Abstract
Background
The cell fate determinant Numb is aberrantly expressed in cancer. Numb is alternatively spliced with one isoform containing an extended proline rich region (PRRL) compared to the other (PRRS). PRRL was recently reported to enhance proliferation of breast and lung cancer cells. However, the importance of Numb alternative splicing in hepatocellular carcinoma (HCC) remain unexplored.
Main Results
We report here that Numb PRRL expression is increased in HCC and is associated with early recurrence and reduced overall survival after surgery. In a panel of HCC cell lines, PRRL generally promotes and PRRS suppresses proliferation, migration, invasion and colony formation. PRRS knockdown leads to increased Akt phosphorylation and c-Myc expression, and Akt inhibition or c-Myc silencing dampens the proliferative impact of Numb PRRS knockdown. In the cell models explored in this study, alternative splicing of Numb PRR isoforms is coordinately regulated by the splicing factor Rbfox2 and the kinase SRPK2. Rbfox2 knockdown causes accumulation of PRRL while SRPK2 knockdown causes accumulation of PRRS. SRPK2 subcellular location is regulated by the molecular chaperone Hsp90, and Hsp90 inhibition or knockdown phenocopies SRPK2 knockdown in promoting accumulation of Numb PRRS. Finally, HCC cell lines that predominately express PRRL are differentially sensitive to Hsp90 inhibition.
Conclusion
Our data suggest that alternative splicing of Numb may provide a useful prognostic biomarker in HCC and is pharmacologically tractable.
Keywords: Numb, alternative splicing, Hsp90, SRPK2, Rbfox2
Hepatocellular carcinoma (HCC) is the fifth most common cancer, and the third most frequent cause of cancer-related death worldwide (1). HCC prognosis is dismal due to lack of effective treatment options and the high rate of postsurgical recurrence. Lack of reliable early diagnostic and screening tests further exacerbates patient survival as more than 70% of HCC is in an advanced stage at diagnosis (2). Therefore, identification of prognostic molecular markers and development of novel therapeutic approaches are critically important.
The Numb gene was originally identified in Drosophila as a key determinant of cell fate. Mammalian Numb homologues are evolutionarily conserved and function in neurogenesis (3). In addition, Numb may also play a role in tumorigenesis and is expressed at high levels in some tumors (4). However, Numb expression and function in HCC have not been explored. Alternative splicing of Numb transcripts produces two major isoforms that differ in the length of their proline-rich region (PRR), due to inclusion (PRRL) or exclusion (PRRS) of a 48 amino-acid insert encoded by exon 12 (3). These isoforms play contrasting roles in regulating cellular functions. Thus, Numb PRRL promoted proliferation but not differentiation in murine embryonic carcinoma cells, whereas Numb PRRS promoted differentiation but not proliferation (5). Recently, global profiling identified Numb exon 12 inclusion as a frequent splicing event in human non-small cell lung cancer and in invasive bladder cancer (6–8). Furthermore, expression of Numb PRRL was increased in tumors compared with normal tissues, and correlated with poor prognosis. A similar phenomenon has been observed in colon and breast cancers (6). In the current study, we investigated the role of Numb and regulation of its alternative splicing in HCC.
MATERIALS AND METHODS
Patients and samples
HCC tissues were obtained with informed consent from patients who underwent curative resection between 2002 and 2003 at the Liver Cancer Institute and Zhongshan Hospital (Fudan University, Shanghai, China), and between 2011 and 2012 at the Beijing 302 Hospital (Beijing, China). Sample collection was approved by the Institutional Review Board of the corresponding institutes and recorded by the National Institutes of Health Office of Human Subjects Research. A total of 241 HCC cases were used for the mRNA array analyses, and the data were deposited into GEO Omnibus with accession number GSE14520 (9). Forty patients who underwent curative hepatectomy were randomly selected for qRT-PCR detection by using patient-matched pairs of tumor (T) and adjacent non-tumor (NT) liver tissues. The initial diagnosis was made based on serological tests and imaging, and was confirmed by histopathologic evaluation.
Cell Line, culture, and transfection
Huh1 and Huh7 cell lines were obtained from the Japanese Collection of Research Bioresources Cell Bank; SK-Hep1 and HEK293A cell lines were purchased from ATCC. Cells were cultured in DMEM (Huh1 and Huh7) or MEM (SK-Hep1) medium supplemented with 10% fetal bovine serum and 2 mmol/L L-glutamine. The UOK171 cell line was established from a clear cell renal cell carcinoma and cultured in DMEM containing 10% FBS. Numb overexpressing Huh7 cells were made by first transfecting the cells with lentiviruses containing the Tet transactivator and selected with blasicidin, and then transfecting the selected cells with lentiviruses containing either Numb PRRL or PRRS followed by selection with puromycin. Numb expression was induced by doxycycline.
RNA interference
siRNAs specific to Numb (SASI_Hs01_00245423, 5′GAUAGUGUUGGUGCAUCA, 3′UGAUGAACCAACGACUAUC), SRPK1, SRPK2, c-Myc, Oct4, Rbfox1, Rbfox2, Rbfox3, Hsp90 and negative control siRNA were purchased from Sigma-Aldrich. Transfection was performed using Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer’s instructions. A final concentration of 20 nM siRNA duplex was used for each transfection.
Immunoprecipitation and western blotting
Cells were lysed and processed as previously described (10). Antibodies specific for Numb, Akt, Akt308, poly ADP ribose polymerase, and caspase 3 were purchased from Cell Signaling Technology, tubulin antibody was from Santa Cruz Biotechnology, and SRPK1 and SRPK2 antibodies were from BD Bioscience. Rabbit anti-Rbfox2 polyclonal antibody was a generous gift from Dr. Sachiyo Kawamoto, Laboratory of Molecular Cardiology, National Heart, Lung, and Blood Institute, NIH.
Quantitative Real-Time Reverse-Transcription Polymerase Chain Reaction (qRT-PCR) Analysis
Total RNA was extracted from cells by using Trizol reagent and was subjected to qRT-PCR. All reactions were performed using a 7500 Real-Time PCR System from Applied Biosystems. Probes used for the Numb PRRL/S analyses were as follows: Hs01105434 (Numb PRRL isoform), Hs01105435 (Numb PRRS isoform) (Invitrogen). Experiments were performed in triplicate. The TaqMan gene assay for 18S RNA and β-actin was used to normalize the relative abundance of mRNA. Other genes were monitored using SYBR Green® (Invitrogen); primers for these genes are listed in the Supporting Materials (Table S2). All templates were amplified using the following standard PCR program: 95 °C for 10 min; 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s.
Immunohistochemistry
An Envision kit (DAKO) was used according to the manufacturer’s instructions. Tissues were embedded in paraffin and cut into 5 μm sections. After deparaffinization, antigen retrieval was performed with sodium citrate buffer in a steamer for 20 min. Endogenous peroxidases were blocked by incubation for 30 min in 0.3% H2O2. After blocking, primary antibodies were applied at 4 °C overnight and then slides were rinsed in wash buffer and incubated with HRP-conjugated appropriate secondary antibodies. Signal was developed using Dako chromogen (DAB/Hydrogen peroxidase) and slides were counterstained with Harris’ hematoxylin (blue nuclear stain). Immunohistochemical labeling intensity was assessed by two experienced pathologists.
Soft agar colony formation and flat colony formation
Cells (7500/dish) were re-suspended in 2.0 ml mixture of 0.7 % low-melting temperature agarose (SeaPlaque® Agarose, Lonza) and 2× DMEM (v:v=1:1), and were loaded in triplicate on top of the solidified bottom agar layer comprised of an equal volume mixture of 0.7 % low-melting temperature agar and DMEM in 6 cm dishes. The cells were incubated at 37 °C, 5 % CO2 for 4 weeks. For flat colony formation assay, cells (1000/dish) were seeded in 6 cm petri dishes and incubated at 37 °C, 5 % CO2 for 10 days. Colonies were visualized by staining with 1 ml of 0.02% crystal violet in 10% neutral buffered formalin for 1 h at room temperature. Colonies comprised of 50 or more cells were photographed using bright-field microscopy at a magnification of ×40/×100 and counted.
Real-time invasion assay
Invasiveness of liver cancer cells was evaluated using the RT-CIM™ system (Acea Biosciences) with the following modifications. The membrane separating upper and bottom chambers was coated with fibronectin prior to the assay. One-day post-transfection or treatment, cells were serum-starved for 16 h before being plated in upper chambers containing serum-free media. The lower chambers contained media with 10% serum (serum was used as attractant). Impedance signals from invasive cells passing through the membrane between the chambers were recorded in real time to provide continuous quantitative assessment of invasion. The assay was performed in a humidified incubator at 37°C and 5 % CO2.
MTT assay
Cell viability was determined using either MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. Briefly, cells were seeded in 96-well plates at 5×103 cells/well and treated with different reagents. Then each well was supplemented with 10 μL MTT (Sigma Aldrich) and incubated for 4 h at 37°C. The medium was then removed, and 100 μL DMSO (Sigma Aldrich) was added to solubilize the MTT reagent. Optical density was read at 490 nm.
Statistical analysis
Statistical analyses were performed using SPSS 19.0 software for Windows. The association of HCC subtypes and clinicopathological characteristics was examined using either Mann-Whitney U-tests or χ2 tests. Student t-tests were used to compare various test groups in colony formation assays, invasion assays, and MTT assays. Kaplan-Meier analysis was used to assess survival. Log-rank tests were used to compare patient survival between subgroups. Data are presented as mean ± SD unless otherwise indicated. Differences were considered to be statistically significant with P values < 0.05.
RESULTS
Numb is more highly expressed in HCC compared to adjacent liver tissues and expression of Numb PRRL correlates with poor prognosis
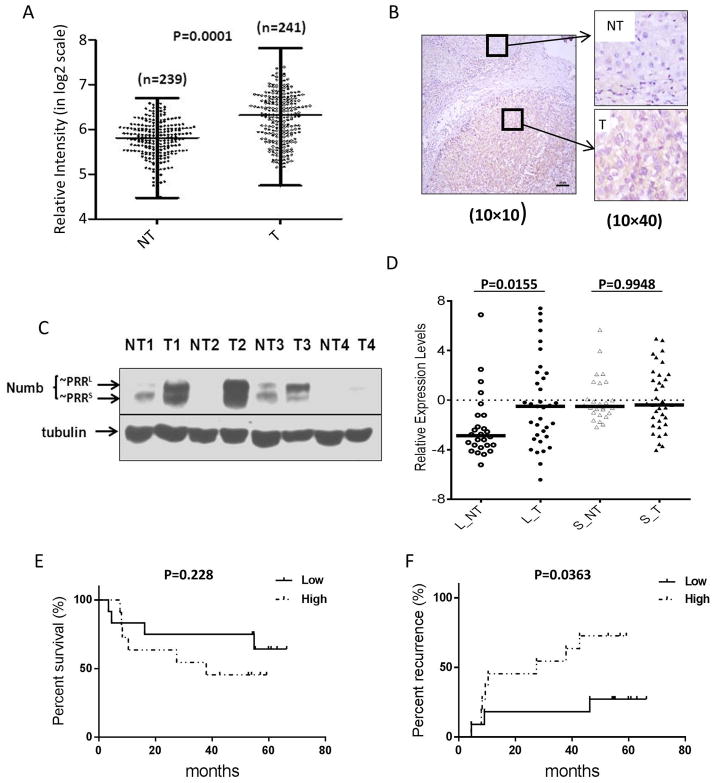
We first examined Numb mRNA expression in tumors and adjacent normal tissues by interrogating a microarray dataset of 241 HCC cases (GEO accession number: GSE14520). Numb expression was significantly up-regulated in tumors compared with matched adjacent non-tumor liver tissues (Figure 1A). Next, we assessed Numb protein expression by immunohistochemistry and western blotting, again finding higher expression in tumors compared to matched non-tumor liver tissues (Figure 1B and 1C).
Figure 1.
Numb expression in HCC patients. A, Total Numb mRNA levels are higher in HCC tumors (T) than in non-tumor liver tissues (NT). Microarray analysis of total Numb mRNA expression levels in patient-matched pairs of tumor and adjacent non-tumor liver tissues from HCC patients (n=241). B, Numb protein expression is elevated in HCC tumor compared to normal tissue. Immunohistochemistry staining of HCC tumor and adjacent liver tissue. C, Numb PRRL is highly expressed in HCC tumors while PRRS is the dominant isoform in non-tumor liver tissue. Western blot detection of Numb PRRL/S isoforms expressed in paired human HCC tumor tissues and adjacent non-tumor liver tissues. D, Numb PRRL but not PRRS expression is higher in HCC tumors. Numb PRRL/S mRNA levels were quantitated by qRT-PCR (n=40). E&F, elevated Numb PRRL expression correlates with reduced survival and more rapid recurrence in HCC patients after surgery (N=23).
Using western blotting, we compared the differential expression of Numb PRRL and PRRS in HCC. While total Numb was elevated in HCC, expression of PRRL (the upper band) was substantially higher than expression of PRRS (Figure 1C). These results were confirmed by qRT-PCR. Among 40 samples of paired HCC and non-tumor liver tissues, we found that the PRRL isoform was expressed at significantly higher levels in tumor (Figure 1D).
To investigate the potential clinical significance of PRRL expression, we examined whether PRRL expression was correlated with patient survival or tumor recurrence after surgery. In the cohort interrogated in our study, there were 23 patients who satisfied the requirements of the analysis (Table S1). Within this subgroup, we observed that patients whose HCC tumors had high PRRL expression (as determined by qRT-PCR) exhibited a trend for worse overall survival, although statistical significance was not reached. However, patients whose tumors had high PRRL expression exhibited a significantly shorter time to recurrence after surgery (median 27.5 months for PRRL high vs 54.9 months for PRRL low, p < 0.05) (Figure 1E, F, Table S3).
Numb PRRL and PRRS isoforms play opposing roles in HCC cell proliferation
The correlation between the expression profiles of Numb PRR isoforms and patient prognosis led us to investigate underlying mechanisms. For this purpose, we searched for HCC cell lines with different Numb PRR expression profiles. We selected Huh1 (expresses mainly PRRL), SK-Hep1 (expresses mainly PRRS), and Huh7 (expresses both PRRL and PRRS) for further studies (Figure S1A).
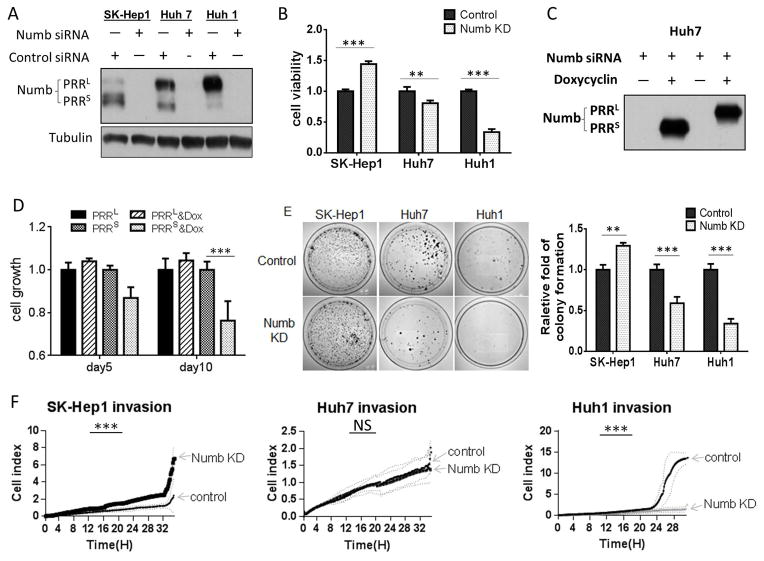
First, we used siRNA to silence Numb expression and we measured cell growth by MTT assay. Efficient Numb knockdown in all cell lines (Figure 2A) led to cell line-dependent effects on cell growth. Thus, while a significant growth increase was observed upon Numb silencing in SK-Hep1, a dramatic growth decrease was seen in Huh1, and a modest growth decrease was seen in Huh7 (Figure 2B). These results correlate with the unique profiles of Numb expression in the three cell lines and are consistent with PRRL promoting and PRRS suppressing the growth of HCC cells in vitro.
Figure 2.
Numb PRRL and PRRS have opposing activities. A, Western blot showing siRNA-mediated knockdown of Numb proteins in HCC cell lines. B, Numb knockdown differentially affects the growth of HCC cells. Numb knockdown was induced by transfecting cells with siRNA. Cell growth was evaluated by MTT assay. C, Exogenous expression of Numb PRRL/S in Huh7 cells. Huh7 cells were stably transduced with lentiviral vectors encoding either Numb PRRL or Numb PRRS, and expression was induced with doxycycline. Endogenous Numb was knocked down with an siRNA targeting the 3′UTR, which is absent in the exogenously introduced constructs. D, Numb PRRS suppresses cell growth. Numb overexpression and knockdown were as described in C, and cell growth was evaluated by MTT assay. E, Numb PRR isoforms differentially affect colony formation. Numb knockdown was achieved by siRNA, and colony formation was performed in petri dishes untreated for cell adhesion. Ten days after seeding, colonies were stained with crystal violet and counted. Assays were performed in triplicate and repeated 3 times. F, Numb PRR isoforms differentially affect HCC cell invasion. Real-time invasion assay in fibronectin-coated transwell inserts. Invaded cells were electronically counted every 10 min and graphed.
To test this hypothesis, we independently overexpressed the two Numb isoforms by engineering stable inducible cell lines derived from Huh7 by transfection with doxycycline-inducible lentiviral vectors expressing either PRRL or PRRS. To minimize effects of endogenously expressed Numb, we silenced its expression with an siRNA targeting the 3′-UTR, which was absent in the transfected constructs (Figure 2C). We found that PRRL over-expression enhanced, albeit not significantly, Huh7 cell proliferation, while PRRS over-expression significantly decreased cell proliferation (Figure 2D). These data are consistent with the effects of endogenous Numb knockdown in the three HCC cell lines and they generally support the opposing influence of PRRL and PRRS on HCC cell growth.
Importantly, cell death did not contribute to this phenotype, since we observed no significant changes in caspase 3 or poly ADP ribose polymerase levels in Huh1 and Huh7 cells when Numb was silenced (Figure S1B), nor did we find any alteration in the number of apototic or necrotic cells (Figure S1C).
Numb PRRL and PRRS isoforms differentially affect HCC colony formation, motility and invasion
Given these results, we wondered whether similar effects could be observed if growth was assessed under anchorage-independent conditions. Colony formation assays carried out in plates untreated for cell attachment showed that Numb knockdown in SK-Hep1 cells dramatically increased the number of colonies. In contrast, colony number significantly decreased in Huh7, and even more so in Huh1, upon Numb knockdown (Figure 2E). The poor ability of Huh1 cells to form colonies on untreated plastic prompted us to examine the impact of Numb knockdown on Huh1 colony growth in soft agarose, where Huh1 cells form robust colonies. Numb knockdown significantly reduced the number and size of these colonies (Figure S1D, E). Together, these results indicate that Numb PRRL promotes while PRRS suppresses anchorage-independent HCC growth.
We used wound-healing and real-time invasion assay to evaluate possible impact of the two Numb PRR isoforms on cell motility and invasiveness. Wound-healing assay showed that Numb knockdown increased the migration of SK-Hep1 cells (Figure S1F, G), while decreasing migration of Huh1 (Figure S1H, I). Likewise, real-time invasion assay documented a nearly 2-fold increase in SK-Hep1 invasion and a more than 10-fold decrease in Huh1 invasion after Numb knockdown. In Huh1 cells, the strong effect of Numb silencing on cell invasiveness was confirmed using two distinct non-overlapping siRNAs (Figure S1K). While the invasiveness of Huh7 cells was not significantly affected (Figure 2F), these data are consistent with our observation that PRRL over-expression in the context of endogenous Numb silencing had no effect on Huh7 invasiveness, while PRRS over-expression moderately but significantly reduced invasiveness in this background (Figure S1J).
Akt activation and c-Myc expression are affected by modulating expression of Numb PRR isoforms
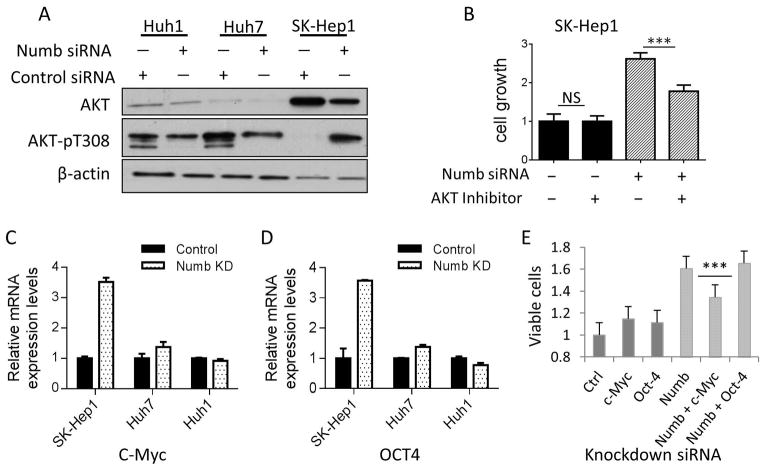
To identify possible mechanism(s) underlying Numb-modulated cell proliferation, we examined the signaling pathways affected by Numb silencing. Western blotting revealed that Akt activation correlated with Numb-dependent modulation of cell growth. Thus, Akt phosphorylation on Thr308 was decreased in Huh1 and Huh7 cells, and increased in SK-Hep1 cells, following Numb knockdown (Figure 3A). Furthermore, the specific Akt inhibitor MK-2206 reduced the increased proliferation in PRRS-expressing SK-Hep1 cells resulting from Numb silencing (Figure 3B).
Figure 3.
Numb-regulated cell growth is mediated by Akt activation and c-Myc expression. A, Numb PRRS knockdown induces Akt phosphorylation on Thr308. Numb knockdown was accomplished with siRNA. Akt protein and phosphorylation on Thr308 were detected by western blot. Actin was used as loading control. B, Akt inhibitor suppresses Numb PRRS knockdown-induced cell growth. SK-Hep1 cells that were transfected with Numb siRNA and treated with Akt inhibitor MK-2206 were cultured for 7 days. Cell proliferation was assessed using MTT assay and expressed as mean cell number on day 7 ± SD (6 wells per condition). C&D, c-Myc and Oct4 mRNA levels are increased in SK-Hep1, Huh7 cell line and decreased in Huh1 cell lines following Numb knockdown. Cells transfected with Numb siRNA were cultured for 3 days. c-Myc and Oct4 mRNA expression were detected using qRT-PCR, and were normalized with the expression levels of actin. Data are expressed as relative expression level (mean ± SD for triplicate samples). E, c-Myc knockdown suppresses Numb PRRS knockdown-induced cell growth. SK-Hep1 cells were transfected with Numb siRNA, together with or without siRNA for c-Myc and Oct4, respectively. Cell growth was measured by MTT assay.
Expression of c-Myc and Oct4 was significantly increased in SK-Hep1 cells upon Numb silencing (Figure 3C & 3D). While Numb knockdown-stimulated SK-Hep1 cell growth was not affected by Oct4 silencing, it was significantly reduced by c-Myc knockdown (Figure 3E). Together, these results are consistent with the possibility that both Akt and c-Myc contribute to Numb-regulated HCC proliferation, particularly when PRRS is the predominant Numb isoform expressed. However, we cannot rule out the influence of cell line variability on these parameters and more work needs to be done to confirm this model.
Numb PRR splicing is reciprocally regulated by Rbfox2 and SRPK2
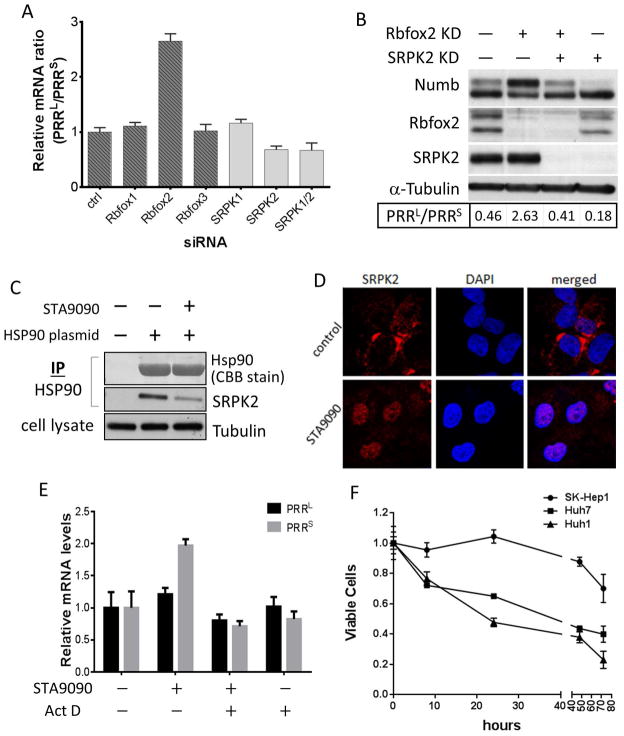
Inclusion of Numb exon 12 in chicken neuronal cells was recently reported to be repressed by Rbfox3, which binds to a conserved UGCAUG sequence in the intron upstream of exon 12 (11). Based on this report, we investigated the possible involvement of the 3 members of the RbFox family of splicing factors in Numb PRR alternative splicing. We found that knockdown of either Rbfox1 or Rbfox3 did not affect the ratio of Numb PRRL to PRRS in both Huh7 and UOK171, a clear cell kidney cancer cell line that, like Huh7, also expresses both Numb PRR isoforms. However, silencing of Rbfox2 dramatically increased the level of Numb PRRL and decreased the level of PRRS, resulting in an elevated ratio of PRRL to PRRS (Figure 4A, darkly shaded bars; Figure S3A).
Figure 4.
Regulation of the alternative splicing of Numb PRR isoforms. A, Rbfox2 silencing increases, while SRPK2 silencing decreases, PRRL/PRRS ratio. Numb mRNA in UOK171 cells with Rbfox or SRPK genes silenced by siRNA was quantitated by qRT-PCR; Numb PRRL/PRRS mRNA levels were calculated and normalized to that in control cells. B, Rbfox2 and SRPK2 function reciprocally. Rbfox2 and SRPK2 in UOK171 were knocked down with siRNA, and the indicated proteins were detected by western blot. C, SRPK2 associates with Hsp90. HEK293A cells were transfected with FLAG-tagged Hsp90, and treated with 1 μM STA9090 for 1 hour. Hsp90 was immunoprecipitated, and SRPK2 was detected by western blot. Hsp90 was stained with Coomassie blue. D, Hsp90 inhibitor induces SRPK2 nuclear translocation. Huh7 cells were treated with 1 μM STA9090 for 40 hrs; SRPK2 was photographed by confocal microscopy. E, Hsp90 inhibitor alters PRRL/PRRS ratio. PRRL and PRRS mRNA levels in Huh7 cells treated with STA9090 and/or actinomycin D were quantitated as above. F, Numb PRRL-expressing HCC are more sensitive to STA9090 than PRRS expressing cells. Indicated HCC cells were treated with 500 nM STA9090 for different times. Cell growth was evaluated by MTT assay.
Another layer of regulation of alternative splicing involves the serine/arginine (SR) protein-specific kinases (SRPKs) that modulate activity and cellular localization of several SR motif-containing splicing factors (12). We examined the role of SRPK1 and SRPK2 in the alternative splicing of Numb exon 12. While SRPK1 knockdown did not affect Numb PRR splicing, SRPK2 knockdown substantially increased the level of PRRS, suggesting that this kinase functions to promote the inclusion of exon 12 (Figure 4A, lightly shaded bars; Figure S3B).
These data indicate that Rbfox2 and SRPK2 exert opposing effects on Numb PRR splicing. To determine whether they work independently or coordinately, we compared the ratio of Numb PRRL and PRRS proteins in cells in which either or both Rbfox2 and SRPK2 were knocked down. In agreement with our earlier data, the PRRL protein level was increased and the PRRS protein level was decreased in cells with reduced Rbfox2 expression, while the opposite outcome was observed in cells with reduced SRPK2 expression (Figure 4B). Interestingly, when expression of Rbfox2 and SRPK2 was simultaneously reduced, the ratio of PRRL to PRRS was restored to a level comparable to that in untreated cells (Figure 4B, Figure S3D), suggesting that SRPK2 and Rbfox2 work in tandem to reciprocally modulate alternative splicing of Numb exon 12 (and, importantly, that additional splicing factors compensate for the simultaneous loss of both proteins).
A number of splicing factors contain SR phosphorylation motifs and are SRPK substrates. Other proteins contain similar motifs but their SRPK-dependent phosphorylation has not been examined. One such protein, RBM6, was recently reported to promote Numb exon 12 inclusion to yield PRRL (13). To determine if RBM6 were downstream of SRPK2, we silenced these two proteins and Rbfox2, either separately or in pairs, and examined the relative abundance of Numb PRRL and PRRS proteins (Figure S3C). Knockdown of RBM6 reduced the ratio of PRRL to PRRS by nearly 67% and countered to some extent the effect of Rbfox2 silencing (27% reversal). However, SRPK2 silencing reduced the PRRL/PRRS ratio by 95% with or without inclusion of RBM6 knockdown, and fully reversed the impact of Rbfox2 silencing (Figure 4B, Figure S3D). Thus, while RBM6 may contribute to Numb PRR splicing in these cells its effect is less dominant than that of SRPK2.
The splicing factor SF2 (SRSF1/ASF) is a known SRPK substrate whose transit from cytoplasm to nucleus depends on phosphorylation of SR motifs (14). Therefore, we investigated whether knockdown of SF2 would phenocopy the effect of SRPK2 silencing on Numb PRR splicing while opposing the effect of Rbfox2 knockdown (Figure S3D). However, SF2 knockdown alone did not alter the PRRL/PRRS ratio and did not counter the effect of Rbfox2 knockdown, ruling out its participation in Numb PRR splicing.
Numb PRR alternative splicing is regulated by the molecular chaperone Hsp90
Given the opposing roles of Numb PRRL and PRRS in regulating cell growth, migration, invasion and colony formation, it is tempting to search for a means of pharmacologically manipulating Numb alternative splicing in cancer cells to increase PRRs or to reduce PRRL. As many kinases, including SRPK1 (15), are dependent on the molecular chaperone Hsp90, we investigated whether SRPK2 may also be an Hsp90 interactor. Immunoprecipitation results indicate that SRPK2 indeed associates with Hsp90 in the cell, and this association can be disrupted by the Hsp90 inhibitor STA9090 (Figure 4C). Using confocal microscopy, we found that, concomitant with its dissociation from Hsp90, SRPK2 markedly relocalized to the nucleus. In contrast, in untreated cells SRPK2 was predominantly cytoplasmic (Figure 4D), suggesting that association with Hsp90 anchors SRPK2 in the cytoplasm.
Next, we determined the impact of STA9090 on Numb PRR splicing. We found that, similar to SRPK2 knockdown, STA9090 increased the abundance of PRRS mRNA and protein relative to PRRL (Figure 4E, Figure S3E), in parallel with STA9090-induced relocalization of SRPK2 to the nucleus (Figure 4D). Importantly, STA9090 did not affect SRPK2 protein stability (Figure S3E), but Hsp90 inhibition did alter the nuclear pattern of protein SR motif phosphorylation (Figure S3F) suggesting that SRPK dissociation from Hsp90 and subsequent localization within the nucleus alters its activity and/or substrate profile. STA9090-induced changes in the mRNA level of Numb PRR isoforms were due to effects on mRNA synthesis rather than turnover, as they were abrogated by actinomycin D (Figure 4E). Finally, a role for Hsp90 in regulating the alternative splicing of Numb PRR isoforms was verified by manipulating Hsp90 expression. When Hsp90 expression was reduced by siRNA-mediated knockdown, the ratio of Numb PRRS to Numb PRRL was increased contemporaneously with SRPK2 nuclear accumulation (Figure S3G & S3H), an effect that was partially reversed by transfection with a plasmid encoding Hsp90 cDNA not targetable by the siRNA (Figure S3H).
Finally, in accordance with their endogenous Numb expression profiles, we found that STA9090 decreased Huh1 and Huh7 proliferation, while minimally affecting growth of SK-Hep1 (Figure 4F & S3I). While increased sensitivity to Hsp90 inhibition could be influenced by factors other than Numb, similarly disparate sensitivity among the three HCC cell lines was not observed upon exposure to other agents, including adriamycin, 5-fluorouracil and AZD8055 (Figure S3J), suggesting that high Numb PRRL expression may uniquely predict for sensitivity to Hsp90-targeted drugs.
DISCUSSION
In searching for the biologic consequences of elevated Numb PRRL expression in HCC tumors, we found that this isoform accelerates proliferation and anchorage-independent growth, and promotes migration and invasion of HCC cells in vitro. In contrast, Numb PRRS suppresses these phenotypes. These functional characteristics of the two PRR splice variants are consistent with our analysis of clinical data indicating that HCC patients whose tumors are characterized by high Numb PRRL expression fared poorly with respect to recurrence after surgery and overall survival. Thus, expression of Numb PRRL may serve as a useful prognostic biomarker for HCC, although this will require further validation.
In chicken neurons, Rbfox3 promotes skipping of Numb exon 12, thus favoring the expression of PRRS (11). The RNA-binding Fox (Rbfox) family consists of three members, Rbfox1-3, with different expression profiles in various tissues and cell types (16). In HCC, we found that Rbfox2, but not Rbfox1 or Rbfox3, regulates Numb PRR alternative splicing. Similar to Rbfox3 in chicken neurons, Rbfox2 promotes skipping of Numb exon 12, as reduced Rbfox2 expression resulted in increased expression of Numb PRRL and decreased expression of Numb PRRS. These results underscore the importance of Rbfox splicing factors in Numb PRR alternative splicing and further emphasize the importance of splicing regulation in cancer (17). Indeed, Rbfox2 is either downregulated or expressed as an inactive variant in ovarian and breast cancers, consistent with the predominance of Numb PRRL in these tumors (18). Such changes in splicing factor expression are not limited to tumors themselves, as Rbfox2 under-expression has been observed in the tumor microenviroment (19).
We found that Numb PRR splicing is further regulated by the SR kinase SRPK2. SRPK2 promotes inclusion of exon 12 and thus favors the expression of Numb PRRL. SRPK2 knockdown increased Numb PRRS and decreased Numb PRRL in HCC, HEK293, and in renal cancer cells. Our data suggest that SRPK2 and Rbfox2 likely function reciprocally to regulate Numb PRR splicing. The mechanism by which SRPK2 affects Numb splicing is a subject of ongoing investigation.
Importantly, we found that Numb PRR splicing can be modulated by the molecular chaperone Hsp90, via its ability to restrict SRPK2 to the cytosol. Hsp90 knockdown or pharmacologic inhibition frees SRPK2 to translocate into the nucleus, thus disrupting its normal function and resulting in increased PRRS, phenocopying the effect of SRPK2 knockdown.
Targeting Hsp90 in cancer is being extensively evaluated in the clinic (20). Like other molecularly targeted agents, Hsp90 inhibitors do not display similar activity in all cancers and predictive biomarkers are urgently needed. Our data raise the possibility that Numb PRRL expression may represent such a biomarker in HCC and perhaps in other cancers. Since Numb PRRL expression also predicts for poor prognosis, evaluation of Hsp90 inhibition as a treatment strategy in HCC patients stratified for Numb PRR isoform expression warrants further clinical investigation.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by the National Natural Science Foundation of China (Grant No. 81302154), by funds from the Intramural Research Program of the U.S. National Cancer Institute, Center for Cancer Research (YYL, WX and LN were supported by grant # ZIA SC 010074; JJ and XWW were supported by grant # Z01-BC010876), and by a National Cancer Institute Career Development Award to WX.
We thank Dr. Sachiyo Kawamoto, Laboratory of Molecular Cardiology, National Heart, Lung, and Blood Institute, NIH, for the generous gift of Rbfox antibodies. We thank Dr. Dom Esposito and Katie Beam of the Protein Expression and Purification Laboratory at the Frederick National Laboratory, NCI for preparing the Numb expressing lentiviruses used in this study. We thank Susan Garfield, Poonam Mannan, and Langston Lim at the CCR Confocal Microscopy Core Facility, Laboratory of Experimental Carcinogenesis, National Cancer Institute, for their help with confocal microcopy. This study was supported by funds from the Intramural Program of the National Cancer Institute (LN).
List of Abbreviations
- HCC
hepatocellular carcinoma
- PRR
proline rich region
- PRRL
PRR long
- PRRS
PRR short
- SR
Serine/arginine
- SRPK
SR protein-specific kinase
- Rbfox
RNA-binding Fox domain containing
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays
- qRT-PCR
quantitative real-time polymerase chain reaction
- siRNA
small interfering RNA
- HSP90
heat shock protein 90
Contributor Information
Yin Ying Lu, Email: luyinying1973@163.com.
Wanping Xu, Email: xuw@mail.nih.gov.
Junfang Ji, Email: jijun@mail.nih.gov.
Dechun Feng, Email: fengdechun@gmail.com.
Carole Sourbier, Email: sourbierc@mail.nih.gov.
Youfeng Yang, Email: youfengy@mail.nih.gov.
Jianhui Qu, Email: qjhzxl@126.com.
Zhen Zeng, Email: zengzhen1970@sina.com.
Chunping Wang, Email: wchping302@yahoo.com.cn.
Xiujuan Chang, Email: anna1chang@msn.com.
Yan Chen, Email: yanchen302@yahoo.com.cn.
Alok Mishra, Email: alokmishra.office@gmail.com.
Max Xu, Email: mxxxuu@gmail.com.
Min-Jung Lee, Email: leemin@mail.nih.gov.
Sunmin Lee, Email: leesun@mail.nih.gov.
Jane Trepel, Email: trepelj@mail.nih.gov.
W. Marston Linehan, Email: linehanm@mail.nih.gov.
Xin Wei Wang, Email: xw3u@nih.gov.
Yongping Yang, Email: yongpingyang@hotmail.com.
Len Neckers, Email: neckersl@mail.nih.gov.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Thomas MB, Jaffe D, Choti MM, Belghiti J, Curley S, Fong Y, Gores G, et al. Hepatocellular carcinoma: consensus recommendations of the National Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol. 2010;28:3994–4005. doi: 10.1200/JCO.2010.28.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan B. Numb--from flies to humans. Brain Dev. 2010;32:293–298. doi: 10.1016/j.braindev.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Li SS. Numb: A new player in EMT. Cell Adh Migr. 2010;4:176–179. doi: 10.4161/cam.4.2.10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verdi JM, Bashirullah A, Goldhawk DE, Kubu CJ, Jamali M, Meakin SO, Lipshitz HD. Distinct human NUMB isoforms regulate differentiation vs. proliferation in the neuronal lineage. Proc Natl Acad Sci U S A. 1999;96:10472–10476. doi: 10.1073/pnas.96.18.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Misquitta-Ali CM, Cheng E, O’Hanlon D, Liu N, McGlade CJ, Tsao MS, Blencowe BJ. Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Mol Cell Biol. 2011;31:138–150. doi: 10.1128/MCB.00709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S, Liu Y, Liu Z, Zhang C, Cao H, Ye Y, Wang S, et al. Transcriptome profiling of a multiple recurrent muscle-invasive urothelial carcinoma of the bladder by deep sequencing. PLoS One. 2014;9:e91466. doi: 10.1371/journal.pone.0091466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zong FY, Fu X, Wei WJ, Luo YG, Heiner M, Cao LJ, Fang Z, et al. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014;10:e1004289. doi: 10.1371/journal.pgen.1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu W, Mimnaugh E, Rosser MF, Nicchitta C, Marcu M, Yarden Y, Neckers L. Sensitivity of mature Erbb2 to geldanamycin is conferred by its kinase domain and is mediated by the chaperone protein Hsp90. J Biol Chem. 2001;276:3702–3708. doi: 10.1074/jbc.M006864200. [DOI] [PubMed] [Google Scholar]
- 11.Kim KK, Nam J, Mukouyama YS, Kawamoto S. Rbfox3-regulated alternative splicing of Numb promotes neuronal differentiation during development. J Cell Biol. 2013;200:443–458. doi: 10.1083/jcb.201206146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giannakouros T, Nikolakaki E, Mylonis I, Georgatsou E. Serine-arginine protein kinases: a small protein kinase family with a large cellular presence. FEBS J. 2011;278:570–586. doi: 10.1111/j.1742-4658.2010.07987.x. [DOI] [PubMed] [Google Scholar]
- 13.Bechara EG, Sebestyen E, Bernardis I, Eyras E, Valcarcel J. RBM5, 6, and 10 Differentially Regulate NUMB Alternative Splicing to Control Cancer Cell Proliferation. Mol Cell. 2013;52:720–733. doi: 10.1016/j.molcel.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 14.Wang HY, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC, Fu XD. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong XY, Ding JH, Adams JA, Ghosh G, Fu XD. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 2009;23:482–495. doi: 10.1101/gad.1752109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ladomery M. Aberrant alternative splicing is another hallmark of cancer. Int J Cell Biol. 2013;2013:463786. doi: 10.1155/2013/463786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, et al. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol. 2009;16:670–676. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- 19.Brosseau JP, Lucier JF, Nwilati H, Thibault P, Garneau D, Gendron D, Durand M, et al. Tumor microenvironment-associated modifications of alternative splicing. RNA. 2014;20:189–201. doi: 10.1261/rna.042168.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neckers L. Heat shock protein 90: the cancer chaperone. J Biosci. 2007;32:517–530. doi: 10.1007/s12038-007-0051-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.