97–100. doi: 10.1111/j.1574-6968.1984.tb00704.x. [DOI] [Google Scholar]
- 10.Barug D, de Groot K. 1985. Effect of the imidazole derivative lombazole on the ultrastructure of Staphylococcus epidermidis and Candida albicans. Microb Agents Chemother 28:643–647. doi: 10.1128/AAC.28.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osumi M. 1998. The ultrastructure of yeast: cell wall structure and formation. Micron 29:207–233. doi: 10.1016/S0968-4328(97)00072-3. [DOI] [PubMed] [Google Scholar]
- 12.Takeo K. 1998. Ultrastructure of fungal plasma membranes as revealed by freeze-replica. Jpn J Med Mycol 39:129–134. doi: 10.3314/jjmm.39.129. [DOI] [PubMed] [Google Scholar]
- 13.Arakawa-Kobayashi S, Kanaseki T. 2004. A study of lipid secretion from the lichen symbionts, ascomycetous fungus Myelochroa leucotyliza and green alga Trebouxia sp. J Struct Biol 146:401–415. doi: 10.1016/j.jsb.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Young ME, Karpova TS, Brügger B, Moschenross DM, Wang GK, Schneiter R, Wieland FT, Cooper JA. 2002. The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol Cell Biol 22:927–934. doi: 10.1128/MCB.22.3.927-934.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malinska K, Malinsky J, Opekarova M, Tanner W. 2004. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J Cell Sci 117:6031–6041. doi: 10.1242/jcs.01493. [DOI] [PubMed] [Google Scholar]
- 16.Walther TC, Brickner JH, Aguilar PS, Bernales S, Pantoja C, Walter P. 2006. Eisosomes mark static sites of endocytosis. Nature 439:998–1003. doi: 10.1038/nature04472. [DOI] [PubMed] [Google Scholar]
- 17.Aguilar PS, Fröhlich F, Rehman M, Shales M, Ulitsky I, Olivera-Couto A, Braberg H, Shamir R, Walter P, Mann M, Ejsing CS, Krogan NJ, Walther TC. 2010. A plasma-membrane E-MAP reveals links of the eisosome with sphingolipid metabolism and endosomal trafficking. Nat Struct Mol Biol 17:901–909. doi: 10.1038/nsmb.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grossmann G, Opekarová M, Malinsky J, Weig-Meckl I, Tanner W. 2007. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J 26:1–8. doi: 10.1038/sj.emboj.7601466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fröhlich F, Christiano R, Olson DK, Alcazar-Roman A, DeCamilli P, Walther TC. 2014. A role for eisosomes in maintenance of plasma membrane phosphoinositide levels. Mol Biol Cell 25:2797–2806. doi: 10.1091/mbc.E13-11-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabeche R, Robuev A, Krogan NJ, Moseley JB. 2014. A Pil1-Sle1-Syj1-Tax4 functional pathway links eisosomes with PI(4,5)P2 regulation. J Cell Sci 127:1318–1326. doi: 10.1242/jcs.143545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walther TC, Aguilar PS, Fröhlich F, Chu F, Moreira K, Burlingame AL, Walter P. 2007. Pkh-kinases control eisosome assembly and organization. EMBO J 26:4946–4955. doi: 10.1038/sj.emboj.7601933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo G, Gruhler A, Liu Y, Jensen ON, Dickson RC. 2008. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J Biol Chem 283:10433–10444. doi: 10.1074/jbc.M709972200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fröhlich F, Moreira K, Aguilar PS, Hubner NC, Mann M, Walter P, Walther TC. 2009. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J Cell Biol 185:1227–1242. doi: 10.1083/jcb.200811081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berchtold D, Piccolis M, Chiaruttini N, Riezman I, Riezman H, Roux A, Walther TC, Lowsith R. 2012. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat Cell Biol 14:542–548. doi: 10.1038/ncb2480. [DOI] [PubMed] [Google Scholar]
- 25.Douglas LM, Konopka JB. 2014. Fungal membrane organization: the eisosome concept. Annu Rev Microbiol 68:377–393. doi: 10.1146/annurev-micro-091313-103507. [DOI] [PubMed] [Google Scholar]
- 26.Malinsky J, Opekarová M, Grossman G, Tanner W. 2013. Membrane microdomains, rafts, and detergent-resistant membranes in plants and fungi. Annu Rev Plant Biol 64:501–529. doi: 10.1146/annurev-arplant-050312-120103. [DOI] [PubMed] [Google Scholar]
- 27.Olivera-Couto A, Graña M, Harispe L, Aguilar PS. 2011. The eisosome core is composed of BAR domain proteins. Mol Biol Cell 22:2360–2372. doi: 10.1091/mbc.E10-12-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ziółkowska NE, Karotki L, Rehman M, Huiskonen JT, Walther TC. 2011. Eisosome-driven plasma membrane organization is mediated by BAR domains. Nat Struct Mol Biol 18:854–856. doi: 10.1038/nsmb.2080. [DOI] [PubMed] [Google Scholar]
- 29.Seger S, Rischatsch R, Philippsen P. 2011. Formation and stability of eisosomes in the filamentous fungus Ashbya gossypii. J Cell Sci 124:1629–1634. doi: 10.1242/jcs.082487. [DOI] [PubMed] [Google Scholar]
- 30.Moreira KE, Schuck S, Schrul B, Frölich F, Mosely JB, Walther TC. 2012. Seg1 controls eisosome assembly and shape. J Cell Biol 198:405–420. doi: 10.1083/jcb.201202097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loibl M, Grossman G, Stradalova V, Klingl A, Rachel R, Tanner W, Malinsky J, Opekarová M. 2010. C terminus of Nce102 determines the structure and function of microdomains in the Saccharomyces cerevisiae plasma membrane. Eukaryot Cell 9:1184–1192. doi: 10.1128/EC.00006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grossmann G, Malinsky J, Stahlschmidt W, Loibl M, Weig-Meckl I, Frommer WB, Opekarová M, Tanner W. 2008. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J Cell Biol 183:1075–1088. doi: 10.1083/jcb.200806035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vangelatos I, Roumelioti K, Gournas C, Suarez T, Scazzocchio C, Sophianopoulou V. 2010. Eisosome organization in the filamentous ascomycete Aspergillus nidulans. Eukaryot Cell 9:1441–1454. doi: 10.1128/EC.00087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brach TS, Kaksonen M. 2011. Assessment of the role of plasma membrane domains in the regulation of vesicular traffic in yeast. J Cell Sci 124:328–337. doi: 10.1242/jcs.078519. [DOI] [PubMed] [Google Scholar]
- 35.Athanasopoulos A, Boleti H, Scazzocchio C, Sophianopoulou V. 2013. Eisosome distribution and localization in the meiotic progeny of Aspergillus nidulans. Fungal Genet Biol 53:84–96. doi: 10.1016/j.fgb.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Murphy ER, Boxberger J, Colvin R, Lee SJ, Zahn G, Loor F, Kim K. 2011. Pil1, an eisosome organizer, plays an important role in the recruitment of synaptojanins and amphiphysins to facilitate receptor-mediated endocytosis in yeast. Eur J Cell Biol 90:825–833. doi: 10.1016/j.ejcb.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Bray DF, Nakamura K, Costerton JW, Wagenaar EB. 1974. Ultrastructure of Chlamydomonas eugametos as revealed by freeze-etching: cell wall, plasmalemma and chloroplast membrane. J Ultrastruct Res 47:125–141. doi: 10.1016/S0022-5320(74)80065-1. [DOI] [PubMed] [Google Scholar]
- 38.Goodenough UW, Heuser JE. 1988. Molecular organization of the cell wall and cell-wall crystals from Chlamydomonas eugametos. J Cell Sci 90:735–750. [Google Scholar]
- 39.Hagen C, Siegmund S, Braune W. 2002. Ultrastructural and chemical changes in the cell wall of Haematococcus pluvialis (Volvocales, Chlorophyta) during aplanospore formation. Eur J Phycol 37:217–226. doi: 10.1017/S0967026202003669. [DOI] [Google Scholar]
- 40.Heuser JE. 2011. The origins and evolution of freeze-etch electron microscopy. J Electron Microsc (Tokyo) 60(Suppl 1):S3–S29. doi: 10.1093/jmicro/dfr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singer SK, Nicolson GL. 1972. The fluid mosaic model of the structure of cell membranes. Science 175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 42.Parchert KJ, Spilde MN, Porras-Alfaro A, Nyberg AM, Northup DE. 2012. Fungal communities associated with rock varnish in Black Canyon, New Mexico: casual inhabitants or essential partners? Geomicrobiol J 29:752–766. doi: 10.1080/01490451.2011.619636. [DOI] [Google Scholar]
- 43.Kabeche R, Baldissard S, Hammond J, Howard L, Mosely JB. 2011. The filament-forming protein Pil1 assembles linear eisosomes in fission yeast. Mol Biol Cell 22:4059–4067. doi: 10.1091/mbc.E11-07-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nečas O, Kopecká M, Brichta J. 1969. Interpretation of surface structures in frozen-etched protoplasts of yeasts. Exp Cell Res 58:411–419. doi: 10.1016/0014-4827(69)90522-9. [DOI] [PubMed] [Google Scholar]
- 45.Miragall F, Rico H, Sentandreu R. 1986. Changes in the plasma membrane of regenerating protoplasts of Candida albicans as revealed by freeze-fracture electron microscopy. J Gen Microbiol 132:2845–2853. [DOI] [PubMed] [Google Scholar]
- 46.Büdel B, Rhiel E. 1987. Studies on the ultrastructure of some cyanolichen haustoria. Protoplasma 139:145–152. doi: 10.1007/BF01282285. [DOI] [Google Scholar]
- 47.Armaleo D, May S. 2009. Sizing the fungal and algal genomes of the lichen Cladonia grayi through quantitative PCR. Symbiosis 49:43–51. doi: 10.1007/s13199-009-0012-3. [DOI] [Google Scholar]
- 48.Misumi O, Matsuzaki M, Nozaki H, Miyagishima S, Mori T, Nishida K, Yagisawa F, Yoshida Y, Kuroiwa H, Kuroiwa T. 2005. Cyanidioschyzon merolae genome. A tool for facilitating comparable studies on organelle biogenesis in photosynthetic eukaryotes. Plant Physiol 137:567–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi Y, Ohnuma M, Kuoiwa T, Tanaka K, Hanaoka M. 2010. The basics of cultivation and molecular genetic analysis of the unicellular red alga Cyanidioschyzon merolae. J Endocytobiosis Cell Res 20:53–61. [Google Scholar]
- 50.Stark MR, Dunn EA, Dunn WSC, Grisdale CJ, Daniele AR, Halstead MRG, Fast NM, Rader SD. 2015. Dramatically reduced spliceosome in Cyanidioschyzon merolae. Proc Natl Acad Sci U S A 112:E1191–E1200. doi: 10.1073/pnas.1416879112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toplin JA, Norris TB, Lehr CR, McDermott TR, Castenholz RW. 2008. Biogeographic and phylogenetic diversity of thermoacidophilic Cyanidiales in Yellowstone National Park, Japan, and New Zealand. Appl Environ Microbiol 74:2822–2833. doi: 10.1128/AEM.02741-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schönknecht G, Chen W, Ternes CM, Barbier GG, Shrestha RP, Stanke M, Bräutigam A, Baker BJ, Banfield JF, Garavito RM, Carr K, Wilderson C, Rensing SA, Gagneul D, Dickenson NE, Oesterhelt C, Lercher MJ, Weber APM. 2013. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339:1207–1210. doi: 10.1126/science.1231707. [DOI] [PubMed] [Google Scholar]
- 53.Selvaratnam T, Pegallapati AK, Montelya F, Rodriguez G, Nirmalakhandan N, Van Voorhies W, Lammers PJ. 2014. Evaluation of a thermo-tolerant acidophilic alga, Galdieria sulphuraria, for nutrient removal from urban wastewaters. Bioresour Technol 156:395–399. doi: 10.1016/j.biortech.2014.01.075. [DOI] [PubMed] [Google Scholar]
- 54.Darienko T, Pröschold T. 2015. Genetic variability and taxonomic revision of the genus Auxenochlorella (Shihira et Krauss) Kulina et Puncocharova (Trebouxiophyceae, Chlorophyta). J Phycol 51:394–400. doi: 10.1111/jpy.12279. [DOI] [PubMed] [Google Scholar]
- 55.Gao C, Wang Y, Shen Y, Yan D, He X, Dai J, Wu Q. 2014. Oil accumulation mechanisms of the oleaginous microalga Chlorella protothecoides revealed through its genome, transcriptomes, and proteomes. BMC Genomics 15:582. doi: 10.1186/1471-2164-15-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sadowska-Deś AD, Dal Grande F, Lumbsch HT, Beck A, Otte J, Hur J, Kim JA, Schmitt I. 2014. Integrating coalescent and phylogenetic approaches to delimit species in the lichen photobiont Trebouxia. Mol Phylogenet Evol 76:202–212. doi: 10.1016/j.ympev.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 57.Peksa O, Škaloud P. 2011. Do photobionts influence the ecology of lichens? A case study of environmental preferences in symbiotic green alga Asterochloris (Trebouxiophyceae). Mol Ecol 20:3936–3948. doi: 10.1111/j.1365-294X.2011.05168.x. [DOI] [PubMed] [Google Scholar]
- 58.Beck A, Friedl T, Rambold G. 1998. Selectivity of photobiont choice in a defined lichen community: inferences from cultural and molecular studies. New Phytol 139:709–720. doi: 10.1046/j.1469-8137.1998.00231.x. [DOI] [Google Scholar]
- 59.VanWinkle-Swift KP, Aubert B. 1983. Uniparental inheritance in a homothallic alga. Nature 303:167–169. doi: 10.1038/303167a0. [DOI] [Google Scholar]
- 60.Nakada T, Misawa K, Nozaki H. 2008. Molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18S rRNA phylogenetic analyses. Mol Phylogenet Evol 48:281–291. doi: 10.1016/j.ympev.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 61.Demchenko E, Mikhailyuk T, Coleman AW, Pröschold T. 2012. Generic and species concepts in Microglena (previously the Chlamydomonas monadina group) revised using an integrative approach. Eur J Phycol 47:264–290. doi: 10.1080/09670262.2012.678388. [DOI] [Google Scholar]
- 62.Harris EH. 2009. The Chlamydomonas sourcebook. I. Introduction to Chlamydomonas and its laboratory use. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 63.Triemer RE, Brown RM. 1976. Ultrastructure of meiosis in Chlamydomonas reinhardtii. Br Phycol J 12:23–44. [Google Scholar]
- 64.Mallet MA, Lee RW. 2006. Identification of three distinct Polytomella lineages based on mitochondrial DNA features. J Eukaryot Microbiol 53:79–84. doi: 10.1111/j.1550-7408.2005.00079.x. [DOI] [PubMed] [Google Scholar]
- 65.Smith DR, Lee RW. 2014. A plastid without a genome: evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiol 164:1812–1819. doi: 10.1104/pp.113.233718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brown DL, Leppard GG, Massalski A. 1976. Fine structure of encystment of the quadriflagellate alga, Polytomella agilis. Protoplasma 90:139–154. doi: 10.1007/BF01276484. [DOI] [PubMed] [Google Scholar]
- 67.Buchheim MA, Sutherland DM, Buchheim JA, Wolf M. 2013. The blood alga: phylogeny of Haematococcus (Chlorophyceae) inferred from ribosomal RNA gene sequence data. Eur J Phycol 48:318–329. doi: 10.1080/09670262.2013.830344. [DOI] [Google Scholar]
- 68.Verni F, Rosati G. 2011. Resting cysts: a survival strategy in protozoa Ciliophora. Ital J Zool 78:134–145. doi: 10.1080/11250003.2011.560579. [DOI] [Google Scholar]
- 69.Takei K, Slepnev VI, Haucke V, De Camilli P. 1999. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol 1:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- 70.Frost A, Perera R, Roux A, Spasov K, Desting O, De Camilli P, Unger VM. 2008. Structural basis of membrane invagination by F-BAR domains. Cell 132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 72.Slepnev VI, Ochoa G-C, Butler MH, De Camilli P. 2000. Tandem arrangement of the clathrin and AP-2 binding domains in amphiphysin 1 and disruption of clathrin coat function by amphiphysin fragments comprising these sites. J Biol Chem 275:17583–17589. doi: 10.1074/jbc.M910430199. [DOI] [PubMed] [Google Scholar]
- 73.Huser S, Suri G, Crottet P, Spiess M. 2013. Interaction of amphiphysins with AP-1 clathrin adaptors at the membrane. Biochem J 450:73–83. doi: 10.1042/BJ20121373. [DOI] [PubMed] [Google Scholar]
- 74.Mason JM, Arndt KM. 2004. Coiled coil domains: stability, specificity, and biological implications. Chembiochem 5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 75.Gallop JL, McMahon HT. 2005. BAR domains and membrane curvature: bringing your curves to the BAR. Biochem Soc Symp 72:223–231. doi: 10.1042/bss0720223. [DOI] [PubMed] [Google Scholar]
- 76.Opekarová M, Caspari T, Tanner W. 1993. Unidirectional arginine transport in reconstituted plasma-membrane vesicles from yeast overexpressing CAN1. Eur J Biochem 211:683–688. doi: 10.1111/j.1432-1033.1993.tb17596.x. [DOI] [PubMed] [Google Scholar]
- 77.Goodenough U, Blaby I, Casero D, Gallaher SD, Goodson C, Johnson S, Lee J, Merchant SS, Pellegrini M, Roth R, Rusch J, Singh M, Umen JG, Weiss Wulan TLT. 2014. The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot Cell 13:591–613. doi: 10.1128/EC.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Konomi M, Fujimoto K, Toda T, Osumi Opekarová MM. 2003. Characterization and behavior of α-glucan synthase in Schizosaccharomyces pombe as revealed by electron microscopy. Yeast 20:427–438. doi: 10.1002/yea.974. [DOI] [PubMed] [Google Scholar]
- 79.Moreira KE, Walther TC, Aguilar PS, Walter P. 2009. Pil1 controls eisosome biogenesis. Mol Biol Cell 20:809–818. doi: 10.1091/mbc.E08-03-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roberts K, Gurney-Smith M, Hills GJ. 1982. The structure of algal cell walls, p 1–40. In Harris JR. (ed), Electron microscopy of proteins, vol 3 Academic Press, London, United Kingdom. [Google Scholar]
- 81.Goodenough UW, Heuser JE. 1985. The Chlamydomonas cell wall and its constituent glycoproteins analyzed by the quick-freeze deep-etch technique. J Cell Biol 101:1550–1568. doi: 10.1083/jcb.101.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Branton D. 1966. Fracture faces of frozen membranes. Proc Natl Acad Sci U S A 55:1048–1056. doi: 10.1073/pnas.55.5.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ziółkowska NE, Christiano R, Walther TC. 2012. Organized living: formation mechanisms and functions of plasma membrane domains in yeast. Trends Cell Biol 22:151–158. doi: 10.1016/j.tcb.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 84.Levin KR, Page E. 1980. Quantitative studies on plasmalemmal folds and caveolae of rabbit ventricular myocardial cells. Circ Res 46:244–255. doi: 10.1161/01.RES.46.2.244. [DOI] [PubMed] [Google Scholar]
- 85.Vaskovicova K, Stradalova V, Efenberk A, Opekarova M, Malinsky J. 2015. Assembly of fission yeast eisosomes in the plasma membrane of budding yeast: import of foreign membrane microdomains. Eur J Cell Biol 94:1–11. doi: 10.1016/j.ejcb.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 86.Steele SD, Miller JJ. 1974. Ultrastructural changes in germinating ascospores of Saccharomyces cerevisiae. Can J Microbiol 20:929–933. doi: 10.1139/m74-142. [DOI] [PubMed] [Google Scholar]
- 87.Olivera-Couto A, Salzman V, Mailhos M, Digman AM, Gratton E, Aguilar PS. 2015. Eisosomes are dynamic plasma membrane domains showing Pil1-Lsp1 heteroligomer binding equilibrium. Biophys J 108:1633–1644. doi: 10.1016/j.bpj.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Buchheim MA, Buchheim JA, Chapman RL. 1997. Phylogeny of Chloromonas (Chlorophyceae): a study of 18S ribosomal RNA gene sequences. J Phycol 33:286–293. doi: 10.1111/j.0022-3646.1997.00286.x. [DOI] [Google Scholar]
- 89.Brown RM, Montezinos D. 1976. Cellulose microfibrils: visualization of biosynthetic and orienting complexes in association with the plasma membrane. Proc Natl Acad Sci U S A 73:143–147. doi: 10.1073/pnas.73.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scazzocchio C, Vangelatos I, Sophianopoulou V. 2011. Eisosomes and membrane compartments in the ascomycetes. Commun Integr Biol 4:64–68. doi: 10.4161/cib.13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alvarez FJ, Douglas LM, Rosebrock A, Konopka JB. 2008. The Sur7 protein regulates plasma membrane organization and prevents intracellular cell wall growth in Candida albicans. Mol Biol Cell 19:5214–5225. doi: 10.1091/mbc.E08-05-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang HX, Douglas LM, Aimanianda V, Latgé J, Konopka JB. 2011. Candida albicans Sur7 protein is needed for proper synthesis of the fibrillar component of the cell wall that confers strength. Eukaryot Cell 10:72–80. doi: 10.1128/EC.00167-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Scholz MJ, Weiss TL, Jinkerson RE, Jing J, Roth R, Goodenough U, Posewitz MC, Gerken HG. 2014. Ultrastructure and composition of the Nannochloropsis gaditana cell wall. Eukaryot Cell 13:1450–1464. doi: 10.1128/EC.00183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weiss TL, Roth R, Goodson C, Vitha S, Black I, Azadi P, Rusch J, Holzenburg A, Devarenne TP, Goodenough U. 2012. Colony organization in the green alga Botryococcus braunii (race B) is specified by a complex extracellular matrix. Eukaryot Cell 11:1424–1440. doi: 10.1128/EC.00184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu H, Schulten K. 2013. Membrane sculpting by F-BAR domains studied by molecular dynamics simulations. PLoS Comput Biol 9:e1002892. doi: 10.1371/journal.pcbi.1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van der Rest ME, Kamminga AH, Nakano A, Anraku Y, Poolman B, Konings WN. 1995. The plasma membrane of Saccharomyces cerevisiae: structure, function, and biogenesis. Microbiol Rev 59:304–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Needham D, Nunn RS. 1990. Elastic deformation and failure of lipid bilayer membranes containing cholesterol. Biophys J 58:997–1009. doi: 10.1016/S0006-3495(90)82444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Drolle E, Kučerka N, Hoopes MJ, Choi Y, Katsaras J, Karttunen M, Leonenko Z. 2013. Effect of melatonin and cholesterol on the structure of DOPC and DPPC membranes. Biochim Biophys Acta 1828:2247–2254. doi: 10.1016/j.bbamem.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 99.Zimmerberg J, Kozlov M. 2006. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol 7:9–19. [DOI] [PubMed] [Google Scholar]
- 100.Patterson GW. 1971. The distribution of sterols in algae. Lipids 6:120–127. doi: 10.1007/BF02531327. [DOI] [Google Scholar]
- 101.Desmond E, Gribaldo S. 2009. Phylogenomics of sterol synthesis: insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biol Evol 1:364–381. doi: 10.1093/gbe/evp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.