Abstract
Senescent cells secrete a combination of factors collectively known as the senescence-associated secretory phenotype (SASP). The SASP reinforces senescence and activates an immune surveillance response but it can also display pro-tumorigenic properties and contribute to age-related pathologies. In a drug screen to find novel SASP regulators, we uncovered the mTOR inhibitor rapamycin as a potent SASP suppressor. Here we report a mechanism by which mTOR controls the SASP by differentially regulating the translation of the MK2/MAPKAPK2 kinase through 4EBP1. In turn, MAPKAPK2 phosphorylates the RNA binding protein ZFP36L1 during senescence, inhibiting its ability to degrade the transcripts of numerous SASP components. Consequently, mTOR inhibition or constitutive activation of ZFP36L1 impairs the non-cell-autonomous effects of senescent cells both in tumour-suppressive and promoting-promoting contexts. Altogether, our results place regulation of the SASP as a key mechanism by which mTOR could influence cancer, age-related diseases and immune responses.
Keywords: mTOR, SASP, senescence, rapamycin, MAPKAPK2, ZFP36L1
INTRODUCTION
Senescence is a stable arrest induced in damaged or ageing cells 1. Senescent cells undergo changes in their transcription, chromatin organization and metabolism 2. They also secrete a group of factors collectively termed the senescence-associated secretory phenotype (SASP), which influence the tumour microenvironment and the homeostasis of aged tissues 3, 4. For example, the SASP of senescent fibroblasts promotes the epithelial to mesenchymal transition (EMT) of epithelial cancer cells 3 while also enhancing their tumorigenic potential 5. In contrast, the SASP can induce senescence 4, 6, 7 and activate the immune system to keep neoplastic cells in check 8. The SASP is regulated transcriptionally by NF-κB and CEBP/β and its expression is sustained by redundant autocrine feedback loops 7, 9-11. In addition, signalling via p38α MAPK 12, and the persistent DNA damage response (DDR) associated with senescence 13 contribute to SASP induction 14. Cells undergoing oncogene-induced senescence (OIS) augment their secretory phenotypes by coordinating protein synthesis and autophagy in the TOR-autophagy spatial coupling compartment (TASCC) 15. Disruption of mTOR recruitment to the TASCC affects IL6 and IL8 production 15. However, it is still unclear how mTOR ultimately regulates this and to what extent mTOR affects the SASP.
mTOR integrates inputs from nutrient and growth signals to control processes like protein and lipid synthesis or autophagy 16. To regulate protein synthesis, mTOR phosphorylates the eukaryotic translation initiation factor 4E (eIF4E)-binding proteins (4EBPs) and S6 kinases (S6K1 and S6K2) 17. While S6K1/2 phosphorylation controls translation initiation and elongation 18, phosphorylation of 4EBPs promotes cap-dependent translation by preventing 4EBP binding to eIF4e 19. Besides controlling overall translation rates, mTOR differentially regulates the translation of transcripts with 5′ terminal oligopyrimidine (TOP) motifs in a 4E-BP dependent fashion 20. Rapamycin-treated mice display a significant increase in life span 21 and resistance to age-related pathologies and mTOR inhibitors are being used for cancer therapy. Elucidating how mTOR regulates the SASP may aid understanding of the aetiology of these complex disorders.
RESULTS
mTOR inhibition prevents the induction of the SASP
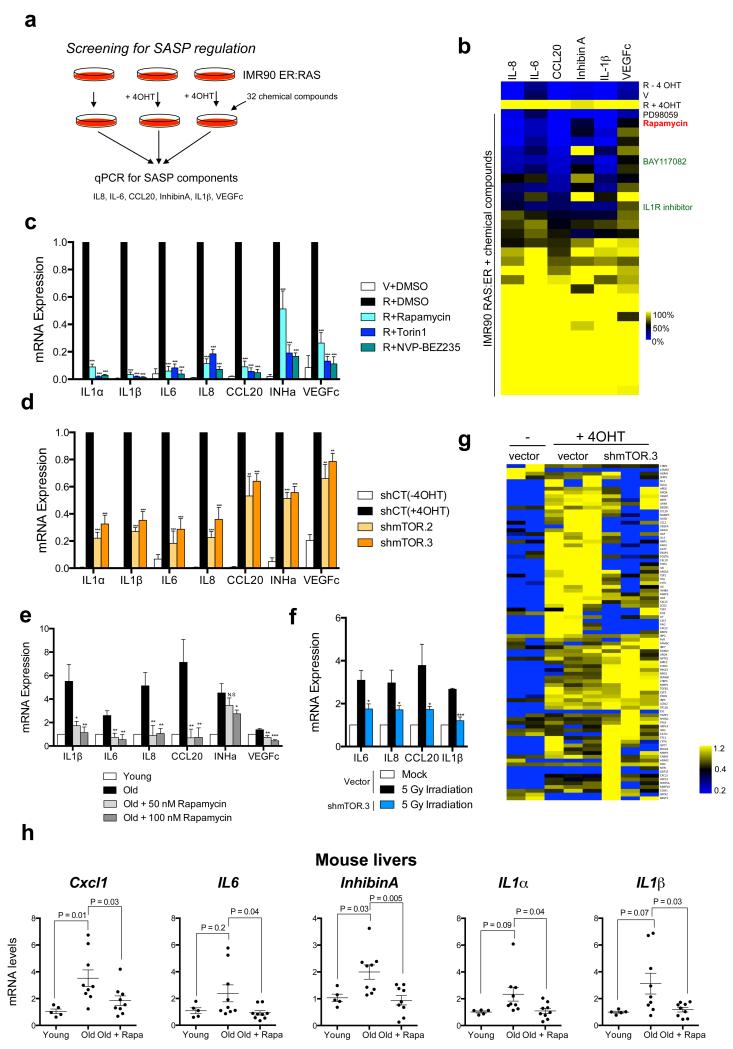
To screen for drugs blunting the induction of the SASP, we used IMR90 cells bearing an ER:RAS fusion protein (IMR90 ER:RAS) as a model of OIS. Treatment of IMR90 ER:RAS cells with 4-hydroxytamoxifen (4OHT) activates RAS, inducing senescence and the SASP 6. The expression of six robustly induced SASP components, which are known to mediate the paracrine effects of senescent cells 6, was used as readout of the screen (Fig 1a). Amongst compounds preventing SASP induction, we identified antagonists of IL1 or NF-κB signalling (Fig 1b, Supplementary Table S1), as previously described 6, 10, 11. We also identified that rapamycin, a natural mTOR inhibitor, prevented the SASP of cells undergoing OIS (Fig 1b). It has been suggested that rapamycin inhibits the SASP 22, 23, but to what extent and how, is unclear. Rapamycin effects were validated using shRNAs targeting mTOR, other mTOR inhibitors (Torin1 and NVP-BEZ235) and additional fibroblast strains (BJ and HFFF2) (Fig 1c-d and Supplementary Fig S1a). mTOR depletion or rapamycin treatment also diminished the SASP during replicative senescence (Fig 1e) and in response to γ-irradiation (Fig 1f). The ability of mTOR inhibition to impair the SASP was also observed at the protein level (Supplementary Fig S1b-c).
Figure 1. A drug screen identifies rapamycin as a regulator of the SASP.
a. Scheme of the screening. b. A drug screen identifies rapamycin as a regulator of the SASP. IMR90 ER:RAS cells were induced with 4OHT to activate ER:RAS and treated with a library of chemical compounds from the day of induction. Seven days after 4OHT induction, the expression of selected SASP components (IL-1β, IL-6, IL-8, CCL20, InhibinA and VEGFc) was analysed by qPCR. c. IMR90 ER:RAS cells were treated as in (b) in the presence of 10 nM rapamycin, 25 nM Torin1, or 0.5 nM NVP-BEZ235. Expression of SASP genes was assessed by qRT-PCR (n=4). V, IMR90 expressing an empty vector; R, IMR90 infected with pLNC-ER:RAS. d. Expression of SASP genes in the indicated IMR90 ER:RAS cells was measured by qRT-PCR 7 days after 4OHT induction (n=3) e. The expression of SASP genes was assessed in young (passage 11) and old (passage 23) IMR90 human fibroblasts treated as indicated for 7 days (n=3). f. mTOR knockdown inhibits the SASP observed in irradiated IMR90 fibroblasts. RNA was collected 8 days after irradiation (5 Gy), (n=3). g. MS analysis shows a global effect of mTOR inhibition on SASP regulation. IMR90 ER:RAS cells (vector or shmTOR) were incubated with DMSO or 4OHT with no FBS. After 6 days, CM was collected and processed for MS. Heatmap represents the global effect of mTOR depletion on the identified secreted factors (see Methods for details). 3 independent experiments are shown; yellow indicates above-mean expression and blue indicates below-mean levels. h. Decreased expression of SASP genes in aged mice treated with rapamycin. Expression of SASP components in livers of young mice (3 months, n=5) and old mice (22 months) either untreated (n=9) or treated (n=9) with rapamycin was measured by qRT-PCR. P values are included. All statistical significances were calculated using two-tailed Student’s t-test, *** P < 0.001; ** P < 0.01; *P < 0.05; n.s, non significant. All error bars represent means ± s.d. n represents number of mice in h and independent experiments in c-f. For raw data, see Supplementary Table 7.
To understand to what extent mTOR regulates the SASP, we analysed the secretome of senescent cells by mass spectroscopy (MS) 25. Amongst the SASP factors (secreted at higher levels in senescent than normal cells) detected by MS, mTOR depletion reduced secretion by at least 20% for half of them (41/78) (Fig 1g and Supplementary Table S2). Inhibiting mTOR with rapamycin, Torin1 or NVP-BEZ235 had similar effects (Supplementary Fig S1d). Importantly, amongst the SASP components downregulated we identified IL6, IL8 and other functionally important factors (Supplementary Table S2) 6, 7, 9.
Since rapamycin extends the lifespan of mice 21, and the ablation of senescent cells improves age-related diseases 26, 27, downregulating the SASP could contribute to the benefits observed in rapamycin-treated old mice. Analyzing liver samples, we observed an upregulation of the SASP during ageing (Fig 1h). Interestingly, 22 months old mice treated with rapamycin from 9 months of age 21 expressed lower levels of the SASP than their untreated age-matched counterparts (Fig 1h). Altogether our results indicate that mTOR regulates the SASP.
mTOR inhibition affects the SASP without reversing the senescence growth arrest
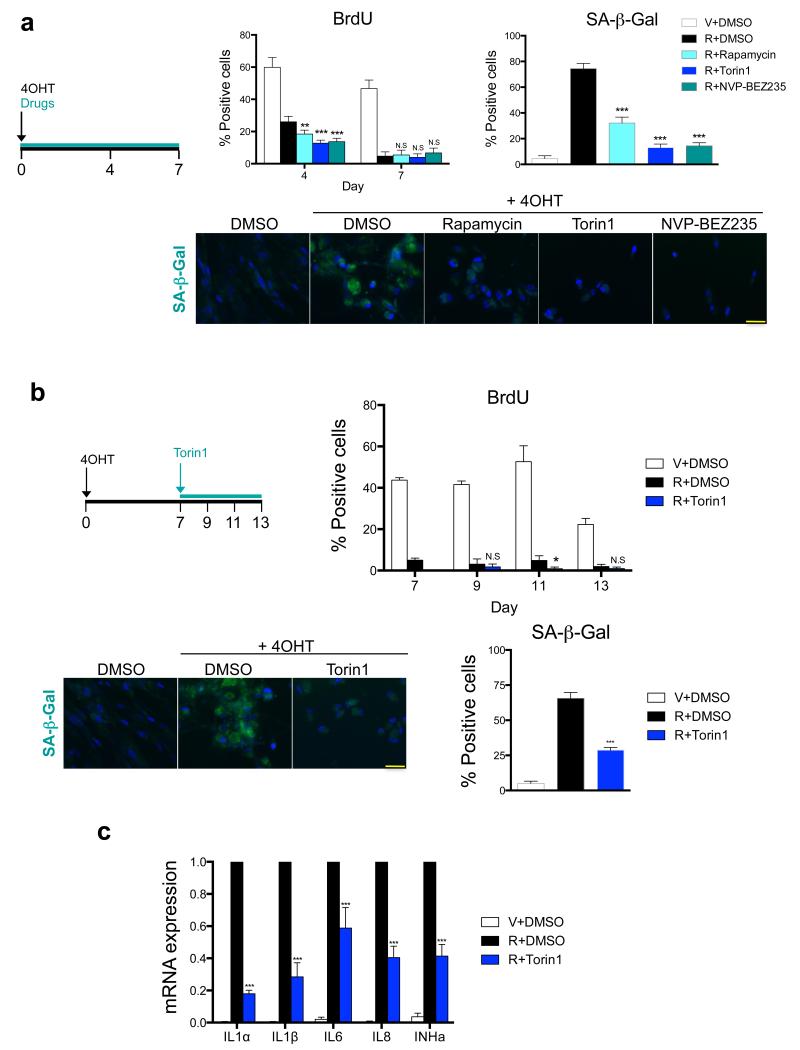
Inhibition of mTOR has been shown to impair the senescence phenotype, but there is conflicting evidence as to whether it also reverses the senescence growth arrest 22, 28, 29. Blocking mTOR signalling in IMR90 ER:RAS cells resulted in fewer SA-β-Gal positive cells and decreased levels of other senescence markers, such as p16INK4a and p21CIP1a. However, mTOR inhibition did not rescue the growth arrest (Fig 2a, Supplementary Fig S2a-c). This may be explained by the well-described antiproliferative effects caused by mTOR inhibition30, 31. In fact, rapamycin significantly decreased the levels of Cyclin D3 in IMR90 ER:RAS senescent cells (Supplementary Fig S2d).
Figure 2. mTOR inhibition impairs the SASP without reversing the senescence growth arrest.
a. mTOR inhibition results in decreased SA-β-Gal activity but cells remain arrested. IMR90 ER:RAS cells were induced to undergo senescence by 4OHT treatment. Cells were treated with the indicated drugs from day 0. BrdU incorporation was measured at day 4 and 7 after induction while SA-β-Gal activity was determined at day 7. Data are mean ± s.d. from n=6 (BrdU) and n=4 (SA-β-Gal) independent experiments. Representative images for SA-β-Gal activity staining are shown. Scale bar, 40 μm. b. Inhibition of mTOR after senescence induction downregulates SA-β-Gal activity without reversing growth arrest. Senescence was induced in IMR90 ER:RAS cells by 4OHT treatment for 7 days. At that stage, senescent cells were treated with 25 nM Torin1. BrdU incorporation was monitored at the indicated times and SA-β-Gal activity was measured at day 13. Data are mean ± s.d. n=4 independent experiments. Representative images for SA-β-Gal activity staining are shown. Scale bar, 40μm. c. Inhibition of mTOR after senescence induction downregulates the SASP. IMR90 ER:RAS cells were treated as in (b). The expression of the indicated SASP components was monitored by qRT-PCR 13 days after 4OHT induction (6 days after adding Torin1). Data are mean ± s.d. from n=3 independent experiments. All statistical significances were calculated using two-tailed Student’s t-test, *** P < 0.001; ** P < 0.01; *P < 0.05; n.s, non significant. For raw data, see Supplementary Table 7.
In the experiments described above, we simultaneously added mTOR inhibitors and 4OHT (Fig 2a). To exclude that SASP downregulation was a consequence of cells not dividing or being prevented to enter into senescence by mTOR inhibition, we conducted experiments in which we treated IMR90 ER:RAS cells with mTOR inhibitors only once OIS was established (Fig 2b). In this setting, Torin1-mediated inhibition of mTOR signalling (Supplementary Fig S2e) and also resulted in downregulation of the SASP and reduced SA-β-Gal staining but did not rescue cell growth (Fig 2b,c and Supplementary Fig S2f). Therefore, mTOR inhibition can dissociate the senescence growth arrest from SASP induction.
Regulation of translation initiation by 4EBP controls the SASP
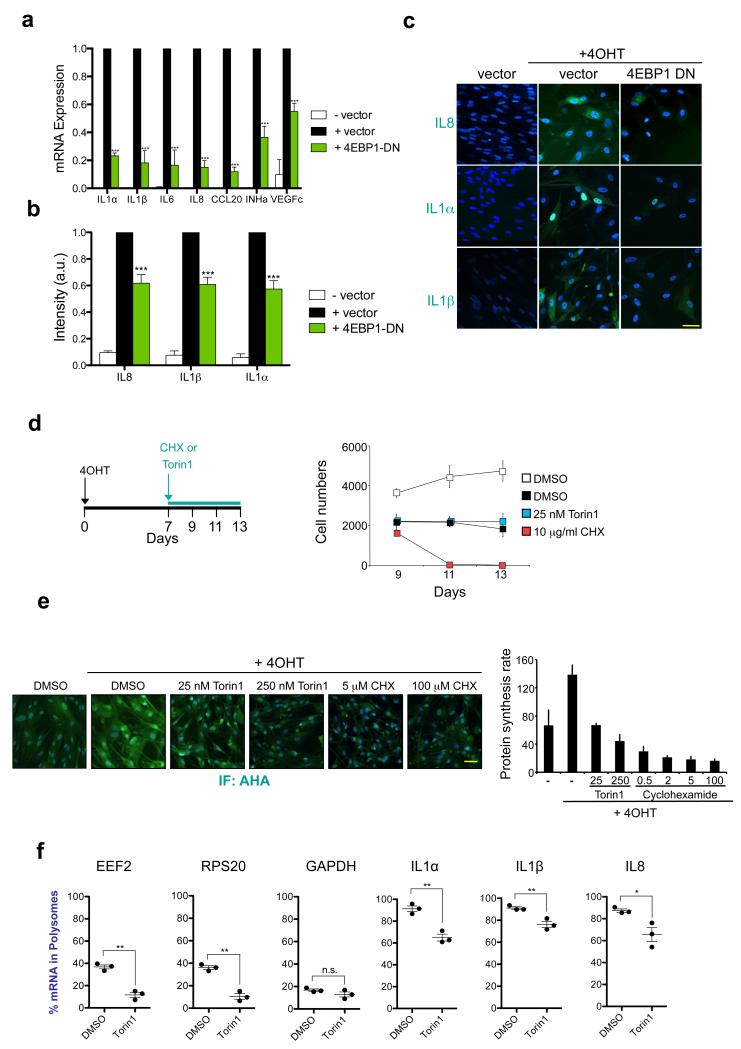
mTOR phosphorylates several downstream effectors, with 4EBPs being one of the best studied. 4EBPs control global translation rates but also differentially regulate the translation of specific mRNAs 20. Interestingly, expression of a dominant negative 4EBP1 mutant, (4EBP1 DN) (Supplementary Fig S3a), inhibited the SASP in response to both OIS and γ-irradiation (Fig 3a-c and Supplementary Fig S3a-b).
Figure 3. 4EBP mediates the regulation of the SASP downstream of mTOR.
a-c. Expression of an eIF4EBP1-4A phosphomutant (4EBP1 DN) that cannot be phosphorylated by mTOR prevents SASP induction. IMR90 ER:RAS cells were infected with a vector expressing 4EBP1 DN (or empty vector). (a) Expression of SASP genes was measured by qRT-PCR 7 days post-induction with 4OHT. Data are mean ± s.d. from n=3 independent experiments. (b) Fold changes of normalised intensity values obtained from SASP components immunostainings (IL8, IL1α and IL1β). Data are mean ± s.d. from n=4 independent experiments. (c) Representatives images from (b). Scale bar, 30μm. d. 7 days after 4OHT induction, IMR90 ER:RAS cells were treated as indicated in the scheme. DAPI staining was used to asses the cell numbers at the indicated times. Data are mean ± s.d. n=6 independent experiments. e. 7 days after 4OHT induction, the global protein synthesis rate in IMR90 ER:RAS cells was measured by using an AHA-based fluorescent assay. Cells were pre-treated for 2h as indicated prior to addition of 100 μM AHA. Fluorescent intensities, reflecting newly synthesized protein, were measured after 30min (DAPI used as a counterstain). (right) Data are mean ± s.d. from n=4 independent experiments. (left) Representative images are shown. Scale bar, 40μm. f. Effect of Torin1 on the distribution of SASP mRNAs in the polysomes of senescent cells. IMR90 ER:RAS cells were incubated for 7 days with 4OHT and then treated with DMSO or 250 nM Torin1 for 3h prior to sucrose gradient fractionation and polysome profiling. The resulting polysome profiles as well as a schematic representation of the position of fractions, monosomes and polysomes are shown in Supplementary Fig. 3c-d. Graphs show the % of the indicated mRNAs present in actively translating polysome fractions. EEF2 and RPS20 transcripts are mTOR canonical targets. GAPDH is insensitive to mTOR inhibition. Data are mean ± s.d. n=3 independent experiments. All statistical significance was calculated using two-tailed Student’s t-test, *** P < 0.001; ** P < 0.01; *P < 0.05; n.s, non significant. For raw data, see Supplementary Table 7.
To exclude that mTOR inhibition affected the SASP by non-specifically blocking translation, we compared the effects of Torin1 and Cyclohexamide (CHX, which blocks translational elongation). While senescent cells treated with Torin1 remained arrested (Fig 3d), CHX treatment triggered rapid cell death. These results (Fig 3d) suggested that the effects caused by mTOR inhibition during senescence were not due to a global translational shut down. To investigate how mTOR inhibition affects the rate of protein synthesis during senescence, we took advantage of a quantitative fluorogenic assay using the methionine analogue L-Azidohomoalanine (AHA) 32. The global translation rate increased during OIS and, in agreement with previous studies20, treatment with Torin1 greatly reduced de novo protein synthesis. However, overall translation was still comparable to that of non-senescent cells. In contrast, CHX almost completely shut down protein synthesis (Fig 3e).
The above results suggest that mTOR and 4EBP1 might control the SASP by regulating the translation of specific mRNA(s). To investigate this, we fractioned ribosomes from senescent cells treated with Torin1 or vehicle for 3 hours (Supplementary Fig S3c). We assessed the distribution of mRNAs in polysome and non-polysome (monosome) fractions (Fig 3f and Supplementary Fig S3d, e). In cells treated with Torin1, the distribution of mRNAs for canonical mTOR targets (e.g. EEF2 or RPS20) shifted almost completely to the ‘monosome’, non-translated fractions (Fig 3f). This was not the case for the mRNA of a housekeeping gene such as GAPDH (Fig 3f). The polysome association of the mRNAs of most SASP components analysed decreased slightly (Fig 3f and Supplementary Fig S3e), consistent with the general effect of Torin1 on translation. Amongst the SASP components analyzed, the mRNAs coding for IL8 and IL1α suffered the biggest drop in polysome association upon Torin1 treatment (Fig 3f). Nonetheless, more than 60 % of the mRNA for all SASP components tested remained associated with polysomes under acute mTOR inhibition in OIS (Fig 3f and Sup Fig S3e), suggesting that mTOR might regulate the translation of other target(s) to control the SASP.
mTOR regulates the SASP by controlling the translation of MAPKAPK2
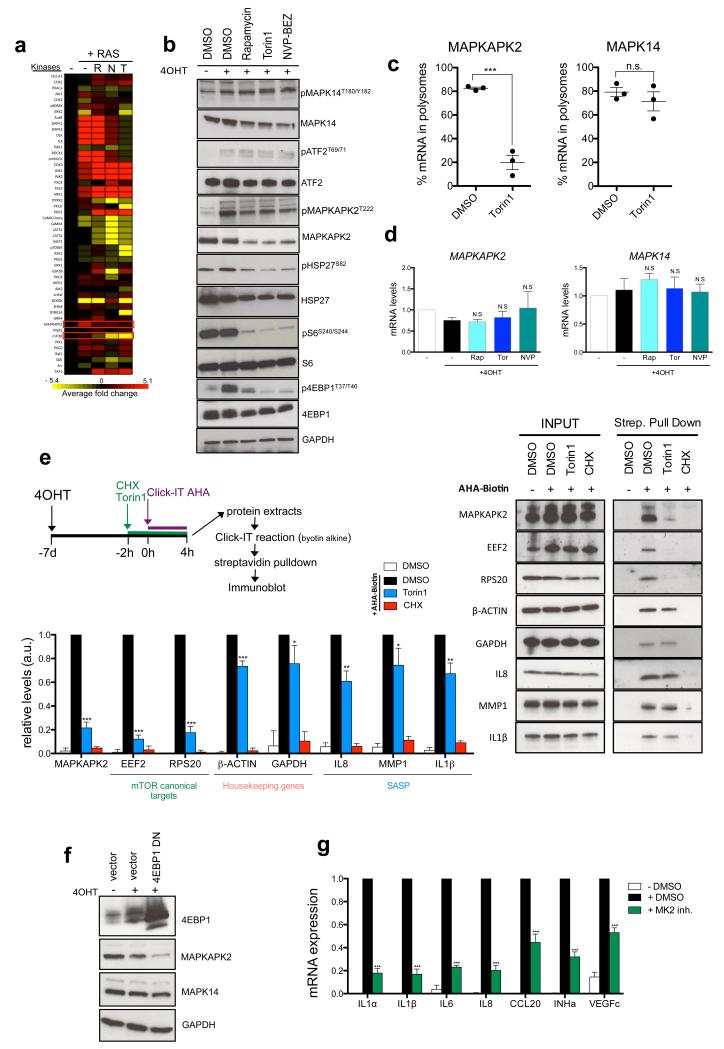
By analyzing phosphoproteomic data using Kinase Substrate Enrichment Analysis (KSEA 33), we identified that MAPKAPK2/MK2 kinase activity increased during senescence but declined upon mTOR inhibition (Fig 4a and Supplementary Table S3). MAPKAPK2/MK2 is located downstream of p38α MAPK 34, a known SASP regulator 12. Immunoblotting confirmed that MAPKAPK2 phosphorylation was blunted upon mTOR inhibition, but it also revealed reduced levels of MAPKAPK2, and to a lesser extent, of p38α MAPK (Fig 4b). However, while p38α activity was not affected by mTOR inhibition, as shown by unchanged phosphorylation of its substrate ATF2, phosphorylation of HSP27 (a MAPKAPK2 substrate) decreased significantly (Fig 4b). As p38α and MAPKAPK2 interact physically, reciprocally controlling their stability 34, decreased MAPKAPK2 could explain the reduced p38α levels.
Figure 4. mTOR regulates the SASP by controlling MAPKAPK2 translation.
a. Phosphoproteomic analysis reveals that mTOR signalling controls MAPKAPK2 kinase activity. IMR90 ER:RAS cells were incubated with DMSO or 4OHT and the indicated drugs for 7 days and further processed for MS analysis. Heatmap shows the enrichment of substrate groups for the different kinases calculated by the KSEA algorithm during senescence and in response to mTOR inhibition. (see Methods for details). The average of 3 independent experiments is shown. R, Rapamycin; T, Torin1; N, NVPBEZ-235. b. IMR90 ER:RAS cells were treated as indicated for 7 days. Immunoblots were performed with the indicated antibodies. c. The association of MAPKAPK2 mRNA with polysomes significantly decreases upon acute mTOR inhibition. Graphs show the percentage of MAPKAPK2 and MAPK14 (encoding for p38α MAPK) mRNAs associated with polysomes (n=3) IMR90 ER:RAS cells were incubated for 7 days with 4OHT and then treated with DMSO or 250 nM Torin1 (3h). d. IMR90 ER:RAS cells were treated as indicated for 7 days. Expression of MAPKAPK2 and MAPK14 was assessed by qRT-PCR (n=3) e. mTOR inhibition strongly impairs de novo translation of MAPKAPK2. After 7 days of 4OHT treatment, IMR90 ER:RAS cells were treated as shown in the scheme. AHA containing proteins were biotinylated and further purified using streptavidin beads. The expression of de novo synthesised proteins was analyzed by immunoblot with the indicated antibodies (right) and quantified (left) (n=3). f. IMR90 ER:RAS cells were infected with a 4EBP1-DN. The expression of 4EBP1, MAPKAPK2 and p38α was measured by immunoblot 7 days after 4OHT addition g.IMR90 ER:RAS cells were treated with 100nM MK2 inhibitor III. The expression of the indicated SASP components was monitored by qRT-PCR 7 days after 4OHT induction (n=3). All statistical significance was calculated using two-tailed Student’s t-test, *** P < 0.001; ** P < 0.01; *P < 0.05; n.s, non significant. All error bars represent means ± s.d. n represents number of independent experiments. For b,e and f representative images from 3 independent experiments are shown. Unprocessed original scans of blots are shown in Supplementary Fig. 9. For raw data, see Supplementary Table 7.
Interestingly, global ribosome profiling have suggested that mTOR can control MAPKAPK2 translation 20, 35. Accordingly, the percentage of MAPKAPK2 mRNA (but not the mRNAs for MAPK14 or the upstream kinase MKK6) bound to polysomes dropped dramatically upon acute mTOR inhibition in OIS (Fig 4c, Supplementary Fig S4a). Since transcript levels for MAPKAPK2 or MAPK14 (that encodes for p38α) remained unchanged when mTOR signalling was blocked (Fig 4d), our results suggested that mTOR specifically regulates MAPKAPK2 translation during senescence.
To confirm this, we analysed MAPKAPK2 translation by giving an AHA-biotin pulse followed by streptavidin pull-down and immunoblotting (Fig 4e and Supplementary Fig S4b). Acute mTOR inhibition decreased de novo synthesis of MAPKAPK2 (~80%) to an extent similar to that observed for mTOR canonical targets. In contrast, de novo translation of different SASP components (IL8, IL1β and MMP1) decreased only by 25-35%, comparable to the effect observed for housekeeping genes such as GAPDH and β-ACTIN (Fig 4e). These differential effects were in contrast with CHX treatment, which similarly suppressed de novo synthesis of all analysed proteins (Fig 4e).
To further address whether mTOR controls MAPKAPK2 translation, we monitored MAPKAPK2 expression and mTOR signalling during OIS. RAS induction quickly activated PI3K/AKT and mTOR signalling, which dropped after 48h-72h due to the activation of a negative feedback loop as reported before 36, (Supplementary Fig S4c). Remarkably, MAPKAPK2 protein levels and mTOR signalling significantly correlated across the analysed time course (Supplementary Fig S4c). In addition, overexpression of a 4EBP1-DN strongly downregulated MAPKAPK2 protein levels (Fig 4f) and pharmacological inhibition or knockdown of MAPKAPK2 prevented SASP induction (Fig 4g, Supplementary Fig S4d). Altogether our results suggest that mTOR controls the SASP via 4EBP1-mediated regulation of MAPKAPK2 translation.
Phosphorylation of ZFP36L1/BRF1 by MAPKAPK2 regulates the SASP
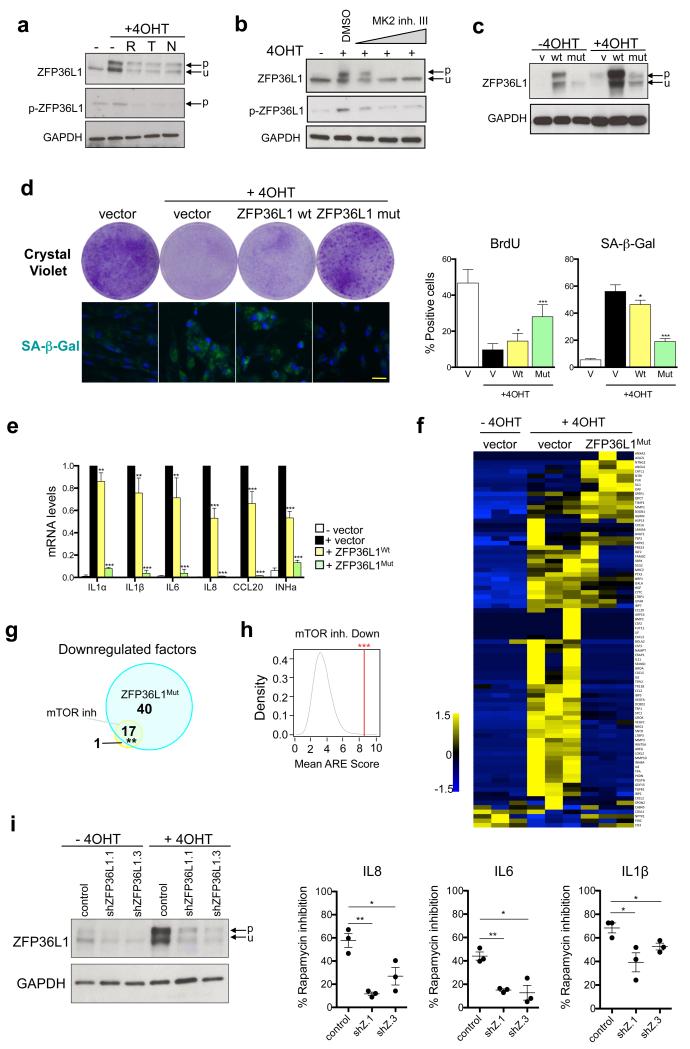
Phosphoproteomics identified that a known MAPKAPK2 substrate, ZFP36L1, was phosphorylated at Serine 54 (S54) during OIS. Importantly, this phosphorylation was impaired upon mTOR inhibition (Supplementary Fig S5a and Supplementary Table S3). ZFP36L1 (also known as Tis11b or BRF1) is a Zn-Finger protein that destabilises mRNAs by binding to AU-rich elements (AREs) in their 3’UTR 37. ZFP36L1 targets mRNAs of several cytokines for degradation 38. Phosphorylation of ZFP36L1 by MAPKAPK2 at S54, S92 and S203 inhibits its binding to mRNAs 39 while stabilizing ZFP36L1 40. The effect of mTOR inhibition on ZFP36L1 phosphorylation during OIS was corroborated using antibodies against ZFP36L1 (which detected a band shifted in senescent cells) and a phospho-specific pZFP36L1S203 antibody 41 (Fig 5a). Importantly, ZFP36L1 phosphorylation during OIS was sensitive to MAPKAPK2 inhibition (Fig 5b and Supplementary Fig S5b). Inhibition of mTOR also affected ZFP36L1 protein levels (Fig 5a, Supplementary Fig S5c), probably as a result of phosphorylation-regulated proteasomal degradation 40 as ZFP36L1 translation did not change upon mTOR inhibition (Supplementary Fig S5d)
Figure 5. MAPKAPK2 phosphorylates ZFP36L1 to regulate the SASP.
a. The effect of mTOR inhibition on ZFP36L1 phosphorylation during OIS was analysed by immunoblot using a ZFP36L1 antibody and a pZFP36L1S203 antibody41. p, phosphorylated; u, unphosphorylated. b. Immunoblot analysis of ZFP36L1 phosphorylation in IMR90 ER:RAS cells treated with MK2 inhibitor III (20, 100 and 200 nM). c-e. IMR90 ER:RAS cells were infected with ZFP36L1 wt or a mutant version (ZFP36L1S54A,S92,S203, ZFP36L1Mut) predicted to not be phosphorylated by MAPKAPK2. (c) Immunoblot analysis of ZFP36L1. (d) (left) Effect on cell proliferation was assessed by CV staining 14 days after 4OHT addition. (right) BrdU incorporation (n=7) and SA-β-Gal activity (n=4) were monitored by IF. Scale bar, 40μm. (e). Expression of SASP components was assessed by qRT-PCR (n=5). f. Indicated IMR90 ER:RAS cells were treated with DMSO or 4OHT in the absence of FBS. After 6 days, CM was collected and processed for MS. Heatmap represents the global effect of ZFP36L1Mut on the identified secreted factors (see Methods for details). 3 independent experiments are shown. g. Venn diagrams representing the overlap between SASP factors downregulated by mTOR inhibitors, shmTOR and ZFP36L1Mut expression. h. The mean ARE score42 for the 18 SASP factors commonly downregulated by shmTOR and mTOR inhibitors (See Sup. Fig 1d) was calculated (red line). Plot denotes the ARE score distribution for 105 random combinations of 18 mRNAs. i. IMR90 ER:RAS cells were infected with 2 shRNAs targeting ZFP36L1 and treated as indicated. (left) Immunoblot analysis of ZFP36L1 expression. (right) The effect of rapamycin treatment on the SASP was assessed by qRT-PCR (n=3). Statistical significance was calculated by using: (d,e and j: Student’s t-test) (g: hypergeometric test), (h: permutation tests). *** P < 0.001; ** P < 0.01; *P < 0.05; n.s, non significant. Error bars represent means ± s.d. n represents number of independent experiments. For a-d and i, data is representative of 3 (c,d,i) or 4 (a,b) independent experiments. Unless otherwise stated, IMR90 ER:RAS cells were induced with 4OHT for 7 days. For unprocessed original scans of blots, see Supplementary Fig. 9. For raw data, see Supplementary Table 7.
Next, we expressed either ZFP36L1 wt or a non-phosphorylatable mutant version (ZFP36L1S54A, S92A, S203A, ZFP36L1Mut) in IMR90 ER:RAS cells (Fig 5c and Supplementary Fig S5d). ZFP36L1Mut behaved as a constitutively active mutant, resistant to inactivation by MAPKAPK2. ZFP36L1Mut expression prevented SASP induction but it also rescued cell proliferation and inhibited senescence (Fig 5d-e, Supplementary Fig S5e-f). ZFP36L1 belongs to a family of RNA-binding proteins that also includes TTP and ZFP36L2. Interestingly, expression of ZFP36L2MUT, and to a lesser extent TTP MUT also compromised SASP induction (Supplementary Fig S5g-i). Although we only identified ZFP36L1 phosphorylation during OIS (Supplementary Table S3), we cannot exclude a similar role for TTP or ZFP36L2 in SASP regulation. Proteomic analysis showed that ZFP36L1Mut downregulates the majority of the SASP (60/89, Fig 5f, Supplementary Table S2). A significant overlap was observed between the factors down-regulated (but not those up-regulated) by mTOR inhibition and by ZFP36L1Mut expression (Fig 5g, Supplementary Fig S5j, k), highlighting a role for ZFP36L1 in mediating SASP regulation by mTOR. In addition, the factors down-regulated upon mTOR inhibition or by ZFP36L1Mut expression presented more AREs in their 3’UTRs than expected or than factors up-regulated by mTOR (Fig 5h, Supplementary Fig S5l), as predicted using ARE score 42. Finally, cells in which ZFP36L1 was knocked down repressed the SASP less efficiently in response to mTOR inhibition (Fig 5i), suggesting that ZFP36L1 mediates SASP regulation by mTOR.
ZFP36L1 is a direct regulator of the SASP
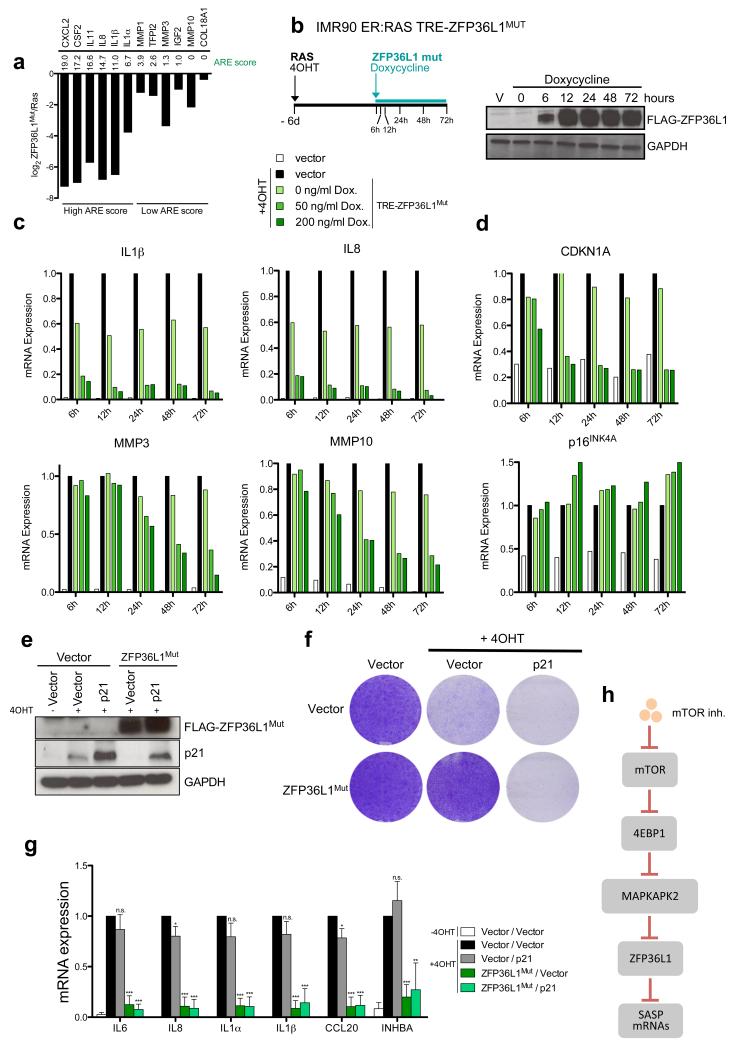
Gene set enrichment analysis (GSEA) of IMR90 ER:RAS cells expressing ZFP36L1Mut confirmed that besides inhibiting the SASP, ZFP36L1Mut also prevented senescence (Supplementary Fig S6a). Consistent with ZFP36L1 targeting mRNAs enriched in AU-rich elements in their 3’UTR, greater downregulation by ZFP36L1Mut correlated with a higher ARE score (Fig 6a). To investigate whether ZFP36L1 regulated the SASP directly, we generated a doxycycline-inducible ZFP36L1Mut construct, and induced its expression after OIS establishment (Fig 6b). Genes with a high ARE score (like IL8 or IL1β) were strongly downregulated as early as 6h after ZFP36L1Mut induction, suggesting a direct event. Downregulation of genes with a lower ARE score (like MMP3 or MMP10) took at least 24-48h, suggesting secondary effects, while expression of SASP components unaffected by ZFP36L1Mut (such as TIMP1) did not change (Fig 6c, Supplementary Fig S6c). Using this system, increased BrdU incorporation was also observed 24 h after ZFP36L1Mut induction (Supplementary Fig S6d).
Figure 6. ZFP36L1 is a direct regulator of the SASP.
a. Downregulation of the indicated genes (shown as log2 of the fold change) when comparing IMR90 ER:RAS cells expressing ZFP36L1Mut versus control IMR90 ER:RAS, as derived from the RNA-Seq data obtained from 3 independent experiments. ARE scores for the genes42 are shown. b. (left) Scheme of the experiment shown in c-d. (right) Expression of FLAG-ZFP36L1Mut was monitored by immunoblot upon treatment with doxycycline (200ng/ml) at the indicated times. TRE, Tetracycline response element. c-d. IMR90 ER:RAS were infected with TRE-FLAG-ZFP36L1Mut (or empty vector) and the reverse transactivator (rtTA-M3). Six days after 4OHT addition, expression of ZFP36L1Mut was induced with the indicated doses of doxycycline. Expression of the indicated transcripts was monitored by qRT-PCR at the indicated times. e-g. IMR90 ER:RAS expressing TRE-FLAG-ZFP36L1Mut (or empty vector) were further infected with a vector encoding the coding sequence of p21 (or empty vector). Six days after incubation with 4OHT, expression of ZFP36L1Mut was induced with 200ng/ml doxycycline for 72h (e). Expression of FLAG-ZFP36L1Mut and p21 was analysed by immunoblot. Data is representative of 3 independent experiments (f) Effect on cell proliferation was assessed by CV staining 14 days after 4OHT addition (8 days after doxycycline addition). Images are representative of 4 independent experiments. (g) Expression of the indicated SASP components was analysed by qRT-PCR (n=3 independent experiments) h. In senescent cells, mTOR signalling promotes translation of MK2 via 4EBP1. Ras activation triggers a signalling cascade that results in MK2 phosphorylation (via p38MAPK). Once phosphorylated, MK2 phosphorylates and inactivates ZFP36L1. If mTOR signalling is blocked, translation of MK2 is strongly compromised. As a consequence, the pool of phosphorylated MK2 is not sufficient to inactivate ZFP36L1. When active, ZFP36L1 binds to the 3’UTRs of many SASP components and promotes their degradation. Statistical significance was calculated using two-tailed Student’s t-test, *** P < 0.001; ** P < 0.01; *P < 0.05; n.s, non significant. Error bars represent means ± s.d. For b-d, data is representative of 2 independent experiments. Unprocessed original scans of blots are shown in Supplementary Fig. 9. For raw data, see Supplementary Table 7.
ZFP36L1 has been shown to target the CDKN1A mRNA (encoding for p21CIP1) 43. Indeed, ZFP36L1Mut induction during OIS resulted in an early and strong downregulation of CDKN1A but not other CDKIs (such as p16INK4a, Fig 6d). This could explain how ZFP36L1Mut rescues growth arrest. To determine if the effect of ZFP36L1Mut on the SASP can be uncoupled from the bypass of the senescence arrest, we expressed a CDKN1A mRNA lacking its UTRs, which is resistant to ZFP36L1Mut –mediated downregulation. Remarkably, restoration of p21CIP1 levels impaired the ability of ZFP36L1Mut to rescue the senescence phenotype (Fig 6e-f, Supplementary Fig S6e-f) but not its ability to downregulate the SASP (Fig 6g). Therefore, ZFP36L1Mut regulates the SASP as part of a direct pathway involving mTOR, 4EBP1 and MAPKAPK2 (Fig 6h).
The mTOR/ ZFP36L1 pathway controls the protumorigenic effects of the SASP
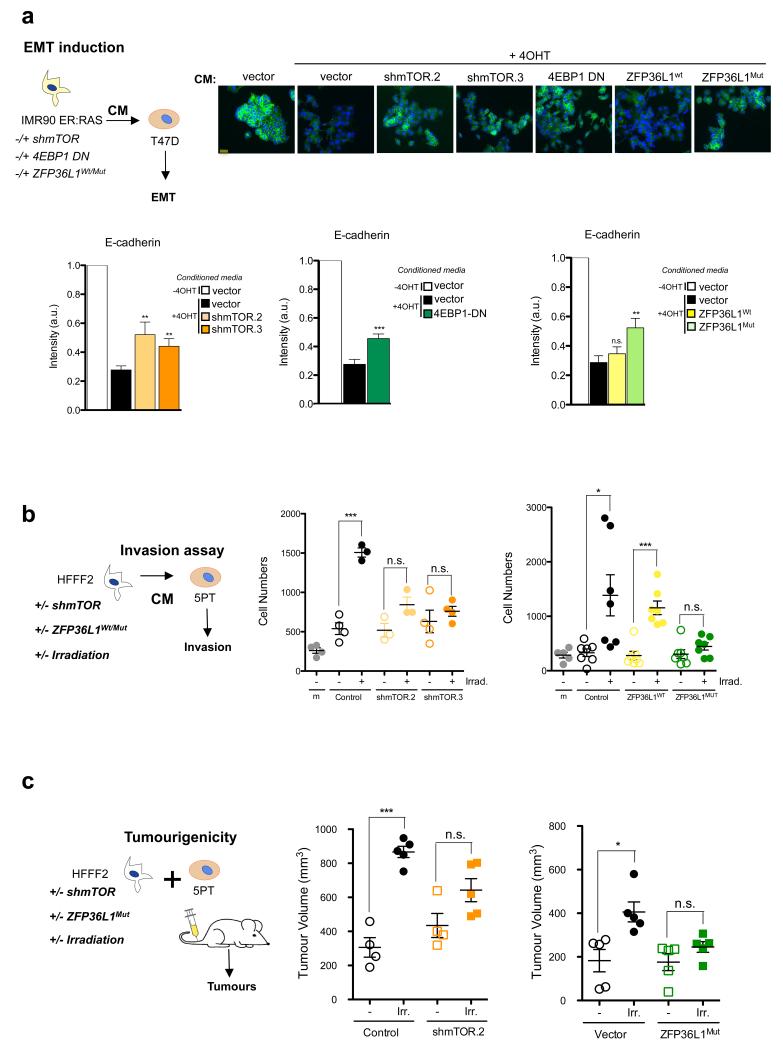
Since inhibiting mTOR only suppresses a subset of SASP components, we investigated whether this was sufficient to affect the paracrine effects of senescence, starting by its pro-tumorigenic abilities. The SASP of senescent fibroblasts can promote EMT, increasing the tumorigenic potential of epithelial cancer cells 3, 5. While the conditioned medium (CM) of senescent fibroblast induced EMT in T47D breast cancer cells, CM of IMR90 ER:RAS in which mTOR was depleted or inhibited did not (Fig 7a, Supplementary Fig S7a-b). A similar result was observed by expressing 4EBP1 DN or ZFP36L1Mut (Fig 7a). Importantly, the CM of cells expressing both ZFP36L1Mut and a non-degradable CDKN1A also failed to promote EMT (Supplementary Fig S7c). Similarly, while CM of fibroblasts that had undergone irradiation-induced senescence enhanced the invasive properties of the squamous cell carcinoma 5PT cell line, fibroblasts with depleted mTOR levels, or expressing ZFP36L1Mut were less capable of doing so (Fig 7b). In nude mice, co-injection of 5PT cancer cells with irradiated, senescent, fibroblasts resulted in increased tumour growth (Fig 7c). In contrast, co-injection with irradiated fibroblasts expressing ZFP36L1Mut or shRNAs against mTOR failed to enhance tumour growth (Fig 7c).
Figure 7. The mTOR/ZFP36L1 pathway regulates the protumorigenic effects of the SASP.
a. For all the indicated conditions in the scheme (top, left), conditioned media (CM) of IMR90 ER:RAS was collected 7 days after 4OHT induction. Breast epithelial tumour T47D cells were cultured in that CM and, 48h after, E-Cadherin expression was monitored by IF. Data are mean ± s.d. from n=4 independent experiments. Fold changes of normalised intensity values (bottom) and representative pictures are shown (top). Student’s T-test was performed to compare the indicated cells to their respective controls. *** P < 0.001; ** P<0.01; n.s., non significant. Scale bar, 40 μm. b. Both mTOR knockdown and expression of ZFP36L1Mut prevent the ability of CM from senescent cells to induce invasion of tumoral cells. HFFF2 fibroblasts were infected with the indicated plasmids and subjected to irradiation (5Gy). Conditioned media was collected 8 days after and tested for their ability to induce invasion of 5PT squamous carcinoma cells. Data are mean ± s.d. from n=4 independent experiments (shMTOR) and n=7 independent experiments (ZFP36L1Wt/Mut) . Student’s T-test was performed to compare the indicated conditions to the control. *** P < 0.001; * P<0.05; n.s., non significant. c. mTOR knockdown and expression of ZFP36L1Mut blunt the protumorigenic effect of the SASP in vivo. Irradiated (senescent) or nonirradiated (non senescent) HFFF2 fibroblasts, infected with the indicated vectors, were co-injected with 5PT squamous carcinoma cells in partially immunocompromised Rag1−/− mice. Tumour volume was measured after 5-6 weeks. Student’s T-test was performed to compare mice injected with irradiated versus nonirradiated fibroblasts for every single condition. n=5 mice per group. Data presented is mean ± s.d. *** P < 0.001; * P<0.05; n.s., non significant. For raw data, see Supplementary Table 7.
mTOR inhibition also blunts the tumour suppressive effects of the SASP
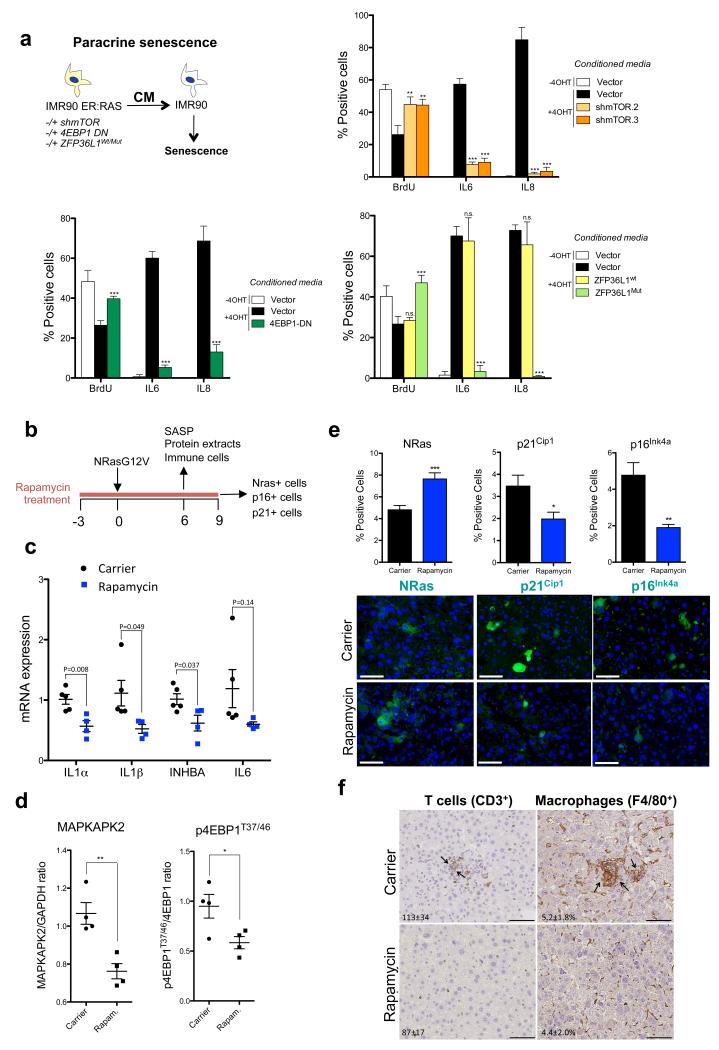
Next we analysed how mTOR inhibition affects the tumour suppressive properties of the SASP. We previously showed that CM of senescent cells can induce ‘paracrine senescence’ in normal cells 6. In contrast, CM of IMR90 ER:RAS cells with depleted mTOR or overexpressing 4EBP1 DN or ZFP36L1Mut (in the presence or absence of CDKN1A) failed to induce paracrine senescence (Fig 8a, Supplementary Fig S8a). mTOR knockdown also diminished the ability of IMR90 ER:RAS cells to induce senescence of normal IMR90 cells when co-cultured (Supplementary Fig S8b).
Figure 8. mTOR inhibition blunts the tumour suppressive effects of the SASP.
a. (top left) Diagram summarizing the paracrine senescence/conditioned media experiments. CM of the indicated cells was collected 7 days after incubation with 4OHT. The effect of CM on BrdU incorporation and SASP induction in IMR90 wt cells was evaluated by IF (n=4 independent experiments). (top right) IMR90 ER:RAS shmTOR (bottom left) IMR90 ER:RAS cells expressing 4EBP1-DN (bottom right) IMR90 ER:RAS cells expressing ZFP36L1wt/ZFP36L1Mut. b-f. Treatment with rapamycin reduces the SASP, senescence and immune clearance in vivo. (b) NrasG12V transposons and a transposase were co-delivered into mouse livers through hydrodynamic injection (Day 0). Mice were treated with carrier or drugs from day −3 (3 days before NrasG12V injection) and sacrificed 6 or 9 days post-injection depending on the experiment. (c) Quantification of the expression of SASP components by qRT-PCR in livers of the indicated mice 6 days after NrasG12V injection. Carrier, n=5 mice; rapamycin, n=4 mice. P values are included. (d) MAPKAPK2 expression and 4EBP1 phosphorylation were analysed by immunoblot (see Supplementary Fig. S8d) and further quantified. MAPKAPK2 levels were normalised to GAPDH expression and 4EBP1 phosphorylation (p4EBP1T37/T46) was referred to normalised 4EBP1. Mice livers were obtained 6 days after NrasG12V induction. (n=4 mice per condition). (e) Quantification of Nras+ (left), p21Cip1+ (centre) and p16Ink4a+ (right) cells on liver sections 9 days after NrasG12V injection (n=5 mice per condition). Representative images of the sections quantified in the top panel are shown in the bottom. Scale bar, 40μm (f) Liver sections were collected 6 days after NrasG12V injection and stained for the CD3 (T cell marker) and F4/80 (macrophage marker). (Carrier n=5 mice, rapamycin n=4 mice; 200x). Arrows indicate clusters of infiltrating immune cells, which suggest clearance of senescence hepatocytes. Quantification of positive cells/mm2 (CD3+ T cells) or % of positive area (F4/80+ macrophages) is included in the images. Scale bar, 50 μm. All statistical significances were calculated using two-tailed Student’s t-test, *** P < 0.001; ** P < 0.01; *P < 0.05; n.s, non significant. All error bars represent means ± s.d. For raw data, see Supplementary Table 7.
Finally, we took advantage of a mouse model in which OIS is triggered in hepatocytes via transposon-mediated transfer of oncogenic Nras (NrasG12V). NrasG12V expression induces senescence and the SASP triggers an immune response that eliminates preneoplastic hepatocytes 44. To investigate whether rapamycin affects the SASP, senescence and immune surveillance, we pre-treated mice with rapamycin for three days before injecting the NrasG12V transposon and treated with rapamycin for nine more days (Fig 8b). Treatment with rapamycin impaired SASP induction (Fig 8c, Supplementary Fig S8c) and correlated with reduced MAPKAPK2 levels (Fig 8d, Supplementary Fig S8d). Nine days after NrasG12V expression, there was a higher percentage of Nras-positive cells but less p21Cip1 and p16Ink4a positive hepatocytes in rapamycin-treated mice (Fig 8e). Since treatment with rapamycin did not affect the number of Nras-positive cells shortly after NrasG12V injection (Supplementary Fig S8e), the most plausible explanation for the differences in Nras+ hepatocytes is that the impaired SASP resulted in decreased immune surveillance and elimination. Consistent with this, we detected reduced infiltration of T cells, B cells, NK cells and macrophages in the livers of mice treated with rapamycin (Fig 8f and Supplementary Fig S8f). To discard that the observed differences were due to rapamycin pre-treatment inhibiting entry into senescence, we treated mice with rapamycin only after senescence was triggered and obtained similar results (Supplementary Fig S8g). Although we cannot discard a direct effect of rapamycin on the immune cells, the above results suggest that mTOR inhibition can control senescence and the SASP during cancer initiation in vivo.
DISCUSSION
The inhibition of mTOR suppresses the SASP by specifically downregulating MAPKAPK2 translation. Since in OIS MAPKAPK2 phosphorylates and inhibits ZFP36L1, an mRNA binding protein involved in ARE-mediated decay that targets SASP components, it follows that mTOR inhibition results in reduced mRNA levels and not only decreased translation of the SASP. Bioinformatic predictions and our experimental results suggest that several SASP components are directly targeted by ZFP36L1. The question remains as to whether SASP components degraded with delayed kinetics are less efficient ZFP36L1Mut targets or a hierarchical model could be proposed in which the downregulation of a few specific SASP components (such as IL8 or IL1β) results in decreased levels of others due to the disruption of autocrine feedback loops. Our results identify an mTOR-dependent pathway required to control the SASP. Whether a similar mechanism operates in non-senescent cells with elevated mTOR signalling, remains to be investigated.
In contrast with mTOR inhibition, a constitutively active ZFP36L1Mut not only prevented SASP induction but also rescued growth arrest. The explanation for their different effects on cell growth is two-fold. mTOR inhibition causes an arrest, partially explained by downregulation of Cyclin D3 31 (Supplementary Fig S2d). In contrast, ZFP36L1Mut can target p21Cip1 for degradation (Fig 6d). Despite these differences, mTOR and ZFP36L1 converge to regulate the SASP as demonstrated by the impaired ability of rapamycin to suppress the SASP in cells lacking ZFP36L1 (Fig 5j).
As most SASP factors used as readouts in our screen are NF-κB targets, it could be argued that our screen was biased to identify regulators of the ‘pro-inflammatory’ SASP. However, we found that a subset of both NF-κB-dependent and -independent SASP components are regulated upon mTOR inhibition or ZFP36L1Mut expression. Importantly, regulating this subset of the SASP is sufficient to impair both its tumour suppressive and tumour promoting functions. Late in cancer or during ageing 45, the pro-inflammatory SASP can be harmful. Therefore, inhibiting mTOR may suppress protumorigenic effects or help to maintain tissue homeostasis by alleviating the chronic, low-grade inflammation observed in aged tissues 45. However the inhibition of mTOR can also compromise senescence and its associated immune surveillance, being potentially detrimental during the early stages of cancer.
How individual SASP components contribute to all these processes and whether the MAPKAPK2/ZFP36L1 pathway described here is involved in the outcome of mTOR inhibition in vivo, requires further investigation. Nevertheless, our findings provide evidence of a critical integration between mTOR and senescence. This may partly explain the reported beneficial effects of rapamycin in cancer and age-related diseases, and may also yield a potential avenue to improve the treatment and/or prevention of these pathologies.
METHODS
Cell culture, retroviral and lentiviral infection
HEK-293T, HEK-293 Gag Pol, IMR-90, BJ, HFFF2, 5PT and T47D cells were obtained from the ATCC and tested for mycoplasma contamination regularly. No cell lines used in this paper are listed in the database of commonly misidentified cell lines maintained by ICLAC and NCBI Biosample. The cell lines have not been authenticated. Cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% foetal bovine serum (Sigma), 1% antibiotic-antimycotic solution (Invitrogen). 5PT cells were cultured in keratinocyte growth medium or KGM (α-MEM medium supplemented with 2,2 g/L Sodium bicarbonate, 10 ng/ml EGF, 5 μg/ml Insulin, 0.4 μg/ml Hydrocortisone, 10% FBS, 1% L-Glutamine and 1% Adenine). Cell number and viability measurements were determined using the Guava Viacount reagent (Millipore) and the Guava Cytometer (Millipore). Retroviral and lentiviral infection were performed as previously described 46, 47.
Chemical compounds and treatments
Chemical compounds and concentrations used in the drug screen are summarized in Supplementary Table S4. Unless stated otherwise, 10nM rapamycin (Tocris), 25nM Torin1 (Tocris), 0.5nM NVPBEZ-235 (LC laboratories), and 100nM MK2 inhibitor III (Calbiochem) were added simultaneously with 125nM 4-hydroxytamoxifen (4OHT) on the day of induction or just after irradiation. In the replicative senescence experiments, cells were treated for 1 week before analysis. In all cases, drugs were re-added every 48h.
Plasmids
pLNC-ER:RAS has been described previously 7,. MSCViresGFP-4EBP1-4A (4EBP1-DN) was obtained from Hans-Guido Wendel (MSKCC, NY), and subsequently subcloned in MSCVpuro retroviral vector. pBabepuro ZFP36L1wt, Flag-ZFP36L1wt, ZFP36L1Mut, Flag-ZFP36L1Mut, Flag-ZFP36L2wt, Flag-ZFP36L2mut, Flag-TTPwt, and Flag-TTPmut were synthesised by Genscript (Piscataway, NJ, USA). To generate a doxycycline-inducible Flag-ZFP36L1Mut transgene, we PCR-amplified it from pBabepuro Flag-ZFP36L1Mut in order to include BstBI and XbaI restriction sites (Fw: 5-CGTTCGAAGCCATGGACTACAAGGACG-3′, Rv: 5-GCTCTAGAGGCTTAGTCATCTGAGATGGAAAGTC-3′) and we then subcloned it into a modified pLenti CMVtight eGFP Puro (w771-1) obtained from Addgene. pGIPZ-based shRNA vectors targeting mTOR were from SIGMA. p21 coding sequence was PCR-amplified using MSCVpuro p21 CDS as a template (Fw: CGGGATCCGCCATGTCAGAACCGGCT, Rv: GCGTCGACGGCTTAGGGCTTCCTCTTG) and subcloned into a pWZLBlast backbone. For de novo generation of ZFP36L1 and MAPKAPK2 miR-E shRNAs, 97-mer oligonucleotides coding for the respective shRNAs 48 were PCR amplified using the primers miRE-Xho-fw and miRE-EcoOligo-rev and cloned into pRRL lentiviral vector. Used RNAi sequences are listed in Supplementary Table S5b.
BrdU incorporation, crystal violet staining and senescence-associated β-galactosidase staining
These methods have been previously described elsewhere 6, 46, 47, 49.
Conditioned medium (CM)
The indicated cells (2.5 × 106) were seeded in a 10cm dish and incubated for 7 days with 125 nM 4OHT in DMEM with 0.5% FBS. The CM was then collected, centrifuged at 4,000 g and filtered through a 0.2 μm pore filter. The resulting CM was mixed with DMEM 40% FBS in a proportion of 3 to 1 to generate CM containing 10% FBS. When CM was collected from cells treated with chemical compounds, the cells were incubated with the drugs until day 4, and at this point, the cells were thoroughly washed with PBS and the media was replaced. The cells were kept drug-free for the next 3 days until CM collection.
Gene expression analysis
Total RNA was extracted using Trizol reagent (Invitrogen) and the RNeasy isolation kit (Qiagen). DNAs were generated using SuperScript II reverse transcriptase (Invitrogen), dNTPs and Random Hexamers. PCR reactions were performed in a Real-Time PCR Detection System (BioRad) using Power SYBR Green Master Mix or TaqMan Universal PCR Master Mix (Applied Biosystems). Expression was normalized to ribosomal protein S14 (RPS14) expression in human cell extracts and to 18S ribosomal RNA or GAPDH in mouse tissues. Primer sets and TaqMan probes used are listed in Supplementary Table S5a. Mice tissue samples were weighed and ground to a fine powder in liquid nitrogen. The RNA was then extracted and processed as described above.
RNA-Seq
RNA-Seq libraries were prepared from 500 ng of total RNA using the Illumina Truseq mRNA stranded library prep kit (Illumina Inc. San Diego, USA) according to the manufacturer’s protocol. Library quality was checked on a Bioanalyser HS DNA chip and concentrations were estimated by Qubit measurement. Libraries were pooled in equimolar quantities and sequenced on a Hiseq2500 using paired end 100 bp reads. At least 40 million reads passing filter were achieved per sample, and they were processed using standard procedures.
Invasion assays
Transwell invasion assays were carried out using 8 μm pore size polycarbonate Transwell filters (Costar) coated with Matrigel diluted 1:2 in α-MEM. The cells that migrated to the lower chamber were trypsinized and counted on a Casy counter (Sharfe System GmbH, Germany) 3 days after plating onto the upper chamber.
Immunofluorescence and high content analysis
IF was performed as previously described 50 using the antibodies and dilutions listed in Supplementary Table S6. Image acquisition was performed using an automated high throughput microscope (IN Cell Analyzer 2000, GE Healthcare). A minimum of 1,000 cells was acquired for each sample per duplicate. High content analysis (HCA) was performed using the IN Cell Investigator software (v 3.2; GE Healthcare), as described elsewhere 6, 47, 50, 51. Briefly, DAPI staining of the nuclei was used to identify cells. The nuclei were segmented using top-hat segmentation, specifying a minimum nucleus area of 100 μm2. To define the cell area, a collar segmentation approach was used with a border of 1 μm around DAPI staining or alternatively, multiscale top-hat was used to detect cytoplasmic intensity for a given staining. Each cell was assigned a nuclear intensity value (and cell intensity value when applicable) for the specific protein being studied. A histogram of the intensity values of all the cells in a sample was produced and this was used to set a threshold filter to determine positive and negative expressing cells. Alternatively, normalized intensity values for a given staining were obtained by subtracting the intensity of the secondary antibody alone (background intensity) to raw intensity values. Normalized intensity values were used to calculate fold changes among different conditions. Error bars and P values were derived from independent biological replicates. The antibodies used for the analysis were validated with robust controls (shRNAs, overexpression or drug inhibition) to assess their specificity. Statistical analysis of IF data was conducted using GraphPad software Prism® (version 6.0b).
De novo protein synthesis experiments
Global protein synthesis was detected by using the Click-iT 2-amino-6-hydroxylaminopurine (AHA) Alexa Fluor 488 Protein Synthesis HCS Assay (Invitrogen) according to the manufacturer’s protocol. Briefly, IRM90 ER:RAS cells were grown in absence or presence of 4OHT for 7 days and then pre-treated for 2h with the indicated drugs (the last 30 min in L-methionine free media in order to deplete methionine reserves). At that point 100 μM Click-iT-labelled AHA was added to the media. 30 mins later cells were washed and fixed. Following incubation with Click-iT-reactive Alexa Fluor 488 dye and DAPI staining, images were acquired and analyzed as previously described for immunofluorescence and high content analysis. The represented intensity values were obtained by subtracting the intensity value of cells that were not grown in presence of AHA (but were incubated with Click-iT-reactive Alexa Fluor 488 dye) from raw intensity values. To analyse nascent synthesis of specific proteins, we also used Click-iT AHA. (Invitrogen) IMR90 ER:RAS cells were grown with 4OHT for 7 days and then pre-treated for 2h with 250 nM Torin1, 10 μg/ml CHX or DMSO (the last 30 min in L-methionine free media in order to deplete methionine reserves). At that point 50 μM AHA was added to media and cells were kept in culture for 4h and then immediately lysed in 1% SDS, 50m M Tris-HCl buffer. After quantification, 600 μg of protein per condition were processed using the Click-iT Protein Analysis Detection Kit (Biotin Alkyne) according to manufacturer’s protocol. Resulting samples, in which AHA has been biotinylated, were loaded into Sephadex SpinTrap G-25 columns (GE Helathcare) in order to get rid of Free Biotin. Afterwards, samples were diluted to 0.5% SDS and further subjected to immunoprecipitation with Streptavidin beads in order to purify biotinylated (de novo synthesized) protein. Samples were eluted with Laemli Buffer and subjected to Western Blotting. As a control we used extracts from untreated cells, which incorporated AHA, but were not incubated with Biotin Alkyne.
Irradiation
To induce SASP induction by ionizing radiation, the indicated cells were γ-irradiated (5Gy) and analyzed at the indicated times.
Immunoblot
Protein extracts from cell lines were processed and analyzed as previously described 47. Tissues were removed and homogenized in lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% w/v Triton X-100) supplemented with complete protease inhibitor cocktail and phosphatase inhibitors (1 mM sodium orthovadanate, 5 mM sodium fluoride and 2 mM β-glycerophosphate). The antibodies and dilutions used are listed in Supplementary Table S6. p-ZFP36L1S203 antibody has been previously described 41.Immunoblots were quantified using ImageJ.
Analysis of mRNA distribution in polysomes
IMR90 ER: RAS fibroblasts (106) were seeded in 10 cm dishes (16 plates) and incubated for 6 days with 125 nM 4OHT. Cells were then treated with vehicle (8 plates) or 250 nM Torin1 (8 plates) for 2h. 10 min before lysis, cells were treated with 100 μg/ml cyclohexamide, washed in ice-cold PBS plus 100 μg/ml cyclohexamide at 4°C and then lysed in polysome lysis buffer (0.3 M NaCl, 15 mM MgCl2, 15 mM Tris-HCl (pH 7.6), 1% Triton X-100, 0.1 U/μL RNAsin (Promega), 100 μg/ml cyclohexamide, 1 μg/mL heparin,) at 4°C. Cells were centrifuged at 10,000 rpm for 10 min at 4°C, and the resulting supernatant was layered onto a 5 mL continuous sucrose gradient (15-50% sucrose in 0.3M NaCl, 15 mM MgCl2, 15 mM Tris-HCl (pH 8), 100 μg/ml cyclohexamide, 1 μg/mL heparin, 0.1 U/μL RNAsin). Gradients were centrifuged at 39,000 rpm for 2 hr in an SW41-Ti rotor at 4°C, and then sampled using a Teledyne ISCO density gradient fractionation system with continuous monitoring at 254 nm. 0.5 mL fractions were collected (20 fractions per sample) and extracted in 3 volumes of Trizol LS reagent (Invitrogen). The resulting 20 fractions (per sample) were pooled into 10 sequential fractions and the RNA extracted with the RNeasy isolation kit (Qiagen). The cDNAs were prepared and PCR reactions were performed as mentioned above. mRNA amounts in each fraction were plotted as a percentage of the total mRNA in the sample.
Secretome characterization
The indicated cells were seeded in two 10 cm dish per condition (4 × 106 cells per plate) with vehicle or 125 nM 4OHT in phenol red free DMEM with no FBS. After 6 days, CM was collected, centrifuge at 4000g and filter through a 0.2um pore filter. The resulting media CM was then concentrated about 100-200 times by ultrafiltration using the vivaspin 20 5,000 MWCO by centrifμgation at 5000g. At this stage, the protein concentration was determined by Pierce 660 nm Protein Assay Reagent (Pierce 1861426). 100 μg of protein for each sample were reduced and alkylated with diothiothreitol and iodoacetamide respectively, then digested overnight with immobilized trypsin at 37°C. Reverse-phase solid phase extraction (RP-SPE) was used to desalt the sample and tryptic peptides were eluted from the column using 50% acetonitrile containing 0.1% formic acid. Peptides were lyophilized in a speed vacuum centrifuge then solubilized in 0.1% trifluoroacetic acid prior to automated loading onto an Acclaim PepMap 100 trap column (100 μm × 2 cm) using an Ultimate 300 RSLC instrument. Peptides were separated on an Acclaim PepMap RSLC analytical column (75 μm × 25 cm) and eluted directly into the nanoelectrospray source of an LTQ-Orbitrap Velosmass spectrometer (ThermoFisher).. Separation was performed using a 120-minute gradient from 5-45% mobile phase B (80% acetonitrile + 0.1% formic acid; mobile phase A 98% water, 2% acetonitrile, 0.1% formic acid). The mass spectrometer was operated in data dependent acquisition mode for top 10 CID acquisitions.
Phospoproteomic analysis
Phosphoproteomic was performed as described previously 52 with some modifications 53. Three 10cm dishes of IMR90 ER:RAS cells were seeded (2 × 106 per plate) per condition. 7 days after 4OHT induction, cells were lysed in urea lysis buffer (8 M Urea, 20 mM HEPES) containing phosphatase inhibitors and, after diluting to <2M Urea, 400 μg proteins were digested using immobilized trypsin overnight at 37°C. The resulting peptides were desalted by RP-SPE in 1cc Oasis cartridges (Waters Corp.) using manufacturer’s instructions with the exception that peptide elution was with 1 mL of 2M glycolic acid, 50% acetonitrile, 5% trifluroracetic acid. Eluted peptides were spiked with 50 μL TiO2 (50% slurry) that was then collected by centrifugation and washed with washing solution (2 M glycolic acid, 50% acetonitrile, 5% trifluoroacetic acid) three times. Phosphopeptides were eluted from TiO2 using ammonium hydroxide and dried in a speed vacuum centrifuge until the time of analysis. Samples were solubilised and separated by nano-LC as per the proteomic analyses, but a top 10 MSA (multi-stage activation) method was utilized for increased coverage for neutral loss phosphoproteomic data acquisition.
Analysis of the proteomics and phosphoproteomics data
Raw files were converted to peak lists (in the Mascot Generic Format) using Mascot Distiller (v2.3.0) and searched using Mascot server (v2.3.01) against the SwissProt Uniprot database (2012_03_10) restricted to the relevant taxonomy. Mass windows for tolerance for MS scans were 10 ppm and 600 mmu for MS/MS. Fixed modification of carbamidomethylation of cysteine and variable modifications of oxidation of methionine and glutamine to pyro-glutamate conversion were permitted. For phosphoproteomic data additional variable modifications of phospho(S/T/Y) were included. Mascot result files were parsed using a Perl script written in house that uses the mascot parser files provided by Matrix Science. Threshold for accepting peptides as being positively identified was set at an expectancy score of 0.05. Pescal was then used to automate the construction of XIC of all peptides identified in an experiment across all samples being compared as described previously 25, 54. Data were normalised by dividing each peptide intensity by the sum of all peptide intensities within a sample. Fold changes were calculated by averaging the normalized intensities of peptides within a sample group and dividing those by the control group. Fold changes were then log transformed prior to calculation of significance using an unpaired t-test. The resulting p-values were corrected for multiple testing using the Benjamini-Hochberg procedure.
Kinase substrate enrichment analysis
KSEA was performed as described before 33. Briefly, an script written in VBA was used to link identified phosphorylation sites to those listed in the phosphoSite database 55 and to calculate values of enrichment by subtracting means of all fold changes to the means of those belonging to predefined kinase-substrate groups. Significance of enrichment was calculated using a Z test.
Heatmaps and ARE score
Secreted proteins were identified as previously described 6, and only those secreted factors increased in RAS expressing cells were further analysed. Proteins measurements were then mean normalised using ‘scale’ function in R and visualised as heat maps using Multi-Experiment viewer (MeV). The significance of observed ARE-score was computed using permutation tests. The observed mean of ARE-score is compared to the distribution of mean values generated by permutation (number of permutations = 100,000). The p-values are estimated as the relative frequency of simulated means greater than the observed mean ARE score of the group. Significance of overlap between groups was computed by hypergeometric test.
In vivo hepatocyte senescence experiments
5 mg/ml Rapamycin stock solution in 100% ethanol was prepared. This stock solution was first diluted in sterile 10% PEG400/8%ethanol and in an equal volume of sterile 10% Tween 80 for a final concentration of 10mg/100ml. Rapamycin (1 mg/kg) was delivered once every 3 days by oral gavage. Treatment started at day −3 or +3 and continued until the animals were sacrified. At day 0 hydrodynamic tail vein injection of a transposon-based Nras expression plasmid together with an expression plasmid for the sleeping beauty 13 transposase 44 was performed. At day 3, 6, 9 or 12 (depending on the experiment) the animals were sacrificed and livers collected. Samples were fixed and subjected to IHC/IF analysis In brief, after deparaffinization of paraffin slides, slides were blocked, incubated with the primary antibody at 8°C over night, washed, incubated with the appropriate secondary antibody for one hour at room temperature, washed, mounted and sealed. Five high power fields were counted on two liver sections from each mouse liver (200X, >200 counted cells per field). No statistical method was used to predetermine sample size. For all animal experiments, only mice in a C57BL/6 background were used (equal gender distribution in randomized groups). 16-20 week old mice were used. For the experiments in which mice were treated with carrier or rapamycin previous to hydrodynamic tail vein injection, 14 carrier-treated mice and 13 rapamycin-treated mice were used. For the experiment in which mice were treated with carrier or rapamycin 3 days after hydrodynamic tail vein injection, 4 mice per group (carrier or rapamycin) were used. The Investigators were not blinded to allocation during experiments and outcome assessment. All mice were maintained under pathogen-free conditions in accordance with the institutional guidelines of the Helmholtz Centre for Infection Research and Hannover Medical School. Experiments have been approved by the German legal authorities.
Samples from rapamycin treated old mice
Mice used in this project were previously described 56. Briefly, all mice were UM-HET3, bred as the progeny of CByB6F1 mothers and C3D2F1 fathers. Encapsulated rapamycin was administered from 9 months of age to mice at doses of 14 parts per million embedded in the food (calculated on the basis of the equivalent mass of non encapsulated rapamycin). The 14-ppm dose has previously been shown to extend lifespan when started at 9 or at 20 months of age 21, 57. Each donor used in this study was 22 months of age when sacrificed. No statistical method was used to predetermine sample size. Only female mice livers were analysed in this report. Mice were equally randomized into different treatment groups. The Investigators were not blinded to allocation during experiments and outcome assessment. These experiments were performed following the guidelines of The National Institute on Aging Interventions Testing Program and the University of Michigan.
Immunohistochemistry
Sections (2μm) of livers (fixed in 4% paraformaldehyde and paraffin-embedded) were stained with the indicated antibodies. Incubation in Ventana buffer and staining was performed on a NEXES immunohistochemistry robot (Ventana Instruments) using an IVIEW DAB Detection Kit (Ventana) or on a Bond MAX (Leica). For quantification of stainings, slides were scanned using a SCN400 slide scanner (Leica) and analyzed using Tissue IA image analysis software (Slidepath, Leica).
Nude mice experiments
106 5PT cells ± 3×106 of the indicated HFFF2 cells were re-suspended in 150 μl of supplement free DMEM; 100 μl of this mix was injected subcutaneously in the flank of partially immunocompromised Rag1−/− mice. Each mice was injected in both flanks. 16-20 weeks old female mice were used for these experiments. 10 mice were used for the shmTOR experiment (Fig 7c, centre) and 10 mice were used for theZFP36L1 experiment (Fig 7c, right). Tumour size was measured using an electronic calliper and calculated using the formula 4π/3 × r3 (radius (r) is calculated from the average diameter, measured as the tumour width and length) 58. Following 5-6 weeks of tumour growth the animals were culled (prior to reaching ethical limits). Animal experiments were performed with institutional review and approval. Animals were only excluded from the analysis due to technical issues with the injection. No statistical method was used to predetermine sample size. Mice were randomized to receive the different injections. The Investigators were not blinded to allocation during experiments and outcome assessment. These experiments were approved and performed in accordance with the institutional guidelines of the University of Southampton (UK).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to T. Rodríguez, A. Nebreda, R. Agami, M. Gaestel, D. Engelberg, G. Wendel, J Campisi, C. Moroni, C. Speck, A. Day and G. Peters for reagents, helpful comments and contributions to this project. R. Miller is funded by NIH Grant U01-AG022303. Core support from MRC funded the research in J. Gil’s laboratory. N Herranz was funded by an EMBO fellowship.
Footnotes
Accession numbers. RNASeq data have been deposited at the Gene Expression Omnibus under the accession number GSE61130.
REFERENCES
- 1.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Salama R, Sadaie M, Hoare M, Narita M. Cellular senescence and its effector programs. Genes Dev. 2014;28:99–114. doi: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 5.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Acosta JC, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol. 2013;15:978–990. doi: 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Acosta JC, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 8.Hoenicke L, Zender L. Immune surveillance of senescent cells--biological significance in cancer- and non-cancer pathologies. Carcinogenesis. 2012;33:1123–1126. doi: 10.1093/carcin/bgs124. [DOI] [PubMed] [Google Scholar]
- 9.Kuilman T, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 10.Jing H, et al. Opposing roles of NF-{kappa}B in anti-cancer treatment outcome unveiled by cross-species investigations. Genes & development. 2011;25:2137–2146. doi: 10.1101/gad.17620611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien Y, et al. Control of the senescence-associated secretory phenotype by NF-{kappa}B promotes senescence and enhances chemosensitivity. Genes & development. 2011;25:2125–2136. doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freund A, Patil CK, Campisi J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. The EMBO journal. 2011;30:1536–1548. doi: 10.1038/emboj.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 14.Rodier F, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narita M, et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011;332:966–970. doi: 10.1126/science.1205407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 18.Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci. 2006;31:342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 20.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iglesias-Bartolome R, et al. mTOR inhibition prevents epithelial stem cell senescence and protects from radiation-induced mucositis. Cell Stem Cell. 2012;11:401–414. doi: 10.1016/j.stem.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorr JR, et al. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature. 2013;501:421–425. doi: 10.1038/nature12437. [DOI] [PubMed] [Google Scholar]
- 24.Acosta JC, Snijders AP, Gil J. Unbiased characterization of the senescence-associated secretome using SILAC-based quantitative proteomics. Methods Mol Biol. 2013;965:175–184. doi: 10.1007/978-1-62703-239-1_11. [DOI] [PubMed] [Google Scholar]
- 25.Cutillas PR, Vanhaesebroeck B. Quantitative profile of five murine core proteomes using label-free functional proteomics. Mol Cell Proteomics. 2007;6:1560–1573. doi: 10.1074/mcp.M700037-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tchkonia T, Zhu Y, van Deursen J, Campisi J, Kirkland JL. Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. J Clin Invest. 2013;123:966–972. doi: 10.1172/JCI64098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pospelova TV, et al. Suppression of replicative senescence by rapamycin in rodent embryonic cells. Cell Cycle. 2012;11:2402–2407. doi: 10.4161/cc.20882. [DOI] [PubMed] [Google Scholar]
- 29.Demidenko ZN, et al. Rapamycin decelerates cellular senescence. Cell Cycle. 2009;8:1888–1895. doi: 10.4161/cc.8.12.8606. [DOI] [PubMed] [Google Scholar]
- 30.Bundscherer A, Vogt T, Kohl G, Landthaler M, Hafner C. Antiproliferative effects of rapamycin and celecoxib in angiosarcoma cell lines. Anticancer research. 2010;30:4017–4023. [PubMed] [Google Scholar]
- 31.Dowling RJ, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci U S A. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casado P, et al. Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci Signal. 2013;6:rs6. doi: 10.1126/scisignal.2003573. [DOI] [PubMed] [Google Scholar]
- 34.Gaestel M. MAPKAP kinases - MKs - two’s company, three’s a crowd. Nat Rev Mol Cell Biol. 2006;7:120–130. doi: 10.1038/nrm1834. [DOI] [PubMed] [Google Scholar]
- 35.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young AR, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross CR, Brennan-Laun SE, Wilson GM. Tristetraprolin: roles in cancer and senescence. Ageing Res Rev. 2012;11:473–484. doi: 10.1016/j.arr.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciais D, et al. Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene. 2004;23:8673–8680. doi: 10.1038/sj.onc.1207939. [DOI] [PubMed] [Google Scholar]
- 39.Maitra S, et al. The AU-rich element mRNA decay-promoting activity of BRF1 is regulated by mitogen-activated protein kinase-activated protein kinase 2. RNA. 2008;14:950–959. doi: 10.1261/rna.983708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciais D, Cherradi N, Feige JJ. Multiple functions of tristetraprolin/TIS11 RNA-binding proteins in the regulation of mRNA biogenesis and degradation. Cellular and molecular life sciences : CMLS. 2013;70:2031–2044. doi: 10.1007/s00018-012-1150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benjamin D, Schmidlin M, Min L, Gross B, Moroni C. BRF1 protein turnover and mRNA decay activity are regulated by protein kinase B at the same phosphorylation sites. Mol Cell Biol. 2006;26:9497–9507. doi: 10.1128/MCB.01099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spasic M, et al. Genome-wide assessment of AU-rich elements by the AREScore algorithm. PLoS Genet. 2012;8:e1002433. doi: 10.1371/journal.pgen.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan FE, Elowitz MB. Brf1 posttranscriptionally regulates pluripotency and differentiation responses downstream of Erk MAP kinase. Proc Natl Acad Sci U S A. 2014;111:E1740–1748. doi: 10.1073/pnas.1320873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang TW, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011 doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 45.Franceschi C, et al. Inflammaging and anti-inflammaging: a systemic perspective on aging and longevity emerged from studies in humans. Mech Ageing Dev. 2007;128:92–105. doi: 10.1016/j.mad.2006.11.016. [DOI] [PubMed] [Google Scholar]
METHODS REFERENCES
- 46.Banito A, Gil J. Induced pluripotent stem cells and senescence: learning the biology to improve the technology. EMBO Rep. 2010;11:353–359. doi: 10.1038/embor.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barradas M, Gil J, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 49.Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc. 2009;4:1798–1806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 50.Banito A, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bishop CL, et al. Primary cilium-dependent and -independent Hedgehog signaling inhibits p16(INK4A) Mol Cell. 2010;40:533–547. doi: 10.1016/j.molcel.2010.10.027. [DOI] [PubMed] [Google Scholar]
- 52.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Montoya A, Beltran L, Casado P, Rodriguez-Prados JC, Cutillas PR. Characterization of a TiO(2) enrichment method for label-free quantitative phosphoproteomics. Methods. 2011;54:370–378. doi: 10.1016/j.ymeth.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alcolea MP, Casado P, Rodriguez-Prados JC, Vanhaesebroeck B, Cutillas PR. Phosphoproteomic analysis of leukemia cells under basal and drug-treated conditions identifies markers of kinase pathway activation and mechanisms of resistance. Mol Cell Proteomics. 2012;11:453–466. doi: 10.1074/mcp.M112.017483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hornbeck PV, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkinson JE, et al. Rapamycin slows aging in mice. Aging Cell. 2012;11:675–682. doi: 10.1111/j.1474-9726.2012.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller RA, et al. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2011;66:191–201. doi: 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Orimo A, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 59.Chrestensen CA, et al. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J Biol Chem. 2004;279:10176–10184. doi: 10.1074/jbc.M310486200. [DOI] [PubMed] [Google Scholar]
- 60.Fridman AL, Tainsky MA. Critical pathways in cellular senescence and immortalization revealed by gene expression profiling. Oncogene. 2008;27:5975–5987. doi: 10.1038/onc.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.