Abstract
Sheath rot complex and seed discoloration in rice involve a number of pathogenic bacteria that cannot be associated with distinctive symptoms. These pathogens can easily travel on asymptomatic seeds and therefore represent a threat to rice cropping systems. Among the rice-infecting Pseudomonas, P. fuscovaginae has been associated with sheath brown rot disease in several rice growing areas around the world. The appearance of a similar Pseudomonas population, which here we named P. fuscovaginae-like, represents a perfect opportunity to understand common genomic features that can explain the infection mechanism in rice. We showed that the novel population is indeed closely related to P. fuscovaginae. A comparative genomics approach on eight rice-infecting Pseudomonas revealed heterogeneous genomes and a high number of strain-specific genes. The genomes of P. fuscovaginae-like harbor four secretion systems (Type I, II, III, and VI) and other important pathogenicity machinery that could probably facilitate rice colonization. We identified 123 core secreted proteins, most of which have strong signatures of positive selection suggesting functional adaptation. Transcript accumulation of putative pathogenicity-related genes during rice colonization revealed a concerted virulence mechanism. The study suggests that rice-infecting Pseudomonas causing sheath brown rot are intrinsically diverse and maintain a variable set of metabolic capabilities as a potential strategy to occupy a range of environments.
Introduction
The increasing global trade activities are the main cause of movement of plant pathogens that continue to threaten modern agriculture. In such scenario, pathogen populations tend to diversify and increase their evolutionary potential as they encounter more favorable conditions and recombination opportunities [1]. This is particularly true for many plant pathogens that colonize the rice (Oryza sativa L.) seed, which include members of the genus Pseudomonas. One of the most common rice-infecting pathogens is Pseudomonas fuscovaginae (Pfv), a seed-borne and seed-transmitted Gram-negative bacterium that causes sheath brown rot and grain discoloration. Several reports have demonstrated epiphytic and endophytic colonization of Pfv in symptomless rice seeds [2–4], suggesting that gene flow is still effective and more research is needed to understand pathogen diversity and distribution. This pathogen was first reported in Hokkaido, Japan in 1976 [2, 5] and soon appeared in many tropical and subtropical rice-growing regions of the world i.e. Latin America [6, 7], Sub-Saharan Africa [8–10], Southeast Asia [11–13], and Australia [14].
Under favorable low temperature conditions, Pfv colonizes the rice sheath, producing brown to reddish brown necrotic lesions. If conditions persist, the lesions can progress toward the panicle, causing seed discoloration and grain sterility [6, 7]. The intrinsic capability of Pfv in colonizing multiple plant tissues as well as its ability to survive as an epiphyte on the seed surface [3] or endophytically in roots, stems, and leaves [4] probably require quite a versatile metabolism. In addition, Pfv has a broad host range among wild and cultivated grasses [3, 15, 16], which also reveals a diverse panel of pathogenicity factors that is worth exploring.
From a taxonomical perspective, the genus Pseudomonas comprises at least two main lineages, the P. aeruginosa lineage and the P. fluorescens lineage. Phylogenetic analysis of 16S rRNA, gyrB, rpoB and rpoD sequences clearly associates Pfv with the P. fluorescens subgroup members [17]. This group appears to be highly diverse, harboring unusual levels of intra-species heterogeneity. Silby and co-workers [18] compared three P. fluorescens genomes (SBW25, Pf0-1, and Pf-5) and found that only 61% of genes were shared among them. Subsequent studies also found high numbers of unique genes when comparative analysis was performed among different genomes of P. fluorescens [19–21].
Apart from Pfv, other rice-infecting Pseudomonas pathogens have been isolated from plant tissues showing sheath brown rot symptoms. In the Philippines, disease surveys on tropical rice ecologies have isolated fluorescent Pseudomonas colonies similar to Pfv [12]. In 1998, two of these colonies, named IRRI 6609 and IRRI 7007, were isolated from rice sheaths collected in Davao and Palawan regions in the month of December when temperatures are usually less than 20°C. Also, the strain S-E1 was isolated in an agronomy trial in Siniloan, Luzon Island during a low temperature spell [12, 22]. Serological, biochemical, and genetic analyses of IRRI 6609, IRRI 7007, and S-E1 together with other Pseudomonas groups revealed that all three strains were closely related to Pfv but were part of a distinct population [12, 22]. Until the taxonomic status of these populations is clarified, we will designate these populations as P. fuscovaginae-like (Pfv-like).
It is well documented that plant pathogenic bacteria evolved a plethora of mechanisms to modulate host environment in order to facilitate colonization [23–25]. Several secretion systems are used to deliver molecules that interact with apoplastic or cytoplasmic plant components [26]. While many Gram-negative bacteria inject type III (T3) effector proteins into host cells [25, 27], other key virulence factors are also widely used such as cell wall-degrading enzymes (CWDEs), phytotoxins, extracellular polysaccharides, and phytohormones, among others [24]. However, only few studies have really addressed the presence of pathogenicity factors in rice-infecting Pseudomonas. For instance, the two N-acyl homoserine lactone (AHL) quorum sensing systems (PfsI/R and PfvI/R) of Pfv have been reported by Mattiuzzo et al. [28] to be involved in virulence. The presence of syringotoxin in Pfv extracts [29, 30] was highlighted as a virulence factor when Batoko and co-workers [31] showed that it may affect plant membrane integrity by inhibiting a membrane-associated H+-ATPase in vitro. Recently, a Pfv mutant screening on Chenopodium quinoa and rice identified additional virulence factors involved in adhesion, phytotoxins, and secretion [32]. At the moment, it is not clear whether other factors may be relevant during the Pseudomonas–rice interaction.
The fact that other Pseudomonas populations are able to infect rice opens the door for comparative analysis to identify a common set of virulence factors and depict the evolutionary context of this group. To better explore Pfv-like genomic composition, structure, and diversity, we obtained shotgun sequences of the two strains from the Philippines (IRRI 6009 and IRRI 7007) and compared their genome assemblies with draft genomes of several Pfv and Pfv-like strains that were recently released [33–35]. We found that Pfv-like strains are closely related to Pfv. Although sequenced strains represent only a fraction of the overall diversity, we showed that Pfv as well as Pfv-like populations are not genetically homogeneous, having acquired high levels of diversification. We redefined the understanding of the Pfv pan-genome and identified a set of common virulence factors that may be important to successfully colonize rice sheath. Interestingly, Pfv and Pfv-like have plastic genomes with a high proportion of strain-specific genes and unique metabolic capabilities. We also defined the core secretome for Pfv and Pfv-like, which showed strong signatures of positive selection that matched with both pathogen lifestyles.
Materials and Methods
Pathogenicity test
The pathogenicity of Pfv-like strains IRRI 6609, IRRI 7007, and S-E1 were performed on the rice cultivars Azucena and Moroberekan using the toothpick method at maximum tillering stage (40–45 days after transplanting). Bacterial cultures were grown for 24 h at 28–30°C on King’s medium B (KB) and suspended in sterile demineralized water [22]. The middle portion of the sheath was pricked with the toothpick dipped in the bacterial suspension with 108 cfu/ml. Plants inoculated with sterile demineralized water served as negative control. The inoculated plants were incubated in a growth chamber at 23°C/18°C day/night temperature with 90% relative humidity and photoperiod 12h/12h (light/dark). Sheath discoloration was observed at 14 days post inoculation (dpi) and, to further confirm the disease, the inoculated plants were kept until maturity stage for observations of grain discoloration.
Pseudomonas genomes and whole-genome alignments
We used different sources to obtain a representative sample of Pfv-like genomes. The genome sequence of the strain S-E1 was downloaded from GenBank [35]. Pfv-like cultures of IRRI 6609 and IRRI 7007 were grown overnight at 28°C. Isolation of genomic DNA was done using Easy-DNA kit (Invitrogen, USA) following the manufacturer’s protocol. Genome sequencing was contracted as service to BGI-Shenzhen (Shenzhen, China), producing 90-bp paired-end reads using Illumina GAIIx technology. Filtered paired-end reads were de novo assembled into contigs and scaffolds using CLC Genomics Workbench 6.5 (CLC bio, Denmark). Alternative assemblies did not result in better outputs so we followed CLC. Gene calling and annotation were performed using JGI/IMG-ER 4 [36]. The genome sequence of the five Pfv strains UPB0736 (Madagascar), CB98818 (China), ICMP 5940 (Japan), and DAR 77795 and DAR 77800 (Australia) were downloaded from GenBank [33–35]. All Pfv (5) and Pfv-like (3) genes were classified according to cluster of orthologous groups (COG) terms [36]. The datasets of the strains ICMP 5940, DAR 77795, and DAR 77800 were re-annotated using RAST [37]. Predicted genes with sizes less than 50 bp were removed from the analysis. Draft genome sequences of Pfv-like strains IRRI 6609 and IRRI 7007 were deposited at GenBank under accession numbers JSYZ00000000 and JTBY00000000. For comparative analysis, we used a set of 79 different Pseudomonas genomes comprising main lineages (S1 Table). Intact prophage prediction and annotation were obtained using the PHAST server [38]. To assess if the prophage is complete or not, we calculated the number of bases, genes, and cornerstone genes and detected the presence of phage-like genes. All intact prophage should have at least a score of 90 by PHAST standards. For secondary metabolite cluster prediction, the antiSMASH 2.0 [39] website was used. In addition, selected genes were BLASTp against the Pseudomonas dataset with E-values less than 1e-20 and with a minimum identity of 20%. These were then visualized using CodaChrome 1.1 [40]. Whole-genome alignment was performed in two steps: first, we reordered contigs of Pfv-like and Pfv genomes against IRRI 6609 using MAUVE 2.3.1 [41] under default parameters. Locally collinear blocks and iterative alignment were arranged using MUSCLE [42]. In the second step, we produced the final alignments of the eight strains with BLASTn and visualized with BLAST Ring Image Generator 0.95 (BRIG) [43]. All alignments had a threshold E-value ≤ 1e-25.
Average nucleotide identity (ANI) and tetranucleotide frequency correlation coefficients (TETRA) analysis
To determine the relatedness among Pfv and Pfv-like strains, average nucleotide identity (ANI) and tetranucleotide frequency correlation coefficients (TETRA) analyses were done using the JSpecies 1.2.1 [44] software under default parameters. For the ANI values, alignment calculation was done using the MUMmer algorithm [45]. The ANI and TETRA matrices were used to construct a pairwise relationship. The heatmap.2 function from the R package gplots was used to build the dendrograms and heatmaps. Furthermore, 79 Pseudomonas species (S1 Table) were compared for ANI using Pfv-like IRRI 6609 as the reference strain. The percentage value cut-off for the ANI and TETRA analyses were >95% and >99%, respectively.
Orthologous gene identification and average amino acid identity (AAI) analysis
Orthologous gene clustering of protein and nucleotide sequences collected from the Pfv and Pfv-like genomes was performed using GET_HOMOLOGUES [46]. Pairwise alignment was done using BLASTall [47] among sequences with a minimum E-value of 1e-5. For the orthologous gene identification, OrthoMCL [48] and COGtriangles [49] algorithms were used to filter and cluster the BLAST results with sequences having a coverage of at least 50% and a minimum identity of 50%. Genes that were not positive for both algorithms were filtered out from the final list. Following this approach, we used all BLAST results to compute the average amino acid identity (AAI) for each comparison and to build the pairwise relationship as stated above.
Secretome prediction and positive selection analysis
The secretome of Pfv and Pfv-like was predicted using 50,066 open reading frames obtained from eight genomes. The presence of secretion signal was predicted using SignalP 4.1 [50] and further filtered for transmembrane domain-containing proteins using TMHMM 2.0 [51]. To predict type III effectors, we used BLASTp against T3DB resource [52]. All the predicted secreted proteins were clustered using GET_HOMOLOGUES [46] with the same parameters mentioned above. The core genes were then extracted for exhaustive Gene Ontology (GO) term search using BLAST2GO [53] and through the Pfam 27.0 [54] and InterPro 48.0 [55] websites. Multiple alignment of the core orthologous genes were generated using TranslatorX [56] with the guidance of the MUSCLE program for the translated protein sequences. Gblocks [57] was then applied to remove spurious alignments. An estimation of the numbers of synonymous (Ks) and nonsynonymous (Ka) substitutions per site was used as parameters to assess selection at molecular level [58, 59]. The higher the ratio (ratio ω = Ka/Ks > 1), the stronger the signal of positive selection between two DNA-coding sequences. Ka/Ks ratios were calculated with the support of KaKs-Calculator 2.0 [58]. The Yn00 model [59] was used for determining the ω values.
Phylogeny based on MLSA
The phylogenetic relationship was evaluated using concatenated nucleotide sequences of rpoB and rpoD genes (S2 Table). Multiple alignment was then performed in MEGA6 [60] using the implementation of the MUSCLE algorithm and with the help of Gblocks to secure the conserved blocks of the alignment [57]. Maximum Likelihood (ML) analysis was carried out to infer the evolutionary relationship of eight Pfv and Pfv-like strains and 26 closely related Pseudomonas species. The E. coli K-12 sub-strain MG1655 was used as outgroup. The ML tree was constructed using RAxML [61], with 1000 bootstrap replicates and GTRGAMMA as the substitution model. FigTree 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/) was used to visualize the tree. To strengthen the phylogenetic analysis, we used concatenated nucleotide sequences of 10 housekeeping genes: acsA, aroE, dnaE, guaA, gyrB, mutL, ppsA, pyrC, recA, and rpoB (S2 Table) on a subsample of species.
Transcript accumulation of Pfv-like genes
The rice cultivar Azucena and the Pfv-like strain IRRI 7007 were used in a time course gene expression experiment. At maximum tillering, the leaf sheath of rice was syringe-infiltrated with Pfv inoculum and sampling points 0, 3, 24, 48, and 72 hours post infection (hpi) were considered. IRRI 7007 was chosen because of its high virulence. For semi-quantitative RT-PCR, the total RNA from the treated rice sheath tissues of Azucena, as well as tissues from mock samples, were obtained using the Trizol method (Invitrogen, USA). Complementary DNA (cDNA) was synthesized from the pooled RNA molecules using SuperScript III and random hexamers (Invitrogen, USA). Primers were designed to target the coding sequences of candidate pathogenicity genes (S3 Table). In addition, two core random genes were selected. RT-PCR was done with the gene-specific primers in a 20ul reaction mix and was performed in a thermal cycler machine (G-storm GS1). We used the 16S gene to normalize the Pfv-like gene expression assessment. This experiment was done in two independent biological replicates with three technical replicates per sample. RT-PCR products were visualized on 1.25% agarose gels.
Result and Discussion
Pfv-like populations are able to infect rice sheath and cause seed discoloration
To reproduce sheath brown rot and grain discoloration symptoms produced by Pfv-like pathogens, we inoculated the rice cultivars Azucena and Moroberekan using the toothpick method (Fig 1A). Phenotypic symptoms caused by the three Pfv-like strains were similar to Pfv as reported in Asia, Africa, and South America [6, 7, 16] which include the appearance of brown to reddish brown discolorations extending to the entire length of the sheath to its inner tissues (Fig 1B and 1C). Similar to the findings made by Cottyn et al. [22], Pfv-like strains were pathogenic on rice and showed variation in virulence spectrum. In order to validate pathogen spread, plants were kept until maturity to assess phenotypic symptoms in the grain. In most cases, panicles that emerged from the inoculated plants were necrotic and produced discolored grains that are often sterile compared with the control plants (Fig 1D and 1E). Fluorescent Pseudomonas colonies similar to Pfv were re-isolated from the infected sheath (data not shown). In addition to previous findings [22], these observations indicate that Pfv-like populations represented by IRRI 6609, IRRI 7007, and S-E1 harbor similar capabilities as that of Pfv to infect the rice host. Although sheath brown rot and seed discoloration phenotype assessment was not the aim of this study, we found that all strains were able to spread successfully across different tissues. Therefore, we decided to investigate the composition of Pfv-like factors contributing to bacterial sheath brown rot at the genomic level.
Fig 1. Infection caused by P. fuscovaginae-like strain IRRI 7007 in O. sativa cv. Azucena.

A) Plants were inoculated at 45 days after transplanting using toothpick method. B) Symptom development along the sheath showing brown necrotic lesions. C) Discolored inner sheath. D) Poorly emerged panicles with brown to dark brown grains. E) Emerged panicles with discolored grains and progressive necrotic stripes at maturity stage.
Genome sequence of Pfv-like IRRI 6609 and IRRI 7007 and other available Pfv draft genomes
To date, the draft genomes of five Pfv and one Pfv-like strains are available in the public domain [33–35], representing a useful resource to consolidate our understanding of this novel pathogen population. To gain insight into the overall genome structure of Pfv-like, we generated high quality draft genomes of IRRI 6609 and IRRI 7007. Genome statistics of the strains IRRI 6609, IRRI 7007, S-E1 (Philippines), UPB0736 (Madagascar), CB98818 (China), ICMP 5940 (Japan), and DAR 77795 and DAR 77800 (Australia) are summarized in Table 1. Among all Pfv-like genomes, the assembly of IRRI 6609 produced the largest genome and the least number of scaffolds. Based on this, we considered IRRI 6609 as our reference genome for further analysis. It is not clear whether the difference in contig number between IRRI 6609 and IRRI 7007 is due to either the intrinsic differences between the strains or unresolved problems with the assembly. However, there is no reason to believe that contamination or low-quality reads are responsible for this difference. Interestingly, draft genomes of Pfv obtained from different sources [33–35] also show variability regarding genome size and number of protein coding genes (Table 1), suggesting that genome structure among groups may be more complex than we previously thought. Although not significant, we detected variation of G+C content between Pfv and Pfv-like genomes, with the Pfv genomes having an average of 61.31% while Pfv-like genomes had 63.12%. The G+C content has a phylogenetic signal over a short evolutionary time period [62], thus suggesting a close relatedness.
Table 1. General features of the eight rice-infecting Pseudomonas draft genomes.
| Pseudomonas fuscovaginae-like | Pseudomonas fuscovaginae | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| IRRI 6609 | IRRI 7007 | S-E1 | CB98818 | UPB0736 | ICMP 5940 | DAR 77795 | DAR 77800 | ||
| Origin a | Philip | Philip | Philip | China | Madag | Aus | Aus | Aus | |
| No. contigs | 79 | 617 | 692 | 263 | 102 | 459 | 482 | 791 | |
| N50 (kbp) | 355.2 | 27.0 | 92.3 | 44.5 | 205.3 | 47.4 | 39.6 | 17.8 | |
| Largest contig size (kbp) | 746.8 | 133.1 | 305.2 | 202.4 | 605.8 | 189.6 | 142.3 | 96.1 | |
| Total size (Mbp) | 7.14 | 6.73 | 6.55 | 6.54 | 6.35 | 6.37 | 6.25 | 5.97 | |
| G+C content (%) | 63.25 | 63.00 | 63.13 | 61.4 | 61.46 | 61.2 | 61.40 | 61.10 | |
| RNA coding genes | 133 | 127 | 113 | 140 | 125 | 56 | 53 | 51 | |
| Protein coding genes | 6,342 | 6,699 | 5,897 | 6,433 | 5,689 | 6,367 | 6,035 | 7,202 | |
a Country names: Philip = Philippines, Madag = Madagascar, and Aus = Australia.
Pfv-like strains are closely related to Pfv
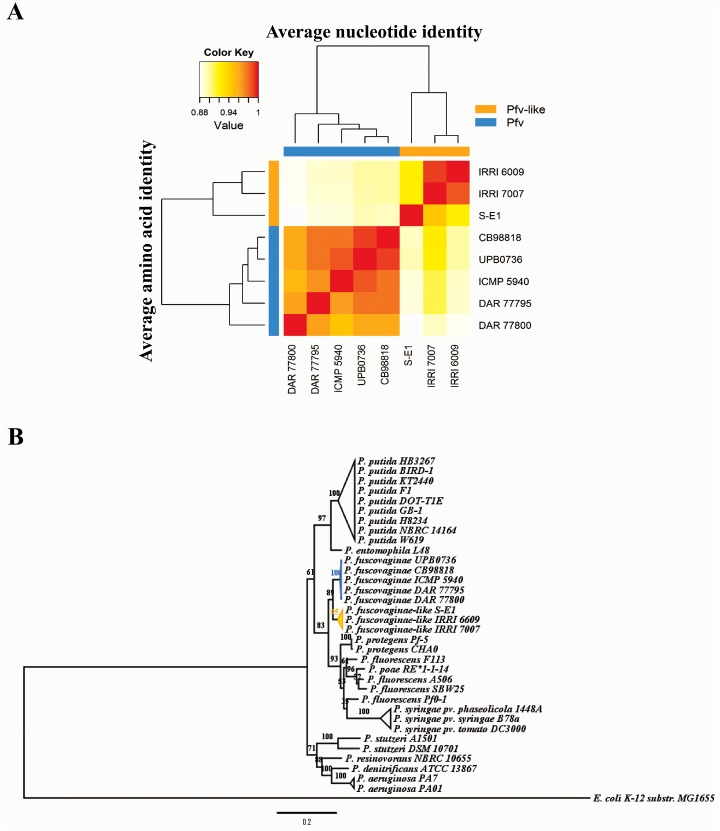
To get insight into the overall homology levels and to evaluate genetic relatedness among Pfv and Pfv-like genomes, we calculated both ANI and AAI values for the eight genomes [44, 63]. We found levels of homology that were inconsistent with a single monophyletic group. In our analysis, the ANI values ranged from 87.95% to 98.65% while the AAI values varied from 87.95% to 99.35% (Fig 2A). We built a distance matrix based on pair-wise ANI-AAI values which identified at least two subgroups within rice-infecting Pseudomonas (Fig 2A). The dendrogram topology clustered the Pfv strains from Madagascar, Japan, China, and Australia separately from those Pfv-like strains from the Philippines. Similar to previous reports [22], IRRI 6609, IRRI7007, and S-E1 appear to be more related to each other. Values that separate Pfv and Pfv-like groups were below the species boundary cut-off of 95% (Fig 2A). This result also correlates with the TETRA analysis that had a species delineation cut-off of ≥ 99% (S1 Fig). Using a one-way comparison approach, we also explored the distribution of homology between IRRI 6609 and 78 closely related Pseudomonas genomes (S1 Table). Results were more consistent with a continuous distribution rather than a clearly defined cluster, with average ANI values ranging from 82.57% for P. stutzeri (n = 6) genomes to 96.13% for Pfv-like (n = 2) genomes (S2 Fig). The data suggest that Pfv-like genomes were more related to Pfv than to any of the other 71 closely related Pseudomonas genomes. We also characterized genome ancestry using a phylogenetic analysis involving 34 Pseudomonas accessions. Using rpoB and rpoD, we found two separated clusters (Fig 2B) that correlated with the homology status of ANI and AAI, in which Pfv and Pfv-like populations share a common ancestor. To further support the phylogenetic relationship of Pfv and Pfv-like strains (Fig 2B), we constructed a robust evolutionary tree using 10 housekeeping genes and observed similar results (S3 Fig). It is clear that species definition in the genus Pseudomonas can be particularly problematic for some of the groups due to the intrinsic diversity in their genomic content [17, 21]. For instance, the P. fluorescens group actually involved multiple species [20, 21] occupying quite diverse ecological niches. Based on the whole genome comparison or phylogenetic inference, we did not find any evidence suggesting that Pfv-like strains may be considered as Pfv sensu stricto. Moreover, Pfv may be part of a species complex composed of different groups including the Pfv-like organisms analyzed in this study. Whether these strains can be considered a novel species or not will need further research efforts.
Fig 2. P. fuscovaginae-like (Pfv-like) strains are closely related to P. fuscovaginae (Pfv).
A) Average nucleotide identity (ANI) and average amino acid identity (AAI) clustering analysis of the eight rice-infecting Pseudomonas draft genomes. Clustering analysis identified two separated groups involving Pfv-like strains (orange) collected in the Philippines and Pfv strains (blue) collected elsewhere. Values scale is depicted in red, orange, yellow, and white colors in ANI (horizontal) and AAI (vertical) pairwise comparison. Value cut-offs with >95% reflect the possibility of same species grouping. The heatmap was generated in the R package gplots using the heatmap.2 function. B) Phylogenetic reconstruction of rice-infecting Pseudomonas and closely related Pseudomonas species using the concatenated housekeeping rpoB and rpoD. Maximum likelihood was used to infer the phylogenetic relationship with bootstrap of 1000 using the RAxML software [61]. Pfv and Pfv-like are highlighted in blue and orange, respectively.
Pfv-like and Pfv harbor high levels of structural polymorphism
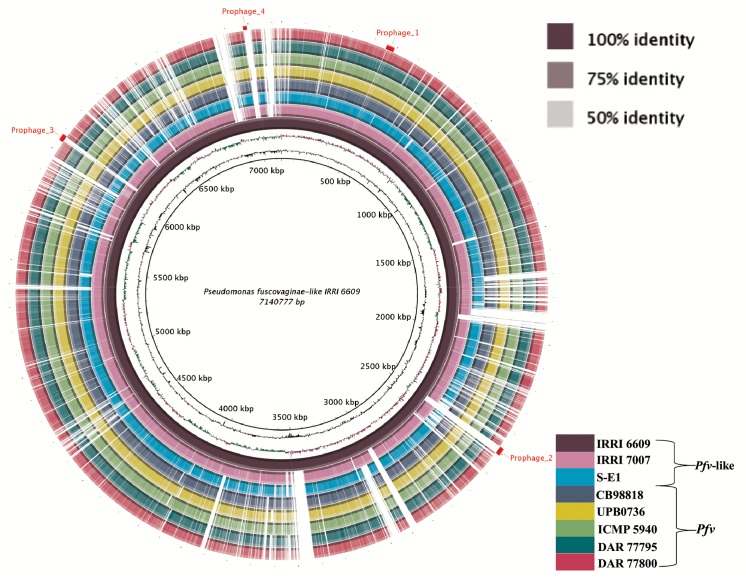
A significant variation in the genomic composition of Pseudomonas groups has been reported recently [20, 64] and previous studies on Pfv have also found important genetic and biochemical variations among strains from different parts of the world [10, 35]. To understand the genome structure of rice-infecting Pseudomonas pathogens, we performed a comparative genomics analysis on eight draft genome datasets (Table 1). Whole genome alignments showed a high level of polymorphism among strains of Pfv and Pfv-like (Fig 3). Many syntenic blocks were interrupted by insertions, deletions, and rearrangements. A closer comparison of the eight genomes in terms of percentage of nucleotide identity and orthologous genes can be found in Table 2. Although Pfv and Pfv-like genomes appear to have many syntenic regions (Fig 3), our structural and gene content assessments illustrate a large degree of genomic diversity in Pfv-like strains. Loper et al. [20] also found a similar pattern when they analyzed members of the P. fluorescens subgroup that are closely related or belonged to the same taxa.
Fig 3. The genome of rice-infecting Pseudomonas harbor high level of structural polymorphism.
Global comparison of eight rice-infecting Pseudomonas draft genomes using BLASTn. The inner most ring corresponds to the genomic position at IRRI 6609. The second and third rings indicate G+C content and G+C skew, respectively. The rest of the rings indicate presence and absence portions of the eight rice-infecting Pseudomonas draft genomes against IRRI 6609. Solid colors represent genomic regions with hits while white spaced represent gaps. P. fuscovaginae (Pfv) and P. fuscovaginae-like (Pfv-like) strains are depicted. Sequence identity is related to color intensity. Also included are locations of four intact prophage insertions found in Pfv-like IRRI 6609 (S4 Fig). The global alignment was visualized using BRIG [43].
Table 2. Nucleotide identity and percentage of orthologous genes obtained in rice-infecting Pseudomonas draft genomes compared to IRRI 6609.
| Species | Strain | Nucleotide Identity (%) a | Percentage of orthologous genes |
|---|---|---|---|
| Pseudomonas fuscovaginae-like | IRRI 6609 | 100 | 100 |
| Pseudomonas fuscovaginae-like | IRRI 7007 | 91.93 | 90.19 |
| Pseudomonas fuscovaginae-like | S-E1 | 86.39 | 80.19 |
| Pseudomonas fuscovaginae | CB98818 | 84.03 | 74.17 |
| Pseudomonas fuscovaginae | UPB7036 | 84.21 | 76.22 |
| Pseudomonas fuscovaginae | DAR 77795 | 83.89 | 71.13 |
| Pseudomonas fuscovaginae | DAR 77800 | 84.52 | 60.56 |
| Pseudomonas fuscovaginae | ICMP 5940 | 84.04 | 71.44 |
a Identity based on BLASTn results
A considerable large proportion of prophage-related gene clusters were found in Pfv and Pfv-like strains (S4 Fig), indicating that they might have played an important evolutionary role in lateral gene transfer and contributed as well in the internal rearrangement of the genomes [65]. Out of the nine putative intact prophage clusters, three were present in more than one genome and five were found to be strain-specific. An overall assessment of prophage sequences in the eight genomes identified 644 phage-related genes, 37 novel non-phage related intact coding sequences, and 10 putative secreted proteins (S4 Fig). We showed that structural variation is consistent with major events of insertion/deletion in Pfv-like. While prophage insertions may not be the only mechanism incorporating foreign genes in the Pfv genetic pool, it is certainly contributing to genetic diversity. The presence of predicted secreted proteins within the inserted fragments may add an additional layer of functional adaptation as observed in other Pseudomonas groups [21].
Pan-genome of Pfv and Pfv-like are highly accessorized
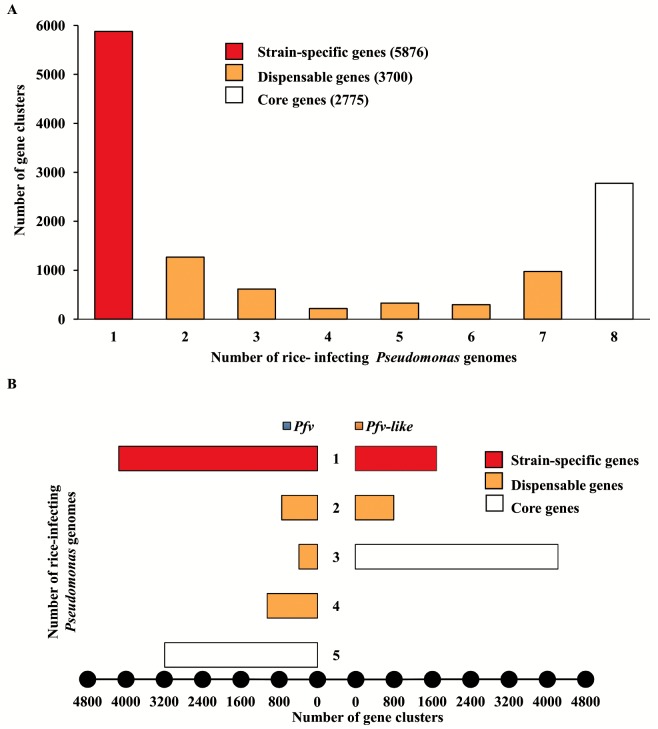
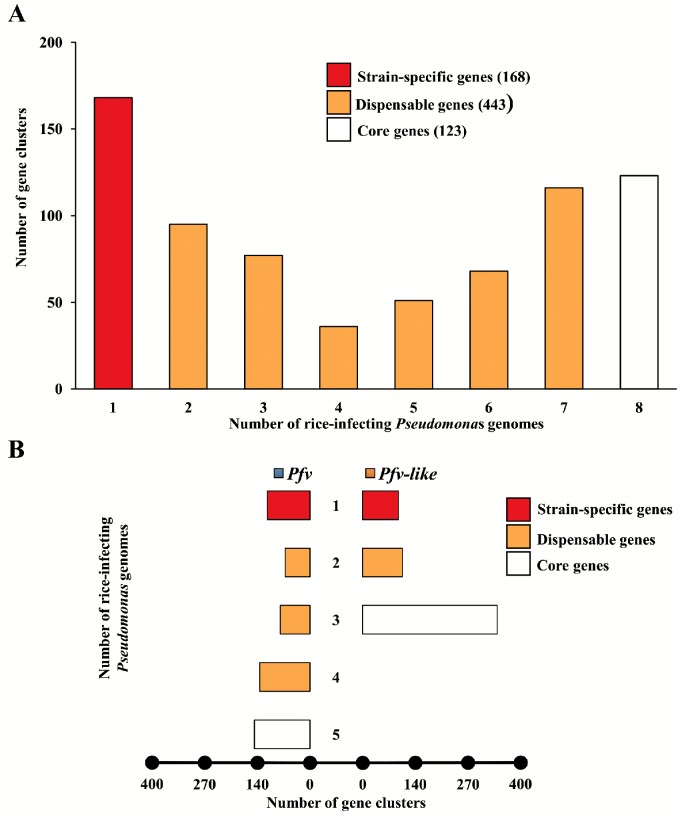
The pan-genome is the overall repository of genes within a species which is further subdivided into core genes shared by all members, dispensable genes shared by more than one member, and strain-specific genes which can be found only in one member [66]. To get insight into the pan-genome of rice-infecting Pseudomonas, we first grouped all 50,664 predicted open reading frames from the eight genomes into 12,351 orthologous gene clusters. Interestingly, the majority of clusters were present in only one of the strains, suggesting that Pfv and Pfv-like have highly accessorized pan-genomes (Fig 4A). It is not likely that the reduced core genome observed in Pfv and Pfv-like is due to poorly assembled draft genomes. All strains showed big genome sizes, high number of coding genes, and random distribution of strain-specific genes located in syntenic regions. For that reason, we predicted that fully assembled Pfv and Pfv-like genomes will produce a slight increase in the core genome but will not change drastically the pattern of strain-specific genes.
Fig 4. The pan-genome of rice-infecting Pseudomonas reveals high proportion of strain-specific genes.
A) Distribution of the 12,351 orthologous gene clusters according to strain-specific genes (only in one genome = 1), dispensable genes (in more than one genome = 2 ≥ x ≤ 7), and core genes (in all genomes = 8). B) Orthologous gene distribution in the P. fuscovaginae (blue) and P. fuscovaginae-like (orange) genomes depicting number of core, dispensable, and strain-specific gene clusters.
To explore the genetic differences within rice-infecting Pseudomonas populations in more detail, we repeated the analysis using Pfv and Pfv-like genomes independently and found that 77% of the 12,351 orthologous clusters were present in Pfv. As expected, the number of strain-specific clusters remained high, which suggests that each strain maintains its own repertoire of genes (Fig 4B). Although Pfv-like have only three strains and share the 54% of the overall orthologous clusters, we identified 1,696 strain-specific clusters in this dataset (Fig 4B). In line with these findings, the functional annotation of Pfv and Pfv-like core genes was also consistent with metabolic versatility (S5 Fig). We observed an enrichment in core genes related to transcription, inorganic ion transport metabolism, and intracellular trafficking between Pfv-like and Pfv. The examined trend suggests that each Pfv-like and Pfv genome is extremely plastic and harbored a battery of genes potentially involved in their adaptation to a number of hosts or different environments. As of the moment, we cannot discard that sampling bias may also contribute to the observed differences.
Accurate estimation of bacterial pan-genome can be highly dependent on the number and diversity of strains involved in the analysis [67]. To estimate if Pfv has an open or close pan-genome, we used a power law regression model [68] to establish the relationship between cluster size and strain number. Our result was consistent with an infinite or open Pfv pan-genome (S6 Fig), in which the number of genes in the pool increased exponentially with each genome added without reaching a clear plateau. The number of core genes was also constantly decreasing with the addition of each new genome (S6 Fig). The inferred pan-genome also correlates with high levels of structural variation (Fig 3), low levels of sequence homology as measured by ANI-AAI analysis (Fig 2A), and high proportion of strain-specific genes (Fig 4A and 4B). Therefore, our findings are not surprising since Pfv appears to be an opportunistic pathogen with a broad host range and mechanism that allow frequent gene exchange in multiple niches. It is likely that Pfv-like groups are also capable to use multiple sources and colonize different environments. A similar situation was described for other Pseudomonas groups that have evolved high levels of genomic plasticity such as P. fluorescens and P. syringae pathovars [19, 21, 64]. Despite the limited understanding about Pfv and Pfv-like life cycle, the analysis of the pan-genome structure is giving us important clues into the biology of these rice pathogens.
Pfv-like has four major secretion apparatus
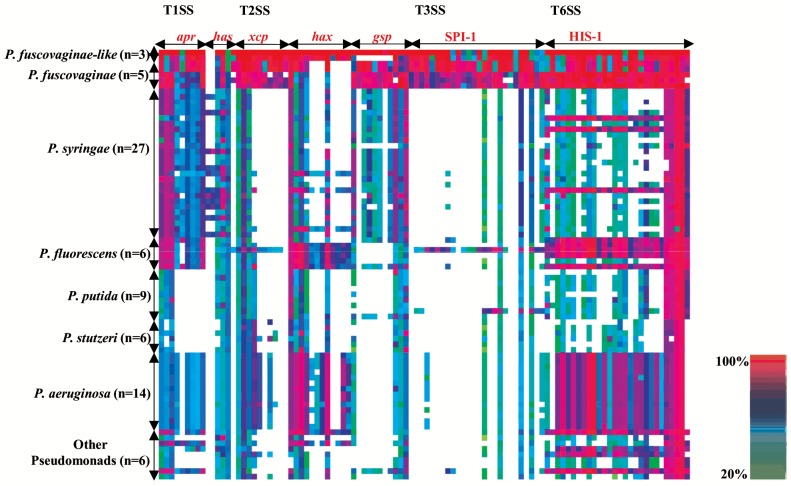
Plant pathogenic bacteria use a combination of secretion systems to modify the surrounding environment and to interact with hosts and other microbes [69]. Using homology pairwise comparison, we investigated the composition and conservation of the secretion apparatus in Pfv-like genomes and found intact Type I (T1SS), Type II (T2SS), Type III (T3SS), and Type VI (T6SS) systems (Fig 5 and S4 Table). All these secretion systems are commonly found in host-associated bacteria and have been reported in Pfv draft genomes [33–35].
Fig 5. Comparative genomic analysis of rice-infecting Pseudomonas secretion apparatus.
Genetic components of T1SS, T2SS, T3SS and T6SS apparatus of P. fuscovaginae-like (Pfv-like) IRRI 6609 was used to compare against 79 closely related Pseudomonas genomes (S1 Table). The apr, has, xcp, hxc, gsp, SPI-1, and HSI-1 are previously characterized gene clusters found within each secretion system. Horizontal axis describes the number of species used for comparison. The rows were sorted by amino acid sequence identity with threshold set at 20%. The heat map was visualized in CodaChrome. Homology range values are shown in bottom right.
We then used a comparative approach to investigate the level of conservation of the Pfv-like secretion apparatus in 79 closely related Pseudomonas genomes, including rice-infecting Pseudomonas. The T1SS, which includes the apr and has clusters [70], was highly conserved across all Pseudomonas genomes with the exception of P. putida and P. stutzeri (Fig 5). This cluster includes an alkaline protease which has a strong homology to arpA [71] and additional genes with predicted exoprotease activities. We found three different T2SS gene clusters in Pfv-like. The xcp cluster showed strong homology with P. aeruginosa [70] while the gsp cluster appears to be conserved within P. syringae genomes [72]. Interestingly, the hxc cluster is only present in Pfv-like strains and has homology to P. fluorescens and P. aeruginosa genomes (Fig 5). Substrates for T2SS, such as CWDE, were also identified in Pfv-like genomes (see below).
Interestingly, Pfv and Pfv-like populations lack the typical hrp/hrc T3SS found in other plant pathogenic Pseudomonas but carry another T3SS family member, the SPI-1 (Salmonella Pathogenicity Island 1) (Fig 5 and S4 Table). Loss-of-function experiments suggest that Salmonella enterica SPI-1 contributes to virulence by secreting type III effectors during human cell colonization [73]. Recent reports that showed SPI-1 in several plant-associated bacteria [21, 33, 74–77] suggested alternative functions outside the mammalian system [78]. Indirect evidence from Arabidopsis thaliana and Nicotiana tabacum showed that S. typhimurium SPI-1 mutants were unable to suppress plant immune response [79, 80]. In our analysis, SPI-1 was poorly conserved in almost 70 Pseudomonas genomes. Only the P. fluorescens strain F113, a plant growth-promoting bacterium, carries an intact SPI-1 cluster [21]. Interestingly, our analysis identified only a few candidate type III effectors in the IRRI 6609 and IRRI 7007 genomes but their role during host colonization is still unclear. Whether SPI-1 is used by Pfv or Pfv-like to deliver unknown type III effectors to interact with rice or with alternative hosts outside the plant kingdom remains to be an area that should be investigated further.
T6SS showed strong homology to the corresponding hcp Secretion Island 1 (HSI-1) [81] encoded in the genome of all 14 P. aeruginosa, 5 out of 6 P. fluorescens, and 3 out of 27 P. syringae strains (Fig 5). This finding is not surprising since T6SS is highly conserved among pathogenic bacteria and has been implicated in multiple functions. For instance, P. aeruginosa uses T6SS to secret the effector protein Tse2 as a component of a toxin-substrate system that mediates interactions between bacteria [82]. Meanwhile, T6SS mutants of the seed-borne pathogen Acidovorax citrulli were impaired in seed-to-seedling transmission in citrus plants [83]. More recently, a Pfv Tn5 mutant of the T6SS showed impaired colonization of rice tissue [32], suggesting that T6SS contributes to pathogenicity. Even though functional data that links Pfv-like secretion system with virulence in rice is still missing, it is likely that each secretion system was acquired independently. This observation indicates a diversity of functional roles that is aligned with the general idea that Pfv-like, similar to Pfv, is capable of colonizing multiple environments.
The core secreted repertoire of Pfv and Pfv-like has signatures of positive selection
Similar to other bacterial pathogens, rice-infecting Pseudomonas are predicted to secrete a number of effector proteins that contribute to disease progression. To investigate the secretion capabilities in Pfv and Pfv-like genomes and to identify putative core and dispensable secretome, we used a combination of prediction tools. We first used SignalP [50] and identified 4,244 proteins that had canonical secretion signals in all the eight genomes. Then, we removed 715 proteins which were predicted to have at least one transmembrane domain, as those may represent membrane-anchored proteins. Using the same approach as described above, we estimated 734 orthologous gene clusters as the overall repertoire of putative secreted proteins in the eight genomes (Fig 6A). Among those, 168 corresponded to strain-specific genes (Fig 6A). We also analyzed the distribution of putative secreted proteins in Pfv and Pfv-like groups independently. Both groups maintained high proportion of unique genes reaching 20.98% for Pfv and 17.05% for Pfv-like (Fig 6B). These findings point out to a unique set of secreted proteins in each rice-infecting Pseudomonas genome consistent with multiple functional capabilities. In the same way, Baltrus et al. [64] found dramatic variation in the number and distribution of effector genes across a range of P. syringae clades from different hosts. Recent analysis of the P. fluorescens F113 strain harbored an unprecedented combination of unique genes related to rhizosphere colonization [21]. Therefore, it can be assumed that some Pseudomonas groups probably evolved to maximize the dispensable secretome as a strategy to occupy a range of environments.
Fig 6. The secretome of rice-infecting Pseudomonas has high proportion of dispensable genes.
A) Distribution of the 729 orthologous gene clusters in the secretome according to strain-specific genes (only in one genome = 1), dispensable genes (in more than one genome = 2 ≥ x ≤ 7) and core genes (in all genomes = 8). B) Orthologous gene distribution in the P. fuscovaginae (blue) and P. fuscovaginae-like (orange) genomes depicting number of core, dispensable, and strain-specific gene clusters.
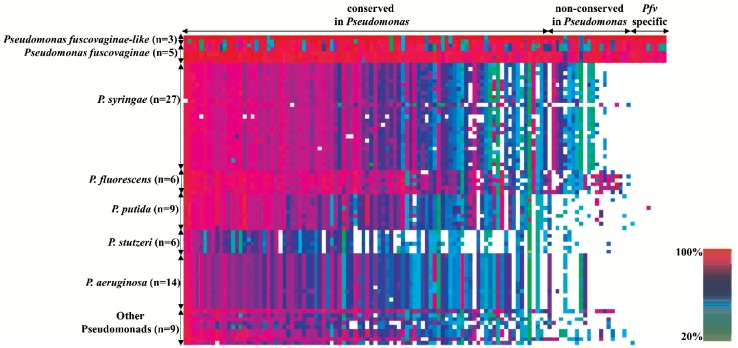
Importantly, we identified 123 core genes that codified for putative secreted proteins in all the rice-infecting Pseudomonas genomes (Fig 6A and 6B, and S5 Table). Most of the candidate core secretome was associated with transport, catalytic, and binding activities (S6 Table), which are the possible molecular functions of the core secreted proteins. To further categorize the level of conservation in other Pseudomonas genomes, we used homology comparison and identified 31 candidate genes that were homologous to known plant pathogenic Pseudomonas (Fig 7). Among these, 13 putative secreted proteins were unique to rice-infecting Pseudomonas strains. Since all strains were isolated from rice and showed exactly the same brown rot and seed discoloration symptoms, this set of genes is likely to play a role during interaction with the host plant. The other 96 secreted proteins were conserved among free living and pathogenic Pseudomonas isolated from a range of eukaryotic hosts (Fig 7). Future research is needed to understand the role of these secreted proteins during interaction with rice.
Fig 7. The core secretome of rice-infecting Pseudomonas harbor unique genes.
Conservation of core secreted proteins from rice-infecting Pseudomonas was evaluated in 79 closely related Pseudomonas genomes (S1 Table). Columns were sorted by averaging the amino acid identity to identify conserved and species-specific proteins using threshold of 20%. Secreted proteins are also classified in: conserved in all Pseudomonas, non-conserved in all Pseudomonas, and Pfv- and Pfv-like-specific. Horizontal axis describes the number of species used for comparison. The heat map was visualized in CodaChrome [40]. Homology range values are shown in bottom right.
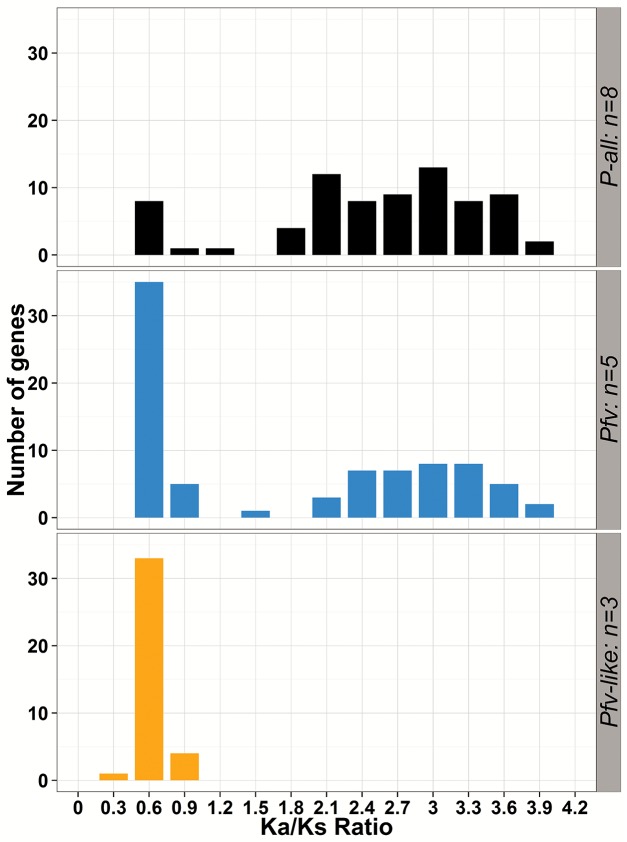
A common feature of effector genes from plant pathogenic microbes is the strong signature of positive selection [84–86]. To characterize the selection pressures underlying the Pfv and Pfv-like core secretome and to identify candidate effector genes, we calculated Ka/Ks ratio using Yn00 [59] on alignments of 123 orthologous loci across the eight genomes. Using a cutoff p-value of 95%, we found that Ka value was greater than Ks (ω = Ka/Ks > 1) in 75 of 123 genes (Fig 8). We obtained ω values ranging from 0.45 to 4.0 (average of 2.4). When we analyzed each group separately, Pfv secretome ω values ranged from 0.37 to 3.76 (average of 1.65) while Pfv-like ranged from 0.14 to 0.8 (average of 0.48) (Fig 8). Among the 75 selected genes, we found high ω values in 3 out of 13 genes that were specific to rice-infecting Pseudomonas (Fig 8). This result shows significant differences in the number of genes under selection between Pfv and Pfv-like. Whether this difference is due to variation in selection pressure or sampling bias between groups is still unknown. Collectively, our findings point out that natural selection is continuously shaping the secreted repertoire of the rice-infecting Pseudomonas, which is a common pattern for other Pseudomonas pathogens [85, 87].
Fig 8. The core secretome of rice-infecting Pseudomonas has signatures of positive selection.
Distribution of Ka/Ks ratio for 123 protein-coding genes, calculated with Yn00 [59] method on rice-infecting Pseudomonas-all (black, P-all, n = 8), P. fuscovaginae (blue, Pfv, n = 5), and P. fuscovaginae-like (orange, Pfv-like, n = 3) datasets. All secreted protein selected on this graph have p-values ≤ 0.01.
Additional features potentially involved in pathogenicity of Pfv-like
To further explore the presence of additional pathogenicity mechanisms within the Pfv-like genome, we identified putative genes involved in secondary metabolism biosynthesis, hormone metabolism, motility, cell adhesion, pilus formation, and cell wall degradation (S3 and S4 Tables). All of the probable secondary metabolite gene clusters of Pfv-like are found in S4 Table. Three gene clusters related to AHL biosynthesis and quorum sensing were identified. Pfv harbors both AHL clusters (PfsI/R and PfvI/R) described in Mattiuzzo et al. [28] and one additional putative novel cluster yet to be characterized (S3 and S4 Tables). Additionally, two classes of siderophores biosynthetic genes were located in the genomes of Pfv-like. The cluster for pyoverdine biosynthesis found in Pfv-like is homologous to the pvd gene cluster from P. aeruginosa [88]. Pyoverdine gives Pseudomonas spp. their fluorescent pigments and is associated with iron acquisition [88]. We also found the acs gene cluster that is associated to achromobactin production [89]. Achromobactin, just like pyoverdine, appears to facilitate iron acquisition as an alternative function. Mutant strains of P. syringae pv. phaseolicola 1448a showed that pyoverdine and achromobactin were not essential in causing halo blight in beans [90]. In addition, the pyoverdine production of P. fluorescens F113 was effective in inhibiting the growth of Pectobacterium atrosepticum in vitro [91]. Whether the production of pyoverdine and achromobactin is important for microbial competition or during host pathogenicity of Pfv-like is still not known.
We found three non-ribosomal peptide (NRP) gene clusters involving at least 69 genes (S3 and S4 Tables). Two of these clusters showed no homology to any NRP cluster in other Pseudomonas species. Moreover, the biosynthetic products could not be identified. We also predicted a gene that codifies for tryptophan 2-monooxygenase, which is an important enzyme in auxin anabolism (S4 Table). Heterologous expression of this gene in A. thaliana promoted susceptibility to the bacterial pathogen P. syringae pv. tomato DC3000 [92]. Pfv-like genome also harbored T4 pili and flagella formation gene clusters which are important in bacterial adhesion to the cell and motility, respectively. In contrast to plant pathogens that cause rotting [93], Pfv-like appeared to have reduced repertoire of CWDE. From the 85 proteins predicted to have carbohydrate-active enzymes, only three harbored a canonical secretion signal (S4 Table). Some of the putative pathogenicity factors found in Pfv-like were also present in Pfv. The tryptophan 2-monooxygenase gene, clusters involved in pilus and flagella formation, biosynthetic clusters of achromobactin and pyoverdine, and the second putative NRP gene cluster were all conserved in Pfv, suggesting similar mechanisms. However, the third quorum sensing, and the two remaining NRP gene clusters appear to be unique to Pfv-like (S4 Table).
Pfv-like induce multiple putative virulence factors during its interaction with rice
Infection of host plants by pathogenic bacteria is a complex process that requires the coordinated expression of many virulence components [94]. To determine the contribution of Pfv-like putative virulence factors during rice colonization, we analyzed the expression of 19 pathogen genes during a time course infection experiment using semi-quantitative RT-PCR. Total RNA was isolated from rice sheaths at 0, 3, 24, 48, and 72 hpi of strain IRRI 7007 in rice cv. Azucena. Selected genes are listed in S4 Table and represents: secretion apparatus T1SS (1), T2SS (3), T6SS (1), and SPI-1 (1); one gene involved in T4 pili formation; one flagella gene; two putative CWDEs; one gene predicted to be involved in putative NRP biosynthesis; six core secreted proteins; and two random core genes present in IRRI 6609 and IRRI 7007 (S3 and S4 Tables).
We detected transcript accumulation in 14 of 19 pathogen genes in at least one of the examined time points (S7 Fig). As expected, the pattern of expression varied considerably among genes. Some genes were constitutively expressed while others were induced as early as 3 hpi or down regulated after infiltration. Interestingly, genes that formed the secretion apparatus and those presumed to be secreted throughout this mechanism were detected at early stages of bacterial infection (S7 Fig). A putative alkaline proteases (PF66_01465), which is known to be secreted by T1SS, was induced at an early time point. In the same way, two genes with predicted CWDE activity (PF66_04809 and PF66_04639) were expressed during the first hours of infection. However, none of the genes representing the T2SS apparatus (PF66_01622, PF66_04896, and PF66_01114) were induced, suggesting that CWDE may be secreted throughout alternative pathways. Two putative methionine aminopeptidase with a predicted T3 secretion signal were expressed in this experiment (S7 Fig) The predicted T3SS effector gene PF66_01486 showed detectable levels at 3 hpi and was down regulated afterwards while PF66_00566 showed constitutive expression (S7 Fig). Recent experimental data showed that T6SS might be important for Pfv pathogenicity [32]. In our experiment, PF66_03771, which is part of T6SS was upregulated during the first three hours. These results may highlight the contribution of at least three secretion systems during the infection of Pfv-like in rice sheath.
The gene involved in membrane adhesion (PF66_01105) was found to be expressed at 3 hpi while the gene involved in locomotive function (PF66_05042) was highly expressed at all time points. Four (PF66_04809, PF66_01042, PF66_04639, and PF66_00996) out of six predicted secreted genes showed very early response during the interaction of rice and the Pfv-like strain IRRI 7007. The gene PF66_00996, which has a putative copper oxidase function, is also specific for Pfv and Pfv-like and has strong signature of positive selection (S7 Fig). Similar to known effector proteins, the induction of candidate core secreted genes during the early stage of infection may suggest the important functional role of PF66_04809, PF66_01042, PF66_04639, and PF66_00996 in Pfv-like pathogenicity. At a later stage (72 hpi), most of the tested genes had low level of expression. Overall, data obtained here revealed that Pfv-like interaction with rice sheath is accompanied by transcript accumulation of genes involved in secretion, adhesion, and trafficking, representing a concerted virulence mechanism to infect rice.
Conclusion
In this study, we focused on understanding the complexity of rice-infecting Pseudomonas genomes that cause sheath brown rot and the underlying mechanisms that might govern its virulence in rice. Rather than genetically homogeneous lineages, Pfv and Pfv-like appear to have an open pan-genome with each isolate representing a very distinct lineage carrying its own repertoire of accessory genes. Structural variation may reflect major events of insertion-deletion in a genome that allows frequent gene exchange. The plasticity in genome content, the overall diversity of metabolic capabilities, and the functional adaptation of its secretome are all consistent with the idea that Pfv and Pfv-like are able to occupy multiple niches. Similar to other Pseudomonas species that show intrinsic genetic variation and continuous distribution of genetic diversity [17, 20, 21], Pfv and Pfv-like appear to represent different phylogenetic groups. However, the phylogenetic status of these strains needs to be analyzed in the context of Pfv meta-population. At the genomic level, Pfv-like harbors many distinct mechanisms that are potentially involved in rice colonization. Many of the genes involved in secretion, adhesion, and trafficking are activated during the early stages of infection, thereby highlighting its potential role during rice pathogenicity. We predicted a conserved set of secreted proteins in the pan-genome of Pfv-like and Pfv, most of which have strong signatures of positive selection. These observations reveal a unique evolutionary pathway of rice-infecting Pseudomonas in the agricultural landscape but also explain partially the ability of Pfv to infect a broad range of host plants outside the rice cropping system. Functional validation of core and strain-specific genes as well as an assessment of phenotypic differences between Pfv and Pfv-like groups will be important to reveal the conserved mechanism of infection and the unique set of metabolic capabilities of these populations. We hope these observations can lead to better strategies of control, monitoring and management of sheath blight rot in the rice plant.
Supporting Information
Clustering analysis differentiates P. fuscovaginae-like (Pfv-like) collected in the Philippines and P. fuscovaginae (Pfv) collected elsewhere. Values scale is depicted in red, orange, yellow, and white colors in TETRA pairwise comparison. Value cut-offs with >99% reflect the possibility of same species grouping. The heatmap was generated in the R package gplots using the heatmap.2 function.
(TIFF)
The middle line in each box plot represents the mean of ANI values for a particular taxonomic group. Also shown in each box plot are the minimum and maximum values. The number of genomes is denoted in brackets. All Pseudomonas genomes used in this analysis are listed in S1 Table.
(TIFF)
Maximum likelihood was used to infer the phylogenetic relationship with bootstrap of 1000 using the RAxML software [61]. Groups Pfv-like and Pfv are highlighted as orange and blue, respectively.
(TIFF)
A) Distribution of intact prophage insertions are color-coded among rice-infecting Pseudomonas strains. Prophage ID is based on PHAST database [38]. All intact prophage have scores >90 [38]. B) Bar plot distribution of the number of genes found within all prophage regions in each of the eight rice-infecting Pseudomonas draft genomes. Non-phage related genes are further classified as secreted and non-secreted.
(TIFF)
Bar plots representing number of genes in each COG category per rice-infecting Pseudomonas-all (black, n = 8), P. fuscovaginae (blue, n = 5) and P. fuscovaginae-like (orange, n = 3).
(TIFF)
Black dots represent standard error; X and Y axes represent genome size and number of genomes, respectively.
(TIFF)
Rice sheath (O. sativa cv. Azucena) infected with P.fuscovaginae-like strain IRRI 7007 was evaluated at 0, 3, 24, 48, and 72 hpi. Gen ID and functional classification are indicated in both sides.
(TIFF)
The 79 Pseudomonas genomes are listed according to species, strains and their corresponding GenBank ID.
(DOCX)
Locus ID and sequence correspond to P. fuscovaginae-like strain IRRI 6609.
(XLSX)
(XLSX)
(DOCX)
Annotation was done using Pfam. Locus ID based on P.fuscovaginae-like IRRI 6609.
(XLSX)
(DOCX)
Acknowledgments
We acknowledge the technical assistance of Fanny Garcia, Pauline Capistrano, Elenita Silab, Ismael Mamiit and Eufrocino Pizarra. We greatly acknowledge Bart Cottyn for the original classification of bacterial cultures.
Data Availability
All genome sequences are deposited at GenBank with accession numbers JSYZ00000000 and JTBY00000000.
Funding Statement
Scientists at the International Rice Research Institute are partially funded by the Global Rice Science Partnership (GRiSP). The program is supported by the Consortium for International Agricultural Research (CGIAR) with headquarters in Montpellier, France. More information can be found at http://www.cgiar.org/. No additional funding source is declared.
References
- 1. McDonald BA, Linde C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol. 2002;40: 349–379. [DOI] [PubMed] [Google Scholar]
- 2. Miyajima K, Tanii A, Akita T. Pseudomonas fuscovaginae sp. nov., nom. rev. Int J Syst Bacteriol. 1983;33: 656–657. [Google Scholar]
- 3. Zeigler RS, Alvarez E. Characteristics of Pseudomonas spp. causing grain discoloration and sheath rot of rice, and associated pseudomonad epiphytes. Plant Dis. 1990;74: 917–922. [Google Scholar]
- 4. Adorada DL, Stodart BJ, Pangga IB, Ash GJ. Implications of bacterial contaminated seed lots and endophytic colonization by Pseudomonas fuscovaginae on rice establishment. Plant Pathol. 2015;64: 43–50. [Google Scholar]
- 5. Tanii A, Miyajima K, Akita T. The sheath brown rot disease of rice plant and its causal bacterium, Pseudomonas fuscovaginae sp. nov. Ann Phytopathol Soc Jpn. 1976;42: 540–548. [Google Scholar]
- 6. Zeigler RS, Alvarez E. Bacterial sheath brown rot of rice caused by Pseudomonas fuscovaginae in Latin America. Plant Dis. 1987;71: 592–597. [Google Scholar]
- 7. Rott P, Honegger J, Notteghem JL, Ranomenjanahary S. Identification of Pseudomonas fuscovaginae with biochemical, serological, and pathogenicity tests. Plant Dis. 1991;75: 843–846. [Google Scholar]
- 8. Autrique A, Maraite H. La pourriture brune de la gaine foliaire du riz causee par Pseudomonas fuscovaginae . FAO Plant Prot B. 1983;31: 94. [Google Scholar]
- 9. Duveiller E, Miyajima K, Snacken F, Autrique A, Maraite H. Characterization of Pseudomonas fuscovaginae and differentiation from other fluorescent Pseudomonads occurring on rice in Burundi. J Phytopathol. 1988;122: 97–107. [Google Scholar]
- 10. Rott P, Notteghem JL, Frossard P. Identification and characterization of Pseudomonas fuscovaginae, the causal agent of bacterial sheath brown rot of rice, from Madagascar and other countries. Plant Dis. 1989;73: 133–137. [Google Scholar]
- 11. Marzukhi H, Ali AH, Hassan S. The presence of a new disease of rice in paddy estates Seberang Perak. Teknologi Padi. 1991;7: 49–52. [Google Scholar]
- 12. Cottyn B, VanOutryve MF, Cerez MT, DeCleene M, Swings J, Mew TW. Bacterial diseases of rice. 2. Characterization of pathogenic bacteria associated with sheath rot complex and grain discoloration of rice in the Philippines. Plant Dis. 1996;80: 438–445. [Google Scholar]
- 13. Xie G. First report of sheath brown of rice in China and characterization of the causal organism by phenotypic tests and biolog. Intl Rice Res Notes. 2003;28: 50–52. [Google Scholar]
- 14. Cother EJ, Stodart B, Noble DH, Reinke R, van de Ven RJ. Polyphasic identification of Pseudomonas fuscovaginae causing sheath and glume lesions on rice in Australia. Australas Plant Path. 2009;38: 247–261. [Google Scholar]
- 15. Duveiller E, Snacken F, Maraite H, Autrique A. First detection of Pseudomonas fuscovaginae on maize and sorghum in Burundi. Plant Dis. 1989;73: 514–517. [Google Scholar]
- 16. Duveiller E, Notteghem JL, Rott P, Snacken F, Maraite H. Bacterial sheath brown rot of rice caused by Pseudomonas fuscovaginae in Malagasy. Trop Pest Manage. 1990;36: 151–153. [Google Scholar]
- 17. Mulet M, Lalucat J, Garcia-Valdes E. DNA sequence-based analysis of the Pseudomonas species. Environ Microbiol. 2010;12: 1513–1530. 10.1111/j.1462-2920.2010.02181.x [DOI] [PubMed] [Google Scholar]
- 18. Silby MW, Cerdeno-Tarraga AM, Vernikos GS, Giddens SR, Jackson RW, Preston GM, et al. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens . Genome Biol. 2009;10: R51 10.1186/gb-2009-10-5-r51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kimbrel JA, Givan SA, Halgren AB, Creason AL, Mills DI, Banowetz GM, et al. An improved, high-quality draft genome sequence of the germination-arrest factor-producing Pseudomonas fluorescens WH6. BMC Genomics. 2010;11: 522 10.1186/1471-2164-11-522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Loper JE, Hassan KA, Mavrodi DV, Davis EW, Lim CK, Shaffer BT, et al. Comparative genomics of plant associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012;8: e1002784 10.1371/journal.pgen.1002784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Redondo-Nieto M, Barret M, Morrissey J, Germaine K, Martinez-Granero F, Barahona E, et al. Genome sequence reveals that Pseudomonas fluorescens F113 possesses a large and diverse array of systems for rhizosphere function and host interaction. BMC Genomics. 2013;14: 54 10.1186/1471-2164-14-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cottyn B, Barrios H, George T, Vera Cruz CM. Characterization of rice sheath rot from Siniloan, Philippines. Intl Rice Res Notes. 2002;27: 39–40. [Google Scholar]
- 23. Alfano JR, Collmer A. Bacterial pathogens in plants: life up against the wall. Plant Cell. 1996;8: 1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abramovitch RB, Anderson JC, Martin GB. Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Bio. 2006;7: 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol. 2006;60: 425–449. [DOI] [PubMed] [Google Scholar]
- 26. Alfano JR. Roadmap for future research on plant pathogen effectors. Mol Plant Pathol. 2009;10: 805–813. 10.1111/j.1364-3703.2009.00588.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444: 567–573. [DOI] [PubMed] [Google Scholar]
- 28. Mattiuzzo M, Bertani I, Ferluga S, Cabrio L, Bigirimana J, Guarnaccia C, et al. The plant pathogen Pseudomonas fuscovaginae contains two conserved quorum sensing systems involved in virulence and negatively regulated by RsaL and the novel regulator RsaM. Environ Microbiol. 2011;13: 145–162. 10.1111/j.1462-2920.2010.02316.x [DOI] [PubMed] [Google Scholar]
- 29. Flamand MC, Pelsser S, Ewbank E, Maraite H. Production of syringotoxin and other bioactive peptides by Pseudomonas fuscovaginae . Physiol Mol Plant P. 1996;48: 217–231. [Google Scholar]
- 30. Ballio A, Bossa F, Camoni L, DiGiorgio D, Flamand MC, Maraite H, et al. Structure of fuscopeptins, phytotoxic metabolites of Pseudomonas fuscovaginae . FEBS Lett. 1996;381: 213–216. [DOI] [PubMed] [Google Scholar]
- 31. Batoko H, d'Exaerde AD, Kinet JM, Bouharmont J, Gage RA, Maraite H, et al. Modulation of plant plasma membrane H+-ATPase by phytotoxic lipodepsipeptides produced by the plant pathogen Pseudomonas fuscovaginae . BBA-Biomembranes. 1998;1372: 216–226. [DOI] [PubMed] [Google Scholar]
- 32. Patel HK, Matiuzzo M, Bertani I, Bigirimana VD, Ash GJ, Hofte M, et al. Identification of virulence associated loci in the emerging broad host range plant pathogen Pseudomonas fuscovaginae . BMC Microbiol. 2014;14: 274 10.1186/s12866-014-0274-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Patel HK, da Silva DP, Devescovi G, Maraite H, Paszkiewicz K, Studholme DJ, et al. Draft genome sequence of Pseudomonas fuscovaginae, a broad host range pathogen of plants. J Bacteriol. 2012;194: 2765–2766. 10.1128/JB.00341-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xie GL, Cui ZQ, Tao ZY, Qiu H, Liu H, Ibrahim M, et al. Genome sequence of the rice pathogen Pseudomonas fuscovaginae CB98818. J Bacteriol. 2012;194: 5479–5480. 10.1128/JB.01273-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ash GJ, Lang JM, Triplett LR, Stodart BJ, Verdier V, Cruz CV, et al. Development of a genomics-based LAMP (Loop-Mediated Isothermal Amplification) assay for detection of Pseudomonas fuscovaginae from rice. Plant Dis. 2014;98: 909–915. [DOI] [PubMed] [Google Scholar]
- 36. Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Pillay M, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42: D560–D567. 10.1093/nar/gkt963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42: D206–D214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou Y, Liang YJ, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39: W347–W352. 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, et al. antiSMASH 2.0-a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41: W204–W212. 10.1093/nar/gkt449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rokicki J, Knox D, Dowell RD, Copley SD. CodaChrome: a tool for the visualization of proteome conservation across all fully sequenced bacterial genomes. BMC Genomics. 2014;15: 65 10.1186/1471-2164-15-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics. 2009;25: 2071–2073. 10.1093/bioinformatics/btp356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5: 113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12: 402 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. P Natl Acad Sci USA. 2009;106: 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5: R12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Contreras-Moreira B, Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microb. 2013;79: 7696–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li L, Stoeckert CJ, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kristensen DM, Kannan L, Coleman MK, Wolf YI, Sorokin A, Koonin EV, et al. A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches. Bioinformatics. 2010;26: 1481–1487. 10.1093/bioinformatics/btq229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8: 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 51. Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6: 175–182. [PubMed] [Google Scholar]
- 52. Wang Y, Huang H, Sun M, Zhang Q, Guo D. T3DB: an integrated database for bacterial type III secretion system. BMC Bioinformatics. 2012;13: 66 10.1186/1471-2105-13-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 54. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40: D290–D301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, et al. InterPro: an integrated documentation resource for protein families, domains and functional sites. Brief Bioinform. 2002;3: 225–235. [DOI] [PubMed] [Google Scholar]
- 56. Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38: W7–W13. 10.1093/nar/gkq291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17: 540–552. [DOI] [PubMed] [Google Scholar]
- 58. Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 2010;8: 77–80. 10.1016/S1672-0229(10)60008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17: 32–43. [DOI] [PubMed] [Google Scholar]
- 60. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30: 2725–2759. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Silvestro D, Michalak I. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 2012;12: 335–337. [Google Scholar]
- 62. Haywood-Farmer E, Otto SP. The evolution of genomic base composition in bacteria. Evolution. 2003;57: 1783–1792. [DOI] [PubMed] [Google Scholar]
- 63. Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. P Natl Acad Sci USA. 2005;102: 2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, et al. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 2011;7: e1002132 10.1371/journal.ppat.1002132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Canchaya C, Fournous G, Brussow H. The impact of prophages on bacterial chromosomes. Mol Microbiol. 2004;53: 9–18. [DOI] [PubMed] [Google Scholar]
- 66. Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15: 589–594. [DOI] [PubMed] [Google Scholar]
- 67. Liang WL, Zhao YB, Chen CX, Cui XY, Yu J, Xiao JF, et al. Pan-genomic analysis provides insights into the genomic variation and evolution of Salmonella Paratyphi A. PLoS One. 2012;7: e45346 10.1371/journal.pone.0045346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". P Natl Acad Sci USA. 2005;102: 13950–13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Records AR. The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol Plant Microbe In. 2011;24: 751–757. [DOI] [PubMed] [Google Scholar]
- 70. Filloux A. Protein secretion systems in Pseudomonas aeruginosa: an essay on diversity, evolution, and function. Front Microbiol. 2011;2: 155 10.3389/fmicb.2011.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Guzzo J, Pages JM, Duong F, Lazdunski A, Murgier M. Pseudomonas aeruginosa alkaline protease evidence for secretion genes and study of secretion mechanism. J Bacteriol. 1991;173: 5290–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. P Natl Acad Sci USA. 2003;100: 10181–10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6: 53–66. [DOI] [PubMed] [Google Scholar]
- 74. Triplett LR, Zhao Y, Sundin GW. Genetic differences between blight-causing Erwinia species with differing host specificities, identified by suppression subtractive hybridization. Appl Environ Microb. 2006;72: 7359–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Alavi SM, Sanjari S, Durand F, Brin C, Manceau C, Poussier S. Assessment of the genetic diversity of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans as a basis to identify putative pathogenicity genes and a type III secretion system of the SPI-1 family by multiple suppression subtractive hybridizations. Appl Environ Microb. 2008;74: 3295–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Correa VR, Majerczak DR, Ammar ED, Merighi M, Pratt RC, Hogenhout SA, et al. The bacterium Pantoea stewartii uses two different type III secretion systems to colonize its plant host and insect vector. Appl Environ Microb. 2012;78: 6327–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Barret M, Egan F, Moynihan J, Morrissey JP, Lesouhaitier O, O'Gara F. Characterization of the SPI-1 and Rsp type three secretion systems in Pseudomonas fluorescens F113. Environ Microbiol Rep. 2013;5: 377–386. 10.1111/1758-2229.12039 [DOI] [PubMed] [Google Scholar]
- 78. Egan F, Barret M, O'Gara F. The SPI-1-like type III secretion system: more roles than you think. Front Plant Sci. 2014;5: 34 10.3389/fpls.2014.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schikora A, Virlogeux-Payant I, Bueso E, Garcia AV, Nilau T, Charrier A, et al. Conservation of Salmonella infection mechanisms in plants and animals. PLoS One. 2011;6: e24112 10.1371/journal.pone.0024112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shirron N, Yaron S. Active suppression of early immune response in tobacco by the human pathogen Salmonella Typhimurium. PLos One. 2011;6: e18855 10.1371/journal.pone.0018855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bleves S, Viarre V, Salacha R, Michel GPF, Filloux A, Voulhoux R. Protein secretion systems in Pseudomonas aeruginosa: a wealth of pathogenic weapons. Int J Med Microbiol. 2010;300: 534–543. 10.1016/j.ijmm.2010.08.005 [DOI] [PubMed] [Google Scholar]
- 82. Hood RD, Singh P, Hsu FS, Guvener T, Carl MA, Trinidad RRS, et al. A type VI secretion system of Pseudomonas aeruginosa targets, a toxin to bacteria. Cell Host Microbe. 2010;7: 25–37. 10.1016/j.chom.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tian Y, Zhao Y, Wu X, Liu F, Hu B, Walcott RR. The type VI protein secretion system contributes to biofilm formation and seed-to-seedling transmission of Acidovorax citrulli on melon. Mol Plant Pathol. 2015;16: 38–47. 10.1111/mpp.12159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Win J, Morgan W, Bos J, Krasileva KV, Cano LM, Chaparro-Garcia A, et al. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell. 2007;19: 2349–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. McCann HC, Rikkerink EHA, Bertels F, Fiers M, Lu A, Rees-George J, et al. Genomic analysis of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLoS Pathog. 2013;9: e1003503 10.1371/journal.ppat.1003503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Guyon K, Balague C, Roby D, Raffaele S. Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum . BMC Genomics. 2014;15: 336 10.1186/1471-2164-15-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Guttman DS, Gropp SJ, Morgan RL, Wang PW. Diversifying selection drives the evolution of the type III secretion system pilus of Pseudomonas syringae . Mol Biol Evol. 2006;23: 2342–2354. [DOI] [PubMed] [Google Scholar]
- 88. Visca P, Imperi F, Lamont IL. Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol. 2007;15: 22–30. [DOI] [PubMed] [Google Scholar]
- 89. Berti AD, Thomas MG. Analysis of achromobactin biosynthesis by Pseudomonas syringae pv. syringae B728a. J Bacteriol. 2009;191: 4594–4604. 10.1128/JB.00457-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Owen JG, Ackerley DF. Characterization of pyoverdine and achromobactin in Pseudomonas syringae pv. phaseolicola 1448a. BMC Microbiol. 2011;11: 218 10.1186/1471-2180-11-218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cronin D, MoenneLoccoz T, Fenton A, Dunne C, Dowling DN, OGara F. Ecological interaction of a biocontrol Pseudomonas fluorescens strain producing 2,4-diacetylphloroglucinol with the soft rot potato pathogen Erwinia carotovora subsp. atroseptica . FEMS Microbiol Ecol. 1997;23: 95–106. [Google Scholar]
- 92. Yin C, Park JJ, Gang DR, Hulbert SH. Characterization of a tryptophan 2-monooxygenase gene from Puccinia graminis f. sp. tritici involved in auxin biosynthesis and rust pathogenicity. Mol Plant Microbe In. 2014;27: 227–235. [DOI] [PubMed] [Google Scholar]
- 93. Charkowski A, Blanco C, Condemine G, Expert D, Franza T, Hayes C, et al. The role of secretion systems and small molecules in soft-rot Enterobacteriaceae pathogenicity. Annu Rev Phytopathol. 2012;50: 425–449. 10.1146/annurev-phyto-081211-173013 [DOI] [PubMed] [Google Scholar]
- 94. Mole BM, Baltrus DA, Dangl JL, Grant SR. Global virulence regulation networks in phytopathogenic bacteria. Trends Microbiol. 2007;15: 363–371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clustering analysis differentiates P. fuscovaginae-like (Pfv-like) collected in the Philippines and P. fuscovaginae (Pfv) collected elsewhere. Values scale is depicted in red, orange, yellow, and white colors in TETRA pairwise comparison. Value cut-offs with >99% reflect the possibility of same species grouping. The heatmap was generated in the R package gplots using the heatmap.2 function.
(TIFF)
The middle line in each box plot represents the mean of ANI values for a particular taxonomic group. Also shown in each box plot are the minimum and maximum values. The number of genomes is denoted in brackets. All Pseudomonas genomes used in this analysis are listed in S1 Table.
(TIFF)
Maximum likelihood was used to infer the phylogenetic relationship with bootstrap of 1000 using the RAxML software [61]. Groups Pfv-like and Pfv are highlighted as orange and blue, respectively.
(TIFF)
A) Distribution of intact prophage insertions are color-coded among rice-infecting Pseudomonas strains. Prophage ID is based on PHAST database [38]. All intact prophage have scores >90 [38]. B) Bar plot distribution of the number of genes found within all prophage regions in each of the eight rice-infecting Pseudomonas draft genomes. Non-phage related genes are further classified as secreted and non-secreted.
(TIFF)
Bar plots representing number of genes in each COG category per rice-infecting Pseudomonas-all (black, n = 8), P. fuscovaginae (blue, n = 5) and P. fuscovaginae-like (orange, n = 3).
(TIFF)
Black dots represent standard error; X and Y axes represent genome size and number of genomes, respectively.
(TIFF)
Rice sheath (O. sativa cv. Azucena) infected with P.fuscovaginae-like strain IRRI 7007 was evaluated at 0, 3, 24, 48, and 72 hpi. Gen ID and functional classification are indicated in both sides.
(TIFF)
The 79 Pseudomonas genomes are listed according to species, strains and their corresponding GenBank ID.
(DOCX)
Locus ID and sequence correspond to P. fuscovaginae-like strain IRRI 6609.
(XLSX)
(XLSX)
(DOCX)
Annotation was done using Pfam. Locus ID based on P.fuscovaginae-like IRRI 6609.
(XLSX)
(DOCX)
Data Availability Statement
All genome sequences are deposited at GenBank with accession numbers JSYZ00000000 and JTBY00000000.