Abstract
Signal transducer and activator of transcription 3 (STAT3) is one of the key players in liver cancer. Increased levels of phosphorylated STAT3 (p-STAT3) have been detected in many cancers including hepatocellular carcinoma (HCC), and are usually associated with a more aggressive phenotype and poor prognosis. In addition to aberrant activation of STAT3, upregulation of total STAT3 was also detected in HCC, for which the underlying mechanisms and significance remain to be fully elucidated. Here we report that a reciprocal regulation exists between miR-197 and the IL-6/STAT3 inflammatory signaling pathway in HCC. We found that IL-6 stimulation increased total STAT3 expression at protein level but not mRNA level in HCC cells, suggesting the existence of post-transcriptional regulation of STAT3. Our study showed that IL-6/STAT3 pathway decreases expression of miR-197 in HCC, which amplifies IL-6/STAT3 pathway and contributes to HCC progression. miR-197 can significantly inhibit HCC growth both in vitro and in vivo. In addition, IL-6/STAT3-induced downregulation of miR-197 in HCC may be via affecting Drosha binding to primary miR-197 (pri-miR-197) and thus reducing mature miR-197 generation. Our study suggests that miR-197 may serve as a potential therapeutic target for interfering with the IL-6/STAT3 inflammatory pathway in HCC.
Keywords: inflammatory cytokine, miRNA, progression, signaling pathway, tumor
Abbreviations
- AFP
α-fetoprotein
- FBS
fetal bovine serum
- HCC
hepatocellular carcinoma
- IL6
interleukin 6
- miR-197
microRNA-197
- p-STAT3
phosphorlated STAT3
- pre-miR-197
precursor miR-197
- pri-miR-197
primary miR-197
- STAT3
signal transducer and activator of transcription 3
- UTR
untranslated region.
Introduction
HCC is one of the most common human malignant tumors in the world and the second leading cause of cancer-related death in China.1 Although clinical treatments have been developed to manage HCC, uncontrolled metastasis and high recurrence always lead to poor prognosis of HCC patients. Therefore, it is urgent to explore the molecular mechanisms underlying tumorigenesis and progression of HCC. Dysregulation of coding and non-coding gene expression was considered to be the main cause of HCC.2-6
STATs, which mostly exist in the cytoplasm, are key transcription factors mediating cytokine and growth factor signaling pathways.7 So far, seven STAT members have been identified in mammals.8-10 After cytokine binding, the receptors are quickly phosphorylated by Jak kinases. Then STATs are phosphorylated, dimerized and translocated into the nucleus where they regulate expression of target genes.11 STAT3, one of the most extensively studied STAT members, has been proven to be constitutively activated in various cancers including HCC.12,13 Activated STAT3 contributes to oncogenesis by promoting cell proliferation, preventing apoptosis and impairing host tumor immunity.14-17 Knockdown of STAT3 protein level by STAT3 antisense oligonucleotide can greatly inhibit cell proliferation and tumorigenic growth of the HCC cell line transplanted in nude mice.18
Constitutive activation of STAT3 plays pivotal roles in the development of many human tumors.19 However, total STAT3 protein is also found to be upregulated in many human tumors including HCC.2,20 The increased amounts of unphosphorylated STAT3 contribute greatly to the development of cancer by driving expression of oncogenes such as MRAS and MET in hTERT-HME1 cells.20 However, the role and mechanism of upregulation of total STAT3 in tumorigenesis and progression remain unclear. Our study shows a reciprocal regulation between miR-197 and the IL-6/STAT3 inflammatory signaling pathway in HCC. IL-6/STAT3 downregulates miR-197 in HCC, thus increasing STAT3 level and amplifying IL-6/STAT3 pathway. miR-197 inhibits HCC growth both in vitro and in vivo, and may serve as a potential therapeutic target in HCC.
Results
STAT3 Protein is Quickly Upregulated in HCC Cells by IL-6 Stimulation
In order to explore the underlying mechanisms of the upregulation of total STAT3 in HCC, we conducted an analysis of the response of HCC cells to IL-6 over a period of time as inflammation is closely related to cancer formation and progression. HCC cell lines HepG2 and QGY-7703 were stimulated with IL-6, the levels of p-STAT3 increased rapidly, peaking at 30 min and returning to low level by 60 min. Surprisingly, the total amount of STAT3 also increased throughout this process (Fig. 1A). Our results showed that not only p-STAT3 but also total STAT3 levels were quickly increased after IL-6 stimulation in HCC cells, which was consistent with previous studies.2,20 An analysis of STAT3 mRNA in cells treated with IL-6 was also performed. Unexpectedly, STAT3 mRNA levels remained unchanged throughout IL-6 stimulation, indicating that IL-6 stimulation rapidly increased STAT3 expression at protein level but not at mRNA level in HCC cells, and suggesting that IL-6-induced upregulation of total STAT3 may be modulated at post-transcriptional level rather than transcriptional level.
Figure 1.
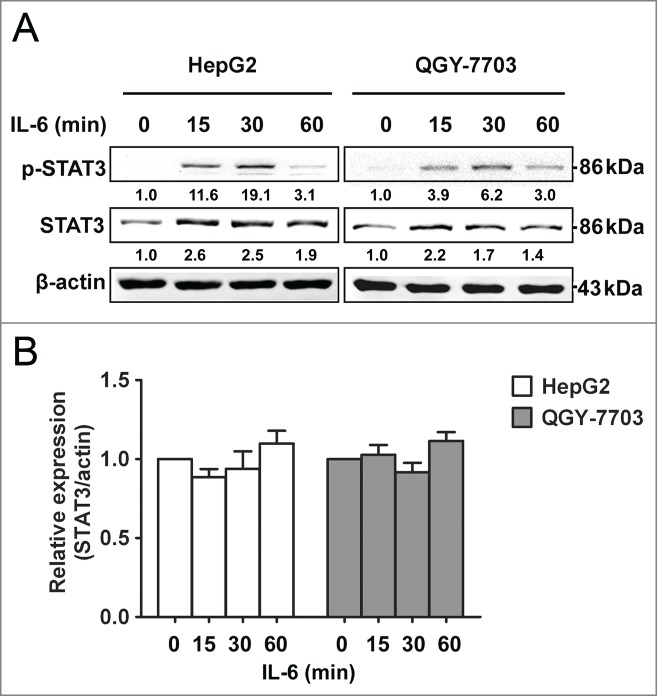
STAT3 protein level is quickly upregulated by IL-6 in HCC cells. (A) Protein expression levels of p-STAT3 and STAT3 were analyzed by Western blotting in HepG2/QGY-7703 cells after being treated with IL-6 (25 ng/mL) as indicated. (B) mRNA expression of STAT3 was measured in HepG2/QGY-7703 cells by qRT-PCR. Data are shown as mean ± s.d. (n = 3) of one representative experiment. Similar results were obtained in at least three independent experiments.
IL-6 Downregulates miR-197, Upregulating STAT3 Protein in HCC
miRNAs are short non-coding RNAs which bind to the 3′UTR of target mRNAs and suppress target expression at post-transcriptional level.21 In order to investigate whether miRNAs were involved in IL-6-induced upregulation of STAT3 in HCC, TaqMan miRNA qPCR array assay was performed to investigate miRNAs expression profile in HepG2 cells treated with IL-6. Many miRNAs were dysregulated by IL-6 stimulation in HepG2 cells (Table S1). Among these miRNAs, 10 miRNAs were mostly reduced by IL-6 (Fig. 2A). To identify the miRNA targeting STAT3 in HCC cells, all these 10 miRNAs were transfected into two HCC cell lines (HepG2 and QGY-7703) respectively, and protein level of STAT3 was detected by Western blotting. Only miR-197 could downregulate protein level of STAT3 (Fig. 2B, C and Fig. S1). These data suggested that STAT3 may be one target of miR-197 in HCC cells. To confirm whether STAT3 was really a target of miR-197, luciferase reporter plasmid with or without wild-type 3′UTR of STAT3 was constructed, and a dual-luciferase reporter assay was employed in HEK293T cells. We found that expression of a luciferase reporter containing 3′UTR of STAT3 was inhibited by cotransfection with miR-197 mimics, whereas reporter plasmid without 3′UTR of STAT3 showed no change in luciferase activity (Fig. 2D).
Figure 2.
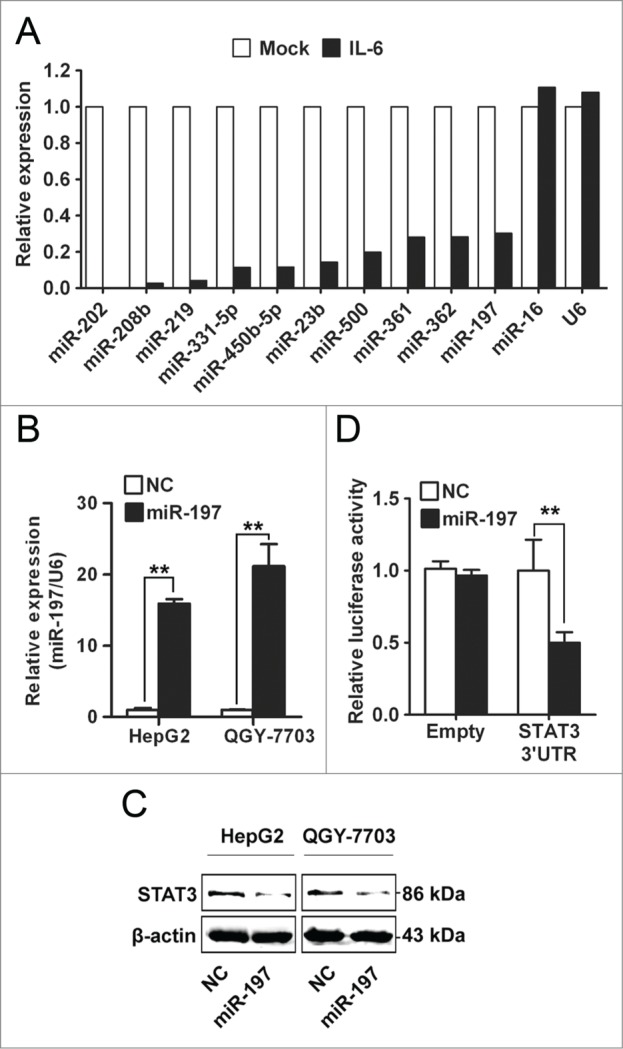
miR-197 is downregulated by IL-6 and targets STAT3 in HCC cells. (A) By TaqMan miRNA qPCR Array, the top 10 downregulated miRNAs in IL-6-treated HepG2 cells were listed, miR-16 and U6 were used as control. (B–C) Expression level of miR-197 was detected by qRT-PCR after transfection with miR-197 mimics (miR-197) or negative control (NC) (B) and STAT3 protein level were detected by Western blotting (C). (D) Firefly luciferase activity was measured in HEK293T cells after co-transfected with pMIR firefly luciferase reporter plasmids with or without 3′UTR of STAT3, pTK-Renilla luciferase plasmids, together with miR-197. Data are shown as mean ± s.d. (n = 3) of one representative experiment. Similar results were obtained in at least three independent experiments. %p < 0.05, %%p < 0.01.
Opposite results were obtained when using antisense oligonucleotides (inhibitors) directed against miR-197 (Fig. S2). In addition, we found that endogenous STAT3 mRNA level in HCC cells was not affected by transfection with miR-197 mimics or miR-197 inhibitors (Fig. S3). These results suggested that IL-6 stimulation could downregulate expression of miR-197, and downregulation of miR-197 in turn upregulated protein level of STAT3 in HCC cells.
Negative Correlation Between miR-197 and Protein Levels of IL-6/STAT3 in HCC Tissues
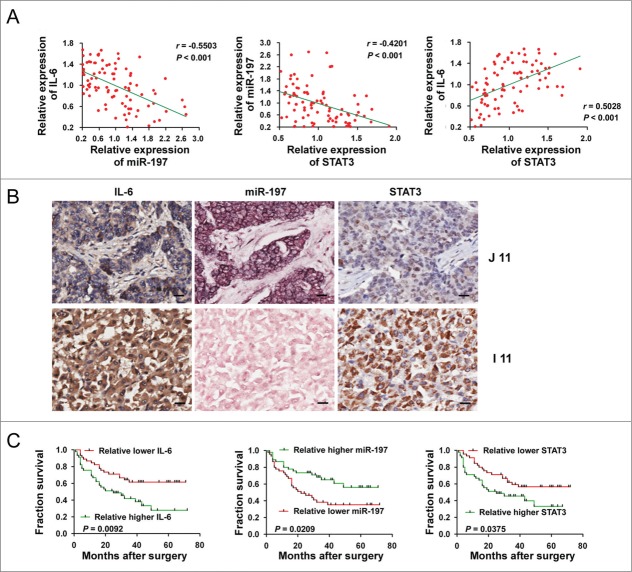
Next, we explored the correlation between miR-197 expression level and protein levels of IL-6 or STAT3 in HCC tissues. In situ hybridization was performed to investigate the expression of miR-197, while immunohistochemical staining was performed to detect the protein expression of IL-6 and STAT3 in patients with HCC by using tissue array. After normalization to the mean expression value, RNA levels of miR-197 and protein levels of IL-6 or STAT3 were analyzed by Pearson's correlation coefficient analysis. Markedly, IL-6 (r = −0.5503, p < 0.001) and STAT3 (r = −0.4201, p < 0.001) protein levels were both inversely correlated with miR-197 expression level in HCC tissues. We also detected that IL-6 was positively related to STAT3 in HCC tissues (r = 0.5028, p < 0.001) (Fig. 3A). ISH of miR-197 and IHC of IL-6 and STAT3 in two representative HCC tissues confirmed negative correlation between miR-197 and protein levels of IL-6/STAT3 in HCC tissues (Fig. 3B).
Figure 3.
Correlation and prognostic significance analyses of miR-197, IL-6 and STAT3 in tissues of HCC patients. (A) A negative Spearman correlation between IL-6 and miR-197, miR-197 and STAT3 and a positive Spearman correlation between IL-6 and STAT3 were found in 90 HCC tissues. (B) Immunohistochemistry staining of STAT3, IL-6 and detection of miR-197 by ISH in two representative HCC tissues (I 11 and J 11). Scale bar = 20 μm. (C) The prognostic significance of IL-6, miR-197 and STAT3 for HCC patients assessed by Kaplan–Meier analyses.
We then analyzed whether low endogenous miR-197 or high IL-6 and STAT3 in HCC tissues correlated with prognosis of HCC patients. As shown in Fig. 3C and Table 1, statistical analysis revealed that patients with high IL-6 (p = 0.0092), STAT3 (p = 0.0375) or low miR-197 (p = 0.0209) had a poorer prognosis.
Table 1.
Univariate analysis of factors correlated with overall survival of HCC patients
| Clinical variables | Case number | HR (95% CI) | p value |
|---|---|---|---|
| miR-197 (lower vs. higher) | 45/45 | 0.49 (0.27–0.90) | 0.0209 |
| IL6 (lower vs. higher) | 45/45 | 2.24 (1.22–4.11) | 0.0092 |
| STAT3 (lower vs. higher) | 45/45 | 1.881 (1.037–3.41) | 0.0375 |
| Sex (male vs. female) | 77/13 | 1.054 (0.45–2.49) | 0.9040 |
| Age (> 55 y vs. ≤ 55 y) | 42/48 | 0.997 (0.55–1.80) | 0.9910 |
| Cirrhosis (yes vs. no) | 32/58 | 0.782 (0.415–1.472) | 0.4460 |
| Tumor size (≥5 cm vs. <5 cm) | 54/36 | 2.087 (1.092–3.992) | 0.0261 |
| Tumor number (>1 vs. 1) | 9/81 | 1.97 (0.879–4.418) | 0.0998 |
| TNM stage (III/IV vs. I/II) | 47/43 | 2.65 (1.40–5.01) | 0.0029 |
| Histological grade (>II vs. I/II) | 36/54 | 1.281 (0.748–2.196) | 0.3670 |
CI: confidence interval; TNM: tumor-node-metastasis staging system.
Collectively, these results suggested that low endogenous miR-197 may be induced by IL-6, and decrease of miR-197 may in turn upregulate STAT3 protein level in HCC. IL-6-induced downregulation of miR-197 in HCC may amplify IL-6/STAT3 pathway, contributing to the progression of HCC. Downregulation of miR-197 may be a prognosis predictor for HCC patients.
miR-197 Inhibits Growth and Invasion of HCC Cells in Vitro through IL-6/STAT3 Signaling Pathway
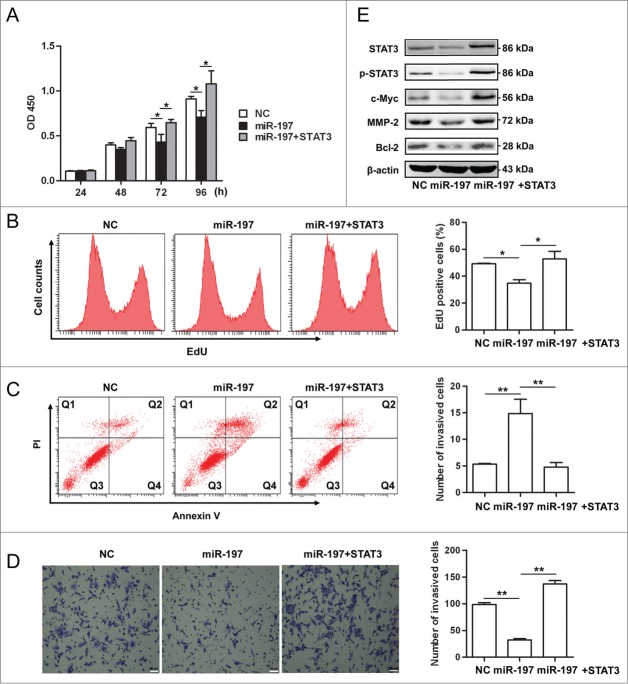
Because our previous study has shown that miR-197 was downregulated in HCC tissues,22 gain-of-function and rescue studies were performed in HCC cells. Overexpression of miR-197 in HepG2 cells significantly suppressed cell proliferation, while, re-expression of STAT3 by transfecting STAT3 cDNA that cannot be targeted by miR-197 in miR-197-tranfected cells rescued this suppression (Figs. 4A, B). Transfection of miR-197 mimics induced apoptosis in HepG2 cells (14.86% versus 5.33% in the control group, p < 0.01), re-expression of STAT3 in miR-197-tranfected cells significantly decreased apoptosis in HepG2 cells (4.8% vs. 14.86% in the miR-197 mimics group, p < 0.01) (Fig. 4C). In matrigel invasion assays, overexpression of miR-197 significantly decreased migration of HepG2 cells (32 versus 98, p < 0.01), re-expression of STAT3 in miR-197-tranfected cells significantly increased migration of HepG2 cells (98 vs. 137, p < 0.01) (Fig. 4D). Moreover, we found that the protein expression levels of c-Myc, Bcl-2 and MMP-2, downstream targets of IL-6/STAT3 signaling pathway, STAT3 and p-STAT3 were decreased in HepG2 cells overexpressing miR-197; re-expression of STAT3 in miR-197-tranfected cells increased the protein expression in these genes (Fig. 4E).
Figure 4.
miR-197 suppresses proliferation, invasion but promotes apoptosis in HepG2 cells through IL-6/STAT3 signaling pathway. (A–B) HepG2 cells growth were measured by CCK-8 analysis (A) or using EdU incorporation (B) after transfection of negative control (NC), miR-197 mimics (miR-197) or miR-197 plus pcDNA-STAT3 which contain STAT3 cDNA that cannot be targeted by miR-197 (miR-197+STAT3). (C) Apoptotic HepG2 cells were analyzed by FACS after they are transfected with NC, miR-197 or miR-197+STAT3. The AnnexinV-positive cells were regarded as apoptotic cells. (D) The invasive ability of HepG2 cells was evaluated by in vitro invasion assays after transfection of NC, miR-197 or miR-197+STAT3. (E) HepG2 cells were transfected with NC, miR-197 or miR-197+STAT3, 48 h later, the expression levels of STAT3, p-STAT3, c-Myc, Bcl−2, and MMP-2 were analyzed by Western blotting. Data are shown as mean ± s.d. (n = 3) of one representative experiment. Similar results were obtained in at least three independent experiments. %p < 0.05, %%p < 0.01, %%%p < 0.001. Scale bar = 50 μm.
In addition, similar results were found in QGY-7703 cells (Fig. S4). Taken together, our data suggested that overexpression of miR-197 suppressed viability and migration of HCC cells possibly through impairing IL-6/STAT3 signaling pathway, thus indicating that downregulation of miR-197 in HCC may contribute to HCC progression by amplifying IL-6/STAT3 pathway.
miR-197 Suppresses Tumor Growth of HCC Xenografts
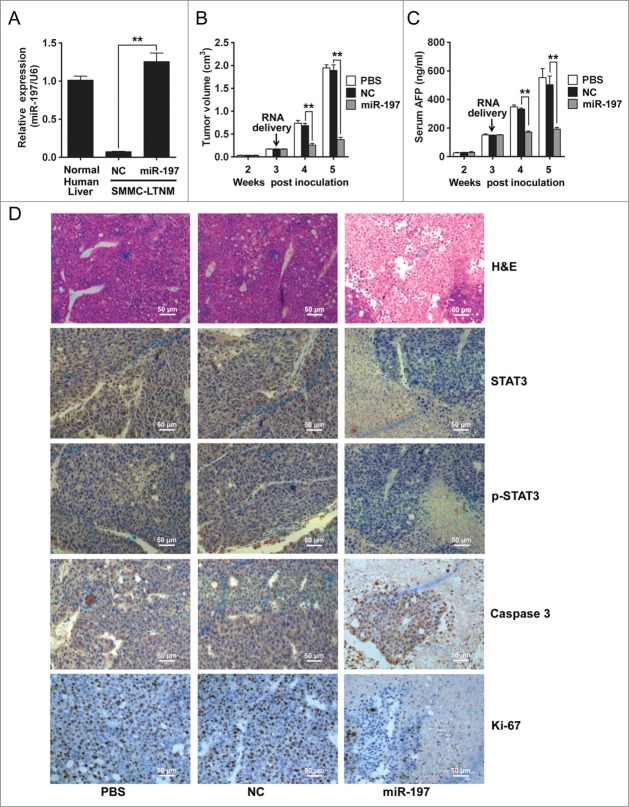
A human HCC-bearing nude mouse model SMMC-LTNM was employed to identify the in vivo effects of miR-197 on HCC growth. As compared to other human HCC-bearing nude mouse model generated by subcutaneously inoculating with HCC cell lines, SMMC-LTNM is more similar to clinical progression of HCC as α-fetoprotein (AFP) is detected with high level in sera of SMMC-LTNM model.22,23 The expression of miR-197 was much lower in SMMC-LTNM tumor tissue when compared to normal human liver, and in vivo intratumoral injection of cholesterol-conjugated miR-197 mimics restored its expression in tumor tissue (Fig. 5A). After intratumoral administration of the cholesterol-conjugated miR-197 mimics, significant reduction in tumor size (Fig. 5B), decrease of serum AFP (Fig. 5C) and down-regulation of STAT3 and p-STAT3, increased apoptosis, suppressed cell proliferation and more severe liver necrosis (Fig. 5D) were observed in SMMC-LTNM-bearing mice. We also found that the protein expression levels of c-Myc, Bcl-2 and MMP-2, downstream targets of IL-6/STAT3 signaling pathway, were decreased in SMMC-LTNM-bearing mice overexpressing miR-197 (Fig. S5), further supporting a potential suppressive effect of miR-197 on HCC.
Figure 5.
miR-197 suppresses HCC growth in vivo. (A) qRT-PCR analysis of miR-197 expression in normal human liver tissue, or SMMC-LTNM tumor tissue two weeks after intratumoral injection of cholesterol-conjugated miR-197 mimics (miR-197) or negative control (NC). (B–C) Two weeks after subcutaneous inoculation of SMMC-LTNM tumor cells, HCC-bearing nude mice were treated by intratumoral injection of cholesterol-conjugated miR-197, NC or PBS. Tumor volume (B), serum AFP levels (C) were shown as indicated in (C). (D) H&E staining and detection of STAT3, p-STAT3, Caspase 3 and Ki-67 by IHC in HCC tissues was performed two weeks after intratumoral injection of cholesterol-conjugated miR-197, NC or PBS. Scale bars, 50 μm. Data are shown as mean ± s.d. (n = 3) of one representative experiment. Similar results were obtained in three independent experiments. %%p < 0.01.
IL-6 Induces Downregulation of miR-197 in HCC by Impairing Binding of Drosha to pri-miR-197
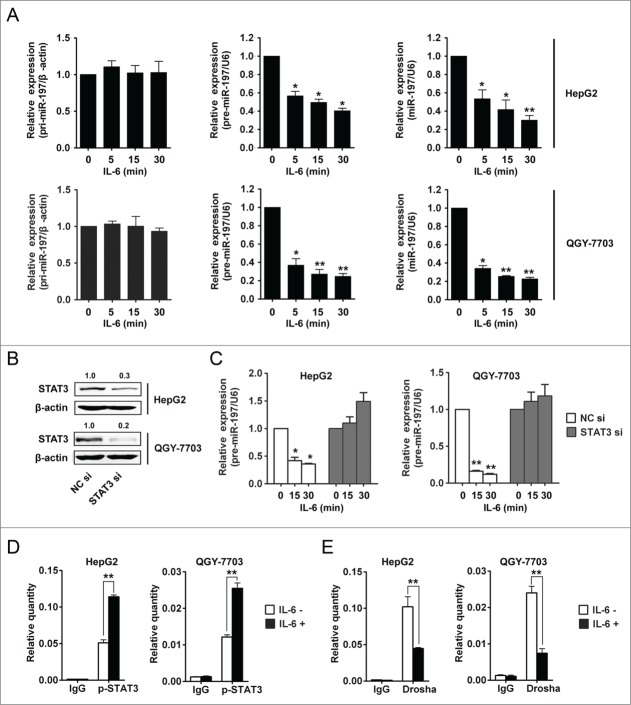
Our results have showed that IL-6 could induce downregulation of miR-197 in HCC cells, but the associated mechanism was not clear. To explore the mechanism underlying the downregulation of miR-197 by IL-6, HepG2 and QGY-7703 cells were employed to detect the expression of pri-miR-197, precursor miR-197 (pre-miR-197) and mature miR-197 after IL-6 treatment at different time points. Our results showed that after IL-6 stimulation, pri-miR-197 expression remained unchanged while pre-miR-197 and mature miR-197 expression reduced significantly (Fig. 6A), suggesting that biogenesis of miR-197 may be regulated at pri-miR-197 to pre-miR-197 level by IL-6 stimulation in HCC cells. Previous studies had demonstrated that maturation of miR-21 could be regulated by transcription factor Smad3 at post-transcriptional level.24 STAT3 is one important transcription factor of IL-6/STAT3 pathway, so we wonder whether STAT3 participated in the generation of miR-197. HepG2 and QGY-7703 cells were transfected with siRNAs specific to human STAT3, and STAT3 protein levels were detected to be significantly inhibited (Fig. 6B). IL-6 was then added to HCC cells after STAT3 interference; pre-miR-197 was detected by qRT-PCR at the indicated time points. The results showed that reduction of pre-miR-197 by IL-6 stimulation disappeared in the cells with STAT3 interference (Fig. 6C). In addition, similar results were obtained for mature miR-197 but not pri-miR-197 (Fig. S6). These data suggested that IL-6-induced downregulation of miR-197 was at the post-transcriptional level and may be STAT3-dependent.
Figure 6.
IL-6 downregulates miR-197 expression through p-STAT3 by disrupting Drosha binding to pri-miR-197 and reducing mature miR-197 generation. (A) pri-miR-197, pre-miR-197 and mature miR-197 were detected by qRT-PCR in HepG2 and QGY-7703 cells after IL-6 treatment (25 ng/mL) as indicated. (B) HepG2 and QGY-7703 cells were transfected with negative control siRNA (NC si) or STAT3 siRNAs (STAT3 si), 48 h later, STAT3 and β-actin were detected by Western blotting. (C) Cells in (B) were treated with IL-6 (25 ng/mL) as indicated, and pre-miR-197 was detected by qRT-PCR and normalized to U6. (D–E) HepG2 cells and QGY-7703 cells were stimulated with or without IL-6 (25 ng/mL) for 5 min, and qRT-PCR analysis of p-STAT3 (D) or Drosha (E) binding to pri-miR-197 after RNA ChIP assay were performed. Data are shown as mean ± s.d. (n = 3) of one representative experiment. Similar results were obtained in at least three independent experiments. %p < 0.05, %%p < 0.01.
In order to explore the mechanism of STAT3-dependent reduction of miR-197 by IL-6 stimulation in HCC cells, RNA-ChIP assay was performed. Because pri-miRNAs are processed to pre-miRNAs in the nucleus, so binding of p-STAT3 to pri-miR-197 was examined. The results showed that the binding of p-STAT3 to pri-miR-197 was increased after IL-6 stimulation in HCC cells (Fig. 6D). Drosha plays an important role in the generation of pre-miRNA,25 so we wonder if IL-6 stimulation exerts its effect via affecting the binding of Drosha to pri-miR-197. Our results showed that the binding of Drosha to pri-miR-197 was decreased after IL-6 stimulation (Fig. 6E). We also detected the binding of STAT3 to pri-miR-197 after IL-6 stimulation in HCC cells; our results showed that there was no binding of STAT3 to pri-miR-197 (Fig. S7). These results indicated that the induction of p-STAT3 binding to pri-miR-197 by IL-6 stimulation affected Drosha binding and then reduced miR-197 generation in HCC cells.
Discussion
It has been reported that constitutive activation of STAT3 is likely to contribute to the development of many human tumors. However, total STAT3 was also found to be upregulated in many human tumors including HCC.2,20 The mechanism for and role of upregulation of total STAT3 in tumorigenesis and progression remain unclear. Our study demonstrates that a reciprocal regulation exists between IL-6/STAT3 pathway and miR-197. Our data suggest that miR-197, by targeting STAT3 protein, can inhibit HCC progression and may serve as a potential therapeutic target for HCC, thus providing insights for aberrant IL-6/STAT3 activation in HCC.
Constitutive STAT3 activation has been linked to oncogenesis in a variety of human tumors. Diverse cytokines and growth factors, including IL-6, EGF and PDGF, activate the JAK/STAT3 signaling pathway.26 STAT3 can also be activated by different oncogenic kinases and viral proteins (e.g. from hepatitis B virus or hepatitis C virus).27 Interestingly, total STAT3 protein is also upregulated in many human tumors including HCC.2,20 Yang et al. reported that total amount of STAT3 continued to increase throughout 32 h after IL-6 treatment in hTERT-HME1 cells, which was attributed to the binding of p-STAT3 to STAT3 promoter. In our study, we found that total STAT3 continued to increase at protein level but not mRNA level in HCC cells during IL-6 treatment, suggesting that post-transcriptional regulation of STAT3 likely exists during IL-6 stimulation. Our results showed that downregulation of miR-197, which was induced by IL-6/STAT3 signaling pathway, in HCC cells could directly target and upregulate STAT3 in HCC tissues and cells. Our results thus reveal one novel regulation mode for IL-6/STAT3 signaling pathway during HCC tumorigenesis.
Yang also reported that increased amounts of unphosphorylated STAT3 contribute importantly to the development of cancer by driving expression of oncogenes such as MRAS and MET in hTERT-HME1 cells. However, our results showed that overexpression of miR-197 in HCC cells downregulated protein levels of STAT3 and p-STAT3 (Fig. 4E, Figs. S4E and S5D), but did not affect the expression of MRAS and MET (Fig. S8), suggesting miR197-mediated regulation of total STAT3 protein in HCC oncogenesis could very well be due to altered p-STAT3 levels as described in previous study.2,18
Tumorigenesis and tumor progression always occurs after accumulation of multistep mutations and is characterized by uncontrolled cell division and survival.28,29 At the same time, environmental conditions and extracellular stimuli exert great effect on these processes by affecting coding genes and non-coding genes.30 Inflammatory microenvironment has been shown to be a strong inducement in many cancers including HCC.31-33 HCC has been regarded as an example of inflammation-related cancer. IL-6 is an important inflammatory cytokine that plays essential roles in HCC development through activating IL-6/STAT3 signaling pathway.34 IL-6 deficient mice were resistant to diethylnitrosamine-induced HCC development,35 and hepatocyte-specific STAT3 knockout significantly impaired the development as well as the tumor growth of HCC.36 Inflammatory stress as IL-6 can alter the expression profile of miRNAs showing tumor suppressive or oncogenic activity.37,38 Therefore, miRNAs may function as mediators of inflammation related tumors. Although the association between IL-6 and malignancy has been recognized for many decades, the role of IL-6 as stimulator of miRNA expression remained to be explored. Our results showed that IL-6 decreased the expression of many miRNAs including miR-197. Moreover, our results demonstrated that miR-197 could significantly inhibit proliferation, invasion and promote apoptosis in HCC cells through impairing IL-6/STAT3 pathway. These data indicate that miR-197 might function as an inflammatory mediator during inflammation-related HCC development.
miR-197 is frequently identified as a potential biomarker in a variety of cancer types including HCC. miRNA expression profiling of oral carcinoma, gastric carcinoma, malignant astrocytoma and HCC revealed the downregulation of miR-197, indicating miR-197 as one potential antitumor biomarker.22,39-41 miR-197 was also found to be upregulated in follicular thyroid cancer and lung cancer and may function as oncogenic biomarker.42,43 However, the role of miR-197 during tumorigenesis of HCC remains largely unknown due to lacking information of target genes. Our data revealed that STAT3 may be the target of miR-197 in HCC cells. Moreover, miR-197 could significantly inhibit HCC growth both in vitro and in vivo, supporting the antitumor effects of miR-197 in HCC. However, the function of miR-197 was much controversial in different cancers. So the role of miR-197 in carcinogenesis of HCC awaits further investigations.
In summary, our study suggests that a reciprocal regulation exists between IL-STAT3 pathway and miR-197 in HCC. We demonstrate that miR-197, by targeting STAT3, inhibits HCC growth both in vitro and in vivo, thus highlighting miR-197 as a potential therapeutic target in HCC.
Materials and Methods
Cell Lines and Human Tissue Specimens
HEK293T as well as human HCC cell lines HepG2 and QGY-7703 were obtained from ATCC. HEK293T cells were cultured in DMEM media containing 10% (v/v) fetal bovine serum (FBS, PAA Laboratories, Pasching, Australia). HepG2 and QGY-7703 cell lines were routinely maintained in RPMI-1640 supplemented with 10% (v/v) FBS at 37°C in a humidified incubator containing 5% CO2. Normal human liver tissues were obtained from distal normal liver tissue of liver hemangioma. Tissue samples were immediately frozen in liquid nitrogen until analysis. Informed consent was obtained from each patient and the study was approved by the ethics committee of Second Military Medical University, Shanghai, China.
Vector Construction and Luciferase Reporter Assay
The vectors used in this study were constructed as described in Supplementary Materials, and luciferase reporter assay was performed as described previously.44
Cell Proliferation, Invasion and Apoptosis Analyses
Cell proliferation, invasion and apoptosis analyses were performed as described in Supplementary Materials.
HCC-bearing Nude Mouse Model and in Vivo Treatment
All animal experiments were conducted according to the National Institute of Health Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of Second Military Medical University, Shanghai. The HCC-bearing male nude mice were prepared and treated as previously described.22 Human HCC-bearing male nude mice with subcutaneous passage of SMMC-LTNM were used for evaluating the antitumor effect of miR-197 mimics in vivo. For preparation of subcutaneous model, 0.2 mL grinded SMMC-LTNM tumor tissue was subcutaneously injected and inoculated. For delivery of cholesterol-conjugated RNA, 10 nmol RNA in 0.1 mL saline buffer was locally injected into the tumor mass once every 3 d for 2 weeks. Tumor size was measured and serum AFP was detected using ELISA as described previously.22
RNA-ChIP Assay
Detail RNA-ChIP assay is described in Supplementary Material. The primers spanning the hairpin of pri-miR-197 are shown in Table S2.
Immunohistochemical Staining (IHC)
IL-6 and STAT3 levels in liver tumors were evaluated by IHC as narrated in Supplementary Materials and the statistical method was described as previously.45
In Situ Hybridization (ISH)
miR-197 level in liver tumors were evaluated by ISH as narrated in Supplementary Materials and the statistical method was described as previously.46
Statistical Analysis
All data are presented as the mean ± standard deviation. Statistical analysis was performed using SPSS 15.0 as detailed in Supplementary Materials.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
The work was supported by Grants from the National Key Basic Research Program of China (2013CB530502, 2010CB911903), the National Natural Science Foundation of China (81230074, 81123006, 30972704, 81222039), and the National 125 Key Project of China (2012ZX10002-014).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69-90; PMID:21296855; http://dx.doi.org/ 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 2.Yang X, Liang L, Zhang XF, Jia HL, Qin Y, Zhu XC, Gao XM, Qiao P, Zheng Y, Sheng YY et al.. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology 2013; 58:158-70; PMID:23389848; http://dx.doi.org/ 10.1002/hep.26305 [DOI] [PubMed] [Google Scholar]
- 3.Hou J, Zhou Y, Zheng Y, Fan J, Zhou W, Ng IO, Sun H, Qin L, Qiu S, Lee JM et al.. Hepatic RIG-I predicts survival and interferon-alpha therapeutic response in hepatocellular carcinoma. Cancer Cell 2014; 25:49-63; PMID:24360797; http://dx.doi.org/ 10.1016/j.ccr.2013.11.011 [DOI] [PubMed] [Google Scholar]
- 4.Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology 2010; 51:836-45; PMID:20041405; http://dx.doi.org/ 10.1002/hep.23380 [DOI] [PubMed] [Google Scholar]
- 5.Feitelson MA, Sun B, Satiroglu Tufan NL, Liu J, Pan J, Lian Z. Genetic mechanisms of hepatocarcinogenesis. Oncogene 2002; 21:2593-604; PMID:11971194; http://dx.doi.org/ 10.1038/sj.onc.1205434 [DOI] [PubMed] [Google Scholar]
- 6.Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology 2010; 52:60-70; PMID:20578129; http://dx.doi.org/ 10.1002/hep.23660 [DOI] [PubMed] [Google Scholar]
- 7.Darnell JE, Jr. STATs and gene regulation. Science 1997; 277:1630-5; PMID:9287210; http://dx.doi.org/ 10.1126/science.277.5332.1630 [DOI] [PubMed] [Google Scholar]
- 8.Zhong Z, Wen Z, Darnell JE Jr. Stat3 and Stat4: members of the family of signal transducers and activators of transcription. Proc Natl Acad Sci U S A 1994; 91:4806-10; PMID:7545930; http://dx.doi.org/ 10.1073/pnas.91.11.4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakraborty A, Tweardy DJ. Stat3 and G-CSF-induced myeloid differentiation. Leuk Lymphoma 1998; 30:433-42; PMID:9711905; http://dx.doi.org/ 10.3109/10428199809057555 [DOI] [PubMed] [Google Scholar]
- 10.Chakraborty A, Tweardy DJ. Granulocyte colony-stimulating factor activates a 72-kDa isoform of STAT3 in human neutrophils. J Leukoc Biol 1998; 64:675-80; PMID:9823774 [DOI] [PubMed] [Google Scholar]
- 11.Zhong Z, Wen Z, Darnell JE Jr. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 1994; 264:95-8; PMID:8140422; http://dx.doi.org/ 10.1126/science.8140422 [DOI] [PubMed] [Google Scholar]
- 12.Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, Forman S, Jove R, Pardoll DM, Yu H. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell 2009; 15:283-93; PMID:19345327; http://dx.doi.org/ 10.1016/j.ccr.2009.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE Jr. Stat3 as an oncogene. Cell 1999; 98:295-303; PMID:10458605; http://dx.doi.org/ 10.1016/S0092-8674(00)81959-5 [DOI] [PubMed] [Google Scholar]
- 14.Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Kloppel G, Yoshimura A, Reindl W, Sipos B, Akira S et al.. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell 2011; 19:456-69; PMID:21481788; http://dx.doi.org/ 10.1016/j.ccr.2011.03.009 [DOI] [PubMed] [Google Scholar]
- 15.Fukuda A, Wang SC, Morris JPt, Folias AE, Liou A, Kim GE, Akira S, Boucher KM, Firpo MA, Mulvihill SJ et al.. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell 2011; 19:441-55; PMID:21481787; http://dx.doi.org/ 10.1016/j.ccr.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li N, Grivennikov SI, Karin M. The unholy trinity: inflammation, cytokines, and STAT3 shape the cancer microenvironment. Cancer Cell 2011; 19:429-31; PMID:21481782; http://dx.doi.org/ 10.1016/j.ccr.2011.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kortylewski M, Xin H, Kujawski M, Lee H, Liu Y, Harris T, Drake C, Pardoll D, Yu H. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell 2009; 15:114-23; PMID:19185846; http://dx.doi.org/ 10.1016/j.ccr.2008.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li WC, Ye SL, Sun RX, Liu YK, Tang ZY, Kim Y, Karras JG, Zhang H. Inhibition of growth and metastasis of human hepatocellular carcinoma by antisense oligonucleotide targeting signal transducer and activator of transcription 3. Clin Cancer Res 2006; 12:7140-8; PMID:17145839; http://dx.doi.org/ 10.1158/1078-0432.CCR-06-0484 [DOI] [PubMed] [Google Scholar]
- 19.Bar-Natan M, Nelson EA, Xiang M, Frank DA. STAT signaling in the pathogenesis and treatment of myeloid malignancies. JAKSTAT 2012; 1:55-64; PMID:24058751; http://dx.doi.org/ 10.4161/jkst.20006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, Stark GR. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res 2005; 65:939-47; PMID:15705894 [PubMed] [Google Scholar]
- 21.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009; 136:642-55; PMID:19239886; http://dx.doi.org/ 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y et al.. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 2011; 19:232-43; PMID:21316602; http://dx.doi.org/ 10.1016/j.ccr.2011.01.001 [DOI] [PubMed] [Google Scholar]
- 23.Li D, Liu X, Lin L, Hou J, Li N, Wang C, Wang P, Zhang Q, Zhang P, Zhou W et al.. MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. J Biol Chem 2011; 286:36677-85; PMID:21878637; http://dx.doi.org/ 10.1074/jbc.M111.270561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 2008; 454:56-61; PMID:18548003; http://dx.doi.org/ 10.1038/nature07086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:21532838; http://dx.doi.org/21447371 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 26.Santos CI, Costa-Pereira AP. Signal transducers and activators of transcription-from cytokine signalling to cancer biology. Biochim Biophys Acta 2011; 1816:38-49; PMID:21447371; http://dx.doi.org/ 10.1016/j.bbcan.2011.03.003 [DOI] [PubMed] [Google Scholar]
- 27.Gelatti U, Donato F, Tagger A, Fantoni C, Portolani N, Ribero ML, Martelli C, Trevisi P, Covolo L, Simonati C et al.. Etiology of hepatocellular carcinoma influences clinical and pathologic features but not patient survival. Am J Gastroenterol 2003; 98:907-14; PMID:12738476; http://dx.doi.org/ 10.1111/j.1572-0241.2003.t01-1-07289.x [DOI] [PubMed] [Google Scholar]
- 28.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science 2013; 339:1546-58; PMID:23539594; http://dx.doi.org/ 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 2012; 338:1622-6; PMID:23258894; http://dx.doi.org/ 10.1126/science.1229164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 2009; 139:693-706; PMID:19878981; http://dx.doi.org/ 10.1016/j.cell.2009.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet 2001; 357:539-45; PMID:11229684; http://dx.doi.org/ 10.1016/S0140-6736(00)04046-0 [DOI] [PubMed] [Google Scholar]
- 32.Naugler WE, Karin M. NF-kappaB and cancer-identifying targets and mechanisms. Curr Opin Genet Dev 2008; 18:19-26; PMID:18440219; http://dx.doi.org/ 10.1016/j.gde.2008.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naugler WE, Karin M. The wolf in sheep's clothing: the role of interleukin-6 in immunity, inflammation and cancer. Trends Mol Med 2008; 14:109-19; PMID:18261959; http://dx.doi.org/ 10.1016/j.molmed.2007.12.007 [DOI] [PubMed] [Google Scholar]
- 34.Taub R. Hepatoprotection via the IL-6/Stat3 pathway. J Clin Invest 2003; 112:978-80; PMID:14523032; http://dx.doi.org/ 10.1172/JCI19974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science 2007; 317:121-4; PMID:17615358; http://dx.doi.org/ 10.1126/science.1140485 [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Lafdil F, Wang L, Park O, Yin S, Niu J, Miller AM, Sun Z, Gao B. Hepatoprotective versus oncogenic functions of STAT3 in liver tumorigenesis. Am J Pathol 2011; 179:714-24; PMID:21684247; http://dx.doi.org/ 10.1016/j.ajpath.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis 2010; 31:37-49; PMID:19955394; http://dx.doi.org/ 10.1093/carcin/bgp272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K et al.. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 2007; 110:1330-3; PMID:17496199; http://dx.doi.org/ 10.1182/blood-2007-03-081133 [DOI] [PubMed] [Google Scholar]
- 39.Scapoli L, Palmieri A, Lo Muzio L, Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M, Carinci F. MicroRNA expression profiling of oral carcinoma identifies new markers of tumor progression. Int J Immunopathol Pharmacol 2010; 23:1229-34; PMID:21244772 [DOI] [PubMed] [Google Scholar]
- 40.Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, Sun L, Ji G, Shi Y, Han Z et al.. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res 2011; 9:824-33; PMID:21628394; http://dx.doi.org/ 10.1158/1541-7786.MCR-10-0529 [DOI] [PubMed] [Google Scholar]
- 41.Yang C, Wang C, Chen X, Chen S, Zhang Y, Zhi F, Wang J, Li L, Zhou X, Li N et al.. Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer 2013; 132:116-27; PMID:22674182; http://dx.doi.org/ 10.1002/ijc.27657 [DOI] [PubMed] [Google Scholar]
- 42.Stokowy T, Eszlinger M, Swierniak M, Fujarewicz K, Jarzab B, Paschke R, Krohn K. Analysis options for high-throughput sequencing in miRNA expression profiling. BMC Res Notes 2014; 7:144; PMID:24625073; http://dx.doi.org/ 10.1186/1756-0500-7-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fiori ME, Barbini C, Haas TL, Marroncelli N, Patrizii M, Biffoni M, De Maria R. Antitumor effect of miR-197 targeting in p53 wild-type lung cancer. Cell Death Differ 2014; 21:774-82; PMID:24488097; http://dx.doi.org/ 10.1038/cdd.2014.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su X, Qian C, Zhang Q, Hou J, Gu Y, Han Y, Chen Y, Jiang M, Cao X. miRNomes of haematopoietic stem cells and dendritic cells identify miR-30b as a regulator of Notch1. Nat Commun 2013; 4:2903; PMID: 24309499; http://dx.doi.org/ 10.1038/ncomms3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen T, Yang M, Yu Z, Tang S, Wang C, Zhu X, Guo J, Li N, Zhang W, Hou J et al.. Small GTPase RBJ mediates nuclear entrapment of MEK1/MEK2 in tumor progression. Cancer Cell 2014; 25:682-96; PMID:24746703; http://dx.doi.org/ 10.1016/j.ccr.2014.03.009 [DOI] [PubMed] [Google Scholar]
- 46.Nilsson S, Möller C, Jirström K, Lee A, Busch S, Lamb R, Landberg G. Downregulation of miR-92a is associated with aggressive breast cancer features and increased tumour macrophage infiltration. PloS One 2012; 7:e36051; PMID:22563438; http://dx.doi.org/ 10.1371/journal.pone.0036051 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.