Abstract
Chromatin boundaries are architectural elements that determine the three-dimensional folding of the chromatin fiber and organize the chromosome into independent units of genetic activity. The Fab-7 boundary from the Drosophila bithorax complex (BX-C) is required for the parasegment-specific expression of the Abd-B gene. We have used a replacement strategy to identify sequences that are necessary and sufficient for Fab-7 boundary function in the BX-C. Fab-7 boundary activity is known to depend on factors that are stage specific, and we describe a novel ∼700-kDa complex, the late boundary complex (LBC), that binds to Fab-7 sequences that have insulator functions in late embryos and adults. We show that the LBC is enriched in nuclear extracts from late, but not early, embryos and that it contains three insulator proteins, GAF, Mod(mdg4), and E(y)2. Its DNA binding properties are unusual in that it requires a minimal sequence of >65 bp; however, other than a GAGA motif, the three Fab-7 LBC recognition elements display few sequence similarities. Finally, we show that mutations which abrogate LBC binding in vitro inactivate the Fab-7 boundary in the BX-C.
INTRODUCTION
Chromosomes of multicellular eukaryotes are subdivided into autonomous domains by special elements called chromatin boundaries or insulators (1–6). The classically defined functions ascribed to boundaries/insulators include an enhancer- or silencer-blocking activity and an ability to bring distant chromosomal DNA segments into close proximity (7–19). Genome-wide chromatin immunoprecipitations (ChIPs) with known insulator proteins together with chromatin conformation capture experiments indicate that insulators are ubiquitous features of chromosomes from flies to human, demarcating distinct chromatin and regulatory domains and helping mediate long-range interactions (20–30).
Although boundaries were discovered 25 years ago, our knowledge about the cis-acting sequences and trans-acting factors that confer activity remains rudimentary. The most thoroughly characterized Drosophila insulator is from the gypsy transposon and consists of reiterated sites for a single DNA binding protein (31, 32). In contrast, endogenous fly insulators are generated by a unique assemblage of proteins on heterogeneous and rather large (>250 bp) sequences (33–40). Moreover, in the few examples that have been studied in detail, this assemblage is a composite of functionally redundant cis/trans elements, with no single element being absolutely essential (3, 37–41). A further complication is the fact that insulators are not autonomous. Instead, their activities depend upon other insulators in the neighborhood (3, 12–15, 30, 42–46). This means that insulators cannot be studied in isolation but, rather, must be analyzed in appropriate experimental contexts.
One context in which boundaries are known to play critical regulatory roles is the Drosophila bithorax complex (BX-C) (47, 48). The BX-C contains three homeotic genes, Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B), which are responsible for specifying the parasegments (PS5 to PS13) that make up the posterior two-thirds of the fly (49, 50). An ∼300-kb regulatory region that is organized into three gene-specific transcriptionally associated regulatory domains (TARDs) orchestrates the parasegment-specific expression of the three homeotic genes. For example, the Abd-B TARD contains four parasegment-specific cis-regulatory domains, iab-5, iab-6, iab-7, and iab-8, which direct Abd-B expression in PS10, PS11, PS12, and PS13, respectively (Fig. 1A) (47, 48).
FIG 1.
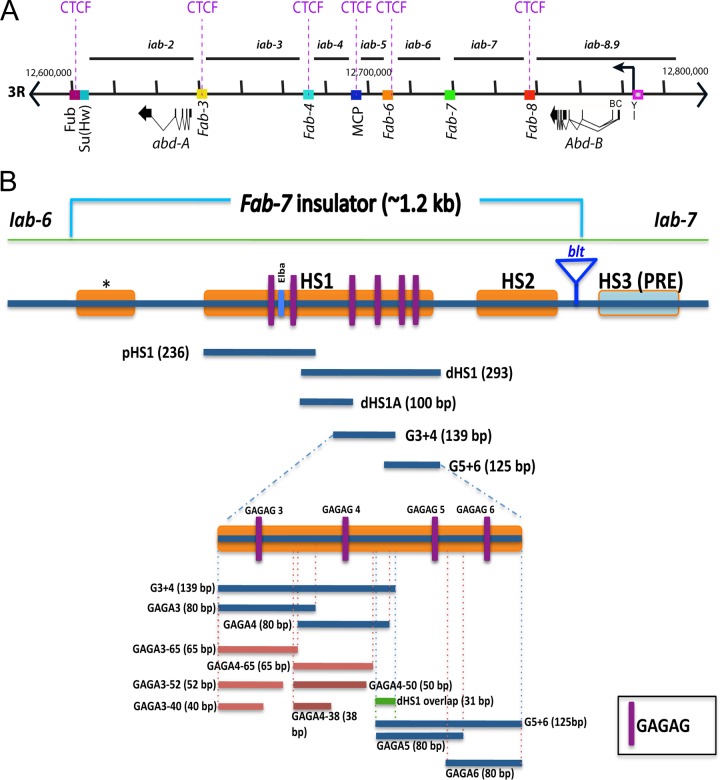
The bithorax complex (BX-C) and Fab-7. (A) Genomic map showing the cis-regulatory regions within the bithorax complex (BX-C) located on the 3R chromosome. The two BX-C genes, abd-A and Abd-B, along with their nine regulatory domains (iab-2 to iab-9) are illustrated. BX-C insulators (20, 40, 52, 92) that are interspersed between the chromosomal domains are depicted as rectangular boxes of different colors. (B) A schematic drawing of the Fab-7 insulator (∼1.2 kb) and the probes used for electrophoretic mobility shift assays (EMSAs). DNase I-hypersensitive regions *, HS1, and HS2 are shown as orange boxes. Within HS1, the binding sites for GAGA factor (GAF) are shown as purple lines (GAGAG). The distal part of dHS1 corresponding to probes G3+4 and G5+6 is enlarged. The names, sizes, and locations of the various probes derived from dHS1 are illustrated.
In order to properly specify PS identity, individual PS-specific cis-regulatory domains in each TARD must be able to function autonomously. Boundaries (Fub, Fab-3 to -8, and MCP) (Fig. 1A) bracketing each cis-regulatory domain confer this autonomous activity. The most thoroughly characterized of these BX-C insulators is Fab-7, which is located between iab-6 and iab-7 (Fig. 1A) (38, 51–54). Deletions of Fab-7 result in a complex mixture of gain- and loss-of-function (GOF and LOF, respectively) phenotypes in PS11, which arise because of cross talk between regulatory elements in the iab-6 and iab-7 regulatory domains (38). Like other fly boundary elements, BX-C insulators also function in enhancer/silencer-blocking transgene assays. In addition to preventing cross talk between adjacent cis-regulatory domains, BX-C insulators must permit interactions between the cis-regulatory domains and their homeotic gene targets (30). For example, the three cis-regulatory domains in the Abd-B TARD (iab-5, iab-6, and iab-7) must be able to bypass one or more insulators to contact the Abd-B promoter (Fig. 1A).
A combination of P-element excisions in BX-C and transgene assays have mapped Fab-7 to a 1.2-kb DNA segment that includes three chromatin-specific major nuclease-hypersensitive sites, the minor site “*” and major sites HS1 and HS2 (Fig. 1B) (38, 53–55). These studies also showed that although Fab-7 is active throughout development irrespective of cell or tissue type, this constitutive activity is generated by subelements whose activity is developmentally restricted. The first hint of developmentally restricted activity came from mutations in the GAGA factor (GAF) binding sites in the largest hypersensitive region, HS1 (Fig. 1B) (56). While mutation in GAGA motifs 1 through 5 (GAGA1–5) weakened the insulator activity of the 1.2-kb Fab-7 element in transgene assays at all stages, mutations in the proximal GAGA motifs, GAGA1 and GAGA2, had an effect only in early embryos. In contrast, mutations in GAGA motifs 3 and 4 had the opposite effect; they weakened insulator activity from midembryogenesis onwards but not in early embryos. Further evidence that HS1 is composed of subelements with developmentally restricted activities came from multimerizing the proximal (p) and distal (d) halves of HS1 (39). pHS1, which spans GAGA sites 1 and 2, has enhancer-blocking activity in early embryos but not thereafter. Conversely, dHS1, which spans GAGA motifs 3 to 6, blocks enhancer-promoter interactions from midembryogenesis onwards, even more efficiently than Fab-7 itself. However, dHS1 had only weak to moderate blocking activity in early embryos. Subsequent experiments showed that the early insulator activity of pHS1 depends in part upon the heterotrimeric Elba factor, which binds to an 8-bp sequence, GGAATAAG, located between GAGA sites 1 and 2 (57, 58). Elba DNA binding activity is detected in early, 0- to 6-h, embryonic nuclear extracts but not in late, 6- to 18-h, nuclear extracts. Moreover, as with pHS1, a multimerized 27-bp oligonucleotide spanning the Elba recognition sequence is sufficient to confer insulator activity in transgene assays in early embryos but not later in development. Elba factor DNA binding and insulator activity is developmentally restricted because midblastula transition genes encode two of the three Elba proteins (57).
Most of the detailed functional analysis of Fab-7 has relied on transgene assay, and these experiments have focused largely, though not exclusively, on sequences from HS1. Consequently, it remains uncertain whether the sequences/developmentally restricted factors implicated in insulator function in transgene assays are important for the proper functioning of Fab-7 in its endogenous context. To address this issue, we developed a BX-C landing platform that can be used to replace Fab-7. Using this platform, we show that the ∼500-bp HS1 sequence is both necessary and sufficient for full insulator function in the BX-C. In contrast, sequences spanning the other Fab-7 hypersensitive sites are not essential for insulator function. While the functioning of the Elba complex in the early insulator activity of the pHS1 subelement has been documented, little is known about the factors that interact with the dHS1 subelement and are responsible for insulator activity later in development. Here, we report the identification of a very large developmentally regulated protein complex, called the late boundary complex (LBC), which binds to three ∼65-bp recognition elements in the late dHS1 region and is required for Fab-7 insulator activity in the context of the BX-C.
MATERIALS AND METHODS
Nuclear extracts.
Nuclear extracts from 0- to 6-h and 6- to 18-h embryos, utilized for electrophoretic mobility shift assays (EMSAs) and size exclusion chromatography, were prepared using methods adopted from previously published procedures (58). For 6- to 18-h extracts, 0- to 12-h embryos from Oregon R were collected from apple juice plates and aged 6 h at room temperature. The final dialysis step described by Aoki et al. (58) was omitted, and the extraction was completed with the final concentration of KCl as 360 mM.
Probes.
Using a 3.35-kb Fab-7 HindIII-to-XbaI fragment inserted in pBluescript as a template, probes pdHS1, G3+4, and G5+6 were obtained by PCR. The following primers were used to generate the probes: pdHS1, chromosome 3R (starting position to ending position) 16899048 to 16899147; pdHS1 forward (pdHS1-F), CATTGGGGCATATCAACGCG; pdHS1 reverse (pdHS1-R), GCTTATATTTACTACTGCACCTGTTCACG; G3+4, chromosome 3R 16899107 to 16899240; G3+4-F, CAAAGAGCGACACGTGAACAG; G3+4-R, GGTGTGCGTGCGGTTCTC; G5+6, chromosome 3R 16899215 to 16899339; G5+6-F, CGTGATAAGAGAACCGCACGC; G5+6-R, CGAACGGCAACTGAATTCCAATC. Sequence numbers are based on the new Drosophila melanogaster Release_6 assembly from the Berkeley Drosophila Genome Project (BDGP).
The remaining probes were obtained with PCR using G3+4 and G5+6 as templates along with the appropriate primers. The exact sequences and locations of these primers are available upon request.
EMSA.
One picomole of probe was 5′ end labeled with [γ-32P]ATP (MP Biomedicals) using T4 polynucleotide kinase (New England BioLabs) in a 50-μl total reaction volume at 37°C for a period of 1 h. Columns packed with Sephadex G-50 fine gel (Amersham Biosciences) were used to separate free ATP from the labeled probes. The volume of the sample eluted from the column was adjusted to 100 μl using deionized water so that the final concentration of the probe was 10 fmol/μl.
Binding reactions were performed in a 20-μl volume consisting of 25 mM Tris-Cl (pH 7.4), 100 mM KCl, 1 mM EDTA, 0.1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 0.03 mg/ml bovine serum albumin, 10% glycerol, 0.25 mg/ml poly(dA-dT) · poly(dA-dT) and 20 μg of protein derived from nuclear extract or an equal volume of 360 mM nuclear extraction buffer. In some samples, unlabeled competitor DNA was included so that the final concentration of the competitor would be in 5- to 100-fold excess. The reaction mixtures containing the 32P-labeled DNA probes were incubated for 30 min at room temperature with or without 20 μg of nuclear extracts derived from 0- to 6-h (early) and 6- to 18-h (late) embryos and loaded onto precleared 4% acrylamide–bis-acrylamide gels in 0.5× Tris-borate-EDTA (TBE)–2.5% glycerol gel. Binding reactions were electrophoresed at 180 V for 3 to 4 h with 0.5× TBE–2.5% glycerol running buffer at 4°C, dried, and imaged using a Typhoon 9410 scanner and Image Gauge software and/or X-ray film.
For supershift experiments, antibodies against different insulator-associated proteins were incubated in the reaction mixtures along with 32P-labeled DNA probes and nuclear extracts for 30 min at room temperature. Supershift experiments were carried out using 1 μl of rat polyclonal antibodies against GAF and E(y)2 or 1 μl of rabbit polyclonal against Mod(mdg4) in each 20-μl reaction mixture. Antibodies were generously donated by Anton Golovnin and Pavel Georgiev [GAF, E(y)2, Mod(mdg4), and CP190], Elissa Lei [Mod(mdg4)], Carl Wu (GAF), and David Gilmour (GAF). One microliter of rat or rabbit serum was used as a control.
Size exclusion chromatography.
Nuclear extracts derived from 6- to 18-h embryos were fractionated using size exclusion chromatography (Superdex 200 16/60 column; GE Healthcare). Molecular mass markers ranging from 1,350 to 670,000 Da (Bio-Rad) were used as gel filtration standards to calculate the partition coefficient and estimate the size of LBC.
Generation of the Fab-7attP50 integration platform.
The strategy to create the Fab-7attP50 landing platform is diagrammed in Fig. S1 in the supplemental material. A donor template composed of a 3.6-kb HindIII fragment spanning the Fab-7 boundary and adjacent iab-7 Polycomb response element (PRE) was generated (from chromosome 3R position 16897271 to position 16900888). Within this fragment an attP minimal element of 50 bp (59) was inserted along with a loxP site (60) near the unique NcoI site at position 16898070. A second loxP site together with an Flp recognition target (FRT) site was inserted at position 16900020, just adjacent to the distal endpoint of the hypersensitive region, HS3, that corresponds to the iab-7 PRE. Note that the FRT sequence contains an XbaI site that is unique in this donor plasmid as well as in the next plasmid, KSattBFLFab7ry, depicted in Fig. S1 in the supplemental material. The donor template was introduced by P-element transformation. One of the third chromosome inserts of this donor was then recombined with the Fab-7 bluetail transposon insertion. A double-strand break for gene conversion at the site of the bluetail insertion was generated by mobilizing the P-elements (see reference 45 for a detailed description of the gene conversion strategy). Out of 58 ry− flies, two individuals had integrated the attP50 sequence along with the two loxP sites. As expected, these individuals are fully wild type (WT). Excision of the Fab-7 was recovered on the basis of its Fab-7 GOF phenotype after a cross with a line expressing Cre recombinase (60).
Integration of modified Fab-7 elements within the Fab-7attP50 platform.
Figure S1 in the supplemental material shows the structure of the KSattBFLFab7ry vector that was used for insulator replacements in the Fab-7attP50 platform (WT replacement is depicted here). All elements were assembled within the KSpBluescript vector. The plasmid contains, in the following order, a full-length attB sequence (59) followed by the same Fab-7 fragment that is deleted in the Fab-7attP50 platform along with the FRT sequence cloned just downstream from the HS3 PRE. The rosy+ gene carried on a 7.2-kb HindIII fragment extracted from the Carnegie 3 vector (61) was chosen because of its non-cell-autonomous behavior (a white+ gene inserted in the BX-C is typically silenced in the eye disk). A loxP sequence was placed at the 3′ end of the rosy+ marker gene.
Integration of the KSattBFLFab7ry plasmid within Fab-7attP50 was achieved by injecting the plasmid into progeny from a cross of Fab-7attP50 males to females carrying a P{y+; nos-ϕC31-nos} transgene inserted onto the X chromosome as a source of integrase (62). These females also carried two third-chromosome balancers, MKRS/TM2. The emerging Go individuals were then crossed to TM2/MKRS flies, and the resulting integrants were recognized on the basis of their ry+ eyes. The ry+ and plasmid sequences were then flipped, introducing a source of flippase (63) and selecting ry− progeny. All stocks are available upon request.
RESULTS
HS1 is necessary and sufficient for Fab-7 insulator activity.
Previous studies on Fab-7 in its endogenous context relied on the bluetail (btl) transposon inserted between HS2 and the iab-7 Polycomb response element (PRE) in HS3 (Fig. 2A) (52, 64). All of the Fab-7 mutations generated by imprecise excision of bluetail had one endpoint at the transposon insertion site and extended proximally through HS2 and into and beyond HS1. Since the smallest deletions with a Fab-7 mutant phenotype had breakpoints in HS1 or removed HS1, we concluded that HS1 must be intact for proper Fab-7 boundary function. However, it was not possible to use this P-element-based mutagenesis to determine whether HS2 is also important for boundary function, nor was it possible to directly manipulate Fab-7 sequences. For this reason, we generated a phiC31-based integration platform (Fab-7attP50) in which the region spanning the minor nuclease-hypersensitive site (*) and three major nuclease-hypersensitive sites (HS1 to HS3) was deleted and replaced by a minimal attP site target for the phiC31 integrase (see Materials and Methods and Fig. S1A in the supplemental material) (45, 62, 65). We then constructed an attB integration plasmid with a rosy+ (ry+) marker and restriction sites for introducing DNAs of interest (see Fig. S1B). Once integrated, the ry+ marker and integration plasmid are inserted within the BX-C between the end of HS3 and the rest of the iab-7 region. These foreign sequences can be removed by an FRT/flipase site-specific recombination system (63).
FIG 2.
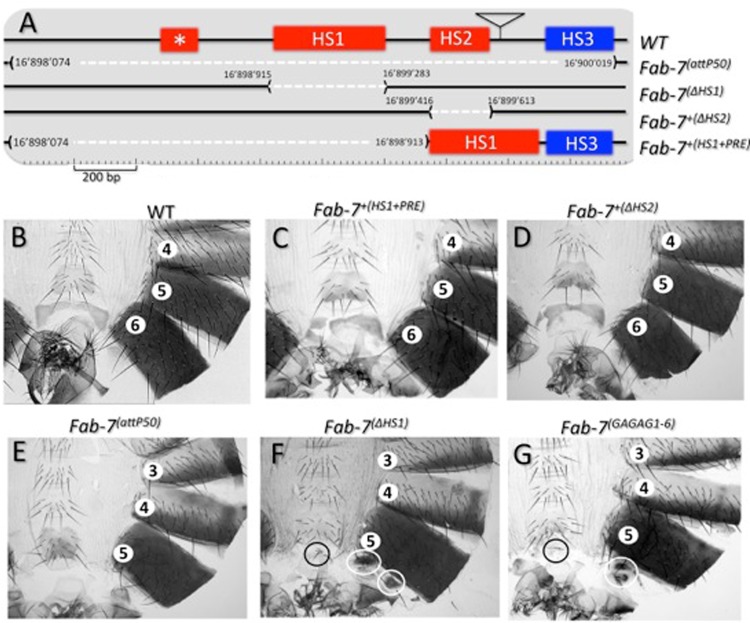
(A) Identifying sequences required for Fab-7 boundary activity in the BX-C. The Fab-7 boundary and abutting PRE (WT) are drawn on top with the four nuclease-hypersensitive regions, *, HS1, HS2, and HS3, as indicated. The location of the bluetail transposon insertion (btl) is indicated by a triangle. Shown in the second line (Fab-7attP50) are the sequences of Fab-7 to the iab-7 PRE that are deleted in the Fab-7attP50 ϕC31 integration platform. Note that this 1,949-bp-long deletion removes all four HS regions as well as flanking DNA sequences (see Materials and Methods). The structures (and coordinates; from release 6 of the BDGP Drosophila genome) of the HS1 and HS2 deletions are indicated in the next two lines [Fab-7ΔHS1 and Fab-7+(ΔHS2)]. The last line in panel A [Fab-7+(HS1+PRE)] shows the structure of the attP50 rescue construct in which only HS1 and HS3 are included. (B to G) Cuticles of the posterior abdominal segments of a wild-type male and of males homozygous for Fab-7+(HS1+PRE) (HS1+ HS3), Fab-7+(ΔHS2), Fab-7attP50, Fab-7ΔHS1, and Fab-7GAGAG1–6, respectively (see Mihaly et al. for a detailed description of cuticular phenotypes [38]). Note that WT males (B) have only six abdominal segments as well as genitalia and analia that are visible at the posterior of each cuticle. The seventh abdominal segments present in embryos and larvae disappear by apoptosis during metamorphosis (93). Because of the GOF transformation of A6 into A7, males homozygous for deletions like Fab-7attP50 (E) that remove both the Fab-7 insulator and the iab-7 PRE have only five abdominal segments (compare panels B and E). While Fab-7+(ΔHS2) males resemble the wild type (compare panels B and D), Fab-7ΔHS1 (F) males exhibit mixed loss- and gain-of-function phenotypes. As expected from a GOF transformation, the A6 segment is significantly reduced in size. On the other hand, the small patches of residual tissues in A6 have a PS10/A5 identity, which is characteristic of a LOF transformation. Fab-7GAGAG1–6 (G) has mutations in GAGA motifs 1 to 6, plus mutations in two GAGAA motifs, as described in the text. Its phenotype is the same as that of Fab-7ΔHS1 (compare panels F and G).
Since the integration platform deletes both the boundary and the HS3 iab-7 PRE, it has a strong Fab-7 gain-of-function phenotype (GOF) in which the sixth abdominal segment (A6) is completely transformed into a copy of A7 (a type 1 Fab-7 phenotype in Mihaly et al. [38]) (Fig. 2E). As expected, reinserting the deleted DNA segment into Fab-7attP50 fully reverts the GOF Fab-7 phenotype, giving rise to wild-type flies (data not shown). Complete rescue is also observed prior to flipping out the ry+ and plasmid sequences, indicating that increasing the distance between iab-6 and its Abd-B target promoter does not by itself impede the activity of the iab-6 cis-regulatory domain.
We next tested the activity of Fab-7 insulators lacking either HS2 or HS1 [Fig. 2A, Fab-7+(ΔHS2) and Fab-7ΔHS1, respectively]. The ΔHS2 insulator appears to be almost fully functional, and most flies homozygous for this deletion look like wild-type flies (Fig. 2D). However, occasionally males were observed that had a single bristle on the sixth sternite. The presence of bristles on the sixth sternite is indicative of a transformation toward A5 (PS11 → PS10) identity, which arises from the spread of Polycomb group (PcG) gene silencing from the inactive iab-7 cis-regulatory domain into iab-6. In addition to the weak expression displayed by ΔHS2, the penetrance of the sternite PS11 → PS10 transformation is also very low, and the frequency of males with a bristle on the sixth sternite is less than 5%. This defect in insulator activity also appears to be tissue specific as we did not observe any trichomes (small hairs found in A5 and more anterior tergites) in the sixth tergite of homozygous ΔHS2 males.
A quite different result is obtained for Fab-7ΔHS1. As shown in Fig. 2F, the ΔHS1 deletion inactivates the Fab-7 boundary, resulting in a mixed GOF/LOF phenotype in PS11/A6 that is the same as that observed for class II deletions generated by imprecise excision of the bluetail transposon (38). On the dorsal side, the sixth tergite is almost completely missing, as expected when iab-7 is ectopically activated in PS11 cells. The small patches of “ A6” tergite remaining (white circles) are covered by trichomes, which means that these cells have a PS10 (A5), not PS11 (A6), identity. On the ventral side, the presence of bristles (Fig. 2F, black circle) on the diminutive sternite indicates that the cells have assumed a PS10 identity. Taken together, these findings show that HS1 is essential for Fab-7 insulator activity, while HS2 is not. This conclusion is supported by an experiment in which we tested whether the HS1 sequence alone is sufficient to rescue the *-HS1-HS2 deletion in Fab-7attP50. As evident from the wild-type abdomen shown in Fig. 2C, the HS1 sequence is not only necessary but also sufficient for full Fab-7 boundary function in an otherwise wild-type background.
A late-stage-specific factor binds to different dHS1 sequences.
Previous studies showed that HS1 is composed of subelements that have developmentally restricted activities. Since an insulator complex, Elba, that functions in early embryos has already been described, we sought to identify factors responsible for HS1 insulator activity from midembryogenesis onwards. Transgene experiments and partial Fab-7 deletions indicate that the late factor(s) must interact with sequences in the dHS1 region of HS1 (Fig. 1). Within dHS1 we found that the proximal half (Fig. 1B, dHS1A) had even stronger late blocking activity in transgene assays than the full-length Fab-7 (when multimerized) but displayed no blocking activity in early embryos (39). In contrast, the distal half (dHS1B) had weak to moderate blocking activity throughout most of development. To identify factors that interact with these sequences we subdivided dHS1 into three overlapping probes, pdHS1A, G3+4, and G5+6 (in Fig. 1B pdHS1A together with G3+4 corresponds to dHS1A, while G5+6 corresponds to dHS1B). The most proximal, pdHS1A, is 100 bp and extends from the proximal end of dHS1 to just before GAGA motif 3. The central probe, G3+4, is 130 bp and spans GAGA motifs 3 and 4. Finally the 125-bp distal probe, G5+ 6, includes all of dHS1B and contains GAGA motifs 5 and 6. We used these probes for electrophoretic mobility shift assays (EMSAs) with nuclear extracts derived from early (0- to 6-h) or late (6- to 18-h) embryos. From our functional studies (39), we anticipated that the late factors would associate primarily with the proximal pdHS1A and/or G3+4 probes. In contrast, G5+6 should differ from pdHS1A and G3+4 in that it would likely be recognized either by continuously expressed factors or by a combination of early and late factors.
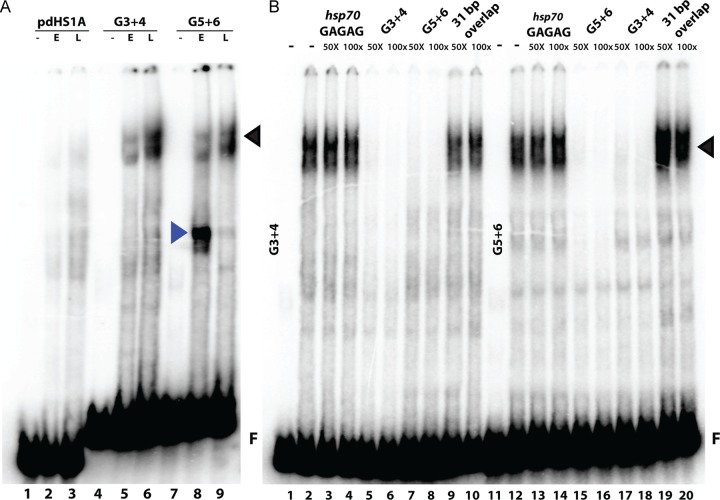
While the proximal pdHS1A probe gave only weak and variable shifts, several quite prominent shifts with distinct stage specificity were reproducibly detected with G3+4 and G5+6 (Fig. 3A). As expected from our enhancer-blocking assays, a shift is observed with early nuclear extracts for probe G5+6. However, the shift of most interest is the very prominent slowly migrating band in nuclear extracts prepared from late embryos. This shift is detected with G3+4 and G5+6 and has the stage specificity expected for late insulator activity as extracts from early embryos generate only a weakly labeled shift (Fig. 3A). Since these two probes give similar shifts, we suspected that they are bound by the same factor(s) in late nuclear extracts. To determine if this is correct, we did cross-competition experiments. As illustrated in Fig. 3B, the G3+4 shift can be competed by excess cold G3+4 or G5+6. Conversely, the late shift detected with G5+6 can be competed with excess cold G5+6 or G3+4. We have called the factor generating this late shift the LBC (late boundary complex).
FIG 3.
Multiple developmental-stage-specific binding activities are observed within the dHS1 region of Fab-7. (A) Gel shift analysis with probes derived from dHS1 of Fab-7 demonstrates stage-specific binding activities. EMSAs were carried out as described in Materials and Methods with 32P-labeled DNA probes. Probes pdHS1A, G3+4, and G5+6 were incubated with nuclear extracts derived from early (E; 0- to 6-h) and late (L; 6- to 18-h) embryos. The identities of the binding activities with early- and late-stage specificity are represented with blue and black arrowheads, respectively. F, free probe. (B) EMSAs with late nuclear extracts were performed with probes G3+4 (lanes 1 to 10) and G5+6 (lanes 11 to 20) in the absence (lanes 1, 2, 11, and 12) or presence (lanes 3 to 10 and 13 to 20) of cold competitor, as indicated above the lanes. A 50- or 100-fold excess of cold competitor was added to the reaction mixes.
As probes G3+4 and G5+6 have a 31-bp overlap, it was possible that this sequence contains the LBC recognition motif. However, this is not the case. The 31-bp sequence gives a shift with early and late nuclear extracts that migrates much more rapidly than the shift observed with probe G3+4 or G5+6 (data not shown). In addition, the 31-bp sequence does not compete the LBC shift generated by probes G3+4 and G5+G6 when it is added in excess (Fig. 3B).
Defining the minimal LBC recognition element.
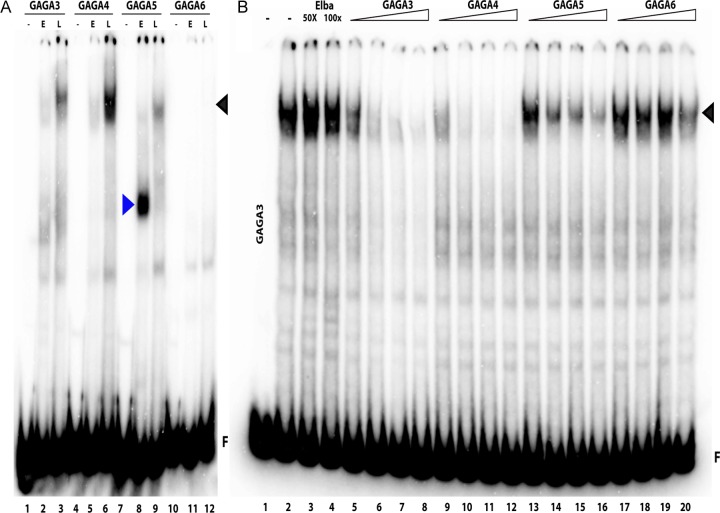
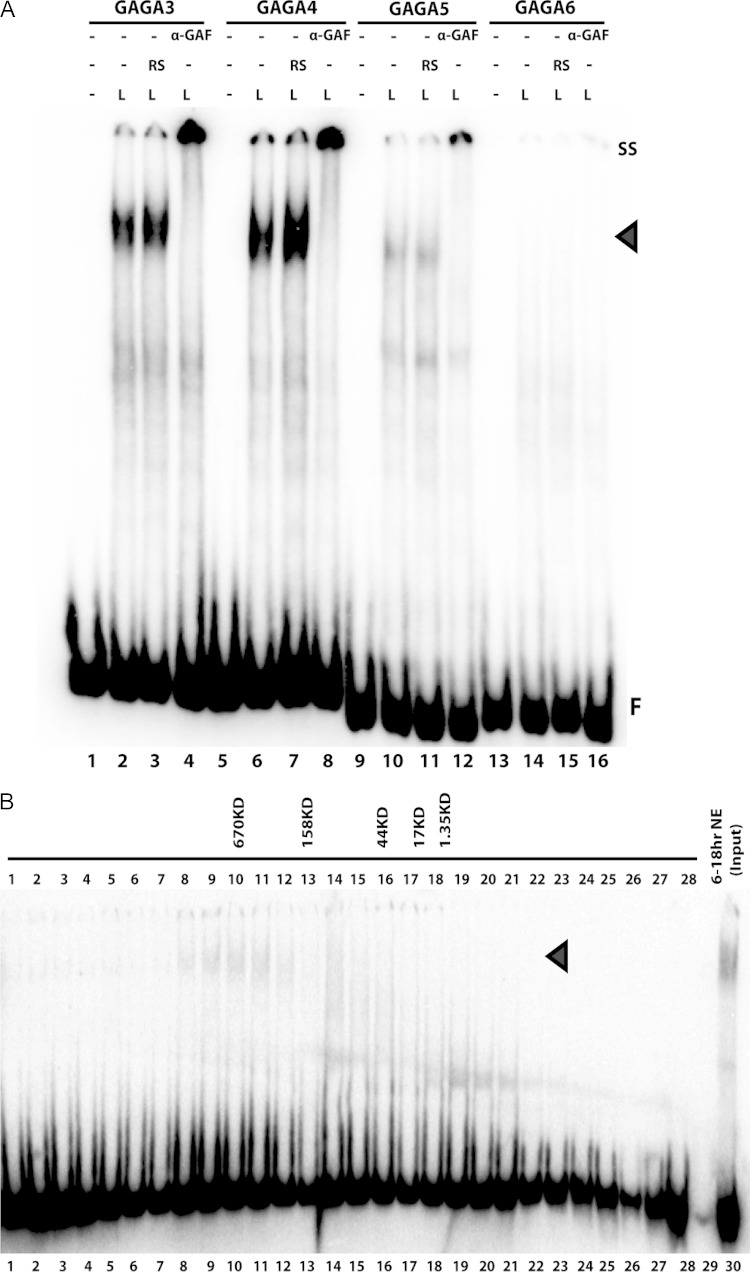
To further localize the LBC recognition sequences in G3+4 and G5+6, we subdivided them into four 80-bp overlapping probes, GAGA3, GAGA4, GAGA5, and GAGA6, so that each spans one of the dHS1 GAGA sites (Fig. 1B). Figure 4A shows that GAGA3, GAGA4, and GAGA5 probes generate the LBC shift, while the probe GAGA6 does not. The yield of the LBC shift differs among the three probes. The most strongly labeled LBC shift is observed with GAGA4 while the GAGA3 shift is intermediate, and the GAGA5 shift is the weakest. Additionally, there are differences in the profiles of LBC shifts generated by the three probes. The GAGA4 shift is typically a composite of three to four closely spaced bands. In contrast, the LBC complexes detected with GAGA3 are enriched for the more slowly migrating GAGA4 shifts, while the complexes detected with GAGA5 usually correspond to the faster-migrating GAGA4 shifts (Fig. 4A).
FIG 4.
Late binding complex (LBC) recognizes sequences associated with the dHS1 GAGA motifs. (A) EMSA was performed with four 80-bp overlapping fragments, GAGA3, GAGA4, GAGA5, and GAGA6. The labeled probes were incubated with early (E; 0- to 6-h) and late (L; 6- to 18-h) nuclear extracts. Binding activities with early-stage (blue) and late-stage (black) specificity are indicated with arrowheads. F, unbound probe. (B) Cross-competition experiments with GAGA3. Using labeled GAGA3 probe, EMSAs with late nuclear extracts were performed in the absence (lanes 1 and 2) or presence (lanes 3 to 20) of increasing amounts of unlabeled cold competitor as indicated above the lanes. The Elba competitor is a 27-bp fragment derived from the pHS1 region of Fab-7. It fails to compete for LBC binding and is used as a negative control. A 5-, 10-, 50-, or 100-fold excess of the cold competitor (69) was added to the reaction mixture. Black arrowhead, LBC; F, free probe.
The differences in the apparent affinities of the LBC for the three ∼80-bp probes are recapitulated in competition experiments. In the experiment shown in Fig. 4B, we challenged LBC-GAGA3 complexes with increasing amounts of cold competitors. Even at a 100-fold excess, a competitor spanning the pHS1 Elba recognition sequence failed to interfere with LBC binding to GAGA3. In contrast, cold GAGA3 or GAGA4 is an efficient competitor. Of the two, GAGA4 appears to be marginally more effective than GAGA3. This small difference in affinity was also observed in competition experiments when the labeled probe was GAGA4 rather than GAGA3 (see Fig. S2A in the supplemental material). As expected, GAGA5 is a less efficient competitor than either GAGA3 or GAGA4 when it is used to challenge LBC binding to GAGA3 (Fig. 4B) or GAGA4 (see Fig. S2A). As seen in Fig. 4B, there is residual LBC binding to GAGA3 even when the GAGA5 sequence is present in 100-fold excess. Interestingly, while we failed to detect an LBC shift with the GAGA6 probe, it competes, albeit poorly, with LBC binding to GAGA3 (compare the GAGA3 LBC shift in the presence of 100-fold excess GAGA6 with the shift in the presence of 100-fold excess of the Elba sequence). Similar results were obtained when GAGA6 competed LBC binding to GAGA5 (see Fig. S2B in the supplemental material).
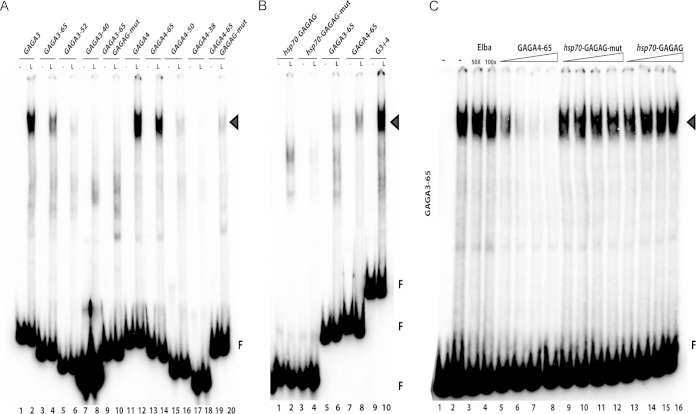
Since the LBC binds with higher affinity to GAGA3 and GAGA4 than to GAGA5 (or GAGA6), these two probes were used to generate a series of terminal truncations to further localize the recognition sequence. We anticipated that this approach would allow us to map the LBC binding site to a smaller ∼20-bp sequence, which we could then further dissect using a series of base pair substitutions. However, as illustrated in Fig. 5A, the LBC requires an unusually long DNA sequence to generate a complex sufficiently stable to give an electrophoretic mobility shift. Of the terminal truncations shown in Fig. 5A, only the 65-bp probes, GAGA3-65 and GAGA4-65, retain substantial LBC binding activity. All of the more extensive truncations of GAGA3 or GAGA4 largely abrogate LBC binding (Fig. 5A). In fact, LBC binding even to the 65-bp probes is noticeably less than that observed for the 80-bp probes (Fig. 5A, compare lanes 2 and 12 with lanes 4 and 14). In spite of this reduced affinity, the two 65-bp probes resemble their larger 80-bp counterparts. First, LBC appears to have greater affinity for GAGA4-65 than for GAGA3-65 (compare lanes 4 and 14). Second, the two 65-bp probes can compete with each other for LBC binding. Third, LBC binding to GAGA3-65 or GAGA4-65 (see Fig. S3A and B in the supplemental material) can also be competed by GAGA3, GAGA4, and GAGA5.
FIG 5.
Identification of the LBC binding sequence. (A) EMSAs were performed with a series of truncations of the 80-bp GAGA3 and GAGA4 probes. For GAGA3, the truncations were 65 bp, 52 bp, and 40 bp in length. For GAGA4, the truncations were 65 bp, 50 bp, and 38 bp in length. The GAGAG motifs within GAGA3 and GAGA4 were mutated to ACAAA to produce two 65-bp fragments, GAGA3-65 GAGAG-mut and GAGA3-65 GAGAG-mut. Each subfragment was incubated with nuclear extracts from late (L; 6- to 18-h) embryos. (B) EMSAs were carried out using a probe, hsp70-GAGAG, that spans a previously characterized GAF binding sequence in the hsp70 promoter (69, 70). The GAGAG sequence within the hsp70 promoter was mutated to TCTCT to generate probe hsp70-GAGAG-mut. (C) Labeled GAGA3-65 was incubated with nuclear extracts from late (6- to 18-h) embryos in the absence (lanes 1 and 2) and presence (lanes 3 to 16) of unlabeled cold competitor as indicated above the lanes. A 5-, 10-, 50-, or 100-fold excess of the cold competitor (from the left to right lane, respectively) of each set was added to the reaction mixture. Arrowheads, LBC; F, free probe.
The GAGA motif is necessary but not sufficient for high-affinity LBC binding.
In addition to the unusual length, sequence comparisons of the probes that generate stable electrophoretic mobility shifts suggest that the LBC recognition properties must be complex. The only sequence in common appears to be the GAF binding motif (66). Since the GAGA motifs in dHS1 are important for late insulator activity in transgene assays, we tested whether LBC binding to GAGA3-65 and GAGA4-65 requires the GAGA motif. Figure 5A shows that mutations in the GAGA3-65 GAGA motif largely disrupt LBC binding, while a residual shift is still observed for GAGA4-65 (Fig. 5A). Mutations in the GAGA motifs of larger probes spanning GAGA3 and/or GAGA4 also reduce LBC binding; however, the effects are somewhat less pronounced than those observed for the two 65-bp probes (data not shown).
Since the GAGA motif appears to be important for LBC binding to the dHS1 probes, we wondered whether it is also sufficient to generate the LBC shift in late nuclear extracts. To investigate this question, we generated probes spanning three previously characterized GAF binding sequences in the hsp70, histones H3 and H4, and alcohol dehydrogenase promoters (67–69). As illustrated for a probe from the hsp70 promoter in Fig. 5B, these GAF binding sequences give shifts in late nuclear extracts that migrate more rapidly than the shift generated by LBC (and seem to correspond to several of the minor shifts often seen with GAGA3). In addition, the hsp70 GAF factor binding sequence is not an effective competitor for LBC binding (Fig. 2B and 5C; see also Fig. S2 and S3 in the supplemental material).
The GAGA factor is a component of the LBC.
Though our experiments indicate that the GAGA motifs in GAGA3-65 and GAGA4-65 are important for LBC binding, a number of findings would seem inconsistent with the involvement of GAF. To begin with, the minimal LBC binding sequence is much larger than that needed for complex formation by GAF. Additionally, several well-characterized GAF binding sequences give shifts that are different from LBC and fail to compete for LBC binding to Fab-7 dHS1 probes. To resolve these apparent discrepancies, we used GAF antibodies in supershift experiments. We also tested Pipsqueak (Psq), which, like GAF, recognizes the GAGA motif (70, 71). We found that Psq antibodies failed to induce a supershift or to block LBC binding (data not shown). In contrast, the GAGA3, GAGA4, and GAGA5 LBC shifts can all be supershifted by a rat polyclonal GAF antibody (Fig. 6A). The LBC supershifts do not appear to be an anomaly of this particular GAF antibody as similar results were obtained when two other independent rabbit polyclonal GAF antibodies were used (data not shown).
FIG 6.
LBC is a 700-kDa complex that contains the GAGA factor (GAF). (A) The probes GAGA3, GAGA4, GAGA5, and GAGA6 of Fab-7 were incubated with late nuclear extracts, without or with either a control rat serum (RS) (lanes 3, 7, 11, and 15) or a rat polyclonal anti-GAF antibody (α-GAF) (lanes 4, 8, 13, and 15). Antibody supershifts (SS) are indicated. (B) EMSA using probe GAGA4 incubated with column fractions derived from late-stage nuclear extracts fractionated by size exclusion chromatography. EMSAs using GAGA3 were used to detect the LBC in column fractions. The samples used in lanes 1 to 28 were incubated with 10 μl of the corresponding column fractions plus rat serum. The 6- to 18-h nuclear extracts (NE) were used as input (lane 30). The sizes of molecular mass markers used as standards are listed above the appropriate column fractions. Arrowheads, LBC; F, free probe (see also Fig. S3 in the supplemental material).
A ∼700-kDa multiprotein complex makes up the late-stage insulator factor.
The findings described in the previous sections indicate that GAF is an important component of the LBC; however, the known properties of this protein would not account for the unusually long and seemingly complex sequences that are required to generate the LBC shift. One explanation for the novel sequence recognition properties of the LBC is that it is a complex of several proteins including GAF. Potentially, this complex could be assembled, perhaps in a stepwise fashion, on LBC probes during the incubation with the nuclear extracts. Alternatively, the complex could exist preassembled in the extract, in which case it would bind to DNA as a complex. To distinguish between these possibilities, we fractionated late nuclear extracts by size exclusion chromatography and then tested the fractions for LBC activity using probe GAGA4. LBC activity was found in fractions 8 to 12, which corresponds to a complex on the order of ∼700 kDa (Fig. 6B; see also Fig. S4 in the supplemental material).
The LBC has components in addition to GAF.
The fact that LBC exists as a preassembled complex in nuclear extracts and exhibits DNA binding properties that differ substantially from those of GAF alone would suggest that this complex must contain additional protein species. To explore this possibility, we used antibodies directed against known insulator proteins for supershift experiments. The antibodies tested included BEAF, CP190, Elba, Enhancer of yellow 2 [E(y)2], Insensitive (Insv), Lola-like, Mod(mdg4), Su(Hw), and Pita (11, 14, 24, 33, 35, 57, 72–82). Like GAF (and Pipsqueak), CP190 and Lola-like have N-terminal BTB protein-protein interaction domains and several C-terminal zinc fingers that mediate DNA binding. Mod(mdg4) also has an N-terminal BTB domain; however, the C-terminal half of the protein is highly variable due to a complex pattern of alternative splicing. The Elba complex and Insv bind DNA via conserved BEN domains, while the Pita protein has a ZAD domain and binds DNA via 10 C2H2 zinc fingers (79, 83). There are two BEAF isoforms, which have different N-terminal DNA binding domains but share a C-terminal interaction domain. Finally E(y)2 is a small, 101-amino-acid protein that is widely conserved. In the case of BEAF, CP190, Elba, Insv, Lola-like, and Pita, we did not observe an LBC supershift, nor were there obvious reductions in the yield of the LBC shift (data not shown). With the caveat that the epitopes recognized by the antibodies might be hidden in the complex, it would appear that these six proteins are not part of the LBC. On the other hand, as shown in Fig. 7, both Mod(mdg4) and E(y)2 antibodies generated an LBC supershift. These findings would suggest that in addition to GAF, Mod(mdg4) and E(y)2 are components of the ∼700-kDa LBC.
FIG 7.
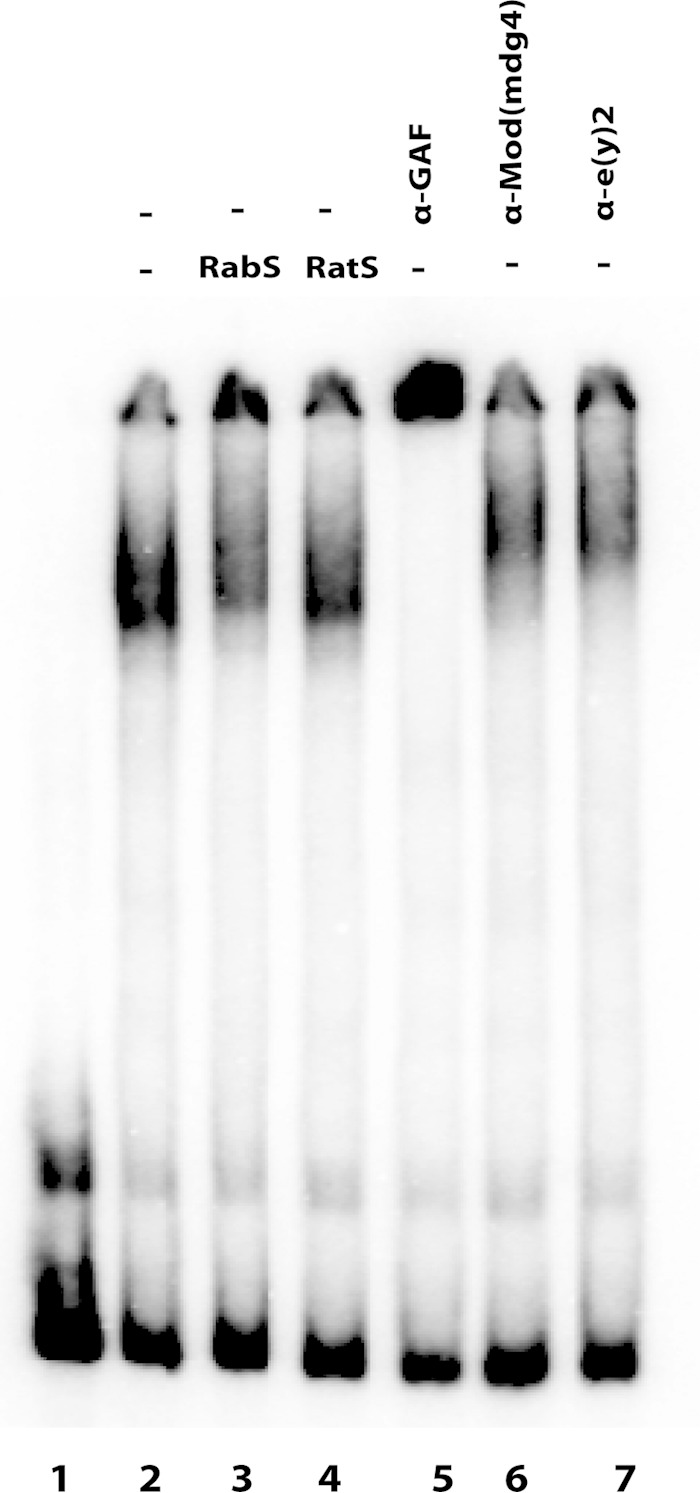
Mod(mdg4) and E(y)2 are also components of the LBC. Probe GAGA4 was incubated with late nuclear extracts, without and with either a control preimmune rabbit serum (RabS) (lane 3), a preimmune rat serum (lane 4), a rat polyclonal anti-GAF antibody (lane 5), a rabbit polyclonal anti-Mod(mdg4) antibody (lane 6), or a rat polyclonal anti-E(y)2 (Enhancer of yellow 2) antibody (lane 7).
Fab-7 GAGA motifs are required for full insulator activity.
Transgene assays have shown that mutations in the GAGA3 and GAGA4 motifs affect the enhancer-blocking activity of Fab-7 during midembryogenesis and in adults (56). The experiments described above suggest that this reduction in late blocking activity is likely due to impaired LBC binding. To explore this possibility further, we used the Fab-7attP50 integration platform to test whether the GAGA motifs in the LBC recognition elements are important for Fab-7 insulator activity in the context of the BX-C. For these experiments we introduced the GAGA motif mutations in a Fab-7 fragment spanning *, HS1, and HS2 (that also includes the HS3 iab-7 PRE). In addition to mutating the three GAGA motifs implicated (directly, as with GAGA3 and GAGA4, or indirectly, as with GAGA5) in LBC binding, we also mutated two other GAGA motifs in the dHS1 sequence. One of these is GAGA6. Though the LBC shift is not observed with the GAGA6 probe, competition experiments indicated that LBC is nevertheless able to bind, albeit weakly, to this region of dHS1. The other is an evolutionarily conserved lower-affinity GAF recognition sequence, GAGAA, located nine nucleotides distal to GAGA4. Finally, to ensure that the phenotypic effects of mutations in the dHS1 GAGA motifs are not masked by the boundary activity of the pHS1 subelement, we mutated the two GAF binding sequences in pHS1 that are known to be important for pHS1 boundary activity in transgene assays plus a low-affinity site at the proximal end of pHS1 (56).
The phenotypic effects of the GAGA motif mutations on Fab-7 boundary activity in the BX-C recapitulate those observed when the entire HS1 sequence is deleted (compare Fig. 2F and G). Like Fab-7ΔHS1 flies, Fab-7GAGA1–6 flies have a mixed GOF/LOF phenotype in PS11/A6. As expected when iab-7 is ectopically activated in PS11 cells, only a residual sixth tergite and sternite remain in Fab-7GAGA1–6 males. The small patch of “A6” tergite tissue (white circle) is covered by trichomes, implying that the surviving cells have a PS10 (A5), not PS11 (A6), identity. Likewise, the residual sternite (black circle) has bristles and thus has a PS10 identity. In addition to demonstrating that the GAGA motifs in HS1 are essential for boundary activity, the fact that mutations which compromise LBC binding to its recognition elements in dHS1 in vitro disrupt boundary function in vivo argues that the LBC is a critical component of the Fab-7 insulator.
DISCUSSION
We have used boundary replacement experiments to identify sequences in Fab-7 that are essential for its boundary functions in the context of the BX-C. We show that the HS1 nuclease-hypersensitive region is necessary for Fab-7 function. In contrast, the nuclease-hypersensitive regions HS2 and also * (48) are not essential. Moreover, even though these sequences are known to contribute to Fab-7 insulator activity in transgene assays (53), they apparently harbor dispensable activities in the context of the BX-C as the HS1 sequence alone is sufficient for full function.
Both transgene assays and partial Fab-7 deletions (39, 56) indicate that HS1 is composed of subelements whose boundary activity is developmentally restricted. Subelements in the proximal half of HS1 (pHS1) confer insulator activity during early development, while those in the distal half (dHS1) function (for the most part) from midembryogenesis through to the adult stage. We have shown previously that the early insulator activity of the pHS1 subelement is conferred, at least in part, by the heterotrimeric Elba complex. Here, we have identified a novel protein complex, the LBC, which binds to subelements in dHS1 that have insulator activity in late embryos and in adult flies. Although a conclusive demonstration that the LBC is responsible for Fab-7 late insulator activity will require the identification of all of the protein components of this complex, several lines of evidence provide a compelling case that this suggestion is correct. The first is the developmental profile. The LBC is enriched in nuclear extracts prepared from late 6- to 18-h embryos, while only little LBC activity is detected in extracts from early 0- to 6-h embryos. Second, the LBC binds to multiple recognition elements in the dHS1 region of HS1. This region of HS1 is known to be responsible for late Fab-7 insulator activity both in the BX-C and in transgene assays (39, 56). Moreover, the pattern of LBC binding to its recognition elements in dHS1 fits with the enhancer-blocking activity of the multimerized dHS1 subfragments, dHS1A and dHS1B. dHS1A, which spans the two higher-affinity LBC elements, GAGA3 and GAGA4, has stronger late blocking activity than the full-length Fab-7, while dHS1B, which contains the two lower-affinity LBC recognition elements, GAGA5 and GAGA6, has only moderate to weak late blocking activity. Third, we found that LBC binding to recognition elements in dHS1 requires a GAGA motif. Consistent with a critical role for LBC in insulator activity, when mutations in the dHS1 GAGA motifs are combined with mutations in the GAGA motifs in pHS1 that are needed for early insulator activity, Fab-7 boundary function in the BX-C is abrogated. Drawing a further connection between the LBC and late insulator activity is the finding that mutations in the LBC GAGA motifs GAGA3 and GAGA4 weaken late insulator activity in transgene assays (56). Fourth, as might be expected from the effects of GAGA motif mutations on DNA binding in vitro and insulator function in vivo, one of the protein components of the LBC is the GAGA factor, GAF. ChIP experiments show that GAF is associated with Fab-7 HS1 sequences in vivo (25, 26). Though there is the caveat that the GAF antibody does not distinguish between GAF in the LBC complex and bulk GAF, this would nevertheless be consistent with the notion that the LBC is bound to the Fab-7 insulator in vivo. The involvement of GAF also fits with other experiments that have implicated this protein in Fab-7 boundary activity (56, 84, 85). However, it was previously thought that the primary function of GAF was to generate a nucleosome-free region that would permit the binding of proteins that function as insulators. The fact that GAF is a component of the LBC would suggest that it likely has a much more intimate connection to insulator activity than previously anticipated. Fifth, further supporting a functional connection between the LBC and late boundary activity is the fact that two other proteins implicated in insulator function, Mod(mdg4) and E(y)2, also appear to be LBC components. Moreover, ChIPs show that Mod(mdg4), like GAF, is associated with Fab-7 HS1 sequences in vivo (25). As GAF and Mod(mdg4) often colocalize at other known or predicted boundaries elsewhere in the genome, the LBC is likely to be important for insulator function on a more global scale.
While our characterization of the LBC is incomplete, what we do know suggests that it is a fascinating complex. In addition to having a surprisingly large minimal binding sequence of ∼65 bp, its DNA sequence recognition properties are unusually flexible. A comparison of the three LBC recognition elements in dHS1 reveals no obvious sequence similarity other than the fact that they share a GAGA motif (plus a preceding AA sequence). In unpublished experiments, we have identified multiple LBC recognition elements (by EMSA) in Fab-7 boundaries from three other Drosophila species, D. yakuba, D. erecta, and D. pseudoobscura. As in D. melanogaster, the LBC recognition elements in each of these boundaries differ from those of their neighbors. Moreover, though the recognition elements in the two D. melanogaster group species, D. yakuba and D. erecta, exhibit a high degree of sequence similarity to the corresponding elements in D. melanogaster, the sequences of the D. pseudoobscura recognition elements have little in common with those in the D. melanogaster group species. While mutations in the GAGA motifs GAGA3 and GAGA4 largely disrupt binding to probes spanning GAGA3 and GAGA4 sites, the presence of a GAGA motif in other contexts is not sufficient for LBC binding. The LBC does not shift well-characterized binding sequences for GAF in the promoters of several fly genes even though these sequences are bound by GAF in nuclear extracts. In addition to not being sufficient for stable binding, we have found that a GAGA motif is not always necessary for LBC binding. The Fab-7 boundaries from D. erecta and D. yakuba have four rather than three LBC recognition elements. In both cases, one of the four elements lacks a GAGA motif (D. Wolle, unpublished data). Further evidence that the sequence recognition properties of the LBC are unusual comes from mutagenesis experiments on GAGA3-65 and GAGA4-65. While mutating the GAGA motifs weakened LBC binding, mutations (12-bp substitutions) elsewhere in the two probes had minimal effect.
Gel filtration experiments show that the LBC does not assemble in a stepwise fashion on DNA but rather exists as a preassembled 700-kDa complex. Our supershift experiments show that there are three different proteins in this complex, GAF, Mod(mdg4), and E(y)2. However, as we were able to test only known insulator proteins, there could be as yet other unidentified or unknown proteins in the complex. Additionally, as the LBC shift consists of several subbands whose yield varies depending upon the probe, there may be adjutant factors that associate stably with the complex only when the LBC binds to a specific element.
The fly has two GAF protein isoforms, GAF519 and GAF581. They share an N-terminal BTB domain and a single internal C2H2 zinc finger but have a distinct C-terminal domain. The zinc finger is responsible for DNA binding, while the N-terminal BTB domain functions in protein-protein interactions. Since the GAF BTB domains assemble into dimers, tetramers, and octomers, it seems likely that the LBC contains two, if not more, GAF proteins (66). Though this would not explain why the minimal recognition element is at least 65 bp in length, it raises the possibility that the LBC could simultaneously interact with several GAGA motifs. In this respect it should be noted that each of the recognition elements in dHS1 has a single GAGA motif. While we have found that the LBC binds independently to each element, our experiments do not exclude the possibility that it binds simultaneously to two or more elements. If this is the case, the LBC could potentially interact with several GAGAG motifs spread out over a sequence extending 120 to 200 bp. However, this would likely require that the DNA wrap around the outside surface of the complex as in the nucleosome. At about six times the mass of a nucleosome, the size of the complex should be more than sufficient to accommodate a DNA sequence of this length. Moreover, there are precedents for wrapping extended recognition sequences around large regulatory complexes. For example, fly PREs (which, like insulators, span large [>200-bp] nuclease-hypersensitive regions) appear to be assembled by wrapping the PRE sequence around a large complex containing DNA binding and Polycomb group proteins (86).
Like GAF, Mod(mdg4) has an N-terminal BTB domain that can self-assemble; however, in contrast to GAF, where the most stable complex appears to be a dimer, the primary Mod(mdg4) multimer is thought to be an octamer (87). Thus, the Mod(mdg4) protein could be one of the more abundant components in the LBC. If this is correct, then it could have a scaffolding role in the complex since the Mod(mdg4) and GAF BTB domains interact with not only themselves but also each other (82). While the BTB domain is common to all Mod(mdg4) isoforms, there are 31 different C-terminal domains. Twenty-nine of these isoforms are predicted to have DNA binding activity. Of these, one isoform has a BED zinc finger domain like the DNA binding domain in the BEAF insulator protein, while another, like the Elba1, Elba2, and Insv insulator proteins, has a BEN DNA binding domain. Two of these isoforms have C-terminal sequences that show no homology to known DNA binding domains. The remaining 27 Mod(mdg4) isoforms have a FLYWCH type Zn finger domain. While DNA binding by Mod(mdg4) isoforms containing one of these FLYWCH Zn fingers has not yet been demonstrated, the Caenorhabditis elegans PEB-1 protein has been shown to bind DNA via its FLYWCH domain (80). Thus, a reasonable expectation is that the Mod(mdg4) protein(s) in the LBC will likely contribute to DNA binding. Since the known Mod(mdg4)-GAF interactions are mediated by the common BTB domain, it is possible that the LBC we detect in 6- to 18-h embryos contains a mixture of several different Mod(mdg4) proteins (which could even vary depending upon the alternative splicing patterns that predominate in cells or tissue types from which the complex is derived). If multiple Mod(mdg4) isoforms are present in the LBC, this could provide an explanation for its unusually flexible sequence recognition properties. Alternatively, the variable domains in one or more of the isoforms may contain signals that mediate preferential incorporation of the isoform(s) into the LBC. Resolving this issue as well as the possible roles of the Mod(mdg4) protein(s) as a scaffolding factor and/or in determining sequence specificity will require the purification of the complex.
The third component of the LBC is E(y)2. This small conserved protein has been shown to interact with the general transcription factor TFIID in flies, while the Saccharomyces cerevisiae counterpart, Sus1, is a component of the SAGA complex (88, 89). In addition to functioning with the transcriptional apparatus, E(y)2 has been shown to interact with the insulator protein Su(Hw). While E(y)2 is not needed for the enhancer-blocking activity of Su(Hw) insulators, it is required to block silencing induced by Polycomb response elements (PREs). Since one of the important functions of Fab-7 as well as of other insulators in the BX-C is to prevent PREs in silenced cis-regulatory domains from turning off their active neighbors, it would be reasonable to think that E(y)2 plays a similar role in the LBC. In fact, this is precisely what happens in the ΔHS1 and GAGA1–6 mutants: the PRE in iab-7 inappropriately silences the iab-6 cis-regulatory domain in a subset of the PS11 cells.
In addition to the unusual sequence recognition properties of the LBC, the other distinguishing feature of the LBC is that its activity is developmentally restricted. It is enriched in nuclear extracts prepared from late embryos, while it is present at only low levels in extracts from early embryos. The Elba factor is the only other known developmentally regulated insulator complex, and its activity is restricted to early embryos because two of the Elba subunits, Elba1 and Elba3, are midblastula transition genes (57, 58). Conceivably, a similar mechanism could account for the late appearance of the LBC activity in nuclear extracts. One potential candidate would be the GAF isoform GAF581. Unlike GAF519, whose message is maternally deposited, expression of GAF581 does not commence until around 6 h (90). While the delayed expression of the GAF581 isoform would fit nicely with the LBC developmental profile, functional studies would argue that if there is an LBC-specific isoform, it would be GAF519, not GAF581 (91). As the two GAF isoforms are subject to stage-specific posttranslational modifications, an alternative possibility is that modification plays a critical role in the assembly of the LBC and the incorporation of the GAF protein. Of course, other components of the LBC components could be responsible for its stage specificity. One possibility would be the expression of specific Mod(mdg4) isoforms that are needed to scaffold the assembly of the LBC complex. There could also be an as yet unidentified stage-specific protein(s) that is needed for complex formation. Further studies should resolve this question.
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert Maeda and Henrik Gyurkovics for insightful discussions. We also thank Eva Favre, Benjamin Barandun, Jorge Faustino, and Gordon Gray for excellent technical assistance. We thank Anton Golovnin, Pavel Georgiev, Elissa Lei, Victor Coreces, Carl Wu, and David Gilmour for the gift of antibodies. We also thank the reviewers for their thoughtful comments.
F.K. acknowledges support by grants from the Donation Claraz, the State of Geneva, and the Swiss National Fund for Research. P.S. acknowledges support from a grant from the NIH (GM043432) and from a grant to the Gene Biology Institute by the Russian Federation Ministry of Education and Science (14.B25.31.0022).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00456-15.
REFERENCES
- 1.Gaszner M, Felsenfeld G. 2006. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 2.Vogelmann J, Valeri A, Guillou E, Cuvier O, Nollmann M. 2011. Roles of chromatin insulator proteins in higher-order chromatin organization and transcription regulation. Nucleus 2:358–369. doi: 10.4161/nucl.2.5.17860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chetverina D, Aoki T, Erokhin M, Georgiev P, Schedl P. 2014. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. Bioessays 36:163–172. doi: 10.1002/bies.201300125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herold M, Bartkuhn M, Renkawitz R. 2012. CTCF: insights into insulator function during development. Development 139:1045–1057. doi: 10.1242/dev.065268. [DOI] [PubMed] [Google Scholar]
- 5.Barkess G, West AG. 2012. Chromatin insulator elements: establishing barriers to set heterochromatin boundaries. Epigenomics 4:67–80. doi: 10.2217/epi.11.112. [DOI] [PubMed] [Google Scholar]
- 6.Ghirlando R, Giles K, Gowher H, Xiao T, Xu Z, Yao H, Felsenfeld G. 2012. Chromatin domains, insulators, and the regulation of gene expression. Biochim Biophys Acta 1819:644–651. doi: 10.1016/j.bbagrm.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kellum R, Schedl P. 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64:941–950. doi: 10.1016/0092-8674(91)90318-S. [DOI] [PubMed] [Google Scholar]
- 8.Kellum R, Schedl P. 1992. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol 12:2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holdridge C, Dorsett D. 1991. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol Cell Biol 11:1894–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sigrist CJ, Pirrotta V. 1997. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147:209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erokhin M, Parshikov A, Georgiev P, Chetverina D. 2010. E(y) 2/Sus1 is required for blocking PRE silencing by the Wari insulator in Drosophila melanogaster. Chromosoma 119:243–253. doi: 10.1007/s00412-009-0253-1. [DOI] [PubMed] [Google Scholar]
- 12.Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P. 2001. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291:495–498. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- 13.Cai HN, Shen P. 2001. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 291:493–495. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- 14.Comet I, Savitskaya E, Schuettengruber B, Negre N, Lavrov S, Parshikov A, Juge F, Gracheva E, Georgiev P, Cavalli G. 2006. PRE-mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev Cell 11:117–124. doi: 10.1016/j.devcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Kyrchanova O, Chetverina D, Maksimenko O, Kullyev A, Georgiev P. 2008. Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements. Nucleic Acids Res 36:7019–7028. doi: 10.1093/nar/gkn781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muller M, Hagstrom K, Gyurkovics H, Pirrotta V, Schedl P. 1999. The Mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics 153:1333–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vazquez J, Muller M, Pirrotta V, Sedat JW. 2006. The Mcp element mediates stable long-range chromosome-chromosome interactions in Drosophila. Mol Biol Cell 17:2158–2165. doi: 10.1091/mbc.E06-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li HB, Muller M, Bahechar IA, Kyrchanova O, Ohno K, Georgiev P, Pirrotta V. 2011. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol Cell Biol 31:616–625. doi: 10.1128/MCB.00849-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujioka M, Sun G, Jaynes JB. 2013. The Drosophila eve insulator Homie promotes eve expression and protects the adjacent gene from repression by polycomb spreading. PLoS Genet 9:e1003883. doi: 10.1371/journal.pgen.1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. 2007. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet 3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith ST, Wickramasinghe P, Olson A, Loukinov D, Lin L, Deng J, Xiong Y, Rux J, Sachidanandam R, Sun H, Lobanenkov V, Zhou J. 2009. Genome wide ChIP-chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev Biol 328:518–528. doi: 10.1016/j.ydbio.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang N, Emberly E, Cuvier O, Hart CM. 2009. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol Cell Biol 29:3556–3568. doi: 10.1128/MCB.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, Stein L, Henikoff S, Kellis M, White KP. 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet 6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz YB, Linder-Basso D, Kharchenko PV, Tolstorukov MY, Kim M, Li HB, Gorchakov AA, Minoda A, Shanower G, Alekseyenko AA, Riddle NC, Jung YL, Gu T, Plachetka A, Elgin SC, Kuroda MI, Park PJ, Savitsky M, Karpen GH, Pirrotta V. 2012. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res 22:2188–2198. doi: 10.1101/gr.138156.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, Tian Y, Shu W, Bo X, Wang S. 2012. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS One 7:e41374. doi: 10.1371/journal.pone.0041374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nora EP, Dekker J, Heard E. 2013. Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? Bioessays 35:818–828. doi: 10.1002/bies.201300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, Gribnau J, Barillot E, Bluthgen N, Dekker J, Heard E. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cleard F, Moshkin Y, Karch F, Maeda RK. 2006. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet 38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- 31.Gdula DA, Gerasimova TI, Corces VG. 1996. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci U S A 93:9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott KC, Taubman AD, Geyer PK. 1999. Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics 153:787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao K, Hart CM, Laemmli UK. 1995. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81:879–889. doi: 10.1016/0092-8674(95)90008-X. [DOI] [PubMed] [Google Scholar]
- 34.Oliver D, Sheehan B, South H, Akbari O, Pai CY. 2010. The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions. BMC Cell Biol 11:101. doi: 10.1186/1471-2121-11-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golovnin A, Biryukova I, Romanova O, Silicheva M, Parshikov A, Savitskaya E, Pirrotta V, Georgiev P. 2003. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development 130:3249–3258. doi: 10.1242/dev.00543. [DOI] [PubMed] [Google Scholar]
- 36.Soshnev AA, Li X, Wehling MD, Geyer PK. 2008. Context differences reveal insulator and activator functions of a Su(Hw) binding region. PLoS Genet 4:e1000159. doi: 10.1371/journal.pgen.1000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vazquez J, Schedl P. 1994. Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J 13:5984–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch F. 1997. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 124:1809–1820. [DOI] [PubMed] [Google Scholar]
- 39.Schweinsberg SE, Schedl P. 2004. Developmental modulation of Fab-7 boundary function. Development 131:4743–4749. doi: 10.1242/dev.01343. [DOI] [PubMed] [Google Scholar]
- 40.Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F. 2000. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127:779–790. [DOI] [PubMed] [Google Scholar]
- 41.Karch F, Galloni M, Sipos L, Gausz J, Gyurkovics H, Schedl P. 1994. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res 22:3138–3146. doi: 10.1093/nar/22.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujioka M, Wu X, Jaynes JB. 2009. A chromatin insulator mediates transgene homing and very long-range enhancer-promoter communication. Development 136:3077–3087. doi: 10.1242/dev.036467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanton J, Gaszner M, Schedl P. 2003. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev 17:664–675. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gohl D, Aoki T, Blanton J, Shanower G, Kappes G, Schedl P. 2011. Mechanism of chromosomal boundary action: roadblock, sink, or loop? Genetics 187:731–748. doi: 10.1534/genetics.110.123752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hogga I, Mihaly J, Barges S, Karch F. 2001. Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol Cell 8:1145–1151. doi: 10.1016/S1097-2765(01)00377-X. [DOI] [PubMed] [Google Scholar]
- 46.Iampietro C, Cleard F, Gyurkovics H, Maeda RK, Karch F. 2008. Boundary swapping in the Drosophila bithorax complex. Development 135:3983–3987. doi: 10.1242/dev.025700. [DOI] [PubMed] [Google Scholar]
- 47.Maeda RK, Karch F. 2006. The ABC of the BX-C: the bithorax complex explained. Development 133:1413–1422. doi: 10.1242/dev.02323. [DOI] [PubMed] [Google Scholar]
- 48.Mihaly J, Barges S, Sipos L, Maeda R, Cleard F, Hogga I, Bender W, Gyurkovics H, Karch F. 2006. Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Development 133:2983–2993. doi: 10.1242/dev.02451. [DOI] [PubMed] [Google Scholar]
- 49.Lewis EB. 1978. A gene complex controlling segmentation in Drosophila. Nature 276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez-Herrero E, Vernos I, Marco R, Morata G. 1985. Genetic organization of Drosophila bithorax complex. Nature 313:108–113. doi: 10.1038/313108a0. [DOI] [PubMed] [Google Scholar]
- 51.Gyurkovics H, Gausz J, Kummer J, Karch F. 1990. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J 9:2579–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galloni M, Gyurkovics H, Schedl P, Karch F. 1993. The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J 12:1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hagstrom K, Muller M, Schedl P. 1996. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev 10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 54.Zhou J, Barolo S, Szymanski P, Levine M. 1996. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev 10:3195–3201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]
- 55.Rodin S, Kyrchanova O, Pomerantseva E, Parshikov A, Georgiev P. 2007. New properties of Drosophila fab-7 insulator. Genetics 177:113–121. doi: 10.1534/genetics.107.075887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. 2004. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168:1371–1384. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aoki T, Sarkeshik A, Yates J, Schedl P. 2012. Elba, a novel developmentally regulated chromatin boundary factor is a hetero-tripartite DNA binding complex. eLife 1:e00171. doi: 10.7554/eLife.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aoki T, Schweinsberg S, Manasson J, Schedl P. 2008. A stage-specific factor confers Fab-7 boundary activity during early embryogenesis in Drosophila. Mol Cell Biol 28:1047–1060. doi: 10.1128/MCB.01622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Groth AC, Olivares EC, Thyagarajan B, Calos MP. 2000. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci U S A 97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siegal ML, Hartl DL. 1996. Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics 144:715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rubin GM, Spradling AC. 1983. Vectors for P element-mediated gene transfer in Drosophila. Nucleic Acids Res 11:6341–6351. doi: 10.1093/nar/11.18.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A 104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Golic KG, Lindquist S. 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 64.Mishra RK, Mihaly J, Barges S, Spierer A, Karch F, Hagstrom K, Schweinsberg SE, Schedl P. 2001. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol Cell Biol 21:1311–1318. doi: 10.1128/MCB.21.4.1311-1318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iampietro C, Gummalla M, Mutero A, Karch F, Maeda RK. 2010. Initiator elements function to determine the activity state of BX-C enhancers. PLoS Genet 6:e1001260. doi: 10.1371/journal.pgen.1001260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adkins NL, Hagerman TA, Georgel P. 2006. GAGA protein: a multi-faceted transcription factor. Biochem Cell Biol 84:559–567. doi: 10.1139/o06-062. [DOI] [PubMed] [Google Scholar]
- 67.Wilkins RC, Lis JT. 1998. GAGA factor binding to DNA via a single trinucleotide sequence element. Nucleic Acids Res 26:2672–2678. doi: 10.1093/nar/26.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weber JA, Gilmour DS. 1995. Genomic footprinting of the hsp70 and histone H3 promoters in Drosophila embryos reveals novel protein-DNA interactions. Nucleic Acids Res 23:3327–3334. doi: 10.1093/nar/23.16.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao J, Benyajati C. 1998. Specific local histone-DNA sequence contacts facilitate high-affinity, non-cooperative nucleosome binding of both adf-1 and GAGA factor. Nucleic Acids Res 26:5394–5401. doi: 10.1093/nar/26.23.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grillo M, Furriols M, Casanova J, Luschnig S. 2011. Control of germline torso expression by the BTB/POZ domain protein pipsqueak is required for embryonic terminal patterning in Drosophila. Genetics 187:513–521. doi: 10.1534/genetics.110.121624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horowitz H, Berg CA. 1996. The Drosophila pipsqueak gene encodes a nuclear BTB-domain-containing protein required early in oogenesis. Development 122:1859–1871. [DOI] [PubMed] [Google Scholar]
- 72.Pai CY, Lei EP, Ghosh D, Corces VG. 2004. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell 16:737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 73.Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RA, Renkawitz-Pohl R, Saumweber H, Renkawitz R. 2007. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J 26:4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilbert MK, Tan YY, Hart CM. 2006. The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics 173:1365–1375. doi: 10.1534/genetics.106.056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kurshakova M, Maksimenko O, Golovnin A, Pulina M, Georgieva S, Georgiev P, Krasnov A. 2007. Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol Cell 27:332–338. doi: 10.1016/j.molcel.2007.05.035. [DOI] [PubMed] [Google Scholar]
- 76.Dai Q, Ren A, Westholm JO, Duan H, Patel DJ, Lai EC. 2015. Common and distinct DNA-binding and regulatory activities of the BEN-solo transcription factor family. Genes Dev 29:48–62. doi: 10.1101/gad.252122.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Faucheux M, Roignant JY, Netter S, Charollais J, Antoniewski C, Theodore L. 2003. batman Interacts with polycomb and trithorax group genes and encodes a BTB/POZ protein that is included in a complex containing GAGA factor. Mol Cell Biol 23:1181–1195. doi: 10.1128/MCB.23.4.1181-1195.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mishra K, Chopra VS, Srinivasan A, Mishra RK. 2003. Trl-GAGA directly interacts with lola like and both are part of the repressive complex of Polycomb group of genes. Mech Dev 120:681–689. doi: 10.1016/S0925-4773(03)00046-7. [DOI] [PubMed] [Google Scholar]
- 79.Maksimenko O, Bartkuhn M, Stakhov V, Herold M, Zolotarev N, Jox T, Buxa MK, Kirsch R, Bonchuk A, Fedotova A, Kyrchanova O, Renkawitz R, Georgiev P. 2015. Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res 25:89–99. doi: 10.1101/gr.174169.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beaster-Jones L, Okkema PG. 2004. DNA binding and in vivo function of C.elegans PEB-1 require a conserved FLYWCH motif. J Mol Biol 339:695–706. doi: 10.1016/j.jmb.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 81.Dorn R, Krauss V. 2003. The modifier of mdg4 locus in Drosophila: functional complexity is resolved by trans splicing. Genetica 117:165–177. doi: 10.1023/A:1022983810016. [DOI] [PubMed] [Google Scholar]
- 82.Melnikova L, Juge F, Gruzdeva N, Mazur A, Cavalli G, Georgiev P. 2004. Interaction between the GAGA factor and Mod(mdg4) proteins promotes insulator bypass in Drosophila. Proc Natl Acad Sci U S A 101:14806–14811. doi: 10.1073/pnas.0403959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dai Q, Ren A, Westholm JO, Serganov AA, Patel DJ, Lai EC. 2013. The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors. Genes Dev 27:602–614. doi: 10.1101/gad.213314.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li M, Belozerov VE, Cai HN. 2010. Modulation of chromatin boundary activities by nucleosome-remodeling activities in Drosophila melanogaster. Mol Cell Biol 30:1067–1076. doi: 10.1128/MCB.00183-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakayama T, Shimojima T, Hirose S. 2012. The PBAP remodeling complex is required for histone H3.3 replacement at chromatin boundaries and for boundary functions. Development 139:4582–4590. doi: 10.1242/dev.083246. [DOI] [PubMed] [Google Scholar]
- 86.Mohd-Sarip A, van der Knaap JA, Wyman C, Kanaar R, Schedl P, Verrijzer CP. 2006. Architecture of a polycomb nucleoprotein complex. Mol Cell 24:91–100. doi: 10.1016/j.molcel.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 87.Bonchuk A, Denisov S, Georgiev P, Maksimenko O. 2011. Drosophila BTB/POZ domains of “ttk group” can form multimers and selectively interact with each other. J Mol Biol 412:423–436. doi: 10.1016/j.jmb.2011.07.052. [DOI] [PubMed] [Google Scholar]
- 88.Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116:75–86. doi: 10.1016/S0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 89.Georgieva S, Nabirochkina E, Dilworth FJ, Eickhoff H, Becker P, Tora L, Georgiev P, Soldatov A. 2001. The novel transcription factor e(y)2 interacts with TAFII40 and potentiates transcription activation on chromatin templates. Mol Cell Biol 21:5223–5231. doi: 10.1128/MCB.21.15.5223-5231.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Benyajati C, Mueller L, Xu N, Pappano M, Gao J, Mosammaparast M, Conklin D, Granok H, Craig C, Elgin S. 1997. Multiple isoforms of GAGA factor, a critical component of chromatin structure. Nucleic Acids Res 25:3345–3353. doi: 10.1093/nar/25.16.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Greenberg AJ, Schedl P. 2001. GAGA factor isoforms have distinct but overlapping functions in vivo. Mol Cell Biol 21:8565–8574. doi: 10.1128/MCB.21.24.8565-8574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bender W, Lucas M. 2013. The border between the ultrabithorax and abdominal-A regulatory domains in the Drosophila bithorax complex. Genetics 193:1135–1147. doi: 10.1534/genetics.112.146340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Foronda D, Martin P, Sanchez-Herrero E. 2012. Drosophila Hox and sex-determination genes control segment elimination through EGFR and extramacrochetae activity. PLoS Genet 8:e1002874. doi: 10.1371/journal.pgen.1002874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.