Abstract
Hedgehog (Hh) proteins function in cell/cell signaling processes linked to human embryo development and the progression of several types of cancer. Here we describe an optical assay of hedgehog cholesterolysis, a unique autoprocessing event critical for Hh function. The assay uses a recombinant FRET-active hedgehog precursor whose cholesterolysis can be monitored continuously in multi-well plates (dynamic range, 4; Z’, 0.7), offering advantages in throughput over conventional SDS-PAGE assays. Application of the optical assay in a pilot small molecule screen produced a novel cholesterolysis inhibitor (apparent IC50, 5×10−6 M) that appears to inactivate hedgehog covalently by a SNAr mechanism.
Proteins in the hedgehog (Hh) family function as cell-cell signaling factors that help govern embryo development, while also having pathogenic effects in multiple types of cancer [1-4]. Hedgehog’s oncogenic activity has galvanized small molecule discovery efforts, resulting in the recent approval of vismodegib (Erivedge) for the treatment of advanced basal cell carcinoma [5]. Vismodegib, like several other Hh inhibitors under clinical study, interferes with transduction of the Hh signal by antagonizing a cell surface receptor called Smoothened [6, 7]. While a major milestone, alternative approaches to combat aberrant Hh activity are worth pursuing for combination therapy and to contend with clinical resistance to vismodegib [8-10]. Here we describe an optical activity assay to monitor Hh cholesterolysis, a crucial yet relatively under-studied autoprocessing reaction characteristic of this important protein family. We demonstrate the assay’s utility with a small molecule screen, resulting in the identification of a novel cholesterolysis inhibitor. .
Cholesterolysis of a hedgehog precursor liberates functional hedgehog ligand (HhN) and couples its incipient carboxyl terminus to cholesterol [11-13] (Figure 1A). At least two chemical steps are involved in the reaction: first, an N-S acyl shift activates the terminal glycine residue of HhN as a thioester; next, the hydroxyl group of substrate cholesterol attacks the thioester, thereby displacing HhN [12]. No cofactors or accessory proteins seem to be required for either step. All catalytic activity resides in the adjacent C-segment of the precursor, referred to as HhC [14][15, 16]. Congenital mutations in the HhC segment of human “sonic” Hh that attenuate cholesterolysis are associated with developmental anomalies of the brain, highlighting the importance of this reaction [17-20].
Figure 1.
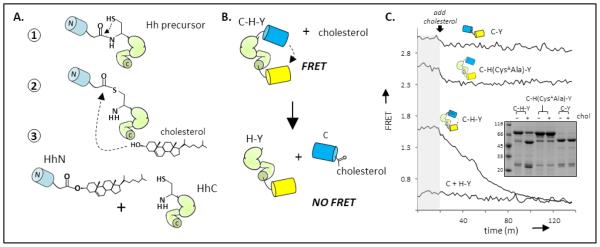
Mechanism and monitoring of Hh cholesterolysis. (A) Reaction pathway. Cholesterolysis of Hh precursor proteins involves an N-S acyl shift at a conserved glycine-cysteine dipeptide (step 1); transesterification to cholesterol (step 2); and dissociation of cholesteroylated Hh ligand (HhN) from the Hh autoprocessing segment, (HhC) (step 3). (B) FRET reporter of cholesterolysis. Construct, abbreviated C-H-Y, contains HhC as an internal fusion to Cyan fluorescent protein and Yellow fluorescent protein. (C) Cholesterolysis assay. FRET from catalytically active C-H-Y, and inactive control constructs,C-Y and C-(CysAAla)H-Y, are monitored before and after cholesterol addition. The kinetic trace of C-H-Y shows FRET loss following cholesterol addition, approaching FRET of a 1:1 mixture of CFP and YFP. Inset Characterization of cholesterolysis reactions by SDS-PAGE followed by Coomassie blue staining. Molecular weights: C-H-Y, 80 kDa; CFP, 27 kDa; H-Y, 52 kDa; C-Y, 48 kDa.
With a view toward increasing the speed of antagonist/agonist screens, as well as enabling structure-function studies of this unusual lipidation event, we sought a continuous cholesterolysis assay suitable for microplate readers. Current means of assaying cholesterolysis include the separation of precursor and products by SDS-PAGE, and more recently by fluorescent polarization [21]. The latter assay, while attractive in many respects, suffers from a low dynamic range and requires refolding of HhC followed by labeling with arsenic-based reagents [21]. The assay system we describe here involves FRET (Förster resonance energy transfer) between soluble recombinant fluorescent proteins genetically fused to an HhC segment (residues 255-471) from the Drosophila melanogaster Hh (Figure 1B). It is analogous to the FRET sensor of Amitai et. al, used to characterize the kinetics of self-cleaving “inteins” [22]. The key construct, termed C-H-Y, has cyan (C) and yellow (Y) fluorescent proteins [23] fused to the amino and carboxyl termini of HhC. Recently we reported that C-H-Y could be used to detect binding of HhC by an irreversible inhibitor of cholesterolysis [24]. Here we pursue the expectation that cholesterolysis of C-H-Y will yield cholesterol-modified CFP and HhC-YFP, with a relatively large change in the FRET ratio (i.e. loss) as the fluorescent proteins physically separate. We calculate the FRET ratio after excitation at 400 nm by dividing the emission intensity at 540 nm, predominately from YFP, by the emission intensity at 460 nm, predominately from CFP. In E. coli, C-H-Y expresses in soluble form, and the precursor remains largely unprocessed owing to the absence of cholesterol. Two control constructs used for the FRET system are a catalytic mutant, C-H(CysAAla)-Y, (residue Cys258Ala) whose cholesterolysis activity is reduced by > 104-fold; and, C-Y, which is identical to C-H-Y except that the HhC domain has been replaced by a short peptide linker. The first control construct, C-H(CysAAla)-Y, can be used to mimic 100% inhibition in screens for antagonists of cholesterolysis; C-Y serves as a control to flag compounds that interact with the fluorescent proteins.
Activity assays using C-H-Y were conducted in Bis-Tris buffer (50 mM) at pH 7.1, the prevailing pH of the endoplasmic reticulum where cholesterolysis seems to occur in the cell [25, 26]. Following earlier work, Triton X-100 was added to a final concentration of 0.4% for the purpose of solubilizing cholesterol [11]. Ethylenediaminetetraacetic acid (5 mM, final) and Tris(2-carboxyethyl)phosphine (4 mM, final) were also included in the reaction buffer to suppress oxidation of two catalytically essential cysteines of HhC [26]. After a 10 minute preincubation with C-H-Y (0.2 µM) in assay buffer at 30 °C, reactions were initiated by the addition of cholesterol from an ethanolic stock. As can be seen in Figure 1C, added cholesterol led to a gradual attenuation of the FRET ratio from C-H-Y, falling from ~1.6 to 0.4. Full spectra were also recorded at selected intervals, which showed time dependent loss of FRET acceptor signal at 540 nm, and gain of FRET donor signal at 460 nm, consistent with donor and acceptor separation. The final FRET value corresponds to that of a 1:1 mixture of C and H-Y (Figure 1C, bottom trace), suggesting that cholesterolysis had proceeded to completion. The extent of cholesterolysis was also confirmed by separating the product mixture with SDS-PAGE (Figure 1C, inset). In addition, the FRET ratio of the catalytic mutant, C-H(CysAAla)-Y, and control C-Y seemed relatively insensitive to added cholesterol (Figure 1C, top traces), as expected.
To substantiate the role of cholesterol as a nucleophile in the C-H-Y reaction, control assays were conducted with C-H-Y using a fluorescent sterol derivative, 25-NBD cholesterol. Separation of the product mixture by denaturing SDS-PAGE, followed by UV illumination of the gel, showed that the fluorophore tracked with liberated CFP (Supporting Figure 2A). Co-migration of 25-NBD cholesterol with denatured CFP is consistent with covalent interaction. No appreciable activity of C-H-Y was apparent in control experiments lacking cholesterol or with the de-oxy cholesterol analogue, 5-cholestene (Supporting Figure 2B, C). The apparent stability of C-H-Y in the absence of added sterol indicates that spontaneous hydrolysis of the internal thioester is slow; our preliminary experiments indicate a rate constant for hydrolysis at 30 °C of ~4×10−6 sec−1 (t ½, 2 days).
To further establish that the observed changes in the FRET ratio with C-H-Y were reporting cholesterolysis activity, we confirmed that steady state kinetics exhibited a saturable dependence on cholesterol concentration. With C-H-Y at 0.2 µM, reactions were initiated by adding cholesterol to concentrations ranging from 5 µM to 350 µM. Assays were carried out in triplicate, and wells containing C-H-Y without cholesterol served as control. Michaelis-Menten plots of initial rates as a function of increasing cholesterol concentration showed saturation behavior (Supporting Figure 3A). Fits of the data to a hyperbolic function (see Supporting Information) yielded an apparent Km for cholesterol of 15 µM (+/− 3), in accord with earlier work [21]. To determine a maximum rate of cholesterolysis, we fit reaction progress curves at saturating cholesterol (350 µM) to a first order exponential, yielding an apparent kcat value of 0.001 sec−1 (t ½ ~ 11 minutes) (Supporting Figure 3B). This value at pH 7.1, the prevailing pH of the ER [25], is remarkably similar to the apparent rate of Hh precursor processing measured by pulse-chase experiments in cultured mammalian cells (0.0006 sec−1) [27]; it is also within range of autocatalytic “protein splicing” reactions brought about by inteins [22, 28-30], which may be ancestral to Hh proteins [14, 31]. Thus, the optical format facilitates kinetic analysis, and affords results that are reproducible and appear physiologically relevant.
To evaluate the assay’s high-throughput compatibility, we carried out a pilot screen aimed at identifying cholesterolysis inhibitors. The compound library consisted of 240 protease inhibitors and their analogues, a selection biased by the target, which acts in a manner analogous to a protease (i.e. peptide bond cleavage). Assays were conducted in 96-well plates with C-H-Y at a final concentration of 0.2 µM in the reaction buffer above. Potential inhibitors were added from DMSO stocks to a final concentration of 40 µM into columns 2-11 of the plate. Control experiments indicated that the final concentration of DMSO (2%, v/v) has a minor effect (<5%) on reaction kinetics (Supporting Figure 4A, B). Prior to cholesterol addition, samples were equilibrated at 30 °C in the plate reader. FRET values calculated during this preincubation proved useful in identifying compounds that interfered optically with the assay output, as well as compounds that were not optically active but could alter the FRET signal from C-H-Y presumably through direct binding [24]. Reactions were initiated with cholesterol (50 µM, or 250 µM, final) and monitored continuously for a period of 1 to 3 h.
Intraplate controls were located in column 1 (minus cholesterol, minus compound) and in column 12, (plus cholesterol, minus compound) (Figure 2A). Data from these wells were used to evaluate assay performance through the Z’ calculation; column 1 used as the “max” and column 2 used as the “min”, according to formulation of Zhang [32]. The Z’ values were consistently above 0.7, indicating a reliable assay. Readings from control wells were also used to flag highly fluorescent compounds whose emission washed out signal from C-H-Y. Anomalously high, stable FRET signals were observed in the presence of compounds that fluoresced at 540 nm, whereas anomalously stable, low FRET signal was apparent in the presence of compounds fluorescing at 460 nm. Compounds in the latter class were more abundant, in accord with similar studies [33]. If emission values at 460 nm were >3 standard deviations from the mean emission value at 460 nm of the control wells, the well data was discarded. A histogram of the 460 nm emission data from compound and control wells is provided in the supporting information (Supporting Figure 5).
Figure 2.
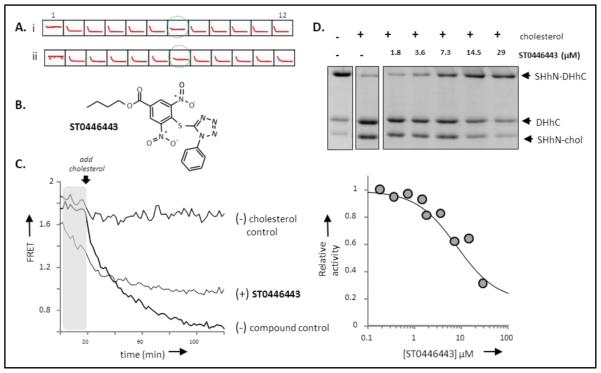
Hh cholesterolysis inhibitor identified by chemical screening. (A) Screening output. Traces of 12 sample wells from a 96 well plate, showing controls, column (1, 12); and compound wells (2-11). Compound in well 7 (green circle) was identified as an potential inhibitor in screens with cholesterol at 60 µM (upper) and 200 µM (lower). (B) Inhibitor structure. TIMTEC chemical identification code ST044643. (C) Effects of ST044643 on C-H-Y. Added ST044643 diminishes FRET from C-H-Y in the pre-incubation period (gray), and limits the extent of reaction compared to the C-H-Y control. (D) Secondary screen of Hh cholesterolysis inhibition by ST044643. Upper: Cholesterolysis activity of chimeric hedgehog precursor, ShhN-DHhC, in the absence and presence of increasing amounts of ST044643. Precursor and products were separated by SDS-PAGE and stained with Coomassie blue. Lower: Relative cholesterolysis activity plotted as a function of increasing ST044643, using data in upper panel. Dose-response curve (solid line) was calculated using an IC50 value of 5 µM.
With optically active compounds flagged, our attention was drawn to compound ST044643 as a potential inhibitor (Figure 2B). Kinetic traces in the presence of ST044643 showed minor changes in the initial cholesterolysis rate, however, the end point FRET ratio seemed high compared with controls (Figure 2C). That elevated end point signal suggested to us that a fraction of the original C-H-Y remained unprocessed; a hypothesis we subsequently confirmed by SDS-PAGE (Supporting Figure 6A). Interestingly, data from the pre-incubation period in the presence of ST044643, but in the absence of cholesterol, indicated that FRET ratio of C-H-Y underwent a noticeable and reproducible quenching (Figure 2C, shaded). No evidence for decomposition of C-H-Y in the presence of ST044643 was found by SDS-PAGE analysis (Supporting Figure 6B). We have observed similar quenching behavior with C-H-Y in the presence of phenylarsine oxide, an active-site covalent inhibitor of cholesterolysis [24]. To explore the specificity of this effect, ST044643 was added to the control construct, C-Y, in which the HhC segment is replaced with a glycine-serine linker; no significant change in the FRET ratio of C-Y was apparent with added ST044643 (p= 0.276, n= 15) (Supporting Figure 6C). Next, we tested whether added ST044643 could alter the FRET from C-H(CysAAla)-Y, which is identical to C-H-Y except that a conserved cysteine residue of HhC (residue 258) is mutated to alanine, rendering it catalytic inert[26]; here, a significant change in the FRET ratio with ST044643 was observed (p < 0.005, n=12). Together these results are consistent with the notion that the ST044643 binding site on C-H-Y maps to the “H” segment, but does not seem to involve this particular catalytically essential cysteine residue (CysA) of HhC.
To further examine the strength and specificity of inhibition by ST044643, the compound was repurchased as a powder from the supplier and used in dose response experiments in a gel-based activity assay. In place of C-H-Y, we used a chimeric Hh precursor, SHhN-DHhC, where the signaling domain from human sonic hedgehog protein (residues 24–197) is fused to the same autoprocessing C-terminal segment of Hh from Drosophila melanogaster [24]. This construct is overexpressed as a soluble, His-tagged protein in E. coli. ST044643 was tested as an inhibitor of SHhN-DHhC cholesterolysis at concentrations ranging from 0.1 µM to 29 µM. The extent of processing was determined by gel densitometry 3 h and 24 h after adding cholesterol (200 µM, final). A representative gel and corresponding dose response curve, which indicate an apparent IC50 value of ~ 5 µM, are shown in Figure 2D. Consistent with covalent inhibition, the extent of cholesterolysis after 3 h and after 24 h in the presence of equivalent concentration of ST044643 were comparable. The IC50 value with SHhN-DHhC also accords with inhibitory strength toward C-H-Y measured in the optical assay. Collectively, these results serve to validate ST044643 as a cholesterolysis inhibitor and further suggest a covalent inhibitory mechanism.
Covalent modification of an active-site cysteine through SNAr (substitution nucleophilic aromatic) is a likely mode of action for ST044643. The pair of nitro groups, along with the ester, all in resonance with the carbon bearing the mercapto tetrazole moiety, activate the phenyl group of ST044643 for SNAr (Supporting Figure 7A). Structurally similar molecules have been reported to inhibit other cysteine-dependent biocatalytic transformations by SNAr [34, 35]. In addition, preliminary mass spectrometry of HhC treated with ST044643 support covalent inactivation by this mechanism (Supporting Figure 7B). Notwithstanding, results of the FRET quenching experiments (above), argue against SNAr, at least to the extent that it requires the CysA residue of HhC. There is a however a second conserved cysteine residue of HhC, termed CysB (residue number 400), that is required for cholesterolysis and located in the active site [14, 26]. To explore the potential involvement of CysB in ST044643 binding, we generated the point mutant, C-H(CysBAsp)-Y, and subjected it to the FRET quenching analysis. Attempts to prepare an Ala point mutant at this position resulted in a protein that expressed poorly. Unlike C-H-Y and C-(CysAAla)H-Y, the FRET ratio of C-H(CysBAsp)-Y mutant seemed unresponsive to added ST044643 (p = 0.162, n=13 ) (Supporting Figure 6C). The unresponsiveness of this mutant is consistent with the notion that CysB is required for interaction with ST044643. Unambiguous identification of the binding site, and the nature of the adduct await more detailed structural analysis. Meanwhile we have begun using ST044643 as a positive control in small molecule screens using larger, more diverse chemical libraries.
To accelerate discovery of a novel class of Hh-directed small molecules that may ultimately develop into effective drug candidates, we devised a continuous, optical assay to monitor Hh cholesterolysis. Agonists of cholesterolysis are expected to increase the concentration of Hh ligand in circulation, with potential use in rescuing Hh mutations linked to birth defects; antagonists have potential use in treating Hh-driven cancers, where Hh is chronically over produced. The FRET assay’s dynamic range, favorable Z’ factor, and ease of use encourage its application to these and to other discovery efforts.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by start-up funds from Binghamton University and by the Office of the Assistant Secretary of Defense for Health Affairs, through the Prostate Cancer Research Program, grant W81XWH-14-1-0155 (BPC). Early stages of this work were supported by grant R01 GM04484 (MB). The Regional NMR Facility (600 MHz instrument) at Binghamton University is supported by the NSF (grant CHE-0922815).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Atwood SX, Chang AL, Oro AE. Hedgehog pathway inhibition and the race against tumor evolution. The Journal of cell biology. 2012;199(2):193–7. doi: 10.1083/jcb.201207140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature reviews. Molecular cell biology. 2013;14(7):416–29. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 3.Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nature medicine. 2013;19(11):1410–22. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- 4.Chen M, Carkner R, Buttyan R. The hedgehog/Gli signaling paradigm in prostate cancer. Expert review of endocrinology & metabolism. 2011;6(3):453–467. doi: 10.1586/EEM.11.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gould SE, et al. Discovery and preclinical development of vismodegib. Expert opinion on drug discovery. 2014;9(8):969–84. doi: 10.1517/17460441.2014.920816. [DOI] [PubMed] [Google Scholar]
- 6.Sharpe HJ, et al. Regulation of the oncoprotein Smoothened by small molecules. Nature chemical biology. 2015;11(4):246–551. doi: 10.1038/nchembio.1776. [DOI] [PubMed] [Google Scholar]
- 7.Wu VM, et al. Small molecule inhibitors of Smoothened ciliary localization and ciliogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(34):13644–9. doi: 10.1073/pnas.1207170109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metcalfe C, de Sauvage FJ. Hedgehog fights back: mechanisms of acquired resistance against Smoothened antagonists. Cancer research. 2011;71(15):5057–61. doi: 10.1158/0008-5472.CAN-11-0923. [DOI] [PubMed] [Google Scholar]
- 9.Stanton BZ, et al. A small molecule that binds Hedgehog and blocks its signaling in human cells. Nature Chemical Biology. 2009;5(3):154–6. doi: 10.1038/nchembio.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrova E, et al. Inhibitors of Hedgehog acyltransferase block Sonic Hedgehog signaling. Nature chemical biology. 2013;9(4):247–9. doi: 10.1038/nchembio.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 1996;274(5285):255–9. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 12.Mann RK, Beachy PA. Novel lipid modifications of secreted protein signals. Annual Review of Biochemistry. 2004;73:891–923. doi: 10.1146/annurev.biochem.73.011303.073933. [DOI] [PubMed] [Google Scholar]
- 13.Steinhauer J, Treisman JE. Lipid-modified morphogens: functions of fats. Current opinion in genetics & development. 2009;19(4):308–14. doi: 10.1016/j.gde.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall TM, et al. Crystal structure of a Hedgehog autoprocessing domain: homology between Hedgehog and self-splicing proteins. Cell. 1997;91(1):85–97. doi: 10.1016/s0092-8674(01)80011-8. [DOI] [PubMed] [Google Scholar]
- 15.Tukachinsky H, et al. Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell reports. 2012;2(2):308–20. doi: 10.1016/j.celrep.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vincent S, et al. Targeting of proteins to membranes through hedgehog auto-processing. Nat Biotechnol. 2003;21(8):936–40. doi: 10.1038/nbt844. [DOI] [PubMed] [Google Scholar]
- 17.Roessler E, et al. The mutational spectrum of holoprosencephaly-associated changes within the SHH gene in humans predicts loss-of-function through either key structural alterations of the ligand or its altered synthesis. Hum Mutat. 2009;30(10):E921–35. doi: 10.1002/humu.21090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Traiffort E, et al. Functional characterization of sonic hedgehog mutations associated with holoprosencephaly. J Biol Chem. 2004;279(41):42889–97. doi: 10.1074/jbc.M405161200. [DOI] [PubMed] [Google Scholar]
- 19.Maity T, Fuse N, Beachy PA. Molecular mechanisms of Sonic hedgehog mutant effects in holoprosencephaly. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(47):17026–31. doi: 10.1073/pnas.0507848102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh S, et al. Sonic hedgehog mutations identified in holoprosencephaly patients can act in a dominant negative manner. Hum Genet. 2009;125(1):95–103. doi: 10.1007/s00439-008-0599-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang SQ, Paulus H. A high-throughput, homogeneous, fluorescence polarization assay for inhibitors of hedgehog protein autoprocessing. J Biomol Screen. 2010;15(9):1082–7. doi: 10.1177/1087057110377498. [DOI] [PubMed] [Google Scholar]
- 22.Amitai G, et al. Modulation of intein activity by its neighboring extein substrates. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(27):11005–10. doi: 10.1073/pnas.0904366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nature biotechnology. 2005;23(3):355–60. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 24.Owen TS, et al. Active site targeting of hedgehog precursor protein with phenylarsine oxide. Chembiochem : a European journal of chemical biology. 2014 doi: 10.1002/cbic.201402421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JH, et al. Noninvasive measurement of the pH of the endoplasmic reticulum at rest and during calcium release. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(6):2997–3002. doi: 10.1073/pnas.95.6.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, et al. Processing and turnover of the Hedgehog protein in the endoplasmic reticulum. The Journal of cell biology. 2011;192(5):825–38. doi: 10.1083/jcb.201008090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guy RK. Inhibition of sonic hedgehog autoprocessing in cultured mammalian cells by sterol deprivation. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(13):7307–12. doi: 10.1073/pnas.97.13.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah NH, et al. Extein residues play an intimate role in the rate-limiting step of protein trans-splicing. Journal of the American Chemical Society. 2013;135(15):5839–47. doi: 10.1021/ja401015p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pereira B, et al. Spontaneous proton transfer to a conserved intein residue determines on-pathway protein splicing. Journal of molecular biology. 2011;406(3):430–42. doi: 10.1016/j.jmb.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood DW, Camarero JA. Intein applications: from protein purification and labeling to metabolic control methods. The Journal of biological chemistry. 2014;289(21):14512–9. doi: 10.1074/jbc.R114.552653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perler FB. Protein splicing of inteins and hedgehog autoproteolysis: structure, function, and evolution. Cell. 1998;92(1):1–4. doi: 10.1016/s0092-8674(00)80892-2. [DOI] [PubMed] [Google Scholar]
- 32.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. Journal of Biomolecular Screening. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 33.Simeonov A, et al. Fluorescence spectroscopic profiling of compound libraries. Journal of medicinal chemistry. 2008;51(8):2363–71. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 34.Groom H, et al. Inhibition of human glutathione transferases by dinitronaphthalene derivatives. Archives of biochemistry and biophysics. 2014:555–556. 71–6. doi: 10.1016/j.abb.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Dahlin JL, et al. PAINS in the Assay: Chemical Mechanisms of Assay Interference and Promiscuous Enzymatic Inhibition Observed during a Sulfhydryl-Scavenging HTS. Journal of medicinal chemistry. 2015;58(5):2091–113. doi: 10.1021/jm5019093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.