Abstract
CRISPR/Cas9 technology provides a powerful system for genome engineering. However, variable activity across different single guide RNAs (sgRNAs) remains a significant limitation. We have analyzed the molecular features that influence sgRNA stability, activity and loading into Cas9 in vivo. We observe that guanine enrichment and adenine depletion increase sgRNA stability and activity, while loading, nucleosome positioning and Cas9 off-target binding are not major determinants. We additionally identified truncated and 5′ mismatch-containing sgRNAs as efficient alternatives to canonical sgRNAs. Based on these results, we created a predictive sgRNA-scoring algorithm (CRISPRscan.org) that effectively captures the sequence features affecting Cas9/sgRNA activity in vivo. Finally, we show that targeting Cas9 to the germ line using a Cas9-nanos-3′-UTR fusion can generate maternal-zygotic mutants, increase viability and reduce somatic mutations. Together, these results provide novel insights into the determinants that influence Cas9 activity and a framework to identify highly efficient sgRNAs for genome targeting in vivo.
Introduction
Genome editing systems are essential for understanding gene function by means of reverse genetics. Zinc finger nucleases (ZFNs) and TALENs have been broadly used to generate short insertion or deletion (indel) mutations1, 2. These techniques have enabled genetic engineering in multiple biological systems through protein-based recognition of DNA, but are limited by variable efficiency and cumbersome assembly. In prokaryotes, the endogenous CRISPR/Cas9 machinery protects against foreign DNA sequences by generating double strand breaks (DSB)3. This targeting system was recently adapted to eukaryotic cells using two components: the endonuclease Cas9 and a single guide RNA (sgRNA) with complementarity to the target DNA4–6. It has been successfully used to induce targeted genetic mutations in several model organisms such as C. elegans, Drosophila, mouse and zebrafish7–10. However, variable activity of different sgRNAs still represents a significant limitation, resulting in inconsistent CRISPR/Cas9 activity. The molecular features that determine sgRNA stability, loading, and targeting in vivo, remain largely unexplored. Recently, studies performed in human and mouse cell lines have identified features shown to modulate CRISPR/Cas9 activity11, 12. In these studies sgRNA activity was indirectly assessed through phenotypic selection. Thus, the effects of extrinsic features such as the microenvironment mediating DSB DNA repair and selection for deleterious mutations are difficult to separate from the effects of intrinsic features of the sgRNA. Indeed one of the strongest features correlating with high sgRNA activity was a depletion of uridines in the sgRNA, which in fact relates to the termination signal of the Pol III transcription machinery used to express the sgRNAs. Circumventing this problem, Gagnon and collaborators targeted over 100 genes in zebrafish embryos, each with a single sgRNA13. They confirmed that a guanine adjacent to the protospacer adjacent motif (PAM) favors cleavage, in agreement with studies in cell lines11, 12, and suggested other sequence features that correlate with targeting efficiency. However, it is unclear how much these features are influenced by the specific genomic loci tested, given that only one sgRNA was evaluated per gene. This could limit the capacity to identify specific features that mediate sgRNA activity, such as stability, Cas9 loading and target recognition. Consequently, the principles underlying effective targeting using CRISPR/Cas9 system in vivo are still largely unknown.
In this study, we analyzed the stability, loading into Cas9 and mutagenic activity of 1280 sgRNAs targeting 128 genes. This revealed that efficient sgRNAs have biased sequence composition that affect CRISPR/Cas9 stability and activity. We compared the efficiency of 640 alternative sgRNAs (truncated, extended and 5′ mismatch containing) increasing the number of target sites in the genome. We have integrated these findings into a predictive model (CRISPRscan.org). Finally, we designed a Cas9 construct that targets mutations preferentially in the germ line, reducing somatic mutations allowing the generation of maternal mutants. Together these findings improve the CRISPR/Cas9 system providing insights into the factors that influence sgRNA activity.
Results
Measuring the activity of >1000 sgRNAs
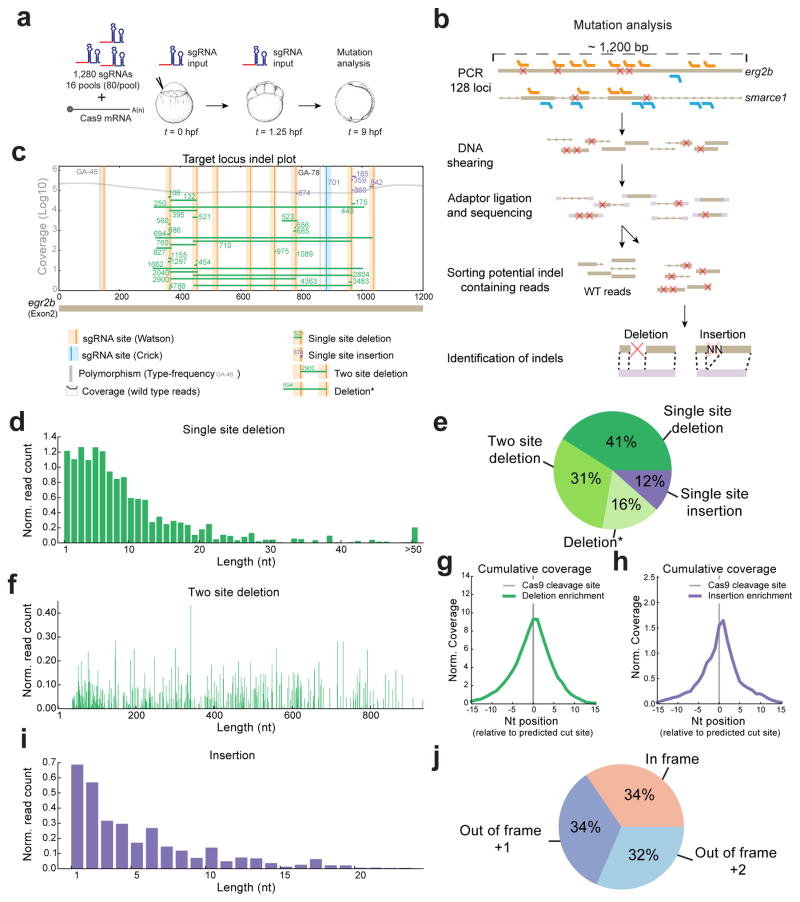
To determine the factors that influence the generation of sgRNA-mediated DNA lesions, we first measured the activity of 1280 sgRNAs targeting 128 different genes in the zebrafish genome. For each gene, we designed a total of 10 sgRNAs falling within a 1.2kb span (Supplementary Fig. 1a), with the majority (96%) targeting exons. We reasoned that in vitro transcribed sgRNAs (Supplementary Fig. 1c, d) would allow us to measure the influence of sgRNA stability, Cas9-loading and target sequence composition independently of transcription rates that might differ in DNA-based libraries typically used in cell culture. To this end, we first injected 16 pools of 80 sgRNAs (targeting 8 loci per pool) together with Cas9-encoding mRNA into zebrafish embryos at 1 cell stage (Fig. 1a). We analyzed insertion-deletion frequency at 9 hpf (hours post fertilization) using high-throughput sequencing, and additionally collected embryos at 0 and 1.25 hpf to measure input sgRNA levels (Fig. 1b, Supplementary Fig. 1b, Supplementary Table 1, Supplementary Data 1 and see Methods). As a control, we sequenced uninjected sibling embryos and discarded 37 target sites presenting polymorphism with the reference zebrafish genome. We measured individual sgRNA activity as the fraction of reads containing indels over the total number of reads in each site. We observed that the vast majority of sgRNAs induced deletions (88%) (Fig. 1c, e). Their activity ranged from 0 to 77%, even among sgRNAs within the same locus, indicating that the wide range of efficiency depends on additional factors that are independent of the targeted locus. Deletions caused by individual sgRNAs had a median length of 7nt, and represented >40% of the events (Fig. 1d and e). Insertions were shorter (median 4nt) and represented 12% of the events (Fig. 1e, i). Because multiple different sgRNAs targeting the same locus were co-injected, we observed a large fraction of deletions spanning two sgRNA target sites (Two site deletion) (31%) with median length 400nt, and as high as 900nt (Fig. 1e and f). Indels had the maximum enrichment at the predicted position of the cleavage site, 3nt upstream of the PAM motif14 and showed no 5′ or 3′ bias (Fig. 1g, h, Supplementary Fig. 1d). Indeed, when we calculated the percentage of indels that maintain the frame or cause frame-shifts, we observed an even distribution of frames after repair (Fig. 1j). Taken together these results indicate that sgRNA induced mutations comprise a wide range of efficiencies causing mainly deletions. This provides a valuable dataset to identify the determinants that influence cleavage efficiency and derive better rules to predict CRISPR/Cas9 activity.
Figure 1. Measuring the activity of >1000 sgRNAs.
a. Schematic diagram illustrating the experimental setup to analyze CRISPR/Cas9 mediated mutations in vivo. 1280 sgRNAs in pools of 80 along with cas9 mRNA were injected into 1-cell stage zebrafish embryos collected after 9 hours of development. In parallel, sgRNA levels at 0 and 1.25 hpf were measured to analyze sgRNA stability and to normalize sgRNA activity.
b. Schematic diagram illustrating the bioinformatics pipeline developed to characterize CRISPR/Cas9 induced mutations in vivo. egr2b and smarce1 are two representative genes among the 128 targeted loci.
c. Schematic representation of deletions and insertions found on the egr2b gene locus with read coverage (grey). Vertical bars represent sgRNA target sites with the solid line indicating the Cas9 cleavage site. Horizontal bars represent deletions (green) and insertions (purple) with the supporting number of reads. Vertical grey bars show polymorphisms.
d. Histogram of deletion lengths induced by single sgRNAs (median of 7 nt).
e. Distribution of sgRNA mutations caused by a single or between two site(s). *Deletion with boundaries that cannot be unambiguously assigned to a single or a two site(s) deletion (see Methods).
f. Histogram of deletion lengths induced by sgRNA pairs (median of 400 nt).
g. Cumulative coverage of mutated reads overlapping single site deletions normalized by WT reads.
h. Cumulative coverage of mutated reads overlapping single site insertions normalized by WT reads.
i. Histogram of insertion lengths induced by single sgRNAs (median of 4 nt).
j. Distribution of frame shifts caused by all CRISPR/Cas9 induced mutations.
Stable sgRNAs are more active, G-rich and A-depleted
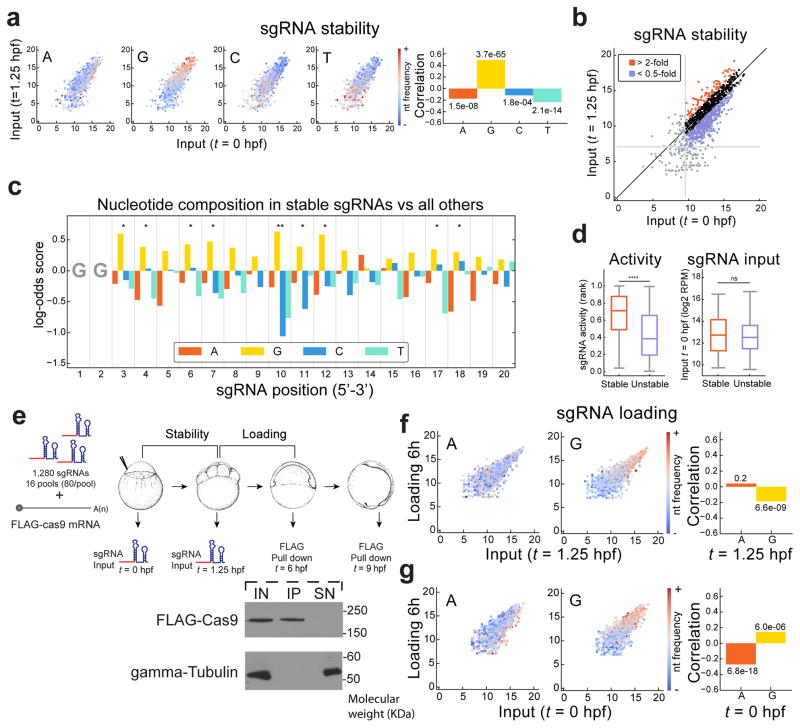
DNA-mediated delivery of sgRNAs typically consists of constitutive expression systems. In contrast, direct delivery of in vitro transcribed sgRNAs allows us to isolate the effects of sgRNA stability and loading into the Cas9 complex. To this end, we compared sgRNA levels at 0 hpf (injected input) and 1.25 hours after injection using high-throughput sequencing (Fig. 1a; see Methods). Analysis of sgRNA nucleotide composition revealed a reproducible and significant enrichment of guanine and depletion of adenine in more stable sgRNAs (p=3.7e-65 and 1.5e-8 respectively; Fig. 2a, c and Supplementary Fig. 2a). Next, we analyzed whether stability could modulate sgRNA activity. We partitioned the sgRNAs into stable and unstable groups by comparing levels in the input vs. 1.25 hpf (Fig. 2b and Supplementary Fig. 2b). While these two groups had similar input levels at 0 hpf, the mutagenic activity of the stable sgRNAs was significantly higher than the other group (p=6.9e-12, Mann–Whitney U test) (Fig. 2d and Supplementary Fig. 2c). Additionally, mutagenic activity and sgRNA levels were significantly correlated (r=0.39 p=2.1e-39, Supplementary Fig. 3a). Thus, sgRNA stability represents an important determinant of sgRNA function.
Figure 2. Stable sgRNAs are more active, G-rich and A-depleted.
a. Biplot of sgRNA levels (log2 RPM) comparing 0 and 1.25 hpf (Experiment shown in Figure 1), colored to indicate the frequencies of the 4 nucleotides in each sgRNA. Corresponding Spearman correlations between nucleotide frequencies and sgRNA stability (ratio of 1.25 hpf to 0 hpf levels) are shown (right), with p values indicated.
b. Biplot illustrating stable and unstable groups of sgRNAs, defined by >2-fold enrichment or depletion between 0 and 1.25 hpf (log2 RPM). sgRNAs with low read-counts (bottom 10%) were excluded (grey lines).
c. Barplot representing the nucleotide composition of the 20% most stable sgRNAs compared to all others. Bars show log-odds scores of nucleotide frequencies for each position in the sgRNA (1 to 20) (G-test: * <0.05, ** <0.01).
d. Box and whisker plots (Box spans first to last quartiles with 1.5x interquartile range distance for whiskers) showing sgRNA activity (left) and the input levels (right) in the stable and unstable sgRNAs. Mann-Whitney U test (**** p< 0.0001, ns: not significant).
e. Diagram illustrating the experiment to analyze the Cas9 loading activity in vivo. One-cell stage embryos were injected with 1280 sgRNAs in pools of 80 along with FLAG-cas9 mRNA (see Methods). sgRNA levels were measured at 0 and 1.25 hpf and immunoprecipitation of FLAG-Cas9 was performed at 6 and 9 hpf to analyze levels of loaded sgRNAs. Analysis of the immunoprecipitation of FLAG-Cas9 at 6 hpf (bottom). 1/50 of the total input (IN), the immunoprecipitation (IP) and the supernatant after the immunoprecipitation (SN) were analyzed by western blot using FLAG and gamma-tubulin (as loading control) antibodies.
f. Biplot of sgRNA levels (log2 RPM) comparing 1.25 hpf and loaded into Cas9 at 6 hpf, colored to indicate the frequencies of A and G in each sgRNA. Corresponding Spearman correlations between nucleotide frequencies and sgRNA stability (ratio of loaded at 6h to 1.25 hpf levels) are shown (right) with p values indicated.
g. Biplot of sgRNA levels comparing 0 hpf and loaded into Cas9 at 6 hpf. (see panel f).
Guanine and cytosine-rich RNAs fold into stable structures. Thus, we hypothesized that sgRNA folding energy could explain the guanine enrichment in stable sgRNAs. To test this, we folded in silico all sgRNAs (including the 80nt tail) to compute their ensemble free energies (EFEs). We observed a significant anti-correlation between sgRNA stability and EFE (r=−0.495, p=2.2e-62) (Supplementary Fig. 3b). sgRNA EFEs also significantly anti-correlated with guanine content (r=−0.475, p=3.8e-57) but cytosine content correlated weakly (r=0.073, p=0.0021), suggesting that guanine enrichment is not a mere consequence of lower folding energy. We postulated that guanines may protect against 5′-directed exonuclease degradation as this enrichment occurred more prominently at the 5′ end of the sgRNA (Fig. 2c and Supplementary Fig. 2a). Guanine-rich sequences can fold into stable non-canonical structures called G-quadruplexes in vivo15. Indeed, a stronger correlation between guanine content and stability was observed when more than 8 guanines were present in the sgRNA, which is the minimal requirement to form a G-quadruplex (Supplementary Fig. 3c). To further support this, we tested a subset of stable and unstable sgRNAs for their ability to fold into G-quadruplex structures using in-line probing16. Notably, 7 out of 9 guanine-rich stable sgRNAs tested were able to form G-quadruplexes compared to none out of 5 unstable sgRNAs (Supplementary Fig. 3d, e and Supplementary Table 2). These results suggest that G-quadruplex formation contributes to sgRNA stability.
Next, we examined whether the loading of sgRNAs into Cas9 was influenced by their nucleotide sequence using a FLAG-Cas9 (Fig. 2e and Supplementary Fig. 3f). We compared the total sgRNA composition at 1.25 hpf with the sgRNAs loaded into FLAG-Cas9, after immunoprecipitation at 6hpf. We observed a small but significant guanine depletion suggesting that Cas9 might slightly disfavor loading guanine-rich sgRNAs (p=6.6e-9; Fig. 2f and Supplementary Fig. 2d). However, this effect was negligible compared to the guanine enrichment observed between 0 and 1.25 hpf (Fig. 2a, c and Supplementary Fig. 2a). Indeed, when we compared loaded sgRNA levels to the input at 0 hpf, we still observed a significant guanine enrichment together with an adenine depletion (p=6.0e-6; Fig. 2g and Supplementary Fig. 2f, g) correlating with a higher level of activity (Supplementary Fig. 2h, i). Preferentially loaded sgRNAs were also slightly enriched for cytosines in several positions (Supplementary Fig. 2d, e). Time course analysis of sgRNAs loaded in to Cas9 at 6 hpf and 9 hpf suggested that sgRNAs are stably retained into Cas9 once they are loaded, since we did not detect a significant difference between time-points (Supplementary Fig. 3g). Together, these results show that sgRNA stability in vivo is strongly influenced by their nucleotide composition, favored by guanines and disfavored by adenines, modulating sgRNA activity.
CRISPR/Cas9 activity is modulated by the sgRNA sequence
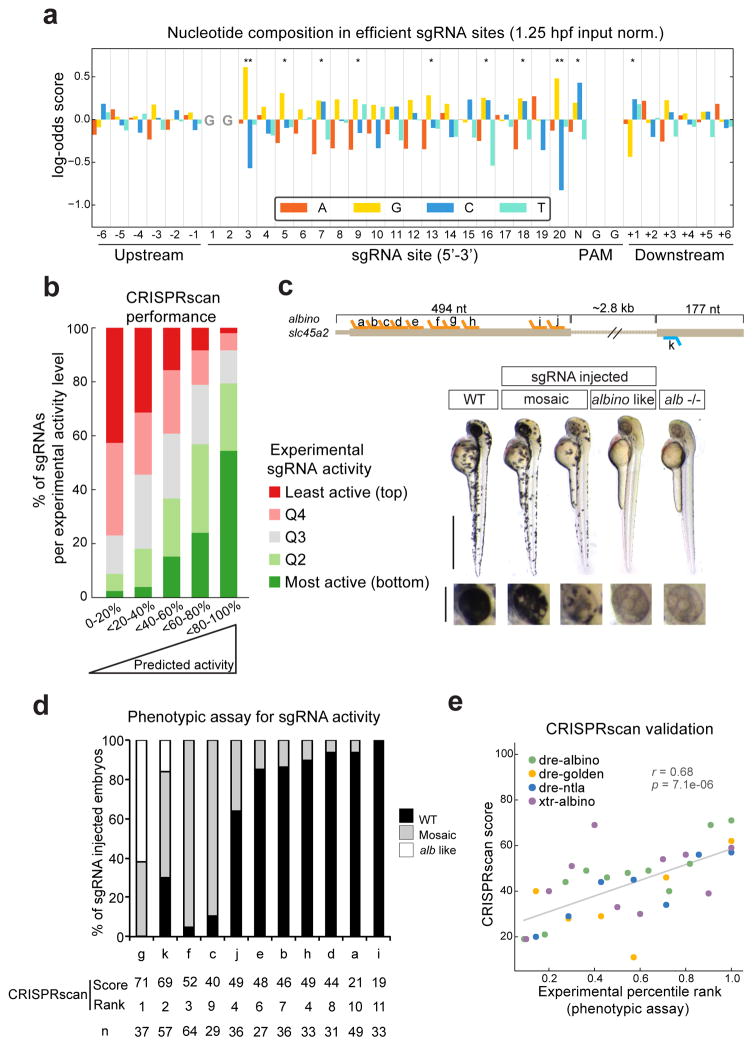
To test whether the mutagenic activity of sgRNAs is influenced by their sequence, we examined the nucleotide composition of efficient sgRNAs for each position in the target site extended by 6 nucleotides upstream and downstream. To exclude the effect of sgRNA stability, we normalized sgRNA activity by its input level at 1.25 hpf. We identified ten positions with significantly different nucleotide distributions in the most efficient sgRNAs (top 20%) (Fig. 3a). First, we observed a strong guanine enrichment in positions 3 and 20 (adjacent to the PAM) of the sgRNA. Both positions were balanced by a strong cytosine depletion. Second, nucleotides distal to the PAM (positions 1–14) were dominated by guanine enrichment, while positions 15–18 were characterized by cytosine enrichment. Thymidine and adenine nucleotides were depleted overall, with the exception of positions 9 and 10. Lastly, outside the sgRNA-binding site we observed (i) a cytosine/guanine enrichment overlapping the first nucleotide of the PAM and (ii) a guanine depletion one nucleotide downstream of the PAM. Thus, specific sgRNA sequences mediate efficient sgRNA-Cas9 target recognition and cleavage.
Figure 3. CRISPR/Cas9 activity is modulated by the sgRNA sequence.
a. Barplot representing the nucleotide composition of the 20% most efficient sgRNA sites (positions 1 to 20 extended by the PAM sequence and 6 nt) compared to all others. Bars show log-odds scores of nucleotide frequencies for each position (G-test: * <0.05, ** <0.01).
b. Performance of linear regression based prediction model (CRISPRscan). sgRNAs were divided into quintiles based on CRISPRscan scores (horizontal axis), then each quintile was evaluated based on their experimentally determined activities (colors indicate five activity levels).
c. Diagram showing 11 sgRNA sites targeting albino exons 1 and 2 used in an independent validation of the prediction model. Phenotypes obtained after the injection of the sgRNAs, showing different levels of mosaicism compared to the wild type (WT) (bottom). Lateral views and insets of the eyes of 48 hpf embryos are shown. Picture of an albino loss of function mutant (−/−) described in White et al.17 (right). (scale bars: 1mm, 0.25mm inset)
d. Phenotypic evaluation of 11 sgRNA targeting albino. Stacked barplots showing the percentage of albino like (white), mosaic (gray) and phenotypically WT (black) embryos 48 hpf after injection. Predicted CRISPRscan scores, ranks and number of embryos evaluated (n) are shown for each sgRNA.
e. Scatter plot showing the correlation between CRISPRscan scores and experimentally measured activities based on all phenotypes used to independently validate CRISPRscan (Panel c and d and Supplementary Fig. 4). Spearman correlation and p value are indicated.
Next, we integrated these observations into a model to predict mutagenic activity based on sgRNA target sequences. We used a randomized logistic regression to select stable features that were the strongest determinants of sgRNA efficiency. Using these selected features, we trained a linear regression model on ranked sgRNA activity normalized by the input at 0 hpf (Supplementary Table 3). The resulting sgRNA scoring method, which we named CRISPRscan, performed strongly when evaluated against our experimental data (r=0.58, p=7.1e-93, Pearson’s correlation between predicted and experimental sgRNA activity). For example, of the top-scoring quintile of sgRNAs (CRISPRscan score > 0.6), 54% were among the most active sgRNAs by experimental evaluation, while only 2% were among the least active (Fig. 3b). To ensure that our prediction model was not over-trained and was generalizable to other sgRNAs, we performed cross-validations (see Methods), which confirmed the performance of our model (r=0.45, sd=0.071). Finally, we independently validated CRISPRscan by determining the efficiency of 35 different sgRNAs targeting the albino (slc45a2), golden (slc24a5) and no-tail (ntla) loci in zebrafish, as well as the albino (slc45a2) locus in Xenopus tropicalis (Fig. 3c, d and Supplementary Fig. 4)17. We observed a significant high correlation between CRISPRscan scores and phenotypic experimental activities (r=0.68, p=7.1e-06) (Fig. 3e). In most cases at least two sgRNAs with the highest CRISPRscan scores were the most active in vivo. Based on the experimental validation, scores above 0.55 identified efficient sgRNAs, while highly efficient ones scored above 0.70. In addition, we tested whether the mutagenic activity of sgRNA-Cas9 complexes was influenced by (i) the accessibility of the chromatin at the targeted locus, and (ii) competition by putative off-target binding sites, but these did not significantly affect CRISPR/Cas9 activity (Supplementary Fig. 5). Together, these results show that sequence composition of the sgRNA and the target strongly influence the mutagenic activity of the CRISPR/Cas9 system and CRISPRscan successfully identifies the most active sgRNAs that mediate mutagenesis in vivo.
Alternative sgRNA formulations have variable activity
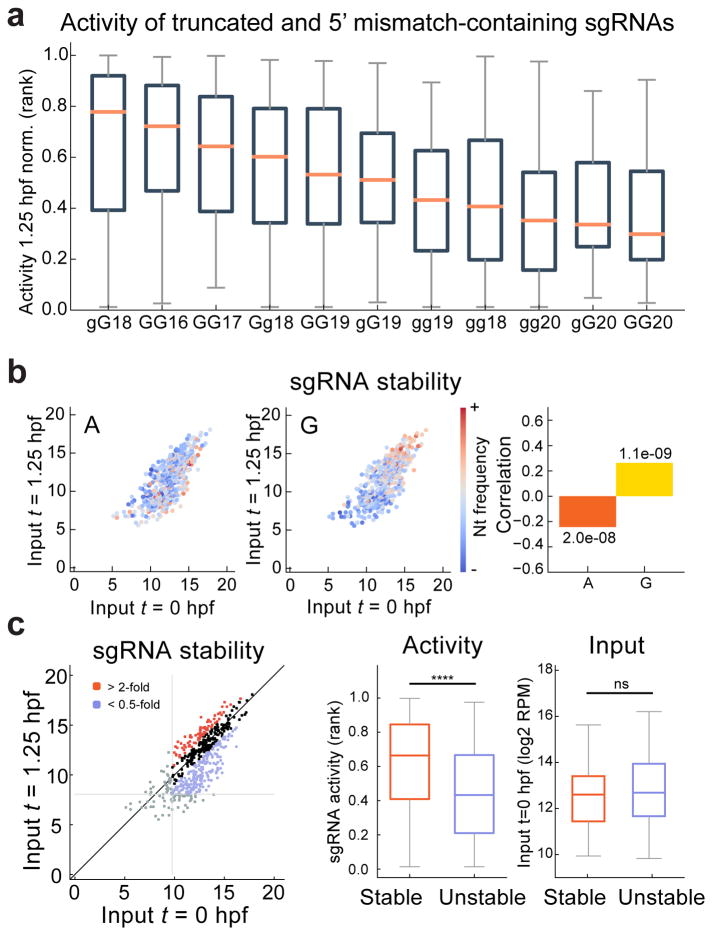
Sequence specificity of the T7 or SP6 promoters restricts 5′ sgRNA sequences to GG or GA, limiting the number of available targets in vivo. To extend the repertoire of potential targeting sites in the genome, we evaluated sgRNAs of various lengths (18–22nt), with up to two nucleotide mismatches in positions 1,2 of the sgRNA (mismatch denoted g); these were termed alternative sgRNAs (Supplementary Fig. 6b). We compared the activity of 640 alternative sgRNAs spanning 11 different sgRNA formulations targeting a total of 64 loci (Supplementary Fig. 6a–c, Supplementary Table 1 and Supplementary Data 2). We observed significantly different activities among them (p=1.0e-9, Kruskal-Wallis H-test). The most efficient alternatives to the canonical sgRNAs were sgRNAs shorter at the 5′ end by 1–2nt, or sgRNAs of canonical length but inducing 1 mismatch in the 5′GG (Fig. 4a). In contrast, longer sgRNAs were less effective, particularly those containing a 22nt binding sequence. The average activity of alternative sgRNAs decreased with sequence variants in the following order: gG18 ~ GG16 ~ GG17 ~ Gg18 > GG19 ~ gG19 > gg19 ~ gg18 > gg20 ~ gG20 > GG20 (Fig. 4a and Supplementary Fig. 6b). Consistent with the stability features described above for canonical sgRNAs, alternative sgRNAs were more stable when enriched in guanine and depleted for adenine, which resulted in higher activity (Fig. 4b, c and Supplementary Fig. 6d). Stability among the different types of alternative sgRNAs was not significantly different (p=0.99, χ2 test). Next, we directly compared the activity of the canonical and alternative sgRNAs targeting the same site in the albino and golden loci (GG18-16nt and GA18 vs Gg18) (Supplementary Fig. 6e–g). Most sgRNAs (8/10) showed no significant difference in the generation of bi-allelic mutation, as quantified by the loss of pigmentation in the F0 injected embryos (Supplementary Fig. 6f, h). Thus, the activity of the most efficient alternative sgRNAs is equivalent to that of canonical sgRNAs. Indeed, we were able to adapt CRISPRscan to predict the activity of Gg18, GG17, and gG18, but not of GG16 (Supplementary Fig. 6i–j). These results suggest that CRISPRscan can infer the efficiency of those sgRNAs increasing by 8-fold (from ~5e6 to ~44e6 sites) the number of potential target sites in the zebrafish genome.
Figure 4. Extending the CRISPR target repertoire with truncated, extended and 5′ mismatch-containing sgRNAs.
a. Boxplot representing the ranked normalized activity of each class of alternative sgRNAs, ordered by median activity. Shorter sgRNAs (GG16 and GG17) and sgRNAs inducing 1 mismatch in the 5′ GG (gG18 and Gg18) are the most active alternatives to the canonical GG18 sgRNA.
b. Biplot of sgRNA levels (log2 RPM) comparing 0 and 1.25 hpf, colored to indicate the frequencies of A and G in each sgRNA. Corresponding Spearman correlations between nucleotide frequencies and sgRNA stability (ratio of 1.25 hpf to 0 hpf levels) are shown (right), with p values indicated.
c. Biplot illustrating stable and unstable groups of sgRNAs, defined by >2-fold enrichment or depletion between 0 and 1.25 hpf (log2 RPM) (left). sgRNAs with low read-counts (bottom 10%) were excluded (grey lines). Box and whisker plots showing sgRNA activity (middle) and the input levels (right) in the stable and unstable sgRNAs. Mann-Whitney U test (**** p< 0.0001, ns: not significant).
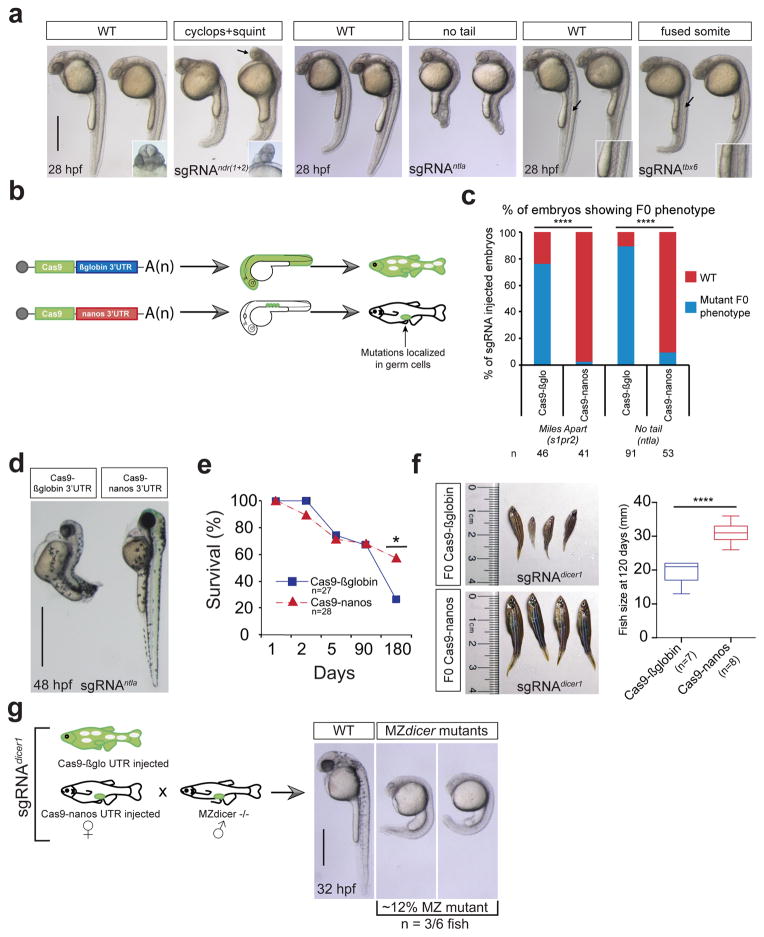
Targeting CRISPR/Cas9 activity to germ cells
CRISPRscan efficiently detects the most active sgRNAs in vivo. However, bi-allelic mutations derived from the use of highly efficient sgRNAs, can result in a lethal phenotype for many essential genes (Supplementary Fig. 4c, Fig. 5a, Supplementary Fig. 7). Concentrating the mutagenic activity of Cas9 to the germ line would (i) minimize lethality due to somatic mutations, and (ii) allow bi-allelic mutations in the germ cells removing the maternal contribution of the targeted gene. To this end we localized CRISPR/Cas9 expression to the germ line by fusing the cas9 ORF to the 3′-UTR of nanos118, 19 (Fig. 5b). To test this method we mutagenized dicer1, ntla and s1pr2, whose zygotic loss of function impairs larval growth, notochord development, and heart development respectively20–23. We compared the activity of Cas9-nanos-3′-UTR (Cas9-nanos) or Cas9-βglobin 3′-UTR (Cas9-βglobin) co-injected into one-cell stage embryos with sgRNAs to target ntla and dicer1 (Fig. 5c–f, Supplementary Fig. 8a). The majority of embryos showed the expected phenotype when injected with Cas9-βglobin but not with Cas9-nanos, suggesting that Cas9-nanos reduces the rate of somatic mutations and embryonic lethality. For example, when targeting the dicer1 locus, the viability and size of Cas9-βglobin injected fish were dramatically reduced and none of the females were fertile (Fig. 5e, f). In contrast, Cas9-nanos injected fish presented normal growth and homozygous mutant germ cells in 50% of the females, resulting in the generation of maternal zygotic dicer mutant embryos24 (Fig. 5g). Quantification of MZdicer mutant embryos revealed that 12% of the germ line was homozygous dicer mutant. Similarly, nlta Cas9-nanos injected fish also resulted in a high rate of germ line transmission for nlta mutations (in 50% of the chromosomes) (Supplementary Fig. 8b). Together, these results show that germ-line targeting of Cas9 can generate MZ and Z mutants for genes whose function is required during embryonic and larval development. Coupled with the increased target site repertoire and optimized sgRNA sequence design rules, these findings provide a valuable characterization of the elements that influence activity of the CRISPR/Cas9 system, with the potential to increase both the efficiency and applicability of CRISPR/Cas9-mediated mutagenesis.
Figure 5. Targeting CRISPR/Cas9 activity to germ cells.
a. Wild-type embryos were injected with a combination of 3 sgRNAs (20 pg each) targeting ntla, tbx6 or ndr1/2 and 100 pg of cas9 mRNA (150 in the case of ndr1/2). Pictures were taken at 28 hpf (lateral view). Arrows are indicating cyclopia (ndr1/2) and fused somites (tbx6). (scale bar: 0.5mm)
b. Schema illustrating the Cas9-nanos 3′-UTR strategy. Nanos 3′-UTR was cloned after Cas9 ORF. Injection of Cas9-nanos will concentrate the expression in the germ cells (green circles).
c. Stacked barplot showing the percentage of coherent F0 phenotype (mutant phenotype) or WT after injection with a combination of 3 sgRNAs (20 pg each) targeting s1pr2 or ntla and using cas9-globin or cas9-nanos mRNA (100 pg). n= number of embryos analyzed. χ2 test (**** p< 0.0001).
d. Individual pictures (lateral view) of 48 hpf old embryos injected with the same sgRNA/Cas9 combinations described in panel c (targeting ntla). (scale bar: 1mm)
e. Survival curve of Cas9-nanos or Cas9-globin injected fishes. χ2 test (* < 0.05) Embryos were injected with the same sgRNA/Cas9 combinations described in panel c (targeting dicer1).
f. Pictures showing fish injected with sgRNA (dicer1) and Cas9-nanos or Cas9-globin 4 months of age. Box and whisker plot showing their size distribution (right). n= number of embryos analyzed. Two-tailed Student’s t test (**** p< 0.0001).
g. Scheme illustrating an F0 out cross between sgRNA (dicer1), Cas9-nanos or Cas9-globin injected female fish and dicer1 −/− mutants males generated by germ line transplantation24. Pictures of 32 hpf MZ dicer1 embryos derived from F0 females injected with Cas9-nanos and sgRNA (dicer1) (right). (scale bar: 0.5mm)
Discussion
Our study provides three insights into the factors that determine the mutagenic activity of CRISPR/Cas9 system in vivo. First, guanine-rich and adenine-depleted sgRNAs are more stable and correspondingly, more mutagenic. While increased stability could be explained by sgRNA folding energy, we also observed that guanine accumulation in the 5′ end resulted in the formation of G-quadruplexes suggesting guanines may protect against 5′-directed exonuclease degradation. These structured sgRNAs could prevent target recognition. However, G-quadruplex forming sgRNA sequences could also benefit from the formation of a G-quadruplex structure on the complementary DNA strand at their targeting site. The presence of this structure in double stranded DNA stabilize R-loops25, which are formed in the CRISPR/Cas9-DNA interaction complexes14. This might also stabilize the sgRNA-Cas9 complex to its cytosine-rich target site consequently increasing the activity.
Second, we uncovered sequence specificity of Cas9/sgRNA targeting that was integrated into a predictive model called CRISPRscan. Consistent with previous studies, we observed a strong guanine enrichment at position 20 of the sgRNA (Fig. 3a)13. Also, this bias was extended by an additional guanine to a GG motif starting at position 1926. Interestingly, this dinucleotide has the second highest coefficient in the CRISPRscan model (Supplementary Table 3). Completing this observation, we find a corresponding depletion in cytosine at position 20 in vivo11. Furthermore, we observed that (i) guanine and cytosine are enriched at the first nucleotide of the PAM sequence, reconciling independent observations by Kuscu et al.27 and Doench et al.11 and (ii) position 3 is strongly enriched for guanine and depleted for cytosine. Our study capitalizes on these data to train a linear regression model to predict sgRNA activity (CRISPRscan). CRISPRscan scores show a stronger correlation with sgRNA activity than previous approaches11, 13, not only in zebrafish but also in X. tropicalis (Supplementary Fig. 9a–d). While folding energy is correlated with stability of the sgRNA, introducing this feature in CRISPRscan did not improve the predictions for 35 experimentally validated sgRNAs. Furthermore, criterion commonly used to select sgRNA site such as GC content within the sgRNA between 40% and 80%12, 28 were outclassed by CRISPRscan (Supplementary Fig. 9e). Finally, analysis of 132 canonical sgRNAs29 recently published by the Burgess laboratory, revealed that CRISPRscan scores were significantly higher (p=2.7e-3, Mann–Whitney U test; Supplementary Fig. 9f) for sgRNAs with higher germline transmission rates. Together, CRISPRscan provides a valuable resource to predict the most efficient sgRNA(s), and will facilitate direct functional screenings in vivo30.
Finally, we proposed two optimizations that increase the number of efficient targets and concentrate mutations to germ cells. Consistent with previous studies31, 32, we observe that truncated sgRNAs are as stable and efficient as canonical sgRNAs in vivo. Conversely, sgRNAs with more than 20nt complementarity to the target were less efficient, consistent with studies in cell lines33 and Drosophila32. Designing efficient alternative sgRNAs (truncated, with 5′ mismatches) increases the number of available target sites up to 8-fold in the zebrafish genome. Such an improvement is also important for precise targeting of short regions in the genome such as small ORFs, or specific DNA and RNA functional elements, such as transcription factor binding sites, or miRNA target sites.
The use of a Cas9-nanos-3′-UTR fusion concentrates mutagenic activity to the germ cells. This reduces lethality due to deleterious phenotypes in somatic tissue and allows the generation of maternal-zygotic mutants in the F1, a process that otherwise requires laborious germ line replacement34. Given that mRNAs representing over 70% of genes are maternally provided to the early embryo35, this method provides an entry point toward characterizing complete loss-of-function phenotypes during embryogenesis.
Together, this resource provides an accessible framework for designing the most efficient sgRNAs for in vivo targeting, particularly in zebrafish and insights into the determinants that mediate sgRNA efficiency.
Online Methods
Target sites design
Ten sgRNA target sites were designed within ~1.2kb locus for 128 loci for a total of 1280 sgRNAs. Among these, 1232 were located in exons, with 1217 targeting coding sequences. Target sites were spaced on average by 81 nt with a minimum distance of 18 nt (Supplementary Fig. 1a) and had a maximum of 10 off-targets with 1 mismatch. Gene annotations from Ensembl 7436 were used.
sgRNA and cas9 mRNA generation
sgRNA DNA template was generated by fill in PCR (Supplementary Fig. 1c and d). Briefly, a 52 nt oligo (sgRNA primer), containing the T7 promoter (Supplementary Table 1 #1), the 20 nt of the specific sgRNA DNA binding sequence and a constant 15 nt tail for annealing, was used in combination with a 80 nt reverse oligo to add the sgRNA invariable 3′ end (tail primer). A 117 bp PCR product was generated following these parameters: 3 minutes at 95°C, 30 cycles of 30 seconds at 95°C, 30 seconds at 45°C and 30 seconds at 72°C, and a final step at 72°C for seven minutes. PCR products were purified using Qiaquick (Qiagen) columns and approximately 120–150 ng of DNA were used as template for a T7 In vitro transcription (IVT) reaction (AmpliScribe-T7-Flash transcription kit from Epicentre) (Supplementary Fig. 1c). In vitro transcribed sgRNAs were DNAse treated and precipitated with Sodium Acetate/Ethanol. Alternative sgRNAs were generated similarly using shorter (50 or 51 nt) and longer (53 or 54nt) sgRNA primers, with 18–22nt complementary to the target. SgRNAs beginning with GA sequences contained the SP6 promoter (Supplementary Table 1 #2) instead of the T7 promoter. A MAXIscript SP6 transcription Kit (Life technologies) was used for SP6 based IVT reactions.
Zebrafish codon-optimized protein from pT3TS-nCas9n (#2656)37 was used in all experiments except for the pull down, where FLAG-Cas9 was employed. A N-terminal 3xFLAG-tag was cloned in pT3TS-nCas9n in the NcoI site. The resulting pT3TS-FLAG-nCas9n (#2722) plasmid is identical to one previously used for a similar experiment in cell lines4, 12. For the cas9-nanos 3′-UTR construct, the nanos 3′-UTR and SV40 late polyA signal was PCR amplified from a previous plasmid pCS2+GFP-nanos 3′UTR18, 19 using two oligos (Supplementary Table 1 #3 #4). The following PCR product was then digested in 3′ with NotI and ligated into the pCS2-nCas9n 37 plasmid previously digested with SnaBI and NotI. The final pCS2-nCas9n-nanos 3′UTR (#2662) (addgene #62542) construct was confirmed by sequencing. Cas9 mRNA was in vitro transcribed from DNA linearized by either NotI (pCS2-nCas9n-nanos 3′UTR) or XbaI (pT3TS-nCas9n and pT3TS-FLAG-nCas9n) using the mMachine SP6 or T3 kit (Ambion), respectively. In vitro transcribed mRNAs were DNAse treated and purified using RNeasy Mini Kit (Qiagen).
RNA injections, large-scale experiments and plasmids
For large-scale experiments, 1280 sgRNAs (Supplementary Table 1) were injected in 16 different cocktails. Independent injections containing 240 pg from 80 sgRNAs targeting 8 genes and 300 pg of cas9 mRNA were carried out at one-cell stage. Five embryos per injection (80 in total) were collected at 1.25 hpf for sgRNA input. Seven embryos per injection were collected at 6 and 9 hpf (102 in total) for the Cas9 loading experiment (see below). Twenty embryos per injection were collected at 9 hpf and DNA was extracted following the HotShot protocol38 with minor modifications. Briefly, a ~1.2 kb PCR product was obtained for each of the 128 loci (Supplementary Table 1) using these parameters: 3 minutes at 95°C, 35 cycles of 30 seconds at 95°C, 30 seconds at 60°C and 2 minutes at 72°C, and a final step at 72°C for seven minutes. Forty non-injected embryos were collected and analyzed to determine possible polymorphisms. PCR products were visualized and quantified on agarose gel (Adobe Photoshop). Next, similar amounts of PCR products per gene were pooled and purified (QIAquick PCR purification. Qiagen). Purified amplicons were sheared to obtain 150 pb DNA products that were used to generate DNA libraries (see below). The same approach was performed for the alternative sgRNAs experiment but using 640 sgRNAs in 8 different injections (Supplementary Table 1).
To compare the efficiency of Cas9-nanos vs Cas9-βglobin in phenotype-analysis experiments and in the CRISPRscan independent validation experiments, 100 pg of cas9 mRNA and 20 or 10 pg of each sgRNA were injected at the one-cell stage, respectively. Phenotypes were analyzed and quantified between 24 and 48 hpf.
FLAG-Cas9 immunoprecipitation
FLAG-Cas9 immunoprecipitation was performed as previously described39 with some modifications. Briefly, 102 FLAG-Cas9 injected embryos were collected at either 6 or 9 hpf and flash frozen in liquid nitrogen. Embryos were lysed in 1 ml of NET-2 buffer (100 mM Tris-HCl pH 7.5, 150 mM NaCl and 0.05% NP-40) supplemented with protease and RNase inhibitor (Roche). A fiftieth (25 μl) of the lysate was collected as an input control and the remaining fraction was added to 100 μl FLAG-M2 magnetic beads (Sigma) previously washed 3 times with NET-2 buffer. Samples were incubated at 4°C for 2h with orbital shaking. After incubation, another 25 μl aliquot was collected from the supernatant as a supernatant control. Beads were then washed 4 times with 1 ml of NET-2 buffer at 4°C. A final 25 ul aliquot was collected during the last wash as a pulled down control. Next, 500 μl of Trizol (Life Technologies) was added to the beads, RNA was purified and used as starting material for sgRNAs cloning (see below). The same protocol was applied to 102 non-injected embryos as negative control. Finally, the input, supernatant and pulled down controls were subjected to SDS–PAGE and analyzed by western blot. Mouse monoclonal ANTI-FLAG® M2 antibody (Sigma-Aldrich F1804) (1:2000) and rabbit polyclonal Anti-gamma antibody (Abcam ab11317) (1:20000) were used according to manufacturers instructions.
Cloning of the sgRNAs
Total RNA was isolated from embryos injected with sgRNAs using TRIzol reagent (Life Technologies). Purified total RNA was then subjected to reverse transcription using the SuperScript III First Strand Kit (Invitrogen), following manufacturer’s protocol, and a primer containing a 3′ sequence complementary to the constant part of the sgRNAs and a 5′ sequence corresponding to part of the Illumina TruSeq Adapter (Supplementary Table 1 #5). The resulting first-strand cDNAs were purified using Agencourt AMPure XP system (Beckman Coulter) following manufacturer’s protocol. cDNAs were dissolved in 10 μL of water and a ssDNA linker was added at their 3′ ends (5′-/5Phos/linker/3InvdT/-3′, where /5Phos/ is a 5′ phosphate and /3InvdT/ is an inverted deoxythymidine; linker in Supplementary Table 1 #6.) using CircLigase ssDNA Ligase (Epicentre), with slight modifications to the manufacturer’s protocol. In brief, reagents were added to 3 μL of dissolved cDNA samples to reach the following final concentrations in a total volume of 10 μL: 1x CircLigase buffer, 0.05 mM ATP, 2.5 mM MnCl2, 10% PEG 6000, 1M betaine, 5 μM ssDNA linker and 50 U CircLigase enzyme. Ligation mixture was then incubated at 60°C for 2 h, 68°C for 1 h and 80°C for 10 min to deactivate the CircLigase. Volume was increased by the addition of 10 μL of water and ligated products were purified using Agencourt AMPure XP system (Beckman Coulter) following manufacturer’s protocol. PCR amplification was performed on the ligated product using Illumina primers (Illumina Small RNA forward primer and Illumina TruSeq reverse index in Supplementary Table 1 #7 #8; indexes 6, 7, 12–16, 19, 23, 25, 27 were used in this study). PCR products were purified on an 8% native polyacrylamide gel and bands corresponding to sgRNA final library size (179 nt) were extracted. DNA was eluted, ethanol precipitated, dissolved in water and sent for sequencing. For input samples, in vitro transcribed sgRNA cocktails used for injection were pooled together, diluted 1/100 and 1 μL of this dilution was combined with total RNA isolated from uninjected embryos prior to reverse transcription. Alternative sgRNA samples and RNA samples isolated from the Cas9 pull-down experiment were treated as mentioned above with few modifications. Briefly, 50 U of RNase I was added during the RNase H treatment prior to AMPure XP purification. Finally, the AMPure XP purification previous to the PCR amplification was substituted by a 10% urea-denaturing PAGE purification step. Bands corresponding to the ligated product size (114 nt) were cut. DNA was then eluted, ethanol precipitated, dissolved in water and PCR amplified as mentioned above.
sgRNA labeling and in-line probing
To produce 5′-end-labeled sgRNAs, purified sgRNA transcripts were dephosphorylated by adding 1 U of antartic phosphatase (New England BIoLabs) to 50 pmol of sgRNA in a final volume of 10 μL containing 50 mM Bis-Propane pH 6.0, 1 mM MgCl2, 0.1 mM ZnCl2 and RNase OUT (20U, Invitrogen). The mixture was incubated for 30 min at 37°C and, then, the enzyme was inactivated by incubation for 5 min at 65°C. Dephosphorylated sgRNAs (5 pmol) were 5′-end-rediolabeled using 3 U of T4 polynucleotide kinase (New England Biolabs) for 1 h at 37°C in the presence of 3.2 pmol of [α-32P]ATP (6000 Ci/mmol; Perkin Elmer). The reaction was stopped by adding formamide dye buffer (95% formamide, 10 mM EDTA, 0.025% bromophenol blue and 0.025% xylene cyanol), and the radiolabeled sgRNA purified by 8% polyacrylamide gel electrophoresis. 5′-end-labeled sgRNAs were visualized by autoradiography and the bands corresponding to the correct sizes were excised from the gel, gel slices shredded and the transcripts eluted for 10 min at 37°C in 300 μL of water. The labeled sgRNAs were then ethanol-precipitated, dried and dissolved in water.
In-line probing was performed to test G-quadruplex formation by sgRNAs as described previously 40. This technique and the conditions used allowed the study of the RNA structure in conditions that either do not (LiCl) or do (KCl) support G-quadruplex formation. Since G-quadruplexes are the only potassium dependent structure (at least at the concentrations used), a sequence can be considered as forming such a structure if a difference in the band patterning is observed40. Briefly, 5′-end-labeled sgRNAs (50 000 cpm), corresponding to trace amount of RNA (<1 nM), was heated at 70°C for 5 min and then slow-cooled to room temperature over 1h in buffer containing 50 mM Tris-HCl pH 7.5 and either in the presence of 100 mM LiCl or KCl in a final volume of 10 μL. After incubation, the final volume of each sample was adjusted to 100 μL to reach final concentrations of 50 mM Tris-HCl pH 7.5, 20 mM MgCl2 and either 100 mM LiCl or KCl. The mixtures were incubated for 40 h at room temperature, ethanol-precipitated and the RNAs dissolved in formamide dye loading buffer (95% formamide, 10 mM EDTA, 0.025% bromophenol blue and 0.025% xylene cyanol). For alkaline hydrolysis, 50 000 cpm of 5′-end-labeled sgRNAs (<1 nM) were dissolved in 5 μL water, 1 μL of 1 N NaOH added and the reactions incubated for 30 sec at room temperature prior to being quenched by the addition of 3 μL of 1 M Tris-HCl pH 7.5. The RNAs were then ethanol-precipitated and dissolved in formamide dye loading buffer. RNase T1 ladders were prepared using 50 000 cpm of 5′-end-labeled RNA (<1 nM) dissolved in of buffer containing 20 mM Tris-HCl pH 7.5, 10 mM MgCl2 and 100 mM LiCl. The mixtures were incubated for 1.5 min at 37°C in the presence of 3 U of RNase T1 (Fisher Scientific), and was stopped by ethanol-precipitation prior to being dissolved in formamide dye loading buffer. The radioactivity of the in-line probing samples and both ladders was calculated, and equal amount in terms of counts per minute of all conditions and ladders of each sgRNA were fractionated on denaturing (8 M urea) 10% polyacrylamide gels. The resulting gels were subsequently dried and visualized by exposure to phosphorscreen. The band intensities for each condition were calculated using the Semi-Automated Footprinting Analysis (SAFA) software. sgRNAs showing a ratio of band intensities (KCl/LiCl) above 2 for a nucleotide in or flanking the guanine-rich sequence were identified as positively folding into a G-quadruplex structure. A previous study showed that a threshold of 2 was associated to guanine-rich sequences folding into active G-quadruplexes in cellulo 41.
Libraries preparation and sequencing of genomic loci
Amplicon quality and concentration were determined by estimating the A260/A280 and A260/A230 ratios by nanodrop. Amplicon integrity and size were confirmed by running an Agilent Bioanalyzer gel. 100 ng of PCR amplicons (1–2kb) were sheared to 120–150bp using a Covaris E210 instrument. Following shearing, a SPRI bead cleanup was performed using Ampure XP SPRI (Beckman Coulter Genomics). A shearing QC was then performed on sheared products using the DNA 1000 Bioanalyzer chip to confirm the target size. Sheared amplicons were then end-repaired and A-tailed and adapters were ligated. Indexed libraries that meet appropriate cut-offs were quantified by both qRT-PCR using a commercially available kit (KAPA Biosystems) and insert size distribution determined with a LabChip GX. Samples with a yield of ≥0.5 ng/ul were used for sequencing. Sample concentrations were normalized to 2 nM and loaded onto Illumina version 3 flow cells at a concentration that yields 170–200 million passing filter clusters per lane. Samples were then sequenced using 75 bp paired-end reads on an Illumina HiSeq 2000 according to Illumina protocols. The 6 bp index is read during an additional sequencing read that automatically follows the completion of read 1. A positive control (prepared bacteriophage Phi X library) provided by Illumina was spiked into every lane at a concentration of 0.3% to monitor sequencing quality in real time. Primary analysis and sample demultiplexing were performed using Illumina’s CASAVA 1.8.2 software suite. Raw reads are publicly accessible in the Sequence Read Archive under SRP059430.
Zebrafish maintenance
Zebrafish wild-type embryos were obtained from natural mating of TU strain of mixed ages (5–18 months) for large-scale experiments except for the Cas9-FLAg pull-down where TU-AB and TLF strains of mixed ages (5–17 months) were used. TU-AB and TLF strains of mixed ages were also used for the other experiments. Selection of mating pairs was random from a pool of 60 males and 60 females allocated for a given day of the month. Fish lines were maintained in accordance with AAALAC research guidelines, under a protocol approved by Yale University IACUC.
Frog husbandry and injections
X. tropicalis were housed and cared for in our aquatics facility according to established protocols that were approved by the Yale Institutional Animal Care and Use Committee (IACUC).
Ovulation was induced and eggs were collected according to established protocols42. Staging of Xenopus tadpoles was done according to Nieuwkoop and Faber43. mRNA was injected into the one-cell or two-cell embryo as previously described44, along with dextran mini-Ruby as a tracer. 500 pg of mRNA of Cas9 and 400 pg of each sgRNA were injected in one-cell stage embryos.
Image acquisition
Embryos were analyzed using a Zeiss Axioimager M1 and Discovery microscopes and photographed with a Zeiss Axiocam digital camera. Images were processed with Zeiss AxioVision 3.0.6. Adult fish were photographed with a Panasonic Lumix DMC-FZ18.
Mapping of “WT” and “mutant” reads
To filter the reads that potentially were mutated by CRISPR/Cas9, all reads were mapped to the zebrafish genome Zv9 using Bowtie2 2.245 not allowing any insertions or deletions and increasing mutation costs with the following options: --end-to-end, --rdg 1000,100, --rfg 1000,100, --mp 12,4. Reads that aligned to the loci were counted as “WT reads” to be used as a reference for sgRNA activity (see below). PCR oligos were filtered from the unmapped pairs (one or both unmapped mate(s)) by trimming the oligo(s). Reads were first mapped to the oligos with Bowtie2 with the following options: --local, -L 4, -a, --score-min G,8,4, --rfg 10,5, --rdg 10,5, --reorder. Then unmapped reads were kept together with trimmed reads if: less than 2 oligos aligned to the read, the oligo aligned within the first 4 nt of the read and the trimmed read was longer than 30 nt. Reads were aligned to the sequence of the 128 loci using gmap 2014.10.2246. The alignments with gmap allowed the detection of distinct mutations on the same read caused by different sgRNAs. Reads aligned as concordant pairs on one of the loci were kept for the rest of the analysis. To call an insertion or a deletion event, a 15 nt match was required on each side of the called mutation in the read alignment.
Characterization of single and two sgRNA mutations
A region centered on the known Cas9 cleavage site (6 nt upstream of the PAM14, extended by 15 nt upstream and downstream was used to attribute mutations to a specific sgRNA target site. A mutation starting or ending in one of the 1280 regions was attributed to that sgRNA and counted as a sgRNA mediated mutation. A mutation starting and ending in two different regions was counted as a “Two site mutation”. A mutation that overlaps a single region was counted as a “Single site mutation”. Otherwise, it was counted as a “mutation” (overlapping more than one region). To analyze length and frame distribution of the mutations, the 1% top outliers were discarded.
Detection of polymorphisms
Reads of the non-injected control were mapped similarly to the sgRNA injected experiment. Mapped reads were then piled up with samtools47 and all variants were detected using bcftools with the following options -mv -P1e-3. A minimum of quality of 5, a 150 read coverage and a 50% frequency were required to call a polymorphism. Target sites overlapping with at least one polymorphism were discarded.
Measuring sgRNA input
Reads were mapped to the sgRNA sequences using Bowtie2 configured to perform local alignments for automatic adapter trimming with increased gap and mismatch costs with the following options: --local, -L 4, -a, --score-min G,8,4, --rfg 10,5, --rdg 10,5. Mapped reads (only primary alignment as defined in the SAM format specifications to ensure that in the rare cases where a sequencing read aligns partially to multiple sgRNAs, only the correct alignment is selected) for each sgRNA were then counted. As sgRNAs were injected by cocktail of 80, counts were normalized by the total number of reads in each cocktail.
Computing sgRNA folding energy
RNA fold from the ViennaRNA package48 was used with the --partfunc parameter to compute the ensemble free energy.
Computing sgRNA activity
To obtain a robust sgRNA raw activity, a minimum of 1000 reads overlapping each target site region was required as well as a minimum of 150 reads in the non-injected controls to assure any polymorphism could be detected. The raw activity was computed as the percentage of “mutated reads” over “WT reads” and “mutated reads” overlapping each sgRNA region as described above. To obtain a robust sgRNA normalized activity measure, the 5% least abundant sgRNAs were removed (based on normalized counts at 0 hpf or 1.25 hpf). This step also reduced the noise level of sgRNA abundances. The normalized sgRNA activity was computed as raw activity divided by the log2 of the sgRNA normalized count. A rank percentile was then computed. The sgRNA with the highest normalized activity received a rank of 1.
sgRNA activity regression model
To build the linear regression model 684 features were included corresponding to the sequence of the sgRNA, the PAM and 6 upstream and downstream nucleotides. These 684 features consist of the following: 140 features representing mononucleotides (4 base identities × 35 nucleotide positions [6nt upstream context + 20nt sgRNA + 3nt PAM + 6nt downstream context]); and 544 dinucleotide features (16 possible dinucleotides × 34 positions). Each sgRNA plus context (35nt total) was represented as a binary vector of length 684 encoding presence (value of 1) or absence (value of 0) for each feature. A randomized logistic regression49, 50 with 500 fold resampling, L1 penalty and 0.3 regularization strength was used to select 91 features that were determinant to classify the top 20% most efficient sgRNAs. A linear regression was then fitted on the 91 features and the ranked normalized sgRNA activities. Cross-validation was performed using the ShuffleSplit method of scikit-learn with 200 iterations.
The canonical model was applied directly on mismatch-containing alternative sgRNAs: correlations between experimental rank and CRISPRscan predicted score for Gg18 (r=0.57, p=0.004) and gG18 (r=0.26, p=0.06) alternatives were observed. To apply the canonical scoring model to shorter sgRNAs, two strategies were explored to account for missing nucleotide positions that are encoded in the GG18 model. An empirical approach evaluating performance was employed where a) index position(s) are omitted from the encoding of shorter sgRNAs and thus not scored; or b) index positions are assigned best-approximation base identities, defined by the next nucleotide downstream (i.e., base X could contribute to either nucleotide position i or i+1 in the canonical encoding) (Supplementary Fig. 6i). Since the biophysical significance of base preferences at each nucleotide position is unclear, these options did not favor any a priori assumptions regarding equivalent nucleotide indices between shorter and canonical-length sgRNAs, but instead allow for a slightly “fuzzy” encoding.
MNase
MNase-seq data were obtained from GEO GSE44269 and analyzed similarly as described in Zhang et al.51. Normalized read coverage was summed over the sgRNA target site regions.
Cas9 distraction factor
Off-target seeds were the number of 5+N+2 nt or 7+N+2 nt (N+2 is the PAM) occurrences found on both strand of the zebrafish genome. For the control, the seed were on the opposite side of the PAM in the sgRNA target.
Code availability
Mutagenesis analysis pipeline is available upon request.
Bioinformatics libraries
Python custom scripts were used to perform the analysis using the Python libraries Matplotlib (matplotlib.org) for plotting, Numpy (numpy.org) and Pandas (pandas.pydata.org) for data mining, Scipy (scipy.org) for statistics, scikit-learn (scikit-learn.org) and Statsmodels (statsmodels.sourceforge.net) for machine learning.
Supplementary Material
Acknowledgments
We thank E. Fleming and H. Codore for technical help; D. Cifuentes, A. Bazzini, M. Lee for discussions and all the members of the Giraldez laboratory for intellectual and technical support; S. Lau, M. Lee, and M. Fernandez-Fuertes for cloning of nanos 3′-UTR and helping with MNase analysis and with pictures of the adult fish, respectively. C. Takacs, M. Lee and K. Divito for manuscript editing. The Swiss National Science Foundation grant P2GEP3_148600 (C.E.V), Programa de Movilidad en Áreas de Investigación priorizadas por la Consejería de Igualdad, Salud y Políticas Sociales de la Junta de Andalucía (M.A.M-M.), NIH grants R21 HD073768, R01 GM103789, R01 GM102251, R01 GM101108 and GM081602 (A.J.G.) and R01 HD081379 (E.M. and M.K.K.) supported this work. M.K.K. is supported by the Edward Mallinckrodt Jr Foundation.
Footnotes
Online CRISPRscan predictions
Available at http://www.crisprscan.org
Author contributions
M.A.M-M., C.E.V. and A.J.G. designed the project, performed experiments, data analysis and wrote the manuscript. J-D. B. performed the sgRNA libraries, G-quadruplex experiments and helped to write part of the manuscript. J.P.F. performed F0 phenotype analysis and Xenopus phenotype analysis experiment with M.A.M-M. E.M. carried out Xenopus injections and phenotype analysis.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 2.Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Molecular therapy: the journal of the American Society of Gene Therapy. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 3.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassett AR, Liu JL. CRISPR/Cas9 and genome editing in Drosophila. Journal of genetics and genomics. 2014;41:7–19. doi: 10.1016/j.jgg.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagnon JA, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huppert JL. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chemical Society reviews. 2008;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 16.Beaudoin JD, Perreault JP. Exploring mRNA 3′-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Res. 2013;41:5898–5911. doi: 10.1093/nar/gkt265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White RM, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell stem cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877–2885. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishima Y, et al. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Current biology. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen JN, et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- 21.Halpern ME, Ho RK, Walker C, Kimmel CB. Induction of muscle pioneers and floor plate is distinguished by the zebrafish no tail mutation. Cell. 1993;75:99–111. [PubMed] [Google Scholar]
- 22.Schulte-Merker S, et al. Expression of zebrafish goosecoid and no tail gene products in wild-type and mutant no tail embryos. Development. 1994;120:843–852. doi: 10.1242/dev.120.4.843. [DOI] [PubMed] [Google Scholar]
- 23.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 24.Giraldez AJ, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 25.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farboud B, Meyer BJ. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics. 2015;199:959–971. doi: 10.1534/genetics.115.175166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 28.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varshney GK, et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015 doi: 10.1101/gr.186379.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shah AN, Davey CF, Whitebirch AC, Miller AC, Moens CB. Rapid reverse genetic screening using CRISPR in zebrafish. Nat Methods. 2015 doi: 10.1038/nmeth.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren X, et al. Enhanced Specificity and Efficiency of the CRISPR/Cas9 System with Optimized sgRNA Parameters in Drosophila. Cell reports. 2014;9:1151–1162. doi: 10.1016/j.celrep.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciruna B, et al. Production of maternal-zygotic mutant zebrafish by germ-line replacement. Proc Natl Acad Sci U S A. 2002;99:14919–14924. doi: 10.1073/pnas.222459999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee MT, Bonneau AR, Giraldez AJ. Zygotic genome activation during the maternal-to-zygotic transition. Annual review of cell and developmental biology. 2014;30:581–613. doi: 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cunningham F, et al. Ensembl 2015. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meeker ND, Hutchinson SA, Ho L, Trede NS. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Bio Techniques. 2007;43:610, 612, 614. doi: 10.2144/000112619. [DOI] [PubMed] [Google Scholar]
- 39.Cifuentes D, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaudoin JD, Jodoin R, Perreault JP. In-line probing of RNA G-quadruplexes. Methods. 2013;64:79–87. doi: 10.1016/j.ymeth.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 41.Beaudoin JD, Perreault JP. 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 2010;38:7022–7036. doi: 10.1093/nar/gkq557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.del Viso F, Bhattacharya D, Kong Y, Gilchrist MJ, Khokha MK. Exon capture and bulk segregant analysis: rapid discovery of causative mutations using high-throughput sequencing. BMC Genomics. 2012;13:649. doi: 10.1186/1471-2164-13-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin): a Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis. Garland Pub; New York: 1994. [Google Scholar]
- 44.Khokha MK, et al. Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn. 2002;225:499–510. doi: 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- 45.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 47.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lorenz R, et al. ViennaRNA Package 2.0. Algorithms for molecular biology: AMB. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. Springer New York Inc; New York, NY, USA: 2001. [Google Scholar]
- 50.Meinshausen N, Buhlmann P. Stability selection. J Royal Stat Society-Series B. 2010;72:417–473. [Google Scholar]
- 51.Zhang Y, et al. Canonical nucleosome organization at promoters forms during genome activation. Genome Res. 2014;24:260–266. doi: 10.1101/gr.157750.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.