Abstract
Coccidioides immitis and Coccidioides posadasii are soil-dwelling fungi and the causative agents of coccidioidomycosis, a mycosis endemic to certain semiarid regions in the Americas. The most common route of infection is by inhalation of airborne Coccidioides arthroconidia. Once a susceptible host inhales the conidia, a transition to mature endosporulated spherules can occur within the first 5 days of infection. For this study, we examined the host response in a murine model of coccidioidomycosis during a time period of infection that has not been well characterized. We collected lung tissue and bronchoalveolar lavage fluid (BALF) from BALB/c mice that were infected with a C. immitis pure strain, a C. immitis hybrid strain, or a C. posadasii strain as well as uninfected mice. We compared the host responses to the Coccidioides strains used in this study by assessing the level of transcription of selected cytokine genes in lung tissues and characterized host and fungal proteins present in BALF. Host response varied depending on the Coccidioides strain that was used and did not appear to be overly robust. This study provides a foundation to begin to dissect the host immune response early in infection, to detect abundant Coccidioides proteins, and to develop diagnostics that target these early time points of infection.
INTRODUCTION
Coccidioidomycosis, more commonly known as valley fever, is a fungal infection caused by inhaling either Coccidioides immitis or Coccidioides posadasii conidia, which are found in the soil in arid to semiarid regions of the Americas. These species share a well-characterized asexual life cycle. During the saprobic cycle, these fungi alternate between two main cell types in the environment: arthroconidia and hyphae. The arthroconidial phase is believed to be the major infectious particle found in the environment. When a susceptible host inhales an arthroconidium, the parasitic cycle begins. During an active parasitic cycle, an arthroconidium can transition to a mature rupturing spherule within 5 days after the initial exposure (1). Once the spherule ruptures and releases endospores, each endospore can develop into a new spherule, and the parasitic cycle can continue exponentially.
It is estimated that 40% of human Coccidioides infections are symptomatic (2). Acute or chronic pulmonary disease is most common manifestation; however, disseminated disease occurs in approximately 1% of Coccidioides infections (2). Certain factors, such as African or Filipino ancestry, immunosuppression, pregnancy, and male gender, increase the risk of a disseminated infection (2, 3). The remaining 60% of human Coccidioides infections are asymptomatic and generally result in clearance or development of asymptomatic lung nodules. Morphological variation is highest during early days of infection, when the inhaled environmental conidia switch to the parasitic lifestyle. However, the host innate immune response in the first 5 days of infection remains an uncharacterized component of the clinical manifestation of disease.
The innate immune and cytokine profile response to coccidioidomycosis has been assessed in a few specific studies. Increases in interferon gamma (IFN-γ), tumor necrosis factor alpha (TNF-α), and interleukin 17 (IL-17) in mononuclear cells from bronchoalveolar lavage fluid (BALF) and in peripheral blood mononuclear cells collected from patients with acute pulmonary coccidioidomycosis have been identified (4). A significant association between lower levels of mannose-binding lectin in serum of patients with active coccidioidomycosis compared to uninfected healthy subjects was described (5). Comparing BALB/c and DBA/2 mouse models of coccidioidomycosis, a greater susceptibility to pulmonary Coccidioides infection in BALB/c mice due to the development of anergy on day 15 postinfection has been reported (6). An acquired suppression of cell-mediated immune reactivity not specific to Coccidioides antigens was also discovered in BALB/c mice (6). In a subsequent study on intranasal infection challenge of 20 arthroconidia of strain Silveira, it was shown that the more resistant DBA/2 mice mounted an IFN-γ-driven response, whereas the more susceptible BALB/c mice manifested a predominant IL-4 response to an infection challenge (7). Expression of these cytokines was measured at 3-day intervals over a 15-day postinfection study, starting on day 3 (7). Increases in IFN-γ and IL-4 were not detected until 12 and 9 days, respectively, postinfection. Further investigation comparing intraperitoneal infection in four inbred mouse strains on days 7 and 14 postinfection indicated nonprotective IL-10 and IL-4 responses to strain RS Coccidioides infections in susceptible C57BL/6 mice (8).
The cytokine response to challenge infections of other human fungal respiratory pathogens has also been examined in murine models of disease. For example, increased transcription of genes that encode IL-2, IL-4, IL-12, and IFN-γ was detected as early as day 3 postinfection in the lungs of C57BL/6 mice challenged with Histoplasma capsulatum (9). IFN-γ is necessary for control of H. capsulatum infection in mice (10). Elevated levels of TNF-α, IL-6, IL-1β, and macrophage inflammatory protein (MIP) were detected during the first 4 days postinfection in BALB/c mice infected with Paracoccidioides brasiliensis (11). The cytokines TNF-α and IL-1α have been shown to be important for leukocyte recruitment and control of infection of the pathogen Aspergillus fumigatus in mice (12, 13). Examination of Cryptococcus neoformans in a C57BL/6 mouse model of infection showed that IL-17α increase is mediated by leukocyte recruitment and activation and IFN-γ production after intratracheal challenge (14).
The goal of this study was to expand upon previous studies of the host innate immune response to pulmonary coccidioidomycosis by examining the first 5 days of infection in a BALB/c mouse model of infection, when the morphological shift from the saprobic to parasitic cycle occurs. Understanding the cytokine expression profile at the onset of infection could reveal new insights into infection dynamics of Coccidioides. For this study, we analyzed the immune responses of mice intranasally infected with arthroconidia from clinical isolates of Coccidioides: a C. immitis pure isolate (2006), a C. immitis hybrid isolate (RS), and a C. posadasii pure isolate (Silveira). The expression profiles of selected cytokines were assessed by real-time reverse transcription-PCR (RT-PCR) of RNA extracted from the mouse lung tissues and a multiplex bead array for proteins present in BALF. Finally, protein expression profiles from BALF during day 5 postinfection were determined using mass spectrometry to identify potential host biomarkers and fungal proteins expressed during pulmonary coccidioidomycosis.
MATERIALS AND METHODS
Mouse inoculations.
Coccidioides immitis isolate 2006 (15), C. immitis hybrid isolate RS (16), and C. posadasii isolate Silveira (17, 18) were used in this study. Female 6- to 8-week-old BALB/c mice were anesthetized with ketamine-xylene and intranasally inoculated with 100,000 arthroconidia suspended in 30 μl phosphate-buffered saline (PBS) as described previously (19). Control mice were inoculated with PBS alone. The mice were housed according to NIH guidelines in a biosafety level 3 animal laboratory. All procedures were approved by the Institutional Animal Care and Use Committee for the University of Arizona. Three mice from each infection group (2006, RS, and Silveira) and two uninfected PBS-treated mice were sacrificed on day 1 through day 5. The lungs were rinsed with 2 ml of phosphate-buffered saline with 0.01 mM EDTA to collect BALF. The right lobe of the lung was harvested and flash frozen in liquid nitrogen. One milliliter of each BALF sample was formalin fixed and underwent a complete cell count using a hemocytometer. The remainder of each BALF sample was filtered using a 0.80-μm Millex-AA syringe filter unit (EMD Millipore, Billerica, MA), and a Thermo Scientific Pierce bicinchoninic acid (BCA) protein assay (Life Technologies, Carlsbad, CA) was used to quantify protein concentrations.
RNA extraction.
Modifications to a previously described protocol were applied to the RNA extraction procedure (20). The whole right lungs were lyophilized for 48 h. The lyophilized lungs were put in 2-ml tubes with lysing matrix D (MP Biomedicals, Santa Ana, CA) ceramic spheres and mixed with 1 ml of TRIsure (Bioline USA, Inc., Taunton, MA). Samples were subjected to bead beating for 2 min and then incubated for an additional 5 min. Chloroform (200 μl) was added, mixed well, and incubated 2 min at room temperature. Samples were centrifuged at maximum speed for 15 min at 4°C. The upper layers were collected and mixed with an equal volume of 80% ethanol (EtOH), and the entire volume was pipetted immediately to RNeasy spin columns (Qiagen, Inc., Valencia, CA). Columns were centrifuged for 1 min, the flowthrough was discarded and washed twice with kit-supplied RPE buffer (Qiagen), and columns were completely dried after the last wash. The spin columns were then transferred to new RNase-free 1.5-ml tubes, 200 μl of RNase free water was added to each spin column, and columns were incubated for 1 min. After that 1-min incubation, the tubes containing the spin columns were centrifuged to obtain nucleic acid. Samples were analyzed with NanoDrop ND-1000 (Thermo Fisher Scientific, Inc., Waltham, MA) and a 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA).
Real-time RT-PCR.
The extracted RNA was treated with an Ambion Turbo DNA-free kit (Life Technologies Corporation, Carlsbad, CA) to remove genomic DNA. The RNA concentration was checked on a NanoDrop ND-1000 to determine quantity. The cDNA was created using the standard procedure of the QuantiTect reverse transcription kit produced by Qiagen (Valencia, CA). The real-time RT-PCR was completed using a SensiFAST SYBR Hi-ROX Mix (Bioline USA, Inc., Taunton, MA) on an ABI7900 system (Applied Biosystems, Waltham, MA). Conditions for real-time RT-PCR were 1 min of polymerase activation at 95°C followed by 40 cycles of 5 s at 95°C, 10 s at 60°C, and 20 s at 72°C. To determine fungal burden, oligomers designed to amplify the Coccidioides glyceraldehyde-3-phosphate dehydrogenase gene (gapdh) were used in a real-time RT-PCR assay (see Table S1 in the supplemental material). To examine cytokine gene expression, PrimeTime qPCR primer assays (Integrated DNA Technologies, Inc., Coralville, IA) of selected genes (see Table S1 in the supplemental material) were used for this study. Change (n-fold) was determined using the 2−ΔΔCT method (21). The mouse gapdh gene was selected as the internal control gene to calculate the relative expression of the tested mouse genes.
Multiplex bead array assay.
The concentrations of selected cytokines (see Table S2 in the supplemental material) for the BALF were examined using a cytokine mouse magnetic 20-Plex panel (Life Technologies, Carlsbad, CA) on a Luminex LX200 system with Luminex xPONENT 3.0 software (Luminex Corporation, Austin, TX). A Thermo Scientific Pierce BCA protein assay (Life Technologies, Carlsbad, CA) was used to quantify protein concentrations. Cytokine concentrations were normalized to the amount of total protein quantified in each sample.
Proteomic analysis.
Equal amounts of protein (20 μg each) were reduced and denatured by boiling for 10 min in loading buffer (Bio-Rad Laboratories, Inc., Hercules, CA) containing 5% β-mercaptoethanol. Each sample was then separated by gel electrophoresis on a 4 to 20% SDS-PAGE gel (Bio-Rad Laboratories, Inc., Hercules, CA) for 45 min at 120 V and stained using Coomassie Plus (Bio-Rad Laboratories, Inc., Hercules, CA). Gel lanes were fully excised into individual bands, destained, washed, dried, and further processed using a previously published method (22). Briefly, each lane fraction was reduced and alkylated using cyclic rehydration/dehydration of the acrylamide gel slabs. Samples were then digested using Trypsin Gold (Promega Corporation, Madison, WI) at 20 ng/ml overnight at 37°C. Peptides were extracted, concentrated to dryness under vacuum, and frozen at −20°C prior to mass spectrometry analysis.
Analyses were conducted on a nanoAcquity ultraperformance liquid chromatography (UPLC) system (Waters Corporation, Milford, MA) coupled to a Thermo LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, Waltham, MA). Each sample fraction was reconstituted in 0.1% formic acid for analysis by using online liquid chromatography coupled to mass spectrometry. Each fraction was reconstituted in 0.1% formic acid and loaded onto a 100-μm-diameter column packed to 100 mm with 3 μm Reprosil Pur C18 AQ resin eluted at 500 nl/minute. Solvents A and B were 0.1% formic acid in water and acetonitrile, respectively. The gradient was 3% B to 40% B in 17 min followed by 40% B to 90% B in 0.5 min, then 90% B for 2 min, and re-equilibration for 10.5 min. The mass spectrometer operated in positive ion mode using a spray voltage of 1.8 kV and a capillary temperature of 200°C. Data were acquired in top-15, data-dependent acquisition mode using a collision voltage of 30 V. Raw mass spectrometry data were searched against a concatenated database from UniprotKB/Swissprot for Mus musculus and UniprotKB/Swissprot+Trembl for C. posadasii isolate Silveira and C. immitis isolate RS using Mascot (version 1.4.1.14; Matrix Science, London, United Kingdom) and X! Tandem (v2010.12.01.1; The GPM). Tandem mass spectra were extracted, and charge state was deconvoluted and deisotoped by Proteome Discoverer 1.4.1.14 (Thermo Fisher Scientific, Waltham, MA). Oxidation (Met), carbamidomethylation (Cys), and pyro-Glu formation (N-terminal Gln or Glu) were specified as variable modifications. A peptide mass tolerance of ±10 ppm, a fragment mass tolerance of ±0.8 Da, and a maximum of two trypsin missed cleavages were allowed. Peptide identifications were accepted at ≥98.0% as specified by the Peptide Prophet algorithm (23) and corrected using Scaffold m correction (Proteome Software Inc., Portland, OR).
Biological concept enrichment analysis.
Biological concept enrichment analysis was performed on the four sets of proteins using the ClueGO v2.1.7 Cytoscape plugin (24). Enrichment was performed with the following Ontologies/Pathway gene sets: GO Biological Process, KEGG, Reactome, and WikiPathways. Advanced Term/Pathway selection options were set at Go Tree interval minimum level 3 and maximum level 8, GO term fusion, and minimum number of genes at five for uninfected and 10 for 2006, RS, and Silveira infected. The kappa score was set at 0.4. A two-sided hypergeometric test was used with Bonferroni step-down correction.
ToppGene functional analysis.
We used the ToppGene functional analysis suite (25) to identify enriched terms in each of our four sets of proteins (uninfected and 2006, RS, and Silveira infected) (false discovery rate [FDR] correction; P < 0.05; gene limits, 1 ≤ n ≤ 2,000). Enriched terms from the biological process ontology were ranked by P value, and only the five most significant terms were retained for interpretation of the host response to Coccidioides infection in BALF.
Statistics.
Statistical analyses were performed using GraphPad Prism version 6.00 for Mac (GraphPad Software, La Jolla, CA) or Scaffold (Proteome Software Inc., Portland, OR) for mass spectrometry-derived data. P values were calculated either by using a two-way analysis of variance (ANOVA) with Tukey's posttest or Kruskal-Wallis test in the case of the proteomics data. A P value of <0.05 was considered statistically significant.
RESULTS
BALB/c mouse infections.
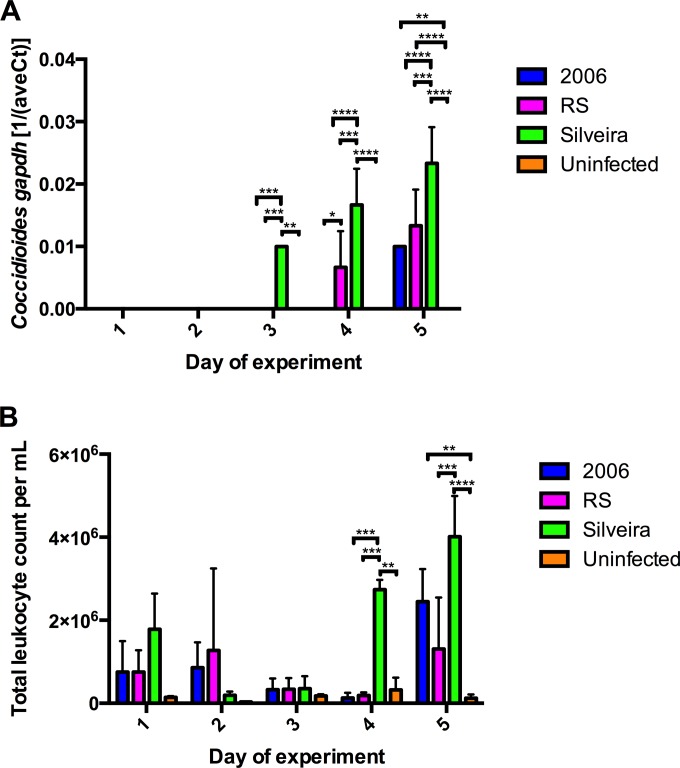
Three mice from each infection group and two mice from the uninfected group were sacrificed on each day postinfection during this 5-day study. Real-time RT-PCR was used to detect Coccidioides gapdh mRNA extracted from the whole right lungs of each mouse to determine a relative fungal burden (Fig. 1A). C. posadasii isolate Silveira gapdh mRNA transcripts were detected on day 3 of infection and beyond (Fig. 1A), whereas C. immitis hybrid isolate RS gapdh mRNA transcripts were not detected until day 4 of infection (Fig. 1A). C. immitis isolate 2006 gapdh mRNA transcripts were not detected until day 5 of infection (Fig. 1A). No Coccidioides gapdh mRNA transcripts were detected in the uninfected mice (Fig. 1A).
FIG 1.
Detection of Coccidioides infection in BALB/c mice. (A) Real-time RT-PCR analysis of Coccidioides gapdh mRNA transcripts in whole right lungs of BALB/c mice above signal background. (B) Total leukocyte count per ml in collected BALF determined by hemocytometer. Error bars indicate standard errors from biological replicates (n = 3 for each infection group; n = 2 for the uninfected group). P values were calculated using two-way ANOVA followed by Tukey's multiple-comparison test. A P value of <0.05 was considered statistically significant, and levels of significance were assigned as follows: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
A total leukocyte count was made from cells present in the BALF to confirm that the mice mounted an inflammatory response to infection (Fig. 1B; Table 1). Non-statistically significant increases were observed for all infection groups compared to the uninfected mice on the first day of infection (Fig. 1B). On day 2 of infection, the 2006 and RS infection groups had non-statistically significant increases of total leukocyte counts compared to the Silveira infection group and uninfected control group (Fig. 1B). On day 3 of infection, all three infection groups had leukocyte counts comparable to that of the uninfected control group. On day 4, however, the Silveira infection group had statistically significant higher number of total leukocytes than the 2006 infection group (P ≤ 0.001), RS infection group (P ≤ 0.001), and uninfected control group (P ≤ 0.01). On day 5, all infection groups had a higher number of leukocytes per ml of BALF than uninfected mice. In agreement with days 1, 3, and 4, the Silveira infection group had the highest number of leukocytes per ml of BALF on average. During necropsy, we observed that mice infected with C. posadasii isolate Silveira suffered from more severe lung damage starting at day 3 of infection than the mice infected with C. immitis isolate RS and C. immitis isolate 2006 (data not shown).
TABLE 1.
Summary statistics for total leukocyte counts per ml of BALF
| Isolate (na) | Mean no. of total leukocyte counts per mlb |
||||
|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | |
| 2006 (3) | 7.53E+05 (0–2.61E+06) | 8.58E+05 (0–2.37E+06) | 3.27E+05 (0–1.0E+06) | 1.25E+05 (0–4.4E+05) | 2.45E+06 (4.95E+05–4.40E+06) |
| RS (3) | 7.50E+05 (0–2.06E+06) | 1.27E+06 (0–6.18E+06) | 3.35E+05 (0–1.01E+06) | 1.85E+05 (0–3.80E+05) | 1.31E+06 (0–4.38E+06) |
| Silveira (3) | 1.78E+06 (0–3.92E+06) | 1.92E+05 (0– 4.17E+05) | 3.53E+05 (0–1.10E+06) | 2.74E+06 (2.15E+06– 3.32E+06) | 4.01E+06 (1.58E+06– 6.45E+06) |
| Uninfected (2) | 1.43E+05 (0–4.28E+05) | 3.25E+04 (0–1.28E+05) | 1.78E+05 (0–5.27E+05) | 3.20E+05 (0–2.99E+06) | 1.23E+05 (0–9.17E+05) |
Number of mice per day per group.
95% confidence intervals are in parentheses.
Real-time RT-PCR analysis of mouse lung tissues.
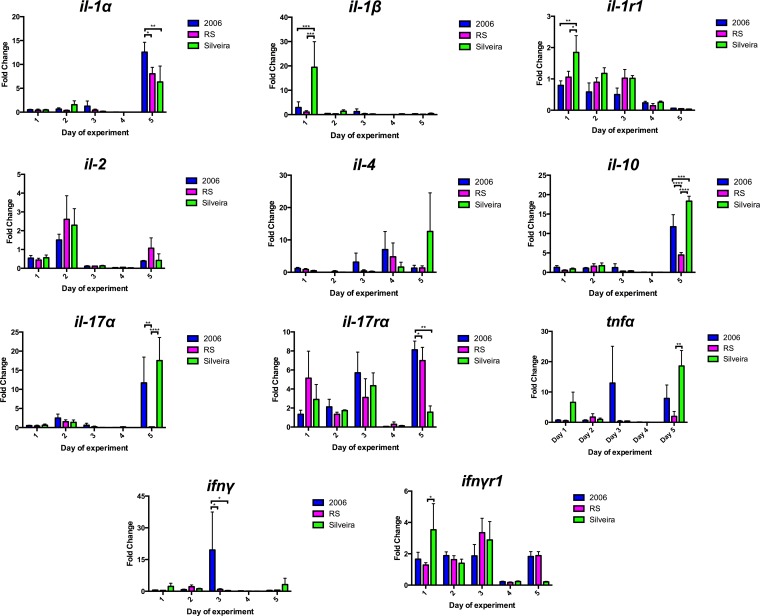
The whole right lungs were harvested from each mouse and underwent real-time RT-PCR analysis to assess transcript levels of pro- and anti-inflammatory cytokines. The cytokine targets and primers used for this study are listed in Table S1 in the supplemental material. We detected a differential cytokine response among the Coccidioides isolates used in this study. Only the Silveira infection group showed a 20-fold increase in the proinflammatory cytokine gene il-1β (Fig. 2). A temporal trend toward an increase in levels of the proinflammatory cytokine gene tnfα varied among the infection groups during the study (Fig. 2). There were also trends that all infection groups shared. On day 1, there was an initial increase of ifnyr1 and il-1r1 for all infection groups, with a significant decrease on days 4 and 5, respectively (Fig. 2). There were transcriptional increases for many of the cytokine genes, including the anti-inflammatory cytokine gene il-10 and the proinflammatory cytokine gene il-1α, analyzed on day 5 of infection for all infection groups. Similar increases were seen in the expression of the proinflammatory cytokine gene il-17α on day 5 for the mice infected with 2006 or Silveira (Fig. 2). No statistically significant increases were noticed for il-17rα before day 5 of infection (Fig. 2). We did observe non-statistically significant increases in changes for other transcripts examined, including il-2 and il-4. Real-time RT-PCR results indicate a differential cytokine response dependent on which Coccidioides isolate was used for infection.
FIG 2.
Real-time RT-PCR analysis of cytokine gene mRNA transcripts in the whole right lungs of BALB/c mice. Error bars indicate standard errors from biological triplicates. P values were calculated using two-way ANOVA followed by Tukey's multiple-comparison test. A P value of <0.05 was considered statistically significant, and levels of significance were assigned as follows: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
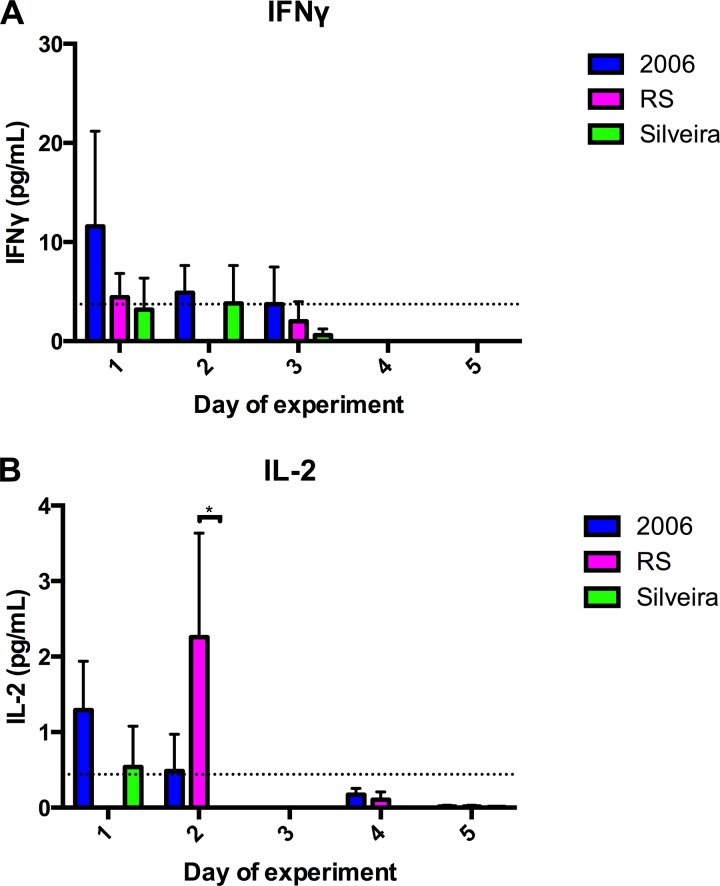
Multiplex bead array analysis of mouse BALF.
The 20 cytokines that were quantified are listed in Table S2 in the supplemental material. Few were detected above the assay's lower limit, as only IFN-γ and IL-2 were detected in the BALF using this method (Fig. 3). The average values of protein concentration for IFN-γ and IL-2 in uninfected control animals are presented in Fig. 3. During the 5 days of infection, day 1 appeared to be the only day in which IFN-γ was at a higher, but not a statistically significantly higher, concentration in the 2006 group than in the uninfected group (Fig. 3A). On day 1 of infection, a non-statistically significant increase of IL-2 was present in the BALF of the 2006 and Silveira infection groups (Fig. 3B). Overall, we did not detect an abundant cytokine response.
FIG 3.
Concentration of cytokines IFN-γ (A) and IL-2 (B) in BALF. Cytokine concentrations were measured using a multiplex bead array and normalized to the amount of total protein quantified in each sample. The dashed line represents the mean for uninfected control animals. Error bars indicate standard errors from biological triplicates. P values were calculated using two-way ANOVA followed by Tukey's multiple-comparison test. A P value of <0.05 (*) was considered statistically significant.
Proteomic analysis of mouse BALF.
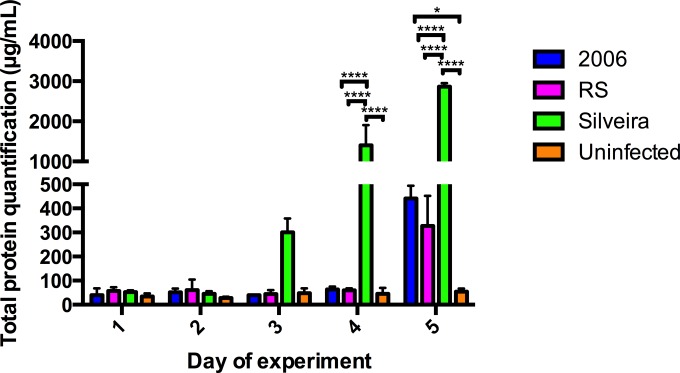
Because we did not detect abundant cytokines in our samples, we employed proteomics analysis on remaining BALF. Overall, total protein concentrations remained below 100 μg/ml in all of the mouse groups until day 3 of infection (Fig. 4). On day 3 of infection, the Silveira-infected mice reached a mean total protein concentration of 300 μg/ml, while other mouse groups remained below 100 μg/ml (Fig. 4). A marked increase in the mean total protein concentration within the BALF of the Silveira infection group on day 4 was statistically significant compared to the 2006 infection group (P ≤ 0.0001), RS infection group (P ≤ 0.0001), and uninfected mouse group (P ≤ 0.0001) (Fig. 4). On day 5, all infection groups had increased levels of total protein in BALF compared to the uninfected mouse group (Fig. 4). As the highest protein concentrations were observed at day 5, we performed an extensive downstream proteomic analysis on these samples.
FIG 4.
Quantifying the concentration (μg/ml) of total proteins present in BALF for downstream proteomics analysis. Error bars indicate standard errors from biological replicates (n = 3 for each infection group; n = 2 for the uninfected group). P values were calculated using two-way ANOVA followed by Tukey's multiple-comparison test (*, P ≤ 0.05; ****, P ≤ 0.0001).
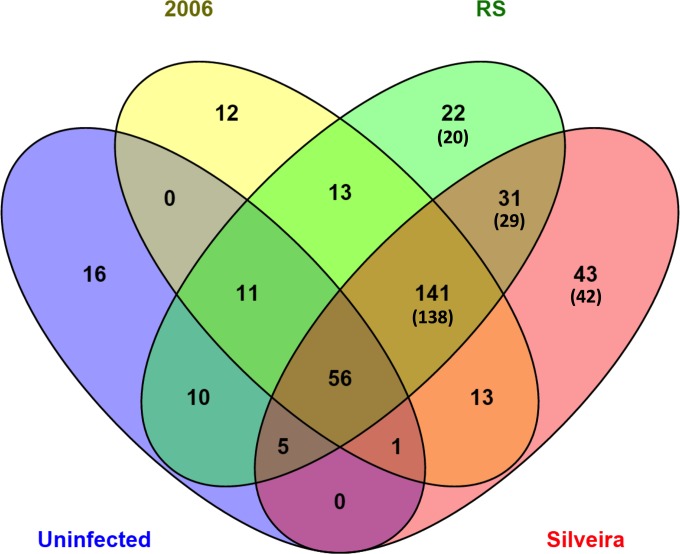
We performed proteomic analysis on the BALF collected on day 5 of the experiment from the nine infected mice (three mice per infection group) and the two mice in the uninfected group. A total of 374 total unique proteins were identified from the proteome of all BALF collected on day 5 of this experiment (Fig. 5; also, see Data Set S1 in the supplemental material). This number includes 366 unique proteins identified overall, as well as eight Coccidioides proteins identified among the infected mice (Fig. 5). Data Set S1 lists the proteins identified in the BALF samples. Of the proteins that we identified, 33 mouse proteins displayed statistically significant differences between the four mouse groups examined in this study (see Data Set S1). Six mouse proteins were significantly differentially abundant in uninfected mice compared to all of the mouse infection groups (Table 2). Two of these, annexin A5 and aminopeptidase N, were increased in all uninfected compared to infected mice. Aminopeptidase N extracellular signaling helps to reduce inflammation, and annexin A5 is a known inhibitor of protein kinase C, thus inhibiting apoptosis and degranulation in healthy mice (26, 27). This suggests activation of cytotoxic activity from mast cells and granulocytes and activation of inflammation and apoptosis in all infected mice. Four of the proteins, α-2-macroglobulin, complement C3, hemopexin, and proteasome subunit, were lower in uninfected mice, and these are associated with general lung damage and inflammation common in all the infected mice (28–31). Twelve proteins were significantly differentially expressed for mice infected with RS compared to the other mouse groups (Table 3). Interestingly, 10 of the 12 were more abundant, including chitinase and iron/heme-associated proteins. For the proteins that were lower in abundance, fibronectin and inter-α-trypsin inhibitor are both biomarkers associated with chronic obstructive pulmonary disease (COPD) and inflammatory processes when found in abundance, suggesting that RS does not induce inflammation via this mechanism (31, 32). Ten significantly differentially expressed proteins were found in mice infected with Silveira compared to the other groups (Table 4). Of the four that are less abundant, carbonic anhydrase 2 and carbonyl reductase (NADPH) 2 suggest a nonprotective response to oxidative stress, whereas complement C5 and lactotransferrin suggest a lack of protective inflammatory response (29, 33–35). For proteins that were more highly expressed, all are associated with damage response, supporting the observation that Silveira causes significant damage to the lung. Interestingly, no unique proteins were differentially abundant for mice infected with isolate 2006 (see Data Set S1).
FIG 5.
Venn diagram comparing and contrasting the number of unique proteins identified in the BALF collected from the day 5 groups using mass spectrometry analysis. A total of 374 unique proteins composed of 366 unique mouse proteins and eight unique Coccidioides proteins were identified. In parentheses are the numbers of mouse proteins that were identified only in a particular group.
TABLE 2.
Mouse proteins significantly differentially expressed in uninfected mice compared to other groups on day 5 of the experiment
| Protein | Gene ID | Abundance |
|---|---|---|
| α-2-Macroglobulin | A2M_MOUSE | Less |
| Aminopeptidase N | AMPN_MOUSE | More |
| Annexin A5 | ANXA5_MOUSE | More |
| Complement C3 | CO3_MOUSE | Less |
| Hemopexin | HEMO_MOUSE | Less |
| Proteasome subunit beta type 6 | PSB6_MOUSE | Less |
TABLE 3.
Mouse proteins significantly differentially expressed in RS-infected mice compared to other groups on day 5 of the experiment
| Protein | Gene ID | Abundance |
|---|---|---|
| Afamin | AFAM_MOUSE | More |
| Apolipoprotein A-I | APOA1_MOUSE | More |
| Chitinase-3-like protein 1 | CH3L1_MOUSE | More |
| Cluster of chitinase-like protein 3 | CHIL3_MOUSE | More |
| Cluster of hemoglobin subunit beta-1 | HBB1_MOUSE | More |
| Ferritin light chain 1 | FRIL1_MOUSE | More |
| Fibronectin | FINC_MOUSE | Less |
| Inter-α-trypsin inhibitor, heavy chain 4 | ITIH4_MOUSE | Less |
| Leukemia inhibitory factor receptor | LIFR_MOUSE | More |
| Polymeric immunoglobulin receptor | PIGR_MOUSE | More |
| Serotransferrin | TRFE_MOUSE | More |
| Triosephosphate isomerase | TPIS_MOUSE | More |
TABLE 4.
Mouse proteins significantly differentially expressed in Silveira-infected mice compared to other groups on day 5 of the experiment
| Protein | Gene ID | Abundance |
|---|---|---|
| Carbonic anhydrase 2 | CAH2_MOUSE | Less |
| Carbonyl reductase (NADPH) 2 | CBR2_MOUSE | Less |
| Ceruloplasmin | CERU_MOUSE | More |
| Complement C5 | CO5_MOUSE | Less |
| Fibrinogen beta chain | FIBB_MOUSE | More |
| Gelsolin | GELS_MOUSE | More |
| Histone H4 | H4_MOUSE | More |
| Inter- α-trypsin inhibitor, heavy chain 4 | ITIH4_MOUSE | More |
| Lactotransferrin | TRFL_MOUSE | Less |
| Thioredoxin | THIO_MOUSE | More |
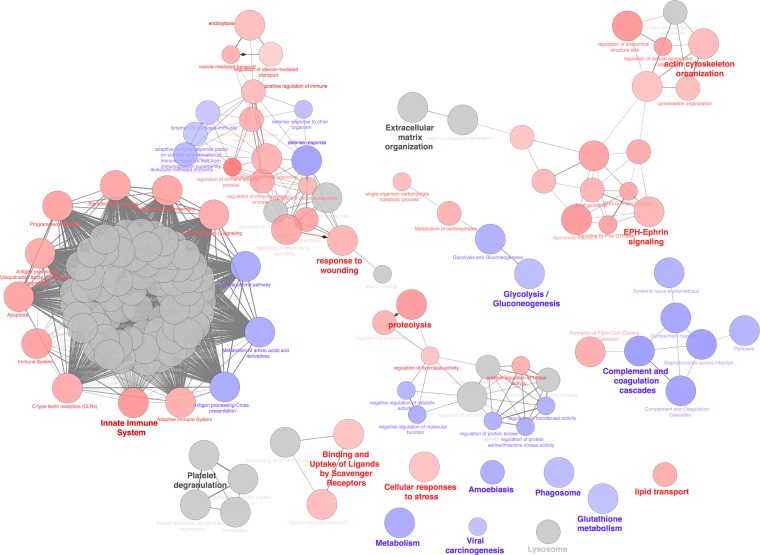
In order to determine representative biology associated with the four sets of proteins, ClueGO was used to perform biological concept enrichment analysis. ClueGO visualizes the functional enrichment results by grouping similar ontologies and networks into overlapping clusters based on similar function and gene membership. The four protein sets were analyzed with ClueGO using the GO Gene Ontology (GO) Biological Process, Kyoto Encyclopedia of Genes and Genomes (KEGG), Reactome, and WikiPathways gene set categories. The enrichment network is visualized in Fig. 6. The red nodes represent ontologies or pathways that show preferential enrichment in the Silveira strain. The blue nodes are those enriched in the RS group of proteins. Interestingly, no category had preferential enrichment for proteins in the 2006 or uninfected protein set. The gray nodes did not have a preferential enrichment in any of the protein sets and represent common terms in all protein sets. The significant ontology/pathway nodes for the Silveira infection group protein set were categories such as innate immune system, proteolysis, cellular response to stress, lipid transport, EPH-ephrin signaling, and actin cytoskeleton organization (Fig. 6). The protein set from the RS infection group had significant node clusters, such as metabolism, phagosome, complement, and coagulation cascades, as well as glutathione metabolism, among others (Fig. 6). A tight cluster of nodes is centered on signaling pathways with a large component of proteasome-related genes (Fig. 6). A cluster of proteasome genes common across all protein sets drive enrichment in this cluster of pathways (Fig. 6).
FIG 6.
Biological concept analysis of the four sets of proteins from day 5. Analysis was performed on the four sets of proteins (uninfected and 2006, RS, and Silveira infected) using ClueGO. The nodes in the diagram represent enriched ontologies or pathways. The red nodes represent ontologies or pathways that show preferential enrichment in the Silveira isolate, and the blue nodes represent those enriched in the RS isolate. The gray nodes do not show preferential enrichment in any one particular protein set. Node size corresponds to significance, as determined by P value. The edges (connections between nodes) represent the statistical association between the nodes based on gene membership and location with gene ontology. The bold node labels represent nodes of the most significant term for cluster of nodes.
ToppGene enrichment analysis was also performed to confirm the results of the protein clustering analysis (see Fig. S1 to S4 and Data Set S2 in the supplemental material). The host proteins identified align with the increase in Coccidioides isolate fungal burden and infection dynamics (2006 ≪ RS ≪ Silveira) we observed in other assays (Fig. 1 and 4). Total proteins assigned to these terms were notably lower in uninfected mice, possibly as a result of a baseline defense response to challenges at the endothelial interface between alveoli and capillaries (see Fig. S4 and Data Set S2). Key terms associated with inflammation were present in all Coccidioides isolate groups (see Fig. S1 to S3 and Data Set S2). As expected, proteins associated with both innate and humoral responses were prioritized for RS and Silveira (defense, inflammatory, and acute inflammatory responses). Proteins secreted in response to wounding were significant in all infection groups (see Fig. S1 to S3 and Data Set S2), confirming our findings on probable lung damage and tissue necrosis (Fig. 4). We detected an increase of platelet degranulation-associated proteins in the 2006 mouse infection group that was not as apparent in any of the other infection groups or uninfected control mice (see Fig. S1 and Data Set S2). Platelet degranulation is an antimicrobial host response to infection that has been observed for other human pathogens, including the fungus Aspergillus fumigatus and multiple bacterial species (36, 37). The high fungal burden of Silveira isolate also likely led to an increase in proteolytic host response (see Fig. S3 and Data Set S2).
Coccidioides proteins identified in BALF.
We also identified Coccidioides proteins present in BALF (Table 5). Three proteins, a Coccidioides serine protease (CIMG_10287/CPSG_04717), a triosephosphate isomerase (CIMG_09361/CPSG_03911), and an uncharacterized protein (CIMG_09001/CPSG_01366), were identified in the BALF collected from all infection groups (Table 5). The Coccidioides serine protease we identified in all of the mouse infection groups is a member of an expanded subtilisin N domain-containing extracellular serine protease family in Coccidioides implicated in host interactions but is currently uncharacterized (38). Orthologous serine proteases have been shown to be virulence factors expressed during infection by other human-pathogenic fungal and bacterial species (38).
TABLE 5.
Coccidioides proteins that were detected in the BALF collected from infected mice on day 5 of infection
| Protein | Gene ORFa | Coccidioides isolateb (nc) |
|---|---|---|
| 40S ribosomal protein S14 | CIMG_04348/CPSG_09614 | RS (1) |
| 60S ribosomal protein L12 | CIMG_04811/CPSG_07687 | Sil (2) |
| Endochitinase 1 | CIMG_02795/CPSG_08657 | RS (1), Sil (3) |
| Gamma-glutamyltranspeptidase | CIMG_05765/CPSG_02828 | Sil (2) |
| Peroxisomal matrix protein | CIMG_05828/CPSG_04764 | RS (2) |
| Serine protease | CIMG_10287/CPSG_04717 | 2006 (3), RS (3), Sil (2) |
| Triosephosphate isomerase | CIMG_09361/CPSG_03911 | 2006 (1), RS (1), Sil (1) |
| Uncharacterized protein | CIMG_09001/CPSG_01366 | 2006 (2), RS (1), Sil (3) |
Gene open reading frame (ORF) name for orthologous loci in both species, in the format C. immitis isolate RS/C. posadasii isolate Silveira.
2006, C. immitis isolate 2006; RS, C. immitis isolate RS; Sil, C. posadasii Silveria.
Number of mice per infection group in which this protein was identified.
Other Coccidioides proteins were found in only one or two infection groups (Table 5). As an example, endochitinase 1 (CIMG_02795/CPSG_08657) was identified from one mouse infected with RS and all three of the mice infected with Silveira (Table 5). It is a member of a well-characterized family of chitinase proteins in Coccidioides that have been shown to be upregulated during the parasitic life cycle (39, 40). CTS1 is also immunogenic and is used as a serodiagnostic antigen for disease (41).
A peroxisomal matrix protein (PMP1) (CIMG_05828/CPSG_04764) was identified in two of the mice infected with RS. A recombinant PMP1 was found previously to be reactive with serum from individuals with both acute and protracted coccidioidomycosis (42). The potential of this protein as a recombinant vaccine candidate was also examined; it evoked protection in two murine models of infection with C. posadasii (42). Thus, our approach was validated by the identification of previously characterized Coccidioides proteins that are known to be associated with the parasitic life cycle.
DISCUSSION
For this study, we sought to examine and characterize the early host response to Coccidioides infection in a BALB/c mouse model of pulmonary coccidioidomycosis at time points not previously characterized. Our focus on BALF was driven by the knowledge that this is a common clinical specimen and that current diagnostics are inadequate for detecting Coccidioides (43). We examined the host response to three different isolates of Coccidioides to assess whether the host responses to the isolates would be similar. We initially anticipated that the three Coccidioides species isolates would have comparable overall inflammatory responses and infection kinetics. In contrast to our expectation, there appeared to be significant differential responses among the isolates used in this study. We discovered that of the three Coccidioides isolates that were used in this study, the C. posadasii isolate Silveira appeared to cause the highest relative fungal burden in mouse lung tissue. This isolate also caused a significantly higher influx of leukocytes as early as day 4 of infection that was not observed in any of the other infection groups on that day. By day 5 of infection, a significant increase in leukocytes was present for all infection groups. Although we do not yet understand the mechanism of pathogenesis and increased fungal growth in the host, the Silveira isolate has been reported to be highly virulent in mice (17, 44).
A differential host response to the three Coccidioides isolates was also observed when host cytokine gene mRNA levels in lung tissue were examined. We discovered a differential and diverse cytokine gene expression profile associated with each Coccidioides isolate. Previous studies have shown that some cytokines, including IFN-γ, can be protective against Coccidioides infection in mice, whereas other cytokines, including IL-4 and IL-10, are not protective against infection (6, 7). Other cytokines, such as TNF-α, IL-1α, and IL-17α, have been shown to be protective in murine models of other mycoses (12–14). The differential cytokine responses involving both probable protective and nonprotective cytokines we observed in our study may associate with the inability of BALB/c mice to respond to Coccidioides infections. We detected few cytokines in the BALF using the multiplex bead array assay.
We quantified various amounts of total protein in BALF, with day 5 having the overall largest amounts of total protein present in infected mice compared to uninfected mice. Increases in total protein concentration and leukocytes are markers for lung injury with other infections (45–47). By these assessments, Silveira appeared to be the most virulent isolate used in this study. Our observation of increasing levels of the total protein during the course of this study was probably due in part to lung damage caused by infection. Because few cytokines were detected, proteomic analysis on the samples collected on day 5 of infection allowed determination of other proteins present. We found differences in the proteomic profile that suggest that lung damage was not the only cause of increased protein concentration. Noninterleukin proteins involved in inflammation, immune signaling, and host defense response proteins varied in abundance for all infection groups compared with uninfected mice. Similar to the cytokine response to infection, the proteome of the BALF varied depending on which isolate was used for infection. The observations that were made in this study will set the foundation for future studies characterizing the host response to different Coccidioides isolates. As well, some proteins and specific host response pathways may be useful host biomarkers for pulmonary coccidioidomycosis.
In addition to finding differential host protein expression profiles, we also identified at least three Coccidioides proteins that have been shown to be associated with the Coccidioides parasitic life cycle by other research groups, confirming that our method of proteomic analysis was able to detect expected proteins. Some of the Coccidioides proteins that we identified are uncharacterized proteins, including a protein (CIMG_09001/CPSG_01366) that was identified in all infection groups. Proteins associated with infections caused by both Coccidioides species are good candidates for further study, as these novel proteins may be necessary for the initiation of the parasitic life cycle and therefore potential targets for diagnostics and/or therapeutics.
Many Coccidioides isolates tested to date cause fatal disease within eight to 12 days with as few as 50 arthroconidia administered intranasally in immunocompetent BALB/c mice (17, 48). We administered 100,000 arthroconidia intranasally to each group and could have potentially overwhelmed the host innate immune response. The data suggest that this was not the case and that this dose is reasonable when dosages administered for other fungal pathogens are considered (49–51). As discussed above, we observed a differential cytokine response to Coccidioides isolates used in the study, and the early innate immune response did not seem overly robust. With regard to the infectious dose in humans, neither the average nor the minimum dosage is known (52). The dose administered to the mice in our study is biologically relevant, as a host could inhale 100,000 or more arthroconidia during an acute environmental exposure. In future studies, it will be worthwhile to assess host cytokine response in a dosage-dependent manner.
The lack of a strong cytokine response to early pulmonary coccidioidomycosis described in this study points to previous studies suggesting host immune system evasion. Circulating Coccidioides antigens have been shown to suppress cell-mediated immune response in BALB/c mice (6, 53, 54). More recently, the virulence factors spherule outer wall glycoprotein (SOWgp) and metalloproteinase 1 (Mep1) were identified and shown to evade the host immune response in a C57BL/6 mouse model of coccidioidomycosis (55–59). The most successful pathogens utilize one or several mechanisms to evade and/or counteract a protective immune response from the host (60–62). Because we did not observe these previously identified protein products among our data, we propose that Coccidioides may utilize additional host immune evasion mechanisms, and these may differ temporally as well as among isolates.
Previous studies compared the virulence of different Coccidioides isolates in a mouse model of infection and found differential virulence among the isolates tested (17, 44). To our knowledge, this is the first study that compares the early cytokine response of BALB/c mice to multiple Coccidioides isolates. All isolates that were used in this study are human clinical isolates with known infectivity. Our data suggest that BALB/c mice have a differential cytokine response dependent on the isolate used for infection. At this time, no differential disease phenotypes have been described for coccidioidomycosis caused by C. immitis compared to C. posadasii. This study does not provide enough evidence to support or negate that claim. However, our findings support the importance of performing additional comparative studies of clinical and even environmental isolates of Coccidioides species. Genomic sequencing has thus far revealed high levels of genetic diversity within and among species of Coccidioides (63). Performing comparative functional studies on a variety of these sequenced isolates could help define the genotypic and phenotypic diversity of the organisms and their relationship to clinical outcomes. As well, gaining a better understanding of the host innate immune response may lead to the development of more sensitive and specific diagnostic tools and effective treatment options.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant K22 AI104801 from the National Institute of Allergy and Infectious Diseases (B.M.B.), an Arizona Biomedical Research Corporation young investigator grant (B.M.B.), and Translational Genomics Research Institute (TGen) startup funds (B.M.B. and P.P.).
We truly appreciate the assistance provided by John Galgiani, Lisa Shubitz, Hien Trinh, and Maria Lourdes Lewis with the mouse experiments and access to ABSL3 for this study. We thank Kylie Sage and Heather Mead for assistance with real-time RT-PCR experiments.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00122-15.
REFERENCES
- 1.Lewis ER, Bowers JR, Barker BM. 2015. Dust devil: the life and times of the fungus that causes valley fever. PLoS Pathog 11:e1004762. doi: 10.1371/journal.ppat.1004762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown J, Benedict K, Park BJ, Thompson GR III. 2013. Coccidioidomycosis: epidemiology. Clin Epidemiol 5:185–197. doi: 10.2147/CLEP.S34434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen C, Barker BM, Hoover S, Nix DE, Ampel NM, Frelinger JA, Orbach MJ, Galgiani JN. 2013. Recent advances in our understanding of the environmental, epidemiological, immunological, and clinical dimensions of coccidioidomycosis. Clin Microbiol Rev 26:505–525. doi: 10.1128/CMR.00005-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nesbit LA, Knox KS, Nguyen CT, Roesch J, Wheat LJ, Johnson SM, Pappagianis D, Chavez S, Ampel NM. 2013. Immunological characterization of bronchoalveolar lavage fluid in patients with acute pulmonary coccidioidomycosis. J Infect Dis 208:857–863. doi: 10.1093/infdis/jit246. [DOI] [PubMed] [Google Scholar]
- 5.Ampel NM, Dionne SO, Giblin A, Podany AB, Galgiani J. 2009. Mannose-binding lectin serum levels are low in persons with clinically active coccidioidomycosis. Mycopathologia 167:173–180. doi: 10.1007/s11046-008-9172-6. [DOI] [PubMed] [Google Scholar]
- 6.Cox RA, Kennell W, Boncyk L, Murphy JW. 1988. Induction and expression of cell-mediated immune responses in inbred mice infected with Coccidioides immitis. Infect Immun 56:13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magee DM, Cox RA. 1995. Roles of gamma interferon and interleukin-4 in genetically determined resistance to Coccidioides immitis. Infect Immun 63:3514–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fierer J, Walls L, Eckmann L, Yamamoto T, Kirkland TN. 1998. Importance of interleukin-10 in genetic susceptibility of mice to Coccidioides immitis. Infect Immun 66:4397–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cain JA, Deepe GS Jr. 1998. Evolution of the primary immune response to Histoplasma capsulatum in murine lung. Infect Immun 66:1473–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou P, Sieve MC, Bennett J, Kwon-Chung KJ, Tewari RP, Gazzinelli RT, Sher A, Seder RA. 1995. IL-12 prevents mortality in mice infected with Histoplasma capsulatum through induction of IFN-gamma. J Immunol 155:785–795. [PubMed] [Google Scholar]
- 11.Gonzalez A, Sahaza JH, Ortiz BL, Restrepo A, Cano LE. 2003. Production of pro-inflammatory cytokines during the early stages of experimental Paracoccidioides brasiliensis infection. Med Mycol 41:391–399. doi: 10.1080/13693780310001610038. [DOI] [PubMed] [Google Scholar]
- 12.Schelenz S, Smith DA, Bancroft GJ. 1999. Cytokine and chemokine responses following pulmonary challenge with Aspergillus fumigatus: obligatory role of TNF-alpha and GM-CSF in neutrophil recruitment. Med Mycol 37:183–194. doi: 10.1046/j.1365-280X.1999.00219.x. [DOI] [PubMed] [Google Scholar]
- 13.Caffrey AK, Lehmann MM, Zickovich JM, Espinosa V, Shepardson KM, Watschke CP, Hilmer KM, Thammahong A, Barker BM, Rivera A, Cramer RA, Obar JJ. 2015. IL-1alpha signaling is critical for leukocyte recruitment after pulmonary Aspergillus fumigatus challenge. PLoS Pathog 11:e1004625. doi: 10.1371/journal.ppat.1004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murdock BJ, Huffnagle GB, Olszewski MA, Osterholzer JJ. 2014. Interleukin-17A enhances host defense against cryptococcal lung infection through effects mediated by leukocyte recruitment, activation, and gamma interferon production. Infect Immun 82:937–948. doi: 10.1128/IAI.01477-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burt A, Dechairo BM, Koenig GL, Carter DA, White TJ, Taylor JW. 1997. Molecular markers reveal differentiation among isolates of Coccidioides immitis from California, Arizona and Texas. Mol Ecol 6:781–786. doi: 10.1046/j.1365-294X.1997.00245.x. [DOI] [PubMed] [Google Scholar]
- 16.Fisher MC, Koenig GL, White TJ, Taylor JW. 2002. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 94:73–84. doi: 10.2307/3761847. [DOI] [PubMed] [Google Scholar]
- 17.Friedman L, Smith CE, Gordon LE. 1955. The assay of virulence of Coccidioides in white mice. J Infect Dis 97:311–316. doi: 10.1093/infdis/97.3.311. [DOI] [PubMed] [Google Scholar]
- 18.Zimmermann CR, Snedker CJ, Pappagianis D. 1994. Characterization of Coccidioides immitis isolates by restriction fragment length polymorphisms. J Clin Microbiol 32:3040–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shubitz LF, Dial SM, Perrill R, Casement R, Galgiani JN. 2008. Vaccine-induced cellular immune responses differ from innate responses in susceptible and resistant strains of mice infected with Coccidioides posadasii. Infect Immun 76:5553–5564. doi: 10.1128/IAI.00885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung D, Barker BM, Carey CC, Merriman B, Werner ER, Lechner BE, Dhingra S, Cheng C, Xu W, Blosser SJ, Morohashi K, Mazurie A, Mitchell TK, Haas H, Mitchell AP, Cramer RA. 2014. ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Pathog 10:e1004487. doi: 10.1371/journal.ppat.1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. [DOI] [PubMed] [Google Scholar]
- 23.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 24.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. 2009. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Bardes EE, Aronow BJ, Jegga AG. 2009. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37:W305-311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuervo AM, Gomes AV, Barnes JA, Dice JF. 2000. Selective degradation of annexins by chaperone-mediated autophagy. J Biol Chem 275:33329–33335. doi: 10.1074/jbc.M005655200. [DOI] [PubMed] [Google Scholar]
- 27.Cowburn AS, Sobolewski A, Reed BJ, Deighton J, Murray J, Cadwallader KA, Bradley JR, Chilvers ER. 2006. Aminopeptidase N (CD13) regulates tumor necrosis factor-alpha-induced apoptosis in human neutrophils. J Biol Chem 281:12458–12467. doi: 10.1074/jbc.M511277200. [DOI] [PubMed] [Google Scholar]
- 28.Scharfstein J. 2006. Parasite cysteine proteinase interactions with alpha 2-macroglobulin or kininogens: differential pathways modulating inflammation and innate immunity in infection by pathogenic trypanosomatids. Immunobiology 211:117–125. doi: 10.1016/j.imbio.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 29.Merle NS, Noe R, Halbwachs-Mecarelli L, Fremeaux-Bacchi V, Roumenina LT. 2015. Complement system part II: role in immunity. Front Immunol 6:257. doi: 10.3389/fimmu.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutra FF, Bozza MT. 2014. Heme on innate immunity and inflammation. Front Pharmacol 5:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vadasz I, Weiss CH, Sznajder JI. 2012. Ubiquitination and proteolysis in acute lung injury. Chest 141:763–771. doi: 10.1378/chest.11-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crosby LM, Waters CM. 2010. Epithelial repair mechanisms in the lung. Am J Physiol Lung Cell Mol Physiol 298:L715-731. doi: 10.1152/ajplung.00361.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McAleer JP, Kolls JK. 2014. Directing traffic: IL-17 and IL-22 coordinate pulmonary immune defense. Immunol Rev 260:129–144. doi: 10.1111/imr.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker D, Prince A. 2011. Innate immunity in the respiratory epithelium. Am J Respir Cell Mol Biol 45:189–201. doi: 10.1165/rcmb.2011-0011RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang XX, Fok KL, Chen H, Chan KS, Tsang LL, Rowlands DK, Zhang XH, Dong JD, Ruan YC, Jiang X, Yu SS, Chung YW, Chan HC. 2012. Lymphocyte CFTR promotes epithelial bicarbonate secretion for bacterial killing. J Cell Physiol 227:3887–3894. doi: 10.1002/jcp.24101. [DOI] [PubMed] [Google Scholar]
- 36.Christin L, Wysong DR, Meshulam T, Hastey R, Simons ER, Diamond RD. 1998. Human platelets damage Aspergillus fumigatus hyphae and may supplement killing by neutrophils. Infect Immun 66:1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeaman MR. 2010. Bacterial-platelet interactions: virulence meets host defense. Future Microbiol 5:471–506. doi: 10.2217/fmb.09.112. [DOI] [PubMed] [Google Scholar]
- 38.Sharpton TJ, Stajich JE, Rounsley SD, Gardner MJ, Wortman JR, Jordar VS, Maiti R, Kodira CD, Neafsey DE, Zeng Q, Hung CY, McMahan C, Muszewska A, Grynberg M, Mandel MA, Kellner EM, Barker BM, Galgiani JN, Orbach MJ, Kirkland TN, Cole GT, Henn MR, Birren BW, Taylor JW. 2009. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res 19:1722–1731. doi: 10.1101/gr.087551.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pishko EJ, Kirkland TN, Cole GT. 1995. Isolation and characterization of two chitinase-encoding genes (cts1, cts2) from the fungus Coccidioides immitis. Gene 167:173–177. doi: 10.1016/0378-1119(95)00654-0. [DOI] [PubMed] [Google Scholar]
- 40.Whiston E, Zhang Wise H, Sharpton TJ, Jui G, Cole GT, Taylor JW. 2012. Comparative transcriptomics of the saprobic and parasitic growth phases in Coccidioides spp. PLoS One 7:e41034. doi: 10.1371/journal.pone.0041034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson SM, Pappagianis D. 1992. The coccidioidal complement fixation and immunodiffusion-complement fixation antigen is a chitinase. Infect Immun 60:2588–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orsborn KI, Shubitz LF, Peng T, Kellner EM, Orbach MJ, Haynes PA, Galgiani JN. 2006. Protein expression profiling of Coccidioides posadasii by two-dimensional differential in-gel electrophoresis and evaluation of a newly recognized peroxisomal matrix protein as a recombinant vaccine candidate. Infect Immun 74:1865–1872. doi: 10.1128/IAI.74.3.1865-1872.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sobonya RE, Yanes J, Klotz SA. 2014. Cavitary pulmonary coccidioidomycosis: pathologic and clinical correlates of disease. Hum Pathol 45:153–159. doi: 10.1016/j.humpath.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 44.Cox RA, Magee DM. 2004. Coccidioidomycosis: host response and vaccine development. Clin Microbiol Rev 17:804–839. doi: 10.1128/CMR.17.4.804-839.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Check IJ, Kidd MR, Staton GW Jr. 1986. Systemic and lung protein changes in sarcoidosis. Lymphocyte counts, gallium uptake values, and serum angiotensin-converting enzyme levels may reflect different aspects of disease activity. Ann N Y Acad Sci 465:407–417. [DOI] [PubMed] [Google Scholar]
- 46.Atochina EN, Beck JM, Preston AM, Haczku A, Tomer Y, Scanlon ST, Fusaro T, Casey J, Hawgood S, Gow AJ, Beers MF. 2004. Enhanced lung injury and delayed clearance of Pneumocystis carinii in surfactant protein A-deficient mice: attenuation of cytokine responses and reactive oxygen-nitrogen species. Infect Immun 72:6002–6011. doi: 10.1128/IAI.72.10.6002-6011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin W, Rong L, Liu Y, Song Y, Li Y, Pan J. 2013. Increased claudin-3, -4 and -18 levels in bronchoalveolar lavage fluid reflect severity of acute lung injury. Respirology 18:643–651. doi: 10.1111/resp.12034. [DOI] [PubMed] [Google Scholar]
- 48.Muhammed M, Feldmesser M, Shubitz LF, Lionakis MS, Sil A, Wang Y, Glavis-Bloom J, Lewis RE, Galgiani JN, Casadevall A, Kontoyiannis DP, Mylonakis E. 2012. Mouse models for the study of fungal pneumonia: a collection of detailed experimental protocols for the study of Coccidioides, Cryptococcus, Fusarium, Histoplasma and combined infection due to Aspergillus-Rhizopus. Virulence 3:329–338. doi: 10.4161/viru.20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dixon DM, Polak A, Walsh TJ. 1989. Fungus dose-dependent primary pulmonary aspergillosis in immunosuppressed mice. Infect Immun 57:1452–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sawyer RT. 1990. Experimental pulmonary candidiasis. Mycopathologia 109:99–109. doi: 10.1007/BF00436790. [DOI] [PubMed] [Google Scholar]
- 51.Wormley FL Jr, Perfect JR, Steele C, Cox GM. 2007. Protection against cryptococcosis by using a murine gamma interferon-producing Cryptococcus neoformans strain. Infect Immun 75:1453–1462. doi: 10.1128/IAI.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nicas M, Hubbard A. 2002. A risk analysis for airborne pathogens with low infectious doses: application to respirator selection against Coccidioides immitis spores. Risk Anal 22:1153–1163. doi: 10.1111/1539-6924.00279. [DOI] [PubMed] [Google Scholar]
- 53.Cox RA. 1988. Immunosuppression by cell wall antigens of Coccidioides immitis. Rev Infect Dis 10(Suppl 2):S415–S418. doi: 10.1093/cid/10.Supplement_2.S415. [DOI] [PubMed] [Google Scholar]
- 54.Cox RA, Kennell W. 1988. Suppression of T-lymphocyte response by Coccidioides immitis antigen. Infect Immun 56:1424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hung CY, Ampel NM, Christian L, Seshan KR, Cole GT. 2000. A major cell surface antigen of Coccidioides immitis which elicits both humoral and cellular immune responses. Infect Immun 68:584–593. doi: 10.1128/IAI.68.2.584-593.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hung CY, Yu JJ, Seshan KR, Reichard U, Cole GT. 2002. A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect Immun 70:3443–3456. doi: 10.1128/IAI.70.7.3443-3456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hung CY, Seshan KR, Yu JJ, Schaller R, Xue J, Basrur V, Gardner MJ, Cole GT. 2005. A metalloproteinase of Coccidioides posadasii contributes to evasion of host detection. Infect Immun 73:6689–6703. doi: 10.1128/IAI.73.10.6689-6703.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johannesson H, Townsend JP, Hung CY, Cole GT, Taylor JW. 2005. Concerted evolution in the repeats of an immunomodulating cell surface protein, SOWgp, of the human pathogenic fungi Coccidioides immitis and C. posadasii. Genetics 171:109–117. doi: 10.1534/genetics.105.040923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hung CY, Xue J, Cole GT. 2007. Virulence mechanisms of Coccidioides. Ann N Y Acad Sci 1111:225–235. doi: 10.1196/annals.1406.020. [DOI] [PubMed] [Google Scholar]
- 60.Deitsch KW, Lukehart SA, Stringer JR. 2009. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol 7:493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chotirmall SH, Mirkovic B, Lavelle GM, McElvaney NG. 2014. Immunoevasive Aspergillus virulence factors. Mycopathologia 178:363–370. doi: 10.1007/s11046-014-9768-y. [DOI] [PubMed] [Google Scholar]
- 62.Reddick LE, Alto NM. 2014. Bacteria fighting back: how pathogens target and subvert the host innate immune system. Mol Cell 54:321–328. doi: 10.1016/j.molcel.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neafsey DE, Barker BM, Sharpton TJ, Stajich JE, Park DJ, Whiston E, Hung CY, McMahan C, White J, Sykes S, Heiman D, Young S, Zeng Q, Abouelleil A, Aftuck L, Bessette D, Brown A, FitzGerald M, Lui A, Macdonald JP, Priest M, Orbach MJ, Galgiani JN, Kirkland TN, Cole GT, Birren BW, Henn MR, Taylor JW, Rounsley SD. 2010. Population genomic sequencing of Coccidioides fungi reveals recent hybridization and transposon control. Genome Res 20:938–946. doi: 10.1101/gr.103911.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.