Abstract
Background: Human exposure to the widespread environmental contaminant mercury is a known risk factor for common diseases such as cancer, cardiovascular disease and neurological disorders through poorly characterized mechanisms. Evidence suggests mercury exposure may alter DNA methylation levels, but to date, the effects in early life on a genome-wide scale have not been investigated.
Methods: A study sample of 141 newborns was recruited in Baltimore, MD, USA and total mercury and methylmercury were measured in cord blood samples. We quantified genome-wide DNA methylation data using CHARM 2.0, an array-based method, and used region-finding analyses to identify concentration-associated differentially methylated regions (DMRs). To test for replication of these identified DMRs in the pilot, or Vanguard, phase of the National Children’s Study (NCS), we compared bisulfite-pyrosequenced DNA at candidate regions from 85 whole cord blood samples with matched first trimester maternal mercury concentration measures.
Results: Total mercury concentration was associated with methylation at DMRs inside ANGPT2 and near PRPF18 genes [false discovery rate (FDR) < 0.05], as well as DMRs near FOXD2 and within TCEANC2 (FDR< 0.1) genes. Methylmercury concentration was associated with an overlapping DMR within TCEANC2 (FDR< 0.05). In NCS replication analyses, methylation levels at three of four cytosine-guanine DNA dinucleotides (CpG sites) within the TCEANC2 DMR were associated with total mercury concentration (P < 0.05), and this association was diminished after adjusting for estimated cell proportions.
Conclusions: Evidence for an association between mercury and DNA methylation at the TCEANC2 region was found, which may represent a mercury-associated shift in cord blood cell composition or a change in methylation within blood cell types. Further confirmatory studies are needed.
Keywords: DNA methylation, mercury exposure, epigenetics, cord blood, methylmercury
Key Messages.
In utero mercury exposure may be related to DNA methylation changes in offspring.
In a genome-wide DNA methylation analysis of 141 cord blood samples, four differentially methylated regions were associated with total or methylmercury concentrations at false discovery rate q-values <0.1.
The region associated with the TCEANC2 gene was validated in the same samples using a second method of DNA methylation detection, and it was replicated in a second independent sample of infants.
Introduction
Environmental and occupational mercury exposure has long been recognized to have severe health consequences, including sensory disturbance, visual field constriction, ataxia, deafness and neurodevelopmental deficits.1,2 Human toxicity to mercury depends on the mercury species, including methylmercury, ethylmercury and inorganic mercury, all of which are encompassed in blood measures of total mercury. Typical exposure is via dietary methylmercury from fish and other seafood.3,4 Toxicity from dietary methylmercury has been observed historically, including the Minamata, Japan and Iraq grain poisoning epidemics, where very high methylmercury in utero exposures caused severe central nervous system damage and death.5–7 Much lower methylmercury in uteroexposures are also associated with subtle, subclinical brain function deficits.8 The mechanisms underlying the early life health effects of mercury exposure are incompletely characterized and suggestions include localized neuronal death, microtubule problems, neuronal migration disruption, oxidative damage and altered brain calcium metabolism.9,10 Given the potential for epigenetic susceptibility to toxicant exposures,11 fetal epigenetic changes resulting from in utero mercury exposure could inform this mechanism of toxicity.
Epigenetic epidemiology research, particularly epigenome-wide association studies (EWAS), interrogates epigenetic changes associated with environmental exposures or disease phenotypes in a population context. Identified epigenetic markers may act directly in disease causality, or they may interact with genetic sequence variation or environmental factors to influence disease. Alternatively, they may be useful biomarkers of previous exposures when the toxicant and metabolites may no longer be observable.11 The prenatal period is a critical window in development and the potential relationship between mercury body burden and DNA methylation at birth may provide an opportunity for epidemiological research with implications concerning central nervous system development.
At present, no genome-scale study has tested the association between DNA methylation and mercury exposure during prenatal development. We performed an epigenome-wide association study of cord blood DNA methylation in 141 newborns to identify regions ofDNA methylation correlated with in utero mercury exposure. We then tested for replication of the identified regional DNA methylation-mercury association results in an independent sample, using a second method of DNA methylation detection.The findings from this genome-scale study enhance our understanding of the relationship between heavy metal exposures and DNA methylation early in fetal development, with potential implications for the usefulness of epigenetic changes as biomarkers of prenatal exposure and potential mediators of health effects.
Methods
Study populations
Discovery sample
Cord blood samples were obtained from newborns at Johns Hopkins Hospital, MD, USA, between November 2004 and March 2005, as part of the previously described cross-sectional Baltimore Tracking Health Related to Environmental Exposures (THREE) study.12 Demographic and limited medical information were obtained from maternal and neonatal medical records. Study clinicians reviewed a 10% random sample of medical record abstractions for accuracy, as previously described.12 The THREE study was reviewed and approved by the Johns Hopkins School of Medicine Institutional Review Board (IRB). A waiver of the need for informed consent was obtained since the study constituted minimal risk, was anonymous and made use of only biological samples that would otherwise be medical waste.
During the Baltimore THREE study time window, there were 609live births. Twelve twin births (24 infants) and 256 births where cord blood was not available were excluded. An additional 41 births had insufficient cord blood samples for methylmercury analyses, leaving 300 infants eligible for the current study. Among the study samples, 187 had umbilical cord blood clots available for DNA extraction, 167 samples yielded sufficient DNA for the methylation assay (5 µg) and 141 samples passed our internal DNA methylation array quality control.13 Samples in the THREE epigenetics set (n = 141) were similar to the samples in the larger THREE exposure study (n = 300) with respect to maternal age, race, mercury exposure, infant sex, gestational age and birthweight (Supplementary Table 1, available as Supplementary data at IJE online).
Replication sample
Maternal matched first and third trimester peripheral blood samples (nwomen = 147), and a subset of corresponding cord blood samples (n = 90) were obtained from the Initial Vanguard Centers of the National Children's Study,14 a pilot study involving mothers and infants recruited from seven different locations across the USA (Duplin County, NC; Queens County, NY; Orange County, CA; Waukesha County, WI; Salt Lake County, UT; Montgomery County, PA; and, a composite location of four adjacent counties in South Dakota and Minnesota). The NCS is an ongoing, large, long-term study of children’s health and development that has previously been described.15,16 The Vanguard phase of the NCS began in 2009, in order to inform the later Main Study, to evaluate the methodologies, feasibility, acceptability and cost of the study. Women were eligible if they were biologically able to become pregnant or were currently pregnant and were aged 18 through 49 years. Participants had up to two pregnancy study visits in the first and third trimesters and a visit with their infants at birth. Demographic information was collected by interview at study visits before or during pregnancy. Race/ethnicity categories of mothers were self-reported as non-Hispanic White, non-Hispanic Black, and all Hispanics including Mexican-Americans and other groups. Due to limited sample size, these were re-categorized as White/non-White for all methylation analyses. Written informed consent was obtained from all NCS participants or their legally authorized representatives and the protocol was approved by the corresponding study centre Institutional Review Board. The infants were born between October 2009 and June 2011. Only material from participants who explicitly consented to use of their biological samples to obtain genetic information was used in these analyses. Of the 90 cord blood samples, 85 had sufficient DNA for replication methylation analyses; however, fewer had available exposure data as described below.
Exposure assessments
In the THREE study samples, total mercury, methylmercury, lead, selenium, copper and omega-3 fatty acids were analysed as previously described.17 Umbilical cord whole blood and serum samples were collected in trace element-free tubes. Whole blood total mercury and lead were measured via inductively coupled plasma mass spectrometry (ICP-MS) and serum concentrations of selenium and copper were measured using inductively coupled plasma dynamic reaction cell mass spectrometry (ICP-DRC-MS) at the Centers for Disease Control and Prevention (CDC) National Center for Environmental Health (CDC/NCEH) laboratory.18 The CDC’s Environmental Health Laboratory was determined not to be participating in human subjects research. The cord serum n-3 polyunsaturated acids, eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) were measured using automated fast gas chromatography at the National Institute for Alcohol Abuse and Alcoholism17 and their levels were summed for analysis (EPA + DHA). The limits of detection (LOD) for exposure concentrations in the THREE study were 0.48 µg/l (methylmercury), 0.33 µg/l (total mercury), 0.25 µg/dl (lead), 5 µg/l (selenium) and 4 µg/dl (copper). Samples with exposure values below the LOD were treated as LOD/√2, except for methylmercury. As there were more than 10% values < LOD for methylmercury, these were replaced with concentrations reported from the laboratory instrument, in order to minimize bias due to left-censoring.
In the NCS Vanguard study, mercury measures in infant cord blood samples were not available. Instead, blood samples collected from pregnant mothers in their first trimester were used for exposure measurement. Blood was collected into CDC-prescreened EDTA tubes that were free of lead, mercury, selenium and copper. Whole blood samples were frozen and shipped on dry ice to the NCS Repository (Fisher Bioservices, Rockville, MD), then stored frozen at vapour phase liquid nitrogen temperatures (between –135 and –190°C) until shipment on dry ice to the CDC laboratory where they were also stored frozen (– 70°C) until they were analysed. The NCEH measured whole blood total mercury, lead and selenium via ICP-MS in the NCS samples as well (LODs respectively: 0.28 total Hg µg/l, 0.25 Pb µg/dl, 5 Se µg/l). Methylmercury was measured via triple-spike isotope dilution with solid-phase microextraction gas chromatography (TSID-SPME-GC) (LOD 0.12 µg/l).19 Of the maternal samples, 77 had available methylmercury measures and corresponding infant DNA methylation data from targeted pyrosequencing assays (see below); 75 had available total mercury data and pyrosequencing DNA methylation data. For cell-type association estimates to explore the potential for confounding (see Statistical methods), 138 mothers had mercury measures and 450K array DNA methylation data (see below).
DNA methylation measures
Genome-scale DNA methylation discovery: CHARM 2.0
DNA methylation in the THREE study samples was measured via the second version of comprehensive high-through put arrays for relative methylation (CHARM 2.0)(Roche NimbleGen, Madison, WI, USA).13 These measures were used to perform genome-scale discovery of candidate differentially methylated regions (DMRs) associated with total mercury and methylmercury concentrations. DNA was isolated from THREE study cord blood clot samples using DNeasy Blood & Tissue kits (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. Of 187 cord blood clot samples, 167 (89.3%) yielded sufficient DNA. After DNA isolation, 5 µg of purified genomic DNA was subjected to shearing, digestion, purification, amplification, labelling and hybridization as previously described,20 generating data on 2.1 million probes covering 5.2 million CpGs, including all CpG islands, CpG island shores, annotated and non-annotated promoters, and microRNA sites. At the sample level, 26 individuals were excluded because observed intensities of < 80% of probes exceeded background levels. At the probe level, sex chromosome probes and those with signals below background in > 25% of samples were removed. The final analysis included 141 samples with 1 569 888 autosomal probes covering 4 254 946 CpGs.
The array hybridization and subsequent processing were performed over 5 days; we addressed this potential batch effect by surrogate variable analysis (SVA) as described in the Statistical methods subsection. Total and methylmercury concentrations did not vary by array hybridization batch (P-valuetotal = 0.58; P-valuemethyl = 0.25).
Pyrosequencing validation of CHARM in THREE samples
DNA methylation, at the candidate DMRs (in or near TCEANC2, ANGPT2, PRPF18 and FOXD2 genes) identified during genome-wide discovery phase as surpassing the significance threshold, were validated via bisulfite pyrosequencing on THREE study samples for which additional bisulfite converted DNA was available (n = 136). MethPrimer software21 was used for assay design. Bisulfite-treated DNA was PCR-amplified using unbiased nested primers (SupplementaryTable 2, available as Supplementary data at IJE online), and pyrosequencing was performed to quantitatively measure DNA methylation using the PSQ HS96 (Biotage, Charlotte, NC, USA). Q-CpG methylation software (Biotage, Charlotte, NC, USA) was used to quantify methylation measurements. Mixtures of Whole Genome Amplified (WGA) Human Genomic DNA: Male (Promega, Fitchburg, WI, USA) via REPLI-g Mini Kit (Qiagen, Valencia, CA, USA) and SSsI-treated WGA DNA were used to construct 0, 25, 50, 75 and 100% methylated control standards.
Gene-specific DNA methylation replication in NCS Vanguard samples
The candidate DMR methylation measures at the ANGPT2, PRPF18, FOXD2 and TCEANC2 genes were measured in the independent sample of NCS Vanguard infants, using pyrosequencing methods as described above. Mixed buffy coat and red blood cell aliquots were derived from cord blood collection bags with citrate phosphate dextrose (CPD) anticoagulant. DNA was isolated using the AgencourtGenefind v2 (Beckman Coulter, Pasadena, CA, USA) on the BiomekNXp laboratory automation workstation according to manufacturer’s instructions. Genomic DNA from each sample was quantified via Picogreen (Invitrogen Life Technologies, Carlsbad, CA, USA). Of 90 cord blood samples, 85 (94%) yielded sufficient DNA for pyrosequencing (200 ng). Genomic DNA from each sample was bisulfite-treated and cleaned using EZ DNA Methylation-Gold Kit (ZymoResearch, Irvine, CA, USA) according to the manufacturer's instructions. Primers for the additional assays are listed in SupplementaryTable 2, available as Supplementary data at IJE online.
Cell type distribution estimation: Genome-scale DNA methylation on the Infinium HumanMethylation450 array
Genome-scale DNA methylation data on NCS Vanguard samples were used to estimate blood cell type distributions per sample. The Infinium HumanMethylation450 assay (Illumina, San Diego, CA, USA) was performed on 147 maternal first and third trimester peripheral blood buffy coat samples and 90 cord blood samples. Agarose gel electrophoresis (1% agarose) was used to check the quality of extracted DNA. Genomic DNA was normalized/aliquotted to 1 µg per sample, and sent to the Johns Hopkins University Center for Inherited Disease Research (CIDR) for bisulfite conversion of genomic DNA with the EZ DNA Methylation-Gold kit (Zymo Research, Irvine, CA, USA) and hybridization to the InfiniumBeadChip. As described below, these data were used to estimate cell type proportions per cord blood sample via a cell type prediction algorithm.22 This algorithm requires reference data from cell type-specific DNA methylation profiles to inform predictions. Cord blood reference samples would be the appropriate input, but no publicly available or proprietary purchaseable cord-blood derived or childhood data are currently available. Instead, we used publicly available cell type-specific DNA methylation profiles from adult men.23
In addition, we estimated cell type proportions in samples from the THREE study, based on CHARM DNA methylation data. This is an incomplete method, since Infinium probes for which (adult male) cell type reference methylation information is available are not highly overlapping with CHARM probe locations. Cell type proportion estimates obtained in this manner were outside previously reported cord blood cell type ranges and were not biologically plausible. These were not included in downstream analyses.
Gene-specific RNA gene expression
Quantitative real-time PCR (qPCR) was used to measure TCEANC2 mRNA expression levels in a subset of the THREEsamples. Fetal cord blood clots were treated with TRIzol (Invitrogen Life Technologies, Carlsbad, CA, USA) and the PureLink RNA Mini Kit (Invitrogen Life Technologies, Carlsbad, CA, USA) was used to isolate RNA, according to the manufacturer’s instructions. cDNA was synthesized using QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA, USA) with random hexamer priming. Samples with sufficient quantities (n = 42) were run for qPCR in triplicates of technical replicates with the Fast SYBR_Green Master Mix (Applied BiosystemsLife Technologies, Carlsbad, CA, USA), on an ABI 7900 Sequence Detection System (Applied Biosystems Life Technologies, Carlsbad, CA, USA).Samples with high qPCR threshold cycle standard deviation between technical replicates (Ct SD > 0.25) were excluded (n = 6). Expression levels of TCEANC2were calculated relative to beta-actin expression. The mercury concentration distributions of 34 THREE participants with final RNA expression results were similar to the discovery methylation population [geometric mean (IQR) methylmercurylevels: 0.92 µg/l (0.62–1.34), geometric mean (IQR) total mercury levels: 1.38 µg/l (0.85–1.68)]. Primer sequences used for the qPCR reactions are described in Supplementary Table 3 (available as Supplementary data at IJE online).
Statistical analyses
Study population demographic and exposure characteristics from the discovery THREE and NCS replication sets were compared using t-tests for continuous data and ANOVA tests for categorical variables. In the THREE samples, Spearman correlations between total and methylmercury concentrations were calculated, as were the bivariate relations between total mercury and selected maternal and infant demographic characteristics.
THREE study CHARM 2.0 DNA methylation data were pre-processed, normalized and analysed for regional associations via ‘bumphunting’.24 In summary, we tested for candidate DMRs associated with total or methylmercury concentration separately by fitting linear models of methylation levels at each probe as a function of log-transformed total or methylmercury concentration levels, adjusted for surrogate variables estimated via SVA.25 Surrogate variables for downstream adjustment of total mercury and methylmercury analyses were calculated separately and their similarity was tested using Spearman correlation tests. ANOVA tests were performed between measured categorical covariates (array scanner, hybridization date, maternal smoking status, BMI, race, age and offspring mercury, copper, selenium, lead, gestational age, birthweight, and sex) and each surrogate variable. Models did not include adjustment for sex, as we removed sex chromosome probes prior to analysis. Next, estimated regression coefficients from the linear models at each probe within regional probe groups pre-defined in CHARM 2.0 array design were smoothed and filtered by a 99.995th percentile cutoff. Region-level statistics representing the DMR area were calculated and family-wise error rate (FWER) and false discovery rate (FDR) q-values for each DMR were estimated based on 1000 permutations under the null. DMRs that passed the empirical FWER cutoff of 0.2 and FDR q-value cutoff of 0.1 were visualized and targeted for validation and replication. Our primary discovery region-finding analysis involved total mercury levels and our secondary analysis used methylmercury levels.
CHARM DNA methylation measures were validated in a subset of the THREE samples at four genes (TCEANC2, ANGPT2, PRPF18 and FOXD2) using pyrosequencing. A Spearman correlation test of the association between CHARM mean methylation levels across the DMR and mean pyrosequencing methylation across four sites assayed was used to assess concordance between DNA methylation methods.
During the replication phase, we analysed gene-specific NCS DNA methylation data from pyrosequencing data at four genes (TCEANC2, ANGPT2, PRPF18 and FOXD2). We fitted linear models at each CpG, predicting DNA methylation as a function of log-transformed total or methylmercury concentration. In addition, exposure quartiles were calculated and a linear trend test across ordinal exposure categories was performed.
To assess functional implications of methylation differences at the TCEANC2 gene, we calculated Spearman’s rank order correlation between DNA methylation and gene expression with respect to each CpG. Gene expression values were transformed to log2 scale.
We performed several analyses in THREE to address potential confounding by measured covariates including sex, race, maternal age, birthweight, gestational age and concentrations of lead, selenium, copper and EPA + DHA. We tested for association between mercury and potential continuous confounders using Pearson correlation tests of log-transformed mercury measures and between mercury and potential binary confounders using t-tests. Similar tests were performed for association between mean methylation at the TCEANC2 DMR and potential confounders. Finally, we estimated the relationships between average DNA methylation for the DMR and mercury concentration by linear regression, adjusting for each confounder individually in addition to the surrogate variables used in our discovery phase analysis.
There is no publicly available cord blood-derived cell-sorted reference panel to use for cell type estimation similar to what has been applied to adult data.23 Instead we considered possible associations with blood cell type based on adult male reference data.23 Four probe sets from this Reinius et al.23 Illumina 450K dataset (cg01109333, cg01986665, cg02270108 and cg02626873) were located inside TCEANC2 DMR. We compared DNA methylation levels at each of these probe sets across the sorted blood cell types (B cells, CD4+ T cells, CD8+ T cells, granulocytes, monocytes, natural killer cells and whole blood) using an ANOVA test. There were six replicates of each cell type.
To assess the relationship between blood cell type proportions per person and total mercury concentration in NCS Initial Vanguard Center study samples, we estimated relative cell type proportions using an epigenetic signature prediction algorithm informed by adult samples22 and calculated Pearson’s correlations between log(total mercury concentration) and predicted cell type proportions. Log(total mercury concentration) was regressed on mean NCS pyrosequencing DNA methylation at theTCEANC2gene both with and without adjustment for predicted granulocyte proportion or all six cell type proportions. The beta estimates and corresponding P-values for total mercury were compared between the models.
Results
Population characteristics
Population univariate descriptive statistics for the discovery Baltimore THREE and NCS Initial Vanguard replication samples used for this study are given in Table 1. Distributions of offspring sex and gestational age between the two studies were similar (P-valuesex = 0.9; P-valuegestational age = 0.9). Younger maternal age and lower birthweight were observed in the THREE sample (P-valueage = 1 × 10−6; P-valuebirth weight = 0.08).
Table 1.
Newborn characteristics of THREE and NCS Vanguard Study samples. Cord blood was used for THREE and whole blood for NCS first-trimester pregnant women
| Characteristics | THREE, n = 141 | NCS, n = 77 |
|---|---|---|
| Offspring gestational age, median (IQR) (days) | 275 (266–282) | 275 (270–281) |
| Offspring birthweight, median (IQR) (g) | 3183 (2794–3566) | 3402 (3102–3691) |
| Offspring male sex (%) | 52 | 51 |
| Cord whole blood concentrations | ||
| Total mercury, median (IQR) (µg/l) | 1.4 (1.0–2.0) | – |
| Methylmercury, median (IQR) (µg/l) | 0.9 (0.6–1.6) | – |
| Cord blood serum concentrations | ||
| Copper, median (IQR) (µg/dl) | 39.7 (28.2–53.4) | – |
| Selenium, median (IQR) (µg/l) | 70.0 (62.0–78.0) | – |
| EPA + DHA, median (IQR) (µg/ml) | 48.1 (40.8–56.9) | – |
| Maternal age, median (IQR) (years) | 24 (20–29) | 28 (25–33) |
| Maternal race (%) | ||
| non-Hispanic White | 23 | 75 |
| non-Hispanic Black | 72 | 6 |
| Asian | 5 | – |
| Other | – | 19 |
| NCS whole blood concentrations | ||
| Total mercury, median (IQR) (µg/l) | – | 0.6 (0.4–1.1) |
| Methylmercury, median (IQR) (µg/l) | – | 0.4 (0.2–0.9) |
| Selenium, median (IQR) (µg/l) | – | 180.5 (165.5–193.3) |
IQR, interquartile range.
Total and methylmercury concentrations were higher in the cord blood sample from THREE compared with the maternal trimester one blood samples in NCS pregnant women (THREE geometric meanTotal Hg = 1.35 µg/l; NCS geometric meanTotal Hg = 0.64 µg/l; P-valueTotal Hg = 0.0013; and THREE geometric meanMethyl Hg = 0.87 µg/l; NCS geometric meanMethyl Hg = 0.41 µg/l; P-valueMethyl Hg = 0.029; see Supplementary Table 4, available as Supplementary data at IJE online). Although the distributions differed, both cohorts only contained two samples with methylmercury levels higher than the US Environmental Protection Agency (EPA) reference dose (RfD) for preventing fetal neurotoxicity (5.8 ug/l) and no samples above the health-based benchmark dose (BMD) (85 ug/l) or benchmark dose lower limit 95% confidence interval (BMDL) (58 ug/l). The THREE study had a higher proportion of non-Hispanic Black whereas the NCS Vanguard sample had a higher proportion of non-Hispanic White individuals (P-valuerace = 1 × 10−16).
Within the THREE sample, total and methylmercury concentrations were highly correlated (R = 0.88, P-value = 2 × 10−16). Total mercury level did not vary by infant sex (P-value = 0.47), gestational age (P-value = 0.25), or birthweight (P-value = 0.34). Total mercury was positively associated with maternal age (P-value = 0.0098) and varied by maternal race (P–value = 9.3 × 10−5). Total mercury was elevated among Asians (mean = 2.9 µg/l), intermediate among non-Hispanic Blacks (1.6 µg/l) and lowest among non-Hispanic Whites (1.2 µg/l).
In all, 19 surrogate variables were estimated in the THREE sample for both total and methylmercury analyses. Spearman correlations between surrogate variables for total and methylmercury were high, ranging from 0.986 to 1.000 with a mean of 0.9964 and standard deviation of 0.0049; 14 of the surrogate variables were related to known covariates including hybridization date, array scanner, maternal race, smoking status and BMI (Supplementary Table 5, available as Supplementary data at IJE online).
Genome-scale discovery
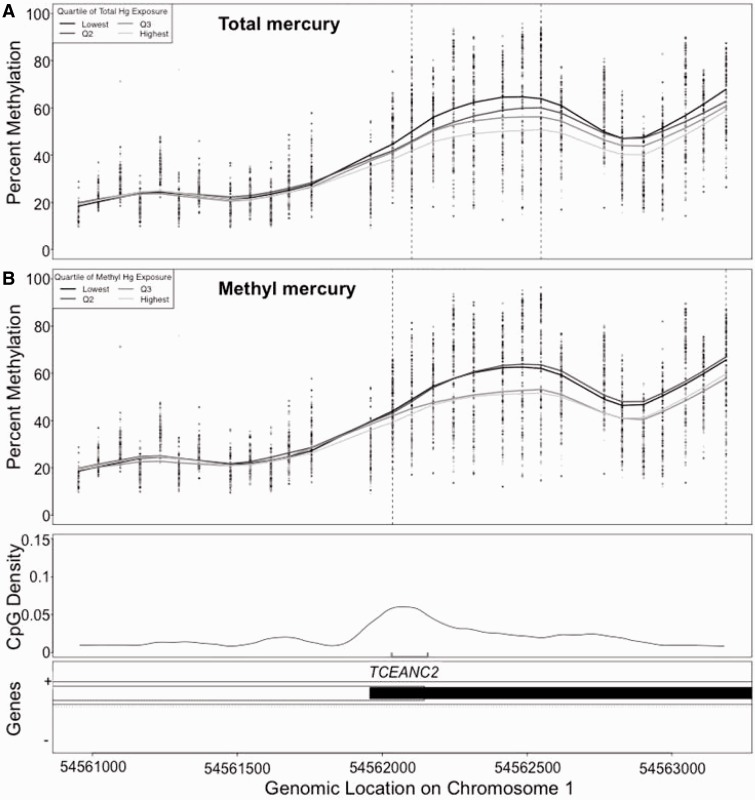
In our primary discovery analyses of total mercury and DNA methylation, four DMRs reached our genome-wide significance thresholds (Table 2). At the DMR in the ANGPT2 gene, encoding angiopoietin 2, DNA methylation was positively associated with total mercury levels though the magnitude of methylation change was modest (Supplementary Figure 1a, available as Supplementary data at IJE online). Mean regional methylation at ANGPT2 in the lowest quartile of total mercury concentration (<1.03 µg/l) was 39.1% (SD: 12.1) and the highest quartile of total mercury concentration (>1.99 µg/l) was 41.4% (SD: 9.4). DNA methylation was inversely associated with total mercury levels in a DMR inside the PRPF18 gene, encoding Pre mRNA Processing Factor 18 (Supplementary Figure 1b, available as Supplementary data at IJE online). Lowest quartile mean (SD) was 72.8% (11.7), highest quartile 59.2% (18.6) and ordinal linear trend test Ptrend = 3.6 × 10−4. In addition, an inverse methylation-total mercury association was observed near the FOXD2 gene, which encodes a transcription factor Forkhead Box D2 (Supplementary Figure 1c, available as Supplementary data at IJE online). Lowest quartile mean (SD) was 53.8% (14.0), highest quartile 43.4% (17.0) and Ptrend = 4.6 × 10−3). Finally, the exonic DMR associated with TCEANC2, Transcription Elongation Factor A (SII) N-Terminal And Central Domain Containing 2, showed DNA methylation inversely associated with total mercury across a region of 446 base pairs spanning seven probe sets on chromosome 1 (Figure 1a). TCEANC2 regional mean methylation in the lowest quartile of total mercury concentration was 56.7% (SD: 14.0) and the highest quartile of concentration had 48.0% (SD: 17.0) methylation (Ptrend = 9.3 × 10−3).
Table 2.
Candidate differentially methylated regions (DMR) associated with total mercury or methylmercury exposure in the THREE study
| Exposure variable | Chr | Nearest gene | FWER | q-value | DMR start position | DMR end position | Location relative to gene | Number of probes | Number of CpGs |
|---|---|---|---|---|---|---|---|---|---|
| Total mercury | 8 | ANGPT2 | 0.03 | 0.045 | 6418036 | 6418808 | Inside intron | 13 | 42 |
| Total mercury | 10 | PRPF18 | 0.05 | 0.045 | 13683914 | 13684474 | Inside intron | 9 | 25 |
| Total mercury | 1 | FOXD2 | 0.13 | 0.078 | 48059831 | 48060319 | Downstream | 8 | 34 |
| Total mercury | 1 | TCEANC2 | 0.19 | 0.079 | 54562102 | 54562548 | Inside exon | 7 | 36 |
| Methylmercury | 1 | TCEANC2 | 0.05 | 0.088 | 54562036 | 54562618 | Inside exon | 16 | 44 |
Chr, chromosome.
Figure 1.
In the THREE study sample, DNA methylation within the TCEANC2 gene was associated with (a) total mercury and (b) methylmercury concentration in µg/l measured in cord blood. Each dot represents the methylation values at each probe for each sample. The smoothed line represents the average methylation curve for each concentration quartile, and demonstrates a dose-dependent inverse association for both total and methylmercury. The black vertical dashed lines define the boundary of the differentially methylated region. The line in the third panel of the plot represents the CpG density in the genomic region. The gray bracket indicates sites assayed in the replication cohort. Quartiles of total mercury are: Q1: [0.233-1.03), Q2: [1.030-1.42), Q3: [1.42-1.99), Q4: [1.99-6.3). Quartiles of methylmercury are: Q1: [0.0849-0.64), Q2: [0.64-0.94), Q3: [0.94-1.69), Q4: [1.69-6.8).
Our secondary discovery analysis using methylmercury resulted in one methylmercury DMR reaching our genome-wide threshold (Table 2). This methylmercury DMR, located in the TCEANC2 gene, was also identified in the total mercury discovery (Figure 1b). Though the remaining three total mercury DMRs (ANGPT2, PRPF18 and FOXD2) did not reach genome-wide significance for methylmercury, the magnitudes and directions of methylation change by methylmercury concentration were consistent with those observed for total mercury (Supplementary Figure 1 d–f, available as Supplementary data at IJE online).
Platform validation
Among 136 samples with CHARM and pyrosequencing data, the Spearman correlation coefficient between DNA methylation measures within the TCEANC2 region was 0.82 (P = 2.2 × 10−16). Pyrosequencing results were also correlated with CHARM results for PRPF16 (ρ = 0.25, P-value = 0.0040) and ANGPT2 (ρ = 0.18, P-value = 0.030). Pyrosequencing methylation was not correlated with CHARM methylation at FOXD2 (ρ = 0.08, P-value = 0.31).
Replication
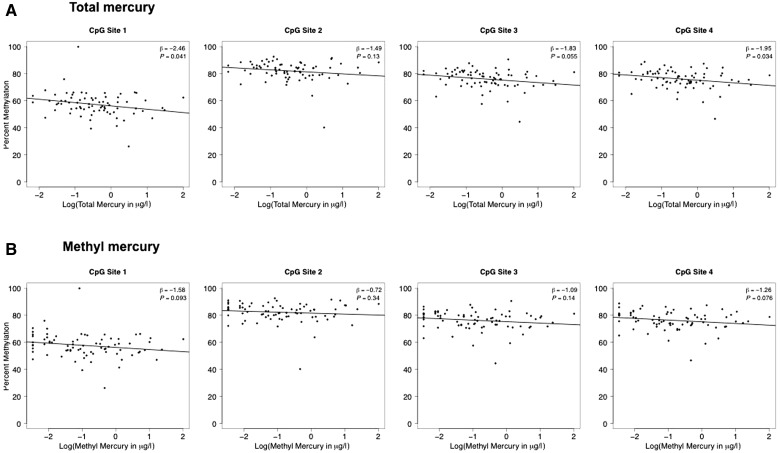
DNA methylation in independent samples from the NCS Vanguard study at four CpGs within the TCEANC2 DMR (red bracket in Figure 1) were inversely associated with both total mercury (Figure 2a) and methylmercury (Figure 2b) concentrations. The direction of association was consistent with the pattern detected during genome-wide discovery pyrosequencing DNA methylation data in NCS Initial Vanguard samples for the ANGPT2, PRPF18 and FOXD2. DMRs were not associated with mercury levels (Supplementary Figure 2, available as Supplementary data at IJE online).
Figure 2.
DNA methylation via bisulfite pyrosequencing in the TCEANC2 gene is associated with total mercury (a) and methylmercury (b) concentration in the NCS replication sample.
Gene expression
TCEANC2 gene expression was not correlated with paired pyrosequencing DNA methylation data from the four TCEANC2 CpGs (Spearman correlation P-valueCpG1 = 0.5; P-valueCpG2 = 0.5; P-valueCpG3 = 0.4; P-valueCpG4 = 0.3; Supplementary Figure 3, available as Supplementary data at IJE online). Mean CHARM DNA methylation across the DMR was also not associated with gene expression (Spearman ρ = –0.08, P–value = 0.65) and expression was not associated with total mercury level (Spearman ρ = –0.15, P-value = 0.38).
Consideration of confounding
Among the potential demographic and exposure confounders examined, race/ethnicity, blood lead and EPA + DHA fatty acid concentrations showed pairwise association with total and methylmercury (P-valuerace-total Hg = 1.8 × 10−3; P-valuerace-methyl Hg = 3.7 × 10−3; P-valuelead-total Hg = 0.079; P-valuelead-methyl Hg = 0.059; P-valueEPA+HDA-total Hg = 3.6 × 10−3; -valueEPA+DHA-methyl Hg = 5.3 × 10−3). Of these, race (non-Hispanic White vs other)and EPA + DHA concentration were also associated with mean DNA methylation at the TCEANC2 DMR (P-valuerace = 7.0 × 10−5; P-valueEPA+DHA = 0.011). After adjusting for race or EPA + DHA, the associations between average methylation in this region and total or methylmercurylevels were attenuated (Table 3).
Table 3.
Change in DMR effect sizes after adjustment for potential confounders
| Model | Total mercury |
Methylmercury |
||
|---|---|---|---|---|
| Change in percent methylation with 10% increase in exposure (95% CI) | P-value | Change in percent methylation with 10% increase in exposure (95% CI) | P-value | |
| Unadjusted | –0.62 (–1.02, –0.23) | 3.1 × 10−2 | –0.61 (–0.93, –0.30) | 2.2 × 10−4 |
| Surrogate variables (SV) adjusteda | –0.79 (–1.31, –0.27) | 3.7 × 10−3 | –0.59 (–0.92, –0.26) | 6.9 × 10−4 |
| SV + sex of baby | –0.79 (–1.31, –0.26) | 3.8 × 10−3 | –0.59 (–0.92, –0.26) | 6.8 × 10−4 |
| SV + maternal age | –0.80 (–1.32, –0.27) | 3.5 × 10−3 | –0.61 (–0.95, –0.28) | 4.7 × 10−4 |
| SV + race (non-Hispanic White vs other) | –0.57 (–1.11, –0.037) | 3.8 × 10−2 | –0.49 (–0.81, –0.16) | 4.6 × 10−3 |
| SV + birthweight | –0.84 (–1.37, –0.32) | 2.2 × 10−3 | –0.61 (–0.94, –0.28) | 4.3 × 10−4 |
| SV + gestational age | –0.83 (–1.35, –0.30) | 2.5 × 10−3 | –0.61 (–0.94, –0.28) | 4.5 × 10−4 |
| SV + lead | –0.78 (–1.30, –0.25) | 4.8 × 10−3 | –0.59 (–0.93, –0.26) | 7.6 × 10−4 |
| SV + selenium | –0.85 (–1.38, –0.33) | 1.9 × 10−3 | –0.64 (–0.97, –0.31) | 2.4 × 10−4 |
| SV + copper | –0.82 (–1.35, –0.29) | 2.8 × 10−3 | –0.63 (–0.96, –0.29) | 3.4 × 10−4 |
| SV + EPA+DHA | –0.74 (–1.28, –0.21) | 7.5 × 10−3 | –0.56 (–0.90, –0.22) | 1.5 × 10−3 |
DMR methylation calculated as mean percent methylation at TCEANC2. Effect sizes reflect 10% increase in cord blood total or methylmercury concentration. Models were estimated in the THREE sample.
aModel reflecting genome-scale discovery analyses with bump-hunter.
DNA methylation at the TCEANC2 DMR does appear to be associated with blood cell type. In the publicly available Reinius et al.23 data containing DNA methylation for six separate blood cell types (B cells, CD4+ T cells, CD8+ T cells, granulocytes, monocytes, natural killer cells and whole blood) from adult blood, methylation among four probe sets at or near the TCEANC2 DMR (chr1: positions 54562102–54562548) varied by cell type (ANOVA: P-valueCpG1 = 7.7 x 10−5; P-valueCpG2 = 0.046; P-valueCpG3 = 0.61; P-valueCpG4 = 3.1 × 10−7; Supplementary Figure 4, available as Supplementary data at IJE online). Associations between mercury concentrations and blood cell type are less clear. In the NCS Vanguard sample, maternal mercury was not correlated with estimated cell type proportions in maternal blood(Supplementary Table 6, available as Supplementary data at IJE online). Maternal mercury concentrations, however, were correlated with estimated cell type proportions of granulocytes and monocytes in child cord blood (based on cell type reference data obtained from adult blood). The effect size for regression of mean methylation at the TCEANC2 gene measured via pyrosequencing in NCS on total mercury concentration was attenuated when adjusting for estimated percent granulocytes or all six cell types (β = –1.9; P-value = 0.03 vs β = –1.2; P = 0.1 for granulocyte adjustment; β = –0.84; P = 0.3 for all cell type adjustment). Again, these cell type proportion estimates were based on adult reference panel data.
Discussion
We identified a genomic region associated with the TCEANC2 gene where DNA methylation in cord blood is inversely associated with total and methylmercury concentrations. This represents the first genome-scale discovery analysis of DNA methylation and mercury concentration in early life, to the best of our knowledge. The TCEANC2 gene region was identified in the predominantly non-Hispanic Black population from Baltimore, MD, using a genome-wide DNA methylation tiling array and mercury concentrations measured in cord blood. Regional mean methylation in the lowest total mercury quartile was 10% higher than in the highest total mercury quartile group. The association between DNA methylation and mercury at this region was replicated in a predominantly non-Hispanic White US sample using gene-specific bisulfite pyrosequencing and mercury concentrations measured in samples from first-trimester women. Replication across independent populations, using a second method of DNA methylation assessment and a different, but related, exposure window, reduces the likelihood of a false-positive result and increases the generalizability of these results to other populations.
The half-life of methylmercury in blood is estimated to be 70 days26 and 70% or more of blood mercury is in the form of methylmercury.27 Therefore blood mercury concentrations measured during trimester one and cord blood represent non-overlapping windows of susceptibility during the in utero period. Regular exposure to mercury is often assumed to result in a ‘pseudo steady state’ mercury body burden; however, in pregnant women dietary exposures are likely to be intermittent and blood mercury concentrations may vary with time.28 Cord blood mercury concentrations exceed maternal blood mercury concentrations at birth29–31 due to the rapid placental transfer of lipophilic methylmercury32 and elevated haematocrit in cord blood,33 underscoring the vulnerability of a developing fetus to even low maternal levels of exposure. In a Japanese population, the mean ratio (SD) of total mercury observed in cord blood to maternal blood was 1.6 (0.3).34 Thus, assuming mercury consumption was at steady state, it would be expected that cord blood concentrations in THREE would be somewhat higher than maternal concentrations in NCS. Mercury processing during pregnancy may vary by racial or ethnic groups, which could be an issue in our study where discovery and replication were performed on different race/ethnicity populations at different points during pregnancy.
Mercury has been associated with altered global and candidate gene-specific DNA methylation in previous laboratory and population studies. Among a population of dental professionals, SEPP1 methylation via bisulfite pyrosequencing was inversely associated with hair mercury concentrations (representing methyl and inorganic mercury exposure) among males (P-value < 0.05).35 In our samples, the CHARM platform did not overlap with SEPP1; 15 CHARM probes were within 50 kb of this gene, but none showed association with methylmercury at a P < 0.05 level. Women undergoing in vitro fertilization (IVF) treatments had a positive association between blood mercury and GSTM1/5 promoter methylation, measured on the Illumina Golden Gate array (P-value = 0.04).36 In our study, there were 19 CHARM probes within 5 kb of the GSTM1 gene and 29 probes within 5 kb of the GSTM5 gene. None were associated with methylmercury at P < 0.05. Primary rat neural stem cells in culture exposed to low levels of methylmercury had reduced global methylation.37 In mouse embryonic stem cells treated with inorganic mercury, DNA hypermethylation at the Rnd2 tissue DMR was observed using the COBRA assay.38 Finally, in our CHARM study, there were 35 probes within 5 kb of the RND2 gene and one was associated with methylmercury exposure at P = 0.0004. The current study expands the genomic coverage of previous work.
The current associations between TCEANC2 methylation and total or methylmercury were robust to several potential confounders including baby’s sex, maternal age, race, gestational age, birthweight, blood lead, selenium and copper concentrations. These results may be confounded by blood cell type composition. Using an adult blood cell reference panel and full-genome Illumina 450 K array data available in the NCS sample, we observed that methylation at the TCEANC2 region varies by estimated blood cell types and that estimated cord blood cell proportions were associated with mercury concentrations. Further, in the NCS sample the association between methylation and mercury in this region was diminished after adjusting for estimated cell type proportions. However, these analyses must be interpreted with caution because it is unknown whether adult reference panels for cell type-specific DNA methylation profiles are appropriate for cord blood. Further work is needed in isolated cell types, using adjustment based on directly measured cord blood cell type proportions or proportions estimated with reference panels from cord blood cell types.
Methylmercury may impair immune function,39 and the developing immune system may be altered by in utero mercury exposure.40 Thus, one possibility is that the observed association between TCEANC2 DNA methylation and mercury concentration may be related in part to mercury-induced changes in blood cell composition. Adjustment for cell type was not possible in our discovery CHARM analyses; however, we applied an SVA that likely accommodates confounding due to cell type. A large proportion of variance in DNA methylation across the genome can be attributed to cell type, and thus a typical surrogate variable obtained from this approach captures cell-type influences.41 The results of our study are consistent with either a biological difference in TCEANC2 methylation within blood cell types, or a biological shift in cell type proportions, which may have implications for immune function.
Whereas the TCEANC2 methylation association with mercury was replicated in the independent NCS sample, we did not observe a relationship between TCEANC2 DNA methylation and gene expression in the small number of available cord blood RNA samples (n = 34). There may be gene expression changes with altered methylation at a different developmental time point or in a different tissue or specific cell type. The TCEANC2 protein product is a little studied transcription elongation factor that is also known as C1orf83. The TCEANC2 DMR spans over 400 base pairs within exon 5 and the 3’UTR. The DMR is 42.2 kb upstream of mir4781 and 42.9 kb upstream of the 5’UTR of TMEM59, a gene functionally implicated in Alzheimer’s disease42 with altered methylation in post-mortem Alzheimer’s disease brain tissue.43The functional consequences of altered methylation at the TCEANC2 region have not yet been determined; however, infants exposed to methylmercury primarily manifest central nervous system effects and do not have altered haematological or immunological function. Further studies are needed to investigate any functional consequences of altered methylation at the TCEANC2 gene in the brain and/or if there are brain regions where altered methylation results from methylmercury. An epigenetic change at a specific point in development could knock processes off course (eg neurological development) in an organ- or time-specific manner; conversely, this mark may a biomarker of exposure and not aetiological in the expression of the mercury-related central nervous system phenotype.
Blood mercury measurements in this exploratory study were of high quality and reliable, as conducted by an experienced laboratory that makes thousands of similar measurements every year. The THREE and NCS study laboratory measurements were done by the same CDC laboratory that carried out the US National Health and Nutrition Examination Survey (NHANES) measurements for methylmercury and total mercury. There is a considerable overlap between mercury concentrations from our two studies and baseline national levels.44 Coastal populations in the USA have elevated blood mercury levels relative to the rest of the USA, associated with regional differences in fish consumption.45 Similarly, we observed higher methylmercury concentrations in the THREE study based out of the coastal city, Baltimore, than the nationwide NCS study. Our results are relevant to methylmercury concentrations measured in the US population.
The current study was limited by cross-sectional biosampling, and repeating the measurements on biosamples collected from the same individuals in the future could demonstrate the persistence or non-persistence of in utero exposure-related DNA methylation marks. In addition, we did not have direct measures or estimates of cell type proportions in the THREE discovery methylation sample because the CHARM methylation platform is not compatible with existing cell-type estimation algorithms. We estimated cell type proportions in the NCS replication sample but, particularly in cord blood, these were limited by: (i) cell type references derived from older, White males;23 and (ii) admixture of red blood cells in the sample (which are more likely to be nucleated in newborns but not in adults). Future work should generate and use cord-derived DNA methylation blood cell type references.
An additional limitation of our study is that brain rather than blood is the target organ for methylmercury toxicity, though DNA methylation was measured in cord blood. However, in a human population context, cord blood is advantageous as a feasible source of epigenomic information at birth. Primary brain tissue at birth is only available using aborted fetuses or stillbirths (which may have genetic abnormalities), or through study of non-human species (which may respond differently to methylmercury). Certainly confirmatory studies in both human epidemiological samples and via different approaches that would allow evaluation of the central nervous system epigenome are warranted. Long-term follow-up in the NCS and future analyses could provide additional information on DNA methylation persistence at the TCEANC2 gene and the utility of these marks as a prenatal exposure biomarker in other studies.
Supplementary Data
Supplementary data are available at IJE online.
Funding
This work was supported by the National Institute for Environmental Health Sciences at the National Institutes for Health (ES017646). The National Children’s Study (NCS) Initial Vanguard Centers from which the samples were collected, the NCS biospecimen team and the NCS repository were supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development at the National Institutes of Health contracts numbers: HHSN27520080033C, HHSN275200503414C, HHSN275200503411C, HHSN275200603416C, HHSN275200503415C, HHSN275200503413C, HHSN275200503410C, HHSN275200503396C, HHSN275201000121U and HHSN275200900010C. Funding for the blood mercury analyses was provided by the National Center for Environmental Health, Centers for Disease Control and Prevention.
Supplementary Material
Acknowledgements
The manuscript was developed by a writing team identified by the National Children’s Study Publications Committee for the purpose of timely sharing of centrally collected NCS data. We acknowledge the contributions of the following Vanguard Centers and principal investigators: Children’s Hospital of Philadelphia, Jennifer Culhane; Mt. Sinai Medical School, Phil Landrigan; South Dakota State University, Bonny Specker; University of California at Irvine, James Swanson and Dean Baker; University of North Carolina at Chapel Hill, Barbara Entwisle and Nancy Dole; University of Utah School of Medicine, Ed Clark; University of Wisconsin, Maureen Durkin.
Conflict of interest: None declared.
References
- 1.Counter SA, Buchanan LH. Mercury exposure in children: a review. Toxicol Appl Pharmacol 2004;198:209–30. [DOI] [PubMed] [Google Scholar]
- 2.Rice KM, Walker EM, Jr, Wu M, Gillette C, Blough ER. Environmental mercury and its toxic effects. J Prev Med Public Health Yebang Uihakhoe chi 2014;47:74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Children’s Exposure to Mercury Compounds. Geneva:WHO, 2010. [Google Scholar]
- 4.Golding J, Steer CD, Hibbeln JR, Emmett PM, Lowery T, Jones R. Dietary predictors of maternal prenatal blood mercury levels in the ALSPAC Birth Cohort Study. Environ Health Perspect 2013;121:1214–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Social Scientific Study Group on Minamata Disease. In the Hope of Avoiding Repetition of Tragedy of Minamata Disease. 1999. http://www.nimd.go.jp/syakai/webversion/SSSGMDreport.html (8 July 2010, date last accessed). [Google Scholar]
- 6.Bakir F, Damluji SF, Amin-Zaki L, et al. Methylmercury poisoning in Iraq. Science 1973;181:230–41. [DOI] [PubMed] [Google Scholar]
- 7.Engleson G, Herner T. Alkyl mercury poisoning. Acta Paediatr 1952;41:289–94. [DOI] [PubMed] [Google Scholar]
- 8.Grandjean P, Weihe P, White RF, et al. Cognitive deficit in 7-year-old children with prenatal exposure to methylmercury. Neurotoxicol Teratol 1997;19:417–28. [DOI] [PubMed] [Google Scholar]
- 9.Castoldi AF, Coccini T, Manzo L. Neurotoxic and molecular effects of methylmercury in humans. Rev Environ Health 2003;18:19–31. [DOI] [PubMed] [Google Scholar]
- 10.Marsh DO, Myers GJ, Clarkson TW, Amin-Zaki L, Tikriti S, Majeed MA. Fetal methylmercury poisoning: clinical and toxicological data on 29 cases. Ann Neurol 1980;7:348–53. [DOI] [PubMed] [Google Scholar]
- 11.Bakulski KM, Fallin MD. Epigenetic epidemiology: Promises for public health research. Environ Mol Mutagen 2014;55:171–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apelberg BJ, Goldman LR, Calafat AM, et al. Determinants of fetal exposure to polyfluoroalkyl compounds in Baltimore, Maryland. Environ Sci Technol 2007;41:3891–97. [DOI] [PubMed] [Google Scholar]
- 13.Lee H, Jaffe AE, Feinberg JI, et al. DNA methylation shows genome-wide association of NFIX, RAPGEF2 and MSRB3 with gestational age at birth. Int J Epidemiol 2012;41:188–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consortium C-DGotPG. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013;45:984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mortensen ME, Hirschfeld S. The National Children's Study: an opportunity for medical toxicology. J Med Toxicol 2012;8:160–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caldwell KL, Pan Y, Mortensen ME, Makhmudov A, Merrill L, Moye J. Iodine status in pregnant women in the National Children's Study and in U.S. women (15-44 years), National Health and Nutrition Examination Survey 2005-2010. Thyroid 2013;23:927–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wells EM, Jarrett JM, Lin YH, et al. Body burdens of mercury, lead, selenium and copper among Baltimore newborns. Environ Res 2011;111:411–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verdon CP, Caldwell KL, Jones RL. Blood Mercury Speciation Performed by HPLC-ICP-DRC-MS. Atlanta, GA: Centers for Disease Control and Prevention Division of Laboratory Sciences Method: 2008. [Google Scholar]
- 19.Sommer YL, Verdon CP, Fresquez MR, et al. Measurement of mercury species in human blood using triple spike isotope dilution with SPME-GC-ICP-DRC-MS. Anal Bioanal Chem 2014;406:5039–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladd-Acosta C, Aryee MJ, Ordway JM, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM). Curr Protoc Hum Genet 2010;20:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002;18:1427–31. [DOI] [PubMed] [Google Scholar]
- 22.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reinius LE, Acevedo N, Joerink M, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One 2012;7:e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaffe AE, Murakami P, Lee H, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol 2012;41:200–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 2007;3:1724–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.al-Shahristani H, Shihab KM. Variation of biological half-life of methylmercury in man. Arch Environ Health 1974;28:342–44. [DOI] [PubMed] [Google Scholar]
- 27.Mortensen ME, Caudill SP, Caldwell KL, Ward CD, Jones RL. Total and methyl mercury in whole blood measured for the first time in the U.S. population: NHANES 2011-2012. Environ Res 2014;134C:257–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartell SM, Ponce RA, Sanga RN, Faustman EM. Human variability in mercury toxicokinetics and steady state biomarker ratios. Environ Res 2000;84:127–32. [DOI] [PubMed] [Google Scholar]
- 29.Jedrychowski W, Perera F, Rauh V, et al. Fish intake during pregnancy and mercury level in cord and maternal blood at delivery: an environmental study in Poland. Int J Occup Med Environ Health 2007;20:31–37. [DOI] [PubMed] [Google Scholar]
- 30.Ong CN, Chia SE, Foo SC, Ong HY, Tsakok M, Liouw P. Concentrations of heavy metals in maternal and umbilical cord blood. Biometals 1993;6:61–66. [DOI] [PubMed] [Google Scholar]
- 31.Stern AH, Smith AE. An assessment of the cord blood:maternal blood methylmercury ratio: implications for risk assessment. Environ Health Perspect 2003;111:1465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kajiwara Y, Yasutake A, Adachi T, Hirayama K. Methylmercury transport across the placenta via neutral amino acid carrier. Arch Toxicol 1996;70:310–14. [DOI] [PubMed] [Google Scholar]
- 33.Doi R, Kasamo M, Ishikawa M, Shimizu T. Factors influencing placental transfer of methylmercury in man. Bull Environ Contam Toxicol 1984;33):69–77. [DOI] [PubMed] [Google Scholar]
- 34.Sakamoto M, Murata K, Kubota M, Nakai K, Satoh H. Mercury and heavy metal profiles of maternal and umbilical cord RBCs in Japanese population. Ecotoxicol Environ Saf 2010;73:1–6. [DOI] [PubMed] [Google Scholar]
- 35.Goodrich JM, Basu N, Franzblau A, Dolinoy DC. Mercury biomarkers and DNA methylation among Michigan dental professionals. Environ Mol Mutagen 2013;54:195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanna CW, Bloom MS, Robinson WP, et al. DNA methylation changes in whole blood is associated with exposure to the environmental contaminants, mercury, lead, cadmium and bisphenol A, in women undergoing ovarian stimulation for IVF. Hum Reprod 2012;27:1401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bose R, Onishchenko N, Edoff K, Janson Lang AM, Ceccatelli S. Inherited effects of low-dose exposure to methylmercury in neural stem cells. Toxicol Sci 2012;130:383–90. [DOI] [PubMed] [Google Scholar]
- 38.Arai Y, Ohgane J, Yagi S, et al. Epigenetic assessment of environmental chemicals detected in maternal peripheral and cord blood samples. J Reprod Dev 2011;57:507–17. [DOI] [PubMed] [Google Scholar]
- 39.Moszczynski P. Mercury compounds and the immune system: a review. Int J Occup Med Environ Health 1997;10:247–58. [PubMed] [Google Scholar]
- 40.Belles-Isles M, Ayotte P, Dewailly E, Weber JP, Roy R. Cord blood lymphocyte functions in newborns from a remote maritime population exposed to organochlorines and methylmercury. J Toxicol Environ Health A 2002;65:165–82. [DOI] [PubMed] [Google Scholar]
- 41.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol 2014;15:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ullrich S, Munch A, Neumann S, Kremmer E, Tatzelt J, Lichtenthaler SF. The novel membrane protein TMEM59 modulates complex glycosylation, cell surface expression, and secretion of the amyloid precursor protein. J Biol Chem 2010;285:20664–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakulski KM, Dolinoy DC, Sartor MA, et al. Genome-wide DNA methylation differences between late-onset Alzheimer's disease and cognitively normal controls in human frontal cortex. J Alzheimers Dis 2012;29:571–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Environmental Protection Agency. Trends in Blood Mercury Concentrations and Fish Consumption Among U.S. Women of Childbearing Age NHANES, 1999-2010. Hyattsville, MD: National Center for Disease Statistics, 2013. [Google Scholar]
- 45.Mahaffey KR, Clickner RP, Jeffries RA. Adult women's blood mercury concentrations vary regionally in the United States: association with patterns of fish consumption (NHANES 1999-2004). Environ Health Perspect 2009;117:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.