Abstract
PPM1D phosphatase, also called wild-type p53-induced phosphatase 1 (Wip1), promotes tumor development by inactivating the p53 tumor suppressor pathway. RBM38 RNA-binding protein, also called RNPC1 and a target of p53, inhibits p53 mRNA translation, which can be reversed by GSK3 protein kinase via phosphorylation of RBM38 at serine 195. Here we showed that ectopic expression of RBM38 increases, whereas knockdown of RBM38 inhibits, PPM1D mRNA translation. Consistent with this, we found that RBM38 directly binds to PPM1D 3' untranslated region (3’UTR) and promotes expression of a heterologous reporter gene that carries PPM1D 3’UTR in a dose-dependent manner. Interestingly, we showed that PPM1D directly interacts with and dephosphorylates RBM38 at serine 195. Furthermore, we showed that PPM1D modulates p53 mRNA translation and p53-dependent growth suppression through dephosphorylation of RBM38. These findings provide evidence that the crosstalk between PPM1D and RBM38, both of which are targets and modulators of p53, plays a critical role in p53 expression and activity.
Keywords: PPM1D, Wip1, RBM38, RNPC1, p53, dephosphorylation, mRNA translation
Introduction
RNA-binding protein RBM38, also called RNPC1, is a target of p53 and E2F1 1, 2. RBM38 is capable of regulating mRNA stability of p21, p73, p63, Mdm2, HuR and GDF15 1–7. RBM38 is found to relieve microRNA repression of several genes, including p21, DDIT4, LATS2, and RBM38 itself 8. RBM38 is also found to regulate alternative splicing of the EPB41 and FGFR2 genes 9, 10. Moreover, p53 expression is regulated by RBM38 via mRNA translation and thus, the mutual regulation between p53 and RBM38 represents a feedback loop in the p53 pathway. Interestingly, the p53-RBM38 loop is regulated by the AKT-GSK3 signaling cascade and phosphorylation of RBM38 at serine 195 by GSK3 converts RBM38 from a repressor to an activator of p53 mRNA translation 5, 11.
PPM1D phosphatase, also called wild-type p53-induced phosphatase 1 (Wip1), is a target of p53 and a member of the PP2C serine/threonine phosphatase family 12. PPM1D has an oncogenic activity through multiple mechanisms, including altered phosphorylation and stability of p53 and Mdm2 13, 14. Consistently, PPM1D is found to be amplified and/or overexpressed in multiple types of human cancer, including breast cancer, neuroblastoma and ovarian cancer 15–18. In addition, the C-terminal truncated version of PPM1D, which is highly expressed in breast and ovarian cancer, impairs p53-dependent genome surveillance system 19, 20. Thus, the p53-PPM1D loop is another major player in the p53 pathway. Here, we identified a crosstalk between two p53 regulators PPM1D and RBM38. We found that RBM38 positively regulates PPM1D expression through mRNA translation. Conversely, PPM1D decreases RBM38 phosphorylation and subsequently its activity in modulating p53 mRNA translation. These findings provide evidence that the reciprocal interplay between PPM1D and RBM38, both of which are targets and modulators of p53, plays a critical role in p53 expression and activity.
Results
PPM1D Expression Is Regulated by RBM38 via mRNA Translation
RBM38 is known to regulate mRNA stability or translation of many p53 related genes via binding to an AU/U-rich element in their 3' UTRs 1, 5, 6, 21. As a major regulator of p53, PPM1D may also be a target of RBM38. To test this, the level of PPM1D protein was determined in HCT116 and MCF7 cells that can inducibly express RBM38 under the control of the tetracycline-regulated promoter. We found that upon RBM38 expression, PPM1D level was increased under normal and DNA-damage-induced conditions (Figure 1A–B). We would like to note that the truncated form of PPM1D, which is encoded by a mutant allele (1349delT:L450X) of the PPM1D gene in HCT116 cells 20, was also highly increased upon ectopic expression of RBM38 (Figure 1A). Since PPM1D is a target of p53, the effect of RBM38 on PPM1D expression was measured in p53−/− HCT116 cells. We showed that the levels of the full-length PPM1D and its truncated version were increased upon ectopic expression of RBM38 in p53−/−HCT116 cells (Figure 1C). Moreover, we found that in p53−/−HCT116 cells, the levels of wild-type PPM1D and its truncated mutant were increased upon expression of non-phosphorylatable S195A and phosphor-mimetic mutant S195D (Fig.1D).
Figure 1. Ectopic expression of RBM38 increases, whereas knockdown of RBM38 decreases, the level of PPM1D protein.
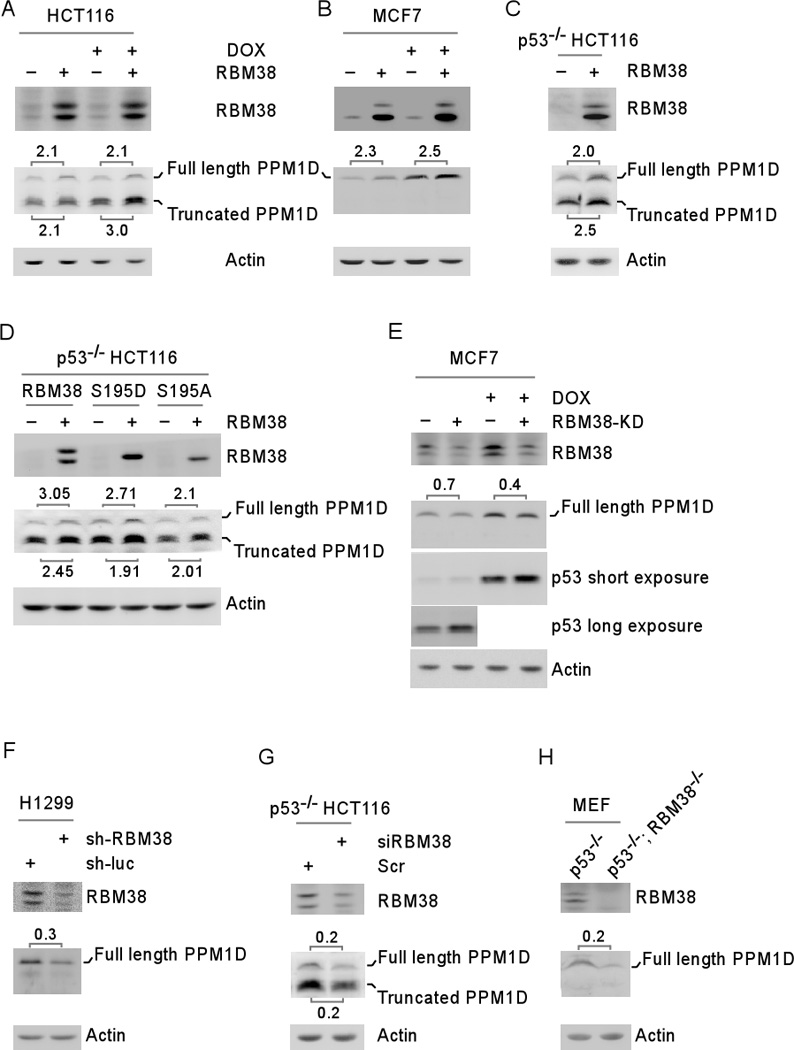
(A–B) The levels of RBM38, PPM1D and actin proteins were measured in HCT116 (A) and MCF7 (B) cells uninduced (−) or induced (+) to express RBM38 for 24 hours, followed by mock-treatment or treatment with doxorubicin (DOX) for 18 h. (C) The levels of RBM38, PPM1D and actin proteins were measured in p53−/− HCT116 cells uninduced (−) or induced (+) to express RBM38 for 24 hours. (D) p53−/− HCT116 cells were uninduced (−) or induced (+) to express HA-tagged wild-type RBM38, S195D, and S195A for 24 h. The levels of RBM38, PPM1D and actin proteins were measured by Western blot analysis. (E) The levels of RBM38, PPM1D, and actin were determined in MCF7 cells uninduced or induced to knock down RBM38 (express a shRNA against RBM38) for 3 days, followed by mock-treatment or treatment with doxorubicin (DOX) for 18 h. (F) The levels of RBM38, PPM1D, and actin were determined in H1299 cells transduced with a lentivirus expressing a control luciferase shRNA (sh-luc) or RBM38 shRNA (sh-RBM38) for 3 days. (G) The levels of RBM38, PPM1D, and actin were determined in p53−/−HCT116 cells transiently transfected with a control or RBM38 siRNA for 3 days. (H) The levels of RBM38, PPM1D, and actin were determined in primary p53−/− and p53−/−; RBM38 −/− MEFs at passage 12.
To determine whether endogenous RBM38 has an effect on PPM1D expression, the level of PPM1D was measured in MCF7 cells uninduced or induced to knock down RBM38. Consistent with the above studies, the level of PPM1D protein was decreased upon RBM38 knockdown at both basal and DNA damage conditions (Figure 1E). As a control, we found that p53 expression was increased (Figure 1E, p53 panel), consistent with our previous studies 5, 21. Additionally, we found that PPM1D expression was decreased in H1299 cells in which RBM38 expression was knocked down with an shRNA targeting RBM38 (Figure 1F) and in p53−/−HCT116 cells in which RBM38 expression was knocked down by an siRNA against RBM38 (Figure 1G). Furthermore, we showed that the level of PPM1D was markedly decreased by RBM38 deficiency in primary RBM38−/−; p53−/− MEFs as compared to that in p53−/− MEFs (Figure 1H).
RBM38 is known to regulate gene expression via post-transcriptional mechanisms. To explore the underlying mechanism by which RBM38 regulates PPM1D, we examined PPM1D mRNA stability and translation. We showed that the levels of PPM1D transcript were not significantly altered in p53−/−HCT116 cells in which RBM38 was overexpressed or knocked down (Figure 2A–B) and in p53-null MEFs in which RBM38 was knocked out (Figure 2C). To examine whether PPM1D mRNA translation is regulated by RBM38, the level of newly synthesized PPM1D protein was measured in 35S-labeled MCF7 cells with or without RBM38 expression. We showed that PPM1D mRNA translation was highly induced (2.38-fold) by ectopic expression of RBM38 in MCF7 cells (Figure 2D). Conversely, we showed that the level of newly synthesized PPM1D protein was markedly decreased by knockdown of RBM38 in MCF7 and H1299 cells (Figure 2E–F).
Figure 2. PPM1D mRNA translation but not mRNA stability is regulated by RBM38.
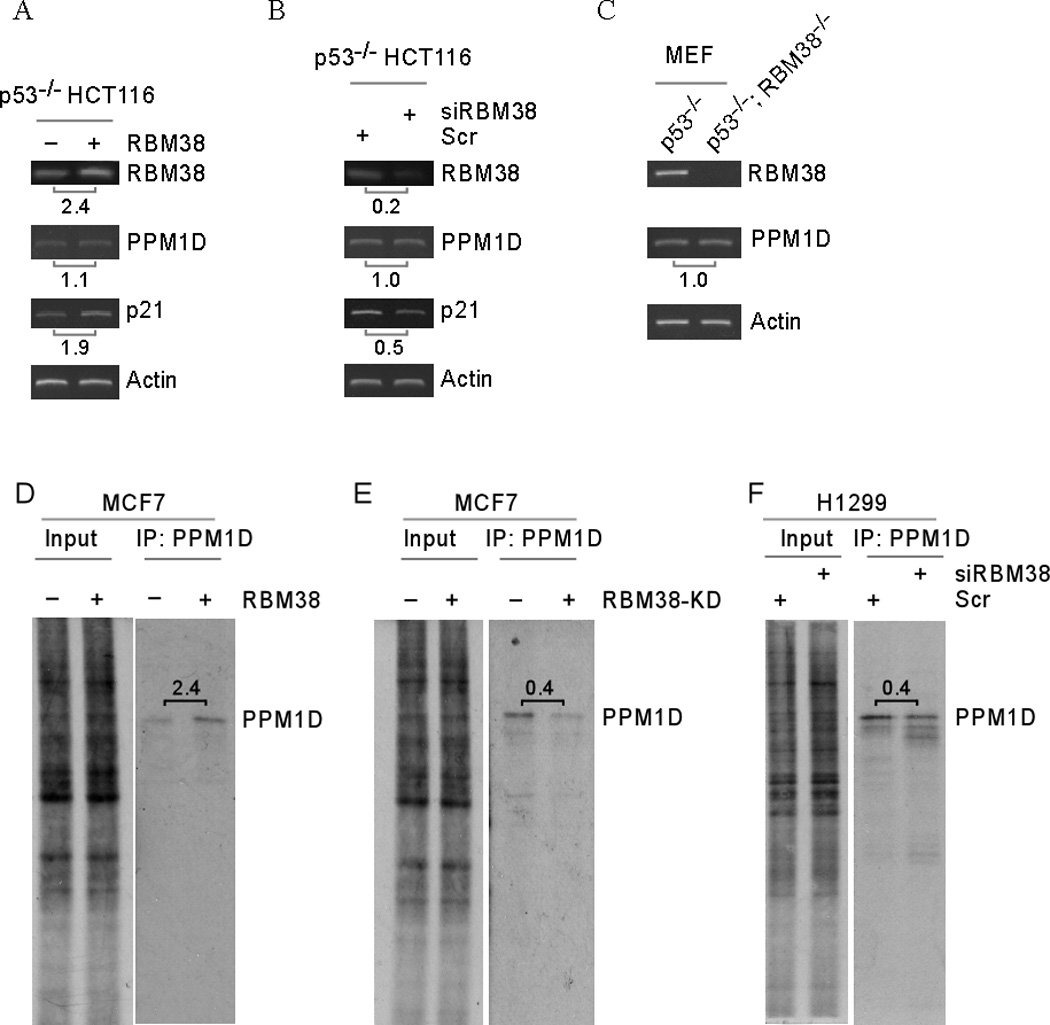
(A) p53−/−HCT116 cells were uninduced (−) or induced (+) to express RBM38 for 24 hours. Total RNAs were purified and used to measure the levels of PPM1D, RBM38, p21 and actin transcripts by semi-quantitative RT-PCR. (B) Total RNAs were purified from p53−/−HCT116 cells transfected with scrambled siRNA or siRNA against RBM38 for 3 days. The levels of PPM1D, RBM38, p21 and actin transcripts were measured by semi-quantitative RT-PCR. (C) Total RNAs were purified from primary p53−/− and p53−/−; RBM38−/− MEFs and used to measure the levels of PPM1D, RBM38 and actin transcripts by semi-quantitative RT-PCR. (D) MCF7 cells were uninduced or induced to express RBM38 for 24 h and then 35S-labeled for 10 min, followed by immunoprecipitation with anti-PPM1D. The immunocomplexes were resolved by SDS-PAGE, and 35S-labeled PPM1D was visualized by autoradiography. (E) The experiment was performed as in (D) with MCF7 cells uninduced or induced to knock down RBM38 for 3 d. (F) The experiment was performed as in (D) with H1299 cells transiently transfected with a control or RBM38 siRNA for 3 days.
Multiple AU- and U-rich elements in PPM1D 3’UTR are recognized by and responsive to RBM38
As a RNA-binding protein, RBM38 may regulate PPM1D expression by binding to PPM1D transcript. To test this, RNA immunoprecipitation (RNA-IP) assay followed by RT-PCR was performed using extracts from MCF7 or p53-null HCT116 cells that were induced to express RBM38. We found that PPM1D mRNA was detectable in RBM38 but not control IgG immunoprecipitates (Figure 3A). As a control, actin transcripts were not found to interact with RBM38 or IgG (Figure 3A). As a positive control, p21 transcripts were also detected in anti-RNPC1 immunoprecipitate (Fig. 3A), consistent with previous reports 1.
Figure 3. Multiple AU- and U-rich elements in PPM1D 3' UTR are bound by and responsive to RBM38.
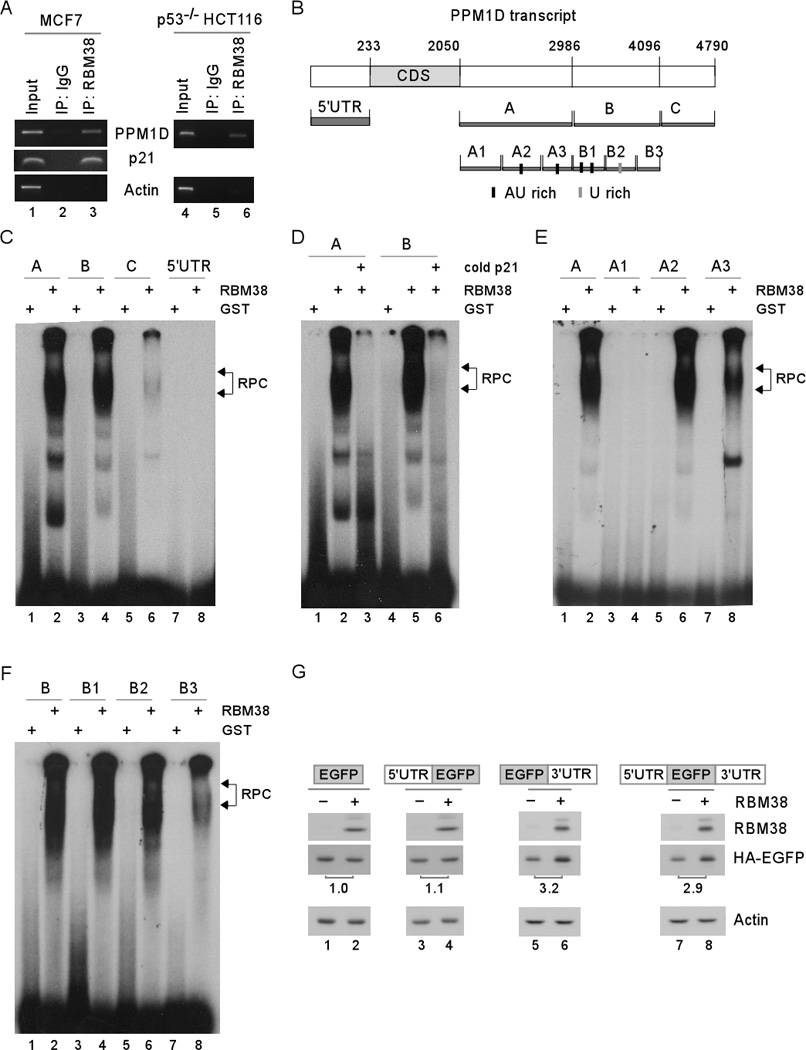
(A) RBM38 interacts with PPM1D transcript. Cell extracts purified from MCF7 and p53-null HCT116 cells, which were induced to express RBM38, were immunoprecipitated with a control IgG or RBM38 antibody. The levels of PPM1D and actin transcripts in RBM38 or IgG immunocomplexes were determined by RT-PCR. (B) Schematic presentation of PPM1D transcript and the location of probes used for REMSA. (C) Probes A and B, but not probes C and 5’ UTR, are bound by RBM38. REMSA was performed by mixing 32P-labeled probe A, B, C, or 5’ UTR with recombinant GST or GST-RBM38 protein. The bracket indicates RNA-protein complexes (RPC). (D) Competition assay was performed by adding an excess amount (50-fold) of unlabeled p21 probe derived from p21 3’UTR to the reaction mix prior to incubation with the 32P-labeled probe A or B. (E) REMSA was performed by mixing recombinant GST or GST-RBM38 protein with 32P-labeled probe A, or A1–3. (F) REMSA was performed by mixing recombinant GST or GST-RBM38 protein with 32P-labeled probe B, or B1–3. (G) PPM1D 3′ UTR is responsive to RBM38. An EGFP expression vector, which contains the coding region (ORF) alone or in combination with PPM1D 5′ and/or 3′UTRs, was co-transfected with RBM38-expressing vector into H1299 cells. The levels of EGFP, RBM38, and actin were analyzed by Western blot analysis. The fold change of EGFP was shown below each lane.
To delineate RBM38-binding site(s) in PPM1D transcript, RNA electrophoretic mobility assay (REMSA) was performed by using radiolabeled probes (probes 5’UTR, A, B, and C) derived from PPM1D 5’ and 3′ UTRs (Figure 3B). We found that recombinant GST-fused RBM38 but not GST alone was able to form a complex with probes A and B (Figure 3C, lanes 2 and 4). However, probe C and probe 5′ UTR showed very weak or no binding with RBM38 (Figure 3C, lanes 6 and 8). In addition, the probe A/B-RBM38 complexes were disrupted by cold p21 probe (Figure 3D, lanes 3 and 6). The p21 probe is derived from p21 3’UTR and known to bind to RBM38 22. To further delineate the RBM38-binding region(s) within probes A-B, several sub-fragments within probe A (A1, A2 and A3) and probe B (B1, B2 and B3) were made for REMSA (Figure 3B). We found that like probe A, probe A2 showed a strong binding to RBM38 whereas probe A3 showed a weak binding to RBM38 (Figure 3E, lanes 6 and 8). However, probe A1 showed no binding to RBM38 (Figure 3E, lane 4). Similarly, we found that probes B1 and B2 showed a strong binding to RBM38 whereas probe B3 showed a weak binding to RBM38 (Figure 3F, lanes 4, 6, and 8).
To determine whether the AU- and U-rich elements are responsible for RBM38 to regulate PPM1D, we generated four EGFP reporters alone or in combination with PPM1D 5' UTR, 3' UTR, or both. We showed that the level of EGFP protein was highly increased by RBM38 as long as the EGFP expression vector carried PPM1D 3' UTR (Figure 3G, lanes 5–8). In contrast, RBM38 had no effect on the level of EGFP expression from a EGFP reporter that carried none or PPM1D 5' UTR (Figure 3G, lanes 1–4).
RBM38 phosphorylation at ser195 is modulated by PPM1D
We showed previously that RBM38, as a p53 target, represses p53 mRNA translation and S195 phosphorylation converts RBM38 from a repressor to an activator of p53 mRNA translation 5, 21. As a protein phosphatase, PPM1D is known to dephosphorylate several proteins in the p53 pathway, including ATM, Chk1, Chk2, p38, Mdm2, and p53 itself 13, 14, 23–25. To evaluate whether PPM1D has effect on RBM38 phosphorylation, the level of S195-phosphorylated RBM38 (p-RBM38) was measured in HCT116 and MCF7 cells treated with PPM1D inhibitor cct007093. We found that upon treatment with the PPM1D inhibitor, the ratio of p-RBM38 vs. underphosphorylated RBM38 was increased in HCT116 and MCF7 in a dose-dependent manner (Figure 4A–B). As a control, the level of p53 protein was also increased (Figure 4A–B), consistent with a previous report 26. As a type 2C phosphatase, PPM1D is relatively insensitive to okadaic acid, a potent inhibitor of the PP1 and PP2A phosphatases. Indeed, we found that the ratio of p-RBM38 vs. RBM38 was not significantly altered in MCF7 cells upon treatment with okadaic acid (Figure 4C). As a control, the level of p53 protein was increased by okadaic acid in MCF7 cells (Figure 4C), consistent with a previous report 27, 28.
Figure 4. Inhibition of PPM1D increases RBM38 phosphorylation at ser195.
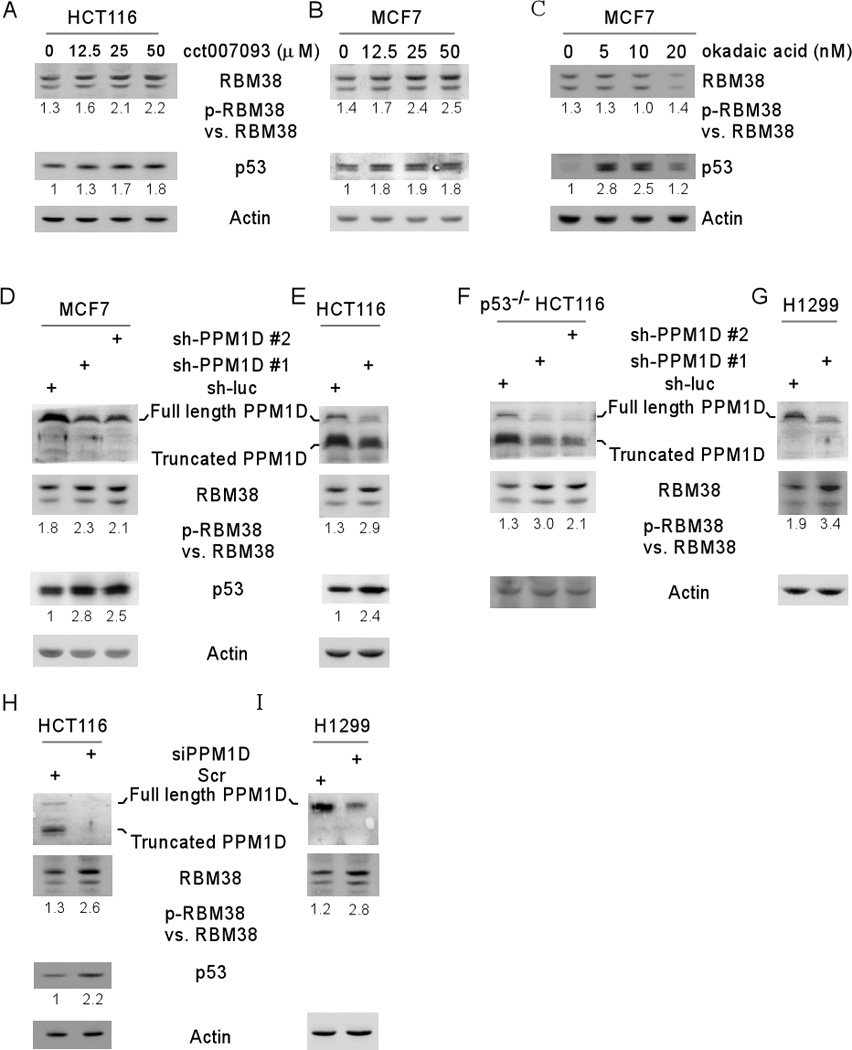
(A–B) The ratio of p-RBM38 vs. RBM38 is increased by a PPM1D inhibitor in a dose-dependent manner. The levels of RBM38, p53, and actin proteins were measured in HCT116 (A) and MCF7 (B) cells mock-treated or treated with PPM1D inhibitor CCT007093 (0–50 µM) for 12 h. (C) Okadaic acid has no effect on RBM38 phosphorylation. The levels of RBM38, p53, and actin proteins were measured in MCF7 cells mock-treated or treated with okadaic acid (0–20 nM) for 12 h. (D–I) Knockdown of PPM1D increases the ratio of p-RBM38 vs. RBM38. The levels of RBM38, p53, and actin proteins were measured in MCF7 (D), HCT116 (E), p53−/− HCT116 (F) and H1299 (G) cells transduced with lentivirus particles expressing a control shRNA, shRNA #1 shRNA #2 against PPM1D for 72 h, or in HCT116 (H) and H1299 (I) cells transduced with siRNA against PPM1D for 72 h. The ratio of p-RBM38 vs. RBM38 was calculated and showed below each lane.
To determine whether endogenous PPM1D is capable of modulating RBM38 phosphorylation, the ratio of p-RBM38 vs. RBM38 was measured in MCF7 and HCT116 cells in which PPM1D was knocked down by two separate shRNAs targeting PPM1D. Similarly, we found that knockdown of PPM1D increased the ratio of p-RBM38 vs. RBM38 in MCF7 and HCT116 cells (Figure 4D–E). As a control, the level of p53 was increased (Figure 4D–E), consistent with a previous study 29. To rule out potential interference from p53, the ratio of p-RBM38 vs. RBM38 was measured in p53−/− HCT116 and H1299 cells and found to be highly increased in these cells upon knockdown of PPM1D (Figure 4F–G). To avoid potential off-target effects, PPM1D was knocked down by another commonly used siRNA20, 30 targeting a different region in PPM1D. Similar results were observed in HCT116 and H1299 cells (Fig. 4H–I).
Conversely, we found that ectopic expression of wild-type PPM1D but not phosphatase-dead PPM1D (D314A) decreased the ratio of p-RBM38 vs. RBM38 in HCT116 and MCF7 cells in a dose-dependent manner (Figure 5A–B). Aspartic acid at codon 314 is necessary for PPM1D phosphatase activity and thus, PPM1D-D314A is phosphatase-dead 23.
Figure 5. PPM1D directly dephosphorylates RBM38.
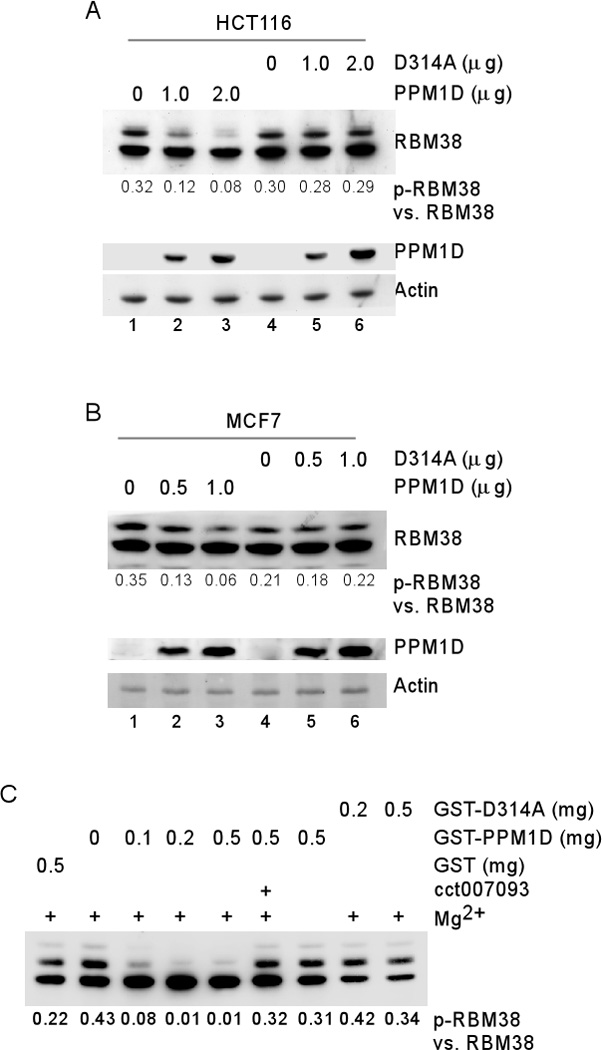
(A–B) Ectopic expression of PPM1D decreases the ratio of p-RBM38 vs. RBM38. HCT116 (A) or MCF7 (B) cells were transiently transfected with various amounts of a pcDNA3 vector expressing wild-type PPM1D or phosphatase dead mutant D314A. 24 h post transfection, cells were induced to express RBM38 for 12h. Cell lysates were harvested for Western blot analysis with antibodies against RBM38, PPM1D or actin. (C) PPM1D directly dephosphorylates RBM38 as measured by in vitro phosphatase assay. RBM38 immunocomplexes from HCT116 cells were treated with various amount of GST, GST-PPM1D, or GST-PPM1D-D314A in the presence or absence of Mg2+ or a PPM1D inhibitor as indicated. The ratio of p-RBM38 vs. RBM38 was measured by Western blot analysis with antibodies against RBM38.
To determine whether PPM1D directly dephosphorylates RBM38 at serine 195, in vitro phosphatase assay was performed using recombinant GST-PPM1D and immuno-purified RBM38. We found that S195-phosphorylated RBM38 was dephosphorylated by PPM1D but not phosphatase-dead PPM1D-D314A in a dose-dependent manner (Figure 5C). Additionally, we found that the activity of PPM1D to dephosphorylate RBM38 was abrogated by PPM1D inhibitor cct007093 or in a buffer lacking Mg2+ (Figure 5C). PPM1D phosphatase activity is Mg2+-dependent 14.
If RBM38 is truly a PPM1D substrate, RBM38 and PPM1D may physically interact with each other. To test this, immunoprecipitation assay was performed with extracts from HCT116 cells in which HA-tagged PPM1D was ectopically expressed. We showed that PPM1D was detected in anti-RBM38 immunocomplexes (Figure 6A). Conversely, p-RBM38 but not underphosphorylated RBM38 was detected in anti-PPM1D (anti-HA) immunocomplexes (Figure 6B). Furthermore, we showed that endogenous p-RBM38 but not underphosphorylated RBM38 in MCF7 cells was detected in anti-PPM1D immunocomplexes (Figure 6C). To further test this, GST pull-down assay was performed and showed that His-tagged RBM38 bound to GST-PPM1D but not GST beads (Figure 6D). Conversely, we showed that His-tagged PPM1D bound to GST-RBM38 but not GST beads (Figure 6E).
Figure 6. RBM38 interacts with PPM1D in vitro and in vivo.
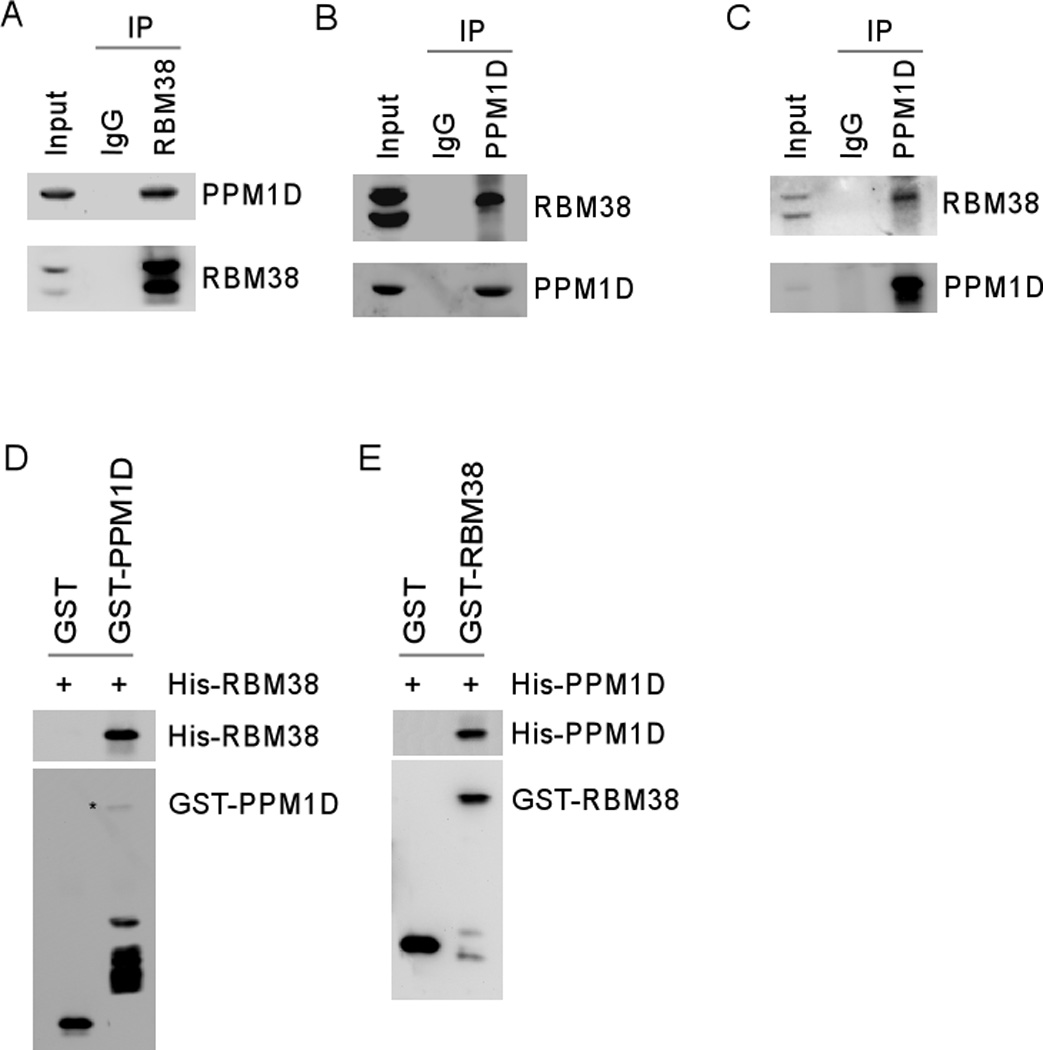
(A) HCT116 cells were transiently transfected with a pcDNA3 vector expressing HA-PPM1D. Cell lysates were treated with RNase A and then immunoprecipitated with a control IgG or antibodies against RBM38, followed by Western blot analysis with antibodies against PPM1D (anti-HA) and RBM38. (B) The experiment was performed as in (A) except that immunoprecipition was performed with α-HA antibody to capture PPM1D immnocomplexes. (C) Lysates purified from MCF7 cells were treated with RNase A and then immunoprecipitated with a control IgG or antibodies against PPM1D, followed by Western blot analysis with antibodies against PPM1D and RBM38. (D) GST pull-down assays were performed with recombinant His-tagged RBM38 and GST-tagged PPM1D. Equal amounts of GST or GST-tagged PPM1D were incubated with His-tagged RBM38 along with glutathione sepharose for 2 h. Complexes were then washed, followed by Western blot analysis using antibody against histidines (anti-omini) or GST. (E) The experiment was performed as in (D) except that His-tagged PPM1D and GST-tagged RBM38 were used.
Knockdown of PPM1D leads to increased p53 mRNA translation in a RBM38-dependent manner
Since S195 phosphorylation converts RBM38 from a repressor to an activator of p53 mRNA translation, we speculate that PPM1D may modulate RBM38 phosphorylation to regulate p53 mRNA translation. To test this, the level of newly synthesized p53 protein was measured in 35S-labeled MCF7 and HCT116 cells upon knockdown of PPM1D. We showed that knockdown of PPM1D led to increased ratios of p-RBM38 vs. RBM38 (Figure 7A–B, left panels). Most importantly, the levels of de novo p53 proteins were highly increased (2.4-fold in MCF7 cells and 2.6-fold in HCT116 cells) (Figure 7A–B, right panels). To determine whether increased phosphorylation of RBM38 is responsible for increased p53 mRNA translation, wild-type and RBM38-null MEFs were used. We found that knockdown of PPM1D led to an increased ratio of p-RBM38 vs. RBM38 (increased from 0.6 to 1.5) (Figure 7C, lanes 1–2) along with increased expression of de novo p53 protein (1.6-fold) (Figure 7C, lanes 9–10). However, in RBM38-null MEFs, the level of de novo p53 protein was not increased (Figure 7C, lanes 11–12). We would like to mention that in RBM38-null MEFs, the steady-state level of p53 was slightly increased (1.3-fold) upon knockdown of PPM1D (Figure 7C, lanes 3–4). This is likely due to the decreased activity of PPM1D, leading to increased expression of p53 directly and/or indirectly via Mdm2.
Figure 7. PPM1D regulates p53 mRNA translation through phosphorylation of RBM38.
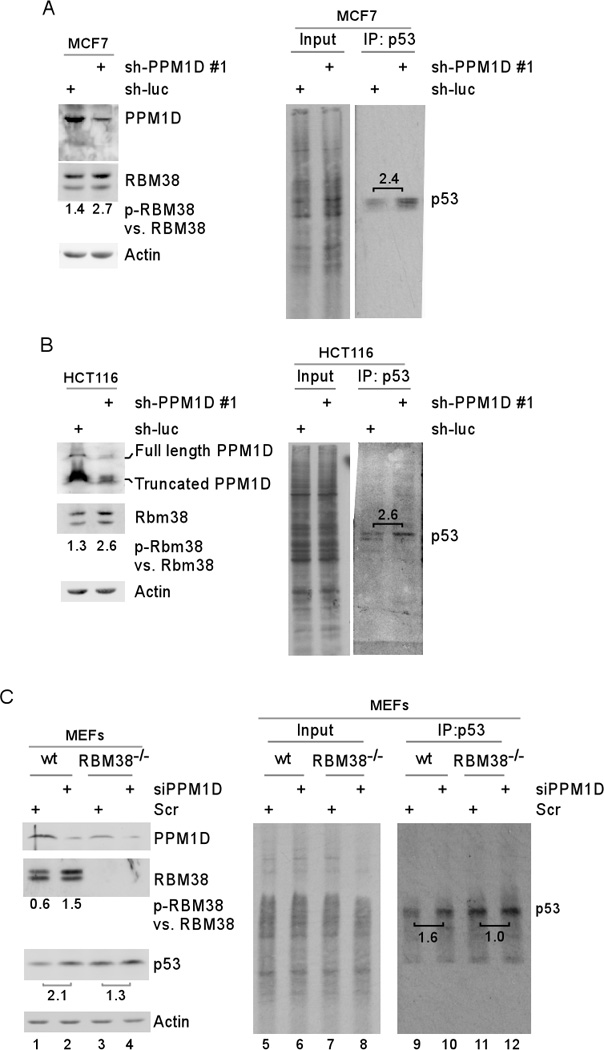
(A–B) Knockdown of PPM1D leads to increased phosphorylation of RBM38 and consequently increased p53 mRNA translation in MCF7 (A) and HCT116 (B) cells. Left panels: cells were transduced with lentivirus particles expressing a control shRNA or shRNA #1 against PPM1D for 72 h. Cell lysates were collected and subjected to Western blot analysis with antibodies against RBM38, PPM1D, or actin. Right panel: Cells treated as in left panel were 35S-labeled for 10 min, followed by immunoprecipitation with anti-p53. The immunocomplexes were resolved by SDS-PAGE and p53 was visualized by autoradiography. The relative level of p53 was measured by densitometry and the relative fold change was shown above each pair. (C) The increase in p53 mRNA translation upon inhibition of PPM1D is RBM38-dependent. Left panels: primary wild-type MEFs and RBM38-null MEFs were transiently transfected with a control or PPM1D siRNA for 72 h. Cell lysates were collected and subjected to Western blot analysis with antibodies against RBM38, PPM1D, p53, or actin. Right panel: Cells treated as in left panel were 35S-labeled for 10 min, followed by immunoprecipitation with anti-p53. The immunocomplexes were resolved by SDS-PAGE and p53 was visualized by autoradiography.
RBM38 knockout diminishes the effect of PPM1D knockdown on growth suppression
Several studies showed that deletion or knockdown of PPM1D or RBM38 promotes p53 expression and subsequently leads to growth suppression5, 31–33. Our studies above showed that the RBM38-PPM1D loop plays a critical role in p53 expression. Thus, the biological function of the RBM38-PPM1D loop in the p53 pathway was measured in MCF7 cells in which PPM1D was knocked down by siRNA along with or without RBM38 knockout via CRISPR-Cas9 technology. We found that knockdown of PPM1D in MCF7 cells led to an increased ratio of p-RBM38 vs. RBM38 along with increased expression of p53 protein (Figure 8A–B, lanes 1–2). Upon inducible knockout of RBM38 by CRISPR-Cas9, the level of PPM1D protein was decreased whereas the level of p53 protein was increased (Figure 8A–B, compare lanes 1 with 3). Interestingly, PPM1D knockdown had very weak effect on the level of p53 expression increased by RBM38 knockout (Fig. 8A–B, compare lanes 3–4). We also found that siRNA-mediated knockdown of PPM1D or RBM38 knockout significantly inhibited cell growth and colony formation (Figure 8C–D). However, PPM1D knockdown had no further effect on colony formation, which was already decreased by RBM38 knockout in MCF7 cells (Figure 8C–D). Thus, the effect of RBM38-PPM1D on colony formation is consistent with the effect of RBM38-PPM1D on p53 expression in MCF7 cells.
Figure 8. RBM38 knockout diminishes the effect of PPM1D knockdown on growth suppression.
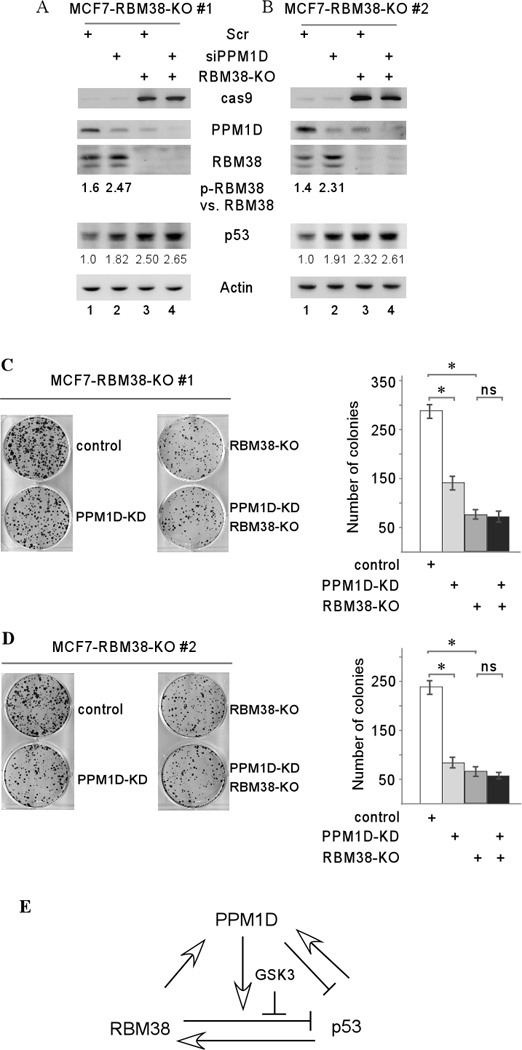
(A–B) Western blots were prepared with extracts from MCF7 cells uninduced or induced to knock out RBM38 for 4 days, in which PPM1D was knocked down by siPPM1D as indicated. The blots were then probed with antibodies against RBM38, PPM1D, p53, or actin. (C–D) Left panels, representative images of the colony formation assay. Colony formation assay was performed with MCF7 cells uninduced or induced to knock out RBM38 for 4 days, in which PPM1D was knocked down by siPPM1D as indicated. Right panels, quantification of colonies shown in the left panels from three separate experiments. An asterisk indicates a difference with statistical significance (P < 0.05). (E) A model for the role of the RBM38-PPM1D loop in the p53 pathway.
Discussion
Defining how p53 activity is regulated is a priority in cancer research because p53 plays a critical role in tumor suppression. RBM38, a target of p53 and a repressor of p53 mRNA translation, is phosphorylated at serine 195 5, 21. Interestingly, S195 phosphorylation converts RBM38 from a repressor to an activator of p53 mRNA translation and GSK3 is found to phosphorylate S195 in RBM38 21. Therefore, it is important to determine whether RBM38 activity is regulated by a phosphatase. Here, we showed that PPM1D physically interacts with p-RBM38 and then dephosphorylates RBM38 at Ser195. Consistent with this, RBM38 phosphorylation was increased upon knockdown of PPM1D and subsequently led to increased p53 mRNA translation. Interestingly, we also found that ectopic expression of RBM38 increased, whereas knockdown of RBM38 decreased, PPM1D expression through mRNA translation. These findings provide evidence that the reciprocal interplay between PPM1D and RBM38, both of which are targets and modulators of p53, plays a critical role in p53 expression and activity (Figure 8E).
As a target of p53, PPM1D is a key modulator of the p53 pathway in response to DNA damage 13, 14, 34. PPM1D dephosphorylates and inhibits several p53 activating kinases, including p38 MAP kinase, which phosphorylates p53 at serines 33 and 46, ATM, which phosphorylates p53 at serine 15, and Chk1 and Chk2, which phosphorylate p53 at serine 20 (Fujimoto et al., 2006, Lu et al., 2005, Oliva-Trastoy et al., 2006,Shreeram et al., 2006, Takekawa et al., 2000 and Yoda et al., 2006). PPM1D also dephosphorylates MDM2 at serine 395, which stabilizes MDM2 and consequently increases p53 ubiquitination and proteasomal degradation 29. Additionally, PPM1D directly dephosphorylates p53 at serine 15 and thus inhibits p53 activity in tumor suppression 14. Here, we found that PPM1D inhibits p53 mRNA translation through dephosphorylation of RBM38 at serine 195. Therefore, we uncovered a novel mechanism by which PPM1D regulates p53 mRNA translation and p53-dependent growth suppression through dephosphorylation of RBM38. Together, we postulated that modulation of the p53-RBM38 loop to suppress p53 mRNA translation is another mechanism by which PPM1D promotes cell survival and transformation in response to DNA damage.
Materials and Methods
Reagents
Anti-p53, anti-PPM1D, anti-GST and anti-histidine were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-mouse p53 (1C12) was purchased from Cell Signaling Technology (Beverly, MA). Anti-HA was purchased from Covance (San Diego, CA). Anti-actin, proteinase inhibitor cocktail, RNase A, protein A/G beads, PPM1D inhibitor (CCT007093) were purchased from Sigma (St. Louis, MO). Anti-RBM38 antibody was made as previously described 1. The Iscript cDNA synthesis kit was purchased from Bio-Rad Laboratories (Irvine, CA). 35S-methionine/cysteine and adenosine-5'-[γ-32P] Triphosphate ([γ-32P] ATP) were purchased from PerkinElmer (Waltham, MA). The Ni-NTA agarose beads were purchased from Biontex (Germany). The glutathione sepharose beads were purchased from Macherey-Nagel (Germany). The EST clone (clone ID 5295521), which contains a full length human PPM1D cDNA, was purchased from OpenBiosystems (Huntsville, AL).
Plasmids
To generate a pcDNA3 vector expressing HA-tagged PPM1D, a fragment encoding the catalytic domain (residues 1–420) of PPM1D was amplified by PCR using full-length PPM1D cDNA as a template with a forward primer, 5′ AAA GGA TCC AAA TGG CGG GGC TGT ACT CGC 3′, and a reverse primer, 5′ AAA CTC GAG TTA CTT GAC TGG TGG TGT AG AAC 3′. The PCR product was inserted into a pcDNA3 vector through BamH1 and XhoI sites and confirmed by sequencing. Phosphatase dead PPM1D 35 was constructed by introducing a D314A mutation. To generate a pcDNA3 vector expressing HA-tagged PPM1D D314A, PCR-based mutagenesis was performed. Specifically, a PCR product was amplified by using PPM1D cDNA as a template with one pair of common primers and one pair of specific primers. The PCR product was inserted into pcDNA3 vector through BamH1 and XhoI sites and confirmed by sequencing. The common primers were the same as the one used for amplifying PPM1D. The specific primers for PPM1D-D314A were a forward primer, 5′ TTG GGG AGT GCT GGA CTT TGG AAT ATG 3′, and a reverse primer, 5′ AAG TCC AGC ACT CCC CAA TAT AAT ATA C 3′.
Histidine (HIS)-tagged or GST-tagged RBM38 expression vectors were generated as previously described 28. To generate vectors expressing GST-tagged PPM1D or HIS-tagged PPM1D, PCR products were amplified with the primers mentioned above using the full-length PPM1D cDNA as a template. These PCR products were inserted into pGEX vector or pcDNA3.1/HisB vector through BamH1 and XhoI sites and confirmed by sequencing.
To generate EGFP expression vector carrying 5′ and/or 3′UTR of PPM1D transcript, a DNA fragment containing EGFP coding region was amplified using pEGFP-N2 vector as template with forward primer, 5′-AAA GGT ACC ATG GTG AGC AAG GGC GAG GAG C −3′, and reverse primer, 5′-AAA GCG GCC GCG TAT GGC TGA TTA TGA TCT AG-3′. The PCR product was digested with KpnI and NotI and cloned into pcDNA3 vector (Invitrogen). The vector was designated as pcDNA3-EGFP. DNA fragment containing 5′ or 3′UTR of PPM1D transcript was amplified using cDNA from H1299 cells as template with forward primer for 5′UTR, 5′-AAA AAG CTT GGG GAA GCG CAG TGC GCA G-3′, and reverse primer for 5′UTR, 5′-AAA GGT ACC GGC CGG CTG GCC GGG ATC C-3′, or forward primer for 3′UTR, 5′-GGG GCG GCC GCA ATG CAT CTG GGA AAT GAG G-3′, and reverse primer for 3′UTR, 5′-GGG TCT AGA AGA TAG TTT ATT TAG GGA CAA AA-3′ respectivly. The PCR products were digested with HindIII and KpnI for PPM1D-5′UTR or NotI and XbaI for PPM1D-3′UTR and cloned into pcDNA3/EGFP vector. The vectors were designated as pcDNA3/PPM1D-5′UTR/ EGFP and pcDNA3/ EGFP/PPM1D-3′UTR. To generate pcDNA3/PPM1D-5′UTR/ EGFP/PPM1D-3′UTR, pcDNA3/PPM1D-5′UTR/ EGFP vector was digested with HindIII and KpnI. The digested DNA fragment containing PPM1D-5′UTR was cloned into pcDNA3/ EGFP/PPM1D-3′UTR.
To generate pcDNA4 vector expressing pcDNA4-Flag-Cas9, pSpCas9 plasmid 36 (Addgene) was cut with ApeI/NotI to generate a DNA fragment encoding 3xFLAG-NLS-Cas9-NLS and then inserted into pcDNA4 vector through BspEI and NotI sites. The plasmid pcDNA4-Flag-Cas9 was confirmed by sequencing.
To generate pBabe-puro vector expressing gRNA, U6-driven sgRNA expression cassette was amplified using pSpCas9 as a template with a forward primer 5′ AAA GAA TTC GAG GGC CTA TTT CCC ATG AT 3′, and a reverse primer 5′ AAA AAG CTT AGA GCC ATT TGT CTG CAG 3′. The PCR product was inserted into pBabe-puro vector through EcoRI and HindIII sites and confirmed by sequencing. RBM38 gRNAs were designed using the CRISPR design tool (crispr.mit.edu) and cloned into the BbsI sites within the sgRNA expression cassette. Two specific gRNAs were used: gRNA #1 ACA CTA CCG ACG CCT CGC TC and gRNA #2 GTA CTT CCT GAG CGA GGC GT. pBabe-puro vectors expressing gRNAs targeting RBM38 were designated as pBabe-U6-sgRBM38 #1 and #2.
Cell culture and cell line generation
MCF7, HCT116, p53−/− HCT116, and H1299 cells were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone) as previously described (Zhang and Chen, 2007). RBM38−/− and p53−/−; RBM38−/− double knockout mouse embryo fibroblasts (MEFs) were generated as described previously 5 and cultured in DMEM supplemented with 10% fetal bovine serum, 55 µM β-mercaptoethanol, and 1× MEM non-essential amino acids solution (Cellgro). MCF7, HCT116, and H1299 cell lines in which RBM38 or HA-tagged RBM38 are inducibly expressed were cultured as previously reported 1, 37. MCF7 cell line in which RBM38 is inducibly knocked down was used as previously reported 1, 37.
Generation of inducible RBM38 knockout cell lines
To generate stable cell lines that inducibly express Cas9, pcDNA4-Flag-Cas9 vector, in which Cas9 expression is driven by CMV promoter along with two tetracycline repressor binding sites, was transfected into MCF7 cells expressing a tetracycline repressor (pcDNA6). The Cas9-expressing cells were selected with zeocin and confirmed by Western blot analysis.
To generate inducible RBM38-KO cell line, pBabe-U6-sgRBM38 #1 or #2 was transfected into MCF7 cells in which Cas9 can be inducibly expressed. The inducible RBM38-KO lines were selected with puromycin and confirmed by Western blot analysis. Inducible RBM38 knockout was achieved by inducing Cas9 expression. For induction, tetracycline (500 ng/mL) was added to culture medium for 4 days.
Western blot analysis, immunoprecipitation and GST-pull down assay
Western blot analysis was performed as previously described 38. Cell lysates suspended in 2X SDS sample buffer were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with indicated antibodies. The immunoreactive bands were visualized by the enhanced chemiluminescence (Pierce) and quantified by densitometry with Software LabWorks (UVP, Upland, CA). Immunoprecipitation assay was performed as previously described 38. Briefly, cells were lysed in 0.2 % Triton lysis buffer (25 mM Tris [pH 7.4], 25 mM NaCl, 0.2 % Triton X-100) supplemented with the proteinase inhibitor cocktail (100 µg/ml) , followed by incubation with 1 µg of antibody or control IgG. The immunocomplexes were brought down by protein A/G beads and subjected to Western blot analysis.
For GST-pull down assay, the recombinant His- or GST-tagged proteins were expressed in bacteria BL21 and purified by using Ni-NTA and glutathione sepharose beads, respectively. 500 ng of recombinant His-tagged proteins and 500 ng of recombinant GST-tagged proteins were incubated in E1A binding buffer (50 mM HEPES, pH 7.6, 50 mM NaCl, 5 mM EDTA, 0.1% Nonidet P-40, and 10% glycerol) for 2 h at 4°C, followed by precipitation with glutathione-sepharose beads. Beads were washed and resuspended in 2x SDS loading buffer and subjected to Western blot analysis.
Probe labeling and RNA Electrophoretic Mobility Shift Assay (REMSA)
All probes were labeled by in vitro transcription using a DNA fragment containing T7 promoter and various region of PPM1D 5′ or 3′UTR. Briefly, 500 ng of purified PCR product was incubated with 50 µCi of α-32P-UTP, 0.5 mM each of NTP (A, G, C), 20 unit of T7 RNA polymerase (Ambion) in 20 µl of reaction at 37°C for 1 h, followed by DNase I (1 unit) treatment for 15 min. The reaction mixture was purified by Sephadex G-50 column to remove unlabeled free nucleotide and the radioactivity of probes was measured by a scintillation counter. REMSA was carried out with a modified protocol as previously described (Zhang et al. 2010). Briefly, 250 nM of RBM38 recombinant protein, 100 µg/ml of yeast tRNA, and 50,000 CPM 32P-labeled RNA probe were mixed in 20 µl of reaction buffer (10 mM Tris-Cl, pH 8.0, 25 mM KCl, 10 mM MgCl2, 1 mM DTT) at 25°C for 25 min. RNA/protein complexes were digested by adding 100 U RNase T1 at 37°C for 15 min and then separated in 7% of native PAGE gel. RNA-protein complexes were visualized by autoradiography.
The primers for generating probe 5′ UTR were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GGG GGA AGC GCA GTG CGC AGG-3′ and 5′-AAA GGT ACC GGC CGG CTG GCC GGG ATC C-3′. The primers for generating probe A were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GAA TGC ATC TGG AAA TGA GG-3′ and 5′-TTC AGC AAC ATG AGG AAC AGT-3′. The primers for generating probe B were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GTG AAC TGT TCC TCA TGT TGC TG-3′ and 5′-TGT GGC ATA AGA CAC ACC GA-3′. The primers for generating probe C were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GAT TCT TGA TCG GTG TGT CTT-3′ and 5′-GGG TCT AGA AGA TAG TTT ATT TAG GGA CAA AA-3′. The primers for generating probe A1 were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GAA TGC ATC TGG AAA TGA GG-3′ and 5′-GTG AAA CAC AGG TCA CTA TG-3′. The primers for probe A2 were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GTT AAT GTT TCT TAG AGC CAA G-3′ and 5′-CAG CAC CAT CTG TTA TTG AAA TAG-3′. The primers for generating probe A3 were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GGC TGC TAA TTC CCA ACA TTT C-3′ and 5′-TTC AGC AAC ATG AGG AAC AGT-3′. The primers for generating probe B1 were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GTG AAC TGT TCC TCA TGT TGC TG-3′ and 5′-GAG ACT CCA TCT CAA ACA AAC-3′. The primers for generating probe B2 were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GCA CTC TGT CAT CCA GGC TAG-3′ and 5′-CCG AGA TCA CAC CAC TGC AC-3′. The primers for generating probe B3 were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GCT CCC TGC AAC CTC TGC CTT C-3′ and 5′-TGT GGC ATA AGA CAC ACC GA-3′.
RNA Interference
Scrambled siRNA (5′ GCA GUG UCU CCA CGU ACU A dTdT 3′), siRNA against RBM38 (5′ CAC CUU GAU CCA GCG GAC UUA dTdT 3′), and siRNA against both mouse and human PPM1D (5′ CGA AAU GGC UUA AGC CGA A dTdT 3′) were purchased from Dharmacon (Chicago, IL). For siRNA transfection, siLentFectTM Lipid Reagent (Bio-Rad) was used according to the user's manual. For lentiviral production, lentivirus vectors (pLKO.1-puro) expressing a shRNA of interest were purchased from Sigma. The sequence for control luciferase shRNA is 5′ CGC TGA GTA CTT CGA AAT GTC 3′. The sequence for RBM38 shRNA is 5′-CAG AAG GAC ACC ACG TTC A-3′. The sequence for PPM1D shRNA#1 is 5′ CCC TTC TCG TGT TTG CTT AAA 3′. The sequence for PPM1D shRNA#2 is 5′ TGG CCT TGT GCC TAC TAA TTC 3′. A lentivirus vector (10 µg) expressing shRNA along with packaging plasmids, pRSV-REV (5 µg), pMDL g/p RRE (5 µg), and VSVG (5 µg), was cotransfected into 293T cells (1 × 107) by ExpressFectTM transfection system (Denville Scientific) according to the user's manual. After 48 h, the supernatant containing shRNA-expressing lentiviruses was filtered and concentrated by ultracentrifugation (28,000 rpm, 4°C, 1 h). The concentrated lentiviral particles were then transduced into cells, followed by puromycin selection (1 µg/ml) for 72 h.
35S metabolic labeling
The metabolic labeling was performed as described 39. Briefly, cells were preincubated in methionine-free DMEM for 1 hr, and then labeled with 100 µCi/ml of 35S-methionine (PerkinElmer) for 10 min. The incorporation of 35S-methionine into newly synthesized proteins was measured by TCA precipitation. 1 × 107 cpm of 35S-labeled lysates was immunoprecipitated with 1.0 µg of anti-PPM1D or anti-p53. The immunocomplexes were resolved in SDS-PAGE gel and then subject to autoradiography.
RNA isolation and RT-PCR analysis
Total RNA was isolated with Trizol reagent. RT-PCR was performed with the Iscript cDNA synthesis kit (Bio-Rad Laboratories) according to the manufacturer's instructions. The PCR program used for amplification was (i) 94°C for 5 min, (ii) 94°C for 30 s, (iii) 56°C for 30 s, (iv) 72°C for 30 s, and (v) 72°C for 10 min. From steps 2 to 4, the cycle was repeated 26 times for actin or 30 times for RBM38 and PPM1D. The primers used to amplify human actin were forward primer, 5' CTG AAG TAC CCC ATC GAG CAC GGC A 3', and reverse primer, 5' GGA TAG CAC AGC CTG GAT AGC AAC G 3'. The primers to amplify mouse actin were forward primer, 5′-CCC ATC TAC GAG GGC TAT-3′, and reverse primer, 5′-AGA AGG AAG GCT GGA AAA-3′. The primers for human PPM1D were forward primer, 5′ GCC AGA ACT TCC CAA GGA AAG 3′, and reverse primer, 5′ GTC TGG TTC AGG TGA CAC CAC 3′. The primers for mouse PPM1D were forward primer, 5′ AAG GTT TCC TCG CCT GTC AC 3′, and reverse primer, 5′ GTG TCA CCT CCA CAG CTC TC 3′. The primers for human RBM38 were forward primer 5′ CTA CCG ACG CCT CGC TCA G 3′, and reverse primer, 5′ CCC AGA TAT GCC AGG TTC AC 3′. The primers for mouse RBM38 were forward primer 5′ GAC GCA TCG CTC AGA AAG T 3′, and reverse primer, 5′ GAG GAG TCA GCC CGT AGG T 3′. The primers for p21 were forward primer, 5′ CCC GTG GAC AGT GAG CAG T 3′, and reverse primer, 5′ CAG CAG GGC AGA GGA AGT A 3′.
RNA-immunoprecipitation (RNA-IP)
RNA-IP was carried out as previously described 1, 40. Briefly, cells (2×107) were lysed with 1 ml of lysis buffer (10 mM HEPES, pH7.0, 100 mM KCl, 10 mM MgCl2, 0.5% NP-40, 1 mM DTT) supplemented with RiboLock Ribonuclease inhibitor (Fermentas) for 30 min on ice, and cell lysates were collected by centrifugation (13,000 rpm, 4°C, 10 min). The RNA-protein immunocomplexes were incubated with 2 µg of anti-RBM38 or isotype control IgG at 4°C for 4 h and brought down by protein G bead. RT-PCR analysis was carried out to examine the RNA-protein interaction.
In vitro phosphatase assay
The assay was performed as described 12. Extracts were collected from HCT116 cells induced to express HA-tagged RBM38 and then immunoprecipitated with anti-HA antibody. The immunocomplexes were resuspended in 50 µl phosphatase buffer (50 mM Tris-HCl (pH 7.5), 30 mM MgCl2, 1 mg/ml bovine serum albumin, 0.05% 2-mercaptoethanol), which was supplemented with purified GST-PPM1D (WT) or GST-PPM1D (D314A). PPM1D inhibitor cct007093 (50 µM) was added as indicated. The mixture was allowed to incubate in a 30°C water bath for 1 h. The phosphorylation status of RBM38 was measured by Western blot analysis with antibody against RBM38.
Colony formation assay
For colony formation assay, 1,000 cells/well were plated in six-well plates. After 15 days, cells were fixed with methanol, stained with Crystal Violet and colonies counted.
Acknowledgement
This work is supported in part by NIH grants CA076069, CA081237, and CA121137.
Footnotes
Authors’ Contribution
Zhang M and Xu E did the experiments; Zhang M, Xu E and Zhang J analyzed the data; Chen X supervised the project and analyzed the data; Zhang M and Chen X wrote the manuscript. All authors read and commented on the draft version of the manuscript and approved the final version.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Shu L, Yan W, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes & development. 2006;20:2961–2972. doi: 10.1101/gad.1463306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feldstein O, Ben-Hamo R, Bashari D, Efroni S, Ginsberg D. RBM38 is a direct transcriptional target of E2F1 that limits E2F1-induced proliferation. Mol Cancer Res. 2012 doi: 10.1158/1541-7786.MCR-12-0331. [DOI] [PubMed] [Google Scholar]
- 3.Xu E, Zhang J, Chen X. MDM2 expression is repressed by the RNA-binding protein RNPC1 via mRNA stability. Oncogene. 2012 doi: 10.1038/onc.2012.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan W, Zhang J, Zhang Y, Jung YS, Chen X. p73 Expression Is Regulated by RNPC1, a Target of the p53 Family, via mRNA Stability. Mol Cell Biol. 2012;32:2336–2348. doi: 10.1128/MCB.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Cho SJ, Shu L, Yan W, Guerrero T, Kent M, et al. Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes & development. 2011;25:1528–1543. doi: 10.1101/gad.2069311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin T, Cho SJ, Chen X. RNPC1, an RNA-binding protein and a p53 target, regulates macrophage inhibitory cytokine-1 (MIC-1) expression through mRNA stability. The Journal of biological chemistry. 2013;288:23680–23686. doi: 10.1074/jbc.M113.480186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho SJ, Jung YS, Zhang J, Chen X. The RNA-binding protein RNPC1 stabilizes the mRNA encoding the RNA-binding protein HuR and cooperates with HuR to suppress cell proliferation. The Journal of biological chemistry. 2012;287:14535–14544. doi: 10.1074/jbc.M111.326827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leveille N, Elkon R, Davalos V, Manoharan V, Hollingworth D, Oude Vrielink J, et al. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nature communications. 2011;2:513. doi: 10.1038/ncomms1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinicke LA, Nabet B, Shen S, Jiang P, van Zalen S, Cieply B, et al. The RNA binding protein RBM38 (RNPC1) regulates splicing during late erythroid differentiation. PloS one. 2013;8:e78031. doi: 10.1371/journal.pone.0078031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Molecular cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang M, Zhang J, Chen XL, Cho SJ, Chen XB. Glycogen synthase kinase 3 promotes p53 mRNA translation via phosphorylation of RNPC1. Genes Dev. 2013;27:2246–2258. doi: 10.1101/gad.221739.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiscella M, Zhang HL, Fan SJ, Sakaguchi K, Shen SF, Mercer WE, et al. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci U S A. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer cell. 2007;12:342–354. doi: 10.1016/j.ccr.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 14.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19:1162–1174. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Natrajan R, Lambros MB, Rodriguez-Pinilla SM, Moreno-Bueno G, Tan DS, Marchio C, et al. Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res. 2009;15:2711–2722. doi: 10.1158/1078-0432.CCR-08-1878. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Yang Y, Peng Y, Austin RJ, van Eyndhoven WG, Nguyen KC, et al. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nature genetics. 2002;31:133–134. doi: 10.1038/ng888. [DOI] [PubMed] [Google Scholar]
- 17.Nannenga B, Lu X, Dumble M, Van Maanen M, Nguyen TA, Sutton R, et al. Augmented cancer resistance and DNA damage response phenotypes in PPM1D null mice. Mol Carcinog. 2006;45:594–604. doi: 10.1002/mc.20195. [DOI] [PubMed] [Google Scholar]
- 18.Tan DS, Lambros MB, Rayter S, Natrajan R, Vatcheva R, Gao Q, et al. PPM1D is a potential therapeutic target in ovarian clear cell carcinomas. Clin Cancer Res. 2009;15:2269–2280. doi: 10.1158/1078-0432.CCR-08-2403. [DOI] [PubMed] [Google Scholar]
- 19.Ruark E, Snape K, Humburg P, Loveday C, Bajrami I, Brough R, et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature. 2013;493:406–410. doi: 10.1038/nature11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleiblova P, Shaltiel IA, Benada J, Sevcik J, Pechackova S, Pohlreich P, et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. The Journal of cell biology. 2013;201:511–521. doi: 10.1083/jcb.201210031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Zhang J, Chen X, Cho SJ, Chen X. Glycogen synthase kinase 3 promotes p53 mRNA translation via phosphorylation of RNPC1. Genes & development. 2013;27:2246–2258. doi: 10.1101/gad.221739.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho SJ, Zhang J, Chen X. RNPC1 modulates the RNA-binding activity of, and cooperate with, HuR to regulate p21 mRNA stability. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkp1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujimoto H, Onishi N, Kato N, Takekawa M, Xu XZ, Kosugi A, et al. Regulation of the antioncogenic Chk2 kinase by the oncogenic Wip1 phosphatase. Cell Death Differ. 2006;13:1170–1180. doi: 10.1038/sj.cdd.4401801. [DOI] [PubMed] [Google Scholar]
- 24.Takekawa M, Adachi M, Nakahata A, Nakayama I, Itoh F, Tsukuda H, et al. p53-inducible wip1 phosphatase mediates a negative feedback regulation of p38 MAPK-p53 signaling in response to UV radiation. The EMBO journal. 2000;19:6517–6526. doi: 10.1093/emboj/19.23.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, et al. Wip1 phosphatase modulates ATM-dependent signaling pathways. Molecular cell. 2006;23:757–764. doi: 10.1016/j.molcel.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Rayter S, Elliott R, Travers J, Rowlands MG, Richardson TB, Boxall K, et al. A chemical inhibitor of PPM1D that selectively kills cells overexpressing PPM1D. Oncogene. 2008;27:1036–1044. doi: 10.1038/sj.onc.1210729. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W, McClain C, Gau JP, Guo XY, Deisseroth AB. Hyperphosphorylation of p53 induced by okadaic acid attenuates its transcriptional activation function. Cancer Res. 1994;54:4448–4453. [PubMed] [Google Scholar]
- 28.Cho SJ, Zhang J, Chen X. RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic acids research. 2010;38:2256–2267. doi: 10.1093/nar/gkp1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu XB, Ma O, Nguyen TA, Joness SN, Oren M, Donehower LA. The Wip1 phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 2007;12:342–354. doi: 10.1016/j.ccr.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 30.Gilmartin AG, Faitg TH, Richter M, Groy A, Seefeld MA, Darcy MG, et al. Allosteric Wip1 phosphatase inhibition through flap-subdomain interaction. Nature chemical biology. 2014;10:181–187. doi: 10.1038/nchembio.1427. [DOI] [PubMed] [Google Scholar]
- 31.Parssinen J, Alarmo EL, Karhu R, Kallioniemi A. PPM1D silencing by RNA interference inhibits proliferation and induces apoptosis in breast cancer cell lines with wild-type p53. Cancer genetics and cytogenetics. 2008;182:33–39. doi: 10.1016/j.cancergencyto.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 32.Pandolfi S, Montagnani V, Penachioni JY, Vinci MC, Olivito B, Borgognoni L, et al. WIP1 phosphatase modulates the Hedgehog signaling by enhancing GLI1 function. Oncogene. 2013;32:4737–4747. doi: 10.1038/onc.2012.502. [DOI] [PubMed] [Google Scholar]
- 33.Wang W, Zhu H, Zhang H, Zhang L, Ding Q, Jiang H. Targeting PPM1D by lentivirus-mediated RNA interference inhibits the tumorigenicity of bladder cancer cells. Brazilian Journal of Medical and Biological Research. 2014;47:1044–1049. doi: 10.1590/1414-431X20143645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, et al. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takekawa M, Adachi M, Nakahata A, Nakayama I, Itoh F, Tsukuda H, et al. p53-inducible Wip1 phosphatase mediates a negative feedback regulation of p38 MAPK-p53 signaling in response to UV radiation. Embo J. 2000;19:6517–6526. doi: 10.1093/emboj/19.23.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Jun Cho S, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, regulates p63 expression through mRNA stability. Proc Natl Acad Sci U S A. 2010;107:9614–9619. doi: 10.1073/pnas.0912594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Chen X. DeltaNp73 modulates nerve growth factor-mediated neuronal differentiation through repression of TrkA. Mol Cell Biol. 2007;27:3868–3880. doi: 10.1128/MCB.02112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonifacino JS. Metabolic labeling with amino acids. Curr Protoc Protein Sci. 2001 doi: 10.1002/0471140864.ps0307s17. Chapter 3: Unit 3 7. [DOI] [PubMed] [Google Scholar]
- 40.Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiriksdottir E, Langel U, et al. Immunoprecipitation of mRNA-protein complexes. Nat Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]


