Abstract
We present the first genome-wide study of recent evolution in Culex pipiens species complex focusing on the genomic extent, functional targets and likely causes of global and local adaptations. We resequenced pooled samples of six populations of C. pipiens and two populations of the outgroup Culex torrentium. We used principal component analysis to systematically study differential natural selection across populations and developed a phylogenetic scanning method to analyse admixture without haplotype data. We found evidence for the prominent role of geographical distribution in shaping population structure and specifying patterns of genomic selection. Multiple adaptive events, involving genes implicated with autogeny, diapause and insecticide resistance were limited to specific populations. We estimate that about 5–20% of the genes (including several histone genes) and almost half of the annotated pathways were undergoing selective sweeps in each population. The high occurrence of sweeps in non-genic regions and in chromatin remodelling genes indicated the adaptive importance of gene expression changes. We hypothesize that global adaptive processes in the C. pipiens complex are potentially associated with South to North range expansion, requiring adjustments in chromatin conformation. Strong local signature of adaptation and emergence of hybrid bridge vectors necessitate genomic assessment of populations before specifying control agents.
Keywords: Culex pipiens, selective sweeps, histones, principal component analysis, population structure, differential selection
1. Introduction
Arms races between antagonistic species have been of longstanding interest to evolutionary biologists [1,2]. Humans have developed unique ways of fighting against competitors, parasites and their vectors through the use of scientific and technological innovations such as application of antibiotics and pesticides, living in artificially designed cities with high hygienic standards and avoidance of high disease-transmission-risk behaviours. The race is, however, far from over, as emphasized by the emergence of new virulent pathogenic strains, evolution of insecticide-resistant vectors and the adaptations of several pathogenic or competitor species for living in cities. Notably, any such adaptation takes place locally at first, even if it does spread to gain global significance eventually.
Here, we present the results of the first genome-wide analysis of population structure and recent natural selection in the Culex pipiens complex, members of which are notorious vectors of West Nile Virus, St. Louise Encephalitis Virus and filariasis worms in the US and worldwide [3–5]. This complex consists of Culex quinquefasciatus (the southern house mosquito) and C. pipiens (the northern house mosquito). Two biological forms (biotypes) C. pipiens f. pipiens and C. pipiens f. molestus have been described within the C. pipiens species based on physiological and ecological differences including the choice of host species, seasonal activity, mating behaviour and preferred habitat [3,6,7]. Hybridization between these two forms and also with C. quinquefasciatus has been reported in certain areas, leading to the rise of bridge vectors transmitting pathogens between birds and humans [3,6,8–10].
We studied six C. pipiens populations and two populations of the closely related species Culex torrentium as an outgroup living within or close to human-inhabited areas in Europe and North America (Moscow and Aleksin, Russia and Sacramento, CA, USA), aiming to investigate two fundamental population genetic aspects of C. pipiens: population structure and natural selection. On population structure, we asked: (i) Does geography, habitat type or biological form mainly determine the organization of genetic variation in Culex? (ii) Do genomic data support genetic isolation and imminent speciation of pipiens and molestus forms, or conversely, do we detect considerable admixture between them? On the matter of natural selection, we asked: (i) Which genes and biological functions are the targets of recent selective sweeps? (ii) To what degree do protein sequence alterations and gene expression changes contribute to adaptation? (iii) What factors are the likely causes of recent sweeps? (iv) Are recent adaptations happening congruently or otherwise in different populations?
2. Material and methods
(a). Mosquito samples
Mosquito samples were taken from urban and suburban areas in Sacramento (CA, USA), Moscow and Aleksin (Central Russia) (table 1).
Table 1.
Samples used in this study and their average genome-wide variability statistics.
| sample | location | habitat | taxonomical identification | no. pooled individuals | average πa | average θa |
|---|---|---|---|---|---|---|
| A1 | Aleksin | urban | C. pipiens f. molestus | 224 | 0.01937 | 0.02030 |
| A4 | Aleksin | suburban | C. pipiens f. pipiens | 132 | 0.02403 | 0.02531 |
| M1 | Moscow | urban | C. pipiens f. molestus | 26 | 0.01821 | 0.01905 |
| M2b | Moscow | suburban | C. torrentium | 28 | 0.01933 | 0.02070 |
| M4b | Moscow | suburban | C. torrentium | 195 | 0.01740 | 0.01820 |
| S1 | Sacramento | urban (males) | C. pipiens f. molestus | 15 | 0.02291 | 0.02354 |
| S2b | Sacramento | suburban (males) | C. pipiens, mixed molestus and pipiens forms | 13 | 0.02347 | 0.02438 |
| S3b | Sacramento | suburban (females) | C. pipiens, mixed molestus and pipiens forms | 64 | 0.02276 | 0.02365 |
aAverage of 10 kb sliding windows. Only positions covered 4–40× were included.
bThe two Moscow suburban samples and the two Sacramento suburban samples were each caught independently at different sites and represent different populations.
(b). Sequencing and mapping to the reference
Genomic DNA was extracted from the pool of mosquitoes collected from each of the eight populations, prepared into separate libraries and sequenced as paired-end 101 bp reads on an Illumina HiSeq machine. Sequenced reads were aligned as pairs using BWA 0.5.7 [11] to the complete C. quinquefasciatus draft genome downloaded from the Broad Institute (see https://www.broadinstitute.org/annotation/genome/culex_pipiens.4/MultiDownloads.html). Reads were allowed up to 12 mismatches throughout the 101 bp per end; they were mapped to the genome and those that did not map uniquely were filtered out. All other BWA alignment parameters were set to default values.
(c). Population genetic analyses
The reads mapping to the mitochondrial cytochrome oxidase subunit I gene (COI) and to the CQ11 microsatellite locus were used to ascertain species and biotype identities of the populations, respectively [12,13].
Fst was calculated for 10 kb sliding windows between each pair of populations according to the methods in [14,15] and averaged across the genome. Maximum-likelihood phylogenetic trees were constructed from sliding windows of non-overlapping 10 kb using RAxML [16]. A custom Python code was used to calculate the percentage of time each two populations were nearest neighbours on the tree. Principal component analysis (PCA) of allele frequencies was done on biallelic positions with coverage 4–40×.
The software package Popoolation v. 1.2.2 was used to estimate measures of variation (π and θ) and Tajima's D from the pooled sequence data [17]. Only positions with coverage 4–40× were used and the minimal legitimate count for the minor allele was set to 2. Synonymous (syn) and non-synonymous (nsyn) polymorphisms were assigned using the same software and the .gff file downloaded from the Broad Institute website.
To detect selective sweeps, we first obtained the allele frequency spectrum (AFS) from the whole genome as the neutral background, and then tried to identify a certain form of skewness in AFS of linked sites, typically associated with selective sweeps [18,19]. The approach in [18,19] has been modified to apply to pooled sequence data [20] and incorporated into the software package Pool-hmm [21]. We ran Pool-hmm in two steps. First, AFS was built based on the whole genome for each sample with coverage 4–40×, θ = 0.02 (based on the Popoolation output, see table 1) and sampling ratio of 20 (5% of positions were used for estimation of AFS). Second, sweep regions were detected separately for each supercontig with the same coverage range as above and transition probability of k = 1 × 10−6 based on the AFS created in the previous step. PCA of Pool-hmm sweep scores was done to compare the broad patterns of genomic selection among the studied populations. Each gene was treated as an observation point and each sample label as an initial variable. We used linear regression to check for potential biases introduced by sequencing coverage variation in calculation of Tajima's D and Pool-hmm scores.
To understand the nature of mutations associated with the sweep events, we did a case study focusing on a 137-kb block (C. quinquefasciatus genome supercontig 3.392: 626-137726) consisting exclusively of 80 histone genes including multiple paralogues of each histone type (H1, H2A, H2B, H3 and H4). The large number of polymorphic sites in H1 genes allowed statistical analysis of association of different types of amino acid changes with certain structural features of the protein. For the polymorphic positions, we cross-examined three structural attributes (domain, secondary structure and solvent accessibility) with three biochemical aspects (addition or removal of proline, the charge difference between the two amino acids, and addition or removal of serine or threonine) using independence tests (χ2 and Fisher's exact). Electrostatic interactions are important in forming the three-dimensional structure of the protein, as well as its DNA-binding function. The reason to include the Ser/Thr category was that they are targets of phosphorylation, which is the best-studied epigenetic modification of H1 [22], although several other types of modification are also reported [23]. We included Pro because, apart from its structural peculiarities, we found tremendous excess conversion from other amino acids in the reference genome to Pro at polymorphic positions (electronic supplementary material, S1).
We examined the functional significance of the sweep genes using three different approaches: Gene Ontology (GO) enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG; see http://www.genome.jp/kegg/) pathway annotations and checking the sweep status of genes with experimentally verified functions reported in the literature. GO enrichment analysis on targets of selection was performed using the online software GOEAST [24]. GO annotations for Culex genes were downloaded from vectorbase.org/biomart. The complete annotation file was used as the default background set. For each population, two enrichment tests were run with different selected gene sets: (i) 200 genes (approx. 1% of the total number of genes in the genome) with highest Pool-hmm scores, and (ii) 200 genes with lowest Tajima's D values. Enrichments with false discovery rate < 0.1 were considered significant. An ANOVA test was done to make sure that gene length did not bias GO enrichment results (electronic supplementary material, S1). The annotated pathways for C. quinquefasciatus were downloaded from the KEGG Pathway database [25]. For each pathway, maximum and average sweep scores were determined and the number of genes with sweep score more than 4 was counted. The number of pathways in which each gene functioned was counted as a proxy for multifunctionality (related to the concept of pleiotropy).
(d). Statistical procedures
All statistical analyses including calculation of descriptive statistics, correlations, independence tests, regressions, ANOVA and PCA were done using SAS v. 9.3 and SAS JMP Pro v. 10.0.0.
More details on the methods can be found in electronic supplementary material, S1.
3. Results
(a). Diversity depends on biotype but population structure is shaped by geography
Whole-genome Illumina resequencing of the eight individually pooled populations resulted in 42× total coverage of the C. quinquefasciatus draft genome, identifying 6.7 M segregating sites among these populations. With more than 461 MB covered by Illumina sequences, this equated to roughly one segregating allele every 69 nucleotides. Table 1 shows average π and θ for 10 kb sliding windows across the genomes of the eight samples. The divergence of our C. pipiens populations from the C. quinquefasciatus reference genome ranged 0.6–1.8%. As expected, C. torrentium populations were more divergent (2.8–3.3%). In accordance with this, about 60–75% and 41–46% of total Illumina reads from C. pipiens and C. torrentium samples mapped onto the reference genome, respectively. The average sequencing depth across the whole covered segments of the genomes was 2–8× for our samples (the lowest ones belonging to C. torrentium). However, we only included the positions with coverages 4–40× in estimation of diversity and selection metrics. The average coverage across those positions in genic regions (whose output was fed into the PCA) ranged 8–19×. Only 7.3% and 5.4% of variance in the observed values of Tajima's D and Pool-hmm score for genes could be explained by coverage differences, respectively (linear regression R2, p < 0.0001). PCA on gene coverages did not produce any patterns similar to the geography-dependent clustering observed for the selection metrics. Thus, coverage did not seem to bias the calculation of population parameters significantly.
Differentiation among populations depended on geographical distance demonstrated by 10 kb sliding window genomic scans of Fst and phylogenetic tree structure (table 2). As expected, the largest distances belonged to C. torrentium versus C. pipiens comparisons. Within the C. pipiens complex, population structure corresponded to geographical proximity for both Russian and American samples. Neither Fst nor phylogenies suggested clustering based on habitat type (urban versus suburban) or biological form (molestus versus pipiens). PCA of allele frequencies mirrored this image (electronic supplementary material, S2). The reference sequence (C. quinquefasciatus) clustered closely with two C. pipiens samples only: A1 and S1 (table 2). The shared genomic regions contributing to this closeness, therefore, seem likely to have originated from recent local admixture rather than ancestral shared polymorphisms between C. pipiens and C. quinquefasciatus.
Table 2.
Population structure in the eight samples demonstrated through pairwise Fst values and phylogenetic frequency of neighbourhood. The lower half of the table reports the average Fst of 10 kb sliding windows in pairwise comparisons. The upper half shows in what percentage of the phylogenetic trees based on 10 kb windows each two populations are nearest neighbours.
| A1 | A4 | M1 | M2 | M4 | S1 | S2 | S3 | Ref | |
|---|---|---|---|---|---|---|---|---|---|
| A1 | − | 15.60 | 12.16 | 0.64 | 0.80 | 7.68 | 7.83 | 8.54 | 8.07 |
| A4 | 0.166 | − | 12.90 | 2.24 | 1.50 | 5.19 | 6.05 | 6.66 | 3.90 |
| M1 | 0.211 | 0.211 | − | 1.32 | 1.10 | 8.66 | 10.59 | 13.05 | 3.41 |
| M2 | 0.487 | 0.457 | 0.497 | − | 78.23 | 0.58 | 0.77 | 0.58 | 2.55 |
| M4 | 0.502 | 0.461 | 0.499 | 0.144 | − | 1.14 | 0.71 | 0.58 | 4.97 |
| S1 | 0.193 | 0.219 | 0.241 | 0.451 | 0.460 | − | 15.29 | 15.72 | 9.18 |
| S2 | 0.176 | 0.201 | 0.228 | 0.430 | 0.427 | 0.143 | − | 18.61 | 5.86 |
| S3 | 0.160 | 0.187 | 0.217 | 0.414 | 0.402 | 0.143 | 0.138 | − | 3.56 |
(b). Positive selection acts on non-coding and coding regions with non-synonymous mutations playing an important role
About 50–65% of the regions targeted by Pool-hmm with high confidence scores in C. pipiens populations coincided with coding sequences of annotated genes. Scanning the genome in 10 kb sliding windows, we found that the windows containing genic sequences were generally more likely to overlap with a sweep region (odds ratio = 1.19, independence χ2 test, p < 0.0001). In all of the C. pipiens samples, the total number of coding sequence polymorphisms per gene, as well as the number of either syn or nsyn polymorphisms, was smaller in sweep regions compared with the rest of the genome, compatible with the purported reduced variation around the sweep site (table 3). On the other hand, the ratio of nsyn/syn sites was always higher in the sweep regions. In C. torrentium samples, the trends were not exactly similar. The higher nsyn/syn ratio was still true; however, the correlation of each type of polymorphism with sweep status was either non-existent or slightly positive.
Table 3.
Coding sequence polymorphisms within and outside sweep regions. Sweep status 0: gene resides in a region not detected by Pool-hmm or detected with a score of less than 4; sweep status 1: gene resides in a region detected by Pool-hmm with a score of greater than or equal to 4; N: number of genes; total: genewise average of all polymorphisms in the coding DNA sequence; syn: genewise average of synonymous polymorphisms; nsyn: genewise average of non-synonymous polymorphisms; values in the correlation columns represent Spearman partial correlation coefficients controlled for gene length; n.s.: correlation not significant at p = 0.05; figures in parentheses: p-value of the correlation (p-value less than 0.0001 where not stated). Repeating the analysis with sweep scores greater than or equal to 2 or 8 as the cut-off (instead of 4) yielded very similar results (not shown). In calculation of the ratio, 0.5 was added to both syn and nsyn counts to avoid division by zero.
| genes with sweep status 0 |
genes with sweep status 1 |
correlation with sweep status 1 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sample | N | total | syn | nsyn | N | total | syn | nsyn | total | syn | nsyn | nsyn/syn ratio |
| A1 | 19 120 | 43.52 | 30.00 | 13.52 | 1186 | 17.50 | 7.94 | 9.56 | −0.1574 | −0.1985 | −0.0743 | 0.2147 |
| A4 | 19 129 | 40.91 | 29.27 | 11.64 | 1177 | 22.51 | 11.81 | 10.70 | −0.1210 | −0.1634 | −0.0263 | 0.1894 |
| M1 | 16 114 | 44.54 | 27.42 | 17.12 | 4192 | 29.90 | 14.93 | 14.97 | −0.1896 | −0.2407 | −0.0990 | 0.2441 |
| M2 | 18 376 | 26.48 | 15.03 | 11.44 | 1930 | 34.75 | 18.72 | 16.03 | 0.0330 | 0.0157 (0.0252) | 0.0563 | 0.0622 |
| M4 | 19 260 | 9.70 | 5.77 | 3.93 | 1046 | 10.23 | 5.81 | 4.42 | n.s. | n.s. | 0.0256 (0.0003) | 0.0202 (0.0040) |
| S1 | 17 713 | 39.48 | 28.62 | 10.87 | 2593 | 19.95 | 11.06 | 8.89 | −0.2221 | −0.2748 | −0.0934 | 0.2655 |
| S2 | 18 055 | 62.80 | 42.74 | 20.06 | 2251 | 30.11 | 16.29 | 13.81 | −0.2390 | −0.2828 | −0.1336 | 0.2647 |
| S3 | 17 510 | 61.82 | 40.12 | 21.70 | 2796 | 30.39 | 16.45 | 13.94 | −0.2766 | −0.3107 | −0.1852 | 0.2478 |
A summary of Tajima's D values and Pool-hmm scores can be found in electronic supplementary material, S3.
(c). Positive selection acts on a wide variety of biological functions in the Culex genome from chromatin organization to insecticide resistance
Table 3 shows the numbers of genes detected as sweep targets in each population. Based on both Pool-hmm and Tajima's D data, the most commonly enriched GO terms were related to chromatin and nucleosome structure and modification (electronic supplementary material, S4). The list of genes contributing to these terms included histones and chromatin remodelling factors (not shown). This was surprising given the well-known conservation of histone sequences, and motivated us to investigate the likely causes of selective sweeps in histones from a structure–function perspective in more detail (§§3e and 4e). Gene length proved not to be a significant confounder in the GO enrichment analysis (electronic supplementary material, S4).
Examination of the Pool-hmm results in the context of the functional pathways annotated for C. quinquefasciatus in the KEGG database demonstrated two important points: first, in every one of the eight populations more than half of the 129 pathways were affected by positive selection as they contained at least one gene with a sweep score more than 4. These pathways encompassed a large variety of functions including but not limited to amino acid biosynthesis, glycosphingolipid biosynthesis, signalling pathways (Notch, Jak-STAT and MAPK) and dorsoventral axis formation. Second, the number of pathways a gene functioned in was not correlated with the number of populations it was selected in nor the strength of selection when it happened.
Finally, we compiled a list of genes that have been shown by gene expression or mutant phenotyping studies to affect specific life-history traits of Culex (such as diapause, autogeny and mating behaviour), confer insecticide resistance or facilitate adaptation to temperature fluctuations. Histones and chromatin remodelling factors, ribosomal proteins, members of the P450 family, chaperonins and heat-shock proteins, vitellogenins and vitellogenin convertase, cadherins, superoxide dismutases and salivary proteins were notable genes with experimentally verified functional roles that were undergoing sweeps in multiple populations (electronic supplementary material, S4, p. 10).
(d). Many specific adaptations in the Culex pipiens genome happen locally
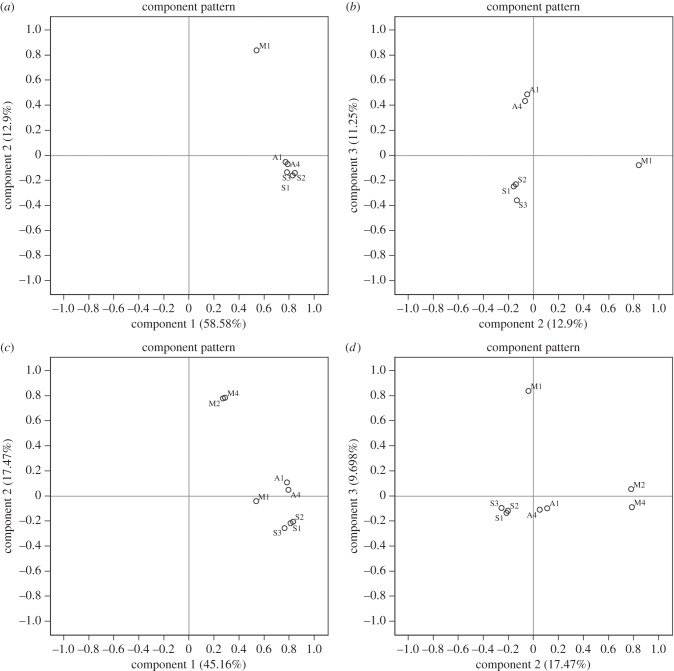
For both Pool-hmm and Tajima's D, we performed PCA once with the six C. pipiens samples only, and a second time with all of eight Culex samples included. The first principal component (PC) was always highly correlated with all of the sample labels and did not separate the samples from each other decisively (figure 1a,c). In the case of the six C. pipiens samples, the second and third PCs demonstrated the local nature of adaptation in the most conspicuous way, producing three distinct clusters containing the samples from Moscow, Aleksin and Sacramento (figure 1b). Including all eight samples, the combination of the second and the third components produced four clusters: one for the C. torrentium samples and three for each of the C. pipiens locations (figure 1d). The results of PCA on Tajima's D values were very similar (not shown).
Figure 1.
The first three PCs of Pool-hmm scores of genes from C. pipiens samples (a,b) or C. pipiens and C. torrentium together (c,d).
(e). Evolution of the conserved: parallel adaptation of histones in Culex pipiens and Culex torrentium
The 137 kb histone block that we examined closely showed reasonably high sweep scores in all of our eight populations. Compared with the non-sweep portions of the genome, this block had lower polymorphism but an increased nsyn/syn ratio (electronic supplementary material, S5), a trend similar to the one demonstrated in table 3 for sweep genes in general. Most of the nsyn polymorphisms occurred at positions where there was no variation among paralogues in the reference, indicating they were bona fide polymorphic sites as opposed to artefacts of mis-mapping onto paralogues (data not shown).
We found the linker histone (H1) to be the most polymorphic among all histone genes, consistent with the fact that it is the least conserved among histones (electronic supplementary material, S5). The basic structure of histone H1 consists of three main domains: a lysine-rich C-terminal domain that binds to linker DNA (the C domain), a central globular domain with a winged helix motif that binds to the nucleosomal DNA (the G domain), and an N-terminal domain whose function is not very well understood (the N domain) [26]. Generally, the globular domain is the most evolutionarily conserved (across taxa and among paralogues) and the N-terminal domain is the most variable [27].
Examination of polymorphic sites in H1 genes confirmed our expectations based on the known evolutionary patterns. The G and N domains showed the lowest and highest propensity for nsyn mutations, respectively (electronic supplementary material, S6, p. 2). The nsyn mutations were quite uncommon in regular secondary structures and in buried residues (electronic supplementary material, S6, pp. 3–4), although these two states tended to coincide with the globular domain, confounding the analysis. Charge-altering mutations were also exceedingly rare in buried residues (electronic supplementary material, S6, p. 7). About 90% of the changes adding or removing Ser/Thr occurred in exposed residues (electronic supplementary material, S6, p. 8) making them potential targets for epigenetic modification.
Polymorphisms converting other amino acids to Pro were vastly overrepresented in the histone block in all of our eight samples (p < 0.0001). We did not test for other amino acid conversions, so there may well be other cases of overrepresentation or underrepresentation in the data that we did not capture. Remarkably, the positions of Pro-permissive mutations in the conserved G domain were much more consistent across populations compared with the N domain. Among the Pro-permissive residues in the G domain, 78.6% showed Pro mutations in multiple populations. By contrast, only 17.5% of Pro-permissive residues in the N domain showed Pro mutations in more than one population (electronic supplementary material, S6, p10). This suggested that Pro mutations in the N domain were mostly neutral or semi-neutral segregating variants in random positions, whereas at least some of the Pro mutations in the G domain happened at specific positions and were probably favoured by selection. Visual inspection of the Pro polymorphisms in G and C domains suggested that almost all of them occurred in irregular parts that connected the regular secondary structures or were located on the domain boundaries.
Generally, nsyn polymorphisms and specifically those involving Pro occurred at similar residues in same-species populations, but were independently positioned when two populations of different species were compared (electronic supplementary material, S7). This verified the expectation of efficient isolation of the C. pipiens and C. torrentium gene pools. Therefore, whatever the evolutionary force behind the overabundance of conversions to Pro might be, it seems to be happening independently and in parallel in C. pipiens and C. torrentium.
4. Discussion
(a). Diversity level and population structure
The three pure biotype molestus populations (A1, M1 and S1) showed reduced variation compared with the one pure biotype pipiens population (A4) (table 1) consistent with previous findings suggesting founder effects during the establishment of molestus populations [28–30]. The dependence of population structure on localities (table 2) agrees with previous reports on US populations [8,30,31] (but also see [32]) but contrasts with the distinct f. molestus versus f. pipiens dichotomy in northern and central Europe [8,29,33].
(b). Mapping efficiency and coverage effects
Theoretical and computational tools are still being developed for pool-seq data analysis [34–36]. It has been proposed that estimates of allele frequencies and population genetic parameters can be improved with increased sequencing depth (up to 20–30×, but not above that [34,36]) and pooling large enough numbers of individuals (about 25 diploid individuals or more [36]). The recommendation of coverage threshold of 20–30× in [34,36] was made to ensure faithful estimates of ‘single-site’ allele frequencies. When diversity or selection parameters are calculated by averaging over 10 kb windows or the length of genes—which are typically several hundred bases long—even lower coverages ought to yield satisfactory results. Accordingly, in a study using simulated data of up to 2% divergence from reference sequence, it has been suggested that direct estimation of population genetic parameters without SNP and genotype calling yields reasonably good results even at low coverages (2–4×) [37]. The methods we used for estimation of diversity and selection strength worked directly on base counts from sequencing reads without any intermediate SNP or genotype calling steps. The coverage in our included genic positions ranged 8–19×, which lay between the two above recommendations. Nevertheless, the relatively low sequencing depth in some regions could have resulted in false negatives in the detection of selection targets, because they were disregarded by our 4–40× filter.
(c). Signature of directional selection in coding sequences
It is well known that some AFS-based metrics of positive selection are sensitive to demography or behave similarly under purifying selection and positive selection (e.g. Tajima's D) [38]. Some false positives might exist among the Pool-hmm hits too, but we are optimistic that most of the detected sweeps are likely to be true positives. Pool-hmm identifies sweep regions based on changes in the AFS regardless of the annotatory features of the alleles; thus, the combination of lower levels of variation and higher ratios of nsyn/syn (table 3) provides independent support for the action of positive selection. Purifying selection would reduce total variation but would also decrease the nsyn/syn ratio. Relaxation of selection would increase the nsyn/syn ratio but would also elevate total variation. Still, alternative scenarios can be envisaged that would produce the kind of pattern we see in our data; for example, a severe decline in population size (a bottleneck) could result in reduced total variation and selection relaxation at the same time. The mathematical models from which the Pool-hmm method was devised were shown through simulations and tests on real data to be relatively robust to several types of demographic changes [18,19]. But unfortunately in the absence of ecological data on our Culex populations, the results of such simulations cannot be confidently extended to them. Further work will be required to disentangle true sweep signals from potential confounders.
A closer look at table 3 reveals that the higher nsyn/syn ratio in sweep regions resulted mainly from depletion of syn mutations. The reason is that in general most of the neutrally segregating variation is syn; so when linked variation is removed from around a sweep site, the reduction in syn variation will be larger. A possible explanation for more abundant nsyn mutations in sweep genes in M2 and M4 when no or a weaker correlation is observed for syn mutations is that a larger proportion of sweeps in C. torrentium were fostered by new mutations (hard sweeps), whereas most of C. pipiens sweeps depended on standing variation (soft sweeps). This scenario seems particularly likely in the case of the M4 population, which has very low levels of standing variation—reflected by smallest number of polymorphic sites in non-swept genes (table 3) and smallest π and θ values (table 1)—providing little raw material for positive selection. Accordingly, M4 shows the lowest number of detected sweep events (table 3). In the Moscow region, populations of C. torrentium have expanded rapidly in the past 10 years [39] indicating that small genetic diversity within M2 and M4 may be owing to founder effect.
An excess of nsyn to syn ‘fixed’ differences (divergence) among multiple taxa is often used as a basis for inference of recurrent positive selection [40–42]. The sites identified by those tests are likely to be the direct targets of selection and emerge owing to positive selection on the nsyn mutations. By contrast, what we present in table 3 is the ratio of nsyn/syn ‘segregating polymorphisms’ (not fixed differences) averaged over the approximately 20 k genes in individual populations, not for single-nucleotide positions across populations or taxa. What we have shown here is that an excess of nsyn/syn ‘segregating polymorphisms’ concurs with selective sweeps.
(d). Principal component analysis on selection metrics as a method of detecting differential selection
PCA across samples on Pool-hmm scores compares the strength of recent adaptive evolution on genes, whereas PCA on Tajima's D captures the contrast between balancing selection (D > 0) and positive or purifying selection (D < 0) against neutral evolution (D = 0) [43]. In either case, the strong correlation of PC 1 with all sample labels meant that most of the genes performed similar functions and were thus selected congruently across the tested populations. This would be expected as our populations all belonged to the same species or closely related ones. On the other hand, small fractions of genes were expected to underlie adaptations unique to each specific population or groups of populations and were supposed to contribute to creation of second, third, fourth and further PCs. In the PCA on all eight samples, the second component separated C. pipiens samples from C. torrentium ones, indicating that the differential selection of genes was driven primarily by species differences (figure 1c). The portion of variance explained by the second component in the eight sample analysis (17.47%) was higher than that explained by any other second or third component, implying that interspecific differences were greater than those caused by geographical isolation of conspecifics.
PC patterns of sweep scores and allele frequencies share a common feature: they are shaped first by species identity and then by geographical distance. Migration between geographically close populations may have contributed to the similarity of allele frequencies and consequently, the detected targets of selection; so do the PCs of Pool-hmm merely reflect population structure? The answer is interestingly NO. First, we need to point out a key difference: in contrast with sweep scores, we do not see a PC1 correlating highly with all of the population labels with allele frequencies. The reason is that we did the PCA only on the polymorphic positions to save on computation time. The majority of genomic positions were fixed for the same allele across all populations and were filtered out. So, for allele frequencies, significant differentiation among populations starts with PC1. PC1 and PC2 of allele frequencies are qualitatively comparable to PC2 and PC3 of sweep scores. Comparing PCA of allele frequencies and Pool-hmm score shows that the order of clustering among the six C. pipiens populations is different between them. PC1 of allele frequencies separates Moscow and Aleksin from Sacramento (electronic supplementary material, S2). Data in table 2 confirm that the allele frequencies of the Moscow and Aleksin populations have more similarity than either do to Sacramento populations. On the contrary, PC1 and particularly PC2 of sweep scores put Aleksin populations closer to Sacramento than Moscow (figure 1). This means that within the C. pipiens species, sweep status does not follow population structure. Besides, Fst between collocal populations is just slightly smaller than Fst between different localities (table 2); for example, Fst between A1 and A4 is 0.166, whereas Fst between A1 and Sacramento populations is in the range of 0.160–0.193. This makes it unlikely that gene flow between collocal populations is sufficiently strong to create similar AFS in them and to lead to corresponding sweep hits. Finally, it should be noted that allele frequency PCA plots were created from biallelic positions (a fraction of polymorphic positions) regardless of gene content; so, presumably most of them came from non-genic parts because only approximately 110 MB out of the approximately 579 MB of the reference genome is genic sequence (including introns), not to mention that polymorphism is expected to be lower in genic sequences on average. On the other hand, Pool-hmm PCA plots used the sweep scores from genic sequences, consisting of monomorphic as well as polymorphic sites. So the two sets of PCA plots represent two potentially overlapping but completely different subsets of genomic positions. Without a formal significance test, it is not possible to statistically disprove or quantitatively evaluate the proposition that demography affects our ‘detected’ selective status. What we can ascertain definitely is that Pool-hmm and Tajima's D do not absolutely follow Fst, phylogenetic neighbourhood or allele frequency PCs. In other words, the variation in neither of the former two can be completely explained by any of the latter three (contribution to the signal of selection is possible but it is never 100%).
We performed PCA on Pool-hmm scores and Tajima's D but it can be as effectively applied to any other selection statistics. There are a great number of indicators of natural selection (including those based on amino acid substitutions, length of haplotype homozygosity, etc.), each optimized to identify certain types of selection and within certain time depths (reviewed, for example, in [44]). PCA will make it possible to use any of them comparatively across populations or taxa to characterize the patterns of differential selection.
From an epidemiological perspective, the dependence of population structure on geographical distribution and strong local signature of adaptations suggest that vector control schemes should be informed by population-specific data rather than presumed global properties of the Culex species complex. For instance, rapid evolution of many insecticide resistance genes indicates that the efficacy of insecticides on each Culex population will have to be tested frequently and on specimens from the same locality with as small gridding as possible.
(e). The special case of histones
Histones are known to be among the most evolutionary conserved genes, although they have been reported to have responded to recent directional selection [45,46]. However, the selective pressures that drive their evolution are not well understood.
Analysis of the biochemical versus structural properties of amino acid residues at polymorphic sites suggested that Pro mutations in the G and C domains of H1 probably acted to modify the orientation of regular structures with respect to each other in space or adjust the rigidity/flexibility of the existing structures without disrupting the basic fold of the protein or its function.
Because seasonal variation in temperature is more substantial in temperate climates compared with tropical or sub-tropical zones, sub-functionalization or neofunctionalization of duplicated genes [47] to accommodate these new environmental conditions seems like a possible scenario for Culex histone evolution. This scenario is corroborated by the heterogeneous distribution of polymorphic sites among paralogues in all of the studied populations (independence χ2, p < 0001; caution: test statistic might have been inflated owing to small number of expected polymorphisms at some loci).
(f). The marks of south to north range expansion
Culex pipiens originated in North Africa and then spread out to colonize other parts of the world [48]. We found certain sweep events that might have helped them adapt to the new environmental conditions (electronic supplementary material, S4).
Heat-shock proteins and chaperonins are crucial for proper protein folding in the cells and also contribute to adaptation to living at high and low temperatures [49,50]. Interestingly, expression of a specific chaperonin component has been reported to be crucial for cold resistance in diapausing members of the Onion Maggot Delia antiqua [51,52]. Positive selection on chaperonins may be a general response to colder climate or bigger seasonal fluctuations in temperature; however, the significantly higher number of sweep genes in C. pipiens compared with C. torrentium makes it more likely to be a specialized adaptation to winter diapause in the colder habitats. Biotype molestus mosquitoes do not undergo diapause during winter, but they have branched off from the pipiens form very recently, so it should not be surprising that they still bear the genomic signatures of diapause-related adaptations.
Chromatin-related factors including histones showed signals of strong sweep in all of the tested populations, and constituted the most significantly enriched GO terms. As major modulators of gene expression, they are known to contribute substantially to adaptation to new environmental challenges [53,54]. Positive selection in several chromatin remodelling genes has been associated with range expansion from tropical to temperate environments in Drosophila [55,56]; it is then plausible to suggest that these genes may have played an equally important adaptive role during the spread of Culex from tropical North Africa to temperate and cold habitats. Interestingly, many regulatory sequences and unannotated sequences have been reported to be highly differentiated between tropical and temperate Drosophila populations, presumably contributing to adaptation to new environmental conditions [56]. Sweeps in histones and chromatin modifiers along with the large proportion of sweeps occurring in non-coding regions emphasize the significance of gene expression regulation as a mechanism of adaptive evolution in Culex.
Supplementary Material
Acknowledgements
We thank Remo Rohs of University of Southern California and Anandasankar Ray of University of California at Riverside for insightful comments about structural properties of histones and physiological significance of sweep hits, Brittany M. Nelms of Lake County Mosquito and Vector Control District for collection of mosquitoes from Sacramento, CA, and John Azizian of University of Southern California for his help in literature survey.
Data accessibility
The Illumina data are available at NCBI under the BioProject PRJNA284197.
Authors' contribution
S.V.N. conceived the project idea, designed the experiments and facilitated the collaboration. H.A. planned and performed the population genetic and statistical analyses and wrote the manuscript. W.K.R. and V.A.S. provided and morphologically identified the mosquito samples from Sacramento and Moscow/Aleksin regions, respectively. S.L. prepared genomic libraries for sequencing. P.L.C. quality-controlled and mapped the sequence reads onto the reference genome and contributed to the population structure tests. All authors edited the manuscript and approved the final content.
Competing interests
The authors declare that this research was conducted in the absence of any competing interests.
Funding
This work was funded by National Institute of Health grant no. GM098741. The work by S.L. was supported by Russian Scientific Foundation grant no. 14-14-00330.
References
- 1.Dawkins R, Krebs JR. 1979. Arms races between and within species. Proc. R. Soc. Lond. B 205, 489–511. ( 10.1098/rspb.1979.0081) [DOI] [PubMed] [Google Scholar]
- 2.Gandon S, Michalakis Y. 2002. Local adaptation, evolutionary potential and host–parasite coevolution: interactions between migration, mutation, population size and generation time. J. Evol. Biol. 15, 451–462. ( 10.1046/j.1420-9101.2002.00402.x) [DOI] [Google Scholar]
- 3.Nelms BM, Macedo PA, Kothera L, Savage HM, Reisen WK. 2013. Overwintering biology of Culex (Diptera: Culicidae) mosquitoes in the Sacramento Valley of California. J. Med. Entomol. 50, 773–790. ( 10.1603/ME12280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arensburger P, et al. 2010. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science 330, 86–88. ( 10.1126/science.1191864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farajollahi A, Fonseca DM, Kramer LD, Kilpatrick AM. 2011. ‘Bird biting’ mosquitoes and human disease: a review of the role of Culex pipiens complex mosquitoes in epidemiology. Infect. Genet. Evol. 11, 1577–1585. ( 10.1016/j.meegid.2011.08.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strickman D, Fonseca DM. 2012. Autogeny in Culex pipiens complex mosquitoes from the San Francisco Bay Area. Am. J. Trop. Med. Hyg. 87, 719–726. ( 10.4269/ajtmh.2012.12-0079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spielman A. 2001. Structure and seasonality of nearctic Culex pipiens populations. Ann. N. Y. Acad. Sci. 951, 220–234. ( 10.1111/j.1749-6632.2001.tb02699.x) [DOI] [PubMed] [Google Scholar]
- 8.Fonseca DM, Keyghobadi N, Malcolm CA, Mehmet C, Schaffner F, Mogi M, Fleischer RC, Wilkerson RC. 2004. Emerging vectors in the Culex pipiens complex. Science 303, 1535–1538. ( 10.1126/science.1094247) [DOI] [PubMed] [Google Scholar]
- 9.Gomes B, et al. 2009. Asymmetric introgression between sympatric molestus and pipiens forms of Culex pipiens (Diptera: Culicidae) in the Comporta region, Portugal. BMC Evol. Biol. 9, 262 ( 10.1186/1471-2148-9-262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornel A, Lee Y, Fryxell RT, Siefert S, Nieman C, Lanzaro G. 2012. Culex pipiens Sensu Lato in California: a complex within a complex? J. Am. Mosq. Control Assoc. 28, 113–121. ( 10.2987/8756-971X-28.4s.113) [DOI] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bahnck CM, Fonseca DM. 2006. Rapid assay to identify the two genetic forms of Culex (Culex) pipiens L. (Diptera: Culicidae) and hybrid populations. Am. J. Trop. Med. Hyg. 75, 251–255. [PubMed] [Google Scholar]
- 13.Hesson JC, Lundström JO, Halvarsson P, Erixon P, Collado A. 2010. A sensitive and reliable restriction enzyme assay to distinguish between the mosquitoes Culex torrentium and Culex pipiens. Med. Vet. Entomol. 24, 142–149. ( 10.1111/j.1365-2915.2010.00871.x) [DOI] [PubMed] [Google Scholar]
- 14.Remolina SC, Chang PL, Leips J, Nuzhdin SV, Hughes KA. 2012. Genomic basis of aging and life-history evolution in Drosophila melanogaster. Evolution 66, 3390–3403. ( 10.5061/dryad.94pv0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jalvingh KM, Chang PL, Nuzhdin SV, Wertheim B. 2014. Genomic changes under rapid evolution: selection for parasitoid resistance. Proc. R. Soc. B 281, 20132303 ( 10.1098/rspb.2013.2303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. ( 10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 17.Kofler R, Orozco-terWengel P, De Maio N, Pandey RV, Nolte V, Futschik A, Kosiol C, Schlötterer C. 2011. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS ONE 6, e15925 ( 10.1371/journal.pone.0015925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, Bustamante C. 2005. Genomic scans for selective sweeps using SNP data. Genome Res. 15, 1566–1575. ( 10.1101/gr.4252305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boitard S, Schlötterer C, Futschik A. 2009. Detecting selective sweeps: a new approach based on hidden Markov models. Genetics 181, 1567–1578. ( 10.1534/genetics.108.100032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boitard S, Schlötterer C, Nolte V, Pandey RV, Futschik A. 2012. Detecting selective sweeps from pooled next-generation sequencing samples. Mol. Biol. Evol. 29, 2177–2186. ( 10.1093/molbev/mss090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boitard S, Kofler R, Françoise P, Robelin D, Schlötterer C, Futschik A. 2013. Pool-hmm: a Python program for estimating the allele frequency spectrum and detecting selective sweeps from next generation sequencing of pooled samples. Mol. Ecol. Resour. 13, 337–340. ( 10.1111/1755-0998.12063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Y, et al. 2010. Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J. Cell Biol. 189, 407–415. ( 10.1083/jcb.201001148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harshman SW, Young NL, Parthun MR, Freitas MA. 2013. H1 histones: current perspectives and challenges. Nucleic Acids Res. 41, 9593–9609. ( 10.1093/nar/gkt700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Q, Wang X-J. 2008. GOEAST: a web-based software toolkit for gene ontology enrichment analysis. Nucleic Acids Res. 36, W358–W363. ( 10.1093/nar/gkn276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–D114. ( 10.1093/nar/gkr988) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brennan RG. 1993. The winged-helix DNA-binding motif: another helix-turn-helix takeoff. Cell 74, 773–776. ( 10.1016/0092-8674(93)90456-Z) [DOI] [PubMed] [Google Scholar]
- 27.Kasinsky HE, Lewis JD, Dacks JB, Ausió J. 2001. Origin of H1 linker histones. FASEB J. 15, 34–42. ( 10.1096/fj.00-0237rev) [DOI] [PubMed] [Google Scholar]
- 28.Byrne K, Nichols RA. 1999. Culex pipiens in London underground tunnels: differentiation between surface and subterranean populations. Heredity 82, 7–15. ( 10.1038/sj.hdy.6884120) [DOI] [PubMed] [Google Scholar]
- 29.Becker N, Jöst A, Weitzel T. 2012. The Culex pipiens complex in Europe. J. Am. Mosq. Control Assoc. 28, 53–67. ( 10.2987/8756-971X-28.4s.53) [DOI] [PubMed] [Google Scholar]
- 30.Kothera L, Godsey M, Mutebi J-P, Savage HM. 2012. A comparison of above-ground and below-ground populations of Culex pipiens pipiens in Chicago, Illinois, and New York City, New York, using 2 microsatellite assays. J. Am. Mosq. Control Assoc. 28, 106–112. ( 10.2987/8756-971X-28.4.106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kothera L, Godsey M, Mutebi J-P, Savage HM. 2010. A comparison of aboveground and belowground populations of Culex pipiens (Diptera: Culicidae) mosquitoes in Chicago, Illinois, and New York City, New York, using microsatellites. J. Med. Entomol. 47, 805–813. ( 10.1603/ME10031) [DOI] [PubMed] [Google Scholar]
- 32.Kent RJ, Harrington LC, Norris DE. 2007. Genetic differences between Culex pipiens f. molestus and Culex pipiens pipiens (Diptera : Culicidae) in New York. J. Med. Entomol. 44, 50–59. ( 10.1093/jmedent/41.5.50) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weitzel T, Collado A, Jöst A, Pietsch K, Storch V, Becker N. 2009. Genetic differentiation of populations within the Culex pipiens complex and phylogeny of related species. J. Am. Mosq. Control Assoc. 25, 6–17. ( 10.2987/08-5699.1) [DOI] [PubMed] [Google Scholar]
- 34.Zhu Y, Bergland AO, González J, Petrov DA. 2012. Empirical validation of pooled whole genome population re-sequencing in Drosophila melanogaster. PLoS ONE 7, e41901 ( 10.1371/journal.pone.0041901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gautier M, et al. 2013. Estimation of population allele frequencies from next-generation sequencing data: pool-versus individual-based genotyping. Mol. Ecol. 22, 3766–3779. ( 10.1111/mec.12360) [DOI] [PubMed] [Google Scholar]
- 36.Ferretti L, Ramos-Onsins SE, Pérez-Enciso M. 2013. Population genomics from pool sequencing. Mol. Ecol. 22, 5561–5576. ( 10.1111/mec.12522) [DOI] [PubMed] [Google Scholar]
- 37.Nevado B, Ramos-Onsins SE, Perez-Enciso M. 2014. Resequencing studies of nonmodel organisms using closely related reference genomes: optimal experimental designs and bioinformatics approaches for population genomics. Mol. Ecol. 23, 1764–1779. ( 10.1111/mec.12693) [DOI] [PubMed] [Google Scholar]
- 38.Nielsen R. 2005. Molecular signatures of natural selection. Annu. Rev. Genet. 39, 197–218. ( 10.1146/annurev.genet.39.073003.112420) [DOI] [PubMed] [Google Scholar]
- 39.Vinogradova EB, Shaikevich EV, Ivanitsky AV. 2007. A study of the distribution of the Culex pipiens complex (Insecta: Diptera: Culicidae) mosquitoes in the European part of Russia by molecular methods of identification. Comp. Cytogenet. 1, 129–138. [Google Scholar]
- 40.Hudson RR, Kreitman M, Aguade M. 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreitman M, Hudson RR. 1991. Inferring the evolutionary histories of the Adh and Adh-dup loci in Drosophila melanogaster from patterns of polymorphism and divergence. Genetics 127, 565–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macpherson JM, Sella G, Davis JC, Petrov DA. 2007. Genomewide spatial correspondence between nonsynonymous divergence and neutral polymorphism reveals extensive adaptation in Drosophila. Genetics 177, 2083–2099. ( 10.1534/genetics.107.080226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabeti PC, et al. 2006. Positive natural selection in the human lineage. Science 312, 1614–1620. ( 10.1126/science.1124309) [DOI] [PubMed] [Google Scholar]
- 45.Malik HS, Henikoff S. 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157, 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berdnikov VA, Bogdanova VS, Rozov SM, Kosterin E. 1993. Geographic patterns of histone HI allelic frequencies formed in the course of Pisum sativum L. (pea) cultivation. Heredity 71, 199–209. ( 10.1038/hdy.1993.125) [DOI] [Google Scholar]
- 47.Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. 2002. Selection in the evolution of gene duplications. Genome Biol. 3, research0008.1–0008.9. ( 10.1186/gb-2002-3-2-research0008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harbach RE. 2011. Classification within the cosmopolitan genus Culex (Diptera: Culicidae): the foundation for molecular systematics and phylogenetic research. Acta Trop. 120, 1–14. ( 10.1016/j.actatropica.2011.06.005) [DOI] [PubMed] [Google Scholar]
- 49.Fujiwara S, Aki R, Yoshida M, Higashibata H, Imanaka T, Fukuda W. 2008. Expression profiles and physiological roles of two types of molecular chaperonins from the hyperthermophilic archaeon Thermococcus kodakarensis. Appl. Environ. Microbiol. 74, 7306–7312. ( 10.1128/AEM.01245-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Somer L, Shmulman O, Dror T, Hashmueli S, Kashi Y. 2002. The eukaryote chaperonin CCT is a cold shock protein in Saccharomyces cerevisiae. Cell Stress Chaperones 7, 47–54. () [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kayukawa T, Chen B, Miyazaki S, Itoyama K, Shinoda T, Ishikawa Y. 2005. Expression of mRNA for the t-complex polypeptide-1, a subunit of chaperonin CCT, is upregulated in association with increased cold hardiness in Delia antiqua. Cell Stress Chaperones 10, 204–210. ( 10.1379/CSC-106R.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kayukawa T, Ishikawa Y. 2009. Chaperonin contributes to cold hardiness of the onion maggot Delia antiqua through repression of depolymerization of actin at low temperatures. PLoS ONE 4, e8277 ( 10.1371/journal.pone.0008277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pecinka A, Mittelsten Scheid O. 2012. Stress-induced chromatin changes: a critical view on their heritability. Plant Cell Physiol. 53, 801–808. ( 10.1093/pcp/pcs044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levine MT, Eckert ML, Begun DJ. 2011. Whole-genome expression plasticity across tropical and temperate Drosophila melanogaster populations from Eastern Australia. Mol. Biol. Evol. 28, 249–256. ( 10.1093/molbev/msq197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levine MT, Begun DJ. 2008. Evidence of spatially varying selection acting on four chromatin-remodeling loci in Drosophila melanogaster. Genetics 179, 475–485. ( 10.1534/genetics.107.085423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolaczkowski B, Kern AD, Holloway AK, Begun DJ. 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187, 245–260. ( 10.1534/genetics.110.123059) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Illumina data are available at NCBI under the BioProject PRJNA284197.