Abstract
Over three quarters of receptor tyrosine kinase (RTK) families are reported to have intracellular localization in response to environmental stimuli. Internalized RTK can bind to non-canonical substrates and affect various cellular processes. Many of the intracellular RTKs exist as fragmented forms that are generated by γ-secretase cleavage of the full-length receptor, shedding, alternative splicing, or alternative translation initiation. Soluble RTK fragments are stabilized and intracellularly transported into subcellular compartments, such as the nucleus, by binding to chaperon or transcription factors while membrane-bound RTKs (full-length or truncated) are transported from the plasma membrane to the endoplasmic reticulum (ER) through the well-established Rab- or clathrin adaptor protein (AP)-coated vesicle retrograde trafficking pathway. Subsequent nuclear transport of membrane-bound RTK can occur via two pathways, INFS and INTERNET, with the former characterized by the release of receptors from ER into the cytosol and the latter by the release of membrane-bound transport from the ER into the nucleoplasm through the inner nuclear membrane. While most non-canonical intracellular RTK signaling is related to transcriptional regulation, there may be other functions that have yet to be discovered. In this review, we summarize the proteolytic processing, intracellular trafficking, and nuclear functions of RTKs and discuss how they promote cancer progression and their clinical implications.
Keywords: Receptor tyrosine kinase, proteolytic cleavage, nuclear translocation, intracellular trafficking
Graphical Abstract

1. Introduction
Receptor protein kinases play critical roles in the regulation of normal biological processes in response to stimuli. However, dysregulation of these kinases, especially when their substrates are also kinases, can further amplify aberrant signaling that leads to diseases such as cancers. Among the receptor protein kinases, receptor tyrosine kinases (RTKs) are thought to be the largest catalytic membrane receptor family, and the development and progression of many human diseases have been attributed to the dysregulation of these RTKs. There are a number of well-characterized and FDA-approved targeted cancer therapies against RTKs, including monoclonal antibodies and small-molecule tyrosine kinase inhibitors (TKIs) [1].
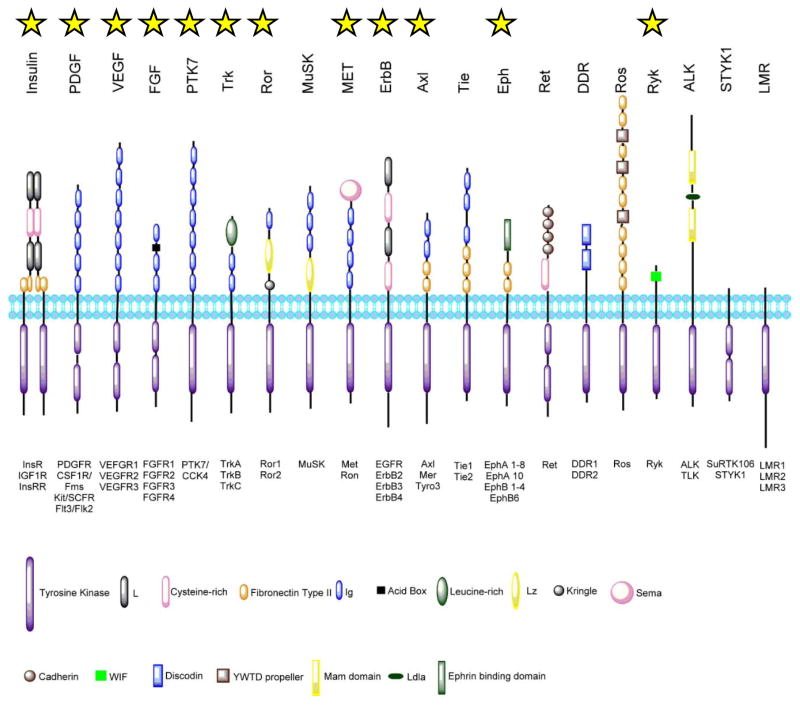
The protein structures, mechanisms of activation, and downstream regulatory pathways of RTKs are highly conserved from C. elegans to humans [2]. In humans, there are 58 known RTKs classified into 20 subfamilies [3]. A prototypical RTK consists of an extracellular ligand-binding domain, a single transmembrane helix domain, a juxtamembrane regulator domain, a cytoplasmic domain containing the tyrosine kinase (TK) domain, and a carboxyl terminal (C-term) domain (Figure 1). Discovered as cell surface proteins, RTKs are membrane receptors that have high affinity for extracellular growth factors, cytokines, and hormones [4]. Overexpression and mutation of RTKs as well as dysregulation of their signaling lead to human diseases, including cancer [5]. While therapeutics against RTKs and their downstream molecules have demonstrated effectiveness in treating cancer, acquired drug resistance and toxicity have also been observed [6, 7]. Thus, investigations are currently underway to further our understanding of RTK signaling toward the development of more effective drugs.
Figure 1. Receptor tyrosine kinase subfamilies.
Among receptor tyrosine kinases, 20 subfamilies are identified in human genome. Those that contain receptor reported as MRIN are marked by a yellow star. The subfamilies are illustrated here with their extracellular domain structures marked accordingly in the key. The intracellular tyrosine kinase domain is shown as purple rectangle. The family members are noted below each subfamily. The lipid bilayer represents the plasma membrane, and the schematic here does not reflect the actual scale. (This figure is modified from the work of Lemmon and Schlessinger [3]).
In the past, a majority of studies have focused on canonical RTK signaling from the cell surface via the following sequential events (Figure 2): (1) The receptor binds with its ligand and then (2) undergoes conformational changes and forms homo- or hetero-oligomers that are essential to activate its kinase activity. The tyrosine kinase domain then undergoes trans autophosphorylation that greatly elevates the receptor’s catalytic activity. (3) The phosphotyrosine residues serve as docking site for cytoplasmic adaptor proteins containing Src homology-2 (SH2) and phosphotyrosine-binding (PTB) domain to form signal transduction complex that determines and initiates the corresponding signaling cascade to regulate cellular processes in response to ligand stimulation [3, 8–13].
Figure 2. Canonical RTK signaling cascade.
The canonical RTK signaling begins with (1) the receptor binding with its ligand. (2) The receptor then undergoes oligomerization and trans auto-phosphorylation. (3) The phosphor-tyrosine residues serve as docking sites for the secondary messenger proteins containing either SH2 and/or PTB domain (green) that are subjected to phosphorylation by the RTK. (4) The secondary messenger proteins recruit and activate downstream proteins (burgundy) which serve as envoys delivering signals into the nucleus to regulate gene transcription.
Interestingly, members of the RTK subfamilies are also present in the nucleus and are referred to as membrane receptor in nucleus or MRIN [14, 15]. Accumulating evidence indicates at least 12 RTK families contain MRINs that exist either as holoreceptor or truncated form with novel non-canonical functions in transcriptional regulation, DNA damage and repair, and cell proliferation and invasion [15–19]. In various cancer types, nuclear RTK expression is associated with poor prognosis [20–23]. Generally, after ligand-induced activation, membrane-bound MRINs are internalized through endocytosis from the cell surface and then transported into the nucleus. Moreover, RTKs can be proteolytically cleaved to release an active RTK fragment that is also transported from the cell membrane to subcellular compartments, including the nucleus.
Drugs that target aberrantly expressed RTKs are designed base on the canonical RTK functions, but they can also lead to drug resistance by inducing RKT internalization. For example, EGFR inhibitors, gefitinib and cetuximab, have both been reported to enhance the levels of nuclear EGFR (nEGFR), which in turn lead to the development of resistance [24–27]. Huang et al. observed increased EGFR in the nucleus of gefitinib-resistant cancer cells and found that nEGFR promotes the transcription of the multidrug resistance efflux transporter breast cancer-resistance protein (BCRP/ABCG2) [25]. Thus, identifying the mechanisms underlying RTK translocation may enhance our understanding of drug resistance. In this chapter, we summarize the proteolytic cleavage processing, mechanisms of nuclear transport, and non-canonical function of RTKs. We mainly focus on members of the ErbB family while briefly discussing other MRINs.
2. Generation of intracellular RTK fragments
Intracellular RTKs do not always exist in the holoreceptor form. Rather, they can be proteolytically cleaved to release an active RTK fragment that is also transported from the cell membrane to subcellular compartments. Cleavage at the transmembrane domain of RTK, which yields an intracellular domain (ICD), can occur by different mechanisms in response to various stimuli [14]. These include caspase-, splicing- and secretase-dependent pathways. Among them, RTK fragments generated by secretase-mediated and splicing are reported to have nuclear localization and functions.
2.1 Caspase-dependent RTK fragment
Many RTKs, including Ret, c-MET, tropomyosin receptor kinase (Trk)-C, anaplastic lymphoma kinase (ALK), ErbB-1/epidermal growth factor receptor (EGFR), ErbB-2/HER-2, and Eph-A4, are subjected to caspase cleavage. Except for EGFR, most of the caspase-dependent intracellular fragments have a direct role in determining the cell fate (reviewed in [28]). Most caspase-mediated RTK cleavage generates detectable fragments that lack the extracellular domain as well as the C-terminal docking sites for downstream signaling proteins. Examples of the caspase-cleaved RTK fragments include intact TK domain of Ret and MET, truncated TK domain of Trk-C, ALK and Eph-A4, and truncated C-term domain of EGFR and ErbB-2. Of these, Ret, MET, Trk-C, and ErbB-2 fragments are also pro-apoptotic factors as they can promote apoptosis without ligand activation [28]. RTK phosphorylation has been shown to inhibit caspase-dependent cleavage [29], suggesting that RTK-addicted cells may rely on not only ligand-stimulated canonical signaling but also inhibition of caspase-related pro-apoptotic factors.
2.2 Alternative splicing RTK variants
Aside from caspase-mediated cleavage, truncated RTKs can also be generated from alternative mRNA splicing. Alternative pre-mRNA splicing is a process that generates mRNA variants of the same gene, yielding structurally and functionally different proteins. Many of the RTK splice variants have distinct functions from the holoreceptor and are also implicated in cancer development [30]. Below we briefly describe some examples of alternative splicing-generated RTK variants.
Both EGFR and ErbB-3 of the ErbB family are known to have alternative splice variants. There are four EGFR splice variants, EGFRvI, II, III, IV, and an in-frame splice variant known as mini-LEEK (mLEEK). Among them, EGFRvIII and mLEEK are known to localize to the nucleus to regulate cellular responses. EGFRvIII is produced by deletion of exons 2–7 of the EGFR gene [31] while mLEEK lacks exons 1 to 23 [32]. Another well-known alternative spliced RTK is TrkA. There are several TrkA splice variants found in neuroblastoma, including TrkAI and TrkAII from exon 9 alternative splicing [33], and TrkAIII from deletion of exons 6, 7, and 9, and extracellular IG-C1 and N-glycosylation domains [34]. Both TrkAI and TrkAIII are reported to affect tumor progression in neuroblastoma cells with TrkAI acting as a tumor suppressor and TrkAIII as an oncogene [35]. In the presence of Hsp90 inhibitor geldanamycin, cytosolic TrkAIII binds to Hsp90, which has been shown to render neuroblastoma cells resistant to geldanamycin–induced apoptosis [35]. TrkAIII also has been reported to localize to the centrosome where it causes genetic instability [35].
2.3 Protease-dependent RTK cleavage
Generally, upon ligand binding, RTKs can undergo multiple proteolytic cleavages in a protease-dependent manner (reviewed in [36]). For example, members of at least 10 RTK subfamilies undergo a series of secretase cleavage after activation to remove the extracellular domain followed by the release of the intracellular domain from the cell membrane, a process that is also known as regulated intramembrane proteolysis (RIP) [37]. α-secretases, e.g., matrix metalloproteinase (MMP) and a disintegrin and metalloproteinase (ADAM), and β-secretases, e.g., aspartic proteases (BACE), are responsible for RTK ectodomain shedding, which releases the extracellular domain of surface membrane RTK by proteolytic cleavage adjacent to the plasma membrane [38].
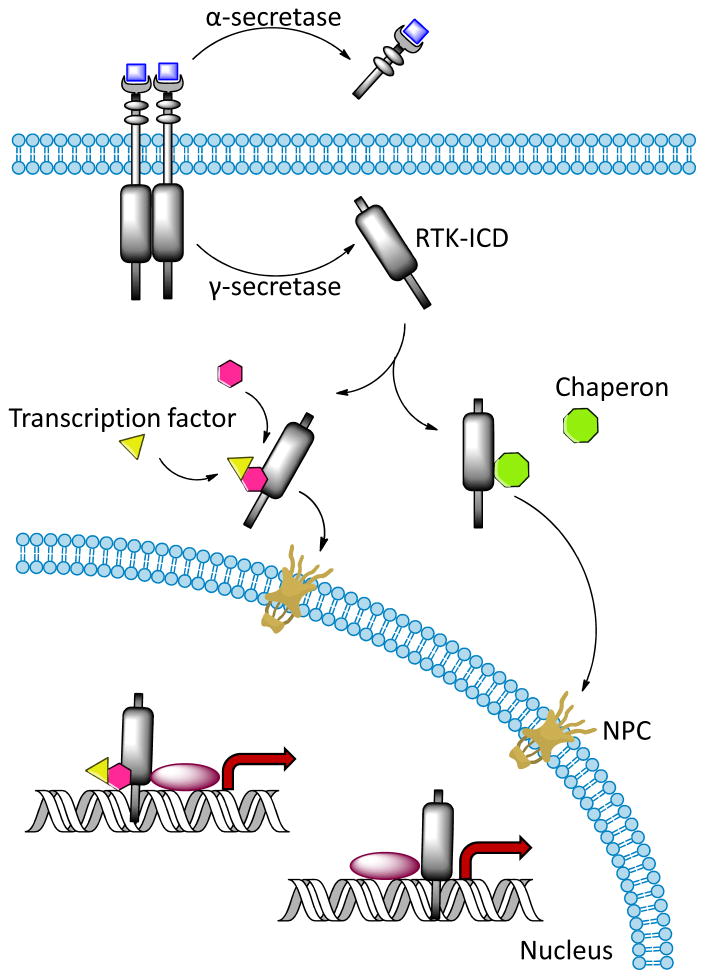
Among these proteases, ADAM-10 and ADAM-17/TACE (tumor necrosis factor-α-converting enzyme) are responsible for the majority of RTK shedding that generates a soluble ectodomain and a membrane-anchored carboxyl-terminal fragment (CTF) [39]. ADAM possesses higher substrate specificity than MMPs and cleaves substrates containing Ala-Val [40–42]. By using a peptide library, Caescu et al. demonstrated that ADAM17 specifically selects for small aliphatic residues immediately downstream of the cleavage site while ADAM10 can tolerate aromatic residues at that position [43]. After shedding, the intramembrane protease complex γ-secretase is responsible for the recognition and intramembrane cleavage of transmembrane proteins to release intracellular protein fragments (reviewed in [44]). The multi-subunit γ-secretase complex contains an enzymatic core comprised of presenilin, presenilin enhancer-2, anterior pharynx defective-1, and nicastrin [45–47]. Nicastrin recognizes and binds to the free amino terminus of the substrates [48], and presenilin cleaves type I single transmembrane proteins, such as RTKs [49]. Previous studies suggested that a conserved structural motif in the transmembrane domain of γ-secretase substrates, such as Notch, β-amyloid precursor protein and Sevenless, may be required for γ-secretase recognition [50–55]. However, so far no conserved primary sequence has been identified. Figure 3 illustrates the cleavage of ligand-bound RTKs by γ-secretase at the juxtamembrane domain to release ICD from the transmembrane domain. The ectodomain of the substrate must be shed prior to γ-secretase binding, and both the transmembrane and cytoplasmic domains must be present for subsequent cleavage.
Figure 3. Protease-dependent RTK-ICD formation and intracellular domain trafficking.
After activation, the RTKs are subjected to α-secretase mediated shedding to free the ectodomain before intracellular domain (ICD) is released into cytoplasm by γ-secretase cleavage. The ICDs are stabilized and guided across the cytoplasm and into nucleus through nuclear pore complex (NPC) by binding to transcription factors or chaperons. ICDs have been reported to act as transcriptional coactivators.
2.3.1 ErbB Family
Of the four members in the ErbB family, the best example of cleavage is that of ErbB-4. ErbB-4 functions in embryogenesis and breast development and its expression is tissue specific (reviewed in [56]). There are two ErbB4 isoforms generated by alternative splicing, Jm-a and Jm-b. The former undergoes secretase cleavage whereas the latter does not [57]. ADMA17/TACE-regulated ectodomain shedding is enhanced when ErbB-4 is bound to heregulin [58, 59] or under protein kinase C (PKC) activation by phorbol 12-myristate 13-acetate (PMA) [60]. This leads to a 120-kDa ectodomain fragment that is released into the extracellular environment, leaving a membrane-bound 80-kDa fragment containing the transmembrane domain and ICD, called m80, which then undergoes γ-secretase cleavage to release the soluble ICD called 4ICD [61, 62]. The cleavable ErbB4 isoform is found in both normal and tumor tissues. In breast cancer, nuclear 4ICD (see later) level is negatively correlated with tumor malignancy [63, 64] and improved response to endocrine therapy but not chemotherapy [65, 66].
Other ErbB receptors also have secretase-dependent fragments. For instance, ErbB-2 ectodomain shedding is mediated by ADAM10 and MMP-1 to generate the p95ErbB2 fragment [67]. Breast tumors resistant to TKIs, such as trastuzumab [68, 69] and lapatinib [70], and anti-oestrogen drug, toremifene [71], also express the p95ErbB2 fragment.
2.3.2 Colony stimulating factor-1 Receptor (CSF-1R)
CSF-1R can be activated by both colony stimulating factor-1 (CSF-1) and interleukin-34 (reviewed in [72]) and is known to regulate the development and maintenance of immune cells, such as monocytes and macrophage [72], as well as regulate neuron progenitor cells, such as myeloid cells [73, 74]. Interestingly, macrophage precursors are better inducers of secretase-mediated CSF-1R cleavage than its ligand CSF-1 [75–80]. In addition, Toll-like receptor-induced Erk activation also promotes secretase-mediated CSF-1R cleavage [81]. During macrophage activation, CSF-1R is downregulated in response to IL-2 and IL-4 stimulation via ADAM17/TACE-mediated shedding, and the cleavage is further enhanced by PMA and lipopolysaccharide [82]. The sequences within the extracellular juxtamembrane region and transmembrane domain of CSF-1R are critical for TACE-mediated shedding and γ-secretase cleavage, respectively [75, 80, 83], but exact cleavage site has not been reported.
2.3.3 Ephrin-B Receptor (Eph-B)
Erythropoietin-producing hepatoma receptors (Eph) family is considered the largest RTK family with 14 members grouped into 2 subfamilies, Eph-A (EphA1-8, Eph-A10), and Eph-B (EphB1-4, EphB6) [84]. Eph receptors are activated by ephrins. Shedding of the Eph-B1 receptor can be blocked by metalloproteinase inhibitor, GM6001, which suggests that the metalloproteinases are involved in its cleavage [85]. The ICD of Eph-B1 generated by γ-secretase cleavage has been speculated to translocate into nucleus via a putative pH-dependent nuclear localization signal in the basic amino acid cluster located within its juxtamembrane domain. However, there is no evidence for Eph-B1 nuclear localization to date [85].
Proteolytic cleavage of Eph-B2 by secretase releases its ectodomain and ICD. Interestingly, the Eph-B2 ectodomain shedding machinery is determined by a specific stimulus that determines the fate of the receptor. Ligand-induced shedding is mainly related to ubiquitinated receptor endocytosis, which is mediated by MMP-2 and MMP-9 [86]. When ligand-induced shedding occurs in the endosome, the process becomes insensitive to GM6001. In contrast, calcium influx- and N-methyl-D-aspartic acid (NMDA)-stimulated Eph-B2 cleavage on the plasma membrane without endocytosis is sensitive to GM6001 and ADAM10 inhibitor [87]. The intramembrane cleavage of Eph-B2 after shedding requires γ-secretase [88].
2.3.4. Vascular Endothelial Growth Factor 1 (VEGFR-1)
Vascular endothelial growth factor receptor (VEGFR) family binds to vascular endothelial growth factor (VEGF) and is responsible for regulating physiological and pathological angiogenesis. VEGFR-1 can also undergo ADAM10- and ADAM 17-mediated shedding to release its soluble ectodomian (sVEGFR1) [89], which has been shown to regulate VEGFR signaling by trapping VEGF. In neural retina, BACE-1-mediated VEGFR-1 shedding is required for VEGF-induced angiogenesis [90] and subsequent γ-secretase cleavage and generation of VEGFR1-ICD [90], which has been reported in leukemic cancer cells [91]. In pregnant women, increasing levels of VEGF and sVEGFR1 are positively correlated with threatened abortion [92].
2.3.5 MET
MET was discovered as TPR-MET oncogene in transformed human osteogenic sarcoma cells [93]. MET overexpression and mutation are found in many types of cancer and contribute to tumor progression, such as cell survival, invasion, and proliferation (reviewed in [94]). Ectodomain shedding of MET correlates with tumor malignant potential in many cancer cells, including non-small cell lung cancer [95, 96]. HGF, PMA, and suramin as well as MET inhibitory antibody have been shown to induce ADAM-10- and ADAM-17-mediated MET ectodomain shedding to produce a membrane–anchored 55 kDa CTF that is found in the nucleus of both normal and cancerous tissues with ADAM-17 being the major protease [97–102]. ADAM-17-mediated MET shedding releases a soluble MET ectodomain that has been shown to promote MET/STAT3 survival signaling in MEK inhibitor-resistant KRAS-mutated colorectal cancer [103]. MET-CTF can further undergo ligand-independent presenilin-dependent γ-secretase cleavage to generate MET-ICD [104].
2.3.6 Tie-1
Tie-1 is an orphan receptor that supports epithelial cell survival [105] and plays important roles in both normal and pathological angiogenesis [106]. In epithelial cells, the metalloproteinase-mediated Tie-1 ectodomain shedding is stimulated by PMA, VEGF, inflammatory cytokines, and shear stress [107–109] to release a 100 kDa extracellular domain and a 45 kDa membrane-anchored CTF. Since Tie-1 forms heterodimers with Tie-2, it has been suggested that the Tie-1 shedding plays an important role in the increased binding of Tie-2 to cartilage oligomeric protein angiopoietin-1 (Ang 1) [110]. Interestingly, Tie-1 shedding to generate CTF is insensitive to metalloproteinase inhibitor TAPI-2 in MCF-7 breast cancer cells [111]. After shedding, Tie-1-CTF then undergoes γ-secretase cleavage that liberates a 43 kDa Tie-1-ICD [110]. Overexpression of Tie-1-ICD has been detected in breast cancer cells, and high levels of Tie-1-ICD in breast and colon tumors were found to correspond to cancer progression [112]. The unique functions of Tie-1 in lymphatic remodeling and maturation [113] and in breast cancer progression raise a question whether the Tie-1 fragments regulate non-canonical signaling pathways. Further studies will be needed to determine that.
2.3.7 Ryk
Unlike most RTKs, receptor-like tyrosine kinase (Ryk) does not have kinase activity due to alterations of the invariant amino acids in the catalytic domain [114, 115]. Ryk contains an extracellular Wnt inhibitory factor domain and has been shown to function as a Wnt co-receptor in axon guidance and pattern formation [116]. In 2008, Lyu et al. reported a 42 kDa Ryk-ICD in the cytosol and nucleus of cells in mice brain section [117]. Using a chimeric Ryk receptor and γ-secretase inhibitor DAPT, they further demonstrated that the secretase cleavage site is located within the transmembrane domain of Ryk and inhibition of γ-secretase leaves a membrane bound ectodomain-shed 45 kDa Ryk fragment.
2.3.8 ICD Formation of Other RTKS
In addition to the RTKs described above, several others have been shown to undergo protease-mediated cleavage, including the insulin receptor (InsR), insulin-like growth factor 1 receptor (IGF-1R), protein-tyrosine pseudokinase 7 (PTK7), and fibroblast growth factor receptor 3 (FGFR-3). Ectodomain shedding of InsR is induced by PKC-activated ADAM17 under PMA treatment to generate InsR-CTF, which is then cleaved by γ-secretase to produce InsR-ICD [118]. Likewise, IGF-1R also relies on PKC-activated shedding and γ-secretase cleavage to release its ICD [119]. PMA also enhances ADAM17- and γ-secretase-mediated proteolytic cleavage of PTK7 to generate two oncogenic PTK7-CTF fragments, PTK7-CTF 1 and PTK7-CTF 2 [120]. For FGFR-3, FGF1-induced shedding does not follow the traditional RIP pathway. While metalloproteinase is not essential for FGFR cleavage to generate ICD, FGFR-3 shedding does require endocytosis [121]. Table 1 lists the secretases known to date that cleave RTKs.
Table 1.
Ectodomain shedding and proteolytic cleavage of RTKs
| RTK | Ectodomain shedding | Ref. | Secretase cleavage | Ref |
|---|---|---|---|---|
| CSF-1R |
ADAM17 Enhanced by CSF, PMA and lipopolysaccharide |
[75, 82, 83] |
γ-secretase Poorly induced by ligand. Induced by PMA, macrophage precursors and Toll-like receptor induced Erk activation |
[75–81, 83] |
| DDR1 | Enhanced by type I collagen | [261] | ||
| Eph-B1 |
MMP Enhanced by PMA, blocked by GM6001 |
[85] | γ-secretase | [85] |
| Eph-B2 |
MMP-2, MMP-9 Mediated by MMP in response to ligand. Insensitive to MMP inhibitor if receptor locates in endosome ADAM10 Mediated by ADAM10 in response to ligand, calcium influx and NMDA stimulation |
[86, 87] | γ-secretase | [87, 88] |
| ErbB-2 |
ADAM10, MMP-1 Enhanced by PMA, trastuzumab, pervanadate and toremifene |
[67, 69, 71] | ||
| ErbB-4 |
ADAM17 Enhanced by binding with heregulin or by PMA-activated PKC |
[58–60] | γ-secretase | [61, 62] |
| FGFR-3 | Enhanced by FGF1 Requires endocytosis | [121] | ||
| IGF-1R | Enhanced by PKC | [119] | γ-secretase | [119] |
| InsR |
ADAM17 Enhanced by PMA |
[118] | γ-secretase | [118] |
| MET |
ADAM10, ADAM17 Enhanced by HGF, PMA, suramin and MET antibody |
[97–102] | γ-secretase | [104] |
| PTK7 |
ADAM17 Enhanced by PMA |
[120] | γ-secretase | [120] |
| Ryk | γ-secretase | [117] | ||
| Tie-1 | Enhanced by PMA, VEGF, inflammatory cytokines and shear stress Enhance binding of its heterodimeric associated Tie-2 receptor to Ang-1 |
[107–110] | γ-secretase | [110] |
| Tie-2 | Enhanced by PMA and VEGF | [262] | ||
| TrkA | Enhanced by NGF | [263, 264] | ||
| VEGFR-1 |
ADAM10, ADAM17 Enhanced by PKC Regulated by the expression of VEGFR-2 and the presence of soluble VEGFR-1 ectodomain BACE-1 Required for VEGF-induced angiogenesis in neural retina |
[89] | γ-secretase | [90] |
| VEGFR-2 |
ADAM17 Enhanced by VEGF-A |
[265] |
3. Intracellular trafficking of MRINs
3.1 Trafficking of membrane-bound MRINs
3.1.1 RTK internalization and endosomal retrograde trafficking to the Golgi and ER
Cell surface receptors are present in many subcellular compartments, including the Golgi apparatus, mitochondria, ER, and nucleus. Upon ligand activation, RTK is rapidly internalized and translocated into the endosomal compartments for signaling, recycling, or degradation by a clathrin-mediated or -independent pathways, depending on the specific coat proteins in the membrane region that form the endocytic vesicles (reviewed in [122]). The endocytic vesicles are then sent to different subcellular compartments based on the associated cargo proteins, such as Rab proteins [123] or clathrin-binding adaptor proteins (AP) [124] (Figure 4).
Figure 4. Endosomal vesicle trafficking of internalized RTK.
RTKs are internalized through either clathrin-dependent or clathrin-independent pathways. In clathrin-dependent endocytosis mechanism, the internalized membrane vesicle is coated with clathrin (green). Meanwhile, caveolin-mediated endocytosis, which is the main clathrin-independent RTK endocytic mechanism, is initiated at the membrane region that contain caveolin-rich lipid raft (purple). The endocytic vesicles from both pathways are sent to early endosome for sorting. Based on the component of coating proteins, the vesicles are then transported to different endosomal components, including recycle endosome, late endosome and trans Golgi network. Several important coating proteins that direct vesicle transport are shown, including Rab proteins, clathrin-dependent adaptor proteins (AP), retromer, syntaxin 6 (Syn 6), and Golgi-associated, gamma adaptin ear containing, ARF binding protein (GGA).
Clathrin-mediated endocytosis (CME) from non-lipid raft membrane domains is the predominant mechanism for RTK internalization. Prior to RTK activation, auto-inhibition of clathrin prevents the recruitment of cytosolic adaptor protein. Upon RTK activation, clathrin recognizes specific posttranslational modifications, such as ubiquitination and acetylation, at the C-terminus of activated RTK [125, 126]. The clathrin complex then recruits cargo-specific adaptors, e.g., AP2, which can also interact with phosphatidylinositol (4,5)-biphosphate (PIP2) to bring in the phospholipids on adjacent plasma membrane to induce conformational changes that promote the formation of clathrin-coated pit through membrane curvature, clathrin polymerization, and internalization of the RTK-containing pit from the plasma membrane [127]. Moreover, CME is a highly selective process that forms only after recognition of the cargo protein sequence by AP. For example, AP2 specifically recognizes the YXXΦ and LL sequence motif ([ED]XXXL[LI]) on the cargo protein (reviewed in [128]). In EGFR, the LL motif is important for AP2 phosphorylation, which further facilitates the interaction between AP2 and EGFR, and subsequent internalization of EGFR via CME [129].
Posttranslationally modified RTKs, such as from ubiquitination, can also be internalized via a clathrin-independent endocytic pathway [130]. There are several clathrin-independent endocytic mechanisms, including phagocytosis, macropinocytosis, and lipid raft-mediated (e.g., caveolin-mediated) endocytosis, and among them, macropinocytosis and lipid raft-mediated endocytosis have been reported for RTK internalization. Macropinocytosis is a growth factor-induced and actin-mediated transient endocytic process that begins from all membranous regions, such as those of the lipid rafts, in larger vesicles containing extracellular fluid and plasma membrane-bound components. Unlike clathrin-dependent endocytosis, caveolae-mediated endocytosis does not require a specific coat protein and is usually associated with lipid raft membrane regions containing caveolin-1 protein (reviewed in [131]). Recently, Boucrot et al. reported a new clathrin-independent endocytic mechanism called endophilin-mediated endocytosis (FEME), which does not require AP2 or clathrin [132] and further demonstrated that FEME uses endophilin-A2 as membrane scissor to release endocytic vesicles [133]. FEME of membrane receptors, including RTKs, e.g., EGFR, MET, VEGFR, PDGFR, IGF-1R, and TrkA, requires ligand activation [132]. FEME of EGFR only occurs under high concentrations of EGF treatment and requires Cbl and CIN85, which are RTK substrates involved in routing activated surface RTKs to the lysosome for degradation, suggesting that this newly identified pathway may be more related to downregulation of canonical RTK signaling [132].
After RTK internalization, receptors are then routed to the early endosomes, where the fate of cargo is determined. In general, RTKs can be degraded or recycled or can undergo retrograde trafficking to the Golgi apparatus (Figure 4). Although it has been reported that in EGFR is recycled after stimulation by high concentrations of ligand in A431 cells, which express high levels of EGFR with 80% of EGFR internalized [134], not all endocytic receptors are subjected to recycling or degradation. Instead, a small portion of them undergoes retrograde transport, which is the influx of protein and lipid from the cell surface to Golgi or from the Golgi to ER. The trans Golgi network (TGN)-targeting coat proteins, including Rab9, syntaxin 6, and GGA, guide the fusion of endosomal vesicle to the TGN [135]. Internalized RTKs, e.g., EGFR [136], c-MET [137], and fibroblast growth factor receptor (FGFR)-1 [138, 139], via CME can also be transported to the TGN for further sorting through coated vesicle transport. However, endocytic RTKs that undergo retrograde transport do not necessarily stop at the TGN as they can be further routed back to the ER and even the nucleus by COPI-mediated retrograde transport as exemplified by the Golgi-to-ER translocation of EGFR [139]. Du et al. recently demonstrated that inhibition of dynein or knockdown of dynein or syntaxin 6 attenuated EGFR accumulations in the Golgi apparatus and nucleus [136]. Their findings indicate that the EGFR detected in these subcellular compartments is indeed from the cell surface.
3.1.2 Nuclear trafficking of MRINs from the ER
After reaching the ER, endocytic RTKs can be further transported into the nucleus via two importin-β-mediated pathways, integral trafficking from the ER to the nuclear envelope transport (INTERNET) and integrative nuclear FGFR-1 signaling (INFS) (Figure 5) [140]. The major difference between the two is that the receptor remains membrane bound and is localized to inner nuclear membrane (INM) before nuclear translocation via INTERNET whereas FGFR-1 becomes a soluble protein after its release from ER or ER-derived membrane vesicle before translocation into the nucleus via INFS [140].
Figure 5. Retrograde and nuclear transport mechanisms of membrane-bound RTKs.
RTKs are transported from Golgi to ER in COPI-coated vesicles via the retrograde pathway. The ER-to-nuclear transport of RTKs is mediated by two pathways, INFS and INTERNET. Via INFS pathway, the RTK is pumped through the Sec61 complex into the cytosol where it binds to cytosolic importin complex and transported into nucleus by the importin-NPC interaction. Via the INTERNET pathway, the RTK is trafficked along ER, translocated from the ONM to INM through the NPC by binding to ER-associated importin, and released from the INM by the Sec61 complex into the nucleoplasm. Nuclear localized RTK interacts with transcription factors and functions as transcriptional regulator.
Both EGFR and ErbB-2 contain a typical NLS that attracts NLS-containing molecules for complex formation with importin-β [141]. It has been shown that EGFR and ErbB-2 are transported from the ER to the outer nuclear membrane (ONM) and then to the nuclear pore complex (NPC), where the receptors enter the INM with the help of importin-β [140, 142]. As demonstrated with digitonin-permeabilized cells, detection of importin-β in the non-nuclear extract suggested that the INTERNET mechanism depends on membrane-associated importin-β to transport EGFR and ErbB-2 from the ONM into the INM [140, 143]. Wang et al. further showed that Sec61β translocon is required for the release of INM-bounded EGFR into the nucleus [144]. Both EGFR and ErbB-2 contain the NLS that interacts with membrane-bound importin-β and translocate into the nucleus via the INTERNET pathway [144]. Given that NLS is conserved among most of RTKs and that nuclear translocation of another NLS-containing RTK, c-MET, also follows the INTERNET pathway (unpublished data), it is likely that INTERNET is a commonly shared mechanism for RTK nuclear trafficking. Further investigation of the nuclear transport mechanism of other RTKs will be required to validate this notion.
Via the INFS pathway, FGFR-1 is released as soluble protein from the ER or ER-derived membrane vesicle into the cytosol through the Sec61 channel. FGFR-1 then associates with importin-β in cytoplasm before being transported into the nucleus [145]. Notably, even though FGFR-1 does not contain a consensus NLS sequence, it can still translocate into the nucleus by association with NLS-containing proteins, such as its NLS-containing ligand, FGF-2 [145].
3.2 Trafficking of RTK-ICD
RTK-ICDs are found in different intracellular compartments, including the mitochondria and nucleus, suggesting that ICDs are also subjected to intracellular transport. To avoid degradation, ICDs can either bind to other proteins, e.g., Hsp90, to enhance their stability [146, 147] or undergo posttranslational modifications by other associated proteins [148]. For example, 4ICD can be SUMOylated by protein inhibitor of activated STAT3 (PIAS3). In addition, the released ICD can bind to transcription factors, such as YAP [149], signal transducer and activator (STAT) [150], and ERβ [151], which may also stabilize the ICD for intracellular trafficking (Figure 3). The nuclear localization sequence (NLS) within the RTK-ICD or its associated proteins binds to adaptor proteins, such as importin-α and importin-β, to facilitate nuclear localization through the NPC [152, 153].
4. Functions of MRINs
4.1 ErbB family
Mutation and overexpression of ErbB receptors (ErbB-1/EGFR, ErbB-2/HER-2/neu, ErbB-3, and ErbB-4) contribute to increased malignancy of solid tumors, including non-small cell lung, breast, and colon cancers, and glioblastoma [154]. The entire ErbB family of receptors is reported to translocate into the nucleus either as holoreceptor or truncated form. In 1994, ErbB-2 was the first RTK reported to have transcriptional activity in the cell’s nucleus. While nuclear localization of EGFR was first described much earlier in 1984 [155], its nuclear functions were not characterized until 2001 [156]. Below, we describe the nuclear functions that have been reported to date for each of the ErbB receptors although more studies are currently ongoing.
4.1.1 ErbB1/EGFR
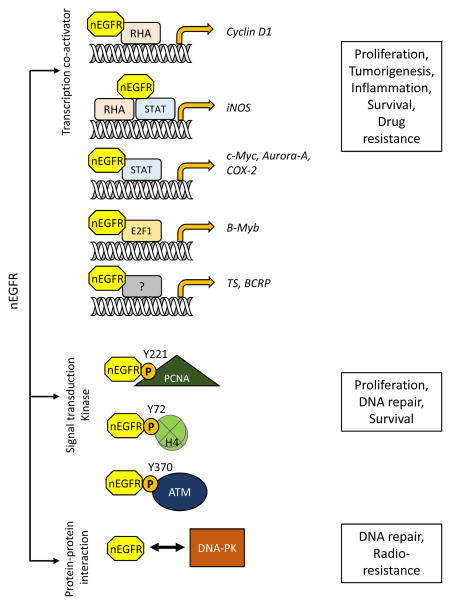
EGFR nuclear translocation is triggered by various stimuli, such as EGF, radiation, anti-cancer drugs, and EGFR antibodies [16, 156, 157]. nEGFR functions as a transcription co-activator by binding to RNA helicase A and MUC1 at the AT-rich response sequence [156, 158–160] to promote cyclin-D1 gene expression. nEGFR also interacts with transcription activators E2F1, STAT3, and STAT5 to upregulate expression of target genes, such as inducible nitric oxide synthase (iNOS) [161], B-Myb [162], Aurora-A [163], cyclooxygenase-2 (COX-2) [164], c-Myc [17], thymidylate synthase (TS) [165], and BCRP [17, 25]. By increasing their expression, nEGFR promotes cell proliferation, inflammation, and genome instability as well as tumor progression.
Several anti-EGFR drugs such as erlotinib, gefitinib, afatinib, and monoclonal antibody cetuximab have been approved by the FDA for cancer treatment [166]. As alluded to above, nEGFR has been shown to contribute to the development of acquired resistance to radio-, chemo-, or targeted therapies [16, 24, 25, 167–169]. For example, phosphorylation of EGFR at Y1101 promotes EGFR nuclear translocation and enhances nEGFR-mediated iNOS and B-Myb transcription in cetuximab-resistant cells [170]. Aside from its role as a transcription co-activator, nEGFR also activates DNA replication and repair. Specifically, nEGFR binds to and phosphorylates proliferation cell nuclear antigen (PCNA) at Y221 residue to stabilize PCNA, which subsequently enhances cell proliferation [171], inhibits DNA mismatch repair, and promotes genome instability [172]. EGFR also interacts with ataxia telangiectasia mutated (ATM) in the nucleus after ionizing radiation and mediates DNA damage response by phosphorylating ATM at Y370 to promote DNA repair and cell survival [18]. nEGFR also phosphorylates histone H4 at Y72, which then modulates K20 methylation of histone H4 to promote DNA synthesis and repair [173]. Moreover, a positive correlation was reported between phosphorylation of DNA-dependent protein kinase (DNA-PK) and nEGFR expression after irradiation. While there is no evidence to suggest that DNA-PK is a direct substrate of nEGFR, these findings point to the importance of nEGFR in DNA damage repair [174–176]. Figure 6 illustrates various functions of nEGFR.
Figure 6. Functions of nuclear EGFR.
Nuclear EGFR (nEGFR) has different roles in regulating cellular fate. (1) nEGFR can function as a transcription co-activator by interacting with different transcription factors, such as RNA helicase A (RHA), STAT, and E2F1, to promote target gene transcription. Elevated levels of these genes result in cell proliferation, tumorigenesis, and inflammation and are also correlated with drug resistance. (2) nEGFR phosphorylates its substrates, such as PCNA, histone H4, and ATM, to promote cell proliferation and DNA repair. (3) nEGFR also interacts with DNA-PK to increase DNA repair, leading to the development of radioresistance.
Besides the nEGFR holoreceptor, some of the EGFR variants are also detected in the nucleus. EGFRvIII and mLEEK have been shown to translocate into the nucleus to regulate cellular responses. In glioblastoma, nuclear EGFRvIII regulates COX-2 and Bcl-XL transcription through its interaction with STAT3 [164] and STAT5b [177]. Nuclear mLEEK has been reported to regulate transcription of ER chaperon protein GRP78 under ER stress [32].
4.1.2 ErbB-2/HER-2/neu
Although ErbB2 itself is not sufficient to induce internalization because it is an orphan receptor that lacks a well-defined ligand, nuclear trafficking of ErbB-2 has been reported via the INTERNET pathway [140, 141, 178]. Ubiquitination of ErbB-2 (reviewed in [179]) or heterodimerization of ErbB-2 with other ErbB members, such as EGFR [180], facilitates its internalization.
Nuclear localization of ErbB2 was first reported more than two decades ago [181]. Similar to EGFR, ErbB-2 can also form a complex with STAT3 to transcriptionally upregulate COX-2 [178], cyclin D1 [182], and ribosomal RNA [183]. It has been shown that the progesterone receptor-induced nuclear ErbB-2/STAT3 complex promotes breast cancer cell growth [182]. In addition, nuclear ErbB-2 also phosphorylates atypical histone macro-H2A1.2 to promote its own gene transcription, resulting in ErbB-2 overexpression and cell proliferation in cancer cells [184]. Aside from transcriptional regulation, nuclear ErbB-2 is reported to phosphorylate cell division cycle protein 2 homolog (Cdc2) Y15, leading to inhibition of Cdc2 and resistance to taxol-induced apoptosis in breast cancer patients [185].
ErbB-2 is also present in nucleus in the truncated form due to alternative translation initiation that generates two ErbB2-CTF variants, 611-CTF and 687-CTF. Interestingly, 611-CTF is a membrane-associated fragment while 687-CTF is not because it lacks the transmembrane domain. Although the ErbB-2 fragments are known to interact with and stabilized by Hsp90, the underlying nuclear trafficking mechanisms are not well understood (reviewed in [179]). Both 611-CTF and 687-CTF are detected in the cytoplasm as well as the nucleus. The 611-CTF variant is hyperactive and more correlated with tumor progression (reviewed in [186]). Studies have indicated that 611-CTF contributes to cell migration and breast cancer cell metastasis by phosphorylating cortactin, a cytoskeletion-binding protein [187], and by promoting transcription of metastasis-related genes, such as MET, MMP1, and IL11, and those that express integrins and Eph receptors [188].
4.1.3 ErbB-3
Due to sequence alteration in its tyrosine kinase domain, the kinase activity of ErbB-3 is lower than that of other ErbB receptors [189]. High expression of nuclear ErbB-3 has been observed in prostate cancer cells as well as in tumor tissue of prostate cancer patients [190–193]. In normal tissues, ErbB-3 is mainly located on the cell surface and in the cytoplasm. Interestingly, immunohistochemistry staining revealed that low nuclear ErbB-3 expression is correlated with cancer cell survival and recurrence [191].
Different from ErbB-1 and ErbB-2, nuclear trafficking of ErbB-3 begins by macropinocytosis instead of clathrin-independent endocytosis [194]. Internalization of ErbB3 is triggered by neuregulin and inefficiently translocated to the lysosome for degradation (reviewed in [195]). Nuclear localization of the ErbB-3 variant (ErbB380kDa) was recently reported by Andrique et al. [196] who demonstrated that nuclear ErbB380kDa, which lacks kinase activity, corresponds to the intracellular domain of the receptor. Although ErbB380kDa may possess little kinase activity, it binds to and regulates the cyclin D1 promoter, which promotes transcriptional activation and cell proliferation [196]. In addition, a 50 kDa nuclear ErbB3 (nuc-ErbB3) variant that regulates transcriptional activity and distribution of Ezrin was reported to contribute to myelination of Schwann and glial cells [197, 198].
4.1.4 ErbB-4
As described in Section 2.3.1, 4ICD is generated by secretase cleavage. The proteolytic processing of ErbB4 is functionally important as 4ICD translocates into nucleus and binds to regulators that modulate transcription, including transcription factors YAP [149], ERβ [151] and STAT5A. In astrocytes, 4ICD forms a complex with corepressor N-CoR and TGF-β-activated kinase 1-binding protein 2 (TAB2), which then translocates into the nucleus to repress gene expression [147]. Nuclear 4ICD has also been reported to regulate β-casein gene expression that is crucial to milk production [199] and fetal lung maturation [150]. In addition, colocalization of 4ICD and HIF-1α has been reported, and their interaction in the nucleus may promote HIF-1α stability and signaling [200].
4.2 Other MRINs
4.2.1 FGFR Family
FGFRs regulate cellular processes including cell proliferation, differentiation, and survival, and thus are important in cancer progression [201]. Traditionally, the FGFR family contains four members–FGFR-1, FGFR-2, FGFR-3 and FGFR-4. Recently, FGFR-5, also known as FGFRL1, was added to this family based on its interaction with FGFR ligands. The conventional FGFR family is highly conserved in sequence homology whereas FGFR-5 lacks the TK domain.
Nuclear translocation of FGFR-1 holoreceptor by ligand stimulation has been reported in many types of tumor tissues and cancer cell lines (reviewed in [202]). Internalization of FGFR-1 is facilitated by the FGF-2-activated heparin sulfate proteoglycan, syndecan 4, via macropinocytosis [203]. Nuclear transport of FGFR-1 occurs via an importin-β-mediated INFS mechanism and has been shown to promote cell proliferation by activating transcription coactivator cAMP response element-binding (CREB)-binding protein to enhance gene transcription (reviewed in [145, 204]). In addition, Hu et al. demonstrated that nuclear FGFR-1 can also translocate into the nucleus from the Golgi directly after protein synthesis [205]. The authors also showed that FGFR-1 phosphorylates ribosome S6 kinase 1 (RSK1), a modification that is critical for RSK1-mediated transcriptional activation of c-Fos, CREB, estrogen receptor, and Iκ Bα as well as histone H3 remodeling [205].
Nuclear FGFR-2 has been shown to regulate mammary gland development through interaction with STAT5. Thus, FGFR-2 is considered to play an important role in mammary tumor growth. Indeed, expression of nuclear FGFR-2 in breast cancer patients is corrected with poor prognosis [206, 207]. FGFR3-ICD, located in both the cytosol and nucleus, is generated by secretase cleavage and released as a soluble protein [121, 208]. FGFR3-ICD is also the only FGFR ICD reported in the nucleus to date. In malignant breast cancer tissues, increased nuclear localization of FGFR3 was observed [209]. More studies are needed to further delineate the physiological and/or pathological role of nuclear FGFR3-ICD.
4.2.2 VEGFR Family
The VEGFR family is composed of three members, VEGFR-1/Flt-1, VEGFR-2/KDR/Flk-1, and VEGFR-3/Flt-4. VEGFR-1 and VEGFR-2 bind to VEGF-A while VEGF-3 binds to VEGF-C and VEGF-D. The biological functions of these three members are distinctively different: VEGFR-1 mainly functions in non-epithelial cells; VEGFR-2 plays an important role in epithelial cells; and VEGFR-3 mediates lymphaogiogenesis (reviewed in [210]).
VEGFR-1 holoreceptor can be found on the membrane, cytoskeleton, and nucleus [211]. Nuclear VEGFR-1 predominantly co-localizes with lamin A/C in the nuclear fraction in breast cancer cells without ligand stimulation and is correlated with cell survival [212]. However, more studies are needed to clarify the function and identify the substrate of nuclear VEGFR-1 holoreceptor. VEGFR1-ICD is generated and detected in the cytoplasm when cells are exposed to pigment epithelium-derived factor (PEDF) in the presence of VEGF. PEDF is also responsible for VEGF-induced VEGFR phosphorylation in addition to VEGFR cleavage [213], suggesting that VEGFR1-ICD generation may be related to downregulation of VEGFR-1 canonical signaling. Nuclear translocation of VEGFR1-ICD has been reported to function in angiogenesis [211, 213], and VEGFR1-ICD without its TK domain can still stimulate normal embryonic development and angiogenesis [214], suggesting that the kinase activity is not crucial for its nuclear function and a possible role of VEGFR1-ICD as a transcription regulator. Further investigation will be required to validate the potential function of VEGFR1-ICD as a transcriptional coactivator.
VEGFR-2 translocates into nucleus under VEGF stimulation in endothelial cells, and has been reported to form a complex with tissue transglutaminase II to mediate this process [215]. Studies have also indicated that nuclear VEGFR2 holoreceptor promotes its own transcription by binding to the transcription factor Sp1 [216] and functions in endothelial wound healing [217]. Still, there may be more functions of nuclear VEGFR-2, pending further investigations.
4.2.3 MET Family
The MET receptor, also known as c-MET or hepatocyte growth factor (HGF) receptor, is responsible for HGF-induced signaling pathways. Overexpression of MET is detected in many cancer types, including non-small cell lung, breast, colorectal, liver, and ovarian cancers. Moreover, overexpression of MET not only correlates to tumor malignancy but also contributes to acquired resistance to EGFR TKIs (reviewed in [94].) In liver SK-HEP-1 cell line, Gomes et al. demonstrated that MET translocates from the plasma membrane to the nucleus in response to HGF stimulation through Gab-1 and importin-β to induce inositol 1,4,5-triphosphate (InsP3) formation in the nucleus. This in turn activates InsP3-dependent calcium release into the nucleoplasm to promote cell proliferation [218]. Androgen deprivation in prostate cancer cells has been reported to induced nuclear accumulation of MET holoreceptor, which contributes to castration-resistant prostate cancer progression through upregulation of SOX9 and activation of the β-catenin signaling pathway [219].
MET intracellular fragments have also been observed in the nucleus. Matteucci et al. reported that nuclear translocation the MET C-terminal fragment, which possesses trans-activating activity that promotes gene transcription in malignant breast cancer cells, does not require ligand stimulation [220, 221]. However, it is not clear from their study whether γ-secretase is essential for the formation of the MET C-terminal fragment. Later, Xie et al. reported their discovery of nuclear MET-ICD in prostate cancer cells and found that its level decreased in response to γ-secretase inhibitor [219]. In addition, based on the molecular weight of the MET-ICD identified, their findings indicate that the MET C-terminal fragment previously described by Matteucci et al. was indeed MET-ICD.
The other member of the MET family, Ron, which binds to hepatocyte growth factor-like protein/macrophage stimulating-protein, is mainly expressed on macrophages and epithelial cells. Similar to MET, overexpression of Ron in various cancer types, including breast, colon and pancreatic cancers, is correlated with poor prognosis and metastasis [222]. In response to serum starvation, Ron translocates into the nucleus, which requires importin and heterodimerization with EGFR. High levels of nuclear Ron have been observed in bladder cancer cells and primary bladder tumors [223]. Hypoxia can also trigger nuclear translocation of Ron to transcriptionally regulate c-JUN and HIF-1α in cancer cells [224]. Moreover, nuclear Ron associates with the chromatin, and potential target genes of nuclear Ron identified are related to several stress-responsive networks [224].
4.2.4 Trk Family
There are three RTKs in this family, TrkA, TrkB and TrkC. Trk is activated by neurotrophins, nerve growth factor, and brain-derived neurotrophic factor, and Trk signaling regulates the development and function of the neural system, including cell survival, proliferation, neural precursors differentiation, growth and patterning of axons and dendrites [225]. Nuclear TrkA has been detected in PC12 rat neuroblastoma cells under NGF stimulation [226] as well as in hepatocytes and activated stellate cells [227]. However, the nuclear function of TrkA is currently unknown.
4.2.5 ROR Family
Receptor tyrosine kinase-like orphan receptor (ROR) is expressed during development in many tissue types and is functionally related to Trk and muscle specific kinase (MuSK). Mutational studies of ROR indicated that it plays important roles in cell migration and cell polarity determination as ROR-deficient mice have defective heart and bone formation (reviewed in [228]). In normal development, ROR is regulated by Wnt5a [229] Interestingly, overexpression of ROR correlates with tumorigenesis [230]. Of the two ROR family members, ROR1 and ROR2, only ROR1 is reported to have nuclear localization, which requires its juxtamembrane domain. In the nucleus, ROR1 functions in cell migration and cytoskeleton regulation [231, 232].
4.2.6 PDGFR Family
There are five members in platelet-derived growth factor receptor (PDGFR) family, PDGFRα, PDGFRβ, colony stimulating factor-1 receptor (CSF-1R)/FMS, KIT/SCFR, and FLT3. The functions and ligands for each member are different. PDGFRα and β are responsible for PDGF-induced cell replication, migration, and survival signaling of myofibroblasts during fibrotic disease pathogenesis [233]. CSF-1R mainly functions in regulating monocytes and macrophages as well as in trophoblast implantation and breast development. KIT is a stem cell factor receptor (SCFR), and the deregulation of KIT is related to mast cell proliferation disease and neoplasms [234]. So far, only PDGFRα and CSF-1R have been reported to translocate into the nucleus. Studies have indicated that nuclear PDGFRα in alveolar fibroblasts plays a role in early embryonic and lung development in mouse model [235, 236]. However, further investigation is needed to determine whether PDGFRα is present in the nucleus in humans and identify its function.
Recently, Barbetti et al. reported nuclear translocation of the CSF-1R holoreceptor and its functions in breast cancer [237]. In this study, they showed that nuclear CSF-1R was present only in breast cancer cells but not in fibroblasts or macrophages. Moreover, they reported a transcriptional regulatory role of nuclear CSF-1R in promoting the expression of CSF-1 and cell proliferation genes, such as CCND1, MYC, and JUN [237]. In addition to the CSF-1R nuclear holoreceptor, CSF-1R ICD is also detected in nucleus but its function is still not well understood. Based on its stimuli, it has been speculated that cleavage of CSF-1R may be important during macrophage activation [81].
4.2.7 Insulin Receptor Family
Members of this family include the insulin-like growth factor 1 receptor (IGF-1R), insulin receptor (InsR), and insulin receptor-related protein (InsRR). In normal tissues, these receptors promote metabolism, cell proliferation, differentiation, and survival. However, there is accumulating evidence to suggest that most cancers highly express these receptors to promote tumorigenesis (reviewed in [238]).
Nuclear IGF-1R has also been detected in many types of cancers with high proliferation rate, including lung, breast, and prostate cancers, and hepatocyte carcinoma [239]. IGF-1-activated IGF-1R can be internalized through either caveolin- or clathrin-dependent endocytosis. However, nuclear translocation of IGF-1R is blocked by clathrin inhibitors but not by caveolin depletion, suggesting that nuclear transport of IGF-1R is CME dependent [239]. Moreover, ligand-dependent SUMOylation of IGF-1R further elevates its nuclear accumulation [240]. Results from ChIP-Seq analysis indicated that nuclear IGF-1R interacts with chromatin, and thus it may also regulate gene transcription [240]. Nuclear translocation of IGF-1R and its association with chromatin can be blocked by IGF-1R inhibitor [239]. IGF-1-induced nuclear InsR/IGF-1R heterodimer has been reported in corneal epithelial cells [241], and the target genes of the nuclear InsR/IGF-1R are involved in cell proliferation and cell cycle control as well as apoptosis. In hepatocyte carcinoma, IGF-1R nuclear translocation increases when cells are treated with gefitinib, and correlated with increased stem cell marker CD133, suggesting that nuclear IGF-1R may be involved in acquired drug resistance in hepatocyte carcinoma [242]. Interestingly, the extracellular α-subunit of IGF-1R (IGF-1Rα) but not the intracellular β-subunit accumulated in the nucleus following IGF treatment in Graves’ disease fibroblasts [243]. While the authors demonstrated that ADAM17 is required for nuclear translocation of IGF-1R, nuclear IGF-1Rα is not the typical ICD fragment observed for other RTKs, and it is not clear why this ligand binding subunit is transported into nucleus instead of the intracellular domain. More studies would be required to understand the intracellular trafficking mechanism of IGF-1Rα.
4.2.8 Eph Family
Because Eph and ephrin are both membrane bound, Eph signaling is activated via cell-cell interaction. However, the signaling outcome differs depending on the cell type and microenvironment [84]. Eph is responsible for embryonic development, disease pathogenesis, and tissue homeostasis.
Cytosolic Eph-B2 ICD (EphB2/CTF2) is generated by γ-secretase cleavage and is reported to phosphorylate and promote tyrosine phosphorylation of NMDA receptor and its cell surface localization [244]. Studies by immunohistochemical staining detected Eph-B2 in the nucleus of prostate cancer cells after demethylation agents, but whether it exists as a holoreceptor or truncated form and what functions are carried out by nuclear Eph-B2 have yet to be determined [245]. Eph-A4 has been reported in both the cytoplasm and nucleus of osteoblastic cells, but the size and function of nuclear Eph-A4 is not known [246].
4.2.9 Ryk Family
Wnt pathway components, Ror and 14-3-3, regulate nuclear localization of Ryk [247]. The function of Ryk signaling is parallel to that of the classical Wnt ligand, Frizzled [248]. Proteolytic cleavage of Ryk by γ-secretase is essential for Wnt-dependent neuronal differentiation [116, 248], and stabilization of Ryk-ICD by Cdc37 is necessary to prevent its degradation. Under Wnt stimulation, Ryk-ICD translocates into nucleus [117, 146] and is essential in neuronal differentiation [117].
4.2.10 PTK Family
PTK7 plays an important role in vertebrate tissue morphogenesis. As mentioned above, PTK7 is cleaved by ADAM17 and γ-secretase. Specifically, PTK-CTF2 has been shown to translocate into the nucleus, which promotes cell proliferation, migration, and colony formation [120]. Studies have indicated that while both PTK7-CTF 1 and 2 reduce the levels of phosphorylated c-Jun, phosphorylation of CREB and ATF1 is enhanced, suggesting that PTK7-CTFs may be involved in CREB/ATF1 regulation via the RAS-ERK pathway [249]. Moreover, PTK7-CTFs are present only in tumor tissues and correlated with cell motility and metastasis in colorectal cancer [250].
4.2.11 TAM Family
The TAM family is composed of Tyro3, Axl, and Mer receptors. Overexpression of members of the TAM family has been reported for many types of human cancer. For example, Axl and Mer are considered to be proto-oncogenes in colon cancer [251]. Growth arrest-specific gene 6 activates TAM receptors to promotes cancer cell proliferation and survival in vitro [252]. Mer contains a conventional NLS, and its glycosylated form has been detected in the nucleus [253]. Long-term ligand exposure induces the production of partially N-glycosylated Mer, which is associated with decreased and increased levels of plasma membrane and nuclear Mer, respectively. Nuclear Mer also exists as a soluble or chromatin-associated form, depending on the extent of its glycosylation [253]. These findings suggest that nuclear Mer may participate in transcriptional regulation in response to ligand stimulation.
5. Conclusion
In this review, we summarize the molecular mechanisms of intracellular RTK formation, trafficking, and their reported functions. Based on the current information, more studies are needed to better characterize the intracellular RTKs. For example, the retrograde trafficking mechanism for both RTK-ICD and membrane-bound RTK is not fully understood, and the underlying molecular machineries are only shown for several receptors. In addition, breakdown of RTKs via the canonical RTK degradation pathway also requires RTK internalization and cytosolic trafficking, and therefore, it may be difficult to distinguish RTK fragments detected in the cytosol from those intended to carry out non-canonical functions in other subcellular compartments. Most of the non-canonical functions are gleaned from studies of the FGFR and ErbB family. While transcriptional regulation seems to be the major function of nuclear RTKs (Table 2), their target genes have not been fully identified.
Table 2.
Functions of MRINs
| RTK | Type | Functions | Ref |
|---|---|---|---|
| RON | holoreceptor |
Transcription regulator Interacts with c-JUN promoter and HIF1α |
[224] |
| EGFR | holoreceptor |
Transcription regulator Interacts with RHA, MUC1, E2F1, STAT3/5 DNA replication Phosphorylates PCNA DNA repair Phosphorylates PCNA, ATM, histone H4 and interacts with DNA-PK |
[17, 18, 25, 156, 158–165, 170–176] |
| EGFR | EGFRvIII |
Transcription regulator Interacts with STAT3/5b |
[164, 177] |
| EGFR | mLEEK |
Transcription regulator Enhances transcription of GRP78 |
[32] |
| ErbB2 | holoreceptor |
Transcription regulator Interacts with STAT3, phosphorylate histone H2A1.2 Anti-apoptosis Phosphorylates Cdc2 |
[178, 182–185] |
| ErbB2 | 611-CTF |
Transcription regulator Enhances metastasis-related genes, e.g., MET, EPH2, MMP1, IL-11 and Integrin Cell mobility Phosphorylates cortactin |
[187, 188] |
| ErbB3 | holoreceptor |
Cell survival Nuclear ErbB3 negatively correlates with cell survival and cancer recurrence |
[191] |
| ErbB3 | 80-kDa |
Transcription regulator Regulates cyclin-D1 promoter |
[196] |
| ErbB3 | 50-kDa |
Transcription regulator Regulates transcription activity and distribution of ezrin |
[198] |
| ErbB4 | 4ICD |
Transcription regulator Interacts with YAP, ERβ, STAT5A, N-CoR, TAB-2, HF1α Regulates β-casein expression |
[147, 149–151, 199, 200] |
| FGFR1 | holoreceptor |
Transcription regulator Interacts with CREB-binding protein Phosphorylates RSK1 |
[145, 204, 205] |
| FGFR2 | holoreceptor |
Transcription regulator Interacts with STAT5 Correlates with poor prognosis in breast cancer |
[206, 207] |
| VEGFR1 | holoreceptor | Cell survival | [212] |
| VEGFR1 | ICD |
Transcription regulator [potential] Promotes angiogenesis |
[211, 213, 214] |
| VEGFR2 | holoreceptor |
Transcription regulator Binds to transcriptional factor Sp1 Wound healing |
[216, 217] |
| MET | holoreceptor |
Cell proliferation Induces InsP3-dependent calcium release |
[218] |
| MET | ICD |
Transcription regulator* Interacts with YAP Cell survival and reporgramming Enhances SOX9 and β-catenin signals Upregulates NANOG protein level |
[219–221] |
| ROR1 |
Transcription regulator Regulates actin cytoskeleton related gene transcription |
[231, 232] | |
| PDGFRα | holoreceptor | Embryonic and lung development [mouse model] | [235, 236] |
| CSF1R | holoreceptor |
Transcription regulator Enhances expression of CSF-1, CCND1, c-Myc and c-JUN |
[237] |
| CSF1R | ICD | Macrophage activation* | [81] |
| IGF1R |
Transcription regulator* Binds to chromatin |
[240] | |
| InsR |
Transcription regulator [Heterodimer of InsR/IGF1R] Promotes cell proliferation and anti-apoptosis Regulates cell cycle |
[241] | |
| Ryk | ICD | Embryonic neuronal differentiation [mouse model] | [117, 146] |
| PTK7 | CTF2 (ICD) | Cell proliferation and migration | [120] |
| PTK7 | CTF1 and 2 (ICD) | Phosphorylates CREB/ATF1 | [249] |
| Mer |
Transcription regulator* Binds to chromatin |
[253] |
Predicted roles.
RTK internalization is a common process in various cell types, but the effects of internalization differ. Normally, in most of cells, RTK internalization is more related to regulation of the magnitude of signaling by either receptor recycling or degradation. However, the internalized RTK also travels through retrograde transport to other subcellular compartments, such as mitochondria and nucleus, where it interacts with non-canonical substrates to regulate the signal specificity. Nuclear RTK is frequently found in highly proliferative tissues. For example, nEGFR is present in regenerating tissues, such as the liver and uterus of pregnant individual, cancer cells, and primary tumor specimens [156, 254–260], and nuclear IGF-1R is detected in many types of highly proliferative cancer [239]. There are also certain stimuli, e.g., ionizing radiation, cisplatin treatment, hypoxia, and some EGFR TKIs, that have been shown to enhance translocation of EGFR into the nucleus, where it promotes aberrant signaling, leading to increased cell proliferation and survival, and resistance to apoptosis and environmental stress [16, 24, 25, 167–169].
It is worthwhile to mention that although a large number of RTKs are internalized, only a small number travel to the nucleus. For example, ~80% of EGFR is internalized [134], but only ~2–6% of EGFR translocates into the nucleus following ligand stimulation [171]. In spite of that, the non-canonical substrates of nuclear RTK are critical regulators, e.g., transcriptional activators and co-activators, which can significantly amplify nuclear RTK signaling. As a result, a few nuclear localized RTK can have a significant impact on cell proliferation and survival.
In this review, we described the translocation of many RTKs into subcellular compartments, especially the nucleus as holoreceptors or intracellular fragments and their roles or substrates in non-canonical RTK signaling, which is much less known compared with the canonical RTK signaling. There is increasing evidence that points to the role of non-canonical RTK signaling in disease progression and therapeutic resistance [16, 21]. Thus, more in-depth investigation on our understanding of ICD formation as well as nuclear trafficking pathways will be required to develop more efficient targeted therapies as the RTK holoreceptors or ICD fragments are not efficiently targeted by current clinically used TKIs.
Acknowledgments
The authors would like to acknowledge the National Breast Cancer Foundation, Inc., National Institutes of Health (RO1 CA109311), and The University of Texas MD Anderson-China Medical University and Hospital Sister Institution Fund for their support.
We apologize to the authors whose original work could not be cited due to space limitation.
Abbreviations
- ADAM
a disintegrin and metalloproteinase
- AP
adaptor proteins
- ATM
ataxia telangiectasia mutated
- BACE
aspartic proteases
- Cdc2
cell division cycle protein 2 homolog
- CME
clathrin-mediated endocytosis
- COPI
coat protein 1
- COX-2
cyclooxygenase-2
- CREB
cAMP response element-binding protein
- CSF-1R
colony stimulating factor-1 receptor
- C-term
carboxyl terminus
- CTF
carboxyl-terminal fragment
- DNA-PK
DNA-dependent protein kinase
- EGFR
epidermal growth factor receptor
- Eph
Erythropoietin-producing hepatoma receptors
- ER
endoplasmic reticulum
- FEME
endophilin-mediated endocytosis
- FGFR
fibroblast growth factor receptor
- HGF
hepatocyte growth factor
- ICD
Intracellular domain
- IGF-1R
insulin-like growth factor 1 receptor
- INFS
integrative nuclear FGFR-1 signaling
- INM
inner nuclear membrane
- iNOS
inducible nitric oxide synthase
- InsP3
inoaitol 1,4,5-triphosphate
- InsR
insulin receptor
- INTERNET
integral trafficking from the ER to the nuclear envelope transport
- mLEEK
mini-LEEK
- MMP
matrix metalloproteinase
- MRIN
membrane receptor in nucleus
- MuSK
muscle specific kinase
- nEGFR
nuclear EGFR
- NGF
neurotrophins-nerve growth factors
- NLS
nuclear localization sequence
- NMDA
N-methyl-D-aspartic acid
- NPC
nuclear pore complex
- ONM
outer nuclear membrane
- PCNA
proliferation cell nuclear antigen
- PDGFR
platelet-derived growth factor receptor
- PEDF
pigment epithelium-derived factor
- PIP2
phosphatidylinositol (4,5)-biphosphate
- PKC
protein kinase C
- PMA
phorbol 12-myristate 13-acetate
- PTK7
protein-tyrosine pseudokinase 7
- RIP
regulated intramembrane proteolysis
- ROR
receptor tyrosine kinase-like orphan receptor
- RSK1
ribosome S6 kinase 1
- RTK
receptor tyrosine kinase
- Ryk
receptor-like tyrosine kinase
- STAT
signal transducer and activator
- TACE
tumor necrosis factor-α-converting enzyme
- TGN
trans Golgi network
- TK
tyrosine kinase
- TKI
tyrosine tyrosine kinase inhibitor
- Trk
tropomyosin receptor kinases
- VEGF
vascular endothelial growth factor
- VEGFR
vascular vascular endothelial growth factor receptor
References
- 1.Keefe DM, Bateman EH. Tumor control versus adverse events with targeted anticancer therapies. Nat Rev Clin Oncol. 2012;9:98–109. doi: 10.1038/nrclinonc.2011.192. [DOI] [PubMed] [Google Scholar]
- 2.Sundaram MV. Canonical RTK-Ras-ERK signaling and related alternative pathways. WormBook. 2013:1–38. doi: 10.1895/wormbook.1.80.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlessinger J. Receptor tyrosine kinases: legacy of the first two decades. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a008912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Templeton AJ, Diez-Gonzalez L, Ace O, Vera-Badillo F, Seruga B, Jordan J, Amir E, Pandiella A, Ocana A. Prognostic relevance of receptor tyrosine kinase expression in breast cancer: A meta-analysis. Cancer Treat Rev. 2014 doi: 10.1016/j.ctrv.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Neal JW, Sledge GW. Decade in review-targeted therapy: successes, toxicities and challenges in solid tumours. Nat Rev Clin Oncol. 2014;11:627–8. doi: 10.1038/nrclinonc.2014.171. [DOI] [PubMed] [Google Scholar]
- 7.Remon J, Moran T, Majem M, Reguart N, Dalmau E, Marquez-Medina D, Lianes P. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in EGFR-mutant non-small cell lung cancer: a new era begins. Cancer Treat Rev. 2014;40:93–101. doi: 10.1016/j.ctrv.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Favelyukis S, Till JH, Hubbard SR, Miller WT. Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat Struct Biol. 2001;8:1058–63. doi: 10.1038/nsb721. [DOI] [PubMed] [Google Scholar]
- 9.Furdui CM, Lew ED, Schlessinger J, Anderson KS. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol Cell. 2006;21:711–7. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 10.Honegger AM, Kris RM, Ullrich A, Schlessinger J. Evidence that autophosphorylation of solubilized receptors for epidermal growth factor is mediated by intermolecular cross-phosphorylation. Proc Natl Acad Sci U S A. 1989;86:925–9. doi: 10.1073/pnas.86.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobb MH, Sang BC, Gonzalez R, Goldsmith E, Ellis L. Autophosphorylation activates the soluble cytoplasmic domain of the insulin receptor in an intermolecular reaction. J Biol Chem. 1989;264:18701–6. [PubMed] [Google Scholar]
- 12.Krauss G. Biochemistry of Signal Transduction and Regulation. Wiley; Hoboken: 2014. p. 845. 1 online resource. [Google Scholar]
- 13.Volinsky N, Kholodenko BN. Complexity of receptor tyrosine kinase signal processing. Cold Spring Harb Perspect Biol. 2013;5:a009043. doi: 10.1101/cshperspect.a009043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carpenter G, Liao HJ. Receptor tyrosine kinases in the nucleus. Cold Spring Harb Perspect Biol. 2013;5:a008979. doi: 10.1101/cshperspect.a008979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YN, Hung MC. Nuclear functions and subcellular trafficking mechanisms of the epidermal growth factor receptor family. Cell Biosci. 2012;2:13. doi: 10.1186/2045-3701-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liccardi G, Hartley JA, Hochhauser D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 2011;71:1103–14. doi: 10.1158/0008-5472.CAN-10-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaganathan S, Yue P, Paladino DC, Bogdanovic J, Huo Q, Turkson J. A functional nuclear epidermal growth factor receptor, SRC and Stat3 heteromeric complex in pancreatic cancer cells. PLoS One. 2011;6:e19605. doi: 10.1371/journal.pone.0019605. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Lee HJ, Lan L, Peng G, Chang WC, Hsu MC, Wang YN, Cheng CC, Wei L, Nakajima S, Chang SS, Liao HW, Chen CH, Lavin M, Ang KK, Lin SY, Hung MC. Tyrosine 370 phosphorylation of ATM positively regulates DNA damage response. Cell Res. 2015 doi: 10.1038/cr.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo HY, Huang YS, Tseng CH, Chen YC, Chang YW, Shih HM, Wu CW. PML represses lung cancer metastasis by suppressing the nuclear EGFR-mediated transcriptional activation of MMP2. Cell Cycle. 2014;13:3132–42. doi: 10.4161/15384101.2014.949212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Traynor AM, Weigel TL, Oettel KR, Yang DT, Zhang C, Kim K, Salgia R, Iida M, Brand TM, Hoang T, Campbell TC, Hernan HR, Wheeler DL. Nuclear EGFR protein expression predicts poor survival in early stage non-small cell lung cancer. Lung Cancer. 2013;81:138–41. doi: 10.1016/j.lungcan.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coleman SJ, Chioni AM, Ghallab M, Anderson RK, Lemoine NR, Kocher HM, Grose RP. Nuclear translocation of FGFR1 and FGF2 in pancreatic stellate cells facilitates pancreatic cancer cell invasion. EMBO Mol Med. 2014;6:467–81. doi: 10.1002/emmm.201302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chioni AM, Grose R. FGFR1 cleavage and nuclear translocation regulates breast cancer cell behavior. J Cell Biol. 2012;197:801–17. doi: 10.1083/jcb.201108077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadzisejdic I, Mustac E, Jonjic N, Petkovic M, Grahovac B. Nuclear EGFR in ductal invasive breast cancer: correlation with cyclin-D1 and prognosis. Mod Pathol. 2010;23:392–403. doi: 10.1038/modpathol.2009.166. [DOI] [PubMed] [Google Scholar]
- 24.Li C, Iida M, Dunn EF, Ghia AJ, Wheeler DL. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009;28:3801–13. doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang WC, Chen YJ, Li LY, Wei YL, Hsu SC, Tsai SL, Chiu PC, Huang WP, Wang YN, Chen CH, Chang WC, Chang WC, Chen AJ, Tsai CH, Hung MC. Nuclear translocation of epidermal growth factor receptor by Akt-dependent phosphorylation enhances breast cancer-resistant protein expression in gefitinib-resistant cells. J Biol Chem. 2011;286:20558–68. doi: 10.1074/jbc.M111.240796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheeler DL, Huang S, Kruser TJ, Nechrebecki MM, Armstrong EA, Benavente S, Gondi V, Hsu KT, Harari PM. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members. Oncogene. 2008;27:3944–56. doi: 10.1038/onc.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brand TM, Iida M, Luthar N, Starr MM, Huppert EJ, Wheeler DL. Nuclear EGFR as a molecular target in cancer. Radiother Oncol. 2013;108:370–7. doi: 10.1016/j.radonc.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 28.Ancot F, Foveau B, Lefebvre J, Leroy C, Tulasne D. Proteolytic cleavages give receptor tyrosine kinases the gift of ubiquity. Oncogene. 2009;28:2185–95. doi: 10.1038/onc.2009.88. [DOI] [PubMed] [Google Scholar]
- 29.Deheuninck J, Goormachtigh G, Foveau B, Ji Z, Leroy C, Ancot F, Villeret V, Tulasne D, Fafeur V. Phosphorylation of the MET receptor on juxtamembrane tyrosine residue 1001 inhibits its caspase-dependent cleavage. Cell Signal. 2009;21:1455–63. doi: 10.1016/j.cellsig.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Bonomi S, Gallo S, Catillo M, Pignataro D, Biamonti G, Ghigna C. Oncogenic alternative splicing switches: role in cancer progression and prospects for therapy. Int J Cell Biol. 2013;2013:962038. doi: 10.1155/2013/962038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ekstrand AJ, Sugawa N, James CD, Collins VP. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc Natl Acad Sci U S A. 1992;89:4309–13. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piccione EC, Lieu TJ, Gentile CF, Williams TR, Connolly AJ, Godwin AK, Koong AC, Wong AJ. A novel epidermal growth factor receptor variant lacking multiple domains directly activates transcription and is overexpressed in tumors. Oncogene. 2012;31:2953–67. doi: 10.1038/onc.2011.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barker PA, Lomen-Hoerth C, Gensch EM, Meakin SO, Glass DJ, Shooter EM. Tissue-specific alternative splicing generates two isoforms of the trkA receptor. J Biol Chem. 1993;268:15150–7. [PubMed] [Google Scholar]
- 34.Tacconelli A, Farina AR, Cappabianca L, Desantis G, Tessitore A, Vetuschi A, Sferra R, Rucci N, Argenti B, Screpanti I, Gulino A, Mackay AR. TrkA alternative splicing: a regulated tumor-promoting switch in human neuroblastoma. Cancer Cell. 2004;6:347–60. doi: 10.1016/j.ccr.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 35.Farina AR, Tacconelli A, Cappabianca L, Cea G, Chioda A, Romanelli A, Pensato S, Pedone C, Gulino A, Mackay AR. The neuroblastoma tumour-suppressor TrkAI and its oncogenic alternative TrkAIII splice variant exhibit geldanamycin-sensitive interactions with Hsp90 in human neuroblastoma cells. Oncogene. 2009;28:4075–94. doi: 10.1038/onc.2009.256. [DOI] [PubMed] [Google Scholar]
- 36.Adrain C, Freeman M. Regulation of receptor tyrosine kinase ligand processing. Cold Spring Harb Perspect Biol. 2014:6. doi: 10.1101/cshperspect.a008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landman N, Kim TW. Got RIP? Presenilin-dependent intramembrane proteolysis in growth factor receptor signaling. Cytokine Growth Factor Rev. 2004;15:337–51. doi: 10.1016/j.cytogfr.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Hayashida K, Bartlett AH, Chen Y, Park PW. Molecular and Cellular Mechanisms of Ectodomain Shedding. The Anatomical Record: Advances in Integrative Anatomy and Evolutionary Biology. 2010;293:925–937. doi: 10.1002/ar.20757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller MA, Meyer AS, Beste MT, Lasisi Z, Reddy S, Jeng KW, Chen CH, Han J, Isaacson K, Griffith LG, Lauffenburger DA. ADAM-10 and -17 regulate endometriotic cell migration via concerted ligand and receptor shedding feedback on kinase signaling. Proc Natl Acad Sci U S A. 2013;110:E2074–83. doi: 10.1073/pnas.1222387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stawikowska R, Cudic M, Giulianotti M, Houghten RA, Fields GB, Minond D. Activity of ADAM17 (a disintegrin and metalloprotease 17) is regulated by its noncatalytic domains and secondary structure of its substrates. J Biol Chem. 2013;288:22871–9. doi: 10.1074/jbc.M113.462267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Black RA, Doedens JR, Mahimkar R, Johnson R, Guo L, Wallace A, Virca D, Eisenman J, Slack J, Castner B, Sunnarborg SW, Lee DC, Cowling R, Jin G, Charrier K, Peschon JJ, Paxton R. Substrate specificity and inducibility of TACE (tumour necrosis factor alpha-converting enzyme) revisited: the Ala-Val preference, and induced intrinsic activity. Biochem Soc Symp. 2003:39–52. doi: 10.1042/bss0700039. [DOI] [PubMed] [Google Scholar]
- 42.Mohan MJ, Seaton T, Mitchell J, Howe A, Blackburn K, Burkhart W, Moyer M, Patel I, Waitt GM, Becherer JD, Moss ML, Milla ME. The tumor necrosis factor-alpha converting enzyme (TACE): a unique metalloproteinase with highly defined substrate selectivity. Biochemistry. 2002;41:9462–9. doi: 10.1021/bi0260132. [DOI] [PubMed] [Google Scholar]
- 43.Caescu CI, Jeschke GR, Turk BE. Active-site determinants of substrate recognition by the metalloproteinases TACE and ADAM10. Biochem J. 2009;424:79–88. doi: 10.1042/BJ20090549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selkoe DJ, Wolfe MS. Presenilin: running with scissors in the membrane. Cell. 2007;131:215–21. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Kimberly WT, LaVoie MJ, Ostaszewski BL, Ye W, Wolfe MS, Selkoe DJ. Gamma-secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2. Proc Natl Acad Sci U S A. 2003;100:6382–7. doi: 10.1073/pnas.1037392100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C. Reconstitution of gamma-secretase activity. Nat Cell Biol. 2003;5:486–8. doi: 10.1038/ncb960. [DOI] [PubMed] [Google Scholar]
- 47.Fraering PC, Ye W, Strub JM, Dolios G, LaVoie MJ, Ostaszewski BL, van Dorsselaer A, Wang R, Selkoe DJ, Wolfe MS. Purification and characterization of the human gamma-secretase complex. Biochemistry. 2004;43:9774–89. doi: 10.1021/bi0494976. [DOI] [PubMed] [Google Scholar]
- 48.Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, LaPlant Q, Ball H, Dann CE, 3rd, Sudhof T, Yu G. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122:435–47. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 49.Xia W, Wolfe MS. Intramembrane proteolysis by presenilin and presenilin-like proteases. J Cell Sci. 2003;116:2839–44. doi: 10.1242/jcs.00651. [DOI] [PubMed] [Google Scholar]
- 50.Lichtenthaler SF, Wang R, Grimm H, Uljon SN, Masters CL, Beyreuther K. Mechanism of the cleavage specificity of Alzheimer’s disease gamma-secretase identified by phenylalanine-scanning mutagenesis of the transmembrane domain of the amyloid precursor protein. Proc Natl Acad Sci U S A. 1999;96:3053–8. doi: 10.1073/pnas.96.6.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tischer E, Cordell B. Beta-amyloid precursor protein. Location of transmembrane domain and specificity of gamma-secretase cleavage. J Biol Chem. 1996;271:21914–9. doi: 10.1074/jbc.271.36.21914. [DOI] [PubMed] [Google Scholar]
- 52.Lichtenthaler SF, Ida N, Multhaup G, Masters CL, Beyreuther K. Mutations in the transmembrane domain of APP altering gamma-secretase specificity. Biochemistry. 1997;36:15396–403. doi: 10.1021/bi971071m. [DOI] [PubMed] [Google Scholar]
- 53.Murphy MP, Hickman LJ, Eckman CB, Uljon SN, Wang R, Golde TE. gamma-Secretase, evidence for multiple proteolytic activities and influence of membrane positioning of substrate on generation of amyloid beta peptides of varying length. J Biol Chem. 1999;274:11914–23. doi: 10.1074/jbc.274.17.11914. [DOI] [PubMed] [Google Scholar]
- 54.Hemming ML, Elias JE, Gygi SP, Selkoe DJ. Proteomic profiling of gamma-secretase substrates and mapping of substrate requirements. PLoS Biol. 2008;6:e257. doi: 10.1371/journal.pbio.0060257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Struhl G, Adachi A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol Cell. 2000;6:625–36. doi: 10.1016/s1097-2765(00)00061-7. [DOI] [PubMed] [Google Scholar]
- 56.Veikkolainen V, Vaparanta K, Halkilahti K, Iljin K, Sundvall M, Elenius K. Function of ERBB4 is determined by alternative splicing. Cell Cycle. 2011;10:2647–57. doi: 10.4161/cc.10.16.17194. [DOI] [PubMed] [Google Scholar]
- 57.Elenius K, Corfas G, Paul S, Choi CJ, Rio C, Plowman GD, Klagsbrun M. A novel juxtamembrane domain isoform of HER4/ErbB4. Isoform-specific tissue distribution and differential processing in response to phorbol ester. J Biol Chem. 1997;272:26761–8. doi: 10.1074/jbc.272.42.26761. [DOI] [PubMed] [Google Scholar]
- 58.Zhou W, Carpenter G. Heregulin-dependent trafficking and cleavage of ErbB-4. J Biol Chem. 2000;275:34737–43. doi: 10.1074/jbc.M003756200. [DOI] [PubMed] [Google Scholar]
- 59.Rio C, Buxbaum JD, Peschon JJ, Corfas G. Tumor necrosis factor-alpha-converting enzyme is required for cleavage of erbB4/HER4. J Biol Chem. 2000;275:10379–87. doi: 10.1074/jbc.275.14.10379. [DOI] [PubMed] [Google Scholar]
- 60.Vecchi M, Baulida J, Carpenter G. Selective cleavage of the heregulin receptor ErbB-4 by protein kinase C activation. J Biol Chem. 1996;271:18989–95. doi: 10.1074/jbc.271.31.18989. [DOI] [PubMed] [Google Scholar]
- 61.Ni CY, Murphy MP, Golde TE, Carpenter G. gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–81. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- 62.Cheng QC, Tikhomirov O, Zhou W, Carpenter G. Ectodomain cleavage of ErbB-4: characterization of the cleavage site and m80 fragment. J Biol Chem. 2003;278:38421–7. doi: 10.1074/jbc.M302111200. [DOI] [PubMed] [Google Scholar]
- 63.Fujiwara S, Hung M, Yamamoto-Ibusuk CM, Yamamoto Y, Yamamoto S, Tomiguchi M, Takeshita T, Hayashi M, Sueta A, Iwase H. The localization of HER4 intracellular domain and expression of its alternately-spliced isoforms have prognostic significance in ER+ HER2- breast cancer. Oncotarget. 2014;5:3919–30. doi: 10.18632/oncotarget.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thor AD, Edgerton SM, Jones FE. Subcellular localization of the HER4 intracellular domain, 4ICD, identifies distinct prognostic outcomes for breast cancer patients. Am J Pathol. 2009;175:1802–9. doi: 10.2353/ajpath.2009.090204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohd Nafi SN, Generali D, Kramer-Marek G, Gijsen M, Strina C, Cappelletti M, Andreis D, Haider S, Li JL, Bridges E, Capala J, Ioannis R, Harris AL, Kong A. Nuclear HER4 mediates acquired resistance to trastuzumab and is associated with poor outcome in HER2 positive breast cancer. Oncotarget. 2014;5:5934–49. doi: 10.18632/oncotarget.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naresh A, Thor AD, Edgerton SM, Torkko KC, Kumar R, Jones FE. The HER4/4ICD estrogen receptor coactivator and BH3-only protein is an effector of tamoxifen-induced apoptosis. Cancer Res. 2008;68:6387–95. doi: 10.1158/0008-5472.CAN-08-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Codony-Servat J, Albanell J, Lopez-Talavera JC, Arribas J, Baselga J. Cleavage of the HER2 ectodomain is a pervanadate-activable process that is inhibited by the tissue inhibitor of metalloproteases-1 in breast cancer cells. Cancer Res. 1999;59:1196–201. [PubMed] [Google Scholar]
- 68.Scaltriti M, Rojo F, Ocana A, Anido J, Guzman M, Cortes J, Di Cosimo S, Matias-Guiu X, Ramon y Cajal S, Arribas J, Baselga J. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J Natl Cancer Inst. 2007;99:628–38. doi: 10.1093/jnci/djk134. [DOI] [PubMed] [Google Scholar]
- 69.Molina MA, Codony-Servat J, Albanell J, Rojo F, Arribas J, Baselga J. Trastuzumab (herceptin), a humanized anti-Her2 receptor monoclonal antibody, inhibits basal and activated Her2 ectodomain cleavage in breast cancer cells. Cancer Res. 2001;61:4744–9. [PubMed] [Google Scholar]
- 70.Xia W, Liu Z, Zong R, Liu L, Zhao S, Bacus SS, Mao Y, He J, Wulfkuhle JD, Petricoin EF, 3rd, Osada T, Yang XY, Hartman ZC, Clay TM, Blackwell KL, Lyerly HK, Spector NL. Truncated ErbB2 expressed in tumor cell nuclei contributes to acquired therapeutic resistance to ErbB2 kinase inhibitors. Mol Cancer Ther. 2011;10:1367–74. doi: 10.1158/1535-7163.MCT-10-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Warri AM, Isola JJ, Harkonen PL. Anti-oestrogen stimulation of ERBB2 ectodomain shedding from BT-474 human breast cancer cells with ERBB2 gene amplification. Eur J Cancer. 1996;32A:134–40. doi: 10.1016/0959-8049(95)00550-1. [DOI] [PubMed] [Google Scholar]
- 72.Nakamichi Y, Udagawa N, Takahashi N. IL-34 and CSF-1: similarities and differences. J Bone Miner Metab. 2013;31:486–95. doi: 10.1007/s00774-013-0476-3. [DOI] [PubMed] [Google Scholar]
- 73.Elmore MR, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, Kitazawa M, Matusow B, Nguyen H, West BL, Green KN. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82:380–97. doi: 10.1016/j.neuron.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stanley ER, Chitu V. CSF-1 receptor signaling in myeloid cells. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Glenn G, van der Geer P. CSF-1 and TPA stimulate independent pathways leading to lysosomal degradation or regulated intramembrane proteolysis of the CSF-1 receptor. FEBS Lett. 2007;581:5377–81. doi: 10.1016/j.febslet.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Downing JR, Roussel MF, Sherr CJ. Ligand and protein kinase C downmodulate the colony-stimulating factor 1 receptor by independent mechanisms. Mol Cell Biol. 1989;9:2890–6. doi: 10.1128/mcb.9.7.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baccarini M, Dello Sbarba P, Buscher D, Bartocci A, Stanley ER. IFN-gamma/lipopolysaccharide activation of macrophages is associated with protein kinase C-dependent down-modulation of the colony-stimulating factor-1 receptor. J Immunol. 1992;149:2656–61. [PubMed] [Google Scholar]
- 78.Dello Sbarba P, Nencioni L, Labardi D, Rovida E, Caciagli B, Cipolleschi MG. Interleukin 2 down-modulates the macrophage colony-stimulating factor receptor in murine macrophages. Cytokine. 1996;8:488–94. doi: 10.1006/cyto.1996.0066. [DOI] [PubMed] [Google Scholar]
- 79.Dello Sbarba P, Rovida E, Caciagli B, Nencioni L, Labardi D, Paccagnini A, Savini L, Cipolleschi MG. Interleukin-4 rapidly down-modulates the macrophage colony-stimulating factor receptor in murine macrophages. J Leukoc Biol. 1996;60:644–50. doi: 10.1002/jlb.60.5.644. [DOI] [PubMed] [Google Scholar]
- 80.Wilhelmsen K, van der Geer P. Phorbol 12-myristate 13-acetate-induced release of the colony-stimulating factor 1 receptor cytoplasmic domain into the cytosol involves two separate cleavage events. Mol Cell Biol. 2004;24:454–64. doi: 10.1128/MCB.24.1.454-464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Glenn G, van der Geer P. Toll-like receptors stimulate regulated intramembrane proteolysis of the CSF-1 receptor through Erk activation. FEBS Lett. 2008;582:911–5. doi: 10.1016/j.febslet.2008.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rovida E, Paccagnini A, Del Rosso M, Peschon J, Dello Sbarba P. TNF-alpha-converting enzyme cleaves the macrophage colony-stimulating factor receptor in macrophages undergoing activation. J Immunol. 2001;166:1583–9. doi: 10.4049/jimmunol.166.3.1583. [DOI] [PubMed] [Google Scholar]
- 83.Vahidi A, Glenn G, van der Geer P. Identification and mutagenesis of the TACE and gamma-secretase cleavage sites in the colony-stimulating factor 1 receptor. Biochem Biophys Res Commun. 2014;450:782–7. doi: 10.1016/j.bbrc.2014.06.061. [DOI] [PubMed] [Google Scholar]
- 84.Gucciardo E, Sugiyama N, Lehti K. Eph- and ephrin-dependent mechanisms in tumor and stem cell dynamics. Cell Mol Life Sci. 2014;71:3685–710. doi: 10.1007/s00018-014-1633-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tomita T, Tanaka S, Morohashi Y, Iwatsubo T. Presenilin-dependent intramembrane cleavage of ephrin-B1. Mol Neurodegener. 2006;1:2. doi: 10.1186/1750-1326-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin KT, Sloniowski S, Ethell DW, Ethell IM. Ephrin-B2-induced cleavage of EphB2 receptor is mediated by matrix metalloproteinases to trigger cell repulsion. J Biol Chem. 2008;283:28969–79. doi: 10.1074/jbc.M804401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Litterst C, Georgakopoulos A, Shioi J, Ghersi E, Wisniewski T, Wang R, Ludwig A, Robakis NK. Ligand binding and calcium influx induce distinct ectodomain/gamma-secretase-processing pathways of EphB2 receptor. J Biol Chem. 2007;282:16155–63. doi: 10.1074/jbc.M611449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barthet G, Dunys J, Shao Z, Xuan Z, Ren Y, Xu J, Arbez N, Mauger G, Bruban J, Georgakopoulos A, Shioi J, Robakis NK. Presenilin mediates neuroprotective functions of ephrinB and brain-derived neurotrophic factor and regulates ligand-induced internalization and metabolism of EphB2 and TrkB receptors. Neurobiol Aging. 2013;34:499–510. doi: 10.1016/j.neurobiolaging.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Raikwar NS, Liu KZ, Thomas CP. N-Terminal Cleavage and Release of the Ectodomain of Flt1 Is Mediated via ADAM10 and ADAM 17 and Regulated by VEGFR2 and the Flt1 Intracellular Domain. PLoS One. 2014;9:e112794. doi: 10.1371/journal.pone.0112794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cai J, Qi X, Kociok N, Skosyrski S, Emilio A, Ruan Q, Han S, Liu L, Chen Z, Bowes Rickman C, Golde T, Grant MB, Saftig P, Serneels L, de Strooper B, Joussen AM, Boulton ME. beta-Secretase (BACE1) inhibition causes retinal pathology by vascular dysregulation and accumulation of age pigment. EMBO Mol Med. 2012;4:980–91. doi: 10.1002/emmm.201101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rahimi N, Golde TE, Meyer RD. Identification of ligand-induced proteolytic cleavage and ectodomain shedding of VEGFR-1/FLT1 in leukemic cancer cells. Cancer Res. 2009;69:2607–14. doi: 10.1158/0008-5472.CAN-08-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Keskin U, Ulubay M, Dede M, Ozgurtas T, Kocyigit YK, Aydin FN, Ergun A. The relationship between the VEGF/sVEGFR-1 ratio and threatened abortion. Arch Gynecol Obstet. 2015;291:557–61. doi: 10.1007/s00404-014-3452-9. [DOI] [PubMed] [Google Scholar]
- 93.Cooper CS, Park M, Blair DG, Tainsky MA, Huebner K, Croce CM, Vande Woude GF. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984;311:29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- 94.Maroun CR, Rowlands T. The Met receptor tyrosine kinase: a key player in oncogenesis and drug resistance. Pharmacol Ther. 2014;142:316–38. doi: 10.1016/j.pharmthera.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 95.Fu L, Guo W, Liu B, Sun L, Bi Z, Zhu L, Wang X, Liu B, Xie Q, Li K. Shedding of c-Met ectodomain correlates with c-Met expression in non-small cell lung cancer. Biomarkers. 2013;18:126–35. doi: 10.3109/1354750X.2012.751455. [DOI] [PubMed] [Google Scholar]
- 96.Athauda G, Giubellino A, Coleman JA, Horak C, Steeg PS, Lee MJ, Trepel J, Wimberly J, Sun J, Coxon A, Burgess TL, Bottaro DP. c-Met ectodomain shedding rate correlates with malignant potential. Clin Cancer Res. 2006;12:4154–62. doi: 10.1158/1078-0432.CCR-06-0250. [DOI] [PubMed] [Google Scholar]
- 97.Galvani AP, Cristiani C, Carpinelli P, Landonio A, Bertolero F. Suramin modulates cellular levels of hepatocyte growth factor receptor by inducing shedding of a soluble form. Biochem Pharmacol. 1995;50:959–66. doi: 10.1016/0006-2952(95)00219-p. [DOI] [PubMed] [Google Scholar]
- 98.Petrelli A, Circosta P, Granziero L, Mazzone M, Pisacane A, Fenoglio S, Comoglio PM, Giordano S. Ab-induced ectodomain shedding mediates hepatocyte growth factor receptor down-regulation and hampers biological activity. Proc Natl Acad Sci U S A. 2006;103:5090–5. doi: 10.1073/pnas.0508156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schelter F, Kobuch J, Moss ML, Becherer JD, Comoglio PM, Boccaccio C, Kruger A. A disintegrin and metalloproteinase-10 (ADAM-10) mediates DN30 antibody-induced shedding of the met surface receptor. J Biol Chem. 2010;285:26335–40. doi: 10.1074/jbc.M110.106435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kopitz C, Gerg M, Bandapalli OR, Ister D, Pennington CJ, Hauser S, Flechsig C, Krell HW, Antolovic D, Brew K, Nagase H, Stangl M, von Weyhern CW, Brucher BL, Brand K, Coussens LM, Edwards DR, Kruger A. Tissue inhibitor of metalloproteinases-1 promotes liver metastasis by induction of hepatocyte growth factor signaling. Cancer Res. 2007;67:8615–23. doi: 10.1158/0008-5472.CAN-07-0232. [DOI] [PubMed] [Google Scholar]
- 101.Chalupsky K, Kanchev I, Zbodakova O, Buryova H, Jirouskova M, Korinek V, Gregor M, Sedlacek R. ADAM10/17-dependent release of soluble c-Met correlates with hepatocellular damage. Folia Biol (Praha) 2013;59:76–86. [PubMed] [Google Scholar]
- 102.Prat M, Crepaldi T, Gandino L, Giordano S, Longati P, Comoglio P. C-terminal truncated forms of Met, the hepatocyte growth factor receptor. Mol Cell Biol. 1991;11:5954–62. doi: 10.1128/mcb.11.12.5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Van Schaeybroeck S, Kalimutho M, Dunne PD, Carson R, Allen W, Jithesh PV, Redmond KL, Sasazuki T, Shirasawa S, Blayney J, Michieli P, Fenning C, Lenz HJ, Lawler M, Longley DB, Johnston PG. ADAM17-dependent c-MET-STAT3 signaling mediates resistance to MEK inhibitors in KRAS mutant colorectal cancer. Cell Rep. 2014;7:1940–55. doi: 10.1016/j.celrep.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 104.Foveau B, Ancot F, Leroy C, Petrelli A, Reiss K, Vingtdeux V, Giordano S, Fafeur V, Tulasne D. Down-regulation of the met receptor tyrosine kinase by presenilin-dependent regulated intramembrane proteolysis. Mol Biol Cell. 2009;20:2495–507. doi: 10.1091/mbc.E08-09-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Puri MC, Rossant J, Alitalo K, Bernstein A, Partanen J. The receptor tyrosine kinase TIE is required for integrity and survival of vascular endothelial cells. EMBO J. 1995;14:5884–91. doi: 10.1002/j.1460-2075.1995.tb00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kaipainen A, Vlaykova T, Hatva E, Bohling T, Jekunen A, Pyrhonen S, Alitalo K. Enhanced expression of the tie receptor tyrosine kinase mesenger RNA in the vascular endothelium of metastatic melanomas. Cancer Res. 1994;54:6571–7. [PubMed] [Google Scholar]
- 107.Yabkowitz R, Meyer S, Black T, Elliott G, Merewether LA, Yamane HK. Inflammatory cytokines and vascular endothelial growth factor stimulate the release of soluble tie receptor from human endothelial cells via metalloprotease activation. Blood. 1999;93:1969–79. [PubMed] [Google Scholar]
- 108.McCarthy MJ, Burrows R, Bell SC, Christie G, Bell PR, Brindle NP. Potential roles of metalloprotease mediated ectodomain cleavage in signaling by the endothelial receptor tyrosine kinase Tie-1. Lab Invest. 1999;79:889–95. [PubMed] [Google Scholar]
- 109.Chen-Konak L, Guetta-Shubin Y, Yahav H, Shay-Salit A, Zilberman M, Binah O, Resnick N. Transcriptional and post-translation regulation of the Tie1 receptor by fluid shear stress changes in vascular endothelial cells. FASEB J. 2003;17:2121–3. doi: 10.1096/fj.02-1151fje. [DOI] [PubMed] [Google Scholar]
- 110.Marron MB, Singh H, Tahir TA, Kavumkal J, Kim HZ, Koh GY, Brindle NP. Regulated proteolytic processing of Tie1 modulates ligand responsiveness of the receptor-tyrosine kinase Tie2. J Biol Chem. 2007;282:30509–17. doi: 10.1074/jbc.M702535200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rees KA, Singh H, Brindle NP. The receptor tyrosine kinase Tie1 is expressed and activated in epithelial tumour cell lines. Int J Oncol. 2007;31:893–7. [PubMed] [Google Scholar]
- 112.Yang XH, Hand RA, Livasy CA, Cance WG, Craven RJ. Overexpression of the receptor tyrosine kinase Tie-1 intracellular domain in breast cancer. Tumour Biol. 2003;24:61–9. doi: 10.1159/000071078. [DOI] [PubMed] [Google Scholar]
- 113.Shen B, Shang Z, Wang B, Zhang L, Zhou F, Li T, Chu M, Jiang H, Wang Y, Qiao T, Zhang J, Sun W, Kong X, He Y. Genetic dissection of tie pathway in mouse lymphatic maturation and valve development. Arterioscler Thromb Vasc Biol. 2014;34:1221–30. doi: 10.1161/ATVBAHA.113.302923. [DOI] [PubMed] [Google Scholar]
- 114.Katso RM, Russell RB, Ganesan TS. Functional analysis of H-Ryk, an atypical member of the receptor tyrosine kinase family. Mol Cell Biol. 1999;19:6427–40. doi: 10.1128/mcb.19.9.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Halford MM, Stacker SA. Revelations of the RYK receptor. Bioessays. 2001;23:34–45. doi: 10.1002/1521-1878(200101)23:1<34::AID-BIES1005>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 116.Yoshikawa S, McKinnon RD, Kokel M, Thomas JB. Wnt-mediated axon guidance via the Drosophila Derailed receptor. Nature. 2003;422:583–8. doi: 10.1038/nature01522. [DOI] [PubMed] [Google Scholar]
- 117.Lyu J, Yamamoto V, Lu W. Cleavage of the Wnt receptor Ryk regulates neuronal differentiation during cortical neurogenesis. Dev Cell. 2008;15:773–80. doi: 10.1016/j.devcel.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 118.Kasuga K, Kaneko H, Nishizawa M, Onodera O, Ikeuchi T. Generation of intracellular domain of insulin receptor tyrosine kinase by gamma-secretase. Biochem Biophys Res Commun. 2007;360:90–6. doi: 10.1016/j.bbrc.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 119.McElroy B, Powell JC, McCarthy JV. The insulin-like growth factor 1 (IGF-1) receptor is a substrate for gamma-secretase-mediated intramembrane proteolysis. Biochem Biophys Res Commun. 2007;358:1136–41. doi: 10.1016/j.bbrc.2007.05.062. [DOI] [PubMed] [Google Scholar]
- 120.Na HW, Shin WS, Ludwig A, Lee ST. The cytosolic domain of protein-tyrosine kinase 7 (PTK7), generated from sequential cleavage by a disintegrin and metalloprotease 17 (ADAM17) and gamma-secretase, enhances cell proliferation and migration in colon cancer cells. J Biol Chem. 2012;287:25001–9. doi: 10.1074/jbc.M112.348904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Degnin CR, Laederich MB, Horton WA. Ligand activation leads to regulated intramembrane proteolysis of fibroblast growth factor receptor 3. Mol Biol Cell. 2011;22:3861–73. doi: 10.1091/mbc.E11-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.El-Sayed A, Harashima H. Endocytosis of gene delivery vectors: from clathrin-dependent to lipid raft-mediated endocytosis. Mol Ther. 2013;21:1118–30. doi: 10.1038/mt.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wandinger-Ness A, Zerial M. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb Perspect Biol. 2014;6:a022616. doi: 10.1101/cshperspect.a022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bonifacino JS. Adaptor proteins involved in polarized sorting. J Cell Biol. 2014;204:7–17. doi: 10.1083/jcb.201310021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Goh LK, Huang F, Kim W, Gygi S, Sorkin A. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J Cell Biol. 2010;189:871–83. doi: 10.1083/jcb.201001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sorkin A, Fortian A. Endocytosis and Endosomal Sorting of Receptor Tyrosine Kinases. In: Wheeler DL, Yarden Y, editors. Receptor Tyrosine Kinases: Structure, Functions and Role in Human Disease. Springer; New York: 2015. pp. 133–161. [Google Scholar]
- 127.Kelly BT, Graham SC, Liska N, Dannhauser PN, Honing S, Ungewickell EJ, Owen DJ. Clathrin adaptors. AP2 controls clathrin polymerization with a membrane-activated switch. Science. 2014;345:459–63. doi: 10.1126/science.1254836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Traub LM, Bonifacino JS. Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb Perspect Biol. 2013;5:a016790. doi: 10.1101/cshperspect.a016790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang F, Jiang X, Sorkin A. Tyrosine phosphorylation of the beta2 subunit of clathrin adaptor complex AP-2 reveals the role of a di-leucine motif in the epidermal growth factor receptor trafficking. J Biol Chem. 2003;278:43411–7. doi: 10.1074/jbc.M306072200. [DOI] [PubMed] [Google Scholar]
- 130.Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, Di Fiore PP, Polo S. Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci U S A. 2005;102:2760–5. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Goh LK, Sorkin A. Endocytosis of receptor tyrosine kinases. Cold Spring Harb Perspect Biol. 2013;5:a017459. doi: 10.1101/cshperspect.a017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Boucrot E, Ferreira AP, Almeida-Souza L, Debard S, Vallis Y, Howard G, Bertot L, Sauvonnet N, McMahon HT. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature. 2015;517:460–5. doi: 10.1038/nature14067. [DOI] [PubMed] [Google Scholar]
- 133.Renard HF, Simunovic M, Lemiere J, Boucrot E, Garcia-Castillo MD, Arumugam S, Chambon V, Lamaze C, Wunder C, Kenworthy AK, Schmidt AA, McMahon HT, Sykes C, Bassereau P, Johannes L. Endophilin-A2 functions in membrane scission in clathrin-independent endocytosis. Nature. 2015;517:493–6. doi: 10.1038/nature14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sorkin A, Krolenko S, Kudrjavtceva N, Lazebnik J, Teslenko L, Soderquist AM, Nikolsky N. Recycling of epidermal growth factor-receptor complexes in A431 cells: identification of dual pathways. J Cell Biol. 1991;112:55–63. doi: 10.1083/jcb.112.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hsu VW, Prekeris R. Transport at the recycling endosome. Curr Opin Cell Biol. 2010;22:528–34. doi: 10.1016/j.ceb.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Du Y, Shen J, Hsu JL, Han Z, Hsu MC, Yang CC, Kuo HP, Wang YN, Yamaguchi H, Miller SA, Hung MC. Syntaxin 6-mediated Golgi translocation plays an important role in nuclear functions of EGFR through microtubule-dependent trafficking. Oncogene. 2014;33:756–70. doi: 10.1038/onc.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Parachoniak CA, Luo Y, Abella JV, Keen JH, Park M. GGA3 functions as a switch to promote Met receptor recycling, essential for sustained ERK and cell migration. Dev Cell. 2011;20:751–63. doi: 10.1016/j.devcel.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Myers JM, Martins GG, Ostrowski J, Stachowiak MK. Nuclear trafficking of FGFR1: a role for the transmembrane domain. J Cell Biochem. 2003;88:1273–91. doi: 10.1002/jcb.10476. [DOI] [PubMed] [Google Scholar]
- 139.Wang YN, Wang H, Yamaguchi H, Lee HJ, Lee HH, Hung MC. COPI-mediated retrograde trafficking from the Golgi to the ER regulates EGFR nuclear transport. Biochem Biophys Res Commun. 2010;399:498–504. doi: 10.1016/j.bbrc.2010.07.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang YN, Lee HH, Lee HJ, Du Y, Yamaguchi H, Hung MC. Membrane-bound trafficking regulates nuclear transport of integral epidermal growth factor receptor (EGFR) and ErbB-2. J Biol Chem. 2012;287:16869–79. doi: 10.1074/jbc.M111.314799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Giri DK, Ali-Seyed M, Li LY, Lee DF, Ling P, Bartholomeusz G, Wang SC, Hung MC. Endosomal transport of ErbB-2: mechanism for nuclear entry of the cell surface receptor. Mol Cell Biol. 2005;25:11005–18. doi: 10.1128/MCB.25.24.11005-11018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lo HW, Ali-Seyed M, Wu Y, Bartholomeusz G, Hsu SC, Hung MC. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J Cell Biochem. 2006;98:1570–83. doi: 10.1002/jcb.20876. [DOI] [PubMed] [Google Scholar]
- 143.Gorlich D, Vogel F, Mills AD, Hartmann E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–8. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 144.Wang YN, Yamaguchi H, Huo L, Du Y, Lee HJ, Lee HH, Wang H, Hsu JM, Hung MC. The translocon Sec61beta localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J Biol Chem. 2010;285:38720–9. doi: 10.1074/jbc.M110.158659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Stachowiak MK, Fang X, Myers JM, Dunham SM, Berezney R, Maher PA, Stachowiak EK. Integrative nuclear FGFR1 signaling (INFS) as a part of a universal “feed-forward-and-gate” signaling module that controls cell growth and differentiation. J Cell Biochem. 2003;90:662–91. doi: 10.1002/jcb.10606. [DOI] [PubMed] [Google Scholar]
- 146.Lyu J, Wesselschmidt RL, Lu W. Cdc37 regulates Ryk signaling by stabilizing the cleaved Ryk intracellular domain. J Biol Chem. 2009;284:12940–8. doi: 10.1074/jbc.M900207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G. Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell. 2006;127:185–97. doi: 10.1016/j.cell.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 148.Sundvall M, Korhonen A, Vaparanta K, Anckar J, Halkilahti K, Salah Z, Aqeilan RI, Palvimo JJ, Sistonen L, Elenius K. Protein inhibitor of activated STAT3 (PIAS3) protein promotes SUMOylation and nuclear sequestration of the intracellular domain of ErbB4 protein. J Biol Chem. 2012;287:23216–26. doi: 10.1074/jbc.M111.335927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hoeing K, Zscheppang K, Mujahid S, Murray S, Volpe MV, Dammann CE, Nielsen HC. Presenilin-1 processing of ErbB4 in fetal type II cells is necessary for control of fetal lung maturation. Biochim Biophys Acta. 2011;1813:480–91. doi: 10.1016/j.bbamcr.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zscheppang K, Dork T, Schmiedl A, Jones FE, Dammann CE. Neuregulin receptor ErbB4 functions as a transcriptional cofactor for the expression of surfactant protein B in the fetal lung. Am J Respir Cell Mol Biol. 2011;45:761–7. doi: 10.1165/rcmb.2010-0179OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zscheppang K, Konrad M, Zischka M, Huhn V, Dammann CE. Estrogen-induced upregulation of Sftpb requires transcriptional control of neuregulin receptor ErbB4 in mouse lung type II epithelial cells. Biochim Biophys Acta. 2011;1813:1717–27. doi: 10.1016/j.bbamcr.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chook YM, Suel KE. Nuclear import by karyopherin-betas: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Peters R. Introduction to nucleocytoplasmic transport: molecules and mechanisms. Methods Mol Biol. 2006;322:235–58. doi: 10.1007/978-1-59745-000-3_17. [DOI] [PubMed] [Google Scholar]
- 154.Roskoski R., Jr The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 155.Gusterson B, Cowley G, Smith JA, Ozanne B. Cellular localisation of human epidermal growth factor receptor. Cell Biol Int Rep. 1984;8:649–58. doi: 10.1016/0309-1651(84)90045-6. [DOI] [PubMed] [Google Scholar]
- 156.Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–8. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 157.Liao HJ, Carpenter G. Cetuximab/C225-induced intracellular trafficking of epidermal growth factor receptor. Cancer Res. 2009;69:6179–83. doi: 10.1158/0008-5472.CAN-09-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Huo L, Wang YN, Xia W, Hsu SC, Lai CC, Li LY, Chang WC, Wang Y, Hsu MC, Yu YL, Huang TH, Ding Q, Chen CH, Tsai CH, Hung MC. RNA helicase A is a DNA-binding partner for EGFR-mediated transcriptional activation in the nucleus. Proc Natl Acad Sci U S A. 2010;107:16125–30. doi: 10.1073/pnas.1000743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Merlin J, Stechly L, de Beauce S, Monte D, Leteurtre E, van Seuningen I, Huet G, Pigny P. Galectin-3 regulates MUC1 and EGFR cellular distribution and EGFR downstream pathways in pancreatic cancer cells. Oncogene. 2011;30:2514–25. doi: 10.1038/onc.2010.631. [DOI] [PubMed] [Google Scholar]
- 160.Bitler BG, Goverdhan A, Schroeder JA. MUC1 regulates nuclear localization and function of the epidermal growth factor receptor. J Cell Sci. 2010;123:1716–23. doi: 10.1242/jcs.062661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lo HW, Hsu SC, Ali-Seyed M, Gunduz M, Xia W, Wei Y, Bartholomeusz G, Shih JY, Hung MC. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–89. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 162.Hanada N, Lo HW, Day CP, Pan Y, Nakajima Y, Hung MC. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol Carcinog. 2006;45:10–7. doi: 10.1002/mc.20147. [DOI] [PubMed] [Google Scholar]
- 163.Hung LY, Tseng JT, Lee YC, Xia W, Wang YN, Wu ML, Chuang YH, Lai CH, Chang WC. Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucleic Acids Res. 2008;36:4337–51. doi: 10.1093/nar/gkn417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lo HW, Cao X, Zhu H, Ali-Osman F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol Cancer Res. 2010;8:232–45. doi: 10.1158/1541-7786.MCR-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kim HP, Yoon YK, Kim JW, Han SW, Hur HS, Park J, Lee JH, Oh DY, Im SA, Bang YJ, Kim TY. Lapatinib, a dual EGFR and HER2 tyrosine kinase inhibitor, downregulates thymidylate synthase by inhibiting the nuclear translocation of EGFR and HER2. PLoS One. 2009;4:e5933. doi: 10.1371/journal.pone.0005933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Köstler W, Zielinski C. Targeting Receptor Tyrosine Kinases in Cancer. In: Wheeler DL, Yarden Y, editors. Receptor Tyrosine Kinases: Structure, Functions and Role in Human Disease. Springer; New York: 2015. pp. 225–278. [Google Scholar]
- 167.Dittmann K, Mayer C, Rodemann HP. Inhibition of radiation-induced EGFR nuclear import by C225 (Cetuximab) suppresses DNA-PK activity. Radiother Oncol. 2005;76:157–61. doi: 10.1016/j.radonc.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 168.Hsu SC, Miller SA, Wang Y, Hung MC. Nuclear EGFR is required for cisplatin resistance and DNA repair. Am J Transl Res. 2009;1:249–58. [PMC free article] [PubMed] [Google Scholar]
- 169.Das AK, Chen BP, Story MD, Sato M, Minna JD, Chen DJ, Nirodi CS. Somatic mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) abrogate EGFR-mediated radioprotection in non-small cell lung carcinoma. Cancer Res. 2007;67:5267–74. doi: 10.1158/0008-5472.CAN-07-0242. [DOI] [PubMed] [Google Scholar]
- 170.Iida M, Brand TM, Campbell DA, Li C, Wheeler DL. Yes and Lyn play a role in nuclear translocation of the epidermal growth factor receptor. Oncogene. 2013;32:759–67. doi: 10.1038/onc.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang SC, Nakajima Y, Yu YL, Xia W, Chen CT, Yang CC, McIntush EW, Li LY, Hawke DH, Kobayashi R, Hung MC. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006;8:1359–68. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- 172.Ortega J, Li JY, Lee S, Tong D, Gu L, Li GM. Phosphorylation of PCNA by EGFR inhibits mismatch repair and promotes misincorporation during DNA synthesis. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1417711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chou RH, Wang YN, Hsieh YH, Li LY, Xia W, Chang WC, Chang LC, Cheng CC, Lai CC, Hsu JL, Chang WJ, Chiang SY, Lee HJ, Liao HW, Chuang PH, Chen HY, Wang HL, Kuo SC, Chen CH, Yu YL, Hung MC. EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev Cell. 2014;30:224–37. doi: 10.1016/j.devcel.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, Chen DJ, Kehlbach R, Rodemann HP. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–9. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 175.Dittmann K, Mayer C, Kehlbach R, Rothmund MC, Peter Rodemann H. Radiation-induced lipid peroxidation activates src kinase and triggers nuclear EGFR transport. Radiother Oncol. 2009;92:379–82. doi: 10.1016/j.radonc.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 176.Dittmann K, Mayer C, Kehlbach R, Rodemann HP. Radiation-induced caveolin-1 associated EGFR internalization is linked with nuclear EGFR transport and activation of DNA-PK. Mol Cancer. 2008;7:69. doi: 10.1186/1476-4598-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Latha K, Li M, Chumbalkar V, Gururaj A, Hwang Y, Dakeng S, Sawaya R, Aldape K, Cavenee WK, Bogler O, Furnari FB. Nuclear EGFRvIII-STAT5b complex contributes to glioblastoma cell survival by direct activation of the Bcl-XL promoter. Int J Cancer. 2013;132:509–20. doi: 10.1002/ijc.27690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang SC, Lien HC, Xia W, Chen IF, Lo HW, Wang Z, Ali-Seyed M, Lee DF, Bartholomeusz G, Ou-Yang F, Giri DK, Hung MC. Binding at and transactivation of the COX-2 promoter by nuclear tyrosine kinase receptor ErbB-2. Cancer Cell. 2004;6:251–61. doi: 10.1016/j.ccr.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 179.Bertelsen V, Stang E. The Mysterious Ways of ErbB2/HER2 Trafficking. Membranes (Basel) 2014;4:424–46. doi: 10.3390/membranes4030424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang SE, Narasanna A, Perez-Torres M, Xiang B, Wu FY, Yang S, Carpenter G, Gazdar AF, Muthuswamy SK, Arteaga CL. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 181.Xie Y, Hung MC. Nuclear localization of p185neu tyrosine kinase and its association with transcriptional transactivation. Biochem Biophys Res Commun. 1994;203:1589–98. doi: 10.1006/bbrc.1994.2368. [DOI] [PubMed] [Google Scholar]
- 182.Beguelin W, Diaz Flaque MC, Proietti CJ, Cayrol F, Rivas MA, Tkach M, Rosemblit C, Tocci JM, Charreau EH, Schillaci R, Elizalde PV. Progesterone receptor induces ErbB-2 nuclear translocation to promote breast cancer growth via a novel transcriptional effect: ErbB-2 function as a coactivator of Stat3. Mol Cell Biol. 2010;30:5456–72. doi: 10.1128/MCB.00012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li LY, Chen H, Hsieh YH, Wang YN, Chu HJ, Chen YH, Chen HY, Chien PJ, Ma HT, Tsai HC, Lai CC, Sher YP, Lien HC, Tsai CH, Hung MC. Nuclear ErbB2 enhances translation and cell growth by activating transcription of ribosomal RNA genes. Cancer Res. 2011;71:4269–79. doi: 10.1158/0008-5472.CAN-10-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li X, Kuang J, Shen Y, Majer MM, Nelson CC, Parsawar K, Heichman KA, Kuwada SK. The atypical histone macroH2A1.2 interacts with HER-2 protein in cancer cells. J Biol Chem. 2012;287:23171–83. doi: 10.1074/jbc.M112.379412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tan M, Jing T, Lan KH, Neal CL, Li P, Lee S, Fang D, Nagata Y, Liu J, Arlinghaus R, Hung MC, Yu D. Phosphorylation on tyrosine-15 of p34(Cdc2) by ErbB2 inhibits p34(Cdc2) activation and is involved in resistance to taxol-induced apoptosis. Mol Cell. 2002;9:993–1004. doi: 10.1016/s1097-2765(02)00510-5. [DOI] [PubMed] [Google Scholar]
- 186.Arribas J, Baselga J, Pedersen K, Parra-Palau JL. p95HER2 and breast cancer. Cancer Res. 2011;71:1515–9. doi: 10.1158/0008-5472.CAN-10-3795. [DOI] [PubMed] [Google Scholar]
- 187.Garcia-Castillo J, Pedersen K, Angelini PD, Bech-Serra JJ, Colome N, Cunningham MP, Parra-Palau JL, Canals F, Baselga J, Arribas J. HER2 carboxyl-terminal fragments regulate cell migration and cortactin phosphorylation. J Biol Chem. 2009;284:25302–13. doi: 10.1074/jbc.M109.001982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pedersen K, Angelini PD, Laos S, Bach-Faig A, Cunningham MP, Ferrer-Ramon C, Luque-Garcia A, Garcia-Castillo J, Parra-Palau JL, Scaltriti M, Ramon y Cajal S, Baselga J, Arribas J. A naturally occurring HER2 carboxy-terminal fragment promotes mammary tumor growth and metastasis. Mol Cell Biol. 2009;29:3319–31. doi: 10.1128/MCB.01803-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc Natl Acad Sci U S A. 2010;107:7692–7. doi: 10.1073/pnas.1002753107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cheng CJ, Ye XC, Vakar-Lopez F, Kim J, Tu SM, Chen DT, Navone NM, Yu-Lee LY, Lin SH, Hu MC. Bone microenvironment and androgen status modulate subcellular localization of ErbB3 in prostate cancer cells. Mol Cancer Res. 2007;5:675–84. doi: 10.1158/1541-7786.MCR-06-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Koumakpayi IH, Diallo JS, Le Page C, Lessard L, Filali-Mouhim A, Begin LR, Mes-Masson AM, Saad F. Low nuclear ErbB3 predicts biochemical recurrence in patients with prostate cancer. BJU Int. 2007;100:303–9. doi: 10.1111/j.1464-410X.2007.06992.x. [DOI] [PubMed] [Google Scholar]
- 192.Offterdinger M, Schofer C, Weipoltshammer K, Grunt TW. c-erbB-3: a nuclear protein in mammary epithelial cells. J Cell Biol. 2002;157:929–39. doi: 10.1083/jcb.200109033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Koumakpayi IH, Diallo JS, Le Page C, Lessard L, Gleave M, Begin LR, Mes-Masson AM, Saad F. Expression and nuclear localization of ErbB3 in prostate cancer. Clin Cancer Res. 2006;12:2730–7. doi: 10.1158/1078-0432.CCR-05-2242. [DOI] [PubMed] [Google Scholar]
- 194.Koumakpayi IH, Le Page C, Delvoye N, Saad F, Mes-Masson AM. Macropinocytosis inhibitors and Arf6 regulate ErbB3 nuclear localization in prostate cancer cells. Mol Carcinog. 2011;50:901–12. doi: 10.1002/mc.20766. [DOI] [PubMed] [Google Scholar]
- 195.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2009;315:683–96. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 196.Andrique L, Fauvin D, El Maassarani M, Colasson H, Vannier B, Seite P. ErbB3(80 kDa), a nuclear variant of the ErbB3 receptor, binds to the Cyclin D1 promoter to activate cell proliferation but is negatively controlled by p14ARF. Cell Signal. 2012;24:1074–85. doi: 10.1016/j.cellsig.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 197.Raabe TD, Deadwyler G, Varga JW, Devries GH. Localization of neuregulin isoforms and erbB receptors in myelinating glial cells. Glia. 2004;45:197–207. doi: 10.1002/glia.10311. [DOI] [PubMed] [Google Scholar]
- 198.Adilakshmi T, Ness-Myers J, Madrid-Aliste C, Fiser A, Tapinos N. A nuclear variant of ErbB3 receptor tyrosine kinase regulates ezrin distribution and Schwann cell myelination. J Neurosci. 2011;31:5106–19. doi: 10.1523/JNEUROSCI.5635-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Williams CC, Allison JG, Vidal GA, Burow ME, Beckman BS, Marrero L, Jones FE. The ERBB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5A nuclear chaperone. J Cell Biol. 2004;167:469–78. doi: 10.1083/jcb.200403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Paatero I, Jokilammi A, Heikkinen PT, Iljin K, Kallioniemi OP, Jones FE, Jaakkola PM, Elenius K. Interaction with ErbB4 promotes hypoxia-inducible factor-1alpha signaling. J Biol Chem. 2012;287:9659–71. doi: 10.1074/jbc.M111.299537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tiong KH, Mah LY, Leong CO. Functional roles of fibroblast growth factor receptors (FGFRs) signaling in human cancers. Apoptosis. 2013;18:1447–68. doi: 10.1007/s10495-013-0886-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Stachowiak MK, Maher PA, Stachowiak EK. Integrative nuclear signaling in cell development--a role for FGF receptor-1. DNA Cell Biol. 2007;26:811–26. doi: 10.1089/dna.2007.0664. [DOI] [PubMed] [Google Scholar]
- 203.Elfenbein A, Lanahan A, Zhou TX, Yamasaki A, Tkachenko E, Matsuda M, Simons M. Syndecan 4 regulates FGFR1 signaling in endothelial cells by directing macropinocytosis. Sci Signal. 2012;5:ra36. doi: 10.1126/scisignal.2002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Dunham-Ems SM, Lee YW, Stachowiak EK, Pudavar H, Claus P, Prasad PN, Stachowiak MK. Fibroblast growth factor receptor-1 (FGFR1) nuclear dynamics reveal a novel mechanism in transcription control. Mol Biol Cell. 2009;20:2401–12. doi: 10.1091/mbc.E08-06-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hu Y, Fang X, Dunham SM, Prada C, Stachowiak EK, Stachowiak MK. 90-kDa ribosomal S6 kinase is a direct target for the nuclear fibroblast growth factor receptor 1 (FGFR1): role in FGFR1 signaling. J Biol Chem. 2004;279:29325–35. doi: 10.1074/jbc.M311144200. [DOI] [PubMed] [Google Scholar]
- 206.Sun S, Jiang Y, Zhang G, Song H, Zhang X, Zhang Y, Liang X, Sun Q, Pang D. Increased expression of fibroblastic growth factor receptor 2 is correlated with poor prognosis in patients with breast cancer. J Surg Oncol. 2012;105:773–9. doi: 10.1002/jso.22120. [DOI] [PubMed] [Google Scholar]
- 207.Martin AJ, Grant A, Ashfield AM, Palmer CN, Baker L, Quinlan PR, Purdie CA, Thompson AM, Jordan LB, Berg JN. FGFR2 protein expression in breast cancer: nuclear localisation and correlation with patient genotype. BMC Res Notes. 2011;4:72. doi: 10.1186/1756-0500-4-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Johnston CL, Cox HC, Gomm JJ, Coombes RC. Fibroblast growth factor receptors (FGFRs) localize in different cellular compartments. A splice variant of FGFR-3 localizes to the nucleus. J Biol Chem. 1995;270:30643–50. doi: 10.1074/jbc.270.51.30643. [DOI] [PubMed] [Google Scholar]
- 209.Zammit C, Barnard R, Gomm J, Coope R, Shousha S, Coombes C, Johnston C. Altered intracellular localization of fibroblast growth factor receptor 3 in human breast cancer. J Pathol. 2001;194:27–34. doi: 10.1002/path.846. [DOI] [PubMed] [Google Scholar]
- 210.Shibuya M. VEGF-VEGFR Signals in Health and Disease. Biomol Ther (Seoul) 2014;22:1–9. doi: 10.4062/biomolther.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Cai J, Jiang WG, Grant MB, Boulton M. Pigment epithelium-derived factor inhibits angiogenesis via regulated intracellular proteolysis of vascular endothelial growth factor receptor 1. J Biol Chem. 2006;281:3604–13. doi: 10.1074/jbc.M507401200. [DOI] [PubMed] [Google Scholar]
- 212.Lee TH, Seng S, Sekine M, Hinton C, Fu Y, Avraham HK, Avraham S. Vascular endothelial growth factor mediates intracrine survival in human breast carcinoma cells through internally expressed VEGFR1/FLT1. PLoS Med. 2007;4:e186. doi: 10.1371/journal.pmed.0040186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Cai J, Chen Z, Ruan Q, Han S, Liu L, Qi X, Boye SL, Hauswirth WW, Grant MB, Boulton ME. gamma-Secretase and presenilin mediate cleavage and phosphorylation of vascular endothelial growth factor receptor-1. J Biol Chem. 2011;286:42514–23. doi: 10.1074/jbc.M111.296590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hiratsuka S, Minowa O, Kuno J, Noda T, Shibuya M. Flt-1 lacking the tyrosine kinase domain is sufficient for normal development and angiogenesis in mice. Proc Natl Acad Sci U S A. 1998;95:9349–54. doi: 10.1073/pnas.95.16.9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Dardik R, Inbal A. Complex formation between tissue transglutaminase II (tTG) and vascular endothelial growth factor receptor 2 (VEGFR-2): proposed mechanism for modulation of endothelial cell response to VEGF. Exp Cell Res. 2006;312:2973–82. doi: 10.1016/j.yexcr.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 216.Domingues I, Rino J, Demmers JA, de Lanerolle P, Santos SC. VEGFR2 translocates to the nucleus to regulate its own transcription. PLoS One. 2011;6:e25668. doi: 10.1371/journal.pone.0025668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Santos SC, Miguel C, Domingues I, Calado A, Zhu Z, Wu Y, Dias S. VEGF and VEGFR-2 (KDR) internalization is required for endothelial recovery during wound healing. Exp Cell Res. 2007;313:1561–74. doi: 10.1016/j.yexcr.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 218.Gomes DA, Rodrigues MA, Leite MF, Gomez MV, Varnai P, Balla T, Bennett AM, Nathanson MH. c-Met must translocate to the nucleus to initiate calcium signals. J Biol Chem. 2008;283:4344–51. doi: 10.1074/jbc.M706550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Xie Y, Lu W, Liu S, Yang Q, Carver BS, Li E, Wang Y, Fazli L, Gleave M, Chen Z. Crosstalk between nuclear MET and SOX9/beta-catenin correlates with castration-resistant prostate cancer. Mol Endocrinol. 2014;28:1629–39. doi: 10.1210/me.2014-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pozner-Moulis S, Pappas DJ, Rimm DL. Met, the hepatocyte growth factor receptor, localizes to the nucleus in cells at low density. Cancer Res. 2006;66:7976–82. doi: 10.1158/0008-5472.CAN-05-4335. [DOI] [PubMed] [Google Scholar]
- 221.Matteucci E, Bendinelli P, Desiderio MA. Nuclear localization of active HGF receptor Met in aggressive MDA-MB231 breast carcinoma cells. Carcinogenesis. 2009;30:937–45. doi: 10.1093/carcin/bgp080. [DOI] [PubMed] [Google Scholar]
- 222.Wagh PK, Peace BE, Waltz SE. Met-related receptor tyrosine kinase Ron in tumor growth and metastasis. Adv Cancer Res. 2008;100:1–33. doi: 10.1016/S0065-230X(08)00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Liu HS, Hsu PY, Lai MD, Chang HY, Ho CL, Cheng HL, Chen HT, Lin YJ, Wu TJ, Tzai TS, Chow NH. An unusual function of RON receptor tyrosine kinase as a transcriptional regulator in cooperation with EGFR in human cancer cells. Carcinogenesis. 2010;31:1456–64. doi: 10.1093/carcin/bgq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Chang HY, Liu HS, Lai MD, Tsai YS, Tzai TS, Cheng HL, Chow NH. Hypoxia promotes nuclear translocation and transcriptional function in the oncogenic tyrosine kinase RON. Cancer Res. 2014;74:4549–62. doi: 10.1158/0008-5472.CAN-13-3730. [DOI] [PubMed] [Google Scholar]
- 225.Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–42. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 226.Moughal NA, Waters C, Sambi B, Pyne S, Pyne NJ. Nerve growth factor signaling involves interaction between the Trk A receptor and lysophosphatidate receptor 1 systems: nuclear translocation of the lysophosphatidate receptor 1 and Trk A receptors in pheochromocytoma 12 cells. Cell Signal. 2004;16:127–36. doi: 10.1016/j.cellsig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 227.Bonacchi A, Taddei ML, Petrai I, Efsen E, Defranco R, Nosi D, Torcia M, Rosini P, Formigli L, Rombouts K, Zecchi S, Milani S, Pinzani M, Laffi G, Marra F. Nuclear localization of TRK-A in liver cells. Histol Histopathol. 2008;23:327–40. doi: 10.14670/HH-23.327. [DOI] [PubMed] [Google Scholar]
- 228.Forrester WC. The Ror receptor tyrosine kinase family. Cell Mol Life Sci. 2002;59:83–96. doi: 10.1007/s00018-002-8407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Minami Y, Oishi I, Endo M, Nishita M. Ror-family receptor tyrosine kinases in noncanonical Wnt signaling: their implications in developmental morphogenesis and human diseases. Dev Dyn. 2010;239:1–15. doi: 10.1002/dvdy.21991. [DOI] [PubMed] [Google Scholar]
- 230.Rebagay G, Yan S, Liu C, Cheung NK. ROR1 and ROR2 in Human Malignancies: Potentials for Targeted Therapy. Front Oncol. 2012;2:34. doi: 10.3389/fonc.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Tseng HC, Lyu PC, Lin WC. Nuclear localization of orphan receptor protein kinase (Ror1) is mediated through the juxtamembrane domain. BMC Cell Biol. 2010;11:48. doi: 10.1186/1471-2121-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Tseng HC, Kao HW, Ho MR, Chen YR, Lin TW, Lyu PC, Lin WC. Cytoskeleton network and cellular migration modulated by nuclear-localized receptor tyrosine kinase ROR1. Anticancer Res. 2011;31:4239–49. [PubMed] [Google Scholar]
- 233.Bonner JC. Regulation of PDGF and its receptors in fibrotic diseases. Cytokine Growth Factor Rev. 2004;15:255–73. doi: 10.1016/j.cytogfr.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 234.Cruse G, Metcalfe DD, Olivera A. Functional deregulation of KIT: link to mast cell proliferative diseases and other neoplasms. Immunol Allergy Clin North Am. 2014;34:219–37. doi: 10.1016/j.iac.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol. 2003;23:4013–25. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Chen L, Acciani T, Le Cras T, Lutzko C, Perl AK. Dynamic regulation of platelet-derived growth factor receptor alpha expression in alveolar fibroblasts during realveolarization. Am J Respir Cell Mol Biol. 2012;47:517–27. doi: 10.1165/rcmb.2012-0030OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Barbetti V, Morandi A, Tusa I, Digiacomo G, Riverso M, Marzi I, Cipolleschi MG, Bessi S, Giannini A, Di Leo A, Dello Sbarba P, Rovida E. Chromatin-associated CSF-1R binds to the promoter of proliferation-related genes in breast cancer cells. Oncogene. 2014;33:4359–64. doi: 10.1038/onc.2013.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Pollak M. The insulin receptor/insulin-like growth factor receptor family as a therapeutic target in oncology. Clin Cancer Res. 2012;18:40–50. doi: 10.1158/1078-0432.CCR-11-0998. [DOI] [PubMed] [Google Scholar]
- 239.Aleksic T, Chitnis MM, Perestenko OV, Gao S, Thomas PH, Turner GD, Protheroe AS, Howarth M, Macaulay VM. Type 1 insulin-like growth factor receptor translocates to the nucleus of human tumor cells. Cancer Res. 2010;70:6412–9. doi: 10.1158/0008-5472.CAN-10-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Sehat B, Tofigh A, Lin Y, Trocme E, Liljedahl U, Lagergren J, Larsson O. SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci Signal. 2010;3:ra10. doi: 10.1126/scisignal.2000628. [DOI] [PubMed] [Google Scholar]
- 241.Wu YC, Zhu M, Robertson DM. Novel nuclear localization and potential function of insulin-like growth factor-1 receptor/insulin receptor hybrid in corneal epithelial cells. PLoS One. 2012;7:e42483. doi: 10.1371/journal.pone.0042483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Bodzin AS, Wei Z, Hurtt R, Gu T, Doria C. Gefitinib resistance in HCC mahlavu cells: upregulation of CD133 expression, activation of IGF-1R signaling pathway, and enhancement of IGF-1R nuclear translocation. J Cell Physiol. 2012;227:2947–52. doi: 10.1002/jcp.23041. [DOI] [PubMed] [Google Scholar]
- 243.Hoa N, Tsui S, Afifiyan NF, Sinha Hikim A, Li B, Douglas RS, Smith TJ. Nuclear targeting of IGF-1 receptor in orbital fibroblasts from Graves’ disease: apparent role of ADAM17. PLoS One. 2012;7:e34173. doi: 10.1371/journal.pone.0034173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Xu J, Litterst C, Georgakopoulos A, Zaganas I, Robakis NK. Peptide EphB2/CTF2 generated by the gamma-secretase processing of EphB2 receptor promotes tyrosine phosphorylation and cell surface localization of N-methyl-D-aspartate receptors. J Biol Chem. 2009;284:27220–8. doi: 10.1074/jbc.M109.048728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Vardi A, Bosviel R, Rabiau N, Adjakly M, Satih S, Dechelotte P, Boiteux JP, Fontana L, Bignon YJ, Guy L, Bernard-Gallon DJ. Soy phytoestrogens modify DNA methylation of GSTP1, RASSF1A, EPH2 and BRCA1 promoter in prostate cancer cells. In Vivo. 2010;24:393–400. [PubMed] [Google Scholar]
- 246.Kuroda C, Kubota S, Kawata K, Aoyama E, Sumiyoshi K, Oka M, Inoue M, Minagi S, Takigawa M. Distribution, gene expression, and functional role of EphA4 during ossification. Biochem Biophys Res Commun. 2008;374:22–7. doi: 10.1016/j.bbrc.2008.06.089. [DOI] [PubMed] [Google Scholar]
- 247.Poh WC, Shen Y, Inoue T. Function of the Ryk intracellular domain in C. elegans vulval development. Dev Dyn. 2014;243:1074–85. doi: 10.1002/dvdy.24159. [DOI] [PubMed] [Google Scholar]
- 248.Inoue T, Oz HS, Wiland D, Gharib S, Deshpande R, Hill RJ, Katz WS, Sternberg PW. C. elegans LIN-18 is a Ryk ortholog and functions in parallel to LIN-17/Frizzled in Wnt signaling. Cell. 2004;118:795–806. doi: 10.1016/j.cell.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 249.Golubkov VS, Strongin AY. Downstream signaling and genome-wide regulatory effects of PTK7 pseudokinase and its proteolytic fragments in cancer cells. Cell Commun Signal. 2014;12:15. doi: 10.1186/1478-811X-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Golubkov VS, Prigozhina NL, Zhang Y, Stoletov K, Lewis JD, Schwartz PE, Hoffman RM, Strongin AY. Protein-tyrosine pseudokinase 7 (PTK7) directs cancer cell motility and metastasis. J Biol Chem. 2014;289:24238–49. doi: 10.1074/jbc.M114.574459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Bosurgi L, Bernink JH, Delgado Cuevas V, Gagliani N, Joannas L, Schmid ET, Booth CJ, Ghosh S, Rothlin CV. Paradoxical role of the proto-oncogene Axl and Mer receptor tyrosine kinases in colon cancer. Proc Natl Acad Sci U S A. 2013;110:13091–6. doi: 10.1073/pnas.1302507110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Verma A, Warner SL, Vankayalapati H, Bearss DJ, Sharma S. Targeting Axl and Mer kinases in cancer. Mol Cancer Ther. 2011;10:1763–73. doi: 10.1158/1535-7163.MCT-11-0116. [DOI] [PubMed] [Google Scholar]
- 253.Migdall-Wilson J, Bates C, Schlegel J, Brandao L, Linger RM, DeRyckere D, Graham DK. Prolonged exposure to a Mer ligand in leukemia: Gas6 favors expression of a partial Mer glycoform and reveals a novel role for Mer in the nucleus. PLoS One. 2012;7:e31635. doi: 10.1371/journal.pone.0031635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Marti U, Hug M. Acinar and cellular distribution and mRNA expression of the epidermal growth factor receptor are changed during liver regeneration. J Hepatol. 1995;23:318–27. [PubMed] [Google Scholar]
- 255.Marti U, Burwen SJ, Wells A, Barker ME, Huling S, Feren AM, Jones AL. Localization of epidermal growth factor receptor in hepatocyte nuclei. Hepatology. 1991;13:15–20. [PubMed] [Google Scholar]
- 256.Kamio T, Shigematsu K, Sou H, Kawai K, Tsuchiyama H. Immunohistochemical expression of epidermal growth factor receptors in human adrenocortical carcinoma. Hum Pathol. 1990;21:277–82. doi: 10.1016/0046-8177(90)90227-v. [DOI] [PubMed] [Google Scholar]
- 257.Lipponen P, Eskelinen M. Expression of epidermal growth factor receptor in bladder cancer as related to established prognostic factors, oncoprotein (c-erbB-2, p53) expression and long-term prognosis. Br J Cancer. 1994;69:1120–5. doi: 10.1038/bjc.1994.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Marti U, Ruchti C, Kampf J, Thomas GA, Williams ED, Peter HJ, Gerber H, Burgi U. Nuclear localization of epidermal growth factor and epidermal growth factor receptors in human thyroid tissues. Thyroid. 2001;11:137–45. doi: 10.1089/105072501300042785. [DOI] [PubMed] [Google Scholar]
- 259.Tervahauta A, Syrjanen S, Syrjanen K. Epidermal growth factor receptor, c-erbB-2 proto-oncogene and estrogen receptor expression in human papillomavirus lesions of the uterine cervix. Int J Gynecol Pathol. 1994;13:234–40. doi: 10.1097/00004347-199407000-00007. [DOI] [PubMed] [Google Scholar]
- 260.Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–48. [PubMed] [Google Scholar]
- 261.Slack BE, Siniaia MS, Blusztajn JK. Collagen type I selectively activates ectodomain shedding of the discoidin domain receptor 1: involvement of Src tyrosine kinase. J Cell Biochem. 2006;98:672–84. doi: 10.1002/jcb.20812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Findley CM, Cudmore MJ, Ahmed A, Kontos CD. VEGF induces Tie2 shedding via a phosphoinositide 3-kinase/Akt dependent pathway to modulate Tie2 signaling. Arterioscler Thromb Vasc Biol. 2007;27:2619–26. doi: 10.1161/ATVBAHA.107.150482. [DOI] [PubMed] [Google Scholar]
- 263.Cabrera N, Diaz-Rodriguez E, Becker E, Martin-Zanca D, Pandiella A. TrkA receptor ectodomain cleavage generates a tyrosine-phosphorylated cell-associated fragment. J Cell Biol. 1996;132:427–36. doi: 10.1083/jcb.132.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Diaz-Rodriguez E, Cabrera N, Esparis-Ogando A, Montero JC, Pandiella A. Cleavage of the TrkA neurotrophin receptor by multiple metalloproteases generates signalling-competent truncated forms. Eur J Neurosci. 1999;11:1421–30. doi: 10.1046/j.1460-9568.1999.00552.x. [DOI] [PubMed] [Google Scholar]
- 265.Swendeman S, Mendelson K, Weskamp G, Horiuchi K, Deutsch U, Scherle P, Hooper A, Rafii S, Blobel CP. VEGF-A stimulates ADAM17-dependent shedding of VEGFR2 and crosstalk between VEGFR2 and ERK signaling. Circ Res. 2008;103:916–8. doi: 10.1161/CIRCRESAHA.108.184416. [DOI] [PMC free article] [PubMed] [Google Scholar]